Abstract
Two major challenges of 3D bioprinting are the retention of structural fidelity and efficient endothelialization for tissue vascularization. We address both of these issues by introducing a versatile 3D bioprinting strategy, in which a templating bioink is deposited layer-by-layer alongside a matrix bioink to establish void-free multimaterial structures. After crosslinking the matrix phase, the templating phase is sacrificed to create a well-defined 3D network of interconnected tubular channels. This void-free 3D printing (VF-3DP) approach circumvents the traditional concerns of structural collapse, deformation and oxygen inhibition, moreover, it can be readily used to print materials that are widely considered “unprintable”. By pre-loading endothelial cells into the templating bioink, the inner surface of the channels can be efficiently cellularized with a confluent endothelial layer. This in-situ endothelialization method can be used to produce endothelium with a far greater uniformity than can be achieved using the conventional post-seeding approach. This VF-3DP approach can also be extended beyond tissue fabrication and towards customized hydrogel-based microfluidics and self-supported perfusable hydrogel constructs.
Keywords: bioprinting, hydrogels, printability, endothelialization, microfluidics
1. Introduction
Vascular networks are essential for supplying tissues with nutrients, oxygen and signalling factors, removing waste products and regulating factors such as temperature and pH. [1] Accordingly, new methods that can rapidly fabricate 3D vascularized constructs would significantly advance studies in tissue repair, cell-based therapeutics, and drug screening. [1–3] One approach that offers clear potential for vascular tissue engineering is 3D bioprinting, which has enabled the controlled, spatial deposition of different biofunctional components, such as cells, extracellular matrix, and biomolecules. For example, co-axial bioprinting can be used to create continuous hollow fibers that can serve as vascular channels, however, this method has not yet been used for the production of 3D interconnected vasculature. [4] An alternative strategy, aimed at building more complex networks, is to directly print structures with interconnected void spaces (pores or channels). [5–10] However, the presence of voids regularly results in structural instability during printing, whereby the top layers cause the lower layers to buckle and deform the underlying pores. [11, 12] This factor has led to the development of alternative print-casting methodologies, in which a vascular template is 3D printed using fugitive inks, such as carbohydrate glass [13] or Pluronic F-127 [14, 15]. The matrix is formed by casting a second material around the printed template, which is then removed to create channels that can be post-seeded with endothelial cells. While this method has yielded impressive results, the use of a typical four step-process (printing, casting, template removal, post-seeding) loses many of the key advantages of single-step 3D printing, such as the speed, simplicity and ability to additively manufacture the matrix component. In particular, post-seeding is an inefficient, poorly controlled step that often results in a non-uniform distribution of cells throughout the templated channels. Ideally, the fugitive ink would be used to seed the endothelial cells in situ, however, carbohydrate glass cannot be used to carry cells, while Pluronic F-127 is known to be highly cytotoxic for cell encapsulation. [16] A very recent study by Dvir and colleagues showed that endothelial cells could be seeded in this way, however, only 2D networks were demonstrated. [17]
Here, we present a versatile bioprinting methodology that successfully overcomes the challenges of structural printability, seeding efficiency/uniformity and 3D interconnected network formation (Figure 1A). This strategy enables, for the first time, the fabrication of 3D complex vascular networks using a single bioprinting step, without the need for material casting or cell post-seeding. We employ two types of bioink, a gelatin-based templating bioink and a photo-crosslinkable matrix bioink, which are printed side-by-side in 3D structures without any void spaces. Derived from collagen, gelatin is a widely used biomaterial in cell culture and tissue engineering, which can undergo a thermoreversible gel-sol transition due to the assembly and disassembly of its triple helix structure. [18–20] Therefore, after photo-crosslinking the matrix bioink component, the printed structure can be incubated at 37 °C to liquefy the gelatin phase and create a network of templated channels. We present a number of key advantages of void-free 3D bioprinting and in-situ endothelialization over conventional methods for additively manufacturing vascular networks (co-axial extrusion, direct lattice printing, print-casting and post-seeding). Notably, we demonstrate: (i) the generation of complex 3D interconnected networks with tubular geometry, rather than the deformed, cuboidal voids present in directly-printed lattices, due to the fact that the bioinks are sequentially deposited on flat layers rather than over voids; (ii) the printing of lattices using low-concentration bioinks that are considered “unprintable” when using direct printing methods, (iii) the engineering of 3D interconnected, vascular networks using cells pre-loaded in the templating bioink (in-situ endothelialization), which provided greater seeding efficiency, uniformity and control compared to post-seeding, and (iv) the construction of customized hydrogel-based microfluidics, including the first example of a 3D printed, self-supported perfusable hydrogel.
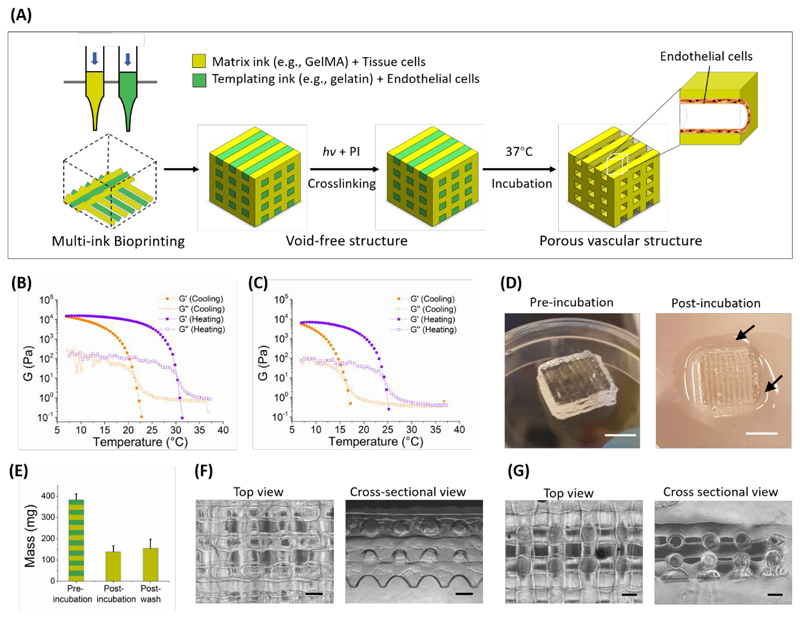
Figure 1.
(A) Schematic of the VF-3DP process, where a biocompatible templating bioink (green) and a matrix bioink (yellow) were printed side-by-side, followed by photo-crosslinking of the matrix phase and 37 °C incubation to release the templating phase. Pre-loading endothelial cells in the templating bioink allowed in-situ endothelialization of the channels. Representative curves of the storage (G’) and loss (G’’) components of the shear modulus (G) were measured for (B) 7.5 wt% gelatin and (C) 7.5 wt% GelMA during cooling and heating temperature sweeps (5 °C min-1). (D) Representative photographs and (E) measured mass of the VF-3DP structure (10 × 10 × 3 mm, 7.5 wt% gelatin as templating bioink and 7.5 wt% GelMA as matrix bioink) before and after incubation. Arrows in the photographs indicate liquified gelatin, while the mass measurements showed a significant loss in mass due to the exuded templating phase. Representative images of (F) the final VF-3DP structure and (G) a directly-printed lattice comparison (7.5 wt% gelatin) showing both the top and cross-sectional views. Scale bars: 5 mm (D), 500 μm (F-G).
2. Results and Discussion
In order to make our approach accessible to bioprinting community, we sought to illustrate our void-free 3D printing (VF-3DP) approach using widely-used biomaterials rather than specialist synthetic systems. Accordingly, we chose to use gelatin and gelatin methacryloyl (GelMA), the latter a semi-synthetic biomaterial that has been used extensively as a photo-crosslinkable extracellular matrix hydrogel for bioprinting and tissue engineering. [20, 21] We synthesized GelMA by methacrylating the lysine sidechain residues of gelatin, and confirmed conjugation using nuclear magnetic resonance (NMR) spectroscopy and a fluoraldehyde assay (degree of functionalization ~80%) (Figure S1). We could form covalently crosslinked GelMA hydrogels in the presence of photo-initiator and light irradiation, either with Irgacure 2959 (I2959) at 365 nm or with lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) at 365 nm or 405 nm (Figure S2). Similar to gelatin, GelMA also exhibits a natural sol-gel transition. We examined this behaviour by performing rheology temperature sweeps between 7 and 37 °C on both 7.5% gelatin and 7.5% GelMA. We observed a sol-gel transition in both cases; however, it should be noted that GelMA had a lower gelation temperature (16 °C) and storage modulus at 7 °C (~6 kPa) than gelatin (21 °C, ~14 kPa) (Figure 1B, 1C). This difference was most likely due to the sidechain modifications in GelMA, which act to sterically hinder the assembly of the triple helix.
Given the thermal setting capability of both gelatin and GelMA, we elected to use a temperature-controlled process to initially set the bioprinted structures. The nozzle was warmed to a temperature that would allow smooth filament extrusion [18], while the print bed was set at 15 °C to ensure rapid gelation after printing. The exact temperature of nozzle was chosen based on the thermal characteristics of the bioink: we printed 7.5 wt% GelMA at 22-23 °C and 7.5 wt% gelatin at 26-27 °C. Given that the sol-gel transition temperature of gelatin is dependent on the concentration (Figure S3), the nozzle temperature was raised for higher weight percentage bioinks, and vice versa (SeeExperimental Section). To illustrate this printing process, we used a standard cubic lattice model (length × width × height of 10 × 10 × 3 mm, slice thickness 300 μm), in which 7.5 wt% gelatin and 7.5 wt% GelMA were printed in alternate parallel lines offset by 500 μm (Movie S1). The structures were structurally stable on the cooled print bed, however, the reversible nature of the sol-gel transition meant that the whole structure rapidly liquified at 37 °C. Therefore, we included 0.05 wt% I2959 within the GelMA bioink, so that we could covalently photo-crosslink the GelMA component of the printed structures (365 nm, ~5 mW cm-2, 5 min). This VF-3DP process replicated the model design well, maintaining excellent structural integrity without any visible holes, bubbles, or defects (Figure 1D).
We next investigated whether we could selectively remove the gelatin component by incubating the printed material at 37 °C. After 30 min incubation, we observed liquid surrounding the base of the structure (Figure 1D), which itself was 37 ± 8% of the mass of the material prior to incubation (Figure 1E). We used a fluoraldehyde assay to confirm that the exuded liquid had a similar level of amines to gelatin (Figure S1B). This analysis indicated that it was predominantly gelatin, rather than GelMA, that was released during the incubation period. When viewing the structure from above, we could clearly observe the regular grid pattern of the remaining GelMA hydrogel (Figure 1F). More importantly, the cross-sectional view revealed an array of well-defined tubular pores formed from the removal of the gelatin template. The orthogonally-aligned hollow channels exhibited a regular cylindrical shape with a cross-sectional diameter of ~450 μm (Figure 1F). The interconnected network of channels in the VF-3DP structure was further demonstrated by adding a dye solution to the structure, which was rapidly taken up (<1 min) into the channel lumen (Movie S2, Figure S4A). This was further confirmed by soaking the structure in a solution of fluorescent beads (0.503 μm in diameter), with fluorescence microscopy clearly showing the tubular lumen pattern and the 3D connectivity when viewed from either above or from the side (Figure S4B). The structure itself presented an initial stiffness of 29.9 ± 3.3 kPa at day 1 (Figure S5A) and remained structurally stable during 15 d of incubation in PBS at 37 °C (Figure S5B).
Such 3D uniform tubular channels are not possible with conventional hydrogel bioprinting: it is traditionally challenging to print voids with a spherical cross-section, moreover, without a supporting template, the weight of the top layers often causes the lower struts to buckle and lead to loss of pore uniformity. We illustrated this concept by directly printing separate lattices of 7.5 wt% gelatin and 7.5 wt% GelMA with the same printing parameters as VF-3DP but without any templating bioink for structural support. This method generated lattices that appeared regular from above, however, cross-sectional imaging revealed deformed or fused pores between adjacent layers (Figure 1G). Moreover, the lattice structure of 7.5 wt% GelMA exhibited some significantly expanded pores from above post UV irradiation (Figure S6). This observation was attributed to oxygen inhibition of the free radical polymerization process, which would result in etching of the areas exposed to air. [11] The VF-3DP process overcomes these issues by using the two hydrogel bioinks to support each other during the layer-by-layer deposition. In essence, this means that each new layer is deposited onto a relatively flat surface and the GelMA photo-crosslinking is performed with an air interface only at the extreme edges of the structure.
While directly printed lattices of 7.5 wt% gelatin showed buckled and deformed features, this bioink could at least be used to generate some form of printed structure. For many bioinks, however, the hydrogel is simply too weak to support structures containing voids or channels. [11, 22] This issue is particularly pertinent for low-concentration hydrogels; the optimal biomaterial for many biological processes, such as angiogenesis, myogenesis, and neurogenesis. [23–26] Therefore, we sought to apply our VF-3DP process as a method for printing low-concentration biomaterials. In particular, we investigated 5 wt% GelMA as the matrix bioink, since other authors have independently concluded that this formulation is not printable by conventional means. [27, 28] We verified that direct printing of 5 wt% GelMA into a lattice structure produced a collapsed structure after the deposition of just two layers (Figure S7A). However, when we used our VF-3DP process with 5 wt% GelMA as the matrix bioink, together with 7.5 wt% gelatin as the templating bioink, we were able to construct a 3D structure of multiple layers (e.g., 10 layers) with excellent fidelity (Figure S7B). We were able to photo-crosslink the GelMA and remove the gelatin phase at 37 °C to obtain a 3D hydrogel lattice made of 5 wt% GelMA. Top-down and cross-sectional imaging confirmed that the lattice contained an interconnected network of uniform tubular channels (Figure S7C).
We next explored whether we could generalize this approach to other photo-crosslinkable hydrogel formulations. For example, hyaluronic acid (HA) is a common extracellular matrix component that can be used for bioprinting and tissue engineering when functionalized with methacrylate or norbornene groups (MeHA and NorHA, respectively). [29, 30] As is the case with GelMA, these two bioinks have been shown to be incompatible with conventional bioprinting protocols at low concentration. [11, 22] Nevertheless, we demonstrated that VF-3DP could be used to assemble 5 wt% MeHA or 5 wt% NorHA cubic lattice structures, when using 7.5 wt% gelatin as a templating bioink (Figure S7D). These results indicated the potential of VF-3DP in enhancing the printability of a range of poorly-printable bioink formulations, including those derived from the extracellular matrix.
We decided, however, to focus on our gelatin-GelMA system for all cell studies. We first assessed the viability of human dermal fibroblasts (HDF) encapsulated in the matrix bioink (7.5 wt% GelMA). LIVE/DEAD™ staining showed a large proportion of living HDFs immediately after printing (Figure S8A). Moreover, image analysis on LIVE/DEAD™ stained constructs showed a consistently high cell viability (~90%) of encapsulated HDF cells over 5 d of culture (Figure S8B). To illustrate dual-cell VF-3DP, we printed a void-free lattice structure with fluorescently-labelled human umbilical vein endothelial cells (HUVECs) and HDF cells in the gelatin and GelMA bioinks, respectively. Fluorescence microscopy, performed immediately after bioprinting, clearly showed the homogeneous distribution of each cell population within their printed boundary but without any gaps or interference between the two phases (Figure S9). These results laid the basis for high-fidelity channel printing and in-situ endothelization. For the latter, we simply included a layer of GelMA around the construct in the bioprinting design in order to block the channel openings, retain the liquified gelatin and allow cells time to adhere to the channel walls. After 6 h, we removed this outer layer to release the liquified gelatin along with any non-adhered cells. This in-situ seeding method produced a remarkably uniform layer of cells adhered to the channel walls, as shown by fluorescence microscopy (Figure 2A, Movie S3) and quantification of the cellularized area (Figure 2B).
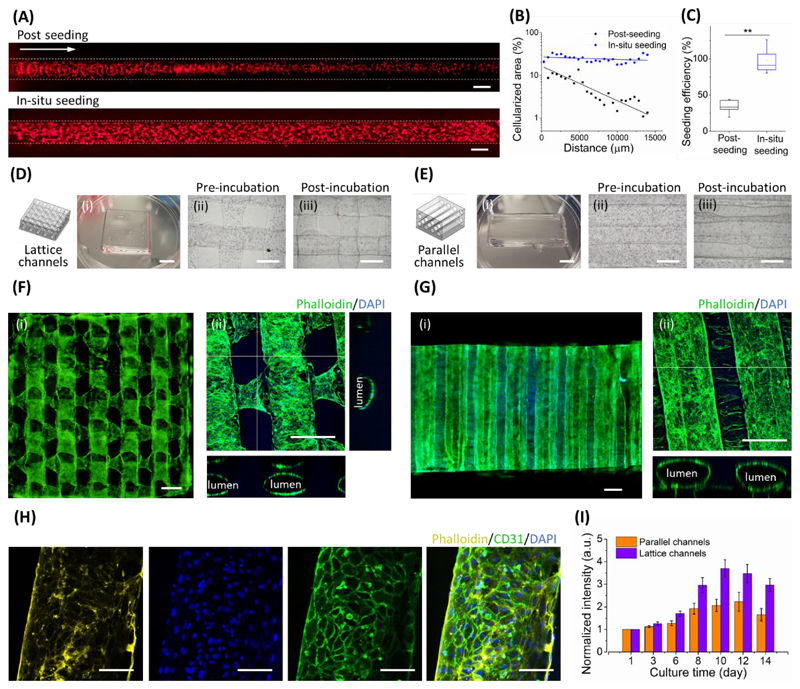
Figure 2.
In-situ endothelization. (A) Representative fluorescence micrographs comparing the conventional post-seeding method with in-situ seeding. RFP-labelled HUVECs (5 × 106 ml-1) were seeded in a single channel, with the white arrow indicating the direction of post-seeding. The white dotted lines denote the approximate width of the cellularized channel. (B) These images were used to assess the uniformity of cell seeding. Note that the metric of cellularized area refers to the observed cell fluorescence in the region-of-interest of the image (i.e. the channel) rather than a measure of absolute seeding density on the cylindrical channel walls. (C) A DNA assay was used to quantitatively assess the efficiency of post-seeding and in-situ seeding. Data shown as mean ± standard deviation from five samples (two-tailed Mann–Whitney test), p≤0.01 (**). VF-3DP was used to assemble HUVEC-laden structures with (D) 3D lattice channels or (E) 3D parallel channels. (i) Photographs were taken after printing, and bright field microscopy was performed (ii) before and (iii) after incubation at 37 °C. (i) Widefield fluorescence microscopy and (ii) confocal fluorescence microscopy was performed on (F) lattice and (G) parallel designs at day 7. These images indicated the widespread formation of endothelialized channels. (H) High-magnification confocal fluorescence microscopy images of immunostained constructs at day 8 indicated full occupation of CD31-positive HUVECs. (I) Metabolic activity of HUVECs in lattice channels (purple bars) and parallel channels (orange bars). Scale bars: 5 mm (D(i), E(i)), 500 μm (A, D(ii-iii), E(ii-iii), F-G), 100 μm (H).
In contrast, conventional post-seeding produced an uneven, gradient distribution with a marked reduction in the number of adhered cells distal to the injection point (Figure 2A, 2B). Moreover, we used a DNA assay to quantify the seeding efficiency, which was considerably higher for in-situ seeding (~97%) than for the post-seeding method (~34%) (Figure 2C). It should be noted that all post-seeding experiments were performed with a cell concentration, media volume and seeding time that matched the corresponding in-situ endothelialization. Moreover, these experiments were performed on straight, relatively large-diameter channels, conditions that are favourable for post-seeding. The non-uniform, low-efficiency cell attachment observed after post-seeding is normally addressed by using multiple injections or a surplus of cells (high cell concentration and/or volume). Such procedures are wasteful (high cell loss), time-consuming and practically challenging, moreover, they provide little control over the seeding process. On the other hand, the cell concentration during seeding could be exploited during in-situ endothelialization (1 - 10 × 106 ml-1) to provide a high degree of control over the number of adhered cells, while maintaining a uniform distribution throughout (Figure S10A, S10B). Furthermore, uniform seeding was also achieved when in-situ endothelialization was performed in channels with varied cross-sectional diameter (Figure S10C, S10D).
To study in-situ endothelization over culture, we used two different geometric designs. The first was the original cubic lattice design, with interconnected channels orthogonally oriented between adjacent layers (Figure 2D), while the second comprised a set of hexagonally-packed parallel channels without any interconnectivity (Figure 2E). HUVECs (5 × 106 cells ml-1) were pre-loaded in 7.5 wt% gelatin as the templating bioink, with 7.5 wt% GelMA used as the matrix bioink. After printing and crosslinking, we transferred the cell-laden constructs to culture medium at 37 °C. At days 1 and 3, we observed the HUVECs still uniformly distributed throughout the channels of both print designs, and importantly, the cells were located on the inside wall of the templated channels (Figure S11, Figure S12). At day 7, the cells had appeared to proliferate to form homogenous, confluent endothelial layers in both the interconnected networks and parallel tubes (Figure 2F, 2G). At day 8, we performed immunostaining that revealed widespread expression of the endothelial cell adhesion marker CD31, which confirmed that the HUVECs had formed a confluent monolayer (Figure 2H, Figure S13, Movie S4). Furthermore, ultrastructural analysis using transmission electron microscopy revealed extensive intercellular junctions, including focal contacts (Figure S14A) and overlapping junctions (Figure S14B). [31] We used an alamarBlue assay to quantitatively assess the metabolic activity of the printed constructs over time. We observed a similar growth trend in both print designs; the metabolic activity increased from day 1, peaked around days 10-12 and then dropped slightly at day 14 (Figure 2I). Interestingly, from day 6 onwards, the growth rate of the cells in the lattice channels was higher than that of the cells in the parallel channels. This observation was attributed to the improved medium penetration in the structures with an interconnected lattice, compared to those with independent channels.
One plausible alternative to VF-3DP would be to post-seed HUVECs onto a directly printed 7.5 wt% GelMA lattice (Figure S15A). However, when we tested this approach we encountered the aforementioned issues with buckling, channel fusion and expanded pores, but also a very non-uniform distribution of HUVECs that resulted in a highly fragmented endothelial layer at day 8 (Figure S15B). Another alternative to VF-3DP would be to post-cast 7.5 wt% GelMA around a pre-printed 7.5 wt% gelatin lattice (Figure S16A). Here, we encountered the additional issue that bubbles formed within the lattice voids, stabilized by surface tension, which led to defective hydrogel casting. With a 1 mm distance between gelatin filaments, bubbles were present in almost every pore (Figure S16B), while increasing the filament separation to 2 mm still resulted in a large number of defects (Figure S16C). Taken together, these failed alternatives illustrated the unique potential of VF-3DP for fabricating uniform, endothelialized tissue constructs.
Aside from vascular tissue engineering, we explored whether the VF-3DP approach could be used to print a hydrogel-based microfluidic device (Figure 3A). Using hydrogels as base materials for microfluidics has recently attracted attention, [32, 33] since they can better support cell migration and tissue remodeling compared to the more common elastomer-based systems. For example, angiogenesis and perivascular interactions have been studied with microvessels formed in degradable hyaluronic acid [34] and collagen [31]. To this end, we printed microfluidic structures using the templating bioink (7.5 wt% gelatin) and matrix bioink (7.5 wt% GelMA) and then sacrificed the former to create well-defined, templated flow channels (Figure S17A). We used a polydimethylsiloxane (PDMS) casing to connect the microfluidic hydrogel to a peristaltic pump for controlled perfusion (Figure 3A, Figure S17B). This process enabled us to create different fluidic patterns, such as a simple S-shaped flow (Figure 3B) or more complex patterns, with the flow path splitting at various circular nodes (Figure 3C). We were even able to use this microfluidic device to perfuse a GelMA lattice structure, fabricated using VF-3DP (Figure 3D). We were able to clearly visualize the channel path by perfusing the system with a fluorescent dye solution (Figure S17C).
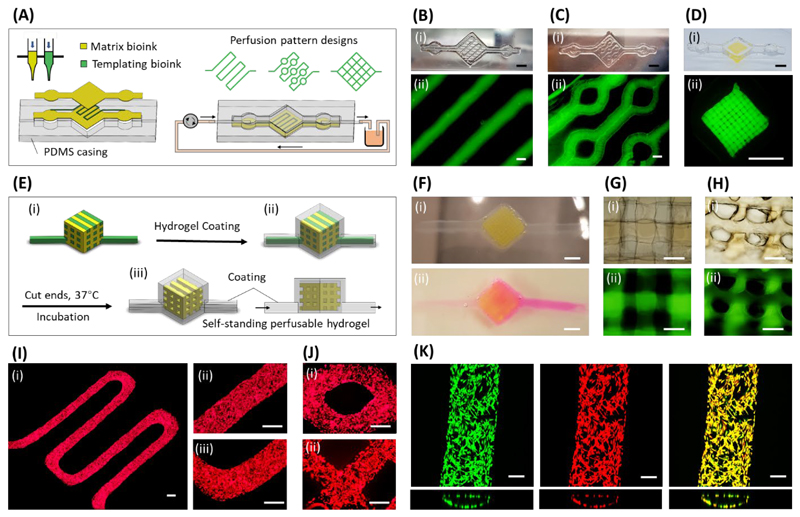
Figure 3.
VF-3DP hydrogel-based microfluidics. (A) Schematic of hydrogel-based microfluidic system with customized perfusion patterns generated using VF-3DP, with a temporary bioink used to template well-defined channels. Representative (i) photographs and (ii) fluorescence microscopy images of (B) an S-shaped single channel, (C) an S-shaped channel with circular joints, and (D) a 3D lattice channel pattern. (E) Schematic of self-standing perfusable 3D constructs generated using VF-3DP, where both the matrix bioink (yellow) and templating bioink (green) were pre-loaded with Ca2+ (87 mM). These doped bioinks were used for (i) the VF-3DP of a lattice structure, which was (ii) dipped into a 1 wt% sodium alginate solution to rapidly form a calcium alginate hydrogel coating. (iii) The two extended ends of the construct could be cut so that the templated bioink could be liquified and released upon incubation. (F) Representative images of the self-standing perfusable construct (i) after alginate coating and (ii) after incubation and dye perfusion. (G-H) (i) Brightfield and (ii) fluorescence microscopy of the self-standing perfusable hydrogel, showing details of the 3D tubular porosity from either (G) top-down or (H) cross-sectional views. (I) Representative images of RFP-labeled HUVECs adhered to the walls of an S-shaped single channel after 12 d of perfusion culture, imaged at (i) low magnification and (ii-iii) high magnification. (J) Representative images of RFP-labeled HUVECs in structures with (i) a circular joint and (ii) lattice channels, imaged after 10 d of perfusion culture. (K) Confocal fluorescence microscopy images of HUVECs after 12 d of perfusion culture. The fluorescence channels show Calcein-AM live stain (green) and RFP-reporter fluorescence (red), with excellent co-localization (yellow). Scale bars: 5 mm (B(i), C(i), D, F), 500 μm (B(ii), C(ii), G, H, I, J), 200 μm (K).
Moreover, we explored whether we could fabricate a self-standing perfusable hydrogel construct, i.e. without any interfacing PDMS structure. GelMA and gelatin bioinks were printed into a VF-3DP cubic structure bearing two extended gelatin handles (Figure 3E). We also pre-supplemented both bioinks with 87 mM CaCl2, which enabled the formation of a thin alginate hydrogel coating over the entire printed structure by immersing it in a 1 wt% solution of sodium alginate. This gelation process was extremely rapid (<1 min) based on the fast diffusion of Ca2+ from the bioinks to the surrounding alginate. We observed excellent co-adhesion between the alginate coating and the internal GelMA lattice, with no visible delamination. This structural integrity was partially achieved by including 5 wt% GelMA within the sodium alginate solution, so that photo-crosslinked covalent bonds could be formed across the interface. [35] After this post-coating step, we could simply cut off the end of the two handles, incubate at 37 °C to remove the gelatin and connect the handles to a syringe pump for perfusion (Figure 3F). Through this method, we achieved a 3D self-standing perfusable hydrogel construct with tubular porosity and no external PDMS interface (Figure 3G, 3H).
Finally, we sought to investigate whether perfusion of culture medium could be used to maintain the viability of in-situ seeded endothelial cells during culture. We used VF-3DP to generate S-shaped single-channel structures connected to a media supply via a PDMS casing (Figure S18). After 12 d of perfusion culture, we observed an endothelial cell layer uniformly distributed across both the straight and turning segments of the channel (Figure 3I). We saw similar results when perfusing VF-3DP structures bearing circular joints or lattice channels for 10 d (Figure 3J). We used Calcein-AM staining, together with the fluorescence of RFP-labelled HUVECs, to visualize a highly viable cell population after 12 d of perfusion culture (Figure 3K). These results are promising in terms of perfusion stability and cell viability; however, further optimization of the seeding, perfusion and culture protocols are required in order to optimize the system towards more confluent, flow-aligned endothelial layers.
3. Conclusion
In conclusion, we have developed a void-free bioprinting strategy that can be used to generate 3D structures with well-defined, uniform, tubular channels, that can be vascularized in situ without the need for post cell seeding. This approach offered multiple benefits over conventional strategies that use a single bioink to directly print porous structures. In terms of print quality, our strategy avoided any lattice buckling, channel fusion or expansion and offered the possibility for interconnected tubular porosity and the capacity for printing low-concentration bioinks, such as 5 wt% GelMA, 5 wt% MeHA and 5 wt% NorHA. Moreover, our demonstration of in-situ cell seeding distinguishes this method from other sacrificial bioprinting strategies, which use templating to create acellular channels that then require post seeding. [13–15,27, 36, 37] In particular, we show that in-situ seeding provides higher efficiency, uniformity and control over cell density, compared to post-seeding methods, and can be used for vascularizing large constructs with 3D interconnected channels, which is challenging for co-axial bioprinting [4]. Our printing approach could also be used to fabricate hydrogel-based microfluidic devices with customized flow patterns. These structures could be integrated with PDMS microfluidics, or alternatively, diffusion-induced gelation could be used to create an alginate-based hydrogel coating and generate perfusable, self-standing 3D constructs. Overall, this void-free bioprinting method represents a technological advance in our ability to print vascularized and perfusable constructs. The simplicity, versatility, and practicality that this method offers will enable it to be applied to a wide range of bioprinting, tissue engineering, and microfluidic applications.
4. Experimental Section
Materials synthesis
Unless otherwise stated, all chemicals were purchased from Sigma-Aldrich, all water used was UltraPure water. GelMA was synthesized by conjugating methacrylate functional groups to gelatin, as described previously. [20] Briefly, gelatin was dissolved in water at 50 °C to a final concentration of 10 wt%. Methacrylic anhydride (MA) was added to the solution dropwise while stirring, at a ratio of 0.6 g MA per 1 g gelatin. After 3 h of reaction at 50 °C, the solution was centrifuged at 3500 g for 3 min and the supernatant, containing the GelMA, was collected and diluted with four volume equivalents of water. The GelMA solution was subsequently dialyzed (12-14 kDa molecular weight cut off) against water at 40 °C for one week, with the water changed twice a day. This was followed by pH adjustment to 7.4 using 1 M NaHCO3 and filter-sterilization using 0.2 μm vacuum filtration. FITC-conjugated GelMA (GelMA-FITC) was synthesized by reacting GelMA with NHS-Fluorescein (Thermo Fisher). Briefly, GelMA was fully dissolved in MES buffer (Thermo Fisher, adjusted to pH 8.1) at 50 °C to a concentration of 10 wt%, followed by addition of 30 mg NHS-fluorescein per 1 g GelMA. The reaction was performed for 3 h at 50 °C in the dark and was subsequently quenched by adding four volume equivalents of water. The product was then dialysed (5 kDa molecular weight cut off) at 40 °C for one week, with the water changed twice a day. GelMA-FITC was freeze-dried and stored at -20 °C before use. Methacrylated HA (MeHA) and norbornene-functionalized HA (NorHA) were kind gifts from the Burdick lab at the University of Pennsylvania. These products were synthesized as previously described. [29, 30] Briefly, MeHA was prepared by esterification between HA (75 kDa, Lifecore) and MA, where 3 molar equivalents of MA were added dropwise to an aqueous 1 wt % HA solution on ice for 6-8 h, while maintaining the pH ~8. After an overnight reaction at 4 °C, the solution was neutralized to pH 7-7.5, followed by dialysis and freeze drying. NorHA was prepared from HA tetrabutylammonium salt (HA-TBA), which was dissolved in DMSO to a concentration of 0.5 wt%. 5-norbornene-2-carboxylic acid (Nor-COOH, 3 eq), 4-dimethylaminopyridine (DMAP, 1.5 eq) and di-tert-butyl dicarbonate (BOC2O, 0.4 eq) were added. The reaction was performed at 45 °C for 24 h, followed by precipitation against acetone, dialysis against water and freeze drying. Final products were stored at -20 °C before use.
Modification degree of GelMA
1H-NMR spectroscopy was used to confirm the successful conjugation of methacrylate groups (Figure S1A), while a fluoraldehyde assay was used to quantify the degree of modification (Figure S1B). Briefly, GelMA samples and gelatin standards were prepared at a concentration range from 0.10-0.75 mg ml-1. These solutions were then thoroughly mixed with fluoraldehyde reagent solution (Thermo Fisher) at a volume ratio of 1:2. Triplicate aliquots of each sample and standard were transferred to an opaque 96-well plate (250 μl per well). The fluorescence intensity was determined at 450 nm with an excitation wavelength of 360 nm using a microplate reader (SpectraMax M5, Molecular Devices).
Rheological characterization
Hydrogel samples were prepared in PBS at the desired concentration. To determine the thermal responsivity of the hydrogels, an oscillation temperature sweep was performed using a rheometer (AR2000, TA Instruments) employing a cone geometry (No. 991569, diameter 40 mm), which corresponded to a sample volume of 600 μl. The temperature was set for a ramp change from 37 °C to 7 °C and then back to 37 °C, at a rate of 5 °C min-1. A consistent strain (1%) and frequency (1.5 Hz) were used throughout the temperature ramp.
Void-free 3D Printing process
The VF-3DP was performed using a multi-nozzle 3D Bioprinter (3D-Bioplotter, EnvisionTEC), where the temperature of both the extrusion nozzle and print bed could be controlled. The prepared bioinks were first incubated at 37 °C for 10 min before loading into an assorted 30 cc cartridge (Nordson EFD) equipped with a 25-gauge needle. The print bed was pre-set to 15 °C, which would trigger the thermal gelation of gelatin-based hydrogels after printing. The nozzle temperature was set at values that would result in smooth gel filament extrusion, as described previously. [18] For instance, the nozzle temperatures used were: 7.5 wt% gelatin (26-27 °C), 7.5 wt% GelMA (22-23 °C), 5 wt% GelMA (19-20 °C). The nozzle temperature for 5 wt% MeHA and 5 wt% NorHA was set at 24 °C, since they exhibit a relatively stable viscosity around this temperature range. In this study, the VF-3DP was mainly realized by printing the templating and matrix bioinks side-by-side in a layer-by-layer fashion. To do so, two identical sliced parts (e.g., 10 × 10 × 3 mm cubic with a slice thickness of 300 μm) were fully overlapped in the bioprinter software (VisualMachines) and assigned with the templating and matrix bioinks, respectively. Meanwhile, the assigned infill printing path for both bioinks were set with an in-line gap of 1 mm and their patterns were shifted 0.5 mm with respect to each other in order to fully fill the printing space. For in-situ endothelialization studies, the matrix bioink was also assigned with a contour path for better cell maintenance during incubation. Normally, a printing speed of 3-6 mm s-1 and an air pressure of 0.7-1.1 bar were used. Unless otherwise stated, 7.5 wt% gelatin was used as the templating bioink. The gelatin was supplemented with 5 × 106 HUVECs per mL for in-situ endothelialization. The matrix bioink comprised different photo-crosslinkable formulations, including 7.5 wt% GelMA, 5 wt% GelMA, 5 wt% MeHA and 5 wt% NorHA. The matrix bioink was always supplemented with 0.05 wt% Irgacure 2959 (I2959). After printing, the void-free hydrogel structure was irradiated with ultraviolet (UV) light (Uvitec, 365 nm, ~ 5 mW cm-2, 5 min) in order to photo-crosslink the matrix phase, followed by incubation at 37 °C to liquify the templating phase. For in-situ endothelialization studies, the printed structures were flipped every 30-60 min after incubation, in order to distribute the HUVECs as uniformly as possible. The structures were then cut off at the edges to open up the channels and allow the release of gelatin. The constructs were incubated in static culture at 37 °C under 5% CO2, with the culture medium changed every two days. All computer-aided design (CAD) files are shown in Figure S19.
Channel feature observation
After incubation, the final VF-3DP constructs were cut vertically, then placed on their side for microscopy (Olympus BX51 and Zeiss Axio Observer). In some cases, GelMA-FITC was used to dope the GelMA bioink at a weight ratio of 1:24 and used as the matrix bioink to better distinguish the hollow channels. The channels were also visualized by dripping food dye or a solution of fluorescent microbeads (diameter of 0.503 μm, plum purple, Bangs Laboratories) onto the construct.
Mechanical properties
Compression testing was performed with the final VF-3DP constructs (designed dimensions: 10 × 10 × 3 mm) using a uniaxial mechanical tester (TA Electroforce 3200) equipped with a 250 g force sensor. Ramp compression at a speed ratio of 0.01 mm s-1 was applied to obtain the stress-strain curve. The compression modulus was calculated within the strain range of 0.1-0.2 mm/mm, ensuring that the linear coefficient of determination (R2) was greater than 0.99.
Perfusable hydrogel-based microfluidics
A PDMS casing with a chamber connected to an inlet and an outlet was used as the holder for the hydrogel-based device. A complete layer of matrix bioink (7.5 wt% GelMA) was first printed as the base (parallel printing patch with a line gap of 0.5 mm), followed by the deposition of both matrix and templating bioinks. In the case of the 3D lattice pattern, a typical VF-3DP process was performed on the base layer using these two bioinks. All patterns had the templating bioink forming a channel connected to the inlet/outlet ports. An additional matrix bioink was subsequently cast in the PDMS chamber to cover the whole hydrogel device. After UV irradiation (365 nm, ~ 5 mW cm-2, 5 min), the whole device was placed at 37 °C to liquify the gelatin, which gradually leached out of the inlet and outlet ports. The perfusion was demonstrated by connecting the device to a peristaltic pump (Ismatec) at flow rates ranging from 0.05-1.0 ml min-1. For perfusion culture of the cell-laden constructs, the hydrogel device together with PDMS casing were covered with glass slides on the top and bottom and clamped with metal clips to avoid leaking. 8-10 mL medium was added in the reservoir and changed every 2-3 days. By connecting the reservoir, the peristaltic pump and the hydrogel microfluidic device into a closed-loop system, the medium level in the reservoir could be used to assess the circulation. No reduction in the level of medium was observed during the culture periods, which indicated smooth watertight perfusion. A perfusion flow rate of 0.05 mL min-1 was used for these cell culture experiments.
Perfusable self-standing hydrogel constructs
The fabrication of perfusable self-standing hydrogel constructs used the standard VF-3DP process and cubic lattice design but with extended inlet and outlet tubes and with both bioinks supplemented with 1 wt% CaCl2 (equivalent to 87 mM Ca2+). The VF-3DP construct was subsequently immersed for 2 min in a solution of 1 wt% sodium alginate and 5 wt% GelMA, with the latter used to bind the alginate coating and the printed matrix, a protocol based on a previous study. [35] After UV crosslinking (365 nm, ~ 5 mW cm-2, 5 min), the ends of the inlet and outlet tubes were cut off to allow the gelatin phase to be removed by incubation at 37 °C. Perfusion was demonstrated by connecting the inlet tube to a syringe pump (Cole-Parmer) at flow rate of 0.4 mL min-1.
Cell culture
Human umbilical vein endothelial cells (HUVECs) were cultured in Endothelial Growth Medium (EGM-2, Lonza), while human dermal fibroblasts (HDF, Sigma) were cultured in high glucose Dulbecco's Modified Eagle Medium (DMEM, Gibco) supplemented with 10 vol% fetal bovine serum (FBS, Thermo Fisher) and 1 vol% penicillin-streptomycin (Thermo Fisher). All cell lines were used at passage 8 or lower.
Cell seeding characterization
To compare the cell seeding efficiency of the conventional post-seeding method and the presented in-situ seeding approach, a single layer of parallel channels were printed. For post-seeding experiments, acellular VF-3DP constructs were printed and the channel ends were cut to release the liquified gelatin. A suspension of RFP-labeled HUVECs (5 × 106 mL-1) with a volume equivalent to the target channel was injected into the channel from one end, followed by incubation for 6 h, during which the construct was carefully flipped upside down every hour. For the in-situ seeding experiments, VF-3DP cellular constructs were printed with varied initial cell concentration (1 × 106, 5 × 106, and 10 × 106 RFP-labeled HUVECs per mL) in the templating bioink. After incubation for 6 h with flipping every hour, the channel ends were cut off to release the liquified gelatin and non-adhered cells. The samples were prepared for DNA quantification by addition of 200 μL of deionized water, homogenization at 20 Hz for 10 min using a TissueLyzer II (QIAGEN), followed by three freeze-thaw cycles from -80°C to room temperature. For the post-seeding control, an equivalent number of cells were transferred, with an unseeded VF-3DP gel, to a tube for lysis and DNA analysis. For the in-situ seeding, uncut VF-3DP cellular constructs were used as a control. A PicoGreen™ dsDNA assay kit (Invitrogen) was used to determine the DNA content of each sample, with the assay performed according to the manufacturer’s protocol. Briefly, PicoGreen™ working solution was added to an equivalent volume of diluted samples, controls or DNA standards in a 96-well plate. After 5 min incubation, the fluorescence emission was measured using a plate reader at 520 nm, with an excitation wavelength of 480 nm. The cell seeding efficiency was calculated by dividing the DNA mass in the samples by the DNA mass in the controls. ImageJ was used to analyze the fluorescence images of seeded cells to estimate the cell occupation and distribution in the channels. Briefly, the cellularized area ratio was calculated by measuring the fluorescence area ratio within the region of interest (the channel) using the Analyze Particles function. By dividing the channels into equivalent segments and acquiring the corresponding cellularized area ratio, the cellularized area ratio along the channel was obtained.
Cell viability
LIVE/DEAD™ staining was conducted by immersing the printed cell-laden construct into calcein-AM/ethidium homodimer-2 (Invitrogen) solution (each at 1 μM in PBS) for 20 min, followed by gentle washing with PBS. Confocal fluorescence microscopy (Leica SP5) was used to capture images of viable and nonviable cells. ImageJ was used to quantify the cell viability, based on the proportion of viable cells to total cells.
Cell proliferation
An alamarBlue assay (Thermo Fisher) was used to determine the metabolic activity of HUVECs in the printed hydrogel constructs over time. Briefly, alamarBlue reagent was mixed with EGM-2 at a volume ratio of 1:9 to form the working solution. At defined time points, cell-laden 3D hydrogel constructs were harvested and then cultured in working solution for 3 h. Triplicate aliquots of supernatant were transferred to a black, clear-bottom 96-well plate (100 μl per well). The fluorescence at 585 nm (excitation wavelength of 570 nm) was measured using a multimode plate reader (EnVision) and used to determine the cell metabolic activity.
Immunostaining
HUVEC-laden constructs were fixed with 4 wt% paraformaldehyde (PFA, Electron Microscopy Sciences) for 1-2 h and then permeabilized with 0.25 vol% Triton-X 100 for 2 h. After blocking with 1 wt% bovine serum albumin (BSA) overnight at 4 °C, the constructs were incubated overnight at 4 °C with primary antibody (5 μg mL-1 CD31 (Thermo Fisher) in 1 wt% BSA solution) and then for 6 h at room temperature with secondary antibody (1 μg mL-1 Alexa Fluor™ 488 (Thermo Fisher) in 1 wt% BSA solution). Optionally, the constructs were subsequently incubated for 1 h with 5 U mL-1 phalloidin solution (Alexa Fluor™ 488 or 647, Thermo Fisher) and then for 15 min with 5 μg mL-1 DAPI solution (Thermo Fisher). Three PBS washes were performed between all incubation steps. All images were taken using widefield fluorescence microscope (Olympus BX51) and confocal fluorescence microscope (Leica SP5).
Transmission electron microscopy
HUVEC-laden constructs were fixed for 1 h in 4 wt% PFA and then 1 h in 2.5 wt% glutaraldehyde (Electron Microscopy Sciences) buffered with 0.1 M sodium cacodylate buffer, pH 7.4. washed with 0.1 M cacodylate buffer and then post-fixed in 2 wt% osmium tetroxide (Electron Microscopy Sciences) for 1 h. The samples were then stained with 1 wt% tannic acid for 1 h and 1 wt% uranyl acetate (Electron Microscopy Sciences) for 2.5 h, followed by dehydration through ascending grades of ethanol (70, 90, 100%) and embedding into epoxy resin. 60 nm films were sectioned and transferred onto 200 mesh Cu grids. 5 nm carbon was coated on the sections to improve conductivity, before imaging using a TITAN 80/300 TEM/STEM with an accelerating voltage at 80 kV.
Supplementary Material
Supporting Information is available from the Wiley Online Library or from the author.
Acknowledgments
The authors thank Miss. D. Čekatauskaitė and Dr. S. Crowder for their help with cell culture, and Dr. A. Nagelkerke for her help with cell transfection in HUVECs labelling. L.O. thanks Prof. J.A. Burdick for the kind gift of modified hyaluronic acid materials. L.O. and M.M.S. acknowledge the financial support from EPSRC Programme Grant “Engineering growth factor microenvironments - a new therapeutic paradigm for regenerative medicine” (EP/P001114/1). J.P.K.A. acknowledges support from the Medical Research Council (MRC) (MR/S00551X/1). M.M.S. acknowledges support from a grant from the UK Regenerative Medicine Platform “Acellular / Smart Materials – 3D Architecture” (MR/R015651/1) and the Wellcome Trust Senior Investigator Award (098411/Z/12/Z).
References
- 1.Novosel EC, Kleinhans C, Kluger PJ. Adv Drug Deliv Rev. 2011;63(4-5):300–311. doi: 10.1016/j.addr.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Osaki T, Sivathanu V, Kamm RD. Curr Opin Biotechnol. 2018;52:116–123. doi: 10.1016/j.copbio.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Place ES, Evans ND, Stevens MM. Nat Mater. 2009;8(6):457–470. doi: 10.1038/nmat2441. [DOI] [PubMed] [Google Scholar]
- 4.Jia W, Gungor-Ozkerimab PS, Zhang YS, Yue K, Zhu K, Liu W, Pi Q, Byambaa B, Dokmeci MR, Shi SR, Khademhosseini A. Biomaterials. 2016;106:58–68. doi: 10.1016/j.biomaterials.2016.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sobral JM, Caridade SG, Sousa RA, Mano JF, Reis RL. Acta Biomater. 2011;7(3):1009–1018. doi: 10.1016/j.actbio.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Roh JD, Nelson GN, Udelsman BV, Brennan MP, Lockhart B, Fong PM, Lopez-Soler RI, Saltzman WM, Breuer CK. Tissue Engineering. 2007;13(11):2743–2749. doi: 10.1089/ten.2007.0171. [DOI] [PubMed] [Google Scholar]
- 7.Zhang YS, Arneri A, Bersini S, Shin SR, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell'Erba V, Bishop C, et al. Biomaterials. 2016;110:45–59. doi: 10.1016/j.biomaterials.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colosi C, Shin SR, Manoharan V, Massa S, Costantini M, Barbetta A, Dokmeci MR, Dentini M, Khademhosseini A. Adv Mater. 2016;28(4):677–684. doi: 10.1002/adma.201503310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan EYS, Yeong WY. International Journal of Bioprinting. 2015;1(1):49–56. [Google Scholar]
- 10.Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue HJ, Ramadan MH, Hudson AR, Feinberg AW. Sci Adv. 2015;1(9):e1500758. doi: 10.1126/sciadv.1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouyang L, Highley CB, Rodell CB, Sun W, Burdick JA. ACS Biomaterials Science & Engineering. 2016;2(10):1743–1751. doi: 10.1021/acsbiomaterials.6b00158. [DOI] [PubMed] [Google Scholar]
- 12.Ribeiro A, Blokzijl MM, Levato R, Visser CW, Castilho M, Hennink WE, Vermonden T, Malda J. Biofabrication. 2017;10(1) doi: 10.1088/1758-5090/aa90e2. 014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DH, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, et al. Nat Mater. 2012;11(9):768–774. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA. Proc Natl Acad Sci U S A. 2016;113(12):3179–3184. doi: 10.1073/pnas.1521342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA. Adv Mater. 2014;26(19):3124–3130. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 16.Khattak SF, Bhatia SR, Roberts SC. Tissue Engineering. 2005;11(5-6):974–983. doi: 10.1089/ten.2005.11.974. [DOI] [PubMed] [Google Scholar]
- 17.Noor N, Shapira A, Edri R, Gal I, Wertheim L, Dvir T. Adv Sci (Weinh) 2019;6(11) doi: 10.1002/advs.201900344. 1900344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouyang L, Yao R, Zhao Y, Sun W. Biofabrication. 2016;8(3) doi: 10.1088/1758-5090/8/3/035020. 035020. [DOI] [PubMed] [Google Scholar]
- 19.Ouyang L, Yao R, Mao S, Chen X, Na J, Sun W. Biofabrication. 2015;7(4) doi: 10.1088/1758-5090/7/4/044101. 044101. [DOI] [PubMed] [Google Scholar]
- 20.Loessner D, Meinert C, Kaemmerer E, Martine LC, Yue K, Levett PA, Klein TJ, Melchels FPW, Khademhosseini A, Hutmacher DW. Nat Protoc. 2016;11(4):727–746. doi: 10.1038/nprot.2016.037. [DOI] [PubMed] [Google Scholar]
- 21.Yue K, Trujillo-de Santiago G, Alvarez MM, Tamayol A, Annabi N, Khademhosseini A. Biomaterials. 2015;73:254–271. doi: 10.1016/j.biomaterials.2015.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ouyang L, Highley CB, Sun W, Burdick JA. Adv Mater. 2017;29(8) doi: 10.1002/adma.201604983. 1604983. [DOI] [PubMed] [Google Scholar]
- 23.Caliari SR, Burdick JA. Nature Methods. 2016;13(5):405–414. doi: 10.1038/nmeth.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YC, Lin RZ, Qi H, Yang Y, Bae H, Melero-Martin JM, Khademhosseini A. Advanced Functional Materials. 2012;22(10):2027–2039. doi: 10.1002/adfm.201101662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costantini M, Testa S, Fornetti E, Barbetta A, Trombetta M, Cannata SM, Gargioli C, Rainer A. Front Bioeng Biotechnol. 2017;5:22. doi: 10.3389/fbioe.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang LS, Chung JE, Chan PP, Kurisawa M. Biomaterials. 2010;31(6):1148–1157. doi: 10.1016/j.biomaterials.2009.10.042. [DOI] [PubMed] [Google Scholar]
- 27.Bertassoni LE, Cardoso JC, Manoharan V, Cristino AL, Bhise NS, Araujo WA, Zorlutuna P, Vrana NE, Ghaemmaghami AM, Dokmeci MR, Khademhosseini A. Biofabrication. 2014;6(2) doi: 10.1088/1758-5082/6/2/024105. 024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin J, Yan M, Wang Y, Fu J, Suo H. ACS Appl Mater Interfaces. 2018;10(8):6849–6857. doi: 10.1021/acsami.7b16059. [DOI] [PubMed] [Google Scholar]
- 29.Khetan S, Guvendiren M, Legant WR, Cohen DM, Chen CS, Burdick JA. Nat Mater. 2013;12(5):458–465. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gramlich WM, Kim IL, Burdick JA. Biomaterials. 2013;34(38):9803–9811. doi: 10.1016/j.biomaterials.2013.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, Lopez JA, Stroock AD. Proc Natl Acad Sci U S A. 2012;109(24):9342–9347. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nie J, Gao Q, Wang Y, Zeng J, Zhao H, Sun Y, Shen J, Ramezani H, Fu Z, Liu Z, Xiang M, et al. Small. 2018;14(45):e1802368. doi: 10.1002/smll.201802368. [DOI] [PubMed] [Google Scholar]
- 33.Lee SH, Shim KY, Kim B, Sung JH. Biotechnol Prog. 2017;33(3):580–589. doi: 10.1002/btpr.2457. [DOI] [PubMed] [Google Scholar]
- 34.Leong JY, Lam WH, Ho KW, Voo WP, Lee MFX, Lim HP, Lim SL, Tey BT, Poncelet D, Chan ES. Particuology. 2016;24:44–60. [Google Scholar]
- 35.Ouyang L, Burdick JA, Sun W. ACS Appl Mater Interfaces. 2018;10(15):12424–12430. doi: 10.1021/acsami.7b19537. [DOI] [PubMed] [Google Scholar]
- 36.Lee V, Lanzi A, Haygan N, Yoo S, Vincent P, Dai G. Cell Mol Bioeng. 2014;7(3):460–472. doi: 10.1007/s12195-014-0340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertassoni LE, Cecconi M, Manoharan V, Nikkhah M, Hjortnaes J, Cristino AL, Barabaschi G, Demarchi D, Dokmeci MR, Yang Y, Khademhosseini A. Lab on a Chip. 2014;14(13):2202–2211. doi: 10.1039/c4lc00030g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.