Abstract
Interface tryptophans are key residues that facilitate the folding and stability of membrane proteins. Escherichia coli OmpX possesses two unique interface tryptophans, namely Trp76, which is present at the interface and is solvent-exposed, and Trp140, which is relatively more lipid solvated than Trp76 in symmetric lipid membranes. Here, we address the requirement for tryptophan and the consequences of aromatic amino acid substitutions on the folding and stability of OmpX. Using spectroscopic measurements of OmpX-Trp/Tyr/Phe mutants, we show that the specific mutation W76 → Y allows barrel assembly > 1.5-fold faster than native OmpX, and increases stability by ~0.4 kcalmol−1. In contrast, mutating W140 → F/Y lowers OmpX thermodynamic stability by ~0.4 kcal mol−1, without affecting the folding kinetics. We conclude that the stabilizing effect of tryptophan at the membrane interface can be position—and local environment—specific. We propose that the thermodynamic contributions for interface residues be interpreted with caution.
Keywords: Outer membrane protein, Interface, Membrane protein folding, Kinetics, Stability, β-Barrel
1. Introduction
Integral β-barrel membrane proteins (OMPs) are major structural determinants of the bacterial outer membrane. Among the array of forces that stabilize a membrane protein in its lipid environment, protein-lipid interactions play a key role in deciding the folded functional form of the mature OMP [1,2]. In turn, strategically placed amino acids in the protein scaffold govern protein-lipid interactions. For example, it is seen that aromatic amino acids cluster largely at the solvent-membrane interface [3], and can contribute significantly to the stability of membrane proteins.
Interface tryptophans play an important role in the folding of several bacterial and mitochondrial OMPs [4–8]. Tryptophan exhibits a membrane depth-dependent contribution in its ability to anchor transmembrane β-barrels. While some studies suggest that tryptophan stabilizes OMPs at the bilayer midplane [9], others show that this contribution is highest at the interface [10]. Further, our study with the model OMP PagP showed that tryptophan is less stabilizing than tyrosine or phenylalanine at the interface [11]. Interestingly, it is the rigid aryl ring, and not the hydrogen bonding, which drives the partitioning of tryptophan to the interface [12]. This observation raises questions on why a metabolically expensive amino acid such as tryptophan is required in an aromatic interface girdle that is otherwise enriched with tyrosines and phenylalanines.
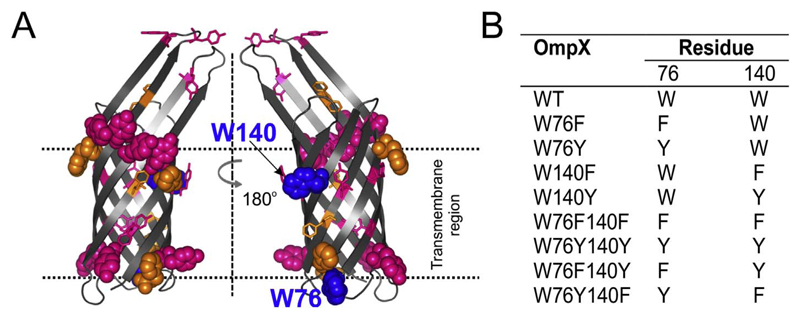
To address the need for tryptophan at the bilayer interface, we carried out a detailed thermodynamic analysis on tyrosine and phenylalanine variants of the E. coli OMP, OmpX. OmpX has two tryptophans at positions 76 and 140. Both tryptophans of OmpX are located near the membrane interface (Fig. 1A); the indole side chains point toward the membrane and can establish non-covalent interactions with the lipid headgroup and hydrocarbon tail. In OmpX, Trp76 forms a part of the lower aromatic girdle, which also contains two phenylalanine and four tyrosine residues (Fig. 1A). Trp140 resides in the upper aromatic girdle, which is more enriched with aromatic residues, particularly tyrosine (eight tyrosine and three phenylalanine) residues, when compared to the lower aromatic girdle (Fig. 1A). OMPs usually contain tryptophan and tyrosine in the aromatic girdles, while phenylalanine shows a distributed occupancy between the interface and the midplane [13]. We have previously observed that Trp140 is more shielded from the solvent than Trp76 [6]. In folded OmpX, Trp140 is likely to be located closer to the midplane when compared with Trp76, in symmetrical bilayer membranes.
Fig. 1.
OmpX has two interface tryptophans. (A) Ribbon diagram of OmpX barrel highlighting phenylalanines (orange), tyrosines (pink), and the two tryptophans (blue) as spheres (interface) or sticks (others). (B) List of the OmpX mutants discussed in this study.
In this study, we generated OmpX mutants where one or both tryptophans were systematically replaced by tyrosine or phenylalanine or both (W76F140Y and W76Y140F) (Fig. 1B). By comparing folding kinetics and equilibrium free energy values, we find that a favorable energetic contribution of an interface tryptophan, when compared to its other aromatic counterparts, can be contextual. Structural plasticity of the extracellular loops of the OmpX family of barrels is required for its binding to host proteins [14]. Hence, we propose that a gain in conformational flexibility at the expense of stability might be required for protein function.
2. Experimental methods
2.1. Protein preparation
OmpX expression, purification, and folding were carried out using reported methods [6,15], and are detailed in the supporting information. The folded protein stock contained 150 μM OmpX, 50 mM DPC (n-dodecylphoshphocholine), 50 mM Tris-HCl pH 9.5 and 400 mM urea. Folded protein stocks were diluted 5-fold to 30 μM protein and 10 mM DPC in all experiments, unless specified otherwise.
2.2. Folding rates in micelles and bicelles
The rate of OmpX folding was determined using far-UV circular dichroism (CD) spectropolarimetry on Jasco J815 CD spectropolarimeter (Jasco Inc., Japan). Briefly, 150 μM unfolded protein in 8 M urea was prepared with 50 mM DPC micelles or DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine): DPC bicelles (q = 0.5) at 25 °C. Additionally, folding rates were also determined in DMPC:DPC bicelles doped with 20% DMPG (1,2-dimyristoyl-sn-glycero-3-(1′-rac-glycerol)). This corresponds to a DMPC:DMPG ratio of 1:0.25 and an overall bicelle q of 0.5. In the presence of 8 M urea, the protein scaffold is unfolded, but OmpX possesses residual secondary structure [16]. Folding was initiated by the rapid 5-fold dilution of the reaction directly in the cuvette. The final protein concentration was 30 μM and urea concentration was 1.6 M. In micelle samples, the final DPC concentration was 10 mM. In bicelles, the final DMPC and DPC concentrations were 2 mM and 4 mM respectively (q = 0.5). Similarly, 2 mM DPC, 3.2 mM DMPC and 0.8 mM DMPG was used for bicelles of q = 0.5, doped with of 20% DMPG. Heat shock [15] was not used in these experiments. The gain in secondary structure was monitored by measuring the change in ellipticity at 215 nm (θ 215), with time. The dead time of the experiment was ~15–20 s. Each data was fitted to a single exponential function to derive the folding rate for the OmpX mutants. The data were then averaged to obtain the mean and standard deviation (s.d.). The endpoint samples were also analyzed on cold SDS-PAGE [15] to confirm that each protein was completely folded (data not shown).
2.3. Equilibrium unfolding using circular dichroism
Equilibrium measurements were monitored using far-UV CD. The folded and unfolded protein stocks were diluted 5-fold in various urea concentrations from 80 mM (1.6 M for folding measurements) to 9.5 M at 0.2 M increments, to promote unfolding and folding, respectively. The system attained equilibrium at 216 h, which was confirmed independently using fluorescence measurements. Far-UV CD wavelength scans were recorded between ~205–260 nm at 25 °C [17]; the data were averaged over three accumulations and corrected for buffer and detergent contributions. The value obtained at 215 nm for each urea concentration was normalized between 0 and 1 to obtain the unfolded fraction (f U) at that urea concentration. Each profile thus obtained was fitted to a two-state linear unfolding model [18,19] provided below:
Here y O is the observed ellipticity at any denaturant concentration D. y F and y U, m F and m U are the intercepts and slopes of the folded and unfolded baselines, respectively. Δ is the equilibrium unfolding free energy, m is the change in accessible surface area upon protein unfolding, R is the gas constant (1.987 cal K−1 mol−1) and T is the temperature in kelvin. The data were fitted globally using a common m value, and an m value of − 1.41 kcal mol−1 M−1 was obtained from the fit. The thermodynamic parameters (Δ and C m) were derived for each independent experiment, and then averaged to obtain the mean and s.d. The complete details are provided in the supporting information.
3. Results
3.1. Tyrosine at position 76 of OmpX increases unassisted barrel folding in DPC micelles
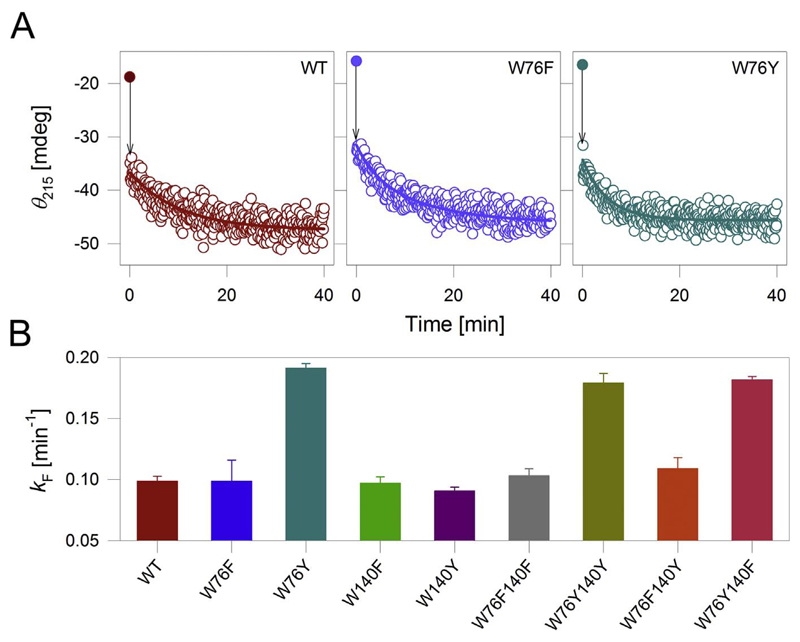
The far-UV circular dichroism (CD) spectrum of OmpX is characteristic of a β-sheet rich protein, with a negative trough centered at 215 nm [17]. First, we measured the rate of folding and insertion (k F) of OmpX, by monitoring the increase in secondary structure content at 215 nm (θ 215) (Fig. 2A, first panel). In urea, OmpX possesses residual structure in its denatured state [16,17]. Further, the burst phase [20,21] of OmpX folding falls within the dead time of our experiment (denoted by the arrow in Fig. 2A). Hence, the k F we obtain represents the gain in β-sheet content as the OmpX β-barrel assembles upon folding [22]. We were able to fit the measured change in θ 215 to a single exponential function to obtain k F (the rate of barrel assembly). In n-dodecylphosphocholine (DPC) micelles, the k F of OmpX-WT is ~0.1 min−1. Next, we measured the k F of the tyrosine and phenylalanine mutants listed in Fig. 1B. The results are shown in Fig. 2B. Surprisingly, we find that introducing tyrosine at position 76 promotes rapid folding of OmpX in DPC micelles. For example, the overall k F of all OmpX Tyr76 mutants, namely W76Y, W76Y140Y, and W76Y140F, is > 1.5-fold faster in DPC (Figs. 2, S1). To assess if similar rates are obtained in lipidic systems, we measured the k F for OmpX in DMPC (1, 2-dimyristoyl-sn-glycero-3-phosphocholine): DPC and 20% DMPG doped isotropic bicelles [23,24] of q = 0.5. Here, the k F of OmpX-WT is ~1.0 min−1 (Fig. S2 and S3). This value is 10-fold higher than what we measure in micelles, which is surprising because OMPs typically show slower folding rates in lipidic systems [25]. Diacylphosphatidylgycerol, which is abundant in native E. coli membranes, is known to slow the unassisted folding rate of OMPs [2]. As expected, when we dope DMPC:DPC bicelles with 20% DMPG, we obtain a 2.5-fold reduction in the k F of OmpX (Figs. S2, S3). As seen in micelles, the k F of the OmpX mutants bearing Tyr76 is higher than OmpX-WT in DMPC:DPC and native-like doped PC:PG bicelles. Additionally, the folding rate is augmented in mutants bearing phenylalanine at position 140 (discussed in detail in Fig. S3). However, as bicelle characteristics are affected in the presence of urea [26], we have limited our studies to DPC micelles. Overall, our results suggest that the rate of unassisted OmpX assembly is considerably accelerated in DPC micelles when tryptophan at position 76 is selectively substituted with tyrosine.
Fig. 2.
OmpX with tyrosine at position 76 exhibits fast folding kinetics in DPC. (A) Folding profiles of select OmpX mutants in DPC micelles monitored using the change in θ 215. Shown here are representative kinetic traces of folding for each mutant in DPC. Note the sudden jump in the θ 215 value (black drop arrow) from 0 min (filled symbol) to 0.017 min. This transition could not be fully captured in our measurements. The data from 0.017 min were fitted to a single exponential function (fits shown as solid lines) to derive the folding rate (k F) summarized in (B). Errors in (B) represent s.d. obtained from 3 to 5 independent experiments. See Figs. S1–S3 for the complete data in micelles and the significance of the measured k F.
3.2. Only one tryptophan affects the thermodynamic stability of OmpX barrel in micelles
Our studies show that tryptophan affects the folding kinetics of OmpX. To assess if the mutations also influence the thermodynamic stability of the barrel, we measured the change in equilibrium free energy (Δ) of OmpX in DPC using chemical denaturation. We monitored the global changes in OmpX structure using far-UV CD post-equilibrium, with urea as the denaturant (Figs. S4–S7). OmpX folded in vesicles is highly stable and resists unfolding even in guanidine [28]; the occurrence of hysteresis precludes measurements of OmpX thermodynamics in lipidic vesicles. Further, urea affects phospholipid hydration [26], which precludes experiments in bicelles. Hence, we carried out all our equilibrium measurements in DPC micelles. Additionally, we were able to achieve path-independent folding of OmpX only in this condition.
We used the total molar ellipticity at 215 nm (ME 215) obtained from far-UV CD for our analysis. Although unfolded OmpX possesses residual structure in urea, the secondary structure content of all the OmpX variants in the folded and unfolded states is similar (Fig. S4). Hence, we assumed that the overall change in the measured accessible surface as OmpX unfolds is unaltered across the mutants. Further, we did not detect intermediates when we analyzed OmpX samples electrophoretically on cold SDS-PAGE [15]. Therefore, we fitted our equilibrium ME 215 data derived from the unfolding profiles to a two-state linear extrapolation model. The common measure of m value (change in accessible surface area) thus derived was − 1.41 kcal mol−1 M−1. Note that the measured m value is low for an ~16 kDa protein such as OmpX (the measured m value for OmpW measured in 12-carbon phosphocholine large unilamellar vesicles is ~4.5 kcal mol−1 M−1 [28]). Such a low value can arise due to residual structure in the unfolded state (see Fig. S4), the use of urea (versus guanidine) as the denaturant, measurements in micellar systems, or a combination of these factors.
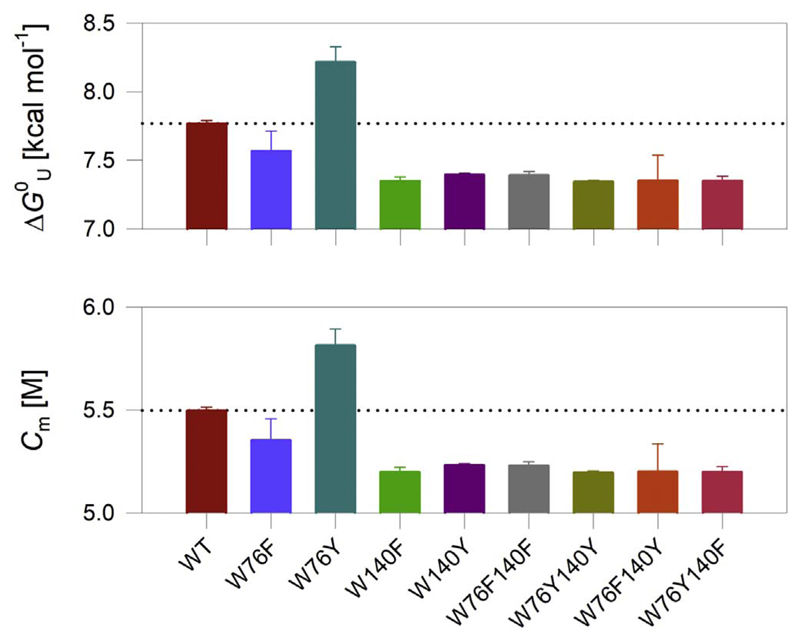
The Δ values measured in DPC micelles are summarized in Fig. 3 (also see Figs. S6, S7). The OmpX-W76Y mutant, which exhibits fast folding kinetics in DPC, also shows high stability in the equilibrium measurements. When compared to OmpX-WT, introducing tyrosine at position 76 increases the barrel stability by > 0.4 kcal mol−1. On the other hand, the presence of phenylalanine at position 76 marginally lowers the stability of OmpX (by < 0.2 kcal mol−1), suggesting that the stabilizing effect we measure at position 76 in micelles, is conferred specifically by tyrosine.
Fig. 3.
Thermodynamic parameters of OmpX mutants in DPC micelles. ME 215 values from equilibrium unfolding measurements were fitted to a two-state linear extrapolation model to obtain the Δ and C m values. We obtained an m value of − 1.41 kcal mol−1 M−1 from a global analysis of the data. Data are mean and s.d. of 2–3 independent experiments. The differences are significant with p-values of 0.19 for W76F, 0.03 for W76Y, and 0.004 for W140F, with respect to the WT protein. The dotted line in each panel represents the value obtained for OmpX-WT, and is shown for easy comparison of the thermodynamic parameters with the mutants. The complete data are presented in Figs. S6, S7.
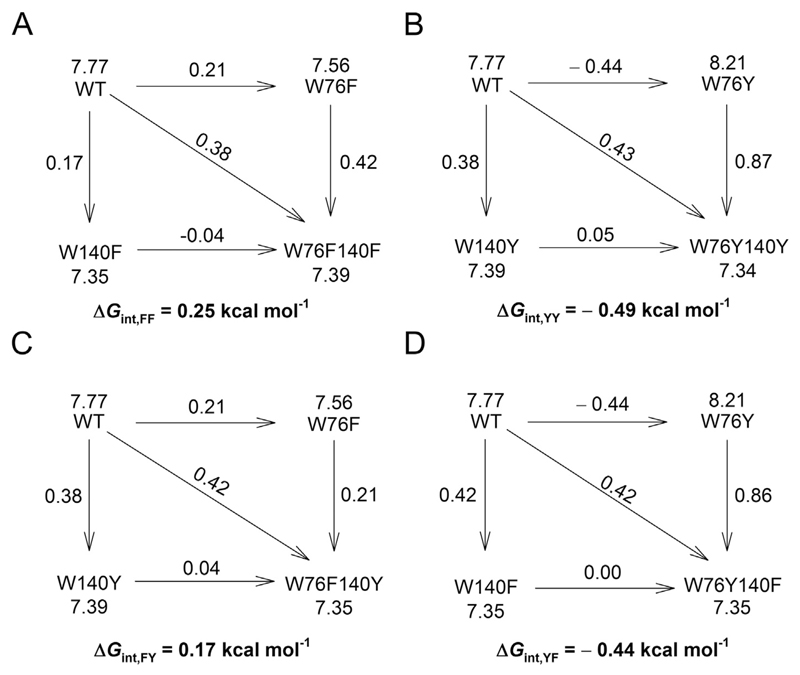
We obtained surprising results when we mutated Trp140. We did not see a pronounced effect in W140 mutants of OmpX in our folding kinetics measurements in DPC (see Fig. 2). Unlike the k F measurements, equilibrium thermodynamic measurements indicate that tryptophan contributes significantly to OmpX stability at position 140. Mutating Trp140 to either phenylalanine or tyrosine destabilizes OmpX by ~0.4 kcal mol−1 (Fig. 3). Furthermore, the stability of OmpX is not rescued when Trp76 is additionally mutated to tyrosine in the W76Y140F and W76Y140Y mutants (Fig. 3). This is also evident from the interaction energies derived from the double mutant cycle (Fig. 4). Additive mutations give an interaction energy (ΔΔGint) of zero. Non-zero values for ΔΔGint indicate that the effect of the mutations is non-additive. Hence, we conclude that tryptophan shows a selective stabilizing contribution of ~0.4 kcal mol−1 at position 140. Equilibrium measurements using tryptophan fluorescence (Figs. S8, S9) and densitometry analysis from electrophoretic mobility on cold SDS-PAGE (Fig. S10) support our conclusions. However, fluorescence measurements were limited to only the single-tryptophan mutants. Electrophoretic mobility measurements exhibited hysteresis. Hence, we have not interpreted these results further due to the limitations outlined above, with these measurements (further details are in Figs. S9, S10).
Fig. 4.
Double mutant free energy values are non-additive. The coupling free energy (ΔΔG int), arising from mutating the two aromatic residues at 76th and 140th in every double mutant is derived using the double mutant cycle [10] and is mentioned below each cycle. It is calculated as: ΔΔG int = ΔG wt − ΔG m1 − ΔG m2 + ΔG m1m2. Here, ΔG wT is the change in free energy (Δ) of OmpX-WT, ΔG m1 and ΔG m2 are the change in free energy of the single mutants, and ΔG m1m2 is the change in free energy of the double mutant. The Δ value for each protein is indicated beside the protein name, and the change in free energy accompanying each mutation is indicated above the arrowhead connecting the two proteins. All values of free energy are in kcal mol−1. The double mutant cycles are presented for (A) W76F140F, (B) W76Y140Y, (C) W76F140Y and (D) W76Y140F.
The results obtained from kinetic and equilibrium measurements were further validated by measuring the thermal stability of OmpX and its variants. OmpX is a highly thermostable protein [15], which shows reversible thermal denaturation but with hysteresis in DPC. In such systems, thermal denaturation can be used to check the stability of the folded OmpX barrel by measurement of the activation energy barrier (E act) separating the folded and unfolded states of the protein [29,30]. In DPC, OmpX unfolds beyond 85 °C [6,17]. In line with previous reports [17], OmpX shows residual secondary structure in its unfolded state (Fig. S11). Hence, we measured the rate of unfolding of OmpX and the mutants at specific temperatures between 87 and 93 °C (see Fig. S12) and generated the Arrhenius plot (Fig. S13A). The E act we obtained is compared in Fig. S13B. Overall, the E act correlates reasonably well with the folding rates and the free energy values. OmpX mutants such as W76Y and W76Y140F that exhibit high k F also show higher E act values, while the values are low for mutants bearing tyrosine at position 140 (Fig. S13). These results suggest that the contribution of aromatic residues to OmpX stability might be independent of the nature of denaturant.
4. Discussion
Well-studied bacterial OMPs such as OmpA and PagP are known to fold in vitro through a multistate pathway, depending on the folding environment [2,22,31]. Here, rapid protein adsorption on membrane occurs within seconds, and is followed by a slower protein insertion and assembly process that occurs in minutes-hours [22]. Further, defects in the bilayer are known to accelerate the OMP folding process [27]. From our study in DPC micelles, we find that the early event in OmpX folding is very rapid, and we are able to explain only the end-state barrel rearrangement process using a simplistic two-state model. It can be argued that the process of adsorption and insertion seen for other similar OMPs also exists for OmpX, but occurs too rapidly for detection. In support of this argument, a previous study that compared the folding efficiency of various OMPs has identified fast folding kinetics (within seconds) for OmpX, while that of OmpA is slower in comparison [2]. We find that the rate of OmpX assembly is accelerated considerably in DPC micelles when the interface tryptophan at residue 76 is specifically replaced with tyrosine. Additionally, the stability of OmpX increases in the OmpX-W76Y mutant. However, we obtain contrasting results at position 140. Here, the stability of OmpX in DPC micelles is lowered even when Trp140 is replaced with another aromatic amino acid. Tryptophan, tyrosine, and phenylalanine differ in their chemical characteristics despite the fact that all three are aromatic residues. For example, tryptophan and tyrosine retain the ability to form hydrogen bonds at the interface with water molecules and the lipid headgroup [13]. The amphipathic nature of the indole side chain of tryptophan allows it to form non-polar interactions, electrostatic N-H…π and C-H…π interactions, as well as hydrogen bonding [32]. On the other hand, tyrosine forms a limited number of interactions when compared with the indole ring. Further, the physical chemistry of the phenylalanine side chain (benzyl ring) does not support strong hydrogen bonding. Hence, we conclude that both the amphipathic chemical nature and aromaticity of tryptophan is necessary at position 140, to retain the stability of OmpX.
Studies of the amino acid distribution pattern in membrane proteins have revealed a clustering of aromatic amino acids at the membrane-protein interface both for transmembrane helical and β-barrel structures [13]. Among the aromatic residues, tryptophan and tyrosine are localized preferentially at the interface, whereas phenylalanine being most hydrophobic of the three aromatics is buried and occurs preferentially closer to the bilayer midplane in membrane proteins [3]. The preferential position of tryptophan in the context of membrane protein stability is still a subject of debate. Different hydrophobicity scales rate tryptophan from being a highly hydrophobic to an amphipathic and moderately hydrophobic amino acid. Energetic contribution of the tryptophan side chain at different membrane depths has been studied for the 8-stranded barrel OmpA [9]. This study indicated a more stabilizing effect of tryptophan when present near the bilayer mid-plane rather than at the solvent-protein interface. Other studies suggest its stabilizing role at the solvent-protein interface where it helps in anchoring the membrane protein to the lipid bilayer ([10], OmpLA). To compare our results from OmpX–DPC systems previous reports, we calculated the transfer free energy predicted for interface aromatics from the well-established Wimley-White interface scale [33] (Table 1). This scale suggests that tryptophan shows a preferential stabilizing effect at the interface, followed by phenylalanine and tyrosine. Interface energetics obtained from several membrane proteins including the β-barrel OmpLA [10] supports a stabilizing role for tryptophan at the interface. In OmpX folded in DPC micelles, the thermodynamic contributions we measure for the two interface positions follows the order Tyr > Trp > Phe at position 76, and Tyr > Phe ≈ Trp at position 140 (Table 1). It is evident from this comparison that tyrosine is expected to have a destabilizing effect of ≥ 1.0 kcal mol−1 over tryptophan (from the Wimley-White scale). However, we obtain a stabilization of > 0.4 kcal mol−1 at position 76. The effective stabilization of ~1.5 kcal mol−1 for Tyr76 is significantly high for a conserved aromatic substitution. We surmise that the high stability we measure for OmpX-W76Y compared to OmpX-WT is specific to this mutant. In DPC micelles, the equilibrium thermodynamic measurements suggest that Trp76 is dispensable for OmpX stability. Hence, tyrosine at residue 76 bears a kinetic contribution to the folding of OmpX. We obtain a stabilizing role for tryptophan only at residue 140. The stability of OmpX is lowered by ~0.4 kcal mol−1 when W140 is mutated to either tyrosine or phenylalanine (see Fig. 3). These observations support a highly regio-specific contribution of tryptophan to OmpX stability in DPC micelles.
Table 1. Energetic cost of Trp/Tyr/Phe mutations of OmpX compared with known scales.
| Residue position in OmpX |
ΔΔ [kcal mol−1]a
|
||||
|---|---|---|---|---|---|
| 76 | 140 | WWint | WWoct | Aroint | OmpX |
| W | W | 0.0 | 0.0 | 0.0 | 0.0 |
| F | W | 0.7 | 0.4 | 0.2 | 0.2 |
| Y | W | 0.9 | 1.4 | 1.1 | − 0.4b |
| W | F | 0.7 | 0.4 | 0.2 | 0.4 |
| W | Y | 0.9 | 1.4 | 1.1 | 0.4 |
calculated for the transfer of tryptophan, tyrosine, or phenylalanine from interface to water (Wimley-White interface scale, WWint [33]), octanol to water (Wimley-White octanol scale, WWoct [40]), and interface to water (aromatic side chain transfer free energy, Aroint [10]).
Residue substitution calculated to be destabilizing from known energetics scales; a stabilizing effect is seen as an exception in the case of OmpX.
Our results reveal an antagonistic behavior of the two tryptophans in OmpX. We and others have previously shown that Trp76 of OmpX faces the lipid milieu and resides at the solvent interface (schematic shown in Fig. 5A) [6,14]. Furthermore, the measurements of tryptophan anisotropy and lifetime, as well as NMR chemical shifts of the indole indicate that Trp76 is also conformationally constrained [6]. It is well known that conformationally constrained residues can increase the free energy of the system by lowering the system entropy [34,35]. In the folded OmpX barrel, tryptophan at residue 76 is surrounded by polar entities such as Asp75, Asn74 and Asn115 in its 5 Å vicinity (Fig. 5A). Hence, we propose that the presence of tyrosine at this position (in OmpX-W76Y) might facilitate barrel rearrangement during OmpX folding by lowering the conformational strain while simultaneously retaining the polar contacts formed by the side chain. We also suggest that favorable packing interactions coupled with the conformational flexibility of the phenolic side chain at this interface will allow tyrosine to increase the stability of OmpX. This could also explain why Tyr76 specifically enhances the folding rate of OmpX in our experiments in DPC micelles.
Fig. 5.
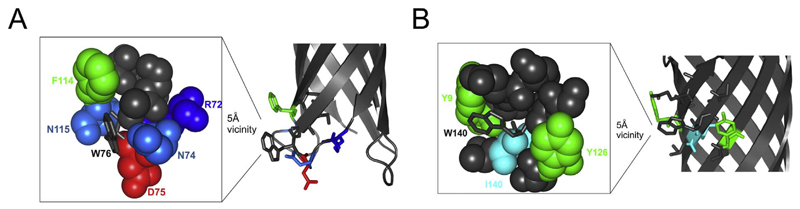
Cartoon representation of OmpX highlighting residues in the vicinity of Trp76 and Trp140. (A) The 5 Å vicinity of Trp76 (shown as grey sticks in the magnified image on the left) is populated with polar residues. Here acidic amino acids are shown as red, basic residues as different shades of blue and the aromatic residue is shown as green. Other apolar residues are shown as grey spheres. (B) The 5 Å vicinity of Trp140 (shown as grey sticks in the magnified image on the left) is populated with aromatic, hydrophobic and polar residues. The vicinity of Trp140 does not contain any charged residues. Here, aromatics are shown as green, hydrophobic as cyan and other apolar residues as grey spheres.
Unlike position 76, which maps to the bilayer interface, we and others have previously shown that residue 140 is relatively more lipid solvated [6,14]. Further, the vicinity of residue 140 is enriched with hydrophobic and aromatic amino acids including Tyr9, Tyr126, and Ile140. Anisotropy and lifetime measurements for Trp140 have established that this residue is lipid solvated and possesses more conformational flexibility than Trp76 [6]. Based on the crystal structure of OmpX, Trp140 is likely to establish electrostatic T-shaped aryl interactions with the two spatially proximal tyrosines in strands β7 and β1 (Fig. 5B). Our previous studies using peptide models have established that tryptophan/tyrosine interactions are energetically more favorable by up to ~1.0 kcal mol−1 than tyrosine/tyrosine or phenylalanine/tyrosine interactions [36]. Based on this observation, we suggest that the measured change in the free energy for the OmpX mutants could be accounted in part by the strengths of aryl interactions in the vicinity of Trp140, which are weakened in the OmpX-W140Y or OmpX-W140F mutants. Furthermore, tryptophan at position 140 is located closer to the bilayer midplane than tryptophan at position 76 in symmetrical membranes as well as in micelles. Such regio-specific contributions could also explain why the free energy changes measured at residue 140 lacks a linear correlation with other well-known scales (see Table 1). Tryptophan and tyrosine residues are usually observed at the lipid-facing interface position in membrane proteins, where they are known to stabilize the protein scaffold [3,5,11,12,37]. Our study reveals that the measure of such contributions of an interface aromatic amino acid cannot be generalized to all membrane proteins. The measured energetics can depend considerably more on the local environment and the model protein system used, than what was believed previously. Finally, we conclude that the tyrosine substitution of tryptophan can bear different effects on the OmpX folding rates and the barrel stability depending upon the position of the residue and the local environment, as both these factors decide the interactions that are formed in the local vicinity.
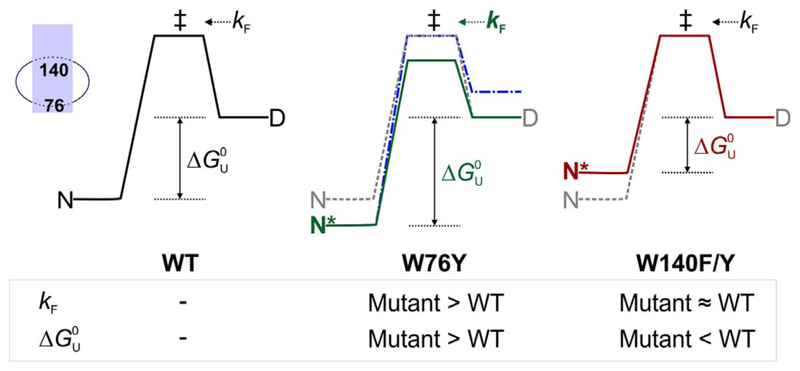
Can the contributions of positions 76 and 140 that we measure in DPC, be used as a means to understand the events during OmpX folding, and the resultant stability in micelle systems? We summarize our hypothesis based on a two-state folding/unfolding model for the OmpX–DPC system, in Fig. 6. Let us assume that OmpX folding from the denatured (D) state to the native (N) state in DPC proceeds via an intermediate (‡), which represents the transition state (Fig. 6, left). The rate of conversion of D → ‡ is k F. The net energetic change between D and N is represented as Δ Introducing tyrosine at position 76 increases the folding rate (Fig. 6, middle; k F now shown in green). Additionally, the mutation lowers free energy of the folded OmpX (now shown as N*) by > 0.4 kcal mol−1 (green solid line, Fig. 6, middle). An alternative explanation could involve a change in D (blue dashed line, Fig. 6, middle panel); we believe that this is less likely, as the residual structure in the unfolded state of OmpX mutants is comparable (see Fig. S4). When Trp140 is mutated, the stability of OmpX is lowered by ~0.4 kcal mol−1 (red solid line, Fig. 6, right), without considerably altering the folding rate. Since the change in free energy of the double mutants W76F140Y and W76Y140F is similar to W140Y and W140F respectively (see Fig. 3), we conclude that the mutations are non-additive (see Fig. 4). The OmpX-W76YW140F mutant is the most stable of the substitutions we studied. The stabilization we observed for OmpX for aromatic residues at positions 76 and 140, respectively, is in the following order: YW > WW > FW > WF ≈ WY ≈ F ≈ YY ≈ FY ≈ FY. Overall, we find that the contribution of Trp76 and Trp140 to OmpX folding and stability is determined by its local environment. A depth-dependent contribution of tryptophan has been reported for a structurally homologous 8-stranded β-barrel OmpA [9]. Hence, the energetic contribution of amino acids, particularly aromatic residues, should be interpreted with caution.
Fig. 6.
Two state diagram depicting possible effects of mutating residues 76 and 140 in an OmpX–DPC micelle assembly. The energy profile for WT (left) and is shown as grey dashed lines in the middle and right profiles. The transition state (‡) bridges the folded (N) and denatured (D) states of OmpX-WT. The measured folding rate k F (D → ‡) increases in W76Y (middle, green, bold) due to a likely change in the height of the transition state (green profile) while it is unaltered in W140F/Y mutations (right). An alternative energy profile where state D changes might also explain the observed increase in k F (middle, blue, dot-dash). The equilibrium folded state of OmpX-W76Y (N*, middle) is more stable than WT, resulting in an overall increase in the Δ of this mutant. OmpX-W140F/Y mutants are destabilized (N*, right, maroon) than WT. The inset (left extreme) shows the positions of residues 76 and 140 in the OmpX-micelle assembly.
Why did the OmpX sequence retain a metabolically expensive amino acid such as tryptophan at position 76, when tyrosine (and phenylalanine) can confer superior barrel folding and stability? This implies that attaining the most stable form is unlikely to be the ultimate aim for β-barrel folding. We reason that a gain in conformational flexibility at the expense of stability might be required for protein function (dynamic association of OmpX with host proteins during invasion) and for tolerance of OMP structures to fluctuations in the outer membrane. High thermodynamic costs associated with the turnover of very stable OMPs could also evolutionarily favor less stable β-barrel structures. Although our present work using OmpX is limited to micelle systems, recent studies on other model proteins such as OmpA [9], PagP [11,38], and OmpLA [38,39] in lipidic systems suggest that the contribution of side chains to the stability of transmembrane β-barrels can be scaffold-dependent. Whether such factors decide the evolutionary selection of all interface residues in membrane protein sequences remains to be established.
Supplementary Material
A detailed experimental section and supporting figures are available free of charge through the journal website. Supplementary data associated with this article can be found in the online version, at https://doi.org/10.1016/j.bbamem.2017.11.003.
Acknowledgment
We thank Ankit Gupta for his invaluable contribution to the data analysis and for critically reading the manuscript. D.C. is supported by senior research fellowships from CSIR, India. R.M. is a Wellcome Trust/DBT India Alliance Intermediate Fellow.
Funding statement
This work was supported by the Department of Biotechnology, Government of India grant BT/HRD/35/02/25/2009 and Science and Engineering Research Board, Govt. of India grant SB/WEA-13/2016 to R.M.
Abbreviations
- CD
circular dichroism
- Cm
midpoint of chemical denaturation
- DMPC, PC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DMPG, PG
1,2-dimyristoyl-sn-glycero-3-(1′-rac-glycerol)
- DPC
n-dodecylphoshphocholine
- Eact
activation energy barrier of protein unfolding
- fU
unfolded fraction
- kF
folding rate
- ME215
total protein molar ellipticity at 215 nm in deg cm2 dmol−1
- OMP
outer membrane protein
- OmpX
outer membrane protein X
- OmpX-WT, WT
wild type OmpX containing tryptophans at positions 76 and 140
- Δ
equilibrium unfolding free energy
- θ215
ellipticity in mdeg, at 215 nm
Footnotes
Author contributions
R.M. designed the research; D.C. performed the experiments; D.C. and R.M. analyzed the data; R.M. wrote the paper with inputs from D.C.
Transparency document
The http://dx.doi.org/10.1016/j.bbamem.2017.11.003 associated with this article can be found, in online version.
Declaration
The authors have no conflict of interest to declare.
References
- [1].Otzen DE, Andersen KK. Folding of outer membrane proteins. Arch Biochem Biophys. 2013;531:34–43. doi: 10.1016/j.abb.2012.10.008. [DOI] [PubMed] [Google Scholar]
- [2].Gessmann D, Chung YH, Danoff EJ, Plummer AM, Sandlin CW, Zaccai NR, Fleming KG. Outer membrane beta-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proc Natl Acad Sci U S A. 2014;111:5878–5883. doi: 10.1073/pnas.1322473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Killian JA, von Heijne G. How proteins adapt to a membrane-water interface. Trends Biochem Sci. 2000;25:429–434. doi: 10.1016/S0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- [4].Hong H, Park S, Jimenez RH, Rinehart D, Tamm LK. Role of aromatic side chains in the folding and thermodynamic stability of integral membrane proteins. J Am Chem Soc. 2007;129:8320–8327. doi: 10.1021/ja068849o. [DOI] [PubMed] [Google Scholar]
- [5].Sanchez KM, Gable JE, Schlamadinger DE, Kim JE. Effects of tryptophan microenvironment, soluble domain, and vesicle size on the thermodynamics of membrane protein folding: lessons from the transmembrane protein OmpA. Biochemistry. 2008;47:12844–12852. doi: 10.1021/bi800860k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chaturvedi D, Mahalakshmi R. Juxtamembrane tryptophans have distinct roles in defining the OmpX barrel-micelle boundary and facilitating protein-micelle association. FEBS Lett. 2014;588:4464–4471. doi: 10.1016/j.febslet.2014.10.017. [DOI] [PubMed] [Google Scholar]
- [7].Gupta A, Zadafiya P, Mahalakshmi R. Differential contribution of tryptophans to the folding and stability of the attachment invasion locus transmembrane beta-barrel from Yersinia pestis . Sci Rep. 2014;4 doi: 10.1038/srep06508. 6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Maurya SR, Mahalakshmi R. Control of human VDAC-2 scaffold dynamics by interfacial tryptophans is position specific. Biochim Biophys Acta. 2016;1858:2993–3004. doi: 10.1016/j.bbamem.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hong H, Rinehart D, Tamm LK. Membrane depth-dependent energetic contribution of the tryptophan side chain to the stability of integral membrane proteins. Biochemistry. 2013;52:4413–4421. doi: 10.1021/bi400344b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].McDonald SK, Fleming KG. Aromatic side chain water-to-lipid transfer free energies show a depth dependence across the membrane normal. J Am Chem Soc. 2016;138:7946–7950. doi: 10.1021/jacs.6b03460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Iyer BR, Zadafiya P, Vetal PV, Mahalakshmi R. Energetics of side-chain partitioning of beta-signal residues in unassisted folding of a transmembrane beta-barrel protein. J Biol Chem. 2017;292:12351–12365. doi: 10.1074/jbc.M117.789446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yau WM, Wimley WC, Gawrisch K, White SH. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- [13].Ulmschneider MB, Sansom MS. Amino acid distributions in integral membrane protein structures. Biochim Biophys Acta. 2001;1512:1–14. doi: 10.1016/S0005-2736(01)00299-1. [DOI] [PubMed] [Google Scholar]
- [14].Vogt J, Schulz GE. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Structure. 1999;7:1301–1309. doi: 10.1016/S0969-2126(00)80063-5. [DOI] [PubMed] [Google Scholar]
- [15].Maurya SR, Chaturvedi D, Mahalakshmi R. Modulating lipid dynamics and membrane fluidity to drive rapid folding of a transmembrane barrel. Sci Rep. 2013;3 doi: 10.1038/srep01989. 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tafer H, Hiller S, Hilty C, Fernandez C, Wuthrich K. Nonrandom structure in the urea-unfolded Escherichia coli outer membrane protein X (OmpX) Biochemistry. 2004;43:860–869. doi: 10.1021/bi0356606. [DOI] [PubMed] [Google Scholar]
- [17].Chaturvedi D, Mahalakshmi R. Methionine mutations of outer membrane protein X influence structural stability and beta-barrel unfolding. PLoS One. 2013;8:e79351. doi: 10.1371/journal.pone.0079351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Agashe VR, Udgaonkar JB. Thermodynamics of denaturation of barstar: evidence for cold denaturation and evaluation of the interaction with guanidine hydrochloride. Biochemistry. 1995;34:3286–3299. doi: 10.1021/bi00010a019. [DOI] [PubMed] [Google Scholar]
- [19].Moon CP, Fleming KG. Using tryptophan fluorescence to measure the stability of membrane proteins folded in liposomes. Methods Enzymol. 2011;492:189–211. doi: 10.1016/B978-0-12-381268-1.00018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Huysmans GH, Radford SE, Baldwin SA, Brockwell DJ. Malleability of the folding mechanism of the outer membrane protein PagP: parallel pathways and the effect of membrane elasticity. J Mol Biol. 2012;416:453–464. doi: 10.1016/j.jmb.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lim SA, Hart KM, Harms MJ, Marqusee S. Evolutionary trend toward kinetic stability in the folding trajectory of RNases H. Proc Natl Acad Sci U S A. 2016;113:13045–13050. doi: 10.1073/pnas.1611781113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kleinschmidt JH, Tamm LK. Folding intermediates of a beta-barrel membrane protein. Kinetic evidence for a multi-step membrane insertion mechanism. Biochemistry. 1996;35:12993–13000. doi: 10.1021/bi961478b. [DOI] [PubMed] [Google Scholar]
- [23].Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chem Rev. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nolandt OV, Walther TH, Grage SL, Ulrich AS. Magnetically oriented dodecylphosphocholine bicelles for solid-state NMR structure analysis. Biochim Biophys Acta. 2012;1818:1142–1147. doi: 10.1016/j.bbamem.2012.01.007. [DOI] [PubMed] [Google Scholar]
- [25].Burgess NK, Dao TP, Stanley AM, Fleming KG. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J Biol Chem. 2008;283:26748–26758. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Barton KN, Buhr MM, Ballantyne JS. Effects of urea and trimethylamine N-oxide on fluidity of liposomes and membranes of an elasmobranch. Am J Physiol. 1999;276:R397–R406. doi: 10.1152/ajpregu.1999.276.2.R397. [DOI] [PubMed] [Google Scholar]
- [27].Kleinschmidt JH, Tamm LK. Secondary and tertiary structure formation of the β-barrel membrane protein OmpA is synchronized and depends on membrane thickness. J Mol Biol. 2002;324:319–330. doi: 10.1016/S0022-2836(02)01071-9. [DOI] [PubMed] [Google Scholar]
- [28].Moon CP, Zaccai NR, Fleming PJ, Gessmann D, Fleming KG. Membrane protein thermodynamic stability may serve as the energy sink for sorting in the periplasm. Proc Natl Acad Sci U S A. 2013;110:4285–4290. doi: 10.1073/pnas.1212527110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jayaraman S, Gantz DL, Gursky O. Kinetic stabilization and fusion of apolipoprotein A-2:DMPC disks: comparison with apoA-1 and apoC-1. Biophys J. 2005;88:2907–2918. doi: 10.1529/biophysj.104.055921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Maurya SR, Mahalakshmi R. Influence of protein-micelle ratios and cysteine residues on the kinetic stability and unfolding rates of human mitochondrial VDAC-2. PLoS One. 2014;9:e87701. doi: 10.1371/journal.pone.0087701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Huysmans GH, Baldwin SA, Brockwell DJ, Radford SE. The transition state for folding of an outer membrane protein. Proc Natl Acad Sci U S A. 2010;107:4099–4104. doi: 10.1073/pnas.0911904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Makwana KM, Mahalakshmi R. Implications of aromatic-aromatic interactions: from protein structures to peptide models. Protein Sci. 2015;24:1920–1933. doi: 10.1002/pro.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- [34].Matthews BW, Nicholson H, Becktel WJ. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc Natl Acad Sci U S A. 1987;84:6663–6667. doi: 10.1073/pnas.84.19.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Brady GP, Sharp KA. Entropy in protein folding and in protein-protein interactions. Curr Opin Struct Biol. 1997;7:215–221. doi: 10.1016/S0959-440X(97)80028-0. [DOI] [PubMed] [Google Scholar]
- [36].Makwana KM, Mahalakshmi R. Asymmetric contribution of aromatic interactions stems from spatial positioning of the interacting aryl pairs in beta-hairpins. Chembiochem. 2014;15:2357–2360. doi: 10.1002/cbic.201402340. [DOI] [PubMed] [Google Scholar]
- [37].Yuen CTK, Davidson AR, Deber CM. Role of aromatic residues at the lipid-water interface in micelle-bound bacteriophage M13 major coat protein. Biochemistry. 2000;39:16155–16162. doi: 10.1021/Bi0016117. [DOI] [PubMed] [Google Scholar]
- [38].Marx DC, Fleming KG. Influence of protein scaffold on side-chain transfer free energies. Biophys J. 2017;113:597–604. doi: 10.1016/j.bpj.2017.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Moon CP, Fleming KG. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proc Natl Acad Sci U S A. 2011;108:10174–10177. doi: 10.1073/pnas.1103979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wimley WC, Creamer TP, White SH. Solvation energies of amino acid side chains and backbone in a family of host-guest pentapeptides. Biochemistry. 1996;35:5109–5124. doi: 10.1021/bi9600153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.