Proinflammatory lipid receptor S1RP3 drives myeloid differentiation in HSC as well as LSC and marks a specific subtype of less-primitive LSC, serving as a promising biomarker and therapeutic target for AML.
Abstract
Acute myeloid leukemia (AML) is a caricature of normal hematopoiesis driven from leukemia stem cells (LSC) that share some hematopoietic stem cell (HSC) programs including responsiveness to inflammatory signaling. Although inflammation dysregulates mature myeloid cells and influences stemness programs and lineage determination in HSCs by activating stress myelopoiesis, such roles in LSCs are poorly understood. Here, we show that S1PR3, a receptor for the bioactive lipid sphingosine-1-phosphate, is a central regulator that drives myeloid differentiation and activates inflammatory programs in both HSCs and LSCs. S1PR3-mediated inflammatory signatures varied in a continuum from primitive to mature myeloid states across cohorts of patients with AML, each with distinct phenotypic and clinical properties. S1PR3 was high in LSCs and blasts of mature myeloid samples with linkages to chemosensitivity, whereas S1PR3 activation in primitive samples promoted LSC differentiation leading to eradication. Our studies open new avenues for therapeutic target identification specific for each AML subset.
Significance:
S1PR3 is a novel regulator of myeloid fate in normal hematopoiesis that is heterogeneously expressed in AML. S1PR3 marks a subset of less primitive AML cases with a distinct inflammatory signature and therefore has clinical implications as both a therapeutic target and a biomarker to distinguish primitive from mature AML.
See related commentary by Yang et al., p. 3.
This article is highlighted in the In This Issue feature, p. 1
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease characterized by impaired myeloid differentiation that is hierarchically organized akin to the normal blood system (1, 2, 3, 4). Leukemia stem cells (LSC) possess properties of self-renewal and lineage differentiation and are responsible for long-term clonal propagation in AML, similar to their normal long-term hematopoietic stem cell (LT-HSC) counterpart that sustains lifelong blood production (2). LSCs are often resistant to standard chemotherapy and are responsible for clinical relapse (2, 5, 6, 7). AML is a cellular hierarchy, driven by LSCs that share many LT-HSC stemness properties but remain impaired for normal myeloid lineage development (3, 4, 7). How myeloid fate networks at the stem cell level become perturbed during leukemogenesis remains poorly understood. Most studies of how hematopoietic stem cells (HSC) choose to differentiate into one lineage versus another have focused on the role of cytokines and the transcriptional factor networks that they engage (8, 9). In parallel, investigations of AML primarily focus on how cytokines and their signaling networks are impaired by AML driver oncogenes (3, 8, 10, 11). However, compelling evidence is emerging that lineage determination is not solely the consequence of lineage-specific cytokine exposure and signaling, but rather that metabolites generated from many pathways can play a role (12). Cellular metabolism is recognized as a hallmark of cancer and is known to be distinct between HSCs and their downstream progenitors (12, 13). We recently showed that sphingolipid composition is diverse across the human hematopoietic hierarchy and uncovered a novel role for sphingolipid metabolism in determining HSC fate (14). Sphingosine-1-phosphate (S1P) is a potent bioactive sphingolipid crucial for systemic health, particularly in disorders associated with inflammation (15, 16, 17). As an obligate ligand for a family of five G-protein–coupled receptors (S1PR1–5), S1P plays pleiotropic roles in cellular proliferation, survival, and migration—processes that are dysregulated in inflammatory diseases (15, 16, 18, 19). Notably, within the murine system, S1P signaling via S1PR1 regulates B lymphopoiesis and neuroinflammation (20). In humans, S1P receptor modulators are used to target immune cells to treat multiple sclerosis, a chronic inflammatory disease (18, 21, 22, 23). Aside from mature lineage cells, inflammation has pleiotropic effects within primitive hematopoietic cells, governing both lineage determination and HSC stemness functions (10, 11). There is also a role in hematopoietic malignancies, although studies exploring inflammation across the individual cells that make up the leukemia hierarchy are limited (11). Aging is also associated with increased inflammation, dysregulated lineage determination, and myeloid skewing, and is a major risk factor for developing AML (24, 25). Some reports suggest that both complex membrane sphingolipids, such as sphingomyelin, that are catabolized for S1P production and levels of S1P itself are dysregulated during aging (26, 27, 28). Overall, the role of S1P signaling in the interplay between inflammation and normal hematopoiesis or in the development of stem cell malignancies like AML is poorly understood.
Dysregulated myeloid cells are the linchpin of inflammation within many chronic human conditions associated with aging, including clonal hematopoiesis and atherosclerotic cardiovascular disease (29, 30, 31). TNFα and IL6 are two key inflammatory cytokines linked to chronic inflammatory diseases, myeloid dysregulation, and stem cell function (11, 23, 32). Moreover, a large body of literature points to NF-κB, originally described as a gatekeeper for inflammatory control of immune cell responses, as a key transcriptional factor in the regulatory network activated by these inflammatory cytokines and other inflammatory inputs in normal and malignant cells of various tissues (11, 33). Importantly, NF-κB is a crucial regulator of inflammatory activation in HSCs and regulates quiescence, proliferation, and survival, particularly upon TNFα stimulation (23). NF-κB was shown to be constitutively active in LSCs from patients with AML (33). At the time, it was anticipated that modulating inflammation through inhibition of some of the key signaling molecules, including TNFα, IL6, and NF-κB, could be exploited therapeutically for treating AML. However, some of these same pathways, for example TNFα via NF-κB, may actually promote differentiation and loss of LSC self-renewal (34). Uncovering novel lineage regulators that interact with such differentiation-inducing inflammatory programs would be pivotal for parsing the functional differences between the different cell types within each AML cell hierarchy and between different patients (3, 35, 36). Such studies could yield novel targets for developing therapeutic approaches focused on driving differentiation of LSCs and blasts. Here, we show that the S1P signaling axis via S1PR3 promotes myeloid differentiation in human HSCs, is dysregulated in AML, and intersects with the TNFα via NF-κB pathway. Furthermore, perturbation of this axis in AML leads to LSC eradication, introducing a previously unreported therapeutic avenue for AML.
Results
S1PR3 Is an Inflammation-Activated S1P Receptor Regulating Lineage Differentiation in Human HSCs
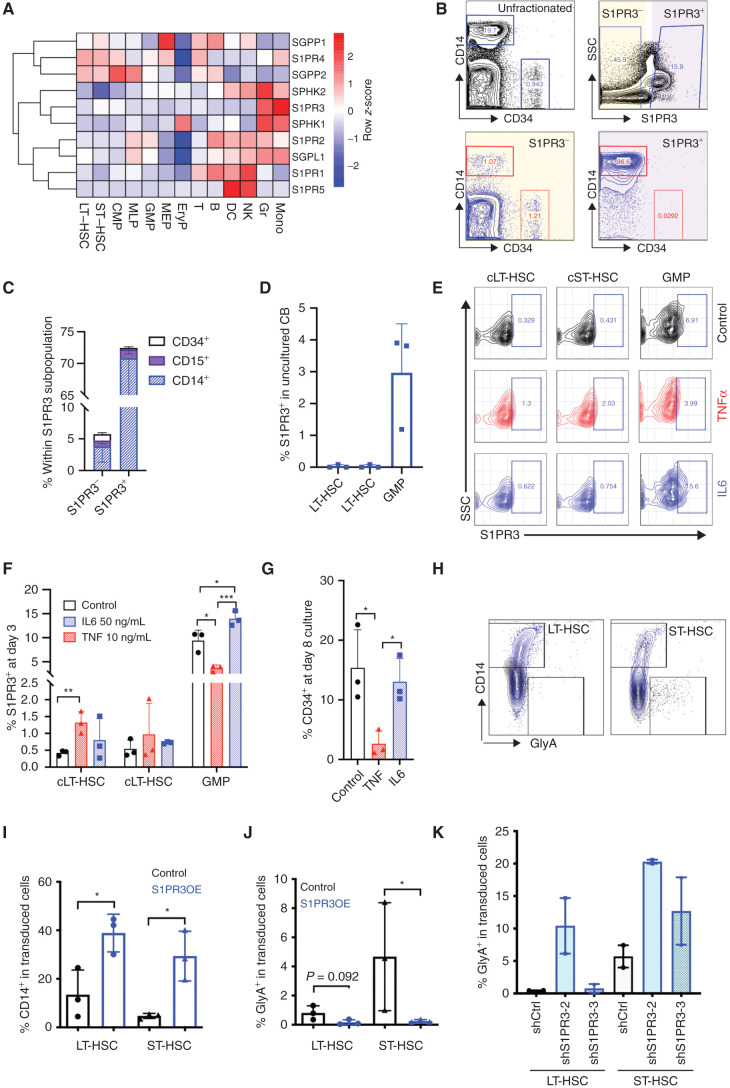
To gain deeper insight into the function of S1P in human hematopoiesis, we undertook gene expression analysis of 10 S1P-related genes, including its five receptors (S1PR1–5) and enzymes involved in its synthesis/degradation across six human hematopoietic stem and progenitor cell (HSPC) populations and seven mature lineages from cord blood (CB; 37). Expression of S1PR1, 3, and 5 was most enriched in specific mature lineages (Fig. 1A). S1PR1 was most highly expressed in T- and B-lymphoid, natural killer (NK), and dendritic cell (DC) populations, whereas S1PR5 was restricted to NK and DC lineages, consistent with published reports primarily in the murine system (16, 20). S1PR3 was of particular interest, as its expression was specific to monocytes and granulocytes, and it had not been previously implicated in myeloid fate specification in either normal or malignant human hematopoiesis. Flow cytometric analysis confirmed that S1PR3 protein was highly expressed on the surface of CD33+ myeloid cells, particularly within the CD14+ subset of mononuclear cells (MNC) where the majority of CD14+ cells showed costaining with S1PR3 (Fig. 1B and C; Supplementary Fig. S1A–S1H). Notably, S1PR3 was expressed at low levels in primitive CD34+ cells and was not detectable in LT-HSC or other HSPC populations except for a very small proportion (∼3%) of granulocyte-monocyte progenitors (GMP; Fig. 1B–D; Supplementary Fig. S1H–S1J). These findings raised the question of whether S1PR3 plays a role in myeloid differentiation.
Figure 1.
S1PR3 is an inflammation-activated S1P receptor regulating lineage differentiation in human HSCs. A, Heatmap of RNA-sequencing gene expression (mean-normalized counts) for 10 S1P genes in 13 human HSPC and mature populations isolated from CB shows myeloid lineage restriction for S1PR3. CMP, common myeloid progenitor; Eryp, erythroid progenitor; MEP, megakaryocyte erythroid progenitor; Gr, granulocyte; MLP, multipotent progenitor; Mono, monocyte. B, Representative flow cytometry plots showing gating scheme for CD14+ myeloid and CD34+ cells from three CB MNCs, in all live cells (black) and after gating for S1PR3− (gray) and S1PR3+ (blue) cells. SSC, side scatter. C, S1PR3 surface expression is absent in CD34+ cells but present in CD14+ monocytes and CD15+ granulocytes shown by comparing the percentage of CD14+, CD15+, and CD34+ cells within S1PR3− and S1PR3+ gated populations. See Supplementary Fig. S1 for additional data, including gating scheme and mean fluorescence intensity quantification of S1PR3 related to B and C. D, Percentage of S1PR3+ in indicated HSPC populations. E, Representative flow cytometry plots showing gating scheme for S1PR3+ in indicated immunophenotypic HSPC populations (see Supplementary Fig. S2A and S2B for culture conditions and gating scheme). cLT-HSC, cultured LT-HSC; cST-HSC, cultured ST-HSC. Percentage of S1PR3+ cells in HSPC populations on day 3 (F) and percentage of CD34+ at day 8 culture (G) in control media or with TNFα or IL6 treatment (n = 3 CB pools). H and I, S1PR3OE alters lineage differentiation in the progeny of LT-HSCs and ST-HSCs in ex vivo culture at day 9 or 10. S1PR3OE, S1PR3 overexpression. H, LT-HSC or ST-HSC progeny was analyzed for CD14 and GlyA, and representative flow cytometry plots for control (black) or S1PR3OE (blue) are shown as overlays. Quantification of CD14+ (I) and GlyA+ (J) populations from H (n = 3 independent CB). K, GlyA expression following lentiviral knockdown of S1PR3 in LT-HSCs and ST-HSCs at 8 or 10 days after transduction (n = 2 CB). *, P < 0.05; **, P < 0.01; and ***, P < 0.001 (unpaired Student t test). Data are mean and SD.
As HSPC subpopulations are primarily quiescent until activated by mitogenic stimulation (14, 38), we asked whether cellular activation alone or with an acute inflammatory stimulus would affect S1PR3 in primitive CB cells. Following 3 days of culture in a low-cytokine media when the majority of LT- and short-term (ST)-HSCs have not yet undergone the first division from quiescence (ref. 38; see Supplementary Fig. S2A and S2B for experimental scheme and HSPC gating scheme), we observed a 3-fold increase in the proportion of S1PR3+ cells within the GMP subset over uncultured GMPs; IL6 further enhanced the percentage of S1PR3+ GMP cells 4.6-fold over uncultured GMPs (Fig. 1D–F). Whereas S1PR3 was not detected on the surface of uncultured LT-HSCs (Fig. 1D; Supplementary Fig. S1I and S1J), TNFα treatment in culture was sufficient to promote expression of S1PR3 on a small but significant population of immunophenotypic cultured LT-HSCs (cLT-HSC; 0.5% S1PR3+ cells in control media vs. 1.3% S1PR3+ with TNFα treatment) despite no significant change in the percentage of cLT-HSCs (Fig. 1D–F; Supplementary Fig. S2C). Moreover, TNFα treatment reduced surface S1PR3 expression on GMPs at 3 days in culture to levels similar to those in uncultured GMPs, and provoked loss of CD34+ cells at 8 days in culture with no significant effects on CD14 differentiation (Fig. 1D–G; Supplementary Fig. S2D and S2E), consistent with the distinct effects of TNFα on LT-HSCs and GMPs previously reported in the murine system (23). In contrast, IL6 treatment tended to increase expression of the CD14 differentiation marker following 8 days of culture (Supplementary Fig. S2E). These data suggest that surface expression of S1PR3 is regulated by proinflammatory cytokines with the potential for influencing stemness and lineage differentiation in response to acute demands.
To determine if S1PR3 activation is sufficient to regulate human HSC function, we utilized a lentiviral system (39) to enforce S1PR3 overexpression (OE) coupled to blue fluorescent protein (BFP) for marking transduced cells. S1PR3OE cells exhibited increased levels of surface S1PR3 in the progeny of LT-HSCs and ST-HSCs by flow cytometry relative to control cells (Supplementary Fig. S3A). S1PR3OE in LT-HSCs and ST-HSCs resulted in enhanced CD14+ myeloid differentiation and decreased GlyA+ erythroid differentiation compared with controls following 9 to 10 days of culture (Fig. 1H–J). Next, we asked what effect perturbing S1PR3 expression in human HSC subpopulations would have on lineage differentiation in culture. Two lentiviral knockdown (KD) constructs to S1PR3 were generated that decreased S1PR3 gene expression to less than 50% of control (shCtrl) in a cell line model (Supplementary Fig. S3B). There was a trend toward enhanced erythroid differentiation with both S1PR3 KD constructs in ST-HSCs, whereas only shS1PR3-2 in LT-HSCs increased GlyA+ expression (Fig. 1K; Supplementary Fig. S3C). However, the level of S1PR3 KD with these constructs did not appear to be sufficient to affect CD14+ myeloid differentiation in vitro (Supplementary Fig. S3D). These data indicate that S1PR3 is a myeloid-associated S1P receptor that regulates lineage differentiation in human HSCs.
S1PR3OE Is Sufficient to Promote Myeloid Differentiation in Human HSPCs In Vitro and In Vivo
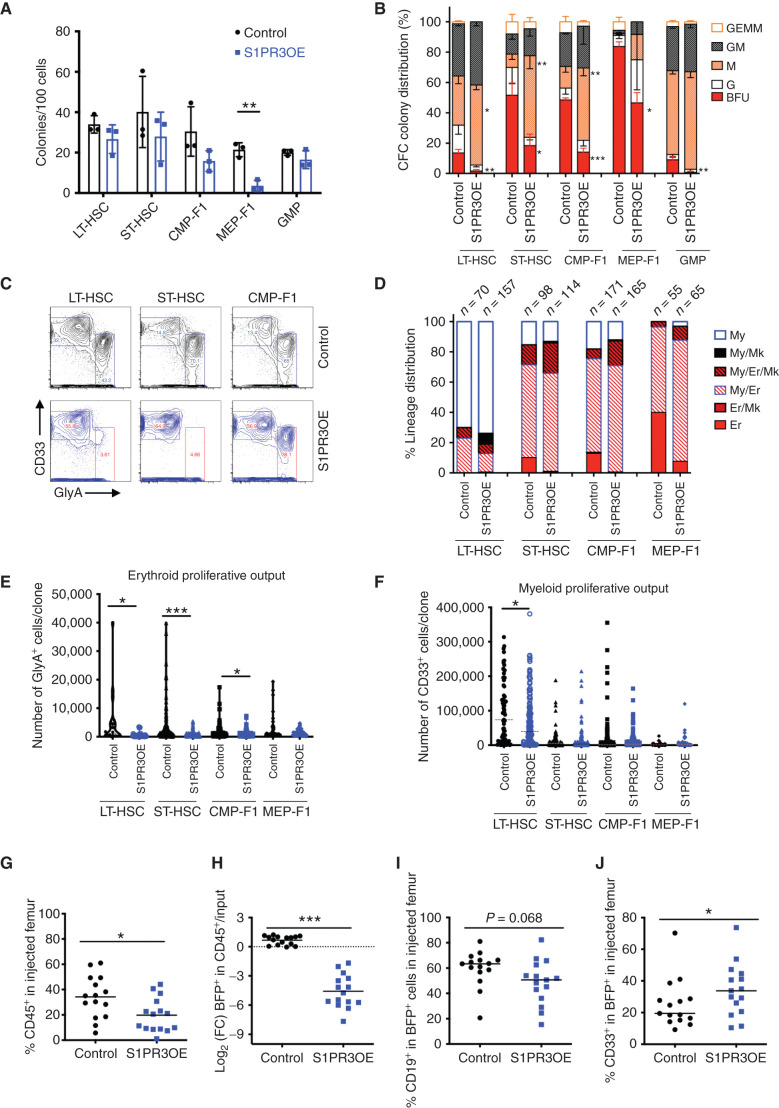
To determine if S1PR3OE was promoting myeloid differentiation in HSPC subpopulations with multilineage differentiation capacity at the single-cell level, transduced (BFP+) LT-HSCs, ST-HSCs, common myeloid progenitor (CMP)-F1, and megakaryocyte erythroid progenitor (MEP)-F1 were isolated 3 days after infection and plated in colony-forming cell (CFC) or single-cell MS5 stromal assays (ref. 14; see Supplementary Fig. S3E and S3F for experimental design and HSPC sorting scheme). S1PR3OE did not significantly decrease clonogenic output by any HSPC population except MEP-F1 (Fig. 2A). However, the colony-type distribution was significantly altered, with an increase in macrophage colonies generated from LT-HSC, ST-HSC, and CMP-F1 populations and a marked decrease of erythroid burst-forming unit (BFU) colonies for all subpopulations relative to controls (Fig. 2B). The alteration in myeloid–erythroid lineage distribution was confirmed by flow cytometry for one representative CFC assay (Fig. 2C). Following single-cell plating on MS5 stroma (40, 41), S1PR3OE reduced unilineage erythroid output (Fig. 2D) as well as total erythroid proliferative output except by MEP-F1 without decreasing cloning efficiency (Fig. 2E; Supplementary Fig. S3G). Total myeloid output was not disrupted except in LT-HSCs (Fig. 2F). The percentage of cells undergoing CD41+ megakaryocytic differentiation was also increased with S1PR3OE (ref. 42; Supplementary Fig. S3H). As S1PR3 KD had a modest effect on erythroid differentiation in liquid culture, we evaluated effects at the single-cell level in LT-HSCs, ST-HSCs, and CMP-F1 in CFC assays. There was no alteration in clonogenic output, but S1PR3 KD with shS1PR3-2 resulted in a modest increase in BFU colonies (Supplementary Fig. S3I and S3J). These data indicate that S1PR3OE is sufficient to promote myeloid differentiation on the single-cell level at the expense of erythroid differentiation in vitro.
Figure 2.
S1PR3OE is sufficient to promote myeloid differentiation in human HSPCs in vitro and in vivo. A, The number of colonies for 100 transduced cells from the indicated populations at 10 days of a CFC assay for control and S1PR3OE lentiviral KD vectors (n = 3). B, Normalized colony distribution showing BFU is significantly decreased with S1PR3OE, and macrophage (M) colonies are significantly increased from all scored cell populations (n = 3 CB). G, granulocyte; GEMM, granulocyte erythrocyte monocyte megakaryocyte; GM, granulocyte monocyte. C, Myeloid (CD33+) versus erythroid (GlyA+) lineage distribution for all pooled cells from one representative assay at day 14 from A. D, Lineage distribution outcomes in single-cell assays performed on MS5 stroma in erythroid–myeloid cytokine conditions. Data for indicated number of cells scored in each condition pooled from three CBs. Er, erythroid; Mk, megakaryocyte; My, myeloid. E and F, Average number of GlyA+ or CD33+ cells from single-cell differentiation assays from D. Human CD45 chimerism (G) and log2 fold change (FC) in BFP (H), which marks transduced cells relative to input. CD19+ B-lymphoid cells in the transduced fraction (I) and CD33+ myeloid cells in BFP+ cells (J) for control or S1PR3OE measured in the injected femur at 4 weeks after transplant in xenotransplantation, with five mice for each condition from three CB. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 (unpaired Student t test). Data are mean and SD except for E and F where data are mean and SEM.
To investigate effects on lineage output in vivo, we xenotransplanted S1PR3OE CD34+CD38− CB cells, containing a mixture of transduced and nontransduced cells, into immunodeficient mice and evaluated engraftment and lineage differentiation after 4 weeks. Compared with controls, S1PR3OE significantly reduced overall human CD45+ engraftment as well as the proportion of BFP+ transduced cells (Fig. 2G and H; Supplementary Fig. S4A–S4C). Mice transplanted with S1PR3OE cells generated grafts with a higher proportion of CD33+ myeloid cells at the expense of CD19+ B-lymphoid cells (Fig. 2I and J). Moreover, the proportion of GlyA+ cells that were BFP+ was lower in S1PR3OE-engrafted mice, indicating that S1PR3OE reduced erythropoiesis in vivo (Supplementary Fig. S4D and S4E). Overall, our findings demonstrate that S1PR3OE promotes myeloid and megakaryocytic differentiation at the expense of the erythroid and lymphoid lineages. These phenotypes are reminiscent of those observed in chronic inflammation–induced myelopoiesis models (22, 42).
S1PR3 Induces a Myeloid Inflammatory Program in HSCs
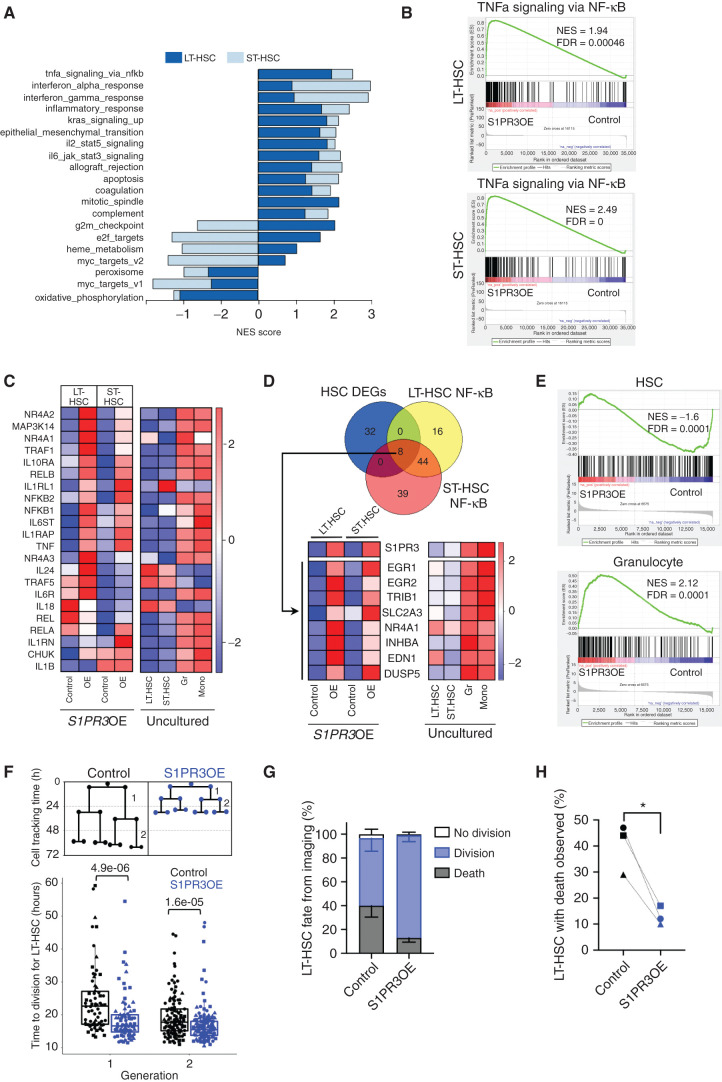
To ascertain the molecular and biological pathways altered by S1PR3OE in human CB HSCs, we performed RNA sequencing (RNA-seq) of LT- and ST-HSCs 3 days after transduction with control or S1PR3OE vectors (Supplementary Fig. S5A and S5B). Gene set enrichment analysis (GSEA) with the hallmark gene sets (FDR q value < 0.01; Fig. 3A; Supplementary Table S1) showed that S1PR3OE upregulated many of the same pathways in both HSC subsets. Mitotic spindle, G2–M checkpoint, and TNFα signaling via NF-κB (Fig. 3B) were the top three enriched gene sets in LT-HSCs relative to controls, suggesting that S1PR3OE induced an activated inflammatory program. Inflammation gene sets were enriched in S1PR3OE ST-HSCs, with genes related to the IFNα response, IFNγ response, and NF-κB being the top three enriched gene sets (Fig. 3A and B; Supplementary Table S1). The differential expression patterns of selected inflammation and NF-κB–related genes in S1PR3OE HSCs compared with controls resembled those of uncultured mature myeloid lineages relative to HSC subpopulations, including TNF and IL6 receptor genes (IL6R or IL6T; Fig. 3C). Notable exceptions were a downregulation of IL1B (22) and upregulation of IL24 (43) in LT-HSCs, highlighting the complex interplay of pro- and anti-inflammatory processes governing human HSC fate. These data could also provide an explanation for why S1PR3OE was not sufficient to induce a de novo AML in xenografts in contrast to the malignancy described in a S1PR3 transgenic mouse model (44). NF-κB and TNFα have been associated with S1P signaling in other contexts (15, 45) and shown to regulate HSC survival and fate decisions (9, 23). Hence, we performed a focused analysis comparing the significantly enriched leading-edge genes from the NF-κB hallmark gene set (Supplementary Table S2) and the genes that were differentially expressed between controls and S1PR3OE HSC subsets (Supplementary Table S3; q < 0.05, fold change = 1). This identified a common set of eight myeloid lineage–associated genes, including genes encoding the known myeloid differentiation factors Early Growth Response 1 and 2 (EGR1/2) and AML-associated Tribbles 1 (TRIB1; refs. 46, 47, 48, 49; Fig. 3D). These results suggest that S1PR3 regulates the well-known ERG–NAB–GFI1 myeloid gene regulatory network (46) in human HSCs. GSEA using a published human hematopoietic gene expression dataset (50) showed that S1PR3OE LT- and ST-HSCs had significant positive enrichment of granulocyte genes and negative enrichment for HSC genes, providing independent confirmation of myeloid lineage induction (Fig. 3E; Supplementary Fig. S5C and S5D). S1PR3OE cells resembled granulocytes with not only strong upregulation of inflammation gene sets but also downregulation of HSC-enriched metabolic gene sets (Supplementary Fig. S5E). If S1PR3OE promotes differentiation via an inflammation-induced emergency hematopoiesis-like mechanism, then S1PR3OE LT-HSCs should exhibit cell-cycle activation and a shorter division time, as has been observed in murine HSCs following treatment with inflammatory cytokines (22). Indeed, live-cell imaging analysis (51) demonstrated that S1PR3OE resulted in faster cell-division kinetics and better survival of LT-HSCs compared with controls (Fig. 3E–H; Supplementary Movie). Overall, this strong induction of inflammatory gene sets upon S1PR3OE together with the upregulation of S1PR3 membrane expression on HSC by TNFα treatment suggests that S1PR3 may function as a downstream potentiator of inflammatory signaling in human HSCs to regulate survival and myeloid fate.
Figure 3.
S1PR3OE induces a myeloid inflammatory program in HSCs. A, Normalized enrichment scores (NES) for the pathways that are significantly different between control and S1PR3OE cells (FDR < 0.01) in LT-HSCs and ST-HSCs (superimposed) following GSEA of the MSigDB v6.2 hallmark gene sets in RNA-seq expression data from three CBs. S1PR3OE strongly upregulates inflammatory pathways in both HSC subsets. B, GSEA plots of TNFα via NF-κB hallmark gene set for LT-HSCs and ST-HSCs. C, Heatmap showing the median expression of selected NF-κB pathway genes, associated with both the canonical and noncanonical signaling pathway, and inflammatory cytokines and receptor genes from the S1PR3OE RNA-seq and uncultured LT-HSCs, ST-HSCs, granulocytes (Gr), and monocytes (Mono). The majority of genes are highly expressed in myeloid lineages and are upregulated with S1PR3OE in the HSC subsets. Of note, a number of inflammation genes exhibit distinct expression patterns in LT-HSCs and ST-HSCs with S1PR3OE, including TRAF5, IL1RL1, IL18, IL24 (an IL10 family member), and IL1B. D, Venn diagram representing the intersection of 40 significantly differentially expressed genes (DEG; FDR < 0.05) in S1PR3OE from both LT- and ST-HSC compared with control and leading-edge genes of the NF-κB gene set in both LT-HSCs and ST-HSCs. The eight genes found in the intersection pointed to upregulation of an EGR1/2-associated myeloid transcriptional network. Heatmaps of the median expression of these eight genes and S1PR3 in control and S1PR3OE at day 3 compared with uncultured LT-HSCs, ST-HSCs, Gr, and Mono are shown. E, GSEA plots for previously published HSC and granulocyte gene sets indicate that S1PR3OE enriches for granulocyte and depletes HSC identity in genes common for LT-HSC and ST-HSC S1PR3OE RNA-seq (see Supplementary Fig. S4C–S4E for individual GSEA plots for LT-HSCs and ST-HSCs and enrichment map of pathways altered by S1PR3OE). F, Quantification of time to division for single cells tracked in the first two generations of control (n = 34)- or S1PR3OE (n = 41)-transduced LT-HSCs beginning at day 3 after transduction by time-lapse imaging (three CB, marked by different symbols). Statistical significance by Mann–Whitney test. G, Distribution of division, death, or no division fate for control (n = 179) or S1PR3OE (n = 198) LT-HSC cells tracked from three independent experiments. H, Percentage of LT-HSCs that died during time-lapse movies for control and S1PR3OE vectors. *, P < 0.05 (paired t test).
S1PR3 Marks a Subset of LSCs in Human AML with a Mature Myeloid State and a Distinct Inflammatory Signature
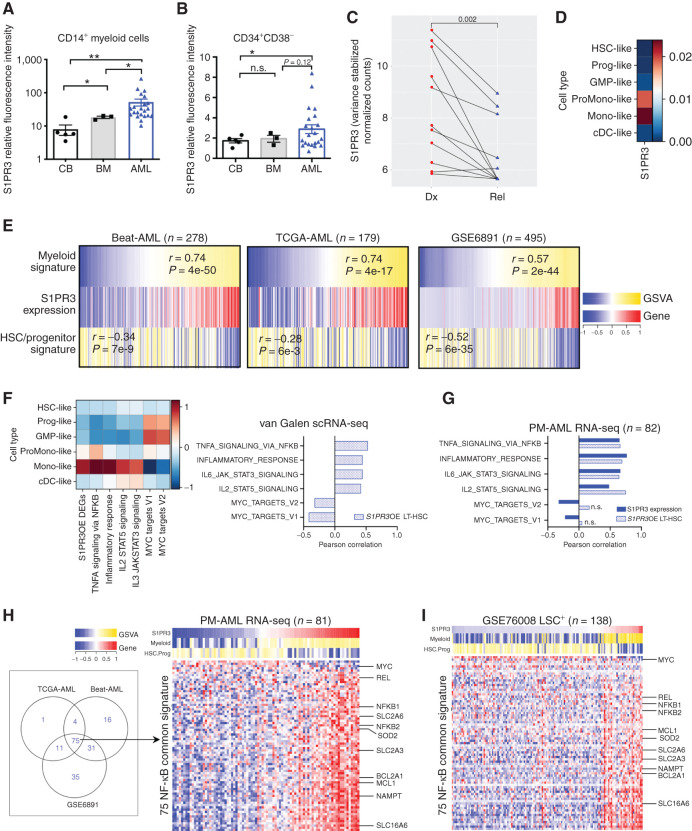
Previous studies have suggested a possible link between inflammation and perturbed myeloid differentiation (30). We therefore investigated the role of S1PR3 in the pathogenesis of AML, a disease of impaired myelopoiesis (3). S1PR3 expression by flow cytometry was higher on CD14+ myeloid and CD34+CD38− primitive cell subsets from patients with AML (n = 22) compared with CB (n = 5) and normal adult bone marrow (n = 3; Fig. 4A and B; Supplementary Table S4), although there was considerable interpatient heterogeneity. In keeping with the observation that AML cells at relapse are typically more primitive than at diagnosis, S1PR3 gene expression was lower in relapsed patient samples compared with paired diagnosis samples (ref. 5; Fig. 4C). Thus, we hypothesized that higher S1PR3 expression may identify less primitive AML cases. We analyzed S1PR3 gene expression in a recently described single-cell RNA-seq (scRNA-seq) dataset (3) across 11,641 malignant cells from 12 patients with AML clustered by their similarity to six comparable populations along the normal bone marrow HSC to myeloid axis [HSC-like, progenitor-like, GMP-like, promonocyte-like, monocyte-like, or conventional DC (cDC)–like; Fig. 4D]. As seen in the normal hierarchy, S1PR3 in AML was enriched exclusively in samples classified as myeloid phenotype (mono-like and promono-like). As this scRNA-seq platform was only able to capture the low abundance of S1PR3 transcript in <0.01% of cells, we analyzed transcriptomes from unfractionated patients with AML generated by RNA-seq or microarray analysis. Gene set variation analysis (GSVA) of the Beat-AML (35), The Cancer Genome Atlas (TCGA)-AML (52), and GSE6491 (53) de novo AML datasets revealed that AML cases with the highest S1PR3 expression were enriched for the myeloid signature and depleted of the HSC/progenitor signature previously defined using the scRNA-seq AML dataset (ref. 3; Fig. 4E; Supplementary Tables S5 and S6). Hereafter, we will refer to these AML cases with high S1PR3 expression and enrichment of the myeloid signature as myeloid-like and the AML cases with low S1PR3 expression and enrichment of the HSC/progenitor signature as stem-like. Together, these findings point to a strong association between S1PR3 expression and myeloid differentiation in both normal hematopoiesis and AML.
Figure 4.
S1PR3 marks a subset of LSCs in human AML with a mature myeloid state and a distinct inflammatory signature. Relative fluorescence intensity of S1PR3 in CD14+ myeloid cells (A) and CD34+CD38− cells (B) calculated as ratio of S1PR3 mean fluorescence intensity to fluorescence minus one controls from human CB MNC (n = 5), bone marrow (BM; n = 3), and AML (n = 22) samples. n.s., not significant; *, P < 0.05; and **, P < 0.01, unpaired Student t test. Data are mean and SD. C, S1PR3 gene expression in paired diagnosis (Dx; red) and relapse (rel, blue) samples from 11 patients previously used to study the origin of AML relapse. Statistical significance by Wilcoxon signed-rank test. D, AUCell analysis across 11,641 malignant cells from 12 patients with AML for S1PR3 gene expression as defined in indicated AML cell types as described in study by van Galen and colleagues (3). Mono-like, monocyte-like; Prog-like, progenitor-like; ProMono-like, promonocyte like. E, Heatmaps depicting S1PR3 gene expression with previously defined myeloid and HSC/progenitor (stem) signatures for patients with de novo AML in the Beat-AML, TCGA-AML, and GSE6891 cohorts. Rho correlation scores and P values for S1PR3 gene expression to the signatures indicated for each cohort showing correlation to the myeloid signature and anticorrelation to the stem signature. F, AUCell analysis across van Galen scRNA-seq data for the set of differentially expressed genes upon S1PR3OE in LT-HSC (S1PR3OE DEGs; Supplementary Table S3) and selected hallmark gene sets upregulated upon S1PR3OE in LT-HSCs with associated Pearson correlation. G, GSVA of S1PR3 gene expression and the set of S1PR3OE DEGs to selected hallmark gene sets upregulated upon S1PR3OE in LT-HSCs in a PM-AML RNA-seq patient cohort. H, Venn diagram of genes that show significant correlation from the NF-κB hallmark gene set with S1PR3 gene expression in diagnostic samples from Beat-AML, TCGA-AML, and GSE6981 patient cohorts yielded 75 common genes. Heatmap of common genes (rows) for the PM-AML patient (columns) cohort shows myeloid-like AMLs have enrichment of NF-κB common genes. S1PR3 gene expression and GSVA correlation values for the myeloid and HSC/progenitor signatures are shown on top of the heatmap. I, Heatmap of 75 genes in the LSC+ subpopulations from GSE76008 indicates a similar relationship of S1PR3 gene expression to enrichment for the set of 75 NF-κB genes as in unfractionated patient samples.
To explore further the role of S1PR3 in human AML, we investigated the S1PR3OE-induced signatures described in Fig. 3 in cohorts of AML samples. AUCell (area under the curve Cell) analysis across 11,641 single malignant cells revealed that the S1PR3OE LT-HSC signature was expressed in 86.5% of cells and most enriched in the mono-like subpopulation of samples of patients with AML (ref. 3; Figs. 3A and 4F). Importantly, the S1PR3OE signature was significantly correlated to a number of inflammatory gene sets, including the NF-κB hallmark gene set, whereas MYC targets were negatively correlated, reminiscent of the phenotype observed in LT-HSCs upon S1PR3OE (Figs. 3A and 4F; Supplementary Table S7). Moreover, GSVA in an independent RNA-seq AML cohort from Princess Margaret (PM) showed strong correlation of both single S1PR3 gene expression and the S1PR3OE signature to a number of inflammatory gene sets that were also induced by S1PR3OE in HSCs (Fig. 4G). These data suggest that the malignancy state in S1PR3-expressing AML cells is congruous with the transcriptional inflammatory state activated by S1PR3OE in LT-HSCs.
We next investigated whether individual components of the S1PR3-mediated inflammatory pathways such as NF-κB were similarly regulated in individual myeloid-like or stem-like AML patients. To this end, we derived a common 75 S1PR3–NF-κB–related gene signature from the Beat-AML (35), TCGA-AML (52), and GSE6491 (53) cohorts (P < 0.05) based on GSVA correlation between the NF-κB hallmark gene set and S1PR3 expression, and validated it in the PM cohort (Fig. 4H; Supplementary Fig. S6A; Supplementary Table S8; clinical data in Supplementary Table S9). Importantly, the S1PR3–NF-κB–related gene signature also varied in a continuum from stem-like to mature-like myeloid states in functionally validated LSC-containing (LSC+) cell fractions (7). Similar to observations in bulk patient samples (Fig. 4H; Supplementary Fig. S6A; Supplementary Tables S5, S6, and S8), myeloid-like LSC+ and LSC− cell fractions were enriched for the common S1PR3–NF-κB signature (Fig. 4I; Supplementary Fig. S6B). The S1PR3–NF-κB signature included genes encoding the NF-κB subunits REL, NFKB1, and NFKB2; the antiapoptotic BCL2 family members MCL1 and BCL2A1; and the energy metabolism/oxidative stress–related genes NAMPT, SOD2, SLC2A3 (GLUT3), SLC2A6, and SLC16A6. MYC was a notable negatively correlated gene, in accordance with downregulation of MYC targets upon S1PR3OE in normal HSCs. These results indicate that NF-κB regulation is heterogeneous among individual patient with AML and raise the possibility that the NF-κB–S1PR3 regulatory loop serves different functions in the LSCs isolated from stem-like patients or myeloid-like patients (11). Moreover, Beat-AML cases with high S1PR3 expression were more resistant to ex vivo treatment with the BCL2 inhibitor venetoclax, consistent with a recent report linking myeloid differentiation in AML and resistance to combination therapy with azacytidine and venetoclax (refs. 54, 55; Supplementary Fig. S6C). Overall, these results demonstrate that S1PR3 expression can classify individual patient with AML into two broad categories: more differentiated and more stem-like cases, each with distinct inflammatory, metabolic, stress response, and cell survival programs.
S1PR3 Selects for Less Functional LSCs and Regulates AML Differentiation via TNFa via NF-kB Signaling
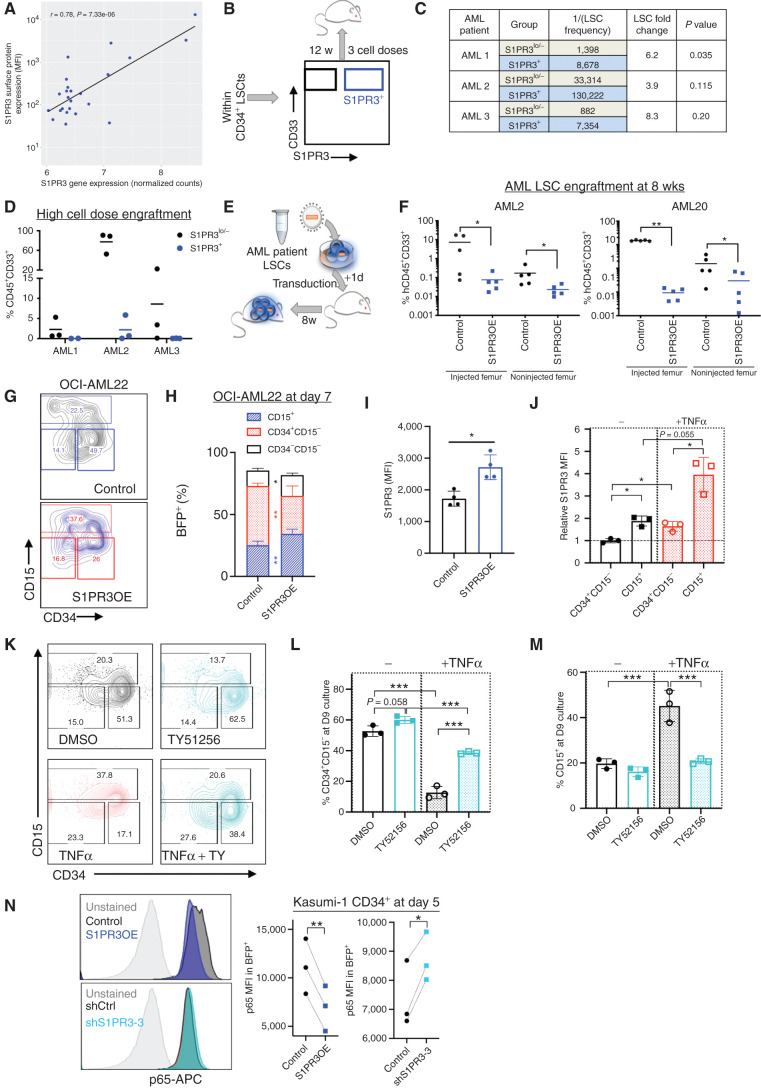
As high S1PR3 expression is associated with a more differentiated phenotype in AML and marks a distinct activated inflammatory state, we evaluated whether S1PR3 surface expression could resolve LSC+ from LSC− cells. We focused on AML samples classified as stem-like. S1PR3 gene expression in LSC+ fractions (n = 24) from 13 patients in the GSE76008 cohort (7) was highly correlated with S1PR3 surface expression measured by flow cytometry (r = 0.78; Fig. 5A; Supplementary Tables S10 and S11). CD34+ LSCs from three of these patients were fractionated based on surface expression of S1PR3 (1%–6% S1PR3+; see Supplementary Fig. S7) and transplanted at limiting dilution into NSG mice (Fig. 5B). S1PR3+ AML cells had reduced engraftment potential at 12 weeks and lower LSC frequency compared with S1PR3low/− cells (Fig. 5C and D). These data show that S1PR3 expression is sufficient to identify a subset of cells with low LSC activity in samples of patients with AML.
Figure 5.
S1PR3 selects for less functional LSCs and regulates AML differentiation via TNFα via NF-κB signaling. A, Correlation of S1PR3 gene expression from the GSE76008 cohort and S1PR3 mean fluorescence intensity (MFI) measured by flow cytometry for LSC+ subpopulations (n = 24) from 13 patients. B, Experimental schematic showing LSC+-containing patient cell fractions (Supplementary Fig. S7) were sorted based on S1PR3 surface expression and transplanted in different cell doses into NSG mice for 12 weeks. C, Table with LSC frequency as calculated by ELDA. D, CD45+CD33+ engraftment of mice injected with the highest cell dose of S1PR3+ (blue) or S1PR3−/lo (black) cells is shown. E, Experimental schematic for lentiviral transduction of control (black) or S1PR3OE (blue) vectors into patient AML LSCs and transplanted into NSG mice for F. F, CD45+CD33+ engraftment of human CD34+CD38− AML cells from two patients in NSG mice at 8 weeks after transplantation in the injected and noninjected femurs. OCI-AML22 is an AML cell line model derived from a patient with AML at relapse where the stem cell fraction is contained in the CD34+ fraction. G, Representative flow cytometry plots for CD15 versus CD34 markers 7 days after transduction (n = 2 experiments, in duplicate). Distribution of indicated immunophenotypic populations (H) and S1PR3 surface expression (MFI; I) for control- and S1PR3OE-transduced OCI-AML22 cells at day 7 after transduction. J, S1PR3 MFI in OCI-AML22 subpopulations normalized to an untreated CD34+CD15− sample 9 days after treatment with 10 ng/mL TNFα (n = 3). K–M, Representative flow cytometry plots and quantitation for CD15 and CD34 markers 9 days after treatment with DMSO or 5 μmol/L TY51256 alone or with 10 ng/mL TNFα in OCI-AML22. N, Representative histogram plots of p65 RelA expression by intracellular flow cytometry and the quantitation of p65 MFI with S1PR3OE or shS1PR3-3 relative to controls in Kasumi-1 CD34+ cells at 5 days after transduction (n = 3) *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Unpaired t test (F, H, and I); one-way ANOVA (J, L, and M); and paired t test (N). Data are mean and SD.
To evaluate whether activation of S1PR3 could represent a therapeutic approach to disrupt LSC function in AML, we transduced AML cells with S1PR3OE and control vectors. S1PR3OE in LSC+ fractions from two AML patient samples virtually abolished leukemic engraftment in xenotransplantation assays (Fig. 5E and F). S1PR3OE in a hierarchical AML model and three primitive AML cell lines with low surface expression of S1PR3 resulted in acquisition of CD15 and loss of CD34 expression in vitro, suggesting that enforced expression of S1PR3 is sufficient to promote myeloid differentiation in AML cells (Fig. 5G–I; Supplementary Fig. S8A–S8E). We found that TNFα enhanced S1PR3 surface expression in both the OCI-AML22 model we derived from a relapse patient (56) and in Kasumi-1 cells, compared with controls (Fig. 5J; Supplementary Fig. S8F). Next, we interrogated the role of endogenous S1PR3 in AML. The S1PR3 antagonist TY51256 restricted myeloid differentiation in both AML cell line models during in vitro culture (Fig. 5K–M; Supplementary Fig. S8G–S8I). Importantly, in OCI-AML22, TY51256 counteracted the myeloid differentiation enacted by TNFα treatment (Fig. 5K–M). Moreover, shRNA-mediated S1PR3 KD with shS1PR3-3 in Kasumi-1 CD34+ cells significantly decreased the CD34−CD15+ and CD34+CD15+ subpopulations at 9 days after transduction (Supplementary Fig. S8J). We also measured the intracellular levels of the p65 RelA subunit of NF-κB following S1PR3OE and KD in Kasumi-1 cells and found that S1PR3OE decreased p65 while shS1PR3-3 increased p65 relative to their respective controls (Fig. 5N). Overall, these findings suggest that S1PR3 is a biomarker that strongly anticorrelates with LSC activity in human AML, engagement of which disrupts LSC function in part by inducing differentiation via the TNFα–NF-κB axis.
Sphingolipid Genes Including S1PR3 Predict Outcomes in Human AML
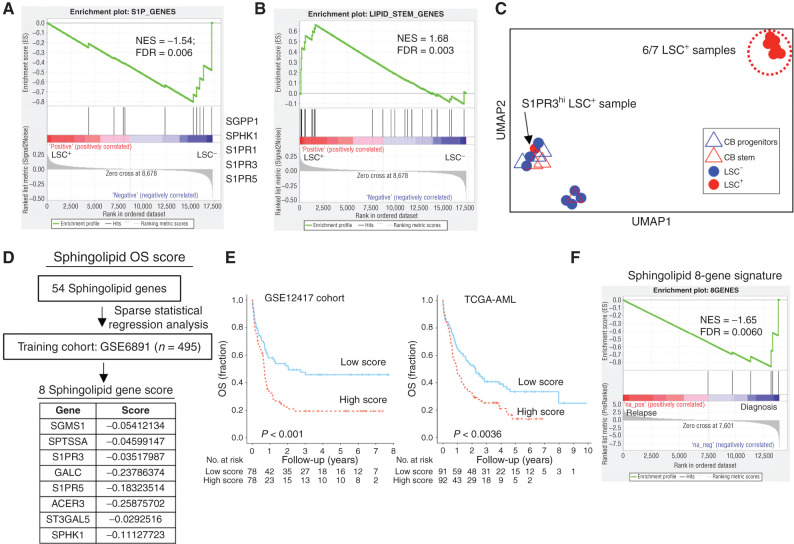
As no specific S1PR3 agonist has been shown to have in vivo efficacy to our knowledge, we asked if there were other possibilities for therapeutic targeting in the sphingolipid pathway. GSEA of functionally defined LSC+ and LSC− subpopulations from samples of patients with AML in the GSE76008 dataset demonstrated enrichment of S1PR3 and other S1P pathway genes only in the LSC− fractions (Fig. 6A). S1PR1, S1PR3, and S1PR5 were in the lowest 10% of analyzed genes expressed in LSC+ fractions (Supplementary Table S12). These findings suggest that downregulation of S1P signaling may be required for LSC maintenance. We previously identified a lipid-stem signature that is distinct between HSC and committed progenitors, and showed that modulation of sphingolipid synthesis alters hematopoietic fate in HSCs (14). This lipid-stem signature was enriched in LSC+ compared with LSC− fractions by GSEA (Fig. 6B). Among the significantly enriched genes in LSC+ samples were the de novo sphingolipid metabolism genes CERS6, CERS5, and SPLTLC2, suggesting that human AML retains features of sphingolipid regulation found in normal HSCs (Supplementary Table S12) and in particular upregulates the biosynthetic pathway generating S1P via ceramide as an intermediate (see Supplementary Fig. S9 for biosynthetic pathway; refs. 14, 17).
Figure 6.
Sphingolipid genes including S1PR3 predict prognosis in human AML. A, GSEA of 10 S1P genes in AML LSC+ and LSC− gene expression data shows enrichment of S1PR1, S1PR3, S1PR5, and S1P kinase (SPHK1) in LSC− samples. See Supplementary Table S12 for LSC ranking. B, GSEA of 23 lipid-stem genes, previously shown to be enriched in HSC subsets relative to committed progenitor populations, in AML LSC+ and LSC− gene expression data shows enrichment in LSC+ samples. C, A UMAP plot for the sphingolipid composition of samples measured by LC/MS spectrometry in functionally defined individual LSC+ (n = 7) and LSC−(n = 7) fractions from 10 patient AMLs as determined by xenotransplantation and normal stem (CD34+CD38−) and progenitor (CD34+CD38+) fractions (CB, n = 3). LSC+ (red circles), LSC− (blue circles), CB stem (red triangles), and CB progenitor (blue triangles). LSC+ subpopulation from AML patient 9, which is high for S1PR3 gene expression, clusters away from the other six LSC+ samples. See Supplementary Fig. S9C–S9G for sphingolipid composition distribution of individual samples and quantification of specific sphingolipid species and Supplementary Table S9 for patient AML data. D, Sparse regression analysis, as described in Ng and colleagues (7), to derive a weighted sphingolipid gene score predictive of OS applied to a large training cohort of 495 patients with AML yielded a set of eight genes with their associated scores, which were then validated on GSE12417 and TCGA-AML cohorts in univariate analysis. These included three genes associated with S1P production and signaling and three genes encoding sphingolipid enzymes. E, Kaplan–Meier estimates of OS based on above- versus below-median values of a weighted sphingolipid gene score. A low score is associated with greater OS. F, GSEA plot for sphingolipid eight-gene OS signature shows negative enrichment in patient samples at relapse.
To determine if sphingolipid levels are dysregulated in LSC+ populations, we profiled the sphingolipidome of LSC+ (n = 7) and LSC− (n = 7) fractions from 10 AML patient samples (7) as well as CB CD34+CD38− (stem) and CD34+CD38+ (progenitor) populations by LC/MS-MS for S1P, sphingosine, ceramides, dihydroceramides (dhCer), hexosylceramides (HexCer; ceramides containing glucose or galactose), and sphingomyelins (Supplementary Fig. S9A–S9H; Supplementary Table S9). Notably, dhCer, HexCer, and S1P levels were significantly different in LSC+ and LSC− samples (Supplementary Fig. S9C–S9E). Clustering samples by sphingolipid composition using uniform manifold approximation and projection (UMAP) showed that six of the seven LSC+ samples were most similar to each other and clustered away from both LSC− samples and CB stem and progenitor populations despite diverse immunophenotypes and molecular subtypes (Fig. 6C; Supplementary Fig. S9B). The seventh LSC+ sample (AML patient 9) that did not cluster with the others showed relatively higher S1PR3 surface and gene expression (see Supplementary Table S12). Thus, we speculated that a differential requirement for sphingolipid metabolism as well as S1PR3 signaling may exist in primitive AML cases (Fig. 4C). Regression analysis (7) of 54 sphingolipid genes including 10 S1P genes trained on survival outcomes in the GSE6891 cohort yielded an eight-gene sphingolipid signature [including S1PR3 (-0.035), S1PR5 (-0.183), and SPHK1 (-0.111)] that was strongly associated with overall survival (OS) in the independent GSE12417 and TCGA-AML cohorts (Fig. 6D and E). This signature was enriched in gene expression profiles from diagnostic relative to paired relapse samples by GSEA (Fig. 6F). These data suggest a link between sphingolipid metabolism and S1P signaling, with stemness state and response to chemotherapy in subsets of patients with AML.
In Vivo S1P Signaling Modulation Disrupts LSC Function in Human AML
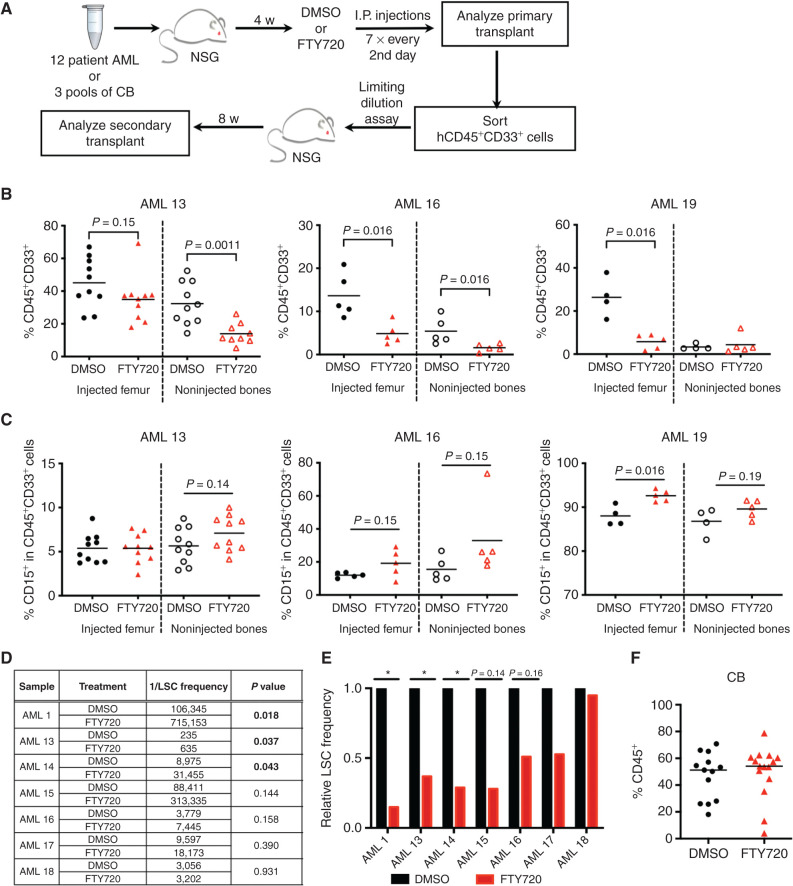
Because LSCs exhibited depletion of S1P signaling genes and higher S1PR3 and S1PR5 levels were associated with better prognosis in our sphingolipid signature, we evaluated the effects of modulating S1P signaling in patients with AML using xenograft assays. FTY720, a prodrug whose phosphorylation generates the S1P mimetic FTY720-phosphate, possesses both agonist and antagonist activities for four S1P receptors including S1PR3. Clinically known as fingolimod, FTY720 is used for treatment of patients with relapse-remitting multiple sclerosis, with the possibility of rapid repurposing for AML (18, 21). Treatment of mice bearing primary AML xenografts with FTY720 resulted in decreased leukemia burden for 3 of 12 patient samples tested (Fig. 7A and B; Supplementary Fig. S10A; Supplementary Table S10). Notably, an interesting trend of modest increase in CD15 expression in the xenografts of the three FTY720 responders (AML patient 1, AML patient 16, and AML patient 19) was observed compared with controls (Fig. 7C) but was not seen in the nonresponders except for AML patient 21 (Supplementary Fig. S10B). Importantly, serial repopulation assays at limiting dilution demonstrated a reduction in LSC frequency following FTY720 treatment in five of the seven AML samples tested (P = 0.018–0.158), including relapsed and treatment-refractory cases (Fig. 7D and E; Supplementary Table S13), demonstrating that FTY720 treatment disrupted LSC activity even without overt immunophenotypic changes in myeloid differentiation markers. Engraftment of CB cells was not affected by FTY720 treatment, suggesting that a therapeutic window exists for targeting of LSCs by FTY720 (Fig. 7F). These data suggest that FTY720 treatment is acting in part as an S1PR3 agonist to promote myeloid differentiation and decrease LSC function in our xenograft studies. In summary, in vivo treatment with the S1P prodrug FTY720 or S1PR3OE can target the disease-sustaining LSCs in AML.
Figure 7.
S1P signaling modulation in vivo targets LSC function in human AML. A–F, Preclinical targeting of the sphingolipid pathway with the S1P prodrug FTY720, clinically known as fingolimod, in samples of patients with AML or CB (n = 3) in vivo. A, Experimental schematic for xenotransplantation. AMLs with >20% engraftment in the injected femur and/or that showed decrease in AML burden were taken through secondary transplant to enumerate LSC frequency in D. B, AML engraftment for each mouse of the three FTY720 (red) responders relative to DMSO control (black), as defined by significant decrease of AML burden at 6 weeks after transplant. AML 13 is a treatment-resistant sample, AML 16 is a relapse sample, and AML 19 is a diagnosis sample. Significance calculated by Mann–Whitney test. See Supplementary Fig. S10 for additional engraftment data. C, Percentage of CD15+ cells in the indicated xenografts. D and E, LSC frequency for indicated AMLs was determined in limiting dilution serial transplantation assays and calculated with ELDA. Mice were considered engrafters if CD45+CD33+ cells >0.5%. The calculated LSC frequency and P value for FTY720 samples relative to DMSO control samples are shown in D, and the relative secondary LSC frequency is shown in E. F, Human CD45+ engraftment of CB following treatment with DMSO or FTY720 at 6 weeks after transplantation.
Discussion
Our study provides direct evidence that S1PR3 governs myeloid commitment in human HSCs and LSCs, and points to S1P signaling modulation as a potential therapeutic approach to target LSC in human AML. S1PR3OE alone was sufficient to induce myeloid differentiation in human HSC and LSC and S1PR3 antagonism or KD-restricted myeloid differentiation in AML. Moreover, S1PR3 marks a subset of LSCs from patients with AML with a distinct inflammatory signature and decreased stemness properties. Thus, S1PR3 could have clinical utility as both a prognostic biomarker and a novel therapeutic target in AML. Although induction of differentiation (e.g., by retinoid agonists) is an effective approach in some AML cases (57) and metabolic processes (6, 58, 59) have been shown to control LSC function, this is the first demonstration to our knowledge of successful human LSC targeting by modulation of a bioactive lipid signaling pathway.
We hypothesize that the dichotomy of S1PR3-high and -low AML patient samples mimics and can be traced to the diverse responses to inflammatory cytokines during stress hematopoiesis that are wired into the hematopoietic hierarchy (22, 23). The S1PR3-mediated promotion of myeloid differentiation at the expense of other lineages, particularly erythropoiesis, and enhanced survival of LT-HSCs are highly reminiscent of TNFα-induced emergency hematopoiesis (23). Thus, S1PR3 plays similar roles in human HSCs to what has been reported for the TNFα–NF-κB axis in the murine system (23). Overall, our data point to S1PR3 as a potentiator of TNFα via NF-κB inflammatory signaling to promote prosurvival and myeloid differentiation in human HSCs. Although TNFα levels are elevated during aging as well as in myeloid malignancies, it remains unclear how this inflammatory environment drives leukemia or which cellular subtypes are susceptible to transformation (11, 25, 30). TNFα has recently emerged as having complex roles in normal hematopoiesis, with different effects on HSC versus myeloid progenitors (23). Notably, we found that acute TNFα treatment was sufficient to upregulate S1PR3 membrane levels in primitive AML cell lines in vitro. In contrast, S1PR3 antagonism was sufficient to counteract the myeloid differentiation enacted by TNFα. A previous study found that S1PR3OE driven by a lysosome M promoter, but not an F4/80 promoter, induced a leukemic transformation in the mouse system, suggesting that the cell of origin for transformation originated from a primitive subpopulation and not from a mature myeloid lineage, although a role for S1PR3 in inflammatory signaling was not identified (44). However, as we found that S1PR3OE in human HSCs was not able to induce a de novo AML in xenografts, other factors such as the microenvironment and species differences need to be considered. We speculate that in a subset of cases of patients with AML, dependent on specific genetic lesions and metabolic states, the NF-κB–S1PR3 feed-forward loop is hijacked to promote prosurvival mechanisms. Although we were able to capture the same inflammatory signatures induced in HSCs by S1PR3OE within samples of patients with AML, whether S1PR3 activity confers enhanced survival to AML cells a priori remains to be explored. Nonetheless, it is intriguing that AML cases with high S1PR3 expression in the Beat-AML cohort were more resistant to venetoclax treatment, and S1PR3OE was sufficient to decrease p65 levels in AML cells in vitro. The nuances around how S1P signaling intersects with inflammation and NF-κB signaling to regulate LSC function are most likely different along the spectrum of stem-like to myeloid-like AML samples and reflective of the differences in canonical and noncanonical NF-κB signaling within LSCs (34). We wonder if the heterogeneity of response to venetoclax is related to such inflammatory wiring. The identification of a distinct NF-κB signature in a subset of S1PR3-high AML cases raises the possibility that different strategies need to be employed for successful targeting of NF-κB in LSCs from stem-like and myeloid-like AML cases.
The FTY720 xenograft study along with our S1PR3OE, KD, and S1PR3 in vitro antagonist studies suggests that FTY720 treatment is acting in part as an S1PR3 agonist to downregulate NF-κB signaling and promote myeloid differentiation to decrease LSC function in stem-like AML. The transcriptome analysis of sphingolipid metabolism in AML and sphingolipidome profiling of LSC+ and LSC− subpopulations hints at connections between S1P signaling, sphingolipid metabolism, and drug response in LSCs beyond S1PR3. Stem-like AML cases may have a different requirement for sphingolipid metabolism and S1P signaling compared with more differentiated myeloid-like cases that appear to be connected to OS and chemotherapy response. Whether chemotherapy selects for preexisting subclones with different sphingolipid metabolism/S1P downstream signaling or whether resistant clones survive by remodeling their underlying metabolism including the sphingolipid network remains to be determined. Importantly, our identification of an eight-gene sphingolipid signature associated with OS that includes S1PR3, S1PR5, and SPHK1 (the kinase that produces S1P) raises the possibility that altering the balance in S1P signaling could be sufficient to disrupt LSC maintenance in a subset of patients with AML. Our data suggest that next-generation S1P modulators approved for multiple sclerosis (21) may have therapeutic efficacy in a subset of patients with AML, although we cannot fully exclude the possibility that FTY720 targets LSCs via factors beyond S1P signaling (60). Our data also provide strong evidence supporting further investigation of recently identified highly potent SPHK1 inhibitors for their effectiveness in targeting LSCs (61, 62, 63). Another approach for therapeutic targeting of LSCs could be direct modulation of sphingolipid metabolic/biosynthetic enzymes such as those in the glycosphingolipid pathway that have recently emerged as regulators of NF-κB (64) and were included in the eight-gene sphingolipid signature. Sphingolipid metabolism has been linked to endoplasmic reticulum stress, autophagy, and inflammatory stress programs in cancer, obesity, and cardiovascular disease, but the mechanisms are ill-defined, especially around drug resistance (14, 15, 65, 66, 67). Our study opens a rich avenue for future mechanistic studies to elucidate the interplay of sphingolipid metabolism and signaling with these potential drug resistance programs in stem cell diseases such as AML. In conclusion, our findings point to a functional role for S1PR3 in myeloid differentiation of LSCs via the TNFα–NF-κB axis and highlight modulation of sphingolipids and their associated signaling pathways as a means to identify and target inflammatory pathways for the treatment of human AML.
Methods
Human Normal and Malignant Hematopoietic Samples
All biological samples were collected with written informed consent according to the procedures approved by the University Health Network (UHN) Research Ethics Board (REB 01-0573-C). Human CB samples were obtained from Trillium and Credit Valley Hospital and William Osler Health Centre, processed as previously described, and stored viably as lineage-depleted (lin−) cells at −80°C or −150°C (14). Normal human bone marrow samples or samples of patients with AML were obtained as viably frozen material from the Leukemia Tissue Bank at PM/UHN. CB MNCs, lin− CB, bone marrow MNCs, and AML patient samples were thawed by dropwise addition of X-VIVO + 50% FCS supplemented with DNase (100 μg/mL final concentration, Roche) and resuspended in PBS + 5% FBS for additional analyses below unless otherwise noted.
S1PR3 Flow Cytometry Analysis and Fluorescence-Activated Cell Sorting
Antibody reagents used throughout this study are listed in Supplementary Table S14. S1PR3 surface protein content was analyzed with an antibody against human S1PR3 (S1P3/EDG-3-PE, 1:30 dilution) in three sets of flow cytometry studies at 1 million cells/mL. Each sample was also analyzed with a panel lacking S1PR3-PE (fluorescence minus one, FMO). FlowJo 9.9 and 10 were used in this study for flow cytometry analysis and for calculation of S1PR3 mean fluorescence intensity (MFI).
1) S1PR3 expressions for CD34+ primitive cells and T (CD3+), B (CD19+), and myeloid (CD33+) cells in fresh human CB MNC samples (n = 4; 2 samples were single CBs from male donors and 2 samples were pools of 6 CB) were analyzed with the following panel on a BD Canto: FITC–anti-CD3, PECy5–anti-CD19, PE–anti-S1PR3, APC–anti-CD33, and APCCy7–anti-CD34. Live/dead discrimination was determined by sytox blue (Thermo Fisher Scientific, S34857).
2) Analysis of S1PR3 on the surface of HSPC populations was performed on three pools of lin− CB with the following panel on a BD LSR II: FITC–anti-CD45RA, PE–anti-S1PR3, APC–anti-CD90, PECy5–anti-CD49f, V450–anti-CD7, BV421–anti-CD10, PECy7–anti-CD38, and APCCy7–anti-CD34. Propidium iodide (BD) was used for live/dead discrimination.
3) Analysis of myeloid subpopulations in CB MNCs (CD15+ or CD14+ cells) and comparison of previously frozen CB MNCs with bone marrow MNCs and AML patient samples were performed by flow cytometry with the following panel on a BD Canto: FITC–anti-CD15, PE–anti-S1PR3, PECy5–anti-CD14, PECy7–anti-CD38, and APCCy7–anti-CD34 or APC–anti-CD34. Live/dead discrimination was determined by sytox blue. Clinical characteristics are listed in Supplementary Table S9. S1PR3 relative fluorescence intensity for each population is the ratio of S1PR3 MFI for the cells stained with S1PR3-PE divided by the FMO.
Lin− CB cells were stained with the following antibodies at a density of 5 × 106 cells/mL (1:50 dilution, unless stated otherwise) for isolation of HSPC subpopulations: FITC–anti-CD45RA, PE–anti-CD90, PECy5–anti-CD49f, V450–anti-CD7, PECy7–anti-CD38 (1:200), BV421–anti-CD10, APCCy7–anti-CD34 (1:200), APC–anti-CD71, and biotin–anti-Flt3. Cells were washed in PBS + 5% FBS and then stained with streptavidin-Qdot605. Cells were washed, resuspended in PBS + 2% FBS and propidium iodide, and FACS-purified on FACS Aria III or Aria Fusion (BD Biosciences), consistently yielding >95% purity. Mature cell populations including B cells, T cells, NK cells, DCs, granulocytes, monocytes, and erythroid progenitors were isolated from CB MNCs for RNA-seq (37).
Cell-surface markers used for each population are listed below.
LT-HSC: CD34+CD38−CD45RA−CD90+CD49f+
ST-HSC: CD34+CD38−CD45RA−CD90−CD49f−
Multipotent progenitor (MLP): CD34+CD38−CD45RA+CD90−
CMP: CD34+CD38+CD10−CD7−CD45RA−Flt3+
CMP-F1: CD34+CD38+CD10−CD7−CD45RA−Flt3+CD71−
GMP: CD34+CD38+CD10−CD7−CD45RA+
MEP: CD34+CD38+CD10−CD45RA−Flt3−
MEP-F1: CD34+CD38+CD10−CD7−CD45RA−Flt3−CD71−
T cell: CD3+CD19−
B cell: CD19+CD3−
NK cell: CD56+CD3−CD19−CD14−
Granulocyte: CD14+CD15+CD3−CD19−
Monocyte: CD14+CD15−CD3−CD19−
DC: CD11c+CD45+
Erythroid cell: CD45−GlyA+CD71+
For sphingolipidome profiling and xenotransplantation, nine samples were selected from the patient cohort previously analyzed for LSC activity (7). Thirty million live cells from 9 patients (Supplementary Table S10) were stained with the following antibodies at a density of 1 × 107 cells/mL: FITC–anti-CD45RA, PE–anti-S1PR3, PECy5–anti-CD33, V450–anti-CD15, PECy7–anti-CD38, V500–anti-CD45 (1:50), APC–anti-CD34, APCCy7–anti-CD3, and BV711–anti-CD19. Cells were washed, resuspended in PBS + 2% FBS and propidium iodide, sorted on CD34 and CD38 surface markers in the CD3−CD19− subpopulations into four fractions on a BD FACSAria Fusion, washed with PBS, and frozen viably.
Lentiviral OE and Knockdown of S1PR3
Lentiviral cloning and VSV-G pseudotyped lentiviral vector particles were produced, and titers calculated as previously described (39). A pLBC2 OE vector for human S1PR3 was generated from a pENTR223_S1PR3_STOP plasmid (a stop codon insertion into HsCD00375638 obtained from PlasmID, DF/HCC DNA Resource Core at Harvard Medical School). The control vector for OE encodes gp91phoxP415H (catalytic inactive gp91phox/CYBB). shRNA sequences to S1PR3 were predicted using the Sherwood algorithm and amplified as previously described (14). shCtrl is a sequence to Renilla that was previously utilized (14, 39). In order to assess shRNA KD efficiency, MOLM13 cells were infected at a multiplicity of infection of 0.3. Transduced cells were sorted for BFP+ expression, and total RNA was isolated and DNAse-treated using the RNeasy Micro Kit (Qiagen, 74004). RNA quality (RNA integrity number > 9) was verified using the Bioanalyzer RNA 6000 Pico Kit (Agilent), and cDNA was synthesized using SuperScript VILO (Thermo Fisher Scientific, 11754050). qPCR was performed on the Roche Lightcycler 480 using Power SYBR Green (Thermo Fisher Scientific, 4367659). All signals were quantified using the ΔCt method and were normalized to the levels of GAPDH.
shRNA sequences to S1PR3 are listed below:
shS1PR3-2: TGCTGTTGACAGTGAGCGCGGTCCCCACTCTTCATCCTCATAGTGAAGCCACAGATGTATGAGGATGAAGAGTGGGGACCATGCCTACTGCCTCGGA
shS1PR3-3: TGCTGTTGACAGTGAGCGACCATCTGGAAAAACAATAAAATAGTGAAGCCACAGATGTATTTTATTGTTTTTCCAGATGGCTGCCTACTGCCTCGGA
Xenotransplantation
All animal experiments were done in accordance with institutional guidelines approved by the UHN animal care committee. Aged-match female or male NSG mice (NOD.Cg PrkdcscidIl2rgtm1Wjl/SzJ; The Jackson Laboratory) 10 to 12 weeks of age were sublethally irradiated with 250 rads 1 day before intrafemoral injection. For CB xenotransplantation experiments, CD34+CD38− cells were transduced with S1PR3OE or control vectors at matching multiplicity of infection in a low-cytokine gene transfer media (38) composed of X-VIVO 10 media, 1% BSA supplemented with 1× Pen/Strep, l-glutamine, and the cytokines SCF (100 ng/mL), Flt3L (100 ng/mL), TPO (50 ng/mL), and IL7 (10 ng/mL) following 18 hours of prestimulation. Cells were collected and transplanted into NSG mice by intrafemoral injection 24 hours after transduction. Cells were also left in culture to assay for transduction efficiency (39) marked by %BFP+ at days 3 and 6 by flow cytometry on a BD Celesta, and the mean of the time points was used to mark initial input. Following xenotransplantation of S1PR3OE into NSG mice for 4 weeks, mice were euthanized and the injected femur and other bones were flushed separately in Iscove's Modified Dulbecco's medium with 5% FBS and human chimerism, and transduced cells marked by BFP expression were assessed by flow cytometry on the BD LSRII with the following: PE–anti-CD19, PE–anti-GlyA or PECy7–anti-GlyA, APC–anti-CD33, PECy5–anti-CD45, APCCy7–anti-CD34, FITC–anti-CD71, and BFP.
For limiting dilution xenotransplantation experiments of patient AMLs fractionated by surface S1PR3 protein expression, AML cells were thawed and stained with the same panel as for isolation of samples for lipid mass spectrometry. Within CD45+CD33+ blasts, CD34+ cells were gated and then isolated for S1PR3+ and S1PR3−/lo cells based on unstained and S1PR3-PE FMO controls. Cells were counted, and three different cell doses based on previous xenotransplantation experiments were injected into NSG mice for 12 weeks and then analyzed for engraftment. The following flow cytometry analysis was performed: FITC–anti-CD15, PE–anti-S1PR3, PECy5–anti-CD45, APC–anti-CD33, PECy7–anti-CD14, and APCCy7–anti-CD34. Mice were considered engrafted if CD33+CD45+ cells >0.05% in the right femur. For S1PR3OE xenotransplantation experiments of human AML patient samples where CD34+ cells were known to contain LSCs, control or S1PR3OE CD34+ cells were FACS-purified, transduced with control or S1PR3OE vectors overnight with previously described AML cytokine culture conditions (39), and then transplanted into NSG mice by intrafemoral injection. Following 8 weeks, right femurs and other leg bones were isolated and analyzed for engraftment by flow cytometry on a BD Celesta with the following: FITC–anti-CD15, PE–anti-S1PR3, V500–anti-CD45, BV786–anti-CD33, PECy5–anti-CD15, APCCy7–anti-CD34, and BFP.
In the FTY720 drug study, cells from 12 patient AMLs depleted of CD3+ T cells using EasySep (Stem Cell Technologies) or three pools of lin− CB (66,000 cells/mouse) were transplanted into NSG mice for 4 weeks (clinical characteristics in Supplementary Table S10). Vehicle control (DMSO) or 1 mg/kg FTY720 (Cayman Chemicals, 10006292, assuming average weight for NSG mouse to be 25 g) was diluted in PBS and administered to mice by i.p. injections every other day (7×) over 2 weeks. On the day after the last injection, mice were sacrificed, bones were isolated, and engraftment was analyzed by flow cytometry on a BD LSR or Celesta. CB engraftment was analyzed with APC–anti-CD33, PECy5–anti-CD45, FITC–anti-CD15, PE–anti-CD19, PE–anti-GlyA, PECy7–anti-CD14, and APCCy7–anti-CD34. AML engraftment (CD45+CD33+ cells) was analyzed with APC–anti-CD45, PECy5–anti-CD33, FITC–anti-CD3, V450–anti-CD15, PECy7–anti-CD14, and APCCy7–anti-CD34. Xenografted AML samples were frozen viably, and those samples that showed decreased engraftment with FTY720 or achieved engraftment over 20% were thawed; CD45+CD33+ cells were isolated using APC–anti-CD45, PECy5–anti-CD33, FITC–anti-CD45, and propidium iodide for serial transplantation assays in NSG mice with the indicated cell doses to assess LSC function at 8 weeks after transplantation (Supplementary Table S13; 9 of 12 samples tested for in the limiting dilution secondary transplantation assays, but AML 19 and AML 21 did not engraft and hence are not listed). Secondary engraftment was assessed by flow cytometry as in the primary transplants, and those mice that had greater than 0.5% CD45+CD33+ engraftment were considered responders. LSC frequency was calculated with ELDA (http://bioinf.wehi.edu.au/software/elda/).
TNFα or IL6 Treatment
Three pools of CB lin− cells were cultured in gene transfer media alone or plus 10 ng/mL human TNFα (Miltenyi Biotec, 130-094-018) or 50 ng/mL IL6 and a portion of cells analyzed on a BD Celesta at 3 days after treatment [FITC–anti-CD45RA, APC–anti-CD90, PECy5–anti-CD49f, Alexa Fluor–anti-CD7, PE-Dazzle–anti-CD38 (1:100), BV605–anti-CD10, APCCy7–anti-CD34, S1PR3-PE, and BV786–anti-CD33] and again at day 8 after treatment (APC–anti-CD90, FITC–anti-CD14, V450–anti-CD15, Alexa Fluor–anti-CD7, BV605–anti-CD10, APCCy7–anti-CD34, S1PR3-PE, and BV786–anti-GlyA).
In Vitro CB HSPC Assays
For in vitro assays, LT-HSCs and ST-HSCs were isolated from three independent pools of CB transduced with indicated OE or KD lentiviral vectors as for xenotransplantation experiments, except cells were prestimulated for 4 hours prior to transduction. For liquid culture, cells were transferred from gene transfer media to a high-cytokine StemPro media starting at day 3 after transduction (Stem Cell Technologies) supplemented with StemPro nutrients (Stem Cell Technologies), l-glutamine (GIBCO), Pen/Strep (GIBCO), human LDL (Stem Cell Technologies, 50 ng/mL), and the following cytokines (all from Miltenyi): SCF (100 ng/mL), Flt3L (20 ng/mL), TPO (100 ng/mL), IL6 (50 ng/mL), IL3 (10 ng/mL), and GM-CSF (20 ng/mL), except EPO (3 units/mL, from Janssen). At day 9 or 10, cells were stained for flow cytometry analysis with PeCy5–anti-CD14 (1:200), APCCy7–anti-CD34 (1:200), and BFP. PE–anti-S1PR3 (1:33) was added into one experiment to confirm enhanced expression. For CFC assays, on day 3 after lentiviral transduction of LT-HSCs, ST-HSCs, CMP-F1, MEP-F1, or GMPs, transduced cells marked by BFP were sorted directly into methylcellulose (cat. no. H4034, Stem Cell Technologies), supplemented with FLT3 Ligand (10 ng/mL) and IL6 (10 ng/mL). Samples were plated onto 35-mm dishes in duplicates, and colonies were allowed to differentiate for 10 days and morphologically assessed for colonies in a blind fashion by a second investigator. At day 14, colonies from replicate plates were pooled and resuspended in PBS + 5% FBS for flow cytometry analysis with PE–anti-Gly, PECy7–anti-CD33, APCCy7–anti-CD34, and BFP. For single-cell eythroid/myeloid/megakaryocyte differentiation assays, on day 3 postlentiviral transduction of LT-HSCs, ST-HSCs, CMP-F1, and MEP-F1, single BFP+ cells were sorted directly onto MS5 (44) stroma in 96-well plates prepared as previously described (28) and cultured for 15 days. Wells containing single cells scored as successfully cloned by eye were detached from MS5 stroma and analyzed by flow cytometry analysis for FITC–anti-CD15, PE–anti-GlyA, APC–anti-CD71, PeCy5–anti-CD14, APCH7–anti-CD41, PECy7–anti-CD33, and BFP on a high-throughput sampler unit of a BD Canto.
Time-lapse Imaging
Time-lapse experiments were conducted at 37°C, with 5% O2 and 5% CO2, on μ-slide VI0.4 channel slides (IBIDI) coated with 20 μg/mL anti-human CD43-biotin antibody (51). BFP+ cells 3 day after lentiviral transduction were sorted and cultured overnight in phenol red–free X-VIVO 10 (Lonza) medium supplemented with BSA (1%), l-glutamine, Pen/Strep, human LDL, and the cytokine cocktail described above before imaging. Brightfield images were acquired every 15 minutes for 4 days using a Nikon-Ti Eclipse equipped with a Hamamatsu Orca Flash 4.0 camera and a 10× CFI Plan Apochromat λ objective (NA 0.45). Single-cell tracking and fate assignment were performed using self-written software as previously described (51). Time to division was calculated using R 3.6.1.
Sphingolipid Quantitation by Mass Spectrometry
To profile the sphingolipid composition of LSC+ and LSC− fraction from samples of patients with AML, LSC+ and LSC− subpopulations (n = 7) were selected from the previously FACS-isolated viably banked samples that contained a minimum of 4 million cells at the time of sort (samples ranged from 4–12 million cells). These were thawed concurrently, and live cells counted, washed twice with PBS, and frozen as cell pellets with viable cells ranging from 1.5 to 5 million cells for LSC− samples and 3.4 to 5.3 million cells for LSC+ samples. The sphingolipid composition of HSPC CB cells (CD34+CD38− and CD34+CD38+ from three pools of lin− CB) was previously reported (14). Subsequent lipid extraction and mass spectrometry of the 14 AML subpopulations and 6 HSPC samples were performed together to allow for direct quantitative comparison of SpL species levels between samples. LC/MS-MS analysis for sphingomyelin species, hexosylceramide species, ceramide species, dihydroceramide species, and sphingoid species was performed by the Lipidomics Facility of Stony Brook University Medical Center on a SCIEX 4000 QTRAP mass spectrometer. Normalization to cellular inorganic phosphate (Pi) was chosen to minimize the potential confounding effects of differences in cellular size and protein content between the profiled populations. Sphingolipid composition for each sample was calculated as proportion of all sphingolipids analyzed and subsequently scaled. Using scanpy (68), a neighborhood graph was calculated with a local neighborhood size of 4, and a UMAP dimensionality reduction was performed.
CB Gene Expression Analysis
LT-HSCs and ST-HSCs from three pools of CB lin− were FACS-purified and transduced as described for in vitro culture assays in gene transfer media. At day 3, 2,000 to 5,300 BFP+ cells were FACS-purified for RNA isolation with a PicoPure kit (Thermo Fisher Scientific, KIT0214). Consistent with our observations that S1PR3OE increases the viability of LT-HSCs over control vectors in live-cell imaging studies (Fig. 2F–H), we were able to isolate only 1,600 to 1,800 BFP+ cells from LT-HSC control samples as opposed to 4,000 to 5,400 BFP+ cells from S1PR3OE samples. Thus, we pooled all control BFP+ LT-HSC cells into one sample for RNA-seq analysis. BFP− LT-HSC cells from control vector transduction were purified from CB1 as an additional LT-HSC control. In total, Nextera libraries generated from 10 ng RNA from five LT-HSC samples (two controls and three S1PR3OE) and six ST-HSC samples (three controls and three S1PR3OE) were subjected to 125 base-pair (bp), paired-end RNA-seq on the Illumina HiSeq 2500, with an average of 50 million reads/sample at the Centre for Applied Genomics, SickKids Hospital. S1PR3OE RNA-seq was aligned using STAR 2.5.2b (69) against GRCh38 and transcript sequences downloaded from Ensembl build 90. Default parameters were used except for the following: “–chimSegmentMin 12 –chimJunctionOverhangMin 12 –alignSJDBoverhangMin 10 –alignMatesGapMax 100000 –alignIntronMax 100000 –chimSegmentReadGapMax parameter 3 –alignSJstitchMismatchNmax 5 -1 5 5.” Counts were obtained using HTSeq v0.7.2 (70). Differential gene expression for the OE of S1PR3 compared with control (mtGP91 and BFP−) for both LT-HSCs and ST-HSCs was performed with DESeq2 1.18.1 using recommended settings. Briefly, the HTSeq counts from the entire RNA-seq cohort were imported into R (3.4.1) and normalized using RLE from DESeq2. To identify differentially expressed genes (DEG), this was followed by a dispersion estimate and a quasi-likelihood negative binomial generalized log-linear model. For visualization, data underwent variance stabilization as recommended by DESeq2. Gene set enrichment was performed using GSEA PreRanked v3.0 (http://www.broad.mit.edu/gsea/) against the hallmark gene set MSigDB v6.2 with recommended settings (Supplementary Table S1).
As independent verification for induction of a myeloid lineage gene expression program upon S1PR3OE, the HSC and granulocyte microarray dataset from GSE24759 (50) was retrieved for pathway and signature analysis using GSEA. Results were visualized as a network using Cytoscape 3.7.1 and EnrichmentMap 3.2.1. AutoAnnotate 1.3.2 automatically labeled pathway modules using most frequent words. HSC and granulocyte gene lists were added as gene sets to the enrichment map, and the significance of overlap with gene sets was calculated using the EnrichmentMap postanalysis integrated hypergeometric test. The CB hierarchy RNA-seq dataset was normalized and preprocessed as noted above. For final visualization, the mean expression of the CB replicates was utilized.
Human AML Gene Expression Analysis
The single-cell correlation analysis of S1PR3 and the S1PR3OE LT-HSC DEGs was performed in a previously published scRNA-seq dataset consisting of 11,641 malignant single cells from 12 patients with AML at diagnosis (3). Counts were scran normalized, and signatures were scored using AUCell with default settings. Signature enrichment was subsequently scaled to facilitate visualization. Four previously published bulk AML gene expression datasets and one dataset described here were used to analyze S1PR3 expression. (i) AML RNA-seq expression read counts and genomic analysis for mutations from the TCGA-AML study (52), consisting of 20,442 genes and 179 patients, were downloaded from the companion website of the original publication (https://tcga-data.nci.nih.gov/docs/publications/laml_2012). Gene expression is defined as log2 [1 + counts per million (CPM)], where the CPM are computed using R package edgeR after filtering and normalizing the raw counts. (ii) The Beat-AML data were downloaded from Supplementary Materials from Tyner and colleagues (35). The CPM table consisting of 22,843 genes and 451 patients, listed in Supplementary Table S9, contained 440 AML samples (280 diagnosis, 23 relapse, 137 not available). We utilized the 280 listed as diagnosis in our analysis here. (iii) GSE6891 (53) is a normalized RNA microarray AML dataset (AFFY HG U133 plus 2) using basic robust multiarray average consisting of 495 patient samples previously analyzed in Ng and colleagues (7). The gene symbols were obtained from R database hgu133plus2.db. Gene expression was calculated using maximum values of multiple probes for each gene, and those genes with SD < 0.01 across samples were filtered, resulting in 17,325 genes. The correlation of genetic mutation data to S1PR3 gene expression was analyzed for any genes reported with mutations in the Beat-AML, TCGA-AML, and GSE6891 cohorts. Briefly, the correlation coefficient and t test P value were calculated for S1PR3 gene expression to any mutation by comparing the two patient groups either containing the genetic mutation or wild-type using two-sample t test. We computed single-sample GSEA (ssGSEA) enrichment scores for the HSC/progenitor and myeloid signatures (Supplementary Table S6) generated from the van Galen dataset (3) and the NF-κB via TNFα hallmark gene set (gene lists in Supplementary Table S5) using the R GSVA_1.30.0 package. The ssGSEA score and gene expression color key represent the normalized values that are equal to centered scores divided by maximum of the absolute centered scores. We analyzed correlations of S1PR3 with the NF-κB genes in these three AML cohorts using R. This resulted in 75 common NF-κB genes correlated to S1PR3 with FDR < 0.05 in the three cohorts (correlation scores, correlation testing P values, and Benjamini and Hochberg FDR are located in Supplementary Table S8). These analyses were also performed on (iv) LSC+/LSC− microarray dataset and (v) an RNA-seq dataset generated from patients from PM described below.
For LSC+/LSC− analysis, normalized RNA microarray (GPL10558 Illumina HumanHT-12 V4.0) data consisting of 138 LSC+ and 89 LSC− samples were obtained from the Gene Expression Omnibus (GEO) database file GSE76008_non-normalized.txt.gz (7). Official gene symbols from the HUGO Gene Nomenclature Committee were retrieved from R database illuminaHumanv4.db. Filtering of duplicated genes (the maximum value was used) resulted in 17,685 genes. For GSVA and NF-κB gene set analysis to S1PR3, we normalized the log2-transformed data using quantile normalization. Following differential gene expression using limma v3.38.3 (71), GSEA was run using default settings against the custom gene sets for S1P-associated or HSC-enriched lipid metabolism–associated genes (ref. 14; SMPD4 was not found in the microarray data, hence we used 22 of 23 genes published previously as the lipid-stem signature) listed in Supplementary Table S12.
The PM-AML RNA-seq dataset consists of 81 AML patient samples (clinical data in Supplementary Table S9), processed in two batches. This dataset only contains patient samples that can engraft in the NSG mouse model. Five patients (90543, 598, 90240, 110484, and 100500) were included in both batches. RNA was extracted from bulk peripheral blood MNCs using the RNeasy Micro Kit (Qiagen Inc.). Libraries were constructed by SMART-Seq (Clontech Inc.). A paired-end, 50 bp flow-cell lane Illumina High seq 2000 yielded an average of 240 million sequence reads aligning to genome per sample at the Genome Sciences Centre, BC Cancer Agency for cohort 1. Cohort 2 was subjected to 125 bp, paired-end RNA-seq on the Illumina HiSeq 2500 with an average of 50 million reads/sample at the Centre for Applied Genomics, SickKids Hospital. Subsequent alignment and normalization were performed as for the S1PR3OE RNA-seq dataset described above. Then R was utilized to perform pathway correlation of S1PR3 and S1PR3OE LT-HSC signature to the indicated hallmark gene sets.
AML paired diagnosis-relapse gene expression analysis for S1PR3 was from RNA-seq data of 12 patient pairs previously described (5).
RNA-seq and ex vivo drug screening data were acquired from Tyner and colleagues (35). Gene expression data were normalized to transcripts per million, and AUC was used from the drug screening data to denote drug tolerance. Diagnostic AML samples were categorized as high S1PR3 and low S1PR3 by a median split on S1PR3 gene expression, and venetoclax tolerance was compared between the groups with a two-tailed t test.
Sparse regression analysis within a set of 54 sphingolipid genes was utilized to derive a weighted sphingolipid gene score applied to a large training cohort of 495 patients with AML (GSE6891). The score was then tested on two independent datasets (GSE12417 and the TCGA-AML cohorts) in univariate analysis.
AML Cell Line Assays
AML cell lines (Kasumi-1, RRID:CVCL_0589; KG-1, RRID:CVCL_0374; ME-1 RRID:CVCL_2110; PL-21 RRID:CVCL_2161; U937, RRID:CVCL_0007; and OCI-AML3, RRID:CVCL_1844) were obtained from the ATCC and cultured in MEM alpha medium (12571-063, GIBCO) supplemented with 20% FBS (F1051, Sigma), 2 mmol/L Pen/Strep (15140-122, GIBCO), and 1% l-glutamine (609-065-EL, Multicell), expanded for <4 passages to generate a stock for subsequent experiments. OCI-AML22 cell line model (56) was derived from the cells of a relapsed PM-AML patient and was cultured in X-Vivo medium (04-380Q, Lonza) supplemented with 20% BIT (9500, Stem Cell Technologies), 1% GlutaMAX (35050061, Thermo Fisher Scientific), 0.2% Primocin (ant-pm-1, Invitrogen), 100 ng/mL SCF (130-096-696, Miltenyi), 10 ng/mL IL3 (130-095-068, Miltenyi), 10 ng/mL TPO (130-095-752, Miltenyi), 10 ng/mL FLT3 Ligand (130-096-474, Miltenyi), and 10 ng/mL G-CSF (130-093-866, Miltenyi). Experiments were conducted from cells within 2 to 3 months of thawing.
For Nanonstring analysis of the LSC104 score (7) from the indicated AML cell lines, RNA from FACS-sorted viable cells was isolated with TRIzol (Invitrogen) or for low cell numbers (<20,000 cells) with PicoPure RNA Isolation Kit (Thermo Fisher Scientific) according to the manufacturer's protocol. The Nanostring nCounter system (NanoString Technologies) was used for multiplexed RNA quantification, and gene expression was analyzed as previously described (7). S1PR3 surface expressions for the AML cell lines were analyzed by flow cytometry on a BD Celesta with the following panel: FITC–anti-CD15, PE–anti-S1PR3, APC–anti-CD38, PeCy5–anti-CD14, APCCy7–anti-CD34, and sytox blue or an FMO panel lacking PE–anti-S1PR3. Statistical analysis of the LSC104 correlation score to S1PR3 MFI was calculated in R (3.6.1).
Cells were spinoculated with 5 μg/mL polybrene (Sigma) at 1,400 × g for 90 minutes for lentiviral transduction. Seventy-two hours after transduction with S1PR3OE, BFP+ cells from the KG-1, Kasumi-1, and ME-1 cell lines were FACS-purified on the Beckman Coulter MoFlo XDP or Sony SH800. For subsequent flow cytometry, BFP+ cells were stained with the following antibodies and analyzed on the BD Celesta flow cytometer: FITC–anti-CD15, PeCy5–anti-CD11b, APCCy7–anti-CD34, and Annexin V–APC (BD). CD34+ cells from Kasumi-1 and OCI-AML22 were isolated on the FACS Aria III or Fusion (BD Biosciences) and transduced with control or S1PR3OE or shCtrl lentiviral vectors, and BFP+ cells were FACS-purified at 72 hours after transduction and analyzed at the indicated time points with FITC–anti-CD15, PE–anti-S1PR3, and APCCy7–anti-CD34 on the BD Celesta. OCI-AML22 or Kasumi-1 CD34+ cells were cultured in 96-well bottom plates with media alone or with 10 ng/mL TNFα (Miltenyi Biotec, 130-094-018) with and without 5 μmol/L S1PR3 antagonist TY5126 (Tocris, 5238) or DMSO control (1:5,000 dilution) and analyzed at days 5 and 9 after treatment by flow cytometry with FITC–anti-CD15, PE–anti-S1PR3, and APCCy7–anti-CD34 on the BD Celesta. For p65 NF-κB flow cytometry analysis, Kasumi-1 CD34+ cells were transduced with indicated lentiviral vectors. Then, BFP+ cells were isolated on the FACS Aria III or Fusion (BD Biosciences) at day 5 after transduction and fixed for intracellular flow cytometry with BD cytoperm/cytofix buffers as described previously (38), stained with APC–anti-p65 overnight at 4°C, and analyzed on a BD Celesta.
Statistical Analyses
GraphPad Prism was used for all statistical analyses except for gene expression and other indicated studies where the analyses were performed with R. Unless otherwise indicated, mean ± SD values are reported in the graphs.
Data Availability
The S1PR3OE RNA-seq data for LT-HSC and ST-HSC (GSE149238) and the PM bulk AML RNA-seq data (GSE156914) were deposited in the GEO. Raw data are available under accession numbers EGAS00001004798 (S1PR3OE) and EGAS00001004792 (AML) in the European Genome-phenome Archive. The human CB 13 population RNA-seq data are available from the authors upon request (37) and includes five HSPC populations at GSE125345. All other data supporting the findings of this study are cited in the Methods, available within the article, or upon request from the authors.
Supplementary Material
Acknowledgments
J.E. Dick is supported by funds from the Princess Margaret Cancer Centre Foundation; Ontario Institute for Cancer Research through funding provided by the Government of Ontario; Canadian Institutes for Health Research grants 130412, 89932, and 154293; International Development Research Centre Ottawa Canada grants 108401 and 109153; Canadian Cancer Society grant 703212; Terry Fox New Frontiers Program Project Grant 1047; University of Toronto's Medicine by Design initiative with funding from the Canada First Research Excellence Fund; and a Canada Research Chair. E. Laurenti is supported by Wellcome grant 107630/Z/15/Z and a core support grant from Wellcome and Medical Research Council (MRC) to the Wellcome - MRC Cambridge Stem Cell Institute. C. Luberto is supported by NIH, NCI grant P01-CA097132. W. Wang was supported by the Swiss Initiative in Systems Biology Transition Postdoc fellowship.
The authors thank the obstetrics units of Trillium Health Partners, Credit Valley and William Osler, the PMH Leukemia Biobank, members of the UHN-SickKids Flow Cytometry Facility, I. Mileva at the Lipidomics Facility of Stony Brook University Cancer Center, K. Golan, Z. Tothova, and T. Hla for support. This work is dedicated to the memory of Lina Obeid. The authors thank all members of the Dick lab for critical feedback.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
#K.B. Kaufmann, W. Wang, and M. Chan-Seng-Yue contributed equally to this article.
Blood Cancer Discov 2021;2:32–53
Authors' Disclosures
K.B. Kaufmann reports personal fees from Deutsche Forschungsgemeinschaft (DFG) during the conduct of the study, as well as a patent for intellectual property around OCIAML22 pending. E. Laurenti reports grants from GlaxoSmithKline outside the submitted work. S.-i. Takayanagi reports personal fees from Kirin outside the submitted work. J.C.Y. Wang reports other from Trillium Therapeutics Inc. outside the submitted work. C. Luberto reports grants from NIH/NCI during the conduct of the study. J.E. Dick reports grants from Canadian Institutes for Health Research, International Development Research Centre Ottawa Canada, Canadian Cancer Society, Terry Fox New Frontiers Program Project, University of Toronto & Medicine by Design initiative with funding from the Canada First Research Excellence Fund, Princess Margaret Cancer Centre Foundation, Ontario Institute for Cancer Research through funding provided by the Government of Ontario, and Canada Research Chair during the conduct of the study; grants from Celgene/Bristol-Myers Squibb and personal fees from Trillium Therapeutics Inc. outside the submitted work; and a patent for OCI-AML-22 cell line pending. No disclosures were reported by the other authors.
Authors' Contributions
S.Z. Xie: Conceptualization, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. K.B. Kaufmann: Resources, investigation, methodology, writing–review and editing. W. Wang: Investigation. M. Chan-Seng-Yue: Data curation, software, formal analysis. O.I. Gan: Resources, investigation. E. Laurenti: Investigation. L. Garcia-Prat: Investigation. S.-i. Takayanagi: Resources. S.W.K. Ng: Data curation, formal analysis. C. Xu: Formal analysis. A.G.X. Zeng: Formal analysis. L. Jin: Methodology. J. McLeod: Methodology. E. Wagenblast: Methodology. A. Mitchell: Resources, methodology. J.A. Kennedy: Data curation. Q. Liu: Investigation. H. Boutzen: Resources. M. Kleinau: Investigation. J. Jargstorf: Investigation. G. Holmes: Investigation. Y. Zhang: Investigation. V. Voisin: Formal analysis, supervision. G.D. Bader: Supervision. J.C.Y. Wang: Supervision, writing–review and editing. Y.A. Hannun: Writing–review and editing. C. Luberto: Investigation, writing–review and editing. T. Schroeder: Supervision. M.D. Minden: Resources, writing–review and editing. J.E. Dick: Supervision, funding acquisition, writing–review and editing.
References
- 1. Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med 2015;373:1136–52. [DOI] [PubMed] [Google Scholar]
- 2. Thomas D, Majeti R. Biology and relevance of human acute myeloid leukemia stem cells. Blood 2017;129:1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Galen P, Hovestadt V, Wadsworth Ii MH, Hughes TK, Griffin GK, Battaglia S, et al. Single-cell RNA-Seq reveals AML hierarchies relevant to disease progression and immunity. Cell 2019;176:1265–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 2016;48:1193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shlush LI, Mitchell A, Heisler L, Abelson S, Ng SWK, Trotman-Grant A, et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature 2017;547:104–8. [DOI] [PubMed] [Google Scholar]
- 6. Ye H, Adane B, Khan N, Sullivan T, Minhajuddin M, Gasparetto M, et al. Leukemic stem cells evade chemotherapy by metabolic adaptation to an adipose tissue niche. Cell Stem Cell 2016;19:23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016;540:433–7. [DOI] [PubMed] [Google Scholar]
- 8. Wilkinson AC, Göttgens B. Transcriptional regulation of haematopoietic stem cells. : Hime G, Abud H, . Transcriptional and translational regulation of stem cells. Dordrecht (the Netherlands): Springer Science+Business Media; 2013. p.187–212. [DOI] [PubMed] [Google Scholar]
- 9. Etzrodt M, Ahmed N, Hoppe PS, Loeffler D, Skylaki S, Hilsenbeck O, et al. Inflammatory signals directly instruct PU.1 in HSCs via TNF. Blood 2019;133:816–9. [DOI] [PubMed] [Google Scholar]
- 10. Ye M, Zhang H, Yang H, Koche R, Staber PB, Cusan M, et al. Hematopoietic differentiation is required for initiation of acute myeloid leukemia. Cell Stem Cell 2015;17:611–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hemmati S, Haque T, Gritsman K. Inflammatory signaling pathways in preleukemic and leukemic stem cells. Front Oncol 2017;7:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol 2014;15:243–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 14. Xie SZ, Garcia-Prat L, Voisin V, Ferrari R, Gan OI, Wagenblast E, et al. Sphingolipid modulation activates proteostasis programs to govern human hematopoietic stem cell self-renewal. Cell Stem Cell 2019;25:639–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature 2014;510:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cartier A, Hla T. Sphingosine 1-phosphate: lipid signaling in pathology and therapy. Science 2019;366:eaar5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hannun YA, Obeid LM. Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol 2018;19:175–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brinkmann V, Davis MD, Heise CE, Albert R, Cottens S, Hof R, et al. The immune modulator FTY720 targets sphingosine 1-phosphate receptors. J Biol Chem 2002;277:21453–7. [DOI] [PubMed] [Google Scholar]
- 19. Golan K, Vagima Y, Ludin A, Itkin T, Cohen-Gur S, Kalinkovich A, et al. S1P promotes murine progenitor cell egress and mobilization via S1P1-mediated ROS signaling and SDF-1 release. Blood 2012;119:2478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blaho VA, Galvani S, Engelbrecht E, Liu C, Swendeman SL, Kono M, et al. HDL-bound sphingosine-1-phosphate restrains lymphopoiesis and neuroinflammation. Nature 2015;523:342–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cohan S, Lucassen E, Smoot K, Brink J, Chen C. Sphingosine-1-phosphate: its pharmacological regulation and the treatment of multiple sclerosis: a review article. Biomedicines 2020;8:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pietras EM, Mirantes-Barbeito C, Fong S, Loeffler D, Kovtonyuk LV, Zhang S, et al. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat Cell Biol 2016;18:607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yamashita M, Passegue E. TNF-alpha coordinates hematopoietic stem cell survival and myeloid regeneration. Cell Stem Cell 2019;25:357–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bruunsgaard H, Pedersen M, Pedersen BK. Aging and proinflammatory cytokines. Curr Opin Hematol 2001;8:131–6. [DOI] [PubMed] [Google Scholar]
- 25. Puchta A, Naidoo A, Verschoor CP, Loukov D, Thevaranjan N, Mandur TS, et al. TNF drives monocyte dysfunction with age and results in impaired anti-pneumococcal immunity. PLoS Pathog 2016;12:e1005368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trabado S, Al-Salameh A, Croixmarie V, Masson P, Corruble E, Feve B, et al. The human plasma-metabolome: reference values in 800 French healthy volunteers; impact of cholesterol, gender and age. PLoS One 2017;12:e0173615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kowalski GM, Carey AL, Selathurai A, Kingwell BA, Bruce CR. Plasma sphingosine-1-phosphate is elevated in obesity. PLoS One 2013;8:e72449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu Z, Zhai G, Singmann P, He Y, Xu T, Prehn C, et al. Human serum metabolic profiles are age dependent. Aging Cell 2012;11:960–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 2017;355:842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abegunde SO, Buckstein R, Wells RA, Rauh MJ. An inflammatory environment containing TNFα favors Tet2-mutant clonal hematopoiesis. Exp Hematol 2018;59:60–5. [DOI] [PubMed] [Google Scholar]
- 31. Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 2017;377:111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kagoya Y, Yoshimi A, Kataoka K, Nakagawa M, Kumano K, Arai S, et al. Positive feedback between NF-kappaB and TNF-alpha promotes leukemia-initiating cell capacity. J Clin Invest 2014;124:528–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guzman ML, Neering SJ, Upchurch D, Grimes B, Howard DS, Rizzieri DA, et al. Nuclear factor-κB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood 2001;98:2301–7. [DOI] [PubMed] [Google Scholar]
- 34. Xiu Y, Dong Q, Li Q, Li F, Borcherding N, Zhang W, et al. Stabilization of NF-kappaB-inducing kinase suppresses MLL-AF9-induced acute myeloid leukemia. Cell Rep 2018;22:350–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cancer Genome Atlas Research Network; Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. García-Prat L, Kaufmann KB, Schneiter F, Voisin V, Murison A, Chan-Seng-Yue M, et al. Dichotomous regulation of lysosomes by MYC and TFEB controls hematopoietic stem cell fate. Blood 2020;136 (Suppl 1):34. [Google Scholar]
- 38. Laurenti E, Frelin C, Xie S, Ferrari R, Dunant CF, Zandi S, et al. CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell 2015;16:302–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kaufmann KB, Garcia-Prat L, Liu Q, Ng SWK, Takayanagi SI, Mitchell A, et al. A stemness screen reveals C3orf54/INKA1as a promoter of human leukemia stem cell latency. Blood 2019;133:2198. [DOI] [PubMed] [Google Scholar]
- 40. Notta F, Zandi S, Takayama N, Dobson S, Gan OI, Wilson G, et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 2016;351:aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Itoh K, Tezuka H, Sakoda H, Konno M, Nagata K, Uchiyama T, et al. Reproducible establishment of hemopoietic supportive stromal cell lines from murine bone marrow. Exp Hematol 1989;17:145–53. [PubMed] [Google Scholar]
- 42. Haas S, Hansson J, Klimmeck D, Loeffler D, Velten L, Uckelmann H, et al. Inflammation-induced emergency megakaryopoiesis driven by hematopoietic stem cell-like megakaryocyte progenitors. Cell Stem Cell 2015;17:422–34. [DOI] [PubMed] [Google Scholar]
- 43. Persaud L, De Jesus D, Brannigan O, Richiez-Paredes M, Huaman J, Alvarado G, et al. Mechanism of action and applications of interleukin 24 in immunotherapy. Int J Mol Sci 2016;17:869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vorbach S, Grunder A, Zhou F, Koellerer C, Jutzi JS, Simoni M, et al. Enhanced expression of the sphingosine-1-phosphate-receptor-3 causes acute myelogenous leukemia in mice. Leukemia 2019;34:721–34. [DOI] [PubMed] [Google Scholar]
- 45. Allende ML, Bektas M, Lee BG, Bonifacino E, Kang J, Tuymetova G, et al. Sphingosine-1-phosphate lyase deficiency produces a pro-inflammatory response while impairing neutrophil trafficking. J Biol Chem 2011;286:7348–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krumsiek J, Marr C, Schroeder T, Theis FJ. Hierarchical differentiation of myeloid progenitors is encoded in the transcription factor network. PLoS One 2011;6:e22649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jin G, Yamazaki Y, Takuwa M, Takahara T, Kaneko K, Kuwata T, et al. Trib1 and Evi1 cooperate with Hoxa and Meis1 in myeloid leukemogenesis. Blood 2007;109:3998–4005. [DOI] [PubMed] [Google Scholar]
- 48. Murphy JM, Nakatani Y, Jamieson SA, Dai W, Lucet IS, Mace PD. Molecular mechanism of CCAAT-enhancer binding protein recruitment by the TRIB1 pseudokinase. Structure 2015;23:2111–21. [DOI] [PubMed] [Google Scholar]
- 49. Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 2006;126:755–66. [DOI] [PubMed] [Google Scholar]
- 50. Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 2011;144:296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Loeffler D, Wehling A, Schneiter F, Zhang Y, Muller-Botticher N, Hoppe PS, et al. Asymmetric lysosome inheritance predicts activation of haematopoietic stem cells. Nature 2019;573:426–9. [DOI] [PubMed] [Google Scholar]
- 52. Cancer Genome Atlas Research Network, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Verhaak RGW, Wouters BJ, Erpelinck CAJ, Abbas S, Beverloo HB, Lugthart S, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica 2009;94:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pei S, Pollyea DA, Gustafson A, Stevens BM, Minhajuddin M, Fu R, et al. Monocytic subclones confer resistance to venetoclax-based therapy in patients with acute myeloid leukemia. Cancer Discov 2020;10:536–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kuusanmäki H, Leppä A-M, Pölönen P, Kontro M, Dufva O, Deb D, et al. Phenotype-based drug screening reveals association between venetoclax response and differentiation stage in acute myeloid leukemia. Haematologica 2020;105:708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Boutzen H, Mandani A, Takayama N, Murison A, Mbong N, Mitchell A, et al. Uncovering non-coding epigenetic determinants that drive stemness programs in human leukaemia stem cells using a hierarchically-organized patient-derived model system. In preparation.
- 57. Wang Z-Y, Chen Z. Acute promyelocytic leukemia: from highly fatal to highly curable. Blood 2008;111:2505–15. [DOI] [PubMed] [Google Scholar]
- 58. Raffel S, Falcone M, Kneisel N, Hansson J, Wang W, Lutz C, et al. BCAT1 restricts alphaKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature 2017;551:384–8. [DOI] [PubMed] [Google Scholar]
- 59. Jones CL, Stevens BM, D'Alessandro A, Reisz JA, Culp-Hill R, Nemkov T, et al. Inhibition of amino acid metabolism selectively targets human leukemia stem cells. Cancer Cell 2018;34:724–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen L, Luo LF, Lu J, Li L, Liu YF, Wang J, et al. FTY720 induces apoptosis of M2 subtype acute myeloid leukemia cells by targeting sphingolipid metabolism and increasing endogenous ceramide levels. PLoS One 2014;9:e103033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Powell JA, Lewis AC, Zhu W, Toubia J, Pitman MR, Wallington-Beddoe CT, et al. Targeting sphingosine kinase 1 induces MCL1-dependent cell death in acute myeloid leukemia. Blood 2017;129:771–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Barth BM, Wang W, Toran PT, Fox TE, Annageldiyev C, Ondrasik RM, et al. Sphingolipid metabolism determines the therapeutic efficacy of nanoliposomal ceramide in acute myeloid leukemia. Blood Adv 2019;3:2598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tan SF, Liu X, Fox TE, Barth BM, Sharma A, Turner SD, et al. Acid ceramidase is upregulated in AML and represents a novel therapeutic target. Oncotarget 2016;7:83208–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chao CC, Gutierrez-Vazquez C, Rothhammer V, Mayo L, Wheeler MA, Tjon EC, et al. Metabolic control of astrocyte pathogenic activity via cPLA2-MAVS. Cell 2019;179:1483–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tam AB, Roberts LS, Chandra V, Rivera IG, Nomura DK, Forbes DJ, et al. The UPR activator ATF6 responds to proteotoxic and lipotoxic stress by distinct mechanisms. Dev Cell 2018;46:327–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mann MJ, Hendershot LM. UPR activation alters chemosensitivity of tumor cells. Cancer Biol Ther 2006;5:736–40. [DOI] [PubMed] [Google Scholar]
- 67. Halbleib K, Pesek K, Covino R, Hofbauer HF, Wunnicke D, Hanelt I, et al. Activation of the unfolded protein response by lipid bilayer stress. Mol Cell 2017;67:673–84. [DOI] [PubMed] [Google Scholar]
- 68. Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 2018;19:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2012;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The S1PR3OE RNA-seq data for LT-HSC and ST-HSC (GSE149238) and the PM bulk AML RNA-seq data (GSE156914) were deposited in the GEO. Raw data are available under accession numbers EGAS00001004798 (S1PR3OE) and EGAS00001004792 (AML) in the European Genome-phenome Archive. The human CB 13 population RNA-seq data are available from the authors upon request (37) and includes five HSPC populations at GSE125345. All other data supporting the findings of this study are cited in the Methods, available within the article, or upon request from the authors.