Abstract
One of the major concerns in the application of nanocarriers in oncology is their scarce penetration capacity in tumoral tissues, which drastically compromises the effctivity. Living organisms as cells and bacteria present the capacity to navigate autonomously following chemical gradients being able to penetrate deeply into dense tissues. In the recent years, the possibility to employ these organisms for the transportation of therapeutic agents and nanocarriers attached on their membrane or engulfed in their inner space have received huge attention. Herein, based on this principle, a new approach to deliver drug loaded nanoparticles achieving high penetration in tumoral matrices is presented. In this case, Escherichia coli (E. coli) bacteria wall is decorated with azide groups, whereas alkyne-strained groups are incorporated on the surface of mesoporous silica nanoparticles loaded with a potent cytotoxic compound, doxorubicin. Both functional groups form stable triazole bonds by click-type reaction allowing the covalent grafting of nanoparticles on living bacteria. Thus, the motility and penetration capacity of bacteria, which carried nanoparticles are evaluated in a 3D tumoral matrix model composed by a dense collagen extracellular matrix with HT1080 human fibrosarcome cells embedded. The results confirmed that bacteria are able to transport the nanoparticles crossing a thick collagen layer being able to destroy almost 80% of the tumoral cells located underneath. These findings envision a powerful strategy in nanomedicine applied for cancer treatment by allowing a homogeneous distribution of therapeutic agents in the malignancy.
Keywords: bacteria motors, mesoporous silica nanoparticles, nanocarriers tumor penetration, nanomedicine
1. Introduction
Conventional antitumoral chemotherapy is based on the administration of highly toxic drugs, which provoke the apparition of systemic toxicity in the host due to their lack of selectivity. After the paramount discovery of the selective accumulation of nanometric devices into tumoral tissues reported by Matsumura and Maeda in 1986,[1] a phenomenon known as enhanced permeation and retention (EPR) effect hundreds of different types of nanocarriers have been developed; from pure inorganic systems as metallic[2] or ceramic particles,[3,4] or organic ones like polymersomes,[5] micelles,[6] liposomes,[7] or polymer nanocapsules[8] to hybrid nanodevices which combines both natures.[9] The capacity to recognize the tumoral cells within the myriad of cells which compose a solid tumor has been provided with the incorporation of targeting moieties on the nanocarrier surface. These moieties bind to specific cell receptors located on the membrane of tumoral cells triggering the particle uptake through a specific receptor-mediated endocytosis mechanism.[10] Thus, theoretically, the nanodevices will be accumulated by EPR effect within the tumoral tissue after their injection in the bloodstream and once there, they will be engulfed by the tumoral cells due to the presence of the targeting groups on their surface. Unfortunately, despite the excellent performance of different nanocarriers in in vitro cell cultures and even in some in vivo models this behavior is not achieved in real clinical situations and only a few nanosystems have reached the market.[11] The reasons of this disappointing outcome are varied. One of them is that tumoral tissues present a dense extracellular matrix, which hampers the particle penetration constraining the therapeutic effect of the nanocarrier only on the tumor periphery.[12] The targeting capacity of the nanocarriers takes place in the nanoscale. Therefore, a proper distribution of the nanocarriers within the malignancy is compulsory to treat the whole tumoral cell population. This scenario is not present when the particles are blocked in the tumor margins.[13] Some research groups have addressed this limitation anchoring proteolytic enzymes on the nanoparticle surface in order to digest the extracellular matrix improving the particle diffusion throughout the malignant tissue.[14,15] Recently, our research group have reported the use of stimuli-responsive polymeric nanocapsules which release collagenase under the mild acidic conditions present in the tumor environment.[16] These capsules were anchored on the surface of drug loaded protocells functionalized with a specific antibody against lung cancer cells, improving their penetration into tumoral tissue and allowing the particle internalization into deep tumoral cells. Other problem which strongly limits the therapeutic efficacy of nanomedicines is the high interstitial pressure usually present in solid tumors caused by their fast growing rate that compresses the surrounding lymphatic vessels blocking the tissue drainage.[17] This pressure makes particle diffusion considerably difficult. Finally, EPR effect is not generally present in all solid tumors but it is limited to a few of them as Kaposi’s sarcoma, being only partially present or completely absent in many of them.[18] Even, EPR effect within the tumor vary during the treatment.
In the last years, the use of living systems as carrier of nanocarriers has been postulated to overcome these limitations.[19] The self-propelled and guiding capacity of cells[20] and bacteria[21] allow them to penetrate living tissues regardless hydrodynamic considerations. Among them, the use of bacteria as drug carriers has received huge attention in anti-tumoral therapy due to their excellent motion capacities.[22] Moreover, these tiny microorganisms are able to move toward gradients in response to small changes in internal stimulus (specific chemicals, temperature, or pH variations), or guided by external stimulus (magnetic field).[23–27] Thus, anaerobes bacteria are able to colonize hypoxic regions into tumoral tissues reaching inner zones of the malignancy, as has been studied in vitro and in vivo.[28–30] Herein, we report the use of Escherichia coli (E. coli) as model of facultative anaerobic bacteria for the transportation of mesoporous silica nanoparticles (MSN) loaded with a potent cytotoxic compound, doxorubicin (Dox). In this work, MSN have been used as nanocarrier in this work due to their excellent biocompatibility and lack of toxicity as has been widely proved elsewhere.[31] Bacteria wall has been decorated with azide groups through the incorporation of an azide-functionalized aminoacid (azide-D-alanine) to the culture media.[32] This aminoacid was incorporated to the bacteria wall through the normal bacteria metabolism and the presence of azide groups on the outer bacteria surface was employed for the covalent attachment of MSN decorated with strained alkynes by the well-known cooper-free azide-alkyne cycloaddition which is compatible with living systems. According to these results, it is established that these modifications do not affect to the bacterial viability.[33,34] Therefore, bacteria were able to transport them into deep zones into a 3D tumoral matrix model composed by a dense collagen extracellular matrix with the HT1080 human fibro-sarcome cells, as tumoral cell line model, embedded within their structure. The homogeneous MSN distribution allowed by the action of bacteria as carriers yielded a significant improvement in the therapeutic effect due to Dox release in comparison with free particles. This proof of concept opens the way to treat solid tumors employing this strategy using nonpathogenic bacteria. Therefore, it would be possible to utilize organisms as nanoparticle carriers capable to follow specific gradients within solid tumors as hypoxic zones or acidic environments improving the distribution of nanomedicines within the malignancy.
2. Results and Discussion
2.1. Synthesis and Characterization of MSN-PEG-DBCO
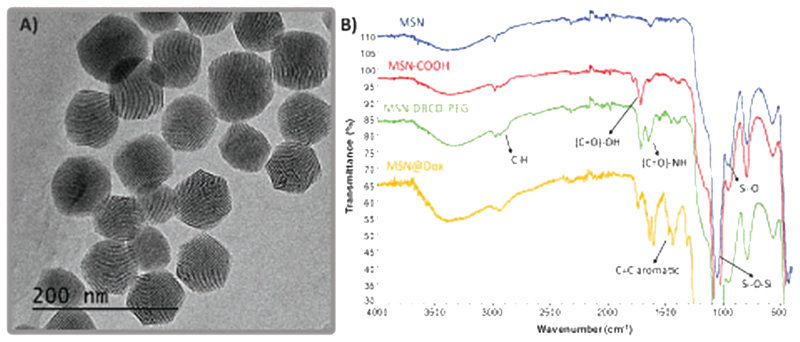
Fluorescent MSN were synthetized by covalently linking the fluorescent dye to the silica network following a modified Stöber method[15] yielding monodisperse nanoparticles with an average diameter of 80-100 nm and a pore size of 2–3 nm. The functionalization of MSN materials was carried out in several steps. First, MSN were functionalized with (3-triethoxysilyl)pro-pylsuccinic anhydride (TESPSA) providing carboxylic groups on the particle surface (MSN-COOH), which allow to bind dibenzylcyclooctyne-PEG4-Amine (DBCO-PEG-NH2) by amide formation via carbodiimide coupling to yield MSN-PEG-DBCO. DBCO is a biorthogonal group that allows the attachment of different biomolecules thanks to the specific reaction with azide derivatives yielding stable triazoles in mild conditions.[35] The hydrophilic polyethylene glycol (PEG) spacer provides a large and flexible connection that minimizes steric hindrance in the ligation of MSN-DBCO to complementary azide-containing molecules and also avoids the aggregation of MSN by steric hindrance.[36] Regarding MSNs degradation, our research group has previously reported that degradability of MSN depends on numerous factors, as nanoparticle diameter, pore type, and surface functionalization.[37] This research work states that 200 nm MSNs dispersed in PBS are partially degraded after a few days in aqueous suspension but the structural integrity of the nanoparticle remains almost invariable for about a week. In addition, the degradation product leached in form of orthosilicic acid is harmless. Dynamic light scattering (DLS) measurements of the obtained MSN-PEG-DBCO revealed an average particle diameter of 91–105 nm (see Figure S1A, Supporting Information). Figure 1A displays the transmission electron microscopy (TEM) image corresponding to MSN-PEG-DBCO sample, showing a nearly hexagonal shape with an average diameter of 100 nm and monodispersed distribution.
Figure 1.
A) TEM micrography corresponding to MSN-PEG-DBCO sample. B) FTIR spectra corresponding to MSN, MSN-COOH, MSN-PEG-DBCO, and MSN@Dox samples.
Fourier transform infrared (FTIR) spectroscopy was employed to verify the successful functionalization with carboxylic groups and DBCO-PEG-NH2, respectively. For pristine MSN, the characteristic absorption bands of the tetrahedron silica structures at 1100 cm−1 (Si–O–Si bending) and 800 cm−1 (Si–O stretching) were clearly observed. After the functionalization with TESPSA, the peaks corresponding to C(=O) –OH stretching vibrations appeared at =1700 cm−1. Finally, the DBCO-PEG on the silica surface was confirmed by the apparition of the characteristic amide band at =1650 cm−1 (N–H stretch) and a slight increase in C–H alkane band (2850–2950 cm−1) (Figure 1B). The organic content of MSN materials was quantified by thermogravimetric analysis (TGA) measurements to follow the incorporation of the organic material (see Figure S1C, Supporting Information). TGA indicates a slight increase in the organic matter after each synthetic step, and Z potential analysis at neutral pH indicates an alteration in the surface charge from −20 mV in the case of naked MSN to −24 mV in MSN-PEG-DBCO. Finally, MSN-PEG-DBCO was incubated with a slight excess of TAMRA-Azide (10 μL of a 1 mg mL−1 solution) in order to evaluate the capacity of these functional groups to attach a biomolecule model, in this case TAMRA fluorophore. The particles which contained the strained alkyne on their surface showed 13-fold higher fluorescence than nonfunctionalized ones, confirming the presence on the nanoparticles surface of DBCO and its reactivity. The amount of DBCO present in the nanoparticles (0.72 μg DBCO-PEG-NH2/mg MSN-PEG-DBCO) was determined from the TAMRA fluorescence obtained for MSN-PEG-DBCO (see Figure S2, Supporting Information).
2.2. Attachment of MSN-PEG-DBCO to Bacteria (Bac-MSN)
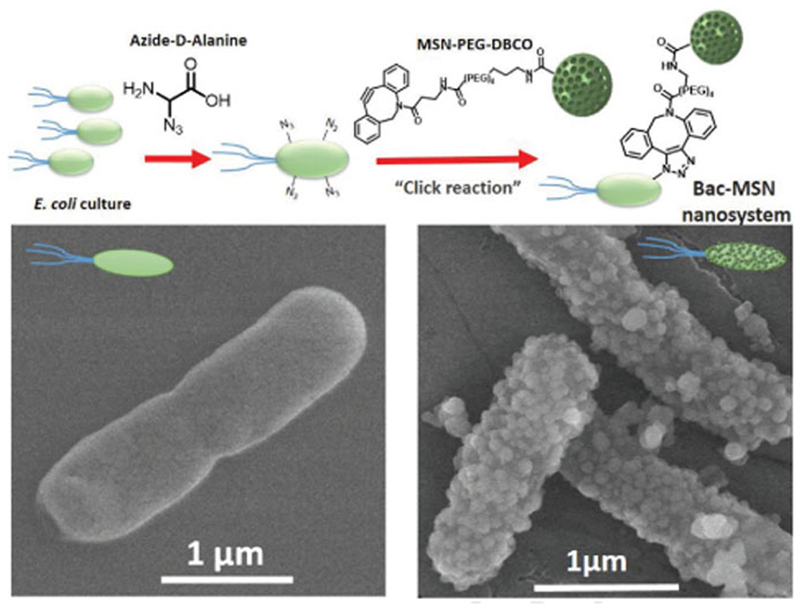
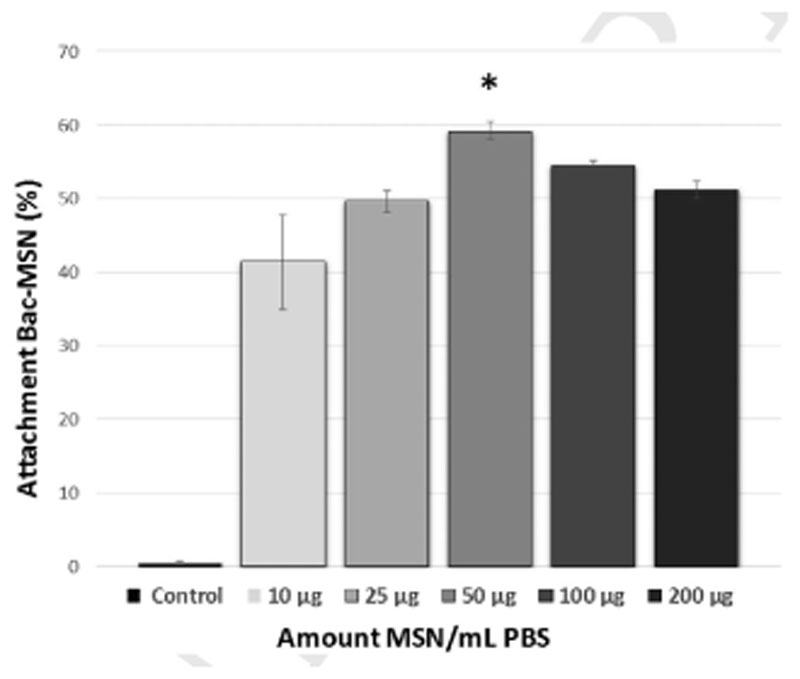
The attachment of a nanoparticle on the surface of a living cell requires the use of chemoselective reactions which can be carried out in mild conditions in the presence of the myriad of compounds that usually constitute the biological fluids. In the recent years, a vast arsenal of these types of reactions, encompassed within the term of bio-orthogonal chemistry, have been developed.[38] Among them, cooper-free strain-promoted azide-alkyne cycloaddition is one of the most widely employed reactions with this purpose thanks to its exquisite selectivity, high yield, and absence of the toxicity caused by the metal which allow its use, even into complex organisms.[39] The first step required for exploiting this chemistry to attach the nanoparticles on the bacteria surface is the incorporation of azide groups on their surface. This requirement was fulfilled growing bacteria in a culture medium enriched with azide-D-alanine. Bacteria incorporates this aminoacid in the structure of peptidoglycans of their cell wall and therefore, it places the azide groups on the bacteria surface. The presence of azide groups on the bacteria wall was evaluated adding DBCO-TAMRA to the cell culture media. Bacteria which were previously incubated with azide-D-alanine exhibited a bright red fluorescence by fluorescence microscopy, whereas bacteria that were cultured without this amino acid did not incorporate the fluorophore and therefore, fluorescence was not visible (see Figure S3, Supporting Information). Once confirmed the presence of azide groups on the bacteria surface, the attachment of MSN-PEG-DBCO was evaluated. Thus, azide-decorated bacteria cultures (O.D. = 0.25) were incubated in the presence of different nanoparticle batches with increasing nanoparticle concentrations for 1.5 h (Figures 2 and 3). After the incubation time, the excess of nanoparticles was removed by washing/centriflugation steps and the attachment of MSN on bacteria wall was determined by flow cytometry analysis (FACS). Bacteria without nanoparticles were used as control. To avoid bacteria growing, each batch was incubated with Paraformaldehyde (3% w/v) overnight previous to FACS analysis. For the detection of bacteria in the cytometry equipment, they were stained with Propidium iodide (PI) as red fluorescence labeling. Nanoparticle attachment to bacteria was evaluated by measuring the green fluorescence (due to FITC-labeled MSN) percentage ratio obtained in the red-positive bacteria (+,+ fluorescence) dot-plots histogram region (see Figures S4–S11, Supporting Information). The results indicate that the highest attachment rate was yielded by the concentration of 50 μg MSN mL−1 (=60%), whereas this amount was reduced when higher nanoparticle concentrations were employed, probably caused by the formation of nanoparticle aggregates in these high concentrated suspensions, as shown in Figure 3. The presence of nanoparticles on bacteria surface was confirmed by scanning electron microscopy (SEM), which showed a homogeneous distribution of nanoparticles in the bacteria wall comparing to bacteria without nanoparticles (Figure 2) and demonstrating that nanoparticles are present in the majority of bacteria (see Figure S12, Supporting Information).
Figure 2.
Upper image shows a schematic procedure of “click reaction” between Bacteria-N3 and MSN-PEG-DBCO nanoparticles. Lower images show SEM micrographs of nude bacteria (left) and bacteria with MSN attached on their surface (right).
Figure 3.
Flow cytometry corresponding to percentage of bacteria which incorporate MSN-PEG-DBCO on their surface depending on the nanoparticle concentration. Bars represent mean ± standard error of the mean (n = 3). *p < 0.05 versus 10 and 25 μg of MSN. (Student’s two-tailed t-test).
2.3. Evaluation of Mobility and Penetration Capacity of Bac-MSN in 3D Nutrient-Rich Collagen Gels
2.3.1. Mobility of Bac-MSN toward Gradient Assay
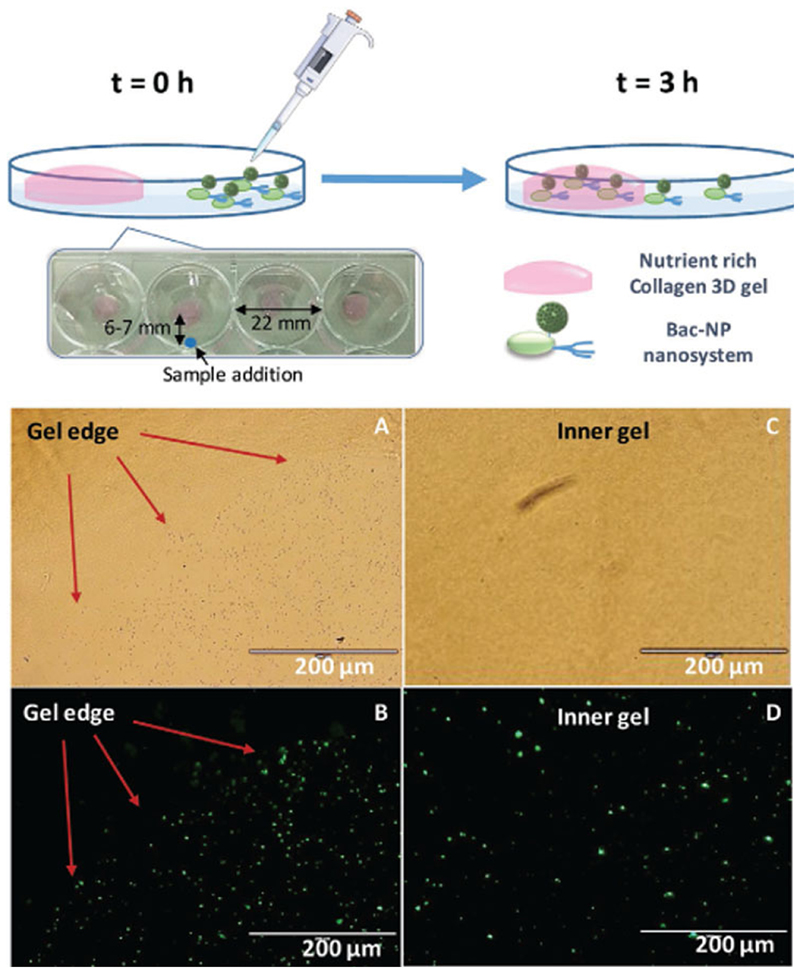
The capacity of bacteria to transport nanosystems to different locations has been previously demonstrated, showing that an anchored nanosystem not affect significantly to the mobility capacity of E. coli comparing with free bacteria.[40] Mobility and penetration capacity of bacteria with MSN attached on their surface (Bac-MSN) was evaluated employing 3D collagen matrices enriched in nutrients that act as bacteria chemoattractors. In this case, the collagen matrix was impregnated with a nutrient media composed by complemented Dulbecco’s modified Eagle’s medium (DMEM) with 10% of fetal bovine serum (FBS) and L-glutamine prepared without antibiotic to preserve the bacteria alive. These gels were placed at a corner of 12-well plates and PBS was added as medium for bacteria migration. Simultaneously, in the opposite corner, 100 μL of a suspension of Bac-MSN was added and incubated at room temperature (RT) in static conditions during 3 h (Figure 4). Thus, thanks to the capacity of bacteria to navigate toward favorable locations, the migration of bacteria to the 3D collagen gel transporting with them the nanoparticles attached were monitored employing fluorescence microscopy, observing fluorescence due to nanoparticles which were present in the collagen gel after the incubation time. Previous to the microscopy observation, gels were washed three times with antibiotic-containing PBS (0.1 mg mL−1 of Levofloxacin) to kill the bacteria and gels were incubated with paraformaldehyde (3% w/v) overnight to freeze the position of the nanoparticles in the gel. The results indicate that when the nanoparticles were transported on bacteria surface, they were present on and inside the gel in higher amount than in the case of free nanoparticles. Figure 4 shows a time comparison of the migration process of Bac-MSN. At the beginning of the experiment (time = 0 h, Figure 4A), gels are completely free of nanoparticles while after 3 h, the high presence of nanoparticles in the whole inner gel in form of green dots was evidently visible by fluorescence microscopy (Figure 4D).
Figure 4.
Top: Motility assay scheme of Bac-MSN in a nutrient-rich collagen 3D gel. Bottom: Microscopy images of a gel edge (left images) with Bac-MSN added at t = 0 h in transmittance A) and fluorescence microscopy B), in which nanoparticles correspond to green dots. Right images show the central area of the same gel at t = 3 h in transmittance C) and fluorescence microscopy D) images.
2.3.2. Penetration of Bac-MSN in 3D Gel Assay
One of the most important properties which can be achieved by self-propelled nanocarriers is their high penetration in tumoral tissues. Bacteria can transport nanoparticles to specific zones as tumoral tissues due to their ability to detect chemical gradients and additionally, they can penetrate deeply into the diseased tissue. This last property is of paramount importance because to achieve a homogeneous nanocarrier distribution in the target tissue is a compulsory requirement for improving the efficacy of nanomedicines. In order to evaluate the penetration of Bac-MSN in a tumoral matrix-mimic model, a 3D collagen gel with tumoral cells embedded within a nutrient-rich organic matrix has been employed. In this case, we have incorporated into the collagen gel HT1080 human fibrosarcome cells because the rheological properties of the resulting gel which contains tumoral cells embedded are close to real tumoral matrix, as has been reported elsewhere.[41] Thus, due to the nutrient guiding capacity of bacteria, the nanoparticles carried by these organisms could reach inner zones of the 3D matrix. It is important to remark that this is a simple model that has been applied only to test the bacteria capacity to transport nanoparticles without losing their motility.
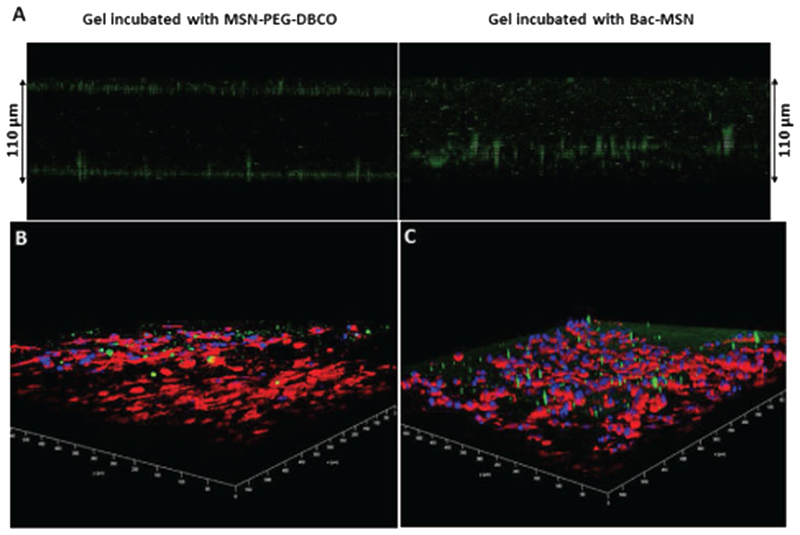
Bac-MSN were placed at the top of collagen gels and gels were incubated during 3 h. Free MSN-PEG-DBCO was employed as a control. After this time, gels were thoroughly washed to remove the particles which did not penetrate the collagen matrix and the particles were visualized employing confocal microscopy. Free particles were accumulated on the gel edges being not capable to penetrate the collagen matrix. Particles attached on the bacteria surface were localized in the whole tissue, which indicate that bacteria were able to penetrate the collagen matrix colonizing the gel, as can be observed in Figure 5A. In order to visualize more properly the location of the particles and the distribution of tumoral cells in the matrix model, HT1080 cells were stained with Phalloidin-Atto 565 that label in red the actin filaments and the cell nuclei were stained with DAPI (blue). Similar to the previous assay, the green dots which correspond to the fluorescein labeled nanoparticles were distributed in the whole tissue only in the case of Bac-MSN (Figure 5C), while they remain on the periphery of the gel in the case of free particles (Figure 5B).
Figure 5.
A) Confocal microscopy Z-axis projection of MSN-PEG-DBCO and Bac-MSN placed on 3D collagen gel. B,C) Confocal microscopy 3D panoramic view of collagen gels which contains HT1080 human fibrosarcoma cells embedded incubated with MSN-PEG-DBCO B) and Bac-MSN C), respectively. Green dots correspond to FITC-labeled MSN, red color corresponds to actin filaments and blue color correspond to cell nuclei.
2.4. Evaluation of Antitumoral Properties of Bac-MSN@Dox in 3D Collagen Tumoral Matrix Model
2.4.1. Doxorubicin Loading into MSN (MSN@Dox) and Bacterial Inhibition Assay
Once the mobility and penetration capacity of Bac-MSN were validated, their capacity to transport antitumoral agents to the tumoral space was tested loading the MSN with a potent cytotoxic drug, Dox, which is widely used in oncology. Its hydrophobicity becomes an ideal drug for the loading into MSN, due to the release from the porous at physiological conditions is slow.[42] However, at mild acidic environment, as is usually present in most of the solid tumors,[43] Dox release from the MSN is around 2.5-fold times, so it has a pH-responsive behavior by itself. Thus, MSN were loaded with Dox by immersion in a highly concentrated Dox solution in PBS (6 mg mL−1), yielding nanoparticles loaded with Dox. The release profiles of Dox were obtained at different pH (7.2 and 5.5, respectively), as are shown in Figure S13A of the Supporting Information, confirming the pH-responsive behavior of these Dox-loaded nanoparticles.
Additionally, the influence of the released Dox in bacteria growth and viability was evaluated exposing them to the Dox released from MSN at 50 µg mL−1 of concentration during the time of the anchoring experiment (4 h). For it, we performed a dilution series (from 10-5 to 10-7) and we pipetted five replicas of 10 µL onto agar plates for each dilution, and then we counted the number of colony forming units (CFU) for the 10-5 dilution, where the number of colonies were statistically representative (between 30 and 300 CFU). Data shown a minor decrease in the bacterial viability of 20.5% (see Figure S13B, Supporting Information). These results agree with other data reported by other research groups which indicate that the presence of Doxorubicin does not affect significantly neither growth nor viability of bacteria.[25]
2.4.2. Evaluation of Antitumoral Potential of Bac-MSN@Dox
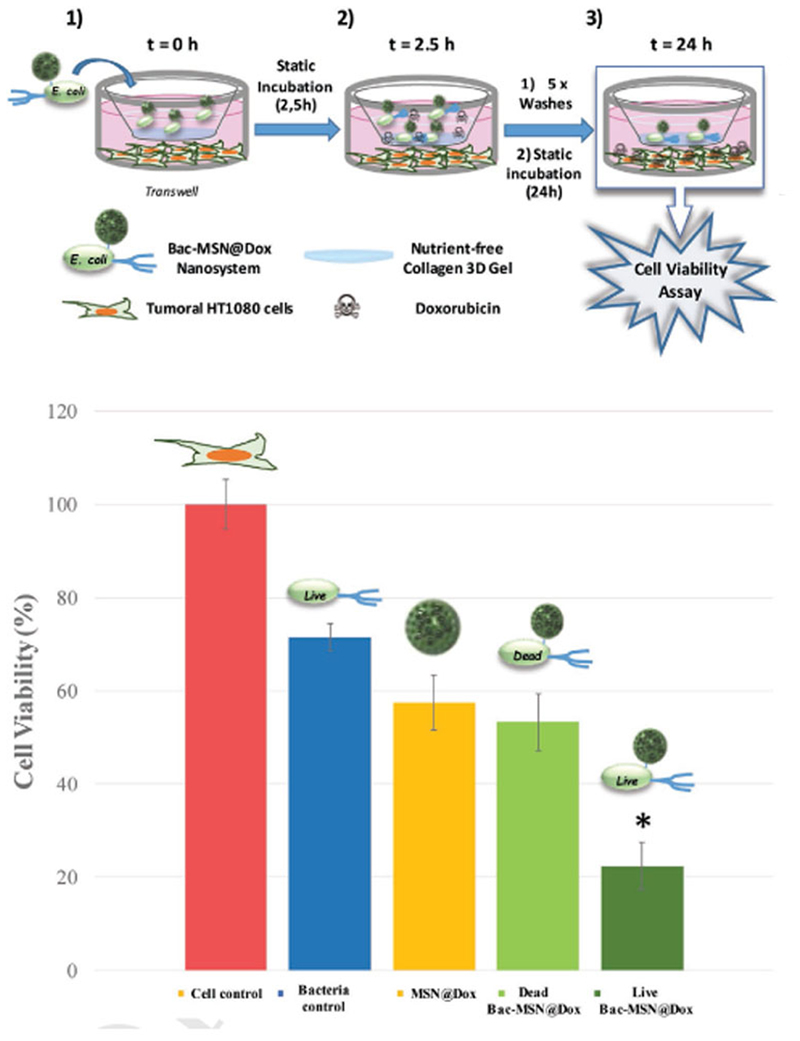
The capacity of bacteria to transport Dox-loaded MSN (Bac-MSN@Dox) was tested employing a homemade tumoral matrix model composed by a Transwell in which a collagen shell of 200–300 µm was grown on the top Transwell insert membrane, while HT1080 cells were seeded at the bottom (see Figure 6). The role of this collagen shell is to act as a barrier which hamper the nanoparticles to reach inner zones. Bac-MSN@Dox (Live) was placed on the top of the collagen gel immersed in PBS and after an incubation time of 2.5 h, the collagen surface was thoroughly washed with PBS to remove the particles which were not able to penetrate into the collagen shell, and the cell culture media at the bottom of the Transwell was replaced and repeatedly washed to eliminate the Dox prematurely released during this time. The cells were incubated for 24 h to let time for Dox release and in the presence of antibiotic in order to kill the bacteria which can interfere in the cell viability evaluation. In this assay, the bacteria role is only to introduce deeply the nanoparticles into the gel. Free nanoparticles are not capable to penetrate into the collagen gel, as was tested in the previous experiments, and therefore, they were rapidly removed from the collagen surface. Cell viability was determined by Alamar blue assay. Different controls were carried out in parallel placing at the top of collagen gel: bacteria alone, MSN@Dox, and Bac-MSN@Dox (Dead) (incubated in PBS with Levofloxacin), and were treated following the same protocol in order to evaluate the influence of each system. Graphic of Figure 6 shows that the addition of bacteria alone provokes a slight decrease in cell viability, which was around 70%. MSN@Dox induced a more pronounced effect due to Dox released by the particles, which remained adsorbed on the collagen surface after the washing steps. This effect is almost the same as that achieved by the MSN@Dox attached on dead bacteria, due to their obvious lack of motility (57% and 53% of cell viability, respectively). Finally, Bac-MSN@Dox exhibited the best performance, yielding a significant tumoral cell mortality being able to destroy almost 80% of the tumoral cells. The presence of fully functional bacteria allows the penetration of the nanocarriers deeply into the collagen matrix resisting the washing steps. The continuous Dox release from the mesoporous silica matrix fixed there induce an efficient antitumoral effect over time.
Figure 6.
Upper image shows the schematic procedure for cytotoxicity experiment: 1) Transwell with HT1080 human fibrosarcoma cells seeded in the lower compartment, and 3D collagen gel covering the upper compartment of the insert. Sample is added in the surface of this gel. 2) After 2.5 h static incubation, bacteria had penetrated and are retained inside the gel. The lower and upper part of the Transwell were abundantly washed and incubated with fresh culture medium. 3) 24 h after incubation, bacteria retained inside the gel have liberated the Doxorubicin loaded into the MSN and it has diffused to the culture medium. Cell viability was measured at this point. Lower graphic show cytotoxicity results for each control and sample. Bars represent mean ± standard error of the mean (n = 3). *p < 0.05 versus Dead Bac-MSN@Dox; MSN@Dox, and Cell control (Student’s two-tailed t-test).
2.5. Evaluation of Immunogenic Response Against the Nanosystem Bac-MSN
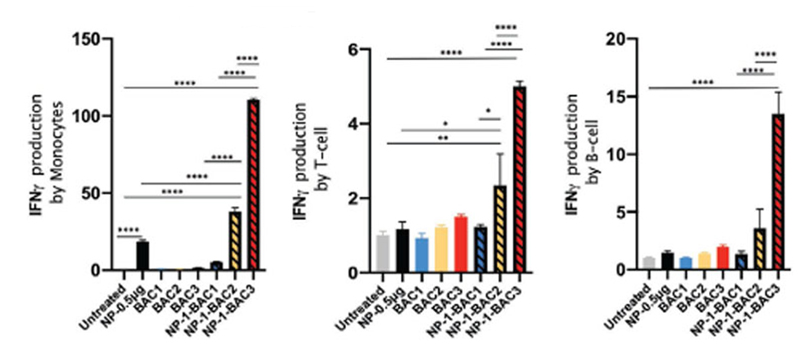
For future biomedical assays, it is very important to evaluate the immune response caused by our nanosystem. In addition to producing toxicity due to the release of Doxorubicin, we could provoke an activation of the immune system that could be a new weapon to achieve better clinical outcomes against the tumor. To elucidate whether our nanodevice (Bac-MSN) induces any immune system response, we detected intracellular Interferon gamma (IFN-γ) production by different immune cells. Mononuclear cells isolated from peripheral blood of a healthy donor were cocultured with our nanodevice at scaling doses. After more than 5 h in presence of Golgi apparatus inhibitor (Monensin), IFN-γ were detected within immune cells by flow cytometry. Serial gating of the different circulating populations (Figure S14, Supporting Information,) showed an ≈100-fold accumulation of IFN-γ in monocytes in response to the highest dose of our nanodevice and ≈20-fold with nanoparticles alone (Figure 7-left). Both T-cells and B-cells showed ≈20 and ≈7-fold less of intra-cellular IFN-γ, suggesting that monocytes are more sensitive, while not significance accumulation was observed in presence of nanoparticles (MSN-PEG-DBCO) alone (Figure 7 center and right). On the other hand, light responses are presented in all populations in presence of high dose of bacterium. Altogether, we observed that our biohybrid nanodevice Bac-MSN (NP-BAC in graphs) produce IFN-γ in a synergic manner in all immune cells, particularly strong retention in monocytes.
Figure 7.
Graphs represent the percentage of IFN-γmeasured in monocytes (left), T lymphocytes (middle), and B lymphocytes (right) by flow cytometry. BAG: 1.91 × 105 cells mL−1; BAC2: 1.91 × 106 cells mL−1; BAC3: 1.91 × 107 cells mL−1. NP-0.5 μg: 0.5 μg. NP-BAC1: 0.05 p,g-1.91 × 105 cells mL−1. NP-BAC2: 0.5 p,g-1.91 × 106 cells mL−1. NP-BAC3: 5 p,g-1.91 × 107cells mL−1. *p < 0.05, **p < 0.01, ****p < 0.0001.
3. Conclusions
In conclusion, a novel strategy to enhance the penetration of drug loaded mesoporous silica nanoparticles in 3D collagen tumoral matrix models through their covalent attachment on the surface of living bacteria have been presented. Bacteria present a remarkable tumor-homing capacity guided by the specific conditions present there, such as the necrotic and hypoxic environment. These tiny organisms have been widely studied for cancer treatment due to their potential capacity to reach even the hardly accessible core of solid tumors.[26]
In this work, bacteria wall was decorated with azide groups to allow the covalent anchorage of mesoporous silica nanoparticles functionalized with strained cycloalkynes click-type reactions. The high chemoselectivity of the azide-alkyne cycloaddition allowed the easy incorporation of nanoparticles loaded with cytotoxic drugs in mild conditions on the surface of living bacteria. The capacity to transport the nanoparticles to deep zones of tumoral matrix was evaluated employing 3D tumor models composed by a dense collagen matrix which contains human fibrosarcoma cells embedded into the organic matrix. The results indicated that nanoparticles carried by bacteria were homogeneously found in the whole tissue, even in deep zones, whereas free nanocarriers were concentrated only in the periphery. This fact allowed an improved release of cytotoxic drugs which caused a significantly higher tumoral cell mortality in comparison with free nanoparticles (80% vs 40%, respectively).
The immune responses obtained in the immunogenic analysis indicate how our nanosystem is able to activate innate and acquire immune cells suggesting a future use for reactivating the intratumoral immune cells which are silenced by tumor microenvironment. Moreover, the capacity of bacteria to transport nanoparticles within dense collagen matrices would be a valuable strategy for improving the efficacy of nanomedicines in the fight against this disease. For a future application in in vivo models, several ways to control the bacterial pathogenicity should be considered, as the employ of attenuated or genetically modified bacteria, the employ of antibiotics after treatment, or the adjust or the bacteria dose.
4. Experimental Section
Materials and Strains
Hexadecyltrimethylammonium bromide (CTAB), Tetraethyl orthosilicate (TEOS), and dry Toluene were purchased from Sigma-Aldrich. Ammonium hydroxide (NH4OH, 28–30%) from Fluka; (3-aminopropyl)triethoxysilane (APTES), TESPSA, and Fluorescein isothiocyanate, isomer I (FITC) were purchased from Abcr; Absolute Ethanol from Panreac; DBCO-PEG-NH2, and (R)-2-Amino-3-azidopropanoic acid hydrochloride (azide-D-alanine) were purchased from Jena Bioscience Gmbh. N,N'-Diisopropylcarbodiimide (DIC), N-Hydroxysuccinimide (NHS), TAMRA-Azide, Ammonium nitrate (NH4NO3), and Dibenzocyclooctyne-PEG4-Fluor 545 (TAMRA-DBCO) from Sigma-Aldrich.
DMEM, Luria-Bertani (LB) medium, Trypsin, FBS, and Antibiotic-Antimycotic (Anti-Anti) 100x from GIBCO. Rat tail Collagen (type I) Life Technologies S.A.; 10X PBS Buffer solution pH = 7.4 from Ambion. Sodium hydroxide (NaOH), Dimethyl sulfoxide (DMSO), Paraformaldehyde, Phalloidin-Atto 565, 4',6-diamidino-2-phenylindole (DAPI), and Doxorubicin hydrochloride (Dox) from Sigma-Aldrich; Alamar blue solution from Invitrogen. E. coli strain Seattle 1946 (ATCC 25 922) was used as a model of Gram negative bacteria. HT1080 human fibrosarcome cell line was obtained from ATCC (ATCC CCL-121).
Synthesis of Mesoporous Silica Nanoparticles
To prepare green fluorescent dye-labeled MSN, 1.5 μL of APTES was added to a solution of 1.5 mg of FITC dissolved in 1 mL of absolute ethanol and was kept under magnetic stirring during 1 h at RT in the dark. Separately, 0.290 g of CTAB was dissolved 150 g in a solution of ammonium hydroxide (NH4OH) 0.22 m and was incubated at 50 °C under magnetic stirring for 1 h in a 250 mL beaker sealed with parafilm. Then, both 3 mL of 0.88 m TEOS in ethanol and FITC-APTES solution were added to the surfactant solution after adjusting the stirring speed to 600 revolutions per minute (rpm). The mixture was incubated at 50 °C under magnetic stirring for 1 h without parafilm, then the solution was aged at 50 °C overnight under static conditions. Subsequently, the nanoparticles were subjected to a hydrothermal treatment at 70 °C for 20 h before being collected by centrifugation and washed three times with water and ethanol, respectively. The ion exchange method was employed for the removal of the surfactant by placing the particles in 500 mL of a solution of 95% ethanol, 5% water, and 10 g of NH4NO3 per liter at 80 °C overnight. The MSNs were washed three times with ethanol and were finally stored in pure ethanol.
Functionalization of the MSNs with DBCO-PEG-NH2 (MSN-PEG-DBCO)
The nanoparticles surface was functionalized in consecutive steps. First, MSNs were functionalized with TESPSA providing functional carboxylic groups (MSN-COOH), which allow the subsequent binding of DBCO-PEG-NH2 trough carbodiimide-mediated coupling to yield MSN-PEG-DBCO. For the obtention of MSN-COOH, 120 mg of dried MSNs were dissolved in 10 mL of anhydrous toluene (99.8%) under N2 atmosphere and gently scattered in an ultrasound bath. After vigorous stirring during 30 min at 80 °C, 18 mg of TESPSA dissolved in 5 mL of anhydrous toluene was added to the mixture. The resulting mixture was stirred overnight at 110 °C under N2. Then, the sample was collected by centriflugation at 13 000 rpm for 1 h and washed three times with toluene and ethanol and dried under vacuum. After that, 15 mg of MSN-COOH was suspended in N,N-dimethylformamide (DMF) and 0.035 mmol of DIC and 0.038 mmol of NHS were added. This mixture was stirred for 30 min under N2 atmosphere at RT. Then, 0.015 mmol of DBCO-PEG-NH2 was added and stirred overnight at RT. The obtained nanoparticles were washed with DMF, H2O, and absolute ethanol, respectively, and were finally stored in absolute ethanol. The resulting samples were named as MSN-PEG-DBCO.
The resulting materials were deeply characterized through different techniques as: DLS to determinate the size distribution and electrophoretic mobility measurements to calculate the values of zeta-potential (Zetasizer Nano ZS equipped with a 633 nm “red” laser); attenuated total reflectance (ATR)-FTIR Thermo Nicolet nexus equipped with a Goldengate attenuated total reflectance device) for the identification of functional organic groups; thermogravimetric and differential thermal analysis (Perkin Elmer Pyris Diamond TG/DTA analyzer, with 5 °C min−1 heating ramps, from R to 600 °C) for the quantification of the loss of organic mass of each sample; and TEM (JEOL JEM 2100 instrument operated at 200 kV, equipped with a CCD Camera (KeenView Camera) to observe the morphology and size of nanoparticles.
Anchoring of MSN-PEG-DBCO to the E. Coli Surface (Bac-MSN)
E. coli strain was routinely grown at 37 °C overnight after inoculation of 20 μL of E. coli into 10 mL of LB medium. This overnight culture was diluted 1:5 with LB medium until obtaining a suspension with optical density (OD600) of 0.25 (2 × 108 cells mL−1). Then, 10 μL of a 10 × 10−3 m of azide-D-alanine solution in DMSO were added to the E. coli suspension and incubated at 37 °C with slight orbital stirring for 1 h. After that, it was centrifuged two times at 8000 rpm for 5 min and suspended with PBS. Afterward, the resulting bacteria suspension was incubated with 50 μL of well-dispersed MSN-PEG-DBCO (1 mg mL−1) suspension in PBS at 37 °C with slight orbital stirring for 1.5 h. Finally, it was centrifuged two times at 8000 rpm for 5 min and suspended with PBS to obtain the Bac-MSN with an ending OD600 of 0.291 (2.33 × 108 cells mL−1).
For the evaluation of the binding efficiency of MSN to Bacteria by fluorescence-activated cell sorter (FACS, Bac-MSN was suspended in a 3% paraformaldehyde solution during 1 h at RT to avoid the growth of bacteria. Then, for the detection of bacteria in the cytometry equipment (FACSCalibur, Becton Dickinson), they were stained with PI as red fluorescence labeling.
This anchoring was also examined by SEM. To observe it, Bac-MSN were suspended with a 3% paraformaldehyde solution for 1.5 h. Then, it was dehydrated in increasing series of water–ethanol solutions (10-100%), and finally air-dried in vacuum. The samples were sputter coated with a layer of gold and examined in a JEOL JSM 7600F
Preparation of 3D Collagen Gels Enriched with Nutrients
Briefly, 2 mL of Rat Tail Collagen type I (3 mg mL−1) and 0.6 mL of complemented DMEM (DMEM medium with 10% FBS and L-Glutamine) were mixed at 0 °C and subsequently 100 μL of a 2 m NaOH solution was added until neutral pH. Then, 0.5 mL of FBS and 1 mL of complemented DMEM were added to the solution at 0 °C. Afterward, 0.5 mL of this mixture was added to the wells of a 24-well plate and incubated at 37 °C at an atmosphere of 5% CO2 for 1 h to promote the collagen gelification. To avoid gel cracking, the edge of each well was encircled with a needle. Finally, 250 μL of complemented DMEM medium was added, and incubated at 37 °C at an atmosphere of 5% CO2 overnight. The resulting collagen gels were employed for the further experiments 1 day after gels formation.
For the collagen gel containing HT1080 human fibrosarcoma cells, the cells were embedded into the collagen matrix following the same procedure by adding 1 mL of complemented DMEM with a suspension of cells (5 × 105 cells mL−1) instead of complemented DMEM alone. As well as above gels, they were used 4 days after for the confocal microscopy (Leica SP2) analysis with the aim of evaluating the bacteria penetration experiment.
In Vitro Bacteria Penetration and Mobility Assay
The previously prepared 3D collagen gels enriched with nutrients were transferred to the corner of a well of a 12-well plate and 0.4 mL of 1X PBS was added to cover it. Straightaway, 100 μL of freshly prepared Bac-MSN sample was added in the other corner of the well (≈6–7 mm), and incubated at RT for 3 h without agitation. Then, the supernatant was removed and gel was washed three times with PBS. This experiment was monitored by EVOS fluorescence microscopy. The same procedure was employed for the gel penetration experiment, but instead of adding the nanosystem on one side of the transwell (mobility assay), it was added over the top of the gel. Then, gels were incubated in a 3% w/v paraformaldehyde solution overnight. Next day, cell-containing collagen gels were stained with 10 μL of Phalloidin-Atto 565 (10 μg mL−1 solution) and 1 μL DAPI (1 μg mL−1 solution) and washed two times with PBS. Thereafter, gels were analyzed by confocal microscopy at different depths.
MSN-PEG-DBCO Loading with Doxorubicin (MSN@Dox) and Effect on Bacterial Viability
2.5 mL of a solution of Dox in PBS (6 mg mL−1) was added to a well-dispersed suspension of MSN-PEG-DBCO in 2 mL of PBS (5 mg mL−1) and were stirred in an orbital shaker overnight at RT. Then, Dox-loaded MSN-PEG-DBCO (MSN@Dox) were centrifuged at 13 000 rpm for 10 min and washed 5 x PBS. Final centrilugation pellet was stored at 4 °C. Loading capacity of nanoparticles was confirmed by Infrared spectroscopy (FTIR) and quantitatively evaluated by TGA and measuring the fluorescence of Dox (λ exc = 480 nm; λ em = 600 nm) liberated along the time (Figures S1C and S13A, Supporting Information).
To evaluate the effect of MSN@Dox on bacterial viability, E. coli strain was routinely grown at 37 °C overnight after inoculation of 10 μL of E. coli into 5 mL of LB medium. This overnight culture was diluted with LB medium until obtaining a suspension with a OD600 of ≈0.25. Then, 50 μL of a MSN@Dox suspension (1 mg mL−1) in PBS were added to the E. coli suspension and incubated at 37 °C with orbital stirring for 4 h (same conditions as in the procedure of anchoring nanoparticles to bacteria). After that, a dilution series of the mixture (from 10−5 to 10−7) was performed and 10 μL of each dilution onto agar plates was pipetted and, thereafter it was incubated at 37 °C overnight. Next day, the number of CFU was determined (Figure S13B, Supporting Information).
Preparation of 3D Collagen Covered Transwell as Tumoral Matrix Model
For this study, the collagen gels were prepared without nutrients in the same way that has been described before, but instead of adding DMEM medium, it was replaced by PBS on each step. Then, 0.2 mL of this mixture was added to each Transwell insert of a 12-well plate and was incubated at 37 °C at an atmosphere of 5% CO2 overnight to promote the gelification, obtaining a gel thickness of 200 μm. At the same time, 20 000 HT1080 human fibrosarcoma cells cm−2 were seeded at the bottom of each well of a 12-well plate with complemented DMEM medium at 37 °C in an atmosphere of 5% CO2.
In Vitro Cytotoxicity Assay in Tumoral Matrix Model
After 24 h, wells of 12-well plate seeded with cells were washed three times with PBS and 1.5 mL of complemented DMEM without Anti-Anti was added. Previously prepared gel-containing transwells were transferred to this 12-well plate with cells, and 100 μL of the corresponding sample suspension (Live Bac-MSN@Dox, Dead Bac-MSN@Dox, MSN@Dox [1 mg mL−1], Bacteria, and PBS control) with a OD600 of 0.239 (1.91 × 108 cells mL−1) were added on top of each gel-containing Transwell insert.
Cells were incubated 2.5 h at 37 °C at 5% CO2 atmospheric concentration, and then both gel-containing Transwell inserts and cell-containing wells were washed five times with PBS to remove the released Dox. Cell medium was refreshed with 1.5 mL of DMEM with Anti–Anti and 100 μL added to the gel-containing inserts. It was incubated at 37 °C at 5% CO2 atmospheric concentration during 24 h.
After 24 h, the cytotoxic capacity of Dox was evaluated with Alamar Blue test. For that, cells supernatant was removed and cells were washed two times with PBS. Then, 1 mL of Alamar Blue solution (10% in DMEM medium) was added to each well, and the gels were incubated at 37 °C at 5% CO2 atmospheric concentration during 2 h. The fluorescence of supernatant was measured at λ ex = 570 nm and λ em = 585 nm using a microplate reader.
Immunogenic Assay
Peripheral blood was used to study IFN-γ production into the main circulating immune cells in response to the bio-hybrid nanodevice. Briefly, mononuclear cells were isolated by density gradient centriflugation method by Ficoll (Histopaque-1077, Sigma-Aldrich). Then, 106 cells per well in a 24-well plate were seeded. One side, 0.5 μg mL−1 was added from a suspension of nanoparticles (MSN-PEG-DBCO) as positive control. On the other side, the same cellular concentration was used to be stimulated at scaling doses of bacteria alone (Bac) and the nanosystem (Bac-MSN) in presence of 2 × 10−6 m mL−1 of Monensin (Sigma-Aldrich), a Golgi apparatus inhibitor for more than 5 h. Finally, intracellular retention of IFN-γ into circulating monocytes, T-cells, and B-cells by FACS following Beckton Dickson’s protocol for intracellular staining with minor modifications (Figure S14, Supporting Information) was analyzed.
For immunophenotype analysis, immune cells were first stained for surface markers with allophycocyanin (APC)-conjugated anti-CD14 (clon MΦP9), and R-phycoerythrin and cyanine dye Cy7 (PECy7)-conjugated anti-CD19 (clon J3-119) and allophycocyanin-H7 (APC)-conjugated anti-CD3 (clon SK7). For IFN-γ detection, R-phycoerythrin (PE)-conjugated anti-IFN-γ(clon GIR-208) was used. All human monoclonal antibodies were purchased from BD Pharmingen, and anti-CD19 from Beckman Coulter. Cell acquisition was performed by flow cytometry (Canto-II, BD Biosciences) equipped with FACSDIVA software (BD, Biosciences) for multiparameter analysis of the data.
Supplementary Material
Supporting Information is available from the Wiley Online Library or from the author.
Acknowledgements
This work was supported by European Research Council, ERC-2015-AdG (VERDI), Proposal No. 694160. The authors wish to thank the ICTS Centro Nacional de Microscopia Electrónica (Spain) and CAI Cytometer and Fluorescence microscopy of the Universidad Complutense de Madrid (Spain) for the assistance and Servier Medical art for the creative commons figures.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
V.M.M. and E.Á. have contributed equally to this work. The manuscript was written through contributions of all the authors. All the authors have given approval to the final version of the manuscript. The authors declare no competing financial interest.
Contributor Information
Alejandro Baeza, Dpto. Materiales y Producción Aeroespacial, ETSI Aeronáutica y del Espacio, Universidad Politécnica de Madrid, Madrid 28040, Spain.
Juana Serrano-Lopez, Experimental Hematology Lab, IIS-Fundación Jiménez Díaz, UAM, Madrid 28040, Spain.
María Vallet-Regí, Dpto. Química en Ciencias Farmacéuticas, Universidad Complutense de Madrid, Instituto de Investigación Sanitaria, Hospital 12 de Octubre i+12, Plaza Ramón y Cajal s/n, Madrid 28040, Spain; CIBER de Bioingeniería, Biomateriales y Nanomedicina, CIBER-BBN, Madrid 28040, Spain.
References
- [1].Matsumura Y, Maeda H. Cancer Res. 1986;46:6387. [PubMed] [Google Scholar]
- [2].Cao-Milán R, Liz-Marzán LM. Expert Opin Drug Delivery. 2014;11:741. doi: 10.1517/17425247.2014.891582. [DOI] [PubMed] [Google Scholar]
- [3].Vallet-Regi M, Rámila A, Del Real RP, Pérez-Pariente J. Chem Mater. 2001;13:308. [Google Scholar]
- [4].Riedinger A, Guardia P, Curcio A, Garcia MA, Cingolani R, Manna L, Pellegrino T. Nano Lett. 2013;13:2399. doi: 10.1021/nl400188q. [DOI] [PubMed] [Google Scholar]
- [5].Oliveira H, Pérez-Andrés E, Thevenot J, Sandre O, Berra E, Lecommandoux S. J Controlled Release. 2013;169:165. doi: 10.1016/j.jconrel.2013.01.013. [DOI] [PubMed] [Google Scholar]
- [6].Talelli M, Barz M, Rijcken CJF, Kiessling F, Hennink WE, Lammers T. Nano Today. 2015;10:93. doi: 10.1016/j.nantod.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Deshpande PP, Biswas S, Torchilin VP. Nanomedicine. 2013;8:1509. doi: 10.2217/nnm.13.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Villegas MR, Baeza A, Vallet-Regí M. Molecules. 2018;23:1008. doi: 10.3390/molecules23051008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].He C, Lu J, Lin W. J Controlled Release. 2015;219:224. doi: 10.1016/j.jconrel.2015.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kobayashi H, Turkbey B, Watanabe R, Choyke PL. Bioconjugate Chem. 2014;25:2093. doi: 10.1021/bc500481x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Venditto VJ, Szoka FC. Adv Drug Delivery Rev. 2013;65:80. doi: 10.1016/j.addr.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Blanco E, Shen H, Ferrari M. Nat Biotechnol. 2015;33:941. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Florence AT. J Controlled Release. 2012;164:115. doi: 10.1016/j.jconrel.2012.03.022. [DOI] [PubMed] [Google Scholar]
- [14].Parodi A, Haddix SG, Taghipour N, Scaria S, Taraballi F, Cevenini A, Yazdi IK, Corbo C, Palomba R, Khaled SZ, Martinez JO, Brown BS, Isenhart L, Tasciotti E. ACS Nano. 2014;8:9874. doi: 10.1021/nn502807n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Villegas MR, Baeza A, Vallet-Regí M. ACS Appl Mater Interfaces. 2015;7 doi: 10.1021/acsami.5b07116. 24075. [DOI] [PubMed] [Google Scholar]
- [16].Villegas MR, Baeza A, Noureddine A, Durfee PN, Butler KS, Agola JO, Brinker CJ, Vallet-Regí M. Chem Mater. 2018;30:112. [Google Scholar]
- [17].Jain RK, Stylianopoulos T. Nat Rev Clin Oncol. 2010;7:653. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nichols JW, Bae YH. J Controlled Release. 2014;190:451. doi: 10.1016/j.jconrel.2014.03.057. [DOI] [PubMed] [Google Scholar]
- [19].Sitti M, Mostaghaci B, Park B-W, Yasa O, Hosseinidoust Z, Singh AV. Adv Drug Delivery Rev. 2016;106:27. doi: 10.1016/j.addr.2016.09.007. [DOI] [PubMed] [Google Scholar]
- [20].Paris JL, La Torre PD, Manzano M, Cabañas MV, Flores AI, Vallet-Regí M. Acta Biomater. 2016;33:275. doi: 10.1016/j.actbio.2016.01.017. [DOI] [PubMed] [Google Scholar]
- [21].Taherkhani S, Mohammadi M, Daoud J, Martel S, Tabrizian M. ACS Nano. 2014;8:5049. doi: 10.1021/nn5011304. [DOI] [PubMed] [Google Scholar]
- [22].Song S, Vuai MS, Zhong M. Infect Agents Cancer. 2018;13:9. doi: 10.1186/s13027-018-0180-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Felfoul O, Mohammadi M, Taherkhani S, De Lanauze D, Xu Y Zhong, Loghin D, Essa S, Jancik S, Houle D, Lafeur M, Gaboury L, et al. Nat Nanotechnol. 2016;11:941. doi: 10.1038/nnano.2016.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Forbes NS. Nat Rev Cancer. 2010;10:785. doi: 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Xie S, Zhao L, Song X, Tang M, Mo C, Li X. J Controlled Release. 2017;268:390. doi: 10.1016/j.jconrel.2017.10.041. [DOI] [PubMed] [Google Scholar]
- [26].Zhou S, Gravekamp C, Bermudes D, Liu K. Nat Rev Cancer. 2018;18:727. doi: 10.1038/s41568-018-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Xie S, Chen M, Song X, Zhang Z, Zhang Z, Chen Z, Li X. Acta Biomater. 2018;78:198. doi: 10.1016/j.actbio.2018.07.041. [DOI] [PubMed] [Google Scholar]
- [28].Yoo J-W, Irvine DJ, Discher DE, Mitragotri S. Nat Rev Drug Discovery. 2011;10:521. doi: 10.1038/nrd3499. [DOI] [PubMed] [Google Scholar]
- [29].Chen F, Zang Z, Chen Z, Cui L, Chang Z, Ma A, Yin T, Liang R, Han Y, Wu Z, Zheng M, Liu C, Cai L. Biomaterials. 2019;214 doi: 10.1016/j.biomaterials.2019.119226. 119226. [DOI] [PubMed] [Google Scholar]
- [30].Suh SB, Jo A, Traore MA, Zhan Y, Coutermarsh-Ott SL, Ringel-Scaia VM, Allen IC, Davis RM, Behkam B. Adv Sci. 2019;6 doi: 10.1002/advs.201801309. 1801309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lu J, Liong M, Li Z, Zink JI, Tamanoi F. Small. 2010;6:1794. doi: 10.1002/smll.201000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].VanNieuwenhze MS, Hughes HV, de Pedro MA, Brown PJ, Cava F, Kuru E, Hall E, Tekkam S, Brun YV. Angew Chem, Int Ed. 2012;51 doi: 10.1002/anie.201206749. 12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sletten EM, Bertozzi CR. Angew Chem, Int Ed. 2009;48:6974. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Prescher JA, Bertozzi CR. Nat Chem Biol. 2005;1:13. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- [35].Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, Bertozzi CR. Proc Natl Acad Sci USA. 2010;107:1821. doi: 10.1073/pnas.0911116107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Guisasola E, Baeza A, Talelli M, Arcos D, Moros M, De La Fuente JM, Vallet-Regí M. Langmuir. 2015;31 doi: 10.1021/acs.langmuir.5b03470. 12777. [DOI] [PubMed] [Google Scholar]
- [37].Paris JL, Colilla M, Izquierdo-Barba I, Manzano M, Vallet-Regí M. J Mater Sci. 2017;52:8761. [Google Scholar]
- [38].Algar WR, Prasuhn DE, Stewart MH, Jennings TL, Blanco-Canosa JB, Dawson PE, Medintz IL. Bioconjugate Chem. 2011;22:825. doi: 10.1021/bc200065z. [DOI] [PubMed] [Google Scholar]
- [39].Koo H, Lee S, Na JH, Kim SH, Hahn SK, Choi K, Kwon IC, Jeong SY, Kim K. Angew Chem, Int Ed. 2012;51 doi: 10.1002/anie.201206703. 11836. [DOI] [PubMed] [Google Scholar]
- [40].Sahari A, Traore MA, Scharf BE, Behkam B. Biomed Micro-devices. 2014;16:717. doi: 10.1007/s10544-014-9876-y. [DOI] [PubMed] [Google Scholar]
- [41].Child HW, Del Pino PA, De La Fuente JM, Hursthouse AS, Stirling D, Mullen M, McPhee GM, Nixon C, Jayawarna V, Berry CC. ACS Nano. 2011;5:7910. doi: 10.1021/nn202163v. [DOI] [PubMed] [Google Scholar]
- [42].Shen J, He Q, Gao Y, Shi J, Li Y. Nanoscale. 2011;3:4314. doi: 10.1039/c1nr10580a. [DOI] [PubMed] [Google Scholar]
- [43].Danhier F, Feron O, Préat V. J Controlled Release. 2010;148:135. doi: 10.1016/j.jconrel.2010.08.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.