Abstract
The chloroplast ATP synthase uses the electrochemical proton gradient generated by photosynthesis to produce ATP, the energy currency of all cells. Protons conducted through the membrane-embedded Fo motor drive ATP synthesis in the F1 head by rotary catalysis. We determined the high-resolution structure of the complete cF1Fo complex by cryo-EM, resolving sidechains of all 26 protein subunits, the five nucleotides in the F1 head, and the proton pathway to and from the rotor ring. The flexible peripheral stalk redistributes differences in torsional energy across three unequal steps in the rotation cycle. Plant ATP synthase is autoinhibited by a β-hairpin redox switch in subunit γ that blocks rotation in the dark.
F-type ATP synthases use the free energy of the membrane potential to synthesize ATP from ADP and inorganic phosphate (1) by rotary catalysis (2, 3). ATP is generated by the tightly coupled action of the catalytic F1 head and the Fo motor in the membrane. F1 consists of three asymmetric αβ heterodimers, which define the catalytic sites (3), and the central stalk of subunits γ and ε, which are attached to the c-ring (4, 5). The Fo motor consists of the c-ring rotor, subunit a and the peripheral stalk. Two aqueous channels in Fo, each spanning half of the membrane, were proposed to conduct protons to and from conserved glutamates in the c-ring to drive rotation (6, 7), and recently observed in mitochondrial ATP synthases (8-10). Rotation of the central stalk, driven by the proton-motive force (pmf) across the membrane, causes sequential conformational changes in the αβ heterodimers, resulting in the synthesis of three molecules of ATP per revolution. The peripheral stalk acts as a stator to prevent unproductive rotation of F1 with the Fo motor.
Green plants, algae and cyanobacteria generate ATP and NADPH by photophosphorylation. The chloroplast ATP synthase (cF1Fo) is located in the stroma lamellae and flat grana end membranes (11). It is spatially separated from the water-splitting photosystem II in the chloroplast grana. In contrast to mitochondrial ATP synthase dimers (12), cF1Fo is monomeric and does not bend the membrane (11). In terms of overall structure and subunit composition, cF1Fo closely resembles the ATP synthases of bacteria.
ATP synthases are fully reversible and can catalyze ATP synthesis or hydrolysis. The catalytic direction depends on the pmf across the membrane and the concentration of ADP and ATP. Most organisms have developed inhibitory mechanisms that block wasteful ATP hydrolysis when the pmf is insufficient to drive ATP synthesis (13, 14). The plant chloroplast ATP synthase has a ~40 amino acid insertion in the γ subunit that attenuates cF1Fo activity in the dark through the formation of a disulfide bond (15, 16), a process known as thiol modulation (17). Without an atomic model of this redox loop, the mechanism of thiol modulation was not understood.
So far, no high-resolution structure of a complete and functional ATP synthase has been available. We determined the structure of the intact spinach chloroplast ATP synthase by cryo-EM, which enabled us to build atomic models of all its 26 subunits.
Overall structure and rotary conformations of cF1Fo
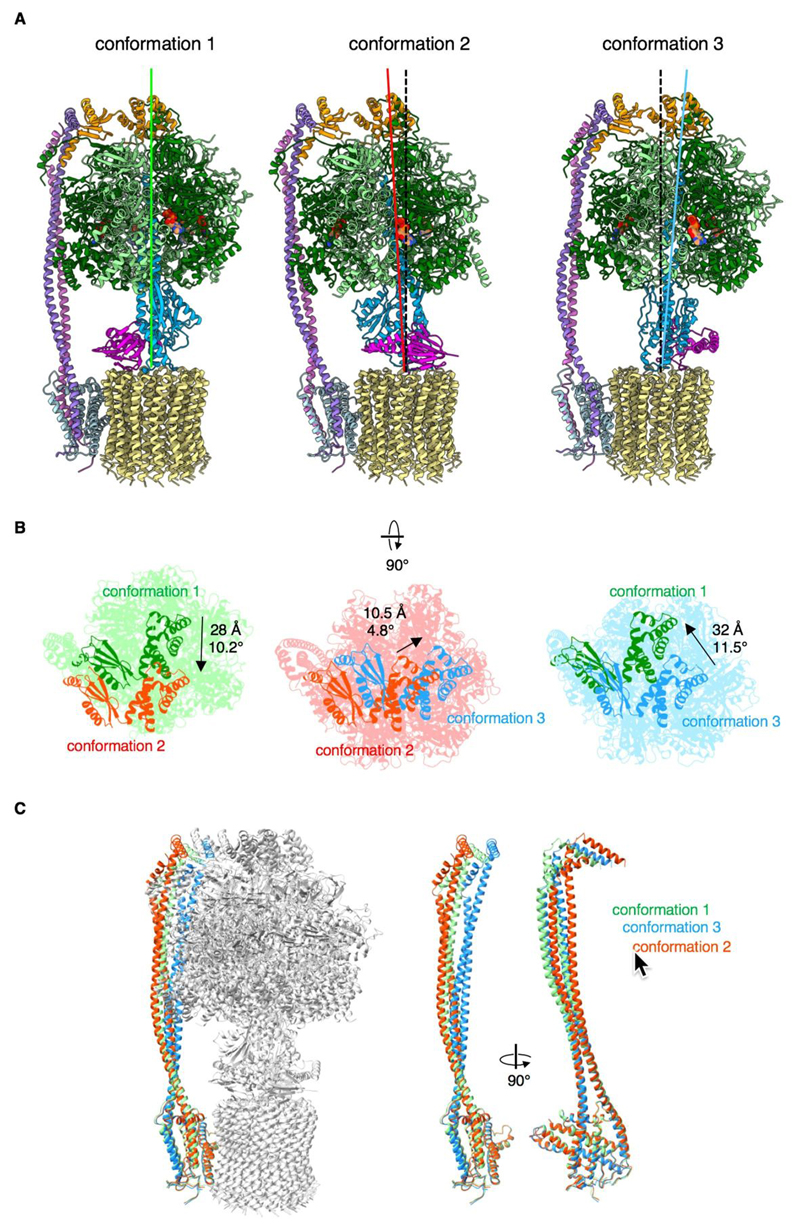
We isolated native cF1Fo from market spinach and reconstituted it into lipid nanodiscs (Fig. S1A). The isolated complex contained all protein subunits, was highly pure and fully autoinhibited (Fig. S1B). When detached from Fo by detergent, F1 was hydrolytically active (Fig. S1B-D). Particles selected from cryo-EM images were sorted into three distinct conformations, each with the rotor arrested in a different rotary state (Fig. S2 and S3). Conformation 1 was the most populated. A 3D reconstruction of particles in this class had an overall resolution of 3.15 Å (Fig. 1A). After masking and local realignment, the F1 head and the membrane-embedded Fo attained average resolutions of 3.0 and 3.4 Å, respectively (Fig. S3C-D, Fig. S4). 3D maps of the less populated conformations 2 and 3 were reconstructed at ~4.5 Å resolution (Fig. S3A-B). The three maps differ in the orientation of the central rotor and represent different resting positions in rotary catalysis (Fig. 2A), similar to what has been observed at lower resolution in E. coli (18) and mitochondrial ATP synthase (19, 20). The three αβ assemblies show the open, loose, and tight conformations of the nucleotide binding pockets (21). In conformation 2 and 3, the cF1 head and peripheral stalk are tilted by ~10° with respect to conformation 1 (Fig. 2). Consequently, the cF1 head and peripheral stalk together perform a precession movement around a central axis, as subunit γ pushes sequentially against each β subunit in the transition between the three rotary states (Movie 1, Movie 2). Simple mechanical models of ATP synthase assume that the 360° turn of the rotor is divided into three equal 120° steps that each result in the production of one ATP. The symmetry mismatch between the 14-fold Fo rotor and the near-threefold F1 head means that the number of c-subunits rotating past subunit a to generate one ATP is not an integer. cF1Fo requires, on average, 4.67 c-subunits, or protons, to produce one ATP. The nearest integral numbers of c-subunits per step would be 4, 5 and 5, equivalent to rotation angles of 103°, 129° and 129°. Surprisingly however, the three conformations are separated by rotations of 103o, 112o and 145o, or 4, 4.4 and 5.6 c-subunits (Fig. S5C). This means that the position of the c-subunits relative to subunit a in the three conformations differs.
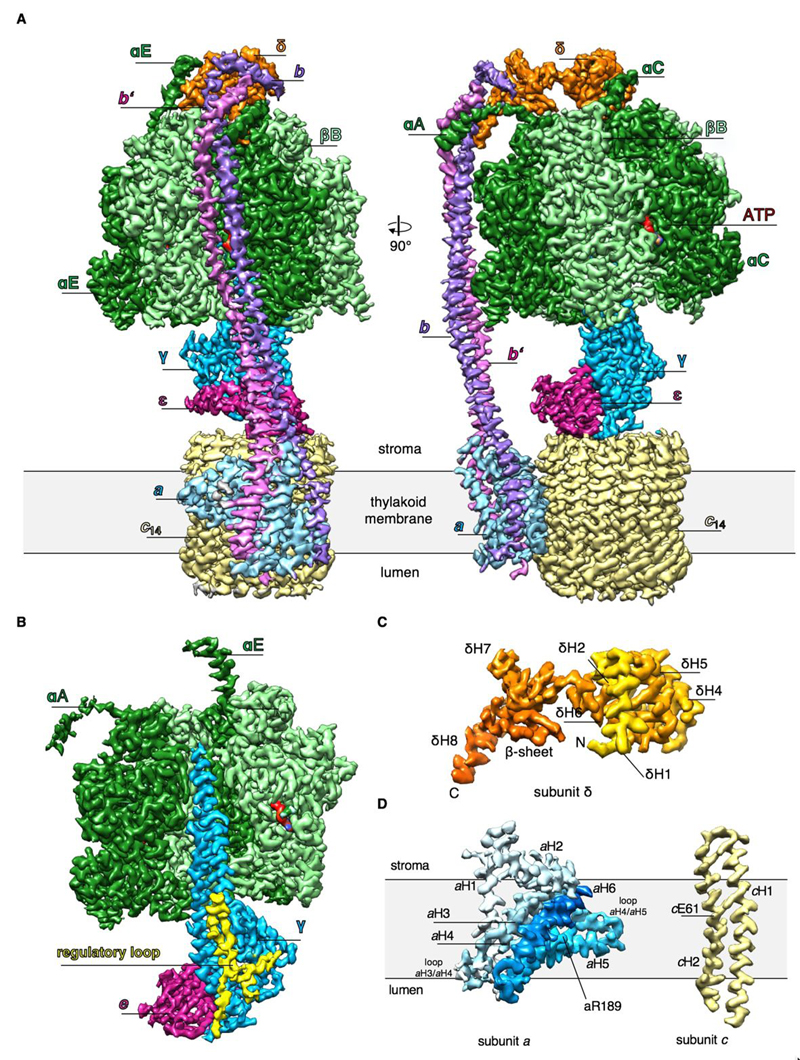
Fig. 1. High-resolution cryo-EM map of the spinach chloroplast cF1Fo ATP synthase.
(A) Surface representation of the overall structure. α subunits, dark green; β subunits, light green; γ, blue; δ, orange; ε, purple; a, light blue; b, violet, b’, pink; c 14, pale yellow; ATP, red. cFo (abb’c 14) is embedded in the thylakoid membrane (grey) while cF1 (α3β3γεδ) extends into the stroma. N-terminal helices of the αA, αC and αE subunits are indicated. (B) Side view of segmented cF1 sub-complex. Subunits βB, αC and δ are omitted for clarity. The plant-specific L-shaped redox-loop of subunit γ is highlighted in yellow. (C) The map density of the two-domain subunit δ is colored from N (yellow) to C-terminus (orange). (D) Segmented maps of membrane-embedded subunits a and c. Subunit a density is colored from light to dark blue to show the N-terminal transmembrane helix H1, the amphipathic helix H2 on the stromal membrane surface and the two membrane-intrinsic helix hairpins H3/H4 and H5/H6. Residues aArg189 and cGlu61 that are essential for pmf-coupled ATP synthesis are indicated.
Fig. 2. Three rotary states of the chloroplast ATP synthase.
(A) 3D classification of pre-aligned cF1Fo particles indicates three different resting states, each with a distinct conformation. In conformation 2, subunits γ (blue) and ε (purple) of the central stalk are rotated by 112° in ATP synthesis direction relative to conformation 1. In conformation 3, the central stalk is rotated further by 103°. The rotation angle from conformation 3 to conformation 1 is 145°. The dashed black line indicates the axis of c 14-ring and central stalk rotation. (B) Top view on the cF1 head. Conformation 2 is tilted by 10.2° with respect to conformation 1, indicated by the shift of subunit δ (green to red). Conformation 3 is tilted by 4.8° with respect to conformation 2 (red to blue) and 11.5° with respect to conformation 1 (blue to green). This movement results in a precession of cF1 relative to cFo during rotary catalysis. (C) Conformation 1, 2 and 3 superimposed and aligned on subunit a. Subunits abb’ are colored by conformation (1, green; 2, red; 3, blue). The peripheral stalk of subunits bb’ is firmly attached to cF1 by subunit δ and bends together with cF1 in rotary conformation 2 and 3 relative to conformation 1.
The free enthalpy ΔG of ATP hydrolysis under physiological conditions in chloroplasts is around -51 kJ/mol (22). Given that ATP synthases operate reversibly close to thermodynamic equilibrium, each proton translocated by cF1Fo contributes -51x3/14 or -10.9 kJ/mol to ATP synthesis. The three observed rotary states indicate energy contributions of -43.7, -48.1 and -61.2 kJ/mol per step. Single-molecule experiments with E. coli ATP synthase have been taken to indicate that energy differences due to symmetry mismatch are stored as a torsional force by the flexible subunit γ (23), whereas the peripheral stalk was thought to be too stiff (24). Our structures of the three rotary cF1Fo states show that the peripheral stalk bends relative to the central axis (Fig. 2; Fig. S5) and thus couples F1 elastically to Fo. The torsion of subunit γ would relax intermittently, each time one molecule of ATP is produced. The bending energy stored in subunits b and b’ is released when the peripheral stalk reverts to its initial conformation after one full rotation, and is thus distributed over all three steps. Acting like an elastic spring, the peripheral stalk evens out the energy minima between the three observed rotational states to optimize ATP synthesis by rotary catalysis.
Structure of the cF1 head with bound nucleotides
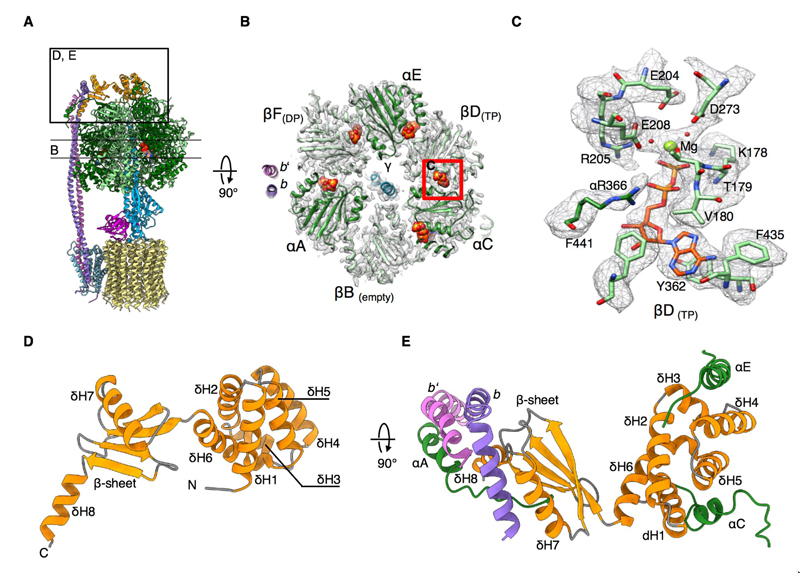
Compared to mitochondrial ATP synthase, detailed information on the structure of the chloroplast complex is limited. In crystal structures of the catalytic subunits, the α3β3 subcomplex is symmetrized and does not contain nucleotides (25, 26). In our structure, the cF1 head is the best-resolved part of the map, with a local resolution of 2.9 Å (Fig. S3C). Sidechains, nucleotides and some water molecules in the nucleotide-binding pockets are readily visible (Fig. 3C). The head is asymmetric with the three αβ pairs in different conformations (Fig. 3B). In crystal structures of mitochondrial F1, one of the catalytic β subunits (βDP) contains ADP, one (βTP) a non-hydrolysable ATP analogue, while the third site (βempty) is unoccupied (3). In our structure, both the βDP and βTP sites contain Mg-ADP (Figure 3B). We isolated cF1Fo without addition of nucleotides, non-hydrolysable substrates or inhibitors. cF1 is most likely in the ADP-inhibited state (27) as a result of hydrolysis during isolation. This conclusion is supported by the absence of phosphate (Pi) density in the binding pockets. Mg-ATP is resolved in the nucleotide-binding sites of the three non-catalytic α-subunits (Fig. 3B).
Fig. 3. Structure of the cF1 head.
(A) Overview of cF1Fo, indicating the section through the nucleotide binding domains (NBDs) in (B) and the position of subunit δ shown in (D) and (E). (B) Cross-section through cF1 as indicated in (A). Each αβ dimer (chains αAβB, αCβD, αEβF) is stalled in a different conformation of the binding-change mechanism (2, 3). Both NBDs of βD and βF (corresponding to βTP and βDP) are occupied by Mg-ADP. βB (corresponding to βempty) is unoccupied. All non-catalytic α subunits bind ATP. (C) Details of the βD NBD with Mg-ADP bound. Mg ions, water molecules and salt bridges involved in Mg coordination are resolved. (D) Two-domain structure of subunit δ. The δ N-terminal domain is a bundle of six α-helices (δH1-δH6). The δ C-terminal domain is a four-stranded mixed β-sheet with two α-helices (δH7, δH8). (E) The δ-subunit connects the peripheral stalk to the F1 head. The N-terminal α-helix bundle interacts in different ways with the N-termini of αC and αE. The N-terminus of αA interacts with the peripheral stalk subunit b’ (pink). The C-terminal domain of δ binds to the kinked C-terminus of b (purple).
The F1 nucleotide-binding sites are highly conserved throughout evolution; the α and β subunits of cF1 respectively share 58 and 67% sequence identity with their mitochondrial counterparts, and the residues forming the nucleotide binding sites are identical (25). A comparison of our cF1 structure to F1 from bovine mitochondria (PDB ID 1BMF) (28) reveals no differences in the nucleotide binding sites (Fig. S6), except the orientation of αArg366, the catalytically essential “arginine finger” that is involved in coordinating the γ-phosphate (3). In the bovine structure this site (βTP) contains the non-hydrolysable ATP analogue AMP-PNP.
Connection of F1 head to the peripheral stalk
The peripheral stalk of cF1Fo consists of subunits δ, b and b’. The δ subunit (called OSCP in mitochondria) connecting the peripheral stalk to the F1 head consists of two domains. The structure of the α-helical N-terminal domain has been determined (29, 30), but the C-terminal domain that binds the peripheral stalk has so far been seen only at low resolution and has not been modelled (18, 20). The C-terminal domain consists of a four-stranded mixed β-sheet and two α-helices (Fig. 3D) and provides a binding platform for the kinked C-terminus of the peripheral stalk subunit b. Remarkably, the fold of this domain is conserved in the peripheral stalk subunit of the A-type ATPase from Thermus thermophilus (31, 32) (Fig. S7). The N-terminal domain of δ is a bundle of six short α-helices that sits in a central position on top of the F1 crown (Fig. 3D). Each of three F1 α subunits binds to δ in a different way. The N-terminal helix of αC (residues α10 to 20) forms an arc that interacts with δH1 and δH5 (Fig. 3E). By contrast, the N-terminal helix of αE (residues α10 to 25) sits vertically next to δH3 and δH4. The N-terminus of αA extends to the far side of the δ-subunit C-terminal domain in a long loop that turns into a short helix (α6 to 18), which interacts with the C-terminal helix of peripheral stalk subunit b’ (Fig. 3E). The peripheral stalk is attached to the F1 head mostly by hydrophobic helix-helix interactions of subunits α, δ, b and b’. With its two-domain structure and central position on the F1 head, subunit δ ensures that only one peripheral stalk can attach to the α3β3 heterotrimer.
Subunits b and b’ are almost entirely α-helical and form a loose right-handed coiled coil (33) that ends just above the membrane surface (Fig. 4A). This arrangement is conserved in all rotary ATPases, including the b-subunit homodimer of the E. coli ATP synthase and the heterodimeric outer stalk of the A/V-type ATPase (18, 31), which is very distantly related (Fig. S7). Near the membrane surface the helices separate and enter the membrane, where they clamp the a-subunit to the c-ring in the Fo subcomplex (Fig. 1A).
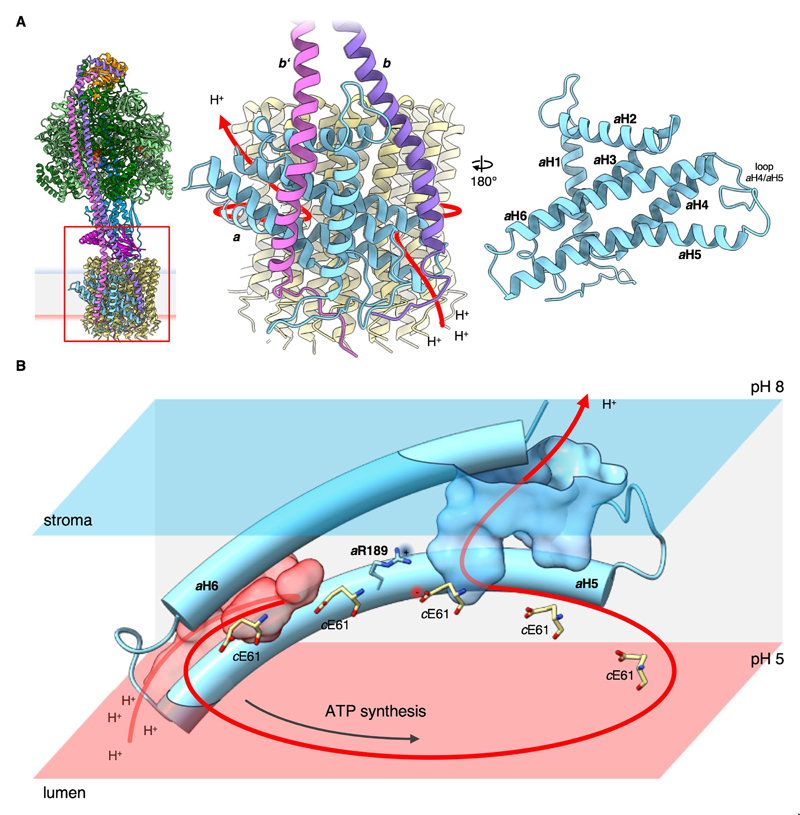
Fig. 4. Proton path through cFo.
(A) Subunits b (violet) and b’ (pink) form a right-handed coiled coil that is separated by the amphipathic H2 of subunit a at the point where b and b’ enter the membrane. (B) The long, membrane-intrinsic hairpin of subunit a helices H5 and H6 follows the curvature of the c 14-ring, forming the lumenal proton entry channel (transparent red) and stromal exit channel (transparent blue). The entry channel conducts protons from the acidic thylakoid lumen to the c-ring glutamate (cE61). After an almost full rotation of the c14-ring, the glutamate encounters the large, hydrophilic exit channel that extends to the stromal membrane surface. Glutamates are deprotonated close to the essential aArg189, which separates the two channels, preventing proton leakage and counteracting the negative charge of the deprotonated glutamate at the subunit a/c 14-ring interface.
Proton translocation through cFo
The proton translocation pathway is formed by the a-subunit and its interface with the c-ring. All 14 subunits of the chloroplast c-ring rotor are equally well resolved (Fig. 1). The crystal structure of an isolated c 14-ring from pea chloroplasts (34) fits the density well. The membrane scaffold protein of the nanodisc is clearly visible as a horizontal belt of two α-helices that wrap tightly around the Fo subcomplex (Fig. S8). Subunit a contains six α-helices H1 to H6 (Fig. 4A, Fig. S9). H1 spans the membrane. H2 is an amphipathic helix on the stromal membrane surface, located between b and b’. H3/H4 and H5/H6 form two long, membrane-intrinsic helix hairpins, as seen also in the mitochondrial and bacterial ATP synthases. The H5/H6 pair forms the interface with the c-ring and is the most conserved part of the subunit.
Two proton channels lead to and from a conserved glutamate in the c-ring (Fig. 4B-C). The proton entrance channel from the thylakoid lumen is a deep cavity, lined by charged and polar residues of a-subunit helices H5 and H6 and the loop connecting H3 and H4 (Fig. S10B). As in the mitochondrial ATP synthase (9, 10), the lumenal channel passes through a narrow gap between the hairpin helices H5 and H6, and ends at the c-ring glutamate receiving the proton. Unlike the tight H5/H6 hairpin, the H3/H4 loop is long, flanked by the TM helices of subunit a and b, and extends to the lumenal surface. The helix segments are highly hydrophobic, but the loop contains many polar residues that line the proton path. The H3/H4 hairpin is characterized by polar residues in all species (Fig. S9), but its sequence is not highly conserved. The only residue in the hairpin conserved across all ATP synthases, aAsn109 on H3, is part of the channel wall (Fig. S10C). The conserved aGln227 in H6 and aAsn193 near aArg189 in H5 contribute to the local hydrophilic environment at the a/c-ring interface. aAsn193 and aGln227 are within hydrogen bonding distance, apparently stabilizing the H5/H6 hairpin. Two charged residues, aGlu198 and aAsp197 on H5, channel protons through the gap between H5 and H6 to the c-ring Glu61 (Fig. S10C). The arrangement of proton-translocating residues in the access channel is different in mitochondria, where a glutamate on H6 (9, 10) appears to take on the role of aAsp197 in chloroplasts. The residue corresponding to aGlu198 in mitochondria is a histidine, which can accept or donate protons. In the E. coli sequence the glutamate and histidine positions are reversed (Fig. S9). A glutamate in one or the other position is essential for function (35). Taken together, the aqueous entrance channel is defined by charged and polar residues of H5, H6 and the H3/H4 loop that funnel protons from the low-pH thylakoid lumen towards the c-ring Glu61. The uncharged, protonated glutamate can partition into the lipid phase. After an almost full rotation of the c-ring, it encounters the proton exit channel and is deprotonated by the high pH of the chloroplast stroma (Fig. 4B-C, Movie 3). The iterative protonation and deprotonation of cGlu61 under physiological conditions enforces the unidirectional rotation of the c-ring.
The exit channel forms a wedge-shaped cavity that extends from the stromal surface to a narrow pocket next to aArg189. The cavity is lined by charged and polar sidechains from H5, H6 and a proline-rich loop between H4 and H5 (Fig. S11B-C). H5 bends to follow the curvature of the c-ring, and polar residues in its amphipathic N-terminal half (Fig. S9) face the c-ring or the stroma. The map density of the essential aArg189 is well-defined and faces the exit channel, as in mitochondrial ATP synthases (9, 10). Overall, the high degree of conservation of this and other features is remarkable considering the long evolutionary distance between mitochondria and chloroplasts of a billion years or more (36). The ~4.5 Å spacing between the aArg189 side chain and nearest c-ring glutamate suggests that it does not form a salt bridge, which would impede ring rotation and tend to arrest the c and a-subunits in the same relative position. This conclusion is supported by the different positions of the c-subunits relative to subunit a in the three conformations (Fig. S5C).
The position of aArg189 between the two proton channels suggests that the positively charged guanidinium group is critical to prevent proton leakage, which would dissipate the pmf (37). Conserved hydrophobic residues in the vicinity of aArg189 (Fig. S9) appear to have a structural role in maintaining the mutual orientation of H5 and H6. aLeu186 and aLeu190 on either side of aArg189 in adjacent helix turns of H5 pack closely against aLeu234 and the conserved aromatic aPhe231 on H6.
The central rotor
The central rotor consists of subunit γ, ε and the c-ring. The γ-subunit contains two long α-helices in a tight, left-handed coiled coil that forms the rotor shaft (Fig. 1B), and their adjacent loops establish most of the close contacts to the c 14-ring. These contacts are mainly mediated by electrostatic interactions with the highly conserved c-subunit loop (Arg41, Gln42, Pro43) (38). Our structure shows that 12 of the 14 c-ring subunits contribute to the firm attachment of the central stalk to the c-ring rotor; similar interactions exist in the ATP synthase of yeast mitochondria, which has a c 10 ring (39). The ε-subunit, which consists of an N-terminal eight-stranded antiparallel β-sandwich and a C-terminal α-helical domain, reinforces the close interaction with the c-ring with its conserved His37 that intercalates between cArg41 and cGln42 of one c subunit. The β-sheet of subunit ε is extended by a loop from the γ subunit. This loop contains the conserved γGlu285 (Fig. S11), which interacts with a cArg41. Mutants of this conserved residue have reduced ATP synthesis capacity due to impaired rotor assembly (40, 41).
In all three observed rotational states of cF1Fo, the C-terminal domain of the ε-subunit forms a horizontal α-helix hairpin next to subunit γ, similar to mitochondrial ATP synthase (4, 20). By contrast, in the E. coli ATPase (18, 42), this hairpin extends along the γ subunit into F1 and interacts with a β subunit, blocking ATP hydrolysis by an effect known as ε inhibition (Kato-Yamada, 1999) (Fig. S12). ε inhibition has also been proposed for chloroplasts (42, 43), but in our structure these helices do not interact with subunit β, suggesting that ε inhibition plays no role in cF1Fo.
Redox regulation of cF1Fo
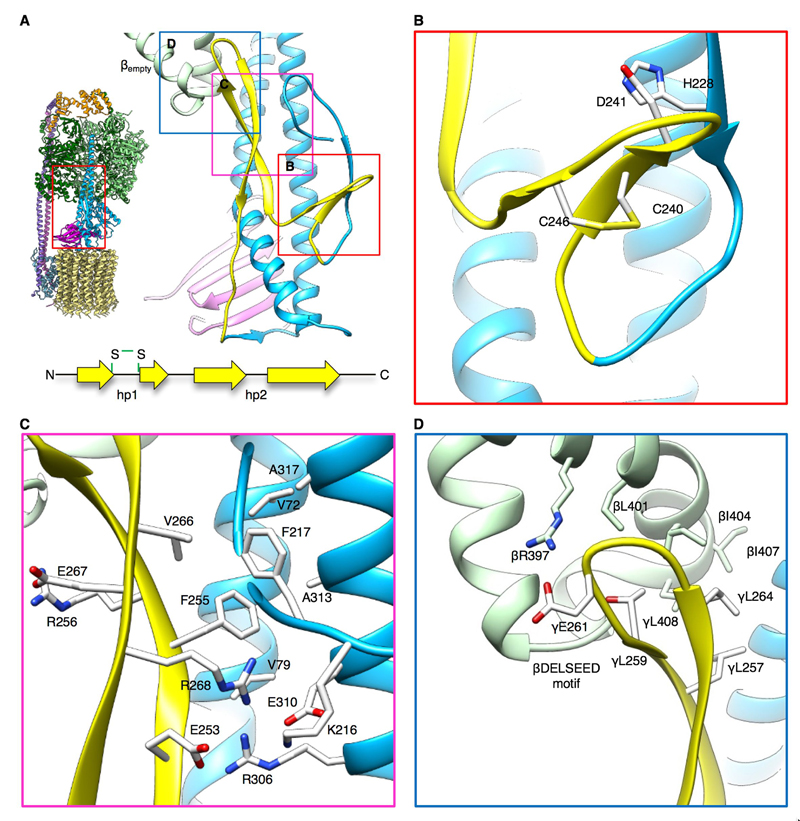
The chloroplast γ-subunit has a conserved ~40 amino acid insertion before the C-terminal α-helix that is thought to work as a redox-controlled inhibitor of ATP hydrolysis (Fig. S13). The insertion comprises two β-hairpins arranged in an L shape between the nucleotide-free β subunit (βempty) and the γ-rotor (Fig. 5A). The short N-terminal hairpin hp1 halfway between the c 14-ring and the F1 head is held in place by an electrostatic interaction between γAsp241 and γHis228, residues conserved in plants. The hp1 arm contains two cysteine residues, γCys240 and γCys246, positioned on opposite strands of the β-hairpin. A disulfide bond is present in our structure between the strands (Fig. 5B), which locks the hairpin together. Previous work suggested a role for these cysteine residues as a redox sensor (16). Thioredoxins and NADPH-dependent thioredoxin reductase (NTRC) serve as electron donors to keep this motif reduced and cF1Fo active under high and low light conditions (44). The redox potential drops during periods of prolonged darkness, when photosynthesis does not sustain the pmf across the thylakoid membrane (45). The formation of the disulfide bond has been proposed to trigger a conformational change that inhibits cF1Fo at night to prevent ATP hydrolysis at low pmf (46). Our structure shows the oxidized, auto-inhibiting conformation of the redox loop.
Fig. 5. Auto-inhibition of cF1Fo by thiol modulation.
(A) Subunit γ of cF1Fo contains a 40-residue insertion that includes two cysteines. The insertion forms two β-hairpins in an L-shaped loop (yellow) that works as an auto-inhibitor of rotation in response to the chloroplast redox potential. (B) The shorter, N-terminal hairpin (hp1) contains the cysteine motif (CDxNGxC, Fig. S11), which forms a disulfide bond under oxidizing conditions. (C) The conserved γF255 of the second hairpin (hp2) interacts with γF217 in a hydrophobic pocket of subunit γ. (D) γE261 in hp2 forms a salt bridge with an arginine in the βempty subunit, blocking rotation. hp2 would clash with the βDELSEED motif if the rotor turns in hydrolysis direction.
The long, slightly twisted C-terminal hairpin hp2 of the redox loop runs vertically along the N-terminal α-helix of subunit γ. It contains charged and polar residues with a conserved phenylalanine (γPhe255) at its center (Fig. S13). The γPhe255 sidechain sits in a hydrophobic pocket formed by valine residues in the coiled coil of subunit γ, in an orthogonal π-π interaction with γPhe217 (Fig. 5C, Movie 4). This interaction, together with electrostatic interactions between γArg268, γArg306, γGlu310, γGlu253 and γLys216, puts hp2 in close contact with the far side of the conserved DELSEED motif (47) of the βempty subunit (Fig. 5D). Reduction of the disulfide bond would destabilize the redox loop and allow for hp2 to be pushed aside when the rotor turns in ATP synthesis direction. Under oxidizing conditions, the disulfide bond and the L-shape of the redox loop re-form. In this position hp2 would clash with the DELSEED motif of the βempty subunit and block rotation of the rotor in ATP hydrolysis direction. In this way, the DELSEED loop functions as a catch for hp2 that blocks rotation in the dark. Although the inhibiting subunit is different, both redox regulation and ε inhibition act on the conserved DELSEED motif of subunit β (βempty and βΤP, respectively; Fig. S12), which moves by more than 10 Å in the process of rotary catalysis (Movie 1).
Concluding remarks
ATP synthases are central components of all membrane-based biological energy conversion systems and the only known macromolecular machines that convert an electrochemical trans-membrane gradient directly into the chemical energy of a covalent bond. ATP generated by photosynthesis is the primary source of biologically useful energy on the planet. ATP synthases of chloroplasts, mitochondria and bacteria all conform to essentially the same building plan and share the same key features, including the α and β subunits of the F1 head, the proton or sodium ion driven c-ring rotor and subunit a with its two ion channels in the Fo motor, as well as the central and peripheral stalks that connect them. Exceptions are the regulatory mechanisms, such as the redox control switch in the central stalk of cF1Fo, which is an adaptation to the day-night cycle of the chloroplasts redox potential. Considering the evolutionary distance between mitochondria and chloroplasts of around 1.5 billion years (36, 48), these key properties are remarkably well conserved. Comparison of the cF1Fo structure to x-ray structures (3) and recent partial cryo-EM structures of mitochondrial ATP synthases (9, 10) reveals that many features are identical, down to the level of individual amino acid sidechains. Evidently, maintaining the structural basis of ATP synthesis is critical to all living organisms; mutants that impair the optimized mechanism of rotary catalysis in any way are strongly selected against. Our high resolution structure informs on all aspects of this mechanism for an intact, functional ATP synthase.
Materials and methods
Isolation of chloroplast ATP synthase from spinach leaves
Preparation of thylakoid membranes from young leaves of market spinach (Spinacia oleracea) and membrane protein solubilization were carried out as described (49). Briefly, cF1Fo was enriched by fractionated ammonium sulfate precipitation. Fractions precipitated from 1.2 M to 1.8 M ammonium sulfate were recovered in 100 ml of Buffer A (30 mM HEPES pH 8.0, 2 mM MgCl2, 0.5 mM Na2 EDTA, 0.1 % (w/v) trans-4-(trans-4’-propylcyclohexyl)cyclohexyl-α-D-maltoside (tPCC-α-M, Glycon, Luckenwalde, Germany) (Hovers et al, 2011). Insoluble material was removed by ultracentrifugation (45,000 x g, 30 min, 4°C). The supernatant enriched in cF1Fo was loaded on a POROS GoPure HQ 50 anion exchange column (Life Technologies, USA) equilibrated with Buffer B (30 mM HEPES pH 8, 50 mM NaCl, 2 mM MgCl2, 0.04% (w/v) tPCC-α-M) and gradually eluted with Buffer C (Buffer B with 1 M NaCl) using an Äkta Explorer chromatography system (GE Healthcare, USA) at 4°C. Hydrolytic activity was assayed as described (20). Samples were analyzed by SDS polyacrylamide gel electrophoresis (SDS-PAGE); the protein concentration was determined by the bicinchoninic acid (BCA) assay (Thermo Fisher/Pierce, Germany).
Reconstitution of cF1Fo into nanodiscs
Expression and purification of MSP2N2 nanodiscs (Addgene, USA) was carried out as described(50). Total membrane lipid was isolated from freshly prepared thylakoid membranes by chloroform/methanol extraction (51). The total lipid concentration was estimated by two-dimensional thin-layer chromatography and iodine vapor staining (52, 53) with phosphatidyl ethanolamine and phosphatidyl glycerol standards (Avanti Polar Lipids, US). MSP2N2, total thylakoid lipid extract and cF1Fo were mixed at a molar lipid:MSP:protein ratio of 400:10:1. After 1 h of incubation, detergent was removed with ~1% (w/w) Bio-Beads™ SM-2 (Bio-Rad Laboratories, USA) and overnight agitation at 4°C. The reconstitution mix was loaded on a 16/300 Superose-6 gel-filtration column equilibrated with Buffer D (Buffer A without detergent) at 4°C using an Äkta Explorer system). Nanodisc-reconstituted cF1Fo elutes at a retention volume of ~12 ml as an almost symmetric peak with a small tailing shoulder of detached cF1. The peak was collected in 0.25 ml fractions. The fraction with the highest protein concentration from the peak front was directly used for cryo-EM sample preparation.
Cryo-EM preparation and electron microscopy
3 µl of cF1Fo at a concentration of 2 mg/ml was applied to freshly glow-discharged Quantifoil R1.2/1.3 grids and plunge-frozen in liquid ethane using a Vitrobot (Thermo Fisher/FEI). Images were recorded in a Titan Krios microscope operated at 300 kV (Thermo Fisher/FEI) on a Falcon III direct electron detector in electron counting mode at a nominal magnification of 75,000x, corresponding to a calibrated pixel size of 1.053 Å. Before data collection, a precise alignment of the pivot points, coma and rotation center was carried out at the electron flux (0.43 e- × pixel-1 × s-1) used to record data. 6,254 dose fractionation movies were recorded using EPU (Thermo Fisher/FEI) over 62 s, corresponding to a total dose of ~25 e-/A2 in a defocus range of -1.5 to -2.5 µm.
Image processing
MotionCor2 was used to correct beam-induced motion and to generate dose-weighted images from movies for initial image processing (54). CTF parameters for each movie were determined with CTFFIND4 (55). 670,614 particle images were automatically selected with RELION-2.1 (56) and extracted with a box size of 350 × 350 pixels. The dataset was cleaned by 2D classification using ISAC (57). Particles were sorted into three classes by 3D classification in RELION-2.1 (58). Final maps were reconstructed in RELION-2.1 from 167,171 (conformation 1), 15,395 (conformation 2) and 14,409 (conformation 3) polished particles. To improve the reconstruction of the membrane region, the F1-rotor (α3β3γε) was subtracted from the extracted particle images (59) and a soft-edged mask around the aδbb’c 14 subcomplex was applied before local realignment. Local resolution was assessed using the built-in routine of RELION-2.1 with an arbitrary B-factor of -150.
Model building and refinement
The structure was built into the EM maps in Coot (60), based on homologous structures where possible, in particular the 3.4 Å X-ray structure of a spinach chloroplast F1-ATPase αβ dimer (PDB ID 1KMH) (26); the NMR solution structure of the E. coli F-ATPase δ subunit N-terminal domain in complex with the α subunit N-terminal 22 residues (PDB ID 2A7U) (29); the γ subunit from the 3.0 Å crystal structure of Caldalkalibacillus thermarum F1-ATPase (PDB ID 5HKK) (61); the solution structure of an ε subunit chimera combining the N-terminal β-sandwich domain from Thermosynechococcus elongatus F1 and the C-terminal α-helical domain from spinach chloroplast F1 (PDB ID 2RQ7) (62); the 3.4 Å crystal structure of chloroplast ATP synthase c 14-ring from Pisum sativum (PDB ID 3V3C) (34). The a, b, and b’ subunits, the C-terminal domain of δ, the redox loop of γ and C and N-termini of the α and β-subunits were built manually de novo. The structure was refined by Phenix real space refinement using Ramachandran restraints (63) followed by manual rebuilding in Coot. MolProbity (64) and EMRinger (65) were used for validation (Table S1). Water-accessible regions of the membrane intrinsic Fo sub-complex were probed by mapping the interior surface using HOLLOW (66). Figures and movies were made with Chimera (67) or ChimeraX (68).
Supplementary Material
The redox loop of cF1Fo. The redox loop (yellow) in the γ subunit of chloroplast ATP synthase (cyan) interacts with a β subunit (light green) to block ATP hydrolysis in the dark. Residues discussed in the text are indicated.
Top view of the interpolation between the three rotary conformations of cF1Fo as in Movie 1. The model was rotated 90° along the x-axis with respect to Movie 1.
Illustration of the proton pathway in through cFo. The long, membrane-intrinsic hairpin of subunit a helices H5 and H6 follows the curvature of the c14-ring, forming the luminal proton entry channel (transparent red) and stromal exit channel (transparent blue). The entry channel conducts protons from the acidic lumen to the c-ring glutamate (cE61). After an almost full rotation of the c14-ring, the glutamate encounters the large, hydrophilic exit channel that extends to the stromal membrane surface. Glutamates are deprotonated close to the essential aArg189, which separates the two channels, preventing proton leakage and counteracting the negative charge of the deprotonated glutamate at the subunit a/c14-ring interface.
Interpolation between the three rotary conformations of cF1Fo. Shown is a morph between the models fitted to the cryo-EM maps of conformation 1, 2 and 3, respectively. The a subunit (light blue) is kept stationary. The interpolation was created with Chimera (67).
One Sentence Summary.
Cryo electron microscopy reveals how oxidation applies the brakes to a proton-powered molecular motor.
Acknowledgements
We thank Marina Amrhein and Thomas Bausewein for their contributions during early stages of the project. We thank Gerhard Hummer for discussions and Niklas Klusch and Bonnie Murphy for reading the manuscript.
Funding
This work was funded by the Max Planck Society, the Collaborative Research Center (CRC) 807 of the German Research Foundation (DFG), and by the Wellcome Trust [WT110068/Z/15/Z].
Footnotes
Author contributions: TM and WK directed the project; AH purified the protein; DJM optimised the high-resolution EM alignment and the data collection procedure; AH and DJM collected cryo-EM data; AH reconstructed the cryo-EM maps; AH and JV built and interpreted the model; AH, JV, TM and WK wrote the paper.
Competing interests: Authors declare no competing interests.
Data and materials availability: The cryo-EM maps have been deposited in the Electron Microscopy Data Bank with accession numbers EMD-4270, EMD-4271 and EMD-4272 for conformation 1, 2 and 3, respectively and EMD-4273 for the masked Fo of conformation 1. Atomic models have been deposited in the Protein Data Bank with accession numbers 6FKF, 6FKH and 6FKI for conformation 1, 2 and 3, respectively.
References
- 1.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemiosmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 2.Boyer PD. The ATP synthase: a splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 3.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 Å resolution of F1 ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 4.Stock D, Leslie AG, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 5.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus . Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- 6.Vik SB, Antonio BJ. A mechanism of proton translocation by F1F0 ATP synthases suggested by double mutants of the a subunit. J Biol Chem. 1994;269:30364–30369. [PubMed] [Google Scholar]
- 7.Junge W, Lill H, Engelbrecht S. ATP synthase: an electrochemical transducer with rotatory mechanics. Trends Biochem Sci. 1997;22:420–423. doi: 10.1016/s0968-0004(97)01129-8. [DOI] [PubMed] [Google Scholar]
- 8.Allegretti M, et al. Horizontal membrane-intrinsic α-helices in the stator a-subunit of an F-type ATP synthase. Nature. 2015;521:237–240. doi: 10.1038/nature14185. [DOI] [PubMed] [Google Scholar]
- 9.Klusch N, Murphy BJ, Mills DJ, Yildiz Ö, Kühlbrandt W. Structural basis of proton translocation in mitochondrial ATP synthase. eLife. 2017;6 doi: 10.7554/eLife.33274. e33274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo H, Bueler SA, Rubinstein JL. Atomic model for the dimeric FO region of mitochondrial ATP synthase. Science. 2017;358:936–940. doi: 10.1126/science.aao4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daum B, Nicastro D, Austin J, McIntosh JR, Kühlbrandt W. Arrangement of photosystem II and ATP synthase in chloroplast membranes of spinach and pea. Plant Cell. 2010;22:1299–1312. doi: 10.1105/tpc.109.071431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies KM, et al. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci USA. 2011;108:14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson PJ, Harris DA. The mitochondrial ATP synthase inhibitor protein binds near the C-terminus of the F1 beta-subunit. FEBS Lett. 1988;299:224–228. doi: 10.1016/0014-5793(88)80832-9. [DOI] [PubMed] [Google Scholar]
- 14.Kato-Yamada Y, et al. Epsilon subunit, an endogenous inhibitor of bacterial F1-ATPase, also inhibits F0F1-ATPase. J Biol Chem. 1999;274:33991–33994. doi: 10.1074/jbc.274.48.33991. [DOI] [PubMed] [Google Scholar]
- 15.Arana JL, Vallejos RH. Involvement of sulfhydryl groups in the activation mechanism of the ATPase activity of chloroplast coupling factor 1. J Biol Chem. 1982;257:1125–1127. [PubMed] [Google Scholar]
- 16.Nalin CM, McCarty RE. Role of a disulfide bond in the gamma subunit in activation of the ATPase of chloroplast coupling factor 1. J Biol Chem. 1984;259:7275–7280. [PubMed] [Google Scholar]
- 17.Junesch U, Gräber P. Influence of the redox state and the activation of the chloroplast ATP synthase on proton-transport-coupled ATP synthesis/hydrolysis. Biochim Biophys Acta. 1987;893:275–288. [Google Scholar]
- 18.Sobti M, et al. Cryo-EM structures of the autoinhibited E. coli ATP synthase in three rotational states. eLife. 2016;5 doi: 10.7554/eLife.21598. e21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou A, et al. Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. eLife. 2015;3 doi: 10.7554/eLife.10180. e10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn A, et al. Structure of a complete ATP synthase dimer reveals the molecular basis of mitochondrial membrane morphology. Mol Cell. 2016;63:445–456. doi: 10.1016/j.molcel.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyer PD. The binding change mechanism for ATP synthase - some probabilities and possibilities. Biochim Biophys Acta. 1993;1140:215–250. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]
- 22.Heineke D, et al. Redox transfer across the inner chloroplast envelope membrane. Plant Physiology. 1991;95:1131. doi: 10.1104/pp.95.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okazaki K, Hummer G. Elasticity, friction, and pathway of gamma-subunit rotation in FoF1-ATP synthase. Proc Natl Acad Sci USA. 2015;112:10720–10725. doi: 10.1073/pnas.1500691112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wächter A, et al. Two rotary motors in F-ATP synthase are elastically coupled by a flexible rotor and a stiff stator stalk. Proc Natl Acad Sci USA. 2011;108:3924–3929. doi: 10.1073/pnas.1011581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groth G, Pohl E. The structure of the chloroplast F1-ATPase at 3.2 Å resolution. J Biol Chem. 2001;276:1345–1352. doi: 10.1074/jbc.M008015200. [DOI] [PubMed] [Google Scholar]
- 26.Groth G. Structure of spinach cloroplast F1-ATPase complexed with the phytopathogenic inhibitor tentoxin. Proc Natl Acad Sci USA. 2002;99:3463–3468. doi: 10.1073/pnas.052546099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carmeli C, Lifshitz Y. Effects of Pi and ADP on ATPase activity in chloroplasts. Biochim Biophys Acta. 1972;267:86–95. doi: 10.1016/0005-2728(72)90140-5. [DOI] [PubMed] [Google Scholar]
- 28.Abrahams JP, et al. The structure of bovine F1-ATPase complexed with the peptide antibiotic efrapeptin. Proc Natl Acad Sci USA. 1996;93:9420–9424. doi: 10.1073/pnas.93.18.9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkens S, Borchardt D, Weber J, Senior AE. Structural characterization of the interaction of the δ and α subunits of the Escherichia coli F1F0-ATP synthase by NMR spectroscopy. Biochemistry. 2005;44:11786–11794. doi: 10.1021/bi0510678. [DOI] [PubMed] [Google Scholar]
- 30.Rees DM, Leslie AGW, Walker JE. The structure of the membrane extrinsic region of bovine ATP synthase. Proc Natl Acad Sci USA. 2009;106:21597–21601. doi: 10.1073/pnas.0910365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee LK, Stewart AG, Donohoe M, Bernal RA, Stock D. The structure of the peripheral stalk of Thermus thermophilus H+-ATPase/synthase. Nat Struct Mol Biol. 2010;17:373–378. doi: 10.1038/nsmb.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart AG, Lee LK, Donohoe M, Chaston JJ, Stock D. The dynamic stator stalk of rotary ATPases. Nat Commun. 2012;3:687. doi: 10.1038/ncomms1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Del Rizzo PA, Bi Y, Dunn SD. ATP synthase b subunit dimerization domain: a right-handed coiled coil with offset helices. J Mol Biol. 2006;364:735–746. doi: 10.1016/j.jmb.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 34.Saroussi S, Schushan M, Ben-Tal N, Junge W, Nelson N. Structure and flexibility of the c-ring in the electromotor of rotary FoF1-ATPase of pea chloroplasts. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0043045. e43045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vik SB, Long JC, Wada T, Zhang D. A model for the structure of subunit a of the Escherichia coli ATP synthase and its role in proton translocation. Biochim Biophys Acta. 2000;1458:457–466. doi: 10.1016/s0005-2728(00)00094-3. [DOI] [PubMed] [Google Scholar]
- 36.Hohmann-Marriott MF, Blankenship RE. Evolution of photosynthesis. Annu Rev Plant Biol. 2011;62:515–548. doi: 10.1146/annurev-arplant-042110-103811. [DOI] [PubMed] [Google Scholar]
- 37.Mitome N, et al. Essential arginine residue of the Fo-a subunit in FoF1-ATP synthase has a role to prevent the proton shortcut without c-ring rotation in the Fo proton channel. Biochem J. 2010;430:171–177. doi: 10.1042/BJ20100621. [DOI] [PubMed] [Google Scholar]
- 38.Vonck J, et al. Molecular architecture of the undecameric rotor of a bacterial Na+-ATP synthase. J Mol Biol. 2002;321:307–316. doi: 10.1016/s0022-2836(02)00597-1. [DOI] [PubMed] [Google Scholar]
- 39.Dautant A, Velours J, Giraud M-F. Crystal structure of the Mg.ADP-inhibited state of the yeast F1c10-ATP synthase. J Biol Chem. 2010;285:29502–29510. doi: 10.1074/jbc.M110.124529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu G, Ortiz-Flores G, Ortiz-Lopez A, Ort DR. A point mutation in atpC1 Raises the redox potential of the Arabidopsis chloroplast ATP synthase γ-subunit regulatory disulfide above the range of thioredoxin modulation. J Biol Chem. 2007;282:36782–36789. doi: 10.1074/jbc.M707007200. [DOI] [PubMed] [Google Scholar]
- 41.Pogoryelov D, et al. Probing the rotor subunit interface of the ATP synthase from Ilyobacter tartaricus . FEBS J. 2008;275:4850–4862. doi: 10.1111/j.1742-4658.2008.06623.x. [DOI] [PubMed] [Google Scholar]
- 42.Cingolani G, Duncan TM. Structure of the ATP synthase catalytic complex (F1) from Escherichia coli in an autoinhibited conformation. Nat Struct Mol Biol. 2011;18:701. doi: 10.1038/nsmb.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richter ML. Gamma–epsilon interactions regulate the chloroplast ATP synthase. Photosynthesis Research. 2004;79:319–329. doi: 10.1023/B:PRES.0000017157.08098.36. [DOI] [PubMed] [Google Scholar]
- 44.Carrillo LR FJ, Cruz JA, Savage LJ, Kramer DM. Multi-level regulation of the chloroplast ATP synthase: the chloroplast NADPH thioredoxin reductase C (NTRC) is required for redox modulation specifically under low irradiance. Plant J. 2016;87:654–663. doi: 10.1111/tpj.13226. [DOI] [PubMed] [Google Scholar]
- 45.Jagendorf AT. Photophosphorylation and the chemiosmotic perspective. Photosynth Res. 2002;73:233–241. doi: 10.1023/A:1020415601058. [DOI] [PubMed] [Google Scholar]
- 46.Ketcham SR, Davenport JW, Warncke K. R. E. McCarty, Role of the gamma subunit of chloroplast coupling factor 1 in the light- dependent activation of photophosphorylation and ATPase activity by dithiothreitol. J Biol Chem. 1984;259:7286–7293. [PubMed] [Google Scholar]
- 47.Tanigawara M, et al. Role of the DELSEED loop in torque transmission of F1-ATPase. Biophys J. 2012;103:970–978. doi: 10.1016/j.bpj.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rand DM, Haney RA, Fry AJ. Cytonuclear coevolution: the genomics of cooperation. Trends in Ecology & Evolution. 19:645–653. doi: 10.1016/j.tree.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 49.Varco-Merth B, Fromme R, Wang M, Fromme P. Crystallization of the c14-rotor of the chloroplast ATP synthase reveals that it contains pigments. Biochim Biophys Acta. 2008;1777:605–612. doi: 10.1016/j.bbabio.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ritchie TK, et al. Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Meth Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 52.Sato N, Tsuzuki M. Carpentier R, editor. Methods in Molecular Biology: Photosynthesis Research Protocols. 2011;684:95–104. doi: 10.1007/978-1-60761-925-3_9. [DOI] [PubMed] [Google Scholar]
- 53.Palumbo G, Zullo F. The use of iodine staining for the quantitative analysis of lipids separated by thin layer chromatography. Lipids. 1987;22:201–205. doi: 10.1007/BF02537303. [DOI] [PubMed] [Google Scholar]
- 54.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Meth. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kimanius D, Forsberg BO, Scheres SHW, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife. 2016;5 doi: 10.7554/eLife.18722. e18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Z, Fang J, Chittuluru J, Asturias Francisco J, Penczek Pawel A. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20:237–247. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bai X-c, Fernandez IS, McMullan G, Scheres SHW. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2 doi: 10.7554/eLife.00461. e00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 61.Ferguson SA, Cook GM, Montgomery MG, Leslie AGW, Walker JE. Regulation of the thermoalkaliphilic F1-ATPase from Caldalkalibacillus thermarum . Proc Natl Acad Sci USA. 2016;113:10860–10865. doi: 10.1073/pnas.1612035113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yagi H, et al. Structural and functional analysis of the intrinsic inhibitor subunit ε of F1-ATPase from photosynthetic organisms. Biochem J. 2010;425:85–94. doi: 10.1042/BJ20091247. [DOI] [PubMed] [Google Scholar]
- 63.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barad BA, et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Meth. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ho BK, Gruswitz F. Generating accurate representations of channel and interior surfaces in molecular structures. BMC Structural Biology. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pettersen EF, et al. UCSF Chimera: a visualisation system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 68.Goddard TD, et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dunn SD, Tozer RG, Zadorozny VD. Activation of Escherichia coli F1-ATPase by lauryldimethylamine oxide and ethylene glycol: relationship of ATPase activity to the interaction of the epsilon and beta subunits. Biochemistry. 1990;29:4335–4340. doi: 10.1021/bi00470a011. [DOI] [PubMed] [Google Scholar]
- 70.Rosenthal P, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 71.Allegretti M, Mills DJ, McMullan G, Kühlbrandt W, Vonck J. Atomic model of the F420-reducing [NiFe] hydrogenase by electron cryo-microscopy using a direct electron detector. eLife. 2014;3 doi: 10.7554/eLife.01963. e01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bartesaghi A, Matthies D, Banerjee S, Merk A, Subramaniam S. Stucture of β-galactosidase at 3.2-Å resolution obtained by cryo-electron microscopy. Proc Natl Acad Sci USA. 2014;111:11709–11714. doi: 10.1073/pnas.1402809111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vonck J, Mills DJ. Recent advances in high-resolution cryo-EM of oligomeric enzymes. Curr Opin Struct Biol. 2017;46:48–54. doi: 10.1016/j.sbi.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 74.Kagawa R, Montgomery MG, Braig K, Leslie AGW, Walker JE. The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 2004;23:2734–2744. doi: 10.1038/sj.emboj.7600293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lau WCY, Rubinstein JL. Subnanometre-resolution structure of the intact Thermus thermophilus H+-driven ATP synthase. Nature. 2012;481:214–218. doi: 10.1038/nature10699. [DOI] [PubMed] [Google Scholar]
- 76.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 77.Rodgers AJW, Wilce MCJ. Structure of the γ-ε complex of ATP synthase. Nat Struct Biol. 2000;7:1051–1054. doi: 10.1038/80975. [DOI] [PubMed] [Google Scholar]
- 78.Tsunoda SP, et al. Large conformational changes of the epsilon subunit in the bacterial F1F0 ATP synthase provide a ratchet action to regulate this rotary motor enzyme. Proc Natl Acad Sci USA. 2001;98:6560–6564. doi: 10.1073/pnas.111128098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The redox loop of cF1Fo. The redox loop (yellow) in the γ subunit of chloroplast ATP synthase (cyan) interacts with a β subunit (light green) to block ATP hydrolysis in the dark. Residues discussed in the text are indicated.
Top view of the interpolation between the three rotary conformations of cF1Fo as in Movie 1. The model was rotated 90° along the x-axis with respect to Movie 1.
Illustration of the proton pathway in through cFo. The long, membrane-intrinsic hairpin of subunit a helices H5 and H6 follows the curvature of the c14-ring, forming the luminal proton entry channel (transparent red) and stromal exit channel (transparent blue). The entry channel conducts protons from the acidic lumen to the c-ring glutamate (cE61). After an almost full rotation of the c14-ring, the glutamate encounters the large, hydrophilic exit channel that extends to the stromal membrane surface. Glutamates are deprotonated close to the essential aArg189, which separates the two channels, preventing proton leakage and counteracting the negative charge of the deprotonated glutamate at the subunit a/c14-ring interface.
Interpolation between the three rotary conformations of cF1Fo. Shown is a morph between the models fitted to the cryo-EM maps of conformation 1, 2 and 3, respectively. The a subunit (light blue) is kept stationary. The interpolation was created with Chimera (67).