Abstract
The importance of peroxisomes for adipocyte function is poorly understood. Herein, we provide insights into the critical role of peroxin 16 (PEX16)-mediated peroxisome biogenesis in adipocyte development and lipid metabolism. Pex16 is highly expressed in adipose tissues and upregulated during adipogenesis of murine and human cells. We demonstrate that Pex16 is a target gene of the adipogenesis “master-regulator” PPARγ. Stable silencing of Pex16 in 3T3-L1 cells strongly reduced the number of peroxisomes while mitochondrial number was unaffected. Concomitantly, peroxisomal fatty acid (FA) oxidation was reduced, thereby causing accumulation of long-and very long-chain (polyunsaturated) FAs and reduction of odd-chain FAs. Further, Pex16-silencing decreased cellular oxygen consumption and increased FA release. Additionally, silencing of Pex16 impaired adipocyte differentiation, lipogenic and adipogenic marker gene expression, and cellular triglyceride stores. Addition of PPARγ agonist rosiglitazone and peroxisome-related lipid species to Pex16-silenced 3T3-L1 cells rescued adipogenesis. These data provide evidence that PEX16 is required for peroxisome biogenesis and highlights the relevance of peroxisomes for adipogenesis and adipocyte lipid metabolism.
Keywords: PEX16, Peroxisome, PPARγ, Adipogenesis, Lipid homeostasis
1. Introduction
The obesity epidemic directed the focus of research on adipose tissue and fat cell development. Adipogenesis is driven by activation of a cascade of genes, especially transcription factors [1–3] with the nuclear receptor peroxisome proliferator-activated receptor gamma (PPARγ) as “master-regulator” [4,5]. PPARγ is essential and sufficient for adipogenesis and regulates or is regulated by genes involved in the adipogenesis-regulatory machinery [6–8]. Adipose tissues contain large numbers of peroxisomes and the number of peroxisomes strongly increases during adipogenesis of 3T3-L1 cells [9,10]. However, the relevance of peroxisomes for adipocyte development and function has been poorly investigated. Peroxisomes have diverse functions including β-oxidation of very long-chain polyunsaturated fatty acids (VLC-PUFAs), α-oxidation of (un)branched-chain FAs [11], biosynthesis of ether lipids and bile acids, and detoxification of H2O2 [12–14], thereby closely interacting with mitochondria. Similar to mitochondria, peroxisomes respond to diverse stimuli, such as cold exposure, thyroid hormonal stimulation and high fat diet with reorganization and fission [15–18].
In mammalian cells, peroxisome biogenesis factor 16 (peroxin 16, PEX16) is required for de novo synthesis of peroxisomes. PEX16 is an integral membrane protein at the ER and in peroxisomes, characterized by two transmembrane domains, with N- and C-termini facing the cytosol [13]. It plays an important role in the early phase of peroxisome de novo synthesis at the ER. Moreover, when integrated into pre-peroxisomes or mature peroxisomes, PEX16 promotes peroxisomal growth by enabling the PEX3-dependent integration of peroxisomal membrane proteins [12,19–22]. Defects in peroxisome assembly are associated with toxic accumulation of VLC-FAs, VLC-PUFAs and branched-chain FAs, culminating in severe developmental and neurological dysfunctions, commonly known as peroxisome biogenesis disorders (PBD) including the fatal Zellweger syndrome [23–25]. Mouse models for PBDs with impaired peroxisomal matrix protein import also suggest a critical role of peroxisomes in adipose tissue function [26,27]. Despite normal food intake, Pex7-knockout mice exhibit strongly reduced adiposity due to deficient peroxisomal ether-lipid synthesis [26]. Another mouse model with adipose tissue-specific loss of PEX5 showed dysfunctional peroxisomes in adipocytes with reduced lipolysis and increased fat mass. This mouse model also exhibited off-target effects impacting central and peripheral nervous system and muscle performance, which could contribute to the adipose tissue phenotype [27]. However, nothing is known about the role of PEX16 in adipose tissue development and function. Our data provide evidence that PEX16-mediated peroxisome biogenesis is crucial for adipogenesis and cellular lipid homeostasis.
2. Methods
2.1. Animal studies
All animal procedures followed the National Institute of Health Guidelines for the Care and Use of Laboratory Animals and were approved by the Austrian Ministry for Science, Research and Economy. All experiments were performed in accordance with these guidelines and regulations. Male C57BL/6 mice (8–10 weeks old) were used for this study. Animals were kept on a 12/12 h light/dark cycle and were fed a chow diet. Tissues were harvested from mice in fed ad-libitum state at room temperature.
2.2. Cell culture, differentiation, lipid staining and quantification
3T3-L1 cells and Cos7 cells were cultured in DMEM (4.5 g/L glucose) containing 10% FBS, 50 units/mL penicillin, 50 μg/mL streptomycin, and 2 mM L-glutamine. 3T3-L1 cells [28], MEFs (Pparγ−/−, Pparγ+/-, kind gift of Evan D. Rosen) [29,30] and Simpson-Golabi-Behmel syndrome (SGBS) cells [28] were differentiated as described elsewhere. Addition of 1 μM rosiglitazone (Cayman Chemical) to the culture medium was used for PPARγ-stimulation, duration and time points are indicated in the figure legends. For rescue experiments with peroxisome-associated ether lipids, 3T3-L1 cells were incubated during the whole differentiation process with differentiation medium containing 10 μM of 1-O-octadecyl-rac-glycerol (18:0-AG, Sigma, B402) or 1-O-hexadecyl-rac-glycerol (16:0-AG, SantaCruz, #sc-205,917), concentrations chosen from [31,32], or vehicle (ethanol). Lipid staining was performed with fixed cells (10% formalin in PBS, 30 min) using Oil red O (ORO; 0.25% in 60% isopropyl alcohol stock solution diluted 3:2 with dH2O) for 45 min. To stimulate lipolysis, cells were incubated with 10 μM isoproterenol for 4 h. Cellular triglyceride content and glycerol release from differentiated 3T3-L1 cells were determined using Infinity Triglyceride Reagent (Thermo). Free FA content was determined using NEFA-HR(2) Kit (WAKO). Values were normalized to total protein content (BCA reagent, Pierce).
2.3. Silencing of Pex16 using short hairpin RNA (shRNA)-containing lentiviral particles
One control non-targeting shRNA lentivirus (ntc, thereafter referred to as control cells) and five shRNA lentiviruses directed against Pex16 (shPex16 1–5) were purchased from Sigma (MISSION® shRNA lentiviral particles NM_145122), see Table S1. 3T3-L1 cells were seeded into 6-well plates 12 h before transduction using 3 × 104 cells/well (around 30% confluence). Cells were infected for 24h with a multiplicity of infection (MOI) of 7.5 in complete medium containing 8 μg/mL polybrene (Sigma). After transduction, the infection medium was replaced with fresh medium and cells were selected with puromycin (3 μg/mL) for 7 days. Silencing efficiency was controlled by quantitative RT-PCR. Only shRNAs #1, #4 and #5 were effective in silencing Pex16. For quantitative RT-PCR analysis and ORO stainings of the 3 efficient silencing constructs #1, #4 and #5 combined see Fig. S1D. Experiments were performed with biological replicates of 3T3-L1 cells silenced with shRNA #4 (referred to as shPex16 in the manuscript), showing the highest silencing efficiency.
2.4. Retroviral expression of Pex16 in Pex16-silenced 3T3-L1 cells
In order to exclude off-target effects, Pex16 was retrovirally re-expressed in Pex16-silenced 3T3-L1 cells. Recombinant retroviruses expressing Pex16 were generated as described elsewhere [33]. Briefly, full-length murine Pex16-coding sequence was amplified by PCR from murine adipose tissue cDNA using Phusion polymerase (Fermentas) and cloned into a murine stem cell virus vector (pMSCV-puro, BD Biosciences Clontech) using BglII/EcoRI restriction sites. Recombinant retroviruses expressing Pex16 were generated by transfection of PhoenixEco packaging cells (cultured in DMEM with 10% FBS in 5% CO2) with pMSCV-Pex16 using Metafectene (Biontex Laboratories GmbH). Empty pMSCV-puro or pMSCV-Pex16 viral particle-containing supernatant was collected 48 h after transfection. Viral supernatants supplemented with 8 μg/mL polybrene were added to ntc or Pex16- silenced 3T3-L1 cells on day −2, 0 and 3 of differentiation for 48 h infections at each time point.
2.5. Silencing of Pex16 in 3T3-L1 adipocytes using small interfering RNA (siRNA)
One universal negative control siRNA (si-ctrl) and three siRNAs directed against Pex16 (si-Pex16) were purchased from Sigma (MISSION® siRNA, NM_145122, ID: SASI_Mm01_00137759 (=siPex16_1); SASI_Mm01_00137760 (=siPex16_2); SASI_Mm01_00137761 (= siPex16_3)). Biological replicates of 3T3-L1 cells (day 5 of differentiation) were used for electroporation (EP) with the Neon® Transfection System (Life Technologies) following the general protocol guidelines. Optimizations to the protocol were the following: Cells were trypsinized and resuspended in pre-warmed growth-medium. Cells were pelleted at 300 ×g for 5 min and washed with PBS. 450,000 cells and 200 nM siRNA were used per 100 μL Neon® EP-tip (program: 1400 V, 20 ms, 2 pulses). The cells were seeded at a density of 450,000 cells/12-well in antibiotic-free DMEM. Medium was changed 24 h after EP to standard growth medium. Silencing efficiency was controlled 48 h, 60 h and 72 h after EP via quantitative RT-PCR (see Fig. 5A).
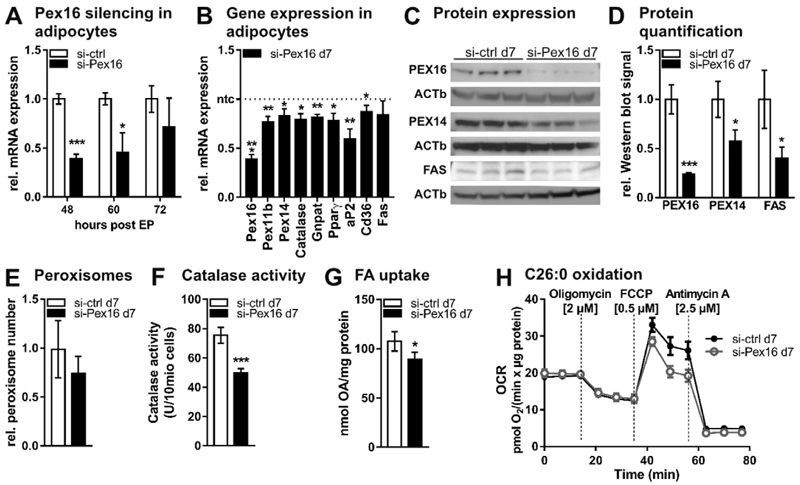
Fig. 5.
Transient silencing of Pex16 in mature 3T3-L1 adipocytes (A)–(H) Transient silencing of Pex16 in mature 3T3-L1 adipocytes (day 5 of differentiation) by electroporation (EP) of 200 nM control siRNA (si-ctrl) or siRNA directed against Pex16 (si-Pex16). (A) RT-PCR analysis of Pex16 expression 48 h, 60 h and 72 h after EP of 3T3-L1 adipocytes with siRNA. (B)–(H) Data analyzed from si-ctrl and si-Pex16 3T3-L1 adipocytes at day 7 (=48 h after EP). (B) RT-PCR analysis of peroxisomal genes (Pex16, Pex11b, Pex14, Catalase, Gnpat), adipogenesis marker genes (Pparγ, aP2) and fatty acid metabolism genes (Cd36, Fas). (C) Protein expression and (D) densitometric analysis of PEX16, PEX14 and fatty acid synthase (FAS). (E) Relative number of peroxisomes counted from ELMI pictures. (F) Catalase activity. (G) 14C–oleic acid (OA) uptake into si-ctrl and si-Pex16 3T3-L1 cells after 30 min incubation. (H) Oxygen consumption rate in the presence of 10 μM hexacosanoic acid (C26:0). Cells were incubated with assay medium containing 10 μM C26:0 45 min prior to and during the measurement with Seahorse XF96 extracellular flux analyzer (n = 4). (A),(B),(D)–(G) Data are presented as mean ± SD (n = 3). (H) Data are presented as mean ± SEM. Statistical significance was calculated using Student’s t-test. *p < 0.05, ** p < 0.01, ***p < 0.001 versus control.
2.6. Protein isolation and Western blot analysis
Cells were harvested for protein analysis in SDS-lysis buffer (50 mm Tris-HCl, pH 6.8, 10% glycerol, 2.5% SDS, 1× protease inhibitor mixture, 1 mM PMSF) and the lysates were digested with benzonase. Tissues were homogenized in RIPA buffer (150 mM Tris-HCl pH 8.0, 50 mM NaCl, 1% TritonX-100, 0.5% Na-Deoxycholate, 0.1% SDS) and incubated on ice for 20 min. After centrifugation at 16000 ×g/10 min/4 °C, intermediate phase was carefully collected into a new Eppendorf tube by puncture of the tube wall with a hot needle. Protein concentrations were determined with the PierceTM BCA Protein Assay Kit (ThermoFisher Scientific, Waltham, MA, US). 70 μg of the cell lysates were subjected to a 10% BisTris gel (NuPAGE, Invitrogen), and gels were blotted to nitrocellulose membranes. The following antibodies were used either in Tris-buffered saline +0.05% Tween20 (TBST) or Phosphate-buffered saline +0.05% Tween20 (PBST): anti-PEX16 (1:500 in 1% milk/TBST, proteintech, #14816-1-AB), anti-ACC1, and anti-FAS (1:1000 in 5% BSA/PBST, Cell Signaling, kit#8335), anti-PEX14 (1:200 in 5% BSA/PBST, SantaCruz, #sc-23197), anti-Catalase (1:1000 in 5% BSA/PBST, Abcam, #ab16731) anti-β-Actin (1:25,000 in 1% milk/PBST, Sigma, #A1978). For chemiluminescent detection, a horseradish peroxidase-conjugated secondary antibody was used (anti-rabbit 1:2000; anti-mouse 1:3000, anti-goat 1:2000, Dako). Amersham ECL Prime (GE Healthcare) or Super Signal West Pico (Thermo Fisher Scientific Inc.) served as substrates. β-Actin (ACTb) or PonceauS staining were used as loading control. Densitometric analysis was performed using ImageJ.
2.7. Human preadipocyte isolation and differentiation
Human subcutaneous adipose tissue was obtained from healthy individuals undergoing lipoaspiration. The detailed method is described in [34]. This study was approved by the Medical University of Vienna’s ethics committee and the General Hospital of Vienna (EK no. 1115/ 2010). All subjects gave written informed consent prior to taking part in the study.
2.8. RNA isolation, reverse transcription, and gene expression analysis
Cellular RNA was isolated using the Total RNA isolation kit (Sigma). RNA from tissue was isolated using TRIzol reagent (Invitrogen). Reverse transcription for cDNA generation was performed using the QuantiTect Reverse Transcription kit (Quiagen). Gene expression was assessed using quantitative RT-PCR as described before [28]. mRNA expression was normalized to TfIIβ or hTBP. For primer sequences and abbreviations, see Table S2 and S3.
2.9. Measurement of catalase activity
Catalase activity was determined as described before [35]. Briefly, undifferentiated or differentiated 3T3-L1 cells (ntc, shPex16, si-ctrl, and si-Pex16) were harvested with trypsin and washed with phosphate buffered saline (PBS). 2 × 107 undifferentiated or 1 × 107 differentiated cells were centrifuged and resuspended in 100 μL PBS. As a standard, concentrations between 0 U and 200 U of catalase from bovine liver (Sigma-Aldrich) solved in 100 μL pure distilled water were used. Cells or catalase-standards (100 μL) were added together with 100 μL 1% Triton X-100 (AppliChem GmbH, Germany) and 100 μL of undiluted 30% hydrogen peroxide (Merck Millipore, Germany) to Pyrex tubes (13 mm diameter, 10 mm height; SciLabware Ltd., UK). Foam height was determined at constant height using a ruler.
2.10. 14C-fatty acid uptake
14C–fatty acid uptake into 3T3-L1 cells (ntc, shPex16, si-ctrl, and si-Pex16) was determined in a 12-well plate. Cells were washed with PBS and pre-incubated for 1 h with 400 μM oleic acid (OA) containing growth medium. Subsequently, medium was changed to growth medium containing 400 μM OA and 0.1 μCi 14C–OA. After 30 min, cells were washed 3× with PBS, lysed for 3 h with SDS-NaOH (0.1% SDS/0.3 M NaOH) and radioactivity was measured by liquid scintillation counting.
2.11. Isolation of mitochondria and peroxisomes from 3T3-L1 cells
Mitochondria/peroxisome fraction (M/P) and cytosol (Cyt) were isolated freshly from ntc and Pex16- silenced 3T3-L1 cells (approx. 10 × 106 cells). Cell pellets were resuspended in 1 mL HES- buffer (20 mM HEPES, 1 mM EDTA, 250 mM sucrose in ddH2O) containing 1× protease inhibitor mixture and 1× phosphatase inhibitor mixture (0.1 M Na-orthovanadate, 50 mM Na-glycerophosphate, 0.5 M Na-fluoride). Cell homogenization was performed using Dounce homogenizer. Cell debris (500 × g, 5 min) and nuclei (1000×g,10min) were removed by centrifugation. Resulting supernatant was centrifuged at 5000 × g for 20 min to pellet mitochondria and peroxisomes (M/P). M/P-fraction was resuspended in 100 μL PBS and cytosolic fraction (100 μL) was used directly for the catalase activity assay.
2.12. Measurement of cellular oxygen consumption rate (OCR) and C26:0- oxidation
Sub-confluent or differentiated 3T3-L1 cells (ntc, shPex16, si-ctrl, and si-Pex16) were plated in Cell-Tak™ coated XF96 polystyrene cell culture microplates (Seahorse Bioscience®) at a density of 20,000 cells per well as indicated in the figure legends. After 24 h, cells were washed and pre-incubated for 30 min in XF assay medium supplemented with sodium pyruvate (1 mM), glutamine (2 mM) and D-glucose (5.5 mM) at 37 °C in air. Oxygen consumption rate (OCR) was subsequently measured every 7 min using an XF96 extracellular flux analyzer (Seahorse Bioscience®). For measurement of cellular hexacosanoic acid (C26:0)-oxidation capacity, ntc and shPex16 3T3-L1 preadipocytes and si-ctrl and si-Pex16 3T3-L1 adipocytes were incubated with 10 μM C26:0/ethanol-containing assay medium 45 min before and during the measurement. Oxygen consumption was normalized to protein content (pmol O2/(min × μg protein)).
2.13. Measurement of cellular ROS production
Intracellular reactive oxygen species (ROS) production was determined using CellROX Deep Red according to the manufacturer’s protocol (Life Technologies). Briefly, differentiated 3T3-L1 adipocytes were incubated for 1 h at 37 °C with 1 μM CellROX, after which cells were harvested and stained with Ghost Dye Red 780 (Tonbo Biosciences) to exclude dead cells. After washing once with PBS, cells (1 × 105) were analyzed by flow cytometry using a Guava Easy Cyte 8 (Merck Millipore, Darmstadt, Germany).
2.14. Lipid extraction and mass spectrometry
Differentiated 3T3-L1 cells were scraped off from a T75-flask (ntc, shPex16) or two 6-wells (si-ctrl, si-Pex16) in PBS and centrifuged at 1200 rpm for 5 min. Total lipids from supernatants and cell pellets were extracted according to the Folch method [36]. Lipid extracts were analyzed by ultra-performance liquid chromatography-quadrupole time of flight-mass spectrometry (UPLC-qTOF-MS) as described elsewhere [37]. In brief, samples were separated using an AQUITY-UPLC system (Waters, Manchester, UK) equipped with a BEH-C18-column, 2.1 × 150 mm, 1.7 μm(Waters). The gradient started from 55% solvent A (methanol/water 1/1, v/v) and 45% solvent B (isopropanol), both containing phosphoric acid (8 μM), ammonium acetate (10 mM) and formic acid (0.1 vol%) and reached 100% solvent B within 32 min at a flow rate of 150 μL/min. A SYNAPT™G1 qTOF HD mass spectrometer (waters) equipped with an ESI source was used for analysis in positive and negative ionization mode. For free FA analysis an 8 μL aliquot of the extract was derivatized using the AMP+ Mass Spectrometry kit (Cayman,No.: 710000) and analyzed by the same method. Data analysis was performed using the Lipid Data Analyzer software [38].
2.15. Sequence analysis
Genome organization of Pex16 was visualized using the UCSC genome browser (NCBI37/mm9). We used custom tracks from preexisting data [39,40] from chromatin-immunoprecipitation followed by sequencing (ChIP-Seq) to identify potential PPAR binding sites in the genomic sequence of Pex16 in inguinal and epididymal WAT derived adipocytes and 3T3-L1 cells on day 6 of differentiation. Sequences of two potential PPARγ target regions downstream the Pex16-transcription start site (R1: 1052–1676 bp, R2: 2160–2554 bp)were selected and subsequently cloned into luciferase reporter vectors.
2.16. Luciferase reporter assays
To test the direct binding of PPARγ to its putative target regions downstream the Pex16-transcription start site (TSS) a luciferase reporter assay was performed. Target sequences R1 (1052–1676 bp downstream Pex16-TSS) and R2 (2160–2554 bp downstream Pex16-TSS) were cloned into luc2- luciferase reporter vector pGL4.26 (Promega). Cos7 cells were transfected with MetafectenePro (Biontex Laboratories GmbH) in 24-well plates according to the manufacturer’s protocol at a ratio of MetafectenePro to DNA 2.5:1 (μl:μg). 200 ng of pGL4.26-R1/R2 or empty vector were co-transfected with 100 ng/well pCMX-PPARγ2 and 100 ng/well pCMX-RXRα expression vectors. In controls, 200 ng of empty pCMX vector were co-transfected. Co-transfection of renilla luciferase reporter vector pGL4.75 in a ratio of 1:50 to the luc2-reporter vectors served as control for varying transfection efficiencies. Luciferase reporter assays were performed 48 h after transfection using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol. Luminescence measurement was done using “Berthold Orion II” luminometer. Luciferase activity was normalized to renilla luciferase activity.
2.17. PPARγ activation assay
Pre-confluent ntc and shPex16 3T3-L1 cells were used for electroporation. Empty pCMX or pCMX-RXRα, pCMX-PPARy2 together with PPRE ×3-TK-luc firefly reporter vector (Addgene, Cambridge, MA, kind gift of Dayoung Oh) were used for EP with the Neon® Transfection System (Life Technologies) following the general protocol guidelines. Additionally, renilla reporter vector pGL4.75 (Promega, Madison, USA) was co-electroporated in all experiments in a ratio of 1:100 to firefly luciferase reporter vectors as a control for varying electroporation efficiencies. Optimizations to the protocol were the following: cells were trypsinized and resuspended in pre-warmed growth medium. Cells were pelleted at 300 ×g for 5 min and washed with PBS. 100,000 cells and a total of 2 μg plasmid DNA per 10 μL Neon® EP-tip were used (program: 1400 V, 20 ms, 1 pulse). 100,000 cells/24-well were seeded in antibiotic-free DMEM supplemented with FBS. Medium was changed to conditioned growth medium (collected after 24 h from ntc or shPex16 3T3-L1 cells at day 7) 18 h after EP and the cells were harvested for luciferase reporter assay 48 h after EP.
2.18. Transmission electron microscopy and cell organelle quantification
For transmission electron microscopy, undifferentiated or differentiated 3T3-L1 cells (ntc, shPex16, si-ctrl, and si-Pex16) were cultured on an Aclar film, fixed in 0.1 M phosphate buffer (pH 7.4) containing 2.5% glutaraldehyde and 2% formaldehyde (2 h), post-fixed in 2% OsO4 (2 h), dehydrated in graded series of ethanol, and embedded in a TAAB epoxy resin. Images of 70 nm sections (stained with lead citrate and platine blue) were taken on a FEI Tecnai G2 20 transmission electron microscope (FEI, Eindhoven, Netherlands) with a Gatan ultrascan 1000 CCD camera (acceleration voltage 120 kV). Peroxisomes and mitochondria were counted in ≥35 images per biological replicate.
2.19. Statistical analysis
If not otherwise stated, results are shown as mean ± S.D. of at least three independent experiments, or results show one representative experiment of three. Statistical analysis was done on all available data. Statistical analysis was performed using the two-tailed Student’s t-test, one-way or two-way ANOVA with a post hoc Tukey’s multiple comparisons test was performed. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. Main effects for the different variables are denoted as § or #, respectively.
3. Results
3.1. Pex16 is a target gene of the adipogenesis “master-regulator” PPARγ
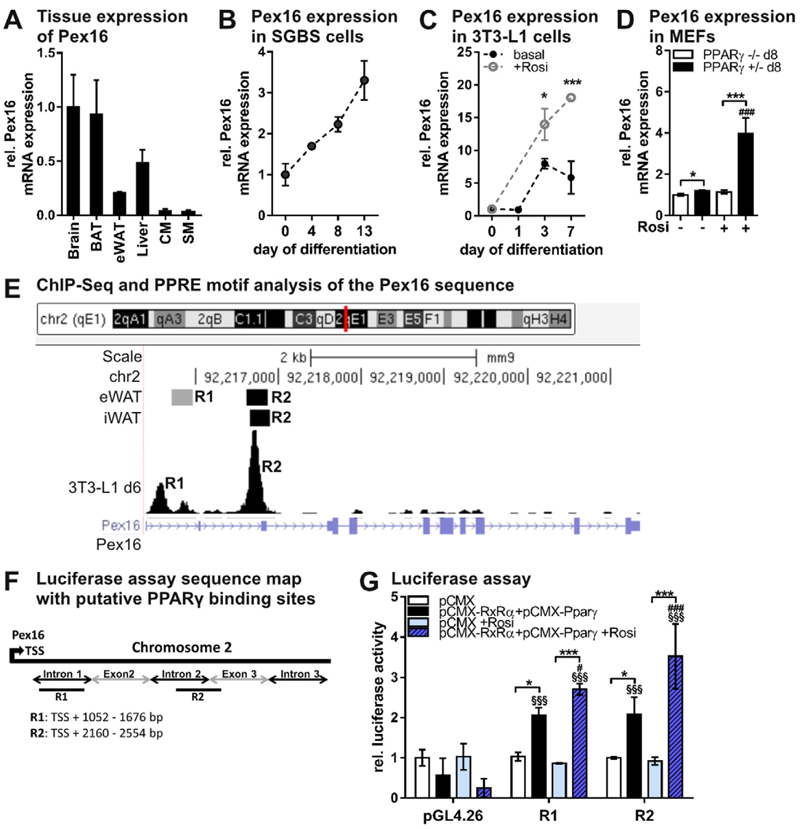
Pex16 is robustly expressed in murine brain, brown adipose tissue (BAT), epididymal white adipose tissue (eWAT), liver, cardiac muscle (CM), and skeletal muscle (SM) (Fig. 1A, Fig. S1A). Further, Pex16 expression strongly increased during adipogenic differentiation of the human SGBS cell line (Simpson-Golabi-Behmel syndrome, Fig. 1B) and human stromal vascular cells (Fig. S1B). During 3T3-L1 cell differentiation, Pex16 expression increased (Fig. 1C) and was even higher upon addition of the PPARγ-agonist rosiglitazone (Fig. 1C), suggesting a potential regulation of Pex16 via PPARγ. To examine whether Pex16 expression is dependent on the presence of PPARγ, we compared Pex16 mRNA expression in mouse embryonic fibroblasts (MEFs) isolated from PPARγ−/− and PPARγ+/−- mice [30] on day 8 after induction of differentiation. Basal Pex16 expression was slightly higher in PPARγ+/−- compared to PPARγ− /− MEFs (Fig. 1D). However, treatment with rosiglitazone increased Pex16 expression in PPARγ+/−- MEFs by 4-fold, while no increase was visible in PPARγ−/− MEFs (Fig. 1D). This increase seems to be independent of adipogenic differentiation as addition of rosiglitazone did not change lipid droplet accumulation (Fig. S1C). To test the potential regulation of Pex16 expression by PPARγ, we explored published ChIP sequencing datasets of global PPARγ binding in murine adipose tissues and 3T3-L1 adipocytes [39, 40] using the UCSC genome browser (http://genome.ucsc.edu). We identified two potential PPARγ binding sites downstream of the Pex16 transcription start site (TSS), referred to as R1 and R2 (Fig. 1E). We next investigated whether these two sequences represent functional binding sites for PPARγ and its cofactor RXRα by luciferase reporter assays with the two putative binding sites (R1: 1052 bp-1676 bp and R2: 2160 bp- 2554 bp downstream of TSS) (Fig. 1F). A functional PPARγ binding to R1 and R2 sequences could be confirmed by increased luciferase activity under basal conditions that was further elevated upon addition of rosiglitazone (Fig. 1G). These results indicate that PPARγ is sufficient to trigger Pex16 expression, and that Pex16 is a functional PPARγ target gene with at least two PPARγ binding sites.
Fig. 1.
Pex16 is a functional target gene of the adipogenesis “master-regulator” PPARγ (A) Pex16 tissue mRNA expression in 8-10-week old male, ad libitum-fed C57BL/6 mice (n = 5). (B) Pex16 mRNA expression inhuman cells (Simpson-Golabi-Behmel syndrome) during adipogenic differentiation (n = 3). (C) Pex16 mRNA expression in3T3-L1 cells during adipogenic differentiation with or without addition of 1 μM PPARγ-agonist rosiglitazone to the culture medium. Student’s t-test (n = 3): *pb 0.05; ***pb 0.001 versus basal. (D) Pex16 mRNA expression in PPARγ−/− and PPARγ+/−- murine embryonic fibroblasts (MEFs) on day 8 of differentiation with or without addition of 1 μM rosiglitazone to the culture medium. (E) Genome organization of Pex16 showing two putative PPARγ binding sites, named R1 and R2, obtained from [39,40], adapted from UCSC genome browser (http://genome.uscs.edu). (F) Map of putative PPARγ binding sites R1 and R2 in the Pex16 sequence depicted in Fig. E, used for the luciferase assay. (G) Putative PPARγ/RXRα binding sites in the Pex16 sequence (R1, R2) were cloned into pGL4.26 luc-2-luciferase reporter vector. Cos7 cells were co-transfected with plasmids encoding luc-2 luciferase, renilla luciferase, and either empty pCMX vector or PPARγ/RXRα expressing vectors. Cells were treated with 1 μM rosiglitazone or DMSO 24 h prior to measurements. Luc2-luciferase activity was measured 48 h after transfection and normalized to renilla luciferase activity. (A)-(D),(G) Data are presented as mean ± SD. (D), (G) Statistical significance was calculated using one-way or two-way ANOVA and subsequent Tukey’s multiple comparisons test (n = 3). *p < 0.05, ** p < 0.01, ***p < 0.001 versus control. Main effects for the different constructs (R1, R2) versus pGL4.26 are denoted as § and for rosiglitazone treatment are denoted as #.
3.2. PEX16-silencing strongly reduces peroxisome number and affects cellular respiration
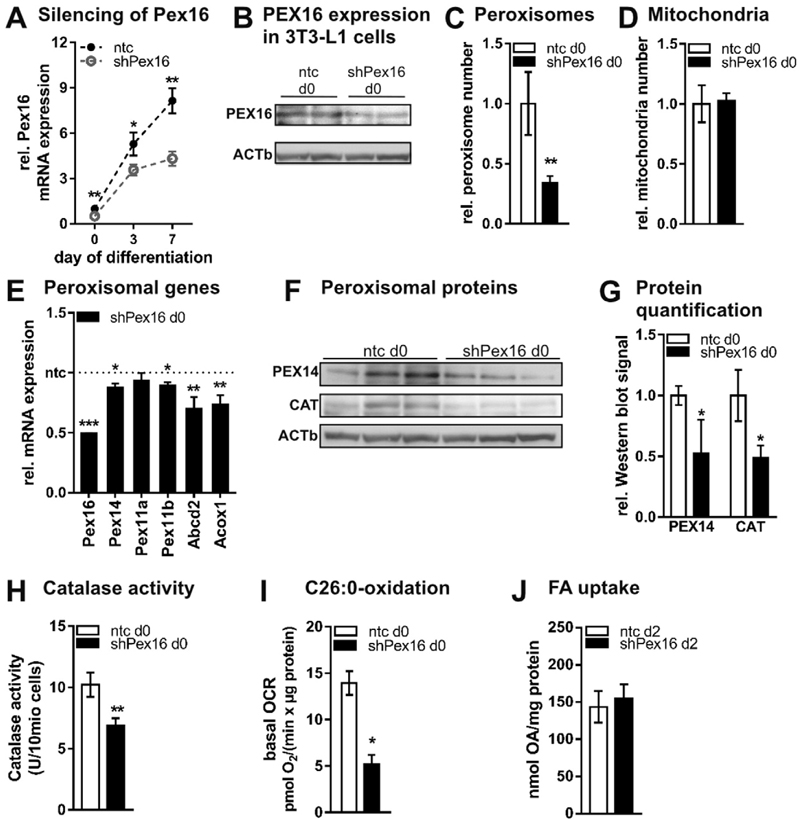
Recently, PEX16 has been postulated as the “master peroxin” for peroxisome de novo synthesis in mammalian cells [19]. To address the function of PEX16 in 3T3-L1 during differentiation, we silenced Pex16 in 3T3-L1 fibroblasts using shRNA-containing lentiviral particles. Stable silencing of ~50% was achieved throughout adipogenic differentiation on mRNA (Fig. 2A, Fig. S1D) and protein level (Fig. 2B, Fig. S1E). In fibroblasts, silencing of Pex16 reduced peroxisome number to one third of control cells (Fig. 2C), while mitochondrial number (Fig. 2D) and morphology (Fig. S1F) were unchanged. Reduced peroxisome number was accompanied by reduced expression of PEX14 (Fig. 2E,F,G), which is involved in peroxisome assembly. Expression of Pex11a and 11b, responsible for peroxisome fission, was unchanged (Pex11a) or moderately reduced (Pex11b) (Fig. 2E), respectively. These data indicate that there is no compensatory up-regulation of peroxisome biogenesis via fission mediated by Pex11 in Pex16-silenced cells. Peroxisomes are a prerequisite for oxidation of certain FAs and hydrogen peroxide detoxification. Upon Pex16-silencing, mRNA expression of the peroxisomal FA importer ATP-binding cassette, sub-family D, member 2 (Abcd2), and acyl-coenzyme A oxidase (Acox1) was reduced (Fig. 2E). Additionally, protein expression and activity of the hydrogen peroxide detoxifying enzyme catalase (CAT) were reduced upon Pex16-silencing (Fig. 2F,G,H). Using Seahorse extracellular flux analyzer, we measured the cellular capacity to oxidize hexacosanoic acid (C26:0), which is generally impaired in peroxisome defects [41–43]. We found C26:0 oxidation strongly reduced in Pex16-silenced fibroblasts (Fig. 2I), which show unchanged FA uptake (Fig. 2J). After differentiating Pex16-silenced fibroblasts to adipocytes (for persistent Pex16 silencing see Fig. S1E, S2A), peroxisome number was still reduced (Fig. S2B). In accordance, expression of genes/proteins involved in peroxisome assembly, FA uptake, oxidation and biosynthesis, as well as detoxification were mostly reduced (Fig. S2C,D). Concomitant with the decreased catalase activity (Fig. S2E), the production of reactive oxygen species (ROS) was increased (Fig. S2F). Importantly, we did not observe any peroxisome defect-associated translocation of peroxisomal proteins to the cytosol in Pex16-silenced 3T3-L1 cells as catalase activity was mainly detected in mitochondrial/ peroxisomal fractions (Fig. S2G), arguing for normal function of remaining peroxisomes in Pex16-silenced cells. Since peroxisomes (in addition to mitochondria) are an important site of oxygen utilization in the cell, we measured the oxygen consumption rate (OCR) of Pex16-silenced and control cells. Pex16-silenced cells showed overall reduced OCR (Fig. S2H), indicating that loss of peroxisomes affects whole cell oxygen utilization.
Fig. 2.
Silencing of Pex16 in 3T3-L1 fibroblasts impairs peroxisome formation and function (A)–(J) Stable silencing of Pex16 in 3T3-L1 fibroblasts. Undifferentiated 3T3-L1 cells were incubated with shRNA-containing lentiviral particles for Pex16 (shPex16) or non-targeting control virus (ntc) followed by antibiotic selection. (B)–(I) Data were collected from confluent fibroblasts on day 0. (A) Pex16 mRNA expression in 3T3-L1 cells during differentiation (n = 3). (B) PEX16 protein expression; one representative replicate is shown. Relative number of (C) peroxisomes and (D) mitochondria counted from ELMI pictures (n = 3). (E) RT-PCR analysis of genes involved in peroxisome assembly (Pex14, Pex11a, Pex11b) and peroxisomal lipid metabolism (Abcd2, Acox1) (n = 3). (F) Expression and (G) densitometric analysis of peroxisomal proteins PEX14 and catalase (CAT) (n = 3). (H) Catalase activity (n = 3). (I) Basal respiration in the presence of 10 μM hexacosanoic acid (C26:0). Cells were incubated with assay medium containing 10 μM C26:0 45 min prior to and during the measurement with Seahorse XF96 extracellular flux analyzer (n = 6). (J) 14C–oleic acid (OA) uptake determined after 30 min incubation on d2 of differentiation (n = 4). (A), (C)–(E), (G), (H), (J) Data are presented as mean ± SD (n ≥ 3). (I) Data are presented as mean ± SEM (n = 4). Statistical significance was calculated using Student’s t-test. *p < 0.05; **p < 0.01; ***p < 0.001 versus ntc.
3.3. Adipogenesis is decreased in Pex16-silenced cells
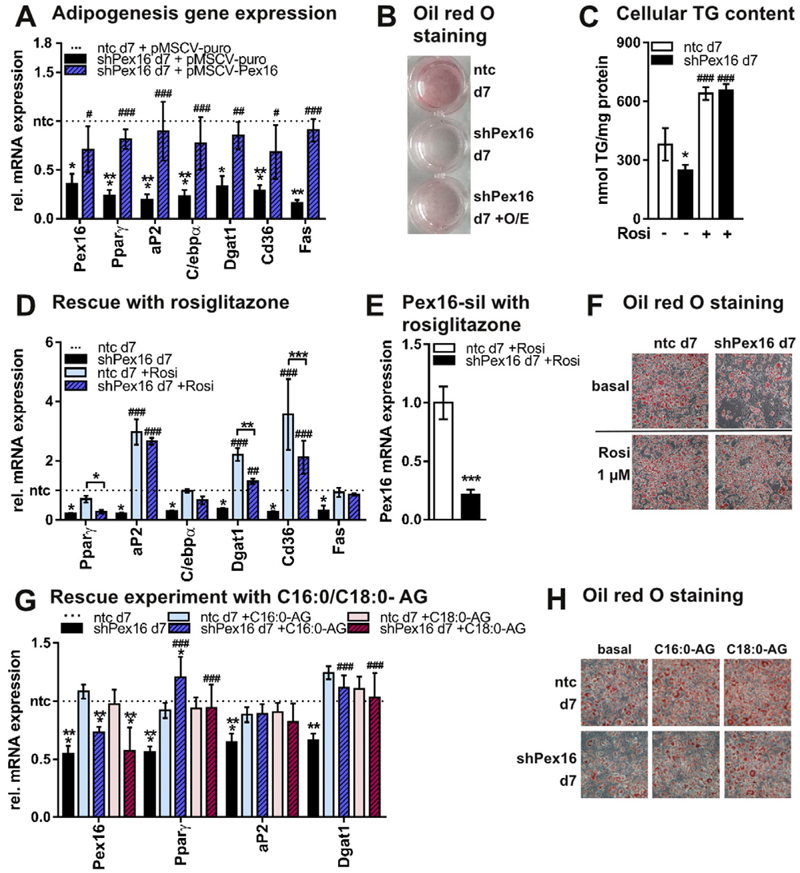
Our data reveal that Pex16 is a PPARγ target gene and expression of Pex16 is highly upregulated during 3T3-L1 adipocyte differentiation. Thus, we were interested how Pex16-silencing and the changes in peroxisome number and activity subsequently impact adipogenesis. Pex16-silencing in 3T3-L1 cells impaired adipogenic differentiation, as corroborated by reduced expression of adipogenesis marker genes (Fig. 3A, Fig. S1D), reduced neutral lipid staining (oil red O, ORO) (Fig. 3B), and reduced triglyceride accumulation (Fig. 3C). This effect could be reversed by retroviral re-expression of Pex16 in Pex16-silenced 3T3-L1 cells (Fig. 3A,B). Additionally, expression of PPARγ and several PPARγ target genes was strongly decreased in Pex16-silenced cells when compared to controls (Fig. 3A, Fig. S1D). Thus, we speculated that reduced PPARγ activity leads to impaired adipogenesis in Pex16-silenced cells and aimed to rescue the differentiation defect of Pex16-silenced cells with a pharmacological agonist of PPARγ. Addition of rosiglitazone during adipogenic differentiation rescued adipogenesis in Pex16-silenced cells, as observed by restored adipogenesis marker and PPARγ target gene expression (Fig. 3D, Fig. S2I), while Pex16 expression was still 70% reduced (Fig. 3E). In addition, ORO staining (Fig. 3F) and cellular TG levels (Fig. 3C) were comparable to controls after treatment of Pex16-silenced cells with rosiglitazone. Since lipids produced by peroxisomes are described to be important for adipogenesis [26,32,44], we used 16:0-alkylglycerol (C16:0-AG) and 18:0-alkyl-glycerol (C18:0-AG) to rescue adipogenesis in Pex16-silenced cells. Both lipids were able to partially restore adipogenesis in Pex16-silenced cells as determined by marker gene expression and ORO staining (Fig. 3G,H). From these data we propose that the reduced number of peroxisomes and the associated effects already observed in Pex16-silenced undifferentiated 3T3-L1 cells subsequently lead to the reduced adipogenesis.
Fig. 3.
PEX16 is required for adipogenesis in 3T3-L1 cells (A)–(H) Data were collected from 3T3-L1 adipocytes atday 7. (A) RT-PCR analysis of adipogenesis marker and PPARγ target genes in ntc and shPex16 3T3-L1 cells on day 7 of differentiation treated with control retrovirus (pMSCV-puro) or Pex16-overexpressing (O/E) retrovirus (pMSCV-Pex16) for 48 h each on day −2, 0 and 3 of differentiation (n = 3). (B) Representative ORO staining of cells described in (A). (C) Cellular triglyceride (TG) content of day 7 ntc and shPex16 3T3-L1 cells, differentiated with or without 1 μM rosiglitazone (n = 3). (D) RT-PCR analysis of adipogenesis marker and PPARγ target genes in ntc and shPex16 3T3-L1 cells differentiated with or without 1 μM rosiglitazone (n = 3). (E) Pex16 mRNA expression in day 7 ntc and shPex16 cells treated with 1 μM rosiglitazone during adipogenesis (n = 3). (F) Representative ORO staining of cells described in (D). (G),(H) Rescue of adipogenesis in shPex16 3T3-L1 cells with ether lipids. (G) RT-PCR analysis of various genes in ntc and shPex16 3T3-L1 cells on day 7 differentiated with or without addition of 10 μM 1-O-hexadecyl-rac-glycerol (C16:0-AG) or 10 μM 1-O-octadecyl-rac-glycerol (C18:0-AG) (n = 3). (H) Representative ORO staining of day 7 ntc and shPex16 3T3-L1 cells from (G). (A), (C), (D), (E), (G) Data are presented as mean ± SD (n ≥ 3). Statistical significance was calculated using Student’s t-test, one-way or two-way ANOVA and subsequent Tukey’s multiple comparisons test (n = 3). *p < 0.05, ** p < 0.01, ***p < 0.001 versus ntc. Main effects of the different treatments versus according untreated samples are denoted as #.
3.4. Pex16-silencing impacts cellular lipid homeostasis
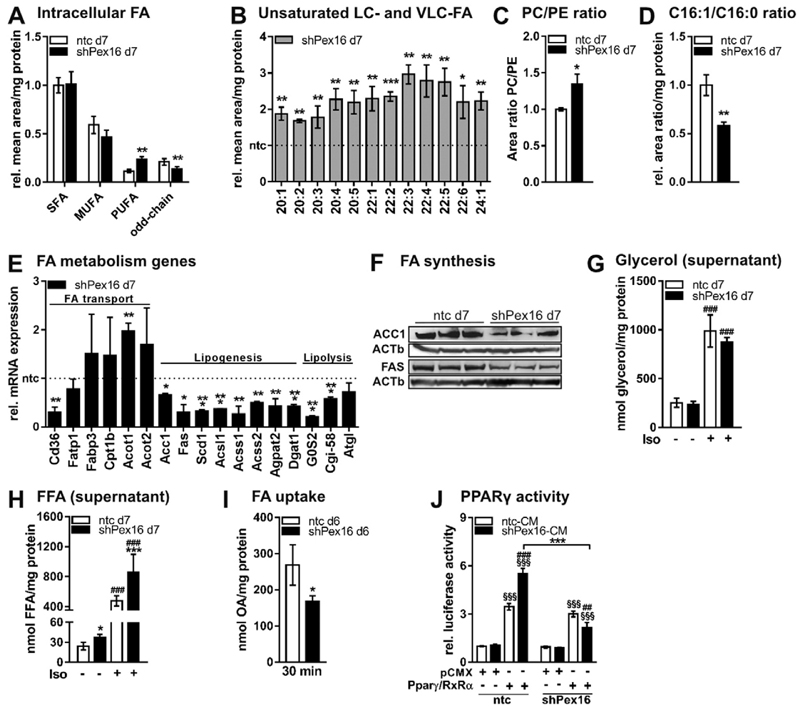
Lipid analysis revealed that silencing of Pex16 in 3T3-L1 cells markedly impacts cellular lipid composition in adipocytes. While saturated (SFAs) and mono-unsaturated fatty acids (MUFAs) were unchanged (Fig. 4A), we found an accumulation of long- and very long-chain (LC/ VLC) MUFAs and PUFAs (Fig. 4A,B) and reduced levels of peroxisomal α-oxidation derived odd-chain FAs (Fig. 4A, Fig. S3A) in Pex16-silenced cells. LC/VLC PUFAs were also strongly increased in triglycerides and phosphatidylcholine (Fig. S3B,C,D). We also found an increased PC/PE ratio in Pex16-silenced cells compared to controls (Fig. 4C, Fig. S3D,E). In addition, the ratio of C16:1 to C16:0 was 50% reduced upon Pex16-silencing (Fig. 4D), suggesting an impaired desaturation of C16:0 to C16:1 via stearoyl-Coenzyme A desaturase 1 (Scd1). Accordingly, Scd1 mRNA expression was 70% decreased in Pex16-silenced cells (Fig. 4E). To investigate which FA metabolism pathways are affected in Pex16-silenced cells, we measured expression of genes involved in FA uptake/transport, lipid biosynthesis, and lipolysis. While expression of the FA import gene Cd36 was 70% reduced upon Pex16-silencing, genes responsible for intracellular FA transport, namely FA transport protein 1 (Fatp1), FA binding protein 3 (Fabp3), and carnitine palmitoyltransferase 1b (Cpt1b) were unchanged (Fig. 4E). Expression of cytosolic (Acot1) and mitochondrial acyl-CoA thioesterases (Acot2), providing free FAs and CoASH, were increased in Pex16-silenced cells (Fig. 4E). Further, mRNA and protein expression of acetyl-CoA carboxylase (Acc1), generating malonyl-CoA from acetyl-CoA as the first step of FA synthesis, were reduced (Fig. 4E,F). Accordingly, expression of FA synthase (Fas), generating FAs such as palmitic acid (C16:0) and stearic acid (C18:0) from malonyl-CoA, was strongly reduced in Pex16-silenced cells on mRNA (Fig. 4E) and protein level (Fig. 4F). Expression of short or long chain acyl-CoA synthetases (Acsl1, Acss1, Acss2), which activate FAs for e.g. biosynthesis of triglycerides or phospholipids, was up to 70% reduced (Fig. 4E). Concomitantly, 1-acylglycerol-3-phosphate O-acyltransferase 2 (Agpat2), generating phosphatidic acid for (phospho-)lipid biosynthesis, and diacylglycerol O-acyltransferase 1 (Dgat1), responsible for the last step of triglyceride biosynthesis, were 50% reduced on mRNA level in Pex16-silenced cells (Fig. 4E), suggesting impaired lipid biosynthesis in Pex16-silenced cells. In contrast to lipid biosynthesis, lipolysis seems not to be affected in Pex16-silenced cells, as mRNA expression of the rate-limiting triglyceride (TG)-hydrolyzing enzyme adipose triglyceride lipase (Atgl) was unchanged and ATGL inhibitor (G0S2) as well as co-activator (Cgi-58) were reduced in Pex16-silenced cells (Fig. 4E). Hence, glycerol levels in the cellular supernatant as readout for lipolysis did not differ between Pex16-silenced and control cells (Fig. 4G). In contrast, free FA levels in the supernatant were strongly increased under basal and lipolysis-stimulated conditions (Fig. 4H). The accumulation of FAs in the supernatant might be due to reduced peroxisomal FA oxidation (Fig. 2I), decreased lipid/TG biosynthesis (Fig. 4E,F), but also caused by impaired FA re-uptake (Fig. 4I). The increased FA concentration in the supernatant of Pex16-silenced cells (Fig. 4H) and the successful rescue with rosiglitazone (Fig. 3D,E,F) made us assume that Pex16-silenced cells release precursors for PPARγ-ligand production into the medium. In control cells, these accumulating FAs should be metabolized by peroxisomes to produce PPARγ ligands. Therefore, we established a luciferase assay measuring PPARγ activity in control and Pex16-silenced fibroblasts in the presence of conditioned medium (CM) from control or Pex16-silenced adipocytes. PPARγ activity was comparable in control and Pex16-silenced fibroblasts when adding CM from control adipocytes. Importantly, control fibroblasts treated with Pex16-silenced CM showed 1.5-fold increased luciferase activity in comparison to treatment with control CM, while no increase was observed in Pex16-silenced cells (Fig. 4J).
Fig. 4.
Lipid homeostasis is altered in Pex16-silenced 3T3-L1 adipocytes (A)-(D) Total lipids were extracted from ntc and shPex16 3T3-L1 cells on day 7 of differentiation and analyzed by UPLC-qTOF-MS.(A) Mean area of total intracellular saturated (SFA), monounsaturated (MUFA), polyunsaturated (PUFA), and odd-chain FAs (n = 3). (B) Profile of unsaturated long-chain (LC) and very long-chain (VLC) FAs (n = 3). (C) Area ratio of phosphatidylcholine (PC) and phosphatidylethanolamine (PE) (n = 3). (D) Ratio of C16:1 to C16:0 FAs (n = 3). (E) RT-PCR analysis of genes (for abbreviations see Table S2) involved inFA transport, lipogenesis and lipolysis inntc and shPex16 3T3-L1 cells on day 7 of differentiation (n = 3). (F) Expression of FA synthesis proteins acetyl-CoA carboxylase (ACC1) and fatty acid synthase (FAS) in ntc and shPex16 3T3-L1 cells on day 7 of differentiation. (G) Glycerol content (n = 3) and (H) free fatty acid (FFA) content (n = 6) in the supernatant of day 7 ntc and shPex16 3T3-L1 cells after 4 h serum-starvation with or without isoproterenol treatment (10 μM). (I) 14C–oleic acid (OA) uptake into ntc and shPex16 3T3-L1 cellson day 6 of differentiation after 30 min incubation. (J) Luciferase assay to assess PPARγ activation in ntc and shPex16 3T3-L1 cells. Preconfluent ntc and shPex16 3T3-L1 cells were co-electroporated with firefly luciferase reporter vector and empty pCMX or PPARγ/RXRα-expressing vectors. Co-electroporation of renilla luciferase encoding vector served as control for varying electroporation efficiencies. After 24 h, medium was changed to CM from day 7 ntc or shPex16 3T3-L1 cells. Firefly luciferase activity was measured 48 h after electroporation and normalized to renilla luciferase activity, (n = 3). (A)–(E), (G)–(J) Data are presented as mean ± SD (n ≥ 3). Statistical significance was calculated using Student’s t-test, one-way or two-way ANOVA and subsequent Tukey’s multiple comparisons test (n = 3). *p < 0.05, ** p < 0.01, ***p < 0.001 versus control. Main effects of the different constructs (pCMX, Pparγ/RxRα) are denoted as § and different treatments are denoted as #.
To clarify whether the changes in lipid homeostasis observed upon stable Pex16-silencing are secondary to impaired adipogenesis, we next silenced Pex16 in mature adipocytes (Fig. 5A–D). Transient silencing of Pex16 in mature adipocytes could not reduce the number of peroxisomes (Fig. 5E), and no changes in supernatant glycerol and FA content (Fig. S4A,B) and cellular lipid profile were detected (Fig. S5A-H). Despite normal adipocyte differentiation (Fig. S4C,D), we found a reduction in peroxisomal, adipogenic and lipogenic gene/protein expression (Fig. 5B–D). Further, catalase activity and FA uptake were decreased (Fig. 5F,G) and C26:0 oxidation showed trends to reduction (Fig. 5H).
These data show that a short term knock-down of Pex16 in mature adipocytes is not efficient in reducing peroxisome number and impacting lipid homeostasis when compared to stable Pex16-silencing from the onset of differentiation.
4. Discussion
This study demonstrates for the first time that Pex16 is a direct and functional PPARγ target gene. In keratinocytes it was already shown that Pex16 expression increased upon incubation with PPARγ agonists [45]. It was also reported that peroxisomes play a key role in regulating the availability of cellular signaling lipids, including ligands for retinoic acid receptors (RARs) and PPARs, which regulate the transcription of genes essential for embryonic development and differentiation of various tissues [25,44,46–48]. Our results strengthen these data, as 3T3-L1 cells stably silenced for Pex16, which have a reduced number of peroxisomes, showed impaired adipogenesis presumably due to reduced PPARγ activation. We draw this conclusion because the differentiation defect of Pex16-silenced 3T3-L1 cells could be rescued by administration of PPARγ-agonist rosiglitazone, by addition of peroxisome-related lipid species, and also by re-expression of Pex16. Our data propose that lipids accumulating in the supernatant of Pex16-silenced cells are potential PPARγ-ligand precursors that can only be used by adipocytes with sufficient number and activity of peroxisomes. It has also been shown that the ratio of phosphatidylcholine (PC) to phosphatidylethanolamine (PE) decreases during adipogenic differentiation [49]. Pex16-silencing increased the PC/PE ratio and might also thereby counteract adipogenesis. In addition, PUFAs such as C22:6, C22:5, C20:5, and C20:4 accumulate in Pex16-silenced 3T3-L1 cells. These PUFAs have all been shown to inhibit the expression of lipogenic enzymes and PPARγ, thereby inhibiting adipocyte differentiation [50,51]. Given that Pex16 expression and the number of peroxisomes markedly increase during adipocyte differentiation [9,10], it is conceivable that peroxisomes are required to metabolize these FAs for final adipocyte differentiation.
Zellweger syndrome, caused by a defect in peroxisome assembly in humans [52], is characterized by an accumulation of VLC-FAs [53] and PUFAs in phosphatidylcholines of fibroblasts [54]. In fact, some of our data resemble this phenotype as Pex16-silenced 3T3-L1 cells, whose peroxisomal number is reduced to one third, accumulate LC/ VLC PUFAs in various lipid classes, including triglycerides and phosphatidylcholine. Additionally, intracellular MUFAs (C20:1, C22:1, and C24:1) accumulated while C16:1 was strongly reduced, leading to unchanged total MUFAs. Further, Zellweger syndrome patients have a defect in mitochondrial electron transport chain and reduced cellular respiration in liver and muscle cells [55]. This might be caused by increased ROS production presumably arising from the defective peroxisomal antioxidant system and defective β-oxidation, as observed in Pex5−/− mice, a mouse model for Zellweger syndrome [56]. In Pex16-silenced cells, we observed reduced catalase activity accompanied by increased production of ROS. In addition to impaired peroxisomal FA oxidation, reduced cellular oxygen consumption rate also indicates diminished mitochondrial β-oxidation. Moreover, Pex16-silenced cells showed an increased concentration of free FAs in the supernatant independent from lipolysis, since Atgl expression and glycerol release as readout for lipolysis were unaffected. Interestingly, genes required for FA activation were down-regulated (Acsl1, Acss1, Acss2) and even more, genes that generate free FAs from activated FAs (Acot1 and Acot2) were upregulated upon Pex16-silencing. These findings propose that Pex16-silenced cells try to reduce the amount of activated FAs as FA oxidation is impaired. Hence, Pex16-silenced cells seem to release FAs into the supernatant. Additionally, we found the uptake of FAs reduced in accordance with decreased Cd36 expression, also suggesting that Pex16-silenced cells try to escape lipotoxicity. On the other hand, the question arises why activated FAs are not re-esterified into triglycerides. This might (amongst others) be due to reduced lysophosphatidic acid availability as Gnpat and Dhrs7b are strongly decreased, but also the expression levels of Agpat and Dgat, responsible for TG synthesis, are reduced.
We have shown that stable Pex16-silencing in fibroblasts revealed profound effects on peroxisome number and activity with concomitant changes in FA oxidation in undifferentiated cells and these effects persisted throughout adipogenic differentiation and strongly influenced adipogenesis and adipocyte lipid profile. Although, transient Pex16-silencing in mature adipocytes showed effects on peroxisomal and lipogenic gene/protein expression and peroxisome activity, no impact on lipid homeostasis was observed. We speculate that these short term changes are not sufficient to lead to measureable modifications of the lipid profile of mature adipocytes which are fully loaded with lipids. Thus, we argue that a reduced peroxisome number and activity throughout adipocyte development is a prerequisite to affect the lipid profile, thereby also reducing PPARγ ligand production and activation [44,48]. Subsequently, adipogenesis is reduced. However, in cells stably silenced for Pex16 throughout differentiation, the attenuated adipogenesis might also contribute or even augment the changes in lipid homeostasis.
These data suggest that PEX16 is required for peroxisome biogenesis during adipocyte development and highlight the relevance of peroxisomes for adipogenesis and cellular lipid metabolism.
Supplementary Material
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbalip.2016.12.009.
Acknowledgements
This work was supportedbyAustrian ScienceFund FWF [grant numbers DK-MCD W1226, P24143, P27108, and P27070]. The authors gratefully acknowledge support from NAWI Graz and BioTechMed Graz. We thank H. Furkan Alkan, Katharina Huber and Wenmin Xia for fruitful discussions and critical review of the manuscript, and Dominique Pernitsch, Thomas Schreiner and Wolfgang Krispel for technical assistance.
Footnotes
Author contribution
D.C.H. and J.G.B.-S. designed the experiments. D.C.H., A.R.P., H.J.P, S.S., C.T.M.-S., D.K., T.O.E., and G.R. performed the experiments together with support of M.B., W.F.G., D.K. and J.G.B.-S. D.C.H. and J.G.B.-S. wrote the manuscript. All authors reviewed the manuscript.
Conflict of interest
The authors declare no potential conflict of interest.
Transparency Document
The Transparency document associated with this article can be found, in online version.
References
- [1].Prokesch A, Hackl H, Hakim-Weber R, Bornstein SR, Trajanoski Z. Novel insights into adipogenesis from omics data. Curr Med Chem. 2009;16(23):2952–2964. doi: 10.2174/092986709788803132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hakim-Weber R, Krogsdam A-M, Jørgensen C, Fischer M, Prokesch A, Bogner-Strauss JG, et al. Transcriptional regulatory program in wild-type and retinoblastoma gene-deficient mouse embryonic fibroblasts during adipocyte differentiation. BMC Res Notes. 2011;4(1):157. doi: 10.1186/1756-0500-4-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Prokesch A, Bogner-Strauss JG, Hackl H, Rieder D, Neuhold C, Walenta E, et al. Arxes: retrotransposed genes required for adipogenesis. Nucleic Acids Res. 2011;39(8):3224–3239. doi: 10.1093/nar/gkq1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Savage DB. PPAR[gamma] as a metabolic regulator: insights from genomics and pharmacology. Expert Rev Mol Med. 2005;7(01):1–16. doi: 10.1017/S1462399405008793. [DOI] [PubMed] [Google Scholar]
- [5].Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nature Reviews, Mol Cell Biol. 2006;7(12):885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- [6].Spiegelman BM, Hu E, Kim JB, Brun R. PPAR gamma and the control of adipogenesis. Biochimie. 1997;79(2–3):111–112. doi: 10.1016/s0300-9084(97)81500-3. [DOI] [PubMed] [Google Scholar]
- [7].Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79(7):1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- [8].Tontonoz P, Hu E, Spiegelman BM. Regulation of adipocyte gene expression and differentiation by peroxisome proliferator activated receptor gamma. Curr Opin Genet Dev. 1995;5(5):571–576. doi: 10.1016/0959-437x(95)80025-5. [DOI] [PubMed] [Google Scholar]
- [9].Novikoff AB, Novikoff PM, Rosen OM, Rubin CS. Organelle relationships in cultured 3T3-L1 preadipocytes. J Cell Biol. 1980;87(1):180–196. doi: 10.1083/jcb.87.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Novikoff AB, Novikoff PM. Microperoxisomes and peroxisomes in relation to lipid metabolism. Ann NY Acad Sci. 1982;386:138–152. doi: 10.1111/j.1749-6632.1982.tb21412.x. [DOI] [PubMed] [Google Scholar]
- [11].Su X, Han X, Yang J, Mancuso DJ, Chen J, Bickel PE, et al. Sequential ordered fatty acid alpha oxidation and Delta9 desaturation are major determinants of lipid storage and utilization in differentiating adipocytes. Biochemistry. 2004;43(17):5033–5044. doi: 10.1021/bi035867z. [DOI] [PubMed] [Google Scholar]
- [12].Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol. 2006;173(4):521–532. doi: 10.1083/jcb.200601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Honsho M, Hiroshige T, Fujiki Y. The membrane biogenesis peroxin Pex16p. Topogenesis and functional roles in peroxisomal membrane assembly. J Biol Chem. 2002;277(46):44513–44524. doi: 10.1074/jbc.M206139200. [DOI] [PubMed] [Google Scholar]
- [14].Islinger M, Cardoso MJR, Schrader M. Be different–the diversity of peroxisomes in the animal kingdom. Biochim Biophys Acta. 2010;1803(8):881–897. doi: 10.1016/j.bbamcr.2010.03.013. [DOI] [PubMed] [Google Scholar]
- [15].Bagattin A, Hugendubler L, Mueller E. Transcriptional coactivator PGC-1alpha promotes peroxisomal remodeling and biogenesis. Proc Natl Acad Sci U S A. 2010;107(47):20376–20381. doi: 10.1073/pnas.1009176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ahlabo I, Barnard T. Observations on peroxisomes in brown adipose tissue of the rat. J Histochem Cytochem. 1971;19(11):670–675. doi: 10.1177/19.11.670. [DOI] [PubMed] [Google Scholar]
- [17].Ishii H, Fukumori N, Horie S, Suga T. Effects of fat content in the diet on hepatic peroxisomes of the rat. Biochim Biophys Acta. 1980;617(1):1–11. doi: 10.1016/0005-2760(80)90218-0. [DOI] [PubMed] [Google Scholar]
- [18].Kramar R, Hüttinger M, Gmeiner B, Goldenberg H. β-oxidation in peroxisomes of brown adipose tissue. Biochim Biophys Acta Lipids Lipid Metab. 1978;531(3):353–356. doi: 10.1016/0005-2760(78)90217-5. [DOI] [PubMed] [Google Scholar]
- [19].Kim PK, Mullen RT. PEX16: a multifaceted regulator of peroxisome biogenesis. Front Physiol. 2013;4:241. doi: 10.3389/fphys.2013.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fransen M, Wylin T, Brees C, Mannaerts GP, Van Veldhoven PP. Human pex19p binds peroxisomal integral membrane proteins at regions distinct from their sorting sequences. Mol Cell Biol. 2001;21(13):4413–4424. doi: 10.1128/MCB.21.13.4413-4424.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yonekawa S, Furuno A, Baba T, Fujiki Y, Ogasawara Y, Yamamoto A, et al. Sec16B is involved in the endoplasmic reticulum export of the peroxisomal membrane biogenesis factor peroxin 16 (Pex16) in mammalian cells. Proc Natl Acad Sci U S A. 2011;108(31):12746–12751. doi: 10.1073/pnas.1103283108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Matsuzaki T, Fujiki Y. The peroxisomal membrane protein import receptor Pex3p is directly transported to peroxisomes by a novel Pex19p- and Pex16p-dependent pathway. J Cell Biol. 2008;183(7):1275–1286. doi: 10.1083/jcb.200806062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Waterham HR, Ebberink MS. Genetics and molecular basis of human peroxisome biogenesis disorders. Biochim Biophys Acta. 2012;1822(9):1430–1441. doi: 10.1016/j.bbadis.2012.04.006. [DOI] [PubMed] [Google Scholar]
- [24].Suzuki Y, Shimozawa N, Orii T, Tsukamoto T, Osumi T, Fujiki Y, et al. Genetic and molecular bases of peroxisome biogenesis disorders. Genet Med. 2001;3(5):372–376. doi: 10.1097/00125817-200109000-00007. [DOI] [PubMed] [Google Scholar]
- [25].Titorenko VI, Rachubinski RA. The peroxisome. J Cell Biol. 2004;164(5):641–645. doi: 10.1083/jcb.200312081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Brites P, Ferreira AS, Ferreira da Silva T, Sousa VF, Malheiro AR, Duran M, et al. Alkyl-glycerol rescues plasmalogen levels and pathology of ether-phospholipid deficient mice. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0028539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Martens K, Bottelbergs A, Peeters A, Jacobs F, Espeel M, Carmeliet P, et al. Peroxisome deficient aP2-Pex5 knockout mice display impaired white adipocyte and muscle function concomitant with reduced adrenergic tone. Mol Genet Metab. 2012;107(4):735–747. doi: 10.1016/j.ymgme.2012.10.015. [DOI] [PubMed] [Google Scholar]
- [28].Bogner-Strauss JG, Prokesch A, Sanchez-Cabo F, Rieder D, Hackl H, Duszka K, et al. Reconstruction of gene association network reveals a transmembrane protein required for adipogenesis and targeted by PPARγ. Cell Mol Life Sci. 2010;67(23):4049–4064. doi: 10.1007/s00018-010-0424-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hansen JB, Petersen RK, Larsen BM, Bartkova J, Alsner J, Kristiansen K. Activation of peroxisome proliferator-activated receptor gamma bypasses the function of the retinoblastoma protein in adipocyte differentiation. J Biol Chem. 1999;274(4):2386–2393. doi: 10.1074/jbc.274.4.2386. [DOI] [PubMed] [Google Scholar]
- [30].Rosen ED, Hsu C-H, Wang X, Sakai S, Freeman MW, Gonzalez FJ, et al. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev. 2002;16(1):22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lodhi IJ, Wei X, Yin L, Feng C, Adak S, Abou-Ezzi G, et al. Peroxisomal lipid synthesis regulates inflammation by sustaining neutrophil membrane phospholipid composition and viability. Cell Metab. 2015;21(1):51–64. doi: 10.1016/j.cmet.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Homan EA, Kim Y-G, Cardia JP, Saghatelian A. Monoalkylglycerol ether lipids promote adipogenesis. J Am Chem Soc. 2011;133(14):5178–5181. doi: 10.1021/ja111173c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pessentheiner AR, Pelzmann HJ, Walenta E, Schweiger M, Groschner LN, Graier WF, et al. NAT8L (N-acetyltransferase 8-like) accelerates lipid turnover and increases energy expenditure in brown adipocytes. J Biol Chem. 2013;288(50):36040–36051. doi: 10.1074/jbc.M113.491324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lindroos J, Husa J, Mitterer G, Haschemi A, Rauscher S, Haas R, et al. Human but not mouse adipogenesis is critically dependent on LMO3. Cell Metab. 2013;18(1):62–74. doi: 10.1016/j.cmet.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Iwase T, Tajima A, Sugimoto S, Okuda K, Hironaka I, Kamata Y, et al. A simple assay for measuring catalase activity: a visual approach. Sci Rep. 2013;3 doi: 10.1038/srep03081. 3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226(1):497–509. [PubMed] [Google Scholar]
- [37].Knittelfelder OL, Weberhofer BP, Eichmann TO, Kohlwein SD, Rechberger GN. A versatile ultra-high performance LC-MS method for lipid profiling. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;951–952:119–128. doi: 10.1016/j.jchromb.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hartler J, Trötzmüller M, Chitraju C, Spener F, Köfeler HC, Thallinger GG. Lipid Data Analyzer: unattended identification and quantitation of lipids in LC-MS data. Bioinformatics. 2011;27(4):572–577. doi: 10.1093/bioinformatics/btq699. [DOI] [PubMed] [Google Scholar]
- [39].Siersbæk MS, Loft A, Aagaard MM, Nielsen R, Schmidt SF, Petrovic N, et al. Genome-wide profiling of peroxisome proliferator-activated receptor γ in primary epididymal, inguinal, and brown adipocytes reveals depot-selective binding correlated with gene expression. Mol Cell Biol. 2012;32(17):3452–3463. doi: 10.1128/MCB.00526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22(21):2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bakkeren JA, Monnens LA, Trijbels JM, Maas JM. Serum very long chain fatty acid pattern in Zellweger syndrome. Clin Chim Acta Int J Clin Chem. 1984;138(3):325–331. doi: 10.1016/0009-8981(84)90140-2. [DOI] [PubMed] [Google Scholar]
- [42].Wanders RJ, van Wijland MJ, van Roermund CW, Schutgens RB, van den Bosch H, Tager JM, et al. Prenatal diagnosis of Zellweger syndrome by measurement of very long chain fatty acid (C26:0) beta-oxidation in cultured chorionic villous fibroblasts: implications for early diagnosis of other peroxisomal disorders. Clin Chim Acta Int J Clin Chem. 1987;165(2–3):303–310. doi: 10.1016/0009-8981(87)90175-6. [DOI] [PubMed] [Google Scholar]
- [43].Schutgens RB, Bouman IW, Nijenhuis AA, Wanders RJ, Frumau ME. Profiles of very-long-chain fatty acids in plasma, fibroblasts, and blood cells in Zellweger syndrome, X-linked adrenoleukodystrophy, and rhizomelic chondrodysplasia punctata. Clin Chem. 1993;39(8):1632–1637. [PubMed] [Google Scholar]
- [44].Lodhi IJ, Yin L, Jensen-Urstad APL, Funai K, Coleman T, Baird JH, et al. Inhibiting adipose tissue lipogenesis reprograms thermogenesis and PPARγ activation to decrease diet-induced obesity. Cell Metab. 2012;16(2):189–201. doi: 10.1016/j.cmet.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Di-Poï N, Tan NS, Michalik L, Wahli W, Desvergne B. Antiapoptotic role of PPARbeta in keratinocytes via transcriptional control of the Akt1 signaling pathway. Mol Cell. 2002;10(4):721–733. doi: 10.1016/s1097-2765(02)00646-9. [DOI] [PubMed] [Google Scholar]
- [46].Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev. 1999;20(5):649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- [47].Lei Z, Chen W, Zhang M, Napoli JL. Reduction of all-trans-retinal in the mouse liver peroxisome fraction by the short-chain dehydrogenase/reductase RRD: induction by the PPAR alpha ligand clofibrate. Biochemistry. 2003;42(14):4190–4196. doi: 10.1021/bi026948i. [DOI] [PubMed] [Google Scholar]
- [48].Lodhi IJ, Semenkovich CF. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 2014;19(3):380–392. doi: 10.1016/j.cmet.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hörl G, Wagner A, Cole LK, Malli R, Reicher H, Kotzbeck P, et al. Sequential synthesis and methylation of phosphatidylethanolamine promote lipid droplet biosynthesis and stability in tissue culture and in vivo. J Biol Chem. 2011;286(19):17338–17350. doi: 10.1074/jbc.M111.234534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wójcik C, Lohe K, Kuang C, Xiao Y, Jouni Z, Poels E. Modulation of adipocyte differentiation by omega-3 polyunsaturated fatty acids involves the ubiquitin-protea-some system. J Cell Mol Med. 2014;18(4):590–599. doi: 10.1111/jcmm.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kim H-K, Della-Fera M, Lin J, Baile CA. Docosahexaenoic acid inhibits adipocyte differentiation and induces apoptosis in 3T3-L1 preadipocytes. J Nutr. 2006;136(12):2965–2969. doi: 10.1093/jn/136.12.2965. [DOI] [PubMed] [Google Scholar]
- [52].Honsho M, Tamura S, Shimozawa N, Suzuki Y, Kondo N, Fujiki Y. Mutation in PEX16 is causal in the peroxisome-deficient Zellweger syndrome of complementation group D. Am J Hum Genet. 1998;63(6):1622–1630. doi: 10.1086/302161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Poulos A, Beckman K, Johnson DW, Paton BC, Robinson BS, Sharp P, et al. Very long-chain fatty acids in peroxisomal disease. Adv Exp Med Biol. 1992;318:331–340. doi: 10.1007/978-1-4615-3426-6_30. [DOI] [PubMed] [Google Scholar]
- [54].Abe Y, Honsho M, Nakanishi H, Taguchi R, Fujiki Y. Very-long-chain polyunsaturated fatty acids accumulate in phosphatidylcholine of fibroblasts from patients with Zellweger syndrome and acyl-CoA oxidase1 deficiency. Biochim Biophys Acta. 2014;1841(4):610–619. doi: 10.1016/j.bbalip.2014.01.001. [DOI] [PubMed] [Google Scholar]
- [55].Trijbels JM, Berden JA, Monnens LA, Willems JL, Janssen AJ, Schutgens RB, et al. Biochemical studies in the liver and muscle of patients with Zellweger syndrome. Pediatr Res. 1983;17(6):514–517. doi: 10.1203/00006450-198306000-00018. [DOI] [PubMed] [Google Scholar]
- [56].Baumgart E, Vanhorebeek I, Grabenbauer M, Borgers M, Declercq PE, Fahimi HD, et al. Mitochondrial alterations caused by defective peroxisomal biogenesis in a mouse model for Zellweger syndrome (PEX5 knockout mouse) Am J Pathol. 2001;159(4):1477–1494. doi: 10.1016/S0002-9440(10)62534-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.