Abstract
The recognition of proteins and their post-translational modifications using synthetic molecules is an active area of research. A common post-translational modification is the phosphorylation of serine, threonine or tyrosine residues. The phosphorylation of proximal tyrosine residues occurs in over 1000 proteins in the human proteome, including in disease-related proteins, so the recognition of this motif is of particular interest. We have developed a luminescent europium(III) complex, [Eu.1]+, capable of the discrimination of proximally phosphorylated tyrosine residues, from analogous mono- and non-phosphorylated tyrosine residues, more distantly-related phosphotyrosine residues and over proximally phosphorylated serine and threonine residues. [Eu.1]+ was used to continuously monitor the phosphatase catalysed dephosphorylation of a peptide containing proximally phosphorylated tyrosine residues.
Keywords: anion sensing, luminescence, europium, phosphorylation, enzyme assay
Introduction
The detection and discrimination of proteins and their post-translational modifications using synthetic molecules is an active area of research.1–4 Traditional approaches for the discrimination of proteins involve the use of enzyme and antibody responses (for example using ELISA),5 which can be cumbersome and expensive. Synthetic molecules may offer advantages in terms of stability, their tunable luminescence properties and their potentially cheaper preparation. One post-translational modification of particular importance is the phosphorylation of serine, threonine and tyrosine residues, which acts as key regulators of protein function and signalling within biological systems.6 The study of the phosphorylation and dephosphorylation of proteins, catalysed by kinases and phosphatases respectively, is of critical importance both for the understanding of biological processes and facilitating drug discovery.
The phosphorylation of tyrosine residues is less common (~1%) compared to that of serine (~90%) or threonine (~4%) residues. However, the phosphorylation of proximal tyrosine residues occurs in over 1000 proteins in the human proteome,7 including in disease-related proteins such as JAK2 and IGFR proteins. Therefore, the development of synthetic probes for the detection of proximal phosphotyrosine (pY) residues within peptides and proteins, with minimal interference from phosphoserine (pS) or phosphothreonine (pT) residues, could be useful for more specific sensing of these proteins. Additionally, such probes could be used for screening of new drugs for the treatment of diseases caused by abnormal phosphorylation.
The detection of phosphorylated amino acid residues within peptides and proteins has been achieved using discrete emissive metal complexes, including Zn(II),8–11 Cu(II),12 Tb(III),13–14 and Eu(III) complexes,15–16 whereby coordination of the phosphorylated residue to the metal complex induces a measurable change in luminescence. However, these probes often show very little sequence specificity or selectivity between phosphorylated amino acid residues. Some sensing selectivity has been achieved for certain phosphorylation patterns, including proximally phosphorylated tyrosine residues. The Hamachi group have worked extensively with Zn(II)-dipicolyamine complexes for the detection of phosphorylated peptides and have utilised selected probes for both end-point phosphatase assays and real-time monitoring.8, 10, 17 Gunning and coworkers prepared a fluorescent Zn(II)–cyclen complex for the detection of proximal di- and triphosphorylated proteins,11 demonstrating its ability to discriminate proteins on a polyacrylamide gel. However, this probe cannot discriminate proximal pY residues (pYpY motifs) from peptides containing a pYXpY motif (where X is any amino acid) nor a pYpT motif,18 with similar spectral responses reported. Increased specificity for a target phosphorylated peptide has been achieved by conjugation of various emissive metal complexes to a designed peptide or protein;19–25 however, this approach involves additional non-trivial synthetic processes.
Herein we report the use of a stable europium(III) complex, [Eu.1]+ (Figure 1a), for the detection of proximally phosphorylated tyrosine residues within a peptide, and demonstrate the probe’s utility in monitoring an acid phosphatase catalysed dephosphorylation reaction in real-time. The use of a discrete luminescent Eu(III) complex for phosphopeptide recognition and monitoring enzyme activity offers several advantages, including line-like emission spectra that permits ratiometric detection, large pseudo Stokes’ shifts and the potential for time-resolved emission spectra, permitting complete removal of background autofluorescence arising from peptide or protein substrates.25–32
Figure 1.
a) Structure of complex [Eu.1]+; b) Different peptides used in this study; c) Cartoon representation of the proposed binding mode of [Eu.1]+ to peptides containing the pYpY motif.
We have previously reported the synthesis33 and application of [Eu.1]+ for the detection of nucleotide polyphosphate anions,34 using the probe to monitor the ratio of ADP/ATP during the kinase catalysed phosphorylation of a serine containing peptide. We now report another application for this Eu(III) complex: the luminescent detection of proximally phosphorylated tyrosine (pYpY) residues within peptides. This spectral response of [Eu.1]+ is distinct compared to mono- and non-phosphorylated tyrosine residues (YpY and YY), proximally phosphorylated serine (pSpS) and threonine (pTpT) residues and more distant phosphorylated tyrosine residues (e.g. pYApY and pYAApY). The excellent discriminatory behaviour of [Eu.1]+ allowed the phosphatase catalysed dephosphorylation of a proximally phosphorylated peptide, ApYpYAA, to be monitored in real-time.
Results and Discussion
Discrimination of proximally phosphorylated tyrosine residues
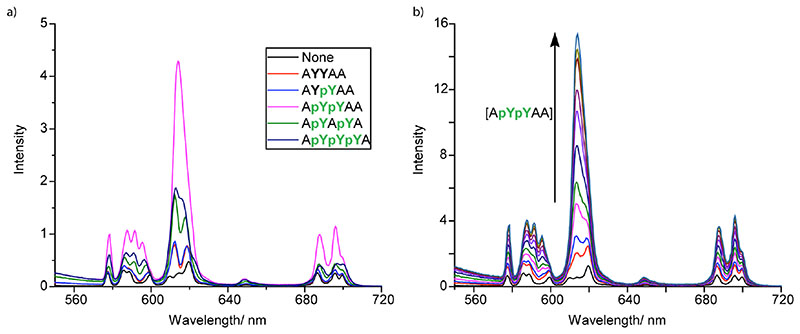
Figure 2a shows the change in emission intensity of [Eu.1]+ in the presence of various phosphotyrosine containing peptides (10 mM), measured in aqueous buffer (10 mM HEPES) at pH 7.0. Addition of the proximally phosphorylated peptide, ApYpYAA, resulted in a striking increase in Eu(III) emission intensity and a pronounced change in spectral form. The emission spectral response was much lower for the other peptides tested, including the analogous mono- and non-phosphorylated peptides, AYpYAA and AYYAA, a proximally triphosphorylated peptide ApYpYpYA, and a diphosphorylated peptide bearing two pY residues separated by a single alanine residue (ApYApYA). The distinctive response to the pYpY containing peptide lead us to explore the use of [Eu.1]+ for the recognition of the pYpY motif over a wider range of different phosphorylated peptides.
Figure 2.
a) Luminescence response of [Eu.1]+ (8 µM) in the presence of different phosphotyrosine containing peptides (10 mM); b) Titration of ApYpYAA (0–8 mM) into [Eu.1]+ (8 µM). Conditions: 10 mM HEPES, pH 7.0, λexc = 330 nm
A titration of the proximally phosphorylated tyrosine peptide, ApYpYAA, into [Eu.1]+ caused a rapid increase in intensity of the ΔJ = 2 emission band centred around 614 nm (Figure 2b). In sharp contrast, titrations of a range of other phosphopeptides resulted in much smaller changes in Eu(III) emission intensity (Figure S1). The high sensing selectivity for the peptide ApYpYAA can be visualised clearly by following the ratiometric change in intensity at 612/599 nm as a function of peptide concentration (Figure 3a). Addition of ApYpYAA (0–8 mM) caused a 10-fold enhancement in the emission intensity ratio, whereas the mono- and non-phosphorylated peptides (AYpYAA and AYYAA, respectively) induced less than 10% increases in the ratiometric signal. Similarly, only minor increases in the emission intensity were observed upon addition of peptides containing two more distantly related phosphotyrosine residues (i, i+2 for ApYApYA, or i, i+3 for pYAApYA). This is significant, as previously reported synthetic probes exhibited limited selectivity between peptides containing differentially located phosphorylated residues.18, 35–37 Importantly, replacing one of the pY residues with a negatively charged glutamic acid residue (ApYEAA or ApYAEA, both purple in Figure 3a), resulted in minimal increases in the intensity ratio at 612/599 nm upon titration of these peptides. These results demonstrate that the presence of a pYpY motif is essential for the distinctive response of [Eu.1]+, rather than there simply being a requirement for two adjacent negatively charged amino acid residues.
Figure 3.
Change in the emission intensity ratio of [Eu.1]+ (8 µM) at 612/599 nm upon incremental addition of: a) various phosphotyrosine (pY) containing peptides; b) proximally diphosphorylated tyrosine, serine and threonine containing peptides. Conditions: 10 mM HEPES buffer, pH 7.0, λexc = 330 nm.
The discrimination of the pYpY motif is likely to arise from both higher affinity binding and a distinctive binding mode to the Eu(III) complex. When the two proximal pY residues are present, it is hypothesised that both phosphate groups coordinate to the Eu(III) metal, causing displacement of the bound water molecule and potentially one of the coordinated quinoline arms, allowing multi-dentate binding (Figure 1c). Evidence for a change in coordination environment of the Eu(III) metal is given by a change in the structure of the hypersensitive ΔJ = 2 band and an increase in the emission lifetime of [Eu.1]+ in the presence of ApYpYAA compared to the Eu(III) complex alone (Table 1). Calculation of hydration state, q, indicates displacement of the bound water molecule from [Eu.1]+ on incubation with the ApYpYAA peptide (Table 1).38 A smaller increase in the emission lifetime of [Eu.1]+ in the presence of the AYYAA and the AYpYAA peptide compared to the Eu(III) complex alone, is also observed, with the non-integral q values indicating formation of ‘partially’ hydrated species on incubation with these peptides. These peptides also induce small changes in the spectral form of the ΔJ = 2 band of [Eu.1]+, but minimal changes in the ΔJ = 1 band, compared to pronounced changes in both bands on incubation with ApYpYAA (Figure S2). This indicates a change in ligand field on incubation with all three peptides, however the different spectral changes, along with the larger change in q in the presence of ApYpYAA indicate a different binding mode. Furthermore, it is reasonable to suggest that this binding mode may only be possible when two proximal pY residues are present in the peptide, as further spacing between pY residues results in less favourable entropy of binding, as seen by the minimal changes in spectral form upon addition ApYApYA and ApYAApY.
Table 1.
Luminescence lifetimes in H2O and D2O (10 mM HEPES, pH 7.0) and calculated q values for [Eu.1]+ (30 µM) alone and in the presence of AYYAA, AYpYAA and ApYpYAA (all 9 mM); λexc = 330 nm, λem = 615 nm.
| peptide | τH2O / ms | τD2O / ms | q a |
|---|---|---|---|
| none | 0.55 | 1.22 | 0.8 |
| AYYAA | 0.68 | 1.30 | 0.5 |
| AYpYAA | 0.72 | 1.21 | 0.3 |
| ApYpYAA | 0.89 | 1.27 | 0.1 |
Values of hydration state, q (20%) are derived using methods in ref. 38.
Next, we examined the ability of [Eu.1]+ to distinguish between peptides bearing proximal pY, pS and pT residues. Titrations of peptides bearing proximally phosphorylated serine and threonine residues (ApSpSAA and ApTpTAA) caused approximately 4-fold enhancements in the 612/599 nm emission intensity ratio of [Eu.1]+, significantly lower than the analogous diphosphotyrosine peptide, ApYpYAA (Figure 3b and Figure S3). Thus, [Eu.1]+ can be used to detect peptides containing proximal pY residues over proximal pS or pT residues. This could be attributed to a change in the orientation of the phosphate groups compared to the peptide backbone and the potential for π-π stacking between the coordinated pY residues and the quinoline arms of the Eu(III) complex. Differences in the binding orientation between the diphosphorylated peptides is indicated by the clear differences in fine splitting of the ΔJ = 1 emission band (Figure S4).39 Additionally, the lower desolvation energy of pY residues compared to pS or pT residues is expected to contribute towards the overall free energy of binding of pYpY containing peptides to [Eu.1]+.
Continuous monitoring of a phosphatase enzyme reaction
Having shown that [Eu.1]+ can distinguish a peptide containing proximal pY residues from other non-phosphorylated, monophosphorylated and diphosphorylated peptides, we wished to utilise the probe to monitor a phosphatase-catalysed dephosphorylation reaction. For this proof-of-concept enzyme reaction we used acid phosphatase, an enzyme which catalyses the hydrolysis of phosphate monoesters, including from pY, pS and pT residues, within the pH range 4.0 – 7.0. The enzyme is therefore expected to remove both the phosphate groups from the ApYpYAA peptide, forming AYYAA via either ApYYAA or AYpYAA.
Before pursuing the enzyme reaction, we modelled the change in Eu(III) emission spectra that might be expected during the enzymatic conversion of peptide ApYpYAA to both the mono- and non-phosphorylated analogues, AYpYAA and AYYAA (Figure S5 and Figure 4, respectively). Titrations of AYpYAA or AYYAA into ApYpYAA, in the presence of [Eu.1]+ resulted in measurable decreases in the intensity of the ΔJ = 2 emission band (Figure 4a and Figure S5a), accompanied by pronounced changes in spectral form. Specifically, the intense single component ΔJ = 2 band, characteristic of the bound diphosphorylated peptide, changed gradually to the less intense, two-component band associated with each of the other peptides. Following the emission intensity at 614 nm (Figure 4b), a decrease in intensity was seen upon addition of AYpYAA or AYYAA to ApYpYAA, which was approximately linear up to 50 % conversion. The simulated dephosphorylation can also be followed ratiometrically, by monitoring the change in the intensity ratio at 614/599 nm (Figure 4c and Figure S5c). A simulation of the second dephosphorylation event, involving conversion of the mono-phosphorylated peptide AYpYAA to the non-phosphorylated peptide AYYAA, was followed in a similar manner, showing minimal changes in emission spectra (Figure S6). Based on these experiments, it was anticipated that in monitoring the enzyme-catalysed dephosphorylation of ApYpYAA, the predominant observation would be the first dephosphorylation, with minor fluctuations in emission intensity being observed for the second dephosphorylation. The byproduct of dephosphorylation is inorganic phosphate, and titration of this species into [Eu.1] +, in the presence of AYYAA, caused a minimal change in emission intensity and spectral form of [Eu.1]+ (Figure S7a). Thus, the accumulation of inorganic phosphate during the enzyme reaction was not expected to interfere with the ability of [Eu.1]+ to signal the dephosphorylation process. Similarly, addition of the acid phosphatase enzyme to [Eu.1]+ also caused negligible changes in the emission spectra of [Eu.1]+ (Figure S7b).
Figure 4.
Model of the phosphatase reaction, by titrating peptide AYYAA (0 – 3.4 mM) into a solution containing ApYpYAA (4 mM initially) and [Eu.1]+ (8 µM), monitoring the change in emission intensity. a) Change in emission spectra of [Eu.1]+; b) Change in the emission intensity at 614 nm over the whole titration (upper) and in the linear range (lower); c) Change in the intensity ratio at 614/599 nm over the whole titration (upper) and in the linear range (lower). Conditions: 10 mM HEPES buffer, pH 7.0, λexc = 330 nm.
Next, we used [Eu.1]+ to monitor the acid phosphatase catalysed dephosphorylation of ApYpYAA in real-time. Incubation of ApYpYAA (8 mM) with acid phosphatase (0.5 units mL-1) resulted in a decrease in the intensity of the ΔJ = 2 band of [Eu.1]+ over time (Figure 5a). A plot of the change in intensity at 614 nm as a function of time revealed a rapid decrease in the emission intensity of [Eu.1]+, with the rate of intensity decrease slowing over time, as expected for the dephosphorylation of the peptide (Figure 5b). In the absence of enzyme, the rate of emission intensity decrease was much slower, and can be attributed to the background hydrolysis of peptide ApYpYAA. As a control, the emission intensity of [Eu.1]+ in the presence of the non-phosphorylated peptide AYYAA remained constant over the time, indicating minimal photobleaching of [Eu.1]+ during the timeframe of the experiment (Figure 5b).
Figure 5.
Luminescence monitoring of the acid phosphatase (0.5 units mL-1) catalysed dephosphorylation of ApYpYAA (8 mM) using [Eu.1]+ (8 µM). a) Change in the emission spectrum of [Eu.1]+ (570–630 nm); b) Change in Eu(III) emission intensity at 614 nm in the presence and absence of enzyme (4 mM ApYpYAA), referenced against the non-phosphorylated peptide AYYAA (4 mM). Conditions: 10 mM HEPES buffer, pH 7.0, λexc = 330 nm.
The magnitude of the observed decrease in Eu(III) emission intensity in the presence of acid phosphatase was lower than that expected based on the titration of either AYYAA or AYpYAA into the ApYpYAA peptide. This could be due to the dephosphorylation reaction not reaching completion in the time frame studied, with some ApYpYAA remaining at the end of the experiment. This is presumably as a result of inhibition of the phosphatase enzyme by the byproduct, inorganic phosphate, a known inhibitor of the enzyme.
Having demonstrated that it was possible to monitor acid phosphatase activity, the reaction was followed at 3 different enzyme concentrations (Figure 6). Pleasingly, the rate of hydrolysis of ApYpYAA increased with increasing concentrations of enzyme. A plot of the initial rates of reaction revealed linear responses over the first 8 minutes (Figure 6b), which would allow for initial enzyme reaction rates to be determined. This demonstrates the utility of [Eu.1]+ for monitoring the acid phosphatase catalysed hydrolysis of proximally phosphorylated tyrosine residues in real-time.
Figure 6.
Monitoring of the acid phosphatase catalysed dephosphorylation of ApYpYAA (4 mM) using [Eu.1]+ (8 µM), using different enzyme concentrations. a) Change in Eu(III) emission intensity at 614 nm over time at the three different enzyme concentrations (0.1, 0.5 and 2.5 units mL-1); b) Initial rates of hydrolysis, showing linear responses over at least the first 8 minutes. Conditions: 10 mM HEPES buffer, pH 7.0, λexc = 330 nm.
Conclusion
A stable luminescent Eu(III) complex, [Eu.1]+, has been shown to discriminate proximally phosphorylated tyrosine residues in peptides under neutral aqueous conditions, from the analogous mono- and non-phosphorylated peptides, as well as differentially located phosphotyrosine residues and proximally phosphorylated serine and threonine residues. The excellent discriminatory behaviour of the Eu(III) complex allowed its application in a continuous-read enzyme assay, wherein the phosphatase catalysed dephosphorylation of a peptide containing a pYpY motif was monitored in real-time, without the need to chemically modify the enzyme or its substrate. Future work will be directed towards the recognition of proximally phosphorylated tyrosine residues in more biologically and therapeutically important peptides and proteins, tracking their dephosphorylation by using more substrate-specific phosphatase enzymes.
Supplementary Material
Acknowledgements
This work was supported by a Wellcome Trust Seed Award (204500/Z/16/Z) and a Royal Society Research Grant. We thank Dr. David Worrall for use of fluorescence spectroscopy facilities.
References
- 1.Collins BE, Anslyn EV. Chem Eur J. 2007;13:4700–4708. doi: 10.1002/chem.200700153. [DOI] [PubMed] [Google Scholar]
- 2.Margulies D, Hamilton AD. Curr Opin Chem Biol. 2010;14:705–712. doi: 10.1016/j.cbpa.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 3.Kubota R, Hamachi I. Chem Soc Rev. 2015;44:4454–4471. doi: 10.1039/c4cs00381k. [DOI] [PubMed] [Google Scholar]
- 4.Hewitt SH, Wilson AJ. Chem Commun. 2016;52:9745–9756. doi: 10.1039/c6cc03457h. [DOI] [PubMed] [Google Scholar]
- 5.Borrebaeck CAK. Immunol Today. 2000;21:379–382. doi: 10.1016/s0167-5699(00)01683-2. [DOI] [PubMed] [Google Scholar]
- 6.Hunter T. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 7.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ojida A, Mito-oka Y, Sada K, Hamachi I. Am Chem Soc. 2004;126:2454–2463. doi: 10.1021/ja038277x. [DOI] [PubMed] [Google Scholar]
- 9.Ojida A, Mito-oka Y, Inoue M-a, Hamachi I. Am Chem Soc. 2002;124:6256–6258. doi: 10.1021/ja025761b. [DOI] [PubMed] [Google Scholar]
- 10.Sakamoto T, Ojida A, Hamachi I. Chem Commun. 2009;0:141–152. doi: 10.1039/b812374h. [DOI] [PubMed] [Google Scholar]
- 11.Kraskouskaya D, Bancerz M, Soor HS, Gardiner JE, Gunning PT. Am Chem Soc. 2014;136:1234–7. doi: 10.1021/ja411492k. [DOI] [PubMed] [Google Scholar]
- 12.Zhang T, Edwards NY, Bonizzoni M, Anslyn EV. Am Chem Soc. 2009;131:11976–11984. doi: 10.1021/ja9041675. [DOI] [PubMed] [Google Scholar]
- 13.Duodu E, Kraskouskaya D, Campbell J, Graca-Lima G, Gunning P. Chem Commun. 2015;51:6675–7. doi: 10.1039/c5cc00679a. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Yang T, Luo J, Yang L, Yao C. Chem Commun. 2015;51:8185–8188. doi: 10.1039/c5cc01056j. [DOI] [PubMed] [Google Scholar]
- 15.Atkinson P, Bretonniere Y, Parker D. Chem Commun. 2004;0:438–439. doi: 10.1039/b313496m. [DOI] [PubMed] [Google Scholar]
- 16.Atkinson P, Murray BS, Parker D. Org Biomol Chem. 2006;4:3166–3171. doi: 10.1039/b608528h. [DOI] [PubMed] [Google Scholar]
- 17.Ojida A, Inoue M-a, Mito-oka Y, Tsutsumi H, Sada K, Hamachi I. Am Chem Soc. 2006;128:2052–2058. doi: 10.1021/ja056585k. [DOI] [PubMed] [Google Scholar]
- 18.Kraskouskaya D, Cabral AD, Fong R, Bancerz M, Toutah K, Rosa D, Gardiner JE, de Araujo ED, Duodu E, Armstrong D, Fekl U, Gunning PT. Analyst. 2017;142:2451–2459. doi: 10.1039/c6an02537d. [DOI] [PubMed] [Google Scholar]
- 19.Anai T, Nakata E, Koshi Y, Ojida A, Hamachi I. Am Chem Soc. 2007;129:6232–6239. doi: 10.1021/ja0693284. [DOI] [PubMed] [Google Scholar]
- 20.Zondlo SC, Gao F, Zondlo NJ. J Am Chem Soc. 2010;132:5619–5621. doi: 10.1021/ja100862u. [DOI] [PubMed] [Google Scholar]
- 21.Pazos E, Golicnik M, Mascarenas JL, Eugenio Vazquez M. Chem Commun. 2012;48:9534–9536. doi: 10.1039/c2cc34958b. [DOI] [PubMed] [Google Scholar]
- 22.Tremblay MS, Zhu Q, Martí AA, Dyer J, Halim M, Jockusch S, Turro NJ, Sames D. Org Lett. 2006;8:2723–2726. doi: 10.1021/ol060614u. [DOI] [PubMed] [Google Scholar]
- 23.Tremblay MS, Lee M, Sames D. Org Lett. 2008;10:5–8. doi: 10.1021/ol701920x. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez-Vera JA, Bouzada D, Bouclier C, Eugenio Vazquez M, Morris MC. Chem Commun. 2017;53:6109–6112. doi: 10.1039/c6cc09948c. [DOI] [PubMed] [Google Scholar]
- 25.Gempf KL, Butler SJ, Funk AM, Parker D. Chem Commun. 2013;49:9104–6. doi: 10.1039/c3cc45875j. [DOI] [PubMed] [Google Scholar]
- 26.Butler SJ, Parker D. Chem Soc Rev. 2013;42:1652–66. doi: 10.1039/c2cs35144g. [DOI] [PubMed] [Google Scholar]
- 27.Weitz EA, Chang JY, Rosenfield AH, Morrow EA, Pierre V. Chem Sci. 2013;4:4052–4060. [Google Scholar]
- 28.Surender EM, Bradberry SJ, Bright SA, McCoy CP, Williams DC, Gunnlaugsson T. Am Chem Soc. 2017;139:381–388. doi: 10.1021/jacs.6b11077. [DOI] [PubMed] [Google Scholar]
- 29.Parker D, Shuvaev S, Starck M. Chem Eur J. 2017;23:9974–9989. doi: 10.1002/chem.201700567. [DOI] [PubMed] [Google Scholar]
- 30.Butler SJ, McMahon BK, Pal R, Parker D, Walton JW. Chem Eur J. 2013;19:9511–7. doi: 10.1002/chem.201301273. [DOI] [PubMed] [Google Scholar]
- 31.McMahon BK, Gunnlaugsson T. J Am Chem Soc. 2012;134:10725–10728. doi: 10.1021/ja300887k. [DOI] [PubMed] [Google Scholar]
- 32.Palmer AJ, Ford SH, Butler SJ, Hawkins TJ, Hussey PJ, Pal R, Walton JW, Parker D. RSC Adv. 2014;4:9356. [Google Scholar]
- 33.Butler SJ. Chem Commun. 2015;51:10879–10882. doi: 10.1039/c5cc03428k. [DOI] [PubMed] [Google Scholar]
- 34.Hewitt SH, Parris J, Mailhot R, Butler SJ. Chem Commun. 2017. [DOI] [PubMed]
- 35.Cabral AD, Murcar-Evans BI, Toutah K, Bancerz M, Rosa D, Yuen K, Radu TB, Ali M, Penkul A, Kraskouskaya D, Gunning PT. Analyst. 2017 doi: 10.1039/c7an00722a. [DOI] [PubMed] [Google Scholar]
- 36.Ishida Y, Inoue M-a, Inoue T, Ojida A, Hamachi I. Chem Commun. 2009:2848–2850. doi: 10.1039/b905814a. [DOI] [PubMed] [Google Scholar]
- 37.Ojida A, Inoue M-a, Mito-oka Y, Hamachi I. Am Chem Soc. 2003;125:10184–10185. doi: 10.1021/ja036317r. [DOI] [PubMed] [Google Scholar]
- 38.Beeby A, Clarkson MI, Dickins SR, Faulkner S, Parker D, Royle L, de Sousa SA, Gareth Williams AJ, Woods M. Chem Soc, Perkin Trans. 1999;2:493–504. [Google Scholar]
- 39.Vonci M, Mason K, Suturina EA, Frawley AT, Worswick SG, Kuprov I, Parker D, McInnes EJL, Chilton NF. Am Chem Soc. 2017;139:14166–14172. doi: 10.1021/jacs.7b07094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.