Summary
During mouse embryonic development, the T-box transcription factor Eomes/Tbr2 is expressed in highly dynamic patterns in various progenitor cell types. Those include the undifferentiated cells of the trophec-toderm, ingressing nascent mesoderm at the primitive streak, and intermediate progenitor cells of the developing cerebral cortex. We generated an EomesGFP-targeted allele to follow the highly dynamic patterns of Eomes expression and to allow for the identification of novel expression domains. We show that our novel allele recapitulates endogenous gene expression at known sites of expression and confirm our results by anti-Eomes immunofluorescent staining. Using this novel allele we were able to identify previously undocumented domains of Eomes expression within the visceral endoderm and at various locations in the developing and adult mouse brain.
Keywords: mouse, Eomesodermin, Tbr2, green fluorescent protein, primitive streak, neuronal progenitors
The T-box family of transcription-factors comprises a group of 17 genes present in all vertebrates. T-box proteins are characterized by the common feature of the evolutionary highly conserved T-box DNA-binding domain, first identified in the founding member of this gene family, brachyury (also known as the T-gene). During development, T-box genes are expressed in highly specific and dynamic patterns, and mutations frequently result in severe phenotypic alterations [reviewed in Naiche et al. (2005)].
In mouse development, the earliest T-box gene to be expressed is Eomesodermin (Eomes, also referred to as Tbr2; Bulfone et al., 1999; Ciruna and Rossant, 1999; Hancock et al., 1999). Eomes is expressed at multiple sites, and loss-of-function studies have revealed crucial functions of Eomes at its various expression domains. Eomes transcripts are initially found in the trophectoderm (TE) of the E3.5 blastocyst and later in its derivative, the extraembryonic ectoderm (ExE). Eomes loss-of-function results in precocious differentiation of TE precursors (Niwa et al., 2005; Russ et al., 2000; Strumpf et al., 2005) that leads to developmental block before the onset of gastrulation. A second expression domain is found at the site of primitive streak (PS) formation, and Eomes is expressed in the nascent mesoderm of the PS from E6.5 until the late streak stage at E7.5 (Ciruna and Rossant, 1999; Hancock et al., 1999; Russ et al., 2000). Within the PS the primary germ layers mesoderm and definitive endoderm (DE) are specified under the influence of signaling molecules [reviewed by Arnold and Robertson (2009) and Tam and Loebel (2007)]. Eomes functions in guiding the epithelial-to-mesenchmymal transition (EMT) of nascent mesoderm and is required for specification of the DE (Arnold et al., 2008a; Russ et al., 2000). From E10.5, Eomes expression is found in neuronal precursor cells of the developing cerebral cortex (Bulfone et al., 1999; Ciruna and Rossant, 1999; Hancock et al., 1999; Russ et al., 2000; Ryan et al., 1998; England et al., 2005). Conditional removal of Eomes from the developing brain results in severe disturbances of cortical development with microcephaly and pronounced behavioral abnormalities (Arnold et al., 2008b; Sessa et al., 2008). Eomes is also transiently expressed in neuronal progenitor cells of the adult dentate gyrus (DG) and in mitral cells of the adult olfactory bulb. The functional relevance of these domains of expression remains to be determined. Finally, Eomes is required during differentiation of T-cell lineages, such as CD8+ effector T cells (Intlekofer et al., 2008; Pearce et al., 2003) and memory cells (Intlekofer et al., 2005), possibly acting in combination with the closely related Tbx21/T-bet.
To monitor highly dynamic expression domains of Eomes during development, we used homologous recombination in embryonic stem (ES) cells to generate an Eomes FP-targeted reporter allele. A reporter cassette for cytoplasmic EGFP expression was inserted into the translational start site of the endogenous Eomes gene, thereby deleting ~500 bp of the Exon 1 coding region (Fig. 1a,b). The loxP-flanked positive selection cassette (PGK.neo) was excised by transient transfection with Cre recombinase (pMCI.Cre, Fig. 1c) and germline chimeras generated following injection of ES cells into E35 blastocysts. To test if the EomesGFP allele acts as a null allele, we intercrossed EomesGFP/+ heterozygous animals and genotyped resulting embryos. As expected, EomesGFP/GFP homozygotes fail to develop beyond early postimplantation stages, and at E7.5, no viable homozygotes were recovered (Fig. 1e) due to the early requirements for Eomes in the TE lineage (Russ et al., 2000). Similarly, using the EomesGFP allele and conditionally excising the second allele in the epiblast via the Sox2.Cre deleter strain (Hayashi et al., 2002) recapitulates the previously reported phenotype (Arnold et al., 2008a). Thus, EomesGFP/CA; Sox2.Cre epiblast cells fail to undergo EMT and to form a distinctive mesodermal cell layer and nascent mesoderm accumulates in the region of the PS (Fig. 1f). Therefore, we conclude that the novel EomesGFP allele acts as a null allele.
Fig. 1.
Generation of the EomesGFP reporter allele. (a) Strategy to introduce EGFP into the first coding Exon 1 of the Eomes locus by homologous recombination in ES cells. A EGFP.pA cassette was placed into the transcriptional start site of the endogenous Eomes locus, followed by a removable, LoxP-flanked (blue arrows) selection cassette (PGK.neo). (b) Drug-resistant ES cell clones were screened by Southern Blot on EcoRV-digested DNA using a 3’ external probe (indicated as red bar in a). (c) Correctly targeted clones were subjected to transient expression of Cre. Individual clones were analyzed by Southern blot to distinguish wildtype allele (wt, 15 kb), targeted allele (T, 7.0 kb), and reporter allele (R, 5.5 kb). (d) PCR genotyping distinguishes wildtype (wt, 302 bp) and EomesGFP reporter allele (R, 373 bp). (e) Genotyping of embryos from EomesGFP/+ intercrosses demonstrates lethality of EomeGFP/GFP embryos prior to E7.5. (f) EomesGFP/CA; Sox2.Cre epiblast-deleted embryos fail to form the mesoderm cell layer and are blocked in gastrulation as shown by cross-sections of E7.5 embryos. E, EagI; H, HindIII; S, SphI; RV, EcoRV; TK, thymidine kinase (negative selection marker).
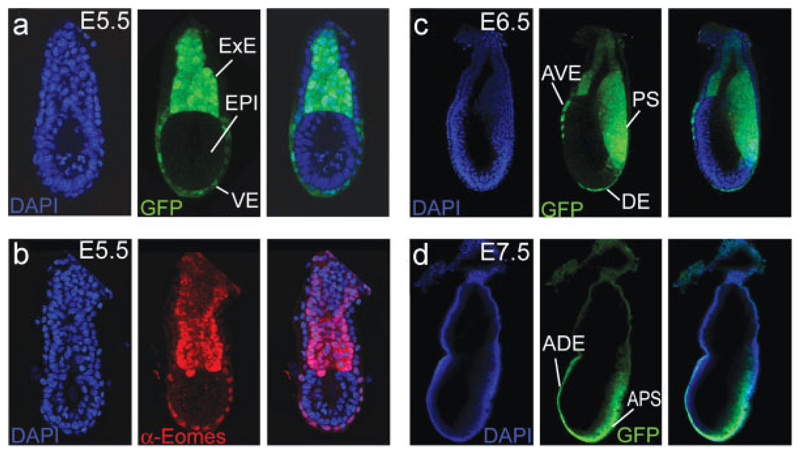
We first visualized EomesGFP expression in embryonic day 5.5 (E5.5) embryos by whole-embryo confocal microscopy (Fig. 2a). GFP was readily visible in the un-differentiated cells of the ExE, the derivative of the TE that gives rise to the embryonic part of the placenta. Surprisingly, we could also detect abundant GFP fluorescence within cells of the visceral endoderm (VE) overlying the epiblast, which in contrast is entirely devoid of GFP expression. To confirm that GFP expression in the VE reflects endogenous Eomes expression, we performed immunofluorescent staining with an anti-Eomes antibody and compared GFP and antibody staining (Fig. 2b). This confirmed the presence of endogenous Eomes protein expression within the VE, albeit at slightly reduced levels when compared to cells of the ExE.
Fig. 2.
EomesGFP expression in the early embryo. (a) At E5.5 EomesGFP expression is found in the extraembryonic ectoderm (ExE) and at reduced levels in the embryonic visceral endoderm (VE). The epiblast (EPI) is devoid of any GFP expression. (b) Immunofluorescence staining with an anti-Eomes antibody at E5.5 confirms the strong expression of endogenous Eomes protein within the ExE and at slightly reduced levels in the VE. (c) After onset of gastrulation at E6.5, EomesGFP is detected in the primitive streak (PS). Additional expression is found in remaining cells of the anterior visceral endoderm (AVE) and in nascent definitive endoderm (DE). (d) During later stages of gastrulation, at E7.5, GFP remains predominantly expressed in the anterior part of the PS (APS) and in cells of the anterior definitive endoderm (ADE). No or only very weak expression is found in the derivatives of the ExE, such as chorion and the ectoplacental cone.
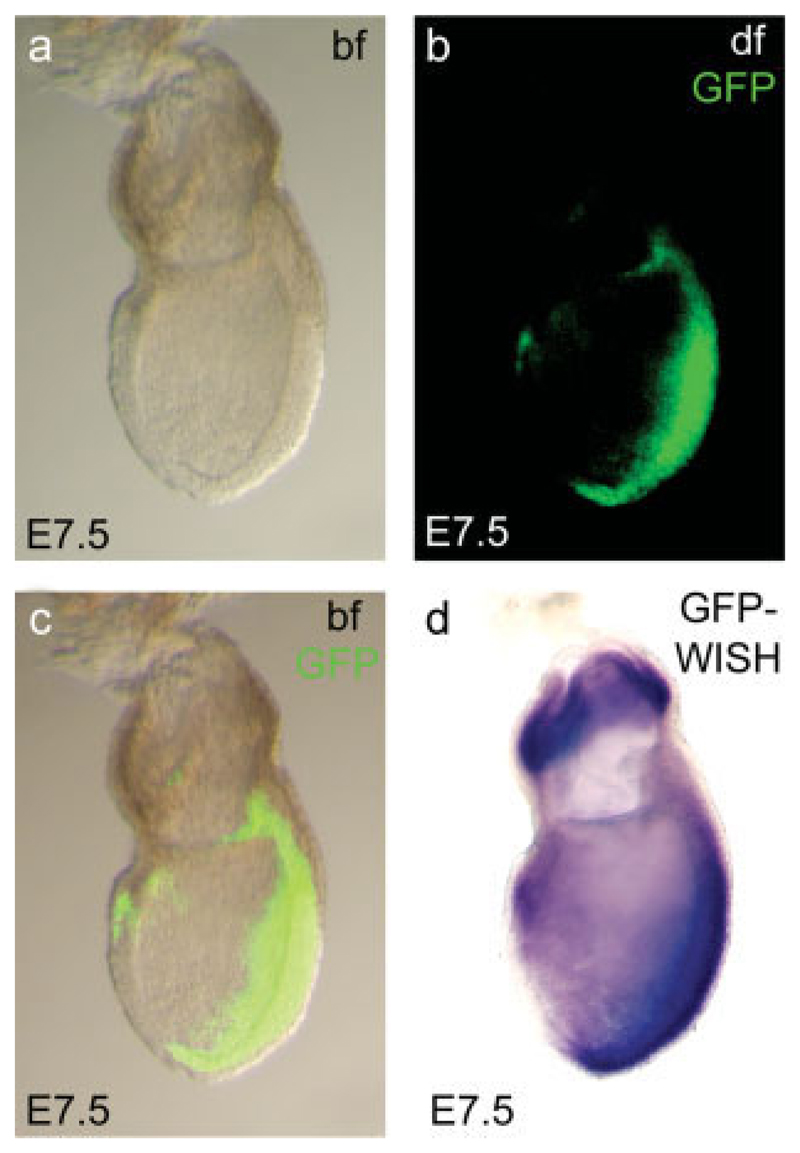
Concomitant with the onset of gastrulation at E6.25 the expression of EomesGFP recapitulates that of the endogenous Eomes RNA in cells of the forming PS (Fig. 2c). PS expression persists until E7.5, predominantly in the region of the anterior PS (Fig. 2d), similar to endogenous RNA (Ciruna and Rossant, 1999). In contrast to RNA, GFP persists in cells of the DE that emerges from the anterior PS (Kwon et al., 2008), most likely reflecting differences in mRNA and GFP stability. Interestingly, the expression of Eomes RNA in cells of the chorion is not recapitulated by GFP-fluorescence similar to results with an Eomes::GFP BAC-transgenic approach (Kwon and Hadjantonakis, 2007) that does not show GFP in the chorion. To investigate if the lack of GFP signal in the chorion is due to insufficient RNA expression, we performed in situ hybridizations on E7.5 embryos using a GFP-specific probe (Fig. 3d) and compared it to the GFP signal from the same embryo (Fig. 3a–c). Surprisingly, despite the absence of GFP fluorescence, we found abundant GFP message levels in the chorion, comparable to that seen in the PS region. We conclude that the lack of GFP activity is due to inappropriate protein folding, or rapid protein degradation in the chorionic region rather than failure to express the reporter gene.
Fig. 3.
Comparison of EomesGFP fluorescence and mRNA expression at late gastrulation. GFP fluorescence can be identified throughout the primitive streak region and in a few cells in the anterior endoderm of the E7.5 late gastrulation stage embryos (a–c). No GFP fluorescence can be detected within the extraembryonic ectoderm (b, c), despite highly abundant mRNA expression as demonstrated by whole mount in situ hybridization (WISH, d). bf, brightfield; df, darkfield.
Eomes is an important regulator of lineage commitment and differentiation of intermediate progenitor cells in the subventricular zone (SVZ) during cortical brain development (Arnold et al., 2008b; Sessa et al., 2008). To examine reporter gene expression of the EomesGFP al-lele in the developing cortex, we analyzed coronal brain sections at E12 and E15. At E12, during early phase of cortical neurogenesis, GFP expression is found in the region of the preplate and within a few cells of the pro-liferative zone, adjacent to the ventricle (Fig. 4c). Merging GFP expression with that of endogenous Eomes protein detected by immunofluorescence staining (Fig. 4b) demonstrates that GFP and Eomes protein overlap in most sites, but that the onset of GFP expression is slightly delayed in comparison to endogenous protein (Fig. 4d). Conversely, the expression of GFP extends more laterally than the endogenous protein. This difference could either be due to an increased protein stability of GFP and/or be a result of differences in the subcellular localization of cytoplasmic GFP and nuclear localization of the endogenous Eomes protein.
Fig. 4.
EomesGFP expression during neurogenesis of the developing cerebral cortex. Coronal sections through the cortical fore-brain regions of E12 (a–d’) and E15 (e–h’) EomesGFP mice demonstrate gross overlap of endogenous Eomes protein expression and GFP. (b, b’) At E12, Eomes protein is found in cells of the pre-plate (arrow) and in few prolifera-tive cells in proximity to the ventricular surface (asterisk). (c) GFP expression extends more lateral (arrows in c’ and d’) than immuno-fluorescent staining. (f, f’) In the E15 cortex, Eomes protein is localized to the nuclei of the basal ventricular zone (VZ) and the sub-ventricular zone (SVZ). (g’, h’) Eomes protein expression is recapitulated by cytoplasmic GFP expression that extends marginally more toward the intermediate zone.
Similarly, at E15 (Fig. 4e–h), Eomes protein and GFP expression show gross overlap in cells of the basal ventricular zone (VZ) and SVZ, the onset of GFP is just marginally delayed, and reduced GFP signals are extended toward the intermediate zone but not beyond the boundary of endogenous Eomes immunofluorescent staining (Fig. 4h,h’). Therefore, this novel EomesGFP reporter allele more faithfully recapitulates endogenous Eomes protein expression than the previously reported Eomes::GFP BAC transgenic mouse, in which GFP expression is found to a large degree in Tbr1+ postmi-totic neurons of the intermediate zone and cortical plate (Kwon and Hadjantonakis, 2007).
We used our novel allele to search for additional expression domains in the adult brain and confirmed sites of GFP expression by simultaneously staining for Eomes protein. In the olfactory bulb, robust expression of both Eomes protein and EomesGFP is found in nuclei of mitral cells and cells of the glomerular layer (Fig. 5a), consistent with the loss of an organized mitral cell layer in Eomes conditional mutants (Arnold et al., 2008b). In addition, the lateral olfactory tract that contains axons of the mitral cells that project to the olfactory cortex shows strong GFP expression (not shown). Coronal sections consistently revealed some Eomes-protein and GFP-positive cells in the subependymal zone, adjacent to the ventricles (Fig. 5b). An additional expression domain has been found in cells of the ventral septum (Fig. 5c) consistent with in situ hybridization analysis in the GEN-SAT atlas (Gong et al., 2003).
Fig. 5.
EomesGFP expression in the adult brain. Comparisons of Eomes immunofluorescent staining and EomesGFP at different locations in the adult (postnatal day 50, P50) mouse brain. (a) In the adult olfactory bulb, Eomes nuclear staining and cytoplasmic GFP expression are found in the mitral cell layer (arrow) and within the granular layer (asterisk). (b) Few Eomes and GFP-positive cells are located in the subepen-dymal zone, adjacent to the ventricles (asterisk) and (c) in the region of the ventral septum. (d) Weak expression of both Eomes protein and GFP expression is found in the internal granular cell layer of the adult cerebellum. (e) In the adult dentate gyrus, Eomes and GFP expression is seen in the subgranular zone, the site of proliferating progenitor cells.
Previous studies reported the expression of Eomes in unipolar brush cells (UBCs) of the internal granular cell layer of the adult cerebellum (Englund et al., 2006; Fink et al., 2006; Fig. 5d). The EomesGFP allele showed similar expression of the reporter gene (Fig. 5e), but the colocalization with endogenous protein was not as obvious compared to other locations. Finally, we examined the DG of the adult hippocampus. Hodge et al. (2008) reported the expression of Eomes in early (type-2) intermediate-stage progenitor cells, a population of transient amplifying cells in the subgranular zone of the adult DG that later gives rise to newly forming granule neurons. Similar to endogenous protein expression, we found EomesGFP in clusters of cells in the subgranular layer (Fig. 5e). Of note, in the report by Hodge et al., only about one-third of Eomes::GFP-positive cells were positive for Eomes protein, but in our mouse line GFP and endogenous protein expression overlap to a significantly higher degree.
In conclusion, we present a novel mouse allele that allows the highly dynamic expression of the critical developmental regulator Eomes to be accurately followed using a GFP reporter. Importantly, this reporter al-lele recapitulates endogenous gene expression more precisely than the previously described Eomes::GFP BAC transgenic line (Kwon and Hadjantonakis, 2007), in spite of the very similar strategy used to insert the reporter gene into Exon 1 of the locus. Most differences could be accounted for by increased copy numbers and/or insertional effects of the transgene that enhance overall GFP expression levels likely resulting in prolonged perdurance of GFP in labeled cells.
The gross differences in detection of endogenous Eomes protein compared to GFP protein expressed from the Eomes::GFP BAC transgenic line suggests that endogenous Eomes is tightly regulated in cells of the embryo and is subject to rapid downregulation at both the RNA and protein level.
We also unambiguously show that in the early mouse embryo, Eomes is expressed within the VE and AVE, critical in imparting the A-P pattern in the pregastrulation stage embryo. This is particularly interesting since we previously demonstrated a failure of the distal VE (DVE) to migrate appropriately in embryos heterozygous for both Eomes and the TGFβ signaling molecule Nodal (Arnold et al., 2008a). There is currently some discrepancy concerning expression of Eomes in the VE. Previous reports suggested VE expression as seen by fluorescent in situ hybridization (Chazaud and Rossant, 2006), DIG-labeled in situ hybridization (Ciruna and Rossant, 1999), and antibody staining (Ralston and Rossant, 2008). However, other reports and our own experiments failed to demonstrate Eomes RNA in the VE expression using DIG-labeled RNA probes (Russ et al., 2000; SJA and EJR, unpublished data), expression of β-galactosidase from an EomesLacZ allele (Russ et al., 2000), or by monitoring GFP activity from the Eomes::GFP BAC reporter transgene (Kwon and Hadjantonakis, 2007). Of note, since the Eomes::GFP BAC reporter transgene does not show GFP expression in the VE, one likely explanation is that the corresponding enhancer elements are located outside the genomic sequences contained on the Eomes::GFP BAC transgene. To determine the potential functional significance of Eomes expression in the VE, it will be important to conditionally delete Eomes exclusively in the VE lineage and examine the resulting phenotypic consequences on axis formation and germ layer specification.
Methods
Generation of the EomesGFP-Targeted Allele
The targeting vector for the generation of an EomesGFP-targeted allele comprises an 8.25-kb HpaI–HpaI fragment of the Eomes locus. An EGFP.pA cassettes from pEGFP-N3 (Clontech) and a LoxP-flanked neomycin resistance cassette were integrated between the SphI site at the translational start site and an EagI site, thereby deleting ~500 bp of the 5′ coding region of Exon 1. The 3′ homology region of the targeting construct was flanked with a pMCI.TK negative selection cassette. Linearized targeting vector was electroporated into CCE ES cells, and neomycin and FIAU resistant ES cell colonies were screened by Southern Blot using a 3′external probe on EcoRV digested DNA (wt allele: 15 kb and targeted allele: 7.0 kb). Correctly targeted clones were transfected with pMCI.Cre and the resulting subclones screened by Southern Blot using a 3′ external probe on EcoRV-digested DNA to detect the 5.5kb neomycin-deleted allele. Two independent correctly targeted ES cell clones were used to generate germline chimeras. F1 progeny and subsequent generations were genotyped by multiplex PCR at 608C annealing temperature with standard conditions to detect wildtype allele (302 bp) and GFP allele (373 bp) using the following primers: forward primer 5′-AAGGAAAGGGGCACCTACAA TC-3′; reverse primer1 5′-AGCACTCTCCAGCGAGTAGAA GTG-3′; reverse primer2 5′-CAGATGAACTTCAGGGTCAG CTTG-3′. The strain was maintained on a mixed genetic background of 129 Sv Ev and C57Bl/6.
Analysis of EomesGFP Reporter Expression
E5.5 to E7.5 embryos were fixed for 12–16 h in 4% PFA/PBS and mounted under coverslips in DAPI-contain-ing mounting medium (Vectashield, H1200, Vector Laboratories) for confocal imaging. Brains were fixed for 12–16 h in 4% PFA/PBS, embedded in 4% agarose/PBS, sectioned on a Leica VT 1000S vibratome at 50 μm, and mounted under coverslips in DAPI-containing mounting medium.
Anti Eomes-antibody staining was performed on whole-mount embryos and 50-μm vibratome sections using a rabbit polyclonal antibody against Eomes/Tbr2 (Chemicon, 1:2,000). Embryos or sections were incubated over night in PBS, 0.1% TritonX-100, 5% goat-serum, and 1:2,000 anti-Eomes antibody, washed three times in PBS, 0.1% TritonX-100, and incubated with the antirabbit Alexa-Fluor 555 secondary antibody (Invi-trogen) for 4 h at 4°C. Sections and embryos were washed three times in PBS, 0.1% TritonX-100, and mounted under coverslips in DAPI-containing mounting medium prior to imaging using epifluorescence or laser scanning confocal microscopy (Zeiss LSM 710 confocal microscope).
In Situ Hybridization and Histological Analysis
In situ hybridization on whole embryos was performed according to standard protocols (Nagy et al., 2003) using a GFP-specific probe. For histology, embryos were postfixed in 4% PFA/PBS, dehydrated through etha-nol series, and embedded in paraffin before sectioning at 8 μm. Hematoxylin and eosin counterstaining was performed according to standard protocols (Nagy et al., 2003).
Acknowledgments
We would like to thank Liz Bikoff for discussion and valuable comments on the manuscript, Carol Paterson and Emily Lejsek for generation of chimeras and animal husbandry, and Thomas Lickiss for assistance with confocal microscopy.
Contract grant sponsors: Wellcome Trust (Programme Grant to EJR), Alexander von Humboldt Foundation (Feodor Lynen-Fellowship to SJA)
Literature Cited
- Arnold SJ, Hofmann UK, Bikoff EK, Robertson EJ. Pivotal roles for eomesodermin during axis formation, epithelium-to-mesen-chyme transition and endoderm specification in the mouse. Development. 2008a;135:501–511. doi: 10.1242/dev.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, Huang GJ, Cheung AF, Era T, Nishikawa S, Bikoff EK, Molnar Z, Robertson EJ, Groszer M. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008b;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, Robertson EJ. Making a commitment: Cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- Bulfone A, Martinez S, Marigo V, Campanella M, Basile A, Quaderi N, Gattuso C, Rubenstein JL, Ballabio A. Expression pattern of the Tbr2 (Eomesodermin) gene during mouse and chick brain development. Mech Dev. 1999;84:133–138. doi: 10.1016/s0925-4773(99)00053-2. [DOI] [PubMed] [Google Scholar]
- Chazaud C, Rossant J. Disruption of early proximodistal patterning and AVE formation in Apc mutants. Development. 2006;133:3379–3387. doi: 10.1242/dev.02523. [DOI] [PubMed] [Google Scholar]
- Ciruna BG, Rossant J. Expression of the T-box gene Eomesoder-min during early mouse development. Mech Dev. 1999;81:199–203. doi: 10.1016/s0925-4773(98)00243-3. [DOI] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Kowalczyk T, Daza RA, Dagan A, Lau C, Rose MF, Hevner RF. Unipolar brush cells of the cerebellum are produced in the rhombic lip and migrate through developing white matter. J Neurosci. 2006;26:9184–9195. doi: 10.1523/JNEUROSCI.1610-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink AJ, Englund C, Daza RA, Pham D, Lau C, Nivison M, Kowalczyk T, Hevner RF. Development of the deep cerebellar nuclei: Transcription factors and cell migration from the rhombic lip. J Neurosci. 2006;26:3066–3076. doi: 10.1523/JNEUROSCI.5203-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Hancock SN, Agulnik SI, Silver LM, Papaioannou VE. Mapping and expression analysis of the mouse ortholog of Xenopus Eo-mesodermin. Mech Dev. 1999;81:205–208. doi: 10.1016/s0925-4773(98)00244-5. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev. 2002;119(1):S97–S101. doi: 10.1016/s0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Hodge RD, Kowalczyk TD, Wolf SA, Encinas JM, Rippey C, Enikolopov G, Kempermann G, Hevner RF. Intermediate progenitors in adult hippocampal neurogenesis: Tbr2 expression and coordinate regulation of neuronal output. J Neurosci. 2008;28:3707–3717. doi: 10.1523/JNEUROSCI.4280-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, Dejong CS, Shin H, Hunter CA, Wherry EJ, Lindsten T, Reiner SL. Anomalous type 17 response to viral infection by CD81 T cells lacking T-bet and eomesodermin. Science. 2008;321:408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, et al. Effector and memory CD81 T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- Kwon GS, Hadjantonakis AK. Eomes::GFP—a tool for live imaging cells of the trophoblast, primitive streak, and telencephalon in the mouse embryo. Genesis. 2007;45:208–217. doi: 10.1002/dvg.20293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon GS, Viotti M, Hadjantonakis AK. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev Cell. 2008;15:509–520. doi: 10.1016/j.devcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. Cold Spring Harbor. 3rd. NY: Cold Spring Harbor Laboratory Press; 2003. Manipulating the mouse embryo: A laboratory manual. [Google Scholar]
- Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Annu Rev Genet. 2005;39:219–239. doi: 10.1146/annurev.genet.39.073003.105925. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Ros-sant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, Di Cioccio CB, Gross DA, Mao CA, Shen H, et al. Control of effector CD81 T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- Ralston A, Rossant J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev Biol. 2008;313:614–629. doi: 10.1016/j.ydbio.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, Wilson V, Evans MJ. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- Ryan K, Butler K, Bellefroid E, Gurdon JB. Xenopus eomesoder-min is expressed in neural differentiation. Mech Dev. 1998;75:155–158. doi: 10.1016/s0925-4773(98)00084-7. [DOI] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blasto-cyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Tam PP, Loebel DA. Gene function in mouse embryogenesis: Get set for gastrulation. Nat Rev Genet. 2007;8:368–381. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]