Abstract
Expanding and reprogramming the genetic code of cells for the incorporation of multiple distinct non-canonical amino acids (ncAAs), and the encoded biosynthesis of non-canonical biopolymers, requires the discovery of multiple orthogonal aminoacyltransfer RNA synthetase/tRNA pairs. These pairs must be orthogonal to both the host synthetases and tRNAs and to each other. Pyrrolysyl-tRNA synthetase (PylRS)/PyltRNA pairs are the most widely used system for genetic code expansion. Here we reveal that the sequences of ΔNPylRS/ΔNPyltRNA pairs (which lack N-terminal domains) form two distinct classes. We show that the measured specificities of the ΔNPylRSs and ΔNPyltRNAs correlate with sequence-based clustering, and most ΔNPylRSs preferentially function with ΔNPyltRNAs from their class. We then identify 18 mutually orthogonal pairs from the 88 ΔNPylRS/ΔNPyltRNA combinations tested. Moreover, we generate a set of 12 triply orthogonal pairs, each composed of three new PylRS/PyltRNA pairs. Finally, we diverge the ncAA specificity and decoding properties of each pair, within a triply orthogonal set, and direct the incorporation of three distinct non-canonical amino acids into a single polypeptide.
Protein translation provides the ultimate paradigm for the encoded cellular synthesis of defined polymer sequences, but natural translation is commonly limited to polymerizing the 20 canonical amino acids. Engineering cellular translation may enable the biosynthesis and evolution of non-canonical biopolymers1,2. However, this will require: (1) strategies for the creation of blank codons, beyond stop codons, that may be assigned to new monomers 3–5; (2) the creation of ribosomes, and other translation factors, with expanded substrate sope6–8; and (3) the creation of a set of aminoacyl-tRNA synthetase (aaRS)/tRNA pairs that are orthogonal with respect to endogenous synthetases and tRNAs in the host organism and mutually orthogonal with respect to each other5,9–12. These pairs must be further engineered to decode distinct blank codons and use unique monomers that are not substrates for other aaRSs. An additional mutually orthogonal pair is required for each new monomer that is uniquely encoded.
Despite exciting progress towards the encoded cellular synthesis of non-canonical biopolymers, the identification of multiple engineered mutually orthogonal aaRS/tRNA pairs that recognize distinct codons and incorporate distinct non-canonical amino acids (ncAAs) remains an outstanding challenge5,9,13,14. Each new ncAA, aaRS, tRNA and codon must function together and be orthogonal to each endogenous amino acid, aaRS, and group of isoacceptor tRNAs and their cognate group of codons. Therefore, for each new ncAA:aaRS:tRNA:codon set, three interactions must be established (ncAA:aaRS, aaRS:tRNA, and tRNA:codon) and 120 interactions (6 x 20 interactions; this analysis counts all isoacceptors for a natural amino acid as one and all codons for an amino acid as one and therefore provides a conservative estimate of the interactions that must be controlled) between the new set and the endogenous translational machinery must be minimized. Moreover, when incorporating more than one ncAA there is the potential for interactions between components of the additional ncAA:aaRS:tRNA:codon sets, and these must also be minimized. Generating ncAA:aaRS:tRNA:codon sets to encode three distinct ncAAs into a polypeptide requires nine specific interactions to be established and minimization of at least 378 specific interactions, including 18 interactions between components of the three sets.
Five pairs (based on Methanococcus janaschii (Mj)tyrosyl-tRNA synthetase (TyrRS)/Mj TyrtRNA, Methanosarcina mazei (Mm) or Methanosarcina barkeri (Mb) pyrrolysyl-tRNA synthetase (PylRS)/Mm PyltRNA, Methanomethylophilus alvus (referred to herein as Alv)PylRS/Alv PyltRNA, Methanococcus maripaludis (Mmp) phosphoseryl-tRNA synthetase (SepRS)/Mj SeptRNA, and Saccharomyces cerevisiae (Sc) tryptophanyl-tRNA synthetase (TrpRS)/Sc TrptRNA) are orthogonal with respect to the endogenous synthetases and tRNAs in Escherichia coli and have been used as part of strategies to incorporate diverse ncAAs into proteins10,15–23. Some combinations of these pairs are mutually orthogonal5,10,12,14 and have been engineered to incorporate two ncAAs into proteins. One report used derivatives of three existing pairs to incorporate ncAAs in response to all three stop codons. This strategy leaves no codon for termination of endogenous genes; since protein termination is essential to cells we anticipate that efficient variants of this strategy will be toxic. Moreover, to obtain a recombinant protein of homogeneous length a protease site had to be introduced and the translation products proteolytically cleaved, as no codon was available for defining the termination of protein synthesis12.
The MmPylRS/Mm PyltRNACUA pair, and closely related MbPylRS/Mb PyltRNACUA pair, are the most widely used systems for genetic code expansion24. These pairs have gained popularity for several reasons: (1) PylRS/PyltRNA pairs are functional across all domains of life and naturally orthogonal to endogenous synthetase/tRNA pairs in both prokaryotic and eukaryotic cells24; (2) the PyltRNA anticodon is not recognized by PylRS and can be mutated to create derivatives of the pair targeted to decode diverse codons14,25; and (3) the active site of PylRS does not recognize canonical amino acids, accepts several non-natural substrates and can be evolved to specifically incorporate a diverse array of ncAAs17,24. The advantages of PylRS/PyltRNA systems for genetic code expansion may also be advantages for non-canonical biopolymer synthesis.
MmPylRS is composed of two domains, an N-terminal domain and a C-terminal domain. The C-terminal domain binds the amino acid substrate and catalyses the aminoacylation of Mm PyltRNA, while the N-terminal domain makes additional contacts to the variable loop of Mm PyltRNA that enhance binding affinity and specificity26,27. Both domains are required to create a functional MmPylRS/Mm PyltRNA pair in E. coli28. Until recently it was widely believed that all functional PylRS/PyltRNA pairs utilized a two domain PylRS, with bacterial PylRS systems encoding each domain on a separate polypeptide29.
A new class of highly active PylRS enzymes - from organisms that encode a PylRS gene lacking an N-terminal domain and do not encode a separate N-terminal domain polypeptide - was recently identified10,19,30 and characterised10. These PylRS enzymes, which we refer to as ΔNPylRSs, function with their cognate PyltRNA, which we term ΔNPyltRNA10. Certain ΔNPylRS/ΔNPyltRNA pairs, or their evolved derivatives, are orthogonal in E. coli, and can be used to incorporate ncAAs10. Moreover, evolved ΔNPylRS/ΔNPyltRNA pairs from M. alvus (Alv), in which the variable loop of Alv ΔNPyltRNA has been expanded, are mutually orthogonal to MmPylRS/Mm PyltRNA10. Subsequent work has shown that derivatives of these pairs are mutually orthogonal in mammalian cells31,32.
Here we identify new ΔNPylRS/ΔNPyltRNA pairs, and reveal that the sequences of ΔNPylRS/ΔNPyltRNA pairs cluster into two distinct classes. Remarkably, the measured specificities of the ΔNPylRSs and ΔNPyltRNAs correlate with the sequence-based clustering, such that class A ΔNPylRSs preferentially function with class A ΔNPyltRNAs, and class B ΔNPylRSs preferentially function with class B ΔNPyltRNAs. We identify 18 mutually orthogonal ΔNPylRS/ΔNPyltRNA pairs. We go on to evolve class A and class B ΔNPyltRNAs to generate a set of 12 triply orthogonal pairs. These pairs are composed of a new MmPylRS/Spe PyltRNA pair, an evolved class A ΔNPylRS/ΔNPyltRNA pair and an evolved class B ΔNPylRS/ΔNPyltRNA pair. We show that these pairs can be engineered to recognize three distinct ncAAs and decode three distinct codons. Finally we show that resulting engineered triply orthogonal pairs can be used to program the cellular incorporation of three distinct ncAAs into a single protein.
Results
Identification of DNPylRS/ΔNPyltRNA pairs
Fifteen ΔNPylRS family members from methanogenic archaea have been reported to date10,30, and for seven of these the cognate ΔNPyltRNA of the pair has been identified in the genome. By performing a HMMER33,34 search for sequence similarity to the C-terminal catalytic domain of MmPylRS (MmPylRSΔ184), we identified two further ΔNPylRS genes (from Methanomassiliicoccales archaeon PtaU1.Bin030 (030) and M. archaeon PtaU1.Bin124),
We used the DNA sequence for each ΔNPylRS gene within its host genome to identify the pyrrolysine gene cluster19, and in four cases we identified new ΔNPyltRNA sequences, in the surrounding sequence. This brought the total number of cognate ΔNPylRS/ΔNPyltRNA pairs to 11(Supplementary Table 1 and 2). All of the identified ΔNPyltRNAs are predicted to fold into clover-leaf structures typical of the previously reported ΔNPyltRNAs from the ΔNPylRS class, sharing the nucleotide loops or bulges in the anticodon stem together with a typical length of D-arm, T-arm, acceptor stem and anticodon stem (Supplementary Fig. 1a).
ΔNPyltRNAs partition into two classes
We analysed the sequence similarity between the 11 ΔNPyltRNA sequences (Supplementary Fig. 2), and performed a hierarchical clustering of the tRNA sequences (Fig. 1a). Interestingly, the ΔNPyltRNA sequences clustered in two groups: one containing 1R26, Alv, H5, G1, and Term which we termed sequence class A, and the other containing Lum1, Lum2, Sheng, 030, Int and RumEn which we termed sequence class B (Fig. 1). Notably, the ΔNPylRSs showed a similar sequence relationship to that of the ΔNPyltRNA sequences and grouped into the same two classes when the clustering was performed on the percentage of protein sequence identity (Supplementary Fig. 3, 4).
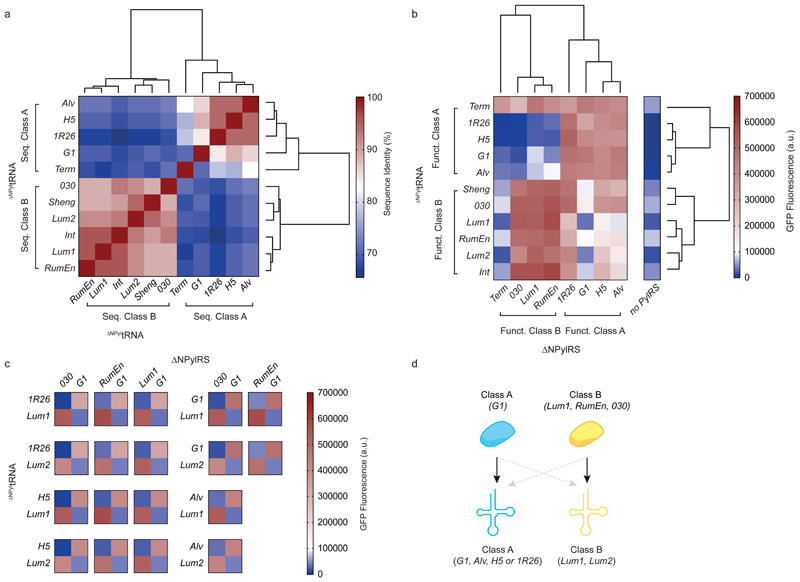
Figure 1. Identifying two classes of ΔNPylRS/ΔNPyltRNA pairs and 18 naturally mutually orthogonal ΔNPylRS/ΔNPyltRNA pairs.
a, Hierarchical clustering of ΔNPyltRNAs with sequence similarity scores converted to Euclidean distance measures and complete linkage clustering (determined using the program R Studio). Percentage sequence identity scores are displayed as a heatmap. The dendrograms resulting from the clustering show the groupings of the ΔNPyltRNAs into two sequence-dependent classes. b, Hierarchical clustering of the indicated ΔNPylRS/ΔNPyltRNACUA pairs on the basis of GFP fluorescence (that is, the resulting expression of GFP(150TAG)His6 in presence of BocK 1) converted to Euclidean distance measures and complete linkage clustering (determined using the program R Studio). Bar charts showing the number of replicates and error bars showing s.d. are provided in Supplementary Fig. 6. All numerical values are provided in Supplementary Table 3. The dendrograms resulting from the clustering show the groupings of the ΔNPyltRNAs and ΔNPylRS into two function-dependent classes. Funct.,functional. c, Heatmaps displaying the in vivo amber suppression data as described in b for the 18 mutually orthogonal pairs. d, Schematic indicating naturally mutually orthogonal ΔNPylRS/ΔNPyltRNA pairs. Class A synthetases and tRNAs are in blue, class B synthetases and tRNAs are in yellow. Black arrows indicate high activity and dashed grey arrows indicate minimal activity corresponding to functional orthogonality.
We identified the nucleotides and predicted secondary structures that are conserved within each class of ΔNPyltRNAs, but differ between sequence class A and sequence class B ΔNPyltRNAs (Supplementary Fig. 1b). These are mostly located in the acceptor stem, T-stem, and T-loop. We hypothesized that these differences might constitute class-specific identity elements, and that some sequence class A and sequence class B DNPylRS/ΔNPyltRNA pairs might, therefore, be mutually orthogonal.
Sequence class B ΔNPyltRNAs contain a cytosine at position 37. Consistent with previous observations35, we found that C37A mutations in sequence class B ΔNPyltRNAs led to increased read through of the amber codon by sequence class B ΔNPylRS/ΔNPyltRNA pairs (Supplementary Fig. 5, Supplementary Table 3). These mutants were used for all further experiments.
Activity, orthogonality and mutual orthogonality of DNPylRS/ΔNPyltRNA pairs
Next we investigated whether the ΔNPylRS/ΔNPyltRNACUA pairs we had identified were active and orthogonal in E. coli. We measured the orthogonality of each tRNA by its ability to produce GFPHis6 from GFP(150TAG)His6 in the absence of any exogenous synthetase. Sequence class A ΔNPyltRNAs led to very low levels of GFP production and sequence class B ΔNPyltRNAs led to low levels of GFP production (Fig. 1b, Supplementary Fig. 6). We concluded that these tRNAs are functionally orthogonal.
To measure the activity and orthogonality of the cognate ΔNPylRS for each ΔNPyltRNA, we introduced each cognate ΔNPylRS/ΔNPyltRNA pair and GFP(150TAG)His6 in cells, along with a known substrate for all previously characterized PylRS enzymes (Nε-((tertbutoxy)carbonyl)-l-lysine (BocK 1)).10 For all sequence class A ΔNPyltRNAs, and the sequence class B ΔNPyltRNAs from 030, Lum1 and RumEn, coexpression of the cognate synthetase in the presence of BocK 1 led to a substantial increase in read through of the amber codon in GFP(150TAG)His6 (Fig. 1b). This indicated that both ΔNPylRS and ΔNPyltRNA are active in E. coli. We observed minimal increase in signal for ShengPylRS, Lum2PylRS or IntPylRS upon addition of BocK 1 (Supplementary Fig. 7, Supplementary Table 3). We concluded that these PylRSs are likely to be either not expressed or not functional in E. coli and we did not consider them further. BRNAPylRS was only weakly active with its cognate ΔNPyltRNA and this tRNA was identical in sequence to the tRNA we identified from 1R26. Since the 1R26PylRS/1R26 ΔNPyltRNA pair was substantially more active than the BRNAPylRS/1R26 ΔNPyltRNA pair we decided to proceed with 1R26PylRS (Supplementary Fig. 7, Supplementary Table 3). ΔNPylRSs for which a cognate tRNA has not been identified from the same host (from Methanonatronarchaeum thermophilum, Methanohalarchaeum thermophilum, MSBL1 archaeon SCGCAAA382A20 or M. archaeon PtaU1.Bin124) were not characterized any further.
Time-of-flight mass spectrometry (TOF-MS) of GFPHis6 produced from GFP(150TAG)His6 in the presence of BocK confirmed that the aaRSs from cognate pairs (AlvPylRS/Alv ΔNPyltRNA, H5PylRS/H5 ΔNPyltRNA, 1R26PylRS/1R26 ΔNPyltRNA, G1PylRS/G1 ΔNPyltRNA, TermPylRS/Term ΔNPyltRNA, 030PylRS/030 ΔNPyltRNA, RumEnPylRS/RumEn ΔNPyltRNA, and Lum1PylRS/Lum1 ΔNPyltRNA) are functionally orthogonal (Supplementary Fig. 8).
We experimentally characterized the activity of all 88 ΔNPylRS/ΔNPyltRNA combinations arising from 11 tRNAs and eight synthetases (Fig. 1b). This provided functional data on the activity of cognate and non-cognate combinations. When clustered on the basis of their relative activity, ΔNPylRSs and ΔNPyltRNAs each form two classes, which we defined as functional class A and functional class B.
Functional class A synthetases were highly active with functional class A ΔNPyltRNAs and most functional class A synthetases showed lower, but measurable, activity when paired with the majority of functional class B ΔNPyltRNAs. Notably, G1PylRS from functional class A was orthogonal with respect to several functional class B ΔNPyltRNAs. Functional class B synthetases, except TermPylRS, were highly active with functional class B ΔNPyltRNAs, but not with most functional class A ΔNPyltRNAs. TermPylRS - a functional class B synthetase - was exclusively selective for its cognate tRNA. Term ΔNPyltRNA was classified as a functional class A ΔNPyltRNA and was a substrate for all ΔNPylRSs tested.
There is striking correspondence between the classes identified on the basis of sequence and the classes identified on the basis of function for both the ΔNPylRSs and ΔNPyltRNAs (Fig. 1a, b and Supplementary Fig. 9). Seven out of eight ΔNPylRSs, and all 11 ΔNPyltRNAs, partition into the same two groups on the basis of sequence and function. We defined consensus class A and class B ΔNPylRSs and ΔNPyltRNAs, which we subsequently referred to as simply class A and class B. Class A contains ΔNPylRSs and ΔNPyltRNAs from Alv, G1, H5 and 1R26, and class B contains ΔNPylRSs and ΔNPyltRNAs from Lum1, Lum2, Sheng, 030, Int and RumEn. We excluded the Term pair (for which the synthetase and tRNA partitioned into different functional classes) from either consensus class and we did not consider this pair further. Thus we proceeded with ten ΔNPyltRNAs and seven ΔNPylRSs.
The correlation between the tRNA classes defined by sequence and the tRNA classes defined by function suggests that the differences in tRNA sequence between each class contain the distinct nucleotide identity elements that specify class-specific synthetase recognition. The preference of class A tRNAs for class A synthetases and the preference of class B tRNAs for class B synthetases will lie within these nucleotide sequences. Similarly, the correlations between synthetase classes defined by sequence, and those defined by function, suggest that the class-specific amino acid sequences within synthetases define class-specific tRNA recognition. The preference of class A synthetases for class A tRNAs and the preference of class B synthetases for class B tRNAs will lie within these amino acid sequences.
Analysis of our data allowed us to identify 18 different combinations of naturally mutually orthogonal DNPylRS/ΔNPyltRNA pairs with good orthogonality and activity. These pairs are composed of G1ΔNPylRS with any class A ΔNPyltRNA and a select class B ΔNPylRS with either Lum1 ΔNPyltRNA or Lum2 ΔNPyltRNA (Fig. 1c, d).
The MmPylRS/MmPyltRNA pair is not orthogonal with respect to many DNPylRSs or ΔNPyltRNAs
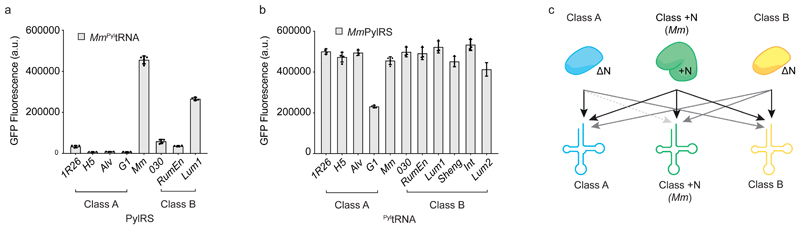
Next we characterized the activity of Mm PyltRNA with each of the seven DNPylRS enzymes, and the activity of MmPylRS with all ten ΔNPyltRNAs (Fig. 2).We found that all of the class B synthetases are functional with Mm PyltRNA while all of the class A synthetases, except 1R26, do not function with Mm PyltRNA (Fig. 2a). We also found that MmPylRS is active with all ΔNPyltRNAs tested, although it is least active with G1 ΔNPyltRNA (Fig. 2b, Supplementary Fig. 10, Supplementary Table 3). These observations define the activity of the MmPylRS/Mm PyltRNA pair with respect to class A and B synthetases and ΔNPyltRNAs. In combination with the activities of class A and B ΔNPylRS/ΔNPyltRNA pairs (Fig. 1), these experiments define the activities we need to engineer to create a set of three mutually orthogonal pairs composed of a class A ΔNPylRS/ΔNPyltRNA pair, a class B ΔNPylRS/ΔNPyltRNA pair, and an MmPylRS/PyltRNA pair (Fig. 2c).
Figure 2. The MmPylRS/Mm PyltRNA pair is not orthogonal with respect to any ΔNPyltRNAs and some ΔNPylRSs.
a, GFPHis6 fluorescence generated by each PylRS with Mm PyltRNACUA resulting from GFP(150TAG)His6 in presence of BocK 1. Each bar represents the mean of four biological replicates ± s.d.. The individual data points are shown as dots. All numerical values are provided in Supplementary Table 3. b, GFPHis6 fluorescence generated by MmPylRS with each PyltRNACUA, amd GFP(150TAG)His6 in presence of BocK 1. Each bar represents the mean of three biological replicates ± s.d. for class B ΔNPyltRNACUAs or four biological replicates ± s.d. for class A ΔNPyltRNACUAs and Mm PyltRNACUA. The individual data points are shown as dots. c, Schematic of the functional interactions between class A and class B ΔNPylRS/ΔNPyltRNA pairs in relation to each other and MmPylRS/Mm PyltRNA. Black arrows indicate high activity, grey arrows indicate low activity and the dashed gray arrow indicates minimal activity corresponding to functional orthogonality.
To discover triply orthogonal PylRS/PyltRNA pairs we needed to create: (1) a tRNA that is a substrate for MmPylRS, while being orthogonal to a class A DNPylRS, a class B DNPylRS, and endogenous synthetases; (2) a tRNA that is only active with class A DNPylRSs while being orthogonal to both MmPylRS, class B DNPylRSs, and endogenous synthetases; and (3) a tRNA that is solely a substrate for class B DNPylRSs while being orthogonal to MmPylRS, class A DNPylRSs, and endogenous synthetases. In our subsequent experiments we addressed each of these challenges in turn.
Discovering class +N +NPyltRNAs that are orthogonal to class A and B DNPylRSs
As Mm PyltRNA functions with both class B DNPylRSs and 1R26PylRS (from class A), it is not compatible with the creation of triply orthogonal PylRS/PyltRNA pairs. We therefore set out to discover PyltRNAs that are both active with MmPylRS, and orthogonal with respect to class A and class B DNPylRSs.
For many PylRS enzymes with an N- and C-terminal domain (denoted class +N herein) the cognate tRNAs (denoted +NPyltRNAs herein) remain unannotated and unknown. We took advantage of existing genome annotations to identify the DNA sequence coding for class +N PylRSs within their host genomes and thereby identified ten +NPyltRNA genes in the surrounding sequence; for six of these sequences we are not aware of previous annotations or reports (Supplementary Table 4). In combination with Mm PyltRNACUA and Mb PyltRNACUA these sequences provided 12 +NPyltRNA sequences for further investigation.
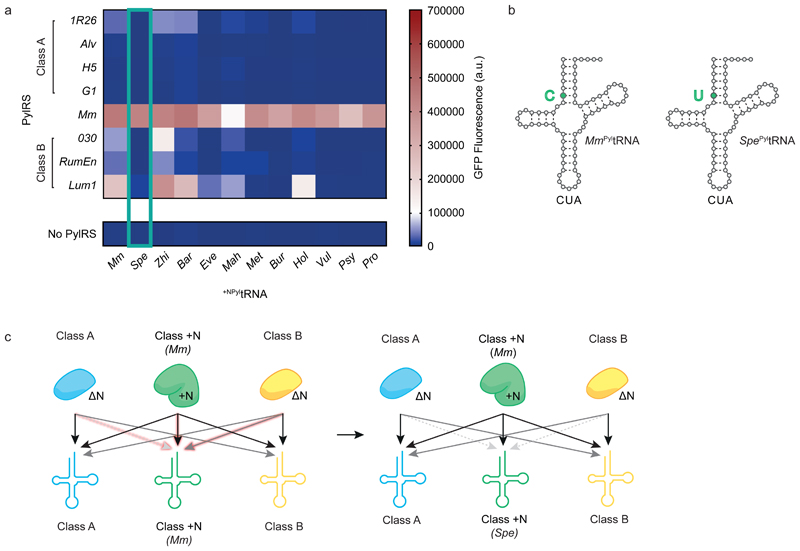
We measured the activity of the 12 +NPyltRNAs with class A and B DNPylRSs, with MmPylRS, or without any PylRS. We discovered six +NPyltRNAs (from Bur, Met, Pro, Psy, Spe and Vul) that were orthogonal to all the DNPylRSs tested, orthogonal with respect to endogenous synthetases and formed active, heterologous pairs with MmPylRS (Fig 3a, Supplementary Fig. 11, Supplementary Table 3).
Figure 3. Identifying triply orthogonal and active Class +N +NPyltRNAs.
a, GFPHis6 fluorescence with indicated +NPyltRNACUA resulting from decoding the amber codon in GFP(150TAG)His6 in the presence of BocK 1. Each heatmap value represents the average of three biological replicates. Bar charts including error bars showing s.d. are provided in Supplementary Fig. 11 and all numerical values are provided in Supplementary Table 3. The data for Spe PyltRNA are outlined in green. b, Clover leaf structure of Mm PyltRNA and Spe PyltRNA. The highlighted C-U point mutation changes a Watson-Crick base pair into a wobble base pair and the Spe PyltRNA is orthogonal to every ΔNPylRS tested. c, Schematic of the progress made towards generating triply orthogonal PylRS/PyltRNA pairs when substituting Mm PyltRNA (left) for Spe PyltRNA (right). Black arrows indicate high activity, grey arrows indicate low activity and dashed grey arrows indicate minimal activity corresponding to functional orthogonality. The interactions of the +NPyltRNA that were manipulated in the experiments shown in a are highlighted.
Among these six heterologous pairs, the MmPylRS/Spe PyltRNA pair was the most active, with similar activity to the native MmPylRS/Mm PyltRNA pair (Fig 3a). The pair produced GFPHis6 from GFP(150TAG)His6 at 80% of the level of wild type GFPHis6 (Supplementary Fig. 12, Supplementary Table 3). Remarkably, Spe PyltRNA only differs from Mm PyltRNA by a single point mutation in the acceptor stem, changing the C-G Watson-Crick base pair to a U-G wobble (Fig. 3b). These experiments established the MmPylRS/Spe PyltRNA pair as the class +N pair for a mutually orthogonal PylRS/PyltRNA triplet (Fig. 3c).
Engineered class A ΔNPyltRNAs are orthogonal to class B DNPylRSs and MmPylRS
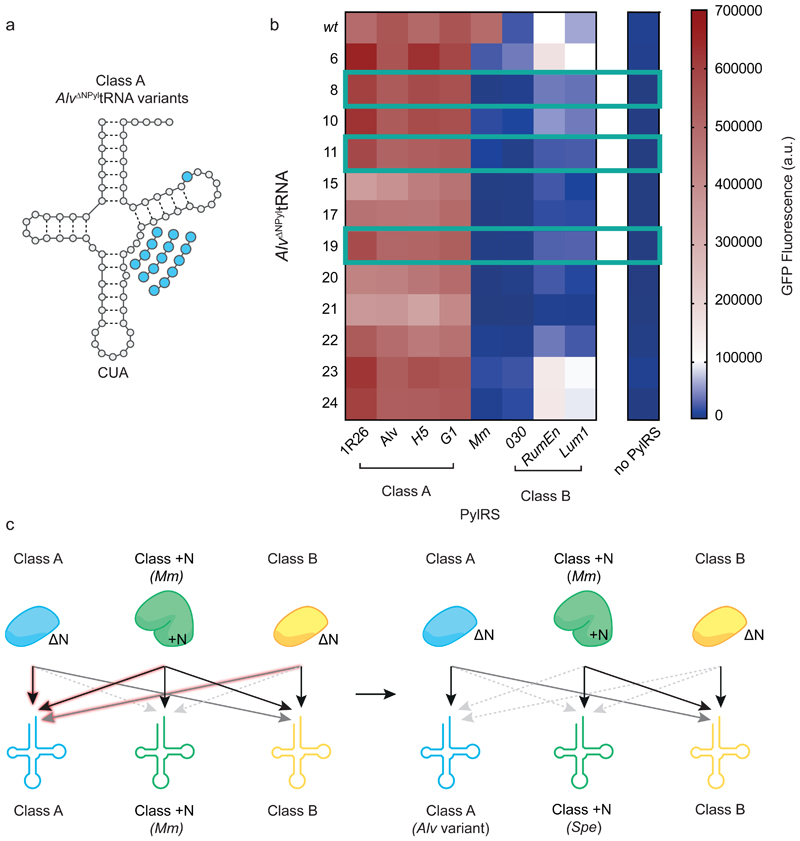
Next we aimed to discover a tRNA that is only active with class A DNPylRSs while being orthogonal to MmPylRS, class B DNPylRSs and endogenous synthetases. We have previously demonstrated that expanding the variable loop of Alv ΔNPyltRNA (a class A ΔNPyltRNA) destroys its activity with MmPylRS without affecting activity with its cognate synthetase, AlvPylRS10.
We tested 12 previously described Alv ΔNPyltRNA variants with expanded variable loops that are active with AlvPylRS (Fig. 4a, Supplementary Fig. 13, Supplementary Table 5)10. All 12 variants showed good activity with class A synthetases, and little or no activity with MmPylRS, class B synthetases or endogenous synthetases (Fig. 4b). We found three variants (Alv ΔNPyltRNA(8), Alv ΔNPyltRNA(11) and Alv ΔNPyltRNA(19)) that had good orthogonality towards both MmPylRS and class B synthetases while remaining highly active with AlvPylRS and other class A synthetases (G1PylRS, H5PylRS and 1R26PylRS) (Fig. 4b). These three Alv ΔNPyltRNA variants are more orthogonal than the parent Alv ΔNPyltRNA with respect to RumEnPylRS, 030PylRS, Lum1PylRS and MmPylRS. These experiments established G1PylRS, AlvPylRS, H5PylRS or 1R26PylRS in combination with Alv ΔNPyltRNA(8), Alv ΔNPyltRNA(11) or Alv ΔNPyltRNA(19) as the class A-derived pairs for a set of triply orthogonal PylRS/PyltRNA pairs (Fig. 4c).
Figure 4. Identifying triply orthogonal and active Class A ΔNPyltRNAs.
a, Clover leaf structure of Alv ΔNPyltRNACUA variants with variable loop expansions and point mutations highlighted in blue. b, GFPHis6 fluorescence resulting from wild type Alv ΔNPyltRNACUA or the indicated Alv ΔNPyltRNACUA variant with each PylRS from cells containing GFP(150TAG)His6 in presence of BocK 1. Each heatmap value represents the average of three biological replicates. Bar charts including error bars showing s.d. are provided in Supplementary Fig. 13 and all numerical values are provided in Supplementary Table 3. Alv ΔNPyltRNA(8)CUA, Alv ΔNPyltRNA(11)CUA and Alv ΔNPyltRNA(19)CUA all showed improved orthogonality to MmPylRS and class B ΔNPylRSs while remaining active with class A ΔNPylRSs (highlighted with a green frame). c, Schematic of the progress made towards generating triply orthogonal PylRS/PyltRNA pairs when substituting wild-type Alv ΔNPyltRNA (left) with selected Alv ΔNPyltRNA variants (right). Black arrows indicate high activity, grey arrows indicate low activity and dashed grey arrows indicate minimal activity corresponding to functional orthogonality. The interactions of Alv ΔNPyltRNA that were manipulated in these experiments are highlighted.
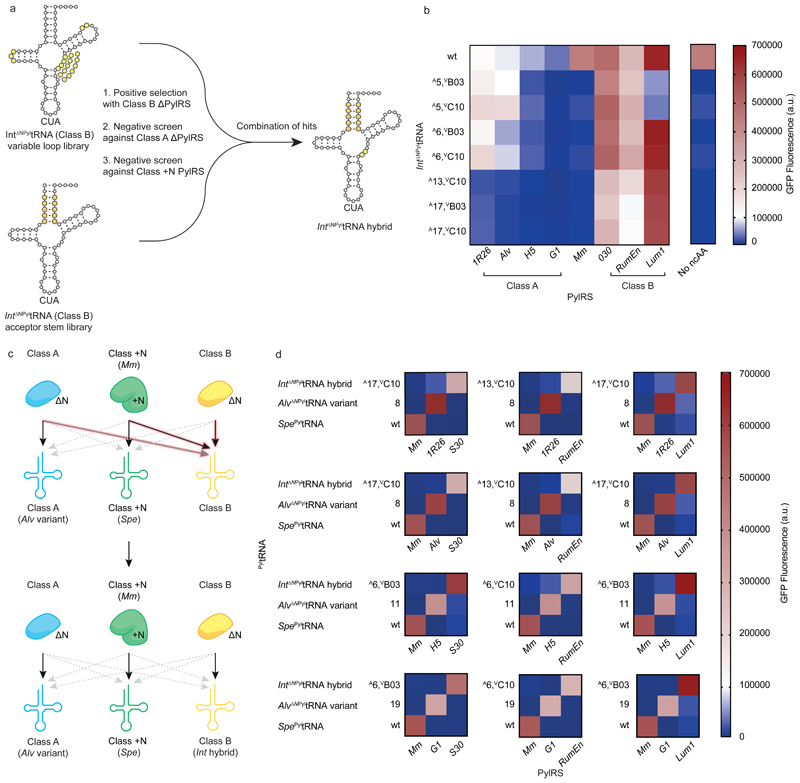
Directed evolution of class B ΔNPyltRNAs orthogonal to class A DNPylRSs and MmPylRS
Class B ΔNPyltRNAs function efficiently with MmPylRS and are active with some class A DNPylRSs. Both of these activities are incompatible with the creation of a set of triply orthogonal PylRS/PyltRNA pairs. We decided to take a convergent approach to evolving a class B ΔNPyltRNA that was active with class B DNPylRSs but did not function with MmPylRS or class A DNPylRSs (Fig. 5a). We focused on Int DNPyltRNA, which has good activity with class B DNPylRSs and lower activity with many class A DNPylRSs than other class B DNPyltRNAs (Fig. 1b).
Figure 5. Identifying triply orthogonal and active Class B ΔNPyltRNAs.
a, Schematic of the evolution of class B Int ΔNPyltRNA. Clover leaf structures of Int ΔNPyltRNACUA libraries are shown with randomized nucleotides highlighted in light yellow (variable loop library) or dark yellow (acceptor stem library). Int ΔNPyltRNACUA hybrids result from combining the best hits from each library. b, GFP fluorescence from GFP(150TAG)His6 in cells containing BocK 1 and Int ΔNPyltRNACUA hybrids with each PylRS. Each heatmap value represents the average of three biological replicates. Bar charts including error bars showing s.d. are provided in Supplementary Fig. 17 and all numerical values are provided in Supplementary Table 3. c, Schematic of the progress made in generating triply orthogonal PylRS/PyltRNA pairs when substituting wild-type Int ΔNPyltRNA (top) with selected Int ΔNPyltRNA hybrids (bottom). Black arrows indicate high activity, grey arrows indicate low activity and dashed grey arrows indicate minimal activity corresponding to functional orthogonality. The interactions of Int ΔNPyltRNA that were manipulated in these experiments are highlighted. d, Heatmap representation of all sets of triply orthogonal PylRS/PyltRNA pairs generated in this study. Each heatmap value represents the average of three biological replicates. Bar charts including error bars showing s.d. are provided in Supplementary Figs. 17 and 18. Each triplet is composed of MmPylRS and Spe PyltRNA, a specific class A ΔNPylRS and an evolved Alv ΔNPyltRNA variant, and a specific class B ΔNPylRS and an evolved Int ΔNPyltRNA hybrid.
We hypothesized that we could abolish any recognition of Int DNPyltRNA by MmPylRS by expanding its variable loop10. We created a variable loop library of Int ΔNPyltRNA mutants in which the length of the variable loop was expanded from three nucleotides to four, five or six randomized nucleotides. In each variable loop library, positions 13, 14, 15, 54 and 55 (in the D-loop and T-loop) (Fig. 5a), which may make contacts with the variable loop in the folded tertiary structure of the tRNA, were also randomized27.
We selected Int ΔNPyltRNA variants that functioned with a class B DNPylRS (030PylRS) and enabled cells to grow on 100 µg ml-1 chloramphenicol in the presence of BocK 1, by facilitating read through of an amber codon at position 111 of a chloramphenicol acetyl transferase reporter, cat(111TAG). Next we performed two negative screens on the selected Int ΔNPyltRNA variants to identify tRNAs that do not function with MmPylRS, a class A synthetase (AlvPylRS) or any endogenous E. coli synthetase. Cells bearing GFP(150TAG) His6, MmPylRS or AlvPylRS, and each Int ΔNPyltRNA variant were provided with BocK 1 and screened for the absence of GFPHis6 expression. This serial positive selection and double negative screen identified two evolved mutants, Int ΔNPyltRNA(VB03) and Int ΔNPyltRNA(VC10), each with single nucleotide insertions into the variable loop. These tRNAs form 030PylRS/Int ΔNPyltRNA pairs which retain two thirds of the activity of the initial 030PylRS/Int ΔNPyltRNA wild type pair, but have much improved orthogonality towards MmPylRS, AlvPylRS and endogenous synthetases (Supplementary Fig. 14, Supplementary Table 6).
The majority of nucleotides that distinguish class A and class B ΔNPyltRNAs are clustered in the acceptor stem (Supplementary Fig. 1b). This observation suggested that the acceptor stem may be an important recognition element in determining tRNA substrate specificity by DNPylRSs. We decided to evolve the acceptor stem of a class B ΔNPyltRNA to discover variants that selectively diminish recognition by class A synthetases without affecting activity with class B synthetases.
We created an acceptor stem library of Int ΔNPyltRNA mutants by randomizing nucleotide positions 3-7 and 61-65 (Fig. 5a). We performed a serial positive selection and double negative screen, analogous to that described for the variable loop library, on the acceptor stem library. Following this process we isolated 11 Int ΔNPyltRNA acceptor stem mutants that retained over 85% of the activity of wild type Int ΔNPyltRNA when paired with 030PylRS (Supplementary Fig. 15, Supplementary Table 7). However, the evolved variants still displayed substantial cross-reactivity with MmPylRS and some activity with AlvPylRS, although both of these were diminished when compared with wild type Int ΔNPyltRNA. One evolved variant, Int ΔNPyltRNA(A17), displayed a level of orthogonality towards both MmPylRS and AlvPylRS that was comparable to Int ΔNPyltRNA(VB03) and Int ΔNPyltRNA(VC10), identified from the variable loop library. All selected variants were orthogonal with respect to endogenous synthetases.
We postulated that combining the mutations isolated from each library might create a tRNA with the desired properties. We therefore transplanted the variable loop extensions from Int ΔNPyltRNA(VB03) and Int ΔNPyltRNA(VC10) into each variant identified from the acceptor stem library, and tested the resulting hybrid tRNAs for activity with 030PylRS, MmPylRS and AlvPylRS. We found that introduction of an extra nucleotide into the variable loop was very effective in removing any cross-reactivity towards MmPylRS, rendering all active hybrids highly orthogonal to MmPylRS (Supplementary Fig. 16, Supplementary Table 3, 8).
We identified four highly active hybrid Int ΔNPyltRNAs (Int ΔNPyltRNA(A5,VB03), Int ΔNPyltRNA(A5,VC10) Int ΔNPyltRNA(A6,VB03) and Int ΔNPyltRNA(A6,VC10)), which retained greater than 80% of wild type activity when paired with 030PylRS and were orthogonal with respect to MmPylRS. However, these four tRNAs still displayed activity with AlvPylRS. Another three tRNAs (Int ΔNPyltRNA(A13,VC10), Int ΔNPyltRNA(A17,VB03) and Int ΔNPyltRNA(A17,VC10)) retained over 50% of wild type activity with 030PylRS and were highly orthogonal with respect to both MmPylRS and AlvPylRS (Fig. 5b, Supplementary Fig. 17). All seven tRNAs were orthogonal with respect to E. coli synthetases, and provided promising candidates for our third tRNA as part of a triply orthogonal PylRS/PyltRNA pair.
Next we investigated the activity of the seven evolved Int ΔNPyltRNAs with the three additional class A synthetases (G1, H5 and 1R26) and two additional class B synthetases (RumEn and Lum1) that were not used for the selection or initial characterization. Overall we characterized 64 combinations of class A synthetases, class B synthetases or MmPylRS with Int ΔNPyltRNA and its selected variants (Fig 5b). From these experiments we identified numerous Int ΔNPyltRNA variants that are orthogonal to specific class A synthetases and MmPylRS while being highly active with a specific class B synthetase (Fig. 5b,c). For example Int ΔNPyltRNA(A6,VB03) is very active with Lum1PylRS (class B) but inactive with MmPylRS or G1PylRS (class A).
Triply orthogonal PylRS/PyltRNA pairs
By combining the observations from our measurements of cognate and non-cognate synthetase/tRNA pair activity (Fig. 1b, Fig. 3a, Fig. 4b and Fig. 5b) we identified 12 sets of triply orthogonal pairs (Fig. 5d, Supplementary Fig. 18). Each triplet is composed of MmPylRS and Spe PyltRNA, a specific class A DNPylRS and an evolved Alv DNPyltRNA, and a specific class B DNPylRS and an evolved Int DNPyltRNA variant.
Incorporating three ncAAs into a protein
The three pairs within a triply orthogonal pair each recognize the same amino acid and decode the amber codon. To use these pairs to encode three distinct ncAAs into a polypeptide, it was necessary to: (1) diverge the active sites of each synthetase to recognize distinct amino acids; and (2) diverge the anticodon of their cognate tRNAs to decode distinct blank codons. We focused on addressing these challenges with the MmPylRS/Spe PyltRNA, Lum1PylRS/Int DNPyltRNA and 1R26PylRS/Alv DNPyltRNA pairs, which are among the most active triply orthogonal pairs (Fig. 5d).
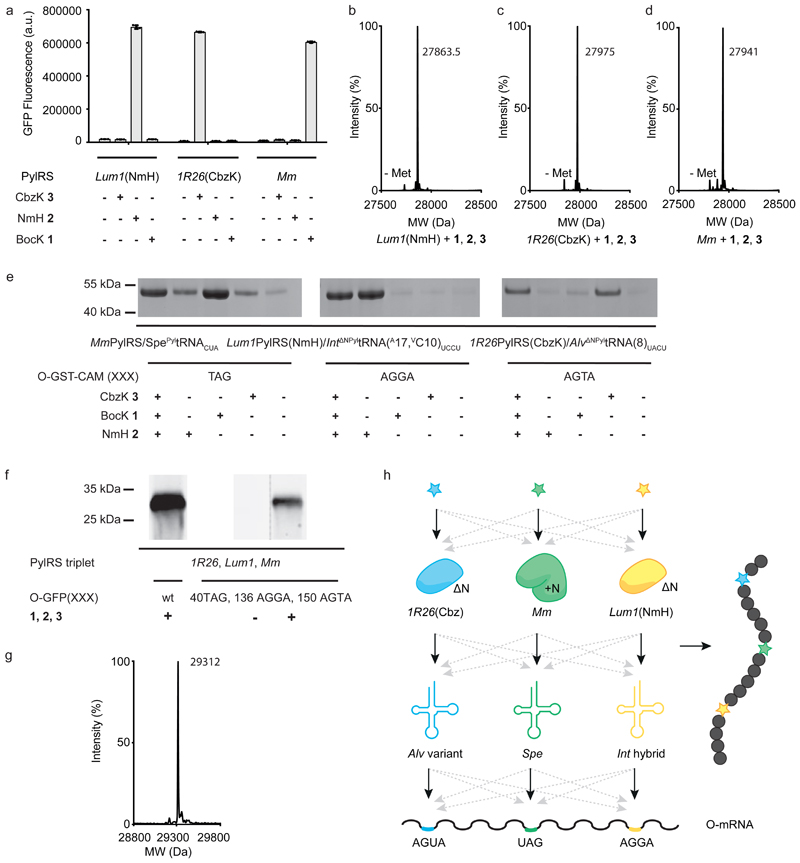
We demonstrated that the active sites of Lum1PylRS, 1R26PylRS and MmPylRS can be diverged to selectively recognize distinct ncAA substrates (Fig. 6a-d). As previously reported, wild type MmPylRS directs the incorporation of BocK 1, but not 3-methyl-lhistidine (NmH 2)32 or N ε -(carbobenzyloxy)-l-lysine (CbzK 3) (Fig. 6a,d) 36. Previous work has shown that five mutations (L121M, L125I, Y126F, M129A and V168F) convert MbPylRS into an aaRS for NmH 2, and that transplanting these mutations into AlvPylRS creates a mutant that directs the incorporation of NmH 2 and excludes BocK 1 32. We transplanted the same mutations into Lum1PylRS, creating Lum1PylRS(NmH), and demonstrated that this enzyme directs the incorporation of NmH 2, but not BocK 1; this shows that that the activity and specificity conferred by these mutations in AlvPylRS can be transplanted to Lum1PylRS. We also showed that Lum1PylRS(NmH) does not direct the incorporation of CbzK 3 (Fig. 6a,b). Finally, we identified a mutant of 1R26, 1R26PylRS(CbzK) that directs the incorporation of CbzK 3, but not NmH 2 or BocK 1 (Fig. 6a,c); the mutations conferring this specificity were discovered in a clone within a laboratory collection of MmPylRS mutants and then transferred to 1R26PylRS.
Figure 6. Diverging amino acid recognition and decoding of triply orthogonal PylRS/tRNA pairs enables the incorporation of three distinct ncAAs into a protein.
a, Each triply orthogonal PylRS or active site mutant, (MmPylRS, Lum1PylRS(NmH) and 1R26PylRS(Cbz)) leads to GFP synthesis from GFP(150TAG)His6 in the presence of its cognate ncAA (2mM), but not with the cognate ncAAs for the other triply orthogonal PylRS. Each bar represents the mean of three biological replicates ± s.d.. The individual data points are shown as dots. All numerical values are provided in Supplementary Table 3. b-d, In the presence of all three ncAAs each PylRS or active site variant selectively incorporates its cognate ncAA in response to the amber stop codon. TOF-MS of nickel-NTA-purified GFP expressed from GFP(150TAG)His6 in the presence of 8 mM BocK 1, 8 mM NmH 2 and 2 mM CbzK 3. (GFP(150BocK)His6 mass predicted: 27,941, mass found: 27,942; GFP(150NmH)His6 mass predicted: 27,864, mass found: 27,863.5; GFP(150CbzK)His6 mass predicted: 27,975, mass found: 27,975) e, Divergence of the triply orthogonal pair derivatives that recognize distinct ncAAs to decode distinct codons. GSTCAM was purified from E. coli containing ribo-Q1, O-GST-CAM(1XXX) (where XXX stands for either TAG, AGGA or AGTA) and the three indicated PylRS/tRNA derivatives. The concentrations of BocK and NmH were 8 mM, while the concentration of CbzK was 2mM. Samples were analysed by SDS–PAGE. This experiment was performed in 3 biological replicates with similar results. Full gels and biological triplicates are shown in Supplementary Fig. 19. f, Incorporation of three ncAAs into a single polypeptide. An anti-Strep western blot following Ni-NTA purification of strepGFPHis6 containing all three ncAAs from O-strepGFP(XX)His6, where XX stands for either wt or the triple mutant: 40TAG, 136AGGA and 150AGTA. E. coli also contained ribo-Q1, MmPylRS/SpePyltRNACUA, Lum1PylRS(NmH)/Int PyltRNA(A17,VC10)UCCU and 1R26PylRS(CbzK)/Alv PyltRNA(8)UACU. ncAAs were used at the following concentrations: 8 mM BocK, 8 mM NmH, 2 mM CbzK). This experiment was performed in three biological replicates with similar results. Full blots and biological triplicates are shown in Supplementary Fig. 20, GFP fluorescence measurements are shown in Supplementary Fig. 21. g TOF-MS of purified strepGFPHis6 purified from cells containing O-strepGFP(40TAG, 136AGGA, 150AGTA)-His6, O-ribo-Q1, MmPylRS/SpePyltRNACUA, Lum1PylRS(NMH)/Int PyltRNA(A17,VC10)UCCU and 1R26PylRS(CbzK)/Alv PyltRNA(8)UACU in presence of 8 mM BocK, 8 mM NmH and 2 mM CbzK. (StrepGFP(40BocK, 136NmH, 150Cbz)His6 mass predicted: 29,314.5, mass found: 29312). h Schematic showing the nine specific functions (three specific ncAA recognitions, three specific aaRS/tRNA recognitions and three specific codon-anticodon interactions) that have been created to generate active pairs (black arrows). The schematic also shows the 18 potential interactions that have been eliminated (dashed grey arrows) to ensure orthogonality between the three pairs. The large number of potential interactions – between the endogenous amino acids, synthetases and tRNAs and the engineered triply orthogonal pairs – that have been eliminated to ensure orthogonality are for clarity. mRNA, messenger RNA.
Next we created MmPylRS/Spe PyltRNACUA, Lum1PylRS(NmH)/Int DNPyltRNA(A17, VC10)UCCU and 1R26PylRS(CbzK)/Alv DNPyltRNA(8)UACU pairs; the synthetase of each pair selectively recognizes its cognate ncAA and the tRNA of each pair contains a CUA, UCCU or UACU anticodon that targets TAG, AGGA or AGTA codons, respectively. We coexpressed all three pairs in a single cell with ribo-Q1 (an orthogonal ribosome that facilitates the decoding of quadruplet codons and amber codons, on its cognate orthogonal message, by tRNAs bearing complementary anticodons5). Cells also contained an OGST(1XXX)CAM gene (glutathione S transferase (GST) linked to calmodulin (CAM) on an orthogonal ribosome binding site (O-rbs); XXX = TAG, AGGA, or AGTA) that was selectively translated by ribo-Q114. Read through of the TAG codon, as judged by production of full length GST-CAM, was observed upon addition of BocK 1 (Fig. 6e). Addition of CbzK 3 or NmH 2 led to low-level read through of the TAG codon; such background incorporation is typically outcompeted in the presence of the cognate pair and ncAA37. These observations are consistent with the MmPylRS/Spe PyltRNACUA pair selectively recognizing BocK 1 and decoding the amber codon. Similarly, we observed read through of the AGGA codon upon addition of NmH 2 but not BocK 1 or CbzK 3(Fig. 6e, Supplementary Fig. 19), and read through of the AGTA codon upon addition of CbzK 3 but not NmH 2 or BocK 1 (Fig. 6e). These observations are consistent with the expected specificity of the Lum1PylRS(NmH)/Int DNPyltRNA(A17, VC10)UCCU and 1R26PylRS(CbzK)/Alv DNPyltRNA(8)UACU pairs. Our data suggest that we have created derivatives of the triply orthogonal pairs that can be expressed in the same cell, recognize distinct ncAAs and decode distinct codons.
Finally, we demonstrated that our new pairs can be used to incorporate three distinct ncAAs into a single protein in response to three distinct codons (Fig. 6f,g,h, Supplementary Figs. 20 and 21). We co-expressed all three pairs and ribo-Q1 in cells also containing O-GFP (40TAG, 136AGGA and 150AGTA) in which translation of a GFP gene containing the target codons is driven from an O-rbs. Full length GFP was produced upon addition of CbzK 3, NmH 2 and BocK 1 to cells (Fig. 6f) and mass spectrometry confirmed the incorporation of all three ncAAs at genetically encoded positions in the protein (Fig. 6g, Supplementary Fig. 22).
Discussion
We have discovered and characterized several new DNPylRS/ΔNPyltRNA pairs, and thereby expanded the diversity of ΔNPylRS/ΔNPyltRNA pairs that are highly active and orthogonal in E. coli. Clustering analysis of our expanded set of DNPylRS and ΔNPyltRNA sequences revealed two distinct classes. These sequence-based classes correlated well with the classes that we independently determined on the basis of function. We demonstrated that class A synthetases function with class A ΔNPyltRNAs, but commonly function poorly with class B ΔNPyltRNAs, and that class B synthetases are naturally orthogonal towards class A ΔNPyltRNAs and function efficiently with class B ΔNPyltRNAs. By characterizing 88 DNPylRS/ΔNPyltRNA combinations we discovered 18 mutually orthogonal DNPylRS/ΔNPyltRNA pairs. These experiments reveal a remarkable divergence in sequence and function between DNPylRS/ΔNPyltRNA pairs.
We discovered an MmPylRS/Spe PyltRNA pair in which Spe PyltRNA is orthogonal to both class A and class B DNPylRSs, but functions efficiently with MmPylRS. We discovered variants of a class A ΔNPyltRNA that are orthogonal to MmPylRS and class B synthetases, but function efficiently with class A synthetases. Similarly, we evolved variants of a class B ΔNPyltRNA that are orthogonal to MmPylRS and certain class A synthetases, but function efficiently with certain class B synthetases. By analysing the specificity of synthetase/tRNA combinations from 248 measurements we identified 12 sets of triply orthogonal pairs: each triplet is composed of the MmPylRS/Spe PyltRNA pair, an evolved class A DNPylRS/ΔNPyltRNA pair and an evolved class B DNPylRS/ΔNPyltRNA pair. Remarkably, each set of three triply orthogonal pairs, in which each pair is orthogonal with respect to each other and to E. coli synthetases and tRNAs (MmPylRS/ Spe PyltRNA, an evolved class A DNPylRS/ΔNPyltRNA, and an evolved class B DNPylRS/ΔNPyltRNA), now equals the number of broadly useful orthogonal pairs in E. coli derived from all other aaRS/tRNA systems. We note that other orthogonal pairs with limited demonstrated utility for ncAA incorporation have also been reported38,39.
We have engineered the active site of the synthetases to generate mutants that recognize distinct ncAAs and exclude others. We have also engineered the pairs to decode distinct codons and demonstrated the incorporation of three distinct ncAAs into a single polypeptide. We note that we have previously reported a scalable strategy for discovering synthetase mutants that use a desired ncAA substrate but exclude other ncAAs40; extensions of this approach, combined with the advances reported herein, should enable an expansion in the diversity of ncAAs that can be incorporated into a single polypeptide. As the anticodon of the pyrrolysyl tRNAs can be altered without destroying aminoacylation by their cognate synthetases25,41(Fig. 6) it may be possible to reprogram the triply orthogonal pairs to incorporate ncAAs in response to other emerging blank codons3,4,42.
Our work enables the co-translational incorporation of three distinct ncAAs into a homogeneous and correctly terminated protein. We anticipate that future work will combine advances in expanding the number of codons available for assignment to non-canonical monomers3–5 with advances in expanding the chemical scope of the ribosome6–8 and the mutually orthogonal systems reported herein to realize the encoded cellular synthesis and evolution of non-canonical biopolymers.
Methods
Identification of ΔNPylRS sequences
We identified PylRS protein sequences homologous to the C-terminal region of MmPylRS or to Desulfitobacterium hafniense (Dh)PylSn by protein HMMER search34 against the UniProtKB database43 using MmPylRSd184 or DhPylSn as the query sequence, respectively, and filtering for expect values below 1 × 10-30. From the identified protein sequences, which contain homology to the C-terminal region of MmPylRS, we eliminated those for which sequence homology to DhPylSn could be found within the same genome. From the remaining protein sequences, which correspond to ΔNPylRSs, we identified those that had not been previously reported.
Identification of PyltRNA sequences
Using the National Center for Biotechnology Information Nucleotide Database we used existing genome annotations to identify the DNA sequence for each PylRS gene within its host genome, and were thus able to identify the genomic region corresponding to the pyrrolysine gene cluster. In the sequences 40 kilobases upstream and downstream of the PylRS gene, ΔNPyltRNA sequences were manually identified by searching for sequence similarity to known ΔNPyltRNA sequences; +NPyltRNA sequences were manually identified by searching for sequence similarity to Mm PyltRNA. tRNA secondary structure prediction was initially performed using RNAstructure 44 and manually curated by inspection and comparison to Mm PyltRNA.
DNA constructs
PylRS and PyltRNA genes were synthesiszd by IDT as gBlock double-stranded DNA fragments. We cloned the genes into pKW vectors by Gibson assembly. PylRS was expressed from a glnS promoter and PyltRNA was expressed from an lpp promoter. PylRS genes were coded for expression in E. coli using the IDT Codon Optimization Tool. We appended the gene for MmPylRS with a sequence encoding a C-terminal Ser(Gly4Ser)4FLAG-tag, while all other PylRS genes were appended with a sequence encoding a C-terminal Ser(Gly4Ser)4His6SerGlyStrep-tag II. We used these plasmids together with pBAD GFP(150TAG)His6 (in which sfGFP containing an amber stop codon at position 150 and a C-terminal His6 tag is expressed from the arabinose promoter of pBAD; GFP refers to sfGFP throughout). We used Gibson cloning to insert each PylRS cassette under constitutive expression from the glnS promoter into pBAD CAT(111TAG) GFP(150TAG)His6 vectors, in which a chloramphenicol acetyl transferase gene containing an amber stop codon at position 111 is under constitutive expression.
To create the plasmid pKW1-Triple for triple ncAA incorporation, PylRS genes were designed to be expressed as a single polycistronic messenger RNA transcript under the control of the glnS promoter, with RBS binding strengths of approximately 10,000 RBS units rationally designed using the RBS Calculator (https://www.denovodna.com/software/)45–49 specifying E. coli K-12 as the host organism. tRNA genes were designed to be expressed as a single polycistronic messengerRNA transcript under the control of the lpp promoter. Sequences between tRNA sequences were designed by manual examination of the E. coli K-12 MG1655 genome using EcoCyc50 and identifying spacer sequences between tRNAs from the same isoacceptor class that are expressed as adjacent tRNA sequences in the same operon. Spacer sequences originating between AlaX and AlaW, and ValU and ValX genes were selected for use. Cassettes containing PylRS and PyltRNA genes were synthesized by IDT as gBlock double-stranded DNA fragments. We cloned the genes into pKW vectors by Gibson assembly.
Library generation
Libraries of Int ΔNPyltRNA with randomized variable loop or acceptor stem sequences were constructed by Golden Gate cloning from a pKW Int ΔNPyltRNA vector using PCR primers listed in Supplementary Table 11 together with restriction enzyme BbsI and T4 DNA ligase. We transformed each library separately into competent E. coli DH10B cells to give library diversities of more than 1 × 108, exceeding the theoretical diversity of 6 × 107 required for complete library coverage.
Selection to identify orthogonal Class A ΔNPyltRNAs
For the variable loop library, we transformed each Int ΔNPyltRNA variable loop library into competent E. coli DH10B cells bearing pBAD IntPylRS CAT(111TAG) GFP(150TAG)His6. We recovered the transformed cells for 1 h at 37°C in 0.5 ml SOB medium supplemented with 8 mM BocK 1. The transformation was plated on LB agar containing 37.5 µg ml-1 spectinomycin, 12.5 µg ml-1 tetracycline and 100 µg ml-1 chloramphenicol. The plates were incubated at 37 °C for 40 h. After incubation, colonies on the plates were washed off and collected in 2XTY buffer and the plasmids were extracted using a DNA midiprep kit (Qiagen). To remove the pBAD IntPylRS CAT(111TAG) GFP(150TAG)His6 plasmid, the extracted DNA was digested with both NcoI restriction endonuclease and T5 exonuclease and re-purified using a PCR purification column. The remaining pKW plasmids were transformed into competent E. coli DH10B cells bearing either pBAD AlvPylRS CAT(111TAG) GFP(150TAG)His6 or pBAD MmPylRS CAT(111TAG) GFP(150TAG)His6. The transformed cells were recovered for 1 h at 37 °C in 0.5 ml SOB medium. The transformation was plated on LB agar containing 37.5 µg ml-1 spectinomycin and 12.5 µg ml-1 tetracycline. The plates were incubated at 37 °C for 20 h. For each library, 1,528 colonies were picked from the plates using a QPix 420 Colony Picking System and inoculated into 190 µl 2XTY-STA (2XTY medium with 75 µg/ml spectinomycin, 25 µg/ml tetracycline and 0.5% l-arabinose) in 96-well microtitre plate format supplemented with 8 mM BocK 1. The plates were incubated at 37°C and 220 r.p.m., and OD600 and GFP fluorescence (λex 485 nm, λem 520 nm) measurements were recorded after 20 h using a SpectraMax i3. Cells from wells with the lowest GFP OD600 -1 ratios were used to inoculate 2XTY medium with 75 µg ml-1 spectinomycin, and the pKW plasmids containing Int ΔNPyltRNA variants were extracted by DNA miniprep and then sequenced. Each hit corresponding to a distinct Int ΔNPyltRNA sequence was cloned into a pKW IntPylRS vector, a pKW AlvPylRS vector, and a pKW MmPylRS vector and rephenotyped with pBAD GFP(150TAG)His6.
For the acceptor stem library, we transformed each Int ΔNPyltRNA acceptor stem library into competent E. coli DH10B cells bearing pBAD IntPylRS CAT(111TAG) GFP(150TAG)His6. We recovered the transformed cells for 1 h at 37°C in 5 ml super optimal broth with catabolite repression (SOC) medium supplemented with 8 mM BocK 1. The transformation was plated on LB agar containing 75 µg ml-1 spectinomycin, 25 µg ml-1 tetracycline and 100 µg ml-1 chloramphenicol. The plates were incubated at 37 °C for 24 h. After incubation, 192 colonies were picked into 1.7 ml 2XTY-STA in a 96-well microtitre plate format supplemented with 8 mM BocK 1 and grown overnight. Plasmids from all fluorescent cultures were extracted by DNA miniprep (Qiagen) and the extracted DNA was digested with both NcoI restriction endonuclease and T5 exonuclease. 1 µl of the digestion products was transformed into chemically competent E. coli DH10B cells bearing either pBAD AlvPylRS CAT(111TAG) GFP(150TAG)His6 or pBAD MmPylRS CAT(111TAG) GFP(150TAG)His6 by heat shock. The transformed cells were recovered for 1 h at 37 °C in 180 µl ml SOC medium, and 10 µl was used to inoculate 180 µl 2XTYSTA in a 96-well microtitre plate format supplemented with 8 mM BocK 1 and grown overnight. Cells from wells with the lowest GFP OD600 ratios were used to inoculate 2XTY medium with 75 µg ml-1 spectinomycin, and the pKW plasmids containing Int ΔNPyltRNA variants were extracted using a DNA miniprep kit (Qiagen) and then sequenced. Each hit corresponding to a distinct Int ΔNPyltRNA sequence was cloned into a pKW IntPylRS vector, a pKW AlvPylRS vector, and a pKW MmPylRS vector and re-phenotyped with pBAD GFP(150TAG)His6.
Measuring the activity and specificity of PylRS/PyltRNACUA pairs with synthetase and tRNA expressed from different plasmids
To measure the activity and specificity of cognate and non-cognate PylRS/PyltRNA combinations, we transformed 0.4 µl pKW PyltRNA plasmids into 8 µl chemically competent E. coli DH10B cells bearing either pBAD GFP(150TAG)His6 or pBAD PylRS GFP(150TAG)His6. We recovered the transformed cells for 1 h at 37 °C and 750 r.p.m. in 180 µl SOC medium in 96-well microtitre plate format. 10 µl of the transformed cells was used to inoculate 180 µl 2XTY-STA in 96-well microtitre plate format, supplemented with or without 8 mM BocK 1. OD600 and GFP fluorescence (λex 485 nm, λem 520 nm) measurements were recorded after 22-28 h incubation at 37 °C and 700 r.p.m. using a Tecan Infinite M200 Pro.
Measuring the activity and specificity of PylRS/PyltRNACUA pairs with synthetase and tRNA expressed from the same plasmid
The same procedure was followed as described above. However, for this expression system both PylRS and PyltRNA were encoded on the same pKW plasmid which was transformed into chemically competent E. coli DH10B cells bearing pBAD GFP(150TAG)His6. For GFP expression, 25 µl transformed cells was inoculated into 500 µl 2XTY-STA in 96-well microtitre plate format, in the presence or absence of 8 mM BocK 1, or 2 mM CbzK 3 or 8 mM NmH 2. Cells were grown for 22-28 h at 750 r.p.m. and 37 °C before 180 µl of each well was transferred to a 96 well plate and GFP fluorescence and OD600 measurements were recorded as described above.
GFP(TAG)His6 expression for mass spectrometry
To express GFP incorporating BocK for mass spectrometry analysis we transformed pKW PylRS/PyltRNA plasmids into competent E. coli DH10B cells bearing pBAD GFP(150TAG)His6. We recovered the transformed cells for 1 h at 37°C in 1 ml SOC medium. The transformation was used to inoculate 20 ml 2XTY-STA supplemented with 8 mM BocK 1 and incubated overnight at 37°C and 220 r.p.m. for 20 h.
The 20 ml culture was pelleted by centrifugation and washed with 2 ml PBS. The cell pellets were resuspended in 1 ml lysis buffer (1X BugBuster Protein Extraction Reagent supplemented with 1X cOmplete protease inhibitor cocktail, 1 mg ml-1 lysozyme and 1 mg ml-1 DNase I) and lysed for 1 h at 25°C with head-over-tail circular rotation. The lysate was clarified by centrifugation (21,000 g; 30 min; 4 °C). GFP was purified by its C-terminal His6 tag using 75 µl Ni-NTA agarose beads and left to bind for 30 min at room temperature. The beads were washed five times with 1 ml PBS supplemented with 10 mM imidazole and eluted in 40 µl PBS supplemented with 250 mM imidazole.
The same procedure was used to assess the active site orthogonality of 1R26PylRS(Cbz)/MaPyltRNA(11)CUA, Lum1PylRS(NmH)/IntPyltRNA(A13,VC10)CUA or MmPylRS/SpePyltRNACUA but all three amino acids were added to the medium (8mM BocK 1, 2 mM CbzK 3, 8 mM NmH 2) simultaneously. The eluted fraction was diluted and analysed by TOF-MS.
Expression and purification of proteins produced from O-GST-CAM(1XXX)His6
To express proteins from O-GST-CaM(1XXX) we co-transformed competent E. coli DH10B cells with pO-GST-CaM(1XXX) (X=TAG, AGGA or AGTA), pKW-Triple MmPylRS/Spe PyltRNACUA, Lum1PylRS(NmH)/Int ΔNPyltRNA(A17,VC10)UCCU , 1R26PylRS(CbzK)/Alv ΔNPyltRNA(8)UACU and pRSF ribo-Q. We recovered the transformed cells for 1 h at 37 °C in 1 ml SOC medium. The transformation was used to inoculate 5 ml 2XTY-KST (2XTY medium with 25 µg ml-1 kanamycin, 75 µg ml-1 spectinomycin, and 12.5 µg ml-1 tetracycline) and incubated overnight (37 °C; 16 h; 220 r.p.m.). 50 µl of the overnight culture was diluted in 5 ml 2XTY-KST containing a combination of the indicated ncAAs (8 mM BocK 1, 8 mM NmH 2 and 2 mM CbzK 3) or none of them and incubated at 37 °C and 220 r.p.m.. At OD600 0.6, 50 µl 1M IPTG was added to a final concentration of 1 mM. After 16 h incubation at 37°C, 220 r.p.m., the cultures were pelleted and washed with 800 µl PBS. The cell pellets were resuspended in 1 ml lysis buffer (1X BugBuster Protein Extraction Reagent supplemented with 1X cOmplete protease inhibitor cocktail) and lysed for 1 h at 25 °C with head over tail rotation. The lysate was clarified by centrifugation (21,000 g; 30 min; 4 °C). GST-containing proteins from the lysate supernatant were left to bind to 60 µl glutathione sepharose beads for 1 h at 25 °C. The beads were washed five times with 800 µl PBS before eluting in 60 µl 20 mM reduced glutathione in PBS at pH 8. Samples were analysed on 4-12% Bis-Tris SDS-PAGE gels, visualized with InstantBlue Coomassie stain and imaged using a ChemiDoc Touch Imaging System.
Expression of GFP containing three ncAAs
To express GFP containing three ncAAs we co-transformed competent E. coli DH10B cells with pO-StrepGFP(40TAG, 136AGGA, 150AGTA)His6, pKW-Triple MmPylRS/Spe PyltRNACUA, Lum1PylRS(NmH)/Int ΔNPyltRNA(A17,VC10)UCCU , 1R26PylRS(CbzK)/Alv ΔNPyltRNA(8)UACU and pRSF ribo-Q1. We recovered the transformed cells for 1 h at 37 °C in 1 ml SOC medium. The transformation was used to inoculate 20 ml 2XTY-KST and incubated overnight (37 °C; 16 h; 220 r.p.m.). 1 ml of the overnight culture was diluted in 50 ml 2XTY-KST containing a combination of the indicated ncAAs (8 mM BocK 1, 8 mM NmH 2 and 2 mM CbzK 3) or none of them and incubated at 37 °C and 220 r.p.m.. At OD600 0.6, 500 µl 1M IPTG was added to a final concentration of 1 mM. After 16 h incubation at 37 °C and 220 r.p.m., the cultures were pelleted and washed with 5 ml PBS. The cell pellets were resuspended in 5 ml lysis buffer (1X BugBuster Protein Extraction Reagent supplemented with 1X cOmplete protease inhibitor cocktail) and lysed for 1 h at 25 °C with head over tail rotation. The lysate was clarified by centrifugation (21,000 g; 30 min; 4 °C). GFP-containing proteins from the lysate supernatant were left to bind to 80 µl Ni-NTA beads for 1 h at 25 °C. The beads were washed five times with 800 µl PBS containing 25 mM imidazole before eluting in 80 µl PBS containing 250 mM imidazole. Samples were analysed by western blot using 4-12% Bis-Tris SDS-PAGE gels, primary antibody rabbit anti-Strep ab76949 (Abcam) and secondary antibody goat anti-rabbit IRDye 800CW (LI-COR).
To obtain GFP for mass spectrometry a 400 ml expression was run. The protein was purified on 250 µl Ni-NTA beads. The purified protein was incubated overnight with 150 µl Strep-Tactin sepharose beads. Beads were washed five times with 500 µl PBS at pH 8 and eluted six times in 75 µl 20 mM desthiobiotin pH 8. Fractions were combined, concentrated and analysed by TOF-MS.
Electrospray ionization mass spectrometry
Denatured protein samples (~10 µM) were subjected to liquid chromatography-mass spectrometry analysis. Briefly, proteins were separated on a BEH C4 UPLC (1.7µm; 1.0 x 100mm; Watewrs) column using a modified nanoAcquity (Waters) to deliver a flow of approximately 50 µl min-1. The column was developed over 20 min with a gradient of acetonitrile (2% vol/vol to 80% vol/vol) in 0.1% vol/volformic acid. The analytical column outlet was directly interfaced via an electrospray ionization source, with a hybrid quadrupole time-of-flight mass spectrometer (Xevo G2, Waters, UK). Data were acquired over an m/z range of 300–2000, in positive ion mode with a cone voltage of 30 V. Scans were summed together manually and deconvoluted using MaxEnt1 (Masslynx; Waters). The theoretical molecular weights of proteins with ncAAs were calculated by first computing the theoretical molecular weight of wild-type protein using an online tool (http://web.expasy.org/protparam/) and then manually correcting for the theoretical molecular weight of ncAAs.
Tandem MS/MS analysis
Proteins were run on 4-12% NuPAGE Bis-Tris gel (Invitrogen) with MES buffer and briefly stained using InstantBlue (Expedeon). The bands were excised and stored in water. Tryptic digestion and tandem MS/MS analyses were performed by K. Heesom (Proteomics Facility, University of Bristol) and, separately, by M. Skehel (Biological Mass Spectrometry and Proteomics Laboratory, Medical Research Council Laboratory of Molecular Biology).
Supplementary Material
Acknowledgements
This work was supported by the Medical Research Council (MRC), UK (MC_U105181009 and MC_UP_A024_1008) and an ERC Advanced Grant SGCR, all to J.W.C. D.L.D was supported by the Boehringer Ingelheim Fonds. We thank Mark Skehel and the MRC-LMB mass spectrometry facility and Kate Heesom at the proteomics facility of the University of Bristol for performing mass spectrometry.
Footnotes
Author contributions D.L.D., J.C.W.W, and J.W.C. designed the project. D.L.D., J.C.W.W, performed experiments. A.T.B. analysed and interpreted MS/MS data. D.L.D., J.C.W.W, and J.W.C wrote the paper with input from A.T.B..
Competing interests: The authors declare no competing interests.
Data availability
Source data for the graphs and heatmaps (for Figs. 1–6, Supplementary Figs. 5-7, 10-18, 21) are provided in Supplementary Table 3. Source data for the gels in Fig. 6 are provided with the paper. All other datasets and material generated or analysed in this study are available from the corresponding author upon reasonable request.
References
- 1.Chin JW. Expanding and reprogramming the genetic code. Nature. 2017;550:53–60. doi: 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- 2.Chin JW. Reprogramming the Genetic Code. Science. 2012;336:428–429. doi: 10.1126/science.1221761. [DOI] [PubMed] [Google Scholar]
- 3.Fredens J, et al. Total synthesis of Escherichia coli with a recoded genome. Nature. 2019;569:514–518. doi: 10.1038/s41586-019-1192-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, et al. A semi-synthetic organism that stores and retrieves increased genetic information. Nature. 2017;551:644–647. doi: 10.1038/nature24659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 6.Schmied WH, et al. Controlling orthogonal ribosome subunit interactions enables evolution of new function. Nature. 2018;564:444–448. doi: 10.1038/s41586-018-0773-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czekster CM, Robertson WE, Walker AS, Söll D, Schepartz A. In Vivo Biosynthesis of a β-Amino Acid-Containing Protein. J Am Chem Soc. 2016;138:5194–5197. doi: 10.1021/jacs.6b01023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dedkova LM, Fahmi NE, Serguei Y, Golovine A, Hecht* SM. Construction of Modified Ribosomes for Incorporation of d-Amino Acids into Proteins. J Am Chem Soc. 2006;45:15541–15551. doi: 10.1021/bi060986a. [DOI] [PubMed] [Google Scholar]
- 9.Neumann H, Slusarczyk AL, Chin JW. De Novo Generation of Mutually Orthogonal Aminoacyl-tRNA Synthetase/tRNA Pairs. J Am Chem Soc. 2010;132:2142–2144. doi: 10.1021/ja9068722. [DOI] [PubMed] [Google Scholar]
- 10.Willis JCW, Chin JW. Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nat Chem. 2018;10:831–837. doi: 10.1038/s41557-018-0052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee A, Xiao H, Schultz PG. Evolution of multiple, mutually orthogonal prolyl-tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli. Proc Natl Acad Sci U S A. 2012;109:14841–14846. doi: 10.1073/pnas.1212454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Italia JS, et al. Mutually Orthogonal Nonsense-Suppression Systems and Conjugation Chemistries for Precise Protein Labeling at up to Three Distinct Sites. J Am Chem Soc. 2019;141:6204–6212. doi: 10.1021/jacs.8b12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterjee A, Sun SB, Furman JL, Xiao H, Schultz PG. A Versatile Platform for Single- and Multiple-Unnatural Amino Acid Mutagenesis in Escherichia coli. Biochemistry. 2013;52:1828–1837. doi: 10.1021/bi4000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K, et al. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat Chem. 2014;6:393–403. doi: 10.1038/nchem.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srinivasan G, James CM, Krzycki JA. Pyrrolysine Encoded by UAG in Archaea: Charging of a UAG-Decoding Specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 16.Krzycki JA. The direct genetic encoding of pyrrolysine. Curr Opin Microbiol. 2005;8:706–712. doi: 10.1016/j.mib.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding Nε acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Brock A, Herberich B, Schultz PG. Expanding the Genetic Code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 19.Borrel G, et al. Unique Characteristics of the Pyrrolysine System in the 7th Order of Methanogens: Implications for the Evolution of a Genetic Code Expansion Cassette. Archaea. 2014;2014 doi: 10.1155/2014/374146. 374146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park H-S, et al. Expanding the Genetic Code of Escherichia coli with Phosphoserine. Science. 2011;333:1151–1154. doi: 10.1126/science.1207203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogerson DT, et al. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog. Nat Chem Biol. 2015;11:496–503. doi: 10.1038/nchembio.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes RA, Ellington AD. Rational design of an orthogonal tryptophanyl nonsense suppressor tRNA. Nucleic Acids Res. 2010;38:6813–6830. doi: 10.1093/nar/gkq521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatterjee A, Xiao H, Yang P-Y, Soundararajan G, Schultz PG. A Tryptophanyl-tRNA Synthetase/tRNA Pair for Unnatural Amino Acid Mutagenesis in E. coli. Angew Chem Int Ed. 2013;52:5106–5109. doi: 10.1002/anie.201301094. [DOI] [PubMed] [Google Scholar]
- 24.Chin JW. Expanding and Reprogramming the Genetic Code of Cells and Animals. Annu Rev Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 25.Elliott TS, et al. Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat Biotechnol. 2014;32:465–472. doi: 10.1038/nbt.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nozawa K, et al. Pyrrolysyl-tRNA synthetase-tRNAPyl structure reveals the molecular basis of orthogonality. Nature. 2008;457:1163–1167. doi: 10.1038/nature07611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki T, et al. Crystal structures reveal an elusive functional domain of pyrrolysyl-tRNA synthetase. Nat Chem Biol. 2017;13:1261–1266. doi: 10.1038/nchembio.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herring S, et al. The amino-terminal domain of pyrrolysyl-tRNA synthetase is dispensable in vitro but required for in vivo activity. FEBS Lett. 2007;581:3197–3203. doi: 10.1016/j.febslet.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang R, Krzycki JA. PylSn and the homologous N-terminal domain of pyrrolysyl-tRNA synthetase bind the tRNA that is essential for the genetic encoding of pyrrolysine. J Biol Chem. 2012;287:32738–32746. doi: 10.1074/jbc.M112.396754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borrel G, et al. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics. 2014;15:679. doi: 10.1186/1471-2164-15-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meineke B, Heimgärtner J, Lafranchi L, Elsässer SJ. Methanomethylophilus alvus Mx1201 Provides Basis for Mutual Orthogonal Pyrrolysyl tRNA/Aminoacyl-tRNA Synthetase Pairs in Mammalian Cells. ACS Chem Biol. 2018;13:3087–3096. doi: 10.1021/acschembio.8b00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beranek V, Willis JCW, Chin JW. An Evolved Methanomethylophilus alvus Pyrrolysyl-tRNA Synthetase/tRNA Pair Is Highly Active and Orthogonal in Mammalian Cells. Biochemistry. 2019;58:387–390. doi: 10.1021/acs.biochem.8b00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Potter SC, et al. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46:W200–W204. doi: 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konevega AL, et al. Purine bases at position 37 of tRNA stabilize codonanticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA. 2004;10:90–101. doi: 10.1261/rna.5142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanagisawa T, et al. Multistep Engineering of Pyrrolysyl-tRNA Synthetase to Genetically Encode Nϵ-(o-Azidobenzyloxycarbonyl) lysine for Site-Specific Protein Modification. Chemistry & Biology. 2008;15:1187–1197. doi: 10.1016/j.chembiol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Wang K, Neumann H, Peak-Chew SY, Chin JW. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat Biotechnol. 2007;25:770–777. doi: 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]
- 38.Ikeda-Boku A, et al. A simple system for expression of proteins containing 3-azidotyrosine at a pre-determined site in Escherichia coli. J Biochem. 2013;153:317–326. doi: 10.1093/jb/mvs153. [DOI] [PubMed] [Google Scholar]
- 39.Anderson JC, et al. An expanded genetic code with a functional quadruplet codon. Proc Natl Acad Sci U S A. 2004;101:7566–7571. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang MS, et al. Biosynthesis and genetic encoding of phosphothreonine through parallel selection and deep sequencing. Nat Methods. 2017;14:729–736. doi: 10.1038/nmeth.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krogager TP, et al. Labeling and identifying cell-specific proteomes in the mouse brain. Nat Biotechnol. 2017;36:156–159. doi: 10.1038/nbt.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang K, et al. Defining synonymous codon compression schemes by genome recoding. Nature. 2016;539:59–64. doi: 10.1038/nature20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Consortium T. U. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2018;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bellaousov S, Reuter JS, Seetin MG, Mathews DH. RNAstructure: web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013;41:W471–W474. doi: 10.1093/nar/gkt290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salis HM. Chapter two - The Ribosome Binding Site Calculator. Methods Enzymol. 2011;498:19–42. doi: 10.1016/b978-0-12-385120-8.00002-4. [DOI] [PubMed] [Google Scholar]
- 47.Espah Borujeni A, Channarasappa AS, Salis HM. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 2013;42:2646–2659. doi: 10.1093/nar/gkt1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Espah Borujeni A, Salis HM. Translation Initiation is Controlled by RNA Folding Kinetics via a Ribosome Drafting Mechanism. J Am Chem Soc. 2016;138:7016–7023. doi: 10.1021/jacs.6b01453. [DOI] [PubMed] [Google Scholar]
- 49.Espah Borujeni A, et al. Precise quantification of translation inhibition by mRNA structures that overlap with the ribosomal footprint in N-terminal coding sequences. Nucleic Acids Res. 2017;45:5437–5448. doi: 10.1093/nar/gkx061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keseler IM, et al. The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2016;45:D543–D550. doi: 10.1093/nar/gkw1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for the graphs and heatmaps (for Figs. 1–6, Supplementary Figs. 5-7, 10-18, 21) are provided in Supplementary Table 3. Source data for the gels in Fig. 6 are provided with the paper. All other datasets and material generated or analysed in this study are available from the corresponding author upon reasonable request.