Abstract
Although sled dogs are one of the most unique groups of dogs, their origin and evolution has received much less attention than many other groups. We applied a genomic approach to investigate their spatiotemporal emergence, by sequencing the genomes of ten modern Greenland sled dogs, a ~9500 year old Siberian dog associated with archaeological evidence for sled technology, and a ~33,000 year old Siberian wolf. We found significant genetic similarity between the ancient dog and modern sled dogs. We detect gene flow from Pleistocene Siberian, but not modern American wolves, to present-day sled dogs. The results indicate that the major ancestry of modern sled dogs traces back to Siberia, where sled dog specific haplotypes of genes that potentially relate to Arctic adaptation were established by 9500 years ago.
Despite decades of studies, consensus is yet to be reached on when and where dogs were first domesticated, and when they were first deliberately used in many of the roles they exhibit today. In Siberia, late Upper Paleolithic artifacts of carved bone, antler and ivory, similar to tools used by modern Inuit for securing dog harness straps, suggest ancient origins of dog sledding (1). Furthermore, archeological findings from Zhokhov Island provide evidence of sled technology and dogs by the Sumnagin Mesolithic Culture ~9-8000 years ago (1–3) (Fig. S1), offering an opportunity to use genomics to further our understanding of early dog domestication and the origin of sled dogs.
We generated nuclear genomes from a dog mandible present at this site (“Zhokhov”, 9.6x coverage), dated to 9524 cal years before present (YBP) (Fig. 1A and Fig. S2) and a Siberian Pleistocene wolf mandible (“Yana”, 4.7x coverage), dated to 33,019.5 cal YBP (Fig. 1A and Fig. S3). In addition, we sequenced 10 modern Greenland sled dog genomes, a dog best described as an indigenous landrace breed, used for hunting and sledging by Inuit. Samples consist of two individuals from each of five geographically diverse localities (Fig. 1A), thus providing a broad representation of the indigenous dog diversity.
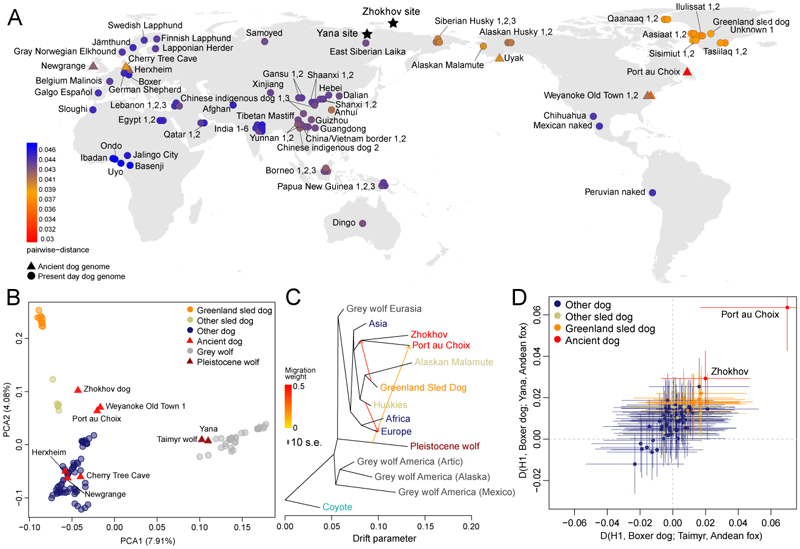
Fig. 1. Geographic location of the samples and overall genetic affinities.
(A) Identity by State pairwise distances between Zhokhov and present-day dogs (Table S1), geographic affiliation of dogs and archaeological sites. Color scale indicates genetic distance between Zhokhov and each sample. Circles and triangles represent modern and ancient dogs, respectively. Stars show Zhokhov and Yana sites. (B) Principal Component Analysis using whole-genome data (2,200,623 transversion sites) on all samples. (C) Treemix admixture graph built using whole genome data (766,082 transversion sites) on a dataset consisting of 66 canids merged into 15 groups according to their geographic location and admixture profile (Table S1 and Fig. S6). Colors indicate main groups as in panel B. Arrows show inferred admixture edges colored by migration weight. (D) D-statistic of the form D(H1, Boxer dog; Taimyr/Yana, Andean fox) testing for Pleistocene wolf gene flow in ancient and modern dogs, testing whether samples share more alleles with Taimyr (x-axis) or Yana (y-axis) wolves when compared to the boxer dog. Color indicates the type of sample in H1. Points show the D-statistic, while horizontal and vertical lines show 3 standard errors for the test with the Taimyr (x-axis) and Yana (y-axis), respectively. The results obtained from both ancient wolves fall along the diagonal, suggesting they are symmetrically related to all dogs.
We analysed our data alongside genomes from 114 geographically and genetically diverse canids (Table S1), using whole-genome pairwise distances, principal component analysis (PCA), Treemix (4) admixture graphs and D-statistics (Fig. 1). Yana appeared alongside wolves (Figs. 1B and 1C), while Zhokhov was found most closely related to dogs. Specifically, Zhokhov was most similar to modern sled dogs (Greenland sled dogs, Alaskan Malamutes, Alaskan and Siberian huskies) and American pre-European-contact dogs (PCDs), best illustrated by the ~2x Port au Choix dog from Maritime Archaic cultural context ~4000 YBP (3). Unsupervised clustering analyses with NGSadmix (4) (Fig. S6) grouped modern domestic dogs into four clusters: African, European, Asian, and sled dogs including Zhokhov. These relationships were confirmed by an admixture graph, where Yana was more closely related to a Pleistocene wolf from Taimyr Peninsula than to modern wolves, whereas Zhokhov represents a lineage that diverged from the ancestor of present-day sled dogs (Figs. 1C and S8-S9). This suggests genetic continuity in Arctic dog breeds for at least the past ~9500 years, setting a lower bound on the origin of the sled dog lineage.
Next, D-statistics indicated an excess of allele sharing between both Yana/Taimyr and PCDs/Zhokhov/sled dogs (Figs. 1D and S14), corroborating previous reports (3, 5). Importantly, this suggests the admixture occurred between Pleistocene wolves and the ancestors of PCDs, sled dogs and Zhokhov.
Previous studies have demonstrated an association between Canine Transmissible Venereal Tumour (CTVT), sled dogs and especially PCDs (3). Here, we evaluated the relationship between Zhokhov, two CTVT genomes (Table S1), dogs and wolves using f3 statistics and phylogenetic analysis. Recent analyses of exome data suggest that CTVT expanded across Eurasia ~6000 years ago (6), thus reducing the likelihood that this transmissible cancer originated in the Americas. In our study, both the phylogenetic analysis (Fig. S9) and f3 statistics (Fig. S10) placed the CTVT genomes closer to PCDs than to sled dogs or Zhokhov. These results imply that (i) the basal dog lineage that led to PCDs (3) occurred in Eurasia ~6000 years ago, and/or (ii) multiple introductions of PCD-like dogs to the Americas.
We employed NGSadmix, admixture analyses and D-statistics (Fig. S6-S8, S11-S15) to evaluate gene flow and shared ancestry between Zhokhov, modern dogs and wolves. We found no significant gene flow between any sled dog (including Zhokhov) and modern American-Arctic wolf populations when compared to the Eurasian wolf (Fig. S15), thus suggesting that gene flow from modern wolves has not contributed to the sled dog gene pool within the past 9500 years. This result was surprising given genetic evidence for post domestication admixture between other wolves and dog breeds (5, 7). Furthermore, ethnographic evidence from Greenland indicates that, at least historically, dog-wolf matings were not uncommon (8). If true, the lack of gene flow from modern American-Arctic wolves into sled dogs implies selection against hybrids.
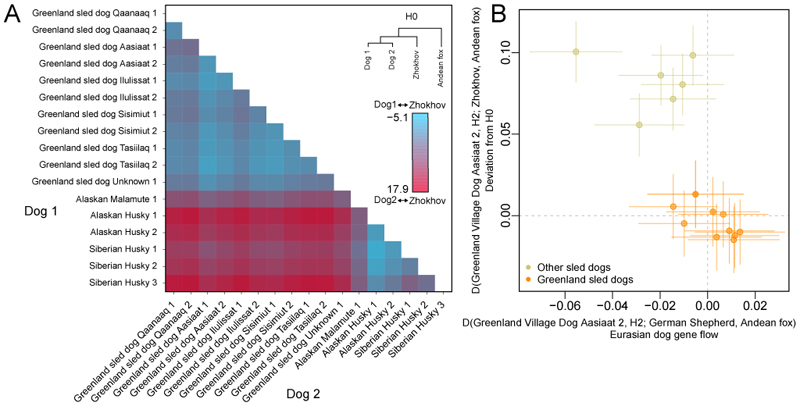
The clustering and admixture results show gene flow between some sled dogs and other modern dog breeds (Fig. 1C, Fig. S6-S8). We further explored this by comparing pairs of sled dogs to Zhokhov using D-statistics (Fig. 2A). While pairs of Greenland sled dogs are symmetrically related to Zhokhov (D~0) indicating a lack of admixture, comparisons involving non-Greenland sled dogs were not always consistent with the null hypothesis of no admixture. D-statistics and admixture analyses (Figs. 2B and S13) indicated that non-Greenland sled dogs carry ancestry from non-sled dogs and that Greenland sled dogs are the least admixed. These results imply that Greenland sled dogs (i) have largely been kept isolated from contact with other dog breeds, and (ii) that their lineage traces more genomics ancestry to Zhokhov-like dogs relative to other dog breeds. Isolation of Greenland sled dogs was supported by inference of their historical effective population size (Fig. S16), which showed Greenland sled dogs had a relatively stable population size until a severe bottleneck ~850 years ago. The timing of the bottleneck is consistent with the colonization of Greenland by Inuit (9), suggesting isolation in Greenland ever since.
Fig. 2. Relationships between Zhokhov and present-day sled dogs (sled dogs).
(A) D-statistics testing the relationships between pairs of sled dogs and Zhokhov. Cell colors indicate the Z-scores obtained from the test D(dog1, dog2; Zhokhov, Andean fox), where dog1 and dog2 are all possible pairs of sled dogs. Comparisons involving pairs of Greenland sled dogs and non-Greenland sled dogs resulted in significant deviations from H0 (|Z|>3). (B) D-statistics showing that sled dogs that are significantly further from Zhokhov compared to Greenland sled dog Aasiaat 2 (y-axis: D(Greenland sled dog Aasiaat 2, H2; Zhokhov, Andean fox)) also show evidence of significant gene flow from other dogs (x-axis: D(Greenland sled dog Aasiaat 2, H2; German shepherd dog, Andean fox)). Points indicate the D-statistic, while horizontal and vertical lines indicate 3 standard errors for the x- and y-axis, respectively. We considered the test to be significant for gene-flow when these lines do not overlap with the dotted line (|Z|>3).
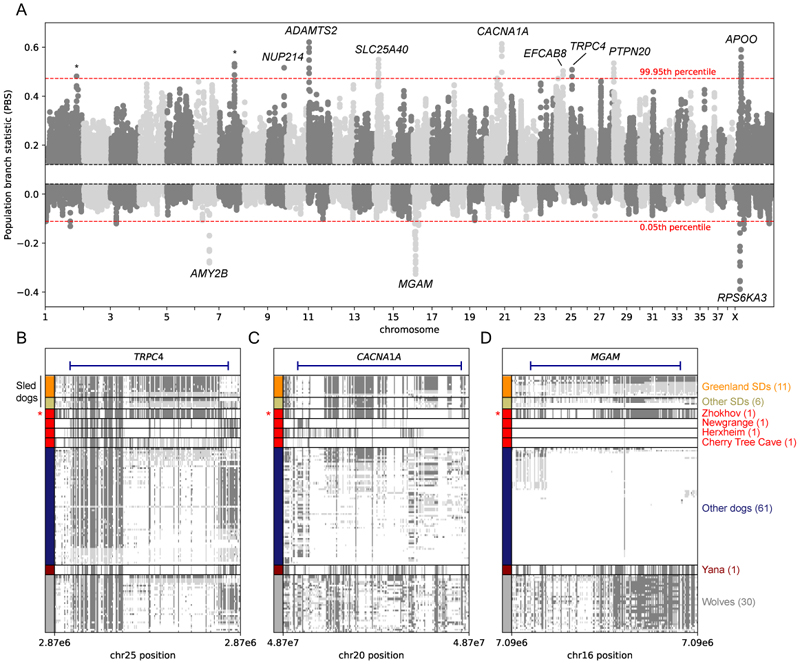
Numerous generations in the Arctic environment and as draft animals may have provided a unique selection pressure on sled dogs. To detect putative signals of positive selection, we used Population Branch Statistics (PBS) (10), to scan for genomic regions highly differentiated in modern sled dogs relative to non-sled dogs (hereafter, other dogs) and wolves. We computed these statistics on modern genomes of 17 sled dogs, 61 other dogs and 30 wolves (Table S1). A sliding window analysis revealed several genomic regions with high PBS values, hinting at selection in sled dogs (Fig. 3A). We took an outlier approach and focused on the most extreme values of the empirical distribution (above 99.95th percentile). For each of these outlier regions (Table S4), we identified overlapping genes and compared haplotypes across samples.
Fig. 3. Adaptation.
(A) Manhattan plot of the PBS values (y-axis) in windows of 100 kilo-base pairs (kb) using a 20 kb slide across chromosomes (x-axis). Data points between the 20th and 80th percentile of the empirical distribution are not plotted and dashed red lines show the 99.95th and 0.05th percentiles. Names of genes within the highest peaks are shown, with asterisks representing no overlap with genes. We note that other genes not displayed in the figure can overlap the outlier regions, a full list can be found in Table S4-5. Haplotype structures for TRPC4 (B), CACNA1A(C) and MGAM (D). Rows represent individuals, columns polymorphic positions in the dog genome. Cells are colored by genotype: dark gray (alternative allele homozygous), light gray (heterozygous) and white (reference allele homozygous). The row height for ancient individuals was increased to facilitate visualization. Zhokhov is highlighted with a red asterisk. SDs is used as an acronym for sled dogs.
Enrichment analysis (4) on genomic regions with high PBS values (above 99.95th percentile) identified three gene ontology terms that were overrepresented (Table S6), namely gamma-aminobutyric acid secretion (GO:0014051, p=0.119), calcium ion import (GO:0070509, p=0.119), and calcium ion transmembrane transport (GO:0070588, p=0.382). To investigate further, we focused on eight genomic regions that are highly differentiated in sled dogs, and three where other dogs differ from sled dogs and wolves (Figs. 3A and S18), and validated the autosomal regions with a cross-population composite likelihood ratio statistic (5) (Fig. S21). In the differentiated regions, we focused on two sets of genes, (i) genes where Zhokhov carries the same haplotype as the modern sled dogs and (ii) genes involved in adaptation to different diets.
TRPC4 is highly differentiated in sled dogs and the putatively selected haplotype bears a striking similarity to Zhokhov (Figs. 3A and B). TRPC4 is a transient receptor potential (TRP) channel protein that plays an important role in vasorelaxation and lung microvascular permeability (11). It is also involved in a temperature sensitivity pathway (12, 13), where it interacts with TRPV2, which is also highly differentiated in sled dogs (99.8th PBS percentile, Table S4 and Fig. S19A), and codes for temperature and potentially pain receptors (14). Several related thermo-TRP sensors in the same pathway - calcium ion transmembrane transport - have been previously reported to be under selection in cold-adapted woolly mammoths (15), which suggests convergent evolution in Arctic adaptation.
Another highly differentiated gene in sled dogs, CACNA1A (Fig. 3A and C), is a calcium channel subunit that plays an essential role in skeletal muscle contraction (16). Further, CACNA1A has been reported to be under positive selection in humans - the Bajau sea nomads (17), where it is involved in hypoxia adaptation (18), indicating a possible role managing exercise-induced hypoxia in sled dogs. Altogether, we hypothesize that the TRPC4, TRPV2 and CACNA1A genes are involved in functions beneficial to physical activity in the Arctic. If so, given that the differentiated haplotypes are also found in Zhokhov (Figs. 3A, B and S19A), any advantages they confer would have been important to dogs in the Arctic ~9500 YBP.
Most domestic dogs are adapted to starch-rich diets via significant increases in AMY2B copy numbers and strong positive selection for a dog-specific MGAM haplotype (19). Consistent with previous findings (20), we observed that sled dogs carry substantially fewer AMY2B copies than other dog breeds (Fig. S20). Interestingly, we also found that MGAM and AMY2B are the regions of the genome with lowest PBS, suggesting high differentiation of other dogs relative to sled dogs and wolves (Fig. 3A). Because negative PBS can arise under different demographic scenarios, we confirmed these observations by computing PBS with other dogs as the focal population (Fig. S18). Indeed, modern sled dogs and Zhokhov are among the only dogs in our dataset that carry the ancestral MGAM haplotype found at high frequency in wolves (Figs. 3C and S18). Therefore, our observations suggest sled dogs do not carry the genetic adaptations to starch rich diets seen in other dog breeds.
In contrast, sled dogs harbor unique haplotypes of genes involved in coping with high intake of fatty acids. SLC25A40, a mitochondrial carrier protein involved in clearing triglycerides from the blood (21), and APOO, an Apolipoprotein gene involved in regulating high levels of fat and fatty acid metabolism (22), are both highly differentiated in sled dogs (Figs. 3A). Interestingly, the derived haplotypes of both genes are absent in Zhokhov, indicating the haplotypes are unique to modern sled dogs and post-date their common ancestors with Zhokhov (Figs. S19B and S19E). As another example of convergent evolution, another gene of the Apolipoprotein family, APOB, is reported to be under selection in polar bears, possibly as a result of adaptation to fat rich diets and clearance of cholesterol from the blood (23). Overall, similar adaptations to high intake of fatty acids have been described in the Inuit and other Arctic human populations (24, 25), thus our observations suggest that sled dogs adapted to a fat rich and starch poor diet, echoing the dietary adaptations of the Arctic human cultures they co-existed with.
Bone composition of polar bears and reindeer consumed at the Zhokhov site indicate an extensive hunting range and transport of large body parts back to camp (26). Further, abundant obsidian tools found at the Zhokhov site reveal movement of obsidian across ~1500 km to the site (3). Together, they indicate significant long-distance travel and transportation of resources, in which dog sledding would have been highly advantageous if not necessary. Putative sled remains and our genomic analyses of a 9500 years old dog from the Zhokhov site indicate that tradition and key genomic variation that define modern sled dogs, were established in the Northeast Asian Arctic over 9500 years ago. Our results imply that the combination of these dogs with the innovation of sled technology facilitated human subsistence since the earliest Holocene in the Arctic.
Supplementary Material
One Sentence Summary.
Specialized Arctic sled dogs form an evolutionarily distinct lineage that was established in Northeast Asia at least 9500 years ago.
Acknowledgments
We thank J. A. Leonard and B. vonHoldt for their input and comments in the conceptualization of this study. We thank the Danish National High-Throughput Sequencing Centre and BGI-Europe for assistance in Illumina data generation. We thank the Danish National Supercomputer for Life Sciences - Computerome (computerome.dtu.dk) for the computational resources to perform the sequence analyses.
Funding
The study is embedded in “The Qimmeq Project” -funded by The Velux Foundations and Aage og Johanne Louis-Hansens Fond, and supported by ArchSci2020 - funded from the European Union's EU Framework Programme for Research and Innovation Horizon 2020 under Marie Curie Actions Grant Agreement No 676154. We thank the Rock Foundation of New York for funding excavations at the Zhokhov and Yana sites in a 15-year-long effort starting 2000. M.-H.S.S. was supported by the Independent Research Fund Denmark (8028-00005B) and NHM Oslo. S.G was supported by Marie Sklodowska-Curie Actions (H2020 655732 - WhereWolf) and Carlsberg (CF14 - 0995). M.d.M.M. was supported by a Formació de personal Investigador fellowship from Generalitat de Catalunya (FI_B01111). V.V.P., E.Y.P. and P.A.N. are supported by the Russian Science Foundation project N 16-18-10265-RNF. T.M.B. was supported by BFU2017-86471-P (MINECO/FEDER, UE), Howard Hughes International Early Career, Obra Social "La Caixa" and Secretaria d’Universitats i Recerca and CERCA Programme del Departament d’Economia i Coneixement de la Generalitat de Catalunya (GRC 2017 SGR 880). M.T.P.G. was supported by a European Research Council grant (ERC-2015-CoG-681396–Extinction Genomics). G.L. and L.A.F. were supported by the ERC (Grant ERC-2013-StG-337574-UNDEAD), and Natural Environmental Research Council (Grants NE/K005243/1 and NE/K003259/1).
Footnotes
Author contributions:
M.-H.S.S., S.G., J.R.-M., M.d.M.M., and M.T.P.G. conceived of the project and designed the research; V.V.P., E.Y.P., P.A.N., A.K.K., V.V.I., and E.W provided archaeological work, logistics and/or ancient collected samples; M.-H.S.S., M.F., S.E.W., M.P.H.-J., R.D., and C.S. coordinated logistics of -and/or provided – modern samples; C.C and M.-H.S.S. conducted the laboratory work; S.G., J.R.-M., M.d.M.M., L.K., L.A.F.F., F.G.V., J.N. and J.A.S.C. conducted the analyses of data with considerable input from M.-H.S.S., B.P., T.S.-P., T.M.-B., A.J.H. and M.T.P.G.; S.G., J.R.-M., M.d.M.M., L.K., L.A.F.F., F.G.V., J.N., J.A.S.C., P.S., M.-H.S.S., T.M.-B., A.J.H. and M.T.P.G interpreted results with considerable input from B.P., T.S.-P., V.V.P., T.R.F., E.U.A.-R., P.D.J., M.M., L.D., G.L., L.B., and Ø.W.; M.-H.S.S., S.G., J.R.-M., M.d.M.M., and M.T.P.G. wrote the paper with input from all other authors.
Competing interests:
Authors declare no competing interests.
Data and materials availability
Raw sequencing data can be accessed at NCBI Short Read Archive with project number PRJNA608847.
References and Notes
- 1.Pitulko VV, Kasparov AK. Ancient Arctic hunters: material culture and survival strategy. Arctic Anthropol. 1996;33:1–36. [Google Scholar]
- 2.Pitulko VV, Kasparov AK. Archaeological dogs from the Early Holocene Zhokhov site in the Eastern Siberian Arctic. Journal of Archaeological Science: Reports. 2017;13:491–515. [Google Scholar]
- 3.Ní Leathlobhair M, Perri AR, Irving-Pease EK, Witt KE, Linderholm A, Haile J, Lebrasseur O, Ameen C, Blick J, Boyko AR, Brace S, et al. The evolutionary history of dogs in the Americas. Science. 2018;361:81–85. doi: 10.1126/science.aao4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.See supplementary materials [Google Scholar]
- 5.Skoglund P, Ersmark E, Palkopoulou E, Dalén L. Ancient wolf genome reveals an early divergence of domestic dog ancestors and admixture into high-latitude breeds. Curr Biol. 2015;25:1515–1519. doi: 10.1016/j.cub.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 6.Baez-Ortega A, Gori K, Strakova A, Allen JL, Allum KM, Bansse-Issa L, Bhutia TN, Bisson JL, Briceño C, Castillo Domracheva A, Corrigan AM, et al. Somatic evolution and global expansion of an ancient transmissible cancer lineage. Science. 2019;365 doi: 10.1126/science.aau9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan Z, Silva P, Gronau I, Wang S, Armero AS, Schweizer RM, Ramirez O, Pollinger J, Galaverni M, Ortega Del-Vecchyo D, Du L, et al. Worldwide patterns of genomic variation and admixture in gray wolves. Genome Res. 2016;26:163–173. doi: 10.1101/gr.197517.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muus B, Salomonsen F, Vibe C. In: Grønlands Fauna. Danish, editor. Gyldendal, Nordisk Forlag; Copenhagen: 1981. [Google Scholar]
- 9.Raghavan M, DeGiorgio M, Albrechtsen A, Moltke I, Skoglund P, Korneliussen TS, Grønnow B, Appelt M, Gulløv HC, Friesen TM, et al. The genetic prehistory of the New World Arctic. Science. 2014;345 doi: 10.1126/science.1255832. 1255832. [DOI] [PubMed] [Google Scholar]
- 10.Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZXP, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiruppathi C, Freichel M, Vogel SM, Paria BC, Mehta D, Flockerzi V, Malik AB. Impairment of Store-Operated Ca2+ Entry in TRPC4-/- Mice Interferes With Increase in Lung Microvascular Permeability. Circ Res. 2002 doi: 10.1161/01.RES.0000023391.40106.A8. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann T, Schaefer M, Schultz G, Gudermann T. Subunit composition of mammalian transient receptor potential channels in living cells. Proc Natl Acad Sci U S A. 2002;99:7461–7466. doi: 10.1073/pnas.102596199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimmermann K, Lennerz JK, Hein A, Link AS, Kaczmarek JS, Delling M, Uysal S, Pfeifer JD, Riccio A, Clapham DE. Transient receptor potential cation channel, subfamily C, member 5 (TRPC5) is a cold-transducer in the peripheral nervous system. Proc Natl Acad Sci U S A. 2011;108:18114–18119. doi: 10.1073/pnas.1115387108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin N, Neeper MP, Liu Y, Hutchinson TL, Lubin ML, Flores CM. TRPV2 is activated by cannabidiol and mediates CGRP release in cultured rat dorsal root ganglion neurons. J Neurosci. 2008;28:6231–6238. doi: 10.1523/JNEUROSCI.0504-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch VJ, Bedoya-Reina OC, Ratan A, Sulak M, Drautz-Moses DI, Perry GH, Miller W, Schuster SC. Elephantid Genomes Reveal the Molecular Bases of Woolly Mammoth Adaptations to the Arctic. Cell Rep. 2015;12:217–228. doi: 10.1016/j.celrep.2015.06.027. [DOI] [PubMed] [Google Scholar]
- 16.Kaja S, van de Ven RCG, van Dijk JG, Verschuuren JJGM, Arahata K, Frants RR, Ferrari MD, van den Maagdenberg AMJM, Plomp JJ. Severely impaired neuromuscular synaptic transmission causes muscle weakness in the Cacna1a-mutant mouse rolling Nagoya. Eur J Neurosci. 2007;25:2009–2020. doi: 10.1111/j.1460-9568.2007.05438.x. [DOI] [PubMed] [Google Scholar]
- 17.Ilardo MA, Moltke I, Korneliussen TS, Cheng J, Stern AJ, Racimo F, de Barros Damgaard P, Sikora M, Seguin-Orlando A, Rasmussen S, et al. Physiological and Genetic Adaptations to Diving in Sea Nomads. Cell. 2018;173:569–580 e15. doi: 10.1016/j.cell.2018.03.054. [DOI] [PubMed] [Google Scholar]
- 18.Wang V, Davis DA, Haque M, Huang LE, Yarchoan R. Differential gene up-regulation by hypoxia-inducible factor-1alpha and hypoxia-inducible factor-2alpha in HEK293T cells. Cancer Res. 2005;65:3299–3306. doi: 10.1158/0008-5472.CAN-04-4130. [DOI] [PubMed] [Google Scholar]
- 19.Axelsson E, Ratnakumar A, Arendt M-L, Maqbool K, Webster MT, Perloski M, Liberg O, Arnemo JM, Hedhammar A, Lindblad-Toh K. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–364. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- 20.Arendt M, Cairns KM, Ballard JWO, Savolainen P, Axelsson E. Diet adaptation in dog reflects spread of prehistoric agriculture. Heredity. 2016;117:301–306. doi: 10.1038/hdy.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenthal EA, Ranchalis J, Crosslin DR, Burt A, Brunzell JD, Motulsky AG, Nickerson DA, NHLBI GO Exome Sequencing Project. Wijsman EM, Jarvik GP. Joint linkage and association analysis with exome sequence data implicates SLC25A40 in hypertriglyceridemia. Am J Hum Genet. 2013;93:1035–1045. doi: 10.1016/j.ajhg.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turkieh A, Caubère C, Barutaut M, Desmoulin F, Harmancey R, Galinier M, Berry M, Dambrin C, Polidori C, Casteilla L, Koukoui F, et al. Apolipoprotein O is mitochondrial and promotes lipotoxicity in heart. J Clin Invest. 2014;124:2277–2286. doi: 10.1172/JCI74668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S, Lorenzen ED, Fumagalli M, Li B, Harris K, Xiong Z, Zhou L, Korneliussen TS, Somel M, Babbitt C, Wray G, et al. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell. 2014;157:785–794. doi: 10.1016/j.cell.2014.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardona A, Pagani L, Antao T, Lawson DJ, Eichstaedt CA, Yngvadottir B, Shwe MTT, Wee J, Romero IG, Raj S, Metspalu M, et al. Genome-wide analysis of cold adaptation in indigenous Siberian populations. PLoS One. 2014;9:e98076. doi: 10.1371/journal.pone.0098076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fumagalli M, Moltke I, Grarup N, Racimo F, Bjerregaard P, Jørgensen ME, Korneliussen TS, Gerbault P, Skotte L, Linneberg A, Christensen C, et al. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science. 2015;349:1343–1347. doi: 10.1126/science.aab2319. [DOI] [PubMed] [Google Scholar]
- 26.Pitulko VV, Ivanova VV, Kasparov AK, Pavlova EY. Reconstructing prey selection, hunting strategy and seasonality of the early Holocene frozen site in the Siberian High\ Arctic: A case study on the Zhokhov site faunal remains, De Long Islands. Environ Archaeol. 2015;20:120–157. [Google Scholar]
- 27.Ramsey CB, Scott M, van der Plicht H. Calibration for archaeological and environmental terrestrial samples in the time range 26-50 ka cal BP. Radiocarbon. 2013;55:2021–2027. [Google Scholar]
- 28.Orlando L, Ginolhac A, Zhang G, Froese D, Albrechtsen A, Stiller M, Schubert M, Cappellini E, Petersen B, Moltke I, Johnson PLF, Fumagalli M, et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499:74–78. doi: 10.1038/nature12323. [DOI] [PubMed] [Google Scholar]
- 29.Ersmark E, Orlando L, Sandoval-Castellanos E, Barnes I, Barnett R, Stuart A, Lister A, Dalén L. Population demography and genetic diversity in the Pleistocene cave lion. Open Quaternary. 2015;1 [Google Scholar]
- 30.Dabney J, Knapp M, Glocke I, Gansauge M-T, Weihmann A, Nickel B, Valdiosera C, García N, Pääbo S, Arsuaga J-L, Meyer M. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci U S A. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allentoft ME, Sikora M, Sjögren K-G, Rasmussen S, Rasmussen M, Stenderup J, Damgaard PB, Schroeder H, Ahlström T, Vinner L, Malaspinas A-S, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 32.Carøe C, Gopalakrishnan S, Vinner L, Mak SST, Sinding M-HS, Samaniego JA, Wales N, Sicheritz-Pontén T, Gilbert MTP. Single-tube library preparation for degraded DNA. Methods Ecol Evol. 2017 [Google Scholar]
- 33.Schubert M, Ermini L, Der Sarkissian C, Jónsson H, Ginolhac A, Schaefer R, Martin MD, Fernández R, Kircher M, McCue M, Willerslev E, et al. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat Protoc. 2014;9:1056–1082. [Google Scholar]
- 34.Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gopalakrishnan S, Castruita JS, Sinding MHS, Kuderna L, Räikkönen J, Petersen B, SIcheritz-Ponten T, Larson G, Orlando L, Marques-Bonet T, Hansen A, et al. The wolf reference genome sequence (Canis lupus lupus) and its implications for Canis spp. population genomics. BMC Genomics. 2017 doi: 10.1186/s12864-017-3883-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 1000 Genome Project Data Processing Subgroup, The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frantz LAF, Mullin VE, Pionnier-Capitan M, Lebrasseur O, Ollivier M, Perri A, Linderholm A, Mattiangeli V, Teasdale MD, Dimopoulos EA, Tresset A, Duffraisse M, et al. Genomic and archaeological evidence suggest a dual origin of domestic dogs. Science. 2016;352:1228–1231. doi: 10.1126/science.aaf3161. [DOI] [PubMed] [Google Scholar]
- 41.Botigué L, Song S, Scheu A, Gopalan S, Pendleton A, Oetjens M, Taravella A, Seregély T, Zeeb-Lanz A, Arbogast R-M, Bobo D. Ancient European dog genomes reveal continuity since the early Neolithic. Nat Commun. 2017;8 doi: 10.1038/ncomms16082. 16082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galinsky KJ, Bhatia G, Loh P-R, Georgiev S, Mukherjee S, Patterson NJ, Price AL. Fast Principal-Component Analysis Reveals Convergent Evolution of ADH1B in Europe and East Asia. Am J Hum Genet. 2016;98:456–472. doi: 10.1016/j.ajhg.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 44.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet. 2011;12:443–451. doi: 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skotte L, Korneliussen TS, Albrechtsen A. Estimating individual admixture proportions from next generation sequencing data. Genetics. 2013;195:693–702. doi: 10.1534/genetics.113.154138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murchison EP, Wedge DC, Alexandrov LB, Fu B, Martincorena I, Ning Z, Tubio JMC, Werner EI, Allen J, De Nardi AB, Donelan EM, et al. Transmissible [corrected] dog cancer genome reveals the origin and history of an ancient cell lineage. Science. 2014;343:437–440. doi: 10.1126/science.1247167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 52.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gopalakrishnan S, Sinding M-HS, Ramos-Madrigal J, Niemann J, Samaniego Castruita JA, Vieira FG, Carøe C, Montero Mde M, Kuderna L, Serres A, et al. Interspecific Gene Flow Shaped the Evolution of the Genus Canis. Curr Biol. 2018;28:3441–3449 e5. doi: 10.1016/j.cub.2018.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Freuchen P. Den rene eskimohund. Danish, editor. Hundevennernes jul - Dansk Kennel Klub. 1943:39–42. [Google Scholar]
- 55.Young SP, Goldman EA. The wolves of North America: Part I. Their history, life habits, economic status, and control. Dover Publications, INC; New York: 1944. [Google Scholar]
- 56.Sinding M-HS, Gopalakrishan S, Vieira FG, Samaniego Castruita JA, Raundrup K, Heide Jørgensen MP, Meldgaard M, Petersen B, Sicheritz-Ponten T, Mikkelsen JB, et al. Population genomics of grey wolves and wolf-like canids in North America. PLoS Genet. 2018;14:e1007745. doi: 10.1371/journal.pgen.1007745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhatia G, Patterson N, Sankararaman S, Price AL. Estimating and interpreting FST: the impact of rare variants. Genome Res. 2013;23:1514–1521. doi: 10.1101/gr.154831.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen H, Patterson N, Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010;20:393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee PH, O’Dushlaine C, Thomas B, Purcell SM. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28:1797–1799. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang G-D, Zhai W, Yang H-C, Wang L, Zhong L, Liu Y-H, Fan R-X, Yin T-T, Zhu C-L, Poyarkov AD, Irwin DM, et al. Out of southern East Asia: the natural history of domestic dogs across the world. Cell Res. 2016;26:21–33. doi: 10.1038/cr.2015.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiedmer M, Oevermann A. A RAB3GAP1 SINE insertion in Alaskan huskies with polyneuropathy, ocular abnormalities, and neuronal vacuolation (POANV) resembling human Warburg micro …. G3: Genes, Genomes. 2016 doi: 10.1534/g3.115.022707. available at https://www.g3journal.org/content/6/2/255.short. [DOI] [PMC free article] [PubMed]
- 62.Decker B, Davis BW, Rimbault M, Long AH, Karlins E, Jagannathan V, Reiman R, Parker HG, Drögemüller C, Corneveaux JJ, Chapman ES, et al. Comparison against 186 canid whole-genome sequences reveals survival strategies of an ancient clonally transmissible canine tumor. Genome Res. 2015;25:1646–1655. doi: 10.1101/gr.190314.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.vonHoldt BM, Cahill JA, Fan Z, Gronau I, Robinson J, Pollinger JP, Shapiro B, Wall J, Wayne RK. Whole-genome sequence analysis shows that two endemic species of North American wolf are admixtures of the coyote and gray wolf. Science Advances. 2016;2:e1501714. doi: 10.1126/sciadv.1501714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Freedman AH, Gronau I, Schweizer RM, Ortega-Del Vecchyo D, Han E, Silva PM, Galaverni M, Fan Z, Marx P, Lorente-Galdos B, Beale H. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 2014;10:e1004016. doi: 10.1371/journal.pgen.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang G-D, Zhai W, Yang H-C, Fan R-X, Cao X, Zhong L, Wang L, Liu F, Wu H, Cheng L-G. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat Commun. 2013 doi: 10.1038/ncomms2814. 1860. [DOI] [PubMed] [Google Scholar]
- 66.Zhang W, Fan Z, Han E, Hou R, Zhang L, Galaverni M, Huang J, Liu H, Silva P, Li P, Pollinger JP. Hypoxia adaptations in the grey wolf (Canis lupus chanco) from Qinghai-Tibet Plateau. PLoS Genet. 2014;10:e1004466. doi: 10.1371/journal.pgen.1004466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, Zody MC, Mauceli E. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. (3) 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 68.Marsden CD, Ortega-Del Vecchyo D, O’Brien DP, Taylor JF, Ramirez O, Vilà C, Marques-Bonet T, Schnabel RD, Wayne RK, Lohmueller KE. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc Natl Acad Sci U S A. 2016;113:152–157. doi: 10.1073/pnas.1512501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Auton A, Rui Li Y, Kidd J, Oliveira K, Nadel J, Holloway JK, Hayward JJ, Cohen PE, Greally JM, Wang J, Bustamante CD, et al. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet. 2013;9:e1003984. doi: 10.1371/journal.pgen.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shannon LM, Boyko RH, Castelhano M, Corey E, Hayward JJ, McLean C, White ME, Abi Said M, Anita BA, Bondjengo NI, Calero J, et al. Genetic structure in village dogs reveals a Central Asian domestication origin. Proc Natl Acad Sci U S A. 2015;112:13639–13644. doi: 10.1073/pnas.1516215112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data can be accessed at NCBI Short Read Archive with project number PRJNA608847.