Abstract
Interactions between RNA-binding proteins (RBPs) and RNAs are critical for cell biology. However, methods to comprehensively and quantitatively assess these interactions within cells were lacking. RNA interactome capture (RIC) employs in vivo UV-crosslinking, oligo(dT) capture and proteomics to identify RNA-binding proteomes. Recent advances have empowered RIC to quantify RBP responses to biological cues such as metabolic imbalance or virus infection. Enhanced (e)RIC exploits the stronger binding of locked nucleic acid (LNA)-containing oligo(dT) probes to poly(A) tails to maximize RNA capture selectivity and efficiency, profoundly improving signal-to-noise ratios. The subsequent analytical use of SILAC and TMT proteomic approaches, together with high-sensitivity sample preparation and tailored statistical data analysis, significantly improves RIC's quantitative accuracy and reproducibility. This optimized approach is an extension of the original RIC protocol. It takes three days plus two weeks for proteomics and data analysis, and will enable the study of RBP dynamics under different physiological and pathological conditions.
Introduction
Development of the protocol
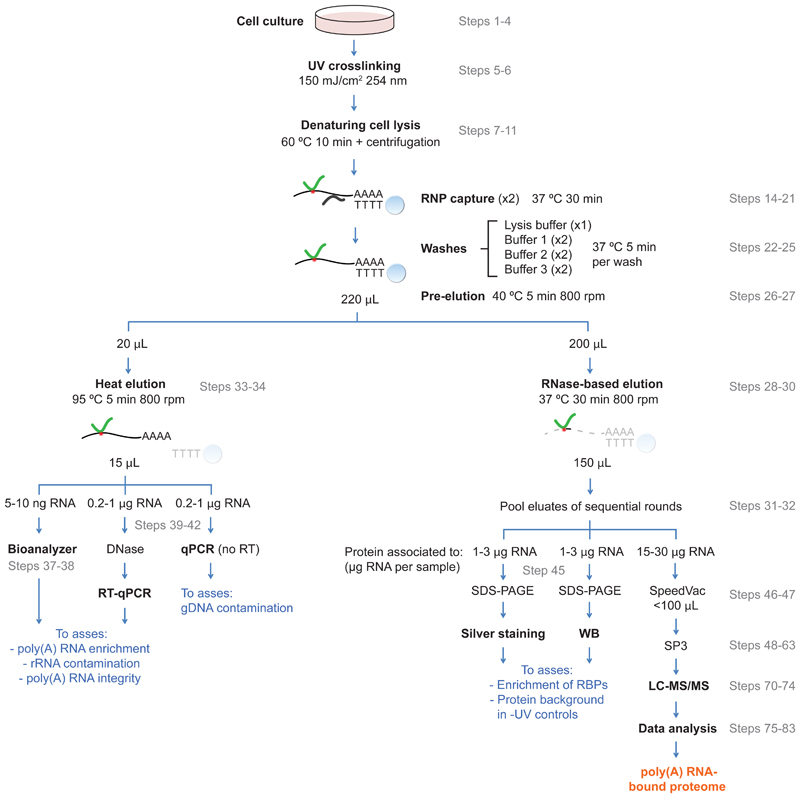
RIC employs irradiation of cultured cells with UV light to trigger crosslinks between protein and RNA interacting at 'zero distance'. This is followed by cell lysis under denaturing conditions, specific isolation of polyadenylated (poly(A)) RNA and its covalently linked proteins using oligo(dT) magnetic beads and stringent washes and proteomic analysis1–3 (Fig. 1). While effective to identify RBPs in multiple cell types1,2,4–7 and organisms8–13, RIC is not readily applicable to comparative analyses aiming to assess the responses of RBPs to physiological and pathological cues. In particular, the original protocol requires a substantial amount of starting material and lacks a specialized proteomics approach and tailored data analysis3. In the last years, several key improvements have empowered RIC to perform comparative analysis efficiently14,15. One of these key advances is the use of an oligo (dT) probe that contains locked nucleic acids (LNAs)14. LNAs are nucleic acid analogues that bear a methylene bridge between the 2'-O and 4'-C atoms of the ribose ring. This modification “locks” oligonucleotides in the optimal conformation for base pairing with complementary strands, leading to a profound increase in the thermal stability of the nucleic acid duplex. By adding LNAs to the probe, it is possible to increase the stringency of the capture and washes, which profoundly depletes the sample of abundant non-poly(A) nucleic acids, such as rRNAs, as well as potential DNA contamination14,16. We describe here this improved variant of RIC that we refer to as enhanced RNA interactome capture (eRIC).
Figure 1. Schematic representation of eRIC.
Cultured cells are exposed to UV light to generate covalent bonds (red dots) between RNA and proteins (green lines) bound at 'zero distance'. Cells are then lysed under denaturing conditions and poly(A) RNAs with their associated proteins are captured using oligo(dT) probes modified with LNAs and coupled to magnetic beads. Extensive washes and a pre-elution in pure water are applied to eliminate contaminant proteins (black lines), as well as contaminating RNA and gDNA. After the pre-elution, the bead suspension containing the captured material is split into two aliquots, which are subjected to either heat or RNase-mediated elution. Heat- and RNase-eluted samples are used for RNA/DNA and protein analyses, respectively.
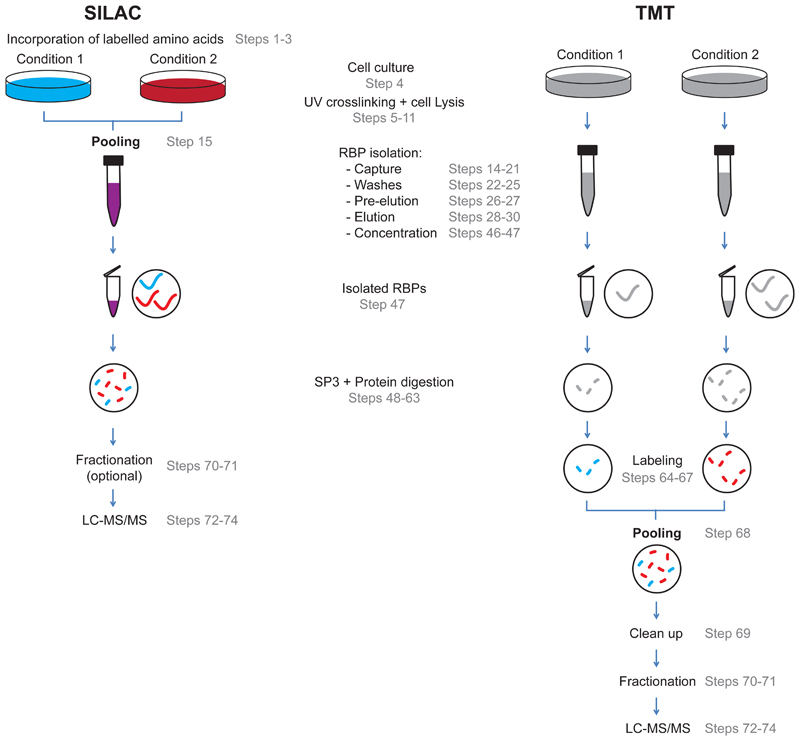
To increase the quantitative power or RIC, we have successfully applied two different proteomic strategies that have already shown their efficacy in proof-of-principle experiments14,15. The first approach exploits the capacity of stable isotope labelling with amino acids in cell culture (SILAC) to reduce technical noise by combining the samples after cell lysis (Fig.2). By mixing the lysates before the oligo(dT) capture, the isolation of poly(A) RNA and the downstream sample preparation for mass spectrometry becomes equally efficient for all the samples15. This, together with the high quantitative power of SILAC17, allows the discovery of even subtle changes in RBP activity15. SILAC allows to parallelize the analysis of up to three samples simultaneously, reducing mass spectrometry run time and improving cross-comparison accuracy when compared to label-free applications. While SILAC has been used in a broad range of cell lines and model systems, it cannot be easily applied to multicellular organisms or to cell types that do not tolerate SILAC reagents or that can only be cultured for a limited time. In such scenarios, it is recommended to employ post-elution peptide labelling techniques, such as isobaric labeling with tandem mass tag (TMT) (Fig.2). TMT labelling has been successfully used in RIC experiments applied to cultured cells and fruit fly embryos10,14, and can virtually be extended to any biological system. Isobaric labelling reagents allow higher level multiplexing with TMT enabling the analysis of up to sixteen samples in one mass spectrometry run. However, the RIC protocol is performed separately for each sample (Fig.2), potentially increasing technical noise. It is also recommended to perform sample fractionation and increase mass spectrometry analysis time to offset the reduction of protein identification rate and maximize proteome coverage. The original RIC protocol3 required a substantial amount of starting material, which is not feasible to obtain in many biological models. To reduce the amount of input material, we have combined eRIC with a recently-developed sample preparation approach, called 'Single-pot solid-phase-enhanced sample preparation' (SP3), that minimizes peptide loss18,19. As a reference, the eRIC workflow combined with SP3 reduces the amount of starting material by ~10 fold, broadening its applicability.
Figure 2. Overview of the proteomic workflow.
Schematic representation of SILAC- (left) and TMT-based (right) comparative eRIC experiments. In the SILAC approach, proteins are labelled 'in cellulo' by incubation of cells with media containing isotopically distinct amino acids. Samples are mixed immediately after cell lysis, so eRIC and downstream sample preparation for proteomics is performed with the combined samples. In the TMT approach, peptide labelling is performed after eRIC elution. Hence, RBP isolation and sample preparation for proteomics are performed individually for each experimental condition.
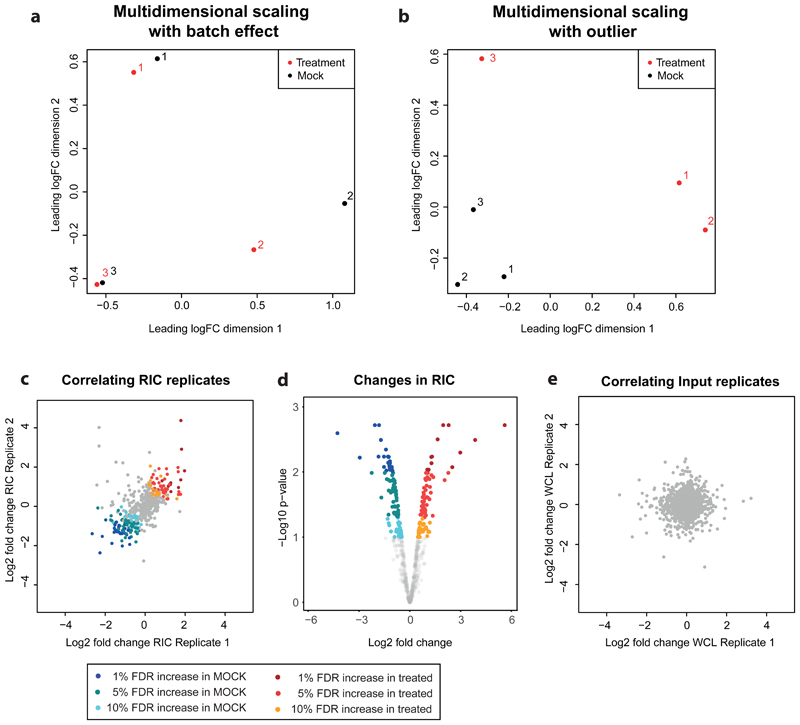
Moreover, the computational workflow used in the past was conceived to compare a UV-irradiated sample versus a non-irradiated control. This comparison yields relatively high fold changes, because the control is largely devoid of proteins. By contrast, the amplitude of the changes in comparisons of two UV-crosslinked samples that have been subjected to different treatments (e.g. uninfected versus infected cells) is expected to be lower, requiring high replicability to gain statistical power. We have implemented an analytical workflow that compiles the ratios between conditions, applies batch effect correction, and uses moderated statistical analysis adjusted for false discovery rate. In addition, our approach employs a complementary semiquantitative analysis to allow the study of proteins with 'zero intensity' values in one or more conditions. These results are problematic, as 'zero' values generate 'zero' or 'infinite' ratios that cannot be, in principle, analyzed statistically10,14,15. This complementary semiquantitative analysis is important because it allows to study RBPs that switch from a dormant to an active RNA-binding state (off/on) and vice versa (on/off). The importance of RBPs with these extreme behaviors is illustrated by IFI16 and IFIT5, whose RNA-binding activities are triggered upon Sindbis virus infection and are indeed critical antiviral host factors15.
Applications and target audience
Here, we describe a modified RIC protocol that is specifically designed to study RBP dynamics on a proteome-wide scale. Comparative RIC studies have recently been applied to profile RBP dynamics during embryo development10, virus infection15 and acute inhibition of alpha-ketoglutarate dependent dioxygenases14, revealing key regulators of these processes. Comparative RIC analyses have broad applications to understand how RBPs regulate RNA metabolism in response to developmental, physiological or environmental cues, including cell division, differentiation, reprogramming or stress. Given the importance of RBPs in disease20–23, this method can also be used to elucidate pathological mechanisms associated with e.g. infection, malignant transformation or metabolic disorders, as well as the modes of action of small molecules. Importantly, recent work has shown that RNA can regulate protein function in response to biological cues, a phenomenon that is referred to here as 'riboregulation'24–27. Comparative RIC analyses have great potential to identify examples of 'riboregulation'26 in a proteome-wide manner. Given that the roles of cellular RNA and RBPs have been greatly expanded24–26,28–30, we foresee an extensive use of comparative RIC analysis not only by RNA biologists, but also by experts in other disciplines such as cell signaling, molecular medicine, development, immunology and virology.
Advantages of (e)RIC and comparison to other methods
eRIC approach described here shares the same advantages with its predecessor3, while adding specific features to efficiently study RBPome dynamics: (1) The improved workflow substantially reduces the required starting material, making it applicable to a broader range of biological systems; (2) UV light promotes RNA-to-protein crosslinks at 'zero' distances and does not induce detectable protein-protein crosslinks with the UV irradiation regime described here2,3,31,32; (3) UV irradiation is applied to living cells, thus providing information on native protein-RNA interactions and their dynamics; (4) The use of an oligo(dT) probe enriches for mRNAs and polyadenylated non-coding RNAs, reducing abundant non-polyadenylated RNAs, (e.g. rRNA and snRNA) and their associated proteins2,14. By depleting these dominant (e.g. ribosome-associated) proteins, RIC allows the detection of medium and low abundant RBPs bound to poly(A) RNA. The use of LNAs in the oligo(dT) probe strongly enhances selectivity14,16,33. (5) DNA is virtually undetectable in eluates of eRIC, even for cell types and experimental settings (e.g. nuclear fractionation) where DNA contamination was found to be problematic. (6) Incorporation of proteomic labelling strategies reduces the technical noise, improves quantification accuracy and reproducibility, while reducing mass spectrometry time. (7) We used a customized data analysis pipeline that allows the identification of subtle differences in RNA binding as well as a complementary semiquantitative approach to deal with 'zero' values and investigate 'on/off' and 'off/on' RBP states14,15. (8) Importantly, RIC has successfully been applied to many organisms and cell lines1,2,4–13,25,34–36, thus offering solid foundations for adaptation to virtually every eukaryotic system.
The original RIC protocol and its enhanced variant (i.e. eRIC) are based on the hybridization of an oligo(dT) probe with poly(A) tails and, thus, can only identify proteins interacting with poly(A) RNA. Recently, other RBP identification methods that also capture non-polyadenylated RNAs have been described37–43. Two approaches employed labelling of nascent RNA transcripts in cells with 5-ethynyluridine (5-EU) and were referred to as RNA Interactome using Click Chemistry (RICK)37 and Click Chemistry-Assisted RNA Interactome Capture (CARIC)38. After UV-mediated crosslinking, cells are lysed, EU residues biotinylated via click chemistry and RNA-proteins conjugates purified with streptavidin-coated beads. While successful at identifying RBPs, these methods present some limitations such as the need of nucleotide incorporation, which is not readily applicable to all samples (e.g. organisms), can introduce biases towards highly transcribed genes and can be toxic44.
2C39 and TRAPP40 build on the realization that silica-based matrixes do not only bind free RNA but also RNA with proteins covalently linked to it. After UV crosslinking, cells are lysed under strong denaturing conditions and RBPs purified using silica columns or beads. Although simple and potent, these approaches demand special considerations to avoid contamination with DNA-interacting proteins, as silica matrices can bind both RNA and DNA40. Of note, eRIC was shown to almost completely prevent genomic (g)DNA contamination even in challenging systems exhibiting large gDNA/poly(A) RNA ratios, such as small cells (e.g. T lymphocytes)14 and isolated nuclei33.
Global purification of RBPs was also achieved based on the physicochemical properties of covalent RNA-protein conjugates41–43. Acid guanidinium thiocyanate-phenol-chloroform extraction (“Trizol”) involves the generation of two phases, with RNA and proteins specifically locating to the aqueous and organic phase, respectively. XRNAX41 and OOPS42 purify RBPs with Trizol, exploiting the property of UV-crosslinked RNA-protein conjugates to migrate to the aqueous-organic interphase. PTex43 is based on the same rationale, but in this case a neutral phenol-toluol extraction is used to remove DNA and membrane contaminants, followed by an acid-phenol extraction.
The main advantage of all the methods based on total RNA purification is that they can be used to study RBPs that interact with non-poly(A) RNA (see below). However, as more than 90% of cellular RNA is composed of rRNA and tRNA, these methods are expected to be biased towards these highly abundant RNA biotypes. mRNA represents only a small fraction of total cellular RNA (~3%) and enrichment of poly(A) RNA with RIC/eRIC have been shown to be instrumental for a comprehensive characterization of poly(A) RNA-bound proteomes. Indeed, RBPomes generated with methods based on total RNA purification lack a significant fraction of RBPs identified by RIC/eRIC in identical systems37,38,40–43. These non-overlapping RBPs may interact exclusively or dominantly with poly(A) RNA.
Limitations of the approach
This protocol will not be suitable to study dynamics of RBPs that (i) shift in RNA preference but not in overall RNA binding; (ii) are not expressed or active in the experimental settings tested; (iii) bind exclusively non-polyadenylated RNA (e.g. prokaryotic RNA); (iv) exhibit a protein-RNA interface that is suboptimal for UV crosslinking (e.g. interaction only with the phosphate-ribose backbone); and (v) do not yield mass spectrometry compatible peptides. (vi) The efficacy of this protocol will also be compromised if the input material is extremely limited (we typically aim for 5-10 x107 cells) or if UV cannot penetrate the sample efficiently (e.g. thick tissue samples).
As described above, RIC/eRIC are biased towards poly(A) RNA and cannot be used to study non-polyadenylated RNAs. Therefore, XRNAX41, OOPS42, PTex43, 2C39 and TRAPP40 are especially useful when focusing on other (or all) classes of cellular RNAs including rRNA, snoRNA, snRNA and pre-mRNA. Indeed, these methods will be the best option to study bacterial systems where RNA (including mRNA) is not polyadenylated. In eukaryotic systems poly(A) RNA is a small fraction of the total RNA and, hence, the amount of starting material required to perform RIC/eRIC is substantially higher than that for total RNA purification approaches. Therefore, if the starting material is extremely limited, total RNA based methods will have higher chances to succeed at identifying RBPs. However, it is important to consider the substantial contribution of non-poly(A) RNA to the proteomic data. In summary, selection of the approach will depend on the specific aim of the project. RIC/eRIC will be the method of choice to study mRNA regulatory processes, while total RNA-based methods will be more appropriate when studying non-poly(A) RNA. Also, the availability of input material will be an important factor to decide between RIC/eRIC and total RNA-based methods. In any case, we consider that both approaches are complementary and can be used in conjunction to generate a complete picture of the cellular RBPome.
Experimental design
Controls
To identify contaminant proteins that may be present in the sample in a UV-independent manner, we recommend including a control sample in which UV irradiation was omitted. If the availability of labels for the proteomic analysis is limited, a separate experiment including this control can be performed (as in 3) prior to the comparative analyses. If samples under study are very similar, a single non-irradiated control of a representative condition can be used. However, when comparing more profoundly different samples (e.g. different cell types, differentiation, long treatments, etc.), we strongly recommend parallelizing one non-irradiated control to each UV-irradiated sample/condition under study. To minimize the incidence of false positives, we typically classify as RBP any protein that is enriched in the UV irradiated over the non-irradiated control with 1% false discovery rate (FDR). For the comparative analysis, the 'treated samples' should be compared to a reference 'untreated' or 'control' condition. For example, cells infected with a virus would be compared to an uninfected control15.
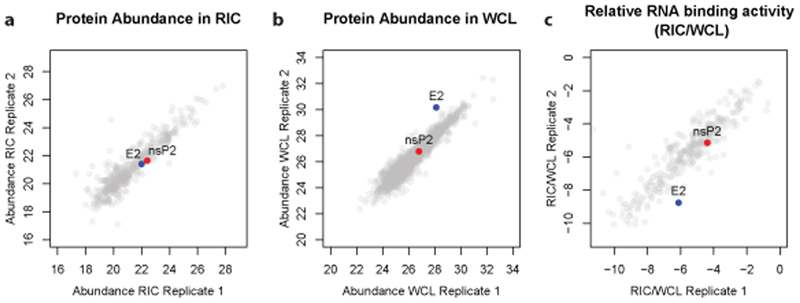
Total proteomic analysis of the cell lysates used as inputs for the eRIC experiment is a very informative addition. The protein intensity ratio between eluates and inputs represents the proportion of protein that crosslinks to RNA upon UV irradiation (crosslinked/total)15,36 (Fig. 3a-c). The eluate/input ratio will be influenced by the amino acid and nucleotide composition, geometry, avidity and duration of the protein-RNA interaction, as these parameters control the ability of a protein to crosslink to RNA25. For example, heterogenous ribonucleoproteins (hnRNPs) establish interactions with RNA that involve multiple high-affinity RNA-binding domains (RBDs). Such long-lived, optimal protein-RNA interactions will give rise to high eluate/input ratios15. Conversely, moderate to low eluate/input ratios are expected for RBPs establishing transitory interactions with RNA (e.g. endonucleases) or when only a subpopulation of the protein engages in RNA binding (e.g. moonlighting RBPs24,25,45). Moreover, we advise that proteins with low eluate/input ratios, which typically appear as outliers in the bottom-left corner of scatter plot (Fig. 3c), should be taken carefully as they may be contaminants or incidental RNA interactors. If interested in a protein falling into this category, we recommend validation with orthogonal methods. The eluate/input ratios have recently been used to model the RNA-binding activities present in large ribonucleoprotein complexes such as the exosome or the ribosome36. Other important applications of the eluate/input ratios are described in the anticipated results section.
Figure 3. Assessment of relative RNA binding by total proteomics and RIC.
a) Scatter plot comparing the protein intensities in the eluates of two replicates of a comparative RIC experiment. Two RBPs with similar abundance are highlighted (E2 in blue and nsP2 in red).b) as (a) but displaying the protein intensity of proteins detected in inputs (total cellular proteome). The two highlighted proteins show a strong difference in protein abundance.c) as(a) but displaying the intensity ratio between RIC eluates and inputs. The comparison highlights that nsP2 displays stronger relative RNA binding than E2. WCL, whole cell lysate.
Selection of the proteomic approach
We have successfully applied two quantitative proteomic approaches to perform comparative RIC analyses: SILAC and TMT. Use of metabolic labeling (SILAC) aids precision and accuracy but possess limited multiplexing. Isobaric labeling such as TMT resolves the multiplexing issue but at the cost of accuracy. There are reviews describing the strength and weaknesses of these approaches46,47. SILAC involves the metabolic incorporation of heavy, medium or light isotopic amino acids into proteins in cultured cells (Fig. 2). For this, cells are grown in media with dialyzed fetal calf serum containing either isotopic variant of (typically) lysine and arginine. When using this approach, isotope incorporation rate must be measured in a pilot experiment. For efficient SILAC-based proteomics, near-complete incorporation rates are required, as lower incorporation rates will compromise the quantification accuracy. For example, if the isotope is incorporated only in 95% of the proteins it would cause on/off changes (i.e. present in condition A and absent in condition B) to be observed as a fold change of 19 (95 divided by 5). As proteins are labelled in cellulo, samples can be mixed immediately after UV crosslinking and cell lysis, and the RNA capture is performed with the combined samples. This reduces technical noise and enhances reliability and accuracy of the protein quantifications. However, the application of SILAC is mostly limited to cultured cells, and specifically to cell types that can be maintained in dialyzed serum. Cells are typically cultured for 5-6 population doublings in SILAC medium to reach high labelling efficiencies (>99%), and thus SILAC is not compatible, in principle, with cells that can only be cultured for a limited time (e.g. certain primary cells). Isobaric reagents such as TMT employ chemical labelling of peptides, and are thus applied after protein elution, concentration and digestion (Fig. 2). Post-elution peptide labelling is compatible with virtually any biological system. The availability of numerous labels allows multiplexing of presently up to sixteen samples in a single run, in contrast to the three labels available for SILAC. The high multiplexing capability simplifies the execution of experiments with larger sample sets, such as those involving multiple treatment conditions or time points. Nevertheless, as peptide labeling is performed further downstream in the protocol, it does not benefit from the early pooling of samples to reduce technical noise and this can compromise the detection of subtle differences.
Moreover, quantification is performed at the ms2 or ms3 level, and thus state-of-the-art mass spectrometers are required for efficient and fast quantification. Quantification and identification with isobaric labelling are performed on the same peptide spectrum thus requiring the mass spectrometry method to strike a balance between identification efficiency and quantification quality. However, the higher 'multiplexing' of TMT reduces mass spectrometry time per sample, allowing the implementation of fractionation to address the identification and quantification loss. Separation of the sample into 3-5 fractions by, for example, high-pH reverse phase StageTip48 or other methods, is thus highly recommended.
Both SILAC and TMT can, in principle, be extended to more conditions than labels available by running a complete experiment in multiple mass spectrometry runs and splitting the conditions between runs. In this case, it is a requisite to run a 'reference' sample (e.g. untreated cells) as a standard in each run, so that ratios between conditions can be robustly generated. In conclusion, we recommend using SILAC when possible, due to its higher robustness, especially when comparing up to three conditions. TMT or similar isobaric tagging strategies will be the approach of choice when using SILAC-incompatible cells, tissues or organisms. It will also be the optimal choice when assessing a large set of conditions. Fractionation is highly recommended when performing TMT experiments.
Although SILAC and TMT provide the highest quantification accuracy and multiplexing capacity, RIC/eRIC can be combined with virtually any quantitative proteomic approach. For example, a recent study has applied label-free quantitation successfully to Saccharomyces pombe36.
DNA vs LNA probes
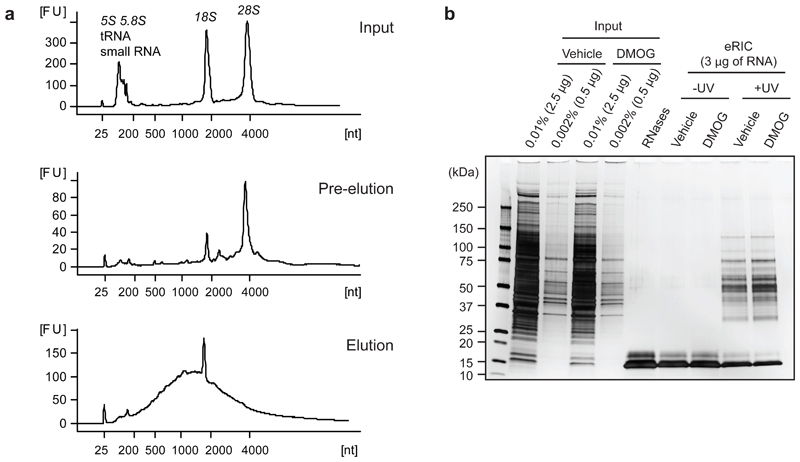
We and others have shown that the use of conventional oligo (dT) DNA capture probes in RIC experiments can lead to contamination with non-poly(A) RNAs such as rRNA. Indeed, depending on the cell type employed and the experimental settings, rRNA can represent from 10% to 30% of the eluted RNA 1,2,14 Proteins that copurify with these abundant non-polyadenylated RNAs (e.g. ribosomal proteins) generate dominant tryptic peptides that mask those from less abundant RBPs. Co-purification of gDNA can also be problematic as it could lead to contamination with DNA-binding proteins or proteins nonspecifically tangled on the chromatin fibers. This can critically impact the quality of RIC experiments when working with nuclear fractions or cells with a low cytoplasm/nucleoplasm ratio such as T lymphocytes. Both contamination with non-polyadenylated RNAs and gDNA can be strongly reduced when using a 20-mer oligo(dT) probe containing locked nucleic acids (LNAs) at every other position (LNA2.T)14. LNAs increase the rigidity of the probe promoting an optimal conformation for hybridization with complementary RNA. The original RIC protocol employs very stringent purification conditions, focused on removing non-covalently linked proteins from the RNA. The increased hybridization strength that the inclusion of LNAs in the oligo(dT) probe provides, allows to additionally maximize the selectivity of the protocol towards poly(A) RNAs. For example, in eRIC the capture of poly(A) RNA and subsequent washes are performed at 37 °C instead of 4°C. Moreover, it includes a 'pre-elution' (final wash) step in water to remove partial hybrids and loosely bound RNAs and DNA14,16 (Fig. 1). The low ionic strength during this pre-elution step destabilizes nucleic acid duplexes, particularly partial hybrids, resulting in efficient removal of rRNA and gDNA (Fig. 4). Here, we describe how to perform comparative RIC studies with the LNA2.T beads (i.e. eRIC), due to the remarkable advantages that this probe adds to the protocol. However, the original DNA oligo(dT) beads have also been successfully used in comparative RIC experiments10,15,36. If using the original DNA probe, apply the experimental design and protocol described here but employ 4 °C during the hybridization step and washes and omit the pre-elution step. Elution can be performed by heating (at 55°C for 3 min) or RNase release as indicated below.
Figure 4. eRIC quality controls.
Poly(A) RNA and associated RBPs were purified from proliferating human leukemia cells (Jurkat) with eRIC and analyzed as follows.a) Representative bioanalyzer profiles of RNA extracted from input material (top), eRIC pre-eluates (middle), and eRIC heat-eluted samples (bottom). rRNAs, tRNAs and small RNAs are predominant in inputs and still present in pre-eluates. eRIC heat-eluted samples display a strong enrichment of RNA species with a size range characteristic of mRNAs. Both 28S rRNA and the small RNAs are undetectable in the heat eluates, although a residual amount of 18S rRNA is consistently observed. [FU] fluorescent units, [nt] length of RNA in number of nucleotides.b) eRIC was performed on Jurkat cells incubated for 6 h with 0.5 mM of the a-ketoglutarate antagonist dimethyloxalylglycine (DMOG) or vehicle (DMSO). Aliquots of the RNase-eluted samples and inputs were analyzed by SDS-PAGE and silver staining. Except for the RNase A and T1 bands (~15 KDa), proteins are only detected in eRIC eluates if cells were UV-irradiated. Protein patterns in untreated and DMOG-treated samples are virtually indistinguishable, which agrees with the proteomic results that revealed a very limited RBP pool (61 proteins out of 721) responding to DMOG14.
Starting material
Incorporation of SP3 together with modifications in the protocol and improvements on mass spectrometry instrumentation have allowed to considerably reduce the starting material required to generate a deep RBPome. For example, we have achieved deep RBPomes from ~50-90 million HeLa and HEK293 cells or ~100-150 million Jurkat cells. For orientation, an optimal eRIC experiment should lead to ~15-30μg of eluted poly(A) RNA per sample to ensure successful downstream proteomic applications. We have noticed that in some cell lines, such as T lymphocytic cells, nuclear RBPs are substantially more abundant than their cytoplasmic counterparts. In such cases, peptide fractionation prior to mass spectrometry greatly improves the identification and quantification of the cytoplasmic RBPs, increasing the dynamic range of the experiment.
RIC and case-specific variations of it have successfully been applied to several unicellular5,8,9,12,34,36 and multicellular organisms8,10,11,35 and it is applicable to tissues13. Starting material and other parameters such as UV light dose must be adapted to the system under study. An excellent starting point is to use the conditions described in the original publications5,8–13,34–36, and to further optimize the protocol with small-scale pilot experiments, if required.
Successive rounds of capture
To maximize poly(A) RNA capture, we perform two sequential rounds of oligo(dT) capture of the same lysate, using 300 of oligo(dT) magnetic beads per round. While two rounds of oligo(dT) capture suffice, in most cases, to capture most poly(A) RNAs from cell extracts, a third round could be required in some instances. To evaluate if the poly(A) RNA depletion has been sufficient, RNA present in each elution round should be measured with e.g. a NanoDrop, separately. Near complete depletion of poly(A) RNA in the lysate leads to a strong reduction in the RNA isolated in the following round of oligo(dT) capture due to the lack of suitable substrate to hybridize with the probe. For example, a third round of oligo(dT) capture typically leads to only 15-10% of the RNA isolated in the first round. In such case, we only recommend performing 2 rounds of capture as the contribution of a third to the proteomic results will be residual.
The efficiency of the oligo(dT) capture can be estimated more accurately using RT-qPCR analysis using primers against selected poly(A) RNAs (e.g. ACTB or GAPDH) in cell lysates before and after the oligo(dT) capture. An efficient eRIC experiment will reduce the levels of a given poly(A) RNA by at least 80%. Conversely, rRNAs are not expected to be depleted.
eRIC optimization
When applying eRIC for the first time to a given cell line, tissue or organism, we recommend performing small-scale experiments (Fig. 1 and Fig. 4). RNA in inputs and eluates can be quality controlled using bioanalyzer and RT-qPCR to assess the proportion of poly(A) RNA versus rRNA. A strong enrichment of poly(A) RNA (e.g. ACTB mRNA) over rRNA (e.g. 18S rRNA) is expected in eluates when compared to inputs (Fig. 4a; see 'anticipated results'). In parallel, RBP isolation can be tested by silver staining (Fig. 4b) or western blotting using antibodies against well-established RBPs and negative controls (e.g. PTBP1 and ACTB, respectively)3. RIC's ability to efficiently isolate RBPs and poly(A) RNA is typically dictated by few key steps that benefit from optimization. These include the i) amount of starting material, ii) the UV crosslinking approach, iii) the cell lysis procedure, and the iv) homogenization of the cell lysate. Optimal starting material is thus fundamental to achieve efficient eRIC experiments. Lack of proteins in eluates despite good quality RNA can often be solved by increasing the number of cells. Still, some cell types (e.g. primary T cells, primary macrophages), give inherently low number of proteins even from large amounts of captured RNA. Conversely, high incidence of contaminant proteins can be caused by excessive protein, RNA and DNA concentration in lysates. We have already estimated the optimal number of cells for HeLa, HEK293 and Jurkat (as indicated in 'Starting material' above). However, cell numbers may need adjustment when working with other cell types. Correct estimation of the starting material becomes more challenging when dealing with multicellular organisms and tissues. In those cases, the starting material will strongly depend on the ability to crosslink protein-RNA interactions with UV light (see below). Improving UV crosslinking efficiency will effectively reduce the input material required; however, excessive UV irradiation can lead to protein and RNA damage.
UV crosslinking can be achieved either exploiting the natural excitability of nucleotide bases at 254 nm (conventional crosslinking; CL)49 or employing nucleotide analogues such as 4-thiouridine (4SU) or 4-thiouracil (4TU) that promote efficient crosslinking between 312 and 365 nm UV light (photoactivatable ribonucleoside-enhanced crosslinking; PAR-CL)50. While CL suffices for most cell lines and is readily applicable to tissues and multicellular organisms, PAR-CL has shown higher performance in budding and fission yeast5,36. Hence, the choice of the crosslinking approach will depend on the model system.
Moreover, UV crosslinking efficiency may differ depending on the properties of the sample. When using cell monolayers, 150 mJ/cm2 of 254 nm UV is a good balance between crosslinking efficiency and lack of RNA damage. However, the optimal dose might vary slightly from cell line to cell line. Conversely, tissues and multicellular organisms may require substantially higher UV irradiation regimens. For example, the optimal UV dose for Drosophila embryos is 4 J/cm2 10 Cell lysates are viscous and one of the key optimization steps is to find the optimal proportion between lysis buffer, biological material and beads. For example, we found that 10 mL of lysis buffer and 300 μL of beads are optimal for selective RBP capture for ~50-90 million HeLa cells or 100-150 million Jurkat cells. Cell lysates are homogenized using a 5 mL syringe and a narrow needle until a waterlike solution is achieved. This step is critical as it promotes the disruption of cellular membranes and the breakage of cellular DNA, reducing lysate viscosity and avoiding the generation of precipitates. If despite of an efficient homogenization, precipitates are observed, increase the lysis buffer volume, homogenize as indicate above and pre-clear the lysate by centrifugation prior to the oligo(dT) capture. See TROUBLESHOOTING.
HPLC and mass spectrometry parameters
For a successful comparative eRIC experiment it is critical to have a suitable proteomic workflow in place. Sample preparation can be performed with any classical technique, including filter-aided sample preparation (FASP)51 or TCA precipitation52. However, to increase sensitivity while reducing the starting material, we employ SP318,19,53. This approach allows efficient removal of detergents and other mass spec-incompatible chemicals, while ensuring efficient protein trypsinization and peptide recovery.
In some experimental settings (see 'selection of the proteomic approach'), peptide fractionation is required to improve protein coverage. In those cases, we apply high pH reversed-phase peptide fractionation54 or similar approaches55,56 prior to LC/MS analysis. The number of fractions can vary in a case-specific manner, but five fractions normally suffice to provide an excellent depth. Peptides are analyzed on a liquid chromatography-tandem mass spectrometry (LC-MS/MS) platform. We used an Ultimate 3000 ultra-HPLC system (ThermoFisher Scientific) coupled to an Orbitrap Elite, QExactive or QExactive plus mass spectrometer (Thermo Fisher Scientific)14,15. However, it is possible to use other equipment with similar characteristics and performance. During LC/MS analysis, each fraction is further separated on low pH gradient of solvent B, the composition of gradient and duration of the separation depend on the complexity of the sample and it should be adjusted for maximum protein identification. Specific mass-spectrometric parameters (such as dynamic exclusion, collision energy, etc) are dependent on the instrument. The current protocol does not require any specific adjustments and can be adapted to virtually any platform optimized for routine protein identification. We recommend monitoring the LC-MS performance between samples and/or fractions by running a blank sample spiked with 20 fmol digested BSA on a 15 min gradient after each sample and/or fraction. BSA digests have characteristic standard chromatographic peaks, which can be used as a proxy to assess the performance of the nanoflow HPLC. Moreover, it will help to clean carryover contaminants between samples.
Statistical data analysis
For robust statistical analysis, it is recommended to generate at least three biologically independent replicates of the complete experimental set, which will include the reference condition (e.g. uninfected cells), treatments (e.g. virus infected cells) and, depending on the experimental design, a negative control (e.g. non-irradiated cells). For more details see 'controls'. Experimental conditions causing subtle variations in the RBPome or with high intrinsic inter-sample variability (e.g. clinical samples), may require higher number of replicates. Peptides are identified and clustered into protein groups and quantified using standard software such as MaxQuant57. Ion counts across samples are summarized and protein intensity ratios between conditions are generated. Significance of the changes is tested using moderated t-test corrected for multiple testing with a Benjamini-Hochberg correction. Software implementation (e.g. Perseus) and customized R packages (Proteus) are available15,32,57,58. In some instances, data normalization and/or batch corrections are required. Analysis of comparative RIC/eRIC experiments are done in two sequential steps. We first compare protein intensities in UV-irradiated and non-irradiated controls and define as RBPs those proteins that are enriched in irradiated over non-irradiated samples with 1% FDR. Secondly, responsive RBPs are identified by comparing the protein intensities of each RBP in crosslinked samples subjected to the different experimental conditions. To define the RBP responses it is critical to avoid normalization against non-irradiated samples, as signal in these controls is low and noisy and provokes artificial distortions of the data when used as normalizer. We typically classify as 'high-confidence' or 'candidate' dynamic RBP any protein that passes the 1% and 10% FDR cut-offs, respectively. Unfortunately, it is not trivial to analyze RBPs with missing ion counts in one experimental condition due to the impossibility to generate a ratio. There are two alternatives to deal which such cases and 'rescue' genuine RBPs (if comparing irradiated vs non-irradiated samples) or responsive RBPs (if comparing two experimental conditions). The first employs a semi-quantitative analysis that takes into consideration the incidence of signal in one condition and lack of signal in the other10,15. Alternatively, it is possible to impute missing values using different computational approaches59. The minimum determination method (Mindet)59 is one of the most commonly used strategies, in which the missing values are replaced by the lowest value detected either globally in the entire dataset or within each sample. Either option has strengths and weaknesses: while the semi-quantitative method does not require the imputation of artificial values, it does not provide ratios to estimate the amplitude of the effect, which may hinder downstream interpretation. Conversely, data imputation provides ratios and thus these RBPs can be included in the statistical analysis. However, imputation methods can introduce artefacts that may affect the statistical analysis. In any case, we recommend applying either strategy.
Complementary analyses
Cellular stimuli often trigger changes at multiple levels, including transcriptomic and proteomic changes. In order to identify the driving factors of the RBP responses we recommend combining comparative RIC analyses with both total proteome and transcriptome analyses. These analyses will provide a snapshot of the protein and RNA landscapes in the experimental conditions, which can be used to interpret the RIC/eRIC results. In other words, the parallel analysis of both protein and RNA abundance is instrumental to determine if any (or both) of these factors contribute to the comparative RIC results10,15. These complementary analyses will be discussed in greater extent in 'anticipated results'.
Material
Reagents
Biological materials
HeLa cells (ATCC, cat. no. CCL-2; https://scicrunch.org/resolver/CVCL_0030)
-
Jurkat cells (DSMZ, cat. no. ACC-282, https://scicrunch.org/resolver/CVCL_0065).
CAUTION: The cell lines used should be regularly checked to ensure they are authentic and are not infected with mycoplasma.
Cell culture reagents
DMEM (Thermo Fisher Scientific, cat. no. 11995065)
DMEM for SILAC (Thermo Scientific, cat. no. 88364)
RPMI 1640 (Thermo Fisher Scientific, cat. no. 21875034)
RPMI-1640 for SILAC (SILANTES, cat. no. 283001300)
Fetal bovine serum (FBS; Thermo Fisher Scientific, cat. no. 10500064)
Glutamine (Gibco, cat. no. G7513)
Penicillin/streptomycin (Sigma-Aldrich, cat. no. P4333)
Dialysed fetal bovine serum (SILANTES, cat. no. 281000900)
Unlabeled L-Arginine HCl (SILANTES, 201004102). C6H14N4O2 •HCl
13C-, 15N-labelled L-Arginine HCl (SILANTES, cat. no. 201604102).
13C6H14 15N4O2 •HCl
• 13C-labelled L-Arginine HCl (SILANTES, cat. no. 201204102). 13C6H14N4O2 •HCl
Unlabeled L-Lysine HCl (SILANTES, cat. no. 211004102). C6H14N2O2•HCl
13C-, 15N-labelled L-Lysine HCl (SILANTES, cat. no. 211604102): 13C6H14 15N2O2 •HCl
2H-labelled D4 L-Lysine 2HCl (SILANTES, cat. no. 211104113). C6 D4H10N2O2 •2HCl. D, deuterium
Sterile filter 0.22 μm pore size (Millipore cat no: SCGVU05RE)
eRIC reagents
Potassium chloride (KCl) (Merck, cat. no. 1.04936.1000)
Potassium dihydrogen phosphate (KH2PO4) (Merck, cat. no. 1.04873.1000)
di-Sodium hydrogen phosphate (Na2HPO4) (Merck, cat. no. 1.06586.0500)
Sodium chloride (NaCl) (Merck, cat. no. 1.06404.1000)
Calcium chloride (CaCl2) (Merck, cat. no. 1.02382.1000)
RNase T1 (Sigma-Aldrich, cat. no. R1003)
RNase A (Sigma-Aldrich, cat. no. R5503)
Complete Protease Inhibitor Cocktail (Roche, cat. no. 11873580001)
Trizma base (Sigma-Aldrich, cat. no. T1503)
-
Lithium dodecyl sulfate (LiDS) (Carl Roth, cat. no. CN25.3)
CAUTION: Harmful if swallowed or inhaled. Can cause serious eye damage. Wear suitable protective equipment, especially when handling as powder.
Lithium chloride (LiCl) (Sigma-Aldrich, cat. no. 62476)
Ethylenediaminetetraacetic acid (EDTA) Disodium Salt, Dihydrate (Merck, cat. no. 324503)
Dithiothreitol (DTT) (Biomol, cat. no. 04020.100)
IGEPAL® CA-630 (Nonidet P-40 (NP-40) replacement) (Sigma-Aldrich, cat. no. I3021)
-
Sodium dodecyl sulfate (SDS) (Serva, cat. no. 20765)
CAUTION: Harmful if swallowed or inhaled. Can cause serious eye damage. Wear suitable protective equipment, especially when handling as powder.
Triton X-100 (Sigma-Aldrich, cat. no. T8787)
-
Sodium deoxycholate (Sigma-Aldrich, cat. no. D6750)
CAUTION: Harmful if swallowed. Causes skin irritation and serious eye irritation. May cause respiratory irritation. Wear dust mask type N95 (US), protective goggles and gloves.
Pierce™ 660nm Protein assay kit (Thermo Scientific, cat. no. 22662)
Ionic Detergent Compatibility Reagent (Thermo Scientific, cat. no. 22663)
Reagents for generation of LNA probes
LNA2.T capture probe: /5AmMC6/+TT+TT+TT+TT+TT+TT+TT+TT+TT+TT (+T: LNA thymidine, T: DNA thymidine) (HPLC purified; Exiqon-Qiagen). The probe bears a primary amine at the 5' end followed by a flexible C6 linker and 20 thymidine nucleotides in which every other nucleotide is an LNA. The required scale of synthesis will depend on the number of samples and capture rounds. A detailed description on this subject is provided in section 'Coupling of the LNA2.T probe to beads'.
Carboxylated magnetic beads M-PVA C11, 50 mg/mL (Perkin Elmer, cat. no. CMG-203)
2-(N-morpholino)ethanesulfonic acid (MES) (Carl Roth, cat. no. 4256.5)
-
N-(3-dimethylaminopropyl)-N'-ethylcarbodiimide hydrochloride (EDC-HCl) (Sigma-Aldrich, cat. no. E7750). CRITICAL Store EDC powder at -20 °C, when required handle it on ice. It is highly hygroscopic, so check that powder is dry before using it.
CAUTION: Toxic on contact with skin. Causes serious eye irritation. Wear protective gloves and eye protection. Very toxic for aquatic environments. Follow suitable disposure procedures
-
≥98% (wt/vol) ethanolamine (Sigma-Aldrich, cat. no. E9508)
CAUTION: Harmful if swallowed, in contact with skin or if inhaled. Causes severe skin burns and eye damage. Wear protective gloves and eye protection. Very toxic for aquatic environments. Follow suitable disposure procedures
Nuclease-free water (Ambion, cat. no. AM9937)
-
Tween® 20 (Carl Roth, cat. no. 9127.2)
CRITICAL Alternatively, oligo(dT) (DNA) probes [Oligo(dT25) magnetic beads (New England Biolabs, cat. no. S1419S)] can be used instead of the LNA2.T probes. If so, use 4 °C instead 37 °C during the capture and washes and omit pre-elution.
Mass spectrometry and SP3 reagents
1M Tetraethylammonium tetrahydroborate (TEAB) (Thermo Fisher Scientific, cat. no. 90114)
-
SP3 purification beads: Speed Bead Magnetic Carboxylate Modified Particles, 15 mL, Azide 0.05%, (Sigma-Aldrich, cat. no. 45152105050250)
CRITICAL Note that these beads are for sample preparation for proteomics and not for poly(A) RNA capture.
CAUTION: Azide is highly toxic, wear gloves and avoid contact with skin.
-
Reduction buffer: Bond-Breaker™ TCEP Solution (Thermo Fisher Scientific, cat. no. 77720)
CAUTION: Causes severe skin burns and eye damage. Wear gloves, clothing and face/eye protection.
-
Chloroacetamide (CAA) (Sigma-Aldrich, cat. no. C0267)
CAUTION: Toxic if swallowed. May cause allergic skin reaction. Wear protective gloves and clothing.
Absolute Ethanol (Sigma-Aldrich, cat. no. 34852)
Acetonitrile (ACN) (Sigma-Aldrich, cat. no. 271004)
Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, cat. no. 34869)
Water LC-MS Grade (Pierce, cat. no. 51140)
Trypsin Gold, MS grade (Promega, cat. no. V5280)
TMT reagents
TMT Isobaric Label Reagent TMT10plex (Thermo Fisher Scientific, cat. no. 90111) Alternatively, duplex TMT, Sixplex TMTTMT11 plex, or TMTpro 16plex can be used according to the number of samples.
OASIS HLB uElution Plate (Waters, cat. no. 186001828BA)
Reagents for eRIC quality assessment
SilverQuest™ Silver staining kit (Invitrogen, cat. no. LC6070)
TURBO DNA-free Kit (Life Technologies, cat. no. 1907)
SuperScript II reverse transcriptase (Invitrogen, cat. no. 18064-014)
SYBR Green PCR Master Mix (Life Technologies, cat. no. 4309155)
Random hexamers (Life Technologies)
Antibodies (Table 1)
qPCR primers for human samples (Table 2)
qPCR primers for mouse samples (Table 3)
(3-Actin and histone H4 are negative controls, the rest correspond to positive controls.
Table 1.
Antibodies employed in Western blot analyses of eRIC eluates β-Actin and histone H4 are negative controls, the rest correspond to positive controls.
| Antigen | Company, catalogue | RRID | Dilution |
|---|---|---|---|
| β-Actin | Sigma-Aldrich, A1978 | AB_476692 | 1:5000 |
| Histone H4 | Abcam, ab10158 | AB_296888 | 1:4000 |
| Polypyrimidine tract binding protein (PTBP1) | Sigma-Aldrich, WH0005725M1 | AB_1843067 | 1:5000 |
| Cold shock domain-containing protein E1 (CSDE1)/UNR | Proteintech, 13319–1-AP | AB_2084902 | 1:5000 |
| Non-POU domain-containing octamer-binding protein (NonO) | Novus Biologicals, NBP1–95977 | AB_11056638 | 1:1000 |
| ELAV-like protein 1 (ELAVL1)/Hu-antigen R (HuR) | Proteintech, 11910–1-AP | AB_11182183 | 1:5000 |
Table 2. qPCR primers for quality assessments of eRIC eluates of human origin.
All sequences from 5' to 3', forward: Fw, reverse: Rv. Note that GAPDH and ACTB forward and reverse primers are designed to target a unique exon of the respective gene and are suitable to quantify both gDNA and cDNA levels.
| Name | Sequence |
|---|---|
| Hm ACTB Fw | GTCATTCCAAATATGAGATGCGT |
| Hm ACTB Rv | GCTATCACCTCCCCTGTGTG |
| Hm GAPDH Fw | CCCCACCACACTGAATCTCC |
| Hm GAPDH Rv | GTACATGACAAGGTGCGGCT |
| Hm 18S rRNA Fw | GAAACTGCGAATGGCTCATTAAA |
| Hm 18S rRNA Fw | CACAGTTATCCAAGTGGGAGAGG |
| Hm 28S rRNA Fw | TTACCCTACTGATGATGTGTTGTTG |
| Hm 28S rRNA Rv | CCTGCGGTTCCTCTCGTA |
Table 3. qPCR primers for quality assessments of eRIC eluates of mouse origin.
All sequences from 5' to 3', forward: Fw, reverse: Rv. Note that GAPDH and and ACTB forward and reverse primers are designed to target a unique exon of the respective gene and are suitable to quantify both gDNA and cDNA levels.
| Name | Sequence |
|---|---|
| Ms Bact Fw | AGAGGGAAATCGTGCGTGAC |
| Ms Bact Rv | CAATAGTGATGACCTGGCCGT |
| Ms GAPDH Fw | AACGACCCCTTCATTGACCT |
| Ms GAPDH Rv | ATGTTAGTGGGGTCTCGCTC |
| Ms 18S rRNA Fw | GCAATTATTCC CCATGAACG |
| Ms 18S rRNA Rv | GGGACTTAATCAACGCAAGC |
| Ms 28S rRNA Fw | AAGCGTTGGATTGTTCACCC |
| Ms 28S rRNA Rv | TCCTCAGCCAAGCACATACA |
Equipment
Class II biosafety cabinet (generic)
Humidified 37 °C, 5% CO2 incubator (generic)
Spectrolinker XL-1500 UV cross-linkers (Spectroline) (254 nm bulbs, BLE-1T155)
Rotator (e.g. Grant-bio, PTR-35)
Conventional incubator without humidification or CO2 control at 37 °C used for the oligo(dT) capture (Aqualytic, Thermostat Cabinet or similar)
SpeedVac concentrator (Thermo Fisher Scientific)
Refrigerated benchtop centrifuge (e.g. Eppendorf, 5415R)
Thermal block (e.g. Eppendorf, cat. no. 5382000015)
500 cm2 dishes (Greiner-BioOne, cat. no. 639160) - for adherent cells
175 cm2 flasks (Falcon, cat. no. 353028) - for suspension cells
150 mm uncoated petri dishes (Greiner Bio-One, 639102) - for suspension cells
Generic metal plate to accommodate 2 x 150 mm petri dishes into the UV crosslinker - for suspension cells
500 mL Steritop Quick Release-GP funnel (Millipore, cat. no. S2GPT05RE)
-
Needle 22G x 1 1/4-inch; Nr. 12, 0.7 mm x 30 mm (BD Microlance, cat. no. 300900) or alternatively Sterican blunt Needle 21G, 0.8 x 22 mm (VWR, cat. no. 720-2562).
CAUTION: handle needles with precaution to avoid harm. We recommend using blunt variants. Dispose the needles in the appropriate sharp bin.
-
Sterican blunt Needle 27G, 0.4 x 25mm (VWR, cat. no. 720-2563)
CAUTION: handle needles with precaution to avoid harm. We recommend using blunt variants. Dispose the needles in the appropriate sharp bin.
Syringes (5 mL; Luer-lock; Medicina, cat. no. IVL05)
DNA LoBind tubes (Eppendorf, 1.5 mL: cat. no. 022431021; 5.0 mL: cat. no. 0030108310; 15 mL: cat. no. 0030122208; 50 mL: cat. no. 0030122232)
Magnetic separation rack, 50 mL (NEB, cat. no. S1507S). For analytical experiments, 12-tube (2 mL) Magnetic separation rack (NEB, cat. no. S1509S) or DynaMag-2 (Invitrogen, cat. no. 123.21D)
NanoDrop spectrophotometer (Thermo Fischer Scientific)
Bioanalyzer with RNA 6000 Pico Kit (Agilent Technologies, cat. no. 50671513 and G2939BA)
Real-time PCR system (e.g. Bio-Rad, CFX96 Touch Real time system, or similar)
Tandem mass spectrometer (e.g. Thermo Scientific Orbitrap Fusion, Q-Exactive, Bruker Impact II or similar)
Software for proteomic data analysis
Protein identification by database searching: Mascot (MatrixScience, http://www.matrixscience.com/distiller_download.html), Sequest (Thermo Fisher Scientific, http://www.selectscience.net/products/sequest-cluster/?prodID=10319#tab-2) or Andromeda60 (via MaxQuant57).
Protein quantification: MaxQuant (https://maxquant.org/)57.
R software (http://www.r-project.org/)
Bioconductor software (http://www.bioconductor.org/), in particular, limma (https://bioconductor.org/packages/release/bioc/html/limma.html)61.
Reagent setup
Cell culture and SILAC reagents
Cell culture media: HeLa cells and Jurkat cells are maintained in DMEM or RPMI-1640, respectively. These media are supplemented with 10% (vol/vol) fetal bovine serum (FBS), glutamine, penicillin/streptomycin, and cells are maintained in a humidified incubator at 37 °C and 5% CO2.
Cell culture media for SILAC: For SILAC, combine medium without arginine and lysine (DMEM for HeLa or RPMI-1640 for Jurkat), with 10% (vol/vol) dialyzed FBS. Prepare one bottle for each SILAC labelling condition by adding either unlabeled arginine and lysine (light medium), 13C L-Arginine HCl and 2HD4 L-Lysine 2HCl (medium labeled medium) or 13C15N L-Arginine HCl and 13C15N L-Lysine HCl (heavy labeled medium). Use 84 mg/L arginine and 146 mg/L lysine for DMEM and 200 mg/L arginine and 40 mg/L lysine for RMPI-1640. Filter the media with a 0.2 μm Sterilefilter and store at 4 °C for up to 3 months.
eRIC reagents
Phosphate-buffered saline (PBS): combine 0.2 g/L KCl, 0.2 g/L KH2PO4, 1.15 g/L Na2HPO4 and 8 g/L NaCl. Prepare, autoclave and store at room temperature for up to 1 year.
1 M DTT stock: prepare 5 mL aliquots in distilled (d)H2O and store at -20 °C for up to 1 year.
1 M Tris-HCl (pH 7.5): dissolve 121 g Trizma base in 800 mL dH2O. Adjust pH to 7.5 with concentrated HCl. Bring volume to 1 L, autoclave and store at room temperature for up to 1 year.
0.5 M EDTA (pH 8.0): dissolve 186.1 g of EDTA disodium salt dihydrate in 700 mL dH2O. Adjust pH to 8.0 with 10 N NaOH. Bring volume to 1 L, autoclave and store at room temperature for up to 1 year.
10% wt/vol LiDS: dissolve 30 g of LiDS in dH2O to a final volume of 300 mL, sterilize by filtration and store at room temperature for up to 1 year.
Lysis buffer: 20 mM Tris-HCl (pH 7.5), 500 mM LiCI, 1 mM EDTA, 5mM DTT and 0.5% (wt/vol) LiDS. Prepare 1 L without DTT, sterilize by filtration and store at room temperature for up to 1 year. Add DTT and Complete protease inhibitor cocktail immediately before usage.
Buffer 1: 20 mM Tris-HCl (pH 7.5), 500 mM LiCI, 1 mM EDTA, 5 mM DTT and 0.1% (wt/vol) LiDS. Prepare 1 L without DTT, sterilize by filtration and store at room temperature for up to 1 year. Add DTT and Complete protease inhibitor cocktail immediately before usage.
Buffer 2: 20 mM Tris-HCl (pH 7.5), 500 mM LiCI, 1 mM EDTA, 5 mM DTT and 0.02% (vol/vol) NP40. Prepare 1 L without DTT, sterilize by filtration and store at room temperature for up to 1 year. Add DTT and Complete protease inhibitor cocktail immediately before usage.
Buffer 3: 20 mM Tris-HCl (pH 7.5), 200 mM LiCI, 1 mM EDTA, 5 mM DTT and 0.02% (vol/vol) NP40. Prepare 1 L without DTT, sterilize by filtration and store at room temperature for up to 1 year. Add DTT and Complete protease inhibitor cocktail immediately before usage.
10x RNase buffer: 100 mM Tris-HCl (pH 7.5), 1.5 mM NaCI and 0.5% (vol/vol) NP-40. Store up to 1 year at 4 °C.
RNase A solution. Prepare 10 mg/mL stock in Tris-HCl (pH 8.0) and 50% (vol/vol) glycerol, aliquot and store at -20 °C.
Heat elution buffer: 20 mM Tris-HCl (pH 7.5) and 1 mM EDTA. Autoclave and store at room temperature for up to 1 year.
RIPA buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% (wt/vol) SDS, 0.5% (wt/vol) sodium deoxycholate, 1% (vol/vol) Triton X-100.
Coupling reagents
50 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer pH 6. Prepare, filter and store at room temperature for several months protected from light by covering the bottle with aluminum foil.
20 mg/mL N-(3-dimethylaminopropyl)-N'-ethylcarbodiimide hydrochloride (EDC-HCl) in MES buffer. Prepare immediately before use.
200 mM ethanoIamine pH 8.5. Add 2.493 mL of 98% (wt/vol) ethanolamine to ~150 mL of dH2O. Adjust pH with 10 N NaOH and bring to 200 mL with H2O. Store at room temperature for several months.
1 M NaCI. Store at room temperature indefinitely.
0.1% (vol/vol) PBS-Tween. Store at room temperature for several months.
Coupling of the LNA2.T probe to beads, Timing 8 h
Commercial DNA oligo(dT) probes used in the original protocol are already coupled to magnetic beads (New England Biolabs, S1419S). Conversely, the LNA-containing probe, LNA2.T, must be coupled to magnetic beads with free carboxylic groups on their surface. Below, we describe the process for coupling the probe to the beads.
Calculate the required amounts of LNA2.T probe and of carboxylated beads. We employ 30 nmol of probe (300 μL of a 100 μM solution) coupled to 15 mg of beads (300 μL of the originaI 50 mg/mL bead suspension) for one round of capture per sample. Note that LNA2.T coupled beads can be reused, at least, up to five times without noticeable loss in performance. We recommend producing enough LNA2.T coupled beads to perform all the biological replicates with the same batch. The following steps show the volumes required to produce LNA2.T-coupled beads for one round of capture in one sample. To obtain final volumes, multiply the indicated volumes of LNA2.T probe and beads by the number of samples and rounds of capture (typically two rounds). The size and number of tubes will depend on the amount of LNA2.T-coupled beads and derived final buffer volumes. We recommend using low-binding tubes throughout the process.
-
Spin down tube with lyophilized LNA2.T probe. Add 1 mL nuclease free water per 100 nmol of probe to obtain 100 μM concentration. Vortex for 30 seconds and spin down briefly.
PAUSE POINT. Probe soIution can be stored at -20 °C at least for several months.
Wash 15 mg of beads (300 μL of 50 mg/mL suspension) three times with 5 volumes of MES buffer pH 6.
Freshly prepare 1.5 mL of a 20 mg/mL solution of the coupling activator EDC-HCl in MES buffer. Transfer a small aliquot (~25 μL) of the EDC solution to a new 1.5 mL tube. This will be used as blank to estimate the coupling efficiency (see below).
Collect the beads from step 3 on a magnet and discard the supernatant. Remove from magnet and reserve.
Combine 1.5 mL of EDC solution with 30 nmol of probe (300 μL of 100 μM probe solution). Transfer a small aliquot (~25 μL) of this probe-EDC solution to a fresh 1.5 mL tube to estimate coupling efficiency (see below).
Add 1.8 mL of probe-EDC solution (step 6) to 15 mg of beads (step 5) and resuspend.
Incubate for 5 h at 50 °C and 800 rpm in a thermal block, occasionally spinning down the liquid that condenses on the lid. Incubate likewise the small aliquots collected for quality control analysis i.e. EDC solution (step 4) and probe-EDC solution (step 6).
Collect the beads with a magnet. Take a small aliquot (~25 μL) of supernatant to estimate coupling efficiency (see below). Set aside the three ~25 μL test samples collected in steps 4, 6 and 9 and keep them at room temperature (15-25 °C).
Wash beads with 1.75 mL of PBS. Repeat wash once.
Incubate with 1.5 mL 200 mM ethanoIamine pH 8.5 for 1 h at 37 °C and 800 rpm to inactivate any residuaI carboxyI residue. Meanwhile estimate coupling efficiency as explained in the next section.
Using a magnet, wash the LNA2.T-coupled beads three times with 1.75 mL of 1 M NaCl.
Collect the beads with a magnet and resuspend in 300 μL of PBS supplemented with 0.1% (vol/vol) Tween-20. If performing the coupling reaction in multiple tubes in parallel, combine them into a single tube of appropriate size to correct for potential coupling differences and subsequent batch effects. PAUSE POINT Coupled beads can be stored at 4°C for at least three months. Supplement with 0.02 % sodium azide as preservative for longer storage.
Estimate the probe concentration in the probe-EDC solution collected before (step 4) and after (step 9) coupling in a NanoDrop device at 260 nm wavelength, using the EDC solution (i.e. without probe) taken in step 4 as a blank. A robust drop on the absorbance after coupling should be observed.
Recycling of probe-coated beads
CRITICAL LNA2.T-coupled magnetic beads can be reused several times (at least five times). Before reusing the beads, the RNase A/T1-resistant poly(A) stretches still associated with the probe must be eluted by heat to release the probe. Beads should also be washed several times for removal of any trace of poly(A) tails and RNases.
CRITICAL Steps 1-4 should be performed immediately after eRIC.
Resuspend 300 μL of the used beads in 400 μL of nucIease-free water and transfer them into a 1.5 mL low binding tube.
Incubate at 95 °C in a thermal block at 800 rpm for 5 min. Proceed and collect the beads immediately after, using a magnet, to prevent the sample from cooling down. Discard supernatant.
Resuspend beads in 5 volumes of water. We recommend pooling the beads used for the different conditions to ensure a homogeneous bead batch for subsequent captures. Wash three times with 5 volumes of water, using the magnet to collect the beads.
Wash the beads three times with 5 volumes of lysis buffer, if the LNA2.T-beads will be used immediately after, or with 5 volumes of PBS supplemented with 0.1% (vol/vol) Tween-20 for longer term storage at 4°C. PAUSE POINT Recycled beads can be stored for at least three months. If required, supplement with 0.02 % sodium azide as preservative for longer storage. Beads can be successfully re-used for at least five times.
Before re-using the beads stored in PBS-Tween-20, wash three times with five volumes of lysis buffer.
Commercial DNA oligo(dT) magnetic beads used in the original RIC protocol can be recycled up to three times. To do so, follow the steps described in 3.
SP3 reagents
100 mM TEAB. Prepare it from a 1M TEAB stock solution in LC/MS-grade H2O immediately before use.
Alkylation buffer. Prepare 500 mM Chloroacetamide (C-IAA) stock solution by dissolving 0.0467 g of CAA in 1 mL of 100 mM TEAB. This must be prepared immediately before use.
Wash buffer 1: 70% (vol/vol) Ethanol (EtOH) (vol/vol) in LC/MS-grade H2O. Prepared fresh and keep at RT for up to 1 week.
Wash buffer 2: 100% Acetonitrile (ACN).
Trypsin digestion buffer: Use freshly prepared 50 mM TEAB in LC/MS-grade H2O for SILAC or 50 mM HEPES pH 8.5 in LC/MS-grade H2O for TMT.
SP3 elution buffer: 2% (vol/vol) DMSO in LC/MS-grade H2O for SILAC or 50 mM HEPES pH 8.5 in LC/MS-grade H2O for TMT. Prepare immediately before use.
Mass spectrometry loading buffer: Prepare 5% (vol/vol) DMSO and 5% (vol/vol) formic acid in LC/MS-grade H2O. Keep at 4 °C for one week.
SP3 purification bead stock preparation
Remove bottle of SP3 purification beads from fridge and keep it at room temperature for 10 minutes.
Mix 40 μL of bead slurry with 160 μL of water (LC/MS grade).
Place tube on a magnetic rack and let the beads settle for 2 minutes. Once beads are fully collected, remove and discard supernatant.
Resuspend the beads in 200 μL of water (LC/MS grade), collect the beads with a magnet and discard the supernatant. Repeat this step twice.
Store SP3 purification beads (10 ug/uL) in 200 μL of water at 4 °C. This stock suffices for up to twenty samples and can be stored for up to one month.
Procedure
CRITICAL If you are using SILAC-based proteomics start at step 1, otherwise go to step 4.
Incorporation of labelled amino acids in adherent or suspension cells Timing: 14 days
-
1
Grow three separate populations of cells for 5-6 passages in SILAC media. Each of the three cell populations should be maintained in media lacking arginine and lysine, supplemented with 10% (vol/vol) dialyzed FBS and one set of isotopic labelled arginine and lysine (i.e. light, medium or heavy amino acids, as described in 'Cell culture and SILAC reagent setup').
CRITICAL STEP: Make sure you do not switch cells from a given isotopic amino acid combination to another e.g. cells growing with heavy arginine and lysine must always be maintained with these heavy amino acids.
-
2
Seed one 6 cm dish with each cell population (i.e. incubated with light, medium or heavy amino acids) and keep them in their respective media overnight at 37 °C and 5% CO2. Lyse the cells with 500 μL of RIPA buffer supplemented with protease inhibitors and confirm isotope incorporation by proteomics as in62,63.
TROUBLESHOOTING.
CRITICAL STEP: It is required a high incorporation rate (optimally >98%) to continue with the experiment.
-
3
Freeze aliquots of each SILAC-labeled cell population with their respective SILAC medium supplemented with 20% (vol/vol) dialyzed FBS and 10% (vol/vol) DMSO. It is possible to initiate any comparative RIC/eRIC experiment from these frozen cell stocks starting from step 4.
PAUSE POINT. Cells can be stored indefinitely in liquid nitrogen.
Cell preparation for eRIC. Timing 1-2 days
For adherent cells follow option A and for suspension cells follow option B. Seeding densities for HeLa and Jurkat cells are provided as a reference. CRITICAL STEP: If using SILAC, labels should be permutated between conditions in different biological replicates to correct for incidental unlabeled contaminants (e.g. keratins from skin) and potential isotope-driven effects.
-
4
-
A
Adherent cells
Seed 1 x 500 cm2 dish per condition with x 107 HeLa cells (or 3 x 150 mm dishes per condition with 3 x 106 cells per dish). If using SILAC you must assign one label to each condition at this stage (e.g. condition A> light; condition B> medium; condition C> heavy). Incubate the cells at 37 °C and 5% CO2 until 80% confluent (usually 1 -2 days) and apply the desired physiological, pathological or pharmacological treatment. TROUBLESHOOTING
-
B
Suspension cells
Seed 3 x 175 cm2 flasks per condition with Jurkat cells at a density of ~0.5 x 106 cells/mL. If using SILAC you must assign one label to each condition. Incubate cells in a humidified incubator at 37 °C and 5% CO2 until reaching a density of 1-1.5 x 106 cells/mL (typically 1-2 days, with 30-45 x 106 cells/175 cm2 flask) and apply the desired physiological, pathological or pharmacological treatment.
-
A
TROUBLESHOOTING
UV irradiation and cell lysis. Timing: 1-2 h
-
5
See Option A if working with adherent cells and Option B if working with suspension cells.
-
A
Adherent cells
Aspirate carefully the culture media of one dish and wash twice with 15 mL of ice-cold PBS. After the second wash tilt the plate on ice for a few seconds to collect remaining PBS and remove it by aspiration. Proceed quickly to the next step.
Remove the lid of the dish and place the dish inside the crosslinker in an appropriate ice chamber at 15-30 cm from the UV source. Irradiate with the dish on ice with 150 mJ/cm2 of 254 nm UV light. Proceed quickly to the next step.
Add 10 mL of ice-cold lysis buffer supplemented with protease inhibitors to each plate and harvest the cells with a cell scraper. Transfer the lysate to a 50 mL conic tube and place it on ice.
-
B
Suspension cells
-
Ensure that cells are monodispersed by pipetting multiple times using a 25 mL pipette. Take an aliquot and test viability (e.g. with trypan blue).
CRITICAL STEP: only proceed if cells show high viability (optimally >90%).
-
Collect 100-130 x 106 living cells by centrifugation at 400 xg for 5min at 4°C. Resuspend in 30 mL of ice-cold PBS and split into two 150 mm uncoated petri dishes with no lids. Proceed quickly to the next step.
CRITICAL STEP: Use uncoated petri dishes to avoid cells attaching to the surface. If cells attach to the uncoated surface, use dishes with low cell binding surfaces instead (e.g. HydroCell, Nunc).
Deposit the petri dishes containing the cells on an ice-cooled metal block and place inside the crosslinker at 15-30 cm from the UV source. Irradiate with 150 mJ/cm2 of 254 nm UV light. Proceed quickly to the next step.
-
Transfer the cell suspension from the petri dishes to a pre-cooled 50 mL conical centrifuge tube and keep on ice. Tilt the dishes on ice for 30 seconds and collect the remaining cell suspension accumulated on the bottom. Combine with the rest in the 50 mL tube and keep on ice.
CRITICAL STEP. Check the dishes with a brightfield microscope. If a large number of cells remains on the dish surface, add 10 mL ice-cold PBS and pipet up and down to recover them. Combine with the rest of the suspension. Repeat if necessary.
Pellet the cells at 400 xg for 5min at 4 °C. Working on ice, discard the supernatant and lyse immediately after with 10 mL of ice-cold lysis buffer supplemented with protease inhibitors. Keep on ice.
-
-
A
CRITICAL STEP: We recommend applying a 300 mJ/cm2 program to the crosslinker immediately before irradiating the cells. This will warm up the bulbs and make UV irradiation cycles more homogeneous. UV crosslinkers typically have a sensor in the bottom-deep part of the chamber. Make sure the ice chamber or metal block does not occlude the sensor. 150 mJ/cm2 UV irradiation typically takes from 30 to 60s. If the irradiation takes longer, make sure the UV sensor is not occluded and all the bulbs are in working order.
-
6
Repeat step 5 with the rest of the dishes/conditions until all samples are processed.
Lysate homogenization. Timing: 60 min
-
7
Working on ice, homogenize the lysate to shear genomic DNA and any remaining cellular structures. Do so by passing each sample 3 times through a 5 mL syringe with a 22-Gauge (0.7 mm diameter) needle. As the volume of the lysate is larger than 5 mL, transfer the lysate from one 50 mL tube to a new one stepwise.
CAUTION: During the homogenization steps, extra care needs to be taken while working with syringes and needles in order to avoid injuries. We recommend the use of blunt needles and syringes with Luer-Lock. Additionally, extra care is recommended to avoid spillage, particularly, when removing the plunger from a syringe.
CRITICAL STEP: Efficient homogenization is critical for successful RIC experiments. As the strength required to pass the lysate through the needle relies on the plunger area, syringes bigger than 5 mL are not recommended. To obtain optimal homogenization, apply as much pressure as possible on the syringe's plunger, releasing the lysate against the wall of the 50 mL tube to avoid the generation of foam. When almost all the lysate has passed through the needle, reduce pressure as this also prevents the generation of foam. Apply all these considerations to the next step.
-
8
Repeat step 7 with a 27-Gauge needle (0.4 mm diameter) until the lysate has lost its viscosity, which typically takes three or more homogenization cycles with the 27-Gauge needle.
TROUBLESHOOTING
-
9
Either snap freeze the samples in liquid nitrogen and store at -80 °C or proceed directly to step 11.
PAUSE POINT: Samples can be stored at -80 °C for up to one month. We recommend avoiding repeated freeze thaw cycles.
CRITICAL STEP: If using SILAC, you can stop at this stage until all the biological replicates are collected. As the different conditions from each replicate will be combined, the number of tubes will thus be reduced (i.e. 1 x 50 mL tube per replicate). Hence, it is feasible to proceed with all 3 replicates in parallel. This parallel processing of replicates will notably reduce technical variation and subsequent batch effects.
RNP capture. Timing: 2-4 hs
-
10
Thaw samples in a 37 °C water bath and continue to the next step immediately after defrosting.
-
11
(optional) Incubate samples at 60 °C for 15 min, quickly cool down on ice and centrifuge at 16000 xg for 5 min and 4 °C to remove any insoluble material. Transfer supernatant to a fresh 50 mL low-binding conical centrifuge tube. NOTE: This step is particularly recommended when lysates look turbid and/or gDNA/protein contaminants had been detected in the eluates of a previous eRIC experiment. If none of these apply to your samples, you can omit this step.
TROUBLESHOOTING
CRITICAL STEP: Incubate simultaneously 1 mL of lysis buffer at 60 °C for 15 min for use in protein quantification in step 13.
-
12
Take 2% of the volume of each sample as 'input' control and store at -80 °C.
-
13
Estimate protein concentration of each sample using an ionic detergent compatible commercial kit and following manufacturer's recommendations.
CRITICAL STEP: Not all protein assays are compatible with the ionic detergent and DTT concentration contained in the lysis buffer. We recommend Pierce 660nm Protein Assay supplemented with Ionic Detergent Compatibility Reagent (IDCR) (Thermo Fisher Scientific). Importantly, prepare the protein standard in the same lysis buffer to correctly calibrate the assay. If step 11 was performed, use an aliquot of lysis buffer incubated at 60 °C for 15 min as blank and to dilute the protein standard, as DTT influences protein quantification and is unstable at high temperatures.
-
14
If optional step 11 was applied, supplement the lysate with 5 mM DTT from 1 M stock to compensate for DTT loss.
-
15
If using SILAC see A, if using other proteomic approach (e.g. TMT) go to B.
-
A
SILAC
Transfer the volume corresponding to an equal amount of protein from each sample into a single 50 mL tube to combine samples. Adjust the combined volume with lysis buffer to 10 mL per sample. CRITICAL STEP: Volumes indicated 'per sample' across the protocol will be multiplied by the number of SILAC samples combined in this step.
-
B
TMT (or other proteomic approaches)
Transfer the volume corresponding to an equal amount of protein of each sample into separate 15 mL tubes. Adjust the volume of each tube to 10 mL with lysis buffer.
-
A
CRITICAL STEP: The amount of protein used is determined by the sample with the lowest protein mass. For example, if sample A=10 mL at 1 mg/mL and sample B=10 mL at 2 mg/mL, 10 mg of protein will be used (corresponding to 10 mL of A and 5 mL of B).
-
16
Add 300 nL/sample of LNA2.T beads to the tube and mix by pipetting up and down (e.g. use 900 μL for SILAC if 3 samples were combined).
-
17
Incubate for 30 min at 37 °C with gentle rotation.
-
18
Collect beads with a magnet in a thermostat cabinet at 37 °C. Wait until the lysate is fully clarified. This should not take more than a few min.
-
19
Transfer supernatant to a new tube (50 mL tube for SILAC and 15 mL tube for TMT) and use it for the second round of capture (step 21).
-
20
Add 5 mL of ice-cold lysis buffer to the LNA2.T beads from step 18. Keep them at 4 °C.
-
21
Repeat steps 16 to 18 for the second round of LNA2.T capture. Proceed with the next steps simultaneously with the beads from the first and second round of capture.
CRITICAL STEP: Once the protocol is successfully implemented for a given cell type and thus the quality of the outcome of each round of capture is known, the beads from the first and second capture can be combined here. Bead combination reduces the number of tubes, which is especially recommended if the experimental design includes numerous conditions. If the beads are combined, double the volumes throughout the protocol from step 22.
-
22
After the second round of capture (step 21), collect the beads and transfer supernatant to a new tube and store at -80 °C for quality control purpose or proceed to a third capture cycle if required (see 'experimental design'). Discard the supernatant if none of these apply. Resuspend the bead pellet from the first and second capture in 10 mL/sample of lysis buffer at room temperature. Incubate for 5 min at 37 °C with gentle rotation in a thermostat cabinet. Collect the beads using a magnet and discard the supernatant.
CRITICAL STEP. For this and the consecutive washes steps, it is essential that the beads are monodispersed and do not clump as this can promote the purification of contaminants. Resuspend the beads by pipetting up and down with a 5 mL pipette. If clumps are observed, resuspend in the following washes gently in 1 mL of the corresponding buffer using a p1000 micropipette with a low binding tip and then add the remaining volume of buffer. The narrower exit of the p1000 tip facilitates clump disruption.
-
23
Resuspend the beads with 10 mL/sample of buffer 1 (kept at room temperature) and incubate for 5 min at 37 °C with gentle rotation. Collect the beads on the magnet and discard the supernatant. Repeat this step once.
-
24
Resuspend the beads with 10 mL/sample of buffer 2 (kept at room temperature) and incubate for 5 min at 37 °C with gentle rotation. Collect the beads on the magnet and discard the supernatant. Repeat this step once.
-
25
Resuspend the beads with 10 mL/sample of buffer 3 (kept at room temperature) and incubate for 5 min at 37 °C with gentle rotation. Collect the beads on the magnet and discard the supernatant. Repeat this step once.
CRITICAL STEP. In the final wash, make sure that the supernatant has been completely removed.
-
26
For the pre-elution step, resuspend the beads in 220 μL/sample of nuclease-free water and transfer to 1.5 mL low-binding tubes. Incubate in a thermal block at 40 °C and 800 rpm for 5 min.
CRITICAL STEP: The pre-elution step should reduce potential contamination with rRNA and gDNA without compromising poly(A) RNA capture when using LNA2.T probe. However, this step is not compatible with the commercial DNA oligo(dT) probe used in the original protocol3. Hence, if using commercial DNA oligo(dT) beads, skip this step.
TROUBLESHOOTING.
-
27
Transfer 20 μL/sample of the bead suspension to a fresh 1.5 mL low- binding tube for RNA and DNA quality control analyses. Capture beads with a magnet and discard the supernatant (which contains eluted contaminants). Add 15 of heat elution buffer and store on ice until step 33.
RNase-based elution for protein analysis. Timing: 60-90 min
-
28
Place the remaining sample from step 26 (200 μL/sample upon removal of 20 μl in step 27) on a magnet rack. Transfer supernatant (which contains eluted contaminants) to a new tube and keep it on ice.
CRITICAL STEP: We recommend keeping this supernatant when adapting the protocol to a new cell type or organism, especially if the yield of captured RNA is low, as it can be used for quality control purposes (see RNA and DNA analysis). No or minimal levels of mRNA should be detected in pre-eluates.
-
29
For RBP elution, resuspend beads in 150 μL/sample of freshly prepared RNase solution (prepare according to the table below) and incubate for 30 min at 37 °C and 800 rpm.
| Component | For 150 μL | For 450 μL* |
|---|---|---|
| Water | 133.65 μL | 401 μL |
| 10x RNase buffer | 15 μL (final: 1x) | 45 μL |
| 1M DTT | 0.75 μL (final: 5 mM) | 2.25 μL |
| 10 mg/mL RNase A | 0.2 μL (final: ~100 U) | 0.6 μL |
| RNase T1 | 0.5 μL (final: ~100 U) | 0.5 μL |
use if 3 SILAC samples were combined
CRITICAL STEP: During this incubation perform steps 33 and 34.
-
30
Collect the beads on a magnetic rack and transfer supernatant to a fresh 1.5 mL low-binding tube.
CRITICAL STEP: captured proteins are now in the supernatant.
-
31
Combine the supernatants of the first and second round of LNA2.T capture (see steps 22-25) if performing a proteomic experiment. Keep them separate if performing a pilot experiment.
-
32
Place samples from step 31 on a magnet to eliminate any remaining traces of beads. Transfer supernatant to a new 1.5 mL low-binding tube.
PAUSE STEP: Eluted proteins can be snap frozen in liquid nitrogen and stored at -80 °C indefinitely or be processed immediately (step 44).
Heat elution for RNA/DNA analysis. Timing: 15 min
-
33
Elute captured RNA from the aliquots taken in step 27 by incubating for 5 min at 95 °C and 800 rpm. Proceed immediately to next step, before samples cool down to prevent recapture of RNA.
-
34
Collect the beads with a magnet and quickly transfer supernatant to a new 1.5 mL low-binding tube. Place the sample again on a magnet to eliminate any remaining traces of beads and transfer the supernatant to a new low-binding 1.5 mL tube.
PAUSE STEP: Samples can be snap frozen in liquid nitrogen and stored at -80 °C for at least one month.
Quality control analyses
RNA and DNA analyses. Timing: 2d
CRITICAL When adapting eRIC to a new biological model, we recommend to analyse RNA and DNA levels on eluates of each round of capture separately.
-
35
Isolate the total RNA from 50 μL of input (step 12) using TRIzol (follow the manufacturer's protocol).
-
36
Determine RNA concentration in samples from step 34 (eRIC heat eluates) and step 35 (purified input RNA) in a NanoDrop device.
Assessment of quality and purity of captured RNA by capillary electrophoresis (bioanalyzer)
-
37
Take 1 μL aliquots of heat eluates (Step 34) and of the purified RNA from inputs (Step 35) and adjust RNA concentration to 5 ng/uL with nuclease free water.
-
38
Run 5 ng (i.e. 1 μL) of each sample in a Bioanalyzer using the Agilent RNA 6000 Pico Kit following manufacturer's indications. See Fig. 4a for an example of the expected outcome.
TROUBLESHOOTING.
Assessment of poly(A) RNA enrichment and gDNA contamination by qPCR
-
39
Mix 0.2-1 μg of RNA from eRIC heat eluates (Step 34) and inputs (Step 35) with TURBO DNase mixture (see below), and incubate for 30 min at 37 °C in a thermal block. eRIC eluates treated with TURBO DNase will be used to measure RNA and will be then referred to as eRIC-RNA. Treat identically and simultaneously an equal amount of eRIC eluates but replacing the DNase by water. These samples will be employed to estimate gDNA contamination and are thus referred to as eRIC-gDNA. It is not required to perform this mock treatment with input samples.
| Component | Reaction | Mock reaction |
|---|---|---|
| RNA | 0.2-1 μg | 0.2-1 μg |
| 10x TURBO DNase buffer | 1 μL | 1 μL |
| TURBO DNase | 1 μL | - |
| Nuclease free water | up to 10 μL | up to 10 μL |
-
40
Add 2 μL of TURBO DNase Inactivation Reagent to eRIC-RNA, eRIC-gDNA and input samples and proceed following manufacturer's indications. CRITICAL STEP: The inactivation reagent is added to the mock reactions to account for any bias it may introduce in the qPCR reaction.
-
41
Using random hexamers, reverse transcribe 5ul of the eRIC-RNA and input samples from previous step. Include a no reverse transcriptase (RT) control to assess the occurrence of gDNA contamination due to an incomplete DNA digestion in step 39. Treat eRIC-gDNA samples from previous step simultaneously and identically but replacing the RT enzyme by water.
-
42
Determine by qPCR the levels of GAPDH mRNA, ACTB mRNA, 18S rRNA and 28S rRNA in eRIC-RNA and inputs. Simultaneously, asses the degree of gDNA contamination in eRIC-gDNA samples using the same GAPDH and ACTB primers. Sequence of primers for human and mouse are provided in 'eRIC quality control reagents' (Table 2 and 3). The qPCR set up is indicated below. The 'anticipated results' section described the expected outcomes of this analysis.
| Component | Amount per reaction |
|---|---|
| SYBR Green PCR Master Mix | 5 μL (final: 1X) |
| Sample diluted 1:50-1:100 | 5 μL |
| 100 μM forward primer | 0.05 μL (final: 0.5 μM) |
| 100 μM reverse primer | 0.05 μL (final: 0.5 μM) |
Use the following thermal cycling parameters:
| Step 1 | 50 °C, 02:00 min |
| Step 2 | 95 °C, 10:00 min |
| Step 3 | 95 °C, 00:15 min |
| Step 4 | 60 °C, 01:00 min |
Perform 40 cycles of steps 3 and 4.
-
43
If RNA/DNA quality controls are satisfactory, combine the RNase-treated eluates of each round of capture (step 32) to perform the protein analysis.
Assessment of RBP capture: 2d
-
44
Take the RNase-treated eluates from step 32. Adjust them to the same RNA concentration determined for heat eluates (step 36) and final volume using 1x RNase solution (see step 32). Use the sample with the lowest concentration as reference.
PAUSE STEP: Samples can be frozen at -20 °C indefinitely.
-
45
Analyze by SDS-PAGE followed by silver staining (Fig.4b) and Western blotting with antibodies against known RBPs (e.g. PTBP1) and negative controls (e.g. ACTB, histone H3). In addition to eluates, load 0.01-0.05% of inputs (step 12) for comparative purposes. Recommended sample amounts for these analyses are provided in Fig.1 and 4. Information of recommended antibodies is provided in Table 1.
CRITICAL STEP: proceed to the next steps only if protein, RNA and DNA quality controls are satisfactory.
TROUBLESHOOTING
Proteomic analysis
Sample concentration. Timing: 60 min
-
46
Add 0.01 volumes of 10% (wt/vol) SDS to the eluates from Step 44 for a final concentration of 0.1% (wt/vol) SDS.
-
47
Concentrate equal amounts of each sample using a SpeedVac at 37 °C until a volume of ~100uL is reached. We recommend performing the SILAC or TMT proteomic analysis with the amount of RNase treated eluate equivalent to 15-30 ug of RNA (inferred from the RNA quantification in step 36; Fig. 1).
CRITICAL STEP: For the complementary total proteome analysis, use material from step 12. Required protein amount depends on mass spectrometry workflow and equipment, but we usually process up to 2 ug for unfractionated samples per mass spec run. More material is usually required if fractionation is applied (20 ug per 10 fractions).
CRITICAL STEP: If the experiment leads to larger quantities of RNA than those indicated above, material excess can be used to perform additional quality control experiments or for validation of candidates from the proteomic approach (see below by Western blotting as in in step 45.
CRITICAL STEP. Avoid concentrating the samples to volumes lower than 50 μL as RBPs can precipitate. If this occurs, supplement the samples with water to reach 100 μL and incubate at 37 °C until precipitate dissolves. PAUSE POINT. Samples can be snap frozen in liquid nitrogen and store at -80 °C.
SP3 sample purification. Timing: 3 hours
-
48
Sample clean up (Steps 48-57). Add 2 μL of 500 mM TCEP (final concentration 10 mM) to the 100 μL of sample from step 47 to disrupt disulfide bonds. Mix by pipetting and incubate at RT for 30 min.
-
49
Add 10.2 μL of 500 mM CAA to alkylate the sulfhydryl groups and prevent re-formation of the disulfide bridges. Mix by pipetting and incubate in the dark at RT for 30 min.
-
50
Add 10 μL of SP3 purification beads (see “SP3 purification bead stock preparation”) to each sample.
-
51
Add 409.1 μL of 100% ACN to obtain a final percentage of 77% (vol/vol) ACN. Incubate for 30 min at RT.
-
52
Collect the beads with a magnet and wait until the supernatant is clear. At this stage, proteins are bound to the SP3 purification beads. You can either keep the supernatant for quality control purpose or discard it if the protocol has already been implemented successfully.
-
53
Keep the tubes on the magnetic rack and add 1 mL of 70% (vol/vol) EtOH [Wash buffer 1]. Incubate for 30 seconds and discard the supernatant. Do not resuspend the beads or take the tube off the magnet.
-
54
Keeping the tubes on the magnet, add 1 mL of 70% (vol/vol) EtOH and incubate for 30 seconds on the magnetic rack. Remove and discard supernatant.
-
55
Keeping the tubes on the magnetic rack, add 1 mL of 100% ACN [Wash buffer 2] and incubate for 15 seconds on the magnetic rack. Remove and discard the supernatant.
-
56
Repeat steps 53-55 seven times.
CRITICAL STEP: It is crucial that at this step, samples are free of any residual detergent, which will negatively affect the nanoflow HPLC system performance and consequentially protein identification.
-
57
Remove any residual ACN. Take the tubes with the beads off the magnetic rack and proceed immediately to the next step.
Trypsin-digestion and peptide elution: Timing: 15-17 hours
-
58
Resuspend the SP3 purification beads in 50 μL (SILAC) or 10 μL (TMT) of Trypsin digestion buffer. Avoid excessive pipetting of the beads as they can stick to the tips, causing protein loss.
CRITICAL STEP: Avoid TEAB when using TMT as primary amines of TEAB can quench TMT peptide labelling (see Trypsin digestion buffer in 'Reagent setup').
-
59
Add 250 ng of sequencing grade trypsin to each sample and incubate in a thermal block at 37 °C and 400 rpm, overnight (~14 hours).
-
60
Collect the beads with a magnet and transfer the supernatant containing the tryptic peptides to a new tube (this is referred to as SP3 eluate 1).
-
61
Take the tube off the magnetic rack, re-suspend the beads with 1 mL 100% ACN, and incubate 5 min at RT. Collect beads with magnet and discard the supernatant.
-
62
Resuspend the beads in 20 μL of SP3 elution buffer. Incubate for 5 min at RT.
CRITICAL STEP: The composition of the SP3 elution buffer for SILAC or TMT approach is different (See 'Reagent setup').
-
63
Collect beads with a magnet making sure the solution becomes clear, carefully collect the supernatant (referred to as SP3 eluate 2), without disturbing the bead pellet, and combine with SP3 eluate 1 (step 60). Mix gently by pipetting up and down. If using TMT, proceed to step 64. If using SILAC (or a label-free approach) proceed directly to step 70 if fractionation is required or to 72 if it is not required.
TMT labelling. Timing: 2 hours
-
64
Select the kit required for your experimental design based on the number of conditions and replicates: i.e. Duplex TMT, Sixplex TMT, TMT10plex, TMT11 plex or TMTpro 16plex for up to 2, 6, 10, 11 or 16 samples, respectively. While we have not tested TMTpro 16plex with this protocol yet, we do not anticipate any incompatibility. Although the process is explained in detail in the manufacturer's manual, we describe it here in the context of our experimental design.
-
65
Dissolve 0.8 mg of each TMT label in 42 μL 100% ACN. One unique label will be used for each sample.
-
66
Add 4 μL of label solution to each sample and incubate for 1 hour at RT.
-
67
Terminate the reaction by adding 5% (wt/vol) hydroxylamine for 15 min at RT.
-
68
Combine equal amounts of each sample.
-
69
Clean up the peptides with an OASIS HLB uElution Plate following manufacturer's indications.
Sample fractionation. Timing: 2 hours
CRITICAL: If using TMT or samples with high dynamic range between nuclear and cytoplasmic proteins (e.g. T lymphocytes), we strongly recommend fractionation.
-
70
Dry the sample completely by vacuum centrifugation. Reconstitute in 20 μL of 20 mM ammonium formate pH 10.0.
-
71
Fractionate samples into 5-6 fractions. We typically perform an offline high pH reverse phase fractionation on an Agilent 1200 Infinity high-performance liquid chromatography system, equipped with a Gemini C18 column (3μm, 110Å, 100x1.0mm2, Phenomenex) or in StageTips54.
CRITICAL STEP: These parameters are provided for guidance and other fractionation regimens or approaches can be used. We do not recommend fractionation at the protein level using SDS-PAGE, as RBPs tend to concentrate at 220-100 KDa and 75-50 KDa, resulting in poor protein spread across fractions. We thus recommend fractionation at peptide level using high pH reversed-phase chromatography 54, isoelectric focusing 56 or ion exchange 55.
Peptide identification and quantification by LC-MS. 1 day
-
72
Dry the sample by vacuum centrifugation. Reconstitute in 20 μL of loading buffer (5% (vol/vol) DMSO and 5% (vol/vol) formic acid).
-
73
We recommend performing a pilot run with 10% of sample to estimate the volume needed for maximum protein identification without overloading the nanoflow HPLC system. This can be estimated based on overflow of the sample peptides to the subsequent blank run with digested BSA (see 'Experimental Design').
-
74
Based on the results from the pilot run, inject the optimal volume into the LC-MS and separate the digested peptide using a typical gradient for the available mass spectrometry instrumentation.
Data analysis. Timing: 1 week
CRITICAL Data analysis can be performed on multiple platforms, including MaxQuant57, R packages or Perseus. Here we describe the workflow based on the R package LIMMA but the workflow will be similar using other tools. An R markdown file, including the code developed to perform analysis for a SILAC labelled experiment15, is provided as Supplementary Software and Supplementary Note.
-
75
Analyze the raw data with software based on database searching that also performs intensity-based quantitation. We used MaxQuant57 (version 1.6.3.4). Define the search database (in our case Uniprot, Human, Organism ID 9606, downloaded on 16/01/13). Define the search parameters: trypsin as protease with full tryptic specificity and a maximum of two missed cleavage sites, carbamidomethyl (C) as a fixed modification, and acetylation (protein N-term, i.e. only the amino terminus of the protein) and oxidation (M) as variable modifications. All other parameters were used as default. By default, MaxQuant will filter the results with a 1 % FDR at both the PSM (peptide to spectrum match) and protein level. If working with multiple fractions per sample, define fraction number in the fraction column before using the 'match between runs' option. For SILAC-based analyses set up the SILAC labels in the 'group specific parameters' tab using the multiplicity option. For isobaric labelling, navigate to the 'Group-specific parameters' tab and select 'Reporter ion MS2' from the 'Type' drop down menu. Then select appropriate kit (e.g. TMT10plex). Consult reference57 for more details.
-
76
Extract the intensities for each peptide from the resulting modificationSpecificPeptide.txt file as in15,32.
-
77
Add the list of reference protein sequences in FASTA format in 'Global parameter-sequence'. Use uniquely mapped peptides and summarize the ion count for each individual sample.
-
78
Perform multidimensional scaling to determine closeness of samples in an unbiased way e.g. plotMDS in LIMMA. Biological samples within the treatment condition should cluster together. Clustering will help to identify technical problems including batch effects and labelling biases.
-
79
Optional: If a batch effect is detected (Fig. 5a-b), we recommend applying a 'batch correction' to the data. This can be incorporated in the R package LIMMA with an appropriate design matrix before linear modeling.
Figure 5. Statistical analysis of proteomic data.
a) Unbiased clustering of data using multidimensional scaling of 3 biological replicates (1-3) and 2 different conditions (Mock in black and Treatment [SINV virus infection] in red). In this case, the clustering of the RIC inputs (WCL) is based on the replicates, rather than experimental conditions, suggesting that the dataset has a batch effect. Batch effects can potentially hide the underlying biological signal and we recommend investigating their cause e.g. assessing if the mean intensity between mass spectrometry runs differ. If the cause of the effect can be identified, a batch correction algorithm can be used after data acquisition.b) Multidimensional scaling as above. In this example, RIC eluates from control (mock) samples cluster together with a clear separation from the treated (infected) samples. No batch correction is necessary in this case. However, the RIC eluate from treated cells in the replicate 3 displays a differential signature when compared to the other 2 replicates. Detailed analysis of the proteomic data can help to elucidate whether this results from a technical problem (e.g. an error estimating the amount of protein prior to mixing the samples).c) Scatter plot displaying the treated/untreated protein ratios from two independent replicates. A good correlation between replicates indicates the high quality of the experiment. Every dot represents a protein and they are colored based on the statistical significance of the change between the two conditions (1%, 5% and 10% FDR). Volcano plot displaying the fold change and p value of the proteins in(c). Every dot corresponds to a protein and they are colored based on the statistical significance of the change between the two conditions (1%, 5% and 10% FDR. as(c) but displaying the ratios in the whole cell lysate (input). No significant changes were observed during the time-course of the treatment, suggesting that the changes observed in eluates are not due to matching changes in protein abundance. Original data was generated in 15 and was re-analyzed here to illustrate the analytical workflow. Mock vs 18hpi with SINV infection of RIC data is displayed in panel a, c and d. Panel b and e represents the matching input proteome.
CRITICAL STEP: A batch effect can be detected because samples cluster not because of the treatment, but due to a technical reason, such as differences between replicates, independent mass spectrometry runs, or labelling performance (Fig. 5a-b). As with any data manipulation, batch effect removal can distort data, hence should be applied only if a clear batch effect is detected by principal component analysis. In our experience, the SILAC light label can introduce batch effects in some instances as exogenous contaminants (e.g. keratins) and unlabeled peptides will only be present in the light channel. This potential bias is minimized by the permutation of the SILAC labels but, if problematic, it can easily be removed by applying the batch effect correction to the SILAC labels.
-
80
Transform the protein intensities into log2 scale.
-
81
Using at least 3 biological replicates, use LIMMA to test whether the fold changes between conditions are different from 'zero' using a moderated t-test.
-
82
Correct the p-values for multiple testing by the Benjamini-Hochberg procedure. Use 1% and 10% FDR proteins to define 'high-confidence' and 'candidate' responsive RBPs. We have recently shown that a large proportion of the 10% FDR responsive RBPs are truly regulated by the cue (in this case, by virus infection)15. Nevertheless, we recommend orthogonal validation experiments when working with 'candidate' responsive RBPs.
-
83
Optional: Proteins that are not detected in all conditions or replicates can lead to 'zero' or 'infinite' ratios, eliminating these proteins effectively from the statistical analysis. As these proteins can represent biologically important on/off and off/on states, we used a semi-quantitative analysis that considers the replicability of these 'zero' and presence of intensity signal in control and treatment 10,15, as implemented in the R markdown provided in the Supplementary Software and Supplementary Note. Alternatively, imputation methods59 that add a value to each 'zero' can be used. The advantages and disadvantages of each option are described in the 'experimental design'.
CRITICAL STEP: The comparison between UV-irradiated and non-irradiated samples is used to discover the repertoire of RBPs present in a given biological system. However, to study the RBP responses it is critical to avoid normalization against non-irradiated samples as signal in these samples is low and noisy and creates artificial distortions of the data when used as normalizer.
-
84
Optional: If complementary proteomic analysis of the input was performed, process the sample as described in Step 47-83 for eluates. Then subtract the log2 fold intensity of the input from the log2 fold intensity of the eluate to estimate eluate/input ratios.
Troubleshooting
Troubleshooting advice can be found in Table 4.
Table 4.
Troubleshooting table.
| Step | Problem | Possible reasons | Solutions |
|---|---|---|---|
| 2 | Incomplete peptide labelling | SILAC incorporation not sufficient | Incubate cells longer in SILAC media. If the cells do not tolerate SILAC reagents, switch to TMT or other quantitative method. |
| 7,8 | Viscous lysate | Incomplete shearing of Chromatiny Too high protein concentration in lysates | Apply more pressure when passing lysate through needle. Repeat homogenization cycles or explore other homogenization strategies5,10; e.g. TissueLyser (Qiagen). Increase lysis volume until lysate reaches a watery consistency. |
| 38, 42, 45 | High contamination with rRNA, gDNA and/or protein | Viscous lysate/Incomplete shearing of chromatin Presence of insoluble chromatin and /or cellular debris Unsatisfactory pre-elution | See “Viscous lysate” in troubleshooting. If skipped, implement optional Step 11. Increase length and/or temperature of pre-elution step, controlling in parallel for poly(A) RNA lost. |
| 38, 42 | RNA degradation | Residual RNase activity in the lysate Excessive UV irradiation | Add RNasin (Promega, N2511) or RNase inhibitors e.g. ribonucleoside vanadyl complex (NEB, S1402S) to the lysis buffer. Note that ribonucleoside vanadyl complexes are incompatible with EDTA. Hence, when working with this RNase inhibitor prepare a lysis buffer without EDTA. Following wash buffers can contain EDTA. Decrease temperature to 4 °C during capture and the first two washes. Remaining washes can be performed at 37 °C due to RNase dilution during previous washes. If eRIC captures substantially more RNA in non-irradiated than UV irradiated samples, further optimize the UV dosage. Make sure the UV sensor is not occluded, which would lead to UV overdose. |
| 38, 42, 45 | Low RNA and protein yield in eluates | Low poly(A) RNA levels per cell Insufficient capture RNA degradation | Increase starting material (number of cells). Increase the length of the capture step, reduce hybridization and/or pre-elution temperature. Increase the number of LNA2.T capture cycles (typically from 2 to 3) and/or the amount of beads to ensure full depletion of poly(A) RNA. See “RNA degradation” in troubleshooting. |
| 45, 74 | Low protein recovery | Insufficient cross-linking | Replace UV bulbs, optimize UV irradiation dose, in particular, for thicker samples or samples containing UV-absorbing pigments. |
| 82-83 | High level of infinite and zero protein ratios in the proteomic analysis | Proteins with low abundance are not identified in all samples | Use more material if MS has not reached saturation. Perform mass spectrometry on a more sensitive mass spectrometer or use more fractions to gain better coverage. If the 'zeros' follow a pattern that resembles a biological regulation (i.e. on/off or off/on states), use a semiquantitative approach or apply suitable data imputation to the dataset. |
| 82 | Few significant proteins or protein responses due to high variability | Batch effect, sample collection or mass spectrometry run | Perform multidimensional scaling analysis to identify the dominant factor of the batch effect. Employ batch correction algorithms to eliminate batch effects, for example by implementing an appropriate design matrix in the LIMMA package before linear modelling. If an experiment is technically inferior due to substantially lower identification and quantification rate (batch effect due to the mass spec run), we recommend first use batch correction and if this does not help, substitute the replicate. |
Timing
Coupling of the LNA2.T probe to beads, 8 h
Comparative eRIC analysis
(Optional) SILAC labelling Step 1-3, 14 days
Cell preparation for RIC: Step 4, 1-2 days
UV irradiation and cell lysis: Step 5-6, 1-2 h
Lysate homogenization: Step 7-9, 1 h
RNP capture: Step 10-26, 2-4 h
RNase based elution: Step 28-32, 60-90 min
Heat based elution: Step 27, 33-34, 15 min
Quality control analyses
RNA and DNA analysis: 35-43, 2 days
Assessment of RBP capture: Step 44-45, 2 days
Proteomic analysis
Sample concentration: Step 46-47, 60 min
Sample preparation by SP3: Step 48-57, 3 h
Trypsin digestion and peptide elution: Step 58-63, 15-17 h
(Optional) TMT labelling Step 64-69, 2 h
(Optional) Sample fractionation: Step 70-71, 2 h
Peptide identification and quantification by LC-MS: Step 72-74, 1 day
Data analysis: Step 75-84, 1 week
Anticipated Results
Initially, RIC was used to identify the RBPs that bind to poly(A) RNA in cells and organisms1,2,4,6,7,9–12,14,15,35,36. RBDmap was derived from RIC and applied to discover the protein regions that directly interact with RNA31,32. The recently developed eRIC protocol boosts RIC selectivity for poly(A) RNA by including locked nucleotides in the oligo(dT) probe14. We exploit here this improved RIC protocol in combination with quantitative proteomics and tailored data analysis10,14,15 to enable system-wide profiling of RBP dynamics in response to biological cues.
A critical aspect of RIC and eRIC is that the eluates should be strongly enriched in intact polyadenylated RNA. To assess whether poly(A) is predominant in eluates and is not degraded, we first use a Bioanalyzer with an RNA 6000 Pico Kit (Fig. 4a). In a successful eRIC experiment, the bioanalyzer profile should show a strong reduction in the rRNA peaks, especially for the 28S rRNA (~4,000 nt), when compared to the inputs (Fig. 4a). Moreover, the eluate RNA profile should show a 'bulge' composed of RNAs with heterogeneous sizes (as expected for mRNAs) that peaks between 500-4,000 nt (Fig. 4a). RNA degradation is highlighted by a strong reduction of the 500-4,000 nt mRNA 'bulge' to smaller RNA fragments (50-500 nt) in eluates.
If eRIC enriches for intact poly(A) RNA based on the bioanalyzer profile, we then utilize RT-qPCR and qPCR for detailed characterization of the isolated RNA pool. We measure the levels of mRNAs (GAPDH and ACTB) and rRNAs (28S and 18S) in inputs and eluates to quantitatively assess the degree of poly(A) RNA enrichment over non-polyadenylated RNA. We expect a 100- and 10-fold enrichment of mRNAs (e.g. GAPDH and ACTB) over 28S and 18S rRNA, respectively2,14. Moreover, if gDNA is depleted in eluates the Ct value in -RT samples should be null or at least 10 Ct higher than those observed in +RT samples2,14. To obtain a system-wide snapshot of the RNA content, inputs and eluates can further be analyzed by RNAseq. This analysis will reveal the relative abundance of each RNA biotype as well as each individual RNA species, providing further contextualization to the eRIC results2.
Once the quality of the isolated RNA has been controlled, it is critical to assess if RBPs have been co-purified efficiently. To test this, we first use silver staining due to its relatively high sensitivity. The eluates of a successful eRIC experiment should yield a complex protein pattern, while the lane corresponding to the non-irradiated control should remain clean (Fig. 4b). Moreover, the protein pattern in eluates should strongly differ from that in inputs, showing the enrichment of a specific subset of proteins. For comparative eRIC analyses, it is important to perform pilot experiments to test if the eluates display similar protein patterns and if individual differences can be spotted. Note that silver staining will only detect the most abundant proteins in eluates, which often play housekeeping roles. Hence, it is not expected to detect massive differences between conditions using silver staining (Fig. 4b)10,15. Therefore, the silver staining analysis is a time-efficient approach to assess the overall performance of eRIC in different experimental settings and can be used to instruct the design of a comparative eRIC experiment.
To further assess the selectivity of eRIC, we also employ Western blotting with antibodies against well-established RBPs (e.g. PTBP1 and/or HNRNPC) and negative controls (e.g. ACTB and histone H4). While both types of proteins should be detected in inputs, only bona fide RBPs should be present in eluates and their presence should be UV-dependent. Detection of non-RBPs such as ACTB in eRIC eluates is a clear indication that further optimization is required (see Troubleshooting).
Only if RNA and protein analyses support the successful implementation/adaptation of eRIC, it is recommended to proceed with the proteomic analysis. Analysis by proteomics of eRIC eluates typically yields 600-1,000 proteins with the workflow and equipment described here. Only proteins within the UV-irradiated samples which are reproducibly enriched over the non-irradiated control should be considered as high-confidence RBPs. As a reference, we only classify proteins as RBPs if they are enriched in the eluates of UV-irradiated samples with 1% FDR. Several RBPomes have been established in different cell lines and species (collected in 25). This super-dataset can be used as reference to quality control the proteomic results. In other words, an RBPome of excellent quality is expected to overlap heavily with previously established RBPomes from the same or closely related cell types. Moreover, both newly and previously established RBPomes are expected to be enriched in similar gene ontology (GO) terms and protein domains, which will indeed be dominated by RNA-related annotation and RNA-binding domains. For example, RNA recognition motif (RRM), K-homology domain (KH), and other RNA-binding domains, should rank amongst the most prevalent domains present in the RBPome1,2,31. By contrast, DNA-binding proteins should be underrepresented, even though specific DNA binders may be endowed with dual DNA/RNA-binding activity and will thus crosslink to RNA (e.g. HNRNPU, EDF1, SUB1). These enrichment analyses can be performed with freely available tools such as DAVID64 (https://david.ncifcrf.gov) and AmiGO 2 (http://amigo.geneontology.org/amigo) as well as dedicated R packages e.g. topGO (https://doi.org/doM0.18129/B9.bioc.topGO).
Comparative eRIC analysis will discover RBPs that associate with RNA differentially under distinct experimental conditions. The number of RBPs regulated by a given cue can vary depending on the strength of the stimulus and the depth of the proteomics analysis, ranging from a dozen to hundreds. We typically classify these proteins as 'high confidence' or 'candidate' dynamic RBPs if passing the 1% or 10% FDR cut-off, respectively (Fig. 5c-d). To confirm that these proteins have differential RNA-binding properties under the selected experimental conditions, we repeat the RIC experiment and analyze few selected dynamic RBPs by Western blotting. Note that, for this validation approach the samples should be processed separately, as described in the TMT workflow. Applying this approach to SINV-infected cells, we validated the differential RNA-binding activity of 11 RBPs (both 1% and 10% FDR) out of 11 proteins tested, supporting the excellent correlation between proteomics and Western blot data15. Nevertheless, the abundance of some RBPs in eRIC eluates can be low, making their detection by Western blotting difficult. For these cases we recommend increasing the amount of sample loaded in the gel and employing ultra-sensitive enhanced chemiluminescent (ECL) reagents (e.g. SuperSignal West Femto Maximum Sensitivity Substrate, Thermo Fisher Scientific). In addition to these experiments, we recommend further validation with orthogonal methods such as polynucleotide kinase assay1,4,5, fluorescence-based assays2,31,65, RIP or CLIP combined with RT-qPCR, CLIPseq-based approaches or localization studies15. To assess the overall impact of protein abundance on eRIC results, we recommend analyzing the whole cell proteome by mass spectrometry (i.e. inputs) (Fig. 3 and 5e-f). By relating the levels of proteins in the inputs and eluates, it is possible to elucidate if the differential association of a given RBP with RNA correlates with matching changes in protein abundance (Fig. 5c vs e). In other words, if there is more protein, more protein is expected to be bound to RNA, and vice versa10,15. For example, protein abundance drives most of the alterations detected in the RBPome throughout fruit fly embryo development10; however, it has minor impact in SINV-infected cells15 (Fig. 5c vs e). Importantly, even under conditions where protein abundance drives the remodeling of the RBPome, it is possible to identify individual RBPs that are apparently regulated in an abundance-independent manner10. This analysis is the first step towards understanding how a given RBP responds to a biological cue.
Alterations in the activity of an RBP can also be triggered by changes in the availability of its RNA targets. Hence, we also recommend performing parallel RNAseq analysis of the inputs and/or eluates of the eRIC experiment. This analysis will reveal if the biological cue causes pervasive, subtle or no changes in RNA abundance. For example, we recently discovered that SINV infection causes a massive degradation of cellular mRNAs15. This phenomenon occurs in parallel with the emergence of the viral RNA, which becomes the dominant mRNA in the cell. Taken together, cellular RNA degradation and accumulation of viral RNA causes deep remodeling of the cellular RBPome. While virus infection has a global impact on RNA availability, other biological cues may cause more subtle differences. In such circumstances, it is possible to utilize RIP or CLIP (e.g. iCLIP66, PAR-CLIP67, eCLIP68 and other variants) data to elucidate if the changes in activity of a given RBP can be explained by matching changes in its target RNAs. For this, it is already possible for many RBPs to use available data such as the eCLIP repository in ENCODE (https://www.encodeproject.org/search/?type=Experiment&status=released&internal_tags=ENCORE&assay_title=eCLIP)69
Other mechanisms may also contribute to RBP control. RBDs are strongly enriched in post-translational modifications (PTMs)31,70, suggesting that PTMs may regulate RNA binding. Therefore, the combination of comparative eRIC studies with proteome-wide methods to profile PTMs can be very powerful to elucidate the contribution of PTMs to RBP function. Similarly, RBPs can assemble into different complexes in response to biological cues. For example, the glutamyl-prolyl tRNA synthase (EPRS) is released from the tRNA multisynthetase complex upon interferon y-induced phosphorylation, and assembles with GAPDH, SYNCRIP and rpL13a forming the GAIT complex71. This newly formed complex regulates translation of specific pro-inflammatory mRNAs. It is thus plausible that differential protein complex assembly has a broader regulatory role of RBP activity than previously anticipated. Hence, protein-protein interaction analyses under the conditions of the eRIC experiment can shed light on the relevance of this regulatory mechanism in RBP function. In summary, the comparative eRIC approach described here can be applied to virtually any eukaryotic system or experimental condition to discover RBPs with condition-specific regulatory roles. By expanding comparative eRIC studies, it will be possible to understand in greater detail how cells and, ultimately organisms, adapt to an ever-changing environment.
Supplementary Material
Editorial Summary.
This protocol extension describes an improved method for global profiling of poly(A) RNA-binding proteins (RBPs) and quantitative analysis of RBP dynamics in response to biological and pharmacological cues using UV crosslinking, capture with LNA-modified oligo-dT probes, and proteomics.
Acknowledgments
We thank Mandy Rettel and Ni Shuai for fruitful discussions on sample preparation for mass spectrometry and data analysis, respectively. A.C. is funded by MRC Career Development Award MR/L019434/1, MRC grant MR/R021562/1, and John Fell Funds from the University of Oxford. W.K. is funded by the Marie Sklodowska-Curie fellowship DLV-842067.
Footnotes
Contributions
J.I.P.-P., M.N., S.M., M.W.H. and A.C. conceived and designed the protocol. J.I.P.-P. and M.N. carried out the experimental work. M.N., W.K. C.E.L. and S.M. performed the proteomic analyses. J.I.P.-P., M.N., S.M., M.W.H., and A.C. performed the data analyses. J.I.P.-P., M.N., W.K., M.W.H. and A.C. wrote the manuscript with input from all authors.
Competing Interests
We declare the authors have no competing interests as defined by Nature Research, or other interests that might be perceived to influence the interpretation of the article.
Data Availability
Raw and processed proteomic data is deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009789. Original data was generated in15 and was re-analyzed here to illustrate the analytical workflow presented in Figure 3 and 5.
Code Availability
An R markdown file, including the code to perform analysis for a SILAC labelled experiment, is provided as Supplementary Software as rmd file and as Supplementary Note including exemplary output in pdf format.
References
- 1.Baltz AG, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 2.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 3.Castello A, et al. System-wide identification of RNA-binding proteins by interactome capture. Nat Protoc. 2013;8:491–500. doi: 10.1038/nprot.2013.020. [DOI] [PubMed] [Google Scholar]
- 4.Kwon SC, et al. The RNA-binding protein repertoire of embryonic stem cells. Nature structural & molecular biology. 2013;20:1122–1130. doi: 10.1038/nsmb.2638. [DOI] [PubMed] [Google Scholar]
- 5.Beckmann BM, et al. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat Commun. 2015;6:10127. doi: 10.1038/ncomms10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao Y, et al. The Cardiomyocyte RNA-Binding Proteome: Links to Intermediary Metabolism and Heart Disease. Cell Rep. 2016;16:1456–1469. doi: 10.1016/j.celrep.2016.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liepelt A, et al. Identification of RNA-binding Proteins in Macrophages by Interactome Capture. Mol Cell Proteomics. 2016;15:2699–2714. doi: 10.1074/mcp.M115.056564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matia-Gonzalez AM, Laing EE, Gerber AP. Conserved mRNA-binding proteomes in eukaryotic organisms. Nature structural & molecular biology. 2015;22:1027–1033. doi: 10.1038/nsmb.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reichel M, et al. In Planta Determination of the mRNA-Binding Proteome of Arabidopsis Etiolated Seedlings. Plant Cell. 2016;28:2435–2452. doi: 10.1105/tpc.16.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sysoev VO, et al. Global changes of the RNA-bound proteome during the maternal-to-zygotic transition in Drosophila. Nat Commun. 2016;7:12128. doi: 10.1038/ncomms12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Despic V, et al. Dynamic RNA-protein interactions underlie the zebrafish maternal-to-zygotic transition. Genome Res. 2017;27:1184–1194. doi: 10.1101/gr.215954.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nandan D, et al. Comprehensive Identification of mRNA-Binding Proteins of Leishmania donovani by Interactome Capture. PLoS One. 2017;12:e0170068. doi: 10.1371/journal.pone.0170068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bach-Pages M, et al. Discovering the RNA-Binding Proteome of Plant Leaves with an Improved RNA Interactome Capture Method. Biomolecules. 2020;10:661. doi: 10.3390/biom10040661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Perri JI, et al. Discovery of RNA-binding proteins and characterization of their dynamic responses by enhanced RNA interactome capture. Nat Commun. 2018;9:4408. doi: 10.1038/s41467-018-06557-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Moreno M, et al. System-wide Profiling of RNA-Binding Proteins Uncovers Key Regulators of Virus Infection. Mol Cell. 2019;74:196–211. doi: 10.1016/j.molcel.2019.01.017. e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogell B, et al. Specific RNP capture with antisense LNA/DNA mixmers. RNA. 2017;23:1290–1302. doi: 10.1261/rna.060798.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 18.Hughes CS, et al. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol Syst Biol. 2014;10:757. doi: 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes CS, et al. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat Protoc. 2019;14:68–85. doi: 10.1038/s41596-018-0082-x. [DOI] [PubMed] [Google Scholar]
- 20.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Castello A, Fischer B, Hentze MW, Preiss T. RNA-binding proteins in Mendelian disease. Trends Genet. 2013;29:318–327. doi: 10.1016/j.tig.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Moore S, Jarvelin AI, Davis I, Bond GL, Castello A. Expanding horizons: new roles for non-canonical RNA-binding proteins in cancer. Curr Opin Genet Dev. 2018;48:112–120. doi: 10.1016/j.gde.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Moreno M, Jarvelin AI, Castello A. Unconventional RNA-binding proteins step into the virus-host battlefront. Wiley Interdiscip Rev RNA. 2018;9:e1498. doi: 10.1002/wrna.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castello A, Hentze MW, Preiss T. Metabolic Enzymes Enjoying New Partnerships as RNA-Binding Proteins. Trends Endocrinol Metab. 2015;26:746–757. doi: 10.1016/j.tem.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19:327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 26.Horos R, et al. The Small Non-coding Vault RNA1-1 Acts as a Riboregulator of Autophagy. Cell. 2019;176:1054–1067. doi: 10.1016/j.cell.2019.01.030. e1012. [DOI] [PubMed] [Google Scholar]
- 27.Choudhury NR, et al. RNA-binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination. BMC Biol. 2017;15:105. doi: 10.1186/s12915-017-0444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiao AL, Slack FJ. RNA-mediated gene activation. Epigenetics. 2014;9:2736. doi: 10.4161/epi.26942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanyi-Nagy R, et al. The RNA interactome of human telomerase RNA reveals a coding-independent role for a histone mRNA in telomere homeostasis. Elife. 2018;7 doi: 10.7554/eLife.40037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumari P, Sampath K. cncRNAs: Bi-functional RNAs with protein coding and non-coding functions. Semin Cell Dev Biol. 2015;47-48:40–51. doi: 10.1016/j.semcdb.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castello A, et al. Comprehensive Identification of RNA-Binding Domains in Human Cells. Mol Cell. 2016;63:696–710. doi: 10.1016/j.molcel.2016.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castello A, et al. Identification of RNA-binding domains of RNA-binding proteins in cultured cells on a system-wide scale with RBDmap. Nat Protoc. 2017;12:2447–2464. doi: 10.1038/nprot.2017.106. [DOI] [PubMed] [Google Scholar]
- 33.Backlund M, et al. Plasticity of nuclear and cytoplasmic stress responses of RNA-binding proteins. Nucleic Acids Res. 2020;48:4725–4740. doi: 10.1093/nar/gkaa256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bunnik EM, et al. The mRNA-bound proteome of the human malaria parasite Plasmodium falciparum. Genome Biol. 2016;17:147. doi: 10.1186/s13059-016-1014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wessels HH, et al. The mRNA-bound proteome of the early fly embryo. Genome Res. 2016;26:1000–1009. doi: 10.1101/gr.200386.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kilchert C, et al. System-wide analyses of the fission yeast poly(A)(+) RNA interactome reveal insights into organization and function of RNA-protein complexes. Genome Res. 2020 doi: 10.1101/gr.257006.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bao X, et al. Capturing the interactome of newly transcribed RNA. Nat Methods. 2018;15:213–220. doi: 10.1038/nmeth.4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang R, Han M, Meng L, Chen X. Capture and Identification of RNA-binding Proteins by Using Click Chemistry-assisted RNA-interactome Capture (CARIC) Strategy. J Vis Exp. 2018 doi: 10.3791/58580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asencio C, Chatterjee A, Hentze MW. Silica-based solid-phase extraction of cross-linked nucleic acid-bound proteins. Life Sci Alliance. 2018;1:e201800088. doi: 10.26508/lsa.201800088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shchepachev V, et al. Defining the RNA interactome by total RNA-associated protein purification. Mol Syst Biol. 2019;15:e8689. doi: 10.15252/msb.20188689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trendel J, et al. The Human RNA-Binding Proteome and Its Dynamics during Translational Arrest. Cell. 2019;176:391–403. doi: 10.1016/j.cell.2018.11.004. e319. [DOI] [PubMed] [Google Scholar]
- 42.Queiroz RML, et al. Comprehensive identification of RNA-protein interactions in any organism using orthogonal organic phase separation (OOPS) Nat Biotechnol. 2019;37:169–178. doi: 10.1038/s41587-018-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urdaneta EC, et al. Purification of cross-linked RNA-protein complexes by phenol-toluol extraction. Nat Commun. 2019;10:990. doi: 10.1038/s41467-019-08942-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burger K, et al. 4-thiouridine inhibits rRNA synthesis and causes a nucleolar stress response. RNA Biol. 2013;10:1623–1630. doi: 10.4161/rna.26214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muckenthaler MU, Rivella S, Hentze MW, Galy B. A Red Carpet for Iron Metabolism. Cell. 2017;168:344–361. doi: 10.1016/j.cell.2016.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altelaar AF, et al. Benchmarking stable isotope labeling based quantitative proteomics. J Proteomics. 2013;88:14–26. doi: 10.1016/j.jprot.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 47.Hogrebe A, et al. Benchmarking common quantification strategies for large-scale phosphoproteomics. Nat Commun. 2018;9:1045. doi: 10.1038/s41467-018-03309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dimayacyac-Esleta BR, et al. Rapid High-pH Reverse Phase StageTip for Sensitive Small-Scale Membrane Proteomic Profiling. Anal Chem. 2015;87:12016–12023. doi: 10.1021/acs.analchem.5b03639. [DOI] [PubMed] [Google Scholar]
- 49.Greenberg JR. Ultraviolet light-induced crosslinking of mRNA to proteins. Nucleic Acids Res. 1979;6:715–732. doi: 10.1093/nar/6.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He C, et al. High-Resolution Mapping of RNA-Binding Regions in the Nuclear Proteome of Embryonic Stem Cells. Mol Cell. 2016;64:416–430. doi: 10.1016/j.molcel.2016.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 52.Niu L, et al. Modified TCA/acetone precipitation of plant proteins for proteomic analysis. PLoS One. 2018;13:e0202238. doi: 10.1371/journal.pone.0202238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sielaff M, et al. Evaluation of FASP, SP3, and iST Protocols for Proteomic Sample Preparation in the Low Microgram Range. J Proteome Res. 2017;16:4060–4072. doi: 10.1021/acs.jproteome.7b00433. [DOI] [PubMed] [Google Scholar]
- 54.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 55.Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krijgsveld J, Gauci S, Dormeyer W, Heck AJ. In-gel isoelectric focusing of peptides as a tool for improved protein identification. J Proteome Res. 2006;5:1721–1730. doi: 10.1021/pr0601180. [DOI] [PubMed] [Google Scholar]
- 57.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11 doi: 10.1038/nprot.2016.136. 23012319. [DOI] [PubMed] [Google Scholar]
- 58.Gierlinski M, Gastaldello F, Cole C, Barton GJ. Proteus: an R package for downstream analysis of MaxQuant output. bioRxiv. 2018 doi: 10.1101/416511. 416511. [DOI] [Google Scholar]
- 59.Lazar C, Gatto L, Ferro M, Bruley C, Burger T. Accounting for the Multiple Natures of Missing Values in Label-Free Quantitative Proteomics Data Sets to Compare Imputation Strategies. J Proteome Res. 2016;15 doi: 10.1021/acs.jproteome.5b00981. 11161125. [DOI] [PubMed] [Google Scholar]
- 60.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 61.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harsha HC, Molina H, Pandey A. Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nat Protoc. 2008;3 doi: 10.1038/nprot.2008.2. 505516. [DOI] [PubMed] [Google Scholar]
- 63.Anantharaman V, Aravind L. The PRC-barrel: a widespread, conserved domain shared by photosynthetic reaction center subunits and proteins of RNA metabolism. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-11-research0061. RESEARCH0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 65.Strein C, Alleaume AM, Rothbauer U, Hentze MW, Castello A. A versatile assay for RNA-binding proteins in living cells. RNA. 2014;20:721–731. doi: 10.1261/rna.043562.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature structural & molecular biology. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Nostrand EL, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Nostrand EL, et al. A Large-Scale Binding and Functional Map of Human RNA Binding Proteins. bioRxiv. 2018 doi: 10.1101/179648. 179648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jarvelin AI, Noerenberg M, Davis I, Castello A. The new (dis)order in RNA regulation. Cell Commun Signal. 2016;14:9. doi: 10.1186/s12964-016-0132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arif A, et al. The GAIT translational control system. Wiley Interdiscip Rev RNA. 2018;9 doi: 10.1002/wrna.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Related links
Key reference(s) using this protocol
- 1.Perez-Perri J, et al. Nat Commun. 2018;9:4408. doi: 10.1038/s41467-018-06557-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Moreno M, et al. Mol Cell. 2019;74:196–211.e11. doi: 10.1016/j.molcel.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sysoev V, et al. Nat Commun. 2016;7:12128. doi: 10.1038/ncomms12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castello A, et al. Cell. 2012;149:1393–406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
Key data used in this protocol
- 5.Garcia-Moreno M, et al. Mol Cell. 2019;74:196–211.e11. doi: 10.1002/wrna.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.This protocol is an extension to. Nat Protoc. doi: 10.1038/nprot.2013.020. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed proteomic data is deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009789. Original data was generated in15 and was re-analyzed here to illustrate the analytical workflow presented in Figure 3 and 5.
An R markdown file, including the code to perform analysis for a SILAC labelled experiment, is provided as Supplementary Software as rmd file and as Supplementary Note including exemplary output in pdf format.