Abstract
Purpose
Chemotherapy drug resistance and relapse of the disease have been the major factors limiting the success of acute myeloid leukemia (AML) therapy. Several factors, including the pharmacokinetics (PK) of Cytarabine (Ara-C) and Daunorubicin (Dnr), could contribute to difference in treatment outcome in AML.
Methods
In the present study, we evaluated the plasma PK of Dnr, the influence of genetic polymorphisms of genes involved in transport and metabolism of Dnr on the PK, and also the influence of these factors on clinical outcome. Plasma levels of Dnr and its major metabolite, Daunorubicinol (DOL), were available in 70 adult de novo AML patients. PK parameters (Area under curve (AUC) and clearance (CL)) of Dnr and DOL were calculated using nonlinear mixed-effects modeling analysis performed with Monolix. Genetic variants in ABCB1, ABCG2, CBR1, and CBR3 genes as well as RNA expression of CBR1, ABCB1, and ABCG2 were compared with Dnr PK parameters.
Results
The AUC and CL of Dnr and DOL showed wide inter-individual variation. Patients with an exon1 variant of rs25678 in CBR1 had significantly higher plasma Dnr AUC [p = 0.05] compared to patients with wild type. Patients who achieved complete remission (CR) had significantly lower plasma Dnr AUC, Cmax, and higher CL compared to patients who did not achieve CR.
Conclusion
Further validation of these findings in a larger cohort of AML patients is warranted before establishing a therapeutic window for plasma Dnr levels and targeted dose adjustment.
Keywords: Acute myeloid leukemia, Daunorubicin, Pharmacokinetics, Genetic factors
Introduction
Acute myeloid leukemia (AML) is an aggressive malignancy of the hematopoietic system associated with relatively poor outcome [1]. Though 80 % of patients achieve complete remission with the current treatment protocol, the 5-year overall survival in younger AML patients ranges from 25 % and is much lower in older patients [2]. Relapse of the disease and resistance to chemotherapy contribute to poor treatment outcome. Daunorubicin (Dnr) has been the choice of anthracycline in the treatment of AML along with Cytarabine (Ara-C) for the past four decades. Altered pharmacokinetics of Dnr and Ara-C may be one of the several reasons for variation in treatment outcome [3, 4].
Pharmacokinetics (PK) of Dnr has been investigated since its use in the induction chemotherapy of AML [5]. Though the plasma as well as intracellular PK analysis of Dnr carried out in a small set of AML patients showed wide variation among the patients studied, these parameters were not significantly associated with clinical outcome [3, 4, 6]. PK models have been developed for Dnr in AML for infants [7], children [8], and adults [9, 10] to identify the sources explaining the wide variation in PK among patients. Genetic variants prevailing in the drug transporter and metabolizing genes can also contribute to inter-individual variation among patients as these variants may alter the expression, structure, and function of the enzymes. Major enzymes involved in metabolism of Dnr are carbonyl reductase 1 (CBR1) and carbonyl reductase 3 (CBR3), which convert Dnr into lesser cytotoxic hydroxyl derivative Daunorubicinol (DOL). Polymorphisms in CBR1 and CBR3 have also been shown to influence disposition and toxicity of doxorubicin in breast-cancer patients [25]. A recent population PK model developed in patients aged less than 21 years with hematological malignancies suggested that polymorphisms in FMO3 and GSTP1 influence the efficacy and toxicity of Dnr [11]. However, the effect of mRNA expression and polymorphisms of Dnr efflux transporters and metabolizing genes on the PK of Dnr in AML patients has not been carried out previously. We carried out a prospective study evaluating the factors which may influence the plasma PK of Dnr and its metabolite Daunorubicinol (DOL) and whether the PK of Dnr and DOL influence the treatment outcome in AML.
Patients and methods
Patients
Seventy adult de novo AML (excluding AML-M3) patients treated with standard induction chemotherapy between 2009 and 2014 at the Department of Hematology, Christian Medical College, Vellore, were enrolled in this study. Bone marrow or peripheral blood samples at diagnosis before the initiation of therapy, as well as samples for pharmacokinetic analysis, were collected after obtaining written informed consent from the patients, and this study was approved by the Institutional Review Board.
Diagnosis of AML was carried out by standard morphology, cytochemistry, and Immunophenotyping techniques. Cytogenetic analysis of bone marrow samples was performed using standard protocols. Karyotypes were reported following the International System of Human Cytogenetic Nomenclature (ISCN) 2005. Patients were classified as good, intermediate, and poor risk cytogenetics groups. FLT3-ITD and NPM1 mutation status at diagnosis were determined by PCR followed by Gene Scan analysis using genomic DNA samples as reported previously [12–14].
Chemicals and reagents
Daunorubicin hydrochloride and Tri Reagent were obtained from Sigma-Aldrich. Daunorubicinol was from Toronto Research Chemicals, Canada. All other reagents and solvents including acetonitrile, chloroform, and n-heptanol were of the high-performance liquid chromatography (HPLC) grade. Ficol-Paque was from GE Healthcare (Uppsala, Sweden).
Pharmacokinetic sampling
Dnr was administered to AML patients on the first three days as intravenous bolus infusion for 1 h (60 mg/m2/day). Peripheral blood was collected (in EDTA tubes) at 0, 0.25, 1, 2, 4, 6, and 24 h on the first day of the induction therapy. Plasma was separated by centrifuging the sample at 4000 rpm for 10 min and stored at −80 °C until analysis.
Estimation of plasma Dnr/DOL levels
Levels of Dnr and DOL in plasma samples were analyzed using high-performance liquid chromatography (HPLC) coupled with fluorescence detector as reported previously [15]. Briefly, plasma samples were treated with 0.2 M sodium (dibasic) phosphate buffer (pH 8.4) using idarubicin as internal standard (100 ng/ml). Processed sample (50 μl) was used for injection. HPLC (LC10 AT, Shimadzu) coupled with fluorescence detector (RF10AXL, Shimadzu) and an autosampler (Bio-Rad AS 100, CA, USA) were used. Phenomenex Luna Cyano column (250 × 4.6 mm × 5 μm) protected with guard column (μBondapak-CN precolumn) was used for separation. The mobile phase was composed of a mixture (35:65 v/v) of acetonitrile and 50 mM monobasic sodium phosphate, pH 4.5. The elution was carried out in an isocratic mode with a flow rate of 0.7 ml/min. Standards (100–1000 ng/ml) for daunorubicin and DOL (100–500 ng/ml) were included in every batch of samples. The retention times of Dnr, DOL, and the internal standard were 8.7, 7.6, and 9.5 min, respectively. The limit of detection (LOD) and limit of quantification (LOQ) for both Dnr and DOL were 1 and 10 ng/ml, respectively. The inter-day and intraday coefficients of variation (CV) were less than 10 % for the concentrations used. Area of Dnr and DOL peaks were normalized with area of IS peak, and amount of Dnr and DOL were obtained by extrapolating the values in the standard graphs of Dnr and DOL. Concentration of Dnr and DOL were expressed as ng/ml. Samples were appropriately diluted and re-analyzed when the concentrations were beyond the upper limit of quantification (ULOQ) of the assay.
Pharmacokinetic analysis
Nonlinear mixed-effects modeling analysis was performed with Monolix (version 4.3.2) using the Stochastic Approximation Expectation–Maximization (SAEM) method. A two-compartment pharmacokinetic model was used to describe each component (Supplemental Fig. 1). The pharmacokinetic parameters estimated included apparent clearance and volume (CL (L/hr) and V (L)) along with the inter-compartmental rate constants (k12 and k21 (1/hr)) for each component. The inter-individual variability of the parameters was assumed to be log normally distributed. A proportional residual error model was used with assumed normal distribution of the residuals. Also, the individual post hoc parameter values were used to estimate the area under the concentration curve (AUC) for Dnr and DOL in plasma.
Pharmacogenetic analysis
Polymorphisms in CBR1 and CBR3 (exons 1 and 3), ABCG2 (Exon 2 and 5) and ABCB1 (exon21 and 26) were screened using direct sequencing or restriction fragment length polymorphism (RFLP) as described previously [16, 17].
RNA expression of CBR1 and ABC transporters
Bone marrow mononuclear cells (MNCs) (collected at diagnosis) were stored in Tri Reagent (Sigma-Aldrich), and RNA was extracted. One microgram of RNA was converted to cDNA using Superscript II (Invitrogen) firststrand cDNA synthesis system. RNA expression of CBR1 (Taqman assay ID Hs00156323_m1), ABCB1 (Taqman assay ID Hs 01067802_m1), and ABCG2 (Taqman assay ID Hs 01053787_m1) were normalized to the expression of the housekeeping gene GAPDH (Taqman assay ID 4352934E). Samples were analyzed in duplicates, and relative expression was calculated using 2-ddCT (where pooled normal control samples were used as reference).
Clinical outcome
Complete remission is defined as bone marrow blasts <5 %; absence of extramedullary disease; absence of blasts with Auer rods; absolute neutrophil count >1.0 × 109/L (1000/μL); platelet count >100 × 109/L (100,000/μL); and independence of red cell transfusions after induction chemotherapy [18]. Induction death is defined as death within 28 days of treatment initiation and before documentation of complete remission. Relapse of the disease after CR is defined as reappearance of leukemic blasts in the peripheral blood or ≥5 % blasts in the bone marrow not attributable to any other cause (e.g., bone marrow regeneration after consolidation therapy).
Statistical analysis
Statistical analysis was performed using GraphPad Prism (California, USA) or SPSS 16.0 (Chicago, USA) software as appropriate. Mann–Whitney U test, Kruskal–Wallis test, and Spearman rank correlation analysis were used as appropriate. A p value of <0.05 was considered statistically significant. The probability of survival was estimated with the use of the product-limit method of Kaplan–Meier for overall survival (OS), disease-free survival (DFS) and was compared using the log-rank test. All survival estimates are reported as ±1SE. All P values were two-sided, with values of 0.05 or less indicating statistical significance. Cox proportional hazard regression was performed for the determination of variables affecting OS and DFS.
Results
Patients
From 2009 to 2014, 70 adult AML patients (who agreed to participate in the study and gave informed consent) undergoing conventional induction chemotherapy comprising of Ara-C and Dnr at our center were enrolled in this study. Patient demographics are summarized in Table 1.
Table 1. Patient demographics (De novo AML (n = 70)).
| Age | 38 years (16–60 years) |
| Sex | Males: 42; females: 33 |
| Cytogenetics risk group | |
| Favorable | 8 (12 %) |
| Intermediate | 47 (68 %) |
| Adverse | 15 (20 %) |
| Classification by WHO | |
| AML with recurrent genetic abnormalities | 9 |
| AML with MDS related features | 34 |
| AML not otherwise specified | 25 |
| NPM1 mutation | 24 (34.28 %) |
| FLT3-ITD | 19 (27.14 %) |
| NPM1 +/FLT 3 -ITD+ | 15 (21.42 %) |
| FLT3-TKD | 6 (8.5 %) |
| WBC | 26,000/cu mm (1100–324,100) |
| Platelet | 44,000/cu mm (4000–242,000) |
| Blast (PB) | 47 % (0–97) |
| Hb | 8.7 mg % (3.6–14.2) |
| LDH | 868 U/l (228–3640) |
| Creatinine | 0.975 mg % (0.37–2.2) |
Plasma pharmacokinetics of Dnr in AML patients
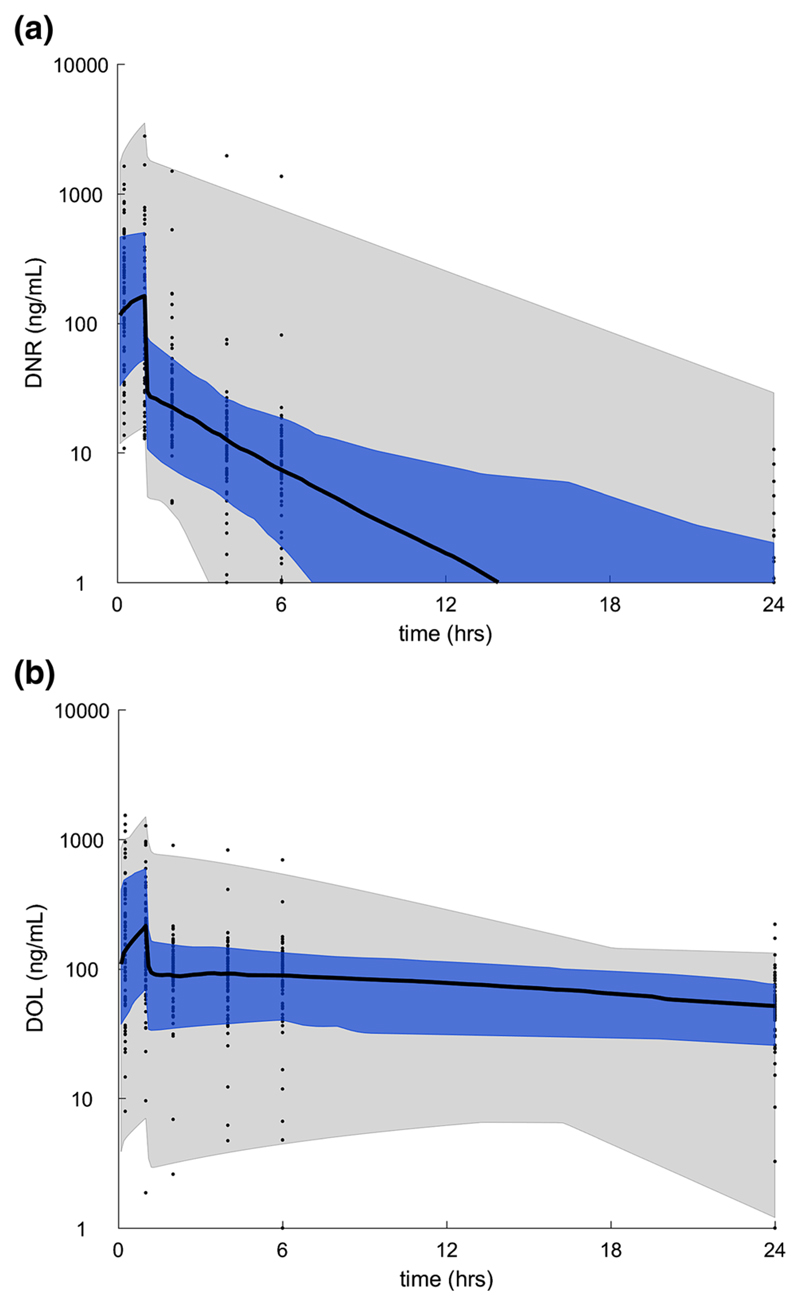
Plasma levels of Dnr and DOL were available in all the 70 patients. Population pharmacokinetics of plasma Dnr and DOL in AML are summarized in Table 2. Wide variation in AUC and clearance of both plasma Dnr and DOL was observed (Supplementary Table 1). The median concentration of DNR was lower than that of DOL in the plasma (Fig. 1a, b).
Table 2. Population pharmacokinetics of plasma Dnr and DOL in AML.
| Parameter | Population mean | rse (%) | Inter-individual variability (CV %) | rse (%) |
|---|---|---|---|---|
| DNR | ||||
| Clearance (L/hr) | 269.8 | 10 | 84 | 18 |
| Volume (L) | 15.0 | 21 | 143 | 21 |
| K12 (1/hr) | 22.4 | 16 | 69 | 43 |
| K21(1/hr) | 0.6 | 9 | 72 | 19 |
| DOL | ||||
| Clearance (L/hr) | 23.6 | 11 | 64 | 32 |
| Volume (L) | 7.6 | 16 | 65 | 42 |
| K12 (1/hr) | 32.0 | 18 | 73 | 39 |
| K21(1/hr) | 0.4 | 10 | 68 | 27 |
| Residual error | Relative error (CV %) | |||
| DNR | 47 | 6 | ||
| DOL | 34 | 6 |
rse relative standard error
Fig. 1.
Plasma Dnr and DOL PK. Plasma concentration/time curve for (a) Dnr and (b) DOL in AML patients receiving a 60 mg/m2 dose of Dnr. The black curve is the median, the blue shaded region is the 25th–75th percentile, and the gray shaded region is the 5th–95th percentile of the population pharmacokinetics
mRNA expression of CBR1 correlates with AUC of plasma DOL
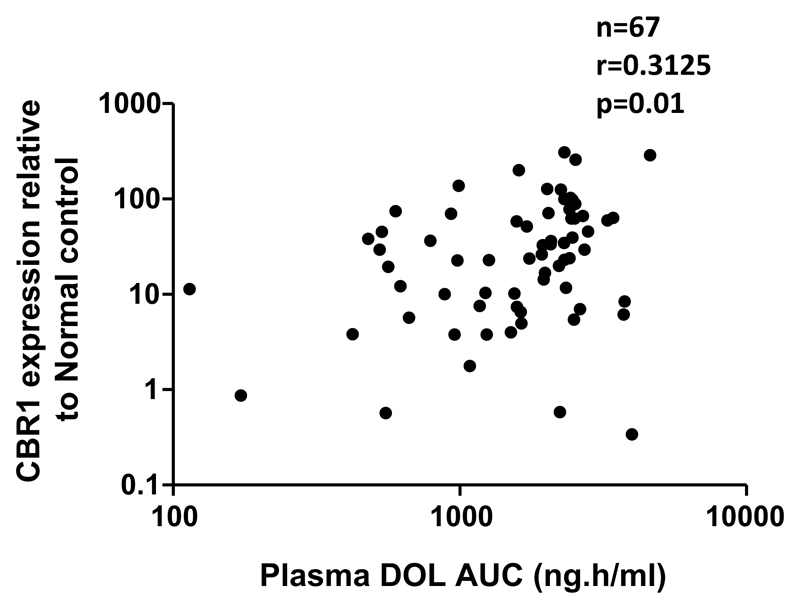
As Dnr efflux transporters and metabolizing genes have been reported to influence the treatment outcome in AML, we evaluated the role of mRNA expression of ABCB1, ABCG2, CBR1, and CBR3 on Dnr PK. Expression of CBR1 showed a positive correlation with plasma DOL AUC (r = 0.3125; p = 0.01) (Fig. 2). However, mRNA expression of ABCB1, ABCG2, and CBR3 did not show significant association with any of the PK parameters (Supplementary Table. 3).
Fig. 2.
CBR1 mRNA expression influences plasma DOL AUC. Patients with higher expression of CBR1 had higher plasma DOL AUC than patients with lower CBR1 expression. Spearman correlation was used to assess the association between CBR1 expression and plasma DOL AUC. mRNA expression of CBR1 was evaluable for 67 patients only and undetectable in three patients
Common polymorphisms in the genes involved in transport and metabolism of Dnr influence pharmacokinetics of Dnr and DOL in AML patients
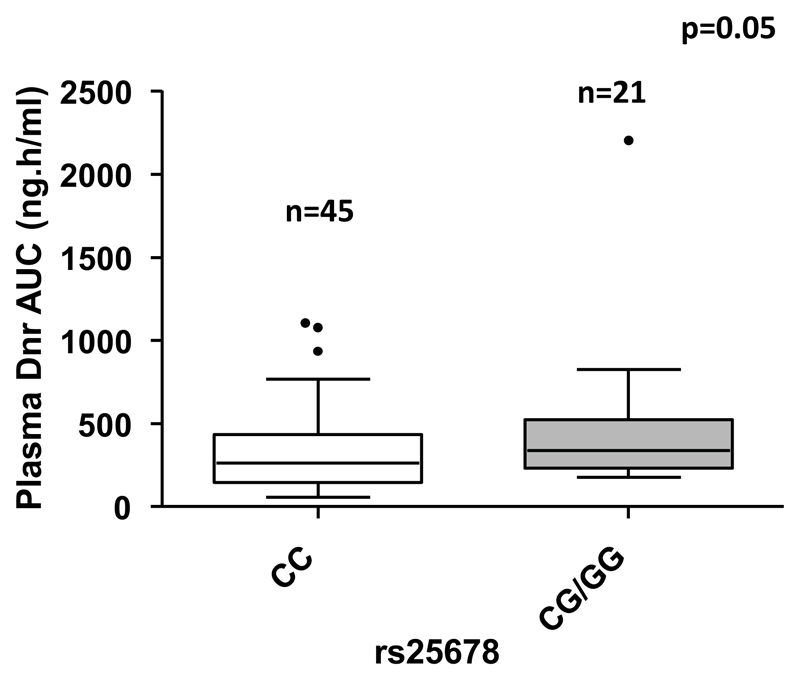
Polymorphisms in efflux transporters and metabolizing genes have been previously shown to influence PK of anthracyclines like doxorubicin in solid tumors. We screened for the known polymorphisms in ABCB1, ABCG2, CBR1, and CBR3, and compared with the PK parameters of Dnr and DOL. The allelic frequencies of common polymorphisms ABCB1, ABCG2, CBR1, and CBR3 are listed in Supplementary Table. 2. Patients with rs25678 variant genotype had significantly higher plasma Dnr AUC [339.4 (179–2203) vs. 262 (56–1106) ng*h/ml; p = 0.05] (Fig. 3). None of the other polymorphisms screened showed any significant association with PK parameters of Dnr and DOL.
Fig. 3.
Influence of CBR1 (rs25678) genetic variant on plasma Dnr exposure. AML patients with CBR1 variant genotype (rs25678) showed higher plasma Dnr AUC than patients with wild type
Daunorubicin PK in AML is not influenced by NPM1 or FLT3-ITD mutations
Various molecular and cytogenetic prognostic markers have been reported to influence the outcome of therapy in AML. We compared Dnr PK parameters in patients carrying mutations in NPM1 or FLT3, with those who did not have these mutations. There was no significant influence of these mutations on the plasma pharmacokinetic parameters of Dnr and DOL (data not shown).
Role of Dnr PK on clinical outcome in AML patients
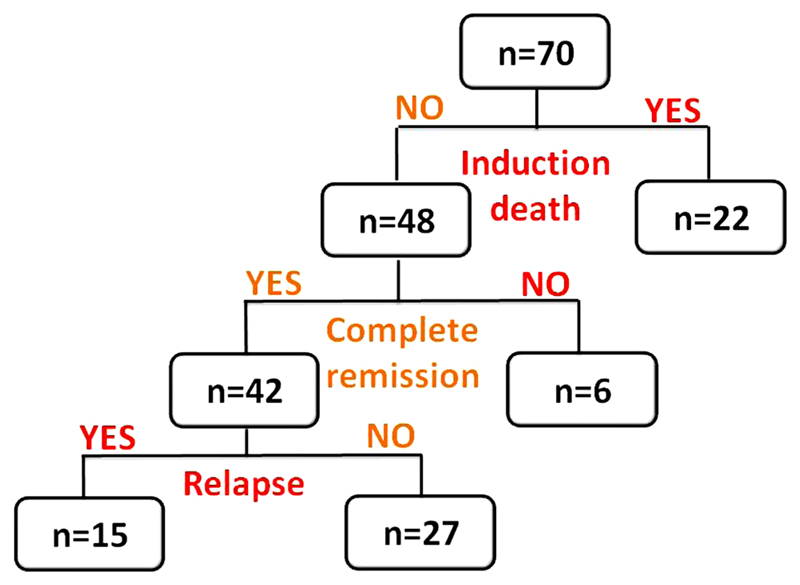
Figure 4 shows the breakdown of AML patients based on clinical events. Induction death occurred in 22 of the 70 patients (31.42 %). Of the remaining 48 patients, 42 (87.5 %) achieved complete remission (CR1). Fifteen of those patients who achieved CR1 relapsed (35.7 %).
Fig. 4.
Flowchart depicting breakdown of AML patients based on clinical events
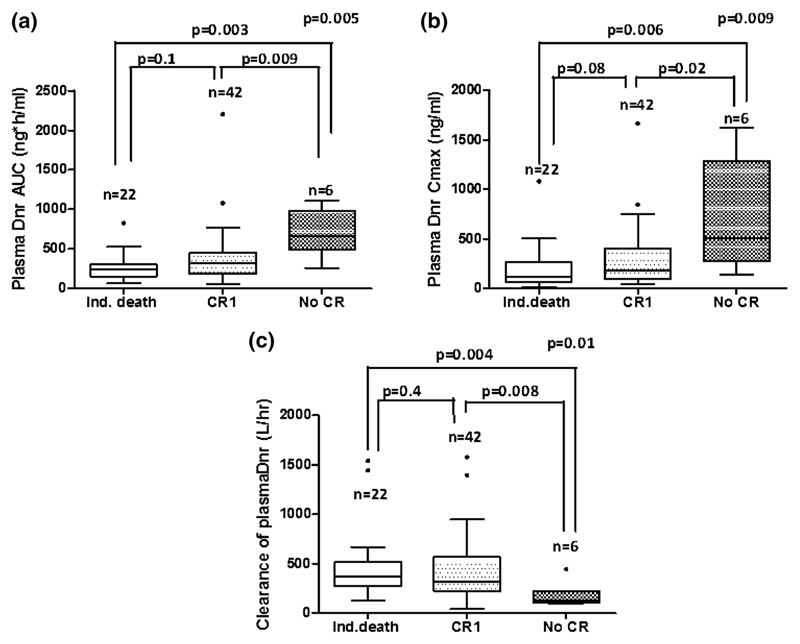
Pharmacokinetic parameters were compared with the clinical outcome such as achievement of CR1 and relapse. Patients achieving CR1 showed significantly lower plasma Dnr AUC (314 ng.h/ml (56–2203) vs. 339.4 ng.h/ml (113–1076); p = 0.005; Fig. 5a), lower Cmax (182 ng/ml (45–1668) vs. 507 ng/ml (135–1627); p = 0.009; Fig. 5b), and higher clearance (321.5 L/hr (41.38–1577) vs. 125 L/hr (93–444); p = 0.01; Fig. 5c) compared to those who did not achieve CR1.
Fig. 5.
Association of Dnr PK with achieving CR1. Patients who achieved CR1 showed significantly a lower plasma Dnr AUC b lower Cmax and c higher clearance compared to those who did not achieve CR1
Discussion
To the best of our knowledge, this is the first study evaluating the influence of RNA expression and genetic variants in the Dnr transporter and metabolism genes, on the pharmacokinetics of Dnr and its major metabolite DOL in adult AML patients.
Inter-individual variation in exposure and clearance of a drug influences the efficacy, toxicity, and ultimately, the treatment outcome of a disease. Similar to the previous reports [3, 9, 10], wide inter-individual variation in the pharmacokinetic parameters of both Dnr and DOL was observed in this study. Dnr levels were lower than DOL in the plasma, which is in line with previous reports [3, 19].
Among numerous oxido-reductase enzymes present in the cell, CBR1 is the most efficient one in the conversion of Dnr to DOL in AML [20]. We had previously shown that CBR1 expression in primary AML samples influences the ex vivo cytotoxicity of Dnr and ex vivo intracellular DOL levels [16]. In the present study, we show that CBR1 expression is positively correlated with plasma AUC of DOL, which is the major metabolite of Dnr known to be causing cardiotoxicity. Inhibiting CBR1 expression has been shown as a strategy to improve the efficacy of daunorubicin in various cancers [21, 22]. CBR1 inhibition has been shown to overcome arsenic trioxide (AS2O3) resistance in AML cells [23] and proposed as a rational target for the development of combination chemotherapy with As2O3 in diverse leukemia including APL.
Polymorphisms in the coding or regulatory regions of the genes encoding drug metabolizing enzymes may have an impact on phase I metabolism of both endogenous and xenobiotic compounds [24]. We had previously shown that a polymorphism in CBR1, rs25678, influenced plasma AUC of Dnr in a small subset of AML patients [16]. We extended this study and screened common polymorphisms in ABCB1, ABCG2, and CBR3 along with CBR1 in a larger cohort of patients. Patients carrying the variant genotype of rs25678 had significantly higher plasma AUC of Dnr. Our finding is consistent with a report on Asian breast-cancer patients where the presence of at least one CBR1 genetic variant (either the synonymous SNP rs25678 or the 3′UTR variant rs9024) was shown to have higher systemic exposure and lower doxorubicin clearance [25].
It has been reported that synonymous SNPs may alter RNA stability, translational efficiency and may contribute to drug disposition. The 3′UTR variant, rs9024, showed similar trend with plasma Dnr AUC, though statistically not significant (p = 0.1635) (data not shown). This polymorphism has been shown to be at the miR-574-5p and miR-921 binding sites [26]. The presence of this polymorphism has shown to alter the binding of miRNA and subsequently affect the protein expression and activity of CBR1 [26]. Two non-synonymous polymorphisms in CBR3 (rs8133502 and rs1056892) are shown to influence doxorubicin disposition and toxicity [27, 28]. None of the CBR3 polymorphisms screened in the present study showed any significant influence on the pharmacokinetics of Dnr and DOL.
There are controversial reports on the role of common polymorphisms in ABCB1 and ABCG2 on drug disposition of various anticancer drugs [29, 30]. We studied the effect of these polymorphisms on pharmacokinetics of Dnr in AML. The presence of non-synonymous polymorphism rs2231142 (G421A) in exon5 of ABCG2 has been shown to reduce protein expression and function of ABCG2 [31, 32]. However, in the present study, none of the ABCB1 and ABCG2 polymorphisms showed any significant association with plasma pharmacokinetics of Dnr. This is consistent with a previous report where there was no significant association with these polymorphisms on doxorubicin pharmacokinetics in breast-cancer patients [33].
When we tested the impact of plasma Dnr pharmacokinetics on treatment outcome in AML, we observed that patients who did not achieve complete remission had higher plasma Dnr AUC and Cmax as well as lower clearance than those who achieved CR. Kokenberg et al. [3] reported that plasma AUC values of Dnr correlated inversely with AUC values of Dnr in white blood cells; however, none of the plasma or cellular levels of Dnr predicted treatment outcome. Previous reports have shown that patients who achieve complete remission had higher intracellular Dnr AUC [19] and higher Dnr concentration after the end of infusion of Dnr [9]. Though we did not have data on the intracellular Dnr AUC, we could extrapolate that the patients who achieved CR had higher intracellular Dnr exposure as these patients had lower plasma Dnr exposure than those who did not achieve CR. These findings need to be validated in a larger cohort of AML patients and potentially help us establish a therapeutic window for plasma Dnr levels and targeted dose adjustment.
Supplementary Material
Electronic supplementary material The online version of this article (doi:10.1007/s00280-016-3166-8) contains supplementary material, which is available to authorized users.
Acknowledgments
This study is supported by DBT Program support grant: BT/01/COE/08/03 and ICMR grant No.50/4/2010/BMS to Dr. Poonkuzhali Balasubramanian SV is a recipient of the senior research fellowship from ICMR. AA and SK are funded by University Grants Commission, Government of India. VM and PB are supported by Wellcome DBT India Alliance (IA/S/11/2500267 and IA/S/15/1/501842).
Footnotes
Author contributions Savitha Varatharajan performed the research, analyzed data, and wrote the paper, John C Panetta performed population pharmacokinetic analysis, Ajay Abraham performed the research, wrote the paper, Sreeja Karathedath: performed the research, Ezhilpavai Mohanan performed the research, Kavitha M.L performed statistical analysis, Nancy Arthur performed cytogenetics analysis, Vivi M Srivastava performed cytogenetics analysis, Sandeep Nemani clinical data accrual, Biju George clinical data accrual, Alok Srivastava clinical data accrual, Vikram Mathews clinical data accrual, administrative support, and wrote the paper, Poonkuzhali Balasubramanian designed study, performed research, analyzed data, and wrote paper.
Conflict of interest We declare that none of the authors have any potential conflicts of interest to disclose.
Ethical approval This study was approved by the Institutional review board (EC Min. No. IRB-8-19-03-2008; IRB Min. No. 7173 dated 09/06/2010. All patients were enrolled in the study after obtaining the written informed consent from the patients. The formats of patient information sheet and informed consent were approved by the institutional review board before the study was initiated.
References
- 1.Döhner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 2.Dombret H, Gardin C. An update of current treatments for adult acute myeloid leukemia. Blood. 2016;127(1):53–61. doi: 10.1182/blood-2015-08-604520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kokenberg E, Sonneveld P, Sizoo W, Hagenbeek A, Löwenberg B. Cellular pharmacokinetics of daunorubicin: relationships with the response to treatment in patients with acute myeloid leukemia. J Clin Oncol Off J Am Soc Clin Oncol. 1988;6(5):802–812. doi: 10.1200/JCO.1988.6.5.802. [DOI] [PubMed] [Google Scholar]
- 4.Speth PA, Linssen PC, Boezeman JB, Wessels HM, Haanen C. Leukemic cell and plasma daunomycin concentrations after bolus injection and 72 h infusion. Cancer Chemother Pharmacol. 1987;20(4):311–315. doi: 10.1007/BF00262582. [DOI] [PubMed] [Google Scholar]
- 5.Greene W, Huffman D, Wiernik PH, Schimpff S, Benjamin R, Bachur N. High-dose daunorubicin therapy for acute nonlymphocytic leukemia: correlation of response and toxicity with pharmacokinetics and intracellular daunorubicin reductase activity. Cancer. 1972;30(6):1419–1427. doi: 10.1002/1097-0142(197212)30:6<1419::aid-cncr2820300602>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 6.Trillet V, Lakhal M, Lang J, Perrin-Fayolle E, Timour Chah Q, Fière D, et al. Cellular pharmacokinetics of daunorubicin: uptake by leukaemic cells in vivo and fate. Eur J Clin Pharmacol. 1985;29(1):127–129. doi: 10.1007/BF00547382. [DOI] [PubMed] [Google Scholar]
- 7.Hempel G, Relling MV, de Rossi G, Stary J, De Lorenzo P, Valsecchi MG, et al. Pharmacokinetics of daunorubicin and daunorubicinol in infants with leukemia treated in the interfant 99 protocol. Pediatr Blood Cancer. 2010;54(3):355–360. doi: 10.1002/pbc.22266. [DOI] [PubMed] [Google Scholar]
- 8.Hempel G, Reinhardt D, Creutzig U, Boos J. Population pharmacokinetics of liposomal daunorubicin in children. Br J Clin Pharmacol. 2003;56(4):370–377. doi: 10.1046/j.1365-2125.2003.01886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogason A, Quartino AL, Lafolie P, Masquelier M, Karlsson MO, Paul C, et al. Inverse relationship between leukaemic cell burden and plasma concentrations of daunorubicin in patients with acute myeloid leukaemia. Br J Clin Pharmacol. 2011;71(4):514–521. doi: 10.1111/j.1365-2125.2010.03894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krogh-Madsen M, Bender B, Jensen MK, Nielsen OJ, Friberg LE, Honoré PH. Population pharmacokinetics of cytarabine, etoposide, and daunorubicin in the treatment for acute myeloid leukemia. Cancer Chemother Pharmacol. 2012;69(5):1155–1163. doi: 10.1007/s00280-011-1800-z. [DOI] [PubMed] [Google Scholar]
- 11.Thompson P, Wheeler HE, Delaney SM, Lorier R, Broeckel U, Devidas M, et al. Pharmacokinetics and pharmacogenomics of daunorubicin in children: a report from the Children’s Oncology Group. Cancer Chemother Pharmacol. 2014;74(4):831–838. doi: 10.1007/s00280-014-2535-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto Y, Kiyoi H, Nakano Y, Suzuki R, Kodera Y, Miyawaki S, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97(8):2434–2439. doi: 10.1182/blood.v97.8.2434. [DOI] [PubMed] [Google Scholar]
- 13.Parihar M, Kumar JA, Sitaram U, Balasubramanian P, Abraham A, Viswabandya A, et al. Cytogenetic analysis of acute myeloid leukemia with t(8;21) from a tertiary care center in India with correlation between clinicopathologic characteristics and molecular analysis. Leuk Lymphoma. 2012;53(1):103–109. doi: 10.3109/10428194.2011.603447. [DOI] [PubMed] [Google Scholar]
- 14.Gupta M, Ashok Kumar J, Sitaram U, Neeraj S, Nancy A, Balasubramanian P, et al. The t(6;9)(p22;q34) in myeloid neoplasms: a retrospective study of 16 cases. Cancer Genet Cytogenet. 2010;203(2):297–302. doi: 10.1016/j.cancergencyto.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 15.Fogli S, Danesi R, Innocenti F, Di Paolo A, Bocci G, Barbara C, et al. An improved HPLC method for therapeutic drug monitoring of daunorubicin, idarubicin, doxorubicin, epirubicin, and their 13-dihydro metabolites in human plasma. Ther Drug Monit. 1999;21(3):367–375. doi: 10.1097/00007691-199906000-00022. [DOI] [PubMed] [Google Scholar]
- 16.Varatharajan S, Abraham A, Zhang W, Shaji RV, Ahmed R, Abraham A, et al. Carbonyl reductase 1 expression influences daunorubicin metabolism in acute myeloid leukemia. Eur J Clin Pharmacol. 2012;68(12):1577–1586. doi: 10.1007/s00228-012-1291-9. [DOI] [PubMed] [Google Scholar]
- 17.Cascorbi I, Gerloff T, Johne A, Meisel C, Hoffmeyer S, Schwab M, et al. Frequency of single nucleotide polymorphisms in the P-glycoprotein drug transporter MDR1 gene in white subjects. Clin Pharmacol Ther. 2001;69(3):169–174. doi: 10.1067/mcp.2001.114164. [DOI] [PubMed] [Google Scholar]
- 18.Cheson BD. Overview of the revised response criteria for acute myelogenous leukemia. Clin Adv Hematol Oncol HO. 2004;2(5):277–279. [PubMed] [Google Scholar]
- 19.Galettis P, Boutagy J, Ma DD. Daunorubicin pharmacokinetics and the correlation with P-glycoprotein and response in patients with acute leukaemia. Br J Cancer. 1994;70(2):324–329. doi: 10.1038/bjc.1994.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plebuch M, Soldan M, Hungerer C, Koch L, Maser E. Increased resistance of tumor cells to daunorubicin after transfection of cDNAs coding for anthracycline inactivating enzymes. Cancer Lett. 2007;255(1):49–56. doi: 10.1016/j.canlet.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 21.Hintzpeter J, Hornung J, Ebert B, Martin H-J, Maser E. Curcumin is a tight-binding inhibitor of the most efficient human daunorubicin reductase–Carbonyl reductase 1. Chem Biol Interact. 2015;5(234):162–168. doi: 10.1016/j.cbi.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 22.Tak E, Lee S, Lee J, Rashid MA, Kim YW, Park J-H, et al. Human carbonyl reductase 1 upregulated by hypoxia renders resistance to apoptosis in hepatocellular carcinoma cells. J Hepatol. 2011;54(2):328–339. doi: 10.1016/j.jhep.2010.06.045. [DOI] [PubMed] [Google Scholar]
- 23.Jang M, Kim Y, Won H, Lim S, Jyothi KR, Dashdorj A, et al. Carbonyl reductase 1 offers a novel therapeutic target to enhance leukemia treatment by arsenic trioxide. Cancer Res. 2012;72(16):4214–4224. doi: 10.1158/0008-5472.CAN-12-1110. [DOI] [PubMed] [Google Scholar]
- 24.Oppermann UC, Maser E. Molecular and structural aspects of xenobiotic carbonyl metabolizing enzymes. Role of reductases and dehydrogenases in xenobiotic phase I reactions. Toxicology. 2000;144(1–3):71–81. doi: 10.1016/s0300-483x(99)00192-4. [DOI] [PubMed] [Google Scholar]
- 25.Lal S, Sandanaraj E, Wong ZW, Ang PCS, Wong NS, Lee EJD, et al. CBR1 and CBR3 pharmacogenetics and their influence on doxorubicin disposition in Asian breast cancer patients. Cancer Sci. 2008;99(10):2045–2054. doi: 10.1111/j.1349-7006.2008.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalabus JL, Cheng Q, Blanco JG. MicroRNAs differentially regulate carbonyl reductase 1 (CBR1) gene expression dependent on the allele status of the common polymorphic variant rs9024. PLoS ONE. 2012;7(11):e48622. doi: 10.1371/journal.pone.0048622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bains OS, Karkling MJ, Lubieniecka JM, Grigliatti TA, Reid RE, Riggs KW. Naturally occurring variants of human CBR3 alter anthracycline in vitro metabolism. J Pharmacol Exp Ther. 2010;332(3):755–763. doi: 10.1124/jpet.109.160614. [DOI] [PubMed] [Google Scholar]
- 28.Fan L, Goh B-C, Wong C-I, Sukri N, Lim S-E, Tan S-H, et al. Genotype of human carbonyl reductase CBR3 correlates with doxorubicin disposition and toxicity. Pharmacogenet Genomics. 2008;18(7):621–631. doi: 10.1097/FPC.0b013e328301a869. [DOI] [PubMed] [Google Scholar]
- 29.Bruhn O, Cascorbi I. Polymorphisms of the drug transporters ABCB1, ABCG2, ABCC2 and ABCC3 and their impact on drug bioavailability and clinical relevance. Expert Opin Drug Metab Toxicol. 2014;10(10):1337–1354. doi: 10.1517/17425255.2014.952630. [DOI] [PubMed] [Google Scholar]
- 30.Lepper ER, Nooter K, Verweij J, Acharya MR, Figg WD, Sparreboom A. Mechanisms of resistance to anticancer drugs: the role of the polymorphic ABC transporters ABCB1 and ABCG2. Pharmacogenomics. 2005;6(2):115–138. doi: 10.1517/14622416.6.2.115. [DOI] [PubMed] [Google Scholar]
- 31.Imai Y, Nakane M, Kage K, Tsukahara S, Ishikawa E, Tsuruo T, et al. C421A polymorphism in the human breast cancer resistance protein gene is associated with low expression of Q141 K protein and low-level drug resistance. Mol Cancer Ther. 2002;1(8):611–616. [PubMed] [Google Scholar]
- 32.Morisaki K, Robey RW, Ozvegy-Laczka C, Honjo Y, Polgar O, Steadman K, et al. Single nucleotide polymorphisms modify the transporter activity of ABCG2. Cancer Chemother Pharmacol. 2005;56(2):161–172. doi: 10.1007/s00280-004-0931-x. [DOI] [PubMed] [Google Scholar]
- 33.Lal S, Wong ZW, Sandanaraj E, Xiang X, Ang PCS, Lee EJD, et al. Influence of ABCB1 and ABCG2 polymorphisms on doxorubicin disposition in Asian breast cancer patients. Cancer Sci. 2008;99(4):816–823. doi: 10.1111/j.1349-7006.2008.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.