Abstract
Expression of programmed death-ligand 1 (PD-L1) is frequently observed in human cancers. Binding of PD-L1 to its receptor PD-1 on activated T cells inhibits anti-tumor immunity by counteracting T cell-activating signals. Antibody-based PD-1-PD-L1 inhibitors can induce durable tumor remissions in patients with diverse advanced cancers, and thus expression of PD-L1 on tumor cells and other cells in the tumor microenviroment is of major clinical relevance. Here we review the roles of the PD-1-PD-L1 axis in cancer, focusing on recent findings on the mechanisms that regulate PD-L1 expression at the transcriptional, posttranscriptional, and protein level. We place this knowledge in the context of observations in the clinic and discuss how it may inform the design of more precise and effective cancer immune checkpoint therapies.
Introduction
The T cell-based immune system has evolved to recognize and destroy aberrant cells, such as pathogen-infected cells and cancer cells. Detection of such aberrant cells occurs through binding of the T cell receptor (TCR) on T cells to peptide-major histocompatibility complexes (MHC) on target cells. However, the outcome of this interaction is to a very large extent controlled by a series of co-stimulatory and co-inhibitory receptors and their ligands (also known as immune checkpoints). By regulating the quantity and functional activity of antigen-specific T cells, these checkpoint pathways play a critical role in limiting tissue damage and maintenance of self-tolerance.
Among all immune checkpoints, the PD-L1-PD-1 pathway has stood out because of its proven value as a therapeutic target in a large number of malignancies. At present, antibodies targeting the PD-L1-PD-1 axis are being evaluated in more than 1,000 clinical trials and have been approved for cancers including melanoma, non-small cell lung cancer (NSCLC), renal cell carcinoma (RCC), Hodgkin’s lymphoma, bladder cancer, head and neck squamous cell carcinoma (HNSCC), Merkel-cell carcinoma, and microsatellite instable-high (MSI-H) or mismatch repair-defi-cient (dMMR) solid tumors. Despite the considerable improvement in patient outcome that has been achieved with PD-L1-PD-1 blockade, durable responses to these therapies are observed in only a minority of patients and intrinsic therapy resistance is common. In some tumor types, expression of PD-L1 on tumor cells and in the tumor microenvironment has been associated with clinical response, highlighting the need for a better understanding of the processes that regulate PD-L1 expression.
In this review, we first discuss the fundamental biology of the PD-L1-PD-1 immune checkpoint. We then describe the promise and limitations of current anti-PD-L1-PD-1 therapies and the relevance of PD-L1 expression in predicting clinical response. Subsequently, we cover the current understanding of the molecular mechanisms that control such PD-L1 expression. In this section we dissect the complex regulatory network that determines PD-L1 levels into five major components that involve (1) genomic aberrations, (2) inflammatory signaling, (3) oncogenic signaling, (4) microRNA-based control, and (5) posttranslational modulation. Finally, we will discuss how this knowledge may guide further research and potentially be used to design more precise and effective cancer immune checkpoint therapies.
The PD-L1-PD-1 Axis: Structure and Function
Programmed cell death protein 1 (PD-1; also called CD279) is one of the co-inhibitory receptors that is expressed on the surface of antigen-stimulated T cells (Ishida et al., 1992). PD-1 interacts with two ligands, PD-L1 (CD274) and PD-L2 (CD273). Expression of PD-L2 is observed on, for instance, macrophages, DCs, and mast cells. PD-L1 expression can be detected on hematopoietic cells including T cells, B cells, macrophages, dendritic cells (DCs), and mast cells, and non-hematopoietic healthy tissue cells including vascular endothelial cells, keratinocytes, pancreatic islet cells, astrocytes, placenta syncytiotrophoblast cells, and corneal epithelial and endothelial cells. Both PD-L1 and PD-L2 can be expressed by tumor cells and tumor stroma. Engagement of PD-L2 at such tumor sites may potentially contribute to PD-1-mediated T cell inhibition (Yearley et al., 2017). However, to date, there is no compelling evidence indicating that antibodies against PD-1, which block binding to both PD-L1 and PD-L2, show higher clinical activity than antibodies against PD-L1. These data are consistent with a model in which PD-L1 is the dominant inhibitory ligand of PD-1 on T cells in the human tumor microenvironment.
Both PD-1 and PD-L1 are type I transmembrane proteins that belong to the immunoglobulin (Ig) superfamily. PD-1 consists of an Ig-V like extracellular domain, a transmembrane domain, and a cytoplasmic domain that harbors two tyrosine-based signaling motifs (Ishida et al., 1992; Zhang et al., 2004). PD-L1 contains an Ig-V and Ig-C-like extracellular domain, a transmembrane domain, and a short cytoplasmic tail that does not contain canonical signaling motifs (Dong et al., 1999; Keir et al., 2008; Lin et al., 2008). Interactions between the extracellular domains of PD-L1 and PD-1 can induce a conformational change in PD-1 that leads to phosphorylation of the cytoplasmic immunoreceptor tyrosine-based inhibitory motif (ITIM) and the immunoreceptor tyrosine-based switch motif (ITSM) by Src family kinases (Gauen et al., 1994; Straus and Weiss, 1992; Zak et al., 2015). These phosphorylated tyrosine motifs subsequently recruit the SHP-2 and SHP-1 protein tyrosine phosphatases to attenuate T cell-activating signals. While traditionally PD-1 engagement was thought to reduce the strength of the TCR signal itself (Chemnitz et al., 2004; Sheppard et al., 2004; Ugi et al., 1994), recent work suggests that the co-stimulatory receptor CD28, rather than the TCR, may be a primary target for dephosphorylation by the SHP2 phosphatase (Hui et al., 2017). In addition to its interaction with PD-1, PD-L1 can also interact with CD80, which may deliver inhibitory signals to activated T cells (Butte et al., 2007; Park et al., 2010). Engagement of PD-1 by PD-L1 alters the activity of T cells in many ways, inhibiting T cell proliferation, survival, cytokine production, and other effector functions (Butte et al., 2007; Chang et al., 1999; Curiel et al., 2003; Dong et al., 1999; Freeman et al., 2000; Keir et al., 2006; Latchman et al., 2004).
While the effects of checkpoint signaling through PD-1 are reasonably well understood, potential “reverse signaling” through PD-L1 has been explored less, possibly because of the absence of canonical signaling motifs in the short cytoplasmic tail of PD-L1. Nevertheless, some evidence has been obtained for a signaling role of the PD-L1 molecule. First, stimulation of PD-L1 by recombinant PD-1 delivers pro-survival signals to tumor cells, resulting in resistance to Fas- or staurosporine-induced apoptosis (Azuma et al., 2008). Moreover, antibody-based PD-L1 targeting reduces mTOR activity and glycolytic metabolism in tumor cells in the absence of T cells (Chang et al., 2015). Finally, PD-L1 can protect tumor cells from the cytotoxic effects of type I and type II interferons and from cytotoxic T lymphocyte (CTL)-mediated cytolysis with no requirement of PD-1 signaling in T cells (Azuma et al., 2008; Gato-Cañas et al., 2017). While it was shown that the cytoplasmic domain of PD-L1 is critically required for this antiapoptotic signaling (Azuma et al., 2008), it remains unclear which intracellular factor(s) participate(s) in such proposed signal transduction. In spite of the above data on potential signaling through PD-L1, the current thinking on the mechanism of action of PD-1 and PD-L1 blocking agents focuses to a very large extent on their effects on the activity of PD-1-expressing T cells.
PD-1 Expression in Tolerance, Infection, and Cancer
The PD-L1-PD-1 axis is a critical determinant of physiological immune homeostasis. PD-1-deficient mice of different genetic backgrounds are prone to develop lupus-like autoimmune disease (Nishimura et al., 1999) or fatal autoimmune cardiomyopathy (Nishimura et al., 2001). Similarly, genetic or antibody-based inhibition of PD-1 accelerates diabetes onset in a non-obese diabetic (NOD) background (Ansari et al., 2003; Wang et al., 2005). Moreover, PD-1-deficient mice have altered thymic T cell education (Nishimura et al., 2000) and PD-L1 blockade has been shown to impair fetomaternal tolerance (Guleria et al., 2005). Collectively these data demonstrate a critical role for PD-1 in immune tolerance at various levels. Upon recognition of antigen, T cells rapidly express PD-1 (Agata et al., 1996), and it has been demonstrated that PD-1 expression on recently activated T cells is involved in modulating the strength of the initial T cell response (Honda et al., 2014). Consistent with a role of PD-1 in suppressing T cell activity, inhibition of the PD-1-PD-L1 axis increases immune responses toward pathogens (Erickson et al., 2012; Lázár-Molnár et al., 2008; Liu et al., 2014; Yao et al., 2009).
Chronic infection and cancer are both characterized by the continued presence of antigen-expressing target cells. Work in mouse models of chronic LCMV infection has provided compelling evidence that in such a setting of chronic antigen encounter, T cells progressively lose effector functions, developing into an exhausted (or dysfunctional) state (Wherry and Kurachi, 2015). Importantly, high expression of PD-1 is one of the defining characteristics of such exhausted T cells (Wherry and Kurachi, 2015). Furthermore, in exhausted T cells, PD-1 has a role in maintenance of the dysfunctional state, both in chronic infection (Barber et al., 2006) and in cancer (Hirano et al., 2005). In human cancers, PD-1-positive T cells in tumors (Gros et al., 2014) and in peripheral blood (Gros et al., 2016) are enriched for tumor reactivity, again consistent with antigen-driven PD-1 expression. However, PD-1 expression can also be induced by other factors, such as IL-10 (Sun et al., 2015) and TGF-b (Park et al., 2016), and “bystander T cells” in mouse tumor models do express intermediate levels of PD-1 (Erkes et al., 2017). Next to its role on T cells, recent data provide evidence for a role of PD-1 on other cell types. In one study, a case was made for a possible role of PD-1 expression on melanoma cells (Kleffel et al., 2015). In addition, blockade of the interaction between PD-1 and PD-L1 has been shown to counteract the activity of immunosuppressive PD-1hi B cells in hepatocellular carcinoma (Zhao et al., 2016). Finally, Gordon et al. (2017) have demonstrated how PD-1 blockade can enhance tumor cell phagocytosis by PD-1+ macrophages. A detailed discussion of PD-1 biology in the context of normal physiology (Sharpe and Pauken, 2018) and T cell exhaustion (Wherry and Kurachi, 2015) is provided elsewhere.
The PD-1-PD-L1 Axis as a Therapeutic Target: Clinical Value and Issues Remaining
Given their role in T cell suppression, PD-1 and PD-L1 have over the past years developed into prominent targets of antibody-based cancer therapeutics. In 2010, the first data were reported on the clinical activity of PD-1-blocking agents in patients with colorectal cancer, melanoma, and renal cell carcinoma, demonstrating an acceptable toxicity profile and providing the first evidence of antitumor activity (Brahmer et al., 2010). Subsequent studies have revealed significant activity of PD-1 blockade in a large variety of cancers, leading to clinical approval for treatment of melanoma (Larkin et al., 2015; Ribas et al., 2015; Robert et al., 2015a, 2015b; Weber et al., 2015, 2017), NSCLC (Antonia et al., 2017; Borghaei et al., 2015; Brahmer et al., 2015; Fehrenbacher et al., 2016; Herbst et al., 2016; Langer et al., 2016; Reck et al., 2016), MSI-H and dMMR cancers (Kok et al., 2017; Le et al., 2017, 2015; Overman et al., 2017), RCC (Motzer et al., 2015), Hodgkin’s lymphoma (Ansell et al., 2015; Chen et al., 2017b), urothelial carcinoma (Balar et al., 2017a, 2017b; Bellmunt et al., 2017; Patel et al., 2018; Powles et al., 2017; Rosenberg et al., 2016; Sharma et al., 2017b), HNSCC (Ferris et al., 2016; Seiwert et al., 2016), Merkel-cell carcinoma (Kaufman et al., 2016), hepatocellular carcinoma (HCC) (El-Khoueiry et al., 2017), and gastric cancer (Fuchs et al., 2017). On the other hand, in tumor types such as non-MSI colorectal cancer, prostate cancer, ovarian cancer, and breast cancer, the clinical activity of anti-PD-1-PD-L1 monotherapy is only modest. Furthermore, even in tumor types for which PD-1-PD-L1 blockade is now an approved therapy such as NSCLC, only a fraction of patients shows objective clinical responses (Table 1).
Table 1. Overview of Approved Indications for PD-1 and PD-L1 Blocking Agents.
| Tumor Type | Target Antibody | Response Rate | Notes | Reference |
|---|---|---|---|---|
| Melanoma | PD-1 - nivolumab | CR 3.3%, PR 28.3% | Checkmate-037. Previously treated, PD-L1+ IHC predictive. | Weber et al., 2015 |
| CR 4%, PR 30% | Checkmate-066. Previously untreated, BRAF wild-type, PD-L1+ IHC predictive. | Robert et al., 2015a | ||
| CR 8.9%, PR 41% | Checkmate-067. Combination with Ipilimumab. | Larkin et al., 2015 | ||
| Hazard ratio 0.65 versus ipilimumab adjuvant | Checkmate 238. Stage III and IV, adjuvant therapy after resection. PD-L1+ IHC predictive. | Weber et al., 2017 | ||
| PD-1 - pembrolizumab | CR 5-6%, PR 27-29% | Keynote-006. Ipilimumab naive. | Robert et al., 2015b | |
| CR 2-3%, PR 19-23% | Keynote-002. Ipilimumab refractory. | Ribas et al., 2015 | ||
| NSCLC | PD-1 - nivolumab | CR 0.7%, PR 19.3% | Checkmate-017. Second line squamous NSCLC, PD-L1+ IHC not predictive. | Brahmer et al., 2015 |
| CR 1.4%, PR 17.8% | Checkmate-057. Second line non-squamous NSCLC, PD-L1+ IHC predictive. | Borghaei et al., 2015 | ||
| PD-1 - pembrolizumab | CR 4%, PR 41% | Keynote-024. PD-L1 > 50%, first line. | Reck et al., 2016 | |
| Hazard ratio 0.53 versus chemotherapy alone | Keynote-021. PD-L1 > 1%, first line, combination with chemotherapy. | Langer et al., 2016 | ||
| ORR 18-19% (PD-L1 > 1%), 29-30% (PD-L1 > 50%) | Keynote-010. PD-L1 > 1%. Previously treated. | Herbst et al., 2016 | ||
| PD-L1 - atezolizumab | OS 12.6 m versus 9.7 m in chemotherapy arm | POPLAR and OAK. Previously treated. PD-L1+ IHC on tumor and IC predictive. | Fehrenbacher et al., 2016 | |
| PD-L1 - durvalumab | Hazard ratio 0.52 versus placebo | PACIFIC. Stage III, durvalumab after chemotherapy. PD-L1+ IHC not predictive. | Antonia et al., 2017 | |
| RCC | PD-1 - nivolumab | ORR 21.5% | Checkmate-025. Progressive patients on or after anti-angiogenic therapies. PD-L1+ IHC not predictive. | Motzer et al., 2015 |
| HNSCC | PD-1 - nivolumab | OS 7.5 m versus 5.1 m for investigator’s choice | Checkmate-141. Patients progressing after platinum-based chemotherapy. PD-L1+ IHC predictive. | Ferris et al., 2016 |
| PD-1 - pembrolizumab | CR 5%, PR 11% | Keynote-012. Patients recurrent on or after chemotherapy. PD-L1+ IHC and IFN-γ-related gene expression predictive. | Seiwert et al., 2016 | |
| Urothelial carcinoma | PD-1 - nivolumab | CR 2.6%, PR 17% | Checkmate-275. Patients progressing after chemotherapy. | Sharma et al., 2017b |
| PD-1 - pembrolizumab | ORR 21% | Keynote-045. Patients progressing after chemotherapy. PD-L1+ IHC not predictive. | Bellmunt et al., 2017 | |
| ORR 28.6% | Keynote-052. Cisplatin-ineligible patients. PD-L1+ IHC predictive. | Balar et al., 2017a | ||
| PD-L1 - atezolizumab | CR 6.7%, PR 16.8% | NCT02951767. Cisplatin-ineligible patients. | Balar et al., 2017b | |
| CR 5.5%, PR 9.4% | NCT02108652. Chemotherapy refractory. PD-L1+ IHC on IC predictive. | Rosenberg et al., 2016 | ||
| PD-L1 - avelumab | CR 5.6%, PR 10.6% (at 6 months follow-up) | JAVELIN solid tumors. Patients failing platinum-based chemotherapy. Weak predictive value for PD-L1+ IHC. | Patel et al., 2018 | |
| PD-L1 - durvalumab | CR 2.7%, PR 14.3% | NCT01693562. Patients that are progressive, ineligible, or have refused platinum-based chemotherapy. PD-L1+ IHC on tumor and IC predictive. | Powles et al., 2017 | |
| Merkel cell carcinoma | PD-L1 - avelumab | CR 11.4%, PR 21.6% | JAVELIN Merkel 200. PD-L1 expression and polyomavirus status not predictive. | Kaufman et al., 2016 |
| Hodgkin’s lymphoma | PD-1 - nivolumab | CR 14%, PR 55% | Checkmate-039 and Checkmate-205. Patients progressing after autologous HSCT and brentuximab vedotin. | Ansell et al., 2015 |
| PD-1 - pembrolizumab | CR 22%, PR 47% | Keynote-087. Relapsed or refractory Hodgkin’s lymphoma. | Chen et al., 2017b | |
| MSI-H and dMMR CRC | PD-1 - nivolumab | CR 2.7%, PR 30% | Checkmate-142. Patients progressing after or ineligible for chemotherapy. | Overman et al., 2017 |
| MSI-H and dMMR solid tumors | PD-1 - pembrolizumab | ORR 39.6% | NCT-01876511. Patients with no satisfactory alternative treatment options. | Le et al., 2015, 2017 |
| HCC | PD-1 - nivolumab | ORR 14.3% | Checkmate-040. Patients progressing after sorafenib. PD-L1+ IHC not predictive. | El-Khoueiry et al., 2017 |
| Gastric cancer | PD-1 - pembrolizumab | Fuchs et al., 2017 | ORR 13.3% in PD-L1 positive | Keynote-059. Disease progression after at least two prior lines of therapy. |
Clinical trials leading to approval, patient subgroups, and predictive value or lack of predictive value of intratumoral PD-L1 expression are indicated where applicable. Abbreviations are as follows: NSCLC, non-small cell lung cancer; RCC, renal cell carcinoma; HNSCC, head and neck squamous cell carcinoma; MSI-H, microsatellite instability high; dMMR, mismatch repair deficient; CRC, colorectal cancer; HCC, hepatocellular carcinoma; CR, complete response; PR, partial response; ORR, overall response rate; OS, overall survival; IHC, immunohistochemistry; IC, immune cells.
The observed heterogeneity in clinical response to PD-1-PD-L1 blockade has led to a major effort to understand which parameters predict clinical response to these therapies. From a conceptual point of view, this question can be divided into two smaller questions. First, is there evidence that the PD-1-PD-L1 inhibitory axis is active in a given tumor? Second, for those tumors in which the PD-1-PD-L1 axis is operational, would blockade of this inhibitory interaction suffice to re-activate the process of tumor cell killing? With respect to the second question, tumor control by T cells is critically dependent on a series of distinct parameters that can be summarized in a “cancer immunogram” that depicts the state of cancer-immune interaction in an individual patient (Blank et al., 2016). In support of this model, the outcome of PD-L1-PD-1 blockade has now been shown to be influenced not only by PD-L1 expression (as discussed in the next section), but also by a series of unrelated parameters that include (1) the “foreignness” of the tumor, (2) the immune status of the patient, (3) the presence and activity of the intratumoral T cell infiltrate, (4) the presence of other inhibitory processes within the tumor, and (5) the sensitivity of tumor cells to tumor-specific T cells at that site.
With respect to the role of tumor foreignness, in most (Goodman et al., 2017; Rizvi et al., 2015; Rosenberg et al., 2016) but not all (Hugo et al., 2016) studies, the number of non-synonymous DNA mutations in human tumors has been shown to correlate with the likelihood of response to PD-1 axis blocking therapies, consistent with the view that the amount of neoantigens influences the likelihood and/or strength of T cell recognition (Schumacher and Schreiber, 2015). While this correlation has now been described for a series of tumor types, the most compelling evidence for a role of “tumor foreignness” has come from the demonstration that MSI-H tumors, which carry an uncommonly high mutational load, respond particularly well to inhibition of the PD-L1-PD-1 pathway as compared to other tumors of the same histology (Kok et al., 2017; Le et al., 2017, 2015).
Several parameters that reflect the general immune status of patients, such as absolute lymphocyte count (ALC), neutrophil to lymphocyte ratio (NLR), and absolute eosinophil count, are also being considered as predictive biomarkers for response to PD-1-PD-L1 blocking therapies. These parameters have previously shown value in predicting response to CTLA-4 blockade. To date, only neutrophil to lymphocyte ratio has shown predictive value in one study that examined response to PD-1-PD-L1 blockade (Bumma et al., 2017). Recently, the frequency of circulating monocytes has also been described as a systemic biomarker to predict response to anti-PD-1 immunotherapy (Krieg et al., 2018).
As a third category of predictive markers, assays that measure accumulation of (clonally expanded) T cells and the activity of such T cells at tumor sites have been shown to be informative. As specific examples, the presence of CD8-positive cells at the tumor invasive margin (Tumeh et al., 2014), clonality of the intratumoral TCR repertoire (Snyder et al., 2017; Tumeh et al., 2014), and a transcriptional signature in which interferon (IFN)-γ-response genes are included (Ayers et al., 2017; Fehrenbacher et al., 2016; Seiwert et al., 2016) all form positive predictive markers. In addition to providing information on intratumoral T cell activity, RNA-seq data have also been utilized to derive a specific transcriptional profile that correlates with lack of response to anti-PD-1 therapies with some of the genes that show increased expression in non-responders, such as IL-10, known to be immunosuppressive (Hugo et al., 2016).
Finally, two classes of predictive markers may be found at the level of tumor cell recognition and tumor cell sensitivity to T cell attack. Specifically, defects in the antigen presentation machinery have been identified in various malignancies (Seliger, 2014). Such defects could lead to both intrinsic and acquired resistance to PD-1-PD-L1 inhibition, and evidence for the latter has already been obtained (Sade-Feldman et al., 2017; Zaretsky et al., 2016). MSI-H tumors should be a useful testing ground to further understand the clinical impact of such alterations, as the beta-2 micro-globulin (B2M) gene, a critical component of the HLA class I antigen presentation pathway, contains microsatellites that are prone to mutation in this class of tumors (Yamamoto et al., 2001). Interestingly, also defects in the IFN-γ receptor pathway have been identified in patients that have developed resistance to PD-1 blocking therapies (Sade-Feldman et al., 2017; Zaretsky et al., 2016). Consistent with this observation, genetic screens have identified inhibition and inactivation of the IFN-γ receptor pathway as a mechanism of resistance of tumor cells to T cell attack, both in vitro (Patel et al., 2017) and in vivo (Manguso et al., 2017). Whether the observed role of the IFN-γ receptor pathway primarily reflects its effect on expression of components of the antigen presentation machinery, or direct cytotoxic or cytostatic effects of IFN-γ receptor signaling in tumor cells, remains to be established.
The Role of PD-L1 Expression in Immunotherapy Response
Expression of PD-L1 within the tumor microenvironment has predictive value for response to monotherapies blocking the PD-L1-PD-1 axis in many studies in melanoma (Robert et al., 2015a; Weber et al., 2017, 2015), NSCLC (Borghaei et al., 2015; Fehrenbacher et al., 2016), and bladder cancer (Rosenberg et al., 2016), but was not predictive in all studies (Motzer et al., 2015; Sharma et al., 2017b). Of note, in certain tumor types, such as bladder cancer (Rosenberg et al., 2016), predictive value is primarily observed for PD-L1 expression on immune infiltrating cells rather than tumor cells, whereas for NSCLC, PD-L1 expression on both immune cells and tumor cells has shown predictive value (Fehrenbacher et al., 2016). These observations have encouraged experimental research to dissect which of the two cell compartments is more relevant in PD-L1-mediated suppression of tumor control. In a first study that aimed to address this issue, Noguchi et al. (2017) took advantage of a murine transplantable sarcoma system in which tumor cells fail to grow in mice treated with PD-1 axis blocking agents. In this setting, selective disruption of PD-L1 on tumor cells resulted in tumor control in most, but not all, animals. Of note, those PD-L1-negative tumors that escaped immune elimination were still sensitive to antibody-mediated PD-L1 blockade, indicating that PD-L1 expression on both cell compartments can play a role under these experimental conditions (Noguchi et al., 2017). In another study, Lau et al. (2017) transplanted either PD-L1-proficient or -deficient tumor cells in either wild-type or PD-L1-deficient mice and observed that absence of PD-L1 from either of the two cell compartments impaired tumor growth, with optimal tumor control being observed when PD-L1 was absent from both cell compartments. In a third report, Juneja et al. (2017) used a similar strategy and implanted either highly immunogenic or mildly immunogenic PD-L1-proficient tumors in either wild-type, PD-L1 plus PD-L2-deficient, or PD-1-deficient mice to dissect once again the role of PD-L1 on the different cell compartments. In these experiments, PD-L1 on non-tumor cells played a dispensable role in case of MC38, the most immunogenic cell line. In contrast, in case of the less immunogenic cell lines, PD-L1 expression on both the tumor and non-tumor compartment contributed to tumor growth (Juneja et al., 2017). Collectively, these data indicate that PD-L1 expression on both cell compartments can play a role, but that the relative importance of both cell compartments is context dependent. In one of these reports (Juneja et al., 2017), the most immunogenic tumor did not rely on non-autonomous PD-L1 expression, but whether this reflects a causal link between immunogenicity and PD-L1 dependency on different cell compartments has not been assessed. Another parameter that is likely to influence the relative role of PD-L1 on tumor cells and stromal cells is its relative expression level on the two cell compartments. In a recent report, in which loss of the PD-L1 3’ UTR was shown to increase the levels of PD-L1, it was observed that tumor cells with increased PD-L1 protein have a greater suppressive capacity in in vitro and in vivo assays (Kataoka et al., 2016). Consistent with these data, overexpression of PD-L1 on tumor cells has been shown to prevent rejection of two highly immunogenic murine cancer cell lines in immunocompetent hosts (Noguchi et al., 2017). Other contextual factors that should be assessed in future work are the relative abundance of PD-L1+ tumor cells and PD-L1+ stromal cells, the specific type(s) of stromal cells that express the PD-L1 protein, and the role that additional suppressive pathways may play.
With the very rapid increase in clinical studies that evaluate combination immunotherapies, it will be important to also understand the value of PD-L1 as a predictive marker in those settings. In melanoma, combination therapy with anti-PD-1 and anti-CTLA-4 results in a numerical increase in progression-free survival relative to anti-PD-1 monotherapy. Strikingly, the added value of CTLA-4 blockade in this study appeared restricted to those patients with tumors that were PD-L1 negative before start of treatment (Larkin et al., 2015), in line with the model that CTLA-4 blocking therapies broaden the tumor-specific T cell response (Kvistborg et al., 2014). Conceivably, increased tumor-specific T cell reactivity induced by CTLA-4 blockade could in this setting lead to IFN-γ-induced PD-L1 expression, at which point blockade of the PD-1-PD-L1 axis would also become valuable.
In spite of the value of PD-L1 staining as a predictive marker in different tumor types, many PD-L1-positive tumors do not respond, and a fraction of PD-L1-negative tumors do respond to PD-L1-PD-1 blockade (Table 2). Clinical response of seemingly PD-L1-negative tumors may be explained by technical issues (e.g., inadequate sampling of the tumor, or insufficient sensitivity of the detection technique that is used), by a potential role of the PD-L1-PD-1 axis outside of the tumor microenvironment, by the dynamic nature of PD-L1 expression, or by immune cell inhibition through PD-L2. Conversely, absence of clinical response in PDL1-positive tumors may be explained by mechanisms described in the prior section, such as loss of tumor cell sensitivity to T cell effector mechanisms, or the presence of additional immunosuppressive mechanisms in the tumor microenvironment, such as TGF-b (Mariathasan et al., 2018). As an alternative explanation, absence of response in PD-L1-positive tumors may occur in settings in which the observed PD-L1 expression is not due to an ongoing immune response but is driven by other mechanisms.
Table 2. Potential Explanations for Discrepencies between Clinical Response to PD-1-PD-L1 Blocking Therapies and Intratumoral PDL1 Expression.
| Clinical Response | ||
|---|---|---|
| PD-L1 expression | Non responding | Responding |
| PD-L1 negative | false negative in assays role of PD-L1 outside of the tumor microenvironment dynamic nature of PD-L1 expression role of PD-L2 | |
| PD-L1 positive | other suppressive mechanism(s) tumor insensitivity to T cells expression of PD-L1 not due to an ongoing immune response |
Given the emerging evidence indicating how PD-L1 levels on different intratumoral cell compartments can influence tumor control and predict treatment response, it is critical to understand how the levels of PD-L1 expression at the cell surface are regulated, and also how this regulation may vary between tumor cells and (different) immune cells. In the next sections, we will discuss the data demonstrating that PD-L1 levels in tumors are regulated in a highly complex manner, with expression being influenced by genomic aberrations, transcriptional control, mRNA stability, oncogenic signaling, and protein stability (Figure 1, Table 3).
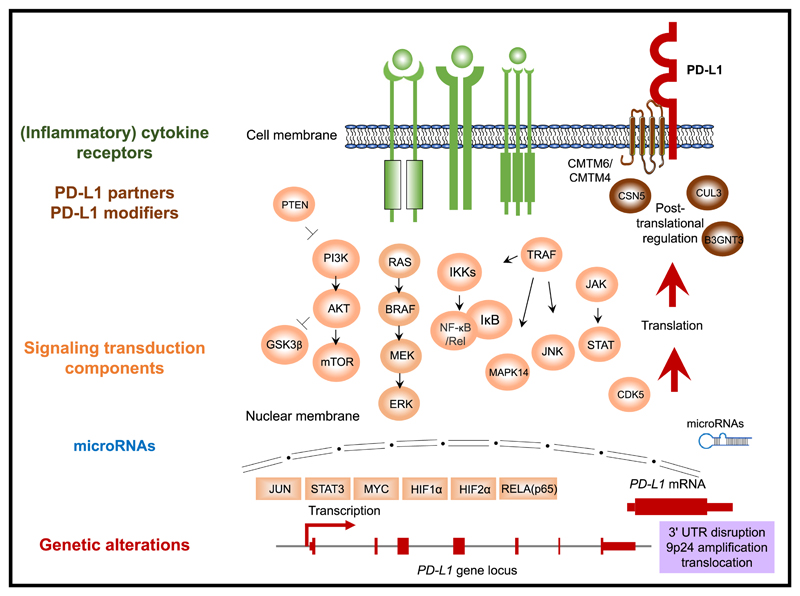
Figure 1. Overview of the Regulatory Mechanisms of PD-L1 Expression.
Regulators are color coded by class. Green, (inflammatory) cytokine receptors; orange, signal transduction components; blue, microRNAs; red, genetic alterations; brown, PD-L1 protein partners and modifiers. Directionality (“up” or “down”) and references are provided in Table 3.
Table 3. Regulators of PD-L1.
| Type | Regulators of PD-L1 | Tissue type |
|---|---|---|
| Inflammatory signaling | IFN-γ | ↑ multiple tumor types (Dong et al., 2002), endothelial cells (Mazanet and Hughes, 2002), glioma (Wintterle et al., 2003), renal tubular epithelial cells (Schoop et al., 2004), colon cancer (Nakazawa et al., 2004), monocytes (Brown et al., 2003), dendritic cells and macrophages (Brown et al., 2003; Kryczek et al., 2008), neutrophils (de Kleijn et al., 2013) |
| IFN-α and IFN-α | ↑ endothelial cells (Eppihimer et al., 2002), monocytes and dendritic cells (Schreiner et al., 2004), melanoma (Garcia-Diaz et al., 2017) | |
| TLR4 | ↑ macrophages (Loke and Allison, 2003), bladder cancer (Qian et al., 2008), monocytes (Huang et al., 2013), dendritic cells (Mezzadra et al., 2017) | |
| TLR3 | ↑ dendritic cells (Pulko et al., 2009), endothelial cells (Cole et al., 2011), neuroblastoma (Boes and Meyer-Wentrup, 2015) | |
| IL-17 | ↑ monocytes (Zhao et al., 2011), prostate and colon cancer (Wang et al., 2017b) | |
| IL-10 | ↑ dendritic cells (Curiel et al., 2003), monocytes (Zhao et al., 2011) | |
| TNF-α | ↑ endothelial cells (Mazanet and Hughes, 2002), myelodysplastic syndrome blasts (Kondo et al., 2010), dendritic cells (Karakhanova et al., 2010), monocytes (Zhao et al., 2011), monocytes (Ou et al., 2012), RCC (Quandt et al., 2014), breast cancer (Lim et al., 2016), prostate and colon cancer (Wang et al., 2017b) | |
| IL-4 | ↑ RCC (Quandt et al., 2014) | |
| IL-1β | ↑ dendritic cells (Karakhanova et al., 2010) | |
| IL-6 | ↑ dendritic cells (Karakhanova et al., 2010; Kil et al., 2017) | |
| IL-27 | ↑ dendritic cells (Karakhanova et al., 2011; Matta et al., 2012), ovarian cancer (Carbotti et al., 2015) | |
| TGF-β | ↑ dendritic cells (Ni et al., 2012; Song et al., 2014), T cells (Baas et al., 2016) ↓ renal tubular epithelial cells (Starke et al., 2007), monocytes (Ou et al., 2012) |
|
| IL-12 | ↑ endothelial cells (Eppihimer et al., 2002),
monocyte-derived macrophages (Xiong et
al., 2014) ↓ THP-1-derived macrophages (Xiong et al., 2014) |
|
| Oncogenic signaling | MYC | ↑ melanoma, NSCLC, HCC, osteosarcoma, leukemia and (mouse) lymphoma (Casey et al., 2016; Atsaves et al., 2017; Wang et al., 2017a) |
| HIF1α | ↑ breast cancer, prostate cancer (Barsoum et al., 2014), mouse myeloid cells (Noman et al., 2014), NSCLC (Guo et al., 2017) | |
| HIF2α | ↑ RCC (Messai et al., 2016) | |
| STAT3 | ↑ lymphoma (Marzec et al., 2008; Atsaves et al., 2017), melanoma (Jiang et al., 2013), HNSCC (Bu et al., 2017) | |
| RELA | ↑ primary monocytes (Huang et al., 2013), melanoma (Gowrishankar et al., 2015), NSCLC (Bouillez et al., 2017), breast cancer (Xue et al., 2017) | |
| MUC1-C | ↑ NSCLC (Bouillez et al., 2017) | |
| JUN | ↑ lymphoma (Green et al., 2012), melanoma (Jiang et al., 2013) | |
| CDK5 | ↑ medulloblastoma (Dorand et al., 2016) | |
| (PTEN loss) PI3K-AKT-mTOR | ↑ glioma (Parsa et al., 2007), dendritic cells (Karakhanova et al., 2010), monocytes (Muthumani et al., 2011), CRC (Song et al., 2013), melanoma (Jiang et al., 2013; Atefi et al., 2014; Gowrishankar et al., 2015), NSCLC (Xu et al., 2014; Lastwika et al., 2016; Zhang et al., 2017b), breast cancer (Mittendorf et al., 2014; Yang et al., 2017) | |
| MEK-ERK | ↑ multiple myeloma (Liu et al., 2007), NSCLC (Sumimoto et al., 2016), melanoma
(Jiang et al., 2013; Liu et al., 2015), bladder cancer
(Qian et al., 2008),
lymphoma (Yamamoto et al.,
2009), dendritic cells (Karakhanova et al., 2010) ↓ melanoma (Atefi et al., 2014), mouse breast cancer (Loi et al., 2016), NSCLC (Liu et al., 2015) |
|
| MAPK14 | ↑ multiple myeloma (Liu et al., 2007), bladder cancer (Qian et al., 2008), lymphoma (Yamamoto et al., 2009), dendritic cells (Karakhanova et al., 2010) | |
| RAS | ↑ NSCLC (Sumimoto et al., 2016; Coelho et al., 2017; Chen et al., 2017a; Guo et al., 2017), mouse lung (Lastwika et al., 2016), bronchial epithelial cells (Chen et al., 2017a) | |
| EGFR | ↑ NSCLC (Akbay et al., 2013; Chen et al., 2015; Lin et al., 2015; Zhang et al., 2016; Lastwika et al., 2016; Coelho et al., 2017), bronchial epithelial cells (Akbay et al., 2013), breast cancer (Li et al., 2016), head and neck cancer (Concha-Benavente et al., 2016) | |
| ALK | ↑ lymphoma (Marzec et al., 2008; Yamamoto et al., 2009), NSCLC (Ota et al., 2015; Koh et al., 2015) | |
| microRNA | miR-513 | ↓ cholangiocytes (Gong et al., 2009; Gong et al., 2010) |
| miR-155 | ↓ dermal lymphatic endothelial cells (Yee et al., 2017) | |
| miR-34a | ↓ AML (Wang et al., 2015), NSCLC (Cortez et al., 2015) | |
| miR-142-5p | ↓ pancreatic cancer (Jia et al., 2017) | |
| miR-93, miR-106b | ↓ pancreatic cancer (Cioffi et al., 2017) | |
| miR-138-5p | ↓ CRC (Zhao et al., 2016) | |
| miR-217 | ↓ laryngeal cancer (Miao et al., 2017) | |
| miR-200 | ↓ NSCLC (Chen et al., 2014; Gibbons et al., 2014), gastric cancer (Xie et al., 2017) | |
| miR-152 | ↓ gastric cancer (Xie et al., 2017) | |
| miR-570 | ↓ gastric cancer (Wang et al., 2013) | |
| miR-17-5p | ↓ melanoma (Audrito et al., 2017) | |
| miR-15a, miR-193a, miR-16 | ↓ malignant pleural mesothelioma (Kao et al., 2017) | |
| miR-20, miR-21, miR-130b | ↑ CRC (Zhu et al., 2014) | |
| miR-197 | ↓ NSCLC (Fujita et al., 2015) | |
| Genetic alteration | amplifications | ↑ Hodgkin’s lymphoma (Green et al., 2010; Roemer et al., 2016), B cell lymphoma (Green et al., 2010; Twa et al., 2014), NSCLC (Ikeda et al., 2016), SCLC (George et al., 2017), squamous cell carcinoma of the oral cavity (Straub et al., 2016), stomach cancer (Cancer Genome Atlas Research Network, 2014) |
| translocations | ↑ mediastinal large B cell lymphoma (Twa et al., 2014) | |
| disruption of 3’ UTR region | ↑ multiple tumor types, e.g., T cell leukemia, T and B cell lymphoma, stomach adenocarcinoma (Kataoka et al., 2016) | |
| Post-translational regulation | CMTM6 | ↑ melanoma, NSCLC, CRC, thyroid cancer, dendritic cells, CML-derived cells, pancreatic cancer, breast cancer (Burr et al., 2017; Mezzadra et al., 2017) |
| CMTM4 | ↑ NSCLC, melanoma, CML-derived cells (Mezzadra et al., 2017) | |
| CDK4 | ↓ cervical cancer, breast cancer cells (Zhang et al., 2017a) | |
| GSK3B | ↓ breast cancer (Li et al., 2016) | |
| B3GNT3 | ↑ breast cancer (Li et al., 2018) | |
| CSN5 | ↑ breast cancer (Lim et al., 2016) |
Up and down arrows are used to annotate positive and negative regulators of PD-L1s, respectively.
Genetic Alterations at the PD-L1 Locus
The two ligands of PD-1, PD-L1 and PD-L2, are located on chromosome 9p24.1, only 42 kilobases apart. Over the past years, both amplifications and translocations have been implicated in increasing the levels of PD-L1 in several types of tumors. 9p copy number amplifications were initially shown to occur in cases of Hodgkin’s lymphoma and mediastinal large B cell lymphoma and were reported to correlate with increased expression of PD-L1 and PD-L2. Interestingly, janus kinase 2 (JAK2), which is involved in transduction of pro-inflammatory signals known to modulate PD-L1 expression (see below), is also located on chromosome 9p and its amplification was suggested to contribute to increased PD-L1 expression by enhancing IFN-γ receptor signaling (Green et al., 2010). In a subsequent study that analyzed 571 cases of mediastinal large B cell lymphoma, genetic aberrations were also observed frequently, with translocations being identified in 20% of the cases and amplification in 29% of the cases (Twa et al., 2014). Furthermore, in-depth analysis of a larger cohort of Hodgkin’s lymphomas showed alterations of the PD-L1 PD-L2 locus in 97% of the cases tested (Roemer et al., 2016). In both of these studies, samples harboring genomic aberrations show increased PD-L1 and PD-L2 expression (Roemer et al., 2016; Twa et al., 2014). Evidence for a functional role of locus amplification in pathogenesis is given by the fact that Hodgkin’s lymphomas respond particularly well to PD-1 blocking agents (Ansell et al., 2015; Armand et al., 2016; Chen et al., 2017b; Younes et al., 2016). It should be noted, though, that these data do not provide conclusive evidence for positive selection of PD-L1, as co-amplification of PD-L2 is generally observed and may also explain the activity of anti-PD-1 therapy. Amplification of the chromosomal portion containing PD-L1, PD-L2, and JAK2 has also been reported in cases of NSCLC, in which they were associated with elevated PD-L1 expression (Ikeda et al., 2016). Similarly, amplifications of 9p24 and focal amplifications of PD-L1 have been described in a small subset of small cell lung cancer patients (George et al., 2017). PD-L1 amplification has likewise been observed in cases of squamous cell carcinoma of the oral cavity (Straub et al., 2016) and in Epstein-Barr virus (EBV)-positive gastric cancer (Cancer Genome Atlas Research Network, 2014).
A series of reports has provided compelling evidence that PD-L1 protein levels are also regulated by microRNAs (miRNA) that bind to the 3’ UTR of the PD-L1 mRNA (reviewed below). In line with a regulatory role of the 3’ UTR of the PD-L1 gene, loss of this gene segment as a consequence of different structural variations has been described in a small fraction of human tumors of diverse histology and correlates with increased PD-L1 expression of those tumors. Furthermore, deletion of the 3’ UTR of the PD-L1 gene by CRISPR Cas9 results in increased PD-L1 mRNA stability in human and murine cells, thereby increasing their resistance to T cell attack in vitro and in vivo (Kataoka et al., 2016). Likewise, in a genetic screen for factors that modulate PD-L1 expression, gene trap vector integrations that result in loss of the 3’ UTR of the PD-L1 gene were enriched in cells with high PD-L1 levels (Mezzadra et al., 2017).
To date, whether tumors with chromosomal aberrations that result in higher PD-L1 expression show an increased response rate to PD-L1-PD-1 blocking agents has not been systematically explored. However, an ovarian cancer patient harboring disruption of the 3’ UTR of PD-L1 was reported as an exceptional responder to PD-1 blockade (Bellone et al., 2018), and chromosomal aberrations of the locus should in future work be explored as a potential biomarker.
Control of PD-L1 Gene Expression through Inflammatory Signaling
Expression of the PD-L1 gene has been shown to be controlled by inflammatory signaling, consistent with the physiological role of the PD-1-PD-L1 axis in suppressing T cell activation. A number of soluble factors produced by immune cells have over the past years been described as inducers of PD-L1. In one of the first reports indicating that PD-L1 could be employed by tumor cells as a defense mechanism against T cell attack (Dong et al., 2002), regulation of PD-L1 by IFN-γ was described for various tumor types, healthy tissues, and immune cells, and this has been extended in further studies (Brown et al., 2003; de Kleijn et al., 2013; Kryczek et al., 2008; Mazanet and Hughes, 2002; Nakazawa et al., 2004; Schoop et al., 2004; Wintterle et al., 2003). IFN-γ is a pro-inflammatory cytokine that is abundantly produced by T cells upon activation and that is also produced by NK cells. Binding of IFN-γ to its receptor results in signaling through the classical JAK-STAT pathway, preferentially through STAT1, thereby inducing increased expression of a series of transcription factors, called the interferon-responsive factors (IRFs) (Platanias, 2005). Of those factors, IRF1 has been shown to play a central role in the IFN-γ-mediated induction of PD-L1 (Lee et al., 2006). IFN-γ is generally considered the most prominent soluble inducer of PD-L1, and expression of PD-L1 may therefore be viewed as a crude measure of local IFN-γ signaling and T cell activity in most settings. It is important to realize though that the effects of IFN-γ on PD-L1 expression can be context dependent. As an example, some evidence has been obtained suggesting that PD-L1 expression on tumor and immune cells can be differentially regulated. In a sarcoma mouse model, treatment of mice with IFN-γ-blocking antibodies largely abrogated PD-L1 expression on tumor cells but only partially reduced PD-L1 levels on tumorassociated macrophages (Noguchi et al., 2017). Importantly, mutations in JAK1 and JAK2 have been described in tumors that were unresponsive to PD-1 blockade despite a high number of somatic mutations. It is plausible that such tumors do not rely on the PD-L1 pathway to escape immune recognition, but have evolved alternative strategies (Shin et al., 2017).
Next to IFN-γ, type I interferons, i.e., IFN-α and IFN-β, can also induce PD-L1 expression in cultured melanoma cells, endothelial cells, monocytes, and dendritic cells (Eppihimer et al., 2002; Garcia-Diaz et al., 2017; Schreiner et al., 2004). Interestingly, exposure to type I interferons has a much greater effect on PD-L2 than on PD-L1 expression in melanoma cells, highlighting their differential regulation. This study systemically interrogated other signaling pathways that may affect IFN signaling in melanoma, and identified that silencing of MAK14, CRKL, and PI3K greatly impairs IFN-γ-mediated PD-L1 expression in these tumor cell types (Garcia-Diaz et al., 2017).
Interferons are not the only inflammatory stimuli that have been linked to PD-L1 expression. For instance, treatment of macrophages (Loke and Allison, 2003), monocytes (Huang et al., 2013), primary bone marrow-derived dendritic cells (Mezzadra et al., 2017), and bladder cancer cells (Qian et al., 2008) with lipopolysaccharide (LPS) leads to increased PD-L1 expression. LPS signals through toll-like receptor (TLR) 4, and the subsequent induction of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) results in induction of type I interferons (Lu et al., 2008), possibly explaining the link between LPS exposure and PD-L1 expression. In addition, it has been demonstrated that inhibition of the NF-kB pathway impairs IFN-γ-mediated induction of PD-L1 in melanoma cells in a context in which NF-kB was not directly stimulated, suggesting a crosstalk between the two pathways (Gowrishankar et al., 2015). Similarly, stimulation of TLR3 with polyinosinic-polycytidylic acid (poly(I:C)) was shown to result in increased PD-L1 expression on dendritic cells (Pulko et al., 2009), endothelial cells (Cole et al., 2011), and neuroblastoma cells (Boes and Meyer-Wentrup, 2015). Whether this effect is directly due to TLR3 triggering or is mediated by an autocrine or paracrine factor that is produced upon TLR3 stimulation was not established. Other inflammatory stimuli that have been shown to induce PD-L1 are IL-17 in monocytes (Zhao et al., 2011) and prostate and colon cancer cells (Wang et al., 2017b), IL-10 in dendritic cells and monocytes (Curiel et al., 2003; Zhao et al., 2011), TNF-a in endothelial cells (Mazanet and Hughes, 2002), myelodysplastic syndrome blast cells (Kondo et al., 2010), dendritic cells, monocytes (Ou et al., 2012; Zhao et al., 2011), RCC (Quandt et al., 2014), prostate, breast cancer (Lim et al., 2016), and also colon cancer cells (Wang et al., 2017b), IL-4 in RCC (Quandt et al., 2014), IL-1b in dendritic cells (Karakhanova et al., 2010), IL-6 in dendritic cells (Karakhanova et al., 2010; Kil et al., 2017), and IL-27 in dendritic cells (Karakhanova et al., 2011; Matta et al., 2012) and ovarian cancer cells (Carbotti et al., 2015). Finally, TGF-b, a molecule that is in general considered an anti-inflammatory cytokine, appears to have a context-dependent effect on PD-L1 expression. Specifically, whereas exposure of cultured monocytes (Ou et al., 2012) or tubular epithelial cells (Starke et al., 2007) to TGF-b represses PD-L1 expression, production of TGF-b by CD8 T cells in an in vivo model of pancreatic islet transplantation was shown to be required for sustained expression of PD-L1 by the same cells (Baas et al., 2016), in line with the observation that TGF-b induces PD-L1 expression in dendritic cells in vitro (Ni et al., 2012; Song et al., 2014). Similarly, IL-12-mediated PD-L1 regulation is also context dependent, as it positively regulates PD-L1 in endothelial cells and monocyte-derived macrophages but represses its expression in macrophages derived from the monocytic cell line THP-1 (Xiong et al., 2014). While the data above indicate that PD-L1 expression can be regulated by a substantial number of inflammatory mediators, in many cases it is unclear whether such induction occurs in an indirect manner, for instance via regulation of IFN production. In addition, more data are required to establish (1) in which cell types these different stimuli do or do not alter PDL1 levels, (2) whether such regulation is also observed in vivo, and (3) what the functional consequences of the altered PD-L1 expression are on local T cell function.
Aberrant Oncogenic Signaling Influences PD-L1 Expression
Alterations in major signaling pathways that control cell survival, proliferation, and differentiation constitute key events in cancer development and maintenance (Hanahan and Weinberg, 2011). In addition to fueling cancer progression in a tumor cell-intrinsic manner, certain oncogenic signaling pathways may also contribute to tumor outgrowth by driving PD-L1 expression, thereby potentially contributing to immune evasion. Findings in this area are of relevance not only because they can highlight mechanisms of PD-L1 gene (dys)regulation, but also because they may offer hypotheses on combinations of immune checkpoint therapies and therapies targeting oncogenic signaling pathways that could be tested clinically. Evidence obtained over the past years demonstrates that oncogenic signaling stemming from altered activity of basal transcription factors, effector components of signaling pathways, and upstream receptors can all influence PD-L1 expression.
A number of oncogenic transcription factors have been identified that directly regulate PD-L1 transcription. Elevated expression of the MYC oncogene contributes to tumorigenesis and is seen in approximately 70% of human cancers (Dang, 2012). Genetic or pharmacological inactivation of MYC has been shown to result in reduced PD-L1 expression in multiple mouse and human tumor cell models including melanoma, NSCLC, leukemia, lymphoma, and HCC (Atsaves et al., 2017; Casey et al., 2016; Wang et al., 2017a). In addition, chromatin immunoprecipitation (ChIP) analysis revealed that MYC binds to the PD-L1 promoter (Casey et al., 2016), indicating that PD-L1 expression can be directly regulated by MYC at the transcriptional level. Consistent with these data, it has been reported that MYC expression positively correlates with PD-L1 expression in NSCLC (Kim et al., 2017). Hypoxia represents a key feature of most tumors, as the expanding tumor mass often outgrows the oxygen supply. Tumors respond to this oxygen deficiency by activating a series of hypoxia-inducible factors (HIFs). As a result of HIF activation, a transcriptional program that promotes angiogenesis and tumor metastasis is initiated (Brown and Wilson, 2004; Liao et al., 2007). In addition, this response can result in increased PD-L1 expression. Specifically, both HIF-1a and HIF-2a have been shown to physically interact with the hypoxia response element (HRE) in the promoter region of PD-L1 (Barsoum et al., 2014; Messai et al., 2016). PD-L1 expression has been shown to be regulated by HIF-1a in mouse melanoma, human breast cancer, prostate cancer, NSCLC cells, and myeloid-derived suppressor cells (MDSCs) (Barsoum et al., 2014; Guo et al., 2017; Koh et al., 2015; Noman et al., 2014), and by HIF-2a in renal cell carcinoma cells (Messai et al., 2016), as shown by perturbation of gene expression. STAT3 is a transcription factor that promotes cell proliferation, metastasis, and survival. It acts downstream of a variety of growth factors and cytokines and is found to be constitutively active in various cancers (Buettner et al., 2002). Multiple studies indicate that active STAT3 can directly act on the promoter of PD-L1 to increase its expression in human lymphoma and HNSCC cells (Atsaves et al., 2017; Bu et al., 2017; Marzec et al., 2008). Similarly, NF-kB, a family of transcription factors, can be activated in cancers by oncogenic mutations and cytokines produced in the inflammatory microenvironment. It has been reported that inhibition of the NF-kB pathway leads to decreased PD-L1 expression in human NK/T cell lymphomas, melanoma cells, and primary monocytes (Bi et al., 2016; Gowrishankar et al., 2015; Huang et al., 2013). The observed binding of RELA (p65; a subunit of NF-kB) to the PD-L1 promoter in NSCLC cells (in the form of a RELA-MUC1-C complex), monocytes, and breast cancer cells suggests that NF-kB can directly regulate PD-L1 transcription (Bouillez et al., 2017; Huang et al., 2013; Xue et al., 2017). AP-1, a dimeric transcription factor consisting of c-Jun, FOS, MAF, or ATF subunits, is strongly oncogenic (Shaulian and Karin, 2002). Tandem candidate AP-1 binding sites were identified in the first intron of PD-L1, which potentially recruit AP-1 components to elevate PD-L1 transcription in Hodgkin’s lymphoma cells (Green et al., 2012). Since EBV-encoded latent membrane protein-1 (LMP1) triggers AP-1 and JAK-STAT activity (Kieser et al., 1997; Chen et al., 2013), EBV infection could conceivably contribute to the increased expression of PD-L1 that is observed in the subset of classical Hodgkin’s lymphoma (CHL) in which no 9p24.1 amplification was detected (Green et al., 2012). In line with this, EBV+ CHLs are characterized by relatively higher PD-L1 H scores than their EBV counterparts, in spite of the similar distribution of 9p24.1 amplification in EBV+ and EBV CHLs (Roemer et al., 2016), although this difference may also be explained by IFN-γ production upon T cell recognition of EBV antigens. In melanoma cells that have developed resistance to BRAF inhibition, constitutive PD-L1 expression (here defined as PD-L1 expression that is not dependent on external stimuli) can be mediated by both JUN and STAT3, as interference with expression of either one blocked PD-L1 expression (Jiang et al., 2013).
As described above, PD-L1 expression can be induced in various cell types by inflammatory cytokines such as IFN-γ (Chen, 2004; Dong et al., 2002; Eppihimer et al., 2002; Keir et al., 2008). Cyclin-dependent kinase 5 (CDK5), a serine-threonine kinase that is highly expressed in medulloblastoma and many other cancers, can regulate PD-L1 expression through its influence on the IFN-γ-signaling pathway. Mechanistically, CDK5 destabilizes IRF2, a competitive repressor of IRF1, by posttranslational modification of the co-repressor IRF2BP2, and this destabilization then leads to IRF1-mediated induction of PD-L1 (Dorand et al., 2016).
The phosphatidylinositol 3-kinase (PI3K) signaling pathway impacts cancer cell survival, proliferation, metabolism, and mobility. The AKT-mTOR cascade that sits downstream of PI3K can be activated by type I and type II interferons and controls interferon-dependent mRNA translation (Kaur et al., 2008; Lekmine et al., 2004), implying a certain level of cooperation between interferon receptor signaling pathway and AKT-mTOR signaling pathway. In line with this, pharmacological inhibition of PI3KAKT signaling suppressed IFN-γ-induced PD-L1 expression, while loss of PTEN (a tumor-suppressor gene that negatively regulates PI3K-AKT signaling) strengthens PD-L1 expression in CRC and NSCLC cells (Song et al., 2013; Zhang et al., 2017b). In addition, the PI3K-AKT pathway can regulate PD-L1 expression in an IFN-γ-independent manner in NSCLC, CRC, glioma, breast cancer, and melanoma cells (Atefi et al., 2014; Lastwika et al., 2016; Parsa et al., 2007; Song et al., 2013; Xu et al., 2014), and in melanoma and breast cancer cells, it was suggested that this regulation occurs at least partially by altering PD-L1 mRNA levels (Jiang et al., 2013; Mittendorf et al., 2014). Moreover, PI3K signaling is critically involved in the increase in PD-L1 expression following EGF stimulation in NSCLC cells, 17b-estradiol treatment of estrogen receptor alpha (ERa)-positive breast cancer cells, CpG oligodeoxynucleotide, or poly(I:C) stimulation of dendritic cells, and HIV infection of APCs (Karakhanova et al., 2010; Lastwika et al., 2016; Muthumani et al., 2011; Yang et al., 2017). Finally, some studies have shown that the influence of PI3K inhibition on PDL1 expression varies substantially across melanoma cells, but the underlying mechanisms have yet to be revealed (Atefi et al., 2014; Gowrishankar et al., 2015).
The MEK-ERK pathway is commonly activated in human cancers, in most cases due to abnormal upstream signaling initiated by amplification or activating mutations in receptor tyrosine kinases, the RAS GTPase, or BRAF (Roberts and Der, 2007). Evidence suggests that MEK-ERK signaling can regulate PD-L1 gene expression through crosstalk with inflammatory signaling. Chemical or genetic suppression of MEK has been shown to inhibit IFN-γ-induced STAT1 phosphorylation and PD-L1 transcription in multiple myeloma cells (Liu et al., 2007). In line with this, activation of MEK-ERK signaling by PMA or by a constitutively active variant of MEK (MEK-DD) increases PD-L1 expression, and this effect can be abolished by MEK inhibition in multiple myeloma cells (Liu et al., 2007; Loi et al., 2016). Similarly, in BRAF mutant melanoma cells that have developed resistance to BRAF inhibition, constitutive PD-L1 expression is elevated in conjunction with the activation of JUN. JUN is a primary target of MAPK signaling and cooperates with STAT3 in transcriptional regulation of PD-L1 expression. It was suggested that suppression of MEK leads to decreased PD-L1 expression through inactivation of JUN and STAT3 in melanoma cells and in several NSCLC cells (Jiang et al., 2013; Sumimoto et al., 2016; Zhang et al., 1999). In addition, the increased PD-L1 expression seen in multiple myeloma, bladder cancer, lymphoma, and dendritic cells upon stimulation by TLR ligands can be abrogated by MEK inhibition (Karakhanova et al., 2010; Liu et al., 2007; Qian et al., 2008; Yamamoto et al., 2009). Moreover, p38 MAPK, an effector in another MAPK signaling cascade that functions in parallel to the MEK/ERK pathway, also positively regulates PD-L1 expression in multiple myeloma, bladder cancer, lymphoma cells, and dendritic cells (Karakhanova et al., 2010; Qian et al., 2008; Yamamoto et al., 2009).
Activation of KRAS, epidermal growth factor receptor (EGFR), and anaplastic lymphoma kinase (ALK) due to genetic alterations or ligand stimulation can drive PD-L1 expression via their downstream effector pathway(s). In some KRAS mutant NSCLC cells, silencing of KRAS by RNAi decreases ERK activation which in turn can suppress PD-L1 expression (Chen et al., 2017a; Sumimoto et al., 2016). In line with this, ectopic expression of mutant KRAS results in increased PD-L1 expression in bronchial epithelial cells (Chen et al., 2017a). Recently, it was shown that RASMEK signaling in part controls expression of PD-L1 in NSCLC cells by modulating the stability of the PD-L1 transcript (Coelho et al., 2017). Activating mutations in EGFR, and also stimulation with EGF, induce PD-L1 expression in bronchial epithelial cells, NSCLC, head and neck cancer (HNC), and breast cancer cells. Consistent with these data, pharmacological inhibition of EGFR reduces constitutive and IFN-γ-induced PD-L1 expression (Akbay et al., 2013; Chen et al., 2015; Concha-Benavente et al., 2016; Lin et al., 2015; Zhang et al., 2016). The increase in PD-L1 expression that occurs as a result of EGFR activation in NSCLC cells can be blocked by rapamycin (Lastwika et al., 2016) and by an ERK inhibitor (Chen et al., 2015), suggesting mTOR- and ERK-dependent regulatory mechanisms. In addition, in different studies in head and neck cancer and NSCLC, evidence was obtained that supports a model in which NF-kB p65, STAT3, and/or JAK2-STAT1 serve as mediators between EGFR signaling and PD-L1 expression (Concha-Benavente et al., 2016; Lin et al., 2015; Zhang et al., 2016).
Finally, oncogenic ALK signaling often results from gene translation or amplification, and the NPM-ALK fusion protein has been shown to promote PD-L1 expression via STAT3 and MEK-ERK in lymphomas (Marzec et al., 2008; Yamamoto et al., 2009). Likewise, in NSCLC cells harboring the EML4-ALK fusion gene, PD-L1 expression is increased by the constitutively active ALK kinase, with ERK, AKT, STAT3, and HIF1a as downstream mediators (Koh et al., 2015; Ota et al., 2015).
While there are now abundant data on the regulation of the PD-L1 gene by a number of signaling pathways that are frequently activated in cancer, a number of aspects remain to be clarified. First, discrepancies with respect to the role of different signaling components are sometimes observed across studies. Specifically, in contrast to the above-mentioned studies that demonstrate that MEK-ERK signaling positively regulates PD-L1 expression, in two other studies MEK inhibitor treatment had no effect or even increased PD-L1 expression in mouse breast cancer, NSCLC, and melanoma cells (Atefi et al., 2014; Liu et al., 2015; Loi et al., 2016). Moreover, whereas the increased PD-L1 expression upon EGFR activation was in one case shown to be mediated by the AKT-mTOR pathway (Lastwika et al., 2016), in a second study it was shown not to depend on AKT, but on ERK (Chen et al., 2015). These inconsistencies may be explained in several ways. First, inhibition of a major signaling node, such as the MEK-ERK or PI3K-AKT pathway, initiates signaling rewiring through feedback loops and pathway crosstalk to compensate for the perturbations (Sun and Bernards, 2014). Depending on both the specific cell or tissue type used and the duration or the intensity of pathway inhibition, the kinetics and specific type of compensatory signaling can vary. Support for this potential explanation is provided by time-course experiments that show that PD-L1 expression can fluctuate during treatment of KRAS mutant NSCLC cells with either a MEK inhibitor or AKT inhibitor (Lastwika et al., 2016; Minchom et al., 2017). In addition, the effect of the same pathway may vary between cell systems depending on the activity of other pathways. As a somewhat more trivial explanation for the observed discrepancies, the small-molecule kinase inhibitors that are frequently used to assess pathway involvement often have multiple targets. When different inhibitors are used to target the same pathway, a different outcome may result from differential off-target effects (Klaeger et al., 2017). As a second topic that deserves further attention, while the relationship between activity of signaling pathways and expression of PD-L1 has now been documented in many cell systems, it is presently unclear whether the increased PD-L1 expression that is observed acts as a positive selection pressure for pathway activation or is a mere epiphenomenon. As an example, MYC activation leads to increased transcription of a plethora of genes, and enhanced expression of only a part of these is likely to form a driving force in oncogenesis.
miRNA-Mediated PD-L1 mRNA Regulation
In normal physiology, miRNAs function as posttranscriptional regulators of gene expression by mediating target mRNA degradation and/or by inhibiting translation. Recently, the role of miRNAs in regulating PD-L1 expression has been revealed. Such regulation can either involve direct binding to the PD-L1 mRNA, or occur indirectly, by influencing the expression of other PD-L1 regulators. miR-513 was the first miRNA identified as a PD-L1 regulator. miR-513 suppresses PD-L1 at the level of translation by direct binding to the 3’ UTR of PD-L1 mRNA. It was shown that IFN-γ suppresses miR-513 expression, and enforced expression of miR-513 blocks IFN-γ-induced PD-L1 expression in cholangiocytes (Gong et al., 2009, 2010). In contrast, miR-155 can be induced by TNFa and IFN-γ and suppresses PD-L1 expression at the protein level by direct binding to the 3’ UTR of PD-L1 in human dermal lymphatic endothelial cells (Yee et al., 2017). Together, miR-513 and 155 can be considered a system to fine-tune PD-L1 expression upon IFN-γ signaling. miR-34a, another miRNA that binds to the 3’ UTR of PD-L1, has been shown to reduce PD-L1 mRNA levels in AML (Wang et al., 2015) and NSCLC cells (Cortez et al., 2015). In NSCLC, it was suggested that p53 suppresses PD-L1 expression via miR-34 (Cortez et al., 2015; Wang et al., 2015). Moreover, miR-142-5p (Jia et al., 2017), miR-93, and miR-106b (Cioffi et al., 2017) in pancreatic cancer, miR-138-5p (Zhao et al., 2016) in CRC, miR-217 (Miao et al., 2017) in laryngeal cancer, miR-200 (Chen et al., 2014; Gibbons et al., 2014; Xie et al., 2017) in NSCLC and gastric cancer, miR-152 (Xie et al., 2017) and miR-570 (Wang et al., 2013) in gastric cancer, miR-17-5p (Audrito et al., 2017) in melanoma, and miR-15a, miR-193a, and miR-16 (Kao et al., 2017) in malignant pleural mesothelioma have all been identified as suppressors of PD-L1 expression. As discussed above, the structural variations in the 3’ region of the PD-L1 gene that are found in adult T cell leukemia-lymphoma, diffuse large B cell lymphoma, and stomach adenocarcinoma (with variable frequencies ranging from 2%–27%) are known to lead to increased PD-L1 mRNA levels. A role of altered miRNA binding in the observed increase in PD-L1 mRNA levels appears plausible (Kataoka et al., 2016). Next to these direct effects, miRNAs can also influence PD-L1 expression in an indirect manner. Specifically, miR-20, miR-21, and miR-130b repress PTEN, which in turn causes increased PD-L1 expression in CRC (Zhu et al., 2014), and miR-197 suppresses PD-L1 expression via its direct action on the CKS1B-STAT3 cascade in NSCLC (Fujita et al., 2015).
PD-L1 Regulation at the Protein Level
Posttranslational regulation is final mechanism by which the level of PD-L1 expression is modulated, and several regulators of the PD-L1 protein have recently been identified. In two large-scale genetic screens in pancreatic cancer cells and CML-derived haploid cells, CMTM6 was identified as a positive regulator of PD-L1 expression (Burr et al., 2017; Mezzadra et al., 2017). Interference with CMTM6 expression reduces both constitutive and IFN-γ-induced PD-L1 protein expression, with no effect on PD-L1 mRNA levels. This control of PD-L1 levels by CMTM6 has to date been observed in all cell systems tested, including melanoma, NSCLC, breast cancer, pancreatic cancer, thyroid cancer, and CRC, and also LPS-stimulated primary dendritic cells. However, the magnitude of CMTM6-mediated PD-L1 regulation does differ substantially across the cell models tested. A modifier genetic screen in CMTM6-deficient cells identified CMTM4 as a second PD-L1 regulator. The variable effect size of CMTM6 modulation across cell systems may therefore potentially be explained by differences in CMTM4 levels, but may also point to additional context dependencies. Mechanistically, CMTM6 was shown to bind to PD-L1 and thereby increase its half-life, presumably by preventing ubiquitination and lysosomal degradation during protein recycling. Mass spectrometric analysis suggests that CMTM6 associates with only a limited number of proteins, adding to its attractiveness as a possible target (Burr et al., 2017; Mezzadra et al., 2017). One aspect of the CMTM6-PD-L1 interaction that deserves further attention is whether CMTM6 has additional roles in this complex. Such roles could involve the clustering of PD-L1, allowing PD-L1 co-localization with other proteins, and/or signal transduction in the PD-L1-expressing cell.
As another proposed mechanism of PD-L1 protein regulation, in a study by Zhang et al. (2017a), it was demonstrated that the level of PD-L1 protein fluctuates during the cell cycle and that the cyclin D-CDK4 cascade participates in the regulation of PD-L1 levels in human cervical cancer and breast cancer cells. CDK4 phosphorylates SPOP, which stabilizes the latter by dissociating it from the ubiquitin E3-ligase complex APC/CCdh1. SPOP is an adaptor protein of Cullin 3 (CUL3), which interacts with and serves as an E3 ubiquitin ligase for PD-L1. Thus, CDK4 functions as a negative regulator of PD-L1 expression by indirectly regulating its ubiquitination (Zhang et al., 2017a). Evidence has also been obtained for a possible interaction between glycogen synthase kinase 3b (GSK3b) and non-glycosylated PD-L1 in breast cancer cells, thereby increasing PD-L1 degradation (Li et al., 2016). In addition, glycosylation of PD-L1 appears to influence its interaction with PD-1. Regulation of such glycosylation has been proposed as an additional level of control, but evidence for physiological relevance is presently limited (Li et al., 2018). Finally, evidence has been obtained indicating that, in addition to the role of NF-kB signaling in transcriptional regulation of PD-L1, NF-kB signaling induces expression of COP9 signalosome 5 (CSN5), resulting in increased PD-L1 protein levels in breast cancer cells through removal of ubiquitin chains (Lim et al., 2016).
Comparison of the effect of the different regulatory processes described in the sections above makes it apparent that the effect size of different PD-L1 regulators varies widely. In addition, the role of a given PD-L1 regulator can vary strongly, both between tissues and between tumors. As a straightforward example, whereas some tumors show a very profound increase in PD-L1 levels upon IFN-γ exposure, only a minor induction is observed in other cases. In the coming years, it will be important to understand under which conditions a specific regulatory process contributes to the PD-L1 levels in human cancers in a physiologically meaningful way.
Concluding Remarks
Immunotherapies that target the PD-L1-PD-1 axis have demonstrated unprecedented efficacy in the treatment of a variety of human cancers. Nevertheless, only a proportion of patients show objective responses to treatment, with only a small fraction experiencing complete responses.
PD-L1 expression has undergone extensive assessment with respect to its value as a predictive biomarker of anti-PD-1-PD-L1 therapies (Pitt et al., 2016; Sharma et al., 2017a). As shown in a number of cancer types, patients with PD-L1-positive tumors have a higher objective response rate and improved progression-free survival and overall survival as compared to PD-L1-negative subgroups (Borghaei et al., 2015; Fehrenbacher et al., 2016; Robert et al., 2015a; Rosenberg et al., 2016; Weber et al., 2015, 2017). However, a non-negligible number of exceptions is observed, both with patients in the PD-L1-negative subgroup still benefiting and with patients with PD-L1-positive tumors displaying intrinsic resistance to therapy (Carbognin et al., 2015). A major goal is therefore to improve our ability to predict response to these agents. Such an improvement will certainly come in part from a better understanding of the activity of other inhibitory mechanisms in individual patients (Blank et al., 2016). At the same time, a better understanding of the activity of the PD-1-PD-L1 axis itself in individual tumors can be expected to contribute. Specifically:
One reason why PD-L1 expression is thought to have information value with respect to outcome upon PD-1-PD-L1 targeting is the fact that PD-L1 expression is frequently driven by IFN-γ exposure. As such, intratumoral PD-L1 expression can be seen as an indirect measure of local T cell activity. A confounder in these analyses, however, is that PD-L1 expression is also regulated by many other mechanisms, thereby reducing its accuracy as a measure of T cell activity. Strategies to distinguish IFN-γ-induced and IFN-γ-independent PD-L1 expression would therefore be of value. A straightforward way to achieve this goal would for instance be the simultaneous measurement of other IFN-γ-induced genes (Seiwert et al., 2016).
As a second issue, the expression of PD-L1 may potentially increase during PD-1-PD-L1 blockade in some patients, thereby reducing the predictive value of a low pre-treatment PD-L1 expression. Specifically, in a case in which PD-L1 blockade reactivates rare tumor-reactive T cells, the resulting local cytokine secretion can induce multiple positive feedback loops: IFN-γ-induced production of CXCL9 and CXCL10 can promote T cell infiltration, and IFN-γ-induced increases in expression of components of the antigen presentation machinery can enhance visibility of tumor cells to T cells. The possible occurrence of such feedforward loops makes it important to understand whether PD-1-PD-L1 blockade is or is not likely to spark enhanced T cell reactivity in an individual tumor.
As a third and slightly related issue, as the ambition should be to understand whether the PD-1-PD-L1 axis is active in the tumor of an individual patient, it is somewhat counterintuitive to solely focus on expression of PD-L1. This issue is all the more important in view of the fact that intratumoral T cells can vary widely in the levels of PD-1 that they express (Thommen and Schumacher, 2018), and the effect of PD-1-PD-L1 axis blockade on T cells with different levels of PD-1 expression are presently ill understood. In addition to the possible joint assessment of expression of PD-L1 and PD-1, it may also be attractive to identify downstream consequences of PD-1 ligation in intratumoral T cells and other PD-1-expressing cells in tumor microenvironment, as a direct strategy to probe the activity of the PD-1-PD-L1 axis.
It is presently unclear whether our increased understanding of PD-L1 regulation will offer novel opportunities for therapeutic intervention. As described above, aberrant cell signaling that results from genetic or epigenetic alterations drive PD-L1 expression in many cancers, and these pathways may be targeted with small molecules to reverse this effect. Whether this will add to the activity of current anti-PD-1-PD-L1 antibodies depends on whether the current agents already fully block checkpoint signaling. Especially at sites of close T cell-tumor cell contact, it would seem possible that newly emerging checkpoint molecules may not be occupied sufficiently fast by antibody to fully prevent ligand binding. By the same token, dual targeting of CMTM6 and PD-L1 may conceivably form a strategy to more efficiently block the PD-1-PD-L1 checkpoint. Specifically, as CMTM6 is constitutively expressed, targeting of this molecule may pro-actively limit IFN-γ-induced adaptive PD-L1 expression by reducing the CMTM6 pool available for PD-L1 stabilization. Finally, the identification of such a bewildering variety of regulatory mechanisms of the PD-L1 checkpoint makes analysis of regulatory pathways that control the expression of other immune checkpoints also a highly attractive research area.
Acknowledgments
We would like to thank Lisette Rozeman, Daniela Thommen, and Christian Blank for valuable discussions. This work was supported by ERC AdG SENSIT (to T.N.S.) and The Cancer Research Institute (CRI) Irvington Postdoctoral Fellowship (to C.S.). We apologize to colleagues whose work was not cited in this review owing to space constraints.
References
- Agata Y, Kawasaki A, Nishimura H, Ishida Y, Tsubata T, Yagita H, Honjo T. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int Immunol. 1996;8:765–772. doi: 10.1093/intimm/8.5.765. [DOI] [PubMed] [Google Scholar]
- Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christen-sen CL, Mikse OR, Cherniack AD, Beauchamp EM, Pugh TJ, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013;3:1355–1363. doi: 10.1158/2159-8290.CD-13-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari MJ, Salama AD, Chitnis T, Smith RN, Yagita H, Akiba H, Yamazaki T, Azuma M, Iwai H, Khoury SJ, et al. The programmed death-1 (PD-1) pathway regulates autoimmune diabetes in nonobese diabetic (NOD) mice. J Exp Med. 2003;198:63–69. doi: 10.1084/jem.20022125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, Schuster SJ, Millenson MM, Cattry D, Freeman GJ, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372:311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonia SJ, Villegas A, Daniel D, Vicente D, Murakami S, Hui R, Yokoi T, Chiappori A, Lee KH, de Wit M, et al. PACIFIC Investigators Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. N Engl J Med. 2017;377:1919–1929. doi: 10.1056/NEJMoa1709937. [DOI] [PubMed] [Google Scholar]
- Armand P, Shipp MA, Ribrag V, Michot JM, Zinzani PL, Kuruvilla J, Snyder ES, Ricart AD, Balakumaran A, Rose S, Moskowitz CH. Programmed death-1 blockade with pembrolizumab in patients with classical Hodgkin lymphoma after brentuximab vedotin failure. J Clin Oncol. 2016;34:3733–3739. doi: 10.1200/JCO.2016.67.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atefi M, Avramis E, Lassen A, Wong DJ, Robert L, Foulad D, Cerniglia M, Titz B, Chodon T, Graeber TG, et al. Effects of MAPK and PI3K pathways on PD-L1 expression in melanoma. Clin Cancer Res. 2014;20:3446–3457. doi: 10.1158/1078-0432.CCR-13-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsaves V, Tsesmetzis N, Chioureas D, Kis L, Leventaki V, Drakos E, Panaretakis T, Grander D, Medeiros LJ, Young KH, Rassidakis GZ. PD-L1 is commonly expressed and transcriptionally regulated by STAT3 and MYC in ALK-negative anaplastic large-cell lymphoma. Leukemia. 2017;31:1633–1637. doi: 10.1038/leu.2017.103. [DOI] [PubMed] [Google Scholar]
- Audrito V, Serra S, Stingi A, Orso F, Gaudino F, Bologna C, Neri F, Garaffo G, Nassini R, Baroni G, et al. PD-L1 up-regulation in melanoma increases disease aggressiveness and is mediated through miR-17-5p. Oncotarget. 2017;8:15894–15911. doi: 10.18632/oncotarget.15213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma T, Yao S, Zhu G, Flies AS, Flies SJ, Chen L. B7-H1 is a ubiquitous antiapoptotic receptor on cancer cells. Blood. 2008;111:3635–3643. doi: 10.1182/blood-2007-11-123141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas M, Besançon A, Goncalves T, Valette F, Yagita H, Sawitzki B, Volk HD, Waeckel-Enée E, Rocha B, Chatenoud L, You S. TGFb-dependent expression of PD-1 and PD-L1 controls CD8 (+) T cell anergy in transplant tolerance. eLife. 2016;5:e08133. doi: 10.7554/eLife.08133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balar AV, Castellano D, O’Donnell PH, Grivas P, Vuky J, Powles T, Plimack ER, Hahn NM, de Wit R, Pang L, et al. First-line pembrolizumab in cisplatin-ineligible patients with locally advanced and unresectable or metastatic urothelial cancer (KEYNOTE-052): a multicentre, single-arm, phase 2 study. Lancet Oncol. 2017a;18:1483–1492. doi: 10.1016/S1470-2045(17)30616-2. [DOI] [PubMed] [Google Scholar]
- Balar AV, Galsky MD, Rosenberg JE, Powles T, Petrylak DP, Bellmunt J, Loriot Y, Necchi A, Hoffman-Censits J, Perez-Gracia JL, et al. IMvigor210 Study Group Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet. 2017b;389:67–76. doi: 10.1016/S0140-6736(16)32455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- Barsoum IB, Smallwood CA, Siemens DR, Graham CH. A mechanism of hypoxia-mediated escape from adaptive immunity in cancer cells. Cancer Res. 2014;74:665–674. doi: 10.1158/0008-5472.CAN-13-0992. [DOI] [PubMed] [Google Scholar]
- Bellmunt J, de Wit R, Vaughn DJ, Fradet Y, Lee JL, Fong L, Vogelzang NJ, Climent MA, Petrylak DP, Choueiri TK, et al. KEYNOTE-045 Investigators Pembrolizumab as second-line therapy for advanced urothelial carcinoma. N Engl J Med. 2017;376:1015–1026. doi: 10.1056/NEJMoa1613683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellone S, Buza N, Choi J, Zammataro L, Gay L, Elvin JA, Rimm DL, Liu Y, Ratner E, Schwartz PE, Santin AD. Exceptional response to pembrolizumab in a metastatic, chemotherapy/radiation resistant ovarian cancer patient harboring a CD274/PD-L1-genetic rearrangement. Clin Cancer Res. 2018 doi: 10.1158/1078-0432.CCR-17-1805. Published online January 19, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi XW, Wang H, Zhang WW, Wang JH, Liu WJ, Xia ZJ, Huang HQ, Jiang WQ, Zhang YJ, Wang L. PD-L1 is upregulated by EBV-driven LMP1 through NF-κB pathway and correlates with poor prognosis in natural killer/T-cell lymphoma. J Hematol Oncol. 2016;9:109. doi: 10.1186/s13045-016-0341-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank CU, Haanen JB, Ribas A, Schumacher TN. CANCER IMMUNOLOGY. The “cancer immunogram”. Science. 2016;352:658–660. doi: 10.1126/science.aaf2834. [DOI] [PubMed] [Google Scholar]
- Boes M, Meyer-Wentrup F. TLR3 triggering regulates PD-L1 (CD274) expression in human neuroblastoma cells. Cancer Lett. 2015;361:49–56. doi: 10.1016/j.canlet.2015.02.027. [DOI] [PubMed] [Google Scholar]
- Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillez A, Rajabi H, Jin C, Samur M, Tagde A, Alam M, Hiraki M, Maeda T, Hu X, Adeegbe D, et al. MUC1-C integrates PD-L1 induction with repression of immune effectors in non-small-cell lung cancer. Oncogene. 2017;36:4037–4046. doi: 10.1038/onc.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer JR, Drake CG, Wollner I, Powderly JD, Picus J, Sharfman WH, Stankevich E, Pons A, Salay TM, McMiller TL, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28:3167–3175. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Wilson WR. Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer. 2004;4:437–447. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
- Brown JA, Dorfman DM, Ma FR, Sullivan EL, Munoz O, Wood CR, Greenfield EA, Freeman GJ. Blockade of programmed death-1 ligands on dendritic cells enhances T cell activation and cytokine production. J Immunol. 2003;170:1257–1266. doi: 10.4049/jimmunol.170.3.1257. [DOI] [PubMed] [Google Scholar]
- Bu LL, Yu GT, Wu L, Mao L, Deng WW, Liu JF, Kulkarni AB, Zhang WF, Zhang L, Sun ZJ. STAT3 induces immunosuppression by upregulating PD-1/PD-L1 in HNSCC. J Dent Res. 2017;96:1027–1034. doi: 10.1177/0022034517712435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8:945–954. [PubMed] [Google Scholar]
- Bumma N, Jeyakumar G, Kim S, Galasso C, Thakur MK, Gadgeel SM, Vaishampayan UN, Wozniak AJ. Neutrophil lymphocyte ratio (NLR) as a predictive biomarker for PD-1/PD-L1 directed therapy in metastatic non-small cell lung cancer (NSCLC) J Clin Oncol. 2017;35 doi: 10.1200/JCO.2017.35.15_suppl.e20633. [DOI] [Google Scholar]
- Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, Lam EYN, Henderson MA, Bell CC, Stolzenburg S, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–105. doi: 10.1038/nature23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butte MJ, Keir ME, Phamduy TB, Sharpe AH, Freeman GJ. Programmed death-1 ligand 1 interacts specifically with the B7-1 costimulatory molecule to inhibit T cell responses. Immunity. 2007;27:111–122. doi: 10.1016/j.immuni.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbognin L, Pilotto S, Milella M, Vaccaro V, Brunelli M, Caliò A, Cuppone F, Sperduti I, Giannarelli D, Chilosi M, et al. Differential activity of nivolumab, pembrolizumab and MPDL3280A according to the tumor expression of programmed death-ligand-1 (PD-L1): sensitivity analysis of trials in melanoma, lung and genitourinary cancers. PLoS ONE. 2015;10:e0130142. doi: 10.1371/journal.pone.0130142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbotti G, Barisione G, Airoldi I, Mezzanzanica D, Bagnoli M, Ferrero S, Petretto A, Fabbi M, Ferrini S. IL-27 induces the expression of IDO and PD-L1 in human cancer cells. Oncotarget. 2015;6:43267–43280. doi: 10.18632/oncotarget.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, Gouw AM, Bay-lot V, Güutgemann I, Eilers M, Felsher DW. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TT, Jabs C, Sobel RA, Kuchroo VK, Sharpe AH. Studies in B7-deficient mice reveal a critical role for B7 costimulation in both induction and effector phases of experimental autoimmune encephalomyelitis. J Exp Med. 1999;190:733–740. doi: 10.1084/jem.190.5.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Qiu J, O’Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemnitz JM, Parry RV, Nichols KE, June CH, Riley JL. SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J Immunol. 2004;173:945–954. doi: 10.4049/jimmunol.173.2.945. [DOI] [PubMed] [Google Scholar]
- Chen L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat Rev Immunol. 2004;4:336–347. doi: 10.1038/nri1349. [DOI] [PubMed] [Google Scholar]
- Chen BJ, Chapuy B, Ouyang J, Sun HH, Roemer MG, Xu ML, Yu H, Fletcher CD, Freeman GJ, Shipp MA, Rodig SJ. PD-L1 expression is characteristic of a subset of aggressive B-cell lymphomas and virus-associated malignancies. Clin Cancer Res. 2013;19:3462–3473. doi: 10.1158/1078-0432.CCR-13-0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, Zhang X, Yi X, Dwyer D, Lin W, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Fang W, Zhan J, Hong S, Tang Y, Kang S, Zhang Y, He X, Zhou T, Qin T, et al. Upregulation of PD-L1 by EGFR activation mediates the immune escape in EGFR-driven NSCLC: implication for optional immune targeted therapy for NSCLC patients with EGFR mutation. J Thorac Oncol. 2015;10:910–923. doi: 10.1097/JTO.0000000000000500. [DOI] [PubMed] [Google Scholar]
- Chen N, Fang W, Lin Z, Peng P, Wang J, Zhan J, Hong S, Huang J, Liu L, Sheng J, et al. KRAS mutation-induced upregulation of PD-L1 mediates immune escape in human lung adenocarcinoma. Cancer Immunol Immunother. 2017a;66:1175–1187. doi: 10.1007/s00262-017-2005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Zinzani PL, Fanale MA, Armand P, Johnson NA, Brice P, Radford J, Ribrag V, Molin D, Vassilakopoulos TP, et al. KEYNOTE-087 Phase II study of the efficacy and safety of pembrolizumab for relapsed/refractory classic Hodgkin lymphoma. J Clin Oncol. 2017b;35:2125–2132. doi: 10.1200/JCO.2016.72.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioffi M, Trabulo SM, Vallespinos M, Raj D, Kheir TB, Lin ML, Begum J, Baker AM, Amgheib A, Saif J, et al. The miR-25-93-106b cluster regulates tumor metastasis and immune evasion via modulation of CXCL12 and PD-L1. Oncotarget. 2017;8:21609–21625. doi: 10.18632/oncotarget.15450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho MA, de Carné Trécesson S, Rana S, Zecchin D, Moore C, Molina-Arcas M, East P, Spencer-Dene B, Nye E, Barnouin K, et al. Oncogenic RAS signaling promotes tumor immunoresistance by stabilizing PD-L1 mRNA. Immunity. 2017;47:1083–1099.e6. doi: 10.1016/j.immuni.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JE, Navin TJ, Cross AJ, Goddard ME, Alexopoulou L, Mitra AT, Davies AH, Flavell RA, Feldmann M, Monaco C. Unexpected protective role for Toll-like receptor 3 in the arterial wall. Proc Natl Acad Sci USA. 2011;108:2372–2377. doi: 10.1073/pnas.1018515108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha-Benavente F, Srivastava RM, Trivedi S, Lei Y, Chandran U, Seethala RR, Freeman GJ, Ferris RL. Identification of the cell-intrinsic and -extrinsic pathways downstream of EGFR and IFNg that induce PD-L1 expression in head and neck cancer. Cancer Res. 2016;76:1031–1043. doi: 10.1158/0008-5472.CAN-15-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, Araujo L, Carbone DP, Shilo K, Giri DK, et al. PDL1 regulation by p53 via miR-34. J Natl Cancer Inst. 2015;108:108. doi: 10.1093/jnci/djv303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curiel TJ, Wei S, Dong H, Alvarez X, Cheng P, Mottram P, Krzysiek R, Knutson KL, Daniel B, Zimmermann MC, et al. Blockade of B7-H1 improves myeloid dendriticcell-mediated antitumor immunity. Nat Med. 2003;9:562–567. doi: 10.1038/nm863. [DOI] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kleijn S, Langereis JD, Leentjens J, Kox M, Netea MG, Koenderman L, Ferwerda G, Pickkers P, Hermans PW. IFN-γ-stimulated neutrophils suppress lymphocyte proliferation through expression of PD-L1. PLoS ONE. 2013;8:e72249. doi: 10.1371/journal.pone.0072249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med. 1999;5:1365–1369. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- Dorand RD, Nthale J, Myers JT, Barkauskas DS, Avril S, Chirieleison SM, Pareek TK, Abbott DW, Stearns DS, Letterio JJ, et al. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science. 2016;353:399–403. doi: 10.1126/science.aae0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, Kim TY, Choo SP, Trojan J, Welling THR, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppihimer MJ, Gunn J, Freeman GJ, Greenfield EA, Chernova T, Erickson J, Leonard JP. Expression and regulation of the PD-L1 immunoinhibitory molecule on microvascular endothelial cells. Microcirculation. 2002;9:133–145. doi: 10.1038/sj/mn/7800123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JJ, Gilchuk P, Hastings AK, Tollefson SJ, Johnson M, Downing MB, Boyd KL, Johnson JE, Kim AS, Joyce S, Williams JV. Viral acute lower respiratory infections impair CD8+ T cells through PD-1. J Clin Invest. 2012;122:2967–2982. doi: 10.1172/JCI62860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkes DA, Smith CJ, Wilski NA, Caldeira-Dantas S, Mohgbeli T, Snyder CM. Virus-specific CD8+T cells infiltrate melanoma lesions and retain function independently of PD-1 expression. J Immunol. 2017;198:2979–2988. doi: 10.4049/jimmunol.1601064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J, Park K, Smith D, Artal-Cortes A, Lewanski C, et al. POPLAR Study Group Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet. 2016;387:1837–1846. doi: 10.1016/S0140-6736(16)00587-0. [DOI] [PubMed] [Google Scholar]
- Ferris RL, Blumenschein G, Jr, Fayette J, Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE, Even C, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016;375:1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs CS, Doi T, Jang RW-J, Muro K, Satoh T, Machado M, Sun W, Jalal SI, Shah MA, Metges J-P, et al. KEYNOTE-059 cohort 1: Efficacy and safety of pembrolizumab (pembro) monotherapy in patients with previously treated advanced gastric cancer. J Clin Oncol. 2017;35 doi: 10.1200/JCO.2017.35.15_suppl.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Yagishita S, Hagiwara K, Yoshioka Y, Kosaka N, Takeshita F, Fujiwara T, Tsuta K, Nokihara H, Tamura T, et al. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol Ther. 2015;23:717–727. doi: 10.1038/mt.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X, et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017;19:1189–1201. doi: 10.1016/j.celrep.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gato-Cañas M, Zuazo M, Arasanz H, Ibañez-Vea M, Lorenzo L, Fernandez-Hinojal G, Vera R, Smerdou C, Martisova E, Arozarena I, et al. PDL1 signals through conserved sequence motifs to overcome interferon-mediated cytotoxicity. Cell Rep. 2017;20:1818–1829. doi: 10.1016/j.celrep.2017.07.075. [DOI] [PubMed] [Google Scholar]
- Gauen LK, Zhu Y, Letourneur F, Hu Q, Bolen JB, Matis LA, Klausner RD, Shaw AS. Interactions of p59fyn and ZAP-70 with T-cell receptor activation motifs: defining the nature of a signalling motif. Mol Cell Biol. 1994;14:3729–3741. doi: 10.1128/mcb.14.6.3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Saito M, Tsuta K, Iwakawa R, Shiraishi K, Scheel AH, Uchida S, Watanabe SI, Nishikawa R, Noguchi M, et al. Genomic amplification of CD274 (PD-L1) in small-cell lung cancer. Clin Cancer Res. 2017;23:1220–1226. doi: 10.1158/1078-0432.CCR-16-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons DL, Chen L, Goswami S, Cortez MA, Ahn Y-H, Byers LA, Lin W, Diao L, Wang J, Roybal J, et al. Regulation of tumor cell PD-L1 expression by microRNA-200 and control of lung cancer metastasis. J Clin Oncol. 2014;32 doi: 10.1200/jco.2014.32.15_suppl.8063. [DOI] [Google Scholar]
- Gong AY, Zhou R, Hu G, Li X, Splinter PL, O’Hara SP, LaRusso NF, Soukup GA, Dong H, Chen XM. MicroRNA-513 regulates B7-H1 translation and is involved in IFN-γamma-induced B7-H1 expression in cholangiocytes. J Immunol. 2009;182:1325–1333. doi: 10.4049/jimmunol.182.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong AY, Zhou R, Hu G, Liu J, Sosnowska D, Drescher KM, Dong H, Chen XM. Cryptosporidium parvum induces B7-H1 expression in cholangiocytes by down-regulating microRNA-513. J Infect Dis. 2010;201:160–169. doi: 10.1086/648589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SR, Maute RL, Dulken BW, Hutter G, George BM, McCracken MN, Gupta R, Tsai JM, Sinha R, Corey D, et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545:495–499. doi: 10.1038/nature22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar K, Gunatilake D, Gallagher SJ, Tiffen J, Rizos H, Hersey P. Inducible but not constitutive expression of PD-L1 in human melanoma cells is dependent on activation of NF-κB. PLoS ONE. 2015;10:e0123410. doi: 10.1371/journal.pone.0123410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O’Donnell E, Chapuy B, Takeyama K, Neuberg D, Golub TR, et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood. 2010;116:3268–3277. doi: 10.1182/blood-2010-05-282780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR, Rodig S, Juszczynski P, Ouyang J, Sinha P, O’Donnell E, Neuberg D, Shipp MA. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clin Cancer Res. 2012;18:1611–1618. doi: 10.1158/1078-0432.CCR-11-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros A, Robbins PF, Yao X, Li YF, Turcotte S, Tran E, Wunderlich JR, Mixon A, Farid S, Dudley ME, et al. PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J Clin Invest. 2014;124:2246–2259. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros A, Parkhurst MR, Tran E, Pasetto A, Robbins PF, Ilyas S, Prickett TD, Gartner JJ, Crystal JS, Roberts IM, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22:433–438. doi: 10.1038/nm.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guleria I, Khosroshahi A, Ansari MJ, Habicht A, Azuma M, Yagita H, Noelle RJ, Coyle A, Mellor AL, Khoury SJ, Sayegh MH. A critical role for the programmed death ligand 1 in fetomaternal tolerance. J Exp Med. 2005;202:231–237. doi: 10.1084/jem.20050019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Li Y, Bai H, Wang J. KRAS mutants to regulate PDL1 expression through NF-κB and HIF-1a pathways in non-small cell lung cancer cells. J Clin Oncol. 2017;35 doi: 10.1200/JCO.2017.35.15_suppl.e20049. [DOI] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Herbst RS, Baas P, Kim DW, Felip E, Pérez-Gracia JL, Han JY, Molina J, Kim JH, Arvis CD, Ahn MJ, et al. Pembrolizumab versus docetaxel for previouslytreated, PD-L1-positive, advancednon-small-celllungcancer (KEYNOTE-010): a randomised controlled trial. Lancet. 2016;387:1540–1550. doi: 10.1016/S0140-6736(15)01281-7. [DOI] [PubMed] [Google Scholar]
- Hirano F, Kaneko K, Tamura H, Dong H, Wang S, Ichikawa M, Rietz C, Flies DB, Lau JS, Zhu G, et al. Blockade of B7-H1 and PD-1 by monoclonal antibodies potentiates cancer therapeutic immunity. Cancer Res. 2005;65:1089–1096. [PubMed] [Google Scholar]
- Honda T, Egen JG, Lüammermann T, Kastenmüller W, Torabi-Parizi P, Germain RN. Tuning of antigen sensitivity by T cell receptor-dependent negative feedback controls T cell effector function in inflamed tissues. Immunity. 2014;40:235–247. doi: 10.1016/j.immuni.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Wen Q, Zhao Y, Gao Q, Bai Y. NF-κB plays a key role in inducing CD274 expression in human monocytes after lipopolysaccharide treatment. PLoS ONE. 2013;8:e61602. doi: 10.1371/journal.pone.0061602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui E, Cheung J, Zhu J, Su X, Taylor MJ, Wallweber HA, Sasmal DK, Huang J, Kim JM, Mellman I, Vale RD. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science. 2017;355:1428–1433. doi: 10.1126/science.aaf1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S, Okamoto T, Okano S, Umemoto Y, Tagawa T, Morodomi Y, Kohno M, Shimamatsu S, Kitahara H, Suzuki Y, et al. PD-L1 is upregulated by simultaneous amplification of the PD-L1 and JAK2 genes in non-small cell lung cancer. J Thorac Oncol. 2016;11:62–71. doi: 10.1016/j.jtho.2015.09.010. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Agata Y, Shibahara K, Honjo T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992;11:3887–3895. doi: 10.1002/j.1460-2075.1992.tb05481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Xi Q, Wang H, Zhang Z, Liu H, Cheng Y, Guo X, Zhang J, Zhang Q, Zhang L, et al. miR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem Biophys Res Commun. 2017;488:425–431. doi: 10.1016/j.bbrc.2017.05.074. [DOI] [PubMed] [Google Scholar]
- Jiang X, Zhou J, Giobbie-Hurder A, Wargo J, Hodi FS. The activation of MAPK in melanoma cells resistant to BRAF inhibition promotes PD-L1 expression that is reversible by MEK and PI3K inhibition. Clin Cancer Res. 2013;19:598–609. doi: 10.1158/1078-0432.CCR-12-2731. [DOI] [PubMed] [Google Scholar]
- Juneja VR, McGuire KA, Manguso RT, LaFleur MW, Collins N, Haining WN, Freeman GJ, Sharpe AH. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med. 2017;214:895–904. doi: 10.1084/jem.20160801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SC, Cheng YY, Williams M, Kirschner MB, Madore J, Lum T, Sarun KH, Linton A, McCaughan B, Klebe S, et al. Tumor suppressor microRNAs contribute to the regulation of PD-L1 expression in malignant pleural mesothelioma. J Thorac Oncol. 2017;12:1421–1433. doi: 10.1016/j.jtho.2017.05.024. [DOI] [PubMed] [Google Scholar]
- Karakhanova S, Meisel S, Ring S, Mahnke K, Enk AH. ERK/p38 MAP-kinases and PI3K are involved in the differential regulation of B7-H1 expression in DC subsets. Eur J Immunol. 2010;40:254–266. doi: 10.1002/eji.200939289. [DOI] [PubMed] [Google Scholar]
- Karakhanova S, Bedke T, Enk AH, Mahnke K. IL-27 renders DC immunosuppressive by induction of B7-H1. J Leukoc Biol. 2011;89:837–845. doi: 10.1189/jlb.1209788. [DOI] [PubMed] [Google Scholar]
- Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, Maeda T, Nagata Y, Kitanaka A, Mizuno S, et al. Aberrant PD-L1 expression through 3’-UTR disruption in multiple cancers. Nature. 2016;534:402–406. doi: 10.1038/nature18294. [DOI] [PubMed] [Google Scholar]
- Kaufman HL, Russell J, Hamid O, Bhatia S, Terheyden P, D’Angelo SP, Shih KC, Lebbé C, Linette GP, Milella M, et al. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: a multicentre, single-group, open-label, phase 2 trial. Lancet Oncol. 2016;17:1374–1385. doi: 10.1016/S1470-2045(16)30364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Sassano A, Dolniak B, Joshi S, Majchrzak-Kita B, Baker DP, Hay N, Fish EN, Platanias LC. Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proc Natl Acad Sci USA. 2008;105:4808–4813. doi: 10.1073/pnas.0710907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keir ME, Liang SC, Guleria I, Latchman YE, Qipo A, Albacker LA, Koulmanda M, Freeman GJ, Sayegh MH, Sharpe AH. Tissue expression of PD-L1 mediates peripheral T cell tolerance. J Exp Med. 2006;203:883–895. doi: 10.1084/jem.20051776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser A, Kilger E, Gires O, Ueffing M, Kolch W, Hammerschmidt W. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J. 1997;16:6478–6485. doi: 10.1093/emboj/16.21.6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kil SH, Estephan R, Sanchez J, Zain JM, Kadin ME, Young JW, Rosen ST, Querfeld C. PD-L1 is regulated by interferon gamma and interleukin 6 through STAT1 and STAT3 signaling in cutaneous T-cell lymphoma. Blood. 2017;130:1458. [Google Scholar]
- Kim EY, Kim A, Kim SK, Chang YS. MYC expression correlates with PD-L1 expression in non-small cell lung cancer. Lung Cancer. 2017;110:63–67. doi: 10.1016/j.lungcan.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Klaeger S, Heinzlmeir S, Wilhelm M, Polzer H, Vick B, Koenig PA, Reinecke M, Ruprecht B, Petzoldt S, Meng C, et al. The target landscape of clinical kinase drugs. Science. 2017;358:358. doi: 10.1126/science.aan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleffel S, Posch C, Barthel SR, Mueller H, Schlapbach C, Guenova E, Elco CP, Lee N, Juneja VR, Zhan Q, et al. Melanoma cell-intrinsic PD-1 receptor functions promote tumor growth. Cell. 2015;162:1242–1256. doi: 10.1016/j.cell.2015.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh J, Jang JY, Keam B, Kim S, Kim MY, Go H, Kim TM, Kim DW, Kim CW, Jeon YK, Chung DH. EML4-ALK enhances programmed cell death-ligand 1 expression in pulmonary adenocarcinoma via hypoxia-inducible factor (HIF)-1a and STAT3. OncoImmunology. 2015;5:e1108514. doi: 10.1080/2162402X.2015.1108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok M, Horlings HM, Snaebjornsson P, Chalabi M, Schumacher TN, Blank CU, Linn SC, Dieren Jv. Profound immunotherapy response in mismatch repair-deficient breast cancer. JCO Precision Oncology. 2017;9:1–3. doi: 10.1200/PO.17.00052. [DOI] [PubMed] [Google Scholar]
- Kondo A, Yamashita T, Tamura H, Zhao W, Tsuji T, Shimizu M, Shinya E, Takahashi H, Tamada K, Chen L, et al. Interferon-gamma and tumor necrosis factor-alpha induce an immunoinhibitory molecule, B7-H1, via nuclear factor-kappaB activation in blasts in myelodysplastic syndromes. Blood. 2010;116:1124–1131. doi: 10.1182/blood-2009-12-255125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg C, Nowicka M, Guglietta S, Schindler S, Hartmann FJ, Weber LM, Dummer R, Robinson MD, Levesque MP, Becher B. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat Med. 2018;24:144–153. doi: 10.1038/nm.4466. [DOI] [PubMed] [Google Scholar]
- Kryczek I, Wei S, Gong W, Shu X, Szeliga W, Vatan L, Chen L, Wang G, Zou W. Cutting edge: IFN-γamma enables APC to promote memory Th17 and abate Th1 cell development. J Immunol. 2008;181:5842–5846. doi: 10.4049/jimmunol.181.9.5842. [DOI] [PubMed] [Google Scholar]
- Kvistborg P, Philips D, Kelderman S, Hageman L, Ottensmeier C, Joseph-Pietras D, Welters MJ, van der Burg S, Kapiteijn E, Michielin O, et al. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8+ T cell response. Sci Transl Med. 2014;6 doi: 10.1126/scitranslmed.3008918. 254ra128. [DOI] [PubMed] [Google Scholar]
- Langer CJ, Gadgeel SM, Borghaei H, Papadimitrakopoulou VA, Patnaik A, Powell SF, Gentzler RD, Martins RG, Stevenson JP, Jalal SI, et al. KEYNOTE-021 investigators Carboplatin and pemetrexed with or without pembrolizumab for advanced, non-squamous non-small-cell lung cancer: a randomised, phase 2 cohort of the open-label KEYNOTE-021 study. Lancet Oncol. 2016;17:1497–1508. doi: 10.1016/S1470-2045(16)30498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastwika KJ, Wilson W, 3rd, Li QK, Norris J, Xu H, Ghazarian SR, Kitagawa H, Kawabata S, Taube JM, Yao S, et al. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res. 2016;76:227–238. doi: 10.1158/0008-5472.CAN-14-3362. [DOI] [PubMed] [Google Scholar]
- Latchman YE, Liang SC, Wu Y, Chernova T, Sobel RA, Klemm M, Kuchroo VK, Freeman GJ, Sharpe AH. PD-L1-deficient mice show that PD-L1 on T cells, antigen-presenting cells, and host tissues negatively regulates T cells. Proc Natl Acad Sci USA. 2004;101:10691–10696. doi: 10.1073/pnas.0307252101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau J, Cheung J, Navarro A, Lianoglou S, Haley B, Totpal K, Sanders L, Koeppen H, Caplazi P, McBride J, et al. Tumour and host cell PD-L1 is required to mediate suppression of anti-tumour immunity in mice. Nat Commun. 2017;8:14572. doi: 10.1038/ncomms14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázár-Molnár E, Gácser A, Freeman GJ, Almo SC, Nathenson SG, Nosanchuk JD. The PD-1/PD-L costimulatory pathway critically affects host resistance to the pathogenic fungus Histoplasma capsulatum. Proc Natl Acad Sci USA. 2008;105:2658–2663. doi: 10.1073/pnas.0711918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Jang BC, Lee SW, Yang YI, Suh SI, Park YM, Oh S, Shin JG, Yao S, Chen L, Choi IH. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-γamma-induced upregulation of B7-H1 (CD274) FEBS Lett. 2006;580:755–762. doi: 10.1016/j.febslet.2005.12.093. [DOI] [PubMed] [Google Scholar]
- Lekmine F, Sassano A, Uddin S, Smith J, Majchrzak B, Brachmann SM, Hay N, Fish EN, Platanias LC. Interferon-gamma engages the p70 S6 kinase to regulate phosphorylation of the 40S S6 ribosomal protein. Exp Cell Res. 2004;295:173–182. doi: 10.1016/j.yexcr.2003.12.021. [DOI] [PubMed] [Google Scholar]
- Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, Khoo KH, Chang SS, Cha JH, Kim T, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016;7:12632. doi: 10.1038/ncomms12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CW, Lim SO, Chung EM, Kim YS, Park AH, Yao J, Cha JH, Xia W, Chan LC, Kim T, et al. Eradication of triple-negative breast cancer cells by targeting glycosylated PD-L1. Cancer Cell. 2018;33:187–201.e10. doi: 10.1016/j.ccell.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D, Corle C, Seagroves TN, Johnson RS. Hypoxia-inducible factor-1alpha is a key regulator of metastasis in a transgenic model of cancer initiation and progression. Cancer Res. 2007;67:563–572. doi: 10.1158/0008-5472.CAN-06-2701. [DOI] [PubMed] [Google Scholar]
- Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu Y, Chang SS, Lin WC, Hsu JM, Hsu YH, et al. Deubiquitination and stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30:925–939. doi: 10.1016/j.ccell.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DY, Tanaka Y, Iwasaki M, Gittis AG, Su HP, Mikami B, Okazaki T, Honjo T, Minato N, Garboczi DN. The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors. Proc Natl Acad Sci USA. 2008;105:3011–3016. doi: 10.1073/pnas.0712278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Cheng J, Yang T, Li Y, Zhu B. EGFR-TKI down-regulates PD-L1 in EGFR mutant NSCLC through inhibiting NF-κB. Biochem Bio-phys Res Commun. 2015;463:95–101. doi: 10.1016/j.bbrc.2015.05.030. [DOI] [PubMed] [Google Scholar]
- Liu J, Hamrouni A, Wolowiec D, Coiteux V, Kuliczkowski K, Hetuin D, Saudemont A, Quesnel B. Plasma cells from multiple myeloma patients express B7-H1 (PD-L1) and increase expression after stimulation with IFN-γamma and TLR ligands via a MyD88-, TRAF6-, and MEK-dependent pathway. Blood. 2007;110:296–304. doi: 10.1182/blood-2006-10-051482. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang E, Ma Z, Wu W, Kosinska A, Zhang X, Möller I, Seiz P, Glebe D, Wang B, et al. Enhancing virus-specific immunity in vivo by combining therapeutic vaccination and PD-L1 blockade in chronic hepadnaviral infection. PLoS Pathog. 2014;10:e1003856. doi: 10.1371/journal.ppat.1003856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Mayes PA, Eastman S, Shi H, Yadavilli S, Zhang T, Yang J, Seestaller-Wehr L, Zhang SY, Hopson C, et al. The BRAF and MEK inhibitors dabrafenib and trametinib: effects on immune function and in combination with immunomodulatory antibodies targeting PD-1, PD-L1, and CTLA-4. Clin Cancer Res. 2015;21:1639–1651. doi: 10.1158/1078-0432.CCR-14-2339. [DOI] [PubMed] [Google Scholar]
- Loi S, Dushyanthen S, Beavis PA, Salgado R, Denkert C, Savas P, Combs S, Rimm DL, Giltnane JM, Estrada MV, et al. RAS/MAPK activation is associated with reduced tumor-infiltrating lymphocytes in triple-negative breast cancer: therapeutic cooperation between MEK and PD-1/PD-L1 immune checkpoint Inhibitors. Clin Cancer Res. 2016;22:1499–1509. doi: 10.1158/1078-0432.CCR-15-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loke P, Allison JP. PD-L1 and PD-L2 are differentially regulated by Th1 and Th2 cells. Proc Natl Acad Sci USA. 2003;100:5336–5341. doi: 10.1073/pnas.0931259100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, Collins NB, Bi K, LaFleur MW, Juneja VR, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547:413–418. doi: 10.1038/nature23270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, Kadel Iii EE, Koeppen H, Astarita JL, Cubas R, et al. TGFb attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzec M, Zhang Q, Goradia A, Raghunath PN, Liu X, Paessler M, Wang HY, Wysocka M, Cheng M, Ruggeri BA, Wasik MA. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1) Proc Natl Acad Sci USA. 2008;105:20852–20857. doi: 10.1073/pnas.0810958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matta BM, Raimondi G, Rosborough BR, Sumpter TL, Thomson AW. IL-27 production and STAT3-dependent upregulation of B7-H1 mediate immune regulatory functions of liver plasmacytoid dendritic cells. J Immunol. 2012;188:5227–5237. doi: 10.4049/jimmunol.1103382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazanet MM, Hughes CC. B7-H1 is expressed by human endothelial cells and suppresses T cell cytokine synthesis. J Immunol. 2002;169:3581–3588. doi: 10.4049/jimmunol.169.7.3581. [DOI] [PubMed] [Google Scholar]
- Messai Y, Gad S, Noman MZ, Le Teuff G, Couve S, Janji B, Kammerer SF, Rioux-Leclerc N, Hasmim M, Ferlicot S, et al. Renal cell carcinoma programmed death-ligand 1, a new direct target of hypoxia-inducible factor-2 alpha, is regulated by von Hippel-Lindau gene mutation status. Eur Urol. 2016;70:623–632. doi: 10.1016/j.eururo.2015.11.029. [DOI] [PubMed] [Google Scholar]
- Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, Logtenberg MEW, Slagter M, Rozeman EA, Hofland I, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549:106–110. doi: 10.1038/nature23669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao S, Mao X, Zhao S, Song K, Xiang C, Lv Y, Jiang H, Wang L, Li B, Yang X, et al. miR-217 inhibits laryngeal cancer metastasis by repressing AEG-1 and PD-L1 expression. Oncotarget. 2017;8:62143–62153. doi: 10.18632/oncotarget.19121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchom A, Thavasu P, Ahmad Z, Stewart A, Georgiou A, O’Brien MER, Popat S, Bhosle J, Yap TA, de Bono J, Banerji U. A study of PD-L1 expression in KRAS mutant non-small cell lung cancer cell lines exposed to relevant targeted treatments. PLoS ONE. 2017;12:e0186106. doi: 10.1371/journal.pone.0186106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittendorf EA, Philips AV, Meric-Bernstam F, Qiao N, Wu Y, Harrington S, Su X, Wang Y, Gonzalez-Angulo AM, Akcakanat A, et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol Res. 2014;2:361–370. doi: 10.1158/2326-6066.CIR-13-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER, et al. CheckMate 025 Investigators Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthumani K, Shedlock DJ, Choo DK, Fagone P, Kawalekar OU, Goodman J, Bian CB, Ramanathan AA, Atman P, Tebas P, et al. HIV-mediated phosphatidylinositol 3-kinase/serine-threonine kinase activation in APCs leads to programmed death-1 ligand upregulation and suppression of HIV-specific CD8 T cells. J Immunol. 2011;187:2932–2943. doi: 10.4049/jimmunol.1100594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa A, Dotan I, Brimnes J, Allez M, Shao L, Tsushima F, Azuma M, Mayer L. The expression and function of costimulatory molecules B7H and B7-H1 on colonic epithelial cells. Gastroenterology. 2004;126:1347–1357. doi: 10.1053/j.gastro.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Ni XY, Sui HX, Liu Y, Ke SZ, Wang YN, Gao FG. TGF-b of lung cancer microenvironment upregulates B7H1 and GITRL expression in dendritic cells and is associated with regulatory T cell generation. Oncol Rep. 2012;28:615–621. doi: 10.3892/or.2012.1822. [DOI] [PubMed] [Google Scholar]
- Nishimura H, Nose M, Hiai H, Minato N, Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–151. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- Nishimura H, Honjo T, Minato N. Facilitation of beta selection and modification of positive selection in the thymus of PD-1-deficient mice. J Exp Med. 2000;191:891–898. doi: 10.1084/jem.191.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura H, Okazaki T, Tanaka Y, Nakatani K, Hara M, Matsumori A, Sasayama S, Mizoguchi A, Hiai H, Minato N, Honjo T. Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science. 2001;291:319–322. doi: 10.1126/science.291.5502.319. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Ward JP, Gubin MM, Arthur CD, Lee SH, Hundal J, Selby MJ, Graziano RF, Mardis ER, Korman AJ, Schreiber RD. Temporally distinct PD-L1 expression by tumor and host cells contributes to immune escape. Cancer Immunol Res. 2017;5:106–117. doi: 10.1158/2326-6066.CIR-16-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noman MZ, Desantis G, Janji B, Hasmim M, Karray S, Dessen P, Bronte V, Chouaib S. PD-L1 is a novel direct target of HIF-1a, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J Exp Med. 2014;211:781–790. doi: 10.1084/jem.20131916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota K, Azuma K, Kawahara A, Hattori S, Iwama E, Tanizaki J, Harada T, Matsumoto K, Takayama K, Takamori S, et al. Induction of PD-L1 expression by the EML4-ALK oncoprotein and downstream signaling pathways in non-small cell lung cancer. Clin Cancer Res. 2015;21:4014–4021. doi: 10.1158/1078-0432.CCR-15-0016. [DOI] [PubMed] [Google Scholar]
- Ou JN, Wiedeman AE, Stevens AM. TNF-α and TGF-β counter-regulate PD-L1 expression on monocytes in systemic lupus erythematosus. Sci Rep. 2012;2:295. doi: 10.1038/srep00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18:1182–1191. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JJ, Omiya R, Matsumura Y, Sakoda Y, Kuramasu A, Augustine MM, Yao S, Tsushima F, Narazaki H, Anand S, et al. B7-H1/CD80 interaction is required for the induction and maintenance of peripheral T-cell tolerance. Blood. 2010;116:1291–1298. doi: 10.1182/blood-2010-01-265975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BV, Freeman ZT, Ghasemzadeh A, Chattergoon MA, Rutebemberwa A, Steigner J, Winter ME, Huynh TV, Sebald SM, Lee SJ, et al. TGFb1-mediated SMAD3 enhances PD-1 expression on antigen-specific T cells in cancer. Cancer Discov. 2016;6:1366–1381. doi: 10.1158/2159-8290.CD-15-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsa AT, Waldron JS, Panner A, Crane CA, Parney IF, Barry JJ, Cachola KE, Murray JC, Tihan T, Jensen MC, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med. 2007;13:84–88. doi: 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, Gartner JJ, Jia L, Steinberg SM, Yamamoto TN, et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537–542. doi: 10.1038/nature23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MR, Ellerton J, Infante JR, Agrawal M, Gordon M, Aljumaily R, Britten CD, Dirix L, Lee KW, Taylor M, et al. Avelumab in metastatic urothelial carcinoma after platinum failure (JAVELIN Solid Tumor): pooled results from two expansion cohorts of an open-label, phase 1 trial. Lancet Oncol. 2018;19:51–64. doi: 10.1016/S1470-2045(17)30900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt JM, Vétizou M, Daillère R, Roberti MP, Yamazaki T, Routy B, Lepage P, Boneca IG, Chamaillard M, Kroemer G, Zitvogel L. Resistance mechanisms to immune-checkpoint blockade in cancer: tumorintrinsic and -extrinsic factors. Immunity. 2016;44:1255–1269. doi: 10.1016/j.immuni.2016.06.001. [DOI] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Powles T, O’Donnell PH, Massard C, Arkenau HT, Friedlander TW, Hoimes CJ, Lee JL, Ong M, Sridhar SS, Vogelzang NJ, et al. Efficacy and safety of durvalumab in locally advanced or metastatic urothelial carcinoma: updated results from a phase 1/2 open-label study. JAMA Oncol. 2017;3:e172411. doi: 10.1001/jamaoncol.2017.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulko V, Liu X, Krco CJ, Harris KJ, Frigola X, Kwon ED, Dong H. TLR3-stimulated dendritic cells up-regulate B7-H1 expression and influence the magnitude of CD8 T cell responses to tumor vaccination. J Immunol. 2009;183:3634–3641. doi: 10.4049/jimmunol.0900974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Deng J, Geng L, Xie H, Jiang G, Zhou L, Wang Y, Yin S, Feng X, Liu J, et al. TLR4 signaling induces B7-H1 expression through MAPK pathways in bladder cancer cells. Cancer Invest. 2008;26:816–821. doi: 10.1080/07357900801941852. [DOI] [PubMed] [Google Scholar]
- Quandt D, Jasinski-Bergner S, Müuller U, Schulze B, Seliger B. Synergistic effects of IL-4 and TNFa on the induction of B7-H1 in renal cell carcinoma cells inhibiting allogeneic T cell proliferation. J Transl Med. 2014;12:151. doi: 10.1186/1479-5876-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe S, et al. KEYNOTE-024 Investigators Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- Ribas A, Puzanov I, Dummer R, Schadendorf D, Hamid O, Robert C, Hodi FS, Schachter J, Pavlick AC, Lewis KD, et al. Pembrolizumab versus investigator-choice chemotherapy for ipilimumab-refractory melanoma (KEYNOTE-002): a randomised, controlled, phase 2 trial. Lancet Oncol. 2015;16:908–918. doi: 10.1016/S1470-2045(15)00083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C, Kalinka-Warzocha E, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015a;372:320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, et al. KEYNOTE-006 investigators Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015b;372:2521–2532. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- Roemer MG, Advani RH, Ligon AH, Natkunam Y, Redd RA, Homer H, Connelly CF, Sun HH, Daadi SE, Freeman GJ, et al. PD-L1 and PD-L2 genetic alterations define classical Hodgkin lymphoma and predict outcome. J Clin Oncol. 2016;34:2690–2697. doi: 10.1200/JCO.2016.66.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, Dawson N, O’Donnell PH, Balmanoukian A, Loriot Y, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman M, Jiao YJ, Chen JH, Rooney MS, Barzily-Rokni M, Eliane JP, Bjorgaard SL, Hammond MR, Vitzthum H, Blackmon SM, et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun. 2017;8:1136. doi: 10.1038/s41467-017-01062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoop R, Wahl P, Le Hir M, Heemann U, Wang M, Wüthrich RP. Suppressed T-cell activation by IFN-γamma-induced expression of PD-L1 on renal tubular epithelial cells. Nephrol Dial Transplant. 2004;19:2713–2720. doi: 10.1093/ndt/gfh423. [DOI] [PubMed] [Google Scholar]
- Schreiner B, Mitsdoerffer M, Kieseier BC, Chen L, Hartung HP, Weller M, Wiendl H. Interferon-beta enhances monocyte and dendritic cell expression of B7-H1 (PD-L1), a strong inhibitor of autologous T-cell activation: relevance for the immune modulatory effect in multiple sclerosis. J Neuroimmunol. 2004;155:172–182. doi: 10.1016/j.jneuroim.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, Heath K, McClanahan T, Lunceford J, Gause C, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016;17:956–965. doi: 10.1016/S1470-2045(16)30066-3. [DOI] [PubMed] [Google Scholar]
- Seliger B. The link between MHC class I abnormalities of tumors, oncogenes, tumor suppressor genes, and transcription factors. J Immunotoxicol. 2014;11:308–310. doi: 10.3109/1547691X.2013.875084. [DOI] [PubMed] [Google Scholar]
- Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017a;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Retz M, Siefker-Radtke A, Baron A, Necchi A, Bedke J, Plimack ER, Vaena D, Grimm MO, Bracarda S, et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol. 2017b;18:312–322. doi: 10.1016/S1470-2045(17)30065-7. [DOI] [PubMed] [Google Scholar]
- Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol. 2018;18:153–167. doi: 10.1038/nri.2017.108. [DOI] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- Sheppard KA, Fitz LJ, Lee JM, Benander C, George JA, Wooters J, Qiu Y, Jussif JM, Carter LL, Wood CR, Chaudhary D. PD-1 inhibits T-cell receptor induced phosphorylation of the ZAP70/CD3zeta signalosome and downstream signaling to PKCtheta. FEBS Lett. 2004;574:37–41. doi: 10.1016/j.febslet.2004.07.083. [DOI] [PubMed] [Google Scholar]
- Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, Grasso CS, Hugo W, Sandoval S, Torrejon DY, et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov. 2017;7:188–201. doi: 10.1158/2159-8290.CD-16-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder A, Nathanson T, Funt SA, Ahuja A, Buros Novik J, Hellmann MD, Chang E, Aksoy BA, Al-Ahmadie H, Yusko E, et al. Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: An exploratory multi-omic analysis. PLoS Med. 2017;14:e1002309. doi: 10.1371/journal.pmed.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Chen D, Lu B, Wang C, Zhang J, Huang L, Wang X, Timmons CL, Hu J, Liu B, et al. PTEN loss increases PD-L1 protein expression and affects the correlation between PD-L1 expression and clinical parameters in colorectal cancer. PLoS ONE. 2013;8:e65821. doi: 10.1371/journal.pone.0065821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Yuan P, Wu H, Chen J, Fu J, Li P, Lu J, Wei W. Dendritic cells with an increased PD-L1 by TNF-β induce T cell anergy for the cytotoxicity of hepatocellular carcinoma cells. Int Immunopharmacol. 2014;20:117–123. doi: 10.1016/j.intimp.2014.02.027. [DOI] [PubMed] [Google Scholar]
- Starke A, Wüthrich RP, Waeckerle-Men Y. TNF-βeta treatment modulates PD-L1 and CD40 expression in proximal renal tubular epithelial cells and enhances CD8 cytotoxic T-cell responses. Nephron, Exp Nephrol. 2007;107:e22–e29. doi: 10.1159/000106506. [DOI] [PubMed] [Google Scholar]
- Straub M, Drecoll E, Pfarr N, Weichert W, Langer R, Hapfelmeier A, Götz C, Wolff KD, Kolk A, Specht K. CD274/PD-L1 gene amplification and PD-L1 protein expression are common events in squamous cell carcinoma of the oral cavity. Oncotarget. 2016;7:12024–12034. doi: 10.18632/oncotarget.7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus DB, Weiss A. Genetic evidence for the involvement of the lck tyrosine kinase in signal transduction through the T cell antigen receptor. Cell. 1992;70:585–593. doi: 10.1016/0092-8674(92)90428-f. [DOI] [PubMed] [Google Scholar]
- Sumimoto H, Takano A, Teramoto K, Daigo Y. RAS-mitogenactivated protein kinase signal is required for enhanced PD-L1 expression in human lung cancers. PLoS ONE. 2016;11:e0166626. doi: 10.1371/journal.pone.0166626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Bernards R. Feedback and redundancy in receptor tyrosine kinase signaling: relevance to cancer therapies. Trends Biochem Sci. 2014;39:465–474. doi: 10.1016/j.tibs.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Sun Z, Fourcade J, Pagliano O, Chauvin JM, Sander C, Kirkwood JM, Zarour HM. IL10 and PD-1 cooperate to limit the activity of tumorspecific CD8+ T cells. Cancer Res. 2015;75:1635–1644. doi: 10.1158/0008-5472.CAN-14-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thommen DS, Schumacher T. T cell dysfunction in cancer. Cancer Cell. 2018;33 doi: 10.1016/j.ccell.2018.03.012. Published online April 9, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twa DD, Chan FC, Ben-Neriah S, Woolcock BW, Mottok A, Tan KL, Slack GW, Gunawardana J, Lim RS, McPherson AW, et al. Genomic rearrangements involving programmed death ligands are recurrent in primary mediastinal large B-cell lymphoma. Blood. 2014;123:2062–2065. doi: 10.1182/blood-2013-10-535443. [DOI] [PubMed] [Google Scholar]
- Ugi S, Maegawa H, Olefsky JM, Shigeta Y, Kashiwagi A. Src homology 2 domains of protein tyrosine phosphatase are associated in vitro with both the insulin receptor and insulin receptor substrate-1 via different phosphotyrosine motifs. FEBS Lett. 1994;340:216–220. doi: 10.1016/0014-5793(94)80141-x. [DOI] [PubMed] [Google Scholar]
- Wang J, Yoshida T, Nakaki F, Hiai H, Okazaki T, Honjo T. Establishment of NOD-Pdcd1-/- mice as an efficient animal model of type I diabetes. Proc Natl Acad Sci USA. 2005;102:11823–11828. doi: 10.1073/pnas.0505497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Li F, Mao Y, Zhou H, Sun J, Li R, Liu C, Chen W, Hua D, Zhang X. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum Genet. 2013;132:641–648. doi: 10.1007/s00439-013-1275-6. [DOI] [PubMed] [Google Scholar]
- Wang X, Li J, Dong K, Lin F, Long M, Ouyang Y, Wei J, Chen X, Weng Y, He T, Zhang H. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal. 2015;27:443–452. doi: 10.1016/j.cellsig.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Wang J, Jia Y, Zhao S, Zhang X, Wang X, Han X, Wang Y, Ma M, Shi J, Liu L. BIN1 reverses PD-L1-mediated immune escape by inactivating the c-MYC and EGFR/MAPK signaling pathways in non-small cell lung cancer. Oncogene. 2017a;36:6235–6243. doi: 10.1038/onc.2017.217. [DOI] [PubMed] [Google Scholar]
- Wang X, Yang L, Huang F, Zhang Q, Liu S, Ma L, You Z. Inflammatory cytokines IL-17 and TNF-α up-regulate PD-L1 expression in human prostate and colon cancer cells. Immunol Lett. 2017b;184:7–14. doi: 10.1016/j.imlet.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JS, D’Angelo SP, Minor D, Hodi FS, Gutzmer R, Neyns B, Hoeller C, Khushalani NI, Miller WH, Jr, Lao CD, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2015;16:375–384. doi: 10.1016/S1470-2045(15)70076-8. [DOI] [PubMed] [Google Scholar]
- Weber J, Mandala M, Del Vecchio M, Gogas HJ, Arance AM, Cowey CL, Dalle S, Schenker M, Chiarion-Sileni V, Marquez-Rodas I, et al. CheckMate 238 Collaborators Adjuvant nivolumab versus ipilimumab in resected stage III or IV melanoma. N Engl J Med. 2017;377:1824–1835. doi: 10.1056/NEJMoa1709030. [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 2015;15:486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintterle S, Schreiner B, Mitsdoerffer M, Schneider D, Chen L, Meyermann R, Weller M, Wiendl H. Expression of the B7-related molecule B7-H1 by glioma cells: a potential mechanism of immune paralysis. Cancer Res. 2003;63:7462–7467. [PubMed] [Google Scholar]
- Xie G, Li W, Li R, Wu K, Zhao E, Zhang Y, Zhang P, Shi L, Wang D, Yin Y, et al. Helicobacter pylori promote B7-H1 expression by suppressing miR-152 and miR-200b in gastric cancer cells. PLoS ONE. 2017;12:e0168822. doi: 10.1371/journal.pone.0168822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong HY, Ma TT, Wu BT, Lin Y, Tu ZG. IL-12 regulates B7-H1 expression in ovarian cancer-associated macrophages by effects on NF-κB signalling. Asian Pac. J Cancer Prev. 2014;15:5767–5772. doi: 10.7314/apjcp.2014.15.14.5767. [DOI] [PubMed] [Google Scholar]
- Xu C, Fillmore CM, Koyama S, Wu H, Zhao Y, Chen Z, Herter-Sprie GS, Akbay EA, Tchaicha JH, Altabef A, et al. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell. 2014;25:590–604. doi: 10.1016/j.ccr.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Chen C, Qi M, Huang Y, Wang L, Gao Y, Dong H, Ling K. Type Ig phosphatidylinositol phosphate kinase regulates PD-L1 expression by activating NF-κB. Oncotarget. 2017;8:42414–42427. doi: 10.18632/oncotarget.17123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Yamashita K, Perucho M. Somatic mutation of the beta2-microglobulin gene associates with unfavorable prognosis in gastrointestinal cancer of the microsatellite mutator phenotype. Gastroenterology. 2001;120:1565–1567. doi: 10.1053/gast.2001.24497. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Nishikori M, Tashima M, Sakai T, Ichinohe T, Takaori-Kondo A, Ohmori K, Uchiyama T. B7-H1 expression is regulated by MEK/ERK signaling pathway in anaplastic large cell lymphoma and Hodgkin lymphoma. Cancer Sci. 2009;100:2093–2100. doi: 10.1111/j.1349-7006.2009.01302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Huang F, Mei J, Wang X, Zhang Q, Wang H, Xi M, You Z. Posttranscriptional control of PD-L1 expression by 17b-estradiol via PI3K/Akt signaling pathway in ERa-positive cancer cell lines. Int J Gynecol Cancer. 2017;27:196–205. doi: 10.1097/IGC.0000000000000875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Wang S, Zhu Y, Luo L, Zhu G, Flies S, Xu H, Ruff W, Broad-water M, Choi IH, et al. PD-1 on dendritic cells impedes innate immunity against bacterial infection. Blood. 2009;113:5811–5818. doi: 10.1182/blood-2009-02-203141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yearley JH, Gibson C, Yu N, Moon C, Murphy E, Juco J, Lunceford J, Cheng J, Chow LQM, Seiwert TY, et al. PD-L2 expression in human tumors: relevance to anti-PD-1 therapy in cancer. Clin Cancer Res. 2017;23:3158–3167. doi: 10.1158/1078-0432.CCR-16-1761. [DOI] [PubMed] [Google Scholar]
- Yee D, Shah KM, Coles MC, Sharp TV, Lagos D. MicroRNA-155 induction via TNF-α and IFN-γ suppresses expression of programmed death ligand-1 (PD-L1) in human primary cells. J Biol Chem. 2017;292:20683–20693. doi: 10.1074/jbc.M117.809053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes A, Santoro A, Shipp M, Zinzani PL, Timmerman JM, Ansell S, Armand P, Fanale M, Ratanatharathorn V, Kuruvilla J, et al. Nivolumab for classical Hodgkin’s lymphoma after failure of both autologous stem-cell transplantation and brentuximab vedotin: a multicentre, multicohort, single-arm phase 2 trial. Lancet Oncol. 2016;17:1283–1294. doi: 10.1016/S1470-2045(16)30167-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak KM, Kitel R, Przetocka S, Golik P, Guzik K, Musielak B, Dömling A, Dubin G, Holak TA. Structure of the complex of human programmed death 1, PD-1, and its ligand PD-L1. Structure. 2015;23:2341–2348. doi: 10.1016/j.str.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wrzeszczynska MH, Horvath CM, Darnell JE., Jr Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol Cell Biol. 1999;19:7138–7146. doi: 10.1128/mcb.19.10.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Schwartz JC, Guo X, Bhatia S, Cao E, Lorenz M, Cammer M, Chen L, Zhang ZY, Edidin MA, et al. Structural and functional analysis of the costimulatory receptor programmed death-1. Immunity. 2004;20:337–347. doi: 10.1016/s1074-7613(04)00051-2. [DOI] [PubMed] [Google Scholar]
- Zhang N, Zeng Y, Du W, Zhu J, Shen D, Liu Z, Huang JA. The EGFR pathway is involved in the regulation of PD-L1 expression via the IL-6/JAK/STAT3 signaling pathway in EGFR-mutated non-small cell lung cancer. Int J Oncol. 2016;49:1360–1368. doi: 10.3892/ijo.2016.3632. [DOI] [PubMed] [Google Scholar]
- Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, Tan Y, Ci Y, Wu F, Dai X, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via Cul3(SPOP) to control cancer immune surveillance. Nature. 2017a;553:91–95. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zeng Y, Qu Q, Zhu J, Liu Z, Ning W, Zeng H, Zhang N, Du W, Chen C, Huang JA. PD-L1 induced by IFN-γ from tumorassociated macrophages via the JAK/STAT3 and PI3K/AKT signaling pathways promoted progression of lung cancer. Int J Clin Oncol. 2017b;22:1026–1033. doi: 10.1007/s10147-017-1161-7. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Xiao X, Wu Y, Wei Y, Zhu LY, Zhou J, Kuang DM. Interleukin-17-educated monocytes suppress cytotoxic T-cell function through B7-H1 in hepatocellular carcinoma patients. Eur J Immunol. 2011;41:2314–2322. doi: 10.1002/eji.201041282. [DOI] [PubMed] [Google Scholar]
- Zhao L, Yu H, Yi S, Peng X, Su P, Xiao Z, Liu R, Tang A, Li X, Liu F, Shen S. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget. 2016;7:45370–45384. doi: 10.18632/oncotarget.9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chen L, Zou L, Yang P, Wu R, Mao Y, Zhou H, Li R, Wang K, Wang W, et al. MiR-20b, -21, and -130b inhibit PTEN expression resulting in B7-H1 over-expression in advanced colorectal cancer. Hum Immunol. 2014;75:348–353. doi: 10.1016/j.humimm.2014.01.006. [DOI] [PubMed] [Google Scholar]