Abstract
Iron is an essential trace element for most organisms. A common way for bacteria to acquire this nutrient is through the secretion of siderophores, which are secondary metabolites that scavenge iron from environmental stocks and deliver it to cells via specific receptors. While there has been tremendous interest in understanding the molecular basis of siderophore synthesis, uptake and regulation, questions on the ecological and evolutionary consequences of siderophore secretion have only recently received increasing attention. In this Review, we outline how eco-evolutionary questions can complement the mechanistic perspective, and help to obtain a more integrated view on siderophores. In particular, we explain how secreted diffusible siderophores can affect other community members, leading to cooperative, exploitative and competitive interactions between individuals. These social interactions in turn can spur co-evolutionary arms races between strains and species, lead to ecological dependencies between them and potentially contribute to the formation of stable communities. In brief, our Review shows that siderophores are much more than just iron carriers: they are important mediators of interactions between members of microbial assemblies and the eukaryotic hosts they inhabit.
Introduction
Like most other organisms, virtually all bacteria need iron as a co-factor in enzymes to catalyze redox reactions involved in fundamental cellular processes such as respiration, DNA synthesis and protection from reactive oxygen species1. Although iron is abundant in the earth crust, its bioavailability is generally low2,3. This is because iron mainly exists in its oxidized ferric Fe3+ state, which is largely insoluble at neutral and basic pH levels, conditions that prevail in many natural habitats. To cope with iron shortage, bacteria have evolved a number of mechanisms to acquire this essential trace element from the environment. These mechanisms include the uptake of iron bound to organic molecules such as citrate or heme, the absorption of iron by membrane-bound uptake systems, and the secretion of siderophores, which are secondary metabolites that scavenge iron from environmental stocks by forming soluble Fe3+-complexes that are then actively taken up via specific receptors4–11.
Almost all known bacterial species produce siderophores, making their secretion the most prevalent mechanism for iron scavenging in the microbial world4–6. Siderophores are a chemically diverse group of secondary metabolites12. There is an extensive body of work on their chemical structures and the molecular mechanisms of their synthesis, export, uptake, and regulation1,5–10,12–14. The enormous interest in the molecular basis of siderophore biology has created an imbalance between what we know on the mechanistic versus the evolutionary and ecological aspects of these vital compounds15. While mechanistic knowledge on siderophores grew immensely, evolutionary questions fell behind, probably because of the obvious physiological function of siderophores, having evolved as a means for bacteria to obtain iron for metabolism1,9,11. Only relatively recently it was recognized that complementary evolutionary questions, which go beyond physiological aspects, can be addressed, and are important to understand evolutionary and ecological dynamics in bacterial communities and the evolution of bacterial pathogens within hosts16–24.
One of these complementary aspects is the question why bacteria have evolved highly diffusible siderophores – a strategy that might successfully solubilize iron, but comes with the problem that siderophores might not find their way back to producers18,24,25? Another complementary aspect is highlighted by the observation that siderophores secreted by one cell can have dramatic fitness consequences for other cells in the microbial community, including members of the same and different species16,17,20,21. For example, siderophores might not only make iron available for the producer, but also to other individuals with a matching receptor, such that the costs and benefits of siderophore production are distributed across different individuals within the population16,17. Alternatively, the binding of iron to siderophores can induce iron starvation in bacteria that lack the matching receptor for uptake20,26. Thus, siderophores seem to be important players in mediating interactions between cells of the same and different species.
The aim of this Review is to elucidate these complementary aspects by summarizing the recent insights gained on the ecology and evolution of siderophores and by linking them to the mechanistic aspects of siderophore production. We will start with an overview on the mechanistic aspects of siderophore synthesis, secretion, uptake, and regulation. Although this topic has been extensively reviewed elsewhere1,6–10,12,13,27, we will briefly treat it here as it is essential for understanding the ecological and evolutionary consequences of siderophore secretion at the community level. We will then move on to specifically elucidating eco-evolutionary aspects by addressing the following four questions: What are the physical consequences of siderophore secretion for individual cells and bacterial groups? Which types of social interactions are driven by siderophores? How do these social interactions impact evolutionary and ecological dynamics in bacterial communities? How can insights into these eco-evolutionary aspects help us to understand bacteria-host interactions, including the evolution of beneficial functions of the microbiota and detrimental effects by pathogens in infections?
A mechanistic primer on siderophores
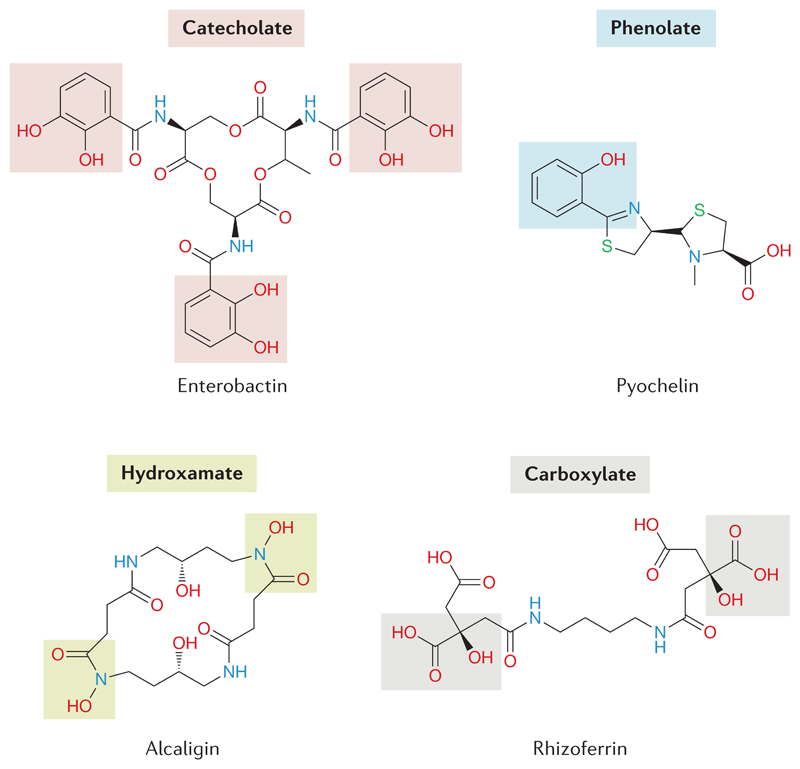
Siderophore (Greek: iron carrier) is a functional term and defines organic ligands that show specificity for iron, enable iron acquisition through specific uptake systems and are regulated in response to iron availability28 (but also see Box 1 for alternative functions of siderophores). Four chemical types of siderophores can be distinguished based on the moieties involved in iron chelation (Figure 1). These are catecholate, phenolate, hydroxamate and carboxylate types of siderophores, whereby mixtures of the various types are also common6,12. Siderophores are typically synthetized by non-ribosomal peptide synthetases (NRPS) or polyketide synthase (PKS) domains that work in concert with NRPS modules. A smaller fraction of siderophores are also produced by pathways that are independent of NRPS and PKS6,29–33. Siderophores synthesis via NRPS is the most common form and consists of a multienzyme assembly line, in which amino acids, as well as carboxy and hydroxy acids are built into a peptidic precursor molecule, which is subsequently modified by either NRPS or other enzymes to form the final siderophore. One such modification includes the formation of heterocycles, such as the thiazoline ring arising through the cyclization of cysteine side chains6,29. The secretion of siderophores is an energy-driven process and mediated by efflux pumps, which belong to different superfamilies depending on the siderophore type they transport6.
Box 1. Alternative functions of siderophores.
Siderophores might have alternative functions in addition to iron scavenging. From an evolutionary point of view, it is not always clear whether these additional functions reflect mere by-products of iron chelation or adaptations in their own right.
Non-iron metal transport
Siderophores can bind metals other than iron, including zinc, manganese, copper and nickel. In some cases, these metal-siderophore complexes do not enter the cell149,150, possibly indicating that their formation is a by-product of iron acquisition. However, there are also cases of siderophores that function as multi-purpose metallophores: for example, Yersinia pestis produces yersiniabactin and this siderophore scavenges zink, cooper and nickel, in addition to iron, for bacterial metabolism151,152,153,154.
Toxic metal sequestration
Heavy metals can be lethal at high concentrations155. Because siderophores typically bind heavy metals without shuttling them into the cell, they can effectively detoxify the local environment150,156. This bioremediation mechanism can be adaptive and specifically upregulated in response to pollution150,157. Moreover, it constitutes a cooperative behaviour as producers detoxify the environment for everybody158.
Signalling
Siderophores can function as signals and thereby regulate their own production, and induce the expression virulence factors and other components of the iron acquisition pathway, whose regulation is linked44. Additionally, siderophores can be cues in inter-species interactions. For example, siderophores produced by one species can stimulate sporulation or induce the production of endogeneous siderophores in another species159,160.
Protection from oxidative stress
Siderophores can offer protection against reactive oxygen species (ROS) created by UV-radiation or antibiotic stressors161–164. For example, pyoverdine can absorb UV-radiation extracellularly and thus reduce ROS-generation161, but it can also chelate ferrous iron that is released from ROS-damaged Fe-S clusters, thereby preventing the iron-catalyzed reaction of H2O2 to harmful hydroxyl radicals within cells164.
Antibiotic activity
Siderophore receptors are gateways into the cytoplasm, and some bacteria exploit this weak spot by deploying sideromycins, a class of antibiotics in which a bactericidal ‘warhead’ is attached to a siderophore138,165. The specificity of sideromycins is largely determined by the prevalence of the respective cognate siderophore receptor, and their activity can be antagonized by siderophores that compete for the same receptor.
Figure 1. Representative examples of the four main chemical classes of siderophores.
Siderophore is a functional term and includes a chemically diverse group of molecules. The four main types are distinguished based on the moieties involved in iron chelation, which entail catecholate, phenolate, hydroxamate and carboxylate functional groups (grey shadings). Siderophores with mixtures of functional groups are also common. Representative siderophore examples include enterobactin (which is produced by Escherichia coli, for example), pyochelin (which is produced by Pseudomonas aeruginosa, for example) featuring a heterocycle (thiazoline) ring, alcaligin (which is produced by Bordetella pertussis, for example) and rhizoferrin (which is produced by Ralstonia pickettii, for example). Note that the same siderophore can be produced by different species18.
The uptake of iron-loaded siderophores differs between Gram-negative and Gram-positive bacteria, mainly because Gram-positive bacteria have no outer membrane across which iron-loaded siderophores must be translocated1,6,10,13,14,27,34,35. In Gram-negative bacteria, a common mechanism involves the specific recognition of the iron-loaded siderophore by a ß-barrel receptor in the outer membrane. After binding the ligand, the receptor undergoes a conformational change, translocating the iron-loaded siderophore into the periplasm. This process is supported by a TonB-complex, which delivers the energy via the proton motive force13,34,35. Transport of the iron-loaded siderophore into the cytosol, where iron reduction occurs, is then typically mediated by an ABC-transporter in the inner membrane13,34,35. In some special cases, the iron is reduced in the periplasm and only the ferrous Fe2+ iron is imported into the cytosol36. In Gram-positive bacteria no outer-membrane receptors are required and siderophores are directly imported by ABC-transporters spanning the cell membrane6,34. The fate of siderophores after the release of iron is not well understood. In some cases, siderophores can be recycled through specific recycling mechanisms 35,37,38, whereas other siderophores undergo hydrolysis to release iron39,40.
The above described siderophore synthesis, release and uptake mechanisms are under tight regulatory control aimed at maintaining iron homeostasis within the cell. The most common key regulator is Fur (ferric uptake regulator), a transcriptional repressor of siderophore synthesis and regulatory activator genes5,41,42. When intracellular iron stores are replenished, Fur binds ferrous iron in the cytosol and this complex then binds to the promoter region of either siderophore synthesis42 or regulatory activator genes43 to block their transcription. Under iron limitation, repression by Fur is released and siderophore synthesis induced. Another level of siderophore regulation occurs when iron-loaded siderophores enter the cell. Here, extracytoplasmic function sigma factors or two-component systems serve as checkpoints, triggering increased siderophore synthesis. This positive feedback loop is interpreted as a signal to cells that iron scavenging by siderophores is a successful course of action6,44,45.
Consequences of siderophore secretion
Siderophore secretion from the perspective of an individual cell
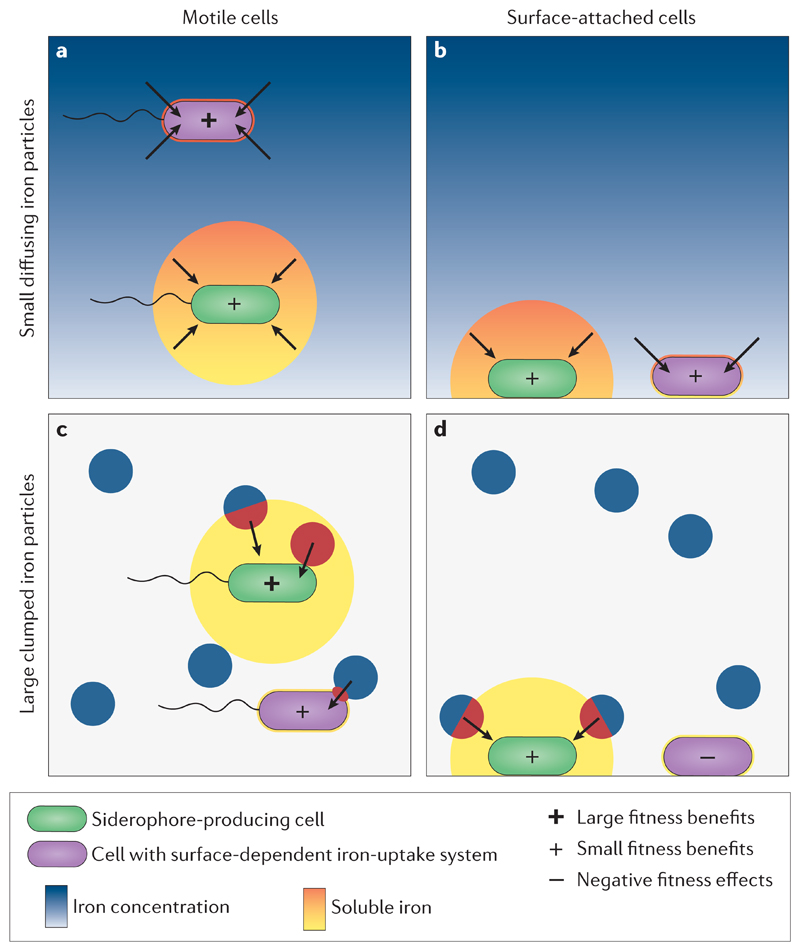
The physical consequence of siderophore secretion is that the molecules can diffuse away, beyond the reach of producers, and might thus not deliver benefits back to producer cells. One key evolutionary question related to the physical consequence of loss through diffusion is why bacteria have evolved diffusible siderophores in the first place and do not solely rely on membrane-bound iron uptake systems, which occur in many taxa46,47. Mathematical models have tackled this question on multiple fronts24,25,48–51. One insight gained from these theoretical analyses is that membrane-embedded systems seem superior over siderophores in planktonic cells living in unstructured environments. This advantage arises because a lack of spatial structure results in high siderophore losses through diffusion, and at the same time leads to a more homogeneous iron distribution, which makes iron easily accessible by membrane-embedded systems (Figure 2a)24,25. When bacteria are attached to surfaces, siderophore secretion enables cells to chelate iron beyond the local pool, which in turn might become rapidly depleted if iron is taken up by membrane-embedded systems (Figure 2b). Yet for single cells without neighbours, most of the additional iron chelated by siderophores is still likely lost due to diffusion and membrane-embedded system might still perform reasonably well24. The key advantage of siderophores seems to kick in when iron resources are clumped (for example, in mineral particles) and bacteria might not necessarily be located near the iron source (Figures 2c+d)24,25,48. In this scenario, siderophores enable increased solubilisation of iron, fostering a more homogenous distribution of this nutrient and making it accessible to both planktonic and sessile cells. Conceivably, membrane-embedded systems perform poorly when iron is clumped, especially when cells are sessile and physically disconnected from the iron source (Figure 2d).
Figure 2. Comparing siderophore-dependent and surface-dependent iron-uptake systems.
Considerations of why bacteria have evolved siderophore-mediated iron scavenging systems (green cells), where siderophores can be lost due to diffusion, as opposed to surface-dependent iron uptake mechanisms (purple cells), where benefits are secured. We consider cases in which iron is either relatively homogenously distributed (as small particles along a blue-shaded gradient) or occurs clumped in large particles (blue circles). Siderophores increase the range across which iron can potentially be sequestered (yellow area) and the amount of iron that is brought into solution (orange-red gradient), but are at the risk of not finding their way back to producers (arrows). The siderophore’s reach of action (yellow area) depends on the viscosity of the environment and the chemical diffusion property of the siderophore. Emojis inside cells depict the relative fitness consequences of the two iron-scavenging strategies. a | When iron is homogenously distributed and cells are motile, surface-dependent iron uptake mechanisms are efficient for iron scavenging, and siderophores unlikely confer an advantage. b | When iron is homogenously distributed, but cells are attached to surfaces, siderophores can access iron beyond the local pool, potentially making it available for bacterial metabolism. However, many siderophore molecules might get lost due to diffusion. c | When iron is clumped and cells are motile, bacteria with surface-dependent iron uptake mechanisms must necessarily make contact with the iron source to be able to take up this nutrient, whereas no direct cellular contact is required with siderophores and more distant iron pools can be accessed. d | When iron is clumped and cells are non-motile, siderophores might be the only way to obtain iron. These considerations suggest that siderophore production is most beneficial when iron resources are clumped and cells are non-motile.
Despite these considerations, siderophore loss due to diffusion can still be substantial and compromise bacterial fitness. One important factor that has been shown to limit loss through diffusion and increase benefits of siderophores is environmental structure, a factor that takes effect when bacteria either live in physically structured habitats (for example, on organic or inorganic particles) or in viscous medium (for example, in the mucus of individuals with cystic fibrosis)49–52. Indeed, a study investigating bacterioplankton17 showed that siderophore-mediated iron scavenging is beneficial in Vibrio spp. attached to organic particles, which represent a structured island in the otherwise highly diffusive marine environment. Another factor that contributes to the control of loss through diffusion is the chemical structure of the siderophore molecule itself18,53,54. A comparative analysis revealed that many marine bacteria produce large amphiphilic siderophores that can, at least partially, be embedded into the cell membrane18, an adaptation that clearly helps to retain siderophores and reduce loss through diffusion in a highly diffusive environment like the open ocean53,54. Although such amphiphilic siderophores can also occur in species living in relatively structured environments such as soil55, they are much rarer in these environments, presumably because the physical structure of the habitat itself limits the diffusion of molecules18.
Siderophore secretion from the perspective of a group
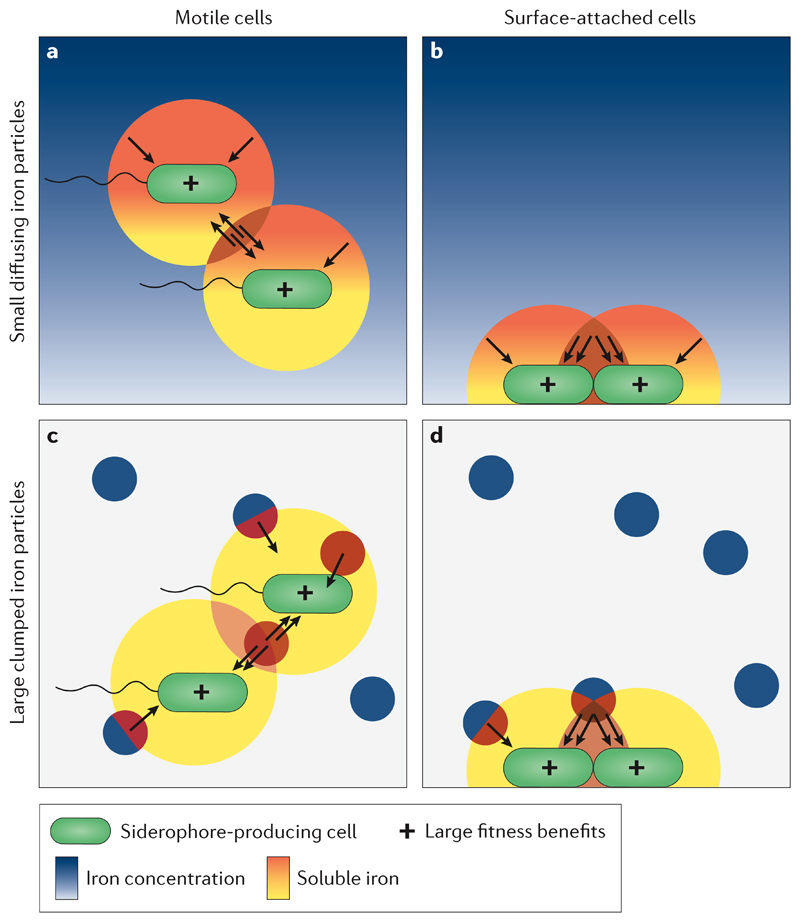
An important feature of bacterial life is that cells are hardly ever alone, but rather grow in groups, often in biofilms attached to surfaces in soil, hosts, and particles in aquatic environments56–58. This has important consequences for the use of siderophores (and other secreted goods), as cells do not necessarily rely on their own siderophores for iron scavenging, but can also benefit from the molecules secreted by their clonemates (Figure 3)59. Siderophore sharing is thus a powerful mechanism to limit the negative consequences of siderophore loss through diffusion, and could be an additional factor that determines whether diffusible siderophores can gain an edge over membrane-embedded iron uptake systems. This effect could be important even in habitats in which iron is relatively homogeneously distributed and cells are motile, as long as local cell density is sufficiently high (Figure 3)25,49,60.
Figure 3. Siderophores can synergistically increase iron uptake when bacteria live in groups.
The problem of siderophore loss due to diffusion can be reduced when cells live in groups. In this scenario neither the diffusion range of siderophores (yellow) nor the amount of iron brought into solution (orange-red gradient) increases per cell, but the probability of iron-loaded siderophores returning to cells increases, because cells can access each other’s siderophores (brown area, double black arrows). Emojis inside cells depict the relative fitness consequences for a cell in the different scenarios. This synergistic effect manifests irrespective of whether the iron is relatively homogenously distributed (a, b, blue-shaded gradient) or occurs in clumped particles (c, d, blue circles) in the environment, or whether cells are motile (a, c) or surface-attached (b, d).
Social interactions and siderophores
Cooperation
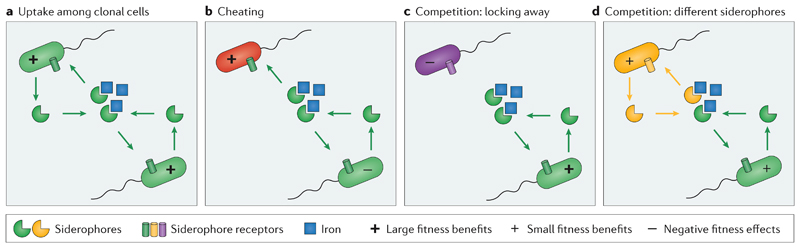
Because secreted molecules can be shared between cells siderophore production is often considered a form of cooperation61. Cooperative behaviours involve an actor generating benefits for another individual, with such interactions having at least partly been selected for this purpose15,62. According to evolutionary theory two forms of cooperation can be distinguished depending on whether the direct (lifetime) fitness consequence for the actor is negative (altruistic cooperation) or positive (mutually beneficial cooperation)62. For both types of cooperation, the inclusive fitness (that is, direct fitness benefits plus indirect benefits of assisting relatives) must be positive in order to be selected for15. These evolutionary aspects have been very well studied for the siderophore pyoverdine produced by Pseudomonas aeruginosa 16, for which both the cost of production63,64 and the benefit to others65 have been demonstrated. The consensus across numerous papers64–67 studying the cooperative nature of this system is that siderophores are public goods and their production typically represents a mutualistic form of cooperation, providing direct benefits to both the producers and the recipients (Fig. 4a). Similar evidence emerged from studies on other siderophores including pyochelin from P. aeruginosa and Burkholderia cenocepacia 68,69, ornibactin from B. cenocepacia 69 and enterochelin from Escherichia coli 70.
Figure 4. Siderophore-mediated social interactions.
The figure shows representative examples of social interactions driven by secreted siderophores (partial circles) that bind insoluble iron (blue squares) and for which specific receptors (cylinders) are required for uptake. Emojis inside cells depict the relative fitness consequences for the interacting partners. a | During cooperation in clonal groups, siderophores are mutually shared and accelerate iron uptake through a common siderophore pool. b | Cheating happens when siderophore non-producers have the matching receptor for uptake and thereby exploit the common pool of siderophores without contributing to it. c | Competition can happen when producers lock iron away from non-producers that lack the matching receptor for heterologous siderophore uptake. d | Another competition scenario involves two species that each produce their specific siderophore. The competitive strength of the species is influenced by the amount and kinetics of siderophore production and the iron affinity of their siderophores.
The evolutionary origin of siderophore secretion is unknown, and a key question is whether the relative importance of self-directed benefits versus indirect benefits to others has changed over evolutionary time. One plausible scenario is that secretion was initially selected for the direct fitness advantages it provides over membrane-bound iron-scavenging systems under the conditions discussed in Fig. 2. Then perhaps later on, regulatory mechanisms evolved that enabled fine-tuned regulation of siderophore production to increase fitness benefits to others. Such adaptations could include siderophore-mediated signalling44 to adjust production levels to the local neighbourhood, siderophore recycling37,71, the specific upregulation of siderophore production to compensate for the presence of non-producers65,66, and the evolution of more diffusible molecules to increasingly benefit others18.
Cheating
Although cooperation can generate substantial fitness benefits for all individuals involved, it might not be evolutionarily stable. The reason is simple: every cooperative act can be exploited by cheaters that save the costs of cooperation but freeride on the cooperative benefits generated by others72,73 (Figure 4b). The tug-of-war between cooperators and cheaters can result in the so-called ‘tragedy of the commons’72,74–76, in which cooperation, although maximising group benefits, collapses due to exploitation by selfish cheaters. These evolutionary concepts were the starting point for a wealth of studies examining interactions between cooperative siderophore producers and non-producers acting as cheaters16,77–81. Again, work on the siderophore pyoverdine from P. aeruginosa took a lead role. This body of work revealed that the collapse of cooperation can indeed occur in certain setups16,82–84, but under many conditions producers are also able to co-exist with non-producers76. This finding spurred interest in understanding the mechanisms that contribute to the maintenance of cooperative siderophore production. One mechanism identified is increased spatial structure, which limits both cell dispersal and siderophore diffusion, and thereby ensures that siderophores are more likely shared between clonemates than cheaters52,67,79,85. In addition, fine-tuned regulatory circuits and recycling mechanisms, which optimise the benefit-to-cost ratio of siderophore investment, also play a role37,71,86,87. Furthermore, genetic architecture constrains cheater success, as found in the case of P. aeruginosa in which pyoverdine non-producers up-regulate the production of the secondary siderophore pyochelin68, and in B. cenocepacia in which ornibactin mutants can no longer produce the receptor required for ornibactin uptake69. Low cell density can also limit the cheaters’ access to siderophores60,70. Finally, negative-frequency dependent selection leads to cheaters only experiencing relative fitness benefits when rare but not when common, as the exploitable pool of siderophores depletes when cooperator frequency declines88,89.
While this work has fundamentally improved our understanding of social interactions between producers and non-producers under laboratory conditions, there is increasing evidence that the observed social patterns also matter in natural systems. Specifically, siderophore non-producers were found in soil20,90, freshwater20, marine17, hospital91 and host-associated19,92 habitats. Moreover, it was demonstrated that in certain cases non-producers can act as cheaters and exploit foreign siderophores17,20,90,93. Although non-producers can evolve for other reasons than cheating (for example, in habitats with high iron availability)94,95, the cumulative evidence suggests that siderophore-mediated cooperation and cheating are important features of bacterial communities.
Competition
The discussed scenarios of cooperation and cheating typically involve interactions among closely related individuals that share the same siderophore system. However, bacteria often live in diverse consortia with interacting strains that likely have different siderophores, each requiring a specific cognate receptor for iron uptake12,18. For these scenarios, evolutionary theory predicts that siderophores can function as competitive agents against other species26,96. The reasoning is straightforward: in an iron-limited environment, siderophores can lock iron away from competitors that do not have the matching receptor for uptake and thereby reserve iron for the cognate producers (Figure 4c). The consequences of locking iron away were found to be particularly dramatic for non-producers, which have no opportunities to enter the competition with their own siderophores and thus experience severe iron starvation20(Figure 4c). If two competing strains both produce their own specific siderophore (Figure 4d) the outcome of the competition can depend on a number of factors, including the amount of siderophores produced, the iron-binding properties of the competing siderophores and the plasticity and temporal dynamics of siderophore production. For example, experiments with Pseudomonas and Burkholderia species showed that the relative differences in iron-binding properties between siderophores indeed determined the outcome of the competition21,97. At the regulatory level, it was found that P. aeruginosa up-regulates siderophore production in competition with Staphylococcus aureus 98, whereas, in competition with B. cenocepacia, P. aeruginosa increased not the overall production of pyoverdine but the rate of production during the early growth phase, which conferred P. aeruginosa a competitive advantage99.
Evolutionary and ecological dynamics
Co-evolutionary arms races
Because social interactions driven by siderophores can have substantial fitness effects, one would predict (based on evolutionary theory) that these interactions should trigger reciprocal evolutionary adaptations in interacting strains or species100. In other words, we would expect evolutionary transitions between the scenarios outlined in Fig. 4. Aspects of these transitions have been well studied in Pseudomonas bacteria and their siderophore pyoverdine. A first and frequently observed transition can occur in populations of siderophore producers, which are invaded by de novo arising non-producer mutants. This invasion induces a shift from cooperation to cheating at the population level (Figure 4a to 4b)68,78,98. Another transition could be induced by siderophore producers that adapt to the presence of cheaters. In P. aeruginosa such adaptations were found to include reduced pyoverdine production82,101, increased investment in alternative iron acquisition mechanisms102, and possibly the evolution of new variants of siderophore-receptor pairs that restrict access of the cheaters to siderophores103. The evolution of siderophore-receptor diversity has attracted a lot of attention, especially in Pseudomonas spp., in which dozens of different pyoverdine-receptor variants exist20,103–107. Although a direct proof that cheating drives pyoverdine-receptor diversification is still lacking, diversification could induce the transition from cheating to locking iron away from non-producers (Figure 4b to 4c). Non-producers could finally close the co-evolutionary cycle by evolving receptors that match the modified siderophores, or by acquiring a matching receptor via horizontal gene transfer. Indeed, natural Pseudomonas spp. isolates from soil and freshwater habitats (non-producers and producers) had a whole array of different pyoverdine receptors, suggesting that bacteria have evolved strategies to exploit multiple pyoverdine types produced by different community members20,108,109 with regulatory mechanisms being in place to selectively up-regulate the corresponding receptors106.
Antagonistic co-evolution should also occur between two species or strains with different siderophore systems(Figure 4d), in which reciprocal siderophore competition could select for increased siderophore production and/or novel siderophore variants with increased iron affinity26 or altered diffusion properties11,18. Moreover, the acquisition of heterologous siderophore receptors through horizontal gene transfer seems to be a common strategy to increase competitive abilities. For example, P. aeruginosa has receptors for the bacterial siderophores enterobactin and ferrioxamine, and a receptor for the fungal siderophore ferrichrome45,110. To add another level of complexity, simultaneous co-evolutionary arms races between different siderophore producers and non-producers in diverse natural communities might offer a potential explanation for why bacteria often produce multiple types of siderophores (for example, Vibrio anguillarum produces anguibactin and vanchrobactin111; P. aeruginosa produces pyovedine and pyochelin112; B. cenocepacia produces ornibactin and pyochelin113). Specifically, the production of different siderophores might increase competitiveness, because it should decrease the likelihood that a competitor can exploit the full siderophore repertoire of a producer. Taken together, these considerations demonstrate that siderophore-mediated iron acquisition strategies are not fixed but in flux, likely driven by the bacterial battle for iron.
Ecological dependencies
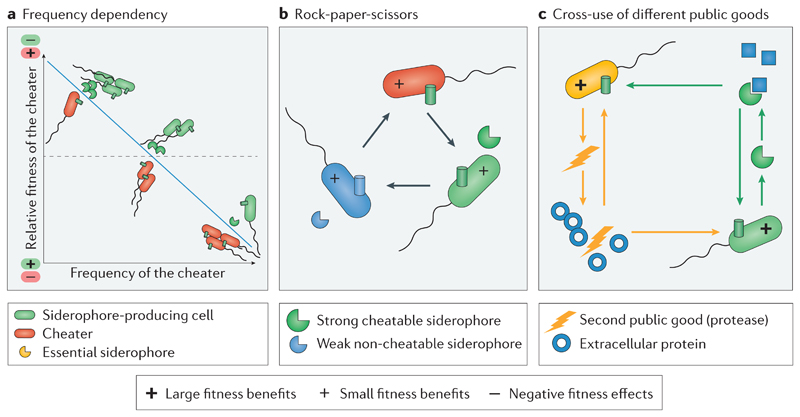
Although cheating provides a short-term benefit to siderophore non-producers, it can also make them dependent on producers in the long run, especially in environments in which siderophores are essential for growth88,89,114. Extreme cases of such ecological dependencies have been described for marine bacteria115: several unculturable species became culturable upon the supplementation of exogenous siderophores. Siderophore-based dependencies typically result in negative frequency-dependent selection (Figure 5a), as the relative fitness of non-producers declines when they become more common in the population, leading to a stable co-existence between donating siderophore producers and dependent non-producers88,89.
Figure 5. Siderophores induce eco-evolutionary dynamics in bacterial communities.
a | If siderophores (yellow partial circles) are essential for growth, siderophore producers (green cells) and non-producing cheaters (red cells) can co-exist in a community through negative frequency-dependent selection. b | Strains or species with different siderophore strategies can engage in non-transitive community dynamics, and thereby co-exist and foster biodiversity. The example depicts interactions between a cheater (red) outcompeting the siderophore producer, which secretes a compatible siderophore (green). This siderophore producer in turn outcompetes the second siderophore producer (blue), secreting a less potent siderophore. This weaker siderophore producer, however, outcompetes the non-producer because it produces a siderophore for which the non-producer lacks the matching receptor. This scenario leads to a rock-paper-scissors dynamic, in which strains chase each other in circles with no overall winner. c | When, in addition to siderophores (green partial circles), a second public good is important for growth (yellow flash, for example, proteases required for extra-cellular protein degradation), specialization could evolve, leading to each strain or species producing only one public good and exchange at the community level. Emojis inside cells depict the relative fitness consequences for the interacting partners.
Non-transitive community dynamics
For simplicity, we have introduced siderophore-mediated social interactions as pairwise interactions between two strains and/or species (Figure 4). This obviously does not capture the diversity of bacterial strategies existing in complex communities that harbour many different types of siderophore non-producers and producers with different siderophore-receptor pairs. From an ecological perspective, the co-occurrence of a diverse set of social strategies could give raise to non-transitive population dynamics, in which the co-existence of strains and species can be fostered without any evolutionary change116–118(Figure 5b). Laboratory studies with experimental communities with either different P. aeruginosa strains97 or P. aeruginosa and B. cenocepacia assemblies21 indeed described such non-transitive population dynamics. In both cases, interacting strains combined the social interactions described in Figs. 4b-d: a strain with a relatively weak siderophore (i) locked iron away from a non-producer, but (ii) was dominated by a strain producing a strong siderophore, which in turn (iii) was exploited by the non-producer. Because each of the strains outcompeted one community member but lost against the other one, there was no overall winner, such that all strains could co-exist. Although these studies provide proofs of concept, the role these non-transitive interactions might have in shaping the composition and stability of natural species assemblies still remains to be explored.
Specialization and cross-use
Although most of the studies referred to in this Review focussed on siderophores alone, we should not forget that bacteria often secrete other compounds, including enzymes (for example, proteases119), biosurfactants120 and other metallophores28, which can also potentially be shared as public goods among cells59,121. Thus, arguments have been raised that siderophore non-producers might only be cheaters with regard to this public good, whereas they are cooperators for other public goods81,122, and that they might exchange their public goods with siderophore producers in a mutually beneficial way94,123(Figure 5c). Such division of labour could eventually lead to mutual dependencies124. Although laboratory studies on genetically engineered or experimentally selected communities demonstrated the benefits of mutualistic cross-feeding interactions involving the exchange of nutrients such as amino acids125–127, evidence for mutualist interactions involving siderophores is scarce. There is, however, one convincing example involving marine bacteria that traded siderophores for fixed carbon with an algal phytoplankton128. The scarcity of mutualistic interactions involving public goods (apart from amino acids) might match theoretical work suggesting that the conditions favouring mutualism between different bacterial strains and/or species are restrictive129. In further support, empirical observations show that competitive interactions predominate over cooperative interactions in bacterial communities130.
Host interactions
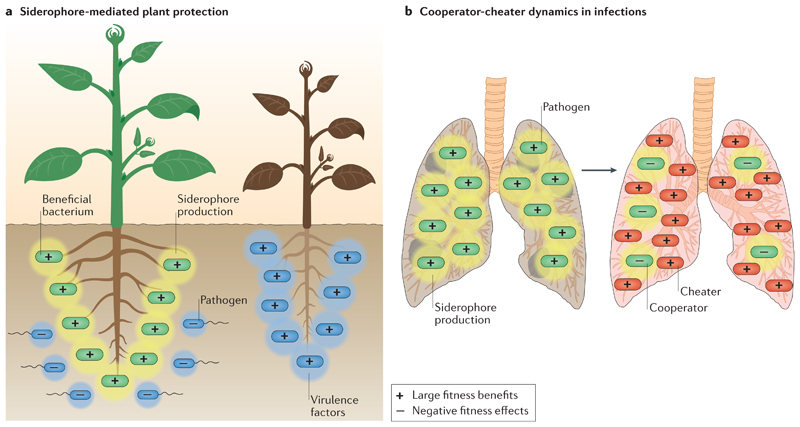
As many bacteria associate with multicellular organisms including plants, animals and humans131, the above described siderophore-mediated social interactions and population dynamics can potentially have consequences for the host, too. The role bacterial siderophores can play for hosts was first recognized in the plant rhizosphere132,133. This habitat harbours diverse microbial communities134, and it was found that siderophores of commensal or mutualistic species can reduce the growth of plant pathogens132,133,135,136. Although the exact mechanism of pathogen suppression still remains to be elucidated, there is evidence that siderophore-mediated competition is involved as described in Figures 4c+d, which leads to plant-protecting bacteria locking iron away from pathogens133 (Figure 6a). Similar mechanisms of pathogen suppression might operate in gut microbiota. For example, a probiotic E. coli strain, which occurs in the human gut, reduced the intestinal colonization by Salmonella enterica subs. enterica serovar Typhimurium through siderophore-mediated iron competition137 and through the targeted antibacterial activity of the siderophore mimic microcin138.
Figure 6. Siderophores and their effects on hosts.
a | Siderophores (yellow shaded zones) from beneficial rhizosphere bacteria (green cells) can protect plants from pathogens (blue cells) and their virulence factors (blue shaded zones), and keep the plants healthy (left green plant). In this scenario, siderophore incompatibility between the plant beneficial bacteria and the pathogen is thought to lead to plant protection. b | Siderophores (yellow shaded zones) of pathogenic bacteria (green cells) can function as virulence factors and damage host tissue and promote pathogen growth (left lung). If siderophore non-producers (red cells) occur or de novo evolve in such infections, they can act as cheaters. If these cheaters can spread the induced cooperator-cheater dynamic can potentially lower the virulence of the infection. Emojis inside cells depict the relative fitness consequences for the interacting partners.
Another set of studies has examined whether siderophore-mediated evolutionary dynamics occur within hosts, especially in the context of infections. Iron is limited within the host as it is bound to proteins such as transferrin and lactoferrin6,9. Thus, pathogens typically have to deploy siderophores to scavenge iron from host tissues, rendering siderophores important determinants of virulence6,9,139. Because of their importance for bacterial growth, it has been suggested that cheating could occur during infections, when de novo evolved non-producers exploit the siderophores of producers, inducing cooperator-cheater dynamics that could steer infections towards lower virulence23,61,140. This scenario has been studied in detail in chronic lung infections with P. aeruginosa in individuals with cystic fibrosis19,22,92,141 (Figure 6b). Over time, the following sequence of evolutionary events were repeatedly observed across patients: infections were established by wildtype producers of the siderophore pyoverdine; mutations in the pyoverdine regulon gave rise to pyoverdine non-producers; pyoverdine non-producers fixed in a high proportion of the patients; mutations accumulated in the pyoverdine receptor genes of non-producers to alleviate the costs of receptor production; and finally the upregulation of the Phu system occurred, a private iron acquisition system extracting iron from host heme molecules. One way to interpret this sequence of events is that non-producers evolved as cheaters, taking over the cooperative population, but because iron is still limited in the lung, siderophore non-producers eventually switched to an alternative membrane-bound iron acquisition system to compensate for the inability to make siderophores. However, it is also important to consider that the lung environment itself changes over time142–144, and the increasing level of anaerobiosis has been proposed as an additional factor inducing a shift in the use of siderophore-bound Fe3+ to heme-bound Fe2+ 145.
Finally, potential cheating in infections is not limited to within-species interactions, as evidenced by a study on inter-species competition in the gut of the nematode Caenorhabditis elegans 146. In this study, the commensal Enterococcus faecalis exploited siderophores produced by the pathogen Staphylococcus aureus. In response to exploitation, S. aureus down-regulated siderophore production, which in turn reduced virulence as a by-product.
Conclusions
Nobel Prize laureate Nikolaas Tinbergen explained in one of his seminal papers that every biological phenomenon can be approached by four different questions, which belong to two main categories: proximate and ultimate questions147. Proximate (how) questions address mechanistic and developmental aspects of the biological phenomenon, whereas ultimate (why) questions address evolutionary (phylogenetic) and adaptive questions. Bacterial siderophores are a perfect example of how this concept of complementary questions can be used to obtain an integrative view on this important class of secreted metabolites. While there was initially a focus on the proximate aspects of siderophores, exploring the mechanistic aspects of synthesis, secretion, uptake and regulation, ultimate questions covering evolutionary and adaptive aspects have caught up over the last decade. The aim of our Review was to elucidate theses ultimate aspects and link them to the mechanistic aspects of siderophore production. The emerging view of this integrative approach is that siderophores are more than just iron carriers: they are important mediators of interactions between members of microbial assemblies and their eukaryotic hosts.
Despite these novel insights there are still fundamental gaps in our knowledge. One obvious shortcoming is that the current research on eco-evolutionary aspects of siderophore production is biased towards Pseudomonas species, and one particular siderophore — pyoverdine. We clearly need additional study systems to verify whether the insights gained from pseudomonads are representative of general siderophore biology, transcending bacterial phylogeny, and habitat and host types. Valuable contributions in this context are the evolutionary studies on aerobactin and vibrioferrin in Vibrio sp.17, enterochelin in E. coli 70, ornibactin and pyochelin in B. cenocepacia 69, and a comparative study across 189 different siderophores18, which could confirm general eco-evolutionary roles of siderophore but also helped to add new elements to the picture. Along a similar line, there is a bias towards studies on simplified laboratory systems. Although these systems offer many advantages as they are tractable and easy to manipulate, we must also embrace more natural systems, studying interactions between more than two strains and species at a multi-dimensional scale and including other social traits in addition to siderophores. Only such integrative approaches will enable us to understand assembly patterns and the functioning (for example, the stability or productivity) of complex microbial consortia. Finally, we might also want to go back in time and think about the evolutionary origins of siderophores. The two-pronged and co-ordinated evolution of a secreted iron-chelating ligand and a receptor for the purpose of iron uptake is an extremely unlikely scenario. A more plausible (yet speculative) possibility is that siderophores have evolved prior to the Great Oxygen Event148 as agents to prevent iron intoxication during an era when iron was highly soluble. Only after the Great Oxygen Event when iron availability became drastically reduced, bacteria might have evolved receptors to retrieve this newly limiting resource. In the light of all these considerations, we believe that there is still much to be discovered and we have so far only explored the tip of the iceberg of siderophore biology.
Acknowledgments
This work was funded by the European Research Council under the Grant Agreement no. 681295 and the Swiss National Science Foundation no. 182499 (both to RK), the German Science Foundation no. KR 5017/2-1 (to JK), and a University Research Priority Program URPP grant (to ÖÖ).
Glossary
- Siderophores
secondary metabolites with high affinity and specificity for iron that function as an organic ligands, serve the purpose of iron acquisition, and are regulated in response to the producer’s need for iron.
- Biofilms
aggregates of microorganisms that are embedded within a self-produced matrix of extracellular polymeric substances and that adhere to each other and/or a surface.
- Cooperation
a social behaviour which provides a benefit to another individual, and which has evolved and/or is currently maintained (at least partly) because of its beneficial effect on the recipient.
- Pseudomonas aeruginosa
a metabolic versatile, ubiquitous, rod-shaped, Gram-negative bacterium that can opportunistically infect plants, animals and humans, and is known for its high intrinsic resistance to antibiotics.
- Public goods
costly resources that not only benefits the producer, but also other members of the population or local community.
- Tragedy of the commons
a situation in which cooperation would be beneficial in the long term, but cooperation breaks down because individuals selfishly pursue their own short-term interests.
- Negative frequency dependent selection
an evolutionary process by which the relative fitness of a phenotype is high when it occurs at low frequency in the population, but decreases as it becomes more common relative to other phenotypes.
- Cheating
exploitation of a cooperative behaviour by an individual that does not cooperate (or cooperates less than its fair share), whereby the cheating individual reaps the benefits of cooperation at the expense of the cooperating individual.
- Competition
a situation that arises when two or more individuals of the same or different species strive for the same limited resource, resulting in immediate costs for all involved individuals.
- Horizontal gene transfer
the transfer of genetic material from one individual to another individual (of the same or different species) that does not involve the vertical transmission of DNA typical for cell division and reproduction.
- Co-evolutionary arms races
evolutionary tug-of-war between competing strains or species, whereby adaptations in one party select for counter-adaptations in the other party.
- Non-transitive population dynamics
population dynamics arising from non-hierarchical circular competitive relationships between species, in which each species is both superior and inferior towards different community members, with no overall winner existing in the population.
- Metallophores
a secondary metabolite with high affinity and specificity for a given metal. It functions as an organic ligand, serves the purpose of acquiring the metal in question, and is regulated in response to metal limitation.
- Division of labour
the division of a task that occurs when cooperating individuals specialize to carry out specific subtasks.
- Cross-feeding
a nutritional interdependence between different strains or species, in which each species feeds on the metabolic products released by the other species.
- Virulence
the damage caused to the host by a parasite or pathogen, measured as the decrease in host fitness.
Footnotes
Author contributions
All authors developed together the concept of the review and wrote the paper.
Competing interests
The authors declare no competing interests.
References
- 1.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 2.Boyd PW, Ellwood MJ. The biogeochemical cycle of iron in the ocean. Nat Geosci. 2010;3:675–682. [Google Scholar]
- 3.Emerson D, Roden E, Twining BS. The microbial ferrous wheel: iron cycling in terrestrial, freshwater, and marine environments. Front Microbiol. 2012;3 doi: 10.3389/fmicb.2012.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerinot ML. Microbial iron transport. Annu Rev Microbiol. 1994;48:743–772. doi: 10.1146/annurev.mi.48.100194.003523. [DOI] [PubMed] [Google Scholar]
- 5.Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 6.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. 2013;13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frawley ER, Fang FC. The ins and outs of bacterial iron metabolism. Mol Microbiol. 2014;93:609–616. doi: 10.1111/mmi.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barber MF, Elde NC. Buried treasure: evolutionary perspectives on microbial iron piracy. Trends Genet. 2015;31:627–636. doi: 10.1016/j.tig.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheldon JR, Heinrichs DE. Recent developments in understanding the iron acquisition strategies of gram positive pathogens. FEMS Microbiol Rev. 2015;39:592–630. doi: 10.1093/femsre/fuv009. [DOI] [PubMed] [Google Scholar]
- 11.Boiteau RM, et al. Siderophore-based microbial adaptations to iron scarcity across the eastern Pacific Ocean. Proc Natl Acad Sci U S A. 2016;113:14237–14242. doi: 10.1073/pnas.1608594113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hider RC, Kong X. Chemistry and biology of siderophores. Nat Prod Rep. 2010;27:637–657. doi: 10.1039/b906679a. [ This is an extensive and comprehensive review on siderophore chemistry, biosynthesis, and transport. ] [DOI] [PubMed] [Google Scholar]
- 13.Faraldo-Gómez JD, Sansom MSP. Acquisition of siderophores in gram-negative bacteria. Nat Rev Mol Cell Biol. 2003;4:105–116. doi: 10.1038/nrm1015. [DOI] [PubMed] [Google Scholar]
- 14.Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 15.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [ This review offers a conceptual overview of social evolution theory as it applies to this review and microorganisms in general. ] [DOI] [PubMed] [Google Scholar]
- 16.Griffin A, West SA, Buckling A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430:1024–1027. doi: 10.1038/nature02744. [ This study demonstrates for the first time that siderophores are a public good in Pseudomonas aeruginosa, and reveals conditions under which cheaters can spread in populations. ] [DOI] [PubMed] [Google Scholar]
- 17.Cordero OX, Ventouras L-A, DeLong EF, Polz MF. Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc Natl Acad Sci U S A. 2012;109:20059–20064. doi: 10.1073/pnas.1213344109. [ This study shows that public good interactions drive the evolution of siderophore-based iron acquisition strategies in Vibrionaceae living on marine particles. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kümmerli R, Schiessl KT, Waldvogel T, McNeill K, Ackermann Habitat structure and the evolution of diffusible siderophores in bacteria. Ecol Lett. 2014;17:1536–1544. doi: 10.1111/ele.12371. [DOI] [PubMed] [Google Scholar]
- 19.Andersen SB, Marvig RL, Molin S, Johansen HK, Griffin AS. Long-term social dynamics drive loss of function in pathogenic bacteria. Proc Natl Acad Sci U S A. 2015;112:10756–10761. doi: 10.1073/pnas.1508324112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butaitė E, Baumgartner M, Wyder S, Kümmerli R. Siderophore cheating and cheating resistance shape competition for iron in soil and freshwater Pseudomonas communities. Nat Commun. 2017;8:414. doi: 10.1038/s41467-017-00509-4. [ This study shows that siderophore-mediated social interactions drive competitive dynamics in soil and freshwater communities of Pseudomonas bacteria. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leinweber A, Fredrik Inglis R, Kümmerli R. Cheating fosters species co-existence in well-mixed bacterial communities. ISME J. 2017;11:1179–1188. doi: 10.1038/ismej.2016.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen SB, et al. Privatisation rescues function following loss of cooperation. eLife. 2018;7 doi: 10.7554/eLife.38594. e38594. [ This study traces the evolutionary trajectory of siderophore production, exploitation, and switch to alternative iron uptake strategies in chronic lung infections of cystic fibrosis patients. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Granato ET, Ziegenhain C, Marvig RL, Kummerli R. Low spatial structure and selection against secreted virulence factors attenuates pathogenicity in Pseudomonas aeruginosa . ISME J. 2018;12:2907–2918. doi: 10.1038/s41396-018-0231-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leventhal GE, Ackermann M, Schiessl KT. Why microbes secrete molecules to modify their environment: the case of iron-chelating siderophores. J R Soc Interface. 2019;16 doi: 10.1098/rsif.2018.0674. [ This theoretical study pins down the conditions under which siderophore secretion is favoured over surface-bound iron uptake systems. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Völker C, Wolf-Gladrow DA. Physical limits on iron uptake mediated by siderophores or surface reductases. Mar Chem. 1999;65:227–244. [Google Scholar]
- 26.Niehus R, Picot A, Oliveira NM, Mitri S, Foster KR. The evolution of siderophore production as a competitive trait. Evolution. 2017;71:1443–1455. doi: 10.1111/evo.13230. [ This theoretical paper shows that siderophores can evolve as competitive agents against other bacteria lacking the cognate receptor required for siderophore uptake. ] [DOI] [PubMed] [Google Scholar]
- 27.Sandy M, Butler A. Microbial iron acquisition: marine and terrestrial siderophores. Chem Rev. 2009;109:4580–4595. doi: 10.1021/cr9002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraemer SM, Duckworth OW, Harrington JM, Schenkeveld WDC. Metallophores and trace metal biogeochemistry. Aquat Geochem. 2015;21:159–195. [Google Scholar]
- 29.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grünewald J, Marahiel MA. Chemoenzymatic and template-directed synthesis of bioactive macrocyclic peptides. Microbiol Mol Biol Rev. 2006;70:121–146. doi: 10.1128/MMBR.70.1.121-146.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donadio S, Monciardini P, Sosio M. Polyketide synthases and nonribosomal peptide synthetases: the emerging view from bacterial genomics. Nat Prod Rep. 2007;24:1073–1109. doi: 10.1039/b514050c. [DOI] [PubMed] [Google Scholar]
- 32.Lamb AL. Breaking a pathogen's iron will: Inhibiting siderophore production as an antimicrobial strategy. Biochim Biophys Acta. 2015;1854:1054–1070. doi: 10.1016/j.bbapap.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carroll CS, Moore MM. Ironing out siderophore biosynthesis: a review of non-ribosomal peptide synthetase (NRPS)-independent siderophore synthetases. Crit Rev Biochem Mol Biol. 2018;53:356–381. doi: 10.1080/10409238.2018.1476449. [DOI] [PubMed] [Google Scholar]
- 34.Krewulak KD, Vogel HJ. Structural biology of bacterial iron uptake. Biochim Biophys Acta. 2008;1778:1781–1804. doi: 10.1016/j.bbamem.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 35.Schalk IJ, Guillon L. Fate of ferrisiderophores after import across bacterial outer membranes: different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways. Amino Acids. 2013;44:1267–1277. doi: 10.1007/s00726-013-1468-2. [DOI] [PubMed] [Google Scholar]
- 36.Ganne G, et al. Iron release from the siderophore pyoverdine in Pseudomonas aeruginosa involves three new actors: FpvC, FpvG, and FpvH. ACS Chem Biol. 2017;12:1056–1065. doi: 10.1021/acschembio.6b01077. [DOI] [PubMed] [Google Scholar]
- 37.Imperi F, Tiburzi F, Visca P. Molecular basis of pyoverdine siderophore recycling in Pseudomonas aeruginosa . Proc Natl Acad Sci U S A. 2009;106:20440–20445. doi: 10.1073/pnas.0908760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones CM, et al. Self-poisoning of Mycobacterium tuberculosis by interrupting siderophore recycling. Proc Natl Acad Sci U S A. 2014;111:1945–1950. doi: 10.1073/pnas.1311402111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin H, Fischbach MA, Liu DR, Walsh CT. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J Am Chem Soc. 2005;127:11075–11084. doi: 10.1021/ja0522027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neumann W, Sassone-Corsi M, Raffatellu M, Nolan EM. Esterase-catalyzed siderophore hydrolysis activates an enterobactin-ciprofloxacin conjugate and confers targeted antibacterial activity. J Am Chem Soc. 2018;140:5193–5201. doi: 10.1021/jacs.8b01042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Escolar L, Pérez-Martín J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Troxell B, Hassan HM. Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front Cell Infect Microbiol. 2013;3:59. doi: 10.3389/fcimb.2013.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leoni L, Orsi N, de Lorenzo V, Visca P. Functional analysis of PvdS, an iron starvation sigma factor of Pseudomonas aeruginosa . J Bacteriol. 2000;182:1481–1491. doi: 10.1128/jb.182.6.1481-1491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamont IL, Beare P, Ochsner U, Vasil AI, Vasil ML. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa . Proc Natl Acad Sci U S A. 2002;99:7072–7077. doi: 10.1073/pnas.092016999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornelis P. Iron uptake and metabolism in pseudomonads. Appl Microbiol Biotechnol. 2010;86:1637–1645. doi: 10.1007/s00253-010-2550-2. [DOI] [PubMed] [Google Scholar]
- 46.Mathew A, Eberl L, Carlier AL. A novel siderophore-independent strategy of iron uptake in the genus Burkholderia . Mol Microbiol. 2014;91:805–820. doi: 10.1111/mmi.12499. [DOI] [PubMed] [Google Scholar]
- 47.Lau CKY, Krewulak KD, Vogel HJ. Bacterial ferrous iron transport: the Feo system. FEMS Microbiol Rev. 2015;40:273–298. doi: 10.1093/femsre/fuv049. [DOI] [PubMed] [Google Scholar]
- 48.Vetter YA, Deming JW, Jumars PA, Krieger-Brockett BB. A predictive model of bacterial foraging by means of freely released extracellular enzymes. Microb Ecol. 1998;36:75–92. doi: 10.1007/s002489900095. [DOI] [PubMed] [Google Scholar]
- 49.Driscoll WW, Pepper JW. Theory for the evolution of diffusible external goods. Evolution. 2010;64:2682–2687. doi: 10.1111/j.1558-5646.2010.01002.x. [DOI] [PubMed] [Google Scholar]
- 50.Allen B, Gore J, Nowak MA. Spatial dilemmas of diffusible public goods. eLife. 2013;2 doi: 10.7554/eLife.01169. e01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobay A, Bagheri HC, Messina A, Kümmerli R, Rankin DJ. Interaction effects of cell diffusion, cell density and public goods properties on the evolution of cooperation in digital microbes. J Evol Biol. 2014;27:1869–1877. doi: 10.1111/jeb.12437. [DOI] [PubMed] [Google Scholar]
- 52.Kümmerli R, Griffin AS, West SA, Buckling A, Harrison F. Viscous medium promotes cooperation in the pathogenic bacterium Pseudomonas aeruginosa . Proc R Soc B. 2009;276:3531–3538. doi: 10.1098/rspb.2009.0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu G, Martinez JS, Groves JT, Butler A. Membrane affinity of the amphiphilic marinobactin siderophores. J Am Chem Soc. 2002;124:13408–13415. doi: 10.1021/ja026768w. [DOI] [PubMed] [Google Scholar]
- 54.Martinez JL, et al. Structure and membrane affinity of a suite of amphiphilic siderophores produced by a marine bacterium. Proc Natl Acad Sci U S A. 2003;100:3754–3759. doi: 10.1073/pnas.0637444100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sidebottom AM, Johnson AR, Karty JA, Trader DJ, Carlson EE. Integrated metabolomics approach facilitates discovery of an unpredicted natural product suite from Streptomyces coelicolor M145. ACS Chem Biol. 2013;8:2009–2016. doi: 10.1021/cb4002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Claessen D, Rozen DE, Kuipers OP, Søgaard-Andersen L, van Wezel GP. Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat Rev Microbiol. 2014;12:115–124. doi: 10.1038/nrmicro3178. [DOI] [PubMed] [Google Scholar]
- 57.Flemming HC, et al. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 58.Flemming HC, Wuertz S. Bacteria and archaea on Earth and their abundance in biofilms. Nat Rev Microbiol. 2019;17:247–260. doi: 10.1038/s41579-019-0158-9. [DOI] [PubMed] [Google Scholar]
- 59.West SA, Diggle SP, Buckling A, Gardner A, Griffin AS. The social lives of microbes. Annu Rev Ecol Evol Syst. 2007;38:53–77. [Google Scholar]
- 60.Ross-Gillespie A, Gardner A, Buckling A, West SA, Griffin AS. Density dependence and cooperation: theory and a test with bacteria. Evolution. 2009;63:2315–2325. doi: 10.1111/j.1558-5646.2009.00723.x. [DOI] [PubMed] [Google Scholar]
- 61.Buckling A, et al. Siderophore-mediated cooperation and virulence in Pseudomonas aeruginosa . FEMS Microbiolol Ecol. 2007;62:135–141. doi: 10.1111/j.1574-6941.2007.00388.x. [DOI] [PubMed] [Google Scholar]
- 62.West SA, Griffin AS, Gardner A. Social semantics: altruism, cooperation, mutualism, strong reciprocity and group selection. J Evol Biol. 2007;20:415–432. doi: 10.1111/j.1420-9101.2006.01258.x. [DOI] [PubMed] [Google Scholar]
- 63.Sexton DJ, Schuster M. Nutrient limitation determines the fitness of cheaters in bacterial siderophore cooperation. Nat Commun. 2017;8:230. doi: 10.1038/s41467-017-00222-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schiessl KT, et al. Individual- versus group-optimality in the production of secreted bacterial compounds. Evolution. 2019;73:675–688. doi: 10.1111/evo.13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weigert M, Kümmerli R. The physical boundaries of public goods cooperation between surface-attached bacterial cells. Proc R Soc B. 2017;284 doi: 10.1098/rspb.2017.0631. 20170631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harrison F. Dynamic social behaviour in a bacterium: Pseudomonas aeruginosa partially compensates for siderophore loss to cheats. J Evol Biol. 2013;26:1370–1378. doi: 10.1111/jeb.12126. [DOI] [PubMed] [Google Scholar]
- 67.Julou T, et al. Cell-cell contacts confine public goods diffusion inside Pseudomonas aeruginosa clonal microcolonies. Proc Natl Acad Sci U S A. 2013;110:12577–12582. doi: 10.1073/pnas.1301428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ross-Gillespie A, Dumas Z, Kümmerli R. Evolutionary dynamics of interlinked public goods traits: an experimental study of siderophore production in Pseudomonas aeruginosa . J Evol Biol. 2015;28:29–39. doi: 10.1111/jeb.12559. [DOI] [PubMed] [Google Scholar]
- 69.Sathe S, Mathew A, Agnoli K, Eberl L, Kümmerli R. Genetic architecture constrains exploitation of siderophore cooperation in Burkholderia cenocepacia . Evolution Letters. 2019 doi: 10.1002/evl3.144. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scholz RL, Greenberg EP. Sociality in Escherichia coli: enterochelin is a private good at low cell density and can be shared at high cell density. J Bacteriol. 2015;197:2122–2128. doi: 10.1128/JB.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kümmerli R, Brown SP. Molecular and regulatory properties of a public good shape the evolution of cooperation. Proc Natl Acad Sci U S A. 2010;107:18921–18926. doi: 10.1073/pnas.1011154107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.MacLean RC. The tragedy of the commons in microbial populations: insights from theoretical, comparative and experimental studies. Heredity. 2008;100:233–239. doi: 10.1038/sj.hdy.6801073. [DOI] [PubMed] [Google Scholar]
- 73.Ghoul M, Griffin AS, West SA. Toward and evolutionary definition of cheating. Evolution. 2014;68:318–331. doi: 10.1111/evo.12266. [DOI] [PubMed] [Google Scholar]
- 74.Hardin G. The tragedy of the commons. Science. 1968;162:1243–1248. [PubMed] [Google Scholar]
- 75.Rankin DJ, Bargum K, Kokko H. The tragedy of the commons in evolutionary biology. Trends Ecol Evol. 2007;22:643–651. doi: 10.1016/j.tree.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 76.Özkaya Ö, Xavier KB, Dionisio F, Balbontin R. Maintenance of microbial cooperation mediated by public goods in single and multiple traits scenarios. J Bacteriol. 2017;199:e00297–00217. doi: 10.1128/JB.00297-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiricny N, et al. Fitness correlates with the extent of cheating in a bacterium. J Evol Biol. 2010;23:738–747. doi: 10.1111/j.1420-9101.2010.01939.x. [DOI] [PubMed] [Google Scholar]
- 78.Dumas Z, Kümmerli R. Cost of cooperation rules selection for cheats in bacterial metapopulations. J Evol Biol. 2012;25:473–484. doi: 10.1111/j.1420-9101.2011.02437.x. [DOI] [PubMed] [Google Scholar]
- 79.Tekwa EW, Nguyen D, Loreau M, Gonzalez A. Defector clustering is linked to cooperation in a pathogenic bacterium. Proc R Soc B. 2017;284 doi: 10.1098/rspb.2017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Becker F, Wienand K, Lechner M, Frey E, Jung H. Interactions mediated by a public good transiently increase cooperativity in growing Pseudomonas putida metapopulations. Sci Rep. 2018;8:4093. doi: 10.1038/s41598-018-22306-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Özkaya Ö, Balbontin R, Gordo I, Xavier KB. Cheating on cheaters stabilizes cooperation in Pseudomonas aeruginosa . Curr Biol. 2018;28:2070–2080. doi: 10.1016/j.cub.2018.04.093. [DOI] [PubMed] [Google Scholar]
- 82.Kümmerli R, et al. Co-evolutionary dynamics between public good producers and cheats in the bacterium Pseudomonas aeruginosa . J Evol Biol. 2015;28:2264–2274. doi: 10.1111/jeb.12751. [DOI] [PubMed] [Google Scholar]
- 83.Vasse M, Torres-Barcelo C, Hochberg ME. Phage selection for bacterial cheats leads to population decline. Proc R Soc B. 2015;282 doi: 10.1098/rspb.2015.2207. 20152207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vasse M, et al. Antibiotic stress selects against cooperation in the pathogenic bacterium Pseudomonas aeruginosa . Proc Natl Acad Sci U S A. 2017;114:546–551. doi: 10.1073/pnas.1612522114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bruce JB, West SA, Griffin AS. Functional amyloids promote retention of public goods in bacteria. Proc R Soc B. 2019;286 doi: 10.1098/rspb.2019.0709. 20190709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kümmerli R, Jiricny N, Clarke LS, West SA, Griffin AS. Phenotypic plasticity of a cooperative behaviour in bacteria. J Evol Biol. 2009;22:589–598. doi: 10.1111/j.1420-9101.2008.01666.x. [DOI] [PubMed] [Google Scholar]
- 87.Ghoul M, et al. Pyoverdin cheats fail to invade bacterial populations in stationary phase. J Evol Biol. 2016;29:1728–1736. doi: 10.1111/jeb.12904. [DOI] [PubMed] [Google Scholar]
- 88.Ross-Gillespie A, Gardner A, West SA, Griffin AS. Frequency dependence and cooperation: theory and a test with bacteria. Am Nat. 2007;170:331–342. doi: 10.1086/519860. [DOI] [PubMed] [Google Scholar]
- 89.Morris JJ. Black Queen evolution: the role of leakiness in structuring microbial communities. Trends Genet. 2015;31:475–482. doi: 10.1016/j.tig.2015.05.004. [ This review explains how secreted and leaky metabolites can spur the evolution of trait loss and dependencies in microbial communities. ] [DOI] [PubMed] [Google Scholar]
- 90.Bruce JB, Cooper GA, Chabas H, West SA, Griffin AS. Cheating and resistance to cheating in natural populations of the bacterium Pseudomonas fluorescens . Evolution. 2017;71:2484–2495. doi: 10.1111/evo.13328. [DOI] [PubMed] [Google Scholar]
- 91.De Vos D, et al. Study of pyoverdin type and production by Pseudomonas aeruginosa isolated from cystic fibrosis patients: prevalence of type II pyoverdine isolates and accumulation of pyoverdine-negative mutations. Arch Microbiol. 2001;175:384–388. doi: 10.1007/s002030100278. [DOI] [PubMed] [Google Scholar]
- 92.Jiricny N, et al. Loss of social behaviours in populations of Pseudomonas aeruginosa infecting lungs of patients with cystic fibrosis. PLoS One. 2014;9:e83124–e83124. doi: 10.1371/journal.pone.0083124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lujan AM, Pedro G, Buckling A. Siderophore cooperation of the bacterium Pseudomonas fluorescens in soil. Biol Lett. 2015;11 doi: 10.1098/rsbl.2014.0934. 20140934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang XX, Rainey PB. Exploring the sociobiology of pyoverdin-producing Pseudomonas . Evolution. 2013;67:3161–3174. doi: 10.1111/evo.12183. [DOI] [PubMed] [Google Scholar]
- 95.Butaitė E, Kramer J, Wyder S, Kümmerli R. Environmental determinants of pyoverdine production, exploitation and competition in natural Pseudomonas communities. Environ Microbiol. 2018;20:3629–3642. doi: 10.1111/1462-2920.14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schiessl KT, Janssen EML, Kraemer SM, McNeill K, Ackermann M. Magnitude and mechanism of siderophore-mediated competition at low iron solubility in the Pseudomonas aeruginosa pyochelin system. Front Microbiol. 2017;8:1964. doi: 10.3389/fmicb.2017.01964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Inglis RF, Biernaskie JM, Gardner A, Kümmerli R. Presence of a loner strain maintains cooperation and diversity in well-mixed bacterial communities. Proc R Soc B. 2016;283 doi: 10.1098/rspb.2015.2682. 20152682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harrison F, Paul J, Massey RC, Buckling A. Interspecific competition and siderophore-mediated cooperation in Pseudomonas aeruginosa . ISME J. 2008;2:49–55. doi: 10.1038/ismej.2007.96. [ This theoretical study shows how competitive interactions between cheaters and cooperators can drive the diversification of siderophores. ] [DOI] [PubMed] [Google Scholar]
- 99.Leinweber A, Weigert M, Kummerli R. The bacterium Pseudomonas aeruginosa senses and gradually responds to interspecific competition for iron. Evolution. 2018;72:1515–1528. doi: 10.1111/evo.13491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dawkins R, Krebs JR. Arms races between and within species. Proc R Soc B. 1979;205:489–511. doi: 10.1098/rspb.1979.0081. [DOI] [PubMed] [Google Scholar]
- 101.Lee W, van Baalen M, Jansen VA. A. Siderophore production and the evolution of investment in a public good: An adaptive dynamics approach to kin selection. J Theor Biol. 2016;388:61–71. doi: 10.1016/j.jtbi.2015.09.038. [DOI] [PubMed] [Google Scholar]
- 102.O'Brien S, Lujan AM, Paterson S, Cant MA, Buckling A. Adaptation to public goods cheats in Pseudomonas aeruginosa . Proc R Soc B. 2017;284 doi: 10.1098/rspb.2017.1089. 20171089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith EE, Sims EH, Spencer DH, Kaul R, Olson MV. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa . J Bacteriol. 2005;187:2138–2147. doi: 10.1128/JB.187.6.2138-2147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Meyer JM, et al. Siderotyping of fluorescent Pseudomonas: molecular mass determination by mass spectrometry as a powerful pyoverdine siderotyping method. Biometals. 2008;21:259–271. doi: 10.1007/s10534-007-9115-6. [DOI] [PubMed] [Google Scholar]
- 105.Lee W, van Baalen M, Jansen VA. A. An evolutionary mechanism for diversity in siderophore producing bacteria. Ecol Lett. 2012;15:119–125. doi: 10.1111/j.1461-0248.2011.01717.x. [ This study shows that siderophores can act as competitive agents in inter-specific competition. ] [DOI] [PubMed] [Google Scholar]
- 106.Sexton DJ, Glover RC, Loper JE, Schuster M. Pseudomonas protegens Pf-5 favours self-produced siderophore over free-loading in interspecies competition for iron. Environ Microbiol. 2017;19:3514–3525. doi: 10.1111/1462-2920.13836. [DOI] [PubMed] [Google Scholar]
- 107.Stilwell P, Lowe C, Buckling A. The effect of cheats on siderophore diversity in Pseudomonas aeruginosa. J Evol Biol. 2018;31:1330–1339. doi: 10.1111/jeb.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jurkevitch E, Hadar Y, Chen Y. Differential siderophore utilization and iron uptake by soil and rhizosphere bacteria. Appl Environ Microbiol. 1992;58:119–124. doi: 10.1128/aem.58.1.119-124.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Champomier-Vergès MC, Stintzi A, Meyer JM. Acquisition of iron by the non-siderophore-producing Pseudomonas fragi . Microbiology. 1996;142:1191–1199. doi: 10.1099/13500872-142-5-1191. [DOI] [PubMed] [Google Scholar]
- 110.Llamas MA, et al. The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa . J Bacteriol. 2006;188:1882–1891. doi: 10.1128/JB.188.5.1882-1891.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lemos ML, Balado M, Osorio CR. Anguibactin- versus vanchrobactin-mediated iron uptake in Vibrio anguillarum: evolution and ecology of a fish pathogen. Environ Microbiol Rep. 2010;2:19–26. doi: 10.1111/j.1758-2229.2009.00103.x. [DOI] [PubMed] [Google Scholar]
- 112.Dumas Z, Ross-Gillespie A, Kümmerli R. Switching between apparently redundant iron-uptake mechanisms benefits bacteria in changeable environments. Proc R Soc B. 2013;280 doi: 10.1098/rspb.2013.1055. 20131055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tyrrell J, et al. Investigation of the multifaceted iron acquisition strategies of Burkholderia cenocepacia . BioMetals. 2015;28:367–380. doi: 10.1007/s10534-015-9840-1. [DOI] [PubMed] [Google Scholar]
- 114.Estrela S, Morris JJ, Kerr B. Private benefits and metabolic conflicts shape the emergence of microbial interdependencies. Environ Microbiol. 2016;18:1415–1427. doi: 10.1111/1462-2920.13028. [DOI] [PubMed] [Google Scholar]
- 115.D'Onofrio A, et al. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol. 2010;17:254–264. doi: 10.1016/j.chembiol.2010.02.010. [ This study shows that unculturable marine bacteria become culturable upon the supplementation of exogenous siderophores, demonstrating the existence of complete ecological dependencies. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kerr B, Riley MA, Feldman MW, Bohannan BJ. M. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [ This study shows (theoretically and empirically) that non-transitive rock-paper-scissor dynamics can maintain species diversity in bacterial communities. ] [DOI] [PubMed] [Google Scholar]
- 117.Allesina S, Levine JM. A competitive network theory of species diversity. Proc Natl Acad Sci U S A. 2011;108:5638–5642. doi: 10.1073/pnas.1014428108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kelsic ED, Zhao J, Vetsigian K, Kishony R. Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature. 2015;521:516–519. doi: 10.1038/nature14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Diggle SP, Griffin AS, Campell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–414. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 120.Xavier JB, Kim W, Foster KR. A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa . Mol Microbiol. 2011;79:166–179. doi: 10.1111/j.1365-2958.2010.07436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Crespi BJ. The evolution of social behavior in microorganisms. Trends Ecol Evol. 2001;16:178–183. doi: 10.1016/s0169-5347(01)02115-2. [DOI] [PubMed] [Google Scholar]
- 122.Brown SP, Taylor PD. Joint evolution of multiple social traits: a kin selection analysis. Proc R Soc B. 2010;277:415–422. doi: 10.1098/rspb.2009.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Driscoll WW, Pepper JW, Pierson LS, Pierson EA. Spontaneous Gac mutants of Pseudomonas biological control strains: cheaters or mutualists. Appl Environ Microbiol. 2011;77:7227–7235. doi: 10.1128/AEM.00679-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Morris JJ, Lenski RE, Zinser ER. The black queen hypothesis: evolution of dependencies through adaptive gene loss. mBio. 2012;3:e00036–00012. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shou W, Ram S, Vilar JM. Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci U S A. 2007;104:1877–1882. doi: 10.1073/pnas.0610575104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Harcombe WR. Novel cooperation experimentally evolved between species. Evolution. 2010;64:2166–2172. doi: 10.1111/j.1558-5646.2010.00959.x. [DOI] [PubMed] [Google Scholar]
- 127.Pande S, Kost C. Bacterial unculturability and the formation of intercellular metabolic networks. Trends Microbiol. 2017;25:349–361. doi: 10.1016/j.tim.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 128.Amin SA, et al. Photolysis of iron-siderophore chelates promotes bacterial-algal mutualism. Proc Natl Acad Sci U S A. 2009;106:17071–17076. doi: 10.1073/pnas.0905512106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Oliveira NM, Niehus R, Foster KR. Evolutionary limits to cooperation in microbial communities. Proc Natl Acad Sci U S A. 2014;111:17941–17946. doi: 10.1073/pnas.1412673111. [ This conceptual study explains why the evolution of cooperative mutual exchange of secreted compounds is constrained in bacterial communities. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Foster KR, Bell T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol. 2012;22:1845–1850. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 131.Koskella B, Hall LJ, Metcalf CJ. E. The microbiome beyond the horizon of ecological and evolutionary theory. Nat Ecol Evol. 2017;1:1606–1615. doi: 10.1038/s41559-017-0340-2. [DOI] [PubMed] [Google Scholar]
- 132.Loper JE, Buyer JS. Siderophores in microbial interactions on plant surfaces. Mol Plant Microbe Interact. 1991;4:5–13. [Google Scholar]
- 133.Raaijmakers JM, et al. Utilisation of heterologous siderophores and rhizosphere competence of fluorescent Pseudomonas spp. Can J Microbiol. 1995;41:126–135. [Google Scholar]
- 134.Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol. 2017;15:579–590. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 135.Kupferschmied P, Maurhofer M, Keel C. Promise for plant pest control: root-associated pseudomonads with insecticidal activities. Front Plant Sci. 2013;4:287–287. doi: 10.3389/fpls.2013.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pieterse CM, et al. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 2014;52:347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- 137.Deriu E, et al. Probiotic bacteria reduce Salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013;14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sassone-Corsi M, et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature. 2016;540:280–283. doi: 10.1038/nature20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Granato ET, Harrison F, Kümmerli R, Ross-Gillespie A. Do bacterial “virulence factors” always increase virulence? a meta-Analysis of pyoverdine production in Pseudomonas aeruginosa as a test case. Front Microbiol. 2016;7:1952. doi: 10.3389/fmicb.2016.01952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Harrison F, Browning LE, Vos M, Buckling A. Cooperation and virulence in acute Pseudomonas aeruginosa infections. BMC Biol. 2006;4:21. doi: 10.1186/1741-7007-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Marvig RL, et al. Within-host evolution of Pseudomonas aeruginosa reveals adaptation. mBio. 2014;5:1–8. doi: 10.1128/mBio.00966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Harrison F. Microbial ecology of the cystic fibrosis lung. Microbiology. 2007;153:917–923. doi: 10.1099/mic.0.2006/004077-0. [DOI] [PubMed] [Google Scholar]
- 143.Callaghan M, McClean S. Bacterial host interactions in cystic fibrosis. Curr Opin Microbiol. 2012;15:71–77. doi: 10.1016/j.mib.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 144.Cutting GR. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet. 2015;16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cornelis P, Dingemans J. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front Cell Infect Microbiol. 2013;3:75. doi: 10.3389/fcimb.2013.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ford SA, Kao D, Williams D, King KC. Microbe-mediated host defence drives the evolution of reduced pathogen virulence. Nat Commun. 2016;7 doi: 10.1038/ncomms13430. 13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tinbergen N. On aims and methods of ethology. Z Tierpsychol. 1963;20:410–433. [Google Scholar]
- 148.Sessions AL, Doughty DM, Welander PV, Summons RE, Newman DK. The continuing puzzle of the great oxidation event. Curr Biol. 2009;19:R567–R574. doi: 10.1016/j.cub.2009.05.054. [DOI] [PubMed] [Google Scholar]
- 149.Braud A, Hoegy F, Jezequel K, Lebeau T, Schalk IJ. New insights into the metal specificity of the Pseudomonas aeruginosa pyoverdine–iron uptake pathway. Environ Microbiol. 2009;11:1079–1091. doi: 10.1111/j.1462-2920.2008.01838.x. [DOI] [PubMed] [Google Scholar]
- 150.Braud A, Geoffroy V, Hoegy F, Mislin GLA, Schalk IJ. Presence of the siderophores pyoverdine and pyochelin in the extracellular medium reduces toxic metal accumulation in Pseudomonas aeruginosa and increases bacterial metal tolerance. Environ Microbiol Rep. 2010;2:419–425. doi: 10.1111/j.1758-2229.2009.00126.x. [DOI] [PubMed] [Google Scholar]
- 151.Bobrov AG, et al. The Yersinia pestis siderophore, yersiniabactin, and the ZnuABC system both contribute to zinc acquisition and the development of lethal septicaemic plague in mice. Mol Microbiol. 2014;93:759–775. doi: 10.1111/mmi.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Perry RD, Bobrov AG, Fetherston JD. The role of transition metal transporters for iron, zinc, manganese, and copper in the pathogenesis of Yersinia pestis . Metallomics. 2015;7:965–978. doi: 10.1039/c4mt00332b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Koh EI, Henderson JP. Microbial copper-binding siderophores at the host-pathogen interface. J Biol Chem. 2015;290:18967–18974. doi: 10.1074/jbc.R115.644328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Robinson AE, Lowe JE, Koh EI, Henderson JP. Uropathogenic enterobacteria use the yersiniabactin metallophore system to acquire nickel. J Biol Chem. 2018;293:14953–14961. doi: 10.1074/jbc.RA118.004483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Giller KE, Witter E, McGrath SP. Toxicity of heavy metals to microorganisms and microbial processes in agricultural soils: a review. Soil Biol Biochem. 1998;30:1389–1414. [Google Scholar]
- 156.Schalk IJ, Hannauer M, Braud A. New roles for bacterial siderophores in metal transport and tolerance. Environ Microbiol. 2011;13:2844–2854. doi: 10.1111/j.1462-2920.2011.02556.x. [DOI] [PubMed] [Google Scholar]
- 157.Hesse E, et al. Ecological selection of siderophore-producing microbial taxa in response to heavy metal contamination. Ecol Lett. 2018;21:117–127. doi: 10.1111/ele.12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.O'Brien S, Hodgson DJ, Buckling A. Social evolution of toxic metal bioremediation in Pseudomonas aeruginosa . Proc R Soc B. 2014;281 doi: 10.1098/rspb.2014.0858. 20140858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Guan LL, Kanoh K, Kamino K. Effect of exogenous siderophores on iron uptake activity of marine bacteria under iron-limited conditions. Appl Environ Microbiol. 2001;67:1710–1717. doi: 10.1128/AEM.67.4.1710-1717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Grandchamp GM, Caro L, Shank EA. Pirated siderophores promote sporulation in Bacillus subtilis . Appl Environ Microbiol. 2017;83:e03293–03216. doi: 10.1128/AEM.03293-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Burke RM, Upton ME, McLoughlin AJ. Influence of pigment production on resistance to ultraviolet irradiation in Pseudomonas aeruginosa ATCC 10145. Ir J Food Sci Technol. 1990;14:51–60. [Google Scholar]
- 162.Achard ME, et al. An antioxidant role for catecholate siderophores in Salmonella . Biochem J. 2013;454:543–549. doi: 10.1042/BJ20121771. [DOI] [PubMed] [Google Scholar]
- 163.Adler C, et al. The alternative role of enterobactin as an oxidative stress protector allows Escherichia coli colony development. PLoS One. 2014;9 doi: 10.1371/journal.pone.0084734. e84734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jin Z, et al. Conditional privatization of a public siderophore enables Pseudomonas aeruginosa to resist cheater invasion. Nat Commun. 2018;9:1383. doi: 10.1038/s41467-018-03791-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Braun V, Pramanik A, Gwinner T, Köberle M, Bohn E. Sidermycins: tools and antibiotics. Biometals. 2009;22:3–13. doi: 10.1007/s10534-008-9199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]