Abstract
Sodium azide is a commonly used cytochrome oxidase inhibitor that leads to translation repression and RNA granule assembly. The global changes in mRNA abundance in response to this stressor are unknown. RGG-motif proteins Scd6 and Sbp1 are translation-repressors and decapping-activators that localize to and affect the assembly of RNA granules in response to sodium azide stress. Transcriptome-wide effects of these proteins remain unknown. To address this, we have sequenced transcriptome of the: a) wild type strain under unstressed and sodium azide stress, b) Δscd6 and Δsbp1 strains under unstressed and sodium azide stress. Transcriptome analysis identified altered abundance of many transcripts belonging to stress-responsive pathways which were further validated by qRT-PCR results. Abundance of several transcripts was altered in Δscd6/Δsbp1 under normal conditions and upon stress. Overall, this study provides critical insights into transcriptome changes in response to sodium azide stress and the role of RGG-motif proteins in these changes.
Keywords: Sodium azide, Transcriptome, RNA granule, SCD6, SBP1, Oxidative stress, RGG-motif proteins
1. Introduction
The cellular response to stress is a feature of all kingdoms of life. The ability of cells to sense and respond to several kinds of environmental stress is essential for survival and evolutionary progress. The ultimate aim of the stress response is to alter the cellular proteome to efficiently combat the stress. This involves upregulating stress-responsive genes and downregulating various energy-consuming pathways, including mRNA translation. Rewiring mRNA abundance, which in turn affects the proteome, is critical for cellular stress response pathways [1–3].
The specific genes and signaling pathways altered in response to stress may vary depending on the type of stressor. A thorough study of the differential network of genes affected by various stressors is key to understand the molecular mechanisms underlying the stress response machinery. Oxidative stress has been implicated in the development and progression of many diseases such as cancer, diabetes, and cardiovascular and neurodegenerative disorders [4–7]. Sodium azide is a well-known oxidative stress-inducing agent that inhibits cytochrome oxidase to impair the mitochondrial electron transport chain and induces oxidative stress [8,9]. Sodium azide has been used to study various biological processes such as the fermentative ability of yeast grown under different oxygen conditions [10]. Interestingly, it has also been used as a weak mutagen in yeast protoplasts [11]. It increases thermo-tolerance of glucose-grown yeast [12] and is routinely used in studies of mRNA translation control since sodium azide treatment leads to general translation repression [13]. Despite the knowledge that sodium azide can inhibit protein and RNA synthesis [14,15], the global impact of sodium azide on mRNA abundance has not been characterized.
Global mRNA translation repression has emerged as an important hallmark of stress responses. The resulting non-translating mRNAs assemble into conserved membraneless mRNA-protein (mRNP) compartments known as RNA granules. Stress granules and P-bodies are well-known examples of RNA granules implicated in translation repression [16,17]. Along with non-translating mRNAs, stress granules and P-bodies are also sites of mRNA storage and decay [16,18]. RNA granule-dynamics are greatly influenced by a number of different categories of proteins, some of which include RNA-binding proteins, translation repressors, and mRNA decay proteins. RNA granules can assemble in response to a wide array of stresses such as glucose deprivation, amino acid starvation, oxidative stress (sodium azide), heat shock, and stationary phase [19–22].
A subset of RGG-motif proteins have been identified as translation repressors that target eIF4G1, a conserved translation initiation factor [23]. Scd6 and Sbp1 are two such proteins with multiple RG−/RGG-repeats. Scd6 is a conserved RGG-motif protein with orthologs identified in most model systems, including humans [24]. Along with the RGG-motif, these two proteins have different RNA binding domains. Scd6 contains an N-terminal Lsm domain, a central FDF-domain, and a C-terminal RGG-motif [24]. The RGG-motif of Sbp1 is flanked by two RRM-domains, one on either side [25]. Both the proteins can affect mRNA stability by activating decapping [26,27]. Deletion of Scd6 and Edc3 (Enhancer Of Decapping 3) leads to a synthetic growth defect [28]. MFA2 mRNA decapping was defective in the absence of both Edc3 and Scd6, suggesting that the two proteins participate in decapping activation. Consistent with this idea, Scd6 co-localizes with Edc3 and Dcp2 in RNA granules such as processing bodies (P-bodies), which are the sites of mRNA decay [23,29]. Sbp1, on the other hand, was identified as a multicopy suppressor of decapping defects in a conditional mutant of Dcp1 and Dcp2 [27]. Sbp1, like Scd6, localizes to P-bodies and perturbation of Sbp1 levels perturbs P-body assembly [27]. Consistent with the presence of RNA-binding domains, mRNAs associate with Scd6 and Sbp1 in vivo [30,31]. As decapping activators, Scd6 and Sbp1 affect transcript abundance by altering mRNA stability, although only MFA2 has been identified as an mRNA target so far. Given that sodium azide treatment induces P-body formation and localization of Scd6 and Sbp1 to P-bodies [23,25], it is likely to modulate the role of Scd6 and Sbp1 in maintaining transcript stability. However, the effect of Scd6 and Sbp1 on the transcriptome under normal and sodium azide treated conditions has not been assessed.
mRNAs bound by the translation repression complex are recruited to mRNP granules and are protected from decay machinery. Consequently, they can be stored for a long period of time through a process known as ‘masking’, an evolutionarily conserved mechanism. For example in worms and flies, certain key mRNAs encoding developmental proteins are stored and protected from translation and decay [18]. Tral, the Drosophila ortholog of Scd6, is involved in masking and storage of nanos mRNA and regulates the Gurken secretory pathway in conjunction with Me31B (Dhh1 in yeast) and Cup. Further, Tral mutants show dysregulated embryonic development [32]. In worms, CAR-1 (Scd6 ortholog), along with CGH-1 (Dhh1 ortholog), is crucial for germline granule maintenance [33]. As such, the deletion of Scd6 and Sbp1 could render certain sequestered mRNAs unprotected, thereby promoting their decay. Investigating mRNA abundance in Δscd6 and Δsbp1 strains can provide insight about specific mRNAs downregulated in response to the loss of masking by translation repression. Such response is likely to be altered upon exposure to stress, for example sodium azide.
Keeping the above in focus, this work aimed to a) understand the impact of sodium azide on the yeast transcriptome and b) identify the transcripts affected by the RGG-motif proteins Scd6 and Sbp1 in response to sodium azide stress. Our results show that sodium azide treatment leads to widespread changes in transcript abundance in S. cerevisiae wild type strain (BY4741). Many functional categories of transcripts with altered expression overlap with those reported in response to other oxidative stresses. We further identified multiple transcripts whose abundance was altered in normal growth conditions in the absence of Scd6 and Sbp1 and in response to sodium azide stress. It is possible that some transcripts identified in this study are direct mRNA targets of Scd6 and Sbp1. In summary, this work provides a detailed insight into the gene expression changes induced by sodium azide stress and also sheds light on the role of Scd6/Sbp1 in maintaining transcript abundance under normal conditions and in response to sodium azide stress.
2. Materials and methods
2.1. Growth conditions
Yeast strains used in this study are listed in Supplementary Table 1. Strains were grown at 30C in yeast extract-peptone (YP) medium supplemented with 2% glucose. For secondary culture, cells were diluted (OD600 ~ 0.1) in YP + 2% glucose. At mid-log phase (OD600 0.4–0.5), cultures were divided into two; one was treated with 0.5% sodium azide (NaN3), and the other with water (control) for 30 min. A 20% NaN3 stock solution was made in water and added to culture to obtain a final concentration of 0.5% NaN3. Post-treatment, cells were harvested by centrifugation and used for further analysis immediately or stored at −80 °C for further experiments.
2.2. Microscopy
The PAB1GFP strain was grown in YP + 2% glucose at 30 °C (until OD600 ~ 0.4–0.5), followed by 0.5% NaN3 treatment for 30 min. Cells were centrifuged at 14,000 rpm for 12 s and pellets were resuspended in 20 μl of supernatant media. A total of 5 μl of the cell suspension was spotted on coverslips for live cell imaging. All images were acquired using a Deltavision Elite microscope system with softWoRx 3.5.1 software (Applied Precision, LLC) and an Olympus 100×, oil-immersion 1.4 NA objective. Exposure time and transmittance settings for the Green Fluorescent Protein (GFP) channel were 0.2 s and 32%, respectively. Images were captured as 512 × 512-pixel files with a CoolSnapHQ camera (Photometrics) using 1 × 1 binning for yeast. All the images were deconvolved using standard softWoRx deconvolution algorithms. ImageJ was used to adjust all images to equalize contrast ranges.
2.3. RNA isolation and analysis
RNA was isolated from two biological replicates for each test sample using the hot acidic phenol protocol described in Current Protocols in Molecular Biology (1996) Unit 13.12 [34]. Briefly, the cell pellet was washed with nuclease free water once and resuspended thoroughly in TES (10 mM TrisCl pH 7.5, 10 mM EDTA and 0.5% SDS). 400 μl acidic phenol pH 4.5 (0.1 M citrate buffer saturated) was added to the suspension and incubated at 65 °C for 60 min with occasional vortexing. Samples were spun at 14,000 rpm for 10 min at 4 °C. The top aqueous layer was transferred to a fresh tube. Subsequently, 400 μl chloroform was added, mixed, and spun as above. The aqueous layer was taken and mixed with 1/10th volume 3 M NaAc pH 5.2 and 2.5 volumes of 100% ethanol. The samples were precipitated at -80 °C overnight, then spun as above and the pellet was washed with 70% ethanol, dried, and resuspended in 50 μl nuclease free water. RNA quality was checked by 1.2% agarose formamide gel electrophoresis. RNA quality and quantity were also assessed using the Agilent Bioanalyzer RNA 6000 Nano kit (Agilent Technologies) and the Qubit HS RNA kit (Thermo Fisher Scientific). Samples were sent to Clevergene for RNA-seq.
2.4. RNA library preparation and sequencing
One μg of total RNA was enriched for mRNA with the NEBNext Poly (A) mRNA magnetic isolation Module (E7490, New England Biolabs, USA) according to the manufacturer's instructions. The protocol uses polyT magnetic beads to enrich mRNA molecules with polyA tails. The polyA enriched mRNA was used for library preparation. The NEBNext Ultra II RNA Library Prep Kit for Illumina (E7770, New England Biolabs, USA) was used for library preparation following the manufacturer's protocol. In brief, enriched mRNA was fragmented and converted to cDNA followed by second-strand synthesis. NEBNext sample purification beads were used to isolate double-stranded cDNA. After end repair, hairpin adapters were ligated to the purified cDNA and the hairpin structure was cleaved using USER enzyme. NEBNext Multiplex Oligos for Illumina (E7335S, New England Biolabs, USA) were used for polymerase chain reaction (PCR) amplification to generate indexed sequencing libraries. The PCR conditions were 98 °C for 1 min for initial denaturation, followed by 7 cycles of 98 °C for 10 s, 65 °C for 75 s, and a final extension at 65 °C for 5 min. The amplicons were purified using NEBNext sample purification beads.
Library quality and quantity was checked using an Agilent Bioanalyzer 2100 with DNA 1000 kits and a Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific), and libraries were sequenced on an Illumina HiSeqX to generate 150 bp paired-end reads.
2.5. Transcriptome analysis
The QC passed reads were mapped to the indexed Saccharomyces cerevisiae reference genome (S288c) using STAR v2 aligner [35]. PCR and optical duplicates were marked and removed using Picard tools. Gene expression level values were obtained as read counts using the featureCounts program [36]. Differential expression analysis was carried out using the edgeR package [37] after normalizing the data based on the trimmed mean of M (TMM) values. After normalization, 248 features (3.76%) were removed from the analysis because they did not have at least 1 counts-per-million in at least two samples.
Enrichment analysis for Biological process, Molecular function, Cellular component and KEGG Pathway analysis was performed using the ClusterProfiler R package [38]. Gene Ontology (GO) and pathway terms with adjusted p-value ≤.05 were considered significant. The GOplot R package was used to visualize GO enrichment results [39]. KEGG pathway visualization was performed by the pathView R Bio-conductor package [40]. The submitted raw data can be accessed here (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145162).
2.6. Reverse transcription and quantitative real-time PCR
For target validation, isolated RNA was first subjected to DNase1 treatment (Thermo, EN0525) by combining 8 μg of total RNA, 4 units of DNase1, and DNase1 Buffer with MgCl2 in a 30 μl reaction. After incubating for 30 min 37 °C, DNase1 was inactivated with 3 μl of 50 mM EDTA for 10 mins at 65 °C. Following DNase1 treatment, RNA quality was checked again using 1.2% agarose formamide gel electrophoresis. DNase1 treated RNA was then used for cDNA synthesis.
One microgram of RNA was used to synthesize cDNA using the RevertAid RT Reverse Transcription Kit (Thermo, K1691) according to the manufacturer's protocol. cDNA was diluted 1:10 and real-time PCR was performed using TB Green™ Premix Ex Taq™ (TaKaRa). For qRTPCR, three technical replicates were assembled with 2 μl cDNA/reaction and 0.5 μM each primer in a BioRad iQ5 Real-Time PCR Detection System. Primers used for qRT-PCR are listed in Supplementary Table 2. The PCR conditions were 95 °C for 12 min for initial denaturation, followed by 30 cycles of 95 °C for 20 s, 46 °C for 30 s, and 72 °C for 30 s. DNA was quantified in every cycle at the extension step. Melt curve acquisition was carried out at 64 °C for 8 s. Ct values were extracted with auto baseline and manual threshold. ΔΔCt method was used to calculate the final log2 FoldChange values, which were then plotted on a box and whisker plot using GraphPad prism 7.0. Significance was calculated by a ratio paired t-test.
3. Results
3.1. RNA sequencing analysis of wild type, Δscd6, and Δsbp1 strains
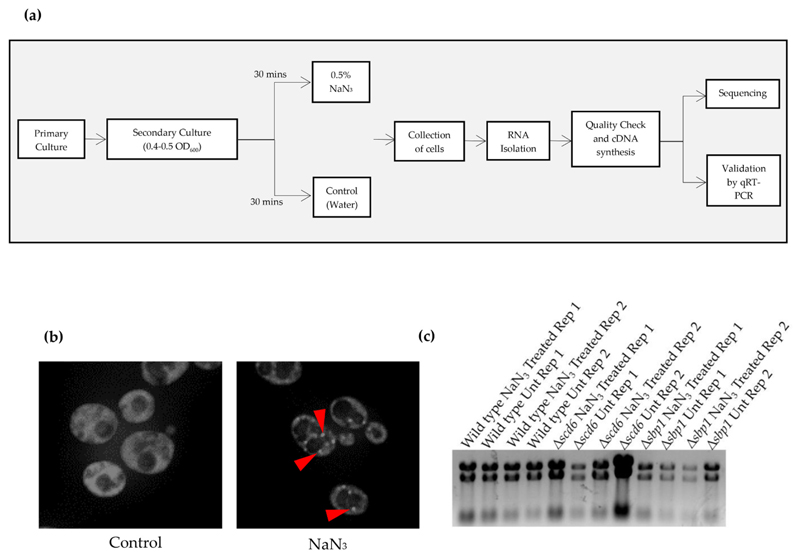
Wild type, Δscd6, and Δsbp1 strains were grown in duplicates (biological replicates) under normal conditions overnight. Secondary cultures were inoculated at 0.1 OD600 and allowed to grow for two generations. Cultures were then divided in half and one sample was treated with 0.5% sodium azide and the other with water, followed by incubation at 30 °C for 30 min (Fig. 1a). To ensure that sodium azide stress was induced under these conditions, a strain expressing Pab1 (a conserved stress granule marker) endogenously tagged with GFP was treated with sodium azide in an identical manner and observed under a fluorescence microscope to visualize stress granule formation. Pab1-GFP foci were induced in cells treated with sodium azide but not in those treated with an equivalent volume of water, confirming that the stress granules were indeed induced by sodium azide treatment (Fig. 1b).
Fig. 1.
(a) Schematic depicting the experimental workflow. Primary yeast cultures were grown for two biological replicates of every strain. For secondary culture, cells were diluted to 0.1 OD600. At mid-log (0.4–0.5 OD600), cells were treated with 0.5% NaN3 for 30 min followed by collection of cells by centrifugation. For control cells, equivalent volume of water was added. RNA was isolated using hot phenol followed by quality check. Sequencing was carried out following cDNA synthesis. Differentially expressed genes were validated by qRT-PCR analysis which was carried out for three technical replicates of each biological replicates; (b) Live cell imaging of cells expressing genomically-tagged PAB1GFP strain treated with NaN3 at the same time as the cells used for isolating the RNA used for RNA-sequencing. Arrows indicate the presence of stress granules; (c) Agarose formamide gel electrophoresis image of the RNA sent for RNA sequencing analysis. Rep 1 and Rep 2 refer to Replicate 1 and Replicate 2, respectively.
RNA was isolated from treated and untreated wild type, Δscd6, and Δsbp1 strains and subjected to denaturing formamide agarose gel electrophoresis (Fig. 1c). PolyA enriched mRNA was used for cDNA synthesis and library preparation. Illumina sequencing was performed on each sample with two biological replicates. Sequencing data quality was checked using FastQC and MultiQC software. The data was checked for base call quality distribution, % bases above Q20, Q30, % GC, and sequencing adapter contamination, and all samples passed the QC threshold (Q30 > 90%). On average, 90.69% of the QC-passed reads aligned to the reference genome. After removing PCR and optical duplicates, gene expression values were obtained as read counts using featureCounts software. Expression similarity between biological replicates was checked by Spearman correlation and the co-efficient value between replicates was > 0.97.
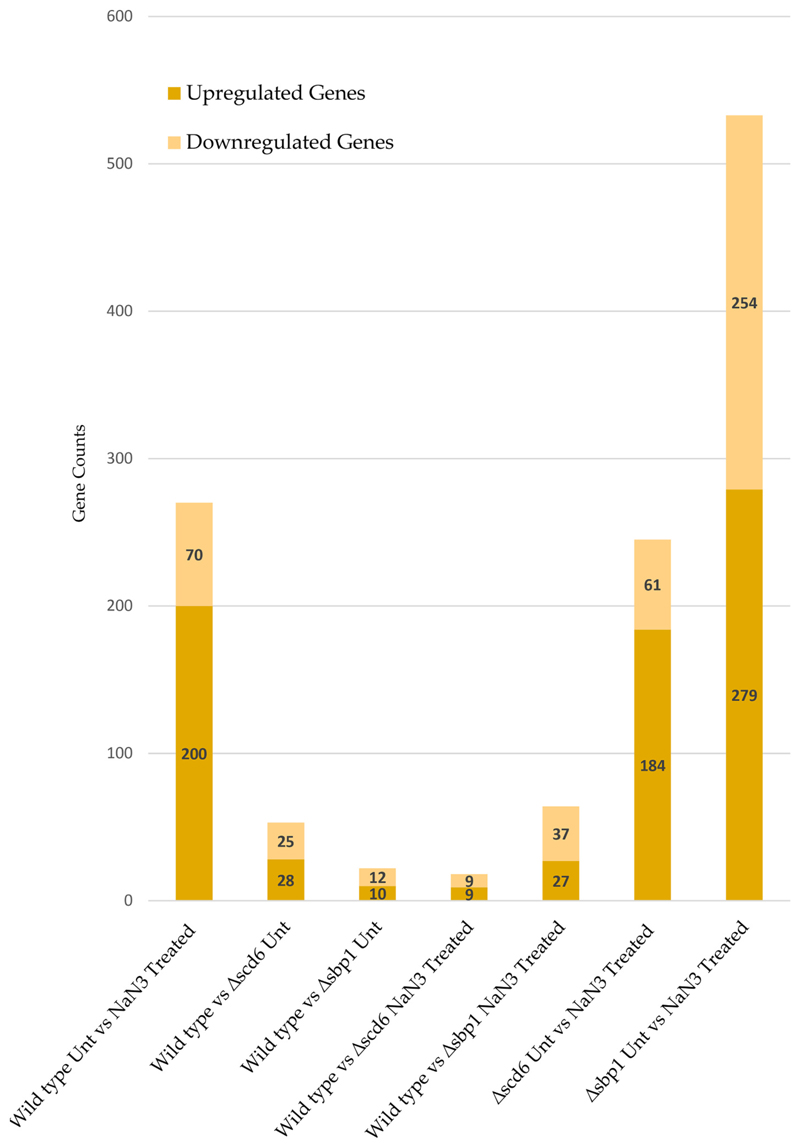
For differential expression analysis, biological replicates were grouped as “Control” and “Test” with the control category listed first in all comparisons (for example: Wild type Unt vs NaN3 Treated considers Wild type Unt as control and NaN3 Treated as test). Genes with an absolute log2 fold change ≤2 and p-value ≥.05 were considered significantly differentially expressed (Fig. 2). A summary of differentially expressed genes is presented as volcano plots and heatmaps (Fig. 3 and Supplementary Figs. 1 & 2).
Fig. 2.
Summary of significantly up and downregulated genes in different conditions.
Fig. 3.
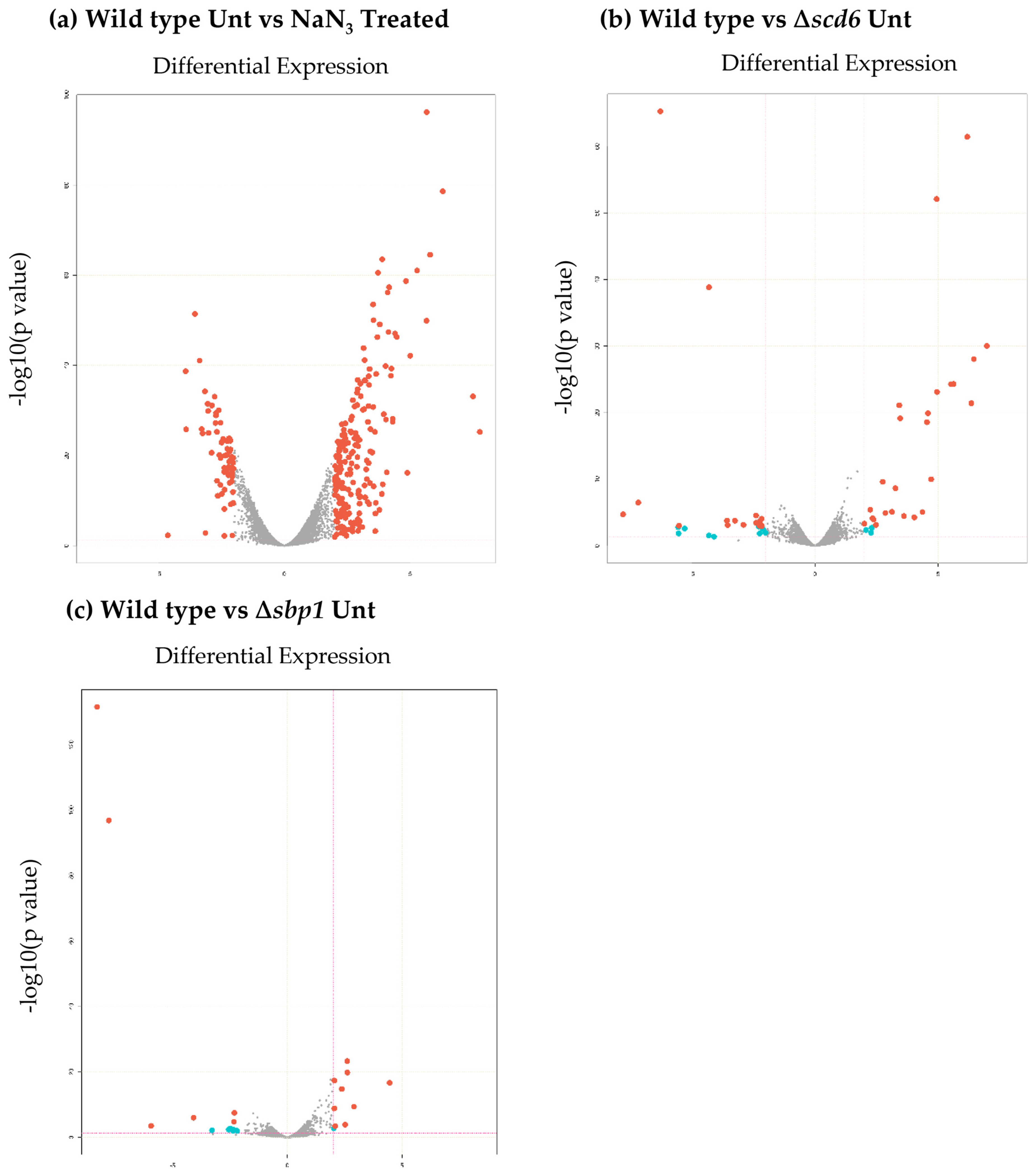
Volcano plot showing changes in transcript levels in: (a) wild type cells upon sodium azide treatment, (b) wild type cells upon deletion of Scd6 and, (c) wild type cells upon deletion of Sbp1. Red dots indicate absolute log2fold change≥2 and FDR/adjusted p-value≤.05. Blue dots indicate log2fold change≥2 and pvalue≤.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Stress-response genes show significant changes in transcript abundance in response to sodium azide treatment
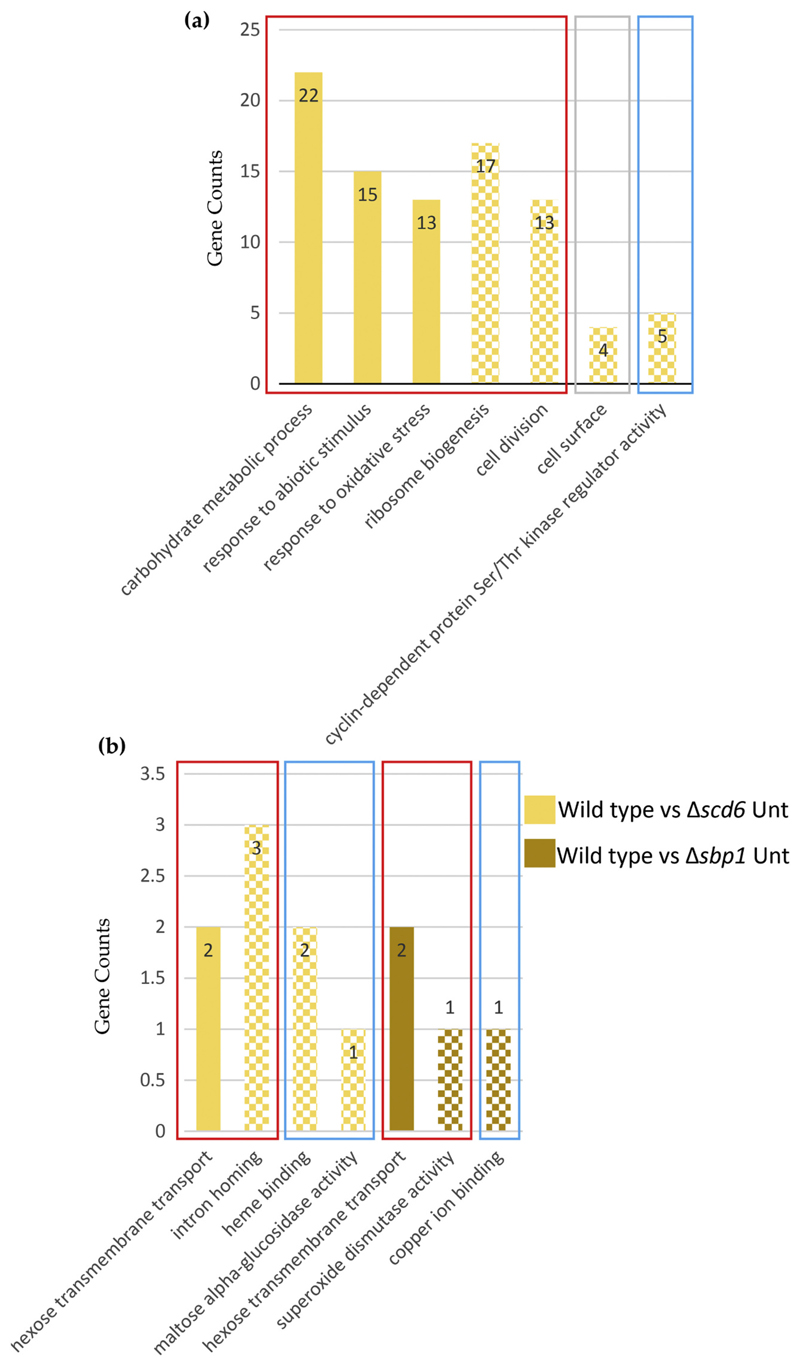
RNA-seq analysis revealed that 270 genes were differentially expressed following sodium azide treatment, of which 200 were upregulated and 70 were downregulated (Fig. 2). Genes belonging to the carbohydrate metabolic process gene ontology (GO) category constituted the largest (22 genes) category of upregulated genes in sodium azide treated wild type cells (Fig. 4a). Two other GO categories upregulated upon sodium azide stress include oxidative stress-responsive genes (11 genes) and abiotic stress-responsive genes (15 genes). This is consistent with the action of sodium azide on cytochrome oxidase and on cellular redox balance. The downregulated genes were prominently represented in the GO category of ribosome biogenesis (17 genes) and cell division (13 genes) (Fig. 4a). The downregulation of ribosome biogenesis genes was consistent with previously reported observations that sodium azide treatment globally represses translation.
Fig. 4.
Gene ontology enrichment analysis of differentially expressed genes in (a) wild type untreated vs treated cells and (b) wild type vs Δscd6/Δsbp1 untreated cells. Solid and pattern fill indicate up and downregulated categories, respectively. Red, blue and grey rectangular boxes represent Biological Processes, Molecular Function and Cellular Component, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Interestingly, two non-coding RNAs HRA1 (Hidden-in-Reading frame Antisense-1) and RUF21 (RNA of Unknown Function 21) are downregulated in wild type cells following sodium azide treatment (Supplementary Fig. 3). Interestingly these RNAs were downregulated in Δsbp1 (Supplementary Fig. 3) cells also following sodium azide treatment. HRA1 is a substrate of RNase P and is speculated to be involved in 18S rRNA maturation [41].
3.3. Transcriptome changes in Δscd6 and Δsbp1 strains
In Δscd6 background as compared to wild type, 53 genes were differentially expressed as compared to wild type under normal conditions; 28 genes were upregulated, and 25 genes were downregulated (Fig. 2). Of the upregulated genes, sugar transporter genes (including hexose transporter HXT7) were prominently represented (Fig. 4b). The downregulated genes belong to different GO categories including intron homing, heme-binding, and alpha-glucosidase activity. Three non-coding RNAs, RUF23, RUF5-1, and RUF5-2, were upregulated upon deletion of SCD6, indicating that Scd6 may play a role in maintaining non-coding RNA levels apart from mRNA levels (Supplementary Fig. 3).
RNA-seq analysis of the Δsbp1 strain as compared to wild type revealed an altered abundance of 22 different transcripts. This included 10 upregulated and 12 downregulated genes (Fig. 2). Interestingly, the sugar transporter gene HXT7, which was upregulated in the absence of Scd6, was also upregulated upon Sbp1 deletion, indicating that both proteins could play a cooperative role in regulating the abundance of HXT7 mRNA. The downregulated genes include GO categories such as superoxide dismutase and copper ion binding (Fig. 4b).
Following sodium azide stress, in Δscd6 strain as compared to wild type, 18 genes were differentially expressed with 9 genes being upregulated and an equal number of genes being downregulated (Fig. 2). GO categories such as chitin deacetylase activity and spermine transmembrane transporter activity were represented in the downregulated genes (Supplementary Fig. 4). Interestingly, GO categories including intron homing and cell division were represented in the upregulated genes (Supplementary Fig. 4). Following sodium azide stress, in Δsbp1 strain as compared to wild type, there was differential expression of 64 genes, with 27 genes being upregulated and 37 genes being downregulated (Fig. 2). It is noteworthy that the intron homing genes (Al1, Al5_beta, and SceI) upregulated in the absence of Scd6 were also upregulated in the absence of Sbp1 upon oxidative stress, although to different extents (Supplementary Fig. 4). This indicated that both the RGG-motif proteins acted on similar mRNAs in this important biological process.
In response to sodium azide treatment in Δscd6 and Δsbp1 backgrounds as compared to untreated, abundance of 245 and 533 transcripts changed respectively (Fig. 2). Genes encoding proteins involved in carbohydrate metabolic process, oxidative and abiotic stress response were significantly enriched in the sodium azide treated Δscd6 and Δsbp1 cells (Supplementary Fig. 4b & c). Downregulation of cellular component genes of cell surface family is a common feature of sodium azide stress response mounted by wild type, Δscd6 and Δsbp1 strains (Fig. 4a, Supplementary Fig. 4b & c). Specifically, different aspartyl proteases like MKC7 and BAR1 were significantly reduced in response to sodium azide. In contrast to this, ribosome biogenesis and cell division related genes did not show any significant change in case of Δscd6 background upon sodium azide stress as compared to untreated cells. In Δscd6 background, genes belonging to cell wall category were downregulated whereas in Δsbp1 background genes belonging to helicase activity category were downregulated (Supplementary Fig. 4b). This is in contrast to the wild type response to sodium azide stress.
3.4. Validation of RNA-seq results by qRT-PCR
To choose a suitable control gene for validating the RNA-seq results, we checked the mRNA levels of three different control genes, namely ACT1, SCR1, and PGK1, under different conditions by qRT-PCR. In absence of stress, both ACT1 and SCR1 genes were not modulated significantly in Δscd6 and Δsbp1 strains (Supplementary Fig. 5). SCR1 was used as a control to validate transcript levels in Δscd6 and Δsbp1 backgrounds under untreated condition; however, in response to sodium azide treatment, SCR1 expression was significantly upregulated when compared with ACT1 and PGK1 (Supplementary Fig. 5). Therefore, PGK1 was chosen as the control for mRNA target validation specific to sodium azide treatment.
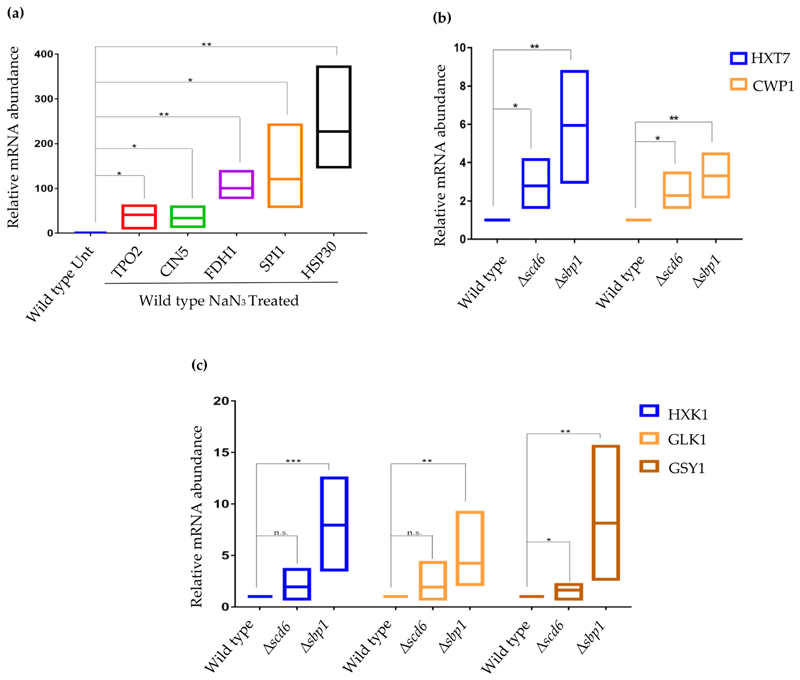
We validated the change in abundance of transcripts that were differentially expressed upon sodium azide treatment in wild type yeast cells. We focused on HSP30, SPI1, TPO2, CIN5, and FDH1. All the tested transcripts were upregulated in wild type cells upon sodium azide treatment (Fig. 5a). A comparison of the changes in transcript abundance measured by RNA-seq and qRT-PCR are presented (Supplementary Fig. 6a).
Fig. 5.
Validation of differentially expressed genes by qRT-PCR analysis in (a) wild type untreated vs NaN3 treated cells and (b) wild type vs Δscd6/Δsbp1 cells and (c) wild type vs Δscd6/Δsbp1 cells. Internal control for (a) and (b and c) are PGK1 and SCR1, respectively (n > 3; *, ** and *** denotes p-value > .05, p-value > .01 and p-value > .001 respectively).
We performed qRT-PCR to validate the RNA-seq results for transcripts differentially expressed in Δscd6. HXT7 and CWP1 transcript levels were validated using SCR1 as the internal control. The abundance of these transcripts increased in Δscd6 (Fig. 5b and Supplementary Fig. 6b). Interestingly, both transcripts also increased in abundance in Δsbp1 (Supplementary Fig. 6b & c). Based on these results, we conclude that Scd6 and Sbp1 reduce the abundance of HXT7 and CWP1 transcripts.
Similarly, to validate RNAseq results for differentially expressed mRNAs in Δsbp1, candidate mRNAs such as HXK1, GLK1, GSY1, and GPH1 were chosen. Two points motivated us to investigate these transcripts. First, RNA-seq results indicated that the levels of these mRNAs were significantly modulated. Second, Sbp1 binds to these transcripts in yeast cells [30], indicating that these changes in transcript level are likely mediated by Sbp1 binding to these mRNAs. The results of qRT-PCR showed increased abundance of these mRNAs (Fig. 5c and Supplementary Fig. 6c). Based on these results, we conclude that the deletion of Sbp1 increases the abundance of HXK1, GLK1, GSY1, and GPH1 transcripts, all of which encode enzymes involved in glucose metabolism.
Changes in transcript levels in Δscd6 and Δsbp1 cells upon sodium azide treatment as compared to wild type treated cells were also validated. HXK1, GPH1, and GSY1 were tested as all three transcripts showed increased abundance in RNA-seq data (Supplementary Fig. 7). We observed that all the tested transcripts showed increased levels by qRT-PCR in the absence of Scd6 and Sbp1 following sodium azide treatment, thus confirming the RNA-seq results. Taken together, these observations validate the RNA-seq results by qRT-PCR.
4. Discussion
Sodium azide represses global translation and induces RNA granule formation within minutes after cells are exposed to the stressor [15]. This in turn, could affect the half lives of many transcripts, thereby contributing to altered gene expression profile. However, the subset of genes affected by sodium azide stress has remained unknown. The present study used RNA sequencing to analyze the change in mRNA levels of yeast cells exposed to sodium azide stress. Our study also evaluated the mRNA abundance changes upon deletion of RGG-motif containing decapping activators such as Scd6 and Sbp1 under normal conditions and in response to sodium azide stress.
Wild type cells showed modulation of many transcripts in response to sodium azide treatment. Thirteen genes were significantly upregulated and belonged to the GO biological process category of ‘response to oxidative stress’ (Fig. 4a and Supplementary Fig. 4b & c). In this category, genes involved in protection against oxidative damage (CTT1, SRX1, CCP1, GAD1, and NCE103) and multi-stress response (HSP30, MTL11, and DDR2) were strongly induced. These results emphasized the role of sodium azide as an agent of oxidative damage to the cells. Cytosolic Catalase T (CTT1) is known for its role in the oxidative stress response. Ctt1 activity is crucial to protect proteins from oxidative damage [42] and CTT1 upregulation is reported in response to a number of stresses such as starvation, osmotic, and oxidative stress [43]. A similar increase in CTT1 mRNA level was observed in our study, highlighting a feature of stress response mounted by yeast cells to counter oxidative damage induced by sodium azide.
Transcripts encoding proteins associated with carbohydrate metabolism were induced in stressed cells (Fig. 4a and Supplementary Fig. 4b & c). This response could fulfil the energy requirements during the stress response. Some of these upregulated genes belong to the glycolytic pathway (TDH1, GPM2, and ENO1) while others were related to closely associated pathways (YIG1, GCY1, GID8, and GPH1). Glycogen and trehalose are the two storage forms of glucose. Our transcriptome analysis also showed upregulation of transcripts (GLC3, IGD1, GSY2, and TSL1) encoding enzymes catalyzing the synthesis of these two polysaccharides. This is in agreement with the reported observation that glycogen and trehalose production are elevated in other stresses [44].
Genes involved in gamma-aminobutyric (GABA) acid metabolism (belonging to the Butanoate pathway) play a role in the oxidative stress response to stressors such as H2O2 and diamide [45]. Diamide depletes glutathione and oxide thiol groups. Two genes of this pathway, GAD1 and UGA2, were upregulated upon sodium azide treatment in wild type, Δscd6 and Δsbp1 cells, indicating their role in the sodium azide stress response. The pentose phosphate pathway (PPP) is also upregulated in response to oxidative stress [46]. SOL4, PGM2, and NQM1 are PPP components upregulated in wild type cells in response to sodium azide stress in our study. This increase would generate NADPH to provide reducing potential and act as a cofactor for glutathione- and thioredoxin-dependent enzymes that play a major role in survival during the oxidative stress response [47].
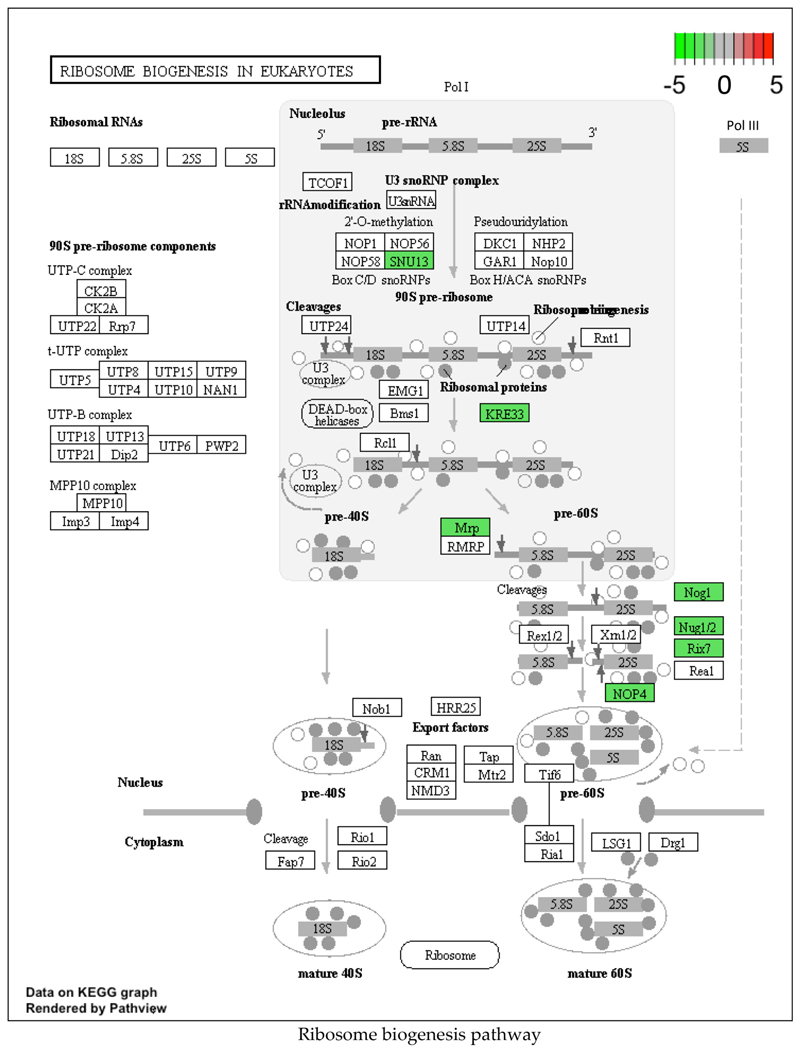
Downregulation of cellular processes that consume a high amount of energy, such as translation and cell cycle progression, is a feature of stress response pathways. We find evidence supporting this in the sodium azide stress response program. In our study, multiple genes in the ribosome biogenesis pathway (SNU13, KRE33, MRP, NOG1, NUG1/2, RIX7, and NOP4) were downregulated following sodium azide treatment (Fig. 6). Ribosome biogenesis is a highly energy-expensive process and defects in it lead to translation repression [48]. Our observation is highly consistent with the report that sodium azide treatment leads to global translation repression and the assembly of cytoplasmic repression foci [49]. Similarly, cell cycle genes such as PCL1, CLN1/2, CLB5/ 6, CDC6, HSL1 were downregulated upon azide treatment (Supplementary Fig. 8a). This is in accordance with the observation that oxidative stress downregulates expression of cell cycle genes involved in the G1 to S transition [50]. Together, the above observations indicate that the response to sodium azide stress is similar to other oxidative stress response pathways. Interestingly, though ribosome biogenesis related genes were downregulated in wild type, Δscd6 and Δsbp1 background upon sodium azide treatment, the cell cycle related target genes were downregulated in wild type and Δsbp1 background but not in Δscd6 cells (Supplementary Fig. 4b & https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145162).
Fig. 6.
Ribosome biogenesis pathway showing genes which are significantly changing upon sodium azide treatment of wild type cells. The scale is set as log2fold change where downregulated genes are shaded green. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Some GO categories were found to be enriched only in the case of deletion of Scd6 and Sbp1 upon sodium azide treatment (Supplementary Fig. 4b & c). This reflects the importance of these proteins in mounting an appropriate cell response to stress.
The cell wall is a major barrier to physical stressors in yeast [51]. Cell wall synthesis and repair enzymes are activated in response to oxidative stress such as H2O2. We capture this behavior in our study in response to sodium azide treatment. Genes involved in cell wall integrity pathways, such as SDP1, MID2, and MTL1, are upregulated upon sodium azide treatment, as has been observed with other oxidative stressors [52].
Many mitogen-activated protein kinase (MAPK) pathway genes are downregulated in response to sodium azide stress, including MSB2, HSL1, BAR1, SPA2, YPS1, and YPS2. This is surprising given that the MAPK pathway is activated upon H2O2-mediated oxidative stress [53]. This observation indicates that although shared metabolic regulators are elicited by sodium azide and H2O2, the signaling pathways activated in response to these two stressors may be distinct.
Downregulation of purine and pyrimidine biosynthesis pathway genes (HPRT1, RNR1, and URA2) likely indicate a stress response mechanism aimed at reducing nucleotide biosynthesis during oxidative stress (Supplementary Fig. 9).
We validated the modulation in the expression levels of HSP30, SPI1, TPO2, CIN5, and FDH1 genes in wild type cells subjected to sodium azide treatment (Fig. 5a). HSP30 and SPI1 were previously reported to be stress-responsive genes, albeit not in response to sodium azide stress. HSP30 is induced by heat shock, ethanol treatment, and weak organic acid. SPI1 is a GPI-anchored cell wall protein (encoded by SPI1 mRNA) induced by stress conditions and contributes to weak acid resistance [54]. FDH1 encodes formate dehydrogenase, and is induced by the presence of formate in the media [55]. TPO2 encodes a spermine-specific polyamine transporter that localizes to the plasma membrane [56]. All of the above genes were not reported to change in response to oxidative stress such as H2O2, indicating that the stress response mediated through these genes may not overlap with the H2O2 stress response, thereby supporting the idea that cells use different sets of proteins to respond to different stressors. In our study, upregulation of these transcripts upon sodium azide treatment indicates their involvement in the sodium azide stress response. The specific contribution of these genes to the sodium azide stress response will be evaluated in future.
RNA binding proteins play a critical role in regulating mRNA fate and abundance. Our report studied the importance of two RNA binding proteins, Scd6 and Sbp1, which are known translation repressors, decapping activators, and RNA granule modulators [23,25,26,57]. Three meiosis/sporulation specific genes were downregulated in Δscd6 including SPO19, SPO23, and AMA1 (Supplementary Table 10). Spo19 and Spo23 are pro-spore and Spo1-associated protein, respectively [58,59]. Ama1 is an activator of the meiotic anaphase-promoting complex, which is required to initiate spore wall assembly during anaphase [60,61]. These observations suggest that Scd6 could be involved in sporulation and/or meiosis related processes, which has not been addressed previously in the literature. DAN1 transcript expression increases in Δscd6 cells. DAN1 is a cell wall mannoprotein expressed under anaerobic conditions and repressed under aerobic conditions [62]. Although the precise role of DAN1 in anaerobic growth is not clear, it is interesting to note that the yeast strain lacking Scd6 is sensitive to oxygen deprivation stress [63]. The mechanisms underlying changes in DAN1 expression level and Δscd6 sensitivity to anaerobic stress is an interesting aspect to study in future. Strikingly, many transcripts encoding proteins of unknown function (PUF) were differentially expressed in the absence of Scd6 (refer to the deposited data, GEO: GSE145162). The functional significance of these changes is currently unclear. Studying the function of Scd6 in detail could reveal the function of genes encoding PUFs. Overall, the transcripts upregulated in Δscd6 formed two clusters (Supplementary Fig. 11). The first cluster contains eight genes with known or putative helicase functions, including YRF1–5 and YRF1–4. The second cluster includes transporter proteins, such as sugar transporter genes like HXT6 and HXT7, and sodium pump ENA2.
The decapping activator Sbp1 binds mRNA, likely in the 5’UTR region [30]. SBP1 deletion upregulated some genes, including HXK1 and HXT7 (Supplementary Fig. 6c), which are normally downregulated in the presence of glucose. Other genes, such as GSY1, GSY2, ALD4, and GLK1 that are downregulated in the presence of glucose were also found to be upregulated when the threshold value of log2FC was lowered to > 1 from > 2. HXK1 and GLK1 transcripts both encode for kinases that phosphorylate glucose in the first step of glycolysis. Expression of both the genes is repressed in the presence of glucose and de-repressed in the presence of non-fermentable carbon sources and galactose [64]. HXK2, a paralog of HXK1, is a predominant hexokinase that promotes repression of HXK1 and GLK1 expression in media containing glucose [65]. It was recently reported that Sbp1 overexpression from a galactose-inducible promoter leads to a growth defect on galactose media [25]. The growth defect could be due to sustained translation repression of HXK1 and GLK1 by Sbp1 overexpression, leading to less availability of non-glucose specific kinases. It would be important to test if the lack of de-repression of HXK1 and GLK1 upon Sbp1 overexpression in galactose media contributes to the growth defect phenotype.
A feature of a subset of differentially regulated mRNAs in Δsbp1 was their association with mitochondria. This included transcripts encoding mitochondrial enzymes like aldehyde dehydrogenase (ALD4) and glyoxylate reductase (GOR1), and mitochondrial membrane proteins like FMP45 and CYC7. The connection between Sbp1 and mitochondria-related transcripts would be an important direction to pursue in future studies.
Genes upregulated in Δsbp1 include stress responsive genes such as DDR2 and HSP12. Interestingly, these genes were also upregulated in wild type cells in response to sodium azide treatment. A similar observation was made with carbohydrate metabolic process genes such as GSY2, GPM2, GPH1, and SOL4. Based on the above observations, it is likely that these genes are repressed by Sbp1 under normal conditions and derepressed in response to sodium azide stress. This de-repression could be orchestrated by post-translational modifications of Sbp1, such as arginine methylation. Interestingly, Sbp1 methylation, which is increased upon glucose deprivation, promotes translation repression and decapping [25].
Comparing the sodium azide stress response in wild type and Δscd6 or wild type and Δsbp1 strains revealed differential expression in several classes of transcripts (Supplementary Fig. 4). In Δscd6 as compared to wild type in azide treatment, transcripts encoding cell cycle category proteins were significantly upregulated (Supplementary Fig. 8b). For example, PCL1/2, CLN1/2, and CLB5/6, which were otherwise down-regulated in the wild type untreated vs treated condition (Supplementary Fig. 8a). Other interesting observation which came out was the differential regulation of MAL32, an indicible protein involved in maltase catabolism. In Δscd6 the abundance of MAL32 transcript reduced significantly as compared to in wild type (Supplementary Fig. 10) but upon treatment of Δscd6 cells with sodium azide, transcript levels got enriched. These observations suggest that Scd6 might play a role in controlling the abundance of different transcripts during azide stress in a different manner. In the case of Δsbp1 sodium azide treated cells as compared to wild type treated cells, one of the subunits of cytochrome C oxidase (COX1) belonging to the GO category of aerobic electron transport chain (Supplementary Fig. 4c) is significantly upregulated (Supplementary Fig. 10). This is interesting given that COX1 mRNA was downregulated in wild type cells following sodium azide treatment (Supplementary Fig. 10). Further, Cox1 is a mitochondriallyencoded protein. Understanding the mechanism underlying differential modulation of mitochondrial transcripts by cytosolic translation repressor proteins will be an important future direction.
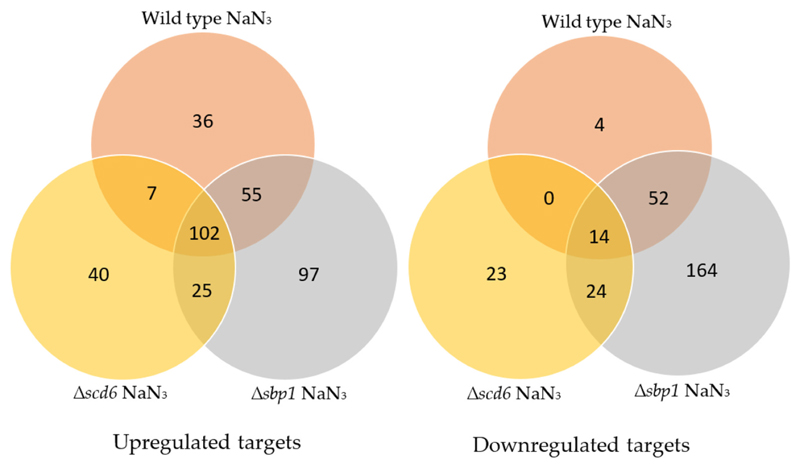
Comparative analysis of transcripts differentially expressed in response to sodium azide stress in wild type, Δscd6, and Δsbp1 strains (Fig. 7) indicated that 120 transcripts change in abundance only in Δsbp1. Similarly, 204 transcripts changed in abundance in Δscd6, but not in wild type or Δsbp1. Based on this, we propose that the absence of Sbp1 or Scd6 affects the transcriptome differentially in response to sodium azide stress. We observe that the number of genes differentially changing between wild type and Δsbp1 cells upon sodium azide treatment is much smaller (Fig. 2) than the wild type (untreated vs treated) and Δsbp1 (untreated vs treated). We interpret this to indicate that global expression level changes are similar in wild type and Δsbp1 backgrounds when azide treatment was given. This also seems to be the case between wild type and Δscd6 background. Our study provides a critical starting point to address the mechanistic details of the sodium azide stress response in yeast. The information provided by this study will allow hypothesis-driven dissection of the sodium azide stress response in general and specifically regarding translation repression and RNA granule assembly. The observations presented here identify the mRNA targets of Scd6 and Sbp1. Interestingly out of all the genes validated, two genes, HXT7 and CWP1, are affected by deletion of both Scd6 and Sbp1, although to different extents (Fig. 5b and Supplementary Fig. 6b & c). It is possible that both proteins act in conjunction to affect these mRNAs. It will be interesting to investigate the effect of Δsbp1Δscd6 double deletion on the commonly affected genes. As discussed in the introduction, since both the proteins can act as decapping activators, it is likely that the upregulated transcripts are a result of increased mRNA stability. Identifying mRNA decay targets of Scd6 and Sbp1 from the upregulated transcripts will be an obvious future endeavor. It is intriguing that the number of downregulated transcripts in the absence of these decapping activators are comparable to the number of upregulated transcripts (Fig. 2). Scd6 orthologs play a role in protection and maintenance of certain developmental mRNAs in Drosophila and C. elegans [24,32,33]. Upon depletion of Scd6 orthologs, the repression complex is hampered, leading to the loss of mRNA-protection in germ granules, making those transcripts susceptible to degradation. In our study, the genes downregulated in Δsbp1 and Δscd6 could be due to a similar loss of mRNA masking mechanisms. Such mRNAs could become substrates for mRNA degradation and hence get downregulated. It is possible that Scd6/Sbp1 could also affect transcription of some of these mRNAs either directly or indirectly. mRNA degradation can affect transcription, and some decay factors have been implicated in directly modulating transcription [66]. Interestingly, Sbp1 (earlier referred to as Ssb1) was reported as a nucleolar protein that can bind snRNAs based on studies using anti-Sbp1 antibodies [67]. However, GFP-tagged Sbp1 is uniformly localized in yeast cells [25]. Whether this is due to differences pertaining to live cell imaging versus fixed cell staining or due to GFP-tagging remains to be addressed. Many RGG-motif proteins, such as Npl3 and Gbp2, are nucleo-cytoplasmic shuttling proteins and are known to affect nuclear processes [23]. The ability of Scd6/Sbp1 to shuttle in and out of the nucleus followed by its impact on the target mRNAs identified in this study will be addressed in our future studies.
Fig. 7.
Venn diagrams showing number of genes that are differentially expressed upon sodium azide treatment in wild type, Δscd6 and Δsbp1 cells.
The following is the supplementary data related to this article.
The data discussed in this publication has been deposited in NCBI's Gene Expression Omnibus [68] and are accessible through GEO accession number: GSE145162 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145162).
Supplementary Data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ygeno.2020.05.001.
Acknowledgements
The authors thank Dr. Rajasekhara Reddy (Clevergene) for help with the analysis. We thank Prof. Dipankar Nandi at the Biochemistry department of IISc for help with the Real-time PCR machine. PIR thanks Dr. Meenal Vyas for providing motivation to initiate this work. MG thanks Raju Roy and Madhulika for helping with the validation of RNA-seq results.
Funding sources
This work was supported by the DBT/Wellcome Trust India Alliance Fellowship [grant number IA/I/12/2/ 500625] awarded to PIR. This work was also supported by the Department of Biotechnology grant (BT/PR13267/BRB/ 10/1444/2015) awarded to PIR. Funding from DBT-IISc partnership program is acknowledged. MG was supported by a University Grant Commission (UGC) fellowship. GP was supported by a fellowship from the Indian Institute of Science.
Footnotes
Declaration of Competing Interest
The authors declare no conflict of interest.
References
- [1].Galluzzi L, Yamazaki T, Kroemer G. Linking cellular stress responses to systemic homeostasis. Nat Rev Mol Cell Biol. 2018;19:731–745. doi: 10.1038/s41580-018-0068-0. [DOI] [PubMed] [Google Scholar]
- [2].Fulda S, Gorman AM, Hori O, Samali A. Cellular stress responses: Cell survival and cell death. Int J Cell Biol. 2010;2010 doi: 10.1155/2010/214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kültz D. Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol. 2005;67:225–257. doi: 10.1146/annurev.physiol.67.040403.103635. [DOI] [PubMed] [Google Scholar]
- [4].Uttara B, Singh AV, Zamboni P, Mahajan R. Oxidative stress and neurodegenerative diseases: A review of upstream and downstream antioxidant therapeutic options. Curr Neuropharmacol. 2009;7:65–74. doi: 10.2174/157015909787602823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liguori I, Russo G, Curcio F, Bulli G, Aran L, Della-Morte D, Gargiulo G, Testa G, Cacciatore F, Bonaduce D. Oxidative stress, aging, and diseases. Clin Interv Aging. 2018;13:757. doi: 10.2147/CIA.S158513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Halliwell B. Oxidative stress and cancer: Have we moved forward? Biochem J. 2007;401:1–11. doi: 10.1042/BJ20061131. [DOI] [PubMed] [Google Scholar]
- [7].Kumar V, Khan AA, Tripathi A, Dixit PK, Bajaj U. Role of oxidative stress in various diseases: Relevance of dietary antioxidants. J Pharm Exp Ther. 2015;4:126–132. [Google Scholar]
- [8].Duncan HM, Mackler B. Electron transport systems of yeast III. Preparation and properties of cytochrome oxidase. J Biol Chem. 1966;241:1694–1697. [PubMed] [Google Scholar]
- [9].Wilson D, Chance B. Azide inhibition of mitochondrial electron transport I. The aerobic steady state of succinate oxidation. Biochim Biophys Acta (BBA)-Bioenerg. 1967;131:421–430. doi: 10.1016/0005-2728(67)90002-3. [DOI] [PubMed] [Google Scholar]
- [10].Brockmann M, Stier T. Steady state fermentation by yeast in a growth medium. J Cell Comp Physiol. 1947;29:1–14. doi: 10.1002/jcp.1030290102. [DOI] [PubMed] [Google Scholar]
- [11].Šilhánková L, Smiovska V, Velemínský J. Sodium azide-induced mutagenesis in Saccharomyces cerevisiae . Mutat Res Fundam Mol Mech Mutagen. 1979;61:191–196. doi: 10.1016/0027-5107(79)90125-8. [DOI] [PubMed] [Google Scholar]
- [12].Rikhvanov EG, Varakina NN, Rusaleva TM, Rachenko EI, Voinikov VK. Sodium azide reduces the thermotolerance of respiratively grown yeasts. Curr Microbiol. 2002;45:0394–0399. doi: 10.1007/s00284-002-3758-x. [DOI] [PubMed] [Google Scholar]
- [13].Amesz W, van der Zeijst B. Azide as inhibitor of protein synthesis in yeast protoplasts. FEBS Lett. 1972;26:165–168. doi: 10.1016/0014-5793(72)80565-9. [DOI] [PubMed] [Google Scholar]
- [14].Jarett L, Hendler RW. 2, 4-Dinitrophenol and azide as inhibitors of protein and ribonucleic acid synthesis in anaerobic yeast. Biochemistry. 1967;6:1693–1703. doi: 10.1021/bi00858a018. [DOI] [PubMed] [Google Scholar]
- [15].Buchan JR, Yoon J-H, Parker R. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae . J Cell Sci. 2011;124:228–239. doi: 10.1242/jcs.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- [17].Ivanov P, Kedersha N, Anderson P. Stress granules and processing bodies in translational control. Cold Spring Harb Perspect Biol. 2019;11 doi: 10.1101/cshperspect.a032813. a032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- [19].Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae . J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Buchan JR, Parker R. Eukaryotic stress granules: The ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shah KH, Zhang B, Ramachandran V, Herman PK. Processing body and stress granule assembly occur by independent and differentially regulated pathways in Saccharomyces cerevisiae . Genetics. 2013;193:109–123. doi: 10.1534/genetics.112.146993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hoyle NP, Castelli LM, Campbell SG, Holmes LE, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rajyaguru P, She M, Parker R. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol Cell. 2012;45:244–254. doi: 10.1016/j.molcel.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Roy D, Rajyaguru PI. Suppressor of clathrin deficiency (Scd6)—An emerging RGG-motif translation repressor, Wiley Interdisc. Rev RNA. 2018;9:e1479. doi: 10.1002/wrna.1479. [DOI] [PubMed] [Google Scholar]
- [25].Bhatter N, Roy R, Shah S, Sastry SP, Parbin S, Iyappan R, Kankaria S, Rajyaguru PI. Arginine methylation augments Sbp1 function in translation repression and decapping. FEBS J. 2019;286:4693–4708. doi: 10.1111/febs.15057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell. 2010;39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Segal SP, Dunckley T, Parker R. Sbp1p affects translational repression and decapping in Saccharomyces cerevisiae . Mol Cell Biol. 2006;26:5120–5130. doi: 10.1128/MCB.01913-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Decourty L, Saveanu C, Zemam K, Hantraye F, Frachon E, Rousselle J-C, Fromont-Racine M, Jacquier A. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc Natl Acad Sci. 2008;105:5821–5826. doi: 10.1073/pnas.0710533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Barbee SA, Estes PS, Cziko A-M, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R. Staufen-and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mitchell SF, Jain S, She M, Parker R. Global analysis of yeast mRNPs. Nat Struct Mol Biol. 2013;20:127–133. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tsvetanova NG, Klass DM, Salzman J, Brown PO. Proteome-wide search reveals unexpected RNA-binding proteins in Saccharomyces cerevisiae . PLoS One. 2010;5 doi: 10.1371/journal.pone.0012671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jeske M, Moritz B, Anders A, Wahle E. Smaug assembles an ATP-dependent stable complex repressing nanos mRNA translation at multiple levels. EMBO J. 2011;30:90–103. doi: 10.1038/emboj.2010.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Snee MJ, Macdonald PM. Bicaudal C and trailer hitch have similar roles in gurken mRNA localization and cytoskeletal organization. Dev Biol. 2009;328:434–444. doi: 10.1016/j.ydbio.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Collart MA, Oliviero S. Preparation of Yeast RNA. John Wiley & Sons; 1993. Vol. ringbou edition. [DOI] [PubMed] [Google Scholar]
- [35].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liao Y, Smyth GK, Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- [37].Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J Integr Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Walter W, Sánchez-Cabo F, Ricote M. GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31:2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
- [40].Luo W, Brouwer C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yang L, Altman S. A noncoding RNA in Saccharomyces cerevisiae is an RNase P substrate. Rna. 2007;13:682–690. doi: 10.1261/rna.460607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jamieson DJ. Oxidative stress responses of the yeast Saccharomyces cerevisiae . Yeast. 1998;14:1511–1527. doi: 10.1002/(SICI)1097-0061(199812)14:16<1511::AID-YEA356>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- [43].Perpiñán E Herrero, Ros Salvador J, Bellí i Martínez G, Català E Cabiscol. Redox Control and Oxidative Stress in Yeast Cells. 2008 doi: 10.1016/j.bbagen.2007.12.004. [DOI] [PubMed] [Google Scholar]
- [44].Parrou JL, Teste M-A, François J. Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: Genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology. 1997;143:1891–1900. doi: 10.1099/00221287-143-6-1891. [DOI] [PubMed] [Google Scholar]
- [45].Coleman ST, Fang TK, Rovinsky SA, Turano FJ, Moye-Rowley WS. Expression of a glutamate decarboxylase homologue is required for normal oxidative stress tolerance in Saccharomyces cerevisiae . J Biol Chem. 2001;276:244–250. doi: 10.1074/jbc.M007103200. [DOI] [PubMed] [Google Scholar]
- [46].Slekar KH, Kosman DJ, Culotta VC. The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J Biol Chem. 1996;271:28831–28836. doi: 10.1074/jbc.271.46.28831. [DOI] [PubMed] [Google Scholar]
- [47].Birben E, Sahiner UM, Sackesen C, Erzurum S, Kalayci O. Oxidative stress and antioxidant defense. World Allergy Org J. 2012;5:9–19. doi: 10.1097/WOX.0b013e3182439613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- [49].Buchan JR. mRNP granules: Assembly, function, and connections with disease. RNA Biol. 2014;11:1019–1030. doi: 10.4161/15476286.2014.972208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chiu J, Tactacan CM, Tan S-X, Lin RC, Wouters MA, Dawes IW. Cell cycle sensing of oxidative stress in Saccharomyces cerevisiae by oxidation of a specific cysteine residue in the transcription factor Swi6p. J Biol Chem. 2011;286:5204–5214. doi: 10.1074/jbc.M110.172973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Klis FM, Mol P, Hellingwerf K, Brul S. Dynamics of cell wall structure in Saccharomyces cerevisiae . FEMS Microbiol Rev. 2002;26:239–256. doi: 10.1111/j.1574-6976.2002.tb00613.x. [DOI] [PubMed] [Google Scholar]
- [52].Rodicio R, Heinisch JJ. Together we are strong—cell wall integrity sensors in yeasts. Yeast. 2010;27:531–540. doi: 10.1002/yea.1785. [DOI] [PubMed] [Google Scholar]
- [53].Lee YM, Kim E, An J, Lee Y, Choi E, Choi W, Moon E, Kim W. Dissection of the HOG pathway activated by hydrogen peroxide in Saccharomyces cerevisiae . Environ Microbiol. 2017;19:584–597. doi: 10.1111/1462-2920.13499. [DOI] [PubMed] [Google Scholar]
- [54].Simoes T, Mira N, Fernandes A, Sá-Correia I. The SPI1 gene, encoding a glycosylphosphatidylinositol-anchored cell wall protein, plays a prominent role in the development of yeast resistance to lipophilic weak-acid food preservatives. Appl Environ Microbiol. 2006;72:7168–7175. doi: 10.1128/AEM.01476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Overkamp KM, Kötter P, van der Hoek R, Schoondermark-Stolk S, Luttik MA, van Dijken JP, Pronk JT. Functional analysis of structural genes for NAD+-dependent formate dehydrogenase in Saccharomyces cerevisiae . Yeast. 2002;19:509–520. doi: 10.1002/yea.856. [DOI] [PubMed] [Google Scholar]
- [56].Tomitori H, Kashiwagi K, Asakawa T, Kakinuma Y, Michael AJ, Igarashi K. Multiple polyamine transport systems on the vacuolar membrane in yeast. Biochem J. 2001;353:681–688. doi: 10.1042/0264-6021:3530681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Poornima G, Shah S, Vignesh V, Parker R, Rajyaguru PI. Arginine methylation promotes translation repression activity of eIF4G-binding protein, Scd6. Nucleic Acids Res. 2016;44:9358–9368. doi: 10.1093/nar/gkw762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, Davis RW, Esposito RE. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- [59].Tevzadze GG, Pierce JV, Esposito RE. Genetic evidence for a SPO1-dependent signaling pathway controlling meiotic progression in yeast. Genetics. 2007;175:1213–1227. doi: 10.1534/genetics.106.069252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Cooper KF, Mallory MJ, Egeland DB, Jarnik M, Strich R. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc Natl Acad Sci. 2000;97:14548–14553. doi: 10.1073/pnas.250351297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Coluccio A, Bogengruber E, Conrad MN, Dresser ME, Briza P, Neiman AM. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae . Eukaryot Cell. 2004;3:1464–1475. doi: 10.1128/EC.3.6.1464-1475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Sertil O, Cohen BD, Davies KJ, Lowry CV. The DAN1 gene of S. cerevisiae is regulated in parallel with the hypoxic genes, but by a different mechanism. Gene. 1997;192:199–205. doi: 10.1016/s0378-1119(97)00028-0. [DOI] [PubMed] [Google Scholar]
- [63].Samanfar B, Omidi K, Hooshyar M, Laliberte B, Alamgir M, Seal AJ, Ahmed-Muhsin E, Viteri DF, Said K, Chalabian F. Large-scale investigation of oxygen response mutants in Saccharomyces cerevisiae . Mol BioSyst. 2013;9:1351–1359. doi: 10.1039/c3mb25516f. [DOI] [PubMed] [Google Scholar]
- [64].Herrero P, Galindez J, Ruiz N, Martinez-Campa C, Moreno F. Transcriptional regulation of the Saccharomyces cerevisiae HXK1, HXK2 and GLK1 genes. Yeast. 1995;11:137–144. doi: 10.1002/yea.320110205. [DOI] [PubMed] [Google Scholar]
- [65].Rodriguez A, Cera Tdl, Herrero P, Moreno F. The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae . Biochem J. 2001;355:625–631. doi: 10.1042/bj3550625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Haimovich G, Medina DA, Causse SZ, Garber M, Millán-Zambrano G, Barkai O, Chávez S, Pérez-Ortín JE, Darzacq X, Choder M. Gene expression is circular: Factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153:1000–1011. doi: 10.1016/j.cell.2013.05.012. [DOI] [PubMed] [Google Scholar]
- [67].Clark MW, Yip M, Campbell J, Abelson J. SSB-1 of the yeast Saccharomyces cerevisiae is a nucleolar-specific, silver-binding protein that is associated with the snR10 and snR11 small nuclear RNAs. J Cell Biol. 1990;111:1741–1751. doi: 10.1083/jcb.111.5.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.