Abstract
Evolution, as we currently understand it, strikes a delicate balance between animals’ ancestral history and adaptations to their current niche. Similarities between species are generally considered inherited from a common ancestor whereas observed differences are considered as more recent evolution. Hence comparing species can provide insights into the evolutionary history. Comparative neuroimaging has recently emerged as a novel subdiscipline, which uses magnetic resonance imaging (MRI) to identify similarities and differences in brain structure and function across species. Whereas invasive histological and molecular techniques are superior in spatial resolution, they are laborious, post-mortem, and oftentimes limited to specific species. Neuroimaging, by comparison, has the advantages of being applicable across species and allows for fast, whole-brain, repeatable, and multi-modal measurements of the structure and function in living brains and post-mortem tissue. In this review, we summarise the current state of the art in comparative anatomy and function of the brain and gather together the main scientific questions to be explored in the future of the fascinating new field of brain evolution derived from comparative neuroimaging.
Introduction
Our brain is the fruit of billions of years of evolution. Evolution, as we currently understand it, strikes a delicate balance between animals’ ancestral history and adaptations to their current niche. Within each generation, discreet changes can occur across phenotypes mostly through genetic recombination (Hirsch 1963). If disadvantageous, these changes are more likely eliminated by natural selection (i.e. survival and reproduction). Accordingly, it is generally assumed that similarities between species are inherited from a common ancestor whereas observed differences are more recent occurrences (Darwin 1859; Fig. 1a). Hence comparing species gives us insight into evolutionary history, and has been applied in multiple fields where precise quantitative measurements are easy to access in large numbers including, for example skeletal structure (e.g. Dutel et al., 2019, see also Fig. 1b) or genetics (e.g. Boffelli et al., 2003; Clifen et al., 2003, see also Fig. 1c)
Fig. 1.
Comparative anatomy as a glimpse at the evolution of species. a) First evolutionary tree (courtesy of © The Complete Work of Charles Darwin Online) as depicted in the 6th edition of the origin of species (Darwin 1859), b) Comparative anatomy of the skeletal structure whereby obvious similarities can be found between a sea lion and a cheetah, suggesting a close common ancestor (Rybczynski et al., 2009, picture taken at the Museum National D’Histoire Naturelle in Paris) c) Example of comparative genetics (limited to the Preferentially Expressed Antigen In Melanoma – PRAME – gene cluster) whereby the evolutionary tree combine within (i.e. interindividual variability in genetics) and between species (comparative genetics) differences (modified from Gibbs et al., 2007). Hue level differences have been coded so that it represents the level of difference with the original phylogenetic branch (in pink). New non-human variations have been coloured in black.
Thus far, however, large numbers of observations and precise quantitative measurements are lacking for the brain across species due to its fragile, ephemeral and complex organisation. While we know a great deal about the evolution of species, the aforementioned difficulty to work with brain data has hampered progress in our understanding of the evolution of the brain. Better understanding evolution will allow for targeted studies with animal models matching the brain mechanisms in the human to its phylogenetic counterpart. Further, it may also help discover neuroprotective mechanisms allowing for resilience to disease in animals. Recent advancements in neuroimaging, with regard to both hardware and software as well as larger cohort datasets are now opening the door to embark on this new adventure of comparative brain evolution.
Brains differ in many respects across species (Haug, 1987; Stephan, 1975; Ariëns Kappers, 1909) and MRI can compare most of these levels digitally at moderate costs (Krubitzer and Kaas, 2005; Mars et al., 2018). With the advent of better hardware and higher-resolution magnetic resonance imaging sequences that allow researchers to characterise many different aspects of the same brain’s structure and function, it has become feasible to compare different species using a non-invasive repeatable multimodal method of investigation (Thiebaut de Schotten, Croxson and Mars, 2019). Another striking advantage of using magnetic resonance imaging for comparative studies is its feasibility to study large cohorts longitudinally as there is no need to sacrifice animals. Thereby, brains can be manipulated, and the effects of aging, training or lesions can be compared not only within but also between species. Finally, the non-invasive nature of the methods facilitates functional studies that better elucidate brain-behaviour interactions. Amongst the most commonly used MRI sequences to probe the structure of the brain are T1-weighted and T2-weighted scans to visualise different brain tissues (i.e. separate grey matter and white matter). Diffusion-weighted imaging (DWI) can be used to estimate microstructural properties within the white matter (Zhang et al., 2012) and to visualise the trajectory of white matter pathways (Basser, Mattiello & Le Bihan, 1994). Other sequences have been tuned to assess myelination (Glasser and Van Essen, 2011; Prasloski et al., 2012; for review see Heath et al., 2018). Using in-vivo recordings, the function of the brain can be assessed by measuring task-related blood oxygen level changes (BOLD; Ogawa et al., 1990; Logothetis et al., 2001) or modelling brain functional dynamics at rest (Fox and Raichle, 2007; Biswal, 2012). Compared to histology, MRI data gives access to a significantly greater number of specimens as it does not require the death (i.e. natural or sacrifice) of animals and allows to acquire complementary information on the structure and function of the brain within the same sample and can even be extended to measures of plasticity mechanisms using longitudinal designs. MRI is conveniently digital and shareable amongst researchers for easier replication of findings. MRI can also be mathematically modified (e.g. log transformation, Donahue et al., 2016) and reanalysed to address novel questions in already collected data (Balezeau et al., 2020). Finally, while acquiring and using MRI data in primates come with challenges with regards to collecting and harmonising data across species (see Milham et al., 2020 for a detailed discussion), MRI allows for the methodologically most similar cross-species comparisons (Thiebaut de Schotten and Zilles, 2019).
In this review, we summarise the emerging field of MRI-based neuroimaging of the primate brain evolution as well as gather the main scientific questions to be explored in the future.
Brain structure
Brain size
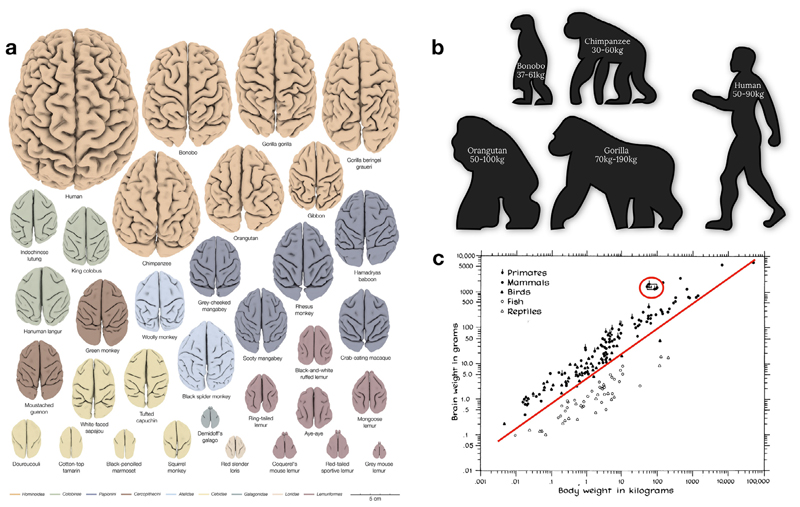
The arguably most apparent and seemingly systematic change that occurred over primate brain evolution is the increase in total brain size relative to body size (Fig. 2). The effect of evolutionary expansion in relation to brain size is, however, not equally distributed across brain structures. This disproportional composition of the brain led to investigations of allometric rules of brain evolution. The size of one region can vary consistently with the size of another structure within the brain (Butler and Hodos, 2005). In this context, allometry refers to the study of the different pace of expansion of brain regions (Montgomery et al., 2016).
Fig. 2.
Relative brain size cross-species comparison. a) 34 three-dimensional digital brain reconstructions from the brain catalogue (Heuer et al., 2019) b) Body size and weight comparison across apes c) Brain and body weight scatter plot comparison (Jerison 1975). Note that the red circle indicates human primates who deviate from the linear relationship existing between body and brain weight.
Allometric changes may provide a window for understanding the adaptations of specific neural systems in response to evolutionary pressure (Willemet, 2019; Finlay et al., 2001). In this regard, evolutionary psychology and neuroscience suggest that allometry arises from evolutionary developmental constraints, as a brain adjustment to optimise its functional organisation (Montgomery 2013; Willemet, 2015a, Montgomery et al., 2016). Along these lines, some authors have argued that similar patterns of allometric slopes across brain regions can be used to identify different grades of evolution. For instance, it has been argued that across different primate species, the prefrontal cortex shows allometric scaling with the visual cortex size. Importantly, this scaling factor is different in apes compared to monkeys (Passingham and Smaers, 2014). This finding suggests that the same developmental constraints happened across all primates, but that a major adaptive change separates monkeys from apes.
Due to the limited availability of tissue, comparative studies often rely on small samples and many studies still rely on antiquated datasets. These datasets often did not delineate different cortical territories with high accuracy, leading to fierce debates (e.g., Passingham and Smaers, 2014; Barton and Venditti, 2013). These limitations impede on our ability to assess within-species diversity accurately and might have biased our understanding of between-species differences. The ability of neuroimaging to acquire data from multiple individuals per species and imaging sequences might allow a more representative parcellation of the brain (Van Essen et al., 2011, 2016 Donahue et al., 2018). Such an approach will benefit allometric studies by providing better quantitative measurements and replicable findings as well as improve the granularity of investigations.
Gyrification
Typically, primates with smaller brains show a smoother, less convoluted brain surface than species with larger brains (see Fig. 2a; Hofmann, 2012; Heuer et al., 2019). The level of convolution of the cortex, also called gyrification index, is easily quantifiable with surface-derived MRI measurements. A convoluted cortex allows for more surface area to be packed into the limited volume within the skull (i.e. linear scaling between surface area and brain volume; Prothero and Sundsten, 1984) providing more space for grey matter cell bodies, white matter connections, and glial cells (Namba and Huttner 2017). The convolution of the cortex (i.e. gyrification) would occur because of an imbalance in the expansion of cortical (i.e. outer layer) and subcortical layers (i.e. inner layer) of the brain (Richman, 1975; adapted by Lui et al., 2011). Computational modelling of an imbalance between inner and outer layer growth success-fully reproduced a folding pattern similar to the mammalian brain (i.e. buckling shell models, Toro and Burnod 2005; Toro 2012; Bayly et al., 2013; Tallinen et al., 2014, 2016; Foubet et al., 2019). Biologically, the cortico-subcortical imbalance would be due to the tangential migration (Lui et al., 2011; Reillo et al., 2011), and the radial intercalation of neurons during development (i.e. pushing of neighbouring neurons in the outer cortical plate aside, Striedter et al., 2015). With evolutionary expansion, a disproportional expression of these biological mechanisms could explain increased cortical folding (Mota and Herculano-Houzel 2015; Amiez et al., 2019). This latter hypothesis partially implies that the dynamic relationship between brain expansion and gyrification during early stages of brain development differ across species. However, investigating such relationships across the brain developmental stages is more likely achievable by means of comparative “longitudinal” imaging during brain development, which is typically be unthinkable with standard histological methods but is feasible with MRI (Rabiei et al., 2017). While models based on the ideas described above are successful in producing random folding patterns, they do not explain why folding patterns show similarities across the brains of the same or even different species (see Fig. 2a).
Hypothetically, similarities in folding patterns could be related to preferences for neurons to migrate in cortical areas (i.e. proliferation hotspots; Retzius, 1896; Kriegstein et al., 2006) and genetically coded. If this assumption is correct, combining genetic measurements with cortical folding patterns derived from neuroimaging in the future should offer some novel insights. Recent evidence already demonstrates a significant relationship between brain surface and genetics in humans in a collaborative cross-laboratory dataset of more than 50,000 participants (Grasby et al., 2020) as well as its distribution over the brain (Valk et al., 2020). Extending genetic brain imaging MRI to other primates will not only validate this work but also shed light on the main brain surface evolutionary mechanisms.
While the placement of proliferation hotspots may as well be determined genetically, other authors pointed out that the mechanical formation of folding leads to a complex stress influencing the stiffness of the cortex (e.g., Foubet et al., 2019). The resulting differences in stiffness might potentially influence the migration of neurons during brain development (Franze, 2013). Therefore, initial folding as proposed by the buckling shell models might be sufficient to create the intricate folding pattern as seen in mammalian cortices, which in turn, may lead to the observed pattern of regional cell composition and neural connections (Heuer and Toro, 2019). Reversely, another hypothesis suggests that the stereotypical pattern of folding would come from the tension applied by the axons on the cortex (i.e. axonal tension hypothesis; Van Essen, 1997). In this theory, tangential forces that are created by the tension along obliquely oriented axonal trajectories induce folds at specific locations. As genetic molecular gradients drive axonal migration during brain development (Krubitzer, 2007; Renier et al., 2017), the future combination of cortical folding estimate, white matter diffusion imaging tractography and genetic measurements across species may reveal a tripartite relationship between these factors.
Sulcal anatomy
A prominent anatomical feature on the primate brain is the presence of folds, or sulci (Fig. 2a). Even though folding patterns may appear to vary greatly, even between individuals of the same species, sul-cal organisation is not at all random, and adheres strongly to a topographical organisation (Petrides, 2012). Anatomically, sulci often constitute borders between cytoarchitectonic areas (White et al., 1997). For instance, across human (Penfield and Boldrey, 1937) and non-human mammal brains (Ferrier, 1873), the central sulcus serves as the border between the motor cortex and the somatosensory cortex. Function-wise, a growing body of work has demonstrated precise relationships between an individual’s local sulcal morphology and the location of functional areas including the sensorimotor cortex (e.g. Zlatkina et al., 2016; Germann et al., 2020), prefrontal cortex (e.g. Loh et al., 2020; Lopez-Persem et al., 2019; Amiez and Petrides, 2018), cingulate cortex (Amiez and Petrides, 2014) and the temporal cortex (Bodin et al., 2018). This robust correspondences with the anatomical-functional organisation of the brain allows for the sulcal organisation to guide our interpretation of neuroimaging data. Especially since brain sulci are still often used as critical anatomical landmarks for navigating the brain during human and non-human primate brain surgeries. Also, most surface-based registration methods use sulci either explicitly (Auzias et al., 2013) or implicitly via geometrical maps such as curvature that are indicators of folding (Robinson et al., 2014).
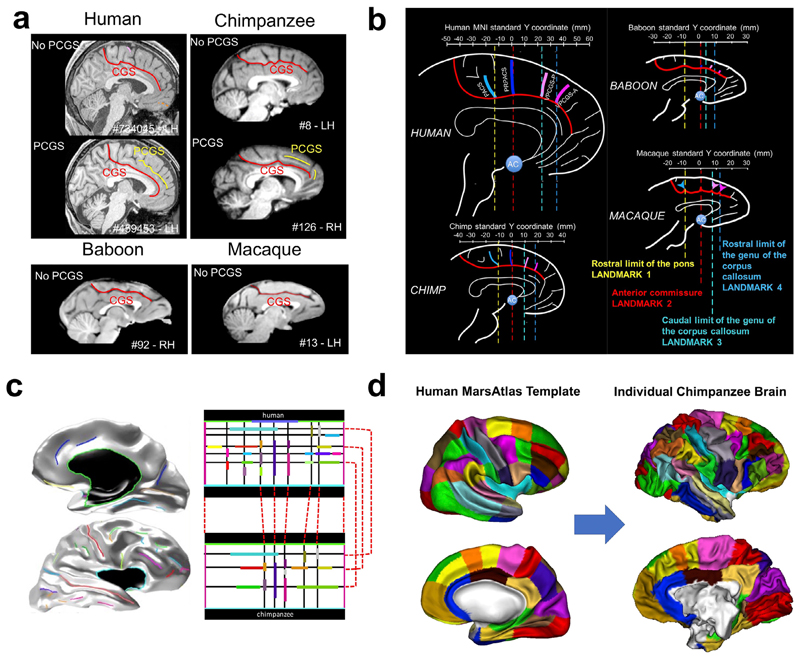
In the primate brain, some brain sulci are conserved across species. These typically include primary sulci such as the central sulcus, superior temporal sulcus, cingulate sulcus, and the calcarine sulcus. The relationship between these primary sulci and the location of anatomical/functional regions appear to be conserved across species. For instance, the somatotopic organisation along the dorsal-ventral extent of the central sulcus, as well as along the rostro-caudal extent of the cin-gulate sulcus appears to be highly conserved across the primate lineage (Procyk et al., 2016; Loh et al., 2018). This indicates that we can potentially anchor the brains of various primate species on the basis of homologous sulcal landmarks, to perform interspecies comparisons on brain structure. In Amiez et al., 2019, this principle has been implemented to reveal the evolutionary trajectories of the medial frontal cortex across macaques, baboons, chimpanzees, and humans (Fig. 3a, 3b). This work demonstrated that, unlike previously thought the paracingulate sulcus is not a human specific feature and could be observed in chimpanzees (Amiez et al., 2019; Fig. 3a). The lateralisation of these sulci, however, is only observed in humans, which suggests further hemispheric specialisation in humans since the last common ancestors to humans and great apes (Croxson et al., 2018). As shown in Fig. 3b, by aligning brains on the basis of common sulci landmarks, the evolutionary changes in the primate medial frontal cortex become apparent and quantifiable.
Fig. 3.
Sulcal anatomy for inter-primate brain comparisons. a) Emergence of the para-cingulate sulcus (PCGS) the primate medial frontal cortex (Amiez et al., 2019 : non-existing in baboons and macaques, but sometimes present for great apes and humans. b) Sulcal landmarks in the primate medial frontal cortex (Amiez et al., 2019). c) Projection of human brain sulci (left) onto a rectangular sulcal model (top right). Correspondences are defined between the human rectangular cortical sulci model and its chimpanzee equivalent (bottom right). d) Application of the model correspondences to map a human surface-based brain atlas onto an individual chimpanzee surface (Coulon et al., 2018).
Inter-species sulcal-based brain alignment can be implemented and tested using model-driven cortical surface matching (Fig. 3c, 3d). For a given species, after building a model of relative positions, orientations, and alignment of sulci in a rectangular domain, any individual cortical surface can be registered to this rectangular model, which leads to an explicit matching of different cortical surfaces based on their sulci (Auzias et al., 2013). For two different species, two different rectangular models can be built, and inter-species sulcal correspondences can then be used to define a homology between the two models (Fig. 3c). This allows for a correspondence between inter-species cortical surfaces, which can be used to compare these species and warp information from one species to the other (Coulon et al., 2018; Fig. 3d).
A key challenge to this approach is the great inter-individual variability of sulci morphology. More work will be necessary to (1) characterise the morphological variability of the various sulci in the primate brain, (2) to determine the relationship between these variations and the localisation of anatomical and functional areas, and lastly, (3) to establish the sulcal homologies between the various species. Such work would only be achievable by means of comparative MRI and data sharing in order to gather enough data to model evolutionary trends (with a minimum of three species; e.g. Balezeau et al., 2020) and quantify variability appropriately (with a minimum of 10 sample per species; Croxson et al., 2018).
Brain connectivity
Brain areas interact in order to orchestrate cognition and behaviour. Short (local) and long (distant) connections link neighbouring and remote brain regions to facilitate these interactions. Importantly, exploring connections informs about the organisational principles of the information processing in the brain (Mars et al., 2018) and is one step closer to explaining the functioning of the brain (Takemura and Thiebaut de Schotten 2020). Therefore, studying the extent to which evolutionary changes in brain structure entail specific differences in brain connectivity is a current agenda in comparative neuroscience. Neuroimaging is ideal for this purpose, as there are many imaging techniques available to elucidate various aspects of connectivity. Hence, connectivity is one of the most developed areas of comparative neuroimaging (Goulas et al., 2014; Miranda-Dominguez et al., 2014; van den Heuvel et al. 2016).
Brain connectivity can be assessed either by reconstructing structural connections (i.e. tractography based on diffusion weighted imaging) or measuring the covariation of activity across brain regions (i.e. the synchronisation of activation derived from functional MRI).
Gross anatomy
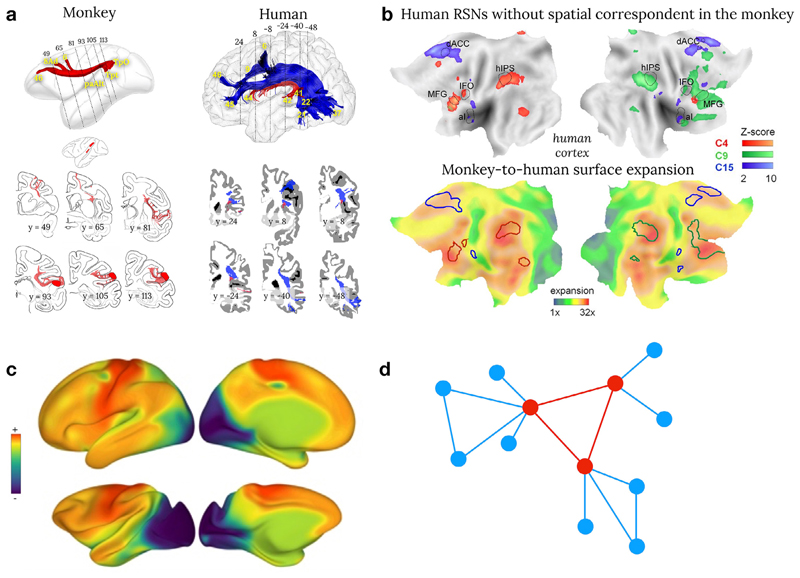
Comparative neuroimaging has revealed gross connectivity differences between primate species (Ardesch et al., 2019; Balsters et al., 2020; Xia et al., 2019). For instance, the inferior fronto-occipital connections (Thiebaut de Schotten et al., 2012; Barrett et al., 2020) and the arcuate fasciculus (see Fig. 4a, Rilling, 2008; Thiebaut de Schotten et al., 2012; Eichert et al., 2018; Barrett et al., 2020; Balezeau et al., 2020) are more prominent in humans than in monkeys. The arcuate fasciculus has sparked interest, in particular, because of its fundamental role in human language processing (Barbeau et al., 2020; Balezeau et al., 2020). In addition, the parietal lobe has been linked to uniquely human functions and the underlying white matter has some prominent structures in humans but also some unique connections in monkeys (Catani et al., 2017). The functional interactions between the frontal and parietal lobes are also more prominent in humans than in macaques (see Fig. 4b, Patel et al., 2015; Mantini et al., 2013; Mars et al., 2011) and might reflect evolutionary trends within the attentional networks (Patel et al., 2015). However, structural connections and functional connectivity are usually examined separately. Comprehensive models of connectivity will require the combination of structural-functional methods to fully grasp the modus operandi of how evolutionary pressure and adaptation might have modified the wiring and the organisational principles of information processing in the brain.
Fig 4.
Brain connectivity cross-species comparison. a) Comparison between post-mortem axonal tracing in monkeys (cases 7&9 modified from Schmahmann and Pandya, 2006) and human in vivo spherical deconvolution tractography. Common anatomical features between human and monkey are reconstructed in red whereas anatomical differences have been coloured in blue (Thiebaut de Schotten et al., 2012) b) Flat maps of the human resting state functional connectivity without correspondence with the monkey (upper row) and its correspondence to cortical expansion maps (Mantini et al., 2013) c) Preliminary comparison of the principal gradient in humans and macaques (see Brain integration section of this paper for a definition of brain gradients ; Xu et al., 2019) d) The rich club organisation of the brain where regions in red are interconnected together and a hub for regions in blue (Bullmore and Sporns 2012).
Connectivity principles
Although species-specific features exist in connectivity, the tendency of two regions to be connected respects several organisational principles across (mammalian) species (Bullmore and Sporns, 2012; Ercsey-Ravasz et al., 2013, Horvat et al., 2016, Goulas et al., 2019, Vértes et al., 2012). In particular, two regions are more likely to display interconnections if they are adjacent to each other (Human: Betzel et al., 2016; Macaque: Kaiser and Hilgetag, 2006). They will also be more connected to each other if they share a similar microstructural composition (Pandya et al., 2015; Barbas, 2015) or connections with the same regions (Song et al., 2014). Recent efforts in human neuroimaging have revealed a seemingly overarching organisation principle (Huntenburg et al., 2018), providing a window of comparison into the features that underlie the spatial arrangement of cortical areas previously reported (Abbie 1940, 1942; Sanides 1962, 1970; Brockhaus 1940; Goulas et al., 2018). While classical studies have focused mainly on cross-species similarities in this overarching organisation scheme (Margulies et al., 2016; Goulas et al., 2019) preliminary evidence suggest these differences might be an evolutionary adaptation (Fig. 4c; Xu et al., 2019).
Since the neural system is costly in energy consumption, one core principle in the neural architecture has to be the minimisation of energy costs (Bullmore and Sporns, 2012). The brain wiring, therefore, can be expected to follow rules that minimise energy costs while maintaining a set of features that are indispensable for efficient brain functioning. The “rich-club organisation” (van den Heuvel and Sporns, 2011) represents a shared feature of brain architecture that fits this description (see Fig. 4d). Within the rich-club, few nodes act as hubs between otherwise segregated nodes and thus, facilitate efficient communication across the entire network (Sporns, 2013). Additionally, hubs tend to interconnect densely with each other. However, the unique properties of hubs come at a high price (van den Heuvel and Sporns, 2011). Cortical regions that represent hubs of the rich-club tend to show high metabolic demand and are vulnerable targets for pathogenic agents (Bullmore and Sporns, 2012; Griffa and Van den Heuvel, 2018). From an evolutionary perspective, the advantages of a rich-club organisation appear to outweigh its drawbacks. Rich-club topology is a common feature amongst various species, ranging from invertebrates to primates (van den Heuvel, Bullmore and Sporns, 2016; Rubinov et al., 2016). Another example includes the comparison of the macroscale organisation of human and macaque connectivity (Goulas et al., 2014; Miranda-Dominguez 2014). However, these promising analyses should be extended to other primates in order to establish their approximate phy-logeny.
Brain function
Brain areas increase their activity when contributing to cognitive function, and this increase is detectable with task-related functional magnetic resonance imaging. The question about comparability between cognitive abilities is debated, for advanced functions such as communication (Mertz et al., 2019) or decision making (Tremblay et al., 2017; Fouragnan et al., 2019) as well as more primary functions such as episodic memory (Croxson et al., 2011; Pause et al., 2013) or even motor cognition (Borra and Luppino 2019). For instance, the superiority of chimpanzees over college students in a working memory task (Inoue and Matsuzawa 2007) is directly related to training (Cook and Wilson 2010) and indeed highlights the issue of comparability of functions across species. A recent alternative has been to compare functional activation related to the free viewing of video during fMRI measurements across primates (Mantini et al., 2012ab; Mantini et al., 2013; Russ and Leopold 2015; Sliwa and Freiwald 2017). However, species differences likely exist in the interpretation of the video limiting the interpretability of such interspecies differences. Therefore, more general features of brain function, such as brain lateralisation and brain integration measures, have been preferred in comparative neuroimaging paradigms.
Brain lateralisation
In order to conserve the speed of brain oscillations across species, a functional reorganisation might have occurred (Buzsaki, Logothetis & Singer, 2013) to compensate for interhemispheric delay related to brain size (Phillips et al., 2015). Accordingly, the inter-hemispheric independence theory suggests that during evolution, the increase in brain size led to increased functional lateralisation in order to avoid excessive conduction delays between the hemispheres (Ringo et al., 1994). Functional processing asymmetries have been derived from the neuroimaging-based study of the corpus callosum (Friedrich et al., 2017; Karolis et al., 2019; Horowitz et al., 2015) and hemispheric asymmetries (Hopkins 2015; Margiotoudi et al., 2019; Eichert et al., 2019; Marie, 2018; Thiebaut de Schotten, 2011; Amiez et al., 2019). Accordingly, an increase in functional lateralisation should be associated with a decrease of corpus callosum size or density as well as an increase in anatomical asymmetries. However, a comprehensive study of functional lateralisation across primate brains is still missing due to the scarcity of appropriate data. Lateralised patterns in tracing studies and cytoarchitectonic maps from macaques and marmosets, for instance, are rarely investigated for understandable ethical and financial reasons. As a consequence, both hemispheres are usually considered as equal limiting this line of research. An HCP-like multimodal neuroimaging approach would enable addressing brain lateralisations at the microarchitecture, connectomics, and functional levels as well as their interdependencies. For instance, in the orbitofrontal cortex, even when both hemispheres are similar at the cytoarchitectonic level (Mackey and Petrides, 2010), rs-fMRI analysis can reveal hemispheric differences in connectivity within the default-mode network (Lopez-Persem et al., 2020), in both humans and macaques. Studying the evolution of brain lateralisation in primate models would benefit from the reuse of MRI data progressively made available thanks to new open data initiatives (Milham et al., 2018, 2020). This will allow to compare larger numbers of species and to disentangle true species differences from individual noise.
Brain integration
The brain processes incoming sensory information (e.g. auditory and visual) along processing streams towards increasingly abstract and integrative, or associative, levels (Pandya and Yeterian 1990). Before comparative neuroimaging, post-mortem studies already suggested that cortical areas related to association processes are enlarged in humans compared to other primate species (Schoenemann, 2006; Van Essen and Dierker, 2007; Hrvoj-Mihic et al., 2013; Hofman, 2014). The pre-frontal lobe has been specifically explored, comparatively, with regards to expansion (Sherwood et al., 2005; Semendeferi et al., 2002, 2001 Petrides et al., 2012; Hofman 2014), cytoarchitecture (Palomero-Gallagher et al., 2013; 2019) and relative scaling of white matter (Smaers et al., 2010; Barrett et al., 2020). Bryant et al. (2019) extended this work to the visual and auditory systems. They investigated the connections of the primary and secondary processing areas of the visual and auditory cortex. In humans, chimpanzees and macaques, the connectivity of the primary visual cortex showed a retinotopic organisation with its association area. However, the primary visual cortex had additional connections to the temporal pole only in humans and chimpanzees. Quite similarly, the primary auditory cortex showed a gradual increase of connection with temporal associative cortex in chimpanzee and humans, but not in the macaque. These results suggest that a gradual expansion of the associative cortex between the auditory and visual cortices must have occurred along the chimp-human phylogenetic lineage. In line with these gradual changes, the advent of advanced MRI analyses has enabled the comparison of a variety of other properties such as the “principal gradient” of connective properties (Margulies et al., 2016; Buckner and Margulies, 2019), which summarise a specific functional connectivity signature distributed across the human brain (Huntenburg et al., 2018).
While the idea of organising the brain in terms of gradients is relatively new in neuroimaging, the concept itself has been evinced across modalities and species for more than 100 years (Vogt and Vogt, 1919; Flechsig 1920; Hopf, 1954a,b, 1955, 1956, 1968a, 1969, 1970b). In a broad perspective, comparative neuroimaging could provide a systematic assessment of the structural variation between cortical areas in multiple species. This could test whether this organisational principle is the basis of functional specialisation and evolution of brain areas, as recently suggested in rodents (Fulcher et al., 2019; Lu et al., 2012) and humans (Waymel et al., 2020).
Overall, these results are encouraging in our endeavour to understand the differences in the structure of the brain and its functions across species. However, it is important to stress that the same network in different species can have different dynamic properties and potentially different functions (Mantini et al., 2013). Consequently, similarities in brain organisation across species should not be considered as entirely equivalent brain functions.
Perspectives & future directions
Imaging the primate evolutionary tree would be a new stepping-stone for neuroscience. Access to more data across species will allow us to model the brains of common ancestors by extrapolating from the wealth of information on commonalities and divergences between species, families, orders, and classes. Having access to all levels of primate entities will allow us to create reference spaces, which in turn may grant better methods for inter- and intra-species comparisons. Ultimately, these developments can help us to form a true ‘neuroecology’ of different brains (Mars and Bryant, in press). In other words, we would be able to understand how a given brain is adapted to fit its environmental niche within the constraints of its evolutionary history.
The resources and methodologies outlined above not only allow for further investigation of primate evolution but can be extended to address crucial questions about similarities and differences in other mammalian species, including humans. The mouse is currently the most commonly used mammalian model in scientific research (Dietrich et al., 2014), and the species for which the most detailed mapping of a wide range of cellular and anatomical brain properties has been obtained. Yet, there is still limited consensus on how primates and rodents differ in terms of their brain structure and connectivity. Employing the MRI-based methodologies outlined above, Balsters et al. (2020) showed an 69-80% overlap in cortico-striatal connectivity fingerprints for humans and macaques compared to a meagre 15% overlap between humans and mice and a 31% overlap between mice and macaques. Given the prevalence of animal models in biomedical research, it is of paramount importance that neuroecology understand the differences between primates, both human and nonhuman, and other species such as rodents.
Whilst primates share some cognitive abilities such as visual perception and motor functions, as well as many emotional processes, their underlying neurobiology may differ. These similarities in function are typically investigated with the assumption that shared traits between primates (i.e. homology) are inherited from a common ancestor—divergent evolution. Most comparative neuroimaging studies investigate closely related species and hence examine divergent evolution. In more distant species, however, another process may have led to the onset of a similar function— “convergent evolution”. This so-called convergent evolution postulates that similar functions in distantly related species evolved independently from each other as a result of evolutionary pressure to adapt to similar environmental or ecological factors (i.e. homoplasy). As a consequence, both evolutionary principles, namely homology and homoplasy, can both lead to structural and/or functional similarities. Supporting evidence for homology and homoplasy is well documented in the field of genetics. For instance, many animal phyla share basic multifunctional regulatory genes such as Pax-6. This gene is involved in the development of light-sensitive cells and initiates eye formation in flies, but also frogs (Altmann et al., 1997; Halder et al., 1995). Despite the eyes of flies and frogs being homoplasies, the initiating gene is homologous. Hence, homology and homoplasy should be considered as complementary in our understanding of brain evolution and can be assessed using an extensive database of primate species only accessible through collaborative neuroimaging.
Another perspective would be to derive the brain of our phylogenetic ancestors (see Kaas, 2011; 2013 for discussion) by registering different species’ brains into a common space Although this might sound implausible, recent preliminary evidence already indicates that it is feasible (Heuer et al., 2020; https://katjaq.github.io/brainscapes). Using this possibility across proximal and more distantly related primate species may offer new insights into brain anatomy across taxonomic families, classes, and orders. Ideally, such endeavours will require the integration of multiple modalities of magnetic resonance imaging with several specimens for each primate species.
Finally, although there is much effort to identify the neural basis of species-defining cognitive functions, less research is devoted to the evolutionary processes through which those functions and their underlying neural adaptations have arisen. Questions about the evolutionary processes imply that an event, such as a genetic mutation or external evolutionary pressure, is responsible for the occurrence of adaptations. In this regard, many species share a common environment and live through competitive or collaborative interactions or in a predator-prey relationship. Therefore, the imaging of the evolutionary tree is accompanied by the intriguing opportunity to investigate the co-evolution of brain structures across interacting species and thus, investigate brain evolution from a novel neuro-ecological perspective.
Acknowledgements
This work was inspired by the #CompMRI meeting in Dusseldorf, Germany (April 11–12 2019), which was supported by the Human Brain Project. The work of RBM is supported by the Biotechnology and Biological Sciences Research Council (BBSRC) UK [BB/N019814/1] and the Netherlands Organization for Scientific Research NWO [452-13-015]. J.S. was supported by a Sir Henry Dale Wellcome Trust Fellowship (105651/Z/14/Z) and IDEXLYON “IMPULSION 2020 grant (IDEX/IMP/2020/14). The Wellcome Centre for Integrative Neu-roimaging is supported by core funding from the Wellcome Trust [203139/Z/16/Z]. MTS has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 818521). EEH was supported by National Science Foundation awards IOS-1457291 and NCS-1631563.
Footnotes
Credit Author Statement
All the authors wrote and revised the manuscript.
Data and code availability
N/A
References
- Abbie AA. The excitable cortex in Perameles, Sarcophilus, Dasyurus, Trichosurus and Wallabia (Macropus) J Comp Neurol. 1940;72(3):469–487. [Google Scholar]
- Abbie AA. Cortical lamination in a polyprotodont marsupial, Perameles nasuta. J Comp Neurol. 1942;76(3):509–536. [Google Scholar]
- Altmann CR, Chow RL, Lang RA, Hemmati-Brivanlou A. Lens Induction by Pax-6 inXenopus laevis. Dev Biol. 1997;185(1):119–123. doi: 10.1006/dbio.1997.8573. [DOI] [PubMed] [Google Scholar]
- Amiez C, Petrides M. Neuroimaging evidence of the anatomo-functional organization of the human cingulate motor areas. Cereb Cortex. 2014;24(3):563–578. doi: 10.1093/cercor/bhs329. [DOI] [PubMed] [Google Scholar]
- Amiez C, Petrides M. Functional rostro-caudal gradient in the human posterior lateral frontal cortex. Brain Structu Funct. 2018;223:1487–1499. doi: 10.1007/s00429-017-1567-z. [DOI] [PubMed] [Google Scholar]
- Amiez C, Sallet J, Hopkins WD, Meguerditchian A, Hadj-Bouziane F, Hamed SB, Petrides M. Sulcal organization in the medial frontal cortex provides insights into primate brain evolution. Nat Commun. 2019;10(1):1–14. doi: 10.1038/s41467-019-11347-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardesch DJ, Scholtens LH, Li L, Preuss TM, Rilling JK, van den Heuvel MP. Evolutionary expansion of connectivity between multimodal association areas in the human brain compared with chimpanzees. Proc Natl Acad Sci. 2019;116(14):7101–7106. doi: 10.1073/pnas.1818512116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariëns Kappers CU. The phylogenesis of the paleocortex and archicortex compared with the evolution of the visual neocortex. Arch Neurol Psychiatry. 1909;4:161–173. [Google Scholar]
- Auzias G, Lefevre J, Le Troter A, Fischer C, Perrot M, Regis J, Coulon O. Model-driven harmonic parameterization of the cortical surface: HIP-HOP. IEEE Trans Med Imaging. 2013;32(5):873–887. doi: 10.1109/TMI.2013.2241651. [DOI] [PubMed] [Google Scholar]
- Balezeau F, Wilson B, Gallardo G, Dick F, Hopkins W, Anwander A, Petkov CI. Primate auditory prototype in the evolution of the arcuate fasciculus. Nat Neurosci. 2020;23(5):611–614. doi: 10.1038/s41593-020-0623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsters JH, Zerbi V, Sallet J, Wenderoth N, Mars RB. Primate homologs of mouse cortico-striatal circuits. Elife. 2020;9:e53680. doi: 10.7554/eLife.53680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas H. General cortical and special prefrontal connections: principles from structure to function. Ann Rev Neurosci. 2015;38:269–289. doi: 10.1146/annurev-neuro-071714-033936. [DOI] [PubMed] [Google Scholar]
- Barbeau EB, Descoteaux M, Petrides M. Dissociating the white matter tracts connecting the temporo-parietal cortical region with frontal cortex using diffusion tractography. Sci Rep. 2020;10 doi: 10.1038/s41598-020-64124-y. 8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RL, Dawson M, Dyrby TB, Krug K, Ptito M, D’Arceuil H, Dell’Acqua F. Differences in frontal network anatomy across primate species. J Neurosci. 2020;40(10):2094–2107. doi: 10.1523/JNEUROSCI.1650-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, Le Bihan D. MR diffusion tensor spectroscopy and imaging. Biophys J. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton RA, Venditti C. Reply to Smaers: Getting human frontal lobes in proportion. Proc Natl Acad Sci USA. 2013;110(22):E3683–3684. doi: 10.1073/pnas.1310334110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly PV, Okamoto RJ, Xu G, Shi Y, Taber LA. A cortical folding model incorporating stress-dependent growth explains gyral wavelengths and stress patterns in the developing brain. Phys Biol. 2013;10(1) doi: 10.1088/1478-3975/10/1/016005. 016005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzel RF, Avena-Koenigsberger A, Goñi J, He Y, De Reus MA, Griffa A, Van Den Heuvel M. Generative models of the human connectome. Neuroimage. 2016;124:1054–1064. doi: 10.1016/j.neuroimage.2015.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal BB. Resting state fMRI: a personal history. Neuroimage. 2012;62(2):938–944. doi: 10.1016/j.neuroimage.2012.01.090. [DOI] [PubMed] [Google Scholar]
- Bodin C, Takerkart S, Belin P, Coulon O. Anatomo-functional correspondence in the superior temporal sulcus. Brain Struct Funct. 2018;223(1):221–232. doi: 10.1007/s00429-017-1483-2. [DOI] [PubMed] [Google Scholar]
- Boffelli D, McAuliffe J, Ovcharenko D, Lewis KD, Ovcharenko I, Pachter L, Rubin EM. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299(5611):1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- Borra E, Luppino G. Large-scale temporo–parieto–frontal networks for motor and cognitive motor functions in the primate brain. Cortex. 2019;118:19–37. doi: 10.1016/j.cortex.2018.09.024. [DOI] [PubMed] [Google Scholar]
- Brockhaus H. Cyto-and myelo-architectonics of the cortex claustralis and the clu-astrum in humans. Journal fur Psychologie und Neurologie. 1940;49:249–348. [Google Scholar]
- Bryant KL, Glasser MF, Li L, Bae JJC, Jacquez NJ, Alarcón L, Preuss TM. Organization of extrastriate and temporal cortex in chimpanzees compared to humans and macaques. Cortex. 2019;118:223–243. doi: 10.1016/j.cortex.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Margulies DS. Macroscale cortical organization and a default-like apex transmodal network in the marmoset monkey. Nat Commun. 2019;10(1):1–12. doi: 10.1038/s41467-019-09812-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. The economy of brain network organization. Nat Rev Neurosci. 2012;13(5):336–349. doi: 10.1038/nrn3214. [DOI] [PubMed] [Google Scholar]
- Butler AB, Hodos W. Comparative Vertebrate Neuroanatomy: Evolution and Adaptation. John Wiley & Sons, Inc; Hoboken, NJ, USA: 2005. [Google Scholar]
- Buzsáki G, Logothetis N, Singer W. Scaling brain size, keeping timing: evolutionary preservation of brain rhythms. Neuron. 2013;80(3):751–764. doi: 10.1016/j.neuron.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catani M, Robertsson N, Beyh A, Huynh V, de Santiago Requejo F, Howells H, Krug K. Short parietal lobe connections of the human and monkey brain. Cortex. 2017;97:339–357. doi: 10.1016/j.cortex.2017.10.022. [DOI] [PubMed] [Google Scholar]
- Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic foot-printing. Science. 2003;301(5629):71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Cook P, Wilson M. Do young chimpanzees have extraordinary working memory. Psychon Bull Rev. 2010;17(4):599–600. doi: 10.3758/PBR.17.4.599. [DOI] [PubMed] [Google Scholar]
- Coulon O, Auzias G, Lemercier P, Hopkins W. Nested cortical organization models for human and non-human primate inter-species comparisons. Proceedings of the 24th Annual Meeting of the Organization for Human Brain Mapping; Singapore. 2018. [Google Scholar]
- Croxson PL, Forkel SJ, Cerliani L, Thiebaut de Schotten M. Structural variability across the primate brain: a cross-species comparison. Cereb Cortex. 2018;28(11):3829–3841. doi: 10.1093/cercor/bhx244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. On the Origin of Species London. John Murray; UK: 1859. p. 62. [Google Scholar]
- Dietrich MR, Ankeny RA, Chen PM. Publication trends in model organism research. Genetics. 2014;198(3):787–794. doi: 10.1534/genetics.114.169714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue CJ, Glasser MF, Preuss TM, Rilling JK, Van Essen DC. Quantitative assessment of prefrontal cortex in humans relative to nonhuman primates. Proc Natl Acad Sci. 2018;115(22):E5183–E5192. doi: 10.1073/pnas.1721653115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue CJ, Sotiropoulos SN, Jbabdi S, Hernandez-Fernandez M, Behrens TE, Dyrby TB, Glasser MF. Using diffusion tractography to predict cortical connection strength and distance: a quantitative comparison with tracers in the monkey. J Neurosci. 2016;36(25):6758–6770. doi: 10.1523/JNEUROSCI.0493-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutel H, Galland M, Tafforeau P, Long JA, Fagan MJ, Janvier P, Herbin M. Neurocranial development of the coelacanth and the evolution of the sarcopterygian head. Nature. 2019;569(7757):556–559. doi: 10.1038/s41586-019-1117-3. [DOI] [PubMed] [Google Scholar]
- Eichert N, Verhagen L, Folloni D, Jbabdi S, Khrapitchev AA, Sibson NR, Mantini D, Sallet J, Mars RB. What is special about the human arcuate fasciculus? Lateralization, projections, and expansion. Cortex. 2018 doi: 10.1016/j.cortex.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichert N, Verhagen L, Folloni D, Jbabdi S, Khrapitchev AA, Sibson NR, Mars RB. What is special about the human arcuate fasciculus? Lateralization, projections, and expansion. Cortex. 2019;118:107–115. doi: 10.1016/j.cortex.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercsey-Ravasz M, Markov NT, Lamy C, Van Essen DC, Knoblauch K, Toroczkai Z, Kennedy H. A predictive network model of cerebral cortical connectivity based on a distance rule. Neuron. 2013;80(1):184–197. doi: 10.1016/j.neuron.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrier D. Experimental researches in cerebral physiology and pathology. West Rid Lunatic Asylum Med Rep. 1873;3:30–96. doi: 10.1136/bmj.1.643.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BL, Darlington RB, Nicastro N. Author’s Response: developmental structure in brain evolution. Behav Brain Sci. 2001;24(2):298–304. [PubMed] [Google Scholar]
- Flechsig P. Anatomie Des Menschlichen Gehirns und Ruckenmarks auf Myelo-genetischer Grundlage. Thieme; Leipzig: 1920. [Google Scholar]
- Fulcher BD, Murray JD, Zerbi V, Wang XJ. Multimodal gradients across mouse cortex. Proc Natl Acad Sci. 2019;116(10):4689–4695. doi: 10.1073/pnas.1814144116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foubet O, Trejo M, Toro R. Mechanical morphogenesis and the development of neocortical organisation. Cortex. 2019;118:315–326. doi: 10.1016/j.cortex.2018.03.005. [DOI] [PubMed] [Google Scholar]
- Fouragnan EF, Chau BK, Folloni D, Kolling N, Verhagen L, Klein-Flügge M, Rush-worth MF. The macaque anterior cingulate cortex translates counterfactual choice value into actual behavioral change. Nat Neurosci. 2019;22(5):797–808. doi: 10.1038/s41593-019-0375-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8(9):700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- Franze K. The mechanical control of nervous system development. Development. 2013;140(15):3069–3077. doi: 10.1242/dev.079145. [DOI] [PubMed] [Google Scholar]
- Friedrich P, Ocklenburg S, Heins N, Schlüter C, Fraenz C, Beste C, Genç E. Callosal microstructure affects the timing of electrophysiological left-right differences. Neuroimage. 2017;163:310–318. doi: 10.1016/j.neuroimage.2017.09.048. [DOI] [PubMed] [Google Scholar]
- Goulas A, Bastiani M, Bezgin G, Uylings HB, Roebroeck A, Stiers P. Comparative analysis of the macroscale structural connectivity in the macaque and human brain. PLoS Comput Biol. 2014;10(3):e1003529. doi: 10.1371/journal.pcbi.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas A, Betzel RF, Hilgetag CC. Spatiotemporal ontogeny of brain wiring. Sci Adv. 2019;5(6):eaav9694. doi: 10.1126/sciadv.aav9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas A, Zilles K, Hilgetag CC. Cortical gradients and laminar projections in mammals. Trends Neurosci. 2018;41(11):775–788. doi: 10.1016/j.tins.2018.06.003. [DOI] [PubMed] [Google Scholar]
- Germann J, Chakravarty MM, Collins DL, Petrides M. Tight coupling between morphological features of the central sulcus and somatomotor body representations: a combined anatomical and functional MRI study. Cereb Cortex. 2020 Mar 14;30(3):1843–1854. doi: 10.1093/cercor/bhz208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Batzer MA. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316(5822):222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Van Essen DC. Mapping human cortical areas in vivo based on myelin content as revealed by T1-and T2-weighted MRI. J Neurosci. 2011;31(32):11597–11616. doi: 10.1523/JNEUROSCI.2180-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasby KL, Jahanshad N, Painter JN, Colodro-Conde L, Bralten J, Hibar DP, Sha-tokhina N. The genetic architecture of the human cerebral cortex. Science. 2020;367(6484) doi: 10.1126/science.aay6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffa A, Van den Heuvel MP. Rich-club neurocircuitry: function, evolution, and vulnerability. Dialogues Clin Neurosci. 2018;20(2):121. doi: 10.31887/DCNS.2018.20.2/agriffa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267(5205):1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Haug H. Brain sizes, surfaces, and neuronal sizes of the cortex cerebri: a stereolog-ical investigation of man and his variability and a comparison with some mammals (primates, whales, marsupials, insectivores, and one elephant) Am J Anat. 1987;180(2):126–142. doi: 10.1002/aja.1001800203. [DOI] [PubMed] [Google Scholar]
- Heath F, Hurley SA, Johansen-Berg H, Sampaio-Baptista C. Advances in non-invasive myelin imaging. Dev Neurobiol. 2018;78(2):136–151. doi: 10.1002/dneu.22552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer K, Gulban OF, Bazin PL, Osoianu A, Valabregue R, Santin M, Toro R. Evolution of neocortical folding: a phylogenetic comparative analysis of MRI from 34 primate species. Cortex. 2019;118:275–291. doi: 10.1016/j.cortex.2019.04.011. [DOI] [PubMed] [Google Scholar]
- Heuer K, Kleineberg M, Dinnage R, Sherwood C, Schwartz E, Langs G, Valabregue R, Santin M, Herbin M, Toro R. A Generative Model For Primate Brain Shapes. 2020 doi: 10.5281/ZENODO.4291032. [DOI] [Google Scholar]
- Heuer K, Toro R. Role of mechanical morphogenesis in the development and evolution of the neocortex. Phys Life Rev. 2019;31:233–239. doi: 10.1016/j.plrev.2019.01.012. [DOI] [PubMed] [Google Scholar]
- Hrvoj-Mihic B, Bienvenu T, Stefanacci L, Muotri AR, Semendeferi K. Evolution, development, and plasticity of the human brain: from molecules to bones. Front Hum Neurosci. 2013;7:707. doi: 10.3389/fnhum.2013.00707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntenburg JM, Bazin PL, Margulies DS. Regul, editor. Large-scale gradients in human cortical organization. Trends Cogn Sci. 2018;22(1):21–31. doi: 10.1016/j.tics.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Hirsch J. Behavior genetics and individuality understood. Science. 1963;142(3598):1436–1442. doi: 10.1126/science.142.3598.1436. [DOI] [PubMed] [Google Scholar]
- Hofman MA. Progress in Brain Research. Vol. 195. Elsevier; 2012. Design principles of the human brain: an evolutionary perspective; pp. 373–390. [DOI] [PubMed] [Google Scholar]
- Hofman MA. Evolution of the human brain: when bigger is better. Front Neu-roanat. 2014;8:15. doi: 10.3389/fnana.2014.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf A. Die Myeloarchitektonik des Isocortex temporalis beim Menschen. J Hirn-forsch. 1954a;1:208–279. [Google Scholar]
- Hopf A. Die Myeloarchitektonik des Isocortex temporalis beim Menschen. J Hirn-forsch. 1954b;1:443–496. [Google Scholar]
- Hopf A. Über die Verteilung myeloarchitektonischer Merkmale in der isokortikalen Schla¨fenlappenrinde beim Menschen. J Hirnforsch. 1955;2:36–54. [Google Scholar]
- Hopf A. Über die Verteilung myeloarchitektonischer Merkmale in der Stirnhirn-rinde beim Menschen. J Hirnforsch. 1956;2(4):311–333. [PubMed] [Google Scholar]
- Hopf A. Photometric studies on the myeloarchitecture of the human temporal lobe. J Hirnforsch. 1968a;10(4):285–297. [PubMed] [Google Scholar]
- Hopf A. Photometric studies on the myeloarchitecture of the human parietal lobe. I Parietal region J Hirnforsch. 1969;11(4):253–265. [PubMed] [Google Scholar]
- Hopf A. Photometric studies on the myeloarchitecture of the human parietal lobe. II. Postcentral region. J Hirnforsch. 1970b;12(1):135–141. [PubMed] [Google Scholar]
- Hopkins WD, Misiura M, Pope SM, Latash EM. Behavioral and brain asymmetries in primates: a preliminary evaluation of two evolutionary hypotheses. Ann N Y Acad Sci. 2015;1359(1):65–83. doi: 10.1111/nyas.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvát S, Gamanuţ R, Ercsey-Ravasz M, Magrou L, Gamanuţ B, Van Essen DC, Kennedy H. Spatial embedding and wiring cost constrain the functional layout of the cortical network of rodents and primates. PLoS Biol. 2016;14(7):e1002512. doi: 10.1371/journal.pbio.1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Matsuzawa T. Working memory of numerals in chimpanzees. Curr Biol. 2007;17(23):R1004–R1005. doi: 10.1016/j.cub.2007.10.027. [DOI] [PubMed] [Google Scholar]
- Jerison HJ. Evolution of the brain and intelligence. Curr Anthropol. 1975;16(3):403–426. [Google Scholar]
- Kaas JH. Neocortex in early mammals and its subsequent variations. Ann N Y Acad Sci. 2011;2011 doi: 10.1111/j.1749-6632.2011.05981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas JH. The Evolution of Brains from Early Mammals to Humans. Wiley Inter-discip Rev Cogn Sci. 2013;4(1):33–45. doi: 10.1002/wcs.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser M, Hilgetag CC. Nonoptimal component placement, but short processing paths, due to long-distance projections in neural systems. PLoS Comput Biol. 2006;2(7):e95. doi: 10.1371/journal.pcbi.0020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martínez-Cerdeño V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7(11):883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- Krubitzer L. The magnificent compromise: cortical field evolution in mammals. Neuron. 2007;56(2):201–208. doi: 10.1016/j.neuron.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Krubitzer L, Kaas J. The evolution of the neocortex in mammals: how is pheno-typic diversity generated? Curr. Opin Neurobiol. 2005;15(4):444–453. doi: 10.1016/j.conb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Logothetis NK, Pauls J, Augath M, Trinath T, Oeltermann A. Neurophys-iological investigation of the basis of the fMRI signal. Nature. 2001;412(6843):150–157. doi: 10.1038/35084005. [DOI] [PubMed] [Google Scholar]
- Loh KK, Procyk E, Neveu R, Lamberton F, Hopkins WD, Petrides M, Amiez C. Cognitive control of orofacial motor and vocal responses in the ventrolateral and dorsomedial human frontal cortex. Proc Natl Acad Sci. 2020;117(9):4994–5005. doi: 10.1073/pnas.1916459117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh KK, Hadj-Bouziane F, Petrides M, Procyk E, Amiez C. Rostro-caudal organization of connectivity between cingulate motor areas and lateral frontal regions. Front Neurosci. 2018;11:753. doi: 10.3389/fnins.2017.00753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Persem A, Verhagen L, Amiez C, Petrides M, Sallet J. The human ven-tromedial prefrontal cortex: sulcal morphology and its influence on functional organization. J Neurosci. 2019;39(19):3627–3639. doi: 10.1523/JNEUROSCI.2060-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Persem A, Roumazeilles L, Folloni D, Marche K, Fouragnan EF, Khalighine-jad N, Rushworth MFS, Sallet J. Differential functional connectivity underlying asymmetric reward-related activity in human and nonhuman primates. Proc Natl Acad Sci. 2020;117(45) doi: 10.1073/pnas.2000759117. 2845262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zou Q, Gu H, Raichle ME, Stein EA, Yang Y. Rat brains also have a default mode network. Proc Natl Acad Sci. 2012;109(10):3979–3984. doi: 10.1073/pnas.1200506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146(1):18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey S, Petrides M. Quantitative demonstration of comparable architectonic areas within the ventromedial and lateral orbital frontal cortex in the human and the macaque monkey brains. Eur J Neurosci. 2010;32:1940–1950. doi: 10.1111/j.1460-9568.2010.07465.x. [DOI] [PubMed] [Google Scholar]
- Mantini D, Corbetta M, Romani GL, Orban GA, Vanduffel W. Evolutionarily novel functional networks in the human brain? J Neurosci. 2013;33(8):3259–3275. doi: 10.1523/JNEUROSCI.4392-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantini D, Hasson U, Betti V, Perrucci MG, Romani GL, Corbetta M, Vanduf-fel W. Interspecies activity correlations reveal functional correspondence between monkey and human brain areas. Nat Methods. 2012;9(3):277–282. doi: 10.1038/nmeth.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margiotoudi K, Marie D, Claidière N, Coulon O, Roth M, Nazarian B, Anton JL. Handedness in monkeys reflects hemispheric specialization within the central sulcus. An in vivo MRI study in right-and left-handed olive baboons Cortex. 2019;118:203–211. doi: 10.1016/j.cortex.2019.01.001. [DOI] [PubMed] [Google Scholar]
- Margulies DS, Ghosh SS, Goulas A, Falkiewicz M, Huntenburg JM, Langs G, Jef-feries E. Situating the default-mode network along a principal gradient of macroscale cortical organization. Proc Natl Acad Sci. 2016;113(44):12574–12579. doi: 10.1073/pnas.1608282113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie D, Roth M, Lacoste R, Nazarian B, Bertello A, Anton JL, Meguerditchian A. Left Brain Asymmetry of the Planum Temporale in a Nonhominid Primate: redefining the Origin of Brain Specialization for Language. Cereb Cortex. 2018;28(5):1808–1815. doi: 10.1093/cercor/bhx096. [DOI] [PubMed] [Google Scholar]
- Mars RB, Eichert N, Jbabdi S, Verhagen L, Rushworth MF. Connectivity and the search for specializations in the language-capable brain. Curr Opin Behav Sci. 2018a;21:19–26. doi: 10.1016/j.cobeha.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars RB, Jbabdi S, Sallet J, O’Reilly JX, Croxson PL, Olivier E, Behrens TE. Diffusion-weighted imaging tractography-based parcellation of the human parietal cortex and comparison with human and macaque resting-state functional connectivity. J Neurosci. 2011;31(11):4087–4100. doi: 10.1523/JNEUROSCI.5102-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz J, Surreault A, van de Waal E, Botting J. Primates are living links to our past: the contribution of comparative studies with wild vervet monkeys to the field of social cognition. Cortex. 2019;118:65–81. doi: 10.1016/j.cortex.2019.03.007. [DOI] [PubMed] [Google Scholar]
- Milham MP, Ai L, Koo B, Xu T, Amiez C, Balezeau F, Croxson PL. An open resource for non-human primate imaging. Neuron. 2018;100(1):61–74. doi: 10.1016/j.neuron.2018.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milham M, Petkov CI, Margulies DS, Schroeder CE, Basso MA, Belin P, Messinger A. Accelerating the evolution of nonhuman primate neuroimaging. Neuron. 2020;105(4):600–603. doi: 10.1016/j.neuron.2019.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Dominguez O, Mills BD, Grayson D, Woodall A, Grant KA, Kroenke CD, Fair DA. Bridging the gap between the human and macaque connectome: a quantitative comparison of global interspecies structure-function relationships and network topology. J Neurosci. 2014;34(16):5552–5563. doi: 10.1523/JNEUROSCI.4229-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota B, Herculano-Houzel S. Cortical folding scales universally with surface area and thickness, not number of neurons. Science. 2015;349(6243):74–77. doi: 10.1126/science.aaa9101. [DOI] [PubMed] [Google Scholar]
- Montgomery SH. The human frontal lobes: not relatively large but still disproportionately important? A commentary on Barton and Venditti Brain Behav Evol. 2013;82(3):147–149. doi: 10.1159/000354157. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Mundy NI, Barton RA. Brain evolution and development: adaptation, allometry and constraint. Proc Royal Soc B: Biol Sci. 2016;283(1838) doi: 10.1098/rspb.2016.0433. 20160433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba T, Huttner WB. Neural progenitor cells and their role in the development and evolutionary expansion of the neocortex. Wiley Interdiscip Rev: Dev Biol. 2017;6(1):e256. doi: 10.1002/wdev.256. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Lee TM, Kay AR, Tank DW. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc Natl Acad Sci. 1990;87(24):9868–9872. doi: 10.1073/pnas.87.24.9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero-Gallagher N, Zilles K, Schleicher A, Vogt BA. Cyto-and receptor architecture of area 32 in human and macaque brains. J Comp Neurol. 2013;521(14):3272–3286. doi: 10.1002/cne.23346. [DOI] [PubMed] [Google Scholar]
- Pandya D, Petrides M, Cipolloni PB. Cerebral cortex: architecture, connections, and the Dual Origin Concept. Oxford University Press; 2015. [Google Scholar]
- Pandya DN, Yeterian EH. Neurobiology of Higher Cognitive Function. The Guiford Press; New York, NY: 1990. Architecture and connections of cerebral cortex: implications for brain evolution and function. [Google Scholar]
- Passingham RE, Smaers JB. Is the prefrontal cortex especially enlarged in the human brain? Allometric relations and remapping factors Brain Behav Evol. 2014;84(2):156–166. doi: 10.1159/000365183. [DOI] [PubMed] [Google Scholar]
- Pause BM, Zlomuzica A, Kinugawa K, Mariani J, Pietrowsky R, Dere E. Perspectives on episodic-like and episodic memory. Front Behav Neurosci. 2013;7:33. doi: 10.3389/fnbeh.2013.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel GH, Yang D, Jamerson EC, Snyder LH, Corbetta M, Ferrera VP. Functional evolution of new and expanded attention networks in humans. Proc Natl Acad Sci. 2015;112(30):9454–9459. doi: 10.1073/pnas.1420395112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield W, Boldrey E. Somatic motor and sensory representation in the cerebral cortex of man as studied by electrical stimulation. Brain. 1937;60:389–440. [Google Scholar]
- Petrides M. Academic Press; 2012. The Human Cerebral cortex: An MRI Atlas of the Sulci and Gyri in MNI Stereotaxic Space. [Google Scholar]
- Petrides M, Tomaiuolo F, Yeterian EH, Pandya DN. The prefrontal cortex: comparative architectonic organization in the human and the macaque monkey brains. Cortex. 2012;48(1):46–57. doi: 10.1016/j.cortex.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Phillips KA, Stimpson CD, Smaers JB, Raghanti MA, Jacobs B, Popratiloff A, Hof PR, Sherwood CC. The corpus callosum in primates: processing speed of axons and the evolution of hemispheric asymmetry. Proc Biol Sci. 2015;282 doi: 10.1098/rspb.2015.1535. 20151535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasloski T, Rauscher A, MacKay AL, Hodgson M, Vavasour IM, Laule C, Mädler B. Rapid whole cerebrum myelin water imaging using a 3D GRASE sequence. Neuroimage. 2012;63(1):533–539. doi: 10.1016/j.neuroimage.2012.06.064. [DOI] [PubMed] [Google Scholar]
- Procyk E, Wilson CR, Stoll FM, Faraut MC, Petrides M, Amiez C. Midcingu-late motor map and feedback detection: converging data from humans and monkeys. Cereb Cortex. 2016;26(2):467–476. doi: 10.1093/cercor/bhu213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prothero JW, Sundsten JW. Folding of the cerebral cortex in mammals. Brain Behav Evol. 1984;24(2–3):152–167. doi: 10.1159/000121313. [DOI] [PubMed] [Google Scholar]
- Rabiei H, Richard F, Coulon O, Lefevre J. Local spectral analysis of the cerebral cortex: new gyrification indices. IEEE Trans Med Imaging. 2016;36(3):838–848. doi: 10.1109/TMI.2016.2633393. [DOI] [PubMed] [Google Scholar]
- Reillo I, de Juan Romero C, García-Cabezas MÁ, Borrell V. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb Cortex. 2011;21(7):1674–1694. doi: 10.1093/cercor/bhq238. [DOI] [PubMed] [Google Scholar]
- Renier N, Dominici C, Erzurumlu RS, Kratochwil CF, Rijli FM, Gaspar P, Chédo-tal A. A mutant with bilateral whisker to barrel inputs unveils somatosensory mapping rules in the cerebral cortex. Elife. 2017;6:e23494. doi: 10.7554/eLife.23494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retzius G. Das Menschenhirn K Buchdruckerei. Stockholm: 1896. [Google Scholar]
- Richman DP, Stewart RM, Hutchinson JW, Caviness VS. Mechanical model of brain convolutional development. Science. 1975;189(4196):18–21. doi: 10.1126/science.1135626. [DOI] [PubMed] [Google Scholar]
- Rilling JK, Glasser MF, Preuss TM, Ma X, Zhao T, Hu X, Behrens TE. The evolution of the arcuate fasciculus revealed with comparative DTI. Nat Neurosci. 2008;11(4):426–428. doi: 10.1038/nn2072. [DOI] [PubMed] [Google Scholar]
- Ringo JL, Doty RW, Demeter S, Simard PY. Time is of the essence: a conjecture that hemispheric specialization arises from interhemispheric conduction delay. Cereb Cortex. 1994;4(4):331–343. doi: 10.1093/cercor/4.4.331. [DOI] [PubMed] [Google Scholar]
- Robinson EC, Jbabdi S, Glasser MF, Andersson J, Burgess GC, Harms MP, Jenk-inson M. MSM: a new flexible framework for Multimodal Surface Matching. Neuroimage. 2014;100:414–426. doi: 10.1016/j.neuroimage.2014.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M. Constraints and spandrels of interareal connectomes. Nat Commun. 2016;7(1):1–11. doi: 10.1038/ncomms13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ BE, Leopold DA. Functional MRI mapping of dynamic visual features during natural viewing in the macaque. Neuroimage. 2015;109:84–94. doi: 10.1016/j.neuroimage.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybczynski N, Dawson MR, Tedford RH. A semi-aquatic Arctic mammalian carnivore from the Miocene epoch and origin of Pinnipedia. Nature. 2009;458(7241):1021–1024. doi: 10.1038/nature07985. [DOI] [PubMed] [Google Scholar]
- Sanides F. Architectonics of the human frontal lobe of the brain. With a demonstration of the principles of its formation as a reflection of phylogenetic differentiation of the cerebral cortex Monographien aus dem Gesamtgebiete der Neurologie und Psy-chiatrie. 1962;98:1. [PubMed] [Google Scholar]
- Sanides F. Evolutionary aspect of the primate neocortex. Proc 3rd Int Congr Primat, Zürich. 1970;1:92–98. [Google Scholar]
- Schmahmann JD, Pandya DN. Fiber Pathways of the Brain. Oxford University Press; New York: 2006. [Google Scholar]
- Schoenemann PT. Evolution of the size and functional areas of the human brain. Annu Rev Anthropol. 2006;35:379–406. [Google Scholar]
- Semendeferi K, Armstrong E, Schleicher A, Zilles K, Van Hoesen GW. Pre-frontal cortex in humans and apes: a comparative study of area 10. Am J Physical Anthropol: Offic Publ Am Assoc Phys Anthropol. 2001;114(3):224–241. doi: 10.1002/1096-8644(200103)114:3<224::AID-AJPA1022>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Lu A, Schenker N, Damásio H. Humans and great apes share a large frontal cortex. Nat Neurosci. 2002;5(3):272–276. doi: 10.1038/nn814. [DOI] [PubMed] [Google Scholar]
- Sherwood CC, Holloway RL, Semendeferi K, Hof PR. Is prefrontal white matter enlargement a human evolutionary specialization? Nat Neurosci. 2005;8(5):537. doi: 10.1038/nn0505-537. [DOI] [PubMed] [Google Scholar]
- Sliwa J, Freiwald WA. A dedicated network for social interaction processing in the primate brain. Science. 2017;356(6339):745–749. doi: 10.1126/science.aam6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaers JB, Schleicher A, Zilles K, Vinicius L. Frontal white matter volume is associated with brain enlargement and higher structural connectivity in anthropoid primates. PLoS One. 2010;5(2):e9123. doi: 10.1371/journal.pone.0009123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HF, Kennedy H, Wang XJ. Spatial embedding of structural similarity in the cerebral cortex. Proc Natl Acad Sci. 2014;111(46):16580–16585. doi: 10.1073/pnas.1414153111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O. Network attributes for segregation and integration in the human brain. Curr Opin Neurobiol. 2013;23(2):162–171. doi: 10.1016/j.conb.2012.11.015. [DOI] [PubMed] [Google Scholar]
- Stephan H. Allocortex. Springer; Berlin: 1975. [Google Scholar]
- Striedter GF, Srinivasan S, Monuki ES. Cortical folding: when, where, how, and why? Ann Rev Neurosci. 2015;38:291–307. doi: 10.1146/annurev-neuro-071714-034128. [DOI] [PubMed] [Google Scholar]
- Takemura H, Thiebaut de Schotten M. Perspectives given by structural connectivity bridge the gap between structure and function. Brain Struct Funct. 2020;225(4):1189–1192. doi: 10.1007/s00429-020-02080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallinen T, Chung JY, Biggins JS, Mahadevan L. Gyrification from constrained cortical expansion. Proc Natl Acad Sci. 2014;111(35):12667–12672. doi: 10.1073/pnas.1406015111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallinen T, Chung JY, Rousseau F, Girard N, Lefèvre J, Mahadevan L. On the growth and form of cortical convolutions. Nat Phys. 2016;12(6):588–593. [Google Scholar]
- Thiebaut de Schotten M, Croxson PL, Mars RB. Large-scale comparative neu-roimaging: where are we and what do we need? Cortex. 2019;118:188–202. doi: 10.1016/j.cortex.2018.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut de Schotten M, Dell’Acqua F, Valabregue R, Catani M. Monkey to human comparative anatomy of the frontal lobe association tracts. Cortex. 2012;48(1):82–96. doi: 10.1016/j.cortex.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Thiebaut De Schotten M, Dell’Acqua F, Forkel S, Simmons A, Vergani F, Murphy DG, Catani M. A lateralized brain network for visuo-spatial attention. Nat Proc. 2011;14:1245–1246. doi: 10.1038/nn.2905. [DOI] [PubMed] [Google Scholar]
- Thiebaut de Schotten M, Zilles K. Evolution of the mind and the brain. Cortex. 2019;118:1. doi: 10.1016/j.cortex.2019.06.002. [DOI] [PubMed] [Google Scholar]
- Toro R. On the possible shapes of the brain. Evol Biol. 2012;39(4):600–612. [Google Scholar]
- Toro R, Burnod Y. A morphogenetic model for the development of cortical convolutions. Cereb Cortex. 2005;15(12):1900–1913. doi: 10.1093/cercor/bhi068. [DOI] [PubMed] [Google Scholar]
- Tremblay S, Sharika KM, Platt ML. Regul, editor. Social decision-making and the brain: a comparative perspective. Trends Cogn Sci. 2017;21(4):265–276. doi: 10.1016/j.tics.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valk SL, Xu T, Margulies DS, Masouleh SK, Paquola C, Goulas A, Eickhoff SB. Shaping Brain structure: Genetic and Phylogenetic Axes of Macro Scale Organization of Cortical Thickness. BioRxiv. 2020 doi: 10.1126/sciadv.abb3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Heuvel MP, Bullmore ET, Sporns O. Comparative connectomics. Trends Cogn Sci. 2016;20(5):345–361. doi: 10.1016/j.tics.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Van Den Heuvel MP, Sporns O. Rich-club organization of the human connectome. J Neurosci. 2011;31(44):15775–15786. doi: 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature. 1997;385(6614):313–318. doi: 10.1038/385313a0. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Dierker DL. Surface-based and probabilistic atlases of primate cerebral cortex. Neuron. 2007;56(2):209–225. doi: 10.1016/j.neuron.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Donahue C, Dierker DL, Glasser MF. Parcellations and connectivity patterns in human and macaque cerebral cortex. Kennedy H, Van Essen DC, Christen Y, editors. Micro-, Meso- and Macro-Connectomics of the Brain Cham (CH) 2016:89–106. [PubMed] [Google Scholar]
- Van Essen DC, Glasser MF, Dierker DL, Harwell J, Coalson T. Parcellations and hemispheric asymmetries of human cerebral cortex analyzed on surface-based atlases. Cereb Cortex. 2012;22:2241–2262. doi: 10.1093/cercor/bhr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vértes PE, Alexander-Bloch AF, Gogtay N, Giedd JN, Rapoport JL, Bullmore ET. Simple models of human brain functional networks. Proc Natl Acad Sci USA. 2012;109(15):5868–5873. doi: 10.1073/pnas.1111738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt C, Vogt O. Allgemeine Ergebnisse unserer Hirnforschung. J Psychol Neurol. 1919;25:279–468. [Google Scholar]
- Waymel A, Friedrich P, Bastian PA, Forkel SJ, De Schotten MT. Anchoring the human olfactory system to a functional gradient. Neuroimage. 2020;216 doi: 10.1016/j.neuroimage.2020.116863. 116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White LE, Andrews TJ, Hulette C, Richards A, Groelle M, Paydarfar J, Purves D. Structure of the human sensorimotor system. I: morphology and cytoarchitec-ture of the central sulcus. Cereb Cortex. 1997;7(1):18–30. doi: 10.1093/cercor/7.1.18. [DOI] [PubMed] [Google Scholar]
- Willemet R. Allometry unleashed: an adaptationist approach of brain scaling in mammalian evolution. PeerJ Preprints. 2019;7:e27872v1. [Google Scholar]
- Willemet R. Commentary: greater addition of neurons to the olfactory bulb than to the cerebral cortex of eulipotyphlans but not rodents, afrotherians or primates. Front Neuroanat. 2015;9:84. doi: 10.3389/fnana.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Fan L, Cheng C, Yao R, Deng H, Zhao D, Jiang T. Interspecies differences in the connectivity of ventral striatal components between humans and macaques. Front Neurosci. 2019;13:623. doi: 10.3389/fnins.2019.00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Nenning KH, Schwartz E, Hong SJ, Vogelstein JT, Fair DA, Langs G. Cross-species functional alignment reveals evolutionary hierarchy within the connectome. Neuroimage. 2019;223 doi: 10.1016/j.neuroimage.2020.117346. 117346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage. 2012;61(4):1000–1016. doi: 10.1016/j.neuroimage.2012.03.072. [DOI] [PubMed] [Google Scholar]
- Zlatkina V, Amiez C, Petrides M. The postcentral sulcal complex and the transverse postcentral sulcus and their relation to sensorimotor functional organization. Eur J Neurosci. 2016;43(10):1268–1283. doi: 10.1111/ejn.13049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
N/A