Abstract
Inducible expression systems represent key modules in regulatory circuit design and metabolic engineering approaches. However, established systems are often limited in terms of applications due to high background expression levels and inducer toxicity. In bacteria, xenogeneic silencing (XS) proteins are involved in the tight control of horizontally acquired, AT-rich DNA. The action of XS proteins may be opposed by interference with a specific transcription factor, resulting in the phenomenon of counter-silencing, thereby activating gene expression. In this study, we harnessed this principle for the construction of a synthetic promoter library consisting of phage promoters targeted by the Lsr2-like XS protein CgpS of Corynebacterium glutamicum. Counter-silencing was achieved by inserting the operator sequence of the gluconate-responsive transcription factor GntR. The GntR-dependent promoter library is comprised of 28 activated and 16 repressed regulatory elements featuring effector-dependent tunability. For selected candidates, background expression levels were confirmed to be significantly reduced in comparison to established heterologous expression systems. Finally, a GntR-dependent metabolic toggle switch was implemented in a C. glutamicum l-valine production strain allowing the dynamic redirection of carbon flux between biomass and product formation.
Keywords: Corynebacterium glutamicum, synthetic biology, metabolic toggle switch, synthetic promoter library, l-valine production, xenogeneic silencing
Inducible expression systems allow the rational and precise control of transcription and are the most frequently used mechanisms for controlling gene expression in synthetic biology and metabolic engineering approaches1, 2. They usually form the first layer in synthetic regulatory circuits allowing for the response to extra- and intracellular stimuli and are applied for the accurate coordination of metabolic fluxes in various microbial production strains1, 3. In many bacterial model strains, for instance in the industrial platform strain Corynebacterium glutamicum, the widely used expression systems Ptac 2, ParaBAD 4 and Ptet 5, 6 are key parts used for circuit design. They are well established but limited in terms of applications due to considerably high background expression levels, heterogeneous inducer uptake, inducer toxicity or high inducer costs2, 7–9. Especially approaches where gene products are toxic to the recipient cell or disturb the central carbon flux are in demand for very stringent control of gene expression8. Besides the set of heterologous expression systems, the repertoire of homologous inducible promoter systems is comparatively small for C. glutamicum 10–15 and only a few promoter libraries were constructed containing either constitutive promoters7, 16, 17 or synthetic, Isopropyl-β-D-1-thiogalactopyranoside- (IPTG-) inducible regulatory elements17.
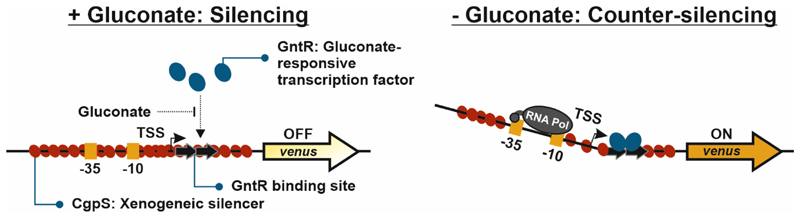
In a recent project, we designed synthetic promoter constructs for C. glutamicum, which are regulated by the mechanisms of xenogeneic silencing and counter-silencing18. Xenogeneic silencing is based on specialized nucleoid-associated proteins, so-called xenogeneic silencers (XS), which preferentially bind to horizontally acquired AT-rich DNA19–21. Known XS proteins are grouped into four classes including H-NS-like XS proteins from proteobacteria22, 23, Lsr2-like proteins of the actinobacteria24, MvaT/U-like XS of gammaproteobacteria of the order Pseudomonodales 25, and Rok found in different Bacillus strains26. The major function of the prophage encoded Lsr2-like XS protein CgpS from C. glutamicum is the control of the lysogenic status of the cryptic prophage element CGP327. CgpS binds to its target promoters and oligomerization of this XS protein leads to the formation of a nucleoprotein complex inhibiting transcription27 (silencing, Figure 1). In our recent study, we inserted the operator site of the regulator of gluconate catabolism GntR at different positions within various CgpS target promoters serving as platforms for the construction of synthetic counter-silencer promoters18. Binding of the transcription factor (TF) GntR to its operator sequence in the absence of gluconate leads to repression of its native target promoter PgntK 28. In contrast, binding of GntR to the synthetic counter-silencer promoter was shown to interfere with the nucleoprotein complex allowing the RNA polymerase to bind and to initiate transcription18 (counter-silencing, Figure 1). Interestingly, GntR binding to different counter-silencing constructs close to the transcriptional start site (TSS) led to counter-silencing, where it would usually cause a block of transcription at classical promoters. These competing effects were observed for several counter-silencing constructs highlighting that the interference of the silencer and the specific transcription factor (GntR) shape promoter activities of the respective targets18.
Figure 1. Schematic overview of the GntR-dependent mechanism of counter-silencing.
Synthetic counter-silencer constructs are based on phage promoters targeted by the xenogeneic silencer CgpS (red ovals). The 15 bp short operator sequence (black arrows) of the regulator of gluconate catabolism GntR (blue ovals)28 was inserted within the silenced promoter regions. In the absence of the effector molecule gluconate, binding of GntR to its operator sequence will interfere with the silencer-DNA complex leading to transcription initiation. Adapted from18.
Appropriate inducible expression systems are key modules for the design of synthetic regulatory circuits such as toggle switches and are frequently applied in metabolic engineering strategies aiming at improving the performance of bacterial production strains 3. Several natural and non-natural biotechnological products such as amino acids, organic acids and alcohols are derived from metabolites from the central carbon metabolism such as the glycolysis and the tricarboxylic acid (TCA) cycle, thereby competing with pathways essential for bacterial growth and physiology29-33. To achieve a redirection of carbon flux towards the molecule of interest, recent rationally engineered production strains are often based on permanent knockouts or knockdowns of genes concerning the central carbon metabolism34-40. This may, however, have detrimental effects on cellular fitness resulting in metabolic imbalances, impaired growth rates and final cell densities, thereby reducing the overall productivity of a fermentation process41. As an alternative strategy for these static engineering approaches, recent synthetic biology studies aimed at the dynamic redirection of carbon flux between cell growth and product biosynthesis by using metabolic toggle switches or other synthetic genetic circuits for the control of metabolic flux29-33, 42-45. For instance, Soma and colleagues constructed a metabolic toggle switch for isopropanol production in Escherichia coli. This synthetic genetic circuit allows switching between growth and production mode by controlling the conditional knockout of the citrate synthase GltA and the simultaneous upregulation of a synthetic isopropanol production pathway, both competing for acetyl-CoA29, 30. Using a comparable circuit architecture, Tsuruno and colleagues constructed a metabolic toggle switch for the redirection of the central carbon flux from glycerol towards 3-hydroypropionic acid31.
In this work, we systematically tested and compared a broad set of published18 and newly constructed synthetic promoters based on the principle of xenogeneic silencing and counter-silencing. The resulting promoter library comprises 44 synthetic promoters regulated by GntR in a gluconate-dependent manner. Comparison to established expression systems revealed very low background expression and good tunability of the resulting constructs. Finally, a GntR-based genetic toggle switch was implemented in C. glutamicum for the dynamic switch between cell growth and l-valine production. By controlling the activity of the pyruvate dehydrogenase complex (PDHC), the redirection of carbon flux towards l-valine biosynthesis was realized.
Results and Discussion
A synthetic promoter library based on the principles of xenogeneic silencing and counter-silencing
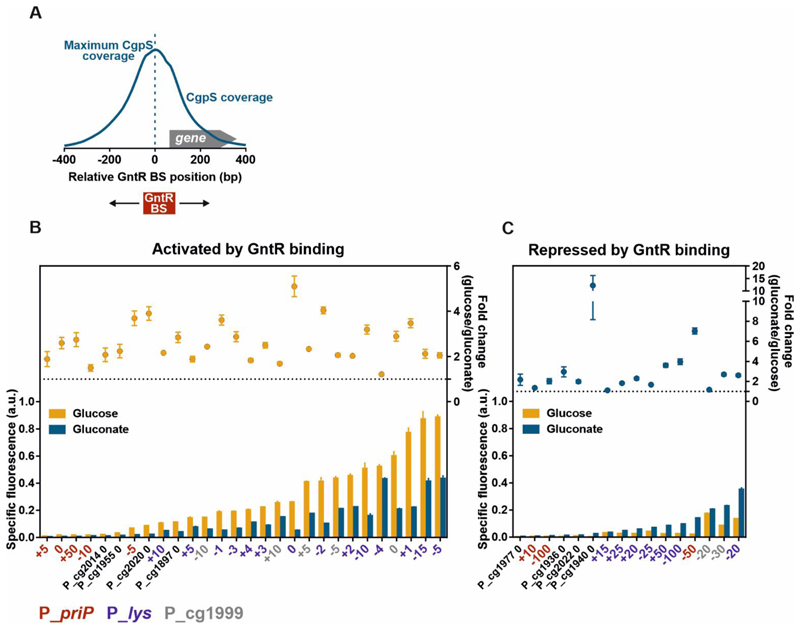
Xenogeneic silencing and counter-silencing represent efficient mechanisms for the tight control of gene expression18. Following a synthetic counter-silencing approach, we recently demonstrated that binding of the TF GntR within promoters silenced by the XS protein CgpS allows their effector-responsive reactivation18. However, depending on the architecture of the phage promoter and the position of the inserted GntR binding site (TATGATAGTACCAAT), counter-silencing promoters strongly vary in minimal and maximal promoter activities18. Here, we systematically tested and compared a broad set of gluconate-dependent, inducible promoters, which are based on the counter-silencing principle. Figure 2 provides an overview of synthetic promoter variants activated by counter-silencing (Figure 2B) or repressed (Figure 2C) by GntR binding (p-values < 0.05) by combining newly generated data (PpriP-based constructs) as well as previously published data18. Promoter activity was measured by means of the fluorescence output (production of the Venus reporter protein) over time18. Constructs activated by GntR-mediated counter-silencing showed a 100-fold range in maximal reporter output (-gluconate; 0.009 to 0.89 a.u. under the applied conditions) with small increment sizes and a large range of non-induced background levels (+gluconate; ranging from 0.005 to 0.43 a.u. under the applied conditions). Fold-change ratios of the individual promoters (-gluconate/+gluconate) ranged from 1.2-fold to 5-fold. As demonstrated in our recent study, highest counter-silencing efficiencies were obtained for constructs in which the GntR binding site was located directly at the position of maximal CgpS binding. The fold change ratios decreased with increasing distance to the CgpS peak18. However, not all synthetic promoter variants were activated by GntR binding. Depending on the core promoter part and the position of the binding, GntR binding led to the repression of gene expression in some cases. These constructs showed a 70-fold range in induced expression strength (+gluconate). Remarkably, several synthetic promoter variants appeared to have very low background expression levels. Lowest background expression levels among constructs repressed by GntR binding were observed for Pcg1977 0, while the minimum background activity of activated counter-silencer constructs was measured for PpriP +5. In C. glutamicum, standard systems used for inducible gene expression are heterologous expression systems like the IPTG inducible promoters Plac, Ptac and Ptrc 2, the arabinose-dependent promoter ParaBAD 4, the anhydrotetracycline inducible promoter Ptet 5, 6 and the heat induced PrPl promoter of bacteriophage λ46. Remarkably, the set of native inducible promoter systems is comparatively small. Described were promoters induced by gluconate namely Pgit1 10, PgntK 11, 12 and PgntP 11, the PmalE promoter induced by maltose10, the propionate inducible PprpD2 promoter13 as well as the amino acid responsive biosensors Lrp-PbrnFE 14 and LysG-PlysE 15. So far, only a few promoter libraries have been constructed containing either constitutive promoters7, 16, 17 or synthetic, IPTG-inducible regulatory elements17.
Figure 2. A synthetic GntR-dependent promoter library based on different CgpS target promoters with inserted GntR binding sites.
The library consists of previously obtained data for different recently described counter-silencer promoters18 as well as additionally constructed promoter variants based on the phage promoter PpriP following the same design approach. A) Schematic overview of a representative phage promoter which is bound by CgpS. The positions of the GntR binding site (BS) were referred to the nucleotide position associated with maximal CgpS binding27. The position directly upstream of this nucleotide was defined as position 0, negative numbers describe upstream and positive numbers downstream positions in relation to the CgpS peak maximum. B/C) Shown are reporter outputs driven by the promoters in the presence (gluconate) and absence (glucose) of the effector molecule after five hours of cultivation and the corresponding fold-change ratios. Promoters are grouped into two sets depending on their response to gluconate availability: activation by GntR binding (counter-silencing) (B) and repression by GntR binding (C) (p-values <0.05). Constructs based on the phage promoter PpriP (red), Plys (violet), or Pcg1999 (grey) are color-coded and indicate the position of the GntR binding site. Cells harbouring the plasmid-based synthetic promoter constructs were grown in CGXII medium supplemented with either 111/100 mM glucose or 100 mM gluconate. Bars represent the means and error bars the standard deviation of at least three biological replicates. Names indicate the platform promoter and the position of the GntR binding site positions. Specific fluorescence values were background corrected by subtracting values of strains harbouring the control plasmid pJC1-venus-term (no promoter in front of venus)74. Specific fluorescence was calculated by dividing the Venus fluorescence signal by the backscatter signal per time point63.
The here presented set of promoters being either activated or repressed by GntR binding provided a gluconate-dependent promoter library covering a broad range of promoter activities which may be useful for synthetic biology or metabolic engineering approaches demanding for tightly controlled and inducible regulation of gene expression. In favor for biotechnological applications, the effector molecule gluconate is a non-toxic, degradable and cost-efficient effector molecule compared to the frequently used inducer IPTG.
XS target promoters permit tight control of gene expression
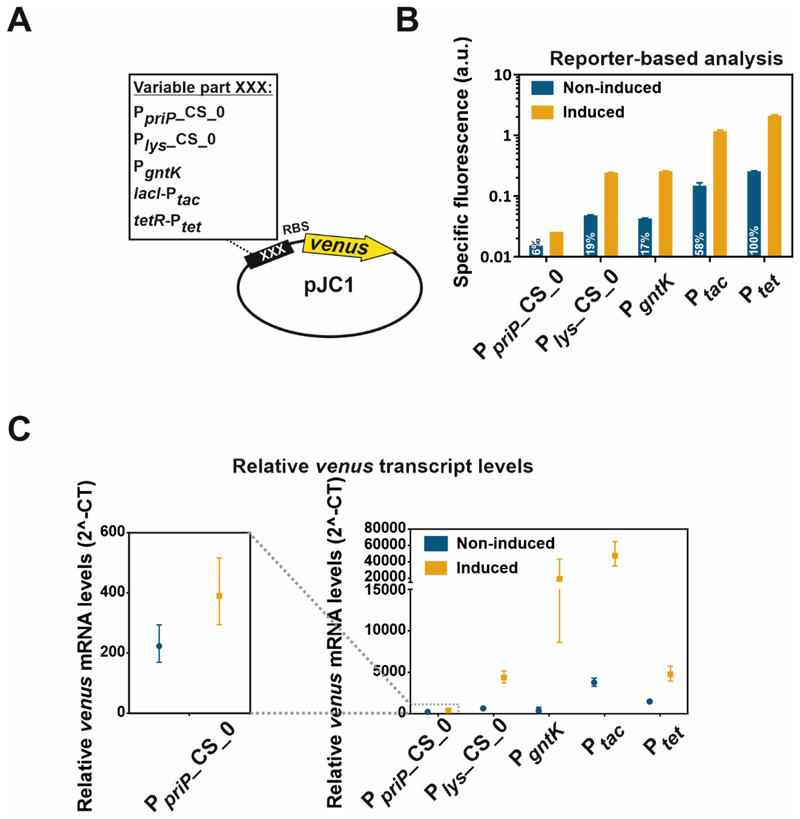
To validate the performance of counter-silencing constructs as expression systems, the synthetic promoters PpriP_CS_0 and Plys_CS_0 were exemplarily compared with the native GntR target promoter PgntK and the well-established expression systems Ptac and Ptet on protein (Venus) and transcript level (venus mRNA). Therefore, all promoters were fused to the reporter gene venus via a consistent linker containing a ribosomal binding site to avoid differences in translation efficiency and were integrated into the plasmid pJC1. Since the genes encoding for the TFs LacI and TetR are not present in the C. glutamicum genome, the lacI and tetR genes were additionally inserted into the respective plasmids (Figure 3A). Analysis of the reporter levels under inducing and non-inducing conditions revealed a wide range of expression levels for the different constructs (Figure 3B). The lowest background level was observed for the PpriP-based counter-silencer (6% of Ptet) which also showed the lowest induced reporter output. The Plys counter-silencer and the native GntR target promoter PgntK showed similar, but inverted response to gluconate availability. Both showed at least 5-fold lower background levels when compared to Ptet emphasizing their potential for application demanding stringent control of gene expression (Figure 3B). Similar ranges were observed on transcript levels using quantitative real-time PCR (qRT-PCR) (Figure 3C). The induced transcript levels of the Ptet construct were surprisingly low compared to the high protein level-dependent reporter outputs. To reduce effects on translation efficiencies, all constructs were based on a consistent sequence containing a conserved ribosomal binding site (RBS; AGGAG47) which was used to link the promoter sequences with the reporter gene venus. However, the 5´ untranslated regions (5´-UTR) differ between the constructs with potential effects on mRNA stability and translation efficiencies48, 49. Furthermore, potentially higher levels of the plasmid-encoded TFs TetR and LacI in comparison to the chromosomally encoded GntR as well as the different effector molecules may impact transcription activation dynamics. Finally, the high stability of fluorescence proteins12, 50 and the time needed for protein synthesis and maturation might explain the high reporter outputs, although most of the mRNA was already degraded.
Figure 3. Comparison of counter-silencing constructs with established expression systems.
A) For the comparison of the counter-silencer constructs Plys_CS_0 and PpriP_CS_018 as well as the native GntR target promoter PgntK18 with the established expression systems Ptac and Ptet, all of the different promoters were cloned into the plasmid pJC1 and fused to the reporter gene venus via a consistent linker containing a ribosomal binding site (RBS; AGGAG47). C. glutamicum wild type cells harbouring the plasmid-based constructs were cultivated in a microtiter cultivation system under inducing and non-inducing conditions depending on the particular promoter construct: PpriP_CS_0 and Plys_CS_0: +: 100 mM glucose, -: 100 mM gluconate; PgntK: +: 100 mM gluconate, -: 100 mM glucose; Ptac: +: 100 mM glucose + 100 µM IPTG, -: 100 mM gluconate + 0 µM IPTG; Ptet: +: 100 mM glucose + 235 nM ATc, -:.100 mM gluconate + 0 nM ATc B) Reporter outputs of the native GntR target promoter PgntK as well as PpriP- and Plys-based counter-silencer constructs (PpriP_CS_0; Plys_CS_0) in comparison to the established expression systems Ptac and Ptet after five hours of cultivation. All strains were pre-cultivated in CGXII containing 100 mM gluconate. Bars show the mean and error bars the standard deviation of specific fluorescence of biological triplicates. Specific fluorescence was calculated by dividing the Venus fluorescence signal by the backscatter signal per time point63. Indicated numbers represent the percentage of the background expression level of Ptet. C) Promoter-derived relative venus transcript levels measured by quantitative real-time PCR after five hours of cultivation under non-inducing and inducing conditions. Symbols represent the means and error bars the range of relative venus mRNA levels measured in biological and technical duplicates. All strains were pre-cultivated under non-inducing conditions.
Nevertheless, Plys_CS_0 and PgntK showed only 44 and 27% of the background level of Ptet, respectively, and background expression of PpriP_CS_0 was even reduced to 15% (Figure 3C), confirming that synthetic counter-silencer promoters represent suitable tools for tightly controlled gene expression.
Tunability of native and synthetic GntR target promoters
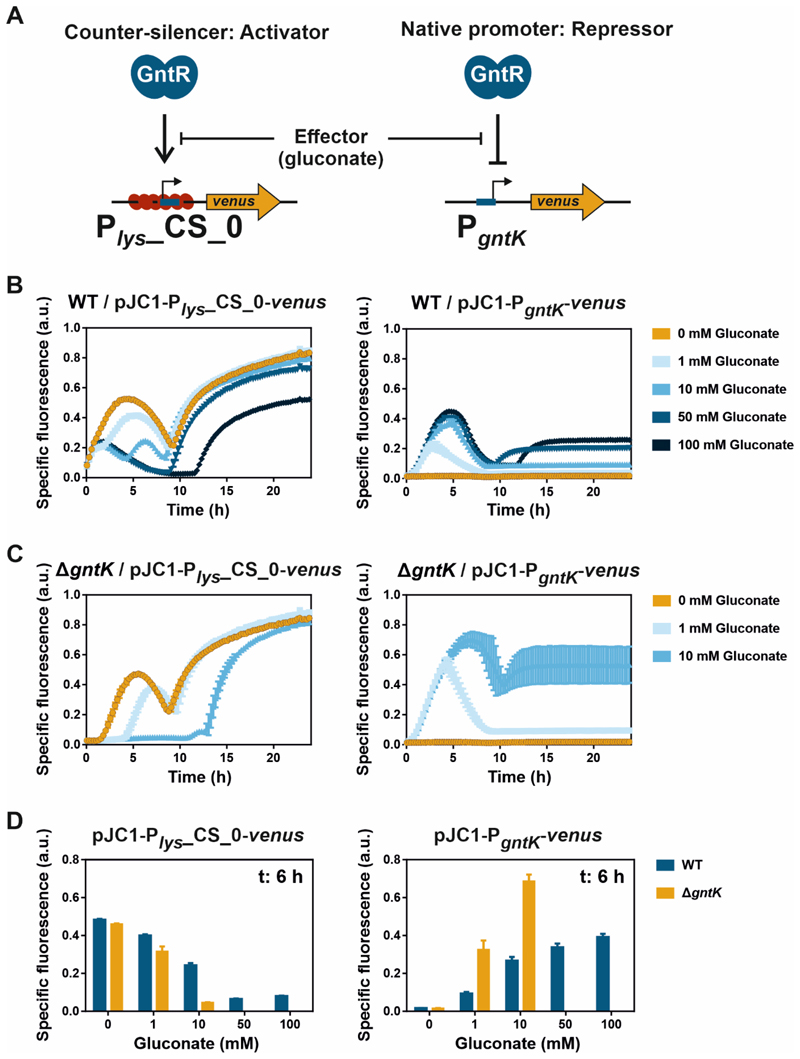
Tunability of promoters, for example by varying the amount of effector molecule, is an important feature for their use as bacterial expression systems in synthetic and biotechnological applications. In this study, the impact of varying gluconate concentrations on temporal dynamics and dynamic ranges of promoter activities of the native GntR target promoter PgntK and the counter-silencer promoter Plys_CS_0 were characterized. Both promoters inversely react to GntR binding and, therefore, to gluconate availability18 (Figure 4). GntR binding within the synthetic counter-silencer promoter Plys_CS_0 leads to promoter activation due to interference between GntR and the CgpS-DNA complex18. In contrast, binding of GntR to the native target promoter PgntK represses transcription initiation28 (Figure 4A).
Figure 4. Tunability of the counter-silencer construct Plys_CS_0 and the native GntR target promoter PgntK.
A) Shown are schematic overviews of both promoter constructs. B/C) Reporter outputs (specific Venus fluorescence) of C. glutamicum wild type (WT) (B) or C. glutamicum strain ΔgntK (C) harbouring the plasmid-based constructs pJC1-Plys_CS_0-venus or pJC1-PgntK-venus. Graphs show the mean and error bars the standard deviation of specific Venus fluorescence of biological triplicates over time. Increasing gluconate concentrations are indicated as shades of blue. D) Bar plots show the reporter outputs (from B and C) after six hours of cultivation. Cells were cultivated in a microtiter cultivation system in CGXII medium supplemented with glucose (100 mM in analysis of pJC1-PgntK-venus and 111 mM for characterization of pJC1-Plys_CS_0-venus) and either no or varying amounts of gluconate as effector. Backscatter and fluorescence were measured at 15 min intervals. Corresponding backscatter values are given in Figure S1. Specific fluorescence was calculated by dividing the Venus fluorescence signal by the backscatter signal per time point63.
Highest Plys_CS_0-derived reporter outputs were reached in the absence of gluconate (0 mM), meaning that the default state in the absence of the effector molecule of this construct is ON. In contrast, maximal PgntK promoter activity was observed in the presence of highest gluconate concentrations (100 mM), demonstrating the inverted response of both systems (Figure 4B). It has to be noted that the drop in fluorescence signal after five to seven hours of cultivation was probably caused by oxygen deprivation in the cultivation system affecting the maturation of the fluorescent protein and not by decreasing and increasing promoter activities. The default state in the absence of gluconate of PgntK promoter activity is OFF and the signal remained stable in this state after cells had entered the stationary phase (Figure 4B). In contrast, in the presence of 100 mM gluconate, Plys_CS_0 promoter activities stayed low during the first 11 hours of cultivations, but reporter outputs strongly increased shortly after cells had entered the stationary phase (Figure S1). At this time point, gluconate was probably fully consumed allowing GntR to bind to the DNA and to interfere with the silencer-DNA complex. Small amounts of supplemented gluconate (1 mM) also led to reduced reporter outputs in the beginning of the cultivation, however, the fluorescence signal already increased after two hours of exponential growth. Increasing amounts of supplemented gluconate (10 and 50 mM) gradually shifted the time point of induction, demonstrating the potential of counter-silencer promoters as expression systems with temporal tunability (Figure 4B). In contrast, a positive correlation between gluconate concentrations and reporter outputs was observed for PgntK (Figure 4B) demonstrating the opportunity of fine-tuning maximal promoter activity.
Upon transport into the cell, gluconate is phosphorylated by the gluconate-specific kinase GntK gaining 6-phosphogluconate which is further metabolized in the pentose phosphate pathway28. To reduce degradation of the effector molecule gluconate, we deleted the gntK gene encoding for the gluconate kinase GntK. Analysis of activities of the counter-silencer promoter Plys_CS_0 in a C. glutamicum ΔgntK deletion strain revealed the gradual temporal shift of the time point of reporter output increase. However, the time periods of low promoter activity were significantly extended and already 10 mM gluconate led to an almost complete repression of Plys_CS_0 promoter activity (Figure 4C, D). On the other hand, constitutively high effector concentrations (ΔgntK) led to more than 2.5-fold higher PgntK reporter outputs in comparison to the wild type (Figure 4C, D). The assumed increased and more stable intracellular gluconate levels were only reduced by dilution effects during cell growth comparable to the use of non-degradable structural sugar analogues like IPTG. However, the higher levels of gluconate can affect cell growth of C. glutamicum ΔgntK (Figure S1), therefore, fine-tuning of effector concentrations must be considered for potential applications of this strain.
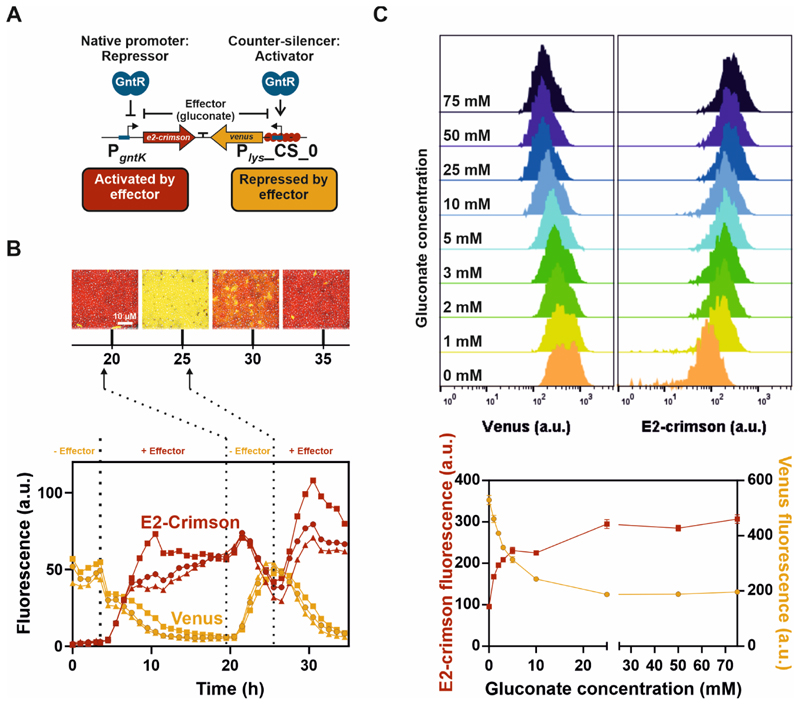
A GntR-based genetic toggle switch - Characterization of its switching dynamics and effector responsiveness
Based on their similar, but inverted response to gluconate availability, the native GntR target promoter PgntK and the synthetic Plys counter-silencing construct (Plys_CS_0) were recently combined in a gluconate-dependent, GntR-controlled genetic toggle switch18. Switching between different expression states was monitored by fusing the counter-silencer promoter Plys_CS_0 to the reporter gene venus and the native GntR target promoter PgntK to the reporter gene e2-crimson (Figure 5A). Previous analysis of the toggle state and its switching dynamics had confirmed its principal functionality. The addition of gluconate led to a switch in reporter outputs from Venus to E2-Crimson, while the removal of gluconate had the inverted effect18. In this study, we characterized the reversibility and robustness of the GntR-dependent toggle system in long-term experiments. Therefore, we cultivated C. glutamicum wild type cells harboring the plasmid-based toggle design in a microfluidic cultivation system starting with continuous supply of CGXII minimal medium containing 100 mM glucose as carbon source. Cultivation in the microfluidic experiment had the advantage that the cells were grown under constant conditions including constant levels of the effector molecule gluconate. The cells had been in a stable Venus-dominant state until the cultivation medium was switched to medium containing 100 mM gluconate after the first 3.5 hours of cultivation. Gluconate interfered with binding of GntR to its operator site leading to PgntK activation and restoring of silencing of Plys_CS_0, visualized by the following switch of the toggle from the Venus- to the E2-Crimson-state within the next eight hours of cultivation. The generally high stability and long half-lives of fluorescent proteins (e.g. GFP ~24h)50 and their expression and maturation rates probably contribute to a certain delay in switching dynamics of the toggle system. This could explain the relatively long period of time required for the complete signal change. In the following cultivation, a stable expression profile was observed demonstrating the robustness of the toggle design. During cultivations, single cells occurred which stayed in one of both toggle states. However, these cells had also stopped growth suggesting that they were in a dormant state. Three consecutive medium switches revealed the dynamic reversibility of the toggle switch (Figure 5B).
Figure 5. Reversibility and graduated responsiveness of the GntR-dependent toggle switch.
A) Schematic overview of the GntR-dependent toggle switch (adapted from18). B) Reversible switch between both reporter outputs. C. glutamicum wild type cells harbouring the plasmid-based toggle were cultivated in a microfluidic cultivation system65 with continuous supply of CGXII medium supplemented with either 100 mM glucose or 100 mM gluconate and analyzed by time-lapse microscopy at 15 min intervals. Cells were pre-cultivated in shaking flasks in the absence of the effector molecule gluconate (100 mM glucose). After the first 3.5 h (left dotted vertical line) of cultivation in the microfluidic chip in the absence of gluconate, the medium supply was switched to CGXII supplemented with 100 mM gluconate. Further switches of effector supply were performed after 19.5 and 25.5 h (dotted vertical lines). The graphs show the background corrected fluorescence (Venus and E2-Crimson) of three independent microcolonies (circles, squares, triangles) over time and images display one representative colony (triangle) after 20, 25, 30 and 35 h of cultivation. C) Graduated response of the GntR-dependent toggle switch to varying amounts of the effector molecule gluconate. Histograms of Venus (left) and E2-crimson fluorescence (right) after five hours of cultivation of C. glutamicum wild type cells harbouring the plasmid-based toggle. Cultivation was performed in a 96 deep-well plate in CGXII medium supplemented with varying amounts of gluconate and glucose (100 mM carbon source in total) and fluorescence values were analyzed by flow cytometry (FACSAria™ III). Gluconate concentrations are given as numbers. Cells were pre-cultivated in shaking flasks in the absence of the effector molecule gluconate (CGXII with 100 mM glucose). The graph shows the means of heights of fluorescence levels of biological triplicates and error bars the standard deviation.
The further characterization of the effect of varying amounts of the effector molecule gluconate revealed a graduated responsiveness of the toggle switch (Figure 5C). Gradually increasing gluconate levels from 0 to 25 mM shifted the toggle output step wisely from Venus towards E2-crimson fluorescence, demonstrating that the GntR-dependent toggle does not show strict bistability and intermediate toggle states exist instead. No further shift was observed at higher effector concentration suggesting that 25 mM gluconate are sufficient to achieve complete switching of the toggle and a saturation of the system (Figure 5C).
Previously, Gardner and colleagues designed a bistable synthetic toggle switch inspired by the genetic switch of bacteriophage λ governing the lysis-lysogenic decision51. This network consists of two repressors, namely Cro and CI, and their corresponding promoters Pr and Prm, each being repressed by the gene product of the other52. Comparably, in the modular synthetic toggle switch variants constructed by Gardner and colleagues, the authors imitated this circuit design by combining the IPTG-dependent LacI-Ptrc expression system with the temperature-sensitive λ repressor (CIts) and its corresponding promoter PLslcon or the anhydrotetracycline (ATc) inducible TetR-PLtetO-1 system51. This design principle was successfully applied as metabolic toggle switch for improving product formation in engineered E. coli strains29-31. However, especially inappropriate TF synthesis and degradation rates and insufficient repressor strengths were discussed as major challenges for toggle stability and functionality51. In contrast to this circuit architecture, the here presented toggle consists of less components and is controlled by only one effector molecule, namely gluconate, and one specific TF, namely GntR. This design is comparably simple but also very robust and allows the use of native TF levels, thereby reducing metabolic burden and circumventing challenges like TF synthesis and degradation rates. The graduated responsiveness of the toggle to varying gluconate concentrations allows the dynamic adjustment of the toggle to different application requirements. Due to the modular circuit architecture, the components of the toggle are independent of each other allowing for tuning of one side of the toggle without affecting the other. However, it has to be noted that the toggle described in this study does not confer memory to the system, but responds solely to the presence or absence of the effector molecule gluconate in a ‘seesaw-like’ mode of action.
Implementation of a metabolic toggle switch for dynamic control of l-valine production
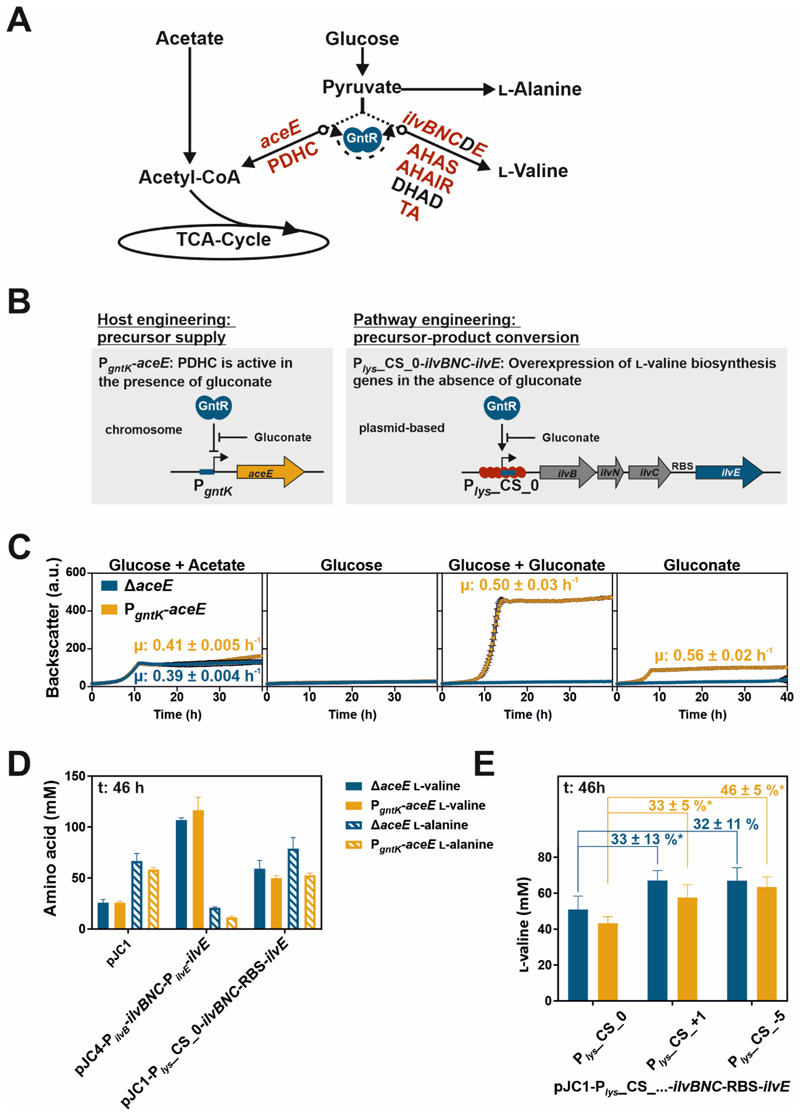
In a next step, the GntR-dependent toggle switch was implemented for the dynamic control of growth and l-valine biosynthesis in C. glutamicum. l-valine, an essential amino acid for human and animals, is used for a variety of application ranging from supplementation in human and animal nutrition, as precursor for herbicides, antibiotics and anti-viral drugs and as compound in cosmetic industries53. l-valine is formed from two molecules of the precursor pyruvate in a pathway comprising four reaction steps catalyzed by acetohydroxyacid synthase (AHAS, ilvBN), acetohydroxyacid isomeroreductase (AHAIR, ilvC), dihydroxyacid dehydratase (DHAD, ilvD) and transaminase B (TA, ilvE) (Figure 6A). The theoretical yield for l-valine production from glucose is one mol l-valine per mol glucose53. Several, previously established l-valine production strains were constructed by engineering the activity of the PDHC, which catalyzes the competing conversion of pyruvate to acetyl-CoA (Figure 6A). Engineering strategies ranged from the deletion of the aceE gene encoding the E1 subunit of the enzyme complex, which leads to a permanent loss of PDHC activity, to a reduction of aceE expression by promoter exchanges35, 37, 54-56. Growth of cells with inactive PDHC is decoupled from l-valine production and depends on costly carbon sources like acetate also reducing biomass yields35, 55. After acetate depletion, glucose is consumed and converted to l-valine35. The dynamic control of precursor supply and of flux through biosynthetic routes, for example by applying synthetic regulatory circuits like the presented GntR-dependent toggle, can represent an alternative strategy to the static permanent knockout approaches. For instance, this design may allow to overcome the growth dependency on acetate.
Figure 6. Application of the GntR-dependent toggle for the dynamic switch between growth and l-valine production.
A) Schematic representation of relevant parts of the central carbon metabolism and the l-valine biosynthesis pathway of C. glutamicum. The GntR-dependent toggle controlled the redirection of the carbon flux allowing for the conversion of pyruvate to either l-valine or acetyl-CoA entering the tricarboxylic acid cycle (TCA-Cycle). aceE encodes for the E1 subunit of the pyruvate dehydrogenase complex (PDHC), which converts pyruvate to acetyl-CoA. l-valine is formed from pyruvate in a four-step reaction pathway catalyzed by acetohydroxyacid synthase (AHAS, ilvBN), acetohydroxyacid isomeroreductase (AHAIR, ilvC), dihydroxyacid dehydratase (DHAD, ilvD), and transaminase B (TA, ilvE). Dynamically controlled genes and their products are highlighted in red. B) Schematic overview of the genetic background of the dynamic l-valine production strain. The native promoter of the aceE gene was replaced by the GntR target promoter PgntK allowing for control of PDHC activity. The l-valine biosynthesis genes were combined in a synthetic operon by fusing ilvE via a linker containing an RBS sequence to the end of the operon ilvBNC. Its expression was controlled by the synthetic GntR-dependent counter-silencer promoter Plys_CS_0. C) Growth of the strain with dynamically controlled aceE expression (PgntK-aceE) in comparison to the previously established ΔaceE strain35, 55. Both strains had been pre-cultivated in CGXII containing 222 mM glucose and 254 mM acetate before they were cultivated in a microtiter cultivation system in medium with either 222 mM glucose and 254 mM acetate, 222 mM glucose, 222 mM glucose and 51 mM gluconate or 51 mM gluconate. Calculated growth rates (µ) are given (colour-coded) when significant growth had been observed. D) l-valine titers of the aceE toggle strain (PgntK-aceE) and the ΔaceE l-valine production strain harbouring either the empty control plasmid pJC1 (pJC1-venus-term74), plasmid-based l-valine biosynthesis genes controlled by the counter-silencer (pJC1-Plys_CS_0-ilvBNC-RBS-ilvE) or the natively regulated variant pJC4-PilvB-ilvBNC-PilvE-ilvE (pJC4-ilvBNCE57). All strains were cultivated in CGXII supplemented with 222 mM glucose and 254 mM acetate. Bar plots represent the l-valine and l-alanine titers of biological triplicates and error bars the corresponding standard deviations after 46 h of cultivation. l-valine and l-alanine titers, glucose concentrations and cell densities (OD600) are shown in Figure S3. Further details are given in Table 1. E) l-valine titers of the aceE toggle strain (PgntK-aceE) and the ΔaceE l-valine production strain harbouring the plasmid-based l-valine biosynthesis genes either controlled by the abovementioned counter-silencer promoter Plys_CS_0 serving as reference or by two different counter-silencer variants (Plys_CS_+1 and Plys_CS_-5) belonging to the strongest promoters of the library shown in Figure 2. Bar plots represent the l-valine titers of biological triplicates and error bars the corresponding standard deviations after 46 h of cultivation. Numbers indicate the percentage increase in produced l-valine titers referred to the strain with l-valine biosynthesis genes under control of the weaker counter-silencer promoter Plys_CS_0 and stars its significance level (*:t-test, p-value < 0.05). All strains were cultivated in CGXII supplemented with 222 mM glucose and 254 mM acetate. Further details are given in Table 1.
For the construction of the toggle-based C. glutamicum l-valine production strain, the native aceE promoter was replaced by the gluconate-dependent promoter PgntK, which is repressed in the absence of gluconate leading to PDHC downregulation (Figure 6B). Growth analysis of the PgntK-aceE strain in comparison to the previously designed ΔaceE deletion strain revealed that both strains did not grow in medium containing glucose as sole carbon source for 40 hours of cultivation (Figure 6C). Both strains grew in medium containing glucose and acetate but showed relatively low final backscatter values (Figure 6C). In contrast, the PgntK-aceE strain reached 3.4-fold higher backscatter values in medium containing 222 mM glucose and 51 mM gluconate in comparison to growth on acetate-glucose medium (molarity of carbon: 254 mM acetate > 51 mM gluconate, factor 1.7) (Figure 6C). The gluconate-induced aceE expression allowed the co-utilization of glucose, as shown by the significantly reduced backscatter values when cells were cultivated in medium containing only 51 mM gluconate (Figure 6C). Remarkably, no differences in cell growth were observed between C. glutamicum PgntK-aceE and wild type cells in minimal medium containing 100 mM gluconate (Figure S2). The costs per kg gluconate are in a comparable range as those for glucose, while acetate represents a more costly carbon source (https://www.sigmaaldrich.com, Table S1A). This is also reflected by the twofold higher costs for carbon sources per litre CGXII medium containing 254 mM acetate and 222 mM glucose (4.91 €) than for medium containing 51 mM gluconate and 222 mM glucose (2.49 €) (Table S1B). Additionally, the last mentioned carbon source composition improved cell growth (3.4-fold) (Figure 6C), highlighting the economic potential of the dynamic gluconate controlled PDHC activity.
Previously applied successful engineering strategies targeting PDHC activity were based on aceE promoter replacements leading to reduced or growth phase-dependent aceE expression and acetate-independent cell growth54, 56. These approaches were either based on weaker, but constitutive mutated promoter variants of dapA 56 or on a growth-regulated promoter of an industrial used L-leucine strain (PCP_2836; C. glutamicum CP)54. However, in contrast to an effector-responsive expression system like the gluconate-dependent promoter PgntK (Figure 4), both regulatory circuits are independent of inducers and cannot be influenced by extracellular supplementation of effector molecules. Thus, fine-tuning of strain performance is constrained, for instance regarding the time point of switching or maximal promoter activities.
In the absence of gluconate, when the PDHC is inactive in the toggle strain, pyruvate is available for l-valine production. Therefore, the counter-silencer Plys_CS_0, as counterpart of the toggle, was used to control the overexpression of the l-valine biosynthesis genes ilvBNCE (combined in a synthetic operon) (Figure 6B). The plasmid-based construct was analyzed in the dynamic PgntK-aceE strain and in the static ΔaceE deletion strain in comparison to the same strains harbouring the empty plasmid pJC1 (pJC1-venus-term) or the previously designed plasmid pJC4-PilvB-ilvBNC-PilvE-ilvE (pJC4-ilvBNCE)57. The plasmid pJC4-PilvB-ilvBNC-PilvE-ilvE containing l-valine biosynthesis genes under control of their native promoters served as reference for strain performance. For comparison of l-valine productivity, all strains were cultivated in CGXII minimal medium containing 222 mM glucose and 254 mM acetate. This cultivation condition fully erased the growth benefit of dynamic aceE and ilvBNCE expression (aceE: OFF state; ilvBNCE: ON state); however, it allowed the comparison of strain performance independent of cell densities. Analysis of the culture supernatant revealed that after 46 hours of cultivation glucose was fully consumed by the production strains, while l-valine titers reached maximal values (Figure S3).
Both platform strains PgntK-aceE and ΔaceE produced comparable amounts of l-valine during cultivations in the absence of gluconate indicating that the PDHC was inactive in the toggle strain and that this redirected carbon flux towards l-valine (Figure 6D, Table 1).
Table 1. Product (l-valine) and by-product (l-alanine) formation of strains PgntK-aceE and ΔaceE.
| Plasmid | Strain | OD600 | l-valine (mM) | l-alanine (mM) | l-valine titer (g*L-1) | Specific titer (g l-valine* g CDW-1) | Yield (g l-valine* g glucose-1) |
|---|---|---|---|---|---|---|---|
| pJC1 | ΔaceE | 45.9 ± 1.0 | 25.9 ± 2.6 | 66.7 ± 6.0 | 3.0 ± 0.31 | 0.26 ± 0.02 | 0.087 ± 0.007 |
| PgntK-aceE | 48.7 ± 1.5 | 25.8 ± 0.9 | 58.3 ± 1.8 | 3.0 ± 0.11 | 0.25 ± 0.02 | 0.076 ± 0.003 | |
| pJC4-P ilvB - ilvBNC -P ilvE - ilvE | ΔaceE | 41.1 ± 0.8 | 106.9 ± 1.7 | 20.8 ± 0.8 | 12.5 ± 0.2 | 1.22 ± 0.01 | 0.313 ± 0.005 |
| PgntK-aceE | 44.3 ± 1.1 | 116.5 ± 10.4 | 11.3 ± 1.0 | 13.7 ± 1.22 | 1.23 ± 0.1 | 0.341 ± 0.03 | |
| pJC1-P lys _CS_0- ilvBNC -RBS- ilvE * | ΔaceE | 41.9 ± 0.9 | 50.9 ± 6.2 | 77.2 ± 5 | 6 ± 0.72 | 0.57 ± 0.08 | 0.149 ± 0.018 |
| PgntK-aceE | 46.6 ± 1.4 | 43.3 ± 3 | 56.6 ± 3.2 | 5.1 ± 0.35 | 0.44 ± 0.02 | 0.127 ± 0.009 | |
| pJC1-P lys _CS_+1- ilvBNC -RBS- ilvE | ΔaceE | 42.2 ± 0.3 | 67.1 ± 4.5 | 68.6 ± 0.6 | 7.9 ± 0.53 | 0.74 ± 0.04 | 0.196 ± 0.013 |
| PgntK-aceE | 45.5 ± 0.6 | 57.7 ± 5.7 | 46.2 ± 3.1 | 6.8 ± 0.67 | 0.59 ± 0.05 | 0.169 ± 0.017 | |
| pJC1-P lys _CS_-5- ilvBNC -RBS- ilvE | ΔaceE | 41.6 ± 1.2 | 67 ± 5.9 | 64.6 ± 2.7 | 7.8 ± 0.69 | 0.76 ± 0.07 | 0.196 ± 0.017 |
| PgntK-aceE | 45.1 ± 0.2 | 63.4 ± 4.6 | 43.3 ± 2.4 | 7.4 ± 0.54 | 0.66 ± 0.05 | 0.186 ± 0.014 |
Corresponding bar plots are shown in Figure 6D and E. Given are the amino acid concentrations, l-valine titers, cell dry weight-(CDW-) specific l-valine titers and l-valine yields after 46 h of cultivation. Means and standard deviations of biological triplicates are listed. pJC1-Plys_CS_0-ilvBNC-RBS-ilvE*: Calculations were based on measurements shown in Figure 6E. The CDW was calculated by the following equation: CDW = OD600 * 0.25 g L-1 76.
The toggle strain PgntK-aceE/pJC1-Plys_CS_0-ilvBNC-RBS-ilvE produced 1.9-fold higher amounts of l-valine than cells harbouring the empty plasmid pJC1 (Figure 6D, Table 1), confirming the importance of increased levels of biosynthesis enzymes. However, in comparison to the previously characterized plasmid pJC4-PilvB-ilvBNC-PilvE-ilvE (pJC4-ilvBNCE)57, l-valine titers were reduced by about 50% in strains ΔaceE and PgntK-aceE. In contrast, pyruvate-derived by-product formation (l-alanine) was 4-5-fold increased (Figure 6D, Table 1). This shift in amino acid concentrations hinted towards limitations in the conversion of pyruvate to l-valine, apparently caused by lower promoter activity of the counter-silencer (Plys_CS_0) compared to the native promoters PilvB and/or PilvE and the resulting lower enzyme levels. The reporter outputs of the synthetic counter-silencer promoters Plys_CS_+1 and Plys_CS_-5 were 3-fold and 3.4-fold higher in their induced state referred to the counter-silencer promoter Plys_CS_0 (Figure 2). Therefore, their applicability in the metabolic l-valine production toggle was characterized in a next step (Figure 6E, Table 1). In comparison to the strains based on the regulatory element Plys_CS_0, both promoter variants improved the product titers of l-valine by about 30-45% (Figure 6E, Table 1), while by-product formation (l-alanine) was reduced by 10-25% (Table 1). Although the l-valine titers did not reach the levels obtained with the native promoters, these results demonstrated the functionality of the modular toggle design as well as the potential for further optimization.
AHAS (ilvBN) is the key enzyme of the l-valine biosynthesis pathway and catalyzed the first reaction step. AHAS is feedback inhibited by branched chain amino acids like l-valine58, 59 and its biosynthesis intermediate α-ketoisovalerate40. In previous AHAS engineering approaches, a genomically encoded, feedback resistant enzyme variant led to increased l-valine production59, suggesting that this reaction step can be a bottleneck for efficient substrate conversion. Consistently, several engineering approaches succeeded in increasing l-valine titers by increasing the gene dosage of l-valine biosynthesis genes by plasmid-based expression35, 57, 59, 60.
In conclusion, the production performance of the here presented toggle strains was not optimal in comparison to previously established l-valine production strains, probably due to lower ilvBNCE transcription levels. However, the achieved increase of l-valine titers by using a stronger counter-silencer promoter demonstrated the potential for improvement of this metabolic toggle. For example, the generation of optimized counter-silencer promoters featuring an increased dynamic range and maximal promoter activity (Figure 2) might provide promising candidates for further toggle strain optimizations.
In the absence of gluconate, both platform strains (PgntK-aceE and ΔaceE) produced comparable amounts of l-valine indicating that the PDHC was inactive in the toggle strain redirecting carbon flux towards l-valine (Figure 6D, Table 1). However, higher cell densities of the C. glutamicum PgntK-aceE strain observed during co-utilization of 222 mM glucose and 51 mM gluconate (Figure 6C) showed on the one hand the potential to overcome growth dependency on costly and inefficient carbon sources like acetate and, on the other hand, the potential to improve substrate to product conversion by accelerating biomass formation. Further in-depth analyses of this effect are indispensable for final evaluation of the dynamically controlled PgntK-aceE strain.
Conclusion
Tightly controlled, inducible promoter systems are key modules in biotechnological and synthetic biological applications and represent the most frequently used principle for controlling gene expression1, 2. In this study, we demonstrated the potential of synthetic counter-silencer promoters as expression system allowing for a more stringent control of gene expression in comparison to well-established systems. The here presented promoter constructs are tightly repressed by CgpS-mediated xenogeneic silencing and induced by GntR-dependent counter-silencing. Synthetic promoter variants were combined in a GntR-dependent bilateral promoter library comprising in total 44 regulatory elements which were repressed (16 promoters) or activated (28 promoters) in the presence of the non-toxic and cheap effector molecule gluconate. These sets of regulatory elements provided a wide range of promoter activities meeting the requirements of various applications.
The effect of varying effector concentrations on the dynamics and timing of promoter activities was analyzed in detail for exemplary chosen synthetic GntR target promoters revealing that counter-silencer constructs are expression systems with temporal tunability.
Furthermore, we performed a comprehensive characterization and application of a GntR-dependent toggle switch allowing for opposed expression of two sets of target genes. This toggle is controlled by only one TF (GntR) and one effector molecule (gluconate) and integrates two promoters displaying an inverse response to gluconate-dependent GntR binding18. Finally, the GntR-dependent toggle was successfully applied as metabolic toggle switch for the dynamic redirection of carbon flux between the growth-related formation of acetyl-CoA and l-valine biosynthesis, both competing for the precursor molecule pyruvate.
In conclusion, our results demonstrated the high potential of XS target promoters for synthetic circuit designs and the establishment of tightly controlled gene expression systems.
Methods
Bacterial strains and cultivation conditions
All bacterial strains and plasmids used in this project are listed in Table S2, S3 and S4. The strain C. glutamicum ATCC 1303261 served as wild type strain. All plasmids and their sequences are available from the corresponding author on request. For cultivations of C. glutamicum strains, brain heart infusion (BHI, Difco Laboratories, Detroit, MI, USA) complex medium was inoculated with a single colony from a fresh BHI agar plate and incubated for 8 to 16 hours. For cultivation of ΔaceE deletion strains as well as cells with dynamically controlled aceE expression (PgntK-aceE), BHI medium was supplemented 85 mM potassium acetate. If not stated otherwise, cultivation steps were performed with 1 - 1.1 ml medium at 30°C, 900 rpm and 75% humidity in sterile 2.2 ml square-shaped V-bottom 96 deep-well plates (VWR, Radnor, PA) covered with a sterile breathable rayon film (VWR, Radnor, PA) in a Microtron incubator shaker (Infors, Bottmingen, Switzerland), in four ml medium in glass tubes at 30°C and 160 rpm or in 15-25 ml medium in round 100 ml glass shaking flasks with two facing baffles covered with a metal cap at 30°C and 120 rpm. For reporter-based assays and analysis of relative venus transcript levels driven by the different expression systems, BHI pre-cultures were used to inoculate a second overnight pre-culture in CGXII minimal medium (buffered medium, initial pH 7)62 supplemented with 25 µg/ml kanamycin and 100 mM sodium gluconate (named gluconate in the following) or 100 mM glucose. For growth analysis of ΔaceE- and PgntK-aceE-based strains, the second overnight pre-culture was performed in CGXII minimal medium containing 222 mM glucose and 254 mM acetate. Subsequently, the second pre-culture was used to inoculate the main culture at a start OD600 of one. For reporter-based assays, the main cultures were grown in CGXII medium with 25 µg/ml kanamycin and 100 mM gluconate, 100/111 mM glucose without or with varying supplemented amounts of gluconate (0, 1, 10, 50, 100 mM), 100 mM glucose with either 100 µM IPTG (induction of Ptac) or 235 nM ATc (induction of Ptet). Growth analysis of the ΔaceE deletion strains as well as cells with dynamically controlled aceE expression (PgntK-aceE) were performed in CGXII minimal medium supplemented with either 222 mM glucose and 254 mM acetate, 222 mM glucose, 222 mM glucose and 51 mM gluconate or 51 mM gluconate. E. coli DH5α was used for plasmid amplification and cells were cultivated in 10-15 ml Lysogeny Broth (LB, 5 g L-1 NaCl, 10 g L-1 tryptone, 10 g L-1 yeast extract) medium in shaking flasks at 37°C and 120 rpm or on LB agar plates at 37°C. If needed, 50 µg/ml kanamycin was added. For long-term storage, single colonies from C. glutamicum and E. coli DH5α strains were cultivated overnight in tubes in four ml BHI or LB medium supplemented with appropriate amounts of antibiotic. Glycerol was added to the culture to a final concentration of 20% and 900 µl of the culture were stored at -80°C.
Microtiter cultivation to monitor cell growth and fluorescence
Analysis of cell growth and reporter-based assays were performed in microliter scale in the BioLector® microcultivation system (m2p-labs, Aachen, Germany)63. Main cultures with a volume of 750 µl (see Bacterial strains and cultivation conditions) and a starting OD600 of one were incubated in 48-well FlowerPlates® (m2p-labs, Aachen, Germany) covered with a sterile breathable rayon film (VWR, Radnor, PA) at 30°C, 1200 rpm and 85% humidity. If needed, Venus fluorescence was measured with an excitation wavelength of 508 nm and emission wavelength of 532 nm (signal gain factor 60). Biomass production was measured as backscattered light intensity of sent light with a wavelength of 620 nm (signal gain factor 20). All samples were measured at 15 min intervals. Arbitrary units (a.u.) of specific fluorescence were calculated by dividing the Venus signal by the backscatter signal per time point63. Growth rates were determined for the exponential growth phase based on the backscatter signal.
Microtiter cultivation of l-valine production strains
For comparison of the productivity of the l-valine production strains ΔaceE and PgntK-aceE harbouring different plasmids, per strain, three single colonies from a fresh BHI agar plate supplemented with 85 mM potassium acetate and 25 µg/ml kanamycin were used to inoculate either tubes containing four ml BHI supplemented with 85 mM potassium acetate (named acetate in the following) and 25 µg/ml kanamycin or 96-deep well plates containing one ml medium per well. After 10 hours of cultivation in tubes at 30°C and 160 rpm or in 96-deep well plates at 30°C, 900 rpm and 75% humidity, cells from the first pre-culture were transferred into fresh CGXII minimal medium (1:10 diluted) supplemented with 222 mM glucose, 254 mM potassium acetate and 25 µg/ml kanamycin. Cultivation was either performed in 100 ml shaking flasks (25 ml culture volume) at 30°C and 120 rpm for 13 hours or in 96 deep-well plates (1.5 ml culture volume) at 30°C, 900 rpm and 75% humidity. . Subsequently, cells were washed with 0.9% NaCl solution and used to inoculate 5-25 ml CGXII containing 222 mM glucose, 254 mM potassium acetate and 25 µg/ml kanamycin. The following cultivation was performed in 96 deep-well plates. Therefore, 1.5 ml culture aliquots were transferred into plates and incubated at 30°C, 900 rpm and 75% humidity. Per sample time point and clone, the complete 1.5 ml culture volume of one well was transferred into a 2 ml reaction tube. Separation of cells and supernatant was performed by centrifugation for 5-30 min at 11,325 x g and 4°C. The supernatant was transferred into a fresh 1.5 ml tube and stored at -20°C before it was used for quantification of l-valine, l-alanine and glucose concentrations.
Cultivation in microfluidic chip devices
For the characterization of reversibility and dynamics of the fluorescence-based GntR-dependent toggle (pJC1-Plys_CS_0-venus-T-PgntK-e2-crimson)18, C. glutamicum wild type cells harbouring the plasmid-based construct were cultivated in an in-house developed microfluidic platform64, 65. Therefore, tubes containing four ml BHI and 25 µg/ml kanamycin were inoculated with a single colony from a BHI agar plate with the same antibiotic concentration and incubated at 160 rpm and 30°C for eight hours. All further incubation steps of this strain were performed with 25 µg/ml kanamycin. One ml of the first pre-culture was transferred to 20 ml CGXII minimal medium containing 100 mM glucose as carbon source and cells were cultivated overnight in a 100 ml shaking flasks at 120 rpm and 30°C. After approximately 16 hours, cells from the second pre-culture were used to inoculate the main culture in 15 ml CGXII containing 100 mM glucose in 100 ml shaking flasks to an OD600 of one. This culture was incubated for 3-4 hours at 30°C and 120 rpm until cells were used to inoculate the microfluidic chip. The experimental setup and chip design was performed as previously described 66. Venus and E2-Crimson fluorescence as well as phase contrast were imaged at 15 min intervals by fully motorized inverted Nikon Eclipse Ti microscope (Nikon GmbH, Düsseldorf, Germany) as described before64, 65, 67. The exposure times for phase contrast was 100 ms, for Venus 200 ms and for E2-Crimson 300 ms. In the microfluidic chip system, cells were cultivated in CGXII medium supplemented with either 100 mM gluconate or 100 mM glucose. A high-precision syringe pump system (neMESYS, Cetoni GmbH, Korbussen, Germany) and disposable syringes (Omnifix-F Tuberculin, 1 ml; B. Braun Melsungen AG, Melsungen, Germany) were used to achieve continuous medium supply (flow rate of 200 nl*min-1) and waste removal during the cultivation. The switches of carbon source supply between gluconate and glucose were performed by changing the syringes and the connecting tubing to ensure an immediate medium switch. The temperature was set to 30°C using an incubator system (PeCon GmbH, Erbach, Germany). Data analysis on colony level was performed using the image-processing package Fiji68 which is based on ImageJ69. Measured fluorescence data were background normalized and plotted with GraphPad prism 7.00 (GraphPad Software, La Jolla. CA. USA).
Flow cytometry to monitor fluorescence state of the GntR-dependent toggle switch
For the characterization of responsiveness of the fluorescence-based GntR-dependent toggle to gradual gluconate concentrations, C. glutamicum wild type cells harbouring the corresponding plasmid (pJC1-Plys_CS_0-venus-T-PgntK-e2-crimson 18) were measured via flow cytometry. Therefore, single colonies from a fresh BHI agar plate were used to inoculate the first pre-culture (tube, four ml BHI supplemented with 25 µg/ml kanamycin) which was incubated for eight hours at 160 rpm and 30°C. Subsequently, one ml of the first main culture were used to inoculate 20 ml CGXII supplemented with 100 mM glucose and 25 µg/ml kanamycin (100 ml shaking flasks) and cells were cultivated overnight (~ 16 hours) at 120 rpm and 30°C. Cells from this second pre-culture were used to inoculate the main culture at a starting OD600 of one. The following cultivation was performed in CGXII medium supplemented with 25 µg/ml kanamycin and varying amounts of gluconate and glucose (100 mM carbon source in total) in 96 deep-well plates (1.5 ml culture volume) at 30°C, 900 rpm and 75% humidity. After five hours of cultivation, flow cytometry analyses was performed on a FACSAria™ III (Becton Dickinson, San Jose, USA) equipped with a blue (Venus, 488 nm excitation) and a yellow-green (E2-crimson, 561 nm excitation) solid state laser. Forward-scatter (FSC) and side-scatter characteristics (SSC) were detected as small and large angle scatters of the 488 nm laser, respectively. Venus fluorescence was detected using a 530/30-nm band-pass filter and E2-crimson fluorescence was measured using a 610/20 band pass filter. FACS-Diva software v8.0.2. was used to adjust and record the measurements. Thresholds on FSC and SSC were used to remove noise. For analysis, C. glutamicum culture samples were diluted to an OD600 of 0.05 in FACSFlow™ sheath fluid buffer (BD, Heidelberg, Germany). Data were analyzed using FlowJo v10.6.2 analysis software (TreeStar, Ashland, USA).
Recombinant DNA work
All standard molecular methods such as DNA restriction, PCR and Gibson assembly were performed according to manufacturer instructions or following previously described standard protocols70, 71. Details on plasmid construction by Gibson assembly are provided in Table S4. DNA sequencing and synthesis of oligonucleotides used for amplification of DNA fragments for plasmid construction (Table S5) and sequencing (Table S6 and S7) were performed by Eurofins Genomics (Ebersberg, Germany). Chromosomal DNA of C. glutamicum ATCC 13032 used as PCR template was prepared as described previously72. PpriP-based counter-silencer promoters were constructed as previously described18.
Construction of strains ΔgntK and PgntK-aceE
For the deletion of the gene gntK encoding the gluconate kinase GntK, C. glutamicum ATCC 13032 wild type cells61 were transformed with the pK19mobsacB-ΔgntK deletion plasmid (Table S4). The plasmid pK19-mobsacB-ΔPaceE-aceE was used to delete the aceE gene and its promoter before the plasmid pK19mobsacB-PgntK-aceE (Table S4) was used for the re-integration of the aceE gene under control of the GntR target promoter PgntK within its native locus. Subsequently, two step homologous recombination and selection was performed as described previously73. Successful integration was verified by sequencing, all primers are listed in Table S7.
Quantitative Real-time PCR (qRT-PCR)
For the comparison of promoter strength of PgntK, Plys_CS_0 and PpriP_CS_0 with the established promoter systems Ptet and Ptac on transcript levels, promoter sequences were cloned into the plasmid pJC1 and fused to the reporter gene venus via a consistent linker containing the conserved ribosomal binding site (AGGAG47). Since LacI and TetR are natively not encoded in the chromosome of C. glutamicum, the corresponding genes were inserted into the respective plasmids. Duplicates of single clones of C. glutamicum wild type cells harbouring the plasmid-based constructs or the control plasmid pJC1-venus-term (no appropriate promoter in front of venus 74) were analyzed. The first BHI pre-culture (see Bacterial strains and cultivation conditions) was used to inoculate the second pre-culture in one ml CGXII medium with either 100 mM gluconate or glucose as carbon source (non-inducing conditions). Cells from the second overnight pre-culture were used to inoculate 1.6 ml of the main culture starting with OD600 of one. All cultivation steps were performed in 96 deep-well plates at 30°C, 900 rpm and 75% humidity. Cells harbouring different constructs were cultivated under non-inducing and inducing conditions depending on the particular promoter construct: PpriP_CS_0 and Plys_CS_0: +: 100 mM glucose, -: 100 mM gluconate; PgntK: +: 100 mM gluconate, -: 100 mM glucose; Ptac (100 mM glucose): +: 100 µM IPTG, -: 0 µM IPTG; Ptet (100 mM glucose): +: 235 nM ATc, -: 0 nM ATc. After five hours of incubation, cells were cooled on ice and harvested by centrifugation (4°C, 10 min, 11,325 x g). Cell pellets were snap-frozen in liquid nitrogen and stored at -80°C until use.
Total RNA was isolated using the Monarch® Total RNA Miniprep Kit (New England Bio Labs, Ipswitch, MA, USA) following the manufacturer protocol. qRT-PCR was performed with 5 ng total RNA and primers listed in Table S8 using the Luna® Universal One-Step RT-qPCR Kit (New England Biolabs, Ipswitch, USA). Measurements of the relative venus expression levels were done in biological and technical duplicates with a qTower 2.2 system (Analytik Jena, Jena, Germany). The chromosomal encoded ddh gene served as reference gene. The software qPCR-soft 3.1 (Analytik Jena, Jena, Germany) was used for obtaining the corresponding Ct-values. Means of relative venus expression levels and the corresponding range were calculated based on the 2-ΔCT method of Livak and Schmittgen75 using the following equations:
Plots in Figure 3C are showing the means of relative venus expression levels and error bars their range. Maximum and minimum of the range was calculated by the following equations:
Quantification of amino acid concentrations
The amino acids l-valine and l-alanine were quantified as ortho-phthaldialdehyde derivatives by using ultra-high performance liquid chromatography (uHPLC) or high performance liquid chromatography (HPLC) with an automatic pre-column derivatization. Derivatives were separated by reverse-phase chromatography using an Agilent 1290 Infinity LC ChemStation or an Agilent 1260 Infinity II LC system (Agilent, Santa Clara, USA) equipped with a fluorescence detector. A gradient of sodium borate buffer (10 mM Na2HPO4, 10 mM Na2B4O7, pH 8.2) and methanol was applied as eluent for the Zorbax Eclipse AAA 3.5 micron 4.6 x 7.5 mm column (Agilent, Santa Clara, USA). Prior to analysis, samples were centrifuged for 5-30 min at 11,325x g and 4°C and were either measured directly or as dilutions of 1:100, 1:400 or 1:1000 in ddH2O.
Determination of the glucose concentration in the culture supernatant
Glucose concentrations in the supernatant were determined with the D-Glucose UV-Test Kit (R-Biopharm, Darmstadt, Germany) according to the manufacturer’s instructions (absorption was measured at 340 nm) with the modification that measurements were performed with only half of the indicated reaction volume. Glucose concentrations were calculated as described in the manual.
Supplementary Material
Acknowledgement
We thank the European Research Council (ERC Starting Grant, grant number 757563) and the Helmholtz Association (grant number W2/W3-096) for financial support.
Footnotes
Author Contributions
J.W. and J.F. designed the experiments. J.W., C.G. and A.W. performed the experiments. J.W. and J.F. wrote the manuscript.
Conflict of Interest
The authors declare no competing financial interest.
References
- 1.Liu X, Yang Y, Zhang W, Sun Y, Peng F, Jeffrey L, Harvey L, McNeil B, Bai Z. Expression of recombinant protein using Corynebacterium glutamicum: progress, challenges and applications. Crit. Rev. Biotechnol. 2019;36:652–664. doi: 10.3109/07388551.2015.1004519. [DOI] [PubMed] [Google Scholar]
- 2.Patek M, Holatko J, Busche T, Kalinowski J, Nesvera J. Corynebacterium glutamicum promoters: a practical approach. Microb. Biotechnol. 2013;6:103–117. doi: 10.1111/1751-7915.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xia PF, Ling H, Foo JL, Chang MW. Synthetic genetic circuits for programmable biological functionalities. Biotechnol Adv. 2019;37 doi: 10.1016/j.biotechadv.2019.04.015. 107393. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Samoun K, Leblon G, Reyes O. Positively regulated expression of the Escherichia coli araBAD promoter in Corynebacterium glutamicum . FEMS Microbiol. Lett. 1999;174:125–130. doi: 10.1111/j.1574-6968.1999.tb13558.x. [DOI] [PubMed] [Google Scholar]
- 5.Lausberg F, Chattopadhyay AR, Heyer A, Eggeling L, Freudl R. A tetracycline inducible expression vector for Corynebacterium glutamicum allowing tightly regulable gene expression. Plasmid. 2012;68:142–147. doi: 10.1016/j.plasmid.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Radmacher E, Stansen KC, Besra GS, Alderwick LJ, Maughan WN, Hollweg G, Sahm H, Wendisch VF, Eggeling L. Ethambutol, a cell wall inhibitor of Mycobacterium tuberculosis elicits L-glutamate efflux of Corynebacterium glutamicum . Microbiology (Reading, U. K.) 2005;151:1359–1368. doi: 10.1099/mic.0.27804-0. [DOI] [PubMed] [Google Scholar]
- 7.Yim SS, An SJ, Kang M, Lee J, Jeong KJ. Isolation of fully synthetic promoters for high-level gene expression in Corynebacterium glutamicum . Biotechnol. Bioeng. 2013;110:2959–2969. doi: 10.1002/bit.24954. [DOI] [PubMed] [Google Scholar]
- 8.Baritugo KA, Kim HT, David Y, Choi JI, Hong SH, Jeong KJ, Choi JH, Joo JC, Park SJ. Metabolic engineering of Corynebacterium glutamicum for fermentative production of chemicals in biorefinery. Appl. Microbiol. Biotechnol. 2018;102:3915–3937. doi: 10.1007/s00253-018-8896-6. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Shang X, Lai S, Zhang G, Liang Y, Wen T. Development and application of an arabinose-inducible expression system by facilitating inducer uptake in Corynebacterium glutamicum . Appl. Environ. Microbiol. 2012;78:5831–5838. doi: 10.1128/AEM.01147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okibe N, Suzuki N, Inui M, Yukawa H. Isolation, evaluation and use of two strong, carbon source-inducible promoters from Corynebacterium glutamicum . Lett. Appl. Microbiol. 2010;50:173–180. doi: 10.1111/j.1472-765X.2009.02776.x. [DOI] [PubMed] [Google Scholar]
- 11.Letek M, Valbuena N, Ramos A, Ordonez E, Gil JA, Mateos LM. Characterization and use of catabolite-repressed promoters from gluconate genes in Corynebacterium glutamicum . J. Bacteriol. 2006;188:409–423. doi: 10.1128/JB.188.2.409-423.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hentschel E, Will C, Mustafi N, Burkovski A, Rehm N, Frunzke J. Destabilized eYFP variants for dynamic gene expression studies in Corynebacterium glutamicum . Microb. Biotechnol. 2013;6:196–201. doi: 10.1111/j.1751-7915.2012.00360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plassmeier JK, Busche T, Molck S, Persicke M, Puhler A, Ruckert C, Kalinowski J. A propionate-inducible expression system based on the Corynebacterium glutamicum prpD2 promoter and PrpR activator and its application for the redirection of amino acid biosynthesis pathways. J. Biotechnol. 2013;163:225–232. doi: 10.1016/j.jbiotec.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 14.Mustafi N, Grünberger A, Kohlheyer D, Bott M, Frunzke J. The development and application of a single-cell biosensor for the detection of L-methionine and branched-chain amino acids. Metab Eng. 2012;14:449–457. doi: 10.1016/j.ymben.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Binder S, Schendzielorz G, Stäbler N, Krumbach K, Hoffmann K, Bott M, Eggeling L. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol. 2012;13:R40. doi: 10.1186/gb-2012-13-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang X, Chai X, Lu X, Li Y, Zhang Y, Wang G, Zhang C, Liu S, Zhang Y, Ma J, Wen T. Native promoters of Corynebacterium glutamicum and its application in L-lysine production. Biotechnol. Lett. 2018;40:383–391. doi: 10.1007/s10529-017-2479-y. [DOI] [PubMed] [Google Scholar]
- 17.Rytter JV, Helmark S, Chen J, Lezyk MJ, Solem C, Jensen PR. Synthetic promoter libraries for Corynebacterium glutamicum . Appl. Microbiol. Biotechnol. 2014;98:2617–2623. doi: 10.1007/s00253-013-5481-x. [DOI] [PubMed] [Google Scholar]
- 18.Wiechert J, Filipchyk A, Hünnefeld M, Gätgens C, Brehm J, Heermann R, Frunzke J. Deciphering the rules underlying xenogeneic silencing and counter-silencing of Lsr2-like proteins using CgpS of Corynebacterium glutamicum as a model. mbio. 2020;11:e02273–02219. doi: 10.1128/mBio.02273-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfeifer E, Hünnefeld M, Popa O, Frunzke J. Impact of xenogeneic silencing on phage-host interactions. J. Mol. Biol. 2019;431:4670–4683. doi: 10.1016/j.jmb.2019.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navarre WW. The impact of gene silencing on horizontal gene transfer and bacterial evolution. Adv. Microb. Physiol. 2016;69:157–186. doi: 10.1016/bs.ampbs.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Will WR, Navarre WW, Fang FC. Integrated circuits: how transcriptional silencing and counter-silencing facilitate bacterial evolution. Curr. Opin. Microbiol. 2015;23:8–13. doi: 10.1016/j.mib.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella . Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 23.Oshima T, Ishikawa S, Kurokawa K, Aiba H, Ogasawara N. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 2006;13:141–153. doi: 10.1093/dnares/dsl009. [DOI] [PubMed] [Google Scholar]
- 24.Gordon BR, Imperial R, Wang L, Navarre WW, Liu J. Lsr2 of Mycobacterium represents a novel class of H-NS-like proteins. J. Bacteriol. 2008;190:7052–7059. doi: 10.1128/JB.00733-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tendeng C, Soutourina OA, Danchin A, Bertin PN. MvaT proteins in Pseudomonas spp.: a novel class of H-NS-like proteins. Microbiology (Reading, U. K.) 2003;149:3047–3050. doi: 10.1099/mic.0.C0125-0. [DOI] [PubMed] [Google Scholar]
- 26.Smits WK, Grossman AD. The transcriptional regulator Rok binds A+T-rich DNA and is involved in repression of a mobile genetic element in Bacillus subtilis . PLoS Genet. 2010;6:e1001207. doi: 10.1371/journal.pgen.1001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfeifer E, Hünnefeld M, Popa O, Polen T, Kohlheyer D, Baumgart M, Frunzke J. Silencing of cryptic prophages in Corynebacterium glutamicum . Nucleic Acids Res. 2016;44:10117–10131. doi: 10.1093/nar/gkw692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frunzke J, Engels V, Hasenbein S, Gatgens C, Bott M. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol. Microbiol. 2008;67:305–322. doi: 10.1111/j.1365-2958.2007.06020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soma Y, Tsuruno K, Wada M, Yokota A, Hanai T. Metabolic flux redirection from a central metabolic pathway toward a synthetic pathway using a metabolic toggle switch. Metab Eng. 2014;23:175–184. doi: 10.1016/j.ymben.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Soma Y, Yamaji T, Matsuda F, Hanai T. Synthetic metabolic bypass for a metabolic toggle switch enhances acetyl-CoA supply for isopropanol production by Escherichia coli . J. Biosci. Bioeng. 2017;123:625–633. doi: 10.1016/j.jbiosc.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 31.Tsuruno K, Honjo H, Hanai T. Enhancement of 3-hydroxypropionic acid production from glycerol by using a metabolic toggle switch. Microb. Cell Fact. 2015;14:155. doi: 10.1186/s12934-015-0342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta A, Reizman IM, Reisch CR, Prather KL. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat. Biotechnol. 2017;35:273–279. doi: 10.1038/nbt.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doong SJ, Gupta A, Prather KLJ. Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli . Proc Natl Acad Sci U S A. 2018;115:2964–2969. doi: 10.1073/pnas.1716920115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becker J, Zelder O, Hafner S, Schroder H, Wittmann C. From zero to hero--design-based systems metabolic engineering of Corynebacterium glutamicum for L-lysine production. Metab Eng. 2011;13:159–168. doi: 10.1016/j.ymben.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Blombach B, Schreiner ME, Holatko J, Bartek T, Oldiges M, Eikmanns BJ. L-valine production with pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum . Appl. Environ. Microbiol. 2007;73:2079–2084. doi: 10.1128/AEM.02826-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Litsanov B, Kabus A, Brocker M, Bott M. Efficient aerobic succinate production from glucose in minimal medium with Corynebacterium glutamicum . Microb. Biotechnol. 2012;5:116–128. doi: 10.1111/j.1751-7915.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blombach B, Schreiner ME, Bartek T, Oldiges M, Eikmanns BJ. Corynebacterium glutamicum tailored for high-yield L-valine production. Appl. Microbiol. Biotechnol. 2008;79:471–479. doi: 10.1007/s00253-008-1444-z. [DOI] [PubMed] [Google Scholar]
- 38.Peters-Wendisch P, Stolz M, Etterich H, Kennerknecht N, Sahm H, Eggeling L. Metabolic engineering of Corynebacterium glutamicum for L-Serine production. Appl. Environ. Microbiol. 2005;71:7139–7144. doi: 10.1128/AEM.71.11.7139-7144.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hüser AT, Chassagnole C, Lindley ND, Merkamm M, Guyonvarch A, Elišáková V, Pátek M, Kalinowski J, Brune I, Pühler A, Tauch A. Rational design of a Corynebacterium glutamicum pantothenate production strain and its characterization by metabolic flux analysis and gnome-wide transcriptional profiling. Appl. Environ. Microbiol. 2005;71:3255–3268. doi: 10.1128/AEM.71.6.3255-3268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krause FS, Blombach B, Eikmanns BJ. Metabolic engineering of Corynebacterium glutamicum for 2-ketoisovalerate production. Appl. Environ. Microbiol. 2010;76:8053–8061. doi: 10.1128/AEM.01710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brockman IM, Prather KLJ. Dynamic metabolic engineering: New strategies for developing responsive cell factories. Biotechnol. J. 2015;10:1360–1369. doi: 10.1002/biot.201400422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farmer WR, Liao JC. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat. Biotechnol. 2000;18:533–537. doi: 10.1038/75398. [DOI] [PubMed] [Google Scholar]
- 43.Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat. Biotechnol. 2012;30:354–359. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- 44.Brockman IM, Prather KLJ. Dynamic knockdown of E. coli central metabolism for redirecting fluxes of primary metabolites. Metab Eng. 2015;28:104–113. doi: 10.1016/j.ymben.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lo TM, Chng SH, Teo WS, Cho HS, Chang MW. A two-layer gene circuit for decoupling cell growth from metabolite production. Cell Syst. 2016;3:133–143. doi: 10.1016/j.cels.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 46.Tsuchiya M, Morinaga Y. Genetic control systems of Escherichia coli can confer inducible expression of cloned genes in coryneform bacteria. Nat. Biotechnol. 1988;6:428–430. [Google Scholar]
- 47.Pfeifer-Sancar K, Mentz A, Rückert C, Kalinowski J. Comprehensive analysis of the Corynebacterium glutamicum transcriptome using an improved RNAseq technique. BMC Genomics. 2013;14:888. doi: 10.1186/1471-2164-14-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evfratov SA, Osterman IA, Komarova ES, Pogorelskaya AM, Rubtsova MP, Zatsepin TS, Semashko TA, Kostryukova ES, Mironov AA, Burnaev E, Krymova E, et al. Application of sorting and next generation sequencing to study 5’-UTR influence on translation efficiency in Escherichia coli . Nucleic Acids Res. 2017;45:3487–3502. doi: 10.1093/nar/gkw1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arnold TE, Yu J, Belasco JG. mRNA stabilization by the ompA 5' untranslated region: two protective elements hinder distinct pathways for mRNA degradation. RNA (New York, N.Y.) 1998;4:319–330. [PMC free article] [PubMed] [Google Scholar]
- 50.Snapp EL. Fluorescent proteins: a cell biologist's user guide. Trends Cell Biol. 2009;19:649–655. doi: 10.1016/j.tcb.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli . Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 52.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat. Rev. Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oldiges M, Eikmanns BJ, Blombach B. Application of metabolic engineering for the biotechnological production of L-valine. Appl. Microbiol. Biotechnol. 2014;98:5859–5870. doi: 10.1007/s00253-014-5782-8. [DOI] [PubMed] [Google Scholar]
- 54.Ma Y, Cui Y, Du L, Liu X, Xie X, Chen N. Identification and application of a growth-regulated promoter for improving L-valine production in Corynebacterium glutamicum . Microb. Cell Fact. 2018;17:185. doi: 10.1186/s12934-018-1031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schreiner ME, Fiur D, Holatko J, Patek M, Eikmanns BJ. E1 enzyme of the pyruvate dehydrogenase complex in Corynebacterium glutamicum: molecular analysis of the gene and phylogenetic aspects. J. Bacteriol. 2005;187:6005–6018. doi: 10.1128/JB.187.17.6005-6018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buchholz J, Schwentner A, Brunnenkan B, Gabris C, Grimm S, Gerstmeir R, Takors R, Eikmanns BJ, Blombach B. Platform engineering of Corynebacterium glutamicum with reduced pyruvate dehydrogenase complex activity for improved production of L-lysine, L-valine, and 2-ketoisovalerate. Appl. Environ. Microbiol. 2013;79:5566–5575. doi: 10.1128/AEM.01741-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Radmacher E, Vaitsikova A, Burger U, Krumbach K, Sahm H, Eggeling L. Linking central metabolism with increased pathway flux: L-valine accumulation by Corynebacterium glutamicum . Appl. Environ. Microbiol. 2002;68:2246–2250. doi: 10.1128/AEM.68.5.2246-2250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eggeling I, Cordes C, Eggeling L, Sahm H. Regulation of acetohydroxy acid synthase in Corynebacterium glutamicum during fermentation of α-ketobutyrate to l-isoleucine. Appl. Microbiol. Biotechnol. 1987;25:346–351. [Google Scholar]
- 59.Elišáková V, Pátek M, Holátko J, Nešvera J, Leyval D, Goergen J-L, Delaunay S. Feedback-resistant acetohydroxy acid synthase increases valine production in Corynebacterium glutamicum . Appl Environ Microbiol. 2005;71:207–213. doi: 10.1128/AEM.71.1.207-213.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sahm H, Eggeling L. D-Pantothenate synthesis in Corynebacterium glutamicum and use of panBC and genes encoding L-valine synthesis for D-pantothenate overproduction. Appl. Environ. Microbiol. 1999;65:1973–1979. doi: 10.1128/aem.65.5.1973-1979.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kinoshita S, Udaka S, Shimono M. Studies on the amino acid fermentation. Part 1. Production of L-glutamic acid by various microorganisms. J Gen Appl Microbiol. 1957;3:193–205. [PubMed] [Google Scholar]
- 62.Keilhauer C, Eggeling L, Sahm H. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 1993;175:5595–5603. doi: 10.1128/jb.175.17.5595-5603.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kensy F, Zang E, Faulhammer C, Tan RK, Büchs J. Validation of a high-throughput fermentation system based on online monitoring of biomass and fluorescence in continuously shaken microtiter plates. Microb. Cell Fact. 2009;8:31. doi: 10.1186/1475-2859-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grünberger A, Paczia N, Probst C, Schendzielorz G, Eggeling L, Noack S, Wiechert W, Kohlheyer D. A disposable picolitre bioreactor for cultivation and investigation of industrially relevant bacteria on the single cell level. Lab Chip. 2012;12:2060–2068. doi: 10.1039/c2lc40156h. [DOI] [PubMed] [Google Scholar]
- 65.Grünberger A, Probst C, Helfrich S, Nanda A, Stute B, Wiechert W, von Lieres E, Nöh K, Frunzke J, Kohlheyer D. Spatiotemporal microbial single-cell analysis using a high-throughput microfluidics cultivation platform. Cytometry, Part A. 2015;87:1101–1115. doi: 10.1002/cyto.a.22779. [DOI] [PubMed] [Google Scholar]
- 66.Grünberger A, Probst C, Heyer A, Wiechert W, Frunzke J, Kohlheyer D. Microfluidic picoliter bioreactor for microbial single-cell analysis: fabrication, system setup, and operation. J. Visualized Exp. 2013:e50560. doi: 10.3791/50560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Helfrich S, Pfeifer E, Krämer C, Sachs CC, Wiechert W, Kohlheyer D, Nöh K, Frunzke J. Live cell imaging of SOS and prophage dynamics in isogenic bacterial populations. Mol. Microbiol. 2015;98:636–650. doi: 10.1111/mmi.13147. [DOI] [PubMed] [Google Scholar]
- 68.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinf. 2017;18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sambrook J, Russel WD. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 71.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 72.Eikmanns BJ, Thum-Schmitz N, Eggeling L, Ludtke KU, Sahm H. Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology (Reading, U K) 1994;140:1817–1828. doi: 10.1099/13500872-140-8-1817. [DOI] [PubMed] [Google Scholar]
- 73.Niebisch A, Bott M. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch. Microbiol. 2001;175:282–294. doi: 10.1007/s002030100262. [DOI] [PubMed] [Google Scholar]
- 74.Baumgart M, Unthan S, Rückert C, Sivalingam J, Grünberger A, Kalinowski J, Bott M, Noack S, Frunzke J. Construction of a prophage-free variant of Corynebacterium glutamicum ATCC 13032 for use as a platform strain for basic research and industrial biotechnology. Appl. Environ. Microbiol. 2013;79:6006–6015. doi: 10.1128/AEM.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, CA, U S) 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 76.Kabus A, Georgi T, Wendisch VF, Bott M. Expression of the Escherichia coli pntAB genes encoding a membrane-bound transhydrogenase in Corynebacterium glutamicum improves l-lysine formation. Appl. Microbiol. Biotechnol. 2007;75:47–53. doi: 10.1007/s00253-006-0804-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.