Abstract
A key step in translational initiation is the recruitment of the 43S preinitiation complex by the cap-binding complex [eukaryotic initiation factor 4F (eIF4F)] at the 5′ end of messenger RNA (mRNA) to form the 48S initiation complex (i.e., the 48S). The 48S then scans along the mRNA to locate a start codon. To understand the mechanisms involved, we used cryo–electron microscopy to determine the structure of a reconstituted human 48S. The structure reveals insights into early events of translation initiation complex assembly, as well as how eIF4F interacts with subunits of eIF3 near the mRNA exit channel in the 43S. The location of eIF4F is consistent with a slotting model of mRNA recruitment and suggests that downstream mRNA is unwound at least in part by being “pulled” through the 40S subunit during scanning.
The recruitment of the 43S preinitiation complex (43S PIC) to the 5′ end of mRNA is a critical step during translation initiation. Eukaryotic initiation factors eIF1, eIF1A, eIF3, and eIF5 and the ternary complex (TC) of eIF2–guanosine 5′-triphosphate (GTP)–methionine initiator transfer RNA (tRNAiMet) bind to the 40S ribosomal subunit to form the 43S PIC. Once assembled, the 43S PIC is recruited to the cap-binding complex eIF4F at the 5′ end of mRNA to form a 48S initiation complex (i.e., the 48S). eIF4F consists of a scaffold protein eIF4G, a 7-methylguanosine (m7G) cap-binding protein eIF4E, and a DEAD-box helicase eIF4A. This complex enhances 43S PIC binding and scanning along the mRNA until the start codon is recognized (1–3). In mammals, the recruitment of 43S PIC to mRNA requires interactions between eIF3 and the eIF4G subunit of eIF4F (4–6).
Mammalian eIF3 is a 13-subunit complex (eIF3a to -m) that coordinates several aspects of translation. It stabilizes the binding of the TC on the 40S and interacts with eIF1, eIF1A, and eIF5, which are involved in fidelity of start-site recognition (7, 8). It also prevents pre-mature association of the ribosomal subunits (9) and regulates recruitment of the 43S PIC to mRNA by interacting directly with eIF4F (4–6). Although structures of mammalian eIF3 have been determined at low resolution (~6 Å) (10), it is not clear how it coordinates these vital functions during translation. Additionally, a fundamental question remains: How does eIF4F and its adenosine triphosphatase (ATPase) cycle promote mRNA recruitment and scanning along mRNA? In particular, how is the activity of eIF4F coordinated with eIF3 in the 48S?
In this work, we use single-particle cryo–electron microscopy (cryo-EM) to determine the structure of a reconstituted human 48S. Our work provides a detailed structure of eIFs and structural insights into how eIF4F interacts with eIF3 as part of the 48S.
In vitro reconstitution of the human 48S and its overall structure
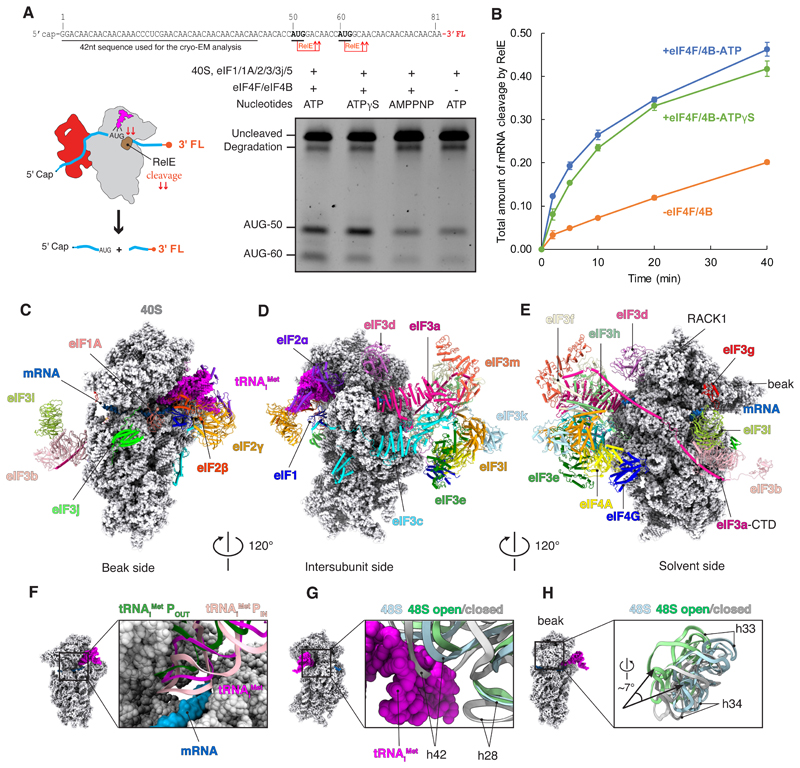
To characterize our purified reconstituted system, we established that mRNA recruitment and scanning follow an eIF4F-dependent pathway. To this end, start-site selection on an m7G capped mRNA was monitored by using the RelE toxin to cleave mRNA in the A site of the 40S subunit when a codon-anticodon interaction forms between tRNAMet and the AUG codon in the P site (11) (Fig. 1, A and B, and fig. S1A). The first AUG codon is preferentially selected in our system, consistent with the scanning model of initiation. We observe efficient start-site selection in the presence of ATP and ATP-g-S. By contrast, adenylyl-imidodiphosphate (AMP-PNP) appreciably reduces start-site selection, which is consistent with our previous work (12). The kinetics of start-site selection is strongly enhanced by eIF4F, indicating that mRNA recruitment and scanning preferentially follow an eIF4F-dependent pathway. To gain insights into the mechanism of mRNA recruitment and scanning, we used single-particle cryo-EM to determine the structure of a 48S complex assembled (with ATP-g-S) on a very similar mRNA but without a start site (Fig. 1A and figs. S2 and S3). An mRNA without a start site was used to capture the 48S at a pathway intermediate after mRNA recruitment but before start-site selection. Because ATP-g-S behaves similarly to ATP in the reconstituted system, we reasoned that this intermediate most likely resembles a scanning intermediate.
Fig. 1. Structure and functional characterization of human 48S complex.
(A) RelE cleavage shows that the efficiency of start-site selection depends on eIF4F (lanes 1 and 4) as well as on ATP hydrolysis (lines 1 and 3). The slowly hydrolysable ATP analog ATPγS (lane 2) has almost the same efficiency of start-site selection when compared with ATP. For the gel, RelE cleavage assay was performed 20 min after the formation of the 48S complex. The degradation band is present in the mRNA prep even in the absence of RelE cleavage (fig. S1A). FL, fluorescent labeled; nt, nucleotide. (B) Kinetic analysis in the presence of eIF4F with ATP or ATPgS, and with ATP absence of eIF4F, shows an eIF4F and ATP-dependent mechanism of scanning under our experimental conditions, and that ATPγS is almost as good as ATP. Error bars indicate SEM. (C to E) Overall structure of 48S shown in different orientations. eIFs are shown in cartoon. tRNAiMet and 40S are represented as magenta and gray spheres. Sugar phosphate backbone of the mRNA is shown as a blue surface. CTD, C-terminal domain. (F) Superposition of the tRNAi Met with the structure of tRNAi Met in the context of 48S in open (POUT) and closed conformation (PIN) (14) shows an intermediate conformation of the anticodon stem loop. (G and H) Superposition of 18S rRNA with the structure of 48S in the open and closed conformations (14) to highlight the changes in their conformation and the swivel movement of the head during scanning.
Although cross-linking with BS3 (materials and methods) did not change the overall structure we obtained (fig. S4), it increased the number of particles containing eIFs and thereby improved resolution. For the analysis of several regions that appeared highly dynamic, we used multibody and focused refinement (fig. S5). The overall resolution of 3.1 Å (figs. S2 and S3) allowed us to identify and place in the maps the previously known structures of the 40S, eIF1, eIF1A, eIF2 (a, b, and g), tRNAiMet, and the octameric structural core of eIF3 as well as its peripheral subunits (b, d, g, i, j) (Fig. 1, C to E; fig. S6; and table S1). These placements were used to segment the maps for detailed model building. Further masked classification on additional unaccounted density yielded a map (fig. S2 and S3) that, despite having a slightly reduced overall resolution of 3.4 Å, improved this additional density. On the basis of our structural and biochemical analysis (fig. S1, B and C) as well as prior data (4–6, 13), we identified this density to be eIF4F (eIF4G, eIF4A, and possibly eIF4E) (Fig. 1, C to E, and fig. S6).
The structure reveals a conformation of the TC that does not involve codon-anticodon base pairing (fig. S7), which is likely to reflect the 48S complex in the process of scanning. The tRNA Met is in a previously unseen orientation, which is intermediate between the previously identified PIN (in which the tRNA is stably base-paired with the start codon in the P site) and POUT (in which the tRNA is not fully inserted into the P site) states (14) (Fig. 1F). Furthermore, the 40S in this complex has an unusual conformation (Fig. 1, G and H). A downward movement of the 40S head when transitioning from the pre- to postscanning state has been described (14, 15), but we observe an additional swivel movement of the head (Fig. 1H). In bacterial initiation, a head swivel changes the position of the tRNAfMet from the P site to between the P and E sites (16). Although the movement is similar, in this case the tRNAiMet remains in the P site. The much looser interaction of the tRNAiMet with mRNA (fig. S7), along with the absence of a start codon in the mRNA, suggests that the structure represents the conformation of the 48S during or just before scanning.
A near-atomic resolution structure of the human eIF3 in the context of 48S
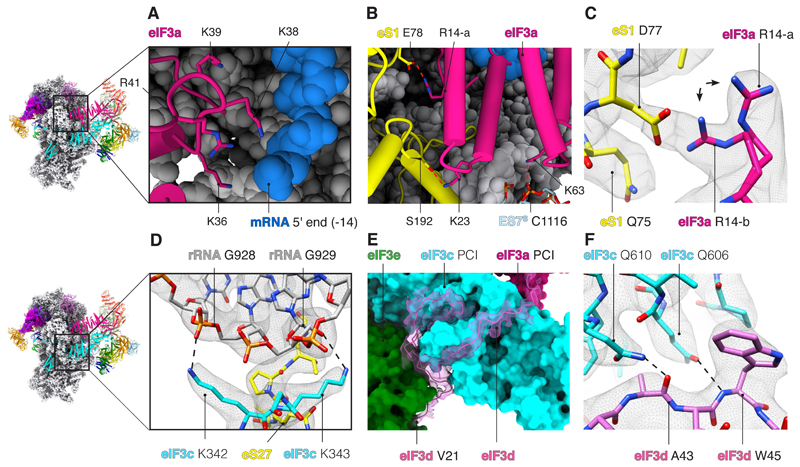
The details of the organization of the 48S and its intramolecular interactions were poorly understood in the absence of a high-resolution structure of the complex. The 43S part of our 48S structure has a resolution of ~3 Å and includes all 13 subunits of eIF3 (Fig. 1 and fig. S8, A to C), most of which could be modeled in atomic detail. As a result, we can under-stand how evolutionarily conserved residues of eIF3 are important for interactions with 18S ribosomal RNA (rRNA) and ribosomal proteins (r-proteins) and rationalize previous biochemical data (17, 18). The subunits eIF3a and eIF3c are universally conserved across eukaryotes. They interact with the 40S as well as mRNA through their RNA binding motifs (Fig. 2, A to D, and table S2) (18). The RNA binding motif of eIF3a (conserved in metazoans) interacts with mRNA near the exit site (Fig. 2A), whereas the RNA binding motif of eIF3c (conserved in eukaryotes) interacts with rRNA on the back of 40S (Fig. 2D and table S2). Additionally, the highly conserved residue Lys63 in eIF3a interacts with rRNA expansion segment 7 (ES7S) near the exit site (Fig. 2B and table S2). The structure also reveals the basis of the interaction of eIF3a and eIF3c with r-proteins (Fig. 2B and table S2). Although the sequence register of our structure differs from that of a previous low-resolution structure of rabbit eIF3 (10), it agrees with the crystal structure of yeast eIF3a and eIF3c (fig. S8, D to H) (19). Furthermore, the amino acid sequence of the region of eIF3a that interacts with r-protein eS1 (table S2) is highly conserved in mammals (fig. S8F). Thus, it is likely that there is also structural conservation between human and rabbit eIF3a. Consistently, some interactions observed between human eIF3a and r-protein eS1 (table S2) have also been partially described in a recent structure of rabbit eIF3 (20).
Fig. 2. Interactions of eIF3 with mRNA and 40S.
(A) Close-up of the mRNA exit site highlighting the interaction of eIF3a (pink) with mRNA (blue sugar-phosphate backbone). K, Lys. (B) eIF3a interacts with ribosomal protein eS1 (yellow) near the exit site. Red dashed lines represent protein-protein or protein-RNA interactions. eIF3a R14-a and K23 interact with eS1 E78 and S192. Furthermore, eIF3a K63 interacts with rRNA ES7S C1116. C, Cys; E, Glu; K, Lys; R, Arg; S, Ser. (C) Close-up of R14 in eIF3a to highlight in two alternative rotamer conformations (R14-a and R14-b) and the interaction of R14-b with D77 of r-protein eS1. D, Asp; Q, Gln. (D) Close-up of the interaction of eIF3c with 18S rRNA on the back of the 40S. G, Gly. (E) eIF3d-NTT (orchid) fitted into the cryo-EM map to highlight the close interactions with PCI domains of eIF3c (cyan surface) and eIF3e (green surface). V, Val. (F) Close-up of the PCI domain of eIF3c to highlight some described interactions with eIF3d-NTT. A, Ala; W, Trp.
Additionally, eIF3d interacts with both 40S and the octameric structural core of eIF3 (Fig. 2, E and F; table S2; and fig S8). The N-terminal tail of eIF3d (eIF3d-NTT), hitherto unseen, interacts with the PCI domain of eIF3e (Fig. 2E), consistent with previous biochemical data (21).
The structure shows that eIF3d interacts with the eIF3 octameric structural core, as well as potentially with eIF4F. The subunit binds to a region of eIF3e that is also involved in binding to eIF4F, which agrees with predicted interaction between eIF3d and eIF4G (4). eIF3d also interacts with eIF3c and probably eIF3a (Fig. 2E and fig. S8B). The eIF3d-NTT loop (residues 33 to 59), which binds to the PCI domain of eIF3c, contains highly conserved residues such as Trp45, which interacts with Gln606 in eIF3c (Fig. 2F and table S2).
eIF1 binds to a mammalian-specific insertion in the eIF3c N-terminal domain
eIF3 coordinates start-site selection by interacting with the fidelity factors eIF1 and eIF5 (7, 8) and prevents premature association between ribosomal subunits (9). The N-terminal domain of eIF3c (eIF3c-NTD) extends toward the decoding center of the ribosome, where it interacts with eIF1 (fig. S9).
Our structure and accompanying bio-chemistry (fig. S9) unexpectedly reveal that a conserved mammalian-specific insertion in eIF3c (fig. S9) is involved in the interaction with eIF1. In yeast, the interaction between eIF3c-NTD and eIF1 occurs through the very N-terminal tail of eIF3c (residues 1 to 63) (7), whereas our structures and biochemical data reveal that this interaction occurs through the C-terminal end of eIF3c-NTD (residues 166 to 287) (fig. S9).
The resolution and completeness of the structure allowed us to build and assign to eIF3c-NTD a cluster of four helices located in a pocket formed by rRNA helices h11, h27, and h44 and ribosomal protein uS15 (fig. S9). The main interaction with 40S occurs through the conserved and charged residues located in helix 4 (fig. S9 and table S2). Because this domain would clash sterically with parts of rRNA from the large subunit involved in the formation of intersubunit bridges B4 and eB11 (fig. S10), it should contribute to the anti-association activity of eIF3 (9). The same density, but at low resolution, has also been observed in yeast, in which it was also tentatively assigned to eIF3c-NTD (7, 14). Thus, the structural basis for the anti-association activity of eIF3c-NTD appears to be evolutionarily conserved among eukaryotes.
eIF3g binds at the mRNA entry site
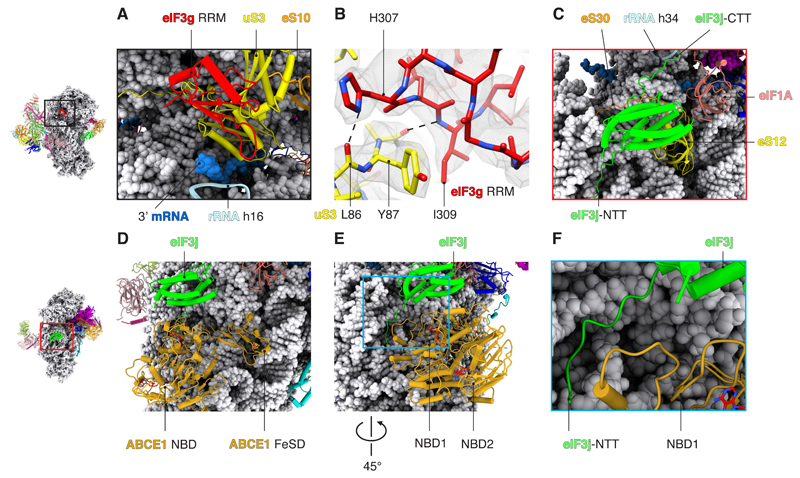
eIF3g contains an RNA-recognition motif (eIF3g-RRM) that is thought to enhance scanning efficiency (22). In a previous low-resolution study, a density corresponding to RRM motifs was observed at the mRNA entry site and assigned to eIF4B (23). However, this density was still present in a 48S complex we assembled without eIF4B (fig. S11). We have now assigned this density to eIF3g-RRM, although we cannot rule out that eIF4B can also bind to this region during the initiation pathway. In our structure, eIF3g-RRM interacts with rRNA h16 and ribosomal proteins uS3 and eS10 (Fig. 3, A and B, and table S2). Considering that it binds at the mRNA entry channel and its possible interaction with backbone of the mRNA, eIF3g-RRM could facilitate the recruitment of the 43S PIC to mRNA, thereby enhancing translational efficiency (22).
Fig. 3. Interactions of peripheral subunits of eIF3.
(A) eIF3g-RNA binding motif viewed from solvent side to highlight its interaction with rRNA in helix 16 and ribosomal proteins uS3 and eS10. (B) Binding interface between eIF3g and the ribosomal protein uS3 at the mRNA entry channel. H, His; I, Ile; L, Leu; Y, Tyr. (C) eIF3j binds to ribosomal proteins eS30 and uS12 near the A site. The C-terminal tail of eIF3j (eIF3j-CTT) interacts with ribosomal rRNA in helix 34. eIF3j-NTT is positioned next to the GTPase binding region of the 40S. (D) Superposition of eIF3j with the structure of ABCE1 (PDB: 5LL6) bound to the 40S subunit (post splitting) (29). (E and F) eIF3j-NTT extends toward the GTPase binding region of the 40S, where the nucleotide binding domain 1 (NBD1) of ABCE1 binds.
The eIF3bgi subcomplex was previously suggested to bind the 40S subunit through an interaction between the eIF3 b-propeller domain and uS4 ribosomal protein (19). A better local resolution in our structure (figs. S5 and S12), together with the identification of the eIF3g-RRM, improves our understanding about the residues in the eIF3bgi sub-complex that interact with the 40S subunit (table S2).
A dual role for eIF3j in start-site selection and recycling
eIF3j binds near the decoding center of the ribosome as predicted (24, 25), where it could coordinate start-site selection. The eIF3j-NTD and helix 5 interact with 40S body through ribosomal proteins eS30, uS12, and 18S rRNA. Additionally, eIF3j is close enough to interact with eIF1A (Fig. 3C and fig. S13). The C-terminal domain of eIF3j bridges the head with the body of 40S by interacting with the rRNA h34 in the mRNA entry latch (a constriction in the mRNA entry channel). This interaction may have a regulatory role by pre-venting the movement of the head of 40S and limiting the closed conformation of the ribosome (12). Furthermore, it might be associated with the known role of eIF3j in preventing leaky scanning (26).
Human eIF3j interacts with 40S in the same location where the mammalian translational auxiliary factor DHX29 has been observed to bind (fig. S14) (10). Thus, the recruitment of DHX29 during the translation of select mRNAs may trigger the release of eIF3j and overcome its possible regulatory role. This interpretation agrees with the unwinding independent role of DHX29 during scanning (27).
The structure also suggests how eIF3j bridges recycling with a new round of translation. The eIF3j-NTT extends toward the conserved GTPase binding region of the ribosome (Fig. 3, D to F), where ABCE1 binds. ABCE1 is an ATPase that is involved in recycling of eukaryotic ribosomes after termination. Although ATP hydrolysis is not required for ribosome splitting, it is required for subsequent release of ABCE1 (28). The structure of ABCE1 is different in the 80S ribosome (presplitting), compared with the one bound just to the 40S subunit (post splitting). Superimposing our structure with structures of ABCE1 in the post-splitting complex (29) places the eIF3j-NTT in a position where it could interact with the nucleotide-binding domain of ABCE1 (Fig. 3, E and F). This model would be consistent with previous studies showing that the eIF3j-NTD facilitates ribosome recycling by ABCE1 (30). The structure suggests that the rotation of the iron-sulfur cluster domain of ABCE1 that occurs after splitting (20, 29) would prevent a steric clash with eIF3j (fig. S14). Thus, eIF3j is likely to be recruited after subunit splitting to prepare the resulting 40S subunit for a new round of initiation. This interpretation agrees with previous data showing co-purification of ABCE1 with components of the 43S PIC, including eIF3j (29). After the release of ABCE1, eIF3 would prevent premature association of 60S.
Location of eIF4F adjacent to eIF3e, -k, and -l near the mRNA exit channel of the 40S subunit
Adjacent to eIF3e, -k, and -l, we see a region that has low local resolution due to flexibility (fig. S15A) that we attribute to eIF4F. Biochemical and genetics studies have indicated that the recruitment of 43S to an mRNA bound to the cap-binding complex and the subsequent scanning process are greatly enhanced by interactions between the middle domain of eIF4G and eIF3 (4–6). Rigid-body fitting of a crystal structure of a partial eIF4G-eIF4A complex from yeast (31) shows close agreement with the density (fig. S15), thus identifying the position of eIF4F in the 48S, consistent with previous biochemical data (4, 5, 32). It is likely that other domains of eIF4G, not visible in this work because of possible flexibility, could also make interactions with nearby subunits such as the eIF3d-NTT and eIF3e, which has been previously suggested by biochemical cross-linking (4).
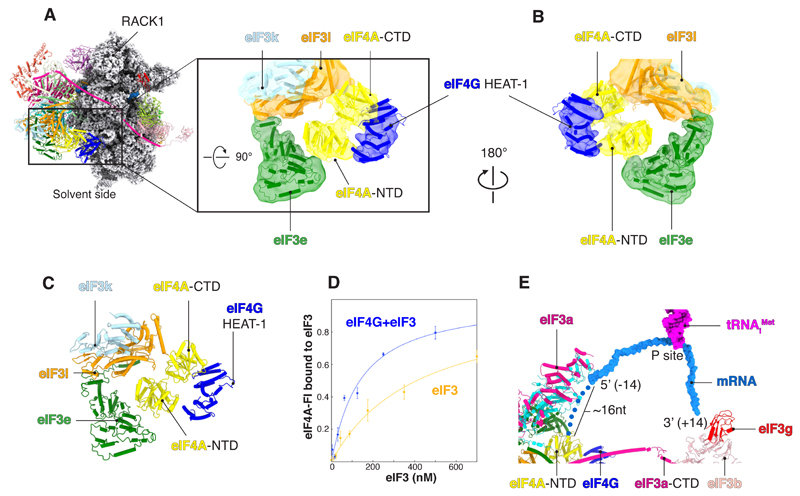
eIF4A binds to eIF3k and -l and eIF3e near the ES6S (Fig. 4, A and C), which agrees with recent cross-linking and immuno-EM data (13). To further validate the interaction between eIF4A and eIF3, we determined the binding affinity between these factors by fluorescence anisotropy. Consistent with our structure, eIF4A alone binds to eIF3, and its affinity is increased in the presence of eIF4G (Fig. 4D and table S3). We also used cross-linking mass spectrometry (XL-MS) to identify interactions between eIF4F and eIF3 in the absence of the 48S (fig. S15). These data are consistent with the interactions observed in our structure.
Fig. 4. Interactions between eIF4F and eIF3 octameric structural core.
(A and B) Rigid-body fitting (correlation = 0.92) of human homology model of eIF4A/eIF4G-HEAT1 into a cryo-EM map filtered to local resolution (6 to 11 Å). (C) eIF4A binds to a pocket formed by eIF3l and eIF3e. (D) Saturation binding curves showing the fraction of eIF4A bound to eIF3 in the presence or absence of eIF4G. Error bars indicate SEM. (E) Surface representation of the mRNA (sugar-phosphate backbone) to highlight the path in the 48S. Blue dots represent a tentative path for the mRNA from the exit site (position −14 from the P site) toward eIF4A-NTD. The tentative path is based on weak unassigned density (fig. S16).
Our map contains unassigned density adjacent to the eIF4G-eIF4A density, in close contact with eIF3k and -l (fig. S17). Although we cannot unambiguously assign it because of the low local resolution, the size and shape of the density are consistent with that of eIF4E. This location of eIF4E agrees well with our XL-MS data, which show that eIF4E interacts with eIF3k and -l (fig. S17), as well as previous proximity-labeling (BioID) data, indicating that eIF4E and eIF3l are in close proximity in live cells (33).
In our structure, eIF4F interacts with the 43S PIC entirely through subunits of eIF3 that are not present in yeast (eIF3e, -k, and -l). The interaction we observe between eIF4A and eIF3k and -l is particularly surprising given that these individual eIF3 subunits are dispensable in Neurospora crassa and Caenorhabditis elegans (34, 35). In addition to a likely interaction between eIF3e and eIF4G, our structure together with biochemical and genetic evidence indicates that substantial redundancy is likely to exist between these interactions. It will, therefore, be important to test this possibility in the future by using double eIF3 subunit knockouts in different organisms. The structure suggests that the interactions that eIF4F makes with other components of the 48S are likely to differ greatly between species, the molecular basis of which will be important to solve with future structures.
The cap-binding complex is positioned at the 5′ end of mRNA relative to the 40S subunit (Fig. 4). Translation complex profile sequencing data indicated that the scanning 48S has 5′-extended footprints upstream but not downstream of the 40S (36, 37), which is consistent with the position of eIF4F in our structure. Nevertheless, we cannot rule out that other conformations of eIF4A may exist during its ATPase cycle and movement along mRNA.
A blind spot in the mRNA
In our structure, the location of eIF4F upstream (to the 5′ end) of the 43S complex is most consistent with mRNA being recruited to the 40S subunit by a slotting mechanism. Direct slotting of mRNA would also be compatible with translation of circular mRNAs (38) as well as initiation on mRNAs containing an internal ribosome entry site. Even during canonical initiation, mRNA is thought to be circularized by the polyadenylate [poly(A)]– binding protein (PABP) interacting with the poly(A) tail at the 3′ end and eIF4F (through eIF4G) at the 5′ end (39), thus favoring a slotting model for mRNA loading into the 40S. Such slotting would require rearrangements in some elements of eIF3 to make the mRNA channel accessible initially; however, eIF3 is known to be dynamic, with various parts of its structure becoming ordered in different states, and in other contexts, the small subunit is known to close upon mRNA binding (14).
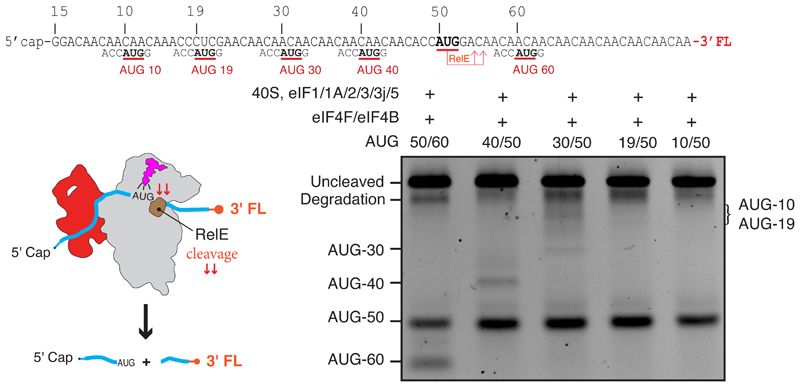
A slotting model of mRNA recruitment would result in a “blind spot” that would preclude recognition of start sites upstream of the location of the P site at the point of recruitment, which would be at least 30 nucleotides from the 5′ end on the basis of our structure (Fig. 4E). We tested for a blind spot in our RelE assay using a series of mRNAs that have a fixed start site at 50 nucleotides from the m7G cap, which would be downstream of the blind spot, and an additional start site located either upstream (10, 19, 30, and 40 nucleotides) or downstream (60 nucleotides). Our data show that in either case, efficient initiation primarily occurs on the first start site that is encountered beyond the blind spot, namely at 50 nucleotides from the m7G cap (Fig. 5). We do observe some initiation at a distance of 40 nucleotides from the m7G cap but very little or none at distances of 30, 19, or 10 nucleotides. This blind spot is consistent with the fact that leaky scanning is observed on mRNAs that contain 5′ untranslated regions (5′UTRs) of less than 32 nucleotides (40). We thus propose that a blind spot of between 30 and 40 nucleotides exists on human mRNAs, which would be compatible with the typical 5′UTR of mRNA in humans, whose median length is 218 nucleotides (41).
Fig. 5. Blind spot for start-site recognition.
RelE cleavage assay (materials and methods) shows a blind spot for start-site selection for mRNAs with a start codon less than 30 to 40 nucleotides from the 5′ end. All five mRNAs tested have an AUG at position 50 just beyond the blind spot and a second AUG at positions downstream (60 nucleotides) or upstream (40, 30, 19, and 10 nucleotides) of it. The main site of cleavage is at AUG 50 in all cases, showing that initiation occurs primarily at the first start codon downstream of the blind spot, and there is little or no cleavage at start codons for 10 to 40 nucleotides.
A model for recruitment and scanning
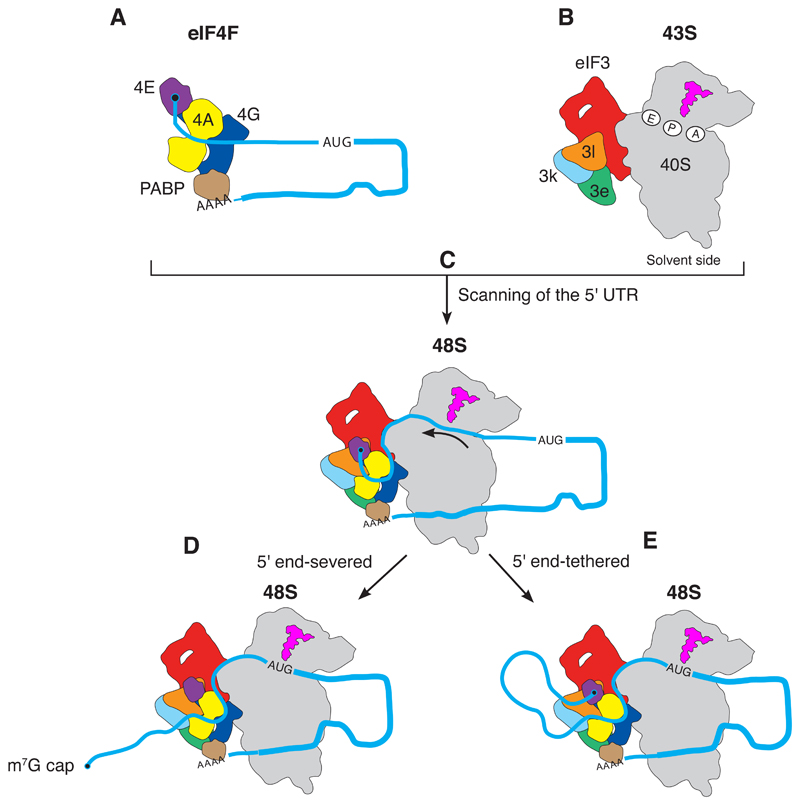
The structural and biochemical data suggest a model in which mRNA is slotted into the 40S just downstream of eIF4F during recruitment (Fig. 6). The location of eIF4A upstream of the 40S subunit (Fig. 4) suggests that the mRNA is likely to be pulled through the channel in the 40S subunit during scanning. The model is economic in terms of rearrangements: Once the 43S PIC is recruited to the cap-binding complex, the entire 48S initiation complex essentially stays intact while the mRNA is pulled through the 40S subunit, until the start codon reaches the P site of the ribosome and triggers subsequent steps in initiation.
Fig. 6. Model for mRNA scanning during canonical translational initiation suggested by the structure.
The eIF4F at the m7G cap at the 5′ end of mRNA (A) recruits the 43S complex of the 40S subunit with initiation factors and initiator tRNAi Met (B) to form the 48S complex (C). eIF4F binds to the eIF3 structural core, which places eIF4E 30 to 40 nucleotides upstream of the P site of the 40S ribosomal subunit. During scanning, the mRNA is pulled through the 40S subunit [indicated by arrow in (C)], until the start codon is reached (D and E). In two alternative scenarios, eIF4F could dissociate from the cap during scanning (D), or it could stay bound resulting in the mRNA forming a loop (E). Although not part of the structure in this work, the mRNA is shown with a poly(A) tail and a PABP interacting with eIF4F to reflect the situation in vivo (39).
The hydrolysis of ATP by eIF4A is required for efficient scanning (Fig. 1A), but precisely how ATP hydrolysis promotes scanning is not clear. It is possible that eIF4F could work as a Brownian ratchet (42) in which the 40S subunit would slide along the mRNA in a stochastic manner, but eIF4F would move unidirectionally along the mRNA to keep up with the 40S in an ATP-dependent manner and act as a pawl or backstop to prevent reverse movement of the 40S subunit. In this model, the ATP would ensure unidirectionality of scanning, but the energy to melt secondary structure would come just from thermal fluctuations and the 40S interaction, consistent with the finding that eIF4F alone is not processive (43, 44). Ribosome proteins uS3, uS4, and uS5 located in the entry channel of the small ribosomal subunit help to unwind mRNA secondary structure during translocation (45). It is, therefore, possible that eIF4A may be exploiting an intrinsic property of the 40S subunit in a similar way during scanning. This model leaves the mRNA entry site free to bind factors known to facilitate translation of mRNAs with extended secondary structure, such as DHX29, which has been observed at the entry site (10). Because there is a second eIF4A binding site on eIF4G, and moreover, eIF4A is present in excess over other components of eIF4F and is known to have additional roles in melting RNA secondary structure (46), it is possible that one or more additional eIF4A molecules could also play a role downstream of the 48S to facilitate translation. Therefore, pulling of the mRNA through the 40S may be only one part of a complex mechanism of unwinding of mRNA secondary structure and scanning. The role of eIF4B also remains unclear and may be part of this process.
eIF4E stimulates the helicase activity of eIF4A (47) and thus likely remains bound to the rest of eIF4F throughout the process (37). From our structure, it is not clear whether the 5′ end of mRNA is released from eIF4E and progressively moves farther away from the 48S PIC during scanning (Fig. 6D). Although the dissociation rate of isolated eIF4E from the m7G cap is quite high (48), its interaction with the rest of eIF4F could enable efficient and rapid rebinding of eIF4E to the 5′ cap (fig. S1, B and C) (49). This would require the 5′UTR mRNA to loop out as it is pulled through the 40S subunit during scanning (Fig. 6E), a possibility also previously suggested (3). Recent evidence to support this model has been provided by 40S selective ribosome footprinting, which indicates that the scanning 48S complex remains tethered to the m7G cap throughout the scanning process in human cells (37).
Some rare mRNAs with unusually short 5′UTRs would place the recruited 43S PIC beyond the initiation codon (6). We propose that these types of mRNAs would likely exploit an alternative recruitment pathway, perhaps occurring in the absence of eIF1 (6). Such mRNAs could possibly require a different role of eIF4F or even be independent of it. Previous work showing eIF4F-dependent translation of mRNAs with very short 5′UTRs propose that eIF4F binds at the entry rather than exit site, followed by dissociation of eIF4E from the cap and threading of mRNA into the 40S subunit through its decoding site (6). For such a model to be compatible with our structure, the eIF4F complex would need to relocate to the opposite side of the 40S at some stage, which we consider unlikely. To resolve these discrepancies, it will be important to determine structures of 48S complexes with other mRNAs, including those with a short 5′UTR.
Outlook
This work reveals the structure of an essentially complete 43S PIC (in the context of 48S PIC) at high resolution as well as its interactions with the cap-binding complex at the 5′ end of mRNA (movie S1). The structure sheds light on several important but hitherto unresolved aspects of initiation, including the mechanism of mRNA recruitment to the 43S PIC and how the position of eIF4F at the mRNA exit channel likely facilitates the process of scanning.
Supplementary Material
Acknowledgments
We thank T. Nakane and V. Chandrasekaran for advice on data processing; A. Hinnebusch, J. Lorsch, R. Hegde, J. Llácer, C. Rae, W. Filipowicz, and N. Sonenberg for helpful comments on the manuscript; and P. Emsley for advice on model building and refinement. The cryo-EM data were collected at the MRC Laboratory of Molecular Biology Electron Microscopy Facility and at the UK national Electron Bio-Imaging Centre (eBIC) (proposal EM17434-62 and EM17434-72, funded by the Wellcome Trust, MRC, and BBSRC). We thank the MRC LMB and eBIC facilities for support with the EM data collection, J. Grimmett and T. Darling for computing, and S. Maslen for assistance with XL-MS.
Funding
J.B.Q. was supported by a FEBS long-term fellowship; V.R. was supported by the UK Medical Research Council (MC_U105184332), a Wellcome Trust Senior Investigator award (WT096570), and the Louis-Jeantet Foundation; C.S.F. was supported by the NIH (grant R01 GM092927).
Data and materials availability
EM maps have been uploaded to the Electron Microscopy Data Bank with accession codes EMD-10775 (48S-40S body), EMD-10772 (48S-40S head), EMD-10769 (48S-eIF3 structural core), EMD-10773 (48S-eIF3bgi), EMD-10774 (48S-eIF2-TC), and EMD-11302 (48S). Protein coordinates have been deposited in the Protein Data Bank (PDB) with IDs 6YBW (48S-40S body), 6YBS (48S-40S head), 6YBD (48S-eIF3 structural core), 6YBT (48S-eIF3bgi), 6YBV (48S-eIFe-TC), and 6ZMW (48S).
Footnotes
Author contributions: M.S. and C.S.F. purified eIFs and performed the binding assay and functional analysis. J.M.S. performed XL-MS. J.B.Q. assembled and biochemically characterized the complex, prepared cryo-EM grids, and performed the cryo-EM analysis. J.B.Q., Y.G., S.K., and V.R. interpreted the structure. J.B.Q. and S.K. interpreted the density to build and refine atomic models. J.B.Q., M.S., C.S.F., and V.R. wrote the manuscript with input from all authors. S.K. made movie S1. V.R. supervised the project.
Competing interests: The authors declare no competing interests.
References and Notes
- 1.Grifo JA, Tahara SM, Morgan MA, Shatkin AJ, Merrick WC. New initiation factor activity required for globin mRNA translation. J Biol Chem. 1983;258:5804–5810. Medline. [PubMed] [Google Scholar]
- 2.Morino S, Imataka H, Svitkin YV, Pestova TV, Sonenberg N. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol. 2000;20:468–477. doi: 10.1128/MCB.20.2.468-477.2000. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villa N, Do A, Hershey JWB, Fraser CS. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J Biol Chem. 2013;288:32932–32940. doi: 10.1074/jbc.M113.517011. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeFebvre AK, Korneeva NL, Trutschl M, Cvek U, Duzan RD, Bradley CA, Hershey JWB, Rhoads RE. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J Biol Chem. 2006;281:22917–22932. doi: 10.1074/jbc.M605418200. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar P, Hellen CUT, Pestova TV. Toward the mechanism of eIF4F-mediated ribosomal attachment to mammalian capped mRNAs. Genes Dev. 2016;30:1573–1588. doi: 10.1101/gad.282418.116. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Obayashi E, Luna RE, Nagata T, Martin-Marcos P, Hiraishi H, Singh CR, Erzberger JP, Zhang F, Arthanari H, Morris J, Pellarin R, et al. Molecular landscape of the ribosome pre-initiation complex during mRNA scanning: Structural role for eIF3c and its control by eIF5. Cell Rep. 2017;18:2651–2663. doi: 10.1016/j.celrep.2017.02.052. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valásek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol Cell Biol. 2004;24:9437–9455. doi: 10.1128/MCB.24.21.9437-9455.2004. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CUT, Pestova TV. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.des Georges A, Dhote V, Kuhn L, Hellen CUT, Pestova TV, Frank J, Hashem Y. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015;525:491–495. doi: 10.1038/nature14891. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreev D, Hauryliuk V, Terenin I, Dmitriev S, Ehrenberg M, Shatsky I. The bacterial toxin RelE induces specific mRNA cleavage in the A site of the eukaryote ribosome. RNA. 2008;14:233–239. doi: 10.1261/rna.693208. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sokabe M, Fraser CS. A helicase-independent activity of eIF4A in promoting mRNA recruitment to the human ribosome. Proc Natl Acad Sci USA. 2017;114:6304–6309. doi: 10.1073/pnas.1620426114. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toribio R, Díaz-López I, Boskovic J, Ventoso I. Translation initiation of alphavirus mRNA reveals new insights into the topology of the 48S initiation complex. Nucleic Acids Res. 2018;46:4176–4187. doi: 10.1093/nar/gky071. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Llácer JL, Hussain T, Marler L, Aitken CE, Thakur A, Lorsch JR, Hinnebusch AG, Ramakrishnan V. Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex. Mol Cell. 2015;59:399–412. doi: 10.1016/j.molcel.2015.06.033. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hussain T, Llácer JL, Fernández IS, Munoz A, Martin-Marcos P, Savva CG, Lorsch JR, Hinnebusch AG, Ramakrishnan V. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell. 2014;159:597–607. doi: 10.1016/j.cell.2014.10.001. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hussain T, Llácer JL, Wimberly BT, Kieft JS, Ramakrishnan V. Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation. Cell. 2016;167:133–144.e13. doi: 10.1016/j.cell.2016.08.074. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aitken CE, Beznosková P, Vlckova V, Chiu W-L, Zhou F, Valášek LS, Hinnebusch AG, Lorsch JR. Eukaryotic translation initiation factor 3 plays distinct roles at the mRNA entry and exit channels of the ribosomal preinitiation complex. eLife. 2016;5:e20934. doi: 10.7554/eLife.20934. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun C, Querol-Audí J, Mortimer SA, Arias-Palomo E, Doudna JA, Nogales E, Cate JHD. Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. Nucleic Acids Res. 2013;41:7512–7521. doi: 10.1093/nar/gkt510. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erzberger JP, Stengel F, Pellarin R, Zhang S, Schaefer T, Aylett CHS, Cimermancic P, Boehringer D, Sali A, Aebersold R, Ban N. Molecular architecture of the 40S⋅eIF1⋅eIF3 translation initiation complex. Cell. 2014;158:1123–1135. doi: 10.1016/j.cell.2014.07.044. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonetti A, Guca E, Bochler A, Kuhn L, Hashem Y. Structural Insights into the Mammalian Late-Stage Initiation Complexes. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.03.061. 107497, Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith MD, Arake-Tacca L, Nitido A, Montabana E, Park A, Cate JH. Assembly of eIF3 Mediated by Mutually Dependent Subunit Insertion. Structure. 2016;24:886–896. doi: 10.1016/j.str.2016.02.024. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuchalová L, Kouba T, Herrmannová A, Dányi I, Chiu W-L, Valásek L. The RNA recognition motif of eukaryotic translation initiation factor 3g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol Cell Biol. 2010;30:4671–4686. doi: 10.1128/MCB.00430-10. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eliseev B, Yeramala L, Leitner A, Karuppasamy M, Raimondeau E, Huard K, Alkalaeva E, Aebersold R, Schaffitzel C. Structure of a human cap-dependent 48S translation pre-initiation complex. Nucleic Acids Res. 2018;46:2678–2689. doi: 10.1093/nar/gky054. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraser CS, Berry KE, Hershey JWB, Doudna JA. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol Cell. 2007;26:811–819. doi: 10.1016/j.molcel.2007.05.019. Medline. [DOI] [PubMed] [Google Scholar]
- 25.Aylett CHS, Boehringer D, Erzberger JP, Schaefer T, Ban N. Structure of a yeast 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex. Nat Struct Mol Biol. 2015;22:269–271. doi: 10.1038/nsmb.2963. Medline. [DOI] [PubMed] [Google Scholar]
- 26.ElAntak L, Wagner S, Herrmannová A, Karásková M, Rutkai E, Lukavsky PJ, Valásek L. The indispensable N-terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1-RRM and with eIF1A in stringent AUG selection. J Mol Biol. 2010;396:1097–1116. doi: 10.1016/j.jmb.2009.12.047. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pisareva VP, Pisarev AV. DHX29 and eIF3 cooperate in ribosomal scanning on structured mRNAs during translation initiation. RNA. 2016;22:1859–1870. doi: 10.1261/rna.057851.116. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gouridis G, Hetzert B, Kiosze-Becker K, de Boer M, Heinemann H, Nürenberg-Goloub E, Cordes T, Tampé R. ABCE1 Controls Ribosome Recycling by an Asymmetric Dynamic Conformational Equilibrium. Cell Rep. 2019;28:723–734e6. doi: 10.1016/j.celrep.2019.06.052. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heuer A, Gerovac M, Schmidt C, Trowitzsch S, Preis A, Kötter P, Berninghausen O, Becker T, Beckmann R, Tampé R. Structure of the 40S-ABCE1 post-splitting complex in ribosome recycling and translation initiation. Nat Struct Mol Biol. 2017;24:453–460. doi: 10.1038/nsmb.3396. Medline. [DOI] [PubMed] [Google Scholar]
- 30.Young DJ, Guydosh NR. Hcr1/eIF3j Is a 60S Ribosomal Subunit Recycling Accessory Factor In Vivo. Cell Rep. 2019;28:39–50e4. doi: 10.1016/j.celrep.2019.05.111. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schütz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: An RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci USA. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yourik P, Aitken CE, Zhou F, Gupta N, Hinnebusch AG, Lorsch JR. Yeast eIF4A enhances recruitment of mRNAs regardless of their structural complexity. eLife. 2017;6:e31476. doi: 10.7554/eLife.31476. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chapat C, Jafarnejad SM, Matta-Camacho E, Hesketh GG, Gelbart IA, Attig J, Gkogkas CG, Alain T, Stern-Ginossar N, Fabian MR, Gingras A-C, et al. Cap-binding protein 4EHP effects translation silencing by microRNAs. Proc Natl Acad Sci USA. 2017;114:5425–5430. doi: 10.1073/pnas.1701488114. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cattie DJ, Richardson CE, Reddy KC, Ness-Cohn EM, Droste R, Thompson MK, Gilbert WV, Kim DH. Mutations in Nonessential eIF3k and eIF3l Genes Confer Lifespan Extension and Enhanced Resistance to ER Stress in Caenorhabditis elegans. PLOS Genet. 2016;12:e1006326. doi: 10.1371/journal.pgen.1006326. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith MD, Gu Y, Querol-Audí J, Vogan JM, Nitido A, Cate JHD. Human-like eukaryotic translation initiation factor 3 from Neurospora crassa. PLOS ONE. 2013;8:e78715. doi: 10.1371/journal.pone.0078715. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Archer SK, Shirokikh NE, Beilharz TH, Preiss T. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature. 2016;535:570–574. doi: 10.1038/nature18647. Medline. [DOI] [PubMed] [Google Scholar]
- 37.Bohlen J, Fenzl K, Kramer G, Bukau B, Teleman AA. Selective 40S Footprinting Reveals Cap-Tethered Ribosome Scanning in Human Cells. Mol Cell. 2020 doi: 10.1016/j.molcel.2020.06.005. Medline. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen L-L, Wang Y, Wong CC, et al. Extensive translation of circular RNAs driven by N6methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/S1097-2765(00)80122-7. Medline. [DOI] [PubMed] [Google Scholar]
- 40.Kozak M. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1991;1:111–115. Medline. [PMC free article] [PubMed] [Google Scholar]
- 41.Leppek K, Das R, Barna M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol. 2018;19:158–174. doi: 10.1038/nrm.2017.103. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spirin AS. How does a scanning ribosomal particle move along the 5′-untranslated region of eukaryotic mRNA? Brownian Ratchet model. Biochemistry. 2009;48:10688–10692. doi: 10.1021/bi901379a. Medline. [DOI] [Google Scholar]
- 43.García-García C, Frieda KL, Feoktistova K, Fraser CS, Block SM. RNA BIOCHEMISTRY. Factor-dependent processivity in human eIF4A DEAD-box helicase. Science. 2015;348:1486–1488. doi: 10.1126/science.aaa5089. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rozen F, Edery I, Meerovitch K, Dever TE, Merrick WC, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/MCB.10.3.1134. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. Medline. [DOI] [PubMed] [Google Scholar]
- 46.Tauber D, Tauber G, Khong A, Van Treeck B, Pelletier J, Parker R. Modulation of RNA Condensation by the DEAD-Box Protein eIF4A. Cell. 2020;180:411–426.e16. doi: 10.1016/j.cell.2019.12.031. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci USA. 2013;110:13339–13344. doi: 10.1073/pnas.1303781110. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slepenkov SV, Korneeva NL, Rhoads RE. Kinetic mechanism for assembly of the m7GpppG.eIF4E.eIF4G complex. J Biol Chem. 2008;283:25227–25237. doi: 10.1074/jbc.M801786200. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee AS, Kranzusch PJ, Doudna JA, Cate JHD. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature. 2016;536:96–99. doi: 10.1038/nature18954. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sokabe M, Fraser CS. Human eukaryotic initiation factor 2 (eIF2)-GTP-Met-tRNAi ternary complex and eIF3 stabilize the 43 S preinitiation complex. J Biol Chem. 2014;289:31827–31836. doi: 10.1074/jbc.M114.602870. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Griffin MA, Davis JH, Strobel SA. Bacterial toxin RelE: A highly efficient ribonuclease with exquisite substrate specificity using atypical catalytic residues. Biochemistry. 2013;52:8633–8642. doi: 10.1021/bi401325c. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brito Querido J, Mancera-Martínez E, Vicens Q, Bochler A, Chicher J, Simonetti A, Hashem Y. The cryo-EM Structure of a Novel 40S Kinetoplastid-Specific Ribosomal Protein. Structure. 2017;25:1785–1794.e3. doi: 10.1016/j.str.2017.09.014. Medline. [DOI] [PubMed] [Google Scholar]
- 53.Götze M, Pettelkau J, Fritzsche R, Ihling CH, Schäfer M, Sinz A. Automated assignment of MS/MS cleavable cross-links in protein 3D-structure analysis. J Am Soc Mass Spectrom. 2015;26:83–97. doi: 10.1007/S13361-014-1001-1. Medline. [DOI] [PubMed] [Google Scholar]
- 54.Russo CJ, Scotcher S, Kyte M. A precision cryostat design for manual and semi-automated cryo-plunge instruments. Rev Sci Instrum. 2016;87:114302. doi: 10.1063/1.4967864. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lomakin IB, Steitz TA. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature. 2013;500:307–311. doi: 10.1038/nature12355. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakane T, Kimanius D, Lindahl E, Scheres SH. Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife. 2018;7:e36861. doi: 10.7554/eLife.36861. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen S, McMullan G, Faruqi AR, Murshudov GN, Short JM, Scheres SHW, Henderson R. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei Z, Zhang P, Zhou Z, Cheng Z, Wan M, Gong W. Crystal structure of human eIF3k, the first structure of eIF3 subunits. J Biol Chem. 2004;279:34983–34990. doi: 10.1074/jbc.M405158200. Medline. [DOI] [PubMed] [Google Scholar]
- 62.Llácer JL, Hussain T, Saini AK, Nanda JS, Kaur S, Gordiyenko Y, Kumar R, Hinnebusch AG, Lorsch JR, Ramakrishnan V. Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition. eLife. 2018;7:e39273. doi: 10.7554/eLife.39273. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmitt E, Panvert M, Lazennec-Schurdevin C, Coureux P-D, Perez J, Thompson A, Mechulam Y. Structure of the ternary initiation complex aIF2-GDPNP-methionylated initiator tRNA. Nat Struct Mol Biol. 2012;19:450–454. doi: 10.1038/nsmb.2259. Medline. [DOI] [PubMed] [Google Scholar]
- 64.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. Medline. [DOI] [PubMed] [Google Scholar]
- 65.Natchiar SK, Myasnikov AG, Kratzat H, Hazemann I, Klaholz BP. Visualization of chemical modifications in the human 80S ribosome structure. Nature. 2017;551:472–477. doi: 10.1038/nature24482. Medline. [DOI] [PubMed] [Google Scholar]
- 66.Casañal A, Lohkamp B, Emsley P. Current Developments in Coot for Macromolecular Model Building of Electron Cryo-microscopy and Crystallographic Data. Protein Sci. 2020;29:1055–1064. doi: 10.1002/pro.3791. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, Adams PD. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D. 2018;74:531–544. doi: 10.1107/S2059798318006551. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 2018;430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. Medline. [DOI] [PubMed] [Google Scholar]
- 69.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. Medline. [DOI] [PubMed] [Google Scholar]
- 70.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache J-P, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, Karcher A, et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482:501–506. doi: 10.1038/nature10829. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V. Structural basis for stop codon recognition in eukaryotes. Nature. 2015;524:493–496. doi: 10.1038/nature14896. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grüner S, Peter D, Weber R, Wohlbold L, Chung M-Y, Weichenrieder O, Valkov E, Igreja C, Izaurralde E. The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation. Mol Cell. 2016;64:467–479. doi: 10.1016/j.molcel.2016.09.020. Medline. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
EM maps have been uploaded to the Electron Microscopy Data Bank with accession codes EMD-10775 (48S-40S body), EMD-10772 (48S-40S head), EMD-10769 (48S-eIF3 structural core), EMD-10773 (48S-eIF3bgi), EMD-10774 (48S-eIF2-TC), and EMD-11302 (48S). Protein coordinates have been deposited in the Protein Data Bank (PDB) with IDs 6YBW (48S-40S body), 6YBS (48S-40S head), 6YBD (48S-eIF3 structural core), 6YBT (48S-eIF3bgi), 6YBV (48S-eIFe-TC), and 6ZMW (48S).