Abstract
Cartilage defects can result in pain, disability, and osteoarthritis. Hydrogels providing a chondroregeneration-permissive environment are often mechanically weak and display poor lateral integration into the surrounding cartilage. This study develops a visible-light responsive gelatin ink with enhanced interactions with the native tissue, and potential for intraoperative bioprinting. A dual-functionalized tyramine and methacryloyl gelatin (GelMA-Tyr) is synthesized. Photo-crosslinking of both groups is triggered in a single photoexposure by cell-compatible visible light in presence of tris(2,2'-bipyridyl)dichlororuthenium(II) and sodium persulfate as initiators. Neo-cartilage formation from embedded chondroprogenitor cells is demonstrated in vitro, and the hydrogel is successfully applied as bioink for extrusion-printing. Visible light in situ crosslinking in cartilage defects results in no damage to the surrounding tissue, in contrast to the native chondrocyte death caused by UV light (365–400 nm range), commonly used in biofabrication. Tyramine-binding to proteins in native cartilage leads to a 15-fold increment in the adhesive strength of the bioglue compared to pristine GelMA. Enhanced adhesion is observed also when the ink is extruded as printable filaments into the defect. Visible-light reactive GelMA-Tyr bioinks can act as orthobiologic carriers for in situ cartilage repair, providing a permissive environment for chondrogenesis, and establishing safe lateral integration into chondral defects.
Keywords: biofabrication, bioglue, bioprinting, cartilage tissue engineering, tissue integration
1. Introduction
Articular cartilage defects are a major problem in the orthopedic field, affecting 36% of athletes and 16% of patients that underwent arthroscopic investigation following pain complaints.[1] These cartilage injuries can cause disability in patients, greatly affecting their quality of life and exerting significant impact on overall healthcare costs. When left untreated, cartilage defects can lead to early-onset osteoarthritis and higher risk of knee replacement surgery.[2] Current treatment options are limited and often result in the formation of fibrotic tissue that exhibits lesser quality than native articular cartilage and increased propensity toward degeneration.[3]
In recent years, injectable hydrogels have risen as promising candidates for cartilage repair. This is due to their highly hydrated microenvironment, which mimics the native extracellular matrix (ECM) and allows effective transfer of various solutes and nutrients.[4] Furthermore, these hydrogels normally provide a biocompatible and/or biodegradable structure that enables cell encapsulation and delivery of bioactive molecules to targeted sites for cartilage regeneration.[5] Bioactive hydrogel formulations show promise as clinically translatable new therapies, with two formulations currently undergoing clinical trials to obtain FDA approval: GelrinC (Regentis Biomaterials), a cell-free hydrogel composed of poly(ethylene glycol)-diacrylate (PEG-DA) and denatured fibrinogen, designed to be photo-crosslinked in situ at the site of the defect; and CARTISTEM (Medipost), which is an injectable product composed an allogeneic human umbilical cord blood-derived mesenchymal stem cells mixed in a hyaluronic acid hydrogel and approved for clinical use in South Korea since 2012.[6]
An ideal hydrogel for cartilage regeneration should be able to mimic 3D environment of cartilage ECM, support the development of neo-cartilage matrix, and integrate with the surrounding native tissue. Moreover, articular cartilage in joints has a specific zonal orientation (superficial, middle, deep, and calcified zones), in which cell morphology, biomarker expression profiles, matrix composition, and mechanical properties vary in a depth-dependent fashion.[7,8] Biofabrication technologies, which enable precise control over the spatial deposition of cells and bioactive cues by means of bioprinting and bioassembly, have emerged as promising strategies to recapitulate such in vivo native architectures,[9–14] for instance by the controlled extrusion of hydrogel-based bioinks via layer-by-layer deposition.[16] Importantly, via controlling the architecture of a printed construct, biofabrication approaches hold great promise to capture salient functions of living tissues and guide the maturation of engineered constructs.[16] In recent years, advancements in the field have led to intraoperative biofabrication, where cell-laden bioinks can be extruded or printed in situ in a surgical setting.[17,18] For example, PEG hydrogels containing chondrocytes have been directly ink-jet printed into the cartilage defect of an osteochondral plug model ex vivo.[19] Similarly, a handheld extrusion printer has been developed to simultaneously deliver mesenchymal stromal cells (MSCs) and gelatin-methacryloyl (GelMA) hydrogels into chondral defects in a single-session surgery.[17] Although promising, both studies demonstrated that the integration of the printed constructs to the native host tissue remained a significant issue,[20] and in general, the in vivo stability and integration of tissue engineered constructs remains a major challenge in the field of cartilage regeneration.[20–22] Importantly, while stable axial integration can be achieved for treatment strategies targeting osteochondral defect repair, i.e., through anchoring in the subchondral bone,[20] to achieve a reliable lateral integration within the chondral region is a particularly daunting task. This type of integration is especially challenging when using hydrogels or bioinks that display optimal cell embedding properties and thus low mechanical properties. This is particularly relevant since cells thrive best in hydrogels with low crosslinking density, and stiffer bioinks have bene demonstrated to limit neo-cartilage deposition.[23] Overcoming the limitation imposed by integrating soft hydrogels in situ particularly relevant in the treatment of joint diseases, as poor interconnection between engineered repair tissue and the surrounding native cartilage is a key cause of failure upon cyclic compressive and shear loading exhibited in healthy synovial joints.[24] Finally, there is also evidence that chondrocyte activity and matrix production and remodeling at a biomaterial–host tissue interface have an effect on lateral integration of the hydrogel.[24] Therefore, cell viability in and around the hydrogel is deemed vital for long-term success.
Another important aspect concerning implant integration when injecting or printing hydrogels in situ is the selection of an appropriate crosslinking mechanism for the material. This can impact the viability of the cells encapsulated within it and the interaction with the surrounding native tissue. Ideally, the crosslinking process should be simple, fast, and safe in a clinical setting. Photo-crosslinkable hydrogels can generally be formed rapidly (within seconds to minutes) and on demand, upon exposure to various light sources in the presence of the appropriate photoinitiators. Moreover, light-based reactions offer facile and accurate spatiotemporal control over the physicochemical properties of hydrogels.[25] As a consequence, these are often common steps in many bioprinting workflows to stabilize printed bioinks and improve the shape fidelity of biofabricated constructs.[9] Thus, engineer such photochemistry can open new possibilities to introduce new functionalities into bioinks.
A commonly used crosslinking mechanism for most photocrosslinkable hydrogels is based on free-radical chain-growth and step-growth polymerization,[26,27] often initiated upon exposure to UV or visible light depending on the selected initiators, such as hydroxy-1-[4-(2-hydroxyethoxy)phenyl]-2-methyl-1-propanone (Irgacure 2959) and lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP). There are however major considerations of using UV light in situ, such as the potential damage to cells in the surrounding native tissue, which may impair the restoration of healthy cellular functionality.
The aim of this study was to develop a hydrogel-based cartilage bioink compatible with in situ delivery that could act as a cell carrier, as well as an adhesive gel that is able to bind to cartilage. Importantly, the activation of these multiple functionalities could be achieved in a single step via a biocompatible one-step photocrosslinking reaction, a process that is compatible with a safe application on native living tissues. GelMA was used as a platform for further modification due to its ability to provide a permissive environment for cell growth and its well-demonstrated applicability for 3D bioprinting applications.[15,28] Herein, GelMA was further derivatized with tyramine moieties (GelMA-Tyr) that can establish covalent bonds with tyrosine residues present in the extracellular matrix of native tissues. Moreover, a visible light photoinitiating system, based on tris(2,2’-bipyridyl)ruthenium(II) chloride and sodium persulfate (Ru/SPS),[29] was employed to assess its potential to initiate both the methacryloyl chain-growth polymerization and dityramine bond formation in a single step. This multifunctional, biofunctionalized hydrogel was characterized as a 3D culture environment for cartilage repair, a bioink for bioprinting and as bioglue for binding to the native cartilage.
2. Results and Discussion
2.1. Synthesis of GelMA-Tyr
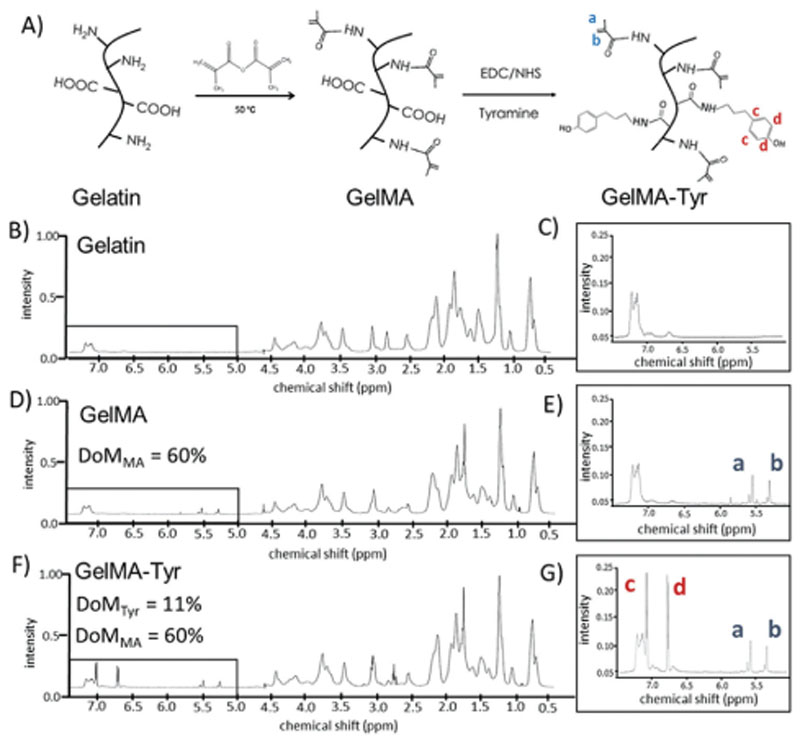
To produce a hydrogel system carrying two photoresponsive functionalities, GelMA-Tyr was successfully synthesized in two steps as shown by the 1H-NMR spectra in Figure 1, and a schematic of the functionalization step is reported in Figure 1A. For GelMA, the degree of modification (DoM) is defined as the percentage of modified lysine groups that are able to react with methacrylic anhydride.[30] Although methacrylic anhydride can also react with carboxyl and hydroxyl groups, it was previously reported that the primary amine groups have highest reaction affinity with the anhydrides.[30] When comparing the 1H-NMR spectra of GelMA to that of gelatin (Figure 1B–E), the presence of proton peaks corresponding to the MA groups can be clearly observed at δ = 2.5–2.6 ppm. The DoM of methacryloylation (DoMMA) is quantified to be 60%, and is in agreement with previously published studies following similar synthesis protocols.[24,31,32] The synthesized GelMA was subsequently grafted with tyramine groups onto the GelMA backbone. In this study, a simple carboxyl-amine coupling reaction was employed where Tyr groups were conjugated via their primary amine to the carboxyl moieties of GelMA. 1H-NMR characterization showed that the reaction was successful as indicated by the presence of extra proton peaks assigned to the Tyr groups at δ = 6.8–7.2 ppm.[33,34] In the context of the tyramination reaction, the DoMTyr was defined as the percentage of modified amino acids containing carboxyl groups (glutamic acid and aspartic acid), and was calculated to be 11% (Figure 1F).[35] Previous reports have demonstrated the derivation of Tyr groups onto pristine gelatin, applying a different carboxyl-amine coupling route, in which the Bolton–Hunter reagent (N-succinimidyl-3-4-hydroxypropionate) was used as the Tyr carrier.[36] This reaction however, targets the accessible primary amines (lysine) of gelatin, and is less suitable in our targeted dual functionalization approach, given that the initial methacryloylation reaction also reacts with the lysine moieties. Furthermore, the carboxyl-targeting tyramination reaction was demonstrated to not affect the methacryloyl groups that were already conjugated onto the gelatin backbone, where the DoMMA remained as 60% for GelMA-Tyr. This is particularly important, as the MA groups are known to be highly functional and might react with the reactants and byproducts generated from the tyramination reaction. The DoMTyr (11%) was deliberately aimed to be lower than DoMMA (60%) by taking into consideration that gelatin already possesses 0.5 mol% native tyrosine groups.[37] Collectively, these results confirm that it is possible to functionalize gelatin in a dual-step reaction, firstly with MA groups and then by subsequent Tyr moieties, where both the chemical modification processes targeted different grafting sites on gelatin, and hence were compatible with each other. Importantly, this allowed for controlled experiments to be accurately performed to study the application of the dual functionalization in the context of biofabrication.
Figure 1. Functionalization process to obtain the dual responsive hydrogels.
A) Schematic representation of the synthesis of GelMA and GelMA-Tyr; representative 1H-NMR spectra of gelatin, GelMA and GelMA-Tyr. B–G) 1H-NMR spectra of the functionalized hydrogels, showing the presence of the characteristic peaks for acrylates (a,b) in the GelMA and GelMA-Tyr groups and for the added phenolic groups in the GelMA-Tyr polymers.
2.2. Physicochemical and Mechanical Characterization of GelMA-Tyr Hydrogels
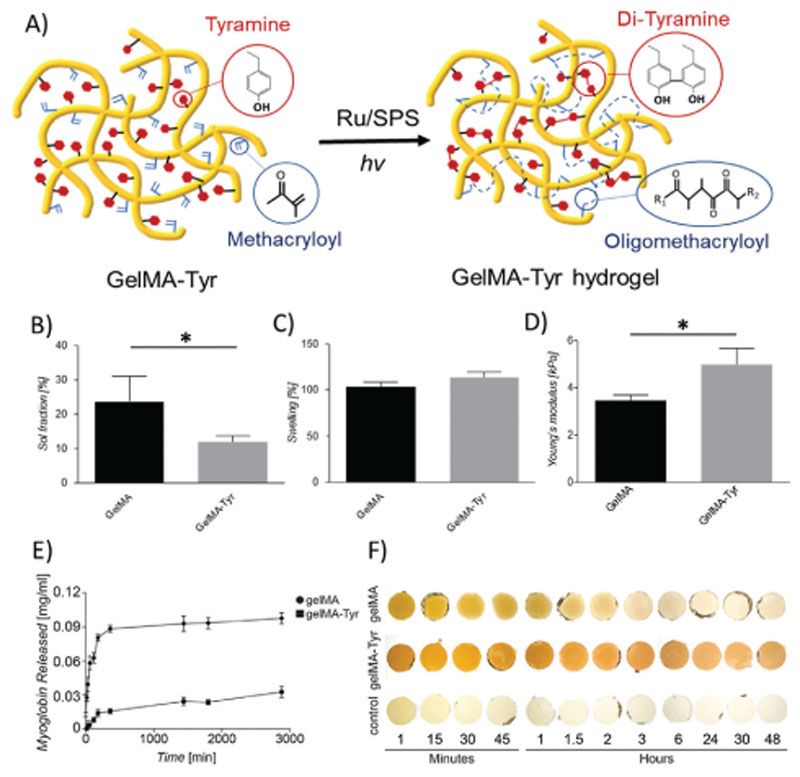
The synthesized GelMA-Tyr macromer was successfully fabricated into hydrogels using the visible light mediated Ru/SPS system (Figure 2A), and was further compared to GelMA hydrogel controls, a well-studied system in biofabrication and common choice in the field of cartilage tissue regeneration.[12,22,38] Optimization of the photoinitiator content to minimize the sol fraction of the fabricated GelMA-Tyr hydrogels led to selection of 0.5 × 10−3 M Ru and5 × 10−3 M SPS as the optimal photoinitiator concentration. Mass loss and swelling studies demonstrated that GelMA-Tyr hydrogels had significantly lower sol fraction value (11.9%, p < 0.05) than GelMA gels (23.6%, Figure 2B). Ru is a transition metal complex and has been characterized to be highly absorptive in the visible light range (ϵ ≈ 14 600 M–1 cm–1 at 450 nm). When irradiated with visible light, Ru2+ becomes photoexcited and then oxidizes into Ru3+, primarily by donating electrons to SPS. After accepting electrons, SPS dissociates into sulfate anions and sulfate radicals. For GelMA, these generated sulfate radicals trigger the chain-growth polymerization of the MA groups, forming nondegradable oligomethacryloyl kinetic chains that crosslink the network together.[24,39] Similarly, these sulfate radicals have been previously shown to also facilitate step-growth thiol-ene polymerization of gelatin-based hydrogels.[31] However, for GelMA-Tyr, it was hypothesized that in addition to the MA chain-growth polymerization, the photo-oxidized Ru3+ could also abstract electrons from the grafted Tyr groups, forming tyrosyl radicals that eventually establish covalent di-tyramine bonds with nearby Tyrmoieties.[36,40,41] This photomediated di-tyramine crosslinking is well characterized and has been previously to fabricate protein hydrogels such as those based on resilin, fibrinogen, and silk.[42–44] Therefore, it is expected that the GelMA-Tyr hydrogels have a higher crosslinking density as the hydrogel network consists of crosslinks in the form of both oligomethacryloyl kinetic chains and di-tyramine bonds (Figure 2A). Interestingly, both the GelMA and GelMA-Tyr gels showed similar swelling behavior (Figure 2C), where the additional di-tyramine crosslinks did not affect the overall water uptake capacity of the GelMA-Tyr hydrogels (Figure S1, Supporting Information). The GelMA-Tyr hydrogels also exhibited a significantly higher mechanical compressive modulus (4.98 kPa) compared to GelMA (3.46 kPa, Figure 2D), which is in agreement with previous studies where hydrogels of higher crosslinking density possess higher mechanical stiffness.[45,46] Moreover, both GelMA and GelMA-Tyr macromers maintained the ability, typical of their common precursor gelatin, to form thermosensitive gels upon cooling at 4 °C, which is then stabilized via photo-crosslinking (Figure S2, Supporting Information). Interestingly, the addition of the tyramine moieties appeared not to modify the susceptibility of the GelMA hydrogel to enzymatic degradation, in presence of collagenase, suggesting that cell mediated remodeling of the gel over time is possible (Figure S3, Supporting Information).
Figure 2. Fabrication and physicochemical and mechanical characterization of the one-step inducible, dual crosslinked hydrogels.
A) Schematic representation of the crosslinking process of the GelMA-Tyr macromer. B) Sol fraction values and C) swelling behavior of the hydrogels as observed via sol–gel analysis; D) compressive young’s modulus of the casted hydrogels. Incorporation of myoglobin into GelMA or GelMA-Tyr hydrogels: E) release profile of myoglobin from GelMA or GelMA-Tyr hydrogels over 3000 min; F) macroscopic images of GelMA and GelMA-Tyr hydrogels incorporated with myoglobin over 48 h.
In addition, Ru-mediated di-tyramine and di-tyrosine crosslinking can be used to induce gelation in pristine proteins (Figure S4, Supporting Information). As such, Tyr groups on the dual functionalized GelMA-Tyr system can bind to any phenolic moieties on proteins (i.e., endogenous tyrosine and tryptophan residues), with potential implications to establish controlled release systems. Hence, the ability of GelMA-Tyr to further covalently immobilize proteins within the hydrogel network was evaluated using myoglobin as a model compound, which is rich in tyrosine groups and exhibits a distinct UV–vis absorbance spectrum, facilitating its detection without the need for further modification. It was shown that in GelMA hydrogels, the incorporated myoglobin displayed a burst release profile where approximately all the myoglobin had leached out of the network within the first 4 h of incubation in phosphate buffered saline (PBS). In contrast, the GelMA-Tyr hydrogels showed a significantly higher retention of myoglobin, where 70% of the initially incorporated protein remained within the hydrogel network after 48 h of incubation (Figure 2E). This result was further confirmed with the macroscopic images, where the dark brown color of myoglobin was retained within the GelMA-Tyr hydrogels throughout the 48 h period (Figure 2F). In comparison, the GelMA gels showed a shift in color from dark brown to clear over the incubation period, as a consequence of the rapid release profile of myoglobin (Figure 2F). Importantly, previous research has demonstrated the incorporation of proteins (lysozyme and α-amylase) into hydrogels via similar di-tyrosine/tyramine crosslinking, and showed that upon release, the functionality of these proteins was preserved. These observations suggest that the linked proteins did not undergo denaturation,[47a] a situation that would facilitate the translation of this light-based system for controlled delivery of bioactive agents. Furthermore, this process is versatile, and can be applied to crosslink or form hydrogels from a wide array of pristine, tyrosine-carrying proteins. To our knowledge, the Ru/SPS system has been applied separately to either crosslink methacryloylated or tyraminated hydrogels biomaterials.[31,32,36,47a,b] Importantly, in this study, this visible light mediated Ru/SPS system was utilized for the first time to crosslink a dual functionalized polymer/macromer, where both the MA chain-growth polymerization and di-tyramine crosslinking occurred concurrently in a single hydrogel system in one photoexposure step. This phenomenon is a unique advantage of using this Ru/SPS system over other conventional photoinitiating systems such as I2959 or LAP.
2.3. Assessment of In Vitro Chondrogenesis
Given that GelMA-Tyr hydrogels were designed for cell delivery to chondral defects, articular chondroprogenitor cells (ACPCs) were encapsulated into the one-step, dual crosslinked gels, where GelMA hydrogels served as a control. Both the GelMA and GelMA-Tyr hydrogels supported high cell viability (>70%) at 1 and 7 days postfabrication (Figure 3A). This result is in accordance to previously published studies where the Ru/SPS concentration (0.5 × 10−3/5 × 10−3 M) used in this study is not toxic to cells,[31,32] as well as to previous work with other lightand enzymatic-activated tyraminated polymers, suggesting no adverse cytotoxicity risk due to undesired interaction with the cell membrane proteins.[49,50] In order to evaluate the functionality of the encapsulated ACPCs, the cell-laden hydrogels were further cultured in chondrogenic differentiation medium for 28 days. It was observed that the cells embedded within the GelMA-Tyr hydrogels offered a permissive environment for the chondrogenic differentiation of ACPCs, as indicated by a significantly higher GAG/DNA value (120 μg μg –1) after 28 days in culture compared to GelMA (52 μg μg –1) (Figure 3B). Surprisingly, the compressive modulus of the cell-laden GelMA hydrogels was significantly higher (28 kPa) than the GelMA-Tyr samples (10 kPa), even though both materials showed similar mechanical properties at the beginning of the culture period (Figure S5, Supporting Information). While the mechanical properties of both gels are relatively low for applications in large tissue defects subjected to continuous mechanical loads, several reinforcing strategies based on the combination or coprinting with stiff thermoplastic support scaffolds have already been reported in the literature to address this common limitation for bioinks.[51–54] Further analysis into the gene expression over the 28 day culture period showed that both collagen type II and type I expression was upregulated in the GelMA and GelMA-Tyr hydrogels (Figure 3D,E). It was also observed that the collagen type II expression is significantly higher in the GelMA-Tyr samples (0.8-fold) after 28 days as compared to GelMA (0.3-fold). There was no significant difference in terms of the collagen type I expression for both 1 and 28 days in the two sample groups. Collagen type II is a well-known marker of native hyaline cartilage while collagen type I is expressed prevalently in fibrocartilage.[55] Therefore, an higher value for the ratio between the expression levels of collagen type II and type I is generally indicative of a chondrogenic phenotype,[55–57] and this indicator was found significantly higher in GelMA-Tyr samples (Figure 3E), albeit still lower than 1. A further analysis on the expression of the superficial zone marker PRG4, a key factor in joint lubrication,[12,58] revealed a higher expression (4-fold) in the GelMA-Tyr samples compared to GelMA (2-fold) after 1 day, but showed a downregulation after 28 days in culture (Figure 3F). Immunohistological analysis confirmed the results of the in vitro biochemical assays, where deposition of glycosaminoglycans (GAGs) (Figure 3G,J), collagen type II (Figure 3H,K) and collagen type I (Figure 3I,L) was observed. Interestingly, albeit collagen I gene expression was relatively high in all samples, at a protein level as detected by immunohistochemistry, this molecule appeared to be less densely present in the neo-cartilage matrix compared to collagen type II, with a higher distribution of collagen type II over collagen type I in both GelMA (5.93 ± 0.95-fold) and GelMA-Tyr (2.27 ± 1.38-fold) (Figure S6, Supporting Information), suggesting the differentiation of ACPCs toward a hyaline cartilage-forming phenotype. Overall, quantification from histological sections provides measurements on the distribution of a given ECM component, in terms of area covered. However, it should be noted that unlike the performed biochemical assays, such measurements do not provide a quantitative measurement of the amounts of produced GAGs or collagens. In the analyzed slides GAG and collagen type II distribution was found to be higher in the GelMA group compared to the GelMA-Tyr group (5.09-fold for the GAGs and 2.07-fold for collagen type II). No significant difference was found for collagen type I in terms of area coverage (Figure S6, Supporting Information). Such results seem to suggest that the extra di-tyramine crosslinking provided by the tyramine moieties at the specific polymer concentration and degree of functionalization tested in this study, on top of that provided by the oligomethacryloyl kinetic chain, may limit the diffusion of neo-secreted ECM moieties. Overall, such inhomogeneous distribution of the neo-synthesized ECM components in the GelMA-Tyr group, which appeared to accumulate prevalently in the pericellular space, may be responsible for the limited stiffening of these hydrogels over the culture time. GelMA hydrogels have been extensively studied as matrices for cartilage regeneration, where cartilage-relevant cells such as articular chondrocytes, nasal chondrocytes, and chondroprogenitor cells have been encapsulated within GelMA gels and showed good chondrogenic differentiation.[59–62a] In this study, the focus is instead placed on the extra Tyr groups grafted onto GelMA, and the influence of formation of di-tyramine bonds during the crosslinking reaction on cellular chondrogenic behavior. Initially, it was hypothesized that both GelMA and GelMA-Tyr hydrogels should support similar levels of cartilage regeneration given that the initial swelling behavior (≈100%) and mechanical properties (4–6 kPa) were similar. Surprisingly, considerable differences between both the sample groups were observed, where although GelMA-Tyr facilitated chondrogenic differentiation of the embedded ACPCs, based on the indication of quantitative markers such as shown by higher GAG/DNA value and collagen type II expression after 28 days, the immunohistological data showed inhomogenous distribution of the neo-cartilage matrix when compared to GelMA. It can be hypothesized that the GelMA-Tyr hydrogels have higher crosslinking density due to the formation of both oligomethacryloyl kinetic chains and di-tyramine crosslinks, which could hinder the diffusion of neo-matrix. In addition, high crosslinking densities have been previously suggested to impact cellular functions such as mitosis and differentiation,[62b–d] and previous studies have shown that introduction of secondary covalent crosslinking into hydrogels inhibited the spreading and differentiation of encapsulated mesenchymal stromal cells.[62c] In this study, the DoMMA was kept constant at 60% for both GelMA and GelMA-Tyr. We anticipate that by reducing the DoMMA for GelMA-Tyr, hydrogels of similar crosslinking density to GelMA can be fabricated, which could be applied to avoid potential drawbacks given by the degree of crosslinking density. Future studies will also focus on covalently incorporating chondrogenic supporting biomolecules into these gels to further enhance the differentiation of the encapsulated cells.[61] Finally, as the tyramination reaction targets carboxyl containing amino acids such as aspartic and glutamic acid, which are respectively a key component of the cell-attachment RGD sequence (arginine– glycine–aspartic acid) and of the collagen-specific GFOGER sequence, it is speculated that the GelMA-Tyr hydrogels possibly possess less cell-adhesive motifs. This reduction in cell-adhesive sites might indeed contribute to the chondrogenic capacity as indicated in previous studies where less spreading area and lower RGD domain density promote higher extent of chondrogenic differentiation.[63] Accordingly, other studies also indicated that conjugating RGD sequences onto alginate or hyaluronan hydrogels inhibited in vitro chondrogenesis.[64,65]
Figure 3. Chondrogenic differentiation of ACPCs encapsulated within GelMA or GelMA-Tyr hydrogels.
A) Cell viability after 1 and 7 days in culture. B) GAG/DNA and C) Young’s modulus of ACPC-laden GelMA or GelMA-Tyr gels after 1 and 28 days in culture. D) Collagen type II, E) Collagen type I, and F) PRG4 gene expression of ACPC-laden GelMA or GelMA-Tyr gels after 1 and 28 days in culture. Histological stainings of ACPC-laden GelMA or GelMA-Tyr gels after 28 days, G,J) safranin-O, H,K) collagen type II, scale bar = 20 μm, and I,L) collagen type 1, scale bar = 40 μm.
2.4. Bioprinting of GelMA-Tyr Hydrogels
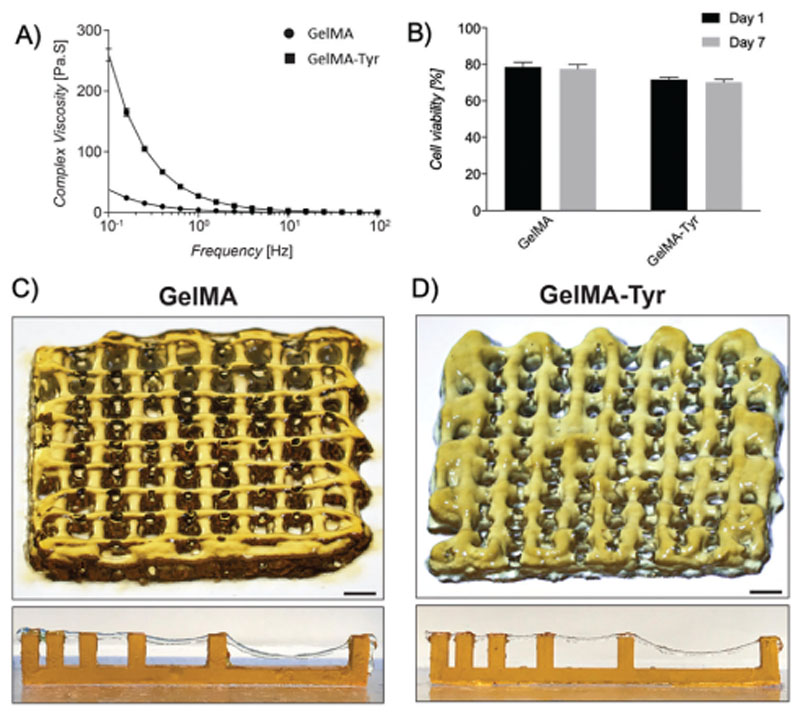
The suitability of GelMA-Tyr as a bioink for extrusion 3D bioprinting was further evaluated. While GelMA has been extensively characterized as a bioink for extrusion bioprinting,[39,66] in this study, we evaluated what effect grafting of Tyr groups onto GelMA had on the printability of the resulting bioink. It was observed that although both materials have shear-thinning properties, GelMA-Tyr displayed a higher complex viscosity compared to GelMA at low shear rates (10−1 to 101 Hz, Figure 4A). This might be due to the conjugated Tyr groups enhancing the overall hydrophobicity of the macromer solution, which results in a higher solution viscosity. It is well documented that hydrophobic effect is an important phenomenon that drives the interaction between proteins which stabilizes the protein conformation.[67] Hence, it is logical that GelMA-Tyr, which is more hydrophobic thanGelMA, has an increased interaction between the macromer chains and thus requires a higher yield stress to facilitate extrusion of the material. Importantly, ACPCs were able to withstand the shear stress during extrusion where cell-laden GelMA or GelMA-Tyr bionks showed high cell viability (>70%, Figure 4B) and sustained metabolic activity (Figure S7, Supporting Information) 1 and 7 days postprinting, demonstrating the suitability of the dual functionalized bioinks for applications in such a bioprinting approach. The diameters of printed hydrogel filaments made of GelMA and GelMA-Tyr were comparable and ranged between about 677 ± 87 μm (highest value observed in GelMA prints) to 280 ± 29 μm (lowest value as observed in GelMATyr prints), when increasing the velocity of the collector plate from3 to 21mms–1 (Figure S8, Supporting Information). Moreover, both GelMA and GelMA-Tyr were able to be extruded into 3D structures which shape was rapidly stabilized by exposure to 450 nm light (Figure 4C,D). Even so, occasional and undesired occlusion of the printed pores could be observed (Figure 4D), possibly due to relaxation of the extruded hydrogel prior to stabilization by photo-crosslinking. Printing resolution could be further improved, for instance through the use of nozzles with finer gauge, as shown via printing grids of GelMA and GelMA-Tyr using a 27G nozzle (Figure S9, Supporting Information), although such choice should be weighed carefully, as recent reports indicated potential detrimental effects in terms of chondrogenic potential of stressed cells sheared through needles with smaller diameters.[68] Finally, the filament collapse test further showed that both GelMA and GelMA-Tyr filaments could easily span over supporting pillars placed at different distances, even bridging 16mmgaps, with no noticeable difference between both bioinks.
Figure 4. Extrusion bioprinting of GelMA-Tyr bioinks.
A) Rheological profile of GelMA and GelMA-Tyr in response to shear rate. B) Cell viability of ACPCs bioprinted within GelMA and GelMA-Tyr constructs. Macroscopic images of extrusion bioprinted C) GelMA and D) GelMA-Tyr constructs, together with a representative image of the filament collapse assay, showing the ability of the printed struts to bridge gaps of 4 mm without noticeable deformation, and up to 16 mm while experiencing sagging. Scale bar = 1 mm.
2.5. Interaction and Integration with Native Cartilage
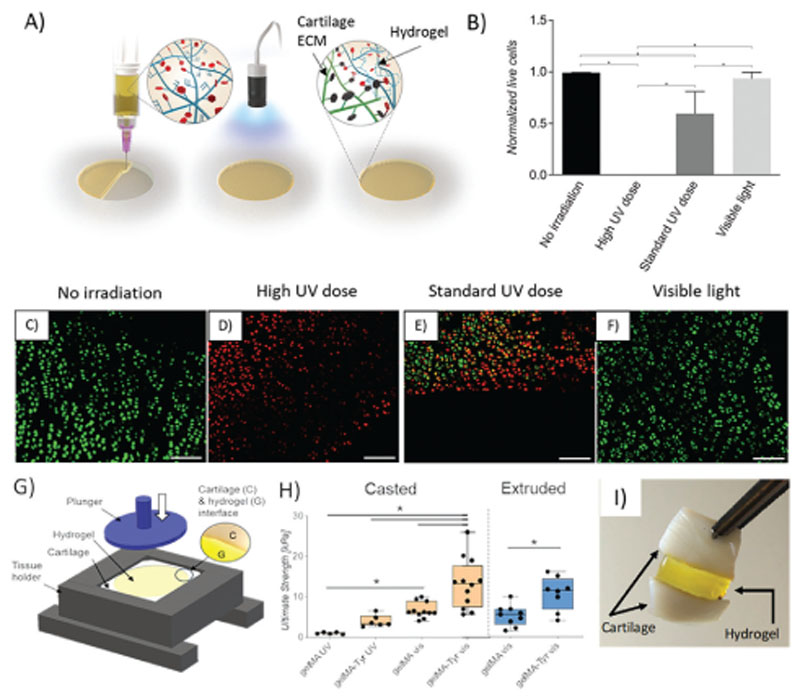
An exciting translational opportunity for photo-crosslinkable hydrogels is their application intraoperatively, either via minimally invasive injections or through advanced in situ bioprinting approaches.[13,69–71] For this purpose, a carrier system in which the cartilage bioink consisting of GelMA-Tyr and cells can be extruded directly into the chondral defect, followed by photoirradiation to crosslink the hydrogel network is envisioned, while, at the same time, exploiting the applied photochemistry to improve lateral integration into the cartilage region (Figure 5A). Such an approach could allow both the delivery of cells to the defect and provide an enhanced integration with the surrounding native cartilage. In terms of photo-crosslinking, most studies have shown that cells can be encapsulated into hydrogels using either UV or visible light irradiation and remain viable and functional after encapsulation.[38,72] For example, UV-A and near UV wavelengths are known to generate reactive oxygen species and free radicals that can indirectly damage DNA.[73,74] In this context, hydrogels with gelation chemistries based on radical initiation have been shown to consume such potentially harmful radicals, thus protecting the embedded cells, and allowing to identify safer crosslinking windows.[38,75] However, in a clinical setting, it can be challenging to confine such photo-crosslinking reactions exclusively to the defect volume. Thus, neighboring healthy tissues, which are not enclosed in such protective hydrogels, may be harmed by harsh light sources. These effects can be more evident for gels based on methacryloyl chemistry (or more broadly on chain growth polymerization) when using photoinitiators which require considerably high irradiation dosages to overcome oxygen inhibition, given by the inherent difficulty of limiting oxygen concentration in an in vivo, intraoperative setting. First, the effect of using UV light or visible light on the surrounding cartilage tissue was thus evaluated. LIVE/DEAD images showed that irradiation of cartilage tissue with UV had a dose-dependent detrimental effect on cell viability, whereas high intensity of visible light irradiation had no effect on cell survival within the native cartilage (Figure 5B–F). It was further observed that a high UV irradiation dosage (36 000 mJ cm–2), used as negative control, resulted in 100% chondrocyte death in the proximity of the exposed defect, whereas 40% cell-death was observed in samples irradiated with standard UV dosage (1800 mJ cm–2) typically used to crosslink GelMA (with I2959) hydrogels in a normoxic environment.[76,77] On the other hand, the percentage of normalized live cells in samples irradiated with visible light (14 400 mJ cm–2) was not statistically different to that of cartilage biopsies that were not photoexposed (Figure 5B). This was also the case for samples crosslinked with the type I initiator LAP 0.1% w/v, able to initiate the acryloyl-based chain polymerization, upon exposure to a 405 nm light source (Figure S10, Supporting Information). In the field of cartilage engineering and bioprinting, GelMA-based bioinks have been extensively studied to encapsulate cartilage relevant cells (chondrocytes, chondroprogenitors, and MSCs), and have been extensively fabricated using the UV and I2959 system. Although several reports have suggested that such UV-based system is not detrimental to embedded cells during the encapsulation process,[39] the results presented here showed that UV irradiation can be harmful on the healthy, native tissue surrounding the gel-filled cartilage defects in vivo. Hence, the visible light-mediated crosslinking system may be more clinically relevant especially if the cell-laden hydrogels are to be administered intraoperatively.
Figure 5.
A) Schematic of intraoperative administration of GelMA-Tyr to the chondral defect. B) Normalized amount of live cells and C–F) LIVE/DEAD images of cartilage biopsies irradiated with UV or visible light. Scale bar = 100 μm. G) Setup of the pushout assay to determine bond-strength. H) Bond-strength of GelMA or GelMA-Tyr administered to the cartilage biopsies as a solution or physically crosslinked gel. I) Cartilage biopsies adhered together using GelMA-Tyr.
A key challenge for the in situ application of cell-laden hydrogels is that of their retention at the target site and the integration within the native tissue. Herein, the adhesion strength of the cell-laden cartilage bioink to the native tissue was evaluated using a custom-made push-out apparatus (Figure 5G), to assess whether the one-step crosslinking of the dual functionalized GelMA-Tyr could endow the bioinks with tissue-adhesive properties. A simple way to deliver hydrogels into a defect site could be that of casting a prepolymer solution. However, the rapid development of bioprinting technologies has opened new opportunities to print hydrogel-based 3D structures, and pattern multiple cell types for instance, with the goal of recreating the zonal organization of native cartilage.[12,22] In the context of zonal cartilage regeneration, gelatin bioinks can be initially dispensed in their solution form when using inkjet printing. On the other hand, with the most widely used extrusion-based bioprinting approaches, most gelatin-based bioinks are delivered at low temperature, where the gelatin is first allowed to physically crosslink through hydrogen bonding, then dispensed taking advantage of its the shear-thinning properties.[78] To model both situations, the bond strength between the resulting hydrogel and the surrounding tissue was evaluated for GelMA and GelMA-Tyr macromers administered to chondral defects created on cartilage explants, either via casting or via extrusion through a nozzle, followed by in situ photoirradiation. When cast, the GelMA-Tyr samples exhibited significantly higher bond strength (13.25 kPa) compared to GelMA (6.7 kPa) upon exposure to visible light, indicating enhanced integration with the surrounding native cartilage (Figure 5H). Bond strength was significantly reduced (≈15fold vs GelMA-Tyr with visible light) in hydrogels crosslinked using a UV 365 nm light source and I2959 photoinitiator. The type-I I2959 photoinitiator was specifically used as a control and it cannot efficiently induce the formation of di-tyramine bonds. Furthermore, when the macromers were extruded as filaments into the defect through a nozzle, the integration of GelMA-Tyr to the native cartilage tissue remained significantly higher compared to GelMA, exhibiting bond strengths of 10.41 ± 4.04 kPa and 5.10 ± 2.51 kPa, respectively (p = 0.0049) (Figure 5H). The adhesive capacity of GelMA-Tyr crosslinked with visible light was also visually highlighted by applying a patch of gel as a glue to link two cartilage biopsies together (Figure 5I). Together, these results indicate that the introduction of Tyr groups to GelMA enhanced binding to the surrounding native cartilage through the di-tyramine crosslinking as hypothesized. Gitten et al. previously reported on using chitosan-based hydrogels crosslinked using genipin or rose bengal to improve the binding between the hydrogel and the cartilage interface.[79] However, such an approach required enzymatic degradation of the cartilage tissue to expose collagen fibers in order to increase the available crosslinking sites at the hydrogel-cartilage interface. Similarly, Broguiere et al. developed factor XIII/transglutaminase crosslinked hyaluronan hydrogels (HA-TG), which showed good stability and adhesiveness to the native cartilage, but also required enzymatic digestion to expose crosslinking sites from the cartilage matrix to achieve a bond strength of 6 kPa, a value 3-times higher than what found for fibrin glue, a standard fixator in cartilage treatments such as autologous chondrocyte implantation,[80] yet 2.5 times lower than what found for Ru/SPS crosslinked GelMA-Tyr. In this study, we showed that the visible light-mediated crosslinking of GelMA-Tyr can be administered to chondral defects safely, without the use of detrimental UV light sources or requiring any modification of the host tissue, i.e., via enzymatic digestion strategies, whereby the resultant GelMA-Tyr hydrogels were still able to bind strongly to the surrounding native cartilage.
3. Conclusion
Visible light crosslinkable GelMA-Tyr hydrogels, in combination with Ru/SPS photoinitiators, have potential for the in situ repair of cartilage defects. In particular, thanks to the dual crosslinking mechanism triggered by Ru/SPS, this hydrogel demonstrates potential for direct injection and integration into damaged cartilage. It is also suitable for bioprinting applications, further enhancing its possibilities for the repair or replacement of complex, patientspecific structures. This versatile system also enables grafting of unmodified proteins onto the bioink backbone through a one-step photoexposure process, potentially enabling the applicability of this ink for controlled protein release and for further applications in biofunctionalization. Overall, the combination of Ru/SPS-mediated visible light crosslinking and dual functionalized (bio)inks could be expanded to a wide range of biocompatible materials and has potential for intraoperative bioprinting applications.
4. Experimental Section
Synthesis of GelMA and GelMA-Tyr
All materials were obtained from Sigma-Aldrich and used without further modification, unless stated otherwise. GelMA was synthesized by adding 0.6 g of methacrylic anhydride per gram of gelatin (type A, from porcine skin, 10 w/v% in PBS), and left to react for 1 h at 50 °C under constant stirring, as previously described.[81] The resultant GelMA solution was then dialyzed against deionized water at 40 °C to remove unreacted methacrylic anhydride and byproducts. For the synthesis of GelMA-Tyr and tyramine-modified gelatin (Gel-Tyr, used as control for the dual functionalization process), tyramine groups were coupled to the carboxyl groups of GelMA (or pristine gelatin) using carbodiimide chemistry. GelMA (10% w/v in (2-(N-morpholino)ethanesulfonic acid), MES buffer) was reacted with 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC, 1.5 × 10−3 mol L–1) and N-hydroxysuccinimide (NHS, 0.75 × 10−3 mol L–1) for 15min at 40 °C, followed by addition of tyramine (36.5 × 10−3 mol L–1), then left to react for another 24 h at 40 °C with gentle stirring. The resultant GelMA-Tyr solution was then dialyzed against deionized water at 40 °C. All purified macromer solutions were sterile filtered (0.22 μm), freeze-dried, and stored at 4 °C.
Nuclear Magnetic Resonance
The DoM was quantified using 1H-proton nuclear magnetic resonance (NMR; Bruker Avance 400 MHz). GelMA, Gel-Tyr, or GelMA-Tyr were dissolved in deuterium oxide (D2O) and analyzed using 1H NMR (300 MHz Bruker Advance DPX-300 spectrometer). For GelMA, DOMMA is defined as the percentage of modified amino acid groups containing primary amines that reacted with methacrylic anhydride. The area of methacryloyl (MA) proton peaks, δ = 2.5–2.6 ppm, was compared to the area of the phenylalanine protons in the gelatin backbone. For Gel-Tyr and GelMA-Tyr, DoM with tyramine (DoMTyr) is defined as the percentage of modified amino acids that contain carboxylic groups. The area of the tyramine (Tyr) proton peaks, δ = 6.8–7.2 ppm, was compared to the peaks corresponding to the phenylalanine groups. The composition of acid-treated porcine skin gelatin was acquired from the gelatin handbook of the gelatin manufacturer institute of America (GMIA).[82] The phenylalanine peaks at δ = 7.2–7.4 ppm were used as reference signals for both the methacryloylation and tyramination, and were calibrated to 10.5 protons as this amino acid’s content in the used gelatin is 2.1%.
Preparation and Characterization of Casted Hydrogel
Lyophilized GelMA and GelMA-Tyr were dissolved at 8% w/v in PBS at 37 °C. Photoinitiators (Ru and SPS) were added to the different macromer solutions at a 0.5 × 10−3/5 × 10−3 M final concentration. Hydrogel discs (6 mm in diameter × 2 mm in height) were created by crosslinking in open air using a visible light lamp for 8 min at 30 mW cm–2. Sol and gel fractions as well as swelling studies were performed as previously described (n = 6).[83] An unconfined uniaxial compression test was performed by applying –20% min–1 strain rate with a dynamic mechanical analyzer (DMA Q800, TA Instruments, The Netherlands). The compression modulus was calculated as the slope of the stress/strain curve in the 10–15% strain range. The degradability of both GelMA and GelMA-Tyr hydrogels was assessed via incubation in a 0.15% w/v solution of collagenase type II in Dulbecco’s modified Eagle medium (DMEM, 31966, Gibco, TheNetherlands), supplemented with 10% v/v heat-inactivated fetal bovine serum (FBS Gibco, The Netherlands), and 1% v/v penicillin and streptomycin (Life Technologies, The Netherlands). Samples (n = 3 per time point) were freeze dried at different time points of incubation (10, 20, 30, 45, and 60min), and the mass loss was measured compared to that of pristine, as-casted hydrogels.
In Vitro Biological Evaluation
All animal-derived materials were obtained from deceased horses donated to science with informed consent from their owner, and in accordance with the Ethical Guidelines of the University Medical Center Utrecht and the Faculty of Veterinary Medicine of Utrecht University. Equine ACPCs were isolated as previously reported,[15] expanded in culture to passage 3 in ACPC expansion medium consisting of DMEM supplemented with 10% v/v FBS, 1% v/v penicillin and streptomycin (Life Technologies, The Netherlands), 1% MEM nonessential amino acids solution (NEAA, Gibco, The Netherlands), and 5 ng mL–1 basic fibroblast growth factor (bFGF, Peprotech, UK). ACPCs were then resuspended in 8% w/v GelMA and GelMA-Tyr hydrogels at a concentration of 20 × 106 cells mL–1, and then irradiated with visible light. Hydrogel samples were cultured in chondrogenic differentiation medium (DMEM, supplemented with 1% insulin–transferrin–selenous acid (ITS+ Premix, Corning, USA), 0.2 × 10−3 M ascorbic acid-2-phosphate, 1% v/v penicillin and streptomycin, 100 × 10−9 M dexamethasone, and 10 ng mL–1 transforming growth factor-b1 (TGF-b1). Medium was refreshed 3 times a week. Cell viability was assessed at day 1 and day 7 using a LIVE/DEAD assay (calcein AM, ethidium homodimer-1, Life Technologies, The Netherlands) (n = 3) and taking images with a confocal laser scanning microscope (Leica SP8X, Leica Microsystems, Germany). Cell laden GelMA and GelMA-Tyr hydrogels were harvested at day 1 and 28 for further analysis. Neocartilage formation was evaluated by sulfated GAGs and DNA quantification (n = 3). GAGs were quantified through a dimethylmethylene blue assay (DMMB, Sigma Aldrich, The Netherlands). DNA content was measured using a Quant-iT PicoGreen dsDNA kit (Life Technologies, The Netherlands). Gene expression was analyzed by quantitative polymerase chain reaction (qPCR) (n = 3). ACPC-laden hydrogels were harvested and mechanically ground in RLT buffer (Qiagen, Germany). The lysate was processed with the RNeasy Mini kit (Qiagen, Germany) in order to isolate mRNA. A Superscript III Platinum SYBR Green One-Step qRT-PCR Kit (Life Technologies, The Netherlands) was used for amplification and cDNA synthesis. The relative expression levels for collagen type I (COL1A1), collagen type II (COL2A1), and proteoglycan 4 (PRG4) were analyzed compared to the housekeeping gene hypoxanthine phosphoribosyltransferase-1 (HPRT1), using primers that have been previously described.[15] Relative expression, Ct, and efficiency values were calculated using the PCR Miner algorithm.[84] For histological analysis, the hydrogel samples were fixed in formalin and embedded in paraffin (n = 3). 5 μm sections were stained to visualize cartilage matrix production via Safranin-O staining for sGAG content and immunohistochemistry for both collagen type I (sc-8784, Santa Cruz Biotechnology, USA) and collagen type II (DSHB, II-II6B3, USA). For the quantitative assessment of the area covered by each ECM component, for each staining, three different slides were selected randomly from the samples (n = 3). Microscopy images were converted to binary images via thresholding with the software ImageJ, and the percentage of the area covered by the staining in pixels was calculated.
Myoglobin-Binding Assay
To test the potential of GelMA-Tyr as a carrier of biomolecules, 10% w/v GelMA and 10% w/v GelMA-Tyr solutions were supplemented with 10 mg mL–1 equine muscle-derived myoglobin and cast into cylindrical samples as previously described (n = 3). For these experiments, the oxidized form of myoglobin (metmyoglobin) was utilized. Myoglobin-free samples were included as controls. After crosslinking, samples were incubated at 37 °C in DPBS to assess protein release over a 48 h time span (after 1, 15, 30, and 45 min and 1, 1.5, 2, 3, 6, 24, 30, and 48 h). At each time point, the hydrogel samples were collected for stereomicroscopy imaging and the media was analyzed with a UV–vis spectrometer (Lambda 35, Perkin Elmer, USA) to quantify the amount of released myoglobin over a wavelength range of λ = 360–460 nm. Myoglobin concentrations were derived from the peak absorbance value at 409 nm of using a standard curve.[85]
Rheometry
The rheological properties of the hydrogel precursor solutions were assessed using a DHR2 rheometer (TA Instruments, The Netherlands). A stainless-steel flat plate (diameter = 40 mm) with a 60 μm plate-to-plate distance was used for all rheological tests. GelMA and GelMA-Tyr solutions were loaded and their complex viscosity was recorded at 21 °C as a function of shear rate (0.01–100 Hz), after two cooling (5min at 4° C) and recovery (5 min at 21 °C) conditioning cycles, at a constant strain of 5% (n = 5).
Hydrogel Printability
The GelMA or GelMA-Tyr macromers were loaded in a pneumatic-driven, extrusion-based printing system (23G stainless-steel nozzle, extrusion pressure between 1.9 and 2.5 bar, printing temperature = 18 °C, 3DDiscovery, regenHU, Switzerland). The effect of increasing the collector velocity (feed rate) from 3 to 21 mm s–1 on the diameter of the printed filaments was assessed, printing several straight lines (n = 3) and measuring their diameter from microscopy images using ImageJ software. In order to assess the printability of the solutions, a filament collapse test was also performed as previously described using photoinitiator-free gel and images were captured using a digital camera to visualize the extent of the spanning filaments (n = 5).[86] These prints were made using a feed rate ranging from 15 to 25 mm s–1. A 5-layered, 10 × 10mmsquare grid with 1 mm interfilament spacing was also printed with each hydrogel blend to assess the stacking ability of the gels (n = 5). In order to stabilize the extruded filaments, the hydrogel solutions were supplemented with 0.5 × 10−3/5 × 10−3 M Ru/SPS and were irradiated during the printing process, and for 5 min postprinting to ensure polymerization of the printed constructs. The printed grids were imaged with a stereomicroscope (Olympus SZ2-ILST, Olympus Corporation, Japan).
Evaluating Hydrogel Blends as Potential Bioinks for Bioprinting
ACPCs were harvested at passage 3 and embedded in GelMA and GelMA-Tyr inks at a density of 20 × 106 cells mL–1. These cell-laden hydrogel solutions were supplemented with 0.5 × 10−3/5 × 10−3 M Ru/SPS. Bioprinting was performed with the previously described extrusion-based system, with the same nozzle and temperature, and under the same visible light crosslinking conditions described in Section 2.7. Printed cylindrical samples (diameter = 5 mm, height = 1 mm) were cultured in ACPC expansion medium for 7 days. Cast controls were fabricated as previously described using the same cell density and crosslinking conditions. Medium was refreshed twice per week. Samples were analyzed for cell viability through a LIVE/DEAD assay and measuring their metabolic activity through a resazurin assay (resazurin salt, Alfa Aesar, Germany) after 1 and 7 days of culture (n = 3).
In Situ Photo-Crosslinking and Effects on the Native Cartilage
To evaluate the impact photoexposure on the tissue surrounding the implanted hydrogel constructs, equine cartilage samples of about 1 cm2 were freshly harvested from fetlock joints postmortem. Using a biopsy punch, 4 mm defects were made in the center of the explants, filled with 8% GelMA-Tyr and photoexposed. Hydrogel-free, nonirradiated explants with a punched chondral defect were used as positive controls. Ru/SPS gels crosslinked with visible light were compared to Irgacure crosslinked with either high (36 000 mJ cm–2, negative control) or a standard (1800 mJ cm–2) UV dose (365 nm, Vilber-Lourmat 144 portable UV-lamp, France) (n = 3). The standard dose represented the minimum necessary to crosslink the hydrogels in normoxic conditions. Cartilage explants were subjected to a LIVE/DEAD assay.
Adhesion of the Bioink to Cartilage
To evaluate the adhesion strength of GelMA and GelMA-Tyr hydrogels to native cartilage tissue, a push-out test was carried out. Cartilage discs of 10 mm in diameter and 1.5–2 mm in thickness were obtained from equine stifle joints. The cartilage disks were fixed in between two custom-made holders, and a cylindrical defect (4 mm) was imparted in the center of the explant using a biopsy punch. GelMA and GelMA-Tyr hydrogel solutions, together with Ru/SPS (0.5 × 10−3/5 × 10−3 M) were cast into the defects at a concentration of 8% gel and exposed to visible light for 10 min (n = 11–12). After incubation in PBS, a mechanical push-out test performed with a custom-made clamp in a Dynamic Mechanical Analyzer (DMA, TA Instruments, USA) was used to measure the adhesion strength between the hydrogels and the native cartilage. A force ramp of 0.1 N min–1 (no preload) was applied until failure. The thickness of each cartilage disk was measured with a digital caliper in order to calculate the interface area. The ultimate pushout stress was calculated by dividing the static force by the interface area (2πrh). As controls, GelMA and GelMA-Tyr hydrogels were also prepared using 0.1% w/v Irgacure 2959 and UV-crosslinked with 1800 mJ cm–2 (n = 5–6). Furthermore, to assess the binding of the bioink in an in situ printing setting, GelMA and GelMA-Tyr bioinks were loaded in a syringe and let undergo thermal gelation at 4 °C (n = 8–9). Subsequently, after being equalized at the printing temperature, the bioinks were extruded (23G nozzle) as filaments to fill the chondral defect, and crosslinked with visible light in presence of Ru/SPS, incubated in PBS and finally subjected to the push-outt tests.
Statistical Analysis
Results were reported as mean ± standard deviation. Statistical analyses were performed using GraphPad Prism 7.0 (GraphPad Software, USA). For the quantitative data, single comparisons were assessed via a Student’s t-test, and multiple comparisons with a oneway ANOVA, followed by post hoc Bonferroni correction to test differences between groups. When normality could not be assumed, nonparametric tests were performed (Mann–Whitney for single comparisons and Kuskal– Wallis for multiple comparisons). Differences were found to be significant when p < 0.05.
Supplementary Material
Supporting Information is available from the Wiley Online Library or from the author.
Acknowledgements
K.S.L. and F.A. contributed equally to this work. The authors would like to thank Anneloes Mensinga and Mattie van Rijen for their support with the qPCR and histological analysis. F.A., P.N.B., J.M., and R.L. acknowledge the funding from the Dutch Arthritis Society (LLP-12 and LLP-22), the European Research Council (grant agreement #647426, 3DJOINT), from the EU-Horizon 2020 research and innovation program under the grant agreement #814444 (MEFISTO), and from the AO Foundation (OCD consortium, ReInforce project). K.S.L. acknowledges funding by New Zealand Health Research Council (Emerging Researcher First Grant – 15/483, Sir Charles Hercus Health Research Fellowship – 19/135) and Royal Society of New Zealand (Marsden Fast Start – MFP-UOO1826). T.B.F.W. acknowledges funding from Royal Society of New Zealand (Rutherford Discovery Fellowship – RDF-UOO1204).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Khoon S. Lim, Christchurch Regenerative Medicine and Tissue Engineering (CReaTE) Group and Medical Technologies Centre of Research Excellence (MedTech CoRE); Department of Orthopaedic Surgery and Musculoskeletal Medicine University of Otago Christchurch 2 Riccarton Ave, Christchurch 8140, New Zealand.
Florencia Abinzano, Department of Orthopaedics and Regenerative Medicine Center University Medical Center Utrecht Utrecht University Heidelberglaan 100, Utrecht 3584 CX, The Netherlands.
Paulina Nuñez Bernal, Department of Orthopaedics and Regenerative Medicine Center University Medical Center Utrecht Utrecht University Heidelberglaan 100, Utrecht 3584 CX, The Netherlands.
Ane Albillos Sanchez, Department of Orthopaedics and Regenerative Medicine Center University Medical Center Utrecht Utrecht University Heidelberglaan 100, Utrecht 3584 CX, The Netherlands.
Pau Atienza-Roca, Christchurch Regenerative Medicine and Tissue Engineering (CReaTE) Group and Medical Technologies Centre of Research Excellence (MedTech CoRE); Department of Orthopaedic Surgery and Musculoskeletal Medicine University of Otago Christchurch 2 Riccarton Ave, Christchurch 8140, New Zealand.
Iris A. Otto, Department of Orthopaedics and Regenerative Medicine Center University Medical Center Utrecht Utrecht University Heidelberglaan 100, Utrecht 3584 CX, The Netherlands
Quentin C. Peiffer, Department of Orthopaedics and Regenerative Medicine Center University Medical Center Utrecht Utrecht University Heidelberglaan 100, Utrecht 3584 CX, The Netherlands
Michiya Matsusaki, Department of Applied Chemistry Graduate School of Engineering Osaka University 2-1 Yamadaoka, Suita, Osaka 565-0871, Japan.
Tim B. F. Woodfield, Christchurch Regenerative Medicine and Tissue Engineering (CReaTE) Group and Medical Technologies Centre of Research Excellence (MedTech CoRE) Department of Orthopaedic Surgery and Musculoskeletal Medicine University of Otago Christchurch 2 Riccarton Ave, Christchurch 8140, New Zealand.
Jos Malda, Department of Orthopaedics and Regenerative Medicine Center University Medical Center Utrecht Utrecht University Heidelberglaan 100, Utrecht 3584 CX, The Netherlands; Department of Clinical Sciences Faculty of Veterinary Medicine Utrecht University Yalelaan 1, Utrecht 3584 CL, The Netherlands.
Riccardo Levato, Levato Department of Orthopaedics and Regenerative Medicine Center University Medical Center Utrecht Utrecht University Heidelberglaan 100, Utrecht 3584 CX, The Netherlands; Department of Clinical Sciences Faculty of Veterinary Medicine Utrecht University Yalelaan 1, Utrecht 3584 CL, The Netherlands.
References
- [1].Flanigan DC, Harris JD, Trinh TQ, Siston RA, Brophy RH. Med Sci Sports Exercise. 2010;42:1795. doi: 10.1249/MSS.0b013e3181d9eea0. [DOI] [PubMed] [Google Scholar]
- [2].a Wluka AE, Ding C, Jones G, Cicuttini FM. Rheumatology. 2005;44:1311. doi: 10.1093/rheumatology/kei018. [DOI] [PubMed] [Google Scholar]; b Mouser VHM, Dautzenberg NMM, Levato R, van Rijen MHP, Dhert W, Malda J, Gawlitta D. ALTEX. 2018;35:65. doi: 10.14573/altex.1704171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hunziker EB. Osteoarthritis Cartilage. 2002;10:432. doi: 10.1053/joca.2002.0801. [DOI] [PubMed] [Google Scholar]
- [4].Vega SL, Kwon MY, Burdick JA. Eur Cells Mater. 2017;33:59. doi: 10.22203/eCM.v033a05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li J, Chen G, Xu X, Abdou P, Jiang Q, Shi D, Gu Z. Regener Bio-mater. 2019;6:129. doi: 10.1093/rb/rbz022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].a Regentis Biomaterials, GelrinC [Google Scholar]; b Medipost, CARTISTEM [Google Scholar]
- [7].Schuh E, Hofmann S, Stok KS, Notbohm H, Müller R, Rotter N. J Tissue Eng Regener Med. 2012;6:e31. doi: 10.1002/term.501. [DOI] [PubMed] [Google Scholar]
- [8].Schon BS, Hooper GJ, Woodfield TBF. Ann Biomed Eng. 2017;45:100. doi: 10.1007/s10439-016-1609-3. [DOI] [PubMed] [Google Scholar]
- [9].Malda J, Visser J, Melchels FP, Jüngst T, Hennink WE, Dhert WJA, Groll J, Hutmacher DW. Adv Mater. 2013;25:5011. doi: 10.1002/adma.201302042. [DOI] [PubMed] [Google Scholar]
- [10].Lim KS, Baptista M, Moon S, Woodfield TBF, Rnjak-Kovacina J. Trends Biotechnol. 2019;37:1189. doi: 10.1016/j.tibtech.2019.04.004. [DOI] [PubMed] [Google Scholar]
- [11].Groen WM, Diloksumpan P, van Weeren PR, Levato R, Malda J. J Orthop Res. 2017;35:2089. doi: 10.1002/jor.23602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mouser VHM, Levato R, Bonassar LJ, D’Lima DD, Grande DA, Klein TJ, Saris DBF, Zenobi-Wong M, Gawlitta D, Malda J. Cartilage. 2017;8:327. doi: 10.1177/1947603516665445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Daly AC, Freeman FE, Gonzalez-Fernandez T, Critchley SE, Nulty J, Kelly D. Adv Healthcare Mater. 2017;6 doi: 10.1002/adhm.201700298. 1700298. [DOI] [PubMed] [Google Scholar]
- [14].Kosiz-Koziol A, Costantini M, Mróz A, Idaszek J, Heljak M, Jaroszewicz J, Kijen´ska E, Szöke K, Frerker N, Barbetta A, Brinchmann JE, ´Swiefiszkowski W. Biofabrication. 2019;11 doi: 10.1088/1758-5090/ab15cb. 035016. [DOI] [PubMed] [Google Scholar]
- [15].Levato R, Webb WR, Otto IA, Mensinga A, Zhang Y, van Rijen M, van Weeren R, Khan IM, Malda J. Acta Biomater. 2017;61:41. doi: 10.1016/j.actbio.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Levato R, Jungst T, Scheurer R, Blunk T, Groll J, Malda J. Adv Mater. 2020 doi: 10.1002/adma.201906423. 1906423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Di Bella C, Duchi S, O’Connell CD, Blanchard R, Augustine C, Yue Z, Thompson F, Richards C, Beirne S, Onofrillo C, Bauquier SH, Ryan SD, Pivonka P, Wallace GG, Choong PF. J Tissue Eng Regener Med. 2018;12:611. doi: 10.1002/term.2476. [DOI] [PubMed] [Google Scholar]
- [18].Murphy SV, De Coppi P, Atala A. Nat Biomed Eng. 2019 doi: 10.1038/s41551-019-0471-7. [DOI] [PubMed] [Google Scholar]
- [19].Cui X, Breitenkamp K, Finn MG, Lotz M, ’lima DDD. Tissue Eng, Part A. 2012;18:1304. doi: 10.1089/ten.tea.2011.0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mancini IAD, Vindas Bolaños RA, Brommer H, Castilho M, Ribeiro A, Van Loon JPAM, Mensinga AM, Van Rijen HP, Malda J, Van Weeren R. Tissue Eng, Part C. 2017;23:804. doi: 10.1089/ten.TEC.2017.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Spiller KL, Maher SA, Lowman AM. Tissue Eng, Part B. 2011;17:281. doi: 10.1089/ten.teb.2011.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Khan IM, Gilbert SJ, Singhrao SK, Duance VC, Archer CW. Eur Cells Mater. 2008;16:26. doi: 10.22203/ecm.v016a04. [DOI] [PubMed] [Google Scholar]
- [23].Mouser VHM, Melchels FP, Visser J, Dhert WJ, Gawlitta D, Malda J. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/3/035003. 035003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].DiMicco MA, Sah RL. J Orthop Res. 2001;19:1105. doi: 10.1016/S0736-0266(01)00037-7. [DOI] [PubMed] [Google Scholar]
- [25].Chung C, Mesa J, Randolph MA, Yaremchuk M, Burdick JA. J Biomed Mater Res, Part A. 2006;77A:518. doi: 10.1002/jbm.a.30660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hoyle CE, Bowman CN. Angew Chem, Int Ed. 2010;49:1540. doi: 10.1002/anie.200903924. [DOI] [PubMed] [Google Scholar]
- [27].Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Bioma-terials. 2009;30:6702. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Idaszek J, Costantini M, Karlsen TA, Jaroszewicz J, Colosi C, Testa S, Fornetti E, Bernardini S, Seta M, Kasarello K, Wrzesien R, Cannata S, Barbetta A, Gargioli C, Brinchman JE, Swieszkowski W. Biofabrication. 2019;11 doi: 10.1088/1758-5090/ab2622. 044101. [DOI] [PubMed] [Google Scholar]
- [29].Bjork JW, Johnson SL, Tranquillo RT. Biomaterials. 2011;32:2479. doi: 10.1016/j.biomaterials.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yue K, Li X, Schrobback K, Sheikhi A, Annabi N, Leijten J, Zhang W, Zhang YS, Hutmacher DW, Klein TJ, Khademhosseini A. Biomaterials. 2017;139:163. doi: 10.1016/j.biomaterials.2017.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bertlein S, Brown G, Lim KS, Jungst T, Boeck T, Blunk T, Tessmar J, Hooper GJ, Woodfield TBF, Groll J. Adv Mater. 2017;29 doi: 10.1002/adma.201703404. 1703404. [DOI] [PubMed] [Google Scholar]
- [32].Lim KS, Klotz BJ, Lindberg GCJ, Melchels FPW, Hooper GJ, Malda J, Gawlitta D, Woodfield TBF. Macromol Biosci. 2019;19 doi: 10.1002/mabi.201900098. 1900098. [DOI] [PubMed] [Google Scholar]
- [33].Lim KS, Roberts JJ, Alves MH, Poole-Warren LA, Martens PJ. J Appl Polym Sci. 2015;132:42142. [Google Scholar]
- [34].Lim KS, Alves MH, Poole-Warren LA, Martens PJ. Biomaterials. 2013;34:7907. doi: 10.1016/j.biomaterials.2013.06.005. [DOI] [PubMed] [Google Scholar]
- [35].Van Hoorick J, Gruber P, Markovic M, Tromayer M, Van Erps J, Thienpont H, Liska R, Ovsianikov A, Dubruel P, Van Vlierberghe S. Biomacromolecules. 2017;18:3260. doi: 10.1021/acs.biomac.7b00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Elvin CM, Vuocolo T, Brownlee AG, Sando L, Huson MG, Liyou NE, Stockwell PR, Lyons RE, Kim M, Edwards GA, Johnson G, McFarland GA, Ramshaw JAM, Werkmeister JA. Biomaterials. 2010;31:8323. doi: 10.1016/j.biomaterials.2010.07.032. [DOI] [PubMed] [Google Scholar]
- [37].Cobbett WG, Kenchington AW, Ward AG. Biochem J. 1962;84:468. doi: 10.1042/bj0840468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bartnikowski M, Bartnikowski NJ, Woodruff MA, Schrobback K, Klein TJ. Acta Biomater. 2015;27:66. doi: 10.1016/j.actbio.2015.08.038. [DOI] [PubMed] [Google Scholar]
- [39].Klotz BJ, Gawlitta D, Rosenberg AJWP, Malda J, Melchels FPW. Trends Biotechnol. 2016;34:394. doi: 10.1016/j.tibtech.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Fancy DA, Denison C, Kim K, Xie Y, Holdeman T, Amini F, Kodadek T. Chem Biol. 2000;7:697. doi: 10.1016/s1074-5521(00)00020-x. [DOI] [PubMed] [Google Scholar]
- [41].Fancy DA, Kodadek T. Proc Natl Acad Sci USA. 1999;96:6020. doi: 10.1073/pnas.96.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Elvin CM, Carr AG, Huson MG, Maxwell JM, Pearson RD, Vuocolo T, Liyou BE, Wong DC, Merrott DJ, Dixon NE. Nature. 2005;437:999. doi: 10.1038/nature04085. [DOI] [PubMed] [Google Scholar]
- [43].Elvin CM, Brownlee AG, Huson MG, Tebb TA, Kim M, Lyons RE, Vuocolo T, Liyou NE, Hughes TC, Ramshaw JAM, Werkmeister JA. Biomaterials. 2009;30:2059. doi: 10.1016/j.biomaterials.2008.12.059. [DOI] [PubMed] [Google Scholar]
- [44].Cui X, Soliman BG, Alcala-Orozco CR, Li J, Vis MAM, Santos M, Wise SG, Levato R, Malda J, Woodfield TBF, Rnjak-Kovacina J, Lim KS. Adv Healthcare Mater. 2020;9 doi: 10.1002/adhm.201901667. 1901667. [DOI] [PubMed] [Google Scholar]
- [45].Kong H-J, Lee KY, Mooney DJ. Polymer. 2002;43:6239. [Google Scholar]
- [46].Hutson CB, Nichol JW, Aubin H, Bae H, Yamanlar S, Al-Haque S, Koshy ST, Khademhosseini A. Tissue Eng, Part A. 2011;17:1713. doi: 10.1089/ten.tea.2010.0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].a Lee F, Chung JE, Kurisawa M. J Controlled Release. 2009;134:186. doi: 10.1016/j.jconrel.2008.11.028. [DOI] [PubMed] [Google Scholar]; b Sando L, Danon S, Brownlee AG, McCulloch RJ, Ramshaw JAM, Elvin CM, Werkmeister JA. J Tissue Eng Regener Med. 2011;5:337. doi: 10.1002/term.318. [DOI] [PubMed] [Google Scholar]
- [48].Elvin CM, Danon SJ, Brownlee AG, White JF, Hickey M, Liyou NE, Edwards GA, Ramshaw JAM, Werkmeister JA. J Biomed Mater Res A. 2010;93:687. doi: 10.1002/jbm.a.32572. [DOI] [PubMed] [Google Scholar]
- [49].Loebel C, Szczesny SE, Cosgrove BD, Alini M, Zenobi-Wong M, Mauck RL, Eglin D. Biomacromolecules. 2017;18:855. doi: 10.1021/acs.biomac.6b01740. [DOI] [PubMed] [Google Scholar]
- [50].Jooybar E, Abdekhodaie MJ, Alvi M, Mousavi A, Karperien M, Dijkstra PJ. Acta Biomater. 2019;83:233. doi: 10.1016/j.actbio.2018.10.031. [DOI] [PubMed] [Google Scholar]
- [51].Bahcecioglu G, Bilgen B, Hasirci N, Hasirci V. Biomaterials. 2019;218 doi: 10.1016/j.biomaterials.2019.119361. 119361. [DOI] [PubMed] [Google Scholar]
- [52].Mouser VHM, Melchels FPW, Visser J, Dhert WJA, Gawlitta D, Malda J. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/3/035003. 035003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Schuurman W, Khristov V, Pot MWP, Van Weeren R, Dhert WJA, Malda J. Biofabrication. 2011;3 doi: 10.1088/1758-5082/3/2/021001. 021001. [DOI] [PubMed] [Google Scholar]
- [54].de Ruijter M, Ribeiro A, Dokter I, Castilho M, Malda J. Adv Health-care Mater. 2019;8 doi: 10.1002/adhm.201800418. 1800418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].O’Driscoll SW, Commisso CN, Fitzsimmons JS. Osteoarthritis Cartilage. 1995;3:197. doi: 10.1016/s1063-4584(05)80054-8. [DOI] [PubMed] [Google Scholar]
- [56].Woodfield TBF, Miot S, Martin I, van Blitterswijk CA, Riesle J. Biomaterials. 2006;27:1043. doi: 10.1016/j.biomaterials.2005.07.032. [DOI] [PubMed] [Google Scholar]
- [57].Schrobback K, Klein TJ, Woodfield TB. Tissue Eng, Part A. 2015;21:1785. doi: 10.1089/ten.tea.2014.0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Jay GD, Waller KA. Matrix Biol. 2014;39:17. doi: 10.1016/j.matbio.2014.08.008. [DOI] [PubMed] [Google Scholar]
- [59].Pahoff S, Meinert C, Bas O, Nguyen L, Klein TJ, Hutmacher DW. J Mater Chem B. 2019;7:1761. doi: 10.1039/c8tb02607f. [DOI] [PubMed] [Google Scholar]
- [60].Mouser VHM, Abbadessa A, Levato R, Hennink WE, Vermonden T, Gawlitta D, Malda J. Biofabrication. 2017;9 doi: 10.1088/1758-5090/aa6265. 015026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Brown GCJ, Lim KS, Farrugia BL, Hooper GJT, Woodfield BF. Macromol Biosci. 2017;17:1700158. doi: 10.1002/mabi.201700158. [DOI] [PubMed] [Google Scholar]
- [62].a Mouser VHM, Levato R, Mensinga A, Dhert WJA, Gawlitta D, Malda J. Connect Tissue Res. 2020;61:137. doi: 10.1080/03008207.2018.1553960. [DOI] [PubMed] [Google Scholar]; b Nam S, Chaudhuri O. Nat Phys. 2018;14:621. [Google Scholar]; c Zhang K, Feng Q, Xu J, Tian F, Yeung KWK, Bian L. Adv Funct Mater. 2018:30. 1701642. [Google Scholar]; d Feng Q, Wei K, Xu Z, Sun Y, Shi P, Li G, Bian L. Biomaterials. 2016;101:217. doi: 10.1016/j.biomaterials.2016.05.043. [DOI] [PubMed] [Google Scholar]
- [63].Li Z, Cao B, Wang X, Ye K, Li S, Ding J. J Mater Chem B. 2015;3:5197. doi: 10.1039/c5tb00455a. [DOI] [PubMed] [Google Scholar]
- [64].Vega SL, Kwon MY, Song KH, Wang C, Mauck RL, Han L, Burdick JA. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03021-5. 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Connelly JT, García AJ, Levenston ME. Biomaterials. 2007;28:1071. doi: 10.1016/j.biomaterials.2006.10.006. [DOI] [PubMed] [Google Scholar]
- [66].Ying G, Jiang N, Yu C, Zhang YS. Bio-Des Manuf. 2018;1:215. [Google Scholar]
- [67].Hong T, Iwashita K, Shiraki K. Curr Protein Pept Sci. 2018;19:746. doi: 10.2174/1389203719666171213114919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Muller M, Ozturk E, Arlov O, Gatenholm P, Zenobi-Wong M. Ann Biomed Eng. 2017;45:210. doi: 10.1007/s10439-016-1704-5. [DOI] [PubMed] [Google Scholar]
- [69].O’Connell CD, Di Bella C, Thompson F, Augustine C, Beirne S, Cornock R, Richards CJ, Chung J, Gambhir S, Yue Z, Bourke J, Zhang B, Taylor A, Quigley A, Kapsa R, Choong P, Wallace GG. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/1/015019. 015019. [DOI] [PubMed] [Google Scholar]
- [70].Li L, Yu F, Shi J, Shen S, Teng H, Yang J, Wang X, Jiang Q. Sci Rep. 2017;7 doi: 10.1038/s41598-017-10060-3. 9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Shen S, Chen M, Guo W, Li H, Li X, Huang S, Luo X, Wang Z, Wen Y, Yuan Z, Zhang B, Peng L, Gao C, Guo Q, Liu S, Zhuo N. Tissue Eng, Part B. 2019;25:187. doi: 10.1089/ten.TEB.2018.0248. [DOI] [PubMed] [Google Scholar]
- [72].Fedorovich NE, Oudshoorn MH, van Geemen D, Hennink WE, Alblas J, Dhert WJA. Biomaterials. 2009;30:344. doi: 10.1016/j.biomaterials.2008.09.037. [DOI] [PubMed] [Google Scholar]
- [73].de Gruijl FR, van Kranen HJ, Mullenders LHF. J Photochem Photobiol, B. 2001;63:19. doi: 10.1016/s1011-1344(01)00199-3. [DOI] [PubMed] [Google Scholar]
- [74].Masuma R, Kashima S, Kurasaki M, Okuno T. J Photochem Pho-tobiol, B. 2013;125:202. doi: 10.1016/j.jphotobiol.2013.06.003. [DOI] [PubMed] [Google Scholar]
- [75].Ruskowitz ER, Deforest CA. ACS Biomater Sci Eng. 2019;5:2111. doi: 10.1021/acsbiomaterials.9b00177. [DOI] [PubMed] [Google Scholar]
- [76].Abbadessa A, Mouser VHM, Blokzijl MM, Gawlitta D, Dhert WJA, Hennink WE, Malda J, Vermonden T. Biomacro-molecules. 2016;17:2137. doi: 10.1021/acs.biomac.6b00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Zhuang P, Ng WL, An J, Chua CK, Tan LP. PLoS One. 2019;14 doi: 10.1371/journal.pone.0216776. e0216776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Billiet T, Gevaert E, De Schryver T, Cornelissen M, Dubruel P. Biomaterials. 2014;35:49. doi: 10.1016/j.biomaterials.2013.09.078. [DOI] [PubMed] [Google Scholar]
- [79].Gittens J, Haleem AM, Grenier S, Smyth NA, Hannon CP, Ross KA, Torzilli PA, Kennedy JG. J Orthop Res. 2016;34:1139. doi: 10.1002/jor.23142. [DOI] [PubMed] [Google Scholar]
- [80].Broguiere N, Cavalli E, Salzmann GM, Applegate LA, Zenobi-Wong M. ACS Biomater Sci Eng. 2016;2:2176. doi: 10.1021/acsbiomaterials.6b00378. [DOI] [PubMed] [Google Scholar]
- [81].Lim KS, Schon BS, Mekhileri NV, Brown GCJ, Chia CM, Prabakar S, Hooper GJ, Woodfield TBF. ACS Biomater Sci Eng. 2016;2:1752. doi: 10.1021/acsbiomaterials.6b00149. [DOI] [PubMed] [Google Scholar]
- [82].Gelatin Manufacturers Institute of America. Gelatin Handbook. GMIA; New York: 2012. [Google Scholar]
- [83].Lim KS, Levato R, Costa PF, Castilho MD, Alcala-Orozco CR, Van Dorenmalen KMA, Melchels FPW, Gawlitta D, Hooper GJ, Malda J, Woodfield TBF. Biofabrication. 2018;10 doi: 10.1088/1758-5090/aac00c. 034101. [DOI] [PubMed] [Google Scholar]
- [84].Zhao S, Fernald RD. J Comput Biol. 2005;12:1047. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Armstrong JPK, Shakur R, Horne JP, Dickinson SC, Armstrong CT, Lau K, Kadiwala J, Lowe R, Seddon A, Mann S, Anderson JLR, Perriman AW, Hollander AP. Nat Commun. 2015;6 doi: 10.1038/ncomms8405. 7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ribeiro A, Blokzijl MM, Levato R, Visser CW, Castilho M, Hennink WE, Vermonden T, Malda J. Biofabrication. 2017;10 doi: 10.1088/1758-5090/aa90e2. 014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.