Abstract
The Reoviridae have double-stranded RNA genomes of 10–12 segments, each in a single copy in the mature virion. The basis of genome segment sorting during virus assembly that ensures each virus particle contains the complete viral genome is unresolved. Bluetongue virus (BTV) NS2 is a single-stranded RNA-binding protein that forms inclusion bodies in infected cells. Here, we demonstrate that the specific interaction between NS2 and a stem-loop structure present in BTV S10 RNA, and phylogenetically conserved in other BTV serotypes, is abolished by mutations predicted to disrupt the structure. Subsequently, we mapped RNA regions in three other genomic segments of BTV that are bound preferentially by NS2. However, structure probing of these RNAs did not reveal secondary structure motifs that obviously resembled the stem- loop implicated in the NS2–S10 interaction. In addition, the specific binding by NS2 to two different viral RNAs was found to occur independently. Together, these data support the hypothesis that the recognition by NS2 of different RNA structures may be the basis for discrimination between viral RNAs during virus assembly.
Keywords: BTV, Bluetongue, NS2, Inclusion body, Reoviridae, Orbivirus, Reovirus, RNA sorting, Packaging
Introduction
The Reoviridae contains viruses with segmented double stranded RNA (dsRNA) genomes. Each mature virus particle contains a single copy of each of 10–12 dsRNA genome segments, depending on the virus (Bellamy et al., 1967; Shatkin et al., 1968). Reassortment of specific genome segments between different isolates infecting the same cell has been well documented, and defective interfering RNA segments packaged instead of a single cognate genome segment have been reported (Ahmed and Fields, 1981; Anzola et al., 1987; Kimura and Black, 1972; Nonoyama and Graham, 1970; Reddy and Black, 1977; Spandidos et al., 1976). Together, these observations have led to the conclusion that genome segments are selectively packaged into virus particles. Analysis of reovirus and rotavirus temperature-sensitive mutants has implicated a viral nonstructural protein present in inclusion bodies as necessary for the packaging of the viral genome (Ramig and Petrie, 1984; Ramig et al., 1978; Taraporewala et. al., 2002). Unlike the rotavirus and reovirus systems, Blue- tongue virus (BTV) inclusion bodies contain only a single major viral nonstructural protein, NS2. In addition to forming inclusion bodies and interacting with structural proteins of the virion, the 41-kDa NS2 protein has been shown to bind ssRNA (Huismans et al., 1987; Thomas et al., 1990). Initial studies indicated that, consistent with its highly charged amino acid sequence, the affinity of NS2 for RNA was relatively nonspecific (Huismans et al., 1987; Taraporewala et al., 2001; Thomas et al., 1990; Uitenweerde et al., 1995). However, evidence from cross linking studies using radiolabeled viral RNA and NS2 produced during virus replication in mammalian cells has shown that there is an interaction between NS2 and viral RNA (Theron and Nel, 1997). These data are supported by subsequent in vitro studies using baculovirus expressed NS2 that confirmed the preferential and specific binding of BTV RNA by NS2 (Lymperopoulos et al., 2003; Markotter et al., 2004). In one of these studies, the region of the smallest segment of genomic RNA (S10) that interacts with NS2 was mapped to a specific RNA domain using purified recombinant protein and viral RNA in electrophoretic mobility shift assays (EMSA) (Lymperopoulos et al., 2003). RNA structure mapping using chemical and enzymatic probes was consistent with the formation of a stem-loop structure in this region. However, mutagenesis experiments aimed at disrupting the predicted structure were not carried out (Lymperopoulos et al., 2003).
In the present study, we demonstrate that destabilization of this stem-loop previously mapped to the 5’-end of the S10 coding sequence abolishes specific RNA binding of S10 by NS2. Subsequently, we employed electrophoretic mobility shift assays (EMSA) to identify the presence of NS2 interacting regions in the plus-strand RNA of three other viral segments; S8, S9 and M6 and carried out RNA structure probing of these regions. Intriguingly, although there is some weak similarity between the structure likely bound by NS2 in M6 with that already described for S10, the structures present in S8 and S9 are quite distinct. Together, these data are consistent with the hypothesis that the NS2–RNA interaction is based on RNA structure but suggest that different RNA structures may be the basis for discrimination between viral RNAs during virus assembly.
Results
Mutational analysis of the specific binding site of NS2 within BTV segment 10
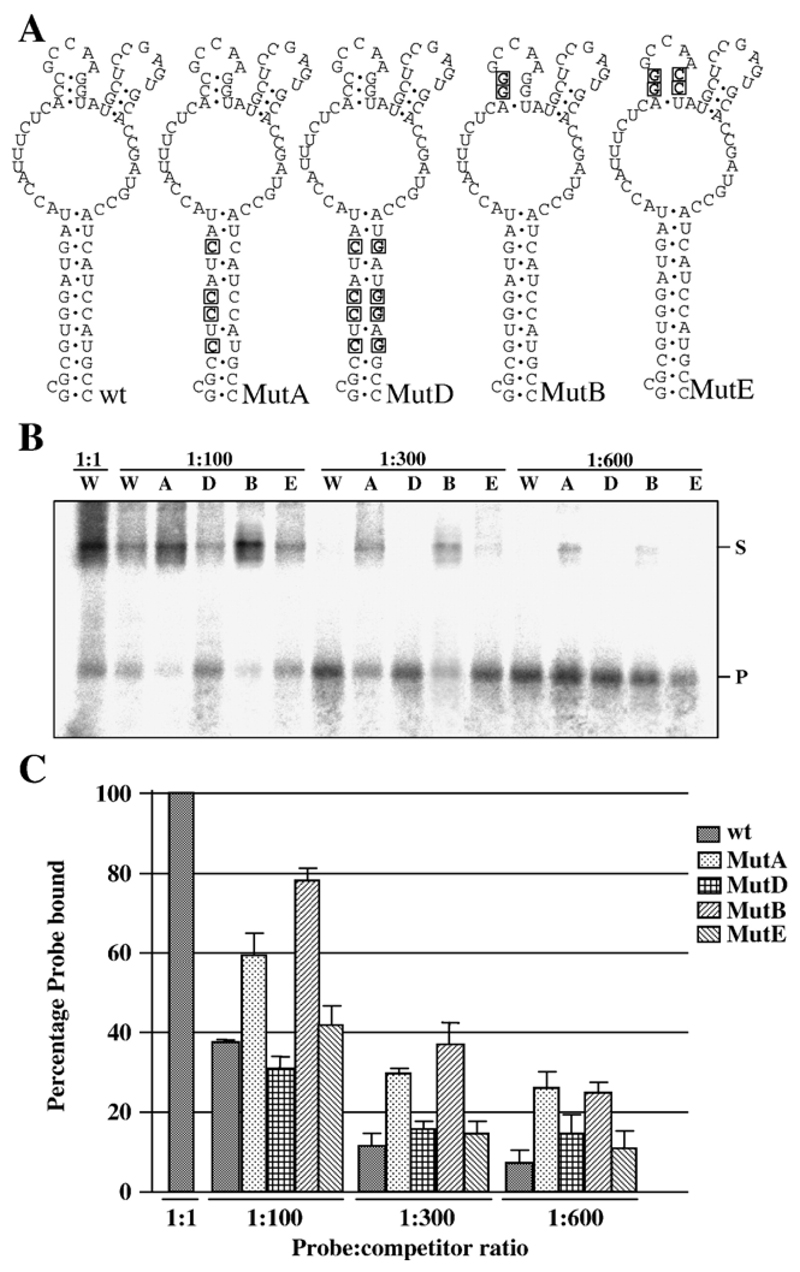
NS2 binds preferentially to a 70-nucleotide sequence located between nucleotides 99 and 169 of BTV S10 plus- strand RNA (Lymperopoulos et al., 2003). To establish whether the interaction of NS2 with this RNA was dependent on RNA secondary structure or primary sequence, we generated mutants designed to disrupt the structure of the RNA, with limited changes in the primary sequence. One mutant was predicted to destabilize the main stem by exchanging four guanines for cytosines (Fig. 1A, MutA). A second mutation was designed to similarly destabilize one of the stems that allow formation of the smaller stem-loops (Fig. 1A, MutB). To confirm that any reduction in binding affinity due to these mutations was attributable to disruption of the RNA structure and not alteration of the primary sequence, compensating mutations designed to restore the structure but not the sequence of the wild-type RNA was also tested (Fig. 1A, MutD and MutE respectively).
Fig. 1. Interaction between NS2 and mutant S10 RNAs.
(A) Mutations introduced into the wild-type (wt) sequence in BTV-10 S10 between nucleotides 99 and 169 are boxed and in bold. Changes were designed to destabilize the major stem (MutA) or a stem supporting a minor loop (MutB). Compensating mutations designed to allow the major stem (MutD) or minor stem (MutE) to reform were also produced. (B) EMSA data using purified NS2 and RNAs shown in panel A. All binding reactions were performed with 1 fmol probe and either 1:1, 1:100, 1:300 or 1:600 molar ratio of probe:competitor as indicated. W, A, D, B and E indicate that competitor RNA was wild type, MutA, MutD, MutB or MutE respectively. Position of free probe (P) and probe complexed with NS2 (S) are indicated. (C) Graph of combined data from three independent EMSA experiments. Error bars indicate the standard error of the mean. Note that second-site mutations that restore the RNA structure but not primary sequence show similar competition to wild-type RNA.
EMSA competition experiments using 32P-labeled S10 RNA as probe and increasing concentrations of each mutant subgenomic RNA as competitor were performed. The amount of NS2 in the binding reaction was adjusted so that 2 fmol of RNA saturated NS2 binding as indicated by some unbound probe in the reaction (Fig. 1B, far left lane). Under these conditions, wild-type unlabeled S10 competitor RNA reduced binding by 63% when present in 100-fold molar excess (Fig. 1C). In contrast, MutA, designed to destroy the major stem of the S10 structure bound by NS2, and MutB, which destabilized one of the minor stems, resulted in only 35% and 20% reductions in the amount of wild-type probe bound respectively. Interestingly, second site mutations designed to restore the structure of the RNA to wild type but preserve the original mutations (MutD and MutB) were as efficient as wild-type RNA in competition experiments. The relative efficiency of competition of the different RNAs was preserved at 300-fold and 600-fold molar excess of competitor RNAs (Fig. 1C).
These experiments support the hypothesis that NS2–S10 interaction is mediated by a structure present in the viral RNA as mutations disrupting this structure reduced the affinity of binding between the RNA and NS2 and restoring the structure restored binding. The difference in binding specificity between wild-type and mutant RNAs was modest but reproducible. The likely involvement of the S10 structure in specific binding of NS2 is strongly supported by phylogenetic covariance analysis. The predicted conformations of the equivalent sequence of nine other serotypes of BTV (BTV-2, -3, -4, -8, -11, -12, -15, -17, -18) are extremely similar, with the stable stem and loop substructures rigorously conserved, despite primary sequence differences (data not shown).
Regions of BTV RNA segments that confer specificity for NS2 recognition
Structures similar to that bound by NS2 in S10 have been predicted in other BTV RNA segments (Lymperopoulos et al., 2003). To test whether these structures are indeed present and involved in NS2 binding, RNA structure mapping and EMSA assays were carried out for three additional RNA segments (S9, S8 and M6). Subfragments of these RNAs were derived and used as competitors against the binding of full-length RNAs in EMSA experiments. To maximize the probability that internal RNA structures were preserved, competitors were designed to overlap over a region of at least 50% of their length. For each segment tested, regions that were preferentially recognized by NS2 were identified.
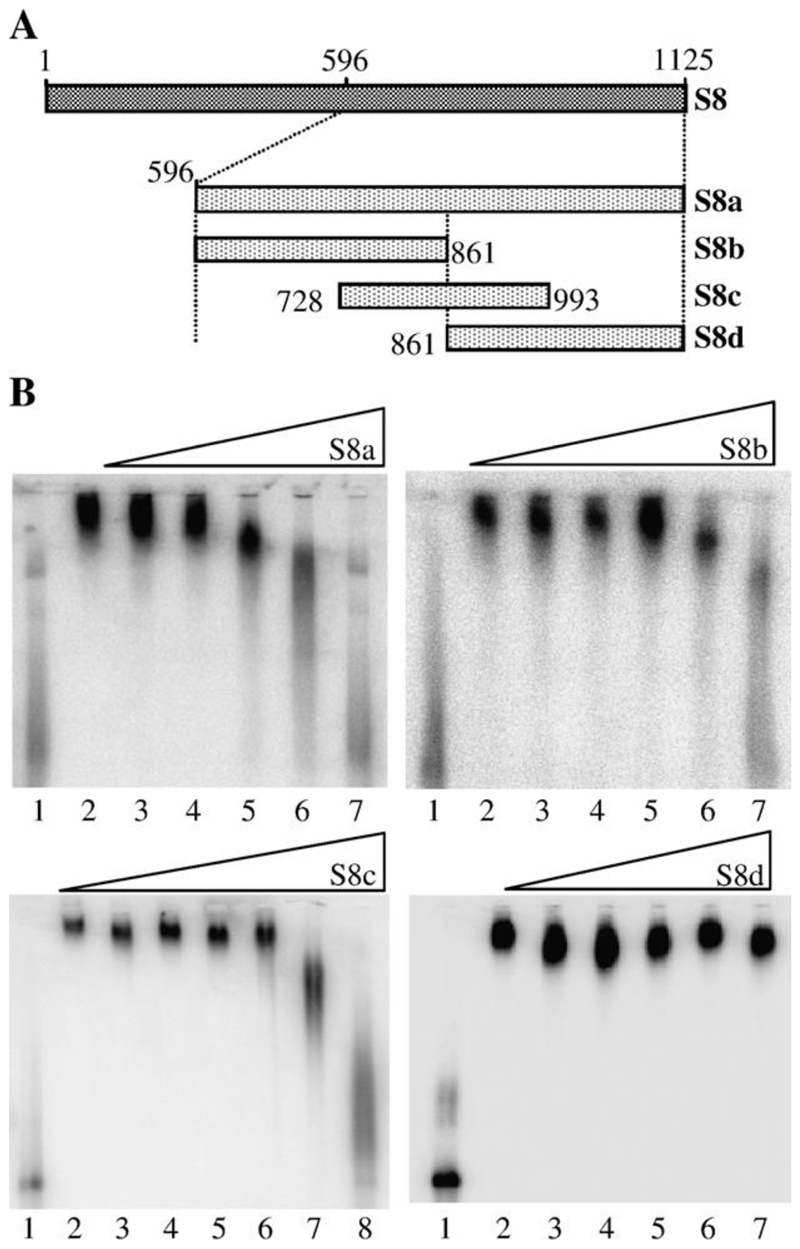
Previous studies by another group have indicated that NS2 has preference for binding nucleotides 619–1124 of the full- length S8 RNA segment (Theron and Nel, 1997). Our studies supported these data and confirmed that the 3’ half of BTV-10 S8 (nt 596–1125, S8a) was as efficient a competitor as the full- length (1–1125) RNA. However, this region is essentially the whole 3’ half of the viral RNA. S8a was further divided into three overlapping subfragments: S8b (nucleotides 596 to 861), S8c (nucleotides 728 to 993) and S8d (nucleotides 861–1125) (Fig. 2A). S8b and S8c were considerably more efficient competitors with full-length S8 RNA for NS2 binding than S8d (Fig. 2B). S8b and S8c each began competing with full-length S8 RNA at less than 1000-fold molar excess, while S8d did not compete even at 10,000-fold excess. S8b and S8c overlap by 140 nt between positions 721 and 861 in the full-length sequence. Therefore, this sequence is implicated in the binding of S8 RNA by NS2.
Fig. 2. Localization of the region of S8 RNA bound by NS2.
(A) Schematic of the truncations in S8 plus-strand RNA used as competitors in EMSA. Four fragments; S8a (nt 596–1125), S8b (nt 596–861), S8c (728–993) and S8d (861–1125) were used. (B) EMSA data. The S8 truncation used as competitor for full-length probe is shown above each panel. Lane 1 contains 1 × 10−9 M 32P- labeled full-length S8 probe only; lane 2, 32P-labeled probe and NS2. For S8a, S8b and S8d, lanes 3–7 contain 32P-labeled probe, NS2 and unlabeled competitor in a molar ratio of probe: competitor of 1:10, 1:100, 1:500, 1:1000 and 1:10,000, respectively. For S8c, lanes 3–8 contain 32P-labeled probe, NS2 and unlabeled competitor in a molar ratio of probe: competitor of 1:10, 1:100, 1:300, 1:600, 1:1000 and 1:10,000 respectively.
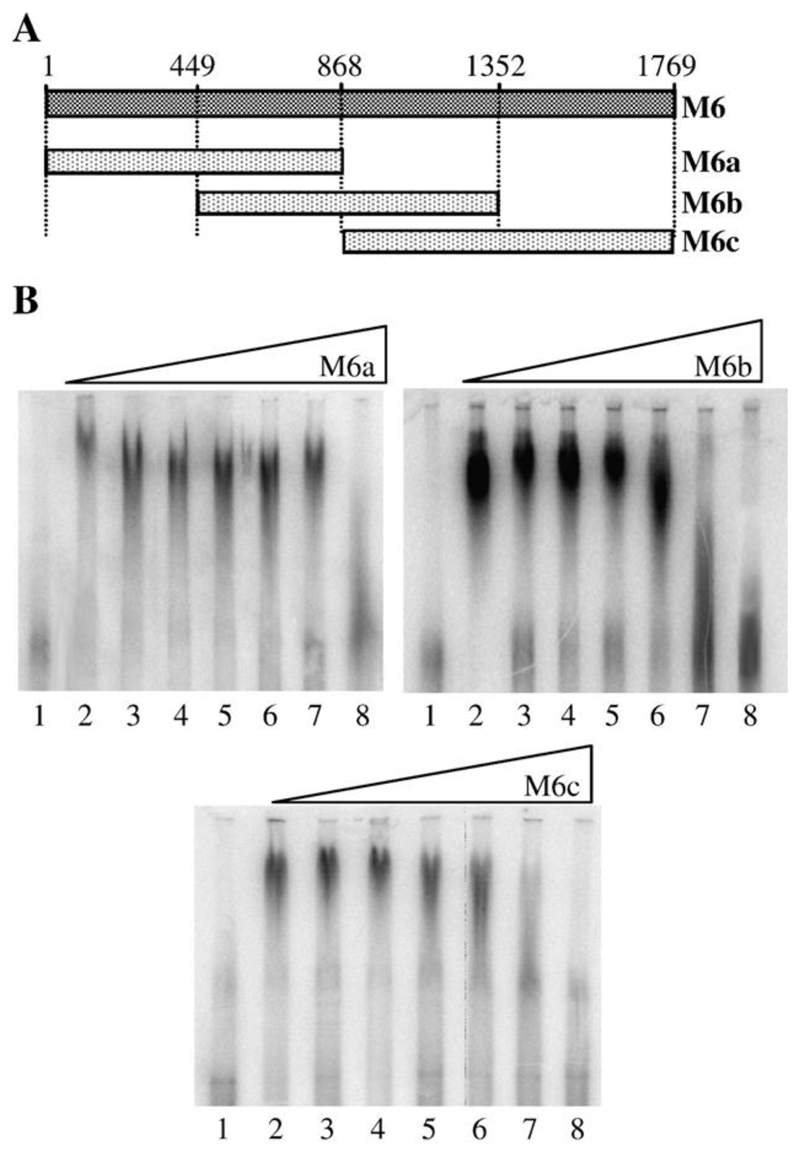
Segment M6 was chosen as a representative of the medium size RNA segments (M4, M5 and M6). As with S8, M6 plus- strand RNA was divided into three overlapping regions of similar length: M6a (nt 1–868), M6b (nt 449–1352) and M6c (nt 868–1769) (Fig. 3A). Subgenomic RNA transcripts were used as competitors for full-length M6 RNA probe in EMSA (Fig. 3B). In these experiments, the NS2-radiolabeled RNA complexes were somewhat diffuse, but it was apparent that M6b and M6c had a similar ability to compete with full-length radiolabeled M6 RNA. In each case, competition was first noted at 600-fold molar excess by the formation of lower molecular weight RNA-NS2 complexes. Competition was clearly evident at 1000-fold molar excess. In contrast, M6a was required in 10,000-fold excess before it could efficiently compete with the wild-type RNA. From these experiments, the NS2 interacting domain of M6 was localized to the overlap region of M6b and M6c (nt 901–1352).
Fig. 3. Localization of the region of M6 RNA bound by NS2.
(A) Schematic of the truncations in M6 plus-strand RNA used as competitors in EMSA. Three fragments, M6a (nt 1–868), M6b (nt 449–1352, M6c (868–1769) were used. (B) EMSA data. The identity of the M6 truncation used as competitor for binding of the full-length probe is shown above each panel. Lane 1 contains 1× 10−9 M 32P-labeled full-length M6 probe only; lane 2, 32P-labeled probe and NS2 and lanes 3–8 contain 32P-labeled probe and NS2 and unlabeled competitor in molar ratio of probe: competitor of 1:10, 1:100, 1:300, 1:600, 1:1000 and 1:10,000 respectively.
The specific interaction of NS2 with BTV S9 and S10 occurs at independent sites
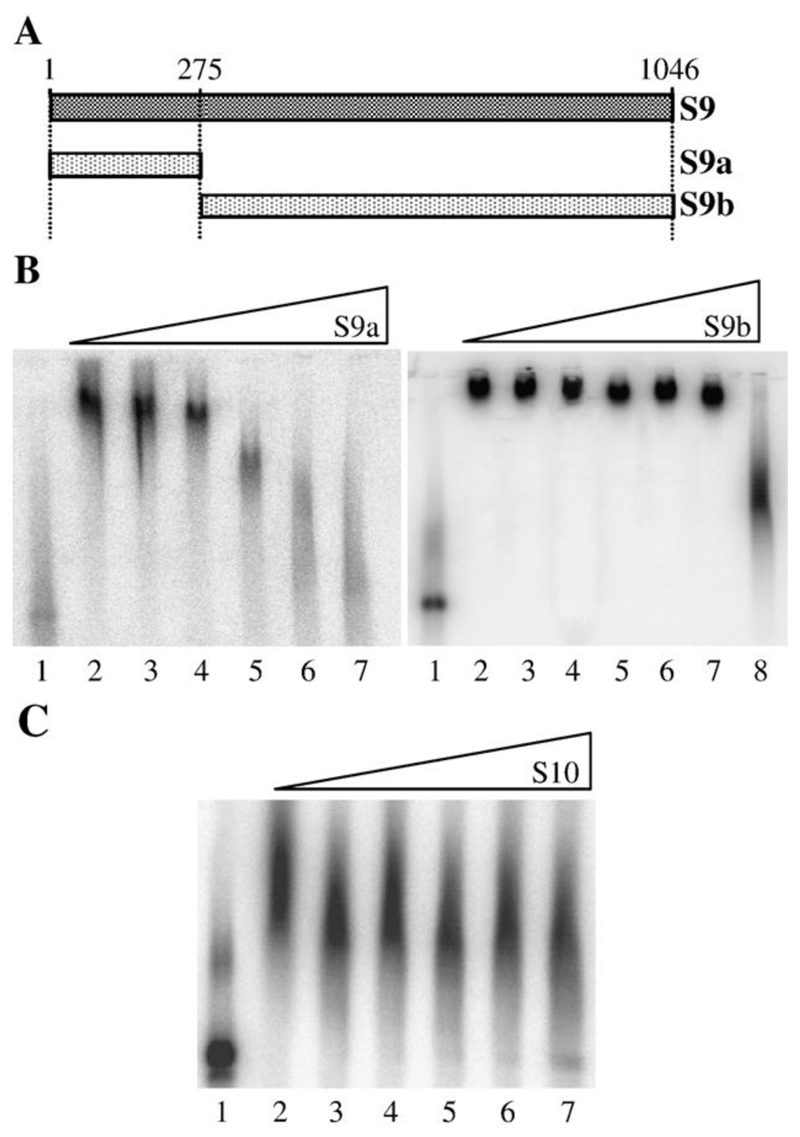
Previous EMSA analysis of S9 indicated that the 273 nt at the 5’-end of the RNA was bound preferentially by NS2 (Lymperopoulos et al., 2003). To confirm that there were no additional sequences in the S9 RNA that were specifically bound by NS2, we generated two S9 subfragments, S9a (nt1–275), containing the known binding site, and S9b (275–1046) (Fig. 4A). In competition experiments, S9a could compete with full-length S9 probe at only 300-fold molar excess, while S9b needed to be in 10,000-fold molar excess (Fig. 4B) before the major complex was disrupted. Therefore, although S9b was three times the length of S9a, its competition efficiency was much lower and equivalent to a nonspecific competitor (poly U; (Lymperopoulos et al., 2003). At this point, we had established that both the S9a and S10 RNAs were able to compete, respectively, with labeled S9 and S10 probes at about a 300-fold molar excess (Figs. 1B, 4B). To assess whether the same site on NS2 was responsible for binding both RNAs, the ability of increasing amounts of unlabeled full-length S10 RNA to compete for binding of NS2 to a full-length labeled S9 probe was tested. As seen in Fig. 4 (panel C), the S10 RNA was found to be a poor competitor for NS2 binding of the S9 RNA. This observation suggests either that there are multiple specific binding sites for viral RNAs in the NS2 complex or that there is a gradient of affinity between NS2 and the RNA of different genomic segments. However, that the S9 and S10 RNAs show similar profiles in competition assays supports more the former hypothesis.
Fig. 4. Noncompetitive binding of S9 and S10 RNA by NS2.
(A) Schematic of the truncations in S9 plus-strand RNA used as competitors in EMSA. Two fragments, S9a (nt 1–275) and S9b (nt 275–1046) were used. (B) EMSA data. The identity of the RNA used as competitor for binding of the full-length probe is shown above each panel. Lane 1 contains 1 × 10−9 M 32P-labeled full-length S9 probe only; lane 2, 32P-labeled probe and NS2 and lanes 3–8 contain 32P- labeled probe and NS2 and unlabeled competitor in molar ratio of probe: competitor of 1:10, 1:100, 1:300, 1:600, 1:1000 and (for S9b) 1:10,000, respectively. (C) In this EMSA, radiolabeled S9 (1 × 10−9 M) was challenged with unlabeled S10 RNA as competitor; lane contents as in panel B.
Structure probing of the specific NS2-binding BTV RNA subfragments
Computer-based sequence searches failed to identify con- served primary sequence motifs within the S8, S9, S10 and M6 RNA sequences (data not shown). Thus, consistent with the specific interaction identified between NS2 and the stem-loop motif present in the S10 RNA (Fig. 1), the interaction between NS2 and the S8, S9 and M6 RNAs is likely to be dependent on RNA structural motifs. Enzymatic and chemical structure probing was performed on those regions of the S9, S8 and M6 RNAs that were bound preferentially by NS2 in EMSA. Probing employed the enzymes RNase U2 and T1 that cleave single-stranded RNA (ssRNA) with a preference for A and G respectively, and CV1 that cleaves double-stranded and stacked RNA. In addition, the chemical probes imidazole and lead acetate were used to specifically cleave ssRNA with little base specificity. Representative examples of the structure mapping gels are shown in Fig. 5, and summaries are provided in Figs. 6–8. 5’-end labeled RNA was subjected to limited digestion with probing reagents prior to resolution by denaturing gel electrophoresis. Although this was a convenient methodology given the large number of RNAs to be probed, it is limited by the resolution of the gels and did not allow us to obtain a complete set of reactivities for all residues in the RNA. Therefore, structure mapping data were used as a measure of the accuracy of mfold predictions for the probed RNA. The structures shown in Figs. 6–8 represent those most consistent with the mapping data.
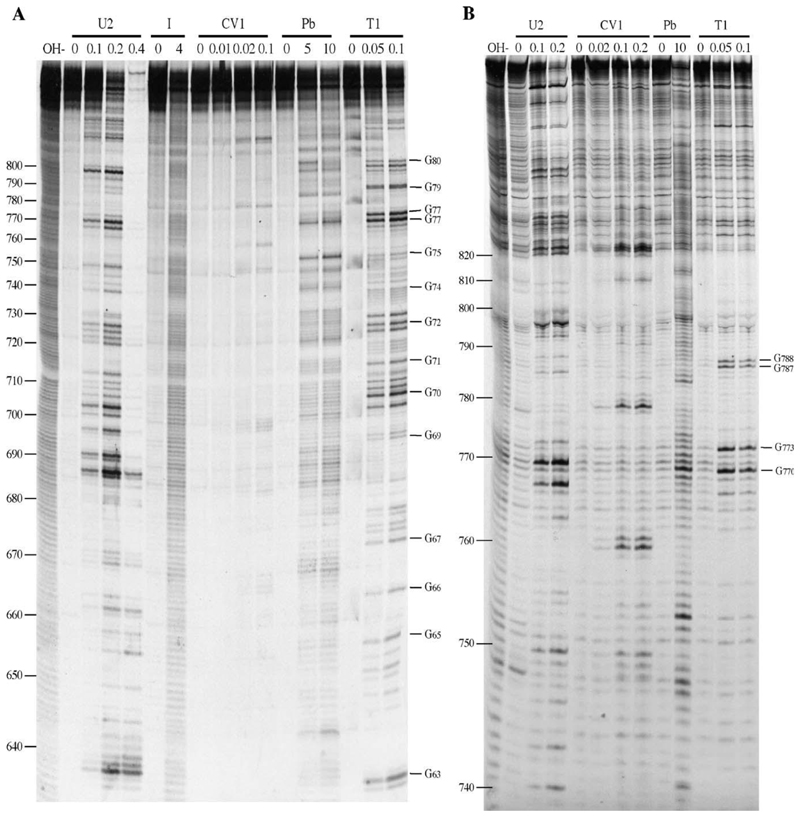
Fig. 5. Enzymatic and chemical structure probing of S8.
Base numbers correspond to those of the S8 cDNA. Products were analyzed on denaturing acrylamide/7M urea gels and data collected from 6%, 10% and 17.5% gels. Enzyme units per reaction (U2, T1), hours of reaction (I [imidazole]) and mM concentration in reaction (Pb [lead]) are shown. OH–, alkaline hydrolysis ladder; R, RNA only. (A) 6% denaturing acrylamide gel showing the results of chemical and enzymatic probing of S8b nt (nt 596–861). (B) As panel A, but structure mapping of S8c (nt 728–993) is shown. RNA structure probing analysis was also carried out on M6 and S9 RNA as detailed in the text
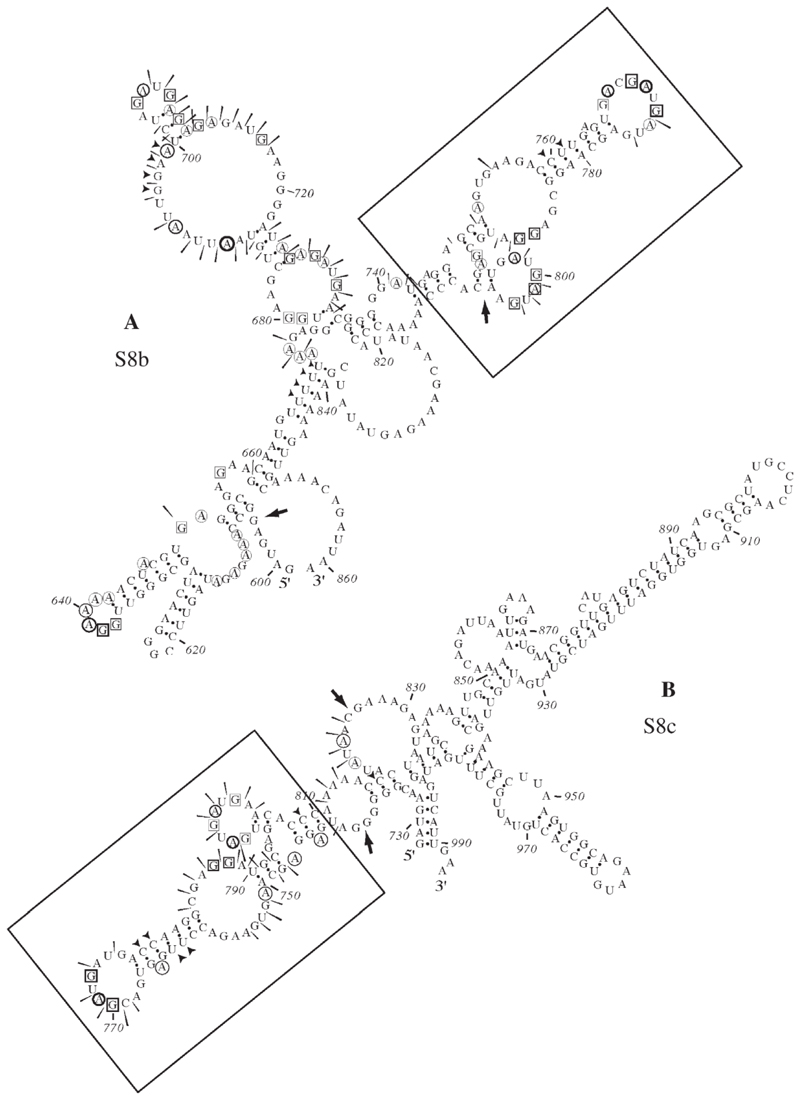
Fig. 6. Summary of RNA structure probing results for S8b and S8c.
Structure probing was carried out on panel (A) S8b (nt 596–861) and (B) S8c (728–993). The sensitivity of bases to the various probes is indicated by a black arrow head (double-stranded RNA—RNase CV1), circles (single-stranded A—RNase U2), boxes (single-stranded G—RNase T1) or a slender arrow (single-stranded RNA—lead acetate). The imidazole probing data, similar to that seen with lead acetate, are omitted for clarity. Large arrows represent regions of the predicted structure where structural information can be assigned. A conserved structure motif shared between these NS2-binding RNAs was identified (boxed).
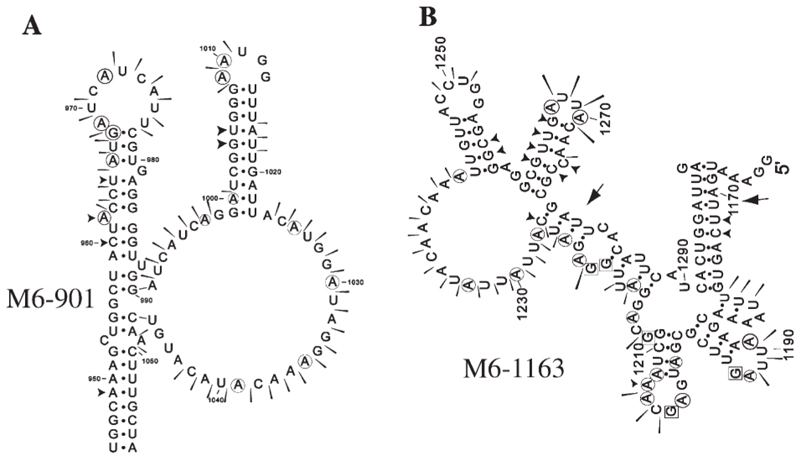
Fig. 8. Summary of RNA structure probing results for M6.
Structure probing was carried out on (A) M6 nt 901–1352 and (B) M6 nt 1163–1458. The sensitivity of bases to the various probes is indicated as in the legend to Fig. 6. The structure probing revealed good correspondence with predicted structures with the exception of M6 (901–1352), where the structure shown is derived from the structure probing results.
Examination of the secondary structures obtained for each RNA did not reveal any regions of conserved structure that could unambiguously define the NS2 binding site. In the case of S8, EMSA experiments predicted that NS2 interactions were dependent on the overlap between S8b (nt 596–861) and S8c (728–993). S8b and S8c were subjected to structure probing and the three primary sequences (596–861, 728–993 and 728– 861) were used as input into the mfold program. From the mfolds, an 82 nt structure present in all three sequences was identified, located in the overlapping region of S8b and S8c (736–818nts). Structure probing data confirmed that the structure of this region is conserved between the two RNAs (Fig. 6 panels A and B). The fact that this structural motif is conserved in vitro and in silico in both S8a and S8b and is predicted (by mfold) to be present in the full-length S8 segment supports the hypothesis that it is involved in the specific recognition of the S8 segment by NS2.
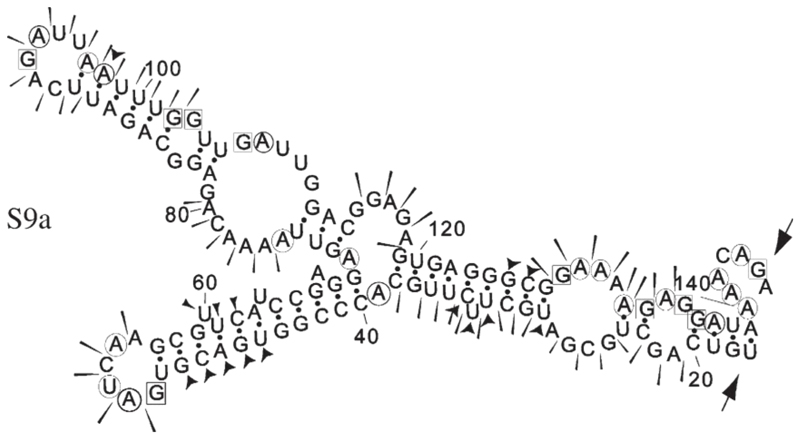
The S9 subgenomic fragment contains a structure, between nucleotides 29 and 127, with some similarity to that previously characterized for S10, although the stems of the smaller loops are much longer and the main loop appears to be constrained by some internal base-pairing (residues 72–74 with 111–113) (Fig. 7). This motif was present in the five most thermodynamically stable mfold predictions and was also retained in mfolds of the complete S9 segment.
Fig. 7. Summary of RNA structure probing results for S9a.
The sensitivity of bases to the various probes is indicated as in the legend to Fig. 6. The probing data matched closely the predicted mfold structure.
The M6 segment was the longest to be tested in competition assays and structure probing. Based on the competition assay, the overlapping region (nucleotides 901 to 1352) of subgenomic fragments M6b and M6c was investigated by structure probing. Due to the limitations in resolution described earlier, two different fragments were probed, 901 to 1352 (M6-901) and 1163 to 1458 (M6-1163). For M6-901, it was evident that the structure probing data did not fit closely to any individual mfold but seemed to be a composite of several predicted structures. For this reason, a model for M6-901 was prepared manually based solely on the structure probing data obtained (Fig. 8A). The model bears some similarity to the NS2 binding site of S10 however like the structure identified in S9, the loop sub- structures are longer and predicted to be more stable. With the M6-1163 RNA, mfolds gave more consistent predictions which closely matched the structure probing data (Fig. 8B). Two regions of the RNA bear similarity to the NS2 binding site of S10, each containing a stable stem leading to a loop from which protrudes two subloops (regions 1168–1213 and 1216–1298).
Discussion
The problem of how BTV, a virus that encodes only 11 proteins, can selectively package 10 different RNA segments into each virion is key to understanding assembly and replication of the virus. Similar problems exist for other viruses with selectively packaged segmented genomes, including other members of the Reoviridae. Previously, we have demonstrated that a region of BTV S10 contains a region of RNA with a defined structure, which is bound by NS2 (Lymperopoulos et al., 2003). In this study, we demonstrate that disruption of this structure by mutagenesis leads to a reduction in the affinity of NS2 for the S10 RNA fragment (Fig. 1). In addition, this effect is reversed if compensating mutations are introduced that recreate the wild-type RNA structure while preserving the original mutations. Furthermore, the MutB mutation was specifically designed because a previous RNase footprinting study (Lymperopoulos et al., 2003) indicated it did not form direct contacts with NS2. Thus, the data indicate that NS2–S10 RNA interactions are based on RNA structure not primary sequence. While our chemical mapping and mutagenesis studies define the likely secondary structure bound by NS2, precise details of the interaction will require structural analysis at higher resolution.
In addition, we have localized specific interactions between NS2 and the plus-strand RNAs of S8, S9 and M6. In each case, it was possible to identify fragments of the viral RNA that competed efficiently with full-length RNA probes for binding NS2 using EMSA competition experiments. These NS2 interacting sequences were located at different positions in each of the viral RNAs. In S9, like the previously characterized S10 sequence, the NS2 binding domain was located towards the 5’-end of the RNA. In contrast, in M6, the NS2 interacting domain was close to the center of the RNA, and in S8, it was closer to the 3’-end. In each case, the sequence bound by NS2 was distinct from the conserved hexanucleotides at the ends of the viral RNA and any other conserved primary sequence features. The RNA domain in S8 that we have identified as interacting with NS2 is consistent with a previous publication that used cross-linking of transcripts to NS2 in cytoplasmic extracts (Theron and Nel, 1997). However, we note that the domain we identify is less consistent with another recent publication that used a filter binding assay and mutant RNA (Markotter et al., 2004). It is likely that the discrepancy is largely due to a difference in interpretation. In the filter binding study, deletions were made in the S8 RNA, and a reduction in binding was correlated with the deleted sequence despite large predicted changes in RNA structure (Markotter et al., 2004). In the studies reported here, we have experimentally mapped the structure of the S8 fragments in order to validate the EMSA data.
In the absence of any conserved primary sequence motif bound by NS2, and the evidence that S10-NS2 interactions are based on RNA structure, we carried out experimental mapping of the structure of S8, S9 and M6 domains bound by NS2. This is the first time that the actual secondary structure of multiple BTV RNA segments has been analyzed and compared experimentally. Intriguingly, while there were structures with weak similarity to that critical for NS2–S10 interactions present in M6 and S9, there were no similar structures mapped in S8. Although our studies cannot rule out a possible common tertiary structure for the RNAs, the data suggest that several distinct structures are adopted by BTV RNAs, and that these structures allow independent interaction with NS2. The observation that S10 RNA, which has previously been demonstrated to interact specifically with NS2 (Lymperopoulos et al., 2003), is a poor competitor of NS2 binding of S9 RNA supports this hypothesis (Fig. 4B). Interestingly, a recent 2.4 Å structure of the RNA- binding domain of BTV NS2 has suggested that the protein forms a decameric complex (Butan et al., 2004). The fact that the remaining C-terminal domain of NS2 does not assemble into multimeric structures (Zhao et al., 1994) and that BTV has 10 different genome segments indicates that a decameric packaging complex is a theoretical possibility. Whether there are also 10 specific monomer–RNA interactions, or whether a smaller number of RNA complex–NS2 interactions are involved in the packaging of BTV RNA will be an exciting area of future research. The RNA-binding assays in this part of the study were all performed under conditions where NS2 was present in excess. Although it was possible to quantify the amount of NS2 that was included in each assay, it was not possible to demonstrate how much of that protein was active for RNA binding. NS2 is certainly not monomeric in these studies as size exclusion chromatography data (not shown) have indicated that the purified NS2 is multimeric in solution even in the absence of RNA. However, intermediate complexes in EMSA gels, for example, in Figs. 1(B–E) and 2 (S8c competition), may indicate that RNA concentration has some effect on the multimerization of NS2. It is interesting to note that one study has reported three different single-stranded RNA-binding domains on NS2 (Fillmore et al., 2002), but the contribution of these sites to the preferential binding of BTV RNA by NS2 is not known.
The wider implications of the specific interaction of BTV NS2 with RNA remain to be resolved. Rotavirus NSP2 has been shown to be an octamer (Jayaram et al., 2002; Taraporewala et al., 1999) but also interacts with NSP5 and NSP6 in inclusion body formation. Similarly, reovirus σNS and μNS act together to form viral inclusions (Becker et al., 2003). Therefore, the inclusions formed by the NS2 of Orbiviruses like BTV are arguably distinct from those formed by other members of the Reoviridae. Thus, it is quite possible that the selective RNA- binding activity associated with BTV NS2 will only be found associated with complexes of proteins in other genera of the Reoviridae family. Nevertheless, based on the accumulating evidence from studies on BTV NS2 (Lymperopoulos et al., 2003; Markotter et al., 2004; Theron and Nel, 1997), including this report, we would predict that similar selective RNA-binding activities will be found associated with the inclusion body complexes of other members of the Reoviridae.
Materials and methods
Virus, cells and purification of recombinant NS2
A recombinant baculovirus expressing BTV NS2 (AcBTV- 10-NS2) was propagated in Spodoptera frugiperda (Sf9) cells grown in Sf900 II media (Invitrogen). Protein expression studies were carried out in Trichoplusia ni cells (Tn5) grown in TC100 media supplemented with 10% fetal calf serum (Invitrogen). Expression, purification and concentration of recombinant NS2 were carried out as described previously (Lymperopoulos et al., 2003).
DNA templates for in vitro transcription
Plasmids pECS8, pECS9, pECS10 and pECM6 contain the full-length cDNA for BTV-10 segments S8, S9, S10 and M6 downstream of the T7 RNA polymerase promoter and upstream of a hepatitis delta ribozyme sequence. These plasmids were used to generate full-length plus-strand RNAs that were used as radiolabeled probes in EMSA. Subgenomic fragments of each RNA were generated by PCR incorporating the T7 promoter sequence at the 5’end of the plus-strand primer. Primers were designed such that the region amplified corresponded to the subfragment of the full-length cDNA that was under test, as detailed in the Results. Mutagenesis of the NS2-binding RNA structure in S10 was carried out using the QuikChange method (Stratagene) with pECS10 according to the manufacturers' instructions. Following mutagenesis, the entire S10 sequence was sequenced to confirm the presence of the mutation and to confirm that there were no other changes in the cDNA.
In vitro transcription and RNA labeling
Transcripts were produced and processed for EMSA experiments as described previously (Lymperopoulos et al., 2003).
RNA-NS2 binding and electrophoretic mobility shift assays (EMSA)
Binding reactions and EMSAs were performed as described previously (Lymperopoulos et al., 2003). For the experiments relating to Fig. 1, 300 ng NS2 and 1 fmol radiolabeled probe was used. For all other experiments, 3 μg of NS2 and 1 fmol 32P labeled RNA probe were routinely used in each EMSA experiment. The reason for these differences is that on preliminary analysis different batches of NS2 had different proportions of active and inactive protein. For quantification of the amount of probe bound by NS2 in the S10 EMSA assays, gels were dried and analyzed using a Molecular Dynamics PhosphorImager. Signals were quantified using ImageQuaNT version 5.0 (Molecular Dynamics). Experiments were repeated three times and the standard error of the mean calculated.
Computational analysis of RNA structures
Predictions of RNA secondary structure were obtained using the mfold programme, version 3.1 (Mathews et al., 1999) on the mfold RNA server of the Rensselaer Polytechnic Institute. Searches for primary sequence motifs were carried out using MEME version 2.2 (Bailey and Elkan, 1994).
Chemical and enzymatic mapping of RNA structure
RNAs for secondary structure mapping were prepared by in vitro transcription as described above. RNA secondary structure probing followed the protocol described (Lymperopoulos et al., 2003).
Acknowledgments
KL would like to thank Christoph Wirblich for technical advice offered during the experiments. This work was partly supported by the Wellcome Trust and NIH grant #R01 AI45000-01AI.
References
- Ahmed R, Fields BN. Reassortment of genome segments between reovirus defective interfering particles and infectious virus: construction of temperature-sensitive and attenuated viruses by rescue of mutations from DI particles. Virology. 1981;111(2):351–363. doi: 10.1016/0042-6822(81)90339-1. [DOI] [PubMed] [Google Scholar]
- Anzola JV, Xu ZK, Asamizu T, Nuss DL. Segment-specific inverted repeats found adjacent to conserved terminal sequences in wound tumor virus genome and defective interfering RNAs. Proc Natl Acad Sci USA. 1987;84(23):8301–8305. doi: 10.1073/pnas.84.23.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Becker MM, Peters TR, Dermody TS. Reovirus sigma NS and mu NS proteins form cytoplasmic inclusion structures in the absence of viral infection. J Virol. 2003;77(10):5948–5963. doi: 10.1128/JVI.77.10.5948-5963.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellamy AR, Shapiro L, August JT, Joklik WK. Studies on reovirus RNA.I. Characterization of reovirus genome RNA. J Mol Biol. 1967;29(1):1–17. doi: 10.1016/0022-2836(67)90177-5. [DOI] [PubMed] [Google Scholar]
- Butan C, Van Der Zandt H, Tucker PA. Structure and assembly of the RNA binding domain of Bluetongue virus non-structural protein 2. J. Biol Chem. 2004;279(36):37613–37621. doi: 10.1074/jbc.M400502200. [DOI] [PubMed] [Google Scholar]
- Fillmore GC, Lin H, Li JK-K. Localization of the single- stranded RNA-binding domains of Bluetongue virus nonstructural protein NS2. J Virol. 2002;76(2):499–506. doi: 10.1128/JVI.76.2.499-506.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huismans H, van Dijk AA, Bauskin AR. In vitro phosphorylation and purification of a nonstructural protein of Bluetongue virus with affinity for single-stranded RNA. J Virol. 1987;61(11):3589–3595. doi: 10.1128/jvi.61.11.3589-3595.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram H, Taraporewala Z, Patton JT, Prasad BV. Rotavirus protein involved in genome replication and packaging exhibits a HIT-like fold. Nature. 2002;417(6886):311–315. doi: 10.1038/417311a. [DOI] [PubMed] [Google Scholar]
- Kimura I, Black LM. The cell-infecting unit of wound tumor virus. Virology. 1972;49(2):549–561. doi: 10.1016/0042-6822(72)90506-5. [DOI] [PubMed] [Google Scholar]
- Lymperopoulos K, Wirblich C, Brierley I, Roy P. Sequence specificity in the interaction of Bluetongue virus non-structural protein 2 (NS2) with viral RNA. J Biol Chem. 2003;278(34):31722–31730. doi: 10.1074/jbc.M301072200. [DOI] [PubMed] [Google Scholar]
- Markotter W, Theron J, Nel LH. Segment specific inverted repeat sequences in Bluetongue virus mRNA are required for interaction with the virus non structural protein NS2. Virus Res. 2004;105(1):1–9. doi: 10.1016/j.virusres.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Nonoyama M, Graham AF. Appearance of defective virions in clones of reovirus. J Virol. 1970;6(5):693–694. doi: 10.1128/jvi.6.5.693-694.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramig RF, Petrie BL. Characterisation of temperature-sensitive mutants of simian rotavirus SA11: protein synthesis and morphogenesis. J Virol. 1984;49:665–673. doi: 10.1128/jvi.49.3.665-673.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramig RF, Mustoe TA, Sharpe AH, Fields BN. A genetic map of reovirus: II. Assignment of the double-stranded RNA-negative mutant groups C, D, and E to genome segments. Virology. 1978;85(2):531–534. doi: 10.1016/0042-6822(78)90459-2. [DOI] [PubMed] [Google Scholar]
- Reddy DV, Black LM. Isolation and replication of mutant populations of wound tumor virions lacking certain genome segments. Virology. 1977;80(2):336–346. doi: 10.1016/s0042-6822(77)80009-3. [DOI] [PubMed] [Google Scholar]
- Shatkin AJ, Sipe JD, Loh P. Separation of ten reovirus genome segments by polyacrylamide gel electrophoresis. J Virol. 1968;2(10):986–991. doi: 10.1128/jvi.2.10.986-991.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos DA, Krystal G, Graham AF. Regulated transcription of the genomes of defective virions and temperature-sensitive mutants of reovirus. J Virol. 1976;18(1):7–19. doi: 10.1128/jvi.18.1.7-19.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraporewala Z, Chen D, Patton JT. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J Virol. 1999;73(12):9934–9943. doi: 10.1128/jvi.73.12.9934-9943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraporewala Z, Chen D, Patton J. Multimers of the Bluetongue virus nonstructural protein, NS2, possess nucleotidyl phosphatase activity: similarities between NS2 and rotavirus NSP2. Virology. 2001;280(2):221–231. doi: 10.1006/viro.2000.0764. [DOI] [PubMed] [Google Scholar]
- Taraporewala Z, Schuck P, Ramig R, Silvestri L, Patton J. Analysis of a temperature-sensitive mutant rotavirus indicates that NSP2 octamers are the functional form of the protein. J Virol. 2002;76(14):7082–7093. doi: 10.1128/JVI.76.14.7082-7093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theron J, Nel LH. Stable protein-RNA interaction involves the terminal domains of Bluetongue virus mRNA, but not the terminally conserved sequences. Virology. 1997;229(1):134–142. doi: 10.1006/viro.1996.8389. [DOI] [PubMed] [Google Scholar]
- Thomas CP, Booth TF, Roy P. Synthesis of Bluetongue virus- encoded phosphoprotein and formation of inclusion bodies by recombinant baculovirus in insect cells: it binds the single-stranded RNA species. J Gen Virol. 1990;71(Pt. 9):2073–2083. doi: 10.1099/0022-1317-71-9-2073. [DOI] [PubMed] [Google Scholar]
- Uitenweerde JM, Theron J, Stoltz MA, Huismans H. The multimeric nonstructural NS2 proteins of Bluetongue virus, African horsesickness virus, and epizootic hemorrhagic disease virus differ in their single-stranded RNA-binding ability. Virology. 1995;209(2):624–632. doi: 10.1006/viro.1995.1294. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Thomas C, Bremer C, Roy P. Deletion and mutational analyses of Bluetongue virus NS2 protein indicate that the amino but not the carboxy terminus of the protein is critical for RNA-protein interactions. J Virol. 1994;68:2179–2185. doi: 10.1128/jvi.68.4.2179-2185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]