Abstract
The tumor suppressor p53 exerts pivotal roles in hematopoietic stem cell (HSC) homeostasis. Mutations of the TP53 gene have recently been described in individuals with clonal hematopoiesis conferring substantial risk of developing blood cancers. In patients with acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS), TP53 aberrations—mutations, deletions, and a combination thereof—are encountered at a constant frequency of approximately 10%. These aberrations affect HSCs transforming them into preleukemic stem cells, pinpointing their central role in leukemogenesis. AML and MDS with TP53 aberrations are characterized by complex chromosomal aberrations. Respective patients experience a dismal long-term outcome following treatment with both intensive and nonintensive regimens including novel agents like venetoclax combinations or even allogeneic HSC transplantation. However, according to the 2016 WHO classification, AML and MDS with TP53 aberrations are still regarded as separate disease entities. On the basis of their common biological and clinical features, we propose to classify AML and MDS with TP53 aberrations as a single, distinct stem cell disorder with a unique genetic make-up, comparable with the WHO classification of “AML with recurrent genetic abnormalities.” This approach will have implications for basic and translational research endeavors, aid in harmonization of current treatment strategies, and facilitate the development of master trials targeting a common deleterious driver event.
Taxonomy of human malignancies has traditionally been performed by histopathologic examination of affected organs and tissues. However, primarily in myeloid neoplasms and consecutively in other malignancies as well, underlying genomic aberrations have increasingly been elucidated and incorporated into classification schemes. The “2016 revision to the WHO classification of myeloid neoplasms and acute leukemia” now includes a multitude of cytogenetic and molecular abnormalities essential for diagnosis, prognostication, and treatment decisions (1).
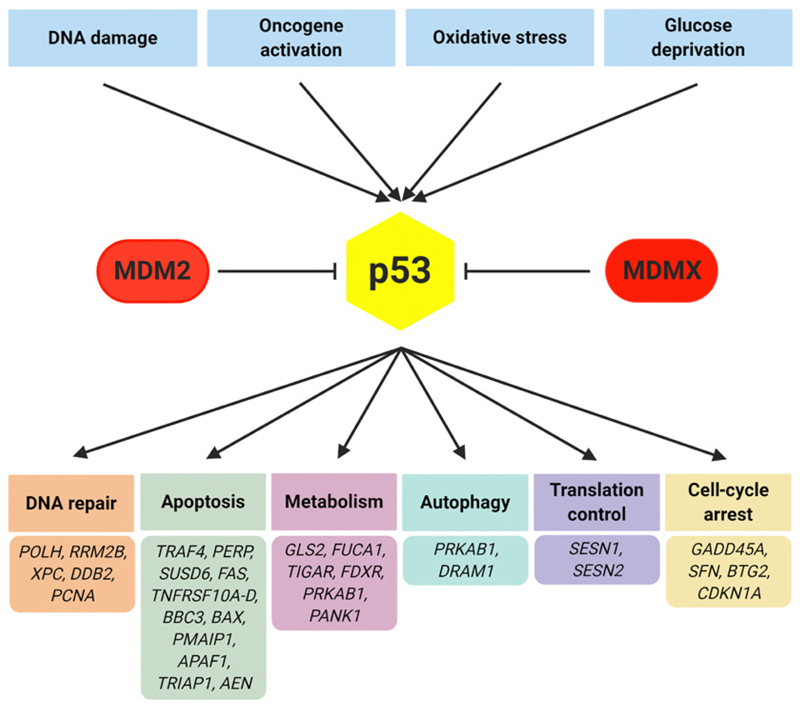
The TP53 tumor suppressor gene encodes a transcription factor involved in an abundance of pivotal cellular functions including DNA damage response (Fig. 1). Importantly, by pursuing these functions, p53 acts in a cell context–specific manner (2). Aberrations of TP53 have been described in more than 50% of human neoplastic disorders and recent work has elucidated their fundamental role in patients with acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS; refs. 3, 4). The data published so far point towards common features of AML and MDS with TP53 aberrations. Here, we focus on this topic that will support genomic classification efforts and aid in harmonization of current treatment strategies as well as the development of master trials.
Figure 1.
The p53 network. Among a multitude of biological processes influencing p53 activity, major inductors and inhibitors are depicted. Once activated, p53 is involved in many different cellular processes. A selection of high-confidence p53 target genes, as recently been described, is shown (50).
p53 in Normal Hematopoiesis
By interacting with the bone marrow microenvironment, hematopoietic stem cells (HSC) sustain a lifelong pool of mature blood cells. Early studies described expression of p53 in human hematopoietic cells and identified its involvement in proliferation, differentiation, and apoptosis (5). Using genetically engineered mice, a fundamental role of p53 for normal HSC function could be elaborated. p53 conveys quiescence of HSCs during steady-state hematopoiesis. In p53-deficient mice, primitive Lin–Sca-1+c-Kit+ cells showed enhanced self-renewal leading to a consistent expansion of this cell compartment. They revealed increased serial replating and repopulating capacity in vitro and in vivo, respectively, indicating the preservation of “stemness” properties (6). In cooperation with oncogenic aberrations like KrasG12D mutations, p53 loss led to indefinite self-renewal of murine HSCs with a propensity to transform into leukemia-initiating cells (7). Intact p53 was also shown to reduce intracellular reactive oxygen species levels, thereby contributing to HSC homeostasis and genetic stability (8). Recent data indicated that p53 is of functional importance for mesenchymal stromal cells (MSC), an essential component of the bone marrow niche. Thus, MSCs derived from p53 knockout mice were defective in supporting growth of cobblestone area-forming cells and hematopoietic progenitors (9).
TP53 Mutations in Clonal Hematopoiesis
Aging is a continuous process induced by exogenous and endogenous cellular toxicities giving rise to persistent DNA damage and, consequently, mutagenesis. In response to such damage, p53 is activated mediating DNA repair, senescence, or apoptosis (10). However, hyperactivity of p53 as induced experimentally or observed in the course of human disorders like Fanconi anemia, is associated with premature aging and/or depletion of tissues with high proliferative activity including the hematopoietic system (11, 12). HSCs, while aging, acquire mutations in various genes including TP53 leading to clonal hematopoiesis of indeterminate potential and age-related clonal hematopoiesis, respectively. Subjects with clonal hematopoiesis have an increased risk of hematologic malignancies, particularly therapy-related myeloid neoplasms (13, 14). Ultra-sensitive sequencing identified clonal hematopoiesis even in younger individuals with an age-dependent increase of TP53 mutations from 50 years on (15). Experimentally, HSCs with TP53 aberrations continuously expand during genotoxic stress, ultimately displacing normal hematopoiesis (6). Intwo recentcase-control studies, genetic profiles of healthy individuals, who developed AML, were compared with age-matched controls, who remained disease free. The pattern of preleukemic mutations differed substantially between both cohorts with TP53 aberrations being among those with the highest HR for leukemia development (16, 17).
Biology of AML and MDS with TP53 Aberrations
Similar to solid neoplasms, different aberrations affecting the TP53 gene have been described in a substantial portion of AML and MDS cases. They comprise allelic losses at a cytogenetic level as well as molecular mutations, insertions, and deletions that are either heterozygous, accompanied by a remaining wild-type allele, or become manifest in a hemi-/homozygous state. As a consequence, p53 haplo-insufficiency with reduced functionality or, alternatively, a complete loss of function due to loss of heterozygosity (LOH) or copy number–neutral LOH may be observed. Furthermore, a dominant-negative effect or even gain of novel functions has been postulated for particular TP53 mutations (2).
In some patients with AML and MDS, TP53 aberrations are of germline origin characterizing the Li-Fraumeni (LF) and LF-like syndromes. Here, myeloid malignancies frequently occur following cytotoxic treatments for a primary disorder pinpointing resistance of mutant HSCs to genotoxic stress (18). In the majority of cases, however, TP53 aberrations are somatically acquired and preferably constitute missense mutations located in the DNA binding domain of the gene. Recent data reveal novel albeit diverse insights into consequences of TP53 aberrations in myeloid neoplasms with respect to loss- or gain-of-function properties. When comparing cells from mice with a TrP53 knockout (k/o) status or exhibiting the murine R172H allele in a hemizygous state, respectively, only TrP53 mutant bone marrow cells showed indefinite serial replating capacity (19). In addition, leukemic cells expressing a “stem cell” signature were observed in the mutant, p53 expressing setting only. Upregulation of Foxh1 as a mediator of mutant p53 was identified and the authors postulated that this TrP53 mutation, corresponding to the human TP53 R175H allele, confers novel gain-of-function properties. These data are supported by observations in mice with either a p53 k/o status or expressing mutant forms of p53 showing marked differences with respect to tumor onset and spectrum (20, 21). In a different approach, TP53 hotspot mutations including R175H and TP53 k/o alleles were generated in myeloid cell lines using CRISPR-Cas9 genome editing. Unexpectedly, TP53 mutations as well as k/o alleles revealed the same oncogenic phenotype including resistance to cytotoxic treatments and displayed a gene expression signature of TP53 inactivation. Therefore, a dominant-negative effect was concluded as the primary force of selection of TP53 mutations in myeloid malignancies (22). In line with those data, loss of p53 function as a mediator of resistance to the BCL2 inhibitor venetoclax was described using a genome-wide CRISPR-Cas9 k/o screen of myeloid cell lines (23).
Clonal cells in AML and MDS are derived from rare leukemia stem cells (LSC) revealing self-renewal and long-term repopulation capacity. Although initial data allocated them to the immature CD34+/CD38– cell compartment, LSCs may also reveal a phenotype indicating transformation at a more mature progenitor cell level (24). The concept of preleukemic HSCs (pLSC) have recently been described in AML. They carry early driver mutations and retain their ability to differentiate into clonal blood cells; however, they are unable to generate leukemia in vivo (25). Transplanting diagnostic AML specimens with clonal TP53 mutations into NSGS mice, it could be shown that these mutations characterize pLSCs. The in vivo data were corroborated by detecting the leukemia-specific TP53 mutation in purified T-lymphocytes from the respective patients (26). Notably, AML specimens with subclonal TP53 mutations display multilineage engraftment potential as well when transplanted into a humanized ossicle mouse model (27). In low- and intermediate-risk MDS, LSCs were identified in the Lin–CD34+CD38–CD90+CD45RA– compartment. In a number of these specimens, LSCs revealed TP53 mutations that were preceded by deletions of chromosome 5q (28). These aberrations were also shown to perturb genomic stability in induced pluripotent stem cells derived from patients with MDS (29). Analyzing highly fractionated stem cell populations of patients with higher risk MDS progressing to secondary AML, TP53 mutations again affected LSCs and persisted throughout the course of the disease (30).
Apart from aberrations in the TP53 gene itself, members of the p53 pathway are frequently affected in myeloid malignancies. MDM2 and MDMX, the latter also known as MDM4, are negative regulators of p53 through binding to its transactivation domain. In addition, MDM2 has intrinsic E3 ubiquitin ligase activity. Over-expression of MDM2 and MDMX could be demonstrated in a high proportion of primary AML specimens with a TP53 wild-type status accompanied by a marked decrease in p21 expression. These patients experienced inferior survial indicating detrimental consequences of a dysfunctional p53 pathway as well (31). However, when analyzing fractionated Lin–CD34+CD38–CD90– stem and Lin–CD34+CD38+CD123+CD45+ progenitor cells from patients with AML, MDMX but not MDM2 expression was significantly higher as compared with age-matched, healthy controls. The data, therefore, provide a scientific basis for investigating MDMX-targeting compounds in AML and related disorders (32).
Clinical Characteristics of Patients with AML and MDS with TP53 Aberrations
TP53 aberrations are encountered in up to 10% of patients with AML and MDS with rising frequencies in older patients and in subtypes like therapy-related myeloid neoplasms and erythroleukemias. They frequently reveal abnormalities of chromosome 5q as well as high cytogenetic complexity or monosomal karyotypes (33, 34). In AML, extensive catastrophic DNA rearrangements, chromothripsis, have recently been linked to TP53 mutations suggesting that copy-number aberrations per se and the extent of genomic imbalances substantially contribute to p53-mediated leukemogenesis (35).
TP53 mutations are well-established adverse risk factors in patients with AML and MDS. Following allogeneic HSC transplantation as the only potentially curative approach, 5-year overall survival (OS) rates are less than 20% (36, 37). Treatment with hypomethylating agents show encouraging response rates although survival is still dismal when compared with TP53 wild-type cases. In a large MDS cohort treated with either azacitidine or decitabine, 5-year OS of TP53-mutated patients was less than 10% (38). In the azacitidine AML-001 trial, median OS was 7.2 months for patients with and 12.0 months for those without TP53 mutations, although this difference was statistically not significant possibly due to low patient numbers (39). Recently, results of the phase Ib study testing the BCL2 inhibitor venetoclax in combination with a hypomethylator showed CR rates for TP53-mutated AML patients of 67%. However, the median OS of 7.2 months was also inferior as compared with that of the total cohort being 17.5 months (40). Preliminary data with the anti-CD47 antibody magrolimab, inducing tumor cell phagocytosis, combined with azacitidine showed CR rates of 78% in TP53-mutated AML and MDS patients with median OS not reached after a median observation period of 6.9 months (41).
In AML, TP53 mutations have already been incorporated into current risk stratification guidelines and claims for a similar approach in MDS have been submitted (34, 38). Using functional classification schemes, it has been shown that different TP53 mutations exert different prognostic impacts in AML (42). However, there is yet conflicting evidence in AML whether mere deletions confer a some-what better prognosis than TP53 mutations or a combination there-of (43, 44). A recent analysis involving >3.000 patients with MDS indicate that a TP53 “multi-allelic state”–mutations combined with deletions or multiple mutations–reveals an extremely adverse outcome (45). The prognostic value of subclonal TP53 mutations and, in particular, the critical clone size threshold, as revealed by next-generation sequencing platforms, has also been regarded controversial in both AMLand MDS patients (33, 39). This might be due to different timepoints when treatments, known to select for TP53-mutated clones, were initiated.
Conclusions and Outlook
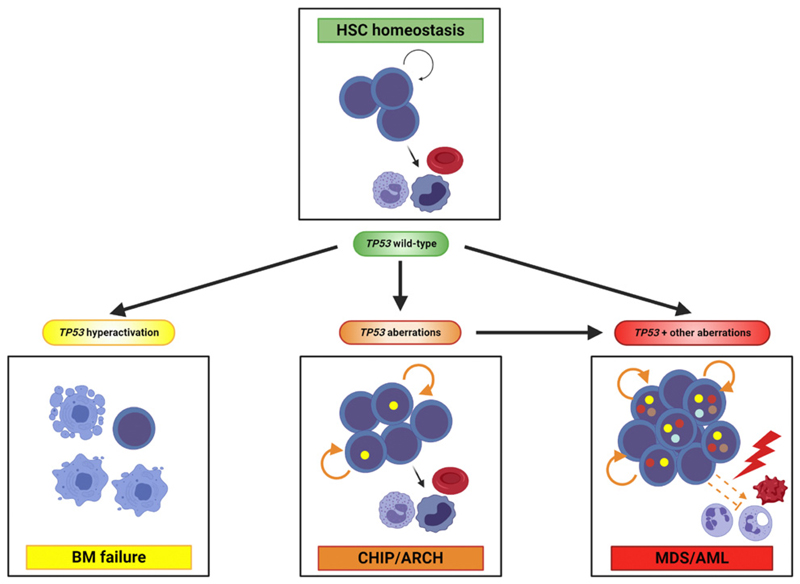
Following acquisition of driver mutations, leukemias may evolve from HSCs or arise from committed progenitors acquiring stem cell abilities (46). As summarized in this report and depicted in Fig. 2, p53 exerts fundamental functions in HSC homeostasis and evidence for the involvement of HSCs in the pathogenesis of AML and MDS with TP53 aberrations is based on both clinical observations and experimental research. In individuals with clonal hematopoiesis, potential precursor states of AML and MDS, mature hematopoietic cells in the bone marrow and peripheral blood show mutations in a variety of genes including TP53 indicating the involvement of pLSCs. Furthermore, single-cell analysis of purified hematopoietic stem and progenitor cells of human AML and MDS specimens revealed TP53 mutations affecting this cell compartment. Transplantation of TP53-mutated AML cells into xenograft mouse models leading to multilineage engraftment further supports this view. Experimentally, murine HSCs with either a TP53-mutant or k/o status are capable of establishing clonal hematopoiesis and, together with cooperating molecular and/or cytogenetic aberrations, frank leukemia.
Figure 2.
p53 hematopoietic stem cell functions under physiologic and pathologic conditions. In normal HSCs, p53 substantially contributes to their homeostasis that constitutes a balance between self-renewal (indicated by circled arrow) and differentiation intomature blood cells. Hyperactivation of p53 following continuousDNA damage, as induced experimentally or observed in patients with Fanconi anemia and other disorders, induces HSC quiescence and apoptosis, ultimately leading to bone marrow failure. Aberrations of TP53 transforms HSCs into pLSCs forming the basis of clonal hematopoiesis of indeterminate potential (CHIP) and age-related clonal hematopoiesis (ARCH). In these cases, their self-renewal capacitymight be increased (indicated by orange circled line). pLSCs with TP53 aberrations may be selected during genotoxic stress, acquire cooperating mutations and transform into LSCs giving rise to MDS or AML. Here, stem cells additionally demonstrate differentiation defects. Importantly, MDS and AML may also develop without overt preleukemic phase. Yellow dots within nuclei represent TP53 aberrations; other colored dots represent cooperating genetic events.
AML and MDS with TP53 aberrations also share clinical features (Table 1). They are observed at a constant frequency of approximately 10% in de novo cases with steep increases in elderly patients and particular leukemia subtypes. Endogenous and exogenous toxicities serve as fundamental cofactors of clonal evolution exerting selective advantage to TP53-mutated cells, ultimately leading to p53-mediated leukemogenesis. Patients with AML and MDS with TP53 aberrations also experience inferior outcomes with low survival rates following both intensive and nonintensive treatments. This might be due to the close association of the TP53 mutational status with highly complex karyotype aberrations representing the most unfavourable parameters in AML and MDS.
Table 1. Common features of AML and MDS with TP53 aberrations.
| TP53 aberrations in AML and MDS affect hematopoietic stem and progenitor cells |
| TP53 aberrations in AML and MDS contribute to genetic instability and are driver events of leukemogenesis |
AML and MDS with TP53 mutations show
|
| Patients with AML and MDS with TP53 aberrations are of older age |
| TP53 aberrations are frequent in therapy-related AML and MDS |
| TP53 aberrations constitute adverse risk parameters in patients with AML and MDS |
Following intensive treatments, patients with AML and MDS with TP53 aberrations show
|
Following nonintensive treatments, patients with AML and MDS with TP53 aberrations show
|
Note: “Intensive treatments” include chemotherapy ± allogeneic hematopoietic stem cell transplantation; “nonintensive treatments” refer to hypomethylating agents (azacitidine and decitabine) ± BCL2 inhibition (venetoclax).
Currently, drugs that specifically target mutant p53 or members of the p53 pathway are under clinical investigation potentially improving the prognosis of these patients. APR-246 is a compound designed to restore the function of mutant p53. In a phase II study, it was combined with azacitidine showing an overall response rate of patients with TP53-mutated AML and MDS of 88% and a median OS of 11.6 months (47). However, a placebo-controlled, double-blind phase III study investigating idasanutlin, an MDM2 inhibitor, in combination with cytarabine, did not show improved CR and OS rates in patients with relapsed and refractory AML (48). This data strengthen preclinical results on the necessity to additionally inhibit MDMX in patients with a TP53 wild-type status (32). Indeed, studies investigating ALRN-6924, a dual MDM2/MDMX inhibitor, are ongoing in patients with AML and MDS (NCT02909972 and NCT03654716).
Classification of human neoplasms is increasingly performed based on biological criteria rather than morphologic features. As early as 1995, AML and MDS with 17p deletion has been regarded “an entity characterized by specific dysgranulopoiesis and a high incidence of P53 mutations” (49). Clinical and experimental data compiled over the past years strongly corroborate such a view. Classification of “AML and MDS with TP53 aberrations” as a distinct, genetically defined stem cell disorder will encourage the development of master trials targeting disease-initiating mutations, thus overcoming the limitations of organ-restricted pathologic approaches. Particularly, in patients with AMLand MDS with TP53 aberrations, we are now facing the advent of such an era with novel compounds specifically targeting mutant p53 or members of a dysfunctional p53 pathway, thus addressing an urgent medical need.
Acknowledgments
We would like to apologize to those authors whose valuable contributions to this field could not be quoted due to space restrictions. Figures 1 and 2 were created with BioRender.com. Work in the laboratories of the authors is supported by “Leukämiehilfe Steiermark” (H. Sill and A. Zebisch) and the Austrian Science Fund FWF (H. Sill, grant number P31430-B26).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 2.Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170:1062–78. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prokocimer M, Molchadsky A, Rotter V. Dysfunctional diversity of p53 proteins in adult acute myeloid leukemia: Projections on diagnostic workup and therapy. Blood. 2017;130:699–712. doi: 10.1182/blood-2017-02-763086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bejar R. Implications of molecular genetic diversity in myelodysplastic syndromes. Curr Opin Hematol. 2017;24:73–8. doi: 10.1097/MOH.0000000000000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivas CI, Wisniewski D, Strife A, Perez A, Lambek C, Bruno S, et al. Constitutive expression of p53 protein in enriched normal human marrow blast cell populations. Blood. 1992;79:1982–6. [PubMed] [Google Scholar]
- 6.Chen S, Wang Q, Yu H, Capitano ML, Vemula S, Nabinger SC, et al. Mutant p53 drives clonal hematopoiesis through modulating epigenetic pathway. Nat Commun. 2019;10:5649. doi: 10.1038/s41467-019-13542-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Z, Zuber J, Diaz-Flores E, Lintault L, Kogan SC, Shannon K, et al. P53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes Dev. 2010;24:1389–402. doi: 10.1101/gad.1940710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung H, Kim MJ, Kim DO, Kim WS, Yoon SJ, Park YJ, et al. TXNIP maintains the hematopoietic cell pool by switching the function of p53 under oxidative stress. Cell Cell Metab. 2013;18:75–85. doi: 10.1016/j.cmet.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Boregowda SV, Krishnappa V, Strivelli J, Haga CL, Booker CN, Phinney DG. Basal p53 expression is indispensable for mesenchymal stem cell integrity. Cell Death Differ. 2018;25:679–92. doi: 10.1038/s41418-017-0004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ou HL, Schumacher B. DNA damage responses and p53 in the aging process. Blood. 2018;131:488–95. doi: 10.1182/blood-2017-07-746396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumble M, Moore L, Chambers SM, Geiger H, Van Zant G, Goodell MA, et al. The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood. 2007;109:1736–42. doi: 10.1182/blood-2006-03-010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceccaldi R, Parmar K, Mouly E, Delord M, Kim JM, Regairaz M, et al. Bone marrow failure in fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11:36–49. doi: 10.1016/j.stem.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science. 2019;366 doi: 10.1126/science.aan4673. eaan4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Busque L, Buscarlet M, Mollica L, Levine RL. Concise review: age-related clonal hematopoiesis: stem cells tempting the devil. Stem Cells. 2018;36:1287–94. doi: 10.1002/stem.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Acuna-Hidalgo R, Sengul H, Steehouwer M, van de Vorst M, Vermeulen SH, Kiemeney LALM, et al. Ultra-sensitive sequencing identifies high prevalence of clonal hematopoiesis-associated mutations throughout adult life. Am J Hum Genet. 2017;101:50–64. doi: 10.1016/j.ajhg.2017.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abelson S, Collord G, Ng SWK, Weissbrod O, Mendelson Cohen N, Niemeyer E, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559:400–4. doi: 10.1038/s41586-018-0317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desai P, Mencia-Trinchant N, Savenkov O, Simon MS, Cheang G, Lee S, et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med. 2018;24:1015–23. doi: 10.1038/s41591-018-0081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulz E, Valentin A, Ulz P, Beham-Schmid C, Lind K, Rupp V, et al. Germline mutations in the DNA damage response genes BRCA1, BRCA2, BARD1 and TP53 in patients with therapy related myeloid neoplasms. J Med Genet. 2012;49:422–8. doi: 10.1136/jmedgenet-2011-100674. [DOI] [PubMed] [Google Scholar]
- 19.Loizou E, Banito A, Livshits G, Ho YJ, Koche RP, Sanchez-Rivera FJ, et al. A gain-of-function p53-mutant oncogene promotes cell fate plasticity and myeloid leukemia through the pluripotency factor FOXH1. Cancer Discov. 2019;9:962–79. doi: 10.1158/2159-8290.CD-18-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–21. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 21.Lavigueur A, Maltby V, Mock D, Rossant J, Pawson T, Bernstein A. High incidence of lung, bone, and lymphoid tumors in transgenic mice overexpressing mutant alleles of the p53 oncogene. Mol Cell Biol. 1989;9:3982–91. doi: 10.1128/mcb.9.9.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boettcher S, Miller PG, Sharma R, McConkey M, Leventhal M, Krivtsov AV, et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science. 2019;365:599–604. doi: 10.1126/science.aax3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nechiporuk T, Kurtz SE, Nikolova O, Liu T, Jones CL, D’Alessandro A, et al. The TP53 apoptotic network is a primary mediator of resistance to BCL2 inhibition in AML cells. Cancer Discov. 2019;9:910–25. doi: 10.1158/2159-8290.CD-19-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vetrie D, Helgason GV, Copland M. The leukaemia stem cell: similarities, differences and clinical prospects in CML and AML. Nat Rev Cancer. 2020;20:158–73. doi: 10.1038/s41568-019-0230-9. [DOI] [PubMed] [Google Scholar]
- 25.Pandolfi A, Barreyro L, Steidl U. Concise review: Preleukemic stem cells: molecular biology and clinical implications of the precursors to leukemia stem cells. Stem Cells Transl Med. 2013;2:143–50. doi: 10.5966/sctm.2012-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lal R, Lind K, Heitzer E, Ulz P, Aubell K, Kashofer K, et al. Somatic TP53 mutations characterize preleukemic stem cells in acutemyeloid leukemia. Blood. 2017;129:2587–91. doi: 10.1182/blood-2016-11-751008. [DOI] [PubMed] [Google Scholar]
- 27.Pabst G, Lind K, Graf R, Zebisch A, Stolzel F, Dohner K, et al. TP53 mutated AML subclones exhibit engraftment in a humanized bone marrow ossicle mouse model. Ann Hematol. 2020;99:653–5. doi: 10.1007/s00277-020-03920-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woll PS, Kjallquist U, Chowdhury O, Doolittle H, Wedge DC, Thongjuea S, et al. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell. 2014;25:794–808. doi: 10.1016/j.ccr.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 29.Hsu J, Reilly A, Hayes BJ, Clough CA, Konnick EQ, Torok-Storb B, et al. Reprogramming identifies functionally distinct stages of clonal evolution in myelodysplastic syndromes. Blood. 2019;134:186–98. doi: 10.1182/blood.2018884338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Kao YR, Sun D, Todorova TI, Reynolds D, Narayanagari SR, et al. Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level. Nat Med. 2019;25:103–10. doi: 10.1038/s41591-018-0267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quintas-Cardama A, Hu C, Qutub A, Qiu YH, Zhang X, Post SM, et al. p53 pathway dysfunction is highly prevalent in acute myeloid leukemia independent of TP53 mutational status. Leukemia. 2017;31:1296–305. doi: 10.1038/leu.2016.350. [DOI] [PubMed] [Google Scholar]
- 32.Carvajal LA, Neriah DB, Senecal A, Benard L, Thiruthuvanathan V, Yatsenko T, et al. Dual inhibition ofMDMXandMDM2as a therapeutic strategy in leukemia. Sci Transl Med. 2018;10 doi: 10.1126/scitranslmed.aao3003. eaao3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prochazka KT, Pregartner G, Rucker FG, Heitzer E, Pabst G, Wolfler A, et al. Clinical implications of subclonal TP53 mutations in acute myeloid leukemia. Haematologica. 2019;104:516–23. doi: 10.3324/haematol.2018.205013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haase D, Stevenson KE, Neuberg D, Maciejewski JP, Nazha A, Sekeres MA, et al. TP53 mutation status divides myelodysplastic syndromes with complex kar-yotypes into distinct prognostic subgroups. Leukemia. 2019;33:1747–58. doi: 10.1038/s41375-018-0351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rucker FG, Dolnik A, Blatte TJ, Teleanu V, Ernst A, Thol F, et al. Chromothripsis is linked to TP53 alteration, cell cycle impairment, and dismal outcome in acute myeloid leukemia with complex karyotype. Haematologica. 2018;103:e17–20. doi: 10.3324/haematol.2017.180497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Middeke JM, Herold S, Rucker-Braun E, Berdel WE, Stelljes M, Kaufmann M, et al. TP53 mutation in patients with high-risk acute myeloid leukaemia treated with allogeneic haematopoietic stem cell transplantation. Br J Haematol. 2016;172:914–22. doi: 10.1111/bjh.13912. [DOI] [PubMed] [Google Scholar]
- 37.Lindsley RC, Saber W, Mar BG, Redd R, Wang T, Haagenson MD, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med. 2017;376:536–47. doi: 10.1056/NEJMoa1611604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bejar R, Lord A, Stevenson K, Bar-Natan M, Perez-Ladaga A, Zaneveld J, et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;124:2705–12. doi: 10.1182/blood-2014-06-582809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dohner H, Dolnik A, Tang L, Seymour JF, Minden MD, Stone RM, et al. Cytogenetics and genemutations influence survival in older patients with acute myeloid leukemia treated with azacitidine or conventional care. Leukemia. 2018;32:2546–57. doi: 10.1038/s41375-018-0257-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DiNardo CD, Pratz K, Pullarkat V, Jonas BA, Arellano M, Becker PS, et al. Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood. 2019;133:7–17. doi: 10.1182/blood-2018-08-868752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sallman DA, Asch AS, Al Malki MM, Lee DJ, Donnellan WB, Marcucci G, et al. The first-in-class anti-CD47 antibody magrolimab (5F9) in combination with azacitidine (AZA) is effective in MDS and AML patients: ongoing phase 1b results. Blood. 2019;134(1):569. [Google Scholar]
- 42.Dutta S, Pregartner G, Rucker FG, Heitzer E, Zebisch A, Bullinger L, et al. Functional classification of TP53 mutations in acute myeloid leukemia. Cancers. 2020;12:637. doi: 10.3390/cancers12030637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stengel A, Kern W, Haferlach T, Meggendorfer M, Fasan A, Haferlach C. The impact of TP53 mutations and TP53 deletions on survival varies between AML, ALL, MDS and CLL: an analysis of 3307 cases. Leukemia. 2017;31:705–11. doi: 10.1038/leu.2016.263. [DOI] [PubMed] [Google Scholar]
- 44.DiNardo CD, Tiong IS, Quaglieri A, MacRaild S, Loghavi S, Brown FC, et al. Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood. 2020;135:791–803. doi: 10.1182/blood.2019003988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernard E, Nannya Y, Yoshizato T, et al. TP53 state dictates genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Blood. 2019;134(1):675. [Google Scholar]
- 46.Passegue E, Jamieson CH, Ailles LE, Weissman IL. Normal and leukemic hematopoiesis: are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc Natl Acad Sci U S A. 2003;100:11842–9. doi: 10.1073/pnas.2034201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sallman DA, DeZern AE, Garcia-Manero G, et al. Phase 2 results of APR-246 and azacitidine (AZA) in patients with TP53 mutant myelodysplastic syndromes (MDS) and oligoblastic acute myeloid leukemia (AML) Blood. 2019;134(1):676. [Google Scholar]
- 48.Konopleva MY, Röllig C, Cavenagh J, Deeren D, Girshova L, Krauter J, et al. A randomized double-blind phase 3 trial of cytarabine with MDM2 inhibitor idasanutlin or placebo in relapsed/refractory acute myeloid leukemia (R/R AML): primary analysis results of the MIRROS study. HemaSphere. 2020;4:EP532. [Google Scholar]
- 49.Lai JL, Preudhomme C, Zandecki M, Flactif M, Vanrumbeke M, Lepelley P, et al. Myelodysplastic syndromes and acute myeloid leukemia with 17p deletion. an entity characterized by specific dysgranulopoiesis and a high incidence of P53 mutations. Leukemia. 1995;9:370–81. [PubMed] [Google Scholar]
- 50.Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017;36:3943–56. doi: 10.1038/onc.2016.502. [DOI] [PMC free article] [PubMed] [Google Scholar]