Abstract
Autophagy is a constitutive and cytoprotective catabolic process. Aberrations in autophagy lead to a multitude of degenerative disorders, with neurodegeneration being one of the most widely studied autophagy-related disorders. While the field has largely been focusing on the cytosolic constituents and processes of autophagy, recent studies are increasingly appreciating the role of chromatin modifications and epigenetic regulation in autophagy maintenance. Autophagy has been implicated in the regulation of neurogenesis, and disruption of neurogenesis in response to psychological stress is a proximal risk factor for development of neuropsychiatric disorders such as major depressive disorder (MDD). In this review, we will discuss the regulation of autophagy in normal neurogenesis as well as during chronic psychological stress, focusing on the epigenetic control of autophagy in these contexts, and also highlight the lacunae in our understanding of this process. The systematic study of these regulatory mechanisms will provide a novel therapeutic strategy, based on the use epigenetic regulators of autophagy to enhance neurogenesis and potentially alleviate stress-related behavioral disorders.
Keywords: autophagy, chromatin, depression, epigenetics, histone, neurogenesis, stress
Introduction
Autophagy (macro-autophagy) is an intracellular, cytoprotective catabolic pathway, in which organelles and cytoplasmic components are recycled via the autophagosome and lysosome. All cells exhibit constitutive autophagy, albeit at differing levels [1]. Dysfunctional autophagy has been implicated in various physiological and pathological conditions [2], in particular the development and progression of neurodegenerative diseases, and much is known about the regulation of autophagy in this context [3]. However, a lesser studied but equally critical role of autophagy is during neurogenesis, which involves the maintenance of stemness and proper differentiation of neural stem cells. Given the fact that environmental influences play a vital role in regulating neurogenesis, and that these can impact behavioral responses to stress and antidepressants [4,5], it becomes all the more important to ask whether autophagy and its regulation, especially during neurogenesis also plays a role in this context. The regulation of gene expression is largely controlled through epigenetic mechanisms, and genes involved in autophagy have also been shown to be regulated in this manner [6]. Epigenetic regulatory modifications alter chromatin states, which render a genomic region amenable or repressive to transcription. These transcription profiles in turn determine crucial processes such as cell fate decisions, differentiation pathways, and responses to stimuli [7,8]. The contribution of epigenetic regulation to autophagy has been largely underappreciated, lesser still, in the context of neurogenesis. This review aims to discuss the epigenetic mechanisms that regulate the expression of autophagy genes and the impact of this regulatory module on normal neurogenesis and neurogenesis in response to psychological stress, which is a prominent proximal risk factor for neurological disorders such as major depressive disorder (MDD) [9]. In addition to understanding the known mechanisms of autophagy regulation in normal and stress conditions, we wish to put forth a novel paradigm of using epigenetic modulators of autophagy in neurogenesis as novel regulators of stress-related behavioral disorders.
Autophagy – mechanisms and key players
The term autophagy (Greek for “Self eating”) was coined by Christian de Duve in the year 1963 and the knowledge in the field has increased exponentially since then. Numerous studies have resulted in the identification of over 100 key players, which carry out various steps in the autophagy pathway, and have implications in development and diseases. Autophagy can be classified into three types namely: (a) Macroautophagy, which involves the formation of a structure called autophagosome which ultimately fuses with the lysosome where the cargo is degraded; (b) Microautophagy, where invagination of the endosomal or lysosomal membrane leads to the engulfing and degradation of target molecules; and (c) Chaperonemediated autophagy, where certain peptide sequences are recognized by and bind to chaperone proteins which deliver them to the lysosome for degradation [1]. Macro-autophagy is the most prevalent and the most extensively studied form of autophagy in cells and herein we use the term “autophagy” to refer to macro-autophagy. In essence, autophagy is a strictly regimented, conserved, pathway that carries out two critical cellular functions: (a) nonselective autophagy, that provides nutrients for cellular processes during starvation and other kinds of stress; and (b) selective autophagy that delivers potentially harmful cytosolic cargo to the lysosome for degradation (reviewed in [1,10–13]).
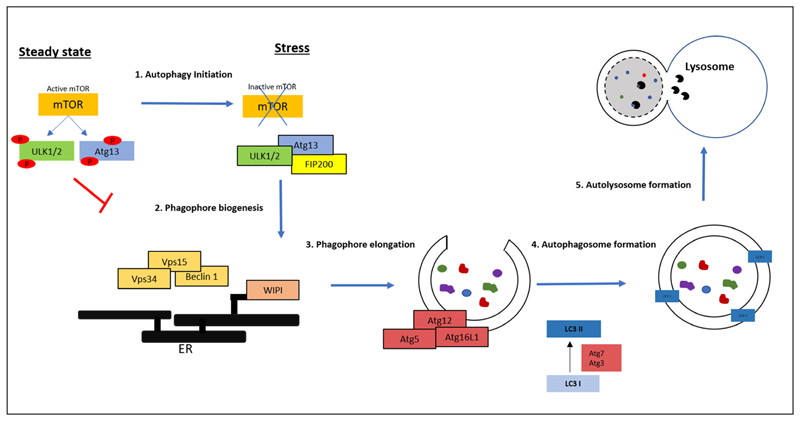
In response to various stressors, autophagy is triggered by the activation of a cascade of signals, which converge onto the nutrient sensor, mechanistic target of rapamycin kinase complex 1, mTORC1. In nutrient-rich conditions, mTORC1 inhibits autophagy by preventing the formation of the autophagosome by phosphorylating key autophagosome components. However, in conditions of starvation and other forms of stress, mTORC1 is inhibited resulting in the activation of autophagy. Figure 1 summarizes the various steps of mammalian autophagy activation and the key players involved therein.
Fig. 1.
Key steps and molecular players in autophagy (Adapted from [1,12,13]): Autophagy initiation (Step 1) involves inactivation of the nutrient sensor mTOR, which in turn activates mammalian ULK1/2 kinases. These kinases form a complex with Atg13 and FIP200, which recruits other autophagy components such as the Beclin1-Vps34-Vps15 complex and initiates phagophore formation from the isolation membrane of the ER (Step 2). This is followed by a cascade of activation and recruitment of other Atg proteins which help in the elongation and maturation of the phagophore, and selection and sequestration of cargo (Step 3). Other Atg proteins are involved in the conversion of LC3 I to LC3 II by phosphatidylethanolamine conjugation. LC3 II is then recruited to the autophagosome which facilitates closure of the autophagosome, and its fusion with the lysosome (Step 4). The cytosolic cargo is then broken down by lysosomal hydrolases (Step 5). Abbreviations: mTOR: mechanistic target of rapamycin, ULK1/2: Unc-51 like autophagy activating kinase 1/2, Atg: Autophagy-related protein, FIP200: FAK family kinase-interacting protein of 200 kDa, Vps 15/34: Vacuolar protein sorting 15/34, WIPI: WD-repeat protein interacting with phosphoinositides, Dfcp1: double FYVE domain-containing protein 1, LC3: Microtubule-associated proteins 1A/1B light chain 3B
Autophagy in neurogenesis
The field of neurogenesis research has come a long way since the days of believing that the adult brain was incapable of generating new neurons. From the first reports of neurogenesis in the adult rodent brain [14], studies in the last few decades have provided unequivocal evidence of neurogenesis in the adult brain using a variety of model systems (reviewed in [15,16]). Adult neurogenesis takes place in the subventricular zone (SVZ) and the subgranular zone (SGZ) of the dentate gyrus in the hippocampus, where neural stem cells (NSCs) and progenitor cells (NPCs) proceed through distinct developmental stages giving rise to functional mature neurons. Multipotent, self-renewing NSCs [17], during the course of neurogenesis, generate type 2 progenitor cells (type2a and type2b cells), which then proliferate into neuroblasts and further differentiate into neurons [16,18]. In addition to neurons, these cells also give rise to glia [19]. NSC-specific markers include Nestin, GFAP (Glial Fibrillary Acidic Protein), and Sox2 [17], although not all cells exhibit similar levels of all the markers [20]. Signaling pathways such as Notch, Wnt, BMP, and Hedgehog have been implicated in the process of adult hippocampal neurogenesis, while transcription factors such as Sox2, NeuroD, FoxO, Tbr2, and Prox1 have been identified as major regulatory players for neurogenesis [16]. Hippocampal neurogenesis has been implicated in the processes of learning, memory and cognitive flexibility [15].
Studies have implicated autophagy in the regulation of neural homeostasis and differentiation and rapidly accumulating data vis-à-vis the role of autophagy in stem cell function suggests that autophagy may be critical not only for the control of self-renewal and proliferation, but also for regulation of differentiation and maintenance of multipotency in neural stem cells [21,22].
One of the most significant reports looking at autophagy in neurogenesis indicated that in the absence of the autophagy gene FIP200, NSCs, NPCs, neuroblasts, and neurons in the hippocampus were dramatically reduced and this was accompanied by p53-dependent apoptosis and cell cycle arrest [23]. Other autophagy genes such as Beclin1, Ambra1, Atg5, Eva1a1 were shown to be highly expressed in the adult SVZ and were required for self-renewal, survival, and differentiation of hippocampal neurons [24–27]. Table 1 summarizes the results of the studies implicating autophagy genes in neurogenesis. A mechanistic insight into how autophagy regulates neurogenesis was demonstrated in a study that showed that autophagy regulated the Notch signaling pathway by degrading Notch1. Autophagy-deficient knockdown cell lines or ATG16L1 hypomorphic mouse models demonstrated hyperactivity of Notch signaling and perturbation of neuronal differentiation resulting in increase in stem cell populations and a delay in brain development [28]. Another interesting study reported that treadmill exercise induced autophagy in the cerebral cortex of mice, which was accompanied by increased neurogenesis [29]. The negative regulator of autophagy mTOR, was also found to regulate embryonic as well as adult neuronal development (reviewed in [30]), however whether mTOR acts by modulating autophagy itself in this context remains to be understood. These studies and more indicate that autophagy, and autophagy genes play an important role in promoting both embryonic and adult neurogenesis.
Table 1. Regulation of neurogenesis by autophagy.
| Gene | Condition | Effect on neurogenesis | References |
|---|---|---|---|
| FIP200 | Conditional deletion | Thinner SVZ, reduced olfactory bulb, decreased NSC proliferation | [23] |
| Ambra1 | Ambra1+/- Isolated NSCs | Reduced proliferation, increased cell death | [24] |
| Ambra1 | Ambra1 -/- Ambra1 -/+ | Reduced neuronal markers, smaller neurospheres, lesser neuronal differentiation (as seen by which markers) | [25] |
| Beclin 1 | Beclin1+/- | Reduced proliferation, increased cell death, smaller neurospheres | [24] |
| Atg5 | Atg5 -/- | Reduced neuronal differentiation | [25] |
| Atg5 | In utero electroporation of shRNA | Decreased neural differentiation, aberrant morphology of cortical neurons | [26] |
| Atg5 | Atg5 -/- NSCs | Delayed neuronal differentiation, transient reduction of spine density | [31] |
| Eva1a1 | Eva1a1 -/- NSCs | Reduced self-renewal, reduced differentiation | [27] |
Neurogenesis and stress
The term “stress” was coined to define the response of an organism to stressors believed to be harmful for health [31]. Since then, numerous studies have successfully dissected the differences between the physiological and psychological aspects of stress and the distinct neurobiological responses of the two [32]. Early life stress, be it physiological or psychological, has been implicated as a major risk factor in the development of psychiatric disorders. Epidemiological and clinical studies demonstrate a strong correlation between childhood adversity, stress and depressive symptoms, more specifically, MDD [9,32,33]. MDD is a highly prevalent debilitating condition, with symptoms including mood fluctuations, anhedonia, weight changes, feelings of worthlessness, and suicidal tendencies among many others [34]. Despite the increasing morbidity and mortality associated with this disease, little is known about the role of stress and the molecular components involved therein, in development of MDD.
Exposure to stress elicits a response which involves the hypothalamic-pituitary-adrenal (HPA) axis that plays a role in adaptation to stress through a neuroendocrine pathway that eventually dictates the behavioral, neural and hormonal response to stress [35]. Hippocampal cells are sensitive to the effects of environmental stress. Studies in animals as well as humans suggest that prolonged stress and glucocorticoid exposure are associated with hippocampal dendritic atrophy, change in hippocampal morphology, and reduction in hippocampal size. These changes pose as a risk factor for the development of neuropsychiatric pathophysiological conditions such as depression, anxiety, post traumatic stress disorder (PTSD), personality disorders etc. [36–40].
The neurogenic theory of depression dictates that depression is triggered by impaired adult neurogenesis, and studies also indicate that increasing hippocampal neurogenesis is sufficient to reduce depressive behavior [41–43]. A seminal study where neurogenesis was disrupted in the mouse hippocampus by x-ray irradiation, revealed that these mice failed to exhibit behavioral effects of antidepressants imipramine and fluoxetine, suggesting that neurogenesis is required for the efficacy of antidepressants [44]. When social defeat stress was used as a paradigm for depressive behavior in rats, neurogenesis–measured by quantifying immature doublecortin positive cells – was found to be reduced; and this reduction could be reversed upon treatment by the antidepressant imipramine [45]. Studies in adult rats have also demonstrated that the thyroid hormone (TH) plays a role in adult hippocampal neurogenesis and that mood disorders associated with hypothyroidism could be a result of impaired neurogenesis [46]. Psychosocial as well as physical stressors have been shown to reduce neurogenesis. Examples of stress include reports of subordination stress in tree shrews and marmosets and social defeat stress in mice resulting in reduced neurogenesis as measured by decreased cell proliferation [37,47–49]. Different chronic stressors exhibit distinct effects on neurogenesis. Neural cell proliferation and differentiation were reduced upon chronic social stress in tree shrews [50] and mice [51]; however, chronic restraint stress was also shown to increase neural cell proliferation in mice [52]. This indicates that while in general, stress results in reduced neurogenesis; this response may exhibit context-dependent, stressor and strain-specific variations.
Autophagy and stress
Autophagy changes were found to be associated with normal stress response mechanisms, and aberrant autophagy was found in neuropsychiatric disorders [53]. Postmortem studies in subjects diagnosed with MDD showed deficits in mTOR signaling which is an autophagy regulatory pathway (see Fig. 1). Also, antidepressants were found to reverse the behavioral effects of chronic stress in an mTOR-dependent manner [54]. Brain-derived neurotropic factor (BDNF) has been shown to maintain neuronal plasticity and reduced BDNF expression in the hippocampus is shown to correlate with major depressive disorders [55]. Recent studies show that the neuroprotective role of BDNF correlates with increased autophagy by upregulating LC3 and inhibiting the Akt/mTOR/p70S6K pathway. Whether the changes in autophagy pathway molecules directly reflect a change in autophagy itself, implicating autophagy in neural homeostasis and stress response, however, still remains to be elucidated [55,56]. A recent study conducted in mice, demonstrated that nicotine alleviates chronic stress-induced behavioral deficits and neuropathological alterations via autophagy regulation. More specifically, treatment with nicotine led to an increase in autophagy marker levels such as Beclin1, LC3, and p62 in mice exposed to mild chronic unpredictable stress (CUS) [57]. While it is increasingly accepted that dysregulated autophagy plays a role in neuropsychiatric disorders, conflicting reports still exist, demonstrating contrasting trends of autophagy in stress disorders. Adverse childhood experiences such as maternal separation (MS) is a well-characterized model for stress which has detrimental long-term behavioral and physiological impacts [58]. A recent report demonstrated different effects of MS in different areas of the rat brains. MS led to decreased autophagy levels in the hippocampus but increased autophagy in the prefrontal cortex [59]; indicating that a context-specific autophagy pathway is crucial to maintain neural homeostasis in different regions of the brain. These variations make it all the more essential to understand the regulation of autophagy itself and the genes involved in response to various cues beyond the cytosolic assembly of autophagy molecules.
Studies have demonstrated a strong correlation between antidepressant action and autophagy activation. Rapamycin, an autophagy activator, itself displays antidepressant activity [studied by the forced swim test (FST) or tail suspension test (TST)], when administered to depression models of mice and rats at sub-chronic doses [60]. In addition to this, classical antidepressants such as lithium, trehalose, tocopherol, amitriptyline, and fluoxetine exhibit increased autophagy. Further investigation of this correlation is required to shed light on the possible causative relationship between increase in autophagy and antidepressant action [61–63]. One is tempted to speculate that autophagy might work in this system by increasing adult hippocampal neurogenesis, which in turn alleviates depressive behavioral symptoms. Despite being increasingly recognized as a debilitating illness that has the potential to affect a significant proportion of the population [64], the treatment paradigms for depressive disorders are far from ideal [65,66]. The latency period for clinical effects of antidepressants is long and is associated with numerous side effects [66,67]. Additionally, a significant percentage of patients do not respond to treatment and are prone to relapse [68]. Alternative treatment modalities targeting genetic markers, cytokine therapies, metabolic pathways, inflammation response have been initiated, however with inconsistent results [69]. Given the crucial role of autophagy in regulating both neurogenesis and stress response, modulation of autophagy could potentially be used as a novel paradigm to rescue the neurogenic disruption brought about by acute and chronic psychological stress with an aim to ameliorate behavioral symptoms.
Regulation of autophagy
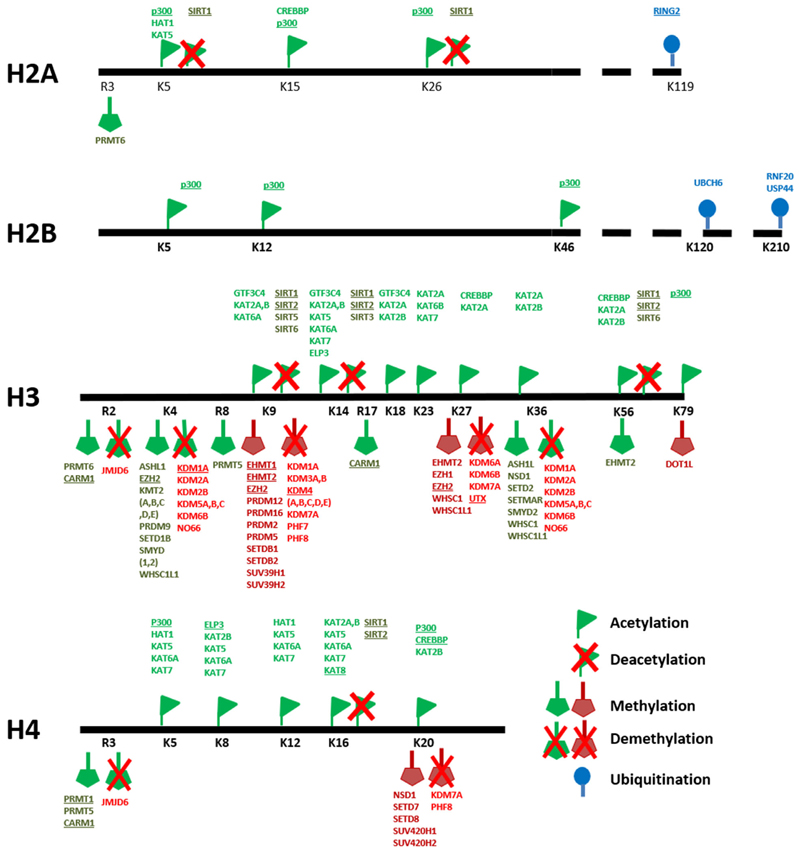
In order to develop autophagy modulation as a novel therapeutic tool to induce neurogenesis and combat behavioral disorders thereof, understanding the process of autophagy itself and its regulation becomes crucial. Autophagy has been considered to be primarily a cytosolic process. The molecules involved in the process of autophagy, and their positive and negative regulators are well understood and have been reviewed elsewhere [70,71]. While these studies have been influential in enhancing our understanding of the mechanism of autophagy under steady state as well as under different stress conditions, a molecular understanding of autophagy regulation is far from clear. A critical layer of regulation of cellular pathways is at the level of chromatin. Epigenetic regulatory mechanisms control gene expression profiles by altering the accessibility of chromatin to the transcriptional machinery making it either conducive or repressive to transcription. Chromatin, made up of nucleosomes, consists of DNA, RNA, and histone proteins, which are modified by chromatin modulators. Maintenance of higher order chromatin is critical for embryogenesis, differentiation, and tissue-specific gene expression patterns which in turn regulate normal development at the cellular and organismal level [72–74]. Various molecules function as readers, writers, or erasers of chromatin modifications to determine a transcriptionally permissive or inhibitory chromatin environment, which regulates transcription states. Figure 2 summarizes key histone modifiers that mark histone tails with activating or repressive modifications to influence transcriptional outcome.
Fig. 2.
Writers and erasers of epigenetic modifications: (Adapted from and reviewed in [7,123,124]). Histone tails are marked by writers of chromatin modifications. These marks include commonly activating modifications such as lysine acetylation (green flags) on H2A, H2B, H3, and H4; lysine and arginine methylation on H3 and H4 (green pentagons) as well as repressive modifications such as lysine and arginine methylation on H3 and H4 (red pentagons). These histone modifications can be removed, and their effects reversed by the action of deacetylases and demethylases (crossed flags and pentagons), which make histone modifications amenable to rapid changes in response to stimuli. Underlined molecules indicate histone modifiers implicated in autophagy regulation.
Global chromatin remodeling takes place during embryogenesis and differentiation and such epigenetic regulatory mechanisms have been shown to be indispensable for normal development [75,76]. Studies have pointed to the presence of a complex interplay of epigenetic modifications which ensure an open chromatin conformation in naïve pluripotent stem cells, with chromatin accessibility being progressively restricted as differentiation proceeds. These modifications include the presence of bivalent domains which are genomic regions marked by both activating as well as repressive chromatin modifications which are present in both stem cells as well as differentiated cells and serve to poise genes for rapid activation upon stimuli [77–79]. Given the central role of epigenetic regulation in cellular pathways, it becomes imperative to understand how a crucial pathway such as autophagy is regulated in this manner. The role of epigenetic regulators in the context of autophagy has been largely neglected [80]. However, the last few years have seen a substantial increase in reports that have implicated transcription factors and epigenetic regulatory mechanisms in governing the autophagy pathway.
Epigenetic control of autophagy
One of the first reports indicating that autophagy was under epigenetic control was conducted in aging yeast [81,82]. Spermidine, a naturally occurring polyamine has been shown to counter oxidative DNA damage, reduce cell necrosis, and enhance longevity. Treatment of yeast cells with spermidine resulted in deacetylation of H3 by suppression of histone acetyl transferases (HATs). This deacetylation resulted in the activation of autophagy genes and induction of autophagy. Depletion of spermidine and other polyamines led to a reduction in autophagy, accumulation of ROS, and ultimately cell necrosis. Similar results were also seen in flies, worms, and humans; indicating that epigenetic activation of autophagy genes played an important conserved role in oxidative stress response [81,82]. Other work has indicated that in response to various cues, autophagy activation is associated with deacetylation of H4K16 and downregulation of the hMOF acetyl transferase [83]. In Drosophila, the histone methyl transferase dG9a was shown to affect starvation sensitivity by affecting autophagy. In the absence of dG9a, flies lost their ability to recycle amino acids by autophagy pathways induced upon starvation, and the autophagy molecule/player ATG8a expression was found to be regulated by dG9a in this system [84]. While G9a was found to perform an activating role in Drosophila, studies in human cells showed a contrasting result. Using inhibitor assays, G9a was found to repress autophagy in HeLa cells as well as in naïve human T cells by binding to the promoters of autophagy genes such as LC3, WIPI, and DOR, and repressing their expression. Upon glucose starvation, G9a was no longer bound to the autophagy gene promoters which led to expression of autophagy genes and activation of the autophagy pathway, indicating that G9a targets distinct loci in different systems which may explain its contrasting effects on autophagy [85]. Another study identified the role of G9a in the autophagy-dependent serine metabolism pathway of human cancer cells, where G9a acts as an activator of the serine-glycine metabolism pathway which in turn sustains the survival of cancer cell lines by repressing autophagy and cell death [86]. These results signify the presence of regulatory epigenetic mechanisms which act in concert, or in opposition, resulting in different transcriptional outcomes in different contexts.
Amino acid starvation in mouse embryonic fibroblasts led to an increase in autophagy and a concomitant decrease in H4K16 acetylation levels, which has been shown to be associated with both active as well as repressed genes [87–89]. On the other hand, glucose starvation resulted in a CARM1-dependent activation of autophagy and lysosomal genes by increase in H3R17 dimethylation [90]. Autophagy induction upon serum starvation was delayed upon EZH2 overexpression in HeLa cells, implicating the Polycomb pathway and the H3K27me3 modification as a negative regulator of autophagy [91]. A recent study was conducted in zebrafish myotubes to identify histone modifications associated with autophagy promoters in normal versus starvation conditions. Under normal conditions, autophagy genes were marked by both activating (H3K4me3) and repressive (H3K9me3) modifications [92]. Such domains, called bivalent domains, are present on developmentally important genes to balance their expression levels and poise these genes for activation upon stimuli [77,93]. The presence of bivalent marks on autophagy genes indicates that epigenetic modifications may be required to balance the levels of autophagy and poise the pathway for induction upon stress. In zebrafish myotubes, upon starvation, H3K4me3 and H3K9me3 marks are reduced from autophagy genes resulting in increased gene expression and an induction of autophagy. A detailed systematic analysis of combinations of chromatin modifications is needed to understand the complex interplay between activating and repressive histone modifications to ultimately lead to precisely balanced transcription of autophagy genes. The histone deacetylase, Sirt1, activates autophagy upon caloric restriction or starvation [94–96]. In HeLa cells, starvation led to a reduction of H2B monoubiquitination on autophagy genes, resulting in activation of autophagy [97]. A recent high-throughput ChIP-seq and RNA-seqbased study conducted in the human HAP1 line (Chronic myeloid leukemia cells), indicated that indeed, upon starvation, transcription of many autophagy genes is increased and this increase is associated with concomitant establishment of activating histone modifications and RNA-Pol II recruitment [98]. Histone demethylases have also been implicated in autophagy regulation. In Drosophila salivary glands, the H3K27 demethylase, dUTX, regulates temporal expression of autophagy genes [99]. Additionally, the H3K36 demethylase KDM4A is found to repress many Atg genes and this function is conserved from yeast to humans [100]. Recent reports in neuroblastoma cells have implicated KDM1A/LSD1 as a regulator of the autophagy gene expression program [101]. While a majority of studies have identified epigenetic regulators of autophagy by using specific inhibitors or knockout/knockdown studies [83,85,99,101], it would be interesting to determine whether these modulators themselves show altered expression under starvation and other stress conditions.
In addition to chromatin modulators, transcription of autophagy genes is also regulated by a variety of transcription factors that bind to autophagy promoters and activate their transcription in response to stimuli. More than 20 transcription factors have now been identified as regulators of autophagy, and their role in autophagy regulation has been discussed in detail elsewhere [102,103]. These studies indicate that a controlled, concerted interplay between various modulators of chromatin and transcription is vital for the regulation of autophagy.
Epigenetic regulation of autophagy in stress
It is now clear that epigenetic modifications play a crucial role in the regulation of autophagy. The implications this regulatory pathway has in stress response, and by extension, neuropsychiatric disorders is gradually being appreciated, as recent studies have reported that epigenetic control of autophagy regulates depressive behavior and antidepressant action. An important report in this field pertains to the HSP90 co-chaperone FKBP51. FKBP51 is a regulator of the glucocorticoid receptor pathway, and in turn, the stress response pathway. In patients, FKPB51 levels are thought to predict antidepressant responses. FKBP51 was found to interact with and activate autophagy genes in response to antidepressant treatment in cell lines, primary cells, and animal models of chronic stress [104–106]. Antidepressant treatment also resulted in an FKBP51-dependent reduction in DNMT1-mediated DNA methylation which led to the activation of numerous genes including autophagy genes as well as BDNF, which is also known to regulate neurogenesis by modulating autophagy [56].
Another interesting but not well-studied aspect of how epigenetic modulation of autophagy genes could help reduce neural stress-related maladaptive responses, involves the autophagic clearance of RNA generated from mobile genetic elements in the brain. Studies in various cell lines indicate that aberrant transcripts generated from retrotransposons and other repeat elements are degraded by autophagy-dependent mechanisms [107]. Stress leads to mis-expression of repeat elements which is associated with correlating epigenetic modifications [108,109]. A study in rats demonstrated that upon acute stress, H3K9me3 (a repressive mark) is increased in the hippocampus and not in other regions of the brain, and this mark could be traced to mobile genetic elements, especially short interspersed nuclear elements (SINEs). This increase in H3K9me3 could no longer be seen upon chronic stress [110]. This presents an interesting paradigm for repeat element regulation upon stress. Acute stress may lead to a wave of transposon expression that is repressed by establishment of the H3K9me3 mark. However, the loss of the repressive mark upon chronic stress points to perhaps the involvement of alternate regulatory pathways. It is tempting to hypothesize that autophagy might play a compensatory role in clearing aberrantly expressed repeat transcripts upon chronic stress, and reduced autophagy in turn leads to increased repeat RNA levels which could then result in altered neurogenesis in chronic stress. It would be interesting to compare epigenetic modifications associated with autophagy genes along with epigenetic profiles present at repeat elements to understand how regulation of autophagy governs stress response by clearing aberrant transcripts.
Given that activating autophagy increases neurogenesis, and increased neurogenesis in turn can counter the detrimental effects of chronic stress, autophagy activating molecules emerge as promising candidates to combat neuropsychiatric disorders. Rapamycin, a potent autophagy activator has been tested as a putative drug for numerous diseases [111]. However, the known risk of developing adverse side effects, such as metabolic defects including hyperglycemia, hyperlipidemia, insulin resistance among others, upon rapamycin treatment makes it a suboptimal candidate as a therapeutic autophagy inducer [112,113]. Combinatorial drug treatments are emerging to be viable and efficient alternatives to single drug treatment regimens, especially, drug therapies involving cytotoxic and epigenetic drugs [114–116]. Altering epigenetic regulatory pathways is being increasingly used to combat various disorders either independently or in conjunction with classical cytotoxic drugs [117]. While the epigenetic regulation of a number of pathways regulating cell fate and lineage commitment has been extensively studied [73,118,119], the transcriptional regulation of autophagy genes is poorly understood. The field of “neuroepigenetics,” deals with the epigenetic regulation of the nervous system and a number of studies in the field have identified chromatin modifications such as DNA methylation and demethylation, histone modifications, and small RNAs as being indispensable for neurogenesis [120–122]. What remains to be understood is whether and how epigenetic regulation of autophagy and neurogenesis are connected. For efficient therapeutic use of epigenetic drugs as modulators of autophagy to combat neuropsychiatric disorders, a systematic approach to understand the epigenetic regulation of autophagy during neurogenesis is essential. This will elucidate the possible link between autophagy and neurogenesis in steady state versus stress conditions.
Conclusions and future perspectives
As the abovementioned reports and more suggest, autophagy plays an indispensable role in neural homeostasis and aberrant autophagy may lead to neuropsychiatric disorders. Understanding the regulation of autophagy, especially in the context of chromatin becomes increasingly important given the ability of chromatin modifications to regulate cellular pathways across generations. This is extremely relevant in today’s scenario where stress is an integral part of most lives and variability in stress response pathways can result in increased or decreased resilience to development of stress disorders. Understanding the upstream regulators of long-term behavioral disorders can facilitate the development of efficient drugs to counter the effects of undesirable stimuli such as stress. It is thus tempting to hypothesize that activation of autophagy using epigenetic modulators (readers, writers, or erasers), can be used therapeutically to increase hippocampal neurogenesis and alleviate depressive behavior. However, as with other regulatory mechanisms, caveats exist and systematic analyses need to be conducted to address the following questions:
What is the epigenetic profile of autophagy genes in neural stem cells and during neural differentiation? What is the correlation between epigenetic marks and the transcriptional profile of autophagy genes under normal conditions as well as under conditions of acute or chronic stress?
Does exposure to acute or chronic stress have distinct effects on autophagy levels? Are they connected to the differences in hippocampal neurogenesis and resulting behavioral responses? The biphasic autophagy theory in the field proposes that autophagy induction in response to stimuli happens in two stages: (a) the initial acute response is independent of transcription and relies on the action of already existing cytosolic autophagy proteins, while; (b) a long-term chronic response requires activation of autophagy genes to elicit an induction. Epigenetic mechanisms of regulation might play a crucial role in this long-term response and need to be further dissected.
What measures can be taken to address the off-target effects of epigenetic drugs to limit the effect primarily to the autophagy pathway and improve the efficiency of these modulators as prospective therapeutic agents?
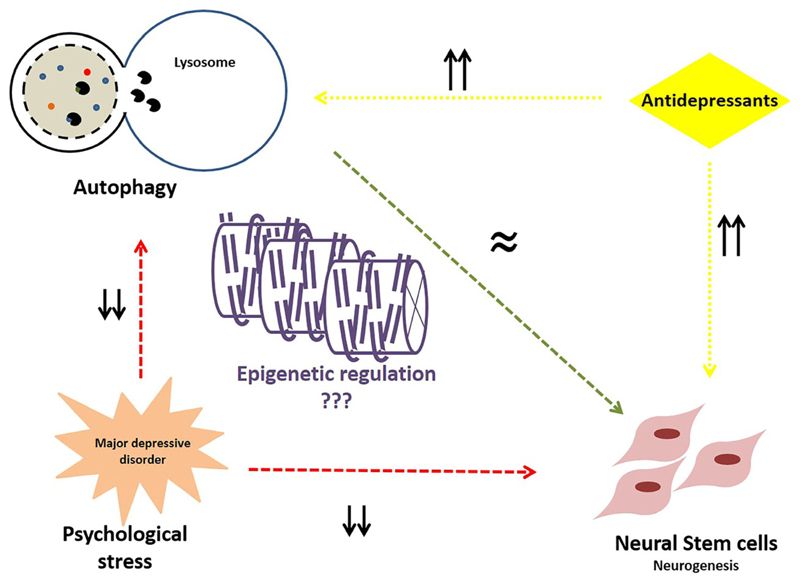
In conclusion, while links exist between autophagy and neurogenesis; neurogenesis and depression; antidepressant response and autophagy; and epigenetic regulation of autophagy in different cellular states (Fig. 3), the mechanistic underpinnings of how these pathways are regulated and interconnected still remain to be dissected. The past few decades have witnessed a multitude of technological breakthroughs that have made it possible to study in detail, the complex connections between environmental cues and the subsequent responses at a cellular and organismal level. This, coupled with the increasing awareness of previously underdiagnosed and suboptimally treated psychological stress disorders, brings to the forefront the relevance of investigating these disorders in the context of perturbed cellular mechanisms. Aberrant autophagy affecting neurogenesis upon exposure to stress is but one of many such pathways that are still underappreciated, but can potentially have tremendous therapeutic benefits in our continuing efforts to combat prevalent disorders and improve the quality of life.
Fig. 3.
Autophagy, neurogenesis, and stress: Precise levels of autophagy are critical for the maintenance of normal neurogenesis (green arrow). In conditions of psychological stress and the resultant neuropsychiatric conditions, neurogenesis as well as autophagy levels in the hippocampus decrease (red arrows). Conversely, antidepressant treatment results in increased autophagy and also hippocampal neurogenesis (yellow arrows). Epigenetic regulatory mechanisms may play a crucial role in maintaining autophagy and in turn, normal neurogenesis, and could be used to ameliorate behavioral disorders arising from exposure to stress.
Acknowledgements
DP is funded by the Department of Science and Technology (DST) INSPIRE Faculty fellowship (DST/INSPIRE/04/2017/001707) and DS is supported by an Intermediate Fellowship from Wellcome Trust DBT India Alliance (IA/I/12/1/500507), and by a grant from the Department of Biotechnology (BT/PR25883/GET/119/105/2017).
Abbreviations
- CUS
chronic unpredictable stress
- DNA
deoxyribonucleic acid
- ESC
embryonic stem cell
- GFAP
glial fibrillary acidic protein
- HAT
histone acetyl transferase
- HPA
hypothalamic pituitary adrenal
- MDD
major depressive disorder
- MS
maternal separation
- NPC
neural progenitor cell
- NSC
neural stem cell
- PTSD
post traumatic stress disorder
- RNA
ribonucleic acid
- SGZ
subgranular zone
- SVZ
subventricular zone
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Author contributions
DP and DS were equally involved in planning writing the review.
References
- 1.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 4.Kuipers SD, Bramham CR, Cameron HA, Fitzsimons CP, Korosi A, Lucassen PJ. Environmental control of adult neurogenesis: from hippocampal homeostasis to behavior and disease. Neural Plast. 2014;2014 doi: 10.1155/2014/808643. 808643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LaDage LD. Environmental change, the stress response, and neurogenesis. Integr Comp Biol. 2015;55:372–383. doi: 10.1093/icb/icv040. [DOI] [PubMed] [Google Scholar]
- 6.Füllgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2013;15:65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- 7.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 8.De Los Angeles A, Ferrari F, Xi R, Fujiwara Y, Benvenisty N, Deng H, Hochedlinger K, Jaenisch R, Lee S, Leitch HG, et al. Hallmarks of pluripotency. Nature. 2015;525:469–478. doi: 10.1038/nature15515. [DOI] [PubMed] [Google Scholar]
- 9.Hammen C. Stress and Depression. Annu Rev Clin Psychol. 2005;1:293–319. doi: 10.1146/annurev.clinpsy.1.102803.143938. [DOI] [PubMed] [Google Scholar]
- 10.Bento CF, Renna M, Ghislat G, Puri C, Ashkenazi A, Vicinanza M, Menzies FM, Rubinsztein DC. Mammalian Autophagy: How Does It Work? Annu Rev Biochem. 2016;85:685–713. doi: 10.1146/annurev-biochem-060815-014556. [DOI] [PubMed] [Google Scholar]
- 11.Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 12.Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhi X, Feng W, Rong Y, Liu R. Anatomy of autophagy: from the beginning to the end. Cell Mol Life Sci. 2018;75:815–831. doi: 10.1007/s00018-017-2657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altman J, Das GD. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J Comp Neurol. 1965;124:319–335. doi: 10.1002/cne.901240303. [DOI] [PubMed] [Google Scholar]
- 15.Gonçalves JT, Schafer ST, Gage FH. Adult neurogenesis in the hippocampus: from stem cells to behavior. Cell. 2016;167:897–914. doi: 10.1016/j.cell.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 16.Kempermann G, Song H, Gage FH. Neurogenesis in the adult hippocampus. Cold Spring Harb Perspect Biol. 2015;7 doi: 10.1101/cshperspect.a018812. a018812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, Song H. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145:1142–1155. doi: 10.1016/j.cell.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lugert S, Basak O, Knuckles P, Haussler U, Fabel K, Götz M, Haas CA, Kempermann G, Taylor V, Giachino C. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell. 2010;6:445–456. doi: 10.1016/j.stem.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Encinas JM, Michurina TV, Peunova N, Park JH, Tordo J, Peterson DA, Fishell G, Koulakov A, Enikolopov G. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell. 2011;8:566–579. doi: 10.1016/j.stem.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gebara E, Bonaguidi MA, Beckervordersandforth R, Sultan S, Udry F, Gijs PJ, Lie DC, Ming GL, Song H, Toni N. Heterogeneity of radial glia-like cells in the adult hippocampus. Stem Cells. 2016;34:997–1010. doi: 10.1002/stem.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casares-Crespo L, Calatayud-Baselga I, García-Corzo L, Mira H. On the role of basal autophagy in adult neural stem cells and neurogenesis. Front Cell Neurosci. 2018;12:339. doi: 10.3389/fncel.2018.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhaliwal J, Trinkle-Mulcahy L, Lagace DC. Autophagy and adult neurogenesis: discoveries made half a century ago yet in their infancy of being connected. Brain Plast. 2017;3:99–110. doi: 10.3233/BPL-170047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C, Liang CC, Bian ZC, Zhu Y, Guan JL. FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nat Neurosci. 2013;16:532–542. doi: 10.1038/nn.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yazdankhah M, Farioli-Vecchioli S, Tonchev AB, Stoykova A, Cecconi F. The autophagy regulators Ambra1 and Beclin 1 are required for adult neurogenesis in the brain subventricular zone. Cell Death Dis. 2014;5:e1403. doi: 10.1038/cddis.2014.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vázquez P, Arroba AI, Cecconi F, De La Rosa EJ, Boya P, De Pablo F. Atg5 and Ambra1 differentially modulate neurogenesis in neural stem cells. Autophagy. 2012;8:187–199. doi: 10.4161/auto.8.2.18535. [DOI] [PubMed] [Google Scholar]
- 26.Lv X, Jiang H, Li B, Liang Q, Wang S, Zhao Q, Jiao J. The crucial role of Atg5 in cortical neurogenesis during early brain development. Sci Rep. 2014;4:6010. doi: 10.1038/srep06010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, Lu G, Hu J, Shen X, Ju J, Gao Y, Qu L, Xia Y, Chen Y, Bai Y. EVA1A/TMEM166 regulates embryonic neurogenesis by autophagy. Stem Cell Rep. 2016;6:396–410. doi: 10.1016/j.stemcr.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, Fleming A, Ricketts T, Pavel M, Virgin H, Menzies FM, Rubinsztein DC. Autophagy regulates Notch degradation and modulates stem cell development and neurogenesis. Nat Commun. 2016;7:10533. doi: 10.1038/ncomms10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He C, Sumpter R, Levine B. Exercise induces autophagy in peripheral tissues and in the brain. Autophagy. 2012;8(10):1548–1551. doi: 10.4161/auto.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garza-Lombo C, Gonsebatt ME. Mammalian target of Rapamycin: its role in early neural development and in adult and aged brain function. Front Cell Neurosci. 2016;10:157. doi: 10.3389/fncel.2016.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selye H. Stress and disease. Science. 1955;122:625–631. doi: 10.1126/science.122.3171.625. [DOI] [PubMed] [Google Scholar]
- 32.Richter-Levin G, Xu L. How could stress lead to major depressive disorder? IBRO Rep. 2018;4:38–43. doi: 10.1016/j.ibror.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kendler KS, Karkowski LM, Prescott CA. Causal relationship between stressful life events and the onset of major depression. Psychiatry Interpers Biol Process. 1999;156:837–841. doi: 10.1176/ajp.156.6.837. [DOI] [PubMed] [Google Scholar]
- 34.Otte C, Gold SM, Penninx BW, Pariante CM, Etkin A, Fava M, Mohr DC, Schatzberg AF. Major depressive disorder. Nat Rev Dis Primer. 2016;2:16065. doi: 10.1038/nrdp.2016.65. [DOI] [PubMed] [Google Scholar]
- 35.Joëls M, Baram TZ. The neuro-symphony of stress. Nat Rev Neurosci. 2009;10:459–466. doi: 10.1038/nrn2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mareckova K, Marecek R, Bencurova P, Klanova J, Dusek L, Brazdil M. Perinatal stress and human hippocampal volume: findings from typically developing young adults. Sci Rep. 2018;8:4696. doi: 10.1038/s41598-018-23046-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe Y, Gould E, McEwen BS. Stress induces atrophy of apical dendrites of hippocampal CA3 pyramidal neurons. Brain Res. 1992;588:341–345. doi: 10.1016/0006-8993(92)91597-8. [DOI] [PubMed] [Google Scholar]
- 38.Magariños AM, McEwen BS, Flügge G, Fuchs E. Chronic psychosocial stress causes apical dendritic atrophy of hippocampal CA3 pyramidal neurons in subordinate tree shrews. J Neurosci Off J Soc Neurosci. 1996;16:3534–3540. doi: 10.1523/JNEUROSCI.16-10-03534.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dranovsky A, Hen R. Hippocampal neurogenesis: regulation by stress and antidepressants. Biol Psychiatry. 2006;59:1136–1143. doi: 10.1016/j.biopsych.2006.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duman RS. Depression: a case of neuronal life and death? Biol Psychiatry. 2004;56:140–145. doi: 10.1016/j.biopsych.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 41.Miller BR, Hen R. The current state of the neurogenic theory of depression and anxiety. Curr Opin Neurobiol. 2015;30:51–58. doi: 10.1016/j.conb.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill AS, Sahay A, Hen R. Increasing adult hippocampal neurogenesis is sufficient to reduce anxiety and depression-like behaviors. Neuropsychopharmacology. 2015;40:2368–2378. doi: 10.1038/npp.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sahay A, Hen R. Adult hippocampal neurogenesis in depression. Nat Neurosci. 2007;10:1110–1115. doi: 10.1038/nn1969. [DOI] [PubMed] [Google Scholar]
- 44.Santarelli L, Saxe M, Gross C, Surget A, Battaglia F, Dulawa S, Weisstaub N, Lee J, Duman R, Arancio O, et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301:805–810. doi: 10.1126/science.1083328. [DOI] [PubMed] [Google Scholar]
- 45.Van Bokhoven P, Oomen CA, Hoogendijk WJG, Smit AB, Lucassen PJ, Spijker S. Reduction in hippocampal neurogenesis after social defeat is long-lasting and responsive to late antidepressant treatment. Eur J Neurosci. 2011;33:1833–1840. doi: 10.1111/j.1460-9568.2011.07668.x. [DOI] [PubMed] [Google Scholar]
- 46.Montero-Pedrazuela A, Venero C, Lavado-Autric R, Fernández-Lamo I, García-Verdugo JM, Bernal J, Guadaño-Ferraz A. Modulation of adult hippocampal neurogenesis by thyroid hormones: implications in depressive-like behavior. Mol Psychiatry. 2006;11:361–371. doi: 10.1038/sj.mp.4001802. [DOI] [PubMed] [Google Scholar]
- 47.Gould E, McEwen BSS, Tanapat P, Galea LA, Fuchs E. Neurogenesis in the dentate gyrus of the adult tree shrew is regulated by psychosocial stress and NMDA receptor activation. J Neurosci Off J Soc Neurosci. 1997;17:2492–2498. doi: 10.1523/JNEUROSCI.17-07-02492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gould E, Tanapat P, McEwen BS, Flugge G, Fuchs E. Proliferation of granule cell precursors in the dentate gyrus of adult monkeys is diminished by stress. Proc Natl Acad Sci USA. 1998;95:3168–3171. doi: 10.1073/pnas.95.6.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yap JJ, Takase LF, Kochman LJ, Fornal CA, Miczek KA, Jacobs BL. Repeated brief social defeat episodes in mice: effects on cell proliferation in the dentate gyrus. Behav Brain Res. 2006;172:344–350. doi: 10.1016/j.bbr.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 50.Czeh B, Michaelis T, Watanabe T, Frahm J, de Biurrun G, van Kampen M, Bartolomucci A, Fuchs E. Stress-induced changes in cerebral metabolites, hippocampal volume, and cell proliferation are prevented by antidepressant treatment with tianeptine. Proc Natl Acad Sci USA. 2001;98:12796–12801. doi: 10.1073/pnas.211427898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferragud A, Haro A, Sylvain A, Velazquez-Sanchez C, Hernandez-Rabaza V, Canales JJ. Enhanced habit-based learning and decreased neurogenesis in the adult hippocampus in a murine model of chronic social stress. Behav Brain Res. 2010;210:134–139. doi: 10.1016/j.bbr.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 52.Snyder JS, Glover LR, Sanzone KM, Kamhi JF, Cameron HA. The effects of exercise and stress on the survival and maturation of adult-generated granule cells. Hippocampus. 2009;19:898–906. doi: 10.1002/hipo.20552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jia J, Le W. Molecular network of neuronal autophagy in the pathophysiology and treatment of depression. Neurosci Bull. 2015;31:427–434. doi: 10.1007/s12264-015-1548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abelaira HM, Reus GZ, Neotti MV, Quevedo J. The role of mTOR in depression and antidepressant responses. Life Sci. 2014;101:10–14. doi: 10.1016/j.lfs.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 55.Lee BH, Kim YK. The roles of BDNF in the pathophysiology of major depression and in antidepressant treatment. Psychiatry Investig. 2010;7:231–235. doi: 10.4306/pi.2010.7.4.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen A, Xiong LJ, Tong Y, Mao M. Neuroprotective effect of brain-derived neurotrophic factor mediated by autophagy through the PI3K/Akt/mTOR pathway. Mol Med Rep. 2013;8:1011–1016. doi: 10.3892/mmr.2013.1628. [DOI] [PubMed] [Google Scholar]
- 57.Xiao X, Shang X, Zhai B, Zhang H, Zhang T. Nicotine alleviates chronic stress-induced anxiety and depressive-like behavior and hippocampal neuropathology via regulating autophagy signaling. Neurochem Int. 2018;114:58–70. doi: 10.1016/j.neuint.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 58.Nishi M, Horii-hayashi N, Sasagawa T, Matsunaga W. General and comparative endocrinology effects of early life stress on brain activity: implications from maternal separation model in rodents. Gen Comp Endocrinol. 2013;181:306–309. doi: 10.1016/j.ygcen.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 59.Liu C, Hao S, Zhu M, Wang Y, Zhang T, Yang Z. Maternal separation induces different autophagic responses in the Hippocampus and prefrontal cortex of adult rats. Neuroscience. 2018;374:287–294. doi: 10.1016/j.neuroscience.2018.01.043. [DOI] [PubMed] [Google Scholar]
- 60.Cleary C, Linde JA, Hiscock KM, Hadas I, Belmaker RH, Agam G, Flaisher-Grinberg S, Einat H. Antidepressive-like effects of rapamycin in animal models: implications for mTOR inhibition as a new target for treatment of affective disorders. Brain Res Bull. 2008;76:469–473. doi: 10.1016/j.brainresbull.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 61.Gulbins A, Schumacher F, Becker KA, Wilker B, Soddemann M, Boldrin F, Müller CP, Edwards MJ, Goodman M, Caldwell CC, et al. Antidepressants act by inducing autophagy controlled by sphingomyelin-ceramide. Mol Psychiatry. 2018;23:2324–2346. doi: 10.1038/s41380-018-0090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang X, Wu H, Jiang R, Sun G, Shen J, Ma M, Ma C, Zhang S, Huang Z, Wu Q, et al. The antidepressant effects of a-tocopherol are related to activation of autophagy via the AMPK/mTOR pathway. Eur J Pharmacol. 2018;833:1–7. doi: 10.1016/j.ejphar.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 63.Hosseinpour-Moghaddam K, Caraglia M, Sahebkar A. Autophagy induction by trehalose: molecular mechanisms and therapeutic impacts. J Cell Physiol. 2018;233:6524–6543. doi: 10.1002/jcp.26583. [DOI] [PubMed] [Google Scholar]
- 64.Murray CJL, Lopez AD. Alternative projections of mortality and disability by cause 1990-2020: Global Burden of Disease Study. Lancet. 1997;349:1498–1504. doi: 10.1016/S0140-6736(96)07492-2. [DOI] [PubMed] [Google Scholar]
- 65.Lépine JP, Briley M. The increasing burden of depression. Neuropsychiatr Dis Treat. 2011 doi: 10.2147/NDT.S19617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kelly K, Posternak M, Jonathan EA. Toward achieving optimal response: understanding and managing antidepressant side effects. Dialogues Clin Neurosci. 2008;10:409–418. doi: 10.31887/DCNS.2008.10.4/kkelly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ferguson JM. SSRI Antidepressant medications: adverse effects and tolerability. Prim Care Companion J Clin Psychiatry. 2001;3:22–27. doi: 10.4088/pcc.v03n0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thase ME. Preventing relapse and recurrence of depression: a brief review of therapeutic options. CNS Spectr. 2006;11(12 Suppl. 15):12–21. doi: 10.1017/s1092852900015212. [DOI] [PubMed] [Google Scholar]
- 69.Cattaneo A, Gennarelli M, Uher R, Breen G, Farmer A, Aitchison KJ, Craig IW, Anacker C, Zunsztain PA, McGuffin P, et al. Candidate genes expression profile associated with antidepressants response in the GENDEP study: differentiating between baseline “predictors” and longitudinal “targets”. Neuropsychopharmacology. 2013;38:377–385. doi: 10.1038/npp.2012.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Y, Liang Z, Gu Z, Qin Z. Molecular mechanism and regulation of autophagy. Acta Pharmacol Sin. 2005;26:1421–1434. doi: 10.1111/j.1745-7254.2005.00235.x. [DOI] [PubMed] [Google Scholar]
- 71.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 73.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 74.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 75.Chen T, Dent SYR. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet. 2014;15:93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cuvier O, Fierz B. Dynamic chromatin technologies: from individual molecules to epigenomic regulation in cells. Nat Rev Genet. 2017;18:457–472. doi: 10.1038/nrg.2017.28. [DOI] [PubMed] [Google Scholar]
- 77.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 78.Puri D, Gala H, Mishra R, Dhawan J. High-wire, act: The poised genome and cellular memory. FEBS J. 2015;282:1675–1691. doi: 10.1111/febs.13165. [DOI] [PubMed] [Google Scholar]
- 79.Vastenhouw NL, Schier AF. Bivalent histone modifications in early embryogenesis. Curr Opin Cell Biol. 2012;24:374–386. doi: 10.1016/j.ceb.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 82.Vellai T. Autophagy genes and ageing. Cell Death Differ. 2009;16:94–102. doi: 10.1038/cdd.2008.126. [DOI] [PubMed] [Google Scholar]
- 83.Füllgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, Hermanson O, Rosenfeld MG, Klionsky DJ, Joseph B. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500:468–471. doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.An PNT, Shimaji K, Tanaka R, Yoshida H, Kimura H, Fukusaki E, Yamaguchi M. Epigenetic regulation of starvation-induced autophagy in Drosophila by histone methyltransferase G9a. Sci Rep. 2017;7:7343. doi: 10.1038/s41598-017-07566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Narvajas AA-M, Gomez TS, Zhang J-S, Mann AO, Taoda Y, Gorman JA, Herreros-Villanueva M, Gress TM, Ellenrieder V, Bujanda L, et al. Epigenetic regulation of autophagy by the methyltransferase G9a. Mol Cell Biol. 2013;33:3983–3993. doi: 10.1128/MCB.00813-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ding J, Li T, Wang X, Zhao E, Choi J-H, Yang L, Zha Y, Dong Z, Huang S, Asara JM, et al. The histone H3 methyltransferase G9A epigenetically activates the serine-glycine synthesis pathway to sustain cancer cell survival and proliferation. Cell Metab. 2013;18:896–907. doi: 10.1016/j.cmet.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou Y, Grummt I. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr Biol. 2005;15:1434–1438. doi: 10.1016/j.cub.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 88.Shogren-Knaak M, Ishii H, Sun J-M, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311 doi: 10.1126/science.1124000. 844 LP-847. [DOI] [PubMed] [Google Scholar]
- 89.Shin H-JR, Kim H, Kim K Il, Baek SH. Epigenetic and transcriptional regulation of autophagy. Autophagy. 2016;12:2248–2249. doi: 10.1080/15548627.2016.1214780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shin HJR, Kim H, Oh S, Lee JG, Kee M, Ko HJ, Kweon MN, Won KJ, Baek SH. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016;534:553–557. doi: 10.1038/nature18014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wei FZ, Cao Z, Wang X, Wang H, Cai MY, Li T, Hattori N, Wang D, Du Y, Song B, et al. Epigenetic regulation of autophagy by the methyltransferase EZH2 through an MTOR-dependent pathway. Autophagy. 2015;11:2309–2322. doi: 10.1080/15548627.2015.1117734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Biga PR, Latimer MN, Froehlich JM, Gabillard J-C, Seiliez I. Distribution of H3K27me3, H3K9me3, and H3K4me3 along autophagy-related genes highly expressed in starved zebrafish myotubes. Biol Open. 2017;6:1720–1725. doi: 10.1242/bio.029090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheedipudi S, Puri D, Saleh A, Gala HP, Rumman M, Pillai MS, Sreenivas P, Arora R, Sellathurai J, Schrøder HD, et al. A fine balance: epigenetic control of cellular quiescence by the tumor suppressor PRDM2/RIZ at a bivalent domain in the cyclin a gene. Nucleic Acids Res. 2015;43:6236–6256. doi: 10.1093/nar/gkv567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, Criollo A, Galluzzi L, Malik SA, Vitale I, et al. The life span-prolonging effect of sirtuin-1 is mediated by autophagy. Autophagy. 2010;6:186–188. doi: 10.4161/auto.6.1.10817. [DOI] [PubMed] [Google Scholar]
- 95.Jeong JK, Moon MH, Lee YJ, Seol JW, Park SY. Autophagy induced by the class III histone deacetylase Sirt1 prevents prion peptide neurotoxicity. Neurobiol Aging. 2013;34:146–156. doi: 10.1016/j.neurobiolaging.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 96.Ou X, Lee MR, Huang X, Messina-Graham S, Broxmeyer HE. SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells. 2014;32:1183–1194. doi: 10.1002/stem.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen S, Jing Y, Kang X, Yang L, Wang DL, Zhang W, Zhang L, Chen P, Chang JF, Yang XM, et al. Histone H2B monoubiquitination is a critical epigenetic switch for the regulation of autophagy. Nucleic Acids Res. 2017;45:1144–1158. doi: 10.1093/nar/gkw1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Peeters JGC, Picavet LW, Coenen SGJM, Mauthe M, Vervoort SJ, Mocholi E, de Heus C, Klumperman J, Vastert SJ, Reggiori F, et al. Transcriptional and epigenetic profiling of nutrient-deprived cells to identify novel regulators of autophagy. Autophagy. 2018;15:1–15. doi: 10.1080/15548627.2018.1509608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Denton D, Aung-Htut MT, Lorensuhewa N, Nicolson S, Zhu W, Mills K, Cakouros D, Bergmann A, Kumar S. UTX coordinates steroid hormone-mediated autophagy and cell death. Nat Commun. 2013;4:2916. doi: 10.1038/ncomms3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang B, Fan X, Ma C, Lei H, Long Q, Chai Y. Downregulation of KDM4A suppresses the survival of glioma cells by promoting autophagy. J Mol Neurosci. 2016;60:137–144. doi: 10.1007/s12031-016-0796-6. [DOI] [PubMed] [Google Scholar]
- 101.Ambrosio S, Sacca CD, Amente S, Paladino S, Lania L, Majello B. Lysine-specific demethylase LSD1 regulates autophagy in neuroblastoma through SESN2-dependent pathway. Oncogene. 2017;36:6701. doi: 10.1038/onc.2017.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Füllgrabe J, Ghislat G, Cho D-H, Rubinsztein DC. Transcriptional regulation of mammalian autophagy at a glance. J Cell Sci. 2016;129:3059–3066. doi: 10.1242/jcs.188920. [DOI] [PubMed] [Google Scholar]
- 103.Pietrocola F, Izzo V, Niso-Santano M, Vacchelli E, Galluzzi L, Maiuri MC, Kroemer G. Regulation of autophagy by stress-responsive transcription factors. Semin Cancer Biol. 2013;23:310–322. doi: 10.1016/j.semcancer.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 104.Zannas AS, Wiechmann T, Gassen NC, Binder EB. Gene - stress - epigenetic regulation of FKBP5: clinical and translational implications. Neuropsychopharmacology. 2016;51:261–274. doi: 10.1038/npp.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gassen NC, Hartmann J, Zschocke J, Stepan J, Hafner K, Zellner A, Kirmeier T, Kollmannsberger L, Wagner KV, Dedic N, et al. Association of FKBP51 with priming of autophagy pathways and mediation of antidepressant treatment response: evidence in cells, mice, and humans. PLoS Med. 2014;11:e1001755. doi: 10.1371/journal.pmed.1001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gassen NC, Fries GR, Zannas AS, Hartmann J, Zschocke J, Hafner K, Carrillo-Roa T, Steinbacher J, Preibinger SN, Hoeijmakers L, et al. Chaperoning epigenetics: FKBP51 decreases the activity of DNMT1 and mediates epigenetic effects of the antidepressant paroxetine. Sci Signal. 2015;8:ra119. doi: 10.1126/scisignal.aac7695. [DOI] [PubMed] [Google Scholar]
- 107.Guo H, Chitiprolu M, Gagnon D, Meng L, Perez-Iratxeta C, Lagace D, Gibbings D. Autophagy supports genomic stability by degrading retrotransposon RNA. Nat Commun. 2014;5:5276. doi: 10.1038/ncomms6276. [DOI] [PubMed] [Google Scholar]
- 108.Hunter RG, McEwen BS, Pfaff DW. Environmental stress and transposon transcription in the mammalian brain. Mob Genet Elem. 2013;3:e24555. doi: 10.4161/mge.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rusiecki JA, Chen L, Srikantan V, Zhang L, Yan L, Polin ML, Baccarelli A. DNA methylation in repetitive elements and post-traumatic stress disorder: a case-control study of US military service members. Epigenomics. 2012;4:29–40. doi: 10.2217/epi.11.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hunter RG, Murakami G, Dewell S, Seligsohn M, Baker MER, Datson NA, McEwen BS, Pfaff DW. Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc Natl Acad Sci USA. 2012;109:17657–17662. doi: 10.1073/pnas.1215810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metab. 2014;19:373–379. doi: 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Salmon AB. About-face on the metabolic side effects of rapamycin. Oncotarget. 2015;20:2585–2586. doi: 10.18632/oncotarget.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Miller RA, Harrison DE, Astle CM, Fernandez E, Flurkey K, Han M, Javors MA, Li X, Nadon NL, Nelson JF, et al. Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell. 2014;13:468–477. doi: 10.1111/acel.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol. 2012;30:679. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- 115.Devita VT, Young RC, Canellos GP. Combination versus single agent chemotherapy: a review of the basis for selection of drug treatment of cancer. Cancer. 2018;35:98–110. doi: 10.1002/1097-0142(197501)35:1<98::aid-cncr2820350115>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 116.Nelander S, Wang W, Nilsson B, She Q, Pratilas C, Rosen N, Gennemark P, Sander C. Models from experiments: combinatorial drug perturbations of cancer cells. Mol Syst Biol. 2008;4:216. doi: 10.1038/msb.2008.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Heerboth S, Lapinska K, Snyder N, Leary M, Rollinson S, Sarkar S. Use of epigenetic drugs in disease: an overview. Genet Epigenetics. 2014;6:9–19. doi: 10.4137/GEG.S12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 119.Lunyak VV, Rosenfeld MG. Epigenetic regulation of stem cell fate. Hum Mol Genet. 2008;17:R28–R36. doi: 10.1093/hmg/ddn149. [DOI] [PubMed] [Google Scholar]
- 120.Covic M, Karaca E, Lie DC. Epigenetic regulation of neurogenesis in the adult hippocampus. Heredity. 2010;105:122–134. doi: 10.1038/hdy.2010.27. [DOI] [PubMed] [Google Scholar]
- 121.Sun J, Sun J, Ming G, Song H. Epigenetic regulation of neurogenesis in the adult mammalian brain. Eur J Neurosci. 2011;33:1087–1093. doi: 10.1111/j.1460-9568.2011.07607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhou H, Wang B, Sun H, Xu X, Wang Y. Epigenetic regulations in neural stem cells and neurological diseases. Stem Cells Int. 2018;2018 doi: 10.1155/2018/6087143. 6087143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 124.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]