Abstract
Polo-like kinase 1 (PLK1) critically regulates mitosis through its dynamic localization to kinetochores, centrosomes and the midzone. The polo-box domain (PBD) and activity of PLK1 mediates its targeting to mitotic structures, but the mechanisms regulating PLK1 dynamics remain poorly understood. Here, we identify PLK1 as a target of the Cullin 3 (CUL3)-based E3 ubiquitin ligase, containing the BTB-adaptor KLHL22, which regulates chromosome alignment and PLK1 kinetochore localization but not PLK1 stability. In the absence of KLHL22, PLK1 accumulates on kinetochores, resulting in activation of the Spindle Assembly Checkpoint (SAC). CUL3-KLHL22 ubiquitinates K492, located within the PBD, leading to PLK1 dissociation from kinetochore phosphoreceptors. Expression of a non-ubiquitinatable PLK1-K492R mutant phenocopies inactivation of CUL3-KLHL22. KLHL22 associates with the mitotic spindle and its interaction with PLK1 increases upon chromosome biorientation. Our data suggest that CUL3-KLHL22-mediated ubiquitination signals degradation-independent removal of PLK1 from kinetochores and SAC satisfaction, which is required for faithful mitosis.
Keywords: mitosis, kinetochore, cullin, CUL3, BTB proteins, KLHL22, kinases, Polo-like kinase 1, PLK1
Introduction
Mitotic division is instrumental for the development of all organisms and defects in its regulatory systems lead to aneuploidy, which is linked to cellular transformation1,2. Polo-like kinase 1 (PLK1) is a critical regulator of mitotic division and an attractive candidate for anticancer drugs3. PLK1 controls chromosome alignment, centrosome function and spindle assembly through its dynamic localization to kinetochores, centrosomes and the midzone4. Downregulation of PLK1 protein or pharmacological inhibition of the PLK1 kinase activity leads to mitotic defects ultimately leading to activation of the Spindle Assembly Checkpoint (SAC) and apoptotic death5,6. However, recent data suggest that a fine balance of PLK1 protein levels and its kinase activity is required for chromosome alignment and faithful mitotic progression7–9. First, PLK1 accumulates at the kinetochores during prometaphase stage to establish kinetochore attachments and subsequently majority of PLK1 protein has to be removed from kinetochores during metaphase to allow for maintenance of stable kinetochore-microtubule interactions, SAC silencing and anaphase onset7–9. The polo-box domain (PBD) and autocatalytic activity of PLK1 have been implicated in its targeting to different mitotic structures, including kinetochores10–13, but the mechanisms regulating PLK1 dynamics remain poorly understood.
Several posttranslational modifications including phosphorylation and acetylation ensure correct expression and activity of key mitotic factors14,15. Moreover, covalent conjugation of ubiquitin to specific lysine residues of target proteins emerged as a critical regulatory signal in cellular physiology, mediating both proteolysis dependent and independent mechanisms16,17. Ubiquitin is attached to a substrate by coordinated cycles of three enzymatic reactions, ubiquitin activation (E1 enzyme), ubiquitin conjugation (E2 enzyme) and ubiquitin ligation (E3 protein ligase). Previous studies in human cells identified critical roles and substrates of Cullin-RING E3 ubiquitin ligases (CRL) in cell proliferation and tumour progression18,19. In particular, CUL3 appears to regulate several aspects of mitosis by assembling E3 ubiquitin ligase complexes with specific BTB (Bric-a-brac/Tramtrack/Broad complex) adaptor proteins20–22.
In this study, we identify PLK1 as a target of the Cullin 3 (CUL3)-based E3 ubiquitin ligase, which in complex with the BTB-adaptor protein KLHL22 regulates chromosome alignment and PLK1 kinetochore localization but not PLK1 stability. In the absence of KLHL22, PLK1 accumulates on mitotic kinetochores, resulting in hyperphosphorylation of its target BubR1, inhibition of stable kinetochore-microtubule interactions and activation of the Spindle Assembly Checkpoint (SAC). CUL3-KLHL22 directly binds PLK1 and ubiquitinates K492, located within the PBD, leading to PLK1 dissociation from kinetochore phosphoreceptors. Expression of a non-ubiquitinatable PLK1-K492R mutant phenocopies inactivation of CUL3-KLHL22. Interestingly, KLHL22 associates with the mitotic spindle and its interaction with PLK1 increases as cells achieve chromosome biorientation. Together, our data suggest that CUL3-KLHL22-mediated ubiquitination signals degradation-independent removal of PLK1 from kinetochores and SAC satisfaction, which is required for faithful progression through mitosis.
Results
CUL3-KLHL22 E3-ligase regulates SAC-dependent chromosome alignment during mitosis
The BTB-Kelch protein KLHL22 interacts with CUL3 (Suppl. Fig. 1a) and is required for mitotic division22, but its molecular function and critical ubiquitination substrates remain unknown. KLHL22 is expressed throughout the cell cycle with levels moderately increasing during mitosis (Suppl. Fig. 1b). Live-cell microscopy of HeLa cells stably expressing the histone marker H2B-mRFP revealed that KLHL22-depleted cells show severe chromosome alignment defects, with 27.9 % (+/- 5 %) of cells dying after a prolonged prometaphase-like arrest without anaphase initiation (Fig. 1a, b). However, unlike CUL3 depletion, KLHL22 RNAi did not lead to an increased occurrence of micronuclei or multinucleated cells (Fig. 1c). To assess whether activation of the Spindle Assembly Checkpoint (SAC) causes the observed prometaphase arrest, KLHL22-depleted cells were arrested in mitosis by addition of the proteasome inhibitor MG132, and the protein mitotic arrest deficient 2 (MAD2) was visualized as a marker for unattached kinetochores23 by immunofluorescence microscopy. In contrast to RNAi-controls, MAD2-positive kinetochores could be detected in 96.7 % of the mitotically arrested KLHL22-depleted cells (Fig. 1d). The presence of unaligned but MAD2-negative kinetochores in CUL3-depleted cells (Suppl. Fig. 1c) could be explained by pleiotropic effects including SAC-slippage24 or incorrect kinetochore attachments, accounting for the fact that CUL3 forms functional complexes with several substrate adaptors22.
Figure 1. The BTB adaptor protein KLHL22 regulates SAC-dependent chromosome alignment during mitosis.
a, HeLa cells expressing H2B-mRFP were treated with the indicated siRNAs for 48 h and analyzed by live-cell microscopy. Stars indicate misaligned and lagging chromosomes. Bar is 10μm. b, The percentage of cells that complete mitosis normally or with a delay from prophase to anaphase (>50 min) or that undergo apoptosis during prometaphase was quantified (n = 50). c, The percentage of cells that undergo normal mitosis, become multinucleated or die during mitosis was quantified (n=50). d, HeLa cells were treated with the indicated siRNAs for 48 h and analyzed by immunofluorescence microscopy for MAD2 and kinetochores. The percentage of prometaphase and metaphase cells that contain one or more MAD2 positive kinetochores was quantified (n = 30). Example images are shown in Suppl. Fig. 1c. e, HeLa cells expressing H2B-mRFP were treated with the indicated mixtures of siRNAs for 48 h and analyzed by live-cell microscopy. The percentage of cells that die in prometaphase was quantified (n = 50). b-d, Bars represent the mean of three independent experiments. Error bars indicate +/-s.d.
Importantly, simultaneous inactivation of KLHL22 and MAD2 rescued the KLHL22 mitotic-cell-death phenotype (Fig. 1e, Suppl. Fig. 1d). KLHL22 depletion did not impede bipolar spindle formation, loading of condensin on chromosomes (data not shown) or kinetochore assembly (Suppl. Fig. 1e-h). However, 57.5 % of the KLHL22-depleted cells failed to form proper metaphase plates in the presence of MG132, compared to 22.1 % of control cells, suggesting that the CUL3-KLHL22 E3-ligase regulates SAC-dependent processes required for efficient chromosome alignment during mitosis.
The CUL3-KLHL22 E3-ligase tagerts PLK1 and regulates its kinetochore localization during mitosis
The KLHL22 RNAi phenotype suggests that CUL3-KLHL22 targets proteins with essential functions during mitosis. To identify these substrates, we probed protein microarrays containing ~8300 human proteins for their binding to AlexaFluor647-labeled KLHL22, and for their in vitro ubiquitination by reconstituted CUL3-Rbx1-KLHL22 complexes in the presence of FITC-labeled ubiquitin. As expected, MBP-KLHL22 interacted with GST-CUL3-Rbx1, but not with GST controls, and reconstituted CUL3-Rbx1-KLHL22 complexes promoted autoubiquitination of GST-CUL3, but not GST- or MBP-tagged negative control substrates (Suppl. Fig. 2). We identified 93 proteins that specifically interacted with MBPKLHL22 and 30 of these were also ubiquitinated by CUL3-Rbx1-KLHL22, suggesting they may be substrates of this E3-ligase (Suppl. Table 1 and 2). Interestingly, in our list of candidates we found a 9- and 6-fold overrepresentation of protein kinases, respectively. Among them was Polo-like kinase 1 (PLK1), which is well known to regulate several processes in mitosis5 including chromosome alignment, centrosome function and spindle assembly through its dynamic localization to kinetochores, centrosomes and the midzone region4. Consistent with the protoarray results, endogenous PLK1, but not PLK2, coimmunoprecipitated with FLAG-tagged KLHL22 in extracts prepared from mitotically synchronized cells (Fig. 2a). Likewise, endogenous KLHL22 was efficiently coimmunoprecipitated with endogenous PLK1 from mitotic-stage cells (Fig. 2b). Accordingly, recombinant KLHL22, but not KLHL21, strongly bound purified PLK1 in vitro and this interaction is established via the Kelch domain of KLHL22 (Fig. 2c, d). Together, these data suggest that PLK1 is indeed a specific and direct target of CUL3-KLHL22.
Figure 2. The CUL3-KLHL22 E3-ligase interacts directly with PLK1 and regulates its kinetochore localization during mitosis.
a, HeLa cells inducibly expressing FLAG-KLHL22 were treated with doxycycline (Dox), synchronized by double thymidine block release and harvested in mitosis by mitotic shake-off. Cell extracts were immunoprecipitated with anti-FLAG antibodies and analyzed by Western blot. The asterisk indicates the band corresponding to PLK2. b, HeLa cells were synchronized as in (a) and extracts were immunoprecipitated with control (IgG IP) or anti-PLK1 (PLK1 IP) antibodies and analyzed by Western blot. c, Recombinant GST or GST fused to full length PLK1 (GST-PLK1) were incubated with recombinant MBP, MBP fused to full length KLHL21 (MBP-KLHL21) or KLHL22 (MBP-KLHL22), and immunoprecipitated using glutathione-sepharose beads. Immunoprecipitates (GST-IP) were washed in low and high salt conditions and analyzed by Coomassie blue staining and Western blot. d, Recombinant GST, GST fused to the Kelch repeats (GST-6K) of KLHL21, KLHL22 or KLHL9 were incubated with recombinant PLK1, immunoprecipitated using glutathionesepharose beads and analyzed by Western blot. e, HeLa cells were treated with the indicated siRNAs for 48 h, arrested in mitosis with Nocodazole for 14 h and analyzed by immunofluorescence microscopy. Bar is 5 μm. f, Quantification of relative intensity ratios of PLK1 : CREST staining on individual kinetochores (from 10 cells) from unsynchronized (as in Suppl. Fig. 3c, light grey) and Nocodazole synchronized cells(as in (e), dark grey). The ratios of control siRNA treated cells were set to 1. g, Quantification of intensity ratios of different kinetochore markers normalized to the corresponding CREST signals in control (light grey) and KLHL22 depleted (dark grey) cells. h, The relative kinetochore intensities of PLK1 were quantified as in (f) from cells treated with indicated siRNAs and Nocodazole (NOC), Taxol (TAX), Monastrol (MON), MG132 or DMSO. f, g, and h, Bars represent the mean of three independent experiments. Error bars indicate +/-s.d.
To understand the physiological consequences of KLHL22-mediated ubiquitination of PLK1, we compared its abundance and subcellular localization in control and CUL3-KLHL22-depleted cells using PLK1-specific antibodies. We were unable to detect significant differences in total cellular levels of PLK1 in the presence or absence of KLHL22 (Fig. 3c). Consistent with published results4,7,9, we observed that kinetochore-associated PLK1 levels were dramatically reduced as chromosomes become bioriented in metaphase (Suppl. Fig. 3a, b). Interestingly, downregulation of KLHL22 or CUL3 led to a striking increase of PLK1 on kinetochores during prometaphase when compared to control siRNA-treated cells (Fig. 2e, f, Suppl. Fig. 3c), while its levels on centrosomes were slightly decreased (Suppl. Fig. 3d, e). In contrast, KLHL22 depletion did not alter the localization of any other kinetochore protein we tested (Fig. 2g and Suppl. Fig. 1e-h). Importantly, PLK1 levels on kinetochores also increased in CUL3- or KLHL22-depleted cells arrested with the microtubule-depolymerizing drug Nocodazole (Fig. 2c, d), excluding the possibility that this effect was indirectly caused by the prolonged prometaphase arrest. Consistent with this notion, downregulation of the previously described CUL3 adaptor KLHL2122 (Fig. 2e, f) or drug-induced arrest in different mitotic stages (Fig. 2h) did not result in PLK1 accumulation on kinetochores. Taken together, we conclude that the CUL3-KLHL22 E3 ligase may act to remove PLK1 from kinetochores before chromosomes achieve biorientation in metaphase, without affecting total PLK1 levels or other subcellular pools of kinetochore proteins.
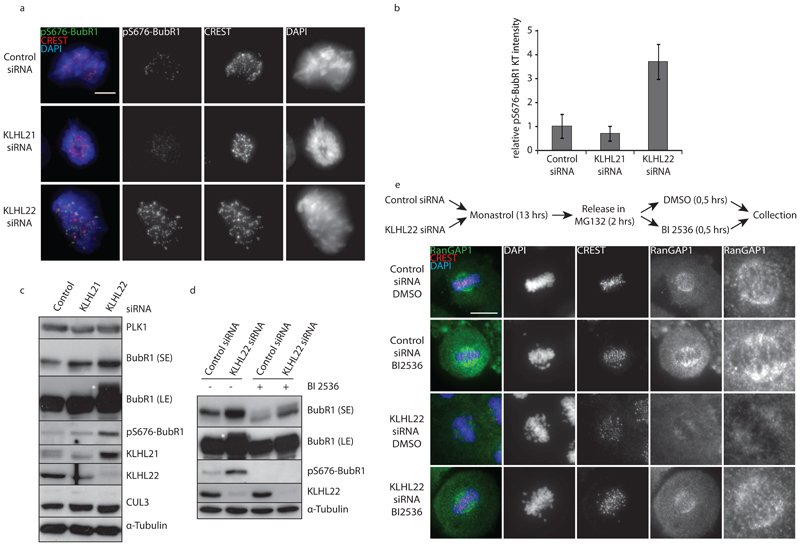
Figure 3. KLHL22 regulates PLK1-mediated phosphorylation of BubR1 and stable KTMT attachments.
a, HeLa cells were treated with the indicated siRNAs for 48 h, arrested in mitosis with Nocodazole for 14 h and analyzed by immunofluorescence microscopy. Bar 5 is μm. b, Quantification of the relative intensity ratios of pS676 BubR1 : CREST on individual kinetochores (n = 10) from (a). Bars represent the mean of three independent experiments. Error bars indicate +/-s.d. c, HeLa cells were treated as in (a), harvested by mitotic shake-off and analyzed by Western blot. d, HeLa cells were treated as in (c), incubated with BI 2536 as indicated and analyzed by Western blot. e, HeLa cells were treated with the indicated siRNAs for 48 h and analyzed by immunofluorescence microscopy. Insets are 3x zooms of the framed region. Bar is 10 μm.
CUL3-KLHL22 regulates maintenance of stable kinetochore-microtubule interactions
In order to test if CUL3-KLHL22 regulates PLK1 activity, we analyzed the phosphorylation state of the kinetochore protein BubR1. PLK1-mediated phosphorylation of BubR1 occurs at several sites and has been correlated with stability of kinetochore-microtubule (KT-MT) interactions7,25. While total, kinetochore-associated BubR1 levels were unaffected (Fig. 2g), inactivation of KLHL22, but not KLHL21 in mitotically-arrested cells led to a 3- to 4-fold augmented phosphorylation of BubR1 on Ser676, consistent with the observed PLK1 increase on kinetochores (Fig. 3a-c). Accordingly, Ser676 phosphorylation of BubR1 was abolished upon treatment with the PLK1 inhibitor BI 25366 (Fig. 3d). In contrast, PLK1 activity-dependent accumulation of γ-tubulin on centrosomes was normal in mitotic cells (Suppl. Fig. 3e). Together, these results suggest that inactivation of CUL3-KLHL22 leads to increased PLK1 levels and sustained kinase activity specifically at kinetochores, leading to increased BubR1 phosphorylation. As BubR1 phosphorylation was reported to regulate stable KT-MT interactions7, we monitored kinetochore localization of RanGAP1, which depends on stable KT-MT interactions26. To this end, we arrested control and KLHL22-depleted cells with Monastrol, and released them into MG132-containing media (Suppl. Fig. 3f) to achieve a highly synchronous population of cells with bioriented chromosomes. In contrast to control-depleted cells, RanGAP1 failed to efficiently localize to kinetochores and mitotic spindles in the absence of KLHL22 (Suppl. Fig. 3f) and this defect could be reversed by treatment with the PLK1 inhibitor BI 2536 ((Fig. 3e). Together these results suggest that in the absence of KLHL22, cells fail to establish stable KT-MT interactions because PLK1 accumulates on kinetochores leading to hyperphosphorylation of BubR1 and, as a consequence, defects in chromosome alignment.
CUL3-KLHL22-mediated ubiquitination of PLK1 within the PBD regulates faithful chromosome alignment during mitosis
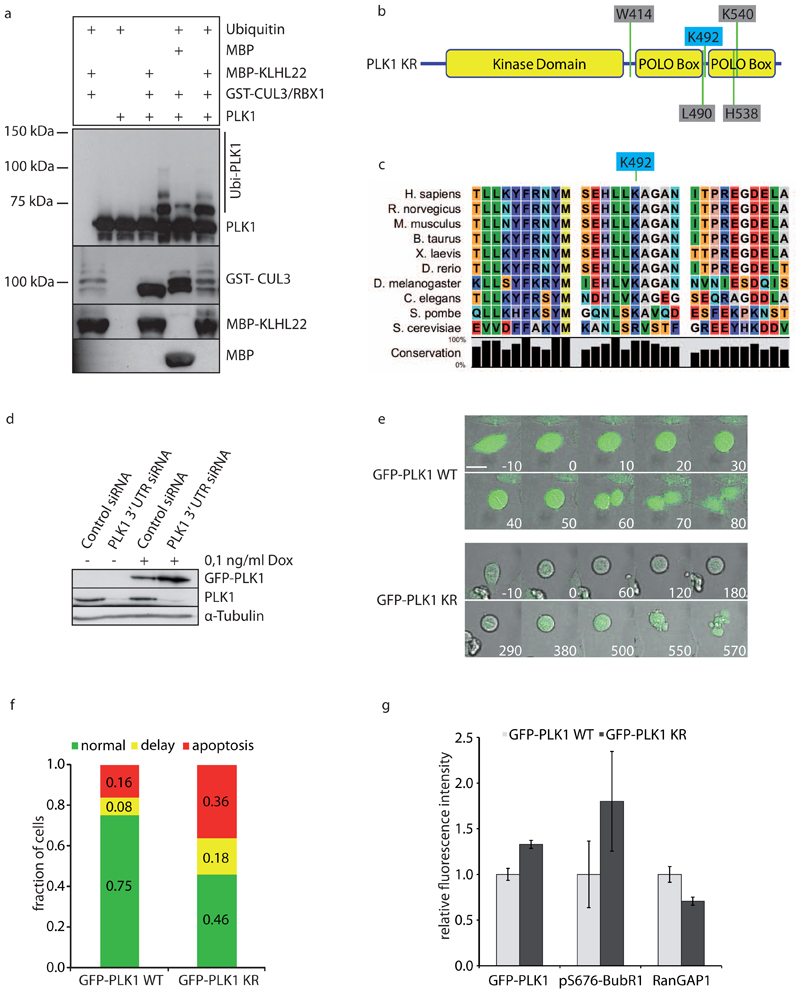
To examine if PLK1 ubiquitination is indeed critical for mitotic progression, we next determined the sites ubiquitinated by CUL3-KLHL22 complexes in vitro. GST-CUL3-Rbx1 complexes purified from Sf9 cells reconstituted with E.coli expressed MBP-tagged KLHL22 were able to mainly mono-ubiquitinate recombinant PLK1, whereas multiple sites were used with lower efficiency (Fig. 4a). Mass spectrometry analysis identified lysine residues 492 (K492) and 19 (K19) as ubiquitin acceptor sites. Interestingly, the lysine 492 resides in the polo box domain (PBD) of PLK1, is conserved from fission yeast to humans (Fig 4b, c) and was confirmed as ubiquitin acceptor site in vivo in two independent proteomic studies27,28.
Figure 4. CUL3-KLHL22-mediated ubiquitination of PLK1 within the PBD domain regulates faithful chromosome alignment during mitosis.
a, Recombinant PLK1 was added to in vitro ubiquitination reactions containing GSTCUL3/RBX1 complexes purified from Sf9 cells. Reactions were incubated for 1 h with MBP or MBP-KLHL22 purified from E. coli and analyzed by Western blot. Note that PLK1 was mainly monoubiquitinated (asterisk) under these conditions, but multiple sites were used with lower efficiency. b, Schematic representation of PLK1. The ubiquitin acceptor lysine K492 is indicated in blue, while residues required for PBD phosphosite targeting are indicated in grey.c, Alignment of PLK1 regions containing the identified ubiquitin acceptor site K492 from different species. d, HeLa cells inducibly expressing GFP-PLK1 WT were treated with doxycycline (+/- Dox) and transfected with control or PLK1 3’UTR siRNA. Cell extracts were analyzed by Western blot. e, HeLa cell lines inducibly expressing GFP-PLK1 wild type (WT) or the KR mutant were transfected with PLK1 3’UTR siRNA and analyzed by live-cell microscopy 24-48 h after transfection. Minutes before and after mitotic entry are indicated. Bar is 10μm. f, Quantification of cells from (e) that complete mitosis (green and yellow) versus cells that undergo apoptosis during prometaphase (red) (total numbers of cells analyzed: nWT=312, nKR=235). For the cells that complete mitosis, the duration from prophase to anaphase was quantified and the fraction of cells displaying a normal (green) or delayed (yellow) anaphase onset was calculated. Delay is defined as ‘duration longer than WT average + 1x standard deviation’. Duration values were quantified from nWT=80, nKR=78 cells. g, Cells expressing GFP-PLK1WT (light grey) or GFP-PLK1KR (dark grey) were transfected with PLK1 3’UTR siRNA, fixed 36 h after transfection and analyzed by immunofluorescence microscopy. GFP-PLK1 was visualized by GFP microscopy, pS676 BubR1, RanGAP1 and CREST were stained using the corresponding antibodies. To quantify kinetochore-associated GFP-PLK1, GFP at individual kinetochores (n=322 kinetochores in total from 3 experiments) was normalized both to cytoplasmic GFP and to the corresponding CREST signal at kinetochores using the following equation: GFP(kinetochore) / GFP(cytoplasm) / CREST(kinetochore). pS676 BubR1 signals of individual kinetochores (n=36 kinetochores from 1 experiment) were normalized both to the corresponding CREST and GFP-PLK1 signal intensities using the following equation: (pS676 BubR1/CREST)/(GFP/CREST), while RanGAP1 intensities on individual kinetochores (n=322 kinetochores in total from 3 experiments) were normalized to CREST staining. Error bars indicate +/-s.d.
Because PBD mediates recruitment to mitotic structures including kinetochores11–13 we tested if ubiquitination by the CUL3-KLHL22 E3-ligase may regulate PBD-mediated PLK1 function at kinetochores. To this end, we generated HeLa cell lines that allow Doxycyclineinducible expression of GFP-fused PLK1 wild type (WT) or a version of PLK1 with K492 mutated to a non-ubiquitinatable arginine residue (KR). Expression of PLK1WT was able to rescue the mitotic defects induced by 3’UTR-directed siRNA targeting endogenous PLK1, including the delay at the metaphase-anaphase transition and induction of apoptosis (Fig. 4d, e, f). In contrast, expression of the PLK1KR mutant resulted in severe mitotic defects leading to a prometaphase delay that was often followed by apoptotic death (36%). Moreover, kinetochore localization of the PLK1KR mutant was augmented and these cells showed increased levels of phosphorylated BubR1 and decreased levels of RanGAP1 on kinetochores (Fig. 4g), thus resembling the KLHL22 RNAi phenotype. These results were confirmed in HeLa cell lines expressing HIS tag-fused PLK1WT or PLK1KR (Fig. 5a, b). As expected, centrosome maturation and bipolar spindle formation were not perturbed under these conditions (data not shown), implying that the PLK1KR mutant is specifically defective for its kinetochore function. Importantly, no significant difference in kinase activity could be detected when comparing PLK1WT and the PLK1KR mutant (Suppl. Fig. 4a). Moreover, the interaction of PLK1 with APC/C activator Cdh1 was not affected by the K492R mutation (Suppl. Fig. 4b). Accordingly, the PLK1KR mutant was efficiently degraded at the end of mitosis with kinetics comparable to PLK1WT or endogenous PLK1 (Suppl. Fig. 4c), implying the K492R mutation does not influence APC/CCdh1-mediated ubiquitination and subsequent proteasomal degradation of PLK1. Together, these results suggest that the accumulation of PLK1 at kinetochores and resulting mitotic defects can be linked to CUL3-KLHL22-mediated ubiquitination of K492 located in the PBD of PLK1.
Figure 5. CUL3-KLHL22-mediated ubiquitination of PLK1 regulates PBD-mediated phospho-interactions at kinetochores and stable KT-MT attachments.
a, HeLa cells inducibly expressing HIS-PLK1 WT or the KR mutant were transfected with PLK1 3’UTR siRNA and analyzed by immunofluorescence microscopy. 5x zooms of the boxed regions are shown on the right side. Note: expression of PLK1KR mutant leads to increase in kinetochore association of PLK1 and decrease in RanGAP1 at kinetochores but not at the nuclear envelope (arrows) in interphase cells. Bar is 5 μm. b, Cells expressing HISPLK1WT (light grey) or HIS-PLK1KR (dark grey) were treated as in (Fig 4 g) and kinetochoreassociated HIS-PLK1 (n=150 kinetochores in total from 3 experiments) and RanGAP1 (n=170 kinetochores in total from 3 experiments) were quantified as in (Fig 4 g). Error bars indicate +/-s.d. c, Model of PLK1 crystal structure43 highlighting the relevant residues of this study. The ubiquitin acceptor lysine K492 is shown in blue, the residues required for phosphopeptide targeting are shown in grey. d, Cells expressing GFP alone, GFP-PLK1WT or GFP-PLK1KR were synchronized in mitosis, immunoprecipitated using GFP-Trap beads and analyzed by Western blot. e, Cells expressing GFP alone, GFP-PLK1WT or GFP-PLK1KR were transfected with either HA alone or HA-KLHL22 and immunoprecipitated using GFPTrap beads and analyzed by Western blot.
The PBD is essential to bring PLK1 to its substrates through phospho-dependent interactions with its target proteins29. Based on structural predictions, K492 is in close proximity to the four key residues involved in phosphosite binding (Trp414, Leu490, His538 and Lys540) (Fig. 4b, Fig. 5c), raising the possibility that ubiquitination by the CUL3-KLHL22 E3-ligase may regulate PBD-mediated interactions of PLK1 at the kinetochores. Thus, we tested the interaction of PLK1WT and PLK1KR with the phosphoreceptor proteins INCENP and BubR1 in mitotically synchronized cells. Remarkably, PLK1KR binding to these kinetochore phosphoreceptors was increased as compared to PLK1WT(Fig. 5d, e). Furthermore, KLHL22 overexpression reduced the interaction between BubR1 and PLK1WT but not PLK1KR(Fig. 5e), suggesting that ubiquitination of K492 inhibits PBD-mediated phospho-interactions of PLK1 at kinetochores.
KLHL22 accumulates at the mitotic spindle and its association with PLK1 increases upon chromosome biorientation
To understand the spatio-temporal regulation of CUL3-KLHL22 E3 ligase, we generated a Doxycycline-inducible cell line expressing GFP-KLHL22 and assayed its localization throughout mitosis. GFP-KLHL22 efficiently interacted with endogenous CUL3 (Suppl. Fig. 5a) and PLK1 (Suppl. Fig. 5b), suggesting that the GFP-fusion does not affect the function of KLHL22 as a CUL3 adaptor in vivo. While GFP-KLHL22 was mostly visible as a diffuse cytoplasmic signal in prophase and prometaphase, it associated with the mitotic spindle as the cells reached chromosome biorientation (Fig. 6a), consistent with the reported localization of CUL330. Interestingly, live video microscopy revealed that GFP-KLHL22 localized to the centrosomes shortly before cells entered anaphase (Fig. 6a, Suppl. Fig. 5c and Suppl. movies). After anaphase onset, GFP-KLHL22 was predominantly associated with the polar microtubules connecting the two opposing centrosomes and gradually diffused into the cytoplasm during telophase. Thus, KLHL22 may use the mitotic spindle compartment to remove PLK1 from kinetochores.
Figure 6. KLHL22 accumulates at the mitotic spindle and its association with PLK1 peaks with chromosome biorentation.
a, Immunofluorescence microscopy was performed using a Doxycycline-inducible GFP-KLHL22 cell line, which was synchronized in mitosis by RO3306 block and release, fixed and stained with CREST and α-tubulin using specific antibodies. GFP-KLHL22 is visualized only by its GFP fluorescence. Images are maximum intensity projections through Z-stacks spanning a total depth of 12 μm in 0.5μm increments. Bar is 10 μm. b, Workflow for synchronizing cells in metaphase and addition of DMSO, the microtubule-stabilizing drug Taxol (TAX) or the microtubule-depolymerizing agent Nocodazole (NOC). Note that Taxol allows MT attachment without tension being exerted on kinetochores, while kinetochores in Nocodacole-treated cells are not attached to MT’s. c, HeLa cells expressing HA-KLHL22 and GFP-PLK1 were treated as outlined in (b), harvested and GFP-PLK1 was immunoprecipitated and analyzed by Western blot. d, HeLa cells expressing HA-KLHL22 were treated as outlined in (b), harvested and endogenous PLK1 was immunoprecipitated and analyzed by Western blot. e, Model for ubiquitination-dependent regulation of PLK1 in mitosis by CUL3/KLHL22. PLK1 (green) localizes to kinetochores (dark blue) and phosphorylates (P) the key SAC component, BubR1 kinase (beige) to control chromosome alignment during prometaphase and metaphase stages. Following the establishment of stable attachments of kinetochores by microtubules (Tension) (black bars), CUL3 (red) together with the substrate specific adaptor protein KLHL22 (light blue) binds and ubiquitinates PLK1 (Ub, yellow) within the PBD domain leading to its dissociation from phosphoreceptor proteins and thereby efficient removal from kinetochores, allowing for silencing of SAC and chromosome segregation. This process may occur dynamically during prometaphase stages to allow for a precise correction mechanism sensing stable MT-KT interactions and microtubule-exerted tension and satisfaction of the SAC.
To understand if the interaction of CUL3-KLHL22 with PLK1 is regulated by chromosome biorientation, we tested if KLHL22-PLK1 binding was influenced by the status of KT-MT interactions. To this end, PLK1 was immunoprecipitated from cells arrested with Monastrol, and released into media containing MG132 to accumulate cells in metaphase. Upon chromosome biorientation, cells were treated with either DMSO, Nocodazole or Taxol, to alleviate kinetochore attachments and/or tension exerted on kinetochores7 (Fig. 6b). As expected, BubR1 was mostly dephosphorylated in DMSO controls whereas upon Taxol or Nocodazole treatment BubR1 was predominantly found in its slow migrating, phosphorylated form. Interestingly, under these conditions the interaction of GFP-PLK1 with HA-KLHL22 was clearly reduced compared to DMSO-treated cells, in which the attached microtubules exert tension across the kinetochores (Fig. 6c). Likewise, interaction of the endogenous PLK1 with HA-KLHL22 was reduced upon Taxol treatment (Fig. 6d). These results suggest that chromosome biorientation and the concomitant structural changes in kinetochores may be a prerequisite for efficient binding of CUL3-KLHL22 to PLK1. This mechanism may thus allow for a spatio-temporal control of PLK1 ubiquitination on K492, which in turn may trigger its dissociation from kinetochores and anaphase onset.
Discussion
In summary, our results strongly suggest that PLK1 is a direct target of the CUL3-KLHL22 E3 ligase complex and that its ubiquitination within the PBD domain regulates PLK1 localization at kinetochores, chromosome alignment, SAC satisfaction and ultimately cell survival during metaphase. Defects in PLK1 ubiquitination result in enhanced interaction with phosphoreceptor proteins and consequently in sustained kinase activity towards its kinetochore substrate BubR1, which in turn interferes with stable KT-MT interactions. PLK1 dissociation from kinetochores has been correlated with bipolar attachment of chromosomes to the mitotic spindle7,9, but the mechanism of its removal was unknown so far. Our data suggest that the CUL3-KLHL22 interaction with PLK1 increases as chromosomes become bioriented, leading to PLK1 ubiquitination and dissociation from kinetochores, thus providing a plausible mechanism of PLK1 removal from kinetochores upon biorientation and consequently satisfaction of SAC signalling (Fig. 6e).
Versatility of functions of CUL3-based ligases
Previous studies established CUL3 as a critical regulator of cellular physiology by assembling E3 ubiquitin ligase complexes with specific BTB adaptor proteins31. CUL3 complexes ubiquitinate and target substrates for proteasomal degradation thereby regulating cellular homeostasis32,33 and development of disease34,35. Importantly, several CUL3 complexes were also shown to regulate cell cycle progression20–22,30.
Our results suggest that the CUL3-KLHL22 E3-ligase ubiquitinates PLK1 at lysine 492 and regulates its localization specifically at kinetochores but does not regulate PLK1 protein stability. Importantly, this site was demonstrated to be ubiquitinyated in vivo in both recent proteomic studies27,28. Although we were unable to demonstrate the specific type of ubiquitin conjugation to PLK1 catalyzed by CUL3/KLHL22 in cells, our in vitro ubiquitintation data strongly suggest that PLK1 is modified by attachment of a mono-ubiquitin. It is also possible that only a small fraction of PLK1, associated with kinetochores specifically when chromosomes achieve bi-orientation, gets modified. This modification is also likely to be very dynamic and quickly reversed by the activity of an unknown DUB. Thus, both the temporal and spatial restriction makes biochemical detection of such modification very difficult with conventional methods. However, in light of the available evidence, and because expression of a PLK1 K492R mutant strikingly mimics the mitotic defects observed upon downregulation of KLHL22 in cells, and conversely overexpression of KLHL22 reduces the interaction of PLK1 and BubR1 in a K492-dependent manner, we believe that CUL3-KLHL22 may directly mono-ubiquitinate this site to promote removal of PLK1 from kinetochores upon bipolar microtubule attachment. Interestingly, other CUL3-based complexes were recently shown to catalyze monoubiquitination36,37. These findings significantly expand the versatility and possible functions of CUL3-based ligases, and implies that they not only target substrates for proteasomal degradation, but also regulate reversible processes by regulating subcellular localization and possibly the activity and/or interactions of substrates in space and time.
Spatio-temporal regulation of CUL3/KLHL22
We demonstrate that KLHL22 associates with the mitotic spindle and its interaction with PLK1 increases upon chromosome biorientation. Interestingly, CUL3 was also demonstrated to associate to mitotic spindle30. These results could provide an insight into how physical attachment of microtubules may covert to a biochemical signal silencing SAC at the kinetochore. While GFP-KLHL22 did not accumulate at kinetochores, it is known that several dynamic kinetochore components are transported towards the spindle poles upon checkpoint inactivation by the dynein/dynactin pathway38–40. As CUL3 was previously reported to interact with components of the dynein-complex30,41, these results could explain how CUL3/KLHL22 uses the microtubule-spindle compartment to ubiquitinate and remove PLK1 from kinetochores upon chromosome biorientation.
CUL3-KLHL22 regulates the kinetochore activity of PLK1 and SAC singalling
We demonstrate that PLK1 ubiquitination specifically regulates its function at kinetochores. This is achieved by ubiquitination of a conserved lysine residue (K492) within its PBD domain. PBD-dependent localization of PLK1 in mitosis is only needed for the function of PLK1 on kinetochores and not on centrosomes13, which explains why no defects in bipolar spindle assembly could be observed upon inactivation of CUL3-KLHL22. These findings thus not only provide surprising insights into the temporal and spatial regulation of PLK1, but also suggest that regulated removal of PLK1 from kinetochores is an important process monitoring chromosome alignment. Current emerging view is that a single pathway may exist linking error correction to SAC activation42. Our data support this notion in that the KLHL22-ubiquitination pathway regulates phosphorylation of BubR1 (Ser 676) by PLK1 and at the same time is sensed by MAD2-dependent SAC mechanism. Importantly, our results demonstrate that both inactivation of KLHL22 and expression of the PLK1-K492R mutant leads to delocalization of the microtubule-kinetochore attachment sensor RanGAP1 protein. Thus, our results suggest that establishment of stable kinetochore-microtubule interactions is tightly regulated consistent with the recently published data8,9 and identify a mechanism leading to satisfaction of SAC and the metaphase to anaphase transition.
Our results suggest that KLHL22-mediated ubiquitination prevents molecular interaction of the PBD with its kinetochore receptor proteins. Thus, the CUL3-ubiquitination pathway may provide a novel mode of substrate regulation by affecting the local concentration of essential enzymatic activities, possibly including other kinases. Since KLHL22 preferentially interacts with a subset of protein kinases in our protoarrays, it is conceivable that CUL3-KLHL22 complexes may also regulate the activity and/or localization of other protein kinases. Interestingly, our previous study demonstrated the role of a different CUL3-based complex in localization of Aurora B kinase21,22. It will therefore be exciting to study if CUL3 complexes more commonly orchestrate localized kinase activities through non-degradative ubiquitination.
Methods
Cell lines
The FLAG-KLHL22 and GFP-KLHL22 cell lines were created using the Flp-IN-T-Rex cell line (Invitrogen). GFP-PLK1 and HIS-PLK1 cell lines were generated using a HeLa FRT/To host cell line kindly provided by U. Kutay (Swiss Federal Institute of Technology, Zurich, Switzerland). The HeLa cell line expressing H2B-RFP44 was kindly provided by D. Gerlich (Swiss Federal Institute of Technology, Zurich, Switzerland). The HeLa cell line expressing tandem of three GFP tags and NLS signal45 was kindly provided by Peter V. Lidsky (Russian Academy of Medical Sciences, Russia). HeLa Kyoto and HEK293T cells were maintained as previously described21. Antibiotics for selection of cell lines were used as follows: Hygromycin (Invitrogen) 100μg/ml, Blasticidin (Invitrogen) 15μg/ml and Puromycin (Sigma Aldrich) 0,5μg/ml. Doxycycline (Sigma) for induction of protein expression was used at 1μg/ml or 0,1 μg/ml.
Chemicals (all Sigma) were used as follows: Taxol (Paclitaxel) 1μM (or as indicated), Nocodazole 2μM, Hydroxyurea 2mM, Thymidine 2mM, MG132 50μM and Monastrol 100μM. The PLK1 inhibitor BI 2536 (Axon Medchem) was used at 100 nM as previously described6 and the general DUB inhibitor PR-619 (TebuBio) was used at 20μM.
cDNAs and siRNAs
cDNAs (RZPD) transfections were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer instructions. siRNA transfections were performed using Oligofectamine (Invitrogen) according to the manufacturer’s instructions at a final concentration of 100 nM siRNA. The following siRNA oligonucleotides (Microsynth) were used: non-silencing control: UUCUCCGAACGUGUCACGU; for KLHL22 3 siRNAs were pooled: GCAACAACGAUGCCGGAUA; CCUAUAUCCUCAAAAACUU; GGACUGGCUCUGUGAUAAA; CUL3: CAACACUUGGCAAGGAGAC; MAD2: GAGUCGGGACCACAGUUUA; for PLK1 3’UUR RNAi 3 siRNAs were pooled: CCAUAUGAAUUGUACAGAA; GGGUUGCUGUGUAAGUUAU; GGGGUUGCUGUGUAAGUUA.
PLK1 mutagenesis
Point mutants of PLK1 were generated using pfu Turbo DNA Polymerase (Stratagene) according to the manufacturer’s instructions. Primers used for mutation of K492R were: GAGCACTTGCTGAGGGCAGGTGCCAAC & GTTGGCACCTGCCCTCAGCAAGTGCTC.
Microscopy
Live-cell microscopy was carried out using a LSM510 Screening Confocal Laser Scanning microscope (Zeiss) or Nikon Eclipse Ti. Immunofluorescence microscopy of fixed cells was performed as previously described21,22 using either a Delta Vision personalDV (Applied Precison) or a Leica DM6000B.
Western Blotting and Immunoprecipitation
Preparation of HeLa cell extracts was described previously21. SDS-PAGE and Western blots were performed according to standard procedures. For immunoprecipitation, antibodies were coupled to Affiprep Protein-A beads (Biorad). FLAG-tagged KLHL22 was immunoprecipitated using anti-FLAG M2-Agarose beads (Sigma). GFP-fused proteins were immunoprecipitated using GFP-Trap agarose beads (Chromotek). Beads were incubated with cell extracts for 2h at 4°C under constant rotation. Prior to elution, beads were washed 5x with TBS-T (20mM Tris pH8, 150mM NaCl, 0.05% Tween20).
For detection of PLK1 interaction with KLHL22 and with the phosphoreceptors, the GFPTrap® coupled to agarose beads (Chromotek) was used according to the manufacturers’ instructions. For the phosphoreceptor-binding experiment, cells were synchronized in mitotic state by Taxol for 13h and subsequently treated with PR-619 for 1h. Expression of PLK1WT and PLK1KR was induced for 24h. To detect the interaction of PLK1 with KLHL22 in response to chromosome biorientation, cells were synchronized in prometaphase by treatment with Monastrol for 16h and released into MG132-containing media for 1.5h to establish metaphase plates. Subsequently, cells were treated with Taxol (10μM) to abrogate microtubule-exerted tension, Nocodazole (10μM) to depolymerize microtubules or solvent control (DMSO) for further 1.5h. 10 μg of PLK1 rabbit polyclonal antibody (Abcam) or IgG control were coupled to protein A beads and used for immunoprecipitations of endogenous PLK1, while GFP-PLK1 was immunoprecipitated using GFP-Trap® beads (Chromotek).
In vitro kinase assay
For in vitro kinase assays GFP-PLK1WT or KR was first immunoprecipitated using GFP-Trap® beads as described above but in the presence of 100 nM Okadaic acid. After the final wash with IP buffer, the beads were washed 3x with kinase buffer (20 mM Tris pH 7.0, 50 mM KCl, 10 mM MgCl2 supplemented with 20 mM ß-glycerophosphate, 1 mM NaOrthovanadate, 2 mM NaF, 1 μg/ml Pepstatin, 1 μg/ml Leupeptin, 1 mM DTT, 1x COMPLETE Protease inhibitors and 100 nM Okadaic acid). 100μg Casein was added as artificial substrate and reactions were started by adding 400 μM ATP and 0.5 MBq γ-32P-ATP.
Protein microarray experiments
For the generation of control protein microarrays, control proteins were spotted at decreasing concentrations onto PATH nitrocellulose slides (Gentel Biosciences) using a Piezoarray platform (Perkin-Elmer). GST-Aurora B was purchased from US Biologicals/Lucerna-Chem and GST-LAPTM5 and GST-xHect were kindly provided by D. Rotin (The Hospital for Sick Children, Toronto, Canada). Ubiquitination microarray screens with CUL3/RBX1/KLHL22 were done in duplicates. For ubiquitination assays, Human ProtoArray PATH V.4 microarray slides (Invitrogen) were rinsed briefly with 0.5 % PBS-T and then blocked at room temperature for 1 h in blocking buffer (50 mM Hepes pH 7.5, 200 mM NaCl, 0.08 % Triton X-100, 25 % Glycerol, 20 mM Glutathione, 1 mM DTT, 1 % BSA). Meanwhile, 10.2 μg of CUL3/RBX1/His-KLHL22 E3 ligase complex was in vitro-neddylated with 2.5 μg human Nedd8 activating enzyme E1 (APPBP1/Uba3), 2.5 μg human E2 (UbcH12), 10 μg human Nedd8, 1x energy regeneration solution (ERS) and 1x Ubiquitination buffer (45 mM Hepes, 2 mM NaF, 0.6 mM DTT) at room temperature for 15 min. The slides were then rinsed gently in 1x Ubiquitination buffer for 5 min following incubation with the reaction mixture (270 μl containing the mixture after CUL3-neddylation and in addition 8 μg FITC-Ubiquitin, 7.5 μg rabbit E1 (UBE1), 7.5 μg rabbit E2 (UbcH5c), 12.5 mM Mg-ATP and 1x Ubiquitination buffer) in the dark for 2 h. Slides were washed 3x in 0.5 % PBS-T for 10 min and then dried by centrifugation at 1000 g at room temperature for 5 min. Slides were scanned using a 488 nm laser on a ProScan Array HT (Perkin-Elmer).
For binding assays, Human ProtoArray PATH V.4 microarray slides were washed, blocked and rinsed gently in 1x Ubiquitination buffer (45 mM Hepes, 2 mM NaF, 0.6 mM DTT) for 5 min following incubation with binding mixture (300 μl 1x Ubiquitination buffer containing 3 μg Alexa647-MBP or Alexa647-MBP-KLHL22 - final concentration: 10 ng/μl) in the dark for 1 h. The slides were washed 2x 5 min and 1x 10 min in 0.1 % PBS-T and then dried by centrifugation at 700 g at room temperature for 5 min. Slides were scanned using a 633 nm laser on a ProScan Array HT (Perkin-Elmer). Binding microarray screens with Alexa647-MBP-KLHL22 were done in duplicates whereas a single binding microarray screen was performed with Alexa674-MBP. Spot fluorescence intensities from the microarray screens were quantified using the ProScan Array HT (Perkin-Elmer) software. Protein Prospector Analyzer (Invitrogen) was used to compare duplicate screens. Proteins were scored as hits if 50% of the pixels of the corresponding spots of both duplicates on both replica slides showed a signal stronger than 2x standard deviation values above background.
Ubiquitination assays and identification of ubiquitinated peptides by mass spectrometry (MS)
For in vitro ubiquitination assays, 1 μg Sf9-purified GST-CUL3/RBX122 mixed with 2 μg MBP or MBP-KLHL22 purified form E. coli was used as E3 ligase. The E3 ligase complexes were mixed with 0.5 μg recombinant PLK1 (Cell Signaling), 15 μg Ubiquitin, 550 ng rabbit E1 (UBE1), 850 ng rabbit E2 (UbcH5b), 12.5 mM Mg-ATP and 1x ubiquitin reaction buffer, and incubated at 30°C for 1 h. All ubiquitination reagents were purchased from LuBio Science Gmbh/Boston Biochem. Reactions were terminated by addition of SDS-containing loading buffer, boiled at 95°C for 5 min and analyzed by Western blot.
For in vitro ubiquitination of PLK1 for MS analysis, the double amount of each reaction component was used in the ubiquitination reaction. After incubation of the reaction at 30°C for 1 h, unreacted free ubiquitin was removed by centrifugation in an Amicon Ultra-15 centrifuge filter (cut-off: 30 kDa; Millipore). 5 mM DTT was added to the eluate and reduction was allowed to proceed at 45°C for 1 h. After cooling the sample, 15 mM Iodoacetamide (Sigma) was added and alkylation was allowed to proceed for 1 h. The sample was digested over night with sequencing grade modified Trypsin (Promega) in the presence of 0.05 % RapiGest (Waters) at 37°C. Prior to MS analysis a microspin column packed with C-18 material (The Nest Group) was used for sample cleanup. Briefly, the sample was acidified with 0.1 % TFA (Promega), loaded onto the reverse phase column, washed 3x with 0.1 % TFA in water, eluted with 0.1 % TFA in 80 % Acetonitrile (Sigma) and dried in a SpeedVac system (Thermo Scientific). Before MS, samples were resuspended in HPLC buffer A (0.1 % formic acid in water) and the peptides were separated on a self-packed 10x 0.075 mm C-18 (Michrom Bioresources) micro-capillary fused silica column (BGB Analytik AG) using a multi-step linear gradient: 2-10 % HPLC buffer B (Acetonitrile) in 1 min; 10-45 % in 120 min; 45-98 % in 3 min generated by an Agilent 1100 series HPLC (Agilent). MS and MS/MS data were acquired on a Thermofinnigan LTQ (Thermofinnigan) using a top three-data-dependent acquisition method. The raw data was converted to the mzXML format46 and analyzed using version 4.3.1 of the Trans Proteomics Pipeline. X!Tandem with the comet score plug-in47 was used for the MS/MS assignment. The protein database used was ipi.HUMAN.v3.41.fasta. The condition files allowed for a variable modification of 114.1 Da at lysine residues, a static modification of 57 Da at cysteine residues and tryptic peptides with a maximum of 2 missed cleavages.
Antibodies
The following antibodies were used in the study: mouse monoclonal BubR1 (BD Biosciences 612502, 1:1000), pSer676 BubR1 (kind gift of E. A. Nigg, University of Basel, 1:???), CREST (Antibodies Incorporated, 15-234, 1:250), rabbit polyclonal CUL321, mouse monoclonal FLAG (Sigma Aldrich F3165, 1:3000), mouse monoclonal GST (Labforce B-14, 1 :4000), rabbit polyclonal HA (Covance HA.11, 1 :3000), rabbit polyclonal KLHL22 and KLHL2122, rabbit polyclonal KLHL9 and 1321, rabbit polyclonal Mad2 (Bethyl Laboratories A300-301A, 1 :5000), mouse monoclonal MBP (Abcam ab49923, 1 :1000), rabbit polyclonal PLK1 (Abcam ab70697, 1:1000), mouse monoclonal PLK1 (Abcam ab17057, 1:500), rabbit polyclonal PLK2 (kind gift of I. Hoffmann, DKFZ Heidelberg, Germany), mouse monoclonal αTubulin (Sigma Aldrich T5169, IF 1:5000, WB 1:10000), rabbit polyclonal γTubulin (Sigma T3559, 1:1000), mouse monoclonal Cyclin A (Sigma C 4710, 1 :2000), mouse monoclonal Cyclin B1 (Santa Cruz GSN1, 1:5000), rabbit polyclonal phospho HistoneH3 (Upstate 06-570, 1:500), mouse monoclonal AuroraB (BD Biosciences 611082/3, 1:250), mouse monoclonal Hec1 (GeneTex, ?? 1:???), rabbit polyclonal Pericentrin (Abcam ???, 1:???), mouse monoclonal GFP (Abcam ?? 1:??), mouse monoclonal RanGAP1 (Invitrogen 330800, 1:250), mouse monoclonal CDH1 (Abcam ab3242, 1:500) and mouse monoclonal HIS (IGBMC antibody facility, clone IG4, 1:250).
Supplementary Material
Acknowledgements
We thank W. Piwko, T. Courtheoux, P. Meraldi, D. Gerlich, A. Smith, O. Pourquié and M. Labouesse for helpful discussions and editing of the manuscript, E.A. Nigg, J.-M. Peters, P. Meraldi, U. Kutay and F. Barr for antibodies, D. Gerlich for cell lines and G. Csucs, J. Kusch, O. Biehlmeier and T. Schwarz from the D-BIOL Light Microscopy Centre for help with microscopy. J.B. was granted an EMBO Short Term Fellowship, and S.M. was funded by ETHZ and the Boehringer Ingelheim Fonds. I.S. was supported by the ETHZ and the Swiss National Science Foundation (SNF), and research in D.R. and M.P.’s laboratories by the Canadian Institute of Health Research (CIHR), the European Research Council (ERC), the SNF and the ETHZ, respectively. Research in I.S. laboratory is supported by the IGBMC, ATIP-AVENIR program from CNRS and INSERM and Sanofi-Aventis.
Footnotes
Authors contribution
J.B., S.M. and T.M. conceived ideas, performed experiments, analysed and interpreted the data. H.S., and K.H. performed bioinformatical analysis of protein microarray hits. A.P. and D.R. collaborated on protein microarrays experiments. P.P. conducted MS analysis of PLK1 ubiquitination sites. M.P. and J.S. collaborated on live video microscopy techniques. M.P. and I.S. conceived ideas. I.S. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Williams BR, Amon A. Aneuploidy: cancer's fatal flaw? Cancer Res. 2009;69(13):5289–5291. doi: 10.1158/0008-5472.CAN-09-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396(6712):643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 3.Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat Rev Drug Discov. 2010;9(8):643–660. doi: 10.1038/nrd3184. [DOI] [PubMed] [Google Scholar]
- 4.Petronczki M, Lenart P, Peters JM. Polo on the Rise-from Mitotic Entry to Cytokinesis with Plk1. Dev Cell. 2008;14(5):646–659. doi: 10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Sumara I, et al. Roles of polo-like kinase 1 in the assembly of functional mitotic spindles. Curr Biol. 2004;14(19):1712–1722. doi: 10.1016/j.cub.2004.09.049. [DOI] [PubMed] [Google Scholar]
- 6.Lenart P, et al. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17(4):304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 7.Elowe S, Hummer S, Uldschmid A, Li X, Nigg EA. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 2007;21(17):2205–2219. doi: 10.1101/gad.436007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maia AR, et al. Cdk1 and Plk1 mediate a CLASP2 phospho-switch that stabilizes kinetochore-microtubule attachments. J Cell Biol. 2012;199(2):285–301. doi: 10.1083/jcb.201203091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu D, Davydenko O, Lampson MA. Polo-like kinase-1 regulates kinetochoremicrotubule dynamics and spindle checkpoint silencing. J Cell Biol. 2012;198(4):491–499. doi: 10.1083/jcb.201205090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee KS, et al. Mechanisms of mammalian polo-like kinase 1 (Plk1) localization: self- versus non-self-priming. Cell Cycle. 2008;7(2):141–145. doi: 10.4161/cc.7.2.5272. [DOI] [PubMed] [Google Scholar]
- 11.Elia AE, et al. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115(1):83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 12.Cheng KY, Lowe ED, Sinclair J, Nigg EA, Johnson LN. The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex. Embo J. 2003;22(21):5757–5768. doi: 10.1093/emboj/cdg558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanisch A, Wehner A, Nigg EA, Sillje HH. Different Plk1 functions show distinct dependencies on Polo-Box domain-mediated targeting. Mol Biol Cell. 2006;17(1):448–459. doi: 10.1091/mbc.E05-08-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2(1):21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 15.Mateo F, Vidal-Laliena M, Pujol MJ, Bachs O. Acetylation of cyclin A: a new cell cycle regulatory mechanism. Biochem Soc Trans. 2010;38(Pt 1):83–86. doi: 10.1042/BST0380083. [DOI] [PubMed] [Google Scholar]
- 16.Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 2005;12(9):1191–1197. doi: 10.1038/sj.cdd.4401702. [DOI] [PubMed] [Google Scholar]
- 17.Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65(15):2397–2406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 19.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6(1):9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 20.Sumara I, Maerki S, Peter M. E3 ubiquitin ligases and mitosis: embracing the complexity. Trends Cell Biol. 2008;18(2):84–94. doi: 10.1016/j.tcb.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Sumara I, et al. A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev Cell. 2007;12(6):887–900. doi: 10.1016/j.devcel.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 22.Maerki S, et al. The Cul3-KLHL21 E3 ubiquitin ligase targets aurora B to midzone microtubules in anaphase and is required for cytokinesis. J Cell Biol. 2009;187(6):791–800. doi: 10.1083/jcb.200906117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274(5285):246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- 24.Sumara I, Peter M. A Cul3-Based E3 Ligase Regulates Mitosis and is Required to Maintain the Spindle Assembly Checkpoint in Human CellS. Cell Cycle. 2007;6(24) doi: 10.4161/cc.6.24.5068. [DOI] [PubMed] [Google Scholar]
- 25.Matsumura S, Toyoshima F, Nishida E. Polo-like kinase 1 facilitates chromosome alignment during prometaphase through BubR1. J Biol Chem. 2007;282(20):15217–15227. doi: 10.1074/jbc.M611053200. [DOI] [PubMed] [Google Scholar]
- 26.Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol. 2004;14(7):611–617. doi: 10.1016/j.cub.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 27.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44(2):325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner SA, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10(10) doi: 10.1074/mcp.M111.013284. M111 013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park JE, et al. Polo-box domain: a versatile mediator of polo-like kinase function. Cell Mol Life Sci. 2010;67(12):1957–1970. doi: 10.1007/s00018-010-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moghe S, et al. The CUL3-KLHL18 ligase regulates mitotic entry and ubiquitylates Aurora-A. Biology Open. 2012;1:82–91. doi: 10.1242/bio.2011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pintard L, Willems A, Peter M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. Embo J. 2004;23(8):1681–1687. doi: 10.1038/sj.emboj.7600186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villeneuve NF, Lau A, Zhang DD. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases. Antioxid Redox Signal. 2010;13(11):1699–1712. doi: 10.1089/ars.2010.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huotari J, et al. Cullin-3 regulates late endosome maturation. Proc Natl Acad Sci U S A. 2012;109(3):823–828. doi: 10.1073/pnas.1118744109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyden LM, et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature. 2012;482(7383):98–102. doi: 10.1038/nature10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan WC, et al. A Cullin3-KLHL20 Ubiquitin ligase-dependent pathway targets PML to potentiate HIF-1 signaling and prostate cancer progression. Cancer Cell. 2011;20(2):214–28. doi: 10.1016/j.ccr.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 36.Hernandez-Munoz I, et al. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci U S A. 2005;102(21):7635–7640. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin L, et al. Ubiquitin-dependent regulation of COPII coat size and function. Nature. 2012;482(7386):495–500. doi: 10.1038/nature10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howell BJ, et al. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155(7):1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varma D, Monzo P, Stehman SA, Vallee RB. Direct role of dynein motor in stable kinetochore-microtubule attachment, orientation, and alignment. J Cell Biol. 2008;182(6):1045–1054. doi: 10.1083/jcb.200710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z, Tulu US, Wadsworth P, Rieder CL. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr Biol. 2007;17(11):973–980. doi: 10.1016/j.cub.2007.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell. 2010;143(6):951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 43.Yun SM, et al. Structural and functional analyses of minimal phosphopeptides targeting the polo-box domain of polo-like kinase 1. Nat Struct Mol Biol. 2009;16(8):876–882. doi: 10.1038/nsmb.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steigemann P, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136(3):473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 45.Belov GA, et al. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J Virol. 2004;78(18):10166–10177. doi: 10.1128/JVI.78.18.10166-10177.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedrioli PG, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol. 2004;22(11):1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 47.MacLean B, Eng JK, Beavis RC, McIntosh M. General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics. 2006;22(22):2830–2832. doi: 10.1093/bioinformatics/btl379. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.