Abstract
In 2013, the “biofabrication window” was introduced to reflect the processing challenge for the fields of biofabrication and bioprinting. At that time, the lack of printable materials that could serve as cell-laden bioinks, as well as the limitations of printing and assembly methods, presented a major constraint. However, recent developments have now resulted in the availability of a plethora of bioinks, new printing approaches, and the technological advancement of established techniques. Nevertheless, it remains largely unknown which materials and technical parameters are essential for the fabrication of intrinsically hierarchical cell–material constructs that truly mimic biologically functional tissue. In order to achieve this, it is urged that the field now shift its focus from materials and technologies toward the biological development of the resulting constructs. Therefore, herein, the recent material and technological advances since the introduction of the biofabrication window are briefly summarized, i.e., approaches how to generate shape, to then focus the discussion on how to acquire the biological function within this context. In particular, a vision of how biological function can evolve from the possibility to determine shape is outlined.
Keywords: biofabrication, bioinks, biological function, regenerative medicine, tissue hierarchy
1. Introduction
The functionality of living tissues is intimately linked to their intricate and highly specialized architecture. Tissues and organs are composed of multiple types of cells and extracellular matrix (ECM) components and, with few exceptions, are infiltrated with vascular and neural networks. The hierarchical spatial arrangement of these elements is paramount to how they interact with each other and, thus, closely orchestrates several processes during embryonic development,[1] in healthy tissue homeostasis, as well as during tissue regeneration.[2] Strategies to generate cell–material constructs that ultimately yield a healthy and mature functional tissue remains a major challenge in the field of regenerative medicine.
With the introduction of additive manufacturing, technologies became available to design and fabricate 3D material scaffolds with unprecedented shape and precision. Although, many of these technologies are potentially harmful for active biological components, including living cells, biological matter can successfully be used as building blocks for the generation of living 3D objects. This constitutes the research field of biofabrication,[3] a revolutionizing toolkit for regenerative medicine that allows cells, biomaterials and bioactive moieties to be precisely combined and patterned into 3D constructs through automated, cell-friendly fabrication methods, such as bioprinting and bioassembly.[4] Indeed, modified extrusion,[5] (stereo)lithographic,[6] inkjet,[7] and laser printing methods[8] are now available for the processing of living cells and can be applied to recreate anatomical parts using medical images as blueprints.[9] Within the past decade there has been a particular focus on techniques and materials compatible with extrusion-based printing due to the high versatility in printing multiple materials, the relative low cost and easy access to the required hardware for this technology.[10] Taken together, biofabrication has gained significant momentum and provides a powerful approach to tackle major hurdles in the generation of engineered living tissues.
As building materials in biofabrication processes, two different types of printable “inks” can generally be distinguished.[11] Firstly, materials that are used to print acellular structures, on which cells are seeded or that can also be used as surgical tools or implants after fabrication, are termed biomaterial inks. Many different materials, including thermoplasts and metal powders, can be processed using a variety of technologies and the process parameters are only restricted by technology and the respective material. Secondly, the printable formulations that contain living cells are termed bioinks.[3] This simple but clear distinction severely limits the number of suitable fabrication technologies as the process must ensure viability of the embedded cells. Therefore, the printing process needs to be performed under physiological conditions and in an aseptic environment. Bioinks are generally aqueous formulations with adjusted rheology that can provide a highly hydrated environment for the encapsulated cells.[11] For this, often formulations of hydrogel precursors are applied that can be crosslinked postfabrication. An important challenge in this context is, however, the counterdirectional effect of polymer concentration on shape fidelity and cytocompatibility. In other words, low polymer content in the bioink results in soft, loosely crosslinked hydrogels after printing that are beneficial to support cell survival, migration and bioactivity, but such formulations do typically result in structures with poor shape fidelity. In contrast, bioinks with higher polymer content and thus higher viscosity can more rapidly be crosslinked into stiffer gels after printing and are in general preferred for the accurate 3D placement and postprinting shape retention,[12] resulting in a printed object that faithfully reproduces its original computer design. However, this comes with the cost of reduced ability of the cells to spread, migrate and colonize the hydrogel matrix with newly synthesized ECM. Additionally, high crosslinking density or polymer content can also hinder the ability of the embedded cells to remodel the hydrogel matrix over time, a process necessary for tissue maturation.
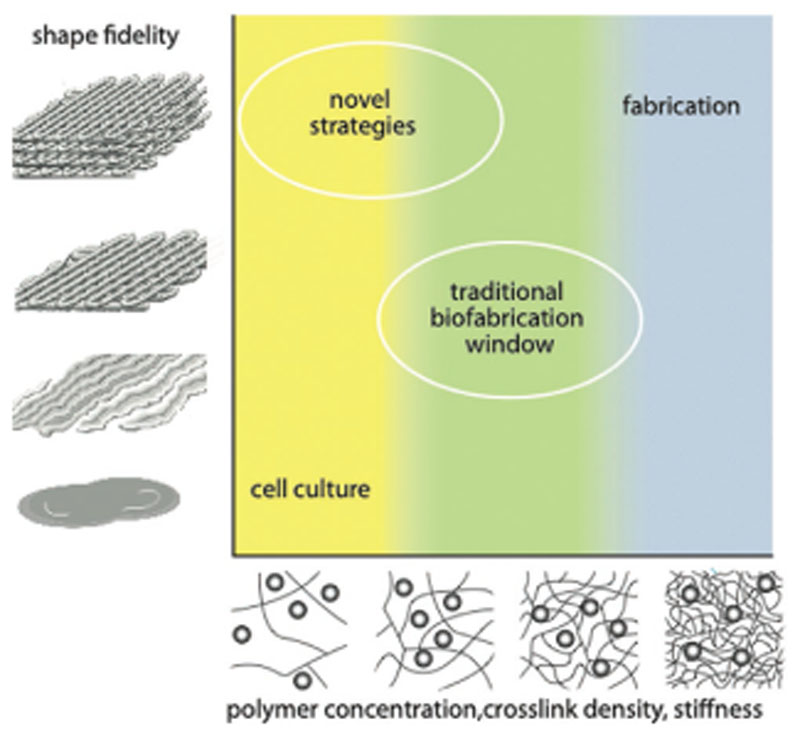
The simultaneous need for these opposing requirements led to the conceptualization of the biofabrication window, the range of material properties suitable both for printability with high shape fidelity and for the support of cell function (Figure 1).[12] This concept has since then widely been appreciated in the biofabrication community.[13–17] Strategies to extend this biofabrication window and allow for printing with good shape fidelity under cytocompatible conditions with as little material content as possible have been an important focus of recent research in the field.
Figure 1.
Schematic representation of the biofabrication window as introduced in 2013. Reproduced with permission.[12] Copyright 2013, Wiley-VCH.
Bioprinted structures are currently being studied as potential transplantable grafts for tissue restoration,[18] as advanced in vitro models to aid the testing of drugs and as potential alternatives to animal experimentation.[19–21] These are used to study tissue development and disease[22] and as components integrated within organ-on-chip devices.[23] While the development of new bioinks and adaptations of existing printing technologies is an important part of current biofabrication research, novel strategies have emerged, introducing alternative approaches to push the boundaries of the biofabrication window. This allows for the fabrication of larger, more sophisticated structures even when softer hydrogels are used and led to significant advances in the generation of 3D constructs displaying salient features of native tissues, such as those of bone,[24] skin,[25] cartilage,[26] cardiac muscle,[27] thyroid,[28] and liver tissue.[29] Despite these promising examples, studies that clearly demonstrate the advantage of bioprinting in achieving 3D cell–material constructs that exhibit, at least to a certain extent, functional characteristics of living tissues are not widespread.
In the present review, we summarize key strategies that have expanded the biofabrication window and that lead to improved control over shape. Building on such advances in material science, the main focus here is on the current and future steps toward mimicking salient functionalities of living tissues, through the creation of hierarchically structured constructs, in particular when using bioinks as building blocks for extrusionbased bioprinting. For this, the impact of bioprinted constructs with preformed spatial organization to facilitate tissue morphogenesis will be critically discussed. We highlight recent and upcoming developments in biofabrication that could influence the next generation of engineered tissues. Finally, we urge that future strategies embrace biological (developmental) processes and integrate them with bioprinting technologies to yield constructs with biological function toward the ambitious goal of printing functional tissues or even entire organs.
2. Recent Progress for Controlling Shape
Among the different subtypes of bioinks,[11] the most commonly used inks for extrusion bioprinting are based on hydrogels, either in the form of gel precursors or as preformed gels. These systems provide a highly hydrated environment that supports the encapsulated cells and generally offer shape retention to maintain the form of the as-printed constructs. During the last decade the biofabrication window has been extended by improving bioink performance and bioprinting techniques. Many excellent reviews discuss the current status of bioink development[16,30–34] and this is beyond the scope of the current review; however, several relevant components for bioinks and the respective crosslinking approaches are listed in Table 1.
Table 1.
Overview of popular natural and synthetic components used in bioinks for extrusion-based bioprinting sorted by the main component. The table summarizes strategies for enabling the printability of common biomaterials using various functionalities and crosslinking approaches.
| Natural | Characteristics/chemical structure | Functionalities/derivates | Crosslinking/fabrication approaches | Remarks/biological activity |
|---|---|---|---|---|
| Polypeptides | ||||
| Gelatin | Denaturated collagen Mainly collagen I Gly-Pro-X (most abundant) Type A (derived from acidic-treated gelatin) → positive-charge at neutral pH Type B (derived from alkali-treated gelatin) → negative-charge at neutral pH Thermoreversible gelation |
Without functionalization[35–37] | Blending with stabilizing component (e.g., alginate, fibrin[37]) Enzymatic crosslinking: transglutaminase[38] tyrosinase[39] Radical (photo-) polymerization |
Biodegradable Cell adhesion motifs are present Moderate biological activity if used alone Common sources: Bovine skin Porcine skin Fish skin |
| Methacrylated[23,40-44]
Allylated[45] Norbornenet[46] Thiolatedt[47,48] Tyraminet[49] Furfury[50] |
Radical (photo-) polymerization | |||
| Collagen type I | Most abundant collagen in human body Gly-Pro-X pH-dependent fibrillogenesis and gelation |
Without functionalization | pH-dependent crosslinking[51–53] photo-crosslinking (e.g., with ribofavin[54]) crosslinked with genipin[55] | Biodegradable Cell adhesion motifs are present Common sources: Rat tail Bovine skin Rabbit skin Calf skin |
| Fibrinogen/fibrin | Fibrous and nonglobular glycoprotein Thrombin (factor IIa) and factor XIIIa can be used to covalently crosslink fibrinogen |
Without functionalization[18,35–37,43,51] | Enzymatic crosslinking (factor IIa, XIIIa and IV) Additionally blended with stabilizing component (e.g., gelatin, hyaluronic acid, and Pluronic F127[37]) | Biodegradable Cell adhesion motifs are present Limited long-term stability (can be prolonged by addition of aprotinin to culture medium) Common sources: Human plasma Bovine plasma Rat plasma |
| Silk/fibroin | Silkworm cocoons Gly-Ser-Gly-Ala-Gly-Ala units[56] Recombinant spider protein GSSAAAAAAAASGPGGYGPE NQGPSGPGGYGPGGP Physical crosslinking by β-sheet crystal formation | Without functionalization | Physical crosslinking[57]
Blended with stabilizing component (e.g., gelatin[57]) Enzymatic crosslinking by tyrosinase[39] |
Biodegradable Poor cell adhesion due to hydrophobic character[58]
Common sources: B. mori silkworm Recombinant silk protein eADF4(C16) mimicking Araneus diadematus silk protein sequences[59] |
| Poly-saccharides | ||||
| Agarose | d-Galactose and 3,6-anhydro-l-galactopyranose Thermoreversible gelation | Without functionalization[60,61] | Physical crosslinking | Biologically inert Biodegradable Often used as sacrifcial material[12] Can be used to modulate the viscosity[60] Common sources: Red algae |
| Alginate | Varying sequences and blocks of β-d-mannuronate (M) and α-l-guluronate (G) Ionic gelation by divalent cations (e.g., Ca[2+]) |
Without functionalization | Physical crosslinking via divalent ions Often blended with, e.g., GelMA, nanocellulose or agarose[40,44,60,62] to improve properties |
Biologically inert Sulfate can bind growth factors, such as FGF, TGF, and HFG Common sources: Brown algae |
| Sulfated[63]
Methacrylated[64] |
Radical (photo-) polymerization | |||
| Characteristics/chemical structure | Functionalities/derivates | Crosslinking/fabrication approaches | Remarks/biological activity | |
| Hyaluronic acid | Glycosaminoglycan (GAG) Units of d-glucuronic acid and N-acetyl-d-glucosamine Forms weak entangled molecular network[65] | Without functionalization[37,66] | Needs modifcation or blending with stabilizing component | Biodegradable Cell adhesion motifs are present Can be used to modulate the viscosity Common sources: Bacteria Bovine vitreous humor Rooster comb |
| Methacrylated[41,67–69]
Phenolic hydroxyl[49] Thiolated[47,48] |
Radical (photo-) polymerization | |||
| Poly(N-isopropylacrylamide)[67]
Adamantane (guest)[68] β-Cyclodextrin (host) Cucurbituril (host)[70] 1,6-diaminohexane (guest) |
Lower critical solution temperature behavior (LCST) Guest–host supramolecular assembly |
|||
| Gellan gum | Tetrasaccharide of repeating units of β-d-glucose, one β-d-glucuronic acid and one α-l-rhamnose[71]
Ionic gelation by mainly divalent cations Thermoreversible gelation |
Without functionalization | Physically crosslinked by cations Blended with stabilizing component (e.g., GelMA[42,72]) | Biodegradable Can be used to modulate the viscosity Common sources: Bacteria |
| Nano-cellulose | Linear linked d-glucose units Cellulose nanocrystals (CNC) Cellulose nanofibers (CNF) Bacterial nanocellulose (BNC) | Without functionalization | Blended with stabilizing and/or bioactive component (e.g., algi-nate[63,73,74]
or hyaluronic acid[62,75]) |
Biodegradable Can be used to modulate the viscosity Common sources: Plants Bacteria |
| Dextran | Branched or linear poly-α-d-glucose | Methacrylatedt[66] | Radical (photo-) polymerization | Biologically inert |
| Chitosan | Progressively deacetylated chitins Linear and random dispersed β-(1-4)-linked d-glucosamine and N-acetyl-d-glucosamine Long gelation time and low mechanical properties[76] | Without functionalization | Blended with stabilizing components (e.g., agarose and alginate[60]) | Biodegradable[77]
improves cell survival[60] cell adhesion properties controlled by N-acetylation groups[78] antibacterial ability[79] Common sources: Chitin shells of seafood (e.g., crabs, shrimps, and prawns) |
| Carboxymethylated[60]
(water soluble) |
||||
| Chondroitin sulfate | Sulfated glycosaminoglycan (GAG) units of N-acetylgalactosamine and glucuronic acid | Methacrylatedt[67] | Radical (photo-) polymerization | Biodegradable Common sources: bovine trachea Shark cartilage |
| Carrageenan | Kappa-carrageenan Ionic gelation mainly by potassium ions iota-carrageenan ionic gelation mainly by calcium ions Thermoreversible gelation |
Without functionalization[80] | Physically crosslinked by cations Blended with a secondary component for covalent polymer network[80]
or nanosilicates[81] |
Kappa-carrageenan mostly used due to its resemblance to natural glycosaminoglycans (GAGs)[82]
Common sources: Red algae |
| Methacrylatedt[83] | Radical (photo-) polymerization | |||
| Characteristics/chemical structure | Functionalities/derivates | Crosslinking/fabrication approaches | Remarks/biological activity | |
| ECM mixtures | ||||
| Matrigel | Solubilized basement membrane preparation Mainly laminin (L-111) and collagen IV, but in total over 1851 unique proteins and over 14 060 unique peptides[84] Temperature-dependent gelation at around RT-37 °C |
Without functionalization | Temperature-triggered gelation | Biodegradable Cell adhesion motifs are present Highly bioactive due to growth factors and vast amount of proteins and peptides No defned composition Batch-to-batch variations Common sources: Engelbreth–Holm–Swarm mouse sarcoma |
| dECM | Decellularized extracellular matrix of a specifc tissue like heart,[85–87]
cartilage,[85] adipose,[85,88,89] aorta,[90] skeletal muscle,[91] and liver[92] Temperature-dependent gelation |
Without functionalization | Temperature-triggered gelation Blended with stabilizing component (e.g., thiolated gelatin, hyaluronic acid, and PEGDA[29]) Vitamin B2-induced UV-A crosslinking[86] | Biodegradable Cell adhesion motifs are present Biodegradable Highly bioactive No defned composition Batch-to-batch variations Common sources: mammalian tissue |
| Synthetic | ||||
| PEG | Poly(ethylene glycol) H(OCH2CH2)nOH Linear or branched | Diacrylatedt[18]
Tetra-acrylatedt[44,48] |
Radical (photo-) polymerization | Biologically inert |
| Methacrylated[61]
Fibrinogen and diacrylate functionalization[18] |
||||
| Succinimidyl valeratet[43] | Amin-carboxylic acid coupling (NHS ester reaction) with, e.g., proteins | |||
| Pluronic | Triblock copolymer of poly(ethylene oxide) Nonionic tenside |
Without functionalization[37] | Temperature-triggered gelation | Biologically inert Often used as sacrifcial material[37] Not suitable for long-term cell culture |
| Diacrylated[93] | Radical (photo-) polymerization | |||
| Additives | ||||
| Nanosilicates | Laponite Na[+] 07[(Mg5.5Li0.3)Si8O20(OH)4]-0.7) |
Without functionalization[83,94] | Physically crosslinked Blended with other bioink components (e.g., kappa-carrageenan and GelMA[95] | Used to modulate the viscosity of an ink to improve printability Can improve cell adhesion and response[96–98] |
| Glycerol | C3H8O3
Viscous and hygroscopic liquid Backbone of many lipids |
Without functionalization | Blended with other bioink components | Can help to induce crosslinking of silk[57]
Can prevent nozzle clogging and used as rheology modifer[37] |
The first strategies to design bioinks involved the chemical modification, or blending of different hydrogels, to control the rheological properties that dictate the printability and shape fidelity of the ink. These efforts resulted in a multitude of applicable systems, including the use of synthetic and natural polymers,[41,67,99] and lay the foundation for improved reproducibility and higher resolutions in bioprinting. As a straightforward, but versatile approach, blending allows to create multimaterial inks, based on mixtures of two or more different materials. In these systems, each component can take on a different function, such as providing suitable biological or chemicophysical properties for bioprinting, including the ability to support a specific cell type or ensuring adequate viscosity for high shape fidelity fabrication. This concept is well exemplified by the inks based on the combination of materials, such as gelatin, fibrinogen, gellan gum, hyaluronic acid (HA), and glycerol[37] (see also Table 1).
Alongside the advances in bioink development, the design of the bioprinting hardware also evolved, offering additional possibilities to enable high shape fidelity prints. Most commercial and custom made bioprinters integrate multiple printheads, enabling the generation of heterocellular and multimaterial 3D constructs. Additionally, many printers incorporate extrusionbased printheads for higher viscosity bioinks, inkjet printheads for low viscosity cell suspensions, heating cores for extruding thermoplastics,[100,101] and recently even melt electrowriting (MEW) functionality within the same platform.[102–104] As such, the potential to converge different printing technologies into a single, hybrid printing process and, therefore, introduce features having spatial resolution spanning a wide range from the nano- to the microscale is becoming more accessible. Coprinting of cell-laden hydrogels and thermoplastic materials via extrusion-based techniques only, or in combination with MEW,[37,105] has been used as an effective approach to adapt the mechanical properties of hybrid constructs to the tissue of interest.[106] As an alternative to thermoplasts to reinforce soft bioinks, stiffer hydrogels,[107] and printable ceramics,[108] have also been used. Apart from the reinforcement and modulation of the construct’s mechanical properties, multimaterial printing can also introduce channels within constructs that can ultimately be perfused and thus would allow for the engineering of larger living structures.[24] Typically, sacrificial materials are being used that can be dissolved after the fabrication process and subsequently be seeded with endothelial cells.
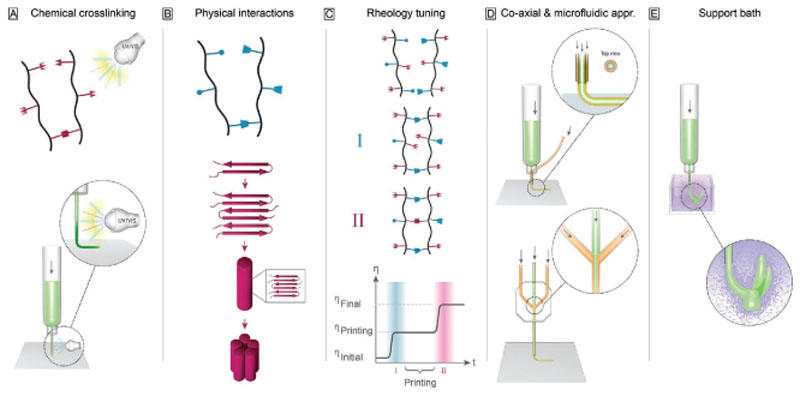
While these hybrid printing methods allow the introduction of structural elements, alternative strategies and further technological developments are necessary to improve shape fidelity of printed structures. In view of this, we here highlight a number of recent important developments that specifically can enhance our ability to create high shape fidelity constructs. These developments can be categorized as methods focusing on bioink design, mainly involving: i) new concepts for chemical crosslinking, ii) hydrogels based on physical interactions beyond ionotropic gelation, and iii) rheological tuning of the ink; as well as methods focusing on the redesign of the printing environment, via the introduction of iv) coaxial and microfluidic printheads, and v) printing into buoyant media as support baths for low-viscosity inks. An overview of the different strategies is depicted in Figure 2 and Table 2 summarizes the advantages and limitations of the fabrication strategies. The evolving strategies will be discussed in more detail in the following chapters. A focus of this chapter is to demonstrate how the recent advantages in the field have helped to extend the biofabrication window and have laid a foundation for tackling new challenges in the field.
Figure 2.
Overview—evolving strategies for controlling shape in bioprinting. A) To enhance chemical crosslinking strategies mainly step growth reactions and the transition from ultraviolet to visible light crosslinking have been applied more frequently. Also, technological advances like in situ photo-crosslinking can improve shape fidelity and broaden the spectrum of applicable materials based on chemical crosslinking strategies. B) In terms of physical interactions, weak bonds, like host–guest interactions and β-sheets, are applied to adjust the rheological properties of the materials. C) Rheological tuning is leading toward a two-step crosslinking utilizing a first step of crosslinking to enable printing with high shape fidelity and a second step to ensure long-term stability enabling to adjust the viscosity of the material to a level that is needed for the different fabrication steps. D) Further technological advances, like coaxial bioprinting and the application of microfluidic approaches in bioprinting, help broadening the spectrum of materials that can be applied for bioprinting. E) Also, printing into support baths is a promising strategy to enable fabrication of more sophisticated structures with less stringent demands on viscoelastic properties of bioinks.
Table 2.
Overview of advantages and limitations of the discussed technological enhancements for extrusion bioprinting.
| Technological advances | Advantages | Limitations |
|---|---|---|
| In situ photo-crosslinking |
|
|
| Coaxial printing |
|
|
| Microfuidic-enhanced printing |
|
|
| Support bath printing |
|
|
2.1. New Concepts for Chemical Crosslinking
2.1.1. Step Growth Reactions
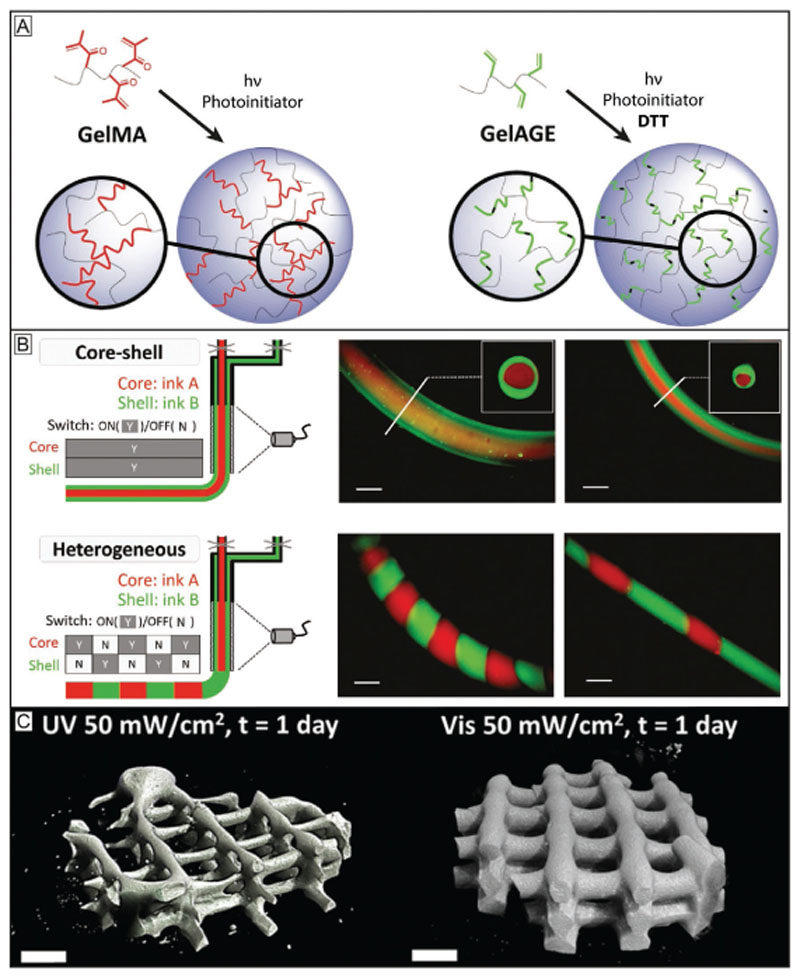
Accurate control over the crosslinking reaction of a bioink is essential to modulate its ultimate mechanical and physical properties, as well as to lead to well defined products that can be obtained under mild, cell-friendly crosslinking conditions. While within the field chain growth-based crosslinking, i.e., the formation of hydrogels by the rapid propagation of active centers through monomers containing multiple carbon–carbon double bonds, has widely been used, step growth systems offer some important advantages. Prominent examples of chain-growth-based system are inks based on acryloyl and vinyl chemistries, such as gelatin methacryloyl (GelMA). Benefits include its stability and straightforward use as no additional crosslinkers are required (Figure 3A). However, the chain growth reaction is prone to oxygen inhibition and leads to a limited control over the number of reacted functionalities resulting in oligo(methacrylamides) and thus also reduced control over mechanical properties and degradation products.[33,109,110] In contrast, thiol–ene step growth reactions, like those based on the modification of gelatin with applied, e.g., to other biopolymers like alginate,[111] or modified hyaluronic acid,[17,100] as well as to synthetic materials, like polyglycidols,[100] and poly(ethylene glycol).[112] This variability and the accompanied improved control over crosslinking render step-growth reactions a suitable tool to expand the biofabrication window toward higher shape fidelity as faster and more homogeneous network formation with more controlled spatiotemporal degradation properties. The difference between the reaction products in chain growth which are less controlled compared to step-growth reactions is schematically depicted in Figure 3A.
Figure 3.
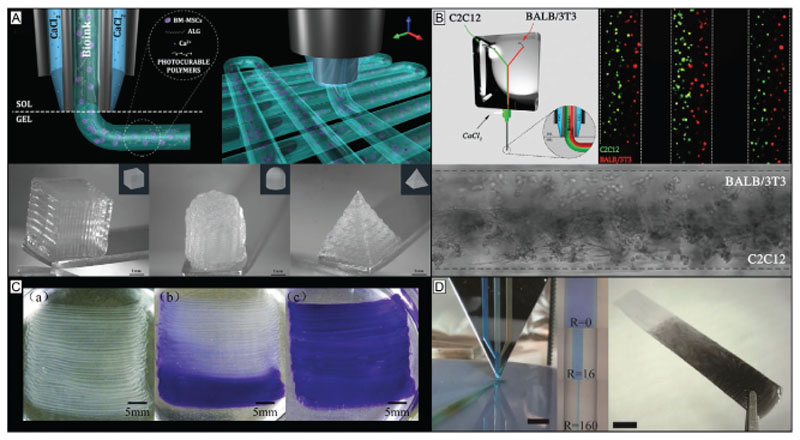
Chemical crosslinking strategies broaden the biofabrication window of inks and improve control over their resulting physicochemical properties. A) The chain growth-based crosslinking, represented here by gelatin methacryloyl (GelMA), is susceptible to oxygen inhibition and lacks precise control over the reaction products and results in undefined oligo(methacrylates) and thus uncontrolled degradation products. In contrast, the step growth reaction, represented here by the thiol–ene reaction between allyl functionalized gelatin (GelAGE) and the crosslinker dithiothreitol (DTT), provides faster reactions and conversation rates, forms more homogeneous networks and is not prone to oxygen inhibition. Reproduced with permission.[45] Copyright 2017, Wiley-VCH. B) In situ photo-crosslinking utilizes photo-crosslinking of materials through a light permeable printing nozzle. This enables printing of low viscosity precursors and the generation of core–shell as well as filaments from different materials. Reproduced with permission.[17] Copyright 2017, Wiley-VCH. C) Visible light crosslinking is a promising strategy to improve photoreactions used in bioprinting. Shown are °CT images of GelMA/collagen constructs crosslinked with UV and Irgacure 2959 (left) or visible light and Ru/SPS (right) at the same conditions. The visible light approach resulted in more homogeneous constructs, while the UV-crosslinked construct exhibited weakly defined lattice structures with gaps and varying filament diameters. Reproduced with permission.[116] Copyright 2016, American Chemical Society.
Rheological tuning is leading toward a two-step crosslinking utilizing a first step of crosslinking to enable printing with high shape fidelity and a second step to ensure long-term stability enabling to adjust the viscosity of the material to a level that is needed for the different fabrication steps.
Further technological advances, like coaxial bioprinting and the application of microfluidic approaches in bioprinting, help broadening the spectrum of materials that can be applied for bioprinting. E) Also, printing into support baths is a promising strategy to enable fabrication of more sophisticated structures with less stringent demands on viscoelastic properties of bioinks.
2.1.2. In Situ Photo-Crosslinking
Besides controlling the chemistry of the crosslinking reaction, shape fidelity can also be tuned through the timing of the crosslinking processes. Therefore, strategies termed in situ photocrosslinking have been introduced as promising approaches to broaden the biofabrication window. In extrusion-based bioprinting, bioinks that crosslink via photoinduced reactions are typically crosslinked after the deposition of the material.[114] This can be performed either by a single crosslinking after the completion of the print,[115] by continuous crosslinking following deposition,[113,114] or by a layer-by-layer crosslinking.[116] All these crosslinking approaches demand initial retention of shape after deposition of the material and thus still need to overcome the challenges described by the biofabrication window concept.
A solution to this is the photo-crosslinking immediately before the extrusion instead of afterward. A first step into this direction was based on aspirating a cell-laden hydrogel precursor (GelMA) into a light-permeable glass capillary, followed by a photocrosslinking. Afterward the strands of crosslinked hydrogel were deposited to produce lattices and stacked-fiber constructs.[117] This approach was refined and continuous printing of different cell-laden inks (GelMA, poly(ethylene glycol) (PEG) diacrylate (PEGDA), methacrylated hyaluronic acid, and norbornene-modified hyaluronic acid) through a photopermeable capillary, which was used as a nozzle, was demonstrated.[17] The length of the capillary that is exposed to the light source and the feed rate of the ink determine the crosslinking density of the ink. This approach permits to print the bioinks applying low extrusion forces and enabling higher cell viability compared to dispensing of pre-photocrosslinked bioinks (>90%).[17] Moreover, it enabled the fabrication of both heterogeneous and hollow filaments when core–shell capillary tubes and multiple inks were used (see Figure 3B). Importantly, it was demonstrated that this strategy can be used for bioinks with viscoelastic properties that cannot be printed with classical extrusion-based approaches.[17]
2.1.3. Visible-Light Photoinitiating Systems
Photo-crosslinking systems are widely used in biofabrication, as they offer rapid and on demand triggering of the polymerization reaction. Early biofabrication approaches have utilized photoinitiators, like Irgacure 2959, that rely on ultraviolet (UV) light to generate radicals, which induce the crosslinking reaction.[72,118,119] As UV light can potentially be harmful for cells,[116,120] the irradiation dose should be limited and several studies have indeed identified irradiation times that permit preservation of cell viability and functionality.[121,122] However, using low intensity UV energies is particularly challenging when using systems characterized by high oxygen inhibition. One possible solution to this is to use materials that rely on step growth reaction with higher conversion rates of functional groups, which generally results in limited oxygen inhibition,[33] leading to shorter exposure times and thus to lower irradiation doses. Another option to reduce the potential risks caused by UV exposure is to use lower energy (visible) light. The different wavelengths generally also require different photoinitiators. For biofabrication purposes a number of UV-A or visible light sensitive photoinitiators have successfully been evaluated, including lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP),[123] eosin Y,[124] and ruthenium (Ru)/ sodium persulfate (SPS) (Ru/SPS).[116] Interestingly, crosslinking of GelMA with visible light in the presence of Ru/SPS yielded higher viability of human articular chondrocytes (HACs) and bone marrow-derived mesenchymal stromal cells (BMSCs) than crosslinking with UV in the presence of Irgacure 2959 (see Figure 3C).[116] In a follow up study comparing GelMA hydrogels photo-crosslinkable at wavelengths between 400 and 450 nm, higher metabolic activity of HACs and higher penetration depths enabling crosslinking of thicker constructs were shown in comparison to initiator systems at 405 nm or in the near-UV range.[125] Due to these advantages, we expect visible-light crosslinking to become a more prominent strategy in the field of bioprinting and help expanding the biofabrication window by preventing the use of potentially harmful irradiation methods and thus making the bioprinting process more cytocompatible.[45] In addition, higher penetration depths can lead to more homogeneous crosslinking especially when bigger construct dimensions are of interest.
2.1.4. Dynamic Covalent Interactions
Given the nature of the extrusion bioprinting process, in which hydrogel-based inks transition from a flowing state in the nozzle to a nonflowing state postprinting, chemistries involving dynamic covalent bonds are offering a promising tool to improve shape fidelity. For example, combination of partially oxidized alginate and gelatin was used to establish a bioink system based on dynamic covalent bonds for extrusion-based bioprinting.[126] The aldehydes resulting from oxidation of the alginate saccharides subunits, in combination with the amino groups of hydroxylysine of gelatin, led to imine formation based on Schiff’s base reactions.[127] These dynamic covalent bonds enabled a cytocompatible gel formation and altered the viscoelastic properties of the material resulting in printable bioinks. Noncovalent crosslinking based on CaCl2 was used to stabilize the constructs after printing for cell culture.[126] Due to the dynamic nature of the bonds, the cell–material interactions can be designed in a way that enables controlling bioink properties like cell migration. This property can be used to design environments which enable cells to reorganize during the maturation process, a useful tool, for example, for researching developments of neuronal network formation. More recently, imine-type dynamic covalent chemistry was also applied for bioink development.[128] Partially oxidized alginate and linear, low molecular weight imine type crosslinkers (alkoxy, semicarbazide, and hydrazide) were used to adjust the viscoelastic properties of the bioinks through dynamic covalent crosslinks (oxime, semicarbazone, and hydrazone). Secondary crosslinking was not necessary to ensure stability over seven days in vitro.[128] Double network formation from dynamic covalent interactions and thiol–ene click photochemistry is another promising approach demonstrating the potential of dynamic covalent interactions for bioink development.[129]
2.2. Physical Interactions beyond Ionotropic gelation
Physical interactions have a dynamic and reversible nature, yielding comparable behavior to dynamic covalent bonds and can be tuned to exhibit shear thinning and recovery behavior. While classic examples among the first bioinks to be developed were based, for instance, on the ionotropic gelation of alginate, more refined and complex chemistries can be used to provide improved control over a wide range of mechanical properties and stability of the crosslinked network. Exploring these chemistries for bioink development resulted in the introduction of weak but highly directed and specific supramolecular interactions like hydrogen bonds or π–π interactions enabling the formation of polymer networks to bioprinting. Such networks can either be based on interactions between noncovalently associated monomers, like polypeptides or via supramolecular interactions, that link covalently bound polymer chains via noncovalent chain interconnections.[130]
2.2.1. Low Molecular Weight Gelators—Polypeptides and Proteins
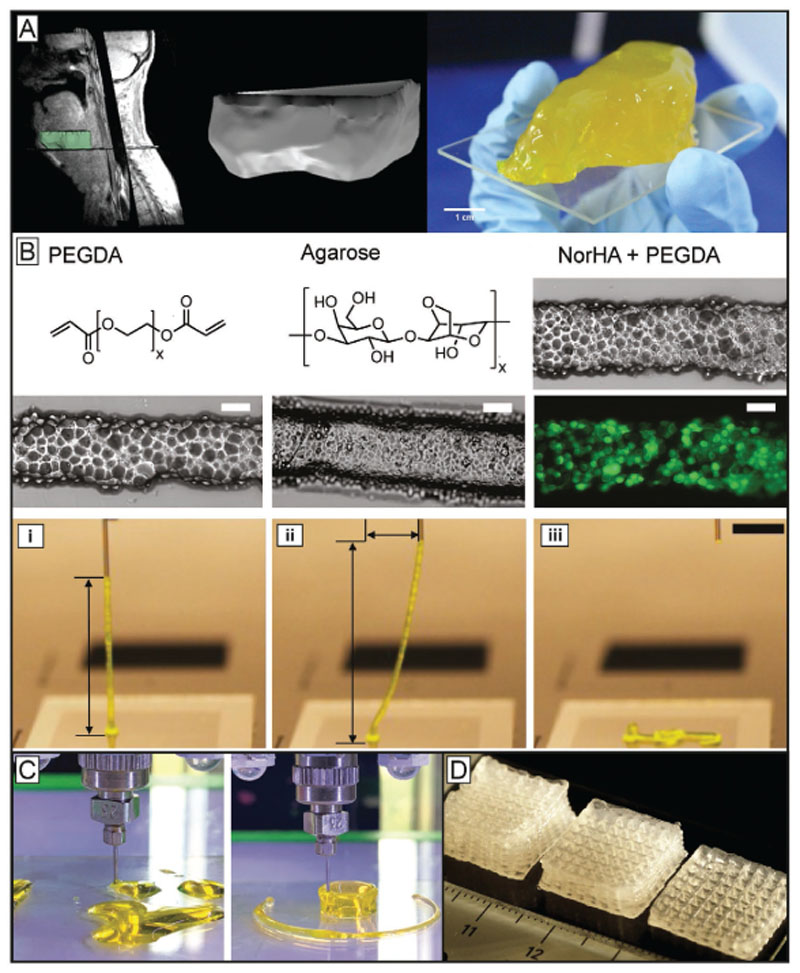
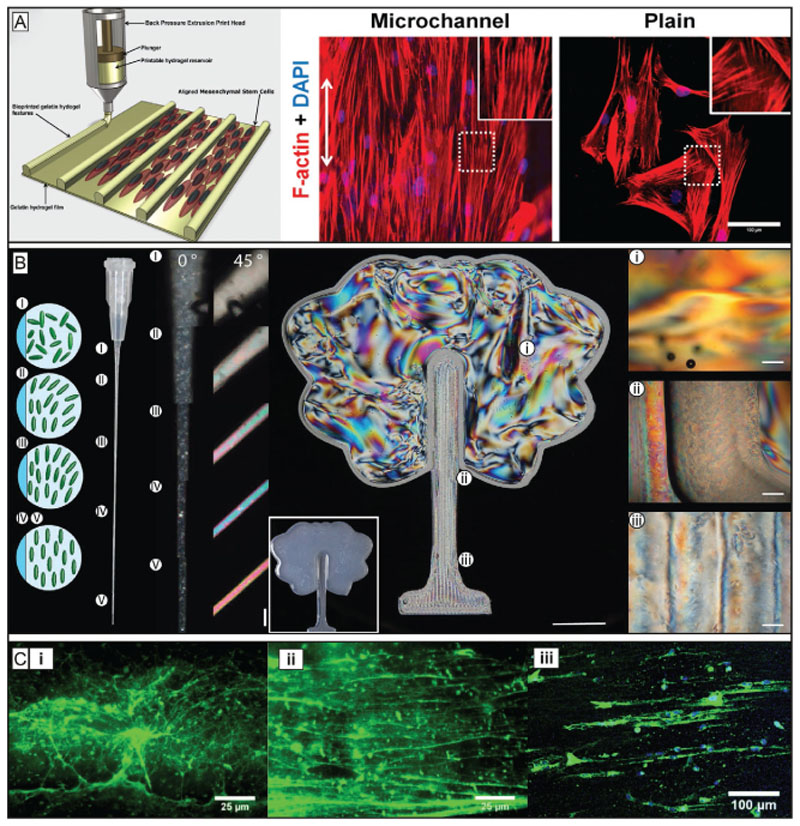
The concept of gels formed by noncovalently bound smaller subunits known from biopolymers like collagen,[131] or elastin,[132] is now also being transferred to synthetic bioinks via polypeptide chemistry.[133] Due to their structure, these materials are biocompatible and biodegradable. Hydrogels can be formed from polypeptide-based materials through different functionalization methods.[134] When stress is applied to these gels, the interaction between the subunits can be destroyed leading to a decrease of material viscosity. After deposition, the materials can recover and thus retain the shape after extrusion. Examples include polyisocyanate hydrogels used as biomaterial inks[135] or oligo-peptide bioinks.[136] Besides polypeptides also proteins are used as physical gels for bioink development. Many native proteins are also able to undergo structural transition that results in the formation of hydrogels. Harnessing the occurrence of such interactions can potentially permit to create shape stable architectures post printing. Silk and silk-like proteins exhibit promising properties as bioink components, since they are nontoxic, low immunogenic, have slow degradation and display shear thinning properties and recovery potential, mainly due to the formation of β-sheet structures during gelation.[137] Recombinant spider silk proteins, for example, can self-assemble into nanofibrillar networks and can, at the right concentration and conditions, form hydrogels based on β-sheet formation. These hydrogels proved to be printable and cytocompatible.[138] They even could be printed without the addition of thickeners and without postprocess crosslinking steps. As silk proteins lack cell binding domains, these needed to be modified with arginine– glycine–aspartic acid (RGD) sequences to improve cell–material interactions. Another option to improve the cell–bioink interaction and to prevent clotting of the nozzle was demonstrated by combining silk fibroin from silk cocoons (Bombyx mori) with gelatin.[39] In situ enzymatic crosslinking by addition of tyrosinase or physical gelation induced by sonication both proved to be suitable crosslinking mechanisms for bioink development. More recently another bioink based on silk fibroin (B. mori) and gelatin was developed (Figure 4A).[57] In contrast to the previous work, glycerol was used to induce physical crosslinking via β-sheet formation. To ensure stability under physiological conditions, the printed constructs were dried for a short period of time and dipped into a glycerol bath to enhance crosslinking of the silk gelatin ink. In general, the physically crosslinking polypeptides and proteins show potential to facilitate the development of new bioinks by already bringing many crucial biomaterial properties with them, such as biocompatibility, degradability and often also bioactivity. Furthermore, they potentially allow chemical or sequential modification of their amino acids to engineer specific mechanical and biochemical properties on demand. Taken together, the approach can help designing bioinks that can expand the biofabrication window toward higher shape fidelity at printing conditions that are more cytocompatible as they can help reducing the shear rates cells are exposed to during extrusion. In addition, properties that are important during maturation like biophysical and biochemical stimuli can be controlled more precisely and thus the materials can be more cell-supportive without sacrificing shape fidelity.
Figure 4.
Physical crosslinking strategies take advantage of reversible interactions to provide bioprinted filaments with structural stability, allowing to broaden the biofabrication window of low viscosity and low polymer density inks, as well as that of colloidal inks. A) From CT data to a printed cheek geometry of cm-scale using silk gelatin inks. To make the physically crosslinked, B. mori derived silk printable, it was mixed with gelatin and glycerol. The additives and the preparation enables the formation of β-sheets, increased the biological activity and helped preventing nozzle clogging. Reproduced with permission.[57] Copyright 2017, Elsevier. B) Colloidal inks from different precursors as deposited strands (scale bar 200 μm) demonstrating viscoelastic properties as proven by the intact filament spanning from the collector to the nozzle tip (i–iii). The approach shows potential to make a broader range of hydrogel systems printable due to its independency from the used crosslinking mechanism. The scale bar represents 5 mm. Reproduced under the terms of the Creative Commons CC-BY 4.0 License (https://creativecommons.org/licenses/by/4.0/).[139] Copyright 2019, The Authors, Published by Wiley-VCH. C) Extruded PEGDA without (left) and with Laponite (right). Reproduced with permission.[98b] Copyright 2019, Wiley-VCH. D) Alginate methylcellulose with Laponite as viscosity enhancer, pointing out the principle of viscosity modulators and their benefits for bioprinting by significantly improving shape fidelity of the constructs. Reproduced with permission.[98a] Copyright 2015, American Chemical Society.
2.2.2. Macromolecular Physical Gels
Noncovalent, supramolecular interactions between covalently bound polymer chains have also been used to develop bioinks based on physical gels. In particular, host–guest interactions have been applied to design bioinks with defined properties.[17,139,140] Blends of HA functionalized with either adamantane or β-cyclodextrin form shear thinning and selfhealing hydrogels based on reversible host–guest complexes between the two functional groups.[141] Given the reversible nature of this bond, irreversible stabilization has also been achieved introducing methacryloyl groups, to enable conventional free radical polymerization. This approach made printing of stable constructs with filament diameters between 100 and 500 μm possible.[17] Besides host–guest interactions, molecular recognition has also been used to improve the printability of bioinks. Alginate was, for example, modified with a peptide (P1; proline-rich peptide domain) and a recombinant, engineered protein (C7) containing cell adhesive domains (RGD).[142] These two components resulted in a weak, physically crosslinked and dispensable gel. It was shown that the damage during the printing process to the cell membrane of 3T3 and hASC encapsulated in the modified alginate compared to nonmodified alginate could be reduced significantly. After extrusion a second crosslinking step via calcium ions stabilized the constructs.
2.2.3. Colloidal Inks
Another approach that enables the decoupling of material printability from its rheological properties, relies on the use of densely packed microgels rather than homogenous hydrogel-forming solutions, also referred as jammed microgels (Figure 4B).[112,139] Here, hydrogel precursors are first processed into stable, crosslinked microgels. This step is followed by the removal of excess water until the increasing physical interactions between the gel particles cause a transition in overall rheological properties from viscosity-dominated to elasticity-dominated viscoelastic behavior. This particle interaction is similar to tightly packed colloidal suspensions known from ceramic processing. The dominance of the interparticle interactions generates so called rest structures that can be disintegrated by external shear stress during printing.[143,144] This is the basis for shear thinning and recovery properties accompanied with a yield stress seen in the jammed microgels. When printed under optimal conditions, it results in intact long filaments (Figure 4Bi–iii) making this approach a promising strategy to generate biomaterial inks and bioinks.
Although only a small number of studies was published on that topic to date, it was shown that this approach is independent of the used hydrogel material, as long as it can be processed into microgels. So far, for the production of microgels thiol–ene crosslinked norbornene-modified hyaluronic acid, photo-crosslinked poly(ethylene glycol) diacrylate, thermosensitive agarose,[139] and photo-crosslinked norbornenemodified poly(ethylene glycol)[112] have been used. However, due to the weak physical interactions between the particles, the printed construct needed to be stabilized in a secondary crosslinking step for long term stability in aqueous solutions. Here, photoinitiated polymerization has been successfully used to crosslink remaining unreacted photoactive groups.[112,139] To prepare bioinks and enable cell-loaded printing, two options to incorporate cells into the biomaterials exist. These are direct encapsulation into the particles or mixing the cells with the microgels. Encapsulation has the advantage of shielding the cells from the shear stress usually experienced during the extrusion. However, to not impede cellular migration and proliferation, the hydrogel needs to be biodegradable,[139] except a spatial cellular confinement is desired, for instance, in drug delivery applications. In contrast, mixing the cells with microgels does not require a biodegradable material to allow spreading and proliferation, since the microporosity of the platform provides sufficient space for the cells.[112] Given the common basis of particle interaction, this approach should be potentially compatible with mixing or separately printing of microgels of different nature or cargo to create multimaterial and hetero-cellular[139] constructs. Taken together, jammed microgels are a promising method to broaden the window of bioprinting, the generalized character still needs to be proved and especially the second crosslinking to stabilize the inks within physiological conditions and reproducible jamming methods still need to be further developed.
2.3. Rheology Tuning
Since the early stage of bioink development, tuning the ink’s rheological properties was and still is a key challenge. It is a highly relevant tool for designing materials that enable high shape fidelity in combination with improved process cytocompatibility and thus help expanding the window of biofabrication. Early approaches were based on increasing polymer concentration or the addition of rheological modifiers like high molecular weight hyaluronic acid or gellan gum to the modified gelatin compromising cytocompatibility.[63,72,145,146] Alternative approaches, like methods to control the crosslinking degree or through addition of nanoscale elements, have been developed to improve shape fidelity while maintaining high cytocompatibility of the bioink and the printing process.
2.3.1. Nanocomposites
Nanosilicates are very prominent additives that, even at very low concentrations, increase shear thinning properties of the precursors they are added to. They can induce self-healing properties and improve shape fidelity. Numerous nanocomposites are used to tune rheological properties of inks and a conclusive discussion is outside the scope of this paper. The interested reader is referred to a list of excellent papers and reviews covering this topic.[16,31,81,147] However, in view of the recent developments for bioprinting a small number of examples is highlighted. The addition of a synthetic nanosilicate clay, called Laponite (Laponite is a trademark of the company BYK Additives Ltd.), to GelMA resulted in improved printability.[94] In addition, the modifications impacted on stiffness and degradation properties of the materials. Laponite was recently also used to adjust the rheological properties of bioinks based on PEGDA, as well as alginate methylcellulose. Besides the exceptional printability of the materials (Figure 4C,D), the alginate methylcellulose-based ink also enabled printing viable human mesenchymal stem cells (hMSC), while the additives showed positive effects on the biological function.[98a,b] Another nanostructure that can be used to tailor printability of inks is nanocellulose. It was, for example, shown that the addition of nanocellulose improved the shape fidelity of alginate-based inks and enabled printing of human nasoseptal chondrocytes although it did impact on their viability.[73]
2.3.2. Pre-Crosslinking Strategies
Pre-crosslinking typically encompasses a stepwise crosslinking procedure that includes two or more different crosslinking methods. For example, inks have been developed that contain linear and branched amine-presenting polyethylene glycol (PEGX; X: succinimidyl valerate),[43] which can act as a crosslinker for macromolecules, like gelatin and fibrinogen via amine-carboxylic acid coupling. A first crosslinking step is performed to adjust the rheological properties of the ink and a second crosslinking step is then introduced to ensure construct stability over extended periods of time and during exposure to physiological conditions. This second step was either chain growth polymerization of GelMA with UV-based initiators, thrombin-based crosslinking of the fibrinogen or EDC (N-(3-dimethylaminopropyl)-N-ethylcarbodiimide)/ NHS (N-hydroxysuccinimide) in terms of gelatin.[43] An additional example of a pre-crosslinking strategy was based on a tyramine-modified hyaluronic acid (HA-Tyr) bioink. Enzymatic crosslinking, mediated by horseradish peroxidase (HRP) and hydrogen peroxide (H2O2), was performed as a first precrosslinking step to adjust the viscoelastic properties of the bioink. This was followed by a second step based on visible light crosslinking in the presence of Eosin Y to crosslink the remaining HA-Tyr components. Furthermore, other approaches involving physical and chemical crosslinking were also demonstrated to be suitable pre-crosslinking strategies,[17,29] emphasizing the potential as a general strategy for rheological tuning of bioinks.
2.4. Coaxial Printing and Microfluidics
2.4.1. Coaxial Printing
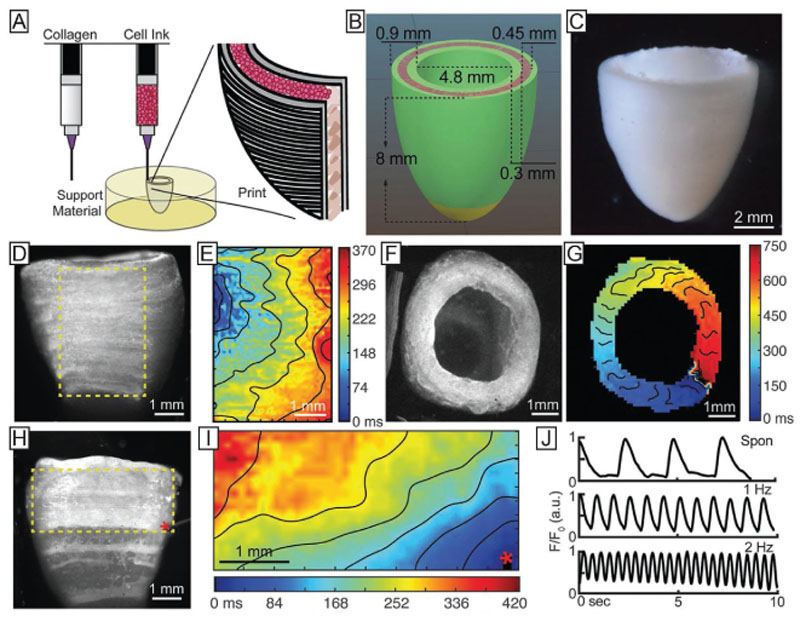
The coaxial extrusion allows the concurrent and concentric deposition of different materials through the use of distinct nozzles assembled in a coaxial fashion resulting in double- or multilayered filaments (Figure 5A,B). It enables printing various materials in a core–shell arrangement to fabricate homogenous or heterogeneous solid or tubular filaments.[17,44,148] In addition, the spatial separation of crosslinker and ink solutions until the very end of the nozzle tip allows the use of lower viscous hydrogel precursors compared to conventional extrusion-based approaches while still maintaining shape fidelity crucial for expanding the biofabrication window. The crosslinker can either be incorporated in the outer compartment and the cell-laden material in the inner one to form solid filaments or vice versa to fabricate hollow filaments. To process medium- or low-viscous inks through a coaxial nozzle, a fast crosslinking mechanism is mandatory to provide the required shape fidelity. The coaxial system has proven to be very suitable for the processing of alginate and its blends, where calcium chloride is used as a crosslinking agent.[18,27,40,149] To enhance its biological activity, alginate is typically blended with more active components. In terms of coaxial printing it can also be coprinted with such components (e.g., collagen, GelMA, and bioactive glass).[27,40,150,151] In addition, coaxially printed, noncrosslinked alginate can also be used as a cytocompatible temporary mechanical support, for example when combined with photocurable bioactive polymers, including GelMA, PEGDAfibrinogen, methacrylated hyaluronic acid and chondroitin sulfate amino ethyl methacrylate.[18,27,40,149,152] Coaxial extrusion can also be used to process materials that exhibit low mechanical stability or long gelation times, such as collagen, Matrigel, fibrin or laminin.[150,153,154] In particular, the coextrusion with mechanically stronger materials can improve their processability while simultaneously increasing the functionality of the resulting constructs.[155,156]
Figure 5.
Coaxial and microfluidic methods are versatile tools to induce rapid crosslinking of extruded filaments. Additionally, using such extrusion devices permit to print multiple materials in a single process, as well as to control the composition and physicochemical properties of the forming hydrogel strands. A) Coaxial bioprinting of an alginate-based bioink (inner compartment) and its crosslinking agent CaCl2 (outer compartment). The rapid crosslinking via calcium ions enabled in situ crosslinking ensuring high shape fidelity printing. Reproduced with permission.[149] Copyright 2016, IOPScience. B) Microfluidic approaches like the utilization of y-junctions in combination with a coaxial nozzle allowed the deposition of strands with parallel aligned multiple cell types, but has also been used to extrude different types of materials. Reproduced with permission.[18] Copyright 2017, Elsevier. C) By coaxially extruding an alginate-based ink through the outer and the crosslinking agent through the inner nozzle, perfusable inks could be produced. Reproduced with permission.[148] Copyright 2015, Elsevier. D) Microfluidic approaches can be used to generate gradients which play crucial roles in biology. Methods like flow focusing (images in the middle) can be also applied to 3D printing and help reducing filament diameters. Reproduced under the terms of the Creative Commons CC-BY 4.0 License (https://creativecommons.org/licenses/by/4.0/).[163] Copyright 2018, The Authors, Published by MDPI.
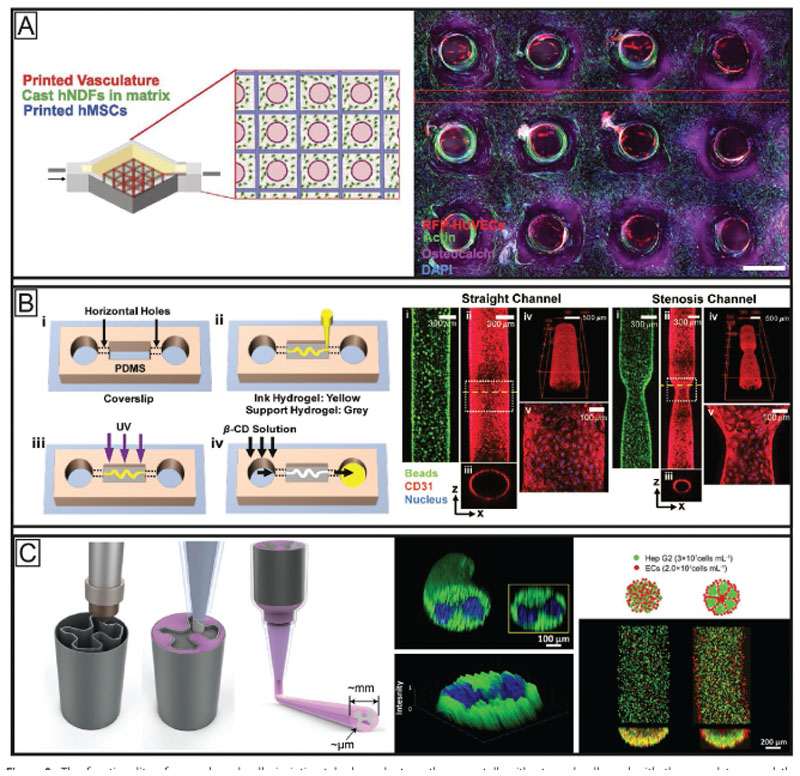
As the supply of sufficient nutrients in larger bioprinted tissue constructs is still a significant challenge,[157–160] coaxial extrusion offers a promising solution as perfusable vascularlike structures can be generated by, for instance, coprinting alginate with a calcium solution for crosslinking (Figure 5C). Here, in one of the first attempts a concentrated paste of alginate (16.7% (w/w)) and aqueous poly(vinyl alcohol) (6% (w/v)) was used to fabricate meter-long hollow fibers.[161] Further, coaxial printing enabled the fabrication of fully perfusable scaffolds with single- or double-walled hollow filaments using other material combinations.[17,44,148] To improve the quality of the extruded tubes, active perfusion of the interior channel with a temporary and mechanically supporting ink, such as Pluronic F127 or gelatin,[44,90,148,162] can prevent the hollow filament from collapsing.
2.4.2. Microfluidic-Enhanced Bioprinting
The microfluidic bioprinting systems can precisely manipulate the behavior of liquids at microscale level and offer a promising solution to accurately control the deposition of, so far, up to seven materials.[18,40,163,164] Each material has its own inlet to a microchannel in a microfluidic chip or nozzle. By merging the different channels, different materials can be mixed together or separately extruded by optimizing the actuation of the syringe pumps. Rapid switching (500 ms) between different inks is possible and the technique also allows for a seamless transition between multiple materials. This renders the approach an interesting tool for creating gradients by varying the spatiotemporal deposition of different materials (Figure 5D). In addition, hydrodynamic focusing of the center flow stream, containing the bioink, can be realized by microfluidic approaches (Figure 5D). By pinching the core bioink flow with at least two side flows, the width of the filament can be reduced.[165] Further, microfluidic-based printheads bear the potential to expand the range of printable materials toward low viscosity bioink by allowing the deposition of multiple materials through a single extruder. A good example are approaches combining coaxial nozzles with microfluidic chips that enable switching between different inks or combining them during printing. Utilizing this approach heterogeneous filaments could be fabricated with a distinct longitudinal interface by using a Y-junction setup.[18,40] Here, the coaxial nozzle concept was used to provide CaCl2 solution for in situ crosslinking through the exterior nozzle and the microfluidic concept to provide up to two different alginate-based bioinks through the interior nozzle (Figure 5B). Furthermore, concentrically layered filaments have been also generated using a microfluidic approach by inserting three needles into a wavy microfluidic channel at different locations. For each needle used, the horizontal microfluidic channel made a vertical turn to allow a vertical and concentric penetration of the respective needle. This setup enabled multiaxial extrusion of up to four different solutions and the fabrication of bi-and tri-layered hollow channels.[166] The cytocompatibility of this approach has been demonstrated by bioprinting alginate-based bioinks containing either fibroblasts or endothelial cells.
2.5. Support Bath and in Gel Printing
The support bath approach (Figure 6) relies on support materials that are placed into a container. Extrusion-based bioprinting is then applied to deposit the desired bioink into this supporting material. The benefit of this key strategy is that the bath provides neutral buoyancy and spatial confinement of the extruded material and allows to print materials that would otherwise not keep their shape. One of the first attempts used a hydrophobic high-density fluid called perfluorotributylamine to support printing a stem cell-laden agarose construct.[167] This first development was then followed by the introduction of hydrogel-based support baths.[51,168,169] Printing into support baths significantly expands the biofabrication window by negating gravity and surface tension,[168] enabling to even process low viscosity inks with a loss modulus (G″) below < 1 Pa (G″ > G′ at a frequency of 1 Hz).[51] The spatial support and confinement of the deposited ink facilitates the fabrication of more delicate and sophisticated structures, which would be difficult or even impossible to fabricate using other extrusionbased bioprinting strategies.[168,169] Besides enhancing the resolution, a support bath can also provide long-term stability in a humid environment, limiting the risk of cell damage during the manufacturing process. This paves the way for long or slow prints and the generation of larger, more clinically relevant constructs and helps expanding the window of biofabrication as it enables more cytocompatible processes without sacrificing shape fidelity (Figure 6A,B).[168,170] But the use of support bath also comes with new challenges. Although the ink does not need to meet the typical requirements for printability, a potential support bath system needs to behave like a non-Newtonian liquid and obey rheological confinements. When shear stress is exerted on the support bath by the movement of a printer nozzle, it has to liquefy locally. After the shear stress caused by the needle movement vanishes, its viscosity needs to increase to trap the deposited ink and support the shape of the printed constructs. Taken together, the benefits of less stringent rheological properties for the dispensed material are associated with the need to accurately tune the properties of the support bath material. In addition to the rheological requirements, a support bath material should also be cytocompatible, easy to remove without leaving significant amounts of residues and not interfere with the crosslinking of the printed materials. To be able to remove the construct safely from the support bath, the ink must undergo a crosslinking step in situ after its deposition and the bath material must allow easy removal of the printed construct without affecting it negatively. Hereby, the crosslinking mechanism of the ink should not interfere with the support bath material itself (or vice versa), to avoid a change in viscoelastic behavior of the bath during printing and the formation of crosslinking artifacts. Furthermore, it is beneficial to have a bath material that allows a controlled transition from solid to liquid state, as offered, for example, by thermoreversible systems, to facilitate the extraction of printed constructs with filigree structures. Collagen I, for instance, can be used as an ink, as it is soluble under acidic conditions and by printing it into a buffered bath the pH-dependent fibrillogenesis and gelation is triggered, resulting in a stable construct that can be harvested as shown in Figure 6A.[53] Another example of a biomaterial class are materials that crosslink with the help of a secondary component, such as alginate with calcium ions or fibrinogen with thrombin. By the addition of the respective crosslinking component into the support bath, the corresponding materials can be printed in liquid-state followed by subsequent in situ crosslinking.[51] Although the development of appropriate support baths is challenging, promising results have been demonstrated. Based on their nature and crosslinking mechanisms, these bath materials can be classified as: i) microparticle-based systems and ii) macromolecule- and nanosilicate-based systems.
Figure 6.
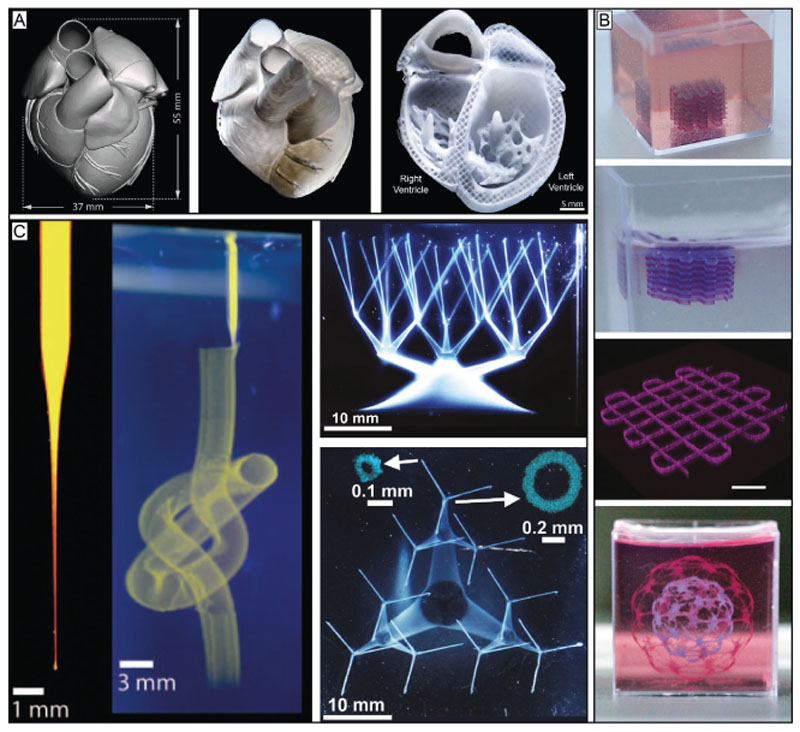
Support bath printing allows to print suspended structures and overhangs, and provides structural stability when using low viscosity bioinks, enhancing control over shape and resolution. A) Based on a CAD-model a construct from collagen I, a material that is usually not printable by itself, was printed using the support bath approach. The support bath, based on gelatin, allowed the deposition of a low viscous collagen ink, utilizing an in situ crosslinking approach, and fabrication of complex structures with high shape fidelity that closely resemble the CAD file templates. Reproduced with permission.[53] Copyright 2019, AAAS. B) The method can also be used to generate filigree multimaterial constructs which could not be printed with classical extrusion-based bioprinting. The alginate-based support bath was enzymatically degraded to gently harvest the fragile objects. Reproduced under the terms of the Creative Commons CC-BY 4.0 License (https://creativecommons.org/licenses/by/4.0/).[88] Copyright 2019, The Authors, Published by Wiley-VCH. C) Carbopol as bath material in combination with a very thin nozzle (50 μm tip) enabled printing of structures with high resolution and challenging shapes. Reproduced with permission.[168] Copyright 2015, The Authors, published by AAAS. Reproduced/modified from ref. [16]. © 2016, The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
2.5.1. Microparticle-Based Systems
Carbopol, a synthetic poly(acrylic acid) with high molecular weight that can be used to make gels consisting of swollen, hydrophilic and spherical-shaped elastic domains with a diameter of ≈7 μm, is one of the first materials that have been successfully utilized as support baths.[171–173] At the right concentration, Carbopol exhibits a yield point and shows excellent self-healing properties. This enables a rapid stabilization of the microparticles after a sudden change in applied shear stress exceeding or undershooting the yield point.[174] Carbopol concentrations between 0.2 and 0.9 wt% did not impede cell migration and proliferation.[175] Although demonstrated in a cell-free approach, Carbopol-based support bath printing (Figure 6C) showed the potential to generate constructs composed of strands with diameters as small as 35–50 μm. This revealed the possibilities offered by the support bath approach and enabled printing of highly complicated structures (Figure 6C).[168,169] Printing cell containing materials into Carbopol allowed the fabrication of objects with a resolution between 100 and 200 μm.[175] A layer thickness of 100 μm could be achieved and even a single-cell ejection along the printing direction by increasing translation speed was possible. A broad range of cells has been printed into Carbopol, including endothelial, epithelial and mesenchymal stem cells with a viability of in average about 94% after 24 h in the gel.[175]
Another common example of granular support baths are those based on so-called fluid or sheared gels,[176] such as agarose or gelatin.[51,177] These microgel-based materials also exhibit rapid self-healing properties postshearing, allowing the quick trapping of extruded inks. Sheared gels from 0.5 wt% agarose particles have been successfully used as support.[170,177] The particle size ranged from 2 to 11 μm and formed a weak, reversible network. It has been demonstrated that an increase of viscosity of the sheared gel correlates with an improved resolution in XY dimensions, but with a decrease in Z dimension. Support baths based on gelatin, which have also been termed freeform reversible embedding of suspended hydrogels (FRESH), included a particle size of ≈25 μm.[53] This enables cell-free print resolution of filaments with diameters of 25 μm. The thermoreversibility of gelatin-based support baths at about 30–40 °C has the advantage of easy removal of the support from the printed construct. However, in the case of using a bioink based on extracted ECM such as Matrigel, a temperature-dependent curing process at 37 °C is required for long-term stabilization of the print. Here, a gelatin-based system would lead to the destruction of delicate structures before they can stabilize. One solution to this issue was demonstrated by supplementing the ink with temperature-independent crosslinking materials, such as fibrinogen or collagen type I.[51] Another option would be to address the problem from the support bath side. Recently another suitable system has been developed, consisting of alginate microparticles and xanthan gum, to support the print and curing of an omental ECM-based bioink.[88] This support setup tolerated a broader range of temperatures and can be enzymatically or chemically degraded to gently harvest the printed object. The used alginate microparticles had a size below 25 μm and enabled printing cellloaded single strands with 100 μm of diameter. Filigree structures, such as a miniaturized hand, heart and spherical network with up to two different materials have been printed (Figure 6B). This setup supported the print of induced pluripotent stem cell (iPSC)-derived cardiac and endothelial cells, as well as human umbilical vein endothelial cells (HUVECs), neonatal cardiomyocytes and fibroblasts with an overall cellular viability of >90%.
2.5.2. Macromolecule- and Nanosilicate-Based Systems
Besides the use of granular materials, there are also support bath concepts that are based on intermolecular guest–host interaction,[169] or Laponite nanoclay.[178–180] In this case the subunits of the support bath are in the molecular scale. For support baths based on guest–host interactions, HA was modified with either adamantane or β-cyclodextrin and then mixed. The reversible supramolecular assembly of the two functional groups through guest–host complexes provided a yield point and thus self-healing properties. This enabled the fabrication of up to 35 μm thick filaments with cell-free printing and structures with ≈600 μm of width with cell-loaded printing.[174] Here, mesenchymal stem cells were printed into support gels containing 3T3 fibroblasts, exhibiting a viability of <90%. Furthermore, the host–guest-modified HA can also include methacrylate groups resulting in a support bath that can be stabilized by a secondary crosslinking step.[174] This allowed the fabrication of perfusable channels, by first extruding an ink as placeholder, followed by a secondary crosslinking step of the support bath and subsequent removal of the ink.[169] In the case of Laponite, the structural components are disk-shaped particles, which release sodium and hydroxide ions in aqueous solutions. This results in areas of positive and negative charge, enabling electrostatic interactions and their arrangement into a colloidal suspension with a yield stress and rapid recovery after exposure to shear stress.[178,181] Depending on the travel speed of the nozzle and Laponite concentration, filaments from gelatin-alginate with a diameter of ≈600 μm could be produced. Taken together, the support bath approach expands the biofabrication window by allowing for the deposition of bioinks in the liquid-phase and thereby the use of low-pressure extrusion-based bioprinting. The humid environment of the bath facilitates long prints by protecting the printed construct from drying out. The buoyancy effect of the support material makes it possible to print delicate and usually mechanically fragile constructs. Moreover, the low initial viscosity of the deposited inks provides more freedom regarding tailoring of local mechanical properties of the biomaterial postprinting. This may be an important step toward improving the imitation of the natural ECM environment.
3. Strategies to Evolve from Shape to Function
3.1. Biological Function in the Context of Biofabrication
A growing number of bioprinting strategies allows for the creation of shapes with intricate architectures and continuously improving resolutions. However, faithfully reproducing the anatomy of a tissue, as permitted by high-shape fidelity bioprinting approaches, does not ensure per se the acquisition of the functionality of the native tissue. Once good control over the spatial deposition of cells and materials is achieved, biological constructs will need to undergo maturation and morphogenesis, which has been defined as an integral part of the bioprinting process.[4] Nonetheless, the achieved widening of the biofabrication window now enables to challenge the core hypothesis of bioprinting that a hierarchical 3D arrangement of cells and (bio-)material/matrix facilitates, accelerates or improves the formation of functional tissue analogues.
However, until now, examples of studies are still limited in which such direct advantage of bioprinting for the evolution of biological function on a tissue level has been demonstrated. One reason for this is that studies focusing on (bio)material development frequently have overlooked this aspect and printing is often shown in proof-of-concept tests, even in cases when conventional fabrication technologies may be sufficient. In other words, an exponential number of contributions to scientific literature and international meetings have included the printing of the materials (with or without cells) without a clear demonstration of an advantage of printing in terms of improvement of the functionality for the ultimate application. This observation is underscored by the fact that there is only a handful of reports that successfully showed the replication or restoration of key functions of specific tissues or organs. One successful example relates to biofabricated ovaries, obtained by including ovary follicle cells in printed gelatin-based scaffolds with cell-instructive architectures that were shown to rescue reproductive capacity, enabling sterilized mice to obtain healthy offspring from natural mating.[182] An additional example of a functional bioprinted structure are the printed spheroids of vascular and thyroid gland cells that were capable of replacing the bioactivity of native thyroid, including its thermoregulatory ability and thyroxine hormone secretion into the systemic circulation, when implanted in mice with a surgically induced hypothyroidism.[28] Also, bioprinted muscle cell-laden hydrogel fibers were demonstrated to mature into homogenously shaped bundles of myofibers, which exhibited contractile function, and could be matured upon ectopic implantation in vivo, showing a more pronounced organization and alignment compared to nonprinted controls.[18]
These few examples show that bioprinting can yield biologically functional constructs. It is however still unclear what the required resolution of the architecture—dictated by the printing process—needs to be and to what extent the fabricated construct can rely on self-organization during the maturation phase. Importantly, based on its ultimate application, the required functionality of the bioprinted structure can be significantly different. Bioprinted tissues for transplantation should ideally recapitulate all functions of the native tissue, either at the time of implantation (i.e., after in vitro culture), or after a maturation period in vivo. The same accounts for in vitro tissue models for diseases, or that are intended to replace animal experiments.
On the other hand, 3D in vitro models that are intended to be used for drug screening, may only need to display a certain salient feature of the replicated biological structure. This concept is well exemplified by the current state-of-the-art of organ-and body-on-a-chip technology, involving soft lithography and replica molding as production processes.[183–185] Such models are often composed by horizontally or vertically aligned microfluidic channels, each representing a tissue compartment, which are interfaced to allow intercompartment cell communication. This geometrical simplification, often displaying planar symmetry, is an elegant and effective way to replicate specific tissue functions, i.e., the barrier function of lung and intestine[183,186] or the invasiveness of tumor cells,[187] and has recently been successfully used to show metabolic interaction between microtissues that are connected via microfluidic channels for drug testing.[188] Nevertheless, they cannot fully recapitulate the hierarchical organization of native tissues. While the exact degree of similarity required for a fully functional engineered tissue is still unclear, there is increasing evidence that controlling shape and architecture of printed constructs in terms of cell and material composition and interaction between the multiple components is essential in order to bridge the gap between engineered and native tissues. The following sections will highlight how cell and tissue functionality can be guided through the biofabrication process, specifically via stimuli derived from: i) the architecture and geometry of the printed structures, ii) the building blocks (bioink) used for printing, and iii) the coordination and cross-talk of multiple cell populations patterned in 3D constructs.
3.2. Design Specifications in Bioprinting to Drive Functional Tissue Maturation
Many studies at the crossroads of regenerative medicine, biomaterials and cell biology have highlighted how different cell types drive tissue morphogenesis and maturation through a combination of cues presented by their extracellular environment.[189] In fact, cells sense and respond to mechanical (e.g., stiffness and viscoelasticity), biochemical (e.g., adhesion molecules and peptides, soluble factors, metabolites) and geometrical cues, including porosity and surface topography. The presentation of such signals to bioprinted cells can be tuned, either during the printing process or through the modification of the hydrogel–bioink composition, effectively enabling a wide array of opportunities to guide cell response and promote the acquisition of the desired tissue functions.
3.2.1. Geometrical Considerations
Porosity
The macroscale architecture of a construct can influence its biological performance and guide functionality of biofabricated tissues. Geometrical cues from scaffolding materials, including surface and 3D topography, shape, size and tortuosity of porous elements, are known to provoke a wide array of responses in cells both in vitro and in vivo, and such reactions greatly differ based on the dimensional scale of these cues. For example, cells can preferentially home or align along topographical features at the micrometer and submicrometer scales, such as nanofibrous elements[190] or periodic grooves and channels,[191] a phenomenon described as contact guidance.[192] At a larger scale, micrometer to submillimeter features play a pivotal role in cell survival and tissue regeneration. For instance, pore sizes in the range of 200–400 μm are fundamental for nutrient diffusion throughout large constructs and for bone infiltration in vivo.[193] Vigorous blood vessel ingrowth and little fibrosis have been observed accompanied with polarization of macrophages toward an M2 phenotype (generally regarded as stimulatory for tissue regeneration) in structures with pores in the 30–40 μm range. In contrast, smaller and larger pores resulted in the formation of fibrotic scar tissue.[194]
Using biofabrication strategies that ensure high shape fidelity and resolution, virtually any convoluted 3D patterns of materials and cells could be created, while providing control over the presentation of topographical and geometrical cues.
In 3D extrusion-based biofabrication processes, strut sizes are typically in the order of 100 μm, and the associated porosity can be accurately tuned from the millimeter scale down to 10 μm, as it can be accurately tuned via the design of the strut-to-strut distance and the laydown angles between each printed layers. Laydown angles and pore geometries have, e.g., shown to greatly influence hepatic cell maturation,[195] stem cell differentiation,[196] macrophage polarization and inflammatory response,[197] and muscle tissue regeneration.[198] Moreover, porosity profiles can be varied across each printed layer. This can, for example, lead to controlled oxygen gradients, mimicking the metabolite concentration across an osteochondral plug.[199]
The phenomena underlying the observed responses are diverse. In some cases cells are directly affected by the physical cues, i.e., via contact guidance, cytoskeletal reorganization or mechanotransduction.[200] This is the case in the responses to strut size and roughness or to attachment to fibers forming different angles at their intersections. However, in many cases the characteristic dimensions of the features of extrusion printed hydrogel structures are too large for cells to sense preferential orientations directly. The selection of a specific geometry only indirectly influences cell behavior, i.e., through favoring cell–cell clustering and via proximity and confinement effects. For example, enhanced maturation of a hepatocytic cell line and improved expression of cytochromes was observed when the cells were seeded onto 0°–60° laydown gelatin constructs, instead of the more common 0–90°. This effect was due to the fact that the narrower angles facilitated cell aggregation at the corners of the pores improving cell aggregation and cell–cell contact, effectively inducing the formation of hepatic spheroids throughout the printed scaffold.[195] Likewise, high densities of chondrocytes bioprinted into porous polycaprolactone (PCL) microchambers were shown to lead to aggregation into spheroids, which grew over time from the depth of the microchamber to its surface, depositing a vertically aligned network of collagen fibers, mimicking the orientation observed in native cartilage.[201]
A combination of pore size and pore geometry can also be exploited to indirectly induce alignment of cells placed within the pore. Printed nerve guides made of GelMA and PEGDA blends, mimicking the orientation of axonal bundles in the spinal cord, were shown to induce the alignment of neurons recruited in vivo and of neural progenitor cells (NPCs) seeded preimplantation, a feature that was not observed when NPCs were injected in a spinal cord injury site simply within a fibrin glue gel.[202] Interestingly, this geometrical cue led to the alignment of astrocyte in the axonal growth direction, ensuring these support cells acted as living scaffold for neural growth, rather than creating an astrocytic scar blocking the regenerative process. These regenerative spinal cord implants led to the recovery of impaired motor function in rat models in vivo.[202]
In extrusion-based printing approaches, due to proximity effects, cells seeded closer to printed filaments tend to align along the filament direction, and in response induce a preferential alignment in neighboring cells, even though the latter are not in direct contact with the printed structure. Taking advantage of this phenomenon, cardiomyocytes placed between relatively large rectangular patterns produced by MEW (200 × 400 μm) were shown to spontaneously align along the main axis of orientation of the pores,[203] in contrast to squared patterns in which cells displayed no preferential alignment. A similar effect was observed when 3D-printed and TGase-crosslinked gelatin patterns were used to confine hMSC or rat-derived cardiomyocytes seeded in between the printed strands (Figure 7A). When cells were placed in between 500 μm spacing, improved alignment and myogenic differentiation were observed, as well as an increment in beating frequency and synchronization.[38]
Figure 7.
Strategies to induce anisotropy within printed constructs can be used to promote cell alignment and therefore maturation of, i.e., muscular and neural structures, as well as to generate 3D constructs with local variations of mechanical properties. A) Cardiomyocytes aligning within confined, printed hydrogel chambers. Reproduced under the terms of the Creative Commons CC-BY 3.0 License (https://creativecommons.org/licenses/by/3.0/).[38] Copyright 2018, The Authors, published by IOP Science. B) Nanocellulose particle alignment tuned by the extrusion speed and shear stress in the printing nozzle, allowing the creation of sophisticated anisotropic hydrogel patterns. Reproduced with permission.[211] Copyright 2017, Wiley-VCH. C) Fluorescently tagged fibrin-based bioinks, showing either: i) random distribution of the fibrin microfibers, when the bioink is printed at a lower printhead velocity, or ii) sub-micrometer scale alignment alongside the printing direction for higher displacement velocity, iii) which eventually drives alignment of bioprinted Schwann cells (actin-dapi staining, in green and red, respectively). Reproduced with permission.[214] Copyright 2018, IOPScience.
Interestingly, tuning of printed pores can be also used to modulate many physical properties of biofabricated constructs, as shown, for instance by bioprinted HCAEC (human coronary artery endothelial cells) in an alginate gel, in which impedance of the overall construct was controlled by pore design.[204] Direct effects on cell behavior could be observed when seeding and adhesion occurs on filaments with sizes comparable with cell dimension, such as micrometer-scale fibers obtained via MEW. MSC adhering to 10° laydown patterns were more likely to elongate along the fibers, whereas cells preferentially followed the curvature at the intersection between fibers printed with 90° laydown angles. This conformation resulted in a mechanical stress that induced nuclear translocation of YAP, and eventually improved osteogenic differentiation.[200]
Besides predesigned pores, the incorporation into bioinks of porogens and their selective removal represents an alternative approach to create room for cells to migrate and reorganize within bioprinted strands. In view of this, biphasic bioinks have been developed. These are, for example, composed of two immiscible components, like GelMA and poly(ethylene oxide) (PEO).[205] Tuning the GelMA/PEO ratio, the PEO component forms spherical phase-separated domains within the GelMA-rich matrix. After photo-crosslinking postprinting, the water-soluble PEO can be readily removed from the 3D matrix, leaving a network of micropores, and can thus facilitate cell stretching and migration within an otherwise dense polymer network. Such approach is beneficial for tumor cluster formation for in vitro models, as well as to aid capillary vessel formation for vascularized constructs, as demonstrated for hepatic carcinoma cells, HUVECs and fibroblastic cells, which were shown to spread and displayed higher proliferation rates compared to when embedded in nonporous gels.[205]
Guided Cell and ECM Orientation
Many tissues owe their functionality to their anisotropic structure. For example, signal transmission across a nerve network, force actuation by bundles of myocytes, ability of bone to withstand mechanical load are all driven by the hierarchical alignment and preferential directionality displayed by cells and their ECM.
Direct alignment of C2C12 myoblast cells along the main axis of bioprinted filaments has been observed and was attributed to the shear during extrusion. This led to maturation of confluent myotubes (enhanced myogenin, MyoD, and α-sarcomeric actin mRNA levels) over 21 days of culture, a phenomenon that was not observed for cells maintained in 2D cultures.[206] In another study using a fibrinogen–PEGDA semisynthetic hydrogel as bioink, confluent sarcomerogenesis and effective myoblastic maturation and stability was also obtained within two weeks. Additionally, by utilizing a microfluidic printing approach, the authors showed the possibility to coprint in the same extruded filament, a myogenic compartment adjacent to a region of fibroblasts known to support myogenesis. An important finding of this work is that adequate myotube formation only occurs when the cell density in the printed filament is high enough to ensure cell–cell contact within the bioink.[18] Cell alignment and myofibers formation was also observed in bioprinted human muscle progenitor cells in fibrin inks, leading to a 82% recovery of the native tibialis anterior muscle function upon implantation in a rodent model.[207]
An alternative strategy to induce cell organization and alignment within hydrogels is the incorporation of anisotropic micro-and nanotopographical cues. For this purpose, paramagnetic ferric oxide nanoparticles have been used either embedded in fragments of electrospun poly(lactic-co-glycolic acid) nanofibers[208] or encapsulated into RGD-functionalized rod-shaped nanogels.[209] These elements were then independently encapsulated together with cells within casted fibrin hydrogels and aligned by applying a magnetic field. Finally, this resulted in a preferred orientation and stretching of fibroblasts, dorsal root ganglia cells, and neurons within the hydrogels. In the case of neurons, cells were also shown to exhibit propagation of calcium signal across the aligned cell network in the preferential direction of alignment, demonstrating an interesting development toward neural network regeneration.
Interestingly, the printing process itself can also be used to induce specific alignment of micro–nanoscale elements. This concept was already demonstrated for the 3D printing of carbon fiber–epoxy resins composites, which yielded the carbon fibers aligned toward the extrusion direction. Interestingly, by applying a rotation of the printhead along its longitudinal axis, the resulting printed filament had the microfibers adopting a random-like and isotropic orientation, following the pattern of rotation.[210] Exploiting this effect can help creating printed constructs with tunable anisotropic/isotropic mechanical properties and topographical cues, even creating regions with diverse properties within the same construct. Potential applications include mimicking tissues such as cartilage and menisci, which have region-dependent mechanical properties, in part determined by ECM anisotropy. Selective alignment of nanoscale elements has also been observed when printing aqueous inks carrying anisotropic cellulose nanocrystal (CNC) suspensions.[211] Interestingly, ordered domains adjacent to areas with random particle orientation were observed within single struts for these materials (Figure 7B). Specifically, this approach created struts with either CNCs aligned toward the extrusion direction only near the outer wall of the filament, or displaying alignment throughout the whole cross-section. This allows to create structures with antagonizing mechanical properties, with highly stretchable filaments (alignment restricted to the surface) and with stiffer ones (fully aligned particles).[211,212] This provides exciting opportunities for recreating tissue interfaces, such as the muscle-tendon transition, in which deformable muscle fibers are connected to high elastic modulus tendon fibers.
Understanding and taking advantage of these phenomena is particularly relevant as many bioinks for extrusion bioprinting contain nanofibrillar elements.[213] Moreover, several ECM-derived materials, such as collagens and fibrins, are even directly composed by nanosized fibers. Bioinks comprising of a blend of alginate and fibrinogen were demonstrated to form a fibrin nano-and microfiber network aligned toward the printing direction, when printed at high displacement velocity of the extrusion nozzle, as visualized by fluorescent staining of the fibrin polymer.[213] This organized structure was observed after crosslinking of the ink in a bath containing CaCl2 and thrombin, and was visualized via fluorescent staining of the fibrin polymer. In turn, such microscale topographical cues were shown to guide the alignment of printed Schwan cells. Finally, once primary neurons were seeded onto these constructs, their newly formed neurites also followed the overall directionality imposed by the printing process (Figure 7C).[214] Along this line of thought, multiwalled carbon nanotubes (mwCNTs) were embedded in alginate and methacrylated collagen (MeCol) as conductive elements for cardiac tissue engineering.[215] In both hydrogels, aligned submicrometer networks of conductive fibrils were observed after printing. The materials were printed alternating the alginate ink as a supportive frame and the MeCol as bioink encapsulating human HCAEC, resulting in improved conductivity of the construct, as well as preferential alignment of the HCAECs in the direction of the nanofibrillar network.[215]
3.2.2. Next Generation Bioinks
As hydrogel-based materials can relatively easily be modified, they can be designed and functionalized with bioactive molecules to guide mechanobiology responses and tune cell behavior. In turn, hydrogels can dynamically respond and adapt to the biosynthetic and catabolic activity of the encapsulated cells. Nevertheless, design requirements greatly vary depending on the target tissue. For further reading on these specific design requirements, we refer to excellent recent reviews.[216–220] However, it is clear that these design requirements are now a next logical step for advancing bioinks toward functional and bioactive bioinks that can, at least partially, orchestrate cellular behavior postprocessing.
Embedded Bioactive Moieties—Peptides, Proteins, and Decellularized ECM
Functionalization with short, integrin-ligand peptides is a well-established strategy to improve cell interaction with otherwise nonadhesive matrices.[221] In biofabrication, grafting of short peptide sequences or full-length proteins selected from the ECM onto the main constituent of the bioink can be exploited. The fine-tuning of physical properties of the hydrogel combined with the presentation of multiple molecular signals, regulate integrin attachment, (stem) cell differentiation, and create a microenvironment in which multiple cell types can thrive, including organoids[222] and heterocellular spheroids.[223] High-throughput screening approaches to evaluate vast libraries of functional peptides have been developed[224,225] and are already applied to hydrogel-based bioink development.[226] Generation of these high-throughput screening platforms can also benefit from biofabrication technology. For example, a library of different ratios of two cellladen bioinks (GelMA and PEG) was generated via microvalve bioprinting to evaluate spreading and proliferation of periodontal ligament stem cells.[227] Further, development of modular bioinks, composed of structural macromolecules that facilitate selective grafting through reactive groups of specific bioactive moieties, including short peptides and full length proteins, has gained increasing attention. This has resulted in a range of bioinks that enable grafting via Michael-type addition[228] and thiol–ene,[100,111,119,140] reactive silane,[229] and succinimidyl ester-amine[43] based reactions.
Engineering the composition of bioinks with bioactive compounds to guide cell response often makes use of cues derived from the native extracellular environment, eventually creating an artificial, and mostly chemically defined, yet simplified ECM substitute. Rather than designing “bottom-up” a bioink composition, a powerful approach is to use natural templates, as already provided by native ECM.[230,231] Printable, cell compatible bioinks have been obtained by decellularizing and solubilizing ECM from adipose tissue, cartilage and cardiac muscle, and each of these bioinks was proven superior to collagen gels in view of their potential to induce differentiation of adipose derived stromal cells, bone-marrow derived stromal cells, and cardiomyocites, respectively.[85,232] In fact, a variety of tissue-specific dECM inks have now been produced from tissues, including liver,[92] brain,[233] skin,[234] skeletal muscle,[91] and cornea.[235]
Importantly, dECM bioinks may be used as allogeneic or even xenogenic materials in tissue engineering applications, since their production and decellularization processes involve harsh enzymatic and detergent treatments, aimed at removing immunogenic residual deoxyribonucleic acid (DNA) or cell membrane antigen fragments. Previous research has also pointed out how ECM composition varies between degenerative and homeostatic processes,[236–239] thus opening interesting avenues for using dECM bioinks from nonhealthy organs to create in vitro models to study tissue pathophysiology. The exact composition of ECM-derived materials, particularly that of less abundant compounds, is not fully defined, although proteomic techniques can aid elucidating part of these components.[240] Nevertheless, such “gray box” mixture of both known and undefined factors has been proven particularly important when aiming to recreate at once complex, multicomponent tissues. For instance, in skin tissue engineering, major challenges revolve around the recreation of nails, melanocytes, hair and sweat glands. Homogenates from dermis of mice were also used as functional component of bioinks, when mixed directly into a gelatin/alginate blend, and then used to print porous constructs embedding epithelial progenitor cells (EPs). The homogenized tissue is rich in BMP-4 (BMP = bone morphogenetic protein) and EGF, which are a major drive toward EP differentiation into functional sweat glands, and was demonstrated to induce the maturation of functional glands, that even restored partial sweating activity upon in vivo implantation in murine burn wound model.[25,241] However, since homogenates, differently from dECM, may still have present immunogens, their compatibility should be carefully ensured prior to implantation in vivo.
An important limitation of dECM bioinks relates to their typically low stiffness, viscoelastic behavior and slow gelation kinetics, which result in limited shape fidelity. Novel technologies such as printing in support baths[88] can offer relevant solutions to this challenge. At the same time, more traditional crosslinking strategies using enzymatic crosslinking, photo-crosslinking, blending with rheology-modifying inks or coprinting with thermoplastic frames are still extensively utilized.[86] For example, to address the problem of controlling the mechanical properties of dECM-based inks, a liver dECM hydrogel has been modified via blending with thiolated gelatin and hyaluronic acid to ensure printability.[29] This bioink also included PEGDA as crosslinker, which could covalently bind to thiols in the viscosity enhancers and in the dECM proteins. Finally, the bioink mixture was further stabilized through multiarm PEG-alkynes and induction of UV-mediated thiol-alkyne crosslinking. By combining different polymer concentrations and 4 or 8 arm alkynes, a library of mechanical properties in a wide range from 0.1 to 20 kPa could be obtained, covering most of the physiological range of soft tissues. With this effort, within the printed material the hepatic differentiation of spheroids composed by three cell populations (hepatocytes, stellate cells, and Kupffer cells) could be enhanced.[29]
Considering the biological advantage of using dECM-derived materials, open challenges for future research in this area include the identification and isolation of components within dECM inks that are responsible for specific cell responses. Elucidating new, key bioactive components could help creating novel, chemically defined bioinks, to facilitate reproducibility and possibly the clinical translation of bioprinted, ECM-derived constructs.
Controlled Degradation
Besides promoting cell attachment, proliferation and differentiation, biomolecule grafting approaches also can control bioink degradation kinetics postprinting, which is paramount for tissue morphogenesis and can also control the phenotype of embedded cells.[242] Peptides and sequences that are the target of specific proteases have for instance been used to print blood vessel-like tubes within 3D hyaluronic acid hydrogels. With such modifications, endothelial cells injected in the vessel lumen could invade the surrounding gel initiating the angiogenic processes.[140] In the context of in vitro cancer models, the improved control of degradation of a matrix, based on gelatin and fibrin components blended with alginate, for the formation of tumor spheroids also resulted in better prediction of resistance to chemotherapy, compared to standard tissue culture.[243]
Another interesting development is to utilize composite bioinks, in which each component is sensitive to specific enzymes or a resorption mechanism, potentially offering two independent mechanisms to modulate the structural and mechanical properties of the polymer network. Recently, a novel dual component bioink was developed, based on a polypeptidesingle strand DNA conjugate.[244] This hydrogel system can be crosslinked through the mixing with short double strand DNA exposing two “sticky” single strand sequences that are complementary for portions of the DNA grafted onto the peptide backbone.[244] As the precursor solution displays low viscosity, this material has been successfully applied in inkjet/drop-ondemand bioprinting. To process such ink also with extrusionbased technologies, strategies such as printing in a support bath would be required. A key advantage of this system is, however, the possibility to modulate its degradation either with proteases, attacking its polymer backbone, or DNase, targeting the crosslinkers, without harming the overall structural stability of the gel. Future developments of this approach could include hydrogels with smart and programmable degradation profile, as suggested using CRISPR as a tool to create cleavable matrices.[245]
An alternative strategy to control the degradation is the addition of exogenously added factors that can degrade the printed structure over time. The feasibility of this concept has been demonstrated with corneal epithelial cells (hCEC) cultured in an alginate/gelatin/collagen blend.[246] Alginate typically does not support the proliferation of the hCECs as it lacks integrinbinding domains and forms a network that is too stiff. However, when sodium citrate, a compound able to disassemble the alginate network over time, is added to the culture, the epithelial cells are capable of proliferating and form spheroids within printed strands.[246]
Spatiotemporal Controlled Release
The bioink can also act as reservoir of bioactive compounds that can be released in a controlled fashion. Incorporation of bioactive molecules, typically growth factors, has been performed by direct grafting onto the hydrogel backbone. Alternatively, growth factor carriers can be mixed into the bioink formulation, either in the form of micro-and nanoparticles, or glycosaminoglycans and native proteins that carry growth-factor binding domains.[247] A protein/growth factor release system based on BMP-2-loaded collagen microfibers BMP has, for example, been included into a GelMA bioink. Loading was promoted by modifying BMP-2 with a collagen binding domain, resulting in an improved osteogenic differentiation of incorporated MSCs.[248] The photocrosslinking reaction of GelMA has also been exploited for its ability to mediate thiol-acrylate crosslinking, for instance to permit attachment of vascular endothelial growth factor (VEGF) mimetic peptides onto the hydrogel.[249] This GelMA with VEGF-like functionality was used to generate an HUVECloaded structure mimicking the Haversian vascular system within a bone-supporting PLA scaffold. Interestingly, this same PLA reinforcing frame was coated with polydopamine carrying BMP-2 mimetic peptides, effectively creating a dual source of morphogenetic signals to induce both osteogenesis and vasculogenesis within the same construct.[249] Growth factor-rich formulations, such as platelet-rich plasma (PRP), are already used in the clinic and could become an important component of future bioinks. As a plasma-derived formulation, PRP can be printed on its own or blended with other hydrogels, such as alginate, potentially creating a patient-specific bioink.[250]
Another approach involves the incorporation of drug-loaded microcarriers or nanoparticles as sustained release vehicles into the bioink. For example, gelatin microspheres have been loaded into alginate bioinks to carry either BMP-2 or VEGF to improve osteoregeneration[251] and vascularization,[252] respectively, upon implantation. An advantage of this strategy is that microcarriers can act both as cell delivery and drug delivery devices.[253,254] Microcarriers offer a high surface area on which cells can attach, spread and proliferate,[255] and can provide a protected microenvironment for the cells.[256] Facilitating cell spreading in 3D has important implications, since mechanosensing, cell adhesion and cytoskeleton rearrangement are key steps to drive (stem) cell differentiation. Since in many stiffer bioinks cells tend to maintain a rounded morphology, such approach can have important implications for tissues in which elongated and spread cells are preferred. This was demonstrated for the printing of bone, where a GelMA bioink, carrying cell-laden polylactide microcarriers, was shown to improve the expression of osteogenic differentiation markers and mineralized ECM deposition from MSCs.[42]
Besides microparticles, nanoparticles from a wide range of materials have also been extensively studied as drug release vehicles. 2D nanoclay particles have been used as versatile growth factor delivery devices. Given their composition and thin disk like structure, these particles display a strong negative charge on their wider side, and a strong positive charge on their perimetral wall, which are beneficial to carry charged biomolecules via electrostatic interactions. Preliminary experiments with Laponite embedded in an alginate/methylcellulose bioink demonstrated the potential for VEGF release in vitro.[98]
Further, micro-and nanocarriers within bioinks have also been studied as gene delivery devices. In particular, complexes of nanohydroxyapatite and plasmid DNA carrying sequences to boost the secretion of TGF-β3 and BMP-2, which are involved in osteogenic differentiation and bone maturation, have been used to directly transfect bioprinted cells and facilitate bonelike tissue formation in vitro and in vivo.[257] This is paving the way for the development of gene-activated bioinks. Nonviral gene carriers were also embedded in pore forming, phase separated alginate-methylcellulose bioinks to modulate the release profiles of plasmid DNA cargo.[258] These inks were printed to create a zonally loaded construct for the engineering of osteochondral plugs.
3.2.3. Replicating Multicompartment and Multicellular Structures
Layering, Gradients, and Interfaces
Since bioprinting is performed in a layer-by-layer fashion, structures that present a variation in material composition in the Z-direction can be fabricated with relative ease. Many living tissues present depthdependence properties and cellular composition that can be thus be simulated with such approach, for instance printing gradient or interfacing different inks for each layer. Following this principle, models of the brain cortex were fabricated by stacking multiple layers of RGD-modified gellan gum bioinks extruded within a flow of their crosslinker (a Dulbecco’s modified Eagle medium (DMEM) or CaCl2 solution) with a coaxial nozzle. When forming a structure in which only the first and last layer were laden with neurons, axonal sprouting could be observed into the cell-free region, in an attempt to bridge the gap within the cell-laden regions and therefore mimicking the projection neurons present in the native brain cortex.[259]
Among multilayered tissues, skin is perhaps one of the most notable examples, due to its naturally layered histological organization, in which a densely packed keratinized epithelial layer is lodged on top of the vascularized dermis, which hosts structures, such as sweat glands and hair follicles. A combination of inkjet-and extrusion-based bioprinting technologies was used to create complex skin grafts, in which human dermal fibroblasts (HDFs) were printed within a skin-derived dECM bioink to create the dermal layer, in which zone-specific ECM was deposited by the embedded cells. This region was then stabilized by inkjet printing of HEK cells on top of it, to form a homogenous cell layer, resulting in improved barrier function and dermal ECM secretion.[234,260] The same bioink was used to print a vascularized patch (ASC + EPCs) on top of the HEK layer, which was further keratinized. When this full multilayer construct was used, accelerated wound closure, re-epithelialization and neo-vascularization were observed upon implantation in vivo in a mice model. Such printed structures are particularly interesting as patches for skin transplantation,[261] but also to study the barrier function of natural skin and for testing drug absorption and toxicity of cosmetics, as an alternative to animal experimentation. Indeed, bioprinted skin grafts have been produced including melanocyte-laden bioinks to introduce pigmentation.[262] Moreover, layering approaches provide opportunities to recreate a wide array of epithelial tissues and the basal lamina that support them, including but not limited to alveolar tissue in the lungs,[263] cornea[264] and urethral epithelium.[265]
Sequential deposition of multilayer structures can also facilitate the recreation of tissues from articulating joints for applications in the field of orthopedics. Articular cartilage displays a depth-dependent architecture and this zonal organization is responsible for cartilage functionality. In the superficial zone, chondrocytes and collagen type II fibers align parallel to the articulating surface and secrete molecules to facilitate joint movement (i.e., lubricin), while toward the deeper zones cells are more dispersed and assume a columnar organization accompanied by collagen fibers oriented perpendicularly to the underlying subchondral bone. Additionally, glycosaminoglycan concentration increases from the surface to the deeper zones, and a calcified cartilage layer is present at the bone–cartilage interface, both contributing to the high mechanical properties of cartilage under compression. Recreating this structure (and therefore the associated mechanical function) holds great promise for the creation of durable cartilage implants to treat degenerative diseases like osteoarthritis. On the one hand, both bioprinted cartilage-specific cells and stem cells demonstrated some ability to deposit hyaline cartilage-like ECM in vivo,[266] as well as calcified cartilage.[267] Within the field of biofabrication, technologies are being developed to print zone-specific bioinks, for instance by depositing subsequent layers of inks laden with a gradient in cell concentration,[268] or with different cartilagederived and bone-marrow derived specific progenitor cells, to recreate the native gradient of glycosaminoglycans (GAGs) and the expression of zonal markers.[269] Further building on these strategies, layering of multiple materials can allow to create osteochondral graft, comprising of closely interconnected bone and cartilage units. This approach is particularly interesting toward the integration of tissues composed of dissimilar materials (i.e., mineralized bone vs softer cartilage). Low-temperature setting calcium phosphate ceramic cement (CPC) were demonstrated possible to print together with a cell-laden alginate–methycellulose bioink. Cell viability was preserved during cement setting and porous triphasic osteochondral constructs were created, composed of i) a bone-mimetic CPC-only region, ii) a calcified-cartilage mimetic region, where MSC-laden bioinks and CPC strands are alternated, and iii) a bionk only region, to simulate articular cartilage. Adhesion between the three compartments is ensured by the middle layer, in which the alternated cement and bioink strands permit the anchoring to the bone and cartilage region respectively.[108] Osteochondral constructs have in general been studied using a wide array of materials, and recently even strategies to print with soft hydrogel both the bone and cartilage compartments have been enabled for instance through printing into supporting, non-Newtonian baths,[177] showing potential to preserve in each region osteoblastic and chondrogenic cell phenotypes, greatly increasing the versatility and array of materials available for this type of application.
Heterocellular Compartments and Cell–Cell Interactions
From the direct connection of myocytes and nerve cells in the neuromuscular junction that regulate muscle activation to the crosstalk between neighboring cells from bone and cartilage in an osteochondral unit that preserves joint homeostasis, in living organisms, the functionality of organs is tightly linked to the continuous interaction of multiple types of tissues. Clearly, different cell types continuously interact and their reciprocal spatial orchestration is a major player in determining the biological functions. Cocultures are therefore widely used in biological research and are especially important to study these interactions, albeit in simplified models. However, the exact location of the different cell types in these coculture models cannot easily be controlled. Recapitulating in vitro tissue interfaces, vascularized organs and the interplay of multiple cell types has therefore been a major challenge in regenerative medicine. The field of biofabrication is now providing powerful tools to approximate to an unprecedented degree of fidelity the heterocellular, hierarchical composition of native tissues.
Moreover, these technologies can be successfully used to create miniaturized tissue models to study pathological processes and the effect of drugs on multitissue platforms on-a-chip. Eventually, integrating multiple cell types and tissues in one single process can pave the way toward the “holy grail” of bioprinting full organs for transplantation, but also to create advanced models for biomedical research to improve our understanding of intertissue interactions in disease and regeneration.[270]
Biofabrication of spatially defined heterocellular structures have already demonstrated improved functionality and the appearance of emergent properties of native tissues and organs, in comparison with simple, nonorganized cocultures. In the field of drug testing, for instance, interfacing multiple biofabricated tissue compartments in organ-on-chip and bodyon-chip devices can permit to observe off-target toxic effects that could not be analyzed in conventional in vitro screening platforms.[270] Importantly, these advanced platforms facilitate the determination of the minimally required complexity in biofabricated models to reduce the gap in functionality between in vitro engineered grafts and native tissues.
The present section will highlight how advanced bioprinting strategies have been successfully employed to recapitulate in vitro living tissue multifaceted functionalities, from the molecular level (biomarker expression in printed cells) to the system level (organ and tissue functions), when precisely patterning multiple cell-types within the same construct. Kang et al. provided a comprehensive demonstration of this potential fabricating jaw and calvaria bone, ear cartilage and skeletal muscle via printing composites of PCL and cellladen gelatin/fibrinogen/hyaluronan/glycerol blends, and demonstrating good shape retention and tissue formation in vivo.[37] Moreover, in the case of the skeletal muscle construct, the PCL support strands also served as functional guides to induce compaction of the printed cells and their alignment in the longitudinal direction of the patterns, in contrast to bioinks printed without the stabilization of the PCL frame, which exhibited randomly oriented cells. This strategy can also be used to create heterocellular constructs and to recapitulate interfaces between different tissues, e.g., for an engineered muscle-tendon unit composed of two connected regions: a tendon compartment, consisting of PCL-reinforced 3T3 cell-laden fibrin strands, adhering to a muscle compartment (fibrin with C2C12 cells, reinforced by an elastomeric polyurethane). Beside the two structural supports that mimicked the tensile properties of tendon and skeletal muscle, the interaction between cells within the two compartments led to the spontaneous emergence of an upregulated expression of focal adhesion markers, typical of native muscle-tendon junctions, solely at the bioprinted interface. Such zonal marker expression was absent in homogenous constructs, when only the muscle bionk was printed, indicating that anatomically accurate cell compartmentalization through bioprinting could aid to better recreate functionally correct tissues.[36] Recently, multimaterial strategies have been used in combination with printing in a support bath, to enable the use of collagen inks as structural components of an engineered cardiac tissue, to support and encase a soft, high cell density-laden fibrinogen ink.[53] With this approach, human embryonic stem cellderived cardiomyocytes, confined by the collagen ink, could be patterned to replicate the shape of a human left ventricle, and exhibited functionality via synchronized contractility with directed propagation of the action potential (Figure 8), as well as thickening of the wall of the construct up to 14% during systole. Moreover, the same printing technology was used to recreate key cardiac blood vessels.[53]
Figure 8.
Bioprinted models of a cardiac ventricle, exhibiting synchronous electroconductive and contractile functions could be fabricated combining multimaterial printing and printing in a support bath, with the incorporation of high density of cardiomyocytes. A) Schematic representation of the printing process, B) construct dimensions, and C) final printed model. D) Calcium imaging of the printed structure and E) spontaneous, directional propagation of the calcium wave, indicating transmission of the action potential across the cardiomyocytes, also shown from a top view of the construct (F,G). H,I) Calcium signal propagation can be observed also after point stimulation, as also J) measured recording transient calcium waves during both spontaneous contraction or induced contractions with stimulation at 1 and 2 Hz. Reproduced with permission.[53] Copyright 2019, AAAS.
Inclusion of Vasculature
The reconstruction of a functional vascular network within engineered tissues is a long-standing challenge and one of the major limitations hampering clinical translation of laboratory-made grafts. In line with this unsolved need, many advanced and proof-of-concept studies to highlight the impact of newly developed printing technologies focused on introducing vascular networks within biofabricated tissues.[271] Additionally, incorporation of tubular networks in bioprinted tissues is highly relevant for a wide array of applications, as many tissues present microtubular structures (kidney, bile ducts, milk ducts, among others). Commonly used bioinks, including gelatin-, collagen- and fibrin-derived, have been optimized to support both vascular cells and tissue-specific cells,[272,273] taking advantage of the spontaneous ability of endothelial cells to reorganize themselves into capillary networks, when embedded in soft, cell adhesive hydrogels. Aiming for vascularized bone engineering a vasculogenesis-supportive bioink, based on gelatin/alginate blends and HUVECs was reinforced via coprinting with a PCL-based reinforcing frame. The adhesion between the reinforcement and the bioink was increased by functionalizing the PCL with polydopamine-modified calcium silicate, and in between each bioink-PCL layer, Wharton Jelly MSCs were patterned via ink-jet printing. Given the permissive environment provided by the gelatin matrix, HUVECs developed their capillary network throughout the construct, and secreted paracrine signals that stimulated osteogenesis from MSCs.[274]
Microcapillaries (typically few μm in diameter), however, are difficult to anastomose to native vessels in vivo, and cannot be readily perfused. Consequently, several approaches aim to print directly tubular networks in the size range of small arteries (<500 μm, conveniently in the range of most printing nozzles). These designed vessels act as templates in which printed or infused endothelial cells will adhere forming a confluent lining in the lumen. In this respect, bioprinting of cell-laden bioinks and sacrificial, water-soluble inks (i.e., thermoreversible hydrogels and carbohydrate glass) is a popular strategy which has been used both for vascularizing engineered tissues,[160] to create vessel-on-chip models to study vascular pathologies and thrombotic processes,[23] or to recapitulate epithelial microtubules, like proximal tubules in the kidney.[275]
Combining multimaterial printing with strategies for vascularization has great potential to improve survivability of large constructs, but also to recreate in vitro the multifaceted cell interactions that occur in vivo between blood vessels, functional tissue (parenchyma) and its supporting interstitial tissue (stroma) (Figure 9A). Kolesky et al. developed an elegant approach to create a vascularized bone construct of clinically relevant size (>1 cm3). First they printed two separate 3D networks: a parenchyma-precursor bioink, composed of MSC-laden gelatin–fibrinogen, and the other with a Pluronics biomaterial ink, as sacrificial template for the vascular network.[24] The interstitial space between the two lattices was filled with a fibroblast-laden gelatin–fibrinogen solution, which rapidly underwent crosslinking upon casting, and that, in turn, promoted also the in situ covalent crosslinking of the parenchyma bioink. Finally, the Pluronic network was removed, and endothelial cells and culture media with osteogenic factors were perfused into the vascular lattice. This precise architecture not only supported osteogenesis by MSC in the parenchyma-competent region, but also allowed to observe emergent interactions between the different cell populations in the three compartments, as the endothelialized vessels became stabilized by adjacent fibroblasts in the interstitium, but also by subpopulations of MSCs that migrated toward the endothelial lining and assumed a pericyte-like function. Such advanced constructs can help to create in vitro complex tissue models, which can aid observing and unravelling mechanisms behind cell interactions during tissue morphogenesis.[24]
Figure 9.
The functionality of parenchymal cells is intimately dependent on the cross-talk with stromal cells and with the vasculature, and the precise patterning of these components can be used to promote tissue maturation. A) Centimeter-scale vascularized bone constructs and bioprinted vascularized bone niche. Reproduced with permission.[24] Copyright 2016, National Academy of Sciences. B) Providing a precise 3D patterning of vascular cells within bioprinted filaments was proven beneficial to mimic healthy liver lobules, and enhance expression of cytochromes in hepatocytic cells, when compared to nonorganized mixing of endothelial cells and hepatocytes within the same bioink. Reproduced with permission.[140] Copyright 2018, Wiley-VCH. C) Free-form endothelialized vessels can be obtained via printing with sacrificial gels and further studied to understand angiogenic sprouting in 3D. Reproduced under the terms of the Creative Commons CC-BY 3.0 License (https://creativecommons.org/licenses/by/3.0/).[281] Copyright 2018, The Authors, published by IOPScience.
Sacrificial templates can also be fabricated using hydrogels that display quick degradation time. For instance, GelMA with different degree of methacryloyl substitution has been used both as structural bioink and a sacrificial material in the same construct. In an osteon model, composed by a quickly degradable MSC and HUVEC-laden GelMA filament, surrounded by more stable, MSC-laden highly substituted GelMA, the cells in the inner sacrificial gel degraded their matrix within a few days of culture, leaving a hollow tubule and started to adhere to the outer wall of the more stable, highly substituted GelMA filaments, effectively forming an endothelialized lumen. Moreover, these stable bioinks were loaded with both silica nanoparticles and covalently bound with VEGF to both enhance osteogenesis and stimulate angiogenic sprouting from the central vessel toward the periphery of the ostial regions of the construct.[276]
Due to the inherent difficulty in creating overhangs when printing soft hydrogels, advanced fabrication techniques are necessary to print convoluted geometries, typical of several native vascular networks (i.e., spiral arteries in the uterine endometrium).[277] Technologies like digital light processing bioprinting have potential to print directly intricated hollow structures and convoluted porosities,[6,288] whereas printing in supporting baths has been used for extrusion-based printing of free-form vessel-like structures. Patient-specific, vascularized heart constructs have been obtained through printing within an alginate-xanthan gum supporting bath, using an momentumderived dECM bioink, with a vascular network printed using a sacrificial gelatin ink.[88] In a different study, spiraling, straight and stenotic vessel-like channels have been printed using a fugitive ink, based on a shear-thinning and reversible supramolecular hydrogel, formed by a blend of cyclodextrin- and adamantane-modified HA. The supporting bath was composed of the same hydrogel precursors, with added norbornene groups to enable irreversible thiol–ene photo-crosslinking, and therefore turning the bath into a 3D matrix from which the fugitive ink can be removed, leaving a stabilized hollow tube. When matrix-metallopeptidase-sensitive crosslinkers were used in the outer matrix, this system allowed for studying cell migration and the invasion of angiogenic sprouts from the printed vessel into the HA matrix, in response to chemokine gradients (Figure 9B).[140] These findings can aid bioink design to establish angiogenic sprouts and capillary network in vitro by bioprinted cells, which have been indicated as promising tools to promote integration of biofabricated constructs within the host vasculature upon implantation in vivo.[278]
Perfusable channels can even be introduced within printed hydrogel filaments, when these are produced as core–shell struts, for instance via coaxial or microfluidic printing.[279] This concept was shown using templating, fugitive alginate as core material and GelMA–PEGTA blends, encapsulating endothelial cells and MSC as shell bioink. With such technology, vessels down to about 500 μm of diameter were fabricated and endothelialized. Perfusion of these systems was possible, even creating several complicated 3D macroarchitectures (i.e., star-shape, woodlog lattice), in which nutrient transport is carried out by the flow within the printed filaments.[44] Likewise, HUVEC-laden GelMA bioinks, blended with a fugitive alginate, have been used to create hierarchical, Janus-like filaments, but also vascularized cardiac patches, in which the endothelial cells line the lumen created within the core of the filament. Direct seeding of cardiomyocytes onto these printed HUVEC structures permitted to create vascularizedlike engineered cardiac tissue, which displayed synchronous beating.[40] Such vascularized cardiac tissues were also successfully used as heart-on-chip devices, characterizing the response of both myocardial and endothelial cells to the cytotoxic anti-tumoral drug doxorubicin.[27]
In line with most extrusion-based bioprinting methods, the resolution of the printed filament reported in the abovementioned studies is still in the range of ≈100–200 μm, to prevent harming cells during extrusion. Higher resolution was achieved with a nozzle-free printing strategy, also employing a similar microfluidic circuit, to feed multiple bioinks into a miniaturized DLP device, for multimaterial digital light photopolymerization. In this way, complex tissue, as shown with proof-of-concept fabrication of a tumor vascular network, a bone-tendon junction and a vascularized muscle-like tissue, can be approached with a spatial resolution of 20 μm, further increasing the level of detail and anatomical accuracy achievable.[280]
Recently, an elegant alternative has been developed to use extrusion technologies to load spatially organized patterns of multiple cell types and microscale vascular structures within a single filament. Instead of coordinating the flow of multiple bioinks from different reservoirs into the printing nozzle, a single, multicompartment cartridge was developed to lodge more than one bioink.[281] As during printing these inks are extruded in parallel from the same nozzle, the produced filament is templated by the geometry and loading pattern in the cartridge. This pre-set organization is then replicated through the longitudinal axis of the bioprinted strut (Figure 9C). In this way, multicompartment filaments were generated, for instance mimicking the shape of the spinal cord, a capillary-laden tube, or reconstructing the cross-section of a liver lobule, with the characteristic hexagonal pattern of hepatocyte-rich areas, separated by blood vessels. With this latter shape, a hexagonal liver lobule model was prepared with HepG2 and endothelial cells, which were then cocultured, and compared to filaments printed with a conventional, single reservoir cartridge, having a single bioink with nonzonally distributed cells. Interestingly, the patterned coculture showed superior maturation of the hepatocytic cells compared to the nonorganized construct, resulting in improved proliferation and ability to respond to the delivery of rifampicin, as a model drug, by increasing the protein expression of cytochrome CYP3A4A, necessary to metabolize the compound.[281] Although the exact mechanism for this improved cell maturation was not identified, this study further highlights the relevance of controlling cell positioning to improve tissue morphogenesis and allow biofabricated constructs to acquire functionalities expressed in native tissues.
Biofabricated In Vitro Models and Microphysiological Systems for Drug Testing
Heterocellular and vascularized constructs, comprising multiple cell-laden compartments have been bioprinted also to study interactions between cells located at distant regions of a tissue, which are fundamental in many pathological processes. In the field of drug testing, for instance, interfacing multiple biofabricated tissue compartments in organ-onchip and body-on-chip devices can permit to observe off-target toxic effects that could not be analyzed in conventional in vitro screening platforms.[270] Additionally, bioprinting technologies, are also used to bioprint directly perfusable constructs into preformed microfluidic chambers, produced by standard soft lithography with poly(dimethylsiloxane) (PDMS). Coupling the potential to print porous living structure with the capability for perfusion and streamlined analysis of the culture fluid, typical of microfluidic chips, proved valuable to study drug toxicity, as demonstrated, for instance on hepatic constructs.[282] Fully 3D-printed on-chip devices can also be obtained via printing of PDMS or thermoplastics to build the main walls of the chip, in which bioinks are patterned and exposed to a (micro)fluidic channel to incorporate fluid flow. Boxes based on PCL have been printed with luer lock adapters to facilitate perfusion, which enclosed two adjacent regions that were printed using bioinks laden with hepatocytes and endothelial cells. This selfcontained liver-on-chip demonstrated improved hepatocyte maturation, with increased albumin and urea secretion, and showed potential as direct platform to test drug induced liver toxicity.[283] Further expanding on this concept, in another study, miniaturized liver and cardiac modules were independently
printed, together with a reinforcing structure made of PCL. By easily handling the PCL supports, the modules could be lodged into traditional PDMS microfluidic chips that were connected in series to create a multitissue on-chip platform.[270] Connecting the liver module, printed with a gelatin-HA bioink laden with liver organoids (in the form of spheroids composed by hepatocytes, stellate cells and Kupffer cells) to the cardiac module (containing a gelatin–fibrinogen bioink with iPSCderived cardiomyocyte spheroids) allowed to study the effect of drugs that can alter cardiac rhythm (in this case, the beating frequency of the cardiac organoids) after they have been metabolized by the liver unit. Such advanced drug testing platform was further extended with the addition of a non-bioprinted lung module, composed of stacked layers of airway stromal mesenchymal cells, bronchial epithelial cells and endothelial cells, sequentially seeded on top of a transwell membrane. When this three-tissue system was exposed to bleomycin, a drug to treat lung cancer and that posed no known specific risk to the heart, unexpectedly the cardiac organoids ceased beating, despite being viable. This response was not observed when bleomycin was injected in the two-tissue liver–heart platform, and further investigation revealed that inflammatory markers (i.e., IL-1β) released from the treated lung organoid were responsible for affecting the function of the cardiac unit.[270]
The ability to capture the interactions between compartments populated by different cell subsets is also of particular interest in the field of modelling cancer biology, in which the 3D niche and surrounding environment of cancer cells play an active role on drug resistances and on the insurgence of metastatic processes. For instance, it has been recently shown that breast cancer cells can recruit cancer-associated fibroblasts located outside the tumor mass, and once these fibroblast reach the tumor, they are used as carriers by the cancerous cells to escape the primary tumor, an event that could initiate metastatic processes.[284] In a bioprinted breast cancer model, a compartment allowing the proliferation of tumorous spheroids was used to observe the inward migration of fibroblasts previously confined in distant region.[285] Such 3D compartmentalized models are particularly useful to study potential therapies that tackled the cancer-associated milieu, especially since many types of tumor cells are known to exhibit an improved resistance to chemotherapeutic drugs and ability to migrate when placed in 3D and coculture environments with stromal cells,[286] but also with macrophages and immune cells.[287] Overall, these advanced platforms and their future refinement, can help to overcome the limitations of currently oversimplified 2D cultures in standard multiwell plates. Moreover, the possibility to create such models with increasing degree of complexity (i.e., amount of cell population, architecture of the tissue) enable to investigate what is the minimally required complexity in biofabricated models to reduce the gap in functionality between in vitro engineered grafts and native tissues.
3.3. Embracing Biological Complexity
The rationale that mimicry of a healthy native tissue through detailed analysis of its geometrical composition will instruct the embedded cells to interact and together act as native tissue, reveals a truly engineering approach to solve complicated problems. As complicated systems are knowable and can be entirely understood by a defined set of rules, they can thus be addressed by recipes derived thereof. Examples for complicated systems are cars, planes or computers. Such systems can be entirely understood by detailed analysis of their components and their behavior can then be fully predicted. In contrast, complex systems are not fully knowable and only partially predictable, since there are too many variables that interact. Also, a detailed analysis of the system components will not result in an entire understanding. Human beings and their interactions are complex. Accordingly, biological systems are complex and we can thus not understand them by applying the same strategy of reductionism that works for complicated systems. However, we can achieve some understanding by watching and studying how the whole system operates, and, particularly when looking at biological systems, how they developed and evolved. In addition, with the constant progress of biomedical research, new underlying mechanisms relevant to complex biological systems are constantly being discovered. As this basic knowledge increases, it cannot be excluded that in the future we will be able to reduce biological and cellular responses to a complicated collection of processes.
For biofabrication and bioprinting, this means that the composition and morphology of mature and functional native tissue can, but does not have to be, a good or even the best starting point for printing. A thorough analysis of the functional interplay, eventually also with crucial interaction partners outside the tissue of interest, as well as a developmental analysis of the tissue, may be equally or even better suited to reveal 3D architecture and composition of (progenitor) cells and matrix components that lead to the formation of a functional tissue analogue.
In vivo, organs originate from the interaction of stem cells and the subsequent growth and remodeling of progenitor organ structures, rather than from the direct combination of separately generated populations of cells. Moreover, some of these progenitor structures only have a temporary role, i.e., the placenta, while others are the starting point for the formation of the organ.[288] Insights in and the application of these biological self-organizing mechanisms underlying the processes of organogenesis and tissue regeneration will be essential for the successful engineering of functional bioprinted structures.
Indeed, already ten years ago, i.e., prior to the rise of biofabrication, developmental engineering was introduced as a fusion of concepts from developmental biology and engineering and proposed as a new paradigm to advance the field of tissue engineering.[289,290] The key feature is an in vitro process design from sequential subprocesses corresponding to consecutive developmental stages in vivo that follow a gradual and coordinated progression of cell differentiation and tissue growth.[289] Special consideration is given to the first stage of the process, since it represents the basis establishing the necessary conditions for all subsequent stages. A proposed route to approach larger, more complex tissues is, furthermore, the use of multicellular developmental modules (intermediate tissues) as reliable, structurally robust building blocks.[289] Although theoretically convincing, developmental engineering has since shown to be also a challenging concept. One of the major obstacles is the fast growing, but still far from complete, knowledge of developmental biology. On the other hand, technological challenges represent major hurdles that have to be overcome, e.g., in order to facilitate the crucial first step of the envisioned process by providing the correct spatial arrangement of cells and materials or enable the assembly of intermediate modules into larger tissue units in a sufficiently controlled manner. Here, biofabrication with its two major routes of bioprinting and automated assembly of cell-containing building blocks, can greatly synergize with the developmental engineering concept in order to enable the rational generation of engineered tissues.
Partially recapitulating physiological processes, structures that can mimic different stages of morphogenesis have already been obtained through the remarkable ability of certain cell types to self-assemble and reorganize into organoids.[291] However, these organoids are relatively small and will not be sufficient to recapitulate the functionality of larger organ and tissue structures in vitro. Bioprinting approaches provide the opportunity to give structure, organization and/or mechanical support to developing cell-laden constructs. One of the few studies so far that has utilized bioprinting within a developmental engineering approach demonstrated the generation of developmentally inspired implants providing templates to instruct organogenesis in vivo. 3D bioprinting of vertebraeshaped mechanically reinforced MSC-laden bioinks was harnessed to engineer anatomically accurate hypertrophic cartilage templates in vitro, which over time in vivo were shown to mature into vascularized bone organs.[101]
Aiming at the further integration of bioprinting technologies and developmental biology principles, here we underscore that the features of bioprinting, such as the rational spatial arrangement and structuring of developing cell–biomaterial constructs, should be synergistically combined with the selfassembly capacity of cells. This could be implemented in two distinct ways (Figure 10). Firstly, bioprinting could impose specific geometrical designs allowing progenitors to be printed into organoid-like structures and generate conditions that induce further self-organization. Merging the resulting structures, allowing them to interact and self-organize, could subsequently lead to organ progenitors with the potential to form larger functional tissue structures. On the other hand, progenitor cell suspensions can self-organize into submillimeter to millimeter-scale structures. These structures that only encompass an intermediate complexity could then be bioprinted into more complex tissue and organ progenitors with geometries and patterns designed to instruct the formation of functional tissue.[288] The fusion capability of multicellular spheroids representing such intermediate tissue modules has been characterized in detail and utilized in proof-of-principle studies for tissue generation.[28,292] The bioprinting of such spheroids or microtissues into either 3D plotted or MEW-generated thermoplastic polymer scaffolds has recently been proposed and demonstrated, facilitating the assembly into larger tissue units.[293,294]
Figure 10.
Possible approaches toward the convergence of bioprinting and self-organization to guide the maturation of bioprinted constructs toward the generation of functional tissues. Inspired by the composition of adult, native tissues, multiple progenitor or differentiated cells can be loaded into bioinks to build tissues or organoids. In this approach, the architecture imposed by the printing process will be templating the cell-driven development of the tissue and its subsequent maturation. Alternatively, specific stem and progenitor cells that possess the ability to autonomously organize into submillimeter to millimeter organoids that exhibit salient tissue features can be used as intermediate building blocks and as bioink components. In both processes, the stimuli provided by the biomaterials, their architecture, and bioactive factors included in the bioinks play key roles for driving the acquisition of native functions.
Bioprinting approaches can thus synergize with the celldriven processes of self-assembly. In fact, this can be achieved through the precise patterning of morphogens and instructive signals, either inspired by developmental or reparative processes or by the native cues present in adult tissues. In more detail, bioprinting can be directly used to guide cell fate via the reproducible: i) printing of geometrical and topographical cues, ii) spatiotemporal presentation of biochemical signals guiding cell differentiation, and iii) coordinated deposition of multiple cell types and materials, to allow to study interactions beyond simple cocultures and observe emergent properties in engineered, heterocellular tissues. Such approaches implemented in bioprinted structures have been shown to boost construct maturation and promote the emergence of specific tissuelevel function or structures that cannot be easily obtained with conventional tissue engineering approaches. We envision that these approaches can be combined or used as starting point for the biofabrication of the next generation of living constructs expressing tissue- and organ-like functionalities (e.g., immune, hormonal, vascular, and nervous functions).
4. Concluding Remarks and Future Perspectives
The recent progress in hydrogel design together with the development of new bioprinting strategies, have introduced effective solutions to extend the biofabrication window, reducing the need to compromise on the use of materials that display satisfactory structural properties, but provide a nonoptimal environment for cells to thrive. Despite this remarkable progress, we are only beginning to tap into the potential of biofabrication in aiding the reconstruction of fully functional living engineered tissues.
Importantly, while our ability to precisely mimic architectural facets of living tissues increases, the extent of resolution that is required in order to achieve fully functional biofabricated constructs, remains unknown. Salient advances in the creation of tissues that exerted native functions in vivo have been achieved with precisely patterned, yet relatively simple architectures.[18,28,182] At the same time, an increasing number of studies is emerging, suggesting improved maturation of biofabricated tissues mimicking native structures, particularly when multiple cell types are distributed in precise areas of the produced constructs.[53,233,281] Bioprinting itself may help answer such fundamental yet elusive questions. High-accuracy and multimaterial printing can permit the generation of models with increasing degree of complexity, in which different combinations of geometrical cues, physicochemical properties and relative positioning of cells and materials can be combined. In order to achieve this, the influence of each variable should be isolated and studied, possibly even converging the field of biofabrication with automated high-throughput analysis, materiomics, and artificial intelligence-driven approaches that are already being introduced for biomaterial research.[295–297] Such systematic research will provide key insights for the design of the next generation of bioprinted constructs, eventually clearing an important step toward clinical translation. Additionally, although most of the expectations of bioprinting are associated with the generation of constructs that can copy or simulate the function of human tissues, alternative nonphysiological elements derived from other disciplines in biology, physics and engineering could also be envisioned and introduced, as recently suggested with the bioprinting of stimuli-responsive materials, nonmammalian cells as sources of metabolites or constructs designed by deterministic chaos principles as platforms to study cell–cell interactions.[298–301] Next generation bioinks, in addition to design criteria centered on rheology and printability, will need to be inspired by substantial input from advances in cell biology and biotechnology. Fundamental lessons learned from embryonic development, mechanobiology, cell differentiation and repair in species with regenerative capacity superior to that of humans,[302,303] will be paramount to guide bioink development, as well as to instruct which architectures and cell patterns to print to boost maturation. Given the dynamic and multifaceted events that determine development and progress of living tissues, other important criteria will be endowing bioprinted structures with the ability to evolve over time and provide different stimuli to the printed cells, for instance via the incorporation of materials in which biological signals and growth factors can be patterned and released on demand.[304] Together with knowledge from developmental biology, the emerging role of organoids as self-developing, miniature functional units of living tissues, and in general of microtissue analogues created by bottom-up assembly of stem cells can provide new opportunities for biofabrication. One possible avenue is to use preformed organoids and embryoid bodies that can be led to self-assemble and thus to produce tissues with high cell content by jamming them into a mold. These dense cellular structures can then be used as suspended bath to print vasculature within such engineered constructs to ensure organoid viability.[305] Furthermore, hybrid printing strategies can be envisioned, in which part of the construct architecture is imposed by the printing process, and at the same time also rely on the ability of stem cells to self-organize into polarized and heterocellular constructs. This can help to recreate microscale functional groups of cells that can otherwise not easily be resolved with current printing techniques.
Certainly, progress in basic biology is necessary and should be given priority in the near future to fulfill the goal of creating transplantable tissues. However, there is also room and need for further technological developments in additive manufacturing approaches. Interestingly, alongside extrusion-based bioprinting, several new and already existing biofabrication techniques are starting to gain more relevance in the field. In the quest to capture tissue complexity, future work can benefit from converging bioprinting with technologies that exhibit such complementary advantages. For instance, methods to manipulate cell suspensions (and even single cells), such as inkjet printing, as well as technologies that resolve features at the nano- and micro-scale, like two photon polymerization and MEW, permit high resolution,[306,307] but struggle to create large constructs with clinically relevant sizes. The combination with extrusion printing can, however, aid the generation of larger engineered tissues with features spanning across different dimensional scales to mimic the hierarchal composition of native tissues or to introduce regions with unique cell patterns. Likewise, tissues generated through the bioassembly of tissue spheroids, which are typically soft and prone to deform due to cell generated forces, can benefit from extrusion printing technologies to provide mechanically reinforcing frameworks,[293] that could possibly facilitate surgical handling and implantation. Additionally, extrusion technologies are limited in the geometries that can be achieved by the use of printed fibers as building blocks and by the need of supports. Recently, lightbased biofabrication technologies, such as stereolithography and digital light projection-based bioprinting have gathered attention for their ability to print convoluted porous geometries, typical of native tissues, and to replicate functional vasculature and microchannels within bioprinted constructs,[6,308] a step often indicated as major bottleneck toward clinical translation. Therefore, more research related to bioinks for light-based printing, as well as the integration with other biofabrication approaches can also be envisioned toward the generation of complex, functional tissues. Moreover, in the perspective of the generation of functional and transplantable tissue and organs, scaling up and regulatory concerns regarding bioprinted cells remain important topics to be tackled in the near future. Likewise, bioprinted constructs used as in vitro models and drug testing platforms will also need to undergo proper validation and clarification from regulatory bodies will be required to determine the exact criteria these models have to meet in order to be used as a complement to, or more desirably, as replacement for animal experimentation.
In view of a shifting focus on the biology of printing, detailed investigations of the impact of printing processes on cells, beyond the assessment of viability alone, will be required, especially for the creation of large grafts that require long printing times. During long printing processes cells are exposed to nonoptimal conditions (i.e., shear stresses and depletion of nutrients). Approaches like printing in support baths could permit supplementation of nutrients in the support material, possibly alleviating the impact of long fabrication times. Further research, can integrate novel technological solutions for the fast creation of living constructs. The recent introduction of volumetric bioprinting (VBP) based on visible light optical tomography, led to the creation of centimeter scale anatomically shaped constructs in less than 30 s,[309] opening new potential avenues for printing with minimal stress on embedded cells. In summary, adequate bioactivity and biofunctionality of printed cells and biofabricated tissues have to be addressed, as postprinting maturation is necessary for the generation of a functional tissue analogue. Accurate bioink design, as well as utilization of advanced cell culture platforms including cocultures, organoids and bioactive cues during the biofabrication process, are fundamental to achieve this objective. This requires the thorough characterization of cell behavior in the long term to ensure the preservation of superior cell functionalities beyond just viability. Biofabrication strategies are reaching a new cornerstone, maturing toward creating tissue grafts with envisioned applications in pharmaceutical industries and clinical therapies. Bioink design should reflect this yearning by investigating beyond the simple proof of concept to show feasibility of printing. Cell functionality is necessary both for in vitro models and to move forward toward the demonstration of applicability of bioprinted constructs as biomedical devices that can eventually be used as clinical solution to repair damaged tissues, and bring researchers a step closer toward the ambitious goal of organ bioprinting.
Acknowledgements
R.L. and T.J. contributed equally to this work. R.L. and J.M. acknowledge the funding from the ReumaNederland (LLP-12 and LLP-22), the European Research Council (Grant Agreement No. 647426, 3DJOINT), and the Horizon 2020 Research and Innovation Program under the Grant Agreement No. 814444 (MEFISTO). R.L and J.M. also gratefully acknowledge the Gravitation Program “Materials Driven Regeneration,” funded by the Netherlands Organization for Scientific Research (024.003.013). T.J., R.S., T.B., and J.G. thank the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) for funding under the Collaborative Research Center SFB/TRR 225—project number 326998133—TRR 225 (subprojects A02, B04, and Z). J.G. further acknowledges the ERC (Grant Agreement No. 617989, Design2Heal) for financial support. The authors thank Daimon Hall (carbonandneon. com) for support with graphical design.
Biographies
 Riccardo Levato is assistant professor in the group of Prof. Jos Malda at the University Medical Center Utrecht. He holds a masters’ degree in biomedical engineering from the Technical University of Milan, and in 2015 he obtained his Ph.D. in the same field of research from the Institute for Bioengineering of Catalonia and Technical University of Catalonia. His research interests include the development of novel biofabrication strategies and cell-instructive biomaterials for applications in regenerative medicine and tissue biomimicry.
Riccardo Levato is assistant professor in the group of Prof. Jos Malda at the University Medical Center Utrecht. He holds a masters’ degree in biomedical engineering from the Technical University of Milan, and in 2015 he obtained his Ph.D. in the same field of research from the Institute for Bioengineering of Catalonia and Technical University of Catalonia. His research interests include the development of novel biofabrication strategies and cell-instructive biomaterials for applications in regenerative medicine and tissue biomimicry.
 Tomasz Jungst studied nanostructural engineering at the University of Würzburg. After that, he did his Ph.D. at the Department for Functional Materials in Medicine and Dentistry. During his Ph.D., he established 3D-printing methods for biofabrication and regenerative medicine. He is currently a postdoctoral fellow in the group of Prof. Jürgen Groll focusing his research on bioprinting techniques and biofabrication of biomimetic vascular grafts and cardiovascular tissues.
Tomasz Jungst studied nanostructural engineering at the University of Würzburg. After that, he did his Ph.D. at the Department for Functional Materials in Medicine and Dentistry. During his Ph.D., he established 3D-printing methods for biofabrication and regenerative medicine. He is currently a postdoctoral fellow in the group of Prof. Jürgen Groll focusing his research on bioprinting techniques and biofabrication of biomimetic vascular grafts and cardiovascular tissues.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Dr. Riccardo Levato, Department of Orthopaedics University Medical Center Utrecht Utrecht University 3584 CX Utrecht, The Netherlands; Department of Clinical Sciences Faculty of Veterinary Medicine Utrecht University 3584 CX Utrecht, The Netherlands
Ruben G. Scheuring, Department of Functional Materials in Medicine and Dentistry and Bavarian Polymer Institute University of Würzburg Pleicherwall 2, 97070 Würzburg, Germany
Prof. Torsten Blunk, Department of Trauma, Hand, Plastic and Reconstructive Surgery University of Würzburg Oberdürrbacher Str. 6, 97080 Würzburg, Germany
Prof. Juergen Groll, Department of Functional Materials in Medicine and Dentistry and Bavarian Polymer Institute University of Würzburg Pleicherwall 2, 97070 Würzburg, Germany
Prof. Jos Malda, Department of Orthopaedics University Medical Center Utrecht Utrecht University 3584 CX Utrecht, The Netherlands; Department of Clinical Sciences Faculty of Veterinary Medicine Utrecht University 3584 CX Utrecht, The Netherlands
References
- [1].Lincoln J, Lange AW, Yutzey KE. Dev Biol. 2006;294:292. doi: 10.1016/j.ydbio.2006.03.027. [DOI] [PubMed] [Google Scholar]
- [2].Rose JC, De Laporte L. Adv Healthcare Mater. 2018;7 1701067. [Google Scholar]
- [3].Moroni L, Boland T, Burdick JA, De Maria C, Derby B, Forgacs G, Groll J, Li Q, Malda J, Mironov VA, Mota C, Nakamura M, Shu W, Takeuchi S, Woodfield TBF, Xu T, Yoo JJ, Vozzi G. Trends Biotechnol. 2018;36:384. doi: 10.1016/j.tibtech.2017.10.015. [DOI] [PubMed] [Google Scholar]
- [4].Groll J, Boland T, Blunk T, Burdick JA, Cho DWW, Dalton PD, Derby B, Forgacs G, Li Q, Mironov VA, Moroni L, Nakamura M, Shu W, Takeuchi S, Vozzi G, Woodfield TBFF, Xu T, Yoo JJ, Malda J. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/1/013001. 013001. [DOI] [PubMed] [Google Scholar]
- [5].Ozbolat IT, Hospodiuk M. Biomaterials. 2016;76:321. doi: 10.1016/j.biomaterials.2015.10.076. [DOI] [PubMed] [Google Scholar]
- [6].Lim KS, Levato R, Costa PF, Castilho MD, Alcala-Orozco CR, Van Dorenmalen KMA, Melchels FPW, Gawlitta D, Hooper GJ, Malda J, Woodfield TBF. Biofabrication. 2018;10 doi: 10.1088/1758-5090/aac00c. 034101. [DOI] [PubMed] [Google Scholar]
- [7].Gao G, Schilling AF, Hubbell K, Yonezawa T, Truong D, Hong Y, Dai G, Cui X. Biotechnol Lett. 2015;37 doi: 10.1007/s10529-015-1921-2. 2349. [DOI] [PubMed] [Google Scholar]
- [8].Mezel C, Souquet A, Hallo L, Guillemot F. Biofabrication. 2010;2 doi: 10.1088/1758-5082/2/1/014103. 014103. [DOI] [PubMed] [Google Scholar]
- [9].Visser J, Peters B, Burger TJ, Boomstra J, Dhert WJA, Melchels FPW, Malda J. Biofabrication. 2013;5 doi: 10.1088/1758-5082/5/3/035007. 035007. [DOI] [PubMed] [Google Scholar]
- [10].Müller M, Becher J, Schnabelrauch M, Zenobi-Wong M. J Visualized Exp. 2013;10:e50632. doi: 10.3791/50632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Groll J, Burdick JA, Cho DW, Derby B, Gelinsky M, Heilshorn SC, Jüngst T, Malda J, Mironov VA, Nakayama K, Ovsianikov A, Sun W, Takeuchi S, Yoo JJ, Woodfield TBF. Biofabrication. 2019;11 doi: 10.1088/1758-5090/aaec52. 013001. [DOI] [PubMed] [Google Scholar]
- [12].Malda J, Visser J, Melchels FP, Jüngst T, Hennink WE, Dhert WJA, Groll J, Hutmacher DW. Adv Mater. 2013;25 doi: 10.1002/adma.201302042. 5011. [DOI] [PubMed] [Google Scholar]
- [13].Nam SY, Park SH. Adv Exp Med Biol. 2018;1064:335. doi: 10.1007/978-981-13-0445-3_20. [DOI] [PubMed] [Google Scholar]
- [14].Kyle S, Jessop ZM, Al-Sabah A, Whitaker IS. Adv Healthcare Mater. 2017;6 doi: 10.1002/adhm.201700264. 1700264. [DOI] [PubMed] [Google Scholar]
- [15].Jessop ZM, Al-Sabah A, Gardiner MD, Combellack E, Hawkins K, Whitaker IS. J Plast Reconstr Aesthetic Surg. 2017;70 doi: 10.1016/j.bjps.2017.06.001. 1155. [DOI] [PubMed] [Google Scholar]
- [16].Chimene D, Lennox KK, Kaunas RR, Gaharwar AK. Ann Biomed Eng. 2016;44 doi: 10.1007/s10439-016-1638-y. 2090. [DOI] [PubMed] [Google Scholar]
- [17].Ouyang L, Highley CB, Sun W, Burdick JA. Adv Mater. 2017;29 doi: 10.1002/adma.201604983. 1604983. [DOI] [PubMed] [Google Scholar]
- [18].Costantini M, Testa S, Mozetic P, Barbetta A, Fuoco C, Fornetti E, Tamiro F, Bernardini S, Jaroszewicz J, Święszkowski W, Trombetta M, Castagnoli L, Seliktar D, Garstecki P, Cesareni G, Cannata S, Rainer A, Gargioli C. Biomaterials. 2017;131:98. doi: 10.1016/j.biomaterials.2017.03.026. [DOI] [PubMed] [Google Scholar]
- [19].Massa S, Sakr MA, Seo J, Bandaru P, Arneri A, Bersini S, Zare-Eelanjegh E, Jalilian E, Cha BH, Antona S, Enrico A, Gao Y, Hassan S, Acevedo JP, Dokmeci MR, Zhang YS, Khademhosseini A, Shin SR. Biomicrofluidics. 2017;11 doi: 10.1063/1.4994708. 044109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Snyder JE, Hamid Q, Wang C, Chang R, Emami K, Wu H, Sun W. Biofabrication. 2011;3 doi: 10.1088/1758-5082/3/3/034112. 034112. [DOI] [PubMed] [Google Scholar]
- [21].Holmes AM, Charlton A, Derby B, Ewart L, Scott A, Shu W. Biofabrication. 2017;9 doi: 10.1088/1758-5090/aa7bbd. 033001. [DOI] [PubMed] [Google Scholar]
- [22].van der Valk DC, van der Ven CFT, Blaser MC, Grolman JM, Wu PJ, Fenton OS, Lee LH, Tibbitt MW, Andresen JL, Wen JR, Ha AH, Buffolo F, van Mil A, Bouten CVC, Body SC, Mooney DJ, Sluijter JPG, Aikawa M, Hjortnaes J, Langer R, Aikawa E. Nanomaterials. 2018;8:E296. doi: 10.3390/nano8050296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang YS, Davoudi F, Walch P, Manbachi A, Luo X, Dell’Erba V, Miri AK, Albadawi H, Arneri A, Li X, Wang X, Dokmeci MR, Khademhosseini A, Oklu R. Lab Chip. 2016;16 doi: 10.1039/c6lc00380j. 4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA. Proc Natl Acad Sci USA. 2016;113 doi: 10.1073/pnas.1521342113. 3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Huang S, Yao B, Xie J, Fu X. Acta Biomater. 2016;32:170. doi: 10.1016/j.actbio.2015.12.039. [DOI] [PubMed] [Google Scholar]
- [26].Groen WM, Diloksumpan P, van Weeren PR, Levato R, Malda J. J Orthop Res. 2017;35 doi: 10.1002/jor.23602. 2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang YS, Arneri A, Bersini S, Shin SR, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell’Erba V, Bishop C, Shupe T, Demarchi D, Moretti M, Rasponi M, Dokmeci MR, Atala A, Khademhosseini A. Biomaterials. 2016;110:45. doi: 10.1016/j.biomaterials.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bulanova EA, Koudan EV, Degosserie J, Heymans C, Pereira FDAS, Parfenov VA, Sun Y, Wang Q, Akhmedova SA, Sviridova IK, Sergeeva NS, Frank GA, Khesuani YD, Pierreux CE, Mironov VA. Biofabrication. 2017;9 doi: 10.1088/1758-5090/aa7fdd. 034105. [DOI] [PubMed] [Google Scholar]
- [29].Skardal A, Devarasetty M, Kang HW, Mead I, Bishop C, Shupe T, Lee SJ, Jackson J, Yoo J, Soker S, Atala A. Acta Biomater. 2015;25:24. doi: 10.1016/j.actbio.2015.07.030. [DOI] [PubMed] [Google Scholar]
- [30].Donderwinkel I, Van Hest JCM, Cameron NR. Polym Chem. 2017;8 4451. [Google Scholar]
- [31].Ashammakhi N, Ahadian S, Xu C, Montazerian H, Ko H, Nasiri R, Barros N, Khademhosseini A. Mater Today Bio. 2019;1 doi: 10.1016/j.mtbio.2019.100008. 100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kim JE, Kim SH, Jung Y. Tissue Eng Regener Med. 2016;13:636. doi: 10.1007/s13770-016-0125-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Van Hoorick J, Tytgat L, Dobos A, Ottevaere H, Van Erps J, Thienpont H, Ovsianikov A, Dubruel P, Van Vlierberghe S. Acta Biomater. 2019;97:46. doi: 10.1016/j.actbio.2019.07.035. [DOI] [PubMed] [Google Scholar]
- [34].Jang J, Park JY, Gao G, Cho DW. Biomaterials. 2018;156:88. doi: 10.1016/j.biomaterials.2017.11.030. [DOI] [PubMed] [Google Scholar]
- [35].Piard C, Baker H, Kamalitdinov T, Fisher J. Biofabrication. 2019;11 doi: 10.1088/1758-5090/ab078a. 025013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Merceron TK, Burt M, Seol YJ, Kang HW, Lee SJ, Yoo JJ, Atala A. Biofabrication. 2015;7 doi: 10.1088/1758-5090/7/3/035003. 035003. [DOI] [PubMed] [Google Scholar]
- [37].Kang HW, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A. Nat Biotechnol. 2016;34:312. doi: 10.1038/nbt.3413. [DOI] [PubMed] [Google Scholar]
- [38].Tijore A, Irvine SA, Sarig U, Mhaisalkar P, Baisane V, Venkatraman S. Biofabrication. 2018;10 doi: 10.1088/1758-5090/aaa15d. 025003. [DOI] [PubMed] [Google Scholar]
- [39].Das S, Pati F, Choi YJ, Rijal G, Shim JH, Kim SW, Ray AR, Cho DW, Ghosh S. Acta Biomater. 2015;11:233. doi: 10.1016/j.actbio.2014.09.023. [DOI] [PubMed] [Google Scholar]
- [40].Colosi C, Shin SR, Manoharan V, Massa S, Costantini M, Barbetta A, Dokmeci MR, Dentini M, Khademhosseini A. Adv Mater. 2016;28:677. doi: 10.1002/adma.201503310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Duan B, Kapetanovic E, Hockaday LA, Butcher JT. Acta Biomater. 2014;10 doi: 10.1016/j.actbio.2013.12.005. 1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Levato R, Visser J, Planell JA, Engel E, Malda J, Mateos-Timoneda MA. Biofabrication. 2014;6 doi: 10.1088/1758-5082/6/3/035020. 035020. [DOI] [PubMed] [Google Scholar]
- [43].Rutz AL, Hyland KE, Jakus AE, Burghardt WR, Shah RN. Adv Mater. 2015;27 doi: 10.1002/adma.201405076. 1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jia W, Gungor-Ozkerim PS, Zhang YS, Yue K, Zhu K, Liu W, Pi Q, Byambaa B, Dokmeci MR, Shin SR, Khademhosseini A. Biomaterials. 2016;106:58. doi: 10.1016/j.biomaterials.2016.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bertlein S, Brown G, Lim KS, Jungst T, Boeck T, Blunk T, Tessmar J, Hooper GJ, Woodfield TBF, Groll J. Adv Mater. 2017;29 doi: 10.1002/adma.201703404. 1703404. [DOI] [PubMed] [Google Scholar]
- [46].Tytgat L, Van Damme L, Van Hoorick J, Declercq H, Thienpont H, Ottevaere H, Blondeel P, Dubruel P, Van Vlierberghe S. Acta Biomater. 2019;94:340. doi: 10.1016/j.actbio.2019.05.062. [DOI] [PubMed] [Google Scholar]
- [47].Serban MA, Scott A, Prestwich GD. Curr Protoc Cell Biol. 2008;40 doi: 10.1002/0471143030.cb1014s40. 10.14.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Skardal A, Zhang J, Prestwich GD. Biomaterials. 2010;31 doi: 10.1016/j.biomaterials.2010.04.045. 6173. [DOI] [PubMed] [Google Scholar]
- [49].Sakai S, Ohi H, Hotta T, Kamei H, Taya M. Biopolymers. 2018;109 doi: 10.1002/bip.23080. e23080. [DOI] [PubMed] [Google Scholar]
- [50].AnilKumar S, Allen SC, Tasnim N, Akter T, Park S, Kumar A, Chattopadhyay M, Ito Y, Suggs LJ, Joddar B. J Biomed Mater Res, Part B. 2019;107:314. doi: 10.1002/jbm.b.34123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue HJ, Ramadan MH, Hudson AR, Feinberg AW. Sci Adv. 2015;1 doi: 10.1126/sciadv.1500758. e1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Rhee S, Puetzer JL, Mason BN, Reinhart-King CA, Bonassar LJ. ACS Biomater Sci Eng. 2016;2 doi: 10.1021/acsbiomaterials.6b00288. 1800. [DOI] [PubMed] [Google Scholar]
- [53].Lee A, Hudson AR, Shiwarski DJ, Tashman JW, Hinton TJ, Yerneni S, Bliley JM, Campbell PG, Feinberg AW. Science. 2019;365:482. doi: 10.1126/science.aav9051. [DOI] [PubMed] [Google Scholar]
- [54].Diamantides N, Wang L, Pruiksma T, Siemiatkoski J, Dugopolski C, Shortkroff S, Kennedy S, Bonassar LJ. Biofabrication. 2017;9 doi: 10.1088/1758-5090/aa780f. 034102. [DOI] [PubMed] [Google Scholar]
- [55].Kim YB, Lee H, Kim GH. ACS Appl Mater Interfaces. 2016;8 doi: 10.1021/acsami.6b11669. 32230. [DOI] [PubMed] [Google Scholar]
- [56].Melke J, Midha S, Ghosh S, Ito K, Hofmann S. Acta Biomater. 2016;31:1. doi: 10.1016/j.actbio.2015.09.005. [DOI] [PubMed] [Google Scholar]
- [57].Rodriguez MJ, Brown J, Giordano J, Lin SJ, Omenetto FG, Kaplan DL. Biomaterials. 2017;117:105. doi: 10.1016/j.biomaterials.2016.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Dhyani V, Singh N. ACS Appl Mater Interfaces. 2014;6 doi: 10.1021/am4060595. 5005. [DOI] [PubMed] [Google Scholar]
- [59].Wohlrab S, Müller S, Schmidt A, Neubauer S, Kessler H, Leal-Egaña A, Scheibel T. Biomaterials. 2012;33 doi: 10.1016/j.biomaterials.2012.05.069. 6650. [DOI] [PubMed] [Google Scholar]
- [60].Gu Q, Tomaskovic-Crook E, Lozano R, Chen Y, Kapsa RM, Zhou Q, Wallace GG, Crook JM. Adv Healthcare Mater. 2016;5 doi: 10.1002/adhm.201600095. 1429. [DOI] [PubMed] [Google Scholar]
- [61].Daly AC, Critchley SE, Rencsok EM, Kelly DJ. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/4/045002. 045002. [DOI] [PubMed] [Google Scholar]
- [62].Nguyen D, Hgg DA, Forsman A, Ekholm J, Nimkingratana P, Brantsing C, Kalogeropoulos T, Zaunz S, Concaro S, Brittberg M, Lindahl A, Gatenholm P, Enejder A, Simonsson S. Sci Rep. 2017;7:658. doi: 10.1038/s41598-017-00690-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Müller M, Öztürk E, Arlov Ø, Gatenholm P, Zenobi-Wong M. Ann Biomed Eng. 2017;45:210. doi: 10.1007/s10439-016-1704-5. [DOI] [PubMed] [Google Scholar]
- [64].Cooper MHHG, Bartolo PJDS. Int J Bioprint. 2019;5:12. doi: 10.18063/ijb.v5i2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Xu X, Jha AK, Harrington DA, Farach-Carson MC, Jia X. Soft Matter. 2012;8 doi: 10.1039/C2SM06463D. 3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Pescosolido L, Schuurman W, Malda J, Matricardi P, Alhaique F, Coviello T, Van Weeren PR, Dhert WJA, Hennink WE, Vermonden T. Biomacromolecules. 2011;12 doi: 10.1021/bm200178w. 1831. [DOI] [PubMed] [Google Scholar]
- [67].Kesti M, Müller M, Becher J, Schnabelrauch M, D’Este M, Eglin D, Zenobi-Wong M. Acta Biomater. 2015;11:162. doi: 10.1016/j.actbio.2014.09.033. [DOI] [PubMed] [Google Scholar]
- [68].Ouyang L, Highley CB, Rodell CB, Sun W, Burdick JA. ACS Biomater Sci Eng. 2016;2 doi: 10.1021/acsbiomaterials.6b00158. 1743. [DOI] [PubMed] [Google Scholar]
- [69].Poldervaart MT, Goversen B, De Ruijter M, Abbadessa A, Melchels FPW, Öner FC, Dhert WJA, Vermonden T, Alblas J. PLoS One. 2017;12 doi: 10.1371/journal.pone.0177628. e0177628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Shim JH, Jang KM, Hahn SK, Park JY, Jung H, Oh K, Park KM, Yeom J, Park SH, Kim SW, Wang JH, Kim K, Cho DW. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/1/014102. 014102. [DOI] [PubMed] [Google Scholar]
- [71].Jansson PE, Lindberg B, Sandford PA. Carbohydr Res. 1983;124:135. [Google Scholar]
- [72].Mouser VHM, Melchels FPW, Visser J, Dhert WJA, Gawlitta D, Malda J. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/3/035003. 35003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Markstedt K, Mantas A, Tournier I, Martínez Ávila H, Hägg D, Gatenholm P. Biomacromolecules. 2015;16 doi: 10.1021/acs.biomac.5b00188. 1489. [DOI] [PubMed] [Google Scholar]
- [74].Martínez Ávila H, Schwarz S, Rotter N, Gatenholm P. Bioprinting. 2016;22:1–2. [Google Scholar]
- [75].Henriksson I, Gatenholm P, Hägg DA. Biofabrication. 2017;9 doi: 10.1088/1758-5090/aa5c1c. 015022. [DOI] [PubMed] [Google Scholar]
- [76].Geng L, Feng W, Hutmacher DW, Wong YS, Loh HT, Fuh JYH. Rapid Prototyping J. 2005;11:90. [Google Scholar]
- [77].Jiang T, James R, Kumbar SG, Laurencin CT. Kumbar SG, Laurencin CT, Deng M. Natural and Synthetic Biomedical Polymers. 1. Elsevier; Oxford, UK: 2014. pp. 91–113. [Google Scholar]
- [78].Freier T, Koh HS, Kazazian K, Shoichet MS. Biomaterials. 2005;26 doi: 10.1016/j.biomaterials.2005.02.033. 5872. [DOI] [PubMed] [Google Scholar]
- [79].Kumar MNVR, Muzzarelli RAA, Muzzarelli C, Sashiwa H, Domb AJ. Chem Rev. 2004;104 doi: 10.1021/cr030441b. 6017. [DOI] [PubMed] [Google Scholar]
- [80].Bakarich SE, Balding P, Gorkin R, Spinks GM, M In Het Panhuis RSC Adv. 2014;4 38088. [Google Scholar]
- [81].Wilson SA, Cross LM, Peak CW, Gaharwar AK. ACS Appl Mater Interfaces. 2017;9 doi: 10.1021/acsami.7b13602. 43449. [DOI] [PubMed] [Google Scholar]
- [82].Mihaila SM, Gaharwar AK, Reis RL, Marques AP, Gomes ME, Khademhosseini A. Adv Healthcare Mater. 2013;2:895. doi: 10.1002/adhm.201200317. [DOI] [PubMed] [Google Scholar]
- [83].Thakur A, Jaiswal MK, Peak CW, Carrow JK, Gentry J, Dolatshahi-Pirouz A, Gaharwar AK. Nanoscale. 2016;8 doi: 10.1039/c6nr02299e. 12362. [DOI] [PubMed] [Google Scholar]
- [84].Hughes CS, Postovit LM, Lajoie GA. Proteomics. 2010;10 doi: 10.1002/pmic.200900758. 1886. [DOI] [PubMed] [Google Scholar]
- [85].Pati F, Jang J, Ha DH, Won Kim S, Rhie JW, Shim JH, Kim DH, Cho DW. Nat Commun. 2014;5 doi: 10.1038/ncomms4935. 3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Jang J, Kim TG, Kim BS, Kim SW, Kwon SM, Cho DW. Acta Biomater. 2016;33:88. doi: 10.1016/j.actbio.2016.01.013. [DOI] [PubMed] [Google Scholar]
- [87].Jang J, Park HJ, Kim SW, Kim H, Park JY, Na SJ, Kim HJ, Park MN, Choi SH, Park SH, Kim SW, Kwon SM, Kim PJ, Cho DW. Biomaterials. 2017;112:264. doi: 10.1016/j.biomaterials.2016.10.026. [DOI] [PubMed] [Google Scholar]
- [88].Noor N, Shapira A, Edri R, Gal I, Wertheim L, Dvir T. Adv Sci; 1900344; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Pati F, Ha DH, Jang J, Han HH, Rhie JW, Cho DW. Biomaterials. 2015;62:164. doi: 10.1016/j.biomaterials.2015.05.043. [DOI] [PubMed] [Google Scholar]
- [90].Gao G, Lee JH, Jang J, Lee DH, Kong JS, Kim BS, Choi YJ, Jang WB, Hong YJ, Kwon SM, Cho DW. Adv Funct Mater. 2017;27 1700798. [Google Scholar]
- [91].Choi YJ, Kim TG, Jeong J, Yi HG, Park JW, Hwang W, Cho DW. Adv Healthcare Mater. 2016;5 doi: 10.1002/adhm.201600483. 2636. [DOI] [PubMed] [Google Scholar]
- [92].Lee H, Han W, Kim H, Ha DH, Jang J, Kim BS, Cho DW. Biomacromolecules. 2017;18 doi: 10.1021/acs.biomac.6b01908. 1229. [DOI] [PubMed] [Google Scholar]
- [93].Müller M, Becher J, Schnabelrauch M, Zenobi-Wong M. Biofabrication. 2015;7 doi: 10.1088/1758-5090/7/3/035006. 035006. [DOI] [PubMed] [Google Scholar]
- [94].Xavier JR, Thakur T, Desai P, Jaiswal MK, Sears N, Cosgriff-Hernandez E, Kaunas R, Gaharwar AK. ACS Nano. 2015;9 doi: 10.1021/nn507488s. 3109. [DOI] [PubMed] [Google Scholar]
- [95].Chimene D, Peak CW, Gentry JL, Carrow JK, Cross LM, Mondragon E, Cardoso GB, Kaunas R, Gaharwar AK. ACS Appl Mater Interfaces. 2018;10 doi: 10.1021/acsami.7b19808. 9957. [DOI] [PubMed] [Google Scholar]
- [96].Thompson DW, Butterworth JT. J Colloid Interface Sci. 1992;151:236. [Google Scholar]
- [97].Bose S, Tarafder S, Bandyopadhyay A. Ann Biomed Eng. 2017;45:261. doi: 10.1007/s10439-016-1646-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].a) Ahlfeld T, Cidonio G, Kilian D, Duin S, Akkineni AR, Dawson JI, Yang S, Lode A, Oreffo ROC, Gelinsky M. Biofabrication. 2017;9 doi: 10.1088/1758-5090/aa7e96. 034103. [DOI] [PubMed] [Google Scholar]; b) Peak CW, Singh KA, Adlouni M, Chen J, Gaharwar AK, editors. Adv Healthcare Mater. 2019;8 doi: 10.1002/adhm.201801553. 1801553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Hockaday LA, Kang KH, Colangelo NW, Cheung PYC, Duan B, Malone E, Wu J, Girardi LN, Bonassar LJ, Lipson H, Chu CC, Butcher JT. Biofabrication. 2012;4 doi: 10.1088/1758-5082/4/3/035005. 35005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Stichler S, Böck T, Paxton N, Bertlein S, Levato R, Schill V, Smolan W, Malda J, Teßmar J, Blunk T, Groll J. Biofabrication. 2017;9 doi: 10.1088/1758-5090/aa8cb7. 044108. [DOI] [PubMed] [Google Scholar]
- [101].Daly AC, Cunniffe GM, Sathy BN, Jeon O, Alsberg E, Kelly DJ. Adv Healthcare Mater. 2016;5 doi: 10.1002/adhm.201600182. 2353. [DOI] [PubMed] [Google Scholar]
- [102].Visser J, Melchels FPW, Jeon JE, Van Bussel EM, Kimpton LS, Byrne HM, Dhert WJA, Dalton PD, Hutmacher DW, Malda J. Nat Commun. 2015;6 doi: 10.1038/ncomms7933. 6933. [DOI] [PubMed] [Google Scholar]
- [103].de Ruijter M, Hrynevich A, Haigh JN, Hochleitner G, Castilho M, Groll J, Malda J, Dalton PD. Small. 2018;14 doi: 10.1002/smll.201702773. 1702773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].de Ruijter M, Ribeiro A, Dokter I, Castilho M, Malda J. Adv Healthcare Mater. 2019;8 doi: 10.1002/adhm.201800418. 1800418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Schuurman W, Khristov V, Pot MW, Van Weeren PR, Dhert WJA, Malda J. Biofabrication. 2011;3 doi: 10.1088/1758-5082/3/2/021001. 021001. [DOI] [PubMed] [Google Scholar]
- [106].Mouser VHM, Abbadessa A, Levato R, Hennink WE, Vermonden T, Gawlitta D, Malda J. Biofabrication. 2017;9 doi: 10.1088/1758-5090/aa6265. 015026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Melchels FPW, Blokzijl MM, Levato R, Peiffer QC, de Ruijter M, Hennink WE, Vermonden T, Malda J. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/3/035004. 035004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Ahlfeld T, Doberenz F, Kilian D, Vater C, Korn P, Lauer G, Lode A, Gelinsky M. Biofabrication. 2018;10 doi: 10.1088/1758-5090/aad36d. 045002. [DOI] [PubMed] [Google Scholar]
- [109].Kloxin AM, Kloxin CJ, Bowman CN, Anseth KS. Adv Mater. 2010;22 doi: 10.1002/adma.200904179. 3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Mũnoz Z, Shih H, Lin CC. Biomater Sci. 2014;2 doi: 10.1039/c4bm00070f. 1063. [DOI] [PubMed] [Google Scholar]
- [111].Ooi HW, Mota C, Tessa Ten Cate A, Calore A, Moroni L, Baker MB. Biomacromolecules. 2018;19 doi: 10.1021/acs.biomac.8b00696. 3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Xin S, Chimene D, Garza JE, Gaharwar AK, Alge DL. Biomater Sci. 2019;7 doi: 10.1039/c8bm01286e. 1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Cui X, Breitenkamp K, Finn MG, Lotz M, ’lima DDD. Tissue Eng, Part A. 2012;18 doi: 10.1089/ten.tea.2011.0543. 1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].O’Connell CD, Di Bella C, Thompson F, Augustine C, Beirne S, Cornock R, Richards CJ, Chung J, Gambhir S, Yue Z, Bourke J, Zhang B, Taylor A, Quigley A, Kapsa R, Choong P, Wallace GG. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/1/015019. 015019. [DOI] [PubMed] [Google Scholar]
- [115].Fedorovich NE, Swennen I, Girones J, Moroni L, Van Blitterswijk CA, Schacht E, Alblas J, Dhert WJA. Biomacromolecules. 2009;10 doi: 10.1021/bm801463q. 1689. [DOI] [PubMed] [Google Scholar]
- [116].Lim KS, Schon BS, Mekhileri NV, Brown GCJ, Chia CM, Prabakar S, Hooper GJ, Woodfield TBF. ACS Biomater Sci Eng. 2016;2 doi: 10.1021/acsbiomaterials.6b00149. 1752. [DOI] [PubMed] [Google Scholar]
- [117].Bertassoni LE, Cardoso JC, Manoharan V, Cristino AL, Bhise NS, Araujo WA, Zorlutuna P, Vrana NE, Ghaemmaghami AM, Dokmeci MR, Khademhosseini A. Biofabrication. 2014;6 doi: 10.1088/1758-5082/6/2/024105. 024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Billiet T, Gevaert E, De Schryver T, Cornelissen M, Dubruel P. Biomaterials. 2014;35:49. doi: 10.1016/j.biomaterials.2013.09.078. [DOI] [PubMed] [Google Scholar]
- [119].Stichler S, Jungst T, Schamel M, Zilkowski I, Kuhlmann M, Böck T, Blunk T, Teßmar J, Groll JJ. Ann Biomed Eng. 2017;45:273. doi: 10.1007/s10439-016-1633-3. [DOI] [PubMed] [Google Scholar]
- [120].Dahle J, Kvam E, Stokke T. J Carcinog. 2005;4:11. doi: 10.1186/1477-3163-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Ruskowitz ER, Deforest CA. ACS Biomater Sci Eng. 2019;5 doi: 10.1021/acsbiomaterials.9b00177. 2111. [DOI] [PubMed] [Google Scholar]
- [122].Fedorovich NE, Oudshoorn MH, van Geemen D, Hennink WE, Alblas J, Dhert WJA. Biomaterials. 2009;30:344. doi: 10.1016/j.biomaterials.2008.09.037. [DOI] [PubMed] [Google Scholar]
- [123].Lin H, Zhang D, Alexander PG, Yang G, Tan J, Cheng AWM, Tuan RS. Biomaterials. 2013;34:331. doi: 10.1016/j.biomaterials.2012.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Shih H, Lin CC. Macromol Rapid Commun. 2013;34:269. doi: 10.1002/marc.201200605. [DOI] [PubMed] [Google Scholar]
- [125].Lim KS, Klotz BJ, Lindberg GCJ, Melchels FPW, Hooper GJ, Malda J, Gawlitta D, Woodfield TBF. Macromol Biosci. 2019;19 doi: 10.1002/mabi.201900098. e1900098. [DOI] [PubMed] [Google Scholar]
- [126].Zehnder T, Sarker B, Boccaccini AR, Detsch R. Biofabrication. 2015;7 doi: 10.1088/1758-5090/7/2/025001. 25001. [DOI] [PubMed] [Google Scholar]
- [127].Sarker B, Papageorgiou DG, Silva R, Zehnder T, Gul-E-Noor F, Bertmer M, Kaschta J, Chrissafis K, Detsch R, Boccaccini AR. J Mater Chem B. 2014;2 doi: 10.1039/c3tb21509a. 1470. [DOI] [PubMed] [Google Scholar]
- [128].Hafeez S, Ooi H, Morgan F, Mota C, Dettin M, van Blitters wijk C, Moroni L, Baker M. Gels. 2018;4:85. doi: 10.3390/gels4040085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Wang LL, Highley CB, Yeh YC, Galarraga JH, Uman S, Burdick JA. J Biomed Mater Res, Part A. 2018;106:865. doi: 10.1002/jbm.a.36323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Seiffert S, Sprakel J. Chem Soc Rev. 2012;41:909. doi: 10.1039/c1cs15191f. [DOI] [PubMed] [Google Scholar]
- [131].Shoulders MD, Raines RT. Annu Rev Biochem. 2009;78:929. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].MacEwan SR, Chilkoti A. Biopolymers. 2010;94:60. doi: 10.1002/bip.21327. [DOI] [PubMed] [Google Scholar]
- [133].Marx V. Nat Methods. 2019;16:365. doi: 10.1038/s41592-019-0399-6. [DOI] [PubMed] [Google Scholar]
- [134].Loo Y, Hauser CAE. Biomed Mater. 2015;11 doi: 10.1088/1748-6041/11/1/014103. 14103. [DOI] [PubMed] [Google Scholar]
- [135].Celikkin N, Padial JS, Costantini M, Hendrikse H, Cohn R, Wilson CJ, Rowan AE, Święszkowski W. Polymers. 2018;10:555. doi: 10.3390/polym10050555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Raphael B, Khalil T, Workman VL, Smith A, Brown CP, Streuli C, Saiani A, Domingos M. Mater Lett. 2017;190:103. [Google Scholar]
- [137].Panwar A, Tan LP. Molecules. 2016;21:685. [Google Scholar]
- [138].Schacht K, Jüngst T, Schweinlin M, Ewald A, Groll J, Scheibel T. Angew Chem, Int Ed. 2015;54 doi: 10.1002/anie.201409846. 2816. [DOI] [PubMed] [Google Scholar]
- [139].Highley CB, Song KH, Daly AC, Burdick JA. Adv Sci. 2019;6 doi: 10.1002/advs.201801076. 1801076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Song KH, Highley CB, Rouff A, Burdick JA. Adv Funct Mater. 2018;28 1801331. [Google Scholar]
- [141].Rodell CB, Kaminski AL, Burdick JA. Biomacromolecules. 2013;14 doi: 10.1021/bm401280z. 4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Dubbin K, Hori Y, Lewis KK, Heilshorn SC. Adv Healthcare Mater. 2016;5 doi: 10.1002/adhm.201600636. 2488. [DOI] [PubMed] [Google Scholar]
- [143].Strivens TA. In: Paint Surf Coatings. Walters K, editor. Elsevier; Amsterdam, The Netherlands: 1999. pp. 550–574. [Google Scholar]
- [144].Moreno R. Adv Appl Ceram. 2012;111:246. [Google Scholar]
- [145].Schuurman W, Levett PA, Pot MW, van Weeren PR, Dhert WJA, Hutmacher DW, Melchels FPW, Klein TJ, Malda J. Macromol Biosci. 2013;13:551. doi: 10.1002/mabi.201200471. [DOI] [PubMed] [Google Scholar]
- [146].Melchels FPW, Dhert WJA, Hutmacher DW, Malda J. J Mater Chem B. 2014;2 doi: 10.1039/c3tb21280g. 2282. [DOI] [PubMed] [Google Scholar]
- [147].Hong S, Sycks D, Chan HFa, Lin S, Lopez GP, Guilak F, Leong KW, Zhao X. Adv Mater. 2015;27 doi: 10.1002/adma.201570182. 4034. [DOI] [PubMed] [Google Scholar]
- [148].Gao Q, He Y, zhong Fu J, Liu A, Ma L. Biomaterials. 2015;61:203. doi: 10.1016/j.biomaterials.2015.05.031. [DOI] [PubMed] [Google Scholar]
- [149].Costantini M, Idaszek J, Szoke K, Jaroszewicz J, Dentini M, Barbetta A, Brinchmann JE, Święszkowski W. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/3/035002. 035002. [DOI] [PubMed] [Google Scholar]
- [150].Mistry P, Aied A, Alexander M, Shakesheff K, Bennett A, Yang J. Macromol Biosci. 2017;17 doi: 10.1002/mabi.201600472. 1600472. [DOI] [PubMed] [Google Scholar]
- [151].Luo Y, Wu C, Lode A, Gelinsky M. Biofabrication. 2013;5 doi: 10.1088/1758-5082/5/1/015005. 015005. [DOI] [PubMed] [Google Scholar]
- [152].Zhu K, Shin SR, van Kempen T, Li YC, Ponraj V, Nasajpour A, Mandla S, Hu N, Liu X, Leijten J, Lin YD, Hussain MA, Zhang YS, Tamayol A, Khademhosseini A. Adv Funct Mater. 2017;27 doi: 10.1002/adfm.201605352. 1605352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Onoe H, Okitsu T, Itou A, Kato-Negishi M, Gojo R, Kiriya D, Sato K, Miura S, Iwanaga S, Kuribayashi-Shigetomi K, Matsunaga YT, Shimoyama Y, Takeuchi S. Nat Mater. 2013;12:584. doi: 10.1038/nmat3606. [DOI] [PubMed] [Google Scholar]
- [154].Yeo MG, Lee JS, Chun W, Kim GH. Biomacromolecules. 2016;17 doi: 10.1021/acs.biomac.5b01764. 1365. [DOI] [PubMed] [Google Scholar]
- [155].Kim G, Ahn S, Kim Y, Cho Y, Chun W. J Mater Chem. 2011;21 6165. [Google Scholar]
- [156].Liu W, Zhong Z, Hu N, Zhou Y, Maggio L, Miri AK, Fragasso A, Jin X, Khademhosseini A, Zhang YS. Biofabrication. 2018;10 doi: 10.1088/1758-5090/aa9d44. 024102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Asakawa N, Shimizu T, Tsuda Y, Sekiya S, Sasagawa T, Yamato M, Fukai F, Okano T. Biomaterials. 2010;31 doi: 10.1016/j.biomaterials.2010.01.105. 3903. [DOI] [PubMed] [Google Scholar]
- [158].Hasan A, Paul A, Vrana NE, Zhao X, Memic A, Hwang YS, Dokmeci MR, Khademhosseini A. Biomaterials. 2014;35 doi: 10.1016/j.biomaterials.2014.04.091. 7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Lee VK, Kim DY, Ngo H, Lee Y, Seo L, Yoo SS, Vincent PA, Dai G. Biomaterials. 2014;35 doi: 10.1016/j.biomaterials.2014.05.083. 8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DHT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Nat Mater. 2012;11:768. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Luo Y, Lode A, Gelinsky M. Adv Healthcare Mater. 2013;2:777. doi: 10.1002/adhm.201200303. [DOI] [PubMed] [Google Scholar]
- [162].Gao G, Park JY, Kim BS, Jang J, Cho DW. Adv Healthcare Mater. 2018;7 doi: 10.1002/adhm.201801102. 1801102. [DOI] [PubMed] [Google Scholar]
- [163].Serex L, Bertsch A, Renaud P. Micromachines. 2018;9:86. doi: 10.3390/mi9020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Liu W, Zhang YS, Heinrich MA, De Ferrari F, Jang HL, Bakht SM, Alvarez MM, Yang J, Li YC, Trujillo-de Santiago G, Miri AK, Zhu K, Khoshakhlagh P, Prakash G, Cheng H, Guan X, Zhong Z, Ju J, Zhu GH, Jin X, Shin SR, Dokmeci MR, Khademhosseini A. Adv Mater. 2017;29 doi: 10.1002/adma.201604630. 1604630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Knight JB, Vishwanath A, Brody JP, Austin RH. Phys Rev Lett. 1998;80 3863. [Google Scholar]
- [166].Attalla R, Puersten E, Jain N, Selvaganapathy PR. Biofabrication. 2019;11 doi: 10.1088/1758-5090/aaf7c7. 15012. [DOI] [PubMed] [Google Scholar]
- [167].Duarte Campos DF, Blaeser A, Weber M, Jäkel J, Neuss S, Jahnen-Dechent W, Fischer H. Biofabrication. 2013;5 doi: 10.1088/1758-5082/5/1/015003. 015003. [DOI] [PubMed] [Google Scholar]
- [168].Bhattacharjee T, Zehnder SM, Rowe KG, Jain S, Nixon RM, Sawyer WG, Angelini TE. Sci Adv. 2015;1 doi: 10.1126/sciadv.1500655. e1500655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Highley CB, Rodell CB, Burdick JA. Adv Mater. 2015;27 doi: 10.1002/adma.201501234. 5075. [DOI] [PubMed] [Google Scholar]
- [170].Cooke ME, Jones SW, ter Horst B, Moiemen N, Snow M, Chouhan G, Hill LJ, Esmaeli M, Moakes RJA, Holton J, Nandra R, Williams RL, Smith AM, Grover LM. Adv Mater. 2018;30 doi: 10.1002/adma.201705013. 1705013. [DOI] [PubMed] [Google Scholar]
- [171].Piau JM. J Non-Newtonian Fluid Mech. 2007;144:1. [Google Scholar]
- [172].Ketz RJ, Prud’homme RK, Graessley WW. Rheol Acta. 1988;27:531. [Google Scholar]
- [173].Carnali JO, Naser MS. Colloid Polym Sci. 1992;270:183. [Google Scholar]
- [174].Moller P, Fall A, Chikkadi V, Derks D, Bonn D. Philos Trans R Soc, A. 2009;367 doi: 10.1098/rsta.2009.0194. 5139. [DOI] [PubMed] [Google Scholar]
- [175].Bhattacharjee T, Gil CJ, Marshall SL, Urueña JM, O’Bryan CS, Carstens M, Keselowsky B, Palmer GD, Ghivizzani S, Gibbs CP, Sawyer WG, Angelini TE. ACS Biomater Sci Eng. 2016;2 doi: 10.1021/acsbiomaterials.6b00218. 1787. [DOI] [PubMed] [Google Scholar]
- [176].Holland S, Tuck C, Foster T. Food Biophys. 2018;13:175. doi: 10.1007/s11483-018-9523-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Moxon SR, Cooke ME, Cox SC, Snow M, Jeys L, Jones SW, Smith AM, Grover LM. Adv Mater. 2017;29 doi: 10.1002/adma.201605594. 1605594. [DOI] [PubMed] [Google Scholar]
- [178].Ding H, Chang RC. Appl Sci. 2018;8:403. [Google Scholar]
- [179].Jin Y, Chai W, Huang Y. Mater Sci Eng, C. 2017;80:313. doi: 10.1016/j.msec.2017.05.144. [DOI] [PubMed] [Google Scholar]
- [180].Jin Y, Compaan A, Chai W, Huang Y. ACS Appl Mater Interfaces. 2017;9 doi: 10.1021/acsami.7b02398. 20057. [DOI] [PubMed] [Google Scholar]
- [181].Jin Y, Liu C, Chai W, Compaan A, Huang Y. ACS Appl Mater Interfaces. 2017;9 doi: 10.1021/acsami.7b03613. 17456. [DOI] [PubMed] [Google Scholar]
- [182].Laronda MM, Rutz AL, Xiao S, Whelan KA, Duncan FE, Roth EW, Woodruff TK, Shah RN. Nat Commun. 2017;8 doi: 10.1038/ncomms15261. 15261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Benam KH, Villenave R, Lucchesi C, Varone A, Hubeau C, Lee HH, Alves SE, Salmon M, Ferrante TC, Weaver JC, Bahinski A, Hamilton GA, Ingber DE. Nat Methods. 2016;13:151. doi: 10.1038/nmeth.3697. [DOI] [PubMed] [Google Scholar]
- [184].Hassell BA, Goyal G, Lee E, Sontheimer-Phelps A, Levy O, Chen CS, Ingber DE. Cell Rep. 2017;21:508. doi: 10.1016/j.celrep.2017.09.043. [DOI] [PubMed] [Google Scholar]
- [185].Maoz BM, Herland A, Fitzgerald EA, Grevesse T, Vidoudez C, Pacheco AR, Sheehy SP, Park TE, Dauth S, Mannix R, Budnik N, Shores K, Cho A, Nawroth JC, Segrè D, Budnik B, Ingber DE, Parker KK. Nat Biotechnol. 2018;36:865. doi: 10.1038/nbt.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Bein A, Shin W, Jalili-Firoozinezhad S, Park MH, Sontheimer-Phelps A, Tovaglieri A, Chalkiadaki A, Kim HJ, Ingber DE. Cell Mol Gastroenterol Hepatol. 2018;5:659. doi: 10.1016/j.jcmgh.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Chen MB, Whisler JA, Fröse J, Yu C, Shin Y, Kamm RD. Nat Protoc. 2017;12:865. doi: 10.1038/nprot.2017.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Satoh T, Sugiura S, Shin K, Onuki-Nagasaki R, Ishida S, Kikuchi K, Kakiki M, Kanamori T. Lab Chip. 2017;18:115. doi: 10.1039/c7lc00952f. [DOI] [PubMed] [Google Scholar]
- [189].Khademhosseini A, Langer R. Nat Protoc. 2016;11 doi: 10.1038/nprot.2016.123. 1775. [DOI] [PubMed] [Google Scholar]
- [190].Zhang L, Chen S, Liang R, Chen Y, Li S, Li S, Sun Z, Wang Y, Li G, Ming A, Yang Y. J Biomed Mater Res, Part A. 2018;106 doi: 10.1002/jbm.a.36507. 3123. [DOI] [PubMed] [Google Scholar]
- [191].Martínez E, Engel E, Planell JA, Samitier J. Ann Anat. 2009;191:126. doi: 10.1016/j.aanat.2008.05.006. [DOI] [PubMed] [Google Scholar]
- [192].Dalby MJ, Hart A, Yarwood SJ. Biomaterials. 2008;29:282. doi: 10.1016/j.biomaterials.2007.09.030. [DOI] [PubMed] [Google Scholar]
- [193].Barba A, Diez-Escudero A, Maazouz Y, Rappe K, Espanol M, Montufar EB, Bonany M, Sadowska JM, Guillem-Marti J, Öhman-Mägi C, Persson C, Manzanares MC, Franch J, Ginebra MP. ACS Appl Mater Interfaces. 2017;9 doi: 10.1021/acsami.7b14175. 41722. [DOI] [PubMed] [Google Scholar]
- [194].Ratner BD. Regener Biomater. 2016;3:107. doi: 10.1093/rb/rbw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Lewis PL, Green RM, Shah RN. Acta Biomater. 2018;69:63. doi: 10.1016/j.actbio.2017.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Ferlin KM, Prendergast ME, Miller ML, Kaplan DS, Fisher JP. Acta Biomater. 2016;32:161. doi: 10.1016/j.actbio.2016.01.007. [DOI] [PubMed] [Google Scholar]
- [197].Almeida CR, Serra T, Oliveira MI, Planell JA, Barbosa MA, Navarro M. Acta Biomater. 2014;10:613. doi: 10.1016/j.actbio.2013.10.035. [DOI] [PubMed] [Google Scholar]
- [198].Neal RA, Jean A, Park H, Wu PB, Hsiao J, Engelmayr GC, Langer R, Freed LE. Tissue Eng, Part A. 2013;19:793. doi: 10.1089/ten.tea.2012.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Di Luca A, Lorenzo-Moldero I, Mota C, Lepedda A, Auhl D, Van Blitterswijk C, Moroni L. Adv Healthcare Mater. 2016;5 doi: 10.1002/adhm.201600083. 1753. [DOI] [PubMed] [Google Scholar]
- [200].Eichholz KF, Hoey DA. Acta Biomater. 2018;75:140. doi: 10.1016/j.actbio.2018.05.048. [DOI] [PubMed] [Google Scholar]
- [201].Daly AC, Kelly DJ. Biomaterials. 2019;197:194. doi: 10.1016/j.biomaterials.2018.12.028. [DOI] [PubMed] [Google Scholar]
- [202].Koffler J, Zhu W, Qu X, Platoshyn O, Dulin JN, Brock J, Graham L, Lu P, Sakamoto J, Marsala M, Chen S, Tuszynski MH. Nat Med. 2019;25:263. doi: 10.1038/s41591-018-0296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Castilho M, Feyen D, Flandes-Iparraguirre M, Hochleitner G, Groll J, Doevendans PAF, Vermonden T, Ito K, Sluijter JPG, Malda J. Adv Healthcare Mater. 2017;6 doi: 10.1002/adhm.201700311. 1700311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Izadifar M, Babyn P, Kelly ME, Chapman D, Chen X. Tissue Eng, Part C. 2017;23:548. doi: 10.1089/ten.TEC.2017.0222. [DOI] [PubMed] [Google Scholar]
- [205].Ying GL, Jiang N, Maharjan S, Yin YX, Chai RR, Cao X, Yang JZ, Miri AK, Hassan S, Zhang YS. Adv Mater. 2018;30 doi: 10.1002/adma.201805460. 1805460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Mozetic P, Giannitelli SM, Gori M, Trombetta M, Rainer A. J Biomed Mater Res, Part A. 2017;105 doi: 10.1002/jbm.a.36117. 2582. [DOI] [PubMed] [Google Scholar]
- [207].Kim JH, Seol YJ, Ko IK, Kang HW, Lee YK, Yoo JJ, Atala A, Lee SJ. Sci Rep. 2018;8 doi: 10.1038/s41598-018-29968-5. 12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Omidinia-Anarkoli A, Boesveld S, Tuvshindorj U, Rose JC, Haraszti T, De Laporte L. Small. 2017;13 doi: 10.1002/smll.201702207. 1702207. [DOI] [PubMed] [Google Scholar]
- [209].Rose JC, Gehlen DB, Haraszti T, Köhler J, Licht CJ, De Laporte L. Biomaterials. 2018;163:128. doi: 10.1016/j.biomaterials.2018.02.001. [DOI] [PubMed] [Google Scholar]
- [210].Raney JR, Compton BG, Mueller J, Ober TJ, Shea K, Lewis JA. Proc Natl Acad Sci USA. 2018;115:1198. doi: 10.1073/pnas.1715157115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Siqueira G, Kokkinis D, Libanori R, Hausmann MK, Gladman AS, Neels A, Tingaut P, Zimmermann T, Lewis JA, Studart AR. Adv Funct Mater. 2017;27 1604619. [Google Scholar]
- [212].Hausmann MK, Rühs PA, Siqueira G, Läuger J, Libanori R, Zimmermann T, Studart AR. ACS Nano. 2018;12 doi: 10.1021/acsnano.8b02366. 6926. [DOI] [PubMed] [Google Scholar]
- [213].Kim H, Jang J, Park J, Lee KP, Lee S, Lee DM, Kim KH, Kim HK, Cho DW. Biofabrication. 2019;11 doi: 10.1088/1758-5090/ab1a8b. 035017. [DOI] [PubMed] [Google Scholar]
- [214].Ning L, Sun H, Lelong T, Guilloteau R, Zhu N, Schreyer DJ, Chen X. Biofabrication. 2018;10 doi: 10.1088/1758-5090/aacd30. 035014. [DOI] [PubMed] [Google Scholar]
- [215].Izadifar M, Chapman D, Babyn P, Chen X, Kelly ME. Tissue Eng, Part C. 2018;24:74. doi: 10.1089/ten.TEC.2017.0346. [DOI] [PubMed] [Google Scholar]
- [216].Li L, Eyckmans J, Chen CS. Nat Mater. 2017;16 doi: 10.1038/nmat5049. 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Madl CM, Heilshorn SC. Annu Rev Biomed Eng. 2018;20:21. doi: 10.1146/annurev-bioeng-062117-120954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Darnell M, Mooney DJ. Nat Mater. 2017;16 doi: 10.1038/nmat4991. 1178. [DOI] [PubMed] [Google Scholar]
- [219].Thomas D, O’Brien T, Pandit A. Adv Mater. 2018;30 doi: 10.1002/adma.201703948. 1703948. [DOI] [PubMed] [Google Scholar]
- [220].Ooi HW, Hafeez S, Van Blitterswijk CA, Moroni L, Baker MB. Mater Horiz. 2017;4 1020. [Google Scholar]
- [221].Huettner N, Dargaville TR, Forget A. Trends Biotechnol. 2018;36:372. doi: 10.1016/j.tibtech.2018.01.008. [DOI] [PubMed] [Google Scholar]
- [222].Schneeberger K, Spee B, Costa P, Sachs N, Clevers H, Malda J. Biofabrication. 2017;9 doi: 10.1088/1758-5090/aa6121. 013001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Zhao H, Chen Y, Shao L, Xie M, Nie J, Qiu J, Zhao P, Ramezani H, Fu J, Ouyang H, He Y. Small. 2018;14 doi: 10.1002/smll.201802630. 1602630. [DOI] [PubMed] [Google Scholar]
- [224].Caiazzo M, Okawa Y, Ranga A, Piersigilli A, Tabata Y, Lutolf MP. Nat Mater. 2016;15:344. doi: 10.1038/nmat4536. [DOI] [PubMed] [Google Scholar]
- [225].Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, Clevers H, Lutolf MP. Nature. 2016;539:560. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- [226].Vega SL, Kwon MY, Song KH, Wang C, Mauck RL, Han L, Burdick JA. Nat Commun. 2018;9:614. doi: 10.1038/s41467-018-03021-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Ma Y, Ji Y, Huang G, Ling K, Zhang X, Xu F. Biofabrication. 2015;7 doi: 10.1088/1758-5090/7/4/044105. 044105. [DOI] [PubMed] [Google Scholar]
- [228].Murphy SV, Skardal A, Atala A. J Biomed Mater Res, Part A. 2013;101(A):272. doi: 10.1002/jbm.a.34326. [DOI] [PubMed] [Google Scholar]
- [229].Echalier C, Levato R, Mateos-Timoneda MAA, Castaño O, Déjean S, Garric X, Pinese C, Noël D, Engel E, Martinez J, Mehdi A, Subra G. RSC Adv. 2017;7 [Google Scholar]
- [230].Choudhury D, Tun HW, Wang T, Naing MW. Trends Biotechnol. 2018;36:787. doi: 10.1016/j.tibtech.2018.03.003. [DOI] [PubMed] [Google Scholar]
- [231].Yu C, Ma X, Zhu W, Wang P, Miller KL, Stupin J, Koroleva-Maharajh A, Hairabedian A, Chen S. Biomaterials. 2019;194:1. doi: 10.1016/j.biomaterials.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Pati F, Cho DW. Methods Mol Biol. 2017;1612:381. doi: 10.1007/978-1-4939-7021-6_27. [DOI] [PubMed] [Google Scholar]
- [233].Yi HG, Jeong YH, Kim Y, Choi YJ, Moon HE, Park SH, Kang KS, Bae M, Jang J, Youn H, Paek SH, Cho DW. Nat Biomed Eng. 2019;3:509. doi: 10.1038/s41551-019-0363-x. [DOI] [PubMed] [Google Scholar]
- [234].Kim BS, Kwon YW, Kong JS, Park GT, Gao G, Han W, Kim MB, Lee H, Kim JH, Cho DW. Biomaterials. 2018;168:38. doi: 10.1016/j.biomaterials.2018.03.040. [DOI] [PubMed] [Google Scholar]
- [235].Park J, Lee K, Kim H, Park S, Wijesinghe RE, Lee J, Han S, Lee S, Kim P, Cho D, Jang J, Kim HK, Jeon M, Kim J. J Biophotonics. 2019:12. doi: 10.1002/jbio.201900098. e201900098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Rittié L. J Cell Commun Signaling. 2015;9:99. doi: 10.1007/s12079-015-0282-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Hsueh MF, Kraus VB, Önnerfjord P. Eur Cells Mater. 2017;34:70. doi: 10.22203/eCM.v034a05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Raghunathan VK, Benoit J, Kasetti R, Zode G, Salemi M, Phinney BS, Keller KE, Staverosky JA, Murphy CJ, Acott T, Vranka J. Acta Biomater. 2018;71:444. doi: 10.1016/j.actbio.2018.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Andreu I, Luque T, Sancho A, Pelacho B, Iglesias-García O, Melo E, Farré R, Prósper F, Elizalde MR, Navajas D. Acta Biomater. 2014;10 doi: 10.1016/j.actbio.2014.03.034. 3235. [DOI] [PubMed] [Google Scholar]
- [240].Skardal A, Smith L, Bharadwaj S, Atala A, Soker S, Zhang Y. Biomaterials. 2012;33 doi: 10.1016/j.biomaterials.2012.03.034. 4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Liu N, Huang S, Yao B, Xie J, Wu X, Fu X. Sci Rep. 2016;6 doi: 10.1038/srep34410. 34410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Madl CM, Lesavage BL, Dewi RE, Dinh CB, Stowers RS, Khariton M, Lampe KJ, Nguyen D, Chaudhuri O, Enejder A, Heilshorn SC. Nat Mater. 2017;16 doi: 10.1038/nmat5020. 1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Dai X, Ma C, Lan Q, Xu T. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/4/045005. 045005. [DOI] [PubMed] [Google Scholar]
- [244].Li C, Faulkner-Jones A, Dun AR, Jin J, Chen P, Xing Y, Yang Z, Li Z, Shu W, Liu D, Duncan RR. Angew Chem, Int Ed. 2015;54 doi: 10.1002/anie.201411383. 3957. [DOI] [PubMed] [Google Scholar]
- [245].English MA, Soenksen LR, V Gayet R, de Puig H, Angenent-Mari NM, Mao AS, Nguyen PQ, Collins JJ. Science. 2019;365:780. doi: 10.1126/science.aaw5122. [DOI] [PubMed] [Google Scholar]
- [246].Wu Z, Su X, Xu Y, Kong B, Sun W, Mi S. Sci Rep. 2016;6 doi: 10.1038/srep24474. 24474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Brown GCJ, Lim KS, Farrugia BL, Hooper GJ, Woodfield TBF. Macromol Biosci. 2017;17 doi: 10.1002/mabi.201700158. 1700158. [DOI] [PubMed] [Google Scholar]
- [248].Du M, Chen B, Meng Q, Liu S, Zheng X, Zhang C, Wang H, Li H, Wang N, Dai J. Biofabrication. 2015;7 doi: 10.1088/1758-5090/7/4/044104. 044104. [DOI] [PubMed] [Google Scholar]
- [249].Cui H, Zhu W, Nowicki M, Zhou X, Khademhosseini A, Zhang LG. Adv Healthcare Mater. 2016;5 doi: 10.1002/adhm.201600505. 2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Faramarzi N, Yazdi IK, Nabavinia M, Gemma A, Fanelli A, Caizzone A, Ptaszek LM, Sinha I, Khademhosseini A, Ruskin JN, Tamayol A. Adv Healthcare Mater. 2018;7 doi: 10.1002/adhm.201701347. 1701347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Poldervaart MT, Wang H, Van Der Stok J, Weinans H, Leeuwenburgh SCG, Oner FC, Dhert WJA, Alblas J. PLoS One. 2013;8 doi: 10.1371/journal.pone.0072610. e72610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Poldervaart MT, Gremmels H, Van Deventer K, Fledderus JO, Öner FC, Verhaar MC, Dhert WJA, Alblas J. J Controlled Release. 2014;184:58. doi: 10.1016/j.jconrel.2014.04.007. [DOI] [PubMed] [Google Scholar]
- [253].Penna C, Perrelli MG, Karam JP, Angotti C, Muscari C, Montero-Menei CN, Pagliaro P. J Cell Mol Med. 2013;17:192. doi: 10.1111/j.1582-4934.2012.01662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Levato R, Mateos-Timoneda MA, Planell JA. Macromol Biosci. 2012;12:557. doi: 10.1002/mabi.201100383. [DOI] [PubMed] [Google Scholar]
- [255].Levato R, Planell JA, Mateos-Timoneda MA, Engel E. Acta Biomater. 2015;18:59. doi: 10.1016/j.actbio.2015.02.008. [DOI] [PubMed] [Google Scholar]
- [256].Jeon O, Lee YB, Hinton TJ, Feinberg AW, Alsberg E. Mater Today Chem. 2019;12:61. doi: 10.1016/j.mtchem.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Cunniffe GM, Gonzalez-Fernandez T, Daly A, Sathy BN, Jeon O, Alsberg E, Kelly DJ. Tissue Eng, Part A. 2017;23:891. doi: 10.1089/ten.tea.2016.0498. [DOI] [PubMed] [Google Scholar]
- [258].Gonzalez-Fernandez T, Rathan S, Hobbs C, Pitacco P, Freeman FE, Cunniffe GM, Dunne NJ, McCarthy HO, Nicolosi V, O’Brien FJ, Kelly DJ. J Controlled Release. 2019;301:13. doi: 10.1016/j.jconrel.2019.03.006. [DOI] [PubMed] [Google Scholar]
- [259].Lozano R, Stevens L, Thompson BC, Gilmore KJ, Gorkin R, Stewart EM, M in het Panhuis. Romero-Ortega M, Wallace GG. Biomaterials. 2015;67:264. doi: 10.1016/j.biomaterials.2015.07.022. [DOI] [PubMed] [Google Scholar]
- [260].Kim BS, Lee JS, Gao G, Cho DW. Biofabrication. 2017;9 doi: 10.1088/1758-5090/aa71c8. 025034. [DOI] [PubMed] [Google Scholar]
- [261].Cubo N, Garcia M, Del Cañizo JF, Velasco D, Jorcano JL. Biofabrication. 2017;9 doi: 10.1088/1758-5090/9/1/015006. 015006. [DOI] [PubMed] [Google Scholar]
- [262].Ng WL, Qi JTZ, Yeong WY, Naing MW. Biofabrication. 2018;10 doi: 10.1088/1758-5090/aa9e1e. 025005. [DOI] [PubMed] [Google Scholar]
- [263].Horvath L, Umehara Y, Jud C, Blank F, Petri-Fink A, Rothen-Rutishauser B. Sci Rep. 2015;5 doi: 10.1038/srep07974. 7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Kim KW, Lee SJ, Park SH, Kim JC. Adv Healthcare Mater. 2018;7 1800398. [Google Scholar]
- [265].Zhang K, Fu Q, Yoo J, Chen X, Chandra P, Mo X, Song L, Atala A, Zhao W. Acta Biomater. 2017;50:154. doi: 10.1016/j.actbio.2016.12.008. [DOI] [PubMed] [Google Scholar]
- [266].Apelgren P, Amoroso M, Lindahl A, Brantsing C, Rotter N, Gatenholm P, Kölby L. PLoS One. 2017;12 doi: 10.1371/journal.pone.0189428. e0189428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].You F, Chen X, Cooper DML, Chang T, Eames BF. Biofabrication. 2019;11 doi: 10.1088/1758-5090/aaf44a. 015015. [DOI] [PubMed] [Google Scholar]
- [268].Ren X, Wang F, Chen C, Gong X, Yin L, Yang L. BMC Musculoskeletal Disord. 2016;17:301. doi: 10.1186/s12891-016-1130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Levato R, Webb WR, Otto IA, Mensinga A, Zhang Y, van Rijen M, van Weeren R, Khan IM, Malda J. Acta Biomater. 2017;61:41. doi: 10.1016/j.actbio.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Skardal A, Murphy SV, Devarasetty M, Mead I, Kang HW, Seol YJ, Zhang YS, Shin SR, Zhao L, Aleman J, Hall AR, Shupe TD, Kleensang A, Dokmeci MR, Jin Lee S, Jackson JD, Yoo JJ, Hartung T, Khademhosseini A, Soker S, Bishop CE, Atala A. Sci Rep. 2017;7 doi: 10.1038/s41598-017-08879-x. 8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Datta P, Ayan B, Ozbolat IT. Acta Biomater. 2017;51:1. doi: 10.1016/j.actbio.2017.01.035. [DOI] [PubMed] [Google Scholar]
- [272].Klotz BJ, Lim KS, Chang YX, Soliman BG, Pennings I, Melchels FPW, Woodfield TBF, Rosenberg AJWP, Malda J, Gawlitta D. Eur Cells Mater. 2018;35:335. doi: 10.22203/eCM.v035a23. [DOI] [PubMed] [Google Scholar]
- [273].Elomaa L, Yang YP. Tissue Eng, Part B. 2017;23:436. doi: 10.1089/ten.teb.2016.0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Chen YW, Shen YF, Ho CC, Yu J, Wu YHA, Wang K, Shih CT, Shie MY. Mater Sci Eng, C. 2018;91:679. doi: 10.1016/j.msec.2018.06.005. [DOI] [PubMed] [Google Scholar]
- [275].Homan KA, Kolesky DB, Skylar-Scott MA, Herrmann J, Obuobi H, Moisan A, Lewis JA. Sci Rep. 2016;6 doi: 10.1038/srep34845. 34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Byambaa B, Annabi N, Yue K, Trujillo-de Santiago G, Alvarez MM, Jia W, Kazemzadeh-Narbat M, Shin SR, Tamayol A, Khademhosseini A. Adv Healthcare Mater. 2017;6 doi: 10.1002/adhm.201700015. 1700015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Pijnenborg R, Vercruysse L, Hanssens M. Placenta. 2006;27:939. doi: 10.1016/j.placenta.2005.12.006. [DOI] [PubMed] [Google Scholar]
- [278].Peak CW, Singh KA, Adlouni M, Chen JA, Carlson MA, Duan B. RSC Adv. 2017;7 29312. [Google Scholar]
- [279].Attalla R, Ling C, Selvaganapathy P. Biomed Microdevices. 2016;18:17. doi: 10.1007/s10544-016-0042-6. [DOI] [PubMed] [Google Scholar]
- [280].Miri AK, Nieto D, Iglesias L, Goodarzi Hosseinabadi H, Maharjan S, Ruiz-Esparza GU, Khoshakhlagh P, Manbachi A, Dokmeci MR, Chen S, Shin SR, Zhang YS, Khademhosseini A. Adv Mater. 2018;30 doi: 10.1002/adma.201800242. 1800242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Kang D, Ahn G, Kim D, Kang HW, Yun S, Yun WS, Shim JH, Jin S. Biofabrication. 2018;10 doi: 10.1088/1758-5090/aac70b. 035008. [DOI] [PubMed] [Google Scholar]
- [282].Bhise NS, Manoharan V, Massa S, Tamayol A, Ghaderi M, Miscuglio M, Lang Q, Zhang YS, Shin SR, Calzone G, Annabi N, Shupe TD, Bishop CE, Atala A, Dokmeci MR, Khademhosseini A. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/1/014101. 014101. [DOI] [PubMed] [Google Scholar]
- [283].Lee H, Cho DW. Lab Chip. 2016;16 doi: 10.1039/c6lc00450d. 2618. [DOI] [PubMed] [Google Scholar]
- [284].Labernadie A, Kato T, Brugués A, Serra-Picamal X, Derzsi S, Arwert E, Weston A, González-Tarragó V, Elosegui-Artola A, Albertazzi L, Alcaraz J, Roca-Cusachs P, Sahai E, Trepat X. Nat Cell Biol. 2017;19:224. doi: 10.1038/ncb3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [285].Jiang T, Munguia-Lopez JG, Flores-Torres S, Grant J, Vijayakumar S, De Leon-Rodriguez A, Kinsella JM. Sci Rep. 2017;7 doi: 10.1038/s41598-017-04691-9. 4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].Wang X, Li X, Dai X, Zhang X, Zhang J, Xu T, Lan Q. Colloids Surf, B. 2018;171:291. doi: 10.1016/j.colsurfb.2018.07.042. [DOI] [PubMed] [Google Scholar]
- [287].Heinrich MA, Bansal R, Lammers T, Zhang YS, Michel Schiffelers R, Prakash J. Adv Mater. 2019;31 doi: 10.1002/adma.201806590. 1806590. [DOI] [PubMed] [Google Scholar]
- [288].Martin I, Malda J, Rivron NC. Curr Opin Organ Transplant. 2019;24:562. doi: 10.1097/MOT.0000000000000679. [DOI] [PubMed] [Google Scholar]
- [289].Lenas P, Moos M, Luyten FP. Tissue Eng, Part B. 2009;15:381. doi: 10.1089/ten.TEB.2008.0575. [DOI] [PubMed] [Google Scholar]
- [290].Lenas P, Moos M, Luyten FP. Tissue Eng, Part B. 2009;15:395. doi: 10.1089/ten.TEB.2009.0461. [DOI] [PubMed] [Google Scholar]
- [291].Clevers H. Cell. 2016;165 doi: 10.1016/j.cell.2016.05.082. 1586. [DOI] [PubMed] [Google Scholar]
- [292].Susienka MJ, Wilks BT, Morgan JR. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/4/045003. 045003. [DOI] [PubMed] [Google Scholar]
- [293].Mekhileri NV, Lim KS, Brown GCJ, Mutreja I, Schon BS, Hooper GJ, Woodfield TBF. Biofabrication. 2018;10 doi: 10.1088/1758-5090/aa9ef1. 024103. [DOI] [PubMed] [Google Scholar]
- [294].McMaster R, Hoefner C, Hrynevich A, Blum C, Wiesner M, Wittmann K, Dargaville TR, Bauer-Kreisel P, Groll J, Dalton PD, Blunk T. Adv Healthcare Mater. 2019;8 doi: 10.1002/adhm.201801326. 1801326. [DOI] [PubMed] [Google Scholar]
- [295].Vasilevich AS, Mourcin F, Mentink A, Hulshof F, Beijer N, Zhao Y, Levers M, Papenburg B, Singh S, Carpenter AE, Stamatialis D, van Blitterswijk C, Tarte K, de Boer J. Front Bioeng Biotechnol. 2018;6:87. doi: 10.3389/fbioe.2018.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Shi J, Song J, Song B, Lu WF. Engineering. 2019;5:586. [Google Scholar]
- [297].Menon A, Póczos B, Feinberg AW, Washburn NR. 3D Print Addit Manuf. 2019;6:181. [Google Scholar]
- [298].Joshi S, Cook E, Mannoor MS. Nano Lett. 2018;18 doi: 10.1021/acs.nanolett.8b02642. 7448. [DOI] [PubMed] [Google Scholar]
- [299].Tognato R, Armiento AR, Bonfrate V, Levato R, Malda J, Alini M, Eglin D, Giancane G, Serra T. Adv Funct Mater. 2019;29 1804647. [Google Scholar]
- [300].Lode A, Krujatz F, Brüggemeier S, Quade M, Schütz K, Knaack S, Weber J, Bley T, Gelinsky M. Eng Life Sci. 2015;15:177. [Google Scholar]
- [301].Trujillo-De Santiago G, Alvarez MM, Samandari M, Prakash G, Chandrabhatla G, Rellstab-Sánchez PI, Byambaa B, Pour Shahid Saeed Abadi P, Mandla S, Avery RK, Vallejo-Arroyo A, Nasajpour A, Annabi N, Zhang YS, Khademhosseini A. Mater Horiz. 2018;5:813. doi: 10.1039/C8MH00344K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].McDermott AM, Herberg S, Mason DE, Collins JM, Pearson HB, Dawahare JH, Tang R, Patwa AN, Grinstaff MW, Kelly DJ, Alsberg E, Boerckel JD. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aav7756. eaav7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [303].Morley CD, Ellison ST, Bhattacharjee T, O’Bryan CS, Zhang Y, Smith KF, Kabb CP, Sebastian M, Moore GL, Schulze KD, Niemi S, Sawyer WG, Tran DD, Mitchell DA, Sumerlin BS, Flores CT, Angelini TE. Nat Commun. 2019;10 doi: 10.1038/s41467-019-10919-1. 3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Shadish JA, Benuska GM, DeForest CA. Nat Mater. 2019;18 doi: 10.1038/s41563-019-0367-7. 1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [305].Skylar-Scott MA, Uzel SGM, Nam LL, Ahrens JH, Truby RL, Damaraju S, Lewis JA. Sci Adv. 2019;5 doi: 10.1126/sciadv.aaw2459. eaaw2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [306].Van Hoorick J, Gruber P, Markovic M, Rollot M, Graulus GJ, Vagenende M, Tromayer M, Van Erps J, Thienpont H, Martins JC, Baudis S, Ovsianikov A, Dubruel P, Van Vlierberghe S. Macromol Rapid Commun. 2018;39 doi: 10.1002/marc.201800181. 1800181. [DOI] [PubMed] [Google Scholar]
- [307].Robinson TM, Hutmacher DW, Dalton PD. Adv Funct Mater. 2019 1904664. [Google Scholar]
- [308].Grigoryan B, Paulsen S, Corbett D, Sazer D, Fortin CL, Zaita A, Greenfield P, Calafat N, Gounley J, Ta A, Johansson F, Randles A, Rosenkrantz JJ, Louis-Rosenberg J, Galie P, Stevens K, Miller J. Science. 2019;364:458. doi: 10.1126/science.aav9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [309].Bernal PN, Delrot P, Loterie D, Li Y, Malda J, Moser C, Levato R. Adv Mater. 2019;31 doi: 10.1002/adma.201904209. 1904209. [DOI] [PubMed] [Google Scholar]