Abstract
Binding protein generation relies on laborious screening cascades that process candidate molecules individually. To break with this paradigm, we developed NestLink, a binder selection and identification technology able to biophysically characterize thousands of library members at once without handling individual clones at any stage of the process. NestLink centers on genetically encoded barcoding peptides, termed flycodes, which were designed for maximal detectability by mass spectrometry and support accurate deep sequencing. We demonstrate that NestLink has the capacity to overcome fundamental limitations of binder generation. Rare binders against an integral membrane protein were identified directly in the cellular environment of a human pathogen. Hundreds of binder candidates were simultaneously ranked according to kinetic parameters. Deep-mining of a nanobody immune repertoire for membrane protein binders was performed entirely in solution without target immobilization. NestLink opens avenues for the selection of tailored binder characteristics directly in tissues or in living organisms.
Introduction
Binding proteins have proven invaluable for a plethora of applications in basic science, diagnostics and therapy. Their generation involves laborious screening cascades that process individual candidate molecules spatially separated from each other with limited throughput. Particularly slow are analyses that require purified binding proteins, such as for the determination of kinetic parameters by surface plasmon resonance (SPR). Recently developed methods that combine next generation sequencing (NGS) and liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) enabled binder identification directly from large ensembles, such as immune repertoires, without the necessity of handling individual clones1-6. Unfortunately, due to extensive sequence homology within binder ensembles, this approach is currently limited by the low number of peptides, which are unambiguously assignable to individual binder sequences7. Furthermore, many of the peptides suffer from low ionization and fragmentation efficiencies, thus hampering binder identification by LC-MS/MS significantly.
Here, we introduce NestLink, a technology that overcomes these inherent limitations by the unprecedented application of genetically encoded barcoding peptides for binding protein identification and characterization. In three diverse applications, we demonstrate that NestLink enables the unambiguous identification and unprecedented high-throughput characterization of thousands of binding proteins without the need to handle individual clones at any stage of the process.
Results
The NestLink principle
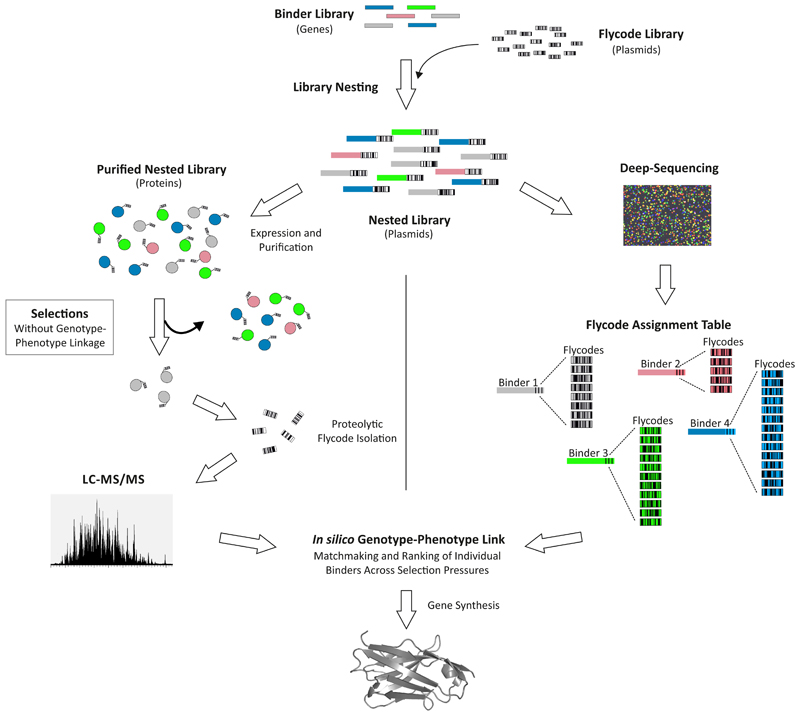
NestLink centers on a diverse library of short peptide barcodes, which are designed for optimal detection by LC-MS/MS and therefore termed flycodes. They are genetically fused to a library of binding proteins in a novel process termed “library nesting” (Fig. 1). The nested library is sequenced by NGS to assign all flycodes to their corresponding binders (Fig. 1, Supplementary Fig. 1). Subsequently, the nested library is expressed as a pool and subjected to selection pressures. Flycodes of selected binders are isolated via sequence-specific proteases and detected via LC-MS/MS (Supplementary Fig. 2). In combination, library nesting, NGS and LC-MS/MS establish an in silico genotype-phenotype linkage, which allows rapid characterization of individual binder properties directly within ensembles.
Figure 1. NestLink overview.
Details are provided in the main text.
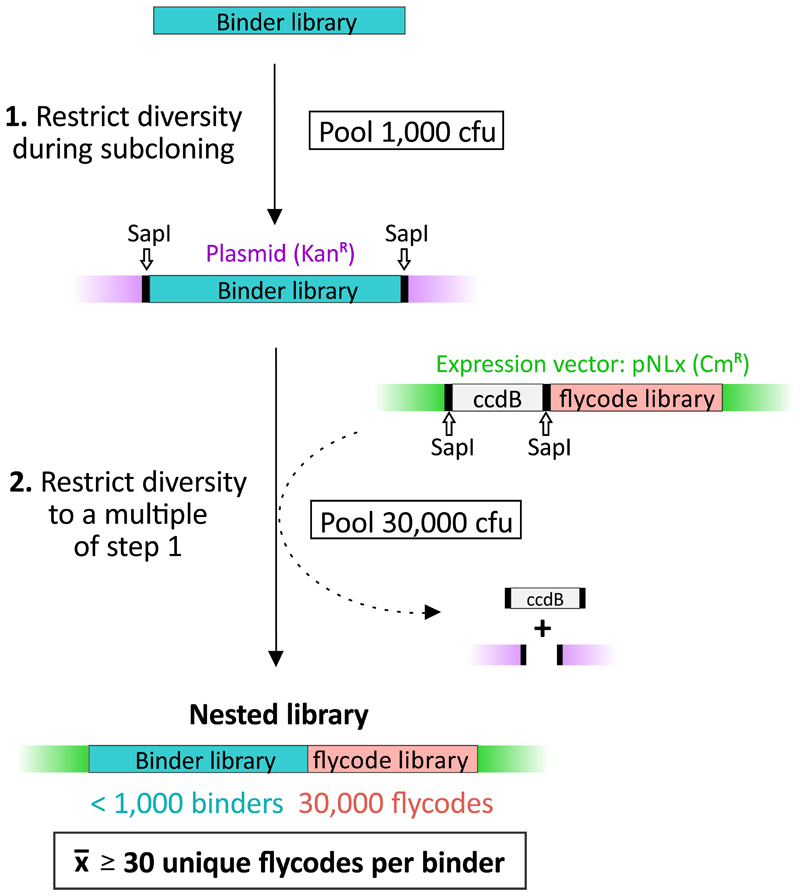
Library nesting
Library nesting links each gene of a binder library in a controlled manner multiple times to unique flycodes (Fig. 2). In a first step, a defined number of bacterial colony forming units (cfu), harboring plasmids that encode binder library members, are pooled for plasmid isolation. This step defines the maximal diversity of the binder library. In a second step, restriction digest and ligation are used to clone the binder library into a plasmid, which harbors the flycode library. Thereby, the binder library and the flycode library are nested. The number of cfu pooled for plasmid isolation in this second step defines the maximal number of flycodes and thus the average number of flycodes per binder. For example, if the binder library size is < 1,000 and 30,000 cfu are pooled after the library nesting step, the number of different flycodes per binder is on average > 30. Importantly, attached flycodes are unique, because the experimental flycode library diversity (≈ 100 Mio) vastly exceeds the total number of flycodes linked via library nesting. Hence, flycodes are unambiguously assignable to library members, which is the basis for unambiguous binder detection via LC-MS/MS. Of note, library nesting avoids PCR amplification and thereby prevents undesired recombination events (Supplementary Fig. 3).
Figure 2. Library nesting.
This example shows how a nested library containing on average 30 flycodes per binder is generated. First, the maximal binder diversity is restricted to 1,000 by isolating 1,000 cfu of bacteria harboring the binder library on a plasmid. This restricted binder pool is cloned into a vector backbone encoding for the flycode library. By isolating 30,000 cfu of the resulting nested library, the maximal number of flycodes is restricted to 30,000 and each binder on average is fused to 30 different flycodes. The large diversity of the flycode library (100 Mio) ensures uniqueness of attached flycodes (30,000 ≪ 100 Mio).
Flycode library design
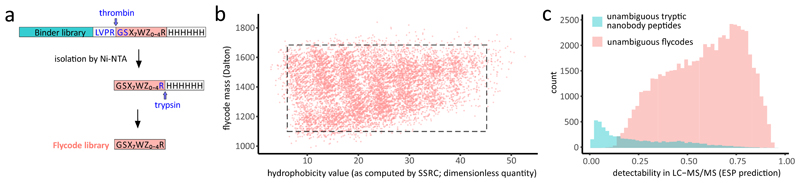
The flycode library is composed of genetically encoded peptide sequences designed for optimal detection via LC-MS/MS upon proteolytic isolation from a protein pool of interest (Fig. 3a). Flycodes are 11-15 amino acids long and contain two randomized regions resulting in a theoretical library diversity of 5.3 x 108. To enable optimal detection of individual flycodes by LC-MS/MS, the library was designed to be maximally diverse in terms of (i) mass-over-charge ratios (m/z) to fall into the optimal m/z-detection window of high-field orbitraps (550-850 m/z), and (ii) hydrophobicity, thus exploiting the full separation capacity of reverse-phase liquid chromatography systems (Fig. 3b, Supplementary Fig. 4a). Flycodes contain an invariant arginine as sole positively charged residue, which supports efficient ionization. The randomized regions are devoid of cysteines and methionines to avoid oxidation and cross-linking, but frequently contain aspartate and glutamate to enhance solubility. Importantly, size exclusion chromatography (SEC) analyses revealed that the attachment of flycodes does not change the oligomeric state of binders (Supplementary Fig. 5). As individual library members are fused redundantly to multiple flycodes, potential negative effects of individual flycodes are averaged out.
Figure 3. Flycode library design and characteristics.
(a) Flycodes are located between a thrombin cleavage site (blue) and a His-tag, which can be cleaved site-specifically by trypsin at the sole positively charged amino acid (blue arginine). “X7” denotes a stretch of seven randomized amino acids, and “Z0-4” represents five distinct sequences of 0-4 amino acids in length. Amino acid compositions are provided in the online methods. (b) Prediction of hydrophobicity (SSRC; Sequence Specific Retention Calculator14) and parent ion mass for 10,000 randomly chosen flycodes. The optimal detection window for the exclusively doubly-charged flycodes is shown as dashed rectangle. (c) Histogram showing the detectability of unambiguously assignable tryptic nanobody peptides (cyan) and unambiguously assignable flycodes (pink) from the same nested library consisting of 3,390 unique nanobodies linked to 59,974 flycodes (see application II, Fig. 5). Peptides are binned according to their ESP prediction value (high ESP values correlate with better detection by LC-MS/MS).
In order to assess the benefits of flycode-mediated protein detection by LC-MS/MS, a nested library comprising 3,390 unique nanobodies linked to 59,974 flycodes (see also application II below) was analyzed in silico to compare the flycodes with the peptides obtained by tryptic digest of the nanobodies (Fig. 3c). The analysis revealed 7.6 fold more flycodes than tryptic nanobody peptides that are unambiguously assignable to a single nanobody of the library. In addition, the enhanced signature peptide (ESP) predictor indicates a high average MS/MS-detectability for flycodes, whereas the majority of tryptic nanobody peptides are predicted to be poorly detected (Fig. 3c)8. A control experiment revealed that redundant tagging of proteins with flycodes allows for highly accurate quantification of proteins (Supplementary note 1, Supplementary Fig. 6 and 7).
Application I: ranking hundreds of off-rates within binder ensembles at once
Biophysical characterization of binder candidates is commonly performed to identify the best binders in terms of stability, affinity and specificity. Such assays often require individually purified candidate molecules (e.g. for the determination of off-rates by SPR), which represents a critical bottleneck in binder screening cascades. NestLink was developed to characterize large numbers of individual binder candidates directly within ensembles, suggesting that it can overcome existing throughput limitations by several orders of magnitude. In order to evaluate this hypothesis, we analyzed how off-rates can be ranked by NestLink.
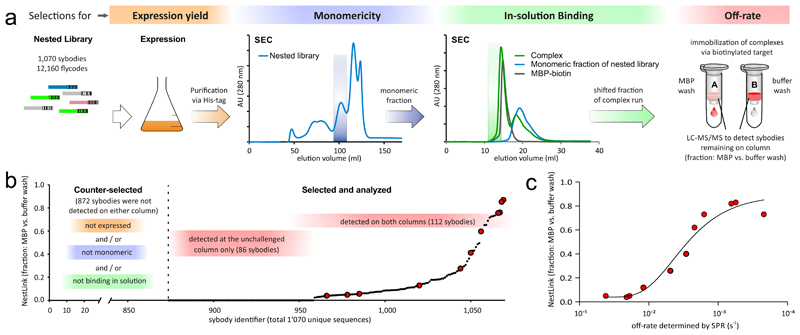
A pool of synthetic nanobodies (sybodies), which was previously selected against maltose-binding protein (MBP) by ribosome display9, was used to construct a nested library as described above. NGS covered the entire sequence length of the nested library with an average redundancy of 451 reads/binder and revealed that it contains 1,070 unique sybodies and 12,160 flycodes (see online methods). The nested library was subsequently expressed as an ensemble in E. coli, purified via His-tag and the monomeric binder candidates were isolated by SEC (Fig. 4a). The monomeric nested library members were mixed with MBP, followed by a second SEC run to isolate MBP-sybody complexes. The complexes were immobilized via biotin previously attached to MBP on two streptavidin-sepharose spin-columns with the aim to identify sybodies exhibiting slow off-rates. To this end, we washed one spin-column with buffer containing a large excess of non-biotinylated MBP, thereby removing binders in an off-rate-dependent manner, while the other column was washed with buffer only. Flycodes of sybodies that remained on the spin-columns were isolated and analyzed by LC-MS/MS (one LC-MS/MS run per spin-column) to determine individual sybody abundances. Of 1,070 nested sybodies, the majority (IDs: 0-872) was not detected on either spin-column, presumably because they were either poorly expressed, not monomeric or not forming a stable complex with MBP (or a combination thereof). Eighty-six sybodies (IDs: 873-958) were only detected on the unchallenged spin-column, suggesting fast off-rates. 112 sybodies (IDs: 959-1,070) were detected on both columns and their summed flycode MS1 intensities were used to determine for each binder the fraction remaining on the challenged column compared to the control column. This allowed ranking of the binders according to their off-rates (Fig. 4b).
Figure 4. Application I: ranking hundreds of binders according to their off-rates.
(a) A pool of 1,070 synthetic nanobodies (sybodies) previously enriched against maltose binding protein (MBP) was linked to 12,160 flycodes, as determined by NGS. The nested library was expressed in E. coli (orange), purified and separated by SEC (blue). The monomeric pool members were mixed with MBP-biotin and the binders co-migrating with the target on SEC were immobilized on two streptavidin-sepharose columns (red). An off-rate selection was performed by washing one column with buffer containing an excess of non-biotinylated target (MBP wash), while the other column was not challenged (buffer wash). Flycodes linked to sybodies that remained on the columns were isolated and analyzed by LC-MS/MS (one run per column). (b) Individual sybodies ranked according to their relative fraction remaining on the MBP-washed column versus the unchallenged column, as determined by the sum of flycode MS1 intensities for each identified binder. (c) Individual sybody genes (red, enlarged data points in (b)) were synthesized, followed by expression, purification and SPR-characterization. The recorded off-rates (x-axis) strongly correlate with the fractions remaining on the columns as determined by NestLink (y-axis).
To validate the NestLink readout, we synthesized the genes of 11 sybodies, expressed and purified them individually and determined their off-rates by SPR for a side-by-side comparison (Fig. 4c and Supplementary Fig. 8). The sigmoidal distribution observed in Figure 4c (R2 = 0.96) shows an excellent correlation between off-rate values determined by SPR and NestLink. NestLink thus overcomes a core bottleneck of binder screening cascades, as it can accurately rank kinetic parameters of hundreds of binders at once without the need to purify individual binders.
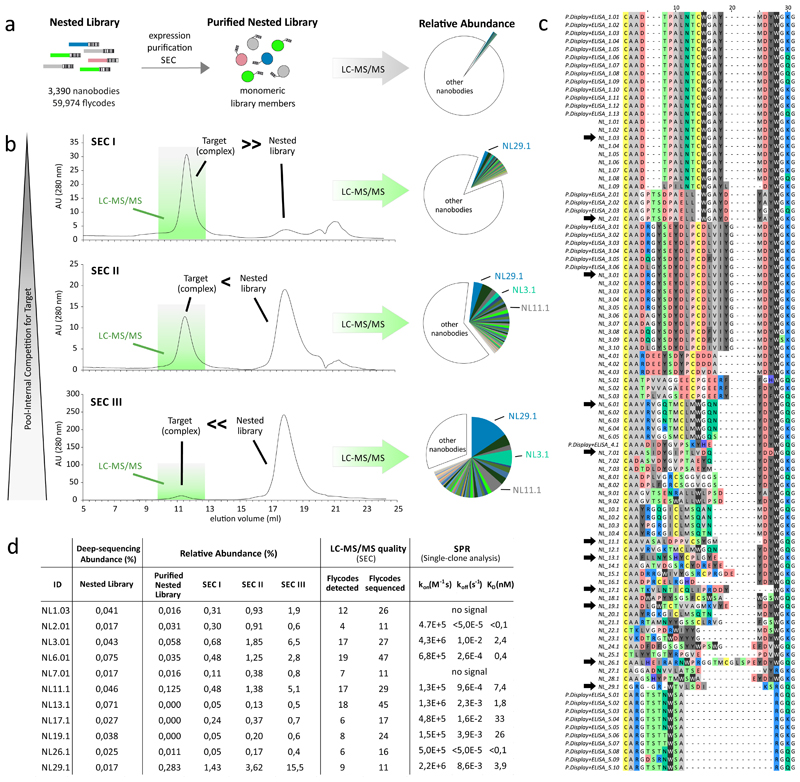
Application II: diversity mining of camelid nanobodies
In an effort to generate nanobodies as crystallization chaperones for the bacterial ABC transporter TM287/28810, 11, we immunized an alpaca with detergent-purified transporter and performed two rounds of phage display, ELISA screening and Sanger sequencing, according to a standard protocol12. Although we sequenced 210 specific ELISA hits, only 33 unique, often nearly identical nanobody sequences belonging to merely 5 binder families were identified.
We suspected that the strong enrichment of certain nanobodies during phage display selection may have caused a heavy overrepresentation of a small number of binders, which were then identified repeatedly by ELISA, thus limiting screening depth. We hypothesized that NestLink can be used to overcome this problem, as it may be suitable to characterize a large number of nanobodies directly cloned from B-cells without the need for phage display, ELISA or Sanger sequencing.
To perform NestLink, we linked 3,390 unique nanobody sequences amplified directly from B-cells of the immunized alpaca to 59,974 flycodes by library nesting (Fig. 5a, Supplementary Fig. 4c). The nested library was purified and all monomeric library members were subsequently mixed with TM287/288 at three different ratios before separation by SEC. Hereby, three distinct levels of pool-internal competition for target binding in solution were achieved (Fig. 5b). Flycodes were isolated from the nanobody-TM287/288 complex fractions of the three SEC runs and from a sample of the purified nested library (selection input). Subsequently, they were analyzed in independent LC-MS/MS runs. The analysis revealed a large number of efficient binders, which gained in relative abundance at the target as a consequence of increasing pool-internal competition. In total, we identified 29 binder families – more than 5-fold the number of families obtained by the conventional workflow (Fig. 5c). A repetition of this experiment using a different LC-MS/MS device proved the robustness of NestLink (Supplementary note 2, Supplementary Fig. 9).
Figure 5. Application II: nanobody selections without target immobilization.
(a) 3,390 unique nanobody sequences from B cells of an alpaca that was previously immunized with the ABC transporter TM287/288, were nested with 59,974 flycodes, followed by expression, purification and SEC separation of the nested pool to isolate monomeric binder candidates. (b) Pool-internal competition for target binding in solution was applied by mixing the nested nanobody pool and TM287/288 at different ratios prior to complex isolation by SEC separation (SEC I – III). Four separate LC-MS/MS runs were performed to analyze the flycodes of the purified nested library as well as of the target bound binders of the three SEC runs. The relative abundance of individual nanobodies within the same LC-MS/MS run was determined according to the summed MS1 intensities of the detected flycodes. The colored slices represent 61 individual nanobodies that gained in relative abundance as a consequence of increased competition for the target. Other nanobodies that did not gain in abundance are collectively represented by the colorless slices. (c) CDR3 alignment of the 61 unique nanobodies identified via NestLink (NL1.01 – NL29.01) or via phage display and extensive ELISA screening using the same immunized animal (P. Display + ELISA_1.01 – 5.10). Black arrows denote clones that were characterized individually by SPR. (d) Comparison of NestLink and SPR results.
The NestLink selection was validated by SPR for 11 individually synthesized and purified nanobodies belonging to 11 different families. Specific binding with affinities down to the picomolar range was observed for 9 nanobodies (Fig. 5d, Supplementary Fig. 10). Two nanobodies did not exhibit target binding in SPR. Of note, the two proteins belong to families that were also identified via the conventional phage display approach, and in spite of strong ELISA signals, target interaction in SPR was not detectable for any member of these families. We therefore concluded that these binders cannot be used for affinity determination by SPR and do not represent false-positive NestLink hits. In agreement with NestLink, four control nanobodies that were well detected in the purified input pool, but not at the target, did not exhibit target binding in SPR.
This NestLink application demonstrates that binding proteins can be identified without the need for target immobilization at any stage of the binder generation process. However, the advantage of full epitope accessibility appears to have contributed only in a minor way to the large number of identified binders, since NestLink also enabled the identification of a significant number of binders from the same pool using the immobilized target (Supplementary note 3, Supplementary Fig. 11). Interestingly, 21 out of these 47 identified nanobodies were distinct from those that were found by the in-solution selection experiment. This suggests that variations of the selection pressure allow to increase the number of identifiable binders.
In summary, these results show that NestLink enables fast and efficient binder identification directly from immunized animals without generating enriched pools by phage display. Hence, redundant analysis of overrepresented clones by ELISA and Sanger-Sequencing can be overcome, allowing for a significantly superior diversity mining capacity than the state of the art method.
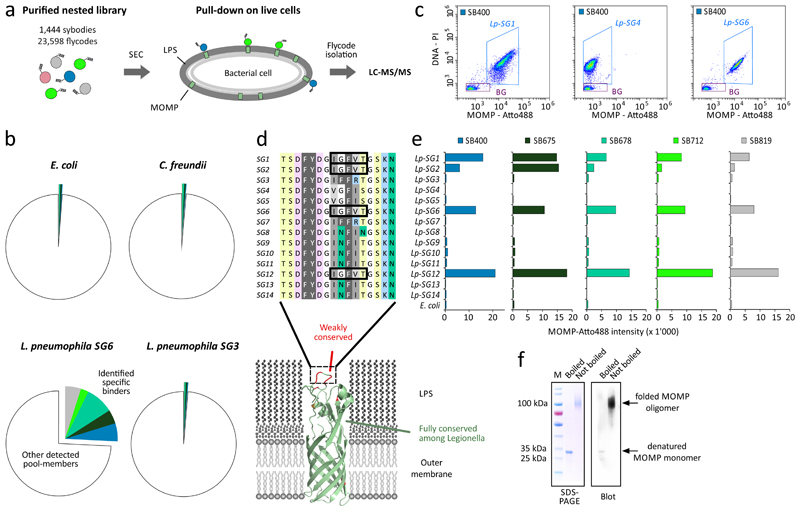
Application III: membrane protein binder identification in the cellular context
Binder generation against cell-surface epitopes of integral membrane proteins on bacterial cells has gained increasing interest for the development of rapid diagnostics and antibiotics against multidrug-resistant bacteria13. Here, we generated sybodies to specifically recognize the major outer membrane protein (MOMP) of Legionella pneumophila serogroup 6 (Lp-SG6) in its native context. To this end, sybodies were selected in vitro against the detergent-purified MOMP. ELISA screening of 576 sybodies revealed 21 unique binders exhibiting specific MOMP interactions in detergent solution. However, flow-cytometry experiments suggested that none of these sybodies recognized the target in the cellular context of Lp-SG6, where MOMP is embedded in a dense layer of lipopolysaccharides (LPS). This indicated that desired binders recognizing MOMP in the cellular context were heavily underrepresented or entirely absent.
We hypothesized that NestLink would be ideally suited to interrogate this binder pool directly in the native cellular context of living Lp-SG6 cells. Using the same binder pool enriched against detergent-purified MOMP, we generated a nested library encoding 1,444 unique sybodies linked to 23,598 flycodes, as determined by NGS. Subsequently, we performed a sybody pull-down with Lp-SG6 or with one of the three control strains Escherichia coli, Citrobacter freundii or Lp-SG3 (Fig. 6a). Captured sybodies were analyzed via their corresponding flycodes by independent LC-MS/MS runs, which allowed monitoring of relative binder abundances on the four bacterial strains. From the initial 1,444 sybodies, 157 passed the pre-selection for monomericity and were unambiguously detected at one or several of the four bacterial strains. Interestingly, five rare sybodies (representing between 0.05-0.22 % of the pool) exhibited a pronounced increase in relative abundance at Lp-SG6 (Fig. 6b). As Lp-SG6 was the only cell type of the pull-down that harbored the MOMP-variant used for the initial in vitro selection, this result confirmed the excellent specificity and sensitivity of NestLink (Fig. 6c).
Figure 6. Application III: specific recognition of an outer membrane protein in the cellular context.
(a) A nested library was constructed from a sybody pool that was previously enriched by in vitro selections against detergent-purified MOMP from Lp-SG6. After isolating monomeric nested library members by SEC, sybodies recognizing MOMP embedded in the outer membrane of Lp-SG6 were selected by a pull-down on intact cells. (b) LC-MS/MS was used to monitor the relative abundance of each sybody on the target cell, as well as on three control strains (E. coli, C. freundii, Lp-SG3). Five specific sybodies exhibiting a high relative abundance on the target cells compared to the control strains are colored. Unspecific sybodies are collectively represented by colorless slices. (c) Flow-cytometry data of Lp-SG1, Lp-SG4 and Lp-SG6 using propidium iodide (PI) for cellular staining and Atto488-labelled sybody SB400 for detection of MOMP. Detection events in the blue-framed gate (5’585, 5’892 and 4’822 events for Lp-SG1, Lp-SG4 and Lp-SG6, respectively) were used to calculate the average MOMP-Atto488 intensity as shown in (e). Flow-cytometry assays were performed once. (d) Alignment of the major non-conserved region of MOMP and illustration of its location on a homology model of the MOMP monomer. MOMP sequences identical to Lp-SG6 are framed. (e) Cell surface binding of the five identified sybodies to an array of Legionella pneumophila serogroups as quantified by flow-cytometry. (f) Coomassiestained SDS-PAGE analysis of MOMP from Lp-SG6 and Western blot detection via SB400. This analysis was performed once.
For further validation, the identified sybodies were individually synthesized, purified, fluorescently labelled and subjected to a flow-cytometry screen, using 15 different Legionella pneumophila serogroups (Fig. 6d and e) and 52 additional bacterial strains as controls (Supplementary Fig. 12). Remarkably, strong cell-surface binding was observed for Legionella pneumophila serogroups 1, 2, 6 and 12, being the only strains with an identical MOMP extracellular region as present in Lp-SG6 (Fig. 6e), which confirms target-binding in the cellular context for all identified sybodies. Since binding to purified MOMP is abolished after heat denaturation of the target, the recognized epitope is likely three-dimensional (Fig. 6f).
In summary, NestLink proved to be highly efficient for the identification of strongly underrepresented binders, which could not be identified using the conventional method of ELISA and flow-cytometry screening. Hence, NestLink is of great value for the identification of binders against challenging membrane protein targets in their native biological context.
Discussion
NestLink processes thousands of binding protein candidates as an ensemble, while generating accurate readouts for individual pool members. This allows for direct binder characterization without laborious handling of individual clones at any stage of the process. In three different applications, we demonstrate the benefits of NestLink compared to state of the art methods. First, NestLink was used to efficiently rank hundreds of binder candidates in a single experiment according to their off-rates. Yeast display allows rank-ordering of binder off-rates as well14, yet without associating the respective binder sequences. Second, NestLink identified a five times larger number of camelid nanobody families that recognize a membrane transporter as compared to classical phage display and ELISA screening. Third, NestLink proved highly effective in deep-mining a pool for rare binders that recognize an outer membrane protein target in its native cellular context, which could not be identified by ELISA and flow-cytometry screening. We wish to emphasize that binder numbers in NestLink refer to unique binder sequences and are thus not merely analogous to the throughput of screens that process individual binder candidates (e.g ELISA), where rare binders can only be identified upon redundant analysis of many identical clones of enriched binders. Therefore, NestLink is ideally suited to discover binding proteins with unique biological properties, which escape identification by current state of the art methods.
NestLink combines NGS and LC-MS/MS in analogy to recently described methods that were used to identify binders from immune repertoires1-6. However, NestLink overcomes previously inherent detection and peptide assignment limitations, as it benefits from a large, controllable number of unambiguously assignable flycodes per binder, which are engineered for optimal detection efficiencies in LC-MS/MS. This leap forward facilitates binder identification and importantly, it uniquely enables in-depth biophysical characterization of binding proteins within large ensembles.
Recently, methods called SMI-Seq15, ProteinSeq16 and IDUP17 were introduced, which employ barcodes for the screening and analysis of protein-protein and protein-ligand interactions. NestLink differs in two fundamental aspects from these existing barcoding methods. Firstly, it employs peptide barcodes and mass spectrometry, whereas existing methods center on DNA barcodes that are quantified by NGS. Secondly, flycodes are attached at the genetic level in a novel high-throughput ensemble process called library nesting, resulting in protein-peptide fusion proteins. The previously described DNA barcodes must be attached to individual proteins via coupling reactions (Supplementary Table 1).
Remarkably, throughput and cost limitations for NestLink are balanced. Illumina MiSeq has a throughput of approximately 20 Mio reads, suggesting that 400,000 flycodes can be sequenced per run at an average read redundancy of 50-fold. This corresponds to maximally 13,000-20,000 binders per nested library, each coupled to 20-30 flycodes on average. Current LC-MS/MS setups can detect several tens of thousands of peptides, corresponding to a few thousand well detected binders per run. Importantly, LC-MS/MS gains in sensitivity upon reduction of sample complexity. Hence, if an input pool is particularly challenging and contains only a tiny fraction of binders passing the applied NestLink selection pressures, LC-MS/MS detects them particularly well. Notably, there are massive economic incentives to render NGS, LC-MS/MS and gene synthesis even more efficient in the near future, from which the NestLink method will greatly benefit.
We wish to stress, that the presented technological principle is not restricted to binders. Rather, it may be applicable to any protein pool analysis that permits a spatial separation of desired from undesired library members. NestLink-type approaches may for example allow efficient identification of flycode-tagged, thermostable G protein-coupled receptor mutants with favorable SEC elution profiles, which are suitable for in vitro drug screening and structural biology. Further, cell-penetrating peptides may be efficiently selected. Antibodies or enzymes with improved stability and aggregation propensities may be identified by NestLink, thereby improving current methods based on phage particles18, 19.
NestLink interconnects genotype and phenotype of library members in silico by matching NGS and LC-MS/MS data. Thereby, it enables binder selections from ensembles in analogy to classical display procedures, such as phage display. However, NestLink operates in the absence of a physical genotype-phenotype linkage and is thus independent of large display particles. This paradigm shift permits for unprecedented, size-dependent selection pressures as we demonstrated by protein selections entirely in solution without target immobilization shown in application II. Consequently, large display particles are no longer preventing size-dependent characterization of binder pools in tissues or in vivo. Hence, NestLink opens avenues to monitor biodistribution, tissue penetration, immunogenicity or serum half-life for thousands of biopharmaceutical drug candidates at once in a single disease-relevant model organism.
Online Methods
Flycode library design
A random experiment was conducted to simulate and visualize in silico the LC-MS/MS detection characteristics of a large number of flycodes using the R environment and the protViz package (https://CRAN.R-project.org/package=protViz)20. The scripts yield a graphical and numerical output enabling characterization of flycode library dispersity in reversed-phase chromatography and mass spectroscopy. Testing a large number of different sets of input parameters that define the flycode lengths, composition of randomized regions and flanking patterns, an optimal flycode library of the sequence “GSX7WZ0-4R” was identified. “GS” corresponds to the C-terminal remainder of a thrombin cleavage site, which enables proteolytic separation from the library of interest. “X7“ corresponds to 7 amino acid positions that encode the following amino acids at their respective frequencies: A: 18 %; S: 6 %, T: 12 %, N: 1 %, Q: 1 %, D: 11 %, E: 11 %, V: 12 %, L: 2 %, F: 1 %, Y: 4 %, W: 1 %, G: 8 %, P: 12%. The constant amino acid “W” was chosen to increase the overall hydrophobicity to the optimal reverse-phase separation range and since a constant amino acid was required at this position for cloning purposes (BfuAI-site). “Z0-4” corresponds to the 5 different combinations “no amino acid”, “L”, “QS”, “LTV” or “QEGG”. The C-terminal “R” was chosen because i) its guanidine group allows for an optimal positive charge stabilization and ii) as it enables efficient separation of the flycode from its C-terminal His-tag by trypsin.
Overall, the flycode library was designed to achieve an even spread of hydrophobicity covering the entire range of typical reverse-phase chromatographic separation powers and an optimal m/z -dispersity that falls within the ideal detection window of high-field orbitraps. All randomized regions are devoid of positively charged residues (K, R, H), such that the N-terminus and the C-terminal arginine render each flycode a well-defined doubly-charged species, which is detectable in the ideal m/z range. We confirmed this assumption also for the gas-phase experimentally and found that more than 99 % of flycode precursor ions correspond to doubly charge species. The omission of positively charged residues is also critical in order to render trypsin a site-specific protease (removal of His-tag, see previous paragraph). Methionine and cysteines were omitted to minimize oxidation events, such as cross-linking via disulfide bonds. Glutamate and aspartate are frequent within the randomized stretch to achieve high library solubility at neutral pH, while still allowing efficient reverse-phase binding in the absence of the negative charge at pH 2 (LC-MS/MS conditions). The flycode library exhibits a theoretical diversity of 5.3 x 108.
Flycode library generation
The flycode library (Fig. 3a) was generated on the basis of the periplasmic expression vector pSb_init9 by standard molecular biology techniques and is designated pNLx (Supplementary Fig. 13). Five vector variants were constructed, designated pNLx-pre1, pNLx-pre2, pNLx-pre3, pNLx-pre4, pNLx-pre5, each encoding one of the five flycode C-terminal sequences, all non-variable regions of pNLx1-5 and two BfuAI-sites for barcode insertion in between the flycode C-terminus and the N-terminal part of the thrombin-site. The oligonucleotide “C GTC ACA TTA ACC TGC TAC TCA AGA GGT AGT xxx xxx xxx xxx xxx xxx xxx TGG CAA GTG CAG GTA TAG AAA CGT” was synthesized using trinucleotides at positions marked with xxx (ELLA Biotech) and encodes the 7 randomized positions at their respective frequencies (see previous section). The flanking sequences allowed for PCR amplification (30 cycles using Q5 high-fidelity polymerase). Restriction by BfuAI, followed by site-directed insertion into pNLx-pre1, pNLx-pre2, pNLx-pre3, pNLx-pre4, pNLx-pre5 resulted in approximately 2x107 clones per construct. Equal mixing of the five sub-libraries resulted in a library size of approximately 1x108 for pNLx. Analysis of NGS data revealed that there is a close match between theoretical and actual amino acid composition in the 7 randomized positions (Supplementary Fig. 4b). The library was prepared for library nesting by the excision of ccdB, followed by agarose gel purification of the linearized vector backbone and gel extraction (Macherey-Nagel).
Application I: ranking hundreds of off-rates within binder ensembles at once
Library nesting
A pool of sybodies with a convex CDR was used for this experiment, which was previously enriched for MBP-binders by three rounds of ribosome display against MBP, as previously described9. After the third round of ribosome display, the recovered sybody pool was amplified by polymerase chain reaction (PCR) using primers 29 and 30 (Supplementary Table 2) and Q5 Polymerase (NEB) according to the manufacturer’s standard reaction conditions, followed by purification of the PCR product by agarose gel-electrophoresis and gel extraction (Macherey-Nagel). The PCR-amplified sybody pool was sub-cloned via BspQI restriction into the FX cloning vector pINITIAL21 containing a kanamycin resistance cassette. After transformation of E. coli MC1061 cells and plating on agar plates containing 50 µg/ml kanamycin and 1 % (w/v) glucose, the diversity of the pool was restricted to 1,200 – 1,500 cfu by scraping off and cultivation of the appropriate number of colonies in LB containing 50 µg/ml kanamycin and 1 % (w/v) glucose, followed by DNA isolation (Macherey-Nagel). Subsequently, the diversity restricted pool was excised from pINITIAL by BspQI, followed by agarose gel purification and gel extraction using a kit (Macherey-Nagel). The purified, diversity restricted pool and the linearized, purified flycode library (see above) were nested by ligation using 1 µg of pNLx and 700 ng of sybody pool and 10 Weiss Units of T4 ligase (ThermoFischer) in a reaction volume of 40 µl for 1 h at 37°C followed by heat inactivation for 10 min an 65°C. The ligation mix was subsequently transformed in 2 x 150 µl electro-competent E. coli MC1061 cells followed by recovery at 37°C in 25 ml SOC medium for 30 min. A small fraction of the recovered cells were distributed on LB-agar plates containing 25 µg/ml chloramphenicol for diversity estimation and larger fractions of varying sizes were used to inoculate several 250 ml over-night LB cultures containing 25 µg/ml chloramphenicol. Based on the cfu dilution series on plates, an over-night culture was inoculated with approximately 12,000 – 15,000 cfu and used for DNA Midi preparation (Macherey-Nagel) and the production of a glycerol stock for storage at -80°C.
Illumina MiSeq sequencing and flycode assignment
The Illumina MiSeq NGS template was prepared as follows: 100 units of SfiI (NEB) were used to digest 25 µg of the prepared pNLx (containing the nested library) in a reaction volume of 200 µl at 50°C for 1.5 h followed by SfiI inactivation by addition of 8 µl of 0.5 M EDTA. The excised linear nested library was isolated by agarose gel purification followed by gel extraction (Macherey-Nagel). Subsequently, 2x332 ng of double-stranded Illumina adaptor oligonucleotides containing compatible sticky ends (previously generated by DNA-synthesis, Supplementary Table 2) were site-specifically ligated to both ends of the linearized nested library (600 ng), thereby avoiding PCR amplification (Supplementary Fig. 3). The ligation product containing two adaptors attached was isolated by agarose gel purification, followed by gel extraction (Macherey-Nagel). The concentration of the ligation product was determined by a NanoDrop 2000c Photospectrometer (Thermo Scientific). The ligation product was mixed with differentially indexed ligation products (originating from unrelated experiments for multiplexed Illumina analysis), to obtain an approximate read redundancy of 50 per flycode for each index, aiming for a total of 10 Mio reads. The molarity of the NGS template mixture was subsequently confirmed using the Tapestation 2200 (Agilent) and adjusted to 4 nM. HT1 hybridization buffer was subsequently used to further dilute the library pool. To generate the sample for clustering, 420 µl of the library at 8 pM was mixed to 180 µl of PHiX (Illumina) at 12.5 pM. The sample was sequenced using a 600cycle v3 Miseq reagent kit for 2 x 300 bp paired-end reads on an Illumina MiSeq Sequencer. 8.4 Gb of data were obtained from a single run with >98% reads passing filters, i.e. 14 Mio passed filter reads that had a mean quality score of 35.
For the relevant index of this experiment, 729,932 raw read pairs were obtained and subsequently preprocessed using Trimmomatic (v0.33, parameters: AVGQUAL:20 MINLEN:100) and Flexbar (v2.5, parameters: --pre-trim-left 4 --pre-trim-right 4). 690,066 high quality read pairs were combined using Flash (v1.2.11, default parameters). 618,049 combined reads were obtained, followed by filtering for read length (611,320 reads were ±5% around observed median read length), for flanking patterns of the flycodes (603,488), for flanking patterns of the sybodies (603,084), for reads without N’s (586,643), for the expected construct lengths (492,580), for sequences without stop codons (484,586) and for sequences with correct flycode endings (482,305). The number of unique flycodes was subsequently determined to be 13,620 (minimum of 4 reads per flycode, uniqueness is defined at the level of identical amino acid sequence). For each of the 13,620 unique flycodes, a consensus of all binders linked to the same flycode sequence (35 binder sequences on average, due to 35-fold read redundancy) was formed at the amino acid (aa) level and scored for filtering. For each aa position of a binder, the relative fraction of the most frequent aa was calculated as follows: #most frequent aa/(#most frequent aa + #second most frequent aa). The consensus score of a binder corresponds to the average relative fraction over all its amino acid positions. Removing flycodes with a consensus score below 0.9 resulted in 12,160 unique flycodes passing the filter, which were linked to 1,070 distinct binders. Hence, flycodes served as unique molecular identifiers (UMIs) to support accurate sequence determination by NGS22, 23. On average, 451 sequences (passing all filtering criteria) were obtained per unique binder. An end-pairing overlap of 62 – 68 bp (depending on the flycode length) allowed acquisition of full-length sybody sequences and not merely CDRs. Based on the NGS analysis, a database for MS/MS ion searches (p1875_db8 (release 2016-07-11) was constructed, which assigns each unique binder sequence (identifier) to a virtual protein consisting of its concatenated unique flycodes (Supplementary Fig. 2).
Expression of nested library and selections
In order to express the nested library in the vector pNLx in E. coli MC1061, the previously generated glycerol stock (see above) was used to inoculate a 37°C overnight pre-culture containing LB and 1 % (w/v) glucose. 2 x 12 ml of saturated pre-culture was used to inoculate 2 x 600 ml of TB containing 25 µg/ml chloramphenicol, followed by induction at an OD600 of 0.6 using 0.05 % (w/v) arabinose for 14 h at 20°C. Cells were harvested by spinning at 5,000 g for 15 min, followed by resuspension in 60 ml of TBS (20 mM Tris-HCl pH 7.5, 150 mM NaCl), 10 mM imidazole pH 8 and a spatula tip of DNase1. The resuspended cells were disrupted using a microfluidizer processor (Microfluidics) at 30,000 lb/in2 and the debris was pelleted by centrifugation at 4,400 g for 30 min. The supernatant was loaded on a 1.5 ml Ni-NTA column (Qiagen), the column was washed by 30 ml of TBS containing 30 mM imidazole pH 8, followed by 3 x 2 ml elution using TBS containing 300 mM imidazole pH 8. The eluted nested library was filtered (0.2 µm syringe filter) and was subjected to a SEC run on an Aekta Purifier (GEHealthcare) system using a HiLoad 16/600 Superdex 200 pg (GE-Healthcare). The nested library members, corresponding to the monomeric binders, were collected and concentrated using centrifugal filters with a 3 kDa cut-off (Amicon Ultra-15) to an absorbance at 280 nm (A280) of 2.0. Please note that the exact binder frequencies within a nested library are unknown and therefore the exact protein concentration can only be estimated. Assuming an average extinction coefficient of 30,000 M-1cm-1 for an average sybody/nanobody-flycode fusion protein, the protein concentration can be estimated using the following equation: Protein conc [M] = A280/30,000. The biotinylated target MBP-biotin at a concentration of 204 µM was prepared as previously described9. Three analogous SEC runs were performed in TBS on a Superdex 200 10/300 (GE-Healthcare). The first sample contained 175 µl of the nested library and 60 µl TBS, the second sample contained 175 µl of the nested library and 60 µl of MBP-biotin and the third sample contained 175 µl TBS and 60 µl of MBP-biotin. Superposition of the three runs allowed collecting those early eluting fractions (3 ml total) of the second run, which contained the library members interacting with MBP in solution. The fractions were split into two equivalent 1.5 ml fractions and 150 µl of streptdavidin-sepharose slurry (Thermo Scientific) was added to each fraction, followed by incubation at 4°C for 2 h. The resins were pelleted by centrifugation (swinging-bucket) for 10 min at 200 g and transferred to two Mini Bio-Spin® Chromatography Columns (Bio-Rad: #732-6207). The columns were drained by centrifugation at 50 g for 5 sec in a table-top centrifuge. Column 1: The resin (75 ul) was resuspended by the addition of 500 µl of TBS containing 10 µM non-biotinylated MBP and incubated for 195 seconds (off-rate selection), followed by draining (5 sec at 50 g). Column 2: was not challenged by MBP but otherwise treated identical to column 1. Both columns were washed immediately by 500 µl TBS.
Flycode isolation and LC-MS/MS
5 µl of a control binder attached to 28 different flycodes of known sequence (NB-control, see below) at an absorbance of 0.05 (280 nm) was added to both columns as an LC-MS/MS standard. The resins were resuspended in 100 µl of buffer TH (20 mM Triethylammonium bicarbonate (TEAB) pH 8.5, 150 mM NaCl, 2.5 mM CaCl2) containing 2.4 units of thrombin (Novagen, #69671-3) and incubated over night at 20°C. The His-tagged flycodes were eluted by centrifugation at 100 g for 10 sec, followed by washing with 2 x 300 µl buffer TH. The flycodes were subsequently pulled down by incubation for 1 h at 20°C with 80 µl Ni-NTA slurry (Qiagen), followed by centrifugation at 800 g for 5 min. The resins were transferred to Mini Bio-Spin® Chromatography Columns (Bio-Rad), washed with 500 µl of buffer TRY (50 mM TEAB pH 8.5, 50 mM NaCl, 2.5 mM CaCl2) and subsequently resuspended in 100 µl buffer TRY containing 0.5 µg trypsin (Promega, #V5113), followed by incubation over night at 37°C. The flycode mixture (severed from His-tags) was eluted from the columns by centrifugation for 30 sec at 100 g, followed by a 100 µl wash (50 mM TEAB pH 8.5, 150 mM NaCl) and addition of 20 µl of 5 % (v/v) TFA and 200 µl of 3 % (v/v) ACN, 0.1 % (v/v) TFA to the elution. The eluted flycode mixture was loaded onto ZipTips (Millipore, #ZTC185960) pre-treated by washing with 200 µl of 60 % (v/v) acetonitrile (ACN), 0.1 % (v/v) trifluoroacetic acid (TFA), 200 µl of methanol and 200 µl of 3 % (v/v) ACN, 0.1 % (v/v) TFA. The ZipTips were washed by 200 µl of 3 % (v/v) ACN, 0.1 % (v/v) TFA, followed by elution with 2 x 40 µl of 60 % (v/v) ACN, 0.1 % (v/v) TFA and lyophilization of the elution and resolubilization of the flycodes in 15 µl of 3 % (v/v) ACN, 0.1 % (v/v) formic acid (FA). For application I, flycodes were analyzed using an Easy-nLC 1000 HPLC system operating in single column mode coupled to an Orbitrap Fusion mass spectrometer (Thermo Scientific). 2 µl of the resuspended flycode solution was injected onto an in-house made capillary column packed with reverse-phase material (ReproSil-Pur 120 C18-AQ, 1.9 µm; column dimension 150 mm x 0.075 mm, Temp. 50°C). The column was equilibrated with solvent A (0.1 % formic acid (FA) in water) and peptides were eluted with a flow rate of 0.3 µl/min using the following gradient: 5 - 20 % solvent B (0.1 % FA in ACN) in 60 min, 20 - 97 % solvent B in 10 min. High accuracy mass spectra were acquired with an Orbitrap Fusion mass spectrometer (Thermo Scientific) using the following parameter: scan range of 300-1500 m/z, AGC-target of 5e5, resolution of 120,000 (at m/z 200), and a maximum injection time of 100 ms. Data-dependent MS/MS spectra were recorded in rapid scan mode in the linear ion trap using quadrupole isolation (1.6 m/z window), AGC target of 1e4, 35 ms maximum injection time, HCD-fragmentation with 30 % collision energy, a maximum cycle time of 3 sec, and all available parallelizable time was enabled. Mono isotopic precursor signals were selected for MS/MS with charge states between 2 and 6 and a minimum signal intensity of 5e4. Dynamic exclusion was set to 25 sec and an exclusion window of 10 ppm. After data collection peak lists were generated using automated rule based converter control24 and Proteome Discoverer 1.4 (Thermo Scientific).
LC-MS/MS data analysis
The two LC-MS/MS runs were aligned in Progenesis QI (Nonlinear Dynamics) with an alignment score of 93.1 %, followed by peak picking with an allowed ion charge of +2 to +5. The fragment spectra with a feature rank-threshold of <5 were exported using deisotoping, charge deconvolution and an ion fragment count limit of 1,000. Mascot 2.5 (Matrix Science) was used for flycode identification by a search against database p1875_db8 (release 2016-07-11, generated as described above) concatenated with an in-house built contaminant database (262 common contaminants). Precursor ion mass tolerance was set to 10 ppm and the fragment ion mass tolerance was set to 0.5 Da. In addition, Scaffold (version Scaffold_4.8.4, Proteome Software Inc.) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were filtered for FDR less than 0.1% by the Peptide Prophet algorithm25 and protein identifications were filtered for FDR less than 1.0% containing at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm26. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters. Scaffold spectrum report was imported into Progenesis QI. The two LC-MS/MS runs were normalized against the spiked reference NB-control (see below) by choosing NB-control as a standard protein (normalization factor = 0.81). The MS1 intensity integrals of all non-conflicting flycode features were summed for each binder. We refer to this sum as “binder abundance”. The ratios between the binder abundances at the two columns were plotted for each individual sybody (Fig. 4b, y-axis).
Single-clone verification by SPR
Based on the NestLink data (Fig. 4b), several sybody genes were chosen that appeared to exhibit different interaction strengths according to the off-rate selection analysis. All chosen genes correspond to sybodies that were detected with at least 2 unique flycodes on the columns (112 passed this filter in total). The sybody genes were synthesized (General Biosystems) and subcloned into pSb_init, followed by expression and purification analogous to the nested library, the only difference being supplementation of the SEC buffer by 0.05 % (v/v) Tween-20. Off-rates were determined in this buffer using a ProteOn™ XPR36 Protein Interaction Array System (Bio-Rad) using biotinylated MBP immobilized on a ProteOn™ NLC Sensor Chip to 1,000 response units (RU). 5 different dilutions of the purified sybodies were applied to the chip for 245 seconds (association phase), followed by dissociation phases of varying lengths. The off-rates were derived from Langmuir fits. Off-rates (x-axis) were plotted against the fractions remaining on the columns as determined by NestLink (y-axis). The data were fitted using equation y = y0 × e-ax, where y corresponds to the fraction remaining, y0 corresponds to the fraction remaining at off-rate of 0 s-1, a is a fitting variable corresponding to the washing time and x corresponds to the off-rate. R2 of the fit was 0.96.
Application II: diversity mining of camelid nanobodies
Immunization of alpaca, phage library preparation and phage display
An alpaca was immunized four times with subcutaneous injections at two week intervals, each time with 200 µg purified TM287/28810 in TBS pH 7.5 containing 0.03% β-DDM. Three additional subcutaneous injections of 200 µg protein were performed at two week intervals. Immunizations of alpacas were approved by the Cantonal Veterinary Office in Zurich, Switzerland (animal experiment licence nr. 188/2011). The nanobody repertoire of the immunized animal served as input for phage display27 and in a separate experiment for NestLink (described in the following paragraphs). Phage display against biotinylated TM287/288 and ELISA screening were performed as previously described9. Of note, phage ELISA revealed that already the non-selected input phage stock exhibited a measurable signal against TM287/288, hence the immune response of this animal was very strong against this antigen.
Library nesting, NGS, expression and purification of nested library
After phage library construction from the B-lymphocytes of the immunized alpaca (without performing phage display), the single-stranded nanobody library was amplified by PCR using Alp-Nb_FX_FW_81 and Alp-Nb_FX_REV_82 (Supplementary Table 2) and GoTaq G2 DNA polymerase (Promega) according to the manufacturer’s standard reaction conditions, followed by purification of the PCR product by agarose gel-electrophoresis and gel extraction using a kit (Macherey-Nagel). The PCR-amplified nanobody pool was subcloned via BspQI restriction into pINITIAL. Nesting of the nanobody pool with the flycode library was performed as described for application I, but using 3,000 – 4,000 cfu of the nanobody pool (pINITIAL) and 60,000 – 80,000 cfu of pNLx after nesting. NGS was performed as described for application I, but with a consensus score cut-off of 0.99. After filtering 59,974 flycodes linked to 3,390 unique nanobodies were obtained, which were used for the generation of the flycode assignment table (p1875_db10 (release 2017-08-18)).
The nested library was expressed and purified as described above, but using 1.5-fold the culture size and two instead of one 1.5 ml Ni-NTA (Qiagen) columns with all buffer volumes adjusted accordingly. Two runs of SEC (HiLoad 16/600 Superdex 200 pg (GE-Healthcare)) were performed to isolate monomeric nested library members, yielding 20 ml solution at an approximate nanobody concentration of 22 µM, assuming an average molar extinction coefficient of the nested library of 30,000 M-1cm-1.
Pool-internal competition binding experiment
Complex formation using the purified nested library and TM287/288 was performed at three different molar ratios of I) 1:2, II) 31:1 and III) 163:1 in 500 µl TBS containing 0.03 % DDM for 1 h at 4°C. The nested library members that bound to TM287/288 in solution were isolated via separate SEC runs (Superdex 200 10/300 increase (GE-Healthcare)) for the three different molar ratios by collecting the appropriate fractions at the elution volume corresponding to the nanobody-TM287/288 complex. Analogously, three additional SEC runs were performed, each analyzing the purified nested library at one of the three quantities that was used for complex formation (described above) but in the absence of the target. For these background runs, the same fractions (as in the runs with the target) at early elution volumes were collected. Nested library members collected in these background runs represent nanobodies that elute at early elution volumes independent of the target.
Flycode isolation, LC-MS/MS and data analysis
Flycodes were individually isolated from 7 different samples: 1) from the purified nested library (200 µl of the monomeric nested library members), 2-4) from the SEC-fractions corresponding to target-nanobody complexes (3 ml of each of the three SEC runs) and 5-7) from the three background SEC runs (3 ml of each of the three SEC runs were collected at the same elution volumes as for the runs isolating target-nanobody complexes). Each sample was spiked with 7 µl of a control binder with 28 known flycodes (NB-control, see below) at an absorbance at 280 nm of 0.052. The 200 µl sample of the purified nested library was diluted to 1.2 ml by TBS for further processing. 100 µl slurry Ni-NTA (Quiagen) was added to each of the 7 different samples, followed by incubation for 2 h at 4°C and pelleting of the resin by centrifugation at 500 g for 5 min. The resins were transferred to Mini Bio-Spin® Chromatography Columns (Bio-Rad: #732-6207) and washed 2 x by 700 µl of buffer Iso (30 % (v/v) isopropanol, 20 mM TEAB, 5 mM imidazole), followed by 3 x 700 µl of buffer TH. The resin was resupsended in 100 µl buffer TH containing 2.4 units of thrombin (Novagen, #69671-3) and incubated over night at room temperature. Subsequently, the resin was washed 5 x by 700 µl buffer TRY containing 10 mM imidazole, followed by elution of the His-tagged flycodes by 2 x 50 µl buffer TRY containing 250 mM imidazole. The eluate was spun through a Microcon filter YM-10 (Amicon, #42407) with a 10 kDa cutoff at 14,000 g at RT. The elution and filtration procedure was repeated by another 2 x 50 µl of the same buffer. Subsequently, 1 µg of trypsin (Promega, #V5113) was added to the flow-through, followed by incubation over night at 37°C. 20 µl of 5 % (v/v) TFA were added to stop the enzymatic digest and the sample was further diluted by addition of 200 µl of 3 % (v/v) ACN, 0.1 % (v/v) TFA. The 7 flycode mixtures were processed by ZipTips (Millipore, #ZTC185960) as described above for application I and analyzed by LC-MS/MS. For application II, flycodes were analyzed by an Easy-nLC 1000 HPLC system operating in trap / elute mode (trap column: Acclaim PepMap 100 C18, 3 µm, 100A, 0.075x20 mm; separation column: EASY-Spray C18, 2 µm, 100A, 0.075x500 mm, Temp: 50°C) coupled to an Orbitrap Fusion mass spectrometer (Thermo Scientific). Trap and separation column were equilibrated with 12 µl and 6 µl solvent A (0.1% FA in water), respectively. 2 µl of the resuspended flycode solution was injected onto the trap column at constant pressure (500 bar) and peptides were eluted with a flow rate of 0.3 µl/min using the following gradient: 5 - 20 % B (0.1 % FA in ACN) in 60 min, 20 - 97 % B in 10 min. High accuracy mass spectra were acquired with an Orbitrap Fusion mass spectrometer (Thermo Scientific) using the following parameter: scan range of 300-1500 m/z, AGC-target of 5e5, resolution of 120,000 (at m/z 200), and a maximum injection time of 100 ms. Data-dependent MS/MS spectra were recorded in the linear ion trap using quadrupole isolation (1.6 m/z window), AGC target of 1e4, 35 ms maximum injection time, HCD-fragmentation with 30 % collision energy, a maximum cycle time of 3 sec, and all available parallelizable time was enabled. Mono isotopic precursor signals were selected for MS/MS with charge states between 2 and 6 and a minimum signal intensity of 5e4. Dynamic exclusion was set to 25 sec and an exclusion window of 10 ppm was used. After data collection, peak lists were generated using automated rule based converter control24 and Proteome Discoverer 1.4 (Thermo Scientific). Two technical replicates were recorded for each sample (total of 14 LC-MS/MS runs).
Two alignments were generated using Progenesis QI: 1) the two LC-MS/MS replicates of the sample representing the purified, monomeric nested library members (alignment score of 94.5 %) and 2) LC-MS/MS runs of the samples corresponding to the collected fractions of the pool-internal competition experiments or their respective background runs (alignment scores between 69.0 and 97.6 %). Note that aligning LC-MS/MS runs was not per se necessary for the NestLink data analysis performed here, but it allowed parallel workflows for similar recordings and thus facilitated data processing in Progenesis QI. Peak picking, peptide filtering and peptide exporting was performed as described for application I (see above). Mascot 2.5 (Matrix Science) was used for flycode identification by two searches (one search for each alignment) against 3 databases per search i) p1875_db10 (release 2017-08-18, generated as described above), ii) p1875_db8 (release 2016-07-11) both have been concatenated with an in-house built contaminant database, and iii) Swissprot database (release 20140403) concatenated with its decoyed entries. Mascot search parameters and processing in Scaffold were analogous to application I (described above). After re-import into Progenesis QI, the LC-MS/MS runs were normalized using the spiked standard NB-control. A normalization factor of 1.00 was obtained for the first alignment and factors between 0.51 – 1.15 were obtained for the second alignment. Note that, normalization was not essential for the analysis performed here, but it served as a control, since extreme normalization factors would hint at inconsistencies in sample preparation. The MS1 intensity integrals of all non-conflicting flycode features were summed for each nanobody in each sample (binder abundance). Binder abundances were averaged between the two technical LC-MS/MS replicates per sample (see above). The “relative abundance” corresponds to the fraction of an individual nanobody abundance relative to the total of all nanobody abundances detected in the same LC-MS/MS run (100 %), thus calculating the relative abundance corresponds to a sample-internal normalization.
Nanobody sequences exhibiting an increase in relative abundance in the following order, 1) purified nested library (input pool), 2) SECI, 3) SEC I, 4) SECIII (target-bound fractions for 2)-4)) and at least 4 detected flycodes on SEC were considered as strong binder hits. Their sequences were extracted from the NGS database and their CDR3 regions were aligned using the alignment tool of the software CLC (Qiagen), followed by editing in Jalview (Fig. 5c). Only nanobodies exhibiting more than 10-fold higher binder abundances in the complex runs compared to the same “shifted” fraction of the background runs (no target, see above), were included in the analysis.
Single-clone verification by SPR
Genes of 11 “binding-nanobodies” (see above alignment) and 4 “negative-control-nanobodies” (detected in the purified nested library, but not at the target) were synthesized (General Biosystems) and subcloned into pSb_init, followed by expression and purification analogous to the nested library. Binding kinetics were determined using a ProteOn™ XPR36 Protein Interaction Array System (Bio-Rad) using biotinylated TM287/288 immobilized on a ProteOn™ NLC Sensor Chip to 1,500 response units (RU) and TBS supplemented with 0.03 % (w/v) DDM. An initial SPR screen was performed at a single concentration of 100 nM for each nanobody. For the “binding-nanobodies”, this screen revealed that two nanobodies (NL2.1 and NL11.1) exhibited off-rates that were too slow to be determined by the ProteOn™ (< 5E-5 s-1) and two nanobodies exhibited significantly higher dissociation constants than 100 nM (NL1.3 and NL7.1). From the 11 purified “binding-nanobodies”, 7 were therefore used for accurate determination of kinetic parameters. To this end, 5 different dilutions of the purified nanobodies were applied to the chip for 245 seconds (association phase), followed by dissociation phases of varying lengths. The data were fitted using the Langmuir method. None of the 4 “negative-control-nanobodies” exhibited a binding signal.
Application III: membrane protein binder identification in the cellular context
Purification of the major outer membrane protein (MOMP) from L. pneumophila serogroup 6
L. pneumophila serogroup 6 strain DSM25182 was grown at 37 °C on BCYE agar (BBL BCYE Agar Base, BD). Single colonies were inoculated in 5 ml liquid BCYE media (Legionella BCYE Growth Supplement, VWR) and grown to stationary phase by shaking overnight at 37 °C. 0.5 ml of the densely grown BYE pre-culture was used to inoculate 500 ml cultures of liquid BYE media (10 g/l yeast extract, 0.25 g/l ferric pyrophosphate, 1 g/l α-ketoglutarate, 0.4 g/l L-cysteine, 7.2 g/l ACES buffer adjusted to pH 6.9) and grown to an OD600 of 0.9. Bacteria were harvested by centrifugation at 4,000 g for 10 min, washed in PBS and centrifuged again at 4,000 g for 10 min.
The MOMP protein was purified from a total of 8 liters of BYE culture according to Gabay et al.28. Briefly, the harvested bacteria were resuspended in lysis buffer (0.1 M sodium acetate, pH 4, 0.45 M CaCl2, 0.45 % Zwittergent 3-14, 10 mM beta-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride), sonicated for 30 s in a sonicating water bath (Elmasonic P) and cooled at 0 °C. Ice-cold absolute ethanol was added dropwise to a final concentration of 20 % ethanol (v/v) and the mixture was stirred for 30 min at room temperature. The preparation was centrifuged at 17,000 g for 10 min and the supernatant was discarded. The pellet was suspended again in lysis buffer and the mixture was sonicated for 30 s using a tip (Branson Sonifier B12), treated with ethanol, and centrifuged at 17,000 g for 10 min. The supernatant, containing MOMP, was collected, treated with ice-cold absolute ethanol to a final concentration of 75 % (v/v) to precipitate proteins, incubated over night at -20 °C and centrifuged at 20,000 g for 35 min. The pellets were suspended in 50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 0.5 % Zwittergent 3-14 and centrifuged at 20,000 g for 35 min to remove insoluble protein. The sample was applied onto two HiTrap FF DEAE columns (GE Healthcare) that were connected in a row and equilibrated with Buffer A (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 0.05 % Zwittergent 3-14). Bound protein was eluted by applying a 50 ml salt gradient of 0.13 M to 1 M NaCl with Buffer B (50 mM Tris-HCl, pH 8.0, 1 M NaCl, 10 mM EDTA, 0.05 % Zwittergent 3-14) on an Aekta Prime. Elution fractions containing MOMP were pooled and treated with ice-cold absolute ethanol to a final concentration of 75 % (v/v), incubated over night at -20 °C and centrifuged at 20,000 g for 35 min to collect precipitated proteins. The pellet was suspended in a minimal volume of 50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 0.5 % Zwittergent 3-14 and centrifuged at 20,000 g for 35 min before injection onto an S200 10/300 (GE-Healthcare) equilibrated with 10 mM Tris, pH 8, 200 mM NaCl, 10 mM EDTA, 0.05 % Zwittergent 3-14. Eluted fractions were analyzed by SDS-PAGE and fractions containing MOMP were pooled, flash-frozen and stored at -80 °C.
Sybody selections against detergent-purified MOMP of Lp-SG6
Purified MOMP of LP-SG6 was biotinylated by EZ-Link™ Sulfo-NHS-LC-LC-Biotin (Thermo Fischer # 21338) at a molar ratio of 1:2.5 at 4°C overnight. Free biotin was removed by SEC in TBS containing 0.03% DDM. The enriched sybody pool was generated using a synthetic nanbody library with a convex CDR3 region9. Briefly, sybodies were selected by performing one round of ribosome display followed by two rounds of phage display. qPCR revealed a 1.5 fold enrichment of the convex Sybody pool against MOMP compared to AcrB as negative control. The enriched sybody pool was subcloned into the pSb_init and single clones were picked for small scale expression and ELISA. ELISA revealed approximately 50 % positive hits. 12 % of the hits exhibited specific ELISA signals for detergent solubilized MOMP and showed only background signals against the negative control AcrB.
Library nesting, NGS, expression and purification of nested library
Library nesting was performed as described for application I. 1,400 – 1,700 cfu and 20,000 – 26,000 cfu were chosen for diversity restriction of sybodies and nested library members, respectively. A consensus score cut-off of 0.99 was used for NGS data filtering analogous to application II. This resulted in a nested library covering 1,444 unique sybodies and 23,598 unique flycodes (database p1875_db9 (release 2017-01-05)). The nested library was expressed and purified as described for application I. Monomeric nested library members (input for pull-down experiment) were selected by a SEC run on an Aekta Purifier (GE-Healthcare) system using a HiLoad 16/600 Superdex 200 pg (GE-Healthcare).
Selection for cell-surface binders by a pull-down experiment
4 x 3 ml of the monomeric nested library members (eluted from SEC at an estimated concentration of 30 µM) were added to 4 individual test tubes each containing 1 ml of either Lp-SG6, Escherichia coli, Citrobacter freundii or Lp-SG3 at an OD600 of 50 in TBS at pH 7.5 supplemented with 0.5 % BSA. All subsequent steps, including LC-MS/MS were carried out independently for the 4 samples. After incubation for 5 min, cells were pelleted by centrifugation at 4,000 g for 10 min, followed by resuspension of the cells in 5 ml PBS at pH 7.5. Pelleting and resuspension was repeated twice to remove low affinity sybodies.
Flycode isolation, LC-MS/MS and data analysis
The pelleted cells were resuspended in 5ml of 100 mM Tris/HCl (pH 7.5), 750 mM NaCl, 2 % (w/v) n-octyl-β-D-glucopyranoside, 50 mM imidazole pH 8.0 containing a pinch of DNaseI and approximately 1 µg of a control binder with 28 known flycodes (NB-control, see below). After 10 min incubation, 20 ml of 6 M GdmCl were added, followed by incubation for 20 min at 20°C. Insoluble components were pelleted by spinning at 4,400 g for 30 min. The supernatant was filtered (0.2 µm syringe filter), followed by the addition of 100 µl slurry Ni-NTA (Qiagen) to the supernatant of each sample and incubation for 2 h at 4°C. The resin was pelleted by centrifugation at 1,500 g for 30 min. The resin was transferred to Mini Bio-Spin® Chromatography Columns (Bio-Rad) for subsequent flycode isolation analogous to application II (see above). For application III, flycodes were analyzed in dublicate by a Waters M-class UPLC system (Waters AG) operating in trap/elute mode coupled to a Q-Exactive HF mass spectrometer (Thermo Scientific). The LC-system were equilibrated with 99% solvent A (0.1% formic acid (FA) in water) and 1% solvent B (0.1% FA in ACN). Trapping of peptides was performed on a Symmetry C18 trap column (5 µm, 75 µm X 250 mm, Waters AG) at 15 µl/min for 30 sec. Subsequently, the peptides were separated using a HSS T3 C18 reverse-phase column (1.8 µm, 75 µm X 250 mm, Waters AG) and the following gradient: 1-40% B in 60 min; 40-98% B in 5 min. The flow rate was constant 0.3 µl/min and the temperature was controlled at 50°C. High accuracy mass spectra were acquired with a Q-Exactive HF mass spectrometer (Thermo Scientific) that was operated in data dependent acquisition mode. A survey scan was followed by up to 12 MS2 scans. The survey scan was recorded using quadrupole transmission in the mass range of 350-1500 m/z with an AGC target of 3E6, a resolution of 120,000 at 200 m/z and a maximum injection time of 50 ms. All fragment mass spectra were recorded with a resolution of 30,000 at 200 m/z, using quadrupole isolation (1.2 m/z window), an AGC target value of 1E5 and a maximum injection time of 50 ms. The normalized collision energy was set to 28%. Dynamic exclusion was activated and set to 30 sec with a mass tolerance of 10 ppm. After data collection, peak lists were generated using automated rule based converter control24 and Proteome Discoverer 1.4 (Thermo Scientific).
Using Progenesis QI, the 8 LC-MS/MS runs of the 4 pull-down samples (2 replicates) were aligned and analyzed as described for application II. Alignment scores of 86.1 - 98.6 % were obtained. Note that aligning LC-MS/MS runs was not per se necessary for the NestLink data analysis performed in application II, but it allowed parallel workflows for similar recordings and thus facilitated data processing in Progenesis QI. Peak picking, peptide filtering and peptide exporting was performed as described for application I (see above). Mascot 2.5 (Matrix Science) was used for flycode identification by a search against database p1875_db9 (release 2017-01-05, generated as described above) concatenated with an in-house built contaminant database. Mascot search parameters and processing in Scaffold were analogous to application I (described above). After re-import into Progenesis QI, the LC-MS/MS runs were normalized using the spiked standard NB-control (normalization factors between 0.97 and 2.34 were obtained). Note that normalization was not essential for the analysis performed here, but it served as a control, since extreme normalization factors would hint at inconsistencies in sample preparation. In analogy to application II, the MS1 intensity integrals of all non-conflicting flycode features were summed for each sybody (binder abundance). The binder abundances were averaged between the two technical LC-MS/MS replicates and each sample was internally normalized by calculating the relative abundance for each sybody.
Single-clone verification by flow-cytometry
5 sybodies exhibiting flycode coverages of more than 20 %, more than 5 unique flycodes detected and 12 – 100 fold higher relative abundances at Lp-SG6 than at any other strain, were chosen for single-clone analysis by flow-cytometry. To this end, the identified sybody genes were synthesized, expressed and purified as described for application I. Subsequently, the sybodies were labelled in the presence of a 1.2 fold molar ratio of AlexaFluor 488-NHS (Alexa488-NHS) and the free dye was removed by dialysis (6,000-8,000 MWCO, Spectra/Por®). Coupling efficiencies were calculated from the absorbance at 280 nm and 488 nm, respectively. The average number of dyes per sybody molecule ranged from 0.8 to 1.1.
For the single-clone verification of cell-surface binding to whole Legionella by flow-cytometry, 14 different Legionella pneumophila serogroups and 50 additional bacterial strains were fixed by glutaraldehyde treatment (strains are listed in Supplementary Fig. 12). To this end, Legionella pneumophila strains were grown in buffered yeast extract (BYE) broth (10 g/L yeast extract, supplemented with Legionella BCYE Growth Supplement from VWR) by shaking at 37 °C. Other bacteria were grown in liquid media according to the strain provider’s specifications (DSMZ, NCTC or ATCC). The bacterial cells were washed three times by centrifugation for 10 min at 4,226 g and resuspension in 20 ml PBS per 200 ml bacteria culture. After the last centrifugation, the cells were resuspended in 10 ml PBS containing 2.5 % glutaraldehyde, vortexed and incubated for two hours at room temperature in the dark. The fixed cells were then washed three times in PBS as described above.
Fixed bacterial strains at a concentration of 100,000 cells/ml were incubated with 0.5 µg/ml of Alexa488 labelled single-clone sybody and 0.5 µg/ml propidium iodide for one hour at room temperature. To test for cell-surface binding to bacterial cells, the samples were analyzed by flow-cytometry using a CytoFLEX (Beckman Coulter), equipped with a 488 nm laser and filter sets of 525/40 (green channel) and 690/50 (red channel). To reduce background noise, a threshold of 400 was used on the green channel and a threshold of 550 was used on the red channel. The analyses were performed at a flow rate of 100 µl/min.
Production of control binder for LC-MS/MS run normalization
Using SapI restriction and ligation, a clone of a convex sybody (Sb_MBP#1) encoded in the vector pINITAL was inserted in the flycode library vector via replacement of the negative selection marker ccdB. After transformation of E. coli MC1061 cells and plating on agar plates containing 25 µg/ml chloramphenicol and 1 % (w/v) glucose, 28 cfu were picked for cultivation as a pool in LB containing 25 µg/ml chloramphenicol and 1 % (w/v) glucose, followed by DNA isolation (Macherey-Nagel) and glycerol stock production. The sample was processed by NGS as described above to determine the sequences of the 28 flycodes. The identified flycodes linked to Sb_MBP#1 were concatenated and formatted as an entry of a mascot search database, appropriate for manual addition to any other NestLink database. Sb_MBP#1 linked to its flycodes was expressed in and purified from E. coli MC1061 as described for nested libraries (see application I).
Supplementary Material
Acknowledgments
We thank Olga Schubert, Roger Dawson and Eric Geertsma for their helpful comments on the manuscript and Saša Štefanić for alpaca immunizations. The authors acknowledge the CRAN and Bioconductor Core teams and in particular Lori Shepherd for making the NestLink package available through Bioconductor. This work was funded by a grant of the Commission for Technology and Innovation CTI (16003.1 PFLS-LS, to MAS), a SNSF Professorship of the Swiss National Science Foundation (PP00P3_144823, to MAS), a SNSF BRIDGE proof of concept grant (20B1-1_175192, to PE) and a BioEntrepreneur-Fellowship of the University of Zurich (BIOEF-17-002, to IZ).
Footnotes
Author Contributions
PE, IZ and MAS developed the conceptual basis of NestLink. PE, BR, CP and MAS designed the flycode library and PE and IZ generated it. PE performed library nesting and selections. NGS was carried out by LO, LP and PE. IZ, FMA, CAJH, DM, HAK and PE performed single-clone analyses via SPR or flow-cytometry. LC-MS/MS was performed and the data was analyzed by PE, BR and CP. PE and MAS wrote the manuscript.
Competing Financial Interests Statement
A patent seeking to protect the NestLink technology has been applied (PCT/EP2017/077816). PE, IZ and MAS are listed as inventors.
Data availability
Mass spectrometry data are available via ProteomeXchange with identifier PXD009301. NGS datasets are available on the European Nucleotide Archive (ENA) under accession number PRJEB25673. The NGS and MS data were handled and annotated using the B-Fabric information management system29 and are available for registered users under project identifiers 1644 and 1875.
Code availability
The custom software used to design the flycode library and to filter and analyze NGS data is available through https://github.com/cpanse/NestLink.
References
- 1.Fridy PC, et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat Methods. 2014;11:1253–1260. doi: 10.1038/nmeth.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheung WC, et al. A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat Biotechnol. 2012;30:447–452. doi: 10.1038/nbt.2167. [DOI] [PubMed] [Google Scholar]
- 3.Sato S, et al. Proteomics-directed cloning of circulating antiviral human monoclonal antibodies. Nat Biotechnol. 2012;30:1039–1043. doi: 10.1038/nbt.2406. [DOI] [PubMed] [Google Scholar]
- 4.Wine Y, et al. Molecular deconvolution of the monoclonal antibodies that comprise the polyclonal serum response. Proc Natl Acad Sci U S A. 2013;110:2993–2998. doi: 10.1073/pnas.1213737110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lavinder JJ, et al. Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc Natl Acad Sci U S A. 2014;111:2259–2264. doi: 10.1073/pnas.1317793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boutz DR, et al. Proteomic identification of monoclonal antibodies from serum. Anal Chem. 2014;86:4758–4766. doi: 10.1021/ac4037679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotham VC, Horton AP, Lee J, Georgiou G, Brodbelt JS. Middle-Down 193-nm Ultraviolet Photodissociation for Unambiguous Antibody Identification and its Implications for Immunoproteomic Analysis. Anal Chem. 2017;89:6498–6504. doi: 10.1021/acs.analchem.7b00564. [DOI] [PubMed] [Google Scholar]
- 8.Fusaro VA, Mani DR, Mesirov JP, Carr SA. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat Biotechnol. 2009;27:190–198. doi: 10.1038/nbt.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmermann I, et al. Synthetic single domain antibodies for the conformational trapping of membrane proteins. Elife. 2018;7 doi: 10.7554/eLife.34317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hohl M, Briand C, Grütter MG, Seeger MA. Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation. Nat Struct Mol Biol. 2012;19:395–402. doi: 10.1038/nsmb.2267. [DOI] [PubMed] [Google Scholar]
- 11.Hohl M, et al. Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc Natl Acad Sci U S A. 2014;111:11025–11030. doi: 10.1073/pnas.1400485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pardon E, et al. A general protocol for the generation of Nanobodies for structural biology. Nat Protoc. 2014;9:674–693. doi: 10.1038/nprot.2014.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storek KM, et al. Monoclonal antibody targeting the β-barrel assembly machine of Escherichia coli is bactericidal. Proc Natl Acad Sci U S A. 2018;115:3692–3697. doi: 10.1073/pnas.1800043115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boder ET, Midelfort KS, Wittrup KD. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu LC, et al. Multiplex single-molecule interaction profiling of DNA-barcoded proteins. Nature. 2014;515:554–+. doi: 10.1038/nature13761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darmanis S, et al. ProteinSeq: high-performance proteomic analyses by proximity ligation and next generation sequencing. PLoS One. 2011;6:e25583. doi: 10.1371/journal.pone.0025583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGregor LM, Jain T, Liu DR. Identification of Ligand-Target Pairs from Combined Libraries of Small Molecules and Unpurified Protein Targets in Cell Lysates. Journal of the American Chemical Society. 2014;136:3264–3270. doi: 10.1021/ja412934t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jespers L, Schon O, Famm K, Winter G. Aggregation-resistant domain antibodies selected on phage by heat denaturation. Nat Biotechnol. 2004;22:1161–1165. doi: 10.1038/nbt1000. [DOI] [PubMed] [Google Scholar]
- 19.Sieber V, Pluckthun A, Schmid FX. Selecting proteins with improved stability by a phage-based method. Nat Biotechnol. 1998;16:955–960. doi: 10.1038/nbt1098-955. [DOI] [PubMed] [Google Scholar]
- 20.Panse C, Trachsel C, Grossmann J, Schlapbach R. specL--an R/Bioconductor package to prepare peptide spectrum matches for use in targeted proteomics. Bioinformatics. 2015;31:2228–2231. doi: 10.1093/bioinformatics/btv105. [DOI] [PubMed] [Google Scholar]
- 21.Geertsma ER, Dutzler R. A versatile and efficient high-throughput cloning tool for structural biology. Biochemistry. 2011;50:3272–3278. doi: 10.1021/bi200178z. [DOI] [PubMed] [Google Scholar]
- 22.Shugay M, et al. Towards error-free profiling of immune repertoires. Nat Methods. 2014;11:653–655. doi: 10.1038/nmeth.2960. [DOI] [PubMed] [Google Scholar]
- 23.Glanville J, et al. Deep sequencing in library selection projects: what insight does it bring? Curr Opin Struct Biol. 2015;33:146–160. doi: 10.1016/j.sbi.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barkow-Oesterreicher S, Turker C, Panse C. FCC - An automated rule-based processing tool for life science data. Source Code Biol Med. 2013;8:3. doi: 10.1186/1751-0473-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 26.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 27.Schenck S, et al. Generation and Characterization of Anti-VGLUT Nanobodies Acting as Inhibitors of Transport. Biochemistry. 2017;56:3962–3971. doi: 10.1021/acs.biochem.7b00436. [DOI] [PubMed] [Google Scholar]
- 28.Gabay JE, Blake M, Niles WD, Horwitz MA. Purification of Legionella pneumophila major outer membrane protein and demonstration that it is a porin. J Bacteriol. 1985;162:85–91. doi: 10.1128/jb.162.1.85-91.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Türker C, et al. B-Fabric: the Swiss Army Knife for life sciences. Proceedings of the 13th International Conference on Extending Database Technology; Lausanne, Switzerland. 2010. 22 March 2010 - 26 March 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.