Abstract
DZIP3/hRUL138 is a poorly characterized RNA binding RING E3-ubiquitin ligase with functions in embryonic development. Here we demonstrate that DZIP3 is a crucial driver of cancer cell growth, migration, and invasion. In mice and zebrafish cancer models, DZIP3 promoted tumor growth and metastasis. In line with these results, DZIP3 was frequently overexpressed in several cancer types. Depletion of DZIP3 from cells resulted in reduced expression of Cyclin D1 and a subsequent G1 arrest and defect in cell growth. Mechanistically, DZIP3 utilized its two different domains to interact and stabilize Cyclin D1 both at mRNA and protein levels. Using an RNA-binding lysine-rich region, DZIP3 interacted with the AU-rich region in 3'UTR of Cyclin D1 mRNA and stabilized it. Using a RING E3-ligase domain, DZIP3 interacted and increased K63-linked ubiquitination of Cyclin D1 protein to stabilize it. Remarkably, DZIP3 interacted with, ubiquitinated, and stabilized Cyclin D1 predominantly in the G1 phase of the cell cycle where it is needed for cell cycle progression. In agreement with this, a strong positive correlation of mRNA expression between DZIP3 and Cyclin D1 in different cancer types was observed. Additionally, DZIP3 regulated several cell cycle proteins by modulating the Cyclin D1-E2F axes. Taken together, this study demonstrates for the first time that DZIP3 employs a unique two-pronged mechanism in its stabilization of Cyclin D1 to drive cell cycle and cancer progression.
Keywords: DZIP3, E3-ubiquitin ligase, Cyclin D1, K63-linked ubiquitination, Oncogene, RNA-binding proteins, RNA-binding RING E3-ligase
Introduction
The human genome encodes more than six hundred E3-ligases and a similar number of RNA binding proteins (RBPs) with a variety of functions in protein and RNA metabolism (1,2). RNA binding RING E3 ligases (RBRLs) are a unique group of a small number of proteins (~20 in number) with a remarkable capability of regulating both RNA and protein metabolism, and also coupling them (3–7). However, the RBRL family proteins remain poorly characterized. Given the role of these proteins in cancer progression and innate immunity (5,6,8), it is crucial to understand the mechanisms by which the RBRLs regulate the turnover of their target RNA and proteins.
The DAZ (Deleted in Azoospermia) family genes are essential for gametogenesis and other developmental processes (9). DAZ family proteins interact with several other zinc finger proteins, including DZIP1, DZIP2 (DZIP1L), and DZIP3. DZIP1 and DZIP1L are centrosomal proteins and have been shown to regulate the Hedgehog signaling pathway, ciliogenesis, and cell cycle (10–12). DZIP3 (DAZ-interacting protein 3) is a poorly characterized RNA-binding RING-H2 ubiquitin E3 ligase, which was originally identified as a Hepatitis B virus interacting protein (13). The RNA binding and E3 ligase functions of DZIP3 were more clearly identified in a recent study where it was shown to interact with HOTAIR (a long noncoding RNA) to mediate ubiquitination and degradation of Ataxin 1 (14). DZIP3 is important for controlling the genes during embryonic development (15,16). DZIP3 interact with CARM1 (an arginine methyltransferase) and act as a transcriptional coactivator of estrogen receptor alpha-responsive genes (17). Although DZIP3 is under-characterized, the studies indicate that it is a multifunctional protein.
In eukaryotic cells, the levels of cell cycle proteins are precisely maintained to ensure an orderly progression of the cell cycle. Dysregulated expression of cell cycle proteins can result in uncontrolled cell growth and cancer. Cyclin D1 plays a critical role in cell cycle progression. Overexpression of Cyclin D1 is a driving feature in a large number of cancer types, including Leukemia, head and neck, breast, non-small cell lung cancer, and prostate (18,19). Multiple mechanisms comprising genomic alterations, post-transcriptional regulation, and post-translational protein stabilization results in Cyclin D1 overexpression leading to a deregulated cell cycle resulting in uncontrolled cell growth and cancer (18–22).
Two E3 ligase complexes, APC/C (Anaphase Promoting Complex/Cyclosome) and Skp/cullin/F-box (SCF) act by increasing K48-linked ubiquitination of cell cycle proteins, and mediate their timely degradation for precise cell cycle progression (23). The cell cycle protein expression is also regulated at mRNA levels by RBPs such as HuR, which binds and stabilizes the mRNA of several cell cycle proteins, including p53, p21, cyclin A, cyclin B1, cyclin E, cyclin D and cdk1 (24–31). Another recently recognized mechanism of regulation of cyclin proteins is by deubiquitinating enzymes such as OTUD7B, USP22, and USP27 in a cell cycle phase-specific manner (32–34). This modification antagonizes the proteasomal degradation of cell cycle proteins, leading to their cell cycle phase specific stabilization. Another layer of regulation is provided by certain E3 ligases, which can increase the K63-linked ubiquitination of specific cell cycle proteins in one particular phase of the cell cycle, resulting in their stabilization (35–37).
In this study, we found that DZIP3 is a novel oncogene with a capacity to drive cancer cells anchorage-independent growth, migration, and invasion. Increased expression of DZIP3 was observed in human cancer patients tumor samples. In agreement, DZIP3 was found to be crucial for cancer progression and metastasis in mice and zebrafish. We show that DZIP3 controls cancer cell growth by regulating the cell cycle and Cyclin D1 stability. DZIP3 utilizes a two-pronged mechanism to positively regulate the expression of Cyclin D1. Firstly, DZIP3 stabilizes the Cyclin D1 transcripts by binding to its 3' untranslated region (UTR), and secondly, DZIP3 interacts and increases K63-linked ubiquitination of Cyclin D1 to stabilize it post-translationally. In addition, DZIP3 controls several of the E2F transcription factor regulated cell cycle and proliferation genes including Cyclin E1, Cyclin A2, CDK1, CDK2, and c-MYC. Taken together, this study identifies DZIP3 as a novel driver of cell cycle and cancer progression by regulating the expression of Cyclin D1 in a unique manner.
Materials and Methods
Cell culture
The cell lines used in the study were obtained from the American Type Culture Collection (ATCC). MCF7, MDA-MB-231, HT-29 (RRID:CVCL_0320), UM-UC3 (RRID:CVCL_1783), HeLa (RRID:CVCL_0030), HEK293, HEK293T (RRID:CVCL_0063) cells were cultured in DMEM medium supplemented with 10% Fetal bovine serum (FBS, Gibco) and penicillin/streptomycin (10,000 units/mL). The cells were tested for mycoplasma contamination routinely (every 2-3 months) using the PCR method. The cell lines were maintained below passage number 20.
Reagents and inhibitors
Cycloheximide (cat #C7698; 100 μg/ml), Puromycin (cat #p8833; 2 μg/ml), Thymidine (cat #T1895; 2 mM), Nocodazole (cat #M1404; 100 ng/ml) PMSF (cat # P7626-5G) were from Sigma. Protease Inhibitor (cat # 11836170001) and phosphatase inhibitors (CAT # 04906845001) were obtained from Roche.
Plasmids, siRNA, and transfection
pRK5-HA-Ubiquitin-K48 (#17605; RRID: Addgene_17604), HA-Ubiquitin (#18712; RRID: Addgene_18712), pRK5-HA-Ubiquitin-K63 (#17606; RRID:Addgene_17606), Rc/CMV Cyclin D1 HA (#8948) plasmids were purchased from addgene. pSG5HA-DZIP3 and pSG5FLAG-DZIP3 were described previously (17). Flag-DZIP3 and Flag-DZIP3 deletion constructs were cloned in gateway cloning vectors as per standard protocol (Invitrogen). For transient knockdown, cells were transfected by electroporation using the Neon transfection system (Invitrogen) and also using INTERFERin (Polyplus) or RNAimax (Invitrogen) as per the manufacturer's instruction. For transient overexpression, Lipofectamine 2000 (Invitrogen) and CALPHOS (Clontech) were used according to the manufacturer's instructions.
CRISPR knockout cells generation
The HEK293T or MCF7 cells were transfected with DZIP3 CRISPR Cas9 (Santacruz; sc-403972) containing a pool of 3 sgRNA's along with DZIP3 HDR plasmids (Santacruz; sc-403972). After 48h, the media was changed, and cells were selected in puromycin (2 µg/mL). The individual colonies were picked, grown, and knockout was evaluated by using western blot analysis.
Western blotting
The cell lysates were prepared in NP-40 (FNN0021, Thermo Fisher Scientific) or Radio-immunoprecipitation assay (RIPA) buffer (20 mM Tris, pH 8.0; 1mM, EDTA; 0.5 mM, EGTA; 0.1% Sodium deoxycholate; 150 mM NaCl; 1% IGEPAL (Sigma); 10% glycerol) supplemented with protease inhibitor cocktail and 1mM PMSF. The western blotting is performed as described previously (38–40).
Cycloheximide chase assay
The cycloheximide chase assay experiments were performed by treating the cells with 100 µg/ml cycloheximide. Cell lysates were prepared at indicated time points and were subjected to western blot analysis with indicated antibodies.
Immunoprecipitation assay
For Immunoprecipitation assays, cells were lysed in NP-40 lysis buffer supplemented with protease inhibitor cocktail and 1 mM PMSF for 20 min at 40C and centrifuged. The supernatant was incubated with the respective antibody at 40 C for 2 h on rotospin followed by incubation with Protein G Dynabeads (Invitrogen, #10004D) for 2 h at 40C. The beads were washed with 1X NP40 and 3X ice-cold PBS. The proteins were eluted from washed beads by boiling for 5 min in 2X SDS gel loading dye and proceeded for immunoblot analysis.
GST pull-down assay
GST pull-down assay was performed according to the methods previously described (39,41). GST or GST-DZIP3 (CCD-RING) proteins were expressed in SoluBL21 (Amsbio), and the proteins were purified on Glutathione Sepharose 4 Fast-Flow beads (GE Healthcare). [35S]-labeled HA-CCND1 protein was in-vitro translated using TnT T7–coupled reticulocyte lysate system (Promega). The GST proteins were incubated with [35S]-labeled HA-CCND1 in 250 ml of NETN-E buffer (50 mm Tris, pH 8.0, 100 mm NaCl, 6 mm EDTA, 6 mm EGTA, 0.5% NP-40, and 1 mm dithiothreitol supplemented with complete mini EDTA-free protease inhibitor cocktail [Roche]) for 2 h at 4°C. Then, GST beads were added, and the mixture was incubated for 30 min at room temperature. The beads were washed with NETN-E buffer five times, boiled with loading buffer, and subjected to SDS-PAGE. The gel was stained with Coomassie Blue and vacuum-dried. The GST or GST-DZIP3 (CCD-RING) was detected by staining with Coomassie Blue, whereas the [35S]-labelled HA-CCND1 was detected in PharosFX imager (Bio-Rad Laboratories).
Colony-forming assay
A total of ~1000 cells in 2 mL cell culture medium were seeded in triplicate in 6-well plates and allowed to grow for 2-3 weeks until the colonies were formed. Cells were fixed in methanol: acetic acid (3:1) for 5 min at room temperature and stained with 0.5% crystal violet for 15 min and washed with water. The images were captured using a digital camera, and colonies were counted.
MTT assay
5000 cells were seeded in triplicate in 25 cm2 flask and allowed to grow for 5 days. Subsequently, at each time point, media was removed, cells were washed with PBS, and were incubated in the dark with MTT (3-[4,5-dimethylthiazolyl-2]-2,5-diphenyltetrazolium bromide) (5 mg/ml, Sigma) for 2-4 h. The dye precipitates were dissolved in DMSO, and the absorbance was measured at 570 nm.
In vitro scratch assay
The cells were grown in six-well plates with a 60-70% confluency and were subjected to a uniform (approximately 1mm in area) scratch with 10ul pipette tip across the width of the well. EVOSTm inverted fluorescence microscope (cell imaging system, Thermo Scientific) was used to measure and photomicrograph the cell migration rate after post wounding at every 24h time duration (0, 24, 48, and 96h).
Immunofluorescence
About 105 cells were seeded on a coverslip. The next day, cells were fixed in 4% paraformaldehyde for 10 min, permeabilized with 0.1% Triton X-100 for 10 min, followed by blocking with 1% BSA for 30 min at RT. Further, cells were incubated with primary antibody for 1 hour (h) at RT, washed thrice with PBS followed by 1 h incubation with Alexa Flour-conjugated secondary antibody. Cells were washed thrice with PBS, mounted (Prolong gold antifade, Invitrogen), air dried, and visualized using a confocal microscope.
Proximity ligation assay
Proximity ligation assay (PLA) was performed using a Duolink in-situ detection (Sigma #DUO92008) kit as per the manufacturer's protocol. The complete protocol is described in supplementary material and methods.
Synchronization of cells
For G1 cell cycle arrest, cells were subjected to serum starvation (DMEM without FBS) for 24 h. For G1/S arrest, MCF7 cells were subjected to double thymidine block (2mM thymidine for 16h followed by a release for 8h and treated again with 2 mM thymidine for 16 h). To arrest HEK293T cells in G1/S phase, cells were cultured in 2 mM thymidine for 16h. For G2/M arrest, cells were cultured in 2mM thymidine for 16h followed by release into medium containing nocodazole (100 ng/ml) for 16 h. The efficiency of synchronization was confirmed by propidium iodide-based cell cycle analysis using flow cytometry.
Cell cycle analysis
For performing cell cycle analysis, the cultured cells were harvested and washed with PBS. The cells were resuspended in PBS containing 2% FBS and fixed by adding 70% ice-cold ethanol dropwise and incubated overnight at 40C. Next day, the cells were washed with 2X PBS and permeabilized with 0.25% Triton x-100 for 15 min. Then, the cells were washed with 1X PBS and resuspended in PBS containing 100 µg RNase A and incubated at 370C for 15 min followed by propidium iodide (PI) staining (10 µg/ml) at RT for 1h. Flow cytometric analysis was performed using BD FACS Calibur. The data was analysed using Cell Quest Pro or FlowJo software (RRID:SCR_008520).
RNA isolation and quantitative real-time PCR
RNA isolation and qRT-PCR was performed as previously described (38). Briefly, total RNA was extracted using Trizol reagent according to the manufacturer's protocols (Invitrogen). 1 μg of RNA was used for reverse transcription using a high capacity DNA reverse transcription kit (Applied Biosystems Cat No: 4368813), and qRT-PCR was performed using Power SYBR green PCR master mix (Applied Biosystems #4367659) according to manufacturer's protocols. The fold-change in expression was calculated by the 2–ΔΔCt methods. mRNA expression profiles were normalized to levels of housekeeping gene Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in each sample The primers used in qRT-PCR are listed in supplementary material and methods.
Ni-NTA pull-down assay
HEK-293T cells were transfected with indicated plasmids. The cells were lysed in NP-40 lysis buffer containing 6M guanidine hydrochloride for 20 min at 4°C. The lysates were clarified at 12,000 g for 15 min. 50 µl of Ni-NTA beads was washed with NP-40 lysis buffer and incubated with cell lysates for 4h at 4°C. The beads were sedimented at 2,000 g, followed by washing with 1x lysis buffer and 4x PBS. The beads were boiled in 50 µl of 2x Lamelli sample buffer and subjected to western blot analysis with indicated antibodies.
Biotin RNA pull-down
Biotinylated RNA prepration: The template PCR fragments were generated using forward primer containing T7 RNA polymerase promoter (TAATACGACTCACTATAGGGAGA). PCR products containing the region of interest were purified for in-vitro transcription. Biotinylated RNA transcripts were generated in a 50 µl reaction containing T7 transcription buffer, 100 mM rNTPs, 40 units RNasin (Cat #N2111; Promega), 10 mM Biotin-14-CTP (cat #16519-016; Thermo Fisher Scientific), 100 ng T7 DNA template and 1 µl of T7 RNA polymerase (Cat #EP0113; 200 u/µl). The reaction mix was incubated at 37 0C for 2 h and RNA was purified using RNA easy spin columns (cat #74104; Qiagen). To remove template DNA on-column DNAse (cat #79254; Qiagen) treatment was performed.
HEK293T cells were transiently transfected with indicated plasmid DNA. Cells were lysed using Polysome Extraction Buffer (20 mM Tris-HCl pH 7.5, 100 mM KCl, 5 mM MgCl2, and 0.5% Nonidet P-40) containing Protease inhibitor cocktail, 1 mM PMSF and 200 u/ml RNase Inhibitor for 10 min at 40C and centrifuged.
500 µg of lysate was diluted in an equal volume of 2x TENT buffer (20 mM Tris-HCl pH 8.0, 2 mM EDTA pH 8.0, 500 mM NaCl, 1% (v/v) Triton X-100) and incubated with 1 µg of biotinylated RNA for 30 min at room temperature (RT). The streptavidin-coated dynabeads were washed thrice with TENT buffer and added to the protein-RNA mix and further incubated on rotospin at RT for 30 min. Finally, the beads were washed thrice with TENT buffer, and the bound proteins were eluted by boiling for 5 min in 2x SDS-sample loading dye and subjected to western blot analysis with indicated antibody.
Human breast cancer tissue microarray, immunohistochemistry, and scoring
Human breast cancer tissue array slides were purchased from US Biomax, Inc. (#BR1008a and #BRM961a). Details of BR1008a is as follows: human breast carcinoma, lymph node metastatic carcinoma, and adjacent normal tissue microarray containing 46 cases of invasive ductal carcinoma, neuroendocrine carcinoma, 3 medullary carcinomas, 40 metastatic carcinomas (4 metastatic carcinomas matched with breast carcinoma), 10 adjacent normal tissue, single core per block. Details of BRM961a are as follows: Human breast carcinoma with matched metastatic carcinoma or breast tissue microarray, containing 48 cases of breast carcinoma, 36 metastatic carcinomas (35 matched with breast carcinoma), 12 matched cancer adjacent or adjacent normal breast tissue, single core per block. DZIP3 immunohistochemical staining is described in supplementary methods and materials.
Mice xenograft and lung metastasis model
The animal work was performed in accordance with a protocol approved by Institutional Animal Care and Use Committee, ILS. 6 to 8 weeks old BALB/C-nude male mice weighing 16-20 g were maintained under specific pathogen-free conditions at the Institutional animal experimental facility. The tumor xenograft model was established by subcutaneous injection of DZIP3 shRNA knockdown and control shRNA UC3 cells suspension under sterile conditions, and the cell number was adjusted to 5×106 cells in PBS containing 50% (V/V) matrigel (BD corning) into right and left flanks respectively (n=7 mice). After eight days, tumor volume was measured every day until the volume reached 1000 mm3 in a group. The mice (n=7) were sacrificed, and tumors were collected for further analysis. For the tumor volume analysis, mean ± SEM tumor volume was plotted (using Excel) for each experimental group.
For lung metastasis study, 2×105 MDA-MB-231 control shRNA and DZIP3 shRNA cells were injected into the lateral tail vein of BALB/C-nude male mice (n=3). 8 weeks after injection, the mice were sacrificed, and the lungs were dissected out to count numbers of tumor nodules and other analysis.
Zebrafish xenograft
The animal work was performed in accordance with a protocol approved by Institutional Animal Care and Use Committee, ILS. The tumor was developed in Zebrafish (Danio rerio) [Tg(fli1:nEGFP)] using the protocol described previously (42). The complete protocol is described in supplementary material and methods.
Migration and Invasion assay
Trypsinized cells were washed with PBS and resuspended in serum-free medium. The cells were plated at a density of 104 per well in the inserts (Costar, 8.0-µm). For Invasion assay, the inserts were coated with 100 µL matrigel (BD Bioscience) before plating cells. The inserts were placed into 24 well plates containing 500 µL DMEM containing fetal bovine serum (chemoattractant). After 24 h of incubation, the cells inside the insert were completely removed by wiping with a cotton swab, and the meshes were fixed and stained with crystal violet. Migration/Invasion was quantitated by manual counting using microsope.
Results
DZIP3 is an important driver for growth, migration, and invasion of cancer cells
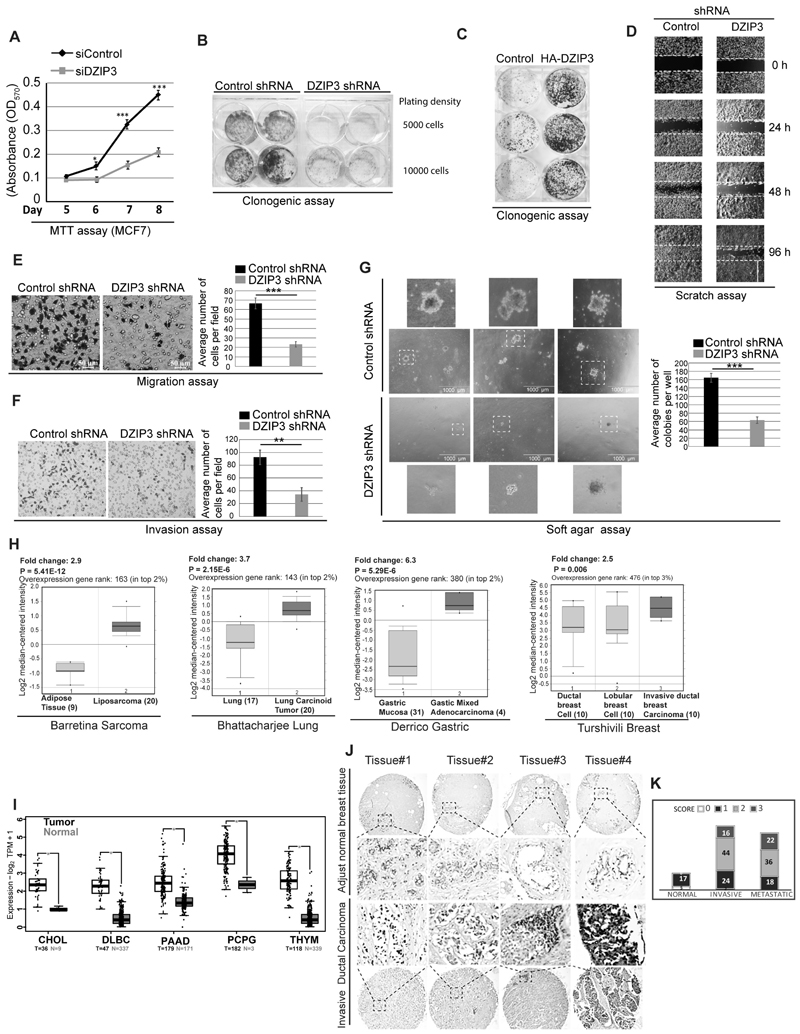
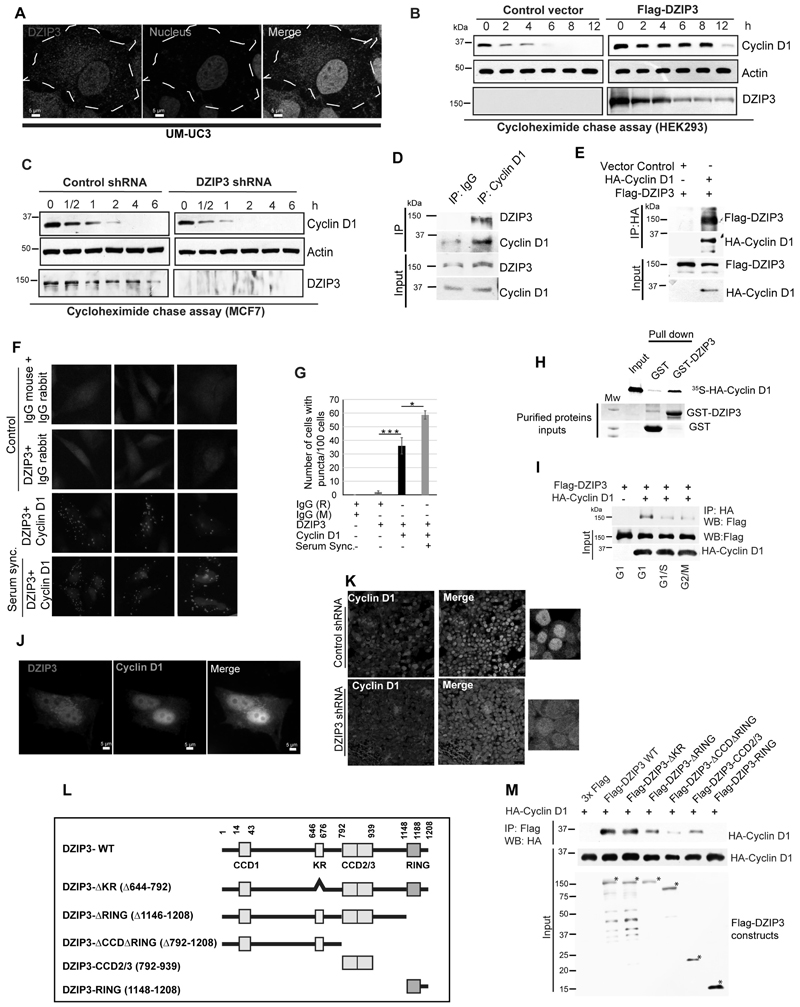
To understand the cellular functions of DZIP3, we generated stable DZIP3 shRNA knockdown cell lines of different origins (Supplementary Fig. S1A). We observed a significant growth defect in the knockdown cells (transient and stable) compared to the control cells in all the cell lines we checked (Fig. 1A; Supplementary Fig. S1B and C). Clonogenic assays are gold standards to assess the oncogenic potential of a single cell to grow into a colony under in vitro conditions. The DZIP3-depleted MCF7 cells were highly attenuated for colony formation as compared to the control cells (Fig. 1B). Next, we tested whether the overexpression of DZIP3 will result in increased cell growth. Indeed, transient overexpression of DZIP3 in HEK293 cells resulted in increased cell growth (Supplementary Fig. S1D) and considerably increased capability to form colonies in clonogenic assays (Fig. 1C). We performed wound healing assays (or scratch assays) to evaluate the migration capacity of DZIP3 knockdown cells. The MCF7 cells depleted of DZIP3 showed a reduction in migration capability and wound closure as compared to control cells (Fig. 1D). In transwell migration assays and matrigel invasion assays, we found significantly reduced migration and invasion of DZIP3 knockdown cells (Fig. 1 E and F). Also, in soft-agar assays a significant decrease in anchorage-independent growth was observed in DZIP3 knockdown cells as compared with the control cells (Fig. 1G). Taken together, the data show that DZIP3 regulates cancer cell growth, migration, and invasion.
Figure 1. DZIP3 drives growth, migration, and invasion of cancer cells.
(A) MTT assays were performed in MCF7 breast cancer cells transfected with control and DZIP3 siRNA. Data is presented as mean ± SD, n=3, *p<0.05, ***p<0.0005.
(B) Clonogenic assay was performed with MCF7 control and DZIP3 stable knockdown cells with an initial plating density of 5000 and 10000 cells.
(C) Clonogenic assay was performed in HEK293 cells transiently transfected with control and HA-DZIP3.
(D) Wound healing or scratch assay was performed to examine migration capacity in control and DZIP3 stable knockdown MCF7 cells.
(E and F) Transwell migration and matrigel invasion assay were performed in UM-UC3 control and DZIP3 stable knockdown cells. Data in bar graph is presented as mean ± SD, n=3, **p<0.005, ***p<0.0005.
(G) Soft agar colony formation assay was performed to check tumorigenicity of UM-UC3 control and DZIP3 stable knockdown cells. Data in bar graph presented as mean ± SD, n=3, ***p<0.0005.
(H) Oncomine analysis of DZIP3 mRNA expression levels in different cancer types (liposarcoma, lung carcinoma, gastric adenocarcinoma, and breast carcinoma) as compared with normal tissues. Fold change and p-values are indicated in the graph.
(I) DZIP3 mRNA expression in tumors and normal control patient samples were evaluated in the TCGA dataset using the GEPIA2 platform. TPM (Transcripts Per Million), Log2FC| Cutoff= 1; p-value Cutoff=0.01
(J and K) (J) Representative images of tissue microarray analysis of DZIP3 protein expression in normal breast tissue (n = 22) and invasive human breast cancer tissue (n = 164) samples. (K) The intensity of staining was scored using a four-tier scale and defined as follows: 0, 0%–20%; 1, weak staining (20%–50%); 2, moderate staining (50%–75%); and 3, strong staining (75%–100%). Based on these scores, the number of tissue samples in each group of tissue array (Normal, Invasive, and Metastatic) having these scores were plotted.
Expression of DZIP3 is increased in several cancer types
Oncomine database (a cancer microarray database, www.oncomine.org) analysis suggests that DZIP3 mRNA levels were significantly increased in several of the cancer types, including sarcoma, lung, gastric, breast, colon, liver, and pancreatic cancers (Fig. 1H; Supplementary Fig. S1E). Next, we analyzed DZIP3 mRNA expression in tumors in the TCGA (The Cancer Genome Atlas) dataset using the GEPIA2 platform (43). DZIP3 levels were found to be significantly higher in tumors as compared to normal control tissues in several cancer types (Fig. 1I).
The levels of DZIP3 expression in human breast carcinomas were investigated using two human tissue microarrays (168 patients cases, 22 adjacent normal controls). The DZIP3 was found to be expressed in low levels (Score-0 or Score-1) in adjacent normal tissues (Fig. 1J and K), and most of the ducts and lobules were devoid of staining in all the cases (Fig. 1J). In breast invasive ductal carcinoma tissues, an intense (Score-2, 88 cases) to very intense staining (score-3, 38 cases) of DZIP3 was observed (Fig. 1J). In some tissues, DZIP3 was exclusively present inside the nucleus, whereas, in most of the tissues, both cytoplasmic and nuclear staining was observed (Fig. 1J). About 126 breast cancer patients cases (metastatic and non-metastatic) out of a total of 168 cases showed increased expression of DZIP3, much above adjacent control tissues (Fig. 1K). The ducts and lobules in these samples were strongly immunoreactive for DZIP3 (Fig. 1K). Altogether, the data suggest that the protein expression of DZIP3 is increased during tumorigenesis.
DZIP3 promotes tumor growth and metastasis in mice and zebrafish
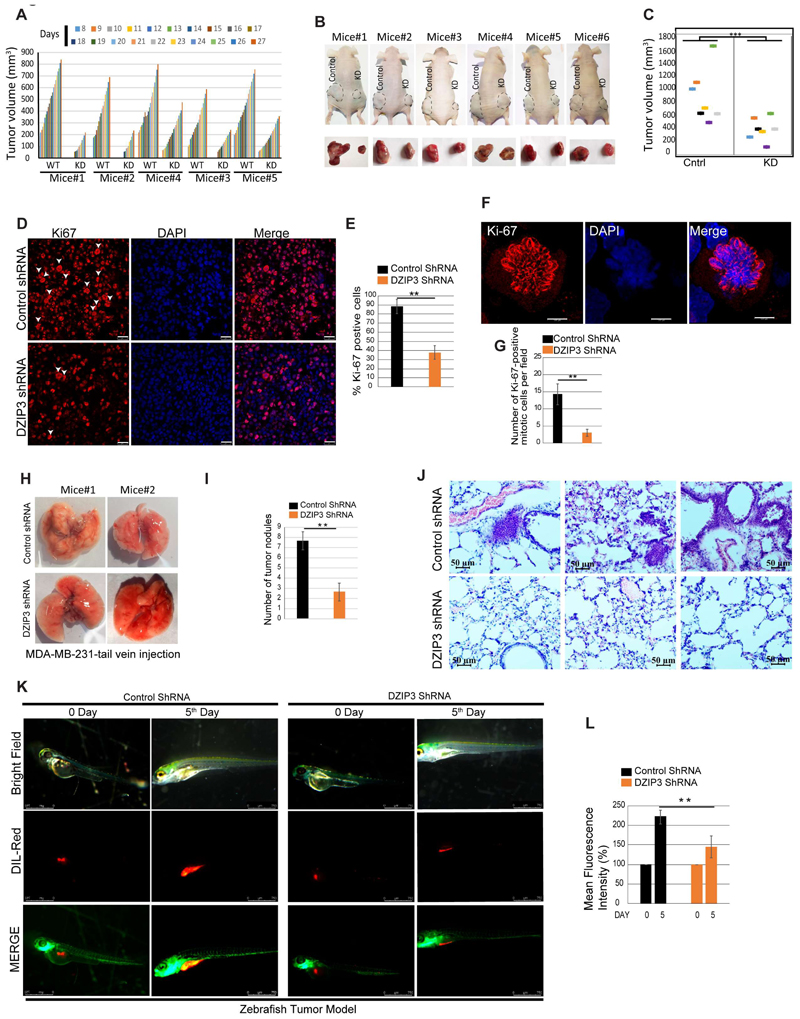
To investigate whether DZIP3 can promote cancer cell growth in vivo, we subcutaneously injected an equal number of control and DZIP3 knockdown UM-UC3 bladder cancer cells in BALB/C-nude mice. To nullify the variations due to innate immunity, we injected both control and knockdown cells in either flank of the same mice. The tumor growth was monitored daily for a period of ~3 weeks and at the termination of the experiment. The growth rate of DZIP3 knockdown tumors was found to be considerably lower than the control tumors (Fig. 2A). In agreement, upon dissection, the volumes of DZIP3 depleted tumors were found to be significantly less than control tumors for all the mice (Fig. 2B and C).
Figure 2. DZIP3 promotes tumor growth and metastasis in animal models.
(A) The graph depicts tumor volumes (mm3) of UM-UC3 control and DZIP3 stable knockdown cells.
(B) Images of tumors formed in the nude mice and pictures of the dissected tumors.
(C) The graph represents the final tumor volume. n=7, mean±SE, ***p<0.0005.
(D and E) (D) Immunohistochemical analysis was performed with the Ki-67 cell proliferation marker in control and DZIP3 stable knockdown tumor sections. (E) The graph represents % of cells with Ki-67 positive staining. >400 cells were analyzed from different sections from different animals, mean ±SD, **p < 0.005. The white arrowhead indicates the Ki-67 positive mitotic cells.
(F and G) (F) Representative images of Ki-67 positive mitotic cells. (G) The graph represents % of Ki-67 positive mitotic cells in control and DZIP3 stable knockdown tumors in IHC's microscopic fields. >10 microscopic fields were analyzed from different sections from 3 different animals, mean ±SD, **p < 0.005
(H-J) (H) Representative images of lungs of mice injected (tail-vein) with MDA-MB-231 control and DZIP3 stable knockdown cell line. (I) The average number of nodules were counted manually. n=3 mean ±SD, **p < 0.005. (J) Representative lung sections showing metastatic foci in control groups.
(K and L) (K) Lateral view of fluorescent transgenic [Tg(fli1:nEGFP)] zebrafish embryos at Day 0 and Day 5 injected with Dil-Red stained MDA-MB-231 control and DZIP3 stable knockdown cells. (L) The tumor growth was assessed by an increase in fluorescence intensity on the 5th day compared to the day of injection. n=10, mean ±SD, **p < 0.005 The quantitation of fluorescence intensity was performed using ImageJ software and represented as % mean fluorescence intensity where day 0 reading was taken as baseline.
The expression of nuclear Ki-67 is strongly associated with tumor cell proliferation, aggressiveness, and growth. Immunohistochemical analysis revealed a significant decrease in nuclear Ki-67 positive cells in DZIP3 knockdown tumors as compared to the controls (Fig. 2D and E). During mitosis, Ki-67 forms peri-chromosomal layer and prevents chromosomes from collapsing into a single chromatin mass (44). Thus, the cells undergoing mitosis can be easily recognized using Ki-67 immunostaining as it gives a floral appearance (Fig. 2F; white arrows, Fig. 2D). We observed a significantly higher number of mitotic cells in control tumors as compared to the DZIP3 knockdown tumors (Fig. 2D-G).
We employed a tail-vein-injection metastasis model to validate the reduced migration and invasion phenotypes of DZIP3 depleted cells. MDA-MB-231 breast cancer cells (control and DZIP3 knockdown) were injected intravenously into the lateral tail vein of nude mice, and lung colonization was evaluated. Although the weight of mice in two groups didn't change significantly, a considerably fewer number of tumor nodules were observed in the lungs of mice that were injected with DZIP3 knockdown cells (Fig. 2H and I). An increased number of metastatic foci were observed in the control group lung sections as compared to the DZIP3 knockdown group (Fig. 2J). The western blot analysis from mouse lung lysates using human-specific antibody confirmed the reduced levels of DZIP3 (and Cyclin D1) in DZIP3 knockdown samples (Supplementary Fig. S2A). These observations suggest that the depletion of DZIP3 reduces the metastatic capacity of the cancer cells.
Next, we used Zebrafish (Danio rerio) [Tg(fli1:nEGFP)] tumor xenograft model (45) to further validate results. Equal number of the control and DZIP3 depleted MDA-MB-231 cells were stained with Dil (1,1'-Dioctadecyl-3,3,3',3'-Tetramethylindocarbocyanine Perchlorate) and injected into perivitelline space of 48-hour post fertilized zebrafish embryos. After 5 days of injection, the primary tumors and metastatic dissemination of cancer cells were documented using a fluorescence microscope. The tumor growth, as measured by fluorescence intensity of primary tumors, was found to be significantly higher in the case of control cells compared to the DZIP3 depleted cells (Fig. 2K and L). The images were captured at the same microscopic settings (intensity, exposure, thresholds) throughout the experiments. At 5 days post-injection, cancer cells showed distal migration from the primary site in the case of control cells but not in the case of DZIP3 knockdown tumors (Supplementary Fig. S2B).
Taken together, the data from different in vivo models suggest that DZIP3 is an important driver of tumor growth and metastasis.
Depletion of DZIP3 results in cell cycle arrest in the G1/S phase
Next, we asked whether the reduced cell growth in vitro and in vivo in DZIP3-depleted cells is due to increased cell death. The Annexin-V/Propidium Iodide double staining of control and DZIP3 knockdown MCF7 cells showed no differences in the apoptotic population (Supplementary Fig. S3A). In an agreement, no difference in Caspase-3 cleavage was observed in cultured cells and xenograft tumors (Supplementary Fig. S3B and C), suggesting that increased apoptotic cell death is not the reason for the reduced proliferation of DZIP3-depleted cells.
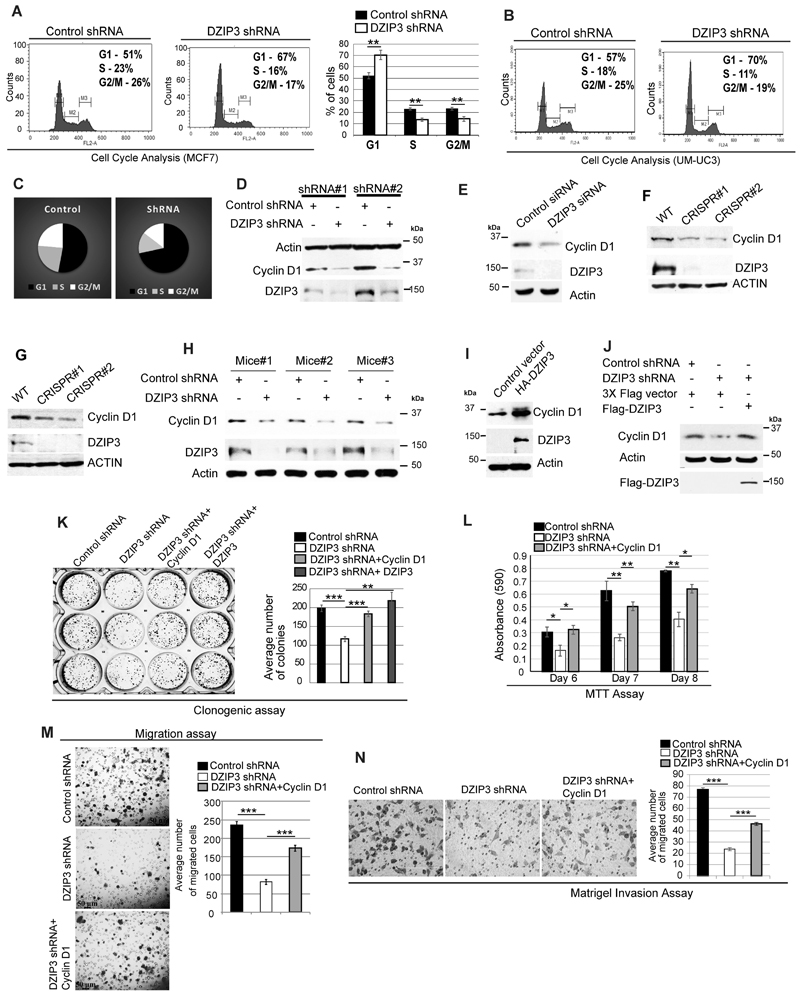
The cell cycle analysis was performed to test whether the defect in growth is due to reduced cell division. The DZIP3 knockdown in MCF7, UM-UC3, and HeLa cells resulted in the disconcerted cell cycle where a significant increase in the percentage of cells in G1 phase and a significant decrease in the percentage of cells in S and G2/M phase was observed in all the cell types tested (Fig. 3A-C; Supplementary Fig. S3D and E). Next, we synchronized the MCF7 cells using the serum starvation method (48 h) and then released the cells in full media to track the progression of the cell cycle (Supplementary Fig. S3F). The control cells progressed to different cell cycle phases normally. However, the knockdown cells were arrested in the G1 phase even after 12 hours of release. Taken together, the data suggest that DZIP3 is required for cell cycle progression, and in its absence, the cells are arrested in the G1 phase of the cell cycle.
Figure 3. DZIP3 controls cell cycle progression and cell growth by regulating the expression of Cyclin D1.
(A) Cell cycle analysis of MCF7 control and DZIP3 stable knockdown cell line. The graph represents the percentage of cells in different cell cycle phases. mean ± SD, n=3, **p<0.005.
(B-C) (B) Cell cycle analysis of UM-UC3 control and DZIP3 stable knockdown cells. (C) Pie charts represent the percentage of cells in different cell cycle phases.
(D-E) Western blot analysis of (D) MCF7 control and DZIP3 shRNA cell lysates probed with indicated antibodies. (E) Control and DZIP3 siRNA transfected MCF7 cell lysates probed with indicated antibodies.
(F-G) Western blot analysis of (F) HEK293T (G) MCF7, control, and DZIP3 CRISPR knockout clones cell lysates probed with indicated antibodies.
(H) Western blot analysis of UM-UC3 control and DZIP3 shRNA tumor lysates probed with indicated antibodies.
(I) Western blot analysis of control and HA-DZIP3 plasmids transfected HEK293 cell lysates probed with indicated antibodies.
(J) Western blot analysis of cell lysates of indicated plasmids transfected MCF7 control and DZIP3 shRNA cells probed with indicated antibodies.
(K) Clonogenic assay was performed with control, DZIP3 stable knockdown cells, and knockdown cells complemented with HA-Cyclin D1 or Flag-DZIP3. The graph depicts the average number of colonies in these conditions. mean ± SE, n=3, **p<0.005, ***p<0.0005.
(L) Cell proliferation was evaluated by performing MTT assay in MCF7 control, DZIP3 stable knockdown, and Cyclin D1 complemented DZIP3 knockdown cells. Data is presented as mean ± SD, n=2, *p<0.05, **p<0.005.
(M) Migration assay was performed with MCF7 control, DZIP3 stable knockdown, and Cyclin D1 complemented DZIP3 knockdown cells. Data are presented as mean ± SE, n=3, ***p<0.0005.
(N) Matrigel invasion assay was performed with UM-UC3 control, DZIP3 stable knockdown, and Cyclin D1 complemented DZIP3 knockdown cells. Data are presented as mean ± SE, n=3, ***p<0.0005.
DZIP3 controls cell growth by regulating the expression of Cyclin D1
The Cyclin D1 drives the G1 to S phase transition (46). Since we observed that DZIP3 depleted cells were arrested in the G1 phase, we examined the levels of Cyclin D1 in different cell lines and xenograft tumors. In all of the cell lines tested (MCF7, UM-UC3, and MDA-MD-231), the expression of Cyclin D1 protein was considerably lower in DZIP3 depleted cells (Fig. 3D; Supplementary Fig. S3G-I). This data was validated using DZIP3 shRNA stable knockdown cells and transient siRNA knockdown cells using two different sets of siRNA and shRNA in 3 different cell lines (Figure 3D and E; Supplementary Fig. S3G-I). Further, DZIP3 was knocked out using the CRISPR-CAS9 technique in two different cell lines. There was a considerable reduction in levels of Cyclin D1 in the DZIP3 knockout clones (Fig. 3F and G).
The levels of Cyclin D1 in DZIP3 knockdown tumors were lower than control tumors (Fig. 3H). Next, to understand whether Cyclin D1 regulation by DZIP3 is direct or is a consequence of growth defect (cause or consequence), we transiently overexpressed DZIP3 in HEK293 cells for a very short duration (6 hours). No growth difference and cell cycle defect were observed in these cells, but the overexpression of DZIP3 was able to strongly stabilize the endogenous Cyclin D1 (Fig. 3I). Taken together, the results suggest that DZIP3 positively regulates the expression of Cyclin D1. To further strengthen this conclusion, we complemented DZIP3 knockdown cells with exogenous DZIP3 (transient expression) and assessed its ability to restore Cyclin D1 protein levels (Fig. 3J). The Cyclin D1 protein levels were restored completely in DZIP3 complemented cells.
An extensive literature suggests that depletion of Cyclin D1 results in reduced growth, migration, and invasion in several cancer types. We also validated these results using scratch assays, migration assays, and invasion assays (Supplementary Fig. S3J-L). To understand whether the growth, migration, and invasion defect observed in DZIP3 depleted cells is due to lower Cyclin D1 expression in these cells, we complemented DZIP3 knockdown cells with Cyclin D1 and performed the assays. The Cyclin D1 complementation significantly rescued the growth defect of DZIP3 knockdown cells in clonogenic assays and MTT assays (Fig. 3K and L). Also, the Cyclin D1 complementation rescued the defective migration and invasion phenotype of DZIP3 knockdown cells (Fig. 3M and N). Taken together, the data suggest that DZIP3 controls the growth and invasion properties of cancer cells by regulating the expression of Cyclin D1.
DZIP3 interacts at 3'UTR of Cyclin D1 mRNA and stabilize it
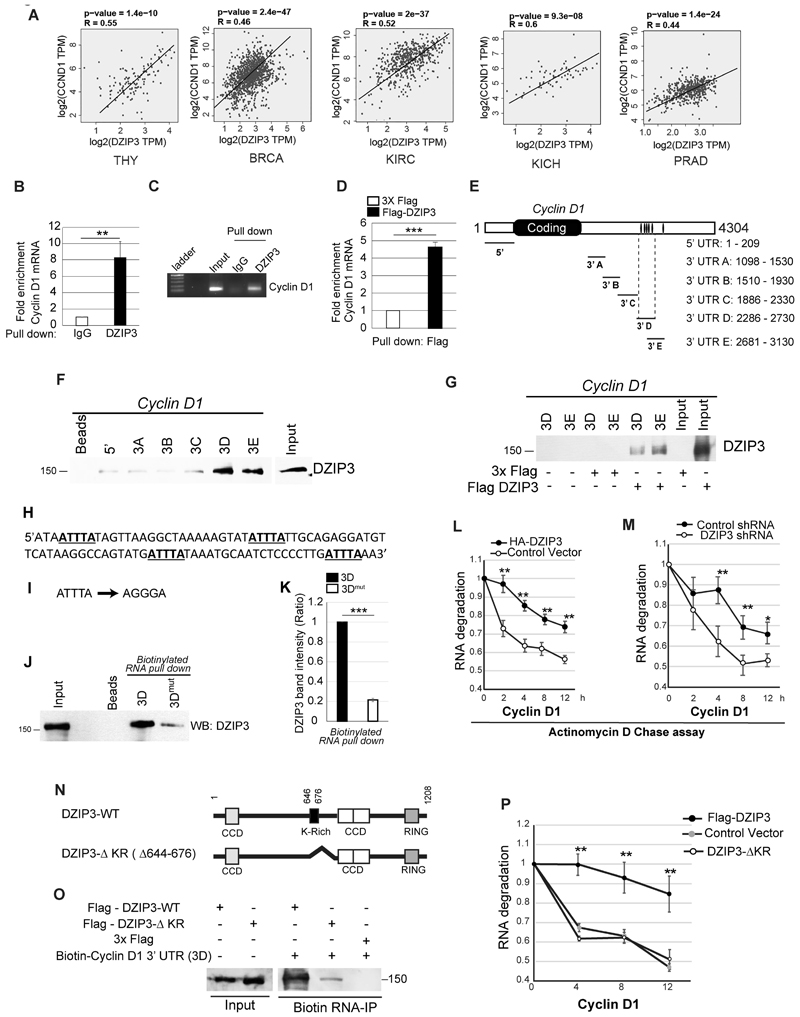
The aberrant overexpression of Cyclin D1 is strongly associated with the pathogenesis and progression of several cancer types (47). So, we analyzed whether the mRNA expression of DZIP3 and Cyclin D1 are correlated in human cancer patients database using GEPIA platform (Gene Expression profiling Interactive analysis, www.gepia.cancer-pku.cn/) (48). We found a strong positive correlation of mRNA expression between DZIP3 and Cyclin D1 in different cancer types, including thymus, breast, kidney, rectum, and prostate (Fig. 4A, Supplementary Fig. S4A), indicating a possible regulation of Cyclin D1 by DZIP3 in cancer patients. However, no significant correlation was found between DZIP3 and Cyclin B1 mRNA expression (Supplementary Fig. S4B).
Figure 4. DZIP3 interacts with Cyclin D1 mRNA and stabilizes it.
(A) The graphs depict the correlation of mRNA expression of DZIP3 and Cyclin D1 in different cancer types. r, Spearman's rank correlation coefficient. TPM (Transcripts Per Million). THY, Thymoma; BRCA, Breast invasive carcinoma; KIRC, Kidney renal clear cell carcinoma; KICH, Kidney Chromophobe; PRAD, Prostate adenocarcinoma
(B and C) (B) qRT-PCR analysis of enrichment of Cyclin D1 mRNA in RNA Immunoprecipitation assay using IgG and DZIP3 antibody. The fold enrichment was calculated by taking IgG values as a baseline. Data is presented as mean ± SD, n=3, **p<0.005. (C) Agarose gel analysis of Cyclin D1 RT-PCR product from the same experiment.
(D) qRT-PCR analysis of enrichment of Cyclin D1 mRNA in RNA Immunoprecipitation assay using Flag antibody. The fold enrichment was calculated by taking control values as a baseline. Data is presented as mean ± SD, n=3, ***p<0.0005
(E) Schematic representation of different RNA fragments of Cyclin D1 mRNA 3'-UTRs and 5'-UTR used in RNA pull-down assays. The black bars in 3'UTR represent the AU-rich elements.
(F) The biotinylated 5'UTR region and 3'UTR's of Cyclin D1 transcripts were incubated with lysates of HEK293T cells followed by pull-down with streptavidin-tagged beads. The pulldown complexes were subjected to western blotting with Flag antibody. Bead control does not contain RNA.
(G) The biotinylated 3'UTR's of Cyclin D1 transcripts were incubated with lysates of 3X Flag vector or Flag-DZIP3 expressing HEK293T cells followed by pull-down with streptavidin-tagged beads. The pull-down complexes were subjected to western blotting with Flag antibody.
(H-K) (H) The nucleotide sequence of the 3D region of 3'UTR of Cyclin D1 mRNA containing four ATTTA sites. (I) The mutation of ATTTA to AGGGA were introduced in all the four sites in indicated 3D region. (J) The biotinylated 3'-UTR of Cyclin D1 -D fragment (3D) wild type and its mutant were incubated with Flag-DZIP3 expressing HEK293T cell lysates followed by pull-down with streptavidin-tagged beads. The pull-down complexes were subjected to western blotting with Flag antibody. (K) Graph depicts the ratio of binding affinity of 3D and 3D-mut regions for Flag-DZIP3 as measured by image J analysis of western blots of biotin-pull down assays (panel J). Data is presented as mean ± SD, n=3, ***p<0.0005
(L and M) (L) Control HA-vector and HA-DZIP3 transiently transfected HEK293T cells and (M) control and DZIP3 shRNA stable knockdown MCF7 cells were serum-starved for 16 h and released into a full medium for 3 h and treated with actinomycin D (5 µg/ml) for indicated time points. The total RNA was isolated and subjected to qRT-PCR with Cyclin D1 specific primers. Data are normalized with GAPDH (transcript levels of GAPDH was not affected by actinomycin D). Data are presented as mean ± SD, n=3, *p<0.05, **p<0.005.
(N) Schematic representation of wild type (WT) and lysine-rich (KR) deletion mutant of DZIP3.
(O) Biotinylated 5' and 3'D UTR's of Cyclin D1 were used for RNA Pull-down experiments with lysates of HEK293T cells transiently transfected with vector control, wild type Flag-DZIP3 and Flag- DZIP3-ΔKR and subjected to western blot analysis with Flag antibody.
(P) Actinomycin D chase assay and qRT PCR assays were performed with HEK293T cells transiently transfected with control, Flag-DZIP3, and Flag DZIP3-ΔKR. Data are presented as mean ± SD, n=3, **p<0.005.
We then investigated the mechanism by which DZIP3 positively controls the expression of Cyclin D1. DZIP3 contains an RNA binding motif called lysine (K)-rich region (or KR motif), a RING domain, and several coiled-coiled domains (CCD) (13) (Supplementary Fig. S4C and D). Previous studies showed that DZIP3 could interact with RNA utilizing the KR motif (13,14). The RNA binding proteins such as HuR (Human antigen R) and AUF1 (AU-rich element RNA-binding protein 1) were shown to regulate the stability of the cyclins mRNA by interacting with AU-rich element (ARE sites) in 3' untranslated regions (3' UTR) (25,26,49). We hypothesized that DZIP3 being RNA binding protein, regulates expression of Cyclin D1 by interacting with its mRNA at 3' UTR and subsequently stabilizing the Cyclin D1 transcripts. To test this hypothesis, first, we performed RNA pull-down assays with either the DZIP3 antibody or control IgG antibody, followed by a real-time qPCR (Fig. 4B) or reverse transcriptase-PCR (Fig. 4C) with Cyclin D1 and GAPDH primers (Supplementary Fig. S4E). Significant enrichment of Cyclin D1 mRNA was observed in RNA pull-down by the DZIP3 antibody compared to IgG control (Fig. 4B and C). No enrichment was observed with GAPDH mRNA (Supplementary Fig. S4E). Similar results were obtained with Flag-DZIP3 overexpressing cells (Fig. 4D; Supplementary Fig. S4F). The data suggest that DZIP3 specifically interacts with Cyclin D1 mRNA.
Next, we performed in vitro biotin RNA pull-down assays with 5' and 3' UTR's RNA fragments of cyclin d1 mRNA (Fig. 4E). The in vitro transcribed RNA fragments were biotinylated and incubated with lysates expressing Flag-DZIP3 or just Flag epitope subjected to pull-down by streptavidin beads. The western blot with DZIP3 showed that DZIP3 interacted specifically and strongly with cyclin D1 3' UTR but very faintly with 5' UTR, and no interaction was observed in bead controls (Fig. 4F). Within 3' UTR of cyclin D1, DZIP3 was bound with 3D (+2286 bp to +2730 bp) and 3E regions (+2681 bp to +3130 bp) (Fig. 4E-G). Thus, DZIP3 specifically interacts with 3' UTR of cyclin D1 mRNA.
Next, we attempted to identify the binding sites of DZIP3 in Cyclin D1 3'UTR (Fig. 4E). Most of the RNA binding proteins control the stability of mRNA via interaction with the AU-rich element (ARE) with the core-binding element "AUUUA" (ATTTA in DNA) (50). Using ARE prediction software (AREsit2; http://rna.tbi.univie.ac.at/AREsite2/welcome), we found four ATTTA elements in 3D region and one such site in 3E region of cyclin D1 3'UTR (Fig. 4E). We mutated these sites in 3D region (ATTTA-> AGGGA) (Fig. 4H and I) and compared them with wild type mRNA for the capability to interact with DZIP3. In biotinylated RNA pull-down assays, the ARE sites mutations dramatically reduced the affinity of 3D region for DZIP3 (Fig. 4J and K), indicating that AUUUA is the binding sites of DZIP3 in 3' UTR of cyclin D1.
In unsynchronized cells and in steady-state conditions, the effect of DZIP3 on total Cyclin D1 mRNA levels was marginal. A slight induction of Cyclin D1 mRNA levels was observed in DZIP3 overexpressing cells (Supplementary Fig. S4G). Similarly, DZIP3 knockdown cells or tumors showed a minor reduction in the Cyclin D1 mRNA amount compared to the control cells (Supplementary Fig. S4H and I). However, unsynchronized cells are in different phases of the cell cycle, which is not an ideal condition to determine the effect of RNA-binding proteins on the stability of cyclins mRNA's (26). To delineate the role of DZIP3 in mRNA stability, we synchronized the cells into the G1 phase by serum starvation and performed the actinomycin D pulse-chase assays to examine the rate of degradation of Cyclin D1 mRNA in the absence and presence of DZIP3. The data showed that the rate of degradation of Cyclin D1 mRNA was significantly less in DZIP3 overexpressing cells compared to the control cells (Fig. 4L). On the other hand, the rate of degradation of cyclin D1 mRNAs was higher in DZIP3 stable knockdown cells compared to the control cells (Fig. 4M). Altogether, these results suggest that DZIP3 interacts and stabilizes cyclin D1 mRNA.
The RNA binding KR motif of DZIP3 is important for its interaction with RNA (13). To validate the specificity of DZIP3-cyclin D1 mRNA interaction and to confirm that the KR motif is important for this interaction, we deleted the KR motif in DZIP3 (Fig. 4N) and compared this protein with the wild-type DZIP3 for its ability to bind and stabilize the cyclin D1 mRNA. In biotin RNA-pulldown assay, the KR motif deleted DZIP3 (DZIP3-ΔKR) was defective in its ability to interact with 3'UTR of cyclin D1 mRNA (Fig. 4O) and also failed to stabilize the cyclin D1 transcripts in actinomycin D chase assays (Fig. 4P). These results are significant as the deletion of a small region (~90 bp) containing the KR motif in the DZIP3 protein (3624 bp gene) crippled its RNA binding and RNA stabilizing potential. The data suggest that the KR motif of DZIP3 plays an important role in DZIP3-cyclin D1 mRNA interaction and stability.
DZIP3 interacts, co-localizes, and stabilizes Cyclin D1 in G1 phase
DZIP3 localizes in both the nucleus and cytoplasm in four different cell lines we tested (Fig. 5A, Supplementary Fig. S5A-C). In the cytoplasm, DZIP3 was found in perinuclear regions as well as in punctuated structures (Fig. 5A, Supplementary Fig. S5A-C). DZIP3 is a bonafide E3-ubiquitin ligase, and several of the E3-ligases are known to modulate the stability of cell cycle proteins (36,51-53). Next, we tested whether DZIP3, in addition to controlling the stability of cyclin D1 mRNA, can modulate the Cyclin D1 protein levels. We synchronized the cells by serum starvation and then performed cycloheximide chase experiments both in DZIP3 overexpressing HEK293 cells (Fig. 5B) and DZIP3-depleted MCF7 cells (Fig. 5C), and monitored the stability of the endogenous Cyclin D1 proteins over a period of time. In the DZIP3 overexpressing cells, Cyclin D1 was well stabilized till 8 h. On the other hand, in control vector-transfected cells, Cyclin D1 was degraded rapidly within 2 h and almost entirely degraded by 6 h (Fig. 5B). In DZIP3 stable knockdown cells, the degradation of the Cyclin D1 proteins was faster as compared to the control cells (Fig. 5C). The data suggest that DZIP3 provides stability to the Cyclin D1 at the protein levels also.
Figure 5. DZIP3 interacts, co-localizes, and stabilizes Cyclin D1.
(A) Representative confocal images of DZIP3 localization in UM-UC3.
(B and C) Western blot analysis of cycloheximide chase experiment performed with (B) HEK293T transiently transfected with indicated plasmids (C) shControl and shDZIP3 MCF7 cells.
(D and E) Immunoprecipitation analysis of the interaction between (D) endogenous DZIP3 and endogenous Cyclin D1 (E) Flag-DZIP3 and HA-Cyclin D1.
(F-G) (F) Representative images of proximity ligation assay performed with unsynchronized and serum synchronized HeLa cells that were transfected with indicated plasmids. (G) The graph represents the number of puncta's present per 100 cells. Data, mean ± SD, n=3, *p<0.05 ***p<0.0005.
(H) GST pull-down assay using purified GST or GST-tagged DZIP3 (CCD-RING domain) and in-vitro-translated and S35- labelled HA-Cyclin D1.
(I) The western blot analysis of co-IP assays performed with HEK293 cells transiently co-transfected with Flag-DZIP3, HA-Cyclin D1, and synchronized in G1 (serum starvation), G1/S (single thymidine block) or G2/M (nocodazole block) phases.
(J) Representative confocal images of DZIP3 and Cyclin D1 protein localization in serum synchronized MCF7 cells.
(K) Representative confocal images of immunofluorescence analysis of Cyclin D1 expression in tumor xenograft sections.
(L) The domain organization map of DZIP3 and its truncated mutants cloned as Flag-tagged proteins.
(M) Co-immunoprecipitation analysis to map the domain interaction between Flag-DZIP3 constructs and HA-Cyclin D1.
To understand how DZIP3 impart stability to Cyclin D1 at protein levels, we first tested whether DZIP3 interacts with Cyclin D1. We performed immunoprecipitation (IP) assays with endogenous as well as overexpressed proteins. In endogenous IP assays, we used IgG and Cyclin D1 antibodies for immunoprecipitation, and the immunoblotting was performed with the DZIP3 antibody (Fig. 5D).
The Cyclin D1 but not IgG antibody was able to pull-down DZIP3, suggesting a specific interaction between DZIP3 and Cyclin D1 (Fig. 5D). Next, we overexpressed HA-Cyclin D1 and Flag-DZIP3 and performed co-IP assay with HA-antibody. A strong interaction between DZIP3 and Cyclin D1 was observed (Fig. 5E). Next, using Flag antibody-tagged beads, we performed pull-down assays with cells overexpressing Flag-DZIP3. We used stringent conditions (6 times NP-40 buffer) for washing the beads to remove weak interactions with DZIP3. The Flag-DZIP3 was eluted from beads using Flag-peptide, and western blotting was performed with the Cyclin D1 antibody. We observed a single band of endogenous Cyclin D1 suggestive of a strong interaction between DZIP3 and endogenous Cyclin D1 (Supplementary Fig. S5D).
Proximity Ligation Assay (PLA) is a powerful quantitative technique that allows in-situ detection of protein-protein interactions with high specificity and sensitivity (54). Since the PLA positive signals (puncta's) are detected only when the two proteins are at close proximities of < 40 nm, such protein interactions are most likely to be the direct interactions (54). In PLA assays, we found a robust increase in the number of PLA positive puncta's in the cells expressing DZIP3 and Cyclin D1, suggestive of direct interactions between DZIP3 and Cyclin D1 (Fig. 5F and G). To further strengthen the direct interaction data, we tested whether purified GST-DZIP3 (CCD-RING domain) directly interact with Cyclin D1 in vitro. For this, we performed GST-pull down assays with purified GST-DZIP3 (CCD-RING domain) and in vitro translated Cyclin D1 in the cell-free milieu. A direct and specific interaction was observed between DZIP3 and Cyclin D1 (Fig. 5H). Taken together, several lines of evidence suggest that DZIP3 directly interacts with Cyclin D1.
Next, we asked whether DZIP3 interacts with Cyclin D1 constitutively or in a cell cycle phase-dependent manner. For this, we overexpressed DZIP3 and Cyclin D1 proteins and then synchronized the cells in the G1 phase, S phase, and G2 phase by using serum starvation, single thymidine block, and nocodazole block, respectively. Serum starvation blocks the cell cycle at the G1 phase, whereas thymidine in the early S phase and nocodazole at the border of the G2 and M phase. The synchronized cells were used in co-IP assays. DZIP3 interacts with Cyclin D1 preferentially during the G1 phase of the cell cycle, suggesting that DZIP3 interacts and stabilizes the Cyclin D1 typically during the G1 phase (Fig. 5I). We also performed the PLA assay in serum synchronized G1 arrested cells. We observed a significant increase in the number of puncta's in synchronized cells than in unsynchronized cells, strengthening our conclusions that DZIP3 interacts with and regulates Cyclin D1 predominantly in the G1 phase of the cell cycle (Fig. 5F and G). This is an interesting finding that DZIP3 interacts with Cyclin D1 only when the latter is required to be stabilized for the cell cycle progression.
Since Cyclin D1 is localized in the nucleus during the G1 phase (55), next, we performed immunofluorescence assays with the G1 phase (serum starvation) synchronized cells to monitor the localization of DZIP3 and Cyclin D1. DZIP3 was found to be completely co-localized with nuclear Cyclin D1 in G1 phase synchronized cells (Fig. 5J). Interestingly, in immunohistochemistry of xenograft tumor tissues, Cyclin D1 showed increased immunoreactivity in the nucleus of the control tumor cells. Whereas considerably less Cyclin D1 nuclear immunostaining was observed in DZIP3 depleted tumors cells (Fig. 5K). The data indicate that DZIP3 is not only required for the stability of Cyclin D1 but might also for its nuclear localization.
CCD and RING domains of DZIP3 are important for its interaction with Cyclin D1
DZIP3 contains three coiled-coiled domain (CCD), a RING domain, and an RNA-binding KR motif (Fig. 5L). To identify the domain/s of DZIP3 required for interaction with Cyclin D1, we generated several DZIP3 domain deletion variants (Fig. 5L). The deletion constructs were co-expressed with HA-Cyclin D1, and co-IP assays were performed. To reduce the effect of DZIP3-mediated stability on Cyclin D1-DZIP3 interaction, in this experiment, we have adjusted the ratio of Cyclin D1 to equalize it in the input, and then the IP was run in the same ratio. There was no apparent change in binding affinity of DZIP3-Cyclin D1 upon KR domain deletion (Fig. 5M). RING domain deletion reduced the interaction, and it was further reduced on the deletion of the CCD2/3 domain (Fig. 5M), suggesting that both domains are important of DZIP3-Cyclin D1 interactions. However, where the RING domain itself cannot interact with Cyclin D1, an interaction was observed between the CCD2/3 domain and Cyclin D1 (Fig. 5M), suggesting that the CCD2/3 domain provides a primary interface for the interaction and the RING domain assist in increasing the interactions.
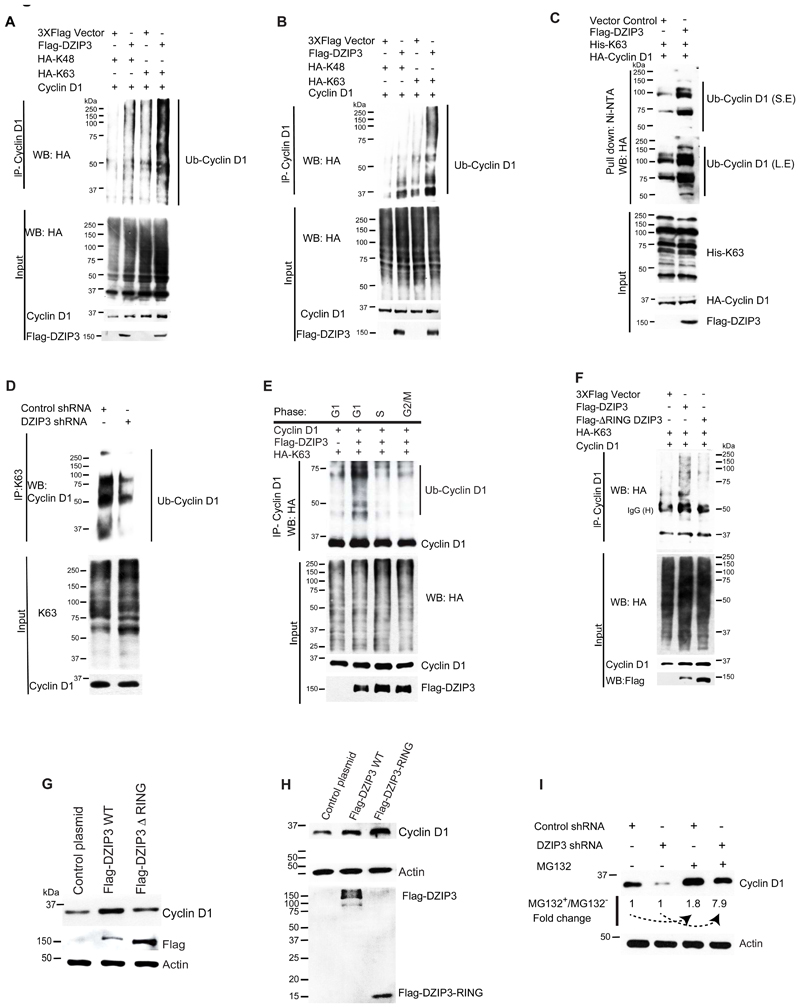
DZIP3 enhances K63-linked ubiquitination of Cyclin D1
DZIP3 is a RING domain-containing E3 ubiquitin ligase (13,14). Next, we examined whether DZIP3 can modulate the ubiquitination status of Cyclin D1. K48-linked ubiquitination of proteins is associated with proteasomal degradation, while K63-linked ubiquitination plays a proteasomal degradation independent role and is shown to be important in signaling, trafficking, sorting, and stabilization of the proteins (36,56–59). DZIP3 and Cyclin D1 were co-expressed in HEK293 cells along with the two ubiquitin variants, HA-K48 and HA-K63. In HA-K48 ubiquitin, all lysine residues were mutated except at amino acid position 48 (K48). In HA-K63, all lysine residues were mutated except at amino acid position 63 (K63). The co-IP assays were performed to understand whether DZIP3 ubiquitinates Cyclin D1. DZIP3 considerably increased the K63-linked ubiquitination of Cyclin D1 with minimally effecting K48-linked ubiquitination (Fig. 6A). Next, to reduce the effect of DZIP3-mediated stability on Cyclin D1-DZIP3 interaction, we repeated the experiment, where we have adjusted the ratio of Cyclin D1 to equalize it in the inputs and then the IP sample are run in the same ratio. Here also, we observed considerably increased K63-linked ubiquitination of Cyclin D1 in the presence of DZIP3 (Fig. 6B).
Figure 6. DZIP3 increases the K63-linked ubiquitination of Cyclin D1 to stabilize it.
(A-B) Western blot analysis of co-IP experiments performed with lysates of HEK293T cells transiently transfected with Flag-DZIP3, Cyclin D1, HA-K48-Ubiquitin and HA-K63-Ubiquitin plasmids (variants of ubiquitin which can only be ubiquitinated at either lysine 48 (K48) or lysine 63 (K63))
(C) Western blot analysis of Cyclin D1 ubiquitination in the absence and presence of DZIP3 by Ni-NTA pull-down assays using transiently transfected plasmid constructs as indicated. His-tagged ubiquitin (His-K63 was mutated at all lysines except at 63 position) was used in these assays. (SE, short exposure; LE, long exposure)
(D) Western blot analysis of endogenous Cyclin D1 ubiquitination in MCF7 Control and DZIP3 knockdown cells. (IP was run in a ratio of adjusted equalized inputs to reduce the artifact of DZIP3 mediated stability).
(E) Western blot analysis of co-IP experiments performed with HEK293T cells transiently transfected with Flag-DZIP3, Cyclin D1, HA-K48-Ubiquitin, and HA-K63-Ubiquitin plasmids, and synchronized in G1 (serum starvation), G1/S (single thymidine block) or G2/M (Nocodazole block) phases. (IP was run in a ratio of adjusted equalized inputs to reduce the artifact of DZIP3 mediated stability).
(F) Western blot analysis of co-IP experiments performed with HEK293T cells transiently transfected with Flag-DZIP3, FLAG ARING DZIP3, Cyclin D1, and HA-K63-Ubiquitin plasmids. (IP was run in a ratio of adjusted equalized inputs to reduce the artifact of DZIP3 mediated stability).
(G) Western blot analysis of Cyclin D1 in HEK293T cells overexpressing Flag-DZIP3 and FLAG ARING DZIP3.
(H) Western blot analysis of Cyclin D1 in HEK293T cells overexpressing Flag-DZIP3 and FLAG-DZIP3-RING.
(I) Western blot analysis of MG132 untreated or treated, shcontrol, and shDZIP3 MCF7 cells lysates probed with indicated antibodies. The imageJ quantification of western band intensity was performed and fold change was depicted, where MG132 untreated samples were used as baseline.
Since in the above experiments, we have performed IP with Cyclin D1 and western blot with HA (ubiquitin), it could be argued that DZIP3 is not directly ubiquitinating Cyclin D1 but possibly one of the Cyclin D1-interacting protein. To eliminate this possibility and also to determine whether DZIP3 directly induce the ubiquitination of Cyclin D1, we performed a classical experiment. The His-K63-Ub (Ubiquitin mutated for all lysines except 63 positions) was co-expressed with Cyclin D1 in the absence and presence of DZIP3. His-K63-Ub was pulled down with Ni-NTA agarose beads and probed with the Cyclin D1 antibody. Since the Ni-NTA affinity pull-down assays were performed in highly denaturing conditions (6M guanidine-HCl), a state in which no protein-protein interaction can exist, any increase in ubiquitination of Cyclin D1 depict direct conjugation of ubiquitin. We found a considerable increase in the direct conjugation of K63-linked ubiquitin of Cyclin D1 (high molecular weight) in the presence of DZIP3 (Fig. 6C), showing that indeed DZIP3 enhances K63-linked ubiquitination of Cyclin D1.
To determine whether DZIP3 controls K63-linked ubiquitination of endogenous Cyclin D1, next, we performed immunoprecipitation with K63-linkage specific polyubiquitin antibody from the lysates of control and DZIP3 knockdown cells. The results demonstrate that upon DZIP3 depletion, K63-linked ubiquitination of endogenous Cyclin D1 is considerably reduced (Fig. 6D). Note that to avoid the artifact of DZIP3-mediated Cyclin D1 stability, we have loaded the IP samples in a ratio that can equalize the Cyclin D1 in inputs. So more IP samples were loaded in the second lane. Still, we found that the ubiquitination was less in DZIP3 knockdown cells suggesting that indeed DZIP3 plays a significant role in K63-linked ubiquitination of Cyclin D1.
DZIP3 interacts with Cyclin D1 in a cell cycle phase-specific manner, predominantly in the G1 phase. So, we asked whether the DZIP3 mediated K63-linked ubiquitination of Cyclin D1 is also phase-specific. For this, we overexpressed DZIP3, Cyclin D1, and HA-K63 and then synchronized the cells in the G1 phase, S phase, and G2 phase. Indeed, we found that DZIP3 mediated K63-linked ubiquitination of Cyclin D1 was increased primarily in the G1 phase (Fig. 6E). Altogether, the results suggest that DZIP3 interactions with Cyclin D1 proteins and subsequent K63-linked ubiquitination are cell cycle phase-specific events.
The RING domain of DZIP3 is important for the K63-linked ubiquitination and stability of Cyclin D1
The RING domain of DZIP3 was shown to be essential for auto-ubiquitination and ubiquitination of other proteins (13,14). The RING domain deleted DZIP3 (Flag-ΔRING DZIP3) variant was found to be inefficient in increasing the K63-linked ubiquitination of the Cyclin D1 proteins (Fig. 6F), suggesting that the RING domain of DZIP3 is absolutely required for enhancing K63-linked ubiquitination of Cyclin D1. Next, we asked whether the RING domain is important for the stability of Cyclin D1. RING-deleted DZIP3 was not able to increase the stability of Cyclin D1 (Fig. 6G), whereas, to our surprise, overexpression of the RING domain itself was sufficient to increase the stability of Cyclin D1 (Fig. 6H). Overall, the data suggest that DZIP3 utilizes its RING domain to increase the K63-linked ubiquitination and stability of the Cyclin D1.
Further, we tested whether the presence of DZIP3 protects from proteasomal degradation. The control and DZIP3 shRNA stable cell lines were treated with MG132, a proteasomal degradation inhibitor (Fig. 6I). The MG132 treated control cells, as compared to untreated cells, showed ~2 fold induction of Cyclin D1, whereas DZIP3 knockdown cells showed ~8 fold induction (Fig. 6I), indicating a considerably higher proteasomal degradation flux of Cyclin D1 in the absence of DZIP3.
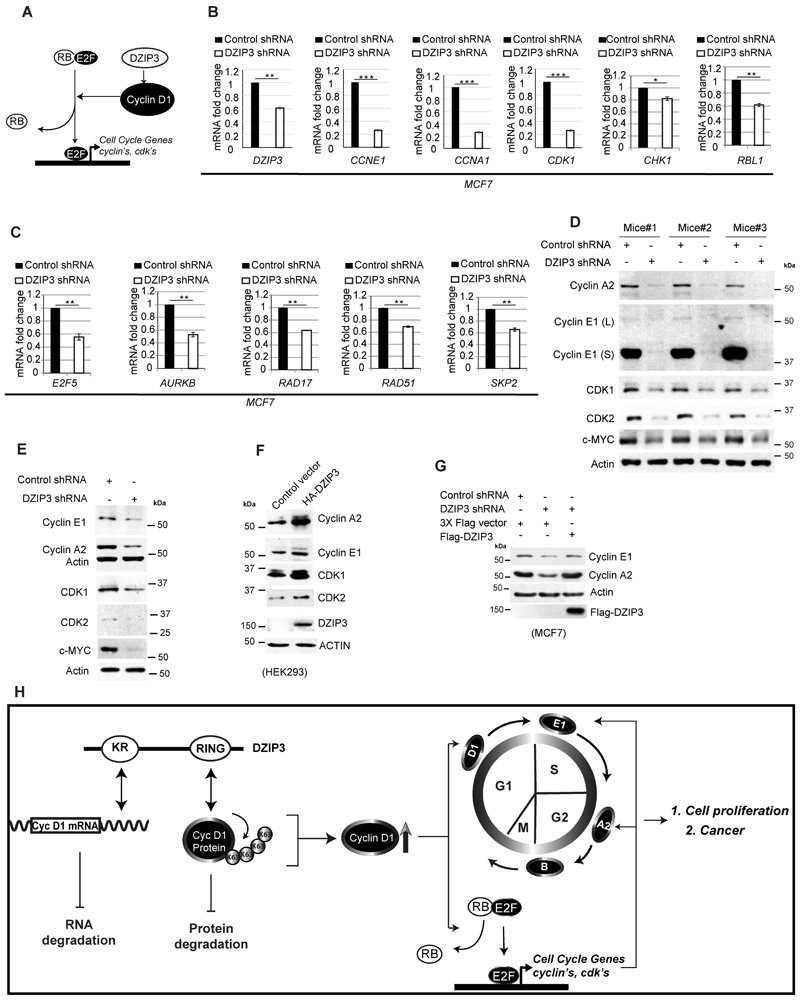
DZIP3 controls E2F target genes
The Cyclin D1-CDK4/6 complex phosphorylates the tumor suppressor retinoblastoma (RB) protein and inactivates it, leading to the release of E2F transcription factors (60). E2F's then activate a large number of genes required for cellular proliferation, including the genes essential for the G1/S phase transition and DNA replication (61) (Fig. 7A). We performed qRT-PCR for several E2F target genes, including CCNE1, CCNA2, CDK1, CHK1, SKP2, RBL1, E2F5, AURKB, RAD17, and RAD52, in control and DZIP3 knockdown cell lines (Fig. 7B and C). The data show that the depletion of DZIP3 significantly reduces the expression of E2F regulated cell proliferation-related genes, including the genes required for cell cycle progression. The expression of some of these genes were also validated in tumors (Supplementary Fig. S6A). In addition, we performed western blotting experiments with few of the E2F regulated cell cycle genes, including Cyclin A2, Cyclin E1, CDK1, CDK2, and c-MYC in control and DZIP3 depleted xenograft tumors and cell lines (Fig. 7D and E, Supplementary Fig. S6B). A marked reduction in expressions of all these essential cell cycle proteins was observed in DZIP3 depleted conditions. In contrast, the overexpression of DZIP3 induced the expression of these proteins (Fig. 7F). Further, the complementation of DZIP3 depleted cells with Flag-DZIP3 restored the levels of Cyclin E1 and Cyclin A2, demonstrating the specificity of the control (Fig. 7G). Taken together, the data show that DZIP3 controls Cyclin D1-E2F axes to regulate cell growth and cell cycle progression.
Figure 7. DZIP3 controls Cyclin D1-E2F axes.
(A) Schematic representation of DZIP3 controlling E2F downstream genes via Cyclin D1.
(B-C) A qRT-PCR analysis to evaluate the expressions of indicated genes from the total RNA of MCF7 control and DZIP3 stable knockdown cells. Data is presented as mean ± SE, n=3, *p<0.05, **p<0.005, ***p<0.0005.
(D) Western blot analysis of UM-UC3 control and DZIP3 stable knockdown tumor xenograft lysates probed with indicated antibodies
(E) Western blot analysis of MCF7 control and DZIP3 stable knockdown cell lysates probed with indicated antibodies
(F) Western blot analysis of HEK293 control and HA-DZIP3 transiently transfected cell lysates with indicated antibodies.
(G) Western blot analysis of MCF7 control and DZIP3 stable knockdown cells transfected with indicated plasmids and probed with indicated antibodies.
(H) Schematic presentation of work presented in this study.
In this study, we found that DZIP3 is a novel regulator of cell cycle progression by controlling the expression of Cyclin D1 in a unique manner. DZIP3 using its RNA binding domain, interacts with AU-rich elements in 3'UTR of Cyclin D1 mRNA to increase its stability (Fig. 7H). Additionally, using its RING domain, DZIP3 increases the K63-linked ubiquitination of Cyclin D1 protein and stabilizes it (Fig. 7H). Predominantly, the interaction and stabilization of Cyclin D1 take place during the G1 phase of the cell cycle, the phase during which Cyclin D1 is required for the progression of the cell cycle (Fig. 7H). By maintaining the expression of Cyclin D1, DZIP3 is an important regulator for E2F-mediated transcription of genes required for proliferation and cell cycle progression (Fig. 7H). In agreement, DZIP3 is necessary for cell cycle progression, cell growth, and cell migration/invasion of cancer cells and thus promotes tumor progression and metastasis (Fig. 7H). Overall, this study identifies DZIP3 as a novel driver of cell cycle and cancer progression.
Discussion
DZIP3 is a developmentally important gene with a capability to suppress differentiation in mouse embryonic stem cells and hematopoietic stem cells (15,16). The mice homozygous for an ENU-induced mutation in the DZIP3 allele exhibit embryonic lethality (http://www.informatics.jax.org/marker/MGI:1917433), suggesting that DZIP3 may be critical for embryonic development. This study, for the first time, underscores the importance and functional relevance of DZIP3 in the progression of cell cycle and cancer.
Cyclin D1 is a critical regulator of cell cycle progression in the G1/S phase, and thus, overexpression or amplification of Cyclin D1 is associated with unregulated cell growth and oncogenic transformations (18–22). A large number of studies identify Cyclin D1 as a protooncogene. This study provides evidence that DZIP3 controls cell cycle and cell growth by controlling the expression of Cyclin D1. DZIP3 is a member of RNA-binding RING E3-ligases (RBRL), a unique class of E3-ligases with the capability to interact with both RNA and protein. The presence of both RNA binding domain and E3-ligase domain in RBRL proteins provides them the capability to perform unique combinatorial spatial and temporal functions. The RBRLs, MEX3B, and DZIP3 were shown to interact with a long noncoding RNA, HOTAIR, to induce K48-linked ubiquitination and degradation of Ataxin-1 and Snurportin-1, respectively (14). Another RBRL, MEX3C, interacts with 3'UTR of HLA-A2 mRNA and causes its RING-dependent degradation (7). One more interesting kind of regulation is mediated by MDM2. MDM2 promotes polyubiquitination and proteasomal degradation of p53. On the other hand, it also binds to p53 mRNA to stabilize it, resulting in a balance of the p53 expression (62). A recent study showed that the mRNA binding activity of TRIM25 induces its ubiquitination capacity suggesting a kind of regulation where the RNA level of one protein can regulate its own proteasomal degradation or degradation of other related proteins (63). In the present study, we revealed another unique kind of regulation by RBRL, where DZIP3 binds with mRNA and protein of the same gene to maintain their stability. The factors involved in phase-specific degradation of cyclins are very well defined; however the factors, which stabilize the cyclins during the specific cell cycle phase, remain elusive. Here, we describe one of such factor and one such mechanism. Further, we also demonstrate that DZIP3, via Cyclin D1, controls the expression of E2F transcription factor regulated genes, including Cyclins (A2 and E1) and CDKs.
Cyclins are regulated at both RNA and protein levels. The HuR protein is known to interact with 3'UTR of pan-cyclin mRNA and impart stability (24–27), whereas AUF1 binds with Cyclin D1 mRNA and increases its degradation (49,64). DZIP3 is similar to HuR in the regulation of the stability of cyclins mRNAs. Several other proteins, including E3 ubiquitin ligases, were shown to interact with cyclin proteins and mediate their degradation or stabilization. The best examples are the APC/C complex and Skp1-Cul1-F-box (SCF) family of E3 ubiquitin ligases. They mediate the degradation of cyclins for the controlled progression of the cell cycle (23). The Cullin-3 mediates ubiquitination and degradation of Cyclin E (65). Hec1 interacts with Cyclin B1 during the early M phase and protects it from degradation by the APC/C complex, however at the late M phase, interaction is broken resulting in the destruction of Cyclin B1 (53). A recent study shows that ERLIN2 stabilizes Cyclin B1 in the G2/M phase by inducing its K63-linked ubiquitination (36). In future studies, it will be interesting to determine whether DZIP3 could cooperate or antagonize activities of some of the known cell cycle regulatory factors to orchestrate Cyclin D1 protein levels during cell cycle progression.
Supplementary Material
Acknowledgments
This work was supported by the DBT/Wellcome Trust India Alliance Fellowship (IA/I/15/2/502071) awarded to Santosh Chauhan. Swati Chauhan is supported by DST WOS-A fellowship (SR/WOS-A/LS-9/2016). Rajeeb Swain is supported by DBT grant (6242-P64/RGCB/PMD/DBT/RJKS/2015). Tor Erik Rusten and Ashish Jain are supported by grants #262652 and #276070 from the Norwegian Research Council. We acknowledge the technical assistance of Mr. Kshitish Rout. We gratefully acknowledge the support of the Institute of Life Sciences central facilities (Imaging, FACS, and Sequencing) funded by the Department of Biotechnology, India.
Footnotes
The authors declare that they have no conflict of interests.
Statement of Significance
Findings show that DZIP3 is a novel driver of cell cycle and cancer progression via its control of Cyclin D1 mRNA and protein stability in a cell cycle phase-dependent manner.
References
- 1.Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol. 2014;21:301–7. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- 2.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–25. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Hildebrandt A, Alanis-Lobato G, Voigt A, Zarnack K, Andrade-Navarro MA, Beli P, et al. Interaction profiling of RNA-binding ubiquitin ligases reveals a link between posttranscriptional regulation and the ubiquitin system. Sci Rep. 2017;7 doi: 10.1038/s41598-017-16695-6. 16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garzia A, Jafarnejad SM, Meyer C, Chapat C, Gogakos T, Morozov P, et al. The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat Commun. 2017;8 doi: 10.1038/ncomms16056. 16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q, Fan L, Hou F, Dong A, Wang YX, Tong Y. New Insights into the RNA-Binding and E3 Ubiquitin Ligase Activities of Roquins. Sci Rep. 2015;5 doi: 10.1038/srep15660. 15660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuniyoshi K, Takeuchi O, Pandey S, Satoh T, Iwasaki H, Akira S, et al. Pivotal role of RNA-binding E3 ubiquitin ligase MEX3C in RIG-I-mediated antiviral innate immunity. Proc Natl Acad Sci U S A. 2014;111:5646–51. doi: 10.1073/pnas.1401674111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cano F, Bye H, Duncan LM, Buchet-Poyau K, Billaud M, Wills MR, et al. The RNA-binding E3 ubiquitin ligase MEX-3C links ubiquitination with MHC-I mRNA degradation. EMBO J. 2012;31:3596–606. doi: 10.1038/emboj.2012.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang L, Malu S, McKenzie JA, Andrews MC, Talukder AH, Tieu T, et al. The RNA-binding Protein MEX3B Mediates Resistance to Cancer Immunotherapy by Downregulating HLA-A Expression. Clin Cancer Res. 2018;24:3366–76. doi: 10.1158/1078-0432.CCR-17-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu XF, Cheng SF, Wang LQ, Yin S, De Felici M, Shen W. DAZ Family Proteins, Key Players for Germ Cell Development. Int J Biol Sci. 2015;11:1226–35. doi: 10.7150/ijbs.11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B, Wang G, Xu X, Yang S, Zhuang T, Wang G, et al. DAZ-interacting Protein 1 (Dzip1) Phosphorylation by Polo-like Kinase 1 (Plk1) Regulates the Centriolar Satellite Localization of the BBSome Protein during the Cell Cycle. J Biol Chem. 2017;292:1351–60. doi: 10.1074/jbc.M116.765438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shigunov P, Sotelo-Silveira J, Stimamiglio MA, Kuligovski C, Irigoin F, Badano JL, et al. Ribonomic analysis of human DZIP1 reveals its involvement in ribonucleoprotein complexes and stress granules. BMC Mol Biol. 2014;15:12. doi: 10.1186/1471-2199-15-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C, Li J, Takemaru KI, Jiang X, Xu G, Wang B. Centrosomal protein Dzip1l binds Cby, promotes ciliary bud formation, and acts redundantly with Bromi to regulate ciliogenesis in the mouse. Development. 2018:145. doi: 10.1242/dev.164236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreft SG, Nassal M. hRUL138, a novel human RNA-binding RING-H2 ubiquitin-protein ligase. J Cell Sci. 2003;116:605–16. doi: 10.1242/jcs.00261. [DOI] [PubMed] [Google Scholar]
- 14.Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4 doi: 10.1038/ncomms3939. 2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De La Fuente R, Baumann C, Viveiros MM. ATRX contributes to epigenetic asymmetry and silencing of major satellite transcripts in the maternal genome of the mouse embryo. Development. 2015;142:1806–17. doi: 10.1242/dev.118927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue D, Aihara H, Sato T, Mizusaki H, Doiguchi M, Higashi M, et al. Dzip3 regulates developmental genes in mouse embryonic stem cells by reorganizing 3D chromatin conformation. Sci Rep. 2015;5 doi: 10.1038/srep16567. 16567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell DJ, Chauhan S, Jimenez-Stinson D, Elliott KR, Tsewang TD, Lee YH, et al. Novel CARM1-Interacting Protein, DZIP3, Is a Transcriptional Coactivator of Estrogen Receptor-alpha. Mol Endocrinol. 2015;29:1708–19. doi: 10.1210/me.2015-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JK, Diehl JA. Nuclear cyclin D1: an oncogenic driver in human cancer. J Cell Physiol. 2009;220:292–6. doi: 10.1002/jcp.21791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sauter ER, Yeo UC, von Stemm A, Zhu W, Litwin S, Tichansky DS, et al. Cyclin D1 is a candidate oncogene in cutaneous melanoma. Cancer Res. 2002;62:3200–6. [PubMed] [Google Scholar]
- 20.Bates S, Peters G. Cyclin D1 as a cellular proto-oncogene. Semin Cancer Biol. 1995;6:73–82. doi: 10.1006/scbi.1995.0010. [DOI] [PubMed] [Google Scholar]
- 21.Callender T, el-Naggar AK, Lee MS, Frankenthaler R, Luna MA, Batsakis JG. PRAD-1 (CCND1)/cyclin D1 oncogene amplification in primary head and neck squamous cell carcinoma. Cancer. 1994;74:152–8. doi: 10.1002/1097-0142(19940701)74:1<152::aid-cncr2820740124>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 22.Tashiro E, Tsuchiya A, Imoto M. Functions of cyclin D1 as an oncogene and regulation of cyclin D1 expression. Cancer Sci. 2007;98:629–35. doi: 10.1111/j.1349-7006.2007.00449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vodermaier HC. APC/C and SCF: controlling each other and the cell cycle. Curr Biol. 2004;14:R787–96. doi: 10.1016/j.cub.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 24.Guo X, Connick MC, Vanderhoof J, Ishak MA, Hartley RS. MicroRNA-16 modulates HuR regulation of cyclin E1 in breast cancer cells. Int J Mol Sci. 2015;16:7112–32. doi: 10.3390/ijms16047112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo X, Hartley RS. HuR contributes to cyclin E1 deregulation in MCF-7 breast cancer cells. Cancer Res. 2006;66:7948–56. doi: 10.1158/0008-5472.CAN-05-4362. [DOI] [PubMed] [Google Scholar]
- 26.Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000;19:2340–50. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kakuguchi W, Kitamura T, Kuroshima T, Ishikawa M, Kitagawa Y, Totsuka Y, et al. HuR knockdown changes the oncogenic potential of oral cancer cells. Mol Cancer Res. 2010;8:520–8. doi: 10.1158/1541-7786.MCR-09-0367. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura H, Kawagishi H, Watanabe A, Sugimoto K, Maruyama M, Sugimoto M. Cooperative role of the RNA-binding proteins Hzf and HuR in p53 activation. Mol Cell Biol. 2011;31:1997–2009. doi: 10.1128/MCB.01424-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci U S A. 2003;100:8354–9. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho SJ, Zhang J, Chen X. RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability. Nucleic Acids Res. 2010;38:2256–67. doi: 10.1093/nar/gkp1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, et al. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000;20:760–9. [Google Scholar]
- 32.Dong L, Yu L, Bai C, Liu L, Long H, Shi L, et al. USP27-mediated Cyclin E stabilization drives cell cycle progression and hepatocellular tumorigenesis. Oncogene. 2018;37:2702–13. doi: 10.1038/s41388-018-0137-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Z, Tan C, Qiu Q, Kong S, Yang H, Zhao F, et al. Ubiquitin-specific protease 22 is a deubiquitinase of CCNB1. Cell Discov. 2015:1. doi: 10.1038/celldisc.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonacci T, Suzuki A, Grant GD, Stanley N, Cook JG, Brown NG, et al. Cezanne/OTUD7B is a cell cycle-regulated deubiquitinase that antagonizes the degradation of APC/C substrates. EMBO J. 2018 doi: 10.15252/embj.201798701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, Li W, Xiao L, Xu J, Chen X, Tang M, et al. Viral oncoprotein LMP1 disrupts p53-induced cell cycle arrest and apoptosis through modulating K63-linked ubiquitination of p53. Cell Cycle. 2012;11:2327–36. doi: 10.4161/cc.20771. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Cai J, Zheng Z, Polin L, Lin Z, Dandekar A, et al. A novel ER-microtubule-binding protein, ERLIN2, stabilizes Cyclin B1 and regulates cell cycle progression. Cell Discov. 2015;1 doi: 10.1038/celldisc.2015.24. 15024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tripathi E, Smith S. Cell cycle-regulated ubiquitination of tankyrase 1 by RNF8 and ABRO1/BRCC36 controls the timing of sister telomere resolution. EMBO J. 2017;36:503–19. doi: 10.15252/embj.201695135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jena KK, Kolapalli SP, Mehto S, Nath P, Das B, Sahoo PK, et al. TRIM16 controls assembly and degradation of protein aggregates by modulating the p62-NRF2 axis and autophagy. EMBO J. 2018:37. doi: 10.15252/embj.201798358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jena KK, Mehto S, Nath P, Chauhan NR, Sahu R, Dhar K, et al. Autoimmunity gene IRGM suppresses cGAS-STING and RIG-I-MAVS signaling to control interferon response. EMBO Rep. 2020 doi: 10.15252/embr.202050051. e50051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehto S, Jena KK, Nath P, Chauhan S, Kolapalli SP, Das SK, et al. The Crohn's Disease Risk Factor IRGM Limits NLRP3 Inflammasome Activation by Impeding Its Assembly and by Mediating Its Selective Autophagy. Mol Cell. 2018 doi: 10.1016/j.molcel.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehto S, Jena KK, Nath P, Chauhan S, Kolapalli SP, Das SK, et al. The Crohn's Disease Risk Factor IRGM Limits NLRP3 Inflammasome Activation by Impeding Its Assembly and by Mediating Its Selective Autophagy. Mol Cell. 2019;73:429–45 e7. doi: 10.1016/j.molcel.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren J, Liu S, Cui C, Ten Dijke P. Invasive Behavior of Human Breast Cancer Cells in Embryonic Zebrafish. J Vis Exp. 2017 doi: 10.3791/55459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA02: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W60. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cuylen S, Blaukopf C, Politi AZ, Muller-Reichert T, Neumann B, Poser I, et al. Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature. 2016;535:308–12. doi: 10.1038/nature18610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letrado P, de Miguel I, Lamberto I, Diez-Martinez R, Oyarzabal J. Zebrafish: Speeding Up the Cancer Drug Discovery Process. Cancer Res. 2018;78:6048–58. doi: 10.1158/0008-5472.CAN-18-1029. [DOI] [PubMed] [Google Scholar]
- 46.Motokura T, Arnold A. Cyclin D and oncogenesis. Curr Opin Genet Dev. 1993;3:5–10. doi: 10.1016/s0959-437x(05)80334-x. [DOI] [PubMed] [Google Scholar]
- 47.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–72. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 48.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Al-Khalaf HH, Colak D, Al-Saif M, Al-Bakheet A, Hendrayani SF, Al-Yousef N, et al. p16(INK4a) positively regulates cyclin D1 and E2F1 through negative control of AUF1. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021111. e21111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bakheet T, Hitti E, Khabar KSA. ARED-Plus: an updated and expanded database of AU-rich element-containing mRNAs and pre-mRNAs. Nucleic Acids Res. 2018;46:D218–D20. doi: 10.1093/nar/gkx975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castro A, Bernis C, Vigneron S, Labbe JC, Lorca T. The anaphase-promoting complex: a key factor in the regulation of cell cycle. Oncogene. 2005;24:314–25. doi: 10.1038/sj.onc.1207973. [DOI] [PubMed] [Google Scholar]
- 52.Higa LA, Yang X, Zheng J, Banks D, Wu M, Ghosh P, et al. Involvement of CUL4 ubiquitin E3 ligases in regulating CDK inhibitors Dacapo/p27Kip1 and cyclin E degradation. Cell Cycle. 2006;5:71–7. doi: 10.4161/cc.5.1.2266. [DOI] [PubMed] [Google Scholar]
- 53.Gui L, Homer H. Hec1-dependent cyclin B2 stabilization regulates the G2-M transition and early prometaphase in mouse oocytes. Dev Cell. 2013;25:43–54. doi: 10.1016/j.devcel.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alam MS. Proximity Ligation Assay (PLA) Curr Protoc Immunol. 2018;123:e58. doi: 10.1002/cpim.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812–21. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- 56.Choi KS, Choi HJ, Lee JK, Im S, Zhang H, Jeong Y, et al. The endothelial E3 ligase HECW2 promotes endothelial cell junctions by increasing AMOTL1 protein stability via K63-linked ubiquitination. Cell Signal. 2016;28:1642–51. doi: 10.1016/j.cellsig.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 57.Lauwers E, Jacob C, Andre B. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J Cell Biol. 2009;185:493–502. doi: 10.1083/jcb.200810114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Erpapazoglou Z, Walker O, Haguenauer-Tsapis R. Versatile roles of k63-linked ubiquitin chains in trafficking. Cells. 2014;3:1027–88. doi: 10.3390/cells3041027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J, Zhang C, Cui J, Ou J, Han J, Qin Y, et al. TRIM45 functions as a tumor suppressor in the brain via its E3 ligase activity by stabilizing p53 through K63-linked ubiquitination. Cell Death Dis. 2017;8:e2831. doi: 10.1038/cddis.2017.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–30. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 61.Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29:409–17. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 62.Cano F, Miranda-Saavedra D, Lehner PJ. RNA-binding E3 ubiquitin ligases: novel players in nucleic acid regulation. Biochem Soc Trans. 2010;38:1621–6. doi: 10.1042/BST0381621. [DOI] [PubMed] [Google Scholar]
- 63.Choudhury NR, Heikel G, Trubitsyna M, Kubik P, Nowak JS, Webb S, et al. RNA-binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination. BMC Biol. 2017;15:105. doi: 10.1186/s12915-017-0444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin S, Wang W, Wilson GM, Yang X, Brewer G, Holbrook NJ, et al. Down-regulation of cyclin D1 expression by prostaglandin A(2) is mediated by enhanced cyclin D1 mRNA turnover. Mol Cell Biol. 2000;20:7903–13. doi: 10.1128/mcb.20.21.7903-7913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singer JD, Gurian-West M, Clurman B, Roberts JM. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 1999;13:2375–87. doi: 10.1101/gad.13.18.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.