Abstract
NLRP3 is an innate immune sensor contributing to the development of different diseases including monogenic autoinflammatory syndromes, gout, atherosclerosis, and Alzheimer’s disease. The molecule sulfonylurea MCC950 is a NLRP3 inflammasome inhibitor with potential clinical utility. However, the mechanism of action of MCC950 remains unknown. Here, we characterize the mechanism of action of MCC950 in both wild-type and autoinflammatory-related NLRP3 mutants, demonstrating that MCC950 closes the ‘open’ conformation of active NLRP3.
The activation of the NOD-like receptor pyrin domain-containing protein 3 (NLRP3) inflammasome is involved in the pathophysiology of numerous non-communicable diseases through the development of sterile inflammation1–5. NLRP3 senses different host-derived sterile signals and oligomerizes with the apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC), forming intracellular signaling hubs that activate caspase-16–9. Caspase-1 proteolytically generates the active forms of a number of pro-inflammatory cytokines from the IL-1 family, and induces pyroptotic cell death by gasdermin D cleavage, resulting in the release of sterile intracellular content, which amplifies host-derived danger signaling6–8. The development of compounds inhibiting IL-1 release started with the identification of the sulfonylurea CP-456,773/CRID3, recently renamed as MCC950, a compound with proven specificity in inhibiting the NLRP3 inflammasome10,11. Different compounds targeting NLRP3 have demonstrated beneficial effects in several murine models of disease, with MCC950 showing a strong inhibitory effect with good pharmacokinetic and pharmacodynamic properties11–15. Despite the selectivity of MCC950 for inhibiting NLRP3 activation, its mechanism of action has not been yet elucidated. In this study, we found that MCC950 is able to modify the active conformation of NLRP3, and that it blocks NLRP3 oligomerization in response to external stimulation or when NLRP3 contains gain-of-function mutations.
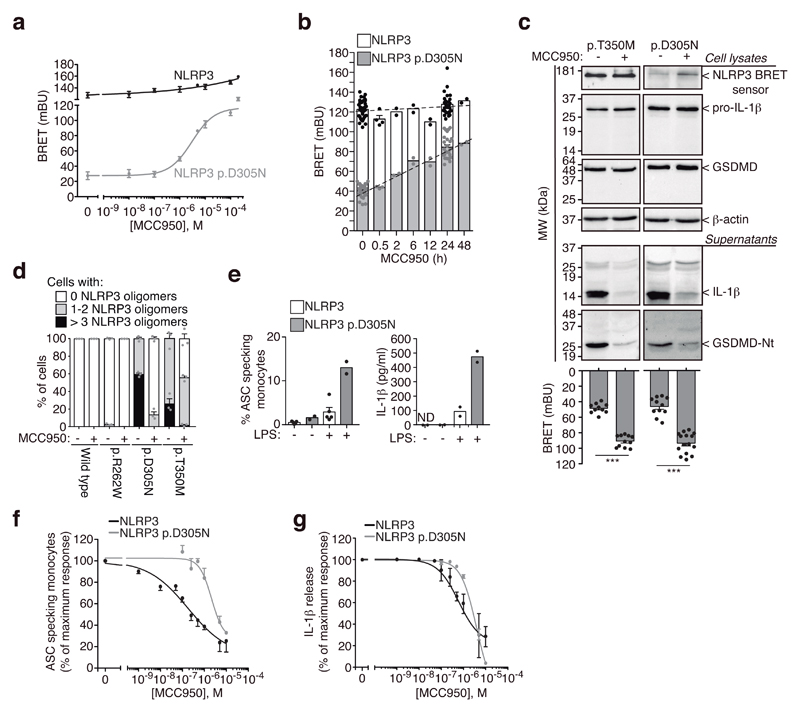
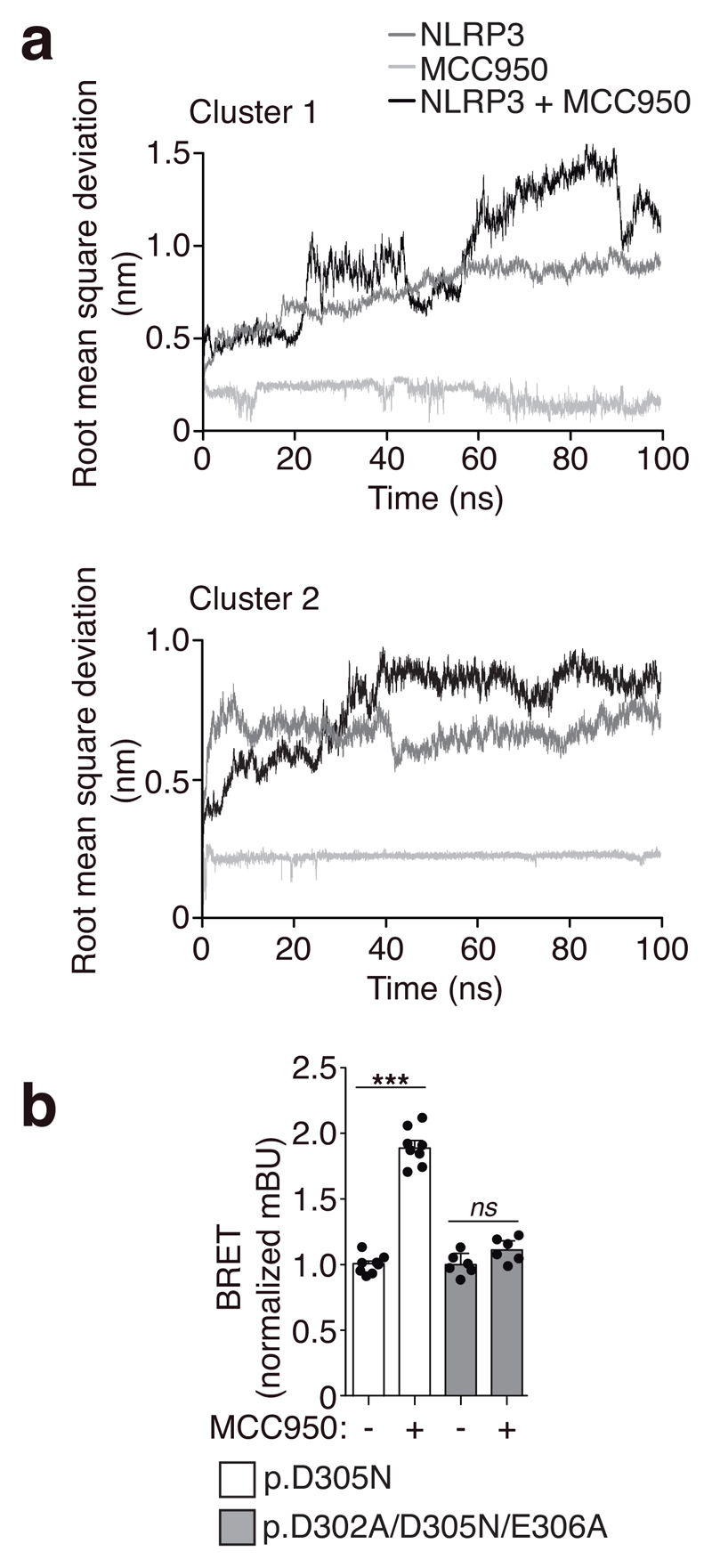
We developed a method to study the molecular conformation of NLRP3 by using bioluminescence resonance energy transfer (BRET)16,17. We found that compared to wild-type NLRP3, the disease-associated mutant p.D305N NLRP3 expressed in HEK293T cells appears to have a distinct conformation (Supplementary Fig. 1a,b), suggesting that the N- and C-terminus of the mutant NLRP3 are separated compared to the wild-type NLRP3, and therefore pathological mutant NLRP3 could be in an ‘open’ conformation. This difference in BRET signal was not due to variations in protein expression (Supplementary Fig. 1c,d) and was also observed in other pathological mutants of NLRP3 (Supplementary Fig. 1c). NLRP3 BRET signal was intramolecular, since a stable signal was found when increased concentration of the sensor was expressed (Supplementary Fig. 1b). The incubation of cells expressing the NLRP3 p.D305N BRET sensor with MCC950 at different doses or over increasing times, resulted in an increase of the BRET signal (Fig. 1a,b). These suggest that MCC950 induced closure of the active NLRP3 p.D305N conformation without changing YFP signals (Supplementary Fig. 1e). MCC950 increased BRET signal from different NLRP3 pathological mutants, and slightly modified the wild-type NLRP3 structure (Supplementary Fig. 1f), suggesting that MCC950 can binds the resting NLRP3. MCC950 effect on NLRP3 structure was reversible (Supplementary Fig. 1g). The MCC950-driven increase in BRET signal from mutant NLRP3 is not necessarily indicative of a structural conformation identical to inactive wild-type NLRP3. To demonstrate that the increase in BRET and the ‘closure’ of the structure are indicative of NLRP3 inhibition, we expressed NLRP3 mutants in immortalized Nlrp3 -/- macrophages, and recorded BRET whilst evaluating NLRP3 function. We found that BRET signal of ectopic mutant NLRP3 in macrophages was similar to that of HEK293T cells (Supplementary Fig. 1h). MCC950 was able to increase NLRP3 BRET signal and in parallel blocked the processing of caspase-1 substrates IL-1β and GSDMD in macrophages (Fig. 1c). We then expressed N-terminal YFP-tagged NLRP3 in HEK293T, and found that gain-of-function NLRP3 mutations resulted in oligomerization (Fig. 1d, Supplementary Fig. 2a). NLRP3 oligomers were able to recruit ASC (Supplementary Fig. 2b), suggesting they present a functional structure. The number of oligomers per cell varied for the three different gain-of-function mutations, with the p.D305N and p.T350M being the mutations showing higher numbers of oligomers per cell when compared to p.R262W (Fig. 1d, Supplementary Fig. 2a). The p.D305N mutation was the strongest inducer of oligomers, correlating with clinical observations that the p.D305N mutation is associated with the most severe form of NLRP3-associated autoinflammatory syndromes5. While MCC950 almost completely disaggregated all p.D305N oligomers, it left >50% of the cells containing at least one p.T350M oligomer (Fig. 1d). Monocytes from individuals carrying the p.D305N NLRP3 mutation released a higher concentration of IL-1β and contained a higher percentage of ASC specks compared to those from healthy donors in response to LPS (Fig. 1e). LPS treatment resulted in an increase of NLRP3 expression in human monocytes (Supplementary Fig. 3a), which is required to induce ASC-specks and the release of IL-1β from cryopyrin-associated periodic syndrome (CAPS) monocytes18. Higher concentration of MCC950 was required to block mutant p.D305N NLRP3 in monocytes from CAPS than to block NLRP3 activation from healthy individuals (Fig. 1f,g). This effect was not due to interference with LPS priming11 (Supplementary Fig. 3b), nor with high expression of NLRP3 or ASC specks in CAPS samples (Supplementary Fig. 3c,d). These suggest that the residue at position 305 could be important for the action of MCC950, which is indeed within the Walker B ATP-hydrolysis motif of NLRP3, an essential motif for NLRP3 activity19. We then performed NLRP3 NACHT homology modeling, followed by blind docking (BD) and molecular dynamics (MD) simulations. Our model systems disclosed the residues within the Walker B motif in both clusters formed an hydrophobic pocket for MCC950 that was stabilized by hydrogen bonds with residues upstream and downstream of this motif (Supplementary Fig. 4a,b), this is compatible with the reversible inhibitory effect of MCC950 (Supplementary Fig. 1g). BD calculations using ADP found the binding amino acids within the Walker A motif and also the distal H522 (Supplementary Fig. 4a). The consensus clusters could represent two potential MCC950 binding spots, that could be further stabilized and alter NLRP3 conformation. 100 ns MD simulations starting from the two clusters found that the binding of MCC950 to both clusters remains stable (Supplementary Fig. 4a-c) and that NLRP3 structure was predicted to be affected by MCC950 binding (Fig. 2a). After 100 ns of MD simulations, cluster 2 residues within Walker B presented a more favorable binding energy to MCC950 (Supplementary Fig. 4d), with F304 establishing stacking interactions with F257 and the aromatic ring of MCC950 (Supplementary Fig. 4b,e). When key amino acids required for Walker B function (D302 and E306 in human, corresponding to D298 and E302 in mouse) were alanine-substituted in the NLRP3 p.D305N background, MCC950 lost its ability to increase the BRET signal (Fig. 2b).
Figure 1. MCC950 closes the conformation of autoinflammatory-associated gain-of-function NLRP3.
a,b, BRET signal in HEK293T cells expressing wild-type or p.D305N YFP-NLRP3-Luc as indicated, incubated for 24 h with different concentrations of MCC950 (a) or for different times with MCC950 (10 μM) (b). Vehicle control data is concentration zero (a) and time zero (b). Centre values represent mean and error bars the SEM; for exact n number see Methods, section Statistics and Reproducibility. c, BRET signal (bottom) and immunoblots for NLRP3, IL-1β, GSDMD and β-actin (top) from immortalized Nlrp3 -/- macrophages expressing YFP-NLRP3-Luc p.T350M or p.D305N, treated for 16 h with doxycycline (1 μg/ml) and LPS (100 ng/ml) alone or in combination with MCC950 (10 μM). Blots are representative of n=2 independent experiments with similar result (full blots are shown in Supplementary Figure 10a) and for BRET assays centre values represent mean and error bars the SEM; for exact n number see Methods, section Statistics and Reproducibility; Mann-Whitney test, two-tailed, ***p<0.0001 (U=0). d, Quantification of the number of mutant NLRP3-YFP oligomers per HEK293T cell incubated or not for 24 h with MCC950 (10 μM). Data are representative of n=3 independent experiments and centre values represent mean and error bars the SEM from 1000-1500 cells. e, Percentage of ASC-specking monocytes identified by time of flight assay (left) and ELISA for IL-1β release (right) from PBMCs isolated from healthy donors (white) and individuals with CAPS carrying the NLRP3 p.D305N (grey) after 4h incubation with LPS (1 μg/ml). Centre values represent mean and error bars the SEM; for exact n number see Methods, section Statistics and Reproducibility. f,g, Inhibition of the percentage of ASC-specking monocytes (f) and IL-1β release (g) from PBMCs after 4h incubation with LPS (1 μg/ml) alone (for CAPS samples) or followed by nigericin (5 μM, 30 min, for healthy individuals) in the presence of different concentrations of MCC950. Centre values represent mean and error bars the SEM; for exact n number see Methods, section Statistics and Reproducibility.
Figure 2. MD simulations of MCC950 targeting the NLRP3 NACHT domain and MCC950 action on the NLRP3 triple walker B mutant.
a, Root mean square deviation during the 100 ns MD for NLRP3 (dark grey), complexed MCC950 (light grey) or NLRP3+MCC950 system (black) models. b, BRET signal in HEK293T cells expressing YFP-NLRP3-Luc p.D305N (white, n= 8 independent experiments) or p.D302A/D305N/E306A (grey, n= 6 independent experiments) incubated for 24 h with MCC950 (10 μM). Centre values represent mean and error bars the SEM; Mann-Whitney test, two-tailed, ***p<0.0001 (U=0) and ns p= 0.2403 (U=10).
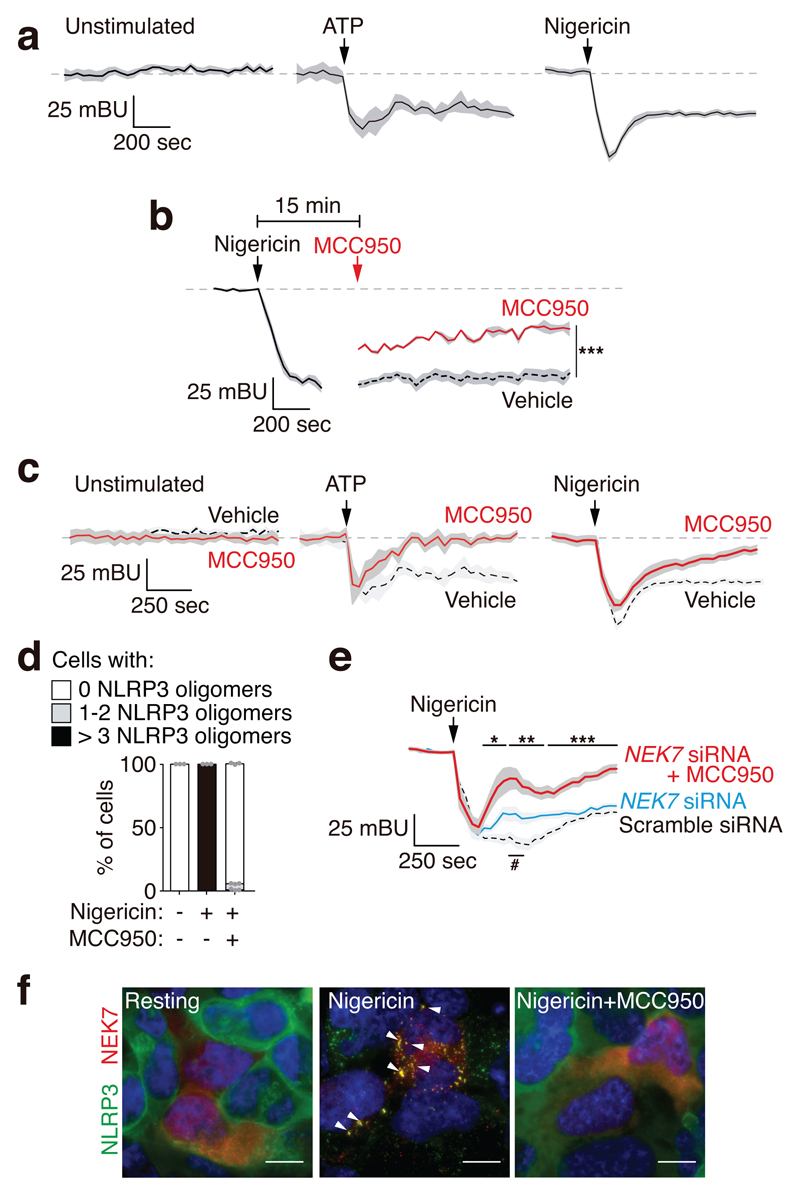
We then stimulated HEK293T cells expressing the wild-type NLRP3 BRET sensor with either nigericin or ATP (the latter in a HEK293T stably expressing P2X7 receptor) and both stimuli decreased NLRP3 BRET signal (Fig. 3a). After stimulation, NLRP3 BRET signal was still intramolecular (Supplementary Fig. 5a). NLRP3-activating stimuli ‘open’ NLRP3 conformation with a separation of the N- and C-terminus that may represent the active NLRP3 conformation, this was confirmed since nigericin induced NLRP3 oligomerization coincided with the stabilization of the ‘open’ structure (Supplementary Fig. 5b). This oligomerization was not due to non-specific aggregation induced by the YFP fluorescent tag, as GFP alone was unable to form oligomers in the cell after nigericin stimulation and NLRP3 without YFP tag was also able to form oligomers after nigericin treatment (Supplementary Fig. 5c). Addition of MCC950 to the wild type NLRP3 during an ‘open’ stable conformation after nigericin stimulation resulted in a higher BRET signal (Fig. 3b). We next incubated HEK293T cells with MCC950 before and during nigericin or ATP (the latter in a HEK293T expressing P2X7 receptor) and MCC950 was able to increase BRET signal after NLRP3 ‘opening’ (Fig. 3c, Supplementary Fig. 5d). MCC950 was also able to block the formation of NLRP3 oligomers in response to nigericin treatment (Fig. 3d, Supplementary Fig. 5e), suggesting that MCC950 forced the ‘open’ active NLRP3 structure into a ‘closed’ conformation that may be unable to self-associate. The inactive NLRP3 structure induced after MCC950 and nigericin treatment is significantly different to the resting NLRP3 structure (Supplementary Fig. 5f). Although both BRET signals indicate a ‘closed’ structure, this does not necessary mean that they have the same conformation. To support this idea, we studied the response of NLRP3 to stimulation subsequent to it being inactivated after nigericin and MCC950 treatment. If this inhibited structure is similar to the resting NLRP3 it should ‘open’ again in response to a second stimulation. The MCC950-nigericin ‘closed’ NLRP3 fails to change its structure in response to a second stimulation (Supplementary Fig. 5g), suggesting that MCC950 could be present in the NLRP3 inactive complex, in a different structure from the resting NLRP3 conformation. The effect of MCC950 on NLRP3 conformation was independent of the PYD domain (Supplementary Fig. 5d) and it was also blocking the activation of a NLRP3 that lacks the LRR domain20, supporting that MCC950 could targets the NACHT domain. As this domain is important for the oligomerization of NLRP3 upon ATP binding and hydrolysis19 and NEK7 is a protein required for the final activation of NLRP321,22, we then performed NEK7 knock-down (Supplementary Fig. 6a). We observed that NEK7 silencing resulted in a potentiation of the MCC950 induced ‘closed’ structure of NLRP3 (Fig. 3e, Supplementary Fig. 6b). While activation of NLRP3 induced the co-localization of NEK7 in NLRP3 oligomers and the co-immunoprecipitation of NEK7 with NLRP3, MCC950 impaired the association of NLRP3 with NEK7 (Fig. 3f, Supplementary Fig. 6c). These data suggest that after decreasing NEK7 expression, MCC950 is able to more efficiently ‘close’ NLRP3 active conformation and that the change induced by MCC950 over the active ‘open’ structure of NLRP3 could disrupt the interaction with NEK7. The accompanying study by Coll et al. shows that MCC950 do not binds NEK7, but directly targets NLRP3 at a site within or close to the Walker B motif and blocks ATP hydrolysis, blocking NLRP3 activation23. Together, these complementary studies demonstrate that MCC950 prevents NLRP3 activation by directly interacting with NLRP3 and affecting Walker B function leading to an inactive NLRP3 conformation.
Figure 3. MCC950 affects NLRP3 conformation upon activation.
a-c, BRET signal for YFP-NLRP3-Luc in HEK293T cells treated or not with nigericin (10 μM) or in HEK293T cells stable expressing P2X7 receptor treated with ATP (3 mM) (a), in the presence or absence of MCC950 (10 μM) incubated 35 min before nigericin or ATP stimulation (red trace in c). In b 15 min after nigericin stimulation, cells were washed, then MCC950 (10 μM) or vehicle was added and BRET was recorded (a representative trace for the initial response to nigericin is included to compare). Mean (black or red line) ±SEM (grey shadow) are shown; for exact n number see Methods, section Statistics and Reproducibility; Mann-Whitney test, two-tailed for each time comparing vehicle with MCC950, ***p<0.0008 (for each time point, exact p and U value are annotated in supplementary datasheet for figure 3). d, Quantification of the number of wild type NLRP3-YFP oligomers per HEK293T cells incubated or not for 30 min with MCC950 (10 μM) and then stimulated or not for 30 min with nigericin (10 μM). Data are representative of n=3 independent experiments and centre values represent mean and error bars the SEM from 1000-1500 cells. e, HEK293T cells transfected with scramble or NEK7 siRNA and then YFP-NLRP3-Luc BRET kinetic was measured before and after nigericin (10 μM) treatment in the absence (blue and dashed trace) or presence of MCC950 (10 μM, red trace). n=11 independent experiments, and mean (blue, red or dashed trace) ±SEM (grey shadow) are shown. Mann-Whitney test, two-tailed for each time comparing vehicle vs MCC950 (*p<0.05; **p≤0.005; ***p≤0.0002) or comparing scramble vs NEK7 siRNA (#p<0.05); for each time point, exact p and U value are annotated in supplementary datasheet for figure 3. f, Representative maximum intensity fluorescence images of HEK293T cells expressing NLRP3-YFP and NEK7 incubated or not for 30 min with MCC950 (10 μM) and then stimulated for another 30 min with nigericin (10 μM). NLRP3, green; NEK7, red; nuclei, blue DAPI; bar 10 μm; arrowheads denote NLRP3 and NEK7 co-localization. Images are representative of n=3 independent experiments with similar result.
In conclusion, our study demonstrates that MCC950 closes the NLRP3 active conformation in pathological gain-of-function NLRP3 mutants or during stimulation of wild-type NLRP3. MCC950 showed different inhibitory effects over distinct pathological NLRP3 mutations, raising the possibility of developing a personalized therapy for individuals with monogenic autoinflammatory syndromes carrying specific mutations where the compound is more effective.
Online Methods
Reagents
Key reagents and their sources were as follows: Escherichia coli LPS serotype 055:B5, nigericin sodium salt; MCC950 (CP-456773), ATP, bovine serum albumin, DAPI and siRNA NEK7 (Mission SiRNA #1 SIHK1408 and #2, SIHK1409) and scrambled siRNA negative control (Mission SiRNA negative control, SIC001) were from Sigma-Aldrich (Madrid, Spain). Ultrapure Escherichia coli LPS serotype 0111:B4 was from Invivogen (California). Coelenterazine-H and Lipofectamine 2000 from Life Technologies (Madrid, Spain). The composition of the physiological buffer used in all experiments to stimulate cells was (in mM): 147 NaCl, 10 HEPES, 13 D-glucose, 2 KCl, 2 CaCl2, and 1 MgCl2; pH 7.4.
Plasmid constructs
The different mutations of human NLRP3 were generated by overlapping PCR to introduce point mutations (p.R262W, p.D305N or p.T350M) or triple mutant (p.D302A/D305N/E306A) (Uniprot #Q96P20 annotation for human NLRP3 was used) and cloned into pcDNA3.1/V5-His TOPO (Life Technologies). Sequencing of all constructs was performed to confirm correct modification and the absence of unwanted mutations. All constructs were fused using overlapping PCR in the N-terminus to YFP for microscopy assays or double tagged with YFP in the N-terminus and Renilla Luciferase (Luc) in the C-terminus to generate the various BRET sensors. NLRP3 single fused to Renilla Luciferase (Luc) in the C-terminus was also produced to use as a control in all BRET assays. They were also cloned into pcDNA3.1/V5-His TOPO and sequenced to confirm correct alignment between tags and the NLRP3 sequence.
Cells and transfections
HEK293T cells (CRL-11268; American Type Culture Collection) were maintained in DMEM:F12 (1:1) (Lonza, Verviers, Belgium) supplemented with 10% fetal calf serum (FCS) (Life Technologies), 2 mM Glutamax (Life Technologies) and 1% penicillin-streptomycin (Life Technologies). HEK293T cells stably expressing the rat P2X7 receptor have previously been described21 and were cultured in F-12 media (Lonza) supplemented with 10% FCS. Lipofectamine 2000 was used for the transfection of HEK293T cells according to the manufacturer’s instructions. After two days of transfection, stable selection of HEK293T clones expressing the different NLRP3 constructs was initiated by supplementing culture media with G418 (2 mg/ml, Acros Organic). After 4 weeks of culture in G418, cell cloning was performed by serial dilution in 96 well plates in the presence of G418 for a further 4-8 weeks. Positive clones were expanded and tested for correct expression by Western blot and fluorescence microscopy. HEK293T cells stably expressing NLRP3 constructions were maintained in DMEM/F12 (1:1) supplemented with 10% FCS, 2 mM Glutamax, and 1% penicillin–streptomycin. All cells were routinely tested for Mycoplasma contamination with a Mycoplasma Detection Kit (Roche). SiRNA were diluted to a final concentration of 2.5 pmol/μl and HEK293T cells stably expressing the wild-type NLRP3 BRET sensor were transfected using Lipofectamine 2000 with 10 pmol of siRNA#1 and 10 pmol of siRNA#2 for 48h.
Construct preparation and transduction of immortalized mouse macrophages Nlrp3 -/-
For doxycycline-inducible expression of NLRP3 BRET sensors in immortalized mouse macrophages we used Tet-ON retroviral system (Clontech #631188). NLRP3 BRET sensors were subcloned into pRETROX Tre3G plasmid (Clontech) using BamHI/NotI sites and transfected using Lipofectamine 2000 into the packaging cell line Gryphon Ampho cell line (Alelle Biotechnology, ABP-RVC-10001). Nlrp3 -/- immortalized mouse macrophages stably expressing the Tet-On 3G transactivator (a kindly gift from Dr. Iva Hafner-Bratkovič, National Institute of Chemistry, Ljubljana, Slovenia) were transduced with different NLRP3 BRET sensors or empty vector encoding retroviruses for two days. Then positive macrophages were selected with puromycin (6 μg/mL) and G418 (1.5 mg/ml).
Bone marrow-derived macrophages
Wild-type C57 BL/6J mice were purchased from Harlan. For all experiments, mice between 8-10 weeks of age bred under SPF conditions were used in accordance with the Hospital Clínico Universitario Virgen Arrixaca animal experimentation guidelines, and the Spanish national (RD 1201/2005 and Law 32/2007) and EU (86/609/EEC and 2010/63/EU) legislation. According to this legislation, no specific procedure approval is needed when animals are sacrificed to obtain biological material. Bone marrow was obtained from leg bones of mice euthanized by CO2 inhalation and bone marrow-derived macrophages (BMDMs) were obtained as described22. BMDMs were primed and stimulated as indicated in the text. Cells were then washed three times with physiological buffer before add the indicated stimulus.
Human samples
Whole peripheral blood samples were collected in EDTA anticoagulated tubes from healthy donors (n= 5), and from 2 individuals with cryopyrin-associated periodic syndrome carrying the NLRP3 p.D305N mutation. The Institutional Review Board of the Hospital Clínico Universitario Virgen de la Arrixaca approved the use of these blood samples. Informed consent was obtained from all individuals enrolled in the study following the principles set out in the WMA Declaration of Helsinki and samples were stored in the Biobanco en Red de la Región de Murcia (PT13/0010/0018) integrated in the Spanish National Biobanks Network (B.000859). Mononuclear cells (PBMCs) were collected using Histopaque-1077 (Sigma-Aldrich) and cultured in RPMI 1640 medium (Lonza) with 10% FCS, 2mM Glutamax and 100 U/ml penicillin-streptomycin. PBMCs from patients were left unstimulated or were stimulated with 1 μg/ml of LPS for 4h at 37ºC in the presence of different concentrations of MCC950 (from 0.1 μM to 10 μM). MCC950 was added 30 min before and during LPS stimulation. To calculate dose inhibition curves for MCC950, PBMCs from healthy donors were similarly treated with LPS and MCC950, but were then stimulated with nigericin (5 μM, 30 min at 37°C).
BRET assays
HEK293T cells or immortalized mouse macrophages expressing the different NLRP3 BRET sensors (wild-type and mutants) were plated on a poly-L-lysine–coated white opaque 96-well plate; after adhesion, cells were incubated with different doses of MCC950 (as stated in the figure legends) or vehicle, washed with PBS with calcium and magnesium, and BRET readings were performed 5 min after the addition of 5 μM coelenterazine-H substrate. BRET signals were detected with two filter settings (Renilla-luciferase (Luc) filter [485 ± 20 nm] and YFP filter [530 ± 25 nm]) at 37ºC using the Synergy Mx plate reader (BioTek) as described before17,23,24. In some experiments, BRET signal was recorded every 35 seconds before and after nigericin or ATP automatic injection for a total of 15 min. For experiments measuring basal BRET signal, a stable signal for 5 min kinetic was recorded and averaged. BRET signal was similar when comparing HEK293T clones stably expressing the different NLRP3 BRET sensor with transient transfections for the same construct. Titration was performed by transfection of different amounts of plasmids encoding YFP-NLRP3-Luc or YFP-NLRP3 p.D305N-Luc (pcDNA empty plasmid was used to have equal amounts of total DNA in all the transfections). Expression of the different sensors was monitored by reading YFP fluorescence in the plate reader or by assessing individual cell relative fluorescence by fluorescence microcopy. Titration of the sensor determines whether the recorded BRET is intra- or inter-molecular. For intramolecular BRET the increase in the BRET sensor expression results in a stable BRET signal while for intermolecular BRET the increase in the BRET sensor expression results in a proportional increase of the BRET signal. The BRET ratio was defined as the difference of the emission ratio 530 nm/485 nm of the BRET sensor minus this ratio of the Luc only tagged NLRP3. Results were expressed in milliBRET units (mBU). The area under curve of the BRET kinetic was measured taking all the points after stimulation (from 315 s), and the data from the MCC950 treatment was normalized to its respective vehicle treated cells in resting, nigericin or ATP treatments independently.
Fluorescence microscopy
Poly-L-lysine coated coverslips (Corning) were used to seed 105 HEK293T cells stably expressing the different NLRP3 constructs tagged with YFP at N-terminus. For NLRP3-NEK7 co-localization experiments stable cell lines were transfected with myc-human NEK7 expressing plasmid (GeneScript) on the coverslips. To study NLRP3 oligomerization without YFP tag, NLRP3-flag or GFP control plasmid were transfected into HEK293T. To analyze wild type or p.D305N NLRP3 oligomerization with ASC a stable HEK293T cell line expressing the desired NLRP3 construction was transfected with ASC. Cells were treated and stimulated as indicated in the figure legends, washed twice with PBS and fixed for 15 min at room temperature with 4% paraformaldehyde, and then were washed three times with PBS. For NEK7, NLRP3 or ASC immunofluorescence, cells were blocked with 2% bovine serum albumin and permeabilized with 0.1% Triton X-100 (Sigma) for 20 min at room temperature. Then cells were stained for 1.5 h at room temperature with the primary monoclonal mouse antibody anti-myc (1:1000, clone 9E10, MA1-980 Thermo Scientific), or the primary monoclonal mouse antibody anti-PYD domain of NLRP3 (1:1000, clone Cryo-2, AG-20B-0014, Adipogen) or the primary monoclonal mouse antibody anti-ASC (1:1000, clone HASC-71, 653902, BioLegend). Cells were washed and then incubated for 1 h at room temperature with anti-mouse IgG fluorescence-conjugated secondary antibody (1:200, Alexa 647 donkey anti-mouse IgG (H+L), A31571, Life Technologies). NEK7, NLRP3 or ASC stained cells or directly fixed cells were washed and nuclei stained with DAPI (1:10000 for 10 min) and coverslips were mounted on slides with mounting medium (S3023, Dako, USA). Images were acquired with a Nikon Eclipse Ti microscope equipped with a 20x S Plan Fluor objective (numerical aperture, 0.45), a 40x S Plan Fluor objective (numerical aperture, 0.60) and a 60x Plan Apo Vc objective (numerical aperture, 1.40) and a digital Sight DS-QiMc camera (Nikon) with a Z optical with spacing of 0.4 μm and 387-nm/447-nm, 472-nm/520-nm, 543-nm/593-nm and 650-nm/668-nm filter sets (Semrock) and the NIS-Elements AR software (Nikon). Images were analyzed with ImageJ (US National Institutes of Health, Bethesda, MD USA).
Quantification of intracellular ASC specks
Following stimulation, PBMCs were fixed with 2% paraformaldehyde and stained for the detection of Apoptosis-associated Speck-like protein containing a Caspase recruitment domain (ASC) specks by Time of Flight Inflammasome Evaluation using the rabbit polyclonal antibody anti-ASC (N-15)-R (sc-22514-R, Santa Cruz, 1:1500) followed by the staining with donkey anti-rabbit alexa-488 antibody (1:1000, Life Technologies) as previously described25. Monocytes were gated using the PE-conjugated anti-human CD14 antibody (61D3, 50-0149-T025, TONBO Biosciences, 1 μl/106 cells) and stained cells were acquired on a FACS Canto cytometer (BD Biosciences). Gating strategy is shown in supplemental figure 7.
Immunoprecipitation and Western blot
BMDMs and immortalized mouse macrophages expressing BRET sensors were lysed in ice-cold lysis buffer (50 mM Tris-HCl pH8.0, 150 mM NaCl, 2% Triton X-100, supplemented with 100 μl/ml of protease inhibitor mixture (Sigma) for 30 min on ice and then were clarified by centrifugation (16,000 g 15 min at 4°C). Cleared cell lysates (800 μg) were incubated at 4 °C overnight with 5 μg of anti-PYD domain of NLRP3 (mouse monoclonal Cryo-2 clone, AG-20B-0014, Adipogen) and 25 μl of Protein A/G magnetic Beads (Pierce Protein A/G Magnetic Beads, Thermo 88802) in a final volume of 500 μl. Immunoprecipitated complexes were washed twice in lysis buffer and eluted in Laemmli buffer (Sigma) after 2 min of incubation. Immortalized mouse macrophages expressing wild type NLRP3-YFP prepared as stated above were treated with doxycycline (1 μg/ml) and ultrapure LPS (100 ng/ml) for 16 h and then stimulated with 10 μM of Nigericin during 30 min in presence or absence of 10 μM of MCC950. Cells were lysed in ice-cold lysis buffer (50 mM Tris-HCl pH8.0, 150 mM NaCl, 5 mM EDTA, 1 % Igepal CA-630, supplemented with 100 μl/ml of protease inhibitor mixture (Sigma) for 30 min on ice and then were clarified by centrifugation (400 g 10 min at 4°C). Cleared cell lysates (1 mg) were incubated at 4 °C overnight with 5 μl (1:100) anti-NEK7 rabbit monoclonal (EPR4900 clone, ab133514, Abcam) with 25 μl of Protein A/G magnetic Beads (Pierce Protein A/G Magnetic Beads, Thermo 88802) in a final volume of 500 μl. Immunoprecipitated complexes were washed twice in lysis buffer and eluted in Laemmli buffer after 2 min of incubation. In the case of immortalized mouse macrophages expressing pathological mutants NLRP3 BRET sensors, MCC950 (10 μM) was added at the same time than with LPS and doxycycline and then, cell supernatants were collected and centrifuged at 300 g for 10 min at 4 ºC to remove detached cells and generate cell-free preparations. Proteins in supernatants were concentrated by centrifugation at 11,200 g for 30 min at 4 °C with a 10 kDa cut-off column (Microcon, Merk-Millipore). Cells lysates, concentrated supernatants and immunoprecipitates were resolved in 4–12% precast Criterion polyacrylamide gels (Biorad) and transferred to nitrocellulose membranes (Biorad) by electroblotting as already described26. Membranes were probed with anti-PYD domain of NLRP3 mouse monoclonal (Cryo-2 clone, AG-20B-0014, Adipogen, 1:1000), anti-NEK7 rabbit monoclonal (EPR4900 clone, ab133514, Abcam, 1:5000), anti-GSDMD rabbit monoclonal (EPR19828, ab209845, Abcam, 1:5000), anti-IL-1β rabbit polyclonal (H-153; sc-7884, 1:1000) and horseradish peroxidase-anti-β-actin (C4; sc-47778HRP, Santa Cruz, 1:10000). HRP-conjugated Protein A (Calbiochem, 539253, 1:8000). Western blot for NLRP3 BRET sensors expression in HEK293T cells were detected from clarified cell lysates obtained as described above for BMDM and using standard procedure for Western blot27 using a polyclonal antibody rabbit anti-GFP (Abcam, ab6556, 1:2500) and HRP-conjugated secondary antibody from GE Healthcare (1:10000). Uncropped immunoblots are shown in supplementary figure 10.
ELISA
Individual culture cell-free PBMCs supernatants were collected, clarified by centrifugation and the concentration of IL-1β was tested by ELISA following the manufacturer’s instructions (R&D Systems).
Quantitative PCR
BMDMs or HEK293T cells where washed twice with PBS before total RNA purification using the RNAesy kit (Qiagen) according to manufacturer’s recommendations and quantified on a nanodrop 2000 (Thermo Fisher). Detailed methods used for qRT-PCR have been described previously26. Briefly, Reverse transcription was realized using iScriptTM cDNA Synthesis kit (BioRad). qPCR was performed in an iQTM 5 Real Time PCR detection System (BioRad) with a SYBR Green mix (Takara) and primers used were obtained from Sigma-Aldrich (KiCqStart® Primers). The presented relative gene expression levels were calculated using the 2-ΔCt method normalizing to Gapdh or HPRT1 expression as endogenous controls.
Human NLRP3 homology modeling (BD) and molecular dynamics (MD)
For the BD and subsequent MD calculations, a homology structural model for the human NACHT NLRP3 was performed using the Phyre2 Protein Fold Recognition Server (http://www.sbg.bio.ic.ac.uk/~phyre2) using the 8 crystalized chains of NLRC4 (PDB: 4KXF) as template and hydrogens and partial charges were added using Gasteiger model with Autodock Tools28. The resulting model system is further refined with the Protein Preparation Wizard as implemented in Maestro (http://www.schrodinger.com), to avoid clashes between residues.
The chemical structures of MCC950 and ADP used in the BD and MD calculations were built up and fully optimized at the B3LYP/ 6-31 + G(d,p) level within the density functional theory (DFT) framework as implemented in Gaussian0929. Additional vibrational calculations were conducted at the same DFT level to verify the nature of the located structures in the potential energy of surface. Partial atomic charges were subsequently computed with the ESP scheme to be used during BD simulations. In our BD approach, multiple independent and parallel docking runs executed on a supercomputer through Blind Docking Server (http://bio-hpc.ucam.edu/achilles) started around geometric centers of all the residues within the selected threshold. A distribution of binding energies and their structural clusters of poses was generated30. The individual docking simulations of MCC950 around the whole protein structure model of NLRP3 and the detailed binding energy calculations were performed with the Autodock Vina docking software28 using default configuration parameters. The size of the grid box for each individual docking was set to extend 120 Å in each direction from the geometric center of each individual docking simulation. The docking score produced by Autodock Vina was taken as the predicted value of the free energy of binding. Only the poses pertaining to the top-ranked clusters in terms of free energy were used for structural and energy analyses. The scoring function from Vina considers the Lennard-Jones term (LJ), hydrogen bonds (H-bonds), electrostatic interactions, hydrophobic stabilization, entropic penalty due to the number of rotatable bonds, and the internal energy of the ligand. Analysis of main interacting residues was performed with PLIP (http://plip.biotec.tu-dresden.de/plip-web) and two dimensional representations with Poseview (http://proteins.plus).
MD simulations were conducted for our NLRP3 model with and without selected pose clusters with the GPU version of Gromacs (http://gromacs.org). For such purpose, topologies were generated by ACPYPE (http://github.com/llazzaro/acpype) with Amber99sb force field parameters (ambermd.org). Molecular systems were solvated in triclinic boxes using periodic boundary conditions and TIP3P water model. Counter ions Na+ were added to neutralize the system. The LINCS algorithm was chosen to constrain covalent bond lengths, and an integration step of 2 fs was applied. As for the electrostatic interactions, calculations were performed by the particle mesh Ewald (PME) method. The Parrinello–Rahman pressure barostat was employed, with a 2.0 ps coupling constant under an isotropic pressure coupling, while the V-rescale (NVT step) and Nosé–Hoover (NPT equilibration and production MD) temperature thermostats were used, with a coupling constant of τ = 0.5. Constant temperature of 300 K and constant pressure of 1 atm were also employed. Steepest Descent algorithm was used in an initial energy minimization, followed by two simulations with position restraints: a 0.1 ns NVT and a 0.9 ns NPT. Subsequently, 100 ns of unrestrained NPT MD simulations were performed for each of the systems, generating the production run from which data were collected.
Statistics and Reproducibility
All data are shown as mean values and error bars represent standard error from the number of independent assays indicated in the figure legend or described below. Raw dot-plot representation of the data could be found in Supplementary Fig. 8 and 9. For two-group comparisons, Mann-Whitney U test was used meanwhile comparisons of multiple groups were analyzed by Kruskal-Wallis test using Prism software (Graph-Pad Software, Inc.). p value is indicated as ***p < 0.001; **p > 0.001 < 0.01; *p > 0.01 < 0.05; p > 0.05 not significant (ns). The exact n number of independent experiments for the different figures were:
For figure 1a n= 6 independent experiments for 0, 0.1 μM, 0.1 mM; n= 4 independent experiments for 0.01 μM, 1 μM, 10 μM, 0.2 mM; and n= 2 independent experiments for 5 μM.
For figure 1b n= 37 independent experiments for NLRP3 and n= 35 for p.D305N NLRP3 at time 0; n= 28 independent experiments for NLRP3 and n= 26 for p.D305N NLRP3 at time 24h; n= 4 independent experiments for NLRP3 and n= 3 at time 0.5h; and n= 2 independent experiments for time 2, 6, 12, and 48h.
For figure 1c n=10 independent experiments for LPS treatment and n=11 independent experiments for LPS+MCC950 treatment for p.T350M NLRP3; n=11 independent experiments for LPS treatment and n=12 independent experiments for LPS+MCC950 treatment for p.D305N NLRP3.
For figure 1e n= 5 healthy individuals and n= 2 individuals carrying the NLRP3 p.D305N mutation for the percentage of ASC specking monocytes (top); and n= 2 individuals healthy and n= 2 individuals carrying the NLRP3 p.D305N mutation for IL-1β release.
For figure 1f n= 3 healthy individuals and n= 2 individuals carrying the NLRP3 p.D305N mutation.
For figure 1g n= 4 healthy individuals and n= 2 individuals carrying the NLRP3 p.D305N mutation.
For figure 3a n= 5 independent experiments for unstimulated and nigericin and n= 4 for ATP stimulation.
For figure 3b n= 3 independent experiments with cell culture triplicates for the vehicle and cell culture duplicates for the MCC950 treatment.
For figure 3c n= 5 independent experiments for MCC950 and MCC950+nigericin and n= 4 for MCC950+ATP stimulation.
Supplementary Material
Acknowledgements
We sincerely thank M.C. Baños and A.I. Gómez for technical assistance with molecular biology and cell culture. We also wish to thank L. Martínez-Alarcón for help with healthy volunteer blood extraction, I. Hafner-Bratkovič (National Institute of Chemistry, Ljubljana, Slovenia) for stable immortalized macrophages lines, and C. Vargas (Hospital Virgen de la Macarena, Sevilla, Spain), E. Ramos and S. Jimenez-Treviño (Hospital Central de Asturias, Oviedo, Spain) and M. Basagaña Torrento (Hospital Universitario Germans Trias i Pujol, Badalona, Spain) for samples of autoinflammatory individuals. We also want to acknowledge the patients and healthy volunteers enrolled in this study and the Biobanco en Red de la Región de Murcia (PT13/0010/0018), which is integrated into the Spanish National Biobanks Network (B.000859) for its collaboration. This research was partially supported by the e-infrastructure program of the Research Council of Norway, and the supercomputer center of UiT - the Arctic University of Norway. The authors thankfully acknowledge the computer resources at CTE-POWER and the technical support provided by Barcelona Supercomputing Center (RES-BCV-2018-3-0008). H.M-B. was supported by Rio Hortega fellowship from Instituto Salud Carlos III (CM14/00008). D.A-B. was supported by Juan de la Cierva postdoctoral fellowship from Ministerio de Economía y Competitividad (FJCI-2014-22041). This work was supported by grants from the Instituto Salud Carlos III-Fondo Europeo de Desarrollo Regional (PI13/00174 to P.P.), the Ministerio de Economia, Industria y Competitividad–Fondo Europeo de Desarrollo Regional (projects no. SAF2017-88276-R to P.P. and CTQ2017-87974-R to H.P-S.), the European Research Council (ERC-2013-CoG 614578 to P.P.), and Fundación Séneca (20859/PI/18 to P.P.).
Footnotes
Author contributions. A.T-A., D.A-B., H.M-B. and C.dT-M. conducted the experiments and interpreted data. J.P.C-C. and H.P-S. conducted blind docking and molecular dynamics simulations. D.A-B. performed structural modeling. J.I.A. coordinated human samples from autoinflammatory individuals. P.P. conceived, designed, and supervised this study, wrote the paper with feedback from all coauthors and provided funding.
Competing financial interests. The authors declare no competing interests.
Data availability. Source data for main figures is presented in Supplementary Source Data files. Supplementary Fig. 8 and 9 present data for main figures 1a,f,g and 3a-c,e as dot-plots to shows data distribution. Supplementary Fig. 10 present uncropped Western blots presented in main and supplementary figures. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.McGettrick AF, et al. Diabetes, Obes Metab. 2013;15:19–25. doi: 10.1111/dom.12169. [DOI] [PubMed] [Google Scholar]
- 2.Heneka MT, et al. Nat Immunol. 2015;16:229–236. doi: 10.1038/ni.3102. [DOI] [PubMed] [Google Scholar]
- 3.Martinon F, et al. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 4.Masters SL, et al. Sci Transl Med. 2011;3:81ps17. doi: 10.1126/scitranslmed.3001902. [DOI] [PubMed] [Google Scholar]
- 5.de Torre-Minguela C, et al. Front Immunol. 2017;8:43. doi: 10.3389/fimmu.2017.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schroder K, Tschopp J. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 7.Rubartelli A, et al. Front Immunol. 2013;4:398. doi: 10.3389/fimmu.2013.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hafner-Bratkovič I, Pelegrín P. Curr Opin Immunol. 2018;52:8–17. doi: 10.1016/j.coi.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Boucher D, et al. J Exp Med. 2018;215:827–840. doi: 10.1084/jem.20172222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laliberte RE, et al. J Biol Chem. 2003;278:16567–16578. doi: 10.1074/jbc.M211596200. [DOI] [PubMed] [Google Scholar]
- 11.Coll RC, et al. Nat Med. 2015;21:248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Primiano MJ, et al. J Immunol. 2016;197:2421–2433. doi: 10.4049/jimmunol.1600035. [DOI] [PubMed] [Google Scholar]
- 13.Jiang H, et al. J Exp Med. 2017;214:3219–3238. doi: 10.1084/jem.20171419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cocco M, et al. J Med Chem. 2017;60:3656–3671. doi: 10.1021/acs.jmedchem.6b01624. [DOI] [PubMed] [Google Scholar]
- 15.Baldwin AG, et al. Cell Chem Biol. 2017;24:1321–1335.e5. doi: 10.1016/j.chembiol.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Compan V, et al. Immunity. 2012;37:487–500. doi: 10.1016/j.immuni.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Martín-Sánchez F, et al. Methods Mol Biol. 2016;1417:159–68. doi: 10.1007/978-1-4939-3566-6_10. [DOI] [PubMed] [Google Scholar]
- 18.Mensa-Vilaro A, et al. Arthritis Rheumatol. 2016;68:3035–3041. doi: 10.1002/art.39770. [DOI] [PubMed] [Google Scholar]
- 19.Duncan JA, et al. Proc Natl Acad Sci. 2007;104:8041–8046. doi: 10.1073/pnas.0611496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hafner-Bratkovič I, et al. Nat Commun. 2018;9:5182. doi: 10.1038/s41467-018-07573-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y, et al. Nature. 2016;530:354–357. doi: 10.1038/nature16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi H, et al. Nat Immunol. 2016;17:250–258. doi: 10.1038/ni.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coll R, et al. Nat Chem Biol. 2019;In press [Google Scholar]
- 21.Virginio C, et al. J Physiol. 1999;519(Pt 2):335–346. doi: 10.1111/j.1469-7793.1999.0335m.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martín-Sánchez F, et al. Cell Death Differ. 2016;23:1219–31. doi: 10.1038/cdd.2015.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Compan V, et al. J Immunol. 2015;194:1261–1273. doi: 10.4049/jimmunol.1301676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martín-Sánchez F, et al. Cell Death Dis. 2017;8:e2984. doi: 10.1038/cddis.2017.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sester DP, et al. J Immunol. 2014;194:455–462. doi: 10.4049/jimmunol.1401110. [DOI] [PubMed] [Google Scholar]
- 26.de Torre-Minguela C, et al. Sci Rep. 2016;6 doi: 10.1038/srep22586. 22586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young MT, et al. Br J Pharmacol. 2006;149:261–268. doi: 10.1038/sj.bjp.0706880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trott O, Olson A. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frisch MJ, et al. Gaussian, Inc; Wallingford, CT: 2016. [Google Scholar]
- 30.Sánchez-Linares I, et al. BMC Bioinformatics. 2012;13:S13. doi: 10.1186/1471-2105-13-S14-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.