Abstract
Controlled conjugation of fluorescent carbon dots (CDs) with DNA and subsequent fabrication of the CDs into an array through hybridization mediated self-assembly in the solution phase is reported. Covalent conjugation of CD with DNA and the subsequent array formation change the mobility of the CD-DNA array in gel electrophoresis and HPLC significantly. Interspatial distance in the CD-DNA array is tuned by the DNA sequence length and maintained at ~8 ± 0.3 nm as revealed by electron microscopy studies. An increase in fluorescence lifetime by ~2 ns was observed for the CD-DNA array compared to a solitary CD, vis-á-vis better imaging prospects of HEK293 cells by the former. Thus, the array displays improved fluorescence and unhindered cell penetration.
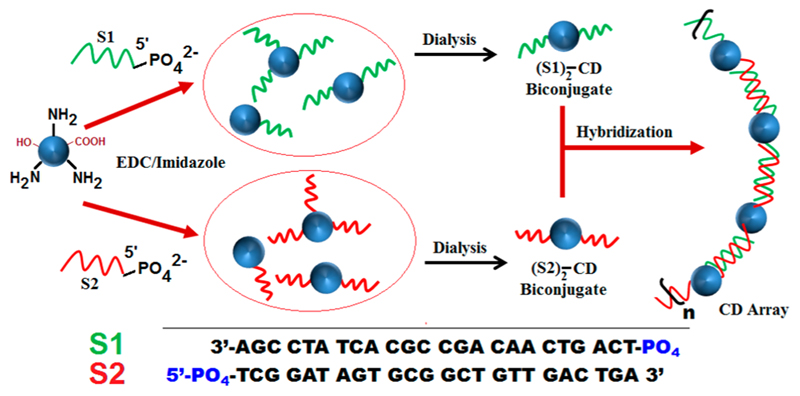
Nanoparticles organized in linear array have numerous interesting applications including energy harvesting, plasmonics, and biosensing.1–3 Metal nanoparticles and quantum dots have been fabricated into arrays mainly by lithographic techniques.4–6 However, so far, very few literature reports have shown the fabrication of arrays from carbon dots (CDs), the latest member of the carbon nanofamily.7–9 The CD arrays were developed through plasma jet, electron beam lithography methods, and others for electrochemical hydrogen generation and potential microarray biosensing. Nonetheless, there is absolutely no report of biomolecule mediated CD array formation. Evidently, there is a lack of general methodology to make CD arrays that may unearth a plethora of interesting applications. For the first time, we report the DNA mediated construction of linear CD array in solution phase based on simple Watson and Crick base pairing that offers precise control over interparticle distance in the array. Reportedly, biomolecules such as DNA and peptide mediated patterning have been applied to fabricate nanoparticles into linear arrays.10–12 In particular, the remarkable base recognition properties of DNA have been taken advantage of to direct selfassembly of nanoparticles.13–15As an obvious consequence, such assemblies that offer temperature dependent tenability are heat labile and reversible.16,17 However, controlled conjugation of nanoparticles to DNA strands presents challenges that are prerequisite for array formation to suppress unwarranted branching, thus reducing fidelity of the bottom-up approach. While conjugation conditions are paramount to achieving the desired nanoparticle–DNA conjugate, wise selection of purification methods takes precedence where controlled reaction conditions often fail to provide the required template comprehensively. Gold nanoparticles, quantum dots (QD), etc., have been conjugated to DNA for nanopatterning by virtue of surface functional groups following simple yet intelligent chemistry and purification technologies.18,19 It remains to be seen whether methods could be engineered to affect patterning of CDs in the nanoscale using DNA as a template. Fluorescent CDs are considered successful substitutes of organic dyes and semiconductor quantum dots due to their easy and inexpensive production, small size (~3–8 nm), high water solubility, surface functionalization, and biocompatibility.20–22 Conse-quently, CDs have found a variety of potential applications in biosensing, imaging technologies, and others.23,24 CDs are being synthesized today from virtually any carbon source and their doping with heteroatoms has resulted in notable optical properties.25,26 Moreover, CDs can be decorated with a variety of functional groups like −COOH, −NH2, and others that provide an excellent opportunity to covalently conjugate them with DNA and peptides.27–29 However, the presence of numerous functional groups on the surface of CDs poses an inevitable challenge as to how to effect a controlled conjugation where one or two single-stranded DNA (ssDNA) or a single peptide could be conjoined to a single CD. Even if such controlled conjugation takes place, isolation and purification of the species for further downstream applications are greatly sought after. Herein, we demonstrate a simple methodology to conjugate amine functionalized CDs with terminal phosphate of ssDNA in a zero linker approach. Reaction conditions of conjugation were carefully optimized to maximize yield of the CD conjoined with two ssDNA of the same sequence. Hybridization-mediated self-assembly of CD-ssDNA conjugate (S1)2-CD and its complementary (S2)2-CD resulted in the creation of a CD array [(S1)2-CD-(S2)2-CD]n (Scheme 1). The distance between adjacent CDs is dictated by the DNA sequence length and hence the number of bases in the DNA. Since the CD array displayed efficient cell penetration and enhanced optical properties, we foresaw potential advantages in its use for imaging studies.
Scheme 1. Scheme for Creation of DNA Templated CD Array through Hybridization.
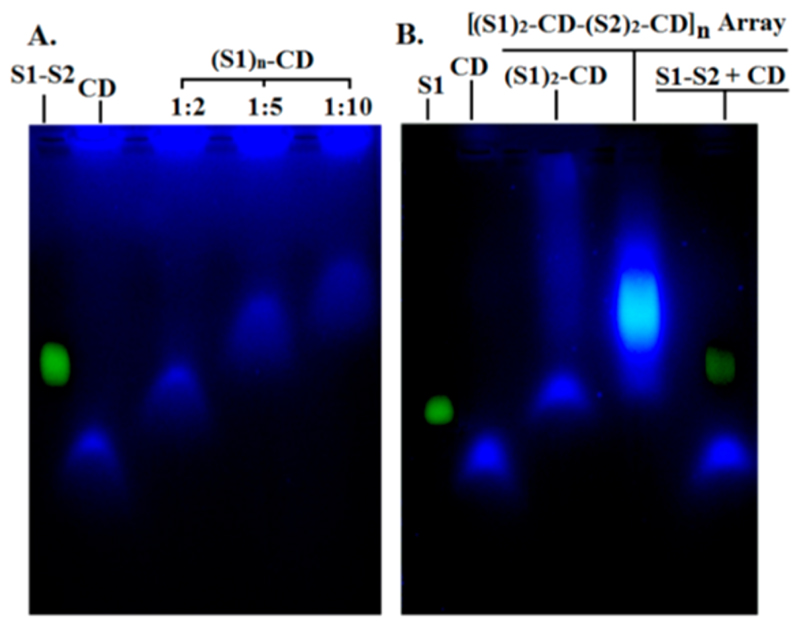
Aqueous soluble CDs were synthesized by low temperature pyrolysis using citric acid and branched polyethylene imine (BPEI) as precursors.30,31 BPEI was used as a capping ligand that presented −NH2 groups as adornment on the CDs having ~4–5 nm diameter. The CDs exhibit a broad band at ~27° having a d-spacing between the (002) planes of 0.3 nm in the powder XRD spectrum indicating weak graphitic crystallinity (Figure S1, SI). Signature FTIR peaks were also observed that confirm the presence of pendant amine functionality on CDs (Figure S2, SI). Estimation of molar concentration of amine groups on the surface of CDs was performed by ninhydrin assay (Figure S3).32 The assay confirms the presence of ~1 nmol −NH2 groups per 1 μg of CD. Considering the abundance of −NH2 groups on CDs and limited yield of the phosphoramidation reaction, a balance was struck to optimize reaction conditions. The ratio of CD to DNA was maintained as 1:2 to minimize multiple conjugation of DNA with the same CD. 5’-PO4 groups of ssDNA (S1) were preactivated with 1-(3-(dimethylamino)propyl)-3-ethylcarbodiimide hydrochloride (eDC·HCI) and 1-methyl imidazole at pH 6 at 37 °C to facilitate coupling with NH2 groups of CDs. A similar reaction was also done with ssDNA (S2) with a separate aliquot of CD. Successful conjugation of S1 and S2 with CDs were confirmed by agarose gel electrophoresis where reduced band mobility with increase in DNA content in the reaction mixture was observed. Evidently, multiple conjugation of DNA strands with several NH2 groups of a single CD reduces gel mobility that justifies the careful optimization of reaction conditions (Figure 1). Crude reaction mixtures of CD-DNA conjugates (1:2) presumably containing (CD-S1)2 and (CD-S2)2 were subjected to multistep dialysis (MWCO-10KDa) to remove the excess of salts, unconjugated DNA, and CDs. The success of biconjugation for the creation of (CD-S1)2 and (CD-S2)2was further confirmed by Reverse Phase-HPLC. A highly reproducible method with Tris.HCl-acetonitrile based buffer in HPLC was developed that enabled the detection of distinct bands corresponding to (S1)2-CD, well separated from other higher-ordered products where multiple (more than two) ssDNA are conjugated to the same CD (Figure S4, SI). Interestingly, this is the first report to demonstrate that HPLC based methods could be useful to characterize and purify DNA-CD conjugates. Purified (CD-S1)2 and (CD-S2)2 were hybridized by heating at 90 °C for 10 min in appropriate buffer solution and gradually cooled to room temperature to obtain CD-DNA array [(S1)2-CD-(S2)2-CD]n in aqueous phase. The annealed CD-DNA conjugates showed limited gel electrophoresis mobility confirming the formation of higher ordered structures. However, the absence of distinct band and smearing of the same indicate wide distribution of products with very high effective molecular weight (Figure 1). Proof of array formation was also obtained indirectly by observing the significant increase in ethidium bromide (EtBr) fluorescence upon hybridization (Figure S5, SI). This confirms the formation of dsDNA and subsequent intercalation of EtBr in the array whose fluorescent intensity in CD solution and in the presence of ssDNA is otherwise subdued.
Figure 1.
A. Agarose gel image showing the mobility of conjugation products of DNA (S1) with CD in various ratios. B. Agarose gel image showing mobility of purified CD array after hybridization.
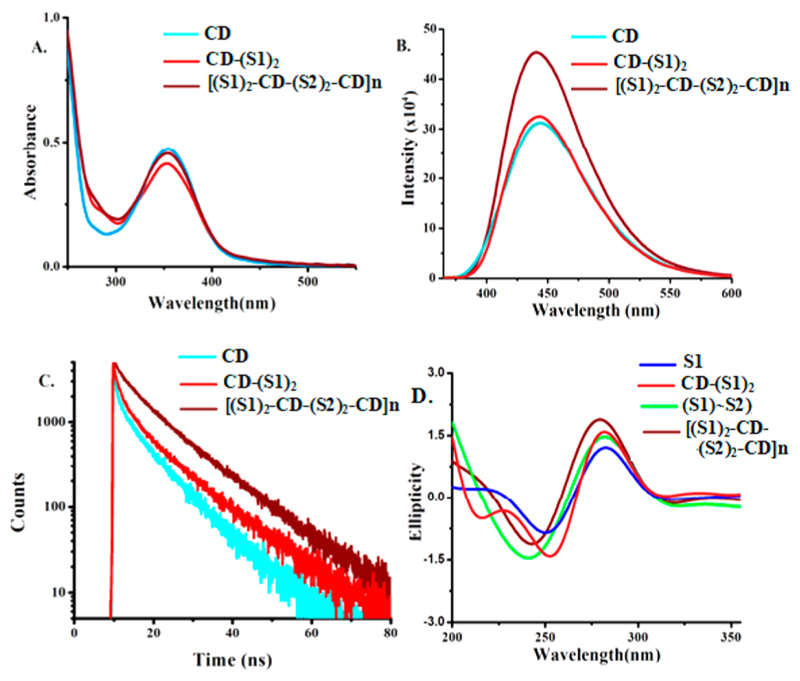
Fluorescence quantum yield of CD and CD-(S1)2 was found to be similar at ~44%, while a slight increase was found for CD array [(S1)2-CD-(S2)2-CD]n at ~48%. Subsequently, optical properties of CDs, CD-DNA conjugates [CD-(S1)2 and CD-(S2)2], and CD array [(S1)2-CD-(S2)2-CD]n were thoroughly investigated. The absorption peak of CD in UV–vis spectra at 350 nm remained consistent for (S1)2-CD, (S2)2-CD, and [(S1)2-CD-(S2)2-CD]n array with the emergence of a shoulder peak at 260 nm due to the presence of DNA (Figure 2A). All the aforementioned samples were excited at 350 nm and emission peaks corresponding to CDs were obtained at 450 nm (Figure 2B) at similar CD concentration. A substantial increase in fluorescence emission intensity was observed for the CD array compared to normal CD solution and CD-DNA conjugates. Time-resolved fluorescence decay was recorded at 400 nm for all the CD, CD-DNA conjugates, and CD-DNA array samples employing time correlated single photon counting (TCSPC) technique with a 375 nm pulsed diode laser (5 MHz) as excitation source (Figure 2C). The excited state lifetime of CD, CD-(S1)2, CD-(S2)2, and [(S1)2-CD-(S2)2-CD]n array were found to be 8.12, 8.13, 8.13, and 10.13 ns, respectively. An increase in lifetime by ~2 ns for [(S1)2-CD-(S2)2-CD]n array is very noteworthy, as it attests to significant fluorescence enhancement. The remarkable fluorescence enhancement is attributed to DNA self-assembly that prevents the CDs from aggregation. Also, steric repulsion between the DNA could play an important role in increasing the colloidal solubility of the CD in the array. In addition to this, Brownian motion of the nanoparticles is also restricted in the assembled CD-DNA array.33,34 Unmistakably, this study has shown that DNA self-assembly could be a reliable source for patterning CDs in aqueous phase that convey added advantages such as improved optical properties. A comparative steady-state spectrum shows negligible loss in fluorescent emission intensity for a period of over eight months for both the CD and CD-DNA array samples stored at 4 °C confirming the solution phase stability of the array (Figure S6, SI).
Figure 2.
A. UV–vis absorption spectra. B. Steady-state fluorescence spectra. C. Time-resolved fluorescence spectra of CD, (S1)2-CD, and [(S1)2-CD-(S2)2-CD]n array. D. Circular dichroism spectra.
S1 and S2 hybridize to form normal B-conformation of DNA that displays a negative peak (~248 nm) and positive peak (~280 nm) in circular dichroism spectra (Figure 2D). The B-conformation and hence Watson and Crick base pairing are also maintained in the [(S1)2-CD-(S2)2-CD]n array. However, the array showed a slight shift in the peak position compared to S1–S2 from 248 to 251 nm for negative peak and 280 to 278 nm for the positive peak with enhanced peak intensity. This is attributed to possible changes in average turn per base resulting from conformational variability upon array formation. Interestingly, loose addition of CD to S1 or S1–S2 results in increase in the intensity of the both negative and positive peaks owing to noncovalent interaction of the DNA bases with the CDs (Figure S7, SI) that results in more compaction of the DNA duplex.35,36 Such interaction was absent in the array, which shows remarkable similarity with S1–S2. This indirectly points toward covalent conjugation of CDs with S1 or S2. Decrease in configurational entropy and increase in sticky end association (additive effect) takes place upon array formation as evident from a slight increase in the DNA melting temperature (T m) (Figure S8, SI). A moderately sharp transition also indicates reduced contribution of terminal dangling ends in the array.37While measurement of chirality and melting transition are indirect proof of array formation, they provide important information regarding the conformational and configuration attributes of the self-assembly of CD-(S1)2 and CD-(S2)2 into the array.
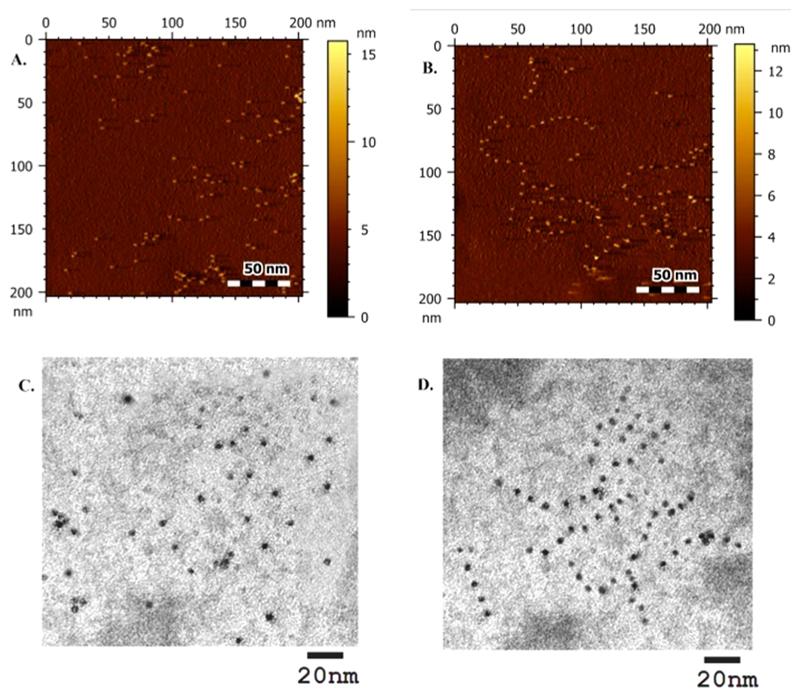
Drop-cast samples of CD and [(S1)2-CD-(S2)2-CD]n array on APS mica were imaged with atomic force microscopy (AFM). CDs displayed narrow size distribution with average size (diameter) of about 4–5 nm (Figure 3A). Arrays are clearly visible with AFM images with definite interparticle distance between CDs maintained at ~8 ± 0.3 nm (Figure 3B and Figure S9, SI). This distance is characteristic of the length of the 24-bases-long S1 and S2 DNA sequences used for array formation (24 × 0.34 ~8.2 nm). Duplex flexibility, contractions in DNA strands and surface van der Waals forces may be the reason for minor nonuniformity of the distance between CDs that affects sample preparation in both AFM and HRTEM. HRTEM correlates well with powder XRD data and showed lattice spacing of CDs as 0.28 nm. Average particle size of CD from HRTEM was found to be similar to AFM data as ~4–5 nm (Figure 3C and Figure S10, SI). The images for [(S1)2-CD-(S2)2-CD]n clearly show the formation of CD arrays (Figure 3D). The distance between two consecutive CDs in the array was estimated to be ~8–8.3 nm in HRTEM (Figure S11, SI). Hybridization defects and different angles of conjugation of the ssDNA to CDs impose conformational strain in the array limiting the length of the array. Nevertheless, electron microscopy images from both AFM and TEM display consistent inter CD distance dictated by the DNA sequence length. Evidently, this opens avenues to tune the array for desired interparticle distance using DNA.
Figure 3.
A and B. AFM image of CD and [(S1)2-CD-(S2)2-CD]n array. C and D. TEM images of CD and [(S1)2-CD-(S2)2-CD]n array.
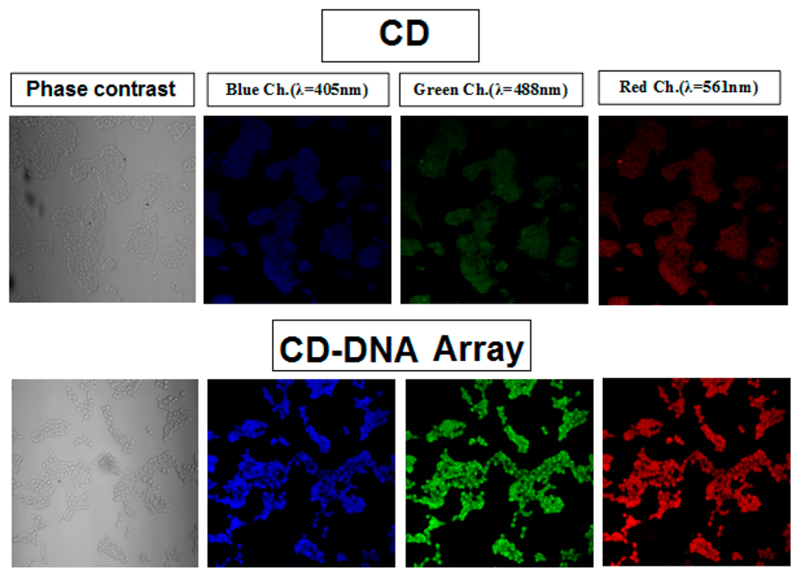
The inherent fluorescence, resistance to photobleaching, high aqueous solubility, improved biocompatibility, and low cytotoxicity of CDs make them valuable candidates for bioimaging applications.38,39 In order to determine whether CDs conjugated to ssDNA in array possessed superior fluorescent properties, they were introduced into HEK293 cells, a common cell line widely used for fluorescence-based imaging and biochemistry. Confocal microscopy revealed improved fluorescence in cells incubated with the CD-arrays compared to the control CDs (Figure 4 and S12, SI), without any increase in toxicity (Figure S13, SI). Thus, the CD-Array functions as an excellent imaging probe with potential to be used for diagnostic applications.
Figure 4.
Confocal images of HEK293 cells incubated with CD and CD-Array.
This study provides a convenient methodology for patterning of CDs where choice of the DNA sequence dictates the selfassembly outcome. Apart from precise control over the inter CD distance in the array, the nanosystem is thermally reversible with accurate knowledge of the dis-assembly temperature. Moreover, one big advantage of using DNA is that, potentially, two different CD/Nanoparticles (having emission in different wavelength) can be patterned in the array in alternate fashion, upon hybridization of two complementary DNA-CD conjugates. This is the first report that confirms controlled DNA functionalization of CDs for fabrication of higher ordered nanostructures potentially serviceable in the field of theranostics, bioimaging, and smart materials, and we forecast an optimistic start to other futuristic applications.
Supplementary Material
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconj-chem.8b00173.
Details of experimental section: Materials and methods, Characterizations, Cell Imaging, MTT assay (PDF)
Acknowledgments
This work was partly supported by a grant to P. D. from Dept. of Biotechnology (DBT), Govt. of India (Project No. BT/PR3444/NNT/28/560/2011). S.K. is thankful to DBT for fellowship. The authors are thankful to IIT Patna, NCCS Pune and SAIF-IIT Bombay for infrastructure and experimental facilities.
Footnotes
Notes
The authors declare no competing financial interest.
References
- (1).Choueiri RM, Galati HT, Klinkova A, Larin EM, Fernandez AQ, Han L, Xin HL, Gang O, Zhulina EB, Rubinstein M, Kumacheva E. Surface patterning of nanoparticles with polymer patches. Nature. 2016;538:79–83. doi: 10.1038/nature19089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Myers BD, Lin QY, Wu H, Luijten E, Mirkin CA, Dravid VP. Size-selective nanoparticle assembly on substrates by DNA density patterning. ACS Nano. 2016;10:5679–86. doi: 10.1021/acsnano.6b02246. [DOI] [PubMed] [Google Scholar]
- (3).Edel JB, Kornyshev AA, Kucernak AR, Urbakh M. Fundamentals and applications of self-assembled plasmonic nanoparticles at interfaces. Chem Soc Rev. 2016;45:1581–96. doi: 10.1039/c5cs00576k. [DOI] [PubMed] [Google Scholar]
- (4).Huang D, Freeley M, Palma M. DNA-mediated patterning of single quantum dot nanoarrays: A reusable platform for single-molecule control. Sci Rep. 2017;7 doi: 10.1038/srep45591. 45591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Nandwana V, Subramani C, Yang YB, Dickert S, Barnes MD, Tuominen MT, Rotello VM. Direct patterning of quantum dot nanostructures via electron beam lithography. J Mater Chem. 2011;21:16859–62. [Google Scholar]
- (6).Palankar R, Medvedev N, Rong A, Delcea M. Fabrication of quantum dot microarrays using electron beam lithography for applications in analyte sensing and cellular dynamics. ACS Nano. 2013;7:4617–28. doi: 10.1021/nn401424y. [DOI] [PubMed] [Google Scholar]
- (7).Camilli L, Jorgensen JH, Tersoff J, Stoot AC, Balog R, Cassidy A, Hornekær L. Self-assembly of ordered graphene nanodot arrays. Nat Commun. 2017;8 doi: 10.1038/s41467-017-00042-4. 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Zhang X, Wang F, Huang H, Li H, Han X, Liu Y, Kang Z. Carbon quantum dot sensitized TiO2 nanotube arrays for photoelectrochemical hydrogen generation under visible light. Nanoscale. 2013;5:2274–78. doi: 10.1039/c3nr34142a. [DOI] [PubMed] [Google Scholar]
- (9).Abuzairi T, Okada M, Mochizuki Y, Poespawati NR, Purnamaningsih RW, Nagatsu M. Maskless functionalization of a carbon nanotube dot array biosensor using an ultrafine atmospheric pressure plasma jet. Carbon. 2015;89:208–16. [Google Scholar]
- (10).Carter JD, LaBean TH. Organization of inorganic nanomaterials via programmable DNA self-assembly and peptide molecular recognition. ACS Nano. 2011;5:2200–05. doi: 10.1021/nn1033983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Si S, Mandal TK. pH-Controlled reversible assembly of peptide-functionalized gold nanoparticles. Langmuir. 2007;23:190–95. doi: 10.1021/la061505r. [DOI] [PubMed] [Google Scholar]
- (12).Li H, Park SH, Reif JH, LaBean TH, Yan H. DNA-templated self-assembly of protein and nanoparticle linear arrays. J Am Chem Soc. 2004;126:418–19. doi: 10.1021/ja0383367. [DOI] [PubMed] [Google Scholar]
- (13).Singh S, Kumari R, Chakraborty A, Hussain S, Singh MK, Das P. Melamine–DNA encoded periodicity of quantum dot arrays. J Colloid Interface Sci. 2016;461:45–49. doi: 10.1016/j.jcis.2015.08.064. [DOI] [PubMed] [Google Scholar]
- (14).Samanta A, Medintz IL. Nanoparticles and DNA – a powerful and growing functional combination in bionanotechnology. Nanoscale. 2016;8:9037–95. doi: 10.1039/c5nr08465b. [DOI] [PubMed] [Google Scholar]
- (15).Bui H, Onodera C, Kidwell C, Tan Y, Graugnard E, Kuang W, Lee J, Knowlton WB, Yurke B, Hughes WL. Programmable periodicity of quantum dot arrays with DNA origami nanotubes. Nano Lett. 2010;10:3367–72. doi: 10.1021/nl101079u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Li Z, Zhu Z, Liu W, Zhou Y, Han B, Gao Y, Tang Z. Reversible plasmonic circular dichroism of Au nanorod and DNA assemblies. J Am Chem Soc. 2012;134:3322–25. doi: 10.1021/ja209981n. [DOI] [PubMed] [Google Scholar]
- (17).Yan Y, Chen JIL, Ginger DS. Photoswitchable oligonucleotide-modified gold nanoparticles: Controlling hybridization stringency with photon dose. Nano Lett. 2012;12:2530–36. doi: 10.1021/nl300739n. [DOI] [PubMed] [Google Scholar]
- (18).Wen Y, McLaughlin CK, Lo PK, Yang H, Sleiman HF. Stable gold nanoparticle conjugation to internal DNA positions: facile generation of discrete gold nanoparticle–DNA assemblies. Bioconjugate Chem. 2010;21:1413–16. doi: 10.1021/bc100160k. [DOI] [PubMed] [Google Scholar]
- (19).Coopersmith K, Han H, Maye MM. Stepwise assembly and characterization of DNA linked two-color quantum dot clusters. Langmuir. 2015;31:7463–7471. doi: 10.1021/acs.langmuir.5b01130. [DOI] [PubMed] [Google Scholar]
- (20).Kargbo O, Jin Y, Ding SN. Recent advances in luminescent carbon dots. Curr Anal Chem. 2014;11:4–21. [Google Scholar]
- (21).Ding C, Zhu A, Tian Y. Functional surface engineering of C-dots for fluorescent biosensing and in vivo bioimaging. Acc Chem Res. 2014;47:20–30. doi: 10.1021/ar400023s. [DOI] [PubMed] [Google Scholar]
- (22).Demchenko AP, Dekaliuk MO. The origin of emissive states of carbon nanoparticles derived from ensemble-averaged and single-molecular studies. Nanoscale. 2016;8:14057–69. doi: 10.1039/c6nr02669a. [DOI] [PubMed] [Google Scholar]
- (23).Huang H, Li C, Zhu S, Wang H, Chen C, Wang Z, Bai T, Shi Z, Feng S. Histidine-derived nontoxic nitrogen doped carbon dots for sensing and bioimaging applications. Langmuir. 2014;30:13542–48. doi: 10.1021/la503969z. [DOI] [PubMed] [Google Scholar]
- (24).Guo Y, Zhang L, Cao F, Leng Y. Thermal treatment of hair for the synthesis of sustainable carbon quantum dots and the applications for sensing Hg2+ . Sci Rep. 2016;6 doi: 10.1038/srep35795. 35795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Prasannan A, Imae T. One-pot synthesis of fluorescent carbon dots from orange waste peels. Ind Eng Chem Res. 2013;52:15673–78. [Google Scholar]
- (26).Wang R, Lu KQ, Tang ZR, Xu YJ. Recent progress in carbon quantum dots: synthesis, properties and applications in photocatalysis. J Mater Chem A. 2017;5:3717–34. [Google Scholar]
- (27).Dimos K. Carbon quantum dots: surface passivation and functionalization. Curr Org Chem. 2016;20:682–95. [Google Scholar]
- (28).Michaelis J, Van der Heden van Noort GJ, Seitz O. DNA-triggered dye transfer on a quantum dot. Bioconjugate Chem. 2014;25:18–23. doi: 10.1021/bc400494j. [DOI] [PubMed] [Google Scholar]
- (29).Medintz IL, Berti L, Pons T, Grimes AF, English DS, Alessandrini A, Facci P, Mattoussi H. A reactive peptidic linker for self-assembling hybrid quantum dot–DNA bioconjugates. Nano Lett. 2007;7:1741–48. doi: 10.1021/nl070782v. [DOI] [PubMed] [Google Scholar]
- (30).Dong Y, Wang R, Li H, Shao J, Chi Y, Lin X, Chen G. Polyamine-functionalized carbon quantum dots for chemical sensing. Carbon. 2012;50:2810–15. [Google Scholar]
- (31).Dong Y, Wang R, Li G, Chen C, Chi Y, Chen G. Polyamine-functionalized carbon quantum dots as fluorescent probes for selective and sensitive detection of copperiIons. Anal Chem. 2012;84:6220–24. doi: 10.1021/ac3012126. [DOI] [PubMed] [Google Scholar]
- (32).Fanizza E, Urso C, Iacobazzi RM, Depalo N, Corricelli M, Panniello A, Agostiano A, Denora N, Laquintana V, Striccoli M, Curri ML. Fabrication of photoactive heterostructures based on quantum dots decorated with Au nanoparticles. Sci Technol Adv Mater. 2016;17:98–108. doi: 10.1080/14686996.2016.1153939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Boal AK, Ilhan F, DeRouchey JE, Thurn-Albrecht T, Russell TP, Rotello VM. Self-assembly of nanoparticles into structured spherical and network aggregates. Nature. 2000;404:746–48. doi: 10.1038/35008037. [DOI] [PubMed] [Google Scholar]
- (34).Vasimalai N, John SA. Protein protected gold nanoparticles as a fluorophore for the highly selective and ultrasensitive determination of bisphenol A in plastic samples. Anal Methods. 2013;5:5515–21. [Google Scholar]
- (35).Sun Y, Harris NC, Kiang CH. Phase Transition and Optical Properties of DNA–Gold Nanoparticle Assemblies. Plasmonics. 2007;2:193–99. doi: 10.1007/s11468-007-9034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Greschner AA, Toader V, Sleiman HF. The role of organic linkers in directing DNA self-assembly and significantly stabilizing DNA duplexes. J Am Chem Soc. 2012;134:14382–89. doi: 10.1021/ja3033197. [DOI] [PubMed] [Google Scholar]
- (37).Kypr J, Kejnovska I, Renciuk D, Vorlickova M. Nucleic Acids Res. 2009;37:1713–25. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Gao X, Ding C, Zhu A, Tian Y. Carbon-dot based ratiometric fluorescent probe for imaging and biosensing of superoxide anion in live cells. Anal Chem. 2014;86:7071–78. doi: 10.1021/ac501499y. [DOI] [PubMed] [Google Scholar]
- (39).Feng T, Ai X, An G, Yang P, Zhao Y. Charge-convertible carbon dots for imaging-guided drug delivery with enhanced in Vivo cancer therapeutic efficiency. ACS Nano. 2016;10:4410–20. doi: 10.1021/acsnano.6b00043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.