Abstract
A group of meiosis-specific proteins, namely budding yeast Spo13, fission yeast Moal, mouse MEIKIN and Drosophila Mtrm, which we call MOKIRs, (meiosis I kinase regulators) are essential for meiotic chromosome segregation. MOKIRs bear no obvious sequence similarity, and yet appear to play functionally conserved roles in regulating meiotic kinases. Meiosis generates haploid gametes from diploid cells through two rounds of chromosome segregation without intervening DNA replication. During meiosis I, homologous chromosomes segregate, reducing chromosome number (reductional division). Conversely, in meiosis II, sister chromatids segregate, similar to mitosis (equational division). Meiosis requires that the cell cycle be re-wired to change both the orientation of kinetochore attachment to microtubules and the timing of sister chromatid cohesion loss. Recent evidence shows that MOKIRs achieve these changes through the spatial and temporal control of key kinases. Here, we review the known roles of MOKIRs and discuss their possible mechanisms of action.
Keywords: MOKIRs, meiosis, Spo13, Moa1, Matrimony, MEIKIN, Polo kinase
1. Introduction
Eukaryotic cells proliferate and divide by one of two different modes. The mitotic cell cycle, which is used to generate the large majority of cell types, involves the separation of a single mother cell into two daughter cells that contain the same chromosomal content as the mothers. To achieve this, mitotic cells first replicate their DNA before segregating sister chromatids to opposite poles. During gametogenesis (see Glossary), however, cells undergo two successive cell divisions without an intervening round of DNA replication, leading to the halving of the chromosomal number. This process is called meiosis.
Glossary.
Chiasmata – the cytological manifestation of reciprocal crossover between DNA molecules from homologous chromosomes. Chiasmata result from homologous recombination and serve to physically link homologs together.
Cohesin – a ring-shaped protein complex formed by the SMC1, SMC3 and a kleisin subunit (such as Rec8), together with accessory subunits. It is thought to topologicially embrace sister chromatids.
DNA replication origins – sites on the DNA where replication is initiated.
Gametogenesis – the process by which a diploid mother cell undergoes meiosis to form haploid male and female germ cells.
Haploinsufficiency – a situation in which a heterozygous loss-of-function mutation of a gene is insufficient to provide normal cellular function.
Holliday junction – a temporary structure formed from two homologous DNA molecules during genetic recombination, which may serve the exchange of genetic information between the two molecules.
Holocentric chromosomes – chromosomes that lack a distinct centromere and thus have multiple microtubule attachment sites along the length of the chromosome.
Kinetochore – a proteinaceous structure that assembles on centromeres and serves as attachment site for microtubules during mitotic and meiotic chromosome segregation
Synaptonemal complex – a proteinaceous structure connecting two homologous chromosomes during meiotic prophase. It serves to facilitate the events of homologous recombination.
Due to the fundamentally different outcomes of mitotic and meiotic cell division, the chromosome segregation machinery has to be adapted in a multitude of ways to facilitate the desired outcome of the cellular division. In the vast majority of organisms studied, meiosis I is considered a specialized division because homologous chromosomes segregate (reductional division). Conversely, mitosis and meiosis II are equational divisions, meaning that sister chromatids are separated to opposite poles. A notable exception to this rule is plants with holocentric chromosomes (see Glossary), in which the typical sequence of reductional and equational division is inverted.[1]
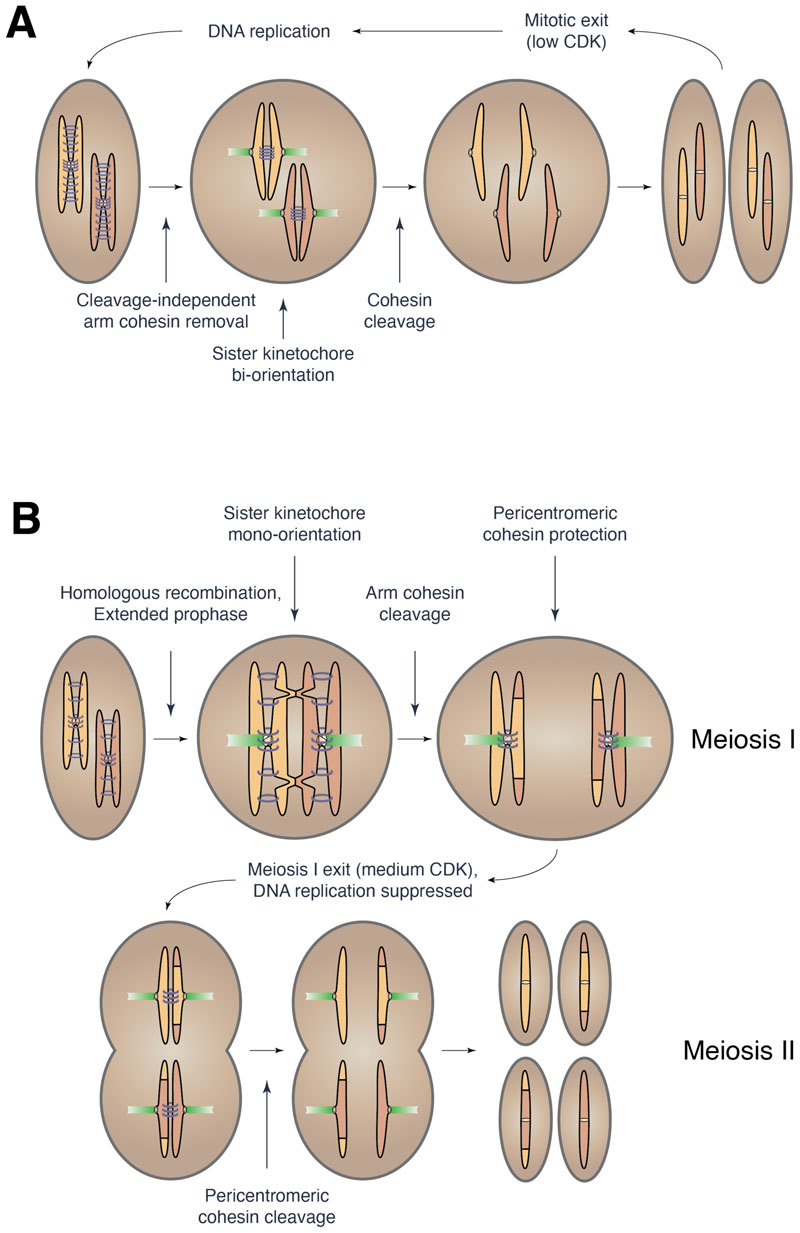
The mitotic and meiotic cell cycles and chromosome segregation machineries differ in five key aspects (Figure 1), the molecular basis of which is largely still elusive.[2] Firstly, in prophase I, homologous recombination creates linkages between homologous chromosomes and these connections are the basis for their successful segregation in meiosis I. Meiotic recombination is a complex process that involves the creation of double strand breaks on chromosomes, the physical linking of homologous chromosomes within a structure called the synaptonemal complex (see Glossary), and the formation and resolution of Holliday junctions (see Glossary). Secondly, the events of meiotic recombination require that meiotic prophase be extended compared to mitotic prophase.[3] This prevents the premature expression of proteins that promote chromosome segregation and thereby ensures that segregation is only initiated once recombination is completed. Thirdly, to segregate sister chromatids in mitosis and meiosis II, sister kinetochores (see Glossary), the proteinaceous structures mediating attachment of chromosomes to microtubules, need to face opposite poles (bi-orientation). In contrast, sister kinetochores must face towards the same pole (mono-orientation) to segregate to the same pole in meiosis I. Fourthly, the cleavage dynamics of cohesin (see Glossary), the protein complex holding sister chromatids together, are altered in meiosis. Cohesin is cleaved on chromosome arms to allow resolution of chiasmata (see Glossary) during anaphase I, which triggers the segregation of homologous chromosomes. However, cohesin in the regions surrounding centromeres (called pericentromeres) is retained until meiosis II to allow faithful segregation of sister chromatids in anaphase II. Finally, the meiotic cell cycle requires the fine-tuning of the activity of cell cycle kinases. Whereas mitotic cells eliminate the activity of cyclin-dependent kinases (CDKs) in anaphase I to promote exit from mitosis and allow re-licensing of DNA replication origins (see Glossary), meiotic cells need to decrease CDK activity enough to drive exit from meiosis I while maintaining some CDK activity to promote an additional round of chromosome segregation and suppress DNA replication.
Figure 1. Comparison of mitotic and meiotic cell cycles.
Mitosis and meiosis need to be specifically adapted to generate two diploid daughter cells or four haploid daughter cells from a single diploid mother cell, respectively. In the mitotic cell cycle (A), replicated sister chromatids attach to opposite spindle poles (bi-orientation). In mammals and Drosophila, cohesin, the protein complex that holds sister chromatids together, is removed from chromosome arms in prophase but retained in the pericentromere, where it resists the microtubule pulling forces to prevent sister chromatid segregation. In anaphase, cohesin is cleaved and sister chromatids move to opposite poles. This is followed by exit from mitosis and re-start of the cell cycle. In meiosis (B), prophase is extended to allow sufficient time for homologous recombination to occur. Recombination generates chiasmata, which link homologous chromosomes. Sister kinetochores are mono-oriented, meaning they face the same pole. When homologous chromosomes become attached by microtubules, chiasmata resist spindle forces and ensure faithful segregation of homologs. Upon anaphase I onset, cohesin is cleaved on chromosome arms to allow the resolution of chiasmata and homolog segregation, but retained in the pericentromere. In the transition from meiosis I to meiosis II, DNA replication is suppressed. In meiosis II, pericentromeric cohesin is cleaved upon sister chromatid bi-orientation, and the meiotic exit programme is consequently initiated.
Both the mitotic and the meiotic cell cycle are driven by fluctuations in the activity of a number of kinases. The transition through interphase and cell division is mainly facilitated by cyclin-dependent kinases (CDKs), which associate with their cyclin co-factors to achieve temporal and spatial specificity for distinct cellular targets. In addition, mitosis and meiosis utilize a select number of kinases to promote cell cycle progression and accurate chromosome segregation (Table 1).[4] These kinases have multifaceted functions and direct chromosome segregation in a multitude of ways, implying that their activity needs to be strictly regulated. Evidence from a range of model organisms indicates that a group of meiosis-specific proteins that are seemingly unrelated in protein sequence control meiotic kinases to execute key features of the meiotic programme. These proteins include Spo13 (budding yeast), Moa1 (fission yeast), Mtrm (Matrimony; Drosophila melanogaster) and MEIKIN (mouse). Here, we review the known functions of these proteins, which we collectively refer to as Meiosis One Kinase Regulators (MOKIRs), and discuss the possibility that they are functionally conserved regulators that orchestrate the action of kinases in meiosis to ensure the accurate segregation of chromosomes.
Table 1. Overview of key meiotic proteins involved in cohesin protection and kinetochore mono-orientation in various model organisms.
| Protein | Budding yeast Saccharomyces cerevisiae | Fission yeast Schizosaccharomyces pombe | Drosophila melanogaster | Mouse |
|---|---|---|---|---|
| Casein kinase 1 | Hrr25 (equivalent to CK1δ isoform) | Cki1 | DoubletimeDbt (CK1ε homolog) | CSNK1 |
| Dbf4-dependent kinase | Cdc7 (with regulatory subunit Dbf4) | Hsk1 (with Dfp1) | Cdc7 (with Chiffon) | CDC7 (with DBF4) |
| Polo kinase | Cdc5 | Plo1 | Polo | PLK1 |
| PP2A B’ subunit | Rts1 | Par1 | Wdb and Wrd | B56 |
| Shugoshin (meiotic cohesin protector) | Sgo1 | Sgo1 | MEI-S332 | Sgo2 |
2. MOKIRs show poor sequence conservation
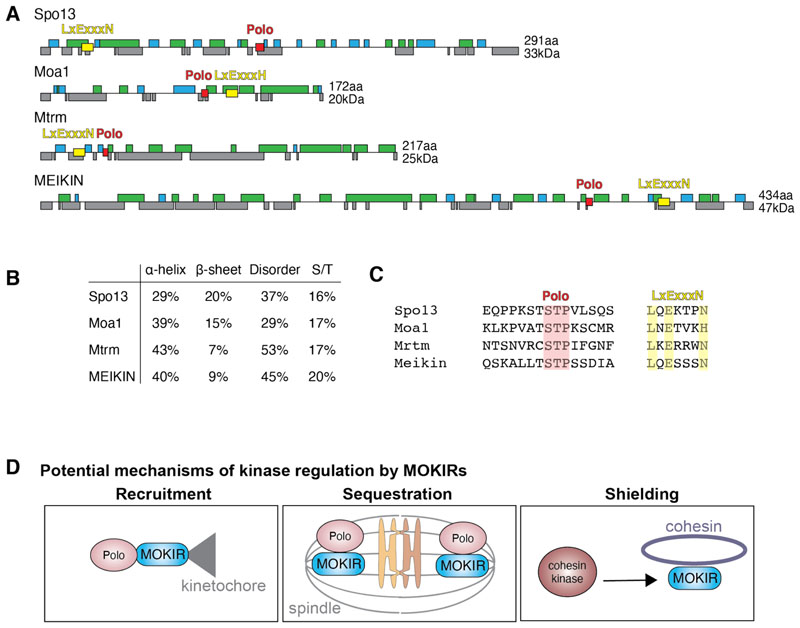
In terms of primary protein sequence, the conservation between the MOKIRs is extremely poor. Accordingly, sequence alignment of the four proteins with Clustal Omega[5] does not identify any conserved regions or domains (data not shown).Nonetheless, these proteins share a number of common features (Figure 2A-C).Generally, they are small in size, ranging from 172 amino acids/20 kDa for Moa1MOKIR to 434 amino acids/47kDa for EIKINMOKIR. Secondary structure analysis with Phyre2[6] shows that they are characterized by large stretches of disordered regions (Figure 2A). The only notable exception is a sterile alpha motif (SAM)-domain found in the C-terminus of MtrmMOKIR, which is required to stabilize its binding to Polo[7,8] (see below). We speculate that a possible explanation for the lack of a clear 3D structure is that the shape of these proteins is largely determined by protein interactions, which could be facilitated by post-translational modifications, as is frequently observed for intrinsically disordered proteins.[9] Indeed, all four proteins carry a large number of serine and threonine residues (Figure 2B), which are targets for phosphorylation. Consistently, both Moa1MOKIR[10] and Spo13MOKIR[11,12] have been found to be phosphorylated.
Figure 2. Structural comparison of MOKIRs and potential mechanisms of kinase regulation.
MOKIRs are largely disordered proteins that show very little sequence conservation amongst each other. (A) We used Phyre2 to model their secondary structure. Green and blue bars represent predicted α-helical domains and β-sheets, respectively. Grey bars indicate disordered regions. The putative Polo-binding site in each protein is highlighted in red and the putative LEN motif for degradation by the APC/C is highlighted in yellow. (B) Table comparing the properties of MOKIRs, as suggested by modelling with Phyre2. Of note is the larger than average content of serine and threonine residues, allowing for large-scale post-translational phosphorylation of these proteins. (C) Alignment of Polo binding site and LEN motifs in MOKIRs. (D) Regulation of kinases by MOKIRs is a common theme and several mechanisms have been proposed. In mouse, budding and fission yeasts, MOKIRs recruit Polo kinase to kinetochores, where they have roles in mono-orientation and, in some cases, cohesin protection. In Drosophila, MtrmMOKIR appears to inhibit active Polo kinase by sequestration to the spindle. In budding yeast, Spo13MOKIR appears to prevent ectopic kinase activity enhancing cohesin cleavage, and localizes to chromosomes in a cohesin-dependent manner. We speculate that Spo13MOKIR may shield cohesin from its kinases, though there is no direct evidence for this. Note that MOKIR interaction with kinases may be inhibitory or activating at specific locations and the three mechanisms above are not mutually exclusive for individual MOKIRs.
The most notable example of the phosphorylation-induced interaction of MOKIRs with their partners is the presence of a Polo-box domain (PBD) binding region (Figure 2C) that requires prior phosphorylation, typically by CDKs, to interact with Polo kinase.[13,14] Indeed, Polo binding through the PBD-binding domain of MOKIRs has been demonstrated in mouse, Drosophila, fission and budding yeasts, and abrogation of the Polo-MOKIR interaction is detrimental for chromosome segregation in all cases.[11,12,15,16] In budding and fission yeasts, and mouse, MOKIRs recruit Polo kinase to kinetochores through the PBD binding region, whereas in Drosophila, this region is important for sequestration of Polo on the spindle (Figure 2D).[11,12,15,16]
Additionally, MOKIRs share a similar degradation motif (Figure 2C). This LxExxxN (short: LEN) motif has been shown to be required for the degradation of both Spo13MOKIR[17] and MtrmMOKIR[18] by the anaphase promoting complex/cyclosome (APC/C) and ensures the degradation of these proteins in anaphase I[17] and at the oocyte-to-embryo transition,[18] respectively. Although failure to degrade Spo13MOKIRin a timely manner does not appear to affect the outcome of meiosis,[17] excess MtrmMOKIR causes developmental defects in embryos.[18] MEIKINmokir also possesses a LEN motif, whereas Moa1MOKIR carries a LxExxxH sequence; however, whether these motifs direct APC/C-mediated degradation of MEIKINMOKIR and Moa1MOKIR remains to be determined.
A recent analysis of MtrmMOKIR orthologs in Drosophila species has provided evidence that MOKIR functionality relies on only short segments of sequence conservation. MtrmMOKIR protein sequences are highly diverse: in the most extreme example, MtrmMOKIR of D. melanogaster and D. grimshawi only share 38.2% overall sequence identity.[8] Even the SAM-domain, which stabilizes the interaction of MtrmMOKIR with Polo,[7] and the SAM proximal region, show only intermediate levels of conservation between these species (46.9% and 57.7%, respectively). Apart from these two regions, and the short PBD-binding region, sequence conservation is very poor, suggesting rapid evolutionary divergence between MtrmMOKIR proteins in Drosophila. Despite this, expression of D. grimshawi MtrmMOKIR rescues the meiotic defects in D. melanogaster carrying a heterozygous mtrm mutation (mtrm is haploinsufficient (see Glossary)).[8] Similarly, artificial targeting of Spo13MOKIR to fission yeast kinetochores rescues the mono-orientation defect observed in moalΔcells.[15] Thus, despite their strong evolutionary divergence at the sequence level, it is possible that MOKIRs have retained similar conserved molecular functions.
3. MOKIRs regulate shugoshin and cohesin kinases to promote cohesin protection
3.1. Cohesin protection requires that cohesin-directed kinase and phosphatase activity is balanced
All MOKIRs appear to ensure the step-wise loss of cohesin, a defining feature of the meiotic chromosome segregation pattern (Figure 3). Cohesin is a ring-shaped complex that topologically links the two newly-duplicated sister chromatids as they are replicated to provide the cohesion that holds them together until the time of their segregation. Cohesin additionally links distant loci of the DNA molecule to provide a structural organisation that impacts on multiple chromosomal processes, including meiotic recombination.[19,20] Although a non-proteolytic cohesin removal way requiring the Wapl protein has been identified,[12–24] the universal trigger for chromosome segregation in both mitosis and meiosis is the proteolytic cleavage of the kleisin subunit of cohesin by separase.[19] During meiosis in the majority of organisms, sister chromatids are held together by cohesin complexes containing the meiosis-specific kleisin Rec8, and it is Rec8 that must be cleaved to trigger chromosome segregation.[25] Rec8-cohesin complexes are cleaved by separase in two steps. First, cohesin on chromosome arms, but not that at pericentromeres, is cleaved at anaphase I onset, resolving chiasmata and triggering segregation of homologous chromosomes. Second, at anaphase II, cohesin persisting at pericentromeres is cleaved to trigger the segregation of sister chromatids.
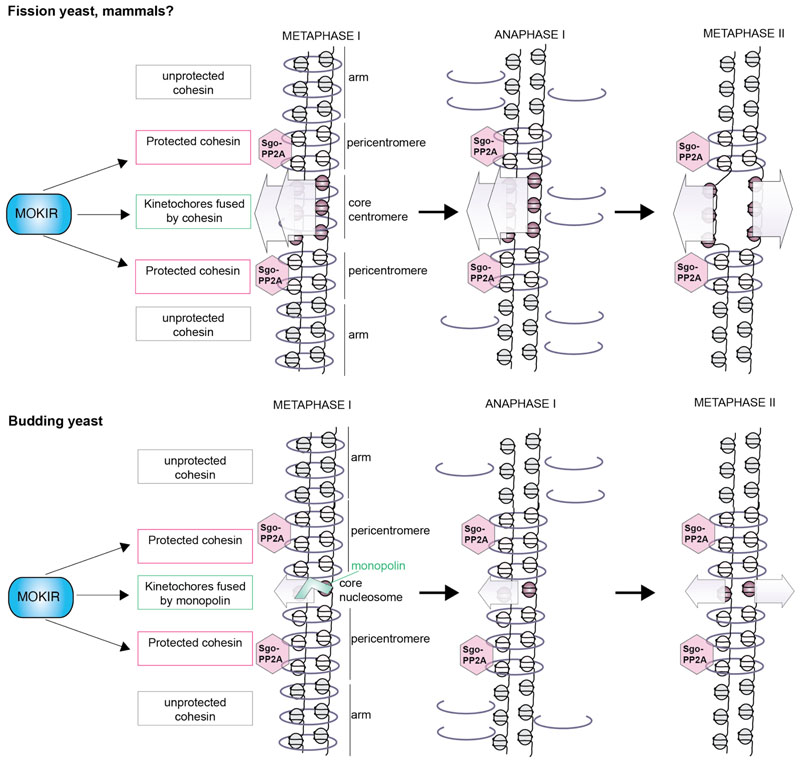
Figure 3. Model for distinct regulation of centromeric domains by MOKIRs.
In fission yeast, the centromeric region is organized such that the core centromere, where the kinetochore assembles, is flanked by surrounding heterochromatin that is cohesin-rich. During meiosis I, Rec8-cohesin is established within the core centromere and builds cohesive links in a Moa1MOKIR-dependent manner. This directs sister kinetochores to attach to microtubules from the same pole. Sister kinetochore mono-orientation relies on recruitment of Plo1Polo kinase to kinetochores; however, the relevant substrates are not known. Meanwhile, Moa1MOKIR promotes the maintenance of cohesin within the pericentromere until meiosis II, potentially in part by ensuring the retention of shugoshin-PP2A within the pericentromeric domain, where cohesin is protected. At anaphase onset, shugoshin protects a specific centromeric pool of cohesin from cleavage, while cohesin cleavage on arms triggers homologous chromosome segregation. Cohesin cleavage in the core centromere has also been proposed to reverse mono-orientation, so that sister kinetochores bi-orient in meiosis II. In budding yeast, the process is similar except that monopolin, rather than cohesin, directs sister kinetochore mono-orientation. In Drosophila, ectopic activity of Polo in mtrm mutants promotes cohesion loss but it is not yet known which domains of cohesin are regulated by MtrmMOKIR and what its impact on sister kinetochore mono-orientation is.
Cohesin cleavage requires the prior phosphorylation of its Rec8 kleisin subunit.[26] Full Rec8 phosphorylation relies on PLK1Polo in mice,[27] Cki1 in fission yeast[28] and casein kinase 1δ(CK1δ), Dbf4-dependent kinase (DDK) and Cdc5Polo in budding yeast,[26,29] although the contribution of Cdc5Polo to cleavage is under debate. [26,29,30] However, our recent data has shown that tethering of Cdc5Polo directly to Rec8 in meiosis can promote loss of cohesion between sister chromatids at anaphase I, suggesting that Cdc5Polo contributes to cohesin removal,[12] although whether it does so through promoting separase-dependent Rec8 cleavage or via the cleavageindependent, Wapl-dependent cohesin removal pathway is unknown. In Drosophila, meiotic cohesin does not contain a Rec8-like subunit. Instead, a meiosis-specific complex containing SMC1 and SMC3 together with the SOLO and SUNN proteins localizes to centromeres, where it persists until metaphase II.[31–33] However, little is known about the phosphorylation state of SOLO-SUNN cohesin in meiosis, and how its removal from chromosomes is regulated.
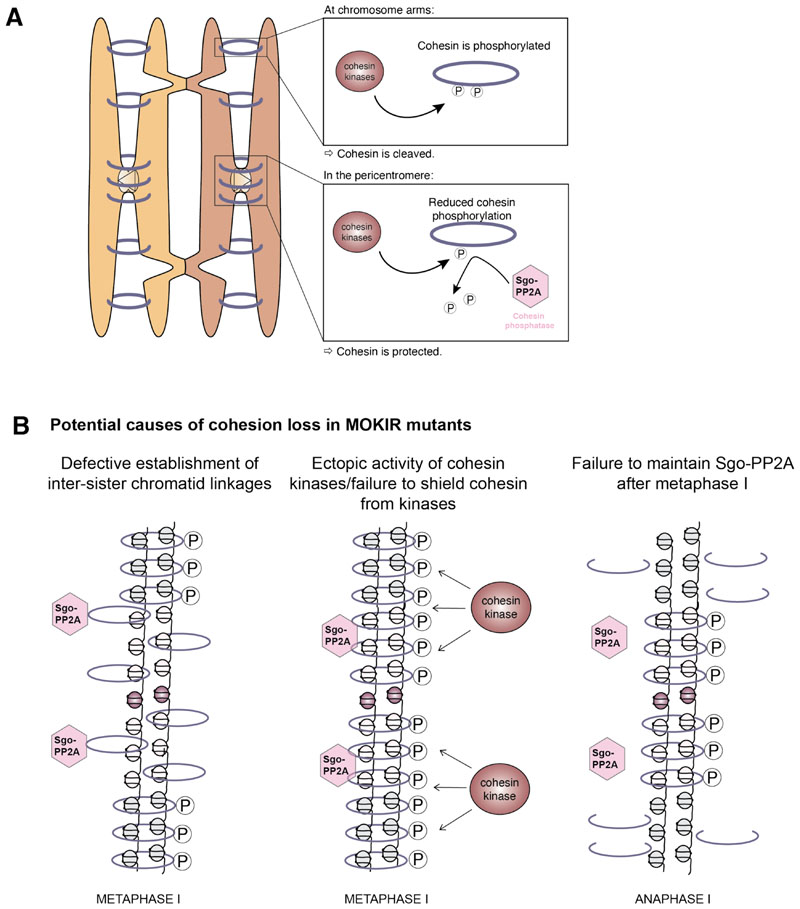
Protection of pericentromeric cohesin is brought about by the pericentromeric adaptor protein Shugoshin (Table 1). Shugoshin specifically localizes to pericentromeres by binding histone H2A that has been phosphorylated by Bub1.[34–39] In mitosis, phosphorylation of human Sgo1 by CDKs promotes its binding to cohesin, and this is required to protect pericentromeric cohesin from its non-proteolytic removal by Wapl.[39,40] Shugoshins also bind to and localize with cohesin during meiosis.[12,41] Whether shugoshins also counteract Wapl during meiosis is not clear, but they are known to protect Rec8 from separase-dependent cleavage in a multitude of organisms, thereby ensuring the maintenance of pericentromeric cohesion until meiosis n.[41–47] To achieve this, shugoshins recruit protein phosphatase 2A (PP2A; Table 1) to pericentromeres by binding of the PP2A B’ regulatory subunit.[48–51] There, PP2A is thought to dephosphorylate cohesin, thus preventing its cleavage by separase. Artificial tethering of PP2A to arm cohesin in fission yeast[51] or budding yeast[52] impairs cohesin cleavage on chromosome arms, highlighting the importance of restricting PP2A activity to the pericentromere. Thus, cohesin protection in the pericentromere requires the careful balancing of cohesin-directed kinase and phosphatase activity; disruption of this balance is likely to interfere with the maintenance of pericentromeric cohesin after metaphase I.
3.2. MOKIRs protect cohesin by regulating shugoshin and the activity of cohesin kinases
MOKIRs are required for the retention of centromeric cohesion in meiosis I in budding yeast, fission yeast, Drosophila and mouse, and may contribute in several different ways (Figure 4).[15,16,53–56] Initial reports of Spo13MOKIR’s involvement in this process suggested that it is required for the maintenance of Sgo1 at budding yeast pericentromeres during metaphase I.[36] Similarly, mouse Sgo2 localisation to pericentromeres is reportedly reduced in Meikin-/- mutants.[15] In fission yeast, however, Sgo1 levels are unaltered in moa1Δ cells.[56] It has been proposed that Moa1MOKIR- Plo1Polo regulates centromeric Bub1 localisation together with the spindle checkpoint kinase Mph1Mps1, since moalΔ mphlΔ double mutants show additive reductions in Bub1 de-localisation, and consequent loss of Sgo1 localization.[56] Still, the fact that moalΔ cells show meiotic cohesion defects despite correctly localising Sgo1 suggests that mechanisms other than Bub1 de-localisation drive cohesion loss in moalΔ cells.
Figure 4. Potential causes of cohesin loss in MOKIR mutants.
(A) Cohesin cleavage requires its prior phosphorylation by cohesin kinases. Although phosphorylation occurs along the length of the chromosome, it is counteracted in the pericentromere by PP2A, which is recruited by the action of the pericentromeric adaptor protein Shugoshin (Sgo1 in yeast). Reduced cohesin phosphorylation in the pericentromere prevents its cleavage by separase in anaphase I. (B) MOKIRs might assist Sgo1-mediated cohesin protection in a number of ways: first, they could ensure the building of inter-sister cohesion; second, they could shield cohesin from the activity of cohesin kinases; and third, MOKIRs may act to retain shugoshin-mediated protection until metaphase II.
Similar to fission yeast, our recent evidence in budding yeast shows that Spo13MOKIR-Cdc5Polo promotes cohesin protection, since artificial tethering of Cdc5Polo to kinetochores, but not to cohesin, mildly enhanced cohesin retention in spo13Δanaphase I cells, although this was not sufficient to provide sister chromatid cohesion.[12] Contrary to initial findings, it is now established that the cohesin protector Sgo1-PP2A is appropriately localized in metaphase I spo13A cells.[12,36,54] Instead, over-activity of the kinases (CK1δ and DDK) that phosphorylate cohesin to promote its cleavage may explain defective cohesion in the absence of Spo13MOKIR, because inhibition of either one of the redundant cohesin kinases CK1δ and DDK in spo13Δcells is sufficient to prevent sister chromatid segregation in anaphase I.[12] Similarly, depletion of Cdc5Polo prevents sister chromatid segregation in spo13Δ though the molecular reasons remain unclear.[12] Importantly, Spo13MOKIR binds all three cohesin kinases directly[11,12] or indirectly,[57] suggesting that it may restrict their activity through direct interactions. Although not required for establishing Sgo1 localization at pericentromeres in meiosis I, at least in budding or fission yeast, MOKIRs may be important to maintain Sgo1 localization: in addition to its function in phosphorylation, CK15 also promotes the permanent removal of Sgo1 upon anaphase I onset in spo13Δ cells.[12] Surprisingly, although artificial tethering of Sgo1 to cohesin reinstates centromeric cohesin in spo13Δ mutants, sister chromatids nevertheless segregate upon anaphase I onset.[12] This is analogous to findings in fission yeast, where loss of Moa1MOKIR interferes with cohesin’s ability to link sister chromatids in the core centromere for sister kinetochore monoorientation (see below).[58] Thus, it is tempting to speculate that the apparent over-activity of kinases disturbs the linkage of sister chromatids in the pericentromere of meiotic budding yeast and fission yeast cells. As described below, MOKIRs in fission yeast and mice are thought to direct the formation of inter-sister cohesive linkages within the core centromere to direct sister kinetochore monoorientation. It is conceivable that Spo13MOKIR plays a similar role in budding yeast, except in this case the cohesive linkages would be in the pericentromere and represent the key linkages that should be protected until meiosis II (Figure 3). Further experiments will however be required to ascertain if this is the case and, if so, whether a similar underlying mechanism is responsible.
Recent evidence from Drosophila supports the notion that kinase inhibition may similarly be the critical function of MtrmMOKIR in cohesin protection. However, in contrast to other organisms, where MOKIRs appear to target Polo to kinetochores (see below), Drosophila MtrmMOKIR may sequester Polo away from chromosomes. In mtrmnull oocytes or oocytes carrying a mutation in the PBD binding region, active Polo kinase is released from the spindle and increased amounts are found on DNA.[16] Since defective centromeric cohesion in mtrm null oocytes is rescued by lowering Polo activity,[16,59] MtrmMOKIR must preserve cohesion by sequestration of Polo on the spindle. How ectopic Polo triggers cohesion loss is not clear, since the cohesin protector MEI-S332Sgo (which is related to the later-discovered shugoshins)[41,47,60] is appropriately localized in metaphase I,[16] similar to spo13Δ and moalΔ mutants in budding yeast and fission yeast, respectively. Instead, MtrmMOKIR may interfere with Polo’s ability to phosphorylate proteins promoting cohesin cleavage. Alternatively, MtrmMOKIR may prevent the premature removal of MEI-S332Sgo in anaphase I by counteracting Polo, which is known to delocalize MEI-S332Sgo from chromosomes in meiosis II.[61] This hypothesis is particularly attractive because such a function of MtrmMOKIR would mirror the role of budding yeast Spo13MOKIR in preventing the CK1δ-mediated removal of Sgo1 in anaphase I.
In summary, regulation of Polo kinase by MOKIRs aids pericentromeric cohesin retention in meiosis I, but other mechanisms are likely to contribute. Indeed, as for the budding yeast example, MOKIRs may regulate other kinases. How ectopic kinase activity results in the premature cleavage of pericentromeric cohesin is not completely clear. However, future studies should address if MOKIRs and their regulated kinases maintain centromeric cohesion beyond meiosis I through mechanisms involving: (1) shugoshin persistence at centromeres during/beyond anaphase I, (2) shielding cohesin from cleavage-promoting phosphorylation by dis-regulated kinases, and (3) altering the types and positions of cohesin-dependent linkages that are established within core centromeres (Figure 2D, Figure 3). These mechanisms are not mutually exclusive and both inhibitory and activating interactions can be envisaged.
4. MOKIRs promote mono-orientation by regulating Polo kinases
4.1. Different organisms have distinct requirements for sister kinetochore monoorientation
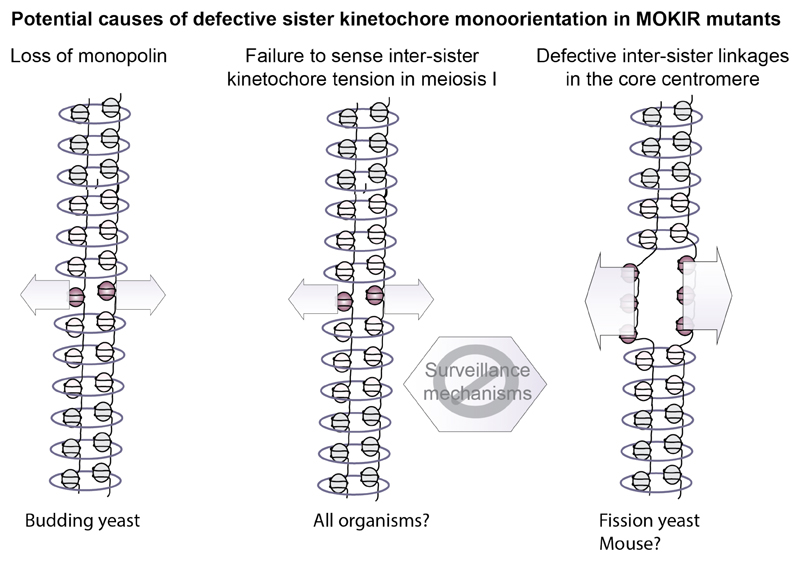
To segregate homologous chromosomes in meiosis I, sister kinetochores need to face towards the same pole, a property referred to as mono-orientation. In budding yeast, Drosophila, maize and mouse, the fusion of sister kinetochores into a single microtubule-binding entity appears to underlie their mono-orientation during meiosis I.[15,62–65] MOKIRs have been implicated in this process in mouse, but it is best understood in budding and fission yeast, where distinct mechanisms have emerged (Figures 3 and 5).
Figure 5. Regulation of mono-orientation by MOKIRs.
The conserved function of MOKIRs is to recruit Polo kinase to kinetochores, and this is important for sister kinetochore mono-orientation in budding yeast, fission yeast and mouse. However, the exact function and targets of Polo have so far remained elusive. In budding yeast, cohesin is largely dispensable for mono-orientation and monopolin is required instead. Consistently, artificial tethering of Cdc5Polo to kinetochores, but not cohesin, forces sister kinetochore co-orientation, even in the absence of monopolin. Thus, a component of the kinetochore, or a kinetochore-associated protein, is the likely target of Cdc5Polo for mono-orientation. Potentially, Cdc5Polo alters surveillance mechanisms to ensure sister kinetochore monoorientation, rather than biorientation. In fission yeast and mouse, Polo is likely to target cohesin, since core cohesion is typically lost in the absence of the respective MOKIR; however, the evidence for centromeric cohesion loss is currently strongest in fission yeast. Whether Drosophila Mtrm also regulates mono-orientation is currently unclear.
4.2. Cohesin is important for sister kinetochore monoorientation in fission yeast
In fission yeast, centromeres comprise a central core, where kinetochores assemble, and flanking pericentromeric heterochromatin, where cohesin is highly enriched and protected during meiosis I. Mono-orientation requires sister chromatid cohesion in the core centromere. This is brought about by cohesin complexes containing the meiosis-specific kleisin subunit Rec8,[66] which are specifically enriched in this region.[58,67] In the absence of Rec8, cohesin containing the alternative kleisin subunit Rad21Scc1, which is typically restricted to the pericentromere, moves into the core centromere; however, mono-orientation is still defective,[68] suggesting a particular requirement for Rec8 in this process. A screen for mono-orientation-defective mutants identified several cohesin regulators, in addition to Moa1MOKIR, which was found to specifically regulate core centromere cohesion.[10,58,68,69] Moa1MOKIR interacts with Plo1Polo and recruits it to kinetochores and Moa1MOKIR mutants unable to bind Plo1Pdo are defective for mono-orientation.[15,56] Although the target of P1o1polo in mono-orientation is still unknown, cohesin is a potential candidate. In moalΔ mutants, defective mono-orientation can be alleviated by artificially linking the centromere sequences of sister chromatids.[58] Thus, in fission yeast, cohesin appears to promote mono-orientation by bringing sister core centromere sequences in close proximity, and this function may require Moa1MOKIR -dependent kinetochore recruitment of Plo1polo. Recent work has suggested that cohesin in the core centromere also directs mono-orientation in mouse.[70] Interestingly, this work argued for a role of separase-dependent cleavage of cohesin in the destruction of mono-orientation and cohesin protection after meiosis I.[70] Collectively, these findings lead to the speculation that the crucial activity of fission yeast and mouse MOKIR-Polo complexes in both monoorientation and cohesion protection might be to alter core cohesion in such a way to make it refractory to separase-dependent cleavage.
4.3. Monopolin directs sister kinetochore monoorientation in budding yeast
In contrast, evidence suggests that sister kinetochore mono-orientation is achieved independently of Rec8 cohesin in budding yeast. Replacement of the meiosis-specific cohesin subunit Rec8 with its mitotic counterpart, Sccl, does not affect mono-orientation[71] and deletion of REC8 causes only a minor mono-orientation phenotype.[72] In budding yeast, mono-orientation is established as a result of sister kinetochore fusion by a dedicated protein complex, called monopolin.[63] Monopolin consists of four subunits: the meiosis-specific kinetochore protein Mam1,[71] the nucleolar proteins Lrs4 and Csm1,[73] and CK16.[74] The monopolin complex forms a V-shaped homodimer,[75] in which the Csml subunits directly interact with the kinetochore protein Dsnl, presumably on sister kinetochores.[75–78] This is thought to fuse sister kinetochores to create a common microtubule-interaction surface.[63,75]Monopolin-induced mono-orientation requires the action of a number of different kinases. First, monopolin-associated CK16 brings the monopolin complex to kinetochores and CK16 kinase activity is required to prevent sister kinetochores splitting in metaphase I.[74] Surprisingly, however, sister chromatid co-segregation in anaphase I is only modestly perturbed when CK16 is inhibited[12] and its activity is dispensable for kinetochore fusion in vitro.[63] Thus, the function of CK16 in monoorientation is still largely elusive. Second, DDK is also required for monopolin association with kinetochores and mono-orientation.[11] Third, Cdc5Polo activity is required for the release of the monopolin subunits Csm1 and Lrs4 from the nucleolus, [79] thus promoting monopolin assembly at kinetochores.[73] Lastly, phosphorylation sites within the monopolin-binding region of Dsn1 that are the targets of CK1δ in vitro [80] are also required for monoorientation, although it is still unknown whether this is the kinase responsible for Dsn1 phosphorylation in vivo.[78]
Although budding yeast apparently uses a monopolin-rather than cohesin-dependent mechanism to direct mono-orientation, a role for Spo13MOKIR is well-established.[54,55] Spo13MOKIR interacts with Cdc5Polo and recruits it to kinetochores, and this interaction promotes monopolin recruitment and mono-orientation in metaphase I-arrested cells. However, a mutant of Spo13MOKIR in which the Cdc5polo interaction site is mutated still largely co-segregates sister chromatids in anaphase I,[12] possibly due to residual Spo13MOKIR -Cdc5Polo interaction in this mutant.[11] Artificial tethering of Cdc5Polo to kinetochores restores co-segregation of sister chromatids in cells lacking Spo13MOKIR[12] and, remarkably, this overrides the requirement for monopolin for mono-orientation.[12] Thus, forcing Cdc5Polo localisation to kinetochores likely does not induce bona fide mono-orientation in budding yeast, although the resulting molecular setup is strikingly similar to that of fission yeast and mouse (see below). Consistently, although Spo13MOKIR is essential for monopolin recruitment to kinetochores,[12,54,55] monopolin is not restored at kinetochores upon tethering Cdc5Polo to kinetochores in the absence of Spo13MOKIR. This suggests that Spo13MOKIR directs monopolin association with kinetochores independently of Cdc5Polo.[12] Furthermore, unlike tethering of Cdc5Polo to kinetochores, tethering to cohesin did not rescue the mono-orientation defect of spo13Δ cells,[12] suggesting that the crucial target of Cdc5polo for mono-orientation in budding yeast may be a kinetochore component. Nevertheless, the finding that Cdc5polo tethering to kinetochores can direct sister chromatid cosegregation independently of monopolin hints at a conserved ancestral mechanism for kinetochore-associated Polo kinases in defining reductional meiosis I segregation.
4.4. MOKIRs play poorly understood roles in sister kinetochore monoorientation
Outside the yeasts, the mechanism of sister kinetochore monoorientation and role for MOKIRs is even less clear. In mouse, MEIKINMOKIR has been shown to be a key regulator of mono-orientation.[15] Although the underlying mechanisms of MEIKINMOKIR-mediated mono-orientation are poorly understood, MEIKINMOKIRrecruits PLK1polo, whose activity is required for mono-orientation, to kinetochores in meiosis I.[15] Notably, both Moa1MOKIR and MEIKINMOKIR bind to kinetochores via the CENP-C kinetochore subunit, providing further evidence to the notion that these proteins are functionally conserved.[15]
In Drosophila, sister centromere fusion similar to that seen in budding yeast has been proposed as a mechanism of mono-orientation.[62] Fusion depends on both centromere cohesion and the activity of protein phosphatase 1, which is thought to counteract the stabilisation of kinetochore-microtubule interactions by Polo and BubRl kinases.[81] Thus, it appears that, in contrast to other model organisms, Drosophila Polo counteracts sister centromere fusion. Whether MtrmMOKIR is important for monoorientation is currently unresolved, though current findings do not rule this possibility out. Homozygous or heterozygous mtrm mutants precociously separate sister centromeres and deletion of a single allele of Polo in mtrm heterozygotes rescues this phenotype,[16] suggesting that MtrmMOKIR primarily acts to counteract the activity of Polo. Indeed, catalytically active Polo relocalizes from the spindle onto the DNA in mtrm null mutants, thereby likely altering the phosphorylation state of Polo targets on chromosomes and the spindle. Although mtrm mutants display a loss of centromere fusion, this is most caused by defective centromeric cohesion.[16] As a consequence, it is currently difficult to assess the status of sister kinetochore orientation in mtrmmutants, thus precluding any conclusions about the mono-orientation functions of MtrmMOKIR and the pool of Polo that it regulates.
Collectively, the above findings argue that the conserved function of MOKIRs is to regulate Polo kinases, either by altering Polo activity or localisation. The MOKIRs Spo13, Moal and MEIKIN recruit Polo to meiosis I kinetochores, where it promotes mono-orientation through mechanisms that are likely to be species-specific, whereas Drosophila MtrmMOKIR appears to exclude Polo from centromeres. Cohesin may be the conserved Polo target in organisms that do not contain monopolin, whereas budding yeast Cdc5polo may target a kinetochore protein or, potentially, the checkpoint and error correction machinery. Understanding the differences between monopolin-dependent and monopolin-independent mechanisms of mono-orientiation may help identify MOKIR-Polo targets in various organisms. Perhaps, monopolin-mediated monoorientation is not feasible in organisms in which centromeric domains are larger than the budding yeast point centromere and in which more than one microtubule attaches to a single kinetochore. Therefore, the MOKIR-Polo system may have been modified throughout the course of evolution to facilitate cohesin-mediated centromere fusion as a substitute for monopolin-dependent mono-orientation.
5. MOKIRs perform specialized functions in budding yeast and Drosophila
Given the strong sequence divergence of MOKIRs, it is not surprising that specialized functions have been reported for a number of these proteins. spo13Δ mutants, for example, only undergo a single meiotic division[82] characterized by a mixture of reductional and equational chromosome segregation.[52,53] This phenotype is not observed in fission yeast moalΔ cells[10] or mouse Meikin-/- oocytes.[15] Little is known about the molecular basis of the altered cell cycle in spo13Δ cells, but it has been observed that deletion of the spindle checkpoint component MAD2 restores the second division in a majority of spo13Δ cells,[53] although chromosome segregation is still defective in spo13Δ mad2Δ strains.[52] How MAD2 deletion promotes a second division in the absence of Spo13MOKIR is unknown, but it has been proposed that the spindle checkpoint-dependent metaphase I delay observed in spo13Δ mutants might cause cells to run out of time to perform a second division.[53] However, monopolin mutants undergo a similar metaphase I delay to spo13Δ cells, but nonetheless appear to biochemically undergo two meiotic divisions and largely form four spores.[54,55,71] arguing against the notion that spo13Δ cells do not have enough time to complete two divisions. Instead, it seems more likely that spo13Δ mutants activate the meiotic exit program already after meiosis I. One of the key characteristics of meiotic exit is the accumulation of the meiosis-specific APC/C activator Ama1, which degrades key meiotic division proteins such as Cdc5Polo and the meiosis-specific transcription factor Ndt80.[3] Indeed, deletion of Amal has been shown to partially restore meiosis II spindle formation in spo13Δ cells,[55] suggesting that premature Amal activation might contribute to early meiotic exit upon Spo13MOKIR loss. Interestingly, the accumulation of Ama1 and a variety of other events occurring during meiotic exit in wild-type cells depends on the activity of CK1δ.[83] Given our findings that Spo13MOKIR restricts CK1δ activity to promote cohesin protection during meiosis I,[12] we speculate that a similar activity of Spo13MOKIR prevents premature meiotic exit.
Although Spo13MOKIR and MtrmMOKIR share a similar LEN degradation motif (Figure 2C), their degradation occurs at different times. Spo13MOKIR is degraded at the onset of anaphase I,[17] but MtrmMOKIR persists until the end of meiosis when it is targeted for proteasomal destruction by the meiosis-specific APC/CCort form.[18] This ensures sufficient Polo activity at the end of meiosis to drive the oocyte-to-embryo transition.[18] Additionally, the inhibitory action of MtrmMOKIR towards Polo is also required at the onset of meiosis, where it ensures that the G2 arrest preceding meiotic entry in oocytes is maintained.[59] In other organisms, pre-division functions of MOKIRs are not known, but, at least in budding yeast, may not be required because Cdc5Polo activity is restricted prior to metaphase I due to active degradation[3] and lack of transcription.[84]
In summary, few specialized functions of MOKIRs, in addition to promoting mono-orientation and cohesin protection, are known, and the mechanisms governing these functions are poorly understood. However, in analogy to the potential function of MOKIRs in cohesin protection, these proteins may control additional meiotic processes through a general property of spatiotemporally restricting the function of meiotic kinases.
6. Conclusions and future perspectives
MOKIRs are key meiotic proteins that almost single-handedly appear to convert many aspects of mitotic chromosome segregation into essential adaptations for meiosis. Yet, their structural features and mechanistic functions are largely elusive. The common denominator for these proteins appears to be their ability to bind and spatially regulate Polo kinases, thus promoting sister kinetochore mono-orientation in meiosis I. Beyond this, MOKIRs seem to act by restricting the activity of meiotic kinases. Although it is tempting to speculate that these proteins directly inhibit meiotic kinases, as has been suggested in the case of MtrmMOKIR and Polo, it is crucial that these kinases still retain some activity in the presence of MOKIRs to perform their essential meiotic functions. One possible explanation to this conundrum is that MOKIRs may target specific pools of a particular kinase, as has been shown recently for MtrmMOKIR.[16] Analysis of the role of post-translational modifications in the regulation of MOKIRs may provide some clues as to how the diverse functions of these proteins are integrated. Moreover, the identification of separation-of-function mutants would greatly aid the study of MOKIRs. It is also essential to determine how MOKIRs are deactivated in meiosis II. Although Spo13MOKIR is degraded in anaphase I, spore viability is similar to wild type in a mutant resistant to degradation,[17] suggesting additional deactivation mechanisms. However, to fully understand the multifaceted functions of MOKIRs, a greater understanding of the meiotic chromosome segregation adaptations in general is required, in particular with regard to the mechanisms of mono-orientation and cohesin regulation. Research on MOKIRs so far has identified them as key rulers of meiotic kinases. The next challenge is to elucidate the identity and role of phosphorylation events during meiotic chromosome segregation and how unruly kinases in MOKIR-deficient cells affect the balance of post-translational modifications in the meiotic cell cycle.
Acknowledgements
We are grateful to Scott R. Hawley for sharing recently published data[16] prior to publication and to A. Jeyaprakash Arulanandam for helpful comments on the manuscript. AM and SG were funded by Wellcome through studentships to SG (096994), a Senior Research Fellowship to ALM (107827) and core funding for the Wellcome Centre for Cell Biology (203149).
References
- [1].Cabral G, Marques A, Schubert V, Pedrosa-Harand A, Schlögelhofer P. Nature Communications. 2014;5:5070. doi: 10.1038/ncomms6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Marston AL, Amon A. Nat Rev Mol Cell Biol. 2004;5:983. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- [3].Okaz E, Argüello-Miranda O, Bogdanova A, Vinod PK, Lipp JJ, Markova Z, Zagoriy I, Novak B, Zachariae W. Cell. 2012;151:603. doi: 10.1016/j.cell.2012.08.044. [DOI] [PubMed] [Google Scholar]
- [4].Marston AL, Wassmann K. Front Cell Dev Biol. 2017;5:109. doi: 10.3389/fcell.2017.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, et al. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. Nat Protoc. 2015;10:845. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bonner AM, Hughes SE, Chisholm JA, Smith SK, Slaughter BD, Unruh JR, Collins KA, Friederichs JM, Florens L, Swanson SK, Pelot MC, et al. Proc Natl Acad Sci USA. 2013;110 doi: 10.1073/pnas.1301690110. E1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bonner AM, Hawley RS. Mol Biol Evol. 2019;36:69. doi: 10.1093/molbev/msy197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wright PE, Dyson HJ. Nat Rev Mol Cell Biol. 2015;16:18. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yokobayashi S, Watanabe Y. Cell. 2005;123:803. doi: 10.1016/j.cell.2005.09.013. [DOI] [PubMed] [Google Scholar]
- [11].Matos J, Lipp JJ, Bogdanova A, Guillot S, Okaz E, Junqueira M, Shevchenko A, Zachariae W. Cell. 2008;135:662. doi: 10.1016/j.cell.2008.10.026. [DOI] [PubMed] [Google Scholar]
- [12].Galander S, Barton RE, Borek WE, Spanos C, Kelly DA, Robertson D, Rappsilber J, Marston AL. Dev Cell. 2019;49:526. doi: 10.1016/j.devcel.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Elia AEH, Cantley LC, Yaffe MB. Science. 2003;299:1228. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- [14].Elia AEH, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. Cell. 2003;115:83. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- [15].Kim J, Ishiguro K-I, Nambu A, Akiyoshi B, Yokobayashi S, Kagami A, Ishiguro T, Pendás AM, Takeda N, Sakakibara Y, Kitajima TS, et al. Nature. 2015;517:466. doi: 10.1038/nature14097. [DOI] [PubMed] [Google Scholar]
- [16].Bonner AM, Hughes SE, Hawley RS. Curr Biol. 2020;1 doi: 10.1016/j.cub.2019.12.027. [DOI] [PubMed] [Google Scholar]
- [17].Sullivan M, Morgan DO. J Biol Chem. 2007;282:19710. doi: 10.1074/jbc.M701507200. [DOI] [PubMed] [Google Scholar]
- [18].Whitfield ZJ, Chisholm J, Hawley RS, Orr-Weaver TL. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001648. e1001648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nasmyth K, Haering CH. Annu Rev Genet. 2009;43:525. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- [20].Yatskevich S, Rhodes J, Nasmyth K. Annu Rev Genet. 2019;53:445. doi: 10.1146/annurev-genet-112618-043633. [DOI] [PubMed] [Google Scholar]
- [21].Challa K, Lee M-S, Shinohara M, Kim KP, Shinohara A. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw034. gkw034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Challa K, Fajish VG, Shinohara M, Klein F, Gasser SM, Shinohara A. PLoS Genet. 2019;15 doi: 10.1371/journal.pgen.1007851. e1007851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Crawley O, Barroso C, Testori S, Ferrandiz N, Silva N, Castellano-Pozo M, Jaso-Tamame AL, Martinez-Perez E. elife. 2016;5:563. doi: 10.7554/eLife.10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Silva MCC, Powell S, Ladstätter S, Gassler J, Stocsits R, Tedeschi A, Peters J-M, Tachibana K. J Cell Biol. 2020;219:977. doi: 10.1083/jcb.201906100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tachibana-Konwalski K, Godwin J, der Weyden Lvan, Champion L, Kudo NR, Adams DJ, Nasmyth K. Genes Dev. 2010;24:2505. doi: 10.1101/gad.605910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Brar GA, Kiburz BM, Zhang Y, Kim J-E, White F, Amon A. Nature. 2006;441:532. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- [27].Kudo NR, Anger M, Peters AHFM, Stemmann O, Theussl H-C, Helmhart W, Kudo H, Heyting C, Nasmyth K. J Cell Sci. 2009;122:2686. doi: 10.1242/jcs.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ishiguro T, Tanaka K, Sakuno T, Watanabe Y. Nat Cell Biol. 2010;12:500. doi: 10.1038/ncb2052. [DOI] [PubMed] [Google Scholar]
- [29].Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W. Dev Cell. 2010;18:397. doi: 10.1016/j.devcel.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Attner MA, Miller MP, Ee L-S, Elkin SK, Amon A. Proc Natl Acad Sci USA. 2013;110:14278. doi: 10.1073/pnas.1311845110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Khetani RS, Bickel SE. J Cell Sci. 2007;120:3123. doi: 10.1242/jcs.009977. [DOI] [PubMed] [Google Scholar]
- [32].Krishnan B, Thomas SE, Yan R, Yamada H, Zhulin IB, McKee BD. Genetics. 2014;198:947. doi: 10.1534/genetics.114.166009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yan R, Thomas SE, Tsai J-H, Yamada Y, McKee BD. J Cell Biol. 2010;188:335. doi: 10.1083/jcb.200904040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tang Z, Sun Y, Harley SE, Zou H, Yu H. Proc Natl Acad Sci USA. 101:18012. doi: 10.1073/pnas.0408600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y. Curr Biol. 15:353. doi: 10.1016/j.cub.2004.12.044. [DOI] [PubMed] [Google Scholar]
- [36].Kiburz BM, Reynolds DB, Megee PC, Marston AL, Lee B, Lee TI, Levine SS, Young RA, Amon A. Genes Dev. 2005;19:3017. doi: 10.1101/gad.1373005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Fernius J, Hardwick KG. PLoS Genet. 2007;3 doi: 10.1371/journal.pgen.0030213. e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kawashima SA, Yamagishi Y, Honda T, Ishiguro K-I, Watanabe Y. Science. 2010;327:172. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- [39].Liu H, Jia L, Yu H. Curr Biol. 2013 doi: 10.1016/j.cub.2013.07.078. [DOI] [PubMed] [Google Scholar]
- [40].Liu H, Rankin S, Yu H. Nat Cell Biol. 2013;15:40. doi: 10.1038/ncb2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kitajima TS, Kawashima SA, Watanabe Y. Nature. 2004;427:510. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- [42].Marston AL, Tham W-H, Shah H, Amon A. Science. 2004;303:1367. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- [43].Katis VL, Gálová M, Rabitsch KP, Gregan J, Nasmyth K. Curr Biol. 2004;14:560. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- [44].Llano E, Gòmez R, Gutiérrez-Caballero C, Herrán Y, Sanchez-Martin M, Vazquez-Quiñones L, Hernandez T, de Alava E, Cuadrado A, Barbero JL, Suja JA, et al. Genes Dev. 2008;22:2400. doi: 10.1101/gad.475308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lee J, Kitajima TS, Tanno Y, Yoshida K, Morita T, Miyano T, Miyake M, Watanabe Y. Nat Cell Biol. 2008;10:42. doi: 10.1038/ncb1667. [DOI] [PubMed] [Google Scholar]
- [46].Rattani A, Wolna M, Ploquin M, Helmhart W, Morrone S, Mayer B, Godwin J, Xu W, Stemmann O, Pendas A, Nasmyth K. elife. 2013;2 doi: 10.7554/eLife.01133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kerrebrock AW, Miyazaki WY, Birnby D, Orr-Weaver TL. Genetics. 1992;130:827. doi: 10.1093/genetics/130.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kitajima TS, Sakuno T, Ishiguro K-I, Iemura S-I, Natsume T, Kawashima SA, Watanabe Y. Nature. 2006;441:46. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- [49].Pinto BS, Orr-Weaver TL. Proc Natl Acad Sci USA. 2017;114:12988. doi: 10.1073/pnas.1718450114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Xu Z, Cetin B, Anger M, Cho US, Helmhart W, Nasmyth K, Xu W. Mol Cell. 2009;35:426. doi: 10.1016/j.molcel.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Gálová M, Petronczki M, Gregan J, Cetin B, Mudrak I, et al. Nature. 2006;441:53. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- [52].Galander S, Barton RE, Kelly DA, Marston AL. Wellcome Open Res. 2019;4:29. doi: 10.12688/wellcomeopenres.15066.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Shonn MA, McCarroll R, Murray AW. Genes Dev. 2002;16:1659. doi: 10.1101/gad.975802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lee B, Kiburz BM, Amon A. Curr Biol. 2004;14:2168. doi: 10.1016/j.cub.2004.12.033. [DOI] [PubMed] [Google Scholar]
- [55].Katis VL, Matos J, Mori S, Shirahige K, Zachariae W, Nasmyth K. Curr Biol. 2004;14:2183. doi: 10.1016/j.cub.2004.12.020. [DOI] [PubMed] [Google Scholar]
- [56].Miyazaki S, Kim J, Yamagishi Y, Ishiguro T, Okada Y, Tanno Y, Sakuno T, Watanabe Y. Genes Cells. 2017;22:552. doi: 10.1111/gtc.12496. [DOI] [PubMed] [Google Scholar]
- [57].Chen Y-C, Weinreich M. J Biol Chem. 2010;285:41244. doi: 10.1074/jbc.M110.155242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sakuno T, Tada K, Watanabe Y. Nature. 2009;458:852. doi: 10.1038/nature07876. [DOI] [PubMed] [Google Scholar]
- [59].Xiang Y, Takeo S, Florens L, Hughes SE, Huo L-J, Gilliland WD, Swanson SK, Teeter K, Schwartz JW, Washburn MP, Jaspersen SL, et al. PLoS Biol. 2007;5 doi: 10.1371/journal.pbio.0050323. e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kerrebrock AW, Moore DP, Wu JS, Orr-Weaver TL. Cell. 1995;83:247. doi: 10.1016/0092-8674(95)90166-3. [DOI] [PubMed] [Google Scholar]
- [61].Clarke AS, Tang TT-L, Ooi DL-Y, Orr-Weaver TL. Dev Cell. 2005;8:53. doi: 10.1016/j.devcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- [62].Goldstein LS. Cell. 1981;25:591. doi: 10.1016/0092-8674(81)90167-7. [DOI] [PubMed] [Google Scholar]
- [63].Sarangapani KK, Duro E, Deng Y, de F, Alves L, Ye Q, Opoku KN, Ceto S, Rappsilber J, Corbett KD, Biggins S, et al. Science. 2014 doi: 10.1126/science.1256729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Li X, Dawe RK. Nat Cell Biol. 2009;11:1103. doi: 10.1038/ncb1923. [DOI] [PubMed] [Google Scholar]
- [65].Chiang T, Duncan FE, Schindler K, Schultz RM, Lampson MA. Curr Biol. 2010;20:1522. doi: 10.1016/j.cub.2010.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Watanabe Y, Nurse P. Nature. 1999;400:461. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- [67].Watanabe Y, Yokobayashi S, Yamamoto M, Nurse P. Nature. 2001;409:359. doi: 10.1038/35053103. [DOI] [PubMed] [Google Scholar]
- [68].Yokobayashi S, Yamamoto M, Watanabe Y. Mol Cell Biol. 2003;23:3965. doi: 10.1128/MCB.23.11.3965-3973.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kagami A, Sakuno T, Yamagishi Y, Ishiguro T, Tsukahara T, Shirahige K, Tanaka K, Watanabe Y. EMBO Rep. 2011;12:1189. doi: 10.1038/embor.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ogushi S, Rattani A, Godwin J, Metson J, Schermelleh L, Nasmyth K. bioRxiv. 2020;106 doi: 10.1016/j.devcel.2021.10.017. 2020.02.06.935171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Toth A, Rabitsch KP, Gálová M, Schleiffer A, Buonomo SB, Nasmyth K. Cell. 2000;103:1155. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- [72].Monje-Casas F, Prabhu VR, Lee B, Boselli M, Amon A. Cell. 2007;128:477. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, Tanaka TU, Nasmyth K. Dev Cell. 2003;4:535. doi: 10.1016/s1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- [74].Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, Mechtler K, Shirahige K, Zachariae W, Nasmyth K. Cell. 2006;126:1049. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- [75].Corbett KD, Yip CK, Ee L-S, Walz T, Amon A, Harrison SC. Cell. 2010;142:556. doi: 10.1016/j.cell.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Corbett KD, Harrison SC. Cell Rep. 2012;1:583. doi: 10.1016/j.celrep.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Sarkar S, Shenoy RT, Dalgaard JZ, Newnham L, Hoffmann E, Millar JBA, Arumugam P. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003610. e1003610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Plowman R, Singh N, Tromer EC, Payan A, Duro E, Spanos C, Rappsilber J, Snel B, Kops GJPL, Corbett KD, Marston AL. Chromosoma. 2019;128:331. doi: 10.1007/s00412-019-00700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, Nasmyth K. Nat Cell Biol. 2003;5:480. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- [80].Ye Q, Ur SN, Su TY, Corbett KD. EMBO J. 2016 doi: 10.15252/embj.201694082. e201694082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wang L-I, Das A, McKim KS. PLoS Genet. 2019;15 doi: 10.1371/journal.pgen.1008072. e1008072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Klapholz S, Esposito RE. Genetics. 1980;96:567. doi: 10.1093/genetics/96.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Arguello-Miranda O, Zagoriy I, Mengoli V, Rojas J, Jonak K, Oz T, Graf P, Zachariae W. Dev Cell. 2017;40:37. doi: 10.1016/j.devcel.2016.11.021. [DOI] [PubMed] [Google Scholar]
- [84].Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. Science. 1998;282:699. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]