Abstract
Two genomes of the closest algal sisters to land plants were sequenced, providing potential evidence that bacterial genes were key in adapting to terrestrial stresses.
Streptophyte algae of the class Zygnematophyceae are the sister lineage to land plants (Embryophyta). A recent paper by Cheng et al.1 reports, for the first time, the genome sequence of two of these algae. The past decade has seen the deciphering and comparative analyses of genomes before and after the water-to-land-transition of plant life (Fig. 1), enabling unprecedented insights into early plant evolution via inference of ancestral states. The genomes of land plants allow for the inference of the state after terrestrialization. Land plants come in two flavours: vascular and non-vascular (the bryophytes). Genomes of vascular plants are abounding. Combining these data with the genomes of bryophytes — moss2, liverwort3 and with the third lineage (hornwort) pending — has revolutionized our understanding of the shared adaptations of embryophytes to living on land. In order to infer the events before and ultimately during terrestrialization, we need data from streptophyte algae. For the three lineages of streptophyte algae that are united with the land plants in the clade Phragmoplastophyta, only a genome for Charophyceae4 has been published, with Coleochaetophyceae and Zygnematophyceae eagerly awaited. Now, the wait for the latter is over. Cheng et al.1 compare the genomes of two Zygnematophyceae with those of other plants and algae, furthering our understanding of the early evolution of the plant lineage.
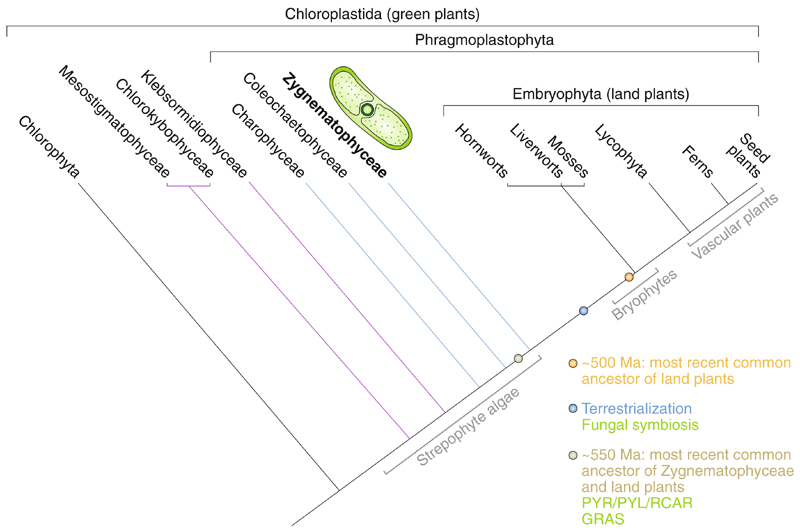
Fig. 1. A simplified cladogram shows the evolution of the green lineage.
The cladogram illustrates the phylogenetic relationships and naming of the organismal lineages discussed in the text. The dark blue dot marks the probable water-to-land-transition of plant life and the earliest land plants; the ochre dot marks the most recent common ancestor of the Z&LP; and the orange dot marks the most recent common ancestor of all land plants. Gains of important genes or features discussed in the text are shown in green.
Genomes of streptophyte algae and bryophytes were long neglected. Currently, it seems that a golden age for genomics on streptophyte diversity is being ushered in. Each new genome allows the development of a more fine-grained understanding of the early steps of land plant evolution. Cheng et al.1 sequenced the genomes of two subaerial species of Zygnematophyceae, Mesotaenium endlicherianum and Spirogloea muscicola, the latter representing a previously undescribed lineage that is probably sister to all other Zygnematophyceae and thus occupies an informative phylogenetic position. Using the genomic information, the authors are able to confirm previous inferences3–6, some of which were based on less robust transcriptomic data, among them the evolutionary gain of the PYR/ PYL/RCAR gene family that, in land plants, encodes abscisic acid (ABA) receptors and the GAI, RGA and SCR (GRAS) family of transcriptional regulators. Both gene families likely provide an advantage to land plants when facing the challenges of living on land. Finding these genes outside of the land plant clade clarifies when, during the course of evolution, the genetic building blocks for adaptive traits arose. GRAS genes are known to be important for the establishment of fungal symbioses as well as the regulation of stress responses. The PYR/ PYL/RCAR-mediated ABA perception is the starting point for the evolutionarily deep-rooted ABA pathway that conveys plant responses to many abiotic stresses6,7. The above-mentioned gains of fungal symbiosis8, as well as the tolerance to drought or desiccation, are thought to be two of the crucial adaptative traits that allowed plants to successfully thrive on land.
The PYR/PYL/RCAR and GRAS genes were apparently gained in the most recent common ancestor (MRCA) of the Zygnematophyceae and land plants (Z&LP). This finding is significant, because the Z&LP form a monophyletic group that excludes other streptophyte algal lineages, such as the Charophyceae (Fig. 1). Hence, these genes are prime candidates for having empowered the land conqueror to overcome the challenges of terrestrialization — they might have protected it from terrestrial stresses and allowed for the gain of nutrients via fungal symbiotic partners8. Moreover, Cheng et al.1 provide data for an interesting coincidence: not only have plant lineages that secondarily lost the ability to engage in fungal symbiosis concomitantly lost the genes for the symbiotic toolkit (including GRAS genes), but such lineages that have secondarily adapted to an aquatic habitat show the same pattern. These data reinforce the importance of plant–fungi symbiosis for terrestrialization brought four decades ago as the ‘mycorrhizal landing’ hypothesis8. But where did these genes come from, and how were they gained? The authors hypothesized that these genes came from soil bacteria and were laterally (horizontally) transferred into the genome of the Z&LP MRCA based on their phylogenetic analyses.
Whole genome duplications (WGDs) are considered drivers of the evolution of plant complexity9. So far, no evidence for WGD had been detected in green algal lineages, including multicellular green algae like Ulva 10 and Volvox, and the morphologically complex streptophyte alga Chara4. Surprisingly, analysis of duplicated blocks suggests that S. muscicola appears to have undergone WGD. Within the green lineage, the propensity to use WGD as a genetic playground for sub- and neofunctionalization of genes was thought to be specific for land plants, but now the finding by Cheng et al.1 extends it to Zygnematophyceae and potentially predates it to the MRCA of Z&LP.
Regardless of their origin, the evolutionary gain of these genes might have been one of those molecular adaptations that allowed the earliest land plants to make use of the terrestrial habitat. The first genomes of Zygnematophyceae are a big leap for the field of early plant evolution. However, the quest for assembling the likely genomic content of the MRCA of Z&LP is far from over. Extant Zygnematophyceae can be as divergent as the whole division of land plants. There is a lot of room for gene gains and losses, as exemplified by the presence of the PYR/PYL/RCAR genes in only one of the two Zygnematophyceae genomes. Furthermore, some features that were likely present in the MRCA of Z&LP are unlikely to be found in extant Zygnematophyceae; such features include the loss of flagellar motility due to the origin of conjugation and the loss of the chassis for plasmodesmata differentiation due to a reduction in morphological complexity. A comparative and complementing approach that includes other streptophyte algal lineages, such as the Coleochaetophyceae, is needed to infer the likely genetic makeup of the earliest land plants.
Footnotes
Competing interests
The authors declare no competing interests.
Contributor Information
Jan de Vries, Email: devries.jan@uni-goettingen.de.
Stefan A. Rensing, Email: stefan.rensing@biologie.uni-marburg.de.
References
- 1.Cheng S, et al. Cell. 2019;179:1057–1067. doi: 10.1016/j.cell.2019.10.019. [DOI] [PubMed] [Google Scholar]
- 2.Rensing SA, et al. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 3.Bowman JL, et al. Cell. 2018;171:287–304. [Google Scholar]
- 4.Nishiyama T, et al. Cell. 2018;12:448–464. [Google Scholar]
- 5.Wilhelmsson PKI, et al. Genome Biol Evol. 2018;9:3384–3397. doi: 10.1093/gbe/evx258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Vries J, et al. Proc Natl Acad Sci USA. 2018;115:E3471–E3480. doi: 10.1073/pnas.1719230115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khandelwal A, et al. Science. 2010;327:546. doi: 10.1126/science.1183672. [DOI] [PubMed] [Google Scholar]
- 8.Pirozynski KA, Malloch DW. Biosystems. 1975;6:153–164. doi: 10.1016/0303-2647(75)90023-4. [DOI] [PubMed] [Google Scholar]
- 9.Van de Peer Y, Mizrachi E, Marchal K. Nat Rev Genet. 2017;18:411–424. doi: 10.1038/nrg.2017.26. [DOI] [PubMed] [Google Scholar]
- 10.De Clerck O. Curr Biol. 2018;28:2921–2933. doi: 10.1016/j.cub.2018.08.015. [DOI] [PubMed] [Google Scholar]