Abstract
Simultaneous binding of molecules by multiple binding partners is known to strongly reduce the apparent dissociation constant of the corresponding molecular complexes and can be used to realize strong, non-covalent molecular interactions. Based on this principle, efficient binding of proteins to DNA nanostructures has been previously achieved by placing several aptamers onto DNA scaffolds within close proximity. Here, we develop an approach for exploring design parameters — such as the geometric arrangement or the nanomechanical properties of the binding sites — using two-dimensional DNA origami-based nanocavities bearing aptamers with known mechanical properties at defined distances and orientations. The origami structures are labelled with barcodes, enabling large numbers of binding cavities to be investigated in parallel, under identical conditions, facilitating a direct and reliable quantitative comparison of their binding yields. We demonstrate that binding geometry and mechanical properties have a dramatic impact on origami-based multivalent binding sites, and that their optimization can be used to significantly improve their effective binding strength.
Keywords: DNA nanotechnology, multivalent binding, scaffolding, in vitro selection
DNA nanotechnology utilizes sequence-programmable self-assembly of DNA molecules for the creation of nanoscale structures and devices1. In particular the DNA origami technique2,3 has enabled the formation of almost arbitrarily shaped molecular assemblies, which can be decorated with functional molecules or nanoparticles with nanoscale precision. Origami structures are made of hundreds of short oligonucleotides, so called “staple strands”, that are hybridized to a long single-stranded “scaffold” strand in a specific manner, which leads to the folding of the scaffold into the desired shape. Each of the staples can, in principle, be further chemically modified or extended, and can thus be regarded as pixels (or, in 3D, voxels) of the resulting structure with a size of ≈ 5 nm.
A wide variety of potential applications for origami structures have already been explored. The capability of placing molecules or nanoparticles at precise distances and into specific geometries allows the biophysical characterization of biomolecular interactions under well-controlled conditions4–6 or the realization of tailored nanophotonic and plasmonic systems composed of optically active components7,8. In particular, origami structures interacting with proteins show great promise for applications in biosensing, theranostics, and biomedical nanorobotics3,9,10.
Functionalization of origami structures has been achieved by covalent modification of staple strands, e.g., with fluorophores, biotin11, or using DNA binding proteins such as zinc finger proteins12 or relaxases13. Alternatively, peptides, proteins or nanoparticles of interest have been covalently attached to DNA linker strands5,14–17, which were then hybridized to complementary staple strand extensions on the origami structures. An attractive approach to couple DNA origami structures with functional protein components is based on the utilization of DNA-antibody conjugates9,18, or protein tags such as the Snap-Tag or Halo-Tag14,19. A different approach that does not require any covalent modification at all is based on DNA aptamers20, which provide a “natural” link between DNA and protein nanotechnology.
However, typical dissociation constants for aptamer-protein interactions are of order Kd=1-100 nM, which often is not strong enough for specific applications. An obvious strategy to improve aptamer-mediated binding of proteins to origami or other DNA structures is to place several aptamers onto the scaffold in close proximity, and thus increase the “local concentration” of binding sites and use multivalency effects21,22. For instance, Yan and coworkers systematically studied the influence of the distance between two thrombin-binding aptamers placed on top of a flat origami sheet on the binding of the protein20, and found an estimated fifty-fold improvement in Kd compared to when using only a single aptamer. The improvement of binding strength via multivalent binding to aptamers arranged on DNA origami scaffolds20,23,24 or also on tile-based DNA or RNA nanostructures20,25,26 were already used in various contexts, for instance for application as anti-coagulants25,27,28.
While in previous work mainly the distance between two aptamers had been controlled with relatively simple DNA scaffolds, DNA nanostructures in principle allow for the spatial modeling of more complex artificial binding sites, in which the geometry of the molecular interactions is precisely controlled. Moreover, the mechanical properties of DNA – with single-stranded (ss) molecules being considerably more flexible than double-stranded (ds) – can be tuned up to a certain degree. Next to geometry, this allows to control the flexibility of the binding site, which is expected to have a profound influence on the overall binding strength6,21,22,29.
Design and assembly of the origami structures
In order to demonstrate and more systematically study these capabilities, we designed origami structures with a central cavity (Figure 1), within which several aptamers can be placed at multiple locations, in different geometric relationships, and with variable flexibilities. Using well-known aptamers for thrombin and streptavidin as examples, we show that these parameters dramatically impact the binding of proteins to the cavities, and that their optimization results in unprecedented binding efficiencies.
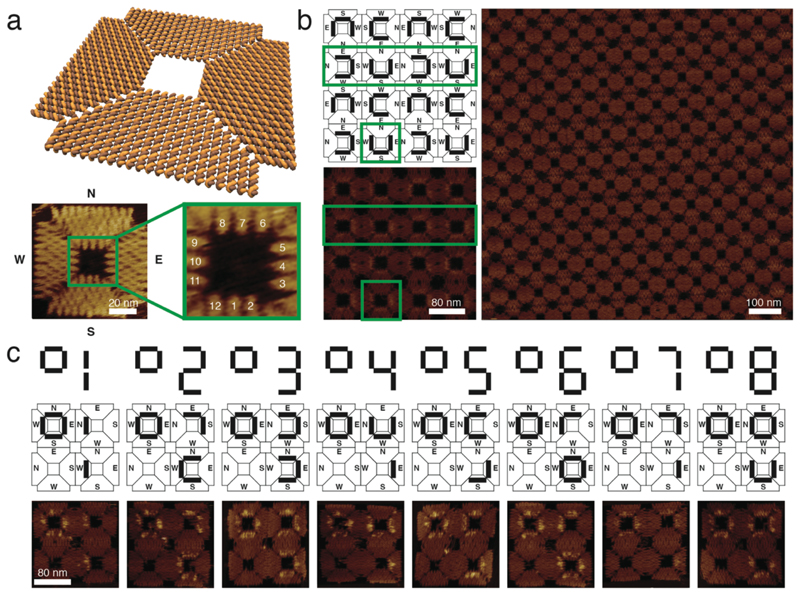
Figure 1. Design of a square-shaped DNA origami structure and its assembly into 2x2 arrays and crystals.
a, Molecular model of the DNA nanostructure. A detailed AFM image shows the origami structure and its cavity. For orientation the outer edges of the structure are addressed with North, South, East and West. The zoomed-in image of the symmetric cavity shows the 12 staples available for modification, and their respective numbers. These staples are used to arrange the DNA aptamers in a controlled orientation and position. b, Scheme and AFM images of a 2D crystal generated from square-shaped DNA origami structures. Connecting origami edges in the way shown in the scheme results in larger and more homogeneous crystals than other connection modes. U-shaped barcodes are used to visualize the connection between monomers in the crystal. Green boxes in the scheme and small AFM image highlight a single structure as well as a line of structures within the crystalline assembly. The large AFM image indicates the size and regularity of the crystals achieved in this way c, Schemes and AFM images of the 2x2 arrays and their respective barcoding. The barcodes represent digital numbers that label uniquely addressable arrays.
Our square-shaped origami platform with an approximate four-fold rotational (C4) symmetry (Figure 1) is based on a design previously used for the assembly of extended DNA origami lattices30. In this design, DNA helices extend from the center in two perpendicular directions (further denoted by North (N) – South (S) and West (W) - East (E)), which facilitates multimerization between origami structures along the helix axis via all four edges, using a combination of sticky and blunt end interactions (Supplementary Fig. 1). The scaffold strand is routed through the structure in a way that creates a symmetric square-shaped cavity in its center, which has a side length of approximately 24 nm (Figure 1a). The cavity is lined with 12 uniquely addressable staples that can be modified with arbitrary DNA aptamers.
In our experiments, we also utilized structures comprising multiple cavities, in particular finite 2 x 2 arrays with four cavities, as well as extended lattices. In spite of the nominal four-fold symmetry of the square structure, we found that multimerization along the N-S and W-E axes only resulted in elongated, one-dimensional assemblies (Supplementary Fig. 1). By contrast, two-dimensional crystal formation proceeded well when connecting the N side of one origami tile to the E side of the neighboring tile and, correspondingly, the W edge to the S edge, which presumably results in a cancellation of the intrinsic twist of the monomers (Figure 1b).
We adopted a hierarchical assembly strategy31 to generate 2 x 2 arrays (Supplementary Figs. 1 and 2) which contained different combinations of aptamers within their four cavities. This allowed us to directly compare the binding yield of different aptamer configurations under identical conditions, within a single experiment. In order to increase throughput, we introduced a barcoding scheme, which let us distinguish between up to eight different 2 x 2 arrays and their single monomers in one AFM experiment (Figure 1c). To this end, between ten and eleven staples on each of the four trapezoids comprising each square were modified with dumbbell hairpins2 to provide contrast during the AFM imaging. The barcode labels were designed in an asymmetric fashion which enabled differentiation between structures which are oriented face up or face down on the mica substrate (for a detailed description of the barcoding scheme see the Supplementary Methods).
Varying aptamer orientation in thrombin-binding cavities
We first investigated the effect of geometry and flexibility of the binding site using thrombin as a model target (Figure 2a). Thrombin has two well-characterized aptamers named TBA132 (orange in the Figure) and HD2233 (green), which target the spatially separated thrombin exosites I and II, respectively (Figure 2b). In one set of experiments, we placed the HD22 aptamer on binding position #1 of the cavity and systematically changed the position of the TBA1 aptamer by attaching it to each of the other 11 available positions (Figure 3a, b). This allowed us to screen different orientations of the two aptamers with respect to each other in 2D.
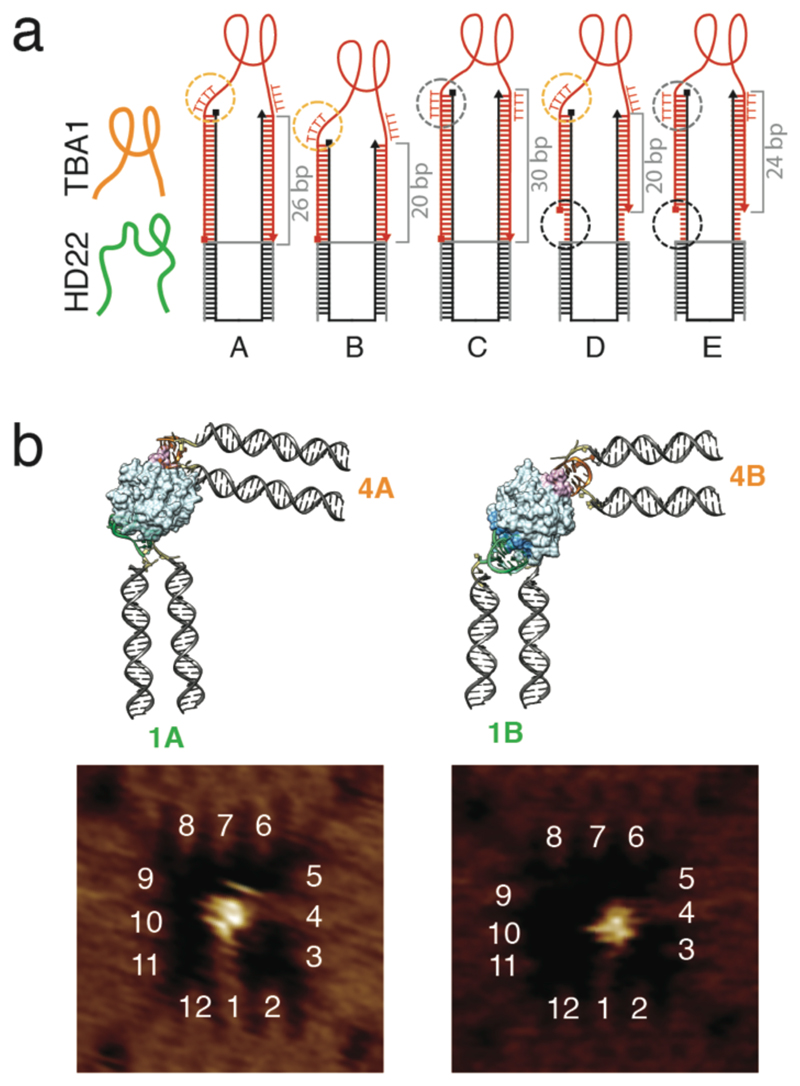
Figure 2. Binding of human thrombin to its two well-characterized aptamers with different origami linkers.
a, Configurations to assess the influence of spacer lengths and flexibilities with HD22 shown in green and TBA1 in orange. The spacers are composed of a rigid double-stranded stem and optional flexible linkers to the origami and the aptamer structure (the staple and aptamer strands are black and red, respectively). The length of the stem is varied from 20 bp to 30 bp. The connection between the stem and the aptamer is made via four thymidines either as a single-stranded (A, B, D) “aptamer linker” (orange circle) or incorporated into the stem (C, E) (grey circle). Cases D and E also have six unpaired nucleotides between the origami structure and the stem to introduce a flexible “stem linker” (black circle). b, Chimera models and AFM images of the cavity with HD22 and TBA1 designed according to case A and B, placed in position 1 and 4 respectively. It can be seen in both AFM images and chimera models that aptamers in case A need to reconfigure themselves to enable binding to their respective sites on thrombin. In the experiments, thrombin and the DNA cavities were first incubated free in solution, followed by deposition and AFM characterization of the nanostructures on mica.
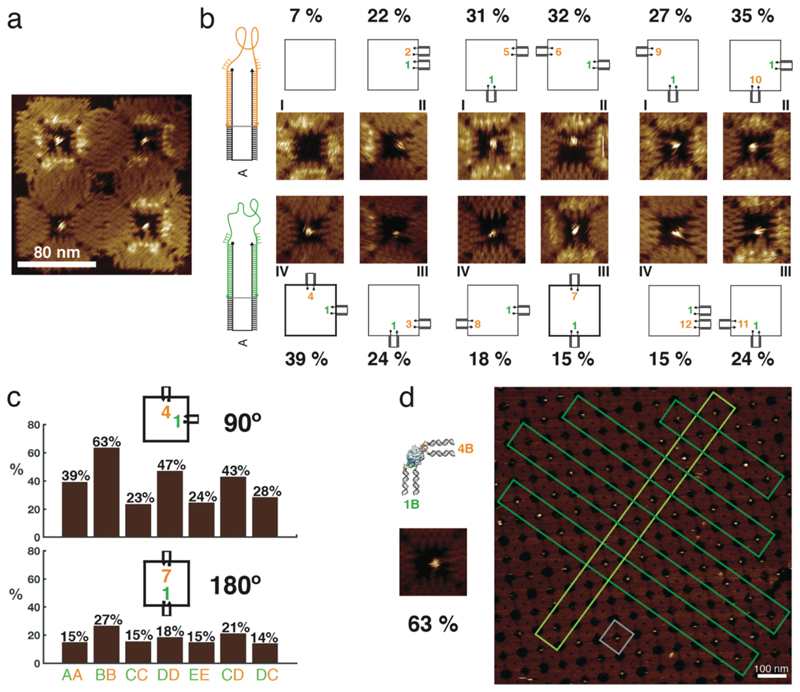
Figure 3. Binding of human thrombin to aptamers in different orientations and in lattices.
a, Example of an AFM image of a 2 x 2 array with thrombin bound in the cavities. Schemes and AFM images of the 11 different configurations of aptamers are shown. HD22 is always in position 1 and the TBA1 position varies in the cavity. All aptamers are attached as in case A. Statistical analysis shows that the 90º (1,4) configuration results in the best binding yield of these configurations. The binding assay for all the configurations was performed in one pot and imaged simultaneously. b, Yield of thrombin binding to its aptamers in 90º (1,4) and 180º (1,7) configuration. The effect of aptamer flexibility at the stem and in the vicinity of the aptamer loop for these configurations are compared. The histogram shows a comparison between the different cases – 90° (1B,4B) has the overall best yield. c, Formation of a 2D crystalline array of the DNA origami structures with the best configuration for binding to thrombin (a scheme and small AFM image of this configuration is shown on the left). The gray box highlights a single structure in the lattice and the dark and light green boxes specify linear arrangements of them within the crystal. All percentages are derived from single AFM experiments to enable direct comparison under identical conditions (n=1, therefore no error bars are given). In all experiments, thrombin and the DNA cavities were first incubated free in solution, followed by deposition and AFM characterization of the nanostructures on mica.
In order to accommodate the target protein in the cavity, the aptamers were attached to the origami frame via DNA spacers, which consisted of a rigid, double-stranded stem and optional flexible linkers between origami and stem (the “stem linker”), and between stem and aptamer (the “aptamer linker”). In the same experiments, we were thus able to test different spacer lengths and flexibilities at the stem and in the vicinity of the aptamer loop to see how these parameters influenced the binding of thrombin to the cavity (Figure 3c and Supplementary Figs. 3, 4, 5, 6).
For instance, in case A (cf. Figure 2a) aptamers were attached to the origami via a 26 bp long, rigid stem, while the aptamers were connected to the stem via a flexible stretch of four (deoxy) thymidines. By contrast, case B had a stem of only 20 bp (shorter by approximately one half-turn of a double helix), which also resulted in a rotation of the aptamer connection with respect to the stem’s helix axis (Supplementary Figs. 7 & 8). A molecular model constructed with Chimera suggests that when the TBA1 aptamer is placed on site 4, in case A the stem would have to bend outward to facilitate binding of the aptamers to the thrombin exosites, while case B would result in a slight inward bending (Figure 2b). Remarkably, the structures expected from these models is confirmed by the AFM images.
To find out the best binding configuration for case A, we compared the results for the 11 different configurations with HD22 fixed at position 1 (Figure 3a). Statistical analysis of over ≈ 200 tiles per configuration showed that an angle between the aptamers of approximately 90° in the cavity plane (position 1 and 4) is the best with a binding yield of 39%. Regardless of the design of the spacer (cases A – E listed in Figure 2a), we found that a 90° configuration (position 1 and 4) always performed better than a corresponding 180° configuration (position 1 and 7) (Figure 3c).
For the analysis and comparison of binding yields, we deliberately focused on single AFM samples, which were prepared from a single batch of origami cavities and proteins. In order to gain statistics, the same sample was imaged at multiple locations. Identification of the barcoded structures then allowed a direct comparison of different binding configurations and spacer designs under identical conditions. We also assessed the reproducibility of the binding yields obtained from different sample preparations and AFM imaging sessions. Here we found a variability on the order of only a few percent (standard error of the mean typically ≤ 3%, cf. Supplementary Fig. 10).
Influence of aptamer linker length and flexibility on binding strength
We further studied the effect of stem length and different flexibilities observed in our data by comparing various corresponding configurations. For instance, the influence of flexibility at the aptamer linker may be assessed by comparing case AA (both aptamers connected via A type spacers) with CC or case DD with EE, respectively. For the 90° configuration, flexibility at the aptamer loop improves the binding yield by about 15-20% in each case. In order to assess the influence of stem linker flexibility, one may compare the results for cases AA and DD, or CC and EE, respectively. While CC and EE have a very similar binding yield, case DD appears to perform better than AA by about 8% (Figure 3c and Supplementary Fig. 3).
Taken together, these results suggest that stem linker flexibility is helpful only when there is also flexibility at the aptamer linker. We also investigated cases of mixed linker flexibilities for the aptamers HD22 and TBA1, respectively. Our results suggest that the mode of attachment of HD22 does not change the binding yield significantly (variations of ≈ 5%). Introducing flexibility into the TBA1 aptamer linker results in a 20% improvement of binding yield, however.
In order to understand these findings on a more detailed geometrical level, we calculated the distance between the positions, where the TBA1 aptamer loop connects to the terminal bases of the stem duplexes (Supplementary Fig. 8). For a stem length of 20 bp (such as in case B), this distance is around 2.7 nm, which is close enough to the optimum distance for the TBA1 aptamer to form (≈ 1.3 nm34), when given enough flexibility by the additional thymidine bases (each thymidine is around 0.4 nm). By contrast, for cases with a 26 bp stem (case AA and CC), this distance is around 4.8 nm, which cannot be compensated by four thymidines alone. As a consequence, mechanical stress will be applied on the aptamer fold, which then results in a reduced binding yield. To further test this hypothesis, we also created a structure with six thymidines for each stem in case A, which indeed improved the binding by about 7% (Supplementary Fig. 8). We also created 2D crystals from square cavities equipped with the best performing aptamer configuration (the 90° 1B-4B configuration), which enabled the arrangement of thrombin proteins into a crystalline pattern at a high yield (Figure 3d).
We wish to note that when using the 2 x 2 arrays, we did not attempt to determine apparent Kd values for the various configurations. As in previous studies20,24, the aptamer binding yield observed in AFM on DNA origami structures is considerably lower than what would be normally expected from the Kd measured on isolated aptamers using other techniques. For instance, with a previously determined Kd ≈ 0.5 nM for the HD22 aptamer, we would naively expect a binding yield of ≈ 100% under our conditions - however, we only observe negligible binding in AFM when using a single HD22 aptamer in the origami cavity (Supplementary Fig. 9). By contrast, the barcoded 2 x 2 arrays enable a direct comparison of different configurations under identical experimental conditions, which allows us to reliably rank the aptamer configurations in terms of binding yields independent of their absolute values, and thus to optimize the binding cavities. This also avoids problems arising from uncertain effective protein concentrations and qualities, which were found to be a source of considerable variability in overall binding yield (> 10% for different protein batches).
Our findings generally correspond well with our expectations for the role of rigidity and flexibility in molecular interactions. As discussed in more detail in the Supplementary Discussion, rigid linkers are preferable, when the distance between the binding partners is well matched and does not require extensive stretching or bending. On the other hand, if the geometry of the binding site does not match the optimum molecular distances for the binding interactions, flexible linkers may facilitate binding, whereas rigid linkers will not. While a rigid binding site with ideal distances would perform even better, a sub-optimal binding site can be improved by introducing flexibility.
Streptavidin-binding origami cavities with four identical aptamers
We next applied our method to streptavidin as another model system and investigated the binding of streptavidin proteins to our aptamer-functionalized square cavities. As in the thrombin case, we first incubated streptavidin and DNA-based cavities free in solution before depositing the nanostructures on mica. Since streptavidin is a tetrameric protein, a total of four DNA aptamers were incorporated into the cavity, each of which was expected to interact with a single monomer. Several laboratories had previously selected aptamers against streptavidin, most of which shared a stem-dumbbell structure as a common secondary structural motif that is surmised to bind specifically to the same site on the protein. A minimal, 40 nt long streptavidin-binding motif (termed streptavidin aptamer SAA1 in the following) was evolved by non-homologous random recombination (NRR) and has a reported Kd of 62 ± 6.6 nM and 105 ± 21 nM with and without flanking primers, respectively35.
We investigated the binding of streptavidin to SAA1 aptamers (without flanking primers) attached to the cavity via 26 bp (case A) and 20 bp (case B) long stems and with 4 nt long aptamer linkers, respectively (Figure 4a). As for the experiments with thrombin, we folded 2 x 2 arrays with their respective barcodes, each representing a different spatial configuration of the four aptamers with different spacers (an example is shown in Figure 4b - Supplementary Figs. 11 to 15 show images of all eight barcodes).
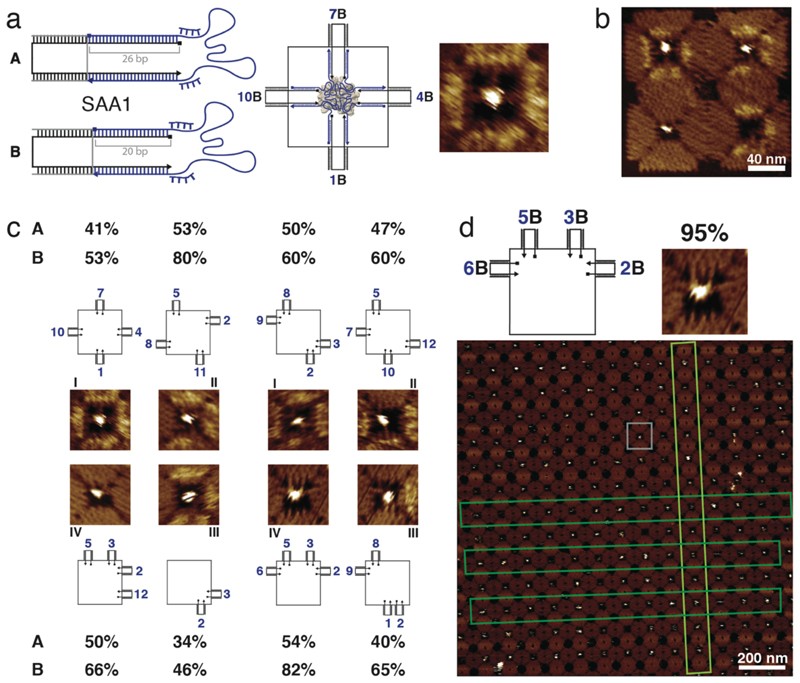
Figure 4. Binding of streptavidin to four identical aptamers in different configurations.
a, Scheme of the streptavidin aptamer SAA1 attached via spacers, each comprised of a 26 bp (case A) or a 20 bp stem (case B) and additional (dT)4 aptamer linkers. On the right, a 2D scheme and a zoomed-in AFM image of the (1,4,7,10) configuration for SAA1 in case B are shown. b, Example of an AFM image of a 2 x 2 array (barcode 3) including different configurations of four SAA1 aptamers in four cavities. c, AFM images and schemes for eight configurations of four SAA1 aptamers binding streptavidin. The binding yields for both long and short stems for these different configurations are given. Typical errors of the percentages are ≤ 3% (standard error of the mean, cf. Supplementary Fig. 15). d, Crystal formation of DNA origami structures with the best configuration (2B,3B,5B,6B) for binding to streptavidin (this configuration is shown in the scheme on top and the small examplary AFM image). In lattice shown in the large AFM image, the gray box highlights a single origami structure and the dark and light green boxes point out linear arrangements of structures within the crystal. In the context of the crystal, we observe a binding yield of ≈ 95%.
Comparison of the results for cases A and B shows that for identical spatial configuration an attachment with a 20 bp long stem always results in a higher binding yield (Figure 4c). As in the thrombin case, the longer distance between the connection points between aptamer and 26 bp stems may result in mechanical stress that disfavors proper folding of the aptamer. When using the short aptamer stem, the binding yield ranged from about 50 up to 80%, which is comparable to what previously had been reported for streptavidin-biotin interactions on origami 11. We also investigated the binding yield for cavities containing only two aptamers, which still resulted in a binding yield of 46 %.
Organizing streptavidin in two-dimensional DNA origami crystals
In the next step, we created two-dimensional DNA origami crystals using cavities equipped with the best-performing SAA1 configuration, which had an observed yield of ≈ 82%, resulting in a DNA lattice-mediated arrangement of streptavidin into an almost crystalline pattern (Figure 4d). Interestingly, with ≈ 95% the binding yield observed in the context of the origami lattice was higher than that for the isolated origami cavities. To verify the replicability of this result, we also generated two-dimensional crystals from origami cavities with a different aptamer configuration. Also in this case we found a higher binding yield for the crystals (again ≈ 95%) in comparison to cavities equipped within 2 x 2 arrays (79%, Supplementary Fig. 16). This result is somewhat surprising as with an inter-origami distance of ≈ 50 nm we would not expect any cooperative action of the neighboring origami cavities within the lattice. Conceivably, the differences are caused by different binding/unbinding kinetics to isolated origami cavities and to origami lattices, which would result in different apparent binding yields for the specific incubation and waiting times used in the experiments. Notably, we do not observe such a difference for lattices of thrombin-binding cavities.
We also performed experiments, in which we first deposited empty DNA origami lattices (equipped with the best performing aptamer configuration) onto a mica substrate, followed by addition of proteins and incubation in situ (Supplementary Fig. 17). We found that in this case the binding yield is slightly reduced compared to DNA lattices incubated in solution, but still significantly higher than for single, solution-incubated origami cavities.
To assess the replicability of the results obtained with streptavidin-binding cavities, we compared binding yields obtained from independent AFM experiments (Supplementary Fig. 17) and also studied the variability within single AFM sessions (Supplementary Fig. 18). A t-student statistical test showed that the differences measured for different configurations are indeed statistically significant, standard errors of the binding yields were found to be ≈ 3% or below. Even though we saw considerable variations among experiments with different protein batches (cf. Supplementary Fig. 16 and 17), the yields and trends were always consistent within the same batch.
Enrichment of optimized aptamer configurations from a small library of DNA structures
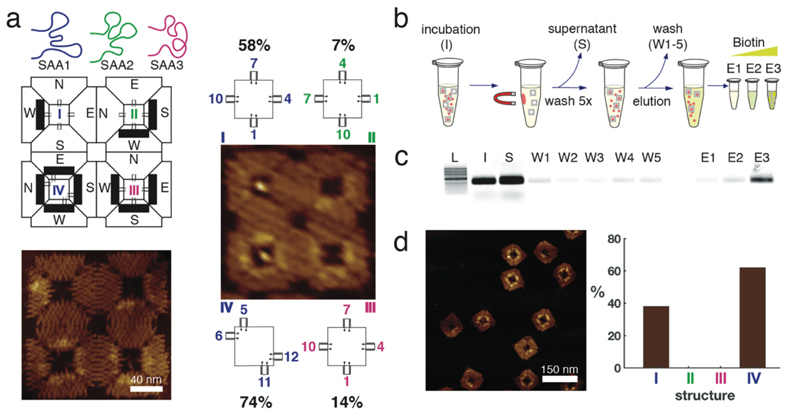
In the long run, it will be interesting to explore whether one can generalize in vitro selection and directed evolution experiments to also address the geometric and mechanical properties of ligand-presenting scaffold structures. As a step towards this goal, we next attempted to “select” cavities with good binding properties from a pool containing a mixture of different aptamer configurations. To this end, we first compared the minimal 40 nt aptamer SAA1 with two other 60 nt long aptamers SAA2 and SAA3 that had previously been selected via micromagnetic separation with reported dissociation constants of 25 nM and 50 nM, respectively36 (Figure 5a). As the binding position of these three aptamers on streptavidin is not known, for comparison we simply chose a symmetric 1,4,7,10 cavity configuration for all of them. Remarkably, even though SAA2 and SAA3 normally are better binders than SAA1, in the context of the aptamer configurations studied on 2 x 2 arrays, SAA1 clearly outperformed the two other aptamers.
Figure 5. Evaluation of the enrichment for the nanostructures with the better binding configuration to streptavidin.
a, Schematic drawing of the three aptamers tested in the 2 x 2 array. Position I and IV have the SAA1 aptamer located in configurations (1,4,7,10) and (5,6,11,12), respectively. Position II and III have SAA2 and SAA3, respectively, in the symmetric configuration (1,4,7,10). AFM images of the array with (on the right) and without (bottom left) streptavidin. The percentages refer to the binding assay done together with different configurations in figure 3. b, scheme of the selection process of the higher affinity configuration using streptavidin coated magnetic particles. On the order of 109 microbeads were incubated with 20 nM nanostructures for 30 min at room temperature (I) and the supernatant (S) was removed using a magnetic separator. Subsequently, beads were washed five times with evolution buffer (W1-W5), and eluted with increasing concentrations of biotin (E1-E3). c, Agarose gel stained with SyBr Gold showing each step of the selection protocol. L, 1Kb ladder NEB, I, initial sample, S, supernatant, W1-W5, washing steps, E1-E3 elution with 0.8, 8 and 80 µM biotin respectively. d, AFM image of the biotin-eluted sample E3. The histogram shows the yield of the nanostructures purified from E3. Nanostructures in position II and III of the 2 x 2 array were not selected by this method. Percentages are derived from single AFM experiments to enable direct comparison under identical conditions (n=1, therefore no errors/error bars are given).
We then tested if we could isolate high affinity multivalent aptamer configurations using a selection protocol based on streptavidin-coated magnetic beads. In our selection experiment individual DNA cavities from the previous experiment were incubated with an excess of streptavidin coated microparticles for 30 minutes, after which unbound nanostructures were removed via magnetic separation (Figure 5b, c). After five washing steps, the origami cavities still bound to the streptavidin beads were eluted by the addition of biotin, which is known to disturb aptamer-binding either through direct competition for the binding site or via induction of a conformational change of the protein36.
Eluted DNA origami cavities were then imaged via AFM and identified via their barcode. Exclusively structures with barcodes corresponding to configurations number I and IV were observed (Figure 5d), which is consistent with the results obtained from the quantitative analysis of binding to the 2 x 2 arrays with all four configurations (Figure 5a).
In other words, the better binding cavities were enriched in this experiment using a single round of affinity-based origami selection. It is conceivable to employ a similar selection strategy to enrich ligands that bind proteins only in a specific and controlled orientation.
Discussion
In conclusion, we have demonstrated how DNA-based scaffold structures can be used to systematically engineer multivalent binding cavities presenting multiple aptamer ligands for a target protein. In contrast to previous work on DNA-based multivalent binders, we not only controlled the distance between the binding partners, but also their orientation and the flexibility of attachment. In the case of the two model aptamer systems used – with aptamers for thrombin and streptavidin -, the interplay of these parameters was shown to result in large variations of effective binding affinity. In the best configurations, we observed a strong increase in binding yield, approaching 95% in the case of streptavidin binding to extended lattices of binding cavities. In our specific implementation of a multivalent binding scaffold, we utilized a flat, symmetric origami structure with a central quadratic cavity, in which the aptamers were presented.
Together with a barcoding scheme for AFM characterization, this structure proved to be versatile and useful for the systematic investigation of geometric and mechanical parameters. In principle, other barcoding schemes could be implemented with a larger address space, and also different readout mechanisms, which should be useful for screening larger numbers of parameters or even selecting multivalent binders de novo.
Our approach need not be restricted to flat two-dimensional cavities, however, but should also be applicable to three-dimensional origami binding pockets5. After an ideal combination of binders and binding parameters has been found, in principle the scaffold structure could be minimized to a much smaller structure, which only preserves the geometry of the binding pocket. This may prove useful in applications, where the relatively large size of standard origami structures is prohibitive.
We have also shown that from a mixture of cavities with different binding geometries of known aptamers, the best binding cavities can be selected. This opens up the exciting possibility that also new binders could be selected that complement and support each other in the context of an origami cavity. Apart from the expected increase in binding affinity and selectivity, this would also allow to select binders that bind to protein targets at specific positions or in specific orientations. Proteins could be fixed in the origami frame with a known aptamer or an antibody at one position, and additional “helper aptamers” could be selected that bind to the protein at an alternative position. With this approach it should also be possible to specifically target active sites of enzymes or allosteric proteins, which would further allow to modulate their function.
Methods
Preparation of scaffold and DNA aptamers
M13mp18 scaffold strand was provided in 100 nM aliquots in water by Florian Praetorius37. Staples were purchased from IDT in 1X TE buffer with a 100 μM concentration. DNA aptamers and modified staples in the cavity were ordered with HPLC or PAGE purification (Supplementary excel file). Details of the sequence designs of the staple strands are included in the Supplementary Information.
2 x 2 Array formation
For 2 x 2 arrays, for each tile reaction mixtures of 50 μl total volume, containing 20 nM scaffold, 140 nM of unmodified staples (a mixture of all staples except barcodes or refills (unmodified staples for the barcode positions), edge staples and the staples in the cavity), 200 nM of either barcodes or their refills for each trapezoid (N, S, E, W), 200 nM to 1 μM of unmodified staples in the cavity, 350 nM of active edge staples for each tile, 100 nM to 180 nM of double hairpin passivation (DHP) edge staples for each side, 1 μM of each modified staple in the cavity which bind to DNA aptamers and 2 μM of each DNA aptamer in 1X TE, 12.5 mM MgCl2 and 5mM KCl (= 1X folding buffer = 1X FB) were annealed using a Thermocycler Nexus X2 (Eppendorf). 2 x 2 arrays folded for streptavidin (STV) have 200 nM of modified staples in the cavity and 2 μM of the corresponding DNA aptamer. Since in the STV case the same aptamer is used at four positions in the cavity we used a lower concentration for the modified staples, so that the aptamers were still present in excess per binding site (i.e., 2 µM/4 > 200 nM). (Note that DNA aptamers should be always added in excess of the modified staples in the cavity in order to ensure complete occupation of all the aptamer binding positions in cavity). First, single DNA origami structures were folded using a temperature ramp which first held temperature at 70°C for 5 min and then decreased from 65°C to 45°C with a rate of 0.1°C per 12 seconds. After individual structures were folded, equal amounts of all the four structures comprising a 2 x 2 array were added to a new reaction tube (200 μl) and mixed properly using a vortex mixer. After mixing, the 200 μl volume was divided into two 100 μl fractions and put back into the Thermocycler. The temperature ramp used for annealing single structures into 2 x 2 arrays started at 55°C and decreased to 20°C with a rate of 0.1°C per 2 min.
2 x 2 Array purification
Folded 2 x 2 arrays were purified at 20°C using 100 kDa Amicon Ultra 0.5 ml filters (UFC5100BK). To wet the filters, first a one-step centrifugation was performed at 10000 rcf for 3 min using 400 μl of 1x TE, 12.5 mM MgCl2 and 5mM KCl (1X FB) buffer solution. Afterwards 30 μl of the 2 x 2 array sample plus 400 μl of 1X FB were added to the filter and centrifuged at 10000 rcf for 3 min. We repeated the same process several times by adding 30 μl aliquots of the 2 x 2 array sample to the same filter until all the sample was purified. Finally, the filter was inserted upside down into a 0.5 ml Amicon tube and centrifuged at 10000 rcf for 3 min to recover the purified arrays. After purification, the sample concentration was determined by measuring its absorbance at 260 nm. We also used a PEG purification protocol adjusted from Stahl et al38 and compared it with spin column filtration (Supplementary Fig. 15). In this case, a 1:1 ratio of sample and precipitation buffer (1XTE, 12.5 mM MgCl2, 5 mM KCl, 10% PEG 8000) were mixed and centrifuged at 20°C with 16000 rcf for 30 min. Afterwards the supernatant was removed and the pellet was resuspended in 1X FB.
2D crystallization of the DNA origami structures
For the formation of 2D crystals, reaction mixtures of 50 μl total volume, containing 40 nM scaffold, 200 nM of unmodified staples (a mixture of all staples except barcodes or refills, edge staples and the staples in the cavity), 200 nM of barcodes or refill stock for each trapezoid (N, S, E, W), 250 nM edge staples for the formation of 2D crystals (N-E and W-S), 200 nM of unmodified staples in the cavity, 1 μM of each modified staple in the cavity which bind to the DNA aptamers (in case of STV 200 nM of each modified staple in the cavity) and 2 μM of each DNA aptamer, all in 1x TE, 12.5 mM MgCl2 and 5mM KCl were annealed using a Thermocycler Nexus X2 (Eppendorf).
Folding of individual structures and 2D crystallization was performed in one pot. The reaction mixture was first held at 70°C for 5 min, then the temperature was ramped from 65°C to 50°C with a rate of 0.1°C per 12 seconds, followed by another temperature ramp from 50°C to 20°C with 0.1°C per 2 min or 0.1°C per 6 min.
Protein binding to the DNA origami structures
Human α-Thrombin (Haematologic Technologies) was diluted in 1X FB to a concentration of 270 nM. Streptavidin from Streptomyces avidinii (Sigma-Aldrich) was diluted in 1X FB to a concentration of 900 nM. For the incubation of 2 x 2 arrays with Streptavidin, 2 x 2 arrays at a concentration of 5.5 nM were mixed with 72 nM Streptavidin in 1X FB and incubated for 30 min to 1 hour at 37°C. For the incubation of α-Thrombin, 2 x 2 arrays at a concentration of 5 nM were mixed with 67 nM α-Thrombin in 1XFB and incubated for 30 min to 1 hour at 37°C. For the incubation of 2D crystals with Streptavidin, DNA origami crystals (25 nM) were mixed with Streptavidin at a concentration of 330 nM in 1X FB and incubated at 37°C for 30 min to one hour. For the incubation of 2D crystals with α-Thrombin, DNA origami crystals (13 nM) were mixed with α-Thrombin at a concentration of 180 nM in 1X FB and incubated at 37°C for 30 min to one hour. After incubation and before imaging, buffer solution was added to adjust the final concentration of DNA origami and proteins to 3 nM and 40 nM, respectively. For the 2D crystals, the final buffer solution has 120 mM to 150 mM NaCl in addition which facilitates the AFM imaging of crystals on mica substrate.
Atomic Force Microscopy
To image the 2 x 2 arrays, 5 μl of the prepared sample after incubation was dropped onto a freshly cleaved mica substrate and 60 μl of 1X FB was added on top for AFM imaging in liquid. Images were collected immediately afterwards. In the case of origami crystals, the whole 50 μl sample which was prepared after incubation was added onto the freshly cleaved mica and imaged immediately.
AFM imaging of the samples was performed using an Asylum Research AFM, Cypher ES (Oxford Instruments, Abingdon, UK). Two types of cantilevers were used for imaging, FASTSCAN-D from BRUKER and BL-AC40TS-C2 from OLYMPUS. In order to image the barcodes, the FASTSCAN-D cantilevers appeared to perform due to their higher resonance frequency. Moreover, it seemed helpful to image with lower scan rates to be able to resolve barcodes nicely. Typically, the scan rate was set to 3 Hz. AFM raw data have been deposited in a data repository.39
Enrichment of scaffolded aptamer configurations using streptavidin microbeads
Selection of origami-scaffolded aptamer configurations was performed using Dynabeads MyOne Streptavidin C1 (ThermoFisher Scientific) with a diameter of 1 μm. 100 μl of magnetic beads were washed three times with Washing Buffer (5 mM Tris-HCl, 0.5 mM EDTA, 1 M NaCl), and two times with 1X FB (1XTE, 12.5 mM MgCl2, 5 mM KCl) in low DNA binding 1.5 ml tubes (Eppendorf) using magnetic separation. Reactions were prepared by adding 5 nM of each origami configuration and 109 microparticles in a total volume of 100 μl of 1X FB. Samples were incubated at RT for 30 min, then placed in the magnetic separator for 1 min, after which the supernatant was carefully removed. Samples were washed 5 times (W1, W2, W3, W4, W5) in 100 μl 1X FB. We briefly incubated the samples with 100 μl of increasing concentrations of biotin (800 nM, 8 μM, and 80 μM) (Sigma-Aldrich) in 1X FB to elute the bound nanostructures from the streptavidin coated beads. Samples were further analyzed under the AFM microscope and using agarose gel electrophoresis.
Gel Electrophoresis
Samples were electrophoresed on 1.0% agarose gels containing 1X TAE and 12.5 mM MgCl2 for 1 hour at 100-V. The gels were stained with SYBR Gold for 25 min and visualized on a Typhoon FLA 9500 laser scanner (GE Healthcare). Gel images were analyzed using ImageJ.
Supplementary Material
Acknowledgements
This work was supported by the European Research Council (grant agreement no. 694410 - AEDNA) and by the Max Planck School Matter to Life. A.A.R. acknowledges support through the TUM Graduate School IGSSE. We thank Enzo Kopperger for the help with 3D modeling of the DNA origami square.
Footnotes
Author Contributions
A.A.R, S.S and F.C.S designed the experiments and wrote the manuscript. A.A.R, S.S and M.T performed the experiments and analyzed the data. AFM measurements were performed by A.A.R.
Competing Interests Statement
The authors declare no competing interests.
Data Availability
The raw data that support the plots within this paper is available in the Zenodo repository via the link: https://doi.org/10.5281/zenodo.3685804
Code Availability
Not applicable.
References
- 1.Jones MR, Seeman NC, Mirkin CA. Nanomaterials. Programmable materials and the nature of the DNA bond. Science. 2015;347 doi: 10.1126/science.1260901. 1260901. [DOI] [PubMed] [Google Scholar]
- 2.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 3.Douglas SM, et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Funke JJ, Dietz H. Placing molecules with Bohr radius resolution using DNA origami. Nat Nanotechnol. 2015;11:47. doi: 10.1038/nnano.2015.240. [DOI] [PubMed] [Google Scholar]
- 5.Sprengel A, et al. Tailored protein encapsulation into a DNA host using geometrically organized supramolecular interactions. Nat Commun. 2017;8 doi: 10.1038/ncomms14472. 14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaw A, et al. Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies. Nat Nanotechnol. 2019;14:184–190. doi: 10.1038/s41565-018-0336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuzyk A, et al. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature. 2012;483:311–314. doi: 10.1038/nature10889. [DOI] [PubMed] [Google Scholar]
- 8.Acuna G, et al. Fluorescence enhancement at docking sites of DNA-directed self-assembled nanoantennas. Science. 2012;338:506–510. doi: 10.1126/science.1228638. [DOI] [PubMed] [Google Scholar]
- 9.Shaw A, et al. Spatial control of membrane receptor function using ligand nanocalipers. Nat Methods. 2014;11:841–846. doi: 10.1038/nmeth.3025. [DOI] [PubMed] [Google Scholar]
- 10.Li S, et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat Biotechnol. 2018;36:258–264. doi: 10.1038/nbt.4071. [DOI] [PubMed] [Google Scholar]
- 11.Voigt NV, et al. Single-molecule chemical reactions on DNA origami. Nat Nanotechnol. 2010;5:200–203. doi: 10.1038/nnano.2010.5. [DOI] [PubMed] [Google Scholar]
- 12.Nakata E, et al. Zinc-finger proteins for site-specific protein positioning on DNA-origami structures. Angew Chem Int Ed. 2012;51:2421–2424. doi: 10.1002/anie.201108199. [DOI] [PubMed] [Google Scholar]
- 13.Sagredo S, et al. Orthogonal Protein Assembly on DNA Nanostructures Using Relaxases. Angew Chem Int Ed. 2016;55:4348–4352. doi: 10.1002/anie.201510313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sacca B, et al. Orthogonal protein decoration of DNA origami. Angew Chem Int Ed. 2010;49:9378–9383. doi: 10.1002/anie.201005931. [DOI] [PubMed] [Google Scholar]
- 15.Grossi G, Dalgaard Ebbesen Jepsen M, Kjems J, Andersen ES. Control of enzyme reactions by a reconfigurable DNA nanovault. Nat Commun. 2017;8:992. doi: 10.1038/s41467-017-01072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogele K, List J, Simmel FC, Pirzer T. Enhanced Efficiency of an Enzyme Cascade on DNA-Activated Silica Surfaces. Langmuir. 2018;34:14780–14786. doi: 10.1021/acs.langmuir.8b01770. [DOI] [PubMed] [Google Scholar]
- 17.Linko V, Eerikainen M, Kostiainen MA. A modular DNA origami-based enzyme cascade nanoreactor. Chem Commun. 2015;51:5351–5354. doi: 10.1039/c4cc08472a. [DOI] [PubMed] [Google Scholar]
- 18.Yamazaki T, Heddle JG, Kuzuya A, Komiyama M. Orthogonal enzyme arrays on a DNA origami scaffold bearing size-tunable wells. Nanoscale. 2014;6:9122–9126. doi: 10.1039/c4nr01598c. [DOI] [PubMed] [Google Scholar]
- 19.Sacca B, Niemeyer CM. Functionalization of DNA nanostructures with proteins. Chem Soc Rev. 2011;40:5910–5921. doi: 10.1039/c1cs15212b. [DOI] [PubMed] [Google Scholar]
- 20.Rinker S, Ke Y, Liu Y, Chhabra R, Yan H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding. Nat Nanotechnol. 2008;3:418–422. doi: 10.1038/nnano.2008.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mammen M, Choi SK, Whitesides GM. Polyvalent Interactions in Biological Systems: Implications for Design and Use of Multivalent Ligands and Inhibitors. Angew Chem Int Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 22.Fasting C, et al. Multivalency as a chemical organization and action principle. Angew Chem Int Ed. 2012;51:10472–10498. doi: 10.1002/anie.201201114. [DOI] [PubMed] [Google Scholar]
- 23.Tintore M, Gallego I, Manning B, Eritja R, Fabrega C. DNA origami as a DNA repair nanosensor at the single-molecule level. Angew Chem Int Ed. 2013;52:7747–7750. doi: 10.1002/anie.201301293. [DOI] [PubMed] [Google Scholar]
- 24.Godonoga M, et al. A DNA aptamer recognising a malaria protein biomarker can function as part of a DNA origami assembly. Sci Rep. 2016;6 doi: 10.1038/srep21266. 21266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rangnekar A, Nash JA, Goodfred B, Yingling YG, LaBean TH. Design of Potent and Controllable Anticoagulants Using DNA Aptamers and Nanostructures. Molecules. 2016;21:202–213. doi: 10.3390/molecules21020202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rangnekar A, et al. Increased anticoagulant activity of thrombin-binding DNA aptamers by nanoscale organization on DNA nanostructures. Nanomedicine. 2012;8:673–681. doi: 10.1016/j.nano.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 27.Krissanaprasit A, et al. Genetically Encoded, Functional Single-Strand RNA Origami: Anticoagulant. Ad Mater. 2019;31 doi: 10.1002/adma.201808262. e1808262. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Y, Qi X, Liu Y, Zhang F, Yan H. DNA-Nanoscaffold-Assisted Selection of Femtomolar Bivalent Human alpha-Thrombin Aptamers with Potent Anticoagulant Activity. Chembiochem. 2019;20:2494–2503. doi: 10.1002/cbic.201900265. [DOI] [PubMed] [Google Scholar]
- 29.Liese S, Netz RR. Influence of length and flexibility of spacers on the binding affinity of divalent ligands. Beilstein J Org Chem. 2015;11:804–816. doi: 10.3762/bjoc.11.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tikhomirov G, Petersen P, Qian L. Programmable disorder in random DNA tilings. Nat Nanotechnol. 2017;12:251–259. doi: 10.1038/nnano.2016.256. [DOI] [PubMed] [Google Scholar]
- 31.Tikhomirov G, Petersen P, Qian L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nature. 2017;552:67–71. doi: 10.1038/nature24655. [DOI] [PubMed] [Google Scholar]
- 32.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection Of Single-Stranded-DNA Molecules That Bind And Inhibit Human Thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 33.Tasset DM, Kubik MF, Steiner W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J Mol Biol. 1997;272:688–698. doi: 10.1006/jmbi.1997.1275. [DOI] [PubMed] [Google Scholar]
- 34.Pica A, et al. Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers. Nucl Acids Res. 2017;45:461–469. doi: 10.1093/nar/gkw1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bittker JA, Le BV, Liu DR. Nucleic acid evolution and minimization by nonhomologous random recombination. Nat Biotechnol. 2002;20:1024–1029. doi: 10.1038/nbt736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh SS, et al. Generation of highly specific aptamers via micromagnetic selection. Anal Chem. 2009;81:5490–5495. doi: 10.1021/ac900759k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kick B, Praetorius F, Dietz H, Weuster-Botz D. Efficient Production of Single- Stranded Phage DNA as Scaffolds for DNA Origami. Nano Lett. 2015;15:4672–4676. doi: 10.1021/acs.nanolett.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stahl E, Martin TG, Praetorius F, Dietz H. Facile and Scalable Preparation of Pure and Dense DNA Origami Solutions. Angew Chem Int Ed. 2014;53:12735–12740. doi: 10.1002/anie.201405991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Original data supporting the findings of the manuscript can be downloaded from. doi: 10.5281/zenodo.3685804. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data that support the plots within this paper is available in the Zenodo repository via the link: https://doi.org/10.5281/zenodo.3685804
Not applicable.