Abstract
Plant pathogenic bacteria cause high crop and economic losses to human societies1-3. Infections by such pathogens are challenging to control as they often arise through complex interactions between plants, pathogens and the plant microbiome4,5. This natural ecosystem is rarely studied experimentally at the microbiome-wide scale, and consequently we poorly understand how taxonomic and functional microbiome composition and the resulting ecological interactions affect pathogen growth and disease outbreak. Here we combine DNA-based soil microbiome analysis with in vitro and in planta bioassays to show that competition for iron via secreted siderophore molecules is a good predictor of microbe-pathogen interactions and plant protection. We examined the ability of 2150 individual bacterial members of 80 rhizosphere microbiomes, covering all major phylogenetic lineages, to suppress the bacterium Ralstonia solanacearum, a global phytopathogen capable of infecting various crops6,7. We found that secreted siderophores altered microbiome-pathogen interactions from complete pathogen suppression to strong facilitation. Rhizosphere microbiome members with growth-inhibitory siderophores could often suppress the pathogen in vitro, in natural and greenhouse soils, and protect tomato plants from infection. Conversely, rhizosphere microbiome members with growth-promotive siderophores were often inferior in competition and facilitated plant infection by the pathogen. Because siderophores are a chemically diverse group of molecules with each siderophore type relying on a compatible receptor for iron uptake8-12, our results suggest that pathogen-suppressive microbiome members produce siderophores the pathogen cannot use. Altogether, our study establishes a causal mechanistic link between microbiome-level competition for iron and plant protection and opens promising avenues to use siderophore-mediated interactions as a tool for microbiome engineering and pathogen control.
Soil-borne pathogens are a global threat for food production1-3. Apart from the damage they inflict, a main problem is that there are very few management strategies available to control soil-borne bacterial phytopathogens. Plant root-associated microbiomes are increasingly seen as a possible driver of natural pathogen resistance and have become a target for innovative strategies aiming at improving crop protection13-15. However, our ability to predict and engineer microbiome function is still very limited and past research splits into two disjointed lines. The first line of studies has used comparative approaches to unravel how microbiome structure and the prevalence of putatively pathogen-suppressive traits correlate with plant health13,16,17. The second line has focused on specific systems to mechanistically test whether factors such as antibiosis, resource competition and activation of plant immunity can affect plant protection18-20. While the former line reveals potentially important candidate traits, species or functions involved in pathogen suppression, it provides limited insights on the underlying causal mechanisms. Conversely, the latter line offers specific mechanistic insights, but it remains often difficult to generalize the results beyond the specific study system. As a result, it still remains largely unclear which bacterial taxa and what type of ecological interactions in the rhizosphere determine disease outcomes by soil-borne pathogens21.
In order to create a predictive framework applicable for disease outcomes for tomato rhizosphere microbiomes, we combined the two above-mentioned lines of research: we first taxonomically characterized 80 tomato rhizosphere microbiomes and then conducted experimental analysis using 2150 representative bacterial members of these microbiomes. In particular, we assessed the interactions of each isolate with the pathogen Ralstonia solanacearum (strain QL-Rs1115), an economically important global pathogen causing damage in more than 200 plants species6,7, both in vitro and in vivo in controlled greenhouse experiments using tomato plants. We then closed the loop by using in vitro interactions as an inference tool to explain covariation in species co-occurrence under natural settings. We hypothesized that competition for iron, through the secretion of siderophores that scavenge iron from the environment, could represent a universal mechanism determining how strongly members of the soil microbiome can suppress the pathogen and protect plants. While other mechanisms including resource competition or antibiosis are probably also involved in bacterial interactions, we reasoned that these mechanisms might be superseded by competition for iron due to its universal importance for bacterial growth. Our reasoning is based on recent studies indicating that siderophore-mediated competition for iron drives eco-evolutionary dynamics in natural and infectious settings22-25. Iron is an essential co-factor for many enzymes, yet its bioavailability is low in most soils, because iron predominantly occurs in its insoluble ferric Fe(III) form26-28. Many bacteria scavenge iron from the environment through the secretion of siderophores, a chemically diverse group of secondary metabolites with high iron affinity23,29. The uptake of iron via iron-loaded siderophores occurs via strain-specific receptors and siderophores can both facilitate and suppress competitors, depending on whether competitors possess the matching receptors for siderophore uptake8-12.
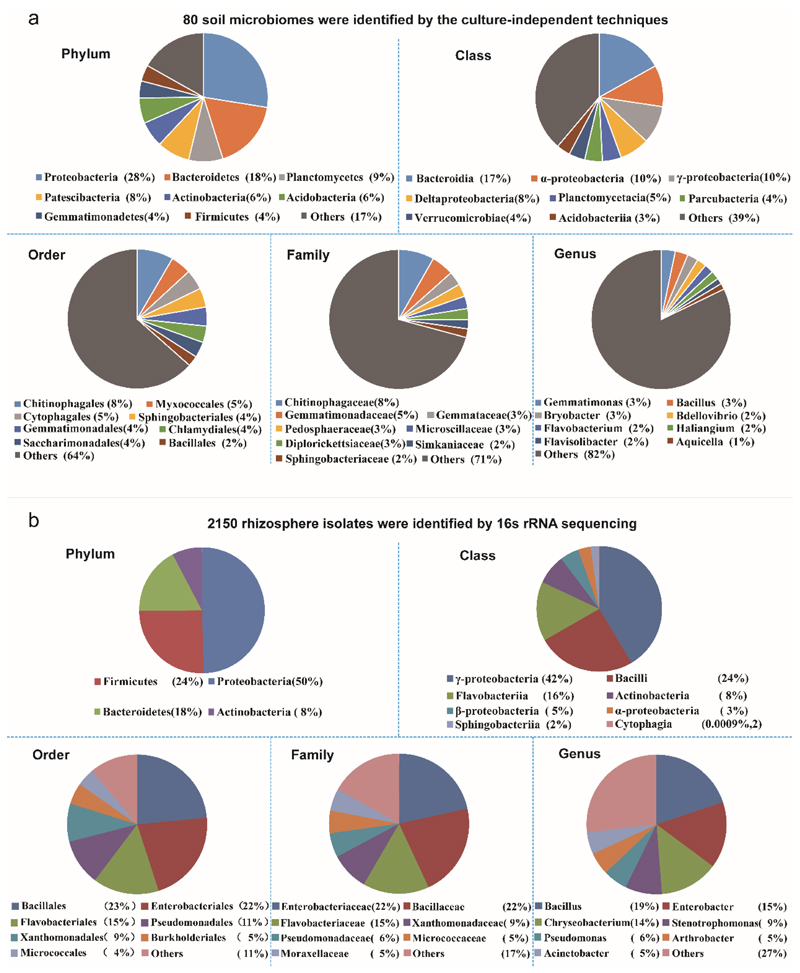
To assess the role of iron competition within the microbiome for plant protection, we first characterized 80 tomato rhizosphere microbiomes (Extended Data Fig. 1a) collected from four different fields in China (Supplementary Table 1), which were all infested by R. solanacearum. The main phyla of these microbiomes included Proteobacteria 28%, Bacteroidetes 18%, Planctomycetes 9%, Patescibacteria 8%, Actinobacteria 6%, Acidobacteria 6% and Firmicutes 4%; in total 293 families and 756 genera. We then randomly isolated and characterized a collection of 2150 culturable bacteria from these samples, which cover four major phylogenetic lineages normally present in plant rhizospheres30 and which were also present in our soil microbiomes (Proteobacteria 50%, Firmicutes 24%, Bacteroidetes 18% and Actinobacteria 8%; in total 35 families and 83 genera, Extended Data Fig. 1b). This collection is thus partly representative of the taxonomic breath of our soil microbiomes.
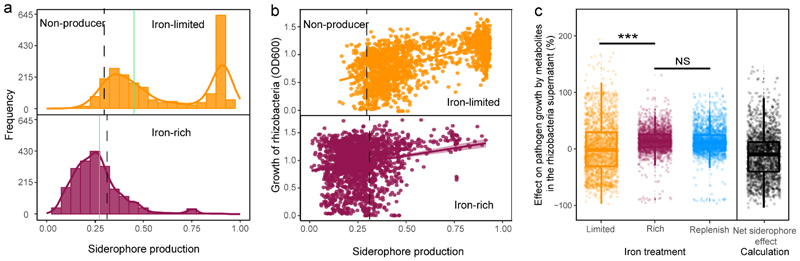
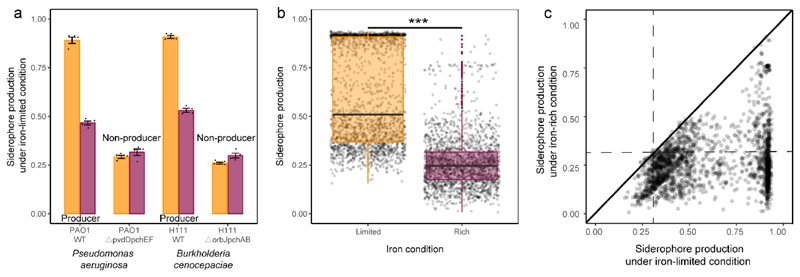
We then used the coulometric CAS (chrome azurol S) assay to estimate the amount of secreted siderophores in the supernatant of all 2150 bacterial isolates. This assay serves as a proxy for siderophore production and revealed that up to 95% of the isolates produced siderophores, given that their CAS values exceeded the ones of siderophore-deficient control strains (Fig. 1a+b). Estimating background CAS-values is important, as this assay also measures the binding activities of other organic iron-binding compounds. When repeating the CAS assay under iron-rich conditions, we found that up to 99% of the siderophore producers upregulated siderophore production under iron-limited as compared to iron-rich conditions (Extended Data Fig. 2). Under iron limitation, siderophore production followed a bimodal distribution with isolates producing either high or low amounts of siderophores (Fig. 1a). Siderophore production (i.e. CAS activity) across all isolates was predictive of bacterial growth under iron-limited condition (R2 = 0.388; n = 2150, Fig. 1b) but barely had an effect under iron-rich condition (R2 = 0.039; n = 2150, Fig. 1b). These results suggest that siderophore production is a widespread trait across the taxa and sampling sites examined here and is important for bacterial growth under iron-limited conditions.
Fig. 1. Siderophore production by rhizosphere bacteria and their growth effects on the plant pathogenic R. solanacearum bacterium.
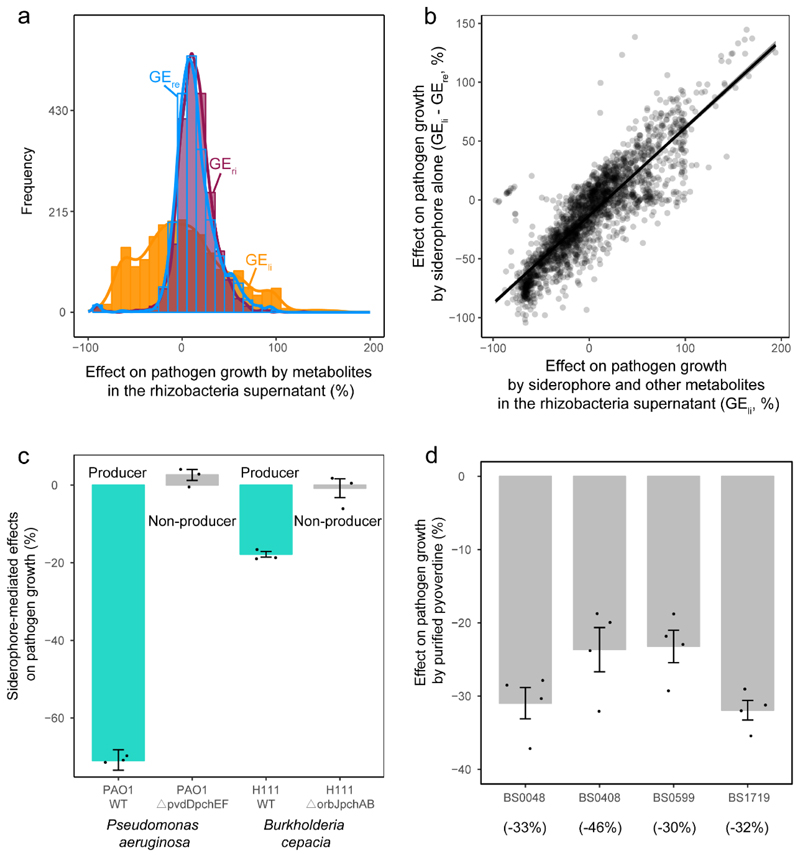
a, Relative siderophore production of the 2,150 rhizosphere isolates (measured in culture supernatants using the CAS assay) under iron-limited (top) and iron-rich (bottom) conditions. The black dashed vertical lines show the average CAS background values measured in the supernatants of two defined siderophore-deficient mutants (non-producers). We used this cut-off line to distinguish background noise from siderophore production. Measuring CAS background is important because this assay also measures the activity of other organic iron-binding compounds in the supernatant. Furthermore, the cut-off can vary slightly between species and the media from which the supernatants were collected (see also Extended Data Fig. 2). The green vertical lines represent siderophore production of our model pathogen R. solanacearum strain QL-Rs1115 (CAS value = 0.462 in iron-limited medium). b, Siderophore production (measured as CAS activity) across all isolates significantly correlated with rhizosphere bacterial growth (determined as the optical density (OD) at 600 nm, OD600) under iron-limited conditions (top), indicating that siderophores are important for growth. This effect was much weaker under iron-rich conditions (bottom). The black dashed vertical lines show the background CAS assay values of defined siderophore non-producers, the yellow and purple lines and the shaded area depict the best-fit trendlines and the 95% confidence interval of the linear regression, respectively. Regression model for iron-limited condition (top): adjusted R 2 = 0.388, F 1,1248 = 1,362 and two-sided P < 2.2 × 10-16; iron-rich condition (bottom): adjusted R 2 = 0.039, F 1,1248 = 87.7 and two-sided P < 2.2 × 10-16 based on a Student’s t-test; n = 2,150 biologically independent rhizobacterial isolates for both. c, The growth effects were measured in supernatants collected under iron-limited (high siderophore concentration + other secreted metabolites), iron-rich (low siderophore concentration + other secreted metabolites) and iron-limited conditions replenished with iron (siderophore effect removed, while the effect of other metabolites is retained). The net effect caused by siderophores alone (right column) was obtained by subtracting the growth effect of the iron-replenished supernatant from the growth effect of the iron-limited supernatant. The values indicate the percentage fold-change in growth. The box plots encompass the 25–75th percentiles, the whiskers extend to the minimum and maximum points, and the midline indicates the median (n = 2,150 biologically independent rhizobacterial isolates). P < 2.2 × 10-16 (left) and P = 0.882 (right) based on an analysis of variance followed by a paired two-sided Student’s t-test. ***P < 0.001; NS, not significant.
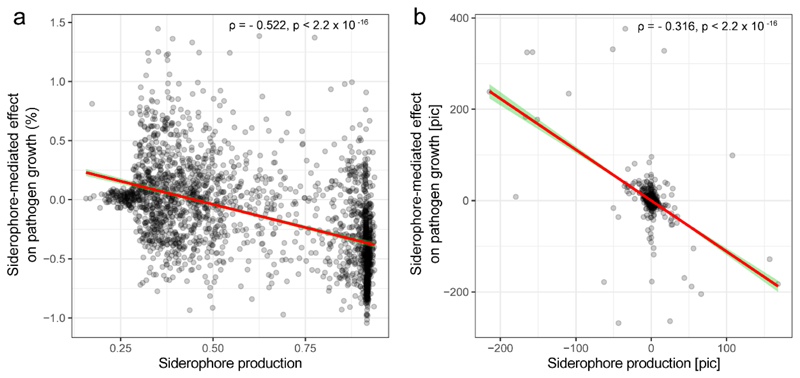
We then assessed the potential of the siderophores from the different isolates to suppress or promote the growth of R. solanacearum (QL-Rs1115). We first applied a baseline treatment by feeding bacterial supernatants collected from iron-rich conditions, which contained little siderophores but presumably other secreted compounds and residual nutrients, to the pathogen. We found that these supernatants had a mild and overall significant stimulatory effect on the growth of R. solanacearum (Fig. 1c and Extended Data Fig. 3a). Conversely, when the pathogen received bacterial supernatants from iron-limited conditions, which contained larger amounts of siderophores, we found much more pronounced growth effects (Fig. 1c and Extended Data Fig. 3a). These effects ranged from almost complete suppression of R. solanacearum growth to high levels of pathogen facilitation. To validate whether the observed differences in growth effects between iron-rich and iron-limited conditions were indeed caused by siderophores and not by other secreted metabolites that might have been up- or down-regulated under iron limitation, we exposed the pathogen to iron-limited supernatants that were replenished with iron to repress iron competition. The addition of iron to these supernatants neutralizes the effect of siderophores but retains the effect of the other secreted metabolites (e.g. antibiotics) on pathogen growth. We found that iron-replenished supernatants had the same (mildly promotive) effects as iron-rich supernatants, suggesting that siderophores are the main drivers of bacterial interactions under iron-limited conditions. Indeed, when estimating the net effect of siderophores (Figure 1c) we found that siderophores explained 76% of the total supernatant-mediated growth effects on R. solanacearum (R2=0.755, n = 2150, p < 0.0001, Fig. 1c and Extended Data Fig. 3b).
To experimentally confirm that siderophores can drive bacteria-pathogen interactions, we grew R. solanacearum in the presence of iron-limited supernatants from two defined siderophore producers (Pseudomonas aeruginosa PAO1 producing pyoverdine and pyochelin; Burkholderia cenocepaciae H111 producing ornibactin and pyochelin) and their isogenic null mutants, which were unable to produce siderophores due to engineered deletions of siderophore-synthesis genes (Extended Data Fig. 3c). We found that the supernatants of the wild type strains (containing siderophores) showed pathogen growth inhibition. Conversely, these inhibitory effects completely disappeared when the pathogen was exposed to the supernatants from the siderophore-deficient isogenic mutants of the two species (Extended Data Fig. 3c). To further confirm the importance of siderophores in mediating inter-species interactions, we isolated and purified the pyoverdines of four different Pseudomonas spp. isolates and exposed the pathogen to these siderophores. We observed that all four siderophores inhibited pathogen growth under iron-limited conditions in a similar way as the iron-limited supernatants did (Extended Data Fig. 3d). Finally, we examined growth and siderophore production of R. solanacearum QL-Rs1115 in monoculture. We found that the pathogen (i) is constrained in its growth in our iron-limited medium (Extended Data Fig. 4a), (ii) up-regulates siderophore production under iron limitation to an intermediate level (CAS-value = 0.462, Fig. 1a, Extended Data Fig. 4b), and (iii) is growth-stimulated when supplemented with its own iron-limited supernatant containing siderophores (Extended Data Fig. 4c). Taken together, these results demonstrate that siderophores can play a key role in mediating interactions between rhizosphere isolates and R. solanacearum QL-Rs1115 under iron-limited conditions. They further suggest that siderophore-mediated growth promotion or inhibition is indicative of whether R. solanacearum has compatible or incompatible receptors for heterologous siderophore uptake, respectively, as it has been described for other bacterial taxa12,22.
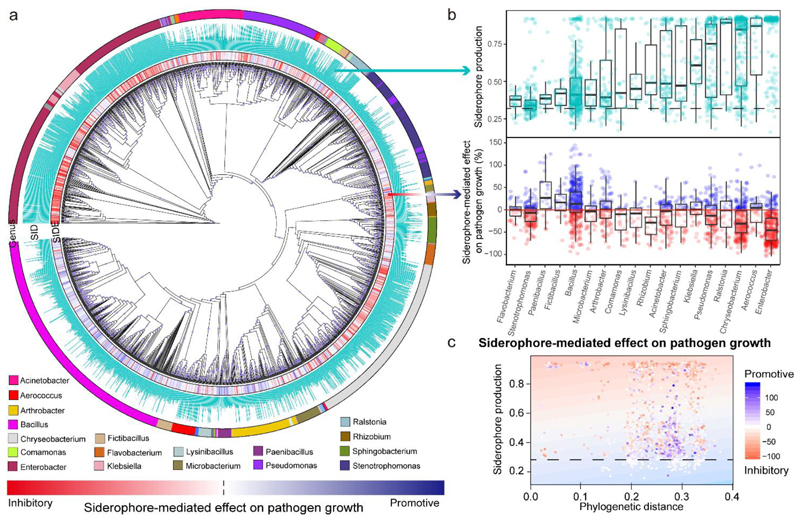
We next examined whether bacterial taxonomic affiliation predict siderophore production and siderophore-mediated effects on the pathogen. We observed a significant phylogenetic signal31 for both traits (Abouheif’s Cmean for siderophore production: 0.262, for siderophore-mediated growth effects: 0.148, p < 0.001 for both traits), which were relatively weak (Cmean = 1 would mean that all the observed variation is explained by phylogeny). This indicates that both siderophore production and siderophore-mediated growth effects vary substantially between related isolates, possibly due to recent change, gain or loss of trait functions. This assertion is supported by our ancestral character state reconstruction analysis showing that high levels of siderophore production and siderophore-mediated growth effects evolved independently multiple times (Supplementary Fig. 1). A focused analysis on the 18 most common genera confirms that both common ancestry and differences within genera contribute to trait variation (Fig. 2b): (a) Certain genera clearly differ from one another in their siderophore production and the extent to which they affect R. solanacearum growth (e.g. compare Enterobacter versus Bacillus); and (b) there is high variation between isolates within genera in the two traits (e.g. Bacillus spp.).
Fig. 2. Taxa identity of rhizobacterial isolates and their phylogenetic distance to R. solanacearum affect siderophore-mediated growth effects on the pathogen.
a, Cladogram depicting the phylogenetic relationship among the 2,150 isolates based on the 16S rRNA gene sequences. The first inner ring depicts the siderophore-mediated growth effects on the pathogen as a heatmap ranging from red (inhibitory) to blue (promotive), the second ring shows relative siderophore production by each isolate and the outer ring shows the most abundant genera (number of isolates greater than 20). SIDE, siderophore-mediated effect on pathogen growth; SID, siderophore production. b, Variation among isolates of the 18 most prevalent genera with regard to siderophore production (top) and siderophore-mediated effect on pathogen growth (bottom). The black dashed line shows the background CAS assay values of siderophore non-producers under iron-limited conditions. Values above and below the red line represent siderophore-mediated promotive and inhibitory effects of rhizobacteria on the pathogen growth. The exact number of bacteria in each genus is shown in Supplementary Table 7. The box plots encompass the 25–75th percentiles, the whiskers extend to the minimum and maximum points, and the midline indicates the median. c, Relationship between the siderophore-mediated effect on pathogen growth, phylogenetic distance between the pathogen and rhizosphere bacteria, and siderophore production in iron-limited conditions. This relationship explained a high proportion of the observed variance (25.6%) in iron-limited conditions (visualized as a heatmap based on the generalized linear model analysis, background). The dotted lines represent the background CAS assay values of defined siderophore non-producers. Each point represents one of the 2,150 isolates and its colour shade indicates the strength of the siderophore-mediated effect on pathogen growth (adjusted R 2 = 0.255, F 1,1248 = 87.7 and two-sided P < 2.2 × 10-16 based on Student’s t-tests for the whole model).
At the quantitative level, we found that R. solanacearum growth inhibition was predicted by additive effects of phylogenetic distance and siderophore production, with the most inhibiting isolates being those that were relatively closely related to the pathogen and that produced high amounts of siderophores (Fig. 2c; whole model R2 = 0.255, n = 2150, Supplementary Table 2). This phylogenetic effect is consistent with ecological theory predicting that resource competition intensifies among more closely related species, as their ecological niches are more likely to overlap32,33. To validate the effect between the level of siderophore production and siderophore-mediated growth effect, we repeated our analysis using phylogenetically independent contrasts, thereby filtering out the variation explained by phylogeny alone. This analysis confirmed that isolates producing high amounts of siderophores were more likely to inhibit R. solanacearum growth than isolates producing low amounts of siderophores (Extended Data Fig. 5).
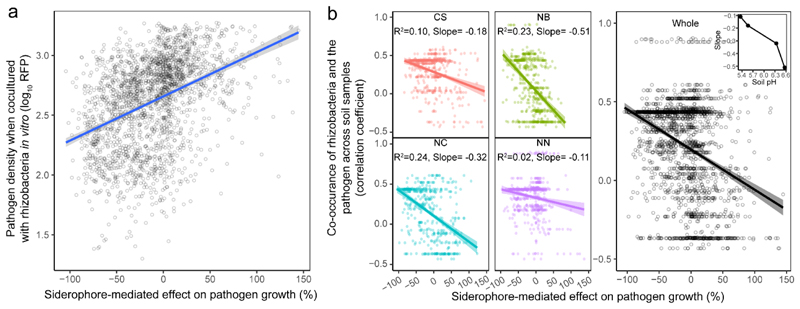
We then examined whether the siderophore-mediated growth effects can predict the outcome of direct competition between each of the 2150 rhizosphere isolates and R. solanacearum in co-cultures. We found that pathogen growth was significantly reduced when competed against taxa producing inhibitory siderophores (R2=0.16, n = 2150, Fig. 3a). To test whether siderophore-mediated growth effects correlate with patterns of co-occurrence between the pathogen and rhizosphere bacteria in natural field conditions, we used 16S rDNA sequencing of the whole microbiome communities and qPCR to estimate co-occurrence patterns of each taxon with the pathogen in each of the 80 soil samples. Thus, for each microbiome we obtained the prevalence of the pathogen and the 2150 taxa. We then correlated these prevalence values across the 80 microbiomes. We found that siderophore-mediated interactions correlated with species co-occurrence in the field: taxa producing inhibitory siderophores tended to co-occur with the pathogen at high density (positive r), while taxa producing promotive siderophores tended to occur at low densities when the pathogen was abundant (negative r, main effect of in vivo coexistence on regression coefficient r with R2 = 0.113, n = 2150, Fig. 3b). These patterns of covariance were strongest at sites with high soil pH values (inset Fig. 3b, Supplementary Table 3), where iron availability is most reduced and siderophores most needed (Fig. 3b inset). Although many other (unknown) factors can influence taxa abundances in the field, our results indicate that siderophore-mediated interactions can be a generalizable mechanism substantially influencing the competitive outcomes and the co-occurrence patterns between the pathogen and taxa isolated from natural rhizosphere microbiomes. Moreover, the power of siderophores to predict species interactions seems to vary with environmental parameters like pH that directly influence the level of iron limitation in soils and we propose that other environmental factors like overall iron content and nutrient availability could have similar effects.
Fig. 3. Siderophore-mediated effects on pathogen growth correlate with R. solanacearum and rhizosphere bacterial abundances in vitro and in vivo under field conditions.
a, Co-culture experiments between the R. solanacearum pathogen and each of the 2,150 rhizosphere isolates show that pathogen density (measured as mCherry fluorescence units) correlates positively with siderophore-mediated growth effects, suggesting that siderophores determine pathogen growth when in direct competition with rhizosphere bacteria. The blue line and grey shaded area depict the best-fit trendline and the 95% confidence interval of the linear regression, respectively (adjusted R 2 = 0.16, n = 2,150 biologically independent rhizobacterial isolates, F 1,1248 = 404.6 and two-sided P < 2.2 × 10-16 based on a Student’s t-test). b, Co-occurrence between the pathogen and rhizosphere bacteria was estimated as the correlation coefficient across the 80 soil samples and plotted against the effect of siderophores on pathogen growth. Exclusively negative correlations were found between these variables, showing that rhizosphere bacteria producing inhibitory siderophores tended to coexist at high densities with the pathogen (positive r), whereas isolates producing promotive siderophores tended to occur at low densities when the pathogen was abundant (negative r). Inset, the strength of this relationship correlated with the soil pH at the four sampling sites (right). Left, the colours represent different sampling sites: CS, Changsha (n = 528 biologically independent rhizobacterial isolates, F 1,526 = 58.72, P < 2.2 × 10-16); NB, Ningbo (n = 526 biologically independent rhizobacterial isolates, F 1,524 = 154.1, P < 2.2 × 10-16); NC, Nanchang (n = 508 biologically independent rhizobacterial isolates, F 1,506 = 157.1, P < 2.2 × 10-16); NN, Nanning (n = 568 biologically independent rhizobacterial isolates, F 1,566 = 14.9, P = 0.0001). The results of linear regression analysis in the left panel are shown as colour-coded best-fit trendlines for each sampling site (the shaded areas depict the 95% confidence interval and adjusted R 2 with the slope of the best-fit trendlines). Right, adjusted R 2 = 0.133 and the shaded area depicting the 95% confidence interval are shown (n = 2,130 biologically independent rhizobacterial isolates, F 1,1228 = 272.7 and two-sided P < 2.2 × 10-16 based on Student’s t-tests for the whole data across four sampling sites).
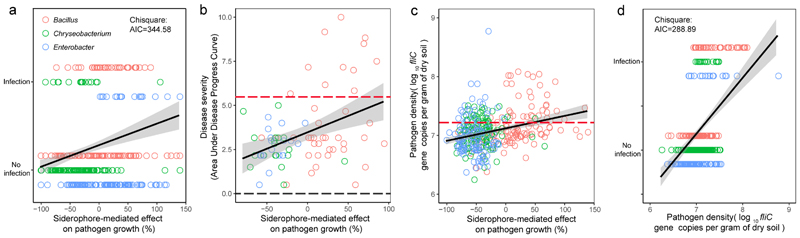
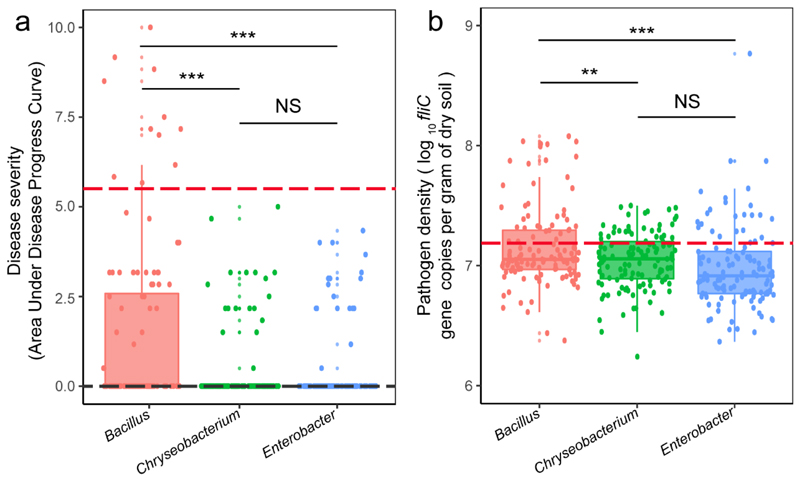
Finally, we tested whether siderophore-mediated interactions can affect the pathogen’s ability to invade the rhizosphere microbiome and cause disease using tomato plants, which were individually pre-colonized by one of 360 randomly selected bacterial isolates belonging to the three most abundant genera (Bacillus: on average promotive, Enterobacter: on average inhibitory, Chryseobacterium: on average inhibitory; 120 isolates tested per genus, Supplementary Table 4). We found that siderophore-mediated interactions were significantly associated with both pathogen growth and plant protection (Fig. 4).
Fig. 4. Siderophore-mediated growth effects predict plant disease outcomes and pathogen load in the tomato plant rhizosphere during greenhouse experiments.
a, The ability of pathogens to infect tomato plants was measured as a binary variable for each of the 360 pathogen–bacteria combinations; infection was scored as successful if at least one of six plants was infected. Successful infection scaled positively with the effect of siderophores on pathogen growth, suggesting that inhibitory siderophores can protect plants from infections (n = 360 biologically independent rhizobacterial isolates and two-sided P = 4.85 × 10-8 based on a X 2 test). b, Disease severity of infected plants (quantified as the area under disease progress curve, AUDPC) correlated positively with the siderophore-mediated growth effect on the pathogen. For this analysis, we only considered the 77 isolates that actually managed to infect plants (adjusted R 2 = 0.149, n = 77 biologically independent rhizobacterial isolates, F 1,75 = 14.34 and two-sided P = 0.0003 based on a Student’s/-test). c, Pathogen densities correlated positively with the siderophore-mediated growth effect on the pathogen, suggesting that the sign of siderophore-mediated effects determine pathogen load in the tomato rhizosphere (adjusted R 2 = 0.105, n = 360 biologically independent rhizobacterial isolates, F 1,358 = 42.92 and two-sided P = 6.16 × 10-9 based on a Student’s t-test). b,c, The red and black dashed lines show the baseline values for the control treatments in which the tomato plants were inoculated with either R. solanacearum alone or sterilized water, respectively. d, Successful infection scaled positively with pathogen densities, suggesting that successful inhibition of pathogen growth is key for disease control (n = 360 biologically independent rhizobacterial isolates and two-sided P = 2.31 × 10-13 based on a X 2 test). a–d, The black lines and grey shaded areas depict the best-fit trendline and the 95% confidence interval of the logistic regression, respectively. AIC, Akaike information criterion.
Specifically, disease incidence and pathogen density were the highest when the pre-inoculated bacterial isolates produced promotive siderophores but dropped when plants were pre-inoculated with bacteria producing inhibitory siderophores (Fig. 4a-c, Extended Data Fig. 6). Crucially, siderophore-mediated effects on infection outcomes were not dependent on the genus (see Supplementary Table 5 for statistical analysis). Moreover, pathogen density correlated positively with disease incidence (Fig. 4d), suggesting that inhibiting pathogen growth plays a key role in disease control. While the associations reported here are all significant, it is important to realize that they only explain a relatively low level of the total variation observed. This suggests that other factors in addition to siderophores also influence infection outcomes. Taken together, our infection experiments match well the growth and co-occurrence patterns observed in vitro and in the field indicating that siderophore-mediated interactions significantly contribute and reliably predict pathogen suppression and disease outcomes in many cases.
In conclusion, our study links global microbiome analyses with high-throughput experimental assays to demonstrate that siderophore-mediated interactions are an overarching mechanism influencing microbiome function in natural field conditions. Our findings set the stage for microbiome-wide ecosystem engineering to suppress pathogens and protect crops. A key component of microbiome engineering or the development of probiotics would be to selectively promote the members of the microbiome with siderophore-inhibitory effects on the pathogen and reduce those with pathogen-promotive siderophores. Great care should be taken to choose species that produce siderophores that cannot be taken up by the pathogen. Our study has identified groups of soil bacteria with such preferable functions, and the next step would be to genetically and chemically identify the pathogen suppressive and promotive siderophores. The siderophore types of all members of the microbiome could form a database against which the siderophore types of any pathogenic bacteria could be mapped, in order to identify the specific members of the microbiome with incompatible siderophores. We propose that such siderophore mismatch analyses could represent a predictive tool for microbiome-mediated pathogen management. An important aspect of such an approach would be to include measures to minimize the risk of resistance evolution by pathogens, for example through the acquisition of heterologous siderophore receptors via horizontal gene transfer34. In this context, it would be important to promote diverse rhizosphere communities that combat the pathogen at multiple fronts, through the secretion of a chemically diverse set of siderophores. It is conceivable that in a scenario where pathogens are confronted with a cocktail of inhibitory siderophores, resistance cannot easily evolve.
Methods
Rhizosphere soil sampling
Rhizosphere soil samples were collected from individual tomato plants located on four geographically distant fields (Supplementary Table 1) that had suffered from bacterial wilt disease for 3-15 years: Changsha (112°58′ E, 28°11′ N), Ningbo (121°67′ E, 29°91′ N), Nanchang (115°51′ E, 28°41′ N), and Nanning (108°21′ E, 22°49′ N). Twenty rhizosphere soil samples (20 tomato plants) were collected from each site. To this end, the excess soil was first gently shaken from the roots and the remaining soil attached to roots was considered as the rhizosphere soil. Each rhizosphere soil sample was then divided into two parts: one for isolating bacteria and the other for extracting DNA for bacterial community sequencing and the quantification of R. solanacearum pathogen densities using qPCR. Soil pH was measured as described in a previous study35, because low soil pH is positively linked to iron bio-availability in the soil36.
Soil DNA extraction
Soil DNA was extracted from 300 mg of soil using Power Soil DNA Isolation Kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) according to the manufacturer’s protocol. DNA quality was checked by running samples on 1% sodium boric acid agarose gels and DNA concentrations were measured with a NanoDrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA). Extracted DNA was stored at -80°C.
Isolation of rhizobacteria
To isolate bacteria, 1 g of rhizosphere soil was mixed with 9 mL MS buffer solution (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 10 mM MgSO4, 0.01% gelatin) in a rotary shaker at 170 rpm at 30 °C for half an hour. After serial dilution in MS buffer solution, 100 μL of diluted soil suspensions were then plated on 1/10 tryptone soya agar (1/10 TSA, tryptone 1.5 g L-1, soytone 0.5 g L-1, 0.5 g L-1 sodium chloride, agar 15 g L-1, pH 7.0). After 48 h of incubation at 30 °C in the dark, 32 isolates were randomly picked per rhizosphere soil sample. Only high dilute samples were used for isolation to prevent potential fungal contaminants. Isolates were then re-streaked on TSA plates for colony purification. Approximately 16.0 % of bacterial isolates failed to grow on the TSA plates for unknown reasons and were omitted from the data set (approximately the same number of isolates failed to grow per each field). The final collection consisted of 2150 bacterial isolates from 80 rhizosphere soil samples. All purified isolates were cultured in 100 μL tryptone soya broth (TSB, liquid TSA) on 96-well microtiter plates at 30 °C with shaking (rotary shaker at 170 rpm) for 18 h before freezing and storing at -80 °C in 15% glycerol.
16S rRNA gene sequencing of rhizobacteria
We used 16S rRNA amplicon sequencing to taxonomically identify all 2150 rhizobacterial isolates. The total genomic DNA was extracted from individual bacterial isolate overnight cultures grown in TSB medium at 30 °C with shaking (170 rpm) using e.Z.N.A. Bacterial DNA kit (OMEGA bio-tek) following the manufacturer’s protocol. The 16S rRNA gene was amplified by PCR using the universal primers F27 (5’-AGA GTT TGA TCA TGG CTC AG-3’) and R1492 (5’-TAC GGT TAC CTT GTT ACG ACT T-3’)37. PCR reactions (25 μL) contained 1 μL of bacterial DNA, 12.5 μL master mix, 1 μL of forward and reverse primer each and 9.5 μL deionized water. PCRs were run as follows: initial denaturation at 95 °C for 5 min, 30 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 1 min 30 and the final extension at 72 °C for 10 min. PCR products were sequenced by Shaihai Songon Biotechnology Co., Ltd, Shanghai Station. The 16S rDNA sequences were identified using NCBI and RDP databases and homologous sequence similarity. To incorporate pairwise phylogenetic distance between the pathogen and each isolate as a covariate into models, the relatedness between rhizobacterial isolates and the pathogen was calculated using pairwise alignment of 16S rDNA sequences using EMBOSS Water38.
Tree construction and phylogenetic analysis of rhizobacterial isolates
To construct and explore phylogenetic relationships between the rhizobacterial isolates and the pathogen, the16S rDNA sequences of all strains (2150) were aligned with MUSCLE. Sequences in the alignment were trimmed at both ends to obtain maximum overlap using the MEGA 7 software, which was also used to construct taxonomic cladograms, and to calculate phylogenetic distances between rhizobacteria and the pathogen39. We constructed a maximum-likelihood (ML) tree, using a General time reversible (GTR) + G + I model, which yielded the best fit to our data set. Bootstrapping was carried out with 100 replicates retaining the gaps. The taxonomic cladogram was created using the iTOL web tool (http://huttenhower.sph.harvard.edu/galaxy/). To allow for further phylogenetic analyses, the tree was midpoint-rooted, multichotomies were randomly resolved into a series of dichotomies, and a small constant (10-6 times the maximum branch length) was added to branches with a length of zero. We first used the revised tree to calculate the phylogenetic signal (Abouheif’s Cmean 31) in the production of siderophores and the siderophore-mediate effects on pathogen growth, using the phyloSignal-function of the phylosignal package40in R. Next, we performed an ancestral character estimation of these two traits. Finally, to test for an effect of phylogeny on the association between siderophore production and siderophore-mediated effects on pathogen growth, we compared the results of a correlation based on the uncorrected trait values with the results of a correlation based on phylogenetically independent contrasts. Both analyses (ancestral character estimation and calculation of phylogenetically independent contrasts) were performed with the ape-package41, using the ace-function (which fits a Brownian motion model by residual maximum likelihood) and the pic-function, respectively.
Measuring the growth of rhizobacteria and their siderophore production
To quantify the growth and siderophore production of each rhizobacterial isolate under iron-limited and iron-rich conditions, we first revived the isolates by transferring 5 μL freezer stocks into new 96-wells plates containing 195 μL TSB per well and cultured bacteria overnight at 30 °C with shaking (rotary shaker set at 170 rpm). We then transferred 10 μL of overnight cultures into new 96-wells plates containing 190 μL MKB medium (K2HPO4 2.5 g L-1, MgSO4·7H2O 2.5 g L-1, glycerin 15 ml L-1, casamino acids 5.0 g L-1, pH 7.2). This medium contains low amounts of residual iron42, but still allows all isolates to grow to a certain extent regardless of their ability to produce siderophores (Fig. 1b). As an iron-rich medium control we supplemented MKB with 50 μM FeCl3. We selected such a high concentration of iron to ensure that iron is no longer a limiting factor for the growth of all the isolates examined. Following 48 h of incubation at 30 °C with shaking (rotary shaker set at 170 rpm), we measured the bacterial growth as maximum density obtained with a plate reader at room temperature (optical density OD at 600 nm; SpectraMax M5, Sunnyvale, CA, USA).
The same samples were then used to quantify rhizobacterial siderophore production. Specifically, we harvested the cell-free supernatant from bacterial cultures by centrifugation (4,000 rpm, 5 min at 4 °C) and filtration (using a 0.22 μm filter). The supernatant was then divided into two parts for measuring i) siderophore production and ii) testing the supernatant effects on R. solanacearum pathogen growth. Siderophore production was assayed using a modified version of the universal chemical assay developed by Schwyn and Neilands43. Briefly, we used the liquid version of the CAS (chrome azurol S) assay, where 100 μl of cell-free supernatant (three biological replicates for all 2150 soil isolates), or deionized water as a control reference, were added to 100 μL CAS assay solution in a 96-well plate. After 2 h of static incubation at room temperature, the optical density (630 nm) of cell-free supernatants (A) and deionized water controls (Ar) was then measured using a plate reader (SpectraMax M5, Sunnyvale, CA, USA) at room temperature. Siderophores induce a color change in the CAS medium, which lowers OD630nm measurements, and siderophore production can thus be quantified using the following formula: 1-A/Ar. Because media components (i.e. organic acids) and other secreted compounds can also bind iron, it is essential to estimate the CAS signal background that is not due to siderophores. We assessed this signal background by using defined siderophore-deficient mutants from two species (Pseudomonas aeruginosa PAO1 △ pvdD △ pchEF and Burkholderia cenocepacia H111 △ orbJ △ pchAB)44,45 and their corresponding wild types using the exact same protocol described above. We then averaged the CAS background signals of the two siderophore-deficient mutants and used it as a cutoff line to distinguish siderophore producers from non-producers among our 2150 soil isolates.
Effect of rhizobacterial supernatants on the growth of R. solanacearum
We used R. solanacearum strain QL-Rs1115 tagged with the pYC12-mCherry plasmid16 as a model pathogen. To explore interactions between this pathogen and the 2150 members of the rhizosphere microbiomes via secreted products (overview of the experimental design is shown in Supplementary Fig. 2), we exposed the pathogen to three different types of supernatant collected from the rhizobacteria. These treatments included: (i) supernatants collected from iron-rich conditions (rhizobacteria grown in iron-rich MKB medium, in which little siderophores are produced but other compounds are secreted, SNri); (ii) supernatants collected from iron-limited conditions (rhizobacteria grown in iron-limited MKB medium, which triggers siderophore production in addition to other secreted compounds, SNli); and (iii) supernatants collected from iron-limited conditions that was subsequently replenished with 50 μM FeCl3 (SNre). This supernatant still contains siderophores, but they are no longer relevant for iron uptake, as iron is available in excess. As a control, we used sterilized water instead of supernatant (SNcontrol). All supernatant assays were carried out in triplicates. Specifically, we inoculated 2 μL overnight culture of the pathogen (adjusted to OD600 = 0.5 after 12 h growth at 30°C with shaking) into 180 μL of fresh MKB medium (10-times diluted, in order to better reflect the effect of the supernatant) and 20 μL of sterile rhizobacterial supernatant. Cultures were then incubated at 30 °C under shaken conditions (rotary shaker set at 170 rpm). Pathogen growth was measured as optical density after 24h of cultivation (OD600; SpectraMax M5 Plate reader). Subsequently, we calculated the rhizobacterial supernatant effect on pathogen growth relative to growth in the absence of supernatants, using the following formula: growth effect GEtreatment = ((SNtreatment / SNcontrol)- 1)*100, where SNtreatment = SNli, SNri, or SNre. For this calculation, we took the average supernatant effects across the three replicates.
Values smaller and greater than zero indicate growth inhibition and facilitation, respectively, expressed as percentage fold-change in growth. From these measures, the net growth effect of siderophores can be measured as GEnet = GEli – GEre.
Control experiments to test whether foreign siderophores affect the growth of R. solanacearum
We performed two control experiments to validate that foreign siderophores affect pathogen growth as shown by the supernatant assays. First, we used the exact same protocol as described above to obtain iron-limited supernatants (in triplicates) from two defined siderophore producers (Pseudomonas aeruginosa PAO1 producing pyoverdine and pyochelin; Burkholderia cenocepaciae H111 producing ornibactin and pyochelin) and their isogenic null mutants, unable to produce siderophores due to engineered deletions of siderophore-synthesis genes (PAO1△pvdD△pchEF and H111△orbJ△pchAB)44,45. We then again followed the above-described protocol to challenge the pathogen with these supernatants to test whether pathogen growth inhibition occurs only with supernatants from the wild type strains, but not with the supernatants from the siderophore-deficient strains.
In a second control experiments, we extracted and purified the siderophores (pyoverdines) from four Pseudomonas isolates from our isolate collection (Supplementary Table 6). These four isolates produce large quantities of pyoverdine, which can be detected via the natural fluorescence of this molecule (relative fluorescence units RFU measured with excitation at 400nm and emission at 460nm) with a microplate reader12. We first revived the 4 isolates by transferring 5 μL freezer stocks into 25-mL conical flask containing 10 mL TSB and cultured bacteria overnight at 30 °C under shaken conditions (rotary shaker set at 170 rpm). We then transferred 1 mL of overnight cultures into 250 mL conical flask containing 100 mL iron-limited medium (MKB). Following 48 h of incubation at 30 °C (shaken at 170 rpm), we extracted the pyoverdines in the supernatants using a standardized method12,46. We then dissolved the purified pyoverdines in 10 mL deionized water and passed it through a filter (0.22 μm) for sterilisation. Finally, the sterile pyoverdine solutions were used to assess their effects on the pathogen growth under iron-limited conditions using the exact same protocol as for the supernatant assay.
Control experiments examining the impact of iron limitation on the growth and siderophore production of R. solanacearum QL-Rs1115
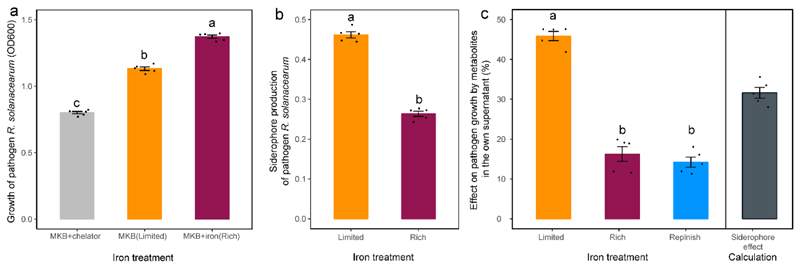
We carried out three additional control experiments to verify that (i) the pathogen R. solanacearum QL-Rs1115 itself is limited by iron in the MKB medium, (ii) that it produces siderophores under iron-limiting conditions, and (iii) that the pathogen is stimulated by its own iron-limited supernatant containing siderophores. We found all these points to hold true (Extended Data Fig. 4).
Prior to these experiments, we inoculated R. solanacearum from freezer stocks into 200 μL TSB medium to grow overnight at 30 °C in 96-well microplates. We then transferred 10 μl of overnight cultures into new 96-wells plates containing 200 μl of one out of three media with five replicates: (i) MKB medium supplemented with 50 μM FeCl3 (high iron availability); (ii) MKB medium (low iron availability); and (iii) MKB medium supplemented with 200 μM 2,2′-dipyridyl, a strong iron chelator (very low iron availability). Following 48 h of incubation at 30 °C with shaking at 170 rpm, we measured the pathogen growth through absorbance at OD600nm with a plate reader (SpectraMax M5, Sunnyvale, CA, USA).
We then used the cultures from the iron-limited (MKB) and iron-rich (MKB+iron) medium to collect sterile supernatants for which we quantified siderophore contents using the CAS assay. For this we used the exact same protocol as described above. Finally, we repeated the supernatant growth assay as reported in Fig. 1c, but this time we provided R. solanacearum with its own supernatants. We did this to test whether the pathogen is stimulated by its own siderophores, being present in the iron-limited, but not the iron-rich and iron-replenished supernatant.
Control experiment testing whether mCherry fluorescence remains stable across a pH gradient
We observed that the pH of MKB medium rises from 7.17 to 7.55 during a 24 hours growth cycle of R. solanacearum. Since we aimed to use the mCherry signal as a proxy for pathogen growth in mixed cultures with the rhizosphere isolates, we tested whether the mCherry fluorescence measured remains stable across a pH range from 6 to 10. Specifically, we took stationary phase cultures of R. solanacearum, and adjusted the pH to 6 (with dilute hydrochloric acid) and to 10 (with sodium hydroxide solution). After allowing the bacterial cultures to adjust to the new pH for 1 hour, we quantified mCherry fluorescence of the original culture (unmanipulated pH) and the pH-adjusted bacterial cultures (excitation: 587nm, emission: 610 nm, using the SpectraMax M5 Plate reader. We found that the mCherry fluorescence intensity was not significantly affected by the pH of the bacterial cultures (Supplementary Fig. 3).
Effect of rhizobacteria on the pathogen growth in co-cultures
In order to test if the siderophores produced by rhizobacteria affect pathogen growth directly, we co-cultured each rhizobacterial isolate with the pathogen in the iron-limited MKB medium that triggers siderophore production. To this end, we first grew all rhizobacterial isolates and R. solanacearum pathogen overnight from 5 μL of freezer stocks at 30 °C in 96-well microplates containing 200 μL of TSB medium. We then co-cultured each rhizobacteria with the pathogen by mixing 10 μL of both overnight cultures into new 96-wells plates containing 200μL iron-limited MKB medium at 30 °C with shaking (170 rpm).
After 24 hours, we quantified pathogen density on the basis of the constitutively expressed mCherry fluorescence signal (excitation: 587nm, emission: 610 nm) using a SpectraMax M5 Plate reader. To control for the auto-fluorescence of rhizobacteria, we also grew each isolate as monoculture in iron-limited MKB medium and subtracted these mono-culture values from the co-culture fluorescence values. This allowed us to correlate pathogen density changes with the siderophore-mediated growth effect for each pathogen-rhizobacterial isolate pair.
Quantification of R. solanacearum densities in the field with qPCR
The R. solanacearum densities in the fields were determined with qPCR using primers targeting the fliC gene, which encodes a flagellar subunit (forward primer: 5′-GAA CGC CAA CGG TGC GAA CT-3′ and reverse primer: 5′-GGC GGC CTT CAG GGA GGT C-3′)47. The qPCR was carried out with the Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, CA, USA). We used SYBR Green I fluorescent dye detection in 20-μl volumes containing 10 μl of SYBR Premix Ex Taq (TaKaRa Bio Inc., Japan), 2 μl of DNA template extracted from rhizosphere soil and 0.4 μl of both forward and reverse primers (10 mM each). The qPCR was performed by initially denaturing step for 30 s at 95 °C with subsequent cycling for 40 times with a 5s denaturizing step at 95 °C. The protocol was followed by a 34s elongation/extension step at 60 °C, and a melt curve analysis for 15 s at 95 °C followed by 1 min at 60 °C and finally for 15 s at 95 °C. Melting curves were obtained based on a standard protocol and used to identify the characteristic peak of PCR product (400 bp)48. Three independent technical replicates were used for each sample.
Estimation of the diversity and the relative bacterial abundances in the field with 16S rRNA amplicon sequencing
To determine the relative abundance of rhizobacterial isolates in the field, we first sequenced the whole bacterial communities of the 80 tomato rhizosphere samples by amplifying the V4 hypervariable regions of the bacterial 16S rRNA gene using the primer pairs 563F (5′-AYT GGG YDT AAA GVG-3 ′) and 802R (5′-TAC NVG GGT ATC TAA TCC-3′)49. PCR reactions (total volume 20 μl) contained 4 μl 5× Fast-Pfu buffer, 2 μl 2.5 mM dNTPs, 0.4 μl of each primer (5 μM), 0.5 μl DNA sample, and 0.4 μl Fast-Pfu polymerase (TransGen Biotech, Beijing, China). PCR amplification was conducted using Applied Biosystems thermal cycler (GeneAmp PCR system 9700, Applied Biosystems, Foster City, CA, USA) and included 30 cycles at 95 °C for 30 s, at 55 °C for 30 s and at 72 °C for 30 s. For each DNA sample, three independent PCRs were performed and triplicate products were pooled to minimize PCR amplification bias. The amplicon products were purified using an AxyPrep PCR Clean-up Kit (Axygen Biosciences, Union City, CA, USA) and sample purities were tested using agarose gel electrophoresis. The concentrations of the purified PCR products were determined with QuantiFluor™-ST (Promega, WI, USA) before subjecting them to 250-nucleotide paired-end sequencing using an Illumina MiSeq platform at Shanghai Biozeron Biotechnology Co., Ltd. The sequence data were processed following the UPARSE pipeline50. Briefly, read pairs from each sample were assembled, low-quality nucleotides (maximal expected error of 0.25) were removed and reads shorter than 200 bp were discarded. After elimination of singletons and chimeras using the UCHIME method51, sequence depth of between 30071~44756 reads were attained for all samples. The soil sample sequences were then clustered using the 16S rDNA sequences of the 2150 culturable rhizobacteria as reference taxa based on 97% similarity threshold. The relative abundance of each identified taxa cluster was then calculated as the percentage of clustered reads of the total sequence reads (30071~44756 reads including also sequences that did not match with 2150 culturable isolates) and analyzed using QIIME52. As we did not find matching sequences for 20 culturable bacterial isolates, these bacterial isolates were removed from further analysis.
Determining rhizobacteria-pathogen co-abundances in the field
We further tested whether siderophore-mediated growth effects could explain co-abundance patterns between the rhizobacteria and R. solanacearum pathogen in the field. To this end, we combined pathogen qPCR and rhizobacterial 16S rRNA data and correlated pathogen densities with the relative abundance of each rhizobacteria across all the 80 tomato rhizosphere samples resulting in 2150 regression coefficients. Positive coefficients indicate that pathogen densities are high when rhizobacterial abundances are also high, whereas negative coefficients indicate that pathogen densities are low when rhizobacterial abundances are high. We then correlated these regression coefficients with siderophore-mediated growth effects on the pathogen.
Establishing a causal link between siderophore-mediated growth inhibition and reduced pathogenicity in the tomato rhizosphere
We carried out a 60-day-long greenhouse experiment to test whether siderophore-mediated growth inhibition in the rhizosphere reduces bacterial wilt disease incidence with tomato. For this experiment, we randomly selected 360 rhizobacteria belonging to the three most abundant genera (120 isolates from Enterobacter, Bacillus and Chryseobacterium taxa each). As iron-limited supernatants from Bacillus had both inhibitory (~1/3 of all isolates) and promotive effects (~2/3 of all isolates), we randomly picked 40 and 80 isolates belonging to the two respective categories. We grew tomato seedlings in 6-well trays filled with growth substrate (100 g of substrate per well; commercially available from Jiangsu-Xingnong Substrate Technology Co., Ltd), which was sterilized with gamma radiation prior to experimentation. Surface-sterilized tomato seeds (Lycopersicon esculentum, cultivar “Micro-Tom”) were germinated on water-agar plates for three days before sowing into 6-well trays. At the three-leaf stage, six plants were independently inoculated with one of the 360 rhizobacterial isolates with a final concentration of 108 CFU per gram of substrate53 (total of 2172 plants). One week later, R. solanacearum was introduced to the roots of all plants at a final concentration of 107 CFU per gram of substrate. In addition, we had two control treatments, including tomato plants treated only with the R. solanacearum (no rhizobacteria) or sterilized water (no rhizobacteria or the pathogen). All plants were grown in a greenhouse with natural temperature variation ranging from 25°C to 35°C and watered regularly with sterile water. Seedling trays were rearranged randomly every three days. Disease progression was monitored every second day after the first disease symptoms appeared. The plant disease incidence was later quantified as Area Under Disease Progress Curve (AUDPC)54 based on disease incidence (percentage of wilted plants), which is frequently used to combine multiple observations of disease progress into a combined single value. The greenhouse experiment was finished forty days after the R. solanacearum inoculation, after which we pooled rhizosphere soil from the six plant replicates, extracted bacterial DNA and quantified R. solanacearum densities using the qPCR protocol as described above.
Statistical analysis
We use generalized linear models (GLM) to test if the production of siderophores and other metabolites affects growth of rhizobacteria directly (co-cultures) and indirectly (supernatants) in iron-limited and iron-rich conditions in the lab and the absolute and relative abundances of the pathogen and rhizobacteria in the field. The relationship between phylogenetic distance, siderophore production and siderophore-mediated effect on the pathogen growth were visualized as heatmap using the R package visreg. Relationships between pathogen and rhizobacterial abundances with siderophore-mediated effects were analyzed using linear regressions. When comparing mean differences between treatments, we used analyses of variance (ANOVA; Duncan’s multiple range tests, Mann–Whitney U tests) and Student’s t-tests where a p-values below 0.05 were considered statistically significant differences. GLMs were also used for analyzing pathogen density and disease incidence data (Area Under Disease Progress Curve; AUDPC) in the greenhouse experiment. To meet assumptions of normality and homogeneity of variance, data was log10-transformed when required. All statistical analyses were carried out using R 3.1.2 program (www.r-project.org).
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. The diversity and taxonomic classification of rhizosphere microbiomes and bacterial isolates.
a. A total of 80 rhizosphere microbiomes were identified by amplifying the V4 hypervariable regions of the bacterial 16S rRNA gene.Eight bacterial groups with highest relative abundances at the phylum, class, order, family, and genus levels are shown in the figure, while groups with relatively low abundances were merged and are presented as one group ‘Others’. b. A total of 2150 rhizosphere isolates were identified by 16s rRNA sequencing and their closest relatives were determined using the NCBI database.Seven bacterial groups with highest relative abundances at phylum, class, order, family, and genus levels are shown in the figure, while groups with relatively low abundances were merged and are presented as one group ‘Others’. In all panels, percentage (%) values in brackets represent the proportion of each bacterial group of the total OTUs (11929 OTUs; 2150 bacterial isolates).
Extended Data Fig. 2. Siderophore production of defined siderophore producers (WT), their isogenic non-producers (deletion mutants), and the 2150 rhizosphere isolates.
a. CAS values of the two defined wild type laboratory strains (producer) and the corresponding siderophore-deficient mutants (non-producer) under iron-limited (yellow) and iron-rich (purple) conditions. We used the background CAS values of the siderophore-deficient mutants as a cutoff value to distinguish background CAS activity from siderophore production. Bars represent the mean±s.d. of the siderophore production, n=4 independent biological replicates (shown as superimposed black dots). b. Mean siderophore production of the 2150 rhizosphere isolates is significantly higher under iron-limited compared to iron-rich conditions. Box plots encompass the 25–75th percentiles, the whiskers indicate outliers, the midline indicates the median and squares the mean (n=2150 biologically independent rhizobacterial isolates. P values were determined based on analysis of variance (ANOVA) followed by two-sided Mann–Whitney U tests. P<2.2×10-16 (*P < 0.05, **P < 0.01, ***P < 0.001)). c. Iron limitation induces siderophore production in up to 99% of all siderophore producers (values fall below the solid black line). Data points that fall on the solid black line, i.e, the diagonal of the square, denote cases where equal amount of siderophores were produced under iron-limited and iron–rich conditions. The black dashed lines represent the background CAS values of the two siderophore non-producers under iron-limited and iron–rich conditions, respectively.
Extended Data Fig. 3. The growth effects of siderophores by rhizosphere bacteria on the plant pathogenic R. solanacearum bacterium.
a. The histogram shows how the growth of the pathogen R. solanacearum was affected by cell-free rhizobacterial supernatants collected from iron-limited (yellow fit, GEli) and iron-rich (purple fit, GEri) media, and when iron-limited supernatants were replenished with iron (blue fit, GEre). b. The panel contrasts GEli (effects mediated by siderophores and other metabolites) with GEli - GEre (effects mediated by siderophores alone). The strong positive correlation demonstrates that growth effects were mainly driven by siderophores and not by other secreted metabolites and residual nutrients. The black line and grey shaded area depict the best-fit trendline and the 95 % confidence interval of the linear regression (adjusted coefficient of determination R2=0.755, n=2150 biologically independent rhizobacterial isolates, F1,1248=6610 and two-side P<2.2×10-16 based on Student’s t-test). c. The siderophore-mediated effects of the wild type strains (producers) on the pathogen were representative of strong (Pseudomonas aeruginosa) and mild (Burkholderia cepacia) inhibition of the pathogen, while the siderophore-deficient isogenic mutants of both species completely lost their inhibitory effect under iron-limited conditions. Bars represent the mean±s.d. of n=3 independent biological replicates (shown as superimposed black dots). d. The growth effect of purified pyoverdines of four Pseudomonas strains on pathogen under iron-limited condition were very similar to the effects of the raw supernatants (shown in brackets on X-axis). Bars represent the mean±s.d. of n=4 independent biological replicates (shown as superimposed black dots over the bars).
Extended Data Fig. 4. R. solanacearum siderophore production and effects on its own growth.
a. Iron deficiency constrains pathogen growth (after 48h growth measured as OD600) in MKB medium (iron-limited) relative to MKB medium supplemented with 50 μM FeCl3 (MKB+iron). Pathogen growth was even further reduced in MKB medium supplemented with 200 μM of the chelator 2,2′-Dipyridyl (MKB+chelator), which mimics the situation where the pathogen is exposed to a heterologous siderophore it cannot use. b. siderophore production of the pathogen R. solanacearum QL-Rs1115 under iron-limited and iron–rich conditions. c. The siderophores produced by the pathogen also promote its own growth under iron-limited conditions. The net effect caused by siderophores alone (right column, black symbols) was obtained by subtracting the growth effect of the iron-replenished supernatant (blue) from the growth effect of the iron-limited supernatant (yellow). Values indicate percentage fold-change in growth. In all panels, bars show the mean±s.d. based on 5 independent biological replicates shown as black dots over the bars) and different lowercase letters above each bar represent significant differences based on analysis of variance (ANOVA) followed by Duncan’s multiple range test (P < 0.05).
Extended Data Fig. 5. The level of siderophore production scales negatively with siderophore-mediated effects on the pathogen growth.
a. The (phylogenetically uncorrected) values of siderophore production and siderophore-mediated effects on pathogen growth are negatively correlated. b. This correlation holds even after applying phylogenetically independent contrasts to both variables, showing that isolates producing high amounts of siderophores are generally more likely to inhibit R. solanacearum growth than isolates producing low amounts of siderophores. In both panels, black dots show values for each rhizosphere isolate (n=2150 biologically independent bacterial isolates). The correlation coefficients and p-values were obtained from Spearman rank correlations that account for the non-normal distribution of the phylogenetically corrected values. The red line and the green shaded area depict the best-fit trendline and the 95 % confidence interval of a linear regression, respectively.
Extended Data Fig. 6. Effect of siderophore-producing rhizosphere bacterial taxa on tomato plant disease incidence and pathogen density.
a. The red dashed line shows the baseline level of disease incidence when soils were not pre-inoculated with rhizobacterial isolates and black dashed line represents results from the negative control treatment, where tomato plants were neither treated with rhizosphere bacteria nor with the pathogen. b. The red dashed line shows the baseline level of pathogen density when soils were not pre-inoculated with rhizobacterial isolates. In (a) and (b), box plots encompass the 25–75th percentiles, the whiskers indicate outliers, the midline indicates the median and squares the mean (n=360 biologically independent rhizobacterial isolates and P values were determined based on analysis of variance (ANOVA) followed by two-sided Mann–Whitney U tests). Significances for pairwise comparisons are (from left to right in) in (a): P=3.92×10-5, P=4.87×10-5 and P=0.933, and in (b): P=0.004, P=0.0001 and P=0.116. NS represents non-significant difference (*P < 0.05, **P < 0.01, ***P < 0.001).
Supplementary Material
Acknowledgements
We thank Bernhard Schmid for insightful comments and suggestions for the manuscript and the students in the class of Re131 who graduated in 2017 from Nanjing Agricultural University for their contributions to this work. This research was financially supported by the National Natural Science Foundation of China (41922053 to Zhong Wei; 41807045 to Tianjie Yang, 31972504 to Yangchun Xu), and the Natural Science Foundation of Jiangsu Province (BK20180527, Tianjie Yang; BK20170085, Zhong Wei). Ville-Petri Friman is supported by the Wellcome Trust [reference no. 105624] through the Centre for Chronic Diseases and Disorders (C2D2) and Royal Society Research Grants (RSG\R1\180213 and CHL\R1\180031) at the University of York. Alexandre Jousset is supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (ALW.870.15.050) and the Koninklijke Nederlandse Akademie van Wetenschappen (530-5CDP18). Rolf Kümmerli is supported by the Swiss National Science Foundation (182499) and the European Research Council under the grant agreement no. 681295. Jos Kramer is supported by the German Science Foundation (DFG; KR 5017/2-1).
Footnotes
Contributions
SG, ZW, XW, ML and XM performed and analyzed most of experiments in lab and SG, ZS, TY, KC performed and analyzed most of experiments in greenhouse. JK performed most of phylogenetic analysis. YX, QS, AJ, VPF and RK contributed intellectual input and helped to interpret data. SG, ZW, AJ, VPF and RK wrote the manuscript. All the authors discussed the results and commented on the manuscript. YX and ZW supervised the study.
Competing interests
The authors declare no competing interests.
Data availability
Raw data of 80 soil samples’ high-throughput sequences (accession numbers SRR8949365-SRR8949444) and 2150 strains’ sequencing data (accession numbers MK823189-MK825338) can be found in the NCBI database. All source data has been deposited to Dryad Digital Repository with the following digital identifier: https://doi.org/10.5061/dryad.p8cz8w9mb.
References
- 1.Fisher MC, et al. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson PK, et al. Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol Evol. 2004;19:535–544. doi: 10.1016/j.tree.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 3.Savary S, et al. The global burden of pathogens and pests on major food crops. Nat Ecol Evol. 2019;3:430–439. doi: 10.1038/s41559-018-0793-y. [DOI] [PubMed] [Google Scholar]
- 4.Andrews JH, Harris RF. The ecology and biogeography of microorganisms on plant surfaces. Annual review of phytopathology. 2000;38:145–180. doi: 10.1146/annurev.phyto.38.1.145. [DOI] [PubMed] [Google Scholar]
- 5.Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant–pathogen interactions. Nature Reviews Genetics. 2010;11:539. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 6.Mansfield J, et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 2012;13:614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poueymiro M, Genin S. Secreted proteins from Ralstonia solanacearum: a hundred tricks to kill a plant. Curr Opin Microbiol. 2009;12:44–52. doi: 10.1016/j.mib.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Niehus R, Picot A, Oliveira NM, Mitri S, Foster KR. The evolution of siderophore production as a competitive trait. Evolution; international journal of organic evolution. 2017 doi: 10.1111/evo.13230. [DOI] [PubMed] [Google Scholar]
- 9.Bruce JB, Cooper GA, Chabas H, West SA, Griffin AS. Cheating and resistance to cheating in natural populations of the bacterium Pseudomonas fluorescens. Evolution; international journal of organic evolution. 2017;71:2484–2495. doi: 10.1111/evo.13328. [DOI] [PubMed] [Google Scholar]
- 10.Butaite E, Kramer J, Wyder S, Kummerli R. Environmental determinants of pyoverdine production, exploitation and competition in natural Pseudomonas communities. Environmental microbiology. 2018;20:3629–3642. doi: 10.1111/1462-2920.14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith EE, Sims EH, Spencer DH, Kaul R, Olson MV. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa. J Bacteriol. 2005;187:2138–2147. doi: 10.1128/JB.187.6.2138-2147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butaite E, Baumgartner M, Wyder S, Kummerli R. Siderophore cheating and cheating resistance shape competition for iron in soil and freshwater Pseudomonas communities. Nature communications. 2017;8:414. doi: 10.1038/s41467-017-00509-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwak MJ, et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat Biotechnol. 2018 doi: 10.1038/nbt.4232. [DOI] [PubMed] [Google Scholar]
- 14.Berendsen RL, Pieterse CM, Bakker PA. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Compant S, Samad A, Faist H, Sessitsch AJJoAR. A review on the plant microbiome: Ecology, functions and emerging trends in microbial application. 2019 doi: 10.1016/j.jare.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei Z, et al. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nature communications. 2015;6:8413. doi: 10.1038/ncomms9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, et al. Facilitation promotes invasions in plant-associated microbial communities. Ecol Lett. 2019;22:149–158. doi: 10.1111/ele.13177. [DOI] [PubMed] [Google Scholar]
- 18.Pieterse CM, et al. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 2014;52:347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- 19.van der Meij A, van Wezel GP, Hutchings MI, Worsley SF. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiology Reviews. 2017;41:392–416. doi: 10.1093/femsre/fux005. [DOI] [PubMed] [Google Scholar]
- 20.Casper BB, Jackson RB. Plant Competition Underground. 1997;28:545–570. [Google Scholar]
- 21.Cordovez V, Dini-Andreote F, Carrión VJ, Raaijmakers JM. Ecology and Evolution of Plant Microbiomes. Annu Rev Microbiol. 2019;73:69–88. doi: 10.1146/annurev-micro-090817-062524. [DOI] [PubMed] [Google Scholar]
- 22.Cordero OX, Ventouras LA, DeLong EF, Polz MF. Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc Natl Acad Sci U S A; 2012. pp. 20059–20064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kummerli R, Schiessl KT, Waldvogel T, McNeill K, Ackermann M. Habitat structure and the evolution of diffusible siderophores in bacteria. Ecol Lett. 2014;17:1536–1544. doi: 10.1111/ele.12371. [DOI] [PubMed] [Google Scholar]
- 24.Andersen SB, Marvig RL, Molin S, Krogh Johansen H, Griffin AS. Long-term social dynamics drive loss of function in pathogenic bacteria. Proc Natl Acad Sci U S A; 2015. pp. 10756–10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barber MF, Elde NC. Buried Treasure: Evolutionary Perspectives on Microbial Iron Piracy. Trends Genet. 2015;31:627–636. doi: 10.1016/j.tig.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 27.Colombo C, Palumbo G, He J-Z, Pinton R, Cesco S. Review on iron availability in soil: interaction of Fe minerals, plants, and microbes. Journal of Soils and Sediments. 2014;14:538–548. [Google Scholar]
- 28.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hider RC, Kong X. Chemistry and biology of siderophores. Natural product reports. 2010;27:637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- 30.Lagos L, et al. Current overview on the study of bacteria in the rhizosphere by modern molecular techniques: a mini–review. Journal of soil science plant nutrition. 2015;15:504–523. [Google Scholar]
- 31.Münkemüller T, et al. How to measure and test phylogenetic signal. Methods in Ecology and Evolution. 2012;3:743–756. [Google Scholar]
- 32.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jousset A, Schmid B, Scheu S, Eisenhauer N. Genotypic richness and dissimilarity opposingly affect ecosystem functioning. Ecol Lett. 2011;14:537–545. doi: 10.1111/j.1461-0248.2011.01613.x. [DOI] [PubMed] [Google Scholar]
- 34.Kramer J, Özkaya Ö, Kümmerli R. Bacterial siderophores in community and host interactions. Nature Reviews Microbiology. 2019;4:019–028. doi: 10.1038/s41579-019-0284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schofield RK, Taylor AW. The Measurement of Soil pH1. Soil Science Society of America Journal. 1955;19:164–167. [Google Scholar]
- 36.Loper JE, Henkels MD. Availability of iron to Pseudomonas fluorescens in rhizosphere and bulk soil evaluated with an ice nucleation reporter gene. Appl Environ Microbiol. 1997;63:99–105. doi: 10.1128/aem.63.1.99-105.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heuer H, Krsek M, Baker P, Smalla K, Wellington EM. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microb. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. P Natl Acad Sci USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keck F, Rimet F, Bouchez A, Franc A. phylosignal: an R package to measure, test, and explore the phylogenetic signal. Ecology and Evolution. 2016;6:2774–2780. doi: 10.1002/ece3.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2018;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 42.HÖfte M, Buysens S, Koedam N, Cornelis P. Zinc affects siderophore-mediated high affinity iron uptake systems in the rhizosphere Pseudomonas aeruginosa 7NSK2 . Biometals. 1993;6:85–91. doi: 10.1007/BF00140108. [DOI] [PubMed] [Google Scholar]
- 43.Schwyn B, Neilands J. Universal chemical assay for the detection and determination of siderophores. Analytical biochemistry. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 44.Ghysels B, et al. The Pseudomonas aeruginosa pirA gene encodes a second receptor for ferrienterobactin and synthetic catecholate analogues. FEMS Microbiology Letters. 2005;246:167–174. doi: 10.1016/j.femsle.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 45.Sathe S, Mathew A, Agnoli K, Eberl L, Kümmerli R. Genetic architecture constrains exploitation of siderophore cooperation in the bacterium Burkholderia cenocepacia. Evolution Letters. 2019;3:144. doi: 10.1002/evl3.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer J-M, et al. Use of Siderophores to Type Pseudomonads: The Three Pseudomonas Aeruginosa Pyoverdine Systems. Microbiology. 1997;143:35–43. doi: 10.1099/00221287-143-1-35. [DOI] [PubMed] [Google Scholar]
- 47.Schonfeld J, Heuer H, Van Elsas JD, Smalla K. Specific and sensitive detection of Ralstonia solanacearum in soil on the basis of PCR amplification of fliC fragments. Appl Environ Microbiol. 2003;69:7248–7256. doi: 10.1128/AEM.69.12.7248-7256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y, et al. A Real-Time PCR Assay for the Quantitative Detection of Ralstonia solanacearum in Horticultural Soil and Plant Tissues. J Microbiol Biotechn. 2010;20:193–201. [PubMed] [Google Scholar]
- 49.Cardenas E, et al. Significant Association between Sulfate-Reducing Bacteria and Uranium-Reducing Microbial Communities as Revealed by a Combined Massively Parallel Sequencing-Indicator Species Approach. Appl Environ Microb. 2010;76:6778–6786. doi: 10.1128/AEM.01097-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 51.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei Z, et al. Efficacy of Bacillus-fortified organic fertiliser in controlling bacterial wilt of tomato in the field. Appl Soil Ecol. 2011;48:152–159. [Google Scholar]
- 54.Jeger MJ, Viljanen-Rollinson SLHJT, Genetics A. The use of the area under the disease-progress curve (AUDPC) to assess quantitative disease resistance in crop cultivars. 2001;102:32–40. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data of 80 soil samples’ high-throughput sequences (accession numbers SRR8949365-SRR8949444) and 2150 strains’ sequencing data (accession numbers MK823189-MK825338) can be found in the NCBI database. All source data has been deposited to Dryad Digital Repository with the following digital identifier: https://doi.org/10.5061/dryad.p8cz8w9mb.