Abstract
Although mouse infection models have been extensively used to study the host response to Mycobacterium tuberculosis, their validity in revealing determinants of human TB resistance and disease progression has been heavily debated. Here, we show that the modular transcriptional signature in the blood of susceptible mice infected with a clinical isolate of M. tuberculosis resembles that of active human tuberculosis disease, with a dominance of a type I IFN response and neutrophil activation and recruitment, together with a loss in B lymphocyte, NK and T cell effector responses. In addition, resistant but not susceptible strains of mice show increased lung B, NK and T cell effector responses in the lung upon infection. Importantly, the blood signature of active disease shared by mice and humans is also evident in latent tuberculosis progressors before diagnosis suggesting that these responses both predict and contribute to the pathogenesis of progressive M. tuberculosis infection.
Tuberculosis (TB) results in over 1.3 million deaths annually1, yet most individuals infected with M. tuberculosis remain asymptomatic. Latent TB infection (LTBI) is defined by an interferon-γ (IFN-γ)-release assay (IGRA) specific for M. tuberculosis antigens, although some patients may have subclinical disease and may progress to active TB2. Protective immune responses against M. tuberculosis include CD4+ T lymphocytes and the cytokines IL-12, IFN-γ, TNF3–6, and IL-17, but these factors do not explain why most individuals control infection, whereas a subset go on to develop active TB. A blood transcriptional signature in active TB patients has implicated type I IFN in TB pathogenesis8–16. Immunological heterogeneity in the blood transcriptome of a cohort of recent TB contacts has been observed, with a small proportion of contacts expressing a persistent blood TB signature and subsequently progressing to active disease (LTBI-progressors)16, suggesting a host response evolving towards active disease16.
How the immune response in blood8,15 reflects that occurring at disease sites is poorly understood, and sampling the latter in humans is prohibitive. The mouse TB model, owing to the richness of genetic and immunological tools available, has been invaluable in defining immune responses in the lung influencing disease outcome after infection4,5,17. However, a global systematic analysis to determine potential common pathways of protection or pathogenesis in different TB mouse models and human disease has not been reported. A role for type I IFN in TB pathogenesis8–16 is supported by mouse TB models6,18 with elevated and sustained levels of type I IFN: (i) infection of particular genetic strains of mice with clinical isolates of M. tuberculosis 19–23; (ii) infection of hosts with genetic mutations in regulators of type I IFN such as Tpl224; IL-17 or ISG1525; (iii) administration of adjuvants, e.g. Poly(I)C7,26; or (iv) viral co-infection27. Whether it is the genetic strain of mouse or the M. tuberculosis pathogen itself which results in an immune response that most resembles human TB is unclear. Although the spectra of human28 and mouse29 TB disease do not completely overlap, comparison of human TB with genetically diverse backgrounds of mice has established points of similarity in their response to M. tuberculosis. Some mouse strains recapitulate key elements of the pathogenesis of human TB disease, at the level of induction of necrotic TB lesions in the lungs29. Whether the global immune response to M. tuberculosis in susceptible mouse strains resembles that of TB in humans is as yet unclear.
Here, we report that the human blood TB type I IFN-inducible signature16,8 is recapitulated in susceptible C3HeB/FeJ mice infected with different strains of M. tuberculosis. Increased expression of granulocyte-associated genes in blood from active TB patients, TB-susceptible mice and LTBI-progressors before TB diagnosis suggested their role in early disease pathogenesis. Conversely, under-abundance of B, NK and effector T cell-signatures in blood from human TB patients16, LTBI-progressors and TB-susceptible mice and yet over-abundance in lungs of M. tuberculosis infected C57BL/6J resistant mice reinforced their role in early disease control. The translationally relevant knowledge dataset presented here on potential pathways of protection and pathogenesis in human TB are easily accessible using an online ShinyApp : https://ogarra.shinyapps.io/tbtranscriptome/
Results
The peak transcriptomic response in M. tuberculosis infected mice
To determine if a mouse blood transcriptional TB signature resembles that of human disease, we tested the human blood modular transcriptional TB signature16 on RNA-Seq data from blood of different genetic inbred strains of mice, C57BL/6J (resistant) and C3HeB/FeJ (susceptible), infected with low and high doses of the M. tuberculosis laboratory strain H37Rv or clinical isolate HN87821,22 (Supplementary Fig.1a-c; Fig. 1, Supplementary Tables 1-3). The human blood TB signature16 was first tested on microarray data from blood of H37Rv infected BALB/c mice at different time-points post-infection, to establish the peak transcriptomic response, where immune signatures were barely detectable at days 14 and 21 post-infection, but most significant by day 138 (Supplementary Fig. 2a; Supplementary Table 3). Analysis of blood microarray data from an independent study30, showed that the blood signature in 129S2 and C57BL/6NCrl mice was again barely detectable at day 14 post H37Rv infection, being observed robustly by day 21, which was the end-point of that study30 (Supplementary Fig. 2a). Upon testing a lung disease modular signature31 on microarray data from lungs of H37Rv infected BALB/c mice, we detected a peak response at day 56 post-infection, only starting to be detected by 28 days post-infection (Supplementary Fig. 2b; Supplementary Table 4). Based on these data we tested the human blood transcriptional TB signature16 and lung disease modular signature31, on the blood and lungs, respectively, from C57BL/6J and C3HeB/FeJ mice infected with HN878, at days 26 to 56 post-infection (Supplementary Fig. 3a and b; Supplementary Tables 3 and 4). The peak response chosen was ca. 42 days post-infection, which best showed a robust signature in blood and lungs from HN878 infected C57BL/6J and C3HeB/FeJ mice (Supplementary Fig. 3a and b; Supplementary Tables 3 and 4; tissues from HN878 infected susceptible mice were harvested after 33-35 days post infection due to excessive pathology).
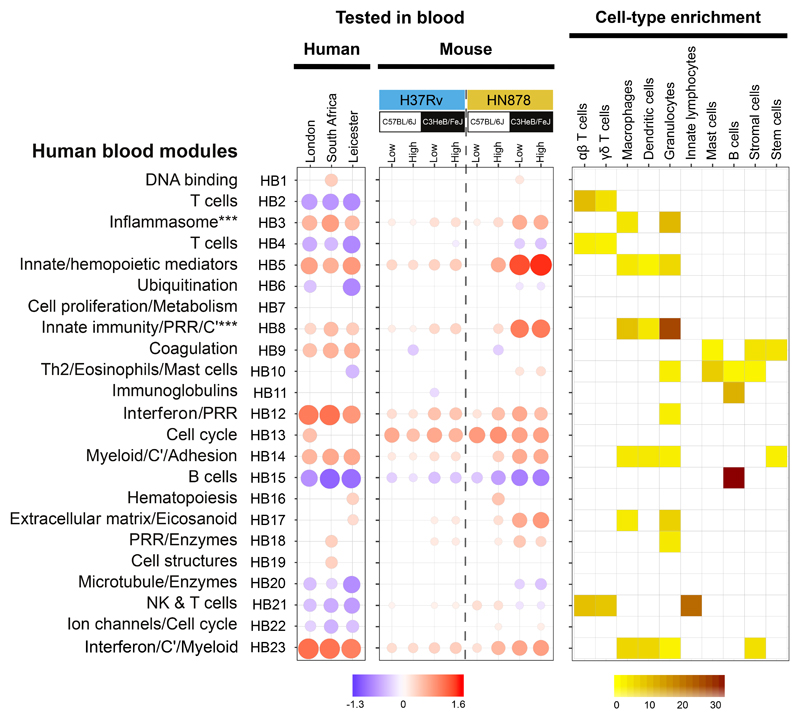
Figure 1. Human TB blood transcriptional signature is preserved in blood of TB susceptible mice.
Blood modules of co-expressed genes derived using WGCNA from human TB datasets in Singhania et al. 201818 are shown for blood RNA-seq datasets from TB patients from London (n=21 biologically independent samples), South Africa (n=16 biologically independent samples) (both compared to London controls; n=12 biologically independent samples) and Leicester (n=53 biologically independent samples) (compared to Leicester controls; n=50 biologically independent samples) (Supplementary Table 2); human blood modules were tested in blood RNA-seq datasets obtained from different genetic strains of mice (C57BL/6J, resistant; C3HeB/FeJ, susceptible) infected with low and high doses of M. tuberculosis laboratory strain H37Rv or the M. tuberculosis clinical isolate HN878 (n=4 biologically independent samples per group for H37Rv infection and n=5 biologically independent samples per group for HN878 infection from one experiment per M. tuberculosis infection as depicted in Supplementary Fig. 1a), compared to their respective uninfected controls (Supplementary Table 3). Fold enrichment scores derived using QuSAGE are depicted, with red and blue indicating modules over- or under-abundant, compared to the controls. Colour intensity of the dots represents the degree of perturbation, indicated by the colour scale. Size of the dots represents the relative degree of perturbation, with the largest dot representing the highest degree of perturbation within the plot. Only modules with fold enrichment scores with FDR p-value < 0.05 were considered significant and depicted here (left and middle panels). Module name indicates biological processes associated with the genes within the module (Supplementary Table 1). C’, complement. PRR, pathogen recognition receptor. Cell-type associated with genes within each module were identified using the mouse cell-type-specific signatures from Singhania et al. 201941 (right panel). Cell-type enrichment was calculated using a hypergeometric test, with only FDR p-value < 0.05 considered significant and depicted here (right panel). Colour intensity represents significance of enrichment..
Blood transcriptional TB signature in mouse and humans
Principal component analysis (PCA) at the peak response depicted distinct global transcriptional signatures in blood of C57BL/6J (resistant) and C3HeB/FeJ (susceptible) mice, infected with low and high doses of H37Rv or HN878, with the largest distance from uninfected mice observed in HN878 infected C3HeB/FeJ mice (Supplementary Fig. 1c). The human blood modular transcriptional TB signature16 was recapitulated in blood of HN878 infected C3HeB/FeJ mice, and high dose HN878 infected C57BL/6J mice (Fig. 1; for annotation see Supplementary Table 1; for genes see Supplementary Tables 2 and 3). Two over-abundant (red) IFN-inducible modules (HB12 and HB23) in blood from TB patients16, showed a graded increase from the C57BL/6J to the C3HeB/FeJ mice infected with low to high dose H37Rv, further increased in C57BL/6J then C3HeB/FeJ mice infected with low to high dose HN878 (Fig. 1). Expression levels of IFN-inducible modules (HB12 and HB23), in blood of HN878 infected C3HeB/FeJ mice most closely resembled the profile in human TB (Fig. 1). Likewise, other over-abundant modules of human TB, including Inflammasome (HB3), Innate/hemopoietic mediators (HB5), Innate immunity/PRR/C’ (HB8) and Myeloid/C’/Adhesion (HB14) modules, were over-abundant in the HN878 infected C3HeB/FeJ mice, and to a lesser extent in high dose HN878 infected C57BL/6J mice (Fig. 1). Under-abundance (blue) of the human TB modules, T cell (HB4) and B cell (HB15)16, was recapitulated in the blood of HN878 infected C3HeB/FeJ mice (Fig. 1). In keeping with this, cellular deconvolution analyses31 of blood RNA-seq data from M. tuberculosis infected mice showed a significant decrease in the percentages of B cell and CD4+ T cell fractions (Supplementary Fig. 1d).
Cell-types associated with each module were identified by comparing cell-type specific gene signatures using the mouse RNA-Seq dataset from ImmGen Ultra Low Input (ULI) (ImmGen Consortium - GSE109125; http://www.immgen.org) as described31, analysed against the mouse gene orthologues within each human blood TB module (Fig. 1, right panel). The cell-type specific enrichment data validated the modular annotation for the blood T cell (HB2, HB4) modules, with enrichment for αβ- and γδ-T cells; the NK and T cell (HB21) module, with enrichment for αβ- and γδ-T cells and innate lymphocytes; and the B cell (HB15) module with enrichment for B cells (Fig. 1). This approach also led to the discovery of previously unappreciated gene signatures, most strikingly, a dominance of granulocyte-associated genes within the Inflammasome (HB3) and Innate immunity/PRR/C’ (HB8) modules (Fig. 1). This set of granulocyte-associated genes was highly expressed in blood from HN878 infected C3HeB/FeJ mice and human TB cohorts (Table 1; Supplementary Table 3 Mouse; Supplementary Table 2 Human). Increased expression of granulocyte-associated genes in blood of HN878 infected C3HeB/FeJ mice was reinforced by data obtained from cellular deconvolution analyses31(Supplementary Fig. 1d).
Table 1.
Granulocyte associated genes within the Inflammasome (HB3, left) and the Innate immunity/PRR/C’ (HB8, right) modules that are over-expressed in the blood of TB patient cohorts from London, South Africa and Leicester, compared to healthy controls (58 out of 87, and 53 out of 92 Granulocyte associated genes, respectively).
| (HB3) Inflammasome*** | |||
|
| |||
| ABCA13 | DMXL2 | KIF1B | NTNG2 |
| AIF1 | EVI2A | LCN2 | OSM |
| APOBR | FAS | LILRB5 | PLA2G4A |
| ASPRV1 | FCGR3B | LPCAT2 | PPP1R3D |
| ATP8B4 | FGL2 | LTF | PRTN3 |
| CAMP | GAS7 | LY96 | RNASEL |
| CASP4 | GPR141 | MARCKS | S100A6 |
| CCR1 | GSN | MCEMP1 | SELL |
| CD177 | HIST1H2BC | MCTP1 | SIGLEC9 |
| CD300A | HP | MEFV | SOCS3 |
| CKAP4 | IL18RAP | MILR1 | TFEC |
| CLEC4A | IL1B | MMP8 | TLR5 |
| CLEC4D | IRAK3 | NAIP | VCAN |
| CLEC4E | KCNJ2 | NCF1 | |
| CLEC5A | KCTD12 | NOD2 | |
|
| |||
| (HB8) Innate immunity/PRR/C'*** | |||
|
| |||
| ACSL1 | FUT7 | MXD1 | REPS2 |
| ALOX5AP | GAB2 | MYBPC3 | RNF19B |
| ANXA3 | GLIPR2 | NCF2 | RRAGD |
| AQP9 | HCAR2 | P2RX1 | S100A11 |
| ARL11 | HRH2 | P2RY13 | S100A8 |
| BCL6 | IFNGR2 | PADI4 | S100A9 |
| BMX | IGSF6 | PGLYRP1 | SIPA1L2 |
| BST1 | ITGAM | PLBD1 | SLC22A4 |
| C1RL | JAML | PPP1R3B | STEAP4 |
| C5AR1 | LITAF | PTPRJ | TIMP2 |
| CHST15 | LRG1 | PYGL | TLR2 |
| CRISPLD2 | LYN | RAB31 | TLR6 |
| ENTPD1 | MMP9 | RALB | |
| FOSL2 | MNDA | RBM47 | |
Host and M. tuberculosis genetic differences drive lung TB signatures
To determine the transcriptional response at the site of infection, RNA-Seq data was obtained from the lungs of the same C57BL/6J and C3HeB/FeJ inbred strains of mice infected with H37Rv or HN878, used for the blood data from Fig. 1 (Supplementary Fig. 1a). PCA depicted distinct global transcriptional signatures for uninfected mice and the different strains of H37Rv or HN878 infected mice, with the largest distance from uninfected controls observed in HN878 infected C3HeB/FeJ mice (Supplementary Fig. 4). The lung transcriptional response depicted a similar but more accentuated difference between the infected and uninfected groups than in blood (Supplementary Fig. 1c and Supplementary Fig. 4).
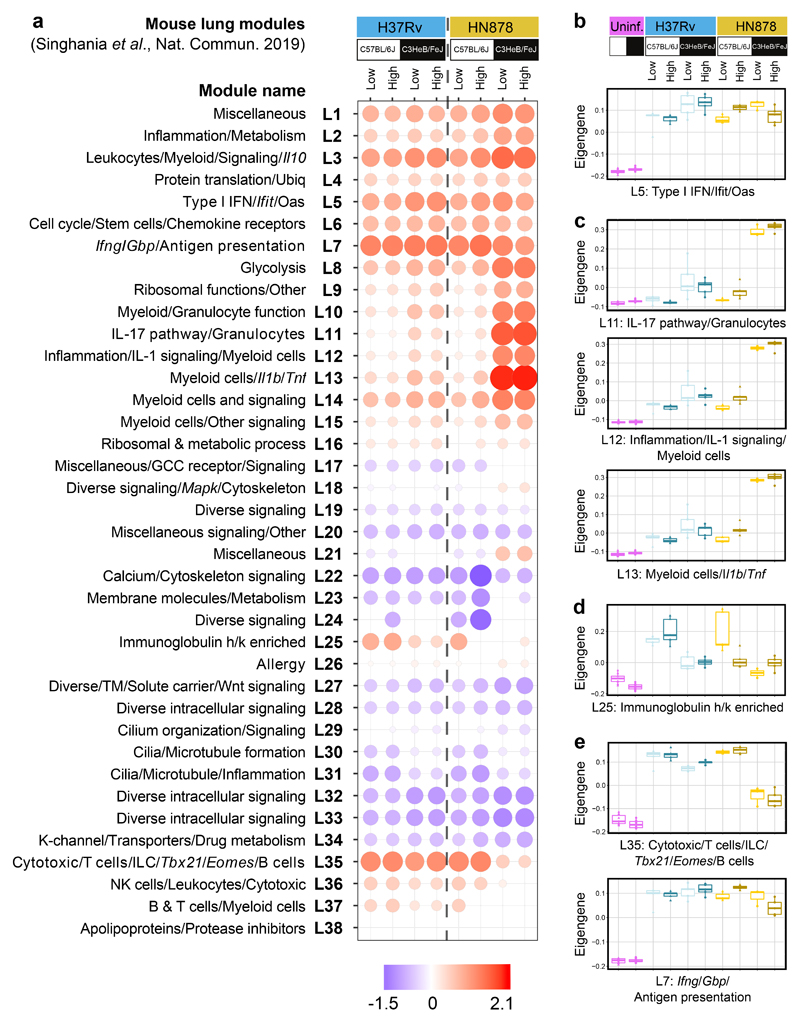
A lung disease modular signature31 was tested on the lung RNA-Seq data from the different groups of infected mice, to identify co-expressed groups of genes across the lung (Fig. 2). The type I IFN/Ifit/Oas (L5) module was over-abundant in the lungs of H37Rv and HN878 infected C57BL/6J and C3HeB/FeJ mice to similar levels, as shown by Eigengene expression (Fig. 2a and b). Six modules (L10 – L15), dominated by an over-abundance of granulocyte, macrophage and myeloid specific genes, including modules with Myeloid/Granulocyte (L10) and IL-17 pathway/Granulocytes (L11) function, showed the highest Eigengene expression in the HN878 infected C3HeB/FeJ mice (Fig. 2a and c; ShinyApp; Supplementary Table 4). Similarly, the Inflammation/IL-1 signalling/Myeloid Cells (L12), Myeloid cells/Il1b/Tnf (L13) and Myeloid cells/Other signalling (L14) modules were also over-abundant in mouse lungs upon M. tuberculosis infection, particularly in the lungs of susceptible HN878 infected C3HeB/FeJ mice (Fig. 2; Supplementary Table 4). Strikingly, an Immunoglobulin h/k module (L25) was over-abundant in the lungs of the C57BL/6J but only minimally in C3HeB/FeJ mice infected with low and high doses of H37Rv, and in the lungs of low dose HN878 infected C57BL/6J mice (Fig. 2a and d; ShinyApp; Supplementary Table 4). However, this Immunoglobulin h/k (L25) module was not changed in the lungs of high dose HN878 infected C57BL/6J mice or C3HeB/FeJ mice (Fig. 2a and d), correlating with these mice showing greater TB susceptibility (Supplementary Fig. 1b). This Immunoglobulin h/k (L25) module was also highly abundant in the lungs of BALB/c mice infected with low dose H37Rv, in keeping with its relatively resistant phenotype (Supplementary Fig. 2b). The Ifng/Gbp/Ag presentation/C’ (L7) and Cytotoxic/T cells/ILC/Tbx21/Eomes/B cells (L35) modules were over-abundant in the lung across both strains of H37Rv or HN878 infected mice (Fig. 2a and e; Supplementary Fig. 3b), and H37Rv infected BALB/c mice (Supplementary Fig. 2b), but less abundant in lungs from HN878 infected C3HeB/FeJ mice (Fig. 2a; Supplementary Fig. 3b), as shown quantitatively by Eigengene profiles (Fig. 2e).
Figure 2. Mouse lung disease modules tested in lungs from diverse mouse TB models.
a, Mouse lung disease modules derived in Singhania et al. 201941 (L1-L38) tested in mouse lung TB samples from different genetic strains of mice (C57BL/6J, resistant; C3HeB/FeJ, susceptible) infected with low and high doses of M. tuberculosis laboratory strain H37Rv or the M. tuberculosis clinical isolate HN878 (n=3 biologically independent samples per group for low dose HN878 infection of C3HeB/FeJ, and n=5 biologically independent samples per group for all other groups as depicted in Supplementary Fig. 1a), compared to their respective uninfected controls (Supplementary Table 4). Red and blue indicate modules over- or under-abundant, compared to the controls. Colour intensity of the dots represents the degree of perturbation, indicated by the colour scale. Size of the dots represents the relative degree of perturbation, with the largest dot representing the highest degree of perturbation within the plot. Only modules with fold enrichment scores with FDR p-value < 0.05 were considered significant and depicted here. GCC, glucocorticoid; K-channel, potassium channel; TM, transmembrane; Ubiq, ubiquitination. b-e, Box plots depicting the module eigengene expression, i.e. the first principal component for all genes within the module, are shown for uninfected (Uninf) and M. tuberculosis infected (Low dose; High dose) C57Bl/6 and C3HeB/FeJ mice, for modules (b) Type I IFN/Ifit/Oas (L5); (c) IL-17 pathway/granulocytes (L11), Inflammation/IL-1 signaling/Myeloid cells (L12), Myeloid cells/Il1b/Tnf (L13); (d) Immunoglobulin h/k enriched (L25); (e) Cytotoxic/T cells/ILC/Tbx21/Eomes/B cells (L35) and Ifng/Gbp/Antigen presentation (L7).
Independent derivation and annotation yielded similar transcriptional modules across all samples from uninfected and M. tuberculosis infected mice, resulting in 27 modules ((ML1 – ML27), Supplementary Fig. 5; Supplementary Tables 5 and 6). The type I IFN/Stat2/Mx1 (ML2) and type I IFN signalling (ML21) modules were similarly over-abundant in the lungs of H37Rv and HN878 infected C57BL/6J and C3HeB/FeJ mice (Supplementary Fig. 5a and b). Over-abundance of modules ML19 and ML27, enriched for Granulocyte/Macrophage specific genes, showed highest Eigengene expression in HN878 infected C3HeB/FeJ mice (Supplementary Fig. 5a and c), confirmed by cell-type specific enrichment analysis (Supplementary Fig. 5a). The Ifng/Gbp/Ag presentation/C’ (ML3) and T cell/NK/ILC/APC/B cell (ML11) modules were over-abundant in lungs from both strains of H37Rv or HN878 infected mice, although significantly less abundant in HN878 infected C3HeB/FeJ mice, as shown quantitatively by Eigengene profiles (Supplementary Fig. 5a and d), and validated by cell-type specific enrichment for T cells, DC, innate lymphocytes (ILC) and B cells (Supplementary Fig. 5a). Thus, two complementary and independently derived modular tools revealed similar transcriptional signatures in the lungs of M. tuberculosis infected susceptible mice, indicating increased type I IFN and granulocyte-associated responses and decreased IFN-γ, NK, T effector and B cell responses (Fig. 2 and Supplementary Fig. 5).
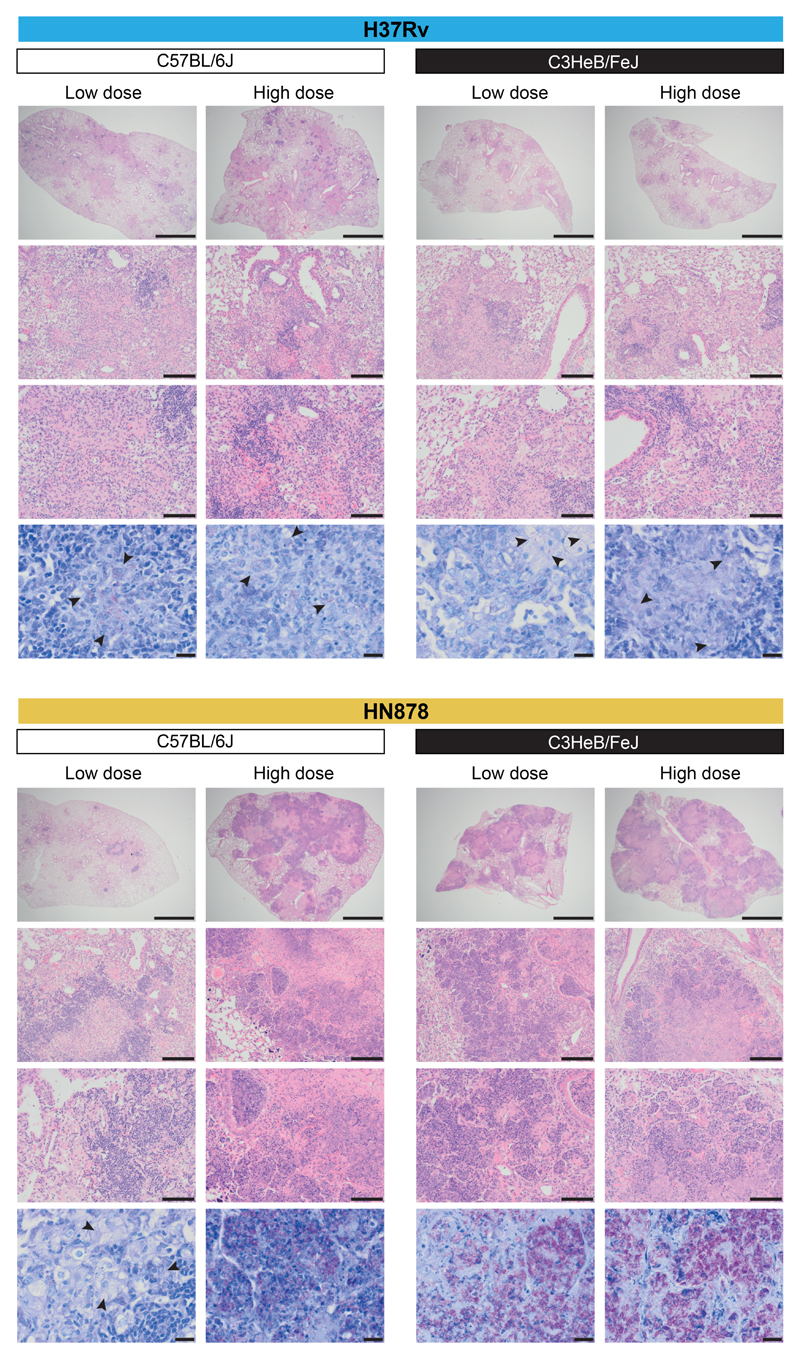
The over-abundance of inflammatory modules associated with granulocytes observed using the two independent modular approaches is in keeping with the more severe inflammation observed by H&E staining in the lungs of HN878 infected C3HeB/FeJ mice and high dose HN878 infected C57BL/6J mice (Fig. 3; Supplementary Fig. 6). This was accompanied by greater numbers of M. tuberculosis bacteria observed in the lungs of these mice by ZN staining (multibacillary infections, Fig. 3; Supplementary Fig. 6).
Figure 3. Histological analysis of mouse lungs from M. tuberculosis infected mice.
Representative photomicrographs of hematoxylin and eosin (H&E) or Ziehl–Neelsen (ZN) stained lung sections from different genetic strains of mice (C57BL/6J, resistant; C3HeB/FeJ, susceptible) infected with low and high doses of M. tuberculosis laboratory strain H37Rv or the M. tuberculosis clinical isolate HN878 (n=2 biologically independent samples per group for H37Rv infection, HN878-infected C57BL/6J mice low dose and HN878-infected C3HeB/FeJ mice high dose, and n=3 biologically independent samples per group for HN878-infected C57BL/6J mice high dose and HN878-infected C3HeB/FeJ mice low dose, from one experiment per M. tuberculosis infection). From top to bottom, scale bar represents 2 mm, 200 μm and 100 μm for H&E staining, 20 μm for ZN staining; arrows locate bacteria.
Degree of preservation of lung modules in human and mouse blood
It is unclear to what extent the airway transcriptional signature is reflected in the blood during M. tuberculosis infection. Certain immune responses across a range of experimental models of disease are well preserved between lung and blood, some not preserved, and others only discernible in blood with prior knowledge from the airway response31. To address this question in TB, the mouse lung modular TB signature (Supplementary Fig. 5a) was tested on the RNA blood samples from the different cohorts of human TB patients and from the different mouse TB models (Supplementary Fig. 7a). The mouse lung modules showed significant preservation in human and mouse blood as assessed by Zsummary scores, indicating the degree of preservation, with scores >10 considered strongly preserved (Supplementary Fig. 7b and c). Type I IFN associated modules (ML2 and ML21) (Supplementary Fig. 5a), were over-abundant in human and mouse blood, being most over-abundant in HN878 infected susceptible mice (Supplementary Fig. 7a). The lung type I IFN/Stat2/Mx1 module (ML2) was the most highly preserved module in human blood (Supplementary Fig. 7b) and the second-most preserved module in mouse blood (Supplementary Fig. 7c) and the type I IFN signalling module (ML21) stood out as the third-most preserved module in both human and mouse blood (Supplementary Fig. 7b and c). The lung Ifng/Gbp/Ag presentation/C’ module (ML3) was weakly over-abundant in the blood of human TB patients and M. tuberculosis infected mice (Supplementary Fig. 7a), albeit to a lesser extent, but highly preserved in both human and mouse blood (Supplementary Fig. 7b and c). The overall increased abundance of the Ifng/Gbp/Ag presentation/C’ module (ML3) was largely attributable to over-expression of genes such as GBP/Gbp genes and complement genes (Supplementary Fig. 7a; Supplementary Tables 2 and 3, and ShinyApp). However, the Ifng gene itself, although upregulated in the blood of M. tuberculosis infected resistant mice, was barely upregulated in the blood of HN878 infected susceptible mice and IFNG was down-regulated in the blood from TB patients (Supplementary Tables 2 and 3, and ShinyApp). The lung Macrophage/Granulocyte modules (ML19 and ML27) and Myeloid cell signalling module (ML10) were also over-abundant in blood of active TB patients and most over-abundant in the blood of HN878 infected susceptible mice (Supplementary Fig. 7a). While lung ML19 and ML10 modules were highly preserved in both human and mouse blood, the ML27 module was only highly preserved in mouse blood and to a much lesser extent in human (Supplementary Fig. 7b and c). Lung modules associated with T, NK and B cells (ML11 and ML13) were over-abundant in the lungs of all relatively resistantmice infected with H37Rv, but to a lesser extent (ML11) or under-abundant (ML13) in HN878 infected susceptible mouse lungs (Supplementary Fig. 5) and blood from all TB cohorts (Supplementary Fig. 7a), ML11 being highly preserved in both human and mouse blood (Supplementary Fig. 7b and c). These findings regarding the preservation of over or under-abundant lung modules in the blood from human TB patients and TB susceptible mouse models (Supplementary Fig. 7), are in keeping with the transcriptional signatures observed on testing human blood TB modules on blood from humans and mouse models of TB (Fig. 1).
Modular gene networks in human versus mouse TB
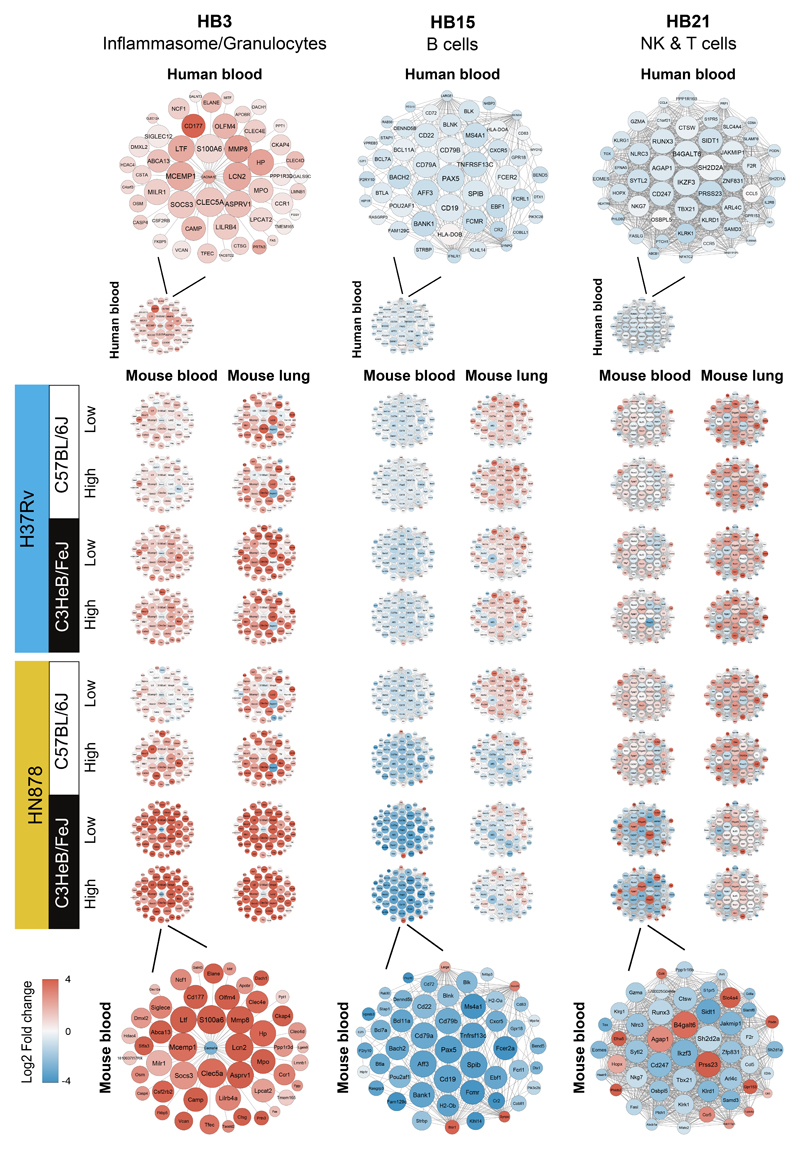
We further interrogated the changes in gene expression of the key modules, HB3, HB15 and HB21, between the blood and lungs of resistant and susceptible mice infected with the different strains of M. tuberculosis, as compared to the human blood. To do so, we examined the expression of top 50 “hub” genes with high intramodular connectivity within the mouse data, on human blood from TB patients, and blood and lungs from mice infected with M. tuberculosis (Fig. 4). In keeping with our current findings that granulocyte specific genes are upregulated within the originally named Inflammasome human blood TB module (HB3)16, granulocyte-specific genes were amongst the 50 “hub” genes within that module (now “Inflammasome/Granulocyte”) (Fig. 4). These granulocyte-specific genes include, Cd177, Elane, Mmp8, Mpo, Ncf1, Camp, Lcn2, S100a6, Ltf (Fig. 4, Supplementary Fig. 8a; ShinyApp), which have been associated with neutrophil recruitment and activation32, were most highly differentially expressed in blood from TB patients and M. tuberculosis infected susceptible mice. Expression of these genes in mouse blood and lungs revealed a graded increase from the C57BL/6J to the C3HeB/FeJ mice infected with low to high dose H37Rv, with a further increase observed in C57BL/6J to the C3HeB/FeJ mice infected with low to high dose HN878 (Fig. 4, Supplementary Fig. 8a). The 50 “hub” genes within Innate immunity/PRR/C’ module (HB8) also showed enrichment for granulocyte-specific genes including Mmp9, Alox5ap, Ncf2, Mxd1, S100a8 and S100a9, also associated with neutrophil activation (Supplementary Fig. 8b), and were most highly expressed in blood from human TB patients and blood and lung from HN878 infected C3HeB/FeJ mice (Fig. 4, Supplementary Fig. 8b; ShinyApp). Increased expression of these neutrophil-specific genes in the lungs of the TB susceptible HN878 infected mice was mirrored by the increased numbers of neutrophils detected in the lungs of these mice by immunohistochemistry (Fig. 5; Supplementary Fig. 6), confirming the H&E data (Fig. 3; Supplementary Fig. 6). Collectively these data support a major role for neutrophils in human TB pathogenesis, similar to the previously reported role for neutrophils in TB susceptible strains of mice33–35.
Figure 4. Gene networks of specific TB modules in human blood from TB patients, and blood and lung from M. tuberculosis infected mice.
Differential expression of genes from human blood modules Inflammasome/Granulocytes (HB3), B cells (HB15) and NK & T cells (HB21) depicting the top 50 “hub” network of genes with high intramodular connectivity found within the mouse data (i.e., mouse genes most connected with all other genes within the module), is shown for data from blood from TB patients (Leicester cohort), and blood and lungs from mice infected with M. tuberculosis, each against their respective controls. An enlarged representative network showing human gene names is shown for human blood (top) and an enlarged representative network showing mouse gene names is shown for blood samples from C3HeB/FeJ mice infected with high dose of HN878 (bottom). Each gene is represented as a circular node with edges representing correlation between the gene expression profiles of the two respective genes. Colour of the node represents log2 foldchange of the gene for human blood TB samples or mouse blood and lung samples from M. tuberculosis infected mice compared to respective controls.
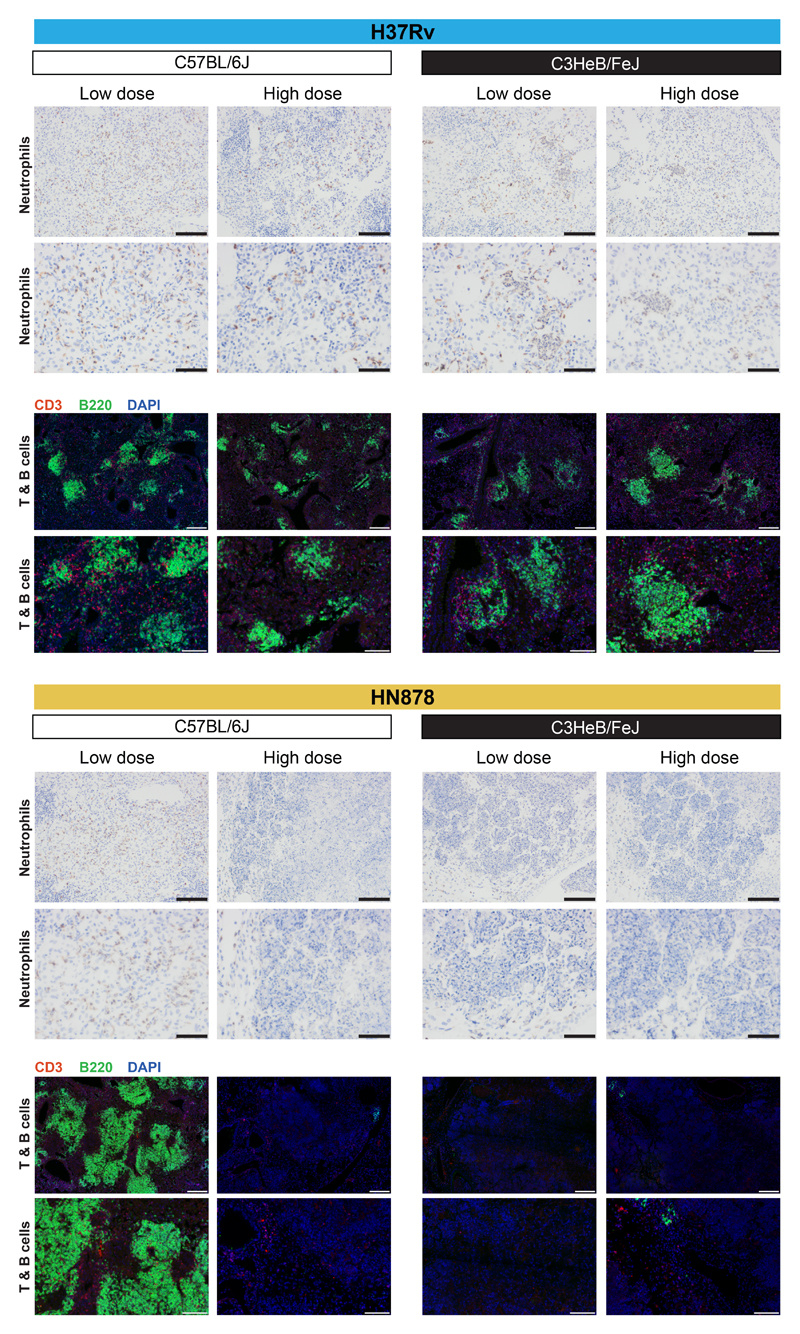
Figure 5. Histological analysis of mouse lungs from M. tuberculosis infected mice for neutrophils, T and B cells.
Representative photomicrographs of lung sections from different genetic strains of mice (C57Bl/6, resistant; C3HeB/FeJ, susceptible) infected with low and high doses of M. tuberculosis laboratory strain H37Rv or the M. tuberculosis clinical isolate HN878 (n=2 biologically independent samples per group for H37Rv infection, HN878-infected C57BL/6J mice low dose and HN878-infected C3HeB/FeJ mice high dose, and n=3 biologically independent samples per group for HN878-infected C57BL/6J mice high dose and HN878-infected C3HeB/FeJ mice low dose, from one experiment per M. tuberculosis infection) depicting neutrophils (2B10, brown) by immunohistochemistry or T (CD3 positive, red) and B (B220 positive, green) cells by immunofluorescence (nuclear staining depicted in blue, DAPI). Scale bar represents 100 μm (top) and 50 μm (bottom) for Neutrophils, 200 μm (top) and 100 μm (bottom) for T & B cells.
The 50 top “hub” genes within the human B cell module (HB15), Cd19, Pax5, Spib, Cd79 and Cd22, were down-regulated in the blood of human TB patients and M. tuberculosis mice (Fig. 4; Supplementary Fig. 8c; ShinyApp). Most of the B cell-specific top “hub” genes were upregulated in the lungs of H37Rv infected mice, but strikingly down-regulated in the lungs of high dose HN878 infected C57BL/6J and C3HeB/FeJ mice (Fig. 4; Supplementary Fig. 8c; ShinyApp). This difference in expression of B cell-specific genes between the lungs of relatively TB resistant and susceptible mouse models, was mirrored by differences in the numbers of B cells detected by B cell-specific immunofluorescent staining of lungs from these mice (Fig. 5; Supplementary Fig. 6). While vastly increased numbers of B cells were observed in the lungs of H37Rv infected mice, with accompanying formation of B cell follicles, these were practically absent in the lungs of C57BL/6J mice infected with high dose HN878 and HN878 infected C3HeB/FeJ mice (Fig. 5; Supplementary Fig. 6). These data support a possible role for B cells in protection against M. tuberculosis infection, as has previously been proposed36,37.
In keeping with the under-abundance of the human blood NK & T cells module (HB21), the top 50 “hub” genes in this module were down-regulated in the blood of patients with active TB (Fig. 4; Supplementary Fig. 8d), as previously reported16. Although upregulated in the blood and lungs of H37Rv infected C57BL/6J and C3HeB/FeJ mice and HN878 infected C57BL/6J mice, the majority of these 50 “hub” genes were down-regulated in the blood and either minimally or not upregulated in the lungs from HN878 infected C3HeB/FeJ mice (Fig. 4; Supplementary Fig. 8d). These included Tbx21, Gzma, Eomes, Cd8a, Nfatc2, Fasl, Nkg7, Klrd1, Klrg1, Ifng and Runx3, reflecting downregulation of effector T and NK cells in the blood of TB patients and HN878 infected C3HeB/FeJ mice (Fig. 4; Supplementary Fig. 8d; ShinyApp). Minimally altered gene expression was mirrored by a decrease in CD3+ T cells in HN878 infected C3HeB/FeJ mouse lungs as shown by immunofluorescence (Fig. 5; Supplementary Fig.6), reflecting an absence of activated effector T cells required for protection against M. tuberculosis infection4–6.
Heatmaps of the top 50 “hub” genes from the human blood TB modules Interferon/PRR (HB12) and Interferon/C’/Myeloid (HB23) demonstrated a large number of genes that were over-expressed in human blood from London and Leicester TB cohorts and were similarly over-expressed in mouse blood from HN878 infected C3HeB/FeJ mice (Supplementary Fig. 9). In contrast, many of these type I IFN-inducible genes in the HB12 module, including Il1rn, Ifit1, Ifit2, Oas2 and Stat2, were not upregulated, or upregulated to a lower extent, in the blood of H37Rv infected C57BL/6J mice, as compared to HN878 infected C3HeB/FeJ mice (Supplementary Fig. 9). The majority of the top 50 “hub” genes from the Interferon/PRR (HB12) and Interferon/C’/Myeloid (HB23) human modules were upregulated in the lungs of all the M. tuberculosis infected mice, with the highest expression observed in the lungs from HN878 infected C3HeB/FeJ mice (Supplementary Fig. 9).
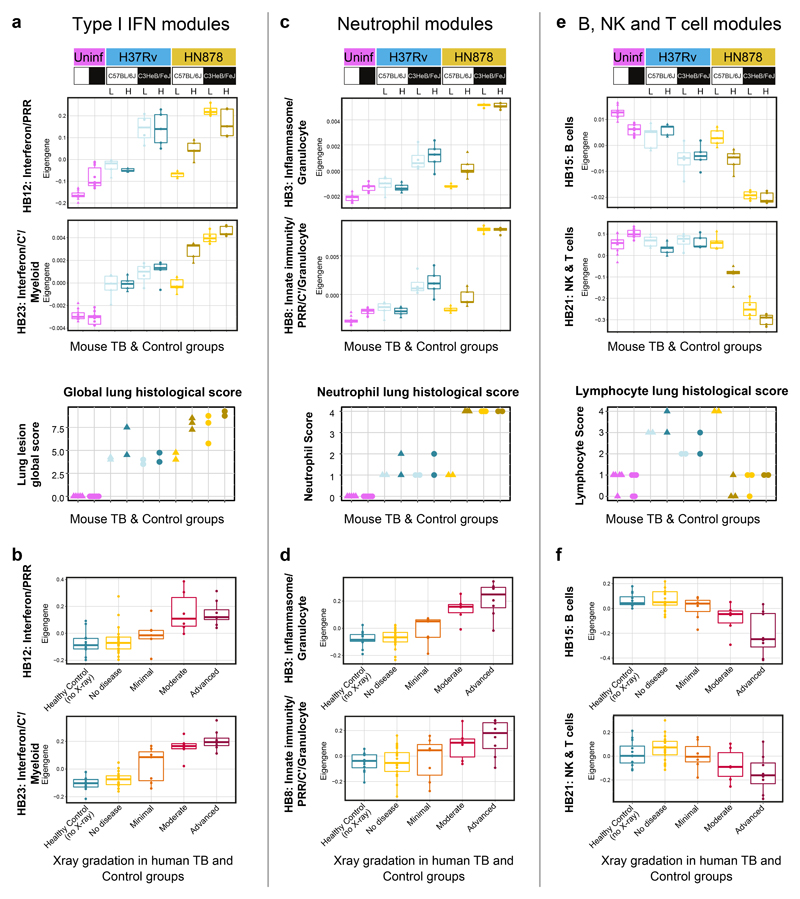
Blood signatures reflect the extent of lung pathology in TB
Correlation between the whole blood TB signature and the extent of lung radiographic burden of human disease has been reported8. A quantitative measure of the transcriptional signature, determined using the molecular distance to health, showed a graded increase in the signature across patients categorised with no disease to minimal, moderate and advanced disease8. Here we show that the extent of the blood modular signatures associated with type I IFN-inducible genes (HB12 and HB23), shown quantitatively by Eigengene expression, positively correlated with the extent of lung pathology assessed by the combined relative lesion burden and percentage of tissue affected scores in the TB mouse models (Fig. 6a). The type I IFN-associated blood modular signature was lowest in the more resistant mouse models of TB increasing with the different levels of lung pathology, peaking in the HN878 infected C3HeB/FeJ mice (Fig. 6a). Similarly, the level of the type I IFN-associated blood modular signatures in human TB, here shown by Eigengene expression, also positively correlated with the radiographic extent of lung disease in patients with different degrees of disease (Fig. 6b). The neutrophil-associated (HB3 and HB8) blood modular signatures, likewise, showed an increased Eigengene expression in the blood of mice in the different TB models correlating with an increased lung neutrophil score (Fig. 6c), and the most severe lung lesions as assessed histopathologically (Fig. 6a). The neutrophil-associated modular blood signature was highest in the HN878 infected C3HeB/FeJ mice correlating with the highest lung neutrophil score (Fig. 6c). Although the neutrophil lung score was similarly high in the high dose HN878 infected C57BL/6J mice, the blood neutrophil-associated modular signature remained low in these mice (Fig. 6c). The blood neutrophil-associated signature in human TB also positively correlated with the radiographic extent of lung disease in TB patients (Fig. 6d), again supporting a role for neutrophils in TB pathogenesis.
Figure 6. Quantitation of specific blood modular signatures against extent of lung pathology in mouse models and human TB.
Box plots depicting the module Eigengene expression for human blood modules Interferon/PRR (HB12) and Interferon/C’/Myeloid (HB23) (a, b), Inflammasome/Granulocytes (HB3) and Innate immunity/PRR/C’/ Granulocytes (HB8) (c, d), B cells (HB15) and NK & T cells (HB21) (e, f), are shown for mouse blood samples from uninfected (Uninf; n = 5 biologically independent samples per group) and M. tuberculosis H37Rv or HN878 infected (L, low dose; H, high dose) C57Bl/6 and C3HeB/FeJ mice (n=3 biologically independent samples per group for low dose HN878 infection of C3HeB/FeJ, and n=5 biologically independent samples per group for all other groups as depicted in Supplementary Fig. 1a) (a, c, e); and for human blood samples from the London TB cohort divided in Healthy Control (no X-ray; n=12 biologically independent samples) and TB patients grouped according to the radiographic extent of disease as No disease (n=21 biologically independent samples), Minimal (n=7 biologically independent samples), Moderate (n = 6 biologically independent samples) or Advanced (n=8, biologically independent samples, described in Berry et al. 20109) (b, d, f). Lung lesion global score (a), neutrophil (c) and lymphocyte (e) scores from H&E stained lung sections are also shown for uninfected (Uninf, n=5 biologically independent samples per group) and M. tuberculosis H37Rv or HN878 infected (L, low dose; H, high dose) C57Bl/6 and C3HeB/FeJ mice (n=2 biologically independent samples per group for H37Rv infection, HN878-infected C57BL/6J mice low dose and HN878-infected C3HeB/FeJ mice high dose, and n=3 biologically independent samples per group for HN878-infected C57BL/6J mice high dose and HN878-infected C3HeB/FeJ mice low dose, from one experiment per M. tuberculosis infection).
In contrast to the increased type I IFN and neutrophil-associated blood modular signatures in TB, the blood B cell (HB15) and NK & T cell (HB21) modular signatures showed a decrease in the blood of M. tuberculosis infected mice showing advanced lung disease, specifically the HN878 infected C3HeB/FeJ mice, and to a lesser extent the high dose HN878 infected C57BL/6J mice (Fig. 6e). This decreased blood signature in advanced disease correlated with a decrease in the lung lymphocyte score, which in the more resistant mice had increased upon infection (Fig. 6e). In human TB, these blood B cell (HB15) and NK & T cell (HB21) modular signatures showed a similar decrease in the blood, inversely correlating with the extent of lung radiographic disease (Fig. 6f).
Modular blood signatures in LTBI-progressors
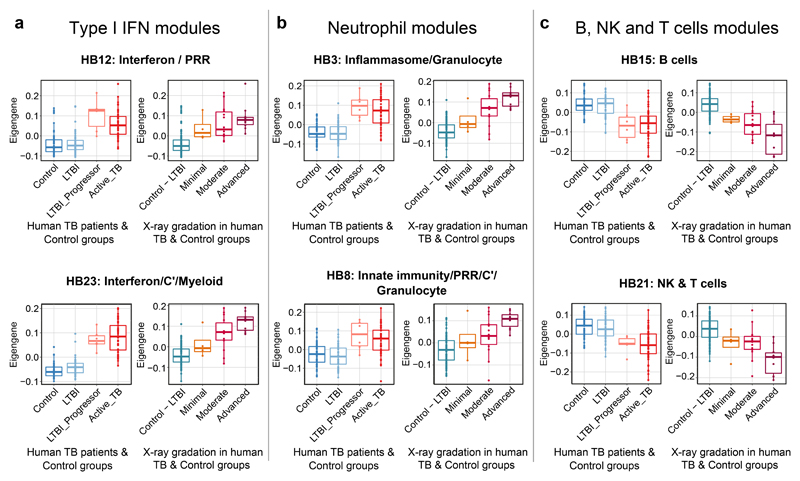
We next set out to determine whether the type I IFN (HB12, HB23), neutrophil (HB3, HB8), B cell (HB15) and NK & T cell (HB21) associated modular signatures, determined in human active TB and susceptible mouse models of TB, could be detected during early M. tuberculosis infection of humans. To this end, we analysed our RNA-Seq data from the blood of recent contacts of active TB patients who were subsequently shown to progress to active TB (LTBI-progressors), active TB patients and healthy controls (IGRA-ve and IGRA+ve contacts who did not progress to TB)16 (Fig. 7).
Figure 7. Quantitation of specific blood modular signatures in blood of healthy controls, LTBI, LTBI-progressors and active TB patients.
Box plots depicting the module Eigengene expression for human blood modules Interferon/PRR (HB12) and Interferon/C’/Myeloid (HB23) (a), Inflammasome/Granulocytes (HB3) and Innate immunity/PRR/C’/ Granulocytes (HB8) (b), B cells (HB15) and NK & T cells (HB21) (c), are shown for human blood samples from the Leicester TB cohort divided in Control (IGRA-ve TB contacts who remained healthy; n=50 biologically independent samples), LTBI (IGRA+ve TB contacts who remained healthy; n=49 biologically independent samples), LTBI_Progressor (TB contacts who developed TB, time point just before the contact was diagnosed with active TB; n=6 biologically independent samples) and Active_TB (patients with active disease; n=53 biologically independent samples) (left panels) or divided in Control – LTBI (IGRA-ve and IGRA+ve TB contacts who remained healthy) or TB patients grouped according to the radiographic extent of disease as Minimal, Moderate and Advanced (right panels; Supplementary Table 7).
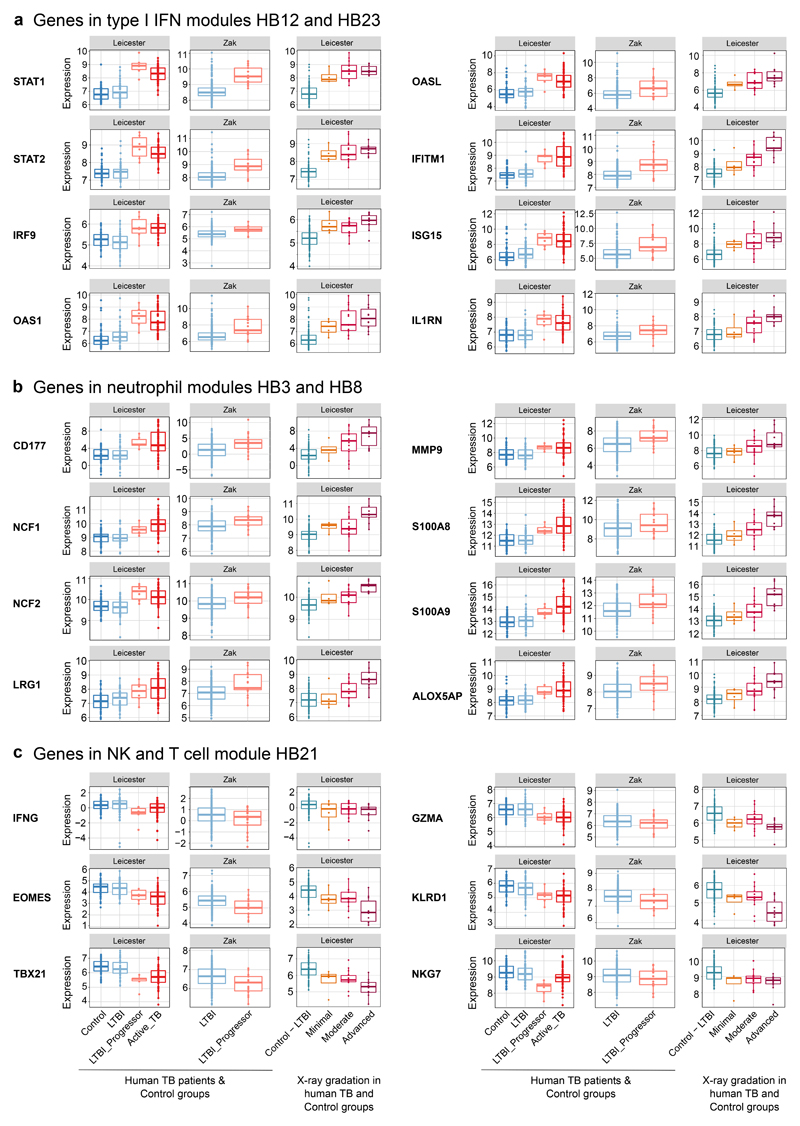
The Interferon/PRR (HB12) and Interferon/C’/Myeloid (HB23) blood modular signatures, shown quantitatively by Eigengene expression, were increased in the blood of LTBI-progressors to the same level as in active TB patients, as compared to healthy controls (Fig. 7a). As shown for the London cohort (Fig. 6b), the type I IFN-associated modular signatures also correlated with the radiographic extent of lung disease in this independent cohort (Fig. 7a). Type I IFN-inducible genes in these modular signatures included STAT1, STAT2, IRF9, OAS1, OASL, IFITM1, ISG15 and IL1RN which were expressed at same level in the blood of LTBI-progressors and active TB patients (Fig. 8a; ShinyApp). Again, the degree of expression of these individual genes positively correlated with the extent of radiographic signs of disease, being already increased in the blood of patients with minimal disease (Fig. 8a). We also observed increased expression of these type I IFN-inducible genes (Fig. 8a) in an independent cohort of LTBI-progressors, as compared to individuals with LTBI who remained healthy38.
Figure 8. Quantitation of IFN, neutrophil and lymphocyte-specific gene expression in blood of healthy controls, LTBI, LTBI-progressors and active TB patients.
Box plots depicting the log2 expression values of selected genes from type I IFN-associated modules HB12 and HB23 (a), neutrophil-associated modules HB3 and HB8 (b) and NK & T cell module HB21 (c) are shown for human blood samples from the Leicester TB cohort divided in Control (IGRA-ve TB contacts who remained healthy; n=50 biologically independent samples), LTBI (IGRA+ve TB contacts who remained healthy; n=49 biologically independent samples), LTBI_Progressor (TB contacts who developed TB, time point just before the contact was diagnosed with active TB; n=6 biologically independent samples) and Active_TB (patients with active disease; n=53 biologically independent samples) (left panels) or divided in Control – LTBI (IGRA-ve and IGRA+ve TB contacts who remained healthy) and TB patients grouped according to the radiographic extent of disease as Minimal, Moderate and Advanced (right panels; Supplementary Table 7). Box plots are also shown for human blood samples of LTBI (non-progressors; n=217 biologically independent samples) and LTBI_Progressor (individuals who developed TB, time point 1 to 180 days before diagnosis; n=17 biologically independent samples) from an independent cohort (GSE79362, Zak et al. 201619) (middle panels).
Strikingly, expression of the neutrophil-associated (HB3 and HB8) modular signatures was also increased to high levels in the blood of LTBI-progressors, to the same level as seen in blood of active TB patients, as compared to healthy controls (Fig. 7b). The extent of these neutrophil associated blood signatures positively correlated with the radiographic signs of lung disease (Fig. 7b). Confirming the contribution of genes associated with neutrophil activation and recruitment, CD177, NCF1, NCF2, LRG1, MMP9, S100A8, S100A9 and ALOX5AP were upregulated in the blood of LTBI-progressors from both cohorts, as compared to controls to a similar level as in the blood of active TB patients, their level of expression correlating with increased signs of radiographic lung disease (Fig. 8b). The increased expression of genes associated with neutrophil activation and recruitment in the blood of TB patients with minimal radiographic signs of disease and LTBI-progressors (Fig. 8b) points to an unappreciated role for neutrophils in early disease.
The expression of the B cell (HB15) and NK & T cell (HB21) associated modular signatures was decreased in the blood of LTBI-progressors to the same extent as in active TB (Fig. 7c), as compared to controls, again correlating with increased radiographic signs of disease (Fig. 7c). Expression of the NK & T cell specific genes IFNG, EOMES, TBX21, GZMA, KLRD1, NKG7, was similarly decreased in the blood of LTBI-progressors in both cohorts, as compared to the healthy controls and in the blood of patients with minimal signs of disease, although further decreased in those with advanced signs of radiographic disease (Fig. 8c). Since T cell and NK cell genes convey protection against M. tuberculosis infection4–6,39,40, their loss may contribute to progression to active TB.
Collectively our findings predict that a dominance of gene expression associated with a type I IFN response and neutrophil activation and recruitment, together with a loss of NK and T cell effector responses, early after infection with M. tuberculosis, may contribute to progression to active TB.
Discussion
Here we show that the IFN-inducible human blood TB transcriptional signature16 is recapitulated in blood from M. tuberculosis HN878 infected TB susceptible C3HeB/FeJ mice, whereas this signature is minimal in blood from M. tuberculosis H37Rv infected resistant C57BL/6J mice. Combining our modular signature data with cell-type specific signatures31 we reveal an increase in neutrophil-associated genes in the blood of TB susceptible mice and TB patients. Genes associated with type I IFN responses and with neutrophil recruitment and activation were increased in LTBI-progressors before diagnosis, suggesting an unappreciated role for neutrophils in early disease. Decreased B, NK and T cell-signatures of human active TB8,16 were observed in the blood of infected TB susceptible mice and LTBI-progressors, whilst upregulated upon infection in the lungs of TB resistant mice, suggesting that their early loss contributes to progression to active TB.
Neutrophils are abundant in the lung lesions of M. tuberculosis infected susceptible mice contributing to TB pathogenesis33,34 whereas lesions of infected resistant mice contain only scattered neutrophils, instead dominated by lymphocytes and macrophages41. M. tuberculosis infected neutrophils have been detected within inflammatory lung granulomas of patients with active TB42,43. We herein reveal low levels of a neutrophil-associated signature in lungs of M. tuberculosis infected C57BL/6J mice, which was maximally increased in HN878 infected susceptible C3HeB/FeJ mice. This was validated by histological analysis, although S100A9 neutrophil staining was lost due to the necrotic nature of the lesions. This led to our discovery of increased expression of neutrophil-associated genes within the over-abundant human TB blood modules, originally annotated as “Inflammasome” and “Innate immunity/PRR/C’”16, which we now rename as “Inflammasome/Granulocyte” and “Innate immunity/PRR/C’/Granulocyte”. Previous studies showed no change by flow cytometry in neutrophil numbers in the blood of active TB patients8, suggesting that the over-abundance of this granulocyte-associated signature of activation and recruitment may be attributable to a subset of activated neutrophils which have circulated to the blood from the lung. Whether these neutrophils are carriers of M. tuberculosis to the blood in human TB, where the bacteria have been recently shown to be detected in early disease44, remains to be investigated. This granulocyte-associated signature was also increased in blood from LTBI-progressors before diagnosis, suggesting a previously unappreciated role for neutrophils in early progression to disease.
The type I IFN-associated signature widely reported in blood of active TB patients8–16 was also present in blood from M. tuberculosis infected mice, with the highest levels observed in the more susceptible models, correlating with more severe lung pathology. This type I IFN-inducible signature resulted from the host genetic background and the dose and strain of M. tuberculosis, possibly explaining differing reports regarding the role of type I IFN in TB pathogenesis18,20–22,24,45,46. The enhanced type I IFN-associated signature in the C3HeB/FeJ mice is in keeping with a recent report that the B6.Sst1S congenic mice carrying the C3H “sensitive” allele of the Sst1 locus that renders them highly susceptible to M. tuberculosis infections47, exhibit markedly increased type I IFN signalling which contributes to their high TB susceptibility via induction of the IL-1 receptor antagonist (IL-1Ra)48. We show that the Il1rn gene expression is increased in mouse blood and lung upon infection, correlating with increasing susceptibility to TB in C3HeB/FeJ mice infected with HN878, a M. tuberculosis strain reported to enhance type I IFN induction and TB pathogenesis21,22. The IL1RN gene was highly expressed in blood from TB patients, but also in the LTBI-progressors, along with other type I IFN-inducible genes such as OAS1, IFITM1 and ISG15, suggesting that type I IFN-inducible genes may contribute to early TB pathogenesis. Genes of the complement cascade were also upregulated in the blood from LTBI-progressors, in keeping with previous reports15,49.
Upregulation of both type I and II IFN have been reported before diagnosis of TB patients15. However, we herein report that in TB patients the IFNG gene itself is down-regulated in the blood, alongside a number of key molecules, including TBX21, EOMES, GZMA, GZMB, NKG7 and KLRD1, suggesting a loss of the protective effector function of T and NK cells5,6,39,40. This decrease was also observed in LTBI-progressors, suggesting that decreased expression of IFNG and other genes associated with effector and cytotoxic functions early after M. tuberculosis infection may contribute to disease progression. This supports reports that IFN-γ, cytotoxic effector molecules and NK cells are important for protection against M. tuberculosis infection in both mouse models and human disease5,6,39,40. In keeping with this, genes associated with effector and cytotoxic NK and T cell responses (Nkg7, Klrd1, Gzma, Gzmb, Tbx21) as well as Ifng were upregulated in the blood and lungs of M. tuberculosis infected TB resistant C57BL/6J mice but drastically reduced in the blood and lungs from HN878 infected susceptible C3HeB/FeJ mice, similarly to in blood from LTBI-progressors and active TB patients. Decreased IFNG expression in the blood of TB patients and Ifng expression in the blood and lungs of susceptible mice parallels the increase in neutrophils, supporting previous reports that IFN-γ regulates neutrophil function35 thus limiting lung inflammation and TB exacerbation.
Our findings of a decrease in the B cell-associated modular expression in the blood of M. tuberculosis infected susceptible mice are in keeping with reports on the reduction in abundance of total B cells in human TB8,40 largely driven by a reduction in circulating naive B cells40. This under-abundance of the B cell-associated module was also observed in blood from LTBI-progressors, although maximal in TB patients and susceptible mice with advanced signs of lung disease. Reduction in peripheral B cells could be due to preferential sequestration of these cells at the site of infection or diminished output of B cells from the bone marrow36,37. Our data support a combination of both, depending on the extent of the disease. The top 50 interacting “hub” genes in the B cell-associated module showed increased expression in the lungs from M. tuberculosis infected resistant mice, but were decreased in lungs from HN878 infected susceptible mice, as verified by histopathology. B cells at the site of infection could be contributing to control of M. tuberculosis infection in the resistant mice as has been proposed36,37.
Using a combination of mouse TB models and human TB cohorts we provide data to suggest that dominance of a type I IFN response and neutrophil activation and recruitment, together with a loss of B cell, NK and T cell effector responses may contribute to the pathogenesis of progressive M. tuberculosis infection. Mouse models of TB have been employed for decades as tools for elucidating mechanisms of host resistance and pathogenesis. While failing to recapitulate many of the features of clinical TB and in several cases protective vaccine responses, they have been remarkably useful in identifying both effector and regulatory responses that have emerged to be important in human infection and disease. The data reported here comparing the host transcriptomic responses of M. tuberculosis infected mice and humans offer further compelling characterization and validation of the mouse model for further mechanistic studies and suggest a peripheral signature associated with progression to clinical disease in TB.
Methods
Experimental animals and ethics
C57BL/6J and C3HeB/FeJ mice were purchased from Jackson Laboratories (Bar Harbour, ME) and housed under barrier conditions in the Animal Biosafety Level 3 (ABSL3) facility at i3S, Porto, Portugal. Experiments were performed in accordance with recommendations of the European Union Directive 2010/63/EU and approved by Portuguese National Authority for Animal Health – Direção Geral de Alimentação e Veterinária. (DGAV-Ref.0421/000/000/2016). Mice were kept with food and water ad libitum and humanely euthanized by CO2 asphyxiation. Every effort was made to minimize suffering. Age matched females were used in experiments.
Mouse models of TB
M. tuberculosis experiments were performed under ABSL-3 conditions. M. tuberculosis H37Rv (laboratory strain) and HN878 (clinical isolate) were grown to midlog phase in Middlebrook 7H9 broth supplemented with 10% oleic acid albumin dextrose complex (Difco), 0.05% Tween 80, and 0.5% glycerol before being quantified on 7H11 agar plates and stored in aliquots at −80°C. Aliquots frozen at −80°C were then thawed (6 aliquots) and quantified, to determine the concentration of the stored inocula. Mice were infected via the aerosol route using an inhalation exposure system (Glas-Col), calibrated to deliver from ∼100 to 1000 CFUs to the lung. The infection dose was confirmed by determining the number of viable bacteria in the lungs of five mice 3 days after the aerosol infection (low dose: ~100-450 CFUs/lung; high dose: ~700-900 CFUs/lung). Infected mice were monitored regularly for signs of illness such as wasting, piloerection and hunching. Mice were euthanized by CO2 inhalation and blood and lung samples from each group were collected from individual mice for RNA isolation, post M. tuberculosis infection at the known peak of the blood transcriptomic response, or in the specific case of the susceptible C3HeB/FeJ mice infected with HN878, when they showed signs of severe illness (Supplementary Fig. 1a). Blood and lung samples from age matched uninfected mice were collected at the same time and used as controls. Lung samples from additional infected mice were collected for bacterial load determination (Supplementary Fig. 1b) or histology (Figs. 3 and 5; Supplementary Fig. 6). Determination of lung bacterial load was performed by plating serial dilution of the organ homogenate on Middlebrook 7H11 agar supplemented with 10% oleic acid albumin dextrose complex plus PANTA to prevent contamination with other infections. CFUs were counted after 3 weeks of incubation at 37°C, and the bacterial load per lung was calculated.
Histopathological analysis of lung samples
Lung tissues from M. tuberculosis infected mice were perfused and fixed in 10% neutral-buffered formalin followed by 70% ethanol, processed and embedded in paraffin, sectioned at 4 μm and stained with hematoxylin and eosin (H&E) or Ziehl-Neelsen (ZN). A single section from each tissue was viewed and scored as a consensus by three board-certified veterinary pathologists (S.L.P., E.H. and A.S.-B.) blinded to the groups (Supplementary Fig. 6). Presence of M. tuberculosis bacilli detected by ZN positive staining was scored as paucibacillary or multibacillary according to their abundance in the tissue. A semi-quantitative scoring (0-4 points) method was devised to assess the following histological features: inflammatory cells (neutrophils, lymphocytes, plasma cells, macrophages), necrosis, pleuritis and fibrosis; using the following scale: 0 = not present, 1 = minimal, 2 = mild, 3 = moderate, and 4 =marked changes. The relative lesion burden scoring (0-5 points) was determined using the following scale: 0= no lesions, 1 = focal lesion, 2 = multiple focal lesions, 3 = one or more focal severe lesions, 4 = multiple focal lesions that are extensive and coalesce, and 5 = extensive lesions that occupy the majority of the lung lobe. The percentage of tissue affected was also scored and the lesion types graded as Type I, Type II and Type III as previously described by Irwin et al. 50. Representative images of each group were acquired on an Olympus BX43 microscope using an Olympus SC50 camera and cellSens Entry software (Ver. 1.18).
Lung lesion global score (Fig. 6a) was calculated by combining the relative lesion burden and the percentage of tissue affected, scored from H&E stained lung sections (Supplementary Fig. 6). Neutrophil and lymphocyte scores for H&E stained lung sections (Supplementary Fig. 6) were plotted in Fig. 6c and 6e, respectively.
Microscopy for neutrophils, T and B cells
Lung sections from M. tuberculosis infected mice were de-waxed and re-hydrated before staining. The neutrophil staining was performed using the automated equipment Ventana Discovery ULTRA. Sections were treated with Protease 1 at 37°C for 8 min for antigen retrieval, incubated with primary antibody Rat anti-mouse 2B10 (in house clone 2B10) at 37°C for 48 min, followed by OmniMap anti-Rt HRP (RUO) at room temperature (RT) for 12 min. For T and B cell staining, sections were microwaved for 23 min (900W) with Citrate Buffer pH6 for antigen retrieval and then incubated with primary antibodies Rabbit monoclonal anti-CD3G (clone ab134096, Abcam) and biotin Rat anti-mouse CD45R/B220 (clone RA3-6B2, BD) for 1h at RT. Sections were then incubated with donkey anti-Rabbit IgG (H+L) secondary antibody Alexa Fluor™ 555 (A-31572, Invitrogen) and Streptavidin, Alexa Fluor™ 488 conjugate (s32354, Invitrogen) for 45 min at RT, followed by DAPI for 15 min at RT and Sudan Black for 20 min at RT. Stained sections were mounted with Tissue-Tek® Glass™ Pertex, examined and scored by two board-certified veterinary pathologists (S.L.P. and A.S.-B). Neutrophils were assessed semi-quantitatively (based on intensity of labelling) as follows: 0 = none present, 1 = low numbers, 2 = moderate numbers and 3 = high numbers. Neutrophil viability was scored as viable (which label with IHC) or necrotic by assessing which subset dominated the stained tissue. Representative images of each group were acquired an Olympus BX43 microscope using an Olympus SC50 camera and cellSens Entry software (Ver. 1.18).
For T and B cell quantification, slides were scanned using the objective magnification of 20x on an Olympus VS120-L100 Slide Scanner. T and B cells were assessed semi-quantitatively (based on positive labelling) as follows: 0 = none present, 1 = very low numbers, 2 = low to moderate numbers, 3 = moderate to high numbers and 4 = very high numbers. The T/B cell ratio (%/%) and presence of follicles with germinal centres were also scored for each slide.
RNA isolation
Blood was collected in Tempus reagent (Life Technologies) at 1:2 ratio. Total RNA was extracted using the PerfectPure RNA Blood Kit (5 PRIME). Globin RNA was depleted from total RNA (1.5–2 μg) using the Mouse GLOBINclear kit (Thermo Fisher Scientific). Lungs were collected in TRI-Reagent (Sigma-Aldrich). Total RNA was extracted using the RiboPure™ Kit (Ambion). All RNA was stored at −80 °C until use.
Quantity and quality of RNA samples
Quantity was verified using NanoDrop™ 1000/8000 spectrophotometers (Thermo Fisher Scientific). Quality and integrity of the total and the globin-reduced RNA were assessed with the HT RNA Assay Reagent kit (Perkin Elmer) using a LabChip GX bioanalyser (Caliper Life Sciences/Perkin Elmer) and assigned an RNA Quality Score (RQS) or RNA 6000 Pico kit (Agilent) using a BioAnalyzer 2100 (Agilent) and assigned an RNA Integrity (RIN) score. RNA with an RQS/RIN >6 was used to prepare samples for microarray or RNA-seq.
Microarray
cRNA was prepared from 200 ng globin-reduced blood RNA or 200 ng tissue total RNA using the Illumina TotalPrep RNA Amplification Kit (Ambion). Quality was checked using an RNA 6000 Nano kit (Agilent) using a BioAnalyzer 2100 (Agilent). Biotinylated cRNA samples were randomized; 1.5 μg cRNA was then hybridized to Mouse WG-6 v2.0 bead chips (Illumina) according to the manufacturer’s protocols.
RNA-Seq
cDNA library preparation: for blood and tissues, total/globin-reduced RNA (200 ng) was used to prepare cDNA libraries using the TruSeq Stranded mRNA HT Library Preparation Kit (Illumina). For cDNA library preparation of FACS sorted cells, total RNA (30–500 pg) was used to prepare cDNA libraries using the NEBNext® Single Cell/Low Input RNA Library Prep Kit NEBNext® Multiplex Oligos for Illumina® #E6609 (New England BioLabs). Quality and integrity of the tagged libraries were initially assessed with the HT DNA HiSens Reagent kit (Perkin Elmer) using a LabChip GX bioanalyser (Caliper Life Sciences/Perkin Elmer). Tagged libraries were then sized and quantitated in duplicate (Agilent TapeStation system) using D1000 ScreenTape and reagents (Agilent). Libraries were normalized, pooled and then clustered using the HiSeq® 3000/4000 PE Cluster Kit (Illumina). The libraries were imaged and sequenced on an Illumina HiSeq 4000 sequencer using the HiSeq® 3000/4000 SBS kit (Illumina) at a minimum of 25 million paired-end reads (75 bp/100 bp) per sample.
Microarray data analysis
Microarray data was processed in GeneSpring GX v14.8 (Agilent Technologies). Flags were used to filter out the probe sets that did not result in a “present” call in at least 10% of the samples, with the “present” lower cut-off of 0.99. Signal values were then normalized using neqc function with default parameters from limma package (v 3.38.3) in R. This function performs background correction, quantile normalization and log2 transformation of intensity signals. For modular fold enrichment analysis, Illumina IDs were converted to Ensembl IDs using both the annotation file available from Illumina and biomaRt package (2.38.0) in R. Next, transcripts were filtered to select the 50% most variable probes across all samples.
RNA-Seq data analysis
Raw paired-end RNA-seq data was subjected to quality control using FastQC (Babraham Bioinformatics) and MultiQC51. Trimmomatic52 v0.36 was used to remove the adapters and filter raw reads below 36 bases long, and leading and trailing bases below quality 25. The filtered reads were aligned to the Mus musculus genome Ensembl GRCm38 (release 86) using HISAT253 v2.0.4 with default settings and RF rna-strandedness, including unpaired reads, resulting from Trimmomatic, using option -U. The mapped and aligned reads were quantified to obtain the gene-level counts using HtSeq54 v0.6.1 with default settings and reverse strandedness. Raw counts were processed using the bioconductor package DESeq255 v1.12.4 in R v3.3.1, and normalized using the DESeq method to remove the library-specific artefacts. Variance stabilizing transformation was applied to obtain normalized log2 gene expression values. Further quality control was performed using principal component analysis, boxplots, histograms and density plots. Differentially expressed genes were calculated using the Wald test in DESeq255. Genes with log2 fold change >1 or <−1 and false discovery rate (FDR) p-value < 0.05 corrected for multiple testing using the Benjamini–Hochberg (BH) method56 were considered significant. Log2 fold changes in mouse blood, mouse lung and human blood datasets (Berry London, Berry South Africa and Leicester: GSE107995), using the top 50 intra-modular genes within selected human blood modules, were represented in heatmaps using the pheatmap package in R (Raivo Kolde (2019). pheatmap: Pretty Heatmaps. R package version 1.0.12.) (Fig. 4; Supplementary Fig. 8 and 9). For lung module generation, and modular fold enrichment, only protein coding genes were considered (Supplementary Fig. 5). PCA plots were generated using prcomp function in R and plotted using ggplot2 package (H. Wickham. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York, 2016.).
Cellular deconvolution
Deconvolution analysis for quantification of relative levels of distinct cell types on a per sample basis was carried out on normalized counts using CIBERSORT57. CIBERSORT estimates the relative subsets of RNA transcripts using linear support vector regression. Mouse cell signatures for 25 cell types were obtained using ImmuCC58 and grouped into 9 representative cell types based on the application of ImmuCC cellular deconvolution analysis to the sorted cell RNA-seq samples from the ImmGen ULI RNA-seq dataset (ImmGen Consortium: GSE109125; http://www.immgen.org) as previously described31,59,60 (Supplementary Fig. 1d).
Module generation
Human blood modules were previously determined in human TB16. Weighted gene co-expression network analysis was performed to identify lung modules using the package WGCNA61 in R. Modules were across all control and infected samples, using log2 RNA-seq expression values. The lung modules were constructed using the 10,000 most variable genes across all lung samples. A signed weighted correlation matrix containing pairwise Pearson correlations between all the genes across all the samples was computed using a soft threshold of β = 22 to reach a scale-free topology. Using this adjacency matrix, the topological overlap measure (TOM) was calculated, which measures the network interconnectedness62 and is used as input to group highly correlated genes together using average linkage hierarchical clustering. The WGCNA dynamic hybrid tree-cut algorithm63 was used to detect the network modules of co-expressed genes with a minimum module size of 20, and deep split = 1. Lung modules were numbered ML1–ML27, and human blood modules previously found in human TB16 were numbered HB1-HB23. An additional “grey” module was identified in lung modules (Supplementary Table 6, module titled NA), consisting of genes that were not co-expressed with any other genes. These grey modules were not considered in any further analysis. To create gene interaction networks, hub genes with the highest intramodular connectivity and a minimum correlation of 0.75 were calculated, with a cut-off of 50 hub genes, and exported into Cytoscape v3.4.0 for visualization.
For checking either human blood modules into mouse data or mouse lung modules into human data, human Ensembl gene ID were translated into Mouse gene ID using BioMart to extract mouse ortholog genes (Supplementary Table 8).
Modular annotation
Lung modules were enriched for biological pathways and processed using IPA (QIAGEN Bioinformatics), Metacore (Thomson Reuters), and a careful manual annotation, by checking cell-type-specific enrichment and individual read counts. Significantly enriched canonical pathways, and upstream regulators were obtained from IPA (top 5). Modules were assigned names based on representative biological processes from pathways and processes from all three methods (Supplementary Table 5 and 6).
Module enrichment analysis
Fold enrichment for the WGCNA modules was calculated using the quantitative set analysis for gene expression (QuSAGE)64 using the bioconductor package qusage v2.4.0 in R, to identify the modules of genes over- or under-abundant in a dataset, compared to the respective control group using log2 expression values. The qusage function was used with n.points parameter set to 215. Only modules with enrichment scores with FDR p-value < 0.05 were considered significant, and plotted using the ggcorrplot function in R. Eigengene profiles, which are representative expression profiles for a given module in aparticular dataset, have been generated using the moduleEigengenes function from the WGCNA package and have plotted using ggplot2 package.
Cell-type-specific enrichment
Cell-type enrichment analysis to identify over-represented cell types in blood and lung modules was performed as previously described31 using a hypergeometric test, using the phyper function in R. p-Values were corrected for multiple testing using the p.adjust function in R, using the BH method, to obtain FDR corrected p-values.
Method for use of online WebApp
An online web application: https://ogarra.shinyapps.io/tbtranscriptome/ accompanies the manuscript to visualize the findings of the study. The app is subdivided into 4 distinct pages that can be accessed through the tabs displayed on the top of the page, with a customized sidebar for user input on each page.
Tab 1: “Expression Table” allows the user to visualize read counts, either as raw counts or log2 normalized expression values, in either the Mouse Blood TB, Mouse Lung TB, Human Blood TB (Leicester, London or South Africa) datasets. Each row represents a different gene, each column a sample in the corresponding dataset. The user can download the dataset into spreadsheet file format.
Tab 2: “Average expression Table” allows the user to visualize the average read counts by group, either as raw counts or log2 normalized expression values, in either the Mouse Blood TB, Mouse Lung TB, Human Blood TB (Leicester, London or South Africa) datasets. Each row represents a different gene, each column a group in the corresponding dataset. The user can download the dataset into spreadsheet file format.
Tab 3: “Gene expression” allows the user to visualize the expression of individual genes, either as raw or log2 normalized expression values, in either the Mouse Blood TB, Mouse Lung TB, Human Blood TB (Leicester, London or South Africa) datasets. Each dot represents the expression value for the chosen gene, in one sample.
Tab 4: “Module profiles” allows the user to visualize the expression profile (EigenGene from WGCNA R package), of a module he can select, either from Human Blood TB Modules (HB1-HB23)16, Mouse Lung TB modules (ML1–ML27) derived de novo in this study, or Mouse Lung Disease modules (L1-L38)31. Each dot represents the EigenGene value for the chosen module, in one sample. A table below the plot displays all genes present within that module.
Supplementary Material
Acknowledgements
We wish to thank S. Hadebe (The Francis Crick Institute UK, now UCT, South Africa) and J. Pitt (MRC NIMR UK) for some of the early mouse M. tuberculosis infections and sample processing leading up to the current study; K. Potempa (MRC NIMR UK) for early analysis of some of the microarray data leading up to the current study. We thank L. Gabryšová (The Francis Crick Institute UK, now Novartis, Basel, Sw) for her intellectual contribution to discussion of the project. We thank X. Wu for her help in organisation of mice for TB experiments. We thank The Francis Crick Institute: Biological Services for breeding and maintenance of the mice used for the early mouse M. tuberculosis infections leading up to the current study; Advanced Sequencing Facility, Bioinformatics and Biostatistics Science Technology Platforms for their contribution to our sequencing processing, and R. Goldstone excellent project management of sequencing and D. Jackson for support of sequencing; and Experimental Histopathology for their excellent work in preparing lung sections for histological analyses.
This whole study was funded by The Francis Crick Institute which receives its core funding from Cancer Research UK (FC001126), the UK Medical Research Council (FC001126), and the Wellcome Trust (FC001126); before that by the UK Medical Research Council (MRC U117565642); and by the European Research Council (294682-TB-PATH). A.O’G., L.M-T., O.T., C.M.G, A.Si., E.S. were supported by The Francis Crick Institute which receives its core funding from Cancer Research UK (FC001126), the UK Medical Research Council (FC001126), and the Wellcome Trust (FC001126); before that by the UK Medical Research Council (MRC U117565642); L.M-T., and A.Si., were additionally funded by the European Research Council (294682-TB-PATH). S.L.P., A.S-B., and E.H. were funded by the Royal Veterinary College and The Francis Crick Institute. K.D.M-B., and A.Sh. were funded by the Intramural Research Program of the National Institutes of Allergy and Infectious Disease. M.S. was funded by FEDER-Fundo Europeu de Desenvolvimento Regional funds through the COMPETE 2020 – Operational Programme for Competitiveness and Internationalisation (POCI), Portugal 2020, and by Portuguese funds through FCT in the frame-work of the project ‘‘Institute for Research and Innovation in Health Sciences’’ (POCI-01-0145-FEDER-007274), and by FCT through Estimulo Individual ao Emprego Científico. K.L.F. and B.C. are funded by FCT PhD scholarships SFRH/BD/114405/2016 and SFRH/BD/114403/2016, respectively. P.H. and R.V. were supported by NIHR Leicester Biomedical Research Centre and the University of Leicester.
Footnotes
Reporting Summary
Further information on research design is available in the Life Sciences Reporting Summary.
Data availability
The materials, data and any associated protocols that support the findings of this study are available from the corresponding author upon request. The RNA-seq datasets have been deposited in the NCBI Gene Expression Omnibus (GEO) database with the primary accession number GSE140945 (TB mouse blood and lung). Publicly available datasets used in this study include GSE107995 (human TB datasets from Singhania et al. 201818) and GSE79362 (human TB dataset from Zak et al. 201619).
Author Contributions. A.O’G. conceived and designed the study with input from L.M-T., M.S., C.M.G., and P.S.R; and lead and co-ordinated the study with help from L.M-T. A.O’G co-wrote the manuscript together with L.M-T. O.T. performed the major part of the bioinformatics analyses with significant input from A.Si. supervised by A.O’G. P.C provided technical bioinformatics support. L.M-T coordinated the logistics of the study. C.M.G. isolated RNA and prepared sequencing libraries from the TB mouse models. E.S. assisted in TB mouse model experiments executed by K.L.F., J.S., B.C., and P.S.R. and designed by A.O’G, L.M-T and M.S. E.S. contributed to all early mouse model TB experiments executed with P.S.R. and L.M-T. supervised by, A.O’G, L.M-T. and M.S. S.L.P., A.S-B, and E.H. performed histopathological analysis and interpretation. K.D.M-B. and A.Sh. provided TB samples from early mouse TB experiments leading up to the current study. M.S., K.D.M-B. and A.Sh., also provided important discussion for the project throughout and critical feedback on the manuscript. R.V. and P.H. provided clinical analysis of TB patients, and also provided critical feedback on the manuscript and important discussion for the project. All co-authors have read, reviewed and approved the manuscript.
Competing Interests. The authors declare no competing interests
References
- 1.WHO. Global Tuberculosis Report 2017. World Health Organization; 2017. [Google Scholar]
- 2.Dowdy DW, Basu S, Andrews JR. Is passive diagnosis enough? The impact of subclinical disease on diagnostic strategies for tuberculosis. American journal of respiratory and critical care medicine. 2013;187:543–551. doi: 10.1164/rccm.201207-1217OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casanova JL, Abel L. Genetic dissection of immunity to mycobacteria: the human model. Annu Rev Immunol. 2002;20:581–620. doi: 10.1146/annurev.immunol.20.081501.125851. [DOI] [PubMed] [Google Scholar]
- 4.Cooper AM, Mayer-Barber KD, Sher A. Role of innate cytokines in mycobacterial infection. Mucosal immunology. 2011;4:252–260. doi: 10.1038/mi.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 6.O'Garra A, et al. The immune response in tuberculosis. Annu Rev Immunol. 2013;31:475–527. doi: 10.1146/annurev-immunol-032712-095939. [DOI] [PubMed] [Google Scholar]
- 7.Mayer-Barber KD, et al. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature. 2014;511:99–103. doi: 10.1038/nature13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry MP, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blankley S, et al. A 380-gene meta-signature of active tuberculosis compared with healthy controls. The European respiratory journal. 2016;47:1873–1876. doi: 10.1183/13993003.02121-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blankley S, et al. The Transcriptional Signature of Active Tuberculosis Reflects Symptom Status in Extra-Pulmonary and Pulmonary Tuberculosis. PloS one. 2016;11:e0162220. doi: 10.1371/journal.pone.0162220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bloom CI, et al. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PloS one. 2013;8:e70630. doi: 10.1371/journal.pone.0070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joosten SA, Fletcher HA, Ottenhoff TH. A helicopter perspective on TB biomarkers: pathway and process based analysis of gene expression data provides new insight into TB pathogenesis. PloS one. 2013;8:e73230. doi: 10.1371/journal.pone.0073230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaforou M, et al. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS medicine. 2013;10:e1001538. doi: 10.1371/journal.pmed.1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maertzdorf J, et al. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun. 2011;12:15–22. doi: 10.1038/gene.2010.51. [DOI] [PubMed] [Google Scholar]
- 15.Scriba TJ, et al. Sequential inflammatory processes define human progression from M. tuberculosis infection to tuberculosis disease. PLoS pathogens. 2017;13:e1006687. doi: 10.1371/journal.ppat.1006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singhania A, et al. A modular transcriptional signature identifies phenotypic heterogeneity of human tuberculosis infection. Nature communications. 2018;9 doi: 10.1038/s41467-018-04579-w. 2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behar SM, Divangahi M, Remold HG. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat Rev Microbiol. 2010;8:668–674. doi: 10.1038/nrmicro2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreira-Teixeira L, Mayer-Barber K, Sher A, O'Garra A. Type I interferons in tuberculosis: Foe and occasionally friend. The Journal of experimental medicine. 2018;215:1273–1285. doi: 10.1084/jem.20180325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carmona J, et al. Mycobacterium tuberculosis Strains Are Differentially Recognized by TLRs with an Impact on the Immune Response. PloS one. 2013;8:e67277. doi: 10.1371/journal.pone.0067277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorhoi A, et al. Type I IFN signaling triggers immunopathology in tuberculosis-susceptible mice by modulating lung phagocyte dynamics. Eur J Immunol. 2014;44:2380–2393. doi: 10.1002/eji.201344219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manca C, et al. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha /beta. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5752–5757. doi: 10.1073/pnas.091096998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manca C, et al. Hypervirulent M. tuberculosis W/Beijing strains upregulate type I IFNs and increase expression of negative regulators of the Jak-Stat pathway. J Interferon Cytokine Res. 2005;25:694–701. doi: 10.1089/jir.2005.25.694. [DOI] [PubMed] [Google Scholar]
- 23.Ordway D, et al. The hypervirulent Mycobacterium tuberculosis strain HN878 induces a potent TH1 response followed by rapid down-regulation. Journal of immunology (Baltimore, Md. : 1950) 2007;179:522–531. doi: 10.4049/jimmunol.179.1.522. [DOI] [PubMed] [Google Scholar]
- 24.McNab FW, et al. TPL-2-ERK1/2 signaling promotes host resistance against intracellular bacterial infection by negative regulation of type I IFN production. Journal of immunology (Baltimore, Md. : 1950) 2013;191:1732–1743. doi: 10.4049/jimmunol.1300146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogunovic D, et al. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science (New York, N. Y.) 2012;337:1684–1688. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antonelli LR, et al. Intranasal Poly-IC treatment exacerbates tuberculosis in mice through the pulmonary recruitment of a pathogen-permissive monocyte/macrophage population. J Clin Invest. 2010;120:1674–1682. doi: 10.1172/JCI40817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Redford PS, et al. Influenza A virus impairs control of Mycobacterium tuberculosis coinfection through a type I interferon receptor-dependent pathway. The Journal of infectious diseases. 2014;209:270–274. doi: 10.1093/infdis/jit424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barry CE, 3rd, et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol. 2009;7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kramnik I, Beamer G. Mouse models of human TB pathology: roles in the analysis of necrosis and the development of host-directed therapies. Seminars in immunopathology. 2016;38:221–237. doi: 10.1007/s00281-015-0538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Domaszewska T, et al. Concordant and discordant gene expression patterns in mouse strains identify best-fit animal model for human tuberculosis. Scientific reports. 2017;7 doi: 10.1038/s41598-017-11812-x. 12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singhania A, et al. Transcriptional profiling unveils type I and II interferon networks in blood and tissues across diseases. Nature communications. 2019;10 doi: 10.1038/s41467-019-10601-6. 2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ley K, et al. Neutrophils: New insights and open questions. Sci Immunol. 2018;3 doi: 10.1126/sciimmunol.aat4579. [DOI] [PubMed] [Google Scholar]
- 33.Dorhoi A, et al. The adaptor molecule CARD9 is essential for tuberculosis control. The Journal of experimental medicine. 2010;207:777–792. doi: 10.1084/jem.20090067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eruslanov EB, et al. Neutrophil responses to Mycobacterium tuberculosis infection in genetically susceptible and resistant mice. Infection and immunity. 2005;73:1744–1753. doi: 10.1128/IAI.73.3.1744-1753.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nandi B, Behar SM. Regulation of neutrophils by interferon-gamma limits lung inflammation during tuberculosis infection. The Journal of experimental medicine. 2011;208:2251–2262. doi: 10.1084/jem.20110919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Achkar JM, Chan J, Casadevall A. B cells and antibodies in the defense against Mycobacterium tuberculosis infection. Immunological reviews. 2015;264:167–181. doi: 10.1111/imr.12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu LL, et al. A Functional Role for Antibodies in Tuberculosis. Cell. 2016;167:433–443.e414. doi: 10.1016/j.cell.2016.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zak DE, et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet. 2016;387:2312–2322. doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng CG, et al. NK cell-derived IFN-gamma differentially regulates innate resistance and neutrophil response in T cell-deficient hosts infected with Mycobacterium tuberculosis. Journal of immunology (Baltimore, Md. : 1950) 2006;177:7086–7093. doi: 10.4049/jimmunol.177.10.7086. [DOI] [PubMed] [Google Scholar]
- 40.Roy Chowdhury R, et al. A multi-cohort study of the immune factors associated with M. tuberculosis infection outcomes. Nature. 2018;560:644–648. doi: 10.1038/s41586-018-0439-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aly S, et al. Oxygen status of lung granulomas in Mycobacterium tuberculosis-infected mice. The Journal of pathology. 2006;210:298–305. doi: 10.1002/path.2055. [DOI] [PubMed] [Google Scholar]
- 42.Gopal R, et al. S100A8/A9 proteins mediate neutrophilic inflammation and lung pathology during tuberculosis. American journal of respiratory and critical care medicine. 2013;188:1137–1146. doi: 10.1164/rccm.201304-0803OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eum SY, et al. Neutrophils are the predominant infected phagocytic cells in the airways of patients with active pulmonary TB. Chest. 2010;137:122–128. doi: 10.1378/chest.09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verma R, et al. A novel high sensitivity bacteriophage-based assay identifies low level M. tuberculosis bacteraemia in immunocompetent patients with active and incipient TB. Clin Infect Dis. 2019 doi: 10.1093/cid/ciz548. [DOI] [PubMed] [Google Scholar]
- 45.Cooper AM, Pearl JE, Brooks JV, Ehlers S, Orme IM. Expression of the Nitric Oxide Synthase 2 Gene Is Not Essential for Early Control of Mycobacterium tuberculosis in the Murine Lung. Infection and immunity. 2000;68:6879–6882. doi: 10.1128/iai.68.12.6879-6882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moreira-Teixeira L, et al. T Cell-Derived IL-10 Impairs Host Resistance to Mycobacterium tuberculosis Infection. Journal of immunology (Baltimore, Md. : 1950) 2017;199:613–623. doi: 10.4049/jimmunol.1601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pichugin AV, Yan BS, Sloutsky A, Kobzik L, Kramnik I. Dominant role of the sst1 locus in pathogenesis of necrotizing lung granulomas during chronic tuberculosis infection and reactivation in genetically resistant hosts. The American journal of pathology. 2009;174:2190–2201. doi: 10.2353/ajpath.2009.081075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ji DX, et al. Interleukin-1 receptor antagonist mediates type I interferon-driven susceptibility to Mycobacterium tuberculosis. bioRxiv. 2018 [Google Scholar]
- 49.Esmail H, et al. Complement pathway gene activation and rising circulating immune complexes characterize early disease in HIV-associated tuberculosis. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:E964–E973. doi: 10.1073/pnas.1711853115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Irwin SM, et al. Presence of multiple lesion types with vastly different microenvironments in C3HeB/FeJ mice following aerosol infection with Mycobacterium tuberculosis. Disease models & mechanisms. 2015;8:591–602. doi: 10.1242/dmm.019570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995:289–300. [Google Scholar]
- 57.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Z, et al. Inference of immune cell composition on the expression profiles of mouse tissue. Scientific reports. 2017;7 doi: 10.1038/srep40508. 40508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heng TS, Painter MW, Immunological Genome Project, C The Immunological Genome Project: networks of gene expression in immune cells. Nature immunology. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida H, et al. The cis-Regulatory Atlas of the Mouse Immune System. Cell. 2019;176:897–912 e820. doi: 10.1016/j.cell.2018.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yip AM, Horvath S. Gene network interconnectedness and the generalized topological overlap measure. BMC bioinformatics. 2007;8:22. doi: 10.1186/1471-2105-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 64.Yaari G, Bolen CR, Thakar J, Kleinstein SH. Quantitative set analysis for gene expression: a method to quantify gene set differential expression including gene-gene correlations. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt660. e170-e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.