Abstract
Therapeutic reinvigoration of tumor-specific T cells has greatly improved clinical outcome in cancer. Nevertheless, many patients still do not achieve durable benefit. Recent evidence from studies in murine and human cancer suggest that intratumoral T cells display a broad spectrum of (dys-)functional states, shaped by the multifaceted suppressive signals that occur within the tumor microenvironment. Here we discuss the current understanding of T cell dysfunction in cancer, the value of novel technologies to dissect such dysfunction at the single cell level, and how our emerging understanding of T cell dysfunction may be utilized to develop personalized strategies to restore antitumor immunity.
Introduction
Tumors harbor genetic alterations resulting from DNA damage, e.g., due to exposure to UV light, tobacco smoke, or deficiencies in DNA repair (Alexandrov et al., 2013; Stephens et al., 2009). These alterations distinguish cancer cells from normal cells, thereby frequently prompting the induction of tumor-reactive T cell responses in both mouse models and cancer patients (Castle et al., 2012; Matsushita et al., 2012; Robbins et al., 2013; van Rooij et al., 2013). While the presence of tumorinfiltrating lymphocytes (TILs), and in particular CD8+ T cells, is a positive prognostic marker in multiple solid tumors (Fridman et al., 2012), these cells fail to effectively eliminate cancer cells (Boon et al., 2006). One reason for this failed immune control is the curtailing of effector functions of infiltrating T cells by a broad spectrum of immunosuppressive mechanisms that are present in the tumor microenvironment (TME) (Chen and Mellman, 2013; Mellman et al., 2011; Schreiber et al., 2011). Among these mechanisms, the upregulation of programmed cell death-1 (PD-1) on T cells has emerged as a major marker of T cell dysfunction. The altered functional state of PD-1+ T cells, termed T cell exhaustion, has originally been described and most extensively studied in murine models of chronic lymphocytic choriomeningitis virus (LCMV) infection (Wherry et al., 2007; Zajac et al., 1998), but ample evidence for it has also been obtained in human infection and cancer (Ahmadzadeh et al., 2009; Baitsch et al., 2011; Day et al., 2006; Trautmann et al., 2006).
The successful reinvigoration of T cell function by blockade of PD-1, or its ligand PD-L1, highlights the importance of the PD-1/ PD-L1 axis in T cell dysfunction (Day et al., 2006). In line with this, antibodies targeting PD-1/PD-L1 have shown impressive activity in multiple cancer types, including melanoma (Robert et al., 2014, 2015), non-small-cell lung cancer (NSCLC) (Borghaei et al., 2015; Brahmer et al., 2015; Fehrenbacher et al., 2016), renal cancer (RCC) (Motzer et al., 2015), urothelial cancer (Balar et al., 2017; Rosenberg et al., 2016), and head and neck squamous cell cancer (HNSCC) (Seiwert et al., 2016). While the objective response rates between 15% and 34% that were observed in these studies signify a clear improvement in patient outcome, the majority of patients still do not respond or do not achieve durable responses to this therapy. Lack of (durable) response is thought to be explained at least in part by the activity of other inhibitory pathways in T cells. Specifically, a simultaneous expression of different inhibitory receptors, so-called immune checkpoints, has been observed on a fraction of T cells and increases with progressive dysfunction (Thommen et al., 2015; Wherry, 2011). Furthermore, it has been found that T cells can differentiate into an exhausted state even in the absence of PD-1 (Legat et al., 2013; Odorizzi et al., 2015). Direct evidence for the role of these additional pathways comes from the observation that T cell subsets expressing certain immune checkpoint combinations display synergistic responses to immunotherapy combinations, compared with anti-PD-1 monotherapy (Fourcade et al., 2010; Sakuishi et al., 2010). As the intratumoral T cell pool is exposed to many distinct immunosuppressive mechanisms, a broad spectrum of dysfunctional T cell states may be expected. Importantly, these states can also be expected to partially diverge from the dysfunctional state of T cells in chronic viral infections, as the microenvironment in tumors will only show a partial overlap with that of chronically infected sites (Figure 1).
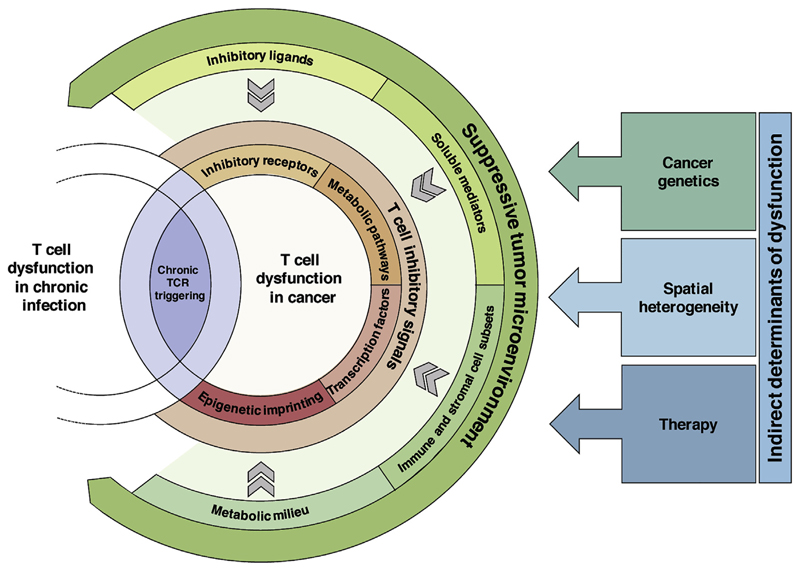
Figure 1. Drivers of T Cell Dysfunction in Cancer.
Dysfunctional T cells in cancer share core exhaustion features with dysfunctional T cells in chronic infection that are at least partially driven by chronic TCR stimulation. The consequences of chronic TCR signaling are further modulated by a multitude of immunosuppressive signals in the TME, including inhibitory ligands, suppressive soluble mediators, cell subsets, and metabolic factors. Strength of these different signals is determined by parameters such as the specific mutations in the cancer cells, spatial gradients in tumor composition, and therapy-induced alterations in the TME. Collectively, the immunosuppressive signals in the TME shape the (dys-)functional state of intratumoral T cells by influencing the expression of inhibitory receptors, changing metabolic pathways, modifying the epigenetic state, and altering their transcription factor profiles.
In this perspective, we review the hallmarks of exhausted T cells in cancer and in chronic viral infection. A better understanding of intratumoral T cell dysfunction should be of value from a fundamental biological perspective, should allow improved patient stratification, and should offer novel avenues for the restoration of intratumoral T cell activity.
Characteristics of Intratumoral T Cell Dysfunction Hallmarks of Exhausted T Cells in Chronic Viral Infection and Cancer
Prolonged exposure of T cells to their cognate antigen results in T cell receptor (TCR) signals that lead to elevated and sustained expression of inhibitory receptors on these cells. In addition, T cells enter a state of dysfunction that is characterized by a hierarchical loss of effector functions and proliferation, as well as distinct transcriptional and metabolic changes. This dysfunctional state was initially described for antigen-specific T cells in chronic murine LCMV infection (Wherry et al., 2007), but a similar state has also been observed in T cells in cancers (Baitsch et al., 2011). Comparison of gene expression profiles derived from T cells in human or murine tumors with those from chronic viral infections has revealed that exhausted T cells in both conditions share many phenotypic and functional characteristics. Of these characteristics, the upregulation of immune checkpoints has been described as one of the hallmarks of T cell exhaustion (Wherry et al., 2007). While transient expression of PD-1 is a characteristic of normal T cell activation, persistent antigen exposure induces a sustained expression of PD-1, which characterizes—and possibly drives—T cell dysfunction (Ahmadzadeh et al., 2009; Baitsch et al., 2011; Wherry, 2011). In addition to PD-1, dysfunctional T cells have been shown to overexpress other inhibitory receptors, including T cell immunoglobulin and mucin domain-3 protein (Tim-3), Lymphocyte-activation gene 3 (Lag-3), Cytotoxic T lymphocyte antigen-4 (CTLA-4), and T cell immunoglobulin and ITIM domain (TIGIT) (Blackburn et al., 2009; Kuchroo et al., 2014). The fraction of T cells that simultaneously express these receptors—and that often also exhibit a high expression level of PD-1—increases during progressive dysfunction, as defined by the gradual loss of effector functions (Blackburn et al., 2008; Schreiner et al., 2016). Analysis of the pattern of inhibitory receptor co-expression on T cells in both chronic hepatitis B virus infection and cancer has revealed a hierarchical expression of these receptors, dominated by the expression of PD-1 (Bengsch et al., 2014; Thommen et al., 2015).
A second feature of exhausted T cells in chronic viral infection is the progressive loss of effector functions, including the secretion of interleukin (IL)-2, tumor necrosis factor (TNF)a, interferon (IFN)g, and b-chemokines (Wherry et al., 2007). Several studies have demonstrated that tumor-infiltrating CD8+ T lymphocytes are also impaired in their production of effector cytokines (Baitsch et al., 2011; Zippelius et al., 2004) in, among others, melanoma (Ahmadzadeh et al., 2009), NSCLC (Thommen et al., 2015), HNSCC (Li et al., 2016), gastric cancer (Lu et al., 2017), and ovarian cancer (Matsuzaki et al., 2010).
Finally, exhausted T cells in chronic LCMV infection display a particular gene expression profile that is distinct from that of naive, effector, or memory T cells and that, in addition to changes in inhibitory receptor expressionand cytokine production, encompasses alterations in transcription factor expression and in pathways involved in chemotaxis, migration, and metabolism (Wherry et al., 2007). The transcriptional profile of CD8+ T cells directed against the Melan-A/MART-1 melanoma antigen isolated from melanoma metastases after vaccination largely overlaps with the exhaustion signature derived from CD8+ T cells during chronic LCMV infection, and contains similar alterations in processes involved in immune response, cell migration, signaling, metabolism, cell cycle, and DNA repair (Baitsch et al., 2011).
Phenotypic and Transcriptional Differences between Dysfunctional T Cells in Cancer and Chronic Viral Infection
Based on the observation that T cells derived from human tumors and chronic viral infection share a number of phenotypic, transcriptional, and functional characteristics, initial studies assumed that the exhausted state of tumor-reactive T cells would be similar to that of virus-specific T cells in chronic infections. More recent work that provides an in-depth analysis of intratumoral T cell dysfunction, however, leads to a model in which the exhaustion signatures during chronic infection and cancer do exhibit clear differences. In particular, whereas the upregulation of inhibitory receptors is seen in both situations, the relative expression of distinct inhibitory receptors appears to differ between tumor- and virus-specific T cells. Specifically, the expression of KLRG-1, CD160, Lag-3, and CTLA-4 varies between melanoma- and EBV-specific T cells, indicating that the upregulation of inhibitory receptors might be context-dependent (Baitsch et al., 2011, 2012).
Transcriptional profiling of T cells in chronic infection has revealed a set of transcription factors that is altered in expression compared with non-exhausted T cells, and thus has been associated with exhaustion (Wherry et al., 2007). While further studies have suggested involvement of a broad number of transcription factors, including NFAT, Eomes, T-bet, Blimp-1, BATF, FoxO1, VHL, and c-Maf, a master regulator that initiates and controls the process of dysfunction has not yet been found (Agnellini et al., 2007; Doedens et al., 2013; Giordano et al., 2015; Kao et al., 2011; Paley et al., 2012; Quigley et al., 2010; Shin et al., 2009; Staron et al., 2014). The understanding of gene regulation in dysfunctional T cells is further complicated by the fact that many of these transcription factors act in a context-specific manner, with a different function in exhausted T cells than in non-exhausted effector or memory T cell subsets (Kao et al., 2011; Paley et al., 2012; Quigley et al., 2010; Shin et al., 2009). One prominent example of this is formed by the T-box transcription factors T-bet and Eomesodermin (Eomes). While T-bet controls normal effector differentiation of CD8+ and CD4+ T cells, it also regulates the non-terminally exhausted progenitor pool in chronic LCMV infection by repressing the expression of PD-1 and other inhibitory receptors. Eomes, in contrast, regulates the protein expression needed for memory formation following acute infection, but is also expressed by the terminally exhausted T cell pool in chronic infection, contributing to the upregulation of inhibitory receptors seen in these cells (Banerjee et al., 2010; Intlekofer et al., 2005; Paley et al., 2012; Zhou et al., 2010). Importantly, while these two transcription factors thus show a reciprocal expression pattern in chronic viral infections, this is different from what is observed in tumor-specific dysfunctional T cells. In the latter situation, T cells do not display high Eomes expression, but rather show both reduced T-bet and Eomes expression over the course of tumor progression, as recently described in a murine liver cancer model (Schietinger et al., 2016). Thus, the role of these two transcription factors in T cell dysfunction may differ between chronic viral infection and tumor outgrowth. It will be of particular interest to assess whether the unresponsiveness to PD-1 blockade that is observed for Eomeshi PD-1hi T cells in chronic infection is also present in highly dysfunctional intratumoral T cells, with their lower level of Eomes expression.
The identification of transcriptional pathways that mediate T cell dysfunction is complicated by the fact that the gene signatures of exhausted T cells show a fair degree of overlap with those of recently activated T cells, consistent with the critical role of TCR signaling in driving both states (Doering et al., 2012; Tirosh et al., 2016). This overlap is also reflected by the large number of immune checkpoints present on exhausted T cells that are transiently expressed during T cell activation, a process that may be interpreted as a protection mechanism against overstimulation (Anderson et al., 2016). Network analysis of exhausted LCMV-specific T cells has, however, revealed that the connectivity patterns of key transcriptional regulators are different between exhausted and memory T cells (Doering et al., 2012). Recent work from the Anderson group described a distinct gene module for dysfunction in murine tumor models that could be separated from that of T cell activation (Singer et al., 2016), and single cell RNA sequencing of intratumoral T cells indicated that these two gene modules are mutually exclusive. Expression of metallothioneins, which regulate intracellular zinc availability, and several zinc-finger transcription factors were highly increased in dysfunctional CD8+ TILs, and targeted deletion of metallothionein 1 and 2 could improve T cell functionality and restore antitumor immunity. Of note, PD-1 and Tim-3 expression remained unchanged upon metallothionein-deletion, indicating that inhibitory receptor expression can be uncoupled from other aspects of T cell dysfunction, and was more likely to reflect T cell activation in this setting.
Tumors are complex ecosystems, defined by the interplay of a large number of cellular and soluble components. Whereas TILs share the core exhaustion signature of virus-specific T cells driven by the persistent antigen exposure, their phenotypic and transcriptional profile is additionally shaped by cell-intrinsic and -extrinsic immunosuppressive factors in the TME that will partially differ from those at sites of chronic infection. These immunosuppressive mechanisms include (1) the expression of inhibitory receptors and their ligands; (2) the recruitment of immunosuppressive cell populations such as regulatory T cells (Tregs) and myeloid-derived suppressor cells; (3) direct repression of T cell function via secretion of suppressive cytokines such as IL-10 and TGFb; (4) direct repression of T cell function via production of metabolites, including adenosine, prostaglandins, and lactate; (5) production of suppressive compounds and enzymes, including nitric oxide synthase, reactive oxygen species and indoleamine-2,3 dioxygenase (IDO); and (6) physiological changes in the TME such as hypoxia, low pH, and the deprivation of nutrients, such as glucose or amino acids (Fridman et al., 2017; Gajewski et al., 2013; Speiser et al., 2016; Zarour, 2016). In the next section, we focus on the role of metabolic factors in T cell dysfunction. For an overview of other immunosuppressive mechanisms in the TME, we refer to the excellent reviews mentioned above.
Impact of the Tumor Metabolism and Genetics on Dysfunctional TIL States
Following TCR triggering, T cells undergo profound metabolic changes to meet the bio-energetic demands associated with cellular proliferation and effector differentiation. Whereas naive T cells largely rely on oxidative phosphorylation (OXPHOS), they switch to the use of aerobic glycolysis upon antigen stimulation, to generate the required ATP (Chang et al., 2013; Pearce and Pearce, 2013). During memory formation, T cells revert back to the use of OXPHOS and become increasingly dependent on mitochondrial fatty acid oxidation (van der Windt et al., 2013; van der Windt and Pearce, 2012). In dysfunctional T cells, the link between antigenic stimulation, and metabolic pathway activation appears altered. In chronic LCMV infection, profound changes in genes related to metabolic pathways have been observed (Wherry et al., 2007). Of note, metabolic dysfunction of virus-specific T cells already develops during the first weeks of infection, before a major loss in effector functions occurs. These bio-energetic alterations include a reduction in glucose uptake and use, dysregulation of mitochondrial energetics, as well as the upregulation of anabolic pathways. Furthermore, it has been shown that PD-1 contributes to these early glycolytic and mitochondrial changes, as PD-1 deficient T cells do not undergo these metabolic changes in the murine LCMV model, and as the key metabolic regulator PPAR-gamma coactivator 1a (PGC-1a) is repressed by PD-1 triggering (Bengsch et al., 2016). Similar observations have been made in a murine tumor model, where enforced expression of PGC-1a led to reprogramming of tumor-specific T cells and improvement of metabolic and effector function (Scharping et al., 2016). While these metabolic changes have been observed in both tumor- and virus-specific T cells, comparison of T cells reactive to the same antigen in either a murine chronic infection model or cancer model revealed a persistent loss of mitochondrial function and mass that was specific to T cells that resided at the tumor site. These mitochondrial alterations were also observed in human CD8+ TILs in HNSCC tumors (Scharping et al., 2016; Siska et al., 2017).
To generate the energy required to exert effector functions, T cells are highly dependent on the availability of nutrients within their microenvironment (Scharping et al., 2016). T cells that fail to upregulate metabolic pathways upon activation, or that are activated under nutrient-poor conditions, have been described to acquire a hyporesponsive phenotype that cannot be reverted by subsequent stimulation (Zheng et al., 2009). The metabolic landscape of tumors is in large part determined by the energetics of cancer cells, because of their high bio-energetic demands for proliferation. The increased utilization of glycolysis and OXPHOS by tumor cells leads to a depletion of essential nutrients, as well as the accumulation of immunosuppressive metabolites such as lactic acid (Vander Heiden et al., 2009). This creates an environment with low glucose, low amino acids, low oxygen, and low pH, which are all considered metabolic checkpoints (Kato et al., 2013; Martinez-Outschoorn et al., 2017; Scharping and Delgoffe, 2016; Wilson and Hay, 2011). Hence, the functionality of T cells will not only be affected by their intrinsic metabolic alterations, but also by the unfavorable metabolic milieu within the TME.
In line with this model, low glucose availability in the TME has been shown to render CD8+ T cells less capable of controlling tumor growth in a mouse sarcoma model (Chang et al., 2015). Similar findings in a mouse melanoma model indicated a role for the glycolytic metabolite phosphoenolpyruvate (PEP) in sustaining Ca2+-NFAT signaling and T cell effector function (Ho et al., 2015). PEP levels are decreased in exhausted T cells, and overexpression of phosphoenolpyruvate carboxykinase (PCK1) to increase PEP production can restore effector functions in CD8+ T cells via metabolic reprogramming. Blockade of the immune checkpoints CTLA-4, PD-1, and PD-L1 in a mouse tumor model has been shown to restore glucose availability in the TME as well as to increase glucose uptake, and to reinvigorate effector functions in CD8+ TILs (Chang et al., 2015). Moreover, this study provided evidence to suggest that PD-L1 blockade may not only target the metabolism of T cells, but also that of tumor cells by directly dampening glucose uptake and glycolysis in the latter. On a more general note, efforts to reduce the metabolic activity of tumor cells may be of potential interest, as an indirect strategy to increase the amount of glucose that can be utilized by T cells.
In addition to glucose, the supply of certain amino acids also impacts the effector functions of intratumoral T cells. First, glutamine is essential for T cell activation and differentiation, but is also often depleted from the TME due to its use as primary fuel for tumor cells (Altman et al., 2016; Carr et al., 2010). Second, tryptophan is lacking in some TMEs due to the secretion of the tryptophan metabolizing enzyme IDO by tumor cells and suppressive immune cells, also leading to an increase in its immunosuppressive catabolite kynurenine (Platten et al., 2012). Kynurenine accumulation in turn promotes the generation of regulatory T cells (Julliard et al., 2014). Third, arginase secretion by myeloid cells leads to depletion of the arginine that is required for T cell activation and proliferation (Lind, 2004; Munder et al., 2009). Next to amino acid deprivation, the pH changes and the loss of cytosolic NAD+ regeneration induced by the lactic acid that is generated as a waste product of tumor cell glycolysis further inhibit T cell function and cytokine production (Choi et al., 2013). In addition, tumor-derived lactic acid has also been shown to induce apoptosis of naive T cells via loss of FAK family-interacting protein of 200 kDa (FIP200), which may further support the ability of tumors to evade immune control (Xia et al., 2017). Neutralization of acidic components in the TME has been shown to boost antitumor immunity in animal models (Pilon-Thomas et al., 2016).
Another metabolic parameter that is closely linked with the above discussed characteristics of the TME is hypoxia. During rapid tumor growth, areas that lack sufficient perfusion develop. The resulting limitation in oxygen availability impacts T cell metabolism, by reducing energy production via OXPHOS and inducing an increased dependency on glycolysis. In different studies, hypoxia has been shown to both increase and impair T cell responses. On the one hand, data from murine chronic infections and tumor models suggest that cytotoxicity of T cells may be enhanced under conditions of hypoxia, leading to more efficient virus and tumor control (Doedens et al., 2013; Gropper et al., 2017). On the other hand, hypoxia induces increased expression and activity of the transcription factor hypoxia-inducible factor 1α (HIF-1α), which promotes the expression of inhibitory receptors on T cells (Doedens et al., 2013), and leads to a reduction in T cell effector functions (Fischer et al., 2007). These seemingly contradictory findings may be explained by a model in which the outcome of hypoxia is determined by the presence or absence of other metabolic checkpoints. In particular the lack of glucose in the TME may restrict the functionality of T cells that rely on glycolysis in an oxygen-low milieu (a detailed review of this topic is provided by Zhang and Ertl, 2016).
Recent studies have provided evidence that classical immune checkpoints can interact with metabolic checkpoints. For instance, the inhibitory receptor CTLA-4 on T cells competes with CD28 for B7 ligands and thereby suppresses the CD28-mediated T cell costimulation that is essential to increase T cell glucose uptake and metabolism (Frauwirth et al., 2002; Parry et al., 2005). Moreover, CTLA-4 ligation has been shown to inhibit mTOR signaling via recruitment of protein phosphatase 2A (PP2A) and subsequent Akt dephosphorylation (Teft et al., 2009; Wlodarchak and Xing, 2016). Activation of PI3K, Akt, and mTOR regulates the switch to anabolic metabolism and subsequent aerobic glycolysis via transcription factors including Myc and HIF-1 (MacIver et al., 2013; Wang et al., 2011). Furthermore, Akt and mTOR are also inhibited by PD-1 ligation (Chemnitz et al., 2004; Wu et al., 2001).
The genetic alterations that cancer cells acquire during tumor development have been shown to sculpt the TME. Such sculpting can either involve repression of T cell recruitment to the tumor site (Nagarsheth et al., 2017; Peng et al., 2015) or inhibition of the functionality of T cells at that site. For instance, activation of the Wnt-b-catenin signaling pathway in melanoma has been found to reduce the recruitment of antigen-presenting cells, particularly CD103+ dendritic cells (DCs). This not only promotes T cell exclusion from the tumor, but also limits the priming and activation of the few T cells that do manage to infiltrate (Spranger et al., 2015). Antitumor T cell responses can also be suppressed by cytokines such as IL-1 and VEGF that are highly abundant in tumors with BRAF mutations (Khalili et al., 2012). In addition, BRAF mutations lead to constitutive activation of the MEK-MAPK pathway, thereby suppressing OXPHOS and enhancing glycolysis (Hall et al., 2013; Haq et al., 2013). Loss of PTEN correlated with reduced T cell-mediated tumor cell killing and effector cytokine secretion. Of note, melanoma patients with tumors that display PTEN loss showed a significantly lower reduction in tumor burden upon PD-1 treatment (Peng et al., 2016). Finally, a recent study investigating longitudinal biopsies of melanoma patients under CTLA-4 and PD-1 blockade observed that, while response to PD-1 blockade was associated with increased expression of genes related to cytotoxicity, antigen processing, and IFNg signaling, therapy resistance correlated with increased VEGFA expression (Chen et al., 2016). VEGF secretion has been found to affect T cell functionality both directly and indirectly, via suppression of DC maturation and recruitment of suppressive cell populations (Gabrilovich et al., 1998; Terme et al., 2013).
Taken together, an increasing amount of evidence has been obtained indicating that intratumoral T cells can adopt a spectrum of dysfunctional states, sculpted by TCR triggering in the context of different micro-environmental factors, such as engagement of inhibitory receptors, presence of immunosuppressive cytokines, and cell populations, as well as metabolic factors. The observation of a series of distinct dysfunctional states provides a strong rationale for the development of individually tailored therapeutic strategies to restore the function of the intratumoral T cell population.
Novel Technologies to Describe T Cell Dysfunction at the Single Cell Level
Capturing T Cell States by Mass Cytometry and Single Cell Sequencing
While the (dys-)functional states of intratumoral T cells show differences between tumors, T cell infiltrates within individual tumors are also highly heterogeneous. This heterogeneity occurs along at least three axes: (1) heterogeneity with respect to the conventional T cell subsets (e.g., CD8+, CD4+ Th1, Th2, Th17, Tregs, γδT cells), (2) heterogeneity with respect to the degree and type of dysfunction of T cells in a given subset, and (3) heterogeneity with respect to the capacity of the TCR of an individual cell to recognize tumor antigen. Because of this heterogeneity, the spectrum of T cell states in a tumor can only be captured appropriately by analyses at the single cell level. Immunologists realized the value of single cell-based analyses long ago, as shown by the development of immunohistochemistry and in particular flow cytometry. These more traditional approaches have over the past few years been complemented by novel approaches that allow one to determine individual cell states of immune cells, stromal cells, and malignant cells in human cancers, such as mass cytometry, single cell sequencing, and highly multiplexed single cell imaging (Figure 2). Mass cytometry allows for quantification of more than 50 readouts at the single cell level by combining metal isotope-labeled antibodies with mass spectrometry-based detection (Bandura et al., 2009; Ornatsky et al., 2010). One drawback of this method is that data are only obtained on a pre-defined set of parameters, thereby precluding completely unbiased analyses. In addition, unlike flow cytometry, isolation of viable cells for downstream (functional) analysis is precluded. Single cell sequencing has the advantage of being truly unbiased. As a downside, the low RNA content of T cells presently precludes the identification of the full transcriptome of cells, and heterogeneity with respect to transcripts that are present at lower levels may be difficult to identify (Chevrier et al., 2017). In addition, heterogeneity at the level of protein translation or protein turnover will be missed.
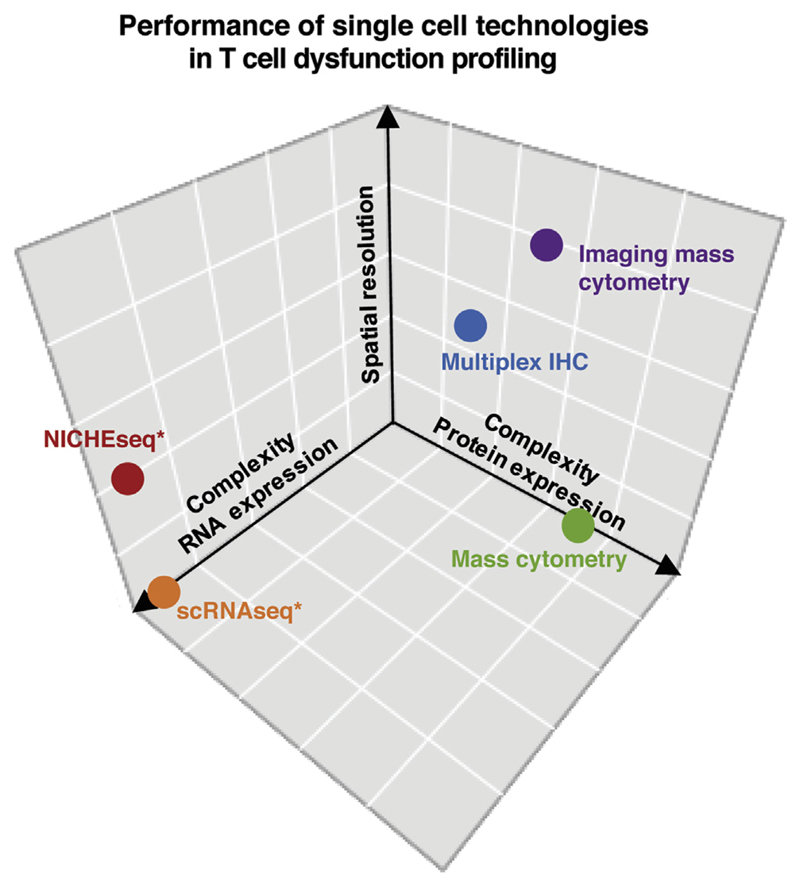
Figure 2. Comparison of Single Cell Technologies to Dissect Intratumoral T Cell Dysfunction.
Technologies that assess T cell dysfunction at the single cell level are required to describe the heterogeneity of functional states within the TME. Current single cell technologies differ in the complexity and spatial resolution they provide. Ability to provide multiparameteric information with respect to RNA and protein expression, and ability to provide spatial information are depicted for the indicated technologies. Asterisks indicate technologies that also provide TCR sequence information and may be combined with functional testing to evaluate tumor reactivity.
Recent work describes the use of these technologies to obtain an in-depth characterization of tumor ecosystems in different cancer types, including breast cancer (Chung et al., 2017), RCC (Chevrier et al., 2017), NSCLC (Lavin et al., 2017), and HCC (Zheng et al., 2017). Single cell sequencing of 11 breast cancer patients revealed three clusters of immune cells: T cells displaying either an exhausted or regulatory phenotype, B cells, and macrophages with mostly an M2 differentiation profile (Chung et al., 2017). Several T cell subclusters could be identified, distinguished by naive, costimulatory, cytotoxic, regulatory, or exhaustion signatures. PD-1 expression was modest in all clusters, but expression of other immune checkpoints such as TIGIT or Lag-3 was observed more frequently, suggesting that these alternative pathways might be important in this tumor type.
A study using single cell mass cytometry compared immune infiltrates in 32 NSCLCs with matched adjacent normal tissue and peripheral blood. Tumors displayed increased numbers of Tregs and decreased numbers of cytolytic T cells compared to normal tissue. PD-1 was expressed on a subset of CD8+ and CD4+ T cells. In addition, changes in the distribution of myeloid cells between tumors, normal lung tissue and peripheral blood were observed (Lavin et al., 2017). A second study, describing the in-depth profiling of immune infiltrates in 73 patients with clear cell RCC by mass cytometry led to the identification of 22 T cell phenotypes and 17 tumor-associated macrophage phenotypes (Chevrier et al., 2017). Among the T cell clusters, seven different clusters of CD8+ T cells that expressed PD-1 were observed. Of note, the co-expression of other inhibitory receptors including Tim-3 and CTLA-4, as well as the expression of costimulatory or proliferation markers, varied greatly between these clusters, which are therefore likely to reflect distinct (dys-)functional states. In patients harboring large numbers of T cell clusters expressing high levels of PD-1 and a marker profile corresponding to intermediate to highly exhausted TILs, other T cell subsets with little or no PD-1 expression were absent, suggesting mutually exclusive T cell states. Furthermore, correlation analyses of the frequencies for each immune cell cluster of each individual tumor were used to unravel relationships between different immune cell subsets. These analyses demonstrated that the presence of a highly exhausted T cell cluster strongly correlated with the presence of both regulatory T cells and PD-L1 and PD-L2 expressing macrophages, indicating that this technology allows the identification of connectivity between cell populations in the TME.
Single cell RNA sequencing of tumor material from 19 melanoma patients yielded profiles of tumor, immune, stromal, and endothelial cells (Tirosh et al., 2016). Intratumoral T cell clusters displayed gene signatures associated with cytotoxicity and T cell exhaustion, suggesting an activation-dependent exhaustion program, as described previously (Fuertes Marraco et al., 2015; Wherry et al., 2007). In line with the data discussed in the previous section, also an activation-independent exhaustion program could be derived. This program was used to further subdivide T cells into ‘‘high-exhaustion’’ and ‘‘low-exhaustion’’ cells, relative to their expression of cytotoxicity genes. Comparison of these high- and low-exhaustion signatures allowed the definition of a core exhaustion signature that consistently identified high-exhaustion cells in most tumors, as well as additional genes that showed variable expression across tumor samples, conceivably reflecting distinct influences of the TME, or treatment-induced effects.
A major advantage of single cell sequencing in the analysis of T cell dysfunction is its truly unbiased nature, making it possible to detect novel markers that are associated with the exhausted T cell state. A recent example of this is the discovery of LAYN as a marker of dysfunctional CD8+ T cells and Tregs in HCC (Zheng et al., 2017). Phenotypic and functional analyses indicated that the expression of LAYN was mutually exclusive with LAG-3 on CD8+ T cells and that overexpression of LAYN impaired IFNγ secretion. Another gene that has repetitively been found to be expressed in exhausted intratumoral T cell clusters, but not in gene expression data from chronic viral infections, is CXCL13 (Baitsch et al., 2011; Tirosh et al., 2016; Zheng et al., 2017). CXCL13 is a chemokine that is critically involved in the recruitment of B cells to lymph nodes and in the formation of germinal centers (Ansel et al., 2000). While its secretion by CD4+ follicular helper T cells has been well described (Crotty, 2011; Kroenke et al., 2012), its role in intratumoral CD8+ T cells is presently unknown.
The combined analysis of transcriptional profiles and TCR sequences of individual cells was recently used to investigate clonal relationships between distinct T cell clusters (Zheng et al., 2017). Comparison of tumor, adjacent normal liver tissue, and blood samples from six patients with HCC revealed a preferential enrichment of two T cell subsets with gene signatures associated with exhausted CD8+ T cells and Tregs in tumor samples. TCR analysis demonstrated a clonal expansion of T cells in these subsets. Pseudotime state transition analysis provided evidence that CD8+ T cells progress from naive via effector memory differentiation to a cytotoxic and finally exhausted T cell state. This conclusion was also supported by sharing of TCRs between the effector memory and cytotoxic T cell clusters, and between the cytotoxic and exhausted T cell clusters. In contrast, CD4+ T helper cells appeared to initially progress from a naive to a T helper cell state and then diverged into either a cytotoxic or exhausted T cell state, with very limited sharing of TCR sequences between the two populations. The use of this type of technology that provides detailed information on the transcriptional or phenotypic profile of distinct T cell states, together with the identification of the TCRs they carry, should greatly increase our understanding of the conditions under which dysfunctional T cell populations develop and which cell states they occupy.
Coupling T Cell State to Tumor Recognition Potential
One of the main limitations of the technologies discussed above is that these approaches do not allow one to couple cell state to the intrinsic capacity of a cell to recognize tumor. To develop novel therapeutic strategies to restore antitumor immunity, it is not only important to achieve a better understanding of the (dys-)functional states that T cells assume, but also how particular dysfunctional states and intrinsic tumor reactivity are connected. Put differently, the functional states of specifically those T cells that carry a tumor-reactive TCR are of most interest, as reactivation of those cells can be expected to have the most profound impact on tumor control. At least four different strategies are currently being developed to address this question. First, peptide-MHC (pMHC) multimers have over the past years been widely used to quantify the effects of cancer immunotherapy on T cell responses to defined antigens (e.g., Kvistborg et al., 2012; van Rooij et al., 2013). The combination of MHC multimer staining with mass cytometry or with single cell sequencing analysis forms a logical next step. Indeed, recent data obtained by mass cytometry of MHC multimer-labeled cells demonstrate feasibility and reveal that intratumoral T cells with the same epitope specificity can display substantial heterogeneity (Fehlings et al., 2017).
As a second strategy to understand tumor reactivity, intratumoral T cells may be expanded to subsequently assess their reactivity, either by co-culture with autologous tumor material (Muul et al., 1987; Topalian et al., 1989; Tran et al., 2008), or by using high-throughput platforms that incorporate tumor-specific epitopes of an individual patient (Gros et al., 2016). With the caveat that proliferative and functional potential may be influenced by T cell exhaustion state, the parallel testing of expanded T cells that are sorted on the basis of phenotypic markers makes it possible to couple T cell differentiation state to tumor reactivity. This approach was successfully used to demonstrate the superior tumor recognition capacity of PD-1+ compared with PD-1 T cells in tumors and peripheral blood of melanoma patients (Gros et al., 2014, 2016). In this approach, the markers used to separate T cell subsets need to be chosen in advance, thereby limiting its value to the analysis of small lists of ‘‘usual suspects.’’ A third approach that does not suffer from this limitation would be to link single cell sequencing data and tumor recognition data by TCR analysis. The identification of TCR sequences in single cell sequencing data allows one to couple the specific state of an intratumoral T cell to the TCR expressed by this cell (Zheng et al., 2017). T cells from the same tumor may conceivably be expanded ex vivo and assessed for tumor reactivity. TCR analysis of those T cells that show or lack tumor reactivity would then enable one to couple tumor recognition potential within a specific cell state, based on the matching of TCR sequences.
A limitation of the above two approaches may be that the functional and proliferative capacity of tumor-infiltrating T cells could be impaired due to their exhausted state, thereby skewing the detection of tumor reactivity in the expanded T cell population. The transfer of TCRab gene pairs identified by single cell TCR sequencing from tumor material into healthy donor peripheral blood T cells overcomes this issue and allows the analysis of tumor reactivity of intratumoral TCR repertoires in a truly unbiased manner. Coupling of this approach with single cell transcriptome analysis should form a fourth and fully unambiguous approach to link tumor recognition potential of a T cell to its transcriptional state. An interesting goal of all these analyses will be to develop algorithms to predict tumor reactivity of T cells with high precision, without any requirement for functional testing.
Spatial Analysis of T Cell States
Signals derived from immune checkpoint ligands on cancer cells, stromal cells, or other immune cells that surround tumorinfiltrating T cells, as well as local gradients of pH, oxygen levels, or suppressive soluble factors, may provide a spatial component to the intratumoral heterogeneity of T cell states. A recent study in melanoma revealed that, rather than the numbers of intratu moral CD8+ T cells, the localization of these cells within the tumor was predictive of response to PD-1 blockade (Tumeh et al., 2014). The same study also observed a predictive effect of PD-1 expression in pretreatment biopsies, if PD-1+ T cells colocalized with PD-L1 expressing cells at the invasive margin. The impact of spatial distribution of intratumoral lymphocyte populations on clinical outcome, and particularly their colocalization with other cell subsets has also been described in other studies. Localization of CD8+ T cells, but not CD4+ or total T cells in proximity to cancer cells correlated with a better overall survival in pancreatic adenocarcinoma (Carstens et al., 2017). Similar findings have been obtained in breast cancer (Heindl et al., 2015; Yuan et al., 2012). Further evidence for the significance of the spatial organization of (PD-1+) lymphocytic infiltrates is provided by recent studies that analyze tertiary lymphoid structures (TLS) in cancer. TLS are present in the large majority of NSCLC, colorectal or ovarian cancers (Di Caro et al., 2014; Goc et al., 2014; Kroeger et al., 2016) and comprise a T cell zone harboring (mostly follicular helper) T cells and DCs, as well as a follicular zone containing B cells, similar to secondary follicles in lymph nodes. The occurrence of TLS is often associated with a favorable prognosis (Dieu-Nosjean et al., 2008; Sautes-Fridman et al., 2016) and frequently correlates with high overall T cell infiltration and lower macrophage infiltration (Lavin et al., 2017; Poschke et al., 2016). T cells in NSCLC tumors harboring many TLS display a more biased TCR repertoire (Zhu et al., 2015), either hinting at a possible role of these structures in promoting clonal expansion of T cells, or at a role of clonally expanded T cells in TLS formation. The former explanation would be consistent with a model in which TLS act as centers where primary or secondary antitumor responses are generated (Fridman et al., 2012). It will be of major interest to investigate how the presence of TLS may have an impact on T cell dysfunction and the response to immunotherapy.
In view of the emerging evidence for a spatial component to T cell heterogeneity, strategies that allow spatial analyses of the T cell state will become increasingly important. Two approaches that have recently been developed—highly multiplexed single cell imaging and single cell sequencing of spatial niches—may be employed to address this question. Immunohistochemistry is commonly used to evaluate the expression of a marker at the single cell level, and to assess the localization of cells carrying this marker within a tissue. The development of multiplex immunofluorescence platforms and novel imaging technologies allow for the detection and quantification of multiple markers on a single cell, or of the spatial organization of cells expressing different markers. While multiplex immunofluorescence imaging is currently limited to the combination of a handful of markers, recent work using imaging mass cytometry demonstrated the simultaneous analysis of 32 parameters in human breast cancer samples (Giesen et al., 2014). Of note, this analysis could not only be used to identify cell populations, but also to detect cell-cell interactions based on protein phosphorylation levels, as well as to distinguish hypoxic and normoxic areas by analysis of gradients of carbonic anhydrase IX.
As a second approach to describe cell states in different niches, Amit and colleagues recently developed technology that combines photoactivatable fluorescent markers, two-photon microscopy, and single cell sequencing, termed NICHE-seq (Medaglia et al., 2017). In situ two-photon irradiation of transgenic mice expressing a photoactivatable fluorescent reporter allowed the identification of cells located at specific tissue regions based on the fluorescent signal that was induced by light exposure. Cells from irradiated and non-irradiated regions could then be sorted and subjected to single cell sequencing, thereby allowing the comparison of transcriptional profiles of cells at specific locations. Development of a similar technology applicable to human material will be of interest.
T Cell Dysfunction and Cancer Immunotherapy Heterogeneity of the PD-1 Expressing T Cell Pool
While the efficacy of antibodies targeting the PD-1/PD-L1 axis has been impressively demonstrated in multiple cancer types, the majority of patients still does not experience durable responses to this therapy. The mechanisms underlying this lack of responsiveness are highly multifactorial (Blank et al., 2016). While in some patients, lack of response to PD-1/PD-L1 blockade can be explained by a scarcity of antigens or by mutations in the antigen-presentation machinery, in part of the patient population lack of responsiveness will likely be due to T cell dysfunction that is not reversible by the sole blockade of the PD-1/PD-L1 axis. For this reason, a better understanding of the relationship between intratumoral T cell state and response to checkpoint blockade forms a major priority.
Evidence from chronic murine infection models indicates that the PD-1 positive CD8+ T cell pool consists of different sub-populations, and that the potential of reinvigoration by PD-1 blockade differs between CD8+ T cell subpopulations that are characterized by distinct expression levels of PD-1 (Blackburn et al., 2008). Specifically, in chronic murine LCMV infection, two subsets of exhausted PD-1+ T cells with distinct responses to PD-1/PD-L1 blockade have been identified. Whereas T cells with high expression of the transcription factor T-bet and intermediate expression of PD-1 (T-bethi PD-1int) can be reinvigorated by in vivo blockade with anti-PD-L1, terminally differentiated T cells with high Eomes and high PD-1 expression (Eomeshi PD-1hi) do not respond (Blackburn et al., 2008; Paley et al., 2012). It is thought that T-bethi PD-1int T cells represent a progenitor T cell subset, that proliferates in response to persisting antigen exposure, and ultimately gives rise to Eomeshi PD-1hi progeny (Paley et al., 2012). The latter cells, which are also characterized by high expression of other inhibitory receptors and loss of cytokine production, accumulate due to the persistent antigen stimulation during ongoing infection. Of note, Eomeshi PD-1hi cells do retain their cytotoxic function, suggesting that while these cells might not be able to completely eliminate the virus, they may play a role in the partial viral control seen during chronic infection.
Two recent studies on chronic LCMV infection identified two PD-1+ antigen-specific T cell populations that differ in their expression of CXCR5 (He et al., 2016; Im et al., 2016). CXCR5 is a chemokine receptor that is normally expressed on B cells and CD4+ follicular helper cells, but that has also been described on a CD8+ T cell subset in a murine model of systemic lupus erythematodes (Kim et al., 2010). The CXCR5-positive and -negative PD-1+ subsets seen in LCMV infection also differ in their expression of other receptors, with higher expression of costimulatory molecules and lower levels of inhibitory receptors on CXCR5+ T cells than on CXCR5- T cells. Of note, also PD-1 expression levels differ between the two populations with an intermediate PD-1 level on CXCR5+ T cells and high PD-1 levels on CXCR5- T cells (Im et al., 2016). The two subsets vary in their functional capacities, with CXCR5+PD-1int T cells secreting higher levels of effector cytokines upon restimulation, compared with the CXCR5 PD-1hi cells (He et al., 2016). Interestingly, in this model, only the CXCR5+PD-1int subset migrates to and resides in lymphoid tissues, and can be reactivated by PD-1 blockade (He et al., 2016; Im et al., 2016). A similar effect has been observed in dysfunctional CD8+ T cells that co-express PD-1 and the transcription factor Tcf1, which also plays an essential role in the generation of the CXCR5+PD-1int subset (Im et al., 2016; Utzschneider et al., 2016). While now firmly established in models of chronic viral infection, it is presently unclear whether the same dichotomy between PD-1 high and low cells might be observed in cancer.
The presence or absence of PD-1 has previously been employed as a strategy to identify tumor-specific T cells in murine cancer and human melanoma and NSCLC (Fernandez-Poma et al., 2017; Gros et al., 2014; McGranahan et al., 2016; Pasetto et al., 2016). However, recent data suggest it will be even more informative to look at the level of PD-1 expression. Specifically, by using co-transfer of tumor-specific and virus-specific T cells in a murine cancer model it was shown that also T cells reactive to viral antigens infiltrate tumor lesions. While the latter cells express PD-1 at low to intermediate levels and retain their functionality, many of the tumor antigen-specific TILs that are found in the same tumors, express high levels of PD-1 and are dysfunctional (Erkes et al., 2017). On a related note, total CD8+ T cells with high PD-1 expression from patients with chronic lymphocytic leukemia display a global dysfunction, which is absent in the CMV-specific T cells that express PD-1 at a lower level (te Raa et al., 2014). Thus, PD-1 expression is not always indicative of dysfunction, and the presence of PD-1 positive virus- or other non-tumor-specific T cells in cancers might reduce the value of global PD-1 expression as a biomarker. At present, it is unclear whether the PD-1int or PD-1hi subset could be a positive or negative predictor of response to anti-PD-1/anti-PD-L1 therapy. While on the one hand tumor reactivity may be enriched in PD-1hi T cells, on the other hand these cells display a highly exhausted state in human cancers (see below) and may be dysfunctional ‘‘beyond repair,’’ as suggested by observations in models of chronic infection (Blackburn et al., 2008; Paley et al., 2012). Additionally, the use of PD-1 as a marker for tumor-specific T cells may be further complicated by the fact that PD-1 expression cannot only be induced by TCR signaling, but also by other factors such as IL-10 (Sun et al., 2015) and TGF-b (Park et al., 2016).
Reactivation of Dysfunctional T Cells by Immunotherapy Combinations
As mentioned above, in addition to PD-1, dysfunctional CD8+ T cells also show (increased) expression of other inhibitory receptors including CTLA-4, Tim-3, Lag-3, and TIGIT, which can bind to their respective ligands expressed by antigen-presenting cells and tumor cells in the TME, thereby potentially impeding T cell functions (Ahmadzadeh et al., 2009; Fourcade et al., 2010; Sakuishi et al., 2010). Co-expression of PD-1 and Lag-3 has been observed on both virus- and tumor antigen-specific T cells (Grosso et al., 2009; Matsuzaki et al., 2010; Wherry et al., 2007). Of note, T cells expressing both receptors are more impaired in effector functions, such as TNF and IFNg secretion, compared with single positive cells. Simultaneous blockade of PD-1 and Lag-3 has been shown to improve T cell responses in chronic infection models and tumor models, compared with anti-PD-1 monotherapy (Blackburn et al., 2009; Matsuzaki et al., 2010), and Lag-3 targeting agents are currently being tested in clinical studies. By the same token, Tim-3 has been shown to be expressed on a subset of PD-1+ NY-ESO-1-specific T cells in advanced melanoma and gastric cancer. Of note, T cells that co-express PD-1 and Tim-3 have substantially higher PD-1 levels and are more dysfunctional than cells with either single or no detectable expression of these receptors (Fourcade et al., 2010; Li et al., 2016; Lu et al., 2017). A study in HNSCC observed that in vitro single-agent PD-1 blockade exerts its effect on PD-1int Tim-3- T cells, but not on PD-1hi Tim-3+ T cells (Li et al., 2016), in line with the previously discussed reactivation of PD-1int T cells that co-express CXCR5 by PD-1 blockade in chronic LCMV infection (He et al., 2016; Im et al., 2016). While the effect of dual PD-1/Tim-3 blockade was not addressed in this work, other studies have suggested that the simultaneous blockade of these two receptors might have additive effects in the reactivation of antigen-specific T cells in chronic infection and cancer in vitro and in vivo (Fourcade et al., 2010; Jin et al., 2010; Lu et al., 2017; Sakuishi et al., 2010). Another inhibitory receptor that is frequently co-expressed with PD-1 is TIGIT, which competes with the costimulatory molecule CD226 for the same ligand, the poliovirus receptor (PVR, CD155) (Bottino et al., 2003; Chauvin et al., 2015). In addition, TIGIT has also been described to bind with lower affinity to CD112 (PVRL2), as well as to disrupt the costimulatory function of CD226 by cis-binding (Johnston et al., 2014; Yu et al., 2009), but the functional impact of these interactions is still unclear. While TIGIT+ CD8+ TILs that co-express PD-1, Tim-3, and Lag-3 show a highly dysfunctional phenotype in murine cancer models (Kurtulus et al., 2015), TIGIT expression by itself does not appear to distinguish PD-1+ T cell subsets with different degrees of dysfunction in melanoma patients. Nevertheless, combined blockade of PD-1 and TIGIT on CD8+ TILs cultured in the presence of autologous tumor did show an additive effect in restoring effector functions (Chauvin et al., 2015).
The most prominent example of combined immune checkpoint blockade that has also reached clinical practice, is the dual blockade of PD-1 and CTLA-4. In contrast to PD-1, which inhibits T cell function by interfering with T cell signaling, CTLA-4 competes with CD28 for binding to CD80/CD86 and thereby inhibits T cell function (Krummel and Allison, 1995; Walunas et al., 2011). The CTLA-4 targeting agent ipilimumab was the first checkpoint inhibitor to demonstrate an improvement in overall survival in patients with advanced melanoma (Hodi et al., 2010; Robert et al., 2011). Treatment of melanoma patients with anti-CTLA-4 leads to a significant broadening of melanoma-reactive T cell responses in peripheral blood, suggesting that at least part of its effect occurs through enhanced T cell priming (Kvistborg et al., 2014). CTLA-4 blockade has also been suggested to target regulatory T cells by either functional inhibition or depletion of this subset (Duraiswamy et al., 2013; Simpson et al., 2013). Together, these observations indicate that the synergistic effect of anti-CTLA-4 and anti-PD-1 in clinical trials (Larkin et al., 2015; Wolchok et al., 2013) is unlikely to occur through the simultaneous targeting of inhibitory molecules on the same cell.
In addition to inhibitory receptors, PD-1+ cells can also express costimulatory molecules such as CD28, which is critical during the activation of naive T cells and enhances cytokine secretion (Bour-Jordan et al., 2011; Watts, 2010). Recently, it was demonstrated that PD-1/PD-L1 signaling suppresses T cell functionality via inactivation of CD28 signaling rather than TCR signaling (Hui et al., 2017). Furthermore, the effect of PD-1 blockade was abrogated by conditional gene deletion of CD28 in a mouse model of chronic infection (Kamphorst et al., 2017), indicating that the CD28/B7 pathway may play a crucial role in the efficacy of anti-PD-1 treatment. CD28, among other costimulatory molecules such as ICOS, is also expressed on the LCMV-specific CXCR5+ PD-1int T cells that mediate response to PD-1 therapy in mouse models (Im et al., 2016).
A synergistic effect of combined targeting of inhibitory and costimulatory molecules in reinvigorating tumor-specific T cells has been implied in other studies. CD137 (or 4-1BB) is a member of the TNFR family that is expressed on recently activated T cells (Pollok et al., 1993). Combination of PD-1 blockade with agonistic anti-CD137 antibodies increases tumor control in murine models, while also enhancing T cell reactivity to tumor antigens as well as effector/memory T cell formation (Chen et al., 2015; Wei et al., 2013, 2014). OX40 is another TNFR family member that is expressed on activated CD4+ T cells and, at lower levels, also on CD8+ T cells after recent TCR engagement (Croft, 2010). OX40 agonists have been shown to promote antitumor immunity in immunogenic mouse tumor models, while failing to exert tumor control in poorly immunogenic tumor models (Kjaergaard et al., 2000; Weinberg et al., 2000). Conceptually, the combination of OX40 agonists with therapies targeting inhibitory receptors on T cells would be an attractive approach to reactivate dysfunctional T cells in such tumors. However, recent data suggest that addition of PD-1 blockade to an OX40 agonist in a vaccination setting can actually abrogate the antitumor effect of OX40 monotherapy, by reducing TIL infiltration and enhancing activation-induced cell death of tumor-reactive CD8+ T cells (Shrimali et al., 2017). It is difficult to predict whether a similar deleterious effect would also take place in human cancer, in which T cell priming may have occurred many years before. However, the data do highlight that it will be crucial to investigate the timing of different immunotherapeutic interventions, to avoid similar negative effects due to T cell overstimulation. As a second word of caution, positive clinical data on immunotherapy combinations are presently still limited, and it is unclear whether the encouraging data obtained in in vitro and in vivo models will be recapitulated in cancer patients. Over the next few years, the results from ongoing early clinical trials that investigate antibodies targeting many of these inhibitory and costimulatory molecules— including additional targets such as GITR, ICOS, and CD27—as monotherapies or in combination with anti-PD-1/PD-L1, should reveal how these agents should be combined and which dysfunctional states might best be targeted by them.
Epigenetic Imprinting of Dysfunction
An additional T cell-intrinsic parameter that may influence the efficacy of immunotherapeutic intervention is the durability of reinvigoration of dysfunctional T cells upon therapy. Observations from chronic viral infection indicate a prominent role for differences in methylation of PDCD1 (encoding PD-1) in sustaining dysfunction. While the PD-1 transcriptional regulatory region is only transiently demethylated in activated T cells, it remains demethylated in exhausted T cells, resulting in prolonged expression of PD-1 (Ahn et al., 2016; Youngblood et al., 2011, 2013). This altered methylation pattern is maintained even after viral clearance to undetectable levels. Recent work reported the existence of low avidity T cell clones within the Melan-A-specific TIL population in human melanoma that are unable to express PD-1 due to continued methylation of the PDCD1 promoter even upon TCR stimulation. In contrast, antigen-specific T cells of high functional avidity within the same TIL population did express PD-1, thereby confirming a link between the strength of TCR signaling, modification of the PDCD1 locus, and PD-1 expression (Simon et al., 2016). When Melan-A-specific TILs were expanded in vitro in the presence of anti-PD-1 antibodies, the T cell repertoire was skewed toward higher avidity T cell clonotypes. In addition to the sustained demethylation of PDCD1, dysfunctional T cells also progressively acquire de novo methylation of genes associated with effector functions during both infections and tumor development, which could not be reverted by PD-1 blockade (Ghoneim et al., 2017). Of note, blockade of these exhaustion-associated methylation programs increased T cell responses and tumor control upon PD-1 blockade in murine tumor models.
On a related note, several reports have recently demonstrated that dysfunctional T cells display specific alterations in chromatin accessibility. Part of this chromatin remodeling is likely driven by the initial antigen stimulation as based on the overlap in chromatin accessibility patterns among effector, memory, and exhausted T cells (Scott-Browne et al., 2016). This notion is further supported by the observation that a fixed state of dysfunction was only induced in tumor-specific T cells, when tumor-specific and bystander T cells were co-transferred in a murine liver cancer model (Schietinger et al., 2016). Thus, micro-environmental factors were not sufficient to induce this fixed exhausted state by themselves, but appear to shape the effects of TCR triggering on intratumoral T cells. Next to the chromatin remodeling patterns that are shared with effector and memory T cells, dysfunctional T cells also display changes in chromatin accessible regions that are specific to these cells. For instance, exhausted T cells in chronic LCMV infection showed additional accessible regions in the PDCD1 locus and lacked several open regions in the IFNG locus that were present in functional T cells (Pauken et al., 2016; Sen et al., 2016). Of note, many of these accessible regions specific to the exhausted state are retained after anti-PD-1 treatment, indicating that this therapy only induces minor epigenetic remodeling (Pauken et al., 2016). In line with this observation, exhausted virus-specific T cells that have regained effector functions upon PD-L1 blockade could not undergo memory development after viral antigen clearance and became dysfunctional again when the infection persisted. A similar observation was made in a murine tumor model, in which both tumor antigen-specific and bystander T cells were adoptively transferred and gene expression as well as chromatin accessibility were compared (Mognol et al., 2017). Only tumor-specific T cells acquired characteristics of dysfunction and, while displaying improved effector functions upon treatment with anti-PD-L1 therapy, these cells showed only very limited alterations in chromatin accessibility patterns.
Interestingly,the imprintingof thisepigeneticstate already starts early during exhaustion, as T cells from premalignant lesions harbor many of the alterations found in exhausted T cells in late-stage tumors or metastases in a murine liver cancer model (Schietinger et al., 2016). This progression from mild exhaustion in early tumor lesions to more severe exhaustion in advanced tumors is reflected by two distinct chromatin states. Whereas the chromatin state of early dysfunctional TILsstill displays some plasticityand can be reprogrammed by PD-1/PD-L1 blockade, the later state is fixed and cannot be altered by anti-PD-1 therapy (Philip et al., 2017). Strikingly, the chromatin accessibility state of PD-1hi CD8+ TILs from human NSCLC was largely overlapping with that of murine late-stage TILs. Hence, also in human tumors PD-1/PD-L1 blockade might only induce transcriptional rewiring due to increased binding of transcription factors to accessible chromatin regions in the more severely dysfunctional T cells, rather than a change in chromatin accessibility patterns themselves. In vitro treatment with histone deacetylase inhibitors has been shown to lead to significantly improved effector functions and memory transition in adoptively transferred exhausted LCMV clone 13-specific T cells (Zhang et al., 2014). Whether this effect is observed on all exhausted T cells and leads to a durable reinvigoration of these cells in combination with PD-1 blockade needs to be further investigated.
Therapy-Induced Resistance and T Cell Dysfunction
Cancer immunotherapy-induced resistance can in some patients be explained by tumor-intrinsic alterations, such as defects in the antigen-presentation machinery or inactivation of the IFNg signaling pathway (Patel et al., 2017; Restifo et al., 1996; Zaretsky et al., 2016). In addition, therapy-induced changes in the functional state of TILs have been suggested to form a cause of resistance to both targeted treatments and immunotherapy. For instance, genomic and non-genomic alterations evolving in BRAF-mutated melanoma upon treatment with MAPK inhibitors co-developed with dysfunction of CD8+ T cells in a subset of resistant tumors. Of note, this often correlated with an enhanced expression of an inflammation signature associated with M2 macrophages that are known to antagonize T cell recruitment and effector function (Hugo et al., 2015). Direct evidence that these changes play a major role in the observed resistance is however currently lacking. Upon adoptive transfer of MART-1 TCR-transduced T cells in patients with advanced melanoma, a change in the functional phenotype of the T cells has been observed at the time of relapse (Ma et al., 2013). Specifically, TCR-transgenic T cells either completely lost their initial cytotoxic activity or acquired a distinct functional profile with secretion of inflammatory cytokines, but lack of cytotoxicity.
Therapy-induced reinvigoration of tumor-specific T cell responses may be expected to in some cases induce the further development of T cell dysfunction. In particular, the IFNg secretion that occurs as a consequence of T cell reactivation may induce immunosuppressive counterbalance mechanisms (Figure 1). In addition, the IFNg-induced increase in MHC expression could also increase the TCR signal that contributes to T cell dysfunction. Recent evidence from a study comparing longitudinal samples from immunocompetent versus immunodeficient murine colon cancer models indicated that immune pressure on tumor cells induced two major tumor escape mechanisms that involved the accumulation of immunosuppressive cell populations, such as Tregs, and the increased expression of multiple inhibitory receptors on T cells (Efremova et al., 2018). The upregulation of alternative immune checkpoints has also been postulated as a possible resistance mechanism upon anti-PD-1 therapy. Data from HNSCC indicated that PD-1 blockade of TILs in vitro as well as in a murine tumor model led to upregulation of Tim-3. Of note, combined blockade of both receptors significantly increased antitumor activity in this model (Shayan et al., 2017). Another study described increased TIGIT expression on NYESO-1-specific CD8+ T cells upon PD-1 blockade in vitro (Chauvin et al., 2015). Upregulation of multiple alternative immune checkpoints including Tim-3, Lag-3, and CTLA-4 upon anti-PD-1 resistance was also observed in a murine lung cancer model (Koyama et al., 2016). Direct evidence of whether this expression of alternative immune checkpoints truly represents a mechanism of resistance, or simply reflects a more activated state of these T cells upon reinvigoration by immunotherapy, is difficult to obtain. However, support for the former model comes from work that assessed resistance to CTLA-4 blockade and radiation in a murine melanoma model (Benci et al., 2016). In these experiments, therapy resistance was accompanied by expression of multiple inhibitory receptor ligands on tumor cells and high inhibitory receptor expression on T cells. While triple or quadruple immune checkpoint blockade combinations were necessary to significantly improve treatment response in resistant tumors, knockout of tumor type I and/or II interferon signaling allowed for response to anti-PD-1 or anti-CTLA-4 monotherapy. To evaluate whether alterations in the dysfunctional state are also important in mediating immunotherapy resistance in cancer patients, studies comparing monotherapy and combination therapy at the start of treatment versus the moment of resistance will be of value.
Conclusions
Dysfunctional tumor-specific T cells display a broad spectrum of cell states that are induced by persistent TCR triggering, but that are additionally influenced by T cell-intrinsic and -extrinsic factors. While traits of their core exhaustion signature are shared with T cells in chronic viral infection, intratumoral T cell dysfunction is further sculpted by the multifaceted immunosuppressive processes ongoing in the TME, which can be expected to deviate from those present at chronically infected sites (Figure 1). This leads to heterogeneous (dys-)functional states between and within tumors, shaped by inhibitory and costimulatory signals from immune, stromal, and cancer cells, but also by gradients of suppressive soluble mediators, metabolic factors, and hypoxia. In order to restore antitumor immunity, an understanding of the processes that drive and maintain these different dysfunctional T cell states is essential. Novel technologies that allow characterization of these cells at the single cell level, in combination with assessment of tumor reactivity and spatial organization within the tumor, will be of value to achieve this goal and thereby support the development of personalized therapeutic strategies to target dysfunctional T cells.
Acknowledgments
This work was supported by a Swiss National Science Foundation grant (P300PB_164755/1) to D.S.T. and ERC AdG SENSIT to T.N.S.
References
- Agnellini P, Wolint P, Rehr M, Cahenzli J, Karrer U, Oxenius A. Impaired NFAT nuclear translocation results in split exhaustion of virus-specific CD8+ T cell functions during chronic viral infection. Proc Natl Acad Sci USA. 2007;104:4565–4570. doi: 10.1073/pnas.0610335104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, Rosenberg SA. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood. 2009;114:1537–1544. doi: 10.1182/blood-2008-12-195792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn E, Youngblood B, Lee J, Lee J, Sarkar S, Ahmed R. Demethylation of the PD-1 promoter is imprinted during the effector phase of CD8 T cell exhaustion. J Virol. 2016;90:8934–8946. doi: 10.1128/JVI.00798-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:749. doi: 10.1038/nrc.2016.114. [DOI] [PubMed] [Google Scholar]
- 6.Anderson AC, Joller N, Kuchroo VK. Lag-3, Tim-3, and TIGIT: Co-inhibitory receptors with specialized functions in immune regulation. Immunity. 2016;44:989–1004. doi: 10.1016/j.immuni.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ansel KM, Ngo VN, Hyman PL, Luther SA, Forster R, Sedgwick JD, Browning JL, Lipp M, Cyster JG. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature. 2000;406:309–314. doi: 10.1038/35018581. [DOI] [PubMed] [Google Scholar]
- 8.Baitsch L, Baumgaertner P, Devevre E, Raghav SK, Legat A, Barba L, Wieckowski S, Bouzourene H, Deplancke B, Romero P, et al. Exhaustion of tumor-specific CD8(+) T cells in metastases from melanoma patients. J Clin Invest. 2011;121:2350–2360. doi: 10.1172/JCI46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baitsch L, Legat A, Barba L, Fuertes Marraco SA, Rivals JP, Baumgaertner P, Christiansen-Jucht C, Bouzourene H, Rimoldi D, Pircher H, et al. Extended co-expression of inhibitory receptors by human CD8 T-cells depending on differentiation, antigen-specificity and anatomical localization. PLoS One. 2012;7:e30852. doi: 10.1371/journal.pone.0030852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balar AV, Galsky MD, Rosenberg JE, Powles T, Petrylak DP, Bellmunt J, Loriot Y, Necchi A, Hoffman-Censits J, Perez-Gracia JL, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet. 2017;389:67–76. doi: 10.1016/S0140-6736(16)32455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 12.Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, Wherry EJ, Reiner SL. Cutting edge: the transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol. 2010;185:4988–4992. doi: 10.4049/jimmunol.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, Cucolo L, Lee DSM, Pauken KE, Huang AC, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167:1540–1554.e12. doi: 10.1016/j.cell.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bengsch B, Johnson AL, Kurachi M, Odorizzi PM, Pauken KE, Attanasio J, Stelekati E, McLane LM, Paley MA, Delgoffe GM, Wherry EJ. Bioenergetic insufficiencies due to metabolic alterations regulated by the inhibitory receptor PD-1 are an early driver of CD8(+) T cell exhaustion. Immunity. 2016;45:358–373. doi: 10.1016/j.immuni.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bengsch B, Martin B, Thimme R. Restoration of HBV-specific CD8+ T cell function by PD-1 blockade in inactive carrier patients is linked to T cell differentiation. J Hepatol. 2014;61:1212–1219. doi: 10.1016/j.jhep.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Blackburn SD, Shin H, Freeman GJ, Wherry EJ. Selective expansion of a subset of exhausted CD8 T cells by alphaPD-L1 blockade. Proc Natl Acad Sci USA. 2008;105:15016–15021. doi: 10.1073/pnas.0801497105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, Betts MR, Freeman GJ, Vignali DA, Wherry EJ. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10:29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blank CU, Haanen JB, Ribas A, Schumacher TN. CANCER IMMUNOLOGY. The ‘‘cancer immunogram’’. Science. 2016;352:658–660. doi: 10.1126/science.aaf2834. [DOI] [PubMed] [Google Scholar]
- 19.Boon T, Coulie PG, Van den Eynde BJ, van der Bruggen P. Human T cell responses against melanoma. Annu Rev Immunol. 2006;24:175–208. doi: 10.1146/annurev.immunol.24.021605.090733. [DOI] [PubMed] [Google Scholar]
- 20.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bottino C, Castriconi R, Pende D, Rivera P, Nanni M, Carnemolla B, Cantoni C, Grassi J, Marcenaro S, Reymond N, et al. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med. 2003;198:557–567. doi: 10.1084/jem.20030788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bour-Jordan H, Esensten JH, Martinez-Llordella M, Penaranda C, Stumpf M, Bluestone JA. Intrinsic and extrinsic control of peripheral T-cell tolerance by costimulatory molecules of the CD28/B7 family. Immunol Rev. 2011;241:180–205. doi: 10.1111/j.1600-065X.2011.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, Aghvanyan A, Turay AM, Frauwirth KA. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J Immunol. 2010;185:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carstens JL, Correa de Sampaio P, Yang D, Barua S, Wang H, Rao A, Allison JP, LeBleu VS, Kalluri R. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat Commun. 2017;8 doi: 10.1038/ncomms15095. 15095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castle JC, Kreiter S, Diekmann J, Lower M, van de Roemer N, de Graaf J, Selmi A, Diken M, Boegel S, Paret C, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72:1081–1091. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 27.Chang CH, Curtis JD, Maggi LB, Jr., Faubert B, Villarino AV, O’Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang CH, Qiu J, O’Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chauvin JM, Pagliano O, Fourcade J, Sun Z, Wang H, Sander C, Kirkwood JM, Chen TH, Maurer M, Korman AJ, Zarour HM. TIGIT and PD-1 impair tumor antigen-specific CD8(+) T cells in melanoma patients. J Clin Invest. 2015;125:2046–2058. doi: 10.1172/JCI80445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chemnitz JM, Parry RV, Nichols KE, June CH, Riley JL. SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J Immunol. 2004;173:945–954. doi: 10.4049/jimmunol.173.2.945. [DOI] [PubMed] [Google Scholar]
- 31.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 32.Chen PL, Roh W, Reuben A, Cooper ZA, Spencer CN, Prieto PA, Miller JP, Bassett RL, Gopalakrishnan V, Wani K, et al. Analysis of immune signatures in longitudinal tumor samples yields insight into bio-markers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016;6:827–837. doi: 10.1158/2159-8290.CD-15-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen S, Lee LF, Fisher TS, Jessen B, Elliott M, Evering W, Logronio K, Tu GH, Tsaparikos K, Li X, et al. Combination of 4-1BB agonist and PD-1 antagonist promotes antitumor effector/memory CD8 T cells in a poorly immunogenic tumor model. Cancer Immunol Res. 2015;3:149–160. doi: 10.1158/2326-6066.CIR-14-0118. [DOI] [PubMed] [Google Scholar]
- 34.Chevrier S, Levine JH, Zanotelli VRT, Silina K, Schulz D, Bacac M, Ries CH, Ailles L, Jewett MAS, Moch H, et al. An immune atlas of clear cell renal cell carcinoma. Cell. 2017;169:736–749.e18. doi: 10.1016/j.cell.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi SY, Collins CC, Gout PW, Wang Y. Cancer-generated lactic acid: a regulatory, immunosuppressive metabolite? J Pathol. 2013;230:350–355. doi: 10.1002/path.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung W, Eum HH, Lee HO, Lee KM, Lee HB, Kim KT, Ryu HS, Kim S, Lee JE, Park YH, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun. 2017;8 doi: 10.1038/ncomms15081. 15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Croft M. Control of immunity by the TNFR-related molecule OX40 (CD134) Annu Rev Immunol. 2010;28:57–78. doi: 10.1146/annurev-immunol-030409-101243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 39.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 40.Di Caro G, Bergomas F, Grizzi F, Doni A, Bianchi P, Malesci A, Laghi L, Allavena P, Mantovani A, Marchesi F. Occurrence of tertiary lymphoid tissue is associated with T-cell infiltration and predicts better prognosis in early-stage colorectal cancers. Clin Cancer Res. 2014;20:2147–2158. doi: 10.1158/1078-0432.CCR-13-2590. [DOI] [PubMed] [Google Scholar]
- 41.Dieu-Nosjean MC, Antoine M, Danel C, Heudes D, Wislez M, Poulot V, Rabbe N, Laurans L, Tartour E, de Chaisemartin L, et al. Long-term survival for patients with non-small-cell lung cancer with intratumoral lymphoid structures. J Clin Oncol. 2008;26:4410–4417. doi: 10.1200/JCO.2007.15.0284. [DOI] [PubMed] [Google Scholar]
- 42.Doedens AL, Phan AT, Stradner MH, Fujimoto JK, Nguyen JV, Yang E, Johnson RS, Goldrath AW. Hypoxia-inducible factors enhance the effector responses of CD8(+) T cells to persistent antigen. Nat Immunol. 2013;14:1173–1182. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doering TA, Crawford A, Angelosanto JM, Paley MA, Ziegler CG, Wherry EJ. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity. 2012;37:1130–1144. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duraiswamy J, Kaluza KM, Freeman GJ, Coukos G. Dual blockade of PD-1 and CTLA-4 combined with tumor vaccine effectively restores T-cell rejection function in tumors. Cancer Res. 2013;73:3591–3603. doi: 10.1158/0008-5472.CAN-12-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Efremova M, Rieder D, Klepsch V, Charoentong P, Finotello F, Hackl H, Hermann-Kleiter N, Lower M, Baier G, Krogsdam A, Trajanoski Z. Targeting immune checkpoints potentiates immunoediting and changes the dynamics of tumor evolution. Nat Commun. 2018;9:32. doi: 10.1038/s41467-017-02424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erkes DA, Smith CJ, Wilski NA, Caldeira-Dantas S, Mohgbeli T, Snyder CM. Virus-specific CD8+ T cells infiltrate melanoma lesions and retain function independently of pd-1 expression. J Immunol. 2017;198:2979–2988. doi: 10.4049/jimmunol.1601064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fehlings M, Simoni Y, Penny HL, Becht E, Loh CY, Gubin MM, Ward JP, Wong SC, Schreiber RD, Newell EW. Checkpoint blockade immunotherapy reshapes the high-dimensional phenotypic heterogeneity of murine intratumoural neoantigen-specific CD8(+) T cells. Nat Commun. 2017;8:562. doi: 10.1038/s41467-017-00627-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J, Park K, Smith D, Artal-Cortes A, Lewanski C, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet. 2016;387:1837–1846. doi: 10.1016/S0140-6736(16)00587-0. [DOI] [PubMed] [Google Scholar]
- 49.Fernandez-Poma SM, Salas-Benito D, Lozano T, Casares N, Riezu-Boj JI, Mancheno U, Elizalde E, Alignani D, Zubeldia N, Otano I, et al. Expansion of tumor-infiltrating CD8+ T cells expressing PD-1 improves the efficacy of adoptive T-cell therapy. Cancer Res. 2017;77:3672–3684. doi: 10.1158/0008-5472.CAN-17-0236. [DOI] [PubMed] [Google Scholar]
- 50.Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, Gottfried E, Schwarz S, Rothe G, Hoves S, et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood. 2007;109:3812–3819. doi: 10.1182/blood-2006-07-035972. [DOI] [PubMed] [Google Scholar]
- 51.Fourcade J, Sun Z, Benallaoua M, Guillaume P, Luescher IF, Sander C, Kirkwood JM, Kuchroo V, Zarour HM. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J Exp Med. 2010;207:2175–2186. doi: 10.1084/jem.20100637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, Elstrom RL, June CH, Thompson CB. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–777. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 53.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 54.Fridman WH, Zitvogel L, Sautes-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14:717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 55.Fuertes Marraco SA, Neubert NJ, Verdeil G, Speiser DE. Inhibitory receptors beyond T cell exhaustion. Front Immunol. 2015;6:310. doi: 10.3389/fimmu.2015.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gabrilovich D, Ishida T, Oyama T, Ran S, Kravtsov V, Nadaf S, Car-bone DP. Vascular endothelial growth factor inhibits the development of dendritic cells and dramatically affects the differentiation of multiple hematopoietic lineages in vivo. Blood. 1998;92:4150–4166. [PubMed] [Google Scholar]
- 57.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghoneim HE, Fan Y, Moustaki A, Abdelsamed HA, Dash P, Dogra P, Carter R, Awad W, Neale G, Thomas PG, Youngblood B. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell. 2017;170:142–157.e19. doi: 10.1016/j.cell.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schuffler PJ, Grolimund D, Buhmann JM, Brandt S, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 60.Giordano M, Henin C, Maurizio J, Imbratta C, Bourdely P, Buferne M, Baitsch L, Vanhille L, Sieweke MH, Speiser DE, et al. Molecular profiling of CD8 T cells in autochthonous melanoma identifies Maf as driver of exhaustion. EMBO J. 2015;34:2042–2058. doi: 10.15252/embj.201490786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goc J, Germain C, Vo-Bourgais TK, Lupo A, Klein C, Knockaert S, de Chaisemartin L, Ouakrim H, Becht E, Alifano M, et al. Dendritic cells in tumor-associated tertiary lymphoid structures signal a Th1 cytotoxic immune contexture and license the positive prognostic value of infiltrating CD8+ T cells. Cancer Res. 2014;74:705–715. doi: 10.1158/0008-5472.CAN-13-1342. [DOI] [PubMed] [Google Scholar]
- 62.Gropper Y, Feferman T, Shalit T, Salame TM, Porat Z, Shakhar G. Culturing CTLs under hypoxic conditions enhances their cytolysis and improves their anti-tumor function. Cell Rep. 2017;20:2547–2555. doi: 10.1016/j.celrep.2017.08.071. [DOI] [PubMed] [Google Scholar]
- 63.Gros A, Parkhurst MR, Tran E, Pasetto A, Robbins PF, Ilyas S, Prickett TD, Gartner JJ, Crystal JS, Roberts IM, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22:433–438. doi: 10.1038/nm.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gros A, Robbins PF, Yao X, Li YF, Turcotte S, Tran E, Wunderlich JR, Mixon A, Farid S, Dudley ME, et al. PD-1 identifies the patient-specific CD8(+) tumor-reactive repertoire infiltrating human tumors. J Clin Invest. 2014;124:2246–2259. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grosso JF, Goldberg MV, Getnet D, Bruno TC, Yen HR, Pyle KJ, Hipkiss E, Vignali DA, Pardoll DM, Drake CG. Functionally distinct LAG-3 and PD-1 subsets on activated and chronically stimulated CD8 T cells. J Immunol. 2009;182:6659–6669. doi: 10.4049/jimmunol.0804211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hall A, Meyle KD, Lange MK, Klima M, Sanderhoff M, Dahl C, Abildgaard C, Thorup K, Moghimi SM, Jensen PB, et al. Dysfunctional oxidative phosphorylation makes malignant melanoma cells addicted to glycolysis driven by the (V600E)BRAF oncogene. Oncotarget. 2013;4:584–599. doi: 10.18632/oncotarget.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, Frederick DT, Hurley AD, Nellore A, Kung AL, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, et al. Follicular CXCR5-expressing CD8+ T cells curtail chronic viral infection. Nature. 2016;537:412–428. doi: 10.1038/nature19317. [DOI] [PubMed] [Google Scholar]
- 69.Heindl A, Nawaz S, Yuan Y. Mapping spatial heterogeneity in the tumor microenvironment: a new era for digital pathology. Lab Invest. 2015;95:377–384. doi: 10.1038/labinvest.2014.155. [DOI] [PubMed] [Google Scholar]
- 70.Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hugo W, Shi H, Sun L, Piva M, Song C, Kong X, Moriceau G, Hong A, Dahlman KB, Johnson DB, et al. Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell. 2015;162:1271–1285. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hui E, Cheung J, Zhu J, Su X, Taylor MJ, Wallweber HA, Sasmal DK, Huang J, Kim JM, Mellman I, Vale RD. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science. 2017;355:1428–1433. doi: 10.1126/science.aaf1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016;537:417–421. doi: 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 76.Jin HT, Anderson AC, Tan WG, West EE, Ha SJ, Araki K, Freeman GJ, Kuchroo VK, Ahmed R. Cooperation of Tim-3 and PD-1 in CD8 T-cell exhaustion during chronic viral infection. Proc. Natl Acad Sci USA. 2010;107:14733–14738. doi: 10.1073/pnas.1009731107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, Park S, Javinal V, Chiu H, Irving B, et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell. 2014;26:923–937. doi: 10.1016/j.ccell.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 78.Julliard W, Fechner JH, Mezrich JD. The aryl hydrocarbon receptor meets immunology: friend or foe? A little of both. Front Immunol. 2014;5:458. doi: 10.3389/fimmu.2014.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, Konieczny BT, Daugherty CZ, Koenig L, Yu K, et al. Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science. 2017;355:1423–1427. doi: 10.1126/science.aaf0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kao C, Oestreich KJ, Paley MA, Crawford A, Angelosanto JM, Ali MA, Intlekofer AM, Boss JM, Reiner SL, Weinmann AS, Wherry EJ. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nat Immunol. 2011;12:663–671. doi: 10.1038/ni.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kato Y, Ozawa S, Miyamoto C, Maehata Y, Suzuki A, Maeda T, Baba Y. Acidic extracellular microenvironment and cancer. Cancer Cell Int. 2013;13:89. doi: 10.1186/1475-2867-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khalili JS, Liu S, Rodriguez-Cruz TG, Whittington M, Wardell S, Liu C, Zhang M, Cooper ZA, Frederick DT, Li Y, et al. Oncogenic BRAF(V600E) promotes stromal cell-mediated immunosuppression via induction of interleukin-1 in melanoma. Clin Cancer Res. 2012;18:5329–5340. doi: 10.1158/1078-0432.CCR-12-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim HJ, Verbinnen B, Tang X, Lu L, Cantor H. Inhibition of follicular T-helper cells by CD8(+) regulatory T cells is essential for self tolerance. Nature. 2010;467:328–332. doi: 10.1038/nature09370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kjaergaard J, Tanaka J, Kim JA, Rothchild K, Weinberg A, Shu S. Therapeutic efficacy of OX-40 receptor antibody depends on tumor immunogenicity and anatomic site of tumor growth. Cancer Res. 2000;60:5514–5521. [PubMed] [Google Scholar]
- 85.Koyama S, Akbay EA, Li YY, Herter-Sprie GS, Buczkowski KA, Richards WG, Gandhi L, Redig AJ, Rodig SJ, Asahina H, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun. 2016;7 doi: 10.1038/ncomms10501. 10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kroeger DR, Milne K, Nelson BH. Tumor-infiltrating plasma cells are associated with tertiary lymphoid structures, cytolytic T-cell responses, and superior prognosis in ovarian cancer. Clin Cancer Res. 2016;22:3005–3015. doi: 10.1158/1078-0432.CCR-15-2762. [DOI] [PubMed] [Google Scholar]
- 87.Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, Crotty S. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J Immunol. 2012;188:3734–3744. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krummel MF, Allison JP. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J Exp Med. 1995;182:459–465. doi: 10.1084/jem.182.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuchroo VK, Anderson AC, Petrovas C. Coinhibitory receptors and CD8 T cell exhaustion in chronic infections. Curr Opin HIV AIDS. 2014;9:439–445. doi: 10.1097/COH.0000000000000088. [DOI] [PubMed] [Google Scholar]
- 90.Kurtulus S, Sakuishi K, Ngiow SF, Joller N, Tan DJ, Teng MW, Smyth MJ, Kuchroo VK, Anderson AC. TIGIT predominantly regulates the immune response via regulatory T cells. J Clin Invest. 2015;125:4053–4062. doi: 10.1172/JCI81187. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 91.Kvistborg P, Philips D, Kelderman S, Hageman L, Ottensmeier C, Joseph-Pietras D, Welters MJ, van der Burg S, Kapiteijn E, Michielin O, et al. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8+ T cell response. Sci Transl Med. 2014;6 doi: 10.1126/scitranslmed.3008918. 254ra128. [DOI] [PubMed] [Google Scholar]
- 92.Kvistborg P, Shu CJ, Heemskerk B, Fankhauser M, Thrue CA, Toebes M, van Rooij N, Linnemann C, van Buuren MM, Urbanus JH, et al. TIL therapy broadens the tumor-reactive CD8(+) T cell compartment in melanoma patients. Oncoimmunology. 2012;1:409–418. doi: 10.4161/onci.18851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, Remark R, Sweeney R, Becker CD, Levine JH, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017;169:750–765.e17. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Legat A, Speiser DE, Pircher H, Zehn D, Fuertes Marraco SA. Inhibitory receptor expression depends more dominantly on differentiation and activation than ‘‘exhaustion’’ of human CD8 T cells. Front Immunol. 2013;4:455. doi: 10.3389/fimmu.2013.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li J, Shayan G, Avery L, Jie HB, Gildener-Leapman N, Schmitt N, Lu BF, Kane LP, Ferris RL. Tumor-infiltrating Tim-3+ T cells proliferate avidly except when PD-1 is co-expressed: evidence for intracellular cross talk. Oncoimmunology. 2016;5:e1200778. doi: 10.1080/2162402X.2016.1200778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lind DS. Arginine and cancer. J Nutr. 2004;134:2837S–2841S. doi: 10.1093/jn/134.10.2837S. discussion 2853S. [DOI] [PubMed] [Google Scholar]
- 98.Lu X, Yang L, Yao D, Wu X, Li J, Liu X, Deng L, Huang C, Wang Y, Li D, Liu J. Tumor antigen-specific CD8+ T cells are negatively regulated by PD-1 and Tim-3 in human gastric cancer. Cell Immunol. 2017;313:43–51. doi: 10.1016/j.cellimm.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 99.Ma C, Cheung AF, Chodon T, Koya RC, Wu Z, Ng C, Avramis E, Cochran AJ, Witte ON, Baltimore D, et al. Multifunctional T-cell analyses to study response and progression in adoptive cell transfer immunotherapy. Cancer Discov. 2013;3:418–429. doi: 10.1158/2159-8290.CD-12-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol. 2017;14:11–31. doi: 10.1038/nrclinonc.2016.60. [DOI] [PubMed] [Google Scholar]
- 102.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matsuzaki J, Gnjatic S, Mhawech-Fauceglia P, Beck A, Miller A, Tsuji T, Eppolito C, Qian F, Lele S, Shrikant P, et al. Tumor-infiltrating NY-ESO-1-specific CD8+ T cells are negatively regulated by LAG-3 and PD-1 in human ovarian cancer. Proc Natl Acad Sci USA. 2010;107:7875–7880. doi: 10.1073/pnas.1003345107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Medaglia C, Giladi A, Stoler-Barak L, De Giovanni M, Salame TM, Biram A, David E, Li H, Iannacone M, Shulman Z, Amit I. Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Science. 2017;358:1622–1626. doi: 10.1126/science.aao4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mognol GP, Spreafico R, Wong V, Scott-Browne JP, Togher S, Hoffmann A, Hogan PG, Rao A, Trifari S. Exhaustion-associated regulatory regions in CD8(+) tumor-infiltrating T cells. Proc Natl Acad Sci USA. 2017;114:E2776–E2785. doi: 10.1073/pnas.1620498114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Munder M, Choi BS, Rogers M, Kropf P. L-arginine deprivation impairs Leishmania major-specific T-cell responses. Eur J Immunol. 2009;39:2161–2172. doi: 10.1002/eji.200839041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Muul LM, Spiess PJ, Director EP, Rosenberg SA. Identification of specific cytolytic immune responses against autologous tumor in humans bearing malignant melanoma. J Immunol. 1987;138:989–995. [PubMed] [Google Scholar]
- 111.Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol. 2017;17:559–572. doi: 10.1038/nri.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Odorizzi PM, Pauken KE, Paley MA, Sharpe A, Wherry EJ. Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J Exp Med. 2015;212:1125–1137. doi: 10.1084/jem.20142237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik MA, Tanner S. Highly multiparametric analysis by mass cytometry. J Immunol Methods. 2010;361:1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 114.Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, Bikoff EK, Robertson EJ, Lauer GM, Reiner SL, Wherry EJ. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 2012;338:1220–1225. doi: 10.1126/science.1229620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Park BV, Freeman ZT, Ghasemzadeh A, Chattergoon MA, Rutebemberwa A, Steigner J, Winter ME, Huynh TV, Sebald SM, Lee SJ, et al. TGFbeta1-mediated SMAD3 enhances PD-1 expression on antigen-specific T cells in cancer. Cancer Discov. 2016;6:1366–1381. doi: 10.1158/2159-8290.CD-15-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Parry RV, Chemnitz JM, Frauwirth KA, Lanfranco AR, Braunstein I, Kobayashi SV, Linsley PS, Thompson CB, Riley JL. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol Cell Biol. 2005;25:9543–9553. doi: 10.1128/MCB.25.21.9543-9553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pasetto A, Gros A, Robbins PF, Deniger DC, Prickett TD, Matus-Nicodemos R, Douek DC, Howie B, Robins H, Parkhurst MR, et al. Tumor- and neoantigen-reactive T-cell receptors can be identified based on their frequency in fresh tumor. Cancer Immunol Res. 2016;4:734–743. doi: 10.1158/2326-6066.CIR-16-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, Gartner JJ, Jia L, Steinberg SM, Yamamoto TN, et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537–542. doi: 10.1038/nature23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, Drake AM, Chen Z, Sen DR, Kurachi M, et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016;354:1160–1165. doi: 10.1126/science.aaf2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Peng D, Kryczek I, Nagarsheth N, Zhao L, Wei S, Wang W, Sun Y, Zhao E, Vatan L, Szeliga W, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527:249–253. doi: 10.1038/nature15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, et al. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6:202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Philip M, Fairchild L, Sun L, Horste EL, Camara S, Shakiba M, Scott AC, Viale A, Lauer P, Merghoub T, et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 2017;545:452–456. doi: 10.1038/nature22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pilon-Thomas S, Kodumudi KN, El-Kenawi AE, Russell S, Weber AM, Luddy K, Damaghi M, Wojtkowiak JW, Mule JJ, Ibrahim-Hashim A, Gillies RJ. Neutralization of tumor acidity improves antitumor responses to immunotherapy. Cancer Res. 2016;76:1381–1390. doi: 10.1158/0008-5472.CAN-15-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Platten M, Wick W, Van den Eynde BJ. Tryptophan catabolism in cancer: beyond IDO and tryptophan depletion. Cancer Res. 2012;72:5435–5440. doi: 10.1158/0008-5472.CAN-12-0569. [DOI] [PubMed] [Google Scholar]
- 126.Pollok KE, Kim YJ, Zhou Z, Hurtado J, Kim KK, Pickard RT, Kwon BS. Inducible T cell antigen 4-1BB. Analysis of expression and function. J Immunol. 1993;150:771–781. [PubMed] [Google Scholar]
- 127.Poschke I, Faryna M, Bergmann F, Flossdorf M, Lauenstein C, Hermes J, Hinz U, Hank T, Ehrenberg R, Volkmar M, et al. Identification of a tumor-reactive T-cell repertoire in the immune infiltrate of patients with resectable pancreatic ductal adenocarcinoma. Oncoimmunology. 2016;5:e1240859. doi: 10.1080/2162402X.2016.1240859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, Julg B, Jesneck JL, Brosnahan K, Imam S, et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med. 2010;16:1147–1151. doi: 10.1038/nm.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Restifo NP, Marincola FM, Kawakami Y, Taubenberger J, Yannelli JR, Rosenberg SA. Loss of functional beta 2-microglobulin in metastatic melanomas from five patients receiving immunotherapy. J Natl Cancer Inst. 1996;88:100–108. doi: 10.1093/jnci/88.2.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, Lin JC, Teer JK, Cliften P, Tycksen E, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C, Kalinka-Warzocha E, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372:320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 132.Robert C, Ribas A, Wolchok JD, Hodi FS, Hamid O, Kefford R, Weber JS, Joshua AM, Hwu WJ, Gangadhar TC, et al. Anti-programmed-death-receptor-1 treatment with pembrolizumab in ipilimumab-refractory advanced melanoma: a randomised dose-comparison cohort of a phase 1 trial. Lancet. 2014;384:1109–1117. doi: 10.1016/S0140-6736(14)60958-2. [DOI] [PubMed] [Google Scholar]
- 133.Robert C, Thomas L, Bondarenko I, O’Day S, Weber J, Garbe C, Lebbe C, Baurain JF, Testori A, Grob JJ, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364:2517–2526. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 134.Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, Dawson N, O’Donnell PH, Balmanoukian A, Loriot Y, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med. 2010;207:2187–2194. doi: 10.1084/jem.20100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sautes-Fridman C, Lawand M, Giraldo NA, Kaplon H, Germain C, Fridman WH, Dieu-Nosjean MC. Tertiary lymphoid structures in cancers: prognostic value, regulation, and manipulation for therapeutic intervention. Front Immunol. 2016;7:407. doi: 10.3389/fimmu.2016.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Scharping NE, Delgoffe GM. Tumor microenvironment metabolism: a new checkpoint for anti-tumor immunity. Vaccines (Basel) 2016;4:E46. doi: 10.3390/vaccines4040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Scharping NE, Menk AV, Moreci RS, Whetstone RD, Dadey RE, Watkins SC, Ferris RL, Delgoffe GM. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity. 2016;45:701–703. doi: 10.1016/j.immuni.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 139.Schietinger A, Philip M, Krisnawan VE, Chiu EY, Delrow JJ, Basom RS, Lauer P, Brockstedt DG, Knoblaugh SE, Hammerling GJ, et al. Tumor-specific T cell dysfunction is a dynamic antigen-driven differentiation program initiated early during tumorigenesis. Immunity. 2016;45:389–401. doi: 10.1016/j.immuni.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 141.Schreiner J, Thommen DS, Herzig P, Bacac M, Klein C, Roller A, Belousov A, Levitsky V, Savic S, Moersig W, et al. Expression of inhibitory receptors on intratumoral T cells modulates the activity of a T cellbispecific antibody targeting folate receptor. Oncoimmunology. 2016;5:e1062969. doi: 10.1080/2162402X.2015.1062969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Scott-Browne JP, Lopez-Moyado IF, Trifari S, Wong V, Chavez L, Rao A, Pereira RM. Dynamic changes in chromatin accessibility occur in CD8(+) T cells responding to viral infection. Immunity. 2016;45:1327–1340. doi: 10.1016/j.immuni.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, Heath K, McClanahan T, Lunceford J, Gause C, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012)an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016;17:956–965. doi: 10.1016/S1470-2045(16)30066-3. [DOI] [PubMed] [Google Scholar]
- 144.Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, Tsao HW, Godec J, LaFleur MW, Brown FD, et al. The epigenetic landscape of T cell exhaustion. Science. 2016;354:1165–1169. doi: 10.1126/science.aae0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shayan G, Srivastava R, Li J, Schmitt N, Kane LP, Ferris RL. Adaptive resistance to anti-PD1 therapy by Tim-3 upregulation is mediated by the PI3K-Akt pathway in head and neck cancer. Oncoimmunology. 2017;6:e1261779. doi: 10.1080/2162402X.2016.1261779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shin H, Blackburn SD, Intlekofer AM, Kao C, Angelosanto JM, Reiner SL, Wherry EJ. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 2009;31:309–320. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shrimali RK, Ahmad S, Verma V, Zeng P, Ananth S, Gaur P, Gittelman RM, Yusko E, Sanders C, Robins H, et al. Concurrent PD-1 blockade negates the effects of OX40 agonist antibody in combination immunotherapy through inducing T-cell apoptosis. Cancer Immunol Res. 2017;5:755–766. doi: 10.1158/2326-6066.CIR-17-0292. [DOI] [PubMed] [Google Scholar]
- 148.Simon S, Vignard V, Florenceau L, Dreno B, Khammari A, Lang F, Labarriere N. PD-1 expression conditions T cell avidity within an antigen-specific repertoire. Oncoimmunology. 2016;5:e1104448. doi: 10.1080/2162402X.2015.1104448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Simpson TR, Li F, Montalvo-Ortiz W, Sepulveda MA, Bergerhoff K, Arce F, Roddie C, Henry JY, Yagita H, Wolchok JD, et al. Fcdependent depletion of tumor-infiltrating regulatory T cells co-defines the effi-cacy of anti-CTLA-4 therapy against melanoma. J Exp Med. 2013;210:1695–1710. doi: 10.1084/jem.20130579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, Nyman J, Sakuishi K, Kurtulus S, Gennert D, et al. A distinct gene module for dysfunction uncoupled from activation in tumor-infiltrating T cells. Cell. 2016;166:1500–1511.e9. doi: 10.1016/j.cell.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Siska PJ, Beckermann KE, Mason FM, Andrejeva G, Greenplate AR, Sendor AB, Chiang YJ, Corona AL, Gemta LF, Vincent BG, et al. Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight. 2017;2 doi: 10.1172/jci.insight.93411. 93411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Speiser DE, Ho PC, Verdeil G. Regulatory circuits of T cell function in cancer. Nat Rev Immunol. 2016;16:599–611. doi: 10.1038/nri.2016.80. [DOI] [PubMed] [Google Scholar]
- 153.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic betacatenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 154.Staron MM, Gray SM, Marshall HD, Parish IA, Chen JH, Perry CJ, Cui G, Li MO, Kaech SM. The transcription factor FoxO1 sustains expression of the inhibitory receptor PD-1 and survival of antiviral CD8(+) T cells during chronic infection. Immunity. 2014;41:802–814. doi: 10.1016/j.immuni.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sun Z, Fourcade J, Pagliano O, Chauvin JM, Sander C, Kirkwood JM, Zarour HM. IL-10 and PD-1 cooperate to limit the activity of tumor-specific CD8+ T cells. Cancer Res. 2015;75:1635–1644. doi: 10.1158/0008-5472.CAN-14-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.te Raa GD, Pascutti MF, Garcia-Vallejo JJ, Reinen E, Remmerswaal EB, ten Berge IJ, van Lier RA, Eldering E, van Oers MH, Tonino SH, Kater AP. CMV-specific CD8+ T-cell function is not impaired in chronic lymphocytic leukemia. Blood. 2014;123:717–724. doi: 10.1182/blood-2013-08-518183. [DOI] [PubMed] [Google Scholar]
- 158.Teft WA, Chau TA, Madrenas J. Structure-Function analysis of the CTLA-4 interaction with PP2A. BMC Immunol. 2009;10:23. doi: 10.1186/1471-2172-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Terme M, Pernot S, Marcheteau E, Sandoval F, Benhamouda N, Colussi O, Dubreuil O, Carpentier AF, Tartour E, Taieb J. VEGFA-VEGFR pathway blockade inhibits tumor-induced regulatory T-cell proliferation in colorectal cancer. Cancer Res. 2013;73:539–549. doi: 10.1158/0008-5472.CAN-12-2325. [DOI] [PubMed] [Google Scholar]
- 160.Thommen DS, Schreiner J, Muller P, Herzig P, Roller A, Belousov A, Umana P, Pisa P, Klein C, Bacac M, et al. Progression of lung cancer is associated with increased dysfunction of T cells defined by coexpression of multiple inhibitory receptors. Cancer Immunol Res. 2015;3:1344–1355. doi: 10.1158/2326-6066.CIR-15-0097. [DOI] [PubMed] [Google Scholar]
- 161.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Topalian SL, Solomon D, Rosenberg SA. Tumor-specific cytolysis by lymphocytes infiltrating human melanomas. J Immunol. 1989;142:3714–3725. [PubMed] [Google Scholar]
- 163.Tran KQ, Zhou J, Durflinger KH, Langhan MM, Shelton TE, Wunderlich JR, Robbins PF, Rosenberg SA, Dudley ME. Minimally cultured tumor-infiltrating lymphocytes display optimal characteristics for adoptive cell therapy. J Immunother. 2008;31:742–751. doi: 10.1097/CJI.0b013e31818403d5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Trautmann L, Janbazian L, Chomont N, Said EA, Gimmig S, Bessette B, Boulassel MR, Delwart E, Sepulveda H, Balderas RS, et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat Med. 2006;12:1198–1202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- 165.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, Danilo M, Alfei F, Hofmann M, Wieland D, et al. T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity. 2016;45:415–427. doi: 10.1016/j.immuni.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 167.van der Windt GJ, O’Sullivan D, Everts B, Huang SC, Buck MD, Curtis JD, Chang CH, Smith AM, Ai T, Faubert B, et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc Natl Acad Sci USA. 2013;110:14336–14341. doi: 10.1073/pnas.1221740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.van der Windt GJ, Pearce EL. Metabolic switching and fuel choice during T-cell differentiation and memory development. Immunol Rev. 2012;249:27–42. doi: 10.1111/j.1600-065X.2012.01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, van Dijk LJ, Behjati S, Hilkmann H, El Atmioui D, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumabresponsive melanoma. J Clin Oncol. 2013;31:e439–442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Walunas TL, Lenschow DJ, Bakker CY, Linsley PS, Freeman GJ, Green JM, Thompson CB, Bluestone JA. Pillars article: CTLA-4 can function as a negative regulator of T cell activation. Immunity. 1994;1:405–413. J. Immunol. 187, 3466–3474. [PubMed] [Google Scholar]
- 172.Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, Green DR. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Watts TH. Staying alive: T cell costimulation, CD28, and Bcl-xL. J Immunol. 2010;185:3785–3787. doi: 10.4049/jimmunol.1090085. [DOI] [PubMed] [Google Scholar]
- 174.Wei H, Zhao L, Hellstrom I, Hellstrom KE, Guo Y. Dual targeting of CD137 co-stimulatory and PD-1 co-inhibitory molecules for ovarian cancer immunotherapy. Oncoimmunology. 2014;3:e28248. doi: 10.4161/onci.28248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wei H, Zhao L, Li W, Fan K, Qian W, Hou S, Wang H, Dai M, Hellstrom I, Hellstrom KE, Guo Y. Combinatorial PD-1 blockade and CD137 activation has therapeutic efficacy in murine cancer models and synergizes with cisplatin. PLoS One. 2013;8:e84927. doi: 10.1371/journal.pone.0084927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Weinberg AD, Rivera MM, Prell R, Morris A, Ramstad T, Vetto JT, Urba WJ, Alvord G, Bunce C, Shields J. Engagement of the OX-40 receptor in vivo enhances antitumor immunity. J Immunol. 2000;164:2160–2169. doi: 10.4049/jimmunol.164.4.2160. [DOI] [PubMed] [Google Scholar]
- 177.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 178.Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 179.Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 180.Wlodarchak N, Xing Y. PP2A as a master regulator of the cell cycle. Crit. Rev. Biochem. Mol Biol. 2016;51:162–184. doi: 10.3109/10409238.2016.1143913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K, et al. Nivolumab plus ipilimumab in advanced melanoma. N. Engl. J Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wu CJ, O’Rourke DM, Feng GS, Johnson GR, Wang Q, Greene MI. The tyrosine phosphatase SHP-2 is required for mediating phosphatidylinositol 3-kinase/Akt activation by growth factors. Oncogene. 2001;20:6018–6025. doi: 10.1038/sj.onc.1204699. [DOI] [PubMed] [Google Scholar]
- 183.Xia H, Wang W, Crespo J, Kryczek I, Li W, Wei S, Bian Z, Maj T, He M, Liu RJ, et al. Suppression of FIP200 and autophagy by tumorderived lactate promotes naive T cell apoptosis and affects tumor immunity. Sci Immunol. 2017;2 doi: 10.1126/sciimmunol.aan4631. eaan4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Youngblood B, Noto A, Porichis F, Akondy RS, Ndhlovu ZM, Austin JW, Bordi R, Procopio FA, Miura T, Allen TM, et al. Cutting edge: prolonged exposure to HIV reinforces a poised epigenetic program for PD-1 expression in virus-specific CD8 T cells. J Immunol. 2013;191:540–544. doi: 10.4049/jimmunol.1203161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Youngblood B, Oestreich KJ, Ha SJ, Duraiswamy J, Akondy RS, West EE, Wei Z, Lu P, Austin JW, Riley JL, et al. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity. 2011;35:400–412. doi: 10.1016/j.immuni.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, Tom I, Ivelja S, Refino CJ, Clark H, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol. 2009;10:48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- 187.Yuan Y, Failmezger H, Rueda OM, Ali HR, Graf S, Chin SF, Schwarz RF, Curtis C, Dunning MJ, Bardwell H, et al. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3004330. 157ra143. [DOI] [PubMed] [Google Scholar]
- 188.Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, Ahmed R. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. 1998;188:2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zarour HM. Reversing T-cell dysfunction and exhaustion in cancer. Clin Cancer Res. 2016;22:1856–1864. doi: 10.1158/1078-0432.CCR-15-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhang F, Zhou X, DiSpirito JR, Wang C, Wang Y, Shen H. Epigenetic manipulation restores functions of defective CD8(+) T cells from chronic viral infection. Mol Ther. 2014;22:1698–1706. doi: 10.1038/mt.2014.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhang Y, Ertl HC. Starved and asphyxiated: how can CD8(+) T cells within a tumor microenvironment prevent tumor progression. Front Immunol. 2016;7:32. doi: 10.3389/fimmu.2016.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, Kang B, Hu R, Huang JY, Zhang Q, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356.e16. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 194.Zheng Y, Delgoffe GM, Meyer CF, Chan W, Powell JD. Anergic T cells are metabolically anergic. J Immunol. 2009;183:6095–6101. doi: 10.4049/jimmunol.0803510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 2010;33:229–240. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhu W, Germain C, Liu Z, Sebastian Y, Devi P, Knockaert S, Brohawn P, Lehmann K, Damotte D, Validire P, et al. A high density of tertiary lymphoid structure B cells in lung tumors is associated with increased CD4+ T cell receptor repertoire clonality. Oncoimmunology. 2015;4:e1051922. doi: 10.1080/2162402X.2015.1051922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zippelius A, Batard P, Rubio-Godoy V, Bioley G, Lienard D, Lejeune F, Rimoldi D, Guillaume P, Meidenbauer N, Mackensen A, et al. Effector function of human tumor-specific CD8 T cells in melanoma lesions: a state of local functional tolerance. Cancer Res. 2004;64:2865–2873. doi: 10.1158/0008-5472.can-03-3066. [DOI] [PubMed] [Google Scholar]