Abstract
Like many organisms, bacteria and archaea have both innate and adaptive immune systems to defend against infection by viruses and other parasites. Innate immunity most commonly relies on endonuclease cleavage of any incoming DNA that lacks a specific epigenetic modification, through a system known as Restriction-Modification. CRISPR-Cas adaptive immunity relies on the insertion of short DNA sequences from parasite genomes into CRISPR loci on the host genome to provide sequence-specific protection. The discovery of each of these systems has revolutionized our ability to carry out genetic manipulations, and, as a consequence, the enzymes involved have been characterized in exquisite detail. In comparison, much less is known about the importance of these two arms of the defence for the ecology and evolution of prokaryotes and their parasites. Here we review our current ecological and evolutionary understanding of these systems in isolation, and discuss the need to study how innate and adaptive immune responses are integrated when they coexist in the same cell.
Introduction
Prokaryotes face infection by a wide range of genetic elements, from lytic viruses to plasmids and integrative elements that can confer fitness benefits. Prokaryotes thus experience selective pressures to defend themselves against parasitic threats, while, if possible, retaining the ability to associate with benign symbionts. Although prokaryotes have a large repertoire of defence systems [1], by far the most widespread defences are Restriction-Modification (RM) and CRISPR-Cas immune systems. RM systems are present in over 90% of sequenced bacterial and archaeal genomes [2], while CRISPR-Cas systems are found in approximately 30-40% of bacterial and >90% of archaeal genomes [3].
The mechanisms of RM and CRISPR immunity have been extensively studied [1], in part due to their value as tools for genetic manipulation. However, the importance of these systems for the ecology and evolution of prokaryotes and their parasites is less well understood. We briefly summarize the mechanistic basis of RM and CRISPR immunity, which has been covered in detail in several recent reviews [4–9]. We discuss the factors governing the distribution of these innate and adaptive immune systems, and their consequences for prokaryotic ecology and evolution. We highlight that despite their frequent co-occurrence [10], most studies have been carried out on individual systems in isolation, and emphasise the need to examine how these systems interact when they coexist in the same cell.
Overview of immune mechanisms in prokaryotes
RM systems: innate immunity based on detecting DNA modification states
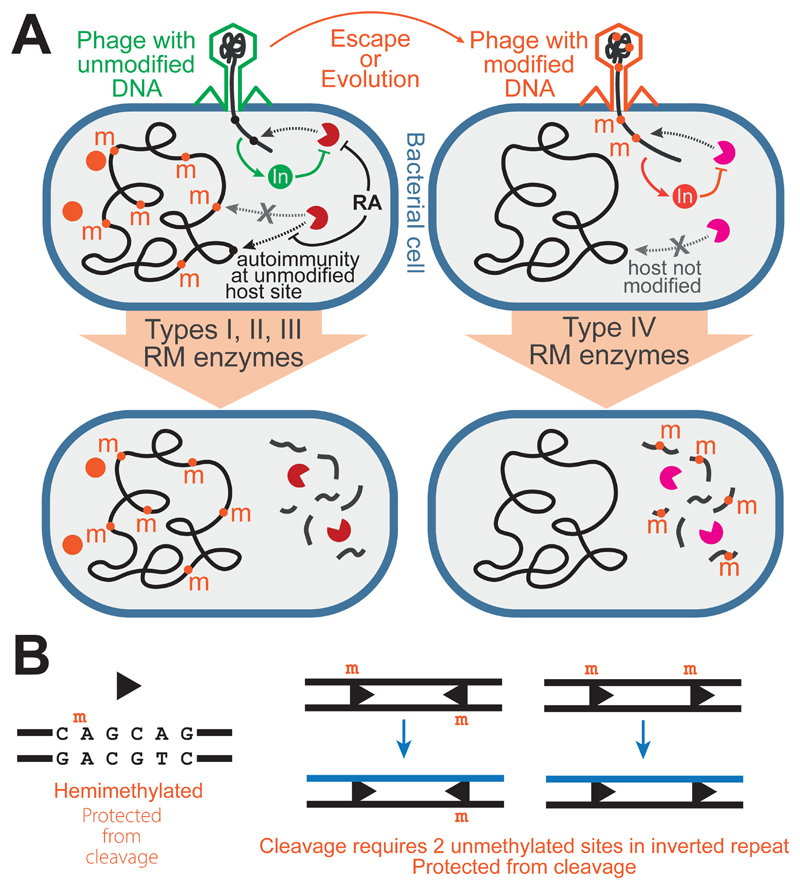
RM systems are innate immune systems that recognise structural features on specific sequences of DNA bases, and target DNA identified as “foreign”. RM systems are divided into Types (I, II, III and IV), largely based on mechanistic properties rather than evolutionary relatedness [2]. Within Types I and III, the relatedness is greater and mechanistic properties more similar compared to Types II and IV. The common feature of Types I, II and III RM systems is specific DNA sequence recognition by an endonuclease activity which triggers a dsDNA break on foreign DNA while self-DNA is protected from cleavage by covalent methylation of the same (cognate) sequences on the host genome producing N6-methyladenine, 5-methylcytosine or N4-methylcytosine (Figure 1A). Type IV enzymes lack methyltransferase activity since they target modified nucleotides on foreign DNA (Figure 1A). Distinguishing self from non-self is thus based on DNA modification (Types I to III) or its absence (Type IV).
Figure 1. Fundamental mechanisms of RM immunity.
(A) Cartoons of bacterial cells infected by phage and the effect of Types I - III (left panel) and Type IV (right panel) RM systems. Types I - III comprise an endonuclease activity (Pacman) and methyltransferase (orange circle). DNA cleavage is targeted to specific sequences (circles) that are protected on the host genome by methylation (m). Appearance of unmethylated sites on the host leads to autoimmunity which is prevented for Type I systems by restriction alleviation (RA). Cleavage can be prevented by phage-encoded inhibitors (In). Type IV systems comprise only an endonuclease. (B) Asymmetric sites (arrowhead) that are only hemimethylated are protected following replication (one daughter DNA shown) by a necessity for interaction between two sites in inverted repeat to activate cleavage.
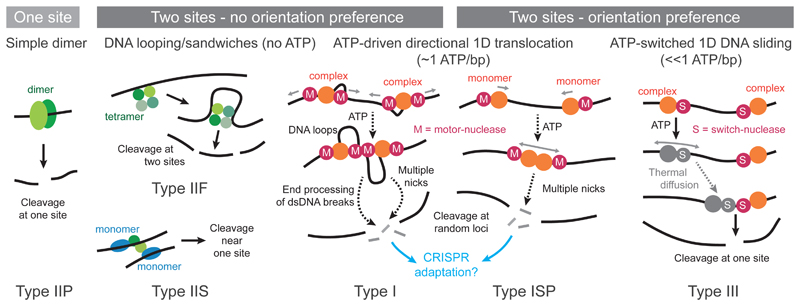
Type I enzymes form multiprotein complexes that undertake both DNA methylation and cleavage. The HsdS subunit provides the recognition site specificity. Target sites are bipartite, comprising semi-specific sequences 3-6 bp separated by a 4-9 bp non-specific spacer. DNA cleavage occurs at distant non-specific sites and requires interaction of two enzyme complexes and ATP hydrolysis [5] (Figure 2). Type ISP enzymes comprise a single polypeptide with recognition, methyltransferase, translocase and endonuclease activities [5].
Figure 2. Mechanisms of DNA cleavage by Type I - III RM enzymes.
The majority of RM enzymes require communication between two target sites to activate cleavage, using either energy-independent DNA looping or ATP-dependent mechanisms.
Type II enzymes are the largest and most diverse group [6]. Classical Type IIP enzymes comprise separate endonuclease and methyltransferase proteins. DNA cleavage occurs within or close to the recognition sites which are 4-8 bp. Several subclasses have the methyltransferase and endonuclease proteins fused as a single polypeptide. Types IIE, IIF and IIS bind two sites and capture a DNA loop to activate cleavage (Figure 2).
Type III enzymes form multiprotein complexes that undertake both DNA methylation and cleavage [2]. Target sites are asymmetric and 5-6 bp, and cleavage occurs ~2.5 DNA turns downstream of one site but requires a pair of recognition sites in inverted repeat and ATP to initiate the reaction [11] (Figure 1B).
Some phages have evolved metabolic pathways to modify bases or have acquired methylation due to avoiding restriction in a cell with a Type I – III RM system. To counter phages carrying these modifications, bacteria evolved Type IV enzymes [4]. These have an endonuclease activity that targets DNA but lack a cognate methyltransferase (Figure 1A). A diverse range of mechanisms have evolved, some of which appear to require interaction with multiple modified sites and some of which require an input of chemical energy (ATP or GTP). Type IV enzymes are the least well-understood at both mechanistic and ecological levels.
CRISPR-Cas, an adaptive immune system
Acquisition of CRISPR-Cas adaptive immunity requires exposure to an MGE. At initial exposure, a “memory” of infection can be recorded on the host DNA by recombining short sequences from the foreign genome. These sequences are then used to detect and destroy the MGE in subsequent infections.
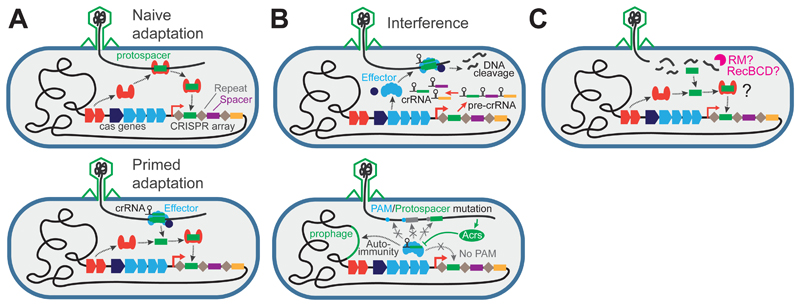
CRISPR arrays are the immunological memory of the CRISPR-Cas immune system, and consist of repeating sequences (repeats, typically 20-40bp) interspersed with variable sequences (spacers, typically 20-40bp) that are complementary to MGE sequences (Figure 3). Cas genes encode proteins responsible for immunity which occurs in three stages: spacer acquisition (often referred to as “adaptation”), expression, and interference [7]. During acquisition, a protein complex including the conserved Cas1 and Cas2 enzymes inserts MGE sequences (protospacers) into the leader end of CRISPR arrays and duplicates the repeats, forming new spacers [12] (Figure 3A). During the expression stage, CRISPR arrays are transcribed and processed into CRISPR RNAs (crRNAs) and loaded onto Cas proteins. Finally, during interference, crRNAs guide Cas effectors towards complementary MGE nucleic acids, triggering cleavage (Figure 3B). Primed adaptation, in which effector recognition triggers further spacer acquisition [13], has been demonstrated in a subset of Type I systems and recently a similar mechanism has been proposed for Type II systems [14] (Figure 3A). To ensure that the system targets only infectious DNA, and not the CRISPR array on the host genome, CRISPR-Cas effector complexes use the sequence flanking the complementary target for discrimination (Figure 4): in a CRISPR array the flanking sequence consists of a CRISPR repeat sequence, whereas targets in an infectious genome are often selected such that they are flanked by the conserved protospacer adjacent motif (PAM) [15,16].
Figure 3. CRISPR-Cas adaptive immunity.
(A) Adaptation is the uptake of MGE sequences by the Cas1-Cas2 complex (orange) into the leader of the CRISPR array. Primed adaptation is facilitated by the effector complex. (B) Interference is the specific recognition and cleavage of an MGE using crRNA processed from the CRISPR array. The process can be blocked by mutation of the PAM and/or protospacer sequences, or by phage-encoded inhibitors (Acrs). Autoimmunity is avoided by an absence of PAMs in the CRISPR array but may occur where foreign sequences are recombined e.g. prophage. (C) Nucleic acid processing by RM enzyme activity or repair nucleases working on stalled replication forks (e.g. RecBCD, [17]) may provide polynucleotide fragments that feed into adaptation.
Figure 4. Polynucleotide cleavage by type I, II and III CRISPR-Cas effectors.
See main text for full details. Polynucleotide cleavage is shown by the orange arrowheads.
CRISPR-Cas systems have been classified into 2 main classes, 6 types and >30 subtypes, based on the phylogeny of Cas1, a conserved spacer acquisition protein, as well as signature genes and gene synteny [18]. However, some of these types and subtypes are relatively rare and concentrated in a few clades [19]. In this review we focus on the more frequent types I (30% of all genomes), II (8%) and III (6%) (Figure 4).
Types I and III belong to Class 1 and encode crRNA-Cas effectors composed of a single crRNA and multiple protein subunits (Figure 4A). Type I systems encode the Cascade ribonucleoprotein complex and a separate ATP-dependent helicase-nuclease Cas3 [20]. Cascade first scans DNA for PAMs. The DNA is then unwound to allow base-pairing between the crRNA and the complementary protospacer (R-loop). Full R-loop zipping recruits Cas3, which cleaves the non-targeted strand within or close to the protospacer. This process in turn provides substrates for spacer acquisition, resulting in more efficient “primed adaptation”, even if targets contain mutations in the protospacer or PAM (Figure 3A)[12]. The majority of Type III systems form Csm or Cmr ribonucleoprotein complexes, which share structural similarities [21] (Figure 4A). These complexes use crRNA to bind complementary RNA transcripts, which triggers Cas10-mediated DNA and Cas7-mediated RNA cleavage activities. Cas10 also produces cyclic oligoadenylates which activate a non-specific RNase activity [22].
Type II systems belong to Class 2, which encode a single effector protein, known as Cas9, which forms a complex with the crRNA and a trans-activating CRISPR RNA, tracrRNA. Cas9 effectors scan DNA for PAMs and form an R-loop with the target DNA sequence. Full complementarity activates the nuclease domains to cut both strands close to the PAM [23,24]. The RNA-guided dsDNA break activity of Cas9 proteins has been widely used for genome editing [9].
Costs and benefits of prokaryotic immune systems
Immune systems commonly carry both fitness costs and benefits which will affect system prevalence (Figure 5). Quantifying these, and understanding how they depend on their environment, can help explain the observed distribution of immune systems in nature.
Figure 5. Summary of balancing factors that can affect immune system prevalence.
Immune systems provide strong benefits in the presence of lytic phage
In the absence of parasites, costs should favour hosts without immune systems; on the other hand, hosts will experience strong selection for immunity when lethal parasites are present. Indeed, bacteria with RM [25] or CRISPR-Cas [26–28] immunity can increase rapidly in frequency in the presence of virulent phages. However, the selective benefits will depend on the level of protection they confer, which varies depending on the system, the phage, and the number and position of sequences targeted [29–34]. In addition, the rate of spacer acquisition can severely limit the benefits of CRISPR-Cas immune systems: if this is low – as is often the case in laboratory culture – the benefits of carrying an adaptive immune system are marginal [35].
Immune systems can be maladaptive in the presence of temperate phages
Temperate phages can replicate through both the lytic and lysogenic cycle. Targeting of incoming phages is beneficial, but targeting integrated prophages leads to immunopathological effects, since cleavage of the phage genome results in a break in the host chromosome (Figure 3B). Indeed, type I and II CRISPR-Cas systems cause cell death when they are programmed to target integrated prophages [36]. Consistent with these findings, carrying a CRISPR immune system was shown to be maladaptive during temperate phage infection of phage-sensitive, but primed, cells (i.e. cells carrying spacers that imperfectly match the temperate phage) [37]. Type III CRISPR-Cas systems, which rely on active transcription (Figure 4), can target phages that replicated through the lytic cycle, whilst tolerating prophages that repress their transcription [38]. However, low levels of transcription of integrated prophages can still lead to a high fitness cost of immunity [39].
RM systems can provide clear benefits in the context of temperate phage infection. During infection with phage λ, experiments with a large panel of RM systems showed that each system favoured lysogeny at the population scale, even though these systems are unable to discriminate between phages that enter the lytic or lysogenic cycle. Instead, this effect was due to RM immunity delaying successful infections until most cells are near to stationary phase, a state in which the probability of lysogeny is greater [32]. This might help explain why temperate phages avoid restriction sites to a lesser extent than lytic ones [40].
Targeting the mobile gene pool entails opportunity costs that can be mitigated
Ideally, immune systems provide protection against parasitic MGEs, but allow for the association with beneficial ones. Yet, immune systems can also limit acquisition of plasmids and prophages, which often confer environment-specific fitness benefits to their hosts [41,42]. Both RM and CRISPR-Cas systems can provide immunity against plasmid conjugation [43,44], transformation [45,46] and transduction [47,48]. While immunity can be beneficial in the presence of costly plasmids [49], both RM and CRISPR-Cas are disadvantageous when targeting beneficial plasmids [44,46,50], causing selection for inactivated RM or CRISPR-Cas systems [50]. When hosts are simultaneously exposed to parasitic and mutualistic elements there may be a trade-off between immunity and access to mutualists.
However, defence systems may discriminate between beneficial and parasitic MGEs. CRISPR-Cas immune systems can do so owing to their high sequence specificity (20-40 nucleotide protospacer targets), which is more limited for restriction enzymes (typically recognition sites of 4-8 nucleotides). Indeed, CRISPR-Cas immunity can lead to elevated levels of generalized transduction, because it protects cells from phage infection but does not cleave encapsulated host DNA of transducing particles [48]. The spacer content of CRISPR arrays can specialize on parasitic sequences over time, through selection and primed adaptation. Indeed, most identified spacers in sequenced genomes are from phages, and a smaller proportion from other MGEs [51].
While RM systems lack the ability to discriminate between beneficial and parasitic MGEs based on their sequence, they will favour MGE exchange among closely related strains over more distantly related ones. Indeed, plasmids are more efficiently transferred among kin than non-kin, a pattern explained partially by shared RM systems [52]. For beneficial MGEs, this preferential transfer among kin is favoured by kin selection, because it allows host cells to restrict MGE benefits to clonemates [53].
Finally, immune systems may use the entry route of MGEs and whether the nucleic acid is single or double stranded for target discrimination. In conjugation and natural transformation, DNA enters the cell single stranded, and ssDNA results in less restriction compared to lytic phage infection [43,54]. CRISPR-Cas systems also acquire spacers preferentially from free dsDNA ends [17], which favors spacer acquisition during dsDNA phage injection [31]. Restriction can also be alleviated when competence is induced [55], and some RM systems even protect DNA entering through natural transformation by dedicated ssDNA methylation [56].
The effect of immunity on MGEs, if consistent over time, could have longer-term consequences on horizontal gene transfer (HGT). RM indeed limits HGT, but specifically among strains bearing non-cognate systems [57]. The role of Type IV systems may be more limited, but this will depend on the epigenetic status of MGEs in the environment, which has not been sufficiently examined. Conclusions vary about CRISPR-Cas effect on long term HGT [58–60]. Some lineages with strong signatures of HGT are depleted in CRISPR-Cas systems [61], but across lineages there are no clear correlations between HGT and CRISPR-Cas activity [59]. Perhaps the transient presence of immune systems in lineages, combined with the high frequency of anti-immune genes born on MGEs [57,58,62,63] obscures these signatures.
Immune systems entail costs linked to activity and self-targeting
Immunity is based on enzyme functionality, which entails a metabolic cost. This cost can be constitutive, due to constitutive enzyme expression [64], or present only upon phage infection [26,64]. For instance, translocation of Type I and III RM enzymes can consume as much as one ATP for each base pair [65], but occurs only upon phage infection. The activity of immunity systems can also affect other cellular processes, for instance through the effect of RM methylation on global epigenetic patterns [66], or CRISPR-Cas interference with DNA repair [67]. The consequences of these pleiotropic effects will depend on the environment.
Immune systems can also present autoimmunity costs. DNA repair and HGT of RM systems create unmodified sites potentially targeted by Types I-III restriction (Figure 1A). Failure to repair DNA damage can result in a new recognition site while an existing methylated site may become demethylated due to repair. Although occurring at low frequency, these events are sufficient to generate toxic dsDNA breaks [68]. When a naïve strain acquires a new RM system by HGT, thousands of unmodified host recognition sites become targets, which can also result in recipient cell death [69]. Whether modified DNA-dependent Type IV RM systems suffer similar autoimmunity issues is unclear (Figure 1A). CRISPR-Cas systems can also cause autoimmune issues by self-targeting (Figure 3B). A small proportion of spacers target loci of the host genome [70,71], which can lead to chromosome dsDNA breaks, growth inhibition and filamentation [72].
Regulation of the expression and activity of immune systems limits their metabolic and autoimmune costs [73]. In Type I RM systems, nuclease activity is downregulated upon translocation events on the host genome but not on invading DNA, a phenomenon called Restriction Alleviation [5]. Host DNA translocation can trigger ClpXP-dependent proteolytic digestion of the nuclease subunit [68,74], or be inefficient compared to efficient translocation/cleavage of foreign DNA [74]. For CRISPR-Cas systems, upregulation of CRISPR-Cas expression frequently occurs following infection [75], through the activation of stress responses or detection of changes in cell metabolism that follow infection. CRISPR-Cas immunity can also be induced by quorum sensing, which anticipates infection by indicating cell densities are high [76,77]. Other mechanisms can also bias spacer acquisition towards foreign sequences. Spacer acquisition is particularly high at stalled replication forks, which are more abundant on foreign DNA, and limited by Chi sites on the chromosome [17] (Figure 3C). The phenomenon of priming in type I systems will then create a positive feedback, amplifying spacer acquisition from previously encountered threats.
In addition, autoimmunity is also limited due to past negative selection of self-targeting. Self-targeting CRISPR-Cas spacers are rare, and they are enriched at the leader end of arrays, suggesting they are recent and strongly selected against [70]. In genomes containing Type II RM systems, restriction site avoidance can also be detected [78]. Chromosomal avoidance is even stronger than observed on phage genomes, suggesting auto-immunity represents a strong selective pressure [40]. Still, some degree of autoimmunity appears an unavoidable trade-off of maintaining efficient immunity. RM systems with higher restriction efficiency also have higher self-restriction [79]. That many RM enzymes must bind two sites to activate cleavage (Figure 2) may be an evolutionary adaptation to prevent autoimmunity where a single unmodified site arises [80], but will also limit immunity. Reliance on sites containing PAMs to limit auto-immunity also limits the choice of spacers available for efficient CRISPR-Cas immunity. On the other hand, the likely absence of auto-immunity in Type IV restriction, at least while modification targets are absent from the host, might explain relatively degenerate sequence context of modification, as it is free to evolve without that trade-off [4].
Selfish behaviour promotes immune system maintenance at a cost to their hosts
Over longer evolutionary timescales, immunity genes are part of the few gene categories to be on average negatively selected [81]. This suggests that costly parasites are not encountered often enough to balance the assorted costs of immune systems, and/or that immunity is not efficient enough (Figure 5). Importantly, long-term immune system maintenance in prokaryotic populations thus requires HGT [82]. HGT decouples immune system fitness from the one of their hosts, allowing them to act as selfish genetic elements as they can spread despite increased costs to the host.
Type II RM systems can exhibit particularly strong selfish behaviour, leading to host killing [83] (Figure 5). In these systems, loss of the M gene eventually leads to toxic dsDNA breaks as methylation patterning is diluted [84]. Even when R and M genes are lost simultaneously (commonly by failed segregation of plasmids carrying the system), endonuclease activity is usually more stable than methyltransferase activity, leading to post-segregational killing similarly to other toxin-antitoxin systems [85]. For the more mechanistically-complex Type I and III RM systems, gene loss does not cause detectable viability problems [68,86]. This may reflect the assembly of these systems into higher order RM machines where the loss of the M genes causes failure of the complete complex (Figure 2), or more stringent control and restriction alleviation. Type II systems thus appear to be the most selfish RM variants. Accordingly, they are also the systems most abundant on MGEs, experiencing frequent HGT [10]. The toxic effects associated with Type II loss do not occur upon entry in a new host due to regulation by associated C protein transcriptional regulators which delay restriction until de novo methylation of the host [87].
Competition between systems can harm or benefit the host: when two RM systems with the same sequence specificity coexist within a cell, each system’s modification protects the host from restriction by the other [88]. Alternatively, when two Type II R genes are regulated by the same C protein, entry of the second RM system causes upregulated restriction activity before the genome is fully methylated, causing cell death [89].
Host death or dormancy can benefit the host in the presence of parasites
Although CRISPR-Cas systems do not have such clear selfish addiction behaviour, several Cas components are evolutionary related to toxins and could arrest growth when induced, similarly to toxin-antitoxin systems [90]. Growth arrest or even death following phage infection can benefit the host population if it stops phages from completing their lytic cycle. Such “Abortive Infection” (Abi) has been demonstrated in some CRISPR-Cas type IF systems [91], and in type VI systems, where RNA targeting leads to growth arrest [92]. Type III systems also activate promiscuous RNAse activity [22,93] (Figure 4), which might also lead to Abi [8]. Competition among RM systems, despite best understood as a selfish behaviour, might similarly benefit host populations if a resident system stops the spread of a more deleterious invasive system.
Because cells engaging in abortive infection stop reproducing, the success of an abortive infection strategy requires the benefits from decreased phage encounters being directed at individuals that are related to the ones paying the cost [94]. However, it has been suggested that dormancy following phage infection might also benefit individual host cells by slowing down metabolism and phage reproduction, giving the host time to mount an immune response [90]. This might particularly be critical to allow spacer integration and CRISPR-Cas adaptive immune response [95].
Overall, bearing an immune system can translate into large benefits when providing defence against parasites, but also presents costs arising from immune activity, targeting the chromosome or beneficial MGEs, or selfish behaviour. Consequently, net fitness effects of immune systems will depend on the balance between all these factors (Figure 5). Moreover, the efficacy of immunity is not a fixed parameter, but evolves as part of an arms race between immune systems and parasites.
Eco-evolutionary dynamics of immune systems and parasites
Parasites escape rapidly from immunity
Parasites commonly escape immunity by genetic or epigenetic mutation of the targeted sequence. For Type I-III RM systems, phage escape occurs through accidental methylation by the host [96], with probabilities ranging from 10-1 to 10-6 per infection [32] depending on relative restriction and modification efficiencies and the number of restriction sites [30]. High probability of phage escape means that the advantage of carrying a RM system is short-lived [25], and other mechanisms of resistance become more relevant after phages overcome restriction [96,97]. Phages can similarly escape CRISPR-Cas immunity by mutating their target sequence or PAM [15] (Figure 3B). If a single site is targeted, type I and II CRISPR-Cas escape by mutation is easier to achieve than for RM systems (commonly targeting multiple sites per MGE) [28]. However, because different bacteria in a population often acquire different spacers, it becomes increasingly hard to overcome CRISPR immunity (discussed below) [98–101]. Escape is even more limited against type III CRISPR-Cas systems because mismatches do not totally suppress interference [102]. Overall, promiscuous immune systems - which can cleave imperfect target sequences - are less susceptible to phage escape. KpnI, a restriction endonuclease that can cleave at non-canonical sequences, confers higher protection against phage than a more specific variant, because it still restricts a fraction of modified phages [103]. However, immune promiscuity also increases self-targeting [79,102], highlighting the trade-off between protection and auto-immunity.
Immune systems can evolve new immune specificities
CRISPR-Cas immune systems rapidly evolve new specificities through spacer incorporation. Type II RM systems are generally more inert: having functionally distinct DNA recognition elements in the separate methyltransferase and endonuclease, evolution of new recognition sites must require convergent evolution. On the other hand, Type I RM systems are particularly adept at evolving new DNA recognition specificity. The specific half-sites of a Type I recognition site are recognised by two target recognition domain (TRD) folds of the HsdS subunit that are separated by a coiled-coil linker that acts as a molecular ruler, setting the non-specific DNA spacer length (Figure 6A). HsdS subunits show structural plasticity: TRDs can be swapped within and between bacteria to generate new recognition sites [104], variation of ±4 amino acids in the first alpha helix of the linker changes the spacer length [105] (Figure 6B), and two half HsdS subunits can dimerise to recognise a palindromic site [106] (Figure 6C). So-called “Shufflons” are Type I operons that exploit HsdS structural plasticity [107] (Figure 6D) and have been identified in many species [108]. They can form replacement recognition subunits using site-specific recombination to “flip” and rearrange HsdS genes, at timescales similar to the ones of spacer acquisition [109].
Figure 6. Generation of diversity in Type I RM systems by genetic recombination of the HsdS DNA recognition subunit.
(A) Computational model of the EcoKI HsdS subunit bound to DNA (PDB:2Y7H, [110]) demonstrating how the target recognition domains (TRDs) and coiled coil region (CCR) allow HsdS to recognise an asymmetric bipartite DNA sequence. (B) Changes in the number of TAEL amino acid repeats in CCR1 of EcoR124I and EcoR124II changes the number of non-specific nucleotides in the spacer. (C) Dimerisation of half HsdS subunits produces a Type I enzyme that recognises a palindrome sequence. (D) Shufflon system. Reversible site-specific inversion between recombination sequences within two inverted hsdS genes produces HsdS subunit that recognise one of two sequences as one of the TRDs is swapped.
Immune specificities can also be acquired through HGT [10,82]. Whole systems can be transferred, but transfer of the subunits encoding specificity can be sufficient, as with hsdS subunits encoded on plasmids [111], or CRISPR array spacers: the recombination of spacers with the phage protospacers they target can even lead to specialized transduction of CRISPR elements [112]. HGT might be a significant factor in generating immune variability, particularly when other mutational pathways are less active [113].
Group-level immunity can counteract parasite escape
If immune hosts differ in target specificity, a phage that overcomes immunity of one host genotype remains sensitive to others. CRISPR-Cas systems often present high diversity in spacer content [26–28,114], that is suggested to lead to ‘distributed immunity’ [115,116]. Diversity benefits were demonstrated by manipulating individual CRISPR-Cas immune clones each with a single targeting spacer. Increasing population-level diversity led to faster extinction of virus populations, which was associated with a reduced evolution and spread of escape mutants [98–101]. These benefits depend on population structure: in spatially structured populations, interactions between clones are strongly reduced, limiting the benefits of diversity [117].
RM immunity diversity could similarly benefit host populations. A rare strain with a different immune specificity should gain a short-term fitness advantage in the presence of phages that escaped restriction from a dominant strain, leading to negative frequency-dependent selection among strains [25]. In S. pneumoniae, a strain bearing an active shufflon generating high levels of diversity appears to have increased resistance to phages compared to ‘locked’ forms not able to undergo phase variation [108,118]. Additionally, phenotypic diversity can arise from variation in the expression of RM systems in a population. Some, particularly Type III, contain repeated sequences that lead to phase variation in ON/OFF expression with subpopulations not expressing any RM function [119]. A recent survey found that 17% of mod genes contain sequence repeats with potential for phase variable expression [120]. Variation in expression of RM immunity has been proposed to benefit host populations because the presence of sensitive hosts would reduce the abundance and weaken the selective pressure for escape phages [121,122]. However, experimental tests of the dynamics and benefits of RM diversity are still lacking.
When multiple systems coexist in a genome, this “within-individual” diversity in specificity also increases the efficacy of targeting, making escape less probable. Bacteria often do not need to encode multiple full-blown RM systems in order to encode multiple specificities, as multiple HsdS subunits can combine with a single HsdM/HsdR complex [111]. Modelling suggests that individual-based diversity might drive population-based diversity: bacteria first accumulate diverse RM systems within cells – each new system providing additional immunity to phages – until phage escape promotes the loss of immunity leading to the evolution of diversity among lineages [123]. Genomes can also encode multiple CRISPR-Cas systems, and multiple spacers targeting a single phage, which results in a strongly reduced probability of escape by point mutation [98,115]. However, phages can still overcome multiple protospacer targeting by insertions, deletions or recombination [29].
Short-term coevolution between parasites and immune systems
The outcome of short-term coevolution depends on the interplay between parasite escape and host response. For CRISPR-Cas systems, the outcome of coevolution depends on the number and diversity of mutations that can be achieved on both sides [97]. The rate of spacer acquisition against escape phages is greatly enhanced by priming [13,124]. P. aeruginosa PA14, which is primed against phage DMS3vir, rapidly generates high spacer diversity, leading to phage extinction [98]. In S. thermophilus, less spacer diversity is generated, and longer-term coevolution can be observed [27,100]. Both immunity and infectivity increase over time as hosts acquire more spacers and phages escape mutations, characteristics of an arms race dynamics. Ultimately, the arms race is asymmetrical [100] because phages are limited by mutation supply whereas hosts can acquire new spacers at low cost [64], and phages go extinct. In natural environments, long-term coevolution can be observed [114,125]. However, CRISPR-Cas immunity can also be lost [126,127], due to loss of spacers or whole systems, or to the inactivation of CRISPR-Cas loci [37,50,126]. CRISPR-Cas loss favours host-phage coexistence if no other resistance mechanism is present [127] or if alternative mechanisms are less efficient in depleting phage [126].
In the case of RM immunity, evidence for short-term arms races has not been observed, although it is possible that the ability of shufflons to rapidly generate diversity leads to short-term coevolution with phages. However, phages cannot accumulate epigenetic modifications and will need to specialize on one RM type at a time (in contrast to the accumulation of escape mutations against CRISPR-Cas targeting) [128], preventing the appearance of generalist phages.
MGEs fight immunity by diverse mechanisms
In the long term, MGEs can avoid immunity by carrying fewer sequences that are recognized by immune systems. Carrying fewer restriction sites than expected by chance increases probability of parasite escape [30,129], and a total absence of target restriction sites explains the resistance of some natural phages to restriction [34]. Restriction avoidance is much rarer against Type I RM target sequences [129], possibly because other strategies can inactivate Type I systems. Some phages avoid Type III RM immunity by carrying all sites in the same orientation, as cleavage requires two inversely-oriented recognition sites [11]. Similar to restriction site avoidance, avoidance of PAM sequences can be detected for some CRISPR-Cas subtypes [130].
Another strategy to avoid immunity is to physically protect nucleic acids. A large variety of chemical modifications have been detected in the nucleic acids of virulent phages. Some common base modifications are C-5-methylcytosine, 5-hydroxylmethylcytosine (HMC), and sugar-derivatives such as glucosyl-HMC [131]. These modifications block the activity of Type I to III RM systems; however, they are targeted by Type IV systems (Figure 1A). Some chemical modifications also inhibit CRISPR-Cas immunity [33,132]. CRISPR-Cas9 is able to cleave methylated DNA [133], but is inhibited by larger modifications [132]. It is not clear how widespread such modifications are in the phage metagenome. Another physical barrier preventing nuclease access to DNA is the production of a nucleus-like structure during infection, which allows escape from type I CRISPR-Cas and RM immunity [134,135], although phages remain sensitive to RNA targeting by type III CRISPR-Cas systems [134]. MGEs can also interfere with host regulation of immunity. Some phages activate host methyltransferases [136] or possess their own [137], while others repress host CRISPR-Cas systems by hijacking host regulators [138]. However, most anti-immune proteins inhibit specific immunity enzymes and are likely to be an important part of phage-immune coevolution.
Type I RM systems are targeted by diverse inhibitors that act through distinct mechanisms, for instance occlusion of restriction sites [47], or competitive inhibition of DNA binding by DNA mimics [62,139]. Anti-restriction proteins against Types II and III systems are not known; the lack of Type II anti-restriction proteins may be due to their mechanistic and structural diversity. Similarly to anti-RM proteins, anti-CRISPR proteins (Acrs) make MGEs able to infect and replicate in hosts with active CRISPR-Cas systems [140]. Acr activity is usually restricted to specific CRISPR-Cas subtypes [141–143]. They can interfere with target DNA recognition or its destruction [140], by associating with Cas proteins and preventing either DNA binding or cleavage. Like RM inhibitors, some Acrs carry negatively charged surfaces that mimic DNA. Additionally, Acrs with enzymatic activity have recently been discovered. For instance, a family of Acrs degrades cyclic nucleotides involved in type III CRISPR-Cas signaling [144].
Acrs bring large benefits to phages in the presence of CRISPR-Cas immune hosts [98]; and costs of expression appear to be very low [145], possibly due to regulated expression. However, Acrs vary in strength and do not totally antagonize CRISPR-Cas activity, requiring cooperation between Acrs to overcome host immunity [146,147]. Because of this cooperative behaviour, Acr phages can also be exploited by non-Acr phages. As this exploitation is costly for Acr phages, it paradoxically increases the competitive fitness of weaker Acrs, less amenable to exploitation [145]. Carrying anti-RM proteins is also likely to benefit MGEs in the presence of restriction [47,62].
Anti-immune strategies impose strong selective pressures on hosts to find alternative ways to defend themselves against phages. A conspicuous evolutionary example is Type IV restriction. Phages bearing chemical modifications conferring RM resistance trigger restriction by Type IV ENases. The best studied is McrBC, conferring immunity against HMC-modified phages [148]. Fitness costs and benefits of Type IV systems have not been studied yet but are likely to depend on the abundance and diversity of modified DNA. Nonetheless, known systems offer a glimpse of multiple rounds of coevolution of restriction and anti-restriction systems: [4,149–151]. No such arms race is known for CRISPR-Cas immunity in response to Acrs. Because Acrs are usually restricted in host range, i.e. they antagonize a specific CRISPR-Cas subtype, switching to another subtype is likely sufficient to respond to Acr presence [152]. Accordingly, several CRISPR-Cas subtypes often coexist within a strain, which could be a way to overcome specific Acrs [19].
Interactions between immune systems
Defence systems are not present in isolation, but often cohabit within genomes, clustered into defence islands [3]. Yet, their interactions have been scarcely studied to date. RM systems can compete with each other [84]; Type IV systems are incompatible with a subset of Type I-III RM systems, as they can target methylated sites and can only coexist when methyltransferases do not create a modification target [4,153]. However, coexisting RM systems can also act in combination [111]. CRISPR-Cas subtypes can also cooperate, for instance type I derived crRNAs can be used by type III machinery, counteracting viral escape from the type I system [154]. Other positive interactions between CRISPR-Cas subtypes are suggested by preferential associations within genomes [19], but remain to be studied in detail.
Cooperation between innate and adaptive immune systems might also be widespread. In Vertebrates, innate and adaptive immunity act in synergy, with each system able to activate the other when detecting a threat. In prokaryotes, CRISPR-Cas and RM have mostly been studied in isolation despite their frequent genomic cooccurrence [10]. In S. thermophilus, the native type II CRISPR-Cas system and a type II RM from Lactococcus work additively, leading to high immunity against phage infection [133]. Both CRISPR-Cas interference and spacer acquisition also work on the methylated escape phage [133]. Two native systems in E. faecalis also work additively against plasmid conjugation [156]. Such additive effect might be enough to prevent MGE escape in many environments, and a simple way to extend the usually transient benefits provided by RM immunity. However, antagonism might also exist as some CRISPR-Cas subtypes are inhibited by DNA modifications [132].
One main challenge to developing CRISPR immunity against phages is spacer acquisition by a cell which is still susceptible to killing. In the same S. thermophilus system, it was demonstrated that restriction promotes spacer acquisition [157] (Figure 3C). Restriction inactivates most incoming phages, providing the CRISPR-Cas acquisition machinery with ‘defective’ phage DNA on which spacer acquisition can proceed [157]. Innate immunity thus allows adaptive immunity to develop, by protecting most hosts from death and increasing the number of cells in which spacer acquisition can proceed. It remains to be seen if such synergy also occurs with other immunity subtypes. How efficiently RM and CRISPR-Cas cooperate could vary between types, for instance depending on the compatibility between the substrates generated by each RM Type, and the ones required by the CRISPR-Cas subtype. For example, Type I and ISP enzymes can liberate short DNA fragments [158,159] that may feed into adaptation (Figure 2). Synergy with RM immunity could be particularly important for naïve spacer acquisition, which can be very inefficient in the absence of priming. The common cooccurrence of CRISPR-Cas systems and RM systems would then increase the spacer acquisition rate, a critical bottleneck for efficient adaptive immunity [35]. It could also allow CRISPR-Cas spacer acquisition to benefit from RM ability to identify and target non-self DNA [73]. Synergy between immunity systems might also be particularly relevant for highly virulent phages. CRISPR-Cas systems confer reduced immunity against these phages, likely because rapid expression of early genes causes damage before spacer acquisition can happen [33,35]. Even inefficient RM immunity (with high rates of phage escape) will increase the probability that some hosts survive and acquire spacers, which might allow CRISPR-Cas immunity to take over.
Conclusions
Immune systems provide defence to prokaryotes against parasites, mostly thanks to their ability to generate high levels of diversity, which is a key element of effective defence against evolving parasites. In response, parasites also present a range of strategies to avoid or fight immunity mechanisms. We are only starting to identify the costs and benefits associated with immune and anti-immune strategies. Current knowledge is primarily based on controlled laboratory experiments with single defence systems. Understanding the costs and benefits and the population and coevolutionary consequences of bacterial immune systems in nature requires future studies that take into account the biotic and abiotic complexity of natural environments (such as interspecific interactions, diverse populations of MGEs, and spatial and social structures) as well as the coexistence of multiple defence mechanisms in the same host genome and the synergistic or antagonistic interactions that exist between them.
Acknowledgements
This work was funded by grants from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (788405 to M.D.S and ERC-STG-2016-714478 -EVOIMMECH to E.R.W.), the BBSRC (BB/L000873 to M.D.S.). and NERC (NE/M018350/1 to E.R.W.).
References
- 1.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nature Reviews Microbiology. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 2.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Research. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koonin EV, Makarova KS, Wolf YI. Evolutionary genomics of defense systems in Archaea and Bacteria. Annual review of microbiology. 2017;71:233–261. doi: 10.1146/annurev-micro-090816-093830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loenen WAM, Raleigh EA. The other face of restriction: modification-dependent enzymes. Nucleic Acids Research. 2014;42:56–69. doi: 10.1093/nar/gkt747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loenen WAM, Dryden DTF, Raleigh EA, Wilson GG. Type I restriction enzymes and their relatives. Nucleic Acids Research. 2014;42:20–44. doi: 10.1093/nar/gkt847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pingoud A, Wilson GG, Wende W. Type II restriction endonucleases—a historical perspective and more. Nucleic Acids Res. 2014;42:7489–7527. doi: 10.1093/nar/gku447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westra ER, Swarts DC, Staals RHJ, Jore MM, Brouns SJJ, van der Oost J. The CRISPRs, They Are A-Changin’: How Prokaryotes Generate Adaptive Immunity. Annu Rev Genet. 2012;46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 8.Hampton HG, Watson BNJ, Fineran PC. The arms race between bacteria and their phage foes. Nature. 2020;577:327–336. doi: 10.1038/s41586-019-1894-8. [DOI] [PubMed] [Google Scholar]
- 9.Swarts DC, Jinek M. Cas9 versus Cas12a/Cpf1: Structure-function comparisons and implications for genome editing. Wiley interdisciplinary reviews. RNA. 2018:e1481. doi: 10.1002/wrna.1481. [DOI] [PubMed] [Google Scholar]
- 10.Oliveira PH, Touchon M, Rocha EPC. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Research. 2014;42:10618–10631. doi: 10.1093/nar/gku734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meisel A, Bickle TA, Kruger DH, Schroeder C. Type III restriction enzymes need two inversely oriented recognition sites for DNA cleavage. Nature. 1992;355:467–469. doi: 10.1038/355467a0. [DOI] [PubMed] [Google Scholar]
- 12.Jackson SA, McKenzie RE, Fagerlund RD, Kieper SN, Fineran PC, Brouns SJ. CRISPR-Cas: Adapting to change. Science. 2017;356:eaal5056. doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- 13.Swarts DC, Mosterd C, van Passel MWJ, Brouns SJJ. CRISPR Interference Directs Strand Specific Spacer Acquisition. PLoS ONE. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nussenzweig PM, McGinn J, Marraffini LA. Cas9 Cleavage of Viral Genomes Primes the Acquisition of New Immunological Memories. Cell Host & Microbe. 2019;26:515–526.e6. doi: 10.1016/j.chom.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage Response to CRISPR-Encoded Resistance in Streptococcus thermophilus. Journal of Bacteriology. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 17.Levy A, Goren MG, Yosef I, Auster O, Manor M, Amitai G, Edgar R, Qimron U, Sorek R. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernheim A, Bikard D, Touchon M, Rocha EPC. Atypical organizations and epistatic interactions of CRISPRs and cas clusters in genomes and their mobile genetic elements. Nucleic Acids Research. 2019;48:748–760. doi: 10.1093/nar/gkz1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs Guide Antiviral Defense in Procaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, Marraffini LA. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell. 2015;161:1164–1174. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kazlauskiene M, Kostiuk G, Venclovas Č, Tamulaitis G, Siksnys V. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science. 2017;357:605–609. doi: 10.1126/science.aao0100. [DOI] [PubMed] [Google Scholar]
- 23.Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, Seidel R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sternberg SH, LaFrance B, Kaplan M, Doudna JA. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levin BR, Antonovics J, Sharma H. Frequency-Dependent Selection in Bacterial Populations. Philosophical Transactions of the Royal Society B: Biological Sciences. 1988;319:459–472. doi: 10.1098/rstb.1988.0059. [DOI] [PubMed] [Google Scholar]
- 26.Westra ER, van Houte S, Oyesiku-Blakemore S, Makin B, Broniewski JM, Best A, Bondy-Denomy J, Davidson A, Boots M, Buckling A. Parasite Exposure Drives Selective Evolution of Constitutive versus Inducible Defense. Current Biology. 2015;25:1043–1049. doi: 10.1016/j.cub.2015.01.065. [DOI] [PubMed] [Google Scholar]
- 27.Paez-Espino D, Sharon I, Morovic W, Stahl B, Thomas BC, Barrangou R, Banfield JF. CRISPR Immunity Drives Rapid Phage Genome Evolution in Streptococcus thermophilus . mBio. 2015;6:e00262–15. doi: 10.1128/mBio.00262-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun CL, Barrangou R, Thomas BC, Horvath P, Fremaux C, Banfield JF. Phage mutations in response to CRISPR diversification in a bacterial population: Strong selection events as host-phage populations establish. Environ Microbiol. 2013;15:463–470. doi: 10.1111/j.1462-2920.2012.02879.x. [DOI] [PubMed] [Google Scholar]
- 29.Watson BNJ, Easingwood RA, Tong B, Wolf M, Salmond GPC, Staals RHJ, Bostina M, Fineran PC. Different genetic and morphological outcomes for phages targeted by single or multiple CRISPR-Cas spacers. Phil Trans R Soc B. 2019;374 doi: 10.1098/rstb.2018.0090. 20180090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pleška M, Guet CC. Effects of mutations in phage restriction sites during escape from restriction–modification. Biology Letters. 2017;13 doi: 10.1098/rsbl.2017.0646. 20170646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Modell JW, Jiang W, Marraffini LA. CRISPR-Cas systems exploit viral DNA injection to establish and maintain adaptive immunity. Nature. 2017;544:101–104. doi: 10.1038/nature21719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pleška M, Lang M, Refardt D, Levin BR, Guet CC. Phage–host population dynamics promotes prophage acquisition in bacteria with innate immunity. Nature Ecology & Evolution. 2018;2:359–366. doi: 10.1038/s41559-017-0424-z. [DOI] [PubMed] [Google Scholar]
- 33.Strotskaya A, Savitskaya E, Metlitskaya A, Morozova N, Datsenko KA, Semenova E, Severinov K. The action of Escherichia coli CRISPR–Cas system on lytic bacteriophages with different lifestyles and development strategies. Nucleic Acids Research. 2017;45:1946–1957. doi: 10.1093/nar/gkx042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korona R, Korona B, Levin BR. Sensitivity of naturally occurring coliphages to type I and type II restriction and modification. Journal of General Microbiology. 1993;139:1283–1290. doi: 10.1099/00221287-139-6-1283. [DOI] [PubMed] [Google Scholar]
- 35.Westra E, Levin B. How important is CRISPR-Cas for protecting natural populations of bacteria against infections with badass DNAs? [Accessed March 31, 2020];2020 doi: 10.1101/2020.02.05.935965. Evolutionary Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar R, Qimron U. The Escherichia coli CRISPR System Protects from Lysogenization, Lysogens, and Prophage Induction. Journal of Bacteriology. 2010;192:6291–6294. doi: 10.1128/JB.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rollie C, Chevallereau A, Watson BNJ, Chyou T, Fradet O, McLeod I, Fineran PC, Brown CM, Gandon S, Westra ER. Targeting of temperate phages drives loss of type I CRISPR-Cas systems. Nature. 2020;578:149–153. doi: 10.1038/s41586-020-1936-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldberg GW, Jiang W, Bikard D, Marraffini LA. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014;514:633–637. doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldberg GW, McMillan EA, Varble A, Modell JW, Samai P, Jiang W, Marraffini LA. Incomplete prophage tolerance by type III-A CRISPR-Cas systems reduces the fitness of lysogenic hosts. Nat Commun. 2018;9:61. doi: 10.1038/s41467-017-02557-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rocha EPC, Danchin A, Viari A. Evolutionary role of restriction/modification systems as revealed by comparative genome analysis. Genome Research. 2001;11:946–958. doi: 10.1101/gr.gr-1531rr. [DOI] [PubMed] [Google Scholar]
- 41.Norman A, Hansen LH, Sorensen SJ. Conjugative plasmids: vessels of the communal gene pool. Philosophical Transactions of the Royal Society B: Biological Sciences. 2009;364:2275–2289. doi: 10.1098/rstb.2009.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obeng N, Pratama AA, van Elsas JD. The Significance of Mutualistic Phages for Bacterial Ecology and Evolution. Trends in Microbiology. 2016;24:440–449. doi: 10.1016/j.tim.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 43.Roer L, Aarestrup FM, Hasman H. The EcoKI Type I Restriction-Modification System in Escherichia coli Affects but Is Not an Absolute Barrier for Conjugation. Journal of Bacteriology. 2015;197:337–342. doi: 10.1128/JB.02418-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marraffini LA, Sontheimer EJ. CRISPR Interference Limits Horizontal Gene Transfer in Staphylococci by Targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berndt C, Meier P, Wackernagel W. DNA restriction is a barrier to natural transformation in Pseudomonas stutzeri JM300. Microbiology. 2003;149:895–901. doi: 10.1099/mic.0.26033-0. [DOI] [PubMed] [Google Scholar]
- 46.Bikard D, Hatoum-Asian A, Mucida D, Marraffini LA. CRISPR Interference Can Prevent Natural Transformation and Virulence Acquisition during In Vivo Bacterial Infection. Cell Host & Microbe. 2012;12:177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 47.Iida S, Streiff MB, Bickle TA, Arber W. Two DNA Antirestriction Systems of Bacteriophage P1, darA, and darB: Characterization of darA-Phages. Virology. 1987;157:156–166. doi: 10.1016/0042-6822(87)90324-2. [DOI] [PubMed] [Google Scholar]
- 48.Watson BNJ, Staals RHJ, Fineran PC. CRISPR-Cas-Mediated Phage Resistance Enhances Horizontal Gene Transfer by Transduction. mBio. 2018;9:e02406–17. doi: 10.1128/mBio.02406-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levin BR. Nasty Viruses, Costly Plasmids, Population Dynamics, and the Conditions for Establishing and Maintaining CRISPR-Mediated Adaptive Immunity in Bacteria. PLoS Genetics. 2010;6:e1001171. doi: 10.1371/journal.pgen.1001171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang W, Maniv I, Arain F, Wang Y, Levin BR, Marraffini LA. Dealing with the Evolutionary Downside of CRISPR Immunity: Bacteria and Beneficial Plasmids. PLoS Genetics. 2013;9 doi: 10.1371/journal.pgen.1003844. e1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shmakov SA, Sitnik V, Makarova KS, Wolf YI, Severinov KV, Koonin EV. The CRISPR Spacer Space Is Dominated by Sequences from Species-Specific Mobilomes. mBio. 2017;8:e01397–17. doi: 10.1128/mBio.01397-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dimitriu T, Marchant L, Buckling A, Raymond B. Bacteria from natural populations transfer plasmids mostly towards their kin. Proceedings of the Royal Society B: Biological Sciences. 2019;286 doi: 10.1098/rspb.2019.1110. 20191110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dimitriu T, Misevic D, Lotton C, Brown SP, Lindner AB, Taddei F. Indirect Fitness Benefits Enable the Spread of Host Genes Promoting Costly Transfer of Beneficial Plasmids. PLoS Biology. 2016;14:e1002478. doi: 10.1371/journal.pbio.1002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lacks SA, Springhorn SS. Transfer of recombinant plasmids containing the gene for DpnII DNA methylase into strains of Streptococcus pneumoniae that produce DpnI or DpnII restriction endonucleases. Journal of Bacteriology. 1984;158:905–909. doi: 10.1128/jb.158.3.905-909.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dwivedi GR, Sharma E, Rao DN. Helicobacter pylori DprA alleviates restriction barrier for incoming DNA. Nucleic Acids Research. 2013;41:3274–3288. doi: 10.1093/nar/gkt024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnston C, Martin B, Granadel C, Polard P, Claverys J-P. Programmed Protection of Foreign DNA from Restriction Allows Pathogenicity Island Exchange during Pneumococcal Transformation. PLoS Pathog. 2013;9:e1003178. doi: 10.1371/journal.ppat.1003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oliveira PH, Touchon M, Rocha EPC. Regulation of genetic flux between bacteria by restriction–modification systems. Proceedings of the National Academy of Sciences. 2016;113:5658–5663. doi: 10.1073/pnas.1603257113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shehreen S, Chyou T, Fineran PC, Brown CM. Genome-wide correlation analysis suggests different roles of CRISPR-Cas systems in the acquisition of antibiotic resistance genes in diverse species. Phil Trans R Soc B. 2019;374 doi: 10.1098/rstb.2018.0384. 20180384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gophna U, Kristensen DM, Wolf YI, Popa O, Drevet C, Koonin EV. No evidence of inhibition of horizontal gene transfer by CRISPR-Cas on evolutionary timescales. [Accessed March 7, 2015];The ISME Journal. 2015 doi: 10.1038/ismej.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Meara D, Nunney L. A phylogenetic test of the role of CRISPR-Cas in limiting plasmid acquisition and prophage integration in bacteria. Plasmid. 2019;104 doi: 10.1016/j.plasmid.2019.102418. 102418. [DOI] [PubMed] [Google Scholar]
- 61.Palmer KL, Gilmore MS. Multidrug-Resistant Enterococci Lack CRISPR-cas. mBio. 2010;1:e00227-10–e00227-19. doi: 10.1128/mBio.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chilley PM, Wilkins BM. Distribution of the ardA family of antirestriction genes on conjugative plasmids. Microbiology. 1995;141:2157–2164. doi: 10.1099/13500872-141-9-2157. [DOI] [PubMed] [Google Scholar]
- 63.Mahendra C, Christie KA, Osuna BA, Pinilla-Redondo R, Kleinstiver BP, Bondy-Denomy J. Broad-spectrum anti-CRISPR proteins facilitate horizontal gene transfer. Nat Microbiol. 2020;5:620–629. doi: 10.1038/s41564-020-0692-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vale PF, Lafforgue G, Gatchitch F, Gardan R, Moineau S, Gandon S. Costs of CRISPR-Cas-mediated resistance in Streptococcus thermophilus . Proc R Soc B. 2015;282 doi: 10.1098/rspb.2015.1270. 20151270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seidel R, Bloom JG, Dekker C, Szczelkun MD. Motor step size and ATP coupling efficiency of the dsDNA translocase EcoR124I. EMBO J. 2008;27:1388–1398. doi: 10.1038/emboj.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Srikhanta YN, Maguire TL, Stacey KJ, Grimmond SM, Jennings MP. The phasevarion: A genetic system controlling coordinated, random switching of expression of multiple genes. Proceedings of the National Academy of Sciences. 2005;102:5547–5551. doi: 10.1073/pnas.0501169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bernheim A, Calvo-Villamañán A, Basier C, Cui L, Rocha EPC, Touchon M, Bikard D. Inhibition of NHEJ repair by type II-A CRISPR-Cas systems in bacteria. Nat Commun. 2017;8:2094. doi: 10.1038/s41467-017-02350-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Makovets S, Doronina VA, Murray NE. Regulation of endonuclease activity by proteolysis prevents breakage of unmodified bacterial chromosomes by type I restriction enzymes. Proceedings of the National Academy of Sciences. 1999;96:9757–9762. doi: 10.1073/pnas.96.17.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Backer O, Colson C. Transfer of the genes for the StyLTI restriction-modification system of Salmonella typhimurium to strains lacking modification ability results in death of the recipient cells and degradation of their DNA. Journal of Bacteriology. 1991;173:1328–1330. doi: 10.1128/jb.173.3.1328-1330.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends in Genetics. 2010;26:335–340. doi: 10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paez-Espino D, Morovic W, Sun CL, Thomas BC, Ueda K, Stahl B, Barrangou R, Banfield JF. Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat Commun. 2013;4:1430. doi: 10.1038/ncomms2440. [DOI] [PubMed] [Google Scholar]
- 72.Vercoe RB, Chang JT, Dy RL, Taylor C, Gristwood T, Clulow JS, Richter C, Przybilski R, Pitman AR, Fineran PC. Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands. PLoS Genet. 2013;9:e1003454. doi: 10.1371/journal.pgen.1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weissman JL, Stoltzfus A, Westra ER, Johnson PLF. Avoidance of Self during CRISPR Immunization. Trends in Microbiology. 2020;28:543–553. doi: 10.1016/j.tim.2020.02.005. [DOI] [PubMed] [Google Scholar]
- 74.Simons M, Diffin FM, Szczelkun MD. ClpXP protease targets long-lived DNA translocation states of a helicase-like motor to cause restriction alleviation. Nucleic Acids Research. 2014;42:12082–12091. doi: 10.1093/nar/gku851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patterson AG, Yevstigneyeva MS, Fineran PC. Regulation of CRISPR–Cas adaptive immune systems. Current Opinion in Microbiology. 2017;37:1–7. doi: 10.1016/j.mib.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 76.Hoyland-Kroghsbo NM, Maerkedahl RB, Svenningsen SL. A Quorum-Sensing-Induced Bacteriophage Defense Mechanism. mBio. 2013;4:e00362–12. doi: 10.1128/mBio.00362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Patterson AG, Jackson SA, Taylor C, Evans GB, Salmond GPC, Przybilski R, Staals RHJ, Fineran PC. Quorum Sensing Controls Adaptive Immunity through the Regulation of Multiple CRISPR-Cas Systems. Molecular Cell. 2016;64:1102–1108. doi: 10.1016/j.molcel.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rusinov I, Ershova A, Karyagina A, Spirin S, Alexeevski A. Lifespan of restriction-modification systems critically affects avoidance of their recognition sites in host genomes. BMC Genomics. 2015;16:1084. doi: 10.1186/s12864-015-2288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pleška M, Qian L, Okura R, Bergmiller T, Wakamoto Y, Kussell E, Guet CC. Bacterial Autoimmunity Due to a Restriction-Modification System. Current Biology. 2016;26:404–409. doi: 10.1016/j.cub.2015.12.041. [DOI] [PubMed] [Google Scholar]
- 80.Halford SE, Welsh AJ, Szczelkun MD. Enzyme-Mediated DNA Looping. Annu Rev Biophys Biomol Struct. 2004;33:1–24. doi: 10.1146/annurev.biophys.33.110502.132711. [DOI] [PubMed] [Google Scholar]
- 81.Iranzo J, Cuesta JA, Manrubia S, Katsnelson MI, Koonin EV. Disentangling the effects of selection and loss bias on gene dynamics. Proc Natl Acad Sci USA. 2017;114:E5616–E5624. doi: 10.1073/pnas.1704925114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koonin EV, Makarova KS, Wolf YI, Krupovic M. Evolutionary entanglement of mobile genetic elements and host defence systems: guns for hire. Nat Rev Genet. 2020;21:119–131. doi: 10.1038/s41576-019-0172-9. [DOI] [PubMed] [Google Scholar]
- 83.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Research. 2001;29:3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Naito T, Kusano K, Kobayashi I. Selfish behavior of restriction-modification systems. Science. 1995;267:897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 85.Mruk I, Kobayashi I. To be or not to be: regulation of restriction-modification systems and other toxin–antitoxin systems. Nucleic Acids Research. 2014;42:70–86. doi: 10.1093/nar/gkt711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.O’Neill M, Chen A, Murray NE. The restriction-modification genes of Escherichia coli K-12 may not be selfish: They do not resist loss and are readily replaced by alleles conferring different specificities. Proceedings of the National Academy of Sciences. 1997;94:14596–14601. doi: 10.1073/pnas.94.26.14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tao T, Bourne JC, Blumenthal RM. A family of regulatory genes associated with type II restriction-modification systems. Journal of Bacteriology. 1991;173:1367–1375. doi: 10.1128/jb.173.4.1367-1375.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kusano K, Naito T, Handa N, Kobayashi I. Restriction-modification systems as genomic parasites in competition for specific sequences. Proceedings of the National Academy of Sciences. 1995;92:11095–11099. doi: 10.1073/pnas.92.24.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nakayama Y, Kobayashi I. Restriction-modification gene complexes as selfish gene entities: Roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proceedings of the National Academy of Sciences. 1998;95:6442–6447. doi: 10.1073/pnas.95.11.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koonin EV, Zhang F. Coupling immunity and programmed cell suicide in prokaryotes: Life-or-death choices. BioEssays. 2017;39:e201600186. doi: 10.1002/bies.201600186. [DOI] [PubMed] [Google Scholar]
- 91.Watson BNJ, Vercoe RB, Salmond GPC, Westra ER, Staals RHJ, Fineran PC. Type I-F CRISPR-Cas resistance against virulent phages results in abortive infection and provides population-level immunity. Nat Commun. 2019;10:5526. doi: 10.1038/s41467-019-13445-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meeske AJ, Nakandakari-Higa S, Marraffini LA. Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature. 2019;570:241–245. doi: 10.1038/s41586-019-1257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang W, Samai P, Marraffini LA. Degradation of Phage Transcripts by CRISPR-Associated RNases Enables Type III CRISPR-Cas Immunity. Cell. 2016;164:710–721. doi: 10.1016/j.cell.2015.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Berngruber TW, Lion S, Gandon S. Evolution of suicide as a defence strategy against pathogens in a spatially structured environment. Ecology Letters. 2013;16:446–453. doi: 10.1111/ele.12064. [DOI] [PubMed] [Google Scholar]
- 95.Jackson SA, Fineran PC. Bacterial dormancy curbs phage epidemics. Nature. 2019;570:173–174. doi: 10.1038/d41586-019-01595-8. [DOI] [PubMed] [Google Scholar]
- 96.Korona R, Levin BR. Phage-mediated selection and the evolution and maintenance of restriction-modification. Evolution. 1993;47:556–575. doi: 10.1111/j.1558-5646.1993.tb02113.x. [DOI] [PubMed] [Google Scholar]
- 97.Gurney J, Pleška M, Levin BR. Why put up with immunity when there is resistance: an excursion into the population and evolutionary dynamics of restriction–modification and CRISPR-Cas. Phil Trans R Soc B. 2019;374:20180096. doi: 10.1098/rstb.2018.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van Houte S, Ekroth AKE, Broniewski JM, Chabas H, Ashby B, Bondy-Denomy J, Gandon S, Boots M, Paterson S, Buckling A, et al. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature. 2016;532:385–388. doi: 10.1038/nature17436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chabas H, Lion S, Nicot A, Meaden S, van Houte S, Moineau S, Wahl LM, Westra ER, Gandon S. Evolutionary emergence of infectious diseases in heterogeneous host populations. PLOS Biology. 2018;16:e2006738. doi: 10.1371/journal.pbio.2006738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Common J, Morley D, Westra ER, van Houte S. CRISPR-Cas immunity leads to a coevolutionary arms race between Streptococcus thermophilus and lytic phage. Philosophical Transactions of the Royal Society B: Biological Sciences. 2019;374:20180098. doi: 10.1098/rstb.2018.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Common J, Walker-Sünderhauf D, van Houte S, Westra ER. Diversity in CRISPR-based immunity protects susceptible genotypes by restricting phage spread and evolution. J Evol Biol. 2020 doi: 10.1111/jeb.13638. jeb.13638. [DOI] [PubMed] [Google Scholar]
- 102.Pyenson NC, Gayvert K, Varble A, Elemento O, Marraffini LA. Broad Targeting Specificity during Bacterial Type III CRISPR-Cas Immunity Constrains Viral Escape. Cell Host & Microbe. 2017;22:343–353.e3. doi: 10.1016/j.chom.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vasu K, Nagamalleswari E, Nagaraja V. Promiscuous restriction is a cellular defense strategy that confers fitness advantage to bacteria. Proceedings of the National Academy of Sciences. 2012;109:E1287–E1293. doi: 10.1073/pnas.1119226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gann AAF, Campbell AJB, Collins JF, Coulson AFW, Murray NE. Reassortment of DNA recognition domains and the evolution of new specificities. Mol Microbiol. 1987;1:13–22. doi: 10.1111/j.1365-2958.1987.tb00521.x. [DOI] [PubMed] [Google Scholar]
- 105.Gubler M, Bickle TA. Increased protein flexibility leads to promiscuous protein-DNA interactions in type IC restriction-modification systems. The EMBO Journal. 1991;10:951–957. doi: 10.1002/j.1460-2075.1991.tb08029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Meister J, MacWilliams M, Hübner P, Jütte H, Skrzypek E, Piekarowicz A, Bickle TA. Macroevolution by transposition: drastic modification of DNA recognition by a type I restriction enzyme following Tn5 transposition. The EMBO Journal. 1993;12:4585–4591. doi: 10.1002/j.1460-2075.1993.tb06147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dybvig K, Sitaraman R, French CT. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proceedings of the National Academy of Sciences. 1998;95:13923–13928. doi: 10.1073/pnas.95.23.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.De Ste Croix M, Vacca I, Kwun MJ, Ralph JD, Bentley SD, Haigh R, Croucher NJ, Oggioni MR. Phase-variable methylation and epigenetic regulation by type I restriction-modification systems. FEMS Microbiology Reviews. 2017;41:S3–S15. doi: 10.1093/femsre/fux025. [DOI] [PubMed] [Google Scholar]
- 109.Croucher NJ, Coupland PG, Stevenson AE, Callendrello A, Bentley SD, Hanage WP. Diversification of bacterial genome content through distinct mechanisms over different timescales. Nature Communications. 2014;5:5471. doi: 10.1038/ncomms6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kennaway CK, Obarska-Kosinska A, White JH, Tuszynska I, Cooper LP, Bujnicki JM, Trinick J, Dryden DTF. The structure of M.EcoKI Type I DNA methyltransferase with a DNA mimic antirestriction protein. Nucleic Acids Research. 2009;37:762–770. doi: 10.1093/nar/gkn988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schouler C, Gautier M, Ehrlich SD, Chopin M-C. Combinational variation of restriction modification specificities in Lactococcus lactis. Molecular microbiology. 1998;28:169–178. doi: 10.1046/j.1365-2958.1998.00787.x. [DOI] [PubMed] [Google Scholar]
- 112.Varble A, Meaden S, Barrangou R, Westra ER, Marraffini LA. Recombination between phages and CRISPR-cas loci facilitates horizontal gene transfer in staphylococci. Nat Microbiol. 2019;4:956–963. doi: 10.1038/s41564-019-0400-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Almendros C, Mojica FJM, Diez-Villasenor C, Guzman NM, Garcia-Martinez J. CRISPR-Cas Functional Module Exchange in Escherichia coli. mBio. 2014;5:e00767–13. doi: 10.1128/mBio.00767-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- 115.Levin BR, Moineau S, Bushman M, Barrangou R. The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR-Mediated Immunity. PLoS Genetics. 2013;9:e1003312. doi: 10.1371/journal.pgen.1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Childs LM, England WE, Young MJ, Weitz JS, Whitaker RJ. CRISPR-Induced Distributed Immunity in Microbial Populations. PLoS ONE. 2014;9:e101710. doi: 10.1371/journal.pone.0101710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pyenson NC, Marraffini LA. Co-evolution within structured bacterial communities results in multiple expansion of CRISPR loci and enhanced immunity. eLife. 2020;9:e53078. doi: 10.7554/eLife.53078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Furi L, Crawford LA, Rangel-Pineros G, Manso AS, De Ste Croix M, Haigh RD, Kwun MJ, Engelsen Fjelland K, Gilfillan GD, Bentley SD, et al. Methylation Warfare: Interaction of Pneumococcal Bacteriophages with Their Host. J Bacteriol. 2019;201:e00370–19. doi: 10.1128/JB.00370-19. /jb/201/19/JB.00370-19.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.De Bolle X, Bayliss CD, Field D, van de Ven T, Saunders NJ, Hood DW, Moxon ER. The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol Microbiol. 2000;35:211–222. doi: 10.1046/j.1365-2958.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 120.Atack JM, Yang Y, Seib KL, Zhou Y, Jennings MP. A survey of Type III restriction-modification systems reveals numerous, novel epigenetic regulators controlling phase-variable regulons; phasevarions. Nucleic Acids Research. 2018;46:3532–3542. doi: 10.1093/nar/gky192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hoskisson PA, Smith MC. Hypervariation and phase variation in the bacteriophage ‘resistome.’. Current Opinion in Microbiology. 2007;10:396–400. doi: 10.1016/j.mib.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 122.Bayliss CD, Callaghan MJ, Moxon ER. High allelic diversity in the methyltransferase gene of a phase variable type III restriction-modification system has implications for the fitness of Haemophilus influenzae. Nucleic Acids Research. 2006;34:4046–4059. doi: 10.1093/nar/gkl568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pagie L, Hogeweg P. Individual-and Population-based Diversity in Restriction-modification Systems. Bulletin of Mathematical Biology. 2000;62:759–774. doi: 10.1006/bulm.2000.0177. [DOI] [PubMed] [Google Scholar]
- 124.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nature Communications. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 125.Laanto E, Hoikkala V, Ravantti J, Sundberg L-R. Long-term genomic coevolution of host-parasite interaction in the natural environment. Nat Commun. 2017;8:111. doi: 10.1038/s41467-017-00158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Meaden S, Capria L, Alseth E, Biswas A, Lenzi L, Buckling A, van Houte S, Westra ER. Transient CRISPR immunity leads to coexistence with phages (Microbiology) [Accessed April 22, 2020];2019 doi: 10.1101/2019.12.19.882027. Available at: [DOI] [Google Scholar]
- 127.Weissman JL, Holmes R, Barrangou R, Moineau S, Fagan WF, Levin BR, Johnson PLF. Immune loss as a driver of coexistence during host-phage coevolution. The ISME Journal. 2018;12:585–597. doi: 10.1038/ismej.2017.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Frank SA. Polymorphism of bacterial restriction-modification systems: the advantage of diversity. Evolution. 1994;48:1470–1477. doi: 10.1111/j.1558-5646.1994.tb02189.x. [DOI] [PubMed] [Google Scholar]
- 129.Rusinov IS, Ershova AS, Karyagina AS, Spirin SA, Alexeevski AV. Avoidance of recognition sites of restriction-modification systems is a widespread but not universal anti-restriction strategy of prokaryotic viruses. BMC Genomics. 2018;19:885. doi: 10.1186/s12864-018-5324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kupczok A, Bollback JP. Motif depletion in bacteriophages infecting hosts with CRISPR systems. BMC Genomics. 2014;15:663. doi: 10.1186/1471-2164-15-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Weigele P, Raleigh EA. Biosynthesis and Function of Modified Bases in Bacteria and Their Viruses. Chem Rev. 2016;116:12655–12687. doi: 10.1021/acs.chemrev.6b00114. [DOI] [PubMed] [Google Scholar]
- 132.Bryson AL, Hwang Y, Sherrill-Mix S, Wu GD, Lewis JD, Black L, Clark TA, Bushman FD. Covalent Modification of Bacteriophage T4 DNA Inhibits CRISPR-Cas9. mBio. 2015;6:e00648–15. doi: 10.1128/mBio.00648-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dupuis M-È, Villion M, Magadán AH, Moineau S. CRISPR-Cas and restriction-modification systems are compatible and increase phage resistance. Nature Communications. 2013;4:2087. doi: 10.1038/ncomms3087. [DOI] [PubMed] [Google Scholar]
- 134.Malone LM, Warring SL, Jackson SA, Warnecke C, Gardner PP, Gumy LF, Fineran PC. A jumbo phage that forms a nucleus-like structure evades CRISPR-Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat Microbiol. 2020;5:48–55. doi: 10.1038/s41564-019-0612-5. [DOI] [PubMed] [Google Scholar]
- 135.Mendoza SD, Nieweglowska ES, Govindarajan S, Leon LM, Berry JD, Tiwari A, Chaikeeratisak V, Pogliano J, Agard DA, Bondy-Denomy J. A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases. Nature. 2020;577:244–248. doi: 10.1038/s41586-019-1786-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Loenen WAM, Murray NE. Modification Enhancement by the Restriction Alleviation Protein (Ral) of Bacteriophage h. Journal of Molecular Biology. 1986;190:11–22. doi: 10.1016/0022-2836(86)90071-9. [DOI] [PubMed] [Google Scholar]
- 137.Murphy J, Mahony J, Ainsworth S, Nauta A, van Sinderen D. Bacteriophage Orphan DNA Methyltransferases: Insights from Their Bacterial Origin, Function, and Occurrence. Appl Environ Microbiol. 2013;79:7547–7555. doi: 10.1128/AEM.02229-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Borges AL, Castro B, Govindarajan S, Solvik T, Escalante V, Bondy-Denomy J. Bacterial alginate regulators and phage homologs repress CRISPR-Cas immunity. Nat Microbiol. 2020;5:679–687. doi: 10.1038/s41564-020-0691-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Walkinshaw MD, Taylor P, Sturrock SS, Atanasiu C, Berge T, Henderson RM, Edwardson JM, Dryden DTF. Structure of Ocr from Bacteriophage T7, a Protein that Mimics B-Form DNA. Molecular Cell. 2002;9:187–194. doi: 10.1016/s1097-2765(02)00435-5. [DOI] [PubMed] [Google Scholar]
- 140.Bondy-Denomy J, Garcia B, Strum S, Du M, Rollins MF, Hidalgo-Reyes Y, Wiedenheft B, Maxwell KL, Davidson AR. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature. 2015;526:136–139. doi: 10.1038/nature15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pawluk A, Amrani N, Zhang Y, Garcia B, Hidalgo-Reyes Y, Lee J, Edraki A, Shah M, Sontheimer EJ, Maxwell KL, et al. Naturally Occurring Off-Switches for CRISPR-Cas9. Cell. 2016;167:1829–1838.e9. doi: 10.1016/j.cell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bhoobalan-Chitty Y, Johansen TB, Di Cianni N, Peng X. Inhibition of Type III CRISPR-Cas Immunity by an Archaeal Virus-Encoded Anti-CRISPR Protein. Cell. 2019;179:448–458.e11. doi: 10.1016/j.cell.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 144.Athukoralage JS, McMahon SA, Zhang C, Grüschow S, Graham S, Krupovic M, Whitaker RJ, Gloster TM, White MF. An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature. 2020;577:572–575. doi: 10.1038/s41586-019-1909-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chevallereau A, Meaden S, Fradet O, Landsberger M, Maestri A, Biswas A, Gandon S, van Houte S, Westra ER. Exploitation of the Cooperative Behaviors of Anti-CRISPR Phages. Cell Host & Microbe. 2020;27:189–198.e6. doi: 10.1016/j.chom.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Landsberger M, Gandon S, Meaden S, Rollie C, Chevallereau A, Chabas H, Buckling A, Westra ER, van Houte S. Anti-CRISPR Phages Cooperate to Overcome CRISPR-Cas Immunity. Cell. 2018;174:908–916.e12. doi: 10.1016/j.cell.2018.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Borges AL, Zhang JY, Rollins MF, Osuna BA, Wiedenheft B, Bondy-Denomy J. Bacteriophage Cooperation Suppresses CRISPR-Cas3 and Cas9 Immunity. Cell. 2018;174:917–925.e10. doi: 10.1016/j.cell.2018.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Raleigh EA, Wilson G. Escherichia coli K-12 restricts DNA containing 5methylcytosine. Proceedings of the National Academy of Sciences. 1986;83:9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dharmalingam K, Goldberg EB. Phage-coded protein prevents restriction of unmodified progeny T4 DNA. Nature. 1976;260:454–456. doi: 10.1038/260454a0. [DOI] [PubMed] [Google Scholar]
- 150.Bair C, Black LW. Exclusion of Glucosyl-Hydroxymethylcytosine DNA Containing Bacteriophages. Journal of Molecular Biology. 2007;366:779–789. doi: 10.1016/j.jmb.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rifat D, Wright NT, Varney KM, Weber DJ, Black LW. Restriction Endonuclease Inhibitor IPI* of Bacteriophage T4: A Novel Structure for a Dedicated Target. Journal of Molecular Biology. 2008;375:720–734. doi: 10.1016/j.jmb.2007.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pausch P, Müller-Esparza H, Gleditzsch D, Altegoer F, Randau L, Bange G. Structural Variation of Type I-F CRISPR RNA Guided DNA Surveillance. Molecular Cell. 2017;67:622–632.e4. doi: 10.1016/j.molcel.2017.06.036. [DOI] [PubMed] [Google Scholar]
- 153.Fukuda E, Kaminska KH, Bujnicki JM, Kobayashi I. Cell death upon epigenetic genome methylation: a novel function of methyl-specific deoxyribonucleases. Genome Biol. 2008;9:R163. doi: 10.1186/gb-2008-9-11-r163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Silas S, Lucas-Elio P, Jackson SA, Aroca-Crevillén A, Hansen LL, Fineran PC, Fire AZ, Sánchez-Amat A. Type III CRISPR-Cas systems can provide redundancy to counteract viral escape from type I systems. eLife. 2017;6:e27601. doi: 10.7554/eLife.27601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bernheim A, Sorek R. The pan-immune system of bacteria: antiviral defence as a community resource. [Accessed December 2, 2019];Nat Rev Microbiol. 2019 doi: 10.1038/s41579-019-0278-2. Available at: http://www.nature.com/articles/s41579-019-0278-2. [DOI] [PubMed] [Google Scholar]
- 156.Price VJ, Huo W, Sharifi A, Palmer KL. CRISPR-Cas and Restriction-Modification Act Additively against Conjugative Antibiotic Resistance Plasmid Transfer in Enterococcus faecalis. mSphere. 2016;1:e00064–16. doi: 10.1128/mSphere.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hynes AP, Villion M, Moineau S. Adaptation in bacterial CRISPR-Cas immunity can be driven by defective phages. Nature Communications. 2014;5:4399. doi: 10.1038/ncomms5399. [DOI] [PubMed] [Google Scholar]
- 158.Kimball M, Linn S. The Release of Oligonucleotides by the Escherichia coli B Restriction Endonuclease. Biochemical and Biophysical Research Communications. 1976;68:585–591. doi: 10.1016/0006-291x(76)91185-2. [DOI] [PubMed] [Google Scholar]
- 159.Chand MK, Nirwan N, Diffin FM, van Aelst K, Kulkarni M, Pernstich C, Szczelkun MD, Saikrishnan K. Translocation-coupled DNA cleavage by the Type ISP restriction-modification enzymes. Nat Chem Biol. 2015;11:870–877. doi: 10.1038/nchembio.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]