Abstract
RNA splicing brings diversity to the eukaryotic proteome. Different spliced variants of a gene may differ in their structure, function, localization, and stability influencing protein stoichiometry and physiological outcomes. Alternate spliced variants of different genes are known to associate with various chronic pathologies including cancer. Emerging evidence suggests precise regulation of splicing as fundamental to normal wellbeing. In this context, infection-induced alternative splicing has emerged as a new pivot of host function, which pathogenic microbes can alter-directly or indirectly-to tweak the host immune responses against the pathogen. The implications of these findings are vast, and although not explored much in the case of pathogenic infections, we present here examples from splicing mediated regulation of immune responses across a variety of conditions and explore how this fascinating finding brings a new paradigm to host-pathogen interactions.
Introduction
Eukaryotic genes get expressed in discontinuous stretches of exons with introns interspersed between them. A matured mRNA involves careful removal of introns and stitching together of exons through a process called RNA splicing. By omitting or retaining specific exons (through alternative splicing), the sequence and therefore, structure, function, and stability of the translated protein products could be altered. More than 90% of expressed human genes undergo alternative splicing, and precise splicing of pre-mRNAs is crucial to normal wellbeing of the cells [1]. Interestingly, a large number of reported exonic mutations linked to various diseases more dramatically affect the splicing efficiency of the exons rather than the ORF itself, underscoring the importance of this process [2–4].
RNA splicing is a complex process, involving assembly of large multi-molecular complexes at splice junctions between introns and exons as well as recognition of several splicing enhancer/silencer elements distributed adjacent to splice junctions which together decide the probability of inclusion or exclusion of an exon in the final mature mRNA [5]. Some of these regulatory elements include “cis” elements like exonic splicing enhancers (ESEs), exonic splicing silencers (ESIs), intronic splicing enhancer (ISEs), intronic splicing silencers (ISSs) and “trans” elements like snRNPs, hnRNPs, SR proteins, and several other accessory proteins.
Alternative splicing is associated with several chronic pathological conditions including neurological disorders, cancers of different types and atherosclerosis [6–23]. Among infectious diseases hijacking and co-opting of the host splicing machinery especially during viral infections including dengue, HIV, Zika etc. is well explored [24–27]. Few recent studies, including one from our group, now suggest that pathogen co-opting of host RNA splicing machinery could be part of a larger scheme of infection mediated perturbation of host response machinery [24, 28]. Thus while one study reports massive alteration in host RNA splicing pattern upon dengue virus infection, we and another group reported global alterations in the RNA splicing pattern upon Mycobacterium tuberculosis or Salmonella and Listeria infection of human macrophages [24, 28, 29]. Here we will briefly review what we know about alternative splicing in immune regulation, factors both intrinsic and extrinsic known to regulate those and explore this new layer of pathogen-mediated regulatory interference on host physiology. We would elaborate on the various implications including the potential for re-strategizing novel therapeutics as well as understanding disease susceptibility and tolerance.
Intrinsic factors regulating alternative splicing during infection and immune activation
Immune activation, which is fundamental to host responses to pathogenic infections, is known to get influenced by alternative splicing. During the activation of T cells, several specific splicing events decide the fate of activation [30–36]. Similarly, in B lymphocytes, several key features are regulated through alternative splicing for example switch between membrane-bound to secreted IgM as well as roles of alternate spliced variants of Oct2 and IL4R, each having distinct immunological consequences [37, 38].
In the cells of innate immunity, signaling downstream to TLRs and other pattern receptors are among the principal mechanisms for pathogen recognition, which elicit inflammatory responses. Several TLRs like TLR2, 3 and 4, rely on the cytosolic adaptor protein MyD88 for activation of NFκB and inflammation [39, 40]. Signaling molecules like IRAK1, Tollip and TFAR6 downstream to the pattern receptors and cytokine receptor also get regulated through alternative splicing [41, 42]. We begin with reviewing some intrinsic mechanisms, specifically reported in the immune cells, which influence alternative splicing during infection or activation.
i). Co-transcriptional regulation
There is a consensus that RNA splicing occurs in parallel to transcription. More recently the role of transcription and factors associated with it has emerged as among the key regulatory components of alternative splicing. For example, the rate of transcription has emerged as a critical gatekeeper for specific splicing event. An excellent set of examples is presented by conditions, which cause pausing of RNA pol II. Pausing of RNA pol II could ensure that the newly transcribed splice junction is utilized thereby retaining the corresponding exon. On the other hand, exon skipping may occur during rapid transcription of genes[43]. An exciting example is activation of T helper 1 (Th1) effector cells, wherein recruitment of RNA Pol II coincides with a genome-wide temporary slowdown in co-transcriptional splicing [44]. Several splicing-associated factors regulate the pausing or rate of transcription. For example, RNA pol II is paused by U2 associated protein Cus2 and only released by the activity of ATPase prp5 which is part of the spliceosome complex [45]. Similarly, an important splicing accessory protein SRSF2 negatively regulates transcription pause of RNA pol II by releasing P-TEFb from the 7SK complex [46]. On the flip side, several transcriptional regulatory factors like TFs, coactivators, transcriptional enhancers, and chromatin remodelers end up impacting RNA splicing [47, 48].
In yet another interesting observation, activated macrophages were shown to maintain distinct pools of chromosome-associated, nucleoplasmic and cytoplasmic messenger RNAs. Out of these, the chromosome associated ones are full-length transcripts but yet to be completely spliced, a feature which is consistent with the concept of cotranscriptional RNA splicing [31]. Macrophage activation being integral to host immune response against a variety of pathogens, the rate of transcription could play a massive role in regulating infection induced alternative splicing and host response. Factors, which could help link rate of transcription with alternative splicing includes the presence of strong or week splice junctions, accessibility of splice junctions due to secondary structures or due to the stoichiometry of splicing accessory proteins which can differentially bind to enhancers or silencers present in the exons or introns [43, 49].
ii). Regulation by splicing factors or accessory proteins
Among factors other than transcriptional kinetics and transcription factors, a large number of splicing factors and accessory proteins involved in splicing play a critical role in regulating alternative splicing. The serine-arginine containing proteins (SR proteins) and hnRNPs are significant regulators of alternative splicing. Functions of SR proteins are regulated through phosphorylation by kinases like CDC2-like kinases (CLKs), dual-specificity tyrosine-regulated kinases (DYRKs) and SR-rich splicing factor protein-kinases (SRPKs) [50–63]. Therefore activity, localization, and concentration of these regulatory proteins can impact on the cellular alternative splicing. Accordingly, these intrinsic factors get modulated during diverse conditions including cellular activation and pathogenic infections. During T cell activation hnRNP U suppresses inclusion of exon 7 in MALT1 transcript, which is needed for TRAF6 recruitment [64]. In macrophages, during TLR activation by pathogen-associated molecular patterns (PAMPs), the production of negatively acting spliced variant of MyD88 gets inhibited by the recruitment of a critical splicing factor SF3a, which then promotes inflammation [41]. In the absence of SF3a, the truncated MyD88 isoform gets increased inhibiting TLR signaling and inflammation [41]. Generation of short isoforms due to alternative splicing as soluble membrane receptors is also known for molecules like TLR4, IL-4R and IL-5R [65–67]. However, in most of these cases, molecular mechanisms are not evident.
While there are cell-intrinsic transcriptional regulators and splicing factors as well as associated accessory proteins and upstream signaling molecules to regulate inclusion or exclusion of specific exons, it is critical to understand whether there are extrinsic factors, which could also alter the inclusion/exclusion of exons from the mature mRNAs? The scope for such a possibility arises through extensive studies involving viral infections and hijacking of the host machinery to help the splicing of viral genes. Interestingly, in several cases, especially those involving viral infections, infection-induced alternative splicing depends on selective uses of intrinsic regulators, thereby suggesting possible crosstalk and cross-regulation between pathogenic factors and host splicing machinery.
Extrinsic factors regulating alternative splicing during infection and immune activation
a). Regulation of alternative splicing during viral infection
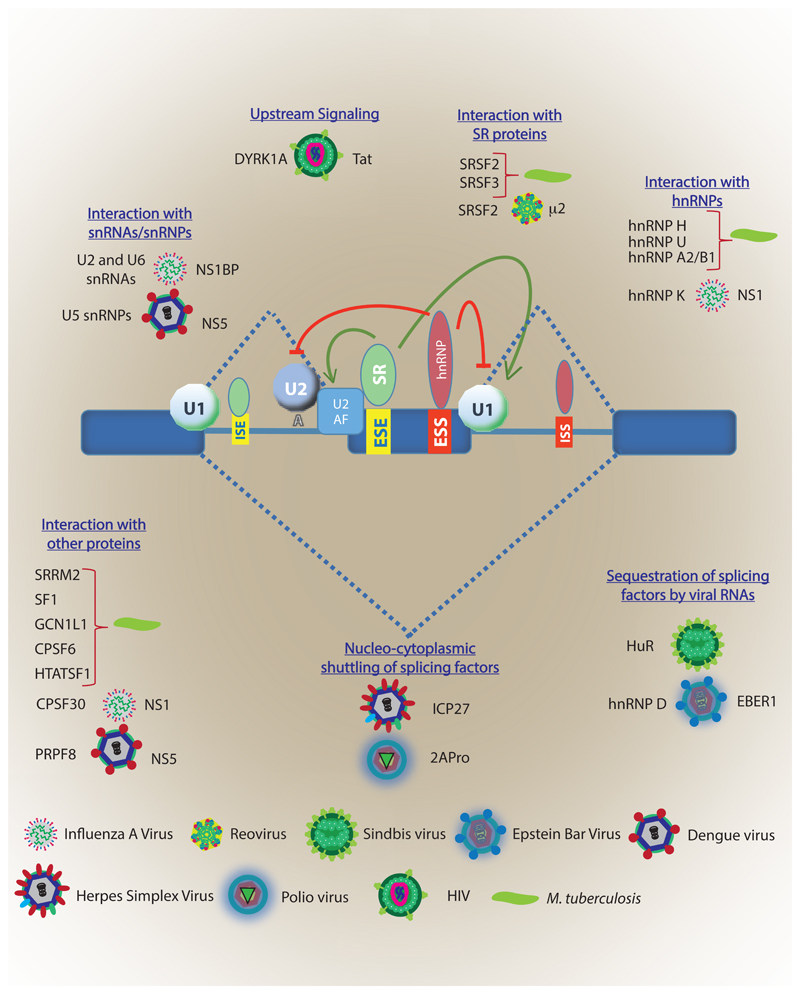
Viral-encoded factors can modulate the complex interplay between cis and trans-regulatory splicing factors thereby facilitating expression of its genes and influencing the defense homeostasis. Such interactions have been well established in papillomavirus, adenovirus and HIV among others [68–71]. An exciting example is presented by the NS1 protein of Influenza A virus, which interacts with the spliceosome complex and blocks the transition to active complex inhibiting the cellular gene expression [24]. NS1 binds to a 30-kDa subunit of CPSF (CPSF30), which is essential for the processing of 3'-end during splicing. Through this interaction, NS1 protein globally inhibits the cellular polyadenylation as well [24]. Another Influenza A protein, NS1-BP, interacts with host hnRNP K and facilitates the splicing of 3 RNA segments of the virus. However, this interaction has a cascading effect on the host RNA splicing as well causing mis-splicing of some of the host genes [72]. Most viruses rely on host splicing machinery to generate multiple proteins from their limited genome. In many cases, viral splice sites match human splice junction consensus sequences, which effectively drive the splicing machinery preferentially towards viral pre-mRNAs that need to get spliced. For example in case of Human parvovirus B19, which harbours an RNA binding motif consensus site (5'-UGUGUG-3') for its intronic splicing enhancer 2 (ISE2) region, utilizes host factor RBM38 for processing of its pre-mRNA during virus replication [73]. Many such examples of viral factors mediated targeting of RNA splicing are depicted in figure 1 [74–79].
Figure 1. Viral and bacterial factors interacting with host RNA splicing machinery.
The key players of the host RNA splicing machinery depicted at the core including snRNAs/snRNPs, SR proteins, hnRNPs and cis-elements like ESE, ESS, ISE, and ISSs. Different pathogens target host RNA splicing through a variety of mechanisms (written in blue). For specific examples mentioned in the figure, for each case, host factors are written at the left, followed by the pathogen and finally the pathogen factors which is known to interact with the corresponding host factors. For cases where interacting partner from the host is not known, like in the case of Mtb, pathogen factors are not explicitly mentioned. ESE: Exonic splicing enhancer, ESS: exonic splicing silencer, ISE: Intronic splicing enhancer, ISS: intronic splicing silencer.
Some RNA viruses replicate within cytoplasmic viral factories independent of splicing. However, their proteins might interact with host splicing factors, modulating splicing for its benefit [79]. For example, mammalian reovirus infection results in a global change in alternative splicing pattern with about 240 alternative splicing events of transcripts frequently involved in regulation of gene expression and RNA metabolism [79, 80]. On the similar line, μ2 protein from T1L reovirus forms a complex with the pre-mRNA splicing factor SRSF2 in nuclear speckles altering the splicing of transcripts for genes involved in RNA processing and maturation and enhances reovirus replication and cytopathic effect [79]. Viruses can also encode proteins, which serve as alternative splicing factors. Like L4-33K of adenoviruses preferentially activates splicing of transcripts with weak 3'-splice sites [81]. In case of HIV-1 infection, the accessory protein Tat directly interacts with TAU RNA and inhibits inclusion of exon 10 in the TAU mRNA in a DYRK1A dependent manner, leading to neurocognitive impairments [82]. Additional mechanisms of viral infection-mediated alteration in host RNA splicing include relocalization of cellular splicing machinery components to the cytoplasm, thereby impacting RNA splicing, like relocalization of hnRNPs to the cytoplasm during VSV infection [83]. Similarly, the expression of several splicing factors is altered during HIV infection, which could impact host RNA splicing through stochiometry of the regulatory factors [84]. Regulation of RNA splicing and overall spliceosome machinery seems fundamental to all such viruses which rely on splicing to generate multiple proteins from their small genomes. As some examples mentioned above shows, they can influence by directly interacting with the core or accessory splicing proteins or by regulating expression, splicing and sub-cellular distribution of splicing proteins.
b). Regulation of alternative splicing during bacterial infection
Unlike the viral infections, not much is known regarding bacterial factors interfering with the host RNA splicing. Nonetheless, there is sufficient, mostly circumstantial evidence now to suggest that bacterial factors may also regulate alternative splicing, either indirectly or direct. In case of Mycobacterium tuberculosis, overexpressing specific secreted proteins in the host macrophage cell line showed coimmunoprecipitation of splicing factors hnRNPU, hnRNP H, hnRNP A2/B1 isoform A2 and SRSF3 with the bacterial protein mtrA [85]. In a more recent study, several mycobacterial proteins like EsxQ, Apa, Rv1827, LpqN, Rv2074, Rv1816 when used as a bait with the macrophage cell lysates, several host RNA splicing proteins like SRSF2, SRRM2, SF1, HTATSF1, GCN1L1, CPSF6 etc. were identified as their interacting partners, suggesting the possibility that mycobacterial proteins could alter host RNA splicing through physically interacting with one or more splicing factors or accessory proteins [86]. Interaction of the virulence factor of Listeria, listeriolysin (LLO) with hnRNP M can specifically impact type IFN response in the host macrophages [87]. All intracellular bacterial pathogens including Mycobacterium, Salmonella, Listeria etc. are known to have different secretory systems, which allow them to secrete virulence factors and perturb host physiology [88]. Through independent studies, it is apparent that bacterial virulence factors can indeed access host nucleus raising the possibility that they can also perturb splicing in the nucleus [89, 90].
A handful of studies now suggest bacterial infection induced alternative splicing of host RNAs [28, 29]. Alternative splicing during intracellular bacterial infections of macrophages is more pervasive than possibly ever imagined. Global changes in the alternative splicing during bacterial infection have been shown in the case of Salmonella and Listeria [29]. A study from our group shows global alterations in the pattern of RNA splicing post Mtb infection of macrophages [28]. Intriguingly, in Mtb-infected macrophages, genes involved in different stages of spliceosome assembly are also regulated at the splicing level, which can directly impact the functioning of splicing machinery. There was also a considerable increase in the expression of truncated/non-translatable variants of several genes, specifically upon virulent infections [28].
As is evident from examples above, perturbation of host RNA splicing during bacterial infections appears as a common occurrence. However currently there is very limited, if any, understanding on involvement/role of specific bacterial factors in regulating host RNA splicing. We assume that work in this area must be rapidly progressing considering its significance.
Physiological impact of infection-induced alternative splicing
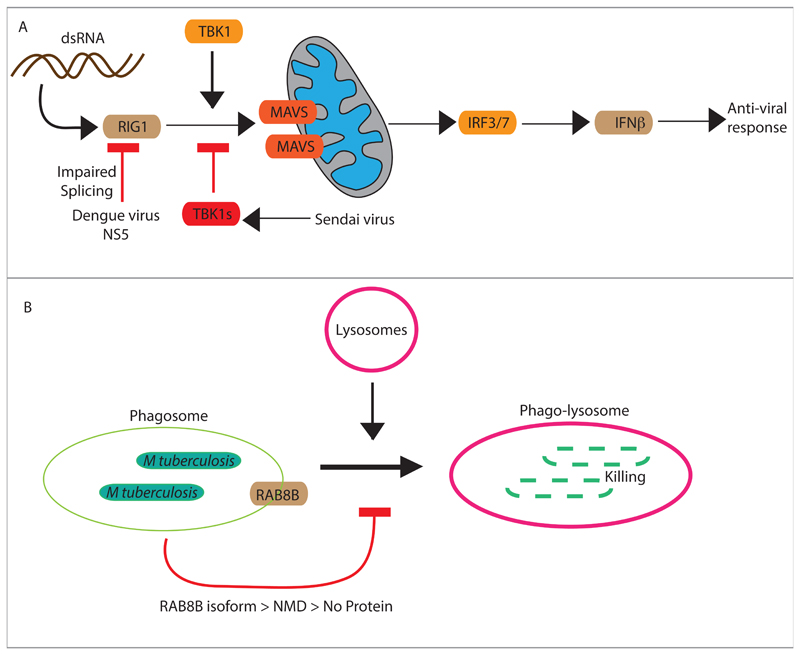
How infection-induced alternative splicing impacts the physiology of host and outcome of infection? During viral infections, host cells recognize viral DNA in cytosol via RIG-I, which initiates downstream signaling via associating with different adapter proteins like mitochondria antiviral-signaling protein (MAVS) and activates TBK1, which is critical for virus-triggered type I IFN signaling [91, 92]. In the condition of some virus infection, like upon Sendai virus infection, TBK1s, an alternative spliced variant, lacking in exon 3-6 gets induced, which binds to RIG-1, and negatively regulates antiviral IFN-β signaling pathway by disrupting the interaction between RIG-I and MAVS [93]. Given the strong anti-viral function of RIG-I, it could be a preferred target by several viral virulent proteins. The recent report on NS5 protein of dengue virus, which targets RIG-I among many other proteins via alternative splicing, clearly underscores this possibility [24]. Among several genes with perturbed splicing patterns during dengue infection, RIG-1 stands out being a part of the innate sensor mechanism against intracellular danger (Fig. 2A). Thus even though the changes in alternative splicing during infections are global, they have highly targeted physiological impact.
Figure 2. Impact of infection-induced alternative splicing on host defense pathways.
A) Regulation of RIG1 mediated anti-viral response by Sendai virus and Dengue virus. By inhibiting RIG1-MAVS interaction, IFNβ expression gets blocked which helps viral survival and replication. B) Phagosome maturation is the integral innate defense mechanism against bacterial pathogens like Mycobacterium tuberculosis. RAB8B is among the several small GTPases, which is required for phagosome maturation and fusion with the lysosome, leading to bacterial killing. However during Mtb infection, due to alternative splicing, RAB8B pre-mRNA is spliced in a way that the resulting RAB8B transcript has a premature stop codon and therefore undergoes non-sense mediated decay. No new RAB8B proteins can, therefore, be synthesized and result in an effective decline in the RAB8B protein levels. The decline in RAB8B protein level helps Mtb escape targeting from the lysosomes.
Macrophage infection of Salmonella and Listeria results in massive alterations in RNA splicing in the host and a large number of genes belonging to immune response functional class specifically undergo alternative splicing [29]. Interestingly, a comparison between alternative splicing events reported upon Listeria or Salmonella and those reported upon Mtb infection shows several common targets, specially for those belonging to critical functional classes like RNA processing, ribosomal proteins, autophagy and immunity, RNA splicing, trafficking and vesicle transport, secretion, translation, immunity to infection etc. [28, 29]. While these two studies highlight the global patterns of alternative splicing during infections, in our recent study, we also establish how the changed splicing products impacts on the host innate defence mechanisms in the macrophages during Mtb infections [28]. In Mtb infected cells, a key regulator of phagosome maturation RAB8B gets spliced in a manner that instead of forming a functional protein the mRNA gets degraded through a process called nonsense-mediated decay [28]. As the RAB8B transcript fails to get translated into a functional protein, it leads to effectively lowered protein level in the cell, hampering the phagosome maturation process (Fig. 2B). While it is difficult to establish at this stage, it is highly likely that increased transcription of RAB8B locus may represent a host response to Mtb infection, whereas alternative splicing to form a truncated matured mRNA reflect the outcome of bacterial intervention in the cellular physiological processes. During Mtb infection of DCs truncated and alternate spliced form of IL12Rβ1 gets induced, which enhances DC migration and Mtb specific CD4+ T-cell activation [94]. In the case of Mycobacterium avium subsp. Paratuberculosis (MAP) infection 46.2% of genes undergo alternative splicing out of which, splice variants of 2 genes related to macrophage maturation (MMD; monocyte to macrophage differentiation-associated) and lysosome function (ADA; adenosine deaminase) results in reduced MAP clearance from the site of infection during the early stage of infection due to failure of macrophage maturation and lysosome function [95]. How RNA splicing gets regulated in such a specific manner constitutes an exciting question and deserves more attention.
It is evident that during global regulation of alternative splicing upon infection, targets do not get randomly selected for altered splicing; instead, they are specially chosen which enhances the chances of pathogen survival. We performed an interesting analysis of RNAseq data from Mtb-infected macrophages on this aspect [96]. We looked at the most dominant isoform, which gets regulated upon infection across the entire family of protein kinases in the host. A large number of kinases, which get dominantly regulated, our analysis revealed, did not express critical functional domains like kinase domain or protein-protein interaction domains like SH2, SH3 and PH domains [96]. It is possible that in the presence of virulence factors from bacterial or viral pathogenic factors, several cryptic or week splice sites get activated, thereby altering the splicing pattern. While the exact mechanisms are probably work in progress, the role of isoform diversity due to the alternative in determining the outcome of infection is established beyond doubt.
Splicing regulators: potential new therapeutics against infectious diseases
Targeting host factors to contain the survival of intracellular pathogens is a non-conventional therapeutic strategy, which has received significant traction over the past two decades. It was based on the realization that certain host factors regulate cellular physiology in a manner that is beneficial to the pathogen. By high throughput screening in cell-based assays, several such host factors have been identified for viral infections like HIV, dengue, West Nile viruses and bacterial pathogens like Mycobacterium tuberculosis, Salmonella sp., etc. [97–100]. Addition of RNA splicing regulators, as among the potential pro-bacterial host factors widens the repertoire of targets for host-directed therapy. While it is too premature to assume and discuss further in this direction, several specific splicing inhibitors are already available, and some of them are also getting tested for chronic diseases like cancer. Mechanisms of alternative splicing during viral infections are relatively more explored, some specific inhibitors are being tested against host splicing factors like SR proteins as well as HIV proteins like Rev and Tat for regulation of splicing during HIV infection [101]. Accordingly, a novel compound which acts on host SRSF10, a splicing factor, impairs HIV replication [102]. There are three major classes of inhibitors used to inhibit RNA splicing: oligonucleotides, RNA binding activators or inhibitors and splicing factor kinase inhibitor [63]. Among these, the most versatile family of splicing regulators are splicing factor kinase inhibitors [63]. Involvement of these kinases bridges the splicing machinery with cellular signaling, potentially explaining, in part, context specificity. However, during pathogenic infections, especially bacterial infections, how cellular signaling could impact the splicing factors remains poorly understood.
Impact of SNPs on isoform diversity and susceptibility/tolerance to infections
A perplexing aspect of most pathogenic infections is the diversity of responses different individuals may have against same pathogenic agent. It is specifically contrasting in the case of Mtb infections since a large number of individuals who get exposed to the bacteria in the endemic countries do not develop active tuberculosis. Only 5-10% of exposed individuals get active disease initially [103]. Remaining population either eliminate the infection or control them where the bacilli stay in a metabolically altered latent phase. Why individuals differ in their susceptibility and tolerance to infection with the same pathogen. For a variety of conditions, genomewide association studies have revealed the association of specific Single nucleotide polymorphisms (SNPs) with specific conditions [104].
As estimated, around ~15% of all point mutations, causing human inherited disorders, disrupt splice-site consensus sequences, particularly at intronic positions [105, 106].
Polymorphisms in high impact exonic splicing enhancers (ESEs) strongly influence the activity of disease-associated genes and modify their association with different pathological conditions [105–107]. Some examples to this include SLC2A2 isoform diversity in Type 2 Diabetes, nmMLCK1 and nmMLCK2 in inflammation and oxidized LDL receptor (LOX-1) in lipid homeostasis and inflammation [107],[108],[109]. In the case of Mtb infections, so far several SNPs are identified which presumably determines the disease susceptibility [110]. Explicit examples are steadily emerging suggesting role of isoform diversity caused due to silent mutations in exonic splice sites and SNPs in intronic sites could play a major role in determining the diversity of host responses against pathogens. For example, a tuberculosis susceptibility locus identified in the intronic region of human ASAP1 gene regulates dendritic cell migration [110]. However, the study does not refer to the effect on RNA splicing. A more definitive report has emerged very recently, showing a polymorphism in IL7RA results in impaired splicing of IL-7Rα and helps protect against tuberculosis[111]. Similarly, through studies on Mendelian susceptibility to mycobacterial disease (MSMD), a splice site mutation in SPPL2A was shown responsible for this susceptibility [112]. While the examples mentioned above report the presence of cis-variants which intrinsically and constitutively impact the alternative splicing, there is no study yet which correlates splice-site variants and their activity upon mycobacterial infection. We believe such studies are poised to emerge rapidly, identifying newer associations for susceptibility and tolerance to infections.
Conclusions
Alternative splicing of transcripts brings unprecedented diversity to the eukaryotic proteome. It serves as a mechanism, which brings dynamism to otherwise static genome in an individual generating diverse isoforms of the same protein with unique functional abilities. Here we have tried to bring examples from the literature which highlight the role alternative splicing could play during pathogenic infection and regulation of immunity as well as inflammation, critical aspects for the well-being of an individual. It is evident through the limited examples discussed here that alternative splicing does impact the outcome of any infection. In several noninfectious pathologies including cancer, such splicing events are explicitly studied and attributed for the pathology. This understanding has led to the development of specific small molecule inhibitors, which targets explicitly certain splicing events and therefore allows for a handle to regulate the cellular outcomes. Finally, as is evident in the final section, several SNPs in the human genomes could impact the alternative splicing pattern rather than impacting the ORFs. These observations highlight the enormous role alternative splicing could play in deciding the individuality whether in the context of physical attributes or tolerance/susceptibility to infectious or chronic diseases. These lines of investigations, however, are very limited in the context of bacterial infections.
Future perspective
Role of alternative splicing in pathogenic infections is slowly but steadily getting recognized. It has opened up an immense possibility for future directions in order to better understand host-pathogen interactions, disease pathogenesis, and therapeutic strategies. While at one level it will be important how perturbed signaling events during various infections impact host RNA splicing, it is also desirable to explore possible pathogenic factors, which could interact with the splicing machinery and alter the host response machinery. Finally, the fact that individuals may vary in terms of the propensity of specific splicing events due to SNPs in coding or non-coding regions, there is unimaginable implications for the development of personalized treatment strategies. Moreover, it will be interesting to test whether one could indeed predict specific disease susceptibility and tolerance based on specific SNPs, which impact the alternative splicing. While several reports do show association between splice-site-specific SNPs and tuberculosis susceptibility, there is no report yet which shows such association for an infection-induced alternative splicing event. It is imperative to note that MSMD is the extreme representation of tuberculosis susceptibility whereas there is a broad spectrum between susceptibility to tolerance. How external factors (like bacterial virulence factors) could impact alternative splicing in an SNP dependent manner could provide resolution to the spectrum of tuberculosis susceptibility.
Altered isoform diversity and the consequent impact on protein domain expression reveal an additional layer of complexity to the host-pathogen interactions during infections [96]. There is a possibility to develop tools to analyze the expression of critical protein domains involved during host defense against bacterial infections and develop novel strategies to integrate this information into deciphering the host response to infection. Adding information about splice-site variations will further personalize the intervention strategies.
While the field of alternative splicing has grown tremendously in the last decade, tools available now in terms of next-generation sequencing and high-end computational capabilities, investigations on the aspects of alternative splicing discussed here, especially in the context of infectious diseases, could yield significant dividends.
Acknowledgements
The research in DK group is supported by the Wellcome-DBT India Alliance Senior Fellowship; Department of Biotechnology, Govt. of India; Science and Engineering Research Board, Govt. of India and Department of Science and Technology, Govt of India. KC is a recipient of a fellowship from Council for Scientific and Industrial Research, Govt. of India and RD receives a fellowship from the Department of Biotechnology, Govt. of India.
References
- [1].Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nature reviews Genetics. 2007;8:749–61. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- [3].Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RK, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347 doi: 10.1126/science.1254806. 1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D, et al. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–4. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- [6].Raj B, Blencowe BJ. Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- [7].Dredge BK, Polydorides AD, Darnell RB. The splice of life: alternative splicing and neurological disease. Nat Rev Neurosci. 2001;2:43–50. doi: 10.1038/35049061. [DOI] [PubMed] [Google Scholar]
- [8].Rizzacasa B, Morini E, Pucci S, Murdocca M, Novelli G, Amati F. LOX-1 and Its Splice Variants: A New Challenge for Atherosclerosis and Cancer-Targeted Therapies. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18020290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Doddapattar P, Gandhi C, Prakash P, Dhanesha N, Grumbach IM, Dailey ME, et al. Fibronectin Splicing Variants Containing Extra Domain A Promote Atherosclerosis in Mice Through Toll-Like Receptor 4. Arterioscler Thromb Vasc Biol. 2015;35:2391–400. doi: 10.1161/ATVBAHA.115.306474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Babaev VR, Porro F, Linton MF, Fazio S, Baralle FE, Muro AF. Absence of regulated splicing of fibronectin EDA exon reduces atherosclerosis in mice. Atherosclerosis. 2008;197:534–40. doi: 10.1016/j.atherosclerosis.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Smith JD, Peng DQ, Dansky HM, Settle M, Baglione J, Le Goff W, et al. Transcriptome profile of macrophages from atherosclerosis-sensitive and atherosclerosis-resistant mice. Mamm Genome. 2006;17:220–9. doi: 10.1007/s00335-005-0099-7. [DOI] [PubMed] [Google Scholar]
- [12].Tan MH, Sun Z, Opitz SL, Schmidt TE, Peters JH, George EL. Deletion of the alternatively spliced fibronectin EIIIA domain in mice reduces atherosclerosis. Blood. 2004;104:11–8. doi: 10.1182/blood-2003-09-3363. [DOI] [PubMed] [Google Scholar]
- [13].Liu F, Dai M, Xu Q, Zhu X, Zhou Y, Jiang S, et al. SRSF10-mediated IL1RAP alternative splicing regulates cervical cancer oncogenesis via mIL1RAP-NF-kappaB-CD47 axis. Oncogene. 2018 doi: 10.1038/s41388-017-0119-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Workenhe ST, Ketela T, Moffat J, Cuddington BP, Mossman KL. Genome-wide lentiviral shRNA screen identifies serine/arginine-rich splicing factor 2 as a determinant of oncolytic virus activity in breast cancer cells. Oncogene. 2016;35:2465–74. doi: 10.1038/onc.2015.303. [DOI] [PubMed] [Google Scholar]
- [15].Tripathi V, Sixt KM, Gao S, Xu X, Huang J, Weigert R, et al. Direct Regulation of Alternative Splicing by SMAD3 through PCBP1 Is Essential to the Tumor-Promoting Role of TGF-beta. Mol Cell. 2016;64:549–64. doi: 10.1016/j.molcel.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Panneerselvam J, Xie G, Che R, Su M, Zhang J, Jia W, et al. Distinct Metabolic Signature of Human Bladder Cancer Cells Carrying an Impaired Fanconi Anemia Tumor-Suppressor Signaling Pathway. J Proteome Res. 2016;15:1333–41. doi: 10.1021/acs.jproteome.6b00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Heinhuis B, Plantinga TS, Semango G, Kusters B, Netea MG, Dinarello CA, et al. Alternatively spliced isoforms of IL-32 differentially influence cell death pathways in cancer cell lines. Carcinogenesis. 2016;37:197–205. doi: 10.1093/carcin/bgv172. [DOI] [PubMed] [Google Scholar]
- [18].Wan Y, Zheng X, Chen H, Guo Y, Jiang H, He X, et al. Splicing function of mitotic regulators links R-loop-mediated DNA damage to tumor cell killing. J Cell Biol. 2015;209:235–46. doi: 10.1083/jcb.201409073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Toker A, Chin YR. Akt-ing up on SRPK1: oncogene or tumor suppressor? Mol Cell. 2014;54:329–30. doi: 10.1016/j.molcel.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Majerciak V, Lu M, Li X, Zheng ZM. Attenuation of the suppressive activity of cellular splicing factor SRSF3 by Kaposi sarcoma-associated herpesvirus ORF57 protein is required for RNA splicing. RNA. 2014;20:1747–58. doi: 10.1261/rna.045500.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Carpenter S, Ricci EP, Mercier BC, Moore MJ, Fitzgerald KA. Posttranscriptional regulation of gene expression in innate immunity. Nat Rev Immunol. 2014;14:361–76. doi: 10.1038/nri3682. [DOI] [PubMed] [Google Scholar]
- [22].Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155:397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nasser NJ, Avivi A, Shafat I, Edovitsky E, Zcharia E, Ilan N, et al. Alternatively spliced Spalax heparanase inhibits extracellular matrix degradation, tumor growth, and metastasis. Proc Natl Acad Sci U S A. 2009;106:2253–8. doi: 10.1073/pnas.0812846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].De Maio FA, Risso G, Iglesias NG, Shah P, Pozzi B, Gebhard LG, et al. The Dengue Virus NS5 Protein Intrudes in the Cellular Spliceosome and Modulates Splicing. PLoS Pathog. 2016;12:e1005841. doi: 10.1371/journal.ppat.1005841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hu B, Huo Y, Yang L, Chen G, Luo M, Yang J, et al. ZIKV infection effects changes in gene splicing, isoform composition and lncRNA expression in human neural progenitor cells. Virol J. 2017;14:217. doi: 10.1186/s12985-017-0882-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liu YC, Kuo RL, Lin JY, Huang PN, Huang Y, Liu H, et al. Cytoplasmic viral RNA-dependent RNA polymerase disrupts the intracellular splicing machinery by entering the nucleus and interfering with Prp8. PLoS pathogens. 2014;10:e1004199. doi: 10.1371/journal.ppat.1004199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Machiels B, Stevenson PG, Vanderplasschen A, Gillet L. A gammaherpesvirus uses alternative splicing to regulate its tropism and its sensitivity to neutralization. PLoS pathogens. 2013;9:e1003753. doi: 10.1371/journal.ppat.1003753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kalam H, Fontana MF, Kumar D. Alternate splicing of transcripts shape macrophage response to Mycobacterium tuberculosis infection. PLoS Pathog. 2017;13:e1006236. doi: 10.1371/journal.ppat.1006236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pai AA, Baharian G, Page Sabourin A, Brinkworth JF, Nedelec Y, Foley JW, et al. Widespread Shortening of 3' Untranslated Regions and Increased Exon Inclusion Are Evolutionarily Conserved Features of Innate Immune Responses to Infection. PLoS genetics. 2016;12:e1006338. doi: 10.1371/journal.pgen.1006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lynch KW. Consequences of regulated pre-mRNA splicing in the immune system. Nat Rev Immunol. 2004;4:931–40. doi: 10.1038/nri1497. [DOI] [PubMed] [Google Scholar]
- [31].Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–90. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Eichelbaum K, Krijgsveld J. Rapid temporal dynamics of transcription, protein synthesis, and secretion during macrophage activation. Mol Cell Proteomics. 2014;13:792–810. doi: 10.1074/mcp.M113.030916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hamilton KS, Phong B, Corey C, Cheng J, Gorentla B, Zhong X, et al. T Cell Receptor-Dependent Activation of mTOR Signaling in T Cells Is Mediated by Carma1 and MALT1, But Not Bcl10. Science Signaling. 2014;7:ra55–ra. doi: 10.1126/scisignal.2005169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lynch KW, Weiss A. A CD45 polymorphism associated with multiple sclerosis disrupts an exonic splicing silencer. J Biol Chem. 2001;276:24341–7. doi: 10.1074/jbc.M102175200. [DOI] [PubMed] [Google Scholar]
- [35].Magistrelli G, Jeannin P, Herbault NB, enoit De Coignac A, Gauchat JF, Bonnefoy JY, et al. A soluble form of CTLA-4 generated by alternative splicing is expressed by nonstimulated human T cells. Eur J Immunol. 1999;29:3596–602. doi: 10.1002/(SICI)1521-4141(199911)29:11<3596::AID-IMMU3596>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- [36].Xu Z, Weiss A. Negative regulation of CD45 by differential homodimerization of the alternatively spliced isoforms. Nat Immunol. 2002;3:764–71. doi: 10.1038/ni822. [DOI] [PubMed] [Google Scholar]
- [37].Arinobu Y, Atamas SP, Otsuka T, Niiro H, Yamaoka K, Mitsuyasu H, et al. Antagonistic effects of an alternative splice variant of human IL-4, IL-4delta2, on IL-4 activities in human monocytes and B cells. Cell Immunol. 1999;191:161–7. doi: 10.1006/cimm.1998.1431. [DOI] [PubMed] [Google Scholar]
- [38].Lillycrop KA, Latchman DS. Alternative splicing of the Oct-2 transcription factor RNA is differentially regulated in neuronal cells and B cells and results in protein isoforms with opposite effects on the activity of octamer/TAATGARAT-containing promoters. J Biol Chem. 1992;267:24960–5. [PubMed] [Google Scholar]
- [39].Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nature immunology. 2001;2:675–80. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- [40].Kawai T, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med. 2007;13:460–9. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- [41].O’Connor BP, Danhorn T, De Arras L, Flatley BR, Marcus RA, Farias-Hesson E, et al. Regulation of toll-like receptor signaling by the SF3a mRNA splicing complex. PLoS Genet. 2015;11:e1004932. doi: 10.1371/journal.pgen.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rao N, Nguyen S, Ngo K, Fung-Leung WP. A novel splice variant of interleukin-1 receptor (IL-1R)-associated kinase 1 plays a negative regulatory role in Toll/IL-1R-induced inflammatory signaling. Mol Cell Biol. 2005;25:6521–32. doi: 10.1128/MCB.25.15.6521-6532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Aslanzadeh V, Huang Y, Sanguinetti G, Beggs JD. Transcription rate strongly affects splicing fidelity and cotranscriptionality in budding yeast. Genome Res. 2018;28:203–13. doi: 10.1101/gr.225615.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Davari K, Lichti J, Gallus C, Greulich F, Uhlenhaut NH, Heinig M, et al. Rapid Genome-wide Recruitment of RNA Polymerase II Drives Transcription, Splicing, and Translation Events during T Cell Responses. Cell Reports. 2017;19:643–54. doi: 10.1016/j.celrep.2017.03.069. [DOI] [PubMed] [Google Scholar]
- [45].Chathoth KT, Barrass JD, Webb S, Beggs JD. A splicing-dependent transcriptional checkpoint associated with prespliceosome formation. Molecular cell. 2014;53:779–90. doi: 10.1016/j.molcel.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, et al. SR Proteins Collaborate with 7SK and Promoter-Associated Nascent RNA to Release Paused Polymerase. Cell. 2013;153:855–68. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, et al. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome research. 2014;24:896–905. doi: 10.1101/gr.171405.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dujardin G, Lafaille C, de la Mata M, Marasco LE, Munoz MJ, Le Jossic-Corcos C, et al. How slow RNA polymerase II elongation favors alternative exon skipping. Mol Cell. 2014;54:683–90. doi: 10.1016/j.molcel.2014.03.044. [DOI] [PubMed] [Google Scholar]
- [50].Aartsma-Rus A, van Ommen GJ. Less is more: therapeutic exon skipping for Duchenne muscular dystrophy. Lancet Neurol. 2009;8:873–5. doi: 10.1016/S1474-4422(09)70229-7. [DOI] [PubMed] [Google Scholar]
- [51].Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: an update. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Coombs TC, Tanega C, Shen M, Wang JL, Auld DS, Gerritz SW, et al. Small-molecule pyrimidine inhibitors of the cdc2-like (Clk) and dual specificity tyrosine phosphorylation-regulated (Dyrk) kinases: development of chemical probe ML315. Bioorg Med Chem Lett. 2013;23:3654–61. doi: 10.1016/j.bmcl.2013.02.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Fukuhara T, Hosoya T, Shimizu S, Sumi K, Oshiro T, Yoshinaka Y, et al. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc Natl Acad Sci U S A. 2006;103:11329–33. doi: 10.1073/pnas.0604616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3:576–83. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- [56].Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol. 2007;3:570–5. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- [57].Mendell JR, Rodino-Klapac LR, Sahenk Z, Roush K, Bird L, Lowes LP, et al. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann Neurol. 2013;74:637–47. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- [58].Spitali P, Rimessi P, Fabris M, Perrone D, Falzarano S, Bovolenta M, et al. Exon skipping-mediated dystrophin reading frame restoration for small mutations. Hum Mutat. 2009;30:1527–34. doi: 10.1002/humu.21092. [DOI] [PubMed] [Google Scholar]
- [59].Szekelyhidi Z, Pato J, Waczek F, Banhegyi P, Hegymegi-Barakonyi B, Eros D, et al. Synthesis of selective SRPK-1 inhibitors: novel tricyclic quinoxaline derivatives. Bioorg Med Chem Lett. 2005;15:3241–6. doi: 10.1016/j.bmcl.2005.04.064. [DOI] [PubMed] [Google Scholar]
- [60].Voit T, Topaloglu H, Straub V, Muntoni F, Deconinck N, Campion G, et al. Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): an exploratory, randomised, placebo-controlled phase 2 study. Lancet Neurol. 2014;13:987–96. doi: 10.1016/S1474-4422(14)70195-4. [DOI] [PubMed] [Google Scholar]
- [61].Yokota T, Nakamura A, Nagata T, Saito T, Kobayashi M, Aoki Y, et al. Extensive and prolonged restoration of dystrophin expression with vivo-morpholino-mediated multiple exon skipping in dystrophic dogs. Nucleic Acid Ther. 2012;22:306–15. doi: 10.1089/nat.2012.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Naryshkin NA, Weetall M, Dakka A, Narasimhan J, Zhao X, Feng Z, et al. Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science. 2014;345:688–93. doi: 10.1126/science.1250127. [DOI] [PubMed] [Google Scholar]
- [63].Bates DO, Morris JC, Oltean S, Donaldson LF. Pharmacology of Modulators of Alternative Splicing. Pharmacol Rev. 2017;69:63–79. doi: 10.1124/pr.115.011239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Meininger I, Griesbach RA, Hu D, Gehring T, Seeholzer T, Bertossi A, et al. Alternative splicing of MALT1 controls signalling and activation of CD4(+) T cells. Nature communications. 2016;7 doi: 10.1038/ncomms11292. 11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Blum H, Wolf M, Enssle K, Rollinghoff M, Gessner A. Two distinct stimulus-dependent pathways lead to production of soluble murine interleukin-4 receptor. Journal of immunology (Baltimore, Md : 1950) 1996;157:1846–53. [PubMed] [Google Scholar]
- [66].Iwami K-i, Matsuguchi T, Masuda A, Kikuchi T, Musikacharoen T, Yoshikai Y. Cutting Edge: Naturally Occurring Soluble Form of Mouse Toll-Like Receptor 4 Inhibits Lipopolysaccharide Signaling. The Journal of Immunology. 2000;165:6682–6. doi: 10.4049/jimmunol.165.12.6682. [DOI] [PubMed] [Google Scholar]
- [67].Tavernier J, Tuypens T, Plaetinck G, Verhee A, Fiers W, Devos R. Molecular basis of the membrane-anchored and two soluble isoforms of the human interleukin 5 receptor alpha subunit. Proceedings of the National Academy of Sciences. 1992;89:7041–5. doi: 10.1073/pnas.89.15.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Biasiotto R, Akusjarvi G. Regulation of human adenovirus alternative RNA splicing by the adenoviral L4-33K and L4-22K proteins. Int J Mol Sci. 2015;16:2893–912. doi: 10.3390/ijms16022893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Graham SV, Faizo AAA. Control of human papillomavirus gene expression by alternative splicing. Virus Res. 2017;231:83–95. doi: 10.1016/j.virusres.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Stoltzfus CM. Chapter 1. Regulation of HIV-1 alternative RNA splicing and its role in virus replication. Adv Virus Res. 2009;74:1–40. doi: 10.1016/S0065-3527(09)74001-1. [DOI] [PubMed] [Google Scholar]
- [71].Wu C, Kajitani N, Schwartz S. Splicing and Polyadenylation of Human Papillomavirus Type 16 mRNAs. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18020366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Thompson MG, Munoz-Moreno R, Bhat P, Roytenberg R, Lindberg J, Gazzara MR, et al. Co-regulatory activity of hnRNP K and NS1-BP in influenza and human mRNA splicing. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04779-4. 2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Ganaie SS, Chen AY, Huang C, Xu P, Kleiboeker S, Du A, et al. RNA Binding Protein RBM38 Regulates Expression of the 11-Kilodalton Protein of Parvovirus B19, Which Facilitates Viral DNA Replication. J Virol. 2018;92 doi: 10.1128/JVI.02050-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Alvarez E, Castello A, Carrasco L, Izquierdo JM. Poliovirus 2A protease triggers a selective nucleo-cytoplasmic redistribution of splicing factors to regulate alternative pre-mRNA splicing. PLoS One. 2013;8:e73723. doi: 10.1371/journal.pone.0073723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Barnhart MD, Moon SL, Emch AW, Wilusz CJ, Wilusz J. Changes in cellular mRNA stability, splicing, and polyadenylation through HuR protein sequestration by a cytoplasmic RNA virus. Cell Rep. 2013;5:909–17. doi: 10.1016/j.celrep.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Bryant HE, Wadd SE, Lamond AI, Silverstein SJ, Clements JB. Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step. J Virol. 2001;75:4376–85. doi: 10.1128/JVI.75.9.4376-4385.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lee N, Pimienta G, Steitz JA. AUF1/hnRNP D is a novel protein partner of the EBER1 noncoding RNA of Epstein-Barr virus. RNA. 2012;18:2073–82. doi: 10.1261/rna.034900.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Qiu Y, Nemeroff M, Krug RM. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA. 1995;1:304–16. [PMC free article] [PubMed] [Google Scholar]
- [79].Rivera-Serrano EE, Fritch EJ, Scholl EH, Sherry B. A Cytoplasmic RNA Virus Alters the Function of the Cell Splicing Protein SRSF2. J Virol. 2017;91 doi: 10.1128/JVI.02488-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Boudreault S, Martenon-Brodeur C, Caron M, Garant JM, Tremblay MP, Armero VE, et al. Global Profiling of the Cellular Alternative RNA Splicing Landscape during Virus-Host Interactions. PLoS One. 2016;11:e0161914. doi: 10.1371/journal.pone.0161914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Tormanen H, Backstrom E, Carlsson A, Akusjarvi G. L4-33K, an adenovirus-encoded alternative RNA splicing factor. J Biol Chem. 2006;281:36510–7. doi: 10.1074/jbc.M607601200. [DOI] [PubMed] [Google Scholar]
- [82].Kadri F, Pacifici M, Wilk A, Parker-Struckhoff A, Del Valle L, Hauser KF, et al. HIV-1-Tat Protein Inhibits SC35-mediated Tau Exon 10 Inclusion through Up-regulation of DYRK1A Kinase. J Biol Chem. 2015;290:30931–46. doi: 10.1074/jbc.M115.675751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Pettit Kneller EL, Connor JH, Lyles DS. hnRNPs Relocalize to the cytoplasm following infection with vesicular stomatitis virus. J Virol. 2009;83:770–80. doi: 10.1128/JVI.01279-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Dowling D, Nasr-Esfahani S, Tan CH, O’Brien K, Howard JL, Jans DA, et al. HIV-1 infection induces changes in expression of cellular splicing factors that regulate alternative viral splicing and virus production in macrophages. Retrovirology. 2008;5:18. doi: 10.1186/1742-4690-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Agrawal AK, Ranjan R, Chandra S, Rout TK, Misra A, Reddy TJ. Some proteins of M. tuberculosis that localise to the nucleus of THP-1-derived macrophages. Tuberculosis (Edinb) 2016;101:75–8. doi: 10.1016/j.tube.2016.07.013. [DOI] [PubMed] [Google Scholar]
- [86].Penn BH, Netter Z, Johnson JR, Von Dollen J, Jang GM, Johnson T, et al. An Mtb-Human Protein-Protein Interaction Map Identifies a Switch between Host Antiviral and Antibacterial Responses. Mol Cell. 2018;71:637–48 e5. doi: 10.1016/j.molcel.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Luo Z, Li Z, Chen K, Liu R, Li X, Cao H, et al. Engagement of heterogeneous nuclear ribonucleoprotein M with listeriolysin O induces type I interferon expression and restricts Listeria monocytogenes growth in host cells. Immunobiology. 2012;217:972–81. doi: 10.1016/j.imbio.2012.01.009. [DOI] [PubMed] [Google Scholar]
- [88].Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol. 2015;13:343–59. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- [89].Sharma G, Upadhyay S, Srilalitha M, Nandicoori VK, Khosla S. The interaction of mycobacterial protein Rv2966c with host chromatin is mediated through non-CpG methylation and histone H3/H4 binding. Nucleic Acids Res. 2015;43:3922–37. doi: 10.1093/nar/gkv261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Yaseen I, Kaur P, Nandicoori VK, Khosla S. Mycobacteria modulate host epigenetic machinery by Rv1988 methylation of a non-tail arginine of histone H3. Nat Commun. 2015;6 doi: 10.1038/ncomms9922. 8922. [DOI] [PubMed] [Google Scholar]
- [91].Sato S, Li K, Kameyama T, Hayashi T, Ishida Y, Murakami S, et al. The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity. 2015;42:123–32. doi: 10.1016/j.immuni.2014.12.016. [DOI] [PubMed] [Google Scholar]
- [92].Deng W, Shi M, Han M, Zhong J, Li Z, Li W, et al. Negative regulation of virus-triggered IFN-beta signaling pathway by alternative splicing of TBK1. J Biol Chem. 2008;283:35590–7. doi: 10.1074/jbc.M805775200. [DOI] [PubMed] [Google Scholar]
- [93].Hu YW, Zhang J, Wu XM, Cao L, Nie P, Chang MX. TANK-Binding Kinase 1 (TBK1) Isoforms Negatively Regulate Type I Interferon Induction by Inhibiting TBK1-IRF3 Interaction and IRF3 Phosphorylation. Front Immunol. 2018;9:84. doi: 10.3389/fimmu.2018.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Robinson RT, Khader SA, Martino CA, Fountain JJ, Teixeira-Coelho M, Pearl JE, et al. Mycobacterium tuberculosis infection induces il12rb1 splicing to generate a novel IL-12Rbeta1 isoform that enhances DC migration. J Exp Med. 2010;207:591–605. doi: 10.1084/jem.20091085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Liang G, Malmuthuge N, Guan Y, Ren Y, Griebel PJ, Guan le L. Altered microRNA expression and pre-mRNA splicing events reveal new mechanisms associated with early stage Mycobacterium avium subspecies paratuberculosis infection. Sci Rep. 2016;6 doi: 10.1038/srep24964. 24964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Kalam HSK, Chauhan K, Fontana MM, Kumar D. Alternate splicing of transcripts upon Mycobacterium tuberculosis infection impacts the expression of functional protein domains. IUBMB Life. 2018 doi: 10.1002/iub.1887. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Jayaswal S, Kamal MA, Dua R, Gupta S, Majumdar T, Das G, et al. Identification of host-dependent survival factors for intracellular Mycobacterium tuberculosis through an siRNA screen. PLoS pathogens. 2010;6:e1000839. doi: 10.1371/journal.ppat.1000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Kumar D, Nath L, Kamal MA, Varshney A, Jain A, Singh S, et al. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140:731–43. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
- [99].Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–6. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- [100].Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–5. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Dlamini Z, Hull R. Can the HIV-1 splicing machinery be targeted for drug discovery? HIV AIDS (Auckl) 2017;9:63–75. doi: 10.2147/HIV.S120576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Shkreta L, Blanchette M, Toutant J, Wilhelm E, Bell B, Story BA, et al. Modulation of the splicing regulatory function of SRSF10 by a novel compound that impairs HIV-1 replication. Nucleic Acids Res. 2017;45:4051–67. doi: 10.1093/nar/gkw1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Russell DG, Barry CE, 3rd, Flynn JL. Tuberculosis: what we don’t know can, and does, hurt us. Science. 2010;328:852–6. doi: 10.1126/science.1184784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Baralle D, Lucassen A, Buratti E. Missed threads. The impact of pre-mRNA splicing defects on clinical practice. EMBO Rep. 2009;10:810–6. doi: 10.1038/embor.2009.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Soukarieh O, Gaildrat P, Hamieh M, Drouet A, Baert-Desurmont S, Frebourg T, et al. Exonic Splicing Mutations Are More Prevalent than Currently Estimated and Can Be Predicted by Using In Silico Tools. PLoS genetics. 2016;12:e1005756. doi: 10.1371/journal.pgen.1005756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Karambataki M, Malousi A, Tzimagiorgis G, Haitoglou C, Fragou A, Georgiou E, et al. Association of two synonymous splicing-associated CpG single nucleotide polymorphisms in calpain 10 and solute carrier family 2 member 2 with type 2 diabetes. Biomed Rep. 2017;6:146–58. doi: 10.3892/br.2016.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Mascarenhas JB, Tchourbanov AY, Fan H, Danilov SM, Wang T, Garcia JG. Mechanical Stress and Single Nucleotide Variants Regulate Alternative Splicing of the MYLK Gene. Am J Respir Cell Mol Biol. 2017;56:29–37. doi: 10.1165/rcmb.2016-0053OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Tejedor JR, Tilgner H, Iannone C, Guigo R, Valcarcel J. Role of six single nucleotide polymorphisms, risk factors in coronary disease, in OLR1 alternative splicing. RNA. 2015;21:1187–202. doi: 10.1261/rna.049890.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Moller M, de Wit E, Hoal EG. Past, present and future directions in human genetic susceptibility to tuberculosis. FEMS Immunol Med Microbiol. 2010;58:3–26. doi: 10.1111/j.1574-695X.2009.00600.x. [DOI] [PubMed] [Google Scholar]
- [111].Lundtoft C, Awuah AA, Guler A, Harling K, Schaal H, Mayatepek E, et al. An IL7RA exon 5 polymorphism is associated with impaired IL-7Ralpha splicing and protection against tuberculosis in Ghana. Genes Immun. 2018 doi: 10.1038/s41435-018-0049-5. [DOI] [PubMed] [Google Scholar]
- [112].Kong XF, Martinez-Barricarte R, Kennedy J, Mele F, Lazarov T, Deenick EK, et al. Disruption of an antimycobacterial circuit between dendritic and helper T cells in human SPPL2a deficiency. Nat Immunol. 2018;19:973–85. doi: 10.1038/s41590-018-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]