Abstract
Chris Hawes had a lively fascination for the immensely complex organization of the endomembrane system, including the process of endocytosis. This is the method by which eukaryotic cells internalise membrane proteins, lipids, carbohydrates, and cell wall enzymes from the cell surface through membrane bound vesicles. Endocytosis occurs progressively, starting with early membrane deformation, scission, and finally the release of the vesicle into the cytoplasm. Next to secretion, endocytosis allows the cell to control the proteome composition of its inner and outer surface membrane and as such, its communication with the outside world. Whereas endocytosis was initially considered theoretically impossible in plants due to their high turgor pressure, it is now established as essential for plant life. Furthermore, endocytosis remains a highly active field of research, both in yeast, animal, and plant model systems. Over the past three decades, the tools and techniques used to visualise, quantify, and characterise endocytosis have resulted in an increasingly higher spatiotemporal understanding of this process. Here we provide a brief history of plant endocytosis research from the time when Chris Hawes was investigating the process, to the current state-of-the-art in the field. We will end this chapter with a discussion on some promising future developments for plant endocytosis research.
A brief history of endocytosis in plants
Chris Hawes had an intense fascination for understanding the intracellular organisation of a cell. Specifically, Chris was intrigued by the immensely complex organization of the endomembrane system, which covers the anterograde trafficking pathways from the endoplasmic reticulum to the Golgi apparatus, to the trans-Golgi network and to the plasma membrane, as well as the retrograde transport route, which recycles material from the Golgi apparatus back to the endoplasmic reticulum. Moreover, he also studied the endocytic pathway, which involves the delivery of material from the cell surface via endosomes and multi-vesicular bodies to the lytic vacuole. He approached his endomembrane research questions largely through imaging, and was very much interested in endocytosis, a process on which he co-authored a book1. Endocytosis is the process by which eukaryotic cells internalise extracellular molecules and parts of their plasma membrane through the formation of endocytic vesicles. This allows cells to regulate the turnover of plasma membrane proteins (cargos), including cell wall enzymes, receptors, and transporters. Endocytosis is progressive, starting with membrane bending leading to the formation of an invagination, to scission, which releases the internalized vesicle into the cytoplasm (Figure 1a).
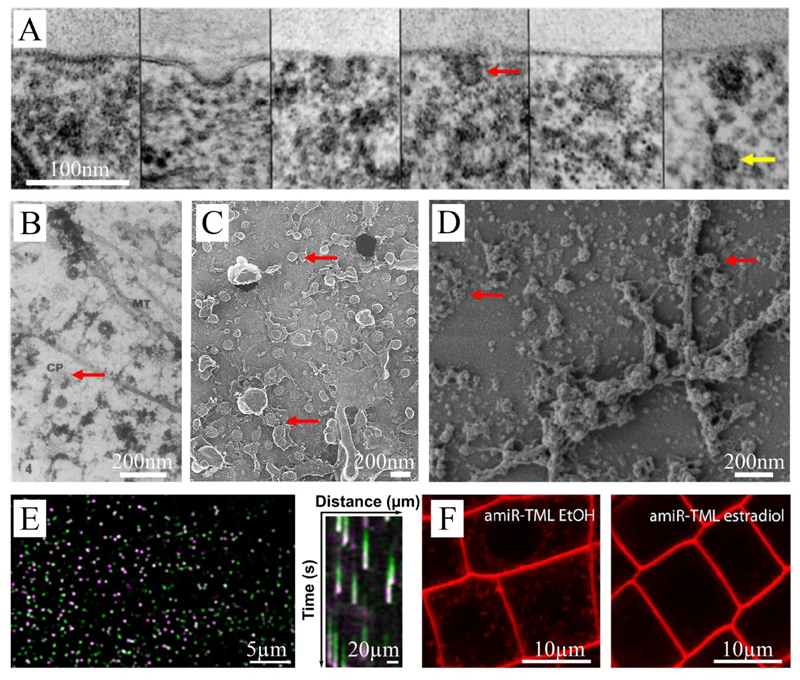
Figure 1. Visualizing endocytosis in plant cells.
(A) Representative transmission electron microscope images of high-pressure frozen, freeze-substituted Arabidopsis root cells. The individual images show subsequent stages of clathrin coated-pit formation (red arrow). Finally, through scission, the vesicle is released into the cytoplasm where it loses its clathrin coat (yellow arrow). The scale bar represents 100 nm. Image adapted and reprinted with permission from 27. (B) Representative transmission electron microscopy image using conventional chemical fixation followed by dry cleaving. Cortical microtubules (MT) and coated pits (CP, red arrow) could be visualized this way. The scale bar represents 200nm. Image adapted and reprinted with permission from 18. (C) Scanning electron microscopy image of Clathrin-coated vesicles from the personal collection of Chris Hawes (unpublished material). The scale bar represents 200nm. (D) A representative scanning electron microscopy image of a metal replica of Arabidopsis protoplasts showing Clathrin-coated pits at the plasma membrane at varying stages of development. The scale bar represents 200 nm. Image courtesy of Madhumitha Narasimhan, Jiri Friml group, IST Austria. (E) Representative spinning disc dual color image (left) and corresponding kymograph (right) of an etiolated Arabidopsis hypocotyl cell expressing TPLATE-GFP (green) and DRP1A-mRFP (magenta). The kymograph visualizes the early arrival of TPLATE and the late arrival of DRP1a at the plasma membrane. The scale bar for the spinning disc image represents 5 μm. The scale bar for the kymograph represents 20 μm. The time plotted in the kymograph totals 120 s. Image courtesy of Jie Wang, Daniel Van Damme group, PSB, Ghent, Belgium. (F) Representative images showing the use of the styryl dye FM4-64 to visualize inhibition of endocytosis. An estradiol-inducible silencing line to reduce the expression of a TPLATE complex subunit (amiR-TML) is either mock treated (EtOH, left) or induced (estradiol, right). In the control condition, the FM-dye is internalized and reaches the early endosomes, whereas this is not the case upon estradiol-mediated silencing of TML. Scale bar is 10 μm. Image adapted and reprinted with permission from 37.
Of the different endocytic mechanisms, clathrin-mediated endocytosis is the most prominent pathway2–4. Clathrin-independent pathways are also present, but do not contribute significantly to endocytic flux2. Clathrin-dependent endocytosis was the focus of Chris’ work during the late 1980s when he was affiliated to the Plant Sciences Department of the University of Oxford. During this time, Chris investigated plant clathrin-coated vesicle formation and the endocytic internalisation of macromolecules, using both electron and fluorescence microscopy5 (Figure 1b and 1c).
To this day, endocytosis remains a highly studied field of research, both in yeast, animal and plant model systems. Over the past three decades, there has been a gradual improvement in the tools and techniques used to visualise, quantify and characterise endocytosis in increasingly higher spatiotemporal resolution. Here we provide a brief history of plant endocytosis research from the time when Chris Hawes was investigating the process, to the current state-of-the-art in the field. We will end this chapter with a discussion on some promising future developments for plant endocytosis research.
The turgor pressure issue
While it is now taken for granted that endocytosis is an essential process in plants, with a key function in diverse processes ranging from nutrient uptake and receptor-mediated signalling, to polarity establishment, it was for a long time considered theoretically impossible in plants due to the high turgor pressure they maintain6. Early work based on theoretical modelling comparing plant to animal cells suggested that endocytosis could not occur in plant cells because the energy required to overcome turgor pressure was greater than could be supplied by the cell6. Later, theoretical work by Dietrich Gradmann and David Robinson similarly argued that based on the known ATP requirements for clathrin-coated pit disassembly, endocytosis would only be possible in plant cells with relatively low turgor, such as root hairs7. Despite the theoretical exclusion of endocytosis in plant cells, various labs were nonetheless interested in its potential. Early work on endocytosis was also encouraged by older studies quantifying the secretory vesicle production in pollen8,9, and the fact that is was later revealed that endocytosis and secretion are balanced processes during pollen growth10. Recent work however points out that shutting down endocytosis in Arabidopsis root cells by preventing uncoating of internalized vesicles does allow secretion to continue and therefore forces an imbalance between both processes11.
One important aspect of endocytosis is the selective uptake of cargo, such as the internalisation of transferrin by the transferrin receptor in animal cells12. However, the existence of the cell wall and lack of markers prevented similar approaches to investigate endocytosis in plants. Early work in the Robinson and Fowke laboratories showed that plant cells could internalise heavy metals such as ferritin or gold-conjugates13–16. Furthermore, clathrin-coated vesicles were identified at the plasma membrane in cell protoplasts using transmission electron microscopy (TEM)16, suggesting that plant cells in an isotonic environment could undergo clathrin-mediated endocytosis. These works provided some of the earliest evidence for endocytosis in plants, although they were not without criticism, which included the use of protoplasts rather than walled cells, and for using potentially toxic heavy metals.
Not long after these studies were published, Chris Hawes and colleagues in Oxford and Dundee, who were similarly interested in endocytosis, had also been working on visualising clathrin-coated pits through electron microscopy approaches. Through an impressive visualisation method involving rapid freezing and deep etching, Hawes and colleagues were able to clearly identify clathrin-coated pits at and near the plasma membrane in carrot suspension cells (Figure 1b-1c)17,18. These images, which are still impressive to this day, provided some of the clearest evidence for the existence of plasma membrane-derived clathrin-coated vesicles in plants. Furthermore, in contrast to earlier theoretical work, a study from Michael Saxton and William Breidenbach at UC Davis proposed that receptor-mediated endocytosis was actually theoretically possible19. These data suggested that endocytosis was a functional process in plants and could occur using fundamentally similar endocytic machinery as described in animals and yeast20.
By the late 1980s and early 1990s many researchers were convinced that endocytosis was a real process in plant cells. Patch clamp experiments in the 1990s also revealed unitary measurements of exo-and endocytosis in plants, similar to what had been shown in other systems21,22. However, at this time clear physiological evidence showing the selective uptake of cargo in plant cells had still not been attained23. In the following decades, further evidence began to emerge irrevocably demonstrating clathrin-mediated endocytosis in plants, largely aided by advances in confocal microscopy and live-cell imaging. The introduction of amphiphilic styryl dyes, which were shown to mark membrane internalisation in a temperature-dependent manner, made a compelling case24,25. Important evidence also came from work in visualising the pattern recognition receptor FLS2, revealing that it undergoes selective internalization and degradation via endocytosis upon stimulation with the bacterial flagellin epitope flg2226. Not long after, even more evidence came from detailed electron microscopy work depicting the formation and maturation of clathrin-coated pits at the plasma membrane in Arabidopsis thaliana cells (Figure 1a), and a role for clathrinmediated endocytosis in the internalization of the PIN auxin efflux carriers27. These, and many other works that followed, were crucial in the establishment of the endocytosis field in plant research.
Clathrin-mediated endocytosis requires adaptor protein complexes
Clathrin does not bind membranes directly3, nor does it recognize the cargo that needs to be internalized. This is achieved through multimeric protein complexes termed adaptor complexes. In addition to several monomeric adaptor complexes, the adaptor protein complex family consists of 5 heterotetramers and functions to transport cargo from one cellular compartment to another28,29. One of those five complexes, adaptor protein complex 2 (AP-2) functions specifically at the plasma membrane30–35. Next to the heterotetrameric AP-2, there is yet another adaptor complex acting specifically in endocytosis at the plasma membrane. This complex is termed the TPLATE complex (TPC) after its founding member, which is recruited to the forming cell plate and to the plasma membrane region surrounding the cell plate insertion site, which appears as a “T” in a single-slice confocal image36. This novel adaptor complex was simultaneously discovered in plants37 and in slime molds, where it was termed, a bit tongue-in-cheek, TSET, referring to its founding member TPLATE, and further consisting of TSPOON, TCUP, TSAUCER and its two TTRAYS38.
TPC/TSET is an evolutionary ancient adaptor complex which emerged in between the COPI complex, which functions in retrograde Golgi-to-ER transport, and the functional diversification of the five AP complexes38. Plants and slime molds have retained this complex, in contrast to yeast and animal cells. Plants however are the only kingdom where this complex is essential for life37,38, indicating that the execution of clathrin-mediated endocytosis in plants occurs fundamentally different from the other model systems. Moreover, a novel degradation pathway was recently discovered in plants, operating between ER-PM contact sites and the vacuole, bypassing the TGN39. Two subunits of TPC (AtEH1/Pan1 and AtEH2/Pan1) play a key role in activating this pathway, and several other TPC and AP-2 subunits as well as clathrin are also implicated. How TPC and AP-2 work together, whether they possess separate roles or differential cargo recognition capacities, and whether this novel degradation pathway is causal to the evolutionary retention of TPC in plants are the subject of ongoing research.
Current tools to study endocytosis in plants: dyes and drugs to visualise endocytosis
In animal systems, Alexa-dye conjugated transferrin (Tf) and epidermal growth factor (EGF) serve as widely used readouts for endocytic internalization. In contrast, no similar tools are available to image endocytosis in live plant cells, although several receptors and corresponding fluorescently labelled ligands have been reported to work40,41. Consequently, by far the most widely used method to visualise and quantify endocytosis rates involves the use of the lipophilic dye FM4-64. FM4-64 only fluoresces when bound to membranes and is readily internalised into root cells through endocytosis, allowing for an approximate visualization and quantification of endocytic flux (Figure 1f)37,42,43. Over long-term incubations (10-60 minutes), FM4-64 labels the trans-Golgi network/early endosome (TGN/EE), multi-vesicular bodies/late endosome (MVB/LE), and eventually the tonoplast, thus revealing the entire endocytic pathway. Importantly, it was initially also through FM4-64 uptake experiments in the Schumacher lab that the plant TGN was discovered to also act as an early endosome44, representing a hub where both the endocytic and secretory pathways converge45.
Another popular method to visualise endocytosis is through the reversible vesicle trafficking inhibitor and fungal toxin brefeldin A (BFA). BFA targets the Sec7 domain in a subclass of ARF-GEFs to disrupt secretion and/or recycling to the plasma membrane46,47. Treatment with BFA causes the accumulation of proteins into an enlarged compartment known as a BFA body, which is composed of TGN/EE and derived vesicles48–50. Interestingly, mutations in the tethering complex TRAPIII can abolish BFA response completely, suggesting a regulatory endosomal function for this complex51,52. As BFA does not strongly inhibit endocytosis, it can be used to accumulate and trap endocytosed membrane proteins. This method has been widely employed in the plant field as a simple method to assess and quantify dynamics of cargo internalisation and secretion or recycling after BFA washout 50,53–57. Use of BFA is not without criticism, and potential concerns were raised by Chris Hawes and colleagues about its tissue-specific effects58. However, due to the convergence of endocytosis and secretion in the TGN/EE, BFA is one of the only tools that plant scientists can use to investigate protein secretion/recycling following endocytosis.
Live-cell visualization of endocytic internalization
One of the major challenges in visualising endocytosis with traditional point-scanning confocal systems is the limited resolution in the z-axis, which results in relatively poor signal-to-noise ratios when imaging at high speeds. An important technique to overcome these limitations is the use of total internal reflection fluorescence microscopy (TIRFM). In this technique the laser light is reflected on the cover slip at a super-critical angle (65–67°), thereby creating a thin evanescent wave that allows selective excitation, and therefore selective imaging, of events at or close to the plasma membrane59. As plant cells have a primary cell wall separating the plasma membrane from the cover slip by more than 100 nm, often a greater penetration depth is needed to clearly image plasma membrane dynamics. Changing the incidence angle of the laser light to sub-critical (59–61°), allows greater penetration depth into the cell which provides improved fluorescent signal60–62. Using TIRFM with sub-critical incident angles has also been referred to as variable-angle epifluorescence microscopy (VAEM).
First popularised by the Bednarek laboratory for use in plants63, TIRFM/VAEM have been effective techniques for visualising the formation and budding of clathrin-coated vesicles from the plasma membrane64–66. Next to TIRFM/VAEM, spinning disc confocal microscopy is also often used to image endocytic events at the plasma membrane, especially if the proteins that are visualized are exclusively localized at the membrane37. Both TIRF and spinning disk microscopy are enabled by very fast and high sensitivity EMCCD and CMOS cameras. These techniques have been used to assess the timing of proteins during plant endocytosis60,67. For example, dual-color live-cell imaging, combined with kymograph visualization can easily discriminate early arriving (e.g. TPLATE) versus late arriving (DRP1A) endocytic players (Figure 1e). Very recently, the temporal resolution of endocytic spinning disc microscopy imaging in Arabidopsis was increased by lowering the temperature of the samples68.
Further developments are focused on adapting quantitative analysis pipelines using automated tracking methods such as TrackMate69 and CmeAnalysis70 for plant endocytosis as was done for yeast and animal model systems. For example, with these and related tools researchers have been able to analyse clathrin-coated vesicle departure times in plants60,71, as well as to quantify membrane-protein dynamics in living plant cells using single-particle tracking72–74. Such analytical tools are increasingly necessary in order to quantitatively assess the spatiotemporal dynamics of endocytosis.
Although confocal microscopy techniques to study endocytosis are now very popular in plants, the diffraction-limited nature of endocytic pits means that ultrastructural insight into plant endocytosis still has its place. Nevertheless, only very recently high resolution images of clathrin coated pits from ‘unroofed’ Arabidopsis protoplasts were obtained (Figure 1d)71. This unroofing process was pioneered by John Heuser’s group and allows a look into the inside of the plasma membrane75, and therefore can provide a unique view of clathrin-coated pits budding from the plasma membrane. Additional images can be found on the Heuser lab website (http://www.heuserlab.wustl.edu/images/galleries/classics/pages/17.shtml). Through their EM analysis, Narasimhan and colleagues report that clathrin-mediated endocytosis may follow a ‘constant curvature’ model and does not require actin for functional endocytic pit progression, providing new insight into how endocytosis in plants differs from other model systems. It will be interesting to perform similar experiments in conditions where endocytosis is perturbed in order to provide new mechanistic insights about plant endocytosis.
Future developments
The rapid pace of improvements in live cell and super-resolution microscopy in the last decade has made future studies in endocytosis especially promising76. One technique with great potential is the combination of TIRFM and structured illumination microscopy (SIM) which enables live-cell super-resolution imaging of clathrin-mediated endocytosis in animal cells77. Whether these techniques can be adapted for use in plants remains an open question and will likely require modifications such as adaptive optics to correct for refractive inhomogeneity inherent to plant samples. Overcoming issues such as autofluorescence and light scattering caused by cell walls remains challenging59, though the recent establishment of live super-resolution photo-activated localization microscopy (PALM) in plant cells suggests that these obstacles can indeed be overcome78.
Alongside new microscopy advancements, the development of more advanced or newer tools and fluorescent labels to study endocytosis beyond traditional FM methods are sorely needed. The creation of a fluorescent chemical analogue of the ligand for BRI1 has been used to visualise the endocytosis and degradation of BRI140, nevertheless its widespread usage is limited by the high cost of production and its relatively weak fluorescent signal. Promising new developments include genetically encoded fluorogen based reporters, such as the fluorescence-activating and absorption-shifting tag (FAST) which can bind exogenously applied fluorogenic chromophores79,80. Such reporters could allow for inducible and selective visualisation of endocytosis.
Conclusions
It is now evident that clathrin-mediated endocytosis does occur in plants and is essential to sustain plant development, yet a clear mechanistically and biophysical understanding of how plant cells undergo endocytosis in a high turgor environment is still unclear. The rapid development in areas such as super-resolution microscopy and live-cell imaging may provide some of the necessary tools and techniques to fully decipher this challenging question and to further explore the field of endocytosis research.
While the majority of research on endocytosis has been undertaken in animal and yeast systems, discoveries in plants have begun to provide interesting insight into how complex membrane trafficking evolved in a broader evolutionarily context71,81,82. For example, new knowledge of how the evolutionarily ancient TPLATE complex functions may reveal key differences in the temporal and spatial co-ordination of endocytosis between plants and other kingdoms.
We believe plant endocytosis remains a highly exciting area of research and are certain that Chris would strongly agree.
Acknowledgements
The authors would like to thank Madhumitha Narasimhan and Jiri Friml (IST, Austria) and Jie Wang (PSB, Ghent, Belgium) for providing unpublished images, and Stefan Hillmer and David Robinson (Heidelberg University) for comments on the manuscript. Research in the Van Damme lab is supported by the European Research Council T-Rex project number 682436.
Reference List
- 1.Hawes CR, Coleman JOD, Evans DE. Endocytosis, exocytosis and vesicle traffic in plants. Vol. 45 Cambridge University Press; 1991. [Google Scholar]
- 2.Bitsikas V, Corrêa IR, Nichols BJ. Clathrin-independent pathways do not contribute significantly to endocytic flux. Elife. 2014;2014:1–26. doi: 10.7554/eLife.03970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 4.Paez Valencia J, Goodman K, Otegui MS. Endocytosis and endosomal trafficking in plants. Annu. Rev. Plant Biol. 2016;67:309–335. doi: 10.1146/annurev-arplant-043015-112242. [DOI] [PubMed] [Google Scholar]
- 5.Oparka KJ, Hawes C. Vacuolar sequestration of fluorescent probes in plant cells: a review. J Microsc. 1992;166:15–27. [Google Scholar]
- 6.Cram WJ. Pinocytosis in plants. New Phytol. 1980;84:1–17. [Google Scholar]
- 7.Gradmann D, Robinson DG. Does turgor prevent endocytosis in plant cells? Plant. Cell Environ. 1989;12:151–154. [Google Scholar]
- 8.Picton JM, Steer MW. Determination of secretory vesicle production rates by dictyosomes in pollen tubes of Tradescantia using cytochalasin D. J Cell Sci. 1981;49:261–272. doi: 10.1242/jcs.49.1.261. [DOI] [PubMed] [Google Scholar]
- 9.Picton JM, Steer MW. A model for the mechanism of tip extension in pollen tubes. J Theor Biol. 1982;98:15–20. [Google Scholar]
- 10.Moscatelli A, Idilli AI. Pollen tube growth: a delicate equilibrium between secretory and endocytic pathways. J Integr Plant Biol. 2009;51:727–739. doi: 10.1111/j.1744-7909.2009.00842.x. [DOI] [PubMed] [Google Scholar]
- 11.Adamowski M, Narasimhan M, Kania U, Glanc M, De Jaeger G, Friml J. A functional study of AUXILIN-LIKE1 and 2, two putative clathrin uncoating factors in arabidopsis. Plant Cell. 2018;30:700–716. doi: 10.1105/tpc.17.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. 1983;97:329–339. doi: 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hübner R, Depta H, Robinson DG. Endocytosis in maize root cap cells. Protoplasma. 1985;129:214–222. [Google Scholar]
- 14.Joachim S, Robinson DG. Endocytosis of cationic ferritin by bean leaf protoplasts. Eur J Cell Biol. 1984;34:212–216. [PubMed] [Google Scholar]
- 15.Tanchak MA, Griffing LR, Mersey BG, Fowke LC. Endocytosis of cationized ferritin by coated vesicles of soybean protoplasts. Planta. 1984;162:481–486. doi: 10.1007/BF00399912. [DOI] [PubMed] [Google Scholar]
- 16.Hillmer S, Depta H, Robinson DG. Confirmation of endocytosis in higher plant protoplasts using lectin-gold conjugates. Eur J Cell Biol. 1986;41:142–149. [Google Scholar]
- 17.Coleman J, Evans D, Hawes C, Horsley D, Cole L. Structure and molecular organization of higher plant coated vesicles. J Cell Sci. 1987;88:35–45. [Google Scholar]
- 18.Hawes C, Martin B. Deep etching of plant cells: cytoskeleton and coated pits. Cell Biol Int Rep. 1986;10:985–992. [Google Scholar]
- 19.Saxton MJ, Breidenbach RW. Receptor-mediated endocytosis in plants is energetically possible. Plant Physiol. 1988;86:993–995. doi: 10.1104/pp.86.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson DG, Depta H. Coated vesicles. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:53–99. [Google Scholar]
- 21.Zorec R, Tester M. Rapid pressure driven exocytosis-endocytosis cycle in a single plant cell: capacitance measurements in aleurone protoplasts. FEBS Lett. 1993;333:283–286. doi: 10.1016/0014-5793(93)80671-g. [DOI] [PubMed] [Google Scholar]
- 22.Homann U, Thiel G. Unitary exocytotic and endocytotic events in guard-cell protoplasts during osmotically driven volume changes. FEBS Lett. 1999;460:495–499. doi: 10.1016/s0014-5793(99)01396-4. [DOI] [PubMed] [Google Scholar]
- 23.Hawes C, Crooks K, Coleman J, Satiat-Jeunemaitre B. Endocytosis in plants: fact or artefact? Plant. Cell Environ. 1995;18:1245–1252. [Google Scholar]
- 24.Bolte S, Talbot C, Boutte Y, Catrice O, Read ND, Satiat-Jeunemaitre B. FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J Microsc. 2004;214:159–173. doi: 10.1111/j.0022-2720.2004.01348.x. [DOI] [PubMed] [Google Scholar]
- 25.Meckel T, Hurst AC, Thiel G, Homann U. Endocytosis against high turgor: intact guard cells of Vicia faba constitutively endocytose fluorescently labelled plasma membrane and GFP-tagged K+-channel KAT1. Plant J. 2004;39:182–193. doi: 10.1111/j.1365-313X.2004.02119.x. [DOI] [PubMed] [Google Scholar]
- 26.Robatzek S, Chinchilla D, Boller T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 2006;20:537–542. doi: 10.1101/gad.366506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, Friml J. Clathrin-Mediated Constitutive Endocytosis of PIN Auxin Efflux Carriers in Arabidopsis. Curr Biol. 2007;17:520–527. doi: 10.1016/j.cub.2007.01.052. [DOI] [PubMed] [Google Scholar]
- 28.Robinson MS. Forty Years of Clathrin-coated Vesicles. Traffic. 2015;16:1210–1238. doi: 10.1111/tra.12335. [DOI] [PubMed] [Google Scholar]
- 29.Hirst J, Barlow L, Francisco GC, Sahlender DA, Seaman MNJ, Dacks JB, Robinson MS. The fifth adaptor protein complex. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Rubbo S, Irani NG, Kim SY, Xu Z-Y, Gadeyne A, Dejonghe W, Vanhoutte I, Persiau G, Eeckhout D, Simon S, Song K, et al. The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis. Plant Cell. 2013;25:2986–97. doi: 10.1105/tpc.113.114058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaksonen M, Roux A. Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2018;19:313–326. doi: 10.1038/nrm.2017.132. [DOI] [PubMed] [Google Scholar]
- 32.Bashline L, Li S, Anderson CT, Lei L, Gu Y. The endocytosis of cellulose synthase in arabidopsis is dependent on μ2, a clathrin-mediated endocytosis adaptin. Plant Physiol. 2013;163:150–160. doi: 10.1104/pp.113.221234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaoka S, Shimono Y, Shirakawa M, Fukao Y, Kawase T, Hatsugai N, Tamura K, Shimada T, Hara-Nishimura I. Identification and dynamics of arabidopsis adaptor protein-2 complex and its involvement in floral organ development. Plant Cell. 2013;25:2958–2969. doi: 10.1105/tpc.113.114082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan L, Hao H, Xue Y, Zhang L, Song K, Ding Z, Botella MA, Wang H, Lin J. Dynamic analysis of Arabidopsis AP2 σ subunit reveals a key role in clathrin-mediated endocytosis and plant development. Development. 2013;140:3826–3837. doi: 10.1242/dev.095711. [DOI] [PubMed] [Google Scholar]
- 35.Kim I, Pan W, Jones SA, Zhang Y, Zhuang X, Wu D. Clathrin and AP2 are required for PtdIns (4, 5) P2-mediated formation of LRP6 signalosomes. J Cell Biol. 2013;200:419–428. doi: 10.1083/jcb.201206096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Damme D, Coutuer S, De Rycke R, Bouget FY, Inzé D, Geelen D. Somatic cytokinesis and pollen maturation in Arabidopsis depend on TPLATE, which has domains similar to coat proteins. Plant Cell. 2006;18:3502–3518. doi: 10.1105/tpc.106.040923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gadeyne A, Sánchez-Rodríguez C, Vanneste S, Di Rubbo S, Zauber H, Vanneste K, Van Leene J, De Winne N, Eeckhout D, Persiau G, Van De Slijke E, et al. The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell. 2014;156:691–704. doi: 10.1016/j.cell.2014.01.039. [DOI] [PubMed] [Google Scholar]
- 38.Hirst J, Schlacht A, Norcott JP, Traynor D, Bloomfield G, Antrobus R, Kay RR, Dacks JB, Robinson MS. Characterization of TSET, an ancient and widespread membrane trafficking complex. Elife. 2014;3:1–18. doi: 10.7554/eLife.02866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang P, Pleskot R, Zang J, Winkler J, Wang J, Yperman K, Zhang T, Wang K, Gong J, Guan Y, Richardson C, et al. Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat Commun. 2019:1–16. doi: 10.1038/s41467-019-12782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irani NG, Di Rubbo S, Mylle E, Van den Begin J, Schneider-Pizoń J, Hniliková J, Šíša M, Buyst D, Vilarrasa-Blasi J, Szatmári A-M, Van Damme D, et al. Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat Chem Biol. 2012;8:583–589. doi: 10.1038/nchembio.958. [DOI] [PubMed] [Google Scholar]
- 41.Ortiz-Morea FA, Savatin DV, Dejonghe W, Kumar R, Luo Y, Adamowski M, Van Begin JDen, Dressano K, De Oliveira GP, Zhao X, Lu Q, et al. Danger-associated peptide signaling in Arabidopsis requires clathrin. Proc Natl Acad Sci. 2016;113:11028–11033. doi: 10.1073/pnas.1605588113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rigal A, Doyle SM, Robert S. Plant Cell. Expansion Springer; 2015. Live cell imaging of FM4-64, a tool for tracing the endocytic pathways in Arabidopsis root cells; pp. 93–103. [DOI] [PubMed] [Google Scholar]
- 43.Jelínková A, Malínská K, Simon S, Kleine-Vehn J, Pařezová M, Pejchar P, Kubeš M, Martinec J, Friml J, Zažímalová E, Petrášek J. Probing plant membranes with FM dyes: Tracking, dragging or blocking? Plant J. 2010;61:883–892. doi: 10.1111/j.1365-313X.2009.04102.x. [DOI] [PubMed] [Google Scholar]
- 44.Dettmer J, Hong-Hermesdorf A, Stierhof Y-D, Schumacher K. Vacuolar H+ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell. 2006;18:715–730. doi: 10.1105/tpc.105.037978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Viotti C, Bubeck J, Stierhof YD, Krebs M, Langhans M, van den Berg W, van Dongen W, Richter S, Geldner N, Takano J, Jürgens G, et al. Endocytic and secretory traffic in Arabidopsis merge in the trans-golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell. 2010;22:1344–1357. doi: 10.1105/tpc.109.072637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh MK, Jürgens G. Specificity of plant membrane trafficking - ARFs, regulators and coat proteins. Seminars in Cell and Developmental Biology. 2018;80:85–93. doi: 10.1016/j.semcdb.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 47.Klausner RD, Donaldson JG, Lippincott-Schwartz J. Brefeldin A: Insights into the control of membrane traffic and organelle structure. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell. 2003;112:219–230. doi: 10.1016/s0092-8674(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 49.Grebe M, Xu J, Möbius W, Ueda T, Nakano A, Geuze HJ, Rook MB, Scheres B. Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr Biol. 2003;13:1378–1387. doi: 10.1016/s0960-9822(03)00538-4. [DOI] [PubMed] [Google Scholar]
- 50.Naramoto S, Otegui MS, Kutsuna N, De Rycke R, Dainobu T, Karampelias M, Fujimoto M, Feraru E, Miki D, Fukuda H. Insights into the localization and function of the membrane trafficking regulator GNOM ARF-GEF at the Golgi apparatus in Arabidopsis. Plant Cell. 2014;26:3062–3076. doi: 10.1105/tpc.114.125880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramírez-Peinado S, Ignashkova TI, van Raam BJ, Baumann J, Sennott EL, Gendarme M, Lindemann RK, Starnbach MN, Reiling JH. TRAPPC13 modulates autophagy and the response to Golgi stress. J Cell Sci. 2017;130:2251–2265. doi: 10.1242/jcs.199521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosquete MR, Worden N, Ren G, Sinclair RM, Pfleger S, Salemi M, Phinney BS, Domozych D, Wilkop T, Drakakaki G. AtTRAPPC11/ROG2: A role for TRAPPs in maintenance of the plant trans-golgi network/early endosome organization and function. Plant Cell. 2019;31:1879–1898. doi: 10.1105/tpc.19.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teh O, Moore I. An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature. 2007;448:493–496. doi: 10.1038/nature06023. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka H, Nodzyński T, Kitakura S, Feraru MI, Sasabe M, Ishikawa T, Kleine-Vehn J, Kakimoto T, Friml J. BEX1/ARF1A1C is required for BFA-sensitive recycling of PIN auxin transporters and auxin-mediated development in arabidopsis. Plant Cell Physiol. 2014;55:737–749. doi: 10.1093/pcp/pct196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richter S, Kientz M, Brumm S, Nielsen ME, Park M, Gavidia R, Krause C, Voss U, Beckmann H, Mayer U, Stierhof Y-D, et al. Delivery of endocytosed proteins to the cell-division plane requires change of pathway from recycling to secretion. Elife. 2014;3:e02131–e02131. doi: 10.7554/eLife.02131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kleine-Vehn J, Leitner J, Zwiewka M, Sauer M, Abas L, Luschnig C, Friml J. Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc Natl Acad Sci. 2008;105:17812–17817. doi: 10.1073/pnas.0808073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Doyle SM, Haeger A, Vain T, Rigal A, Viotti C, Łangowska M, Ma Q, Friml J, Raikhel NV, Hicks GR, Robert S. An early secretory pathway mediated by GNOM-LIKE 1 and GNOM is essential for basal polarity establishment in Arabidopsis thaliana. Proc Natl Acad Sci. 2015;112:E806–E815. doi: 10.1073/pnas.1424856112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson DG, Langhans M, Saint-Jore-Dupas C, Hawes C. BFA effects are tissue and not just plant specific. Trends Plant Sci. 2008;13:405–408. doi: 10.1016/j.tplants.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 59.Grossmann G, Krebs M, Maizel A, Stahl Y, Vermeer JEM, Ott T. Green light for quantitative live-cell imaging in plants. J Cell Sci. 2017 doi: 10.1242/jcs.209270. [DOI] [PubMed] [Google Scholar]
- 60.Johnson A, Vert G. Single Event Resolution of Plant Plasma Membrane Protein Endocytosis by TIRF Microscopy. Front Plant Sci. 2017;8:1–11. doi: 10.3389/fpls.2017.00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wan Y, Ash WM, Fan L, Hao H, Kim MK, Lin J. Variable-angle total internal reflection fluorescence microscopy of intact cells of Arabidopsis thaliana. Plant Methods. 2011;7:27. doi: 10.1186/1746-4811-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He K, Marsland R, Upadhyayula S, Song E, Dang S, Capraro BR, Wang W, Skillern W, Gaudin R, Ma M, Kirchhausen T. Dynamics of phosphoinositide conversion in clathrin-mediated endocytic traffic. Nature. 2017;552:410–414. doi: 10.1038/nature25146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Konopka Ca, Bednarek SY. Variable-angle epifluorescence microscopy: A new way to look at protein dynamics in the plant cell cortex. Plant J. 2008;53:186–196. doi: 10.1111/j.1365-313X.2007.03306.x. [DOI] [PubMed] [Google Scholar]
- 64.Ito E, Fujimoto M, Ebine K, Uemura T, Ueda T, Nakano A. Dynamic behavior of clathrin in Arabidopsis thaliana unveiled by live imaging. Plant J. 2012;69:204–216. doi: 10.1111/j.1365-313X.2011.04782.x. [DOI] [PubMed] [Google Scholar]
- 65.Konopka CA, Backues SK, Bednareka SY. Dynamics of Arabidopsis dynaminrelated protein 1C and a clathrin light chain at the plasma membrane. Plant Cell. 2008;20:1363–1380. doi: 10.1105/tpc.108.059428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Konopka CA, Bednarek SY. Comparison of the dynamics and functional redundancy of the Arabidopsis dynamin-related isoforms DRP1A and DRP1C during plant development. Plant Physiol. 2008;147:1590–1602. doi: 10.1104/pp.108.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fujimoto M, Arimura SI, Ueda T, Takanashi H, Hayashi Y, Nakano A, Tsutsumi N. Arabidopsis dynamin-related proteins DRP2B and DRP1A participate together in clathrincoated vesicle formation during endocytosis. Proc Natl Acad Sci. 2010;107:6094–6099. doi: 10.1073/pnas.0913562107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J, Mylle E, Johnson A, Besbrugge N, De Jaeger G, Friml J, Pleskot R, Van Damme D. High temporal resolution reveals simultaneous plasma membrane recruitment of TPLATE complex subunits. Plant Physiol. 2020 doi: 10.1104/pp.20.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW. TrackMate: An open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 70.Aguet F, Antonescu CN, Mettlen M, Schmid SL, Danuser G. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev Cell. 2013;26:279–291. doi: 10.1016/j.devcel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Narasimhan M, Johnson A, Prizak R, Kaufmann WA, Tan S, Casillas-Pérez B, Friml J. Evolutionarily unique mechanistic framework of clathrin-mediated endocytosis in plants. Elife. 2020;9 doi: 10.7554/eLife.52067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X, Li X, Deng X, Luu D-T, Maurel C,, Lin J. Single-molecule fluorescence imaging to quantify membrane protein dynamics and oligomerization in living plant cells. Nat Protoc. 2015;10:2054. doi: 10.1038/nprot.2015.132. [DOI] [PubMed] [Google Scholar]
- 73.Zhang X, Cui Y, Yu M,, Lin J. Single-Molecule Techniques for Imaging Exo-Endocytosis Coupling in Cells. Trends in Plant Science. 2019;24:879–880. doi: 10.1016/j.tplants.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 74.Wang L, Li H, Lv X, Chen T, Li R, Xue Y, Jiang J, Jin B, Baluška F, Šamaj J, Wang X, et al. Spatiotemporal Dynamics of the BRI1 Receptor and its Regulation by Membrane Microdomains in Living Arabidopsis Cells. Mol Plant. 2015;8:1334–1349. doi: 10.1016/j.molp.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 75.Heuser J, Evans L. Three-dimensional visualization of coated vesicle formation in fibroblasts. J Cell Biol. 1980;84:560–583. doi: 10.1083/jcb.84.3.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baranov MV, Olea RA, van den Bogaart G. Chasing Uptake: Super-Resolution Microscopy in Endocytosis and Phagocytosis. Trends Cell Biol. 2019;29:727–739. doi: 10.1016/j.tcb.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 77.Li D, Shao L, Chen BC, Zhang X, Zhang M, Moses B, Milkie DE, Beach JR, Hammer JA, Pasham M, Kirchhausen T, et al. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 2015;349:6251. doi: 10.1126/science.aab3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Platre MP, Bayle V, Armengot L, Bareille J, del Mar Marquès-Bueno M, Creff A, Maneta-Peyret L, Fiche JB, Nollmann M, Miège C, Moreau P, et al. Developmental control of plant Rho GTPase nano-organization by the lipid phosphatidylserine. Science. 2019;364:57–62. doi: 10.1126/science.aav9959. [DOI] [PubMed] [Google Scholar]
- 79.Jullien L, Gautier A. Fluorogen-based reporters for fluorescence imaging: a review. Methods Appl Fluoresc. 2015;3:42007. doi: 10.1088/2050-6120/3/4/042007. [DOI] [PubMed] [Google Scholar]
- 80.Li C, Mourton A, Plamont MA, Rodrigues V, Aujard I, Volovitch M, Le Saux T, Perez F, Vriz S, Jullien L, Joliot A, Gautier A. Fluorogenic Probing of Membrane Protein Trafficking. Bioconjug Chem. 2018;29:1823–1828. doi: 10.1021/acs.bioconjchem.8b00180. [DOI] [PubMed] [Google Scholar]
- 81.Barlow LD, Dacks JB. Seeing the endomembrane system for the trees: Evolutionary analysis highlights the importance of plants as models for eukaryotic membrane-trafficking. Semin. Cell Dev Biol. 2018;80:142–152. doi: 10.1016/j.semcdb.2017.09.027. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y, Persson S, Hirst J, Robinson MS, van Damme D, Sánchez-Rodríguez C. Change your Tplate, change your fate: Plant CME and beyond. Trends in Plant Science. 2015;20:41–48. doi: 10.1016/j.tplants.2014.09.002. [DOI] [PubMed] [Google Scholar]