Abstract
The mutualistic symbiosis between forest trees and ectomycorrhizal fungi (EMF) is among the most ubiquitous and successful interactions in terrestrial ecosystems. Specific species of EMF are known to colonize specific tree species, benefitting from their carbon source, and in turn, improving their access to soil water and nutrients. EMF also form extensive mycelial networks that can link multiple root-tips of different trees. Yet the number of tree species connected by such mycelial networks, and the traffic of material across them, are just now under study. Recently we reported substantial belowground carbon transfer between Picea, Pinus, Larix and Fagus trees in a mature forest. Here, we analyze the EMF community of these same individual trees and identify the most likely taxa responsible for the observed carbon transfer. Among the nearly 1,200 EMF root-tips examined, 50%–70% belong to operational taxonomic units (OTUs) that were associated with three or four tree host species, and 90% of all OTUs were associated with at least two tree species. Sporocarp 13C signals indicated that carbon originating from labelled Picea trees was transferred among trees through EMF networks. Interestingly, phylogenetically more closely related tree species exhibited more similar EMF communities and exchanged more carbon. Our results show that belowground carbon transfer is well orchestrated by the evolution of EMFs and tree symbiosis.
Keywords: mycorrhiza, Sebacina, host genetics, stable isotopes, temperate forest
1. Introduction
Forest soils commonly contain low levels of bioavailable nutrients, and most trees rely on symbiosis with ectomycorrhizal fungi (EMF) for their nutrition (Grayston, Vaughan, & Jones, 1997; Johnson, Wilson, Bowker, Wilson, & Miller, 2010; Perry, Bell, & Amaranthus, 1992; Wallander, Göransson, & Rosengren, 2004). Symbiosis between trees and EMF is prevalent in boreal and temperate zones (Martin, Kohler, Murat, Veneault-Fourrey, & Hibbett, 2016), where individual trees may be colonized by tens to hundreds of different EMF taxa (Bahram, Polme, Koljalg, & Tedersoo, 2011; Bruns, 1995). EMF compete among each other for two main resources: host-derived carbon and soil-derived mineral nutrients. The availability of the two resources affects EMF community structure in space and time (Bruns, 1995). Climate, soil chemistry, depth, moisture, and nutrient availability (Alday, Martínez de Aragón, de-Miguel, & Bonet, 2017; Bruns, 1995; Castano et al., 2018; McGuire, Allison, Fierer, & Treseder, 2013; Pena et al., 2017; Rosenstock, Berner, Smits, Kram, & Wallander, 2016), as well as biotic interactions (Gehring, Sthultz, Flores-Renteria, Whipple, & Whitham, 2017; Lovelock, Andersen, & Morton, 2003; Mujic, Durall, Spatafora, & Kennedy, 2016; Peay, Kennedy, & Bruns, 2011; Wu, Nara, & Hogetsu, 1999), have all been observed to influence EMF community composition. Still, the crucial drivers maintaining the diversity and the ecological niches of these EMF taxa remain largely unknown. Similar to other relationships between plants and nonplant organisms, a positive correlation has been found between EMF diversity and host tree diversity (Gao et al., 2015; Kernaghan, Widden, Bergeron, Légaré, & Paré, 2003; Nguyen et al., 2016). EMF affect ecosystem function (Högberg & Read, 2006), but the extent to which the taxonomic diversity of fungal populations affects a specific function is poorly defined (Hazard & Johnson, 2018).
Ectomycorrhizal fungi exhibit varying degrees of host specificity, defined as the rate at which a specific EMF species associates with a particular host species across a large geographical range (Wolfe & Pringle, 2012). EMFs with low host specificity are under strong competition (Lang, Seven, & Polle, 2011) and are commonly among the most abundant EMF taxa in mixed forests (Horton & Bruns, 1998). In addition to trees having associations with many different EMF taxa, EMF individuals (defined as a genet with a contiguous mycelium) have the ability to associate with different tree species simultaneously (Van Der Heijden & Horton, 2009). Common mycorrhizal networks can connect multiple plant hosts belowground across spatial scales of cm to tens of meters (Beiler, Durall, Simard, Maxwell, & Kretzer, 2010; Nara, 2006; Selosse, Richard, He, & Simard, 2006; Simard et al., 2012; Toju, Guimaraes, Olesen, & Thompson, 2015). In such cases, single fungal genets may be connected to many mycorrhizal root tips of different trees - although the actual connection is hard to prove (Beiler et al., 2010; Nara, 2006). Allospecific trees of natural mixed-species forests may thus be interlinked through common mycorrhizal networks, forming “guilds” (Pickles et al., 2017; Simard et al., 1997). Net carbon transfer between trees is influenced by EMF morphology (Agerer, 2001) including contact-, short-distance and long distance exploration types, the latter potentially facilitating belowground C transfer on larger scales.
We have recently shown that carbon is transferred bidirectionally among mature forest trees belonging to taxonomically distant taxa (Klein, Siegwolf, & Körner, 2016). This carbon transfer was identified via δ13C signals after 5 years of Free Air CO2 Enrichment (FACE) of 37 m tall Picea abies tree crowns. After ruling out alternative explanations to the 13C label transfer, the trafficking of carbon was attributed to the EMF hyphae networks. Unlike root anastomosis, such an EMF-based carbon transfer can bridge plant phylogenetic barriers. However, common mycorrhizal networks have so far only been shown to nurse conspecific seedlings with carbon assimilates (Brownlee, Duddridge, Malibari, & Read, 1983; Dickie, Guza, Krazewski, & Reich, 2004; Simard et al., 1997). Furthermore, the EMF species responsible for the observed transfer remain unknown.
Here, we examined the patterns of EMF species specificity in the same mixed forest in which the aforementioned 13C transfer was observed, utilizing the specific tree individuals examined previously, and evaluating the level of EMF species sharing among Picea and other species as a potential explanation for the observed patterns in carbon transfer. In addition, we examined whether the similarity in EMF community composition between allospecific trees was correlated to carbon transfer from labelled Picea. We hypothesized that phylogenetically related tree species have more similar EMF communities and, as a consequence, greater carbon transfer.
2. Materials and Methods
2.1. Study site
The study was conducted in a diverse mixed forest 12 km southwest of Basel, Switzerland (47°33′N, 7°36′E, 550 m a.s.l). The site was dominated by 110 to 120-year-old trees belonging to 14 taxa of which Picea abies (Norway spruce), Larix decidua Mill. (European larch), Pinus sylvestris L. (Scots pine), and Abies alba Mill (silver fir). represent conifers and Fagus sylvatica L. (beech), Quercus petraea (Matt.) Liebl. (sessile oak), Carpinus betulus L. (hornbeam), Tilia platy-phylla Scop. (large-leaved lime), Prunus avium L. (wild cherry), Fraxinus excelsior L. (ash), Acer campestre L (field maple). and a few others, represent deciduous angiosperms. Our study included the species Picea abies, Fagus sylvatica, Pinus sylvestris and Larix decidua (Figure 1). Crown heights were 30–40 m, forming a closed tree canopy. The soil was a shallow, 30 cm deep, silty-loamy rendzina on calcareous bedrock. The climate was mild temperate, with mean January and July temperatures of 2.1°C and 19.1°C, respectively and mean temperature during the growing season (May–September) of 14.7°C. Mean annual precipitation was ~900 mm.
Figure 1.
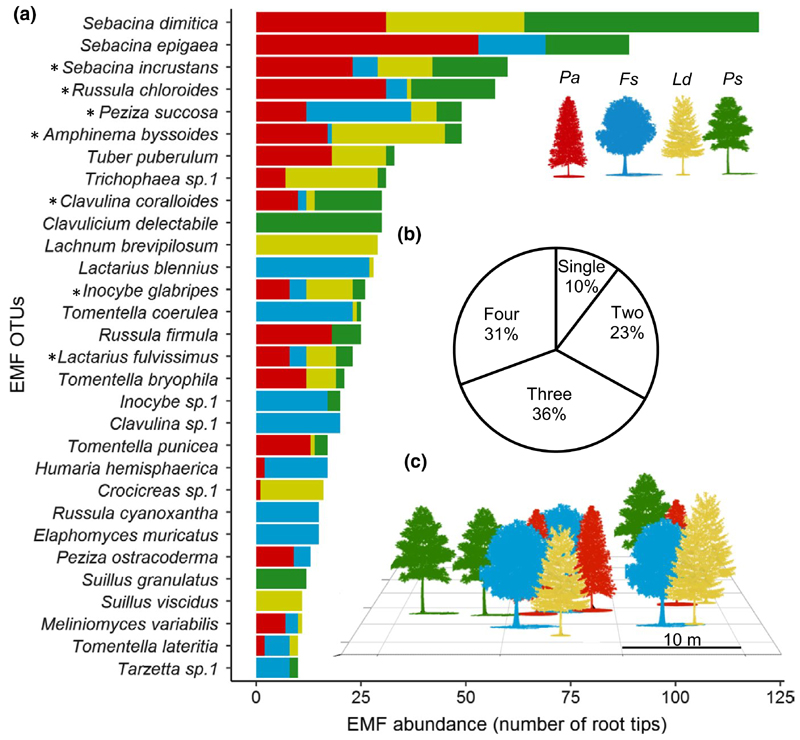
The observed abundance of ectomycorrhizal fungi (EMF) species on root tips of four tree species populating the forest plot near Basel, Switzerland (Picea abies (Pa), Pinus sylvestris (Ps), Larix decidua (Ld) and Fagus sylvatica (Fs); n = 3). (a) Bar colours indicate the tree species from which the roots originated (only top 30 OTUs are displayed). Asterisks denote EMF associations with all four tree species. (b) Proportion of root tips associated with EMF species found on one, two, three, or four different host tree species. (c) Schematic presentation of the distribution of the four tree species in the mixed forest plot
2.2. CO2 labelling
Between 30 July 2009 and 30 October 2014, five 37–40 m tall Picea abies individuals were equipped with a free air CO2 enrichment system (FACE). Since the 1–2 t of food quality CO2 released daily over the canopy carried a constant 13C isotope signal (δ13C = −30‰, compared with ca. −8‰ in the ambient air), FACE permitted tracing of the carbon flows in trees and into the soil, besides confirming the efficiency of the FACE system (Mildner, Bader, Leuzinger, Siegwolf, & Körner, 2014). CO2 release (and hence, labelling) was confined to tree crowns, i.e., at 20–37 m aboveground, without downward CO2 -flow and thus, preventing uncontrolled CO2 label transfer to the soil surface (confirmed by δ13C in understory vegetation; five similarly tall Picea trees away from the labelled trees served as controls under unlabelled, ambient CO2. The FACE system was based on a web of porous tubes, woven into the tree crowns using a 45 m high canopy crane (Klein, Siegwolf, et al., 2016).
2.3. Fine root sampling and DNA extraction
Lateral fine roots of Picea abies, Fagus sylvatica, Pinus sylvestris, Larix decidua trees were sampled on 20–24 September 2016, 2 years after discontinuation of FACE (Klein, Siegwolf, et al., 2016). Lateral fine roots were sampled at 1–4 m distance from stems they belong to, at about 10 cm soil depth; tree species was controlled by following fine roots to the stems they emanated from (and by sequence analysis of roots; detailed below). We excavated roots from a total of 12 individual trees: three labelled Picea abies individuals (n = 3) and nine allospecific, tall individuals of canopy forming non-Picea neighbour species Fagus sylvatica (n = 3), Pinus sylvestris (n = 3), Larix decidua (n = 3). All 12 trees were sampled using the same approach. Six different terminal root branchlets were sampled per tree at ~60º azimuthal intervals: 2–4 root branchlets from the overlapping zone between roots of each labelled Picea and its non-Picea neighbour, and 2–4 root branchlets from the distal side. For each root branch-let we recorded the distance and orientation to the source tree. Sampled roots were stored in an ice box and washed free from attached soil particles on a 1 mm sieve under tap water within 24 hr. Washed fine roots were placed in petri dishes with a thin layer of unchlorinated tap water and stored at 4°C. Within 1–3 days, each root sample was placed over a 1 cm grid, to allow for unbiased sampling of 30 root tips, regardless of their size, form, and colour. Root tips were picked from each root sample using a dissecting scope and tweezers that were flame-sterilized between each picking. EMF tips were placed in prefilled 96 well plates with 18 µl of Sigma XNAT DNA isolation buffer (Sigma-Aldrich) and then stored at 4°C, until DNA extraction.
2.4. Fungal sporocarp sampling and analysis
During August to November 2010 and 2011, 2 years into the 5.5-year FACE experiment period, fruit bodies (sporocarps) were collected from around labelled and unlabelled (control) Picea abies trees. Root spheres of 6 m diameter were specified around each Picea tree, and all the epigeous (aboveground), visible sporocarps were collected (Mildner et al., 2014). The sporocarps were frozen and stored at −20°C until September 2016 when genetic identification and further preparation for carbon isotope analysis were implemented. At least one representative of each sporocarp type was chosen, tissue pieces of 1 mm size were placed in the same prefilled 96 well plates as described above and stored in 4°C until DNA extraction.
2.5. DNA extraction
DNA was extracted with the Extract-N-Amp Tissue PCR kit (SIGMA XNAT2-1KT) according to the manufacturer’s standard protocol, with the following modification: extractions were performed in a volume of 18 µl extraction buffer instead of 100 µl. The 96-well plates were incubated for a minimum of 10 min at room temperature, incubated for 4 min at 95°C using a thermo-cycler and then mixed with the neutralizing inhibitor solution (15 µl) and stored at 4°C.
2.6. PCR amplification and DNA isolation
From each fine root sample (sampled on 20–24 September 2016), we amplify fungal DNA region from the 30 root tips collected. In addition, we amplified four root tips from each root sample with plant primers to verify our assessment of tree species identify. PCR reactions were performed using 1.25 units Dream Taq DNA polymerase (Thermo Scientific) in 25 µl reaction volumes with 1 µl DNA extract, 0.2 mM dNTPs, and 0.5 µl primer. For fungal identification, we used the internal transcribed spacer (ITS) region primers ITS1F (Table S1; Gardes & Bruns, 1993) and ITS4 (Table S1; White, Bruns, Lee, & Taylor, 1990). For the tree species identification, we used TrnL-F (Table S1) and TrnL-R (Table S1; Taberlet, Gielly, Pautou, & Bouvet, 1991). PCR reactions for both sets of primers were performed as follows: initial 1 min at 94°C followed by 35 cycles of 30 s 94°C, 30 s 51°C, and 1 min 72°C final cycle with 8 min 72°C. A total of 6 µl of the PCR products were run on 1.5% agarose gel with 6 µl of SYBR safe for 40 min. If PCR resulted in several bands or gave a weak signal in the agarose gel, higher or lower amount of DNA template was used for the PCR reaction. PCR reaction samples with one clear band were purified by isopropanol-ethanol precipitation, and the DNA pellet was dried in the dark at room temperature. DNA was dissolved with 10 µl H2O and 10 µM ITS4 primer or Trnl-F, vortexed and then stored at –20°C until sequencing.
2.7. Sequencing and data analysis
Amplified purified DNA from root tips and sporocarps was Sanger sequenced by Macrogen (EZ-seq; Macrogen Europe) unidirectionally with primers ITS4 (fungi) and TrnL-F (trees). Sporocarps collected near control trees were sequenced by the Biological Services Department, Weizmann Institute of Science, Rehovot, Israel. From the 2,160 root tips collected, all PCR products were run on agarose gel. In root samples with <50% clear bands, only the first 16 samples (two plate rows) with clear bands were sent for sequencing instead of all 30. A total of 1,698 root tips, from six different roots per tree and a minimum of 138 root tips per tree (except for a single Larix individual with 96 root tips) were sent for sequencing. Electropherograms of raw sequences were checked manually and only high quality portions of the sequence, at least 100 bp long, were used for analysis (short sequences (45) and low quality sequences (other 45) were eliminated). Verifying the root origin using the TrnL tree DNA region indicated 93% of the root tips were from the suspected tree species (124 root tips were eliminated, belonging to a single root from each of three Larix trees and three roots from two Pinus trees. Sequences (from 1,500 root tip) were clustered locally using the sequencher 5.3 software (Gene Codes Corp.) with minimum match percentage 95% and minimum overlap of 100 bp. Consensus sequences were assigned operational taxonomic unit (OTU) names (when possible, at the species level) by manually editing ambiguous bases and queried in sequences databases at PlutoF (unite, blastn 2.4.0) (Altschul et al., 1997). Consensus sequences (~600 bp) with no species level of 90% or higher were identified for the Genus level only. Normalization of the data for statistical tests was facilitated by first combining the six root samples for each tree, and those trees which were represented by more than 87 root tips were rarified to 87 selected root tips (1,015 root tips remaining in total). Finally, for further analysis of the overlapped EMF species, OTUs which were found only on one tree were removed (863 root tips remaining, Table S2). Mycorrhizal status of sporocarps was assigned according to Tedersoo, May, & Smith (2010). Representatives sequences from each taxa were submitted to GenBank in a bulk accession under the accession SUB6629846 (MN970689 - MN970812).
2.8. Sporocarps δ13C
Each sporocarp was measured for the δ13C signature derived from the 13CO2 tree labelling during FACE. Sporocarp material was oven-dried at 60°C for 48 hr and kept dehydrated until analysis. Unground tissue pieces of 1 mg were mounted onto a combustion module equipped with auto-sampler (ECS 4010, Costech Analytical). The CO2 gas product was analyzed with a 13C cavity ring down analyzer (G2131i, Picarro), which was directly interfaced to the combustion module. Results are expressed as part per thousand (‰) deviations from the international carbon isotope standard. Three replicates from each biological sample were measured and an internal standard (homogeneous Spirulina powder with a δ13C of −29.2‰) was used every nine samples.
2.9. Fine roots δ13C
Fine roots for δ13C analysis were sampled in the meeting regions where root spheres of two individual trees from different taxa spatially overlapped and carbon transfer has been recently observed (Klein, Siegwolf, et al., 2016). Briefly, root branchlets of non-Picea neighbour (Fagus, Larix and Pinus) were sampled at the end of the FACE experiment from the labelled and control plot (during 10 and 27 March 2015). Tree-tree C transfer was calculated by the 13C depleted fine roots (n = 4) from unlabelled trees overlapping with root spheres of labeled Picea, relative to the average of δ13C from control root branchlets of the same tree species. Fine roots’ δ13C of Larix (δ13C −27.5‰) from the nonlabelled plot was estimated from the average differences between the trunk and roots’ values of the other species and in agreement with δ13C reported for other Larix species in the literature (Ouimette, Guo, Hobbie, & Gu, 2013). The isotope analysis was performed at the stable isotope facility at Paul Scherrer Institute, Villigen, Switzerland, as previously described.
2.10. Evolutionary relationships of tree taxa
Evolutionary relationship was deduced from the TrnL-F region. Electropherograms of raw sequences were checked manually and only high quality portions of the sequence, at least 100 bp long, were used in the analysis. Sequences (Picea = 89, Pinus = 36, Larix = 55, Fagus = 67) from three individuals per species, were clustered locally using the sequencher 5.3 software (Gene Codes Corp.) with minimum match percentage 95% and minimum overlap of 100 bp. Consensus sequences of the single cluster (Pinus = 36, Fagus = 67) or two clusters (Larix = 19,22 and Picea = 28,58) in every tree species were aligned (ClustalW) and the evolutionary history was inferred using the UPGMA method. The optimal tree with the sum of branch length = 0.54 was used. The evolutionary distances were computed using the maximum composite likelihood method (Tamura, Nei, & Kumar, 2004) and are in the units of the number of base substitutions per site from the four species. All ambiguous positions were removed for each sequence pair. The final data set included a total of 569 positions. The percentage of replicate phylogenetic trees in which the associated taxa clustered together in bootstrap test (Felsenstein, 1985) was >99% or higher in all the branches except between Picea and Larix with only 81%. Evolutionary analyses were conducted in MEGA X platform (Institute of Molecular Evolutionary Genetics, Pennsylvania State University, University Park, USA).
2.11. Statistics
We used R (R Core Team, 2018) and the interface RStudio (R Core Team, 2018) for statistical analysis and EMF networks’ prediction. Levene, ANOVA (p < .05), shapiro and post-hoc Tukey’s test (p < .05) tests were calculated for the different 13C levels measured sporocarps. Species accumulation curves for EMF communities were constructed using the rarefy function of the vegan R package version 2.4 (Oksanen et al., 2013) on the data set obtained before the trimming of EMF taxa only found in on one tree. Differences in EMF community composition between trees were visualized with nonmetric multidimensional scaling (NMDS) (Three dimensions, stress = 0.11). Ellipses were drawn around the 95% confidence interval for the centroid (the standard error multiplied by the Chi-squared distribution) of all samples from a particular tree species. Examination of whether the EMF communities of different tree species were significantly different was performed by permutation multivariate analysis of variance (PERMANOVA; Adonis function of the vegan R package). Multiple comparisons Correction was done by the false discovery rate correction (Benjamini & Hochberg, 1995). Further examination of the relative difference between tree species’ pairings was conducted by averaging Bray-Curtis similarity values for all individual tree parings in the 12 tree data set and then comparing average dissimilarity values between different tree species’ pairings (using the diversity function in vegan R package); significant differences in community similarity between different tree species pairings were determined by ANOVA and post-hoc Tukey tests. The significant amount of carbon transfer was tested by t test (p < .05), of Ps roots (n = 4), Fs roots (n = 4), and of Ld roots (n = 3; ignoring one extreme value). A positive relation (y = 4.4x−0.28), between EMF community similarity (1- Bray Curtis) (n = 6) and tree-tree C transfer (n = 4) was tested by ANOVA (p < .05), one single extreme value was eliminated form Ld carbon transfer data and from the community similarity of Ps data.
3. Results
3.1. Ectomycorrhizal fungi are predominantly associated with multiple tree hosts
We analyzed 1,185 root tips from 12 individuals of four tree species (three individuals of Picea abies, Pinus sylvestris, Larix decidua and Fagus sylvatica) and assigned the fungi colonizing them to 101 operational taxonomic units (OTUs) at the species level. We rarified the EMF community on each tree (compositing six root samples) to 87 root tips (yielding 1,015 root tips and 99 OTUs), and subsequently eliminated all EMF OTUs occurring on one individual tree only (leaving 56 OTUs remaining, comprised 863 root tips or 85% of all identified root tips). Species accumulation curves (Figure S1) for the four tree species nearly saturated at the sampling intensity achieved but also indicated that we were not able to fully capture the EMF diversity from each tree species. The genus Sebacina was most abundant, with S. dimitica, S. epigaea and S. incrustans jointly representing 24% of all EMF root tips (Figure 1). The six most abundant EMF OTUs, the aforementioned three Sebacina, followed by Russula chloroides, Peziza suc-cosa, and Amphinema byssoides, constituted 41% of all identified EMF root tips (Figure 1). EMF diversity was similar for all four trees species with 33–50 OTUs collected per tree species, with median Shannon-diversity index values of 2.25–2.50; yet, rarefaction and Shannon’s index indicated that the Larix EMF community was slightly less diverse than that of the other three species (Figure S1). Among the 56 OTUs found on more than one individual tree, more than 70% of the OTUs found on Larix, Picea, and Pinus were associated with three or all four tree host species, while <50% of the EMF OTUs found on Fagus were (Figure S2). 90% of the identified OTUs, representing 90% of all sampled root tips, were found in more than one host species (Figure 1b). Eight OTUs were common to all four tree species (of which, seven OTUs occurring on >10 root tips are presented in Figure 1). The EMF which were associated with all four tree species were Sebacina incrustans, Russula chloroides, Peziza succosa, Amphinema byssoides, Clavulina coralloides, Inocybe glabripes, Lactarius fulvissimus, and Hymenogaster sp.1 (Figure 1a). Overall, EMF species with a single host species were minor in number and abundance.
3.2. Genetically identified ectomycorrhizal fungal sporocarps contain labelled carbon from Picea
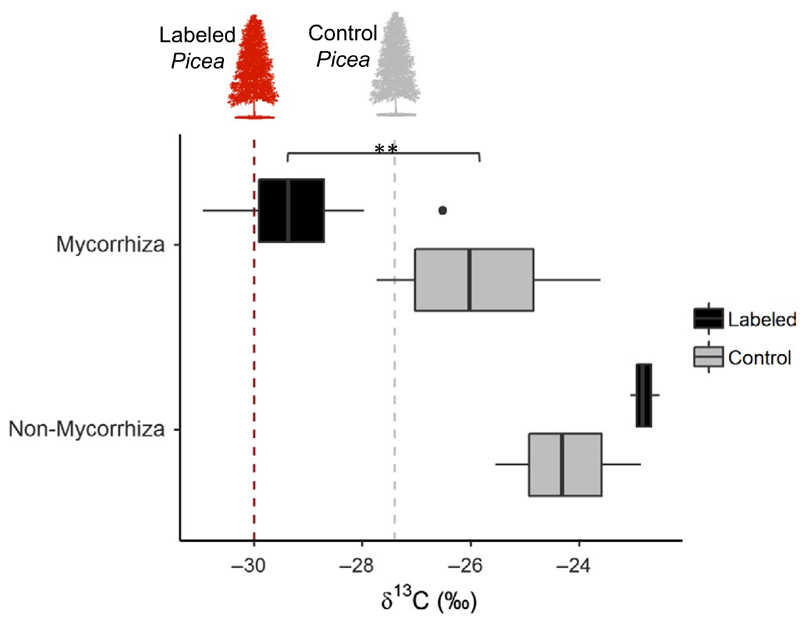
Ectomycorrhizal sporocarps collected from the area under labelled trees had δ13C signatures of −26.5 ± 0.1 to −30.9 ± 0.1, which were significantly more depleted in 13C compared with −23.6 ± 0.2 to −27.7 ± 0.2 from sporocarps collected near control trees (one-way ANOVA, F(3,18) = 28.9 p < .001, Tukey HSD, p = .001; Figure 2, Figure S3). Among the 12 different sporocarps collected under and around labelled trees, three were from EMF OTUs common to 3–4 tree species: Clavulina coralloides (δ13C –29.9 ± 0.1), Russula chloroides (δ13C –29.7 ± 0.1) and Lactarius fulvissimus (δ13C –29.4 ± 0.1; Figure 1a, Figure 2 & Figure S3). Congeneric EMF sporocarps from Lactarius deterrimus (δ13C –25.3 ± 0.2) and Russula nobilis (δ13C –27.7 ± 0.2) collected in the vicinity of unlabelled Picea trees had markedly less depleted δ13C signals. Most of the other shared EMF species did not produce above-ground and visible sporocarps. While ectomycorrhizal sporocarps collected near labelled trees were significantly more depleted in δ13C than EMF sporocarps collected from unlabelled trees, nonectomycorrhizal (primarily saprotrophic) sporocarps did not show a δ13C difference between labelled and unlabelled areas (Tukey HSD, p = .52).
Figure 2.
13C isotopic signature of fungal sporocarps collected near 13C labelled and control (unlabelled) trees following 2 years of continuous labelling of Picea abies tree crowns. Sporocarps were collected 3–6 m from the tree stems. Sporocarps were both morphologically and genetically identified, and analyzed for isotopic composition. Red and grey Picea trees represent the root δ13C values of labelled and control trees, respectively. Box plot (whisker) presenting the median, upper and lower quartiles and outliers of the Mycorrhiza labelled (n = 12), control (n = 4) and the non-Mycorrhiza labelled (n = 3), control (n = 3). Asterisk represent significant differences (p < .001) [Colour figure can be viewed at wileyonlinelibrary.com]
3.3. Ectomycorrhizal community composition is more similar between more closely related tree species
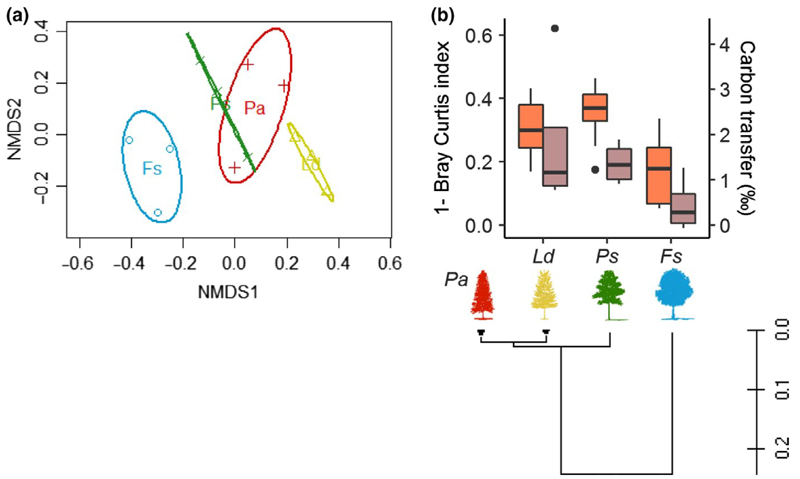
PERMANOVA analysis indicated that EMF community composition was significantly different in all possible species pairings (p < .003), except for Picea-Larix (p < .12) and Picea-Pinus (p < .6) (Table S3). Ordination analysis indicated that the EMF community of Fagus differed significantly from that of the other three tree species, and the EMF community colonizing Larix was more different from Pinus and Picea than the EMF communities on Pinus or Picea were from one another (Figure 3a). Average community dissimilarity (Bray-Curtis index) between Fagus and all other species were highest, and Picea and Fagus were significantly less similar than either Picea and Larix or Picea and Pinus (Figure S4); average community dissimilarity also indicated that Picea and Pinus EMF communities were more similar to each other than either were to Larix EMF communities.
Figure 3.
EMF community similarity by neighbouring tree hosts at the plot level. (a) Relative EMF community differences between tree species and within species, represented by nonmetric multidimensional scaling (NMDS, stress = 0.11). Each point represents the EMF community of each tree. Ellipses are drawn around the 95% confidence interval for the centroid. (b) Relationship between the level of EMF community similarity (1- Bray Curtis index, orange), the level of carbon transfer (13C signal, purple) among Picea and the non-Picea trees and evolutionary relationship of tree taxa. EMF community data were normalized according to the procedures described in the Methods. δ13C was measured in trees neighbouring the labelled Picea relate to control trees from the same species (Picea abies (Pa), Pinus sylvestris (Ps), Larix decidua (Ld) and Fagus sylvatica (Fs). The optimal phylogenetic tree with the sum of branch length = 0.495 is shown, and drawn to scale [Colour figure can be viewed at wileyonlinelibrary.com]
3.4. The magnitude of belowground carbon transfer is correlated with EMF community similarity
We tested whether the similarity in EMF composition between tree species’ pairs was correlated with tree-tree C transfer, measured by the 13C depletion in roots from unlabelled trees overlapping with root spheres of labelled Picea individuals. Based on the evolutionary relationship of tree taxa, the gymnosperm Picea is closely related to the gymnosperms Larix and Pinus, and more distant from Fagus. In correlation with these relationships, EMF community similarity, measured as 1- Bray Curtis dissimilarity, were markedly higher between Picea – Larix and between Picea – Pinus than between Picea – Fagus (Figure 3b). The amount of carbon transfer was significantly higher between Picea and Pinus than that between Picea and Fagus (Figure 3b; t test p < .05) ignoring extreme values (presented as outlier points; more than 1.5 times the interquartile range, inclusive median; n = 3–4), again correlating with the trees’ evolutionary history. Plotting EMF community similarity (1- Bray Curtis) against tree-tree C transfer yielded a positive relation (y = 4.4x–0.28; R 2 = .99), which was significant when the two most extreme values (Figure 3b, outlier points) were eliminated (ANOVA, p < .05, F = 764.45, n = 3–4 roots).
3.5. Ectomycorrhizal networks may interlink multiple tree host across a mixed forest
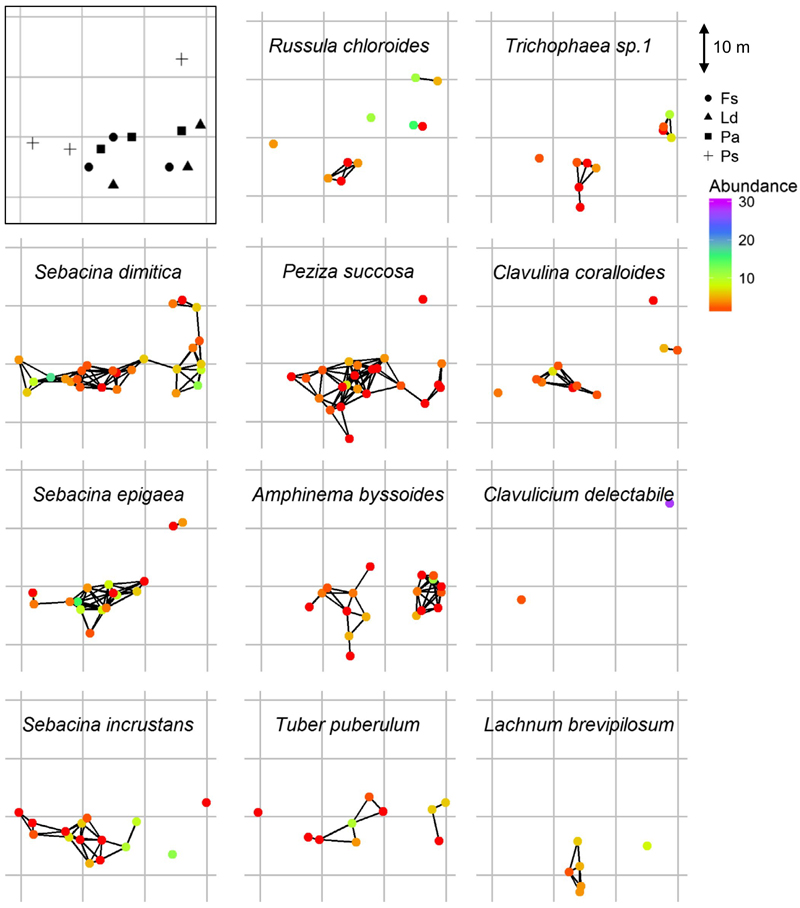
Transfer of labelled carbon through EMF hyphae at our mixed forest site was found to be most effective in a radius of <6 m from tree trunks (Mildner et al., 2014). Based on the spatial distribution of each EMF OTUs in the study plot and a 6 m threshold for connectivity, we predicted a carbon transfer coverage in the forest plot (Figure 4). The most abundant EMF OTU in our study, Sebacina dimitica, was the only EMF OTU to produce a plot-wide network (Figure 4). Interestingly, predicted networks typically consisted of proximal locations with low numbers (e.g., the network of Peziza succosa), whereas locations with high abundance were mostly disconnected from a network (e.g., the network of Russula chloroides). The observed networks shed light on the potential distribution and the possible carbon paths for each EMF species, offering a unique glimpse into the “wood-wide-web” (Simard et al., 1997) of carbon transfer.
Figure 4.
Predicted spatial EMF networks for the most abundant 11 OTUs in the mixed forest plot near Basel, Switzerland. Black frame (top left) represent schematic presentation of the distribution of the four tree species in the mixed forest plot. Colours indicate the species abundance at each location. Lines connect locations where an EMF species was identified 6 m or less from the nearby location. The 6 m threshold is based on the distance dependence of observed isotopic carbon transfer for the studied trees on site (Mildner et al., 2014)
4. Discussion
We investigated patterns of EMF-host species associations to explore the potential role of common mycorrhizal networks in transferring significant amounts of carbon among adult trees belonging to different taxa, as was observed by Klein, Siegwolf, et al. (2016). We found an extremely high level of EMF ‘species’ sharing between tree species. In fact, association with multiple host trees was widespread among EMF taxa. At least eight different EMF OTUs were common to the four tree taxa, and 90% of all EMF OTUs from root tips were associated with at least two host tree species (Figure 1), and 90% of all identified mycorrhizal root tips belonged to OTUs which were observed on at least two tree species. These findings are congruent with those of a recent large scale study of host specificity that found that only approximately 10% of the EMFs are host specific (van der Linde et al., 2018). The basidiomycota Sebacina dimitica, Sebacina epi-gaea and Sebacina incrustans OTUs were particularly widespread and “promiscuous” in our forest plot (Figure 1). The OTUs of Sebacina were common to all tree species (Figure S5), although they were less abundant on Fagus.
Belowground carbon transfer via EMF was observed from Picea trees to roots and stems of neighbouring trees from different taxa. EMF sporocarps collected near labelled trees had significantly depleted δ13C signals compared to EMF sporocarps collected further away from labelled trees (nonmycorrhizal (primarily saprotrophic) fungal sporocarps lacked labelling altogether; Figure 2). Reliance on analysis of sporocarps alone, however, is problematic: at least a third of all the EMF ‘species’ do not commonly produce prominent sporocarps (van der Linde et al., 2018), including Sebacina, and the timing of sporocarp production changes with the microclimate of the specific year (Alday et al., 2017). Our data show that the isotopically labelled carbon assimilated by the canopy of Picea trees, which was detected in roots and stems of neighbouring trees, is being transferred through certain EMF and was not yet detected in nonmycorrhizal fungi.
Despite the overall low degree of EMF host specificity, host association patterns were apparent in EMF communities. EMF community composition significantly changed depending on associated plant phylogeny and, perhaps as a function of this, also correlated with carbon transfer. While many EMF species are host generalists (Horton & Bruns, 1998), host preferences also play a role, for example among conifers (Glassman et al., 2015; van der Linde et al., 2018). We expected the differences in EMF community composition between trees to be a function of the trees’ phylogenetic distance. In our temperate mixed forest site, we sampled the EMF community of four tree species vary in their degree of relatedness. The variation between the Fagus (Fagaceae) EMF communities and that of the other tree species (Pinaceae) was consistently greater than the differences among Pinus, Larix and Picea (Figure S4). Rates of carbon transfer from labelled Picea, measured as isotopic depletion of roots, appeared to match EMF community differences, with Fagus receiving the least carbon and Pinus getting more (though not significantly) than Larix (Figure 3b), with a significant relation among the three groups.
Four lines of evidence support a direct involvement of EMF in the belowground carbon transfer: (a) Canopy, stem, root, and fine root δ13C signals showed belowground movement of labelled carbon (longterm FACE experiment; (Klein, Bader, et al., 2016; Klein, Siegwolf, et al., 2016) from stem to stem, via root-EMF-networks, and not via respired CO2; (b) Trees receiving labelled carbon from labelled Picea individuals shared, to a large extent, similar EMF community as the labelled Picea individuals (Figure 3); (c) EMF sporocarps collected near labelled trees showed significantly depleted δ13C signals compared to EMF sporocarps collected further away from labelled trees and this was only observed in EMF sporocarps, not in non-EMF fungal sporocarps (Figure 2), (d) Carbon exchange between Picea and the non-Picea tree species was greater between tree species that shared more similar EMF communities (Figure 3b). In agreement with previous work (Pickles et al., 2017; Simard et al., 1997; Wagg, Bender, Widmer, & van der Heijden, 2014) these findings provide insight into the hidden belowground carbon networks, which connect trees from different taxa through single or multiple EMF species.
Common mycorrhizal networks may be composed of a single connected genet of EMF fungus or anastomosed clusters of genets interlinked though mycelial connections (Beiler et al., 2010). The resolution of our molecular markers does not allow for inferences of genet identity within species. But even if it were established that genetically identical EMF colonize Picea donor trees and allospecific receiver trees, this would not prove that the clonal connection was intact. A single EMF genotype may be observed in geographically distant, unconnected locations (Croll et al., 2008). The structure of mycorrhizal networks can vary a great deal across a mixed forest (Kennedy, Izzo, & Bruns, 2003) with unknown consequences for carbon transfer. This patchy distribution of EMF species can be dynamic at the scale of years, affected by root distribution and root density (Pickles et al., 2010). The different signal level of δ13C found in the EMF sporocarps (Figure 2), together with the predicted carbon transfer networks (Figure 4), support the hypothesis of multiple, and complex, networks which shuttle carbon and probably other elements among the trees.
Ectomycorrhizal fungis display a wide variation in host specificity (Horton & Bruns, 1998). The ability to associate with multiple host plant species increases the potential habitat for EMF, while specialization can be an advantage if it provides greater physiological compatibility with the host (Bruns, Bidartondo, & Taylor, 2002) or if it diminishes the probability of indirectly transferring resources to competing plant species (Molina, Massicotte, & Trappe, 1992). Recent analysis of the genus Russula found evidence for host switching between conifers and angiosperms (Looney, Ryberg, Hampe, Sanchez-Garcia, & Matheny, 2016). In agreement with these findings, we found the genus Russula to be the second most abundant genus associated with the four different hosts (Figure S5). An example of host expansion triggered by past presence of other tree species in the stand, was shown for Suillus subaureus (Lofgren, Nguyen, & Kennedy, 2018). Nevertheless, in our study, we found Suillus granulatus and Suillus viscidus exclusively on roots of the two conifers, Pinus and Larix respectively (Figure 1a). On the other hand, Clavulina coralloides, which we found associated with Pinus only (Figure 1a), has been observed to associate with both spruce and birch (Tedersoo, Kõljalg, Hallenberg, & Larsson, 2003). In addition to specialization upon host plant species, EMFs vary considerably in their degree of specialization of abiotic, especially edaphic, conditions (Johnson et al., 2010; Peay et al., 2015; Tedersoo et al., 2003). Interestingly, a link between the level of specificity and ecological strategy can be explained by the low mycorrhizal community overlap between early and late successional plant species (Taudiere et al., 2015). The shift from specialist to a generalist EMF and vice versa can influence the EMF guilds and the optimal belowground networks in the forest.
Ectomycorrhizal morphology may be an important factor influencing their role in belowground carbon transfer. In the context of mycorrhizal exploration types (Agerer, 2001), the most abundant EMF genera that we observed shared among the different tree species (Figure S5) were contact- and short-distance types. However, long-distance exploration type (e.g., in Suilus), which were found in the two gymnosperms Larix and Pinus (Figure 1) have also been observed to mediate belowground carbon transfer (Teste, Simard, Durall, Guy, & Berch, 2010). Sebacina species, which belong to the most abundant genus in our forest (Figure 1a) are typically of the short-distance exploration type (Oberwinkler et al., 2013; Urban, Weiss, & Bauer, 2003; Weiss, Waller, Zuccaro, & Selosse, 2016), and have been shown to mediate C transfer between trees and orchids (Selosse, Weiss, Jany, & Tillier, 2002; Yagame, Orihara, Selosse, Yamato, & Iwase, 2012). Russula chloroides, which was associated with the four different hosts (Figure 1), forms short- and medium-distance mycelia (Agerer, 2006), and has been implicated as a likely intermediary for an orchid to receive carbon from neighbouring EMF trees (Girlanda et al., 2006). EMFs that form rhizomorphs have been shown to promote carbon transfer in larger distance and between different species (Teste et al., 2010). However, rhizomorph connections are generally rare amongst EMF species (Finlay & Read, 1986), and we found taxa that are known to form rhizomorphs to be infrequent in the overlapping EMF species (Figure 1). In summary, the high level of overlapping EMF species observed here might suggest that the short distance exploration strategy dominates the below-ground carbon networks in the studied tree assembly, with less prominent contributions by long distance hyphal connections, which are more host specific.
The specific form in which trees exchange carbon remains unknown. Belowground carbon transfer by EMF was found to facilitate a bidirectional transfer among trees at this site (Klein, Siegwolf, et al., 2016). Plants provide to the symbiotic fungi carbon in the form of hexoses (Shachar-Hill et al., 1995), but recently, cross-kingdom lipid transfer was also demonstrated between arbuscular mycorrhizal fungi and a plant host (Jiang et al., 2017). Different EMF species displayed substantial variation in δ13C (Figure 2), which may be explained by the different compounds being transferred and various metabolic pathways being utilized (Deveau, Kohler, Frey-Klett, & Martin, 2008). Analyses combining stronger isotopic labelling together with advanced analytical identification (allowing both identification of potential transfer compounds and measurement of their 13C signatures) are needed to understand the carbon transfer mechanism and the exact organic compounds being exchanged between the EMF and the host tree.
Elevated CO2 levels can alter mycorrhizal community composition in the field (Gryta, Debaud, Effosse, Gay, & Marmeisse, 1997), although the precise effects on the symbiotic interactions are not yet clear: Under limiting CO2 levels, symbiosis can play a more beneficial role (Zhang, Ziegler, Han, Trumbore, & Hartmann, 2015), but a number of studies have observed elevated CO2 levels to increase carbon allocation to EMF (Drigo et al., 2010; Hyvonen et al., 2007) reviewed by Terrer et al. (2018). At our site, the FACE experiment induced increased carbon transfer to belowground sinks such as fine root production of Picea trees (Klein, Bader, et al., 2016; Mildner et al., 2014). Adjustment to this new condition probably depends on both symbiosis partners (host trees and EMF symbionts), and soil nutrient availability. It is possible that carbon transfer from labelled (CO2 enriched) Picea to nearby allospecific trees through the EMF network was increased by the higher CO2 supply. The amount of carbon transfer observed by Klein, Siegwolf, et al. (2016) may thus represent a high estimate for transfer rates compared to a situation in which the entire forest experience the same future CO2 concentrations. Nevertheless, the current study, in conjunction with Klein, Siegwolf, et al. (2016), provide compelling evidence for the important role of EMF in transferring significant amounts of carbon among forest trees belonging to different taxa. Connectedness can be an advantage, particularly for sharing resources related to environmental changes (Pickles & Simard, 2017). The mechanisms and dynamics of this belowground carbon transfer through common EMF networks, as well as its repercussions for patterns of tree species assemblages in forests, and their effects on the ecosystem carbon balance, are now under investigation.
Supplementary Material
Additional supporting information may be found online in the Supporting Information section.
Acknowledgements
The authors thank M. Mildner (University of Basel) for collecting the sporocarps, M. Wilhelm (Basel) for identifying the sporocarps’ species, Professor Hakan Wallander (Lund University, Sweden) for assistance with molecular EMF identification, O. Lapidot (WIS) for assistance with R programing of predicted mycorrhizal networks, C. Ziv (ARO Volcani center) and H. Shemesh (Tel Hai College) for their useful comments on early drafts of this paper. TK wishes to thank the Merle S. Cahn Foundation and the Monroe and Marjorie Burk Fund for Alternative Energy Studies, Mr. and Mrs. Norman Reiser, together with the Weizmann Center for New Scientists, and the Edith & Nathan Goldberg Career Development Chair; Estate of Helen Nichunsky and the Benoziyo Endowment Fund for the Advancement of Science; and The Weizmann - Angel Faivovich Foundation for Ecological Research. The forest CO2 enrichment facility near Basel was funded by the Swiss Science foundation (projects 31003A, 14753/1 to CK.), the Swiss Federal Office of the Environment, and the University of Basel. Research was funded in part by the European Research Council grant 849740 RHIZOCARBON given to TK. IR is supported by the Sustainability and Energy Research Initiative Ph.D. Fellowship.
Funding information
Weizmann Center for New Scientists, and the Edith & Nathan Goldberg Career Development Chair; Merle S. Cahn Foundation; Estate of Helen Nichunsky and the Benoziyo Endowment Fund for the Advancement of Science; The Weizmann -Angel Faivovich Foundation for Ecological Research; Mr. and Mrs. Norman Reiser; Swiss Federal Office of the Environment; Swiss Science foundation, Grant/Award Number: projects 31003A, 14753/1 to C.K.; The European Research Council, 849740 RHIZOCARBON to TK; Monroe and Marjorie Burk
Footnotes
Author Contributions
I.R., and C.K. performed the sampling in the field research site managed by C.K. DNA extraction and PCR were performed by I.R., and N.R. Analyses were performed by all authors and coordinated by T.K. I.R. wrote the manuscript with contributions from all authors.
Data Availability Statement
Data supporting the results will be archived in an appropriate public repository.
References
- Agerer R. Exploration types of ectomycorrhizae. Mycorrhiza. 2001;11(2):107–114. doi: 10.1007/s005720100. [DOI] [Google Scholar]
- Agerer R. Fungal relationships and structural identity of their ectomycorrhizae. Mycological Progress. 2006;5(2):67–107. doi: 10.1007/s11557-006-0505-x. [DOI] [Google Scholar]
- Alday JG, Martínez de Aragón J, de-Miguel S, Bonet JA. Mushroom biomass and diversity are driven by different spatio-temporal scales along Mediterranean elevation gradients. Scientific Reports. 2017;7:45824. doi: 10.1038/srep45824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahram M, Polme S, Koljalg U, Tedersoo L. A single European aspen (Populus tremula) tree individual may potentially harbour dozens of Cenococcum geophilum ITS genotypes and hundreds of species of ectomycorrhizal fungi. FEMS Microbiol Ecology. 2011;75(2):313–320. doi: 10.1111/j.1574-6941.2010.01000.x. [DOI] [PubMed] [Google Scholar]
- Beiler KJ, Durall DM, Simard SW, Maxwell SA, Kretzer AM. Architecture of the wood-wide web: Rhizopogon spp. genets link multiple Douglas-fir cohorts. New Phytologist. 2010;185(2):543–553. doi: 10.1111/j.1469-8137.2009.03069.x. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- Brownlee C, Duddridge J, Malibari A, Read D. The structure and function of mycelial systems of ectomycorrhizal roots with special reference to their role in forming inter-plant connections and providing pathways for assimilate and water transport. In: Atkinson D, Bhat KKS, Coutts MP, Mason PA, Read J DJ, editors. Tree root systems and their mycorrhizas. Dordrecht: Springer; 1983. pp. 433–443. [Google Scholar]
- Bruns TD. Thoughts on the processes that maintain local species diversity of ectomycorrhizal fungi. Plant and Soil. 1995;170(1):63–73. doi: 10.1007/BF02183055. [DOI] [Google Scholar]
- Bruns TD, Bidartondo MI, Taylor DL. Host specificity in ectomycorrhizal communities: What do the exceptions tell us? Integrative and Comparative Biology. 2002;42(2):352–359. doi: 10.1093/icb/42.2.352. [DOI] [PubMed] [Google Scholar]
- Castaño C, Lindahl BD, Alday JG, Hagenbo A, Martínez de Aragón J, Parladé J, Bonet JA. Soil microclimate changes affect soil fungal communities in a Mediterranean pine forest. New Phytologist. 2018 doi: 10.1111/nph.15205. [DOI] [PubMed] [Google Scholar]
- Croll D, Wille L, Gamper HA, Mathimaran N, Lammers PJ, Corradi N, Sanders IR. Genetic diversity and host plant preferences revealed by simple sequence repeat and mitochon-drial markers in a population of the arbuscular mycorrhizal fungus Glomus intraradices. New Phytologist. 2008;178(3):672–687. doi: 10.1111/j.1469-8137.2008.02381.x. [DOI] [PubMed] [Google Scholar]
- Deveau A, Kohler A, Frey-Klett P, Martin F. The major pathways of carbohydrate metabolism in the ectomycorrhizal basidiomycete Laccaria bicolor S238N. New Phytologist. 2008;180(2):379–390. doi: 10.1111/j.1469-8137.2008.02581.x. [DOI] [PubMed] [Google Scholar]
- Dickie IA, Guza RC, Krazewski SE, Reich PB. Shared ectomycorrhizal fungi between a herbaceous perennial (Helianthemum bicknellii) and oak (Quercus) seedlings. New Phytologist. 2004;164(2):375–382. doi: 10.1111/j.1469-8137.2004.01177.x. [DOI] [PubMed] [Google Scholar]
- Drigo B, Pijl AS, Duyts H, Kielak AM, Gamper HA, Houtekamer MJ, Kowalchuk GA. Shifting carbon flow from roots into associated microbial communities in response to elevated atmospheric CO2. Proceedings of the National Academy of Sciences. 2010;107(24):10938–10942. doi: 10.1073/pnas.0912421107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein JJE. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Finlay R, Read D. The structure and function of the vegetative mycelium of ectomycorrhizal plants: II. The uptake and distribution of phosphorus by mycelial strands interconnecting host plants. New Phytologist. 1986;103(1):157–165. doi: 10.1111/j.1469-8137.1986.tb00604.x. [DOI] [Google Scholar]
- Gao C, Zhang YU, Shi N-N, Zheng Y, Chen L, Wubet T, Guo L-D. Community assembly of ectomycorrhizal fungi along a subtropical secondary forest succession. New Phytologist. 2015;205(2):771–785. doi: 10.1111/nph.13068. [DOI] [PubMed] [Google Scholar]
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes–application to the identification of mycorrhizae and rusts. Molecular Ecology. 1993;2(2):113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Gehring CA, Sthultz CM, Flores-Renteria L, Whipple AV, Whitham TG. Tree genetics defines fungal partner communities that may confer drought tolerance. Proceedings of the National Academy of Sciences. 2017;114(42):11169–11174. doi: 10.1073/pnas.1704022114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girlanda M, Selosse MA, Cafasso D, Brilli F, Delfine S, Fabbian R, Perotto S. Inefficient photosynthesis in the Mediterranean orchid Limodorum abortivum is mirrored by specific association to ectomycorrhizal Russulaceae. Molecular Ecology. 2006;15(2):491–504. doi: 10.1111/j.1365-294X.2005.02770.x. [DOI] [PubMed] [Google Scholar]
- Glassman SI, Peay KG, Talbot JM, Smith DP, Chung JA, Taylor JW, Bruns TD. A continental view of pine-associated ectomycorrhizal fungal spore banks: A quiescent functional guild with a strong biogeographic pattern. New Phytologist. 2015;205(4):1619–1631. doi: 10.1111/nph.13240. [DOI] [PubMed] [Google Scholar]
- Grayston S, Vaughan D, Jones D. Rhizosphere carbon flow in trees, in comparison with annual plants: The importance of root exudation and its impact on microbial activity and nutrient availability. Applied Soil Ecology. 1997;5(1):29–56. doi: 10.1016/S0929-1393(96)00126-6. [DOI] [Google Scholar]
- Gryta H, Debaud JC, Effosse A, Gay G, Marmeisse R. Fine-scale structure of populations of the ectomycorrhizal fungus Hebeloma cylindrosporum in coastal sand dune forest ecosystems. Molecular Ecology. 1997;6(4):353–364. doi: 10.1046/j.1365-294X.1997.00200.x. [DOI] [Google Scholar]
- Hazard C, Johnson D. Does genotypic and species diversity of mycorrhizal plants and fungi affect ecosystem function? New Phytologist. 2018;220(4):1122–1128. doi: 10.1111/nph.15010. [DOI] [PubMed] [Google Scholar]
- Högberg P, Read DJ. Towards a more plant physiological perspective on soil ecology. Trends in Ecology & Evolution. 2006;21(10):548–554. doi: 10.1016/j.tree.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Horton TR, Bruns TD. Multiple-host fungi are the most frequent and abundant ectomycorrhizal types in a mixed stand of Douglas fir (Pseudotsuga menziesii) and bishop pine (Pinus muricata) New Phytologist. 1998;139(2):331–339. doi: 10.1046/j.1469-8137.1998.00185.x. [DOI] [Google Scholar]
- Hyvonen R, Agren GI, Linder S, Persson T, Cotrufo MF, Ekblad A, Wallin G. The likely impact of elevated [CO2], nitrogen deposition, increased temperature and management on carbon sequestration in temperate and boreal forest ecosystems: A literature review. New Phytologist. 2007;173(3):463–480. doi: 10.1111/j.1469-8137.2007.01967.x. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Wang W, Xie Q, Liu NA, Liu L, Wang D, Wang E. Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science. 2017;356(6343):1172–1175. doi: 10.1126/science.aam9970. [DOI] [PubMed] [Google Scholar]
- Johnson NC, Wilson GW, Bowker MA, Wilson JA, Miller RM. Resource limitation is a driver of local adaptation in mycorrhizal symbioses. Proceedings of the National Academy of Sciences. 2010;107(5):2093–2098. doi: 10.1073/pnas.0906710107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy P, Izzo A, Bruns T. There is high potential for the formation of common mycorrhizal networks between understorey and canopy trees in a mixed evergreen forest. Journal of Ecology. 2003;91(6):1071–1080. doi: 10.1046/j.1365-2745.2003.00829.x. [DOI] [Google Scholar]
- Kernaghan G, Widden P, Bergeron Y, Légaré S, Paré D. Biotic and abiotic factors affecting ectomycorrhizal diversity in boreal mixed-woods. Oikos. 2003;102(3):497–504. doi: 10.1034/j.1600-0706.2003.12415.x. [DOI] [Google Scholar]
- Klein T, Bader MKF, Leuzinger S, Mildner M, Schleppi P, Siegwolf RT, Körner C. Growth and carbon relations of mature Picea abies trees under 5 years of free-air CO2 enrichment. Journal of Ecology. 2016;104(6):1720–1733. doi: 10.1111/1365-2745.12621. [DOI] [Google Scholar]
- Klein T, Siegwolf RT, Körner C. Belowground carbon trade among tall trees in a temperate forest. Science. 2016;352(6283):342–344. doi: 10.1126/science.aad6188. [DOI] [PubMed] [Google Scholar]
- Lang C, Seven J, Polle A. Host preferences and differential contributions of deciduous tree species shape mycorrhizal species richness in a mixed Central European forest. Mycorrhiza. 2011;21(4):297–308. doi: 10.1007/s00572-010-0338-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofgren L, Nguyen NH, Kennedy PG. Ectomycorrhizal host specificity in a changing world: Can legacy effects explain anomalous current associations? New Phytologist. 2018;220(4):1273–1284. doi: 10.1111/nph.15008. [DOI] [PubMed] [Google Scholar]
- Looney BP, Ryberg M, Hampe F, Sanchez-Garcia M, Matheny PB. Into and out of the tropics: Global diversification patterns in a hyperdiverse clade of ectomycorrhizal fungi. Molecular Ecology. 2016;25(2):630–647. doi: 10.1111/mec.13506. [DOI] [PubMed] [Google Scholar]
- Lovelock CE, Andersen K, Morton JB. Arbuscular mycorrhizal communities in tropical forests are affected by host tree species and environment. Oecologia. 2003;135(2):268–279. doi: 10.1007/s00442-002-1166-3. [DOI] [PubMed] [Google Scholar]
- Martin F, Kohler A, Murat C, Veneault-Fourrey C, Hibbett DS. Unearthing the roots of ectomycorrhizal symbioses. Nature Reviews Microbiology. 2016;14(12):760–773. doi: 10.1038/nrmicro.2016.149. [DOI] [PubMed] [Google Scholar]
- McGuire KL, Allison SD, Fierer N, Treseder KK. Ectomycorrhizal-dominated boreal and tropical forests have distinct fungal communities, but analogous spatial patterns across soil horizons. PLoS ONE. 2013;8(7):e68278. doi: 10.1371/journal.pone.0068278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildner M, Bader M-K-F, Leuzinger S, Siegwolf RT, Körner C. Long-term 13C labeling provides evidence for temporal and spatial carbon allocation patterns in mature Picea abies. Oecologia. 2014;175(3):747–762. doi: 10.1007/s00442-014-. [DOI] [PubMed] [Google Scholar]
- Molina R, Massicotte H, Trappe JM. Specificity phenomena in mycorrhizal symbioses: community-ecological consequences and practical implications. In: Allen MF, editor. Mycorrhizal functioning: An integrative plant-fungal process. New York, NY: Chapman and Hall; 1992. pp. 357–423. [Google Scholar]
- Mujic AB, Durall DM, Spatafora JW, Kennedy PG. Competitive avoidance not edaphic specialization drives vertical niche partitioning among sister species of ectomycorrhizal fungi. New Phytologist. 2016;209(3):1174–1183. doi: 10.1111/nph.13677. [DOI] [PubMed] [Google Scholar]
- Nara K. Ectomycorrhizal networks and seedling establishment during early primary succession. New Phytologist. 2006;169(1):169–178. doi: 10.1111/j.1469-8137.2005.01545.x. [DOI] [PubMed] [Google Scholar]
- Nguyen NH, Williams LJ, Vincent JB, Stefanski A, Cavender-Bares J, Messier C, Kennedy PG. Ectomycorrhizal fungal diversity and saprotrophic fungal diversity are linked to different tree community attributes in a field-based tree experiment. Molecular Ecology. 2016;25(16):4032–4046. doi: 10.1111/mec.13719. [DOI] [PubMed] [Google Scholar]
- Oberwinkler F, Riess K, Bauer R, Selosse M-A, Weiss M, Garnica S, Zuccaro A. Enigmatic sebacinales. Mycological Progress. 2013;12(1):1–27. doi: 10.1007/s11557-012-0880-4. [DOI] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara R, Wagner H. Package ‘vegan’. 2013;2(9) [Google Scholar]
- Ouimette A, Guo D, Hobbie E, Gu J. Insights into root growth, function, and mycorrhizal abundance from chemical and isotopic data across root orders. Plant and Soil. 2013;367(1–2):313–326. doi: 10.1007/s11104-012-1464-4. [DOI] [Google Scholar]
- Peay KG, Kennedy PG, Bruns TD. Rethinking ectomycorrhizal succession: Are root density and hyphal exploration types drivers of spatial and temporal zonation? Fungal Ecology. 2011;4(3):233–240. doi: 10.1016/j.funeco.2010.09.010. [DOI] [Google Scholar]
- Peay KG, Russo SE, McGuire KL, Lim Z, Chan JP, Tan S, Davies SJ. Lack of host specificity leads to independent assortment of dipterocarps and ectomycorrhizal fungi across a soil fertility gradient. Ecology Letters. 2015;18(8):807–816. doi: 10.1111/ele.12459. [DOI] [PubMed] [Google Scholar]
- Pena R, Lang C, Lohaus G, Boch S, Schall P, Schöning I, Polle A. Phylogenetic and functional traits of ectomycorrhizal assemblages in top soil from different biogeographic regions and forest types. Mycorrhiza. 2017;27(3):233–245. doi: 10.1007/s00572-016-0742-z. [DOI] [PubMed] [Google Scholar]
- Perry DA, Bell T, Amaranthus M. In: Mycorrhizal fungi in mixed-species forests and other tales of positive feedback, redundancy and stability. Cannell MGR, Malcolm DC, Robertson PA, editors. Oxford, UK: Special publications of the British Ecological Society; 1992. pp. 151–179. [Google Scholar]
- Pickles BJ, Genney DR, Potts JM, Lennon JJ, Anderson IC, Alexander IJ. Spatial and temporal ecology of Scots pine ectomycorrhizas. New Phytologist. 2010;186(3):755–768. doi: 10.1111/j.1469-8137.2010.03204.x. [DOI] [PubMed] [Google Scholar]
- Pickles BJ, Simard SW. Mycorrhizal networks and forest resilience to drought. In: Johnson NC, Gehring C, Jansa J, editors. Mycorrhizal Mediation of Soil - Fertility, Structure, and Carbon Storage. Amsterdam: Elsevier; 2017. pp. 319–339. [Google Scholar]
- Pickles BJ, Wilhelm R, Asay AK, Hahn AS, Simard SW, Mohn WW. Transfer of 13C between paired Douglas-fir seedlings reveals plant kinship effects and uptake of exudates by ectomycorrhizas. New Phytologist. 2017;214(1):400–411. doi: 10.1111/nph.14325. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2018. Retrieved from https://www.R-project.org. [Google Scholar]
- R Core Team. RStudio: Integrated development environment for R (Version 1.2.1335) Boston, MA: RStudio, Inc; 2018. Retrieved from http://www.rstudio.com/100. [Google Scholar]
- Rosenstock NP, Berner C, Smits MM, Kram P, Wallander H. The role of phosphorus, magnesium and potassium availability in soil fungal exploration of mineral nutrient sources in Norway spruce forests. New Phytologist. 2016;211(2):542–553. doi: 10.1111/nph.13928. [DOI] [PubMed] [Google Scholar]
- Selosse M-A, Richard F, He X, Simard SW. Mycorrhizal networks: Desliaisons dangereuses? Trends in Ecology & Evolution. 2006;21(11):621–628. doi: 10.1016/j.tree.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Selosse M-A, Weiss M, Jany JL, Tillier A. Communities and populations of sebacinoid basidiomycetes associated with the achlorophyllous orchid Neottia nidus-avis (L.) LCM Rich. and neighbouring tree ectomycorrhizae. Molecular Ecology. 2002;11(9):1831–1844. doi: 10.1046/j.1365-294X.2002.01553.x. [DOI] [PubMed] [Google Scholar]
- Shachar-Hill Y, Pfeffer PE, Douds D, Osman SF, Doner LW, Ratcliffe RG. Partitioning of intermediary carbon metabolism in vesicular-arbuscular mycorrhizal leek. Plant Physiology. 1995;108(1):7–15. doi: 10.1104/pp.108.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard SW, Beiler KJ, Bingham MA, Deslippe JR, Philip LJ, Teste FP. Mycorrhizal networks: Mechanisms, ecology and modelling. Fungal Biology Reviews. 2012;26(1):39–60. doi: 10.1016/j.fbr.2012.01.001. [DOI] [Google Scholar]
- Simard SW, Perry DA, Jones MD, Myrold DD, Durall DM, Molina R. Net transfer of carbon between ectomycorrhizal tree species in the field. Nature. 1997;388(6642):579. doi: 10.1038/41557. [DOI] [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17(5):1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences. 2004;101(30):11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taudiere A, Munoz F, Lesne A, Monnet A-C, Bellanger J-M, Selosse M-A, Richard F. Beyond ectomycorrhizal bipartite networks: Projected networks demonstrate contrasted patterns between early- and late-successional plants in Corsica. Frontiers in Plant Science. 2015;6:881. doi: 10.3389/fpls.2015.00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedersoo L, Kõljalg U, Hallenberg N, Larsson KH. Fine scale distribution of ectomycorrhizal fungi and roots across substrate layers including coarse woody debris in a mixed forest. New Phytologist. 2003;159(1):153–165. doi: 10.1046/j.1469-8137.2003.00792.x. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, May TW, Smith ME. Ectomycorrhizal lifestyle in fungi: global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza. 2010;20(4):217–263. doi: 10.1007/s00572-009-0274-x. [DOI] [PubMed] [Google Scholar]
- Terrer C, Vicca S, Stocker BD, Hungate BA, Phillips RP, Reich PB, Prentice IC. Ecosystem responses to elevated CO2 governed by plant-soil interactions and the cost of nitrogen acquisition. New Phytologist. 2018;217(2):507–522. doi: 10.1111/nph.14872. [DOI] [PubMed] [Google Scholar]
- Teste FP, Simard SW, Durall DM, Guy RD, Berch SM. Net carbon transfer between Pseudotsuga menziesii var. glauca seedlings in the field is influenced by soil disturbance. Journal of Ecology. 2010;98(2):429–439. doi: 10.1111/j.1365-2745.2009.01624.x. [DOI] [Google Scholar]
- Toju H, Guimaraes PR, Jr, Olesen JM, Thompson JN. Below-ground plant-fungus network topology is not congruent with above-ground plant-animal network topology. Science Advances. 2015;1(9):e1500291. doi: 10.1126/sciadv.1500291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban A, Weiss M, Bauer R. Ectomycorrhizas involving sebacinoid mycobionts. Mycological Research. 2003;107(Pt 1):3–14. doi: 10.1017/S0953756202007116. [DOI] [PubMed] [Google Scholar]
- Van Der Heijden MG, Horton TR. Socialism in soil? The importance of mycorrhizal fungal networks for facilitation in natural ecosystems. Journal of Ecology. 2009;97(6):1139–1150. doi: 10.1111/j.1365-2745.2009.01570.x. [DOI] [Google Scholar]
- van der Linde S, Suz LM, Orme CDL, Cox F, Andreae H, Asi E, Bidartondo MI. Environment and host as large-scale controls of ectomycorrhizal fungi. Nature. 2018;558(7709):243–248. doi: 10.1038/s41586-018-0189-9. [DOI] [PubMed] [Google Scholar]
- Wagg C, Bender SF, Widmer F, van der Heijden MG. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proceedings of the National Academy of Sciences. 2014;111(14):5266–5270. doi: 10.1073/pnas.1320054111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallander H, Göransson H, Rosengren U. Production, standing biomass and natural abundance of 15N and 13C in ectomycorrhizal mycelia collected at different soil depths in two forest types. Oecologia. 2004;139(1):89–97. doi: 10.1007/s00442-003-1477-z. [DOI] [PubMed] [Google Scholar]
- Weiss M, Waller F, Zuccaro A, Selosse MA. Sebacinales– one thousand and one interactions with land plants. New Phytologist. 2016;211(1):20–40. doi: 10.1111/nph.13977. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications. 1990;18(1):315–322. [Google Scholar]
- Wolfe BE, Pringle A. Geographically structured host specificity is caused by the range expansions and host shifts of a symbiotic fungus. The ISME Journal. 2012;6(4):745–755. doi: 10.1038/ismej.2011.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Nara K, Hogetsu T. Competition between ectomycorrhizal fungi colonizing Pinus densiflora. Mycorrhiza. 1999;9(3):151–159. doi: 10.1007/s005720050300. [DOI] [Google Scholar]
- Yagame T, Orihara T, Selosse MA, Yamato M, Iwase K. Mixotrophy of Platanthera minor, an orchid associated with ectomycorrhiza-forming Ceratobasidiaceae fungi. New Phytologist. 2012;193(1):178–187. doi: 10.1111/j.1469-8137.2011.03896.x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ziegler W, Han X, Trumbore S, Hartmann H. Plant carbon limitation does not reduce nitrogen transfer from arbuscular mycorrhizal fungi to Plantago lanceolata. Plant and Soil. 2015;396(1–2):369–380. doi: 10.1007/s11104-015-2599-x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.