Abstract
Regeneration of lost body parts is essential to regain the fitness of the organism for successful living. In the animal kingdom organisms from different clades exhibit varied regeneration abilities. Hydra is one of the few organisms that possess tremendous regeneration potential, capable of regenerating complete organism from small tissue fragments or even from dissociated cells. This peculiar property has made this genus one of the most invaluable model organisms for understanding the process of regeneration. Multiple studies in Hydra led to the current understanding of gross morphological changes, basic cellular dynamics and the role of molecular signalling such as the Wnt signalling pathway. However, cell-to-cell communication by cell adhesion, role of extracellular components such as extracellular matrix (ECM), nature of cell types that contribute to the regeneration process need to be explored in-depth. Additionally, roles of developmental signalling pathways need to be elucidated to enable more comprehensive understanding of regeneration in Hydra. Further research on cross communication among extracellular, cellular and molecular signalling in Hydra will advance the field of regeneration biology. Here, we present a review of the existing literature on Hydra regeneration biology and outline the future perspectives.
Keywords: Hydra, Regeneration, Morphallaxis, Extracellular matrix, Signalling and Pattern formation
1. Introduction
Replacement of lost body parts or tissues is an extraordinary phenomenon. The demonstration of the ability to regenerate missing parts after amputation in an animal by Trembley is the earliest record of this phenomenon. He called this animal Hydra, after a Greek mythological serpent which can regenerate multiple heads upon decapitation. This initiated a great interest in search for animals with regeneration abilities by biologists. Regeneration of damaged or diseased tissue or body part is important for successful survival of the organism. Therefore, it has fascinated the field of biomedical science for its potential applications and has led to the emergence of regenerative medicine.
Occurrence of regeneration process varies in different animal phyla (Table 1). In certain animals, ability of regenerating the whole organism is observed and in many others it is restricted to specific organs or tissue types. Whole body regeneration ability is evident from the earliest divergent multicellular organisms such as sponges (Ayling 1983; Borisenko et al. 2015). Members of the phylum Cnidaria, which are early divergent animals with defined body column and cell types similar to bilaterians, also exhibit regeneration ability (Schmid and Tardent 1971; Bossert et al. 2013). Especially the fresh water polyp Hydra exhibits tremendous regeneration capacity and it is the earliest record of animal regeneration (Trembley 1744). In Bilateria, members of basal phylum Platyhelminthes which belong to free living flatworm groups possess the whole organismal regeneration ability (Egger et al. 2007; Ritter and Congdon 1900; Stevens and Boring 1905; Monti 1900; Graff 1882; Dalyell 1814; Ruhl 1927; Spallanzani 1769). Until now there is no evidence of whole body regeneration in Ecdysozoa (Arthropoda and Nematoda) (Bely and Nyberg 2010). However, regeneration of limb has been well studied in Arthropoda and this ability varies across the taxa with good regeneration abilities observed in crustaceans (Bohn 1970; Minelli et al. 2013). In annelids, regeneration of anterior and posterior parts has been shown with varying capacity while the regeneration potential is completely absent in some animals like leeches (Bely 2006; Hyman 1940). In Mollusca, siphonophores of bivalves (Meyer and Byers 2005) and arms of cephalopods (Tressler et al. 2014) can regenerate and this ability is not conserved to the same extent in all the members of the phylum. Echinoderms are interesting invertebrates which can regenerate most complex organs/body-parts and this ability is well conserved in the phylum (Cuénot 1948; Goss 1969; Hyman 1955; Carnevali 2006).
Table 1. Animals with diverse ability to regenerate complex tissues across the animal phyla.
| NON-CHORDATES | Sub-clade | Animal | Whole-organism | body part/Structure |
|---|---|---|---|---|
| Porifera | Homoscleromorpha | Oscarella lobularis | yes | |
| Demospongiae | Halisarca dujardini | yes | ||
| Calcaria | Sycon cilatum | yes | ||
| Leucosolenia complicata | yes | |||
| Ctenophora | Tentaculates | Mnemiopsis leidyi | yes | |
| Vallicula multiformis | yes | |||
| Cnidaria | Hydrozoa | Hydra | yes | |
| clytia hemispherica | no | partial medusa | ||
| Anthozoa | Nematostella | yes | ||
| Scyphozoa | Aurelia aurita | only juvenile medusa | ||
| Cubozoa | Carybdea marsupialis | polyp stage | ||
| Platyhelminthes | Trematoda | Macrostomum lignano | yes | |
| Monocelis | yes | |||
| Schmidtea mediterranea | yes | |||
| Dugesia japonica | yes | |||
| Girardia tigrina | yes | |||
| Arthropoda | ||||
| Crustacea | Decapoda | Menippe mercenaria | no | chela |
| Chelicerata | Arachnids | Latrodectus variolus | no | limb |
| Hexapoda | Insecta | Gryllus bimaculatus | no | limb |
| Hexapoda | Insecta | Tribolium castaneum | no | limb |
| Hexapoda | Insecta | Drosphila | no | wing imaginal discs of larva |
| Annelida | Oligochaeta | Pristina leidy | yes | |
| Lumbriculus sp | yes | |||
| Polychaeta | Chaetopterus variopedatus | yes | ||
| Myxicola aesthetica | yes | |||
| Mollusca | Cephalopoda | Sepia officinalis | no | limb |
| Loligo pealei | no | limb | ||
| Argonauta | no | limb | ||
| Octopus sp. | no | limb | ||
| Bivalvia | Mesodesma mactroides | no | inhalent siphon | |
| Echinodermata | Asteroidea | Leptasterias hexactis | yes | |
| Asterias rubens | yes | |||
| Coscinasterias muricata | yes | |||
| Asterina gibbosa | no | arm | ||
| Ophiuroidea | Amphiura filiformis | no | arm, pyloric caeca and cardiac stomach | |
| Crinoidea | Antedon mediterranea | no | arm | |
| Holothuroidea | Holothuria glaberrima | no | appedages and visceral organs | |
| Echinoidea | Paracentrotus lividus | no | external appendages | |
| Lytechinus variegates | larva | |||
| Strongylocentrotus purpuratus | larva | |||
| HEMICHORDATA | ||||
| Enteropneusta | Ptychodera flava | no | proboscis | |
| CHORDATA | ||||
| Tunicata | Ascidiacea | Botrylloides violaceus | yes | |
| Ascidiacea | Ciona intestinalis | no | oral siphon, branchial sac and pharynx | |
| Cephalochordata | Leptocardii | Branchiostoma lanceolatum | posterior and anterior body parts | |
| Pisces | Actinopterygii | Danio rerio | no | heart, lens, fins |
| Sternopygus macrurus | no | tail | ||
| Amphibia | Urodela | Notophthalmus viridescens | limb, tail, spinal cord, lens, jaw and apex of heart | |
| Cynops pyrroghaster | limb, tail, spinal cord, lens, jaw and apex of heart | |||
| Ambystoma mexicanum | limb, tail, spinal cord, lens, jaw and apex of heart | |||
| Anura | Xenopus laevis | juvenile limb | ||
| Reptilia | ||||
| Squmata | Anoles caolinesis | no | tail | |
| Mammals | Dama dama | no | antlers | |
| Cervus sp. | no | antlers | ||
| Rangifer tarandus | no | antlers | ||
| Mus musculus | no | digit tips in fetal and juvenile stages | ||
| Homo sapiens | no | finger tips of children |
Chordates show different degrees of regeneration potential across the phylum. Among these, urochordates, especially ascidians such as Botrylloides violaceus (complete body) (Berrill 1951; Brown et al. 2009) and Ciona intestinalis (partial body) (Dahlberg et al. 2009; Auger et al. 2010) display considerable regeneration capacities. Within the Pisces, selected members were reported to exhibit regeneration (Unguez 2013; Goss 1969; Broussonet 1786) and recent studies in zebrafish have demonstrated the ability to regenerate different organs and fins (Johnson and Weston 1995). In Amphibia, salamanders such as newt and axolotl can regenerate lost limb (a relatively a complex structure) and this property is very unique and not reported in any other vertebrates (Spallanzani 1769; Goss 1969). Noticeable regeneration power has been demonstrated in lizard tail (Reptilia) and they are the closest animals to mammals that exhibit significant replacement potential of complex tissue (structure) (Lozito and Tuan 2017; Simpson 1964; Bellairs and Bryant 1985; Bryant and Bellairs 1967; Etheridge 1967). Aves (Stone and Rubel 2000; Cotanche 1987) and Mammals (Iismaa et al. 2018) possess very poor regeneration abilities and can regenerate relatively simple tissue types in their adult life. They cannot regenerate more complex structures such as appendages. An interesting exception is found in many species of deer which can regenerate their antlers (Goss 1983).
The regeneration ability was lost in different animals which are closely related or belonging to sister clades of organisms which exhibit this property. Many hypotheses such as adaptive, epiphenomenal, proximate causes etc. have been proposed to explain the loss of regeneration ability. However, most of these cannot be applied universally and thus provide no satisfactory explanation (Goss 1963; Wagner and Misof 1992; Goss 1992).
Among the mentioned animals with varied regeneration capacities only a few representative members have gained popularity. Invertebrate model systems such as Hydra and planaria are widely used for understanding the whole organism regeneration. Whereas zebrafish and axolotl are used as vertebrate model systems to understand the regeneration potential limited to few structures or organs. Apart from the regeneration abilities (Hydra and planaria) and phylogenetic closeness to mammals (zebrafish and axolotl) these also have other major advantages as mentioned below.
Established protocols and ease of maintaining in controlled laboratory conditions.
Amenable to genetic manipulations using transgenic technology and siRNA mediated gene expression knockdown.
Availability of genome, transcriptome and other molecular information.
Concerted efforts and sharing of reagents by active research groups.
In this chapter, we aim to review the current understanding of the regeneration process in Hydra, the oldest regeneration model system. We believe it is important to consolidate the recent findings and compare them in light of present understanding in other regenerative model systems which will also help us identify future directions for the studies to unravel the complex process of regeneration. Especially, in vivo model systems are ideal due to limited success of research focused majorly on stem cell based regenerative therapies. Further, since tissue regeneration is a complex phenomenon that depends on different cell types and extracellular factors/niche, it is important to investigate regeneration at an organism level. Previous reports have provided significant insights into the organismal regeneration. These studies suggest that the formation of signalling centre is a critical step in the regeneration process. Such signalling centres called ‘organizers’ were reported in axis induction of Hydra and newt embryo. The organizer comprises of a group of cells that emit signals and regulate the patterning of neighbouring tissue. However, the cells that contribute to formation of signalling centre and the process of evoking tissue pattering required for regeneration remain elusive. Due to these reasons, organisms such as Hydra and planaria which can exhibit complete organismal regeneration serve as important model systems.
2. Hydra
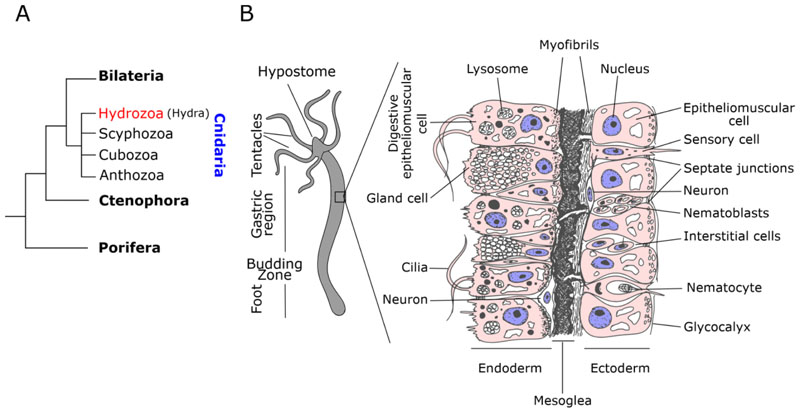
Hydra is a fresh water living organism and belongs to the class Hydrozoa of phylum Cnidaria (Marques and Collins 2004; Campbell 1987). Cnidaria is a sister group to bilateria and an early divergent phylum comprising multicellular animals with tissue level organization (Fig. 1A). Members of the phylum are radially symmetrical and evolved with two germ layers. Animals from this phylum share bilaterian cell types such as neurons and muscular epithelial cells (Fig. 1B). These properties have positioned the members of these phylum as favourite models to understand the molecular evolution determining organized body plan and evolution of cell types conserved in bilateria.
Figure 1. Phylogenetic position of Hydra and general morphology.
A)Phylogenetic relation of basal metazoans and different classes of phylum Cnidaria. B) General body plan and histology cartoon depiction with germ layers and cell types.
Hydra is a diploblast possessing two layers of the cells which form a cylindrical body containing a gastric lumen. These polyps are among the earliest animals to have evolved a defined body plan and has an oral-aboral axis with the oral end consists of a dome shaped hypostome (mouth) being surrounded by a ring of 5-6 tentacles which help them in feeding. The body column of a polyp can be divided into a gastric region, a budding region and a peduncle region. The aboral end of the polyp is called basal disk which can secrete mucus that helps in the attachment of Hydra to various substrates (Hoffmeister and Schaller 1985). As seen in Figure 1B, the cell layers are mainly constituted of epithelial cells with either ectodermal or endodermal stem cell origin. The epithelial cells have myofibrils at the base, due to which they are called as epitheliomuscular cells (Hess et al. 1957). These myofibrils are oriented perpendicular to each other in ectoderm and endoderm. This organization of myofibrils helps in contraction or elongation of the polyp (Passano and McCullough 1964). Endodermal cells are ciliated and have phagocytic capabilities while the ectodermal cells secrete a glycocalyx layer on the outermost side of the cells to protect the polyp from environment (Hess et al. 1957). Apart from epithelial cells, Hydra also possess multipotent interstitial stem cells located in the interstitial spaces of epithelial cells. These stem cells differentiate into cells with special functions such as gland cells, mucus cells, neuronal cells, nematocytes and gametes (oocytes or sperms) (Campbell and David 1974). The ectoderm and endoderm are divided by an acellular mesoglea which is essentially the extracellular matrix (ECM) (Sarras Jr et al. 1993).
3. Gross morphological changes during Hydra vulgaris regeneration
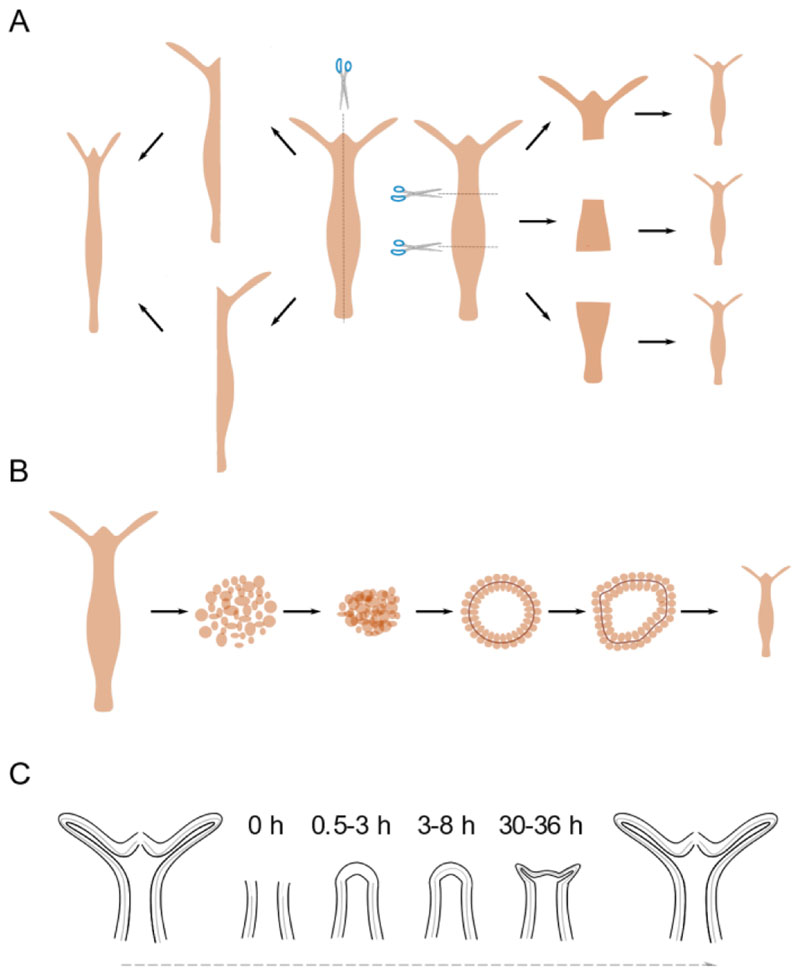
Hydra can regenerate missing body parts upon transverse or longitudinal amputation (Fig. 2a). The Hydra polyps are also capable of regenerating from re-aggregated cells. These polyps when dissociated into single cells can reorganize and regenerate into a whole polyp when these cells are pelleted (Fig. 2b). The cells reorganize into a lumen within first 12 h with ectodermal cells outside and endodermal cells facing inside the lumen (Technau and Holstein 1992; Gierer et al. 1972). This reorganization (possibly) arises from differential cell adhesion between ectodermal and endodermal cells (Technau and Holstein 1992). This process is followed by establishment of head organizer de novo within 18-30 h (Sato et al. 1990). First appearance of the head and tentacle structures can be seen by 48-72 h and finally the whole adult Hydra body forms within 4 to 7 days (Technau and Holstein 1992).
Figure 2. Regeneration capacities of Hydra vulgaris.
The ability of Hydra to regenerate when dissected in various manners and the morphological changes observed are shown. A) Hydra can regenerate missing parts upon both transverse and longitudinal dissection. B) Regeneration from dissociated and reaggregated cells. C) Kinetics of gross morphological changes during Hydra head regeneration.
In case of regeneration in response to amputation, the site of injury responds by re-organization of the epithelial cells to close the wound within an hour. This is then followed by lack of any gross morphological changes until 30 h post amputation. After 30-36 h, small tentacle buds begin to emerge from the regenerating tip beginning the process of morphological differentiation of cells. The emergence of the tentacles happens over the next 24 h with the completion of the whole process taking place by 72 h, where the tentacles are mature enough to catch the prey. On the other hand, basal disk regeneration is completed within 30-36 h (Hoffmeister and Schaller 1985). During regeneration wound healing is the initial step towards the morphological changes leading to major the cellular reorganization. This requires remodelling of ECM to facilitate tissue morphogenesis.
4. ECM dynamics during axis patterning and regeneration
ECM in animals has varied functions like cell-adhesion, mechanical support, cell-signalling, scaffolding for other extracellular molecules etc. The transition of unicellular organism to multicellular organisms requires ECM as a conduit for integration of various information between cells to function together. Recent studies have shown that the basic ‘Toolkit’ required to form the basal membrane coincided with emergence of multicellularity. ECM components have been found to be encoded in eukaryotes as early as the unicellular choanoflagellate Monosiga brevicollis. More recently, it was reported that a unicellular organism named Ministeria vibrans belonging to a filozoan clade of filasterea which diverged earlier to Choanozoa contains a gene similar to collagen IV (Grau-Bove et al. 2017). This premetazoan origin of collagen type IV indicates that it is possibly the most primitive form of collagens and possibly other components of ECM evolved from this. Earliest phylum reported to have a complete functional set of ECM components and related proteins is Cnidaria. The fact that ECM components expanded their repertoire from phylum Cnidaria onwards indicates their eumetazoan specific functions. Hence, studying cnidarian ECM can shed light on the role of ECM components in the organization of the body plans of early multicellular organisms.
ECM has been reported to play a crucial role in regulating cell fate and tissue morphology in multiple organisms. The chemical and physical properties of ECM can influence cell behaviour and fate. It was reported that solely by varying the stiffness properties ECM, the differentiation fate of mesenchymal stem cells could be altered (Engler et al. 2006). The regulation of cell fate by ECM occurs as a function of its interaction with ECM specific cell receptors such as integrins which in turn transmit the information either through chemical signal transduction or mechanically through force distribution via actomyosin networks (Mao and Baum 2015). ECMs can also regulate tissue morphogenesis by controlling and sequestering morphogenic ligands including BMP (Nistala et al. 2010). These capabilities of ECM can therefore make them an important player in ‘sculpting’ an organism during embryogenesis. A recent study showed that during Drosophila embryogenesis, ECM components in the basement membrane are differentially deposited to form a morphogen-like gradient during tissue elongation (Crest et al. 2017). This creates an anisotropic resistance to isotropic tissue expansion and hence helps in shaping organs by creating a mechanical imbalance. Another study in sea urchins showed how spatio-temporal incorporation of the hygroscopic chondroitin sulphate proteoglycan causes swelling of ECM which creates hydrostatic pressure for folding of epithelial sheet during embryonic invagination (Lane et al. 1993). The branching morphogenesis in mice salivary gland has been shown to be regulated by fibronectin deposition which signals the tightly packed epithelial cells to lose the e-cadherin contacts and help in conversion of cell-cell adhesions to cell— matrix adhesions (Sakai et al. 2003).
In mammalian systems, ECM has been reported to be crucial for regeneration and sustaining cell proliferation and differentiation post injury. Mammals possess very limited potency to regenerate their lost or injured tissues. Skeletal muscles are among the few tissues that mammals can regenerate and it was found that upon muscle degeneration, it leaves behind a hull of basement membrane ECM which acts as a scaffold to facilitate myofiber fusion (Vracko and Benditt 1972). In mammals, early stages of skeletal muscle regeneration are associated with downregulation of laminin and collagen type I to facilitate de-differentiation of satellite cells to form myoblasts (Gulati et al. 1983). Fibronectin, Hyaluronic acid and Tenascin-C are shown to be up-regulated during the same period. Another study showed that by altering the ECM stiffness of a 3 day old mouse, it was possible to restore the ability to regenerate heart which is lost after day 1 (Notari et al. 2018). Certain species of mammals are reported to have an astonishing capability to regenerate. For example, the African spiny mouse (Acomys sp.) which can loose and regenerate up to 60% of its dorsal surface including hair, follicle, skin, sweat glands etc. Here, It was shown that laminin and collagen type I molecules are down-regulated while the cells de-differentiate and later upregulated during cellular differentiation (Seifert et al. 2012). Another study which looked at the regeneration of skeletal muscle of tibialis anterior in Acomys found that it has higher collagen type VI in the regenerating tissue as compared to Mus (Maden et al. 2018). This difference renders a softer biomechanical property to the Acomys tissue which is believed to be important to achieve a faster, scar-free regeneration with higher macrophage infiltration, low expression of inflammatory and fibrotic genes at the site of injury. A recent study has demonstrated that fibronectin in the niche was necessary for maintaining a functional muscular stem cell population capable of regenerating lost muscular tissue. The reconstitution of fibronectin levels in aged stem cell niche is enough to remobilize and restore functional muscular stem cells (Lukjanenko et al. 2016).
In axolotls, skeletal muscle regeneration follows similar regulation of collagen type I, laminins, fibronectin, hyaluronic acid and tenascin-C as in mammals in the early phase of regeneration to allow de-differentiation of satellite cells (Mailman and Dresden 1976; Tassava et al. 1996; Contreras et al. 2009). In Xenopus tadpoles, it has been reported that Hyaluronic acid plays an important role in regeneration of tail bud after amputation (Contreras et al. 2009). Adequate level of hyaluronic acid in the regenerating tail bud is required for sustaining mesenchymal cell proliferation.
Studies in zebrafish also indicate a similar role of ECM in regeneration. Fin regeneration studies indicate an early upregulation of fibronectin, hyaluronic acid and tenascin-C which seems to be a conserved mechanism across different organisms in the vertebrates (Contreras et al. 2009). Additionally, Hapln1a, hyaluronan and proteoglycan aggrecan are also upregulated during the process. On the other hand, laminin levels do not show any appreciable changes during fin regeneration in zebrafish. Fibronectin has been shown to be necessary for the heart regeneration and is shown to be expressed by epicardial cells. The cardiomyocytes starts to express itgb3 receptors which can respond to presence of fibronectin in the site of injury for a successful heart regeneration (Wang et al. 2013). A study on the caudal fin regeneration indicate a crucial role of collagen type IV and specific matrix remodellers from the MMP and TIMP family of proteins (Bai et al. 2005).
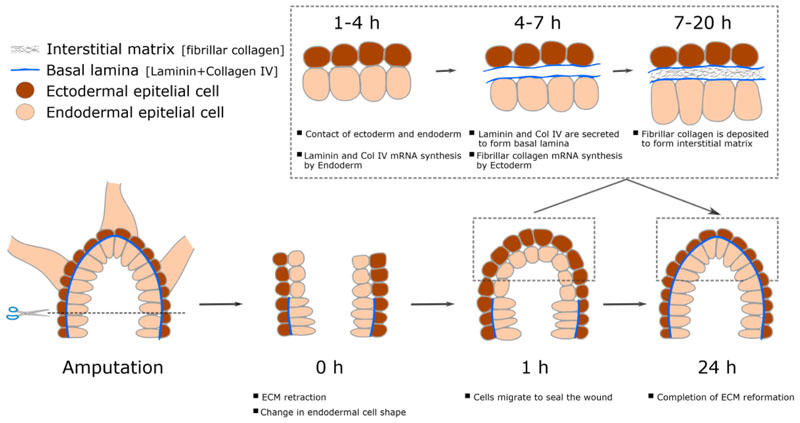
Hydra mesoglea is 0.5 - 2 μm thick depending upon its position in the body. The mesoglea is a trilaminar structure dividing the two epithelia (Davis and Haynes 1968). The epithelial cells are in direct contact with the basal lamina which sandwiches a thick central layer of interstitial matrix. The mesoglea also has trans-mesogleal pores which enable inter-epithelial cellular interactions via cell processes running through them (Shimizu et al. 2008). ECM secretory dynamics have been studied using the Hydra regeneration model. It is known that post amputation there is a retraction of Mesoglea, and its restoration is required for initiation of regeneration. Post-amputation, loss of ECM is followed by sealing of wound which causes direct contact of ectoderm and endoderm cells, leading to change of morphology of these cells from being cuboidal or columnar to flattened (Fig. 3) (Shimizu et al. 2008; Shimizu et al. 2002). During regeneration, within 3 hours, all the components of ECM are actively transcribed by their respective cells and then by 7 hours components of the basal lamina - endoderm secretes laminin and HyCol IV such that basal lamina is formed and hence leading to regaining the normal cuboidal morphology of the cells. Contact of the basal lamina with receptors of the ectodermal cells results in secretion of components of the interstitial matrix and hence leading to complete formation of mesoglea within 20 hrs post amputation (Sarras Jr 2012). During the regeneration of re-aggregated cells, it is reported that mesoglea formation takes place immediately after reorganization of ectoderm and endoderm cells into a lumen (Technau and Holstein 1992). The mesogleal components are first secreted by 12-17 hrs of aggregation and the maturation of the mesogleal structure is reported to be completed by 48-72 hrs (Sarras Jr et al. 1993). The morphogenetic processes required for regeneration of reaggregated cells succeeds the mesoglea formation and the formation of a mature mesoglea is an absolute necessity for success of regeneration (Sarras Jr et al. 1993). It has also been reported that disruption of any of this process either by knockdown of the ECM components or by inhibiting their biosynthesis chemically or by preventing ECM-receptor interaction, the process of regeneration is arrested (Sarras Jr et al. 1993). Therefore, it seems very plausible that the important role of ECM in regulating regeneration and other tissue function evolved very early during the evolution. As can be noted above, the precise mechanistic role of ECM during regeneration in more complex organisms is unclear despite strong evidence showing its importance. A highly regenerative model system such as Hydra with its less complicated tissue system and signalling networks could be crucial to shedding light to ECM mediated regulation. Studies focussing on biomechanical aspects of ECM in Hydra could unravel various aspects of their role in physiological regulation and how it evolved to perform more complex function in a system such as human.
Figure 3. Cellular and structural changes during head regeneration in Hydra .
ECM is retracted immediately upon decapitation followed by change in endodermal cell shape. This is succeeded by wound closure and secretion of ECM components completing the early stages of head regeneration.
5. Cellular basis of regeneration
Regeneration in animals can be either for homeostatic cell replacement like the replacement of blood cells or for reparative replacement wherein a damaged tissue or organ needs to be regenerated. In the context of this chapter, discussion will be limited to the latter type. Regeneration can be broadly divided into two types: -
-
a)
Epimorphosis is a type of regeneration involving active cell proliferation for the completion of regeneration. This process can further be divided into two types i.e., blastema-mediated or compensatory regeneration. In blastema-mediated regeneration, repair of lost tissue or organ proceeds by recruitment of either pre-existing progenitor cells in the vicinity of the injury or by de-differentiation of differentiated cells from the vicinity. These cells then start proliferating to form a mass of heterogeneous undifferentiated cells at the regenerating tip called blastema. Once a necessary number of cells are amassed, the cells in blastema start to differentiate and initiate the process of morphogenesis similar to that seen during embryogenesis. This process then culminates with restoration of the lost tissue either completely or partially depending upon the organism e.g. planaria, axolotls etc.(Alvarado 2006). In the compensatory regeneration, the regeneration proceeds without any blastema formation or requirement of stem cells. In this process, differentiated cells from the vicinity are recruited to site of injury and proliferate to replace the lost tissue. Liver regeneration is a prime example of compensatory regeneration (Michalopoulos and DeFrances 1997).
-
b)
Morphallaxis is a type of regeneration hallmarked by absence of cellular proliferation. The existing tissue is re-patterned to replace the lost tissue. This kind of regeneration is frequently observed in lower invertebrate organisms such as Hydra (Lenhoff et al. 1986; Trembley 1744).
Epimorphic type of regeneration is the most prevalent type of regeneration across the animal kingdom. It would be wrong to assume dividing cells as the sole contributors towards the new tissue during regeneration. There exist possibilities of contribution from trans-differentiated and de-differentiated cells which might contribute to a new population of dividing and differentiating cells (Jopling et al. 2011). Hence to understand how regeneration process is regulated, it becomes important to dissect the origins of cells involved as a source of new cells required for regeneration upon specific injury signals. In planaria, multiple studies have shown that a continuously dividing population of cells called neoblasts contribute to formation of new tissue (Newmark and Sanchez Alvarado 2000). These cells are clonal and pluripotent in nature; have the capability to rescue regeneration deficit planarians to regenerate with as few as 3-5 cells (Wagner et al. 2011). These clonogenic neoblasts are recruited to site of injury where they proliferate into a mass of undifferentiated cells to form blastema. The blastema cells then undergoes differentiation process for patterning the lost tissue (Newmark and Sanchez Alvarado 2000). In vertebrate systems, blastema formation doesn’t consist in entirely of one type of clonogenic pluripotent cells like neoblasts. There seems to have a pre-determined heterogeneous group of cells having unipotent or multipotent capabilities. For example, in Xenopus the muscle regeneration is solely dependent on dormant population of satellite cells found adjacent to muscle fibres (Le Grand and Rudnicki 2007). These are the muscle stem cells which upon activation, differentiate into muscle fibres. In axolotl, it was shown using lineage tracing experiments that muscular satellite cells and Schwann cells are found to be unipotent and were responsible for muscular and Schwann cells respectively; while, dermis cells of mesodermal origin contribute to a diverse repertoire of cartilage and connective tissues (Kragl et al. 2009). In the zebrafish caudal fin regeneration model, it was shown that most of the cell types required for regenerating the fin were derived from specific unipotent progenitors and the same is the case during the fin development during embryogenesis (Tu and Johnson 2011).
In Hydra, regeneration is manifested in two different modes depending on the region of amputation. Any amputation away from mid-gastric region will exhibit morphallactic mode of regeneration. In this mode, the wounded tissue undergoes wound healing by bringing together the ectodermal and endodermal cells which is initiated by the endodermal cells from severed edge in a fashion similar to blastopore closure (Takaku et al. 2005). This is then followed by cellular re-arrangements and differentiation required for regeneration without any proliferation. It has also been reported that there is no involvement of interstitial stem cells or their derivatives towards the regeneration process and hence only requiring epithelial stem cells for the whole process (Sugiyama and Fujisawa 1978). On the other hand, it was reported that head regeneration after a mid-gastric cut proceeds through a process similar to the epimorphosis (Chera et al. 2009). Upon amputation, there are detectable levels of activation of apoptotic pathway in interstitial cells such as neurons, nematocytes etc. present near the area of injury. These cells start secreting Wnt3a in a yet unknown fashion. These molecules of Wnt ligand then activate the Wnt signalling in nearby interstitial stem cells and induce them to do compensatory cell proliferation. This proliferation however does not form a very large mass of cells like in blastema. Simultaneously, the epithelial stem cells which are located at the apical end starts secreting Wnt3a and assume the role of head organizer after which follows the process of regeneration as seen in morphallactic regeneration (Galliot and Chera 2010).
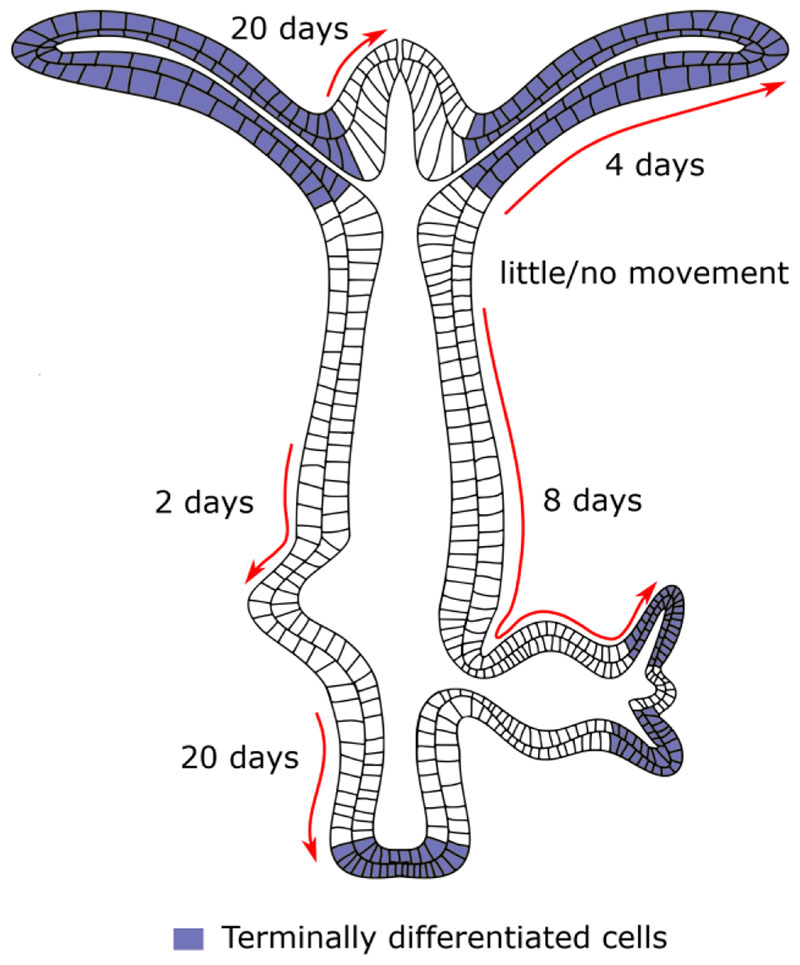
Several studies have shown in Hydra that certain strains of Hydra do not exhibit any signs of aging or senescence while other strains can be induced to senesce upon induction of gametes (Martínez and Bridge 2012; Kaliszewicz 2018). The immortality in Hydra is attributed to the ability of Hydra to maintain a steady state of shedding of cells and cell proliferation. The stem cells in the body column divide continuously either to replenish themselves or differentiate into specialized cells. The epithelial cells divide roughly every 3-4 days and interstitial cells divide every 1.5 days (Martínez and Bridge 2012). These cells produce differentiated cells which are pushed to either end of the body column and are sloughed off. The differentiated cells proceed to move towards their destination depending on the origin of the cells. They move to the basal disk if the cells originate from the peduncle region, the tentacles if the originate from region just below the tentacle ring or are pushed to form new bud if the cells originate from region above the budding region and below the tentacle ring. The cells moving towards their respective destinations have variable rate of movement depending on their position. Cells at the base of the tentacle takes on an average of 4 days to reach the tip of the tentacles and being pushed off. The cells at the base of the hypostome and the at the peduncle reach their respective destination on 20 days and the cells in the body column takes about 2-8 days to reach the budding zone and to bud depending on their position with respect to the bud (Fig. 4) (Campbell 1967). In conclusion, it is evident that specific cell types play important role in regeneration and tissue homeostasis.
Figure 4. Cellular dynamics in Hydra .
The different cell movements that take place during maintenance of the Hydra body plan are depicted here. Cells undergo continuous proliferation and movement in both directions along the oral-aboral axis. The empty cells denote proliferative cells and the blue coloured cells are terminally differentiated cells. The number of days taken for cells to reach their respective destinations such as buds or the hypostome and peduncle termini are shown. The arrows depict the direction of cell movement.
6. Molecular Signalling
For the development of a zygote into an animal, there are seven major signalling pathways that act in a concerted manner and organize the proliferating cells into the final morphology. These different signalling pathways have been classified based on either ligands or signal transducers involved. These are, Wnt/Wingless (Wg), transforming growth factor-β (TGF-β)/BMPs, Notch, RTK, Hedgehog (Hh), JAK-STAT [signal transducers and activators of transcription] pathway) and Nucleic acid receptor/Hormone receptor mediated signalling (Barolo and Posakony 2002; Gerhart 1999). All the signalling pathways work towards a unified goal which is to create axes and break symmetry in the developing embryo. From early metazoans like sponges that have a single oral-aboral axis to mammals that have multiple axes like dorso-ventral (D-V), anterior-posterior (A-P), left-right (L-R) planes, these pathways are critical in the patterning of all organisms. During the process of development, these signalling pathways act in a spatio-temporally regulated manner to regulate various cellular functions like cell division, migration, differentiation, organization into tissues, recycling cellular components, enabling cells to respond to other signalling pathways and apoptosis (Sanz-Ezquerro et al. 2017). These developmental signalling pathways are also reactivated when an organism must undergo the process of regeneration. The reformation of the body axes and proper patterning of lost tissues is a result of the cumulative action of these pathways.
6.1. Injury response
When there is loss of tissues, the first event to occur is the activation of injury mediated signalling response. In multiple organisms such as planaria (Petersen and Reddien 2009), axolotl (Bryant et al. 2017), newt and salamander (Yokoyama 2008), it has been reported that the earliest responses upon wounding were essential for initiation of the cascade of developmental signalling needed to regenerate lost structures. In Hydra, following a mid-gastric amputation, the initial ROS signalling responses upregulated by H2O2 were shown to activate multiple pathways. Majority of these are immune response genes and it has also been shown that multiple signalling pathways such as STK, Pi3K, ERK 1–2, and MAPK pathways that are a part of injury induced responses are necessary for the formation of the head organizer (Arvizu et al. 2006; Tischer et al. 2013) (Fig. 6).
Figure 6. Dynamic expression of signalling molecules during Hydra head regeneration.
Components of multiple signalling pathways that are reported to be differentially regulated across the regeneration time course have been depicted. The signalling pathways shown are Injury responses, TGF-β signalling pathway members and Wnt signalling pathway members. The regeneration timeline is depicted above the table. The shaded region for each gene denotes the time when mRNA expression of the respective molecule has been recorded.
6.2. Wnt signalling
The Wnt signalling pathway is highly conserved in evolution and is reported to play roles in proliferation, cell polarity, cell fate determination, migration and apoptosis during the development of both invertebrates and vertebrates (Miller 2002; Loh et al. 2016). This pathway is essentially a short-range communication system that uses the Wnt lipid-modified glycoproteins and has roles in both axis patterning (canonical) and cell-polarity (non-canonical) via two distinct signalling cascades (Alexandre et al. 2014). The canonical pathway is activated when the Frizzled receptors are bound by the Wnt ligands and the coreceptor Lrp5/6 is ligated to it. This leads to inhibition of the GSK3-β kinase which prevents phosphorylation of β-catenin resulting in its stabilization and nuclear localization. Nuclear β-catenin binds to the Tcf/Lef transcription factors and activates the Wnt target genes resulting in determination of cell fate. The non-canonical component of Wnt signalling is often β-catenin independent and via multiple intracellular effector proteins, plays a role in planar cell polarity (PCP), convergent extension (CE) and directed cell migration (Loh et al. 2016).
Wnt signalling is thought to predate the “Hox patterning system” and is possibly therefore the original symmetry breaking mechanism and primary axis organizer (Larroux et al. 2007; Martindale 2005). Traditionally, Wnt has been considered as a posterior organizer signal although there exists a debate due to the expression of few Wnt targets that play a role in sensory epithelium differentiation in the oral region of pre-bilaterian organisms (Nichols et al. 2006; Ryan et al. 2013).
The different components of the Wnt signalling pathway are differentially present in various animals across the animal phyla. Even in basal metazoans like sponges, it has been observed that the multiple Wnt ligands express in a polarized manner across the larvae from the anterior to posterior extremes (Windsor Reid et al. 2018; Adamska et al. 2007; Adamska et al. 2010). In the ctenophore Mnemiopsis leidyi, it was observed that the Wnt ligands were expressed in the aboral pole, tentacles and the apical organ indicating their role in polarized pattern formation (Pang et al. 2010). In Nematostella, the presence of multiple Wnt ligands and their conservation at sequence level has been shown (Rigo-Watermeier et al. 2012). In Nematostella, the Wnt/β-catenin signalling is important for patterning the oral-aboral axis during the development of the organism using a cross-talk with the Hox patterning system (Leclere et al. 2016; DuBuc et al. 2018).
In bilaterians, deuterostomes (echinoderms to vertebrates), Wnt signalling has been shown to play a role during early embryogenesis in anterior-posterior patterning (Schubert and Holland 2013; Darras et al. 2018). Similar roles have been demonstrated in protostome clades such as Arthropoda (Drosophila, Tribolium, Gryllus and Achaearanea)(Nusslein-Volhard and Wieschaus 1980; Bolognesi et al. 2008; Miyawaki et al. 2004; McGregor et al. 2008). Wnt is also known to regulate the caudal gene and in turn the Hox patterning system in mouse and zebrafish (Chawengsaksophak et al. 2004; Shimizu et al. 2005). The Hox patterning system is another set of genes conserved from cnidarians important in axis patterning (Ryan et al. 2006; Shenk et al. 1993; Gauchat et al. 2000; Chourrout et al. 2006; Naito et al. 1993; Schummer et al. 1992; Reddy et al. 2015).
Besides playing a role in embryonic development, the Wnt/β-catenin signalling also plays a posteriorizing role in the process of homeostasis and regeneration. In sponges, Wnt signalling was found to be important for regeneration (Windsor Reid et al. 2018). In planaria, it has been observed that the Wnt/β-catenin signalling elicited by wounding is necessary for posterior tissue patterning (Petersen and Reddien 2008, 2009). While the canonical Wnt signalling is necessary for proper posterior regeneration, even anterior regeneration has been shown to require nuclear translocation of β-catenin and signalling through unknown mechanisms (Sureda-Gomez et al. 2016). Among the vertebrates, different Wnt ligands have been shown to act in the regulation of the regeneration process. In zebrafish tailfin regeneration, Wnt/β-catenin signalling acts upstream of FGF-signalling and is necessary for the cell proliferation involved in blastema formation (Kawakami et al. 2006). During heart regeneration in adult zebrafish and juvenile mice, Wnt signalling was observed to be induced upon cardiomyocyte injury and necessary to switch between fibrosis and relatively scar-free regeneration (Ozhan and Weidinger 2015). Wnt signalling is active and multiple Wnt ligands express throughout adulthood in urodele tails. Such an expression pattern suggests multiple roles in urodele tail regeneration as well (Caubit et al. 1997). In axolotl, which can regenerate whole appendages, Wnt signalling has been shown to be necessary (Kawakami et al. 2006). Multiple targets and effectors of the Wnt signalling pathway were found to be differentially regulated and were implicated in the lizard tail regeneration (Hutchins et al. 2014).
In Hydra, the Wnt signalling pathway was identified to be a key player in the organization of head, bud formation and head regeneration in Hydra (Hobmayer et al. 2000; Bode 2003; Broun et al. 2005). A total of 11 Wnt ligands have been identified and six of them have been shown to be a part of the organizer centre of the Hydra hypostome (Lengfeld et al. 2009) (Fig. 5). There were also autoregulatory mechanisms discovered to localise the Wnt expression to the hypostome (Nakamura et al. 2011). Systemic activation of the Wnt signalling pathway results in ectopic tentacle formation all over the body column of Hydra, turning the whole organism into a head like structure confirming its role in head patterning (Broun et al. 2005). The regulation of Hox genes by Wnt signalling was observed in Hydra and these were also upregulated during the process of head regeneration (Reddy et al. 2015; Schummer et al. 1992) (Fig. 5). Multiple components of Wnt signalling have been shown exhibit dynamic expression pattern during Hydra head regeneration (Petersen et al. 2015) (Fig. 6).
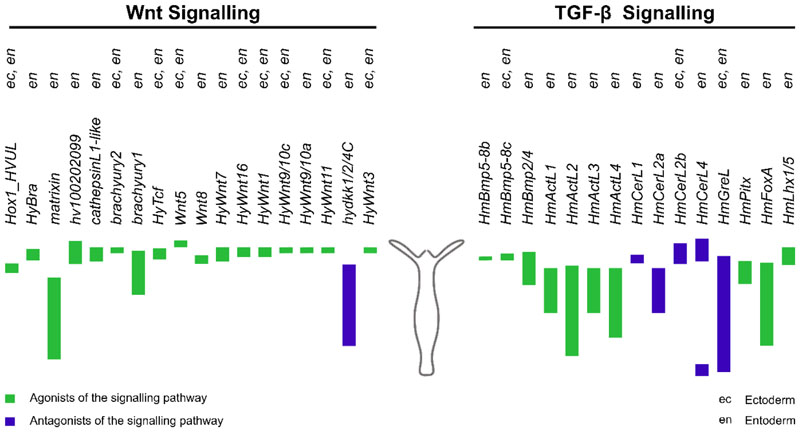
Figure 5. Expression profile of Wnt and TGF-beta superfamily signalling components along the Hydra body axis.
Molecular components that have been shown to be regulated by Wnt (left) and TGF-β (right) signalling pathways have been depicted here. The coloured bars represent the localisation of gene expression along the body column of the Hydra polyp. Green bars represent molecules that play a positive regulatory role and the blue bars represent negative regulators of the pathways. The expression of the genes in the ectoderm and endoderm of Hydra has been denoted as ec and en respectively.
6.3. TGF-beta superfamily signalling
The TGF-β superfamily is a very large group of molecules that play a vital role in development of metazoans. The canonical form of signal transduction is through Ser-Thr kinases to intracellular mediator class of Smad proteins. The superfamily of ligands consists of more than 30 members including TGF-βs, BMPs, GDFs, Activin and Nodal (Feng and Derynck 2005; Kitisin et al. 2007). The effector Smad proteins are classified into 3 classes, the R-Smads, Co-Smads and the Anti-Smads based on their structure and function. The R-Smads including Smad-1, 2, 3, 5 and 8 are the receptor regulated Smads and are directly phosphorylated by binding of the Activins, BMPs and TGF-β ligands. The co-Smads or the common Smads include one member, the Smad 4 and are required for all distinct pathways as they form heteromeric complexes with R-Smads and translocate to the nucleus for target gene activation.
The anti-Smads include the Smad 6 and 7 classes and inhibit signalling by the R-Smads and co-Smads by stably associating with the receptors (Kawabata and Miyazono 1999).
The components of the TGF-β signalling pathway are conserved across metazoans starting from placozoans (Huminiecki et al. 2009). The members of the TGF-β superfamily are morphogens generating gradients and playing an important role in both development of body axes and tissues patterning. Nodal and BMPs initiate regulatory loops in the zygote to form the first embryonic axis. While the Wnt signalling pathway plays an important role in the formation of the antero-posterior axis, the TGF-β members play a critical role in the development of the dorso-ventral (D-V) axis of the organism (Wu and Hill 2009). Components of the TGF-β pathway have been identified in the ctenophore, Mnemiopsis leidyi, and play a role in morphogenesis but not in early axis patterning (Pang et al. 2011). TGF-β members specify the dorsal regions of a developing embryo in both invertebrate (arthropods) and vertebrate (Xenopus and zebrafish) systems (De Robertis and Kuroda 2004; O’Connor et al. 2006).
In different classes of echinoderms, there are studies reporting TGF-β members playing a role in regeneration of the arms (Bannister et al. 2005; Patruno et al. 2003; Patruno et al. 2002). In the zebrafish heart regeneration, TGF-β signalling pathway plays a role in switching between the scar-based repair to cardiomyocyte-based regeneration (Chablais and Jaźwińska 2012; Choi et al. 2013; Dogra et al. 2017). The TGF-β/ALk4/Smad3 pathway has been shown to play important role in the spinal cord development during regeneration (Casari et al. 2014). TGF-β has been shown to play a role in cell proliferation, activation of the BMP and ERK signalling cascades and reversible prevention of wound epithelium resulting in the formation of regeneration structures (Ho and Whitman 2008; Beck et al. 2003). This signalling pathway is also critical for initiation and control of limb regeneration in axolotl (Levesque et al. 2007).
Multiple studies have elucidated the role of BMP signalling in body patterning and regeneration in Hydra. A BMP5-8 homolog was identified in Hydra and was shown to play a role both in tentacle formation and foot patterning during regeneration (Reinhardt et al. 2004) (Fig. 5). A chordin-like protein has been reported to play a conserved role in the formation of the head organiser during budding and regeneration (Rentzsch et al. 2007) (Fig. 5). The Nodal signalling pathway via Pitx in Hydra has been shown to play a role in breaking the radial symmetry during the process of budding and allow for the organizer pathways to form a bud on one side of the body column (Watanabe et al. 2014) (Fig. 5). This suggests a possible role in organizer formation during head regeneration. This is further supported by a report of multiple TGF ligands being downregulated during regeneration at transcript level and the upregulation of Smads (Petersen et al. 2015) (Fig. 6).
6.4. Notch signalling
The Notch signalling pathway is a mediator of cell-cell communication in a juxtracrine manner and plays a crucial role in embryonic development. Like other signalling pathways, it controls cell proliferation, differentiation and cell fate determination during embryonic development in spatio-temporal manner (Liao et al. 2016). A very important function of the Notch signalling pathway is the ability to form boundaries within developing tissues. The Notch signalling pathway is one of the simplest with very few components in its core network. The canonical Notch signalling pathway consists of a Notch transmembrane receptor that contains a Notch intracellular domain (NICD) and is activated by transmembrane ligands like Delta and Serrate/Jagged on neighbouring cells. This leads to the cleavage of the NICD which moves to the nucleus and regulates transcription of target genes (Kopan 2012). The lack of an amplification mechanism in this signalling pathway makes the stoichiometry and the signalling strength, which can be modulated by post-translational modifications, important for manifestation of the effect of the signalling pathway. Such dosage-dependent action and consequences have been studied in Drosophila, mice and human diseases (Andersson et al. 2011). Across the animal kingdom, this pathway has been shown to play a role in the specification of germ layers and differentiation of cells into specific subtypes (Shi and Stanley 2006). Notch signalling has a well characterized role in the development of nervous system and this is conserved from sponges (Richards et al. 2008; Richards and Degnan 2012). In Nematostella, disruption of the Notch signalling pathway affects the development of neurons and cnidocytes and indicates a conserved ancient origin for the multifunctional sensory cells/neurons (Marlow et al. 2012). In amphioxus embryos, Notch signalling was shown to be co-opted into the anterior-posterior boundary patterning of the developing somites (Onai et al. 2015). The role of the Notch signalling pathway during early mouse embryonic development in axial mesoderm formation, ectodermal anterior-posterior patterning and neural tube formation has also been established (Souilhol et al. 2015).
Notch signalling components (like delta-1) were among the genes that have been shown to be upregulated in a translation independent manner, early in the regeneration of planarians (Wenemoser et al. 2012). This is necessary in maintaining the midline identity of the planaria (Sasidharan et al. 2017). Notch signalling is necessary for zebrafish tail-fin regeneration in the formation of a blastema (Münch et al. 2013; Grotek et al. 2013). In addition to the tail-fin, Notch signalling has been shown to be critical for the proliferation of cardiomyocytes and atrial to ventricular trans-differentiation during heart regeneration (Zhao et al. 2014; Zhang et al. 2013; Raya et al. 2003). In the Xenopus tadpole it was shown that the Notch signalling pathway, acting downstream of BMP signalling, is required and positively correlates with regenerative capacity (Beck et al. 2003).
The Notch signalling pathway is also conserved in Hydra and was shown to govern differentiation of cells originating from the interstitial stem cell lineage of the polyps (Kasbauer et al. 2007). It also plays an important role in determining the boundaries in Hydra polyps. These boundaries include the ones between tentacles and the head region and between the newly formed bud and the parent polyp (Munder et al. 2013). The process of boundary formation between tissues/cells is an important part of development and determines the morphogenetic processes. The Notch target is expressed just before the foot cells begin to differentiate at the boundary between the bud and the parent polyp. This signalling was necessary for the completion of the bud development and detachment from the parent polyp (Munder et al. 2010). The Notch signalling pathway in Hydra is necessary for the formation of the Wnt3a expressing head organizer during the regeneration of Hydra. Further, inhibition of Notch signalling led to the loss of boundary between head and tentacle and repression of the head formation. This finally ended in non-regenerating tips because of perturbed spatio-temporal expression of head and tentacle specific genes and networks (Munder et al. 2013).
6.5. RTK signalling
The receptor tyrosine kinases (RTKs) are autocatalytic receptors that function by phosphorylating various tyrosine residues. All RTKs consist of an extracellular ligand binding domain, a transmembrane domain and a cytoplasmic tyrosine kinase domain which harbours serine, threonine and tyrosine residues that can be phosphorylated. When the ligand binds, the receptors dimerize or undergo allosteric transitions leading to activation. The phosphorylation of the receptors recruits other cytoplasmic adaptor proteins via multiple interaction domains. The signals are then transmitted via a network of cytoplasmic proteins to the nucleus resulting in activation of various target genes (Volinsky and Kholodenko 2013).
In planarians, the FGF signalling and Wnt signalling circuits are juxtaposed and regulate the anterior-posterior axis patterning (Scimone et al. 2016). Among other growth factors, the FGFs have been shown to play a critical role in regeneration in few organisms. EGFR-3 was shown to be important for the asymmetric cell divisions that take place in planarian neoblasts upon near-lethal irradiation and was important for regeneration of these animals by regulating the size of the blastema produced (Fraguas et al. 2011; Lei et al. 2016). This signalling was also necessary for proper gut regeneration and homeostasis in planarians (Barberán et al. 2016). During the process of homeostasis of fins in adult zebrafish
, the blastema markers including shh, msxb and mkp3 are expressed at low levels indicating that regenerative processes are important for the maintenance of the fins in these animals. Removal of Fgfs resulted in drastic atrophy of all fin types and showed a critical role for these growth factors in preservation of dermal bone, joint structures and all supporting tissues (Wills et al. 2008). Fgfs are also necessary for the vascularisation of the regenerating heart in zebrafish (Scimone et al. 2016). The FGFs were also shown to play a role in lizard tail regeneration (Fraguas et al. 2011; Lei et al. 2016).
Sponges have 9 members belonging to the RTK family (Suga et al. 2001). In Hydra, 15 RTKs belonging to eight families have been identified, including 3 ephrin family members (Reddy et al. 2011). In Hydra, an FGFR called kringelchen has been shown to express at the boundary between parent polyp and bud. Additionally, multiple components of the FGFR signalling pathway have been shown to be essential for bud detachment by modulating the actin cytoskeleton at that location (Sudhop et al. 2004). Homologs of VEGF and FGF have been identified in Hydra and VEGF has been shown to have roles in bud formation and regeneration indicating a conserved role in tube formation and branching morphogenesis (Krishnapati and Ghaskadbi 2013). HyEph1 has been shown to be upregulated during the later stages of regeneration during which its role in boundary formation could be necessary (Tischer et al. 2013). The members of RTK signalling pathway in Hydra and their expression during regeneration have been characterized, however, their functional significance has not been established yet.
7. Conclusion and Future perspectives
Studies on the early morphological changes during Hydra head regeneration have revealed changes in cell shape and cell-cell contacts established between ecto- and endo-dermal cells. However, the significance of these cell-cell contacts remains elusive. Therefore, a focused approach is required to analyse the molecular mechanism underlying this cell-cell contact during early stages of regeneration.
Another important component that appears to play crucial role in regeneration is the ECM. It retracts immediately after amputation facilitating the contact of outer ectoderm and inner endoderm. Additionally, experiments perturbing the ECM components indicate its role in the regeneration process. The questions that still remain unanswered are, what are the changes occurring in ECM in response to damage and how these changes are read or communicated to neighbouring cells? Such studies will also be important in understanding the molecular mechanism of wound healing and tissue repair in higher organisms.
Formation of a signalling centre (organizer) is important for successful regeneration. The nature of cell types that are competent and form the organizer is not understood. This needs to be addressed to gain insight into the cellular contribution of regeneration process.
The role of signalling pathways such as the Wnt and the TGF-β superfamily are relatively well characterized. At the same time molecular regulation of other developmental signalling pathways such as hedgehog, Hippo etc. are not studied. Understanding the role of these signalling mechanisms and their cross talk will prove invaluable towards elucidation of the regeneration process.
The signalling pathways lead to transcriptional changes in the cell required for both developmental and regenerative processes. To bring about these changes, there are multiple epigenetic regulators that are recruited by or recruit transcription factors to target regions. Very few studies have investigated the role of histone modifiers, modifications and chromatin remodellers in the process of regeneration. The conservation of histones in Hydra (Reddy et al. 2017) and Hydractinia (Török et al. 2016) has been established. Since the histones are conserved their modification machinery, at least in part may also be conserved. Therefore, Hydra can be used as a model organism to study the epigenetic mechanism of regeneration and identify differences with other organisms that cannot regenerate. This will facilitate in identifying the presence or absence of epigenetic blockade in animals with no regeneration ability.
In Hydra, head regeneration process is well studied whereas regeneration of foot is not addressed extensively. Comparing the head and foot regeneration process might provide clues towards a common molecular mechanism necessary for the regeneration process.
Acknowledgements
The authors would like to thank Prof. Sanjeev Galande for discussions and suggestions & Dr. Apurva Barve for reviewing this chapter. PCR is supported by Early Career Fellowship of the Wellcome Trust-DBT India Alliance (IA/E/16/1/503057). AG is supported by a fellowship from the Council of Scientific and Industrial Research, India. MU is supported by fellowship from the University Grants Commission (UGC), India. The final publication is available at https://link.springer.com/chapter/10.1007%2F978-3-030-23459-1_12.
References
- Adamska M, Degnan SM, Green KM, Adamski M, Craigie A, Larroux C, Degnan BM. Wnt and TGF-beta expression in the sponge Amphimedon queenslandica and the origin of metazoan embryonic patterning. PLoS One. 2007;2(10):e1031. doi: 10.1371/journal.pone.0001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamska M, Larroux C, Adamski M, Green K, Lovas E, Koop D, Richards GS, Zwafink C, Degnan BM. Structure and expression of conserved Wnt pathway components in the demosponge Amphimedon queenslandica. Evol Dev. 2010;12(5):494–518. doi: 10.1111/j.1525-142X.2010.00435.x. [DOI] [PubMed] [Google Scholar]
- Alexandre C, Baena-Lopez A, Vincent JP. Patterning and growth control by membrane-tethered Wingless. Nature. 2014;505(7482):180–185. doi: 10.1038/nature12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado AS. Planarian regeneration: its end is its beginning. Cell. 2006;124(2):241–245. doi: 10.1016/j.cell.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138(17):3593. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- Arvizu F, Aguilera A, Salgado LM. Activities of the protein kinases STK, PI3K, MEK, and ERK are required for the development of the head organizer in Hydra magnipapillata. Differentiation. 2006;74(6):305–312. doi: 10.1111/j.1432-0436.2006.00078.x. [DOI] [PubMed] [Google Scholar]
- Auger H, Sasakura Y, Joly JS, Jeffery WR. Regeneration of oral siphon pigment organs in the ascidian Ciona intestinalis. Dev Biol. 2010;339(2):374–389. doi: 10.1016/j.ydbio.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayling AL. Growth and Regeneration Rates in Thinly Encrusting Demospongiae from Temperate Waters. Biol Bull. 1983;165(2):343–352. doi: 10.2307/1541200. [DOI] [PubMed] [Google Scholar]
- Bai S, Thummel R, Godwin AR, Nagase H, Itoh Y, Li L, Evans R, McDermott J, Seiki M, Sarras MP., Jr Matrix metalloproteinase expression and function during fin regeneration in zebrafish: analysis of MT1-MMP, MMP2 and TIMP2. Matrix biology. 2005;24(4):247–260. doi: 10.1016/j.matbio.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Bannister R, McGonnell IM, Graham A, Thorndyke MC, Beesley PW. Afuni, a novel transforming growth factor-beta gene is involved in arm regeneration by the brittle star Amphiura filiformis. Dev Genes Evol. 2005;215(8):393–401. doi: 10.1007/s00427-005-0487-8. [DOI] [PubMed] [Google Scholar]
- Barberán S, Fraguas S, Cebrià F. The EGFR signaling pathway controls gut progenitor differentiation during planarian regeneration and homeostasis. Development. 2016;143(12):2089. doi: 10.1242/dev.131995. [DOI] [PubMed] [Google Scholar]
- Barolo S, Posakony JW. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 2002;16(10):1167–1181. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- Beck CW, Christen B, Slack JM. Molecular pathways needed for regeneration of spinal cord and muscle in a vertebrate. Dev Cell. 2003;5(3):429–439. doi: 10.1016/s1534-5807(03)00233-8. [DOI] [PubMed] [Google Scholar]
- Bellairs AdA, Bryant S. Autotomy and regeneration in reptiles. Biology of the Reptilia. 1985;15(5):301–410. [Google Scholar]
- Bely AE. Distribution of segment regeneration ability in the Annelida. Integr Comp Biol. 2006;46(4):508–518. doi: 10.1093/icb/icj051. [DOI] [PubMed] [Google Scholar]
- Bely AE, Nyberg KG. Evolution of animal regeneration: re-emergence of a field. Trends Ecol Evol. 2010;25(3):161–170. doi: 10.1016/j.tree.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Berrill NJ. REGENERATION AND BUDDING IN TUNICATES. Biological Reviews. 1951;26(4):456–475. doi: 10.1111/j.1469-185X.1951.tb01207.x. [DOI] [Google Scholar]
- Bode HR. Head regeneration in Hydra. Dev Dyn. 2003;226(2):225–236. doi: 10.1002/dvdy.10225. [DOI] [PubMed] [Google Scholar]
- Bohn H. Interkalare Regeneration und segmentale Gradienten bei den Extremitäten von Leucophaea-Larven (Blattaria): II. Coxa und Tarsus. Developmental biology. 1970;23(3):355–379. doi: 10.1016/0012-1606(70)90104-1. [DOI] [PubMed] [Google Scholar]
- Bolognesi R, Farzana L, Fischer TD, Brown SJ. Multiple Wnt genes are required for segmentation in the short-germ embryo of Tribolium castaneum. Curr Biol. 2008;18(20):1624–1629. doi: 10.1016/j.cub.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisenko IE, Adamska M, Tokina DB, Ereskovsky AV. Transdifferentiation is a driving force of regeneration in Halisarca dujardini (Demospongiae, Porifera) PeerJ. 2015;3:e1211. doi: 10.7717/peerj.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert PE, Dunn MP, Thomsen GH. A staging system for the regeneration of a polyp from the aboral physa of the anthozoan Cnidarian Nematostella vectensis. Dev Dyn. 2013;242(11):1320–1331. doi: 10.1002/dvdy.24021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun M, Gee L, Reinhardt B, Bode HR. Formation of the head organizer in hydra involves the canonical Wnt pathway. Development. 2005;132(12):2907–2916. doi: 10.1242/dev.01848. [DOI] [PubMed] [Google Scholar]
- Broussonet M. Observations sur la régénérations de quelques parties du corps des poissons. Hist d I’Acad Roy des Sciences. 1786 [Google Scholar]
- Brown FD, Keeling EL, Le AD, Swalla BJ. Whole body regeneration in a colonial ascidian, Botrylloides violaceus. J Exp Zool B Mol Dev Evol. 2009;312(8):885–900. doi: 10.1002/jez.b.21303. [DOI] [PubMed] [Google Scholar]
- Bryant DM, Sousounis K, Payzin-Dogru D, Bryant S, Sandoval AGW, Martinez Fernandez J, Mariano R, Oshiro R, Wong AY, Leigh ND, Johnson K, Whited JL. Identification of regenerative roadblocks via repeat deployment of limb regeneration in axolotls. npj Regenerative Medicine. 2017;2(1):30. doi: 10.1038/s41536-017-0034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant SV, Bellairs AdA. Tail regeneration in the lizards Anguis fragilis and Lacerta dugesii. Journal of the Linnean Society of London, Zoology. 1967;46(310):297–305. [Google Scholar]
- Campbell R, David CN. Cell Cycle Kinetics and Development of Hydra Attenuata: II. Interstitial Cells. Journal of cell science. 1974;16(2):349–358. doi: 10.1242/jcs.16.2.349. [DOI] [PubMed] [Google Scholar]
- Campbell RD. Tissue dynamics of steady state growth in Hydra littoralis. II. Patterns of tissue movement. Journal of Morphology. 1967;121(1):19–28. doi: 10.1002/jmor.1051210103. [DOI] [PubMed] [Google Scholar]
- Campbell RD. A new species of Hydra (Cnidaria: Hydrozoa) from North America with comments on species clusters within the genus. Zoological Journal of the Linnean Society. 1987;91(3):253–263. [Google Scholar]
- Carnevali MC. Regeneration in Echinoderms: repair, regrowth, cloning. Invertebrate Survival Journal. 2006;3(1):64–76. [Google Scholar]
- Casari A, Schiavone M, Facchinello N, Vettori A, Meyer D, Tiso N, Moro E, Argenton F. A Smad3 transgenic reporter reveals TGF-beta control of zebrafish spinal cord development. Dev Biol. 2014;396(1):81–93. doi: 10.1016/j.ydbio.2014.09.025. [DOI] [PubMed] [Google Scholar]
- Caubit X, Nicolas S, Le Parco Y. Possible roles for Wnt genes in growth and axial patterning during regeneration of the tail in urodele amphibians. Dev Dyn. 1997;210(1):1–10. doi: 10.1002/(SICI)1097-0177(199709)210:1<1::AID-AJA1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Chablais F, Jaźwińska A. The regenerative capacity of the zebrafish heart is dependent on TGFβ signaling. Development. 2012;139(11):1921. doi: 10.1242/dev.078543. [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K, de Graaff W, Rossant J, Deschamps J, Beck F. Cdx2 is essential for axial elongation in mouse development. Proc Natl Acad Sci USA. 2004;101(20):7641–7645. doi: 10.1073/pnas.0401654101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chera S, Ghila L, Dobretz K, Wenger Y, Bauer C, Buzgariu W, Martinou J-C, Galliot B. Apoptotic cells provide an unexpected source of Wnt3 signaling to drive hydra head regeneration. Developmental cell. 2009;17(2):279–289. doi: 10.1016/j.devcel.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Choi W-Y, Gemberling M, Wang J, Holdway JE, Shen M-C, Karlstrom RO, Poss KD. <em>In vivo</em> monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development. 2013;140(3):660. doi: 10.1242/dev.088526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourrout D, Delsuc F, Chourrout P, Edvardsen RB, Rentzsch F, Renfer E, Jensen MF, Zhu B, De Jong P, Steele RE. Minimal ProtoHox cluster inferred from bilaterian and cnidarian Hox complements. Nature. 2006;442(7103):684. doi: 10.1038/nature04863. [DOI] [PubMed] [Google Scholar]
- Contreras EG, Gaete M, Sánchez N, Carrasco H, Larraín J. Early requirement of Hyaluronan for tail regeneration in Xenopus tadpoles. Development. 2009;136(17):2987–2996. doi: 10.1242/dev.035501. [DOI] [PubMed] [Google Scholar]
- Cotanche DA. Regeneration of the tectorial membrane in the chick cochlea following severe acoustic trauma. Hear Res. 1987;30(2-3):197–206. doi: 10.1016/0378-5955(87)90136-5. [DOI] [PubMed] [Google Scholar]
- Crest J, Diz-Muñoz A, Chen D-Y, Fletcher DA, Bilder D. Organ sculpting by patterned extracellular matrix stiffness. Elife. 2017;6:e24958. doi: 10.7554/eLife.24958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuénot Li. Anatomie, éthologie et systématique des échinodermes. Traité de zoologie. 1948;11:1–363. [Google Scholar]
- Dahlberg C, Auger H, Dupont S, Sasakura Y, Thorndyke M, Joly JS. Refining the Ciona intestinalis model of central nervous system regeneration. PLoS One. 2009;4(2):e4458. doi: 10.1371/journal.pone.0004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalyell J. Observations on Some Interesting Phenomena in Animal Physiology. Edinburgh, Archibald Constable: Exhibited by Several Species of Planariae Illustrated by Coloured Figures of Living Animals. 1814 [Google Scholar]
- Darras S, Fritzenwanker JH, Uhlinger KR, Farrelly E, Pani AM, Hurley IA, Norris RP, Osovitz M, Terasaki M, Wu M, Aronowicz J, Kirschner M, Gerhart JC, Lowe CJ. Anteroposterior axis patterning by early canonical Wnt signaling during hemichordate development. PLoS Biol. 2018;16(1):e2003698. doi: 10.1371/journal.pbio.2003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LE, Haynes JF. An ultrastructural examination of the mesoglea of Hydra. Zeitschrift fur Zellforschung und Mikroskopische Anatomie. 1968;92(2):149–158. doi: 10.1007/BF00335643. [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra D, Ahuja S, Kim H-T, Rasouli SJ, Stainier DYR, Reischauer S. Opposite effects of Activin type 2 receptor ligands on cardiomyocyte proliferation during development and repair. Nature Communications. 2017;8(1):1902. doi: 10.1038/s41467-017-01950-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBuc TQ, Stephenson TB, Rock AQ, Martindale MQ. Hox and Wnt pattern the primary body axis of an anthozoan cnidarian before gastrulation. Nature Communications. 2018;9(1):2007. doi: 10.1038/s41467-018-04184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Gschwentner R, Rieger R. Free-living flatworms under the knife: past and present. Development genes and evolution. 2007;217(2):89. doi: 10.1007/s00427-006-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Etheridge R. Lizard caudal vertebrae. Copeia. 1967:699–721. [Google Scholar]
- Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- Fraguas S, Barberan S, Cebria F. EGFR signaling regulates cell proliferation, differentiation and morphogenesis during planarian regeneration and homeostasis. Dev Biol. 2011;354(1):87–101. doi: 10.1016/j.ydbio.2011.03.023. [DOI] [PubMed] [Google Scholar]
- Galliot B, Chera S. The Hydra model: disclosing an apoptosis-driven generator of Wnt-based regeneration. Trends in cell biology. 2010;20(9):514–523. doi: 10.1016/j.tcb.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Gauchat D, Mazet F, Berney C, Schummer M, Kreger S, Pawlowski J, Galliot B. Evolution of Antp-class genes and differential expression of Hydra Hox/paraHox genes in anterior patterning. Proceedings of the National Academy of Sciences. 2000;97(9):4493–4498. doi: 10.1073/pnas.97.9.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart J. 1998 Warkany lecture: signaling pathways in development. Teratology. 1999;60(4):226–239. doi: 10.1002/(SICI)1096-9926(199910)60:4<226::AID-TERA7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Gierer A, Berking S, Bode H, David CN, Flick K, Hansmann G, Schaller H, Trenkner E. Regeneration of hydra from reaggregated cells. Nature New Biology. 1972;239(91):98–101. doi: 10.1038/newbio239098a0. [DOI] [PubMed] [Google Scholar]
- Goss RJ. Adaptive growth. Lagos: 1963. [Google Scholar]
- Goss RJ. Principles of regeneration. Elsevier; 1969. [Google Scholar]
- Goss RJ. Deer antlers: regeneration, function and evolution. Academic Press; 1983. [Google Scholar]
- Goss RJ. The evolution of regeneration: adaptive or inherent? Journal of Theoretical Biology. 1992;159(2):241–260. doi: 10.1016/s0022-5193(05)80704-0. [DOI] [PubMed] [Google Scholar]
- Graff L. VON. 1882. Monographie der Turbellarien. I. Rhabdocoelida Ak der Wiss zu Berlin. 1882:442. [Google Scholar]
- Grau-Bove X, Torruella G, Donachie S, Suga H, Leonard G, Richards TA, Ruiz-Trillo I. Dynamics of genomic innovation in the unicellular ancestry of animals. Elife. 2017;6 doi: 10.7554/eLife.26036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotek B, Wehner D, Weidinger G. Notch signaling coordinates cellular proliferation with differentiation during zebrafish fin regeneration. Development. 2013;140(7):1412–1423. doi: 10.1242/dev.087452. [DOI] [PubMed] [Google Scholar]
- Gulati AK, Reddi A, Zalewski A. Changes in the basement membrane zone components during skeletal muscle fiber degeneration and regeneration. The Journal of cell biology. 1983;97(4):957–962. doi: 10.1083/jcb.97.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess A, Cohen A, ROBSON EA. Observations on the structure of Hydra as seen with the electron and light microscopes. Journal of Cell Science. 1957;3(43):315–326. [Google Scholar]
- Ho DM, Whitman M. TGF-beta signaling is required for multiple processes during Xenopus tail regeneration. Dev Biol. 2008;315(1):203–216. doi: 10.1016/j.ydbio.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobmayer B, Rentzsch F, Kuhn K, Happel CM, von Laue CC, Snyder P, Rothbacher U, Holstein TW. WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature. 2000;407(6801):186–189. doi: 10.1038/35025063. [DOI] [PubMed] [Google Scholar]
- Hoffmeister S, Schaller HC. A new biochemical marker for foot-specific cell differentiation in hydra. Wilhelm Roux’s archives of developmental biology. 194(8):453–461. [Google Scholar]
- Huminiecki L, Goldovsky L, Freilich S, Moustakas A, Ouzounis C, Heldin CH. Emergence, development and diversification of the TGF-beta signalling pathway within the animal kingdom. BMC Evol Biol. 2009;9:28. doi: 10.1186/1471-2148-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins ED, Markov GJ, Eckalbar WL, George RM, King JM, Tokuyama MA, Geiger LA, Emmert N, Ammar MJ, Allen AN, Siniard AL, et al. Transcriptomic analysis of tail regeneration in the lizard Anolis carolinensis reveals activation of conserved vertebrate developmental and repair mechanisms. PLoS One. 2014;9(8):e105004. doi: 10.1371/journal.pone.0105004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman LH. Aspects of regeneration in annelids. The American Naturalist. 1940;74(755):513–527. [Google Scholar]
- Hyman LH. The invertebrates: Echinodermata, the coelomate bilateria. Vol. 4 McGraw-Hill; New York: 1955. [Google Scholar]
- Iismaa SE, Kaidonis X, Nicks AM, Bogush N, Kikuchi K, Naqvi N, Harvey RP, Husain A, Graham RM. Comparative regenerative mechanisms across different mammalian tissues. NPJ Regen Med. 2018;3:6. doi: 10.1038/s41536-018-0044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Weston JA. Temperature-sensitive mutations that cause stage-specific defects in Zebrafish fin regeneration. Genetics. 1995;141(4):1583–1595. doi: 10.1093/genetics/141.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Boue S, Belmonte JCI. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nature reviews Molecular cell biology. 2011;12(2):79. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- Kaliszewicz A. Sex ratio patterns and trade-off between sexual and asexual reproduction in the brown hydra. Freshwater Science. 2018;37(3):551–561. [Google Scholar]
- Kasbauer T, Towb P, Alexandrova O, David CN, Dall’armi E, Staudigl A, Stiening B, Bottger A. The Notch signaling pathway in the cnidarian Hydra. Dev Biol. 2007;303(1):376–390. doi: 10.1016/j.ydbio.2006.11.022. [DOI] [PubMed] [Google Scholar]
- Kawabata M, Miyazono K. Signal transduction of the TGF-beta superfamily by Smad proteins. J Biochem. 1999;125(1):9–16. doi: 10.1093/oxfordjournals.jbchem.a022273. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Rodriguez Esteban C, Raya M, Kawakami H, Marti M, Dubova I, Izpisua Belmonte JC. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006;20(23):3232–3237. doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitisin K, Saha T, Blake T, Golestaneh N, Deng M, Kim C, Tang Y, Shetty K, Mishra B, Mishra L. Tgf-Beta signaling in development. Sci STKE. 2007;2007(399):cm1. doi: 10.1126/stke.3992007cm1. [DOI] [PubMed] [Google Scholar]
- Kopan R. Notch signaling. Cold Spring Harb Perspect Biol. 2012;4(10) doi: 10.1101/cshperspect.a011213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 460(7251):60. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- Krishnapati LS, Ghaskadbi S. Identification and characterization of VEGF and FGF from Hydra. Int J Dev Biol. 2013;57(11-12):897–906. doi: 10.1387/ijdb.130077sg. [DOI] [PubMed] [Google Scholar]
- Lane MC, Koehl M, Wilt F, Keller R. A role for regulated secretion of apical extracellular matrix during epithelial invagination in the sea urchin. Development. 1993;117(3):1049–1060. doi: 10.1242/dev.117.3.1049. [DOI] [PubMed] [Google Scholar]
- Larroux C, Fahey B, Degnan SM, Adamski M, Rokhsar DS, Degnan BM. The NK homeobox gene cluster predates the origin of Hox genes. Curr Biol. 2007;17(8):706–710. doi: 10.1016/j.cub.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Le Grand F, Rudnicki MA. Skeletal muscle satellite cells and adult myogenesis. Current opinion in cell biology. 2007;19(6):628–633. doi: 10.1016/j.ceb.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclere L, Bause M, Sinigaglia C, Steger J, Rentzsch F. Development of the aboral domain in Nematostella requires beta-catenin and the opposing activities of Six3/6 and Frizzled5/8. Development. 2016;143(10):1766–1777. doi: 10.1242/dev.120931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K, Thi-Kim Vu H, Mohan RD, McKinney SA, Seidel CW, Alexander R, Gotting K, Workman JL, Sanchez Alvarado A. Egf Signaling Directs Neoblast Repopulation by Regulating Asymmetric Cell Division in Planarians. Dev Cell. 2016;38(4):413–429. doi: 10.1016/j.devcel.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengfeld T, Watanabe H, Simakov O, Lindgens D, Gee L, Law L, Schmidt HA, Ozbek S, Bode H, Holstein TW. Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev Biol. 2009;330(1):186–199. doi: 10.1016/j.ydbio.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Lenhoff SG, Lenhoff HM, Trembley A. Hydra and the birth of experimental biology, 1744: Abraham Trembley’s Mémoires concerning the polyps. Boxwood Pr. 1986 [Google Scholar]
- Levesque M, Gatien S, Finnson K, Desmeules S, Villiard E, Pilote M, Philip A, Roy S. Transforming growth factor: beta signaling is essential for limb regeneration in axolotls. PLoS One. 2007;2(11):e1227. doi: 10.1371/journal.pone.0001227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao BK, Jorg DJ, Oates AC. Faster embryonic segmentation through elevated Delta-Notch signalling. Nat Commun. 2016;7:11861. doi: 10.1038/ncomms11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh KM, van Amerongen R, Nusse R. Generating Cellular Diversity and Spatial Form: Wnt Signaling and the Evolution of Multicellular Animals. Dev Cell. 38(6):643–655. doi: 10.1016/j.devcel.2016.08.011. [DOI] [PubMed] [Google Scholar]
- Lozito TP, Tuan RS. Lizard tail regeneration as an instructive model of enhanced healing capabilities in an adult amniote. Connect Tissue Res. 2017;58(2):145–154. doi: 10.1080/03008207.2016.1215444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukjanenko L, Jung MJ, Hegde N, Perruisseau-Carrier C, Migliavacca E, Rozo M, Karaz S, Jacot G, Schmidt M, Li L. Loss of fibronectin from the aged stem cell niche affects the regenerative capacity of skeletal muscle in mice. Nature medicine. 2016;22(8):897. doi: 10.1038/nm.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden M, Brant JO, Rubiano A, Sandoval AGW, Simmons C, Mitchell R, Collin-Hooper H, Jacobson J, Omairi S, Patel K. Perfect chronic skeletal muscle regeneration in adult spiny mice, Acomys cahirinus. Scientific reports. 2018;8(1):8920. doi: 10.1038/s41598-018-27178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailman ML, Dresden MH. Collagen metabolism in the regenerating forelimb of Notophthalmus viridescens: synthesis, accumulation, and maturation. Developmental biology. 1976;50(2):378–394. doi: 10.1016/0012-1606(76)90159-7. [DOI] [PubMed] [Google Scholar]
- Mao Y, Baum B. Tug of war—the influence of opposing physical forces on epithelial cell morphology. Developmental biology. 2015;401(1):92–102. doi: 10.1016/j.ydbio.2014.12.030. [DOI] [PubMed] [Google Scholar]
- Marlow H, Roettinger E, Boekhout M, Martindale MQ. Functional roles of Notch signaling in the cnidarian Nematostella vectensis. Dev Biol. 2012;362(2):295–308. doi: 10.1016/j.ydbio.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Collins AG. Cladistic analysis of Medusozoa and cnidarian evolution. Invertebrate Biology. 2004;123(1):23–42. [Google Scholar]
- Martindale MQ. The evolution of metazoan axial properties. Nat Rev Genet. 2005;6(12):917–927. doi: 10.1038/nrg1725. [DOI] [PubMed] [Google Scholar]
- Martínez DE, Bridge D. Hydra, the everlasting embryo, confronts aging. International Journal of Developmental Biology. 2012;56(6-7-8):479–487. doi: 10.1387/ijdb.113461dm. [DOI] [PubMed] [Google Scholar]
- McGregor AP, Pechmann M, Schwager EE, Feitosa NM, Kruck S, Aranda M, Damen WG. Wnt8 is required for growth-zone establishment and development of opisthosomal segments in a spider. Current Biology. 2008;18(20):1619–1623. doi: 10.1016/j.cub.2008.08.045. [DOI] [PubMed] [Google Scholar]
- Meyer JJ, Byers JE. As good as dead? Sublethal predation facilitates lethal predation on an intertidal clam. Ecology Letters. 2005;8(2):160–166. [Google Scholar]
- Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276(5309):60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- Miller JR. The Wnts. Genome Biol. 2002;3(1):reviews3001. doi: 10.1186/gb-2001-3-1-reviews3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minelli A, Boxshall G, Fusco G. Arthropod biology and evolution: molecules, development, morphology. Springer Science & Business Media. 2013 [Google Scholar]
- Miyawaki K, Mito T, Sarashina I, Zhang H, Shinmyo Y, Ohuchi H, Noji S. Involvement of Wingless/Armadillo signaling in the posterior sequential segmentation in the cricket, Gryllus bimaculatus (Orthoptera), as revealed by RNAi analysis. Mech Dev. 2004;121(2):119–130. doi: 10.1016/j.mod.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Monti R. Milan R. istituto lombardo; 1900. La rigenerazione nelle planarie marine. [Google Scholar]
- Münch J, González-Rajal A, de la Pompa JL. Notch regulates blastema proliferation and prevents differentiation during adult zebrafish fin regeneration. Development. 2013;140(7):1402. doi: 10.1242/dev.087346. [DOI] [PubMed] [Google Scholar]
- Munder S, Kasbauer T, Prexl A, Aufschnaiter R, Zhang X, Towb P, Bottger A. Notch signalling defines critical boundary during budding in Hydra. Dev Biol. 2010;344(1):331–345. doi: 10.1016/j.ydbio.2010.05.517. [DOI] [PubMed] [Google Scholar]
- Munder S, Tischer S, Grundhuber M, Buchels N, Bruckmeier N, Eckert S, Seefeldt CA, Prexl A, Kasbauer T, Bottger A. Notch-signalling is required for head regeneration and tentacle patterning in Hydra. Dev Biol. 2013;383(1):146–157. doi: 10.1016/j.ydbio.2013.08.022. [DOI] [PubMed] [Google Scholar]
- Naito M, Ishiguro H, Fujisawa T, Kurosawa Y. Presence of eight distinct homeobox - containing genes in cnidarians. FEBS letters. 1993;333(3):271–274. doi: 10.1016/0014-5793(93)80668-k. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Tsiairis CD, Ozbek S, Holstein TW. Autoregulatory and repressive inputs localize Hydra Wnt3 to the head organizer. Proc Natl Acad Sci U S A. 2011;108(22):9137–9142. doi: 10.1073/pnas.1018109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark PA, Sanchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol. 2000;220(2):142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- Nichols SA, Dirks W, Pearse JS, King N. Early evolution of animal cell signaling and adhesion genes. Proc Natl Acad Sci U S A. 2006;103(33):12451–12456. doi: 10.1073/pnas.0604065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nistala H, Lee-Arteaga S, Smaldone S, Siciliano G, Carta L, Ono RN, Sengle G, Arteaga-Solis E, Levasseur R, Ducy P. Fibrillin-1 and-2 differentially modulate endogenous TGF-β and BMP bioavailability during bone formation. The Journal of cell biology. 2010;190(6):1107–1121. doi: 10.1083/jcb.201003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notari M, Ventura-Rubio A, Bedford-Guaus SJ, Jorba I, Mulero L, Navajas D, Martí M, Raya Á. The local microenvironment limits the regenerative potential of the mouse neonatal heart. Science advances. 2018;4(5):eaao5553. doi: 10.1126/sciadv.aao5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- O’Connor MB, Umulis D, Othmer HG, Blair SS. Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development. 2006;133(2):183–193. doi: 10.1242/dev.02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai T, Aramaki T, Inomata H, Hirai T, Kuratani S. On the origin of vertebrate somites. Zoological Lett. 2015;1:33. doi: 10.1186/s40851-015-0033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozhan G, Weidinger G. Wnt/beta-catenin signaling in heart regeneration. Cell Regen (Lond) 2015;4(1):3. doi: 10.1186/s13619-015-0017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang K, Ryan JF, Baxevanis AD, Martindale MQ. Evolution of the TGF-beta signaling pathway and its potential role in the ctenophore, Mnemiopsis leidyi. PLoS One. 2011;6(9):e24152. doi: 10.1371/journal.pone.0024152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang K, Ryan JF, Mullikin JC, Baxevanis AD, Martindale MQ, Program NCS. Genomic insights into Wnt signaling in an early diverging metazoan, the ctenophore Mnemiopsis leidyi. EvoDevo. 2010;1(1):10. doi: 10.1186/2041-9139-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passano L, McCullough C. Co-ordinating systems and behaviour in Hydra: I. Pacemaker system of the periodic contractions. Journal of Experimental Biology. 41(3):643–664. doi: 10.1242/jeb.42.2.205. [DOI] [PubMed] [Google Scholar]
- Patruno M, McGonnell I, Graham A, Beesley P, Carnevali MC, Thorndyke M. Anbmp2/4 is a new member of the transforming growth factor−β superfamily isolated from a crinoid and involved in regeneration. Proceedings of the Royal Society of London B: Biological Sciences. 2003;270(1522):1341–1347. doi: 10.1098/rspb.2003.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patruno M, Smertenko A, Carnevali MC, Bonasoro F, Beesley P, Thorndyke M. Expression of transforming growth factor β-like molecules in normal and regenerating arms of the crinoid Antedon mediterranea: immunocytochemical and biochemical evidence. Proceedings of the Royal Society of London B: Biological Sciences. 2002;269(1502):1741–1747. doi: 10.1098/rspb.2002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 2008;319(5861):327–330. doi: 10.1126/science.1149943. [DOI] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. A wound-induced Wnt expression program controls planarian regeneration polarity. Proc Natl Acad Sci U S A. 2009;106(40):17061–17066. doi: 10.1073/pnas.0906823106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen HO, Hoger SK, Looso M, Lengfeld T, Kuhn A, Warnken U, Nishimiya-Fujisawa C, Schnolzer M, Kruger M, Ozbek S, Simakov O, Holstein TW. A Comprehensive Transcriptomic and Proteomic Analysis of Hydra Head Regeneration. Mol Biol Evol. 2015;32(8):1928–1947. doi: 10.1093/molbev/msv079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya Á, Koth CM, Büscher D, Kawakami Y, Itoh T, Raya RM, Sternik G, Tsai H-J, Rodríguez-Esteban C, Izpisúa-Belmonte JC. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proceedings of the National Academy of Sciences. 2003;100(suppl 1):11889. doi: 10.1073/pnas.1834204100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PC, Bidaye SS, Ghaskadbi S. Genome-wide screening reveals the emergence and divergence of RTK homologues in basal Metazoan Hydra magnipapillata. J Biosci. 2011;36(2):289–296. doi: 10.1007/s12038-011-9065-6. [DOI] [PubMed] [Google Scholar]
- Reddy PC, Ubhe S, Sirwani N, Lohokare R, Galande S. Rapid divergence of histones in Hydrozoa (Cnidaria) and evolution of a novel histone involved in DNA damage response in hydra. Zoology (Jena) 2017;123:53–63. doi: 10.1016/j.zool.2017.06.005. [DOI] [PubMed] [Google Scholar]
- Reddy PC, Unni MK, Gungi A, Agarwal P, Galande S. Evolution of Hox-like genes in Cnidaria: Study of Hydra Hox repertoire reveals tailor-made Hox-code for Cnidarians. Mech Dev. 2015;138(Pt 2):87–96. doi: 10.1016/j.mod.2015.08.005. [DOI] [PubMed] [Google Scholar]
- Reinhardt B, Broun M, Blitz IL, Bode HR. HyBMP5-8b, a BMP5-8 orthologue, acts during axial patterning and tentacle formation in hydra. Dev Biol. 2004;267(1):43–59. doi: 10.1016/j.ydbio.2003.10.031. [DOI] [PubMed] [Google Scholar]
- Rentzsch F, Guder C, Vocke D, Hobmayer B, Holstein TW. An ancient chordin-like gene in organizer formation of Hydra. Proc Natl Acad Sci U S A. 2007;104(9):3249–3254. doi: 10.1073/pnas.0604501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards GS, Degnan BM. The expression of Delta ligands in the sponge Amphimedon queenslandica suggests an ancient role for Notch signaling in metazoan development. EvoDevo. 2012;3(1):15. doi: 10.1186/2041-9139-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards GS, Simionato E, Perron M, Adamska M, Vervoort M, Degnan BM. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Curr Biol. 2008;18(15):1156–1161. doi: 10.1016/j.cub.2008.06.074. [DOI] [PubMed] [Google Scholar]
- Rigo-Watermeier T, Kraft B, Ritthaler M, Wallkamm V, Holstein T, Wedlich D. Functional conservation of Nematostella Wnts in canonical and noncanonical Wnt-signaling. Biol Open. 2012;1(1):43–51. doi: 10.1242/bio.2011021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter WE, Congdon EM. On the inhibition by artificial section of the normal fission plane in Stenostoma, vol 2. vol 6. The Academy. 1900 [Google Scholar]
- Ruhl L. Regenerationserscheinungen an Rhabdocoelen. Zool Anz. 1927;72:160–175. [Google Scholar]
- Ryan JF, Burton PM, Mazza ME, Kwong GK, Mullikin JC, Finnerty JR. The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: evidence from the starlet sea anemone, Nematostella vectensis. Genome Biol. 2006;7(7):R64. doi: 10.1186/gb-2006-7-7-R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JF, Pang K, Schnitzler CE, Nguyen AD, Moreland RT, Simmons DK, Koch BJ, Francis WR, Havlak P, Program NCS, Smith SA, et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342(6164):1242592. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature. 2003;423(6942):876. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- Sanz-Ezquerro JJ, Munsterberg AE, Stricker S. Editorial: Signaling Pathways in Embryonic Development. Front Cell Dev Biol. 2017;5:76. doi: 10.3389/fcell.2017.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarras MP., Jr Components, structure, biogenesis and function of the Hydra extracellular matrix in regeneration, pattern formation and cell differentiation. International Journal of Developmental Biology. 2012;56(6-7-8):567–576. doi: 10.1387/ijdb.113445ms. [DOI] [PubMed] [Google Scholar]
- Sarras MP, Jr, Zhang X, Huff JK, Accavitti MA, John PS, Abrahamson DR. Extracellular matrix (mesoglea) of Hydra vulgaris: III. Formation and function during morphogenesis of hydra cell aggregates. Developmental biology. 1993;157(2):383–398. doi: 10.1006/dbio.1993.1143. [DOI] [PubMed] [Google Scholar]
- Sasidharan V, Marepally S, Elliott SA, Baid S, Lakshmanan V, Nayyar N, Bansal D, Sánchez Alvarado A, Vemula PK, Palakodeti D. The <em>miR-124</em> family of microRNAs is crucial for regeneration of the brain and visual system in the planarian <em>Schmidtea mediterranea</em> Development. 2017;144(18):3211. doi: 10.1242/dev.144758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Bode HR, Sawada Y. Patterning processes in aggregates of hydra cells visualized with the monoclonal antibody, Ts19. Developmental biology. 1990;141(2):412–420. doi: 10.1016/0012-1606(90)90395-y. [DOI] [PubMed] [Google Scholar]
- Schmid V, Tardent P. The reconstitutional performances of the Leptomedusa Campanularia jonstoni. Marine Biology. 1971;8(2):99–104. [Google Scholar]
- Schubert M, Holland LZ. The Wnt Gene Family and the Evolutionary Conservation of Wnt Expression. 2013 [Google Scholar]
- Schummer M, Scheurlen I, Schaller C, Galliot B. HOM/HOX homeobox genes are present in hydra (Chlorohydra viridissima) and are differentially expressed during regeneration. The EMBO journal. 1992;11(5):1815–1823. doi: 10.1002/j.1460-2075.1992.tb05233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Cote LE, Rogers T, Reddien PW. Two FGFRL-Wnt circuits organize the planarian anteroposterior axis. Elife. 2016;5 doi: 10.7554/eLife.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert AW, Kiama SG, Seifert MG, Goheen JR, Palmer TM, Maden M. Skin shedding and tissue regeneration in African spiny mice (Acomys) Nature. 2012;489(7417):561. doi: 10.1038/nature11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk MA, Bode HR, Steele RE. Expression of Cnox-2, a HOM/HOX homeobox gene in hydra, is correlated with axial pattern formation. Development. 1993;117(2):657–667. doi: 10.1242/dev.117.2.657. [DOI] [PubMed] [Google Scholar]
- Shi S, Stanley P. Evolutionary origins of Notch signaling in early development. Cell Cycle. 2006;5(3):274–278. doi: 10.4161/cc.5.3.2396. [DOI] [PubMed] [Google Scholar]
- Shimizu H, Aufschnaiter R, Li L, Sarras MP, Jr, Borza D-B, Abrahamson DR, Sado Y, Zhang X. The extracellular matrix of hydra is a porous sheet and contains type IV collagen. Zoology. 2008;111(5):410–418. doi: 10.1016/j.zool.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H, Zhang X, Zhang J, Leontovich A, Fei K, Yan L, Sarras MP. Epithelial morphogenesis in hydra requires de novo expression of extracellular matrix components and matrix metalloproteinases. Development (Cambridge, England) 2002;129:1521–1532. doi: 10.1242/dev.129.6.1521. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Bae YK, Muraoka O, Hibi M. Interaction of Wnt and caudal-related genes in zebrafish posterior body formation. Dev Biol. 2005;279(1):125–141. doi: 10.1016/j.ydbio.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Simpson SB., Jr Analysis of Tail Regeneration in the Lizard Lygosoma Laterale. I. Initiation of Regeneration and Cartilage Differentiation: The Role of Ependyma. J Morphol. 1964;114:425–435. doi: 10.1002/jmor.1051140305. [DOI] [PubMed] [Google Scholar]
- Souilhol C, Perea-Gomez A, Camus A, Beck-Cormier S, Vandormael-Pournin S, Escande M, Collignon J, Cohen-Tannoudji M. NOTCH activation interferes with cell fate specification in the gastrulating mouse embryo. Development. 2015;142(21):3649–3660. doi: 10.1242/dev.121145. [DOI] [PubMed] [Google Scholar]
- Spallanzani L. An essay on animal reproductions. T. Becket and PA de Hondt [Google Scholar]
- Stevens N, Boring A. Regeneration in polychrerus caudatus.Part I. Observations on living material. Journal of Experimental Zoology. 1905;2(3):335–346. [Google Scholar]
- Stone JS, Rubel EW. Cellular studies of auditory hair cell regeneration in birds. Proceedings of the National Academy of Sciences. 2000;97(22):11714–11721. doi: 10.1073/pnas.97.22.11714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhop S, Coulier F, Bieller A, Vogt A, Hotz T, Hassel M. Signalling by the FGFR-like tyrosine kinase, Kringelchen, is essential for bud detachment in Hydra vulgaris. Development. 2004;131(16):4001–4011. doi: 10.1242/dev.01267. [DOI] [PubMed] [Google Scholar]
- Suga H, Katoh K, Miyata T. Sponge homologs of vertebrate protein tyrosine kinases and frequent domain shufflings in the early evolution of animals before the parazoan-eumetazoan split. Gene. 280(1-2):195–201. doi: 10.1016/s0378-1119(01)00784-3. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Fujisawa T. Genetic analysis of developmental mechanisms in Hydra. II. Isolation and characterization of an interstitial cell-deficient strain. Journal of cell science. 1978;29(1):35–52. doi: 10.1242/jcs.29.1.35. [DOI] [PubMed] [Google Scholar]
- Sureda-Gomez M, Martin-Duran JM, Adell T. Localization of planarian beta-CATENIN-1 reveals multiple roles during anterior-posterior regeneration and organogenesis. Development. 2016;143(22):4149–4160. doi: 10.1242/dev.135152. [DOI] [PubMed] [Google Scholar]
- Takaku Y, Hariyama T, Fujisawa T. Motility of endodermal epithelial cells plays a major role in reorganizing the two epithelial layers in Hydra. Mechanisms of development. 2005;122(1):109–122. doi: 10.1016/j.mod.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Tassava RA, Nace JD, Wei Y. Extracellular matrix protein turnover during salamander limb regeneration. Wound Repair and Regeneration. 1996;4(1):75–81. doi: 10.1046/j.1524-475X.1996.40113.x. [DOI] [PubMed] [Google Scholar]
- Technau U, Holstein TW. Cell sorting during the regeneration of Hydra from reaggregated cells. Developmental biology. 1992;151(1):117–127. doi: 10.1016/0012-1606(92)90219-7. [DOI] [PubMed] [Google Scholar]
- Tischer S, Reineck M, Soding J, Munder S, Bottger A. Eph receptors and ephrin class B ligands are expressed at tissue boundaries in Hydra vulgaris. Int J Dev Biol. 2013;57(9-10):759–765. doi: 10.1387/ijdb.130158ab. [DOI] [PubMed] [Google Scholar]
- Török A, Schiffer PH, Schnitzler CE, Ford K, Mullikin JC, Baxevanis AD, Bacic A, Frank U, Gornik SG. The cnidarian Hydractinia echinata employs canonical and highly adapted histones to pack its DNA. Epigenetics & chromatin. 2016;9(1):36. doi: 10.1186/s13072-016-0085-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trembley A. Mémoires pour servir à l’histoire d’un genre de polypes d’eau douce, à bras en forme de cornes. Par A. Trembley. Chez Jean & Herman Verbeek. 1744 [Google Scholar]
- Tressler J, Maddox F, Goodwin E, Zhang Z, Tublitz NJ. Arm regeneration in two species of cuttlefish Sepia officinalis and Sepia pharaonis. Invert Neurosci. 2014;14(1):37–49. doi: 10.1007/s10158-013-0159-8. [DOI] [PubMed] [Google Scholar]
- Tu S, Johnson SL. Fate restriction in the growing and regenerating zebrafish fin. Developmental cell. 2011;20(5):725–732. doi: 10.1016/j.devcel.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unguez GA. Electric fish: new insights into conserved processes of adult tissue regeneration. The Journal of Experimental Biology. 2013;216(13):2478. doi: 10.1242/jeb.082396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinsky N, Kholodenko BN. Complexity of receptor tyrosine kinase signal processing. Cold Spring Harb Perspect Biol. 2013;5(8):a009043. doi: 10.1101/cshperspect.a009043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vracko R, Benditt EP. Basal lamina: The scaffold for orderly cell replacement: Observations on regeneration of injured skeletal muscle fibers and capillaries. The Journal of cell biology. 1972;55(2):406–419. doi: 10.1083/jcb.55.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332(6031):811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Misof BY. Evolutionary modification of regenerative capability in vertebrates: a comparative study on teleost pectoral fin regeneration. J Exp Zool. 1992;261(1):62–78. doi: 10.1002/jez.1402610108. [DOI] [PubMed] [Google Scholar]
- Wang J, Karra R, Dickson AL, Poss KD. Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Developmental biology. 2013;382(2):427–435. doi: 10.1016/j.ydbio.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Schmidt HA, Kuhn A, Hoger SK, Kocagoz Y, Laumann-Lipp N, Ozbek S, Holstein TW. Nodal signalling determines biradial asymmetry in Hydra. Nature. 2014;515(7525):112–115. doi: 10.1038/nature13666. [DOI] [PubMed] [Google Scholar]
- Wenemoser D, Lapan SW, Wilkinson AW, Bell GW, Reddien PW. A molecular wound response program associated with regeneration initiation in planarians. Genes Dev. 2012;26(9):988–1002. doi: 10.1101/gad.187377.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills AA, Kidd AR, Lepilina A, Poss KD. Fgfs control homeostatic regeneration in adult zebrafish fins. Development. 2008;135(18):3063. doi: 10.1242/dev.024588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windsor Reid PJ, Matveev E, McClymont A, Posfai D, Hill AL, Leys SP. Wnt signaling and polarity in freshwater sponges. BMC Evol Biol. 2018;18(1):12. doi: 10.1186/s12862-018-1118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MY, Hill CS. Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16(3):329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Yokoyama H. Initiation of limb regeneration: the critical steps for regenerative capacity. Dev Growth Differ. 2008;50(1):13–22. doi: 10.1111/j.1440-169X.2007.00973.x. [DOI] [PubMed] [Google Scholar]
- Zhang R, Han P, Yang H, Ouyang K, Lee D, Lin Y-F, Ocorr K, Kang G, Chen J, Stainier DYR, Yelon D, Chi NC. In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature. 2013;498:497. doi: 10.1038/nature12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Borikova AL, Ben-Yair R, Guner-Ataman B, MacRae CA, Lee RT, Burns CG, Burns CE. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proceedings of the National Academy of Sciences. 2014;111(4):1403. doi: 10.1073/pnas.1311705111. [DOI] [PMC free article] [PubMed] [Google Scholar]