Abstract
Metadata is essential in proteomics data repositories and crucial to interpret and reanalyze the deposited datasets. For every proteomics dataset we should capture at least three levels of metadata: (i) dataset description, (ii) the sample to data files related information, and (iii) standard data file formats (e.g. mzIdentML, mzML or mzTab). While the dataset description and standard data file formats are supported by all ProteomeXchange partners; the information regarding the sample to data files is mostly missing. Recently members of the European Bioinformatics Community for Mass Spectrometry (EuBIC) have created an open-source project called Sample to Data file format for Proteomics (https://github.com/bigbio/proteomics-metadata-standard/) to enable the standardization of sample metadata of public proteomics dataset. Here, the project is presented to the proteomics community and we call for contributors including researchers, journals, and consortiums to provide feedback about the format. We believe this work will improve reproducibility, facilitate the development of new tools dedicated to proteomics data analysis.
Keywords: sample metadata, experimental design, proteomics, multiomics, data repositories, proteomeXchange, open data, reproducibility, data reanalysis, bioinformatics, standards
Introduction
Proteins are the executors of the function encoded by a cell’s genome (1). From protein expression, post-translational modifications, interactions, or even cellular localization; the diversity of biological questions that proteomics studies can answer is immense. This universe is composed of a variety of analytical and bioinformatics methods that enable us to answer each specific question. By June 2020, the PRIDE database (2) stores over 1900 phospho-proteomics, 280 crosslinking, and 120 protein-protein interactions among distinct types of studies (Supplementary Note 1). Most of those experiment designs involve (i) generation of protein samples relevant to the biological hypotheses or phenomena explored; (ii) protein separation by liquid chromatography (LC); (iii) protein digestion using an enzyme; (iv) chromatographic separation of the proteolytic peptides; (v) mass spectrometric (MS) analysis; and (vi) searching a protein sequence collection to identify and quantify proteins based on the LC-MS information (3). The proteomics community has explored in detail the experimental design, including the impact of technical and biological replicates in the statistical significance of the final results (especially in differential expression studies) (4, 5). However, the representation and dissemination of experimental design including the sample related information is still mostly an unexplored field in proteomics.
In 2015, Griss et al. (6) highlighted that the lack of complete metadata in public repositories and datasets made it difficult to reproduce the original results, and therefore limited public data reuse. Then, the analyzed samples must be well-characterized. It is not sufficient to know, for example, that a patient had a certain tumor. It is equally important to know the tumor stage, the tumor’s known molecular characteristics, as well as any possible pre-treatments (6). How to best capture an experimental design for better reuse, reproducibility, and understanding of the original results of a proteomics experiment? How can we represent in a file format or data model the complexity and variety of proteomics experiments? These remain open questions.
Discussion
Metadata standards within a scientific domain provide uniformity and consistency in the way researchers share their results with others. In proteomics, HUPO-PSI (the Human Proteome Organization Proteomics Standard Initiative) has created a set of standard file formats to store mass spectra, peptide evidences, expression values, and/or protein-protein interaction information, among other proteomics data types (7). These file formats not only contain the data (e.g. spectra) in a standardized representation but also information in a standardized representation but also additional metadata related to analysis tools, and settings (for example, search engine scores used to select specific peptide evidence. However, the sample metadata and their relationship with the data files are still missing. The mzML and mzTab file formats have specific sections to annotate sample information, integrating the experimental design in the data files. Unfortunately, the instrument and analysis software providers poorly annotate these data. Part of the problem could be that the sample description is not provided by the researchers running the mass spectrometers and/or that this level of information is not requested by the bioinformatics tools. As a result, the proteomics experimental design and sample related information are missing or stored in very diverse ways and formats. For example, the CPTAC consortium (https://cptac-data-portal.georgetown.edu/) provides for every dataset a set of excel files with the information on each sample (e.g. https://cptac-data-portal.georgetown.edu/study-summary/S048) including tumor size, origin but also how every sample is related to a specific raw file (e.g. instrument configuration parameters). As a resource routinely re-analyzing public datasets, ProteomicsDB (8), captures for each sample in the database a minimum number of properties to describe the sample and the related experimental protocol such as tissue, digestion method or instrument (e.g. https://www.proteomicsdb.org/#projects/4267/6228).
For every proteomics dataset we should capture at least three levels of metadata: (i) dataset description, (ii) the sample to data files related information; and (iii) standard data file formats (e.g. mzIdentML, mzML, or mzTab). The general description includes a piece of minimum information to describe the study: title, description, date of publication, type of experiment (e.g. http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD016060.0-1&outputMode=XML). The standard data files captured all the metadata associated with the dataset including search engine settings, scores, workflows, configuration files, among others. All ProteomeXchange partners mandate this information for each dataset. However, the information regarding the sample and its relation to the data files (Figure 1) is mostly missing. These three levels of metadata are combined in the well-established data formats ISA-TAB (https://www.isacommons.org/) (9) or MAGE-TAB (10), which are used in other omics fields. In both data formats, a tab-delimited file is used to annotate the metadata about the sample and link it to the corresponding data file(s) (sample to data file format - SDRF). Both data formats encode the properties and sample attributes as columns, and each row represents a sample in the study. However, a careful review of an existing proteomics dataset annotated in ISA-TAB (11) makes clear that not only a file format is needed, but most importantly, general guidelines about what information should be encoded in proteomics data repositories. The lack of guidelines to annotate information like disease stage, cell line code, or organism part; and the analytical information labeling channels (e.g. TMT, SILAC) or instrument configurations makes the data representation incomplete to understand the original experiment, reproduce the results or perform a re-analysis. If the information of the fraction, labeling, or enrichment method is not annotated, the reuse and reproduce of the original results will be challenging.
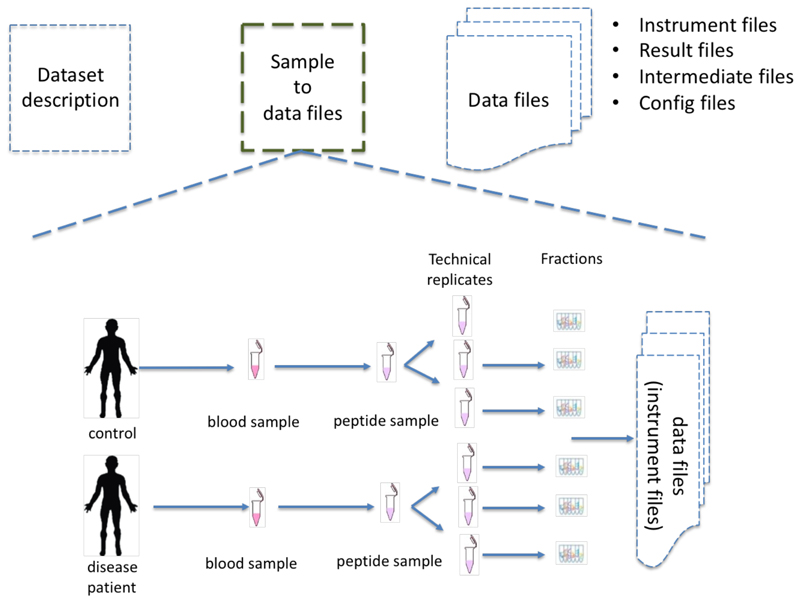
Figure 1.
Different levels of experimental design metadata: (i) dataset description: including sample and data protocols, instruments, dataset submitter (ii) the sample to data files related information; (iii) data files including standard file formats (e.g. mzIdentML, mzML or mzTab). The sample to data metadata information should capture organism, disease, cell type but also information of the analytical method: instrument, fractions, labeling channels (e.g. TMT, SILAC).
Recently members of the European Bioinformatics Community for Mass Spectrometry (EuBIC -https://eubic-ms.org/) have created an open-source project called Sample to Data file format for Proteomics (https://github.com/bigbio/proteomics-metadata-standard/) to enable the standardization of sample metadata on public proteomics datasets. The project aims to extend the sample to the data file format (Sample and Data Relationship Format - SDRF) from MAGE-TAB to represent mass spectrometry-based proteomics experiments. In summary, SDRF is a tab-delimited format that describes the sample characteristics and the relationships between samples and data files. It begins by describing the samples and finishes with the names of the data files generated from the analyses of the experimental results (Figure 2).
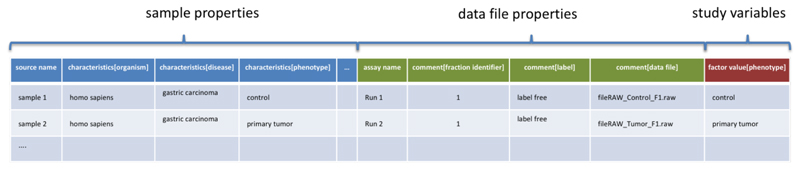
Figure 2.
Sample to the data file format (SDRF). SDRF is a tab-delimited format that describes the sample characteristics and the relationships between samples and data files.
The file format contains three different sections:
-
-
The sample metadata including organism, disease, organism part (https://github.com/bigbio/proteomics-metadata-standard/tree/master/sample-metadata).
-
-
The raw file properties that include information about the instrument, labeling applied, fraction number, mass spectrometry analyzer, fragmentation method.
-
-
The study variables (factor values), which are the variables understudy in the dataset (e.g. phenotype)
To differentiate each section, three different prefixes are used: characteristics (sample property), comment (data file property), and factor value (the study variable).
This file format complements existing submissions formats in ProteomeXchange such as submission summary file and the standard data formats such as mzIdentML, mzTab, or mzML. All the properties are expressed as ontology terms including the name of the properties. A list of supported ontologies is described on the home page of the project and can be extended using pull requests in GitHub or direct contact with the community. The current proposal is compatible with other omics types such as transcriptomics (e.g. GTEX SDRF file -https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-5214/) which will make easier the annotation of multi-omics studies. Also, the file format is compatible with the sample characteristics file format required by the EBI BioSample database (https://www.ebi.ac.uk/biosamples/docs/references/sampletab#SCD). These guidelines define a set of rules and templates that enable the representation of a variety of proteomics experiments, ranging from differential expression datasets to protein-protein interaction studies. This full compatibility with both resources (ArrayExpress and BioSamples) will enable us to perform multi-omics submissions and re-analysis of existing public data.
We are calling to biologists, mass spectrometrists, and researchers in proteomics to contribute with this initiative and provide feedback on the file format, including new use cases and proteomics approaches that have not been modeled in the data format/data model yet. We aim to standardize experimental design annotations, including the definition of the related ontologies and minimum metadata to represent a proteomics experiment, and promote active discussions and interactions around sample metadata annotations.
Researchers can contribute in the following way:
-
-
By providing feedback on the ongoing efforts including new use cases or improvements to the file format and guidelines. Researchers can contribute using GitHub issues (https://github.com/bigbio/proteomics-metadata-standard/issues), pull-requests, or by email using the google group: multiomics-data-annotation-group@googlegroups.com.
-
-
Reviewing existing annotated examples (https://github.com/bigbio/proteomics-metadata-standard/tree/master/annotated-projects) and provide feedback about them.
-
-
For bioinformaticians and software developers: implementation of pipelines and tools to convert from their sample metadata formats to SDRF or extend existing tools to read SDRF and perform automatic or semi-automatic reanalysis of public datasets.
We have recently joined the format to HUPO-PSI projects (http://psidev.info/SRDF), providing an additional forum for discussions and promote the formal adoption of the format by ProteomeXchange partners. We believe this work should improve reproducibility, facilitate the development of new tools dedicated to proteomics data analysis, and facilitate collaborations.
Supplementary Material
Acknowledgement
YPR is supported by the Wellcome Trust, grant number 208391/Z/17/Z. We would like to acknowledge everyone involved in the EuBIC-MS community.
References
- 1.Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER, 3rd, Kalocsay M, Jane-Valbuena J, Gelfand E, Schweppe DK, Jedrychowski M, Golji J, et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell. 2020;180(2):387–402 e16. doi: 10.1016/j.cell.2019.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Perez E, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eriksson J, Fenyo D. Modeling experimental design for proteomics. Methods Mol Biol. 2010;673:223–30. doi: 10.1007/978-1-60761-842-3_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goeminne LJE, Gevaert K, Clement L. Experimental design and data-analysis in label-free quantitative LC/MS proteomics: A tutorial with MSqRob. J Proteomics. 2018;171:23–36. doi: 10.1016/j.jprot.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Karp NA, Spencer M, Lindsay H, O'Dell K, Lilley KS. Impact of replicate types on proteomic expression analysis. J Proteome Res. 2005;4(5):1867–71. doi: 10.1021/pr050084g. [DOI] [PubMed] [Google Scholar]
- 6.Griss J, Perez-Riverol Y, Hermjakob H, Vizcaino JA. Identifying novel biomarkers through data mining-a realistic scenario? Proteomics Clin Appl. 2015;9(3-4):437–43. doi: 10.1002/prca.201400107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deutsch EW, Orchard S, Binz PA, Bittremieux W, Eisenacher M, Hermjakob H, Kawano S, Lam H, Mayer G, Menschaert G, Perez-Riverol Y, et al. Proteomics Standards Initiative: Fifteen Years of Progress and Future Work. J Proteome Res. 2017;16(12):4288–4298. doi: 10.1021/acs.jproteome.7b00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samaras P, Schmidt T, Frejno M, Gessulat S, Reinecke M, Jarzab A, Zecha J, Mergner J, Giansanti P, Ehrlich HC, Aiche S, et al. ProteomicsDB: a multi-omics and multi-organism resource for life science research. Nucleic Acids Res. 2020;48(D1):D1153–D1163. doi: 10.1093/nar/gkz974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez-Beltran A, Maguire E, Sansone SA, Rocca-Serra P. linkedISA: semantic representation of ISA-Tab experimental metadata. BMC Bioinformatics. 2014;15(Suppl 14):S4. doi: 10.1186/1471-2105-15-S14-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rayner TF, Rocca-Serra P, Spellman PT, Causton HC, Farne A, Holloway E, Irizarry RA, Liu J, Maier DS, Miller M, Petersen K, et al. A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB. BMC Bioinformatics. 2006;7:489. doi: 10.1186/1471-2105-7-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau E, Cao Q, Ng DC, Bleakley BJ, Dincer TU, Bot BM, Wang D, Liem DA, Lam MP, Ge J, Ping P. A large dataset of protein dynamics in the mammalian heart proteome. Sci Data. 2016;3 doi: 10.1038/sdata.2016.15. 160015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.