Abstract
The deoxy sugar L-fucose is frequently found as a glycan constituent on and outside living cells, and in mammals it is involved in a wide range of biological processes including leukocyte trafficking, histo-bloodgroup antigenicity, and antibody effector functions. The manipulation of fucose levels in those biomedically important systems may provide novel insights and therapeutic leads. However, despite the large established sequence diversity of natural fucosidases, so far very few enzymes have been characterized. We explored the diversity of the α-L-fucosidase-containing CAZY family GH29 by bio-informatic analysis, and by the recombinant production and exploration for fucosidase activity of a subset of 82 protein sequences that represent the family’s large sequence diversity. After establishing that most of the corresponding proteins can be readily expressed in E. coli, more than half of the obtained recombinant proteins (57% of the entire subset) showed activity towards the simple chromogenic fucosylated substrate 4-nitrophenyl α-L-fucopyranoside. Thirty-seven of these active GH29 enzymes (and the GH29 subtaxa that they represent) had not been characterized before. With such a sequence diversity-based collection available, it can easily be used to screen for fucosidase activity towards biomedically relevant fucosylated glycoproteins. As an example, the subset was used to screen GH29 members for activity towards the naturally occurring sialyl-Lewis x-type epitope on glycoproteins, and several such enzymes were identified. Together, the results provide a significant increase in the diversity of characterized GH29 enzymes, and the recombinant enzymes constitute a resource for the further functional exploration of this enzyme family.
Keywords: Fucosidase, GH29, glycoengineering, glycosylhydrolase
Introduction
Fucose is usually found at the glycan termini on mammalian protein and lipid glycoconjugates, where it is either linked α1–2 to galactose or α1–3/4/6 to N-acetylglucosamine (GlcNAc). Over the last decades, it has become widely appreciated that fucosylated glycoconjugates are often involved in molecular protein and cellular interactions, for example in host-microbe interactions (Pickard et al. 2014; Pickard and Chervonsky 2015), immunological processes (Shields et al. 2002; Ali et al. 2008), and in fertilization and development (Shi and Stanley 2003; Pang et al. 2011); several of these have recently been reviewed (Schneider et al. 2017). In many of these biological processes, the fucose residue is crucial for mediating the molecular interaction. Among other things, fucosylated glycans are involved in the leukocyte homing cascade. The removal of fucose from sialyl-lewis x (SLex) epitopes which cover the densely glycosylated P-selectin glycoprotein ligand 1 (PSGL-1), abolishes the rolling phenotype of leukocytes on activated endothelium (Ali et al. 2008). The absence of the core fucose (i.e. fucose linked to the innermost GlcNAc) on the conserved Fc glycan of Immunoglobulin G (IgG) significantly increases the affinity between IgG and the Fc gamma receptor IIIA (FcγRIIIA) on macrophages and Natural Killer (NK) cells. As a result, the corresponding antibody-dependent cellular cytotoxicity (ADCC) effector function is elicited up to a hundredfold more efficiently (Shields et al. 2002; Shinkawa et al. 2003).
The removal of fucose from glycoconjugates is catalyzed by fucose hydrolases (fucosidases). Given the widespread occurrence of fucosylated ligands in nature, it is not surprising to find a wealth of fucose-hydrolyzing enzymes throughout all domains of life. Most of the currently known fucose hydrolases (EC3.2.1) are classified into two major families in the Carbohydrate Active Enzymes (CAZy) database: GH29 and GH95 (Lombard et al. 2014). However, two new smaller families were recently added: GH139, involving α-2-O-methyl-L-fucosidase activity, and GH141, involved in the release of fucose that is substituted at O-3 with 2-O-methyl-α-D-xylose, as it occurs in the plant cell wall polysaccharide rhamnogalacturonan II (Ndeh et al. 2017). Furthermore, β-fucosidase activity has been described in families GH1 and GH30 (Lombard et al. 2014).
Family GH95 currently contains over 2400 entries and its experimentally characterized members generally have a rather narrow substrate specificity. They all act on fucose that is linked α1-2 to galactose, although α1-2–galactosidase activity towards arabinoxylans has also been reported (Rogowski et al. 2015). GH95 enzymes catalyze the hydrolysis of fucose through an inverting mechanism, resulting in the inversion of the anomeric configuration (Katayama et al. 2004; Nagae et al. 2007). So far no GH95 enzymes have been found in animals. In contrast, the largest fucosidase family, GH29, has over 4700 entries in the CAZy database with members throughout all domains of life. They act through a retaining mechanism and have a broader substrate specificity, including hydrolysis of fucose that is linked α1-3/4/6 to GlcNAc. Given that such linkages are often found on mammalian glycans and that fucosylation may influence the molecular interactions of the glycoprotein involved, GH29 fucosidases are likely to be useful tools for studying the biological impact of the removal of fucose. However, despite the large number of sequences in the CAZy family GH29, only few enzymes have been functionally characterized so far. This discrepancy has been further exacerbated since the onset of whole genome sequencing. New homologous sequences are being discovered rapidly, but true enzymatic characterization typically requires case-by-case studies (Cantarel et al. 2009). To further complicate matters for the discovery of novel and biomedically interesting fucosidases, many of the available sequences are merely derived from in silico predictions. Despite regular updates of the CAZy database and careful entry annotation, there is no guarantee that an annotated domain represents a foldable and functional protein. We therefore set out to map the complexity of the GH29 enzyme family by exploring its primary sequence diversity, to generate a representative subset of proteins that cover the major taxa in this family, and for their miniaturized expression in a microbial system. Here, we show that such a library can indeed be used to delineate those taxa that contain active fucosidases, and to identify those with substrate specificity towards naturally occurring N-glycans and fucosylated macromolecules.
Results
Towards a representative subset of the GH29 family
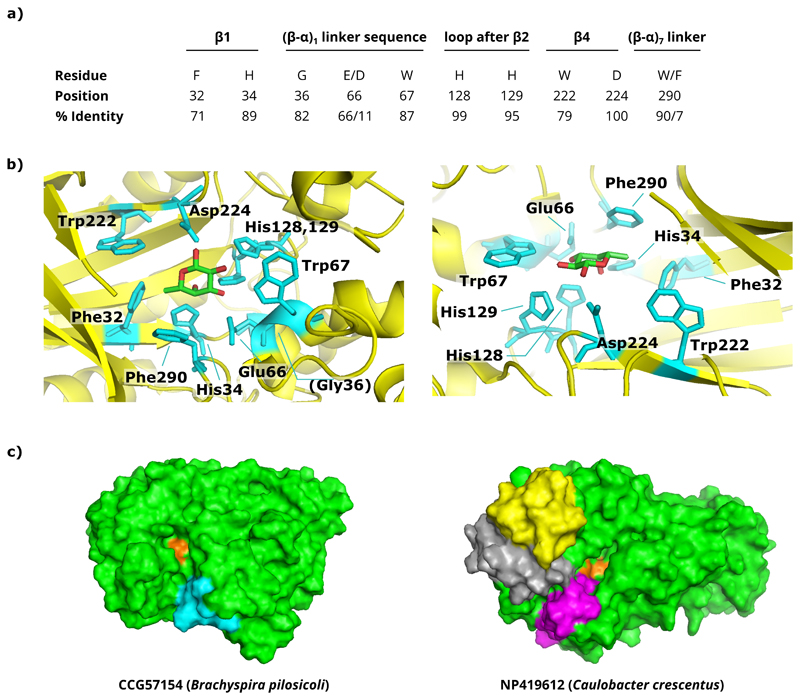
To assess the diversity of the GH29 family, all available GH29 protein sequences (n=705 at the initiation of the project in 2013) and human FUCA2 homologues were extracted from GenBank (also see Supplementary Methods), after which only non-redundant sequences of at least 250 amino acids were retained, based on the sum of the length of the conserved fold-determining structural elements (Wierenga 2001). A redundancy threshold of 92% protein sequence identity was used as a compromise between retaining as much sequence variation as possible while eliminating highly similar sequences. After multiple sequence alignments and the generation of a guide tree, the sequence diversity between the obtained clusters was compared. The catalytic nucleophile (aspartate) is located at position 224 in the protein structure of the Thermotoga maritima (Tm) fucosidase (PDB: 1HL8) (Sulzenbacher et al. 2004) and is completely conserved across the family, but as expected from earlier reports, the position of the general acid/base glutamate is not (Lammerts van Bueren et al. 2010; Shaikh et al. 2013). Other residues that resided in the catalytic pocket were also relatively well-conserved throughout GH29, including a consensus FxHxG motif in β-sheet 1, an (E/D)W sequence in a helical linker sequence between α1 and β1, two adjacent histidine residues just following the second β-sheet, the WxD motif in β4 containing the nucleophilic Asp, and a Trp residue in loop 7 (or a Phe in case of T. maritima fucosidase) (Figure Ia). Guided by the available protein structures of GH29 enzymes and the identification of conserved residues in or near the catalytic pocket (Figure Ib), we then focused on larger insertions or deletions in the loops that surround the catalytic pocket because these are most likely to affect the substrate specificity (Figure Ic).
Figure I. Conserved residues in and surrounding the catalytic pocket of GH29 proteins.
a) Overview of the conserved residues surrounding the catalytic site. The residue positions are based on those in the crystallized T. maritima fucosidase (PDB: 1HL8). The percentage of sequence identity (% identity) over our final GH29 collection that we have analyzed is also given for each residue. b) The position of these residues is highlighted on the crystal structure of the T. maritima fucosidase (left: top view; right: side view). In this structure, the enzyme is complexed with β-L-fucose (PDB: 1ODU). c) Visualization of an example where differences in loop regions from the multi-sequence alignments were identified. Shown are the modelled structures for two fucosidases (and the clusters they represent): CCG57154 from Brachyspirapilosicoli (left) and NP419612 from Caulobacter crescentus (right). The catalytic pocket is shown in orange, and regions where loops were longer as compared to neighboring clusters are highlighted in cyan for CCG57154, or yellow, gray and magenta for NP419612. 3D models were built using the iterative threading assembly refinement (I-TASSER) platform.
The diverse sequence tree was thoroughly pruned by sequentially clustering the sequences based on the presence of larger insertions or deletions surrounding the catalytic pocket and removing entries within each cluster that were highly similar. This manual process was repeated until each cluster contained only one or a few remaining sequences. Finally, a collection of 82 sequences was obtained that largely represents the loop diversity around the catalytic pocket of the GH29 scaffold and therefore likely captures much of the available substrate specificities. Most of these sequences were from bacterial origin (93%), next to five eukaryotic fucosidases and one Archaeal fucosidase (a complete overview can be found in Supplementary Table SI, visually represented in Supplementary Figure S1).
Recombinant expression of GH29 enzymes in E. coli
Despite the availability of many annotated protein sequences in dedicated databases such as CAZy and GenBank, such annotations are usually merely based on sequence similarity. However, no information can be inferred regarding the ability of the corresponding genes to be expressed as stable, folded proteins. Even more, a considerable number of fucosidases consist of multiple protein domains, which led us to investigate whether the fucosidase-annotated domain within the GH29 family will on its own provide functional proteins when expressed recombinantly. This was investigated using the well-characterized GH29 fucosidase of Thermotoga maritima (TmF), of which the annotated fucosidase domain in GenBank does not span the entire protein structure.The catalytic domain of the Tm fucosidase is organized around a (β/α)8 TIM-like (triosephosphate isomerase) barrel at the N-terminus (AA 1-359), a fold that is relatively common among GH enzymes (Henrissat et al. 1995). At the C-terminus, the enzyme also contains a non-conserved domain of unknown function, which is organized in a β-sandwich fold. The GenBank-annotated GH29 domain stretches to AA 401, which is somewhat odd as this is about halfway this C-terminal domain. Our results indicated that in fact the entire gene sequence is required to obtain a functional enzyme, as the expression of the (β/α)8 TIM-like barrel or the annotated GenBank domain (AA 1-401) did not result in a soluble active fucosidase (Supplementary Figure S2). Therefore, the full-length gene sequences were used for generating the expression cassettes of our GH29 collection, excluding only predicted signal sequences and/or predicted N-terminal transmembrane regions (Supplementary Table SII). The corresponding codon-optimized DNA sequences were synthesized and cloned into an expression vector to allow intracytoplasmatic recombinant expression as His6-tagged fusions in E. coli BL21 AI™ cells from the araBAD promoter. This yielded higher expression success rates than IPTG-based induction in BL21(DE3) cells. One caveat of the used Gateway cloning vector (pDEST17) is the inclusion of a linker (LESTSLYKKAGSGS) in between the N-terminal His-tag and ORF, yet we did not encounter issues regarding the soluble expression and activity of the Tm fucosidase.
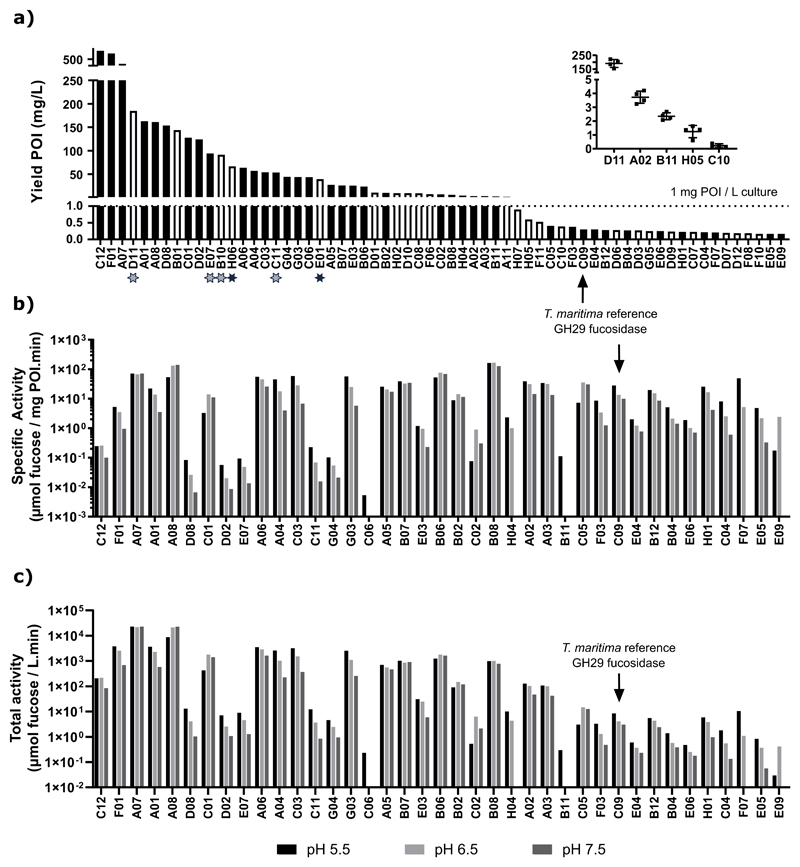
The expression level of each enzyme in the total bacterial lysate was quantified by spiking in a dilution series of a His6-tagged reference protein (in-house produced peptide-N-glycanase from Flavobacterium meningosepticum, PNGaseF) into the samples prior to separation by SDS-PAGE. Both the recombinant fucosidase enzymes and the reference protein were then detected using an infrared fluorescent western blot with an anti-His6-tag antibody. The approach yielded a good linear correlation between the amount of reference protein and the fluorescence signal, establishing a standard curve internal to each gel. This allowed for the use of linear regression to determine the level of fucosidase expression in the total lysates, expressed as nanograms of protein of interest (POI) per microgram of total protein in the lysate (Supplementary Figure S3). From these values, the expression yields were extrapolated to milligrams POI per liter of shake-flask E. coli culture (Figure IIa). As such, the level of expression could be quantified for 63 proteins (77% of our entire collection). The remaining proteins were expressed below the limit of quantification (i.e. less than 150 μg/L). Yields of at least 1 mg per liter of culture were obtained for nearly half of the recombinant proteins (38 proteins, 46%) (Figure IIa). Three proteins were expressed at very high level (>300 mg/L): fucosidases C12 from Bacteroides thetaiotaomicron (834 mg/L) of which the structure has been determined (PDB: 2WVS), F01 (Akkermansia muciniphila, 398 mg/L) and A07 (Leadbetterella byssophila, 322 mg/L).
Figure II. Quantitative protein expression and activity for the GH29 proteins in our collection.
a) Overview of the constructs with a protein expression above 150 μg per liter of culture. For nearly half of our collection (46%), yields of at least 1 mg/L of culture could be obtained. The insert demonstrates the limited variability of the quantification method used over five biological replicates of GH29 proteins with varying degrees of expression. Open bars represent members for which no activity on 4-NPF could be detected, and stars mark those fucosidases which are active on SLex-type α1-3 branched fucose (as also seen in Figure IV). b) Specific activities, determined as the amount of fucose (μmol) that is released per milligram of the protein of interest (POI) and per minute in bacterial total protein lysates. The proteins were sorted according to the expression levels shown in panel a, from highest to lowest. Proteins for which no 4-NPF hydrolysis could be detected or active proteins with expression levels below 150 μg/ml were left out of this panel. c) Total fucosidase activity that can be obtained per liter of shake flask E. coli culture for members of our collection. As a reference, the previously characterized Thermotoga maritima GH29 fucosidase is also highlighted in each panel.
To assess the robustness of this methodology, the quantification method was repeated for three additional biological replicates of five selected sequences with varying levels of expression. For samples with relatively high expression levels (> 2mg/L), the coefficient of variation (CV) was 15% at most, although the CV increased in samples with lower expression levels.
The majority of the GH29 subset are active α-L-fucosidases
Most of the proteins in our collection could be readily expressed in E. coli, but the next step was to determine whether the expressed proteins actually represented active enzymes that are able to remove fucose from glycoconjugates. Considering that the diversity of fucosylated glycoconjugates in nature is very high, the enzymes were first tested for their ability to hydrolyze the chromogenic substrate 4-nitrophenyl α-L-fucopyranoside (4-NPF). The assays were performed at 30°C with a buffer pH of 5.5, 6.5 and 7.5. The total enzyme input ranged between 0.1 to 200 μg per reaction, depending on the required dilution that provided a 4-nitrophenolate (4-NP) absorbance signal within the linear range of the assay. For each recombinant enzyme, the specific fucosidase activity was determined in the clarified E. coli lysates, expressed as the amount of fucose (in μmol) that was released per minute and per milligram of GH29 protein in the cleared lysates. In this way, specific fucosidase activity data was obtained for 44 enzymes, representing 54% of our library (Figure IIb). Using the well-characterized fucosidase from Thermotoga maritima as a reference, we identified fourteen enzymes with a higher specific activity towards 4-NPF at pH 5.5 or pH 7.5 (28 and 10 μmol of fucose per mg fucosidase per minute for the Tm fucosidase, respectively), and twenty with a higher activity at pH 6.5 (13 μmol/(mg.min) for TmF). The highest specific activity was obtained for the following constructs: a human fucosidase (FUCA1, enzyme B08 in our collection) at pH 5.5 (162 μmol of fucose per mg fucosidase per minute), H03 at pH 6.5 (Opitutus terrae, 198 μmol/(mg.min)) and A08 at pH 7.5 (Paenibacillus thiaminolyticus,138 μmol/(mg.min)). As compared to TmF, these specific activities are much higher, up to nearly sixfold at pH 5.5 and up to around 12 to 14-fold at more neutral pH.
We also determined the total fucosidase activity in the prepared lysates (extrapolated per liter of E. coli culture) and found that many enzymes yielded much higher productivity of fucosidase than the one from T. maritima (Figure IIc). Two constructs were especially superior in this regard, namely the strains expressing fucosidases A07 (fucosidase from Leadbetterella byssophila) and A08 (Paenibacillus thiaminolyticus). As compared to the third-best performer, these respectively yielded six- and twofold more total fucosidase activity at pH5.5, which increases to eightfold and 33-fold at higher pH (6.5 and 7.5 respectively). As the fucosidase with the highest specific activity displayed only moderate expression levels (fucosidase B08, 6 mg/L), it ranked only 11th out of 39 enzymes for which the total activity could be determined. Thirty-nine of the activeenzymes in our subset, including some with the highest specific activities and/or expression levels, were as yet uncharacterized.
Fucosidase activity on naturally occurring N-glycans
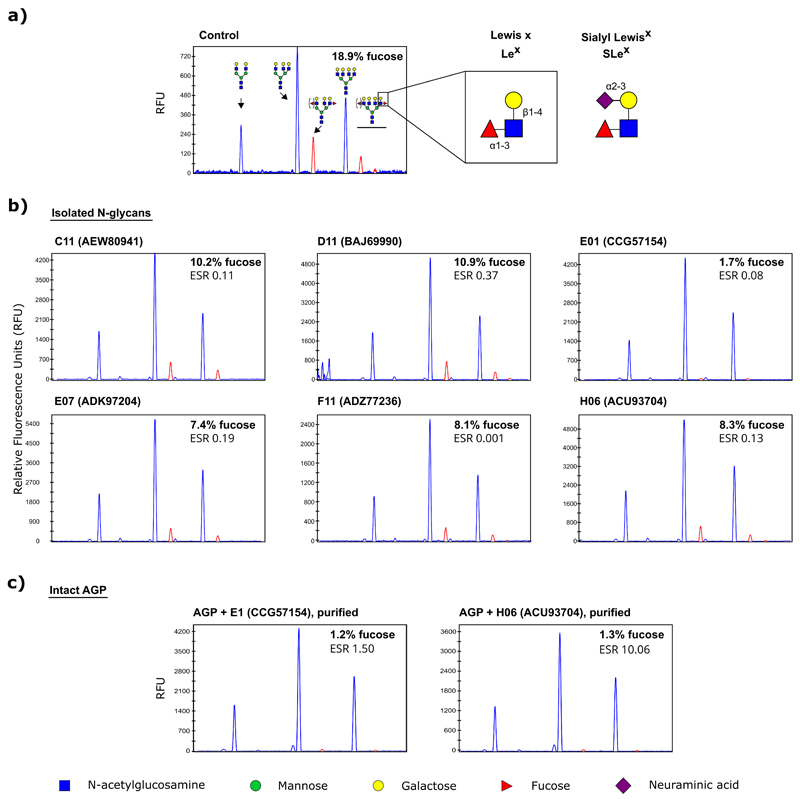
Having established this recombinant GH29-family resource, we then decided to explore the obtained library for activity towards naturally occurring glycoproteins. The human α-1 acid glycoprotein (AGP) was chosen as a test case, as it contains complex, multi-antennary N-glycans carrying the immunologically important SLex antigen. First, the AGP N-glycan composition was determined using capillary electrophoresis combined with laser-induced fluorescence (CE-LIF). AGP N-glycans were confirmed to be highly sialylated with a relative occurrence of bi-, tri-, and tetra-antennary structures of 21%, 51% and 28%, respectively. Branch fucosylation occurred only on tri- and tetra-antennary glycans, and represented about 19% of the total relative peak heights, while core fucosylation was not observed (Supplementary Figure S4). These results are in line with AGP N-glycan structures previously reported in the literature (Van Dijk et al. 1995).
Next, the fucosidase-containing bacterial lysates were screened for activity towards AGP N-glycans. All enzymes of our collection were included for the screening, as the lack of activity towards 4-NPF does not a priori exclude potential activity towards more complex substrates. The samples were incubated either with the native glycoprotein, or with its N-glycans which were first released with PNGaseF and isolated using solid-phase extraction. Six enzymes were identified with branch-fucosidase activity against the isolated sialylated N-glycans (Figures III and IV). As the screening was performed using fixed volumes of lysate, we later determined the enzyme-substrate weight ratios (ESR) of the positive hits, which usually lay between 0.08–0.37 except for F1, which was much lower (0.001).Interestingly, only two of these hits, the fucosidases from Brachyspirapilosicoli and Capnocytophagaochracea (E01 and H06, GenBank identifiers CCG57154 and ACU93704), were also able to defucosylate the N-glycans on intact AGP, which was confirmed after immobilized metal affinity chromatography (IMAC) based purification of the recombinant proteins from the E. coli lysates (Figure III and Supplementary Figure S5). All of the six hits, except for the fucosidase of Bifidobacterium longum (D11, GenBank BAJ69990), were enzymes for which no prior experimental evidence was available regarding substrate or linkage specificity.
Figure III. Defucosylation of AGP N-glycans by certain GH29 members in our collection.
a) CE-LIF profiles of an untreated control or b) after overnight incubation of isolated α1 acid glycoprotein (AGP) N-glycans with a clarified E. coli lysate containing fucosidases C11, D11, E1, E7, F11 or H6 at pH 5.5 (see Supplementary Figure S4 for the composition of these glycans). Each of these enzymes were able to, at least in part, remove the branch α1-3 fucose as found on the multiantennary N-glycans. c) Two of these hits were able to act on the AGP N-glycans while they were still attached to the glycoprotein. In all experiments, the glycans were treated with sialidase after fucosidase treatment and prior to CE-LIF analysis to simplify quantification of the branch fucose levels. Peaks containing branch fucose are highlighted in red. ESR: enzyme-substrate ratio (weight).
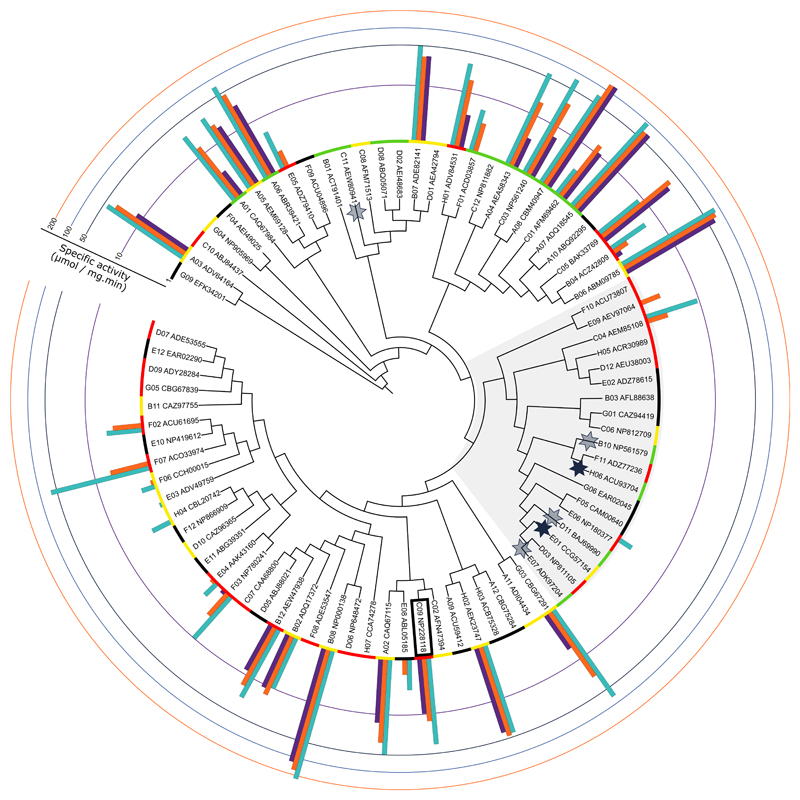
Figure IV. Annotation of the expression yields and specific 4-NPF hydrolysis activity onto the GH29 subset guide tree.
The expression yields are categorized with color codes where green = above 50 mg/L; yellow = between 1 and 50 mg/L; red = below 1 mg/L; and black = below 150 μg/ml. Specific activities at three pH conditions are shown in a log scale where light blue bars represent pH 5.5; orange bars pH 6.5; and purple bars pH 7.5. Proteins with activity towards branch α1-3 fucoses on N-glycans from the human α1-acid glycoprotein are shown with a star, where shaded stars represent activity only on glycans that were isolated from AGP and full stars activity also on the intact glycoprotein. The T. maritima fucosidase is boxed, and the shaded area corresponds to the GH29-B subfamily.
Annotating all of the activity data (both for 4-NPF and AGP N-glycans) onto the guide tree reveals that active members can generally be found throughout the family’s sequence diversity, although highly active members do seem to be confined in clusters (Figure IV). There is a large subdivision of GH29 members where no to very little activity towards 4-NPF was detected, yet interestingly five of the enzymes with activity towards the branch fucose on AGP N-glycans reside in this cluster (active members are indicated with a star in Figure IV). Even more, none of the enzymes with activity towards the α1-3 linked fucose residues on the N-glycans of AGP were able to release fucose from the chromogenic substrate 4-NPF. These findings reveal that the substrate specificity within the GH29 family is indeed determined by a similar coding sequence and reflect the known subdivision of the GH29 family into two subfamilies: enzymes with a substrate specificity that is restricted to α1-3/4-linked fucoses (GH29-B, shaded region in Figure IV) and enzymes with a reportedly broad substrate specificity that, among others, show activity towards fucose that is linked to chromogenic substrates such as 4-NPF (GH29-A) (Ashida et al. 2009; Sakurama, Tsutsumi, et al. 2012).
Discussion
Given the importance of many fucosylated glycoconjugates in various molecular interactions, the possibility of modulating these glycans is a highly attractive outlook both for fundamental research and for developing novel biomedical applications. Therefore, we performed a functional characterization of the CAZy GH29 family, in order to de-orphan many of its members by exploring the sequence diversity within this family. This resulted in a representative subset of 82 members, of which at least 63 proteins could be readily expressed in a bacterial expression system, allowing to determine their activity on a synthetic fucosylated substrate. It is appropriate to point out that, although synthetic substrates such as 4-NPF are regularly used for verifying fucosidase activity, by no means do these serve as universal substrates for all fucosidase enzymes, and no information can be inferred on the possible activity towards other fucosylated conjugates. Nonetheless, we found that over half of the family members in our collection (57%) were able to release fucose from this substrate. Combined with quantitative expression data, the specific activity towards 4-NPF could be calculated for each of these fucosidases. As compared to the well-characterized fucosidase from T. maritima, twenty-one enzymes were identified with up to an order of magnitude higher specific activity towards 4-NPF, especially at physiological pH, and with much better productivity in E. coli.
Encouraged by these results, we then demonstrate the use of the obtained family-comprehensive GH29 subset for identifying GH29 family members with substrate specificities towards naturally occurring N-glycans and glycoproteins that are of biomedical interest. As an example, we targeted the α1-3 branch fucoses in terminal SLex epitopes. These are involved in leukocyte trafficking through interactions with selectins. Several methods for inhibiting leukocyte rolling by interfering with these interactions have already been investigated in order to reduce leukocyte-induced tissue damage, for example in skin inflammation (Schön et al. 2002; Zollner et al. 2007) or reperfusion injury (Kubes et al. 1995). Interestingly, most GH29 enzymes that showed activity towards the SLex substrate were contained within a separate clade in the diversity tree. This clade contains several enzymes which have previously been described to belong to the GH29-B subfamily (Sakurama, Fushinobu, et al. 2012; Sakurama, Tsutsumi, et al. 2012; Shaikh et al. 2013), and which are able to hydrolyze α1-3/4 linked fucose, but not 4-NPF. These findings are thus consistent with the current knowledge on the subdivision of GH29 in two subfamilies with their corresponding substrate specificities. However surprisingly, we found that only a few of those enzymes were able to act on SLex-modified N-glycans while still linked to a protein. This is of course critical if such enzymes are to be used to defucosylate SLex in native biomedical samples.
The experimental findings described here were further compared with the available knowledge in the Uniprot database, where protein sequences are classified as either ‘Unreviewed’ (records that await full manual annotation) or ‘Reviewed’ (containing information extracted from literature and curator-evaluated computational analysis). Within our subset, reviewed experimental evidence was only available for three enzymes and the data here confirms that these sequences correspond to active fucosidases (B08, human FUCA1; C09, Tm fucosidase; and C12, from Bacteroides thetaiotaomicron). We provide experimental evidence for fucosidase activity or lack thereof on two substrates (4-NPF and SLex) for 63 E. coli expressable, previously uncharacterized GH29 members, sampling the entire GH29 diversity. To date, the CAZy database contains 35 characterized GH29 enzymes. Our study considerably enhances the knowledge on this family as our sequences comprehensively sample the entire family’s diversity.
The subset of fucosidase sequences and their expression and activity characteristics described here will be useful to direct future efforts for protein structure determination towards branches of the GH29 family tree that are currently not represented in the Protein Data Bank (Berman et al. 2000), especially given the strong biomedical interest in this family. New crystal structures of GH29 members have recently emerged (Summers et al. 2016; Kovalová et al. 2019), and novel applications involvingnaturally occurring or mutated GH29 members were described over the last years as tools for the biomedical engineering of glycoproteins (Li et al. 2017; Tsai et al. 2017).
In conclusion, this work likely represents the most comprehensive experimental exploration of GH29 so far and provides a broadly applicable starting point for identifying additional enzymes with activity towards biomedically relevant fucosylated glycans.
Materials and methods
Sequence extraction, alignment and visualization
Sequences from the CAZy GH29 family and 1000 homologues to the human fucosidase FUCA2 (NP_114409) were extracted from GenBank using a custom Perl-based script (Supplementary Methods) and basic local alignment search tool (BLAST). Multiple sequence alignments of large datasets (>100 sequences) were generated using the MUltiple Sequence Comparison by Log- Expectation (MUSCLE) algorithm in the Molecular Evolutionary Genetics Analysis software (MEGA) version 6 (Edgar 2004; Tamura et al. 2013). Multiple sequence alignments of smaller datasets were generated using Clustal throughout the refinement of sequence selection (Larkin et al. 2007). The removal of redundant sequences and visualization of the alignments was performed in the Jalview desktop application (Waterhouse et al. 2009). Associated guide trees of the alignments were generated using the Neighbor-Joining algorithm in MEGA 6, and these were visualized and further annotated on the Interactive tree of life (iTOL) web-based tool (Letunic and Bork 2016). Structural protein information was obtained through the Protein Data Bank (PDB) of the Research Collaboratory for Structural Bioinformatics (Berman et al. 2000) and visualized using PyMOL (Schrödinger). For all 82 GH29 protein sequences in our collection, 3D models were built using the iterative threading assembly refinement (I-TASSER) platform (Roy et al. 2010; Yang and Zhang 2015). All the software and tools described in this section are freely available for academic use.
In silico processing of the protein sequences
The prediction of signal peptides and possible N-terminal transmembrane helices was done using SignalP 4.1 (Petersen et al. 2011) and the TMHMM server v2.0 (Krogh et al. 2001), respectively. If present, these regions were removed. The corresponding DNA gene sequences were codon-optimized for expression in E. coli and ordered synthetically with an adapter sequence at both termini to allow efficient cloning into the pDEST17 expression vector (Thermo Fischer Scientific) using the restriction enzymes NdeI and XhoI (New England Biolabs) (N-terminal adapter: 5’–CAGTCATATGTCGTACTACC ATCACCATCA CCATCACCTC GAATCAACAA GTTTGTACAA AAAAGCAGGC TCTGGATCC–3’ ; C-terminal adapter: 5’–GTTTAAACCT CGAGATATCT–3’). These sequences include restriction sites (shown in italics), the start and stop codons (bold), and the attB1 site of the destination vector (underlined and in italics). The gene constructs also included an N-terminal 6xHis-tag (underlined) to enable detection of protein expression in total bacterial lysates using Western blot and to allow for Immobilized Metal Affinity Chromatography (IMAC) purification. The ordered nucleotide sequences are provided in the Supplementary Files.
Molecular cloning and general cultivation techniques
Restriction enzyme and ligation-based molecular cloning was performed using standard laboratory techniques and plasmids were propagated in E. coli MC1061 or DH5α strains. A feature map of the expression vector is provided in Supplementary Figure S6. The cultures were grown in liquid lysogeny broth (LB: 10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl) or on agar plates (LB + 15 g/L agar) at 37°C. Antibiotic selection was applied where required using 50 μg/ml kanamycin and/or 100 μg/ml carbenicillin (Duchefa Biochemie). Recombinant protein expression was done in BL21-AI™ strains (Thermo Fischer Scientific) in either LB or Terrific Broth (Sigma Aldrich) under constant antibiotic selection. The nucleotide sequence of each of the final constructs was verified by Sanger sequencing at the VIB Genomics Core.
Recombinant protein expression and detection
Protein expression in E. coli from a 30 ml culture grown in shake flasks was initiated when the culture OD600 had reached a value between 0.8 and 1, by supplementing the medium with L-arabinose to a final concentration of 0.2%. Cultures were harvested after 16h of growth (28°C while shaking at 225 rpm), the cells pelleted and resuspended in 4 ml of lysis buffer per gram wet cell weight (25 mM Tris-HCl pH 7.5, 200 mM NaCl, 5 mM MgCl2, 100 μg/ml chicken egg white lysozyme and 0.1% Triton X-100). Finally, the cells were lysed by sonication and the soluble fractions isolated by centrifugation (15 minutes at 20,000). Microbial pellets or soluble lysate fractions were flash-frozen in liquid nitrogen and stored at –20°C until further analysis. Total protein concentrations were determined using the bicinchoninic acid (BCA) assay as recommended by the manufacturer (Pierce™ BCA Protein Assay Kit, Thermo Fischer) and protein samples were analyzed after separation on Tris-glycine sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels containing 10 or 12% polyacrylamide. After electrophoresis, the proteins were visualized by staining with Coomassie brilliant blue or first blotted onto a nitrocellulose membrane for western blot (WB) analysis using a Pierce™ Power Blotter (Thermo Fischer) according to the manufacturer’s instructions. Proteins carrying a polyhistidine tag were detected with an anti-His6 antibody conjugated to DyLight 800™ (Rockland, usually diluted 1:20,000 to 50 ng/ml unless otherwise stated), and visualized using a LI–COR® Odyssey western Blot Detection System (Westburg). Data processing was done in the LI–COR® Image Studio™ Lite software application.
Quantification of fucosidase expression
The amount of fucosidase in a given sample was determined using quantitative WB. Depending on the signal intensity observed in preparatory experiments, the total protein loading for the bacterial lysates varied from 0.1 to 50 μg per lane of an SDS-PAGE gel. The signal was quantified relative to a dilution series of an in-house produced and His-tagged peptide-N-glycanase from Flavobacterium meningosepticum (PNGaseF), which was spiked into the samples as an internal standard, allowing linear regression for the unknown sample concentrations in each gel (Supplementary Figure S3). The dilution of the fluorophore-conjugated anti-His6 antibody varied from 1:5,000 to 1:100,000 (1 μg/ml stock) during optimization, and final experiments were done using a dilution of 1:15,000 as this maximized the signal-to-noise ratio (STR). For selected samples, additional biological replicates were used to determine the variation in protein expression.
Activity assay on 4-nitrophenyl α-L-fucopyranoside (4-NPF) and quantification
Activity assays on the chromogenic 4-NPF substrate were performed in a reaction volume of 80 μl containing up to 200 μg of total protein of the cleared lysates. Dilution of the lysate was performed as needed to ensure that the signal of product formation was within the dynamic range of the assay for each enzyme. The reaction further contained 2.5 mM of 4-NPF in 50 mM buffer (MES at pH 5.5 or 6.5; or HEPES at pH 7.5) supplemented with protease inhibitors (cOmplete™ Protease Inhibitor Cocktail without EDTA, Roche). After incubation (30 minutes at 30°C), the reaction was stopped by adding 120 μl of 0.5 M of carbonate/bicarbonate pH 9.6, and the absorbance was measured at 415 nm on an iMark™ microplate reader (Bio-Rad). The obtained absorbance was corrected using a negative control lysate from an untransformed strain, which was applied in the same concentration as the sample. The amount of fucose that was released was inferred by linear regression by including a dilution series of 4-nitrophenol on each plate, with concentrations ranging from 0-1 mM (1 mM corresponds to 80 nmol per well). For selected enzymes, each with varying degrees of activity towards 4-NPF, the assay was repeated with up to four biological replicates. Data processing and visualization was done in Microsoft Excel 2013 and GraphPad Prism 7.
Solid-phase extraction of the N-glycans from α-acid glycoprotein (AGP)
Human AGP (1 mg/ml obtained from Sigma-Aldrich, cat. nr G9885) was supplemented with 10% (v/v) denaturation buffer (0.5% SDS, 40 mM DTT) and incubated for 10 minutes at 98°C. After cooling on ice, 10% (v/v) reaction buffer was added (0.5 M sodium phosphate, 10% IGEPAL® CA-630, pH 7.5), followed by the addition of 77 IUB mU ofPNGaseF (in-house produced as described earlier (Vanderschaeghe et al. 2010)). After 6 hours, another 46.2 IUB mU of PNGaseF was added and the samples further incubated overnight at 37°C. The next day, samples were quenched with ultrapure water (Milli Q, MQ) and the N-glycans extracted using Extract-Clean SPE Carbo Columns (Altech). The columns were first washed three times with 80% acetonitrile (ACN) supplemented with 0.1% trifluoroacetic acid (TFA) and three times with MQ before applying the sample under gravitational flow. The resin was washed five times with MQ before elution with 25% ACN supplemented with 0.05% TFA. The obtained solution was dried in a Savant™ SpeedVac™ Concentrator (Thermo Fischer Scientific) coupled to a vapor trap. Finally, the obtained glycans were resuspended in one-tenth of the starting volume in MQ.
General N-glycan analytics
The isolation and analysis of N-glycans were done on as described previously (Laroy et al. 2006). In short, glycoproteins were immobilized on 96 well plates containing a hydrophobic immobilon-P PVDF membrane (Merck) and their N-glycans released using 15.4 IUB mU/μl of PNGaseF in 10 mM Tris-acetate pH 8.3. Before analysis on an ABI3130 Genetic Analyzer (Applied Biosystems), the N-glycans were derivatized with 8-aminopyrene-1,3,6-trisulfonic acid (APTS, 10 mM final concentration) by reductive amination and excess label was removed by size exclusion chromatography over Sephadex G10 resin (GE Healthcare). As external reference for the glycan electrophoretic mobility, isolated N-glycans from bovine pancreas Ribonuclease B (RNaseB) and a dextran ladder were included. The dextran ladder was obtained by partial heat-hydrolysis of dextran (Mw 9,000-11,000) isolated from Leuconostoc mesenteroides, resulting in a heterogeneous mixture of glucose oligomers that differ in length by one glucose unit. The data were visualized with the Genemapper software (Applied Biosystems), and relative abundances of each peak were calculated based on the peak heights (in relative fluorescence units, RFU). The obtained chromatograms were aligned and processed using Inkscape 0.91 after they were exported in the Scalable Vector Graphics (SVG) format.
Exoglycosidase digests were performed to obtain information on the structure of AGP N-glycans, interpreted in light of the established N-glycan biosynthesis pathways in humans. The exoglycosidases used in this project were Arthrobacter ureafaciens α2,3/6/8-sialidase (in house produced, 40 mU/reaction), Glyko® β1-4-galactosidase from Streptococcus pneumoniae (Prozyme, 0.4 mU), β-N-acetyl-hexosaminidase from Jack Beans (Prozyme, 10 mU), α1-2/3/4/6-fucosidase from bovine kidney (Prozyme, 0.44 mU), α1-3/4-fucosidase from almond meal (Prozyme, 0.2 mU), and α1-2/3/6-mannosidase from Jack Beans (Sigma, 20mU). All digests were executed on APTS-labeled glycans that were subjected to clean-up, and performed overnight in 20 mM ammonium acetate pH 5.2 at 37°C.
Purification of fucosidases E01 and H06
After protein expression and lysis of the obtained E. coli cells, the cleared lysates could be directly loaded onto an Äkta chromatography system for protein purification (GE Healthcare). The lysis buffer was supplemented with 20 mM imidazole to reduce aspecific binding to the column (HisTrap HP 5 ml, GE Healthcare) for immobilized metal affinity chromatography (IMAC). The column was equilibrated with 20 mM NaH2PO4, 0.5 M NaCl and 20 mM imidazole at pH 7.5 before sample loading and then washed after sample loading for 5 column volumes with 8% elution buffer (100% elution buffer contained 20 mM NaH2PO4, 20 mM NaCl, 400 mM imidazole at pH 7.5). Sample elution was done using 100% of the elution buffer and the obtained fractions were subjected to SDS-PAGE followed by WB. Fractions containing the protein of interest were pooled and concentrated using Amicon® Ultra Centrifugal Filter Units with a 3 kDa molecular weight cutoff membrane (Millipore) before size exclusion chromatography on a Superdex200 HiLoad 16/600 (GE Healthcare), which was equilibrated with 25 mM Tris-HCl pH 7.5 and 150 mM NaCl. All purification steps were performed at 4°C, and all buffers were filter-sterilized (0.22 μm). The obtained fractions were analyzed using SDS-PAGE, relevant fractions were pooled (Supplementary Figure S5) and the protein concentration of the final solution was determined using the BCA assay.
Identification of branch fucosidase activity
Incubations of fucosidases with AGP or N-glycans derived thereof were done overnight at 37°C in a reaction volume of 28 μl containing 20 μl of the bacterial lysate or a purified fucosidase solution, 1x cOmplete™ Protease Inhibitor Cocktail without EDTA, substrate, and either 20 mM Tris-HCl pH 7.5 or 20 mM ammonium acetate pH 5.2. The substrate was either intact AGP (10 μg) or N-glycans derived from this amount of AGP by prior PNGaseF release and isolation by solid-phase extraction. The N-glycans of intact AGP were then released and labeled, whereas pre-isolated glycans could be APTS-labeled immediately. In each case, sialic acids were removed from the final glycan preparations prior to CE-LIF analysis to simplify the quantification of branch fucose levels, as sialylated glycans tend to elute very close to each other in CE-LIF, and as sialylation is partial, resulting in many more glycan species than after de-sialylation.
Supplementary Material
Acknowledgments
This work was supported by a PhD Fellowship by the Flanders Agency for Innovation and Entrepreneurship [IWT, grant 111466 to H.G.] and the European Research Council [ERC-2013-CoG-616966 to N.C.].
Abbreviations
- 4-NP
4-nitrophenolate
- 4-NPF
4-nitrophenyl-α-L-fucopyranoside
- ADCC
antibody-dependent cellular cytotoxicity
- AGP
alpha-1 acid glycoprotein
- APTS
8-aminopyrene-1,3,6-trisulfonic acid
- BLAST
basic local alignment search tool
- CAZy
carbohydrate-active enzymes
- CE-LIF
capillary electrophoresis–laser-induced fluorescence
- CV
coefficient of variation
- ESR
enzyme-substrate ratio
- FcγRIIIA
Fc gamma receptor IIIA
- GH
glycoside hydrolase
- GlcNAc
N-acetylglucosamine
- Ig(G)
immunoglobulin (G)
- IMAC
immobilized metal affinity chromatography
- I-TASSER
iterative threading assembly refinement
- MUSCLE
multiple sequence comparison by log-expectation
- N-glycan
asparagine-linked glycan
- NK
natural killer cell
- PDB
protein data bank
- PNGaseF
peptide-N-glycanase from Flavobacterium meningosepticum
- POI
protein of interest
- PSGL-1
P-selectin glycoprotein ligand-1
- SLex
sialyl-Lewis x
- TIM
triosephosphate isomerase
- Tm
Thermotoga maritima
- TmF
Thermotoga maritima GH29 fucosidase
References
- Ali S, Jenkins Y, Kirkley M, Dagkalis A, Manivannan A, Crane IJ, Kirby JA. Leukocyte Extravasation: An Immunoregulatory Role for α-l-Fucosidase? J Immunol. 2008;181(4):2407–2413. doi: 10.4049/jimmunol.181.4.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, Kumagai H, Katayama T, Yamamoto K. Two distinct α-l-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 2009;19(9):1010–1017. doi: 10.1093/glycob/cwp082. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Callebaut I, Fabrega S, Lehn P, Mornon JP, Davies G. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc Natl Acad Sci U S A. 1995;92(15):7090–7094. doi: 10.1073/pnas.92.15.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Sakuma A, Kimura T, Makimura Y, Hiratake J, Sakata K, Yamanoi T, Kumagai H, Yamamoto K. Molecular Cloning and Characterization of Bifidobacterium bifidum 1,2-α-l-Fucosidase (AfcA), a Novel Inverting Glycosidase (Glycoside Hydrolase Family 95) J Bacteriol. 2004;186(15):4885–4893. doi: 10.1128/JB.186.15.4885-4893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalová T, Koval T, Benešová E, Vodicková P, Spiwok V, Lipovová P, Dohnálek J. Active site complementation and hexameric arrangement in the GH family 29; a structure-function study of α-l-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus. Glycobiology. 2019;29(1):59–73. doi: 10.1093/glycob/cwy078. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kubes P, Jutila M, Payne D. Therapeutic potential of inhibiting leukocyte rolling in ischemia/reperfusion. J Clin Invest. 1995;95(6):2510–2519. doi: 10.1172/JCI117952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerts van Bueren A, Ardèvol Albert, Fayers-Kerr J, Luo B, Zhang Y, Sollogoub M, Blériot Y, Rovira C, Davies GJ. Analysis of the Reaction Coordinate of α-l-Fucosidases: A Combined Structural and Quantum Mechanical Approach. J Am Chem Soc. 2010;132(6):1804–1806. doi: 10.1021/ja908908q. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinforma Oxf Engl. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Laroy W, Contreras R, Callewaert N. Glycome mapping on DNA sequencing equipment. Nat Protoc. 2006;1(1):397–405. doi: 10.1038/nprot.2006.60. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhu S, Ma C, Wang L-X. Designer α 1,6-Fucosidase Mutants Enable Direct Core Fucosylation of Intact N-Glycopeptides and N-Glycoproteins. J Am Chem Soc. 2017;139(42):15074–15087. doi: 10.1021/jacs.7b07906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(D1):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae M, Tsuchiya A, Katayama T, Yamamoto K, Wakatsuki S, Kato R. Structural Basis of the Catalytic Reaction Mechanism of Novel 1,2-α-L-Fucosidase from Bifidobacterium bifidum. J Biol Chem. 2007;282(25):18497–18509. doi: 10.1074/jbc.M702246200. [DOI] [PubMed] [Google Scholar]
- Ndeh D, Rogowski A, Cartmell A, Luis AS, Baslé A, Gray J, Venditto I, Briggs J, Zhang X, Labourel A, et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature. 2017;544(7648):65–70. doi: 10.1038/nature21725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang P-C, Chiu PCN, Lee C-L, Chang L-Y, Panico M, Morris HR, Haslam SM, Khoo K-H, Clark GF, Yeung WSB, et al. Human Sperm Binding Is Mediated by the Sialyl-Lewisx Oligosaccharide on the Zona Pellucida. Science. 2011;333(6050):1761–1764. doi: 10.1126/science.1207438. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Pickard JM, Chervonsky AV. Intestinal Fucose as a Mediator of Host-Microbe Symbiosis. J Immunol. 2015;194(12):5588–5593. doi: 10.4049/jimmunol.1500395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514(7524):638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC, Baslé A, Morland C, Day AM, Zheng H, et al. Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun. 2015;6:7481. doi: 10.1038/ncomms8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurama H, Fushinobu S, Hidaka M, Yoshida E, Honda Y, Ashida H, Kitaoka M, Kumagai H, Yamamoto K, Katayama T. 1,3-1,4-α-l-Fucosynthase That Specifically Introduces Lewis a/x Antigens into Type-1/2 Chains. J Biol Chem. 2012;287(20):16709–16719. doi: 10.1074/jbc.M111.333781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurama H, Tsutsumi E, Ashida H, Katayama T, Yamamoto K, Kumagai H. Differences in the Substrate Specificities and Active-Site Structures of Two α-L-Fucosidases (Glycoside Hydrolase Family 29) from Bacteroides thetaiotaomicron. Biosci Biotechnol Biochem. 2012;76(5):1022–1024. doi: 10.1271/bbb.111004. [DOI] [PubMed] [Google Scholar]
- Schneider M, Al-Shareffi E, Haltiwanger RS. Biological functions of fucose in mammals. Glycobiology. 2017;27(7):601–618. doi: 10.1093/glycob/cwx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön MP, Krahn T, Schön M, Rodriguez M-L, Antonicek H, Schultz JE, Ludwig RJ, Zollner TM, Bischoff E, Bremm K-D, et al. Efomycine M, a new specific inhibitor of selectin, impairs leukocyte adhesion and alleviates cutaneous inflammation. Nat Med. 2002;8(4):366–372. doi: 10.1038/nm0402-366. [DOI] [PubMed] [Google Scholar]
- Shaikh FA, Lammerts van Bueren A, Davies GJ, Withers SG. Identifying the Catalytic Acid/Base in GH29 α-l-Fucosidase Subfamilies. Biochemistry. 2013;52(34):5857–5864. doi: 10.1021/bi400183q. [DOI] [PubMed] [Google Scholar]
- Shi S, Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc Natl Acad Sci U S A. 2003;100(9):5234–5239. doi: 10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Meng YG, Weikert SHA, Presta LG. Lack of Fucose on Human IgG1 N-Linked Oligosaccharide Improves Binding to Human FcγRIII and Antibody-dependent Cellular Toxicity. J Biol Chem. 2002;277(30):26733–26740. doi: 10.1074/jbc.M202069200. [DOI] [PubMed] [Google Scholar]
- Shinkawa T, Nakamura K, Yamane N, Shoji-Hosaka E, Kanda Y, Sakurada M, Uchida K, Anazawa H, Satoh M, Yamasaki M, et al. The Absence of Fucose but Not the Presence of Galactose or Bisecting N-Acetylglucosamine of Human IgG1 Complex-type Oligosaccharides Shows the Critical Role of Enhancing Antibody-dependent Cellular Cytotoxicity. J Biol Chem. 2003;278(5):3466–3473. doi: 10.1074/jbc.M210665200. [DOI] [PubMed] [Google Scholar]
- Sulzenbacher G, Bignon C, Nishimura T, Tarling CA, Withers SG, Henrissat B, Bourne Y. Crystal Structure of Thermotoga maritima α-l-Fucosidase - Insights into the catalytic mechanism and the molecular basis for fucosidosis. J Biol Chem. 2004;279(13):13119–13128. doi: 10.1074/jbc.M313783200. [DOI] [PubMed] [Google Scholar]
- Summers EL, Moon CD, Atua R, Arcus VL. The structure of a glycoside hydrolase 29 family member from a rumen bacterium reveals unique, dual carbohydrate-binding domains. Acta Crystallogr Sect F Struct Biol Commun. 2016;72(Pt 10):750–761. doi: 10.1107/S2053230X16014072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai T-I, Li S-T, Liu C-P, Chen KY, Shivatare SS, Lin Chin-Wei, Liao S-F, Lin Chih-Wei, Hsu T-L, Wu Y-T, et al. An Effective Bacterial Fucosidase for Glycoprotein Remodeling. ACS Chem Biol. 2017;12(1):63–72. doi: 10.1021/acschembio.6b00821. [DOI] [PubMed] [Google Scholar]
- Van Dijk W, Havenaar EC, Brinkman-Van Der Linden ECM. α 1-Acid glycoprotein (orosomucoid): pathophysiological changes in glycosylation in relation to its function. Glycoconj J. 1995;12(3):227–233. doi: 10.1007/BF00731324. [DOI] [PubMed] [Google Scholar]
- Vanderschaeghe D, Szekrényes A, Wenz C, Gassmann M, Naik N, Bynum M, Yin H, Delanghe J, Guttman A, Callewaert N. High-throughput profiling of the serum N-glycome on capillary electrophoresis microfluidics systems: toward clinical implementation of GlycoHepatoTest. Anal Chem. 2010;82(17):7408–7415. doi: 10.1021/ac101560a. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga RK. The TIM-barrel fold: a versatile framework for efficient enzymes. FEBS Lett. 2001;492(3):193–198. doi: 10.1016/S0014-5793(01)02236-0. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43:W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollner TM, Asadullah K, Schön MP. Targeting leukocyte trafficking to inflamed skin - still an attractive therapeutic approach? Exp Dermatol. 2007;16(1):1–12. doi: 10.1111/j.1600-0625.2006.00503.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.