Summary
Lonicera japonica is a wide-spread member of the Caprifoliaceae (honeysuckle) family utilized in traditional medical practices. This twining vine honeysuckle is also a much-sought ornamental, in part due to its dynamic flower coloration, which changes from white to gold during development.
The molecular mechanism underlying dynamic flower coloration in L. japonica was elucidated by integrating whole genome sequencing, transcriptomic analysis, and biochemical assays.
Here, we report a chromosome-level genome assembly of L. japonica, comprising nine pseudo-chromosomes with a total size of 843.2 Mb. We also provide evidence for a whole genome duplication event in the lineage leading to L. japonica, which occurred after its divergence from Dipsacales and Asterales. Moreover, gene expression analysis not only revealed correlated expression of the relevant biosynthetic genes with carotenoid accumulation, but also suggested a role for carotenoid degradation in L. japonica’s dynamic flower coloration. The variation of flower color is consistent with not only the observed carotenoid accumulation pattern, but also with the release of volatile apocarotenoids that presumably serve as pollinator attractants.
Beyond novel insights into the evolution and dynamics of flower coloration, the high-quality L. japonica genome sequence also provides a foundation for molecular breeding to improve desired characteristics.
Keywords: Caprifoliaceae (honeysuckle), Lonicera japonica, genome, flower coloration, carotenoids, carotenoid cleavage dioxygenase (CCD)
Introduction
Caprifoliaceae (the honeysuckle family) contains more than 800 species, including vines, shrubs, and small trees. A representative species of this family is Lonicera japonica, a perennial, evergreen, twining vine cultivated worldwide as an ornamental plant owing to its numerous, sweet-smelling, double-tongued flowers. L. japonica is known as golden-and-silver honeysuckle because its flowers change color, from white to gold, during their development (Figure 1A). Medicinal uses for this species are recorded in the pharmacopeia of China, Japan, and the United States (Schierenbeck, 2004; He et al., 2013). For thousands of years, L. japonica dried flower buds have been used in traditional Chinese medicine to treat fever and influenza, and the annual market for these dried flower buds is approximately 10 million kilograms, with sales of more than 2 billion yuan (CNY) in China (Shang et al., 2011). In addition, L. japonica is an important antiviral used to treat the SARS coronavirus (Wu et al., 2004; Liu et al., 2006; Shang et al., 2011; He et al., 2013), influenza A viruses (Zhou et al., 2015), the H1N1 flu virus (Ko et al., 2006), and Enterovirus 71 (Li et al., 2018a). It has further value as a component of healthy food and beverages (Zhang et al., 2016; Yang et al., 2019).
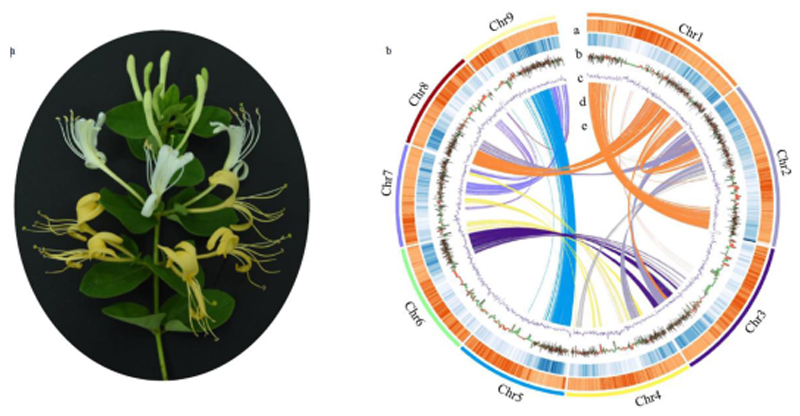
Figure 1.
L. japonica flower morphology, genome features, and synteny information. A. L. japonica dynamic flower coloration from white to yellow. B. a) Transposable elements, b) gene density, c) upregulated (red) and downregulated (green) genes from the GB (green bud) to GF (golden flower) stage, d) GC content, e) synteny blocks of paralogous sequences related to the whole genome duplication event.
Flower color is a vital trait in the plant kingdom that is largely determined by chemically distinct pigments, including carotenoids, flavonoids and/or alkaloids (Grotewold, 2006; Tanaka et al., 2008). Carotenoids were first discovered during the 19th century and have aroused wide interest as medicines, food colorants and nutritional products (Schmidt-Dannert et al., 2000; Laura et al., 2009). More than 700 different carotenoids have been characterized (Britton et al., 1995; Britton et al., 2004). These have diverse and important functions, such as attracting pollinators (Brouillard, 1988; Grotewold, 2006) and providing photoprotection, and they are precursors of the phytohormones abscisic acid (ABA) and strigolactone (Griffiths et al., 1955; Green & Durnford, 1996; Nambara & Marion-Poll, 2005), as well as vitamin A (Hirschberg, 2001; Adrian et al., 2012). Not surprisingly then, carotenoid biosynthesis has been a subject of intense study (Ruiz-Sola & Rodriguez-Concepcion, 2012). Moreover, it is well-known that their degradation is often catalyzed by carotenoid cleavage dioxygenases (CCDs), which are responsible for the synthesis of smaller apocarotenoids (Sandmann et al., 2006; Ruiz-Sola & Rodriguez-Concepcion, 2012). As an important component of photosynthetic organisms, carotenoids are found in all plastids, especially in chloroplasts and chromoplasts. In chloroplasts, carotenoids are involved in photosynthesis; whereas, in chromoplasts, they are considered to be secondary metabolites, endowing flowers and fruits with their distinct coloration (Giuliano et al., 1993; Hirschberg, 2001). However, the molecular regulation of carotenoids in coloration is not well understood.
During L. japonica flower development, the flower color changes from white to yellow in a short time frame. Notably, this interesting dynamic phenomenon occurs during the course of individual flower development, unlike the color differences found between distinct cultivars of chrysanthemum (Ohmiya et al., 2006), Narcissus pseudonarcissus (Li et al., 2018b) and Brassica napus (Zhang et al., 2015). Given that the primary pigments in L. japonica flowers are carotenoids (Fu et al., 2013), the striking developmental color change must be due to generation and degradation of these metabolites. While L. japonica has been a subject of numerous transcriptomic studies, these have focused on investigating the biosynthesis of pharmaceutically active natural products (He et al., 2013; Zhang et al., 2016; Rai et al., 2017). Research bearing on the molecular mechanisms of the distinct color change during flower development has not been reported.
Here, we assembled a chromosome-level genome for L. japonica using a combination of NGS (next-generation sequencing), ONT (Oxford Nanopore Technologies), and Hi-C technologies. Combined analyses of the L. japonica genome and transcriptomic data from six different flower stages revealed the molecular mechanisms underlying carotenoid production and degradation in flower coloration. In particular, functional characterization indicates that carotenoid cleavage dioxygenases (LjCCD4 and LjCCD1b) play important roles in flower coloration by cleaving lutein, β-carotene, and 10'-apo-β-carotenal to produce colorless (C27-apocarotenoids and C14-dialdehyde) and volatile C13-apocarotenoids.
Materials and Methods
Plant materials
L. japonica plants were cultivated in the Beijing medicinal plant garden at IMPLAD (Institute of Medicinal Plant Development) (40°N and 116°E), Beijing, China. All of the samples were collected from an individual L. japonica, whose IMPLAD germplasm registration number is 10107428 (http://www.cumplag.cn). Young leaves (~2 cm width) were collected to extract high-quality DNA for Illumina and ONT sequencing. At each of the different stages of flower development/coloration (i.e., juvenile bud, green bud, white bud, silver flower, golden flower and tawny withering flower) the corollas, androecium, pistil, and calyx were manually collected, excluding subtending bract and bracteole. Flowers at the same stage from individual honeysuckle plant were pooled and divided into three samples. These samples were immediately frozen in liquid nitrogen, and then utilized for RNA sequencing, qRT-PCR analysis, and examination of carotenoid content. Replicates were obtained from separate clonal plants.
ONT sequencing and genome assembly
High-quality L. japonica genomic DNA fragments (>20 kb) were selected using BluePippin and used to construct long-read libraries in the ONT platform (https://nanoporetech.com). The libraries were sequenced using GridION X5 with 22 nanopore flow cells (v9.4.1) and the SQK-LSK108 sequencing kit. Base calling of the raw nanopore reads was performed using the Oxford Nanopore base caller Guppy (v1.8.5) with default parameters. ONT reads were corrected and trimmed using CANU (v1.7) (Koren et al., 2017). Corrected ONT reads were directly assembled using SMARTdenovo (https://omictools.com/smartdenovo-tool) (Schmidt et al., 2017). The assembled contigs were polished three times by Pilon (v1.22), using Illumina short reads. The Embryophya odb 10 dataset was employed to estimate the completeness of the genome assembly using Benchmarking Universal Single-Copy Orthologs (BUSCO; v3) (Simao et al., 2015).
Chromosomal genome assembly and chromatin interactions using Hi-C technology
Young L. japonica leaves were fixed in 1% formaldehyde for crosslinking. Cells were lysed using a Dounce homogenizer and digested using Hind III restriction enzyme. DNA ends were filled and labelled with biotin, and the filled-in Hind III sites were ligated to form Nhe I sites. Complexes containing the biotin-labelled ligation products were purified and sheared, and the biotinylated Hi-C ligation products were pulled down and used to construct Illumina sequencing libraries (Belton et al., 2012). Hi-C Pro was used to validate paired-end reads (Servant et al., 2015). Draft contigs were clustered and ordered into chromosomes in accordance with intrachromosomal interactions estimated by LACHESIS (Burton et al., 2013). Validated paired-end reads were also used to calculate interchromosomal interactions to analyze chromosome territories. The contigs were further independently assembled into scaffolds using SLR (Luo et al., 2019) and SALSA (Ghurye et al., 2017), respectively, to validate the accuracy of the LACHESIS assembly.
Genome annotation
RepeatModeler (v1.0.9), LTR_Finder (v1.0.6), and LTR_retriever were used to identify and trace the repeat elements in the L. japonica genome, as previously described (VanBuren et al., 2018; Xu et al., 2018). Repeat sequences in the L. japonica genome were masked, and coding genes were predicted with the MAKER (v2.31.9) pipeline, integrating transcript and protein sequences from the de novo assembly of L. japonica RNA-Seq (Cantarel et al., 2008). Noncoding RNAs and small RNAs were annotated by alignment to Rfam and miRNA databases using INFERNAL (v1.1.2) and BLASTN, respectively (Kalvari et al., 2018).
Genome evolution analysis
The amino acid sequences of all proteins from L. japonica and 12 other angiosperms were clustered into orthologous groups using BLASTP and OrthoMCL (v 2.0.9), and an MCL inflation of 1.5 was used as the cluster granularity setting (Li et al., 2003). A maximum likelihood phylogenetic tree of single-copy genes from L. japonica and the 12 other angiosperms was constructed using the RAxML package (v 8.1.13) with the best-scoring protein substitution model and 1000 bootstrap replicates (Stamatakis, 2006). Divergence times between the 13 plant species were estimated by the program r8s (Sanderson, 2003), using reference speciation times of 42–52 million years ago (MYA) for the divergence between O. sativa and Z. mays, 98–117 MYA for that between A. thaliana and P. trichocarpa, and 173–199 MYA for that between A. trichopoda and V. vinifera. CAFÉ (v 3.1) was used to identify expansions and contractions of gene families following divergence predicted by the phylogenetic tree with a probabilistic graphical model (De Bie et al., 2006). The CoGe pipeline (https://genomevolution.org/coge/) was used to perform comparative genomic analysis among L. japonica, C. nankingense (Song et al., 2018), L. sativa (Reyes-Chin-Wo et al., 2017), and C. canephora (Denoeud et al., 2014), in order to identify paralogous and orthologous gene pairs and to calculate the synonymous substitutions per synonymous site (K S) values for L. japonica – C. nankingense, L. japonica – L. sativa, L. japonica – C. canephora, L. japonica – L. japonica, C. nankingense – C. nankingense, L. sativa – L. sativa, and C. canephora – C. canephora. The maximum DAGChainer distance between two matches was 120kb. Syntenic blocks between L. japonica and pre-γ AEK chromosomes (Murat et al., 2017) as well as V. vinifera chromosomes (Jaillon et al., 2007) were identified using MCScanX (Wang et al., 2012).
Carotenoid content analysis
Frozen flowers were ground into powder with a Mixer Mill MM400 (Retsch GmbH, Haan, Germany) operated at 30 Hz. For carotenoid isolation, 0.1 g samples of powdered flowers were dissolved in 4 mL methanol that contained 200 μL 60% (m/V) potassium hydroxide (KOH), and the mixture was incubated at 60 °C for 20 min. After cooling, the mixture was then extracted with 50% ethyl ether in petrol ether. After centrifugation, the upper phase was collected and evaporated, and the residue resuspended in methanol. Total carotenoid content was determined by spectrophotometry at 445 nm. Samples at each stage were prepared in triplicate (Tian et al., 2017).
Individual carotenoids were quantified via UPLC analysis with a Thermo Ultimate 3000 system equipped with an Acquity UPLC BEH C18 column (1.7 μm, 100 × 2.1 mm) at 30 °C. The mobile phases were acetonitrile containing 0.1% formic acid (A), and water containing 0.1% formic acid (B). The separation was performed at a flow rate of 0.3 mL min-1 with the following gradient: 0–5 min, linear increase from 10% A to 50% A; 5–8 min, linear increase from 50% A to 90% A; 8–10 min, linear increase from 90% A to 100% A; 10–30 min, sustained at 100% A; 30–31 min, back to 10% A. Carotenoids were detected by UV-VIS absorption at 440 nm.
RNA sequencing and gene expression analysis
Flowers from different developmental stages were collected, and total RNA was extracted using an RNeasy Plus Mini Kit (Qiagen, Germany). Then, mRNA isolation, fragmentation, first strand synthesis, second strand synthesis, and purification were performed using a TruSeq RNA Library Prep Kit v2 (Illumina, USA). The libraries were sequenced using the Illumina NextSeq 500 platform. The raw sequencing data were trimmed using Trimmomatic (v0.39) (Bolger et al., 2014), and the clean data were de novo assembled using Trinity (v2.8.3) for genome annotation (Haas et al., 2013). Fragments per kilobase of exon model per million reads mapped (FPKM) values for RNA-Seq reads were calculated with HISAT2 (v2.0.5) and Cufflinks (v2.2.1) (Trapnell et al., 2012). Highly co-expressed gene modules were inferred from the FPKM values at different flower stages (FPKM > 10) using weighted gene co-expression network analysis (WGCNA), an R package (Langfelder & Horvath, 2008). The WGCNA network was constructed using a minimal module size of 30, an unsigned topological overlap matrix type, a soft power of 17, and a merge cut height of 0.25. WGCNA module eigengene values were calculated and associated with carotenoid contents at different flower stages.
To investigate core gene expression at different flower stages, first strand cDNA was synthesized using the PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara). Primers designed by Primer Premier (v 6) are listed in Table S1. The qPCR experiments were performed in triplicate using TB Green Premix Ex Taq (Takara) in a Bio-Rad CFX96 Real-Time system. LjActin was the reference gene, and the relative expression levels were calculated by the 2–ΔΔCt method from Ct values.
Functional identification of LjCCDs
A phylogenetic tree of CCD peptide sequences from different species was constructed by MEGA 6.0 using the maximum-likelihood method with the Jones-Taylor-Thornton model and 1,000 bootstrap replicates (Tamura et al., 2013). LjCCD4 and LjCCD1b sequences were cloned into the pET32a vector, and then expression constructs were transformed into BL21 (DE3). Recombinant E. coli cells were cultured in Luria-Bertani (LB) liquid medium with 50 μg mL-1 ampicillin, and induced at 18°C and 130 rpm for 24 hours with 0.3 mM IPTG. Crude enzyme was prepared and used for in vitro enzymatic assays. Catalytic reactions, with a total volume of 200 µL, were carried out in 100 mM HEPES (pH 8.0) and 1 mM FeSO4 buffer, with 100 μM substrate and 60 μL crude lysate. After overnight incubation at 30°C, the reaction mixture was extracted, and products were analyzed by UPLC and LC-MS.
LC-MS analysis
Qualitative analyses of carotenoid content were carried out by LC-MS using an Agilent Technologies 1290 Infinity II and 6545 Q-TOF, with the atmospheric pressure chemical ionization (APCI) ion source operated in positive ion mode (mass range: 100–1000 m/z). An Acquity UPLC BEH C18 column (1.7 μm, 100 × 2.1 mm) was used at 30 °C, with water containing 0.1% formic acid (A) and methanol containing 0.1% formic acid (B) as the mobile phases. Separation was performed at a flow rate of 0.3 mL min-1 with the following gradient: 0–0.5 min, 60% B; 0.5–8 min, linear increase from 60% to 100% B; 8–23 min, sustained at 100% B. The drying gas was 350 °C and 5 L min-1, and the sheath gas was 350 °C and 11.0 L min-1. The nebulizer was set to 40 psi, and the VCap was set to 4000 V. Data were analyzed with MassHunter (version B.07.00).
GC-MS analysis
A solid phase micro extraction fiber assembly (65 μm PDMS/DVB, Supelco, USA) was used to investigate volatile cleavage products. Gas above the reaction mixture was exposed to the fiber for 30 min at 60–100 °C and then analyzed by GC-MS with an Agilent 5977A Mass Detector coupled to an Agilent 7890B GC system. The temperature programs used were as follows: for detection of β-ionone, the oven was kept at 50 °C for 5 min and then increased at 25 °C min-1 to 340 °C with a helium carrier gas flow of 1.0 mL min-1; and for detection of 3-OH-β-ionone and 3-OH-α-ionone, the initial temperature of 40 °C was held for 3 min and then increased at 5 °C min-1 to 250 °C with a helium carrier gas flow of 1.5 mL min-1. Electron impact (EI) was set at an ion source potential of 70 eV. The data were analyzed with MassHunter (version B.07.00).
Results
Genome sequencing and assembly of L. japonica
The size of the L. japonica genome was estimated to be 887.15 Mb by 17 k-mer distribution analysis, with an extremely high heterozygosity of 1.27% (Methods S1, Figure S1). We sequenced 78.8 Gb of ONT reads with an N50 of 26.6 kb from 22 flow cells using the GridION X5 platform. The longest ONT read was 149.4 kb, and the genome coverage was approximately 90 × (Tables S2, S3). Due to the low quality of the ONT raw reads, we first corrected the sequenced reads using overlaps (CANU correction stage) and obtained 31.5 Gb of corrected reads with an N50 of 37 kb, which were assembled by SMARTdenovo (Methods S1, Table S4). A contiguous assembly with a contig N50 of 2.1 Mb was produced, but BUSCO estimation showed only 57.1% completeness for this initial assembly. The assembled contigs were then polished three times using Illumina short reads, and the polished genome was found to have 94.7% BUSCO completeness with a 92.1% DNA mapping rate, indicating a high degree of completeness for the genome assembly (Table S4).
In total, 561,004,504 raw paired-end reads were produced during construction of the Hi-C sequencing library, and 202,254,107 valid interaction reads were captured using HiC-Pro. This enabled 84.4% of the draft genome to be anchored into 9 pseudo-chromosomes (2n=18) by the LACHESIS package, and the strong intra-chromosomal interaction signal suggested that the Hi-C assembly was of high quality (Figure S2, S3, S4). The final chromosome-level genome assembly of L. japonica was 843.2 Mb with an N50 of 84.4 Mb. Whole-chromosomal interactions were assessed using the Hi-C interaction matrix in L. japonica. As expected, Hi-C interactions within chromosomes were more frequent than inter-chromosomal interactions (Figure S5). Nevertheless, the inter-chromosomal interactions strongly indicated that four chromosomes, Chr2, Chr6, Chr7, and Chr9, were closely associated with each other, presumably due to their physical proximity in the nucleolar body (Figure S5).
Genome annotation and whole genome duplication
Transposable elements (TEs) account for approximately 58.2% of the L. japonica genome (Figure 1B, Table S5), and 45.6% of these TEs are long terminal repeat (LTR) elements (Methods S1, Table S6). The intact LTR-RTs in L. japonica were estimated to have an average insertion time of 1.4 MYA, with the burst of LTR/Gypsy insertions occurring earlier than that of LTR/Copia, suggesting that these recent LTR insertions might have made an important contribution to rearrangement of L. japonica chromosomes (Figure S6). The L. japonica genome was predicted to contain 33,939 coding gene loci after masking repeat elements (Methods S1, Figure 1B). Orthologs for 91% of the eukaryotic BUSCOs were found in the L. japonica annotation (Table S5), indicating that the annotated genome is largely complete. In total, 81.74% of the RNA-Seq reads from stems, leaves, and different flower stages could be accurately aligned to the coding genes of L. japonica using HISAT2. In addition, annotation of numerous SSRs (simple sequence repeats), rRNAs, and microRNAs provided valuable molecular markers for honeysuckle breeding (Tables S7, S8, S9). MIR2911 from L. japonica, which directly targets influenza A viruses in an example of cross-kingdom regulation (Zhou et al., 2015), was accurately localized to three different TE regions in the genome (Table S10).
Orthologous protein groups from 13 angiosperms were delineated, yielding a total of 21,650 orthologous groups that included 499,064 genes. Evaluation of gene family expansion and contraction indicated that 3,133 gene families expanded and 7,397 gene families contracted in the L. japonica lineage (Methods S2, Figure S7). Among the L. japonica expanded families, functional analysis indicated enrichment of gene ontologies (GO) such as catalytic activity (GO: 0003824) and metabolic process (GO: 0008152), suggesting possible expansion of secondary metabolism (Table S11). On the basis of phylogenetic analysis of 169 single-copy genes shared over these 13 species, L. japonica showed the closest relationship to Asteraceae species, Chrysanthemum nankingense and Lactuca sativa, with a divergence time of approximately 87 MYA estimated based on reference speciation times of 42–52 MYA for the divergence between Oryza sativa and Zea mays, 98–117 MYA for that between Arabidopsis thaliana and Populus trichocarpa, and 173–199 MYA for that between Amborella trichopoda and Vitis vinifera (Methods S2, Figure 2A).
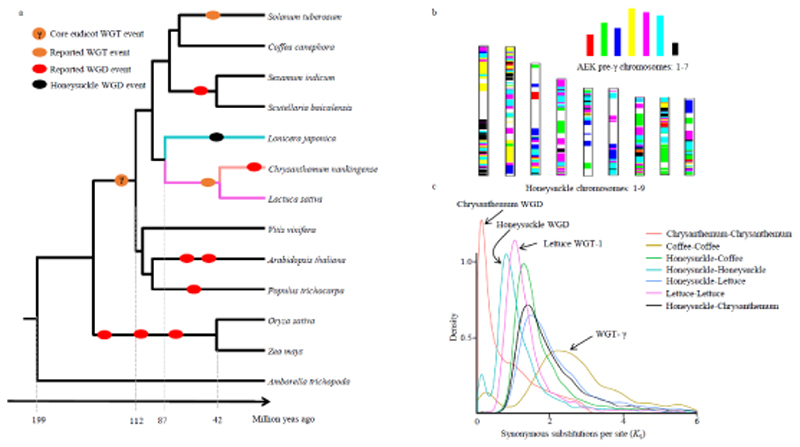
Figure 2.
L. japonica phylogeny and whole genome duplication events. a). Phylogenetic tree based on single copy genes from 13 plant species showing divergence times and reported and identified whole genome duplicatin (WGD) and whole genome triplication (WGT) events. γ represents the gamma triplication event. b). Comparison of the L. japonica genome with the ancestral eudicot karyotype (AEK). Syntenic AEK blocks are painted onto L. japonica chromosomes. C. The synonymous substitutions per synonymous site (K S) distributions of orthologous and paralogous genes among L. japonica, C. nankingense, L. sativa and C. canephora. See text for details.
Comparisons of the L. japonica genome structure with those of the reconstructed ancestral eudicot karyotype (AEK) (pre-gamma triplication, γ) and V. vinifera suggested that significant chromosome rearrangements occurred after the speciation of L. japonica (Figures 2B, S8A). Covering 30.9% of the genome, 4,983 paralogous sequence pairs were identified in L. japonica. The large paralogous regions identified in the intra-genome comparison in L. japonica suggest that a whole genome duplication event occurred during its evolutionary past (Figures 1B, S9). Syntenic genomic blocks found in L. japonica and V. vinifera confirmed that the WGD event occurred after the divergence between these two species (Methods S2, Figures S8B, S8C). The K S distribution for paralogous genes in L. japonica syntenic blocks validated a WGD peak with an approximate K S value of 0.8 (Figure 2C). K S age distributions were also constructed for the paranomes of the C. nankingense, L. sativa and Coffea canephora genomes, as well as for the orthologs between these species and L. japonica. The K S distribution of C. canephora indicated a whole genome triplication (WGT-γ) event. In addition, previously reported WGD in the C. nankingense genome (Song et al., 2018) and WGT-1 in the L. sativa genome (Reyes-Chin-Wo et al., 2017) were assumed to correspond to the peaks at the approximate K S values of 0.1 and 1.0, respectively (Methods S2, Figure 2C). Comparing the estimates of divergence time and the average K S peak values in the K S age distributions of L. japonica with those of C. nankingense, L. sativa, and C. canephora, we inferred the mean synonymous substitutions per site per year to be 7.7e-9 for Dipsacales and Asterales (Table S12). The WGD event in (the ancestor of) L. japonica is hence dated around 51 MYA, after its divergence from Asteraceae.
Carotenoid accumulation at different stages of flower development
The total carotenoid content of six different flower stages in L. japonica, namely the juvenile bud (JB), green bud (GB), white bud (WB), silver flower (SF), golden flower (GF) and tawny withering flower (TWF), were quantified via spectrophotometry analysis (Methods S3). Carotenoid levels were found to first decrease and subsequently dramatically increase with the maturity of the flowers; in particular, the carotenoid level tripled from the SF to GF flower stage (Figure 3C). Furthermore, individual carotenoids were quantified via UPLC separation with spectroscopic detection and comparison to authentic samples (Methods S3). Obvious chromatographic peaks were observed at retention times of 11.09 and 28.18 min, corresponding to lutein and β-carotene, respectively (Figure S10). Interestingly, lutein levels continuously decrease throughout flower development. By contrast, while levels of β-carotene first dropped at the GB stage, this then gradually accumulated at increasing levels from the WB to GF stages, in sync with the variation in total carotenoid content.
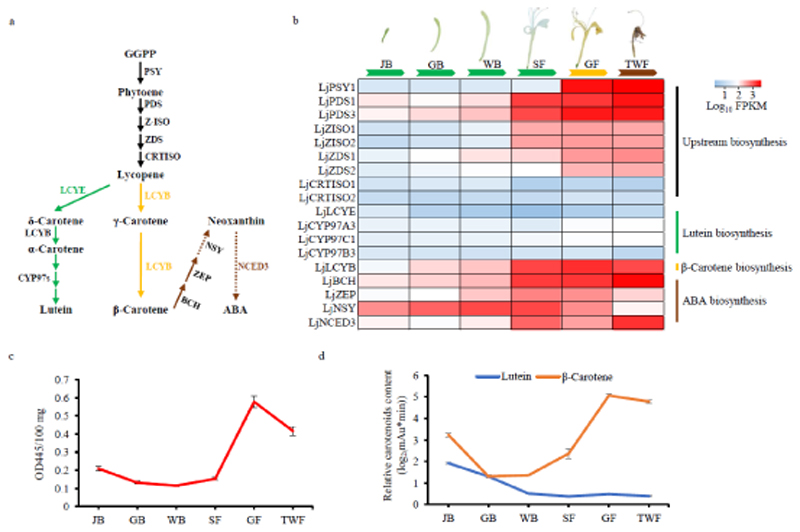
Figure 3.
Expression of core carotenoid biosynthesis pathway genes and carotenoid content variation at six flower developmental stages. a). Schematic of carotenoid metabolism. b). Heat map for the expression of core carotenoid metabolism genes at different flower developmental stages. c). Graph depicting the variation in total carotenoids. d). Graph depicting variation of the two main carotenoids, β-carotene and lutein. Juvenile bud, JB. Green bud, GB. White bud, WB. Silver flower, SF. Golden flower, GF. Tawny withering flower, TWF. The dotted arrow for NSY represents one of the key steps for neoxanthin biosynthesis. The dotted arrow for NCED3 represents one of the key steps in ABA metabolic pathway. Error bars represent standard deviation obtained from three biological repeats.
A total of 13,891 genes were highly expressed (FPKM>10) at different flower stages and these were clustered into 15 modules using WGCNA. Module-trait relationships showed that the 221 gene ‘coral1’ module was highly correlated with carotenoid content (r=0.88, P=0.03) (Figure S11). Functional annotation of selected genes in ‘coral1’ and ‘firebrick4’ modules revealed enrichment of metabolic process (GO:0008152) GO terms, including aromatic compound biosynthetic process (GO:0019438) and regulation of gene expression (GO:0010468) (Figure S12).
Evolution and expression analysis of carotenogenesis genes
Carotenoid biosynthesis has essentially been completely elucidated in higher plants, as shown in Figure 3A. First, two molecules of (E,E,E)-geranylgeranyl diphosphate (GGPP) are condensed to form phytoene by phytoene synthase (PSY). Phytoene then undergoes sequential transformations catalyzed by phytoene desaturase (PDS), 15-cis-ζ-carotene isomerase (Z-ISO), ζ-carotene desaturase (ZDS), and carotenoid isomerase (CRTISO) to produce lycopene. Here, using a genome-wide strategy to identify homologs of known genes from A. thaliana, we identified five LjPSY, three LjPDS, two LjZISO, two LjZDS, and two LjCRTISO upstream carotenoid biosynthesis genes in L. japonica (Table S13). In particular, there are five LjPSY genes in L. japonica; whereas, there are only one, three, and three PSY genes in A. thaliana, C. canephora, and V. vinifera, respectively (Figure S13). Genome synteny analysis showed that the duplication of LjPSY1 and LjPSY3 might be a product of the honeysuckle WGD event, as these fall into two large syntenic blocks on L. japonica chr2 and chr4 that correspond with a homologous block on grape chr4 (Figure S13).
PSY is thought to be the rate-limiting enzyme in carotenoid skeleton production (Hirschberg, 2001). In contrast to the single AtPSY gene found in A. thaliana, this enzyme has been expanded into a small family in L. japonica. Notably, LjPSY1 shares 83.8% amino acid identity with AtPSY, and LjPSY1 mRNA levels in GF and TWF flower stages were 100-fold higher than those measured in four earlier stages. A similar expression pattern was observed with two PDS members (Figure 3B). The protein sequences of LjPDS1, LjPDS2, and LjPDS3 share high identities of 70.1%, 74.0%, and 80.0% with AtPDS1, AtPDS2, and AtPDS3, respectively (Figure S14). LjPDS1 and LjPDS3 mRNA levels were significantly higher during SF to TWF stages than during the first three flower developmental stages. These results are of particular interest for understanding overall carotenoid accumulation and also coloration, as this initial step in carotenoid biosynthesis transform the colorless phytoene to the reddish lycopene.
Lycopene is diverted into one of two further branches of carotenoid biosynthesis by competing transformations to δ-carotene and γ-carotene, catalyzed by lycopene ε-cyclase (LCYE) and lycopene β-cyclase (LCYB), respectively. δ-Carotene is further cyclized by LCYB en route to the formation of lutein, which is further transformed by reactions catalyzed by members of the CYP97 family of cytochromes P450. γ-Carotene is further cyclized to β-carotene by LCYB. Here, we identified three CYP97 family members, along with single copies of LCYE and LCYB in the L. japonica genome. Consistent with the observed accumulation of β-carotene (Figure 3D), LjLCYB showed extremely high expression during the SF to TWF flower stages, and significant co-expression with LjPSY1, LjPDS1, and LjPDS3, as indicated by observed FPKM values that exceeded 500 (Pearson correlation, r > 0.85). In contrast, LjLCYE, LjCYP97A3, LjCYP97C1, and LjCYP97B3, which participate in the alternative lutein branch of carotenoid metabolism, were expressed at much lower levels (Figure 3B), consistent with the low lutein levels observed across all flower stages (Figure 3D).
Abscisic acid (ABA) plays critical roles in plant development, as well as in responses to biotic and abiotic stress. NCED3, a 9-cis-epoxycarotenoid cleavage dioxygenase (NCEDs), is essential for ABA biosynthesis. In the L. japonica genome, five NCED family members were identified, and the protein sequence of LjNCED3 exhibited 81.3% identity with AtNCED3, which has been shown to have a crucial role in ABA biosynthesis (Luchi et al., 2001). Interestingly, LjNCED3 mRNA levels increased during SF to TWF stages of L. japonica flower development, suggesting a role for ABA in withering, which is characteristic of the last (TWF) stage (Figure 3B).
The expression of these core carotenoid metabolism genes was further analyzed by qPCR (Figure S15), and results found to be consistent with the RNA-Seq data (Figure 3B). Evolutionary analyses of carotenogenesis genes reveal the potential impact of a honeysuckle WGD event on carotenoid biosynthesis. The gene expression patterns and carotenoid content analyses further suggest a primary role for β-carotene accumulation in the dynamic flower coloration of L. japonica.
Evolution and functional identification of carotenoid cleavage dioxygenases in flower coloration
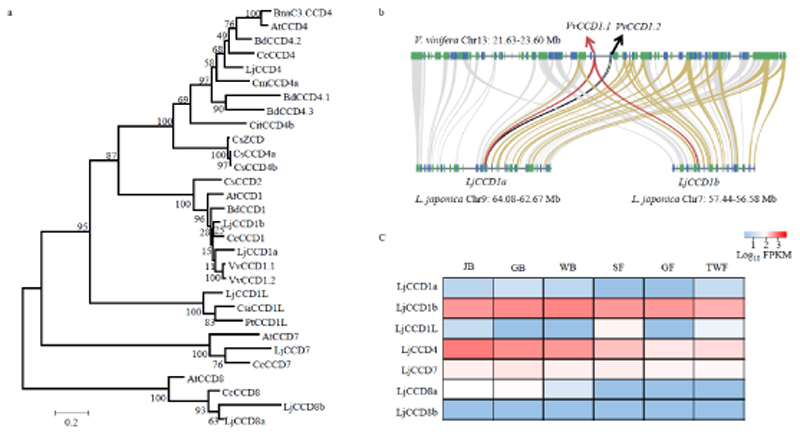
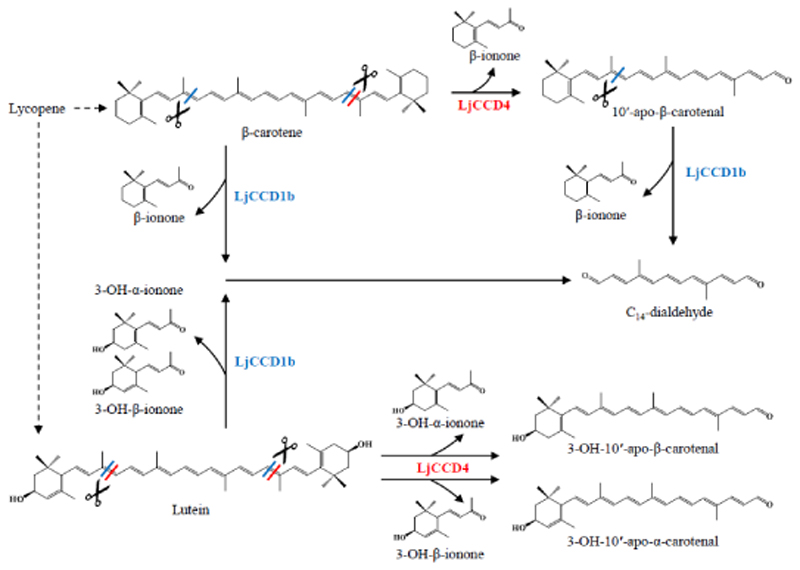
Carotenoid cleavage dioxygenases (CCDs) can be further divided into sub-families according to their substrate preference and cleavage positions (Walter & Strack, 2011). A genome-wide analysis of L. japonica led to the identification of seven CCDs, which fall into five sub-families; with two in CCD1, one in CCD1L, one in CCD4, one in CCD7 and two in CCD8 (Figure 4A). There are two V. vinifera CCD1 duplicates, VvCCD1.1 and VvCCD1.2, which are tandem repeats on Chr13 of the grape genome and share extremely high amino acid sequence identity (97%) (Figure 4B). VvCCD1.2 can catalyze symmetric cleavage at the 9,10 (9′, 10′) double bonds of β-carotene and at the 5,6 (5', 6') double bonds of lycopene (Mathieu et al., 2005; Lashbrooke et al., 2013). Comparative genomic analysis revealed that the genomic segment containing VvCCD1 shows collinearity with two L. japonica segments on Chr7 and Chr9 that contain two LjCCD1 genes, LjCCD1a and LjCCD1b, with amino acid sequence identities of 80% and 85%, respectively (Figure 4B). The apparent collinearity between these two segments suggests duplication of LjCCD1 originating from the honeysuckle WGD event. Comparative analysis of L. japonica and C. canephora indicates that the CCD4 gene is conserved and syntenic between chr7 of L. japonica and chr8 of C. canephora, although the flanking regions appear to have undergone rearrangements after separation of the L. japonica and C. canephora lineages (Figure S16). Most of the CCD family members were not expressed during L. japonica flower development, except for LjCCD1b and LjCCD4 (Figure 4C). LjCCD1b showed stable expression across all transcriptome samples, which indicated a potential function for LjCCD1b in the degradation of carotenoids in L. japonica. By contrast, LjCCD4 mRNA was most abundant at the JB stage, had significantly decreased by later GB and WB stages, and showed no detectable expression in the latest stages (Figure S15, Table S14). This expression pattern is correlated with the observed decreases in carotenoid content.
Figure 4.
Evolution and gene expression analysis of CCDs. a). Phylogenetic analysis of the CCD family, with LjCCD4 indicated in red. BnaC3.CCD4 (Brassica napus, KP658825), AtCCD4 (Arabidopsis thaliana, AT4G19170), BdCCD4.2 (Buddleja davidii, KX374548), CcCCD4 (Coffea canephora, Cc08_g05610), CmCCD4a (Chrysanthemum morifolium, EU334432), BdCCD4.1 (B. davidii, KX374547), BdCCD4.3 (B. davidii, KX374549), CitCCD4b1 (Citrus clementina, Ciclev10028113m.g), CsZCD (Crocus sativus, AJ489276), CsCCD4a (C. sativus, EU523662), CsCCD4b (C. sativus, EU523663), CsCCD2 (C. sativus, KJ541749), AtCCD1 (A. thaliana, AT3G63520), BdCCD1 (B. davidii, KX816559), CcCCD1 (C. canephora, Cc02_g28080), CsiCCD1L (Camellia sinensis, XP_028124847), PtCCD1L (Populus trichocarpa, XP_024449234), AtCCD7 (A. thaliana, AT2G44990), CcCCD7 (C. canephora, Cc02_g24750), AtCCD8 (A. thaliana, AT4G32810), CcCCD8 (C. canephora, Cc08_g09540), VvCCD1.1 (Vitis vinifera, GSVIVT01032103001), VvCCD1.2 (Vitis vinifera, GSVIVT01032110001). b. Synteny analysis between L. japonica and V. vinifera. The block containing VvCCD1.1 in Vitis corresponds with two blocks in L. japonica containing LjCCD1a and LjCCD1b. c). Heat map for the expression of CCDs at different flower developmental stages. Juvenile bud, JB. Green bud, GB. White bud, WB. Silver flower, SF. Golden flower, GF. Tawny withering flower, TWF.
To explore putative cleavage activity for LjCCD4 and LjCCD1b, the coding sequences were cloned into pET32a for recombinant E. coli expression, and activity assays were performed with crude enzyme preparations (Table S15). β-carotene, lutein, and 10'-apo-β-carotenal were tested as potential substrates. LC-MS analyses demonstrated that LjCCD4 and LjCCD1b can convert these carotenoids into different products (Figure 5). LjCCD4 cleaved the 9,10 (or 9',10') double bond of β-carotene and lutein, and the resulting products were identified as 10'-apo-β-carotenal, and 3-OH-10'-apo-β-carotenal ([M+H-H2O]+: m/z 375.2692) and 3-OH-10'-apo-α-carotenal ([M+H]+: m/z 393.2796), respectively (Figures S17, S18). LjCCD1b mediated cleavage of β-carotene and lutein at the 9,10 (9',10') double bonds and the cleavage of 10'-apo-β-carotenal at the 9,10 double bond, and the resulting products were similarly identified as C14-dialdehydes. In addition, the volatile co-products β-ionone, 3-OH-α-ionone, and 3-OH-β-ionone were also detected via GC-MS analysis (Figures S19, S20, S21).
Figure 5.
LjCCD4 and LjCCD1b catalyzed cleavage of β-carotene, lutein, and 10'-apo-β-carotenal. The 9,10 (9',10') double bonds cleavage sites for LjCCD1b are showed with a blue diagonal line (/) through the carotenoid backbone. The red diagonal line (/) through the carotenoid backbone represents the 9,10 (or 9',10') double bonds cleavage sites for LjCCD4.
Discussion
The L. japonica genome sequence reported here will be an important genetic resource, whose utility is demonstrated here by clarification of the mechanisms underlying the intriguing dynamic changes in flower coloration. The chromosome-level genome assembly further cements L. japonica’s status as a model honeysuckle. L. japonica is one of the most important medicinal plants in traditional Chinese medicine because its flower buds contain phenylpropanoids, terpenoids, flavonoids, and fatty acids with putative bioactivity, and the genome sequence reported herein is expected to contribute to understanding the biosynthesis of these natural products.
The L. japonica genome has been reported to have greater than 1% heterozygosity, presenting a significant challenge for genome assembly (Xin et al., 2019). In this study, we took advantage of the longer read lengths offered by ONT sequencing technology, which has been advantageous in the assembly of other plant genomes, such as Solanum pennellii (Schmidt et al., 2017) and Chrysanthemum nankingense (Song et al., 2018). Indeed, the long sequencing reads lead to a contig assembly with an N50 of 2.1 Mb and a BUSCO completeness estimate of 94.7%, an improvement over what has been reported for plant genomes assembled with other sequencing technologies. Polyploidization plays important roles in plant adaption to environment and plant genome evolution (Van de Peer et al., 2017), and L. japonica was estimated here to have a WGD event that occurred ~51 MYA. The phylogenetic tree suggests that this WGD event is distinct from the whole genome triplication event of Asterales (Reyes-Chin-Wo et al., 2017). The honeysuckle genome also shared the ancestral hexaploidization WGD-γ event, thus, the L. japonica genome has undergone at least two rounds of genome duplication events. However, since L. japonica has only nine chromosomes, extensive chromosomal fusions and re-arrangements must have occurred subsequent to these WGD events.
The dynamic color change from white to gold in L. japonica flowers has been reported to be caused by variations in carotenoid content (Fu et al., 2013). However, the molecular mechanism underlying this coloration has been unclear. Quantitative analysis revealed that β-carotene levels dramatically increase in the white to golden flower stages, which is accompanied by a contrasting drop in lutein levels. To investigate this accumulation pattern, the expression of carotenoid biosynthesis genes was analyzed by coupling the high-quality genome sequence generated here with RNA-Seq studies of flower development. This led to several significant observations. First, we found correlated expression of β-carotene biosynthetic genes, in particular LjPSY1, LjPDS1, LjPDS2, and LjLCYB, with total carotenoid levels as the flower color changed from white to yellow. More specifically, mRNA levels for LjPDS1 and LjPDS2 significantly increased between the WB and SF flower stages, consistent with the accompanying increase in β-carotene. However, LjPSY1 mRNA did not accumulate until the GF stage. Thus, we speculate that phytoene consumption resulting from LjPDS1 and LjPDS2 catalysis during the SF stage may produce positive feedback that increases LjPSY1 expression to provide additional metabolic flux into carotenoid biosynthesis. Expansion of the LjPSY family via whole genome duplication or recent tandem repeat events may have played a role in functionally enhancing carotenoid content (Gallagher et al., 2004). Second, from the SF to TWF stage, we detected significant expression of ABA biosynthetic genes, such as LjBCH, LjZEP, LjNSY, and LjNCED3, suggesting that ABA may be involved in flower withering. Third, we observed low mRNA levels for LjLCYE and the LjCYP97s, which are involved in a distinct metabolic branch that leads to lutein biosynthesis. This is consistent with the lack of lutein accumulation observed in L japonica flower development. Thus, combined genomic and transcriptomic analysis of L. japonica flowers indicates important roles for β-carotene biosynthesis in the change from white to gold flower coloration, as well as for ABA in withering.
Surprisingly, the overall carotenoid levels decrease early in flower development (i.e., between the JB and GB stages), with lutein levels continuing to fall throughout development. Thus, an enzyme that catalyzes carotenoid degradation seems to be required. The CCD family generally catalyzes carotenoid cleavage, generating colorless, shorter products (Ahrazem et al., 2017). Notably, the CCD4 sub-family has previously been associated with plant fruit and flower coloration. For example, high CCD4 expression is correlated with white-flowered chrysanthemum cultivars (Ohmiya et al., 2006), white-fleshed potato cultivars (Campbell et al., 2010), white-fleshed peach cultivars (Brandi et al., 2011), three different colors of mandarin fruits (Rodrigo et al., 2013), white-flowered Brassica napus lines (Zhang et al., 2015), and low-carotenoid-accumulating (“Cavendish”) banana cultivars (Buah et al., 2016). CmCCD4a and StCCD4 silencing showed that they contribute to carotenoid catabolism. Biochemical characterization further showed that CmCCD4a cleaves β-carotene at the 9,10 (9',10') double bond and StCCD4 cleaves β-carotene at the 9,10 (or 9',10') double bond (Huang et al., 2009; Bruno et al., 2015). Moreover, CCD1 sub-family members showed carotenoid and apocarotenoid cleavage activity that was correlated with emission of volatiles (Schwartz et al., 2001; Schmidt et al., 2006; Floss et al., 2008; Garcia-Limones et al., 2008). Here, we found the expression pattern of a L. japonica CCD4 family member (LjCCD4) is consistent with a role in carotenoid degradation. Specifically, LjCCD4 mRNA is most abundant at the JB stage, with significant levels still observed in the GB and WB stages. Moreover, we showed that LjCCD4 catalyzes cleavage of the 9,10- (or 9',10'-) double bond in lutein and β-carotene, which are the major carotenoid pigments. Subsequently, LjCCD1b cleaves 10'-apo-β-carotenal to produce colorless C14-dialdehyde. These findings indicate that LjCCD4 and LjCCD1b might be responsible for the carotenoid degradation observed during L. japonica flower development. Notably, this oxidative cleavage also yields the C13-apocarotenoids β-ionone, 3-OH-α-ionone, and 3-OH-β-ionone, which are considered to be volatile signals in plant-pollinator interactions, suggesting a further physiological role for this cleavage activity.
Conclusion
This study reports a chromosome-level genome assembly for L. japonica, the first for the Dipsacales order. The genome sequence represents a valuable genetic resource for molecular breeding and provides insight into the evolution of the honeysuckle family. Here, genomic analysis was also combined with transcriptome sequencing to elucidate the molecular mechanisms underlying the intriguing dynamic flower coloration in golden-and-silver honeysuckle, which can now be confidently assigned to the biosynthesis and degradation of carotenoids by particular enzymatic genes.
Supplementary Material
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 31700264), National Key R&D Program of China (2019YFC1711103), CAMS Innovation Fund for Medical Sciences (CIFMS) (Grant No. 2017-I2M-1-009), and the State Scholarship Fund (CSC no. 201808110110). Z.L. is funded by a postdoctoral fellowship from the Special Research Fund of Ghent University (BOFPDO2018001701). Z.L. is funded by a postdoctoral fellowship from the Special Research Fund of Ghent University (BOFPDO2018001701). Y.V.d.P. acknowledges funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No 833522).
Footnotes
Author contributions
X.P., Z.X. and J.S. designed and coordinated the study. Z.X. assembled and annotated the genome. Z.X. and Z.L. conducted WGD analysis. X.P., Y.T., R.G., Y.H., L.H., and C.H. performed the experiments and analyzed the data. X.P., Z.X., W.S., R.J.P., M.X., Y.V.d.P., and J.S. wrote and revised the manuscript.
Conflict of interest statement
The authors declare no conflict of interest.
Data availability
The raw data of genome and transcriptome sequencing reported in this paper have been deposited in the Genome Sequence Archive (Wang et al., 2017) in BIG Data Center (Members, 2019), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number CRA001975 that are publicly accessible at http://bigd.big.ac.cn/gsa. The assembled genome and gene structures have been deposited in the Genome Warehouse in National Genomics Data Center under the accession number GWHAAZE00000000 with the BioProject ID (PRJCA001719) and the BioSample ID (SAMC097356), which is publicly accessible at https://bigd.big.ac.cn/gwh.
References
- Adrian A, Muhammad J, Mattia M, Mark B, Martina V, Peter B, Sandro G, Harro B, Peter B, Salim AB. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science. 2012;335(6074):1348–1351. doi: 10.1126/science.1218094. [DOI] [PubMed] [Google Scholar]
- Ahrazem O, Diretto G, Argandona J, Rubio-Moraga A, Julve JM, Orzaez D, Granell A, Gomez-Gomez L. Evolutionarily distinct carotenoid cleavage dioxygenases are responsible for crocetin production in Buddleja davidii . J Exp Bot. 2017;68(16):4663–4677. doi: 10.1093/jxb/erx277. [DOI] [PubMed] [Google Scholar]
- Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58(3):268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi F, Bar E, Mourgues F, Horvath G, Turcsi E, Giuliano G, Liverani A, Tartarini S, Lewinsohn E, Rosati C. Study of 'Redhaven' peach and its white-fleshed mutant suggests a key role of CCD4 carotenoid dioxygenase in carotenoid and norisoprenoid volatile metabolism. BMC Plant Biol. 2011;11:24. doi: 10.1186/1471-2229-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton G, Liaaen-Jensen S, Pfander H. Carotenoids, Vol. 1A: Isolation and Analysis. Birkhäuser; Basel, Switzerland: 1995. [Google Scholar]
- Britton G, Liaaen-Jensen S, Pfander H. Carotenoids: handbook. Birkhäuser; Basel, Switzerland: 2004. [Google Scholar]
- Brouillard R. Flavonoids and flower colour. In: Harborne JB, editor. The Flavonoids: Advances in Research since 1980. Boston, MA: Springer US; 1988. pp. 525–538. [Google Scholar]
- Bruno M, Beyer P, Al-Babili S. The potato carotenoid cleavage dioxygenase 4 catalyzes a single cleavage of beta-ionone ring-containing carotenes and non-epoxidated xanthophylls. Arch Biochem Biophys. 2015;572:126–133. doi: 10.1016/j.abb.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Buah S, Mlalazi B, Khanna H, Dale JL, Mortimer CL. The Quest for Golden Bananas: Investigating Carotenoid Regulation in a Fe'i Group Musa Cultivar. J Agric Food Chem. 2016;64(16):3176–3185. doi: 10.1021/acs.jafc.5b05740. [DOI] [PubMed] [Google Scholar]
- Burton JN, Adey A, Patwardhan RP, Qiu R, Kitzman JO, Shendure J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. 2013;31(12):1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R, Ducreux LJ, Morris WL, Morris JA, Suttle JC, Ramsay G, Bryan GJ, Hedley PE, Taylor MA. The metabolic and developmental roles of carotenoid cleavage dioxygenase4 from potato. Plant Physiol. 2010;154(2):656–664. doi: 10.1104/pp.110.158733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, Holt C, Sanchez Alvarado A, Yandell M. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18(1):188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22(10):1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- Denoeud F, Carretero-Paulet L, Dereeper A, Droc G, Guyot R, Pietrella M, Zheng C, Alberti A, Anthony F, Aprea G, et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science. 2014;345(6201):1181–1184. doi: 10.1126/science.1255274. [DOI] [PubMed] [Google Scholar]
- Floss DS, Schliemann W, Schmidt J, Strack D, Walter MH. RNA interference-mediated repression of MtCCD1 in mycorrhizal roots of Medicago truncatula causes accumulation of C27 apocarotenoids, shedding light on the functional role of CCD1. Plant Physiol. 2008;148(3):1267–1282. doi: 10.1104/pp.108.125062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Li H, Li L, Yu H, Wang L. Reason of flower color change in Lonicera japonica . Scientia Silvae Sinicae. 2013;49(10):155–161. [Google Scholar]
- Gallagher CE, Matthews PD, Li F, Wurtzel ET. Gene duplication in the carotenoid biosynthetic pathway preceded evolution of the grasses. Plant Physiol. 2004;135(3):1776–1783. doi: 10.1104/pp.104.039818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Limones C, Schnabele K, Blanco-Portales R, Luz Bellido M, Caballero JL, Schwab W, Munoz-Blanco J. Functional characterization of FaCCD1: a carotenoid cleavage dioxygenase from strawberry involved in lutein degradation during fruit ripening. J Agric Food Chem. 2008;56(19):9277–9285. doi: 10.1021/jf801096t. [DOI] [PubMed] [Google Scholar]
- Ghurye J, Pop M, Koren S, Bickhart D, Chin CS. Scaffolding of long read assemblies using long range contact information. BMC Genomics. 2017;18(1):527. doi: 10.1186/s12864-017-3879-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Bartley GE, Scolnik PA. Regulation of carotenoid biosynthesis during tomato development. Plant Cell. 1993;5(4):379–387. doi: 10.1105/tpc.5.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BR, Durnford DG. The chlorophyll-carotenoids proteins of oxygenic photosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1996;47(1):685–714. doi: 10.1146/annurev.arplant.47.1.685. [DOI] [PubMed] [Google Scholar]
- Griffiths M, Sistrom WR, Cohen-Bazire G, Stanier RY. Function of Carotenoids in Photosynthesis. Nature. 1955;176(4495):1211–1214. doi: 10.1038/1761211a0. [DOI] [PubMed] [Google Scholar]
- Grotewold E. The genetics and biochemistry of floral pigments. Annu Rev Plant Biol. 2006;57:761–780. doi: 10.1146/annurev.arplant.57.032905.105248. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Xu X, Li Y, Li C, Zhu Y, Yan H, Sun Z, Sun C, Song J, Bi Y, et al. Transcriptome analysis of buds and leaves using 454 pyrosequencing to discover genes associated with the biosynthesis of active ingredients in Lonicera japonica Thunb. PLoS ONE. 2013;8(4):e62922. doi: 10.1371/journal.pone.0062922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg J. Carotenoid biosynthesis in flowering plants. Curr Opin Plant Biol. 2001;4(3):210–218. doi: 10.1016/s1369-5266(00)00163-1. [DOI] [PubMed] [Google Scholar]
- Huang FC, Molnar P, Schwab W. Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J Exp Bot. 2009;60(11):3011–3022. doi: 10.1093/jxb/erp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449(7161):463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, Bateman A, Finn RD, Petrov AI. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018;46(D1):D335–D342. doi: 10.1093/nar/gkx1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HC, Wei BL, Chiou WF. The effect of medicinal plants used in Chinese folk medicine on RANTES secretion by virus-infected human epithelial cells. Journal of Ethnopharmacology. 2006;107(2):205–210. doi: 10.1016/j.jep.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrooke JG, Young PR, Dockrall SJ, Vasanth K, Vivier MA. Functional characterisation of three members of the Vitis vinifera L. carotenoid cleavage dioxygenase gene family. BMC Plant Biol. 2013;13:156. doi: 10.1186/1471-2229-13-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laura A, Alvarez-Parrilla E, Gonzalez-Aguilar GA. Fruit and vegetable phytochemicals: Chemistry, nutritional value and stability. John Wiley & Sons; 2009. [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Huang Y, Sun M, Ji H, Dou H, Hu J, Yan Y, Wang X, Chen L. Honeysuckle-encoded microRNA2911 inhibits Enterovirus 71 replication via targeting VP1 gene. Antiviral Res. 2018a;152:117–123. doi: 10.1016/j.antiviral.2018.02.015. [DOI] [PubMed] [Google Scholar]
- Li X, Tang D, Du H, Shi Y. Transcriptome Sequencing and Biochemical Analysis of Perianths and Coronas Reveal Flower Color Formation in Narcissus pseudonarcissus . Int J Mol Sci. 2018b;19(12):4006. doi: 10.3390/ijms19124006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang M, He L, Li YP, Kang YK. Chinese herbs combined with Western medicine for severe acute respiratory syndrome (SARS) Cochrane Database Syst Rev. 2006;(1):CD004882. doi: 10.1002/14651858.CD004882.pub2. [DOI] [PubMed] [Google Scholar]
- Luchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis . Plant J. 2001;27(4):325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- Luo J, Lyu M, Chen R, Zhang X, Luo H, Yan C. SLR: a scaffolding algorithm based on long reads and contig classification. BMC Bioinformatics. 2019;20(1):539. doi: 10.1186/s12859-019-3114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu S, Terrier N, Procureur J, Bigey F, Gunata Z. A carotenoid cleavage dioxygenase from Vitis vinifera L.: functional characterization and expression during grape berry development in relation to C13-norisoprenoid accumulation. J Exp Bot. 2005;56(420):2721–2731. doi: 10.1093/jxb/eri265. [DOI] [PubMed] [Google Scholar]
- Members BIGDC. Database Resources of the BIG Data Center in 2019. Nucleic Acids Res. 2019;47(D1):D8–D14. doi: 10.1093/nar/gky993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat F, Armero A, Pont C, Klopp C, Salse J. Reconstructing the genome of the most recent common ancestor of flowering plants. Nature Genetics. 2017;49(4):490–496. doi: 10.1038/ng.3813. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- Ohmiya A, Kishimoto S, Aida R, Yoshioka S, Sumitomo K. Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 2006;142(3):1193–1201. doi: 10.1104/pp.106.087130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai A, Kamochi H, Suzuki H, Nakamura M, Takahashi H, Hatada T, Saito K, Yamazaki M. De novo transcriptome assembly and characterization of nine tissues of Lonicera japonica to identify potential candidate genes involved in chlorogenic acid, luteolosides, and secoiridoid biosynthesis pathways. J Nat Med. 2017;71(1):1–15. doi: 10.1007/s11418-016-1041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Chin-Wo S, Wang Z, Yang X, Kozik A, Arikit S, Song C, Xia L, Froenicke L, Lavelle DO, Truco MJ, et al. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat Commun. 2017;8:14953. doi: 10.1038/ncomms14953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo MJ, Alquezar B, Alos E, Medina V, Carmona L, Bruno M, Al-Babili S, Zacarias L. A novel carotenoid cleavage activity involved in the biosynthesis of Citrus fruit-specific apocarotenoid pigments. J Exp Bot. 2013;64(14):4461–4478. doi: 10.1093/jxb/ert260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Sola MA, Rodriguez-Concepcion M. Carotenoid biosynthesis in Arabidopsis: a colorful pathway. Arabidopsis Book. 2012;10:e0158. doi: 10.1199/tab.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19(2):301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- Sandmann G, Romer S, Fraser PD. Understanding carotenoid metabolism as a necessity for genetic engineering of crop plants. Metab Eng. 2006;8(4):291–302. doi: 10.1016/j.ymben.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Schierenbeck KJCRiPS. Japanese honeysuckle (Lonicera japonica) as an invasive species; history, ecology, and context. Critical Reviews in Plant Sciences. 2004;23(5):391–400. [Google Scholar]
- Schmidt-Dannert C, Umeno D, Arnold FH. Molecular breeding of carotenoid biosynthetic pathways. Nat Biotechnol. 2000;18(7):750–753. doi: 10.1038/77319. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Kurtzer R, Eisenreich W, Schwab W. The carotenase AtCCD1 from Arabidopsis thaliana is a dioxygenase. J Biol Chem. 2006;281(15):9845–9851. doi: 10.1074/jbc.M511668200. [DOI] [PubMed] [Google Scholar]
- Schmidt MH, Vogel A, Denton AK, Istace B, Wormit A, van de Geest H, Bolger ME, Alseekh S, Mass J, Pfaff C, et al. De Novo Assembly of a New Solanum pennellii Accession Using Nanopore Sequencing. Plant Cell. 2017;29(10):2336–2348. doi: 10.1105/tpc.17.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Zeevaart JA. Characterization of a novel carotenoid cleavage dioxygenase from plants. J Biol Chem. 2001;276(27):25208–25211. doi: 10.1074/jbc.M102146200. [DOI] [PubMed] [Google Scholar]
- Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, Heard E, Dekker J, Barillot E. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang X, Pan H, Li M, Miao X, Ding H. Lonicera japonica Thunb.: ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. J Ethnopharmacol. 2011;138(1):1–21. doi: 10.1016/j.jep.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Song C, Liu Y, Song A, Dong G, Zhao H, Sun W, Ramakrishnan S, Wang Y, Wang S, Li T, et al. The Chrysanthemum nankingense Genome Provides Insights into the Evolution and Diversification of Chrysanthemum Flowers and Medicinal Traits. Mol Plant. 2018;11(12):1482–1491. doi: 10.1016/j.molp.2018.10.003. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 2008;54(4):733–749. doi: 10.1111/j.1365-313X.2008.03447.x. [DOI] [PubMed] [Google Scholar]
- Tian X, Ji J, Wang G, Jin C, Jia C, Li Z. Cloning and Functional Characterisation of Carotenoid Cleavage Dioxygenase 4 from Wolfberry. Transactions of Tianjin University. 2017;23:62–69. [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Mizrachi E, Marchal K. The evolutionary significance of polyploidy. Nat Rev Genet. 2017;18(7):411–424. doi: 10.1038/nrg.2017.26. [DOI] [PubMed] [Google Scholar]
- VanBuren R, Wai CM, Ou S, Pardo J, Bryant D, Jiang N, Mockler TC, Edger P, Michael TP. Extreme haplotype variation in the desiccation-tolerant clubmoss Selaginella lepidophylla . Nat Commun. 2018;9(1):13. doi: 10.1038/s41467-017-02546-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MH, Strack D. Carotenoids and their cleavage products: Biosynthesis and functions. Natural Product Reports. 2011;28(4):663–692. doi: 10.1039/c0np00036a. [DOI] [PubMed] [Google Scholar]
- Wang Y, Song F, Zhu J, Zhang S, Yang Y, Chen T, Tang B, Dong L, Ding N, Zhang Q, et al. GSA: Genome Sequence Archive *. Genomics Proteomics Bioinformatics. 2017;15(1):14–18. doi: 10.1016/j.gpb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CY, Jan JT, Ma SH, Kuo CJ, Juan HF, Cheng YS, Hsu HH, Huang HC, Wu D, Brik A, et al. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc Natl Acad Sci U S A. 2004;101(27):10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin T, Zhang Y, Pu X, Gao R, Xu Z, Song J. Trends in herbgenomics. Sci China Life Sci. 2019;62(3):288–308. doi: 10.1007/s11427-018-9352-7. [DOI] [PubMed] [Google Scholar]
- Xu Z, Xin T, Bartels D, Li Y, Gu W, Yao H, Liu S, Yu H, Pu X, Zhou J, et al. Genome Analysis of the Ancient Tracheophyte Selaginella tamariscina Reveals Evolutionary Features Relevant to the Acquisition of Desiccation Tolerance. Mol Plant. 2018;11(7):983–994. doi: 10.1016/j.molp.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Yang B, Zhong Z, Wang T, Ou Y, Tian J, Komatsu S, Zhang L. Integrative omics of Lonicera japonica Thunb. Flower development unravels molecular changes regulating secondary metabolites. J Proteomics. 2019;208:103470. doi: 10.1016/j.jprot.2019.103470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Liu C, Wang Y, Yao X, Wang F, Wu J, King GJ, Liu K. Disruption of a CAROTENOID CLEAVAGE DIOXYGENASE 4 gene converts flower colour from white to yellow in Brassica species. New Phytol. 2015;206(4):1513–1526. doi: 10.1111/nph.13335. [DOI] [PubMed] [Google Scholar]
- Zhang L, Long Y, Fu C, Xiang J, Gan J, Wu G, Jia H, Yu L, Li M. Different gene expression patterns between leaves and flowers in Lonicera japonica revealed by transcriptome analysis. Frontiers in plant science. 2016;7:637. doi: 10.3389/fpls.2016.00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Li X, Liu J, Dong L, Chen Q, Liu J, Kong H, Zhang Q, Qi X, Hou D, et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015;25(1):39–49. doi: 10.1038/cr.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of genome and transcriptome sequencing reported in this paper have been deposited in the Genome Sequence Archive (Wang et al., 2017) in BIG Data Center (Members, 2019), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number CRA001975 that are publicly accessible at http://bigd.big.ac.cn/gsa. The assembled genome and gene structures have been deposited in the Genome Warehouse in National Genomics Data Center under the accession number GWHAAZE00000000 with the BioProject ID (PRJCA001719) and the BioSample ID (SAMC097356), which is publicly accessible at https://bigd.big.ac.cn/gwh.