Abstract
The T-box transcription factor eomesodermin (Eomes) has been implicated as an important component in germ layer induction and patterning in vertebrate embryos. In the mouse, Eomes is essential for development of the trophectoderm lineage and Eomes loss-of-function mutants arrest at implantation. Here, we have used a novel Eomes conditional allele to test Eomes functions in the embryo proper. Eomes-deficient embryos express both Fgf8 and its downstream target Snail at normal levels but surprisingly fail to downregulate E-cadherin. Eomes functional loss thus efficiently and profoundly blocks EMT and concomitant mesoderm delamination. Marker analysis as well as fate-mapping and chimera studies demonstrate for the first time that Eomes is required for specification of the definitive endoderm lineage. We also describe developmental abnormalities in Eomes/Nodal double heterozygotes, and demonstrate that these phenotypes reflect Eomes and Nodal interactions in different tissue sites. Collectively, our experiments establish that Eomes is a key regulator of anteroposterior axis formation, EMT and definitive endoderm specification in the mouse.
Keywords: Eomesodermin, Nodal, Axis formation, EMT, E-cadherin, Endoderm specification, Mouse
Introduction
The coordinated activities of multiple signalling pathways including TGFβ/Nodal, Wnt and Fgfs govern specification of the embryonic axes and regulate germ layer formation during gastrulation in the early mouse embryo (reviewed by Beddington and Robertson, 1999; Tam and Loebel, 2007). Shortly after implantation reciprocal interactions between cells of the three primary cell lineages, namely the trophectoderm (TE), epiblast (the progenitors of the embryo proper) and visceral endoderm (VE) establish the initial proximodistal (PD) axis. The anteroposterior (AP) axis emerges gradually as a result of subsequent cell movements. Thus, a specialized population of VE cells induced at the distal tip migrates towards the future anterior side of the embryo to form the anterior visceral endoderm (AVE). The AP axis then becomes evident when a discrete population of proximal epiblast cells is induced to form mesoderm and initiates primitive streak (PS) formation on the posterior side of the embryo (reviewed by Beddington and Robertson, 1999; Lu et al., 2001; Tam and Loebel, 2007). Over the next 12-24 hours the PS gradually elongates to reach the distal tip of the epiblast. Fate-mapping studies have shown that cells delaminating from the streak proximally give rise to extra-embryonic mesoderm. The lateral plate and paraxial mesoderm emerge from intermediate levels, whereas cells situated in the anterior primitive streak (APS) give rise to the axial mesendoderm precursors that in turn give rise to the anterior definitive endoderm (ADE) and prechordal plate mesoderm (PCP), as well as the progenitors of the node and its derivative the notochord.
We and others have previously shown that graded Nodal/Smad2 signals govern patterning of the AP axis in mouse. Thus, Nodal expressed in the epiblast activates Smad2 in the VE and promotes formation of the AV E (Waldrip et al., 1998; Brennan et al., 2001). The subsequent migration of the AVE is also dependent on Nodal signals derived from the epiblast (Norris et al., 2002) (reviewed by Tam and Loebel, 2007). During gastrulation, patterning of the mesoderm and endoderm along the proximodistal axis of the PS is regulated by graded levels of Nodal activity (Vincent et al., 2003; Ben-Haim et al., 2006). Highest levels of Nodal/Smad2 signalling is necessary to specify APS derivatives, that selectively give rise to the definitive endoderm (DE) and PCP mesoderm (Vincent et al., 2003; Dunn et al., 2004). Importantly, specification of DE progenitors also depends on transcriptional partnerships with Smad4 (Chu et al., 2004) and FoxH1 (Hoodless et al., 2001; Yamamoto et al., 2001).
The T-box transcription factor eomesodermin (Eomes) has been implicated as an important component in germ layer induction and patterning (Ryan et al., 1996; Ryan et al., 2000; Russ et al., 2000; Bruce et al., 2003; Bjornson et al., 2005). Initially identified in Xenopus as an early ‘panmesodermal’ marker gene, Eomes was shown to be necessary and sufficient for mesoderm induction (Ryan et al., 1996). As for Activin/Nodal ligands, forced expression of Eomes in animal caps induces a wide range of mesodermal marker genes in a dose-dependent fashion and can dorsalize ventral mesoderm (Ryan et al., 1996). Eomes overexpression in zebrafish results in ectopic expression of organizer genes and induction of secondary body axes, consistent with a role for maternal Eomes in regulating organizer formation (Bruce et al., 2003). Collectively, these zebrafish experiments suggest that Eomes may not be required for mesoderm formation per se, but rather plays an essential role in endoderm specification. Eomes together with GATA5 and the homeodomain-protein Bonnie and Clyde is known to activate expression of the essential Sox transcription factor casanova, necessary for endoderm formation (Bjornson et al., 2005). Thus, in lower vertebrates, Eomes appears to act downstream of Nodal to regulate development of both the mesoderm and endodermal cell lineages.
In mouse, Eomes transcripts are initially expressed in the TE at the blastocyst stage and become confined to the extra-embryonic ectoderm (ExE) following implantation. Eomes expression is subsequently induced within the proximal posterior epiblast prior to overt streak formation. During gastrulation, Eomes transcripts are restricted to the PS and nascent mesoderm, and become confined to the APS coincident with the formation of the morphological node, before being abruptly downregulated (Ciruna and Rossant, 1999; Hancock et al., 1999; Russ et al., 2000). Eomes is essential for development of the TE lineage and Eomes loss-of-function mutants arrest at implantation (Russ et al., 2000; Strumpf et al., 2005). In tetraploid chimeras, Eomes-deficient ES cells give rise to limited mesoderm but the development of these embryos is severely disturbed, making it difficult to characterize discrete tissue defects (Russ et al., 2000).
To further explore Eomes functions within the embryo proper, we have generated an Eomes conditional allele. The well-described Sox2.Cre strain (Hayashi et al., 2002) was used to delete Eomes selectively in epiblast derivatives. Eomes is not required for initial AP patterning or induction of mesoderm markers. However, nascent mesoderm fails to delaminate and migrate away from the primitive streak. The failure of mesoderm migration is associated with an inability to efficiently downregulate E-cadherin and undergo an epithelium-to-mesenchyme transition (EMT), but surprisingly the known E-cadherin upstream regulator Fgf8 and its downstream target Snail are expressed at normal levels. Strikingly, in the absence of Eomes expression in the epiblast, the embryo entirely lacks definitive endoderm. As for Smad2 (Tremblay et al., 2000), in chimeras, Eomes- null ES cells can efficiently contribute to the mesoderm lineage, but are entirely excluded from the DE. We also describe developmental abnormalities in Eomes/Nodal double heterozygotes. A subset of embryos arrest early owing to a failure to rotate the initial PD axis; others form an AP axis but fail to specify APS derivatives. Additionally, some are viable postnatally. We show these different phenotypes reflect Eomes and Nodal interactions at different tissue sites. Collectively our experiments demonstrate that Eomes plays pivotal roles during AP axis formation, EMT and DE specification.
Materials and Methods
Generation of Eomes conditional and null alleles
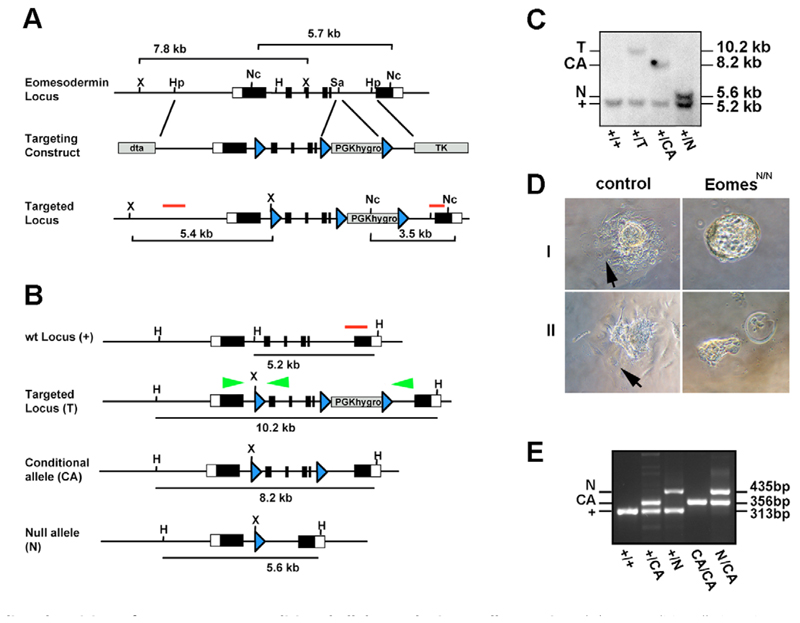
The targeting vector comprised a 8.25 kb HpaI-HpaI fragment of the Eomes locus, with a single LoxP and XbaI site integrated in the HindIII site of intron 1 and a LoxP flanked PGK.hygro selection cassette integrated in the SacI site of intron 5 (Fig. 1). Both homology regions were flanked with negative selection cassettes (PGK.DTA and pMCI.TK). Linearized targeting vector was electroporated into CCE ES cells and drug-resistant ES cell colonies were screened by Southern blot using a 3′external probe on NcoI-digested DNA (wild-type allele 5.7 kb; targeted allele 3.5 kb). The presence of the single LoxP sites was confirmed using an internal probe on Xbal-digested DNA. To generate both the conditional and null alleles, correctly targeted clones were transfected with pMCI.Cre, and the resulting subclones screened by Southern blot using a 3′external probe on HindIII-digested DNA. Three independent correctly targeted ES cell clones carrying the conditional allele were used to generate germline chimeras. F1 progeny were genotyped by Southern blots and thereafter animals were genotyped by PCR. PCR genotyping was performed as multiplex PCR at 59°C annealing temperature with standard conditions to detect wild-type (313 bp), floxed (356 bp) and null configuration (435 bp) using the following primers: forward primer 5′-CCTTACTTCCTAAGGCACACCTGTAGC-3′; reverse primer1 5′-AGCTCAAACCGACTTCTTTCTGCTC-3′; reverse primer2 5′-AATATGTCCTCAGGGAGATGATGACTC-3′. To generate Eomes mutant ES cells, correctly targeted ES cells were transiently transfected with Cre to generate the null configuration and subjected to a second round of targeting and Cre excision.
Fig. 1.
Cre mediated excision of a new Eomes conditional allele results in a null mutation. (A) To conditionally inactivate Eomes, exons 2-5 encoding the Tbox domain were flanked with LoxP sites (blue arrows) and a LoxP-flanked selectable marker (PGK.hygro) was introduced in intron 5-6. ES cell clones were screened by Southern blot on Ncol-digested DNA and the presence of the 5′ LoxP site confirmed on Xbal-digested DNA as indicated. (B) Correctly targeted clones were subjected to transient expression of Cre-recombinase. (C) HindIII digested DNA was analysed by Southern blot to detect various configurations. Green arrowheads represent the primer-sites for PCR genotyping. (D) Cultured blastocysts from Eomes N/+ intercrosses were analysed after 72 hours. Whereas wild-type and Eomes N/+ heterozygous blastocysts develop trophoblast outgrowths with typical giant cells (arrows), Eomes N/N blastocysts fail to form outgrowths (I), or display severely reduced numbers of giant cells (II). (E) Multiplex PCR genotyping of tail DNA from mice at weaning age using the primers indicated in B showing viability of Eomes CA/CA and Eomes CA/N animals. H, HindIII; Hp, HpaI; Nc, Ncol; Sa, Sad; X, Xbal.
Mouse strains, genotyping and generation of chimeric embryos
All mutant mouse strains were maintained on a mixed genetic background of mostly 129SvEv and C57Bl/6. The Nodal LacZ (Collignon et al., 1996), Noda PEE (Vincent et al., 2003), Wnt3 alleles (Liu et al., 1999), ROSA26 gene-trap line (Friedrich and Soriano, 1991), ROSA26R reporter strain (Soriano, 1999), Sox2.Cre (Hayashi et al., 2002) and T.Cre (Perantoni et al., 2005) transgenic strains have been described previously and genotyped accordingly. Blastocysts were recovered from matings of ROSA26 males to CD1 outbred females, injected with 12-14 ES cells, transferred into E2.5 pseudopregnant foster females and embryos recovered at appropriate time-points for lacZ staining.
In situ hybridization, lacZ staining, histology, scanning electron microscopy and immunofluorescence staining
In situ hybridization on whole embryos and paraffin sections were performed according to standard protocols (Nagy et al., 2003). Standard probes for Afp, brachyury, Cer1, cripto (Cfc1), E-cadherin (Cdh1), Eomes, Fgf8, Foxa2, Hex, Mixl1, Mlcv, Otx2, Shh, Snail, Spc4 (Pcsk6), Spry2, Tbx6, Uncx4.1 and Wnt3 were used. lacZ staining was performed as described (Nagy et al., 2003). For histology, embryos were post-fixed in 4% PFA, dehydrated through ethanol series and embedded in paraffin before sectioning at 8 μm. Hematoxylin and Eosin counterstaining was performed according to standard protocols. Embryos for scanning electron microscopy were dissected in DMEM + 10% FCS, fixed overnight [2.5% glutaraldehyde, 2% paraformaldehyde, 0.1% picric acid in 100 mM phosphate (pH 7.0)], transversally bisected, post-fixed for 1.5 hours in 1% osmium/100 mM phosphate (pH 7.0) and subsequently processed according to standard protocols. Images were taken on a JEOL JSM 6390 SEM. Immunofluorescence staining of whole embryos was performed as described (Ciruna and Rossant, 2001) using anti-E-cadherin antibody (Sigma, F3648) at a 1:500 dilution and Alexa-Fluor 488 secondary antibody (Invitrogen, A11006) at a 1:1000 dilution. Embryos were mounted under coverslips in DAPI-containing mounting medium (Vectashield, H1200, Vector Laboratories) for confocal imaging.
Trophoblast outgrowth assay and primitive streak explants
E3.5 blastocysts were collected from Eomes N/+ intercrosses and individually cultured in DMEM containing 15% FCS on untreated tissue culture dishes for 72 hours before assessment and subsequent PCR genotyping. Primitive streak explants from individually genotyped embryos were cultured for 72 hours as described (Ciruna and Rossant, 2001) and stained for E-cadherin expression.
Results
Generation and characterisation of a novel Eomes conditional allele
Eomes loss-of-function mutant embryos arrest shortly after implantation owing to defective TE development (Russ et al., 2000; Strumpf et al., 2005). To study Eomes requirements at later stages of development, we generated a conditional allele (Eomes CA) by flanking exons 2-5 encoding the T-box DNA-binding domain with LoxP sites (Fig. 1A-C). To confirm that the deletion eliminates Eomes function we mated Eomes CA/+ males to Sox2.Cre females. The Sox2.Cre transgene is active in the female germline and in this context functions as a general deleter (Hayashi et al., 2003; Vincent and Robertson, 2003). Eomes N/+ offspring were then intercrossed. As expected, no phenotypically normal Eomes N/N embryos were recovered at E7.5 (Table 1). Cultured blastocysts from Eomes N/+ intercrosses were also assayed for their ability to form trophoblast outgrowths. After 72 hours in culture, as described previously (Russ et al., 2000; Strumpf et al., 2005), Eomes N/N mutant blastocysts either failed to attach and form a trophoblast outgrowth, or, at best, formed a rudimentary outgrowth with few trophoblast giant cells (Fig. 1D). Thus, Cre-mediated excision results in Eomes loss of function and recapitulates the null phenotype. To confirm that the LoxP sites do not compromise Eomes expression, we intercrossed Eomes CA/+ and Eomes N/+ animals. Both Eomes CA/CA and Eomes CA/N animals are fully viable and fertile (Fig. 1E).
Table 1.
Lethality of EomesN/N embryos at E7.5
| EomesN/+ X EomesN/+ intercross | |||||
|---|---|---|---|---|---|
| Empty deciduae | Eo+/+ | EoN/+ | EoN/N | Total | |
| 8 | 9 (24%) 25% | 28 (76%) 50% | 0 (0%) 25% | 37 | |
Loss of Eomes function in the epiblast disrupts mesodermal cell migration and DE formation
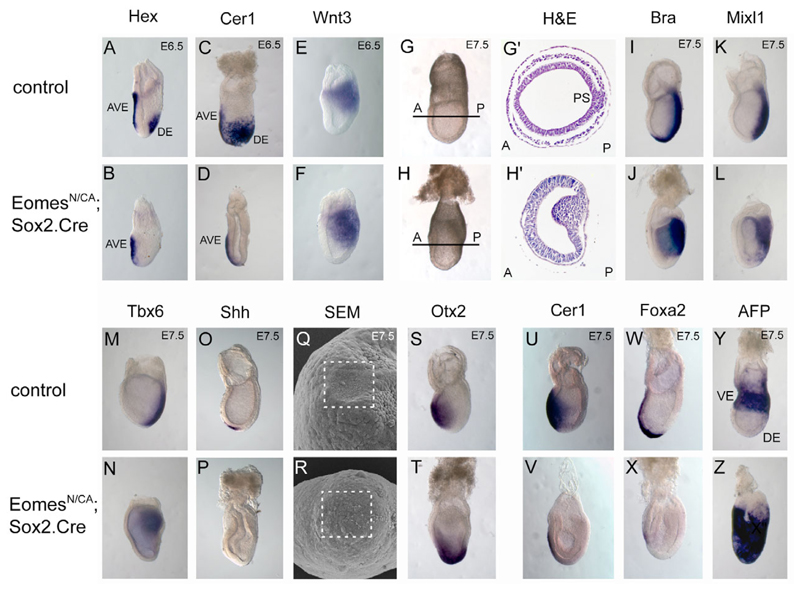
To inactivate Eomes in the embryo proper we made use of the paternally inherited Sox2.Cre transgene, specifically expressed in the inner cell mass (ICM) from early implantation stages onward (Hayashi et al., 2002). Eomes N/+ ; Sox2.Cre males were crossed to Eomes CA/CA females and embryos collected from E6.5-E7.5. At E6.5 Eomes N/CA; Sox2.Cre embryos (hereafter called Eomes-deficient embryos) are phenotypically indistinguishable from control littermates. Analysis by whole-mount in situ hybridization (WISH) shows that initial AP patterning is unaffected by loss of Eomes. Thus, the AVE forms normally, as visualized by expression of Hex and Cer1 (Fig. 2A-D). Similarly, posterior marker genes such as Wnt3 (Fig. 2E,F) and brachyury (Fig. 2I,J) are expressed normally.
Fig. 2.
Epiblast-specific Eomes deletion blocks gastrulation at the primitive streak stage. (A-F) Whole-mount in situ hybridisation with molecular markers for the AVE (Hex and Cer1) and posterior epiblast (Wnt3) at E6.5 reveal correct AP axis specification in Eomes N/CA; Sox2.Cre mutant embryos. (G-H’) At E7.5, Eomes epiblast mutants show a distinct posterior thickening of the epiblast and lack the mesodermal tissue layer as seen in histological sections. (I,J) Posterior (brachyury/Bra) and (K-N) intermediate (Mixl1, Tbx6) streak markers are expressed in Eomes epiblast mutants, but the node fails to form, as revealed by the lack of Shh expression (O,P). (Q,R) Consistent with this, scanning electron microscopy shows lack of a morphological node at the distal tip of mutant embryos (boxed areas indicate the node forming region). (S,T) Instead, the Otx2 expression domain is expanded in Eomes mutants. (U-X) At E7.5 expression of the DE marker Cer1 and the AME marker Foxa2 are lost and (Y,Z) expression of the VE marker AFP fails to become localized to the extra-embryonic region.
From E7.0, Eomes mutant embryos could be identified by their irregular shape resulting from an abnormal thickening of the PS (Fig. 2G,H). In contrast to wild-type epiblast cells that are induced to form mesoderm, undergo EMT and migrate away from the streak towards more lateral and anterior regions of the embryo (Fig. 2G′), mutant embryos show a striking accumulation of cells with a mesenchymal morphology in the PS and a complete absence of mesodermal cells between the VE and the epiblast at E7.5 (Fig. 2H′). Furthermore the epiblast becomes irregularly thickened and expands into the amniotic cavity from E7.5 onwards. By late headfold stages, mutant embryos contain multiple cystic epiblast invaginations and entirely lack a discrete layer of mesoderm (data not shown).
Brachyury and Bmp4, posterior and extra-embryonic mesoderm markers, respectively, are strongly expressed on the posterior side of mutant embryos (Fig. 2I,J and data not shown). Tbx6 and Mixl1, intermediate PS markers are expressed (Fig. 2K-N) at slightly reduced intensities. However brachyury expression fails to extend anteriorly (Fig. 2J) and mutant embryos completely lack expression of markers of APS derivatives, such as Cer1, Shh and Foxa2 (Fig. 2O,P,U-X). This is not simply due to a failure to establish anterior identity, because the Otx2 expression pattern demonstrates that the neuroectoderm develops normally (Fig. 2S,T). However, the Otx2 expression domain normally confined to the anterior region of the embryo is shifted towards the distal tip in Eomes epiblast mutant embryos. Similarly, Hex and Cer1 are expressed normally in the AVE at E6.5, but the corresponding expression domains that normally mark nascent DE are absent (Fig. 2A-D). To evaluate whether the absence of DE markers simply reflects delayed development, we analysed endoderm markers at later time points. At E7.5 in wild-type embryos Foxa2 is expressed in the node and anterior mesendoderm (AME), whereas Cer1 marks cells of the ADE (Fig. 2U,W). Both expression domains were undetectable in mutant embryos (Fig. 2V,X), demonstrating the complete absence of DE. Consistent with a failure of expression of APS and DE markers, scanning electron microscopy analysis shows that mutant embryos fail to develop a morphological node (Fig. 2Q,R).
By E7.5 in wild-type embryos, the VE is displaced proximally into the extra-embryonic region by DE cells emerging from the distal epiblast that migrate anteriorly and proximally. To investigate the origins of endoderm in Eomes mutants, we performed an in vivo fate-mapping experiment. The ROSA26R lacZ reporter allele (Soriano, 1999) was introduced into the Eomes CA/CA background. In embryos recovered from matings between Eomes N/+ ; Sox2.Cre males and Eomes CA/CA ; ROSA26R/R females, lacZ expression is activated in the epiblast lineage. At E7.75 in wild-type embryos, the outermost layer of lacZ-positive cells corresponds to the DE (Fig. 3A-A"). In marked contrast in Eomes mutants the endodermal layer is devoid of lacZ activity and thus represents VE that fails to be displaced proximally (Fig. 3B-B"). Consistent with this interpretation, Afp-positive VE cells overlie only the extra-embryonic tissues in control embryos, whereas Afp is expressed uniformly in the endoderm in Eomes mutants (Fig. 2Y,Z). Collectively, these experiments demonstrate that Eomes functional loss disrupts specification of the DE.
Fig. 3.
Eomes mutant epiblast cells fail to contribute to the endoderm layer. Fate-mapping analysis of the epiblast via Sox2.Cre and the ROSA26R reporter allele show that the outer endodermal layer (arrows) overlying the embryo of (A-A”) E7.75 control embryos is entirely derived from ROSA26-positive epiblast cells. (B-B”) Eomes N/CA; Sox2.Cre; ROSA26R/+ mutant epiblast cells fail to contribute to the endoderm layer (arrows). Occasionally, single lacZ-positive cells are found in the endodermal layer, most probably owing to perdurance of Cre expressed in the early ICM from the Sox2.Cre transgene.
Eomes is essential for downregulation of E-cadherin and EMT
In Eomes-deficient embryos mesoderm induction is initiated but cells fail to migrate away from the streak and accumulate on the posterior side of the embryo (Fig. 4A). This phenotype is highly reminiscent of that caused by an inability to downregulate the epithelial cell-adhesion molecule E-cadherin (Ciruna and Rossant, 2001). E-cadherin is robustly expressed throughout the epiblast, but is specifically lost in mesoderm cells within the PS during gastrulation. Fgf8-mediated activation of the transcriptional repressor Snail (Ciruna and Rossant, 2001), acts directly to silence E-cadherin transcription (Batlle et al., 2000; Cano et al., 2000). Additionally, E-cadherin protein degradation decreases expression levels (Zohn et al., 2006). We analysed E-cadherin expression in mutant embryos by immunofluorescence staining. Interestingly, the distinctive tissue mass that accumulates at the site of the PS maintains E-cadherin expression at levels similar to those in the epiblast and overlying VE (Fig. 4B). The persistence of E-cadherin could potentially reflect continued transcription and/or the lack of protein degradation. To examine these possibilities, we analysed Fgf8, Snail, Spry2 and E-cadherin transcripts in E7.5 wild-type and mutant embryos. Intriguingly, both Fgf8 and its targets Snail (Ciruna and Rossant, 2001) and Spry2 (Nutt et al., 2001) are expressed robustly in the PS of mutant embryos (Fig. 4C,D). Nonetheless, E-cadherin transcription is not downregulated (Fig. 4D).
Fig. 4.
Eomes-deficient epiblast exhibits normal Fgf8/Snail expression but fails to downregulate E-cadherin. (A) Scanning electron microscopy of transverse sections through E7.5 control and Eomes mutant embryos shows the mesenchymal morphology of cells accumulating at the primitive streak of mutant embryos. Eomes mutants are devoid of a mesodermal cell layer. (B) Immunofluorescence staining using an anti-E-cadherin antibody in E7.5 Eomes mutant embryos reveals failure of E-cadherin downregulation at the PS stage. Whereas mesoderm of wild-type embryos is devoid of E-cadherin (arrowhead), the distinctive tissue mass in mutants retains E-cadherin. (C,D) Fgf8 and Snail are expressed at appropriate sites and with normal intensity in Eomes N/CA; Sox2.Cre embryos when analysed by whole-mount in situ hybridisation (C) or in situ hybridisation on sections (D). The mutant PS shows overlapping expression of E-cadherin and Snail, whereas in wild-type controls the expression domains are mutually exclusive. (C) The Fgf target Spry2 is widely expressed in Eomes mutants. (E) Wild-type and Eomes mutant primitive streak explant cultures show indistinguishable migration behaviour and efficiently downregulate E-cadherin in migrating cells. Broken lines indicate the border of E-cadherin-positive explants.
Next, to evaluate whether loss of Eomes results in a generalized cell migration defect we performed explant experiments. Primitive streak regions dissected from mutant and wild-type embryos were cultured for 72 hours on fibronectin-coated dishes as described previously (Ciruna and Rossant, 2001; Zohn et al., 2006). Surprisingly, Eomes-deficient explants were indistinguishable from wild type and showed efficient migration behaviour, associated with downregulation of E-cadherin (Fig. 4E). Thus, under these in vitro culture conditions Eomes-deficient epiblast cells are responsive to Fgf8/Snail signalling, downregulate E-cadherin, and undergo EMT. Thus, the local environment in mutant embryos contributes to the cell migration defects.
Eomes is activated in the posterior epiblast prior to overt streak formation and is maintained in the primitive streak until E7.5, when expression is abruptly downregulated. To delete Eomes from the PS, we made use of the recently described T.Cre deleter strain (Perantoni et al., 2005), in which Cre, expressed from the streak enhancer of the brachyury gene, is activated in nascent mesoderm from day 6.5 onwards. In this case, we recovered Mendelian numbers (7/26; 27%) of T.Cre; EomesN/CA offspring. Thus, we conclude that Eomes functional activity is only transiently required in the posterior epiblast at peri-gastrulation stages to allow nascent mesoderm to efficiently undergo EMT.
Cell autonomous Eomes functions required for cell migration and DE specification
Do EMT and migration defects reflect cell-autonomous Eomes requirements? To evaluate this possibility, we examined the developmental capabilities of Eomes mutant ES cells. Homozygous Eomes null (Eomes N/N) ES cells (Fig. 5A) were injected into ROSA26 blastocysts (Fig. 5B). lacZ staining of chimeric embryos at E7.5 reveals that lacZ-negative Eomes-deficient ES cells efficiently colonize the epiblast (Fig. 5D-G) but only a few mutant cells become integrated into nascent mesoderm. Interestingly, Eomes-deficient cells fail to contribute to the DE at E7.5. Rather, these lacZ-negative cells accumulate within the posterior PS region, frequently forming tissue protrusions into the amniotic cavity, similar to those observed in Eomes epiblast mutant embryos (Fig. 5E,F).
Fig. 5.
Cell-autonomous Eomes requirements for cell migration from the primitive streak and endoderm specification. (A) Eomes-deficient ES cells were generated by retargeting the remaining wild-type allele in Eomes N/+ heterozygous ES cells. (B) Eomes N/N ES cells were introduced into ROSA26LacZ blastocysts, as depicted and chimeric embryos analysed histologically at E7.5 (C-G) and E9.5 (H-N). (C-G) /acZ-negative Eomes N/N ES cells (counterstained with Eosin) are found randomly distributed in the epiblast at E7.5 (C′,D-G) but fail to exit the PS and accumulate in cell masses in the amniotic cavity. (H) Highly chimeric embryos at E9.5 are developmentally delayed and show a relative paucity of mesoderm derivatives, resulting in gross abnormalities affecting the heart, somites, axis elongation and embryonic turning. (I-N) Eomes N/N mutant ES cells fail to contribute to definitive endoderm derivatives and cannot be found within the gut tube at any level (arrows in H-N). (N) Chimeric embryos occasionally exhibit posterior neural tube duplications (arrowheads).
We also analysed chimeric embryos at later stages. At E9.5, all embryos, irrespective of the degree of ES cell contribution, displayed a graded distribution of lacZ-negative cells along the AP axis, with increased numbers of mutant cells found in more posterior regions (Fig. 5H-M). Histological analysis showed that Eomes mutant cells contribute to all derivatives of mesodermal and ectodermal origin, but even in embryos where the entire posterior region is derived from mutant ES cells, only lacZ-positive wild-type cells contribute to the gut tube (Fig. 5I-M).
Embryos with a high contribution (>80%) of mutant cells were developmentally delayed and exhibited pleiotropic defects, including disturbed somitogenesis, heart-looping abnormalities and neural tube closure defects (Fig. 5H). Occasionally, we observed neural tube duplications in posterior regions, reminiscent of observations from Fgfr1 mutant chimera experiments (Ciruna et al., 1997) (Fig. 5N). However, in the previous report the duplicated neural tubes consisted entirely of Fgfr1-mutant cells, whereas here the tissue comprises a mix of wild-type and mutant cells.
Eomes and Nodal function cooperatively to pattern the AP axis
Nodal mutants lack Eomes expression in the epiblast and expression in the TE is not maintained (Brennan et al., 2001; Guzman-Ayala et al., 2004). Nodal and Eomes are both required in the epiblast for specification of APS derivatives (Vincent et al., 2003) (Fig. 2). To investigate Eomes and Nodal genetic interactions, we intercrossed Eomes N/+ and Nodal LacZ/+ animals. Genotyping embryos at different developmental stages revealed three different phenotypes (Table 2). Approximately one-third of the double heterozygotes are severely affected and arrest around gastrulation. Another category of mutants gastrulate, but show severe truncations of the anterior body axis, heart abnormalities and die around E9.5. Finally, roughly a third of double heterozygous mutants develop normally and are adult viable.
Table 2.
Different time-points of lethality of EomesN/+; NodalLacZ/+ embryos
| EomesN/+ × NodalLacZ/+ intercross | |||||
|---|---|---|---|---|---|
| Age | Eo+/+; Nodal+/+ | EoN/+; Nodal+/+ | Eo+/+; NodalLacZ/+ | EoN/+; NodalLacZ/+ | Total |
| E7.5 | 32 (22%) | 31 (21%) | 49 (33%) | 32 (32%) | 144 |
| E9.5 P20 | 35 (24%) | 39 (26%) | 50 (33%) | 21 (17%) | 148 |
| P20 | 25 (28%) | 28 (31%) | 29 (33%) | 7 (8%) | 89 |
The most severely affected class of Eomes N/+; Nodal LacZ/+ embryos display visible abnormalities beginning around E7.0. These embryos fail to form a PS and often develop pronounce constrictions between the embryonic and extra-embryonic regions of the conceptus (Fig. 6P), a disturbance often associated with defective AP axis formation. At E6.5, Nodal LacZ expression is normally restricted to the posterior epiblast but in the mutants we found Nodal LacZ broadly expressed throughout the proximal epiblast (Fig. 6A,B). We also examined extracellular antagonists expressed by the AVE that regulate Nodal activities and position the Nodal expression domain (Beck et al., 2002; Brennan et al., 2001; Perea-Gomez et al., 2002). Mutant embryos express Cer1 and Hex at the distal tip. Thus, the AVE is correctly induced but fails to migrate anteriorly (Fig. 6C-F). Consequently, posterior markers genes, including cripto, brachyury and Eomes itself are broadly expressed throughout the entire proximal part of mutant embryos (Fig. 6G-L). However, Eomes and Nodal expression levels are unaffected in double heterozygotes (Fig. 6A,B,I,J,O,P; data not shown).
Fig. 6.
Severely affected Eomes N/+; NodalLacZ/+ double heterozygotes fail to rotate the PD axis. (A,A’) Nodal expression normally confined to the posterior epiblast at E6.5, (B,B’) persists throughout the entire proximal epiblast of Eomes N/+ ; Nodal LacZ/+ double heterozygous embryos. (C-F) AVE-markers (Hex and Cer) remain localized to the distal tip of double heterozygous mutant embryos. (G-L) Consequently posterior marker genes (cripto, Eomes and brachyury) are expressed throughout the proximal epiblast. (M,N) Spc4 is expressed at normal levels in the ExE of mutant embryos, which fails to be displaced proximally. (O-P’) From late gastrulation stages onwards, double heterozygous mutants show gross disturbances of germ layer formation, and massive cell accumulations of mesenchymal cells within in the amniotic cavity (arrowhead in P′, lacZ staining) and severe constrictions between the embryonic and extra-embryonic regions of the embryo (arrows in P). (Q-T) At E7.5, the mesoderm marker Fgf8 is widely expressed in mutants, whereas Cer1, which marks newly formed DE, fails to be expressed.
Nodal signalling thresholds within the epiblast control the formation as well as the migration of the AV E (Norris et al., 2002). One possibility is that Nodal signalling might be compromised in double heterozygous embryos, owing to decreased activities of the convertases Furin and PACE4/Spc4 synthesized within the ExE (Constam and Robertson, 2000; Beck et al., 2002; Guzman-Ayala et al., 2004; Ben-Haim et al., 2006). We detect wild-type Spc4 expression levels in the ExE of E6.5 mutant embryos, but expression is symmetric owing to the absence of extra-embryonic mesoderm formation (Fig. 6M,N). By late gastrulation stages, double heterozygous embryos are grossly disorganised, frequently show tissue accumulation within the amniotic cavity and lack visible germ layers (Fig. 6O,P). Mesoderm markers such as Fgf8 (Fig. 6Q,R) or Snail (data not shown) are expressed at E7.5 but DE markers are absent (Fig. 6S,T).
A second class of Eomes/Nodal double heterozygotes establish a normal AP axis, initiate gastrulation, but fail to specify the APS. At E7.0 Hex expression, marking the midline DE (Fig. 7A,B) and Cer1, marking the majority of nascent DE, are both greatly reduced (Fig. 7C-D′). Brachyury is normally expressed in the nascent mesoderm as well as the newly forming notochord. In mutant embryos, the brachyury expression domain is truncated because of the lack of notochord specification (Fig. 7E,F). Likewise, Foxa2 expression in axial midline tissues is decreased and fails to extend to the anteriormost regions (data not shown). At E8.0, Shh is normally expressed in the node and the developing axial midline tissues. Double heterozygotes display greatly reduced or completely lack Shh expression in axial tissues, but the node-expression domain remains intact (Fig. 7G-H′). Signalling cues from the APS and its derivatives guide and maintain anterior structures. As shown by the loss of Otx2 expression in the anterior neurectoderm at E8.5 (Fig. 7I,J), Eomes / Nodal heterozygous mutants lack the ability to correctly specify the midline axial mesendoderm, and subsequently fail to maintain anterior identity. By E9.5, mutants lack head structures rostral to the otic placodes and exhibit heart and LR patterning abnormalities (Fig. 7K-N), owing to the loss of APS and midline structures.
Fig. 7.
A second category of Eomes N/+; Nodal LacZ/+ double heterozygous mutants fails to specify the anterior primitive streak and exhibit partial axis duplications. A proportion of compound heterozygous embryos correctly specify the AP axis, but show reduced expression of DE markers (A,B) Hex and (C-D’) Cer1 at E6.5 and E7.5, respectively. Anterior axial midline tissues are lost, as seen by (E,F) truncated brachyury expression anterior to the PS (asterisk) and (G-H’) absence of Shh (arrows indicate remaining expression in the node). (H,H’) Head folds are present at E8.0 in mutant embryos (asterisks), but (I,J) anterior neural identity and Otx2 expression is lost by E8.5. (K-N) At E9.5 double heterozygous mutants exhibit severe anterior truncations, and various degrees of heart defects, ranging from (L) looping defects (arrowheads) to (N) severe malformations with cardia bifida, while posterior development remains grossly unaffected. (O,P) A proportion of Eomes N/+ ; Nodal LacZ/+ compound heterozygous mutants show duplications of the node and notochord, as revealed by Shh expression marking both node and notochord, or (Q-R′) staining for the Nodal LacZ expression in the node. Embryos with node duplications lack Nodal expression in the left lateral plate mesoderm (arrows).
Additionally, development of the node at E8.5 was severely disturbed in a subset of mutant embryos. In some cases, Shh and Nodal LacZ expression domains were enlarged (data not shown) and also complete node duplications accompanied by formation of an accessory notochord were observed (Fig. 7O-R′). As a result, the floor plate of the neural tube is expanded, secondarily causing increased spacing between the rows of somites at E9.5 (Fig. 7N). In the most severely affected embryos, we observed more complete duplications of the posterior body axis, including additional rows of somites (data not shown).
To distinguish abnormalities caused by reduced Eomes in the epiblast or the ExE, Nodal LacZ/+;Sox2.Cre males and Eomes CA/CA females were intercrossed to specifically reduce Eomes in the epiblast. Embryos with anterior axis truncations were present at low frequencies, but we failed to recover the severely affected embryos. Thus, the APS defects are associated with Nodal and Eomes co-expression in the epiblast, while early arrest and AP patterning defects reflect Eomes requirements in the ExE.
Wnt3 regulates Nodal expression levels in the epiblast via a Tcf/Lef-dependent 5′ PEE enhancer element (Ben-Haim et al., 2006). To investigate whether Eomes influences Nodal expression levels indirectly via modulating Wnt3 activities, we crossed Eomes N/+ and Wnt3 N/+ animals, and examined embryos at E9.5. No phenotypic abnormalities were observed in a large panel of Eomes N/+ ; Wnt3 N/+ double heterozygous embryos (n=19). Similarly, we recovered Mendelian numbers of viable Eomes N/+; Nodal ΔPEE/ΔPEE offspring from crosses between Eomes N/+; Nodal ΔPEE/+ mice. Collectively, these data argue that Nodal/Eomes signals patterning the APS are not relayed via the Wnt3 pathway.
Discussion
Genetic studies have identified several T-box transcription factors as key regulators of vertebrate development (reviewed by Naiche et al., 2005; Showell et al., 2004). Depending on the context of conserved DNA binding sites, different T-box transcription factors can either activate or repress target genes. Partnerships between different T-box family members and/or other transcription factors may also contribute to functional specificity (reviewed by Naiche et al., 2005). Owing to this cooperativity and partially overlapping expression patterns, functions of individual family members remain ill defined. Eomes, the first T-box gene to be expressed in the developing mouse embryo, is essential in the trophectoderm lineage (Russ et al., 2000; Strumpf et al., 2005). Eomes acts downstream of Cdx2 to maintain and expand the TS cells in the early post-implantation embryo (Niwa et al., 2005; Strumpf et al., 2005). Additionally Eomes is expressed in the posterior epiblast at pre-gastrulation stages, and maintained for a relatively brief 36 hours interval during gastrulation and elongation of the primitive streak. Here, we examined Eomes function in epiblast derivatives via a conditional inactivation strategy. We demonstrate that Eomes expression in the posterior epiblast is essential for EMT and delamination of nascent mesoderm. Second, Eomes plays a crucial role in specification of the APS and its derivatives, namely the DE cell lineage.
Previous work has shown that Nodal-mediated reciprocal signalling between the epiblast and the ExE is responsible for local mesoderm induction on the prospective posterior pole of the embryo (Brennan et al., 2001) (reviewed by Lu et al., 2001). Fgf family members also control the process of EMT. Fgfr1 receptor activation in response to Fgf4 and Fgf8 expressed by nascent mesoderm in turn activates expression of Snail, a Zn-finger transcriptional repressor that silences E-cadherin transcription (Batlle et al., 2000; Cano et al., 2000) and results in rapid loss of E-cadherin from the adherens junctions, allowing efficient cell delamination and migration. Here, we demonstrate that Eomes-deficient epiblast cells can efficiently activate these signalling cascades and express nascent mesoderm markers, including Wnt3 and brachyury. Strikingly, however, nascent mesoderm fails to undergo EMT and cannot migrate away from the primitive streak. In contrast to Fgfr1, Fgf8 and Snail-deficient embryos, which display reduced mesoderm migration (Ciruna and Rossant, 2001; Sun et al., 1999; Carver et al., 2001), Eomes functional loss efficiently and profoundly blocks EMT. Moreover, Fgf8 and Snail transcripts are robustly expressed in Eomes-deficient cells, strongly suggesting that Fgf signals function normally. The most obvious possibility is that Eomes, together with Snail, cooperatively regulates E-cadherin expression. However, this simple scenario seems unlikely because we failed to identify T-box-binding sites upstream of the E-cadherin promoter region. Eomes may be required to activate Snail transcriptional partners or alternatively Eomes could play a role in epigenetic reprogramming. Perhaps the Snail binding site controlling downregulated E-cadherin expression may be inaccessible in mutant epiblast cells. Consistent with this suggestion, Eomes inactivation at later stages in the primitive streak with the T.Cre deleter fails to disrupt development. Thus, we have identified a crucial timeframe for Eomes function in the early pre-streak epiblast tissue.
Efficient mesoderm delamination also depends on rapid degradation of E-cadherin protein. Interestingly, recent studies demonstrate that E-cadherin protein persists in p38IP mutant embryos (Zohn et al., 2006) and EMT is impaired. Nonetheless, gastrulation proceeds with reduced efficiency, and results in the formation of a well-patterned body axis with abundant mesodermal derivatives, including somites (Zohn et al., 2006). Unlike Eomes mutant explants showing normal migration behaviour in vitro, explants of p38-deficient primitive streak regions fail to migrate in vitro. Overall the p38IP phenotype shares minimal common features with the characteristics of Eomes mutant embryos described here. Components of the p38 pathway are thus unlikely to function as Eomes targets. Collectively, our experiments place Eomes as a novel upstream regulator of EMT in the early mouse embryo.
During gastrulation cells within the epiblast that ingress at the anterior primitive streak intercalate into the overlying visceral endoderm, giving rise to the DE. Chimera studies have uncovered important requirements for Foxh1 and Smad2, downstream effectors of the Nodal pathway, for specification of the DE lineage (Tremblay et al., 2000; Hoodless et al., 2001). Here, we observe in chimeric embryos that Eomes-deficient cells surrounded by wild-type neighbouring nascent mesoderm cells can delaminate and go on to form mesodermal derivatives. Consistent with this Eomes mutant ES cells have been shown to form mesodermal tissues in teratomas (Russ et al., 2000). However, as for Smad2-and Foxh1-deficient ES cells, Eomes mutant ES cells fail to contribute to the DE cell lineage. The present experiments thus demonstrate for the first time that Eomes is essential for endoderm formation in the mouse.
In zebrafish, Eomes expressed in the marginal blastomeres directly interacts with the Nodal targets Faust/Gata5 and the homeodomain protein Bon to assemble transcriptional complexes that activate Cas, a Sox family member that acts as the key endodermal determinant (Bjornson et al., 2005). Thus, it is known that Nodal and Eomes cooperatively govern DE specification (Bjornson et al., 2005) (reviewed by Stainier, 2002). Homologues of Bon and Cas have not been identified in mouse. It will be important to identify Eomes partners and target genes that control endoderm specification in the mouse.
The present study establishes that Nodal and Eomes interact genetically. Thus, double heterozygous embryos display distinctive developmental abnormalities at two different time points (summarized in Fig. 8). The first category of double heterozygous embryos phenocopy those with decreased levels of Nodal signalling owing to targeted deletion of the intronic autoregulatory enhancer (Norris et al., 2002) or defective expression of convertases by the ExE (Beck et al., 2002). Nodal produced by the epiblast maintains Eomes-positive trophoblast stem cells within the ExE (Guzman-Ayala et al., 2004) and Bmp4 expressed by this tissue in turn induces Wnt3 activity and amplifies Nodal activity in the epiblast (Ben-Haim et al., 2006). Reduced Eomes and Nodal expression levels probably compromises these regulatory feedback loops that coordinate early development of the ExE and epiblast.
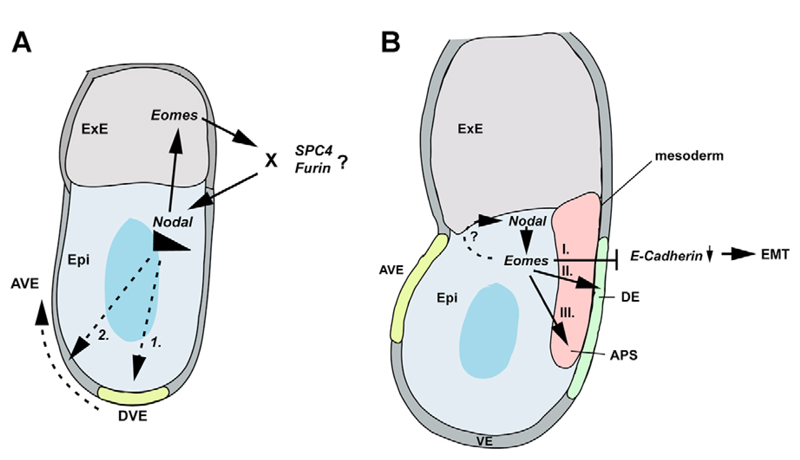
Fig. 8.
Model of Eomes action in gastrulation. (A) At pregastrula stages, Eomes and Nodal expression are restricted to the ExE and epiblast (Epi), respectively. Nodal signalling is required first to induce formation of the DVE, which requires elevated Nodal signalling-levels for migration to the anterior side. A positive-feedback loop between Eomes in the ExE and Nodal in the epiblast is necessary to maintain high Nodal signalling thresholds required for DVE migration. Although Nodal signalling may directly regulate Eomes in the ExE, the requirement for Eomes in maintaining Nodal levels may be indirect possibly via regulation of levels of pro-protein convertases Furin and/or SPC4 in the ExE. (B) During gastrulation, Eomes expression is initiated in the posterior proximal epiblast and the PS, where it overlaps with Nodal. Eomes participates in the transcriptional control of E-cadherin downregulation, essential for cells to traverse the PS and delaminate as mesoderm and endoderm. Additionally, Eomes is required to specify the DE lineage and acts cooperatively with Nodal signalling to activate target genes responsible for APS specification.
A second class of double heterozygous mutant embryos selectively lack APS progenitors and some display axis duplications. Selectively lowering Eomes in the epiblast results in a similar phenotype. The promoter region of the Xenopus Eomes gene contains an activin responsive element (Ryan et al., 2000), consistent with the idea that Nodal and Eomes are coordinately regulated. However, the mouse Eomes promoter lacks the conserved cluster of Foxh1/Smad-binding sites (data not shown). The 5′ Nodal proximal epiblast enhancer (PEE) contains Tcf/Lef binding sites, and a number of potential T-box half binding sites. These Tcf/Lef sites are highly conserved across a broad spectrum of vertebrates, including humans, rodents, cow and armadillo (Ben-Haim et al., 2006). However, by contrast, the PEE associated T-box half sites are not conserved (data not shown) and therefore unlikely to be essential for controlling Nodal/Smad2 signalling. Nodal/Eomes interactions in the epiblast are probably indirect. In zebrafish the Lim-domain gene Lmx1 is a Nodal target (Watanabe et al., 2002) known to co-regulate cerberus expression (Yamamoto et al., 2003). Lim-domain proteins have previously been shown to assemble with T-box proteins (Krause et al., 2004). As in zebrafish (Bjornson et al., 2005), Eomes/Nodal pathways may cooperatively regulate specification of APS progenitors. Alternatively, Nodal may regulate Eomes activity via a post-translational mechanism as in T-bet-mediated control of T helper progenitor cell differentiation. In this case, phosphorylation regulates T-bet interactions with Gata3 and prevents DNA binding in a pathway that represses development of Th2 cells (Hwang et al., 2005). Similar mechanisms may regulate T-box activities controlling cell fate decisions in the early embryo. Future experiments aim to identify Eomes transcription partners and downstream targets governing EMT and endoderm specification in the mouse.
Acknowledgments
We thank Carol Paterson and Emily Lejsek for generation of chimeras, George Trichas and Shankar Srinivas for help with confocal imaging, Mike Shaw for help with SEM, Richard Copley for help with sequence analysis, Mark Lewandoski for the T.Cre mice, and Lee Niswander, Rolf Kemler and Hans-Henning Arnold for probes. S.J.A. is the recipient of a Feodor Lynen Fellowship from the Alexander von Humboldt-Foundation. This work was supported by a Programme Grant from the Wellcome Trust.
References
- Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De, Herreros A. The transcription factor snail is a repressor of E- cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Beck S, Le Good JA, Guzman M, Ben Haim N, Roy K, Beermann F, Constam DB. Extraembryonic proteases regulate Nodal signalling during gastrulation. Nat Cell Biol. 2002;4:981–985. doi: 10.1038/ncb890. [DOI] [PubMed] [Google Scholar]
- Beddington RS, Robertson EJ. Axis development and early asymmetry in mammals. Cell. 1999;96:195–209. doi: 10.1016/s0092-8674(00)80560-7. [DOI] [PubMed] [Google Scholar]
- Ben-Haim N, Lu C, Guzman-Ayala M, Pescatore L, Mesnard D, Bischofberger M, Naef F, Robertson EJ, Constam DB. The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev Cell. 2006;11:313–323. doi: 10.1016/j.devcel.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Bjornson CR, Griffin KJ, Farr GH, Terashima A, Himeda C, Kikuchi Y, Kimelman D. Eomesodermin is a localized maternal determinant required for endoderm induction in zebrafish. Dev Cell. (3) 2005;9:523–533. doi: 10.1016/j.devcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS, Robertson EJ. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–969. doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- Bruce AE, Howley C, Zhou Y, Vickers SL, Silver LM, King ML, Ho RK. The maternally expressed zebrafish T-box gene eomesodermin regulates organizer formation. Development. 2003;130:5503–5517. doi: 10.1242/dev.00763. [DOI] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu GC, Dunn NR, Anderson DC, Oxburgh L, Robertson EJ. Differential requirements for Smad4 in TGFbeta-dependent patterning of the early mouse embryo. Development. 2004;131:3501–3512. doi: 10.1242/dev.01248. [DOI] [PubMed] [Google Scholar]
- Ciruna BG, Rossant J. Expression of the T-box gene Eomesodermin during early mouse development. Mech Dev. 1999;81:199–203. doi: 10.1016/s0925-4773(98)00243-3. [DOI] [PubMed] [Google Scholar]
- Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- Ciruna BG, Schwartz L, Harpal K, Yamaguchi TP, Rossant J. Chimeric analysis of fibroblast growth factor receptor-1 (Fgfr1) function: a role for FGFR1 in morphogenetic movement through the primitive streak. Development. 1997;124:2829–2841. doi: 10.1242/dev.124.14.2829. [DOI] [PubMed] [Google Scholar]
- Collignon J, Varlet I, Robertson EJ. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature. 1996;381:155–158. doi: 10.1038/381155a0. [DOI] [PubMed] [Google Scholar]
- Constam DB, Robertson EJ. SPC4/PACE4 regulates a TGFbeta signaling network during axis formation. Genes Dev. 2000;14:1146–1155. [PMC free article] [PubMed] [Google Scholar]
- Dunn NR, Vincent SD, Oxburgh L, Robertson EJ, Bikoff EK. Combinatorial activities of Smad2 and Smad3 regulate mesoderm formation and patterning in the mouse embryo. Development. 2004;131:1717–1728. doi: 10.1242/dev.01072. [DOI] [PubMed] [Google Scholar]
- Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- Guzman-Ayala M, Ben-Haim N, Beck S, Constam DB. Nodal protein processing and fibroblast growth factor 4 synergize to maintain a trophoblast stem cell microenvironment. Proc Natl Acad Sci USA; 2004. pp. 15656–15660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock SN, Agulnik SI, Silver LM, Papaioannou VE. Mapping and expression analysis of the mouse ortholog of Xenopus Eomesodermin. Mech Dev. 1999;81:205–208. doi: 10.1016/s0925-4773(98)00244-5. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev. 2002;119(1):S97–S101. doi: 10.1016/s0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Tenzen T, McMahon AP. Maternal inheritance of Cre activity in a Sox2Cre deleter strain. Genesis. 2003;37:51–53. doi: 10.1002/gene.10225. [DOI] [PubMed] [Google Scholar]
- Hoodless PA, Pye M, Chazaud C, Labbe E, Attisano L, Rossant J, Wrana JL. FoxH1 (Fast) functions to specify the anterior primitive streak in the mouse. Genes Dev. 2001;15:1257–1271. doi: 10.1101/gad.881501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005;307:430–433. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- Krause A, Zacharias W, Camarata T, Linkhart B, Law E, Lischke A, Miljan E, Simon HG. Tbx5 and Tbx4 transcription factors interact with a new chicken PDZ-LIM protein in limb and heart development. Dev Biol. 2004;273:106–120. doi: 10.1016/j.ydbio.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A. Requirement for Wnt3 in vertebrate axis formation. Nat Genet. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- Lu CC, Brennan J, Robertson EJ. From fertilization to gastrulation: axis formation in the mouse embryo. Curr Opin Genet Dev. 2001;11:384–392. doi: 10.1016/s0959-437x(00)00208-2. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating The Mouse Embryo: A Laboratory Manual. 3 Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Annu Rev Genet. 2005;39:219–239. doi: 10.1146/annurev.genet.39.073003.105925. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Norris DP, Brennan J, Bikoff EK, Robertson EJ. The Foxh1- dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development. 2002;129:3455–3468. doi: 10.1242/dev.129.14.3455. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Dingwell KS, Holt CE, Amaya E. Xenopus Sprouty2 inhibits FGF-mediated gastrulation movements but does not affect mesoderm induction and patterning. Genes Dev. 2001;15:1152–1166. doi: 10.1101/gad.191301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perantoni AO, Timofeeva O, Naillat F, Richman C, Pajni-Underwood S, Wilson C, Vainio S, Dove LF, Lewandoski M. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development. 2005;132:3859–3871. doi: 10.1242/dev.01945. [DOI] [PubMed] [Google Scholar]
- Perea-Gomez A, Vella FD, Shawlot W, Oulad-Abdelghani M, Chazaud C, Meno C, Pfister V, Chen L, Robertson E, Hamada H, et al. Nodal antagonists in the anterior visceral endoderm prevent the formation of multiple primitive streaks. Dev Cell. 2002;3:745–756. doi: 10.1016/s1534-5807(02)00321-0. [DOI] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- Ryan K, Garrett N, Mitchell A, Gurdon JB. Eomesodermin, a key early gene in Xenopus mesoderm differentiation. Cell. 1996;87:989–1000. doi: 10.1016/s0092-8674(00)81794-8. [DOI] [PubMed] [Google Scholar]
- Ryan K, Garrett N, Bourillot P, Stennard F, Gurdon JB. The Xenopus eomesodermin promoter and its concentration-dependent response to activin. Mech Dev. 2000;94:133–146. doi: 10.1016/s0925-4773(00)00300-2. [DOI] [PubMed] [Google Scholar]
- Showell C, Binder O, Conlon FL. T-box genes in early embryogenesis. Dev Dyn. 2004;229:201–218. doi: 10.1002/dvdy.10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stainier DY. A glimpse into the molecular entrails of endoderm formation. Genes Dev. 2002;16:893–907. doi: 10.1101/gad.974902. [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Sun X, Meyers EN, Lewandoski M, Martin GR. Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 1999;13:1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PP, Loebel DA. Gene function in mouse embryogenesis: get set for gastrulation. Nat Rev Genet. 2007;8:368–381. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- Tremblay KD, Hoodless PA, Bikoff EK, Robertson EJ. Formation of the definitive endoderm in mouse is a Smad2-dependent process. Development. 2000;127:3079–3090. doi: 10.1242/dev.127.14.3079. [DOI] [PubMed] [Google Scholar]
- Vincent SD, Robertson EJ. Highly efficient transgene- independent recombination directed by a maternally derived SOX2CRE transgene. Genesis. 2003;37:54–56. doi: 10.1002/gene.10226. [DOI] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Hayashi S, Norris DP, Robertson EJ. Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev. 2003;17:1646–1662. doi: 10.1101/gad.1100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldrip WR, Bikoff EK, Hoodless PA, Wrana JL, Robertson EJ. Smad2 signaling in extraembryonic tissues determines anterior-posterior polarity of the early mouse embryo. Cell. 1998;92:797–808. doi: 10.1016/s0092-8674(00)81407-5. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Rebbert ML, Andreazzoli M, Takahashi N, Toyama R, Zimmerman S, Whitman M, Dawid IB. Regulation of the Lim-1 gene is mediated through conserved FAST-1/FoxH1 sites in the first intron. Dev Dyn. 2002;225:448–456. doi: 10.1002/dvdy.10176. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Meno C, Sakai Y, Shiratori H, Mochida K, Ikawa Y, Saijoh Y, Hamada H. The transcription factor FoxH1 (FAST) mediates Nodal signaling during anterior-posterior patterning and node formation in the mouse. Genes Dev. 2001;15:1242–1256. doi: 10.1101/gad.883901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Hikasa H, Ono H, Taira M. Molecular link in the sequential induction of the Spemann organizer: direct activation of the cerberus gene by Xlim-1, Xotx2, Mix.1, and Siamois, immediately downstream from Nodal and Wnt signaling. Dev Biol. 2003;257:190–204. doi: 10.1016/s0012-1606(03)00034-4. [DOI] [PubMed] [Google Scholar]
- Zohn IE, Li Y, Skolnik EY, Anderson KV, Han J, Niswander L. p38 and a p38-interacting protein are critical for downregulation of E- cadherin during mouse gastrulation. Cell. 2006;125:957–969. doi: 10.1016/j.cell.2006.03.048. [DOI] [PubMed] [Google Scholar]