Abstract
While the cerebral cortex is organized into six excitatory neuronal layers, it is unclear whether glial cells show distinct layering. Here, we developed a high-content pipeline, the Large-area Spatial Transcriptomic (LaST) map, which can quantify single-cell gene expression in situ. Screening 46 candidate genes for astrocyte diversity across the mouse cortex, we identified superficial, mid, and deep astrocyte identities in gradient layer patterns that were distinct from those of neurons. Astrocyte layer features, established in early postnatal cortex, mostly persisted in adult mouse and human cortex. Single cell RNA sequencing and spatial reconstruction analysis further confirmed the presence of astrocyte layers in the adult cortex. Satb2 and Reeler mutations that shifted neuronal post-mitotic development were sufficient to alter glial layering, indicating an instructive role for neuronal cues. Finally, astrocyte layer patterns diverged between mouse cortical regions. These findings indicate that excitatory neurons and astrocytes are organized into distinct lineage-associated laminae.
Introduction
Cortical structure has classically been described in terms of six excitatory neuronal layers, generated sequentially during early development (1). In addition, the cortex is regionally specialized into areas responsible for motor, sensory and cognitive functions (1). Although neurons are known to be a diverse population, astrocytes, which comprise about 50% of brain cells, are generally regarded as functionally homogeneous and interchangeable. Might glia also be regionally specialized to confer added cortical organizational complexity? Gray matter astrocytes generally support central nervous system (CNS) structure, nutrition, regulation of blood flow, the blood-brain barrier, synapse formation and activity (2). While several studies show functional diversity of astrocytes (3–7) with implications for regional neural circuit activity or survival (8), precise molecular features of glial spatial organization underlying cortical architecture remain unclear.
Single-cell spatial transcriptomic approaches can provide unprecedented detail on cellular diversity (9), and have been used to investigate the spatial organization of the mammalian brain (10–12). While some approaches produce highly multiplexed gene expression information, imaging and high-content data analysis restrict their application to relatively small tissue areas. Thus, they leave open the important question of how spatial transcriptomics can be applied to screen large tissue areas, including archival tissues and human organs, in a single-cell and quantitative manner to demonstrate gene expression gradients across regions.
Here we developed a method for automated quantitative high-content screening of in situ single-cell gene expression in the mammalian brain. We screened 46 candidate genes for astrocyte spatial organization against known patterns for neurons in the cerebral cortex. Surprisingly, while astrocytes expression patterns showed laminar and area organization such glial layers did not correspond to the six excitatory neuronal layers, indicating higher order complexity of cortical architecture.
Results
Automated pipeline to map single cell cortical gene expression in situ
To map single-cell gene expression across large tissue areas at scale, we automated multiplexed branched-DNA single-molecule fluorescent in situ hybridization (RNAScope smFISH, Advanced Cell Diagnostics) with favorable signal-to-noise ratio (13), immunohistochemistry (IHC), fast confocal imaging, cellular segmentation and quantification on standard whole mouse brain cryo-sections (Fig 1A). We used spinning disk confocal microscopy, to image whole tissue sections at high XY and Z resolution, then modified a high-content screening workflow software (Harmony, Perkin Elmer) to quantify RNA transcripts per cell. We manually segmented cells into brain regions based on reference atlases for gene expression analysis (see Extended Methods) to generate quantitative 2D tissue maps showing spatial single cell regulation of gene expression.
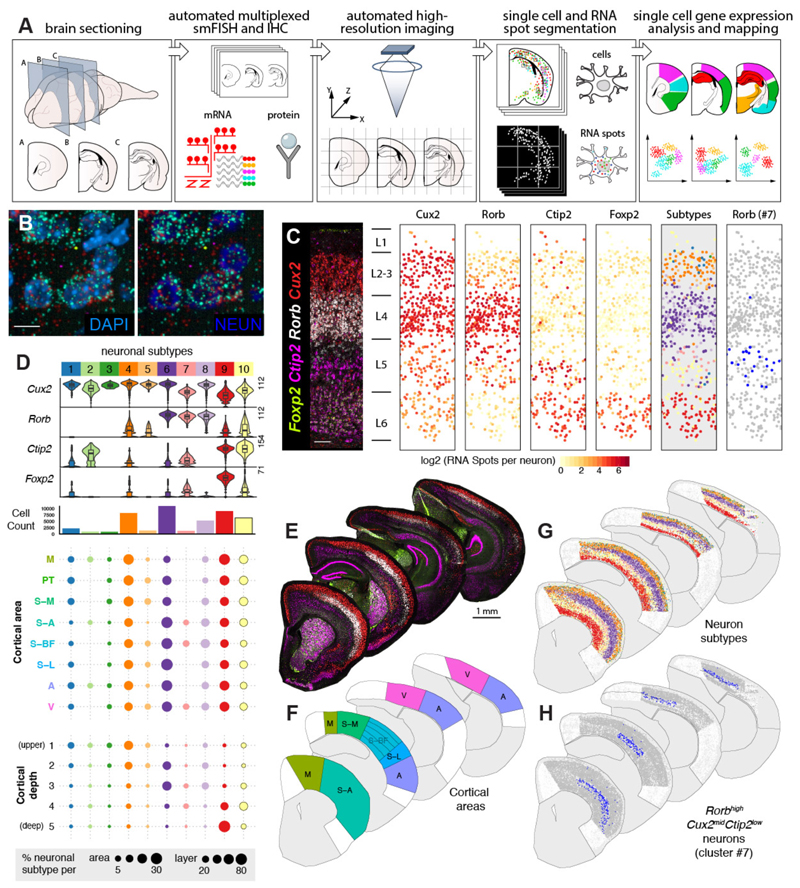
Figure 1. LaST map pipeline for mapping cortical neuronal subtypes in situ.
A) Design of automated spatial transcriptomic pipeline.
B) High resolution imaging of large tissue areas. Shown are 40X z-projection images of Rorb + L4 neurons in the P14 mouse barrel cortex.
C) Automated mapping of layer neuron marker expression and layer neuron subtypes in the mouse barrel cortex. Automatically identified single neurons are plotted as solid circles and colored according to expression (middle panels) or subtype classification (right panels).
D) Identification of neuronal subtypes based on unbiased classification of single cell level smFISH data. tSNE and hierarchical clustering of 46,888 cortical neurons yielded 10 subpopulations. (Top) Violin plots show single cell expression profiles of clusters, highest RNA spot count per cell are shown on the left. (Middle) Histograms showing total number of cells per cluster. (Bottom) Spatial distribution of clusters across cortical areas and five normalized cortical depth bins, shown as percentage of total neurons in given area/depth bin (bottom).
E-H) Single cell mapping of cortical neuron subtypes: (E) Low magnification images of P14 hemisections from four different anatomical levels, (F) broad cortical areas included in the analysis, (G) maps of 10 major neuronal populations, and (H) spatial distribution of area-restricted L5 Rorb high Cux2 mid Bcl11b low neurons.
n = 1 mouse, 10 tissue sections independently imaged. Scalebars: (B) 10 µm, (C) 100 µm, (E) 1 mm.
Abbreviations: M, motor, S-A, anterior- somatosensory, S-M, medial-somatosensory, S-BF, somatosensory barrel, S-L, somatosensory-lateral, PT, parietal, A, auditory, V, visual.
LaST map confirms neuron excitatory layering and reveals novel neuronal architecture in mouse postnatal cortex
We first validated our Large area Spatial Transcriptomic (LaST) pipeline by mapping expression of known cortical layer neuron markers for layers 2-4 (L2-4, Cux2), L4 (Rorb), L5-6 (Bcl11b) and L6 (Foxp2) (1) (Fig 1B,C) in the mouse brain. We performed automated six-color imaging of 10 anterior-posterior sections across the entire P14 cortex (Fig 1E, F), totaling 2,216 fields of view and ~300,000 images collected under 25 hours of continuous acquisition. We used machine-learning based texture analysis combined with NeuN IHC (neuronal nuclei and cytoplasm) and DAPI staining to identify single neuron soma on max-z projection images, while also ruling out doublets/triplets (Supplementary Figure 1, Extended Methods). This approach validated that our technique accurately identified known layer- specific single-cell and layer-specific neuronal gene expression patterns (Fig 1C) with robust reproducibility (Supplementary Figure 2).
Having generated a quantitative map of known cortical neuron gene expression, we tested whether we could identify and/or discover neuronal subtypes solely from smFISH data in an unbiased manner. We analysed transcript counts from 46,888 neurons across select broad cortical areas using a modified dimensionality reduction and hierarchical clustering approach (Extended Methods, Sup Table 1). Our analysis identified 10 distinct major clusters based on the expression of the four markers above (Fig 1D) and could robustly detect low expression differences across neurons (Supplementary Figure 3,4,5). These clusters primarily represented layer-enriched neuron populations present across multiple cortical areas (Fig 1C, D, G;, e.g. Bcl11b high Foxp2 high cluster #9 in L6). In addition, we captured heterogeneity within cortical layers, exemplified by clusters of Cux2 high Rorb neg/low (#1,3-5) and Cux2 high Rorb high (#8) within L2-3. We could also accurately distinguish area-specific differences such as Rorb high Cux2 high neurons (#6 and 8) enriched in L4 of sensory areas and Cux2 mid Bcl11b mid neurons (#2, likely an interneuron subtype, Supplementary Figure 6, (14)) in L1-3 and L5 of somatosensory, motor and auditory cortex (full area maps shown in Supplementary Figure 7). Finally, we found a novel population of Rorb high Cux mid Bcl11b low neurons (#7) restricted to L5 of anterior somatosensory, barrel and visual cortex at P14 and P56 (Fig 1C, H; Supplementary Figure 6). To confirm that Rorb and Bcl11b expression segregates amongst neuronal subtypes in L5, we examined available cortical single cell transcriptome data (15). Consistently, our survey showed that Rorb and Bcl11b expression segregate amongst molecular subtypes of L5 neurons in the adult visual cortex (Supplementary Figure 6). Taken together, these results demonstrate that LaST map can be used to validate regional qualitative and quantitative predictions about gene expression in mammalian brain and that it can serve as a tool to sensitively identify novel/diversified cell populations in situ.
Laminar gene expression patterns are characteristic of cortical gray matter astrocytes
It is generally accepted that astrocytes in the superficial subpia (L1) and deep L6 show white matter astrocyte characteristics (16). We next tested whether gray matter astrocytes in L2-5 are molecularly homogeneous, or alternatively, segment into multiple distinct layers? First, to generate candidate gene lists, we manually dissected the upper (L2-4) and deep (L5-6) layers of the somatosensory cortex from P14 astrocyte reporter Aldh1L1-GFP transgenic mice (17) and performed RNA-seq profiling of FACS-purified astrocytes versus whole cortex (Fig 2A). The expression pattern of whole cortical tissue showed that the microdissection accurately captured upper and deep layers. Astrocyte markers were highly enriched in FACS-purified GFP+ cells over GFP-negative cells and whole cortex tissue as expected; moreover, most neuronal layer markers were not distinctly expressed amongst cortical astrocytes (Supplementary Figure 8).
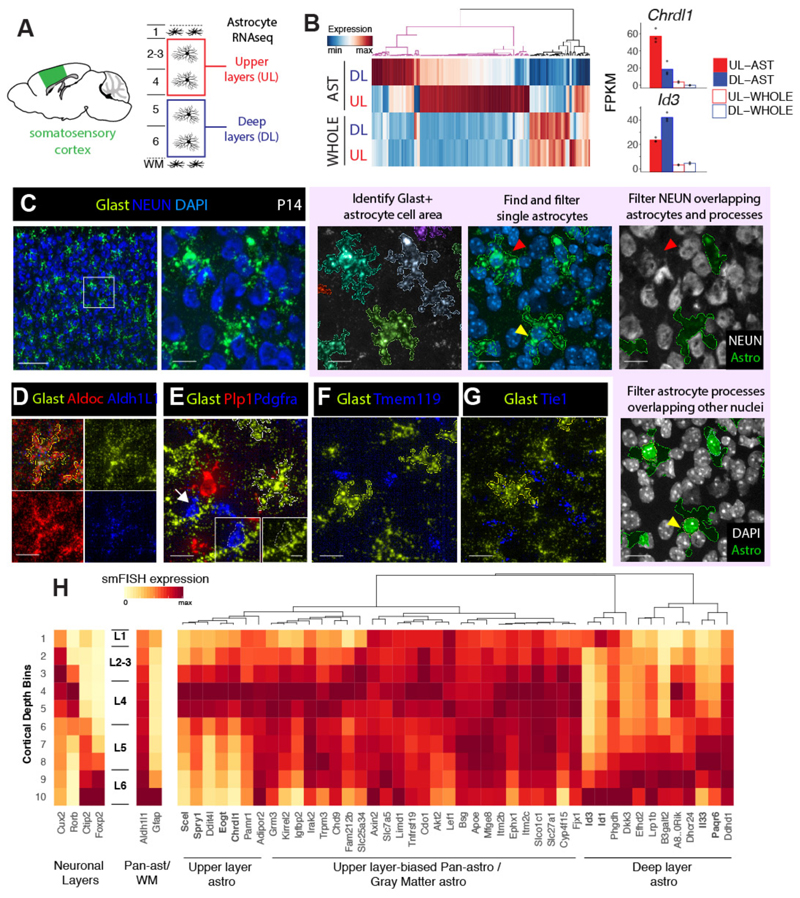
Figure 2. Novel layer expression differences amongst cortical gray matter astrocytes revealed via RNAseq and LaSTmap.
A) Diagram showing somatosensory cortical areas and layers used for laminar gray matter astrocyte RNAseq expression profiling.
B) Novel molecular heterogeneity of layer astrocytes identified by RNAseq. 159 differentially expressed genes between upper and deep layer gray matter astrocytes (FDR<0.05 and expression threshold of FPKM>5) were detected. Bar plots show mean expression (n=3 mice for astrocytes, 2 mice for whole cortex).
C) Identification of astrocyte gene expression with Glast (Slc1a3) smFISH. The astrocyte cell area is segmented from the Glast signal and nuclei are segmented from DAPI. Single astrocytes are selected using morphological and intensity filters, overlapping neurons and non-astrocyte nuclei are excluded.
D-G) Glast is a specific marker of astrocytes. Solid outlines indicate the cell areas of identified single astrocytes.
H) Screening candidate layer astrocyte genes with LaSTmap identifies laminar expression patterns in situ. Tile heatmaps show average single cell gene expression binned across cortical depth in the P14 somatosensory cortex (n =2 pooled biological replicates across multiple tissue sections). Upper and deep layer genes with astrocyte-specific expression are marked in bold. n = 2 mice independently assayed, 3 tissue sections imaged per replicate.
Scalebars: (C, left panel) 100 µm, (C, other panels) 25 µm, (D-G) 25 µm, (E, inset) 10 µm.
This analysis identified 159 candidate genes with a differing expression between the upper and deep layer astrocytes (Fig 2B). While most genes showed moderate-layer enrichment, many also showed layer and astrocyte-enriched expression in published cortical transcriptome datasets (Supplementary Figure 9). We next validated layer astrocyte gene expression in situ. To distinguish astrocyte-specific gene expression by smFISH, we examined markers for astrocytes (Aldh1l1, AldoC, Glast), neurons (Syt1, NEUN), oligodendrocytes (Pdgfra, Plp), microglia (Tmem119) and endothelia (Tie1) (17). As shown in Fig 2C-G, while Glast mRNA showed low expression in Pdgfra+ oligodendrocyte precursors, high Glast levels exclusively marked astrocytes. Glast mRNA (but not AldoC or Aldh1L1) filled astrocyte soma and main processes, permitting clear delineation of astrocyte cell borders by image texture analysis.
To avoid double counting, we selected non-overlapping single astrocytes by filtering cells based on size, DAPI (single-nuclei) and Glast intensity (Extended Methods, Supplementary Figure 10 and 11). In addition, we filtered astrocytes and their processes that overlap with neuronal soma and non-astrocyte nuclei in z-projection images (Fig 2C, Extended Methods). These measures ensured our smFISH approach counted single cell astrocyte-specific gene expression in situ in the context of a large tissue area screen.
To validate candidate layer-associated astrocyte genes in situ, we first analyzed the P14 somatosensory barrel cortex. We selected the top 46 genes from RNA-seq that showed the highest differential enrichment between cortical layers and/or astrocyte specificity (Fig 2H, Sup Table 2). Of these, we confirmed layer enriched expression patterns or rejected genes that showed subtle variations based on quantitative findings in situ. For example, 48% (22/46) showed an expression pattern biased –but not restricted-- to upper layers (e.g., Tnfrsf19, Mfge8), and 15% (7/46) showed enrichment in all gray matter astrocytes across L2-6 (e.g., Igfbp2, Kirrel2). Another third (13%, 6/46) showed expression in astrocytes + neurons and were excluded (e.g., Ddit4l, B3galt2, Supplementary Figure 12). In contrast, eight genes clearly distinguished upper versus deep layer astrocytes in the somatosensory cortex (Fig 2H). As shown (Fig 3A-C, Supplementary Figure 13), expression of the BMP antagonist Chordin-like 1 (Chrdl1) (18) was expressed in L2-4 astrocytes while Interleukin-33 (Il-33) (19) was enriched in L5-6. In distinction, Sciellin (Scel), a component of epithelial cornified envelopes (20), was expressed in mid-cortical (~L4) layers and transcriptional repressors Id1 and Id3 occupied deep (L5-6) and marginal (L1) layers (21). These findings show that subsets of cortical gray matter astrocytes show laminar spatial gene expression heterogeneity.
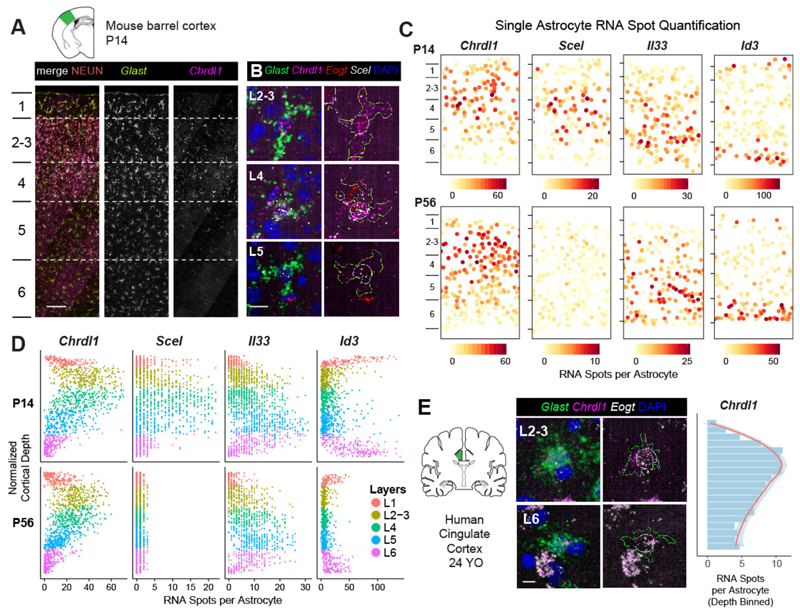
Figure 3. Astrocytes show broad expression gradients across cortical depth and diverge from neuronal layers.
A-B) Upper layer astrocyte enrichment of Chrdl1, Scel and Eogt. Images show the mouse barrel cortex at P14 (A) and close-ups of astrocytes across layers (B). Astrocyte cell areas are marked with yellow outlines and nuclei are marked with white outlines.
C) Astrocytes are organized into superficial, mid and deep layers across the cortical gray matter. Single astrocyte expression maps in the barrel cortex at P14 (top panels) and P56 (bottom panels). Astrocytes are plotted as solid circles and colored quantitively for RNA spot counts per gene per cell.
D) Single astrocyte quantification of layer astrocyte markers across cortical depth in the barrel cortex at P14 and P56.
E) Upper layer astrocyte enrichment of Chrdl1 expression in the adult human cingulate cortex. Quantification of depth binned average single astrocyte expression shown on the right.
n=1 mouse per timepoint, 3 tissue sections imaged independently per gene panel (A-D) and 3 human brains independently assayed, 1-2 sections imaged per case (E). Scalebars: (A) 100 µm, (B, E) 10 µm.
Astrocyte layers diverge from neuronal laminae
To determine whether glial layers matched excitatory neurons we directly compared the spatial expression patterns of the 8 top layer astrocyte markers against layer neuron markers in P14 somatosensory barrel cortex in situ. We found that astrocyte layer genes generally fell into three spatial bins comprising superficial, mid and deep laminae. As shown (Fig 3A-D, Supplementary Figure 13), Chrld1, Scel, Eogt, Spry1, Paqr6 and Il33 were expressed in patterns that spanned multiple neuronal layers. Astrocyte Chrdl1 expression peaked in L2-4, with low levels in L1 and L5. Scel expression was highest in L4-5, with an upper boundary in mid L2-3. Il33 expression peaked in L5, with low levels in L4 and L6. Such patterns were not artifacts of astrocyte size or Glast expression level differences across layers (Supplementary Figure 14) and were highly reproducible with expression levels similar to that of layer neuron markers (Supplementary Figure 15). The layer astrocyte patterns were also validated with immunohistochemistry (Supplementary Figure 16). Moreover, analysis of the P56 mouse brain indicated persistence of these patterns, except for the mid-layer Scel expression into adulthood (Fig 3D). Indeed, we found that the upper layer astrocyte Chrdl1 expression was conserved in the adult human brain (Fig 3E, Supplementary Figure 17).
While we found certain similar features between astrocytes in deep cortical layers and white matter (WM), most cortical astrocyte layer genes were specific to gray matter. L6 astrocytes expressed high levels of Gfap, Id1 and Id3, similar to astrocytes in subcortical WM and L1-subpia (Supplementary Figure 13) (7). Conversely, Il33 was enriched in L5- 6 gray matter astrocytes but absent from WM, showing that deep layer astrocyte identity is distinct from WM. Together, these findings indicate that astrocytes are organized into multiple layers across the cortical gray matter and that several glial laminae are dissimilar to classic neuronal layers (Fig 6E). Moreover, such patterns are evident from P14, persist until adulthood and are conserved in the human cortex.
Figure 6. Astrocyte arealization across the cortex.
A-C) Single cell mapping of astrocyte gene expression across cortical areas. Maps show the (A) P14 cortical areas used in the analysis, (B) single astrocyte expression of Scel and (C) Chrdl1 across the dorsoventral and rostrocaudal extent of the cortex. Astrocytes are plotted as solid circles and colored quantitively for RNA spot counts per gene per cell.
Arrowheads indicate the restriction of Scel expression to sensory areas. Abbreviations as in Fig 1.
D) Astrocyte layers are distinct across cortical areas. Smoothened tile plots showing the quantification of neuronal Rorb expression and astrocyte expression of Scel, Chrdl and Il33 across the cortical areas and layers
E) Diagrams showing the divergent layer heterogeneity of astrocytes. Our study identifies superficial (sAS), mid (mAS) and deep (dAS) layer astrocyte subtypes through cortical gray matter in the postnatal cortex and confirms white matter like (wlAS) properties of L6 astrocytes.
F) 3D model showing astrocyte area and layer heterogeneity. Astrocyte layering is regionally specialized across the dorsoventral and rostrocaudal extent of the cortex. n = 1 mouse, 10 tissue sections independently imaged.
Astrocyte layers are observed in unbiased single cell transcriptomic data
To independently verify astrocyte layers, we examined the molecular heterogeneity of cortical astrocytes in a single-cell RNA-seq (scRNA-seq) study (Fig 4A). The single cell astrocyte transcriptomic database was generated using a modified Smart-Seq2 protocol (23). Unsupervised clustering with the Louvain method identified two major astrocyte subtypes in the P56 cortex (Fig 4C). These astrocyte clusters, designated AST2 and AST3, were distinguished by the expression of Unc13c and Agt, respectively (Fig 4C), and were found to be largely intermixed throughout cortical layers (23). To reconcile our results with these findings, we examined the expression pattern of our cardinal layer astrocyte markers in the scRNA-seq data. As shown (Fig 4D), our layer astrocyte markers were expressed in subpopulations of cells in the single astrocyte transcriptome data, but not in a cluster-associated way. These observations suggest that, while layer astrocyte gene expression patterns can be detected in an unbiased transcriptomic study of cortical astrocytes, there might be multiple axes of astrocyte heterogeneity (i.e. laminar and non-laminar) in the cortex.
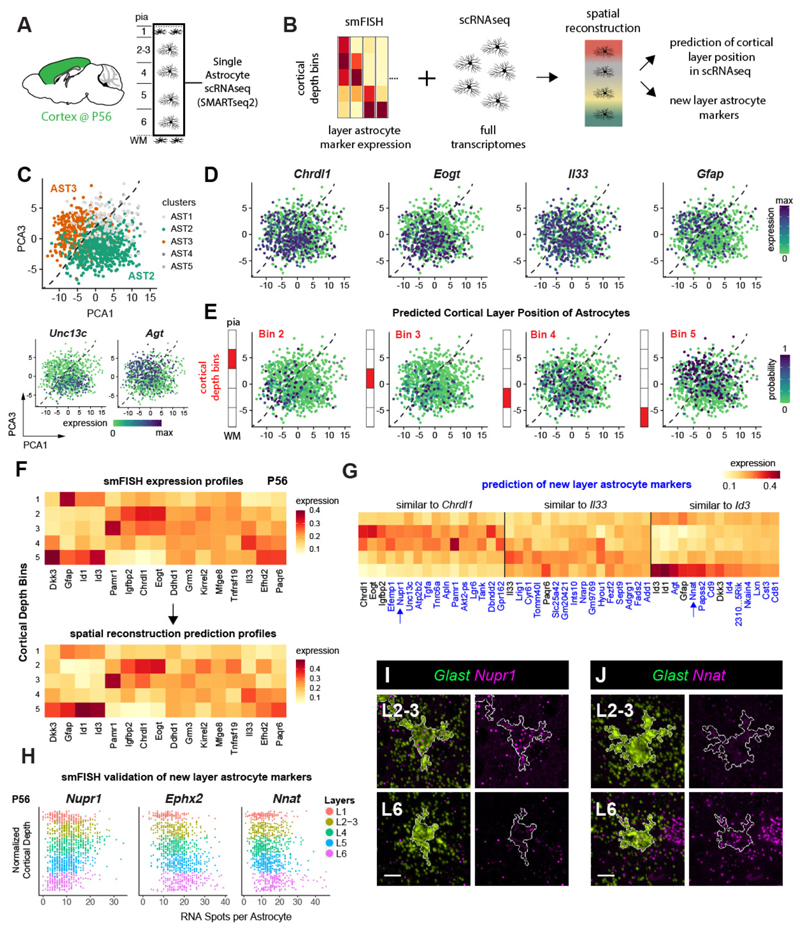
Figure 4. Spatial reconstruction of astrocyte layers from single cell transcriptome data.
A-B) Diagrams showing cortical areas and layers used for single astrocyte RNAseq expression profiling (A) and strategy for spatial gene expression reconstruction (B). C) Astrocyte clusters in scRNA-seq data visualized using PCA plots (PC1: 3% PC2: 0.77%. of variance). Dashed lines indicate the border between the major clusters AST2 and AST3 (colors, top). Bottom plots show the expression of cluster markers (log2 Smart-seq2 read counts). n = 2 independent experiments.
D) Astrocyte layer markers express in subpopulations of astrocytes in scRNA-seq data but not in a cluster-specific manner. PCA plots are shown.
E) Predicted astrocyte layering across scRNA-seq data as a new axis of heterogeneity. PCA plots (X-axis is PC 1, Y-axis is PC 3) indicate the probability of cell assignment (color) to the cortical depth bins as diagrammed.
F) The smFISH reference of 16 layer astrocyte markers used for the reconstruction (top) and the predicted output of the reconstruction(bottom). Expression scaled to sum to 1 across bins (Y-axis).
G) New candidate layer astrocyte genes predicted by the spatial reconstruction model. The expression pattern of top 10 new layer astrocyte genes (X-axis) that are most similar to Chrdl1, Il33 and Id3 are shown. Expression scaled to sum to 1 across bins (Y-axis). H-J) Validation of three new candidate layer astrocyte genes using LaSTmap smFISH. Single astrocyte quantification across cortical depth in the barrel cortex at P56 (n=3 pooled tissue sections from one replicate per timepoint) (H). Close-up images of astrocytes. White outlines mark astrocyte cell areas (I,J). n=2 mice independently assayed, 3 sections imaged.
Scalebars: (I,J) 10 µm.
To determine whether astrocyte layering represents an axis of heterogeneity in the single cell transcriptome data, we then mapped the scRNA-seq data to cortical layers using the smFISH data as a spatial reference (Fig 4B). To infer the cortical layer position of cells profiled in scRNA-seq, we employed a Bayesian model (22) using 16-layer astrocyte markers profiled with LaST map smFISH as a reference. The spatial reconstruction allowed us to assign cells to different layers with high accuracy (Fig 4F, Supplementary Figure 18) and distinguish astrocyte layers in the single cell transcriptome data (Fig 4E). The two cortical astrocyte subtypes identified by unsupervised clustering of scRNA-seq were both located across multiple layers (Fig 4E, Supplementary Figure 19), consistent with the observation that they are spatially intermixed (23). We also found that scRNA-seq clusters derived using the alternative SC3 method showed a similar non-layer specific distribution (Supplementary Figure 19). Taken together, these results suggest that cortical astrocytes show at least two axes of molecular diversity: 1) layer-independent heterogeneity detected by unsupervised clustering of scRNA-seq data and 2) layer-dependent heterogeneity demonstrated by our smFISH-based approach and the spatial reconstruction of scRNA-seq data.
Spatial reconstruction of single astrocyte transcriptomes identifies new layer astrocyte genes
To expand the repertoire of astrocyte layer markers, we used spatial reconstruction of single cell transcriptomes (22). This analysis yielded 125 genes with layer-associated expression features. Layer-associated astrocyte candidate genes comprised on average 19% percent of transcripts per cell and 8% of expressed genes (Supplementary Figure 18, Kruskal–Wallis FDR-corrected p-value < 0.05); of these, 2/3 were predicted to exhibit upper and deep layer astrocyte expression (similar to Chrdl1 and Il33), while the remaining third of genes exhibited WM-like layer profiles (similar to Id3, Fig 4G, Supplementary Figure 18). We validated three of these predictions with smFISH. As shown (Fig 4H-J), the expression of the transcriptional regulator Nuclear Protein 1 (Nupr1) peaked in L2-4 astrocytes, while the enzyme Epoxide Hydrolase 2 (Ephx2) and the proteolipid Neuronatin (Nnat) showed higher expression in deep layer L5/6 astrocytes. Thus, interrogation of scRNA-seq data provides additional insight into the degrees of astrocyte layer heterogeneity in cerebral cortex.
Neuronal specification directly or indirectly determines astrocyte laminar gene expression
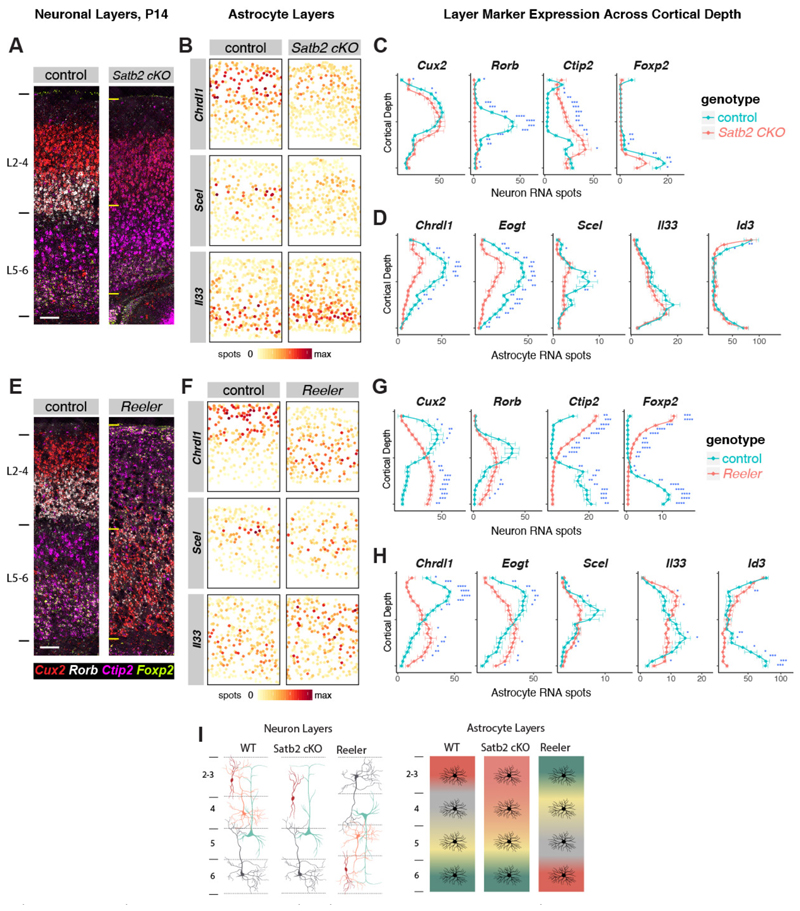
During cortical development, neuronal layers are established along a radial glial scaffold prior to astrogenesis (P0-7 (24)). To investigate whether neuronal factors regulate astrocyte layer organization, we first examined mice that lack the chromatin remodeling factor Satb2, which is required to specify superficial post-mitotic callosal neuron identity (25) Satb2 conditional knockout mice (Satb2 cKO, Emx1-cre x Satb2 floxed/floxed) are deficient in L2-4 neuron gene expression, lack callosal axon projections and ectopically express markers of L5-6 neurons in superficial cortical layers (26,27).
We verified that at P14, L4 neurons (Rorb, Pamr1) are absent in Satb2 cKO while L2-3 neurons show some ectopic deep layer gene expression (Bcl11b) (Fig 5A, C, Supplementary Figure 20). As a consequence, we found complete loss of Scel expression in astrocytes, while upper layer astrocyte genes Chrdl1 and Eogt were significantly reduced (Fig 5B, D). Deep layer astrocyte genes (Il33, Id1, Id3) were unaffected. These findings indicate that post-mitotic L4-specific neuron identity is necessary for the generation of superficial layer astrocyte identity (Fig 5I). Next, to assess deep layer astrocyte identity in relation to neurons, we used Reeler (Reln) mice, deficient for secreted Reelin, that show an inversion of radial glial polarity and neuronal layers (28). In Reln mice excitatory neurons are specified correctly but fail to migrate to their proper layers, yielding an “inside-out” cortex before birth (29). As expected (30), at P14 the expression pattern of superficial (Cux2) and deep laminar markers (Bcl11b, Foxp2) were inverted while L4 neurons (Rorb) formed aberrant clusters across cortical depth (Fig 5E-G). In addition, we found that superficial and deep astrocyte layers were inverted (Fig 5F-H). Consistent with the L4 neuron phenotype, Scel+ astrocytes were aberrantly located across cortical depth. Together, these findings suggest that cortical neuronal cues are involved in regulation of astrocyte layer development (Fig 5I).
Figure 5. Evidence that post-mitotic neuronal cues establish astrocyte layer identities.
A-D) Satb2 cKO mice show defects in upper layer neuron and astrocyte identity.
A) Images showing aberrant upper neuronal layers in the Satb2 cKO barrel cortex at P14.
B) Single astrocyte maps of layer astrocyte marker gene expression.
C-D) Quantification of cortical depth binned layer neuronal (C) and astrocyte (D) marker expression in cKO vs control.
E-H) Reeler mice show inversion of neuronal and astrocyte layers.
E) Images showing neuronal layer inversion in the Reln-/- barrel cortex at P14.
F) Single astrocyte maps of layer astrocyte marker gene expression.
G-H) Quantification of cortical depth binned layer neuronal (G) and astrocyte (H) marker expression in cKO vs control.
I) Diagrams depicting layer neuron and astrocyte changes in Satb2 cKO and Reln-/- mice.
n= 3 mice per genotype independently assayed, 5 tissue sections from each replicate imaged.
Scalebars: 100 µm. All data represent mean ± s.d. : Two-tailed Student’s t-tests were used with *P < 0.05,**P < 0.01, ***P < 0.001.
Astrocyte layer characteristics vary between cortical areas
Are astrocyte layers distinct or similar between functional cortical areas? To investigate astrocyte arealization, we quantified patterns of top layer astrocyte markers across the P14 cortex. As shown (Fig. 6), astrocyte layer gene expression varied significantly across dorsoventral and rostrocaudal cortex. Scel expression showed enrichment in L4-5 astrocytes in the somatosensory areas but was absent from the medial motor and caudal visual cortex (Fig 6A, B), a pattern that correlates closely with L4 neurons (Fig 6D). In a different pattern, Chrdl1 showed dorsoventral enrichment in L1-4 astrocytes throughout the cortex (Fig 6C). In contrast, deep layer astrocyte marker expression was enriched in medial and lateral areas across the dorsoventral axis (Il33, Id3, Fig 6D, Supplementary Figure 21, 22). Thus, astrocytes show both laminar and area heterogeneity across the cortex. Collectively, our observations on the regional heterogeneity of cortical astrocytes and their induction by neuronal signals show that glial laminae are a fundamental biomarker characteristic of cortical layer and area architecture (Fig 6E, F).
Discussion
Our findings indicate LaSTmap as a robust technique to validate single cell gene expression in situ as well as discovery of combinatorial patterns indicative of cell diversity. For example, our findings indicate a novel Rorb, Bcl11b L5 neuronal sub-type in visual cortex, as well as molecular diversity of cortical gray matter astrocytes. While a prior study indicated morphological and gene expression differences in gray matter versus white matter/L6 cortical astrocytes (7), a surprising biological finding of our study is that cortical gray matter astrocytes are organized into laminae that are distinct from classic layers of excitatory neurons. Moreover, our study identified distinct gray matter astrocyte sub-types within gray matter L2-5. Single-cell quantification of astrocyte layer genes uniquely showed peaks and troughs of expression across the superficial-to-deep cortex, as well as differences across functionally distinct cortical areas. As prior studies show that region-restricted astrocyte mRNA and protein expression are predictors of functions tailored to support of local neural circuits (8,31,32), astrocyte laminar genes indicate potential additional localized functions. Indeed, a recent study showed that expression of Chrdl1 in upper layer astrocytes is essential for local synapse regulation and regulation of developmental plasticity (33).
By introducing an in silico spatial reconstruction approach, we identified and validated further markers of layer-restricted astrocytes in situ. This indicates that identification of cell type- and region-specific markers by LaSTmap can used to interrogate other datasets for insights into diversified function, which could play into sophisticated layer-associated neural circuit activity (34). Interestingly, in a companion analysis, genes from cortical astrocytes self-organized into two classes with largely excitatory or inhibitory synapse character across layers (23). Together, these findings suggest that astrocyte laminae include a further axis of heterogeneity in terms of interactions at the synapse level in excitatory or inhibitory circuits.
International collaboratives such as the Human Cell Atlas and BRAIN initiative have generated large repositories of gene expression data, and a logical next step is to map back such expression patterns in situ within developing tissues and/or mature organs. However, spatial transcriptomic analysis in human tissue presents a particular scaling challenge. LaSTmap provides a pipeline to map quantitative gene expression in single cells across large mouse and human tissue areas. It uses commercially available probes and does not require special tissue preparation or bespoke imaging equipment. LaSTmap showed reproducibility across multiple tissue samples/sections and should be adaptable for a range of other tissues to obtain regional/qualitative and cellular/quantitative information from ISH reintegrated into 2D tissue maps. Here we focused on image acquisition from sections representing the mouse cerebral cortex and we show that use of LaSTmap is feasible in human cortex for gene expression analysis at the single cell level. Future elaboration with tissue warp integration would enable in silico 3D map construction (35).
Analysis of cortical lamination mutants suggested that neuronal cues are required, directly or indirectly, to establish astrocyte laminae, particularly in L4 where neurons and mid-layer astrocyte layers do match most closely. While results in Reeler mice are consistent with astrocyte layering following that of excitatory neuronal layering, we cannot rule out that a cortical ‘pre-pattern’ instructs astrocyte precursors to respond to neuronal signals or other mediator cells involved in astrocyte specification. Indeed, astrocyte-encoded gradient patterns are typical of cells that have undergone polarization in response to a morphogen, and upstream regulation of this process is a subject for further study.
Together, our findings indicate the diversification of astrocytes in layer patterns that diverge from excitatory neurons, indicating a composite “neuroglial” cortical architecture with higher order complexity. Our study has revealed markers for astrocyte cortical layers that could provide insight into specialized functions in development and/or disease (36).
Supplementary Material
Acknowledgments
We are grateful to Ben Barres and Sarah Teichmann for helpful discussions and comments. We are grateful to Barry Lynch and Xiao-Jun Ma for advice on RNAScope, as well as Richard Sawkins and James Hutt for technical support on imaging. We are also grateful to Eric Olson (SUNY Upstate University) for providing Reeler mice and Ralph Marcucio (UCSF) for providing the Satb2-flox mice.
Footnotes
Author Contributions
O.A.B. and D.H.R. conceived the study. O.A.B. planned the experiments. O.A.B., T.B. and S.H. performed histology and imaging experiments. O.A.B. performed the image analysis. K.P. and J.S. contributed to the histology and imaging pipeline. D.P. and O.A.B. analyzed neuron gene expression data to identify subtypes. A.B. wrote the slideSegmenter software. A.M.H.Y. and M.F.P. provided human tissue. O.A.B. and L.B.H. performed layer astrocyte purification. O.A.B. and R.K. analyzed RNASeq data. A.M., M.B. and M.G.H. generated the astrocyte scRNA-seq data. V.K. and M.H. analyzed the scRNA-seq data and performed spatial reconstruction analysis. K.S. and S.M. supported mouse work and genotyping. O.A.B. and D.H.R. wrote the manuscript with feedback from all authors.
Competing financial interests
The authors declare no competing financial interests.
Fellowships
The authors were supported by the Life Sciences Research Fellowship and the Howard Hughes Medical Institute (O.A.B.), the Wellcome Trust (T.B.), NIHR Academic Clinical Fellowship and a Wellcome Trust PhD for Clinicians Fellowship (A.M.H.Y.) and Stichting Alzheiemer Onderzoek (A.M.).
Grants
The study was supported by the Paul G. Allen Foundation Distinguished Investigator Program (E.M.U. and D.H.R.), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (D.H.R., D.G. and G. C.), BRAIN initiative (1U01 MH105991 to D.G.) and National Institute of Health (1R01 MH109912 to D.G.; P01NS08351 to D.H.R.), National Institute of Health Research and the European Union Seventh Framework (to P.H.), NINDS Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691 to G.C.), Wellcome Trust core support (M.H., O.A.B.), European Research Council (281961 to M.G.H.), Fonds Wetenschappelijk Onderzoek (G066715N and 1523014N to M.G.H.), Stichting Alzheimer Onderzoek (S#16025 to M.G.H.) and VIB Institutional Support and Tech Watch funding (to M.G.H.), Howard Hughes Medical Institute and the Wellcome Trust (to D.H.R.).
References
- 1.Molyneaux BJ, Arlotta P, Menezes JRL, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007 Jun;8(6):427–37. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 2.Freeman MR, Rowitch DH. Evolving concepts of gliogenesis: a look way back and ahead to the next 25 years. Neuron. 2013 Oct 30;80(3):613–23. doi: 10.1016/j.neuron.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, et al. Application of a Translational Profiling Approach for the Comparative Analysis of CNS Cell Types. Cell. 2008 Nov;135(4):749–62. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chai H, Diaz-Castro B, Shigetomi E, Monte E, Octeau JC, Yu X, et al. Neural Circuit- Specialized Astrocytes: Transcriptomic, Proteomic, Morphological, and Functional Evidence. Neuron. 2017 Jul; doi: 10.1016/j.neuron.2017.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.John Lin C-C, Yu K, Hatcher A, Huang T-W, Lee HK, Carlson J, et al. Identification of diverse astrocyte populations and their malignant analogs. Nat Neurosci. 2017 Feb 6; doi: 10.1038/nn.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boisvert MM, Erikson GA, Shokhirev MN, Allen NJ. The Aging Astrocyte Transcriptome from Multiple Regions of the Mouse Brain. Cell Rep. 2018 Jan 2;22(1):269–85. doi: 10.1016/j.celrep.2017.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanjakornsiripan D, Pior B-J, Kawaguchi D, Furutachi S, Tahara T, Katsuyama Y, et al. Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nat Commun. 2018 Apr 24;9(1):1623. doi: 10.1038/s41467-018-03940-3. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molofsky AV, Kelley KW, Tsai H-H, Redmond SA, Chang SM, Madireddy L, et al. Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature. 2014 May 8;509(7499):189–94. doi: 10.1038/nature13161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lein E, Borm LE, Linnarsson S. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science. 2017 Oct 6;358(6359):64–9. doi: 10.1126/science.aan6827. American Association for the Advancement of Science. [DOI] [PubMed] [Google Scholar]
- 10.Moffitt JR, Hao J, Bambah-Mukku D, Lu T, Dulac C, Zhuang X. High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc Natl Acad Sci USA, National Acad Sciences. 2016 Dec 13;113(50):14456–61. doi: 10.1073/pnas.1617699113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah S, Lubeck E, Zhou W, Cai L. In Situ Transcription Profiling of Single Cells Reveals Spatial Organization of Cells in the Mouse Hippocampus. Neuron. 2016 Oct 19;92(2):342–57. doi: 10.1016/j.neuron.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, et al. Three- dimensional intact-tissue sequencing of single-cell transcriptional states. Science. 2018 Jun 21; doi: 10.1126/science.aat5691. eaat5691, American Association for the Advancement of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Battich N, Stoeger T, Pelkmans L. Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat Methods. 2013 Oct 6;10(11):1127–33. doi: 10.1038/nmeth.2657. [DOI] [PubMed] [Google Scholar]
- 14.Nikouei K, Muñoz-Manchado AB, Hjerling-Leffler J. BCL11B/CTIP2 is highly expressed in GABAergic interneurons of the mouse somatosensory cortex. J Chem Neuroanat. 2016 Jan;71:1–5. doi: 10.1016/j.jchemneu.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016 Feb;19(2):335–46. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sosunov AA, Wu X, Tsankova NM, Guilfoyle E, McKhann GM, Goldman JE. Phenotypic heterogeneity and plasticity of isocortical and hippocampal astrocytes in the human brain. J Neurosci. 2014 Feb 5;34(6):2285–98. doi: 10.1523/JNEUROSCI.4037-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008 Jan 2;28(1):264–78. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakayama N, Han CE, Scully S, Nishinakamura R, He C, Zeni L, et al. A novel chordin- like protein inhibitor for bone morphogenetic proteins expressed preferentially in mesenchymal cell lineages. Dev Biol. 2001 Apr 15;232(2):372–87. doi: 10.1006/dbio.2001.0200. [DOI] [PubMed] [Google Scholar]
- 19.Vainchtein ID, Chin G, Cho FS, Kelley KW, Miller JG, Chien EC, et al. Astrocyte-derived interleukin-33 promotes microglial synapse engulfment and neural circuit development. Science. 2018 Feb 1; doi: 10.1126/science.aal3589. eaal3589, American Association for the Advancement of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Champliaud MF, Baden HP, Koch M, Jin W, Burgeson RE, Viel A. Gene characterization of sciellin (SCEL) and protein localization in vertebrate epithelia displaying barrier properties. Genomics. 2000 Dec 1;70(2):264–8. doi: 10.1006/geno.2000.6390. [DOI] [PubMed] [Google Scholar]
- 21.Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends in Cell Biology. 2003 Aug;13(8):410–8. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 22.Halpern KB, Shenhav R, Matcovitch-Natan O, Tóth B, Lemze D, Golan M, et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017 Feb 6; doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Batiuk MY, Martirosyan A, Voet T, Ponting CP, Belgard TG, Holt MG. Molecularly distinct astrocyte subpopulations spatially pattern the adult mouse brain. bioRxiv. 2018 May 10; 317503, Cold Spring Harbor Laboratory. [Google Scholar]
- 24.Ge W-P, Miyawaki A, Gage FH, Jan Y-N, Jan LY. Local generation of glia is a major astrocyte source in postnatal cortex. Nature. 2012 Apr 19;484(7394):376–80. doi: 10.1038/nature10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Fariñas I, Grosschedl R, et al. Satb2 Regulates Callosal Projection Neuron Identity in the Developing Cerebral Cortex. Neuron. 2008 Feb;57(3):364–77. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 26.McKenna WL, Ortiz-Londono CF, Mathew TK, Hoang K, Katzman S, Chen B. Mutual regulation between Satb2 and Fezf2 promotes subcerebral projection neuron identity in the developing cerebral cortex. Proc Natl Acad Sci USA, National Acad Sciences. 2015 Sep 15;112(37):11702–7. doi: 10.1073/pnas.1504144112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leone DP, Heavner WE, Ferenczi EA, Dobreva G, Huguenard JR, Grosschedl R, et al. Satb2 Regulates the Differentiation of Both Callosal and Subcerebral Projection Neurons in the Developing Cerebral Cortex. Cereb Cortex. 2015 Oct;25(10):3406–19. doi: 10.1093/cercor/bhu156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartfuss E. Reelin signaling directly affects radial glia morphology and biochemical maturation. DEVELOPMENT. 2003 Oct 1;130(19):4597–609. doi: 10.1242/dev.00654. [DOI] [PubMed] [Google Scholar]
- 29.Kwan KY, Sestan N, Anton ES. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. DEVELOPMENT. 2012 May;139(9):1535–46. doi: 10.1242/dev.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyle MP, Bernard A, Thompson CL, Ng L, Boe A, Mortrud M, et al. J Comp Neurol. 11. Vol. 519. Wiley Subscription Services, Inc., A Wiley Company; 2011. Aug 1, Cell-type-specific consequences of Reelin deficiency in the mouse neocortex, hippocampus, and amygdala; pp. 2061–89. [DOI] [PubMed] [Google Scholar]
- 31.Cui Y, Yang Y, Ni Z, Dong Y, Cai G, Foncelle A, et al. Astroglial Kir4.1 in the lateral habenula drives neuronal bursts in depression. Nature. 2018 Feb 14;554(7692):323–7. doi: 10.1038/nature25752. Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- 32.Kelley KW, Ben Haim L, Schirmer L, Tyzack GE, Tolman M, Miller JG, et al. Kir4.1-Dependent Astrocyte-Fast Motor Neuron Interactions Are Required for Peak Strength. Neuron. 2018 Apr 18;98(2):306–7. doi: 10.1016/j.neuron.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanco-Suarez E, Liu T-F, Kopelevich A, Allen NJ. Astrocyte-Secreted Chordin-like 1 Drives Synapse Maturation and Limits Plasticity by Increasing Synaptic GluA2 AMPA Receptors. Neuron. 2018 Dec 5;100(5):1116–1132. doi: 10.1016/j.neuron.2018.09.043. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marshel JH, Kim YS, Machado TA, Quirin S, Benson B, Kadmon J, et al. Cortical layer– specific critical dynamics triggering perception. Science. 2019 Jul 18; doi: 10.1126/science.aaw5202. eaaw5202, American Association for the Advancement of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karreman MA, Mercier L, Schieber NL, Solecki G, Allio G, Winkler F, et al. Fast and precise targeting of single tumor cells in vivo by multimodal correlative microscopy. J Cell Sci. 2016 Jan 15;129(2):444–56. doi: 10.1242/jcs.181842. The Company of Biologists Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Velmeshev D, Schirmer L, Jung D, Haeussler M, Perez Y, Mayer S, et al. Single-cell genomics identifies cell type-specific molecular changes in autism. Science. 2019 May 17;364(6441):685–9. doi: 10.1126/science.aav8130. American Association for the Advancement of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.