Abstract
Chromatin topology is intricately linked to gene expression, yet its functional requirement remains unclear. Here, we comprehensively assessed the interplay between genome topology and gene expression using highly rearranged chromosomes (balancers) spanning ~75% of the Drosophila genome. Using transheterozyte (balancer/wild-type) embryos, we measured allele-specific changes in topology and gene expression in cis, whilst minimizing trans effects. Through genome sequencing, we resolved eight large nested inversions, smaller inversions, duplications, and thousands of deletions. These extensive rearrangements caused many changes to chromatin topology, including long-range loops, TADs and promoter interactions, yet these are not predictive of changes in expression. Gene expression is generally not altered around inversion breakpoints, indicating that mis-appropriate enhancer-promoter activation is a rare event. Similarly, shuffling or fusing TADs, changing intra-TAD connections and disrupting long-range inter-TAD loops, does not alter expression for the majority of genes. Our results suggest that properties other than chromatin topology ensure productive enhancer-promoter interactions.
Keywords: chromatin organization, chromosomal rearrangements, gene expression, genome topology, TADs, allele-specific Hi-C, enhancers, transcription, embryogenesis
Introduction
Complex patterns of gene expression are controlled by enhancer elements, which can be located close to, or far from (in genomic distance) their target genes1–3. How regulatory information is conveyed across such distances is a long-standing, poorly understood question4. In recent years, chromatin topology i.e. the three-dimensional conformation of DNA into complex topologies, has been suggested to play a key role in bringing enhancers into spatial proximity with their target genes5. The genome is organized into topologically associating domains (TADs), which represent contiguous genomic segments with a higher frequency of interactions within them than between them6–9. TADs are thought to create regulatory environments that facilitate enhancer function, and insulate promoters from ectopic activation by enhancers in a neighboring TAD. Evidence for this comes from the disruption of individual TAD boundaries in cis 7,10–14. For example, a genomic inversion overlapping the boundary of the Epha4 containing TAD leads to limb defects due to new interactions between Epha4 enhancers and the Wnt6 gene10. Such enhancer adoption15 or hijacking16 due to structural rearrangements affecting TADs has also been observed in cancer16–20. For example, in medulloblastoma, structural rearrangements facilitate new enhancer-promoter interactions and the activation of proto-oncogenes GFI1 and GFI1B 16.
While these individual examples indicate that changing genome topology can have strong effects on gene expression, other studies suggest a more moderated role. For example, fusing or scrambling chromosomes in yeast has little effect on gene expression21,22, perhaps due to the predominantly promoter-proximal regulation in yeast. In Drosophila, where enhancers are comparatively more distal, deletions23 or engineered inversions within three testis-specific gene clusters24 had little impact on gene expression, although their impact on chromatin topology was not assessed. Similarly, a series of increasingly large deletions overlapping a TAD boundary in the mouse HoxD locus had little effect on limb bud expression25. It was only with larger deletions (> 40 kb) that expression changes and ectopic interactions across the TAD boundary were observed25. Perhaps even more striking, depletion of CTCF26,27 and cohesin28–30 proteins in trans led to a very dramatic reduction in TAD insulation for the majority of TADs, yet this had only moderate effects on gene expression26,28,29. This may reflect the inherent difference between weakening TAD boundaries due to protein depletion in trans 27,29,31, compared to completely altering TAD structure due to rearrangements in cis 10–12. Much more extensive genetic manipulations are needed to resolve the functional role of genome topology in enhancer-promoter communication.
To more systematically assess the functional relationship between chromatin topology and gene expression, we took advantage of the highly rearranged nature of balancer chromosomes in Drosophila melanogaster. We sequenced the genome of a “balancer” line, whose second and third chromosomes contain eight large nested inversions, several smaller inversions, duplications and thousands of deletions. The functional impact of these genetic perturbations on genome topology and gene expression was assessed in cis in heterozygote embryos, using allele-specific Hi-C and RNA-seq. Despite major changes in genome organization, only a few hundred genes have moderate changes in expression, indicating that only a subset of genes are sensitive to changes in their topology. Rearrangements that alter TAD boundaries or reshuffle TADs, for example, impact the expression of only a subset of the associated genes, suggesting that enhancer hijacking is a rare event. Genetic variants that resize TADs, that cause intra-TAD changes in promoter interactions, or that break long-range inter-TAD loops are generally not correlated with changes in gene expression. Although gene expression at distinct loci is influenced by topology, our more global data suggests that this is not generalizable – the expression of many genes appear resistant to rearrangements within their regulatory domain.
Results
Drosophila balancer chromosomes as a source of highly rearranged chromosomes
Balancers are highly rearranged chromosomes that carry multiple nested inversions, suppressing genetic recombination between homologous chromosomes during meiosis. They were generated roughly 60 years ago31,32 by combining a recessive lethal mutation with inversions, often generated sequentially by X-ray irradiation to increase the balancer’s capacity to suppress recombination across large stretches of the chromosome33. Balancers are therefore typically homozygous lethal, and maintained in trans to a non-balancer homologous chromosome. Here, we used a ‘double balancer’ line carrying rearranged chromosomes for both the second (CyO31) and third (TM332) chromosomes, together covering 76% of the Drosophila genome (Fig. 1a). We crossed the double balancer to an isogenic wild-type line (Supplementary Fig. 1a; Methods). Trans-heterozygous adults from the F1 generation were backcrossed to the wild-type parental line and this pool of N1 embryos was used for all embryonic experiments (Supplementary Fig. 1a,b; Methods). Importantly, this ensures that the N1 generation is devoid of homozygous balancer embryos, which are embryonic lethal and therefore likely to exhibit indirect effects on gene expression. Allele-specific chromosome conformation capture (Hi-C, Capture-C) and RNA-seq were used to measure changes in chromatin organization and gene expression from both chromosomes. This design thereby facilitates a direct comparison between genome topology and gene expression in cis – while minimizing trans effects.
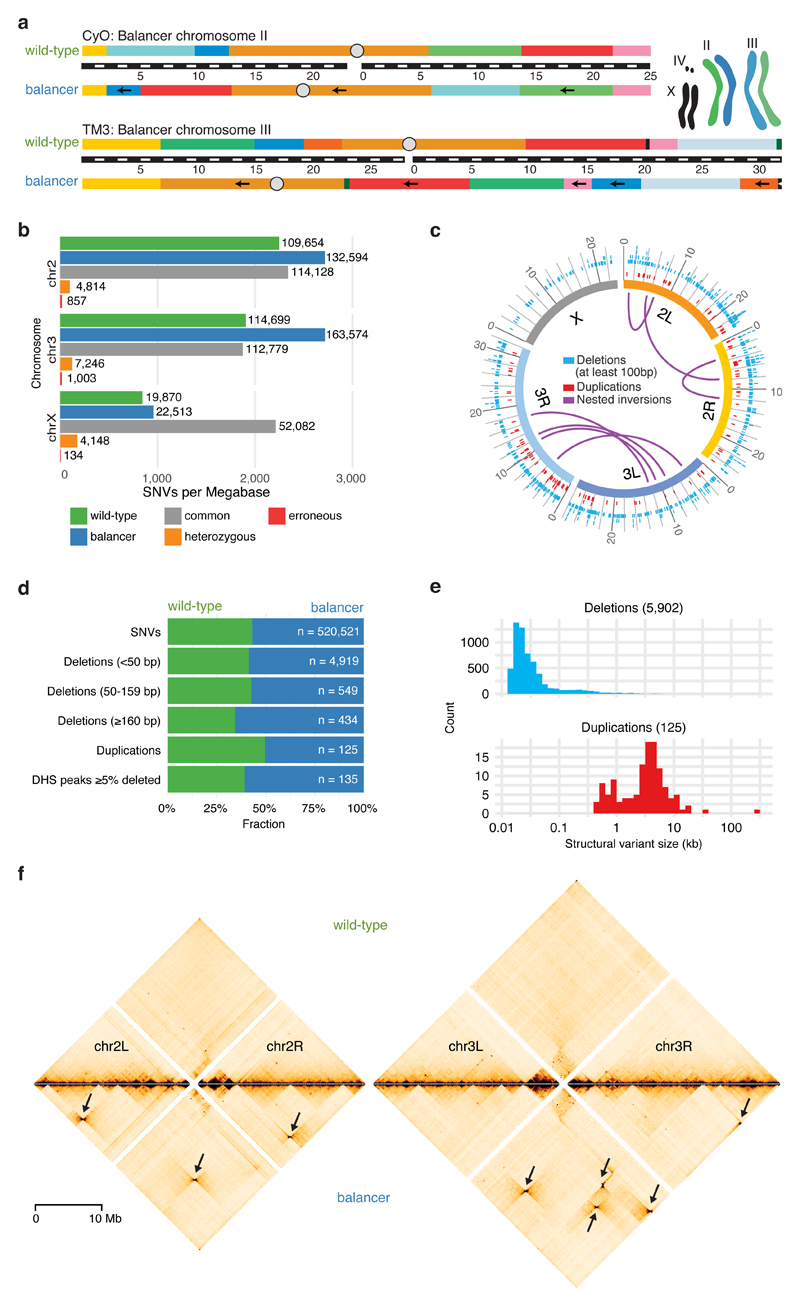
Figure 1. Genomic variation in balancer and wild-type chromosomes.
a, Drosophila melanogaster balancer chromosomes CyO and TM3 carry multiple nested inversions. Arrows indicate segments on the balancer chromosomes that have opposite orientation compared to wild-type. Black bar with white dashes indicates chromosome size (megabases). Cartoon depicts the genotype of the heterozygous (F1) fly line used. b, Number of Single Nucleotide Variants (SNVs) specific to wild-type (green), balancer (blue), common to both haplotypes (grey), heterozygous (orange) or erroneous (red), per megabase for each chromosome. c, Circos plot representing the distribution of structural variants (deletions >100 bp (blue), duplications (red), and nested inversions (purple)) in balancer and wild-type chromosomes. Variants common to both are omitted. d, Fraction of SNVs, deletions (<50 bp, 50-159 bp, ≥160 bp), duplications, and DNase I hypersensitive peaks (DHS, deleted by ≥5% of their length) specific to wild-type (green) or balancer (blue) haplotypes. e, Size distributions of allele-specific deletions (blue) and duplications (red). f, Allele-specific Hi-C contact maps for wild-type (top) and balancer (bottom) chromosomes, relative to the reference assembly (dm6). Bowtie-shaped contacts (arrows) correspond to the nested inversions characteristic of balancer chromosomes.
To identify Single Nucleotide Variants (SNVs) and structural variants (Methods), the DNA of F1 and wild-type adults was sequenced using mate pair34 and whole-genome sequencing. We detected 761,348 SNVs on chromosome 2 and 3 compared to the reference genome, of which 38.9% are balancer-specific and 29.5% wild-type specific (Fig. 1b). An allele-specific SNV occurs on average every 210 bp, allowing sequencing reads to be efficiently assigned to the balancer or wild-type haplotypes. We integrated three approaches (paired-end, split read and read depth35) to identify structural variants with high confidence (Methods), obtaining 6,180 small (15-49 bp), 687 medium (50-159 bp) and 434 large (0.16-5.2 kb) deletions, in addition to 122 tandem duplications (from 400 bp to 33 kb) (Fig. 1c,d,e). This is in line with prior Drosophila population studies, which identified polymorphic duplications to be on average larger but less numerous than deletions36,37. The accuracy of structural variant predictions was confirmed by PCR on randomly selected loci for medium and large deletions (validation rates: 24/25 (medium size), 49/50 (large), Table S1).
The two balancer chromosomes contain eight large nested inversions (Fig. 1c), whose approximate cytological locations were characterized by karyotyping38. Using mate pair sequencing, we mapped their breakpoints to base-pair resolution in 14 out of 16 cases (Fig. 1c, Table S2). Most of the newly formed junctions resulted in a deletions (in one case ~17.5 kb) followed by small duplication (four cases, up to 281 bp), as observed in humans39. Only one inversion has a precise re-ligation. We confirmed all breakpoints using two independent approaches. First, recent independent sequencing of the second40 and third41 balancer chromosomes precisely match our breakpoints in 12 cases. Second, we performed allele-specific Hi-C on the pool of N1 embryos collected at 4 to 8 hours after egg lay (stages 8-11) (Methods; Table S3). After separating the reads from each haplotype, Hi-C contact maps were generated for both the balancer and wild-type chromosomes. The inversions are visible as strong signals off the diagonal with a characteristic “bowtie” shape when mapped to the reference genome (Fig. 1f). The location of the breakpoints perfectly recapitulates the expected karyotype information. In addition to these large inversions, we identified a 38 kb inversion and three non-tandem duplications on the balancer chromosomes, including a 258 kb inverted duplication (Supplementary Fig. 2a,b).
Genomic perturbations accumulate in balancer chromosomes
As the double balancer is only viable in a heterozygous state, there is little selective pressure to eliminate deleterious mutations. Balancer chromosomes also suppress recombination in the broad vicinity of the inversion breakpoints, as recently confirmed40,41. The combination of these two properties means that balancers act as mutational sinks, accumulating deleterious structural variants and SNVs over time. This is exactly what we observe for the double balancer line: the balancer chromosomes have ~1.3 times more allele-specific SNVs compared to wild-type chromosomes (Fig. 1d). Many balancer-specific SNVs are not found in a well-studied wild population (the Drosophila Genetic Reference Panel (DGRP)36,42, with a lower fraction (86.1%) of balancer-specific SNVs present in DGRP lines compared to the wild-type strain (92.1%). Balancer chromosomes also contain more deletions, especially larger ones, compared to their wild-type homologs (ratio 1.9 for deletions ≥160bp, Fig. 1d). Balancer chromosomes have therefore accumulated many genetic variants not present in homozygous viable lines.
Many structural variants could affect functional elements. For example, deletions impact more DNase I hypersensitive sites (DHS) at matching developmental stages43 in the balancer chromosomes than the wild-type (82:53) (Fig. 1d, Supplementary Fig. 2c). Similarly, structural variants impact more protein coding genes on balancer chromosomes. The breakpoints of the large nested inversions, for example, disrupt genes in 12 out of 16 cases, including Glut4EF (a transcription factor involved in developmental patterning and morphogenesis) and p53 (tumor suppressor). A 17.5 kb deletion at breakpoint chr3R:20.32 Mb removes CG42668 (a predicted sterol-binding protein) in the balancer line (Supplementary Fig. 2d,e), while a 258 kb inverted duplication increases the copy number of 31 genes, and disrupts one gene (CG31886) that exists in two truncated copies (Supplementary Fig. 2a,b).
Balancer chromosomes therefore contain many structural variants affecting both regulatory and coding regions. These chromosomes thereby provide a rich resource of genomic rearrangements, including inversions, duplications and deletions, which can be used to systematically assess the functional impact of changes in chromatin organization on gene expression.
Genome rearrangements effect expression of a small proportion of genes
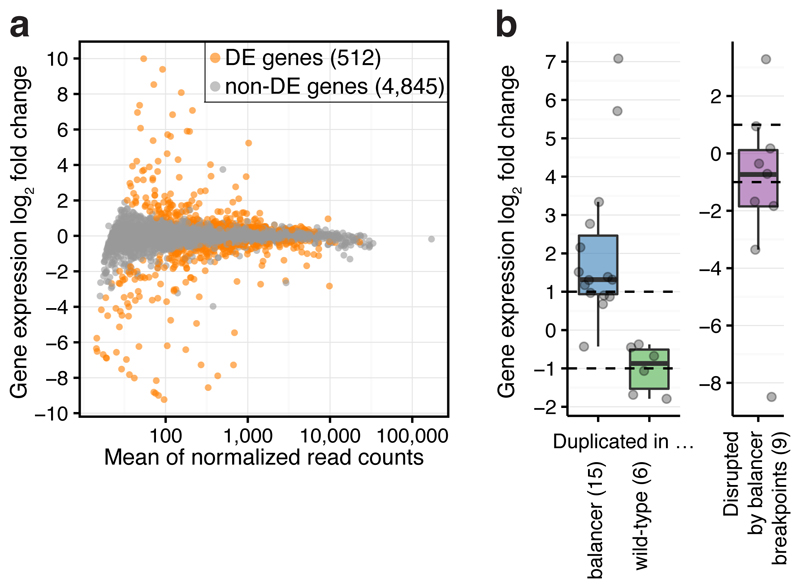
To assess the impact of the extensive changes in genomic structure on gene expression, we performed allele-specific RNA-seq on N1 embryos at 6 to 8 hours after egg lay (stages 10-11; Table S4). To control for possible effects of maternally deposited RNA, embryos were obtained from crosses performed in both directions (Supplementary Fig. 1a, Methods). Sequencing reads were separated based on haplotype-specific SNVs and tested for allele-specific expression (Methods). Of the 5,357 testable genes (on chromosome 2 and 3) with sufficient allele-specific reads, 512 (9.6%) had significant (5% FDR) differential expression between the balancer and wild-type haplotypes, 343 (6.4%) of which had >1.5 fold change (Fig. 2a). Differentially expressed genes have no specific enrichment for biological process or function (Methods).
Figure 2. The impact of chromosomal rearrangements on gene expression.
a, Chromosomal rearrangements generally have a modest effect on gene expression: MA-plot of genome-wide differential gene expression between wild-type and balancer haplotypes. 512 differentially expressed (DE) genes indicated in orange (two-sided Wald test, 5% FDR). b, Change in gene expression (log2 fold change) for testable genes fully duplicated (n=21) or disrupted by balancer breakpoints (n=9). Points, individual genes; center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; dashed lines, expectation for duplications (two-fold increase or decrease).
We confirmed the near absence of trans effects, i.e. the expression of genes on chromosome 3 does not depend on the presence of a chromosome 2 balancer, and vice versa (Methods). The differential expression of only 99/5,981 genes (1.66%) could be explained by trans effects (Supplementary Fig. 3a). While higher than expected from biological noise (0.45%, Supplementary Fig. 3b), this percentage is notably smaller than the total fraction of differentially expressed genes in the F1 generation (6.1%).
Copy-number variants (e.g. duplications or deletions) that fully contain a gene provide a clear prediction of the impact of structural rearrangements on gene expression, and serve as a positive control. Reassuringly, the majority of fully duplicated genes display the expected two-fold change in allelic expression (Fig. 2b), demonstrating the sensitivity of the data. For example, in the context of a 258 kb balancer-specific duplication, the majority of expressed genes have the expected two-fold increase in allelic expression (Supplementary Fig. 2a). This trend is also observed for genes with a partial duplication or deletion, but to a lesser extent as expected (Supplementary Fig. 3c). In total, 50 of the 512 differentially expressed genes (42/343 genes with >1.5 fold change) are likely explained by copy-number variants. An additional 45 of the 343 differentially expressed genes with >1.5 fold change have aberrant transcriptional starts, most likely caused by the insertion of a transposable element in their vicinity. This signal can cause haplotype-imbalanced read counts, which likely explains their differential expression.
In summary, the large diversity of structural variants between the balancer and wild-type chromosomes has moderate effects on gene expression with ~10% (512/5,357) of genes affected on the balancer chromosome. Of these, a relatively small proportion (~18%, 95/512) can be explained by genetic variation directly impacting the genes themselves. In the subsequent analysis we examine if changes in chromatin topology can explain the expression changes of the remaining ~80% of genes.
Chromosomal rearrangements that affect TAD size have little effect on gene expression
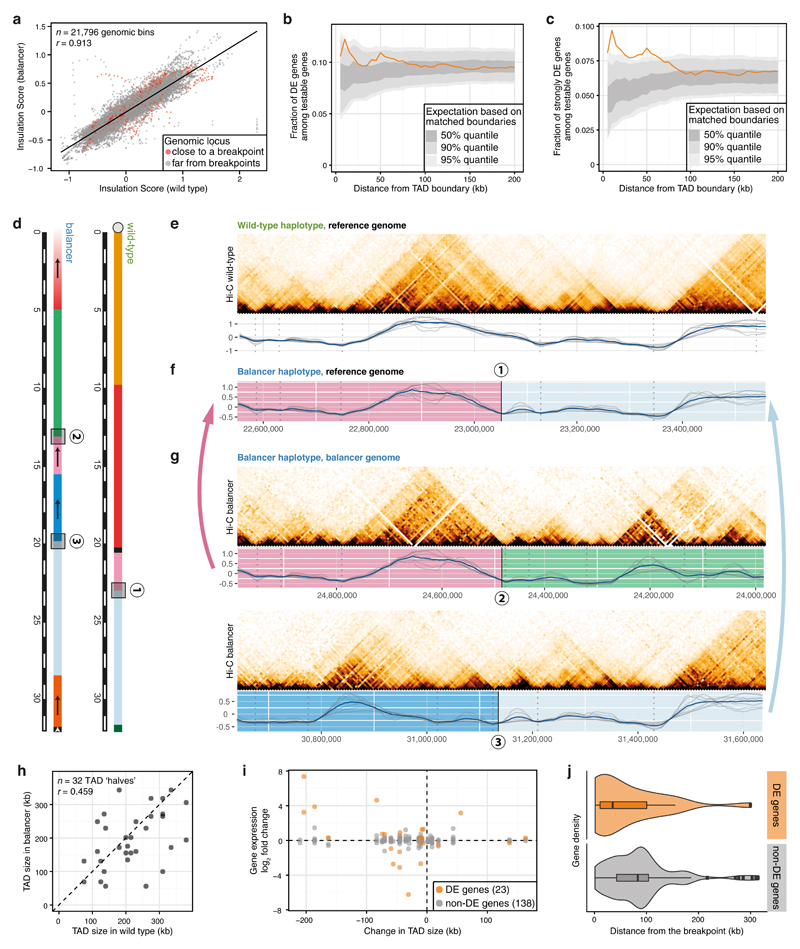
To assess the impact of genome rearrangements on chromatin topology, we used allele-specific Hi-C data and estimated the location of TAD boundaries in each haplotype using insulation scores (IS)44, a normalized measure of Hi-C contacts between a window upstream and downstream (Methods). IS profiles are globally highly correlated between biological replicates for each haplotype (Supplementary Fig. 4a), and between the wild-type and balancer chromosomes (Fig. 3a), indicating that structural variants have a minor impact on chromatin structure genome-wide. TAD boundaries were defined in both haplotypes as the local minima in IS profiles. On chromosomes 2 and 3, we identified 771 TADs in the wild-type and 761 in the balancer haplotypes, with a median size of 125 kb.
Figure 3. Extensive changes in TADs have limited impact on gene expression.
a, Insulation scores (averaged across different window sizes) are highly correlated between wild-type and balancer haplotypes, even close to inversion breakpoints (red). Pearson correlation coefficients (r) indicated. b-c, Fraction of all differentially expressed (DE) genes (b), or those with >1.5 fold-change (c) at varying distances from 182 perturbed TAD boundaries (orange). 50%, 90% and 95% percentiles (grey ribbons) are shown from randomly sampled matched boundaries. d, Schematic of the chr3R:23.05Mb breakpoint in the wild-type (1) and genomic sequences on each side of the breakpoint in the balancer haplotype (2 and 3). e, Top: wild-type Hi-C contact map of a 1 Mb region around the breakpoint (1) from (d). Bottom: wild-type insulation scores for 7 window sizes (50 to 195 kb; grey lines), average profile (blue) and TAD boundaries (grey dotted lines). f, Balancer insulation score profiles lifted over to the wild-type assembly for the region shown in (e). g, As in (e), but for breakpoints (2) and (3) in the balancer haplotype. h, Changes in TAD size due to inversions. Pearson correlation coefficients (r) indicated. i, Changes in TAD size do not correlate with changes in gene expression (log2 fold change) of DE-genes or non-DE genes. j, DE-genes (orange, 23 genes) in disrupted TADs are significantly closer to the inversion breakpoints than non-DE genes (grey, 138 genes). Center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers. p = 0.017, two-sided Kolmogorov-Smirnov test.
Comparing the location of TAD boundaries revealed that ~12% are lost (or shifted by >25 kb) in either haplotype (Methods): 96 out of 767 wild-type boundaries are lost in the balancer, while 86 are gained. Differentially expressed genes are moderately enriched within ±10 kb of perturbed TAD boundaries (Fig. 3b, Supplementary Fig. 4b; Methods): 12.2% (29/237) of testable genes within ±10 kb of a perturbed boundary are differentially expressed genes, compared to 8.5% (155/1,821) around non-perturbed boundaries (p = 0.047, two-sided binomial test). The effect increases for genes with >1.5 fold change: 9.7% (23/237) of differentially expressed genes compared to 4.9% (90/1,821) (Fig. 3c, Supplementary Fig. 4c, p = 0.0023). There is therefore a significant enrichment of differentially expressed genes within 10 kb of a perturbed boundary, however the actual fraction of affected genes is low (only ~10-12% of genes within 10 kb, and ~6% (29/512) of all differentially expressed genes).
We next assessed the effect of inversion breakpoints on TAD size by separating balancer breakpoints falling inside a TAD (disrupting it) from those located close to TAD boundaries. The resulting 16 breakpoints (14 from the large nested inversions, two from a 38 kb inversion) that are located away (>13 kb) from a TAD boundary result in resized (shuffled) TADs that typically still use the existing boundaries (Fig. 3d-g). In fact, the vast majority of TADs (88%) have unchanged boundaries despite drastic rearrangements of the genome. The location of TAD boundaries therefore seems mostly driven by the sequence of the boundary region itself, rather than by the sequence or expression status within the TAD.
Although the position of the boundaries has not changed, the size of these shuffled TADs often changed dramatically (Fig. 3h). The breakpoints split the 16 disrupted TADs into 32 ‘halves’, which are rearranged differently in the balancer. In 5 cases, the ‘halves’ are in a TAD over 50 kb bigger, and in 10 cases over 50 kb smaller in the balancer. 443 genes are located in the 16 resized TADs, of which 322 carry allele-specific SNVs (161 of these genes are expressed, while the remainder have little or no expression at these embryonic stages). Remarkably, these changes in TAD size are not correlated with changes in gene expression (Fig. 3i). 23 genes (out of 161) are differentially expressed (p = 0.040, two-sided Fisher’s exact test), only 16 of which to >1.5 fold change (p = 0.067, two-sided Fisher’s exact test). Interestingly, none of the inactive genes within shuffled TADs were ectopically expressed, suggesting that expressed genes may be more sensitive to changes in their topology.
The expression of the majority of genes within shuffled TADs is not affected by changes in their regulatory environments. To explore this further we examined the location of differentially expressed genes with respect to the breakpoints: differentially expressed genes within disrupted TADs are significantly closer (median distance 35 kb) to inversion breakpoints compared to non-differentially expressed genes (Fig. 3j, p = 0.017, two-sided Kolmogorov-Smirnov test). Therefore, changing the regulatory context within a TAD by inversions that essentially swap TAD parts, leads to significant changes in the expression of some genes that tend to be close to the breakpoint, presumably due to altered enhancer-promoter interactions. However, it is important to note that only a fraction of genes (12/38 expressed genes, 0/34 lowly expressed/inactive genes) within 35 kb of a breakpoint change expression. In 6 of the 16 TADs, it is the gene closest to one side of the breakpoint that is differentially expressed (Supplementary Fig. 4d), suggesting mis-regulation due to new local positioning close to an enhancer. However, in the other 10 cases there are one or more unaffected genes closer to the breakpoint than a differentially expressed gene, indicating that proximity alone is not sufficient to activate all gene’s expression. In addition, the absolute expression level of differentially expressed genes within disrupted TADs is not significantly different from non-differentially expressed genes (Supplementary Fig. 4e, p=0.09, two-sided Wilcoxon test). Therefore, although productive enhancer-promoter interactions can be formed de novo, for example as seen in the 38 kb inversion (discussed below), the majority of genes are not mis-regulated. This suggests selectivity in many enhancer-promoter interactions that cannot be explained by regulatory distance alone.
Changes in gene expression are correlated with local changes in genome topology
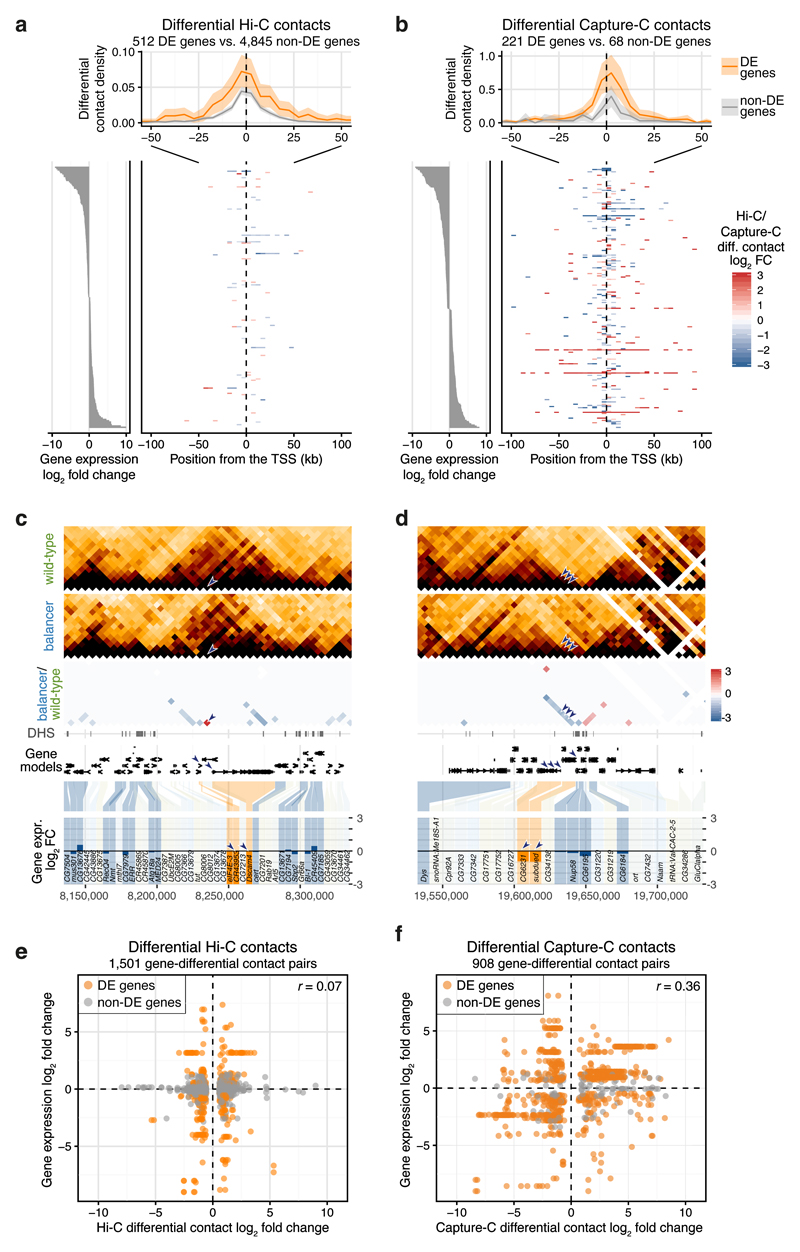
To explore the impact of changes in intra-TAD contacts, putative enhancer-promoter interactions, on gene expression, we performed Capture-C to generate high-resolution views of promoter topologies. Libraries were generated on the same pool of N1 embryos as Hi-C, using probes designed to hybridize equally to the two haplotypes and capture promoters of 221 differentially expressed genes and 68 non-differentially expressed ‘control’ genes (Tables S5, S6, Methods). Differential interactions were defined by comparing the interaction count of balancer vs. wild-type haplotypes using both Hi-C and Capture C data (Methods). Among 216,066 tested pairwise interactions between 5 kb Hi-C bins, 4,329 had significant differential contacts, not explained by overlapping CNVs. Among the 59,605 tested interactions from the Capture-C viewpoints, 931 had differential contacts at the level of individual restriction fragments. Neighboring differential fragments within 1 kb were clustered to form 445 differentially contacted regions.
We first focused on differentially expressed genes and plotted the differential contact density from their transcription start sites (TSS) (Fig. 4a,b). Genes with differential expression generally have twice as many differential contacts from their promoters: differentially expressed genes have on average 0.48 differential Hi-C contacts within ± 100 kb from their TSS, while non-differentially expressed genes have 0.20 (p = 0.006, two-sided Kolmogorov-Smirnov test, Fig. 4a). Similarly with Capture-C; differentially expressed genes have on average 3.59 differential contacts within ± 100 kb from their TSS, while non-differentially expressed genes have 1.46 (p = 0.033, Fig. 4b). This effect is more pronounced for genes with >1.5 fold change in expression (Supplementary Fig. 5a,b). The effect vanishes at larger distances (>50 kb). Genes with changes in their expression therefore have a small, but significant, increase in local (≤50 kb) differential contacts with their promoters.
Figure 4. Changes in promoter contacts and their relationship to gene expression.
a-b, Top: positional distribution of differential Hi-C (a) or Capture-C (b) contacts originating from Transcription Start Sites (TSS) of differentially expressed (DE) (orange) and non-DE (grey) genes. All genes are aligned by their TSS and transcribed to the right. 95% confidence bands (shaded ribbons) estimated using bootstrapping. Bottom right: heatmap of differential contact (Hi-C, log2 fold change) for 200 randomly sampled DE-genes (a) or for all captured DE-genes (Capture-C) (b). Bottom left: expression (log2 fold change) of genes shown in the heatmap. c-d, Downregulated genes (CR43953 and Dscam4 in (c); CG6231 and subdued in (d)) have differential Hi-C contacts, either increasing, red (c) or decreasing, blue (d) proximity in the balancer. Top to bottom: wild-type and balancer Hi-C contact maps, log2 fold change before normalization (balancer/wild-type; red/blue), DNase hypersensitive sites (DHS), gene models and differential gene expression (balancer/wild-type, log2 fold change). Differentially expressed (DE-) genes in orange, non-DE genes in blue and non-tested genes (lowly expressed or lacking SNVs) in grey. Arrowheads highlight differential contacts and the affected genes. e-f, Hi-C (e) and Capture-C (f) differential contacts (log2 fold change) are plotted against changes in expression (log2 fold change) of all genes with a TSS in the two interacting genomic bins. DE-genes highlighted in orange. Pearson correlation coefficient (r) indicated.
Interestingly, such differential interactions from the promoters of differentially expressed genes often involve the promoters of other differentially expressed genes, an observation that occurs more often than expected by chance (Supplementary Fig. 5c). For 14 out of 18 such pairs of differentially expressed genes linked by a differential contact, the direction of change is the same for both genes’ expression (Supplementary Fig. 5d, p = 0.03, two-sided binomial test, Table S7). The promoters of Dscam4 and the long non-coding RNA CR43953, for example, are in proximity in the balancer, but not the wild-type (40-fold increased Hi-C contact), which is associated with a strong down-regulation of both genes’ expression in the balancer (Fig. 4c). Conversely, three differential contacts between the promoter of subdued (a chloride channel) and multiple promoters of CG6231 (a predicted transmembrane transporter), located 10-20 kb away (Fig. 4d) are reduced on the balancer chromosome, which is associated with a ~two-fold decrease in both genes’ expression. The concordance in the changes between both genes expression and their promoter interactions suggests co-regulation of gene pairs, perhaps in a transcriptional hub type of conformation.
Changes in genome topology are not predictive of changes in gene expression
As shown above, genes with changes in their expression have a small but significant enrichment for differential promoter contacts. To test whether the converse is true, i.e. if changes in three-dimensional proximity (Hi-C contacts) are predictive of changes in gene expression, we extracted differential contacts that had a TSS at either of the two contacting loci. Of the 4,329 differential Hi-C contacts (after filtering for CNVs), 1,063 (24.6%) are associated with a promoter of a testable gene. Focusing on these promoters with differential contacts reveals no correlation between promoter contact frequency (Hi-C fold change) and gene expression (Fig. 4e). Only 25% (265/1,063) of differential contacts at promoters are associated with a change in gene expression. Similarly with Capture-C – although the majority (76%) of tested promoters were differentially expressed genes, differential promoter contacts are not correlated with differential gene expression (Fig. 4f, Supplementary Fig. 5b,e), indicating that this observation is likely not due to limited resolution. Rather, there are many differential contacts at promoters that do not correlate with changes in gene expression.
Taken together, our results indicate that differential gene expression is globally correlated with local changes in genome topology (or contacts), as observed at individual loci: genes that change in expression generally have twice as many differential contacts at their promoters compared to non-differentially expressed genes. However, going in the other direction, changes in genome topology at promoters are not globally correlated with changes in gene expression: 75% (798/1,063) of all promoter contacts at testable genes (differential promoter contacts) can change with no measurable effect on gene expression. These results highlight an inherent robustness within regulatory landscapes, where many changes in genome topology, even at promoters, are buffered at the level of changes in gene expression.
Loss of long-range chromatin loops has little impact on gene expression
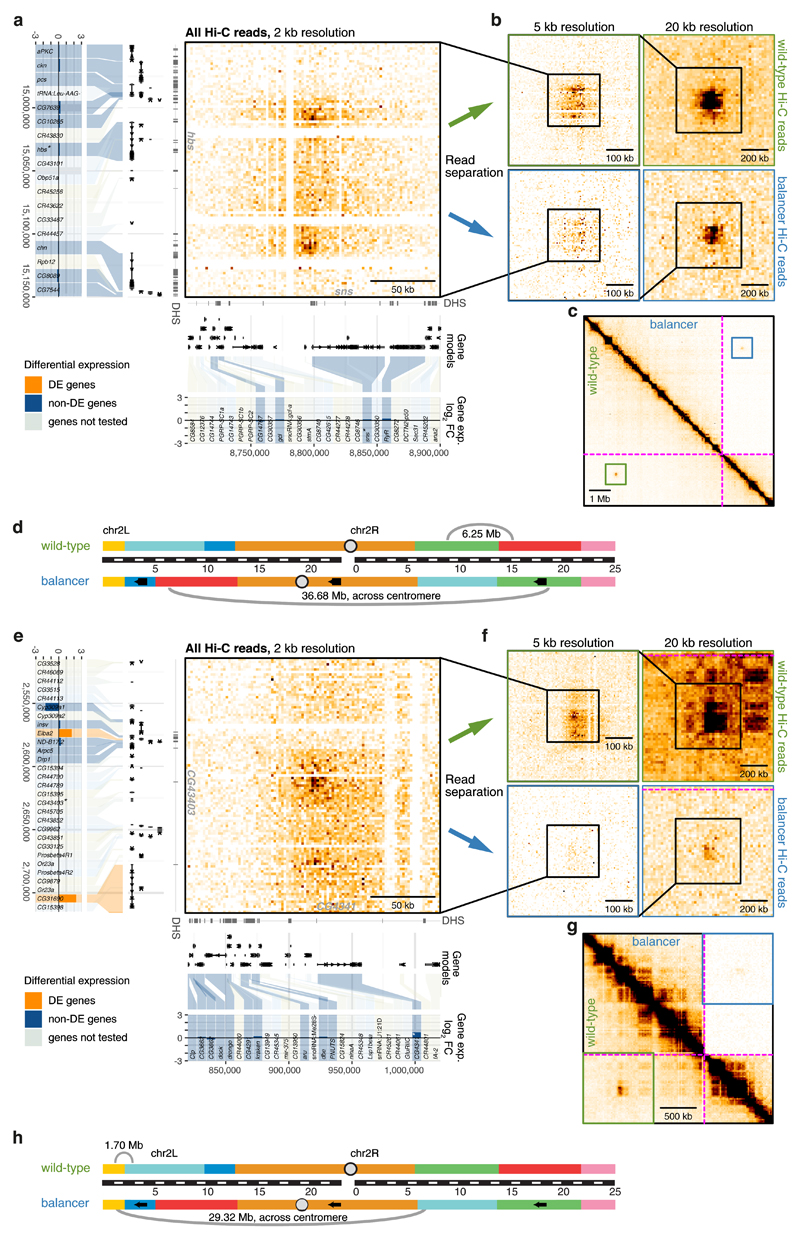
Long-range inter-TAD loops that span multiple TADs have been observed in many species including Drosophila 9. They typically involve co-regulated active genes, e.g. sns and hbs in Drosophila 9, or inactive Polycomb repressed genes45. Six such long-range chromatin loops span across the nested inversion breakpoints, allowing us to assess if genome reorganization affects long-range looping and how this impacts gene expression. In two cases (sns/hbs, Wbp2/Nufip) the loop is still present (although at reduced frequencies for sns) despite changes in the genes’ genomic context and relative distance, while in the other four cases the loop is either lost or severely diminished in balancer chromosomes. For example, a long-range loop forms between the sns and hbs loci, separated by 6.25 Mb on the wild-type chromosome (Fig. 5a-d). On the balancer, these genes are separated by over 36.68 Mb (~20% of the size of the total Drosophila genome) spanning a centromere, and yet remarkably the looping interaction is still present (although at roughly half the interaction frequency), and there is no significant impact on the surrounding genes expression.
Figure 5. Loss of long-range chromatin loops has little impact on gene expression.
a, e, Hi-C contact map (2 kb resolution, all Hi-C reads) showing a long-range loop between hbs (left) and sns (bottom) (a) or CG43403 (left) and CG4341 (bottom) (e). The location of DNase hypersensitive sites (DHS), gene models, and differential gene expression (balancer/wild-type; log2 fold change) are shown. Differentially expressed (DE) genes in orange, non-DE genes in blue and non-tested genes (lowly expressed or lacking SNVs) in grey. b, f, Zoomed-in wild-type (top) and balancer (bottom) Hi-C contact maps at 5 kb (left) and 20 kb (right) resolution showing a slight (b) or strong (f) decrease in the strength of long-range loops. c, g, Zoomed-out Hi-C contact map showing the location of the long-range loops between sns and hbs (c) and CG4341 and CG43403 (g) in the wild-type (bottom left) and balancer (top right) haplotypes. The location of inversion breakpoints is indicated by purple dotted lines. d, h, Cartoon depicting the location, and distances, of both loops on chromosome 2 in wild-type (top) and balancer (bottom) haplotypes with respect to the inversion breakpoints.
In two other cases the long-range loops normally span ~2 Mb (CG4341/CG43403 and eIF4E-4/PGRP-LA) but are now separated by over 28 Mb in the balancer, which results in a severe reduction in looping frequency (Fig. 5e-h). Interestingly, this decrease does not affect the interacting genes’ expression (e.g. CG4341/CG43403, Fig. 5e-h). Two genes located further away from the looping region (Elba2 and CG31690) have elevated expression on one side, while no gene’s change expression on the other. Similarly, the long-range loops between Nufip/kug (normally separated by 1.25 Mb) and Wbp2/kug (7.15 Mb) are separated by distances of 14.83 Mb and 20 Mb in the balancer, respectively (Supplementary Fig. 6). In both cases this leads to a severe diminishment of the looping interactions, but this only seems to impact the expression of CG10960, a gene located close to Wbp2 (Supplementary Fig. 6c,d). There is no significant difference in Wbp2, Nufip or kug’s expression (or other genes) despite the strong reduction in the associated looping interaction.
These inversions have enabled us to disentangle long-range looping interactions from gene expression. In some cases the loop has a remarkable ability to form despite huge changes in genomic distance, suggesting that the underlying mechanism that brings these loci together can still function. In other cases the inversion has broken (or severely diminished) the frequency of long-range interactions, and interestingly this has no effect on the interacting genes’ expression. This indicates that long-range loop formation can be uncoupled from the associated genes’ expression suggesting no direct causal link between the two.
Discussion
Individual examples of genomic rearrangements that alter TAD structure46 indicate that this can lead to mis-expression at specific loci10,11,16–18,20. These studies typically started with a phenotype, a structural variant leading to a developmental defect or cancer cells that have gained a selective advantage, and worked backwards to explain the misexpression in the context of TAD structure. The balancer system may also have some selection bias as the original inversions were viable, before a lethal mutation and more x-ray induced inversions were added. As they are homozygous lethal, balancers are always maintained in trans, and can therefore accommodate extensive recessive lethal rearrangements with loss- or gain- of regulatory interactions. This system thereby allowed us ask more generally if changes in chromatin topology can predict changes in gene expression, within the context of embryonic development.
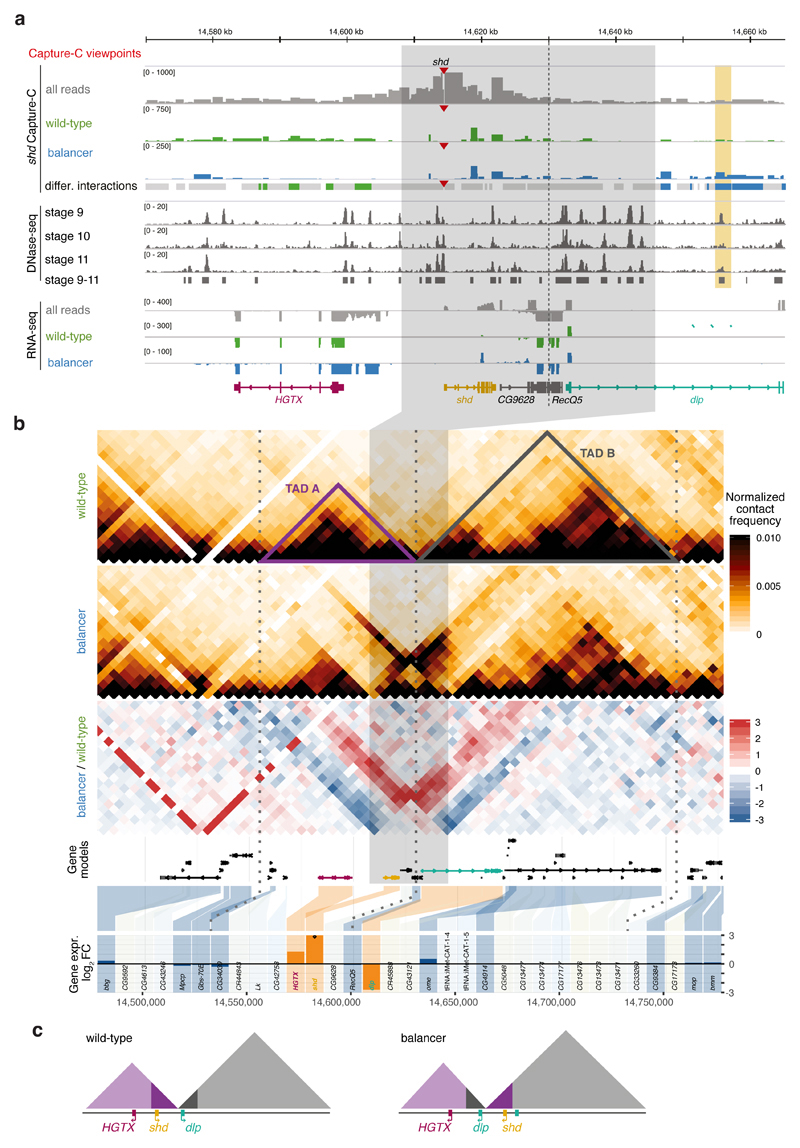
A 38 kb inversion that overlaps a TAD boundary provides a good example (Fig. 6). Of the seven genes with allelic coverage within the affected TADs, only three are differentially expressed: HGTX (Nkx6), shd and dlp (all essential embryonic genes). The dlp gene is directly disrupted by the inversion, while HGTX is likely impacted by a transposable element. The only gene whose differential expression (over-expression in the balancer) is likely caused by a change in regulatory landscape is shd. The shd promoter normally establishes 3D contacts across the left inversion breakpoint within TAD-A, as observed by Capture-C (Fig. 6a). These contacts are lost in the balancer and new ones formed with regions overlapping a DHS43 originally located in TAD-B (orange highlight, Fig. 6a), suggesting that new enhancer-promoter interactions lead to shd misexpression. However, such enhancer adoption did not occur for the other four genes within the newly formed TADs. This is not unique to this breakpoint; in all 16 inversion breakpoints that disrupt TADs, only a fraction of genes change expression, and it’s generally not the closest gene (Supplementary Fig. 4d). This indicates that the activity of many regulatory elements is resistant to topological changes in their regulatory environment.
Figure 6. Chromatin organization and expression around a 38kb inversion.
a, Top to bottom: Capture-C tracks (all reads (grey), wild-type-specific (green), and balancer-specific (blue) reads) for the shd viewpoint (red triangle) in the vicinity of a balancer-specific inversion in chr3L (grey highlight). Dashed vertical line indicates the TAD boundary. Differential Capture-C contacts stronger in wild-type (green), balancer (blue), or not significantly changed (grey) are highlighted. DHS signal at stages 9-11, RNA expression (all reads and allele-specific) and gene models shown underneath. A differential contact is formed between the shd promoter and a DHS (vertical orange rectangle) in the balancer haplotype, observed in two independent Capture-C experiments. b, Top to bottom: Wild-type and balancer Hi-C contact maps, log2 fold change before normalization (balancer/wildtype, red/blue), gene models, and differential gene expression (balancer/wildtype, log2 fold change). Differentially expressed (DE) genes in orange, non-DE genes in blue and non-tested genes (lowly expressed or lacking SNVs) in grey. Dotted vertical lines indicate TAD boundaries. c, Model of the inversion and associated changes in TAD structure and position of three DE-genes in the balancer haplotype.
Our results, obtained from genetic rearrangements in cis, complement recent findings depleting CTCF26,27 and cohesin28–30 proteins in trans, where the majority of TADs were reduced yet hundreds, rather than thousands of genes changed expression26,28,29. This raises a number of interesting questions: what is the role of TADs in gene regulation? What are the regulatory differences between genes that are sensitive to, or resistant to, changes in their topology? Taken together, our results highlight an apparent uncoupling between gene expression and 3D genome organization and suggests that there must be properties, in addition to genome topology, that facilitate productive enhancer-promoter interactions.
Online Methods
Drosophila lines
To obtain the large number of embryos required for high resolution Hi-C and Capture-C at specific embryonic stages, we setup a big cross between the two haplotypes. To obtain a large number of virgin wild-type female flies, we used a ‘virginizer’ line, w[1118]/(P{hs-hid}Y). We first made this line isogenic by back-crossing the w[1118]/(P{hs-hid}Y) line for at least 18 generations in single pair mattings to ensure maximal homozygosity. The resulting isogenic virginizer line was amplified to large amounts. To obtain females, vials containing embryos and larvae were placed in a 38°C water bath for 1 h. This resulted in the heat-shock induced expression of the pro-apoptotic gene hid on the Y-chromosome, leading to the death of all males. The only adults that eclose from these vials are female and therefore virgins. The virginizer line will thereafter be referred to as “wild-type” (+/+; +/+).
Adults from the wild-type isogenic line were crossed to the “double balancer” fly line (w; If/CyO; Sb/TM3,Ser) as follows (Supplementary Fig. 1a):
Cross 1: Male F0 (If/CyO; Sb/TM3) x Female F0 (+/+; +/+)
The F1 offspring is composed of 4 genotypes in equal proportions:
(If/+; Sb/+); (If/+; +/TM3); (+/CyO; +/TM3); (+/CyO; Sb/+)
Cross 2 (back-cross): Male F1 (+/CyO; +/TM3) x Female F0 (+/+; +/+)
The N1 offspring (N1 pat) is composed of 4 genotypes in equal proportions:
(+/+; +/+); (+/+; +/TM3); (+/CyO; +/+); (+/CyO; +/ TM3)
We also generated the reciprocal cross (yielding N1 mat offspring) with Female F1 (+/CyO; +/TM3) x Male F0 (+/+; +/+).
The F0 (+/+; +/+) and F1 (+/CyO; +/TM3) adults were used for whole-genome and mate pair sequencing. The pool of N1 embryos was used for RNA-seq (N1 pat and N1 mat), Hi-C (N1 pat) and Capture-C (N1 pat) experiments. Additional controls were included for RNA-seq experiments (see below and Supplementary Fig. 1b).
Embryo collections
Freshly hatched adults were placed in embryo collection vials with standard apple cap plates. After three 1 h pre-lays, the flies were allowed to lay for 2 h, after which the embryos were aged to the appropriate time-point. The embryos were then dechorionated using 50% bleach, and washed alternately with water and PBS + 0.1% Triton X-100. The embryos used for RNA sequencing were directly snap-frozen in liquid nitrogen. The embryos used for Hi-C and Capture-C experiments were covalently crosslinked in 1.8% formaldehyde for 15 min at room temperature and stored at -80°C.
Whole-genome and mate-pair sequencing
Genomic DNA from F0 flies (+/+; +/+) and double balancer flies (F1; i.e. +/CyO; +/TM3) was isolated by grinding them in liquid nitrogen, followed a 30 min incubation at 65°C in lysis buffer (100 mM Tris-HCl, pH 7.5, 100 mM EDTA, 100 mM NaCl, 1% SDS). The sample was then treated with RNaseA to degrade the RNA, before routine DNA extraction by phenol-chloroform and precipitation with ethanol.
The genomic DNA from these F0 and F1 samples was used to generate whole-genome sequencing libraries with an insert size of about 700 bp. The samples were sequenced on the Illumina MiSeq (300 bp paired-end), yielding a total of 146 M and 68 M read pairs respectively, which corresponds to coverages of ~200x and ~100x.
The same F0 and F1 samples were used to generate mate pair DNA libraries using the Nextera Mate Pair Sample Preparation Kit (Illumina) as previously described47. The samples were multiplexed and sequenced on the Illumina HiSeq2000 (100 bp paired-end). Nextera adapter contaminations were cleaned afterwards using NextClip version 1.3.1, yielding a total of 44 M and 73 M read pairs.
SNV and small indel calling
Both whole-genome (WGS) and mate pair sequencing data were mapped to dm6 using bwa mem48 version 0.7.15. SNV and short indel calling was performed using FreeBayes49 version v0.9.21-19 on the WGS data of both samples simultaneously and with disabled population priors. The results were filtered with vcflib (https://github.com/vcflib/vcflib) based on a quality value of 30 or higher, a minimum of at least two reads carrying the allele to the right and to the left end, and on the fact that the allele was seen on at least two reads mapping in each direction. The command used was: vcffilter -f 'QUAL > 29 & QUAL / AO > 2 & SAF > 1 & SAR > 1 & RPR > 1 & RPL > 1' -s. We further normalized indel variants, removed multi-allelic variants, and decomposed multi-nucleotide substitutions (which are reported as haplotype blocks by FreeBayes) into SNVs using vt normalize50. We finally removed contigs other than chromosome 2, 3, and X, obtaining a total of 860,095 SNVs in addition to 158,564 small indels.
Deletion calling
We applied Delly51 version 0.7.2 on the WGS data of the F0 and F1 samples simultaneously and applied an extensive filtering procedure to reduce the number of false positive calls. From the initial 10,421 deletion calls, 5,150 dropped out that were not flagged as QC PASS, were not on the main chromosomes (2, 3, or X), had a mapping quality value of less than 60 or did not match one of the expected genotypes (i.e. balancer-specific, wild-type-specific and common, which together constituted >90% of the calls). Furthermore, we required a minimum number of supporting read pairs for reference and alternative allele combined, namely 40 read pairs for imprecise Delly calls or 25 split reads for precise Delly calls. Next, we developed a dynamic read depth ratio filter that was applied to only large heterozygous calls (i.e. balancer- or wild-type-specific ones). To this end, the read count within the predicted deletion was normalized by the summed read count in size-matched intervals flanking the locus (two flanking intervals, half the deletion size each) and these values were compared (absolute difference) between the two samples. We required a minimum difference in this read depth ratio between samples with different allele counts, where this threshold adapts with structural variant size in a way that large structural variants need to show a clearer difference in read depth ratio between samples than small structural variants (we required that abs(ratio.bal – ratio.vrg) < 1.25/(deletion.basepairs)0.2 + 0.5). To give an example, this filter removed a number of obviously false calls above 100 kb. Deletions were overlapped with a mappability map to classify them into high- (at least 50% of the structural variant is in a uniquely mappable region) or low-confidence loci. This resulted in four call sets: 3,072 high confidence calls less than 50 bp, 737 high confidence calls between 50-159 bp range, 395 high confidence large calls (≥160 bp), and 75 low confidence large calls (≥160 bp).
Finally, we merged Delly deletion calls with small deletions called by FreeBayes (which underlie aforementioned filtering criteria) and chose a lower size cutoff of 15 bp. During the merging process FreeBayes calls were given priority over matching Delly calls (based on 50% reciprocal overlap criterion). The final data set (referred to as “deletions” in the main text) contains 8,340 deletions on chromosomes 2, 3 and X, namely 7,114 deletions below 50 bp, 756 deletions in the 50-159 bp range, and 470 large deletions (≥160 bp). Of these, 6,180, 687 and 434, respectively, were located on chromosomes 2 and 3. Taking only the allele-specific ones, 4,919 deletions below 50 bp, 549 deletions in the 50-159 bp range, and 434 large deletions (≥160 bp) remained.
We compared these balancer deletion calls to deletions present in the DGRP lines (inbred lines generated from wild isolates). We found that the deletions present in the DGRP line tend to be smaller on average compared to the balancer chromosomes. Besides, 50% of the small (20-50 bp) wild-type-specific deletions are found in the DGRP lines versus 40% for the balancer-specific ones. This is in line with our overall findings that the double balancer chromosomes accumulate mutations not naturally present in the wild.
PCR validation of deletion calls
To validate deletion calls we performed PCR on randomly selected loci in three categories: 25 loci of 50-159 bp in size, 25 loci above 159 bp with high confidence, and 25 loci above 159 bp with low confidence. We designed primers using a lab-internal extension to Primer352. The PCR reactions (under standard conditions) were performed on genomic DNA extracted from F1 (+/CyO; +/TM3) and F0 (+/+; +/+) adult flies to visualize a shift in the size of the PCR product. In the size range 50-159 bp, 24 out of 25 loci validated, 24/25 loci validated for high confidence calls of ≥160 bp, and 25/25 loci validated for low-confidence calls, yielding an estimated FDR of 2.66%. After weighting by the number of deletion calls in each of these categories, we can estimate an FDR of 3.75%.
Duplication calling and filtering
Delly51 version 0.7.5 was run in tandem duplication mode and supplied with both mate pair and WGS libraries for F0 and F1 samples simultaneously. Duplication calls were initially filtered by the quality PASS criteria and by their combined genotypes, which were required to be heterozygous in the F1 sample. We did not require homozygosity in the F0 sample because many homozygous tandem duplications are wrongly classified as heterozygous. This misclassification is a known issue of the classifier function (according to the author of Delly), and due to the fact that reference sequence overlapping the breakpoint of homozygous tandem duplication remains contiguous. For all remaining 352 calls we generated QC plots that contained a total read-depth track, a mappability track, and, importantly, a B-allele frequency measured at SNV positions around the predicted locus. These plots allowed us to sort out false positives, leaving 122 manually curated high-quality tandem duplications.
Aside from tandem duplications, we inspected the B-allele frequency ratio across the genomes and unravelled 3 non-tandem duplications of 4.3 kb, 10.4 kb and 258 kb size. The final set (referred to as “duplications” in the main text) contains 125 duplications.
Refinement of inversion breakpoints on the balancer chromosomes
Approximate breakpoint locations were estimated from an initial inspection of the balancer-specific Hi-C plots and narrowed down to ~10 kb. To identify the exact breakpoints we ran Delly51 version 0.7.1 in inversion and translocation mode (because chromosomes are split into p- and q-arms some intra-chromosomal rearrangements will be reported as translocation calls) on both samples, using both mate pair and WGS libraries separately. As translocation calls are typically very noisy, we only searched the regions of interest for calls that connect the expected loci. Starting from Hi-C-based coordinates, we scanned a region of 200 kb around each breakpoint and identified 14 out of 16 rearrangement junctions in the mate pair data, 12 of 14 additionally in the paired-end data. A single inversion could not be found as one of its breakpoints is likely outside the known reference genome. As exact breakpoints of these structural variants underlie an inevitable alignment uncertainty, we manually determined the exact base position of the breakpoints that best matches the read mapping. Interestingly, Miller et al. 40,41 unravelled the exact breakpoints of multiple balancer chromosomes including TM3 shortly after this analysis and our results match perfectly to their findings. Also they were not able to resolve the last inversion, for which we provide a “best guess” according to IGV inspection and a search for dangling read pairs (i.e. where only one of the reads maps). These results are summarized in Table S2. At one breakpoint (chr3_6) we observed a 17.5 kb balancer-specific deletion that not was identified in the previous characterization of the balancer chromosome TM341. Note that we found no heterozygous SNVs in the putative deletion region.
We manually validated smaller (non-nested) Delly inversion calls against Hi-C contact maps, and took into further consideration one 38 kb inversion which was supported by our Hi-C data.
Fly crosses for RNA-seq experiments
RNA-seq was performed on Drosophila embryos of the N1 generation at 6-8 h after egg lay (stages 10-11) (Supplementary Fig. 1; see Methods/”Fly Lines”). The genotypes of the Drosophila embryos cannot be phenotypically distinguished at this early developmental stage; hence the resulting libraries consist of a pool of four different genotypes where 25% of chromosomes 2 and 3 are balancer chromosomes (CyO and TM3, respectively) and 75% wild-type. As we anticipated a bias from maternally deposited mRNAs (i.e. an excess of RNA from the female parental line), we implemented two critical steps to overcome this problem: First, we manually removed unfertilized eggs among the embryos prior to library preparation, which likely contribute to the biggest part of maternal mRNAs. Second, we collected fly embryos from parents where the N1 backcross was set up in both directions: N1 pat (using F1 males and wild-type parental females) as well as N1 mat (using F1 females and wild-type parental males). The impact of maternal mRNAs for any given gene will act in opposite directions in both samples, allowing us to control for such effects.
To determine the impact of trans effect, we used adult fly heads from single-balancer lines (used in Supplementary Fig. 3a). These lines are based on the same F0 wild-type line as the double balancer line and denoted F1 CyO and F1 TM3 (for chromosome 2 balanced and chromosome 3 balanced, respectively). In parallel, adult fly heads from the double-balancer F1 line were collected (denoted F1CyO;TM3, used in Supplementary Fig. 3b). In addition, we collected female-only fly heads from the F1 cross (denoted F1 female) to assess the biological noise between two lines that both carry the CyO and TM3 balancer chromosomes but have originated from a different cross (comparing F1 female and F1CyO;TM3).
RNA-seq experiments
For RNA isolation, a pool of approximately 100 embryos or 20 adult fly heads was homogenized in TRIzol®LS (Life Technologies) with a Cordless Motor for Pellet Mix and pestels (VWR) on ice. RNA was extracted according to the manufacturer’s instructions, and the remaining DNA digested with RNase-free DNase I (Roche) for 30min. The RNA solution was purified a second time using Agencourt RNAClean XP beads (Beckman Coulter).
Strand-specific RNA-seq was performed from 1 μg of total RNA using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB) according to the manufacturers’ recommendations. RNA-seq was performed on embryos from the N1 pat and N1 mat lines, each in two biological replicates. The samples were multiplexed and sequenced on the Illumina NextSeq500 (150 bp paired-end).
We also generated RNA-seq libraries for the controls mentioned above: four adult fly lines (F1 CyO, F1 TM3, F1 CyO;TM3, and F1 female, each in two biological replicates) sequenced on the Illumina HiSeq2000 (100 bp paired-end) or NextSeq (150 bp paired-end). An overview of the sequencing depth is provided in Table S4.
Differential RNA-seq analysis
The RNA-seq data was mapped to the Drosophila melanogaster reference genome dm6, and the reads separated into the haplotypes based on overlap with haplotype-tagging SNVs. A custom script based on pysam (https://github.com/pysam-developers/pysam) detects such overlaps and integrates information across all SNVs within a read pair. This way, read pairs with conflicting information (i.e. containing both balancer- and wild-type-tagging SNVs) can be filtered; this fraction was below 0.2% of read pairs in all cases. In the embryonic RNA-seq data set (2x144 bp reads after demultiplexing), ~28% of read pairs could be assigned to haplotypes (see Table S4). We then counted fragments per haplotype per gene using HTSeq-counts53.
We tested genes for differential expression by inserting these haplotype-specific counts for all four replicates (2x N1 pat, 2x N1 mat) into a matrix and supplying it to DESeq254. Our experimental design, i.e. examining RNA-seq from embryos obtained from the reciprocal crosses, takes care of any potential influences of maternally deposited mRNAs. DESeq2 could thereby test for differential expression against a 25% ratio, with the multiple replicates (from the reciprocal crosses). Genes were filtered for a minimum number of reads (average of 50 fragments from both haplotypes per gene per sample) and by chromosome (only chromosomes 2 and 3 considered). The resulting p-values were corrected using fdrtool55, identifying 512 differentially expressed genes (two-sided Wald test, 5% FDR) and 4,845 non-differentially expressed genes. The remaining 9,053 genes were not tested for differential expression due to low number of reads, but were further divided into 4,989 lowly expressed separable genes (for which at least one RNA-seq read could be haplotype-separated) and 4,064 unseparable genes. If the former, lowly expressed genes, did change in their expression (i.e. were activated in the balancer) due to rearrangements, we would be able to detect them.
To assess whether the 512 differentially expressed genes are enriched in Gene Ontology terms, we compared them to all 5,357 testable expressed genes using the PANTHER Overrepresentation Test (http://pantherdb.org/tools/compareToRefList.jsp). We found no significant enrichment (Bonferroni-corrected p-value < 0.05, Fisher's exact test) for any of the annotation datasets used: PANTHER GO-Slim Molecular Function, Biological Process, or Cellular Component, indicating that the differentially expressed genes have heterogenous functions.
Controlling for trans effects in the RNA-seq data
To control for potential trans effects (Supplementary Fig. 3a,b), we generated three F1 generations of adult flies with different genotypes: one line with only chromosome 2 balanced (CyO), one with only chromosome 3 balanced (TM3), and one carrying both balancer chromosomes (double balancer, or normal F1 cross; see also Methods/”Fly crosses for RNA-seq experiments”). Note that in this design, allele separation is only feasible on the balanced chromosomes themselves. Apart from two haplotypes (balancer or wild type), we also considered two balancer genotypes (single or double-balancer). We then tested the interaction term between haplotype and balancer genotype on gene expression, using the DESeq2 formula ~ Haplotype + Balancer.Genotype + Haplotype:Balancer.Genotype. For example, in the case of CyO, we asked if the differential expression (balancer vs. wild type) of a gene is significantly different when using the CyO cross (which does not contain TM3) compared to using the double balancer cross (which contains TM3). Biological noise was estimated by comparing the new F1 double balancer cross (F1 CyO;TM3) to a (female) F1 female line with the same interaction approach, asking whether old vs. new sequencing data significantly changes differentially expressed genes. Here, due to lower sequencing depth, we reduced threshold for a minimum number of reads to 30 fragments.
Assessing the overlap of Copy Number Variants (CNVs) with genes
We required at least one exon of a gene to overlap a CNV (deletion or duplication). Additionally (with the exception of Fig. 2b and Supplementary Fig. 3c), for deletions in the wild type and duplications in the balancer, we required the gene to show an increased expression in the balancer comparing to the wild type. Conversely, for deletions in the balancer and duplications in the wild type, we required the gene to show a decreased expression in the balancer.
Capture-C primer design
As Capture-C viewpoints we used DpnII restriction fragments which satisfy the following criteria in the context of SNVs and small indels:
length ≥124 bp,
restriction sites (recognition sequences) at both ends are not disrupted in either of the haplotypes,
no restriction sites are created inside the fragment in either of the haplotypes.
We also imposed additional criteria on the two candidate 120 bp probes starting from the restriction sites and continuing into the restriction fragment. The following criteria had to be satisfied by both of the probes:
GC-content in the range of [0.25, 0.65],
≤20 repeat-masked basepairs,
no secondary BLAT alignment of score ≥10,
no more than 1 bp of allele-specific SNVs,
no allele-specific small indels.
We incorporated the shared SNVs and shared small indels into the probe sequences. We also resolved the possible allele-specific SNVs in a way that the probe has an allele different than the wild-type and different than the balancer, and no new restriction sites are created. All the viewpoints and probe sequences are given in Table S6.
In total, we designed 314 Capture-C viewpoints, manually assigned to 328 genes, namely: 226 differentially expressed genes, 69 non-differentially expressed genes, and 33 genes non-tested. Some of the viewpoints were assigned to more than one gene (Table S6). Seven of the viewpoints overlapped allele-specific SNVs, and were therefore excluded from any analysis. The remaining 307 Capture-C viewpoints are assigned to 321 genes, namely: 221 differentially expressed genes, 68 non-differentially expressed genes, and 32 genes non-tested.
Hi-C and Capture-C library preparation
Drosophila embryos were collected at 4-8 h after egg lay (stages 8-11) from a backcross of male double balancer flies (F1; i.e. +/CyO; +/TM3) with female F0 flies (+/+; +/+), i.e. N1 pat. Embryos were fixed and nuclei extracted as described previously56. For Hi-C, three aliquots of 30x106 nuclei in 2 biological replicates were used for each 3C template preparation. The Hi-C libraries were prepared as previously described9, using DpnII as restriction enzyme. The final library was prepared using the NEBNext Ultra DNA Library Prep Kit for Illumina according to the manufacturer’s instructions from at least 1 μg of DNA. The libraries were sequenced on the Illumina HiSeq2000 (100 bp paired-end).
The Capture-C libraries were prepared as previously described57, using DpnII as restriction enzyme and performing the ligation step “in nucleus”. The template was then processed with the Roche Nimblegen SeqCap EZ Library SR system and a double capture strategy. The libraries were sequenced on the Illumina NextSeq (150 bp paired-end).
Hi-C and Capture-C data processing
Hi-C sequencing reads were mapped onto the Drosophila melanogaster reference genome dm6, considering both reads from a read pair separately, using bwa mem48 with options -E50 -L0 -5. After merging the paired reads, we annotated them with haplotypes based on SNVs. Using pairsamtools (https://github.com/mirnylab/pairsamtools), we formed Hi-C pairs in pairsam format, selected only linear-linear (LL) or rescued chimeric-linear (CX) pair types, and sorted and de-duplicated them. Reads were then separated according to their haplotype, removing supplementary alignments, splitting read pairs into separate BAM files, and filtering and processing them using HiCExplorer58 using hicBuildMatrix with options --restrictionSequence GATC --danglingSequence GATC --binSize 5000 --skipDuplicationCheck. The resulting contact matrices were summed across two biological replicates and normalized by iterative correction using hicCorrectMatrix with options --filterThreshold -1.5 5, taking only chromosomes 2, 3, 4, X and Y.
To obtain balancer contact maps in the balancer genome assembly, we relied on the separate BAM files obtained by splitting read pairs. We used CrossMap59 with a custom chain file to convert them from the reference assembly to the balancer assembly. We then processed them using HiCExplorer as described above.
Capture-C sequencing reads were processed in the same manner, up to and including filtering them using HiCExplorer. We further processed the filtered reads using CHiCAGO60 to obtain contact count tracks in restriction fragment resolution for each replicate, haplotype and viewpoint.
Topologically Associated Domain (TAD) calling
To call TADs from Hi-C data, we used an approach based on the TAD separation score, using hicFindTADs from HiCExplorer with default options. We calculated TAD separation score profiles for 7 different window sizes, ranging from 50 kb to 195 kb. The TAD-separation score is calculated as the average Z-score of all Hi-C contacts between an adjacent window upstream and an adjacent window downstream. We then averaged these profiles into an aggregate insulation score (IS) profile. To ensure that the profile incorporates the proper genomic context around rearrangement junctions, we used the balancer genome assembly to calculate the IS profile for balancer chromosomes, and the reference assembly for wild-type chromosomes. We then converted the balancer profile to the reference assembly.
To compare the insulation score profiles between replicates and haplotypes (Supplementary Fig. 4a), we processed the Hi-C data from two biological replicates separately, and for each replicate down-sampled the wild-type haplotype reads to match the number of reads of the balancer haplotype.
Differential TAD analysis
As described above, from the wild-type Hi-C contact map we identified 767 wild-type TAD boundaries (separating 771 TADs) in chromosomes 2 and 3. In the same manner we identified 757 balancer TAD boundaries (separating 761 TADs) in these two chromosomes from the balancer Hi-C contact map in the balancer genome assembly. We then converted the balancer TAD boundaries to the reference genome assembly to compare these two sets of boundaries. To account for the difficulty in accurately calling TAD boundaries, we compared them against boundaries called with a less stringent p-value threshold (using hicFindTADs with option --thresholdComparisons 0.1) and allowed the boundaries to be shifted by up to 25 kb between haplotypes. This identified 671 matched TAD boundaries, 96 wild-type-specific ones and 86 balancer-specific ones.
In addition to allele-specific TAD boundaries, inversion breakpoints on the balancer chromosomes also shuffle TADs, causing them to be resized (Fig. 3h). Out of the 16 disrupted TADs, 5 also have an allele-specific boundary on one side: three balancer-specific (change of boundary up to 136 kb), and two wild-type-specific (change up to 65 kb). While analysing these TADs, we considered their wild-type boundaries and the set of 161 genes with TSSs located within these boundaries. In the balancer, 15 of the considered genes (2 of which are differentially expressed genes) are across a balancer-specific boundary (outside the disrupted TAD), while 8 genes (2 of which are differentially expressed genes) located within the disrupted TADs were not considered due to being located across a wild-type-specific boundary in the wild type haplotype.
Differential Hi-C contact analysis
Differential Hi-C analysis was performed on contact maps before normalization and before summing biological replicates. For each replicate and haplotype, we fitted a distance-decay trend using the R package locfit61 and provided the fitted values as normalization factors in DESeq254. We compared the haplotypes for all 453,423 Hi-C contacts within 100 kb which were not separated by large nested inversions. Contacts with low average count numbers were excluded in the independent filtering procedure, leaving 216,066 Hi-C contacts to be tested. Out of those, 5,297 were differential (two-sided Wald test, 10% FDR). We excluded differential contacts that had at least 1 kb overlap with CNVs at either of the interacting 5 kb bins, leaving 4,329 high-confidence differential Hi-C contacts. Of those, 4,299 had an absolute fold change >1.5 (i.e. fold change >1.5 or <⅔, equivalent to absolute log2 fold change > log2(1.5)) between the two haplotypes.
Differential Capture-C contact analysis
The reference genome dm6 was in silico digested into restriction fragments using sequence GATC recognized by the restriction enzyme DpnII. The restriction fragments were further filtered in the context of SNVs, small indels and structural variants, removing:
restriction fragments having a disrupted restriction site (different sequence than the recognition sequence) at either end in either of the haplotypes,
restriction fragments having an additional restriction site created in either of the haplotypes.
We considered jointly the Capture-C interactions originating from the 307 viewpoints that did not overlap structural variants. We compared the haplotypes for all 561,958 Capture-C contacts within 100 kb from the viewpoint, which were not separated by large nested inversions. Contacts with low average count numbers were excluded in the independent filtering procedure, leaving 59,605 Capture-C contacts to be tested. Out of those, 984 were differential (two-sided Wald test, 5% FDR). We excluded differential contacts to restriction fragments whose restriction site overlaps a deletion or duplication (but allowed smaller deletions and duplications that were fully contained between the restriction sites). This resulted in 935 differential Capture-C contacts, of which we used 931 that had an absolute fold change >1.5.
Correlating differential contacts and differential gene expression
We aligned and oriented the genes by their 5’-most transcription start site, and considered the differential Hi-C contacts between the TSS and the surrounding genomic region. When considering Capture-C data viewpoints associated to multiple genes, we took them into account multiple times, each time in association with a different gene. Averaging within differentially expressed genes and non-differentially expressed genes separately, the average number of differential contacts from the TSS to a given relative location was plotted.
Genomic features overlap
We used DNase hypersensitive sites (DHS) at the same stages of embryogenesis from62, merging the peaks from stages 9-11 and converting the resulting dataset to the reference genome dm6. Promoters were defined by taking all annotated TSS from FlyBase release FB2015_02 and extending them by ±1 kb. Enhancers were combined from CAD63 and mesoderm CRMs64 converted to dm6. Distal enhancers were defined as DHS overlapping an enhancer and not overlapping any promoters.
For the purpose of overlapping with other genomic features (Supplementary Fig. 5c), differential Hi-C contacts between two genomic bins were contributing twice, with each of the bins serving as either viewpoint or the other end. For the same purpose, differential Capture-C contacts were reduced into 445 clusters, by extending the interacting restriction fragments by ±1 kb and merging the overlapping ones for each viewpoint. Permutation tests were performed by keeping the viewpoints fixed, and randomly shuffling the distances to the other ends of the interactions. The sign of these distances was also randomly flipped. The 95% confidence intervals and p-value were derived from the overlaps obtained for 1,000 random shuffles.
Statistics
We performed differential gene expression, differential Hi-C and differential Capture-C analyses using the two-sided Wald test integrated in DESeq254. For differential gene expression, we corrected the resulting p-values by re-estimating the variance of the null model using fdrtool55. All the other statistical tests are discussed in the context of the analysis for which they were applied, in the corresponding Methods subsections above.
Reporting summary
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
Supplementary Material
Editorial summary.
Systematic analysis of highly rearranged balancer chromosomes in Drosophila shows that only a restricted subset of genes change expression in response to extensive topological changes.
Acknowledgments
We thank all members of the Furlong lab for discussions and comments on the manuscript. We thank Matthew Davis, Hilary Gustafson, David Garfield, Tobias Rausch and Sebastian Waszak for useful discussions and suggestions at the various stages of the project. This work was technically supported by the EMBL Genomics Core facility, with specific thanks to Rajna Hercog for WGS library preparation. This work was financially supported by a FRM grant (AJE20161236686) to Y.G.-H., EMBL International Ph.D. Programme to S.M, an EU Horizon 2020 Marie Skłodowska-Curie grant (708111) to A.J., ERC Starting Grant (336045) to J.O.K and ERC advanced grant DeCRyPT (787611) to E.E.F.
Footnotes
Author Contributions
Y.G.-H. and E.E.M.F. designed the study. Y.G.-H., A.J., S.M., J.K. and E.E.M.F. analyzed the results. Y.G.-H., A.J., S.M. and E.E.M.F. wrote the manuscript. Y.G.-H. performed all experiments, except the mate pair library performed by R.R.V. S.M. performed SNV and structural variant calling and RNA-seq analysis. A.J. performed Hi-C and Capture-C data analysis. Y.G.-H., A.J. and S.M. contributed equally to the study. All authors discussed the results and commented on the manuscript.
Declaration of Interests
The authors declare no competing financial interests.
Data Availability
All raw data, which consists of 75 demultiplexed files, were submitted to ArrayExpress (https://www.ebi.ac.uk/arrayexpress/browse.html) under accession numbers: E-MTAB-7510 (whole-genome and mate-pair sequencing), E-MTAB-7512 (Hi-C), E-MTAB-7513 (Capture-C), E-MTAB-7511 (RNA-seq). The Hi-C contact maps, RNA-seq read counts and other processed data are available on the Furlong lab web page, http://furlonglab.embl.de/data. Custom code used for the analysis is available at https://github.com/ajank/balancer-paper.
References
- 1.Bulger M, Groudine M. Functional and Mechanistic Diversity of Distal Transcription Enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine M. Transcriptional Enhancers in Animal Development and Evolution. Curr Biol. 2010;20:R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 4.Furlong EEM, Levine M. Developmental enhancers and chromosome topology. Science. 2018;361:1341–1345. doi: 10.1126/science.aau0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao SSP, et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sexton T, et al. Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Lupiáñez DG, et al. Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franke M, et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature. 2016;538:265–269. doi: 10.1038/nature19800. [DOI] [PubMed] [Google Scholar]
- 12.Tsujimura T, et al. A Discrete Transition Zone Organizes the Topological and Regulatory Autonomy of the Adjacent Tfap2c and Bmp7 Genes. PLOS Genet. 2015;11:e1004897. doi: 10.1371/journal.pgen.1004897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narendra V, et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347:1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lettice LA, et al. Enhancer-adoption as a mechanism of human developmental disease. Hum Mutat. 2011;32:1492–1499. doi: 10.1002/humu.21615. [DOI] [PubMed] [Google Scholar]
- 16.Northcott PA, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flavahan WA, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hnisz D, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Northcott PA, et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547:311–317. doi: 10.1038/nature22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weischenfeldt J, et al. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat Genet. 2017;49:65–74. doi: 10.1038/ng.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Y, et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science. 2017;355:eaaf4791. doi: 10.1126/science.aaf4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao Y, et al. Creating a functional single-chromosome yeast. Nature. 2018;560:331. doi: 10.1038/s41586-018-0382-x. [DOI] [PubMed] [Google Scholar]
- 23.Lee H, et al. Effects of Gene Dose, Chromatin, and Network Topology on Expression in Drosophila melanogaster. PLOS Genet. 2016;12:e1006295. doi: 10.1371/journal.pgen.1006295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meadows LA, Chan YS, Roote J, Russell S. Neighbourhood Continuity Is Not Required for Correct Testis Gene Expression in Drosophila. PLOS Biol. 2010;8:e1000552. doi: 10.1371/journal.pbio.1000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodríguez-Carballo E, et al. The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev. 2017;31:2264–2281. doi: 10.1101/gad.307769.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nora EP, et al. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 2017;169:930–944.e22. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Splinter E, et al. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao SSP, et al. Cohesin Loss Eliminates All Loop Domains. Cell. 2017;171:305–320.e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarzer W, et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 2017;551:51–56. doi: 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wutz G, et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 2017;36:3573–3599. doi: 10.15252/embj.201798004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oster II. A new crossing-over supressor in chromosome 2 effective in the presence of heterologous inversions. DIS. 1956;30:145. [Google Scholar]
- 32.Tinderholt V. New mutants report. DIS. 1960;34:53–54. [Google Scholar]
- 33.Ashburner M, Golic KG, Hawley RS. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- 34.Korbel JO, et al. Paired-End Mapping Reveals Extensive Structural Variation in the Human Genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet. 2013;14:125–138. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- 36.Mackay TFC, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zichner T, et al. Impact of genomic structural variation in Drosophila melanogaster based on population-scale sequencing. Genome Res. 2013;23:568–579. doi: 10.1101/gr.142646.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindsley DL, Zimm GG. The Genome of Drosophila Melanogaster. Academic Press; 1992. [Google Scholar]
- 39.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller DE, et al. The Molecular and Genetic Characterization of Second Chromosome Balancers in Drosophila melanogaster. G3 Genes Genomes Genet. 2018;8:1161–1171. doi: 10.1534/g3.118.200021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller DE, Cook KR, Arvanitakis AV, Hawley RS. Third Chromosome Balancer Inversions Disrupt Protein-Coding Genes and Influence Distal Recombination Events in Drosophila melanogaster. G3 Genes Genomes Genet. 2016;6:1959–1967. doi: 10.1534/g3.116.029330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang W, et al. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 2014;24:1193–1208. doi: 10.1101/gr.171546.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas S, et al. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biol. 2011;12:R43. doi: 10.1186/gb-2011-12-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramírez F, et al. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun. 2018;9:189. doi: 10.1038/s41467-017-02525-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogiyama Y, Schuettengruber B, Papadopoulos GL, Chang J-M, Cavalli G. Polycomb-Dependent Chromatin Looping Contributes to Gene Silencing during Drosophila Development. Mol Cell. 2018;71:73–88.e5. doi: 10.1016/j.molcel.2018.05.032. [DOI] [PubMed] [Google Scholar]
- 46.Spielmann M, Lupiáñez DG, Mundlos S. Structural variation in the 3D genome. Nat Rev Genet. 2018;19:453–467. doi: 10.1038/s41576-018-0007-0. [DOI] [PubMed] [Google Scholar]
- 47.Mardin BR, et al. A cell-based model system links chromothripsis with hyperploidy. Mol Syst Biol. 2015;11:828. doi: 10.15252/msb.20156505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv13033997 Q-Bio. 2013 [Google Scholar]
- 49.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. ArXiv12073907 Q-Bio. 2012 [Google Scholar]
- 50.Tan A, Abecasis GR, Kang HM. Unified representation of genetic variants. Bioinformatics. 2015;31:2202–2204. doi: 10.1093/bioinformatics/btv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rausch T, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Untergasser A, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- 56.Bonn S, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 57.Davies JOJ, et al. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat Methods. 2016;13:74–80. doi: 10.1038/nmeth.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramírez F, et al. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun. 2018;9:189. doi: 10.1038/s41467-017-02525-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao H, et al. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinforma Oxf Engl. 2014;30:1006–1007. doi: 10.1093/bioinformatics/btt730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cairns J, et al. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 2016;17:127. doi: 10.1186/s13059-016-0992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loader C. locfit: Local Regression, Likelihood and Density Estimation. 2013.
- 62.Thomas S, et al. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biol. 2011;12:R43. doi: 10.1186/gb-2011-12-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cusanovich DA, et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature. 2018;555:538–542. doi: 10.1038/nature25981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EEM. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data, which consists of 75 demultiplexed files, were submitted to ArrayExpress (https://www.ebi.ac.uk/arrayexpress/browse.html) under accession numbers: E-MTAB-7510 (whole-genome and mate-pair sequencing), E-MTAB-7512 (Hi-C), E-MTAB-7513 (Capture-C), E-MTAB-7511 (RNA-seq). The Hi-C contact maps, RNA-seq read counts and other processed data are available on the Furlong lab web page, http://furlonglab.embl.de/data. Custom code used for the analysis is available at https://github.com/ajank/balancer-paper.