Abstract
Our understanding of the human genome has continuously expanded since its draft publication in 2001. Over the years, novel assays have allowed us to progressively overlay layers of knowledge above the raw sequence of A’s, T’s, G’s, and C’s. The reference human genome sequence is now a complex knowledge base maintained under the shared stewardship of multiple specialist communities. Its complexity stems from the fact that it is simultaneously a template for transcription, a record of evolution, a vehicle for genetics, and a functional molecule. In short, the human genome serves as a frame of reference at the intersection of a diversity of scientific fields. In recent years, the progressive fall in sequencing costs has given increasing importance to the quality of the human reference genome, as hundreds of thousands of individuals are being sequenced yearly, often for clinical applications. Also, novel sequencing-based assays shed light on novel functions of the genome, especially with respect to gene expression regulation. Keeping the human genome annotation up to date and accurate is therefore an ongoing partnership between reference annotation projects and the greater community worldwide.
Keywords: human, genome, annotation, genes, variants, regulatory elements
History of the Human Genome and its Annotation
The history of the sequencing and annotation of the human genome is marked by breathtaking acceleration after a prolonged theoretical inception. Many components of the human genome were discovered long before its base pairs were read through astute experimental design.When the DNA was finally readable, these abstract concepts were mapped onto actual sequences, creating a multilayered annotation linking sequence to phenotype.
Defining Concepts
The concept of genes evolved from theoretical consideration to molecular components (54). In 1866, Gregor Mendel published his laws of genetics (97), and three years later, Friedrich Miescher isolated nucleic acids (31). The term gene itself was coined as early as 1909 by Wilhelm Johannsen (79, 130) to designate the characteristics of the gametes that affect the resulting organism. Even though geneticists did not know the exact molecule involved, statistical analyses of inheritance patterns allowed them to determine that genes were stored in a linear fashion and to start computing genetic maps of gene proximity (143).
It was only in the mid-twentieth century that the experiments of Avery et al. (8) (1944) and Hershey & Chase (65) (1952) demonstrated the role of DNA in carrying genetic information. Once the role of DNA was proven, genes became physical components. Protein-coding genes could be characterized by the genetic code, which was determined in 1965 (109, 135), and could thus be defined by the open reading frames (ORFs). However, exceptions to Francis Crick’s central dogma of genes as blueprints for protein synthesis (30) were already being uncovered: first tRNA (27) and rRNA (87) and then a broad variety of noncoding RNAs (38).
The genome also provides mechanisms to regulate when and where genes are expressed, thus refning their phenotypic effects. In 1939, Conrad Hal Waddington (161) coined the term epigenetics to designate the study of cell type differentiation (67). In 1970, John Gurdon (61) demonstrated that differentiation did not involve changes to DNA, raising the question of how a multicellular organism, whose genome is (nearly) identically replicated across all cells, could express a wide diversity of cell types, tissues, and so on. Epigenetics thus became the study of information conserved across mitosis and not carried by the DNA sequence. Confusingly, the term later came to additionally (and simultaneously) refer to the study of non-Mendelian inheritance across generations (45, 70).
The control mechanism of gene expression levels was illuminated by François Jacob and colleagues through the discovery of the lac operon (78), and a model of gene expression regulation was produced: a promoter sequence upstream of the gene to recruit polymerase and operator sequences to recruit transcription factors. Farther away from the promoter, enhancers were found—first in viruses in 1981 (13, 59) and then in eukaryotes in 1983 (9, 55, 98)—to affect transcriptional output at the promoter regardless of distance or orientation.
The genome contains functional regions relevant to its integrity. Centromeric regions, for example, are necessary to recruit the kinetochores to ensure proper separation of chromatin during mitosis, to keep sister chromatids together ahead of mitosis (10), and finally to ensure their own rapid replication during S phase (145). Telomeric regions have long been interpreted to protect the ends of chromosomes, but our understanding of their function is still evolving (133).
Finally, a large amount of the genome is derived from transposable elements. In 1953, Barbara McClintock (95) published the first observation of genes moving in the genome. It was later discovered that transposable elements correspond to repeated sequences that are able to copy themselves within a cell’s genome.
Reading the Genome
Shortly after the discovery of the importance of nucleic acids, the first (RNA) genome was sequenced in 1976 (47), and sequencing methods were refined to allow for large-scale data production (131). Despite this rapid progress, characterizing the entire sequence of a large genome was still a complex and costly endeavor due to the necessity of collecting genetic linkage maps (101), which meant that the first eukaryotic genome (of yeast) was published only in 1996 (58).
As genome sequencing technology improved, one obvious challenge was to sequence the human genome (155). Despite all the technical obstacles, as early as 1985, scientists such as Robert Sinsheimer at the University of California, Santa Cruz (UCSC), started discussing the feasibility of sequencing the human genome (28). This idea gathered support, and in 1988, a joint project of the US National Institutes and Health and Department of Energy was created to sequence the human genome over a period of 15 years, around which parallel efforts in China, France, Germany, Great Britain, and Japan rallied. The project continued slowly, sequencing less than 15% of the genome over the next 11 years, until the competition of the Celera Corporation created uncertainty about the availability of the sequence and spurred a significant ramping up of resources and processes, leading to the back-to-back release of two draft sequences on June 26, 2000 (76, 157).
Mapping Old Concepts to the New Sequence
As soon as the sequence of the human genome could be read, the community set out to associate all the earlier concepts inferred through indirect observation onto actual nucleotide sequences and motifs.
Gene annotation
Before the sequencing of the entire human genome, fragmentary data were already being collected into reference resources that became the foundation of bioinformatics (140). These were naturally focused on the protein products of genes. In 1965, Margaret Oakley Dayhoff created the first bioinformatic database, the Atlas of Protein Sequence and Structure, and in 1971, Bernstein et al. (15) created the Protein Data Bank (PDB). These resources were followed in the early 1980s by nucleotide sequence databases, such as GenBank and the European Molecular Biology Laboratory (EMBL) Data Library, which would later become EMBL-Bank and then the European Nucleotide Archive. These data collections were enriched by annotation databases such as Swiss-Prot (1986), which later became UniProt, and then domain-specific genomic annotations such as RepBase (1992), AceDB (1995), FlyBase (1997), and WormBase (2001). To collect sequence polymorphisms, the dbSNP reference database was created in 1999 (134), establishing an unambiguous assignment of identifiers to single-nucleotide polymorphisms (SNPs), known as reference SNP IDs (rsIDs).
As soon as the raw genomic sequence was released, various teams competed to annotate the gene loci. The initial methods developed fell into three broad categories. The first consisted of ab initio methods such as GENSCAN (20), which used contemporary biological knowledge of transcription, translation, and splicing to build computational models that looked for signals of these processes in the genomic sequence itself and required no additional input. The second was gene annotation methods such as SGP2 (113), SLAM (3), and TWINSCAN (48), which built computational models that leveraged knowledge of patterns of sequence-level conservation among species to identify protein-coding genes subject to purifying selection. The third was to take experimental data from one or more sources of sequenced cDNA and expressed sequence tag (EST) libraries, held in the International Nucleotide Sequence Database Collaboration (INSDC) databases (81), and curated annotation from expert databases such as Swiss-Prot (now UniProt) (154).
Manual and automated annotation
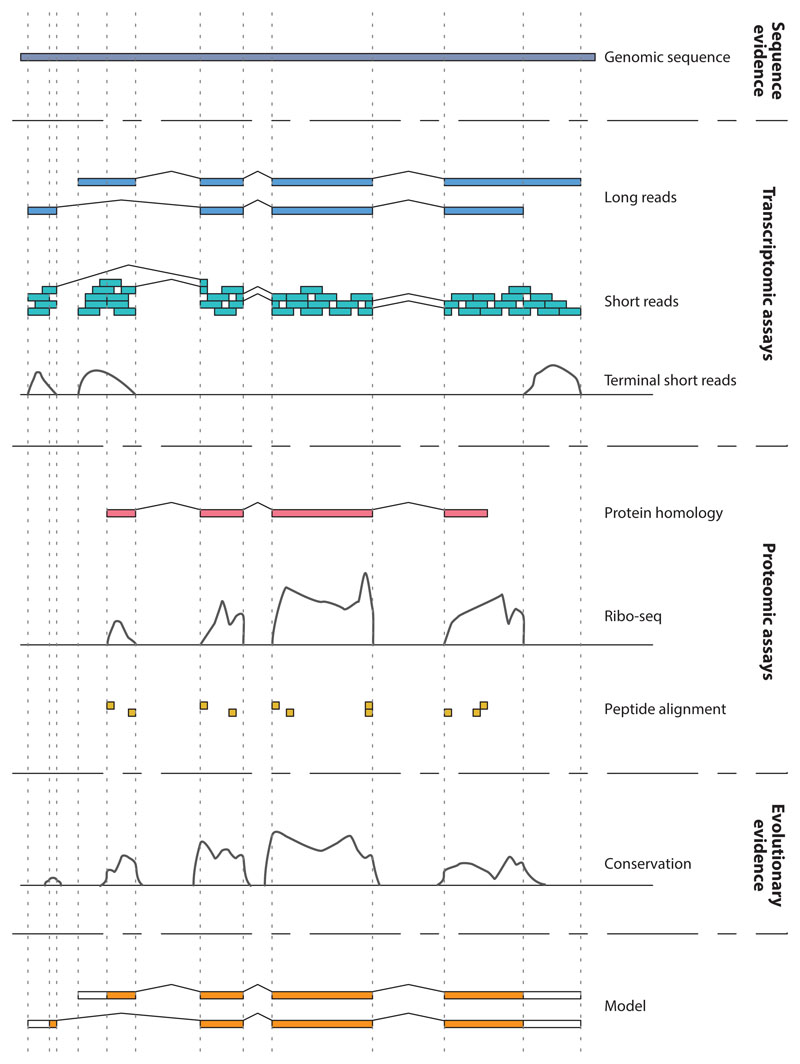
While some evidence-based approaches, such as Ensembl and UCSC genes, were purely computational, both the RefSeq group at the National Center for Biotechnology Information (NCBI) (110) and the Human and Vertebrate Analysis and Annotation (HAVANA) group (49) [initially at the Wellcome Sanger Institute, now merged into Ensembl at EMBL’s European Bioinformatics Institute (EMBL-EBI)] employed manual annotation approaches to complement automated annotation methods. Manual approaches not only require annotators to examine all alignments that are used to create gene and transcript models but also allow them to take into account any orthogonal data, including critical reading of the available literature, to determine the best representation of a gene feature (for a summary, see Figure 1). The manual approach is thus able to give a highly sensitive and specific annotation at the cost of speed. Indeed, the full manual first-pass annotation of the human reference genome by the HAVANA group took approximately 13 years.
Figure 1. Gene annotation process.
Gene annotation uses diverse orthogonal data types to determine first the structure and then the most likely functional class of the transcript and gene locus. Long transcriptomic data aligned to the reference genome identify the overall exon-intron structure of the transcript, while short RNA sequencing reads give confidence to the annotation of precise intron/exon boundaries and extensions at the ends of the transcripts (5′ and 3′ untranslated regions), especially where coverage from longer reads is low. Some transcript structures may be annotated entirely based on RNA sequencing data, again where coverage from longer reads is low. Terminal short-read data sets help define the 5′ and 3′ ends of transcripts, which is important from both a structural and functional point of view; where the termini of a transcript can be identified with confidence, lending certainty of the structural annotation, the annotators gain greater confidence in their determination of functional annotation. The presence of high-quality proteomic data and evidence of the evolutionary conservation of coding sequence informs the annotation of coding potential.
In 2006, the human Encyclopedia of DNA Elements (ENCODE) Genome Annotation Assessment Project (EGASP) compared automated annotation pipelines with HAVANA manual annotation of the ENCODE pilot regions, representing 1% of the human genome (40). This study revealed that, while the best automated annotation pipelines were broadly successful in identifying manually annotated protein-coding gene loci, all methods failed to reproduce the manually determined transcript exon–intron structures, particularly where alternatively spliced transcripts were identified (60). Although far more laborious, the manual annotation provides a detailed review of each edge case and the opportunity to select the evidence relevant to each locus. Manually encoding an algorithm to handle each and every exception would be less cost-effective than directly editing these occurrences in a database. It is, however, conceivable that recent developments in machine learning will enable a computer to devise such knowledge automatically, in which case existing manual gene annotations will prove an invaluable training data set.
Notwithstanding the general adoption of these two reference sets for gene annotation, additional approaches to gene annotation continue to be developed. For example, as well as the automated gene annotation methods that use one or two sources of data, methods such as AUGUSTUS (66) and Maker (21) have been developed that integrate multiple sources of data, including other gene predictions and data from RNA sequencing (RNA-seq). Though these approaches could be used to annotate the human genome, their stated role is to support gene annotation for genome projects with substantially less data and attention to annotation than the human genome.
Advances in transcriptome sequencing
The emergence of new transcript sequencing technologies has supported new approaches for detecting genes and transcripts along the human genome (see Table 1). The first of these next-generation sequencing technologies, RNA-seq (163), was based on Solexa (12) (now Illumina) sequencing and provided significantly higher depth (i.e., more sequenced molecules) than Sanger cDNA reads but with much shorter reads. While the length of reads for the technology has extended from approximately 30 bases in early versions to a maximum of approximately 250 bases today (and a general practical application of approximately 100 bases), the shorter length of reads compared with INSDC cDNA data hampers their assembly into full-length transcripts, which can be several kilobases long.
Table 1. Evidence relevant to the annotation of different types of genes.
| Biotype | Transcription data (INSDC, RNA-seq, PacBio, ONT) | Terminal transcription data (CAGE, RAMPAGE, polyA-seq) | Protein homology data (UniProt) | Protein experimental data (MS, ribo-seq) | Conservation data (PhyloCSF, PhastCons, GERP) | RNA secondary structure data (Infernal) | External expert database (miRBase, Rfam, IMGT) |
|---|---|---|---|---|---|---|---|
| Protein coding | Yes | Yes | Yes | Yes | Yes | No | No |
| lncRNA | Yes | Yes | No | No | No | No | No |
| sRNA | Yes | No | No | No | Yes | Yes | Yes |
| Pseudogene | Noa | Noa | Yes | No | Yes b | No | No |
| IG/TR | No | No | No | Yes | No | No | Yes |
This table illustrates the evidence types generally used by manual annotators in the Ensembl team to determine the correct structure and function of a transcript model. Protein-coding genes require transcriptomic evidence to define structure and terminal transcription data sets to define transcript start and end coordinates. Homology with UniProt and proteomics data informs or validates the decision to assign a transcript or locus the protein-coding biotype—that is, to decide whether a functional protein is encoded. Similarly, evolutionary conservation of sequence and of protein-coding potential also informs this decision. Decisions about protein-coding genes do not generally use RNA secondary structure or other expert databases, although they may be consulted on a case-by-case basis. The annotation of lncRNAs utilizes the same transcriptomic data sets as protein-coding genes; however, the absence of protein homology, experimental proteomics data, and conservation is a key determinant in choosing not to annotate a transcript as protein coding. For sRNAs, transcriptomic data sets, conservation data, RNA secondary structure data, and expert external databases are utilized. Pseudogenes are annotated based solely on their homology to annotated protein sequences, although transcriptomic data are used to support the transcribed pseudogene biotypes. IG/TR gene segments are annotated on the basis of protein experimental data and homology to IG/TR sequences from the IMGT database. Abbreviations: CAGE, cap analysis gene expression; GERP, Genomic Evolutionary Rate Profiling; IG, immunoglobulin; IMGT, International Immunogenetics; Infernal, Inference of RNA Alignment; INSDC, International Nucleotide Sequence Database Collaboration; lncRNA, long noncoding RNA; miRBase, MicroRNA Database; MS, mass spectrometry; ONT, Oxford Nanopore Technologies; PacBio, Pacific Biosciences; PhyloCSF, Phylogenetic Codon Substitution Frequencies; polyA-seq, polyA sequencing; RAMPAGE, RNA annotation and mapping of promoters for the analysis of gene expression; ribo-seq, ribosome profiling; RNA-seq, RNA sequencing; sRNA, small RNA; TR, T cell receptor.
For nontranscribed pseudogenes only; transcribed pseudogenes may be supported by these data.
While pseudogenes are not conserved over large evolutionary distances, known artifacts in the whole-genome alignments on which conservation detection is based permit their identification with care.
This problem was exposed by the RNA-seq Genome Annotation Assessment Project (RGASP) (139), a recapitulation and extension of the EGASP exercise that focused on RNA-seq data. RGASP showed that no method achieved the same level of quality as automated annotation pipelines using Sanger-sequenced INSDC data sets in EGASP. Despite the development of new methods such as StringTie (119) and improvements in the pioneers of RNA-seq transcript assembly such as Cufflinks (151), the fundamental difficulty in assigning short reads to longer transcripts that are subject to alternative splicing with the required resolution appears to be insurmountable (88, 150, 164).
Sequencing technologies generating longer reads, such as Roche’s 454 pyrosequencing (94); Pacific Biosciences’ Single Molecule, Real-Time (SMRT) sequencing (39); and Oxford Nanopore sequencing (99), can aid the reconstruction of transcripts. The latter two methods are still relatively new, but their read length and coverage depth hold the promise of solving the problem of accurately identifying transcript structures. While none of these methods produce reads with the same low error rate as Sanger-sequenced cDNAs, when polished by consensus generation (128), RNA-seq data (146), and variation data (170), they can be used in an equivalent manner for gene annotation by both automated and manual approaches. Combined with intron-spanning RNA-seq reads to validate splice sites with base-pair resolution, they promise to revolutionize transcript annotation in the near future.
Protein-coding genes
Protein-coding genes were the best-understood class of gene features prior to the sequencing of the human genome, with the Swiss-Prot and RefSeq databases providing genome-free curation of protein and gene sequences, respectively. Despite this foreknowledge, the total number of protein-coding genes is still being debated (see the sidebar titled First Surprise: How Many Protein-Coding Genes Are There in the Human Genome?). Frequently, short ORFs are found to be transcribed, suggesting the existence of uncharacterized proteins (see the sidebar titled Second Surprise: Lilliput Genes). In some cases, the evidence from nonreference databases aligns to genomic regions that do not contain an intact coding sequence. Such inconsistencies arise either from sequencing errors in the reference sequence or from natural polymorphisms. Genuine loss-of-function variants in the human reference sequence have been identified at a range of allele frequencies, with some gene regions containing very rare alleles on the reference sequence that were initially thought to be nonfunctional pseudogenes. To correct these inconsistencies, the Genome Reference Consortium (GRC) has supplemented many affected regions with patches and representations of alternative alleles to allow the functional copies of protein-coding genes to be captured in the total gene set (132).
Pseudogenes
Pseudogenes are predominantly duplicate copies of genomic loci that share sequence similarity with their functional parent copy but lack protein-coding potential due to the presence of disruptive mutations such as frame shifts and premature stop codons. Pseudogenes are classified according to the biological processes that led to their creation as (a) processed pseudogenes, which are created by retrotransposition of mRNA from functional protein-coding loci back into the genome, or (b) duplicated or unprocessed pseudogenes, which are created by the complete or partial duplication of functional genes; a third and distinct category is (c) unitary pseudogenes, which are created by loss-of-function mutations in ancestral functional protein-coding genes (116). Pseudogenes are of interest not only because of the insights they can provide into these processes but also because their shared homology with functional protein-coding parent genes can inform interpretation of the alignment of transcriptomic data to the genome. In addition, pseudogenes are a substrate on which evolution can occasionally act to create novel function; for example, the long noncoding RNA (lncRNA) responsible for X inactivation arose from a duplicated or unprocessed pseudogene (37) (see also the sidebar titled Third Surprise: Win Some, Lose Some).
Noncoding RNA
The human genome is pervasively transcribed, with the vast majority of the bases in the reference human genome represented in transcriptomic data sets (34, 41). The resulting transcripts that do not belong to protein-coding genes are usually divided by length (141).
Small-RNA genes are conventionally characterized by the fact that they are shorter than 200 base pairs, do not encode polypeptides, and possess secondary structures that are important to their function. They are generally identified in the genome by (a) homology to sequences of known genes both within the same species and between species; (b) the presence of a known secondary structure; (c) the presence of paired changes in sequence or covariance that preserve structure (105); and, more recently, (d) the presence of small-RNA sequences detected experimentally. Small RNAs are often found in large numbers in the human genome; for example, the approximately 2,000 microRNAs generate massive diversity in their targets through sequence differences in the mature microRNAs, while the U6 small nuclear RNAs have more than 1,300 copies of essentially the same sequence. While the numbers of potential genes may be large,many loci encode nonfunctional (or pseudogenic) copies of the small RNA, and discriminating between the functional and nonfunctional copies remains a problem (36, 86). However, the development of computational methods combined with manual curation and literature review of expert small-RNA databases holds the potential to achieve greater resolution of gene classes where the biology is better understood and where experimental data provide sufficiently comprehensive coverage. The functions of many small RNAs have been very well characterized, and both germline and somatic variation have been linked to disease. As such, it is as important to obtain a full representation of functional small-RNA loci in the reference genome as it is for protein-coding genes.
lncRNAs are a class of transcripts that, by definition, are more than 200 bases in length, frequently extending to tens of thousands of kilobases. Unlike small RNAs, lncRNAs lack known RNA secondary structures, although there are considerable ongoing efforts to investigate whether functional and/or structural motifs can be identified and used to inform the annotation and classification of lncRNAs. lncRNAs generally show little cross-species conservation at the sequence level, although they more commonly show conservation of their position in syntenic regions of the genomes.
Large numbers of lncRNA loci have been identified in the reference annotation catalogs (approximately 18,000 in Ensembl/GENCODE and 15,000 in RefSeq). Even larger catalogs have been created by transcript reconstruction for RNA-seq data (69), and resources that collate other individual catalogs reach even greater numbers of lncRNAs—LNCipedia, for example, contains approximately 49,000 high-confidence loci (160)—although different resources have different criteria for annotation, making direct comparison difficult. Given the rate of discovery of new lncRNA loci in both RNA-seq and long transcriptomic data sets, it is unlikely that these figures represent the final tally.
Some lncRNAs have been clearly demonstrated to be functional. The X-inactive specific transcript (XIST) locus, for example, is an essential component of the X-inactivation process (122). While only a few lncRNA loci have been characterized to the same depth as XIST, more lncRNAs, such as XIST and HOTAIR, have been implicated in the regulation of epigenetic modifications (129) as well as other processes, such as the regulation of transcription (62). lncRNAs such as HOTAIR and MALAT1 have been implicated in disease (84, 171), and while the mechanism for their involvement is frequently unclear, they may serve as useful markers for prognosis via the monitoring of expression levels (127).
Repetitive regions and transposable elements
A large proportion of the human genome consists of repetitive sequences. Transposable elements make up the largest category, covering approximately 45% of the genome, and possess the innate ability to move around the genome (112). The vast majority of transposable elements (approximately 90%) are retrotransposons, which are initially transcribed from DNA to an RNA intermediate before being copied back to DNA by reverse transcriptase enzymes (29). The DNA copy is then inserted back into the genome in a new position, often far from the original locus. Long terminal repeat and long interspersed nuclear element (LINE) retrotransposons encode the reverse transcriptase enzymes that catalyze their creation, but short interspersed nuclear element (SINE) retrotransposons do not. DNA transposons do not utilize an RNA intermediate and instead are excised from the genome and reinserted via the activity of a transposase enzyme. As with the retrotransposons, some classes of DNA transposons encode their own transposases, while others do not and rely on the presence of other transposons for their mobility. The remaining repeat sequences comprise microsatellites, which are very short DNA sequences (typically 5 or fewer bases in length) repeated many times; larger minisatellites (10–60 bases in length); and satellite DNA, such as alpha- and beta-satellite DNA, which forms the main component of centromeres and heterochromatin. Repeat sequences are identified in the genome on the basis of sequence similarity to curated repeat libraries by computational methods such as RepeatMasker (148; http://www.repeatmasker.org).
Polymorphisms
SNPs are characterized by their alleles and the shared flanking sequences, and mapping them to the genome is therefore a matter of performing a sequence search in the genome (24). Since the human reference genome is composed of sequences from a few donors, largely from the anonymous RPCI-11 donor (111), the scientific community endeavored to enrich it with common polymorphisms sampled across wide populations. In some cases, the GRC has added the sequences of alternative haplotypes for highly variable regions of the genome, such as the major histocompatibility complex and leukocyte receptor cluster. Large surveys such as the International HapMap Project (75) and the 1000 Genomes Project (1) have further enriched our knowledge of the genome with short polymorphisms as well as structural variants. These maps have provided researchers with allele frequencies across populations as well as linkage information. Once they are annotated onto the genome, interpreting the functional impact of variants is very much an open research question; however, this process is sensitive to the reference annotations used for genes, regulatory features, repeats, and so on (82). This increasing reliance on annotations for biomedical applications in particular is a driver for current efforts to ensure that annotations are both complete and stable.
GRCH38: The Human Genome and its Current State of Annotations
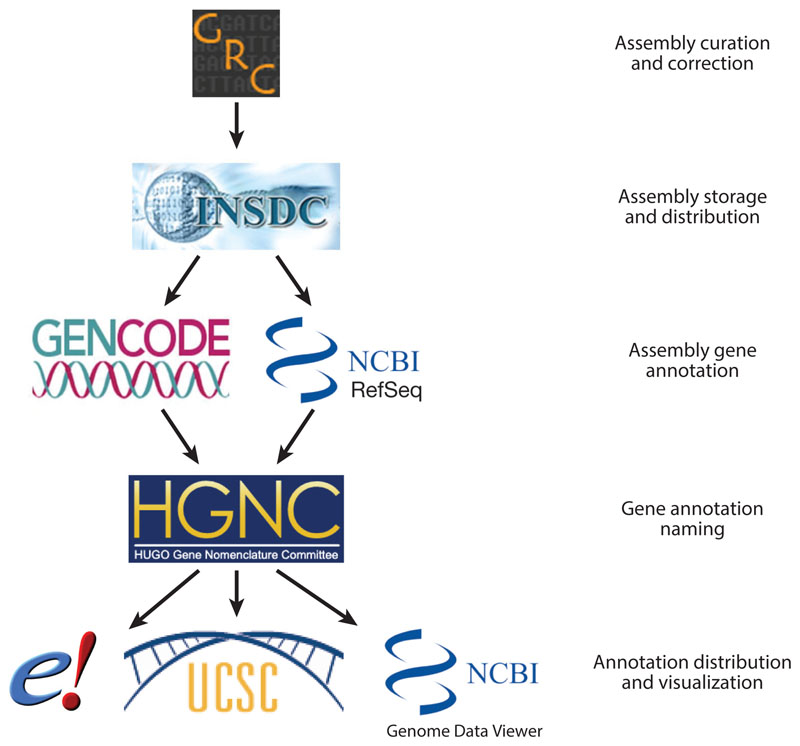
The current official GRCh38 genome assembly and its annotations are a corpus of public knowledge that is kept up to date and accurate under the stewardship of multiple specialist bodies across the world, as illustrated in Figure 2. The GRC (25), a collaboration among five institutes, defines the official genome build sequence and hence lends its name to the assembly. It is responsible for improving the human reference genome assembly, correcting errors, and adding sequences to ensure that it provides the best representation of the human genome to meet basic and clinical research needs. Every time a release or an update is ready, it submits the sequence to the INSDC (81), which freely distributes the sequence via three international nucleotide archives in Japan, Europe, and the United States.
Figure 2. Organizations that support the GRC assembly and its gene annotations.
Abbreviations: e!, Ensembl Project; GRC, Genome Reference Consortium; HGNC, Human Genome Organisation (HUGO) Gene Nomenclature Committee; INSDC, International Nucleotide Sequence Database Collaboration; NCBI, National Center for Biotechnology Information; UCSC, University of California, Santa Cruz.
Once the raw sequence is available, it is vital to assign known elements to it, so that past research, mapped to a previous genome assembly, is not rendered meaningless by a change in the coordinates. This process ensures the backward compatibility of the new build with past research. Human genes are annotated in parallel by two consortia: GENCODE (49) and RefSeq (110). This two-pronged effort serves to stimulate research by providing a point of comparison. These two annotations are regularly compared, producing the Consensus Coding Sequence (CCDS) annotation (125). To mitigate the confusion that could be created by the use of two different reference annotations, the Human Genome Organisation (HUGO) Gene Nomenclature Committee (HGNC) (18) is responsible for assigning common gene names and symbols to both annotations. Variants are separately mapped to the genome by dbSNP (131) and Ensembl (172). All of these annotations are then freely distributed via public genomic databases and browsers, particularly Ensembl, the UCSC Genome Browser (63), and the NCBI Map Viewer (167).
An accurate representation of the gene content of the human genome is of great importance both for supporting research in genome biology and as a foundation for the interpretation of genetic variation in the clinic. Given the relative inaccuracy of even the best automated methods and the chance (or even likelihood) that any error in gene annotation could be propagated into an error in the clinic, the two sets of gene annotations that are generally utilized as a reference are predominantly manually created and maintained on one hand by the Ensembl group in collaboration with the GENCODE consortium (formally known as the Ensembl/GENCODE annotation) and on the other by the RefSeq group.
GRCH39 and Beyond: Future Challenges of Human Genome Annotation
The concept of the reference human genome is changing with the creation of the Human Pangenome Reference Consortium (https://humanpangenome.org), which plans to complete several hundred high-quality haplotype-resolved human assemblies representing populations around the world. These genomes will be collected and presented in a graph-based pangenome structure to best represent human genetic variation. The pangenome and extracts of it representing individual human genomes will be the substrate for future genome annotation and analysis.
The Genome as a Template for Transcription
Despite tremendous progress since the publication of the draft genome, the identification and characterization of transcribed regions of the genome are still moving targets, as we learn more about the subtleties of transcriptions. Thus, annotations are continuously being enriched with subtle new features revealed through novel assays.
Converging on a final list of protein-coding genes
New genes are being regularly detected thanks to a combination of better computational methods to generate and rank targeted lists for manual review (92) and a growing and diverse corpus of transcriptomic and proteomic data sets that cover an expanding number of human cell types and tissues (for an example, see Figure 3), experimental resources also employed by gene annotation resources such as the Comprehensive Human Expressed Sequences (CHESS) catalog (120). While the use of such resources is clearly of tremendous importance in the discovery of new protein-coding genes, the total number of protein-coding genes in reference catalogs is converging on stability, as illustrated in Figure 4. At the same time, many protein-coding annotations are being removed as well. For the most part, this removal happens as an older annotation is reevaluated in the light of better functional, evolutionary, transcriptomic, proteomic, and human variation data on a case-by-case basis. When a locus that was previously annotated as protein coding is found on review to lack the expected level of evidence for a protein-coding gene, its classification will be updated (44).
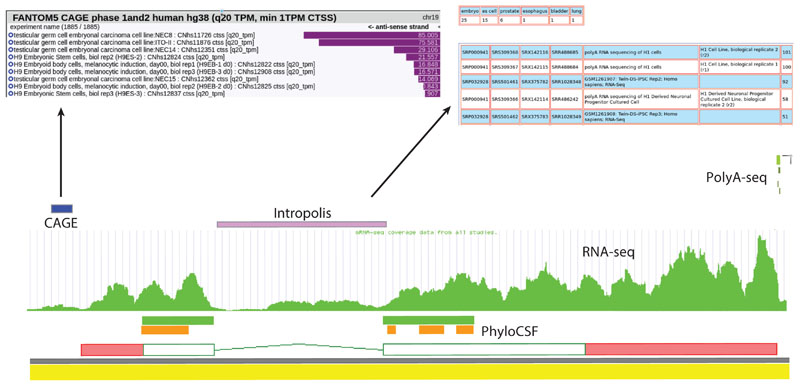
Figure 3. A locus whose identification was possible only through the analysis of recent orthologous data types.
The locus lacks any support from transcript evidence deposited in INSDC databases, and as such, it is not represented in any reference annotation database. Only by identifying the intersection of PhyloCSF data (to identify conserved protein-coding potential), RNA-seq data (to provide evidence of transcription and tissue specificity), Intropolis RNA-seq-supported intron-spanning reads (to provide evidence for precise split junctions and support tissue specificity from other datasets), CAGE data (to define transcript 5′ ends and tissue specificity support), and polyA-seq data (to define transcript 3′ ends and tissue specificity support) could a correctly splicing transcript model be built and the correct coding sequence added. Given the expectation of conservation, protein-coding genes identified by this annotation process were also annotated in mouse to provide an additional check on their validity Abbreviations: CAGE, cap analysis gene expression; INSDC, International Nucleotide Sequence Database Collaboration; PhyloCSF, Phylogenetic Codon Substitution Frequencies; polyA-seq, polyA sequencing; RNA-seq, RNA sequencing.
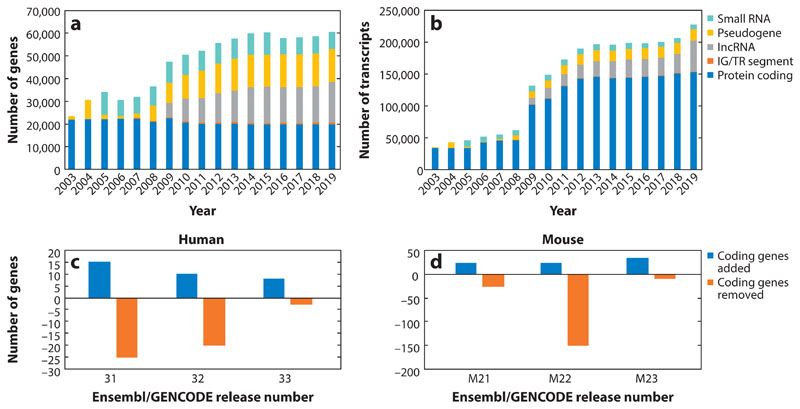
Figure 4. Progress in the annotation of gene loci in Ensembl/GENCODE.
(a) The number of protein-coding genes annotated has generally fallen over time but appears to be generally stable in recent years. The number of pseudogene loci increased rapidly during the annotation of the whole genome (2007–2012) and has maintained slow growth subsequently, while the number of lncRNA experienced a similar pattern of increase but continues to rise. Small-RNA locus totals are generally stable, only changing when there is a significant update to their automated annotation pipeline, and the relatively few IG and TR segments have remained broadly stale since their initial annotation. (b) The number of transcripts continues to increase over time, particularly for protein-coding genes and lncRNA loci, and given the availability of high-quality long-read data sets, this trend is expected to continue. (c,d) The changes to protein-coding gene counts underlying the relatively stable headline totals for human and mouse, respectively, in three recent Ensembl/GENCODE annotation releases. Protein-coding genes were both added and removed in every human and mouse release, with a total of 33 additions and 48 removals in human and 80 additions and 188 removals in mouse, suggesting that the final gene annotation for protein-coding genes has not yet been settled. Abbreviations: IG, immunoglobulin; lncRNA, long noncoding RNA; TR, T cell receptor.
Converging on a definition of protein-coding genes
Given the clear benefit of removing uncertainty from the annotation of protein-coding genes in the reference genome, significant efforts have been made to achieve convergence among the major reference databases, such as the CCDS project being carried out by RefSeq, Ensembl/GENCODE, UCSC, UniProt, and the HGNC (126). While these cross-database exercises have made great strides toward achieving the goal of convergence, they have also revealed some of the remaining gaps in our knowledge, particularly questions on the very definition of a protein-coding gene.
Specifically, new evidence has shown low-level transcription and translation across the genome, although this may not have a role in cellular physiology. The depth of transcriptomic data available allows us to identify a greater number of transcribed regions of the genome. At the same time, new techniques such as ribosome profiling (ribo-seq) provide direct evidence of translation (via the proxy of interaction between ribosome and transcript), demonstrating that translation is perhaps more promiscuous than previously thought (72) (see also the sidebar titled Fourth Surprise: Coding Noncoding RNA?).
Additional methods are therefore required to discriminate functional protein-coding loci from other transcribed and translated regions. Current approaches rely on better determination of evolutionary conservation to provide additional confidence in protein-coding potential, but this precludes the annotation of genuinely emergent functional coding genes (80, 144). A similar class of putative protein-coding genes is those that have clear evidence of transcription, and sometimes translation as well, but have activity restricted exclusively or predominantly to a disease state. Cancer–testis (CT) antigen genes such as GAUGE family members display these characteristics of protein-coding genes but lack evolutionary conservation, and we have no understanding of the role they play in normal cellular function. They are potentially important targets for immunotherapy (57) and demand inclusion in the reference gene catalogs; however, their existence suggests that further subclassification of protein-coding genes is required to capture the functional diversity within the group.
A relatively small number of protein-coding genes have been thoroughly investigated in direct experimental assays to establish their function, although approximately 87% have been detected in high-confidence proteomic experiments (53) (see also the sidebar titled Second Surprise: Lilliput Genes). As such, the determination of protein-coding potential still requires identifying signals of purifying selection on the coding sequence of protein-coding genes (92). While this is partly due to the lack of available primary data—for example, from embryonic or developmental tissue, or subregions of organs such as the brain—other approaches are needed to validate at least the protein-coding potential of a locus, if not its function. One such approach is to raise antibodies against all putative protein-coding loci and use them to detect proteins in a variety of assays, including western blots and immunohistochemistry (153). The latter can be useful in giving hints to function via determination of tissue and subcellular localization. Furthermore, the generation of the antibody itself creates a reagent that can be used in other assays, such as coimmunoprecipitation to identify protein–protein interactions. Similarly, large-scale testing of protein–protein interactions via assays, such as yeast two-hybrid systems, can also provide additional validation for the functional potential of a coding locus.
Transcript annotation
Virtually all protein-coding gene loci are alternatively spliced, meaning that they are transcribed into a variety of transcripts that each include only a subset of the ex-ons at their locus (64, 106). There is frequently disagreement on whether some or all transcript isoforms of a locus are biologically relevant (17) or only one is important (152). One view is that almost all alternative splicing is created by stochastic events during transcription and splicing, creating biologically inert transcripts that could be considered noise (96). Relatively few alternatively spliced transcripts have been functionally characterized. Historically, several approaches have been used to quantify the expression levels of transcripts within a gene, including reverse transcription PCR, ESTs, and microarrays, but RNA-seq has much greater throughput than reverse transcription PCR and ESTs and outperforms microarrays in its throughput, sensitivity, identification of DNA variation, dynamic range, and lack of reliance on existing annotation (174). However, long transcriptomic data may now be used for quantification (149, 169) and may overtake RNA-seq in quantification for the same reasons of length and connectivity that will enable it to do so in transcript annotation.
RNA-seq quantification of individual transcripts suggests that some are persistently more highly expressed than others. However, function has been demonstrated in alternatively spliced transcripts that have long been dismissed as noise, such as isoforms that retain intronic sequence even in their mature forms and those predicted to be targeted by nonsense-mediated decay (NMD) (see the sidebar titled Fifth Surprise: Nonsense-Mediated Decay—Transcription’s Autocorrect). It must be acknowledged that we currently lack the biological understanding to accurately determine whether an individual transcript is functional and, if so, what its function is. However, in the absence of experimental characterization, features such as evolutionary conservation and a high expression level of alternatively spliced transcripts can be strong indicators of functional potential, and a lack of conservation and low expression suggest the opposite. However, while transcripts that do not display these features may be functional either by encoding an alternative protein or by having a regulatory effect, some transcripts may not be functionally important in their own right or even in the context of contributing to or buffering the overall transcriptional output of the gene. Annotation should accept this and seek to add information regarding function and proxies to function (both positive and negative) to transcripts as it emerges.
Read-through genes
Read-through or chimeric transcripts share exonic overlap with two or more loci on the same strand (56). These transcripts were first identified in INSDC data, but the increased sequencing depths of second- and third-generation sequencing technologies (102, 124) may make them more readily identifiable, particularly where genes lie close to one another on the same strand. While some read-through transcripts contain ORFs that span the coding sequence of all the loci they overlap, many others do not and are predicted to be subject to NMD. Read-through transcripts tend to be weakly expressed relative to the protein-coding loci they connect, and although they are clearly not technical sequencing artifacts, it remains unclear what functional role they play (if any) in either increasing protein diversity or regulating the expression of the loci they overlap.
Defining long noncoding genes
One of the difficulties for the annotation or description of genomic features in a world where long and deep transcriptomic data sets are readily available is the increase in the identification of novel transcripts that connect previously independent gene loci. For protein-coding genes where the functional region (the coding sequence) is readily identifiable, this presents less of a problem and can be mitigated by the identification and tagging of transcripts that read through between more than one locus. For long noncoding RNA genes, it is much more of a problem; their functionality is less well understood, both in general terms and regarding which parts of these transcripts are functional effectors. Thus, if novel transcripts connect two annotated loci, it is hard to determine whether the true locus was originally fragmented or whether merging them together is wrong. Incorrectly merging annotations has downstream ramifications for analyses such as locus-level expression quantification. This problem is also compounded in lncRNAs by their generally low and tissue-specific expression, which means that it is often difficult to use orthogonal data such as expression levels derived from RNA-seq to join or break apart loci.
The Genome as a Vehicle of Genetics
From initial surveys of polymorphisms across the general population, targeted projects are now attempting to annotate the functional relevance of variants, especially in a medical context. Thus, large patient cohorts were consented for research by projects such as the International Cancer Genome Consortium (74), the Cancer Genome Atlas (104), Pan-Cancer Analysis of Whole Genomes (22), and the UK Biobank (4). In some cases, this sample collection is integrated into patient care strategies, as in Genomics England and other national initiatives (138). These studies can be analyzed via an array of approaches, ranging from genome-wide association studies, as stored in the NHGRI-EBI GWAS Catalog (19) for common diseases, to individual and familial case studies for rare variants, such as Online Mendelian Inheritance in Man (OMIM) (5), ClinVar (89), ClinGen (35), Orphanet (115), and Deciphering Developmental Disorders (33).
When scaling up to cohorts of millions of patients, it becomes increasingly important to eliminate even occasional artifacts to reduce false positive discoveries. For example, one avoidable source of bias occurs when mapping short sequencing reads to the haploid reference genome. Indeed, reads with the alternate allele of a variant map fractionally less often than reads of the same genomic location with the reference allele. To eliminate this bias, new bioinformatic tools use graph structures to map these short reads to an augmented graph genome that contains the reference as well as all known variants (114). It is likely that, in the future, the human reference genome will be such a graph genome.
Storing the genome as a graph would also cleanly resolve the issue of annotating the segments of immunoglobulin and T cell receptor genes, which is problematic even for the International Immunogenetics specialist reference database (91). These loci are brought together during V(D)J recombination in developing lymphocytes during B and T cell maturation. As a result of this combinatorial operation, there is significant structural variation among the individual lymphocytes within an individual, and it is therefore difficult to provide a meaningful consensus annotation of that region.
The Genome as a Functional Molecule
The last frontier of genomic annotation remains the gene regulatory system, as this system is necessary for the expression of a gene and could even be included in the definition of the gene (54). When considered as a molecule, the genome has many dynamic yet reproducible characteristics that can be assayed (50). From the larger to the smaller scale, it is possible to measure, for example, chromatin loops, chromatin accessibility, histone marks, transcription factor binding, shape, and DNA modifications.As with gene expression,detecting patterns requires assaying these properties across a large number of tissues, cell types, and conditions; hence, large consortia such as the Roadmap Epigenomics Mapping Consortium (14), ENCODE (41), and BLUEPRINT (2), brought together with others under the umbrella of the International Human Epigenome Consortium (142), are currently collecting a substantial array of tissue- or cell type–specific measurements. This multiomic approach to functional genomics was dubbed epigenomics (not to be confused with the already overloaded term of epigenetics). In parallel, several assays can test for regulatory effects using either natural (108) or engineered (117) sequence variation.
Despite the plethora of assays and measurements, converting the classical definition of enhancers into genomic or epigenomic terms is still a matter of intense discussion (52, 83, 118, 136, 159), as no strong distinguishing pattern emerges: Their positions relative to genes are highly variable, their sequences are extremely diverse, their activity in the cell is transient, they are weakly evolutionarily conserved, and their mechanisms of action are not fully understood. Even when a regulatory effect is measured, there is no consensus as to where exactly a regulatory element starts and ends along the genome. For this reason, currently available genome-wide annotations (6, 173). rely largely on indirect evidence of regulatory activity, although direct validation can be performed on selected sites (158). Recent experimental technologies suggest that it may soon be possible to measure regulatory effects on a large scale across the entire genome and across cell types (51, 52, 73, 156), shedding new light on the nature of regulatory elements.
The Genome as a Frame of Reference for Scientific Communication
In effect,the human genome reference sequence is now more than a molecular measurement; itisa frame of reference that the biomedical community uses to connect its knowledge. For example, the HGNC gene symbols are used consistently from basic research to patient genetic reports. From an explosion of independent resources after the initial release of the human genome sequence, we are now observing a consolidation and standardization of the field, such that these resources will gradually form a consistent annotation of the sequence. After years of parallel work, the teams behind RefSeq and Ensembl/GENCODE are now cooperating within the Matched Annotation from NCBI and EMBL-EBI (MANE) project (43) to facilitate mappings from one system to the other. Similarly, Ensembl/GENCODE is collaborating with UniProt on the Gene Integration with Function,Taxonomy,and Sequence (GIFTS) resource (42),and UniProtis collaborating with the Protein Data Bank in Europe (PDBe) on the Structure Integration with Function, Taxonomy, and Sequence (SIFTS) resource (32).
Genomic variants,however,are currently referenced using multiple nomenclatures,which have positives and negatives. As early as 1993, a standard gene-based nomenclature was proposed (11) that would later become the Human Genome Variation Society notation. This approach, however, produces several ambiguous edge cases that hamper exact determination (68).
As the impact of genomics, and biology in general, has expanded to social and economic matters, greater attention has been paid to estimating and mitigating the consequences of sharing annotations. Whereas the academic field generally subscribes to open science to accelerate discovery—for example, in the Fort Lauderdale statement (165)—private companies and lawmakers have tended to prioritize data protection, for the sake of intellectual property as well as personal privacy [e.g., the Health Insurance Portability and Accountability Act in the United States (7) and the General Data Protection Regulation in the European Union (121)]. Respectful of the trust of human donors, the scientific community is currently developing secure methods to exchange knowledge and data without compromising individuals’ ethical and legal rights. Thus, the Global Alliance for Genomics and Health (16) is implementing software solutions such that data do not need to be copied across servers, let alone across territorial boundaries. Instead, computational analysis tasks will be distributed across data centers. Depending on the contractual and legal context, each analysis returns only summary statistics (which are not patient identifiable) or employs adequate encryption. To ensure the usefulness of this infrastructure work, efforts are ongoing to standardize the content available, for example, with respect to data quality or access rights.
Conclusion
Nearly 20 years since its first public draft release, the annotated human genome sequence has reached adulthood and has become a mature reference that the scientific community, in both academia and industry, relies on intensively. In its initial years, many definitions had to be set, refined, and tested, and subsequent iterations led to highly variable annotations. However, under the stewardship of multiple organizations, it is gradually reaching stability, and it now offers a framework to support the consolidation of knowledge around gene sequences, gene regulatory networks, variants, population structure, and evolution.
Nevertheless, the genome sequence is far from retirement, as many of the novel uncharted aspects are regularly brought to light through better experimentation. While the list of protein-coding genes is converging to a fixed set, the definition of noncoding genes has yet to be settled. Regulatory elements and their interactions with genes are even more elusive. Genetic variation across the world’s population is not represented by today’s reference assembly, and the next major release will probably encode a collection of haplotypes. Finally, the human genome reference annotation community is now accountable for its ethical, legal, and societal impact on the world and is taking concrete steps to ensure that everyone benefits from the spectacular advances in the field.
First Surprise: How Many Protein-Coding Genes are there in the Human Genome?
Estimates of the number of human protein-coding genes made around the initiation of the Human Genome Project were as high as 100,000, although the numbers fell quite rapidly over the following decade and a half to reach equilibrium at around 20,000 (26, 46, 77), a figure that is frequently stated to this day. More recently, even lower counts of approximately 19,000 have been predicted (44), and it is this figure that more closely refects the number of functional protein-coding genes in the reference genome sequence found in the RefSeq and Ensembl/GENCODE catalogs, although other resources, such as CHESS, predict more (120). It might also surprise those making some of the higher predictions that the number of protein-coding genes in the human reference genome sequence is approximately 2,500 lower than the equivalent number for the mouse reference genome sequence produced by identical manual annotation workflows. However, despite the aspirations of the Human Genome Project and con-fdent predictions that a final protein-coding gene count is close, we do not yet have the definitive number (see Figure 4).
Second Surprise: Lilliput Genes.
One class of loci that suggests our current coding catalogs may be more incomplete than we believe are the small ORFs (smORFs), which are very short proteins that can still play an important functional role at the level of the cell or organism. While reference protein-coding gene catalogs do contain smORFs, including STRIT1 (107), those that have been found either show very strong signals of conservation or purifying selection or have been comprehensively biochemically characterized, and there remains the possibility that (potentially a very large number of) smORFs with weaker signals of conservation remain unannotated.
One feature that is frequently described as a subclass of smORFs is upstream ORFs (uORFs), which lie in the 5′ untranslated regions of protein-coding loci. uORFs regulate the ability of the translating ribosome to access the translation initiation site of the primary coding sequence of a transcript (100). Again, there has been little functional characterization of uORFs, but where such data exist, they generally support the role of the uORFs as purely regulatory with the proteins they encode, although there are exceptions (137). Regulatory uORFs are not currently included in reference annotation sets even where they have been characterized.
Third Surprise: Win Some, Lose Some.
In a few rare cases, pseudogenes can evolve novel functionality. For example, transcripts originating from the PTENP1 pseudogene of the functional human PTEN locus function as microRNA decoys, regulating the binding of microRNAs to the 3′ untranslated region of the parent locus (123). A prerequisite for novel gain of function is that a pseudogene be transcribed, and although there is an increasing volume of evidence for the transcription of both duplicated and retrotransposed pseudogenes, there is very little experimental evidence that gain of novel function can occur at the transcript level.
Fourth Surprise: Coding Noncoding RNA?
By definition, lncRNAs do not encode functional proteins; however, their translation is more complex than this fact suggests. In experiments to identify evidence for translation, such as proteomics (mass spectrometry) and ribo-seq, signals of translation are frequently detected (71, 85, 166). Transcripts from lncRNAs clearly come into contact with ribosomes, and indeed, lncRNA loci have been demonstrated to be regulated via the act of translation and the NMD process that requires translation (147). Similarly, computational methods to identify regions of lncRNAs with protein-coding potential (e.g., from cross-species conservation) also find evidence that some lncRNAs could encode proteins (162). However, when subject to expert review, lncRNAs with a signal of protein-coding potential almost always fail to reach the standards required for reference annotation sets, with the signals explicable as technical or biological noise (168). Where a transcript or locus annotated as a lncRNA does pass the threshold for annotation as encodinga functional protein,both the transcript and locus are switchedinthe protein-coding biotype—that is, they are changed from being considered lncRNAs to being considered protein-coding genes, and as such, there is currently no recognition of protein-coding lncRNA genes. This may be challenged by the future discovery of protein-coding transcripts that possess demonstrable function at the transcript level. While the literature seems to suggest the existence of such loci, so far no examples have passed the threshold for inclusion in reference annotation sets.
Fifth Surprise: Nonsense-Mediated Decay—Transcription’s Autocorrect.
The NMD cellular mRNA surveillance pathway detects mRNAs with premature termination codons (PTCs) and promotes their degradation. NMD is a highly efficient mechanism for the cell to eliminate mRNAs, thus buffering the biological consequences of irregular splicing. While the precise mechanisms of NMD are complex and not fully understood, a PTC is defined as a stop codon that lies more than 50–55 base pairs upstream of a splice site, with an exon junction complex (EJC) deposited on it during the splicing process. If the EJC is fewer than 50 bases downstream of the PTC, the ribosome will have progressed sufficiently to displace the EJC from the mRNA, and NMD will not be initiated unless additional EJCs lie further downstream (103).
The function of NMD in degrading PTC-containing transcripts that are potentially damaging to the cell is well established, but a role in the active posttranscriptional regulation of genes has recently been identified. The first cases to be identified were genes encoding splicing factors of the SRSF family involved in the regulation of alternative splicing (90). These splicing factors were shown to autoregulate by directing their own splicing to produce SRSF2 transcripts sensitive to NMD when SRSF2 concentration is elevated. The functional link between alternative splicing and NMD provides a mechanism for fne-tuning gene expression, and this mechanism has been described as regulated unproductive splicing and translation (RUST) and alternative splicing coupled NMD (AS-NMD). Variants that affect these mechanisms have been shown to cause disease; for example, a variant in the PTC-containing exon of an NMD transcript in SNRPB was shown to dysregulate the splicing of the exon, causing cerebro-costo-mandibular syndrome (93), while variants promoting the inclusion of a poison exon in SCN1A cause Dravet syndrome (23).
Acknowledgements
We thank Eloise Stapleton and Emily Steed for their careful proofreading. This work was funded by the Wellcome Trust (WT108749/Z/15/Z), the National Human Genome Research Institute (2U41HG007234), and the European Molecular Biology Laboratory.
Footnotes
Disclosure Statement
P.F. is a member of the scientific advisory boards of Fabric Genomics Inc. and Eagle Genomics Ltd.
Contributor Information
Daniel R. Zerbino, Email: zerbino@ebi.ac.uk.
Adam Frankish, Email: frankish@ebi.ac.uk.
Paul Flicek, Email: ficek@ebi.ac.uk.
Literature Cited
- 1.1000 Genomes Proj. Consort. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams D, Altucci L, Antonarakis SE, Ballesteros J, Beck S, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nature. 2012;30:224–26. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- 3.Alexandersson M, Cawley S, Pachter L. SLAM: cross-species gene finding and alignment with a generalized pair hidden Markov model. Genome Res. 2003;13:496–502. doi: 10.1101/gr.424203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen NE, Sudlow C, Peakman T, Collins R. UK Biobank data: come and get it. Sci Transl Med. 2014;6:224ed4. doi: 10.1126/scitranslmed.3008601. [DOI] [PubMed] [Google Scholar]
- 5.Amberger JS, Bocchini CA, Scott AF, Hamosh A. OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019;47:D1038–43. doi: 10.1093/nar/gky1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–61. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Annas GJ. HIPAA regulations—a new era of medical-record privacy? N Engl J Med. 2003;348:1486–90. doi: 10.1056/NEJMlim035027. [DOI] [PubMed] [Google Scholar]
- 8.Avery OT, MacLeod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med. 1944;79:137–58. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banerji J, Olson L, Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33:729–40. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- 10.Barra V, Fachinetti D. The dark side of centromeres: types, causes and consequences of structural abnormalities implicating centromeric DNA. Nat Commun. 2018;9 doi: 10.1038/s41467-018-06545-y. 4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beaudet AL, Tsui L-C. A suggested nomenclature for designating mutations. Hum Mut. 1993;2:245–48. doi: 10.1002/humu.1380020402. [DOI] [PubMed] [Google Scholar]
- 12.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–38. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 13.Benoist C, Chambon P. In vivo sequence requirements of the SV40 early promoter region. Nature. 1981;290:304–10. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–48. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Jr., Brice MD, et al. The Protein Data Bank: a computer-based archival file for macromolecular structures. Eur J Biochem. 1977;80:319–24. doi: 10.1111/j.1432-1033.1977.tb11885.x. [DOI] [PubMed] [Google Scholar]
- 16.Birney E, Vamathevan J, Goodhand P. Genomics in healthcare: GA4GH looks to 2022. 2017 doi: 10.1101/203554. bioRxiv 203554. [DOI] [Google Scholar]
- 17.Blencowe BJ. The relationship between alternative splicing and proteomic complexity. Trends Biochem Sci. 2017;42:407–8. doi: 10.1016/j.tibs.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Braschi B, Denny P, Gray K, Jones T, Seal R, et al. Genenames.org: the HGNC and VGNC resources in 2019. Nucleic Acids Res. 2019;47:D786–92. doi: 10.1093/nar/gky930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 21.Campbell MS, Holt C, Moore B, Yandell M. Genome annotation and curation using MAKER and MAKER-P. Curr Protoc Bioinform. 2014;48:4.11.1–39. doi: 10.1002/0471250953.bi0411s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Res. Netw. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carvill GL, Engel KL, Ramamurthy A, Cochran JN, Roovers J, et al. Aberrant inclusion of a poison exon causes Dravet syndrome and related SCN1A-associated genetic epilepsies. Am J Hum Genet. 2018;103:1022–29. doi: 10.1016/j.ajhg.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Cunningham F, Rios D, McLaren WM, Smith J, et al. Ensembl variation resources. BMC Genom. 2010;11:293. doi: 10.1186/1471-2164-11-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Church DM, Schneider VA, Graves T, Auger K, Cunningham F, et al. Modernizing reference genome assemblies. PLOS Biol. 2011;9:e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clamp M, Fry B, Kamal M, Xie X, Cuff J, et al. Distinguishing protein-coding and noncoding genes in the human genome. PNAS. 2007;104:19428–33. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark BF. The crystal structure of tRNA. J Biosci. 2006;31:453–57. doi: 10.1007/BF02705184. [DOI] [PubMed] [Google Scholar]
- 28.Collins FS, Morgan M, Patrinos A. The Human Genome Project: lessons from large-scale biology. Science. 2003;300:286–90. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- 29.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crick FH. On protein synthesis. Symp Soc Exp Biol. 1958;12:138–63. [PubMed] [Google Scholar]
- 31.Dahm R. Friedrich Miescher and the discovery of DNA. Dev Biol. 2005;278:274–88. doi: 10.1016/j.ydbio.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 32.Dana JM, Gutmanas A, Tyagi N, Qi G, O’Donovan C, et al. SIFTS: updated Structure Integration with Function, Taxonomy and Sequences resource allows 40-fold increase in coverage of structure-based annotations for proteins. Nucleic Acids Res. 2019;47:D482–489. doi: 10.1093/nar/gky1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deciphering Dev. Disord. Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–38. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dolman L, Page A, Babb L, Freimuth RR, Arachchi H, et al. ClinGen advancing genomic datasharing standards as a GA4GH driver project. Hum Mut. 2018;39:1686–89. doi: 10.1002/humu.23625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doucet AJ, Droc G, Siol O, Audoux J, Gilbert N. U6 snRNA pseudogenes: markers of retrotrans-position dynamics in mammals. Mol Biol Evol. 2015;32:1815–32. doi: 10.1093/molbev/msv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–55. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- 38.Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919–29. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 39.Eid J, Fehr A, Gray J, Luong K, Lyle J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;23:133–38. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 40.ENCODE Proj. Consort. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.ENCODE Proj. Consort. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eur. Bioinform. Inst. GIFTs. European Bioinformatics Institute. 2020 https://www.ebi.ac.uk/gifts.
- 43.Eur. Bioinform. Inst. MANE (Matched Annotation between NCBI and EBI) European Bioinfor-matics Institute. 2020 https://www.ensembl.org/info/genome/genebuild/mane.html.
- 44.Ezkurdia I, Juan D, Rodriguez JM, Frankish A, Diekhans M, et al. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum Mol Genet. 2014;23:5866–78. doi: 10.1093/hmg/ddu309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Felsenfeld G. A brief history of epigenetics. Cold Spring Harb Perspect Biol. 2014;6:a018200. doi: 10.1101/cshperspect.a018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fields C, Adams MD, White O, Venter JC. How many genes in the human genome? Nat Genet. 1994;7:345–46. doi: 10.1038/ng0794-345. [DOI] [PubMed] [Google Scholar]
- 47.Fiers W, Contreras R, Duerinck F, Haegeman G, Iserentant D, et al. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976;260:500–7. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- 48.Flicek P, Keibler E, Hu P, Korf I, Brent MR. Leveraging the mouse genome for gene prediction in human: from whole-genome shotgun reads to a global synteny map. Genome Res. 2003;13:46–54. doi: 10.1101/gr.830003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–73. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furey TS. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat Rev Genet. 2012;13:840–52. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gasperini M, Andrew J, Hill AJ, McFaline-Figueroa JL, Martin B, et al. crisprQTL mapping as a genome-wide association framework for cellular genetic screens. 2018 doi: 10.1101/314344. bioRxiv 314344. [DOI] [Google Scholar]
- 52.Gasperini M, Tome JM, Shendure J. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nat Rev Genet. 2020;21:292–310. doi: 10.1038/s41576-019-0209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaudet P, Michel PA, Zahn-Zabal M, Britan A, Cusin I, et al. The neXtProt knowledgebase on human proteins: 2017 update. Nucleic Acids Res. 2017;45:D177–82. doi: 10.1093/nar/gkw1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, et al. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17:669–81. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- 55.Gillies SD, Morrison SL, Oi VT, Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983;33:717–28. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- 56.Gingeras TR. Implications of chimaeric non-co-linear transcripts. Nature. 2009;461:206–11. doi: 10.1038/nature08452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gjerstorff MF, Andersen MH, Ditzel HJ. Oncogenic cancer/testis antigens: prime candidates for immunotherapy. Oncotarget. 2015;6:15772–87. doi: 10.18632/oncotarget.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, et al. Life with 6000 genes. Science. 1996;274:546–67. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 59.Gruss P, Dhar R, Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. PNAS. 1981;78:943–47. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guigó R, Flicek P, Abril JF, Reymond A, Lagarde J, et al. EGASP: the human ENCODE Genome Annotation Assessment Project. Genome Biol. 2006;7(Suppl. 1):S2.1–31. doi: 10.1186/gb-2006-7-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gurdon JB. Nuclear transplantation and the control of gene activity in animal development. Proc R Soc Lond B. 1970;176:303–14. doi: 10.1098/rspb.1970.0050. [DOI] [PubMed] [Google Scholar]
- 62.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haeussler M, Zweig AS, Tyner C, Speir ML, Rosenbloom KR, et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019;47:D853–58. doi: 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, et al. GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hershey AD, Chase M. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J Gen Physiol. 1952;36:39–56. doi: 10.1085/jgp.36.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoff KJ, Stanke M. Predicting genes in single genomes with AUGUSTUS. Curr Protoc Bioinform. 2019;65:e57. doi: 10.1002/cpbi.57. [DOI] [PubMed] [Google Scholar]
- 67.Holliday R. Epigenetics: a historical overview. Epigenetics. 2006;1:76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]
- 68.Holmes JB, Moyer E, Phan L, Maglott D, Kattman BL. SPDI: data model for variants and applications at NCBI. 2019 doi: 10.1101/537449. bioRxiv 537449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Horsthemke B. A critical view on transgenerational epigenetic inheritance in humans. Nat Commun. 2018;9 doi: 10.1038/s41467-018-05445-5. 2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–79. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Inoue F, Ahituv N. Decoding enhancers using massively parallel reporter assays. Genomics. 2015;106:159–64. doi: 10.1016/j.ygeno.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Int. Cancer Genome Consort. International network of cancer genome projects. Nature. 2010;464:993–98. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Int. HapMap 3 Consort. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Int. Hum. Genome Seq. Consort. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 77.Int. Hum. Genome Seq. Consort. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 78.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–56. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 79.Johannsen W. Elemente der exakten Erblichkeitslehre. Jena, Ger: Fischer; 1909. [Google Scholar]
- 80.Kaessmann H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010;20:1313–26. doi: 10.1101/gr.101386.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karsch-Mizrachi I, Nakamura Y, Cochrane G, Int. Nucleotide Seq. Database Collab The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2012;40:D33–37. doi: 10.1093/nar/gkr1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Katsila T, Potamias G, Patrinos GP, Swertz MA. A review of tools to automatically infer chromosomal positions from dbSNP and HGVS genetic variants. In: Lambert CG, Baker DJ, Patrinos GP, editors. Human Genome Informatics: Translating Genes into Health. Cambridge, MA: Academic; 2018. pp. 133–56. [Google Scholar]
- 83.Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, et al. Defining functional DNA elements in the human genome. PNAS. 2014;111:6131–38. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim J, Piao HL, Kim BJ, Yao F, Han Z, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50:1705–15. doi: 10.1038/s41588-018-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, et al. A draft map of the human proteome. Nature. 2014;509:575–81. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kozomara A, Birgaoanu M, Griffths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–62. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kurland CG. Molecular characterization of ribonucleic acid from Escherichia coli ribosomes: I. Isolation and molecular weights. J Mol Biol. 1960;2:83–91. [Google Scholar]
- 88.Lagarde J, Uszczynska-Ratajczak B, Carbonell S, Pérez-Lluch S, Abad A, et al. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat Genet. 2017;49:1731–40. doi: 10.1038/ng.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Landrum MJ, Kattman BL. Clin Var at five years: delivering on the promise. Hum Mut. 2018;39:1623–30. doi: 10.1002/humu.23641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–29. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 91.Lefranc MP, Giudicelli V, Duroux P, Jabado-Michaloud J, Folch G, et al. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 2015;43:D413–22. doi: 10.1093/nar/gku1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–82. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lynch DC, Revil T, Schwartzentruber J, Bhoj EJ, Innes AM, et al. Disrupted auto-regulation of the spliceosomal gene SNRPB causes cerebro-costo-mandibular syndrome. Nat Commun. 2014;5 doi: 10.1038/ncomms5483. 4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McClintock B. Induction of instability at selected loci in maize. Genetics. 1953;38:579–99. doi: 10.1093/genetics/38.6.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Melamud E, Moult J. Stochastic noise in splicing machinery. Nucleic Acids Res. 2009;37:4873–86. doi: 10.1093/nar/gkp471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mendel JG. Versuche über Pfanzenhybriden. Verh Naturforsch Ver Brünn. 1866;4:3–47. [Google Scholar]
- 98.Mercola M, Wang XF, Olsen J, Calame K. Transcriptional enhancer elements in the mouse im-munoglobulin heavy chain locus. Science. 1983;221:663–65. doi: 10.1126/science.6306772. [DOI] [PubMed] [Google Scholar]
- 99.Mikheyev AS, Tin MM. A first look at the Oxford Nanopore MinION sequencer. Mol Ecol Resour. 2014;14:1097–102. doi: 10.1111/1755-0998.12324. [DOI] [PubMed] [Google Scholar]
- 100.Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20:8635–42. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Murray JC, Buetow KH, Weber JL, Ludwigsen S, Scherpbier-Heddema T, et al. A comprehensive human linkage map with centimorgan density. Cooperative Human Linkage Center (CHLC) Science. 1994;265:2049–54. doi: 10.1126/science.8091227. [DOI] [PubMed] [Google Scholar]
- 102.Nacu S, Yuan W, Kan Z, Bhatt D, Rivers CS, et al. Deep RNA sequencing analysis of readthrough gene fusions in human prostate adenocarcinoma and reference samples. BMC Med Genom. 2011;4:11. doi: 10.1186/1755-8794-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–99. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 104.Natl. Cancer Inst. The Cancer Genome Atlas Project. National Cancer Institute. 2020 https://www.cancer.gov/tcga. [Google Scholar]
- 105.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–35. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nellore A, Jaffe AE, Fortin JP, Alquicira-Hernández J, Collado-Torres L, et al. Human splicing diversity and the extent of unannotated splice junctions across human RNA-seq samples on the Sequence Read Archive. Genome Biol. 2016;17:266. doi: 10.1186/s13059-016-1118-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–75. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nica AC, Dermitzakis ET. Expression quantitative trait loci: present and future. Philos Trans R Soc Lond. 2013;368 doi: 10.1098/rstb.2012.0362. 20120362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nirenberg M, Leder P, Bernfield M, Brimacombe R, Trupin J, et al. RNA codewords and protein synthesis, VII. On the general nature of the RNA code. PNAS. 1965;53:1161–68. doi: 10.1073/pnas.53.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–45. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Osoegawa K, Mammoser AG, Wu C, Frengen E, Zeng C, et al. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 2001;11:483–96. doi: 10.1101/gr.169601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pace JK, Feschotte C. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–32. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Parra G, Agarwal P, Abril JF, Wiehe T, Fickett JW, Guigó R. Comparative gene prediction in human and mouse. Genome Res. 2003;13:108–17. doi: 10.1101/gr.871403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Paten B, Novak AM, Eizenga JM, Garrison E. Genome graphs and the evolution of genome inference. Genome Res. 2017;27:665–76. doi: 10.1101/gr.214155.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pavan S, Rommel K, Mateo Marquina ME, Höhn S, Lanneau V, et al. Clinical practice guidelines for rare diseases: the Orphanet database. PLOS ONE. 2017;12:e0170365. doi: 10.1371/journal.pone.0170365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pei B1, Sisu C, Frankish A, Howald C, Habegger L, et al. The GENCODE pseudogene resource. Genome Biol. 2012;13:R51. doi: 10.1186/gb-2012-13-9-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 118.Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. Enhancers: five essential questions. Nat Rev Genet. 2015;14:288–95. doi: 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–95. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pertea M, Shumate A, Pertea G, Varabyou A, Brietwieser FP, et al. CHESS: a new human gene catalog curated from thousands of large-scale RNA sequencing experiments reveals extensive transcriptional noise. Genome Biol. 2018;19:208. doi: 10.1186/s13059-018-1590-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Phillips M. International data-sharing norms: from the OECD to the General Data Protection Regulation (GDPR) Hum Genet. 2018;137:575–82. doi: 10.1007/s00439-018-1919-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–78. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 123.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–38. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Prakash T, Sharma VK, Adati N, Ozawa R, Kumar N, et al. Expression of conjoined genes: another mechanism for gene regulation in eukaryotes. PLOS ONE. 2010;5:e13284. doi: 10.1371/journal.pone.0013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, et al. The Consensus Coding Sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–23. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pujar S, O’Leary NA, Farrell CM, Loveland JE, Mudge JM, et al. Consensus Coding Sequence (CCDS) database: a standardized set of human and mouse protein-coding regions supported by expert curation. Nucleic Acids Res. 2018;46:D221–28. doi: 10.1093/nar/gkx1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Reis EM, Nakaya HI, Louro R, Canavez FC, Flatschart AV, et al. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene. 2004;23:6684–92. doi: 10.1038/sj.onc.1207880. [DOI] [PubMed] [Google Scholar]
- 128.Rhoads A, Au KF. PacBio sequencing and its applications. Genom Proteom Bioinform. 2015;13:278–89. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Roll-Hansen N. The holist tradition in twentieth century genetics. Wilhelm Johannsen’s genotype concept. J Physiol. 2014;592:2431–38. doi: 10.1113/jphysiol.2014.272120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. PNAS. 1977;74:5463–67. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen HC, et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017;27:849–64. doi: 10.1101/gr.213611.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shay JW, Wright WE. Telomeres and telomerase: three decades of progress. Nat Rev Genet. 2019;20:299–309. doi: 10.1038/s41576-019-0099-1. [DOI] [PubMed] [Google Scholar]
- 134.Sherry ST, Ward M, Sirotkin K. dbSNP—database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–79. [PubMed] [Google Scholar]
- 135.Söll D, Ohtsuka E, Jones DS, Lohrmann R, Hayatsu H, et al. Studies on polynucleotides, XLIX. Stimulation of the binding of aminoacyl-sRNA’s to ribosomes by ribotrinucleotides and a survey of codon assignments for 20 amino acids. PNAS. 1965;54:1378–85. doi: 10.1073/pnas.54.5.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Spivakov M. Spurious transcription factor binding: non-functional or genetically redundant? BioEssays. 2014;36:798–806. doi: 10.1002/bies.201400036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, et al. Translation from the 5′ untranslated region shapes the integrated stress response. Science. 2016;351 doi: 10.1126/science.aad3867. aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stark Z, Dolman L, Manolio TA, Ozenberger B, Hill SL, et al. Integrating genomics into healthcare: a global responsibility. AmJ Hum Genet. 2019;104:13–20. doi: 10.1016/j.ajhg.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Steijger T, Abril JF, Engström PG, Kokocinski F, RGASP Consort et al. Assessment of transcript reconstruction methods for RNA-seq. Nat Methods. 2013;10:1177–84. doi: 10.1038/nmeth.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Stevens H. Life Out of Sequence: A Data-Driven History of Bioinformatics. Chicago: Univ. Chicago Press; 2013. [Google Scholar]
- 141.Storz G. An expanding universe of noncoding RNAs. Science. 2002;296:1260–63. doi: 10.1126/science.1072249. [DOI] [PubMed] [Google Scholar]
- 142.Stunnenberg HG, Int. Hum. Epigenome Consort. Hirst M. The International Human Epigenome Consortium: a blueprint for scientific collaboration and discovery. Cell. 2016;167:1145–49. doi: 10.1016/j.cell.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 143.Sturtevant H. The linear arrangement of six sex-linked factors in Drosophila as shown by their mode of association. J Exp Zool. 1913;14:43–59. [Google Scholar]
- 144.Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–46. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tanaka TU, Clayton L, Natsume T. Three wise centromere functions: see no error, hear no break, speak no delay. EMBO Rep. 2013;14:1073–83. doi: 10.1038/embor.2013.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tang AD, Soulette CM, van Baren MJ, Hart K, Hrabeta-Robinson E, et al. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. 2018 doi: 10.1101/410183. bioRxiv410183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tani H, Torimura M, Akimitsu N. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLOS ONE. 2013;8:e55684. doi: 10.1371/journal.pone.0055684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tarailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinform. 2009;25:4.10.1–14. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- 149.Tardaguila M, de la Fuente L, Marti C, Pereira C, Pardo-Palacios FJ, et al. SQANTI: extensive characterization of long-read transcript sequences for quality control in full-length transcriptome identification and quantification. Genome Res. 2018;28:396–411. doi: 10.1101/gr.222976.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tilgner H, Jahanbani F, Blauwkamp T, Moshref A, Jaeger E, et al. Comprehensive transcriptome analysis using synthetic long-read sequencing reveals molecular co-association of distant splicing events. Nat Biotechnol. 2015;33:736–42. doi: 10.1038/nbt.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–15. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tress ML, Abascal F, Valencia A. Alternative splicing may not be the key to proteome complexity. Trends Biochem. Sci. 2017;42:98–110. doi: 10.1016/j.tibs.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 154.UniProt Consort. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–15. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.US Dep. Energy. History of the Human Genome Project. Human Genome Project Information Archive: 1990–2003. 2019 https://web.ornl.gov/sci/techresources/HumanGenome/project/hgp.shtml. [Google Scholar]
- 156.van der Wijst MGP, Brugge H, de Vries DH, Deelen P, Swertz MA, et al. Single-cell RNA sequencing identifies cell type-specific cis-eQTLs and co-expression QTLs. Nat Genet. 2018;50:493–97. doi: 10.1038/s41588-018-0089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 158.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Volders PJ, Anckaert J, Verheggen K, Nuytens J, Martens L, et al. LNCipedia 5: towards a reference set of human long non-coding RNAs . Nucleic Acids Res. 2019;47:D135–39. doi: 10.1093/nar/gky1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Waddington CH. Introduction to Modern Genetics. London: Allen & Unwin; 1939. [Google Scholar]
- 162.Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Weirather JL, de Cesare M, Wang Y, Piazza P, Sebastiano V, et al. Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis. F1000Res. 2017;6:100. doi: 10.12688/f1000research.10571.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wellcome Trust. Sharing data from large-scale biological research projects: a system of tripartite responsibility. Rep., Wellcome Trust; London: 2003. https://www.sanger.ac.uk/legal/assets/fortlauderdalereport.pdf. [Google Scholar]
- 166.Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–87. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 167.Wolfsberg TG. Using the NCBI Map Viewer to browse genomic sequence data. Curr Protoc Hum Genet. 2011;69:18.5.1–25. doi: 10.1002/0471142905.hg1805s69. [DOI] [PubMed] [Google Scholar]
- 168.Wright JC, Mudge J, Weisser H, Barzine MP, Gonzalez JM, et al. Improving GENCODE reference gene annotation using a high-stringency proteogenomics workflow. Nat Commun. 2016;7 doi: 10.1038/ncomms11778. 11778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wyman D, Balderrama-Gutierrez G, Reese F, Jiang S, Rahmanian S, et al. A technology-agnostic long-read analysis pipeline for transcriptome discovery and quantification. 2019 doi: 10.1101/672931. bioRxiv 672931. [DOI] [Google Scholar]
- 170.Wyman D, Mortazavi A. TranscriptClean: variant-aware correction of indels, mismatches and splice junctions in long-read transcripts. Bioinformatics. 2019;35:340–42. doi: 10.1093/bioinformatics/bty483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Xue X, Yang YA, Zhang A, Fong KW, Kim J, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35:2746–55. doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yates AD, Achuthan P, Akanni W, Allen J, Allen J, et al. Ensembl 2020. Nucleic Acids Res. 2020;48:D682–88. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zerbino DR, Wilder SP, Johnson N, Juettemann T, Flicek PR. The Ensembl regulatory build. Genome Biol. 2015;16:56. doi: 10.1186/s13059-015-0621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLOS ONE. 2014;9:e78644. doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]