Abstract
Background
The protein kinase Target Of Rapamycin (TOR) is a nexus for the regulation of eukaryotic cell growth. TOR assembles into one of two distinct signalling complexes, TOR complex 1 (TORC1) and TORC2 (mTORC1/2 in mammals), with a set of largely non-overlapping protein partners. (m)TORC1 activation occurs in response to a series of stimuli relevant to cell growth, including nutrient availability, growth factor signals and stress, and regulates much of the cell’s biosynthetic activity, from proteins to lipids, and recycling through autophagy. mTORC1 regulation is of great therapeutic significance, since in humans many of these signalling complexes, alongside subunits of mTORC1 itself, are implicated in a wide variety of pathophysiologies, including multiple types of cancer, neurological disorders, neurodegenerative diseases and metabolic disorders including diabetes.
Methodology
Recent years have seen numerous structures determined of (m)TOR, which have provided mechanistic insight into (m)TORC1 activation in particular, however the integration of cellular signals occurs upstream of the kinase and remains incompletely understood. Here we have collected and analysed in detail as many as possible of the molecular and structural studies which have shed light on (m)TORC1 repression, activation and signal integration.
Conclusions
A molecular understanding of this signal integration pathway is required to understand how (m)TORC1 activation is reconciled with the many diverse and contradictory stimuli affecting cell growth. We discuss the current level of molecular understanding of the upstream components of the (m)TORC1 signalling pathway, recent progress on this key biochemical frontier, and the future studies necessary to establish a mechanistic understanding of this master-switch for eukaryotic cell growth.
Keywords: mTORC1, nutrient sensing, GATOR complex, TSC complex, Rag GTPases, Rheb
Introduction
The Target Of Rapamycin (TOR) was first identified in Saccharomyces cerevisiae by Hall and colleagues through a genetic screen for mutants resistant to the antifungal activity of the drug Rapamycin (Heitman et al., 1991). The Hall group went on to show that there are two yeast TOR genes, both of which encode kinases (Kunz et al., 1993; Helliwell et al., 1994), and the mammalian homolog mTOR was discovered and published by four groups independently thereafter (Brown et al., 1994; Chiu et al., 1994; Sabatini et al., 1994; Sabers et al., 1995). Since these seminal studies, (m)TORC1 has emerged as the key driver of cell biogenesis throughout the eukaryotes: (m)TORC1 activity has been shown to be stimulated by growth factor signaling and nutrients such as glucose and amino acids, whereas under conditions of stress, such as starvation, energy or oxygen deficiency, or DNA damage, (m)TORC1 activity is robustly supressed (Laplante and Sabatini, 2012).
The core components of (m)TORC1 are (m)TOR, its catalytic core, regulatory-associated protein of mTOR (RAPTOR; Kog1 in yeast) (Kim et al., 2002; Loewith et al., 2002), and mammalian lethal with Sec13 8 (mLST8; Lst8 in yeast) (Roberg et al., 1997; Chen and Kaiser, 2003; Kim et al., 2003). Proline-rich Akt substrate of 40 kDa (PRAS40) and DEP domain-containing partner of TOR (DEPTOR) are additional components which complete the roster, although these proteins are inhibitors of (m)TOR kinase activity (Vander Haar et al., 2007; Oshiro et al., 2007; Sancak et al., 2007; Peterson et al., 2009). When activated, (m)TORC1 phosphorylates substrates with roles in stimulating ribosome biogenesis, protein and lipid synthesis, and inhibition of autophagy (Laplante and Sabatini, 2012; Kennedy and Lamming, 2016). Many of these proteins feature a TOR signaling (TOS) motif, characterized by alternating hydrophobic and acidic residues, which is specifically recruited by RAPTOR (Schalm and Blenis, 2002; Nojima et al., 2003; Schalm et al., 2003).
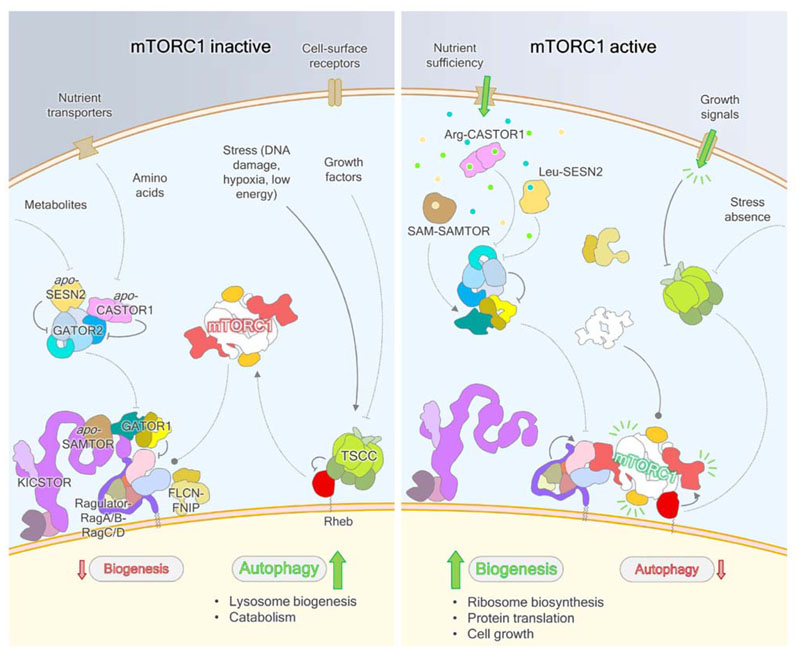
(m)TORC1 activity is understood to be principally controlled by the nucleotide state of three small GTPases. Full mTORC1 activation requires its recruitment to the lysosome by heterodimeric Rag GTPases (Sancak et al., 2008), whereupon GTP-bound Rheb GTPases allosterically activate the kinase (Long et al., 2005; Sato et al., 2009) (Fig. 1).
Figure 1. Schematic of mTORC1 regulation in response to multiple stimuli.
In the absence of the appropriate signals, mTORC1 is not localized to the lysosome by RagA/B-RagC/D and therefore not allosterically activated by Rheb. Inhibitory GAPs for the RagA/B and Rheb GTPases – GATOR1 and TSCC, respectively – act on their targets, while FLCN-FNIP activates RagC/D. In the presence of amino acids and key metabolites, SESN2 and CASTOR1 dissociate from GATOR2, enabling it to inhibit GATOR1 through an as-yet incompletely described mechanism, while SAMTOR dissociates from GATOR1-KICSTOR and may enable recognition of GATOR1 by GATOR2. In the absence of external stresses, such as DNA damage, low cellular energy status and reactive oxygen species production, as well as positive growth factor signaling, TSCC-mediated Rheb inhibition is relieved by posttranslational modifications of the integrator complex, which likely lead to dispersion of TSCC from the lysosome surface. Both the Rag heterodimer and Rheb GTPases can now cooperatively activate mTORC1 to stimulate biogenesis.
Signals are relayed to mTORC1 through these small GTPases, whose nucleotide binding states are regulated by a series of important protein complexes. The TSC complex (TSCC) acts as a GTPase activating protein (GAP) for Rheb, employing a GAP domain within the TSC2 subunit (Castro et al., 2003; Garami et al., 2003; Inoki et al., 2003; Saucedo et al., 2003; Stocker et al., 2003; Tee et al., 2003; Zhang et al., 2003). Upstream regulation of the Rag GTPases is more baroque, featuring at least four large protein complexes including: GATOR1, a GAP for RagA/RagB/Gtr1 (Neklesa and Davis, 2009; Bar-Peled et al., 2013; Panchaud et al., 2013a); FLCN-FNIP (Lst4-Lst7 in yeast), a GAP for RagC/RagD/Gtr2 (Péli-Gulli et al., 2015; Petit et al., 2013; Tsun et al., 2013); GATOR2, an inhibitor of GATOR1 (Bar-Peled et al., 2013; Panchaud et al., 2013a, 2013b); and, KICSTOR, which is thought to regulate the GATOR complexes (Peng et al., 2017; Wolfson et al., 2017). The LAMTOR/Ragulator complex is thought to be a guanine nucleotide exchange factor (GEF) for the RagA/B GTPases in mammals (Bar-Peled et al., 2012), however GEFs for Rheb and RagC/D are yet to be identified. The current consensus understanding within the field is that mTORC1 is only active when both the Rag and Rheb GTPase activation pathways are fully activated, neither being sufficient in isolation.
Recent technological advances have enabled structural and mechanistic characterization of many technically challenging complexes such as mTORC1, (m)TORC2, Ragulator-Rag, and most recently GATOR1 and GATOR1-Rag, shedding increasing light on this key signaling pathway. However, many of the most important upstream components, including the TSCC, GATOR2 and KICSTOR complexes, remain either only partially or completely uncharacterised at a molecular level. Here we provide a survey of the current understanding within the field, first covering the nutrient sensing pathway leading to the Rag GTPases and (m)TORC1 recruitment, and then the plethora of signals communicated through Rheb to (m)TORC1 activation. Finally, we detail the remaining gaps that must be addressed before the complete picture can emerge.
Diverse, independent and ambivalently conserved cytoplasmic sensor proteins detect and signal nutrient insufficiency through the Rag pathway
While it has long been known that (m)TOR responds to nutrient availability (Blommaart et al., 1995; Hara et al., 1998), the direct inputs to (m)TORC1 signaling remained unclear. However, several human proteins are now recognized as key metabolite sensors for the mTORC1 pathway. These include SESN1 and SESN2 (Chantranupong et al., 2014; Parmigiani et al., 2014; Peng et al., 2014), CASTOR1 (Chantranupong et al., 2016), and SAMTOR (Gu et al., 2017), and may also include SLC38A9 (Jung et al., 2015; Rebsamen et al., 2015; Wang et al., 2015) and the vacuolar (v)-ATPase (Zoncu et al., 2011) which have been shown to affect mTORC1 activity in response to amino acid availability. Homologs of only the SESNs have been detected by sequence analysis in other higher eukaryotes such as C. elegans and D. melanogaster, and the known sensor proteins in general are only present in few fungal species, potentially representing species-specific nutritional dependencies (Wolfson and Sabatini, 2017). A great deal of work needs to be done at the periphery of the (m)TORC1 pathway to discover exactly where and how the first steps in nutrient sensing take place, as this is one of its least well-understood aspects. Recent structural and functional studies of the known elements of the pathway have successfully yielded mechanistic insights into how some of these proteins link metabolite detection to mTORC1 signaling.
Sestrins
The Sestrin/SESN proteins were first identified several years before being linked to mTORC1 signaling (Velasco-Miguel et al., 1999; Budanov et al., 2002; Peeters et al., 2003), in a study demonstrating that expression of SESN1 and SESN2 strongly inhibited phosphorylation of the well-characterized mTORC1 substrates ribosomal protein S6 kinase 1 (S6K1) and eukaryotic initiation factor 4E binding protein 1 (4EBP1) (Budanov and Karin, 2008). SESN2 interacts with GATOR2 (Chantranupong et al., 2014; Parmigiani et al., 2014; Kim J S et al., 2015), with the interaction conditionally strengthened by both amino acid deprivation (Chantranupong et al., 2014) and DNA damage-induced stress (Parmigiani et al., 2014). Ectopic SESN2 expression prevents mTORC1 localization to the lysosome in the presence of amino acids (Parmigiani et al., 2014; Peng et al., 2014), while reducing GATOR1-GATOR2, and increasing GATOR1-RagB, association (Kim J S et al., 2015), hence acting as a negative regulator of mTORC1 signaling. Experimentation with individual amino acids led to the discovery that the interaction between SESN2 and GATOR2 is leucine-dependent (Wolfson et al., 2016), and intrinsically coupled to its interaction with GATOR2, and consequently to mTORC1 regulation, as the expression of a GATOR2 binding-deficient mutant, SESN2S190W, does not restore leucine sensitivity to SESN1-3 deficient cells. It has alternatively been suggested to act as a guanine nucleotide dissociation inhibitor (GDI) for RagA (Peng et al., 2014) however subsequent structural studies have not supported this hypothesis. It may be worthwhile considering, in the face of newer observations, that Peng and colleagues did not consider the regulation of SESN2 by amino acids, and therefore SESN2 GDI activity may belong to the apo-protein; however, in this instance their observation between a direct SESN2-RagA/B interaction has not been recapitulated by other groups, and therefore RagA/B-GDI activity remains unproven.
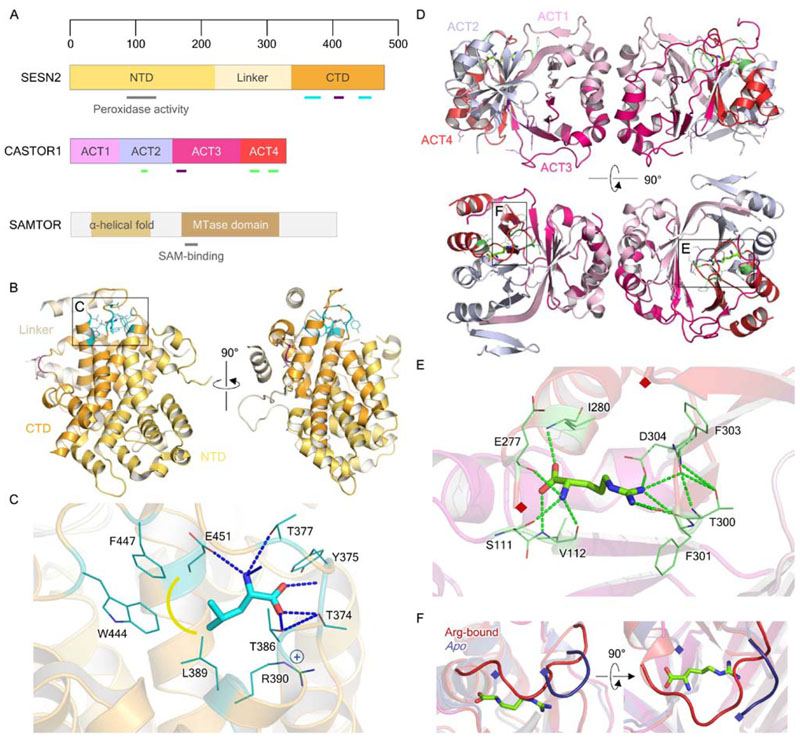
Crystal structures of SESN2 (Kim H et al., 2015; Saxton et al., 2016b) proved particularly revealing. SESN2 shares a common fold with the carboxymucolactone decarboxylase (CMD) protein family, possessing globular CMD-like domains at either terminus, separated by a partially disordered linker (Fig. 2A). CMD family members are commonly homodimers, and therefore SESN2 represents an intramolecular “dimer” of two CMD-like domains, with the N- and C- terminal domains (NTD and CTD, respectively) juxtaposed around a twofold pseudosymmetry axis (Fig. 2B). SESN2 coordinates a single leucine molecule in a pocket in the CTD (Fig. 2C), adjoined by part of the linker, which packs against the side of the pocket. Saxton and colleagues identify an inter-helix loop in the CTD, containing three important threonine residues (T374, T377 and T386), which closes over the binding pocket and is responsible for providing specificity for leucine binding. This closure is thought to relay a conformational change from the binding pocket to the putative GATOR2 interaction surface, of which two residues, D406 and D407, were identified as necessary for GATOR2 recruitment (Kim H et al., 2015; Saxton et al., 2016b). Further mechanistic detail of this interaction remains elusive however, given the lack of any substantial structural or mechanistic understanding of the action of GATOR2.
Figure 2. Structural biology of the nutrient sensors SESN2, CASTOR1, and SAMTOR.
A. Domain organization of SESN2 and CASTOR1 and predicted domain organization of SAMTOR. Black and gray outlines represent structurally resolved or predicted regions, respectively, while colored and gray bars reflect specific features with known or putative functions, respectively. B. Crystal structure of leucine-bound SESN2 (PDB ID: 5DJ4), with bound leucine shown as cyan in stick representation. Leucine- and GATOR2-interacting residues are shown in teal and purple respectively, as highlighted in A. C. Leucine binding pocket of SESN2, as inset in B. Polar contacts are indicated by blue dashed lines, and hydrophobic interactions by yellow. D. Crystal structures of the arginine-bound, and apo, CASTOR1 homodimer (Arg-bound PDB ID: 5I2C; apo: 5GT8), with bound arginine shown as light green in stick representation. Arginine- and GATOR2-interacting residues are shown in green and purple respectively, as shown in A. E. Arginine binding pocket of CASTOR1, as inset in D. Polar contacts are indicated by bright green dashed lines. The loop which closes over the CASTOR1 binding pocket has been omitted for clarity between residues 269-276, indicated by maroon diamonds. F. Comparison of ACT4 residues 270-280 between the arginine-bound and apo CASTOR1, which form a loop over the bound arginine. Residues 275-279 are unobserved in the apo CASTOR1 crystal structure, indicating disorder.
Taken together, the functional and structural data suggest that SESNs act as negative regulators of mTORC1 signaling by inhibiting the function of GATOR2 in the apo state. SESNs have also been suggested to constitute a switch between biogenesis and autophagy given the observation that ULK1, a kinase controlling the induction of autophagy, is able to stimulate phosphorylation of SESN2 either directly or indirectly (Kimball et al., 2016). It seems likely that several stress signals are integrated through the SESNs, and thus they may represent an important, multifaceted sensor upstream of not only mTORC1, but related signaling circuits as well.
CASTORs
The CASTOR proteins (originally GATSL2 and 3) were identified as through their activity as binding partners of the GATOR2 components WDR24, WDR59 and MIOS (Huttlin et al., 2015), although only subsequent biochemical verification of these interactions linked them to the mTORC1 pathway, with GATSL3 renamed CASTOR1, for cellular arginine sensor for mTORC1, and GATSL2 CASTOR2 (Chantranupong et al., 2016). CASTOR1 and 2 homo- and heterodimerize with one another, with the CASTOR1 homodimer interacting with GATOR2 most strongly. CASTORs contain tandem aspartate kinase, chorismate mutase and TyrA (ACT) domains (Fig. 2A), which have an evolutionarily conserved function in binding small molecules. In this case the CASTOR1-GATOR2 interaction is arginine-dependent, with arginine supressing the association of CASTOR1 with GATOR2 to sense arginine upstream of mTORC1. CASTOR2 binding to GATOR2, on the other hand, was amino acid-insensitive; thus, CASTOR2 may serve to retain responsivity of mTORC1 to nutrient signals through CASTOR1 by damping the initial activation of the kinase, affecting the properties of signal feedback as opposed to direct amino acid detection.
The crystal structure of the arginine-bound CASTOR1 homodimer (Gai et al., 2016b; Saxton et al., 2016a; Xia et al., 2016) revealed that each monomer contains four ACT domains (Fig. 2D), and arginine binds to each monomer in a deep cleft at the intramolecular ACT2/4 interface (Fig. 2E). Mutation of critical residues D304, which coordinates the arginine guanidinium group, and S111, which binds the free amine, resulted in complete loss of arginine binding, as well as constitutive GATOR2 binding and strong mTORC1 inhibition even in the presence of arginine. Residues Y118, Q119 and D121 were identified as necessary for the GATOR2 interaction (Saxton et al., 2016a; Gai et al., 2016b) and further noted to form patches on the same side of the CASTOR1 homodimer, which are distinct surfaces to the arginine binding pocket (Gai et al., 2016b). A loop comprising residues 270-280 sits over the arginine binding site and ‘locks’ the bound amino acid in place, however is disordered in the apo-state (Guo and Deng; PDB ID: 5GT8) (Fig. 2F). This observation supports the hypothesis that this key loop mediates a conformational switch between the apo- and occupied states, allosterically modulating the GATOR2 binding site.
It is noticeable that the proposed mechanism is reminiscent of that implied for the SESNs, although the means by which this small conformational change affects the GATOR1-GATOR2 interaction remains unknown. Intriguingly, the binding of the CASTOR1 homodimer to GATOR2 is not competitive with SESN2: both proteins can bind the GATOR2 complex simultaneously. WDR24 and SEH1L are known to form a minimal complex sufficient for interaction with SESN2 (Parmigiani et al., 2014), and these GATOR2 components, with the addition of MIOS, were also identified as sufficient to bind CASTOR1 (Chantranupong et al., 2016; Gai et al., 2016b). It will remain for future structural and mechanistic studies of the GATOR2 complex and interacting proteins to unpick this knot, however the available evidence would imply that signal integration appears likely to occur at the GATOR2 binding step.
SAMTOR
SAMTOR, originally C7orf60, has been recently defined an S-adenosylmethionine sensor upstream of mTOR in mammals (Gu et al., 2017). Sequence analysis of SAMTOR suggests the presence of a class I Rossman fold, typically found in methyltransferases (Fig. 2A), which has been corroborated by in vitro binding assays establishing micromolar affinity binding coefficients for both S-adenosylmethionine (SAM) and S-adenosylhomocysteine (SAH). SAM binding appears to constitute a physiologic signal of methionine sufficiency, and overexpression of even wild type SAMTOR is sufficient to inhibit mTORC1 signaling, implying that the apo-form represents the negative regulator, as for the SESN and CASTOR proteins. SAMTOR is thought to interact with the GATOR1 and KICSTOR complexes simultaneously (Gu et al., 2017; Wolfson et al., 2017), possibly through a tripartite interface given that all three proteins are required for any interaction. Furthermore, SAM/SAH binding reduces the association of SAMTOR with GATOR1-KICSTOR, analogously to the way in which leucine and arginine disrupt the interaction with GATOR2 of SESN2 and CASTOR1 respectively. Further development of the exact mechanisms by which SAMTOR achieves its function must await structural studies of the protein and its complexes.
Further nutrient sensors – an established paradigm
The presence of sensors specific for leucine and arginine in mammalian cells likely reflect their metabolic importance. For example, leucine is one of two amino acids which are completely ketogenic, and is known to potently activate muscle protein synthesis (Grabacka et al., 2016; Norton and Layman, 2006); additionally, arginine is the precursor of nitric oxide, ornithine and urea, all key molecules at the intersections of several critically important metabolic pathways (Morris, 2006). It appears highly likely that further nutrient sensors feeding into the (m)TORC1 signaling pathway await discovery given the great diversity in nutrient signals to be taken account of and the lack of orthologs of even the most well-established sensors (Wolfson and Sabatini, 2017). Although it is likely evolutionarily redundant to encode sensors for isoleucine and valine, the other branched-chain amino acids, it is tempting to speculate that specific sensors may have evolved for more metabolically unique amino acids such as histidine, and other key nutrients. Those discovered to date are diverse in structure, but follow an established paradigm by interacting with further proteins within the pathway in the case of insufficiency rather than sufficiency of the nutrient in question. This has the evolutionary benefit of allowing a small number of conserved complexes to trivially integrate a much larger number of diverse repressive inputs, enabling sensitivity while retaining responsivity in downstream signaling.
Nutrient signaling inhibits (m)TORC1 recruitment to the lysosome by controlling the nucleotide state of the RagA/B GTPases
As our understanding of mTORC1 signaling in response to specific stimuli increases, inputs such as the levels of individual metabolites to the integrator complexes are being recognized as increasingly important (Wolfson and Sabatini, 2017). These complexes, comprising GATOR1 and GATOR2 (Dokudovskaya et al., 2011; Bar-Peled et al., 2013; Panchaud et al., 2013a), and KICSTOR (Peng et al., 2017; Wolfson et al., 2017) constitute a network of integrators which filter the available chemical inputs to produce discrete on-or-off signals to the (m)TORC1 machinery. Though these complexes remain incompletely structurally characterized, a developing body of work is enabling us to understand how signals are integrated and communicated through Rag A/B.
The GATOR complexes
GATOR complexes 1 (GATOR1) and GATOR2, for GAP Activity Toward Rags, are key evolutionarily conserved signal integration and transduction complexes that control the nucleotide state of the RagA/B GTPases. They were first identified and characterized in S. cerevisiae, as Seh1-associated complex (SEAC) inhibiting TORC1 (SEACIT) and SEAC activating TORC1 (SEACAT) respectively (Dokudovskaya etal., 2011). GATOR1 is a negative regulator of (m) TORC1 signaling, whereas GATOR2 inhibits GATOR1 in turn, thereby acting as a positive regulator. SEACIT/ GATOR1 consists of three proteins, Sea1/Iml1, Npr2 and Npr3; while SEACAT/GATOR2 comprises five, Seh1, Sec13, Sea2/Rtc1, Sea3/Mtc5 and Sea4. Whereas Sea1-Sea4 were previously uncharacterised, Npr2 and Npr3 were previously annotated with roles in autophagy, and shown to form an evolutionarily conserved heterodimer (Neklesa and Davis, 2009; Wu and Tu, 2011). Neklesa and colleagues also provided the first lines of evidence that the human homologs, Npr2-like (NPRL2) and NPRL3, act as repressors of mTORC1. Subsequently, orthologous mammalian complexes were identified and categorised, with GATOR1 containing Iml1 homolog DEPDC5, NPRL2 andNPRL3; and GATOR2, consisting of SEC13, SEH1L, Sea2 homolog WDR24, Sea3 homolog WDR59, and Sea4 homolog MIOS (Bar-Peled et al., 2013). Both the human and S. cerevisiae GATOR1 complexes possess evolutionarily conserved GAP activity toward the Gtr1/RagA/B GTPase, while GATOR2 inhibits the activity of GATOR1 through an as-yet-undetermined mechanism (Bar-Peled et al., 2013; Panchaud et al., 2013a).
Bioinformatic analysis of the S. cerevisiae proteins revealed predicted architectures commonly found in vesicle coatomer and tethering proteins (Lee and Goldberg, 2010; Faini et al., 2013; Balderhaar and Ungermann, 2013), and these architectures are largely conserved in the mammalian homologs, especially in GATOR2 components (Dokudovskaya etal., 2011) (Fig. S1A). Sec13/SEC13 and Seh1/SEH1L are both composed of six-bladed β-propellers (Fig. S1B and 1C, respectively) that commonly serve as sites of protein-protein interactions (Xu and Min, 2011), and are subunits directly shared with coatomers and nuclear membrane complexes. The structure of S. cerevisiae Sec13 was first solved at high resolution as part of a Sec13-Sec31 COPII subcomplex (Fath et al., 2007), before both yeast and human forms were later resolved as part of numerous nuclear pore complex (NPC)-related (Hsia et al., 2007; Brohawn and Schwartz, 2009; Nagy et al., 2009; Stuwe et al., 2015; Kelley et al., 2015) and COPII-related (Whittle and Schwartz, 2010; Bharucha et al., 2013; Zanetti et al., 2013) subcomplexes. Seh1, like Sec13, has been consistently resolved at high resolution as part of NPC-related subcomplexes (Brohawn et al., 2008; Debler et al., 2008; Stuwe et al., 2015). WDR24, WDR59 and MIOS are notably also predicted to contain β-propeller domains at their N-termini, separated from C-terminal RING domains, commonly associated with E3 ubiquitin ligases (Metzger et al., 2012). WDR59 is predicted to contain a RING finger and WD domain-containing proteins and DEAD-like helicases (RWD) domain following the β-propeller, which are a common architectural motif observed in kinetochore proteins (Schmitzberger and Harrison, 2012). A structural modeling approach was used to derive an architecture for the GATOR complex based on bioinformatic predictions and interactivity assays (Algret et al., 2014), however the lack of homologous structures for many of the GATOR components render this model highly speculative. Nonetheless in the proposed model the β-propellers of Seh1, Sec13, Sea2 and Sea4 form a cluster, similar to that seen in COPI and COPII vesicle coat complexes (Fath et al., 2007; Lee and Goldberg, 2010). This exchange with the membrane trafficking machinery raises the possibility that mTORC1 is also regulated by signals originating in nucleocytoplasmic transport and vesicle transport processes through the Rag GTPase axis (Panchaud et al., 2013b).
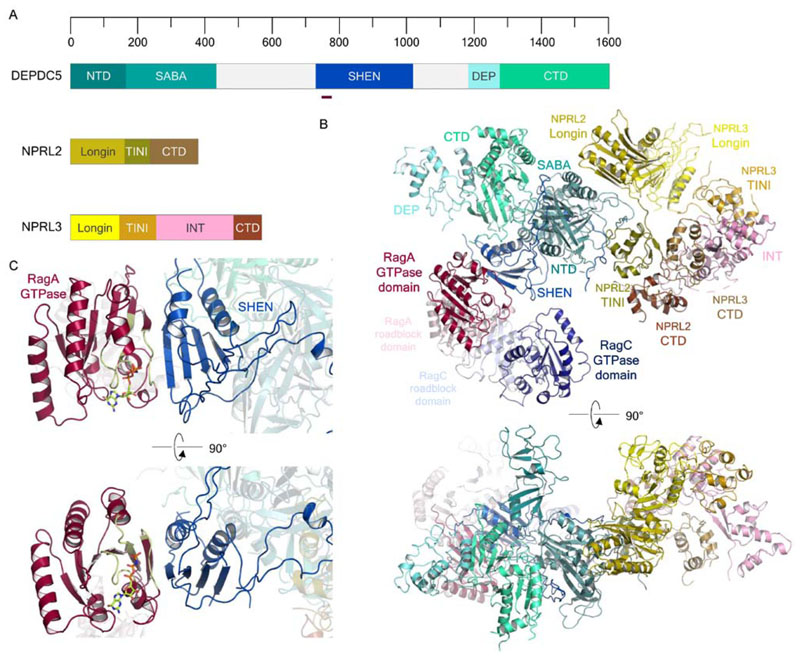
While GATOR2 remains structurally uncharacterised, the structure of the mammalian GATOR1 complex was recently resolved by cryo-EM (Shen et al., 2018), revealing the architecture of each GATOR1 component (Fig. 3A) and the structure of a stable complex with the Rag GTPases (Fig. 3B). NPRL2 and NPRL3 form a pseudosymmetric heterodimer with their N-terminal longin domains and CTDs, interacting together with the DEPDC5 Structural Axis for Binding Arrangement (SABA) domain. The DEPDC5 Steric Hindrance for Enhancement of Nucleotidase activity (SHEN) domain interacts with GTP analog-bound RagA proximal to the nucleotide binding pocket (Fig. 3C), however the interaction captured in the structure does not appear to be responsible for stimulation of GTP hydrolysis. Kinetic assays for GTP hydrolysis suggest that a weaker interaction mediates true GAP activity, and this is thought to be performed by NPRL2-NPRL3 (Shen et al., 2018, 2017). GATOR1 is thus presumed to interact with the Rag heterodimer in two ways: the ‘inhibitory’ conformation, captured in the resolved cryo-EM structure, which binds to GTP-bound RagA with higher relative affinity but insufficient to activate GTP hydrolysis, and a lower-affinity interaction dependent on the NPRL2-NPRL3 heterodimer that stimulates GTPase activity. NPRL2 and NPRL3 form a heterodimer in part using their N-terminal longin domains (Zhang D et al., 2012; Shen et al., 2018). It is intriguing that a recent structure of the Chaetomium thermophilum Mon1-Ccz1-Ypt7 complex highlighted that the Mon1-Ccz1 GEF contacts its cognate GTPase Ypt7 using one face of a conserved longin domain heterodimer (Kiontke et al., 2017). The conservation of this domain architecture in GTPase-regulator interactions suggests that the longin domains of NPRL2-NPRL3 may well bind the Rag GTPases in a similar way. Studies of the NPRL2-NPRL3-Rag heterodimer interaction are required to provide insights into the catalytic GAP mechanism employed by GATOR1, and hence enable us to understand exactly how these GATOR1-Rag interactions differ to regulate the GTPases. The physiologic relevance of the inhibitory binding mode is proposed to be to dampen mTORC1 activation in response to sustained nutrient sufficiency, although such bimodal activity has not previously been observed between a GTPase and its GAP. It is important to note that NPRL2 and NPRL3 are believed to constitute the GATOR2 binding site of GATOR1 (Shen et al., 2018). Since NPRL2 and NPRL3 are also proposed to constitute the weakly binding GAP for the Rag GTPases, it is plausible that GATOR2 inhibits GATOR1 GAP activity by sterically hindering binding of NPRL2-NPRL3 to RagA/B.
Figure 3. Structural biology of the GATOR1 complex.
(A) Domain organization of DEPDC5, NPRL2 and NPRL3. The dark red bar indicates the region of the DEPDC5 SHEN domain which interacts with RagA. (B) Cryo-electron microscopy structure of the mammalian GATOR1-RagA-RagC complex (PDB ID: 6CES; EMDB: EMD-7464). (C) Focused view of the DEPDC5SHEN-RagAGTPase domain interaction, highlighting the end-to-end β-strand interaction of the two domains and its proximity to the RagAGTPase nucleotide binding pocket. The RagA P loop, switch I and II regions are colored pale green. Bound GTP is shown as light green in stick representation.
Mutations in the mammalian GATOR1 genes have been recently linked to focal epilepsies and familial cortical dysplasia, previously not considered to have an underlying genetic basis, supporting a multifaceted role for these proteins in multiple cellular processes (Baldassari et al., 2016; Ricos et al., 2016). Detailed characterization of the interactions between mammalian GATOR2 and GATOR1 will likely provide some insights into the associated pathophysiologies, since unlike in S. cerevisiae, the mammalian complexes do not appear to form a stable octamer, instead performing their functions as independent complexes dynamically associating in mammalian cells (Bar-Peled et al., 2013; Panchaud et al., 2013b).
KICSTOR
The KICSTOR complex is also a key component of the RagA/B control pathway, responsible for the localization of the Rag-GAP GATOR1 to its GTPase substrates. It consists of the proteins KPTN, ITFG2, C12orf66, and SZT2, whose initial letters give the complex its name, which were identified as DEPDC5 interactors and form a distinct complex (Wolfson et al., 2017), while independently SZT2 was found as part of the SESN2 interactome (Peng et al., 2017). The integrity of KICSTOR depends on the largest subunit, SZT2; this component is also responsible for the interaction of KICSTOR with GATOR1, as well as GATOR2, although the latter interaction is indirect (Wolfson et al., 2017). Knockout studies targeting SZT2 impair the localization of GATOR1 to lysosomes, but not that of GATOR2 or the Rag GTPases. The SZT2 component appears to be necessary for coordinated GATOR1 and GATOR2 binding, and is necessary for GATOR1-dependent inactivation ofmTORC1 on the lysosome, as its loss results in constitutive mTORC1 activity and lysosomal localization (Wolfson et al., 2017; Peng et al., 2017).
SZT2 is the largest subunit of KICSTOR, at 3432 residues long, however it exhibits very little homology with any other known proteins or domains (Fig. S2A). Bioinformatic predictions indicate a possible von Willebrand factor (vWA)-like domain toward the N terminus, which is commonly associated with cell-cell adhesion (Whittaker and Hynes, 2002). ITFG2 and KPTN, both much smaller proteins, are both predicted to have almost completely β-stranded architecture, also featuring putative adhesion motifs: ITFG2 with two atypical FG-GAP motifs frequently found in α-integrins (Springer, 1997); and, KPTN with a VCBS motif found in some fungal and bacterial adhesion proteins (Wenter et al., 2010). The FG-GAP repeats as found in integrin α-subunits form a β-propeller (Xiong et al., 2001), consistent with the prediction for the structure of ITFG2. C12orf66, on the other hand, was predicted to be almost completely α-helical, with a putative coiled-coiled region at its N terminus followed by a domain of unknown function (DUF) with predominantly α-helical architecture. The crystal structure of the C-terminal DUF from the Mus musculus C12orf66 homolog (PDB ID: 2GNX) (Fig. S2B) revealed a helical bundle architecture for this domain, similar to that of the classical longin domain. The structure is reminiscent of SNARE proteins, which form helical bundles at membrane contact/fusion sites, and notably form a subfamily of longin domain-containing proteins (De Franceschi et al., 2014; Daste et al., 2015), but no further structural information is currently available.
Mutations in SZT2 has been linked to epilepsy (Frankel et al., 2009; Basel-Vanagaite et al., 2013), and genetic aberrations in KPTN and C12orf66 have been linked to brain malformations (Baple et al., 2014; Mc Cormack et al., 2015; Pajusalu et al., 2015). Consistent with the notion that these abnormalities are associated with mTORC1 hyperactivation and GATOR1 inactivation (Baldassari et al., 2016; Baulac, 2016), KICSTOR clearly plays a role in the GATOR1-dependent repression of mTORC1 signaling. SZT2 contains several key regions that enable interaction with GATOR1 and GATOR2 respectively (Peng et al., 2017), and more recent immunoprecipitation data suggests that a SZT2-DEPDC5 interaction occurs in the absence of other GATOR components (Shen et al., 2018). It has previously been noted that the GATOR proteins exhibit architectural similarity to vesicle coat proteins as well as the related vesicle tethering complexes of the CORVET, HOPS and CATCHR families (van der Kant et al., 2015; Balderhaar and Ungermann, 2013). Structural studies of the GATOR1-GATOR2-KICSTOR complex will be required to understand how KICSTOR organizes the GATOR complexes on the lysosome. Further biochemical and genetic studies on individual KICSTOR components themselves are urgently needed; they might reveal as-yet-unknown functions in the growth pathway, or it may turn out that binding to KICSTOR is simply a convenient means of recruiting GATOR1 to its cognate compartment, with the complex performing another lysosomal-specific task.
Recruitment of the (m)TORC1 assembly to the lysosome or vacuole is accomplished in the absence of inhibitory nutrient signaling, and through incompletely understood control of the RagC/D GTPase
In the activated state of the cellular growth pathway, (m) TORC1 must be recruited to its cognate compartment, colocalizing it with its activator Rheb. This role is performed by the Rag GTPases, which are activated by nucleotide exchange and hydrolysis, in the case of Rag A/B and C/D respectively, and their own recruitment to the lysosome or to the vacuole in yeast. While FLCN-FNIP (Petit et al., 2013; Tsun et al., 2013; Péli-Gulli et al., 2015) and the Ragulator complex are known to be the key regulators promoting this step, the underlying signals for activation of the Rag GTPases remain obscure in comparison to those inhibiting them.
FLCN-FNIP
Just as the GATOR complexes control RagA/B, Folliculin (FLCN) has been identified as the GAP controlling the nucleotide state of RagC/D. Unlike GATOR1 however, this renders it is a positive regulator of (m)TORC1 signaling, given the mismatched nucleotide requirements of the Rag heterodimer. It was first discovered as a novel truncated gene from a cohort of patients with Birt-Hogg-Dubé (BHD) syndrome (Nickerson et al., 2002), associated with multiple pathophysiologies such as renal cancer and impaired skin development (Schmidt and Linehan, 2018). Subsequent work established that FLCN forms functional complexes with folliculin-interacting proteins 1 (FNIP1) and FNIP2, and furthermore that FNIP1 is a substrate, and binding partner, of AMPK, with phosphorylation resulting in downregulated mTOR signaling (Baba et al., 2006; Hasumi et al., 2008). The link between FLCN activity and mTORC1 signaling led to the discovery that FLCN and both FNIP1 and 2 are direct interacting partners of the Rag GTPases in mammalian cells (Petit et al., 2013; Tsun et al., 2013) and in yeast (Péli-Gulli et al., 2015). An obligate FLCN-FNIP complex is necessary for the Rag interaction, since RagA and RagC coimmunoprecipitate with FLCN only when FNIP2 is coexpressed, and this interaction is strengthened during nutrient starvation and resulted in inhibition of (m)TORC1 activity (Tsun et al., 2013; Péli-Gulli et al., 2015). Endogenous FLCN-FNIP1/2 and Lst4-Lst7 complexes exhibit lysosomal or vacuolar localization during nutrient deficiency, respectively (Petit et al., 2013; Tsun et al., 2013; Péli-Gulli et al., 2015), and once there FLCN-FNIP2 acts as the GAP for RagC/D.
FLCN and FNIP both contain conserved N-terminal longin and C-terminal Differentially Expressed in Normal cells and Neoplasia (DENN) domains (Fig. S3A), which are highly represented in Rab GEFs (Zhang D et al., 2012). The C-terminal DENN domain forms part of a larger so-called DENN module, which also comprises an N-terminal longin domain found in FLCN and the FNIPs, as well as the S. cerevisiae homologs. The crystal structure of the human FLCN C-terminal DENN domain (Fig. S3B) revealed the molecular structure of the core, or c-DENN, and downstream (d-)DENN subdomains (Nookala et al., 2012). Notably, FNIP1 and FNIP2 display a more divergent relationship to the prototypical DENN module architecture, encoding large, predicted unstructured, insertions within both the N-terminal longin and C-terminal c-DENN and d-DENN domains. To work around this, the structure of the Kluyveromyces lactis Lst4 N-terminal longin domain was solved (Pacitto et al., 2015), as this domain does not feature any disordered regions compared to the mammalian FNIPs. The structure revealed a classic longin domain architecture, which is the first subdomain of the complete DENN module (Fig. S3C), and which is also observed in the GATOR1 components NPRL2 and NPRL3. Unfortunately structures of the complete complex are as yet unavailable, while the details of its interaction with the Rag GTPases are keenly awaited.
Which signals converge on FLCN-FNIP to control its activity remains unclear. Although there is data implying that FLCN-FNIP is a GAP for RagC/D (Tsun et al., 2013) and that FLCN interacts stably with RagA (Petit et al., 2013), the localization of the FLCN-FNIP complexes, and their binding partners, remains incompletely understood. How FLCN-FNIP complexes are recruited to the lysosome or vacuole surface under nutrient-deficient conditions, and what events occur upon nutrient supplementation which cause their dispersion from these organellar membranes, remain open questions. A more recent study on S. cerevisiae Lst4 and Lst7 characterized TORC1 phosphorylation sites within Lst4 – within an intra-DENN loop insertion – which is involved in the release of the Lst4-Lst7 complex from the vacuolar membrane in response to nutrient repletion (Péli-Gulli et al., 2017). Given this insight, a similar feedback mechanism may occur in mammalian cells, where posttranslational modifications impinging on the intra-DENN regions of FNIP could cause dispersal from the lysosome under conditions of nutrient sufficiency. The known interaction with AMPK might well also imply a close connection to cellular energy state, and therefore the integration of metabolic signals directly. It is intriguing to note that a growing body of work also supports the intrinsic role of FLCN and its associated interacting proteins in regulating lysosome positioning in response to cellular metabolism (Starling et al., 2016; Dodding, 2017). These studies suggest that the association of FLCN-FNIP complexes with the lysosome during nutrient starvation promotes the formation of contacts between the lysosome and peri-nuclear membranes in a manner dependent on Rab34 and Rab-interacting lysosomal protein (RILP). Clearly multiple signals converge on FLCN-FNIP, and it is likely that characterization of the dynamic intra-DENN regions will highlight upstream regulators. Structural studies of the entire FLCN-FNIP-Rag assembly will be required to provide a definitive mechanism of Rag regulation by the FLCN-FNIP heterodimer, and may highlight how FLCN-FNIP cooperates with related pathways in the coordination between cellular metabolism and lysosome positioning (Dodding, 2017).
The Rag GTPase-Ragulator assembly
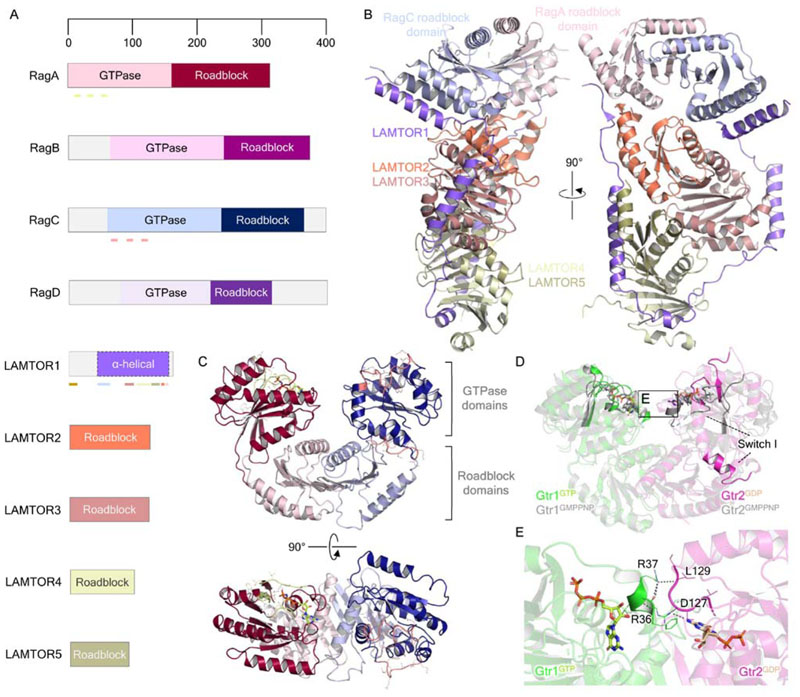
The Rag GTPases and their associated complex, ‘Ragulator’, are the essential platform for (m)TORC1 recruitment to the lysosome. The RagA and RagB genes were first identified as a novel subfamily of Ras-related GTPases of unknown biological function (Schürmann et al., 1995). Sequence alignment then identified RagA/B as the mammalian homologs of yeast Gtr1 (Bun-Ya et al., 1992). Yeast two-hybrid experiments isolated two more candidate Rags, which were homologous to each other as well as yeast to Gtr2, and bound RagA/B in vivo using C-terminal roadblock domains unique to the Rag subfamily (Fig. 4A) similarly to the binding of Gtr1 to Gtr2 (Nakashima et al., 1999; Sekiguchi et al., 2001). The discovery of these proteins, RagC and RagD, completed the Rag family.
Figure 4. Structural biology of the small GTPases Rag and the Ragulator complex.
(A) Domain organization of the Rag GTPases and Ragulator complex components LAMTOR1-5. Pale green lines under RagA indicate, from N- to C terminus, the P-loop (residues 14-21), switch I (residues 39-45) and switch II (residues 60-67) regions. Pink lines under RagC similarly indicate the P-loop (residues 68-75), switch I (residues 91-99) and switch II (residues 114-122) regions. The brown bar at the LAMTOR1 N terminus indicates the palmitoylation and myristoylation motifs, while colored bars reflect the regions of interaction with other Ragulator-Rag components, colored according to the interacting proteins. (B) Crystal structure of Ragulator in complex with the Rag GTPase roadblock domain heterodimer (PDB ID: 6EHP). (C) Structure of the Rag GTPase heterodimer from the cryo-EM structure of the GATOR1-Rag GTPase complex (PDB ID: 6CES; EMDB: EMD-7464). RagA-bound GTP is shown as light green in stick representation. The P loop, switch I and II regions of RagA and RagC are colored as highlighted in A. (D) Comparison of the crystal structures of Gtr1GMPPNP-Gtr2GMPPNP (PDB ID: 3R7W) and Gtr1GTP-Gtr2GDP (PDB ID: 4ARZ), highlighting the conformational change of the Gtr2 switch I region. E. Gtr1-Gtr2 GTPase domain interface, as inset in D. Gtr1 residues R36 and R37 make direct contacts with the neighboring Gtr2 nucleotide binding pocket and the bound GDP itself. Polar contacts are indicated by dark gray dashed lines.
In yeast, the Gtr1 and Gtr2 GTPases were shown to associate with Ego1, Ego3 and more recently Ego2 (Dubouloz et al., 2005; Gao and Kaiser, 2006; Powis et al., 2015) in an analogous complex to Ragulator named the EGO complex (EGOC). However, given the low sequence conservation between the Ego proteins and those from humans, the complex was not recognized as a Ragulator equivalent until structural data became available. The Rag GTPases were only later connected to the (m)TORC1 pathway (Kim et al., 2008; Sancak et al., 2008) when Drosophila melanogaster homologs of RagA/B/Gtr1 and RagC/D/Gtr2 were identified through an RNA interference screen targeting GTPases whose knockdown prevented amino acid-dependent phosphorylation of S6K, a well-known (m)TORC1 target (Burnett et al., 1998; Hara et al., 1998). RagA and RagC regulate mTORC1 in an amino acid-sensitive manner; RagA/B, when GTP-bound, and RagC/D, when GDP-bound, recruit RAPTOR, a core (m)TORC1 component (Sancak et al., 2008), thereby recruiting the kinase complex to late endosomes and lysosomes. None of the Rag proteins contain any lipid modification motifs that would result in their lysosomal localization, and therefore five proteins, MP1, p14, p18, HBXIP and C7orf59, which form a lysosomally-localized complex and which interact with the Rag GTPases, were found to be required to recruit them to their cognate compartment (Bar-Peled et al., 2012;Sancak et al., 2010). The protein p18 was shown to be the key determinant of lysosome localization by virtue of lipid modifications within its N-terminal region (Bar-Peled et al., 2012; Nada et al., 2009). Though MP1 and p14 were previously annotated with other functions (Teis et al., 2002), to reflect the regulation of the Rags and mTORC1 by these proteins, the pentameric complex was termed ‘Ragulator’ and the gene names were renamed LAMTOR1 (p18), LAMTOR2 (p14), LAMTOR3 (MP1), LAMTOR4 (HBXIP) and LAM-TOR5 (C7orf59) for Late Endosomal/Lysosomal Adaptor, MAPK and mTOR activator/regulator (Bar-Peled et al., 2012).
In addition to being a scaffold, Ragulator acts as the GEF for RagA/B, accelerating the release of both GDP and GTP from this subunit of the Rag heterodimer (Bar-Peled et al., 2012). A GEF for RagC/D has yet to be identified, representing a key gap in our understanding of these proteins. Presence of amino acids decreases the amount of Ragulator coimmunoprecipitating with RagB; furthermore, this interaction is strengthened when the Rags are in a nucleotide-free state, as the addition of GTP to an in vitro binding assay weakened the interaction (Bar-Peled et al., 2012). The interaction between Ragulator and the Rag heterodimers has been suggested to be dependent on the v-ATPase (Zoncu et al., 2011), which is thought to transmit intra-lysosomal amino acid signals to the Rags by affecting Ragulator GEF activity. Intriguingly, several other lysosomal membrane proteins have been considered to regulate the Rag GTPase pathway through Ragulator, including putative arginine sensor SLC38A9 (Jung et al., 2015; Rebsamen et al., 2015; Wang et al., 2015), the BLOC-1 related complex (BORC) (Pu et al., 2015), Niemann-Pick C1 (NPC1) (Castellano et al., 2017), and potentially TMEM55B (Hashimoto et al., 2018) and TMEM127 (Deng et al., 2018). Studies of SLC38A9 suggest that presence of amino acids, arginine in particular (Wang et al., 2015) are transmitted through SLC38A9-v-ATPase to the Rag GTPases (Jung et al., 2015; Rebsamen et al., 2015; Wang et al., 2015). It may be the case that stimulation of v-ATPase activity regulates Ragulator-Rag in response to fusion of lysosomes with incoming nutrient-rich endosomes, thus enabling signaling to mTORC1 from within the lysosome. Additionally, NPC1 has been implicated in cholesterol-dependent signaling to mTORC1 through SLC38A9 (Castellano et al., 2017) and TMEM55B and TMEM127 appear to support the stability of the Ragulator-v-ATPase assembly (Hashimoto et al., 2018; Deng et al., 2018); thus, there are likely to exist more peripheral membrane sensors which can regulate mTORC1 through the Rag GTPases. BORC has been implicated in regulating lysosome positioning in concert with amino acid signaling through Ragulator-Rag (Filipek et al., 2017; Pu et al., 2017) and also lysosome-autophagosome fusion during autophagy (Jia et al., 2017), thus representing another link to lysosome positioning and function alongside the Rab-RILP connection to FLCN-FNIP (Starling et al., 2016). A structural dissection of Ragulator interactions with its associated machinery on the lysosome will certainly shed light on the mechanisms by which the complex communicates to the Rag GTPases from multiple peripherally associated proteins.
The structures of LAMTOR2, LAMTOR3 and LAMTOR5 were the first to be characterized prior to the discovery of the Ragulator complex (Kurzbauer et al., 2004; Cui et al., 2008; Lunin et al., 2004; Qian et al., 2005; Garcia-Saez et al., 2011), revealing roadblock folds for the individual LAMTOR3 and LAMTOR5 monomers, and the formation of MP1/p14 heterodimers by edge-to-edge juxtaposition of the roadblock domains. The structure of S. cerevisiae Ego3 provided evidence that the EGOC and Ragulator components are structurally conserved, despite substantial sequence divergence (Kogan et al., 2010; Zhang et al., 2012), exhibiting a dimeric architecture very similar to that of LAMTOR2-LAMTOR3. The crystal structure of Gtr1-Gtr2 confirmed that these GTPases heterodimerize using their roadblock domains analogously (Gong et al., 2011). Given the notion that roadblock domains, like the structurally similar longin domains, are recurring architectures in GTPase-interacting proteins (Levine et al., 2013) the Ragulator-Rag complex serves as an extended interaction hub for the nutrient-sensing machinery, including the GAPs GATOR1 and FLCN-FNIP, peripheral lysosomal proteins such as v-ATPase and SLC38A9, and (m)TORC1. Indeed, crystal structures of the yeast EGOC and mammalian Ragulator-Rag complex support this view (Powis et al., 2015; Mu et al., 2017; Su et al., 2017; Yonehara et al., 2017; Zhang et al., 2017; de Araujo et al., 2017). The pentameric Ragulator complex exhibits an architecture in which heterodimers of LAMTOR2-LAM-TOR3 and LAMTOR4-LAMTOR5 are encircled by LAM-TOR1, which adopts an extended conformation with short regions of partial α-helicity making contacts with the other Ragulator members. The yeast EGOC structure, containing an Ego2-Ego3 heterodimer, and the LAMTOR1 homolog Ego1, demonstrated a remarkably similar architecture, with Ego1 overlapping considerably with LAMTOR1, and Ego2 and Ego3 overlapping with LAMTOR5 and LAMTOR2 respectively. The heptameric Ragulator-Rag complex structure showed Rag C-terminal roadblock domains binding to the LAMTOR2-LAMTOR3 heterodimer (de Araujo et al., 2017; Su et al., 2017; Yonehara et al., 2017) (Fig. 4B), thus building a tower of three roadblock domain-mediated heterodimers tied together by the extended LAMTOR1 subunit, and presenting the GTPase domains for interaction with RAPTOR. It is important to note that when the Rag heterodimer binds Ragulator, the N-terminal portion of LAMTOR1 encompassing residues 47-64 is resolved, forming an α-helix which packs against the RagC roadblock domain, whereas in the Ragulator complex alone this region of LAMTOR1 was disordered (de Araujo et al., 2017; Yonehara et al., 2017). Thus, LAMTOR1 forms a flexible tether for the remainder of the Ragulator complex and the Rag GTPases on the lysosome surface.
The GATOR1-Rag structure has provided the first glimpse of the Rag GTPase domains within the heterodimer (Shen et al., 2018) (Fig. 4C). Although the resolution of the cryo-EM structure was insufficient to accurately identify the RagC-bound ligand, it is thought to be GDP as RagC contains a S75N mutation which induces preferential GDP binding (Shen et al., 2018). A notable difference between both GTPase domains is the conformation of switch I, which in RagC appears to have extended and broken the central β-sheet as seen in RagA. This dramatic structural rearrangement is reminiscent of the Gtr1GTP-Gtr2GDP structure, in which GDP-bound Gtr2 displays an extremely similar switch I conformation (Jeong et al., 2012). In comparison to a Gtr1GMPPNP-Gtr2GMPPNP structure (Gong et al., 2011), the Gtr2GDP switch I region extends into an α-helix away from the nucleotide binding pocket with a concomitant global rotation (Fig. 4D) establishing a small novel interface between the GTPase domains whereby Gtr1 residues directly contact the Gtr2 nucleotide binding pocket and bound nucleotide itself (Fig. 4E). It is of particular interest to note that such an observation may support the recent notion that members within the Rag GTPase heterodimer exhibit negative cooperativity toward one another (Shen et al., 2017). Kinetic analyses of nucleotide binding suggest that the binding of a GTP molecule to one Rag establishes a dominant, conformation-driven, effect on the nucleotide binding pocket of the partner Rag, reducing affinity for GTP and stimulating GTP hydrolysis in the partner active site such that the heterodimer exists stably in a singly-GTP-bound state. Perhaps there exists a novel interface between the RagA-RagC GTPase domains, not captured in the recent cryo-EM structure, which may facilitate such cooperative behavior.
Overall, the Ragulator-Rag structures reveal a long molecular surface comprising three pairs of roadblock domain heterodimers, over which other protein complexes like the GAPs GATOR1 and FLCN-FNIP, as well as mTORC1, can bind. These structures, however, do not shed light on the mechanism by which Ragulator acts as a GEF for the Rags, although some insights from the study by Su and colleagues may reveal an unusual GEF mechanism (Su et al., 2017). These authors used constitutively active or inactive Rag heterodimers and used hydrogen-deuterium exchange-mass spectrometry to probe the dynamics of the RagA and RagC P loop residues, finding that Ragulator appears to modulate the dynamics of the RagA P loop specifically, as RagC did not exhibit spectral differences in the presence or absence of Ragulator. A hypothesis for this unusual GEF activity has been proposed in which GDP/GTP binding and conformational changes of the switch I region cause corresponding changes in the surrounding β-sheet, where the GTPase domain contacts the roadblock domain; the extended structure of LAMTOR1 may imply that its dynamic association with the other Ragulator components and Rag GTPases could regulate GTPase activity based on the interactions between individual roadblock domain heterodimers, as well as the interactions between Ragulator and lysosomal components including v-ATPase (Cherfils, 2017). Finally, the dynamic changes in the GTPase domain during nucleotide cycling must be implicitly linked to RAPTOR binding, however a molecular understanding of this interaction has not yet been detailed. It is clear that static structural determination methods will not be sufficient to understand the regulation of the Rag GTPases in atomic detail.
Extracellular stimuli and survival signals control cell growth through GTPase activation and suppression of Rheb, which acts as the activator of (m)TORC1 catalysis
Whereas recruitment of (m)TORC1 to the lysosome is controlled in response predominantly to signals corresponding to the intracellular nutrient and energy state, its activation once there is controlled by RhebGTP in response to a wide variety of external and survival signals feeding into a single point of control. Several studies in the past few years have greatly improved our understanding of the structure of mTORC1 as well as how the Rag and Rheb GTPases recruit and activate the kinase, respectively (Kim et al., 2008; Long et al., 2005; Sancak et al., 2008; Sato et al., 2009). Together, these GTPases constitute a coincidence detector which cooperate synergistically to robustly induce mTORC1 activity and fail to do so when either is only singly activated under physiologic conditions (Durán and Hall, 2012; Groenewoud and Zwartkruis, 2013).
The TSC complex
The integration of the majority of external and survival signals feeding into the (m)TORC1 pathway occurs through the TSC complex (TSCC), composed of tuberous sclerosis complex protein 1 (TSC1) and TSC2, also known as hamartin and tuberin, and TBC1D7, which is the key signal integrator and negative regulator upstream of Rheb. The genomic loci encoding TSC1 and TSC2 were first mapped upon identification of their linkage to the autosomal dominant disease Tuberous Sclerosis Complex, after which these genes were named (Fryer et al., 1987; Kandt et al., 1992). TSC2 contains a domain with homology to Rap 1-GAP (van Slegtenhorst et al., 1997), and forms a complex in vivo with TSC1, mediated by predicted coiled-coil domains at the TSC1 C terminus and the TSC2 N terminus (van Slegtenhorst et al., 1998; Nellist et al., 1999) (Fig. 5A). The functions of the TSC1-TSC2 complex were largely mysterious until studies investigating the Drosophila homologs provided a link between TSC1-TSC2 and cell growth, specifically downstream of the insulin receptor (Gao and Pan, 2001; Potter et al., 2001; Tapon et al., 2001). This identification of this regulatory mechanism, shown using the human proteins, unequivocally identified TSC1-TSC2 as inhibitors of the mTORC1 signaling pathway (Inoki et al., 2002; Tee et al., 2002), specifically by virtue of TSC2 GAP activity toward the small GTPase Rheb (Castro et al., 2003; Garami et al., 2003; Inoki et al., 2003; Saucedo et al., 2003; Stocker et al., 2003; Tee et al., 2003;Zhang et al., 2003). A third subunit of TSCC, TBC1D7, was later discovered and shown to be a constitutive member of the complex (Dibble et al., 2012). Unlike other TBC proteins, which are generally known to act as GAPs for the Rab family of GTPases (Fukuda, 2011), TBC1D7 does not appear to have GAP activity relevant to the Rheb signaling axis upstream of mTORC1, instead likely increasing the sensitivity and responsivity of the entire complex to environmental signals. Chromatographic analyses suggest that TSCCs also contains multiple copies of TSC1 and TSC2, and it is postulated that including multiple copies of each constituent protein may increase the sensitivity of the complex to allosteric regulation (Hoogeveen-Westerveld et al., 2012). As more recent structural studies appear to suggest, the presence of TBC1D7 may support higher-order oligomerisation of TSC1 and TSC2.
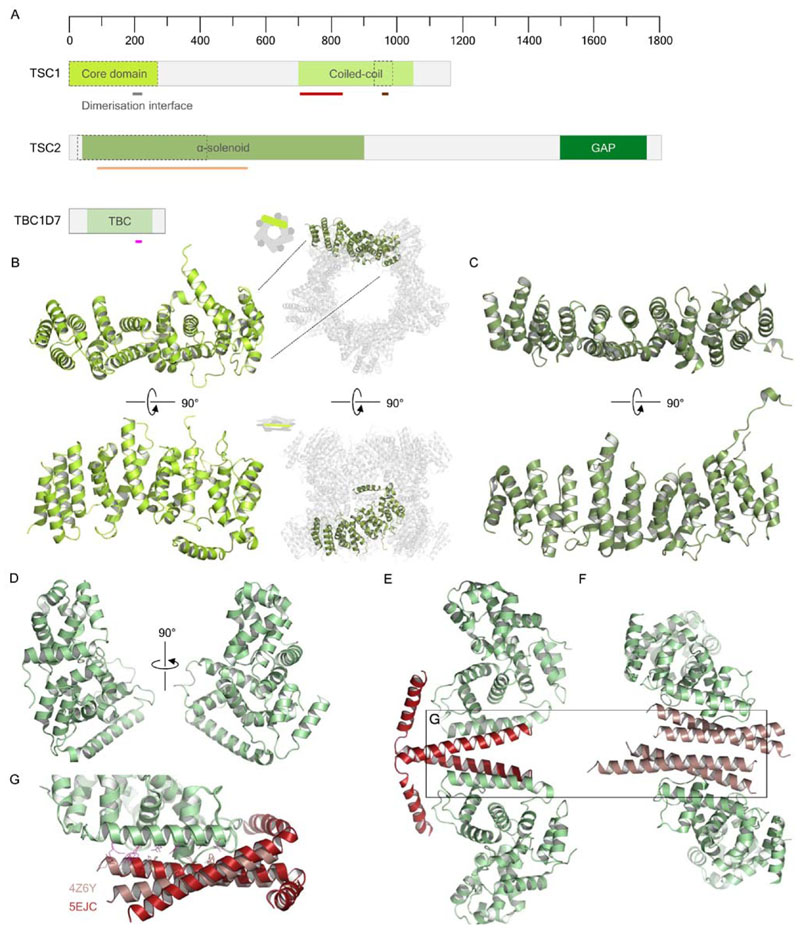
Figure 5. Structural biology and predicted domains of TSCC.
(A) Predicted domain organization of TSC1, TSC2 and TBC1D7. Black and black-dashed outlines indicate structurally resolved proteins or fragments, respectively, while gray outlines represent predicted regions. Grey bars reflect specific features with putative functions. The crimson bar in the TSC1 coiled-coil region denotes a TSC2 interaction site. Similarly, the light orange bar in the TSC2 α-solenoid denotes a TSC1-interacting region. The pink bar in TBC1D7 represents the TSC1-interacting helix. (B) Crystal structure of the S.pombe TSC1 core domain monomer, alongside the decamer observed in each crystal form (PDB ID: 4KK0). (C) Crystal structure of the C. thermophilum TSC2 N-terminal α-solenoid fragment (PDB ID: 5HIU). (D) Crystal structure of the deposited, but as-yet unpublished, TBC1D7 crystal structure (PDB ID: 3QWL). (E) Crystal structure of a TBC1D7-TSC1 coiled coil complex (PDB ID: 4Z6Y), indicating a dimer-of-dimers structure. (F) Crystal structure of TBC1D7-TSC1 coiled coil complex (PDB ID: 5EJC), indicating a helical-bundle architecture formed between dimerization of two TSC1 coiled coil fragments and one TBC1D7 molecule. (G) Superposition of the TBC1D7-TSC1 coiled coil complex structures, highlighting the similarity in binding conformation of the two different TSC1 coil fragments. TSC1-interacting residues are shown in pink, as highlighted in A, while TBC1D7-interacting residues are colored per complex.
The large, complex nature of the TSC1 and TSC2 proteins has largely impeded structure determination, especially in contrast to the smaller TBC1D7 subunit (Fig. 5A). Insights to TSC1 and TSC2 structure first came from a characterization of the N-terminal domain of S. pombe TSC1 (Sun et al., 2013), dubbed the core domain (Fig. 5B). This region of S. pombe TSC1 is homologous to residues 1-266 of human TSC1, notably lacking a region homologous to the first ~90 residues of the yeast protein. These authors found that the core domain is exclusively α-helical, as predicted by bioinformatic analyses, and forms a solenoid built of HEAT repeats. It is worth noting that Sun and colleagues obtained crystals with one of two different space groups, and that in both cases the asymmetric unit contained a higher-order TSC1 oligomer. In one crystal form, ten monomers form two juxtaposed pentameric rings, ultimately forming a ‘face-to-face’ decamer (Fig. 5B). The driver for such an assembly appears to be an overhanging helix from each core domain, which makes contacts with neighboring domains. Though such an arrangement may be a consequence of crystal packing, and thus its physiologic relevance is questionable, the structure may indicate a role for the TSC1 N terminus in homooligomerisation. The core domain is considered essential for the stability of TSC1 (Hoogeveen-Westerveld et al., 2010; Mozaffari et al., 2009); if a function of this region was to enable higher-order oligomer formation, such an assembly may protect TSCC subunits from degradation. More recently, a crystal structure was solved of a stable N-terminal TSC2 fragment from C. thermophilum (Zech et al., 2016) (Fig. 5C). This TSC2 N-terminal fragment, like the S. pombe TSC1 core domain, forms an α-solenoid constituting nine HEAT repeats. This fragment of TSC2 was also shown to be sufficient to pull down the full-length, and C-terminal half of, TSC1, and these interacting regions are conserved in the human proteins (Zech et al., 2016).
Contributing to a greater understanding of TSCC architecture, crystal structures have been solved of the smaller subunit TBC1D7 complexed with coiled-coil fragments of TSC1 (Gai et al., 2016a; Qin et al., 2016), alongside a TBC1D7 apo structure deposited in the PDB by Guan et al. (PDB ID: 3QWL). The apo structure (Fig. 5D) reveals an architecture highly conserved with other TBC domain proteins (Fischer et al., 2016; Fukuda, 2011; Park et al., 2011), with little conformational difference in comparison to the TSC1-complexed structures. Both TBC1D7-TSC1 crystal structures highlight a conserved interaction interface, with a long helix from TBC1D7 packing against the TSC1 coiled-coil to form a three-helix bundle. Both groups crystallized the same TSC1 region – residues 939-992 – with near full-length TBC1D7, either residues 19-293 (Qin et al., 2016) or 21-293 (Gai et al., 2016a). In the complex structure solved by Gai and colleagues, two TBC1D7 molecules and two TSC1 molecules form a dimer of dimers, with the full length of the TSC1 subunit visible, forming a parallel coiled-coil where each helix kinks at G973 and folds back onto each TBC1D7 subunit (Fig. 5E). In contrast, the complex solved by Qin et al. shows two TBC1D7 and four TSC1 molecules per asymmetric unit, whereby each TBC1D7 complexed with two TSC1 molecules, forming a triple helical bundle (Fig. 5F). In this structure, the last ~15 residues of each TSC1 molecule – which formed the kinked helix in the crystal structure by Gai et al. – were not visible. In both cases, one TBC1D7 molecule was able to interact simultaneously with two TSC1 coiled-coil fragments, and alignment of these domains highlights structural similarity (Fig. 5G). As supported by analytical ultracentrifugation data from both studies (Gai et al., 2016a; Qin et al., 2016), the exact stoichiometry of the TBC1D7-TSC1 coiled-coil interaction appears to be dependent on the ratio between the two molecules, and this may underlie physiologic activity of TBC1D7 in regulation TSCC oligomerisation.
Whereas it appears that specific metabolites signal to mTORC1 through the Rag GTPase axis, stress conditions such as hypoxia, DNA damage stress, ER stress, and growth factor and survival signaling, are filtered through to the Rheb GTPase through TSCC by post-translational modification (Kwiatkowski, 2003; Huang and Manning, 2008; Tomasoni and Mondino, 2011; Demetriades et al., 2016). TSCC is phosphorylated by many kinases, each of which contributes either activating or inhibiting modifications under various environmental conditions, including S6K1, cyclin-dependent kinase 1 (CDK1), AMPK and Akt, which is itself activated by mTORC2 (Huang and Manning, 2009). Phosphorylation has been shown to cause dispersion of TSC2 from membrane-localized TSC1 punctae to the cytosol (Cai et al., 2006), in a manner dependent on interaction with 14-3-3 proteins. Notably, TSC2 has already been shown to interact with 143-3 proteins when phosphorylated, and that this interaction disrupts the function of the TSC1-TSC2 complex (Nellist et al., 2003;Shumway et al., 2003). Overall, it appears that the localization of the TSC2 subunit is the main determinant of TSCC-mediated Rheb inhibition (Demetriades et al., 2016). Another example of this localization dependency includes hypoxia-dependent regulation through REDD1 (Brugarolas et al., 2004), which disrupts cytosolic 14-3-3-bound TSC2 and enables subsequent TSC2 localization to the lysosome where it can inhibit Rheb (DeYoung et al., 2008). Future structural work capturing the TSCC assembly and TSC2-Rheb structure will provide molecular insights into the activity of this crucially important but as-yet incompletely understood mTORC1 regulator, and provide a rational basis for understanding the pathogenicity of mutations in human TSC1 and TSC2, associated with diseases such as TSC and sporadic cases of lymphangioleiomyomatosis (Kwiatkowski, 2003; Rosset et al., 2017).
Rheb GTPase
In almost all eukaryotes, RhebGTP is an essential coactivator of (m)TORC1, required for catalytic activity. It was first identified during mRNA screens of rat hippocampal tissue for genes induced after electroconvulsive stimulation (Yamagata et al., 1994). The protein identified was noted to be a Ras GTPase homolog, and was named Ras Homolog Enriched in Brain, although it has since been discerned that Rheb is ubiquitous in all tissues across the body (Saito et al., 2005). It was subsequently confirmed that Rheb is farnesylated at a C-terminal CAAX motif (Clark et al., 1997) (Fig. 6A). The subcellular localization of Rheb has been debated, with many sources arguing for a distribution between endomembrane compartments including the ER, Golgi, mitochondria, and late endosomes and lysosomes (Takahashi et al., 2005; Buerger et al., 2006; Hanker et al., 2010; Parmar and Tamanoi, 2010). Despite a lack of clear experimental evidence pointing to a defined localization pattern, Rheb is known to function on endomembranes in the mTORC1 pathway as a potent activator of the kinase (Long et al., 2005; Sato et al., 2009) and is the physiologic target of TSC2 (Castro et al., 2003; Garami et al., 2003; Inoki et al., 2003; Saucedo et al., 2003; Stocker et al., 2003; Tee et al., 2003; Zhang et al., 2003).
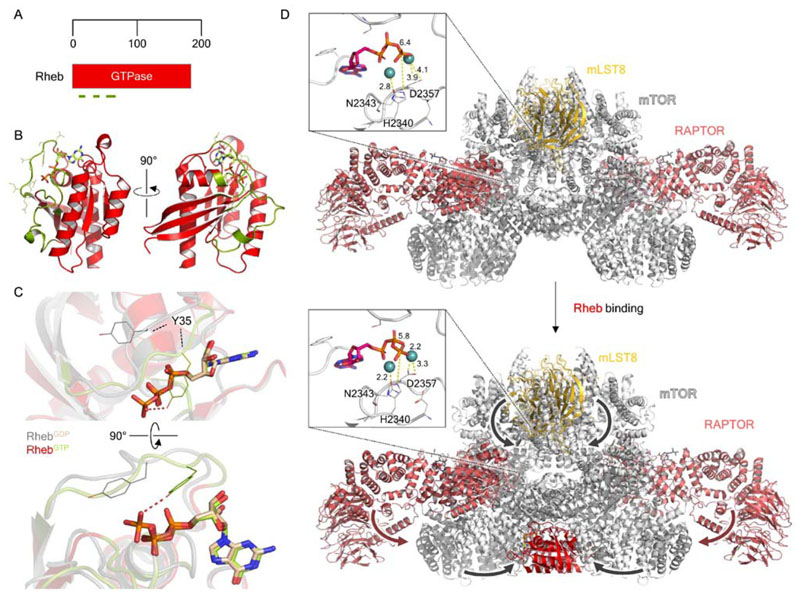
Figure 6. Structural biology of the small GTPase Rheb and the Rheb-dependent activation of mTORC1.
(A) Domain organization of Rheb. Olive green lines indicate, from N- to C terminus, the P-loop (residues 12-20), switch I (residues 33-41) and switch II (residues 6379) regions. (B) Crystal structure of GTP-bound Rheb (PDB ID: 1XTS), with bound GTP shown as light green in stick representation. The P loop, switch I and II regions are shown in olive green, as highlighted in A. (C) Focused view of the nucleotide binding pocket, highlighting the conformational difference between key GTP binding residue Y35 between the GDP- and GTP-bound states. (D) Comparison of the cryo-EM structures of mTORC1 alone (top; PDB ID: 6BCX; EMDB: EMD-7087) and mTORC1-Rheb (bottom; PDB ID: EMD-7086). Major conformational movements which occur upon Rheb binding, such as the N-HEAT rotation and kinase N- lobe compaction, are indicated by arrows. The mTOR kinase active sites are shown as insets in each, with the Mg2+ -coordinating residues N2343 and D2357, and catalytic residue H2340, shown. Interatomic distances labeled in Å indicate that in Rheb-bound mTORC1 these key residues shift by < 1 Å toward the bound ATP.
Kinetic studies of Rheb revealed that the protein is unique compared to other Ras superfamily GTPases in that it exhibits very low basal GTPase activity, probably due to lacking a conserved catalytic glycine residue (G12 in Ras), which is replaced by an arginine. Crystal structures of Rheb complexed with GTP (Fig. 6B) and GDP revealed that the protein undergoes very small, select conformation changes involving switch I (residues 33-41) and switch II (residues 63-79) (Yu et al., 2005). Notably, these structures reveal that the glycine-to-arginine mutation thought to result in low GTPase activity cannot alone be used to explain this difference in activity. Several differences from the canonical GTPase motifs account for these unusual kinetics. Q64, which is required for polarization/activation of the water molecule that initiates nucleophilic attack, is displaced by > 4 Å from the counterpart RasGTP structure, due to an unusual conformation of switch II, and furthermore its sidechain is buried in a hydrophobic pocket. Y35 also prevents GTP dissociation by closing over the nucleotide binding pocket (Fig. 6C), coordinating the phosphates more tightly than in Ras and Rap (Mazhab-Jafari et al., 2012).
Rheb has been observed to maintain a high proportion within the GTP state in within cells (Im et al., 2002). The molecular features which enable this behavior thus prepare for de-inhibition by TSC2 to lead to rapid activation of mTORC1, thereby consolidating the function of Rheb as a potent activator of mTORC1. It is known that the TSC2 GAP domain utilizes an “asparagine-thumb” mechanism for the activation of Rheb GTPase activity, similar to that of the Rap GTPase and its GAP (Marshall et al., 2009; Scrima et al., 2008), and a structure of the TSC2 GAP-Rheb complex is sorely needed to enable us to understand more about how this unique GTPase-GAP pair functions to control the activation of mTORC1. The GEF for Rheb, if one exists, remains one of the most significant gaps in our knowledge, but it is notable that Rheb is found dispersed throughout the endomembrane compartments (Parmar and Tamanoi, 2010), and it has been postulated that Rheb activation occurs at the Golgi, whereupon it then shuttles during maturation to late endosomes and lysosomes where it can activate mTORC1 (Groenewoud and Zwartkruis, 2013). This would also rationalise the slow GTP hydrolysis and release, as it would be required for Rheb to survive translocation to lysosomes in GTP-state. More recent evidence suggests that microspherule protein 1 (MCRS1) retains Rheb on the lysosome surface in the presence of amino acids, thus coordinating the localization of functional Rheb with the Rag signaling axis (Fawal et al., 2015). Given the pleiotropic effects observed upon Rheb knockout in multiple cell lines and mouse models (Heard et al., 2014), more work is required to pin down the exact pathways by which Rheb is activated and localized in proximity to mTORC1.
Architecture and activation of mTORC1
The structure of the mTORC1 core, consisting of an mTOR mLST8 “FATKIN” unit, was solved in 2013 by Yang and colleagues, providing the first structural data on the kinase and enabling a molecular-level understanding of kinase inhibition by rapamycin (Yang et al., 2013). Being one of the mammalian PI3K-related protein kinases (PIKKs), mTOR was known by sequence homology to contain a FRAP, ATM and TRAPP (FAT) domain, a catalytic kinase domain and a C-terminal ‘FATC ’ domain found only in PIKKs (Bosotti et al., 2000). The crystal structure revealed that the kinase domain exhibited a classical bilobed structure, just as in the PI3K family, and the PIKK-specific FAT domain encircles the kinase domain, raising the possibility that interaction of regulatory proteins with this region will allosterically impact the kinase domain structure as well. Notably, however, the kinase N-lobe features a large insertion corresponding to the FKBP12-rapamycin binding (FRB) domain (Chen et al., 1995; Chiu et al., 1994; Vilella-Bach et al., 1999) specific to mTORC1 among the PIKKs. More recently, cryo-EM structures have revealed the architecture of the mTORC1 complex at higher resolution (Aylett et al., 2016; Baretić et al., 2016; Yang et al., 2017, 2016). These structures revealed that mTORC1 is indeed dimeric, with each monomer in the complex interacting with its partner using the N-terminal HEAT repeat domains, which were truncated in the earlier crystal structure (Yang et al., 2013), and with Raptor binding the juncture between the kinase domains (Imseng et al., 2018).
Cross-linking of mTORC1 in the presence of RhebGTPgS and 4EBP1, another well-known mTORC1 substrate (Burnett et al., 1998), yielded the cryo-EM structure of mTORC1 bound to Rheb (Yang et al., 2017). The structure of mTORC1 corresponds well to the previously determined structures (Aylett et al., 2016; Baretić et al., 2016; Yang et al., 2016), enabling the identification of the key mTORC1 domains, and verifying the tripartite dimerization interface across Raptor and the M- and N-HEAT regions. Density corresponding to Rheb was observed at the N-terminal portions of the N- HEAT, M-HEAT and FAT domains, thereby resulting in a four-way interface where Rheb switch I residues bound the M-HEAT solenoid and FAT domain, and switch II formed contacts with all three mTORC1 domains. A mechanistic understanding for the allosteric activation of mTORC1 is now available: Rheb binds to mTOR with its switch I and II regions, which have been previously observed crystallographically to undergo nucleotide-dependent conformational changes (Yu et al., 2005). Rheb binding causes a rotation of the mTORC1 N-HEAT solenoid toward the M-HEAT and FAT domains, where N-HEAT-FAT interactions are formed (Fig. 6D). These in turn reorganise the N-lobe of the kinase domain to a catalytically-primed conformation: the catalytic and Mg2+ binding residues are brought into closer proximity with the terminal phosphate of ATP (Fig. 6D, insets). The binding of Rheb to mTORC1 exhibits enforced cooperativity, since a singly-bound Rheb would result in a highly unfavorable dimerization interface; mTORC1 is therefore robustly activated by the binding of two Rheb GTPases per kinase complex, which causes a global rearrangement culminating in the priming of the active site for catalysis. This final step represents the culmination of the combined input of the entire pathway into a final binary “on/off” decision: to grow.
Concluding remarks
The control of the (m)TORC1 growth on/off switch at the juncture of so many key signaling pathways (Fig. 7) is a highly complex system evolved to reduce the cacophony of information available to the cell to the answer to a simple question: “Is it a good time to grow”?. A great deal of recent progress has been made on unpicking the mechanisms of this “Heath Robinson” contraption, revealing how key features of the process function at a molecular level, as in the case of the recent explosion of structural data for Rag/Ragulator and (m) TORC1, and how input reduction has been achieved, for the GATOR2-interacting nutrient sensors for example, and in the unexpectedly complete isolation of the Rag and Rheb pathways from one another. There have also been many completely unexpected observations, which have thrown up new questions remaining to be understood in time, such as the dual Rag binding sites of GATOR1, hinting at new mechanisms by which responses are tuned, and the proliferation of longin domains, suggesting evolutionary expansion from a simpler precursor pathway.
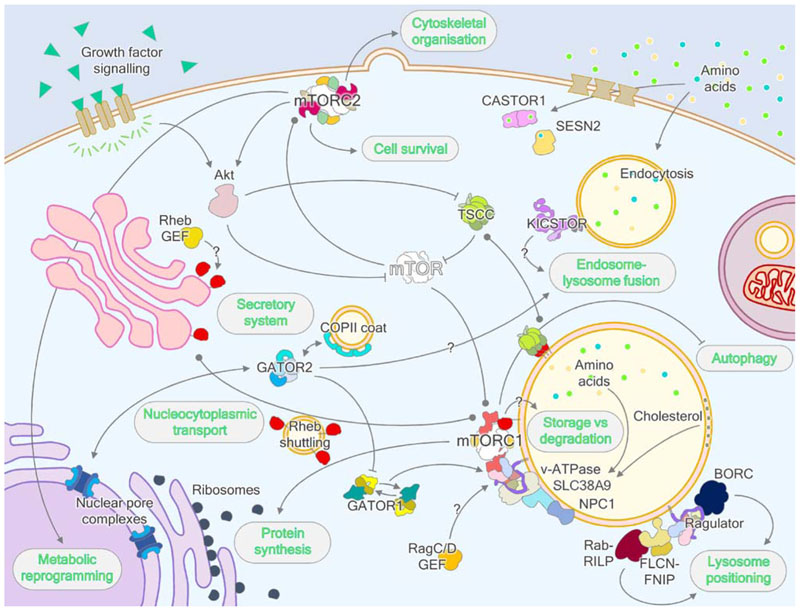
Figure 7. Wider regulation of eukaryotic cell metabolism by mTORC1.
Besides the well-characterized regulation of protein synthesis and autophagy by mTORC1, mTOR also forms a plasma membrane-localized, functionally segregated and rapamycin-insensitive complex called mTORC2, which controls cytoskeletal organization pathways and coordinates cell survival with cellular metabolism. mTORC2 is also a key activator of protein kinase Akt, which inhibits TSCC and therefore activates mTORC1 through Rheb. On the lysosome, the Ragulator complex interacts with several lysosomally-localized proteins, including the v-ATPase, SLC38A9, NPC1 and BORC complex, coordinating wider metabolism and lysosomal biogenesis and positioning to mTORC1 signaling. Additionally, the FLCN-FNIP complex also regulates lysosome positioning through interaction with Rab-RILP complexes. The GATOR2 complex interchanges subunits with the NPC and COPII vesicle coat, and alongside KICSTOR, which also features similar architectures, may have roles in lysosome-endosome and/or lysosome-autophagosome tethering and fusion. As recently determined, GATOR1 exhibits two modes of interaction with RagA/B and this interplay may endow the Rag signaling axis with unique properties. Finally, the Rag and Rheb GTPases both feature GAPs, and there is evidence supporting the role of Ragulator as a RagA/B GEF, however GEFs have not been conclusively identified for RagC/D or Rheb. The Rheb-GEF may localize to the Golgi, which is postulated to be the site of Rheb activation.
The molecular mechanisms of (m)TOR regulation are now tantalisingly close to being understood. Progress remains quite unevenly distributed, however; the recruitment and activation complexes themselves are now becoming relatively well described, only the mechanism of recruitment of (m)TORC1 by Rag/Ragulator and the substrate bound structures of (m)TORC1 eluding our grasp, and discussion moving toward low-level subjects such as kinetics and active site mechanisms, whereas the lack of structural detail on the less proximal parts of the pathway, the TSCC, FLCN-FNIP and GATOR2 complexes, hampers our understanding still. There is great cause for hope, however, given the pace of recent progress, that this highly important and medically relevant pathway will emerge from the scattered pieces to yield a clear picture over the next few years of scientific endeavor.
Supplementary Material
Electronic supplementary material Supplementary material is available in the online version of this article at http://dx.doi.org/10.1007/s11515-018-1501-7 and is accessible for authorized users.
Acknowledgments
This work was supported by the Wellcome Trust and the Royal Society through a Sir Henry Dale Fellowship (206212/Z/17/Z) to CHSA.
Footnotes
Compliance with ethics guidelines
Kailash Ramlaul and Christopher H. S. Aylett declare that they have no conflict of interest.
References
- Algret R, Fernandez-Martinez J, Shi Y, Kim SJ, Pellarin R, Cimermancic P, Cochet E, Sali A, Chait BT, Rout MP, Dokudovskaya S. Molecular architecture and function of the SEA complex, a modulator of the TORC1 pathway. Mol Cell Proteomics. 2014;13(11):2855–2870. doi: 10.1074/mcp.M114.039388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylett CHS, Sauer E, Imseng S, Boehringer D, Hall MN, Ban N, Maier T. Architecture of human mTOR complex 1. Science. 2016;351(6268):48–52. doi: 10.1126/science.aaa3870. [DOI] [PubMed] [Google Scholar]
- Baba M, Hong SB, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, Esposito D, Gillette WK, Hopkins RF, 3rd, Hartley JL, Furihata M, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci USA. 2006;103(42):15552–15557. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassari S, Licchetta L, Tinuper P, Bisulli F, Pippucci T. GATOR1 complex: the common genetic actor in focal epilepsies. J Med Genet. 2016;53(8):503–510. doi: 10.1136/jmedgenet-2016-103883. [DOI] [PubMed] [Google Scholar]
- Balderhaar HJ, Ungermann C. CORVET and HOPS tethering complexes- coordinators of endosome and lysosome fusion. J Cell Sci. 2013;126(Pt 6):1307–1316. doi: 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- Baple EL, Maroofian R, Chioza BA, Izadi M, Cross HE, Al-Turki S, Barwick K, Skrzypiec A, Pawlak R, Wagner K, Coblentz R, et al. Mutations in KPTN cause macrocephaly, neurodevelopmental delay, and seizures. Am J Hum Genet. 2014;94(1):87–94. doi: 10.1016/j.ajhg.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340(6136):1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150(6):1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baretić D, Berndt A, Ohashi Y, Johnson CM, Williams RL. Tor forms a dimer through an N-terminal helical solenoid with a complex topology. Nat Commun. 2016;7 doi: 10.1038/ncomms11016. 11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Hershkovitz T, Heyman E, Raspall-Chaure M, Kakar N, Smirin-Yosef P, Vila-Pueyo M, Kornreich L, Thiele H, Bode H, Lagovsky I, et al. Biallelic SZT2 mutations cause infantile encephalopathy with epilepsy and dysmorphic corpus callosum. Am J Hum Genet. 2013;93(3):524–529. doi: 10.1016/j.ajhg.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulac S. mTOR signaling pathway genes in focal epilepsies. Prog Brain Res. 2016;226:61–79. doi: 10.1016/bs.pbr.2016.04.013. [DOI] [PubMed] [Google Scholar]
- Bharucha N, Liu Y, Papanikou E, McMahon C, Esaki M, Jeffrey PD, Hughson FM, Glick BS. Sec16 influences transitional ER sites by regulating rather than organizing COPII. Mol Biol Cell. 2013;24(21):3406–3419. doi: 10.1091/mbc.E13-04-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem. 1995;270(5):2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- Bosotti R, Isacchi A, Sonnhammer EL. FAT: a novel domain in PIK-related kinases. Trends Biochem Sci. 2000;25(5):225–227. doi: 10.1016/s0968-0004(00)01563-2. [DOI] [PubMed] [Google Scholar]
- Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322(5906):1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Schwartz TU. Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice. Nat Struct Mol Biol. 2009;16(11):1173–1177. doi: 10.1038/nsmb.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369(6483):756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/ TSC2 tumor suppressor complex. Genes Dev. 2004;18(23):2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134(3):451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov AV, Shoshani T, Faerman A, Zelin E, Kamer I, Kalinski H, Gorodin S, Fishman A, Chajut A, Einat P, Skaliter R, et al. Identification of a novel stress-responsive gene Hi95 involved in regulation of cell viability. Oncogene. 2002;21(39):6017–6031. doi: 10.1038/sj.onc.1205877. [DOI] [PubMed] [Google Scholar]
- Buerger C, DeVries B, Stambolic V. Localization of Rheb to the endomembrane is critical for its signaling function. Biochem Biophys Res Commun. 2006;344(3):869–880. doi: 10.1016/j.bbrc.2006.03.220. [DOI] [PubMed] [Google Scholar]
- Bun-Ya M, Harashima S, Oshima Y. Putative GTP-binding protein, Gtr1, associated with the function of the Pho84 inorganic phosphate transporter in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12(7):2958–2966. doi: 10.1128/mcb.12.7.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA. 1998;95(4):1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai SL, Tee AR, Short JD, Bergeron JM, Kim J, Shen J, Guo R, Johnson CL, Kiguchi K, Walker CL. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol. 2006;173(2):279–289. doi: 10.1083/jcb.200507119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano BM, Thelen AM, Moldavski O, Feltes M, van der Welle REN, Mydock-McGrane L, Jiang X, van Eijkeren RJ, Davis OB, Louie SM, Perera RM, Covey DF, Nomura DK, Ory DS, Zoncu R. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science. 2017;355(6331):1306–1311. doi: 10.1126/science.aag1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro AF, Rebhun JF, Clark GJ, Quilliam LA. Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J Biol Chem. 2003;278(35):32493–32496. doi: 10.1074/jbc.C300226200. [DOI] [PubMed] [Google Scholar]
- Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, Wang T, Harper JW, Gygi SP, Sabatini DM. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell. 2016;165(1):153–164. doi: 10.1016/j.cell.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantranupong L, Wolfson RL, Orozco JM, Saxton RA, Scaria SM, Bar-Peled L, Spooner E, Isasa M, Gygi SP, Sabatini DM. The Sestrins interact with GATOR2 to negatively regulate the aminoacid-sensing pathway upstream of mTORC1. Cell Reports. 2014;9(1):1–8. doi: 10.1016/j.celrep.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EJ, Kaiser CA. LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J Cell Biol. 2003;161(2):333–347. doi: 10.1083/jcb.200210141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci USA. 1995;92(11):4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J. Encoding Allostery in mTOR Signaling: The Structure of the Rag GTPase/Ragulator Complex. Mol Cell. 2017;68(5):823–824. doi: 10.1016/j.molcel.2017.11.027. [DOI] [PubMed] [Google Scholar]
- Chiu MI, Katz H, Berlin V. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc Natl Acad Sci USA. 1994;91(26):12574–12578. doi: 10.1073/pnas.91.26.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark GJ, Kinch MS, Rogers-Graham K, Sebti SM, Hamilton AD, Der CJ. The Ras-related protein Rheb is farnesylated and antagonizes Ras signaling and transformation. J Biol Chem. 1997;272(16):10608–10615. doi: 10.1074/jbc.272.16.10608. [DOI] [PubMed] [Google Scholar]
- Cui Q, Sulea T, Schrag JD, Munger C, Hung MN, Naïm M, Cygler M, Purisima EO. Molecular dynamics-solvated interaction energy studies of protein-protein interactions: the MP1-p14 scaffolding complex. J Mol Biol. 2008;379(4):787–802. doi: 10.1016/j.jmb.2008.04.035. [DOI] [PubMed] [Google Scholar]
- Daste F, Galli T, Tareste D. Structure and function of longin SNAREs. J Cell Sci. 2015;128(23):4263–4272. doi: 10.1242/jcs.178574. [DOI] [PubMed] [Google Scholar]
- de Araujo MEG, Naschberger A, Fürnrohr BG, Stasyk T, Dunzendorfer-Matt T, Lechner S, Welti S, Kremser L, Shivalingaiah G, Offterdinger M, Lindner HH, et al. Crystal structure of the human lysosomal mTORC1 scaffold complex and its impact on signaling. Science. 2017;358(6361):377–381. doi: 10.1126/science.aao1583. [DOI] [PubMed] [Google Scholar]
- De Franceschi N, Wild K, Schlacht A, Dacks JB, Sinning I, Filippini F. Longin and GAF domains: structural evolution and adaptation to the subcellular trafficking machinery. Traffic. 2014;15(1):104–121. doi: 10.1111/tra.12124. [DOI] [PubMed] [Google Scholar]
- Debler EW, Ma Y, Seo HS, Hsia KC, Noriega TR, Blobel G, Hoelz A. A fence-like coat for the nuclear pore membrane. Mol Cell. 2008;32(6):815–826. doi: 10.1016/j.molcel.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Demetriades C, Plescher M, Teleman AA. Lysosomal recruitment of TSC2 is a universal response to cellular stress. Nat Commun. 2016;7 doi: 10.1038/ncomms10662. 10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Qin Y, Srikantan S, Luo A, Cheng ZM, Flores SK, Vogel KS, Wang E, Dahia PLM. The TMEM127 human tumor suppressor is a component of the mTORC1 lysosomal nutrientsensing complex. Hum Mol Genet. 2018;27(10):1794–1808. doi: 10.1093/hmg/ddy095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22(2):239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. TBC1D7 is athird subunitofthe TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47(4):535–546. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodding MP. Folliculin- A tumor suppressor at the intersection of metabolic signaling and membrane traffic. Small GTPases. 2017;8(2):100–105. doi: 10.1080/21541248.2016.1204808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokudovskaya S, Waharte F, Schlessinger A, Pieper U, Devos DP, Cristea IM, Williams R, Salamero J, Chait BT, Sali A, Field MC, et al. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.006478. M110.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19(1):15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Durán RV, Hall MN. Regulation of TOR by small GTPases. EMBO Rep. 2012;13(2):121–128. doi: 10.1038/embor.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faini M, Beck R, Wieland FT, Briggs JAG. Vesicle coats: structure, function, and general principles of assembly. Trends Cell Biol. 2013;23(6):279–288. doi: 10.1016/j.tcb.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Fath S, Mancias JD, Bi X, Goldberg J. Structure and organization of coat proteins in the COPII cage. Cell. 2007;129(7):1325–1336. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Fawal MA, Brandt M, Djouder N. MCRS1 binds and couples Rheb to amino acid-dependent mTORC1 activation. Dev Cell. 2015;33(1):67–81. doi: 10.1016/j.devcel.2015.02.010. [DOI] [PubMed] [Google Scholar]
- Filipek PA, de Araujo MEG, Vogel GF, De Smet CH, Eberharter D, Rebsamen M, Rudashevskaya EL, Kremser L, Yordanov T, Tschaikner P, Fürnrohr BG, et al. LAMTOR/Ragulator is a negative regulator of Arl8b- and BORC-dependent late endosomal positioning. J Cell Biol. 2017;216(12):4199–4215. doi: 10.1083/jcb.201703061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer B, Lüthy K, Paesmans J, De Koninck C, Maes I, Swerts J, Kuenen S, Uytterhoeven V, Verstreken P, Versées W. Skywalker-TBC1D24 has a lipid-binding pocket mutated in epilepsy and required for synaptic function. Nat Struct Mol Biol. 2016;23(11):965–973. doi: 10.1038/nsmb.3297. [DOI] [PubMed] [Google Scholar]
- Frankel WN, Yang Y, Mahaffey CL, Beyer BJ, O’Brien TP. Szt2, a novel gene for seizure threshold in mice. Genes Brain Behav. 2009;8(5):568–576. doi: 10.1111/j.1601-183X.2009.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer AE, Chalmers A, Connor JM, Fraser I, Povey S, Yates AD, Yates JR, Osborne JP. Evidence that the gene for tuberous sclerosis is on chromosome 9. Lancet. 1987;1(8534):659–661. doi: 10.1016/s0140-6736(87)90416-8. [DOI] [PubMed] [Google Scholar]
- Fukuda M. TBC proteins: GAPs for mammalian small GTPase Rab? Biosci Rep. 2011;31(3):159–168. doi: 10.1042/BSR20100112. [DOI] [PubMed] [Google Scholar]
- Gai Z, Chu W, Deng W, Li W, Li H, He A, Nellist M, Wu G. Structure of the TBC1D7-TSC1 complex reveals that TBC1D7 stabilizes dimerization of the TSC1 C-terminal coiled coil region. J Mol Cell Biol. 2016a doi: 10.1093/jmcb/mjw001. mjw001. [DOI] [PubMed] [Google Scholar]
- Gai Z, Wang Q, Yang C, Wang L, Deng W, Wu G. Structural mechanism for the arginine sensing and regulation of CASTOR1 in the mTORC1 signaling pathway. Cell Discov. 2016b;2(1):16051. doi: 10.1038/celldisc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol. 2006;8(7):657–667. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- Gao X, Pan D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 2001;15(11):1383–1392. doi: 10.1101/gad.901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garami A, Zwartkruis FJT, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11(6):1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- Garcia-Saez I, Lacroix FB, Blot D, Gabel F, Skoufias DA. Structural characterization of HBXIP: the protein that interacts with the anti-apoptotic protein survivin and the oncogenic viral protein HBx. J Mol Biol. 2011;405(2):331–340. doi: 10.1016/j.jmb.2010.10.046. [DOI] [PubMed] [Google Scholar]
- Gong R, Li L, Liu Y, Wang P, Yang H, Wang L, Cheng J, Guan KL, Xu Y. Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev. 2011;25(16):1668–1673. doi: 10.1101/gad.16968011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabacka M, Pierzchalska M, Dean M, Reiss K. Regulation of ketone body metabolism and the role of pparα. Int J Mol Sci. 2016;17(12):E2093. doi: 10.3390/ijms17122093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenewoud MJ, Zwartkruis FJT. Rheb and Rags come together at the lysosome to activate mTORC1. Biochem Soc Trans. 2013;41(4):951–955. doi: 10.1042/BST20130037. [DOI] [PubMed] [Google Scholar]
- Gu X, Orozco JM, Saxton RA, Condon KJ, Liu GY, Krawczyk PA, Scaria SM, Harper JW, Gygi SP, Sabatini DM. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science. 2017;358(6364):813–818. doi: 10.1126/science.aao3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanker AB, Mitin N, Wilder RS, Henske EP, Tamanoi F, Cox AD, Der CJ. Differential requirement of CAAX-mediated posttranslational processing for Rheb localization and signaling. Oncogene. 2010;29(3):380–391. doi: 10.1038/onc.2009.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273(23):14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Shirane M, Nakayama KI. TMEM55B contributes to lysosomal homeostasis and amino acid-induced mTORC1 activation. Genes Cells. 2018 doi: 10.1111/gtc.12583. [DOI] [PubMed] [Google Scholar]
- Hasumi H, Baba M, Hong SB, Hasumi Y, Huang Y, Yao M, Valera VA, Linehan WM, Schmidt LS. Identification and characterization of a novel folliculin-interacting protein FNIP2. Gene. 2008;415(1-2):60–67. doi: 10.1016/j.gene.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard JJ, Fong V, Bathaie SZ, Tamanoi F. Recent progress in the study of the Rheb family GTPases. Cell Signal. 2014;26(9):1950–1957. doi: 10.1016/j.cellsig.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253(5022):905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- Helliwell SB, Wagner P, Kunz J, Deuter-Reinhard M, Henriquez R, Hall MN. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol Biol Cell. 1994;5(1):105–118. doi: 10.1091/mbc.5.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogeveen-Westerveld M, Exalto C, Maat-Kievit A, van den Ouweland A, Halley D, Nellist M. Analysis of TSC1 truncations defines regions involved in TSC1 stability, aggregation and interaction. Biochim Biophys Acta. 2010;1802(9):774–781. doi: 10.1016/j.bbadis.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Hoogeveen-Westerveld M, van Unen L, van den Ouweland A, Halley D, Hoogeveen A, Nellist M. The TSC1-TSC2 complex consists of multiple TSC1 and TSC2 subunits. BMC Biochem. 2012;13(1):18. doi: 10.1186/1471-2091-13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia KC, Stavropoulos P, Blobel G, Hoelz A. Architecture of a coat for the nuclear pore membrane. Cell. 2007;131(7):1313–1326. doi: 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412(2):179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem Soc Trans. 2009;37(Pt 1):217–222. doi: 10.1042/BST0370217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, et al. The bioplex network: A systematic exploration of the human interactome. Cell. 2015;162(2):425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im E, von Lintig FC, Chen J, Zhuang S, Qui W, Chowdhury S, Worley PF, Boss GR, Pilz RB. Rheb is in a high activation state and inhibits B-Raf kinase in mammalian cells. Oncogene. 2002;21(41):6356–6365. doi: 10.1038/sj.onc.1205792. [DOI] [PubMed] [Google Scholar]
- Imseng S, Aylett CH, Maier T. Architecture and activation of phosphatidylinositol 3-kinase related kinases. Curr Opin Struct Biol. 2018;49:177–189. doi: 10.1016/j.sbi.2018.03.010. [DOI] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17(15):1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4(9):648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Jeong JH, Lee KH, Kim YM, Kim DH, Oh BH, Kim YG. Crystal structure of the Gtr1p(GTP)-Gtr2p(GDP) protein complex reveals large structural rearrangements triggered by GTP-to-GDP conversion. J Biol Chem. 2012;287(35):29648–29653. doi: 10.1074/jbc.C112.384420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R, Guardia CM, Pu J, Chen Y, Bonifacino JS. BORC coordinates encounter and fusion of lysosomes with autophagosomes. Autophagy. 2017;13(10):1648–1663. doi: 10.1080/15548627.2017.1343768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Genau HM, Behrends C. Amino Acid-Dependent mTORC1 Regulation by the Lysosomal Membrane Protein SLC38A9. Mol Cell Biol. 2015;35(14):2479–2494. doi: 10.1128/MCB.00125-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandt RS, Haines JL, Smith M, Northrup H, Gardner RJ, Short MP, Dumars K, Roach ES, Steingold S, Wall S, Blanton SH, et al. Linkage of an important gene locus for tuberous sclerosis to a chromosome 16 marker for polycystic kidney disease. Nat Genet. 1992;2(1):37–41. doi: 10.1038/ng0992-37. [DOI] [PubMed] [Google Scholar]
- Kelley K, Knockenhauer KE, Kabachinski G, Schwartz TU. Atomic structure of the Y complex of the nuclear pore. Nat Struct Mol Biol. 2015;22(5):425–431. doi: 10.1038/nsmb.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Lamming DW. The mechanistic target of rapamycin: the grand conductor of metabolism and aging. Cell Metab. 2016;23(6):990–1003. doi: 10.1016/j.cmet.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110(2):163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11(4):895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10(8):935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, An S, Ro SH, Teixeira F, Park GJ, Kim C, Cho CS, Kim JS, Jakob U, Lee JH, Cho US. Janus-faced Sestrin2 controls ROS and mTOR signalling through two separate functional domains. Nat Commun. 2015;6(1) doi: 10.1038/ncomms10025. 10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Ro SH, Kim M, Park HW, Semple IA, Park H, Cho US, Wang W, Guan KL, Karin M, Lee JH. Sestrin2 inhibits mTORC1 through modulation of GATOR complexes. Sci Rep. 2015;5(1) doi: 10.1038/srep09502. 9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR, Gordon BS, Moyer JE, Dennis MD, Jefferson LS. Leucine induced dephosphorylation of Sestrin2 promotes mTORC1 activation. Cell Signal. 2016;28(8):896–906. doi: 10.1016/j.cellsig.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke S, Langemeyer L, Kuhlee A, Schuback S, Raunser S, Ungermann C, Kümmel D. Architecture and mechanism of the late endosomal Rab7-like Ypt7 guanine nucleotide exchange factor complex Mon1-Ccz1. Nat Commun. 2017;8 doi: 10.1038/ncomms14034. 14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan K, Spear ED, Kaiser CA, Fass D. Structural conservation of components in the amino acid sensing branch of the TOR pathway in yeast and mammals. J Mol Biol. 2010;402(2):388–398. doi: 10.1016/j.jmb.2010.07.034. [DOI] [PubMed] [Google Scholar]
- Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73(3):585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- Kurzbauer R, Teis D, de Araujo MEG, Maurer-Stroh S, Eisenhaber F, Bourenkov GP, Bartunik HD, Hekman M, Rapp UR, Huber LA, Clausen T. Crystal structure of the p14/MP1 scaffolding complex: how a twin couple attaches mitogen-activated protein kinase signaling to late endosomes. Proc Natl Acad Sci USA. 2004;101(30):10984–10989. doi: 10.1073/pnas.0403435101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski DJ. Rhebbing up mTOR: new insights on TSC1 and TSC2, and the pathogenesis of tuberous sclerosis. Cancer Biol Ther. 2003;2(5):471–476. doi: 10.4161/cbt.2.5.446. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Goldberg J. Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats. Cell. 2010;142(1):123–132. doi: 10.1016/j.cell.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Daniels RD, Wong LH, Gatta AT, Gerondopoulos A, Barr FA. Discovery of new Longin and Roadblock domains that form platforms for small GTPases in Ragulator and TRAPP-II. Small GTPases. 2013;4(2):62–69. doi: 10.4161/sgtp.24262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10(3):457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15(8):702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- Lunin VV, Munger C, Wagner J, Ye Z, Cygler M, Sacher M. The structure of the MARK scaffold, MP1, bound to its partner, p14. A complex with a critical role in endosomal map kinase signaling. J Biol Chem. 2004;279(22):23422–23430. doi: 10.1074/jbc.M401648200. [DOI] [PubMed] [Google Scholar]
- Marshall CB, Ho J, Buerger C, Plevin MJ, Li GY, Li Z, Ikura M, Stambolic V. Characterization of the intrinsic and TSC2-GAP-regulated GTPase activity of Rheb by real-time NMR. Sci Signal. 2009;2(55) doi: 10.1126/scisignal.2000029. ra3. [DOI] [PubMed] [Google Scholar]
- Mazhab-Jafari MT, Marshall CB, Ishiyama N, Ho J, Di Palma V, Stambolic V, Ikura M. An autoinhibited noncanonical mechanism of GTP hydrolysis by Rheb maintains mTORC1 homeostasis. Structure. 2012;20(9):1528–1539. doi: 10.1016/j.str.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Mc Cormack A, Sharpe C, Gregersen N, Smith W, Hayes I, George AM, Love DR. 12q14 Microdeletions: Additional Case Series with Confirmation of a Macrocephaly Region. Case Rep Genet. 2015:192071. doi: 10.1155/2015/192071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci. 2012;125(Pt 3):531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SMJr. Arginine: beyond protein. Am J Clin Nutr. 2006;83(2):508S–512S. doi: 10.1093/ajcn/83.2.508S. [DOI] [PubMed] [Google Scholar]
- Mozaffari M, Hoogeveen-Westerveld M, Kwiatkowski D, Sampson J, Ekong R, Povey S, den Dunnen JT, van den Ouweland A, Halley D, Nellist M. Identification of a region required for TSC1 stability by functional analysis of TSC1 missense mutations found in individuals with tuberous sclerosis complex. BMC Med Genet. 2009;10(1):88. doi: 10.1186/1471-2350-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Z, Wang L, Deng W, Wang J, Wu G. Structural insight into the Ragulator complex which anchors mTORC1 to the lysosomal membrane. Cell Discov. 2017;3:17049. doi: 10.1038/celldisc.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nada S, Hondo A, Kasai A, Koike M, Saito K, Uchiyama Y, Okada M. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J. 2009;28(5):477–489. doi: 10.1038/emboj.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy V, Hsia KC, Debler EW, Kampmann M, Davenport AM, Blobel G, Hoelz A. Structure of a trimeric nucleoporin complex reveals alternate oligomerization states. Proc Natl Acad Sci USA. 2009;106(42):17693–17698. doi: 10.1073/pnas.0909373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima N, Noguchi E, Nishimoto T. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics. 1999;152(3):853–867. doi: 10.1093/genetics/152.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neklesa TK, Davis RW. A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS Genet. 2009;5(6):e1000515. doi: 10.1371/journal.pgen.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellist M, Goedbloed MA, Halley DJJ. Regulation of tuberous sclerosis complex (TSC) function by 14-3-3 proteins. Biochem Soc Trans. 2003;31:587–591. doi: 10.1042/bst0310587. [DOI] [PubMed] [Google Scholar]
- Nellist M, van Slegtenhorst MA, Goedbloed M, van den Ouweland AM, Halley DJ, van der Sluijs P. Characterization of the cytosolic tuberin-hamartin complex. Tuberin is a cytosolic chaperone for hamartin. J Biol Chem. 1999;274(50):35647–35652. doi: 10.1074/jbc.274.50.35647. [DOI] [PubMed] [Google Scholar]
- Nickerson ML, Warren MB, Toro JR, Matrosova V, Glenn G, Turner ML, Duray P, Merino M, Choyke P, Pavlovich CP, Sharma N, et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dubé syndrome. Cancer Cell. 2002;2(2):157–164. doi: 10.1016/s1535-6108(02)00104-6. [DOI] [PubMed] [Google Scholar]
- Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K, Hara K, Tanaka N, Avruch J, Yonezawa K. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J Biol Chem. 2003;278(18):15461–15464. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- Nookala RK, Langemeyer L, Pacitto A, Ochoa-Montaño B, Donaldson JC, Blaszczyk BK, Chirgadze DY, Barr FA, Bazan JF, Blundell TL. Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol. 2012;2(8):120071. doi: 10.1098/rsob.120071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton LE, Layman DK. Leucine regulates translation initiation of protein synthesis in skeletal muscle after exercise. J Nutr. 2006;136(2):533S–537S. doi: 10.1093/jn/136.2.533S. [DOI] [PubMed] [Google Scholar]
- Oshiro N, Takahashi R, Yoshino K, Tanimura K, Nakashima A, Eguchi S, Miyamoto T, Hara K, Takehana K, Avruch J, Kikkawa U, et al. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem. 2007;282(28):20329–20339. doi: 10.1074/jbc.M702636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacitto A, Ascher DB, Wong LH, Blaszczyk BK, Nookala RK, Zhang N, Dokudovskaya S, Levine TP, Blundell TL. Lst4, the yeast Fnip1/2 orthologue, is a DENN-family protein. Open Biol. 2015;5(12):150174. doi: 10.1098/rsob.150174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajusalu S, Reimand T, Õunap K. Novel homozygous mutation in KPTN gene causing a familial intellectual disability-macrocephaly syndrome. Am J Med Genet A. 2015;167A(8):1913–1915. doi: 10.1002/ajmg.a.37105. [DOI] [PubMed] [Google Scholar]
- Panchaud N, Péli-Gulli MP, De Virgilio C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci Signal. 2013a;6(277) doi: 10.1126/scisignal.2004112. ra42. [DOI] [PubMed] [Google Scholar]
- Panchaud N, Péli-Gulli MP, De Virgilio C. SEACing the GAP that nEGOCiates TORC1 activation: evolutionary conservation of Rag GTPase regulation. Cell Cycle. 2013b;12(18):2948–2952. doi: 10.4161/cc.26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Jin W, Woo JR, Shoelson SE. Crystal structures of human TBC1D1 and TBC1D4 (AS160) RabGTPase-activating protein (RabGAP) domains reveal critical elements for GLUT4 translocation. J Biol Chem. 2011;286(20):18130–18138. doi: 10.1074/jbc.M110.217323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar N, Tamanoi F. The Enzymes. Elsevier; 2010. Rheb G-Proteins and the Activation of mTORC1; pp. 39–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmigiani A, Nourbakhsh A, Ding B, Wang W, Kim YC, Akopiants K, Guan KL, Karin M, Budanov AV. Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Reports. 2014;9(4):1281–1291. doi: 10.1016/j.celrep.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters H, Debeer P, Bairoch A, Wilquet V, Huysmans C, Parthoens E, Fryns JP, Gewillig M, Nakamura Y, Niikawa N, Van de Ven W, Devriendt K. PA26 is a candidate gene for heterotaxia in humans: identification of a novel PA26-related gene family in human and mouse. Hum Genet. 2003;112(5-6):573–580. doi: 10.1007/s00439-003-0917-5. [DOI] [PubMed] [Google Scholar]
- Péli-Gulli MP, Raucci S, Hu Z, Dengjel J, De Virgilio C. Feedback Inhibition of the Rag GTPase GAP Complex Lst4-Lst7 Safeguards TORC1 from Hyperactivation by Amino Acid Signals. Cell Reports. 2017;20(2):281–288. doi: 10.1016/j.celrep.2017.06.058. [DOI] [PubMed] [Google Scholar]
- Péli-Gulli MP, Sardu A, Panchaud N, Raucci S, De Virgilio C. Amino Acids Stimulate TORC1 through Lst4-Lst7, a GTPase-Activating Protein Complex for the Rag Family GTPase Gtr2. Cell Reports. 2015;13(1):1–7. doi: 10.1016/j.celrep.2015.08.059. [DOI] [PubMed] [Google Scholar]
- Peng M, Yin N, Li MO. Sestrins function as guanine nucleotide dissociation inhibitors for Rag GTPases to control mTORC1 signaling. Cell. 2014;159(1):122–133. doi: 10.1016/j.cell.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Yin N, Li MO. SZT2 dictates GATOR control of mTORC1 signalling. Nature. 2017;543(7645):433–437. doi: 10.1038/nature21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137(5):873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol. 2013;202(7):1107–1122. doi: 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Huang H, Xu T. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell. 2001;105(3):357–368. doi: 10.1016/s0092-8674(01)00333-6. [DOI] [PubMed] [Google Scholar]
- Powis K, Zhang T, Panchaud N, Wang R, De Virgilio C, Ding J. Crystal structure of the Ego1-Ego2-Ego3 complex and its role in promoting Rag GTPase-dependent TORC1 signaling. Cell Res. 2015;25(9):1043–1059. doi: 10.1038/cr.2015.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J, Keren-Kaplan T, Bonifacino JS. A Ragulator-BORC interaction controls lysosome positioning in response to amino acid availability. J Cell Biol. 2017;216(12):4183–4197. doi: 10.1083/jcb.201703094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J, Schindler C, Jia R, Jarnik M, Backlund P, Bonifacino JS. BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell. 2015;33(2):176–188. doi: 10.1016/j.devcel.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Zhang Q, Wang X, Zeng L, Farooq A, Zhou MM. Structure of the adaptor protein p14 reveals a profilin-like fold with distinct function. J Mol Biol. 2005;347(2):309–321. doi: 10.1016/j.jmb.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Qin J, Wang Z, Hoogeveen-Westerveld M, Shen G, Gong W, Nellist M, Xu W. Structural Basis of the Interaction between Tuberous Sclerosis Complex 1 (TSC1) and Tre2-Bub2-Cdc16 Domain Family Member 7 (TBC1D7) J Biol Chem. 2016;291(16):8591–8601. doi: 10.1074/jbc.M115.701870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebsamen M, Pochini L, Stasyk T, de Araujo MEG, Galluccio M, Kandasamy RK, Snijder B, Fauster A, Rudashevskaya EL, Bruckner M, Scorzoni S, et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;519(7544):477–481. doi: 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricos MG, Hodgson BL, Pippucci T, Saidin A, Ong YS, Heron SE, Licchetta L, Bisulli F, Bayly MA, Hughes J, Baldassari S, et al. Epilepsy Electroclinical Study Group. Mutations in the mammalian target of rapamycin pathway regulators NPRL2 and NPRL3 cause focal epilepsy. Ann Neurol. 2016;79(1):120–131. doi: 10.1002/ana.24547. [DOI] [PubMed] [Google Scholar]
- Roberg KJ, Bickel S, Rowley N, Kaiser CA. Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae by SEC13, LST4, LST7 and LST8. Genetics. 1997;147(4):1569–1584. doi: 10.1093/genetics/147.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosset C, Netto CBO, Ashton-Prolla P. TSC1 and TSC2 gene mutations and their implications for treatment in Tuberous Sclerosis Complex: a review. Genet Mol Biol. 2017;40(1):69–79. doi: 10.1590/1678-4685-GMB-2015-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78(1):35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, Abraham RT. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270(2):815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- Saito K, Araki Y, Kontani K, Nishina H, Katada T. Novel role of the small GTPase Rheb: its implication in endocytic pathway independent of the activation of mammalian target of rapamycin. J Biochem. 2005;137(3):423–430. doi: 10.1093/jb/mvi046. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320(5882):1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatin DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25(6):903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sato T, Nakashima A, Guo L, Tamanoi F. Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein. J Biol Chem. 2009;284(19):12783–12791. doi: 10.1074/jbc.M809207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5(6):566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- Saxton RA, Chantranupong L, Knockenhauer KE, Schwartz TU, Sabatini DM. Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature. 2016a;536(7615):229–233. doi: 10.1038/nature19079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Knockenhauer KE, Wolfson RL, Chantranupong L, Pacold ME, Wang T, Schwartz TU, Sabatini DM. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science. 2016b;351(6268):53–58. doi: 10.1126/science.aad2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalm SS, Blenis J. Identification of a conserved motif required for mTOR signaling. Curr Biol. 2002;12(8):632–639. doi: 10.1016/s0960-9822(02)00762-5. [DOI] [PubMed] [Google Scholar]
- Schalm SS, Fingar DC, Sabatini DM, Blenis J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr Biol. 2003;13(10):797–806. doi: 10.1016/s0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- Schmidt LS, Linehan WM. FLCN: The causative gene for Birt-Hogg-Dube syndrome. Gene. 2018;640:28–42. doi: 10.1016/j.gene.2017.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitzberger F, Harrison SC. RWD domain: a recurring module in kinetochore architecture shown by a Ctf19-Mcm21 complex structure. EMBO Rep. 2012;13(3):216–222. doi: 10.1038/embor.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann A, Brauers A, Massmann S, Becker W, Joost HG. Cloning of a novel family of mammalian GTP-binding proteins (RagA, RagBs, RagB1) with remote similarity to the Ras-related GTPases. J Biol Chem. 1995;270(48):28982–28988. doi: 10.1074/jbc.270.48.28982. [DOI] [PubMed] [Google Scholar]
- Scrima A, Thomas C, Deaconescu D, Wittinghofer A. The Rap-RapGAP complex: GTP hydrolysis without catalytic glutamine and arginine residues. EMBO J. 2008;27(7):1145–1153. doi: 10.1038/emboj.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel Gproteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001;276(10):7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- Shen K, Choe A, Sabatini DM. Intersubunit Crosstalk in the Rag GTPase Heterodimer Enables mTORC1 to Respond Rapidly to Amino Acid Availability. Mol Cell. 2017;68(3):552–565.e8. doi: 10.1016/j.molcel.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Huang RK, Brignole EJ, Condon KJ, Valenstein ML, Chantranupong L, Bomaliyamu A, Choe A, Hong C, Yu Z, Sabatini DM. Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes. Nature. 2018;556(7699):64–69. doi: 10.1038/nature26158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumway SD, Li Y, Xiong Y. 14-3-3beta binds to and negatively regulates the tuberous sclerosis complex 2 (TSC2) tumor suppressor gene product, tuberin. J Biol Chem. 2003;278(4):2089–2092. doi: 10.1074/jbc.C200499200. [DOI] [PubMed] [Google Scholar]
- Springe r TA. Folding of the N-terminal, ligand-binding region of integrin alpha-subunits into a beta-propeller domain. Proc Natl Acad Sci USA. 1997;94(1):65–72. doi: 10.1073/pnas.94.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starling GP, Yip YY, Sanger A, Morton PE, Eden ER, Dodding MP. Folliculin directs the formation of a Rab34-RILP complex to control the nutrient-dependent dynamic distribution of lysosomes. EMBO Rep. 2016;17(6):823–841. doi: 10.15252/embr.201541382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5(6):559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- Stuwe T, Correia AR, Lin DH, Paduc h M, Lu VT, Kossiakoff AA, Hoelz A. Nuclear pores. Architecture of the nuclear pore complex coat. Science. 2015;347(6226):1148–1152. doi: 10.1126/science.aaa4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su MY, Morris KL, Kim DJ, Fu Y, Lawrence R, Stjepanovic G, Zoncu R, Hurley JH. Hybrid Structure of the RagA/C-Ragulator mTORC1 Activation Complex. Mol Cell. 2017;68(5):835–846. doi: 10.1016/j.molcel.2017.10.016. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Zhu YJ, Wang Z, Zhong Q, Gao F, Lou J, Gong W, Xu W. Crystal structure of the yeast TSC1 core domain and implications for tuberous sclerosis pathological mutations. Nat Commun. 2013;4(1) doi: 10.1038/ncomms3135. 2135. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Nakagawa M, Young SG, Yamanaka S. Differential membrane localization of ERas and Rheb, two Ras-related proteins involved in the phosphatidylinositol 3-kinase/mTOR pathway. J Biol Chem. 2005;280(38):32768–32774. doi: 10.1074/jbc.M506280200. [DOI] [PubMed] [Google Scholar]
- Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001;105(3):345–355. doi: 10.1016/s0092-8674(01)00332-4. [DOI] [PubMed] [Google Scholar]
- Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tuberous sclerosis complex-1 and-2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci USA. 2002;99(21):13571–13576. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13(15):1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- Teis D, Wunderlich W, Huber LA. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev Cell. 2002;3(6):803–814. doi: 10.1016/s1534-5807(02)00364-7. [DOI] [PubMed] [Google Scholar]
- Tomasoni R, Mondino A. The tuberous sclerosis complex: balancing proliferation and survival. Biochem Soc Trans. 2011;39(2):466–471. doi: 10.1042/BST0390466. [DOI] [PubMed] [Google Scholar]
- Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52(4):495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kant R, Jonker CTH, Wijdeven RH, Bakker J, Janssen L, Klumperman J, Neefjes J. Characterization of the mammalian CORVET and HOPS complexes and their modular restructuring for endosome specificity. J Biol Chem. 2015;290(51):30280–30290. doi: 10.1074/jbc.M115.688440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Slegtenhorst M, de Hoogt R, Hermans C, Nellist M, Janssen B, Verhoef S, Lindhout D, van den Ouweland A, Halley D, Young J, Burley M, et al. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277(5327):805–808. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- van Slegtenhorst M, Nellist M, Nagelkerken B, Cheadle J, Snell R, van den Ouweland A, Reuser A, Sampson J, Halley D, van der Sluijs P. Interaction between hamartin and tuberin, the TSC1 and TSC2 gene products. Hum Mol Genet. 1998;7(6):1053–1057. doi: 10.1093/hmg/7.6.1053. [DOI] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9(3):316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Velasco-Miguel S, Buckbinder L, Jean P, Gelbert L, Talbott R, Laidlaw J, Seizinger B, Kley N. PA26, a novel target of the p53 tumor suppressor and member of the GADD family of DNA damage and growth arrest inducible genes. Oncogene. 1999;18(1):127–137. doi: 10.1038/sj.onc.1202274. [DOI] [PubMed] [Google Scholar]
- Vilella-Bach M, Nuzzi P, Fang Y, Chen J. The FKBP12-rapamycin-binding domain is required for FKBP12-rapamycin-associated protein kinase activity and G1 progression. J Biol Chem. 1999;274(7):4266–4272. doi: 10.1074/jbc.274.7.4266. [DOI] [PubMed] [Google Scholar]
- Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, Wang T, et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347(6218):188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenter R, Hütz K, Dibbern D, Li T, Reisinger V, Plösche r M, Eichacker L, Eddie B, Hanson T, Bryant DA, Overmann J. Expression-based identification of genetic determinants of the bacterial symbiosis ‘Chlorochromatium aggregatum’. Environ Microbiol. 2010;12(8):2259–2276. doi: 10.1111/j.1462-2920.2010.02206.x. [DOI] [PubMed] [Google Scholar]
- Whittaker CA, Hynes RO. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell. 2002;13(10):3369–3387. doi: 10.1091/mbc.E02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JRR, Schwartz TU. Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat. J Cell Biol. 2010;190(3):347–361. doi: 10.1083/jcb.201003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson RL, Chantranupong L, Wyant GA, Gu X, Orozco JM, Shen K, Condon KJ, Petri S, Kedir J, Scaria SM, Abu-Remaileh M, et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature. 2017;543(7645):438–442. doi: 10.1038/nature21423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson RL, Sabatini DM. The Dawn of the Age of Amino Acid Sensors for the mTORC1 Pathway. Cell Metab. 2017;26(2):301–309. doi: 10.1016/j.cmet.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Tu BP. Selective regulation of autophagy by the Iml1-Npr2-Npr3 complex in the absence of nitrogen starvation. Mol Biol Cell. 2011;22(21):4124–4133. doi: 10.1091/mbc.E11-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Wang R, Zhang T, Ding J. Structural insight into the arginine-binding specificity of CASTOR1 in amino acid-dependent mTORC1 signaling. Cell Discov. 2016;2(1):16035. doi: 10.1038/celldisc.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin alpha Vbeta3. Science. 2001;294(5541):339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2(3):202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Sanders LK, Kaufmann WE, Yee W, Barnes CA, Nathans D, Worley PF. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J Biol Chem. 1994;269(23):16333–16339. [PubMed] [Google Scholar]
- Yang H, Wang JLM, Chen X, Huang M, Tan D, Dong MQ, Wong CCL, Wang J, Xu Y, Wang HW. 4.4 Å Resolution Cryo-EM structure of human mTOR Complex 1. Protein Cell. 2016;7:878–887. doi: 10.1007/s13238-016-0346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Jiang X, Li B, Yang HJ, Miller M, Yang A, Dhar A, Pavletich NP. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature. 2017;552(7685):368–373. doi: 10.1038/nature25023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP. mTOR kinase structure, mechanism and regulation. Nature. 2013;497(7448):217–223. doi: 10.1038/nature12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonehara R, Nada S, Nakai T, Nakai M, Kitamura A, Ogawa A, Nakatsumi H, Nakayama KI, Li S, Standley DM, Yamashita E, Nakagawa A, Okada M. Structural basis for the assembly of the Ragulator-Rag GTPase complex. Nat Commun. 2017;8(1) doi: 10.1038/s41467-017-01762-3. 1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Li S, Xu X, Li Y, Guan K, Arnold E, Ding J. Structural basis for the unique biological function of small GTPase RHEB. J Biol Chem. 2005;280(17):17093–17100. doi: 10.1074/jbc.M501253200. [DOI] [PubMed] [Google Scholar]
- Zanetti G, Prinz S, Daum S, Meister A, Schekman R, Bacia K, Briggs JAG. The structure of the COPII transport-vesicle coat assembled on membranes. eLife. 2013;2 doi: 10.7554/eLife.00951. e00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech R, Kiontke S, Mueller U, Oeckinghaus A, Kümmel D. Structure of the Tuberous Sclerosis Complex 2 (TSC2) N Terminus Provides Insight into Complex Assembly and Tuberous Sclerosis Pathogenesis. J Biol Chem. 2016;291(38):20008–20020. doi: 10.1074/jbc.M116.732446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Iyer LM, He F, Aravind L. Discovery of Novel DENN Proteins: Implications for the Evolution of Eukaryotic Intracellular Membrane Structures and Human Disease. Front Genet. 2012;3:283. doi: 10.3389/fgene.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Péli-Gulli MP, Yang H, De Virgilio C, Ding J. Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the ego complex to activate TORC1. Structure. 2012;20(12):2151–2160. doi: 10.1016/j.str.2012.09.019. [DOI] [PubMed] [Google Scholar]
- Zhang T, Wang R, Wang Z, Wang X, Wang F, Ding J. Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1. Nat Commun. 2017;8(1) doi: 10.1038/s41467-017-01567-4. 1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5(6):578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334(6056):678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.