Abstract
Cerebrospinal fluid (CSF) is a vital liquid, providing nutrients, signalling molecules, and clearing out toxic byproducts from the brain. The CSF is produced by the choroid plexus (ChP), a protective epithelial barrier that also prevents free entry from the blood. Here, we establish human ChP organoids with a selective barrier and CSF-like fluid secretion in self-contained compartments. We show that this in vitro barrier exhibits the same selectivity to small molecules as in vivo, and that ChP-CSF organoids can predict CNS permeability of novel compounds. The transcriptomic and proteomic signature of ChP-CSF organoids reveal a high degree of similarity to in vivo. Finally, the intersection of single cell transcriptomics and proteomic analysis uncovers key human CSF components produced by previously unidentified specialized epithelial subtypes.
The polarised, secretory epithelium of the choroid plexus (ChP) plays a central role in brain development and function by actively secreting the cerebrospinal fluid (CSF) and forming the blood-CSF barrier (B-CSF-B) (1). The CSF is a colorless fluid that is rich in nutrients, hormones, and signaling molecules that are vital for brain function (2–4). The B-CSF-B, similar to the blood-brain barrier (BBB), prevents toxic substances in the circulation from reaching the brain (5); it directly contacts the CSF rather than the brain parenchyma and is formed by the ChP epithelial monolayer (5, 6). The BBB and B-CSF-B together make up the CNS barrier and prevent entry of most therapeutic molecules into the brain.
Since the ChP lies deep within the brain, study of ChP function has represented a challenge. In particular, the cellular makeup of the ChP is not well defined. Although there are reports of ChP epithelial subtypes referred to as “dark” and “light” cells due to their observed density in transmission electron microscopy (EM) (7), whether heterogeneous subtypes of epithelial cells exist in this tissue is still unclear. Since the ChP is surrounded by brain tissue and vasculature, it has not been possible to examine the ChP in isolation and understand which of the many roles of the ChP are carried out by which cells and which CSF factors are filtered from the blood versus made de novo by the ChP.
Cerebral organoids can model the human developing brain with remarkable fidelity (8–10). Recent work by Sakaguchi et al. demonstrated the ability to generate dorsal identities, including ChP and hippocampus (11). However, none of the key functions of the ChP (barrier formation or CSF production) were demonstrated. Thus, we are still in need of an authentic in vitro model of the ChP (12). To better understand the development of the human ChP, the B-CSF-B, and the synthesis of CSF, we have established ChP organoids. ChP organoids reliably and reproducibly develop ChP epithelium with polarised cells that actively secrete a colorless fluid with properties closely resembling the CSF. The CSF-like fluid is enclosed and isolated from the surrounding media modeling two fundamental properties of the ChP: CSF secretion and barrier formation. With this model we are now able to elucidate which components of the CSF are specifically generated by the ChP, and assign distinct roles to newly identified ChP epithelial subtypes.
Generation of ChP organoids from human PSCs
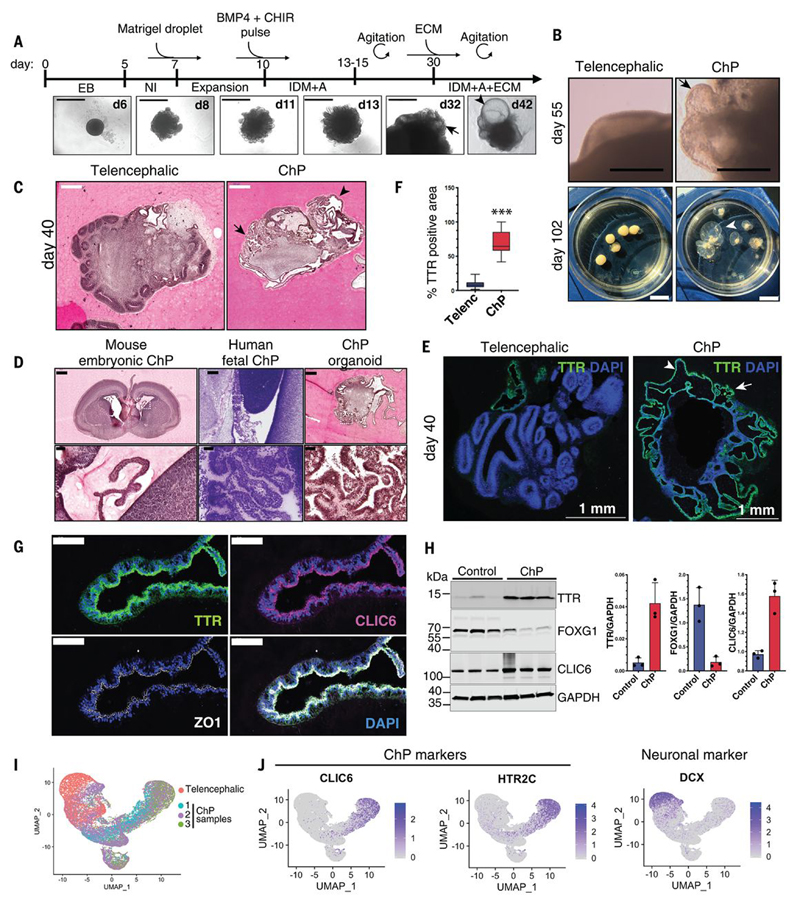
Cerebral organoids are characterized by self-organisation of complex tissue architectures similar to the developing mammalian brain (8–10). Recent protocols describe predominantly forebrain, telencephalic identities (13, 14) which also includes ChP epithelium (Fig. S1A) (9, 13). In order to develop an in vitro model of human ChP with accurate architecture and function, we established a protocol based upon the cerebral organoid method. In vitro derivation of ChP cells using the dorsalising factor Bmp4, critical for the development of ChP epithelial cells both alone (15) and in combination with the Wnt-activator molecule CHIR (11) has been reported before. To promote ChP fate in cerebral organoids, Bmp4 and CHIR were given as a pulse (Fig. 1A, Fig. S1B). At day 14, undirected telencephalic organoids developed large, rounded neuroepithelial lobes whereas Bmp4/CHIR treated organoids generated elongated neuroepithelial tissues (Fig. S1C), consistent with the progressive elongation of the ChP from the neuroepithelium of the dorsal midline in vivo (1, 16, 17). ChP organoids appeared almost entirely enriched in cuboidal epithelium, in contrast to forebrain organoids (Fig. 1B, 1C). A striking feature of ChP organoids was the later development of fluid-filled compartments or ‘cysts’ containing a colorless liquid, rarely present in untreated organoids (Fig. 1B, Fig. S1D). Comparison of histological sections of ChP organoids with human and mouse embryonic ChP showed a resemblance in complexity and organization between the human and the organoid samples (Fig. 1D).
Figure 1. Generation of human ChP organoids with self-contained fluid-filled compartments.
(A) Protocol timeline with images of H9 round seeded ChP organoids over time. Arrow indicates emerging ChP epithelium and arrowhead the later fluid-filled compartment. Scale bar: 1000μm. (B) Comparison of H1 ChP and telencephalic organoids (day 55, scale bar:1mm and day 102, scale bar:1cm). Arrow indicates ChP epithelium, arrowhead points to fluid-filled cavity. (C) H&E stained sections of H1 telencephalic and ChP organoids (day 40). Arrow indicates convoluted ChP epithelium, arrowhead points to fluid-filled cavity. Scale bar:500μm. (D) Histological sections of E18.5 mouse embryonic brain (H&E stained), human fetal ChP at 15 post-conception weeks (Nissl stained, from BrainSpan Atlas of the Developing Human Brain (44)), and H1 human ChP organoid at day 40 (H&E stained). Scale bar: 500μm, 50μm for magnification. (E) Representative confocal images of H1 telencephalic and ChP organoids at day 40 stained for TTR and nuclei (DAPI, blue). Arrow indicates convoluted TTR-positive ChP epithelium, arrowhead points to ChP epithelium surrounding fluid-filled cavity. Scale bar: 1mm. (F) Quantification of percentage of TTR-positive area over total organoid area in n=4 independent H9 batches (3-4 organoids per batch, day 30, 40, 48). ***P<0.00001, Mann-Whitney test. (G) Representative confocal images of ChP epithelium from H1 organoid (day 40) stained for TTR, CLIC6 and ZO1. Scale bar:50μm. (H) Immunoblots from telencephalic control, and ChP organoids probed for TTR, FOXG1, CLIC6, and loading control GAPDH. Quantification of immunoblots show band intensity normalized for GAPDH (n=3 independent H9 batches collected at day 75, 73 and 68, in separate lanes). (I) UMAP dimensional reduction plot of the combined samples (telencephalic organoids, and three ChP organoids), showing overlap among ChP organoid cells. (J) Feature plots showing enrichment of ChP markers CLIC6 and HTR2C in ChP cells, and of neuronal marker DCX in telencephalic organoid cells.
In vivo, ChP epithelium starts as a pseudostratified epithelium, followed by an intermediate columnar stage, and maturing into a highly folded cuboidal epithelium (1, 17, 18). ChP organoids similarly developed a pseudostratified neuroepithelium with TTR positive areas by day 28 (Fig. S1E). By approximately day 40, ChP organoids displayed more columnar and cuboidal epithelia, increasingly enriched in TTR and the cell-cell junction marker ZO1, with only sparse neuroepithelial SOX2-positive cells (Fig. 1E, Fig. S1E). Cells surrounding fluid-filled compartments also stained positive for TTR and displayed polarized apical staining for AQP1 (Fig. S1F, Fig. S1G). Quantification of TTR-positive regions showed enrichment in ChP organoids (Fig. 1F). We detect high levels of CLIC6, a highly specific marker of the ChP (19) (Fig. 1G, Fig. 1H), as well as a number of other ChP related proteins (PLEC, APOE, PLTP, IGFBP7, CA2) with lower levels of the telencephalic marker Foxg1 and other neuronal markers (DCX, GAP43) (Fig. 1H, Fig. S3A). Cortical Tbr2 intermediate progenitors and HuC/D neurons were also largely reduced in early ChP organoids and almost completely absent in mature ChP organoids compared to controls (Fig. S3B). Together, these data demonstrate a reliable and reproducible generation of ChP tissue in vitro, as well as fluid-filled compartments, from different lines of human pluripotent stem cells and with two different seeding methods (Fig. 1F, Fig. S2A, S2B, S2C, S2D, S2E, S2F).
ChP organoids develop epithelial and stromal components and recapitulate the human in vivo transcriptomic signature
To further investigate the identity of ChP organoids, we performed single cell RNA-sequencing (scRNA-seq), sampling two organoids from three separate batches of ChP organoids and two telencephalic organoids as control (Fig. 1I-J). Combined analysis revealed that cells from different ChP organoids/batches were intermingled and largely separate from those of telencephalic organoids (Fig. 1I), suggesting reproducibility across batches. Furthermore, although telencephalic organoid cells expressed neuronal markers, such as DCX, ChP cells expressed ChP markers, such CLIC6 and HTR2C (20) (Fig. 1J).
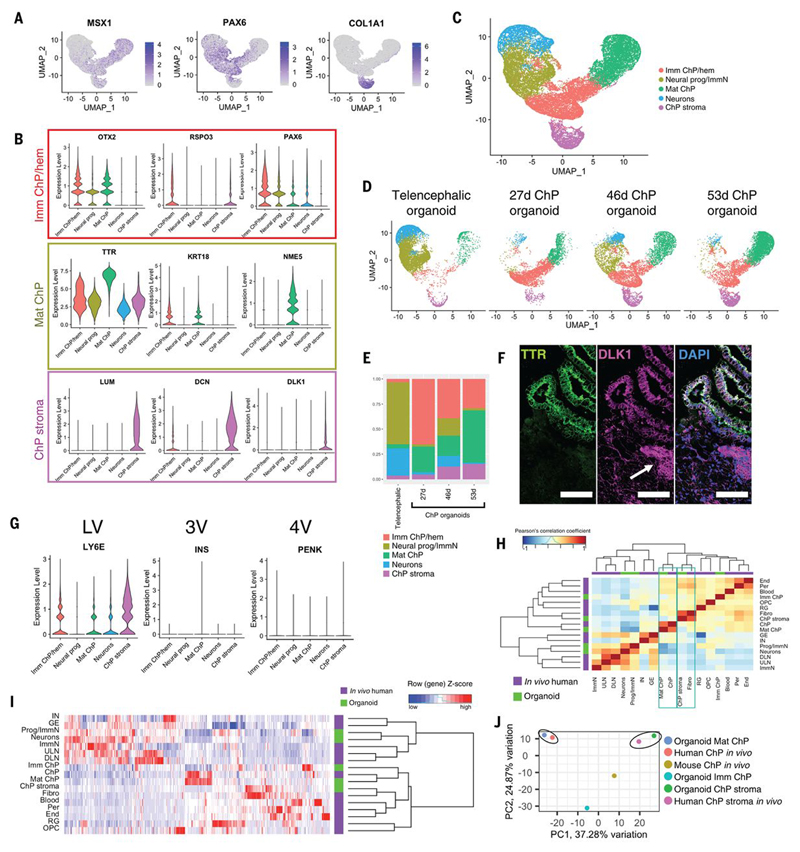
Cluster analysis revealed five major cell types: two major telencephalic clusters consisting of neural progenitors and neurons, and three major ChP clusters. Further examination of marker genes revealed an immature ChP cluster expressing early regulatory factors involved in ChP development including MSX1, OTX2, and R-spondin 3 (RSPO3). This cluster also expressed the progenitor marker PAX6, which was also expressed in the neural progenitor cluster of telencephalic organoids (Fig. 2A-B) owing to their common developmental origin. The third cluster of the ChP organoids expressed numerous mesenchymal markers including the extracellular matrix component COL1A1 (Fig. 2A), DCN, LUM, and DLK1 (Fig. 2B). These markers allowed us to assign three major identities to ChP organoids: immature ChP/hem, mature ChP epithelium, and ChP stroma (Fig. 2C). Further analysis also revealed a consistent and progressive enrichment over time in the mature ChP cluster (Fig. 2D-E). Immunostaining highlighted both stromal and epithelial cells (Fig. 2F, Fig. S3C), and examination of regional ChP markers (4, 20) revealed the ChP organoids to be of lateral ventricle identity rather than third or fourth ventricle (Fig. 2G) consistent with their telencephalic origin.
Figure 2. ChP organoids closely recapitulate transcriptomic signature of human.
in vivo tissue.(A) Feature plots showing enrichment of genes involved in ChP development (MSX1, PAX6) and stromal marker COL1A1. (B) Violin plots of scRNA-seq analysis showing expression levels of ChP immature/hem (OTX2, RSPO3, PAX6), mature (TTR, KRT18, NME5) and stromal markers (LUM, DCN, DLK1). (C) UMAP plot showing scRNA-seq clusters of combined samples. (D) UMAP plots separated by samples showing progressive enrichment in ChP populations in ChP organoids compared to 55 day old mixed telencephalic identity organoids. (E) Stacked bar chart showing relative proportion of clusters in each sample. (F) Confocal images of ChP organoids positive for TTR and DLK1 in an adjacent stromal population (arrow). Scale bar:100μm. (G) Violin plots showing expression levels of regional ChP markers for lateral (LV, LY6E), third (3V, INS) and fourth (4V, PENK) ventricle. (H) Heatmap and dendrogram of Pearson’s correlation coefficients across identified in vivo human dorsal telencephalon (21) and organoid clusters showing high similarity between in vivo and in vitro ChP clusters (green outline). (I) Heatmap and dendrogram of unbiased hierarchical clustering based on the 1000 most variable genes between organoid single cell clusters and in vivo human brain single cell clusters. (J) PCA plot of scRNA-seq ChP clusters from mouse embryo, human fetal dorsal telencephalon, and ChP organoids, revealing higher similarity of ChP organoid clusters to human than to mouse ChP.
Comparison with a published scRNA-seq dataset of developing human brain (21) revealed a high correlation between organoid and in vivo ChP clusters, and between organoid and in vivo stromal clusters (Fig. 2H, Fig. 2I). Further comparison to both human in vivo ChP and developing mouse ChP from scRNA-seq of mouse embryos (22) revealed higher correlation with human ChP than with mouse (Fig. 2J, Fig. S3D). Overall, these data demonstrate that in vitro derived ChP organoids closely recapitulate the cell composition and transcriptomic signature of the human ChP in vivo.
ChP organoids form a tight barrier and recapitulate in vivo CNS permeability to small molecules
To test the functional role of ChP organoids in the formation of a selective barrier, we first assessed expression of tight junction markers such as claudins and occludin (OCLN). scRNA-seq revealed the presence of several of the claudins (CLDN1, CLDN3, CLDN5) (Fig. 3A) as well as other tight junction components OCLN, ZO1 (TJP1), ZO2 (TJP2), and PDZ proteins PATJ (INADL) and MPDZ (Fig. 3B). We observed apical localization of CLDN1, CLDN3, CLDN4 and CLDN5 in the ChP epithelium (Fig. 3C). Low levels of CLDN2 were also observed in ChP organoid epithelium (Fig. S4) as well as the presence of OCLN (Fig. S4). Electron micrographs also revealed tight junctions, as well as primary cilia, extensive microvilli, multi-vesicular bodies and numerous extracellular vesicles (Fig. 3D). We directly assessed barrier function by examining the entry of fluorescently labeled dextrans of varying molecular weights, which were completely excluded from the organoid (Fig. 3E, Fig. 3F).
Figure 3. ChP organoids form a tightly sealed barrier.
(A) Violin plots showing expression levels of CLDN1, CLDN3 and CLDN5 in organoid cell clusters identified by scRNA-seq. (B) Feature plots showing enrichment in tight junction proteins TJP1 and TJP2, OCLN, INADL and MPDZ in ChP immature and mature clusters. (C) Representative images of H1 ChP organoids (day 40) staining positive for TTR, CLDN1, CLDN3, CLDN4 and CLDN5. Scale bar:50μm. (D) Electron micrographs showing tight junctions (TJ), microvilli (upper, middle panel), cilia (C), multivesicular bodies (MVB) and extracellular vesicles (Ex). Scale bar:1μm. (E) Bright field images of ChP organoids with clear, fluid-filled compartments despite incubation for 2h with 647-Alexafluor 10kDa dextran (blue tint). (F) Fluorescent intensity in media and organoid fluid after 2h incubation with 70kDa Oregon green-dextran, 10kDa 647-Alexafluor dextran and 3-5kDa FITC-dextran.
To test for more selective permeability, we applied therapeutically relevant small molecules and assayed their permeability by NMR analysis (Fig. 4A, Table S1). As a proof of principle, we tested dopamine (Dopa) and its precursor, Levodopa (L-Dopa), since these are similar molecules with very different permeability in vivo. Dopa does not enter the brain in vivo, whereas L-Dopa is actively transported in to the CSF via the LAT-1 transporter (23, 24). We incubated the organoids with Dopa, and L-Dopa in the presence of Carbidopa, a small molecule that prevents conversion of L-Dopa into Dopa (25). After 2h, both compounds were detectable in the media but only L-Dopa was detectable in the organoid internal fluid demonstrating that our model exhibited the same selectivity as in vivo (Fig. 4B, Fig. S5). LAT-1 was expressed on the ChP organoid epithelium (Fig. 4C-D), explaining the proper transport of L-Dopa in our assay. We also detected expression of efflux pump P-glycoprotein 1 (Pgp/MDR1) throughout the apicobasal axis (Fig. 4C). Pgp localization in ChP has been somewhat controversial (26); however, it is reported to have a role in pumping molecules from the CSF to the blood (26, 27). These findings would suggest the Pgp is expressed in the human ChP. Additionally, ChP efflux transporters MRP1 and MRP4 (26, 28) were localized on ChP organoid epithelium (Fig. 4D, Fig. 4E). These results suggest that ChP organoids express transporters that are critical for the correct function of the B-CSF-B in human brain.
Figure 4. ChP organoids predict CNS permeability of small molecules.
(A) Schematic of CNS drug permeability measurement in ChP organoids. (B) Top: NMR spectra of organoid fluid upon application of Dopa, L-Dopa or no drug (Ctrl) measured after 2h incubation. Bottom: Quantifications of above NMR spectra in media and organoid fluid. (C) Representative images of ChP organoids stained for TTR, LAT1, Pgp. (D) Immunoblot for ChP transporters MRP1, LAT1, CLIC6, and secreted protein IGF2, and the loading control GAPDH of ChP organoid lysates (day 10 to day 31). (E) Confocal images of ChP organoids stained for transporters MRP4 and MRP1. Scale bar:50μm. (F) Relative quantifications of NMR spectra of bupropionyl, methotrexate and vincristine in media and organoid fluid after 2h. (G) Scatter plot with linear regression showing correlation (R2=0.9921; Slope=1.004) between in vivo CSF/plasma ratio (K p,uu,CSF) and in vitro organoid fluid/media ratio (K m,uu,Org.fluid) of unbound drugs (Table S2). In contrast to other drugs shown, Sephin 1 (red dot) in vivo CSF/plasma measurement is the reported value from mice (32). (H) Relative quantifications of NMR spectra of Sephin 1 in media and organoid fluid after 2h. (I) Relative quantifications of NMR spectra of BIA 10-2474 in media and organoid fluid after 2h. (J) Time course analysis of the ratio of BIA 10-2474 and bupropionyl in organoid fluid to media at 2h, 12h, 24h and 72h. (All drugs were tested in n=3 independent experiments; Table S1).
We next tested whether the system could quantitatively predict drug permeability. We tested bupropionyl, an antidepressant known to readily cross the BBB (29), and two chemotherapeutic compounds, methotrexate and vincristine, which poorly cross the BBB due to the actions of efflux transporters MRP1/4 and Pgp, respectively (30, 31). As expected, bupropionyl was readily detectable in the organoid internal fluid after 2h, while methotrexate and vincristine were completely excluded from the internal fluid (Fig. 4F, Fig. S6, S7). We found high correlation between in vitro fluid/media ratios and the in vivo CSF/plasma ratios reported in the literature for the same drugs (R2=0.9921) (Fig. 4G, Table S2). The values corresponded remarkably well (as indicated by a slope of 1.004), indicating that this in vitro system can qualitatively and quantitatively recapitulate the permeability of drugs reported in vivo.
To test whether ChP organoids could predict CNS permeability of novel drugs, we turned to Sephin 1, an inhibitor of the regulatory subunit PPP1R15A of protein phosphatase 1 (32). Clinical trials for this promising candidate have recently begun but data for this molecule in humans is not yet available. Sephin 1 was detectable in organoid internal fluid, indicating that this compound could also cross the human CNS barrier (Fig. 4H, Fig. S6). Notably, however, the permeability in the human organoid model was much lower than that described in mice (Fig. 4G). This may indicate potential species-related differences in the CNS permeability of this compound, though an in vitro difference cannot be excluded.
Issues related to the pharmacokinetic profile of drugs account for most clinical trial failures (33). One example involved the compound BIA-10-2474, a fatty acid amide hydrolase inhibitor that was in development for treatment of chronic pain and multiple sclerosis (34, 35). Phase I clinical trials were halted when trial participants exhibited severe neurotoxicity, with one fatality (34, 36). Investigations hypothesized toxic drug accumulation in the brain that was not evident in any animal model tested (34, 36, 37). However, the exact cause remains unknown. We compared the pharmacokinetic profile of BIA-10-2474 to a safe compound, bupropionyl, which crosses the barrier but has not been shown to accumulate. Similar to bupropionyl, BIA-10-2474 crossed the ChP epithelial barrier after 2h incubation (Fig. 4I, Fig. S8). However, compared to the bupropionyl, which stabilized to baseline levels after 24h, BIA-10-2474 continued to accumulate in the organoid fluid (Fig. 4J). These data are consistent with the theory that BIA-10-2474 accumulated in the brains of the trial participants.
ChP organoids secrete a CSF-like fluid highly similar to in vivo CSF
The other key function of the ChP is production of CSF. We detected expression of transporters involved in CSF secretion including the water channel aquaporin 1 (AQP1) and the key enzymes for generation of the gradient driving CSF secretion, CA2 and CA12, (38) (Fig. 5A). Staining revealed apical localization of AQP1, similar to in vivo (Fig. 5B). Consistent with the unique role of ChP in the transport of folate and vitamin C, both vital for brain function (39, 40), we also detected expression of specific transporters involved in the trafficking of these nutrients: SLC23A2 (vitamin C) and SLC46A2 (folate) (Fig. 5A).
Figure 5. ChP organoids secrete human CSF proteins and progressively mature to a postnatal state.
(A) Top: violin plots showing expression levels of water transporter AQP1 and CSF secretion enzymes CA2 and CA12 in scRNA-seq clusters. Bottom: feature plots showing enrichment in SLC23A2 (vitamin C transporter) and SLC46A1 (folate transporter) in scRNA-seq clusters. (B) Confocal images of H1 ChP organoid at day 40 and in vivo ChP from a mouse embryo (E18.5) stained for Aqp1 and TTR. Scale bar:50μm. (C) gProfileR (45) analysis of proteins detected in iCSF from at least 2 ChP organoid samples, showing significant (P<0.05) enrichments for GO categories cellular component (GO:CC), molecular function (GO:MF) and biological process (GO:BP), KEGG, REAC, WP, HP databases. (D) Color-coded heatmap showing relative abundance (emPAI values) of proteins detected in organoid iCSF from at least two organoid batches (5 batches of H9, 6 batches of H1 and one iPSCs batch, Table S3) and with a mean emPAI ≥ in organoids. Corresponding values are shown for the media, the mean of the 12 organoid samples, and the mean of 3 in vivo CSF samples: human adult CSF, bovine fetal CSF and embryonic mouse CSF (E12.5-13.5) (Table S4). (E) Venn diagram of all proteins detected with an emPAI ≥ in any sample. Proteins were assigned to each group in which they were detectable at any level. (F) Venn diagram of proteins detected in at least 2 iCSF samples and with an emPAI ≥ in at least one. Overlap is shown for proteins detected in datasets of human adult CSF (46), pediatric CSF from healthy controls (47), and human embryonic CSF (48). (G) Color-coded heatmap of protein emPAI level showing abundant iCSF proteins (emPAI ≥ in early d32-d58 or late d68-d146 stage organoids) shared with in vivo samples (split into developing and adult). Proteins are highlighted and color-coded according to the earliest stage human in vivo CSF dataset in which they are detected from the following published datasets: human adult CSF (46), pediatric CSF (47) and embryonic CSF (48). (H) emPAI heatmap of proteins reproducibly (more than one iCSF sample) and abundantly (emPAI ≥ in early d32-d58 or late d68-d146 stage organoids) detected in iCSF but not in media, and shared with at least one in vivo sample, which are considered de novo ChP-derived CSF proteins.
To investigate whether the colourless fluid inside ChP organoids could represent an in vitro CSF-like fluid (iCSF), we performed mass spectrometry analysis of extracted fluid (5 batches of H9, 6 batches of H1 and 1 batch of iPSCs IMR-90), compared to 3 different in vivo samples (human adult telencephalic CSF, mouse embryonic CSF at E13.5, and fetal bovine CSF) (Table S3-S4) and media. We detected 1,618 proteins in ChP organoid iCSF, 851 of which were reliably detected in more than one iCSF sample (Supplementary Data 1). Gene Ontology Analysis revealed enrichment in GO:CC categories “extracellular vesicle” and “extracellular space”, and GO:BP categories “export from cell” and “secretion from cell” (Fig. 5C). The most abundant proteins in the iCSF largely overlapped with proteins present in vivo (Fig. S9A). Indeed, of the 50 most abundant proteins detected in organoid iCSF, 49 were also detected in in vivo samples (Fig. 5D). Comparison of all the proteins detected revealed a large overlap between iCSF and in vivo samples (Fig. 5E, Fig. S9B). We could detect clinically relevant biomarkers, for example APOE, Insulin-like growth factor binding protein 7 (IGFBP7), Serpin Family F Member 1 (SERPINF1), and Niemann-Pick disease type C2 protein (NPC2) (Table S5).
To ascertain the similarity to in vivo, we examined proteins detected reproducibly in in vivo samples but not in organoid iCSF (Fig. S9C) as these could be factors the organoid failed to produce. Proteins detected exclusively in the in vivo CSF samples showed enrichment in GO:CC “blood microparticle”, GO:BP “platelet degranulation” and GO:MF “Oxygen carrier activity” (Fig. S9D). This suggests these are factors coming from the blood, either through transport across the B-CSF-B or as collection artifacts through blood contamination. These would not be expected to be present in ChP organoids as they lack a vascular system and blood, and hence represent the ChP in isolation.
ChP-CSF organoids progressively mature over time
ChP organoids progressively expressed more mature markers (Fig. 2D-E). To test whether the composition of iCSF also matured over time, we compared proteins in iCSF to published datasets of normal human embryonic, pediatric, and adult CSF (Supplementary Data 2). We observed overlap with all three human age ranges, including a number of genes shared with the more mature timepoints (Fig. 5F). A number of proteins in iCSF changed abundance over time, with a marked shift between the day 58 and 68 time points (Fig. S9A). We further analyzed early (abundant before day 60) and late (abundant after day 60) proteins (Fig. S10A). Whereas early abundant proteins were overlapping with embryonic and fetal in vivo samples and datasets, late abundant proteins overlapped more with pediatric and adult stages (Fig. 5G).
We next examined factors known to exhibit changes in CSF levels over time (Fig. S10B). Abundant extracellular matrix proteins (laminin, fibronectin, and collagens) were shared with human embryonic CSF at the earliest time points (Fig. 5G, Fig. S10B). IGF2 is an important signaling factor in developing CSF (2). We observed a number of IGF factors present in iCSF, enriched in the REAC category “Regulation of Insulin-like Growth Factor” (Fig. 5C). IGF2 was highly abundant with levels dropping off at the latest stage tested of 146 days (Fig. S10B, Fig. S10C) closely matching the pattern in vivo, where IGF2 was detected in developing embryonic, fetal, and pediatric CSF (Fig. 5G).
We next examined proteins whose expression should increase in postnatal and adult CSF. We observed a number of lipoproteins in organoid iCSF, such as APOE, clusterin (CLU), APOD and phospholipid transfer protein (PLTP), which increased over time and were shared with adult in vivo CSF (Fig. S10B). Similarly, we observed marked presence of lipid droplets at later stages (Fig. S10D). Finally, we observed a number of other markers increasing with time, including retinol binding protein 1 (RBP1), ceruloplasmin (CP), and complement proteins (C1R and C4B) (Fig. S10B). Together, these results suggest that late stage organoids (older than 60 days) recapitulate functionality of pediatric and even adult ChP, with production of more mature CSF-like fluid by 100 days.
Identification of the tissue source of key CSF components
Organoids represent tissues in isolation, lacking cell types such as vasculature and an immune system. Although this can be a limitation, we viewed this as a strength when examining secretion of a biological fluid such as CSF. This isolation can be leveraged to reveal factors from external sources, such as surrounding brain tissue or filtered from the blood, versus those made de novo by the ChP itself. To explore which CSF factors may be filtered, rather than made de novo, we examined iCSF proteins also present in fresh media (Fig. S10E). Many of these factors were also shared with in vivo CSF samples, suggesting these may be filtered into the CSF also in vivo.
CSF composition may also be influenced by surrounding brain tissue. We therefore examined organoid fluid from telencephalic organoids of mixed identity, which would include ChP. The fluid largely overlapped that of pure ChP organoids (Fig. S10F), suggesting CSF composition is primarily determined by ChP secretion. However, a number of proteins were specific to mixed identity organoids, 11 of which were shared with in vivo samples (Fig. S10G). These may represent bona fide brain-derived factors such as secretogranin-1 (CHGB), a neuroendocrine secretory protein, and NDRG2, an astrocyte expressed protein implicated in neurite outgrowth. The vast majority of proteins (1,589 of 1,618) were not shared with media, suggesting de novo ChP secretion (Fig. 5H). Overall, these findings suggest that CSF composition is largely dictated by ChP secretion, with a subset of components being filtered from the blood and produced by surrounding brain tissue.
Merging proteomic and transcriptomic data reveals cell-type specific secretion and identifies novel ChP epithelial subtypes
To identify specific ChP cell types responsible for secretion of CSF components, we examined expression in scRNA-seq of key factors present in iCSF. The developmental signaling factor IGF2 was expressed in the ChP stroma (Fig. S11A), but also later within the mature ChP epithelium (Fig. S11B, Fig. S11C). More mature factors were specifically expressed within the mature ChP epithelium (Fig. S11A, S11D), while biomarkers exhibited varying expression patterns across ChP clusters (Fig. S11E). These findings provide insight into which cells within the ChP produce key secreted proteins.
Although several secreted proteins were expressed broadly among mature ChP cells (i.e. PLTP, CP) others such as IGF2, COL1A1, and C1R exhibited more restricted expression (Fig. S11A), suggesting the existence of ChP epithelial subtypes involved in secretion of specific CSF components. We therefore performed sub-clustering of the mature ChP cluster and identified four prominent clusters (Fig. 6A). Differential gene expression analysis of these clusters revealed a cluster expressing markers of ChP epithelium (HTR2C and CLDN5), but also high levels of mitochondrial genes (Fig. 6B-C, Fig. S12A-B). The second most abundant cluster instead showed marked down-regulation of mitochondrial genes, but upregulation of microtubule cytoskeletal and ciliary assembly factors (Fig. 6B-C).
Figure 6. Identification and molecular characterization of ChP epithelial subtypes.
(A) UMAP plot showing subclustering of the mature ChP epithelial cluster identified by scRNA-seq. (B) Heatmap of top 10 differentially up-regulated transcripts in the four subclusters identified, and the major GO terms enriched for each cluster. (C) Dot plot showing average expression and percent of cells expressing the displayed enriched genes identified by scRNA-seq. (D) Electron micrographs showing dark (D) and light (L) cells on the ChP epithelium in organoids. Arrowhead points to basal body in a light cell. Scale bar:40μm. (E) Representative confocal images of H1 ChP organoid (day 82) stained for mitochondrial marker CARD19, ciliated cell marker FOXJ1, and TTR. Scale bar:100μm. (F) Confocal images of H1 ChP organoid (day 46) stained for Foxj1, Arl13b (cilia) and TTR. Scale bar:50μm and 100μm (zoom-in:20μm). (G) Representative confocal images of H1 ChP organoid (day 82) stained for alpha-smooth muscle actin (α-SMA), transgelin (TAGLN) and TTR. Scale bar:100μm.
Some studies of ChP ultrastructure reported the existence of “dark” and “light” cells (7), but nothing is known about their molecular identities. Dark cells, or “mitochondria-rich cells”, have also been observed in other secretory organs (41). These cells also lack cilia. We therefore hypothesized that the two most abundant clusters of the ChP epithelium may be dark and light epithelial cells with opposite expression of mitochondrial and ciliary genes. Electron micrographs revealed abundant dark cells with interspersed light cells with a ciliary basal body (Fig. 6D). We observed nonhomogeneous expression of the mitochondrial marker CARD19, the ciliary transcription factor FOXJ1 and the ciliary markers ARL13B and CCDC67 in subsets of cells (Fig. 6E, Fig. 6F, Fig. S12C). These results provide a first look at the molecular and cell biological hallmarks of dark and light ChP epithelial cells.
Closer examination of differential gene expression in the third cluster of ChP epithelial cells revealed markers typical of myoepithelial cells (Fig. 6B, Fig. 6C, Fig. S12A-B), a cell type described in secretory glands. These cells express contractile actin and myosin, similar to smooth muscle cells (α-SMA), yet are bona fide epithelial cells expressing markers such as keratin 17 (KRT17) Fig. 6B-C). Furthermore, staining for α-SMA and transgelin (TAGLN), revealed their expression in a subset of cells within the epithelium (Fig. 6G). Finally, we observed a cluster of cells expressing markers of DNA replication and cell cycle (Fig. 6B-C, Fig. S12A-B), possibly indicating dividing cells. Indeed, we could observe occasional dividing PH3+ cells in mature ChP epithelium, decreasing over time (Fig. S12D). These findings suggest the presence of a limited number of dividing mature epithelial cells, as well as contractile cells, which had not been described in the ChP and may help promote CSF secretion, similar to secretory glands elsewhere in the body.
We next took an unbiased approach to explore cell type-specific secretion by looking at overlapping genes between mass spectrometry analysis and scRNA-seq (Fig. 7A, Supplementary Data 3). We examined specific genes of interest in this context, such as IGF2, which was restricted to dark cells of the ChP epithelium (Fig. 7B, Fig. S13A), consistent with its staining pattern (Fig. 7C). The expression of more mature marker RBP1 in dark cells (Fig. 7A), was also further confirmed by immunostaining (Fig. S13B). Finally, a number of biomarkers were specifically produced by epithelial subtypes, including SERPINF1, IGFBP7, PARK7, and APOE (Fig 7A, Fig. S13C). Overall, these findings provide insight into specific cell types and their secretions within the ChP epithelium.
Figure 7. Identification of novel factors secreted by distinct ChP epithelial subtypes.
(A) Dot plot showing average expression and percent of cells expressing factors overlapping between iCSF detected proteins and differentially expressed genes within the four subclusters (Supplementary Data 3). (B) Violin plot showing expression of IGF2 in ChP epithelial subtypes identified by scRNA-seq. (C) Representative confocal images of H9 ChP organoid (day 50) stained for IGF2, ZO1, TTR and DAPI. Scale bar:50μm. (D) Color-coded heatmap showing proteins reproducibly (detected in more than one iCSF sample) and abundantly (emPAI ≥ in early d32-d58 or late d68-d146 stage organoids) detected in iCSF but not in media or in in vivo developing mouse or bovine CSF or in adult human CSF. Highlighted and color-coded accordingly are proteins detected in published datasets of human embryonic (48) or pediatric CSF (47), and novel factors predicted to be secreted (GeneCards database). (E) Violin plots showing expression levels of secreted proteins identified exclusively in organoid iCSF: LGALS3BP, MDK and PRAP1. (F) Representative confocal images of H1 ChP organoid (day 56) stained for LGALS3BP, DCN, TTR and DAPI. Scale bar:100μm.
Identification of CSF proteins secreted by the developing human ChP
ChP organoids provide a model system for developing human ChP and CSF. We therefore focused on iCSF-unique factors. As none of the in vivo samples tested here were from developing human CSF, any CSF components unique to the ChP organoids could in fact represent either in vitro-specific or human-specific developmental factors. We identified 24 proteins that were reproducibly abundant in ChP iCSF but absent in mouse and bovine developing CSF as well as adult human CSF (Fig. 7D). We then queried published datasets of human developing CSF from fetal and pediatric stages, which revealed some overlap with known human developmental CSF factors whereas others had not been observed before but are predicted to be secreted proteins (Fig. 7D, Fig. 7E).
One human-specific developmental secreted protein stood out, LGALS3BP, as it has also been identified in outer radial glial progenitors of the human fetal brain (42). We therefore examined the expression of this factor further and found it to be present in neural progenitors (Fig. S13D) but more abundant in the ChP (Fig. S13E), specifically in dark, myoepithelial, and dividing cells, but less in light cells (Fig. 7E). Immunohistochemistry also revealed abundant, broad staining throughout the ChP epithelium (Fig. 7F). These findings highlight a number of promising candidate signaling proteins that may play important roles in human brain development.
Discussion
We present ChP organoids for the investigation of development and function of the human ChP. Using this in vitro organoid model, we were able to demonstrate the two key functions of the ChP: barrier formation and CSF secretion. ChP organoids are selective to highly similar small molecules. This model matches the selectivity of the human blood-CNS barrier in vivo, and it can predict CNS permeability of both known and novel drug compounds.
There is an increasing demand for more effective CNS drugs. New drugs too often progress to clinical trial before failing for lack of efficacy, inability to cross in to the CNS, or limited translatability from animal models (43). ChP organoids could be exploited for preclinical testing to identify new modes of drug entry into the CNS.
We further show that the fluid secreted by ChP organoids represents an authentic biological fluid, separate from the media, and exhibiting a high degree of similarity to CSF in vivo. The organoids and CSF-like fluid also mature over time, reaching a state highly similar to postnatal stages and even adulthood. This would enable the study of disease-related biomarkers, whose functions are still not well understood. The ChP is relatively understudied, but it is garnering increasing attention due to its important roles in development and diseases such as neurodegeneration.
Finally, we have leveraged the isolated nature of ChP organoids to identify and molecularly describe novel ChP epithelial cell types and to identify their specific CSF secretions. With this model we were able to disentangle CSF components that are specifically made by the ChP, from those transported across or generated by the surrounding brain tissue. This revealed a number of key developmental factors and human-specific signaling proteins that may play key roles in brain development and homeostasis. We are hopeful that with this information in hand, further studies can begin to shed light on the vital functions of the ChP in brain development and disease.
Materials and Methods Summary
Cerebral and ChP organoid culture conditions
Stem Cell Technologies Cerebral Organoid kit (catalogue n. 08570, 08571) reagents were used for the generation of cerebral and ChP organoids. For ChP patterning, 3μM CHIR and 20ng/ml BMP4 were added in maturation media on day 10 until day 17. Detailed methods for organoid differentiation protocol are included in the supplementary material.
Immunostaining and immunoblotting
Organoids were fixed in 4% PFA overnight at 4°C then moved to 30% sucrose buffer. Organoids were then embedded in gelatin and sectioned as previously described (49). After blocking and permeabilisation, sections were incubated overnight with primary antibody. Detailed list of antibodies used is included in the supplementary material. Images were acquired using a Zeiss LSM 780 confocal microscope (Carl Zeiss) and prepared using Fiji (NIH). Detailed methods for sample preparation and imaging by electron microscopy (EM) are included in the supplementary material. For immunoblotting, organoids were homogenised in RIPA buffer. Protein samples were loaded into a SDS-PAGE. Membranes were imaged using a Li-COR Odyssey CLx Infrared Imaging System. Detailed methods for preparation of cytosol-enriched fractions are included in the supplementary material.
Single cell RNA sequencing
Single cell dissociation was performed by pooling two organoids for each condition: 55-day H9 telencephalic organoids, 27-day H1 ChP, 46-day H1 ChP, and 53-day H1 ChP. Dissociated cells were resuspended in 0.04% BSA in PBS to load 16,000 cells on the 10X Chromium system (10X Genomics). Detailed methods for single cell RNA sequencing, bioinformatic analysis and comparison with in vivo datasets are included in the supplementary material.
Samples for mass spectrometry and NMR analysis
iCSF was collected using a pulled glass microcapillary attached to filter and tubing using controlled suction. In vivo samples were hCSF, human telencephalic CSF (Caltag-Medsystem), bovine fetal CSF (bCSF, BioIVT), and embryonic mouse CSF from E12.5-13.5 telencephalic ventricles. Detailed methods for mass spectrometry analysis, NMR analysis and comparison between in vivo and in vitro drug permeability are included in the supplementary material.
Supplementary Material
One Sentence Summary.
Choroid plexus organoids predict CNS drug permeability and reveal human CSF proteins produced by specialized cell types
Acknowledgments
The authors would like to thank members of the Lancaster lab for helpful feedback and discussions, particularly A. Philips and I. Kelava for technical and intellectual input. We also thank the Light Microscopy and NMR facilities of the MRC Laboratory of Molecular Biology, and P. Pritchett and T. Stevens for help with bioinformatics.
Funding
This work was supported by the Medical Research Council (MC_UP_1201/9) and the European Research Council (ERC STG 757710).
Footnotes
Author contributions: L.P. designed and conducted experiments, analysed data, and wrote the manuscript. C.B. performed NMR experiments and analysed data. J.C. performed electron microscopy and analysed data. F.B. performed mass spectrometry and analysed data. M.S. planned and analysed mass spectrometry. M.A.L. designed and supervised the project, analysed data, and wrote the manuscript.
Competing interests: The authors have filed a patent based on the ChP organoid method.
Data and materials availability
scRNA-seq data is available at ChPOrg.cells.ucsc.edu and on NCBI GEO (GSE150903). All other data is available in the main text or the supplementary materials. H9, H1, and IMR90-4 cells are available from WiCell under a material transfer agreement with WiCell.
M.A.L. and L.P. are inventors on patent application #GB1900930.7 submitted by UKRI that covers choroid plexus organoids.
References
- 1.Lun MP, Monuki ES, Lehtinen MK. Development and functions of the choroid plexus-cerebrospinal fluid system. Nat Rev Neurosci. 2015;16:445–457. doi: 10.1038/nrn3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehtinen MK, Walsh CA. Neurogenesis at the Brain-Cerebrospinal Fluid Interface. Annu Rev Cell Dev Biol. 2011;27:653–679. doi: 10.1146/annurev-cellbio-092910-154026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silva-Vargas V, Maldonado-Soto AR, Mizrak D, Codega P, Doetsch F. Age-Dependent Niche Signals from the Choroid Plexus Regulate Adult Neural Stem Cells. Cell Stem Cell. 2016;19:643–652. doi: 10.1016/j.stem.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Lun MP, Johnson MB, Broadbelt KG, Watanabe M, Kang Y-j, Chau KF, Springel MW, Malesz A, Sousa AMM, Pletikos M, et al. Spatially Heterogeneous Choroid Plexus Transcriptomes Encode Positional Identity and Contribute to Regional CSF Production. J Neurosci. 2015;35:4903–4916. doi: 10.1523/JNEUROSCI.3081-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghersi-Egea JF, Strazielle N, Catala M, Silva-Vargas V, Doetsch F, Engelhardt B. Molecular anatomy and functions of the choroidal blood-cerebrospinal fluid barrier in health and disease. Acta Neuropathol. 2018;135:337–361. doi: 10.1007/s00401-018-1807-1. [DOI] [PubMed] [Google Scholar]
- 6.Redzic Z. Molecular biology of the blood-brain and the blood-cerebrospinal fluid barriers: Similarities and differences. Fluids Barriers CNS. 2011;8:3. doi: 10.1186/2045-8118-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dohrmann GJ, Bucy PC. Human choroid plexus: a light and electron microscopic study. J Neurosurg. 1970;33:506–16. doi: 10.3171/jns.1970.33.5.0506. [DOI] [PubMed] [Google Scholar]
- 8.Lancaster MA, Renner M, Martin C, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pașca SP. The rise of three-dimensional human brain cultures. Nature. 2018;553:437–445. doi: 10.1038/nature25032. [DOI] [PubMed] [Google Scholar]
- 11.Sakaguchi H, Kadoshima T, Soen M, Narii N, Ishida Y, Ohgushi M, Takahashi J, Eiraku M, Sasai Y. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat Commun. 2015;6:1–11. doi: 10.1038/ncomms9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fame RM, Lehtinen MK. Emergence and Developmental Roles of the Cerebrospinal Fluid System. Dev Cell. 2020;52:261–275. doi: 10.1016/j.devcel.2020.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lancaster MA, Corsini NS, Wolfinger S, Gustafson EH, Phillips AW, Burkard TR, Otani T, Livesey FJ, Knoblich JA. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol. 2017;35:659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giandomenico SL, Mierau SB, Gibbons GM, Wenger LMD, Masullo L, Sit T, Sutcliffe M, Boulanger J, Tripodi M, Derivery E, et al. Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat Neurosci. 2019;22:669–679. doi: 10.1038/s41593-019-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe M, Kang Y-J, Davies LM, Meghpara S, Lau K, Chung C-Y, Kathiriya J, Hadjantonakis A-K, Monuki ES. BMP4 Sufficiency to Induce Choroid Plexus Epithelial Fate from Embryonic Stem Cell-Derived Neuroepithelial Progenitors. J Neurosci. 2012;32:15934–15945. doi: 10.1523/JNEUROSCI.3227-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Currle DS, Cheng X, Hsu C, Monuki ES. Direct and indirect roles of CNS dorsal midline cells in choroid plexus epithelia formation. Development. 2005;132:3549–3559. doi: 10.1242/dev.01915. [DOI] [PubMed] [Google Scholar]
- 17.Lehtinen MK, Bjornsson CS, Dymecki SM, Gilbertson RJ, Holtzman DM, Monuki ES. The Choroid Plexus and Cerebrospinal Fluid: Emerging Roles in Development, Disease, and Therapy. J Neurosci. 2013;33:17553–17559. doi: 10.1523/JNEUROSCI.3258-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liddelow SA. Development of the choroid plexus and blood-CSF barrier. Front Neurosci. 2015;9:1–13. doi: 10.3389/fnins.2015.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sathyanesan M, Girgenti MJ, Banasr M, Stone K, Bruce C, Guilchicek E, Nairn A, Williams K. A molecular characterization of the choroid plexus and stress-induced gene regulation. Transl Psychiatry. 2012;2:e139–9. doi: 10.1038/tp.2012.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dani N, Herbst RH, Habib N, Head J, Dionne D, Nguyen L, McCabe C, Cui J, Shipley FB, Jang A, et al. A cellular and spatial map of the choroid plexus across brain ventricles and ages. bioRxiv. 2019 doi: 10.1016/j.cell.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-radji MA, Di Lullo E, Haeussler M, Sandoval-espinosa C, Liu SJ, Velmeshev D, et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566 doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duelli R, Enerson BE, Gerhart DZ, Drewes LR. Expression of Large Amino Acid Transporter LAT1 in Rat Brain Endothelium. J Cereb Blood Flow Metab. 2000;20:1557–1562. doi: 10.1097/00004647-200011000-00005. [DOI] [PubMed] [Google Scholar]
- 24.Uchino H, Kanai Y, Kim DOK, Wempe MF, Chairoungdua A, Morimoto E, Anders MW, Endou H. Transport of Amino Acid-Related Compounds Mediated by L-Type Amino Acid Transporter 1 (LAT1): Insights Into the Mechanisms of Substrate Recognition. Mol Pharmacol. 2002;61:729–737. doi: 10.1124/mol.61.4.729. [DOI] [PubMed] [Google Scholar]
- 25.Durso R, Evans JE, Josephs E, Szabo G, Evans B, Fernandez HH, Browne TR. Variable Absorption of Carbidopa Affects Both Peripheral and Central Levodopa Metabolism. J Clin Pharmacol. 2000;40:854–860. doi: 10.1177/00912700022009585. [DOI] [PubMed] [Google Scholar]
- 26.Rao Vallabhaneni V, Dahlheimer Julie L, Bardgett Mark E, Snyder Abraham Z, Finch Rick A, Sartorelli Alan C, P-W D. Choroid plexus epithelial expression of MDR1 P glycoprotein and multidrug resistance-associated protein contribute to the blood - cerebrospinal-fluid drug-permeability barrier. 1999;96:3900–3905. doi: 10.1073/pnas.96.7.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ek CJ, Wong A, Liddelow SA, Johansson PA, Dziegielewska KM, Saunders NR. Efflux mechanisms at the developing brain barriers: ABC-transporters in the fetal and postnatal rat. Toxicol Lett. 2010;197:51–59. doi: 10.1016/j.toxlet.2010.04.025. [DOI] [PubMed] [Google Scholar]
- 28.Uchida Y, Zhang Z, Tachikawa M, Terasaki T. Quantitative targeted absolute proteomics of rat blood-cerebrospinal fluid barrier transporters: comparison with a human specimen. J Neurochem. 2015;134:1104–1115. doi: 10.1111/jnc.13147. [DOI] [PubMed] [Google Scholar]
- 29.Summerfield SG, Zhang Y, Liu H. Examining the Uptake of Central Nervous System Drugs and Candidates across the Blood-Brain Barrier s. J Pharmacol Exp Ther. 2016;358:294–305. doi: 10.1124/jpet.116.232447. [DOI] [PubMed] [Google Scholar]
- 30.Angelov L, Doolittle ND, Kraemer DF, Siegal T, Barnett GH, Peereboom DM, Stevens G, Mcgregor J, Jahnke K, Lacy CA, et al. Blood-Brain Barrier Disruption and Intra-Arterial Methotrexate-Based Therapy for Newly Diagnosed Primary CNS Lymphoma: A Multi-Institutional Experience. J Clin Oncol. 2009;27:3503–3509. doi: 10.1200/JCO.2008.19.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zylber-Katz E, Gomori JM, Schwartz A, Lossos A, Bokstein F, Siegal T. Pharmacokinetics of methotrexate in cerebrospinal fluid and serum after osmotic blood-brain barrier disruption in patients with brain lymphoma. Clin Pharmacol Ther. 2000;67:631–641. doi: 10.1067/mcp.2000.106932. [DOI] [PubMed] [Google Scholar]
- 32.Das I, Krzyzosiak A, Schneider K, Wrabetz L, Antonio MD, Barry N, Sigurdardottir A, Bertolotti A. Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. 2015;348:239–243. doi: 10.1126/science.aaa4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong CHIH, Siah KWEI, Lo AW. Estimation of clinical trial success rates and related. Biostatistics. 2019;20:273–286. doi: 10.1093/biostatistics/kxx069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaur R, Sidhu P, Singh S. What failed BIA 10-2474 Phase I clinical trial? Global speculations and recommendations for future Phase I trials. J Pharmacol Pharmacother. 2016;7:120–126. doi: 10.4103/0976-500X.189661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otrubova K, Ezzili C, Boger DL. The Discovery and Development of Inhibitors of Fatty Acid Amide Hydrolase (FAAH) Bioorg Med Chem Lett. 2012;21:4674–4685. doi: 10.1016/j.bmcl.2011.06.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butler D, Callaway E. Scientists in the dark after fatal French clinical trial. Nature. 2016;529:236–264. doi: 10.1038/nature.2016.19189. [DOI] [PubMed] [Google Scholar]
- 37.Bird SM, Bailey RA, Sl M, Senn S. Statistical issues in first-in-human studies on BIA 10-2474: neglected comparison of protocol against practice. Pharm Stat. 2017;16:100–106. doi: 10.1002/pst.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kida E, Palminiello S, Golabek AA, Walus M, Wierzba-bobrowicz T, Rabe A, Albertini G, Wisniewski KE. Carbonic Anhydrase II in the Developing and Adult Human Brain. 2006;65:664–674. doi: 10.1097/01.jnen.0000225905.52002.3e. [DOI] [PubMed] [Google Scholar]
- 39.Wollack JB, Makori B, Ahlawat S, Koneru R, Picinich SC, Smith A, Goldman ID, Qiu A, Cole PD, Glod J, et al. Characterization of folate uptake by choroid plexus epithelial cells in a rat primary culture model. J Neuroch. 2008;104:1494–1503. doi: 10.1111/j.1471-4159.2007.05095.x. [DOI] [PubMed] [Google Scholar]
- 40.Ulloa V, Saldivia N, Ferrada L, Salazar K, Martínez F, Silva-a C, Magdalena R, Oviedo MJ, Montecinos H, Torres-vergara P, et al. Basal Sodium-Dependent Vitamin C Transporter 2 polarization in choroid plexus explant cells in normal or scorbutic conditions. Sci Rep. 2019;9:1–13. doi: 10.1038/s41598-019-50772-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roy A, Al-bataineh MM, Pastor-Soler NM. Collecting Duct Intercalated Cell Function and Regulation. Ren Physiol. 2015;10:305–324. doi: 10.2215/CJN.08880914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pollen AA, Nowakowski TJ, Chen J, Leyrat AA, West JA, Kriegstein AR, Chen J, Retallack H, Sandoval-espinosa C, Kriegstein AR. Molecular Identity of Human Outer Radial Glia during Cortical Development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicolas J-M. Blood-Brain Barrier in Drug Discovery: Optimizing Brain Exposure of CNS Drugs and Minimizing Brain Side Effects for Peripheral Drugs. 2015 [Google Scholar]
- 44.Miller JA, Ding S, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:191–198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dayon L, Galindo AN, Wojcik J, Cominetti O, Corthésy J, Oikonomidi A, Henry H, Kussmann M, Migliavacca E, Bowman GL, et al. Alzheimer disease pathology and the cerebrospinal fluid proteome. Alzheimers Res Ther. 2018;10:1–12. doi: 10.1186/s13195-018-0397-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo L, Ren H, Zen H, Gong Y, Ma X. Proteomic analysis of cerebrospinal fluid in pediatric acute lymphoblastic leukemia patients: a pilot study. Onco Targets Ther. 2019;12:3859–3868. doi: 10.2147/OTT.S193616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zappaterra MD, Lisgo SN, Lindsay S, Gygi SP, Walsh CA, Ballif BA. A comparative proteomic analysis of human and rat embryonic cerebrospinal fluid. J Proteome Res. 2007;6:3537–3548. doi: 10.1021/pr070247w. [DOI] [PubMed] [Google Scholar]
- 49.Lancaster M, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-seq data is available at ChPOrg.cells.ucsc.edu and on NCBI GEO (GSE150903). All other data is available in the main text or the supplementary materials. H9, H1, and IMR90-4 cells are available from WiCell under a material transfer agreement with WiCell.
M.A.L. and L.P. are inventors on patent application #GB1900930.7 submitted by UKRI that covers choroid plexus organoids.