Summary
The use of alternative translation initiation sites enables production of more than one protein from a single gene, thereby expanding cellular proteome. Although several such examples have been serendipitously found in bacteria, genome-wide mapping of alternative translation start sites has been unattainable. We found that the antibiotic retapamulin specifically arrests initiating ribosomes at start codons of the genes. Retapamulin-enhanced Ribo-seq analysis (Ribo-RET) not only allowed mapping of conventional initiation sites at the beginning of the genes but, strikingly, it also revealed putative internal start sites in a number of Escherichia coli genes. Experiments demonstrated that the internal start codons can be recognized by the ribosomes and direct translation initiation in vitro and in vivo. Proteins, whose synthesis is initiated at an internal in-frame and out-of-frame start sites, can be functionally important and contribute to the ‘alternative’ bacterial proteome. The internal start sites may also play regulatory roles in gene expression.
Introduction
A broader diversity of proteins with specialized functions can augment cell reproduction capacity, optimize its metabolism, and facilitate survival in the ever-changing environment. However, the fitness gain acquired by making a new protein is counterbalanced with the cost of expanding the size of the genome, a conundrum particularly onerous in bacteria whose genomes are highly streamlined.
Different strategies can be used for diversifying proteome without expanding genome size. For instance, ribosomes may initiate at a unique start codon of an open reading frame (ORF), but due to programmed ribosomal frameshifting or stop codon readthrough, some of them may produce a polypeptide whose sequence deviates from that encoded in the main ORF. Such recoding events lead to generation of more than one protein from a single gene (Atkins et al., 2016; Baranov et al., 2015).
Another possible way for producing diverse polypeptides from a single ORF is the utilization of alternative internally located start codons. Although translation of the majority of the bacterial genes is initiated at a unique TIS, designated herein as primary (pTIS), several examples of genes with an additional internal TIS (iTIS) have been uncovered by detecting additional polypeptide products during the purification of the primary protein (reviewed in (Meydan et al., 2018)). In these genes, translation initiated at the pTIS results in production of the full-length (primary) protein, while ribosomes that initiate translation at the in-frame iTIS synthesize an alternative, N-terminally truncated polypeptide. Such primary and alternative proteins may have related but specialized functions. The products of inframe internal initiation at several bacterial genes have been reported to participate in various cellular functions ranging from virulence, to photosynthesis, or antibiotic production among others (reviewed in (Meydan et al., 2018)). In very few known cases, iTIS directs translation in a reading frame different from the primary ORF (D'Souza et al., 1994; Feltens et al., 2003; Yuan et al., 2018).
Most of the known examples of translation of a protein from an iTIS have been discovered serendipitously. Although several computational algorithms can predict the pTIS of many bacterial ORFs (Gao et al., 2010; Giess et al., 2017; Makita et al., 2007; Salzberg et al., 1998), iTIS prediction remains by far more challenging and has not even been pursued in most of those studies. The recent advent of new mass-spectrometry-based approaches have allowed the identification of N-terminal peptides of a range of proteins expressed in bacteria (Bienvenut et al., 2015; Impens et al., 2017), including some whose translation was likely initiated at an iTIS. However, the success of the available techniques for identifying such proteins is intrinsically restricted by the stringent requirements for the chemical properties, size, and abundance of the peptides that can be detected by mass-spectrometry. Therefore, the majority of the functional iTISs in the genomes likely remain overlooked.
Ribosome profiling (Ribo-seq), based on deep sequencing of ribosome protected mRNA fragments (“ribosome footprints”), allows for genome-wide survey of translation (Ingolia et al., 2009). Ribo-seq experiments carried out with eukaryotic cells pre-treated with the translation initiation inhibitors harringtonine (Ingolia et al., 2011) and lactimidomycin (Gao et al., 2015; Lee et al., 2012) or with puromycin (Fritsch et al., 2012), showed specific enrichment of ribosome footprints at or near start codons of ORFs and facilitated mapping of TISs in eukaryotic genomes. These studies also revealed active translation of previously unknown short ORFs in the 5’ UTRs of many genes and identified several TISs within the genes that were attributed to leaky scanning through the primary start sites (Lee et al., 2012). Analogous studies, however, have been difficult to conduct in bacteria because of the paucity of inhibitors with the required mechanism of action. An inhibitor useful for mapping start sites should allow the assembly of the 70S translation initiation complex at a TIS but must prevent the ribosome from leaving the start codon. Unfortunately, most of the ribosomal antibiotics traditionally viewed as initiation inhibitors do not satisfy these criteria. Recently, tetracycline (TET), an antibiotic that prevents aminoacyl-tRNAs from entering the ribosomal A site and commonly known as an elongation inhibitor (Cundliffe, 1981), was used in conjunction with Ribo-seq to globally map pTISs in the E. coli genome (Nakahigashi et al., 2016). Although TET Ribo-seq data successfully revealed the pTISs of many of the actively translated genes, identification of iTISs was not feasible in that work because of the substantial number of footprints generated by elongating ribosomes. Furthermore, because TET can potentially bind to the ribosome at every round of elongation cycle, when the A-site is temporarily empty, it is impossible to distinguish whether the footprints within the ORFs represented elongating ribosomes or those initiating translation at an iTIS (Nakahigashi et al., 2016).
Here we show that retapamulin (RET), an antibiotic of the pleuromutilin family, exclusively stalls ribosomes at the start codons of the ORFs. Brief pretreatment of E. coli cells with RET dramatically rearranges the distribution of ribosomes along the ORFs, confining the ribosomal footprints obtained by Riboseq to the TISs of the genes. Strikingly, the application of the Ribo-seq/RET approach to the analysis of bacterial translation revealed that more than many E. coli genes contain actively used iTISs. In vitro and in vivo experiments confirmed initiation of translation at some of the discovered iTISs and show that internal initiation may lead to production of proteins with distinct functions. Our data show that initiation at alternative start sites is widespread in bacteria and reveal the possible existence of a previously cryptic fraction of the proteome.
Results
RET arrests the initiating ribosome at the start codons
Pleuromutilin antibiotics, including clinically-used semi-synthetic RET, bind in the peptidyl transferase center (PTC) of the bacterial ribosome, hindering the placement of the P- and A-site amino acids and thus preventing peptide bond formation (Figure S1A and S1B) (Davidovich et al., 2007; Poulsen et al., 2001; Schlunzen et al., 2004). In vitro studies have shown that presence of fMet-tRNA and RET in the ribosome are not mutually exclusive (Yan et al., 2006). Therefore, we reasoned that RET may allow the assembly of the 70S initiation complex at the start codon, but by displacing the aminoacyl moiety of the initiator fMet-tRNA and interfering with the placement of an elongator aminoacyl-tRNA in the A site, it could prevent formation of the first peptide bond.
The results of polysome analysis were compatible with RET being a selective translation initiation inhibitor, because treatment of E. coli cells with high concentrations of the drug, 100-fold over the minimal inhibitory concentration (MIC), rapidly converted polysomes into monosomes (Figure S1C). We then used toeprinting analysis ((Hartz et al., 1988) to test whether RET captures ribosomes at start codons. When model genes were translated in an E. coli cell-free system (Shimizu et al., 2001), addition of RET stalled ribosomes exclusively at the ORFs start codons (Figure 1A, ‘RET’ lanes), demonstrating that this antibiotic readily, and possibly specifically, inhibits translation initiation. In contrast, TET, which was used previously to map TISs in the E. coli genome (Nakahigashi et al., 2016), halted translation not only at the translation initiation sites but also at downstream codons of the ORFs (Figure 1A, ‘TET’ lanes), confirming that this inhibitor interferes with both initiation and elongation of translation (Orelle et al., 2013).
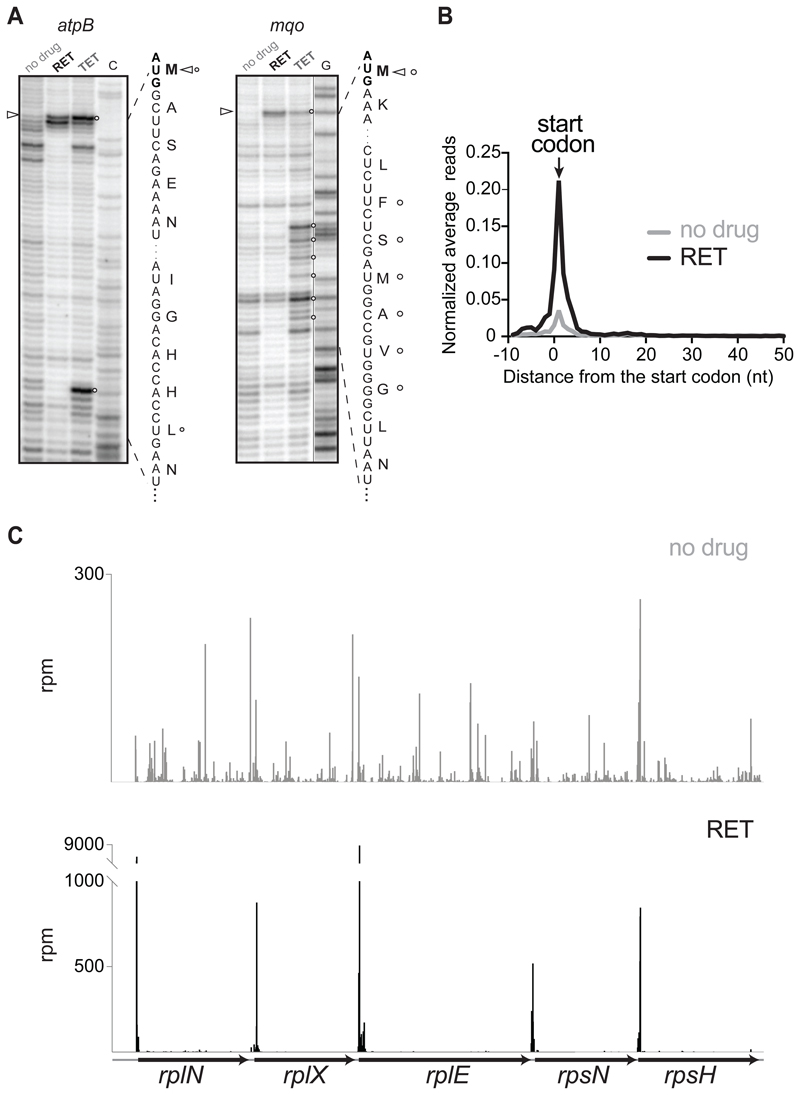
Figure 1. RET specifically arrests ribosomes at translation initiation sites.
(A) Toeprinting analysis showing retapamulin (RET) (triangles)- and tetracycline (TET) (circles)- induced translation arrest sites during cell-free translation of two model E. coli genes. ‘C’ and ‘G’ indicate sequencing lanes. Nucleotide and encoded amino acid sequences are shown.
(B) Metagene analysis plot representing normalized average relative density reads in the vicinity of the annotated start codons of the genes of E. coli cells treated or not with RET.
(C) Ribosome footprints density within the spc operon in cells treated or not with RET.
See also Figures S1.
The outcomes of the polysome- and toeprinting analyses, along with the structural data showing the incompatibility of the nascent protein chain with RET binding (Figure S1B), encouraged us to assess whether RET would enable the use of Ribo-seq for mapping translation start sites in bacterial cells. Even a brief (2 min) exposure of the ΔtolC derivative of the E. coli strain BW25113 to a 32-fold MIC of RET nearly completely halts protein synthesis (Figure S1D). However, in Ribo-seq experiments we exposed cells for 5 min to a 100-fold MIC of RET to ensure that elongating ribosomes complete translation of even long or slowly-translated ORFs prior to cell harvesting. Analysis of the Ribo-seq data showed that the RET treatment led to a striking ribosome redistribution. The occupancy of the internal and termination codons of the expressed genes was severely reduced compared to that of the untreated control, whereas the ribosome density peaks at the start codons dramatically increased (Figure 1B and 1C). Although a generally similar trend can be observed in the metagene analysis of the Ribo-seq data in the RET- (this paper) and TET-treated cells (Nakahigashi et al., 2016), the startcodons peak in the TET experiments is smaller and broader compared to the peak of the RET-stalled ribosomes (Figure S1E and S1F), reflecting a higher potency of RET as initiation inhibitor. Filtered by fairly conservative criteria (see STAR Methods), Ribo-seq data revealed distinct peaks of ribosome density at the annotated start codons (pTISs) of 991 out of 1153 (86%) E. coli genes expressed in the BW25113. The magnitude of the start codon peaks at the remaining 14% of the translated genes did not pass our threshold criteria (see STAR Methods) possibly reflecting changes in gene expression due to the RET treatment.
Taken together, our in vitro and in vivo results showed that RET acts as a specific inhibitor of translation initiation locking the ribosomes at the start codons, and in combination with Ribo-seq can be used for mapping the pTISs of the majority of actively translated genes in bacterial genomes. We named the Riboseq/RET approach ‘Ribo-RET’.
Ribo-RET unmasks initiation of translation at internal codons of many bacterial genes
Even though the majority of the ribosome footprints in the Ribo-RET dataset mapped to annotated pTISs, we also observed peaks at certain internal codons (Figure 2A). Hypothetically, the presence of internal Ribo-RET peaks could be explained by elongating ribosomes paused at specific sites within the ORF. Nonetheless, this possibility seems unlikely, since no substantial Ribo-RET peak was detected even at the most prominent programmed translation arrest site in the E. coli genome within the secM ORF (Nakatogawa and Ito, 2002) (Figure S1G). Similarly implausible was the origin of the internal RET peaks from context-specific elongation arrest observed with some other antibiotics (Kannan et al., 2014; Marks et al., 2016) because biochemical (Dornhelm and Hogenauer, 1978) and structural (Davidovich et al., 2007) data strongly argue that RET cannot bind to the elongating ribosome (Figure S1B). We therefore concluded that the Ribo-RET peaks at internal sites within ORFs must represent ribosomes caught in the act of initiating translation.
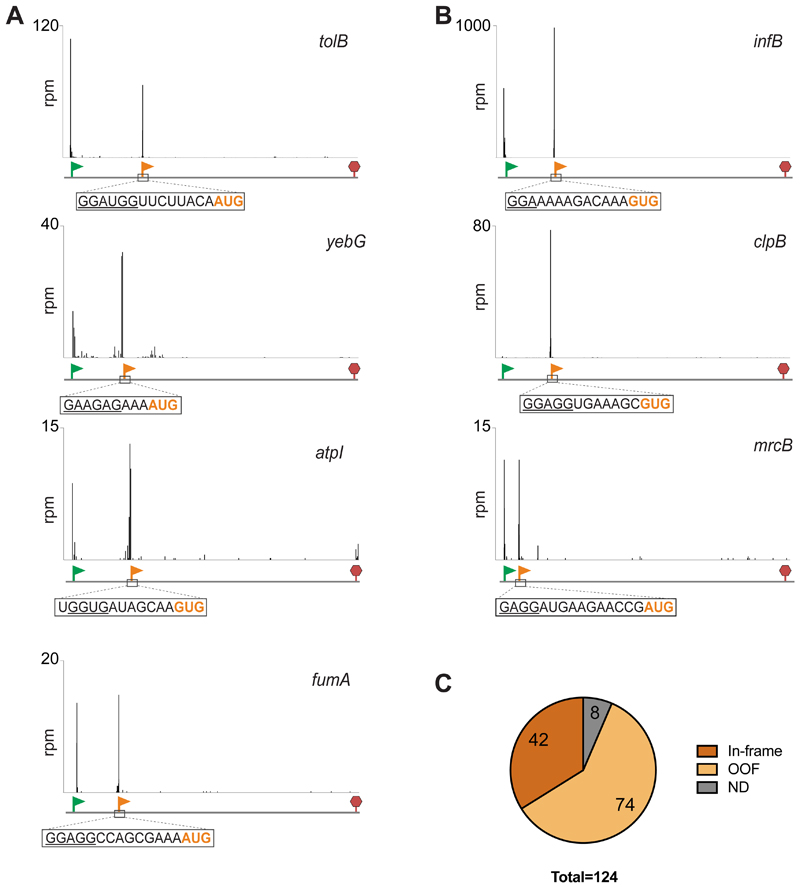
Figure 2. Ribo-RET reveals the presence of iTISs in many bacterial genes.
(A) Examples of Ribo-RET profiles of E. coli genes with newly detected iTISs. The annotated pTISs are marked with green flags and stop codons are shown with red stop signs; orange flags show the iTISs. iTIS start codons are highlighted in orange and the SD-like sequences are underlined.
(B) Ribo-RET profiles of infB, clpB, mrcB, the three E. coli genes where iTISs had been previously characterized.
(C) The iTISs common between the E. coli BW25113 and BL21 strains. ND (not determined): the internal Ribo-RET peaks not associated with the known start codons.
See also Table S1.
Three E. coli genes, infB, mcrB and clpB, were previously reported to encode two different polypeptide isoforms due to the iTIS presence: translation of the full-size protein is initiated at the pTIS while the shorter isoform is expressed from an iTIS (Broome-Smith et al., 1985; Park et al., 1993; Plumbridge et al., 1985). The Ribo-RET profile of these genes showed well-defined and highly-specific ribosome density peaks (Figure 2B) at the known iTISs, thereby verifying the utility of Ribo-RET for mapping iTISs in bacterial genes.
Among the E. coli BW25113 genes expressed in our conditions, we identified 239 iTIS candidates. To further expand the systematic identification of iTISs in E. coli genes, we applied the Ribo-RET approach to the ΔtolC derivative of the E. coli strain BL21, a B-type strain which is genetically distinct from the K-strain BW25113 (Grenier et al., 2014; Studier et al., 2009). Ribo-RET analysis identified 620 iTISs in the BL21 strain. Of these, 124 iTISs were common between the two strains (Table S1). While a notably higher number of iTIS in the BL21 remains somewhat puzzling, it may be related to the fact that more genes are expressed in this strain in comparison with BW25113 (1990 genes with the identified pTISs in BL21 strain vs 1554 such genes in BW25113), and to the sequence variations between the strains, as a result of which 244 BL21-specific iTISs did not have a perfect sequence match in the BW25113 strain. We limited our subsequent analysis to 124 iTISs conserved between the two strains, among which, 42 directed translation in frame with the main gene, whereas start codons of 74 iTISs were out of frame relative to the main ORF; for 8 iTISs the start site was not assigned (Figure 2C and Table S1). In the following sections we consider the first two classes separately.
Internal translation initiation sites that are in frame with the main ORF
The in-frame iTISs exploit various initiator codons that have been shown previously to be capable of directing translation initiation in E. coli (Chengguang et al., 2017; Hecht et al., 2017), although similar to the pTISs, the AUG codon is the most prevalent (Figure 3A). A SD-like sequence could be recognized upstream of many of the in-frame internal start codons (Table S1).
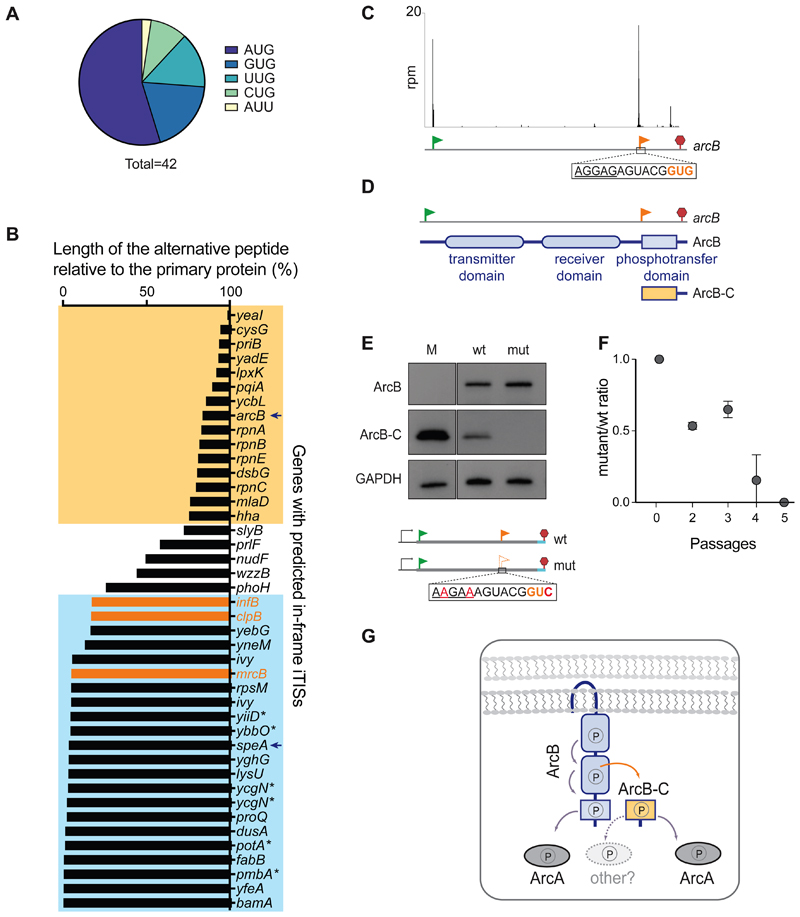
Figure 3. In-frame internal initiation can generate functional N-terminally truncated proteins.
(A) The frequency of various putative start codons at the in-frame iTISs. (B) The relative length of the predicted alternative proteins, products of internal initiation, in comparison with the main protein. The known examples of genes with in-frame iTISs (Figure 2B) are in orange. The genes with the iTISs located within the 3’ or 5’ quartile of the gene length are boxed in yellow or blue, respectively. Asterisks show genes with pTIS re-annotation proposed based on TET-assisted Ribo-seq, (Nakahigashi et al., 2016). Arrows point at arcB and speA genes further analyzed in this work.
(C) Ribo-RET profile of arcB showing ribosomal density peaks at the pTIS (green flag) and the iTIS (orange flag).
(D) Schematics of the functional domains of ArcB. The putative alternative ArcB-C protein would encompass the phosphotransfer domain.
(E) Western blot analysis of the C-terminally 3XFLAG-tagged translation products of the arcB gene expressed from a plasmid in E. coli cells. Inactivation of iTIS by the indicated mutations (mut) abrogates production of ArcB-C. Lane M: individually-expressed marker protein ArcB-C-3XFLAG (see STAR Methods).
(F) Functional iTIS in arcB facilitates growth under low oxygen conditions. BW25113 ΔarcB E. coli cells, expressing from a plasmid either wt arcB or mutant arcB with inactivated iTIS, were co-grown in low oxygen conditions and the ratio of mutant to wt cells was analyzed (see Start Methods for details). Error bars represent deviation from the mean (n=2).
(G) The phosphorelay across the wt ArcB domains results in the activation of the response regulator ArcA (Alvarez et al., 2016). Diffusible ArcB-C could amplify the signal capabilities of the ArcBA system and/or enable cross-talk with other response regulators.
See also Figure S2.
Initiation at an in-frame iTIS would generate an N-terminally truncated form of the primary protein. The sizes of candidate proteins expressed from in-frame iTISs range from 6 to 805 amino acids in length (Table S1). Although the locations of in-frame iTISs are highly variable, the majority are clustered close to the beginning of the gene or are within the 3’ terminal quartile of the ORF length (Figure 3B). We examined the iTIS of the arcB gene as a 3’-proximal start site representative and that of speA as an example of the 5’-proximal iTIS.
A protein with a putative specialized function is translated from the 3’-proximal iTIS of the arcB gene
The gene arcB encodes the sensor kinase ArcB of the two-component signal transduction system ArcB/A that helps bacteria to sense and respond to changes in oxygen concentration (Alvarez and Georgellis, 2010) (Figure 3C and 3D). The ArcB protein consists of the transmitter, receiver and phosphotransfer domains (Figure 3D). Under microaerobic conditions, ArcB undergoes a series of phosphorylation steps that eventually activate the response regulator ArcA that controls expression of nearly 200 genes (Alvarez and Georgellis, 2010; Salmon et al., 2005). The C-terminal ArcB-C domain is the ultimate receiver of the phosphoryl group within the ArcB membrane-anchored protein and serves as the phosphoryl donor for ArcA (Alvarez et al., 2016).
The Ribo-RET data showed a strong ribosome density peak at an iTIS in arcB, with the putative start codon GUG located precisely at the 5’ boundary of the segment encoding the ArcB-C domain (Figure 3C, D). A similarly located iTIS can be found in the arcB gene of several bacterial species (Figure S2B). Initiation of translation at the arcB iTIS could generate a diffusible ArcB-C polypeptide, detached from the membrane-bound ArcB kinase (Figure 3D). To test this possibility, we introduced the 3xFLAG-coding sequence at the 3’ end of the arcB gene, expressed it from a plasmid in E. coli cells and analyzed the protein products.
Expression of the tagged arcB resulted in the simultaneous production of the full-size ArcB and of a smaller protein with apparent molecular weight (MW) of 14 kDa, consistent with that of the FLAG-tagged ArcB-C (Figures 3E and S2A). Disruption of the iTIS by synonymous mutations did not affect the synthesis of the full-length ArcB but abrogated that of ArcB-C (Figure 3E) confirming that the ArcB-C polypeptide is produced via initiation of translation at the arcB iTIS. Previous in vitro experiments showed that the isolated ArcB-C domain could serve as a phosphoryl acceptor and donor for the ArcB-catalyzed phosphorylation reactions (Alvarez and Georgellis, 2010), suggesting that a self-standing ArcB-C protein is likely functional in vivo. In agreement with this possibility, under micro-aerobic conditions E. coli cells with the operational arcB iTIS win over the mutant in which the iTIS is disrupted by synonymous mutations (Figures 3F and S2C). Diffusible ArcB-C may either facilitate the operation of the ArcB-ArcA signal transduction pathway or could enable a cross-talk with other signal transduction systems (Figure 3G).
The expression of ArcB-C from the arcB iTIS is apparently quite efficient because E. coli and Salmonella enterica Ribo-seq datasets show a notable upshift in the ribosome density at the arcB codons located downstream from the iTIS (Baek et al., 2017; Kannan et al., 2014; Li et al., 2014) (Figure S2D-S2G). Curiously, the average ribosome occupancy of the arcB codons before and after the iTIS vary under different physiological conditions (Figure S2F and S2G), suggesting that utilization of the arcB pTIS and iTIS could be regulated.
Another remarkable example of in-frame 3’-proximal iTISs is found in the homologous rpnA-E genes of E. coli, encoding nucleases involved in DNA recombination (Kingston et al., 2017). Each of the rpn genes show Ribo-RET peaks at iTISs that appear to be their major initiation sites (Figure S2H) under the growth conditions of our experiments. Curiously, the polypeptide expressed from the rpnE iTIS is 98% identical to the product of the ypaA gene (Figure S2I), revealing a possible distinct functionality of the alternative products of the rpn gene family.
5’-proximal iTIS gene may generate differentially-targeted proteins
The speA gene encodes arginine decarboxylase (SpeA), an enzyme involved in polyamine production (Michael, 2016). SpeA has been found in the E. coli cytoplasmic and periplasmic fractions (Buch and Boyle, 1985) and was reported to be represented by two polypeptide isoforms, SpeA-74, with an apparent MW of 74 kDa, and a smaller one of ~ 70 kDa, SpeA-70, suggested to be a co-secretional maturation product of the full-length SpeA-74 (Buch and Boyle, 1985; Wu and Morris, 1973). Our analysis, however, revealed two Ribo-RET peaks in the speA ORF: one corresponding to the annotated pTIS and the second one mapped to an iTIS at codon Met-26 (Figure S3A). Initiation of translation at the pTIS and iTIS of speA would generate the 73,767 Da and 71,062 Da forms of SpeA, respectively, arguing that the SpeA-70 isoform is generated due to initiation of translation at the speA iTIS. In support of this conclusion, the peptide (M)SSQEASKMLR, which precisely corresponds to the N-terminus of the short SpeA isoform defined by Ribo-RET, can be found in the database of the experimentally-identified E. coli N-terminal peptides (Bienvenut et al., 2015).
It was suggested that SpeA-74 is targeted to the periplasm due to the presence of a putative N-terminal secretion signal sequence (Buch and Boyle, 1985). A segment of this signal sequence would be missing in the SpeA-70 isoform confining the shorter polypeptide to the cytoplasm (Figure S3B). Therefore, utilization of the 5’-proximal iTIS of speA could change compartmentalization of the encoded protein. The 5’-proximal iTISs identified in some other E. coli genes encoding secreted proteins (e.g. bamA, ivy or yghG), may serve similar (Figure S3C). Analogous strategy for targeting polypeptide isoforms to different cellular compartments has been described for few other bacterial proteins (reviewed in (Meydan et al., 2018)).
Six of the 5’-proximal iTISs (marked by asterisks in Figure 3B) were detected by TET Ribo-seq and suggested to represent incorrectly annotated pTISs (Nakahigashi et al., 2016). Lack of the Ribo-RET peaks at the annotated pTIS of some of these genes (Table S1) is generally consistent with this proposal suggesting that assignment of the pTISs could be reassessed. However, such conclusion should be drawn cautiously because the utilization of the upstream pTIS could depend on growth conditions.
Conservation analysis of in-frame iTISs
We analyzed alignments of bacterial genes homologous to the E. coli genes where internal in-frame start sites were detected by Ribo-RET. Sequence logos and codon conservation plots indicated preservation of in-frame potential initiation sites and locally enhanced synonymous site conservation at phoH, speA, yebG, yfaD and yadD (Figure S4A-S4E). However, it remains to be seen whether these conserved regions are relevant to promoting iTIS usage or simply represent unrelated sequence requirements of these genes. The other iTISs identified by Ribo-RET in the E. coli genome show a lower degree of evolutionary conservation indicating that many of them could be species- or strain-specific.
Ribo-RET identified iTISs that are out of frame relative to the main ORF
Only two examples of a bacterial ORF nested in an alternative frame within another ORF had been previously described: comS within srfAB in B. subtilis and rpmH within rnpA in Thermus thermophilus (reviewed in (Meydan et al., 2018)). Our Ribo-RET analysis showed the presence of 74 OOF iTISs common between the examined E. coli BW25113 and BL21 strains. (Figure S5A). The location of the OOF iTISs within the host genes varies significantly; the peptides generated by translation initiated at the OOF iTISs would range in size from 2 to 84 amino acids (Figures 4A, S5A and S5B).
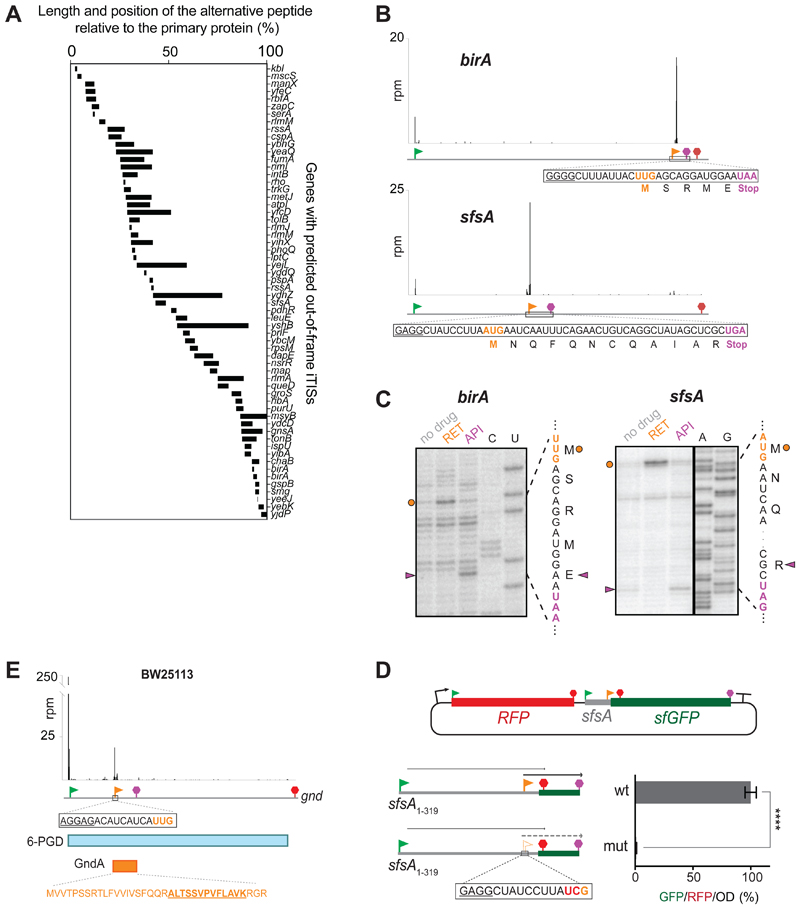
Figure 4. OOF iTISs revealed by Ribo-RET can direct initiation of translation.
(A) The length and location of the alternative OOF protein-coding segments relative to the main ORF.
(B) Ribo-RET profiles of birA and sfsA genes with putative OOF iTISs. The iTIS OOF start codon and the corresponding stop codon are indicated by orange flag and stop sign, respectively. The sequences of the alternative ORFs are shown. (C) Toeprinting analysis reveals that RET arrests ribosomes at the start codons (orange circles) of the alternative ORFs within the birA and sfsA genes; termination inhibitor Api137 arrests translation at the stop codons (purple arrowheads) of those ORFs. The nucleotide and amino acid sequences of the alternative ORFs are shown. Sequencing lanes are indicated.
(D) In the cell, translation is initiated at the sfsA OOF iTIS. Schematics of the RFP/sfGFP reporter plasmid. The rfp and sf-gfp genes are co-transcribed. The sfGFP-coding sequence is expressed from the iTIS (orange flag) and is OOF relative to the sfsA pTIS start codon (green flag). The first stop codon in-frame with the pTIS (red) and the stop codon in-frame with the iTIS (orange) are indicated. The bar graph shows the change in relative green fluorescence in response to the iTIS start codon mutation (mut). The values represent the mean ± standard deviation from technical replicates (n=6). Two-tailed unpaired t-test. (E) Ribo-RET snapshots of the gnd gene revealing the putative location of the start codon of the alternative ORF. The amino acid sequence of the alternative ORF product GndA is shown in orange; the tryptic peptide identified by mass spectrometry (Yuan et al., 2018) is underlined.
See also Figure S5 and Table S1.
We tested two OOF iTIS candidates (found within the birA and sfsA genes) for their ability to direct initiation of translation. Initiation of translation at the OOF UUG internal start site (overlapping the birA gene Leu300 codon) would yield a 5-amino acid long peptide, while translation initiated at the OOF AUG of the sfsA gene (overlapping the Leu95 codon of the main ORF) would generate a 12 amino acid peptide (Figure 4B). When the full-size sfsA and birA genes were translated in vitro, addition of RET resulted in the appearance of toeprint bands not only at the pTISs of the corresponding genes (Figure S5C and S5D) but also at the OOF iTIS start codons (Figures 4C, lanes ‘RET’, orange dots). The addition of the translation termination inhibitor Api137 (Florin et al., 2017) to the reactions generated toeprint bands at the stop codons of the OOF ORFs, indicating that the ribosomes not only bind to the OOF start sites but do translate the entire alternative ORFs (Figure 4C, lanes ‘API’, magenta triangles).
We then examined whether the alternative ORF in the sfsA gene is translated in vivo. For this purpose, we engineered a dual RFP/GFP reporter, where translation of the gfp gene is initiated at the sfsA OOF iTIS (Figure 4D). E. coli cells carrying such reporter construct actively expressed the GFP protein (Figure 4D, right panel), whereas gfp expression was abolished when the internal AUG start codon was mutated to UCG (Figure 4D). This result demonstrates that the OOF iTIS in sfsA is utilized for initiation of translation in the E. coli.
An independent validation of the functional significance of one of the Ribo-RET identified OOF iTISs came from a recent study aimed at characterizing E. coli proteins activated by heat-shock (Yuan et al., 2018). In that work, the sequence of one of the identified tryptic peptides mapped to the -1 frame of the gnd gene, although the location of the start codon from which translation of the alternative protein (named GndA) would initiate remained ambiguous (Yuan et al., 2018). Our Ribo-RET data not only validated those findings, but also suggested that translation of GndA initiates most likely at the UUG codon, which is preceded by a strong SD sequence (Figure 4E).
Expression of functional alternative proteins may be highly specialized since most of the OOF iTISs identified in the E. coli genome are not conserved. The strongest example that exhibits near-threshold significance of the OOF iTIS conservation is that of the tonB gene (Figure S4F). Furthermore, the internal initiation at this site is apparently sufficiently strong to be observed as an upshift of the local ribosome density in the Ribo-seq data collected from E. coli cells not treated with RET (Figure S4F).
Start-Stop sites may modulate translation of the primary gene
Among the 74 OOF iTIS candidates, Ribo-RET revealed 14 unique sites where the start codon is immediately followed by a stop codon, and thus we called them start-stops (Table S1). Although start-stops have been identified in the 5’ UTRs of some viral and plant genes where they likely play regulatory functions (Krummheuer et al., 2007; Tanaka et al., 2016), operational start-stops have not been reported within the bacterial genes.
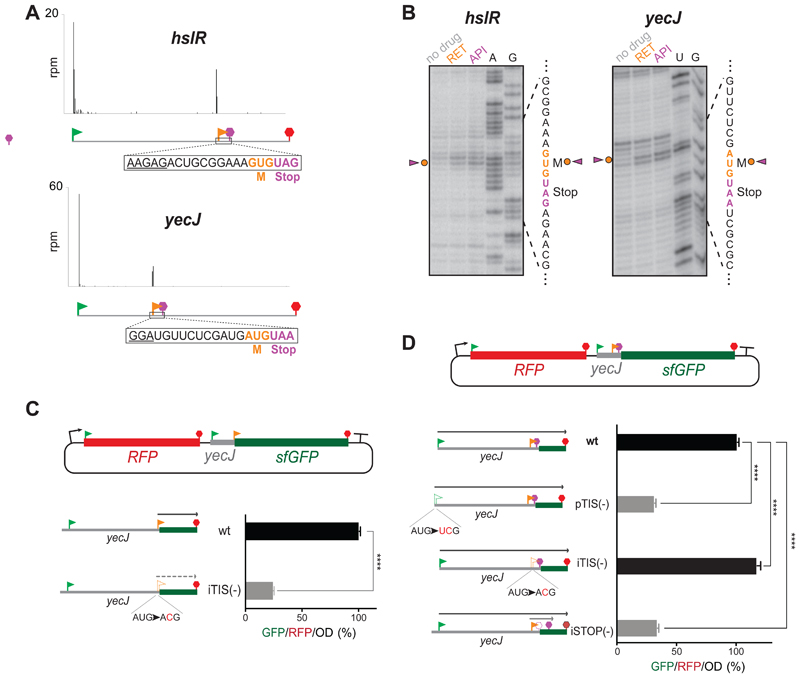
We selected the identified start-stops within two genes, yecJ and hslR (Figure 5A), for further analysis. Consistently, in vitro studies, carried out using the full-length yecJ or hslR genes, showed that in both cases addition of initiation inhibitor RET or termination inhibitor Api137 caused the appearance of coincidental toeprint bands (either one of the inhibitors is expected to stall the ribosome at the initiation codon of the start-stop site) (Figure 5B). Thus, the iTISs of the start-stops nested in yecJ and hslR genes can direct ribosome binding. For in vivo analysis we fused the gfp reporter gene, devoid of its own start codon, immediately downstream from the AUG codon of the yecJ iTIS (stripped from its associated stop codon) (Figure 5C). GFP fluorescence derived from the resulting construct was readily detectable as long as the initiator codon of the start-stop site was intact, but was significantly reduced when this AUG codon was mutated to ACG (Figure 5C). These results demonstrated that the start codon of the yecJ start-stop site is operational in vivo.
Figure 5. Start-Stops within E. coli genes.
(A) Representative Ribo-RET profiles revealing start-stops. The SD-like sequences are underlined.
(B) Toeprinting analysis shows ribosomes stalled at the start codons of the start-stop sites in response to the presence of initiation (RET) and termination (API) inhibitors. The start and stop codons of the start-stop sites are indicated by orange and purple characters, respectively.
(C) The start codon of the yecJ start-stop can direct initiation of translation in vivo. sfGFP expression in the RFP/sfGFP reporter is directed by the start codon of the yecJ start-stop (orange flag). The relative translation efficiency was estimated by measuring GFP/RFP/OD (%) ratio. The expression of sf-gfp is severely abrogated by a mutation that disrupts the start-stop initiation codon [iTIS (-)]. The values represent the standard deviation from the mean in technical replicates (n=3). Two-tailed unpaired t-test.
(D) Start-stop impacts expression of the yecJ gene. The expression of the YecJGFP chimeric protein is controlled by yecJ pTIS (green flag) (gfp sequence is in 0-frame relative to pTIS). The reporter expression increases by ~16% when the start codon of the start-stop site is disrupted by a mutation [(iTIS(-)]. Mutating the stop codon of the start-stop site expands the length of the translated OOF coding sequence and results in severe inhibition of the main frame translation. The error bars represent standard deviation from the mean in technical triplicates (n=3). Two-tailed unpaired t-test.
See also Figure S5 and Table S2.
We surmised that functional OOF start-stops may carry out regulatory functions, possibly affecting the expression efficiency of the protein encoded in the main ORF. To test this hypothesis, we examined whether the presence of a functional start-stop affects the expression of the main ORF that hosts it. For that, we prepared a reporter construct where the gfp coding sequence was placed downstream of the yecJ start-stop but in frame with the yecJ pTIS (Figure 5D). Mutational analysis verified that expression of the YecJ-GFP fusion protein was directed by the yecJ pTIS (Figure 5D, wt vs. pTIS(-) bars). Notably, when the start codon of the OOF start-stop was inactivated by mutation, the efficiency of expression of the YecJ-GFP reporter increased by approximately 16% (Figure 5D, wt vs. iTIS(-) bars). These results demonstrate that the presence of the active start-stop site within the yecJ gene attenuates translation of the main ORF, indicative of its possible regulatory function.
Interestingly, mutating the stop codon of the yecJ start-stop, that should lead to translation of a 14-codon internal ORF originating at the yecJ OFF iTIS, significantly reduced the expression of the YecJ-GFP reporter by ~ 3-fold (the iSTOP(-) construct in Figure 5D). This result shows that active utilization of some of the OOF iTISs could significantly attenuate the expression of the main ORF whereas the position of the corresponding stop codon could modulate this effect.
Ribo-RET reveals TISs outside of the known annotated coding sequences
The ability of Ribo-RET to reveal the cryptic sites of translation initiation makes it a useful tool for identifying such sites located not only within the genes, but outside of the annotated protein coding regions. We have detected 6 upstream in-frame TISs (uTISs) in the E. coli strain BW25113 and 36 uTISs in the BL21 strain that would result in N-terminal extensions of the encoded proteins (Table S2). For one gene (potB), we did not observe any Ribo-RET peak at the annotated pTIS (Figure S6A), suggesting that either its start site has been mis-annotated or that the annotated pTIS is activated under growth conditions different from those used in our experiments. For several other genes (e.g. yifN), we detected Ribo-RET signals for both the annotated pTIS and the uTIS, indicating that two isoforms may be expressed (Figure S6B).
We also detected 41 TISs, common between the two analyzed strains, outside of the annotated genes likely delineating the translation start sites of unannotated short ORFs (Table S2) (analyzed in detail in another study (Weaver, 2019)). Although analysis of such ORFs was beyond the scope of our work, the ability to detect such ORFs underscores the utility of Ribo-RET as a general tool for the genome-wide identification of translation start sites in bacteria.
Discussion
Genome-wide survey of TISs in two E. coli strains revealed translation initiation not only at the known start codons of the annotated genes, but also int eh intergenic regions and, importantly, at over 100 mRNA sites nested within the currently recognized ORFs. Proteins, whose synthesis is initiated as such sites, may constitute a previously obscure fraction of the proteome and may play important roles in cell physiology. In addition, initiation of translation at internal codons may play regulatory role by influencing the efficiency of expression of the main protein product.
Mapping of the cryptic translation start sites was possible due to the action of RET as a highly-specific inhibitor of translation initiation, arresting ribosomes at the mRNA start codons. It is this specificity of RET action that makes it possible to utilize the antibiotic for confidently charting not only the TISs at the beginning of the protein-coding sequences and also for mapping initiation-competent codons within the ORFs. Other antibiotics that exclusively bind to the initiating ribosomes, could also be explored for mapping TISs in bacteria (Weaver, 2019).
Our experiments were confined to the E. coli strains. However, we expect that the drug would exhibit a similar mode of action in other bacterial species. RET has limited activity against Gram-negative species, partly due to the active efflux of the drug (Jones et al., 2006). Therefore, in our experiments with we needed to use the E. coli strains lacking the TolC component of the multi-drug efflux pumps. Newer broad-spectrum pleuromutilins (Paukner and Riedl, 2017), could be likely used even more efficiently for mapping TISs in both Gram-positive and Gram-negative bacterial species.
Ribo-RET revealed the presence of internal start codons in over a hundred E. coli genes, dramatically expanding the number of putative cases of internal initiation in bacteria of which, before this work, only a few examples had been known (Meydan et al., 2018). Our findings suggest that inner-ORF initiation of translation is a much more widespread phenomenon. Although in most cases we have little knowledge about the possible functions of the alternative polypeptides encoded in the bacterial genes, one can envision several general scenarios:
-
1)
In-frame internal initiation generates a protein isoform with a distinct function. One such example is the ArcB-C polypeptide expressed from the iTIS within the arcB gene.
-
2)
The isoform expressed from an in-frame iTIS could partake in the hetero-complex formation with the main protein (reviewed in (Meydan et al., 2018)). Primary proteins encoded by some of the iTIS-containing E. coli genes (e.g. slyB, nudF, lysU and wzzB) are known to form homodimers and thus, could be candidates for the formation of heterodimers with their N-terminally truncated isoforms.
-
3)
Translation from the in-frame 5’-proximal iTIS (e.g. speA, bamA, ivy or yghG) can alter proteins compartmentalization. Similarly, some of the iTISs identified in eukaryotic mRNAs and attributed to the ‘leaky scanning’ may alter the subcellular localization of the alternative polypeptides (Kochetov, 2008; Lee et al., 2012).
-
4)
Because protein stability significantly depends on the nature of the N-terminal amino acid (Dougan et al., 2010), utilization of an alternative start site may alter the protein’s half-life.
-
5)
The utilization of the OOF iTISs may generate polypeptides with structure and function unrelated to those of the main protein.
The significance of some of the cryptic initiation sites, particularly the OOF iTISs, may reside in their regulatory role. In particular, the discovered start-stop sites within the E. coli genes may be utilized by the cell for fine-tuning the expression of proteins encoded in the host ORFs. However, such possible mechanism is likely subtle, because no significant change in the ribosome density before and after the start-stop site in the Ribo-seq data collected with the untreated cells during fast-growth was observed.
It is also likely that not all of the Ribo-RET-identified iTISs directly benefit bacterial cell and a number of them could simply represent unavoidable noise of imprecise start codon recognition by the ribosome. Furthermore, Ribo-RET peak shows only the potential of a codon to be used as a translation start site: by arresting the ribosomes at a pTIS while allowing the elongating ribosomes to run off the mRNAs, RET treatment leads to the generation of ribosome-free mRNAs, thereby allowing ribosome binding to the newly exposed putative iTISs. Nevertheless, several lines of evidence argue that many of the Ribo-RET-identified iTISs are recognized by the ribosomes even in the untreated cells: i) for some genes (e.g. arcB) an increase in ribosome density downstream of the identified iTIS can be seen in the Ribo-seq data collected with the untreated cells; ii) we have experimentally demonstrated the functionality of iTISs in several genes (e.g. arcB, sfsA); iii) the expression of an OOF ORF within the gnd gene was confirmed by proteomics (Yuan et al., 2018).
The mechanisms that control the relative utilization of pTISs and iTISs could operate at the level of translation, via modulating the activity of pTIS and iTIS, or at the level of transcription: some of the experimentally-mapped transcription sites map reside between the pTIS and iTIS in some genes (see Table S1).
Besides iTISs, our Ribo-RET data revealed a number of the translation initiation sites outside of the annotated genes. Most of those sites delineate previously uncharacterized short genes. Proteins encoded in such ORFs may further expand the cryptic bacterial proteome (Storz et al., 2014; Weaver, 2019), whereas translation of the other ORFs could play regulatory roles.
In conclusion, by mapping translation initiation landscape in bacteria Ribo-RET unveils the hidden fraction of the bacterial proteome and offers insights into gene regulatory mechanisms.
Star Methods
Bacterial strains
Ribo-seq experiments were performed in two E. coli strains: the K12-type strain BW25113 (lacI q, rrnB T14, ΔlacZWJ16, hsdR514, ΔaraBAD AH33, ΔrhaBAD LD78) that was further rendered ΔtolC (called previously BWDK Kannan, 2012 #81}) and the B-type strain, BL21, (F-, ompT, gal, dcm, lon, hsdSB(rB-mB-)[malB +]K-12 (λS) and was also rendered ΔtolC by recombineering (Datsenko and Wanner, 2000). For that, the kanamycin resistance cassette was PCR-amplified from BW25113 tolC::kan strain from the Keio collection (Baba et al., 2006) using the primers #P1 and P2 (Table S3). The PCR fragment was transformed into BL21 cells (NEB, #C2530H) carrying the Red recombinase expressing plasmid pKD46. After selection and verification of the BL21 tolC::kan clone, the kanamycin resistance marker was eliminated as previously described (Datsenko and Wanner, 2000). In the subsequent sections of STAR Methods we will refer to BW25113(ΔtolC) strain as ‘K’ strain and to BL21(ΔtolC) as ‘B’ strain.
Reporter plasmids were expressed in the E. coli strain JM109 (endA1, recA1, gyrA96, thi, hsdR17 (rk–, mk+), relA1, supE44, Δ(lac-proAB), [F´ traD36, proAB, laqIqZΔM15]) (Promega, #P9751).
Metabolic labeling of proteins
Inhibition of protein synthesis by RET was analyzed by metabolic labeling. Specifically, the B strain cells were grown overnight at 37°C in M9 minimal medium supplemented with 0.003 mM thiamine and 40 μg/mL of all 19 amino acids except methionine (M9AA-Met). Cells were diluted 1:200 into fresh M9AA-Met medium and grown at 37°C until the culture density reached A600~0.2. Subsequent operations were performed at 37°C. The aliquots of cell culture (28 µL) were transferred to Eppendorf tubes that contained dried-down RET (Sigma-Aldrich, #CDS023386). The final RET concentration ranged from 1x MIC to 32x MIC (0.06 μg/mL to 2 μg/mL). After incubating cells with antibiotic for 3 min, the content was transferred to another tube containing 2 µL M9AA-Met medium supplemented with 0.3 μCi of L-[35S]-methionine (specific activity 1,175 Ci/mmol) (MP Biomedicals). After 1 min incubation, 30 µL of 5% trichloracetic acid (TCA) was added to the cultures and this mixture was pipetted onto 35 mm 3MM paper discs (Whatman, Cat. No. 1030-025) pre-wetted with 25 µL of 5% TCA. The discs were then placed in a beaker with 500 mL 5% TCA and boiled for 5 min. TCA was discarded and this step was repeated one more time. Discs were rinsed in acetone, air-dried and placed in scintillation vials. After addition of 5 ml of scintillation cocktail (Perkin Elmer, Ultima Gold, #6013321) the amount of retained radioactivity was measured in a Scintillation Counter (Beckman, LS 6000). The data obtained from RET-treated cells were normalized to the no-drug control.
The time course of inhibition of protein synthesis by RET was monitored following essentially the same procedure except that antibiotic was added to a tube with the cells and 28 µL aliquots were withdrawn after specified time and added to tubes containing 2 µL M9AA-Met medium supplemented with 0.3 μCi of L-[35S]-methionine. The rest of the steps were as described above.
Ribo-seq experiments
The Ribo-seq experiments were carried out following previously described procedures (Becker et al., 2013). The overnight cultures of E. coli grown in LB medium at 37°C were diluted to A600~0.02 in 100 mL of fresh LB media sterilized by filtration and supplemented with 0.2% glucose. The cultures were grown at 37°C with vigorous shaking to A600~0.5. RET was added to the final concentration of 100X MIC (12.5 μg/mL for the K strain or 5 μg/mL for the B strain) and incubated for 5 min (K strain) or 2 min (B strain). No antibiotic was added to the control nodrug cultures. Cells were harvested by rapid filtration, frozen in liquid nitrogen, cryo-lysed in 650 µL of buffer containing 20 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 100 mM NH4Cl, 5 mM CaCl2, 0.4% Triton X100, 0.1% NP-40 and supplemented with 65 U RNase-free DNase I (Roche, #04716728001), 208 U SUPERasexIn™ RNase inhibitor (Invitrogen, #AM2694) and GMPPNP (Sigma-Aldrich, #G0635) to the final concentration of 3 mM. After clarifying the lysate by centrifugation at 20,000 g for 10 min at 4°C samples were subjected to treatment with ~450 U MNase (Roche, #10107921001) per 25 A260 of the cells for 60 min. The reactions were stopped by addition of EGTA to the final concentration of 5 mM and the monosome peak was isolated by sucrose gradient centrifugation. RNA was extracted and run on a 15% denaturing polyacrylamide gel. RNA fragments ranging in size from ~28 to 45 nt were excised from the gel, eluted and used for library preparation as previously described (Becker et al., 2013). Resulting Riboseq data was analyzed using the GALAXY pipeline (Kannan et al., 2014). The reference genome sequences U00096.3 (BW25113, ‘K’ strain) and CP001509.3 (BL21, ‘B’ strain) were used to map the Ribo-seq reads. The first position of the P-site codon was assigned by counting 15 nucleotides from the 3’ end of the Riboseq reads. The Ribo-seq datasets were deposited under accession number GSE1221129.
Metagene analysis
The genes with the read counts ≥100 in both control and RET-treated samples were used for metagene analysis of K and B strains. The published tetracycline Ribo-seq data (Nakahigashi et al., 2016) were used to generate the corresponding metagene plot. The genes separated by less than 50 bp from the nearest neighboring gene were not included in the metagene analysis in order to avoid the ‘overlapping genes’ effects.
For every nucleotide of a gene, normalized reads were calculated by dividing reads per million (rpm) values assigned to a nucleotide by the total rpm count for the entire gene including 30 nt flanking regions. The metagene plot was generated by averaging the normalized reads for the region spanning 10 nt upstream and 50 nucleotides downstream of the first nucleotide of the start codon.
Computational identification of translation initiation sites
The assignment of RET peaks to the start codons was performed using the algorithm provided in Supplemental Information. Specifically, we searched for a possible start codon (AUG, GUG, CUG, UUG, AUU, AUC) within 3 nucleotides upstream or downstream of the Ribo-RET peak. All other codons associated with an internal RET peak were considered as “non-start” codons (Table S1).
For assessing whether Ribo-RET peaks in K strain match the annotated start codons in the genes expressed under no-drug conditions, we calculated the percentage of genes whose rpkm values were t100 in the no-drug conditions and whose corresponding pTIS Ribo-RET peak values were >1 rpm. More stringent criteria were used for identification of alternative Ribo-RET peaks (rpm >5). If the Ribo-RET peak matched an annotated TIS, it was classified as pTIS (Classification I, Scheme I and Table S1). “Tailing peaks” (peaks within 10 nt downstream and upstream of the start codon) around the pTIS were considered as “near-annotated TIS” and merged with the pTISs after removing duplicates. All pTISs prior to duplicate removal are provided in Table S1 (the ‘pTISs’ tabs). The Ribo-RET peaks within coding regions were considered in Classification II and were assigned as inframe or out-of-frame iTISs depending on the position of the likely start codon (Table S1, the ‘iTISs’ tabs). Finally, the RET peaks outside of the coding regions were considered either as N-terminal extensions or unannotated ORFs (Classification III and Table S2). The criteria for each classification are detailed in Schemes I, II and III in Supplementary Information.
Construction of ArcB - expressing plasmids
The plasmids carrying the wt arcB gene (pArcB) or its mutant variant pArcB(mut) (G1947A, G1950A, G1959C) were generated by Gibson assembly (Gibson et al., 2009). The PCR-generated fragments covering the length of wt or mutant arcB genes or of the ArcB-C coding arcB segment were introduced into NcoI and HindIII-cut pTrc99A plasmid. Three PCR fragments used for the assembly of wt arcB plasmid were generated by using primer pairs P3/P4, P5/P6 and P7/P8 (Table S3). To construct the pArcB(mut) plasmid, the PCR fragments were generated by using primer pairs P3/P9, P7/P8 and P10/P11. The plasmid pArcBC expressing exclusively C-terminal domain of ArcB was prepared by acquiring the ArcB-C coding sequence as a gBlock (fragment #12 in Table S3) an dintroducing it into NcoI and HindIII-cut pTrc99A plasmid. All the plasmids were verified by Sanger sequencing of the inserts. The plasmids were introduced in the E. coli BW25113 or BW25113 (ΔarcB) strains.
Western blot analysis of the FLAG-tagged ArcB
The BW25113 cells carrying either pArcB or pARcB(mut) plasmids (or the pArcBC plasmid encoding the marker ArcB-C segment of ArcB) were grown overnight at 37°C in LB medium supplemented with ampicillin (final concentration of 50 μg/mL). The cultures were diluted 1:100 into 5 mL LB/ampicillin medium supplemented with 0.01 mM of isopropyl-β-D-1-thiogalactopyranoside (IPTG) and grown at 37°C until culture density reached A600~0.5. The cultures were harvested by centrifugation. Cells were resuspended in 300 μL of B-PER™ Bacterial Protein Extraction Reagent (Thermo Fisher, #78248) and centrifuged at 16,000 g for 10 min. Ten μL of the cell lysate were loaded on TGX 4-20% gradient gel (Bio-Rad, #4561096). Resolved proteins were transferred to a PVDF membrane using PVDF transfer pack (Bio-Rad, #1704156) by electroblotting (Bio-Rad Trans-Blot SD Semi-Dry Transfer Cell, 10 min at 25 V). Membrane was blocked by incubating in TBST (50 mM M Tris [pH 7.4], 150 mM NaCl, and 0.05% Tween-20) containing 5% non-fat dry milk and probed with Anti-FLAG M2-Peroxidase (Sigma-Aldrich, #A8592) and anti-GAPDH antibodies (Thermo Fisher, #MA5-15738-HRP) at 1:1000 dilution in TBST. The blot was developed using Clarity Western ECL Substrate (Bio-Rad, #170-5060) and visualized (Protein Simple, FluorChem R).
Growth competition under low oxygen conditions of cells expressing wild type or mutant arcB genes
Dependence of micro-aerobic cell growth on expression of arcB was initially verified by co-growing E. coli BW25113(ΔarcB) cells transformed with the empty vector pTrc99A or the vector carrying wt arcB gene (pArcB). Overnight cultures, grown in LB medium supplemented with 100 µg/mL of ampicillin, were diluted 1:100 into fresh LB supplemented with 100 µg/mL of ampicillin and 10 µM of IPTG, grown to A600~0.5 and mixed in the proportion to provide equal number of pTrc99A-and pArcB cells. Plasmids from the mixed “0 passage” sample were isolated and stored. The 0 passage mix culture was diluted 1:1000 into two 14 ml culture tubes containing each 12.5 ml of fresh LB/amplicillin/IPTG medium. Tubes were tightly capped and grown vertically with no shaking at 37°C. Cell sedimentation was avoided by the slow rotation (~ 40 rpm) of a small magnet placed at the bottom of the tubes. After 24 h, cultures were diluted 1:1000 into tubes with fresh medium, while the rest of the cells were used for isolation of the total plasmid (“passage 1” sample). The same procedure was carried out for two more passages (passage 2 and 3). To assess the relative representation of cells with pTrc99A or pArcB the total plasmid from each of the passages 0-3 was linearized with HindIII. The 4176 bp pTrc99A DNA and 6570 bp pArcB DNA bands were resolved by agarose electrophoresis (Figure S2C).
This same low-oxygen experimental set-up was used for the growth competition of BW25113(ΔarcB) cells expressing wt or mutant arcB, from pArcB or pArcB(mut), respectively. In this case, 5 passages were performed (passages 0-5) and, instead of isolating plasmids from the cultures, cells from 500 µL aliquots of passages 0, 2, 3, 4, and 5 were collected and stored. After completing the passages, cells were resuspended in 200 µL H2O, boiled for 10 min to lyse the cells and 1 µL of the lysate was used to PCR-amplify the segment of the arcB gene encompassing the iTIS region (primers P52 and P53, Table S3). The resulting PCR fragments were purified and subjected to capillary sequencing. The ratio of cells carrying wt and mutant arcB (that carried the G1947A, G1950A, G1959C mutations) genes was estimated by comparing the height of the sequencing peaks corresponding to the position G1959C.
Toeprinting assay
The DNA templates for toeprinting, were prepared by PCR amplification for the respective genes from the E. coli BW25113 genomic DNA. The following primer pairs were used for amplification of specific genes: atpB: P13/P14; mqo: P15/P16; birA: P18/P19; hslR: P30/P31; yecJ: P33/P34. Two point mutations were generated in sfsA in order to change the stop codon of the alternative ORF from TGA to TAG because in the PURE transcription-translation system, the termination inhibitor Api137 arrest termination at the TAG stop codon with a higher efficiency (Florin et al., 2017). This was achieved by first amplifying segments of the sfsA gene using pairs of primers P22/P23 and P24/P25 and then assembling the entire mutant sfsA sequence by mixing the PCR products together and re-amplifying using primers P26/P27. Toeprinting primer P17 was used with the atpB and mqo templates. Primers P20, P28, P32 and P36 were used for analysis of ribosome arrest at pTIS of birA, sfsA, hslR and yecJ templates, respectively. Primers P21, P29, P33 and P37 were used for the analysis of ribosome arrest at the iTISs of birA, sfsA, hslR and yecJ templates, respectively.
Transcription-translation was performed in 5 µL reactions of the PURExpress system (New England Biolabs, #E6800S) for 30 min at 37°C as previously described (Orelle et al., 2013). Final concentration of RET, tetracycline (Fisher scientific, #BP912-100) or Api137 (synthesized by NovoPro Biosciences, Inc.) was 50 μM. The primer extension products were resolved on 6% sequencing gels. Gels were dried, exposed overnight to phosphorimager screens and scanned on a Typhoon Trio phosphorimager (GE Healthcare).
Polysome analysis
For the analysis of the mechanism of RET action, the overnight culture of the K strain was diluted 1:200 in 100 mL of LB medium supplemented with 0.2% glucose. The culture was grown at 37°C with vigorous shaking to A600 ~0.4 at which point RET was added to the final concentration of 100X MIC (12.5 µg/mL) (control culture was left without antibiotic). After incubation for 5 min at 37°C with shaking, cultures were transferred to pre-warmed 50 mL tubes and cells were pelleted by centrifugation in a pre-warmed 37°C Beckman JA-25 rotor at 8,000 rpm for 5 min. Pellets were resuspended in 500 µL of cold lysis buffer (20mM Tris-HCl, pH7.5, 15 mM MgCl2), transferred to an Eppendorf tube and frozen in a dry ice/ethanol bath. Tubes were then thawed in an ice-cold water bath and 50 µL of freshly prepared lysozyme (10 mg/ml) was added. Freezing/thawing cycle was repeated two more times. Lysis was completed by addition of 15 µL of 10% sodium deoxycholate (Sigma, #D6750) and 2 µL (2U) of RQ1 RNase-free DNase (Promega, #M610A) followed by incubation on ice for 3 min. Lysates were clarified by centrifugation in a table-top centrifuge at 20,000 g for 15 min at 4°C. Three A260 of the lysate were loaded on 11 ml of 10%-40% sucrose gradient in buffer 20 mM Tris-HCl, pH 7.5, 10mM MgCl2, 100 mM NH4Cl2, 2 mM E-mercaptoethanol. Gradients were centrifuged for 2 h in a Beckman SW-41 rotor at 39,000 rpm at 4°C. Sucrose gradients were fractionated using Piston Gradient Fractionator (Biocomp).
Construction of the reporter plasmids
The RFP/GFP plasmids were derived from the pRXG plasmid, kindly provided by Dr. Barrick (University of Texas). The vector was first reconstructed by cutting pRXG with EcoRI and SalI and re-assembling its backbone with 2 PCR fragments amplified from pRXG using primer pairs P38/P39 (rfp gene) and P40/P41 (sf-gfp preceded by a SD sequence). The resulting plasmid (pRXGSM), had RFP ORF with downstream SpeI sites flanking the SD-containing sf-GFP ORF. To generate pRXGSM-sfsA plasmids, sfsA sequences were PCR amplified from E. coli BW25113 genomic DNA with primers P42/P43, for the wt gene, or P42/P44, for the mutant variant, and assembled with the SpeI-cut pRXGSM plasmid. To generate the pRXGSM-yecJ reporter plasmids, the pRXGSM was cut with SpeI and assembled with each of the PCR fragments generated using the following primer pairs: P45/P46 (iTIS-wt plasmid), P45/P47 (iTIS(-)); P45/P48 (pTIS-wt), P45/P50 (pTIS-iTIS(-)). pRXGSM-yecJ-pTIS(-) and pRXGSM-yecJ-pTIS-iStop(-) plasmids were generated by site directed mutagenesis of the pRXGSM-yecJ-pTIS-wt plasmid using primers P49 and P51, respectively.
Fluorescence and cell density measurements
E. coli JM109 cells carrying the reporter plasmids were grown overnight in LB medium supplemented with 50 μg/mL kanamycin (Fisher Scientific, #BP906-5). The cultures were then diluted 1:100 into fresh LB medium supplemented with kanamycin (50 μg/mL) and grown to A600 ~0.5-0.8. The cultures were diluted to the final density of A600 ~0.02 in fresh LB/kanamycin (50 μg/mL) medium supplemented with 0.1 mM IPTG and 120 μL were placed in the wells of a clear flat bottom 96 well microplate (Corning, #353072). The plates were placed in a Tecan Infinite M200 PRO plate reader, where they were incubated at 37°C with orbital shaking (duration: 1000 sec; amplitude: 3 mm), and measurements of optical density (at 600 nm), ‘green fluorescence’ (excitation: 485 nm; emission: 520 nm) and ‘red fluorescence’ (excitation: 550 nm and emission: 675 nm) were acquired in real time.
Evolutionary conservation analysis
Protein sequences of the genes of interest were extracted from Ecogene database (Zhou and Rudd, 2013). Homologs for each gene were obtained by performing a tblastn search against the nr database. Briefly, the nr database was downloaded on 19/01/2018 to a local server and tblastn searches were performed for each gene (parameters -num_descriptions 1000000 -num_alignments 1000000 -evalue 0.0001). Only those tblastn hits which share a sequence identity of at least 45% with the query sequence and whose length is at least 75% of the query sequence were retained. Hits that contain in-frame stop codons were also discarded. Alignments for each gene of interest were generated by first translating the nucleotide sequences to protein sequences, aligning the protein sequences using Clustal-Omega (Sievers et al., 2011) and then back-translating the aligned protein sequence to their corresponding nucleotide sequence using T-coffee (Notredame et al., 2000). To analyze the conservation of internal start codons for each candidate gene, a 45-nucleotide region containing the internal start codon and 7 codons on either side of the start codon was extracted from each alignment. Sequence logos were built using this region of alignment and visualized to assess the conservation of the internal start codon. In order to determine if there is purifying selection at synonymous positions in the alignment, Synplot2 was used (Firth, 2014). Synplot2 was applied to each alignment (window size – 15 codons) and the resulting plots were visualized to assess the degree of synonymous site variability in the region of internal start codon.
Analysis of the conservation of arcB internal start site
Bacterial orthologs of arcB were retrieved from OrtholugeDB. Coordinates of histidine-containing phosphotransfer domain (HPT domain) were determined with HMMSEARCH (Wistrand and Sonnhammer, 2005) using the model 0051674 retrieved from Superfamily database version 1.74 (Wilson et al., 2009). For predicting internal initiation site, 40 nt long fragments were extracted containing 12 codons upstream of the predicted beginning of HPT domain and 1 codon downstream. Potential SD-aSD interactions were estimated by scanning the fragment with aSD 5'-ACCUCCU-3' using a ΔG threshold of -8.5. The presence of in-frame initiation codons (AUG, GUG, UUG) was checked. If initiation codon was found closer than 15 nt from SD-aSD sequence, the gene was reported to have inframe iTIS. After removing redundancy for the strains of the same species, internal in-frame iTISs were identified for 26 bacterial species. Maximum Likelihood (ML) tree for 26 arcB sequences was computed using ETE command line tools by executing the command: “ete3 -w standard_fasttree -a arcB_protein_seq_in.fas -o ete_output” (Huerta-Cepas et al., 2016). Final figure (Figure S2B) was produced with function PhyloTree from ETE3 toolkit (Huerta-Cepas et al., 2016).
Supplementary Material
Acknowledgements
We thank T. Florin for help with Ribo-seq experiments, G. Storz, J. Weaver and A. Buskirk for sharing their unpublished results and useful suggestions regarding the manuscript, J.E. Barrick for providing the pRXG plasmid, D. Georgellis for advice with some experiments, Y. Polikanov and N. Aleksashin for help with some figures. This work was supported by the grant from the National Science Foundation MCB 1615851 (to ASM and NV-L). PVB is supported by SFIHRB-Wellcome Trust Biomedical Research Partnership, grant no. 210692/Z/18/Z. AEF is supported by Wellcome Trust grant no. 106207.
Footnotes
Author Contributions
Conceptualization: S.M., J.M., N.V.-L., and A.S.M.; Methodology: S.M., N.V.-L., and A.S.M.; Software and formal analysis: J.M., V.S., P.V.B., A.E.F., T.M., and A.K.; Investigation: S.M. and D.K., Writing: S.M., N.V.-L., and A.S.M.; Supervision, Project Administration and Funding Acquisition: N.V.-L., and A.S.M.
Declarations of Interest
The authors declare no competing interests.
References
- Alvarez AF, Barba-Ostria C, Silva-Jimenez H, Georgellis D. Organization and mode of action of two component system signaling circuits from the various kingdoms of life. Environ Microbiol. 2016;18:3210–3226. doi: 10.1111/1462-2920.13397. [DOI] [PubMed] [Google Scholar]
- Alvarez AF, Georgellis D. In vitro and in vivo analysis of the ArcB/A redox signaling pathway. Methods Enzymol. 2010;471:205–228. doi: 10.1016/S0076-6879(10)71012-0. [DOI] [PubMed] [Google Scholar]
- Atkins JF, Loughran G, Bhatt PR, Firth AE, Baranov PV. Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 2016;44:7007–7078. doi: 10.1093/nar/gkw530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek J, Lee J, Yoon K, Lee H. Identification of unannotated small genes in Salmonella . G3 (Bethesda) 2017;7:983–989. doi: 10.1534/g3.116.036939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov PV, Atkins JF, Yordanova MM. Augmented genetic decoding: global, local and temporal alterations of decoding processes and codon meaning. Nat Rev Genet. 2015;16:517–529. doi: 10.1038/nrg3963. [DOI] [PubMed] [Google Scholar]
- Becker AH, Oh E, Weissman JS, Kramer G, Bukau B. Selective ribosome profiling as a tool for studying the interaction of chaperones and targeting factors with nascent polypeptide chains and ribosomes. Nat Protoc. 2013;8:2212–2239. doi: 10.1038/nprot.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienvenut WV, Giglione C, Meinnel T. Proteome-wide analysis of the amino terminal status of Escherichia coli proteins at the steady-state and upon deformylation inhibition. Proteomics. 2015;15:2503–2518. doi: 10.1002/pmic.201500027. [DOI] [PubMed] [Google Scholar]
- Broome-Smith JK, Edelman A, Yousif S, Spratt BG. The nucleotide sequences of the ponA and ponB genes encoding penicillin-binding protein 1A and 1B of Escherichia coli K12. Eur J Biochem. 1985;147:437–446. doi: 10.1111/j.1432-1033.1985.tb08768.x. [DOI] [PubMed] [Google Scholar]
- Buch JK, Boyle SM. Biosynthetic arginine decarboxylase in Escherichia coli is synthesized as a precursor and located in the cell envelope. J Bacteriol. 1985;163:522–527. doi: 10.1128/jb.163.2.522-527.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chengguang H, Sabatini P, Brandi L, Giuliodori AM, Pon CL, Gualerzi CO. Ribosomal selection of mRNAs with degenerate initiation triplets. Nucleic Acids Res. 2017;45:7309–7325. doi: 10.1093/nar/gkx472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundliffe E. Antibiotic Inhibitors of Ribosome Function. In: Gale EF, Cundliffe E, Reynolds PE, Richmond MH, Waring MJ, editors. The Molecular Basis of Antibiotic Action. London, New York, Sydney, Toronto: John Willey & Sons; 1981. pp. 402–545. [Google Scholar]
- D'Souza C, Nakano MM, Zuber P. Identification of comS a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis . Proc Natl Acad Sci USA. 1994;91:9397–9401. doi: 10.1073/pnas.91.20.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Bashan A, Auerbach-Nevo T, Yaggie RD, Gontarek RR, Yonath A. Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity. Proc Natl Acad Sci USA. 2007;104:4291–4296. doi: 10.1073/pnas.0700041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornhelm P, Hogenauer G. The effects of tiamulin, a semisynthetic pleuromutilin derivative, on bacterial polypeptide chain initiation. Eur J Biochem. 1978;91:465–473. doi: 10.1111/j.1432-1033.1978.tb12699.x. [DOI] [PubMed] [Google Scholar]
- Dougan DA, Truscott KN, Zeth K. The bacterial N-end rule pathway: expect the unexpected. Mol Microbiol. 2010;76:545–558. doi: 10.1111/j.1365-2958.2010.07120.x. [DOI] [PubMed] [Google Scholar]
- Feltens R, Gossringer M, Willkomm DK, Urlaub H, Hartmann RK. An unusual mechanism of bacterial gene expression revealed for the RNase P protein of Thermus strains. Proc Natl Acad Sci USA. 2003a;100:5724–5729. doi: 10.1073/pnas.0931462100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth AE. Mapping overlapping functional elements embedded within the protein-coding regions of RNA viruses. Nucleic Acids Res. 2014;42:12425–12439. doi: 10.1093/nar/gku981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin T, Maracci C, Graf M, Karki P, Klepacki D, Berninghausen O, Beckmann R, Vazquez-Laslop N, Wilson DN, Rodnina MV, Mankin AS. An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome. Nat Struct Mol Biol. 2017;24:752–757. doi: 10.1038/nsmb.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch C, Herrmann A, Nothnagel M, Szafranski K, Huse K, Schumann F, Schreiber S, Platzer M, Krawczak M, Hampe J, Brosch M. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 2012;22:2208–2218. doi: 10.1101/gr.139568.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, Yang Z, Wang Y, Jing L. Identifying translation initiation sites in prokaryotes using support vector machine. J Theor Biol. 2010;262:644–649. doi: 10.1016/j.jtbi.2009.10.023. [DOI] [PubMed] [Google Scholar]
- Gao X, Wan J, Liu B, Ma M, Shen B, Qian SB. Quantitative profiling of initiating ribosomes in vivo. Nat Methods. 2015;12:147–153. doi: 10.1038/nmeth.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Giess A, Jonckheere V, Ndah E, Chyzynska K, Van Damme P, Valen E. Ribosome signatures aid bacterial translation initiation site identification. BMC Biol. 2017;15:76. doi: 10.1186/s12915-017-0416-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier F, Matteau D, Baby V, Rodrigue S. Complete Genome Sequence of Escherichia coli BW25113. Genome Announc. 2014;2:e01038–14. doi: 10.1128/genomeA.01038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- Hecht A, Glasgow J, Jaschke PR, Bawazer LA, Munson MS, Cochran JR, Endy D, Salit M. Measurements of translation initiation from all 64 codons in E. coli . Nucleic Acids Res. 2017;45:3615–3626. doi: 10.1093/nar/gkx070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Serra F, Bork P. ETE 3: reconstruction analysis, and visualization of phylogenomic data. Mol Biol Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impens F, Rolhion N, Radoshevich L, Becavin C, Duval M, Mellin J, Garcia Del Portillo F, Pucciarelli MG, Williams AH, Cossart P. N-terminomics identifies Prli42 as a membrane miniprotein conserved in Firmicutes and critical for stressosome activation in Listeria monocytogenes . Nat Microbiol. 2017;2:17005. doi: 10.1038/nmicrobiol.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RN, Fritsche TR, Sader HS, Ross JE. Activity of retapamulin (SB-275833), a novel pleuromutilin, against selected resistant gram-positive cocci. Antimicrob. Agents Chemother. 2006;50:2583–2586. doi: 10.1128/AAC.01432-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K, Kanabar P, Schryer D, Florin T, Oh E, Bahroos N, Tenson T, Weissman JS, Mankin AS. The general mode of translation inhibition by macrolide antibiotics. Proc Natl Acad Sci USA. 2014;111:15958–15963. doi: 10.1073/pnas.1417334111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston AW, Ponkratz C, Raleigh EA. Rpn (YhgA-Like) Proteins of Escherichia coli K-12 and their contribution to RecA-independent horizontal transfer. J Bacteriol. 2017;199:e00787–16. doi: 10.1128/JB.00787-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochetov AV. Alternative translation start sites and hidden coding potential of eukaryotic mRNAs. Bioessays. 2008;30:683–691. doi: 10.1002/bies.20771. [DOI] [PubMed] [Google Scholar]
- Krummheuer J, Johnson AT, Hauber I, Kammler S, Anderson JL, Hauber J, Purcell DF, Schaal H. A minimal uORF within the HIV-1 vpu leader allows efficient translation initiation at the downstream env AUG. Virology. 2007;363:261–271. doi: 10.1016/j.virol.2007.01.022. [DOI] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci USA. 2012;109:E2424–2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita Y, de Hoon MJ, Danchin A. Hon-yaku: a biology-driven Bayesian methodology for identifying translation initiation sites in prokaryotes. BMC Bioinformatics. 2007;8:47. doi: 10.1186/1471-2105-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks J, Kannan K, Roncase EJ, Klepacki D, Kefi A, Orelle C, Vazquez-Laslop N, Mankin AS. Context-specific inhibition of translation by ribosomal antibiotics targeting the peptidyl transferase center. Proc Natl Acad Sci USA. 2016;113:12150–12155. doi: 10.1073/pnas.1613055113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meydan S, Vazquez-Laslop N, Mankin AS. Genes within genes in bacterial genomes. In: Archaea GStorz, Papenfort K., editors. Regulating with RNA in Bacteria. Washington, DC: ASM Press; 2018. pp. 133–154. [Google Scholar]
- Michael AJ. Biosynthesis of polyamines and polyamine-containing molecules. Biochem J. 2016;473:2315–2329. doi: 10.1042/BCJ20160185. [DOI] [PubMed] [Google Scholar]
- Miller MJ, Wahba AJ. Chain initiation factor 2. Purification and properties of two species from Escherichia coli MRE 600. J Biol Chem. 1973;248:1084–1090. [PubMed] [Google Scholar]
- Nakahigashi K, Takai Y, Kimura M, Abe N, Nakayashiki T, Shiwa Y, Yoshikawa H, Wanner BL, Ishihama Y, Mori H. Comprehensive identification of translation start sites by tetracycline-inhibited ribosome profiling. DNA Res. 2016;23:193–201. doi: 10.1093/dnares/dsw008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Orelle C, Carlson S, Kaushal B, Almutairi MM, Liu H, Ochabowicz A, Quan S, Pham VC, Squires CL, Murphy BT, Mankin A S. Tools for characterizing bacterial protein synthesis inhibitors. Antimicrob Agents Chemother. 2013;57:5994–6004. doi: 10.1128/AAC.01673-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Kim KI, Woo KM, Seol JH, Tanaka K, Ichihara A, Ha DB, Chung CH. Site-directed mutagenesis of the dual translational initiation sites of the clpB gene of Escherichia coli and characterization of its gene products. J Biol Chem. 1993;268:20170–20174. [PubMed] [Google Scholar]
- Paukner S, Riedl R. Pleuromutilins: potent drugs for resistant bugs-mode of action and resistance. Cold Spring Harb Perspect Med. 2017;7:a027110. doi: 10.1101/cshperspect.a027110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge JA, Deville F, Sacerdot C, Petersen HSA, Cenatiempo Y, Cozzone A, Grunberg-Manago M, Hershey JW. Two translational initiation sites in the infB gene are used to express initiation factor IF2 alpha and IF2 beta in Escherichia coli . EMBO J. 1985;4:223–229. doi: 10.1002/j.1460-2075.1985.tb02339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen SM, Karlsson M, Johansson LB, Vester B. The pleuromutilin drugs tiamulin and valnemulin bind to the RNA at the peptidyl transferase centre on the ribosome. Mol Microbiol. 2001;41:1091–1099. doi: 10.1046/j.1365-2958.2001.02595.x. [DOI] [PubMed] [Google Scholar]
- Salmon KA, Hung SP, Steffen NR, Krupp R, Baldi P, Hatfield GW, Gunsalus RP. Global gene expression profiling in Escherichia coli K12: effects of oxygen availability and ArcA. J Biol Chem. 2005;280:15084–15096. doi: 10.1074/jbc.M414030200. [DOI] [PubMed] [Google Scholar]
- Salzberg SL, Delcher AL, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlunzen F, Pyetan E, Fucini P, Yonath A, Harms JM. Inhibition of peptide bond formation by pleuromutilins: the structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin. Mol Microbiol. 2004;54:1287–1294. doi: 10.1111/j.1365-2958.2004.04346.x. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Wolf YI, Ramamurthi KS. Small proteins can no longer be ignored. Annual Review Biochem. 2014;83:753–777. doi: 10.1146/annurev-biochem-070611-102400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Daegelen P, Lenski RE, Maslov S, Kim JF. Understanding the differences between genome sequences of Escherichia coli B strains REL606 and BL21 (DE3) and comparison of the E. coli B and K-12 genomes. J Mol Biol. 2009;394:653–680. doi: 10.1016/j.jmb.2009.09.021. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Sotta N, Yamazumi Y, Yamashita Y, Miwa K, Murota K, Chiba Y, Hirai MY, Akiyama T, Onouchi H, Naito S, et al. The Minimum Open Reading Frame, AUG-Stop, Induces Boron-Dependent Ribosome Stalling and mRNA Degradation. Plant Cell. 2016;28:2830–2849. doi: 10.1105/tpc.16.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver JS, Mohammad F, Buskirk Al, Storz G. Identifying small proteins by ribosome profiling with stalled initiation complexes. mBio. 2019 doi: 10.1128/mBio.02819-18. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M, Chothia C, Gough J. SUPERFAMILY-sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 2009;37:D380–386. doi: 10.1093/nar/gkn762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistrand M, Sonnhammer EL. Improved profile HMM performance by assessment of critical algorithmic features in SAM and HMMER. BMC Bioinformatics. 2005;6:99. doi: 10.1186/1471-2105-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WH, Morris DR. Biosynthetic arginine decarboxylase from Escherichia coli Purification and properties. J Biol Chem. 1973;248:1687–1695. [PubMed] [Google Scholar]
- Yan K, Madden L, Choudhry AE, Voigt CS, Copeland RA, Gontarek RR. Biochemical characterization of the interactions of the novel pleuromutilin derivative retapamulin with bacterial ribosomes. Antimicrob. Agents Chemother. 2006;50:3875–3881. doi: 10.1128/AAC.00184-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, D'Lima NG, Slavoff SA. Comparative membrane proteomics reveals a nonannotated E. coli heat shock protein. Biochemistry. 2018;57:56–60. doi: 10.1021/acs.biochem.7b00864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Rudd KE. EcoGene 3.0. Nucleic Acids Res. 2013;41:D613–624. doi: 10.1093/nar/gks1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.