Abstract
The ability of the skin to grow in response to stretching has been exploited in reconstructive surgery1. Although the response of epidermal cells to stretching has been studied in vitro 2,3, it remains unclear how mechanical forces affect their behaviour in vivo. Here, we develop a mouse model in which the consequences of stretching on skin epidermis can be studied at single-cell resolution. Using a multidisciplinary approach that combines clonal analysis with quantitative modelling and single-cell RNA-seq, we show that stretching induces skin expansion by creating a transient bias in the renewal activity of epidermal stem cells (SC), while a second subpopulation of basal progenitors remains committed to differentiation. Transcriptional and chromatin profiling identifies how cell states and gene regulatory networks are modulated by stretching. Using pharmacological inhibitors and mouse mutants, we define the step-by-step mechanisms that control stretch-mediated tissue expansion at single-cell resolution in vivo.
To shape tissue architecture, cells are subject to mechanical forces arising intrinsically through the cytoskeleton; by cell-cell adhesion; and from the microenvironment through interaction with the extracellular matrix (ECM)4,5. Cells sense and respond to these cues via the integrated activation of different signalling pathways. This process of mechanotransduction eventually leads to changes in cell shape, gene expression and cell fate6. Although studies have shown that mechanotransduction can dictate cell behaviour in vitro 7, the underlying mechanisms that allow organs to sense and mediate mechanical cues in vivo are only beginning to be revealed.
As the first barrier against the environment, the skin is highly exposed to mechanical stress. The skin must resist and respond to physical insults, as well as to adapt its shape and size to ensure vital barrier functions8.
Mechanical stretch-mediated tissue expansion is a procedure commonly used in plastic surgery to generate extra skin to repair birth defects, remove scars, or for breast reconstruction1. In this procedure, an inflatable “skin expander” is inserted underneath the skin and inflated, causing the expansion of the overlaying skin1. During the course of expansion, an excess of cells must be produced. But, do all proliferative cells respond equally to stretch, or do subpopulations respond differentially? How is mechanosensation linked to gene transcription, and which transcription factors relay mechanical stress to control expansion?
Results
Hydrogel induces mouse skin expansion
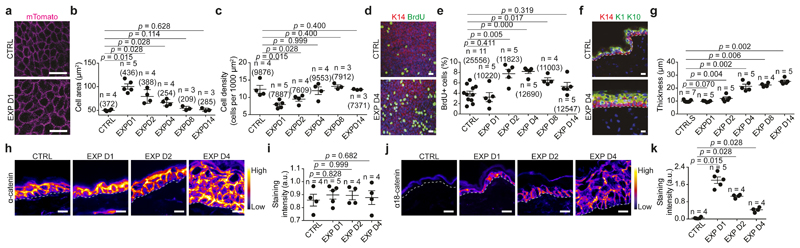
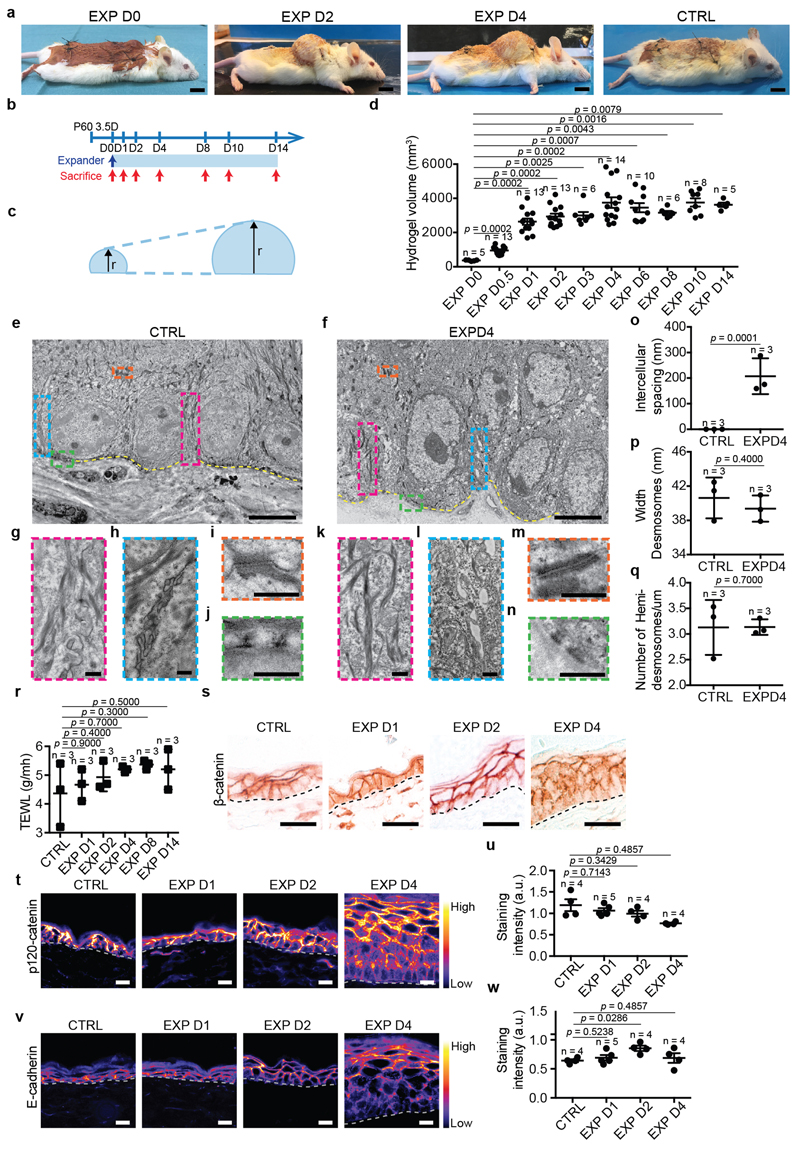
To study the cellular and molecular mechanisms that regulate stretch-mediated expansion in vivo, we established a mouse model that mimics the procedure used in humans in which a self-inflating hydrogel is introduced underneath the skin. The hydrogel is designed to inflate to a predefined shape and size9. The 4ml hydrogel devices expanded rapidly, reaching their maximum volume after 1 day (Extended Data Fig. 1a-d). During the first two days following expansion, the cell area was increased and density decreased, consistent with stretch. After 4 days, these parameters returned to their homeostatic values, indicating the production of new tissue. At day (D)2, BrdU incorporation increased two-fold; and after D4, BrdU incorporation decreased progressively, reaching control levels by D14 (Fig. 1a-e).
Figure 1. Inflated hydrogel mediates skin expansion.
a, Membranous signal (pink), from PFA-perfused Rosa26-mT/mG mice. Scale bar, 15 urn. b, Area of basal cells measured from a. c, Basal cell density. Number of nuclei per 1000 μm2 (5 different independent areas of 40,000 μm2 per mouse). d, Immunostaining for K14 (red), BrdU (green) and Hoechst for nuclei (blue) on whole mount epidermis. Scale bar, 20 μm. e, BrdU positive cells. f, Immunostaining for K14 (red), K1, K10 (green) and Hoechst for nuclei (blue) on tissue sections. Scale bar, 20 μm. g, Tissue thickness, 3 independent measurements per at least 2 sections per mouse. h, j, Adherens junctions (AJ) component a-catenin (h) and the a18 tension sensitive from of a-catenin (α18-cathenin) (j) colour-coded for signal intensity with ImageJ. Protein expression is visualized as a colour gradient going from black to yellow, with black as indicator of no expression and yellow as indicator of maximal expression. Scale bar, 10 μm i, k, Average integrated density signal for α-catenin (i) and α18-cathenin (k). Each data point is the average of 5 measurements per mouse. f, h, j, Dashed line indicate the basal lamina. b, c, e, g, i, k, In parentheses the number of cells and n=number of mice. Two-tailed Mann–Whitney test, mean per mouse + s.e.m.
To investigate whether the differentiation rate was affected, we assessed the production of Keratin 1 (K1+) and Keratin 10 (K10+) suprabasal cells following expansion. From D4 on, we observed an increase in the number of K1+/K10+ suprabasal layers, demonstrating that stretch-mediated proliferation couples renewing divisions with differentiation (Fig. 1f,g).
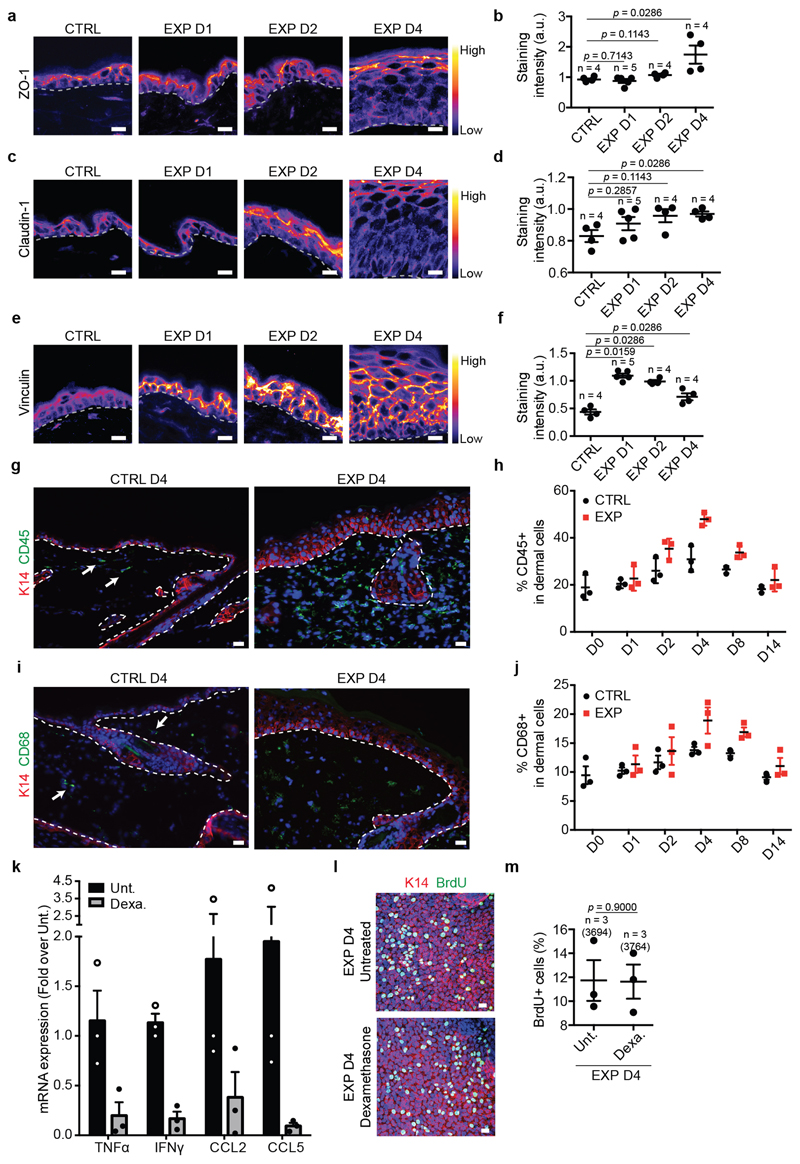
During morphogenesis and in in vitro cell culture, stretch is commonly associated with cell-cell junction rearrangements10. Transmission electron microscopy (TEM) showed that stretch induced intercellular spacing and thicker keratin bundles. Desmosomes and hemidesmosomes remained unchanged (Extended Data Fig. 1e-q). Despite cellular remodelling, the integrity of the skin barrier was maintained, as assessed by trans-epithelial water loss (TEWL) (Extended Data Fig. 1r). Moreover, the expression of adherens junctions and tight-junction proteins was unchanged (Fig.1h,i, Extended Data Fig. 1s-w and 2a-d). However, following expansion, the tension-sensitive epitope of alpha-catenin (a18-catenin)11 was increasingly accessible, and vinculin expression was enriched at the adhesion sites, showing that adherens junctions are remodelled following stretch (Fig. 1j,k and Extended Data Fig. 2e,f).
Although inflammation occurred following expansion, blocking inflammation by dexamethasone administration did not decrease proliferation (Extended Data Fig. 2g-m), suggesting that inflammation is not essential to mediate cell proliferation.
Stretching promotes SC renewal
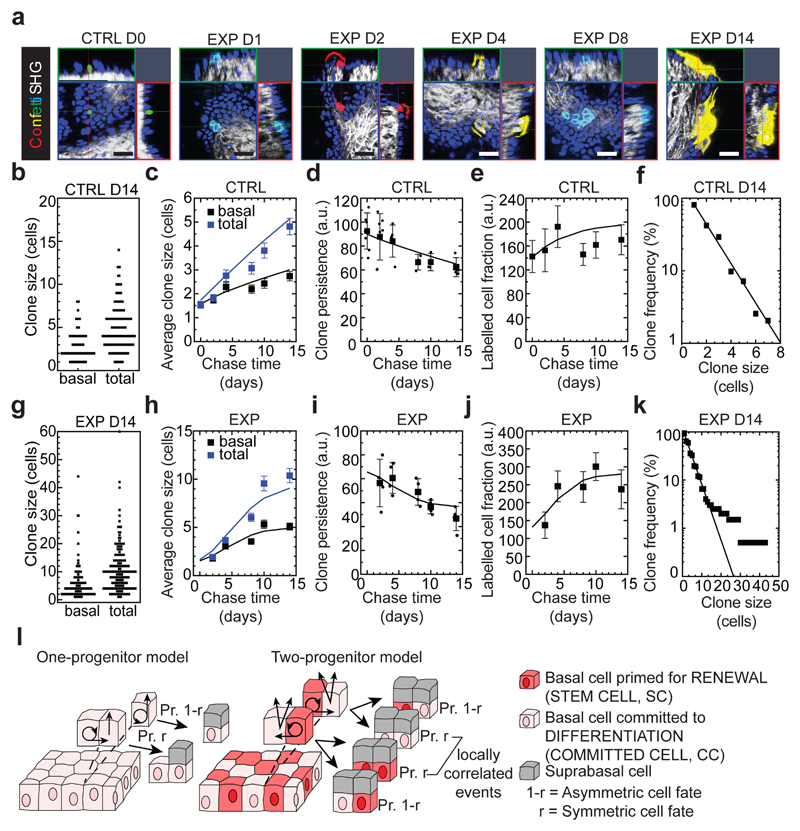
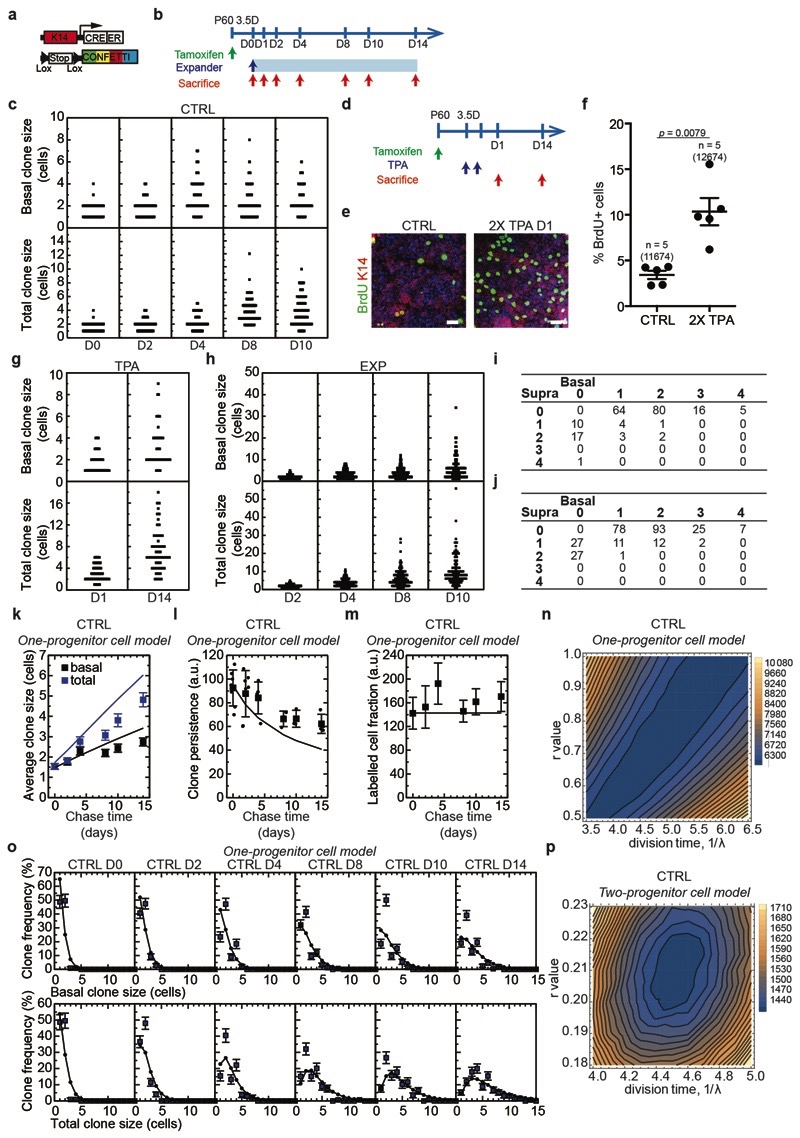
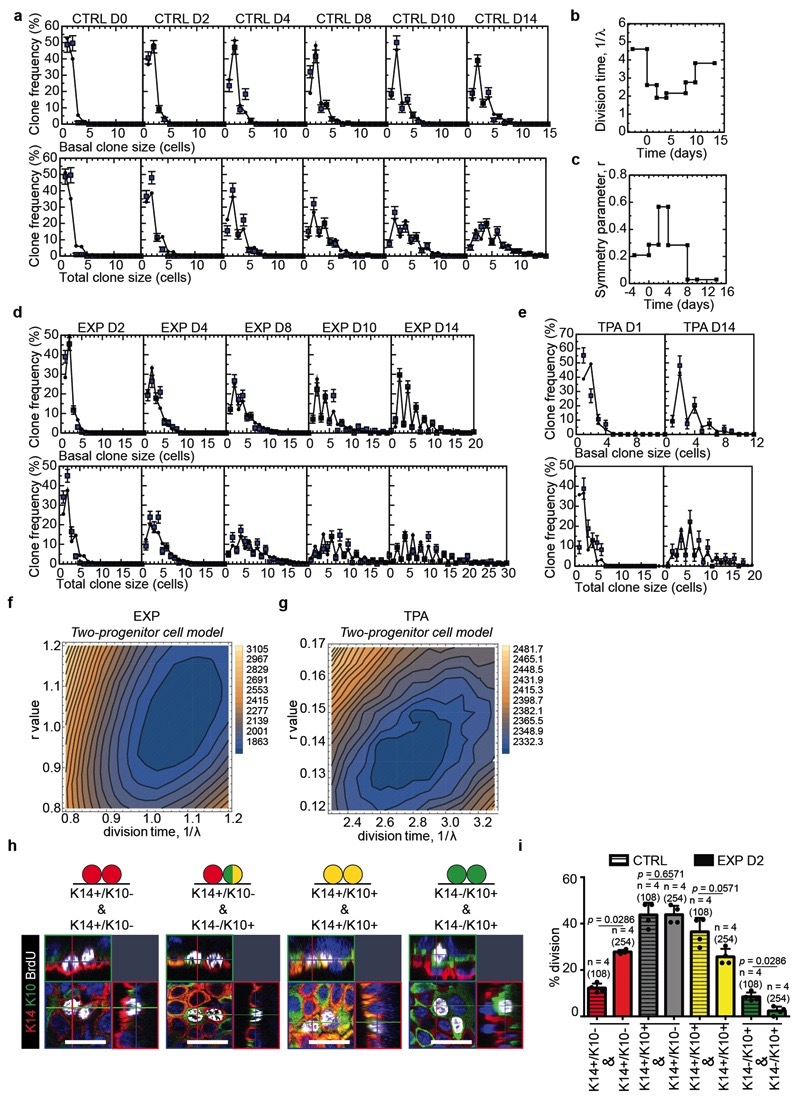
To define the fate dynamics of epidermal cells during stretch-mediated skin expansion, we performed clonal analysis on K14CREER/Rosa-Confetti mice. As described previously12,13,14, the steady increase in basal (and total) clone size over the two week time-course was compensated by a decrease in clone persistence so that the total labelled cell fraction remained constant over time. Further, the clone size distributions showed an exponential-like dependence (Fig. 2a-f and Extended Data Fig. 3a, b), consistent with the dynamics of a single equipotent cell population maintained through population asymmetric self-renewal, as found in other compartments of skin epidermis12,13,14,15.
Figure 2. Clonal analysis of epidermal SC during stretch-mediated skin expansion.
a, K14CREER-RosaConfetti clones (n=4 independent experiments). Second Harmonic Generation (SHG) visualizes the collagen fibers (white). 7AAD for nuclei (blue). Scale bars, 50 μm. b-k, Clonal analysis in control (CTRL) and expansion (EXP) conditions. (b,g), Distribution of clone sizes at D14 based on basal and total cell number, (c,h), average clone size based on basal (black) and total (blue) cell content, (d,i), clone persistence, (e,j), average labelled cell fraction, and (f,k), cumulative clone size distribution at D14 showing an approximate exponential size dependence (lines). b-f, D0: 115 clones from n=7 mice; D2: 175 clones from n=7 mice; D4: 136 clones from n=5 mice; D8: 159 clones from n=3 mice; D10: 146 clones from n=3 mice; D14: 195 clones from n=4 mice. g-k, D2: 231 clones from n=4 mice; D4: 197 clones from n=4 mice; D8: 199 clones from n=4 mice; D10: 157 clones from n=4 mice; D14: 199 clones from n=4 mice. l, Schematic showing the cellular organization in a one-progenitor model and the proposed two-progenitor model of back skin interfollicular epidermis. In the two-progenitor model, the epidermis contains renewing stem cells (SC), committed cells (CC), and suprabasal cells. In homeostasis, stem cells divide at an average rate λ. With probability 1—r, this division results in asymmetric fate outcome, leading to the replacement of the partner committed cell, which in turn is lost through terminal division and stratification from the basal layer. The remaining divisions lead to the correlated loss and replacement of renewing cells through symmetric cell divisions. c-f, h-k, Mean + s.d. c, d, e, h, i, j, Points show data and lines the results of a two-progenitor model (Supplementary Note).
During stretch-mediated expansion, the average clone size was increased by 3.4 fold in the basal and 6.8 fold in the total cell content, some two-fold larger than control conditions. However, in this case, the expansion in basal clone size was not compensated by a decrease in clone persistence (Fig. 2g-k and Methods), indicating that tissue expansion does not simply increase proliferation, but also changes the balance between renewal and differentiation.
To understand the changes that take place in cell fate during expansion, we first considered the homeostatic control condition. Despite its exponential form, detailed inspection of the clone size distribution revealed evidence of a statistically significant bias towards clones bearing an even number of basal cells (65±5% at D14), a striking feature that was also present in the total clone size distribution (57±4% at D14) (Fig. 2b). Remarkably, this feature was greatly enhanced in 12-O-Tetradecanoylphorbol-13-acetate (TPA) treated (Extended Data Fig. 3d-g) and expansion conditions (Fig. 2g, Extended Data Fig. 3h-j), both for basal (80±11% and 84±6%, respectively, at D14) and total (78±11% and 81±6%, respectively, at D14) clone size, suggesting that its origin did not derive from statistical noise or synchrony of division, but may lie in a niche-like organization of tissue. To develop this idea, we considered an arrangement in which back skin IFE comprised a mosaic of “units”, each of which plays host to two basal cells organized in a stem/progenitor celllike hierarchy with one basal cell belonging to a renewing (SC) compartment and the other a progenitor cell committed to differentiation through terminal division and stratification (Fig. 2l and Supplementary Note). To ensure the prevalence of even cell numbers in clones, we posited that the terminal division and stratification of progenitor cells is compensated predominantly by the asymmetric division of SC within the same unit. Further, to account for the expansion of clones beyond two basal cells, we proposed that correlated cell loss and replacement must also occur between neighbouring units. From a fit to the clonal data, we found that this model could predict the distributions of basal and total clone size with a division rate of once per 4.6 days, while some 4 out of 5 divisions of renewing cells result in asymmetric fate outcome, a figure resonant with that found in other skin compartments based on a model of intrinsic fate choice12–16. Notably, we found that a “one-progenitor” model based on the correlated differentiation and divisions of neighbours17 could not reproduce the fine features of the data (Fig. 2l, Extended Data Fig. 3k-p and Supplementary Note).
In common with control and TPA data, the distribution of clone sizes during expansion also showed convergence towards an exponential size dependence (Fig. 2k). We therefore questioned whether a minimal adjustment of the homeostatic model could predict the clone dynamics. Taking as an input the measured proliferation rate based on BrdU incorporation (Fig. 1e), we found that a continuous adjustment of fate imbalance could account for the average clone size increase, as well as the striking enrichment of even-sized clones, providing evidence that progenitor heterogeneity is conserved during expansion (Extended Data Fig. 4a-g and Supplementary Note).
To further challenge the two-progenitor model and the transient changes in cell fate following skin expansion, we marked cells during division using a BrdU single pulse-chase assay. We assessed the fate of proliferative cells during stretch, discriminating between daughter cells biased for renewal (K14+/K10-) and those committed to differentiation (K14+/K10+). We found that stretching increased the proportion of BrdU+/K14+/K10- cells, consistent with the model, demonstrating that expansion promotes an imbalance of the renewing population towards cell duplication (Extended Data Fig. 4h,i).
Molecular features related to stretch
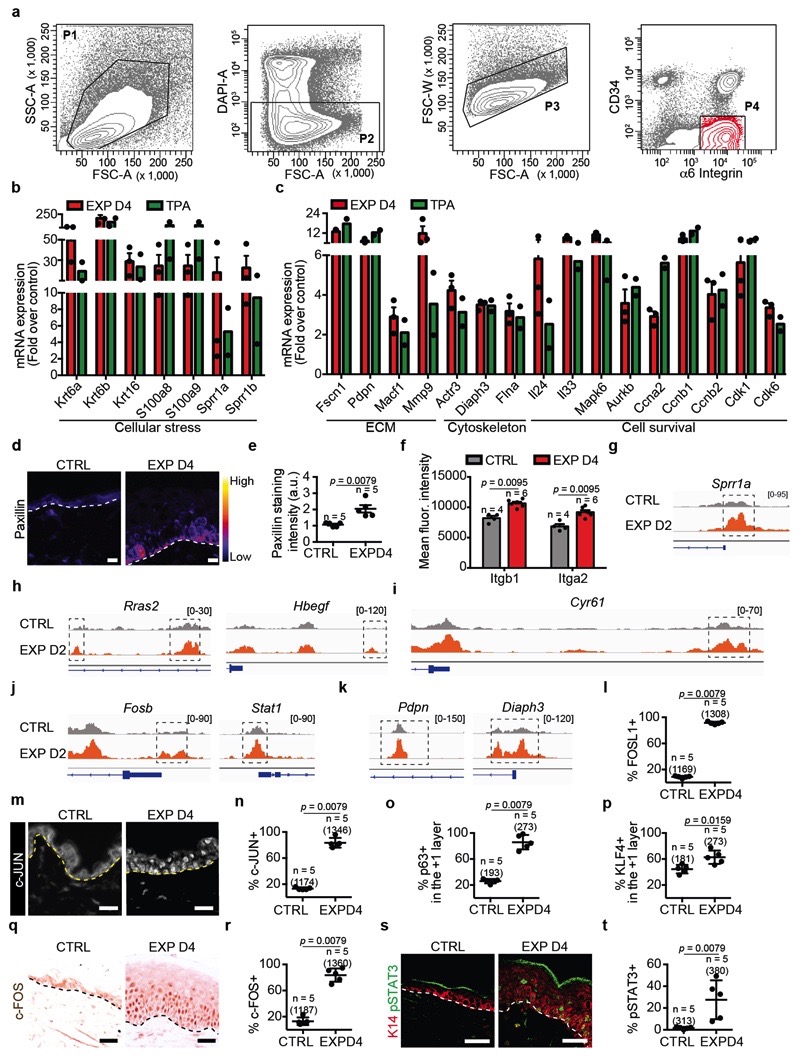
To define the changes in gene expression following expansion, we performed microarray analysis on FACS isolated basal cells in different conditions (Extended Data Fig. 5a). Genes upregulated both in expanded and TPA conditions comprised those regulating cell cycle, DNA replication, cell survival and cytoskeleton remodelling (Extended Data Fig. 5b,c). Many of these genes were also found to be upregulated during wound healing18, suggesting that they represent a common transcription programme linked to cellular stress and proliferation.
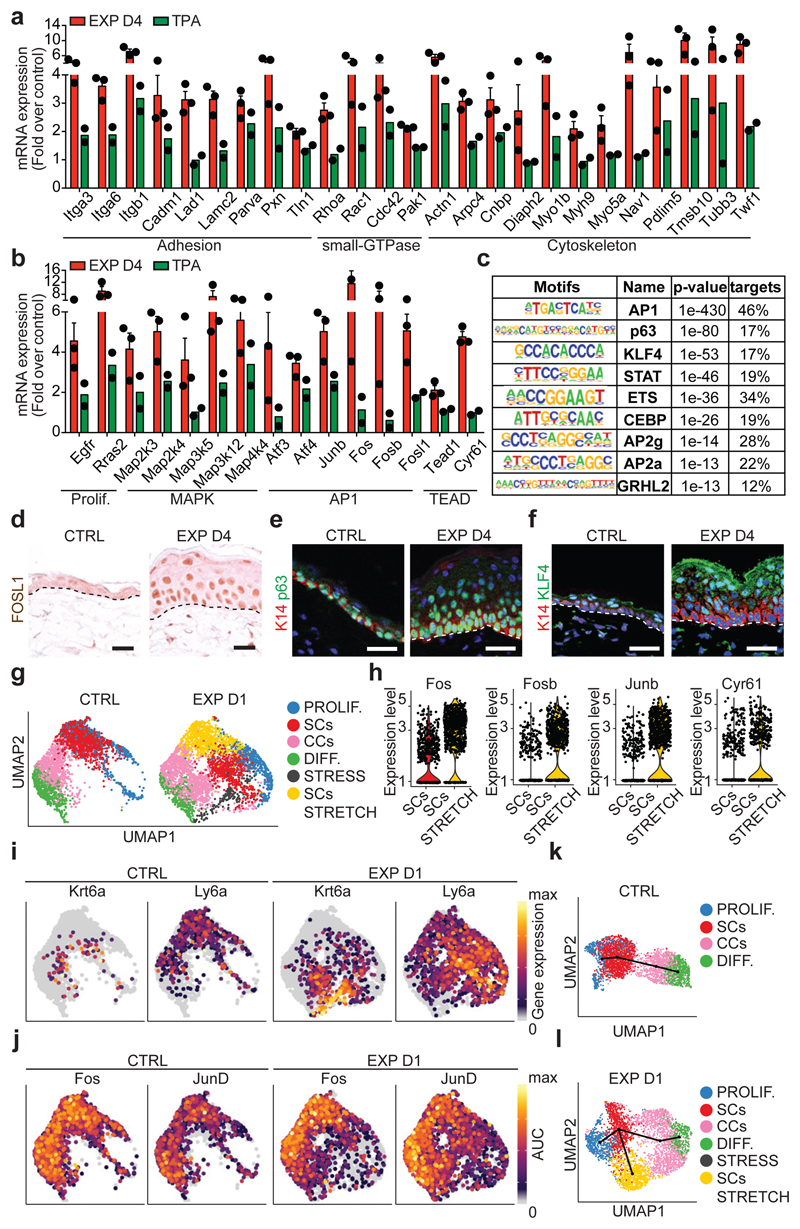
Genes that were preferentially upregulated in expanded conditions revealed enrichment for those related to cell-cell and cell-ECM adhesion, small GTP-ase, regulators of the actomyosin cytoskeleton (Fig. 3a), and genes regulating proliferation, including Egfr, Ras, mitogen-activated protein kinase (MAPK), Activator Protein 1 (AP1) transcription factors (TFs) (Junb, Fos, Fosb, Fosl1), and the YAP/TEAD pathway (e.g. Tead1 and the canonical YAP target gene Cyr61) (Fig. 3b). Consistently, we found changes in the expression of some of these adhesion and cytoskeleton proteins by immunostaining and FACS analysis (Extended Data Fig. 5d-f).
Figure 3. Transcriptional and chromatin remodelling associated with stretch-mediated skin expansion.
a, b, mRNA expression of genes upregulated in EXP D4 (n=3) compared to TPA (n=2). Bars are mean with s.e.m. of the fold change over the average value of the CTRL (n=3). c, TF motifs enriched in the ATAC-seq peaks that were upregulated in EXP D2 compared to CTRL (n=3262 target sequences, 46200 background sequences) as determined by Homer analysis using known motif search. d, Immunohistochemistry for FOSL1. e, f, Immunofluorescence for p63 (e) or KLF4 (f) in green, K14 (red) and nuclei with Hoechst (blue). d-f, Dashed lines delineate the basal lamina. Scale bar, 20 μm. n=3 independent experiments. g, Uniform Manifold Approximation and Projection (UMAP) graphic of the clustering analysis for the CTRL (n=3142 cells) and EXP D1 (n=3756 cells) IFE single-cell RNA-seq projected on an integrated embedding of the dataset. h, Violin plot of the indicated genes in EXP D1 in the SCs (n=700 cells) and SCs STRETCH (n=801 cells) clusters, see Source Data. i, UMAP plot coloured by normalized gene expression values for the indicated genes in the CTRL and EXP D1 IFE. Gene expression is visualized as a colour gradient going from grey to yellow, with grey as indicator of no expression (i.e. expression below the 50th percentile across each respective sample) and yellow as indicator of maximal expression. j, UMAP plots coloured by the degree of regulon activation for TFs differentially activated (AUC rank-sum test FDR corrected p-value < 0.05) in the different conditions. Colour scaling represents the normalized AUC value of target genes in the regulon being expressed as computed by SCENIC. k, l, Lineage trajectories (black lines) computed using Slingshot. i-l, CTRL (n=3142 cells) and EXP D1 (n=3756 cells).
To unravel changes in the chromatin landscape associated with expansion, we made use of transposase-accessible chromatin using sequencing (ATAC-seq) to identify the chromatin regions that were remodelled at D2 following expansion. To define the TFs associated with chromatin remodelling, we performed motifs discovery analyses on the chromatin-remodelled regions. The most frequent motifs associated with open chromatin regions corresponded to AP1, p63, Stat, Ets, Cepb, Ap2a and Grhl2 (Fig. 3c, Extended Data Fig. 5g-k and Supplementary Table 1 and 2). Immunostaining confirmed overexpression of FOSL1, c-FOS and c-JUN of the AP1 TF family, p63, pSTAT3 and Klf4 in the basal and early suprabasal cells (Fig. 3d-f and Extended Data Fig. 5l-t). These results show that stretch-mediated skin expansion is regulated by the EGFR/Ras/MAPK pathway, leading to the activation of Jun/Fos19 TFs, as well as TFs, such as p63, that mediate epidermal stem cell renewal20, and TFs associated with differentiation such as Cebp21, Klf422 and Grhl223, allowing skin expansion while maintaining skin barrier functions.
scRNA-seq during stretching
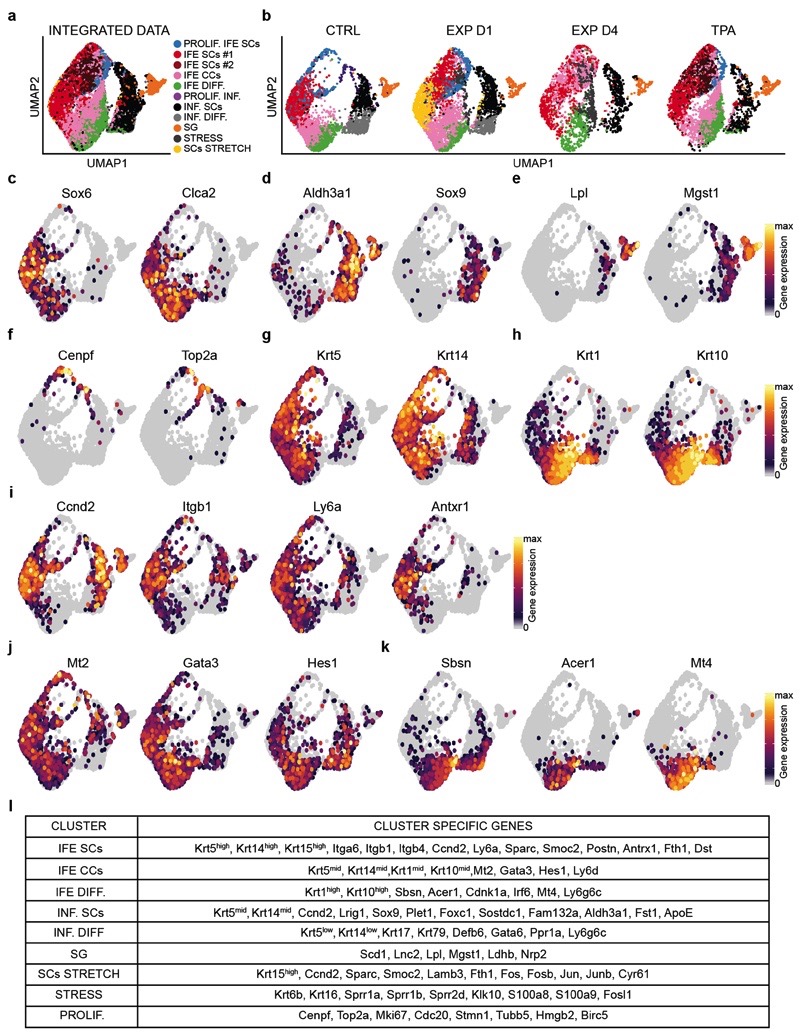
To assess whether all basal cells respond equally to stretch, we performed single-cell RNA sequencing (scRNA-seq) on FACS-isolated cells enriched for basal IFE, infundibulum (INF), sebaceous gland (SG) and basal upper hair follicle cells in control (CTRL) (n=4659) conditions, after TPA treatment (n=4342), and following expansion (EXP D1: n=4934 and EXP D4: n=2716) (Extended Data Fig. 5a). Using graph-based clustering, we identified different subpopulations based on marker gene expression. These different clusters were found in all conditions (Extended Data Fig. 6a-l), and were similar to the clusters previously described by Kasper and colleagues24,25.
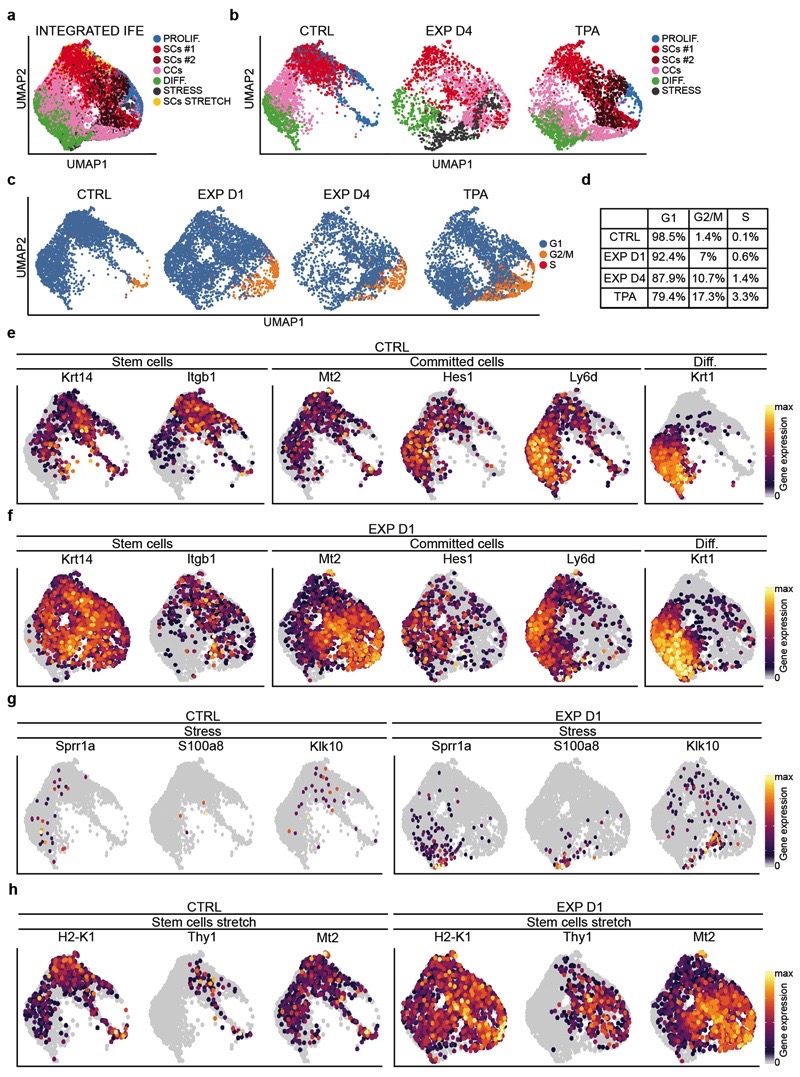
Based on the clonal analysis, we first questioned whether the scRNA-seq data supported the existence of proliferative heterogeneity in the IFE. When only IFE cells were analysed in control, we identified a population of undifferentiated cells with a stem cell (SC)-like signature, a population of proliferative basal committed cells (CCs), co-expressing basal and early differentiation markers such as Krt1 and Krt10, and a population of non-cycling cells expressing differentiation markers (Fig. 3g). The same IFE populations were found in all conditions; although the proliferative activity of SCs and CCs were proportionately increased in expansion and TPA conditions (Fig. 3g, Extended Data Fig. 7a-f and Supplementary Table 3).
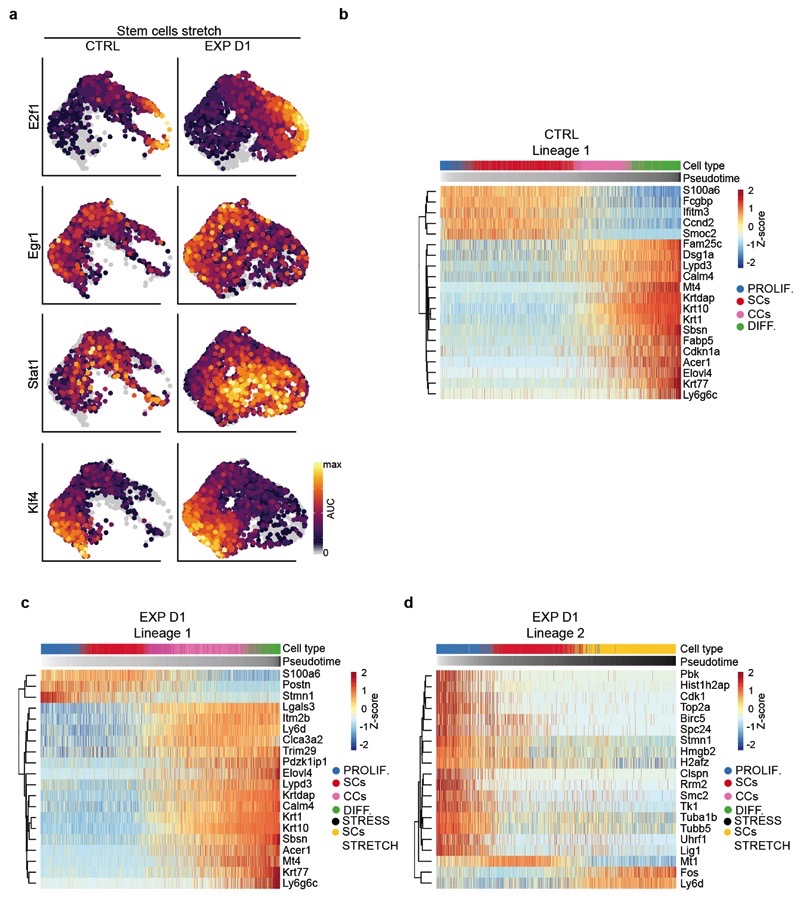
In addition, new cell states expressing genes associated with stress and hyperproliferation (e.g. Krt6a, Sprr1a, S100a8, Klk10) were found in expansion (Fig. 3g and Extended Data Fig. 7g). Importantly, the stem cell-like “stress” cluster (SCs STRETCH) appearing at EXP D1, identified by the expression of basal markers (e.g. Krt14, Itgb1) and higher expression of Ly6a, H2K1, Thy1 and Mt2 (Fig. 3g-i and Extended Data Fig. 7h), presented increased activity of TFs regulating proliferation and immediate early genes (e.g. E2f1 and Egr1) as well as AP1 TFs (e.g. Fos, Junb, Jund), inflammation (e. g. Stat1, Stat3) and commitment/differentiation (e.g. Klf4), as analysed by SCENIC analysis26 (Fig. 3j and Extended Data Fig. 8a and Supplementary Table 4-6), suggesting that only a fraction of basal cells respond transcriptionally to mechanical stress.
To determine the lineage trajectories between the different subpopulations, we performed pseudotemporal ordering of cells using Slingshot27 in CTRL and EXP D1. In all conditions, we found a trajectory starting from undifferentiated basal SCs, passing through the progenitor state, and terminating with the most differentiated cells (Fig. 3k,l and Extended Data Fig. 8b). In EXP D1, we resolved another differentiation pathway involving progression through the “stress states”, delineating the different responses to mechanical cues (Fig. 3l and Extended Data Fig. 8c,d). Together, these data show that SCs present a rapid and profound response to stretch.
Mechanosensing at adherens junctions
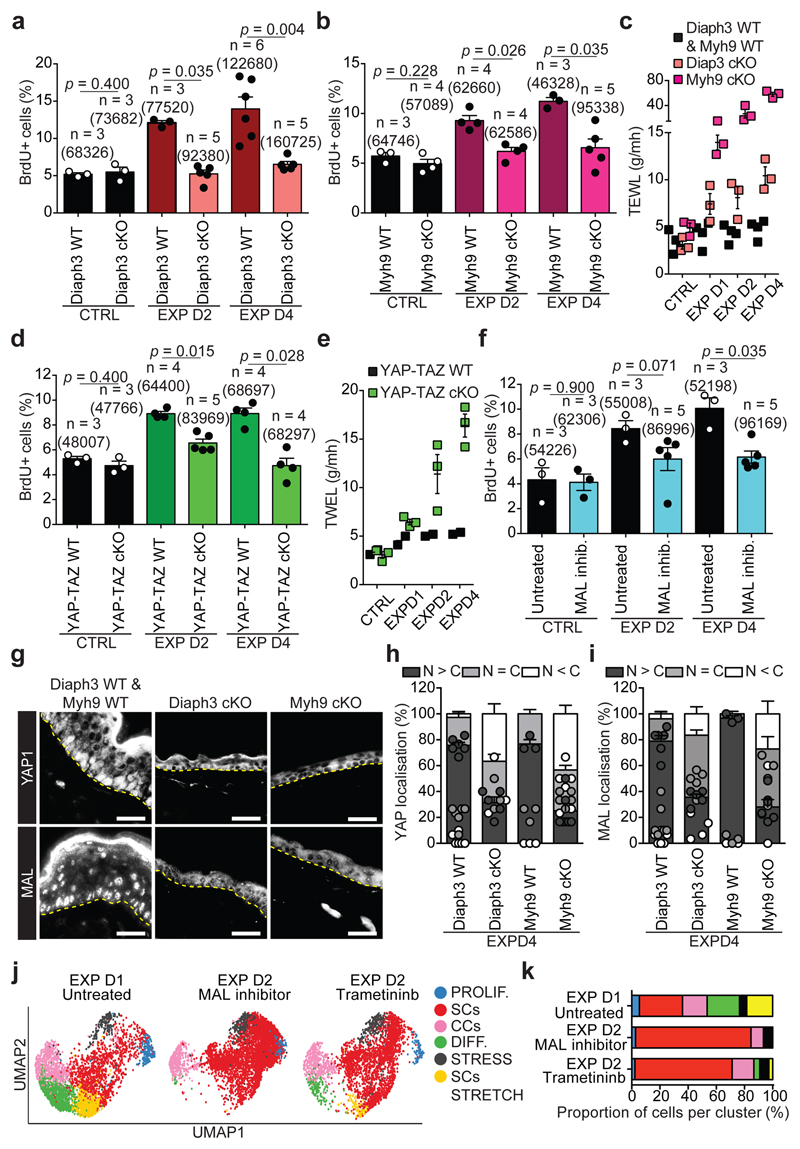
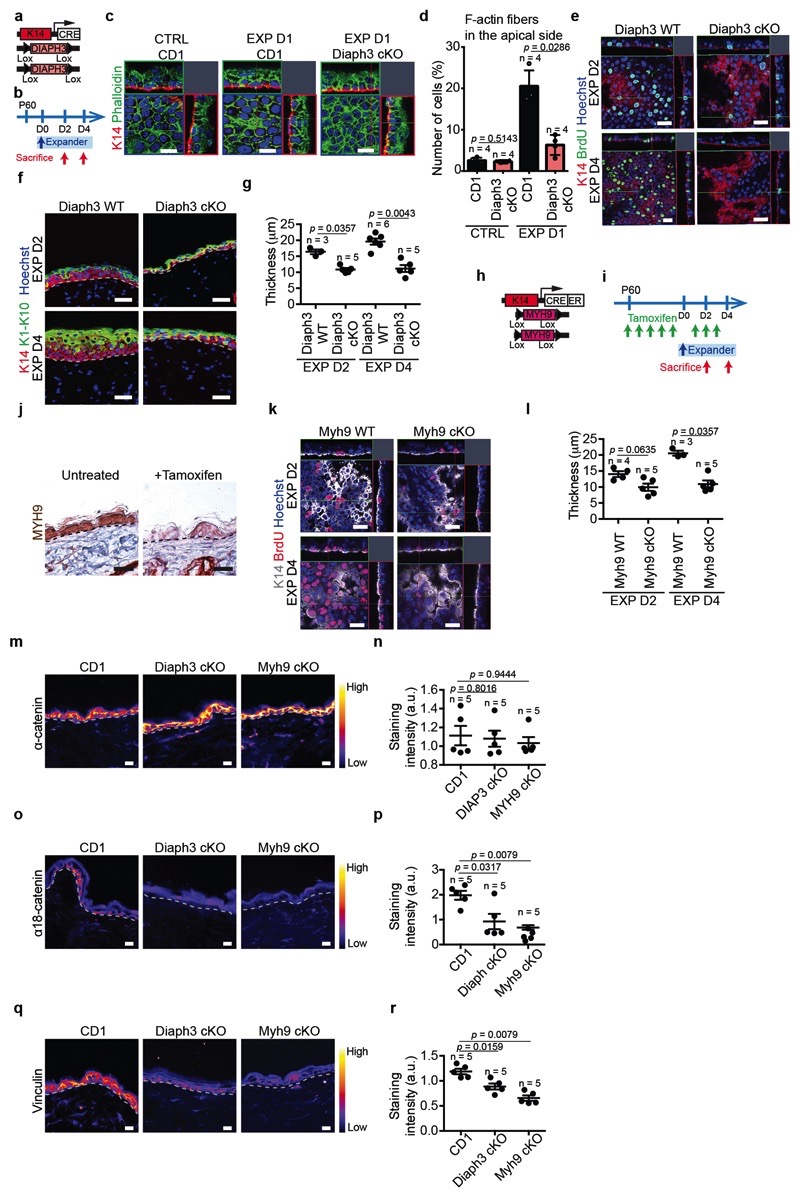
The results of TEM and molecular analysis suggested that stretching induces remodelling of the cytoskeleton, as well as an increase at the transcription level of several genes encoding remodellers of the actomyosin cytoskeleton, such as Diaph2 and Diaph3, and formin-like proteins involved in actin regulation28 (Fig. 3a, Extended Data Fig. 1e-g,k and Extended Data Fig. 5c,k). To test whether rearrangements of actin organisation are essential to mediate the response to stretch, we studied the consequence of Diaph3 deletion (Diaph3 cKO). The increase in the number of cells presenting F-actin structures on their apical surface at EXP D1 was reduced and no increase in cell proliferation following stretch was observed in Diaph3 cKO, (Fig. 4a and Extended Data Fig. 9a-g).
Figure 4. Molecular regulation of stretch-mediated skin expansion.
a, BrdU positive cells in CTRL and EXP in Diaph3 WT or cKO mice. b, BrdU positive cells in CTRL and EXP in Myh9 WT and cKO mice. c, Trans-epithelial water loss (TEWL) measurements from n=2 Diaph3 WT and n=2 Myh9 WT mice (black), n=3 Diaph3 cKO mice (pink) and n=3 Myh9 cKO mice (violet). d, BrdU positive cells in CTRL and EXP in YAP-TAZ WT or cKO mice. e, TEWL measurements from n=2 YAP-TAZ WT mice (black) and n=3 YAP-TAZ cKO mice (green). f, BrdU positive cells in untreated or treated animals with the MAL inhibitor in CTRL and EXP. g, Immunostaining for YAP1 (top) and MAL (bottom) on skin sections of Diaph3 cKO and Myh9 cKO mice at EXP D4. The CTRL images are from a Diaph3 WT (top) mouse and a Myh9 WT (bottom) mouse. Dashed lines delineate the basal lamina. Scale bar, 20 μm. h, i, Quantification of YAP1 (h) and MAL (i) subcellular localization, presented as mean and s.e.m., related to g. N>C, more protein in nucleus than in cytoplasm, N=C, similar level of protein in nucleus as in cytoplasm, N<C, less protein in nucleus than in cytoplasm (n=150 cells per condition). j, UMAP computed on the integrated dataset coloured fort the different cellular clusters in IFE single-cell RNA-seq. n=3869 cells EXP D1 Untreated, n=4762 cells EXP D2 MAL inhibitor, n=3254 cells EXP D2 Trametininb. k, Percentage of the different cellular clusters in j. a-f, Mean + s.e.m. n=number of mice. Total number of cells analysed indicated in parentheses. a, b, d, f, Two-tailed Mann–Whitney test. c, e, Every data point represents the average of 30 individual subsequently recorded measurements at the probe head.
Mechanical stretch has been shown to promote the phosphorylation of Myosin II, relaying mechanotransduction in cell lines in vitro2 and in the Drosophila wing disc29. Consistently, following conditional ablation of Myh9, a key subunit of Myosin IIA (Myh9 cKO), proliferation was not increased in response to stretch (Fig. 4b and Extended Data Fig. 9k,l).
Since basal cells sense stretching by remodelling the traction at adherens junctions, we assessed whether Diaph3 and Myh9 cKO prevent adhesion remodelling. The accessibility of the alpha-catenin tension-sensitive epitope and vinculin staining were not increased following stretching in Diaph3 and Myh9 cKO (Extended Data Fig. 9m-r). Importantly, the deletion of Diaph3 and Myh9 following expansion resulted in the incapacity of the epidermis to adapt to stretch, causing a barrier defect (Fig. 4c).
MEK/ERK/AP1 regulate expansion
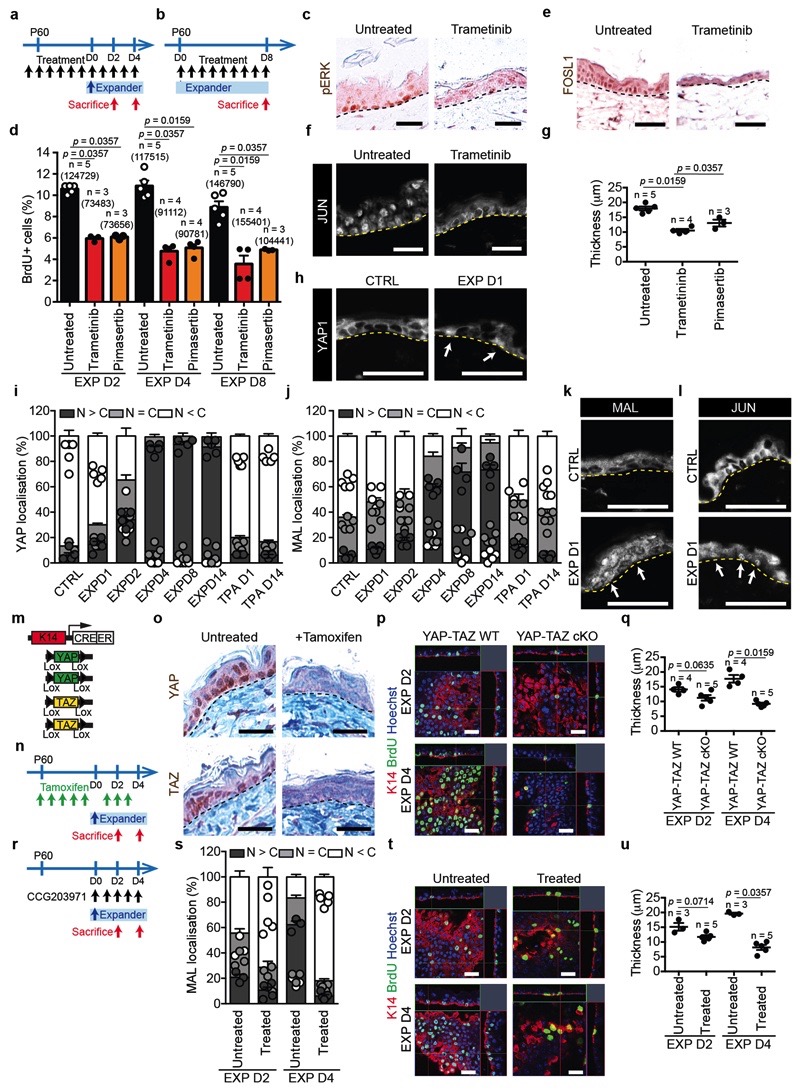
Previous studies demonstrated that in vitro mechanical stretching induces proliferation by activation of MEK (mitogen-activated protein kinase kinases) signalling30, and that ERK is activated on stress fibers in a myosin II-dependent manner31. To assess whether pharmacological inhibition of MEK/ERK pathway could impair the cellular behaviour following skin stretching, we treated mice with MEK inhibitors Trametinib and Pimasertib. Both inhibitors reduced pERK, and resulted in the decrease of cell proliferation and the number of differentiated cells induced by stretching (Extended Data Fig. 10a-g), demonstrating the importance of MEK/ERK/AP1 signalling on the regulation of stretch response in vivo.
YAP and MAL regulate skin stretching
Previous studies have shown that the yes-associated protein 1 (YAP1), the downstream co-effector of Hippo signalling, and Megakaryoblastic leukemia/myocardin-like 1 (MKL1 or MRTFA or MAL), the co-partner of Serum response factor (SRF), are induced in response to mechanical stimuli32,33. In the skin, YAP-TAZ and SRF pathways are essential for epidermal development and wound repair, but are dispensable for homeostasis34,35. In vitro studies have demonstrated the role of MAL/SRF in the survival and differentiation of keratinocytes cultured on micropatterned surfaces36, and YAP as a driver of proliferation in stretched cultured cells in vitro 37.
To test whether and when YAP and MAL were activated in vivo, we assessed their subcellular localization at different times during expansion. While YAP and MAL were localized preferentially in the cytoplasm in the control, they gradually translocated to the nucleus of basal cells during expansion (Extended Data Fig. 10h-k). These data reveal that YAP and MAL are activated immediately following stretching, but only in a subset of basal cells, similar to AP1 family members (Extended Data Fig. 10l).
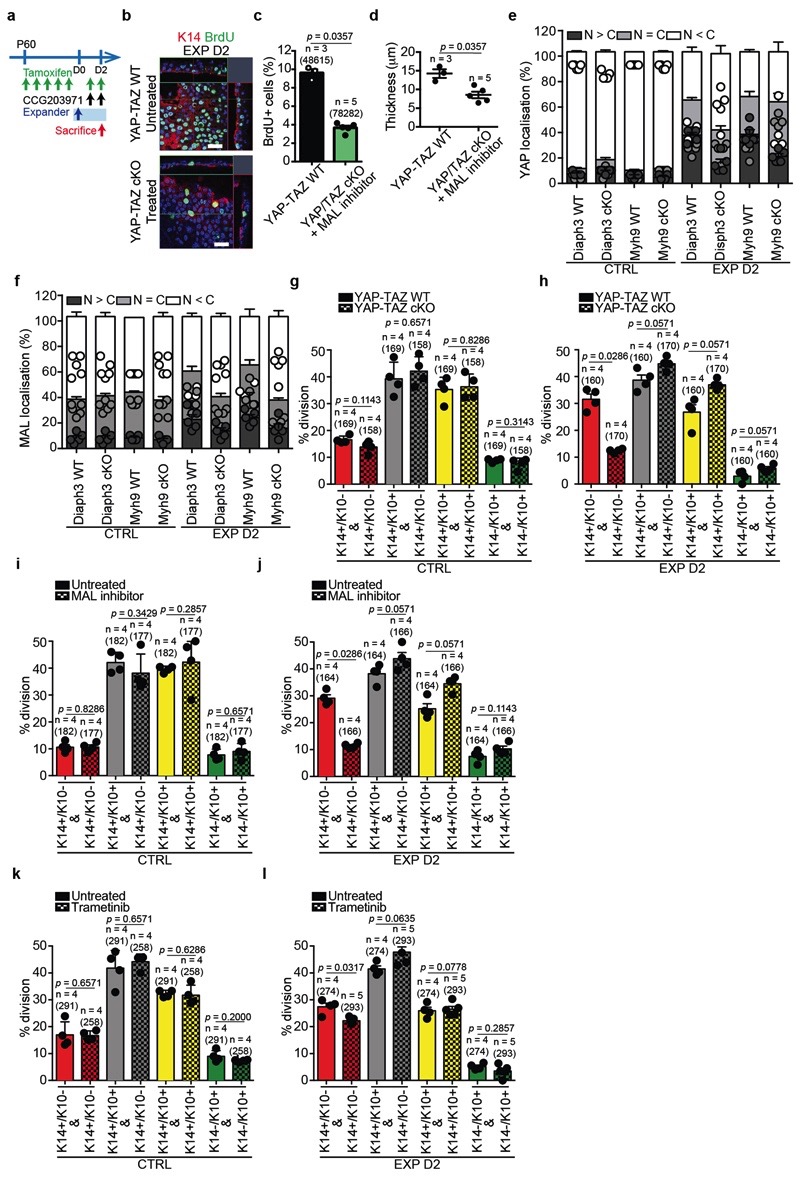
To define the functional role of YAP, we induced deletion of Yap1 and Taz specifically in the epidermis (YAP-TAZ cKO) and assessed their role during stretch. A significant decrease in BrdU incorporation was observed already by D2 in YAP-TAZ cKO epidermis, as well as a reduction in epidermal thickness (Fig. 4d and Extended Data Fig 10m-q). Additionally, the deletion of YAP-TAZ following hydrogel expansion caused a barrier defect (Fig. 4e).
To assess the role of MAL activity, we treated the animals immediately after surgery with the pharmacological MAL/SRF inhibitor CCG2039738, and found a significant decrease of BrdU incorporation and epidermal thickness at D4 (Fig. 4f and Extended Data Fig. 10r-u). Interestingly, inhibition of both pathways blocked completely the response to stretch (Extended Data Fig. 11a-d).
As Diaph3 and Myh9 control adhesion remodelling upon stretching, we tested whether they act upstream of YAP and MAL. In Diaph3 and Myh9 cKO epidermis, YAP and MAL were not translocated into the nucleus following stretch (Fig. 4g-i and Extended Data Fig. 11e,f), demonstrating the essential role of Diaph3 and Myh9 in regulating canonical mechanotransducers. Altogether, these experiments demonstrate that mechanical stretch couples cytoskeletal contractility with the nuclear effectors of mechanotransduction and stem cell activation in vivo.
scRNA-seq after MEK and MAL inhibition
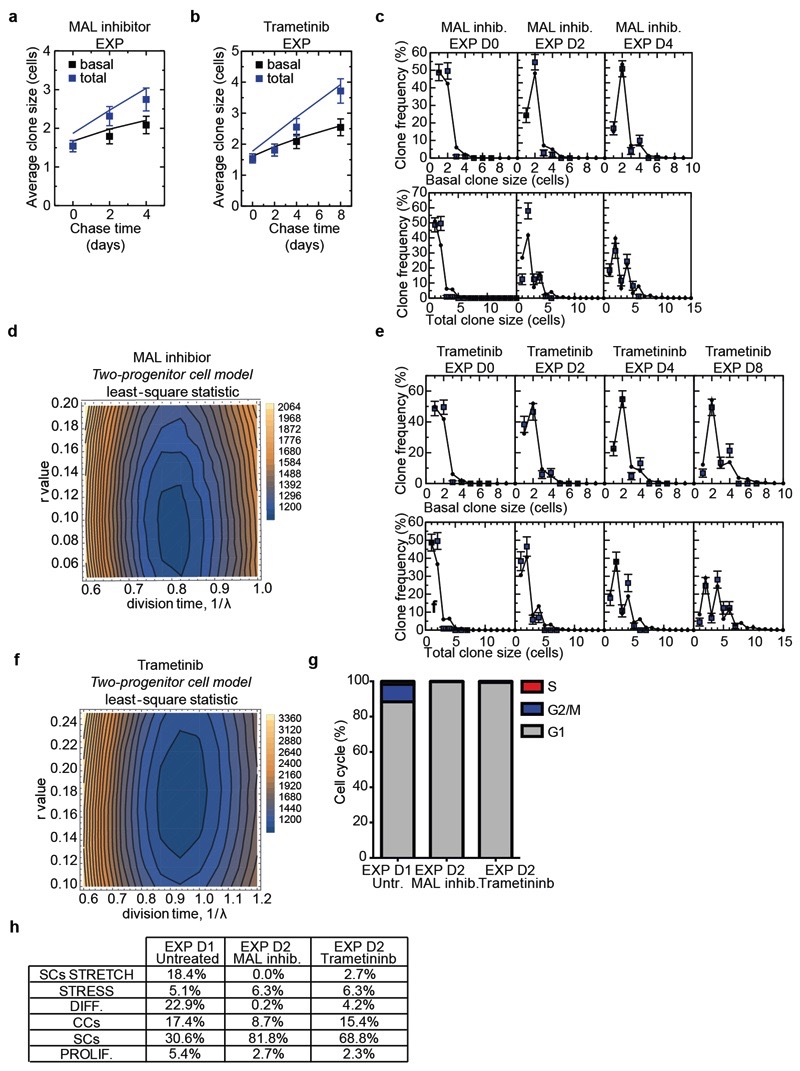
To define the consequences of the inhibition of the signalling pathways that control stretch-induced proliferation on the cellular heterogeneity and SC dynamics in vivo, we performed short-term lineage tracing of basal cells using BrdU pulse-chase combined with immunohistochemistry. The results showed a decrease in renewing divisions (BrdU+/K14+/K10-) in the YAP-TAZ cKO animals and upon inhibition of MAL/SRF or MEK/ERK/AP1 (Extended Data Fig. 11g-l). In addition, short-term genetic lineage tracing in the presence of MAL/SRF or MEK/ERK/AP1 inhibition showed suppression of the increase of clone size following skin expansion (Extended Data Fig. 12a-f). scRNA-seq at D2 following expansion in the presence of the MEK/ERK/AP1 and MAL/SRF inhibitors showed that, while the inhibition of both pathways led to a similar decrease in cell proliferation, the proportion of cells in different states differed between treatments. Cellular heterogeneity was relatively well conserved following the inhibition of the MEK/ERK/AP1 pathway, while the MAL-SRF inhibitor diminished the abundance of SC with stress characteristics and decreased the heterogeneity of basal cells (Fig. 4j,k, Extended Data Fig. 12g,h and Supplementary Table 7-9), suggesting that while MEK/ERK/AP1 pathway controls only proliferation induced by stretching, the MAL/SRF pathway controls cellular heterogeneity in the basal layer and stretch sensing.
Discussion
Here, we have dissected the cellular and molecular spatio-temporal mechanisms that control the behaviour of epidermal stem cells in response to mechanical stretch. Quantitative modelling of clonal data suggests that proliferation and differentiation of basal cells in back skin epidermis is spatially and temporally linked, as has been suggested during homeostasis17,39. During stretch-mediated skin expansion, a transient shift of SC fate towards renewal allows the basal population to expand, while differentiation is maintained. These results were supported by scRNA-seq, which revealed the existence of different cell states within the basal layer, identifying the molecular signature of stem and committed cells in the mouse back skin, and highlighting the heterogeneous response of basal cells to stretching.
Mechanistically, we found that regulators of the actomyosin cytoskeleton, including formin-like proteins and non-muscle myosin, are essential for sensing skin stretching in vivo, and act upstream of canonical mechanotransduscers, such as YAP and MAL. Interestingly, the same signalling pathways are activated during embryonic pancreas development40 and in the fly notum upon mechanical compression41, suggesting that these signalling pathways and transcriptional regulators play a conserved role in mechanotransduction across the animal kingdom and between embryonic development and adult tissue regeneration.
Methods
Mouse strains
K14CREER 42 and K14CRE 43 transgenic mice were kindly provided by E. Fuchs (Rockefeller University). Rosa-Confetti 44 mice were provided by H. Clevers (Hubrecht Institute). YAPfl/fl and TAZfl/fl mice45 were obtained from Georg Halder (KU Leuven) who received them from Randy Johnson at the MD Anderson Cancer Center, Houston. DIAPH3fI/fI 46 mice were produced by Fadel Tissir (UCLouvain) and MYH9fI/fI 2 mice were a kindly gift from Sara Wickstrom (University of Helsinki). Rosa26-mT/mG 47 mice were provided form Isabelle Migeotte (Université Libre de Bruxelles). Mice colonies were maintained in a certified animal facility in accordance with European guidelines. The experiments were approved by the local ethical committee (CEBEA) under protocols #604 and #605. The study is compliant with all relevant ethical regulations regarding animal research.
Expander experiments
Mice were anesthetized (5% xylazine 10% ketamine in PBS) and the dorsal skin was disinfected with 10% Iso-Betadine (Meda Pharma), an incision was created in the most caudal part of the dorsal skin to minimize any tension in the wound as the expansion progressed and to maximise the distance from the access wound, to the location of the expander. A subcutaneous pocket was created with forceps and a 4ml Hemispher Self-inflating tissue expander (Osmed™) was placed in the most rostral part under the dorsal skin in proximity of the neck. Stiches were used to close the subcutaneous pocket, to limit the hydrogel movement and to close the access incision. All analyses were performed in the area on the top of the dome of the hemisphere induced by the skin expander, since this region experiences the highest strain. The hemispherical architecture of the hydrogel has, by its geometric nature, a higher degree of mechanical load at the apex of the hemisphere with isotropic stress48 and only the skin overlying the very upper part of the hydrogel was studied.
Sample size, randomization and blinding
The sample size was chosen based on previous experience in the laboratory, for each experiment to yield high power to detect specific effects. No statistical methods were used to predetermine sample size. The experiments were not randomized. All animals used were of similar age (60-90 days after birth) and between 28 and 33g of weight. The investigators were not blinded to allocation during experiments and outcome assessment.
Skin whole-mount
For skin whole-mount confocal microscopy on induced K14CREER-RosaConfetti mice, pieces of entire skin of around 1 cm2 were cleared with Sca/eCUBIC-1 and Sca/eCUBIC-2 solutions as described in49. The pieces were fixed overnight in 4% paraformaldehyde, incubated 3 days at 37°C in Sca/eCUBIC-1 solution and incubated at room temperature (RT) in Sca/eCUBIC-2 solution for at least 2 days before their analysis. Counterstaining of nuclei was performed with 7AAD (1/1000, Thermo Fisher Scientific). Images were acquired at LiMiF http://limif.ulb.ac.be/ (Université Libre de Bruxelles, Faculté de Médecine, Campus Erasme) on a LSM780NLO confocal system fitted on an Observer Z1 inverted microscope (Zeiss, Iena, Germany) equipped with a Chameleon Vision II 690-1064 nm multiphoton laser (Coherent Europe, Utrecht, The Netherlands) using a Plan Apochromat 20x/0,8 dry objective or LD C Apochromat 40x/1.1 water immersion objective (Zeiss, Iena, Germany). Spectral images were acquired in “lambda mode” with a single excitation wavelength at 920 nm and a GaAsp spectral detector (Zeiss). Images 512 by 512 pixels were acquired as a z-stack across the thickness of the object. Fluorochromes were separated by linear unmixing using ZEN2012 software (Zeiss). Settings were kept identical for all conditions – this does not apply to the depth (z stack) that was adjusted individually to encompass the region of interest. Single plane images were displayed using Zen2012 (Black Edition) software (Zeiss) and exported as uncompressed TIFF images.
Clonal induction in K14CREER-RosaConfetti mice
For lineage tracing experiment, K14CREER-RosaConfetti mice were induced at 2 months of age with Tamoxifen (Sigma) (0.1 mg/g diluted in 10% vol/vol Ethanol and sunflower seed oil, Sigma) by intra-peritoneal (IP) injection.
Inducing YAP-TAZ and MYH9 deletion
For YAP-TAZ deletion in K14CREER YAPfl/fl-TAZ fl/fl mice and for MYH9 deletion in K14CREER MYH9fl/fl mice, animals received a daily dose of 2,5mg of tamoxifen (Simga-Aldrich), for 30g of body weigh, diluted in a mixture of 10% ethanol (VWR) 90% sunflower oil (Sigma-Aldrich) by IP administration, for 5 consecutive days and every other day after surgery.
Mouse treatments
For TPA treatments, TPA (200 μl of 0.02 mg/ml solution in acetone) was administered daily to shaved mouse back skin for 2 days. To inhibit MEK1/2 activity, mice were treated with Trametinib 2mg per kg body weight by daily oral gavage, Pimasertib 20mg per kg body weight daily by oral gavage for at least 7 consecutive days before analysis or as indicated in the figures. To inhibit MAL activity, mice received drug treatment via IP injection daily using 100mg CCG203971 (Cayman Chemical) per kg body weight for 2 or 4 days as indicated in the figures. For the dexamethasone experiments, dexamethasone powder (Sigma) was resuspended at 1 mg ml-1 in ethanol 100% and diluted 5 × in sterile PBS. The mice were injected intraperitoneally once per day at the dose of 1 mg kg-1. The treatment started 2 days before the surgery to place the expander and was sustained until the end of the experiment.
Monitoring hydrogel expansion, skin growth, quantification of clone size and persistence
To measure the increase in the hydrogel volume, the height or radius of the expander (r) was measured every day (D) from D0 to D14 and the volume calculated as the volume of half sphere with the formula V = 2/3πr3.
To measure basal cell area, whole-mount tissues were obtained from Rosa26-mT/mG mice that were fixed by PFA perfusion. Images were acquired by two-photon confocal microscopy (40x objective, z=1 μm). Basal cells were identified based on the orthogonal view and if their basal side was in direct contact with the signal of the second harmonic generation. Cell area was measured using ImageJ after manually drawing the contour of each cell revealed by the fluorescence on the membrane on the Rosa26-mT/mG mice in the x-y plan.
To define the clone persistence, hair follicles were used as landmarks to calibrate the degree of expansion. During expansion, the total number of hair follicles is not changed, implying that, for an area expansion by a factor ε, the hair follicle density must fall by a factor l/ε. If the area a of each hair follicle remains unchanged during the expansion, for a patch of tissue of area A containing hair follicles spanning an area AHF, the fractional increase in area during the expansion is given by where is the corresponding area of hair follicles in the control animal. Then, with an average of n clones in an area A, the clonal fractional change in clonal persistence is given by εn/n0, where n0 represents the corresponding clone number in the control animals. For clonal persistence, large areas of tissue (at least 1 cm2 per animal) were acquired using the 25x objective and the number of basally attached clones was quantified on the two-photon confocal images in a fixed area of 4mm2 across the different time points.
To quantify the size of the clone, whole-mounts obtained from K14CREER-RosaConfetti mice were imaged by two-photon confocal microscopy. Orthogonal view was used to see in 3 dimensions RFP, YFP, mCFP or nGFP positive cells and quantify the number of basal and total cells per clone.
Cells were considered as basal when their basal side was in direct contact with the signal of the second harmonic generation.
Epidermal whole-mount and immunostaining
Almost the entire back skin was dissected from the animal, the fat was removed by mechanical scraping with a scalpel and the remaining tissue was cut in pieces of around 2 cm2 and incubated in PBS/EDTA (20mM) on a rocking plate at 37°C for 90 minutes. Epidermis was separated from the dermis using forceps as an intact sheet and washed 2 times with PBS. Pieces of epidermis were prefixed in 4% paraformaldehyde for 1 hour at RT. Epidermis was rinsed 2 times with PBS for 5 min and conserved in PBS with 0.2% azide at 4°C. For immunofluorescence staining, entire pieces of epidermis were incubated in blocking buffer (1% BSA, 5% horse serum, 0.8% Triton in PBS) for 3 hours at RT on a rocking plate (100 rpm). The samples were incubated in primary antibody with anti-Integrinβ4 (rat, 1:200, BD Biosciences) overnight at 4°C. Samples were then washed 3 times in PBS with 0.2% tween during 1 hour and incubated in appropriate secondary antibodies diluted 1:400 in blocking buffer for 1 hour at RT on the rocking plate. For BrdU staining, samples were incubated in HCl 1M at 37°C for 35 min, washed with PBS 0.2% tween, stained with anti-BrdU (rat, 1:200, Abcam) in blocking buffer and with appropriate secondary antibody. The following secondary antibodies were used: anti-rat conjugated to AlexaFluor488 (Molecular Probes), to Rhodamine Red-X or to Cy5 (Jackson Immuno Research). Alexa488 or Alexa633 conjugated phalloidin (Thermo Fisher Scientific) was used 1:200 in blocking buffer to visualize F-actin microfilaments. Nuclei were stained in Hoechst solution diluted 1:5000 for 30 min and mounted in DAKO mounting medium supplemented with 2.5% Dabco (Sigma). All confocal images were acquired at RT with a LSM780 confocal system fitted on an AxioExaminer Z1 upright microscope equipped with C-Apochromat 40x/1.1 or Plan Apochromat 25x/0.8 water immersion objectives (Zeiss, Iena, Germany). Optical sections 512 x 512 pixels were collected sequentially for each fluorochrome. The data sets generated were merged and displayed with the ZEN2012 software (Zeiss).
Histology and Immunostaining on sections
Skin epidermis was embedded in OCT and kept at -80°C. Sections of 6 μm were cut using a CM3050S Leica cryostat (Leica Mycrosystems). After fixation in 4% paraformaldehyde for 10 minutes at RT, tissues were washed 3 times in PBS for 5 min and incubated in blocking buffer (1% BSA, 5% Horse serum, 0.2% Triton in PBS) for 1h at RT. Primary antibodies were incubated overnight at 4°C. Sections were rinsed 3 times in PBS and incubated with secondary antibodies and Hoechst in blocking buffer for 1h at RT. Sections were again washed three times with PBS. The following primary antibodies were used: anti-K14 (chicken, 1:20000, custom batch, Thermo Fischer); anti-K1 (rabbit, 1:4000, Covance/IMTEC, PRB-165P); anti-K10 (rabbit, 1:4000, Covance/IMTEC, PRB-159P); anti-c-JUN (rabbit, 1:200; Proteintech, 24909-1-AP); anti-p63 (rabbit, 1:100 Abcam, ab124762); anti-KLF4 (rabbit, 1:100 Abcam, ab129473); anti-pSTAT3 (rabbit, 1:100 Cell signaling, 9145); anti-YAP1 (rabbit, 1:100 Proteintech, 13584-1-AP); anti-MAL (rabbit, 1:100, Sigma, HPA030782); anti-α-catenin (rabbit, 1:1000 Sigma, C2081); anti-α18-catenin (rat monoclonal, 1:20000) was a generous gift form Akira Nagafuchi, Kumamoto University; anti-Vinculin (mouse, 1:200 Millipore MAB3574); anti-p120-catenin (rabbit, 1:1000 Invitrogen PA5-82545); anti-E-cadherin (rat, 1:500, ebioscience 14-3249-82); anti-ZO-1(rabbit, 1:200 Invitrogen 61-7300); anti-Claudin-1 (rat, 1:100 Thermo Scientific 51-9000); anti-Paxillin (rabbit, 1:100 Abcam 32084); anti-CD45 (rat, 1:500, BD Biosciences 553081); anti-CD68 (rabbit, 1:100 Abcam ab125212).
The following secondary antibodies were used diluted to 1:400: anti-mouse, anti-rat, anti-rabbit, anti-chicken conjugated to Alexa Fluor 488 (Molecular Probes), to rhodamine Red-X (Jackson Immunoresearch) or to Cy5 (Jackson Immunoresearch). Nuclei were stained in Hoechst solution (1:2000) and slides were mounted in DAKO mounting medium supplemented with 2.5% Dabco (Sigma). All images of section immunostaining were acquired using the Axio Imager M1 Microscope, the AxioCamMR3 or MrC5 camera and using the Axiovision software (Carl Zeiss). Acquisitions were performed at RT using 20x numerical aperture (NA) 0.4 (Carl Zeiss).
Immunofluorescence intensity measurements
To quantify the intensity of the immunostaining for proteins at the adherens junctions (α-catenin, α18-catenin, β-catenin, p120-cathenin) and Paxillin, ImageJ was used to measure the integrated density, a well-established method of measuring fluorescence intensity that accounts for differences in area50 of the signal in the basal layer of the skin (labelled by K14). To quantify the integrated density of the signal of the tight junctions proteins ZO-1 and Claudin-1, the measurements were taken on the upper suprabasal layers. Images are shown with the pseudo-colour “Fire” from ImageJ after they have been contrasted equally and background was uniformly removed with Adobe Photoshop.
Immunohistochemistry
For staining on paraffin sections, 4μm paraffin sections were deparaffinized and rehydrated. Antigen unmasking was performed for 20 min at 98°C in citrate buffer (pH 6) using the PT module. Endogenous peroxidase was blocked using 3% H2O2 (Merck) in methanol for 10 min at RT. Endogenous avidin and biotin were blocked using the Endogenous Blocking kit (Invitrogen) for 20 min at RT. Nonspecific antigen blocking was performed using blocking buffer. Rabbit anti- YAP (1:200, Santa Cruz Biotechnology, sc-15407), rabbit anti- TAZ (1:100, Sigma- Aldrich, HPA007415), rabbit anti-cFOS (1:100, Proteintech, 26192-1-AP), rabbit anti-MYH9 (1:500, Sigma, HPA001644), rabbit anti-pERK (1:200, Cell signaling, 4370S) were incubated overnight at 4°C. For anti-FOSL1 (1:500, Santa Cruz Biotechnology, sc-28310) and anti-β-catenin (mouse, 1:1000 Abcam ab6301) tissue were also blocked with Mouse on Mouse (M.O.M.™) Blocking Reagent (Vector Laboratories). Anti- rabbit biotinylated with blocking buffer, anti-mouse biotinylated with blocking buffer, Standard ABC kit, and ImmPACT DAB (Vector Laboratories) were used for the detection of horseradish peroxidase (HRP) activity. Slides were then dehydrated and mounted using SafeMount (Labonord).
Proliferation experiments
For BrdU experiments, mice were injected with a single IP injection of BrdU (50 mg/kg in PBS) at different time points and sacrificed 4 hours after. For the quantification at least an area of 1,5 mm2 per animal was analysed with Zen2012 (Black Edition) software (Zeiss) to determine the percentage of BrdU positive cells.
Trans-epithelial water loss measurement
Trans-epithelial water loss (TEWL) was measured on back skin of isoflurane-sedated mice using a TEWA meter TM210 (Courage and Khazaka, Cologne, Germany). Every measurement represents the average of 30 individual subsequently recorded measurements at the probe head.
Transmission electron microscopy
The skin was cut into pieces of approximately 1cm2 and pre-fixed 2 hours at RT in the fixative solution composed of 4% paraformaldehyde (Sigma-Aldrich) and 2.5% of electron microscopy grade glutaraldehyde (Sigma-Aldrich) diluted in 0,1M cacodylate buffer pH 7,4 (Sigma). Subsequently the pieces were cut again to reach an approximate size of 1mm2 and immersed in the fixative solution at 4°C overnight. After washing 6 times for 10 minutes in 0,1M cacodylate buffer on ice under slow rotation, the samples were post-fixed for 2 hours in 2% osmium tetroxide and again washed 6 times for 10 minutes in water. Samples were then stained with 0,5% uranyl acetate overnight at 4°C in the dark. The following day the samples were washed 6 times for 10 minutes in water and incubated with lead aspartate (Walton’s lead aspartate composed of 0,02M lead nitrate in 0,03M sodium aspartate pH 5,5) for 45 minutes at 60°C in the dark. After the samples were washed in water (6 x 10 mins) and dehydrated in a graded ethanol series of 50, 70 and 90% on ice (20 minute steps) and 100% ethanol (3 x 20 mins) before being placed in propylene oxide (2 x 20 minutes). The samples were infiltrated in a mixture of 2:1 Propylene oxide/epoxy resin (Agar) for 1 hour at RT and in a mixture of 1:1 Propylene oxide/epoxy resin for 1 hour at RT before being left overnight in 1:2 Propylene oxide/epoxy resin mixture with open lid to allow evaporation of propylene oxide. The next morning, samples were placed in 100% epoxy resin that was changed every 2 hours and embedded at the end of the day in 100% fresh resin. They were incubated overnight at 45°C and at 60°C for 2 days to allow polymerization. Samples were processed into 50-70nm ultrathin sections using a Leica ultracut UCT ultra-microtome. Micrographs were taken in a JEOL JEM1400 transmission electron microscope equipped with an Olympus Quemesa 11 Mpxl camera at 80 kV accelerating voltage.
Dissociation of epidermal cells, cell sorting and microarray analysis
The mice were shaved with an electric shaver, the back skin was dissected, and the fat and underlying subcutis were removed with a scalpel. The samples were placed in 0.25% Trypsin (Gibco, Thermo Fisher Scientific) in DMEM-Dulbecco’s modified Eagle’s medium (Gibco, Thermo Fisher Scientific) and 2mM EDTA (Thermo Fisher Scientific) overnight at 4°C. The epidermis was separated from the dermis and the trypsin was neutralized by adding DMEM medium supplemented with 2% Chelex Fetal Calf Serum (FCS). Samples were filtrated on 70 and 40μm filter (Falcon). Single cells suspension were incubated in 2% FCS/PBS with primary antibodies for 30 min on ice, protected from the light, with shaking every 10 min. Primary antibodies were washed with 2% FCS/PBS and cells incubated for 30 min in APC-conjugated streptavidin (BD Biosciences), on ice, with shaking every 10 min. Living epidermal cells were gated by forward scatter, side scatter and negative staining for Hoechst dye. Basal IFE and infundibulum cells were stained using PEconjugated anti-α6-integrin (clone GoH3, 1:200, ebioscience) or FITC- conjugated anti-α6-integrin (clone GoH3, 1:200, ebioscience) and bulge cells were stained with biotinylated CD34 (clone RAM34; 1:50, BD Biosciences). Basal cells from the interfollicular epidermis were targeted using CD34 negative and α6 integrin positive gating. The geometric mean fluorescence intensity of Itgβl (FITC-coniugated anti-β1-integrin, 1:100, BD Bioscience 555005) and Itga2 (PE-coniugated anti-α2-integrin, 1:100, BD Bioscience 553858) was measured on the α6-integrinhigh/CD34neg population.
Fluorescence-activated cell sorting analysis was performed using FACSAria I at high pressure (70 psi) and FACSDiva software (BD Biosciences). Sorted cells (300 cells per sample) were harvested directly in 45μl of lysis buffer (20 mM DTT, 10 mM Tris.HCl ph 7.4, 0.5% SDS, 0.5μg/μl proteinase K). Samples were then lysed at 65°C for 15 minutes and frozen. RNA isolation, amplification and microarray were performed in the Functional Genomics Core, Barcelona. cDNA synthesis, library preparation and amplification were performed as described in51. Microarrays were then performed on Mouse Genome 430 PM strip Affymetrix array. The data were normalized using RMA algorithm. The entire procedure was repeated in three technical independent samples for the control and the EXP D4 condition and in two technical independent samples for the TPA treatment. Genetic signatures were obtained by considering genes presenting a fold change greater or smaller than 2 or -2, respectively in both biological replicates.
RNA extraction and quantitative PCR
Fresh-frozen pieces of mouse skin were reduced to powder with a mortar and RNA extraction was then carried out with the RNAeasy Microkit from Quiagen according to the manufacturer’s protocol. Purified RNA was used to synthesize the first-strand complementary DNA using SuperScript II (Invitrogen) with random hexamers (Roche). Quantitative PCR analyses were carried out with Light Cycler 96 (Roche). Primers used: TNFα-5’: ACCACGCTCTTCTGTCTACT, TNFα-3’: AGGAGGTTGACTTTCTCCTG, IFNγ-5’: TCAAGTGGCATAGATGTGGAA, IFNγ-3’: CACTCGGATGAGCTCATTGA, CCL2-5’: CAGGTCCCTGTCATGCTTCT, CCL2-3’: GTCAGCACAGACCTCTCTCT, CCL5-5’: ACCATGAAGATCTCTGCAGC, CCL5-3’:TGAACCCACTTCTTCTCTGG. Normalizers: HPRT-5’: GCAGTACAGCCCCAAAATGG, HPRT-3’: TCCAACAAAGTCTGGCCTGT, Gapdh-5’: CGTGTTCCTACCCCCAATGT, Gapdh-3’: GTGTAGCCCAAGATGCCCTT.
ATAC sequencing
For ATAC-seq, 100000 sorted basal cells were collected in 1 ml PBS supplemented with 3% FBS at 4°C. Cells were centrifuged and cell pellets were resuspended in 100 μl lysis buffer (TrisHCl 10 mM, NaCl 10 mM, MgCl2 3 mM, Igepal 0.1%) and centrifuged at 500g for 25min at 4°C. Supernatant was carefully discarded and nuclei were resuspended in 50 μl reaction buffer (Tn5 transposase 2.5 μl, TD buffer 22.5 μl, from Nextera DNA sample preparation kit, Illumina, and 25 μl H20). The reaction was performed at 37 °C for 30 min and was stopped by adding 5 μl clean up buffer (NaCl 900 mM, EDTA 300 mM). DNA was purified using the MiniElute purification kit (QIAGEN) following the manufacturer’s protocol. DNA libraries were PCR amplified (Nextera DNA Sample Preparation Kit, Illumina), and size selected from 200 to 800 bp (BluePippin, Sage Sciences), following the manufacturer’s recommendations.
ATAC-seq analysis
Adaptor sequences were removed with TrimmomaticPE using options ‘HEADCROP:10 CROP:70 ILLUMINACLIP:adaptor.file:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:60’. ATAC-seq paired-end reads were then aligned to mouse genome Grcm38 using Bowtie2 (version 2.2.6)52 using options -X 2000-fr-very-sensitive-no-discordant-no-unal-no-mixed -non-deterministic’. Mitochondrial reads, reads from unmapped or random counting and reads with a mapping quality < 20 were removed using SAMtools53. Duplicate reads were removed by Picard tools (http://broadinstitute.github.io/picard/).
Peak calling was performed on each individual sample by MACS2 (version 2.1.0.20151222)54 using options ‘-f BAMPE -g mm –nomodel–call-summits -B –SPMR’, q-value 0.0001. Peaks from the different subpopulations were merged for downstream analysis.
Reads counts of each merged peak for each individual sample were calculated by HTSeq-count55 using options ‘-f bam -r pos -m intersection-nonempty’. These counts were normalized for one million mapped reads in merged peaks and fold-change was calculated compared to control. Peaks were associated to genes with GREAT software56 with the following parameters: 5.0 kb in proximal upstream, 1.0 kb in proximal downstream and 100.0 kb in distal. For most of the analysis, only peaks annotated to at least one gene were kept.
Differential peaks are defined as peaks having at least a twofold change compared to control and being called peak in the expanded condition and contain at least 3 reads per million. De novo motif search was performed using findMotifsGenome.pl program in HOMER software57 searching for motifs of 6 to 12 bp in a region of 500 bp around the peak center.
Single-cell RNA sequencing
Single basal cells from the back skin were FACS sorted (Integrin alpha 6 high, Cd34 negative) after dissociation. 6000 cells were loaded onto each channel of the Chromium Single Cell 3’ microfluidic chips (V2-chemistry, PN-120232, 10X Genomics) and barcoded with a 10X Chromium controller according to the manufacturer’s recommendations (10X Genomics). RNA from the barcoded cells was subsequently reverse transcribed, followed by amplification, shearing 5’ adaptor and sample index attachment. The Libraries were prepared using the Chromium Single Cell 3’ Library Kit (V2-chemistry, PN-120233, 10X Genomics) and sequenced on an Illumina HiSeq 4000 (paired-end 100bp reads).
Single-cell transcriptomic data analysis
Sequencing reads were aligned and annotated with the mm10-1.2.0 reference data set as provided by 10X Genomics and demultiplexed using CellRanger (version 2.1.1) with default parameters. Further downstream analyses were carried out individually for each of the four samples (CTRL, EXP.D1, EXP.D4 and TPA). Quality control was performed using the scater R package (version 1.10.0)58. For each sample, all of the cells passed the following criteria: had more than 4000 UMI counts, showed expression of more than 1500 unique genes and had less than 10% UMI counts belonging to mitochondrial sequences. Read counts were normalized using deconvoluted cell-specific size factors with the scran R package (version 1.10.2)59. Cell-cycle phases were assigned using the cyclone function from scran60. Dimensional reduction and clustering were performed using the Seurat R package (version 3.1.1). A PCA for each sample was calculated using the scaled expression data of the most variable genes (identified as outliers on a mean/variability plot, implemented in the FindVariableGenes function from Seurat). To remove unwanted sources of variability caused by differences in cycling stages but retaining the distinction between cycling and non-cycling cells, the differences between S-phase and GS/M-phase scores (determined by cyclone) were regressed out during scaling (by setting the vars.to.regress parameter of the ScaleData function in Seurat). UMAP61 calculation and graph-based clustering were done for each sample using the appropriate functions from Seurat (default parameters) with the respective PCA results as input. Different values of the resolution parameter were tested for the clustering. We set the final resolution to 0.6 (after testing a range from 0.3-1.2) first in the control sample, and then applied the same parameters to the other samples. Given that the obtained clustering sensitivity for a given resolution is dependent on the number of cells of that subpopulation in each respective sample, we swept over the same range of resolutions for the other samples, to assure the presence/absence of described clusters in all samples. This resolution best reflected the biological heterogeneity that emerged from the lineage tracing and the analysis of the clonal data, identifying a progenitor population (characterized by intermediate levels of Krt5, Krt14, Krt1 and Krt10) that emerged as an intermediate cluster between the stem cells (that expressed high levels of Krt14, Krt15 and integrins) and the differentiated population (highly enriched in Krt1 and Krt10). A Wilcoxon rank sum test was used to define marker genes for each cluster. Benjamini-Hochberg FDR correction for potential cluster marker genes across all samples (n=10,651) using the p.adjust method in R and only markers expressed in at least 25% of cells of the cluster, having an average log-fold change of at least 0.25 were reported. Differentially expressed genes and regulons between the CTRL SCs and EXP D1 STRETCH SCs and SCs clusters were computed as two-sided Wilcoxon ranked sum tests using the FindMarkers function in Seurat and p-values were FDR adjusted for the total number of comparisons (n=542 and n=571 respectively). Pseudotime ordering of cells was calculated using slingshot (version 1.1.0)27. We checked the robustness of the resulting trajectories by performing the analysis on PCA and UMAP reductions and also on downsampled subsets of the data to half the dataset size, these different reductions and permutations did not affect the described trajectories. Gene regulatory network analysis was performed using SCENIC26 with default parameters. Differentially activated regulons for each cluster were determined by performing a Wilcoxon rank sum test on the normalized regulon AUC values of cells in the various clusters, p-values were FDR-adjusted using the Benjamini-Hochberg method and regulons with an adjusted p-value less than 0.05 were considered differentially activated. For visualization purposes we integrated the CTRL, EXP D1, EXP D2 and TPA samples using the Seurat package’s standard CCA-MNN based data integration workflow. Feature selection was performed using the FindVariableFeatures function from Seurat with default parameters, selecting the 2000 most variable genes. Canonical correlation analysis (CCA) followed by integration anchors selection was then performed on the selected features using the FindIntegrationAnchors function from Seurat, taking the first 20 dimensions from the CCA into account, as described by the Seurat authors62. These anchors were then used to integrate the data with the IntegrateData function.
To compare the single-cell RNA-seq data from the treated EXP D2 samples (EXP D2-Trametinib and EXP D2-MAL inhibitor), data integration was performed with the EXP D1 sample following the approach implemented in Seurat62. The samples with the treatments were sequenced at a different time point than the EXP D1 sample and had in general lower UMI count distributions. Therefore, the UMI counts matrix of the EXP D1 sample was first downsampled, using the downsampleMatrix function from the DropletUtils package (https://bioconductor.org/packages/release/bioc/html/DropletUtils.html), such that its total number of counts was the same as the average of the total number of counts of the treatment samples. QC filtering and pre-processing was then performed as described before for each individual sample prior to integration. Further downstream analyses such as dimensionality reduction and clustering were all performed as described before using the integrated data. Cell-cycle phase prediction and differential expression analysis were performed on the uncorrected data to avoid interfering with the underlying distribution assumptions of these statistical methods. Cluster annotation was mainly performed by comparing the overlap of the new clusters of the integrated data with the original cell type annotations of the EXP D1 sample and by studying the expression of known marker genes. Differentially expressed genes and regulons between the EXP D1 SCs and the EXP D2 SC clusters were computed as two-sided Wilcoxon ranked sum tests using the FindMarkers function in Seurat and p-values were FDR adjusted for the total number of comparisons (n=13,786 and n=758 respectively). Subpopulations that are not associated with the IFE (infundibulum and sebaceous gland) were removed for the final analysis.
Statistical analysis
Two-tailed Student’s t-test, two-tailed Mann–Whitney U-test and Wilcoxon signed-rank test were performed using GraphPad Prism version 7.00 for Mac (GraphPad Software). Bar graphs and dot plots were generated by GraphPad Prism and show mean ± s.e.m. unless otherwise indicated.
Extended Data
Extended Data Figure 1. A mouse model of mechanical stretch-mediated skin expansion.
a, Representative photographs of mice with the skin expander immediately after surgery at day (D) D0, D2, D4 and in control (CTRL) condition. Scale bar, 10 mm. The device was implanted on the back skin of the animals, close to the neck where the rigidity of the proximate cervical spines allows the hydrogel to stretch the skin during the inflation of the expander. Control mice were operated upon similarly but without introducing the hydrogel. b, Timeline of the experiment. CD1 mice were operated to place the expander and followed over time. c, Scheme showing the growth of the hydrogel. The arrows indicate the radius of the hemisphere. d, Hydrogel volume (measured by the height of the hydrogel and calculated as the volume of a hemisphere, see Methods, n=5 D0, n=13 D0.5, n=13 D1, n=13 D2, n=7 D3, n=13 D4, n=10 D6, n=6 D8, n=8 D10, n=5 D14 mice). en, Transmission electron microscopy (TEM) of ultrathin sections of control (e, g, h, i, j) and expanded (f, k, l, m, n) epidermis. In e and f, dashed yellow lines denote dermal-epidermal boundary and boxed area in pink, cerulean, orange and green are shown at higher magnification respectively in g and k, h and l, i and m, j and n. Scale bar, 5 μm. g, k, Keratin bundles. h, l, Ultrastructural analysis of cell-cell adhesion. i, m, Desmosomes. j, n, Hemidesmodomes. o, Quantification of the intercellular spacing on images as in h and l. Wilcoxon signed-rank test, two-sided. p, Quantification of the width of the desmosomes as in i and m. q, Quantification of the width of the number of hemidesmosomes per μm in j and n. r, Trans-epithelial water loss (TEWL) measurements from n=3 CD1 mice in CTRL and at different time point during expansion. s, Immunohistochemistry for the adherens junctions (AJ) component β-catenin, n=3 independent experiments. t, v, Representative images of AJ component p120-catenin (t) and E-cadherin (v) colour-coded for the signal intensity with ImageJ. Protein expression is visualized as a colour gradient going from black to yellow, with black as indicator of no expression and yellow as indicator of maximal expression. Scale bar, 10 μm. u, w, Quantification of the average integrated density signal for p120-catenin (u) and E-cadherin (w). Each data point is the average of 3 sections per mouse (n=3 mice per condition). o-q, The quantifications are made on n=3 different animals per condition on 10 different samples per mouse and represented as mean + s.e.m. g-n, Scale bar, 500 nm. d, p, q, r, u, w, Two-tailed Mann–Whitney test, mean + s.e.m.
Extended Data Figure 2. Adhesion remodelling and inflammatory response during stretch-mediated skin expansion.
a, c, e, Representative images of the tight junction (TJ) components ZO-1 (a) and Claudin-1 (c) and of Vinculin (e) colour-coded for the signal intensity with ImageJ. Protein expression is visualized as a colour gradient going from black to yellow, with black as indicator of no expression and yellow as indicator of maximal expression. Scale bar, 10 μm b, d, f, Quantification of the average integrated density signal for ZO-1 (b), Claudin-1 (d) and Vinculin (f). The number of mice per condition is indicated. g, i, Immunostaining for K14 (red), inflammatory cells stained with CD45 (g) and macrophages stained with CD68 (i) (green) and Hoechst for nuclei (blue) on tissue sections. Scale bar, 10 μm. White arrows indicate positive cells, n=3 independent experiments. h, j, Percentage of CD45 (h) and CD68 (j) positive cells on the total dermal cells quantified based on the nuclear staining, n=3 mice per condition, mean per mouse + s.e.m. k, mRNA expression analysis for the indicated gene in Untreated (Unt., black) skin and skin treated with Dexamethasone (Dexa., grey). Fold change is expressed compared to one Unt. sample, n=3 mice per condition, mean per mouse + s.e.m. l, Maximum intensity projection of confocal pictures showing immunostaining for K14 (red), BrdU (green) and Hoechst for nuclei (blue) 4 hours following BrdU administration on whole mount epidermis. Scale bar, 10 μm. m, Proportion of basal cells that are BrdU positive (n=3,694 cells counted from 3 mice for Untreated and n=3,764 cells from 3 mice for the Dexamethasone treatment). a, c, e, g, i, Dashed lines indicate the basal lamina. b, d, f, m, Two-tailed Mann–Whitney test, mean per mouse + s.e.m.
Extended Data Figure 3. Clonal analysis of epidermal SC during homeostasis, TPA treatment and stretch-mediated skin expansion.
a, Genetic labelling strategy used to trace K14 IFE SC in the back skin during homeostasis and stretch-mediated tissue expansion. b, Timeline of the experiment. K14CREER-RosaConfetti mice were induced with Tamoxifen at 2 months of age and operated upon 3.5 days after to place the expander. The samples were collected 0, 1, 2, 4, 8, 10 and 14 days after surgery. c, Raw distribution of clone size taken from mouse back skin under normal homeostatic conditions (CTRL) at different time points based on basal (top) and total (bottom) cell number. Note that times are calibrated so that the “day 0” time-point is acquired 3.5 days after Tamoxifen injection, requiring effective chase times to be calibrated accordingly, see (b). D0: 115 clones from n=7 mice; D2: 175 clones from n=7 mice; D4: 136 clones from n=5 mice; D8: 159 clones from n=3 mice; D10: 146 clones from n=3 mice. d, Time line of the experiment to perform clonal tracing upon TPA treatment. K14CREER-RosaConfetti mice were induced with Tamoxifen at 2 months of age and after 3.5 days topically treated with 12-0-Tetradecanoylphorbol-13-acetate (TPA) for 2 consecutive days. The samples were collected 1 and 14 days after treatment. e, Maximum intensity projection of representative confocal pictures showing immunostaining for K14 (red) and BrdU (green) following BrdU administration on whole mount epidermis form mice treated with TPA or with vehicle (CTRL). Hoechst nuclear staining in blue. Scale bar, 20 μm. f, Percentage of BrdU positive cells in control and mice treated with TPA at D1 (n = 5). Two-tailed Mann–Whitney test, mean + s.e.m. g, Raw distribution of clone size taken from mouse back skin during TPA treatment (TPA) based on basal (top) and total (bottom) cell number. D1: 85 clones from n=4 mice; D14: 54 clones from n=5 mice. h, Raw distribution of clone size taken from mouse back skin under stretch-mediated tissue expansion (EXP) at different time points based on basal (top) and total (bottom) cell number. As with control, note that times are calibrated so that the “day 0” time-point is acquired 3.5 days after Tamoxifen injection, requiring effective chase times to be calibrated accordingly. D2: 231 clones from n=4 mice; D4: 197 clones from n=4 mice; D8: 199 clones from n=4 mice; D10: 157 clones
from n=4 mice. i, Table showing the abundance (raw counts) of clones by their basal and suprabasal cell composition from the CTRL D2 condition (i.e. 5 days post-induction), n= 203 clones from 7 mice. j, Table showing the abundance (raw counts) of clones by their basal and suprabasal cell composition from the EXPD2 condition, n= 283 clones from 4 mice. k, Fit of the one-progenitor model to the average size of persisting clones in control conditions based on the basal (black) and total (blue) cell content. Points show data and lines are the results of the fit to a one-compartment model (see Methods). l-o, Fit to the one-progenitor cell model. Clone persistence (l), labelled cell fraction (m), and the distribution of basal (upper) and total (lower) clone size (o). Points show data and lines are the results of the fit to a one-progenitor model. k, l, m, o, D0: 115 clones from n=7 mice; D2: 175 clones from n=7 mice; D4: 136 clones from n=5 mice; D8: 159 clones from n=3 mice; D10: 146 clones from n=3 mice; D14: 195 clones from n=4 mice. n, p, Sensitivity analysis of the model fits depicted as a map of the total square-differences of the experimental basal/total clone size data and the respective model predictions as a function of the average division time, 1λ, and the degree of imbalance towards stem cell loss/replacement, r, (see Methods). Panels (n) shows the results of one-progenitor model and the CTRL data, (p) shows the results of two-progenitor model and the CTRL data. These results show both the enhanced accuracy of the two-progenitor model over the one-progenitor model, despite involving the same number of fit parameters. k-m, Mean + s.d. o, Mean + s.e.m.
Extended Data Figure 4. Fit of the data to the two-progenitor model.
a, Fit of the model to the clone size distribution under homeostatic control conditions. Note that, with 1/λ = 4.6 days and r = 0.21, the model faithfully reproduces both the exponential-like clone size distribution and the predominance of clones bearing an even number of basal and total cell numbers. Mean + s.e.m. D0: n=115 clones from 7 mice; D2: n=175 clones from 7 mice; D4: n=136 clones from 5 mice; D8: n=159 clones from 3 mice; D10: n=146 clones from 3 mice; D14: n=195 clones from 4 mice. b, Change of division time (1λ) during stretch-mediated expansion as parameterised from the measured rate of BrdU incorporation, Fig. 1e. c, Change in the probability of symmetric division (parameter, r) during stretch-mediated skin expansion obtained from a fit of the two-compartment model to the clone size data (for details, see Supplementary Note). d, Corresponding fit of the two-compartment model to the clone size distribution during stretch-mediated expansion. The model accurately reproduces both the exponential-like clone size distribution and the predominance of clones bearing an even number of basal and total cell numbers. Notably, the sharp increase in even-sized clones at long times can only be recovered by limiting the frequency of renewing divisions well below that of the control value. Mean + s.e.m. D2: n=231 clones from 4 mice; D4: n=197 clones from 4 mice; D8: n=199 clones from 4 mice; D10: n=157 clones from 4 mice. e, Fit of the model to the clone size distribution at D14 under TPA treatment. Note that with 1/λ = 2.3 days and r = 0.15, the model faithfully reproduces both the exponential-like clone size distribution and the predominance of clones bearing an even number of basal and total cell numbers. Mean + s.e.m. D1: n=85 clones from 4 mice; D14: n=54 clones from 5 mice. f, g, Sensitivity analysis of the model fits depicted as a map of the total square-differences of the experimental basal/total clone size data and the respective model predictions as a function of the average division time, 1/λ, and the degree of imbalance towards stem cell loss/replacement, r, (see Methods). Panels (f) shows the results of the results of two-progenitor model and the EXP data, and (g) shows the results of two-progenitor model and the TPA data. For the EXP data (f), we have imposed the measured relative variation of the proliferation rate (as inferred from BrdU incorporation) (Fig. 1e and panel (b)) and an inferred relative variation of the r parameter as obtained from a model fit (c), while the two parameters in panel (f) represent variation in the net rates. h, Representative orthogonal confocal sections immunostained for K14 (red), K10 (green) following short-term BrdU (white) incorporation identifying cells biased for renewal (K14+/K10-), cells primed for differentiation (K14+/K10+) and differentiated cells (K14-/K10+). i, Percentage of the type of divisions in CTRL (108 divisions from n=4 mice) and EXP D2 (254 divisions from n=4 mice) based on short-term BrdU tracing and staining as in h. Two-tailed Mann–Whitney test, mean + s.e.m.
Extended Data Figure 5. Genetic signature of TPA treated and expanded epidermis.
a, Representative FACS plots showing the strategy used to isolate basal cells. Single living cells were gated by debris exclusion (P1), DAPI exclusion (P2), doublet elimination (P3) and basal IFE Integrin-α6high CD34neg cell were sorted (P4). n=10 independent experiments. b, c, mRNA expression of genes that were upregulated in basal cells at EXP D4 (n = 3) and in cells treated with TPA (n = 2). These genes are related to a generic stress signature (b), regulating ECM remodeling and cytoskeleton, important for cell survival and cell cycle (c). Bars are mean with s.e.m. d, Representative images of Paxillin immunostaining color-coded for the signal intensity with ImageJ. Protein expression is visualized as a color gradient going from black to yellow with black as indicator of no expression and yellow as indicator of maximal expression. Scale bar, 10 μm e, Quantification of the average integrated density signal for Paxillin as in d. Each data point is the average of 3 sections per mouse (n = 3 mice per condition). f, Geometric mean fluorescence intensity for the indicated integrin in CTRL (grey, n = 4 mice) and EXP D4 (red, n = 6 mice) from FACS analysis of basal IFE Integrin-α6high CD34neg cells. g-k, ATAC-seq profiles showing increasing accessibility of chromatin regions that are specifically remodeled during mechanical expansion (CTRL in grey and EXP D2 in orange). l, Quantification of the number of cells FOSL1+ in the basal layer related to Fig. 3d. m, Immunostaining on skin sections for c-JUN (white) in control and EXP D4. n, Quantification of the number of cells c-JUN+ in the basal layer related to m. o, Quantification of the number of cells p63+ in the +1 layer related to Fig. 3e. p, Quantification of the number of cells KLF4+ in the +1 layer related to Fig. 3f. q, Immunohistochemistry on paraffin sections for c-FOS in control and EXPD4. Scale bar, 20 μm. r, Quantification of the number of cells c-FOSL+ in the basal layer related to q. s, Immunofluorescence on tissue sections
for pSTAT3 in green and K14 (red) to identified the epidermis. Scale bar, 20 μm. t, Quantification of the number of cells positive for pSTAT3 in the basal layer related to s. l, n, o, p, r, t, 3 sections quantified per n = number of mice and total number of cells indicated in parentheses d, m, q, s, Dashed lines delineate the basal lamina. e, f, l, n, o, p, r, t, Two-tailed Mann–Whitney test, Mean + s.e.m.
Extended Data Figure 6. Single-cell RNA sequencing clustering analysis.
a, Integrated Uniform Manifold Approximation and Projection (UMAP) graphic representation of the CTRL, EXP D1, EXP D4 and TPA single-cell RNA-seq data, showing the graph-based clustering results annotated by cell type. The proliferating IFE stem cells (PROLIF. IFE SCs) are in light blue, the IFE stem cells cluster are in red (IFE SCs#1) and dark red (IFE SCs#2), the IFE committed cells (IFE CCs) cluster is in pink and the differentiated IFE cells (IFE DIFF.) are in green. The differentiated cells from the infundibulum (INF. DIFF.) are in grey, the stem cells of the infundibulum (INF. SCs.) are in black, the proliferating cells of the infundibulum (PROLIF. INF.) are in plum and the sebaceous gland cluster (SG) is in orange. The IFE stress cells (STRESS) are in dark grey and the cluster of stem cells stretch (SCs STRETCH) in yellow. n=16651 cells. b, UMAP of the different samples (CTRL, EXP D1, EXP D4, TPA) using the same integrated projection. n=4659 cells CTRL, n= 4934 cells EXP D1, n= 2716 cells EXP D4, n= 4342 cells TPA. c-k, UMAP plot of the CTRL sample colored by normalized gene expression values for genes identifying the IFE (c) versus infundibulum (d), the sebaceous gland (e) and the proliferating cells (f). Undifferentiated (g) and more differentiated cells (h) in the IFE identified the SCs cluster (i), the CCs cluster (j) and the differentiated stage (k). Gene expression is visualized as a color gradient going from grey to yellow with grey as indicator of no expression (i.e. expression values below or equal to the 50th percentile for that sample) and yellow as indicator of maximal expression. c-k, n=16651 cells. l, Table showing the specific marker genes used to annotate the different clusters.
Extended Data Figure 7. Single-cell RNA sequencing clustering analysis on the IFE cells.
a, Integrated UMAP graphic representation of the IFE cells in CTRL, EXP D1, EXP D4 and TPA
single-cell RNA-seq data, showing the graph-based clustering results annotated by cell type. The proliferating stem cells (PROLIF.) are in light blue, the stem cells clusters are in red and dark red (SCs#2), the committed cells (CCs) cluster is in pink and the differentiated cells (DIFF.) are in green. The stress cells (STRESS) are in dark grey and the cluster of stem cells stretch (SCs STRETCH) in yellow. n=12747 cells. b, UMAP of the different samples (CTRL, EXP D1, EXP D4, TPA). c, Predicted cell-cycle phases assigned using the cyclone function from scran tool and visualized in the UMAP. Cells in G1 are in light blue, cells in G2/M are in orange and cells in S phase are in red. b-d, n=3142 cells CTRL, n=3756 cells EXP D1, n=2145 cells EXP D4, n=3704 cells TPA. d, Percentage of cells in the different cycling phase calculated on the total number of cells. e-h, UMAP plot colored by normalized gene expression values for the indicated gene and in the indicated sample. Gene expression is visualized as a color gradient going from grey to yellow with grey as indicator of no expression and yellow as indicator of maximal expression. n=3142 cells CTRL, n=3756 cells EXP D1.
Extended Data Figure 8. Pseudotime analysis for single-cell RNA sequencing.
a, UMAP plots coloured by the degree of regulon activation for TFs differentially activated (AUC rank-sum test FDR corrected p-value < 0.05) in the different conditions. Colour scaling represents the normalized AUC value of target genes in the regulon being expressed as computed by SCENIC. b, Heatmap representation of the top 20 gene expression changes along the inferred pseudotime trajectory computed with Slingshot for the CTRL IFE. c, Heatmap representation of the top 20 gene expression changes along the inferred pseudotime homeostatic trajectory computed with Slingshot for the EXP D1 IFE. d, Heatmap representation of the top 20 gene expression changes along the inferred pseudotime trajectory computed with Slingshot characterising the stress state for the EXP D1 IFE. b-d, Columns represent cells ordered by their position along the pseudotime trajectory; rows represent genes whose expression profiles show highest correlation (FDR-correted p-value < 0.01) with the pseudotime variable, calculated using a generalized additive model (GAM). The colour scaling of the cells represents the normalized expression value of a gene in a particular cell, scaled by Z-score. a-d, n= 3142 cells CTRL, n= 3756 cells EXP D1.
Extended Data Figure 9. Cell contractility in stretch-mediated tissue expansion.
a, Scheme of the genetic strategy to delete Diaph3 in the epidermis. b, Protocol to delete Diaph3 during stretch-mediated tissue expansion. c, Orthogonal views of confocal analysis of immunostaining for K14 (red) marking basal cells and Phalloidin (green) to visualize F-actin and Hoechst for nuclei (blue) in whole mounts of IFE in CTRL from a CD1 mouse, EXP D1 from a CD1 or KI4CRE DIAPH3fl/fl (Diaph3 cKO) mouse. Scale bar, 10 μm. d, Percentage of cells with F-actin fibers in the apical side of basal cells related to c (n = 4 mice per condition). e, Orthogonal views of confocal analysis of immunostaining for K14 (red) marking basal cells, BrdU (green) and Hoechst for nuclei (blue) in whole mounts of IFE from K14CRE DIAPH3fl/+ (Diaph3 WT) and KI4CRE DIAPH3fl/fl (Diaph3 cKO) mice during expansion. Scale bar, 20 μm. Epidermal Diaph3 cKO were born at a Mendelian ratio and did not present obvious pathological phenotypes. n=3 independent experiments. f, Immunostaining for the basal marker K14 (red) and the suprabasal markers K1 and K10 (green) in Diaph3 WT and Diaph3 cKO mice in EXP D2 and EXP D4. Scale bar, 20um. g, Epidermal thickness of Diaph3 WT and Diaph3 cKO mice in EXP D2 and EXP D4 (three measurements taken with ImageJ on two sections per mouse, n = at least 3 mice for the different conditions). h, Scheme of the genetic strategy to delete Myh9 in the epidermis. i, Protocol to delete Myh9 during stretch-mediated tissue expansion. j, Immunohistochemistry for MYH9 in untreated and Tamoxifen induced K14CREER MYH9fl/fl mice. Scale bar, 20um. n=3 independent experiments. k, Orthogonal views of confocal analysis of immunostaining for K14 (white), BrdU (red) and Hoechst for nuclei (blue) in whole mounts of IFE in Myh9 WT and Myh9 cKO mice during expansion. Scale bar, 20 [jm. 1, Epidermal thickness of Myh9 WT and Myh9 cKO mice in EXP D2 and EXP D4 (three measurements taken with ImageJ on two sections per mouse, n = at least 3 mice for the different conditions). m-r, Analysis of adherens-junctions in Diaph3 cKO and Myh9 cKO mice. m, o, q, Representative images of adherens junction (AJ) component α-catenin (m), the α18 tension sensitive form of α-catenin (α18-cathenin) (o) and Vinculin (q), colour-coded for the signal intensity with ImageJ. Protein expression is visualized as a colour gradient going from black to yellow, with black as indicator of no expression and yellow as indicator of maximal expression. Dashed lines indicate the basal lamina. Scale bar, 10 μm. n, p, r, Quantification of the average integrated density signal for α-catenin (m), α18-cathenin (o) and Vinculin (q). Each data point is the average of 3 sections per mouse (n=5 mice per condition). d, g, l, n, p, r, Two-tailed Mann–Whitney test, mean + s.e.m.
Extended Data Figure 10. MEK/ERK/AP1, YAP-TAZ and MAL/SRF regulate stretch-mediated proliferation.
a, b, Protocol for Trametinib or Pimasertib treatment in CD1 mice operated to place the expander and scarified at D2, D4 (a) and D8 (b) after surgery. c, Immunohistochemistry for pERK on paraffin sections of epidermis form CD1 mice untreated or treated with the indicated drug at EXP D2. d, Quantification of the proportion of BrdU positive cells during expansion at the indicated time point in CD1 mice untreated or treated with Trimatenib or Pimasertib (n=at least 3 mice per condition as indicated, total number of cells analyzed indicated in parentheses). e, f, Immunohistochemistry for FOSL1 (e) and immunofluorescence for JUN (f) on sections of epidermis from CD1 mice untreated or treated with the indicated drug at EXP D2. g, Epidermal thickness measured with ImageJ on tissue sections at EXP D8 in CD1 mice untreated or treated with the indicated drug (n=5 mice untreated, n=4 mice Trametinib, n=3 mice Pimasertib, 3 measurements on at least 2 sections per mouse). h, Immunostaining (white) for YAP1 on skin sections in the control and in EXP D1. White arrows indicate nuclear localization. i, Quantification of YAP1 subcellular localization, bars and error bars represent the mean and s.e.m. Nuclear (N) > Cytosplasm (C), more YAP1 in nucleus than in cytoplasm, N = C, similar level of YAP1 in nucleus than in cytoplasm, N < C, less YAP1 in nucleus than in cytoplasm (n=150 cells for all samples except n=120 for EXP D8). j, Quantification of MAL subcellular localization, presented as mean and s.e.m. N > C, more MAL in nucleus than in cytoplasm, N = C, similar level of MAL in nucleus than in cytoplasm, N < C, less MAL in nucleus than in cytoplasm (n=150 cells for all samples except n=120 for EXP D8). k, l, Immunostaining (white) for MAL (k) and JUN (l) on skin sections in the control and in EXP D1. White arrows indicate nuclear localization. m, n, Scheme of the genetic strategy to delete YAP-TAZ in the epidermis (m) and protocol to delete YAP and TAZ in stretch-mediated tissue expansion (n). o, Immunohistochemistry for YAP (top) and TAZ (bottom) in K14CREER YAP-TAZfl/fl mice before and after Tamoxifen administration. p, Orthogonal views of confocal analysis of immunostaining for K14 (red) marking basal cells, BrdU (green) and Hoechst for nuclei (blue) in whole mounts of IFE in YAP-TAZfl/fl (YAP-TAZ WT) or K14CREER YAP-TAZfl/fl (YAP-TAZ cKO) mice at the indicated time point following expansion. q, Epidermal thickness of YAP-TAZ WT and YAP-TAZ cKO mice in EXP D2 and EXP D4 (three measurements taken with ImageJ on two sections per mouse, n=at least 4 mice per condition). r, Protocol to inhibit MAL with the CCG203971 small molecule during stretch-mediated tissue expansion. s, Quantification of MAL subcellular localisation in EXP D2 and EXP D4 mice treated or not with the MAL inhibitor. N > C, more MAL in nucleus than in cytoplasm, N = C, similar level of MAL in nucleus than in cytoplasm, N < C, less MAL in nucleus than in cytoplasm (n=150 cells per condition). Data are presented as mean and s.e.m. t, Orthogonal views of confocal analysis of immunostaining for K14 (red) marking basal cells, BrdU (green) and Hoechst for nuclei (blue) in whole mounts of IFE in mice treated with the MAL inhibitor or with vehicle control (untreated) at the indicated time point following expansion. u, Epidermal thickness of CD1 mice in EXP D2 and EXP D4 treated or not with the MAL inhibitor (three measurements taken with ImageJ on two sections per mouse, n=3 for untreated mice, n=5 for treated animals). c, e, f, h, k, l, o, p, t, Scale bar, 20μm. n=3 independent experiments. c, e, f, h, k, l, o, Dashed lines delineate the basal lamina. d, g, q, u, Two-tailed Mann–Whitney test, mean + s.e.m.
Extended Data Figure 11. Pathways associated with stretch-mediated tissue expansion.
a, Protocol used to delete YAP and TAZ and to inhibit MAL with CCG203971 treatment in K14CREER YAP-TAZfl/fl mice in EXP D2. b, Orthogonal views of immunostaining for K14 (red) to mark basal cells, BrdU (green) and Hoechst for nuclei (blue) on whole mounts of IFE in YAP-TAZ WT untreated mice and YAP-TAZ cKO mice treated with the MAL inhibitor at 2 days after the expander placement. Scale bar, 20 μm. c, Proportion of BrdU positive cells in untreated YAP-TAZ WT mice (48615 cells from 3 mice) and in YAP-TAZ cKO mice treated with the MAL inhibitor (78282 cells from 5 mice). Two-tailed Mann–Whitney test, mean + s.e.m. d, Epidermal thickness of YAP-TAZ WT untreated (n=3) and YAP-TAZ cKO treated with the MAL inhibitor in EXP D2 (n=5), three measurements taken with ImageJ on two sections per mouse. Two-tailed Mann–Whitney test, mean + s.e.m. e, f, Quantification of YAP1 (e) and MAL (f) subcellular localization, presented as mean and s.e.m. in CTRL and EXP D2. N > C, more protein in nucleus than in cytoplasm, N = C, similar level of protein in nucleus than in cytoplasm, N < C, less protein in nucleus than in cytoplasm (n=150 cells per condition). g, h, Percentage of the type of divisions in CTRL (g) and EXP D2 (h) in YAP-TAZ WT mice and YAP-TAZ cKO mice based on the short-term BrdU tracing and staining as in Extended Data Fig. 4h. i, j, Percentage of the type of divisions in CTRL (i) and EXP D2 (j) in Untreated mice and with MAL inhibitor based on the short-term BrdU tracing and staining as in Extended Data Fig. 4h. k, l, Percentage of the type of divisions in CTRL (k) and EXP D2 (l) in Untreated mice and with Trametinib based on the short-term BrdU tracing and staining as in Extended Data Fig. 4h. g-l, The number of counted divisions is indicated in parenthesis from n=number of mice. Two-tailed Mann–Whitney test, mean + s.e.m.
Extended Data Figure 12. Single-cell data analysis after MEK and MAL inhibition.
a, Average size of persisting clones in mice treated with MAL inhibitor during expansion, based on the basal (black) and total (blue) cell content. Points show data and lines denote the results from the fit to the two-compartment model (see main text and Methods). D0: n=115 clones from 7 mice; D2: n=86 clones from 3 mice; D4: n=83 clones from 3 mice. b, Average size of persisting clones in mice treated with Trametinib during expansion, based on the basal (black) and total (blue) cell content. Points show data and lines denote the results from the fit to the two-compartment model (see main text and Methods). D0: n=115 clones from 7 mice; D2: n=84 clones from 3 mice; D4: n=80 clones from 4 mice; D8: n=81 clones from 3 mice. c, Fit of the model to the clone size distribution during expansion upon MAL inhibition with 1/λ = 3.8 days and r = 0.08. D0: n=115 clones from 7 mice; D2: n=86 clones from 3 mice; D4: n=83 clones from 3 mice. d, Least-square values indicate the sensitivity of the fit parameters in (c). e, Fit of the model to the clone size distribution during expansion upon Trametininb treatment with 1/λ = 4.3 days and r = 0.17. D0: n=115 clones from 7 mice; D2: n=84 clones from 3 mice; D4: n=80 clones from 4 mice; D8: n=81 clones from 3 mice. f, Least-square values indicate the sensitivity of the fit parameters in (e). g, Predicted cell-cycle phases assigned using the cyclone function from scran tool of EXP D1 Untreated IFE, EXP D2 IFE treated with the MAL inhibitor and EXP D2 IFE treated with Trametinib. Cells in G1 are in grey, cells in G2/M are in blue and cells in S phase are in red. The percentage of cells in the different cycling phases is calculated on the total number of cells. h, Table showing the values of the percentage of the different cellular clusters in Figure 4j,k. a, b, Mean + s.d. c, e, Mean + s.e.m.
Supplementary Material
Acknowledgements
The authors acknowledge the animal house facility from ULB (Erasme campus), the ULB genomic core facility (F. Libert and A. Lefort), J.-M. Vanderwinden and the LiMiF (www.limif.ulb.ac.be) for the help with confocal microscopy, Thomas Van Brussel (KU Leuven) for help with 10X genomics, Barbara Gilbert and Wim Declercq (Ghent University) and Pauline Vieugue for helping with the TWEL assay, Aurelie De Groote (Univerite Libre de Bruxelles) for performing mice perfusion, and colleagues who provided reagents mentioned in the text. Single-cell RNA-sequencing was performed at the Brussels Interuniversity Genomics High Throughput core (www.brightcore.be) and the Genomics Core Leuven. C.B. is an investigator of WELBIO. M.A. is supported by a long-term postdoctoral fellowship of the HFSPO (LT000380/2015-L) and a FNRS fellowship. B.D.S. is supported by a Royal Society EP Abraham Research Professorship and a Wellcome Trust Senior Investigator Award (098357/Z/12/Z). S.H. is supported by a long-term fellowship of the HFSPO (LT000092/2016-L). B.D.S and S.H. acknowledge core funding to the Gurdon Institute from the Wellcome Trust (092096) and CRUK (C6946/A14492). A.S., J.V.H. and T.V. are supported by KU Leuven SymBioSys, Stiching Tegen Kanker and FWO postdoctoral fellowship number 12W7318N and Marie Sklodowska-Curie fellowship number 12O5617N. F.T. is a research Director at the FNRS. This work was supported by the FNRS, TELEVIE, the PAI programme, a research grant from the Fondation contre le Cancer, the ULB foundation, the foundation Bettencourt Schueller, the foundation Baillet Latour and a consolidator grant of the European Research Council (ERC-EXPAND, 616333).
Footnotes
Data availability
Data associated with this study have been deposited in the NCBI Gene Expression Omnibus under accession numbers GSE126231, GSE126734 and GSE146637 respectively for the microarray, ATAC-seq and single-cell RNA-seq. Data supporting the findings of this study are available within the article (and its Supplementary Information files). Source data behind Figures 1-4 and Extended Data Figures 1-12 are available within the manuscript files.
Code availability
Custom computer code and algorithm used to generate results that are reported in the paper are available within the article (and its Supplementary Information files) and from the corresponding authors on reasonable request. The code used for the modeling of the clonal data has been deposited in GitHub (available at https://github.com/BenSimonsLab/Aragona_Nature_2020).
In relation with the single-cells analysis, sequencing reads were preprocessed using cutadapt (version 1.13, https://pypi.org/project/cutadapt/), alignments were generated using STAR (version 2.5.2b, https://github.com/alexdobin/STAR) and transcript counts were generated using HTSeq (version 0.6.0, https://pypi.org/project/HTSeq/). Quality control and analysis was performed using the scater (version 1.8.0, https://bioconductor.org/packages/scater/) and Seurat (version 2.3.3, https://github.com/satijalab/seurat) R packages. Normalization was performed using the scran R package (version 1.8.4, https://bioconductor.org/packages/scran/). Gene regulatory network analysis was performed using pySCENIC (version 0.9.3, https://github.com/aertslab/pySCENIC). Pseudotime trajectory analysis was performed using the Slingshot (version 1.1.0, https://bioconductor.org/packages/slingshot/) R packages.
Author Contribution
M.A. and C.B. designed the experiments. M.A., B.D.S. and C.B. performed data analysis. M.M., A.S., J.V.H. and T.V. performed scRNA-seq and analysis. S.H. helped with data analysis. S.D., S.G and G.L. helped with experiments and animal follow-up. Y.S. and B.S. performed ATACseq analysis. P.B and K.V. performed TEM. F.T. contributed with genetic tools. M.A., B.D.S. and C.B. wrote the manuscript. All authors read and approved the final manuscript.
Author Information
Reprints and permission information is available at www.nature.com/reprints.
Competing interests
The authors declare no competing financial interests.
References
- 1.Zollner AM, Holland MA, Honda KS, Gosain AK, Kuhl E. Growth on demand: reviewing the mechanobiology of stretched skin. Journal of the mechanical behavior of biomedical materials. 2013;28:495–509. doi: 10.1016/j.jmbbm.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le HQ, et al. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nature cell biology. 2016;18:864–875. doi: 10.1038/ncb3387. [DOI] [PubMed] [Google Scholar]
- 3.Nava MM, et al. Heterochromatin-Driven Nuclear Softening Protects the Genome against Mechanical Stress-Induced Damage. Cell. 2020;181:800–817.e822. doi: 10.1016/j.cell.2020.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iskratsch T, Wolfenson H, Sheetz MP. Appreciating force and shape-the rise of mechanotransduction in cell biology. Nature reviews Molecular cell biology. 2014;15:825–833. doi: 10.1038/nrm3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeGoff L, Lecuit T. Mechanical Forces and Growth in Animal Tissues. Cold Spring Harbor perspectives in biology. 2015;8:a019232. doi: 10.1101/cshperspect.a019232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirby TJ, Lammerding J. Emerging views of the nucleus as a cellular mechanosensor. Nature cell biology. 2018;20:373–381. doi: 10.1038/s41556-018-0038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vining KH, Mooney DJ. Mechanical forces direct stem cell behaviour in development and regeneration. Nature reviews Molecular cell biology. 2017;18:728–742. doi: 10.1038/nrm.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanpain C, Fuchs E. Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science (New York, NY) 2014;344 doi: 10.1126/science.1242281. 1242281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obdeijn MC, Nicolai JP, Werker PM. The osmotic tissue expander: a three-year clinical experience. Journal of plastic, reconstructive & aesthetic surgery: JPRAS. 2009;62 doi: 10.1016/j.bjps.2007.12.088. 12191222. [DOI] [PubMed] [Google Scholar]
- 10.Wickstrom SA, Niessen CM. Cell adhesion and mechanics as drivers of tissue organization and differentiation: local cues for large scale organization. Current opinion in cell biology. 2018;54:89–97. doi: 10.1016/j.ceb.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. alpha-Catenin as a tension transducer that induces adherens junction development. Nature cell biology. 2010;12 doi: 10.1038/ncb2055. 533542. [DOI] [PubMed] [Google Scholar]
- 12.Clayton E, et al. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 13.Mascre G, et al. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature. 2012;489:257–262. doi: 10.1038/nature11393. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez-Danes A, et al. Defining the clonal dynamics leading to mouse skin tumour initiation. Science (New York, NY) 2016;536:298–303. doi: 10.1126/science.aao4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim X, et al. Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Science (New York, NY) 2013;342:1226–1230. doi: 10.1126/science.1239730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rompolas P, et al. Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science (New York, NY) 2016;352:1471–1474. doi: 10.1126/science.aaf7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mesa KR, et al. Homeostatic Epidermal Stem Cell Self-Renewal Is Driven by Local Differentiation. Cell stem cell. 2018;23:677–686.e674. doi: 10.1016/j.stem.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aragona M, et al. Defining stem cell dynamics and migration during wound healing in mouse skin epidermis. Nature communications. 2017;8 doi: 10.1038/ncomms14684. 14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 20.Botchkarev VA, Flores ER. p53/p63/p73 in the epidermis in health and disease. Cold Spring Harbor perspectives in medicine. 2014;4 doi: 10.1101/cshperspect.a015248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez RG, et al. C/EBPalpha and beta couple interfollicular keratinocyte proliferation arrest to commitment and terminal differentiation. Nat Cell Biol. 2009;11:1181–1190. doi: 10.1038/ncb1960. [DOI] [PubMed] [Google Scholar]
- 22.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 23.Hopkin AS, et al. GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS Genet. 2012;8:e1002829. doi: 10.1371/journal.pgen.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joost S, et al. Single-Cell Transcriptomics Reveals that Differentiation and Spatial Signatures Shape Epidermal and Hair Follicle Heterogeneity. Cell systems. 2016;3:221–237.e229. doi: 10.1016/j.cels.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joost S, et al. Single-Cell Transcriptomics of Traced Epidermal and Hair Follicle Stem Cells Reveals Rapid Adaptations during Wound Healing. Cell reports. 2018;25:585–597.e587. doi: 10.1016/j.celrep.2018.09.059. [DOI] [PubMed] [Google Scholar]
- 26.Aibar S, Gonzalez-Blas CB. SCENIC: single-cell regulatory network inference and clustering. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Street K, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rottner K, Faix J, Bogdan S, Linder S, Kerkhoff E. Actin assembly mechanisms at a glance. 2017;130:3427–3435. doi: 10.1242/jcs.206433. [DOI] [PubMed] [Google Scholar]
- 29.Duda M, et al. Polarization of Myosin II Refines Tissue Material Properties to Buffer Mechanical Stress. Dev Cell. 2019;48:245–260.e247. doi: 10.1016/j.devcel.2018.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang K, et al. YAP and ERK mediated mechanical strain-induced cell cycle progression through RhoA and cytoskeletal dynamics in rat growth plate chondrocytes. Journal of orthopaedic research: official publication of the Orthopaedic Research Society. 2016;34 doi: 10.1002/jor.23138. 11211129. [DOI] [PubMed] [Google Scholar]
- 31.Hirata H, et al. Actomyosin bundles serve as a tension sensor and a platform for ERK activation. EMBO reports. 2015;16:250–257. doi: 10.15252/embr.201439140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panciera T, Azzolin L, Cordenonsi M, Piccolo S. Mechanobiology of YAP and TAZ in physiology and disease. Nature reviews Molecular cell biology. 2017;18:758–770. doi: 10.1038/nrm.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Posern G, Treisman R. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends in cell biology. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Rognoni E, Walko G. The Roles of YAP/TAZ and the Hippo Pathway in Healthy and Diseased Skin. 2019;8 doi: 10.3390/cells8050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luxenburg C, Pasolli HA, Williams SE, Fuchs E. Developmental roles for Srf, cortical cytoskeleton and cell shape in epidermal spindle orientation. Nature cell biology. 2011;13:203–214. doi: 10.1038/ncb2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Connelly JT, et al. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nature cell biology. 2010;12:711–718. doi: 10.1038/ncb2074. [DOI] [PubMed] [Google Scholar]
- 37.Aragona M, et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 38.Whitson RJ, et al. Noncanonical hedgehog pathway activation through SRF-MKL1 promotes drug resistance in basal cell carcinomas. Nat Med. 2018;24:271–281. doi: 10.1038/nm.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miroshnikova YA, et al. Adhesion forces and cortical tension couple cell proliferation and differentiation to drive epidermal stratification. Nature cell biology. 2018;20:69–80. doi: 10.1038/s41556-017-0005-z. [DOI] [PubMed] [Google Scholar]
- 40.Mamidi A, et al. Mechanosignalling via integrins directs fate decisions of pancreatic progenitors. Nature. 2018;564:114–118. doi: 10.1038/s41586-018-0762-2. [DOI] [PubMed] [Google Scholar]
- 41.Moreno E, Valon L, Levillayer F, Levayer R. Competition for Space Induces Cell Elimination through Compaction-Driven ERK Downregulation. Current biology: CB. 2018 doi: 10.1016/j.cub.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasioukhin V, Bauer C, Degenstein L, Wise B, Fuchs E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell. 2001;104:605–617. doi: 10.1016/s0092-8674(01)00246-x. [DOI] [PubMed] [Google Scholar]
- 44.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 45.Xin M, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Damiani D, Goffinet AM, Alberts A, Tissir F. Lack of Diaph3 relaxes the spindle checkpoint causing the loss of neural progenitors. Nature communications. 2016;7 doi: 10.1038/ncomms13509. 13509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis (New York, NY: 2000) 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 48.Tepole AB, Gart M, Purnell CA, Gosain AK, Kuhl E. The Incompatibility of Living Systems: Characterizing Growth-Induced Incompatibilities in Expanded Skin. Annals of biomedical engineering. 2016;44:1734–1752. doi: 10.1007/s10439-015-1467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Susaki EA, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 50.Ellis SJ, et al. Distinct modes of cell competition shape mammalian tissue morphogenesis. Nature. 2019;569:497–502. doi: 10.1038/s41586-019-1199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonzalez-Roca E, et al. Accurate expression profiling of very small cell populations. PloS one. 2010;5:e14418. doi: 10.1371/journal.pone.0014418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England) 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nature biotechnology. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38 doi: 10.1016/j.molcel.2010.05.004. 576589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCarthy DJ, Campbell KR, Lun AT, Wills QF. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics (Oxford, England) 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lun AT, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome biology. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scialdone A, et al. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods (San Diego, Calif) 2015;85:54–61. doi: 10.1016/j.ymeth.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 61.Becht E, McInnes L. Dimensionality reduction for visualizing single-cell data using UMAP. 2018 doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 62.Stuart T, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.